
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 19 August 2020
Sec. Bacteria and Host
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00419
This article is part of the Research Topic Bacterial Effectors as Drivers of Human Disease: Models, Methods, Mechanisms View all 16 articles
 Jiyao Gan1,2
Jiyao Gan1,2 Nichollas E. Scott2
Nichollas E. Scott2 Joshua P. M. Newson2†
Joshua P. M. Newson2† Rachelia R. Wibawa1,2
Rachelia R. Wibawa1,2 Tania Wong Fok Lung2†
Tania Wong Fok Lung2† Georgina L. Pollock1
Georgina L. Pollock1 Garrett Z. Ng3
Garrett Z. Ng3 Ian van Driel3
Ian van Driel3 Jaclyn S. Pearson1,4
Jaclyn S. Pearson1,4 Elizabeth L. Hartland1,4*
Elizabeth L. Hartland1,4* Cristina Giogha1,4*
Cristina Giogha1,4*During infection, Salmonella species inject multiple type III secretion system (T3SS) effector proteins into host cells that mediate invasion and subsequent intracellular replication. At early stages of infection, Salmonella exploits key regulators of host intracellular vesicle transport, including the small GTPases Rab5 and Rab7, to subvert host endocytic vesicle trafficking and establish the Salmonella-containing vacuole (SCV). At later stages of intracellular replication, interactions of the SCV with Rab GTPases are less well defined. Here we report that Rab1, Rab5, and Rab11 are modified at later stages of Salmonella infection by SseK3, an arginine N-acetylglucosamine (GlcNAc) transferase effector translocated via the Salmonella pathogenicity island 2 (SPI-2) type III secretion system. SseK3 modified arginines at positions 74, 82, and 111 within Rab1 and this modification occurred independently of Rab1 nucleotide binding. SseK3 exhibited Golgi localization that was independent of its glycosyltransferase activity but Arg-GlcNAc transferase activity was required for inhibition of alkaline phosphatase secretion in transfected cells. While SseK3 had a modest effect on SEAP secretion during infection of HeLa229 cells, inhibition of IL-1 and GM-CSF cytokine secretion was only observed upon over-expression of SseK3 during infection of RAW264.7 cells. Our results suggest that, in addition to targeting death receptor signaling, SseK3 may contribute to Salmonella infection by interfering with the activity of key Rab GTPases.
Salmonella enterica Typhimurium (S. Typhimurium) is a common foodborne pathogen that imposes a significant financial burden on healthcare systems in both industrialized and developing countries (Majowicz et al., 2010). Upon ingestion of contaminated food or water, Salmonellae that survive the acidic gastric environment colonize the gut by invading both phagocytic and non-phagocytic cells (Clark et al., 1994; Smith, 2003; Geddes et al., 2007; Muller et al., 2012). Salmonella pathogenicity island 1 (SPI-1) and Salmonella pathogenicity island 2 (SPI-2) both encode type III secretion systems (T3SSs) that deliver virulence effector proteins into host cells that facilitate Salmonella pathogenesis. These two T3SSs are regulated in a spatio-temporal dependent manner, and are responsible for delivering two distinct cohorts of effectors into the host cell cytosol to hijack host physiology (LaRock et al., 2015). The SPI-1-encoded T3SS is activated initially, inducing inflammation and host cytoskeletal rearrangements to promote invasion of the bacteria into epithelial cells, while cells such as macrophages internalize Salmonella by phagocytosis. Following internalization, the SPI-2-encoded T3SS is activated to establish a membrane-bound replicative niche termed the Salmonella-containing vacuole (SCV). The SCV subverts the canonical endolysosomal pathway to facilitate intracellular bacterial survival and replication (Ramsden et al., 2007; Jennings et al., 2017).
SseK1, SseK2, and SseK3 are three SPI-2 translocated effectors which show high amino acid sequence similarity (84, 83, and 80% respectively) to the T3SS effector NleB1 from enteropathogenic Escherichia coli (EPEC) (Kujat Choy et al., 2004; Brown et al., 2011; Li et al., 2013; Pearson et al., 2013). NleB1 is a novel glycosyltransferase that blocks apoptotic cell death during EPEC infection by modifying conserved arginine residues with N-acetylglucosamine (GlcNAc) within death domain containing proteins, including FADD and TRADD (Li et al., 2013; Pearson et al., 2013; Scott et al., 2017). The post-translational modifications are termed Arg-GlcNAcylation and are only mediated by members of the NleB1 effector protein homolog family including the SseKs (El Qaidi et al., 2017; Gunster et al., 2017; Esposito et al., 2018; Park et al., 2018; Newson et al., 2019). Similar to NleB1, expression of SseK1 or SseK3, but not SseK2, suppresses NF-κB activation during infection, although these effectors exhibit different Arg-GlcNAcylation profiles on infected host cells (Gunster et al., 2017; Newson et al., 2019). Our group recently showed that overexpression of SseK1 and SseK3 resulted in broadened substrate specificity, suggesting that authentic host targets of these effectors need to be identified under native expression conditions during infection (Newson et al., 2019). When expressed at endogenous levels during Salmonella infection, SseK1 preferentially modifies the death domain of TRADD, whereas SseK3 targets death domains in the death receptors, TRAILR and TNFR1 (Newson et al., 2019). Interestingly, both SseK2 and SseK3 localize to the Golgi during infection; while SseK1 localizes to the cytosol (Gunster et al., 2017). Such observations suggest SseK2 and SseK3 may have uncharacterized Golgi-localized targets.
In this study, we sought to identify Golgi-associated proteins that are modified by SseK3 during S. Typhimurium infection. Using a mass spectrometry-based approach to examine membrane-enriched fractions of infected macrophages, we identified several Rab GTPases as targets of SseK3, including Rab1, Rab5, and Rab11. Notably, we found SseK3 modified arginine residues in the switch II region of Rab1, and that ectopic expression of either SseK2 or SseK3, but not SseK1, blocked host protein secretion. However, only SseK3 appeared to have the capacity to block host protein secretion during Salmonella infection. Collectively, these results suggest that SseK3 may contribute to Salmonella infection by GlcNAcylating selected Rab GTPases.
The bacterial strains used in this study are listed in Table 1. All bacteria were grown in Luria-Bertani (LB) broth with shaking at 200 rpm in 37°C incubator in the presence of streptomycin (50 μg/ml), kanamycin (100 μg/ml), or ampicillin (100 μg/ml) when necessary.
The plasmids and primers used in this study are listed in Tables 2, 3 respectively. All plasmid extractions were performed using QIAGEN QIAprep Miniprep Kit (Qiagen, Valencia, CA). DNA restriction digests were applied according to manufacturer's instructions (New England Biolabs, Ipswich, MA). PCR products, restriction digest products and DNA from agarose gels were purified using the Wizard SV Gel and PCR Clean-Up system (Promega, Madison, WI). Digested inserts and plasmids were ligated with a 6:1 molar ratio at 4°C overnight using T4 DNA Ligase system in accordance with manufacturer's instructions (Promega, Madison, WI). The pF_TRE-SEAP ligation product was transformed into DH5α cells and other resultant ligation products were transformed into XL1-Blue cells. The pF_TRE-SEAP plasmid was sequenced with pFTRE-F/R and other plasmids were extracted and sequenced with sequencing primer pair p3xFlag-Myc-CMV-24F/p3xFlag-Myc-CMV-24R.
The p3xFlag-Rab1a vector was constructed by amplifying RAB1A from human cDNA using primer pair Rab1aF/Rab1aR, which was then double digested with EcoRV and BamHI and ligated into p3xFlag-Myc-CMV-24 plasmid that had been digested with same restriction enzymes to generate an N-terminal 3xFlag fusion to Rab1a. p3xFlag-Rab5a and p3xFlag-Rab5b were constructed in a similar manner by amplifying RAB5A or RAB5B from human cDNA using primer pair Rab5aF/Rab5aR or Rab5bF/Rab5bR respectively, which were then double digested with HindIII and BamHI and ligated into p3xFlag-Myc-CMV-24. p3xFlag-Rab5c was constructed by amplifying RAB5C from human cDNA using primer pair Rab5cF/Rab5cR, which was then double digested with EcoRV and BamHI and ligated into p3xFlag-Myc-CMV-24. p3xFlag-Rab11b was constructed by amplifying RAB11B from human cDNA using primer pair Rab11bF/Rab11bR, which was then double digested with HindIII and BamHI and ligated into p3xFlag-Myc-CMV-24. p3xFlag-AnkX was constructed by amplifying ankX from Legionella pneumophila Philadelphia 1 genomic DNA using primer pair AnkXF/R, double digested with SalI and BamHI and ligated into p3xFlag-Myc-CMV-24 plasmid that has been digested with same restriction enzymes to generate an N-terminal 3xFlag fusion to AnkX. The pF_TRE-SEAP was constructed by amplifying SEAP from pSEAP vector using primer pair SEAPF/SEAPR, which was subsequently double digested with BclI and NheI and ligated into pF_TRE3G_PGK puro that has been digested with same restriction enzymes.
All site-directed mutagenesis was performed using QuikChange II Site-Directed Mutagenesis Kit according to manufacturer's protocol. p3xFlag-Rab1aR74A was generated with primer pair Rab1aR74A−F/Rab1aR74A−R using p3xFlag-Rab1a vector as a template. Rab1aR82A was generated with primer pair Rab1aR82A−F/Rab1aR82A−R using p3xFlag-Rab1a vector as a template. Rab1aR111A was generated with primer pair Rab1aR111A−F/Rab1aR111A−R using p3xFlag-Rab1a vector as a template. Rab1aR74AR82A and Rab1aR74AR111A were generated with primer pair Rab1aR82A−F/Rab1aR82A−R and Rab1aR111A−F/Rab1aR111A−R respectively using p3xFlag-Rab1aR74A vector as a template. Rab1aR82AR111A was generated with primer pair Rab1aR111A−F/Rab1aR111A−R using p3xFlag-Rab1aR82A vector as a template. Rab1aR74AR82AR111A was generated with primer pair Rab1aR111A−F/Rab1aR111A−R using p3xFlag-Rab1aR74AR82A vector as a template. The sequences of resulting plasmids were confirmed by sequencing using primer pair p3xFlag-Myc-CMV-24F/p3xFlag-Myc-CMV-24R. The pEGFP-C2-SseK1DXD, pEGFP-C2-SseK2DXD and pEGFP-C2-SseK3DXD vectors were generated using primer pairs SseK1AAA−F/R, SseK2AAA−F/R and SseK3AAA−F/R respectively. All resultant plasmids following PCR reactions were digested with DpnI at 37°C overnight and then transformed into XL1-Blue competent cells.
RAW264.7 cells were seeded to 24 well plates at a concentration of 3 × 105 cells per well 1 day before infection. 10 ml LB broths containing appropriate antibiotic were inoculated with Salmonella strains and incubated at 37°C overnight with shaking at 180 rpm. On the day of infection, the OD600 readings of the overnight culture were read and used to estimate bacterial counts. Cells were then infected at a multiplicity of infection (MOI) of 10. 24 well plates were centrifuged at 1500 rpm for 5 min at room temperature to promote and synchronize infection. Infected cells were incubated at 37°C, 5% CO2 for 1 h. Culture media was replaced with media containing 100 μg/ml gentamicin (Pharmacia, Washington, USA), and cells were incubated at 37°C, 5% CO2 for a further 1 h. Culture media was replaced with media containing 10 μg/ml gentamicin, and cells were incubated at 37°C, 5% CO2 for a further 18 h.
Infected cells were washed three times in ice-cold PBS, then collected by scraping in PBS containing cOmplete™ EDTA-free protease inhibitor cocktail (Roche), on a bed of ice. Cells were centrifuged at 4000 rpm for 5 min at 4°C, and the supernatant was carefully removed via aspiration using a vacuum pump. Cells were resuspended in 1 ml of ice-cold lysis buffer (20 mM HEPES (pH 7.5), 100 mM KCl, 2.5 mM MgCl2, 100 mM sucrose, 10% PhosSTOP™ (Sigma-Aldrich), and cOmplete™ EDTA-free protease inhibitor cocktail (Roche). Cells were mixed thoroughly by pipette and incubated on ice for 10 min. Cell lysates were centrifuged at 13000 rpm for 5 min at 4°C, and the supernatant was removed by vacuum-aided aspiration as previously. Pelleted material was resuspended in lysis buffer containing 10% digitonin, mixed thoroughly by pipette and incubated on ice for 1 h with intermittent vortexing. Cell lysates were centrifuged at 13000 rpm for 5 min at 4°C, and the supernatant was collected as the digitonin-soluble membrane fraction, while the remaining pellet was collected as the digitonin-insoluble fraction.
Insoluble membrane pellets were resuspended in 8M urea in 100 mM ammonium bicarbonate and protein concentration determined using a BCA assay. 2.5 mg of protein from each sample type, ΔsseK12 and ΔsseK123, were reduced with 10 mM dithiothreitol for 1 h then alkylated with 50 mM chloroacetamide for a further 1 h in the dark. Samples were digested with Lys-C (1/200 w/w) for 3 h at RT, samples diluted to <2M urea and digested with trypsin (1/100 w/w) overnight at RT. Digested samples were acidified to a final concentration of 0.5% formic acid and desalted with 50 mg tC18 SEP-PAK (Waters corporation, Milford, USA) according to the manufacturer's instructions. Briefly, tC18 SEP-PAKs were conditioned with buffer B (80% acetonitrile, 0.1% formic acid), washed with 10 volumes of Buffer A* (0.1% trifluoroacetic acid, 2% acetonitrile), sample loaded, column washed with 10 volumes of Buffer A* and bound peptides eluted with buffer B then dried.
Peptide affinity purification was accomplished according to the protocol of Scott et al. (2017) for peptide based Arg-GlcNAc enrichment. Briefly, aliquots of 100 μl of Protein A/G plus Agarose beads (Santa Cruz, Santa Cruz CA) were washed three times with 1 ml of immunoprecipitation buffer (IAP, 10 mM Na2HPO4, 50 mM NaCl, 50 mM MOPS, pH 7.2) and tumbled overnight with 10 μg of anti-Arg-GlcNAc antibody (ab195033, Abcam) at 4°C. Coupled anti-Arg-GlcNAc beads were then washed three times with 1 ml of 100 mM sodium borate (pH 9) to remove non-bound proteins and cross-linked for 30 min by gently rotating with 20 mM Dimethyl Pimelimidate (Thermo Fisher Scientific) in 100 mM HEPES, pH 8.0. Cross-linking was quenched by washing beads with 200 mM ethanolamine, pH 8.0, three times then rotating the beads in an additional 1 ml 200 mM ethanolamine, pH 8.0 for 2 h at 4°C. Beads were washed three times with IAP buffer and used immediately. Purified peptides were resuspended in 1 ml IAP buffer and the pH checked to ensure compatibility with affinity conditions (~pH7.2). Peptide lysates were then added to the prepared cross-linked anti-Arg-GlcNAc antibody beads and rotated for 3 h at 4°C. Upon completion of the incubation, antibody beads were spun down at 3000 G for 2 min at 4°C and the unbound peptide lysates collected. Antibody beads were then washed six times with 1 ml of ice-cold IAP buffer and Arg-GlcNAc peptides eluted using two rounds of acid elution. For each elution round, 100 μl of 0.2% trifluoroacetic acid was added and antibody beads allowed to stand at room temperature with gentle shaking every minute for 10 min. Peptide supernatants were collected and desalted using C18 stage tips (Rappsilber et al., 2007) before analysis by LC-MS.
Purified peptides prepared were re-suspend in Buffer A* and separated using a two-column chromatography set up composed of a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μl/min for 5 min and infused into an Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer (Thermo Fisher Scientific) at 300 nl/minute via the analytical column using a Dionex Ultimate 3000 UPLC (Thermo Fisher Scientific). 125 min gradients were run altering the buffer composition from 1% buffer B to 28% B over 90 min, then from 28% B to 40% B over 10 min, then from 40% B to 100% B over 2 min, the composition was held at 100% B for 3 min, and then dropped to 3% B over 5 min and held at 3% B for another 15 min. The Lumos™ Mass Spectrometer was operated in a data-dependent mode automatically switching between the acquisition of a single Orbitrap MS scan (120,000 resolution) every 3 s and Orbitrap HCD for each selected precursor (maximum fill time 100 ms, AGC 2*105 with a resolution of 30,000 for Orbitrap MS-MS scans).
Identification of proteins and Arg-glycosylated peptides was accomplished using MaxQuant (v1.5.3.30) (Cox and Mann, 2008). Searches were performed against the Mouse (Uniprot proteome id UP000000589—Mus musculus, downloaded 18-05-2016, 50306 entries) and Salmonella Typhimurium SL1344 (Uniprot proteome id UP000008962- Salmonella Typhimurium SL1344, downloaded 18-05-2016, 4,657 entries) proteomes with carbamidomethylation of cysteine set as a fixed modification. Searches were performed with trypsin cleavage specificity allowing 2 mis-cleavage events and the variable modifications of oxidation of methionine, N-Acetylhexosamine addition to arginine (Arg-GlcNAc) and acetylation of protein N-termini. The precursor mass tolerance was set to 20 parts-per-million (ppm) for the first search and 10 ppm for the main search, with a maximum false discovery rate (FDR) of 1.0% set for protein and peptide identifications. To enhance the identification of peptides between samples the Match Between Runs option was enabled with a precursor match window set to 2 min and an alignment window of 10 min. For label-free quantitation, the MaxLFQ option within Maxquant (Cox et al., 2014) was enabled in addition to the re-quantification module. The resulting protein group output was processed within the Perseus (v1.4.0.6) (Tyanova et al., 2016) analysis environment to remove reverse matches and common protein contaminates prior. For LFQ comparisons missing values were imputed using Perseus. Enrichment analysis of Arg-GlcNAcylated targets was undertaken using a Fisher exact test within Perseus. Data was exported into the R framework for visualization using ggplot2. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015082.
HEK293T cells were seeded in a 10 cm plate and grown to 60% confluency before being transfected with 1.2 μg pF_TRE-SEAP, 2.0 μg pCMVΔR8.2 and 0.8 μg pCMV-VSV-G. Transfected cells were kept in a tissue culture incubator under 5% CO2 at 37°C for 24 h before the tissue culture media was replaced. The supernatant containing packaged virus was collected 48 h post transfection and then filtered through a 0.45 μm filter. HeLa229 cells were seeded in a 6 well plate and grown to 50% confluency before infection. 5 μg/ml Polybrene (Sigma) was added to virus containing media. DMEM GlutaMAX (Gibco) supplemented with 10% (v/v) Fetal Bovine Serum (FBS, RBS Thermo scientific) was mixed with virus containing media in a 1:1 ratio before being added to HeLa229 cells which were then centrifuged at 1,500 rpm for 30 min at room temperature. Infected cells were incubated for 48 h and then the tissue culture medium was replaced with DMEM GlutaMAX (Gibco) supplemented with 10% (v/v) Fetal Bovine Serum (FBS, RBS Thermo scientific) containing 5 μg/ml puromycin (Sigma). Infected cells were selected with 5 μg/ml puromycin for 3 passages before use.
HEK293T, RAW264.7, HeLa229 and HeLa229 SEAP cells were cultured in T75 cm2 or T25 cm2 tissue culture flasks (Corning) in DMEM GlutaMAX (Gibco) supplemented with 10% (v/v) Fetal Bovine Serum (FBS, RBS Thermo scientific) in a tissue culture incubator under 5% CO2 at 37°C. All cell lines were kept and passaged to maximum of 40 times. HeLa229 cells, HeLa229 SEAP cells and HEK293T cells were split when cells were grown to 80 to 90% confluency. When splitting cells, cells were washed two times with 1 × PBS followed with 0.4 ml for T25 cm2 flask or 1.0 ml for T75 cm2 flask of 0.05% Trypsin-EDTA solution (Gibco, Life Technologies) treatment for 1 min at 37°C. 0.05% Trypsin-EDTA solution treated cells were resuspended with 10 ml of DMEM media supplemented with 10% FBS. RAW264.7 cells were washed two times with 1 x PBS and detached from tissue culture flasks with cell scrapers and diluted in fresh DMEM media supplemented with 10% FBS.
Cells were collected and placed on ice, 1xKal-B cell lysis buffer supplemented with protein inhibitors [50 mM Tris-HCl pH 7.4, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 10 mM NaF, 1 mM PMSF, 2 mM Na3VO4, and 1 x EDTA-free Complete protease inhibitor mixture (Roche)] was used to lyse harvested cells. Collected samples were placed on ice for 30 min, with pipetting up and down every 10 min to complete lysis cells. Cell lysates were pelleted at 13,000 rpm for 10 min. 4 × Bolt® LDS sample buffer (Life Technologies) and DTT (Astral scientific) at a final concentration of 50 mM were added to supernatants of samples and boiled at 70°C for 10 min. Boiled samples were loaded on Bolt® 4–12% Bis-Tris Plus gels (Life Technologies) and electrophoresis was performed according to manufacturer's instructions. Proteins were then transferred to nitrocellulose membranes using iBlot® nitrocellulose transfer stacks (Life Technologies) following manufacturer's protocol. Transferred nitrocellulose membranes were then blocked with 5% (w/v) skim milk in 1 x TBST buffer (20 mM Tris, 50 mM NaCl, 0.1% (v/v) Tween-20, pH 8.0) at room temperature with shaking at 60 rpm for 1 h. Blocked membranes were washed with 1 x TBST buffer 3 times, 5 min for each wash, before being probed with primary antibodies as required at 4°C, shaking at 60 rpm overnight. Primary antibodies are used as follows: mouse monoclonal anti-GFP (1:2000 dilution) (Roche, Basel, Switzerland), mouse monoclonal anti-Flag M2-HRP (1:2000 dilution, Sigma-Aldrich, St Louis, MO), or mouse monoclonal anti-β-actin (1:5000 dilution, Sigma-Adrich, St Louis, MO), or rabbit monoclonal anti-ArgGlcNAc (1:2000 dilution, Abcam, Cambridge, UK). All primary antibodies were diluted in 5% bovine serum albumin (Sigma-Aldrich) in 1 x TBST buffer. Membranes were then washed 3 times, for 5 min each, in 1 × TBST at room temperature with shaking at 60 rpm. Secondary antibodies, if required, were then incubated on membranes at room temperature with shaking at 60 rpm for 1 h. Secondary antibodies used in this study were horseradish peroxidase (HRP) conjugated anti-mouse (PerkinElmer), or HRP conjugated anti-rabbit (Bio-Rad). All secondary antibodies were diluted at 1:3000 with 5% bovine serum albumin (Sigma-Aldrich) in 1 × TBST buffer. Probed membranes were washed 7 times, for 5 min each, with 1 × TBST at room temperature with shaking at 60 rpm before being developed using ECL Prime Western blotting reagent (Amersham Bioscience) according to manufacturer's instructions. Immunoblots were developed using the Amersham Imager 680 blot and gel imager (GE Healthcare).
HEK293T cells were seeded in 24 well plates (Greiner Bio-One) at a concentration of 1 × 105 cells per well on a coverslip for immunofluorescence experiments or at 2 × 105 cells per well for secreted embryonic alkaline phosphatase assays. For immunoprecipitation, HEK293T cells were seeded in 10 cm cell culture dishes (Greiner Bio-One) at 4 × 106 cells per dish. Cells were seeded 1 day before transfection. On the day of transfection, FuGENE 6 Transfection Reagent (Roche) was added into Opti-MEM® I (1x) in GlutaMAX(TM)-I (Gibco, Life Technologies) and incubated for 5 min at room temperature before mixed with relevant plasmids according to manufacturer's instructions. Transfection mixtures were incubated for 20 min before added to seeded cells. Transfected cells were placed in 37°C incubator with 5% CO2 for 18 h incubation before harvesting for other experiments.
Harvested HEK293T cells were collected using 1 × KalB buffer supplemented with protein inhibitors as described above. Samples were placed on ice for 30 min, with pipetting up and down every 10 min, for complete cell lysis. Cell debris was pelleted at 13,000 rpm for 10 min at 4°C. Anti-Flag® M2 Magnetic Beads (Sigma-Aldrich) were washed 3 times, for 5 min each with 1 × Kal-B buffer before being loaded with cell lysates. Cell lysates were incubated with the beads with rotation at 4°C overnight. Flag-tagged proteins were eluted with 80 μl of 150 μg/ml Flag peptide (Sigma-Aldrich) with rotation at 4°C for 30 min. Eluted samples were processed for immunoblotting as described above.
For transfection on HEK293T cells, pSEAP vector was used to co-transfect HEK293T cells with vectors expressing AnkX, SseKs or their catalytic mutants following the transfection method described above. 16 h post transfection or infection, 200 μl DMEM GlutaMAX (Gibco) media was added to each well to replace old media and the 24 well plate was put back at 37°C with 5% CO2 for a further 8 h incubation. 24 h post transfection or infection, supernatants and cells lysates were collected for processing using Phospha-LightTM SEAP Reporter Gene Assay System (ThermoFisher Scientific) following manufacturer's instructions. Processed samples were plated on a 96-well white flat bottom plate and read via CLARIOstar Plus (BMG LABTECH).
RAW264.7 cells were seeded in 24 well plates at a concentration of 1 × 105 cells per well for cytometric bead array assay 1 day before infection. HeLa229 SEAP cells were seeded in 24 well plates at a concentration of 1 x 105 cells per well 1 day before experiment. Salmonella strains were inoculated in LB broth with relevant antibiotics 1 day before infection and grown at 37°C with shaking at 200 rpm overnight. On the day of infection, Salmonella strains were sub-cultured at a ratio of 1:100 in fresh LB broth with relevant antibiotics and grown at 37°C with shaking at 200 rpm for 3.5 h. OD600 of different sub-cultures were recorded using a spectrophotometer (SPECTRONICTM 200, Thermo ScientificTM) to estimate bacterial density. Sub-cultured Salmonella was diluted 10-fold in DMEM GlutaMAX (Gibco) media and added into seeded cells at a multiplicity of infection (MOI) of 10. The plates were centrifuged at 1,500 rpm for 5 min to synchronize the infection and incubated at 37°C with 5% CO2 for 30 min to allow for invasion. The cells were then washed twice with 1 x PBS and incubated with fresh DMEM GlutaMAX (Gibco) media supplemented with 100 μg/ml gentamicin and further incubated at 37°C with 5% CO2 for 1 h. After 1 h incubation, the cells were washed twice with 1 x PBS and incubated with fresh DMEM GlutaMAX (Gibco) media supplemented with 10 μg/ml gentamicin, and where necessary, a final concentration of 1 mM IPTG was added into the media. Infected cells were incubated at 37°C with 5% CO2 for various timepoints. For infection on HeLa229 SEAP cells, the media was replaced and 100 ng/ml doxycycline (Sigma) was used to induce SEAP expression where necessary; for cytometric bead array assays, the media was replaced on infected cells at 16 h post infection, and supernatants were collected at 20 or 24 h post infection when required.
Infection of HeLa229-SEAP and RAW264.7 cells with derivatives of Salmonella SL1344 was carried out as described above. Infected cells were collected for enumeration of bacteria at 2 and 24 h post infection. Infected cells were washed twice in PBS before being lysed for 5 min in 250 μl 0.1% Triton X-100 before been scraped and collected. Cell lysates were serially diluted in PBS, and then plated on LB agar plates containing 50 μg/ml streptomycin. Plated LB agar plates were incubated at 37°C overnight. Salmonella colonies were enumerated, and the colony-forming units (CFU) were calculated for each time point. Fold replication for each Salmonella strain was determined by dividing CFU at 24 h post infection by CFU at 2 h post infection.
Cells grown on coverslips were fixed with 4% paraformaldehyde in 1 × PBS for 10 min at room temperature at required time points following transfection. Cells were permeabilized using 0.2% Triton X-100 in PBS for 3 min, and then blocked with 3% BSA in 1 × PBS for 1 h. Primary antibodies were diluted at 1:200 in 3% BSA in 1 × PBS and cells were stained for 1 h at room temperature. Primary antibodies used for immunofluorescence were: monoclonal anti-Flag M2 (Sigma-Aldrich), polyclonal anti-Golgin 97 (Abcam, ab84830). The cells were then washed 3 times with 1 × PBS and incubated with fluorophore-conjugated secondary antibodies as required and supplemented with Hoechst stain (Sigma-Aldrich) for 1 h at room temperature in the dark. All secondary antibodies were diluted 1:2000 and Hoechst stain was diluted 1:4,000 in 3% BSA in 1 × PBS. Secondary antibodies used in this study were: Alexa Fluor 568 and Alexa Fluor 633 (Thermo Fisher Scientific). Samples were then washed 3 times with 1 × PBS and mounted using Prolong Gold mounting medium (Life Technologies). Confocal imaging was performed using the Olympus FV1200 Confocal.
HeLa229 cells seeded onto coverslips were infected with derivatives of Salmonella SL1344. Samples were fixed with 4% paraformaldehyde in 1 × PBS for 10 min at room temperature at 20 h post infection and prepared for immunofluorescence analysis as described above. Primary antibodies used in this experiment were: polyclonal anti-Golgin 97 (Abcam, ab84830), and anti-Salmonella CSA-1 (BacTrace, 5310-0322); secondary antibodies used were: Alexa Fluor 488 and Alexa Fluor 568 (Thermo Fisher Scientific).
RAW264.7 cells were infected with S. Typhimurium as above, with cell culture media replaced at 16 h post infection to focus on cytokine secretion at late stages of infection. Supernatants from RAW264.7 cells infected with various Salmonella strains were collected at 20 or 24 h post infection for analysis. Samples were processed for analysis using Cytometric Bead Array (CBA) Flex Sets (BD Biosciences) according to manufacturer's instructions. Samples were analyzed on the BD LSRFortessaTM cell analyzer.
Using a mass spectrometry-based approach on total cell lysates, we previously identified TNFR1 and TRAILR as targets of SseK3 during Salmonella infection of RAW264.7 cells (Newson et al., 2019). However, given that SseK3 exhibits strong Golgi localization during Salmonella infection (Gunster et al., 2017), we postulated that SseK3 may also target Golgi-associated membrane proteins. To test this, we enriched membrane fractions from Salmonella-infected RAW264.7 cells at 20 h post infection, and digested these to produce peptides that were then immunoprecipitated using an antibody that recognizes Arg-GlcNAc and then analyzed by mass spectrometry. To identify the Arg-GlcNAcylated targets of SseK3 we analyzed the peptides immunoprecipitated from RAW264.7 cells infected with S. Typhimurium ΔsseK12 and compared these to RAW264.7 cells infected with S. Typhimurium ΔsseK123. This approach revealed Arg-GlcNAcylation of previously identified SseK3 targets, TRAILR (Tnfrsf10b) and TNFR1 (Tnfrsf1a) (Newson et al., 2019), as well as novel host and bacterial protein Arg-GlcNAcylation events (Figure 1A, Supplementary Tables 1, 2). Consistent with the Golgi localization of SseK3, the Arg-GlcNAcylated targets identified were enriched for GO terms associated with Golgi-related biological processes including “ER to Golgi vesicle– mediated transport”, “Golgi vesicle transport” and “Retrograde transport, endosome to Golgi” (Figure 1B, Supplementary Table 3). Among the Golgi-related targets identified were several Rab GTPases, including Rab1, Rab5, and Rab11 (Figure 1A, Supplementary Tables 1, 2).
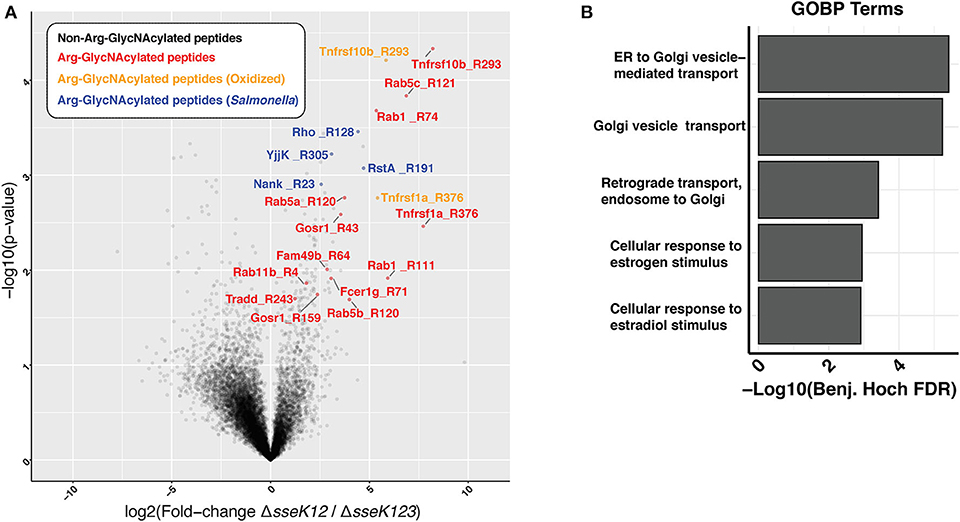
Figure 1. Enrichment of peptides Arg-GlcNAcylated by SseK3 derived from the insoluble fraction of Salmonella-infected RAW264.7 cell lysates. Arg-GlcNAcylated peptides were enriched and immunoprecipitated from the insoluble fraction of RAW264.7 cells infected with S. Typhimurium SL1344 ΔsseK12 or S. Typhimurium SL1344 ΔsseK123. (A) The volcano plot depicts the mean ion intensity peptide ratios of ΔsseK12 vs. ΔsseK123 plotted against the -logarithmic student t-test p-values from biological triplicate experiments. Arg-GlcNAcylated peptides with a fold change > 2 and a p < 0.05 are highlighted. Peptides are labeled by their gene names followed by the location of the Arg-GlcNacylated arginine. Arg-GlcNAcylated peptides from the host cell are highlighted in red; Methionine oxidized Arg-GlcNAcylated peptides from host cell are highlighted in yellow; Arg-GlcNAcylated peptides from Salmonella are highlighted in blue. (B) Bar chart of enrichment analysis of GO-terms (Biological processes) associated with Arg-GlcNAcylated peptides compared to all observed peptides from immunoprecipitation experiments. Fisher's exact enrichment analysis demonstrates the over-representation of Golgi-associated processes with Arg-GlcNAcylated proteins in the insoluble membrane immunoprecipitation.
Rab GTPases are master regulators of intracellular vesicle transport. Rab5 and Rab11 are regulators of early endosomes and recycling endosomes respectively; while Rab1 mediates vesicle transport from the ER to Golgi and can be found on Golgi membranes (Zhen and Stenmark, 2015; Prashar et al., 2017). To confirm that Rabs are modified by SseK3, we co-transfected HEK293T cells with 3xFlag-tagged Rab1a, Rab5a/b/c or Rab11b and GFP tagged SseK3, and then performed anti-Flag immunoprecipitation and subsequent immunoblot analysis using Arg-GlcNAc antibodies. GFP or the catalytically inactive glycosyltransferase motif mutant GFP-SseK3DXD were used as controls. We found that SseK3 does modify human Rab1a (Figure 2A), Rab5a/b/c (Figure 2B) and Rab11b (Figure 2C) with GlcNAc in transfected mammalian cells.
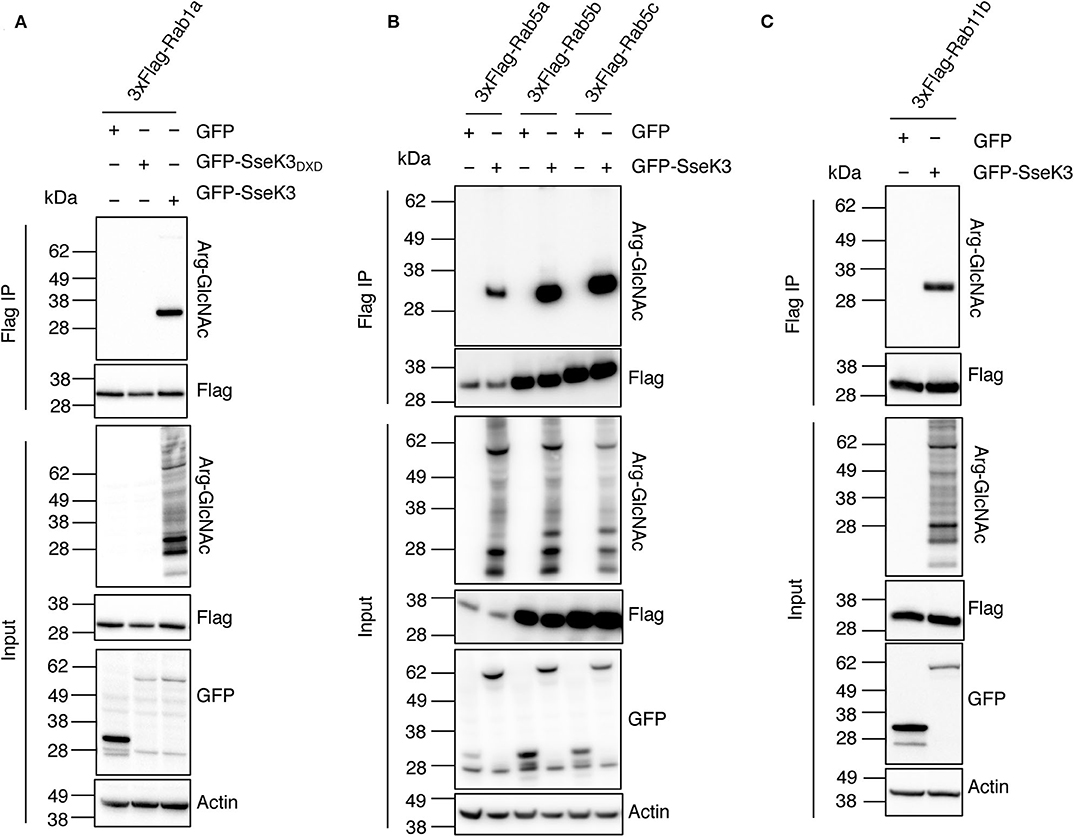
Figure 2. SseK3 modifies Rab1, Rab5, and Rab11 in co-transfected HEK293T cells. pEGFP-C2, pEGFP-C2-SseK3 or pEGFP-C2-SseK3DXD were co-transfected with either p3xFlag-Rab1a (A), p3xFlag-Rab5a/b/c (B), or p3xFlag-Rab11b (C) into HEK293T cells before 3xFlag-tagged proteins were immunoprecipitated. Input and immunoprecipitate (IP) were subjected to immunoblot analysis with anti-ArgGlcNAc, anti-Flag M2-HRP, anti-GFP or anti-β-actin antibodies. Representative immunoblots of at least 3 independent experiments.
Our mass spectrometry analysis revealed three different SseK3-mediated modification sites within Rab1 (corresponding to Arg74, Arg82, and Arg111 in Rab1a), whereas SseK3 modified single arginine residues in Rab5 and Rab11 respectively (Figure 1A, Supplementary Tables 1, 2). Strikingly, two of the Rab1 modification sites were located in the 74RTITSSYYR82 peptide within the catalytic switch II region (Figure 3A). In contrast, the Rab5 and Rab11 modification sites were not located in this region, occurring on Arg120 in the third α-helix and Arg4 at the N-terminus respectively, and the roles of these residues in Rab activity are unknown. The switch II region of Rab1 is a hotspot for post-translational modifications by different bacterial effectors to regulate Rab1 activity (Muller et al., 2010; Mukherjee et al., 2011; Wang et al., 2018). The Rab switch II region, in addition to the switch I region is involved in nucleotide binding, and shift from being unfolded in the GDP-bound state to adopting well-defined conformations in the GTP-bound state to allow for Rab interactions with host effector proteins (Zhen and Stenmark, 2015). Given the importance of the switch II region, we focussed on Rab1a modification by SseK3. To confirm the SseK3 modification sites within Rab1a, we mutated the arginine residues of interest to alanines, which cannot be GlcNAcylated. Individual 3xFlag-tagged Rab1 mutants were co-expressed in HEK293T cells with GFP-tagged SseK3 and then subjected to Flag immunoprecipitation and immunoblot analysis. We found mutation of the arginine residues individually did not significantly impact Arg-GlcNAcylation of Rab1 by SseK3 (Figure 3B). Mutating two residues at a time had a modest effect on blocking modification and mutating all of Arg74, Arg82, and Arg111 resulted in complete abrogation of Rab1a Arg-GlcNAcylation, suggesting SseK3 modifies each of these three arginine residues (Figure 3B). Rab1aR74AR82A was less efficiently modified by SseK3 compared to the other double site mutants, suggesting SseK3 may preferentially modify these two arginine residues located within the critical switch II region of Rab1a (Figure 3B). We then examined the intracellular localization of 3xFlag-tagged Rab1a arginine mutants by confocal microscopy. 3xFlag-tagged Rab1a or arginine mutants were expressed in HEK293T cells before the cells were stained with anti-golgin-97 antibodies for immunofluorescence analysis. 3xFlag-Rab1a showed a staining pattern consistent with Golgi localization (Supplementary Figure 1). All other mutants showed similar localization patterns, other than those containing a mutation of Arg82 (Supplementary Figure 1). All of 3xFlag-Rab1aR82A, 3xFlag-Rab1aR74AR82A, 3xFlag-Rab1aR82AR111A and 3xFlag-Rab1aR74AR82AR111A were instead expressed throughout the cell (Supplementary Figure 1).
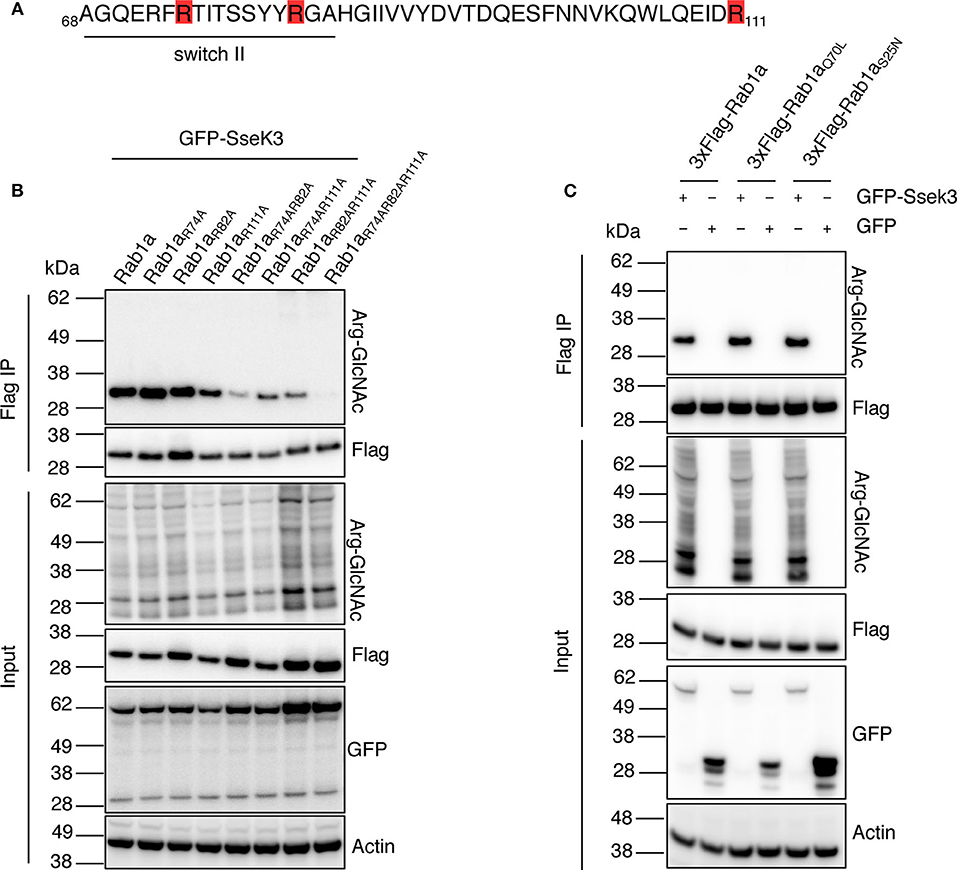
Figure 3. SseK3 modifies Arg74, Arg82, and Arg111 within Rab1a and has no preference for GTP-bound or GDP-bound Rab1a. (A) Amino acid sequence of Rab1a containing the arginine residues modified by SseK3 (highlighted in red) and the Rab1 switch II region (underlined). (B) GFP-SseK3 was co-expressed with 3xFlag tagged Rab1a arginine site-directed mutants in HEK293T cells. Flag-immunoprecipitation was performed on the cell lysates with subsequent immunoblot analysis using anti-ArgGlcNAc, anti-Flag M2-HRP, anti-GFP or anti-β-actin antibodies. Representative immunoblots of at least 3 independent experiments. (C) GFP or GFP-SseK3 together with 3xFlag tagged Rab1a or Rab1a nucleotide binding state mutants (active-state mimetic Rab1aQ70L, or constitutively inactive GDP-bound Rab1aS25N) were co-expressed in HEK293T cells by transfection. 3xFlag tagged proteins were immunoprecipitated for immunoblot analysis with anti-ArgGlcNAc, anti-Flag M2-HRP, anti-GFP or anti-β-actin antibodies. Representative immunoblots of at least 3 independent experiments.
Rab GTPases cycle between a GTP-bound active state and GDP-bound inactive state to mediate different steps of vesicle trafficking. The Rab switch regions undergo major conformational changes depending on the nucleotide binding state (Pfeffer, 2005). These changes may influence the ability of effectors to bind and modify the Rabs. For example, the Legionella glucosyltransferase effector, SetA preferentially modifies GDP-bound Rab1 (Wang et al., 2018), and an endogenous Rab1 regulator, TAK1, also preferentially phosphorylates the GDP-bound form of Rab1 (Levin et al., 2016). Thus, we explored whether SseK3 exhibited a preference for GTP-bound or GDP-bound Rab1a. A constitutively inactive 3xFlag-tagged GDP-bound Rab1aS25N (Nuoffer et al., 1994) or active-state mimetic Rab1aQ70L (Tisdale et al., 1992) were co-transfected with GFP-SseK3 into HEK293T cells before being subjected to Flag-immunoprecipitation and immunoblot analysis. No significant difference in Arg-GlcNAc modification was observed among Rab1a, Rab1aS25N or Rab1aQ70L (Figure 3C). Thus, modification of Rab1a by SseK3 was independent of the Rab1a nucleotide binding state. In this way, SseK3 functions differently in comparison to the endogenous Rab1 regulator TAK1, or the glucosyltransferase bacterial effector, SetA.
We next investigated the cellular localization of Rab1a in the presence of the SseKs. GFP, GFP-SseK1, GFP-SseK2, GFP-SseK3 or their catalytically inactive mutants were co-expressed with 3xFlag-tagged Rab1 in HEK293T cells before the cells were stained with anti-golgin-97 antibodies for immunofluorescence analysis. 3xFlag-Rab1a showed a Golgi-associated staining pattern in all samples tested, while both GFP and GFP-SseK1 were expressed throughout the cell (Figure 4A). Consistent with previous findings, GFP-SseK3 also localized to the Golgi in HEK293T cells (Figure 4A) (Gunster et al., 2017). Furthermore, we observed that GFP-SseK3DXD also localized to the Golgi with 3xFlag-Rab1a, indicating localization was independent of Arg-GlcNAcylation activity (Figure 4A).
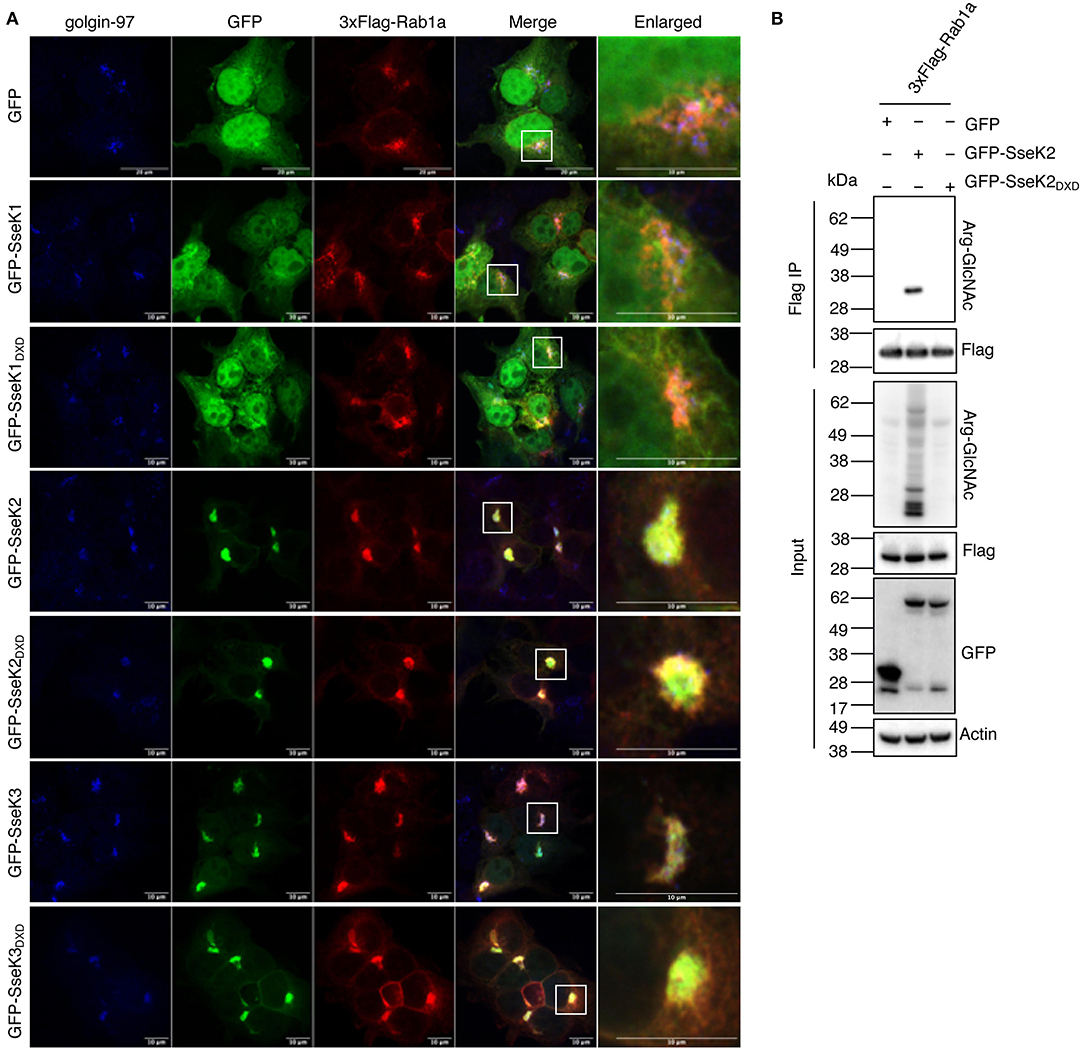
Figure 4. SseK2 modifies Rab1a and both SseK2 and SseK3 co-localize with Rab1a at the Golgi. (A) The intracellular localization pattern of 3xFlag-Rab1a was examined in the presence of GFP, GFP-SseK1, GFP-SseK2, GFP-SseK3 and their catalytically inactive mutants by confocal microscopy in transfected HEK293T cells. Anti-golgin-97 and anti-Flag antibodies were used to identify the Golgi and recombinant 3xFlag-Rab1a respectively. White boxes highlight Golgi that are shown enlarged. Representative immunofluorescence fields of at least 3 independent experiments (B) SseK2 also modifies Rab1a with Arg-GlcNAc. pEGFP-C2, pEGFP-C2-SseK2 or pEGFP-C2-SseK2DXD were co-transfected with p3xFlag-Rab1a into HEK293T cells before 3xFlag-tagged proteins were immunoprecipitated. Input and immunoprecipitate (IP) were subjected to immunoblot analysis with anti-ArgGlcNAc, anti-Flag M2-HRP, anti-GFP or anti-β-actin antibodies.
SseK2 also localizes to the Golgi during Salmonella infection (Gunster et al., 2017), and may therefore share targets with SseK3. We found that SseK2 and Rab1a also co-localized at the Golgi, and this was also independent of SseK2 catalytic activity (Figure 4A). This suggested that SseK2 may also target Rab1a for Arg-GlcNAcylation. Flag-immunoprecipitation and immunoblot analysis confirmed Arg-GlcNAcylation of 3xFlag-Rab1a by GFP-SseK2 in co-transfected HEK293T cells (Figure 4B).
Inhibition of Rab1 can lead to disruption of the Golgi (Dong et al., 2012). In our co-transfection studies we did not observe Golgi disruption (Figure 4A). However, we also examined the Golgi in cells that were not overexpressing Rab1, and found that after 20 h of infection with wild type S. Typhimurium SL1344, 15% of infected cells showed Golgi disruption, but this was independent of SseK2 or SseK3 (Supplementary Figure 2).
As Rab1 mediates vesicle transport from the ER to Golgi early in the secretory pathway, we explored the functional consequences of Arg-GlcNAcylation on Rab1 activity by testing whether the SseK family of proteins inhibited host protein secretion. We employed secreted embryonic alkaline phosphatase (SEAP) as a reporter to examine the activity of the secretory pathway in transfected cells. AnkX, a Legionella effector which significantly inhibits SEAP secretion (Mukherjee et al., 2011), was adopted as a positive control for the assay. Vectors expressing 3xFlag, 3xFlag-AnkX, GFP, GFP-SseK1, GFP-SseK2, GFP-SseK3 or their catalytically inactive DXD motif mutants were co-transfected into HEK293T cells with a SEAP expressing vector. As expected, expression of 3xFlag-AnkX significantly inhibited the secretion of SEAP in comparison to cells expressing 3xFlag only (Figure 5A). Expression of either GFP-SseK2 or GFP-SseK3 significantly inhibited the secretion of SEAP compared to GFP or GFP-SseK1 expressing cells (Figure 5A). Catalytically inactive SseK2 and SseK3 also partially inhibited the secretion of SEAP; however, this was not to the level of inhibition mediated by active SseK2 and SseK3 (Figure 5A).
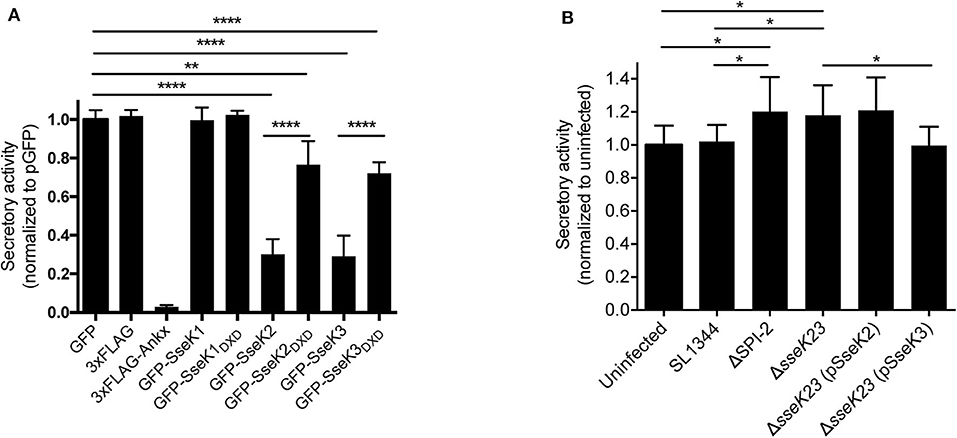
Figure 5. SseK3 inhibits the secretion of alkaline phosphatase in transfected HEK293T cells and during Salmonella infection. (A) pSEAP was co-transfected with mammalian expression vectors expressing GFP, 3xFlag, 3xFlag-AnkX, GFP-SseK1, GFP-SseK2, GFP-SseK3 or their catalytic mutants into HEK293T cells. Supernatants and cell lysates were then analyzed for alkaline phosphatase activity. Secretory activity was calculated as alkaline phosphatase activity in the supernatants divided by total alkaline phosphatase activity, which includes alkaline phosphatase activity in both supernatants and cell lysates. These were then normalized to the GFP expressing sample. Results are mean + SD of three independent experiments performed in duplicate. ****P < 0.0001, **P < 0.01; unpaired, two-tailed t-test. (B) HeLa229 cells stably expressing SEAP were infected with various Salmonella SL1344 strains. The cell culture media was replaced at 16 h post infection, and infection allowed to proceed for a further 8 h. Supernatants and cell lysates were analyzed for alkaline phosphatase activity at 24 h of infection. Secretory activity was calculated as alkaline phosphatase activity in the supernatants divided by total alkaline phosphatase activity, normalized to uninfected cells. Results are mean + SD of three independent experiments performed in duplicate. *P < 0.05; unpaired, two-tailed t-test.
We next explored whether SseK2 and SseK3 inhibited host protein secretion during S. Typhimurium SL1344 infection. A SEAP expressing HeLa229 reporter cell line was constructed and infected with derivatives of S. Typhimurium before a SEAP assay was performed. To analyse protein secretion at a timepoint when SseK2 and SseK3 are likely to be active, the cell media was changed at 16 h post infection, and infection allowed to proceed for a further 8 h before SEAP analysis at 24 h post infection. Wild type S. Typhimurium SL1344-infected cells showed similar levels of SEAP secretion compared to uninfected cells. Complex manipulation of host signaling pathways occurs during infection, thus SL1344 infection may simultaneously activate and interfere with host cell protein secretion. In support of this, modest increases in SEAP secretion were observed from cells infected with either ΔSPI-2 or ΔsseK23 Salmonella strains compared to cells infected with S. Typhimurium SL1344 or uninfected cells (Figure 5B). However, SEAP secretion returned to the levels observed for SL1344-infected cells only upon complementation of ΔsseK23 with SseK3, and not SseK2 when over-expressed from a plasmid (Figure 5B). Hence, although both SseK2 and SseK3 robustly inhibited the secretion of SEAP when transfected into cells, we were unable to confirm a role for SseK2 in inhibition of the host cell secretory pathway in the context of infection, and SseK3 had only a modest impact on SEAP secretion during Salmonella infection. To control for bacterial numbers in these experiments, we examined intracellular replication of S. Typhimurium SL1344 and its derivatives and found that compared to wild type SL1344, only the SPI-2 mutant showed significantly impaired replication in the HeLa229 SEAP cell line (Supplementary Figure 3A).
Given their potential effect on host cell protein export, we next explored whether SseK2 or SseK3 inhibited cytokine secretion during Salmonella infection. RAW264.7 cells were infected with wild type S. Typhimurium SL1344 or mutant derivatives for 16 h before the cell culture media was changed and then supernatants were collected and analyzed for cytokine levels at 20 and 24 h post infection using a cytometric bead array. Replication of the strains at 24 h post infection was also examined, with only the SPI-2 mutant showing impaired replication in RAW264.7 cells (Supplementary Figure 3B). Compared to uninfected cells, S. Typhimurium SL1344 infection resulted in increased cytokine secretion at both 20 and 24 h post infection (Figure 6). In contrast to the SEAP assay, ΔSPI-2 infection of RAW264.7 cells resulted in less IL-1α, IL-6 and GM-CSF cytokine secretion compared to wild type-infected samples at 24 h of infection and no significant differences were observed between wild type SL1344 and ΔsseK23 infected cells (Figure 6). A reduction in IL-1α and GM-CSF secretion was observed when SseK3 was over-expressed in the ΔsseK23 mutant compared to SL1344-infected cells, but not when compared to ΔsseK23-infected cells (Figure 6). In summary, during infection SseK3 appeared to have a greater effect on Rab1-dependent host protein secretion than SseK2, but overall SseK3 had only a marginal influence on the secretion levels of some cytokines when overexpressed.
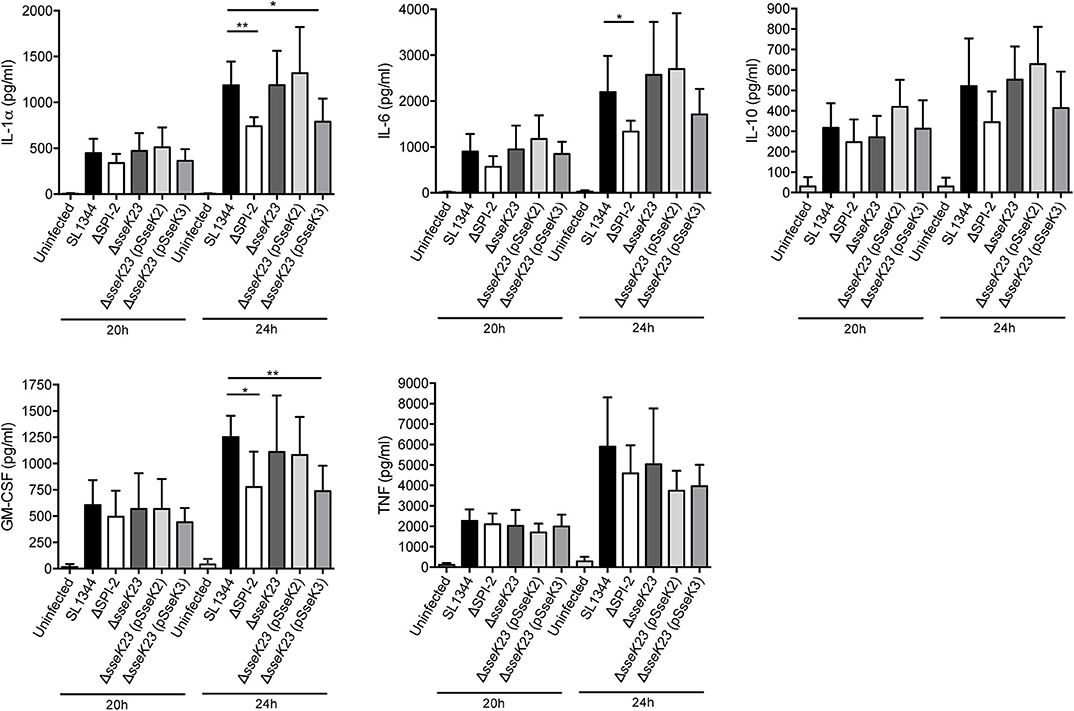
Figure 6. SseK3 does not inhibit the secretion of cytokines during Salmonella infection of RAW264.7 cells. Supernatants from RAW264.7 cells infected with wild type, ΔSPI-2, ΔsseK23, ΔsseK23 (pSseK2), or ΔsseK23 (pSseK3) S. Typhimurium SL1344 strains were collected for cytometric bead array analysis. After 16 h of infection, the cell culture media was changed, and supernatants were collected at 20 or 24 h post infection. Concentrations of cytokines in supernatants were determined based on fluorescence intensities from PE conjugated beads by flow cytometry compared to a standard curve and represented as pg/ml. Results are mean + SD of three independent experiments performed in duplicate. *P < 0.05, **P < 0.01; unpaired, two-tailed t-test.
Rab GTPases are well known for their role in mediating endocytosis and exocytosis as well as other intracellular membrane trafficking events. Many bacterial pathogens including Salmonella hijack Rab-dependent pathways to facilitate infection (Spano and Galan, 2018). For example, maturation of the SCV requires participation of several key intracellular vesicle transport regulators, including Rab5, Rab7, and Rab11 (Knodler and Steele-Mortimer, 2003; Brumell and Grinstein, 2004; Smith et al., 2005). Upon invasion, Salmonella modulates Rab recruitment to the SCV in a SPI-1-dependent manner (Smith et al., 2007). Rab5 is recruited to the SCV by the SPI-1 effector SopB, which is a phosphoinositide phosphatase (Mallo et al., 2008), while another SPI-1 effector, SopE, functions as a guanine exchange factor (GEF) for Rab5 and promotes the formation of GTP-bound active Rab5 on the SCV (Mukherjee et al., 2001). The enhanced retention of active Rab5 on the SCV is hypothesiszed to promote fusion with early endosomes and prevent trafficking to mature lysosomes (Parashuraman and Mukhopadhyay, 2005; Madan et al., 2008). Rab11 is also recruited to early SCVs following Salmonella invasion, and is involved in SCV maturation but is not essential for replication (Smith et al., 2005, 2007). Many studies of Rab manipulation by Salmonella focus on early time points of infection before the complete repertoire of SPI-2 effectors are expressed and translocated into host cells. Less is known about the impact of Salmonella on the host endosomal pathway at later stages of infection, although two SPI-2 effectors, SopD2 and GtgE are known to target Rabs including Rab32. SopD2 functions as a GTPase activating protein (GAP) for Rab32 (Spano et al., 2016) while GtgE is a cysteine protease that cleaves Rab32 (Spano and Galan, 2012, 2018; Wachtel et al., 2018). Together, these effectors inhibit recruitment of Rab32 to the SCV and subsequent Rab32-mediated control of replication (Spano and Galan, 2012; Spano et al., 2016).
In this study, we identified Rab1, Rab5, and Rab11 as host targets of the SPI-2 effector, SseK3 during Salmonella infection. These targets were identified by mass spectrometry and confirmed by immunoblot of ectopically expressed Rabs immunoprecipitated from HEK293T cells co-expressing SseK3. Using a SEAP reporter assay we found SseK3 impaired host protein secretion in transfected cells and modestly reduced secretion levels during Salmonella infection, suggesting Arg-GlcNAcylation of Rab1 by SseK3 at least partially blocked Rab1 activity. However, SseK3 did not appear to reduce the secretion of selected cytokines during Salmonella infection when expressed and translocated at native levels. Interestingly, inactive SseK3DXD and SseK2DXD also partially inhibited SEAP secretion in transfected cells, but the reason for this is unclear.
While this study was under review, another group also reported that Rab1 is Arg-GlcNAcylated by SseK3 (Meng et al., 2020). In contrast to our work, Meng et al. concluded that SseK3 did inhibit cytokine secretion during infection, while the impact of SseK2 on Rab1 and host protein secretion was not examined (Meng et al., 2020). Whereas Meng et al. performed infections with a triple mutant of S. Typhimurium SL1344 lacking all of sseK1, sseK2 and sseK3, we used a double sseK2/sseK3 mutant for our studies. Given that the strains we used retained SseK1, which together with SseK3 can inhibit inflammatory signaling through TNFR1 (Gunster et al., 2017; Newson et al., 2019), differences in the results observed by Meng et al. compared to our study could be due to altered cytokine expression levels due to the presence or absence of SseK1 (Meng et al., 2020). Thus, the SEAP reporter was a more direct measure of the ability of SseK3 to block the host cell secretory pathway as its expression was not influenced by inflammatory signaling. Although a previous study aimed at discovering Salmonella effector proteins that interact with host exocytic pathway failed to identify SseK2 and SseK3 as inhibitors of SEAP secretion, (Perrett and Zhou, 2013), this may have been due to insufficient expression of SseK2 and SseK3, which was not determined.
Two Rab1-targeting SPI-2 effectors, SseF and SseG, were recently reported to inhibit host autophagy during infection by abolishing Rab1 activation, indicating Rab1 is targeted by Salmonella once the SCV is established (Feng et al., 2018). It is not surprising that Salmonella employs multiple effectors to modulate different Rab1-related host cell events, considering intracellular pathogens need to counteract multiple host cell defense pathways. Previous studies on Legionella provide a good example of how intracellular pathogens utilize different effectors to modulate Rab1, even with contradictory impacts. During Legionella infection, spatio-temporal regulation of Rab1 activity is achieved largely via several post-translational modifications. For example the Legionella Dot/Icm effector SidM/DrrA AMPylates Tyr80 of Rab1 to retain it in the GTP-bound active state; while this post-translational modification is reversed by another effector, SidD (Muller et al., 2010; Mukherjee et al., 2011; Tan and Luo, 2011). Another Legionella effector, AnkX modifies Ser79 with a phosphocholine moiety and this modification is eliminated by a dephosphorylcholinase effector, Lem3 (Tan et al., 2011). Furthermore, a Legionella glucosyltransferase effector SetA modifies Thr75 of Rab1 with a glucose molecule and thus limits GTPase activity (Wang et al., 2018). Interestingly, most sites on Rab1 targeted for post-translational modification by different bacterial effectors, as well as the endogenous Rab1 regulator TAK1 (Levin et al., 2016), are located within the 74RTITSSYYR82 peptide of the switch II region, highlighting the susceptibility of this region to attack and/or regulation. Notably, we found SseK3 modified three different arginine residues in Rab1a, Arg74, Arg82 and Arg111, two of which were located within the Rab1 switch II region. Meng et al. identified the same SseK3-mediated Arg-GlcNAc modification sites within Rab1, and an additional modification at Arg72 when SseK3 was expressed from a multi-copy plasmid during S. Typhimurium infection (Meng et al., 2020).
We did not directly examine the activity of Arg-GlcNAc modified Rab1 in this study, however Meng et al. reported that SseK3-modified Rab1 had reduced GTPase activity, impaired interaction with binding partners and the membrane cycling of Rab1 was also perturbed (Meng et al., 2020). Our observations that SseK3 modified Rab1 regardless of its nucleotide binding state was also supported by Meng et al. (2020) and suggests that Arg-GlcNAc modified Rabs may also be present in the soluble fraction of S. Typhimurium-infected cells, as GDP-bound Rabs are not membrane bound (Zhen and Stenmark, 2015; Prashar et al., 2017). Indeed, we previously identified Arg-GlcNAc modified peptides from Rab1 and Rab5 in total cell lysates from RAW246.7 cells infected with S. Typhimurium ΔsseK12, although the modified Rab peptides were either not detected in all three biological replicates or were detected at lower levels compared to TNFR1 and TRAILR2 (Newson et al., 2019).
Similar to Meng et al., we found that SseK3 and SseK2 exhibited Golgi-associated localization with Rab1a, but that this was independent of Arg-GlcNAc activity. SseK3 localized to the cis-Golgi independently of its catalytic motif, with localization mediated by a polybasic region within SseK3 that binds phospholipids (Meng et al., 2020). However, whereas Meng et al. reported that ectopic expression of SseK3 caused fragmentation of the Golgi (Meng et al., 2020), we did not observe altered Golgi morphology when either SseK2 or SseK3 were translocated at native levels during S. Typhimurium infection. Thus the observations by Meng et al. could be artifacts of SseK3 over-expression, making the physiological relevance of SseK3-mediated Golgi disruption during infection unclear (Meng et al., 2020). This may also suggest that Rab1 is not a primary target of SseK3 during infection, or that levels of non-modified Rab1 are still sufficient to maintain Golgi structure. Notably, no host or pathogen factors have been reported to reverse Arg-GlcNAcylation (Scott et al., 2017). As such, the impact of Arg-GlcNAc modification on intracellular vesicle transport could still be significant over time even when only a small portion of target protein is modified. Further work is required to examine the effect of SseK3 on Rab5 or Rab11 function, as they may be preferred targets of SseK3 during S. Typhimurium infection. Interestingly, Meng et al. did not report Rab5 or Rab11 as significant targets of SseK3 during infection. Discrepancies between the targets of SseK3 identified may be due to the different approaches and cell lines used. Our Arg-GlcNAc pulldowns were performed on insoluble fractions of infected RAW246.7 cells, while Meng et al. identified modified proteins in cell lysates from transfected or infected HEK293T cells, or infected HeLa, iBMDM or MEF cells (Meng et al., 2020).
In addition to Rab1, Rab5, and Rab11, we identified several Salmonella proteins that were Arg-GlcNAcylated by SseK3 during infection, including the transcription termination factor Rho and a two-component regulatory system factor, RstA. The consequences of these modifications have not been investigated here, however it is worthwhile noting that NleB of enterohaemorrhagic and enteropathogenic E. coli and C. rodentium also have intra-bacterial Arg-GlcNAcylation activity, and this impacts bacterial survival in oxidative stress conditions (El Qaidi et al., 2020). NleB1, SseK1, and SseK3 also perform auto-Arg-GlcNAcylation, and this is required for activity against death domain protein targets (Xue et al., 2020).
In summary, we found that the Salmonella glycosyltransferase effector SseK3 modified Rab1, Rab5, and Rab11 during Salmonella infection. SseK3 targeted critical arginine residues in the switch II region of Rab1, thereby influencing host cell protein secretion during infection. Hence, in addition to its role in blocking death receptor signaling (El Qaidi et al., 2017; Gunster et al., 2017; Newson et al., 2019), SseK3 may modify the host cell secretome during the later stages of Salmonella infection.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JG designed and performed experiments and wrote the manuscript. NS and JN designed and performed mass spectrometry experiments. RW, TW, and GP made reagents and GN, ID, and JP provided experimental advice. CG and EH conceptualized and supervised the study and wrote the manuscript. All authors provided critical feedback and edited the manuscript.
This work was supported by National Health and Medical Research Council of Australia (NHMRC) project grants awarded to EH (APP1098826) and NS (APP1100164). JG was supported by a China Scholarship Council-University of Melbourne Ph.D. Scholarship. NS was supported by an Overseas (Biomedical) Fellowship (APP1037373). CG was supported by a Victoria Fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Nathaniel Brown (University of British Columbia) and Richard Strugnell (University of Melbourne) for bacterial strains and Craig Roy (Yale University) and John Silke (Walter and Eliza Hall Institute) for plasmids. We thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute at The University of Melbourne for the support of mass spectrometry analysis. The authors acknowledge Monash Micro Imaging, Monash University, for the provision of instrumentation, training and technical support. The use of a cytometer was supported by the Melbourne Cytometry Platform, The University of Melbourne.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00419/full#supplementary-material
Brown, N. F., Coombes, B. K., Bishop, J. L., Wickham, M. E., Lowden, M. J., Gal-Mor, O., et al. (2011). Salmonella phage ST64B encodes a member of the SseK/NleB effector family. PLoS ONE 6:e17824. doi: 10.1371/journal.pone.0017824
Brumell, J. H., and Grinstein, S. (2004). Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7, 78–84. doi: 10.1016/j.mib.2003.12.005
Clark, M. A., Jepson, M. A., Simmons, N. L., and Hirst, B. H. (1994). Preferential interaction of Salmonella typhimurium with mouse Peyer's patch M cells. Res. Microbiol 145, 543–552. doi: 10.1016/0923-2508(94)90031-0
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., and Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526. doi: 10.1074/mcp.M113.031591
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. doi: 10.1038/nbt.1511
Dong, N., Zhu, Y., Lu, Q., Hu, L., Zheng, Y., and Shao, F. (2012). Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029–1041. doi: 10.1016/j.cell.2012.06.050
El Qaidi, S., Chen, K., Halim, A., Siukstaite, L., Rueter, C., Hurtado-Guerrero, R., et al. (2017). NleB/SseK effectors from Citrobacter rodentium, Escherichia coli, and Salmonella enterica display distinct differences in host substrate specificity. J. Biol. Chem. 292, 11423–11430. doi: 10.1074/jbc.M117.790675
El Qaidi, S., Scott, N. E., Hays, M. P., Geisbrecht, B. V., Watkins, S., and Hardwidge, P. R. (2020). An intra-bacterial activity for a T3SS effector. Sci. Rep. 10:1073. doi: 10.1038/s41598-020-58062-y
Esposito, D., Gunster, R. A., Martino, L., El Omari, K., Wagner, A., Thurston, T. L. M., et al. (2018). Structural basis for the glycosyltransferase activity of the Salmonella effector SseK3. J. Biol. Chem. 293, 5064–5078. doi: 10.1074/jbc.RA118.001796
Feng, Z. Z., Jiang, A. J., Mao, A. W., Feng, Y., Wang, W., Li, J., et al. (2018). The Salmonella effectors SseF and SseG inhibit Rab1A-mediated autophagy to facilitate intracellular bacterial survival and replication. J. Biol. Chem. 293, 9662–9673. doi: 10.1074/jbc.M117.811737
Geddes, K., Cruz, F., and Heffron, F. (2007). Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog. 3:e196. doi: 10.1371/journal.ppat.0030196
Gunster, R. A., Matthews, S. A., Holden, D. W., and Thurston, T. L. M. (2017). SseK1 and SseK3 type III secretion system effectors inhibit NF-kappaB signaling and necroptotic cell death in salmonella-infected macrophages. Infect. Immun. 85:17. doi: 10.1128/IAI.00242-17
Hoiseth, S. K., and Stocker, B. A. (1981). Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239. doi: 10.1038/291238a0
Jennings, E., Thurston, T. L. M., and Holden, D. W. (2017). Salmonella SPI-2 Type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22, 217–231. doi: 10.1016/j.chom.2017.07.009
Kagan, J. C., Stein, M. P., Pypaert, M., and Roy, C. R. (2004). Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199, 1201–1211. doi: 10.1084/jem.20031706
Knodler, L. A., and Steele-Mortimer, O. (2003). Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4, 587–599. doi: 10.1034/j.1600-0854.2003.00118.x
Kujat Choy, S. L., Boyle, E. C., Gal-Mor, O., Goode, D. L., Valdez, Y., Vallance, B. A., et al. (2004). SseK1 and SseK2 are novel translocated proteins of Salmonella enterica serovar typhimurium. Infect. Immun. 72, 5115–5125. doi: 10.1128/IAI.72.9.5115-5125.2004
Kupz, A., Guarda, G., Gebhardt, T., Sander, L. E., Short, K. R., Diavatopoulos, D. A., et al. (2012). NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8(+) T cells. Nat. Immunol. 13, 162–169. doi: 10.1038/ni.2195
LaRock, D. L., Chaudhary, A., and Miller, S. I. (2015). Salmonellae interactions with host processes. Nat. Rev. Microbiol. 13, 191–205. doi: 10.1038/nrmicro3420
Levin, R. S., Hertz, N. T., Burlingame, A. L., Shokat, K. M., and Mukherjee, S. (2016). Innate immunity kinase TAK1 phosphorylates Rab1 on a hotspot for posttranslational modifications by host and pathogen. Proc. Natl. Acad. Sci. U.S.A. 113, E4776–E4783. doi: 10.1073/pnas.1608355113
Li, S., Zhang, L., Yao, Q., Li, L., Dong, N., Rong, J., et al. (2013). Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature 501, 242–246. doi: 10.1038/nature12436
Madan, R., Krishnamurthy, G., and Mukhopadhyay, A. (2008). SopE-mediated recruitment of host Rab5 on phagosomes inhibits Salmonella transport to lysosomes. Methods Mol. Biol. 445, 417–437. doi: 10.1007/978-1-59745-157-4_27
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O'Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Mallo, G. V., Espina, M., Smith, A. C., Terebiznik, M. R., Aleman, A., Finlay, B. B., et al. (2008). SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J. Cell Biol. 182, 741–752. doi: 10.1083/jcb.200804131
Meng, K., Zhuang, X., Peng, T., Hu, S., Yang, J., Wang, Z., et al. (2020). Arginine GlcNAcylation of Rab small GTPases by the pathogen Salmonella Typhimurium. Commun. Biol. 3:287. doi: 10.1038/s42003-020-1005-2
Mukherjee, K., Parashuraman, S., Raje, M., and Mukhopadhyay, A. (2001). SopE acts as an Rab5-specific nucleotide exchange factor and recruits non-prenylated Rab5 on Salmonella-containing phagosomes to promote fusion with early endosomes. J. Biol. Chem. 276, 23607–23615. doi: 10.1074/jbc.M101034200
Mukherjee, S., Liu, X., Arasaki, K., McDonough, J., Galan, J. E., and Roy, C. R. (2011). Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106. doi: 10.1038/nature10335
Muller, A. J., Kaiser, P., Dittmar, K. E., Weber, T. C., Haueter, S., Endt, K., et al. (2012). Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe 11, 19–32. doi: 10.1016/j.chom.2011.11.013
Muller, M. P., Peters, H., Blumer, J., Blankenfeldt, W., Goody, R. S., and Itzen, A. (2010). The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949. doi: 10.1126/science.1192276
Newson, J. P. M., Scott, N. E., Yeuk Wah Chung, I., Wong Fok Lung, T., Giogha, C., Gan, J., et al. (2019). Salmonella effectors SseK1 and SseK3 target death domain proteins in the TNF and TRAIL signaling pathways. Mol. Cell. Proteomics 18, 1138–1156. doi: 10.1074/mcp.RA118.001093
Nuoffer, C., Davidson, H. W., Matteson, J., Meinkoth, J., and Balch, W. E. (1994). A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol. 125, 225–237. doi: 10.1083/jcb.125.2.225
Parashuraman, S., and Mukhopadhyay, A. (2005). Assay and functional properties of SopE in the recruitment of Rab5 on Salmonella-containing phagosomes. Meth. Enzymol. 403, 295–309. doi: 10.1016/S0076-6879(05)03025-9
Park, J. B., Kim, Y. H., Yoo, Y., Kim, J., Jun, S. H., Cho, J. W., et al. (2018). Structural basis for arginine glycosylation of host substrates by bacterial effector proteins. Nat. Commun. 9:4283. doi: 10.1038/s41467-018-06680-6
Pearson, J. S., Giogha, C., Ong, S. Y., Kennedy, C. L., Kelly, M., Robinson, K. S., et al. (2013). A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 501, 247–251. doi: 10.1038/nature12524
Perrett, C. A., and Zhou, D. (2013). Salmonella type III effector SopB modulates host cell exocytosis. Emerg. Microbes Infect. 2:e32. doi: 10.1038/emi.2013.31
Pfeffer, S. R. (2005). Structural clues to Rab GTPase functional diversity. J. Biol. Chem. 280, 15485–15488. doi: 10.1074/jbc.R500003200
Prashar, A., Schnettger, L., Bernard, E. M., and Gutierrez, M. G. (2017). Rab GTPases in Immunity and Inflammation. Front. Cell. Infect. Microbiol. 7:435. doi: 10.3389/fcimb.2017.00435
Ramsden, A. E., Holden, D. W., and Mota, L. J. (2007). Membrane dynamics and spatial distribution of Salmonella-containing vacuoles. Trends Microbiol. 15, 516–524. doi: 10.1016/j.tim.2007.10.002
Rappsilber, J., Mann, M., and Ishihama, Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906. doi: 10.1038/nprot.2007.261
Scott, N. E., Giogha, C., Pollock, G. L., Kennedy, C. L., Webb, A. I., Williamson, N. A., et al. (2017). The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J. Biol. Chem. 292, 17337–17350. doi: 10.1074/jbc.M117.805036
Smith, A. C., Cirulis, J. T., Casanova, J. E., Scidmore, M. A., and Brumell, J. H. (2005). Interaction of the Salmonella-containing vacuole with the endocytic recycling system. J. Biol. Chem. 280, 24634–24641. doi: 10.1074/jbc.M500358200
Smith, A. C., Heo, W. D., Braun, V., Jiang, X., Macrae, C., Casanova, J. E., et al. (2007). A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J. Cell Biol. 176, 263–268. doi: 10.1083/jcb.200611056
Smith, J. L. (2003). The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J. Food Prot. 66, 1292–1303. doi: 10.4315/0362-028X-66.7.1292
Spano, S., and Galan, J. E. (2012). A Rab32-dependent pathway contributes to Salmonella typhi host restriction. Science 338, 960–963. doi: 10.1126/science.1229224
Spano, S., and Galan, J. E. (2018). Taking control: Hijacking of Rab GTPases by intracellular bacterial pathogens. Small GTPases 9, 182–191. doi: 10.1080/21541248.2017.1336192
Spano, S., Gao, X., Hannemann, S., Lara-Tejero, M., and Galan, J. E. (2016). A Bacterial Pathogen Targets a Host Rab-Family GTPase Defense Pathway with a GAP. Cell Host Microbe 19, 216–226. doi: 10.1016/j.chom.2016.01.004
Stewart, S. A., Dykxhoorn, D. M., Palliser, D., Mizuno, H., Yu, E. Y., An, D. S., et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. doi: 10.1261/rna.2192803
Tan, Y., Arnold, R. J., and Luo, Z. Q. (2011). Legionella pneumophila regulates the small GTPase Rab1 activity by reversible phosphorylcholination. Proc. Natl. Acad. Sci. U.S.A. 108, 21212–21217. doi: 10.1073/pnas.1114023109
Tan, Y., and Luo, Z. Q. (2011). Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475, 506–509. doi: 10.1038/nature10307
Tisdale, E. J., Bourne, J. R., Khosravi-Far, R., Der, C. J., and Balch, W. E. (1992). GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119, 749–761. doi: 10.1083/jcb.119.4.749
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. doi: 10.1038/nmeth.3901
Wachtel, R., Brauning, B., Mader, S. L., Ecker, F., Kaila, V. R. I., Groll, M., et al. (2018). The protease GtgE from Salmonella exclusively targets inactive Rab GTPases. Nat. Commun 9, 44. doi: 10.1038/s41467-017-02110-1
Wang, Z., McCloskey, A., Cheng, S., Wu, M., Xue, C., Yu, Z., et al. (2018). Regulation of the small GTPase Rab1 function by a bacterial glucosyltransferase. Cell Discov 4:53. doi: 10.1038/s41421-018-0055-9
Xue, J., Pan, X., Peng, T., Duan, M., Du, L., Zhuang, X., et al. (2020). Auto arginine-GlcNAcylation is crucial for bacterial pathogens in regulating host cell death. Front. Cell. Infect. Microbiol 10:197. doi: 10.3389/fcimb.2020.00197
Yamamoto, M., Okuyama, M., Ma, J. S., Kimura, T., Kamiyama, N., Saiga, H., et al. (2012). A cluster of interferon-gamma-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 37, 302–313. doi: 10.1016/j.immuni.2012.06.009
Keywords: Salmonella enterica, Rab, glycosyltransferase, protein secretion, host-pathogen interaction
Citation: Gan J, Scott NE, Newson JPM, Wibawa RR, Wong Fok Lung T, Pollock GL, Ng GZ, van Driel I, Pearson JS, Hartland EL and Giogha C (2020) The Salmonella Effector SseK3 Targets Small Rab GTPases. Front. Cell. Infect. Microbiol. 10:419. doi: 10.3389/fcimb.2020.00419
Received: 07 April 2020; Accepted: 08 July 2020;
Published: 19 August 2020.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Stephanie Rochelle Shames, Kansas State University, United StatesCopyright © 2020 Gan, Scott, Newson, Wibawa, Wong Fok Lung, Pollock, Ng, van Driel, Pearson, Hartland and Giogha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth L. Hartland, ZWxpemFiZXRoLmhhcnRsYW5kQGh1ZHNvbi5vcmcuYXU=; Cristina Giogha, Y3Jpc3RpbmEuZ2lvZ2hhQGh1ZHNvbi5vcmcuYXU=
†Present address: Joshua P. M. Newson, Department of Biology, Institute of Microbiology, ETH Zurich, Zurich, Switzerland
Tania Wong Fok Lung, Department of Pediatrics, Columbia University at the Columbia University Medical Center, New York, NY, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.