
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 11 August 2020
Sec. Clinical Microbiology
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00354
This article is part of the Research Topic Bacterial Vaginosis, a Model of True Polymicrobial Infections: Genetics, Evolution, Clinical and Socio-Clinical Implications View all 14 articles
A healthy female genital tract harbors a microbiome dominated by lactic acid and hydrogen peroxide producing bacteria, which provide protection against infections by maintaining a low pH. Changes in the bacterial compositions of the vaginal microbiome can lead to bacterial vaginosis (BV), which is often associated with vaginal inflammation. Bacterial vaginosis increases the risk of acquiring sexually transmitted infections (STIs) like human immunodeficiency virus (HIV) and affects women's reproductive health negatively. In pregnant women, BV can lead to chorioamnionitis and adverse pregnancy outcomes, including preterm premature rupture of the membranes and preterm birth. In order to manage BV effectively, good diagnostic procedures are required. Traditionally clinical and microscopic methods have been used to diagnose BV; however, these methods require skilled staff and time and suffer from reduced sensitivity and specificity. New diagnostics, including highly sensitive and specific point-of-care (POC) tests, treatment modalities and vaccines can be developed based on the identification of biomarkers from the growing pool of vaginal microbiome and vaginal metabolome data. In this review the current and future diagnostic avenues will be discussed.
Bacterial vaginosis (BV), or vaginal dysbiosis, is one of the most common vaginal conditions associated with aberrant changes in the vaginal microbiome (VMB) (van de Wijgert et al., 2014). Bacterial vaginosis is characterized by a reduction of the resident lactic-acid producing Lactobacillus spp. and an overgrowth of anaerobic bacteria (Hillier et al., 1993; Fredricks et al., 2005; Ravel et al., 2013). This condition poses a major public health concern for women of reproductive age, for their offspring and for their partners as BV is associated with adverse reproductive health outcomes such as pelvic inflammatory disease, miscarriage, preterm birth and may also lead to an increased risk of human immunodeficiency virus (HIV) acquisition and transmission (Hay et al., 1994; Ness et al., 2005; Sha et al., 2005a; Atashili et al., 2008; van de Wijgert et al., 2008).
Bacterial vaginosis can be diagnosed clinically by using the Amsel's criteria (Amsel et al., 1983) and in the laboratory using the Nugent scoring system (Nugent et al., 1991). Amsel's criteria were published in 1983 for use in clinical settings and at least three of four criteria need to be met before being classified as BV positive (Amsel et al., 1983). The Nugent scoring system is a refined version of the grading criteria introduced by Spiegel et al. (1983), where Gram-stained vaginal smears are evaluated for the presence/absence and quantity of specific bacterial morphotypes with a scoring system ranging from 0 to 10 (Nugent et al., 1991).
Although both methods have been widely used worldwide for almost three decades and are considered the “gold standard” of BV diagnosis, these methods are not free of limitations. For example, both methods are often subject to interobserver variability as the assessment of the diagnostic criteria depends on the observer's skill and experience (Klebanoff et al., 2004; Modak et al., 2011). Inaccurate BV results may lead to misdiagnosis and delays in treatment, which eventually puts women at risk for adverse reproductive health sequelae (Allsworth and Peipert, 2007; Modak et al., 2011). Misdiagnosis may also occur because of the unavailability of specific diagnostic tools in e.g., resource-limited settings and the deviation from strict diagnostic criteria by clinicians (Chavoustie et al., 2017). The complex etiology of BV contributes to inaccurate diagnosis, especially in asymptomatic carriers, and subsequently to poor treatment and clinical outcomes. The continuous development of accurate, easy-to-use point-of-care (POC) tests for BV is crucial, particularly in resource-limited settings.
The major drawbacks for the development of new diagnostic assays for BV are the lack of a uniform case definition for BV and that the etiology of BV remains poorly understood (Forsum et al., 2005; Muzny and Schwebke, 2016). In recent years, the advancement of molecular and high-throughput sequencing technologies [e.g., next-generation sequencing (NGS)] has revealed that BV is a multifactorial condition influenced by social, epidemiological, microbiological and host factors (Muzny and Schwebke, 2016; Muzny et al., 2020). Also, evidence suggests that the presence of a polymicrobial biofilm offers protection for many BV-associated bacteria against hydrogen peroxide (H2O2), antibiotics and host immunity (Swidsinski et al., 2005, 2008, 2015; Patterson et al., 2007) and these factors should be considered when designing a POC test. The focus of this review includes factors that complicate the diagnosis of BV, currently available diagnostic avenues and alternative approaches as potential diagnostic avenues for the diagnosis of BV.
The diagnosis of BV is complicated by the lack of consensus over what is defined as BV, the natural variation of the VMB in women of different racial backgrounds and the elusive polymicrobial etiology of BV. Several risk factors have been identified in the pathogenesis of BV, such as age, socioeconomical status, antibiotic usage, sexual behavior and ethnicity (Brumley, 2012; Singh et al., 2015; Ranjit et al., 2018). Lactobacillus bacteria are traditionally linked with a healthy VMB, but L. iners may be more pathogen than friend. These factors remain the reason that still no one method can accurately diagnose BV.
Historically, “non-specific bacterial vaginitis” was defined by Gardner and Dukes (1954) as an “infection” caused by a single etiological agent, Haemophilus vaginalis (now renamed as Gardnerella vaginalis), based on the observation that this bacterium was isolated from 92% (127/138) of women with “non-specific bacterial vaginitis.” In the following year, the authors argued that the direct inoculation of G. vaginalis in women “free of the disease” resulted in clinical manifestations of BV and pure cultures of G. vaginalis could be recovered from these women (Gardner and Dukes, 1955). However, in the very same study, this argument was not true in the majority of the tested “disease-free” women (77%; 10/13), and positive cultures were not obtained from these women (Gardner and Dukes, 1955). In confirmation of these findings, studies based on molecular and culture techniques have shown that G. vaginalis can be found in the vaginal tract of sexually inexperienced women and of women without BV (Nugent score 0–3) (Aroutcheva et al., 2001; Fethers et al., 2012; Balashov et al., 2014; Schwebke et al., 2014a; Janulaitiene et al., 2017). Consequently, the “single etiological agent” theory was not widely accepted (Hickey and Forney, 2014).
To address the controversy of the “single etiological agent” concept, alternative definitions of BV suggested that this condition is of polymicrobial nature. There are several proposed polymicrobial hypotheses under debate, mainly to answer three questions: (i) whether BV is initiated by the establishment of an “early colonizer species” like virulent strains of Gardnerella spp. and Prevotella bivia, which create a favorable vaginal environment to facilitate the adherence and the growth of the “secondary colonizer” species like Atopobium vaginae or Megasphaera type I; (ii) whether BV is initiated by the introduction of polymicrobial communities of BV-associated bacteria; or (iii) whether the displacement of Lactobacillus spp. happens before the establishment of “early colonizer species” or polymicrobial BV communities (Srinivasan and Fredricks, 2008; Schwebke et al., 2014b; Muzny et al., 2018, 2019). In addition, it is also unclear whether BV is caused by de novo polymicrobial biofilm formation or by the transmission of polymicrobial biofilms through sexual activity (Verstraelen and Swidsinksi, 2019). While there is some evidence supporting each hypothesis, these hypotheses explaining the polymicrobial BV theory have not come to a consensus yet. In support of the first question above, a study by Pybus and Onderdonk (1997) suggested that during the onset of BV, Gardnerella spp. may provide a favorable environment by producing amino acids to promote the growth of other BV-associated bacteria (e.g., P. bivia). Muzny et al. (2018) showed that the mean relative abundance of four “key bacteria” (i.e., P. bivia, Gardnerella spp., A. vaginae and Megasphaera type I) is significantly increased zero to four days before the onset of BV, indicating their role during the initiation of BV. However, relative abundances of certain Gardnerella spp. are not always associated with BV as no significant association was found between relative abundances of G. leopoldii and Nugent scoring category by Hill et al. (2019). Gardnerella leopoldii is a recently-identified species within the genus Gardnerella and is part of 13 genomic species found within the genus through whole genome sequence analysis and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis (Vaneechoutte et al., 2019). With regards to the second question above, Srinivasan and Fredricks (2008) have hypothesized that BV is triggered when the polymicrobial communities of BV-associated bacteria are introduced into the vaginal environment, which may lead to simultaneous displacement of lactobacilli. In support of this hypothesis, there is evidence that some strains of BV-associated bacteria (Bacteroides spp., Enterococcus faecium, G. vaginalis, Mobiluncus spp., and Peptostreptococcus spp.) are capable of inhibiting the growth of lactobacilli (Nagy et al., 1991; Kelly et al., 2003). Furthermore, according to a study by Swidsinski et al. (2014), the polymicrobial community of BV-associated bacteria could be transmitted by means of a polymicrobial biofilm (“clue cells,” characteristic vaginal epithelial cells coated with coccobacillary bacteria). This hypothesis, however, is contradicted by the studies that suggest other BV-associated bacteria are not as virulent as Gardnerella spp. in terms of cytotoxicity, adherence and biofilm formation (Patterson et al., 2010; Machado et al., 2013a; Alves et al., 2014).
Despite multiple molecular and genomic studies being conducted at species or microbiome level, there is no consensus on the group of bacterial species that may directly cause BV (Fethers et al., 2012; Srinivasan et al., 2012; Chen et al., 2018; Muzny et al., 2018). Srinivasan et al. (2012) performed broad-range 16S ribosomal RNA (rRNA) gene PCR and pyrosequencing to describe the composition and diversity of the VMB in women with BV, diagnosed either by Amsel's criteria or Nugent scoring. The results revealed that women with BV have a heterogeneous VMB that is not dominated by a single taxon and no species is universally present in all of BV-positive women (Srinivasan et al., 2012). It is supported by the findings of Chen et al. (2018) that significant differences in the bacterial composition of the BV microbiome were observed among BV groups defined by Amsel's criteria or Nugent scoring.
Vaginal inflammation is another controversial feature in BV. Some studies have reported that there was no statistically significant difference between any of the median concentrations of the pro-inflammatory cytokines [interleukin 6 (IL-6), IL-10, IL-12] tested for in BV-positive women vs. controls, and that high levels of prolidase and sialidase have led to the cleavage of the vaginal immunoglobulin A (IgA) and IgM (Cauci et al., 1998; Weissenbacher et al., 2010). A recent study using a mouse coinfection model also found that neither Gardnerella spp. or Prevotella bivia induced vaginal inflammation (Gilbert et al., 2019). In contrast, other studies have provided evidence that vaginal inflammation might well be present in BV and that specific BV-associated bacteria other than Gardnerella spp. and Prevotella spp. may induce proinflammatory responses (Sturm-Ramirez et al., 2000; Hedges et al., 2006; Nikolaitchouk et al., 2008; Anahtar et al., 2015; Jespers et al., 2017; Gardner et al., 2019; Gilbert et al., 2019). A few examples include a study by Sturm-Ramirez et al. (2000), in which women with BV had higher odds of having high levels of IL-1β or a tumor necrosis factor alpha (TNF-α) (adjusted odds ratio: 4.17; 95% confidence interval: 1.69–10.48; p = 0.002) and a study by Jespers et al. (2017), in which statistically significant increased concentrations of IL-12 were observed in women with incident BV. Similar observations were reported regarding other types of proinflammatory cytokines (IL-1α, IL-8, IL-36γ) (Pivarcsi et al., 2005; Nikolaitchouk et al., 2008; Gardner et al., 2019). Accordingly, a conceptual model was recently proposed by Muzny et al. (2020) in order to explain these observations. According to this model, epithelial cells and immune cells during a “healthy state” contribute to homeostasis by producing anti-inflammatory cytokines in response to low levels of cytokines produced by epithelial cells, whereas during early colonization by Gardnerella spp. and Prevotella bivia, sialidase is produced to induce a mucosal barrier disruption and allow the evasion of immune responses (Lewis et al., 2012; Gilbert et al., 2019; Muzny et al., 2020). Ultimately, secondary colonizers like A. vaginae and Sneathia spp. may join the polymicrobial biofilm and elicit the production of proinflammatory cytokines (Libby et al., 2008; Anahtar et al., 2015; Muzny et al., 2020). However, more research needs to be done to elucidate exact mechanisms of how the host-microbiota interactions contribute toward BV etiology.
Bacterial vaginosis has a complex pathogenesis and etiology and a disruption in the vaginal microflora is thought to be the main contributor to the altered vaginal environment and the associated clinical symptoms (Muzny and Schwebke, 2016). However, up to 50% to 75% of BV cases could be asymptomatic, complicating the diagnosis of BV and raising more questions on the etiology of this condition (Klebanoff et al., 2004; Coleman and Gaydos, 2018).
Traditionally, the vaginal environment of “healthy” women (referring to women who do not have adverse reproductive health outcomes and do not show clinical symptoms) were thought to be protected and maintained by hydrogen peroxide (H2O2)-producing Lactobacillus spp. via the production of H2O2, lactic acid and bacteriocins (Klebanoff et al., 1991; Zhou et al., 2004; O'Hanlon et al., 2011; Ravel et al., 2011). A lack or a decrease in the number of Lactobacillus spp. was therefore considered to indicate vaginal dysbiosis and this assertion formed the basis for the Nugent scoring system, which regards the lack of the Lactobacillus morphotype and the abundance of Gram-variable rods and cocci (G. vaginalis and Bacteroides spp. morphotypes) as BV (Nugent score of 7–10; Klebanoff et al., 1991; Nugent et al., 1991; Hillier et al., 1993). Indeed, a large cross-sectional study in the US by Ravel et al. (2011) showed that the VMB of 396 “healthy” women [representing four different ethnic groups (white, black, Hispanic, and Asian)] consisted of five “community state types” (CSTs), in which four CSTs were dominated by four Lactobacillus species each (L. crispatus, L. gasseri, L. iners and L. jensenii) (72.2%; 286/396). However, women with more heterogeneous CST (dominated by strict BV-associated anaerobic bacteria such as Prevotella spp., Dialister spp., Atopobium spp., Gardnerella spp., Megasphaera spp., Peptoniphilus spp., Sneathia spp., Eggerthella spp., Aerococcus spp., Finegoldia spp., and Mobiluncus spp.) were asymptomatic and therefore thought to still maintain a “healthy” vaginal environment (Ravel et al., 2011). This type of VMB was also observed in several other studies (Zhou et al., 2004; Hyman et al., 2005; Fettweis et al., 2014; Gautam et al., 2015). The reason why women with this CST can still maintain “healthy” vaginal environments is yet unknown, but it is possible that anaerobic bacteria like Atopobium spp., Leptotrichia spp. and Megasphaera spp. may substitute an ecological role in the absence of Lactobacillus spp. by e.g., producing lactic acid (Zhou et al., 2004). This substitution of an ecological role by anaerobic bacteria may partially explain why there is a high percentage of asymptomatic BV and may also contribute to the misdiagnosis of BV (Hickey et al., 2012). Therefore, with more information coming to light, the diagnosis of BV should not be focused on only the abundance of lactobacilli and other anaerobic bacteria.
The diagnosis of BV could be complicated by the fact that the vagina is a dynamic ecosystem that undergoes a natural fluctuation in the composition of the VMB throughout a woman's life, which is influenced by menstrual cycle, progesterone and estradiol levels, glycogen content in the vaginal epithelium, vaginal pH and immune responses (Brotman et al., 2010; Thoma et al., 2011; Gajer et al., 2012; Jespers et al., 2017; De Seta et al., 2019; Gliniewicz et al., 2019). At birth, it is believed that the VMB is established when the neonate is exposed to the vaginal tract of her mother during vaginal delivery or to the skin bacteria during Caesarian section (Dominguez-Bello et al., 2010). During this time, the VMB of the neonate delivered through the vaginal tract resembles that of her mother, dominated by either Lactobacillus spp., Prevotella spp. or Sneathia spp. (Dominguez-Bello et al., 2010). When the neonate gets older, the vaginal epithelium becomes thinner (i.e., contains a lower glycogen content) and the vaginal pH becomes neutral due to the decreased number of Lactobacillus spp. producing lactic acid (Farage and Maibach, 2006a). These physiological changes trigger the established VMB to undergo a compositional change from a Lactobacillus spp.-dominated VMB to a VMB dominated by strict anaerobes (e.g., Bacteroides fragilis) or enteric bacteria (e.g., Escherichia coli; Hammerschlag et al., 1978a,b). The vaginal pH and the number of Lactobacillus spp. seem to recover in adolescents and in women of reproductive age as the vaginal epithelium thickens (i.e., the glycogen level rises) under the estrogen influences (Eschenbach et al., 2000; Alvarez-Olmos et al., 2004; Farage and Maibach, 2006b). At this time (reproductive age), the VMB can undergo a natural fluctuation in composition during the menstrual cycle—some women may have a more resilient VMB while others have more extensive fluctuation over short periods of time (Gajer et al., 2012).
The role of ethnicity and race in affecting patterns of the vaginal microbiome have been considered because of the higher prevalence of BV in African-American and Mexican-American women (51.5 and 32.1%, respectively) in comparison to white, non-Hispanic women (23.2%; Alcendor, 2016). Some women (especially of African ethnicity) may also harbor a more heterogeneous VMB consisting of strict anaerobes like Anaerococcus spp., Atopobium spp., Corynebacterium spp., Finegoldia spp., Gardnerella spp., Megasphaera spp., Prevotella spp. and Streptococcus spp., along with Lactobacillus spp. (Zhou et al., 2007; Gajer et al., 2012; Fettweis et al., 2014). This type of VMB is similar to the VMB observed in postmenopausal women, in which the numbers of Lactobacillus spp. are reduced but those of strict anaerobes like Anaerococcus spp., Atopobium spp., Finegoldia spp., Gardnerella spp., Prevotella spp. and Streptococcus spp. are increased (Brotman et al., 2018; Gliniewicz et al., 2019).
The VMB is typically characterized by the dominance of a single or a few Lactobacillus species such as L. crispatus, L. gasseri, L. jensenii, L. iners and to a lesser extent species such as L. acidophilus, L. brevis, L. delbrueckii, L. fermentum, L. mucosae, L. paracasei, L. plantarum, L. reuteri, L. rhamnosus and L. vaginalis are also VMB colonizers (Antonio et al., 1999; Pavlova et al., 2002; Tärnberg et al., 2002; Ravel et al., 2011). If characterization of the VMB is considered as part of the diagnostic criteria for BV, understanding the relevance and functional role of different Lactobacillus species is needed. The identification of different types of lactobacilli in the VMB may be relevant to understand the stability of the VMB and the host's susceptibility to pathogens (Spear et al., 2011). Lactobacillus iners has been detected in the VMB of both BV positive and BV negative women, raising questions about the role of L. iners in the etiology of BV (Macklaim et al., 2011; Petrova et al., 2017). While the detection of G. vaginalis and A. vaginae have received the most attention in the etiology of BV, L. iners has been suggested as an unusual suspect in the etiology of BV and might play a critical role in the diagnosis of BV (Vaneechoutte, 2017).
The predominance of L. iners in an intermediate VMB or in symptomatic and asymptomatic BV might indicate its role in homeostasis and in promoting a lactobacilli-dominated microbial community in an altered vaginal environment (Ferris et al., 2004; Jakobsson and Forsum, 2007; Lambert et al., 2013). High levels of L. iners in BV may also refer to its genetic composition enabling optimal adaptation and survival in altered vaginal environments (Petrova et al., 2017). For instance, functional analysis in cases of BV revealed the expression of genes for the breakdown of glycogen, mannose and maltose (Macklaim et al., 2013). Shipitsyna et al. (2013) highlighted the importance of L. iners after finding L. iners and G. vaginalis as the predominant species in intermediate cases of BV. Another finding in the same study was the decline in the abundance of L. iners in cases of BV as opposed to levels in healthy women. The depletion of the abundance of L. iners together with the possibility that the predominance of L. iners may shift an intermediate VMB further toward dysbiosis contribute to the value of L. iners in the diagnosis of BV (Verstraelen et al., 2009; Petrova et al., 2017).
In the study by Shipitsyna et al. (2013), the qualitative detection of L. iners (negative result) was determined to have a low sensitivity in the prediction of BV (7% sensitivity and 86% specificity) with also little discriminatory power between positive and negative BV cases. Microscopy techniques such as Gram-staining may also lead to false negative results due to different isolates of L. iners presenting with different bacterial morphologies than that previously described (De Backer et al., 2007; Petrova et al., 2017). Thus, far, the most reliable method to include L. iners in the diagnosis of BV would be a quantitative method, including sequencing of the 16S rRNA gene that has previously enabled a comparison of prevalence, bacterial load and relative abundance of BV associated bacteria (Shipitsyna et al., 2013). Together with the relative abundance of L. iners, different ethnic groups with known differences in the abundance of L. iners in the VMB should also be considered when BV is suspected. Especially in cases involving Black African and African-American women where L. iners has been strongly associated with symptomatic and asymptomatic cases of BV (Ravel et al., 2011; Jespers et al., 2012; Srinivasan et al., 2012; Mitchell et al., 2013; Vaneechoutte, 2017).
Bacterial vaginosis is clinically characterized by Amsel's criteria or laboratory diagnosed by the Nugent score (Bautista et al., 2017). Clinical laboratories identify changes in the vaginal environment through microscopic examination and vaginal swab culture (Hong et al., 2016). Nugent scoring involves Gram-staining of vaginal smears and has been suggested as the gold standard in the diagnosis of BV in comparison to Amsel's criteria, which is based on non-quantifiable, non-reproducible clinical symptoms only (Chawla et al., 2013; Amegashie et al., 2017; Antonucci et al., 2017; Coleman and Gaydos, 2018). The diagnosis of BV is made on quantification of the Gram-stained microorganisms, classifying these organisms based on different vaginal morphotypes as well as the identification of clue cells (part of the Amsel's criteria) which could be laborious and requiresskilled personnel (Nugent et al., 1991; Chawla et al., 2013; Antonucci et al., 2017).
Amsel's criteria involves saline microscopy and has been improved over time to include the presence of a thin watery homogenous discharge, elevated vaginal pH (>4.5), the presence of more than 20% of clue cells (vaginal epithelial cells) and a fishy odor after the addition of 10% potassium hydroxide to vaginal secretions (“whiff test”) for a positive BV diagnosis (Amsel et al., 1983; Eschenbach et al., 1988; Mohammadzadeh et al., 2014). In comparison to Nugent scoring, the sensitivity and specificity for Amsel's criteria ranges from 37% to 70% and 94% to 99%, respectively and with moderate reproducibility (Schwebke et al., 1996; Sha et al., 2005b). Previous studies determined that 37% to 54% of women with an intermediate Nugent scoring had BV according to the Amsel's criteria (Taylor-Robinson et al., 2003; Bradshaw et al., 2005). A combination of Amsel's criteria and Nugent scoring may be beneficial for an accurate diagnosis of BV due to an assessment on both clinical symptoms and microbial morphology.
Another diagnostic method based on Gram-stained vaginal smears, the Ison-Hay classification criteria was described in 2002, which allows simplified grading and characterization of the vaginal microflora based on the amount of lactobacilli morphotypes compared to Gardnerella spp. morphotypes (Ison and Hay, 2002; Chawla et al., 2013). A vaginal microflora with a Nugent score of 4 to 6, known as an intermediate score or intermediate microflora is classified in grade II by the Ison-Hay criteria (Amegashie et al., 2017; Antonucci et al., 2017). An intermediate microflora was initially thought as a transitional step between a normal vaginal microflora and BV, or vice versa, but remains an uncharacterized category and is a challenge in the diagnosis of BV due to unknown clinical implications (Verhelst et al., 2005; Menard et al., 2008). Microscopy may be desired by some clinicians above other laboratory tests such as molecular assays due to a shorter turn-around time; however, the identification of different morphotypes is subjective and diagnosis may be influenced by individual skills and experience (Chawla et al., 2013; Antonucci et al., 2017). The involvement of species such as A. vaginae, Ureaplasma spp. and Mycoplasma spp., that cannot be detected by using Gram staining techniques or Nugent score, subsequently decreases the sensitivity of the Nugent score and warrants the need of a confirmatory or an additional molecular test to measure other etiological agents in the diagnosis of BV (Menard et al., 2008; Haggerty et al., 2009).
Several POC diagnostic assays exist to diagnose BV, such as saline microscopy, wet mount microscopy, chromogenic tests such as the OSOM® BVBlue® and the VGTest™ ion motility spectrometry (IMS). The VGTest™ IMS determines the levels of the malodorous biogenic amines associated with BV, whereas the OSOM® BVBlue® test detects elevated levels of the sialidase enzyme to diagnose BV (Madhivanan et al., 2014; Blankenstein et al., 2015). Rapid assays detecting the presence of proline amino peptidase in BV have also demonstrated high levels of specificity and sensitivity (Madhivanan et al., 2014). The combination of testing for vaginal pH and the whiff test is also suggested as a simple and inexpensive POC test for especially resource-limited settings (Madhivanan et al., 2009). The development of nucleic acid amplification tests (NAATs) such as the BD Affirm™ VPIII test can identify and differentiate between organisms associated with vaginitis and should be investigated to be used as a POC test (Cartwright et al., 2013).
Syndromic management is based on the identification of a combination of symptoms presented during a clinical examination (Shrivastava et al., 2014). These symptoms may be easily recognized signs associated with infection and known bacterial pathogens (Shrivastava et al., 2014). A diagnosis is made by a healthcare provider within a short time, without sophisticated skills and laboratory tests (Altini, 2006). Syndromic management of sexually transmitted infections (STIs) and BV has its benefits, especially in resource-limited settings where invasive procedures and laboratory tests are not available. These benefits include the amendment of immediate treatment on the patient's first visit to a clinic, (reducing further transmission of the disorder), it's widely accessible, education is provided, and no laboratory tests are needed (World Health Organization, 2007; Shrivastava et al., 2014). The success of syndromic management relies on accurate information including the sexual history of the patient and a thorough clinical examination (National Department of Health (South Africa), 2015). Bacterial vaginosis is diagnosed based on vaginal discharge syndrome (VDS) and is treated with a stat dose of metronidazole (National Department of Health (South Africa), 2015).
Despite the advantages that have been reported with syndromic management of STIs and BV, the shortfall of this approach includes not detecting asymptomatic STIs (that may be under treatment) and a poor positive predictive value where antibiotic susceptibility testing is not available, possibly resulting in the overuse of antibiotics (Garrett et al., 2018). In resource-limited settings, there are limited opportunities for routine surveillance, complicating the treatment and resolution of infections. Deviation from standard management guidelines by healthcare workers and the habit of relying on their own clinical judgement also contributes to insufficient treatment of vaginal infections (Leitich et al., 2003; Tann et al., 2006).
Syndromic management may lead to the misdiagnosis of BV and treatment failure and high rates of recurrencemay contribute to antibiotic resistance (Bostwick et al., 2016). A study conducted in South Africa investigating the prevalence of asymptomatic BV amongst a group of HIV unaffected pregnant women observed that 43% of BV positive (severe BV as indicated by a Nugent score of 9–10) women were asymptomatic (Joyisa et al., 2019). The high number of asymptomatic BV cases complicates syndromic management. Diagnosis and effective treatment of BV in pregnant women are warranted to avoid the development of pregnancy complications as well as the acquisition of HIV and other STIs. To diagnose and treat asymptomatic BV, low-cost but sensitive and specific diagnostic models for resource-limited countries are needed in addition to a proposed shift from syndromic to diagnostic management (Shrivastava et al., 2014; Garrett et al., 2018).
Rapid and accurate molecular methods targeting the nucleic acid have changed the diagnosis of BV and also provided new insight into our understanding of BV. The report that Gardnerella vaginalis is not just one species and does not only comprise one strain type but that several genomic species can be distinguished in the Gardnerella genus (Vaneechoutte et al., 2019) has emphasized how limited our understanding of bacterial vaginosis is and how the use of current and advanced technologies will shape our understanding of this condition in the future. Next generation technologies, including genomics, metabolomics, proteomics and immunomics shed light on the functional and immune processes during healthy, intermediate and BV states, that can ultimately contribute to the development of diagnostic tests, treatment and prevention strategies. The fourth industrial revolution brings about artificial intelligence and the possibility of diagnosing complex syndromes, like BV, using machine learning algorithms.
Given the limitations of Amsel's clinical criteria and microscopy for the diagnosis of BV, the development of molecular assays is an attractive approach for the diagnosis of BV because of the ability to identify and quantify multiple fastidious microorganisms (Adzitey et al., 2013). As described by Coleman and Gaydos (2018), an example of a molecular technique used in the diagnosis of BV is the direct probe assay (BD Affirm™ VPIII probe assay) that is commercially available (Becton Dickinson, Sparks, MD) and the Bacterial Vaginosis/Vaginitis panel developed by Quest diagnostics. Direct probe assays are rapid and can detect multiple indicator organisms in a single sample but may require high concentrations of the targeted bacteria for a reliable result. The detection of only G. vaginalis in direct probe assays is not a specific marker for BV (thus cannot be used to diagnose BV), as this species may be present in healthy women. The specific target used in these probe assays should be verified as 13 genomic species have been identified in the genus Gardnerella (Vaneechoutte et al., 2019). Other clinical indicators such as Amsel's criteria could be used in conjunction with probe assays to improve diagnostic accuracy and ensure reliable diagnosis (Spiegel, 1991; Fredricks et al., 2005; Zozaya-Hinchliffe et al., 2010; Janulaitiene et al., 2017). Direct probe assays designed for the detection of G. vaginalis might be more specific for the diagnosis of BV in symptomatic patients than in asymptomatic patients. A sensitivity and a specificity of 90% and 97%, respectively have been reported when compared to microscopy and Nugent scoring (94% and 81%, respectively; Coleman and Gaydos, 2018). Nucleic acid amplification tests are another molecular approach that is proven to have a higher diagnostic accuracy (20% to 25%) over direct probe assays, especially in populations with varying prevalence of infective vaginitis (Cartwright et al., 2013). Commercially available NAATs as indicated in Table 1 include the NuSwab® quantitative multiplex PCR assay, SureSwab BV DNA quantitative real-time PCR assay, BD Max vaginal panel and the BV multiplex assay with sensitivity ranging from 90.5% to 96.7% (the latter for symptomatic women) and specificity from 85.8% to 95% compared to Amsel's criteria and Nugent score as previously reported in detail by Coleman and Gaydos (2018). Figure 1 summarizes the main diagnostic tests and/or approaches that are currently available in the diagnostic arena for BV detection and approaches that could be developed for wider use in BV diagnostics.
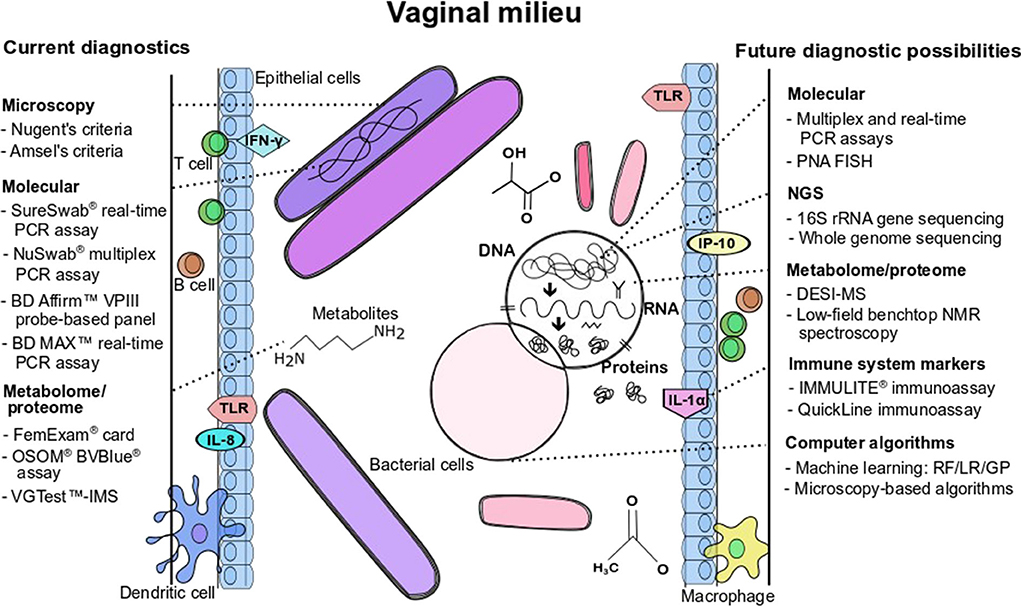
Figure 1. Current diagnostics and future possibilities for BV detection and/or BV pathogenesis characterization. Figure annotations in acronym or abbreviated form are: DESI-MS, desorption electrospray ionization-mass spectrometry; GP, genetic programming; IFN, interferon; IL, interleukin; LR, logistic regression; NMR, nuclear magnetic resonance; PCR, polymerase chain reaction; PNA-FISH, peptide nucleic acid fluorescence in situ hybridization; RF, random forest; TLR, toll-like receptor; VGTest-IMS, VGTest-ion mobility spectrometry.
Nucleic acid amplification tests are dependent on the selection of specific indicator organisms; however, multiplexed NAATs allow the detection of multiple indicator organisms in a single NAAT reaction (Adzitey et al., 2013). Not only is the selection of indicator organisms, but also the inclusion of the quantitative value that would indicate an accurate BV diagnosis, a challenge. Indicator species such as G. vaginalis and A. vaginae can also be commensals of a healthy VMB (Zozaya-Hinchliffe et al., 2010; Redelinghuys et al., 2017). The value of molecular quantification in the diagnosis of BV was investigated by Menard et al. (2008) who, similar to other studies, investigated the bacterial load to determine cut-off values for both G. vaginalis and A. vaginae (Wang and Su, 2014; Redelinghuys et al., 2017). Molecular quantification also enabled the characterization of an intermediate microflora corresponding to a BV-type vaginal microflora with high levels of G. vaginalis and A. vaginae (Bradshaw et al., 2006; Menard et al., 2008). Cut-off values for G. vaginalis and A. vaginae might improve BV diagnosis using molecular techniques; however, previous attempts to determine cut-off values most specific and sensitive for the diagnosis of BV have either taken individual species into consideration or a combination of the two species because of the suggested synergism (Redelinghuys et al., 2017). Consensus has not been reached due to variables such as different populations, study design and co-infection status such as HIV and pregnancy. As suggested by Redelinghuys et al. (2017), before a potential cut-off value can be used, it should first be validated with additional sets of data and in different study populations.
Commercially available molecular diagnostic panels for the diagnosis of BV include the detection of the Lactobacillus genus, but do not allow an extensive differentiation of species. Current diagnostic panels only allow differentiation of L. crispatus and L. jensenii (Table 1), but other studies have shown that L. vaginalis and L. iners also have been found in a population of African women with a low Nugent score (Jespers et al., 2015). In another study L. vaginalis was detected in 8% of Belgian women with BV (Jespers et al., 2012). Different species of the Lactobacillus genus interact differently with the vaginal environment, which may suggest that each species may contribute to the development of BV through a different mechanism (Jespers et al., 2015).
A VMB dominated by L. iners has been characterized as an unstable, transitional VMB and associated with vaginal dysbiosis (Borgdorff et al., 2016; Amabebe and Anumba, 2018; van Houdt et al., 2018). Janulaitiene et al. (2017) found that a healthy VMB was dominated by multiple Lactobacillus species, in contrast to BV positive samples where L. iners was the most frequently detected species, alone or in combination with L. crispatus. Other findings indicated that this particular species of Lactobacillus has been found in women with and without BV, indicating the challenge of using L. iners as a marker of BV (Zozaya-Hinchliffe et al., 2010).
The same concept may apply to G. vaginalis as marker of BV which has four different clades that each possess unique genetic markers and virulence factors (Ahmed et al., 2012). Clades 1 and 3 have been found more commonly among women with BV in contrast to clade 2, which has been associated with an intermediate vaginal microflora (Balashov et al., 2014; Coleman and Gaydos, 2018). Multiple clades have also been associated with BV, suggesting that a polyclonal G. vaginalis might be a risk for BV (Balashov et al., 2014). Genotypic differences between G. vaginalis strains could explain different clinical phenotypes such as with asymptomatic BV or the detection of G. vaginalis in healthy women (Ahmed et al., 2012). It is also of importance to take into consideration that carriers with a dense colonization of G. vaginalis, as seen with a high Nugent score, may remain asymptomatic for BV (Nugent et al., 1991; Ahmed et al., 2012). Due to the challenges of using a single species as a marker of BV and the shortcomings of using microscopy for identification of the VMB in BV, characterization of the VMB and its diversity may play an important role in the diagnosis of BV.
A study utilizing 16S rRNA PCR with clone analysis identified greater bacterial diversity in women with BV in contrast to women without BV, which is expected because of complete Lactobacillus dominance in a healthy VMB (Fredricks et al., 2005). This molecular technique enabled the identification of 35 unique bacterial species in women with BV by using the bacterium specific 16S rRNA gene (Fredricks et al., 2005). The question remains as to what the clinical significance of the identified species is, because species such as Atopobium vaginae and Gardnerella vaginalis, were more frequently detected in women with BV while other bacteria such as Peptostreptococcus spp., were less frequently detected in BV (Fredricks et al., 2005).
Molecular techniques utilizing the 16S rRNA gene for taxonomic identification also led to the identification of unculturable and fastidious microorganisms such as BV-associated bacterium 1 (BVAB1), BVAB2, and BVAB3 that were found to be specific indicators for BV (Fredricks et al., 2005). This technique also enabled a better understanding of bacterial morphotypes identified using Gram stain analysis (Srinivasan et al., 2013). Molecular amplification and sequencing of the 16S rRNA gene has its limitations, including the introduction of potential bias by broad-range amplification of the bacterial 16S rRNA gene (Winsley et al., 2012; Twin et al., 2013). Differences in taxa between studies can also be attributed to different sequencing approaches and to universal PCR primer mismatches to certain bacteria such as Bifidobacterium and G. vaginalis (Verhelst et al., 2004). Although the development of molecular techniques has revealed more bacterial species that form part of the pathophysiology of BV, threshold values and the appropriate number of marker bacterial species included on these panels to remain sensitive, specific and cost effective remain to be determined.
Development of a high-throughput technology such as next generation sequencing (NGS) allows the identification of multiple microorganisms that may be present in low abundance in a clinical sample (Hong et al., 2016). Traditional molecular approaches such as targeted Sanger sequencing of the 16S rRNA gene has enabled clinical identification of microorganisms in a polymicrobial sample when compared with traditional culturing methods (Salipante et al., 2013). However, high-throughput sequencing technologies such as targeted metagenomics on an NGS platform have the advantage of simplified identification of multiple microorganisms in a single sequencing approach, compared to interpreting multiple superimposed Sanger sequencing reads (Salipante et al., 2013).
To date, NGS techniques have not been extensively applied in the diagnosis of BV and therefore this section will mainly focus on the potential of the application in the diagnosis and treatment of BV. Next generation sequencing continues to improve the understanding of the complex pathology of BV by enabling the complete and accurate characterization of the diverse VMB associated with BV (Budd et al., 2015). Metagenomic analysis have indicated that bacterial communities associated with BV are highly diverse, either dominated by G. vaginalis, L. iners and/or combinations of anaerobic bacteria with or without Lactobacillus species in varying proportions (McKinnon et al., 2019). Deep sequencing of the bacterial 16S rRNA gene enabled multiple new bacterial associations with BV, providing new information on the possible role of Atopobium vaginae as indicator organism (Fredricks, 2011; Srinivasan et al., 2012; van de Wijgert et al., 2014).
NGS techniques can provide information on the relative proportion and absolute number of all incident bacterial species in the VMB (Ravel et al., 2011; Bostwick et al., 2016). The targeted NGS approach could identify mixed vaginal infections associated with BV and enables a comprehensive investigation of the VMB including the involvement of antibiotic resistance genes (Mullany, 2014). Targeted NGS has been evaluated by Bostwick et al. (2016) as an approach to diagnose and manage BV in clinical practice. The 16S rRNA sequencing approach was evaluated in a case of recurrent BV, where treatment with metronidazole was unsuccessful and a VMB with 56% of anaerobes was identified. In a second case, the patient immediately received treatment with metronidazole based on a positive diagnosis based on Amsel's criteria. The patient developed recurrent BV symptoms with a mixed BV VMB, a Candida albicans co-infection and antibiotic resistance. This patient had a VMB dominated by L. crispatus and L. iners. The use of broad-spectrum antibiotic treatment can induce shifts in the VMB from a healthy VMB often to a L. iners transitional VMB (Mayer et al., 2015). In the third clinical case, NGS results indicated a VMB composed of BVAB, L. iners and mixed anaerobic bacteria in a patient that was BV positive by Amsel's criteria and with no G. vaginalis present. The possibility of mixed bacterial infection is important to consider with the diagnosis of BV. With the NGS approach, the re-emergence of bacterial infection can also be studied, which may aid in the understanding of recurrent BV (Mayer et al., 2015; Bostwick et al., 2016).
Commercial tests that employ cost-effective, short-read 16S rRNA sequencing include the Ion Torrent 16STM Metagenomics kit (Thermo Fisher scientific, Waltham, MA, USA) in comparison to the Illumina 16S rRNA metagenomic sequencing (Illumina, San Diego, USA), which has been widely used for research purposes (Malla et al., 2019). Next-generation 16S rRNA sequencing has not been utilized in clinical microbiology practice in resource-limited laboratories because of sequencing cost, procedural challenges to prepare the sequencing libraries and the complexity of analysis (Salipante et al., 2013). Various laboratories have evaluated 16S rRNA sequencing for diagnostic purposes and developed analysis pipelines to characterize multiple hypervariable regions of the 16S rRNA gene (Barb et al., 2016; Watts et al., 2017; Culbreath et al., 2019). However, algorithms specifically for the diagnosis of BV is not yet commercially available. Similar to quantitative methods, the cut-off percentage of bacterial abundance related to clinical significance should also be determined. Standardization is needed in terms of the terminology of microbial communities, the methodologies to describe them, and standardization across sample site should also be considered. Targeted sequencing on the NGS platform has similar shortcomings as the 16S rRNA NAATs described earlier. To overcome limited identification of bacteria on species level, long-amplicon PCR-based approaches of the full 16S rRNA gene (~1,500 base pairs) with the Oxford Nanopore Technologies can be considered for accurate characterization of microbial communities and analysis of antibiotic resistance gene islands (Cusc et al., 2018; Malla et al., 2019).
As an alternative to 16S rRNA sequencing, whole genome sequencing (WGS) is another approach to investigate for the diagnosis and management of BV. Whole genome sequencing has the potential to generate a large amount of data from a single isolate, including species, strain type, virulence, antibiotic resistance and other information for outbreak and case management (Besser et al., 2018). Whole genome sequencing has been suggested to be used for microbial epidemiology, the surveillance of pathogens and outbreaks and for the prediction of antibiotic resistance (Jackson et al., 2019). Whole genome sequencing or targeted metagenomics (16S rRNA) might be useful for the diagnosis of BV in clinical cases where an intermediate VMB has been identified [Nugent score finding (score 4–6)]. As an intermediate VMB is proposed to represent a transitional state of the VMB to BV or vice versa, NGS techniques can be used to monitor slight changes in terms of abundance and composition of the VMB. The development of NGS technologies is ongoing and is suggested to be highly applicable in the future, where personalized medicine approaches will be possible for the treatment of specific bacterial infections (Punina et al., 2015; Jackson et al., 2019). The personalized medicine approach might also be applicable in BV because of a lack of consensus on the structure and composition of the VMB in this complex condition.
Proteomics is the study of the collection of proteins produced by the host and microbiome that embody the functional activity of the entire bionetwork (Peters et al., 2019). Metabolomics is the study of low molecular weight compounds (<1,500 Da) that are produced as substrates or by-products of enzymatic reactions in response to stimuli in a biological system (Peters et al., 2019). Compared to other components in an ecosystem, the metabolome may be the component that best represent the phenotype of that ecosystem (Guijas et al., 2018). Nonetheless, both proteomics and metabolomics are useful in studying host-microbiome interactions on a functional level and could be used to elucidate the pathogenesis of disease (Peters et al., 2019). Newer, high-throughput technologies have created the opportunity to screen and study metabolites and proteins on a large scale. High-throughput mass spectrometry (MS) and nuclear magnetic resonance (NMR) techniques have been used to explore the vaginal milieu under BV conditions and identify proteins or metabolites as biomarkers of this condition (Yeoman et al., 2013; McMillan et al., 2015; Ferreira et al., 2018; Parolin et al., 2018).
Although characteristic for its polymicrobial nature, the BV microbiome is typically associated with the production of specific metabolites, such as cadaverine, putrescine, tyramine and succinate, which have been linked to the “fishy” or amine odor and increased vaginal pH characteristic of BV (Yeoman et al., 2013; McMillan et al., 2015; Pruski et al., 2017). Lactic acid, for example, forms part of the metabolome signature of a healthy VMB (Stafford et al., 2017). Information on the proteome of cervical-vaginal fluid (CVF) is limited, but the protein content of CVF has been shown to differ between a BV microbiome and a healthy VMB (Zegels et al., 2009; Cruciani et al., 2013; Ferreira et al., 2018). Proteins associated with the immune response are either exclusive to BV or show elevated levels of expression (Cruciani et al., 2013; Ferreira et al., 2018). Distinctive metabolites and proteins could therefore be used as biomarkers of a healthy or BV microbiome, irrespective of the dominant bacterial species. A factor which complicates the diagnostic value of metabolomics and proteomics is that it could prove difficult to link the metabolites to the species producing them, which may or may not be essential for treatment purposes.
The evolution of diagnostics research has seen the field shift to evaluating a variety of combination criteria for the detection of BV. Laghi et al. (2014) investigated the microbiome and metabolome of women affected by BV. In a group of women treated with a placebo, this study found that high ratios of lactate to BV-associated metabolites and lactobacilli to BV-associated bacteria were likely to experience spontaneous remission in the absence of treatment, indicating that these may not have been true cases of BV. Another study suggested the coupling of acetate, malonate and nicotinate, quantified with proton (1H) NMR, with the VMB species Atopobium spp., M. hominis and Prevotella spp., quantified with qPCR, as diagnostic criteria for BV (Vitali et al., 2015). With the aid of liquid chromatography mass spectrometry (LC-MS), McMillan et al. (2015) have found that higher levels of 2-hydroxyisovalerate (2 HV) and gamma-hydroxy-butyrate (GHB), and reduced levels of lactate and tyrosine, were sensitive and specific for BV. In a validation cohort of 45 pregnant women, 2 HV: tyrosine ratios proved to be the most specific (94%) and sensitive (89%) for BV diagnosis (AUC = 0.946). The combination of a vaginal pH level of more than 4.5 with the detection of specific polyamines has been proposed as diagnostic criteria for BV diagnosis and to assess whether treatment is necessary (Watson and Reid, 2018). The use of high-throughput technologies to screen for biomarkers is still in its infancy (mostly regarding the ease of data analysis) and most of the proposed biomarkers have yet to be validated in different resource-limited settings where they could be implemented as POC tests for BV.
The number of available POC tests that have metabolites or proteins incorporated is limited. The commercial OSOM® BVBlue® test (Gryphus Diagnostics, LLC, Knoxville, TN) is a chromogenic rapid diagnostic test that is based on the detection of activity of the sialidase enzyme, an enzyme that is produced by BV-related bacteria such as Bacteroides spp. (Myziuk et al., 2003). Compared to Nugent's criteria, this test has a sensitivity and specificity of around 90% and 96%, respectively; compared to the Amsel criteria the sensitivity and specificity of this test range between 50% and 88%, and between 91% and 100%, respectively (Myziuk et al., 2003; Bradshaw et al., 2005). Although it is reported that a statistically significant correlation exists between a positive BVBlue® test and a raised vaginal pH, it has been found that combining these two criteria for the diagnosis of BV might improve either the sensitivity or specificity of the BVBlue® test, but at the same time could impair the alternative measure (Bradshaw et al., 2005). Another POC test, the FemExam® (Litmus Concepts, Santa Clara, CA) test, is similar to the Amsel's criteria as it measures vaginal pH and amine activity (West et al., 2003). The test is based on a two-card system where card 1 measures vaginal pH and the presence of trimethylamine, where card 2 measures the activity of proline iminopeptidase in G. vaginalis (West et al., 2003). West et al. (2003) reported a sensitivity of 91% and a specificity of 62% when the combined test results of the two cards were compared to Nugent's criteria. Both the BVBlue® and FemExam® tests are rapid, easy to perform and objective in comparison to the Amsel and Nugent's criteria. The sensitivity and specificity of these POC tests may be affected or compromised by the polymicrobial nature of BV and the wide spectrum of metabolites and proteins associated with its microbiome. Also, different study populations may harbor varying ratios of species to metabolites and/or proteins, which may or may not be produced in different concentrations.
Direct desorption electrospray ionization mass spectrometry (DESI-MS) analysis could be developed as a POC test as this technique was proven to be successful in identifying mucosal metabolite profiles from swabs that were collected in a clinical setting (Pruski et al., 2017). This technique could be used to explore the mucosal metabolite signature(s) associated with BV and help delineate the bacterial-host interactions at the vaginal mucosa during this condition. Another technology that could be optimized for the POC is low-field benchtop NMR spectroscopy, which involves the use of instruments that operate at frequencies below 100 MHz (Percival et al., 2019). Percival et al. (2019) successfully showed that LF NMR could detect and quantify biomarkers of type 2 diabetes in urine. This study therefore demonstrated the feasibility of the technology to do a biochemical screening of human biofluids.
Although several developing countries rely on the syndromic management of BV based on a vaginal discharge syndrome, Mlisana et al. (2012) reported that vaginal discharge and other symptoms are poor predictors of BV with a sensitivity of 10% and a specificity of 94.4%. This study found that asymptomatic women with one or more active STIs had subclinical inflammation and that these women may have increased levels of genital inflammation similar to those of women with symptomatic infections. Bacterial vaginosis has been associated with a range of upregulated genital pro-inflammatory cytokines (Masson et al., 2014; Kyongo et al., 2015) and women with BV may have an increase in the number of HIV target cells in the genital tract with higher expression levels of immune activation markers (Thurman et al., 2015; Gosmann et al., 2017).
The profiling of immune mediators in the genital tract of different groups of women has revealed variation in these profiles according to age, pregnancy status, gestational age and geographical region (Donders et al., 2003; Kyongo et al., 2015). The polymicrobial nature of BV complicates the assignment of a standard signature of immune mediators to a condition that is associated with divergent microbiome profiles. Different species or microbiome profiles may be associated with different immune mediators and/or different levels of inflammation and the risk of acquiring HIV. For example, L. crispatus has been associated with significantly lower levels, and G. vaginalis, A. vaginae and P. bivia with significantly higher levels, of the cytokines interleukin (IL)-1α, IL-1β and IL-12p70 (Kyongo et al., 2015). Women with a high-diversity VMB profile with abundant Prevotella spp. have been reported to have higher levels of interferon (IFN)-γ and IL-1β in cervicovaginal lavages (Anahtar et al., 2015). Also, young women with diverse VMBs dominated by anaerobes other than Gardnerella spp. have been reported to have elevated numbers of activated mucosal CD4+ T cells and an over four-fold higher risk of HIV acquisition compared to women with VMBs dominated by L. crispatus (Gosmann et al., 2017). In South African adolescent girls aged 16 to 22 years, Lennard et al. (2018) characterized the VMB and found three distinct microbiome subtypes, one of which was associated with extreme genital inflammation. The authors suggested that the inflammatory state associated with the BVAB1-dominated subtype may be chronic and that this subtype can be predicted with a sensitivity of 80% and specificity of 88%, based on a Nugent score of ≥9. The high sensitivity and specificity of high Nugent scores to predict this microbiome subtype might explain why high Nugent scores are representative of certain microbiomes and their associated symptoms but not of all VMB signatures. The Nugent scoring diagnostic system is therefore limited in the sense that it has to represent a complex and diverse VMB in terms of its immune mediator profile and bacterial species dominance. Considering the relationship between the VMB and its immune complement, the immune system is another avenue that could be explored for the development and expansion of BV diagnostics.
Microbiome profiles that represent dysbiosis may not always be detectable with conventional diagnostics. High-throughput technologies have enabled the characterization of the VMB on a grand scale and different bacterial species and microbiome profiles that have been associated with inflammation could therefore be easily characterized and used as indirect markers of an inflammatory state. However, characterizing the microbiome may reveal dysbiosis, but it is not an exhaustive approach to indicate the level of inflammation of the genital tract and therefore the risk of acquiring HIV and other STIs and developing reproductive complications (Mlisana et al., 2012; Masson et al., 2018). Consequently, the polymicrobial nature of BV and its association with divergent immune mediator profiles better support the use of direct markers of inflammation, such as immune mediators, as a diagnostic avenue. Compared with normal microflora, BV is often associated with varying levels of the immune mediators IL-1α, IL-1β, IL-6, IL-12(p70), IL-8, interferon gamma-induced protein (IP)-10, tumor necrosis factor (TNF)-β and secretory leukocyte protease inhibitor (SLPI) (Masson et al., 2014, 2016; Kyongo et al., 2015; Jespers et al., 2017).
In cervical lavage (CVL) samples, Masson et al. (2016) identified that the pro-inflammatory cytokine interleukin (IL)-1β and the chemokine interferon-γ-induced protein (IP)-10 could potentially be used at the POC to identify women with STIs or BV. This study reported that, according to STI and/or BV status, a model that comprised IL-1β (direct relationship between its concentration and BV) and IP-10 (inverse relationship between its concentration and BV) could correctly classify 75% of women with a sensitivity of 77% and a specificity of 72%. With the inclusion of another cytokine, IL-1α (direct relationship between its concentration and BV), the fit of the model was significantly improved (p = 0.0001) and the model correctly classified 76% of women with a sensitivity of 72% and specificity of 81%. The performance of the three cytokine biomarkers was validated by Masson et al. (2018) in a cohort of women who were recruited irrespective of vaginal symptoms and who were from four different geographical regions in Africa. The diagnostic model performed best with lateral vaginal swabs as genital specimen, correctly classifying 76% of women based on their STI, BV or intermediate VMB status with a sensitivity and specificity of 86% and 64% respectively. This study found that vaginal pH alone had a low specificity (61%) for classifying women according to their STI or VMB status. However, when vaginal pH was combined with the cytokine biomarkers, the accuracy of the diagnostic test was improved and 82% of cases were correctly identified (sensitivity of 86% and specificity of 64%). Interestingly, the authors also found that by combining pH with only IL-1α, the diagnostic model also correctly classified 82% of cases with a sensitivity and specificity of 86% and 68% respectively. Several other studies support the inverse relationship between IP-10 and BV and the direct relationship between BV and IL-1α and IL-1β (Deese et al., 2015; Kyongo et al., 2015; Jespers et al., 2017).
Immune mediators that are associated with a BV profile could be incorporated into diagnostic tests such as automated chemiluminescent enzyme immunoassays or lateral flow-based immunoassays (POC diagnostic assay). Kunze et al. (2016) measured the concentrations of IL-6 in amniotic fluid and TNF-α in vaginal secretions with the IMMULITE® system (Siemens Healthcare, Erlangen, Germany), an automated chemiluminescent enzyme immunoassay, and the QuickLine rapid test (Milenia Biotec, Gießen, Germany), a lateral flow immunoassay. A comparison between the two assays revealed a strong correlation for each marker with BV, with a Spearman correlation coefficient of 0.88 (p < 0.0001) for IL-6 and 0.86 (p < 0.0001) for TNF-α. The IMMULITE® system offers sensitive biomarker quantification, including an inflammation panel. The IMMULITE® 1,000 system is a small benchtop immunoanalyzer that is easy to use with a low cost of operation (Zirath et al., 2017). Chaemsaithong et al. (2015) compared the concentrations of IL-6 and IP-10 in amniotic fluid determined by standard enzyme-linked immunosorbent assay (ELISA) and a lateral flow-based immunoassay and reported that the results of the POC test strongly correlated with concentrations as determined by ELISA.
Although Amsel's criteria and the Nugent scoring system are considered as the “gold standard” for BV diagnosis, problems still exist because of the “interobserver variability” and the fact that the “intermediate vaginal microbiome” do not necessarily indicate disease progression to BV or vice versa (van de Wijgert et al., 2014). The advances in machine learning and its application in other fields have been followed by attempts to apply computer algorithms in BV diagnosis (Baker et al., 2014; Beck and Foster, 2014, 2015; Carter et al., 2014; Song et al., 2017; Jarvis et al., 2018).
Computer algorithms could potentially have a wide range of applications that may help clinicians and researchers to search for models that identify features relevant to BV diagnosis, to assess relative bacterial abundance data (qPCR) to diagnose BV, or to analyze bacterial morphotypes on microscope images for more accurate Nugent scoring results (Beck and Foster, 2014, 2015; Carter et al., 2014; Song et al., 2017; Jarvis et al., 2018). One of the first attempts to apply machine learning algorithms in BV diagnosis was done by Beck and Foster (2014), where the authors first grouped the correlations in microbial relative abundance data from studies by Ravel et al. (2011) and Srinivasan et al. (2012) and built different classification models (based on Amsel's criteria or Nugent scoring) using three different types of machine learning algorithms [“genetic programming” (GP), “logistic regression” (LR) and “random forest” (RF)]. The classification models using RF and LR obtained accuracy values between 90% and 95% when the models classified potential diagnostic features and bacterial groups associated with Nugent scoring, whereas the models using GP obtained slightly lower accuracy values (above 80%; Beck and Foster, 2014). However, the identified diagnostic features differed between all three machine learning algorithms and a technical difficulty was encountered when the models were deconstructed to identify which diagnostic features influenced the classification accuracy (Beck and Foster, 2014). These observations led to a follow-up study in 2015 where the changes in the accuracy values change were observed when each diagnostic feature was sequentially added to the classification models created by RF and LR (Beck and Foster, 2015). Instead of grouping the VMB according to microbial relative abundances, features were all ranked according to their importance in each classification model (Beck and Foster, 2015). The results revealed that a relatively consistent decrease in classification accuracy was observed as the RF classification model feature ranking decreases, while a more uneven decrease was observed for the LR model feature rankings, suggesting that the RF classification may be more useful in predicting BV (Beck and Foster, 2015). The top 15 important features for BV classification models based on Nugent scoring included several BV-associated bacteria (Prevotella spp., G. vaginalis, Dialister spp., A. vaginae, Megasphaera spp., Eggerthella spp., Sneathia spp., Peptoniphilus spp.) and clinical features like vaginal pH and “clue cells” (Beck and Foster, 2015).
Computer algorithms can also be incorporated into a molecular diagnostic tool to assess the relative abundance data of BV-associated bacteria for diagnosing BV. In a recent study conducted by Jarvis et al. (2018), a novel qPCR-based diagnostic tool (that detects ten BV-associated bacterial species and four Lactobacillus species) with a diagnostic machine learning algorithm (CLS2.0q) was developed. Evaluation of this molecular diagnostic tool with 172 women (149 symptomatic and 23 asymptomatic) resulted in a sensitivity of 93% and a specificity of 90% when compared with Nugent scoring (Jarvis et al., 2018). Although this molecular diagnostic tool currently requires further validation and improvements to become useful in clinical settings, it could be handy in POC settings.
Lastly, computer-based algorithms could be used to automate BV diagnosis based on microscopy results. In a study by Song et al. (2017), different approaches (“Bacteria Regions Segmentation,” “Overlapping Bacteria Clumps Splitting” and “Bacteria Morphotypes Learning”) were introduced to automate BV diagnosis based on the Nugent scoring system. The automated BV diagnosis procedures were as follows: (i) the selection of images from a slide; (ii) the generation of the superpixels by grouping pixels of different regions of bacteria; (iii) the calculation of superpixel contrast at the region level; (iv) the segmentation of “bacteria regions” by “saliency cut”; (v) the splitting of overlapping bacterial clumps; (vi) the classification of bacterial morphotypes through a machine learning algorithm; and (vii) BV diagnosis using the Nugent scoring system (Song et al., 2017). The experimental results using 105 vaginal smears revealed a sensitivity of 58.3% and a specificity of 87.1%, and the authors presumed the reason for low sensitivity was due to an inability of the algorithm to split heavily overlapped bacteria (Song et al., 2017). However, when the images were manually selected by human readers (experts) before the superpixel generation, sensitivity improved by up to 75% (Song et al., 2017). Again, this approach requires more research and improvements before it can be used in clinical settings, but it could be beneficial in future as it provides a more objective analysis of the vaginal smears and prevents “interobserver variability.”
Bacterial vaginosis can be diagnosed microscopically with the presence of “clue cells,” one of the four components of Amsel's criteria, but it was only in 2005, when Swidsinski et al. (2005) described the adherent biofilms found on these squamous vaginal epithelial cells. A spatial organization of bacteria associated with the vaginal epithelium shed new light on the etiology of BV but also led to the definition of BV as a synergistic polymicrobial syndrome with not only G. vaginalis playing an important role (Holst, 1990; Sobel, 2000; Swidsinski et al., 2005). Bacterial vaginosis is associated with a polymicrobial biofilm formed by BV-associated G. vaginalis and other BV-associated bacteria including A. vaginae, Mobiluncus mulieris and Prevotella bivia (Castro and Cerca, 2015; Castro et al., 2016, 2019; Hardy et al., 2016). This polymicrobial biofilm might be one of the reasons for the high recurrence rate of BV due to the protection of bacteria against H2O2, lactic acid, bacteriocins and antibiotics commonly used for treatment (such as metronidazole and tinidazole), but also provides resistance against the host immune system (e.g., prevent macrophage phagocytosis or chemotaxis; Al-Mushrif et al., 2000; Patterson et al., 2007; Alves et al., 2014; Swidsinski et al., 2015; Castro et al., 2019).
Biofilms composed mainly of G. vaginalis have the ability to adhere to vaginal epithelial cells, even in the presence of L. crispatus, and have also shown to benefit from the colonization of other BV-associated bacteria (A. vaginae, M. mulieris, P. bivia, Fusobacterium nucleatum; Machado et al., 2013a,b; Gottschick et al., 2017). Moreover, the adherence of G. vaginalis has been found to increase in the presence of L. iners, which could be due to the weak protective action exhibited by L. iners, although L. iners and L. crispatus have shown similar inhibitory effects against the adherence of BV-associated bacteria (Machado et al., 2013b). This investigation on the adherence of L. iners and G. vaginalis on vaginal epithelial cells confirmed that L. iners does not exhibit an antagonistic effect against G. vaginalis and suggested that the two species may be tolerant toward each other in the vaginal environment (Machado et al., 2013b). Such findings are important to fully understand biofilm formation and structure if the investigation thereof would be used for diagnostic and monitoring purposes and might especially be important in the adjustment of treatment in cases of a high recurrence rate.
Gardnerella vaginalis is known as the initial colonizer in the biofilm formation in BV, accounting for 60% of the bacterial composition. Initial colonization involves the attachment and formation of a biofilm (Machado and Cerca, 2015; Hardy et al., 2016). Transcriptomic analysis of G. vaginalis gene expression (e.g., virulence genes such as vaginolysin and sialidase) in biofilms indicated gene-regulated processes to result in a protected form of bacterial growth with high virulence and low metabolic activity (Castro et al., 2017, 2019). Gene expression of G. vaginalis transcripts encoding antimicrobial resistance proteins might be of particular interest, since a polymicrobial biofilm has been suggested to have greater antibiotic tolerance in contrast to a mono-species biofilm (Castro et al., 2019). This phenotype of G. vaginalis is another mechanism that may contribute to the recurrent nature of BV and may be a potential biomarker for biofilm formation and potential target for treatment remedies; however, extensive investigation on how such gene expression is influenced still needs to be done.
Both G. vaginalis and A. vaginae are accepted as important bacterial constituents of the BV biofilm, although single colonization of A. vaginae has not been found to initiate biofilm formation. Research findings highlighted higher bacterial loads when both species were present in a biofilm, in contrast to biofilms of only G. vaginalis, highlighting the synergistic relationship of these two species in biofilm formationto create favorable conditions for optimal growth and survival (Hardy et al., 2015). Such a synergistic relationship could also play a role in increased antibiotic resistance in BV and it has been demonstrated that different A. vaginae strains have different susceptibility profiles in vitro to metronidazole and secnidazole (Mendling et al., 2019). With the known increased recurrence rate of BV, antiseptics such as dequalinium chloride (DQC) (Fluomizin®) has been accepted as an alternative treatment for BV. This compound has antimicrobial activity against different pathogens (aerobic and anaerobic bacteria), especially against G. vaginalis and A. vaginae and has no known safety concerns and mechanisms of resistance (Weissenbacher et al., 2012).
Microbial communities, such as the VMB, is known as reservoirs for numerous antimicrobial resistance genes. Collectively these genes harbored in a microbial community is called the resistome, which can be studied by means of functional metagenomics and targeted (PCR-based) metagenomics, including real-time PCR and sequence-based metagenomics (Penders et al., 2013). Characterization of the resistome allows the investigation of present antimicrobial resistance (AMR) genes in a microbial environment, which might be useful in the investigation of the VMB and biofilms in BV for optimal treatment remedies. Although resistome research has mostly been done with the gut microbiome, and only aerobic biofilm samples of wastewater have been tested, the resistome might be an area of research that may give more insight into antibiotic resistance associated with BV (van Schaik, 2015; Relman and Lipsitch, 2018; Tian et al., 2019).
Another area that may be relevant to investigate in BV is microbiome-drug interactions, more recently termed pharmaco-microbiomics. In pharmaco-microbiomics, individual variation in drug response can be addressed by characterizing the composition of microbial communities and identify the chemical mechanisms in these microbiomes to understand drug metabolism (Guthrie and Kelly, 2019). A relationship has already been established between the VMB and levels of antiretrovirals (ARVs), for instance, in women with a VMB consisting of G. vaginalis and other anaerobic bacteria lower levels of tenofovir were detected than in women with a Lactobacillus-dominating microbiome (Donahue Carlson et al., 2017; Klatt et al., 2017). The finding was that a non-Lactobacillus microbiome rapidly depleted tenofovir before target cells could actively convert it to a pharmacologically active drug (Klatt et al., 2017). Therefore, tenofovir would be less effective in women with a non-Lactobacillus or BV type of microbiome. Besides the investigation of the metabolism of ARVs by microbial communities, the question is if such an effect would be suspected with the treatment of BV. Research regarding the metabolism of metronidazole or other antimicrobial drugs by the VMB has not been done to establish the efficacy of drugs with a BV type of microbiome. Moreover, research regarding the metabolism of drugs by biofilms in BV might contribute to additional treatment remedies or alteration of current treatment remedies such as initial drug concentration and mode of delivery.
For an optimal treatment approach for BV, the composition and structure of a biofilm can be investigated by using the fluorescence in situ hybridization (FISH) method followed by visualization with fluorescence microscopy, as described by Swidsinski et al. (2005) and Machado et al. (2013a). The presence and adherence of both G. vaginalis and A. vaginae can be investigated by using peptide nucleic acid fluorescence in situ hybridization (PNA FISH), which includes a specific PNA probe specifically selecting for the bacterial species of interest followed by DAPI staining to quantify cells in a mono- and dual-species biofilm (Freitas et al., 2017; Castro et al., 2019). A combination of confocal laser scanning microscopy (CLSM) and FISH has been recommended by Castro et al. (2019) to investigate the spatial distribution of a bacterial population and different architectures of the tested dual-species biofilms. Hardy et al. (2015) developed a PNA probe specific for A. vaginae (AtolTM1 probe), with a 67% sensitivity and 89.4% specificity as tested on clinical samples as compared to quantitative PCR (qPCR). The PNA FISH and qPCR can be used to determine the bacterial load of G. vaginalis and A. vaginae on vaginal epithelial cells, which is relevant in the prediction of the presence of a bacterial biofilm in BV (Hardy et al., 2015). Methods such as PNA FISH may be useful in cases of recurrent treatment failure, where it can be used to detect changes in biofilm composition and simultaneously monitor treatment efficacy. This method may therefore rather form part of a personalized treatment or precision medicine approach, especially in cases of high recurrence of BV with unsuccessful attempts of depletion of the biofilm structure in BV, and not as a method for standard diagnostic purposes.
The polymicrobial nature of BV necessitates the use of diagnostic tests that are based on combination criteria. Part of the challenge lies in determining which combination criteria are sensitive and specific enough as diagnostic criteria for BV; the other challenge is to develop cost-effective diagnostic tests, which could preferably be used at the POC. Although there is a complex interplay between vaginal pH and the concentration of different bacterial species, it is evident that vaginal pH, specifically higher than 4.5, improves the performance of diagnostic tests when combined with other components in CVF.
The efficient and accurate detection of vaginal dysbiosis has always been plagued by factors such as the difference in biomarker levels across populations (e.g., bacterial species) and sample type variations (Kyongo et al., 2015; Masson et al., 2018). Many studies discussed in this review have highlighted the potential of different combination criteria with biomarkers beyond the genetic level to improve BV diagnostics. If the purpose of diagnosis is to treat, the question should be asked whether a perfect equilibrium exists between the VMB and all its related components. That is, does bacterial concentration translate into corresponding levels of metabolites, proteins and inflammatory markers? Nonetheless, the continual reduction in operational costs of high-throughput technologies provide the opportunity to study the vaginal milieu with a systems biology approach on a large scale to map and link potential biomarkers. This review does not necessarily suggest the replacement of diagnostic tools currently available for BV but does highlight the limitations of these tools and calls for the expansion of the BV diagnostics field by exploring the vast array of diagnostic opportunities discussed here.
In many resource-limited settings, however, POC tests for BV are either not available or simply too expensive for routine diagnostic use and healthcare practitioners have to rely on syndromic management of vaginal discharge syndrome. It is therefore imperative that the development and evaluation of new diagnostic tests must include both a cost- and health-benefit analysis in various settings, especially where expensive instrumentation is required. The risk profiles of different populations for adverse sequelae of BV infection, such as increased risk for HIV infection and poor pregnancy outcomes should guide diagnostic test selection. In such at-risk populations, we have to ask the question—what is the cost of cost?
All authors contributed to concept design and layout of the manuscript. MR, JG, and HJ wrote the body of the manuscript and MK contributed as senior author. All authors listed approved the manuscript for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adzitey, F., Huda, N., and Ali, G. R. R. (2013). Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. Biotechnology 3, 97–107. doi: 10.1007/s13205-012-0074-4
Ahmed, A., Earl, J., Retchless, A., Hillier, S. L., Rabe, L. K., Cherpes, T. L., et al. (2012). Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J. Bacteriol. 194, 3922–3937. doi: 10.1128/JB.00056-12
Alcendor, D. J. (2016). Evaluation of health disparity in bacterial vaginosis and the implications for HIV-1 acquisition in African American women. Am. J. Reprod. Immunol. 76, 99–107. doi: 10.1111/aji.12497
Allsworth, J. E., and Peipert, J. F. (2007). Prevalence of bacterial vaginosis: 2001-2004 national health and nutrition examination survey data. Obstet. Gynecol. 109, 114–120. doi: 10.1097/01.AOG.0000247627.84791.91
Al-Mushrif, S., Eley, A., and Jones, B. M. (2000). Inhibition of chemotaxis by organic acids from anaerobes may prevent a purulent response in bacterial vaginosis. J. Med. Microbiol. 49, 1023–1030. doi: 10.1099/0022-1317-49-11-1023
Altini, L. (2006). Syndromic management of sexually transmitted infections. South African Pharm. J. 73:16.
Alvarez-Olmos, M. I., Barousse, M. M., Rajan, L., Van Der Pol, B. J., Fortenberry, D., Orr, D., et al. (2004). Vaginal lactobacilli in adolescents: presence and relationship to local and systemic immunity, and to bacterial vaginosis. Sex. Transm. Dis. 31, 393–400. doi: 10.1097/01.OLQ.0000130454.83883.E9
Alves, P., Castro, J., Sousa, C., Cereija, T. B., and Cerca, N. (2014). Gardnerella vaginalis outcompetes 29 other bacterial species isolated from patients with bacterial vaginosis, using in an in vitro biofilm formation model. J. Infect. Dis. 210, 593–596. doi: 10.1093/infdis/jiu131
Amabebe, E., and Anumba, D. O. C. (2018). The vaginal microenvironment: the physiologic role of Lactobacilli. Front. Med. 5:181. doi: 10.3389/fmed.2018.00181
Amegashie, C. P., Gilbert, N. M., Peipert, J. F., Allsworth, J. E., Lewis, W. G., and Lewis, A. L. (2017). Relationship between nugent score and vaginal epithelial exfoliation. PLoS ONE 12:e0177797. doi: 10.1371/journal.pone.0177797
Amsel, R., Totten, P. A., Spiegel, C. A., Chen, K. C. S., Eschenbach, D., and Holmes, K. K. (1983). Nonspecific vaginitis: diagnostic criteria and microbial and epidemiologic associations. Am. J. Med. 74, 14–22. doi: 10.1016/0002-9343(83)91112-9
Anahtar, M. N., Byrne, E. H., Doherty, K. E., Bowman, B. A., Yamamoto, H. S., Soumillon, M., et al. (2015). Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 42, 965–976. doi: 10.1016/j.immuni.2015.04.019
Antonio, M. A., Hawes, S. E., and Hillier, S. L. (1999). The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180, 1950–1956. doi: 10.1086/315109
Antonucci, F., Mirandola, W., Fontana, C., and Fontana, C. (2017). Comparison between Nugent's and Hay/Ison scoring criteria for the diagnosis of bacterial vaginosis in WASP prepared vaginal samples. Clin. Investig. (Lond.) 07, 89–93. doi: 10.4172/Clinical-Investigation.1000116
Aroutcheva, A. A., Simoes, J. A., Behbakht, K., and Faro, S. (2001). Gardnerella vaginalis isolated from patients with bacterial vaginosis and from patients with healthy vaginal ecosystems. Clin. Infect. Dis. 33, 1022–1027. doi: 10.1086/323030
Atashili, J., Poole, C., Ndumbe, P. M., Adimora, A. A., and Smith, J. S. (2008). Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS 22, 1493–1501. doi: 10.1097/QAD.0b013e3283021a37
Baker, Y. S., Agrawal, R., Foster, J. A., Beck, D., and Dozier, G. (2014). “Detecting bacterial vaginosis using machine learning,” in Proceedings of the 2014 ACM Southeast Regional Conference (Kennesaw, GA: ACM).
Balashov, S. V., Mordechai, E., Adelson, M. E., and Gygax, S. E. (2014). Identification, quantification and subtyping of Gardnerella vaginalis in noncultured clinical vaginal samples by quantitative PCR. J. Med. Microbiol. 63, 162–175. doi: 10.1099/jmm.0.066407-0
Barb, J. J., Oler, A. J., Kim, H.-S., Chalmers, N., Wallen, G. R., Cashion, A., et al. (2016). Development of an analysis pipeline characterizing multiple hypervariable regions of 16S rRNA using mock samples. PLoS ONE 11:e0148047. doi: 10.1371/journal.pone.0148047
Bautista, C. T., Wurapa, E. K., Sateren, W. B., Morris, S. M., Hollingsworth, B. P., and Sanchez, J. L. (2017). Association of Bacterial Vaginosis with chlamydia and gonorrhea among women in the US army. Am. J. Prev. Med. 52, 632–639. doi: 10.1016/j.amepre.2016.09.016
Beck, D., and Foster, J. A. (2014). Machine learning techniques accurately classify microbial communities by bacterial vaginosis characteristics. PLoS ONE 9:e87830. doi: 10.1371/journal.pone.0087830
Beck, D., and Foster, J. A. (2015). Machine learning classifiers provide insight into the relationship between microbial communities and bacterial vaginosis. Bio. Data Min. 8:23. doi: 10.1186/s13040-015-0055-3
Besser, J., Carleton, H. A., Gerner-Smidt, P., Lindsey, R. L., and Trees, E. (2018). Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin. Microbiol. Infect. 24, 335–341. doi: 10.1016/j.cmi.2017.10.013
Blankenstein, T., Lytton, S. D., Leidl, B., Atweh, E., Friese, K., and Mylonas, I. (2015). Point-of-care (POC) diagnosis of bacterial vaginosis (BV) using VGTestTM ion mobility spectrometry (IMS) in a routine ambulatory care gynecology clinic. Arch. Gynecol. Obstet. 292, 355–362. doi: 10.1007/s00404-014-3613-x
Borgdorff, H., Armstrong, S. D., Tytgat, H. L. P., Xia, D., Ndayisaba, G. F., Wastling, J. M., et al. (2016). Unique insights in the cervicovaginal Lactobacillus iners and L. crispatus proteomes and their associations with microbiota dysbiosis. PLoS ONE 11:e0150767. doi: 10.1371/journal.pone.0150767
Bostwick, G. D., Hunt, C. A., Parker, L. R., Mcdyer, D., Woody, J., and Budd, W. (2016). Utility of next-generation sequencing in managing bacterial vaginosis: examples from clinical practice. J. Women's Heal. Care 5:1000322. doi: 10.4172/2167-0420.1000322
Bradshaw, C. S., Morton, A. N., Garland, S. M., Horvath, L. B., Kuzevska, I., and Fairley, C. K. (2005). Evaluation of a point-of-care test, BVBlue, and clinical and laboratory criteria for diagnosis of bacterial vaginosis. J. Clin. Microbiol. 43, 1304–1308. doi: 10.1128/JCM.43.3.1304-1308.2005
Bradshaw, C. S., Tabrizi, S. N., Fairley, C. K., Morton, A. N., Rudland, E., and Garland, S. M. (2006). The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J. Infect. Dis. 194, 828–836. doi: 10.1086/506621
Brotman, R. M., Ravel, J., Cone, R. A., and Zenilman, J. M. (2010). Rapid fluctuation of the vaginal microbiota measured by Gram stain analysis. Sex. Transm. Infect. 86, 297–302. doi: 10.1136/sti.2009.040592
Brotman, R. M., Shardell, M. D., Gajer, P., Fadrosh, D., Chang, K., Silver, M. I., et al. (2018). Association between the vaginal microbiota, menopause status, and signs of vulvovaginal atrophy. Menopause 25, 1321–1330. doi: 10.1097/GME.0000000000001236
Brumley, J. (2012). Testing a model of bacterial vaginosis among black women. (PhD Dissertation). University of South Florida. Available online at: https://scholarcommons.usf.edu/etd/3995 (accessed 16 December 2019).
Budd, W. T., Harwich, M., Bostwick, D. G., Meyers, G., Dilts, J., O'Hanlon, K., et al. (2015). Metagenomics analysis using next generation sequencing of vaginal samples from community practices in the US. MOJ Cell Sci. Report 2:20. doi: 10.15406/mojcsr.2015.02.00020
Carter, J., Beck, D., Williams, H., Foster, J., and Dozier, G. (2014). GA-Based selection of vaginal microbiome features associated with bacterial vaginosis. Genet. Evol. Comput. Conf. 2014, 265–268. doi: 10.1145/2576768.2598378
Cartwright, C. P., Lembke, B. D., Ramachandran, K., Body, B. A., Nye, M. B., Rivers, C. A., et al. (2013). Comparison of nucleic acid amplification assays with BD affirm VPIII for diagnosis of vaginitis in symptomatic women. J. Clin. Microbiol. 51, 3694–3699. doi: 10.1128/JCM.01537-13
Cartwright, C. P., Lembke, B. D., Ramachandran, K., Body, B. A., Nye, M. B., Rivers, CA., et al. (2012). Development and validation of a semiquantitative, multitarget PCR assay for diagnosis of bacterial vaginosis. J. Clin. Microbiol. 50, 2321–2329. doi: 10.1128/JCM.00506-12
Castro, J., and Cerca, N. (2015). BV and non-BV associated Gardnerella vaginalis establish similar synergistic interactions with other BV-associated microorganisms in dual-species biofilms. Anaerobe 36, 56–59. doi: 10.1016/j.anaerobe.2015.10.008
Castro, J., Franca, A., Bradwell, K. R., Serrano, M. G., Jefferson, K. K., and Cerca, N. (2017). Comparative transcriptomic analysis of Gardnerella vaginalis biofilms vs. planktonic cultures using RNA-seq. npj Biofilms Microbiomes 3:3. doi: 10.1038/s41522-017-0012-7
Castro, J., Machado, D., and Cerca, N. (2016). Escherichia coli and Enterococcus faecalis are able to incorporate and enhance a pre-formed Gardnerella vaginalis biofilm. Pathog. Dis. 74:7. doi: 10.1093/femspd/ftw007
Castro, J., Machado, D., and Cerca, N. (2019). Unveiling the role of Gardnerella vaginalis in polymicrobial bacterial vaginosis biofilms: the impact of other vaginal pathogens living as neighbors. ISME J. 13, 1306–1317. doi: 10.1038/s41396-018-0337-0
Cauci, S., Monte, R., Driussi, S., Lanzafame, P., and Quadrifoglio, F. (1998). Impairment of the mucosal immune system: IgA and IgM cleavage detected in vaginal washings of a subgroup of patients with bacterial vaginosis. J. Infect. Dis. 178, 1698–1706. doi: 10.1086/314505
Chaemsaithong, P., Romero, R., Korzeniewski, S. J., Dong, Z., Yeo, L., Hassan, S. S., et al. (2015). A point of care test for the determination of amniotic fluid interleukin-6 and the chemokine CXCL-10/IP-10. J. Matern. Neonatal Med. 28, 1510–1509. doi: 10.3109/14767058.2014.961417
Chavoustie, S. E., Eder, S. E., Koltun, W. D., Lemon, T. R., Mitchell, C., Nyirjesy, P., et al. (2017). Experts explore the state of bacterial vaginosis and the unmet needs facing women and providers. Int. J. Gynaecol. Obstet. 137, 107–109. doi: 10.1002/ijgo.12114
Chawla, R., Bhalla, P., Chadha, S., Grover, S., and Garg, S. (2013). Comparison of Hay's criteria with Nugent's scoring system for diagnosis of bacterial vaginosis. Biomed. Res. Int. 2013, 365194. doi: 10.1155/2013/365194
Chen, H. M., Chang, T. H., Lin, F. M., Liang, C., Chiu, C. M., Yang, T. L., et al. (2018). Vaginal microbiome variances in sample groups categorized by clinical criteria of bacterial vaginosis. BMC Genomics 19:876. doi: 10.1186/s12864-018-5284-7
Coleman, J. S., and Gaydos, C. A. (2018). Molecular diagnosis of bacterial vaginosis: an update. J. Clin. Microbiol. 56, e00342–e00318. doi: 10.1128/JCM.00342-18
Cruciani, F., Wasinger, V., Turroni, S., Calanni, F., Donders, G., Brigidi, P., et al. (2013). Proteome profiles of vaginal fluids from women affected by bacterial vaginosis and healthy controls: outcomes of rifaximin treatment. J. Antimicrob. Chemother. 68, 2648–2659. doi: 10.1093/jac/dkt244
Culbreath, K., Melanson, S., Gale, J., Baker, J., Li, F., Saebo, O., et al. (2019). Validation and Retrospective clinical evaluation of a quantitative 16S rRNA gene metagenomic sequencing assay for bacterial pathogen detection in body fluids. J. Mol. Diagnostics. 21, 913–923. doi: 10.1016/j.jmoldx.2019.05.002
Cusc,ó, A., Catozzi, C., Viñes, J., Sanchez, A., and Francino, O. (2018). Microbiota profiling with long amplicons using Nanopore sequencing: full-length 16S rRNA gene and the 16S-ITS-23S of the rrn operon. F1000 Res. 7:1755. doi: 10.12688/f1000research.16817.2
De Backer, E., Verhelst, R., Verstraelen, H., Alqumber, M. A., Burton, J. P., Tagg, J. R., et al. (2007). Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol. 7:115. doi: 10.1186/1471-2180-7-115
De Seta, F., Campisciano, G., Zanotta, N., Ricci, G., and Comar, M. (2019). The vaginal community state types microbiome-immune network as key factor for bacterial vaginosis and aerobic vaginitis. Front. Microbiol. 10:2451. doi: 10.3389/fmicb.2019.02451
Deese, J., Masson, L., Miller, W., Cohen, M., Morrison, C., Wang, M., et al. (2015). Injectable progestin-only contraception is associated with increased levels of pro-inflammatory cytokines in the female genital tract. Am. J Reprod. Immunol. 74, 357–367. doi: 10.1111/aji.12415
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Donahue Carlson, R., Sheth, A. N., Read, T. D., Frisch, M. B., Mehta, C. C., Martin, A., et al. (2017). The female genital tract microbiome is associated with vaginal antiretroviral drug concentrations in human immunodeficiency virus-infected women on antiretroviral therapy. J. Infect. Dis. 216, 990–999. doi: 10.1093/infdis/jix420
Donders, G. G., Vereecken, A., Bosmans, E., and Spitz, B. (2003). Vaginal cytokines in normal pregnancy. Am. J Obstet. Gynecol. 189, 1433–1438. doi: 10.1067/S0002-9378(03)00653-7
Eschenbach, D. A., Hillier, S. L., Critchlow, C., Stevens, C. E., DeRouen, T., and Holmes, K. K. (1988). Diagnosis and clinical manifestations of bacterial vaginosis. Am. J. Obstet. Gynecol. 158, 819–828. doi: 10.1016/0002-9378(88)90078-6
Eschenbach, D. A., Thwin, S. S., Patton, D. L., Hooton, T. M., Stapleton, A. E., Agnew, K., et al. (2000). Influence of the normal menstrual cycle on vaginal tissue, discharge, and microflora. Clin. Infect. Dis. 30, 901–907. doi: 10.1086/313818
Farage, M., and Maibach, H. (2006a). Lifetime changes in the vulva and vagina. Arch. Gynecol. Obstet. 273, 195–202. doi: 10.1007/s00404-005-0079-x
Farage, M. A., and Maibach, H. I., (eds.). (2006b). The Vulva: Anatomy, Physiology, and Pathology. New York, NY: CRC Press.
Ferreira, C. S. T., Da Silva, M. G., De Pontes, L. G., dos Santos, L. D., and Marconi, C. (2018). Protein content of cervicovaginal fluid is altered during bacterial vaginosis. J. Low Genit. Tract. Dis. 22, 147–151. doi: 10.1097/LGT.0000000000000367
Ferris, M. J., Masztal, A., Aldridge, K. E., Fortenberry, J. D., Fidel, P. L. Jr., and Martin, D. H. (2004). Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect. Dis. 4:5. doi: 10.1186/1471-2334-4-5
Fethers, K., Twin, J., Fairley, C. K., Fowkes, F. J., Garland, S. M., Fehler, G., et al. (2012). Bacterial vaginosis (BV) candidate bacteria: associations with BV and behavioural practices in sexually-experienced and inexperienced women. PLoS ONE 7:e30633. doi: 10.1371/journal.pone.0030633
Fettweis, J. M., Brooks, J. P., Serrano, M. G., Sheth, N. U., Girerd, P. H., Edwards, D. J., et al. (2014). Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 160, 2272–2282. doi: 10.1099/mic.0.081034-0
Forsum, U., Hallen, A., and Larsson, P. G. (2005). Bacterial vaginosis–a laboratory and clinical diagnostics enigma. APMIS 113, 153–161. doi: 10.1111/j.1600-0463.2005.apm1130301.x
Fredricks, D. N. (2011). Molecular methods to describe the spectrum and dynamics of the vaginal microbiota. Anaerobe 17, 191–195. doi: 10.1016/j.anaerobe.2011.01.001
Fredricks, D. N., Fiedler, T. L., and Marrazzo, J. M. (2005). Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353, 1899–1911. doi: 10.1056/NEJMoa043802
Freitas, A. C., Chaban, B., Bocking, A., Rocco, M., Yang, S., Hill, J. E., et al. (2017). The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non-pregnant women. Sci. Rep. 7:9212. doi: 10.1038/s41598-017-07790-9
Gajer, P., Brotman, R. M., Bai, G., Sakamoto, J., Schutte, U. M., Zhong, X., et al. (2012). Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 4:132ra152. doi: 10.1126/scitranslmed.3003605
Gardner, H. L., and Dukes, C. D. (1954). New etiologic agent in nonspecific bacterial vaginitis. Science 120, 853. doi: 10.1126/science.120.3125.853
Gardner, H. L., and Dukes, C. D. (1955). Haemophilus vaginalis vaginitis: a newly defined specific infection previously classified non-specific vaginitis. Am. J. Obstet. Gynecol. 69, 962–976. doi: 10.1016/0002-9378(55)90095-8
Gardner, J. K., Łaniewski, P., Knight, A., Haddad, L. B., Swaims-Kohlmeier, A., and Herbst-Kralovetz, M.M. (2019). Interleukin-36γ is elevated in cervicovaginal epithelial cells in women with bacterial vaginosis and in vitro after infection with microbes associated with bacterial vaginosis. J. Infect. Dis. 221, 983–988. doi: 10.1093/infdis/jiz514
Garrett, N. J., Osman, F., Maharaj, B., Naicker, N., Gibbs, A., Norman, E., et al. (2018). Beyond syndromic management: opportunities for diagnosis-based treatment of sexually transmitted infections in low- and middle-income countries. PLoS ONE 13:e0196209. doi: 10.1371/journal.pone.0196209
Gautam, R., Borgdorff, H., Jespers, V., Francis, S. C., Verhelst, R., Mwaura, M., et al. (2015). Correlates of the molecular vaginal microbiota composition of African women. BMC Infect. Dis. 15:86. doi: 10.1186/s12879-015-0831-1
Gaydos, C. A., Beqaj, S., Schwebke, J. R., Lebed, J., Smith, B., Davis, T. E., et al. (2017). Clinical validation of a test for the diagnosis of vaginitis. Obstet. Gynecol. 130, 181–189 doi: 10.1097/AOG.0000000000002090
Gilbert, N. M., Lewis, W. G., Li, G., Sojka, D. K., Lubin, J. B., and Lewis, A. L. (2019). Gardnerella vaginalis and Prevotella bivia trigger distinct and overlapping phenotypes in a mouse model of bacterial vaginosis. J. Infect. Dis. 220, 1099–1108. doi: 10.1093/infdis/jiy704
Gliniewicz, K., Schneider, G. M., Ridenhour, B. J., Williams, C. J., Song, Y., Farage, M. A., et al. (2019). Comparison of the vaginal microbiomes of premenopausal and postmenopausal women. Front. Microbiol. 10:193. doi: 10.3389/fmicb.2019.00193
Gosmann, C., Anahtar, M. N., Handley, S. A., Farcasanu, M., Abu-Ali, G., Bowmanet, B. A., et al. (2017). Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity 46, 29–37. doi: 10.1016/j.immuni.2016.12.013
Gottschick, C., Deng, Z. L., Vital, M., Masur, C., Abels, C., Pieper, D. H., et al. (2017). The urinary microbiota of men and women and its changes in women during bacterial vaginosis and antibiotic treatment. Microbiome 5:99. doi: 10.1186/s40168-017-0305-3
Guijas, C., Montenegro-Burke, J. R., Warth, B., Spilker, M. E., and Siuzdak, G. (2018). Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 36, 316–320. doi: 10.1038/nbt.4101
Guthrie, L., and Kelly, L. (2019). Bringing microbiome-drug interaction research into the clinic. EBioMedicine 44, 708–715. doi: 10.1016/j.ebiom.2019.05.009
Haggerty, C. L., Totten, P. A., Ferris, M., Martin, D. H., Hoferka, S., Astete, S. G., et al. (2009). Clinical characteristics of bacterial vaginosis among women testing positive for fastidious bacteria. Sex. Transm. Infect. 85, 242–248. doi: 10.1136/sti.2008.032821
Hammerschlag, M. R., Alpert, S., Onderdonk, A. B., Thurston, P., Drude, E., McCormack, W. M., et al. (1978a). Anaerobic microflora of the vagina in children. Am. J. Obstet. Gynecol. 131, 853–856. doi: 10.1016/S0002-9378(16)33130-1
Hammerschlag, M. R., Alpert, S., Rosner, I., Thurston, P., Semine, D., McComb, D., et al. (1978b). Microbiology of the vagina in children: normal and potentially pathogenic organisms. Pediatrics 62, 57–62.
Hardy, L., Jespers, V., Abdellati, S., De Baetselier, I., Mwambarangwe, L., Musengamana, V., et al. (2016). A fruitful alliance: the synergy between Atopobium vaginae and Gardnerella vaginalis in bacterial vaginosis-associated biofilm. Sex. Trans. Inf. 92, 487–491. doi: 10.1136/sextrans-2015-052475
Hardy, L., Jespers, V., Dahchour, N., Mwambarangwe, L., Musengamana, V., Vaneechoutte, M., et al. (2015). Unravelling the bacterial vaginosis-associated biofilm: a multiplex Gardnerella vaginalis and Atopobium vaginae fluorescence in situ hybridization assay using peptide nucleic acid probes. PLoS ONE 10:e0136658. doi: 10.1371/journal.pone.0136658
Hay, P. E., Lamont, R. F., Taylor-Robinson, D., Morgan, D. J., Ison, C., and Pearson, J. (1994). Abnormal bacterial colonisation of the genital tract and subsequent preterm delivery and late miscarriage. BMJ 308, 295–298. doi: 10.1136/bmj.308.6924.295
Hedges, S. R., Barrientes, F., Desmond, R. A., and Schwebke, J. R. (2006). Local and systemic cytokine levels in relation to changes in vaginal flora. J. Infect. Dis. 193, 556–562. doi: 10.1086/499824
Hickey, R. J., and Forney, L. J. (2014). Gardnerella vaginalis does not always cause bacterial vaginosis. J. Infect. Dis. 210, 1682–1683. doi: 10.1093/infdis/jiu303
Hickey, R. J., Zhou, X., Pierson, J. D., Ravel, J., and Forney, L. J. (2012). Understanding vaginal microbiome complexity from an ecological perspective. Transl. Res. 160, 267–282. doi: 10.1016/j.trsl.2012.02.008
Hilbert, D. W., Smith, W. L., Chadwick, S. G., Toner, G., Mordechai, E., Adelson, M. E., et al. (2016). Development and validation of a highly accurate quantitative real-time PCR assay for diagnosis of bacterial vaginosis. J. Clin. Microbiol. 54, 1017–1024. doi: 10.1128/JCM.03104-15
Hill, J. E., Albert, A. Y. K., and the Vogue Research G.roup (2019). Resolution and cooccurrence patterns of Gardnerella leopoldii, G. swidsinskii, G. piotii, and G. vaginalis within the vaginal microbiome. Infect. Immun. 87, e00532–e00519. doi: 10.1128/IAI.00532-19
Hillier, S. L., Krohn, M. A., Rabe, L. K., Klebanoff, S. J., and Eschenbach, D. A. (1993). The normal vaginal flora, H2O2-producing lactobacilli, and bacterial vaginosis in pregnant women. Clin. Infect. Dis. 16, S273–S281. doi: 10.1093/clinids/16.Supplement_4.S273
Holst, E. (1990). Reservoir of four organisms associated with bacterial vaginosis suggests lack of sexual transmission. J. Clin. Microbiol. 28, 2035–2039. doi: 10.1128/JCM.28.9.2035-2039.1990
Hong, K. H., Hong, S. K., Cho, S. I., Ra, E., Han, K. H., Kang, S. B., et al. (2016). Analysis of the vaginal microbiome by next-generation sequencing and evaluation of its performance as a clinical diagnostic tool in vaginitis. Ann. Lab. Med. 36:441. doi: 10.3343/alm.2016.36.5.441
Hyman, R. W., Fukushima, M., Diamond, L., Kumm, J., Giudice, L. C., and Davis, R. W. (2005). Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U.S.A. 102, 7952–7957. doi: 10.1073/pnas.0503236102
Ison, C. A., and Hay, P. E. (2002) Validation of a simplified grading of Gram stained vaginal smears for use in genitourinary medicine clinics. Sex. Transm. Infect. 78, 413–415. doi: 10.1136/sti.78.6.413
Jackson, C., Gardy, J. L., Shadiloo, H. C., and Silva, D. S. (2019). Trust and the ethical challenges in the use of whole genome sequencing for tuberculosis surveillance: a qualitative study of stakeholder perspectives. BMC Med. Ethics. 20:43. doi: 10.1186/s12910-019-0380-z
Jakobsson, T., and Forsum, U. (2007). Lactobacillus iners: a marker of changes in the vaginal flora? J. Clin. Microbiol. 45, 3145. doi: 10.1128/JCM.00558-07
Janulaitiene, M., Paliulyte, V., Grinceviciene, S., Zakareviciene, J., Vladisauskiene, A., Marcinkute, A., et al. (2017). Prevalence and distribution of Gardnerella vaginalis subgroups in women with and without bacterial vaginosis. BMC Infect. Dis. 17:394. doi: 10.1186/s12879-017-2501-y
Jarvis, J. P., Rains, D., Kradel, S. J., Elliott, J., Diamond, E. E., Avaniss-Aghajani, E., et al. (2018). Diagnosing bacterial vaginosis with a novel, clinically-actionable molecular diagnostic tool. J. Appl. Microb. Res. 1, 01–08. doi: 10.1101/334177
Jespers, V., Kyongo, J., Joseph, S., Hardy, L., Cools, P., Crucitti, T., et al. (2017). A longitudinal analysis of the vaginal microbiota and vaginal immune mediators in women from sub-Saharan Africa. Sci. Rep. 7:11974. doi: 10.1038/s41598-017-12198-6
Jespers, V., Menten, J., Smet, H., Poradosu, S., Abdellati, S., Verhelst, R., et al. (2012). Quantification of bacterial species of the vaginal microbiome in different groups of women, using nucleic acid amplification tests. BMC Microbiol. 12:83. doi: 10.1186/1471-2180-12-83
Jespers, V., van de Wijgert, J., Cools, P., Verhelst, R., Verstraelen, H., Delany-Moretlwe, S., et al. (2015). The significance of Lactobacillus crispatus and L. vaginalis for vaginal health and the negative effect of recent sex: a cross-sectional descriptive study across groups of African women. BMC Infect. Dis. 15:115. doi: 10.1186/s12879-015-0825-z
Joyisa, N., Moodley, D., Nkosi, T., Talakgale, R., Sebitloane, M., Naidoo, M., et al. (2019). Asymptomatic bacterial vaginosis in pregnancy and missed opportunities for treatment: a cross-sectional observational study. Infect. Dis. Obstet. Gynecol. 2019, 1–7. doi: 10.1155/2019/7808179
Kelly, M. C., Mequio, M. J., and Pybus, V. (2003). Inhibition of vaginal lactobacilli by a bacteriocin-like inhibitor produced by Enterococcus faecium 62-6: potential significance for bacterial vaginosis. Infect. Dis. Obstet. Gynecol. 11, 147–156. doi: 10.1080/10647440300025513
Klatt, N. R., Cheu, R., Birse, K., Zevin, A. S., Perner, M., Noel-Romas, L., et al. (2017). Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science 356, 938–945. doi: 10.1126/science.aai9383
Klebanoff, M. A., Schwebke, J. R., Zhang, J., Nansel, T. R., Yu, K. F., and Andrews, W. W. (2004). Vulvovaginal symptoms in women with bacterial vaginosis. Obstet. Gynecol. 104, 267–272. doi: 10.1097/01.AOG.0000134783.98382.b0
Klebanoff, S. J., Hillier, S. L., Eschenbach, D. A., and Waltersdorph, A. M. (1991). Control of the microbial flora of the vagina by H2O2-generating lactobacilli. J. Infect. Dis. 164, 94–100. doi: 10.1093/infdis/164.1.94
Kunze, M., Klar, M., Morfeld, C. A., Thorns, B., Schild, R. L., Markfeld-Erol, F., et al. (2016). Cytokines in noninvasively obtained amniotic fluid as predictors of fetal inflammatory response syndrome. Am. J Obstet. Gynecol. 215, e1–e96. doi: 10.1016/j.ajog.2016.01.181
Kyongo, J. K., Crucitti, T., Menten, J., Hardy, L., Cools, P., Michiels, J., et al. (2015). Cross-sectional analysis of selected genital tract immunological markers and molecular vaginal microbiota in sub-Saharan African women, with relevance to HIV risk and prevention. Clin. Vaccine Immunol. 22, 526–538. doi: 10.1128/CVI.00762-14
Laghi, L., Picone, G., Cruciani, F., Brigidi, P., Calanni, F., Donders, G., et al. (2014). Rifaximin modulates the vaginal microbiome and metabolome in women affected by bacterial vaginosis. Antimicrob. Agents Chemother. 58, 3411–3420. doi: 10.1128/AAC.02469-14
Lambert, J. A., John, S., Sobel, J. D., and Akins, R. A. (2013). Longitudinal analysis of vaginal microbiome dynamics in women with recurrent bacterial vaginosis: recognition of the conversion process. PLoS ONE 8:e82599. doi: 10.1371/journal.pone.0082599
Leitich, H., Bodner-Adler, B., Brunbauer, M., Kaider, A., Egarter, C., and Husslein, P. (2003). Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am. J. Obstet. Gynecol. 189, 139–147. doi: 10.1067/mob.2003.339
Lennard, K., Dabee, S., Barnabas, S. L., Havyarimana, E., Blakney, A., Jaumdally, S. Z., et al. (2018). Microbial composition predicts genital tract inflammation and persistent bacterial vaginosis in South African adolescent females. Infect. Immun. 86:e00410–17. doi: 10.1128/IAI.00410-17
Lewis, W. G., Robinson, L. S., Perry, J., Bick, J. L., Peipert, J. F., Allsworth, J. E., et al. (2012). Hydrolysis of secreted sialoglycoprotein immunoglobulin A (IgA) in ex vivo and biochemical models of bacterial vaginosis. J. Biol. Chem. 287, 2079–2089. doi: 10.1074/jbc.M111.278135
Libby, E. K., Pascal, K. E., Mordechai, E., Adelson, M. E., and Trama, J. P. (2008). Atopobium vaginae triggers an innate immune response in an in vitro model of bacterial vaginosis. Microbes Infect. 10, 439–446. doi: 10.1016/j.micinf.2008.01.004
Machado, A., and Cerca, N. (2015). Influence of biofilm formation by Gardnerella vaginalis and other anaerobes on bacterial vaginosis. J. Infect. Dis. 212, 1856–1861. doi: 10.1093/infdis/jiv338
Machado, A., Jefferson, K. K., and Cerca, N. (2013a). Interactions between Lactobacillus crispatus and bacterial vaginosis (BV)-associated bacterial species in initial attachment and biofilm formation. Int. J. Mol. Sci. 14, 12004–12012. doi: 10.3390/ijms140612004
Machado, A., Salgueiro, D., Harwich, M., Jefferson, K. K., and Cerca, N. (2013b). Quantitative analysis of initial adhesion of bacterial vaginosis-associated anaerobes to ME-180 cells. Anaerobe 23, 1–4. doi: 10.1016/j.anaerobe.2013.07.007
Macklaim, J. M., Fernandes, A. D., Di Bella, J. M., Hammond, J. A., Reid, G., and Gloor, G. B. (2013). Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome 1:12. doi: 10.1186/2049-2618-1-12
Macklaim, J. M., Gloor, G. B., Anukam, K. C., Cribby, S., and Reid, G. (2011). At the crossroads of vaginal health and disease, the genome sequence of Lactobacillus iners AB-1. Proc. Natl. Acad. Sci. U.S.A. 108, 4688–4695. doi: 10.1073/pnas.1000086107
Madhivanan, P., Krupp, K., Hardin, J., Karat, C., Klausner, J. D., and Reingold, A. L. (2009). Simple and inexpensive point-of-care tests improve diagnosis of vaginal infections in resource constrained settings. Trop. Med. Int. Health 14, 703–708. doi: 10.1111/j.1365-3156.2009.02274.x
Madhivanan, P., Krupp, K., Li, T., Ravi, K., Selezneva, J., Srinivas, V., et al. (2014). Performance of BVBlue rapid test in detecting bacterial vaginosis among women in Mysore, India. Infect. Dis. Obstet. Gynecol. 2014, 908313. doi: 10.1155/2014/908313
Malla, M. A., Dubey, A., Kumar, A., Yadav, S., Hashem, A., and Abd_Allah, E. F. (2019). Exploring the human microbiome: the potential future role of next-generation sequencing in disease diagnosis and treatment. Front. Immunol. 9:2868. doi: 10.3389/fimmu.2018.02868
Masson, L., Arnold, K. B., Little, F., Mlisana, K., Lewis, D. A., Mkhize, N., et al. (2016). Inflammatory cytokine biomarkers to identify women with asymptomatic sexually transmitted infections and bacterial vaginosis who are at high risk of HIV infection. Sex. Transm. Infect. 92, 186–193. doi: 10.1136/sextrans-2015-052072
Masson, L., Barnabas, S., Deese, J., Lennard, K., Dabee, S., Gamieldien, H., et al. (2018). Inflammatory cytokine biomarkers of asymptomatic sexually transmitted infections and vaginal dysbiosis: a multicentre validation study. Sex. Transm. Infect. 95, 5–12. doi: 10.1136/sextrans-2017-053506
Masson, L., Mlisana, K., Little, F., Werner, L., Mkhize, N. N., Ronacher, K., et al. (2014). Defining genital tract cytokine signatures of sexually transmitted infections and bacterial vaginosis in women at high risk of HIV infection: a cross-sectional study. Sex. Transm. Infect. 90, 580–587. doi: 10.1136/sextrans-2014-051601
Mayer, B. T., Srinivasan, S., Fiedler, T. L., Marrazzo, J. M., Fredricks, D. N., and Schiffer, J. T. (2015). Rapid and profound shifts in the vaginal microbiota following antibiotic treatment for bacterial vaginosis. J. Infect. Dis. 212, 793–802. doi: 10.1093/infdis/jiv079
McKinnon, L. R., Achilles, S. L., Bradshaw, C. S., Burgener, A., Crucitti, T., Fredricks, D. N., et al. (2019). The evolving facets of bacterial vaginosis: implications for HIV transmission. AIDS Res. Hum. Retroviruses 35, 219–228. doi: 10.1089/aid.2018.0304
McMillan, A., Rulisa, S., Sumarah, M., Macklaim, J. M., Renaud, J., Bisanz, J. E., et al. (2015). A multi-platform metabolomics approach identifies highly specific biomarkers of bacterial diversity in the vagina of pregnant and non-pregnant women. Sci. Rep. 5:14174. doi: 10.1038/srep14174
Menard, J., Fenollar, F., Henry, M., Bretelle, F., and Raoult, D. (2008). Molecular Quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin. Infect. Dis. 47, 33–43. doi: 10.1086/588661
Mendling, W., Palmeira-de-Oliveira, A., Biber, S., and Prasauskas, V. (2019). An update on the role of Atopobium vaginae in bacterial vaginosis: what to consider when choosing a treatment? A mini review. Arch. Gynecol. Obstet. 300, 1–6. doi: 10.1007/s00404-019-05142-8
Mitchell, C., Balkus, J. E., Fredricks, D., Liu, C., McKernan-Mullin, J., Frenkel, L. M., et al. (2013). Interaction between lactobacilli, bacterial vaginosis-associated bacteria, and HIV Type 1 RNA and DNA genital shedding in U.S. and Kenyan women. AIDS Res. Hum. Retroviruses 29, 13–19. doi: 10.1089/aid.2012.0187
Mlisana, K., Naicker, N., Werner, L., Roberts, L., van Loggerenberg, F., Baxter, C., et al. (2012). Symptomatic vaginal discharge is a poor predictor of sexually transmitted infections and genital tract inflammation in high-risk women in South Africa. J. Infect. Dis. 206, 6–14. doi: 10.1093/infdis/jis298
Modak, T., Arora, P., Agnes, C., Ray, R., Goswami, S., Ghosh, P., et al. (2011). Diagnosis of bacterial vaginosis in cases of abnormal vaginal discharge: comparison of clinical and microbiological criteria. J. Infection Dev. Countries 5, 353–360. doi: 10.3855/jidc.1153
Mohammadzadeh, F., Dolatian, M., Jorjani, M., and Alavi Majd, H. (2014). Diagnostic value of Amsel's clinical criteria for diagnosis of bacterial vaginosis. Glob. J. Health Sci. 7, 8–14. doi: 10.5539/gjhs.v7n3p8
Mullany, P. (2014). Functional metagenomics for the investigation of antibiotic resistance. Virulence 5, 443–447. doi: 10.4161/viru.28196
Muzny, C. A., Blanchard, E., Taylor, C. M., Aaron, K. J., Talluri, R., Griswold, M. E., et al. (2018). Identification of key bacteria involved in the induction of incident bacterial vaginosis: a prospective study. J. Infect. Dis. 218, 966–978. doi: 10.1093/infdis/jiy243
Muzny, C. A., Laniewski, P., Schwebke, J. R., and Herbst-Kralovetz, M. M. (2020). Host-vaginal microbiota interactions in the pathogenesis of bacterial vaginosis. Curr. Opin. Infect. Dis. 32, 59–65. doi: 10.1097/QCO.0000000000000620
Muzny, C. A., and Schwebke, J. R. (2016). Pathogenesis of bacterial vaginosis: discussion of current hypotheses. J. Infect. Dis. 214, S1–S5. doi: 10.1093/infdis/jiw121
Muzny, C. A., Taylor, C. M., Swords, W. E., Tamhane, A., Chattopadhyay, D., Cerca, N., et al. (2019). An updated conceptual model on the pathogenesis of bacterial vaginosis. J. Infect. Dis. 220, 1399–1405. doi: 10.1093/infdis/jiz342
Myziuk, L., Romanowski, B., and Johnson, S. C. (2003). BVBlue test for diagnosis of bacterial vaginosis. J. Clin. Microbiol. 41, 1925–1928. doi: 10.1128/JCM.41.5.1925-1928.2003
Nagy, E., Petterson, M., and Mårdh, P.-A. (1991). Antibiosis between bacteria isolated from the vagina of women with and without signs of bacterial vaginosis. APMIS 99, 739–744. doi: 10.1111/j.1699-0463.1991.tb01253.x
National Department of Health (South Africa) (2015). Sexually Transmitted Infections Management Guidelines 2015. Pretoria: National Department of Health (South Africa). Available online at: https://health-e.org.za/wp-content/uploads/2015/06/STIguidelines3-31-15cmyk.pdf (accessed December 16 2019).
Ness, R. B., Kip, K. E., Hillier, S. L., Soper, D. E., Stamm, C. A., Sweet, R. L., et al. (2005). A cluster analysis of bacterial vaginosis-associated microflora and pelvic inflammatory disease. Am. J. Epidemiol. 162, 585–590. doi: 10.1093/aje/kwi243
Nikolaitchouk, N., Andersch, B., Falsen, E., Strömbeck, L., and Mattsby-Baltzer, I. (2008). The lower genital tract microbiota in relation to cytokine-, SLPI- and endotoxin levels: application of checkerboard DNA-DNA hybridization (CDH). APMIS 116, 263–277. doi: 10.1111/j.1600-0463.2008.00808.x
Nugent, R. P., Krohn, M. A., and Hillier, S. L. (1991). Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J. Clin. Microbiol. 29, 297–301. doi: 10.1128/JCM.29.2.297-301.1991
O'Hanlon, D. E., Moench, T. R., and Cone, R. A. (2011). In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infect. Dis. 11:200. doi: 10.1186/1471-2334-11-200
Parolin, C., Foschi, C., Laghi, L., Zhu, C., Banzola, N., Gaspari, V., et al. (2018). Insights into vaginal bacterial communities and metabolic profiles of Chlamydia trachomatis infection: positioning between eubiosis and dysbiosis. Front. Microbiol. 9:600. doi: 10.3389/fmicb.2018.00600
Patterson, J. L., Girerd, P. H., Karjane, N. W., and Jefferson, K. K. (2007). Effect of biofilm phenotype on resistance of Gardnerella vaginalis to hydrogen peroxide and lactic acid. Am. J. Obstet. Gynecol. 197, e171–e170. doi: 10.1016/j.ajog.2007.02.027
Patterson, J. L., Stull-Lane, A., Girerd, P. H., and Jefferson, K. K. (2010). Analysis of adherence, biofilm formation and cytotoxicity suggests a greater virulence potential of Gardnerella vaginalis relative to other bacterial-vaginosis-associated anaerobes. Microbiology 156, 392–399. doi: 10.1099/mic.0.034280-0
Pavlova, S. I., Kilic, A. O., Kilic, S. S., So, J. S., Nader-Macias, M. E., Simoes, J. A., et al. (2002). Genetic diversity of vaginal lactobacilli from women in different countries based on 16S rRNA gene sequences. J. Appl. Microbiol. 92, 451–459. doi: 10.1046/j.1365-2672.2002.01547.x
Penders, J., Stobberingh, E. E., Savelkoul, P. H., and Wolffs, P. F. (2013). The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 4:87. doi: 10.3389/fmicb.2013.00087
Percival, B. C., Grootveld, M., Gibson, M., Osman, Y., Molinari, M., Jafari, F., et al. (2019). Low-field, benchtop NMR spectroscopy as a potential tool for point-of-care diagnostics of metabolic conditions: validation, protocols and computational models. High-Throughput 8:2. doi: 10.3390/ht8010002
Peters, D. L., Wang, W., Zhang, X., Ning, Z., Mayne, J., and Figeys, D. (2019). Metaproteomic and metabolomic approaches for characterizing the gut microbiome. Proteomics 19:1800363. doi: 10.1002/pmic.201800363
Petrova, M. I., Reid, G., Vaneechoutte, M., and Lebeer, S. (2017). Lactobacillus iners: friend or foe? Trends Microbiol. 25, 182–191. doi: 10.1016/j.tim.2016.11.007
Pivarcsi, A., Nagy, I., Koreck, A., Kis, K., Kenderessy-Szabo, A., Szell, M., et al. (2005). Microbial compounds induce the expression of pro-inflammatory cytokines, chemokines and human β-defensin-2 in vaginal epithelial cells. Microbes Infect. 7, 1117–1127. doi: 10.1016/j.micinf.2005.03.016
Pruski, P., Macintyre, D. A., Lewis, H. V., Inglese, P., Correia, G. D. S., Hansel, T. T., et al. (2017). Medical swab analysis using desorption electrospray ionization mass spectrometry: a noninvasive approach for mucosal diagnostics. Anal. Chem. 89, 1540–1550. doi: 10.1021/acs.analchem.6b03405
Punina, N. V., Makridakis, N. M., Remnev, M. A., and Topunov, A. F. (2015). Whole-genome sequencing targets drug-resistant bacterial infections. Hum. Genomics 9:19. doi: 10.1186/s40246-015-0037-z
Pybus, V., and Onderdonk, A. B. (1997). Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J. Infect. Dis. 175:406. doi: 10.1093/infdis/175.2.406
Ranjit, E., Raghubanshi, B. R., Maskey, S., and Parajuli, P. (2018). Prevalence of bacterial vaginosis and its association with risk factors among nonpregnant women: a hospital based study. Int. J. Microbiol. 2018:8349601. doi: 10.1155/2018/8349601
Ravel, J., Brotman, R. M., Gajer, P., Ma, B., Nandy, M., Fadrosh, D. W., et al. (2013). Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1, 29. doi: 10.1186/2049-2618-1-29
Ravel, J., Gajer, P., Abdo, Z., Schneider, G. M., Koenig, S. S., McCulle, S. L., et al. (2011). Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U.S.A. 108, 4680–4687. doi: 10.1073/pnas.1002611107
Redelinghuys, M., Ehlers, M., Bezuidenhoudt, J., Becker, P., and Kock, M. (2017). Assessment of Atopobium vaginae and Gardnerella vaginalis concentrations in a cohort of pregnant South African women. Sex. Transm. Infect. 93, 410–415. doi: 10.1136/sextrans-2016-052883
Relman, D. A., and Lipsitch, M. (2018). Microbiome as a tool and a target in the effort to address antimicrobial resistance. Proc. Natl. Acad. Sci. U.S.A. 115, 12902–12910. doi: 10.1073/pnas.1717163115
Salipante, S. J., Sengupta, D. J., Rosenthal, C., Costa, G., Spangler, J., Sims, E. H., et al. (2013). Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS ONE 8:e65226. doi: 10.1371/journal.pone.0065226
Schwebke, J. R., Flynn, M. S., and Rivers, C. A. (2014a). Prevalence of Gardnerella vaginalis among women with lactobacillus-predominant vaginal flora. Sex. Trans. Inf. 90, 61–63. doi: 10.1136/sextrans-2013-051232
Schwebke, J. R., Hillier, S. L., Sobel, J. D., McGregor, J. A., and Sweet, R. L. (1996). Validity of the vaginal Gram stain for the diagnosis of bacterial vaginosis. Obstet. Gynecol. 88, 573–576. doi: 10.1016/0029-7844(96)00233-5
Schwebke, J. R., Muzny, C. A., and Josey, W. E. (2014b). Role of Gardnerella vaginalis in the pathogenesis of bacterial vaginosis: a conceptual model. J. Infect. Dis. 210, 338–343. doi: 10.1093/infdis/jiu089
Sha, B. E., Chen, H. Y., Wang, Q. J., Zariffard, M. R., Cohen, M. H., and Spear, G. T. (2005b). Utility of Amsel criteria, Nugent score, and quantitative PCR for Gardnerella vaginalis, Mycoplasma hominis, and Lactobacillus spp. for diagnosis of bacterial vaginosis in human immunodeficiency virus-infected women. J. Clin. Microbiol. 43, 4607–4612. doi: 10.1128/JCM.43.9.4607-4612.2005
Sha, B. E., Zariffard, M. R., Wang, Q. J., Chen, H. Y., Bremer, J., Cohen, M. H., et al. (2005a). Female genital-tract HIV load correlates inversely with Lactobacillus species but positively with bacterial vaginosis and Mycoplasma hominis. J. Infect. Dis. 191, 25–32. doi: 10.1086/426394
Shipitsyna, E., Roos, A., Datcu, R., Hallen, A., Fredlund, H., Jensen, J. S., et al. (2013). Composition of the vaginal microbiota in women of reproductive age - sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS ONE 8:e60670. doi: 10.1371/journal.pone.0060670
Shrivastava, S. R., Shrivastava, S., and Ramasamy, J. (2014). Utility of syndromic approach in management of sexually transmitted infections: public health perspective. Doc. Head. J. Coast. Life Med. 2, 7–13. doi: 10.12980/JCLM.2.20143D237
Singh, H. O., Singh, A., Dhole, T. N., and Sumitra, N. (2015). Factor associated to bacterial vaginosis in non-pregnant women of North Indian population. J. Biotechnol. Biomater. 05:195. doi: 10.4172/2155-952X.1000195
Sobel, J. D. (2000). Bacterial vaginosis. Annu. Rev. Med. 51, 349–356. doi: 10.1146/annurev.med.51.1.349
Song, Y., He, L., Zhou, F., Chen, S., Ni, D., Lei, B., et al. (2017). Segmentation, splitting, and classification of overlapping bacteria in microscope images for automatic bacterial vaginosis diagnosis. IEEE J Biomed Health Inform 21, 1095–1104. doi: 10.1109/JBHI.2016.2594239
Spear, G. T., Gilbert, D., Landay, A. L., Zariffard, R., French, A. L., Patel, P., et al. (2011). Pyrosequencing of the genital microbiotas of HIV-seropositive and -seronegative women reveals Lactobacillus iners as the predominant Lactobacillus Species. Appl. Environ. Microbiol. 77, 378–381. doi: 10.1128/AEM.00973-10
Spiegel, C. A. (1991). Bacterial vaginosis. Clin. Microbiol. Rev. 4, 485–502. doi: 10.1128/CMR.4.4.485
Spiegel, C. A., Amsel, R., and Holmes, K. K. (1983). Diagnosis of bacterial vaginosis by direct gram stain of vaginal fluid. J. Clin. Microbiol. 18, 170–177. doi: 10.1128/JCM.18.1.170-177.1983
Srinivasan, S., and Fredricks, D. N. (2008). The human vaginal bacterial biota and bacterial vaginosis. Interdiscip. Perspect. Infect. Dis. 2008:750479. doi: 10.1155/2008/750479
Srinivasan, S., Hoffman, N. G., Morgan, M. T., Matsen, F. A., Fiedler, T. L., Hall, R. W., et al. (2012). Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS ONE 7:e37818. doi: 10.1371/journal.pone.0037818
Srinivasan, S., Morgan, M. T., Liu, C., Matsen, F. A., Hoffman, N. G., Fiedler, T. L., et al. (2013). More than meets the eye: associations of vaginal bacteria with Gram stain morphotypes using molecular phylogenetic analysis. PLoS ONE 8:e78633. doi: 10.1371/journal.pone.0078633
Stafford, G. P., Parker, J. L., Amabebe, E., Kistler, J., Reynolds, S., Stern, V., et al. (2017). Spontaneous preterm birth is associated with differential expression of vaginal metabolites by lactobacilli-dominated microflora. Front. Physiol. 8:615. doi: 10.3389/fphys.2017.00615
Sturm-Ramirez, K., Gaye-Diallo, A., Eisen, G., Mboup, S., and Kanki, P. J. (2000). High levels of tumor necrosis factor-α and interleukin-1β in bacterial vaginosis may increase susceptibility to human immunodeficiency virus. J. Infect. Dis. 182, 467–473. doi: 10.1086/315713
Swidsinski, A., Loening-Baucke, V., Mendling, W., Dorffel, Y., Schilling, J., Halwani, Z., et al. (2014). Infection through structured polymicrobial Gardnerella biofilms (StPM-GB). Histol. Histopathol. 29, 567–587. doi: 10.14670/HH-29.10.567
Swidsinski, A., Loening-Baucke, V., Swidsinski, S., and Verstraelen, H. (2015). Polymicrobial Gardnerella biofilm resists repeated intravaginal antiseptic treatment in a subset of women with bacterial vaginosis: a preliminary report. Arch. Gynecol. Obstet. 291, 605–609. doi: 10.1007/s00404-014-3484-1
Swidsinski, A., Mendling, W., Loening-Baucke, V., Ladhoff, A., Swidsinski, S., Hale, L. P., et al. (2005). Adherent biofilms in bacterial vaginosis. Obstet. Gynecol. 106, 1013–1023. doi: 10.1097/01.AOG.0000183594.45524.d2
Swidsinski, A., Mendling, W., Loening-Baucke, V., Swidsinski, S., Dorffel, Y., Scholze, J., et al. (2008). An adherent Gardnerella vaginalis biofilm persists on the vaginal epithelium after standard therapy with oral metronidazole. Am. J. Obstet. Gynecol. 198, e91–e97. doi: 10.1016/j.ajog.2007.06.039
Tann, C. J., Mpairwe, H., Morison, L., Nassimu, K., Hughes, P., Omara, M., et al. (2006). Lack of effectiveness of syndromic management in targeting vaginal infections in pregnancy in Entebbe, Uganda. Sex. Transm. Infect. 82, 285–289. doi: 10.1136/sti.2005.014845
Tärnberg, M., Jakobsson, T., Jonasson, J., and Forsum, U. (2002). Identification of randomly selected colonies of lactobacilli from normal vaginal fluid by pyrosequencing of the 16S rDNA variable V1 and V3 regions. APMIS 110, 802–810. doi: 10.1034/j.1600-0463.2002.1101106.x
Taylor-Robinson, D., Morgan, D. J., Sheehan, M., Rosenstein, I. J., and Lamont, R. F. (2003). Relation between Gram-stain and clinical criteria for diagnosing bacterial vaginosis with special reference to Gram grade II evaluation. Int. J. STD AIDS 14, 6–10. doi: 10.1258/095646203321043183
Thoma, M. E., Gray, R. H., Kiwanuka, N., Wang, M. C., Sewankambo, N., and Wawer, M. J. (2011). The natural history of bacterial vaginosis diagnosed by gram stain among women in Rakai, Uganda. Sex. Transmitted Dis. 38, 1040–1045. doi: 10.1097/OLQ.0b013e3182275499
Thurman, A. R., Kimble, T., Herold, B., Mesquita, P. M. M., Fichorova, R. N., Dawood, H. Y., et al. (2015). Bacterial vaginosis and subclinical markers of genital tract inflammation and mucosal immunity. AIDS Res. Hum. Retroviruses 31, 1139–1152. doi: 10.1089/aid.2015.0006
Tian, Z., Liu, R., Zhang, H., Yang, M., and Zhang, Y. (2019). Developmental dynamics of antibiotic resistome in aerobic biofilm microbiota treating wastewater under stepwise increasing tigecycline concentrations. Environ. Int. 131:105008. doi: 10.1016/j.envint.2019.105008
Twin, J., Bradshaw, C. S., Garland, S. M., Fairley, C. K., Fethers, K., and Tabrizi, S. N. (2013). The potential of metatranscriptomics for identifying screening targets for bacterial vaginosis. PLoS ONE 8:e76892. doi: 10.1371/journal.pone.0076892
van de Wijgert, J. H. H. M., Borgdorff, H., Verhelst, R., Crucitti, T., Francis, S., Verstraelen, H., et al. (2014). The vaginal microbiota: what have we learned after a decade of molecular characterization? PLoS ONE 9:e105998. doi: 10.1371/journal.pone.0105998
van de Wijgert, J. H. H. M., Morrison, C. S., Cornelisse, P. G., Munjoma, M., Moncada, J., Awio, P., et al. (2008). Bacterial vaginosis and vaginal yeast, but not vaginal cleansing, increase HIV-1 acquisition in African women. J. Acquir. Immune Defic. Syndr. 48, 203–210. doi: 10.1097/QAI.0b013e3181743936
van Houdt, R., Ma, B., Bruisten, S. M., Speksnijder, A. G. C. L., Ravel, J., and de Vries, H. J. C. (2018). Lactobacillus iners-dominated vaginal microbiota is associated with increased susceptibility to Chlamydia trachomatis infection in Dutch women: a case-control study. Sex. Transm. Infect. 94, 117–123. doi: 10.1136/sextrans-2017-053133
van Schaik, W. (2015). The human gut resistome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:e0087. doi: 10.1098/rstb.2014.0087
Vaneechoutte, M. (2017). Lactobacillus iners, the unusual suspect. Res. Microbiol. 168, 826–836. doi: 10.1016/j.resmic.2017.09.003
Vaneechoutte, M., Guschin, A., Van Simaey, L., Gansemans, Y., Van Nieuwerburgh, F., and Cools, P. (2019). Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int. J. Syst. Evol. Microbiol. 69, 679–687. doi: 10.1099/ijsem.0.003200
Verhelst, R., Verstraelen, H., Claeys, G., Verschraegen, G., Delanghe, J., Van Simaey, L., et al. (2004). Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis. BMC Microbiol. 4:16. doi: 10.1186/1471-2180-4-16
Verhelst, R., Verstraelen, H., Claeys, G., Verschraegen, G., Van Simaey, L., De Ganck, C., et al. (2005). Comparison between Gram stain and culture for the characterization of vaginal microflora: definition of a distinct grade that resembles grade I microflora and revised categorization of grade I microflora. BMC Microbiol. 5:61. doi: 10.1186/1471-2180-5-61
Verstraelen, H., and Swidsinksi, A. (2019). The biofilm in bacterial vaginosis: implications for epidemiology, diagnosis and treatment: 2018 update. Curr. Opin. Infect. Dis. 32, 38–42. doi: 10.1097/QCO.0000000000000516
Verstraelen, H., Verhelst, R., Claeys, G., De Backer, E., Temmerman, M., and Vaneechoutte, M. (2009). Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol. 9:116. doi: 10.1186/1471-2180-9-116
Vitali, B., Cruciani, F., Picone, G., Parolin, C., Donders, G., and Laghi, L. (2015). Vaginal microbiome and metabolome highlight specific signatures of bacterial vaginosis. Eur. J. Clin. Microbiol. Infect. Dis. 34, 2367–2376. doi: 10.1007/s10096-015-2490-y
Wang, K.-D., and Su, J.-R. (2014). Quantification of Atopobium vaginae loads may be a new method for the diagnosis of bacterial vaginosis. Clin. Lab. 60, 1501–1508. doi: 10.7754/Clin.Lab.2014.131121
Watson, E., and Reid, G. (2018). Metabolomics as a clinical testing method for the diagnosis of vaginal dysbiosis. Am. J. Reprod. Immunol. 80:e12979. doi: 10.1111/aji.12979
Watts, G. S., Youens-Clark, K., Slepian, M. J., Wolk, D. M., Oshiro, M. M., Metzger, G. S., et al. (2017). 16S rRNA gene sequencing on a benchtop sequencer: accuracy for identification of clinically important bacteria. J. Appl. Microbiol. 123, 1584–1596. doi: 10.1111/jam.13590
Weissenbacher, E. R., Donders, G., Unzeitig, V., Martinez de Tejada, B., Gerber, S., Halaska, M., et al. (2012). A comparison of dequalinium chloride vaginal tablets (Fluomizin®) and clindamycin vaginal cream in the treatment of bacterial vaginosis: a single-blind, randomized clinical trial of efficacy and safety. Gynecol. Obstet. Invest. 73, 8–15. doi: 10.1159/000332398
Weissenbacher, T., Walter, C., Mylonas, I., Scholz, C., Gingelmaier, A., and Friese, K. (2010). Interleukin-6, interleukin-10 and interleukin-12 in vaginal fluid from women with bacterial vaginosis. Arch. Gynecol. Obstet. 281, 77–80. doi: 10.1007/s00404-009-1072-6
West, B., Morison, L., van der Loeff, M. S., Gooding, E., Awasana, A. A., Demba, E., et al. (2003). Evaluation of a new rapid diagnostic kit (FemExam) for bacterial vaginosis in patients with vaginal discharge syndrome in the Gambia. Sex. Transm. Dis. 30, 483–489. doi: 10.1097/00007435-200306000-00003
Winsley, T., Dorst, J. M., van, Brown, M. V., and Ferrari, B. C. (2012). Capturing greater 16S rRNA gene sequence diversity within the domain bacteria. Appl. Environ. Microbiol. 78, 5938–5941. doi: 10.1128/AEM.01299-12
World Health Organization (2007). Module 2 Introducing STI Syndromic Case Management. Geneva. Available online at: https://apps.who.int/iris/bitstream/handle/10665/43275/9241593407_mod2_eng.pdf;jsessionid=00099EDAE67E12CBFC8DEAC334AB7296?sequence=3 (accessed September 15 2019).
Yeoman, C. J., Thomas, S. M., Miller, M. E. B., Ulanov, A. V., Torralba, M., Lucas, S., et al. (2013). A multi-omic systems-based approach reveals metabolic markers of bacterial vaginosis and insight into the disease. PLoS ONE 8:e56111. doi: 10.1371/journal.pone.0056111
Zegels, G., Van Raemdonck, G. A. A., Coen, E. P., Tjalma, W. A. A., and Van Ostade, X. W. M. (2009). Comprehensive proteomic analysis of human cervical-vaginal fluid using colposcopy samples. Proteome Sci. 7:17. doi: 10.1186/1477-5956-7-17
Zhou, X., Bent, S. J., Schneider, M. G., Davis, C. C., Islam, M. R., and Forney, L. J. (2004). Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150, 2565–2573. doi: 10.1099/mic.0.26905-0
Zhou, X., Brown, C. J., Abdo, Z., Davis, C. C., Hansmann, M. A., Joyce, P., et al. (2007). Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1, 121–133. doi: 10.1038/ismej.2007.12
Zirath, H., Schnetz, G., Glatz, A., Spittler, A., Redl, H., and Peham, J. R. (2017). Bedside immune monitoring: an automated immunoassay platform for quantification of blood biomarkers in patient serum within 20 minutes. Anal. Chem. 89, 4817–4823. doi: 10.1021/acs.analchem.6b03624
Keywords: bacterial vaginosis, female genital tract, diagnostics, vaginal microbiome, vaginal metabolome, vaginal inflammation
Citation: Redelinghuys MJ, Geldenhuys J, Jung H and Kock MM (2020) Bacterial Vaginosis: Current Diagnostic Avenues and Future Opportunities. Front. Cell. Infect. Microbiol. 10:354. doi: 10.3389/fcimb.2020.00354
Received: 17 December 2019; Accepted: 09 June 2020;
Published: 11 August 2020.
Edited by:
Mario Vaneechoutte, Ghent University, BelgiumReviewed by:
Hans Verstraelen, Ghent University, BelgiumCopyright © 2020 Redelinghuys, Geldenhuys, Jung and Kock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marleen M. Kock, bWFybGVlbi5rb2NrQHVwLmFjLnph
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.