- 1Medical Microbiology, UMC Utrecht, Utrecht University, Utrecht, Netherlands
- 2GSK, Siena, Italy
- 3Interfaculty Institute of Microbiology and Infection Medicine, University of Tübingen, Tübingen, Germany
- 4Department of Medical Microbiology and Infection Prevention, Amsterdam University Medical Center, University of Amsterdam, Amsterdam, Netherlands
- 5Netherlands Reference Laboratory for Bacterial Meningitis, Amsterdam University Medical Center, Amsterdam, Netherlands
Antigen-presenting cells (APCs) are present throughout the human body—in tissues, at barrier sites and in the circulation. They are critical for processing external signals to instruct both local and systemic responses toward immune tolerance or immune defense. APCs express an extensive repertoire of pattern-recognition receptors (PRRs) to detect and transduce these signals. C-type lectin receptors (CLRs) comprise a subfamily of PRRs dedicated to sensing glycans, including those expressed by commensal and pathogenic bacteria. This review summarizes recent findings on the recognition of and responses to bacteria by membrane-expressed CLRs on different APC subsets, which are discussed according to the primary site of infection. Many CLR-bacterial interactions promote bacterial clearance, whereas other interactions are exploited by bacteria to enhance their pathogenic potential. The discrimination between protective and virulence-enhancing interactions is essential to understand which interactions to target with new prophylactic or treatment strategies. CLRs are also densely concentrated at APC dendrites that sample the environment across intact barrier sites. This suggests an–as yet–underappreciated role for CLR-mediated recognition of microbiota-produced glycans in maintaining tolerance at barrier sites. In addition to providing a concise overview of identified CLR-bacteria interactions, we discuss the main challenges and potential solutions for the identification of new CLR-bacterial interactions, including those with commensal bacteria, and for in-depth structure-function studies on CLR-bacterial glycan interactions. Finally, we highlight the necessity for more relevant tissue-specific in vitro, in vivo and ex vivo models to develop therapeutic applications in this area.
Introduction
A large variety of bacterial species, collectively called the microbiome, lives in and on the human body. Most of these species have beneficial effects on human health, but opportunistic pathogens are also frequently present. In case of barrier defects or (temporary) immune system impairments, these microbes can enter tissues with the risk of causing local or, upon further dissemination, systemic infections. Fortunately, the human immune system is well-equipped with a wide range of innate pattern-recognition receptors (PRRs) that sense specific constituents or microbe-associated molecular patterns of bacteria. Among these PRRs, four different families are currently distinguished (Takeuchi and Akira, 2010). Nucleotide-binding oligomerization domain-like receptors and retinoic acid-inducible gene-I-like receptors (RLRs) are the cytoplasmic sensors of a cell, whereas Toll-like receptors (TLRs) are expressed in endosomes and on the cell surface (Takeuchi and Akira, 2010). The family of C-type lectin receptors (CLRs) include both transmembrane and soluble extracellular proteins (Brown et al., 2018). On tissue-resident antigen-presenting cells (APCs), engagement of PRRs including CLRs triggers local cytokine production, which is important to attract other immune cells such as neutrophils from the circulation to the site of infection and clear invading microbes (Janela et al., 2019; Sparber et al., 2019). Moreover, CLR-induced APC activation is instrumental to instruct adaptive immunity, including T cell polarization through production of cytokines and expression of co-stimulatory molecules, antibody production as well as immunological memory formation (Brown et al., 2018). The induction of these adaptive responses is indispensable for protection from reinfection and key for efficacy of vaccines. Therefore, a better understanding of CLR-bacterial glycan interactions could aid the development of therapeutic applications and vaccines.
CLRs are specialized in recognition of exposed sugar residues or sugar motifs present on self as well as non-self structures (Hoving et al., 2014; Yan et al., 2015). Recognition of specific glycans occurs through one or more carbohydrate recognition domains (CRDs) in a Ca2+-dependent manner. CLRs expressed on APCs can be divided based on their topology as type I and type II transmembrane proteins; type I receptors are characterized by the N-terminus pointing out of the cell and multiple CRDs, whereas type II receptors have their N-terminus directed toward the cytoplasm and an extracellular C-terminus that contains a single CRD (van Kooyk, 2008). Receptors of both groups have a stalk region, a transmembrane region and an intracellular domain with or without a signaling motif. Within type I and type II receptors, CLRs can additionally be categorized based on conserved amino acid motifs in their CRDs that determine their glycan specificity and Ca2+ coordination. CLRs with an EPN (Glu-Pro-Asn) amino acid motif in their CRD, such as DC-SIGN (CD209), langerin (CD207) and mannose receptor (MR, CD206), preferentially bind glycans with equatorial 3- and 4-hydroxyl groups such as mannose, fucose, and N-acetylglucosamine (GlcNAc) residues. On the other hand, CLRs with a QPD (Gln-Pro-Asp) motif preferentially bind glycans with axial 4-hydroxyl groups such as galactose and N-acetylgalactosamine (GalNAc) terminated glycans (Drickamer, 1992). In humans, most CLRs possess the EPN motif, with the exception of Macrophage Galactose-type C-type lectin (MGL, CD301), which possesses the QPD motif (Drickamer and Taylor, 2015). Despite the shared structural features of their CRDs, CLRs display considerable variation in overall structure, cellular expression profiles and signal transduction cascades. These differences have important consequences for the specific contributions of CLRs to antimicrobial immunity, since they strongly affect ligand specificity and the induced immune responses, which can either support host defense or allow immune escape. The role of CLRs in host defense against fungal, viral and mycobacterial infections has recently been reviewed by others (Liu et al., 2017; Shiokawa et al., 2017; Bermejo-Jambrina et al., 2018). Instead, our review focuses on recent findings on the importance of CLRs in recognition of and responses to bacterial pathogens by APCs. In addition, we highlight tools and technologies used for the identification of new interactions, and discuss challenges in the choice of appropriate cell model systems and in the translation of in vitro to in vivo studies. We conclude our review with possible applications of the gathered knowledge for the development of new CLR targeting strategies in vaccines or CLR blocking to counter bacterial immune evasion (Lang et al., 2011; Wamhoff et al., 2019).
Recognition of Bacterial Glycans by Tissue-Resident APCs
The bacterial cell wall is essential for bacterial survival; it defines bacterial cell shape, is critical to sequester ions for bacterial homeostasis and serves as a scaffold for proteins and glycopolymers, to name but a few important features (Silhavy et al., 2010; Dorr et al., 2019). As such, it is much more than a structure that provides resistance to physical stress or harmful environmental factors. In fact, the bacterial cell wall and all its associated structures provide an important interface for direct sensing and communication with the environment, including the host. Despite considerable differences in overall cell wall composition between Gram-positive and -negative bacteria, both classes of bacteria decorate their cell wall with glycans. The best studied examples are capsular polysaccharides, lipopolysaccharide (LPS) and peptidoglycan. Capsular polysaccharides and LPS are effective vaccine antigens when conjugated to protein carriers (glycoconjugate vaccines), whereas proteins in the peptidoglycan biosynthesis pathways are proven targets of antibiotics (Schneider and Sahl, 2010; Rappuoli, 2018). However, bacteria produce a much broader array of glycan structures, which are incorporated in glycolipids, proteins, flagella and glycopolymers (Tytgat and Lebeer, 2014). All these structures are potential ligands for CLRs, and considerable insight into specific molecular interactions has been made the past decades (Prado Acosta and Lepenies, 2019). Importantly, these studies have revealed that interactions between bacterial glycans and CLRs do not always support host defense. Instead, bacteria can exploit CLR interactions for immune evasion, resulting in subversion of host defense responses and increased morbidity. Consequently, detailed molecular and functional insight into bacterial glycan recognition by CLRs is critical to distinguish beneficial from detrimental interactions and inform the development of new treatment or prophylactic strategies.
The functional consequences of CLR engagement are determined by the encountered bacterial ligands but also on the location in the body where the interaction occurs. First, different tissues are populated with specific APC subtypes, which can be phenotypically distinguished from each other by presence of specific immunological markers (Bigley et al., 2015; Alculumbre and Pattarini, 2016; Gunawan et al., 2016; Alcantara-Hernandez et al., 2017; Collin and Bigley, 2018). Second, the local microenvironment provides specific signals to induce APC tissue adaptation, resulting in different receptor expression profiles and migratory capacities of similar APC subtypes in different tissues (Lundberg et al., 2013; Alcantara-Hernandez et al., 2017; Collin and Bigley, 2018). Here, we summarize and discuss specific interactions between CLRs and bacterial glycans (Figure 1), categorized by tissue as relevant for site of bacterial entry.
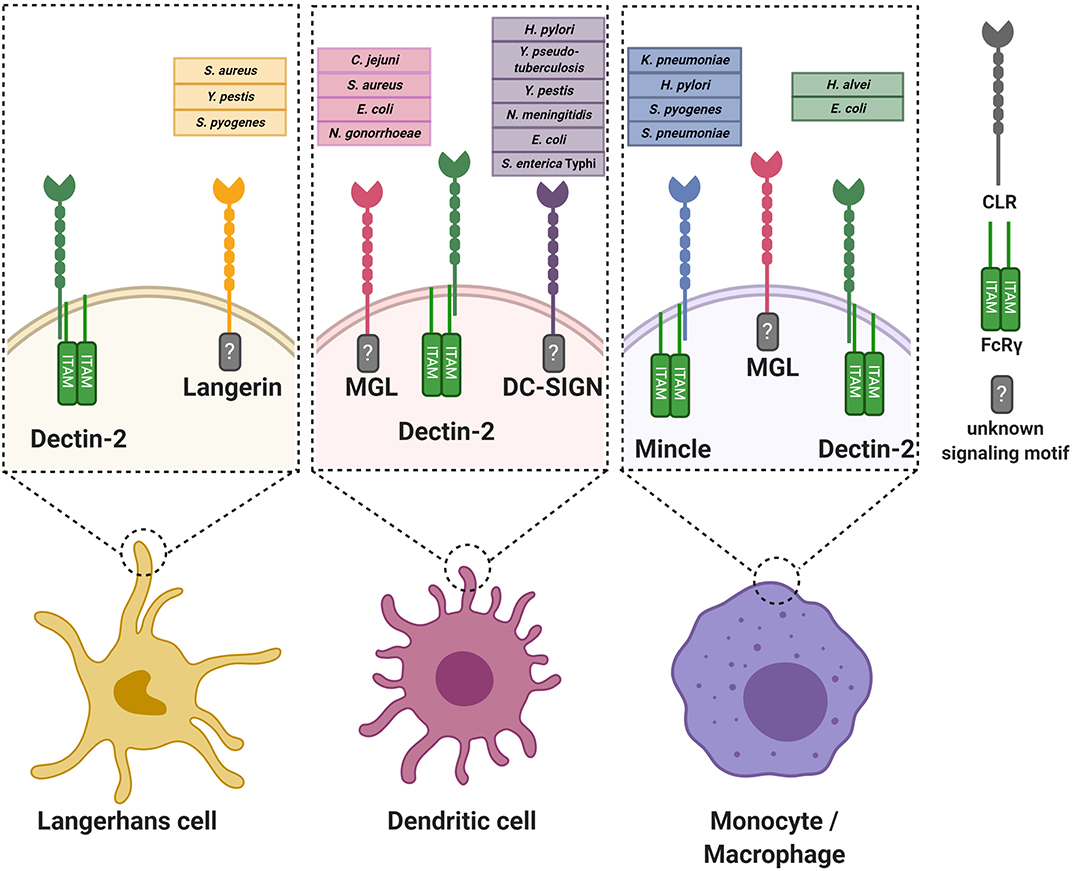
Figure 1. Graphical overview of the discussed C-type lectin receptors on various antigen-presenting cells. For every receptor, the known interacting bacterial species are indicated. Where known, the intracellular signaling motif or associated signaling adaptor molecule is stated. For simplicity, the oligomerization state of the CLRs is not incorporated in the Figure. FcRγ, Fc receptor gamma chain.
Skin
The skin represents the largest organ of the body and is colonized by a plethora of microorganisms (Byrd et al., 2018). Immune cells of the skin constantly interact with microbes and their products, even deeper within the tissue, without causing infection or inflammation (Nakatsuji et al., 2013). However, two common skin resident species, Staphylococcus aureus and Streptococcus pyogenes, are also frequent causes of skin infections (Cardona and Wilson, 2015). Skin infections caused by these pathogens are diverse in their presentation and pose a risk for life-threatening systemic infections. Both these skin-tropic pathogens, but not other more abundant species, such as Staphylococcus epidermidis and Staphylococcus lugdunensis, interact with the CLR langerin (CD207) as confirmed by experiments using recombinant human langerin as well as ectopically-expressing langerin cell lines (van Dalen et al., 2019a,b). Langerin is exclusively expressed on human Langerhans cells (LCs), which are a subclass of myeloid cells that are abundantly present in the skin epidermis. LCs form an important first line of defense against pathogens (Doebel et al., 2017; Deckers et al., 2018). Although most studies focus on skin LCs, their localization is not restricted to the skin; they are also present in mucosal tissues and other epithelial linings, for example foreskin, cervical mucosal tissue, tonsils, tongue and the upper respiratory tract (Patterson et al., 2002; van der Vlist et al., 2011).
Langerin is expressed as a trimer through oligomerization of the neck region, and its cytoplasmic tail contains a putative proline-rich signaling motif (Valladeau et al., 2000; Stambach and Taylor, 2003; Tateno et al., 2010) (Table 1). Based on binding profiles of recombinant human langerin to individual microbes, glycan arrays, langerin has specificity for sulfated and mannosylated glycans as well as β-glucans, which it binds in a Ca2+-dependent manner (de Jong et al., 2010b; Feinberg et al., 2010, 2011; Hanske et al., 2017). However, these particular glycans are unlikely to be involved in the interaction with S. pyogenes or S. aureus, as they are not produced by these bacterial species. Although the ligand of langerin on S. pyogenes has not been identified yet (van Dalen et al., 2019b), human pharyngeal LCs have been observed to interact with S. pyogenes (Reed et al., 1994). On S. aureus, the langerin ligand depends on the gene tarS as confirmed by binding studies comparing wild-type and isogenic mutant strains (van Dalen et al., 2019a). TarS encodes a glycosyltransferase that attaches a conserved β1,4-linked N-acetylglucosamine (β1,4-GlcNAc) moiety on the surface glycopolymer wall teichoic acid (WTA) (Brown et al., 2012), which is an important structural component of the Gram-positive bacterial cell wall (Brown et al., 2013). Recognition of WTA β1,4-GlcNAc increases cytokine production of Th1- and Th17-skewing cytokines by in vitro-generated LCs. Correspondingly, epicutaneous infection with S. aureus producing β1,4-GlcNAc increases skin inflammation in human langerin transgenic mice (van Dalen et al., 2019a). The use of langerin transgenic mice was required since the murine langerin homolog does not recognize S. aureus (van Dalen et al., 2019a), which is of interest as S. aureus is not a mouse commensal. Species-specificity for this interaction may therefore reflect the extended co-evolution of S. aureus with the human host. Approximately 30% of the S. aureus strains are able to co-decorate WTA with the accessory modification α1,4-GlcNAc, which requires the glycosyltransferase TarM (Xia et al., 2010). Langerin does not interact with α1,4-GlcNAc-modified WTA, but co-expression of α1,4-GlcNAc and β-GlcNAc decreased interaction with langerin, suggestion a possible strategy to evade detection by langerin (van Dalen et al., 2019a) Recently, a third WTA glycosyltransferase, TarP, was identified in 5–10% of S. aureus strains (Gerlach et al., 2018). TarP attaches β1,3-GlcNAc to WTA (Gerlach et al., 2018), but it is currently unknown whether or how this modification impacts interaction with langerin or responses by LCs. Several SNPs in the langerin CRD alter the ligand specificity of langerin (Feinberg et al., 2013). These SNPs impair S. aureus recognition and uptake by langerin-expressing cells, thereby providing a potential increase in disease susceptibility (van Dalen et al., 2019b). Intriguingly, interaction with S. pyogenes was much less affected by these SNPs. Although preliminary, these data seem to suggest that such genetic variation does not affect overall susceptibility to bacterial infections (van Dalen et al., 2019b).
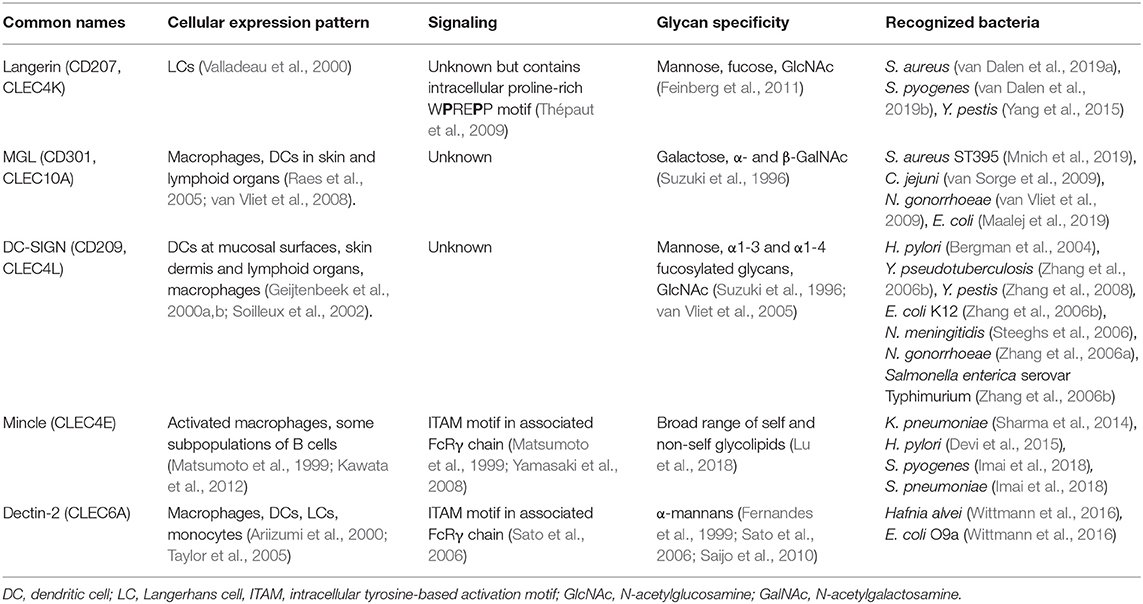
Table 1. Overview of human CLRs and their characteristics discussed in this review, and their characteristics.
Another bacterium that can enter the body through the skin is the Gram-negative pathogen Yersinia pestis, the cause of plague. Infections with Y. pestis can occur after a bite from an infected flee, which delivers the bacterium directly into the tissue, past the protective layer of stratum corneum. Through interaction with langerin, Y. pestis can be phagocytosed by LCs (Yang et al., 2015). In addition to langerin, Y. pestis also interacts with Dendritic-cell-specific intracellular adhesion molecule-3-grabbing non-integrin (DC-SIGN, CD209) (Zhang et al., 2008), which is expressed on dendritic cells (DCs), but not LCs, in mucosal linings, skin dermis, and lymphoid organs and macrophages (Geijtenbeek et al., 2000a,b; Soilleux et al., 2002). DC-SIGN is expressed as a tetramer on the cell surface, with each monomer consisting of a single CRD, neck region and an intracellular domain (van Kooyk and Geijtenbeek, 2003). Although triggering of DC-SIGN alone does not induce DC responses, activation of DC-SIGN modulates signaling cascades induced by other PRRs (Gringhuis et al., 2009). DC-SIGN selectively recognizes oligosaccharides containing high mannose residues but also fucosylated structures in a Ca2+-dependent manner and tetramerization of the receptor favors the recognition of closely-spaced mannose residues on the target molecule (Mitchell et al., 2001; Feinberg et al., 2005; Yu et al., 2009). For Y. pestis, the interaction with langerin and DC-SIGN occurs through the core oligosaccharide of LPS and strains that are genetically-engineered to cover the LPS core are defective in DC and LC invasion (Zhang et al., 2008; Yang et al., 2015). Lectin-dependent phagocytosis seems to favor dissemination of Y. pestis by facilitating migration of bacteria to the lymph nodes (Yang et al., 2015, 2019). However, additional infection experiments in knock-out mice are required to specifically link the observed phenotype to the langerin or DC-SIGN mouse homolog (Zhang et al., 2008; Yang et al., 2015, 2019).
Skin macrophages and DCs, but not LCs, express the CLR Macrophage Galactose-type C-type lectin (MGL, CD301) (Raes et al., 2005; van Vliet et al., 2008). MGL is a homotrimeric receptor that interacts in a Ca2+-dependent manner with galactose and terminal GalNAc residues through the QPD motif in its CRD (Suzuki et al., 1996; Marcelo et al., 2014; Tanaka et al., 2017). Recently, a specific lineage of S. aureus (ST395) was shown to interact with MGL, as based on binding experiments with recombinant human MGL (Mnich et al., 2019). This interaction depends on the unique WTA structure of this S. aureus lineage, which is composed of a glycerol-phosphate (GroP) backbone (as opposed to ribitol-phosphate in all other described S. aureus lineages) decorated with α-N-acetylgalactosamine (α-GalNAc) (Winstel et al., 2013, 2014). WTA α-GalNAc expression by wild-type S. aureus confers binding to human monocyte-derived dendritic cells (moDCs) and induces proinflammatory cytokine production compared to S. aureus strains genetically modified to lack α-GalNAc expression (Mnich et al., 2019). It remains to be determined whether the observed responses correspond to the responses of primary human macrophages or DCs isolated from skin. This knowledge is highly relevant to our understanding of the skin immune detection of these S. aureus strains.
In addition to interaction with langerin, S. pyogenes cell wall components also activate Macrophage inducible C-type lectin (Mincle). Mincle is modestly expressed under homeostatic conditions but significantly upregulated on resident and attracted myeloid cells and neutrophils upon infection or sterile inflammation in murine and human skin (Iborra et al., 2016; Kostarnoy et al., 2017). Increased expression results from increased transcriptional activity induced by inflammatory cytokines such as TNF-α, IL-6, and IFN-γ (Matsumoto et al., 1999). Enhanced surface expression and phagocytic capacity is also conferred by the formation of heteromeric complexes of Mincle with other CLRs such as macrophage C-type lectin (MCL) (Lobato-Pascual et al., 2013). Despite the presence of an EPN motif in its CRD, which usually confers specificity for mannan, all Mincle ligands identified so far contain a lipid moiety in addition to the glycan, suggesting that glycolipids are the molecular signature required for Mincle recognition (Lu et al., 2018). Mincle induces cellular activation through coupling with the Fc receptor common γ chain (FcRγ), which contains an intracellular ITAM signaling motif (Matsumoto et al., 1999; Yamasaki et al., 2008). Mincle interacts with glycolipid antigens from S. pyogenes, specifically the lipophilic components monoglucosyldiacylglycerol (MGDG) and diglycosyldiacylglycerol (DGDG) (Imai et al., 2018), which constitute membrane anchors for lipoteichoic acid (LTA) in the bacterial cell envelope. Intriguingly, MGDG activates DCs via Mincle, resulting in antigen-induced IL-17 production from CD4+ T cells, whereas DGDG prevents MGDG-induced cellular activation through the same receptor (Imai et al., 2018). Abrogation of Mincle-mediated S. pyogenes detection impairs production of pro-inflammatory cytokines, resulting in increased bacteremia and mortality in a mouse model of systemic infection (Imai et al., 2018). Mincle-mediated protection depends on induction of interferon-γ by a specialized lineage of immature myeloid cells, which in turn requires TLR2-induced IL-6 production by these same cells (Matsumura et al., 2019). As MGDG and DGDG are a product of the same biosynthetic pathway, controlled expression of these different lipid anchors may allow S. pyogenes to escape Mincle detection.
Overall, these examples highlight that APC subsets in the skin express distinct CLR repertoires that affect the molecular interaction and immunological responses upon bacterial infection. As DCs, macrophages and LCs are located at different anatomical depths, it is likely that the depth of infection is an important determinant for the outcome of infection. We believe this represents an important area of research to understand pathogenesis of bacterial skin infections (Quaresma, 2019).
Genital Tract
DCs and LCs are both present at barrier sites of the genital tract. Neisseria gonorrhoeae is a Gram-negative pathogenic bacterium and the cause of the sexually-transmitted disease gonorrhea. It can express different lipooligosaccharide (LOS) variants that correlate with altered disease states, i.e. active disease is associated with N. gonorrhoeae variant C (Schneider et al., 1991). N. gonorrhoeae variants A, B, and C only differ in their LOS glycosylation, expressing terminal GlcNAc, galactose and GalNAc, respectively (van Vliet et al., 2009). Using these three well-defined N. gonorrhoeae variants, recombinant soluble CLR-constructs and human moDCs, it was discovered that N. gonorrhoeae variant C interacts with MGL, whereas N. gonorrhoeae variant A is detected by DC-SIGN. Engagement of MGL on moDCs by N. gonorrhoeae variant C shifts DC cytokine secretion and subsequent T helper differentiation toward Th2 in a co-culture system compared to N. gonorrhoeae variants A and B (Zhang et al., 2006a; van Vliet et al., 2009). The interaction with MGL is thought to benefit bacterial survival as a shift toward Th2 is considered to be a non-protective response against N. gonorrhoeae infection. So far, no interactions of LCs with bacteria relevant to genital tract infections or sexually-transmitted diseases have been studied.
Gastrointestinal Tract
The stomach is a hostile environment due to its very low pH. In fact, it was assumed for a long time that this was a sterile site of the body. However, Helicobacter pylori, a Gram-negative bacterium and cause of ulcers and stomach cancer, has evolved unique strategies to survive and colonize the gastric mucosa. Intriguingly, H. pylori expresses phase-variable LPS and LPS variants containing Lewis epitopes interact with DC-SIGN (Bergman et al., 2004). Functional in vitro experiments using human moDCs and H. pylori strains with and without Lewis antigen epitopes on their LPS, demonstrated that engagement of DC-SIGN blocks Th1 development (Bergman et al., 2004). This response is assumed to contribute to long-lasting colonization of H. pylori in a large proportion of the population. In addition to DC-SIGN, H. pylori also affects immune responses through Mincle as based on experiments with PMA-differentiated THP-1 cells, in which Mincle expression was strongly upregulated after exposure to H. pylori. Furthermore, Mincle expression decreases production of pro-inflammatory cytokines while concordantly increasing the production of anti-inflammatory IL-10 in response to H. pylori (Devi et al., 2015). Computational modeling studies indicate that Mincle could interact with LPS of H. pylori containing Lewis-antigens (Devi et al., 2015), but additional studies are required to confirm the interaction and relevance of H. pylori-Mincle interaction at a molecular level.
In contrast to the stomach, the intestines are colonized by a wealth of different bacterial species. Multiple interactions between pathogenic enteric bacteria and CLRs have been discovered. For example, DC-SIGN interacts with the core LPS, especially the GlcNAc residues, of many Gram-negative enteric pathogens, including Salmonella enterica serovar Typhimurium, E. coli K12 and Yersinia pseudotuberculosis (Zhang et al., 2006b). The interaction between Y. pseudotuberculosis and DC-SIGN promotes systemic dissemination and further infection in mice (He et al., 2019). For the other interactions the impact on infection is awaiting further studies. MGL is targeted by at least two gastrointestinal bacteria, Campylobacter jejuni and E. coli R1, through their LOS terminal GalNAc residues and, in case of C. jejuni, through expression of glycosylated proteins (van Sorge et al., 2009; Maalej et al., 2019). MGL targeting by C. jejuni suppresses IL-6 production by moDCs compared to isogenic C. jejuni mutant strains that lack N-glycan protein modifications (van Sorge et al., 2009). Finally, the CLR Dendritic-cell-associated C-type lectin-2 (Dectin-2/CLEC6A) interacts with mannosylated O-antigen of LPS from the nosocomial bacterial pathogens Hafnia alvei and E. coli O9a (Wittmann et al., 2016). Engagement of Dectin-2 by H. alvei mannosylated LPS enhances TLR4-induced cytokine responses of murine bone marrow-derived DCs (BM-DCs) as demonstrated using BM-DCs from Dectin-2 knockout mice (Wittmann et al., 2016). Dectin-2-induced signaling also synergizes with TLR4 responses in human myeloid cells and is dependent on Syk (Wittmann et al., 2016). These data suggest that Dectin-2 could have an important role in sensing bacteria that express mannosylated structures on their surface. The effect of Dectin-2 engagement for the outcome of infection requires further investigation.
Respiratory Tract
The respiratory tract is continuously exposed to airborne microorganisms, including potential pathogens. The distribution and density of APC subsets differs along the respiratory tract. For example, LCs are much more abundant in the upper respiratory tract but are scarce when descending toward the lung (van der Vlist et al., 2011). On the other hand, subsets of DCs, which vary in expression of surface receptors and therefore likely in function, are observed within specific lung microenvironments (Patel and Metcalf, 2018). More precise and comprehensive information on CLR expression along the respiratory tract would be helpful to further interpret results from studies demonstrating that CLRs can protect from bacterial pneumoniae. Specifically, Mincle was shown to have a critical role in protection from pneumonia caused by Klebsiella pneumoniae and Streptococcus pneumoniae. For K. pneumoniae, no bacterial ligand was identified but the increased susceptibility to pneumonia is associated with an increased bacterial burden and exaggerated hyperinflammation (Sharma et al., 2014). The increased bacterial burden may be explained by defects in neutrophil-mediated clearance, as Mincle-deficient neutrophils show an impaired uptake of non-opsonized bacteria and impaired formation of neutrophil-extracellular traps (Sharma et al., 2014, 2017). A similar increase in pneumonia as well as mortality was observed in Mincle-knockout vs. wild-type mice challenged intrapulmonary with S. pneumoniae serotype 19F. The Mincle ligand of S. pneumoniae was identified as the LTA anchor moiety glucosyl-diacylglycerol (Glc-DAG), which is the same ligand as MGDG identified in S. pyogenes (Imai et al., 2018). Engagement of Mincle was not sufficient to protect mice from invasive pneumococcal disease as caused by S. pneumoniae serotype 3, which is more aggressive compared to serotype 19F as this strain causes bacteremia in addition to pneumonia (Behler-Janbeck et al., 2016). These results suggest that Mincle may contribute to local lung immunity against S. pneumoniae but is unable to contain bacteria once in the blood stream (Behler-Janbeck et al., 2016). However, it is very difficult to draw firm conclusions on the role of Mincle in pneumococcal infection, since the strains were not quantified for expression of the Mincle ligand. In addition, the two strains are genetically quite diverse, which likely alters the molecular context in which the Mincle ligand is expressed. As discussed above, this could lead to co-engagement of additional cellular receptors, leading to a different outcome of Mincle activation. Therefore, additional studies are required to fully unravel the role of Mincle in anti-pneumococcal lung immunity.
Nasopharynx
Neisseria meningitidis can colonize the nasopharynx of healthy individuals but is also a common cause of invasive disease, most notably sepsis and meningitis. Dependent on the molecular composition of N. meningitidis LPS, the strain can interact with DC-SIGN with important consequences for uptake and induction of adaptive immunity. Truncation of N. meningitidis LPS (ΔlgtB; representative of immunotype L6) results in enhanced uptake by human moDCs through DC-SIGN and induction of anti-bacterial Th1 responses, as assessed by in vitro co-cultures with human primary cells (Steeghs et al., 2006). Immunotype L6 is rarely found among clinical isolates, suggesting that interaction with DC-SIGN is unfavorable for N. meningitidis survival (Steeghs et al., 2006). From an alternative perspective, immunotype L6 LPS may be used as a powerful vaccine adjuvant as it induces a favorable Th profile compared to unfavorable Th2 type responses with the currently approved human adjuvant alum.
CLR-Mediated Transcompartmental Sampling
The studies described so far all involve interactions between CLRs and pathogenic or nosocomial bacterial species within tissues. However, interactions with commensal members of the microbiome are also likely to occur (Li T. H. et al., 2019). APCs at barrier sites such as the skin, gut or lung do not only respond to bacteria that have penetrated the epithelial barrier, but also have the capacity to actively sample the environment through extension of dendrites across the physical barriers (Rescigno et al., 2001; Niess et al., 2005; Jahnsen et al., 2006; Sung et al., 2006; Kubo et al., 2009; Thornton et al., 2012; Yoshida et al., 2014). CLRs are concentrated at the tips of APC protrusions and are able to trigger localized antigen uptake (Baranov et al., 2014), suggesting a critical role for CLRs in transcompartmental antigen sensing and uptake. The biological significance of this uptake process was demonstrated in a mouse model of staphylococcal scalded skin syndrome (SSSS), which depends on S. aureus exfoliating toxin (ET) (Ouchi et al., 2011). Patch-immunization induces specific cellular and humoral immunity that protects mice from SSSS which depends on the presence of LCs (Ouchi et al., 2011). Importantly, antigen-uptake through patch-immunization does not induce local inflammation or tissue damage (Ouchi et al., 2011). As such, non-invasive application is being explored for development of vaccines against infectious agents (Zheng et al., 2018). Investigations in this area may reveal a role for CLR-mediated APC transcompartmental uptake and activation in maintaining tissue homeostasis at barrier sites.
The Impact of Biological Variation on CLR-Bacterial Interactions
In many of the studies described above, new interactions were identified using a representative bacterial strain of the species of interest and a single growth condition. Biologically-relevant interactions may be missed using such a limited set of experimental conditions. First, the bacterial target may only be expressed in a specific environment or under specific growth condition as a result of regulation. We have recently observed this for the interaction between MGL and Staphylococcus lugdunensis (Mnich et al., 2019). The gene encoding the bacterial glycosyltransferase TagN, responsible for the incorporation of GalNAc on WTA (Winstel et al., 2013, 2014), was identified in the S. lugdunensis genome, but no MGL binding to this bacterial strain was observed (Mnich et al., 2019). In contrast, constitutive expression of S. lugdunensis tagN from a plasmid did confer MGL binding (Mnich et al., 2019). These results seem to indicate lack of tagN expression in S. lugdunensis under our experimental growth conditions. Second, the target may only be present in a restricted subset or lineage from that specific species. Again, we can take the example of MGL interaction with GalNAc-decorated WTA, which is exclusive for the S. aureus ST395 lineage (Mnich et al., 2019). Overall, the glycan expression profile or repertoire on the bacterial surface after in vitro growth may not be representative for the in vivo situation. Variation between strains from a single species can be estimated based on genetic studies, but, as illustrated with the example of S. lugdunensis above, the presence of a gene does not guarantee expression under the used experimental conditions. In addition to genetic sequences, information on conditions that induce the gene or glycan biosynthesis cluster of interested can be estimated by transcriptional data from a wide range of growth conditions (Mader et al., 2016).
Even when a CLR-bacterial interaction is identified in vitro, translation to mouse models is often challenging as no murine homologs exist (e.g., DC-SIGN) or have vastly different glycan-binding profiles compared to the human CLR homolog (Hanske et al., 2016; van Dalen et al., 2019a). As an example of the latter, comparison of the glycan specificity using a bacterial glycan array for recombinantly-expressed murine and human langerin revealed a different binding pattern, in which murine langerin recognizes a broad set of bacterial glycans, but the binding pattern of human langerin is much more restricted (Hanske et al., 2017). Therefore, results from mouse infection models where CLRs are implicated should be interpreted with care with regard to translation of the results for human infection.
Multivalency and Epitope Density in CLR-Ligand Interactions
CLRs not only differ from many other receptors due to their specificity for glycans, but also in their preference for binding multivalent ligands and densely-expressed epitopes (Dam and Brewer, 2010). Indeed, carbohydrate-protein interactions are usually of low affinity, which is overcome by multivalent display of the receptor and/or the ligand (Lepenies et al., 2013). In addition, multivalency and epitope density contribute to discrimination between self and non-self and are not just based on the identity of the glycan, since similar glycan epitopes are often found in pathogens and the host (Medzhitov and Janeway, 2002; Dam and Brewer, 2010). Indeed, data from sugar competition assays for several receptors revealed that glycans carrying multiple carbohydrate epitopes have higher ability to bind to these receptors than monosaccharides (Hsu et al., 2009). Studies that have systematically probed the effect of multivalency and configuration on binding capacity and cellular activation have been conducted for DC-SIGN and MGL (Eriksson et al., 2014; Li R. E. et al., 2019). Overall, higher glycan density increases lectin affinity and kinetics of binding, which is likely explained by one of the following concepts. The first is known as the ‘binding and sliding' mechanism, in which lectins constantly bind to and dissociate from the glycan epitope (Dam and Brewer, 2008). By increasing the number of glycan epitopes along the surface, the lectin-epitope complex becomes longer and of higher affinity. This theory can be applied to soluble lectins such as galectins (Dam and Brewer, 2008). In the second concept, lectins containing multiple CRDs align to allow simultaneous binding of multiple epitopes on a multivalent ligand in a face-to-face interaction (Dam and Brewer, 2008). This concept is proposed for the binding mechanism of DC-SIGN, which forms a tetramer on the surface of DCs and thereby increases affinity for its ligands mannan and gp120 (Mitchell et al., 2001; Bernhard et al., 2004). Additionally, DC-SIGN becomes organized in nanoscale clusters within lipid rafts, which further supports ligand binding and internalization by DCs (Cambi et al., 2004; Neumann et al., 2008). Finally, it was suggested that for the interaction between highly mannosylated structures and DC-SIGN additional secondary binding sites on the CRD are present that further increase the binding affinity of multivalent epitopes (Mitchell et al., 2001). Knowledge of binding and activation requirements have been exploited to target anti-cancer vaccines to DC-SIGN (Li R. E. et al., 2019). Similar targeting strategies could be applied to other CLRs, which may provide the opportunity to target different APC subsets, such as LCs (Porkolab et al., 2018; Neuhaus et al., 2019; Wamhoff et al., 2019).
Interaction between CLRs and other PRR Family Members
Bacteria express a specific combination of PRR ligands. Complementary, PRRs have a specific distribution pattern among different APC subsets. Consequently, engagement of the same CLR can have a different immunological outcome as a result of different co-stimulation in that particular cell type. For example, engagement of MGL by S. aureus ST395 enhances IL-6 and IL-12 production by human moDCs (Mnich et al., 2019), whereas engagement of MGL by C. jejuni glycosylated proteins dampens cytokine production by the same cell type (van Sorge et al., 2009). Since it is known that MGL crosslinking does not induce cellular activation (van Vliet et al., 2013; Heger et al., 2018), these contrasting outcomes may result from differential TLR engagement for S. aureus (activating predominantly TLR2; Takeuchi et al., 2000) and C. jejuni (activating predominantly TLR4; Rathinam et al., 2009). A similar situation of differential co-stimulation may contribute to the different outcomes of langerin targeting by S. aureus and Y. pestis. In this example, langerin-mediated S. aureus recognition induces production of Th1/Th17 cytokines, which is generally accepted to promote S. aureus clearance (van Dalen et al., 2019a), whereas Y. pestis exploits langerin to gain entry into LCs to facilitate dissemination to the lymph nodes (Yang et al., 2015). The exact molecular mechanism underlying these differential outcomes of langerin engagement by these two pathogens is currently not known. It is however intriguing to speculate that langerin targeting modulates intracellular signaling of co-triggered TLRs or possibly alters intracellular routing resulting in different antigen presentation. In the LC context, it is important to realize that these cells appear to express a restricted repertoire of TLRs compared to DCs, with consensus on absence of TLR4, TLR8, and TLR9 (Flacher et al., 2006; van der Aar et al., 2007). Consequently, LCs show no or little response to classical bacterial PAMPs such as LPS (Flacher et al., 2006; van der Aar et al., 2007).
For some CLRs, modulation of signaling pathways from other PRRs is well-characterized. For example, the intracellular signaling pathways of DC-SIGN and TLR4 converge in response to fucose-containing LPS from H. pylori, which skews immune responses from Th1 to Th2 (Bergman et al., 2004; Gringhuis et al., 2014). The molecular details for Th2 skewing have been identified and result from activation of the atypical NF-kB family member Bcl-3 by DC-SIGN, which represses TLR-induced proinflammatory cytokine expression (Gringhuis et al., 2014). This opens up the possibility for therapeutic intervention; addition of anti-DC-SIGN blocking antibodies allow TLR activation to proceed normally, switching T cell responses toward Th1 (Gringhuis et al., 2014), which is thought to enhance pathogen clearance. Another example is the CLR Dectin-2, which interacts with TLR4 signaling after mannosylated LPS (man-LPS) stimulation (Wittmann et al., 2016). In contrast to BM-DCs from wild-type mice, BM-DCs from Dectin-2 and TLR4 knockout mice are unable to enhance IL-10 and TNFα production after stimulation with Man-LPS, suggesting synergy between these two PRRs (Wittmann et al., 2016). These examples illustrate the importance of studying CLR responses in the context of intact bacteria vs. purified components and also in the relevant APC subset to most closely resemble the complex biology of bacteria-APC interaction.
Tools and Challenges in Screening for New Interactions
The described examples of CLR interaction with bacterial pathogens at different barrier sites highlight the importance of this PRR subfamily for pathogen detection and the instruction of appropriate downstream immune responses. As such, it is critical to identify new interactions to improve our understanding of immune tolerance, host defense and disease pathogenesis and apply this knowledge toward the development of new treatment and vaccines. Screening for new interactions between bacterial glycans and CLRs is challenging for multiple reasons. From a biological perspective, we are faced with an overwhelming diversity of glycans that are produced by bacterial species. For example, our gut microbiome contains an estimated 1013 − 1014 bacterial cells (Sender et al., 2016a,b), which produce a plethora of carbohydrate-active enzymes (CAZymes; Cantarel et al., 2009). Unfortunately, it is currently still challenging to predict glycan structures based on genome sequences. In addition, bacterial glycan expression is further regulated by specific environmental conditions in the gut. Interaction with neighboring bacteria may also affect the glycan composition, for example through secretion of glycan-hydrolyzing enzymes that modify (surface-associated) glycans from competitor bacteria. Screening for new interactions among this large diversity of glycans with different CLRs and elucidating their function requires a well-developed tool box. We will discuss developed tools below, as well as the advantages and disadvantages for each strategy.
CLRs can be recombinantly expressed as soluble extracellular domains, which facilitates screening of a diverse range of bacteria. Commercially available recombinant CLRs, both human and mouse, are often expressed as extracellular domains linked to a tag, such as IgG Fc (Hsu and Mahal, 2009; Maglinao et al., 2014; Mayer et al., 2018). Due to the physical nature of IgG Fc, the soluble CLR-fusion proteins are expressed as dimers, which increases avidity of the CLR-glycan interaction compared to single soluble domains. However, the dimeric presentation does not reflect the natural arrangement of several CLRs, such as MGL, langerin and DC-SIGN, as trimers or tetramers on the cell surface (Feinberg et al., 2005, 2010; Jegouzo et al., 2013). In addition, Fc-binding proteins are expressed by several bacterial species (Sidorin and Solov'eva, 2011; Nordenfelt and Bjorck, 2013), such as S. aureus and S. pyogenes, and likely by other species, which further challenges screening for new interactions with these specific human pathogens due to high non-specific background binding. The use of non-Fc-based tags (such as Strep-tag II; Hanske et al., 2016), orientation of the Fc-tagged constructs on arrays or on protein A/G-coated ELISA plates are possible solutions to prevent non-specific Fc-mediated binding (Chen et al., 2008). Alternatively, lectins are spotted on an array, which can then be exposed to bacterial components or whole bacterial cells (Hsu et al., 2006). This platform is able to produce a specific bacterial fingerprint and can assess dynamic changes to the bacterial glycan coat (Hsu et al., 2006). The mentioned platform used well-defined plant lectins to probe the bacterial glycan profile. A similar platform consisting of mammalian CLRs would be a very valuable tool to identify new CLR-bacteria interactions. Finally, CLRs can be ectopically expressed on cell lines, potentially as GFP-reporter constructs (Imai et al., 2018). This approach conserves the natural multimerization state of the receptor, but usually provides only a limited window in which specific interactions can be observed. Furthermore, GFP-reporter cell lines will only identify activating ligands, whereas CLR antagonists remain undetected. Additionally, since not all CLRs contain intrinsic signaling capacity, they need to be expressed as chimeric constructs for example with the CD3ζ chain, which may alter their properties in unknown ways (Imai et al., 2018).
With the aforementioned approaches, specific CLRs can be screened for interactions using intact bacteria. Alternatively, CLR constructs are used to screen glycan arrays (Rillahan and Paulson, 2011; Geissner et al., 2019), which display isolated bacterial glycans. This approach may benefit from the multivalent glycan display, which increases avidity of the interaction, allowing for identification of low affinity interactions. However, isolation of glycans from bacteria can be challenging due to the presence of labile groups or modifications that are lost during sample preparation (Lewis et al., 2004; Edgar et al., 2019). This has sparked the development of synthetic carbohydrate chemistry (Adamo, 2017; van der Es et al., 2017; Zhang and Ye, 2018), which allows the generation of libraries consisting of stable and well-defined glycan structures. In addition, these defined structures enable structure-function studies and crystallography (Gerlach et al., 2018). Crystallography with synthetic glycans is a useful approach once a CLR-glycan interaction is established but due to the vast number of glycosylated bacterial structures it does not allow for a comprehensive approach to screen for new glycan interactions across the entire bacterial kingdom. Complementary use of all mentioned tools is therefore essential to identify new interactions between bacteria and host CLR.
Model Systems to Assess Functional Consequences of CLR Interaction
The above-mentioned studies have provided insight into the functional consequences of CLR-bacteria interactions. However, experimental work with APCs is not without technical and logistical challenges. First and foremost, the transcriptional profiles of APCs from the same subset can differ significantly depending on the tissue where they are isolated from, as cells receive specific signals from their surrounding microenvironment (Lundberg et al., 2013). Consequently, APCs should ideally be isolated from the tissue of interest and be related to the infection route of the bacterium. However, isolation of primary APCs is not feasible for every tissue or for every laboratory due to limited availability or accessibility to human tissues. Even if the relevant tissues are obtained, it is challenging to isolate a suitable number of immature cells for experiments, due to limited cell numbers and (partial) induction of APC maturation by the isolation procedure (Botting et al., 2017). Therefore, APCs differentiated from blood monocytes or CD34+ cord blood cells by a specific cocktail of cytokines, have become widely-used models to study APC- and CLR-induced responses. Using these differentiation strategies, APCs with an immature phenotype can be obtained and differentiated into mature APCs in a controlled manner. In addition to human primary cells, the use of cell lines that can be differentiated in distinct APCs subsets, such as the myeloid CD34+ MUTZ3 cell line (Masterson et al., 2002; Santegoets et al., 2006), have yielded more insight into CLR-mediated interaction and responses (de Jong et al., 2010a; van Dalen et al., 2019a). The advantages of this cell line-based approach are the absence of donor-to-donor variability, independence on donor-derived material and amenability of these cells to CRISPR/Cas9 genetic manipulation, which makes these cells suitable to address the role of specific molecules in immunological interactions. Another possibility to generate APCs is through induced pluripotent stem cells (iPSC) derived from fibroblasts, which offers an unlimited cell source from the same individuals and the possibility to generate patient-specific APCs with genetic defects (Choi et al., 2009, 2011; Yanagimachi et al., 2013; Ackermann et al., 2015). Although these approaches are understandably widely applied for ease of use and quantity of cells, transcriptomics studies have made clear that all in vitro differentiated cell and cell lines are distinct from primary cells (Lundberg et al., 2001; Harman et al., 2013). For example, MUTZ3-derived LCs express several TLRs that are not expressed by primary LCs (Lundberg et al., 2013). Therefore, data obtained with differentiated primary cells, cell lines and iPSC should be interpreted with care, and experiments should be reproduced in more physiologically relevant primary cells, ex vivo tissue models or suitable in vivo models.
Fortunately, in recent years, several ex vivo and 3D tissue models have been developed. Human skin explants have been used to study the interaction between different cell populations and common skin pathogens, including S. aureus (Schaudinn et al., 2017; Olaniyi et al., 2018). This approach is currently the most relevant model to study the interaction of CLRs with bacteria in their natural environment, since the explanted tissue contains all relevant cell populations. Since not all laboratories have access to human skin, an alternative approach is the use of designed skin equivalents consisting of either epidermis alone or together with underlying dermis. Progress is made to repopulate these organotypic skin models with specific skin-resident immune cells (Pupovac et al., 2018), for example CD34+ progenitor cells that differentiate into LCs when co-seeded with human keratinocytes (Regnier et al., 1998). DCs can also be repopulated in the skin equivalent model by introducing human moDCs in between layers of keratinocytes and fibroblasts (Chau et al., 2013). Not only for skin, but also for lung tissue, 3D tissue models have been generated to study molecular pathogenesis of bacterial infections, such as S. aureus toxin-mediated lung pathology relevant for pneumonia (Mairpady Shambat et al., 2015). Also here it was possible to repopulate the model with functional human moDCs (Mizoguchi et al., 2017).
The development of organoid technology has brought about a surge in the use of 3D models, also for studies in the area of infectious diseases (Schutgens and Clevers, 2020). There are now a wide variety of tissues and organs that can be grown from human pluripotent stem cells (hPSCs) with a specific cocktail of growth factors including lung, gut, stomach and even brain organoids (Clevers, 2016). Since organoids consist of all cellular components of the representative organ, they provide opportunities to study bacteria-host interactions in a more physiologically-relevant setting as compared to cell lines. For example, gastric organoids have been used to study host responses to H. pylori (Bartfeld et al., 2015). Despite many advantages, organoids have limitations such as the absence of blood vessels and immune cells, which are crucial aspects of host defense against infections. The development of immune-competent organoids models (Bar-Ephraim et al., 2020) will provide an opportunity to study interactions between CLR and bacteria in the fully differentiated context of different human tissues. Overall, all immunocompetent 3D tissue models can be useful tools to gain insight into bacterial-cell interactions and the influence of the specific environmental context on the functional outcomes of this interactions, i.e., influence of cell-cell communication.
Conclusion and Future Outlook
From their first discovery, CLRs have been studied mainly for their role in anti-fungal and anti-viral immunity, with research on CLR-bacterial interactions lagging behind. Our overview of bacterial-CLR interactions clearly highlights the importance of specific CLRs in anti-bacterial immunity, but also provides examples of pathogens such as Y. pestis that exploit CLR interaction to enhance virulence or survival. Studies have generally focused on the role of CLRs in the detection of bacterial pathogens. However, APCs also actively sample the environment across intact tissue barriers, suggesting an important role in immune homeostasis that has yet to be elucidated. A major challenge in the identification of new interactions of CLRs with bacterial species in the microbiome is the vast diversity of bacterial glycan structures, in combination with technical challenges that have to be overcome to close this knowledge gap. In this review, we have discussed advantages and disadvantages of currently available approaches. Applications of CLR-bacterial interactions include the development of targeting agents for vaccine delivery to specific CLRs on APC subsets, that can help boost an effective adaptive immune and memory response. In this regard, the infectious diseases field should take advantage of the progress in the area of cancer vaccinology, where studies have shown the benefit of in vivo targeting of cancer antigens to APCs to enhance anti-tumor immunity (Hossain and Wall, 2019). Overall, the knowledge gained from studies on bacterial-CLR interactions could therefore not only shed light on the role in immune defense or pathogenicity but also be highly relevant to the translation of vaccine applications.
Author Contributions
MM, RD, and NS wrote, revised, and approved the manuscript. MM prepared the figure and table. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the European Union's Horizon 2020 research H2020-MSCA-ITN (No. 675106 coordinated by Dr. Fabio Bagnoli, GSK, Siena, Italy). RD was supported by the European Molecular Biology Organization and acknowledges infrastructural support by the Cluster of Excellence Controlling Microbes to Fight Infections (EXC 2124) of the German Research Foundation.
Conflict of Interest
MM is a Ph.D. fellow who is enrolled in the Infection and Immunity Ph.D. program, part of the Graduate School of Life Sciences at Utrecht University, and participates in a postgraduate studentship program at GSK.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Figure was created with Biorender.com.
References
Ackermann, M., Liebhaber, S., Klusmann, J. H., and Lachmann, N. (2015). Lost in translation: pluripotent stem cell-derived hematopoiesis. EMBO Mol. Med. 7, 1388–1402. doi: 10.15252/emmm.201505301
Adamo, R. (2017). Advancing homogeneous antimicrobial glycoconjugate vaccines. Acc. Chem. Res. 50, 1270–1279. doi: 10.1021/acs.accounts.7b00106
Alcantara-Hernandez, M., Leylek, R., Wagar, L. E., Engleman, E. G., Keler, T., Marinkovich, M. P., et al. (2017). High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity 47, 1037–1050.e1036. doi: 10.1016/j.immuni.2017.11.001
Alculumbre, S., and Pattarini, L. (2016). Purification of human dendritic cell subsets from peripheral blood. Methods Mol. Biol. 1423, 153–167. doi: 10.1007/978-1-4939-3606-9_11
Ariizumi, K., Shen, G. L., Shikano, S., Ritter, R., Zukas, P. 3rd, Edelbaum, D., et al. (2000). Cloning of a second dendritic cell-associated C-type lectin (dectin-2) and its alternatively spliced isoforms. J. Biol. Chem. 275, 11957–11963. doi: 10.1074/jbc.275.16.11957
Baranov, M., Ter Beest, M., Reinieren-Beeren, I., Cambi, A., Figdor, C. G., and van den Bogaart, G. (2014). Podosomes of dendritic cells facilitate antigen sampling. J. Cell Sci. 127, 1052–1064. doi: 10.1242/jcs.141226
Bar-Ephraim, Y. E., Kretzschmar, K., and Clevers, H. (2020). Organoids in immunological research. Nat. Rev. Immunol. 20, 279–293. doi: 10.1038/s41577-019-0248-y
Bartfeld, S., Bayram, T., van de Wetering, M., Huch, M., Begthel, H., Kujala, P., et al. (2015). In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148, 126–136. e126. doi: 10.1053/j.gastro.2014.09.042
Behler-Janbeck, F., Takano, T., Maus, R., Stolper, J., Jonigk, D., Tort Tarres, M., et al. (2016). C-type lectin mincle recognizes glucosyl-diacylglycerol of streptococcus pneumoniae and plays a protective role in pneumococcal pneumonia. PLoS Pathog 12:e1006038. doi: 10.1371/journal.ppat.1006038
Bergman, M. P., Engering, A., Smits, H. H., van Vliet, S. J., van Bodegraven, A. A., Wirth, H. P., et al. (2004). Helicobacter pylori modulates the T helper cell 1/T helper cell 2 balance through phase-variable interaction between lipopolysaccharide and DC-SIGN. J. Exp. Med. 200, 979–990. doi: 10.1084/jem.20041061
Bermejo-Jambrina, M., Eder, J., Helgers, L. C., Hertoghs, N., Nijmeijer, B. M., Stunnenberg, M., et al. (2018). C-Type lectin receptors in antiviral immunity and viral escape. Front. Immunol. 9:590. doi: 10.3389/fimmu.2018.00590
Bernhard, O. K., Lai, J., Wilkinson, J., Sheil, M. M., and Cunningham, A. L. (2004). Proteomic analysis of DC-SIGN on dendritic cells detects tetramers required for ligand binding but no association with CD4. J. Biol. Chem. 279, 51828–51835. doi: 10.1074/jbc.M402741200
Bigley, V., McGovern, N., Milne, P., Dickinson, R., Pagan, S., Cookson, S., et al. (2015). Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J. Leukoc. Biol. 97, 627–634. doi: 10.1189/jlb.1HI0714-351R
Botting, R. A., Bertram, K. M., Baharlou, H., Sandgren, K. J., Fletcher, J., Rhodes, J. W., et al. (2017). Phenotypic and functional consequences of different isolation protocols on skin mononuclear phagocytes. J. Leukoc. Biol. 101, 1393–1403. doi: 10.1189/jlb.4A1116-496R
Brown, G. D., Willment, J. A., and Whitehead, L. (2018). C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 18, 374–389. doi: 10.1038/s41577-018-0004-8
Brown, S., Santa Maria, J. P. Jr., and Walker, S. (2013). Wall teichoic acids of Gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336. doi: 10.1146/annurev-micro-092412-155620
Brown, S., Xia, G., Luhachack, L. G., Campbell, J., Meredith, T. C., Chen, C., et al. (2012). Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 18909–18914. doi: 10.1073/pnas.1209126109
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi: 10.1038/nrmicro.2017.157
Cambi, A., de Lange, F., van Maarseveen, N. M., Nijhuis, M., Joosten, B., van Dijk, E. M., et al. (2004). Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J. Cell Biol. 164, 145–155. doi: 10.1083/jcb.200306112
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Cardona, A. F., and Wilson, S. E. (2015). Skin and soft-tissue infections: a critical review and the role of telavancin in their treatment. Clin. Infect. Dis. 61(Suppl. 2), S69–S78. doi: 10.1093/cid/civ528
Chau, D. Y., Johnson, C., MacNeil, S., Haycock, J. W., and Ghaemmaghami, A. M. (2013). The development of a 3D immunocompetent model of human skin. Biofabrication 5:035011. doi: 10.1088/1758-5082/5/3/035011
Chen, M. L., Adak, A. K., Yeh, N. C., Yang, W. B., Chuang, Y. J., Wong, C. H., et al. (2008). Fabrication of an oriented Fc-fused lectin microarray through boronate formation. Angew. Chem. Int. Ed. Engl. 47, 8627–8630. doi: 10.1002/anie.200803377
Choi, K. D., Vodyanik, M., and Slukvin, I. I. (2011). Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nat. Protoc. 6, 296–313. doi: 10.1038/nprot.2010.184
Choi, K. D., Vodyanik, M. A., and Slukvin, I. I. (2009). Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J. Clin. Invest. 119, 2818–2829. doi: 10.1172/JCI38591
Clevers, H. (2016). Modeling development and disease with organoids. Cell 165, 1586–1597. doi: 10.1016/j.cell.2016.05.082
Collin, M., and Bigley, V. (2018). Human dendritic cell subsets: an update. Immunology 154, 3–20. doi: 10.1111/imm.12888
Dam, T. K., and Brewer, C. F. (2008). Effects of clustered epitopes in multivalent ligand-receptor interactions. Biochemistry 47, 8470–8476. doi: 10.1021/bi801208b
Dam, T. K., and Brewer, C. F. (2010). Lectins as pattern recognition molecules: the effects of epitope density in innate immunity. Glycobiology 20, 270–279. doi: 10.1093/glycob/cwp186
de Jong, M. A., de Witte, L., Santegoets, S. J., Fluitsma, D., Taylor, M. E., de Gruijl, T. D., et al. (2010a). Mutz-3-derived Langerhans cells are a model to study HIV-1 transmission and potential inhibitors. J. Leukoc. Biol. 87, 637–643. doi: 10.1189/jlb.0809577
de Jong, M. A., Vriend, L. E., Theelen, B., Taylor, M. E., Fluitsma, D., Boekhout, T., et al. (2010b). C-type lectin Langerin is a beta-glucan receptor on human Langerhans cells that recognizes opportunistic and pathogenic fungi. Mol. Immunol. 47, 1216–1225. doi: 10.1016/j.molimm.2009.12.016
Deckers, J., Hammad, H., and Hoste, E. (2018). Langerhans cells: sensing the environment in health and disease. Front. Immunol. 9:93. doi: 10.3389/fimmu.2018.00093
Devi, S., Rajakumara, E., and Ahmed, N. (2015). Induction of mincle by helicobacter pylori and consequent anti-inflammatory signaling denote a bacterial survival strategy. Sci. Rep. 5:15049. doi: 10.1038/srep15049
Doebel, T., Voisin, B., and Nagao, K. (2017). Langerhans cells - the macrophage in dendritic cell clothing. Trends Immunol. 38, 817–828. doi: 10.1016/j.it.2017.06.008
Dorr, T., Moynihan, P. J., and Mayer, C. (2019). Editorial: bacterial cell wall structure and dynamics. Front. Microbiol. 10:2051. doi: 10.3389/fmicb.2019.02051
Drickamer, K. (1992). Engineering galactose-binding activity into a C-type mannose-binding protein. Nature 360, 183–186. doi: 10.1038/360183a0
Drickamer, K., and Taylor, M. E. (2015). Recent insights into structures and functions of C-type lectins in the immune system. Curr. Opin. Struct. Biol. 34, 26–34. doi: 10.1016/j.sbi.2015.06.003
Edgar, R. J., van Hensbergen, V. P., Ruda, A., Turner, A. G., Deng, P., Le Breton, Y., et al. (2019). Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides. Nat. Chem. Biol. 15, 463–471. doi: 10.1038/s41589-019-0251-4
Eriksson, M., Serna, S., Maglinao, M., Schlegel, M. K., Seeberger, P. H., Reichardt, N. C., et al. (2014). Biological evaluation of multivalent lewis X-MGL-1 interactions. Chembiochem 15, 844–851. doi: 10.1002/cbic.201300764
Feinberg, H., Guo, Y., Mitchell, D. A., Drickamer, K., and Weis, W. I. (2005). Extended neck regions stabilize tetramers of the receptors DC-SIGN and DC-SIGNR. J. Biol. Chem. 280, 1327–1335. doi: 10.1074/jbc.M409925200
Feinberg, H., Powlesland, A. S., Taylor, M. E., and Weis, W. I. (2010). Trimeric structure of langerin. J. Biol. Chem. 285, 13285–13293. doi: 10.1074/jbc.M109.086058
Feinberg, H., Rowntree, T. J., Tan, S. L., Drickamer, K., Weis, W. I., and Taylor, M. E. (2013). Common polymorphisms in human langerin change specificity for glycan ligands. J. Biol. Chem. 288, 36762–36771. doi: 10.1074/jbc.M113.528000
Feinberg, H., Taylor, M. E., Razi, N., McBride, R., Knirel, Y. A., Graham, S. A., et al. (2011). Structural basis for langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J. Mol. Biol. 405, 1027–1039. doi: 10.1016/j.jmb.2010.11.039
Fernandes, M. J., Finnegan, A. A., Siracusa, L. D., Brenner, C., Iscove, N. N., and Calabretta, B. (1999). Characterization of a novel receptor that maps near the natural killer gene complex: demonstration of carbohydrate binding and expression in hematopoietic cells. Cancer Res. 59, 2709–2717.
Flacher, V., Bouschbacher, M., Verronese, E., Massacrier, C., Sisirak, V., Berthier-Vergnes, O., et al. (2006). Human Langerhans cells express a specific TLR profile and differentially respond to viruses and Gram-positive bacteria. J. Immunol. 177, 7959–7967. doi: 10.4049/jimmunol.177.11.7959
Geijtenbeek, T. B., Kwon, D. S., Torensma, R., van Vliet, S. J., van Duijnhoven, G. C., Middel, J., et al. (2000a). DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100, 587–597. doi: 10.1016/S0092-8674(00)80694-7
Geijtenbeek, T. B., Torensma, R., van Vliet, S. J., van Duijnhoven, G. C., Adema, G. J., van Kooyk, Y., et al. (2000b). Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100, 575–585. doi: 10.1016/S0092-8674(00)80693-5
Geissner, A., Reinhardt, A., Rademacher, C., Johannssen, T., Monteiro, J., Lepenies, B., et al. (2019). Microbe-focused glycan array screening platform. Proc. Natl. Acad. Sci. U.S.A. 116, 1958–1967. doi: 10.1073/pnas.1800853116
Gerlach, D., Guo, Y., De Castro, C., Kim, S. H., Schlatterer, K., Xu, F. F., et al. (2018). Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity. Nature 563, 705–709. doi: 10.1038/s41586-018-0730-x
Gringhuis, S. I., den Dunnen, J., Litjens, M., van der Vlist, M., and Geijtenbeek, T. B. (2009). Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat. Immunol. 10, 1081–1088. doi: 10.1038/ni.1778
Gringhuis, S. I., Kaptein, T. M., Wevers, B. A., Mesman, A. W., and Geijtenbeek, T. B. (2014). Fucose-specific DC-SIGN signalling directs T helper cell type-2 responses via IKKepsilon- and CYLD-dependent Bcl3 activation. Nat. Commun. 5:3898. doi: 10.1038/ncomms4898
Gunawan, M., Jardine, L., and Haniffa, M. (2016). Isolation of human skin dendritic cell subsets. Methods Mol. Biol. 1423, 119–128. doi: 10.1007/978-1-4939-3606-9_8
Hanske, J., Aleksic, S., Ballaschk, M., Jurk, M., Shanina, E., Beerbaum, M., et al. (2016). Intradomain allosteric network modulates calcium affinity of the C-type lectin receptor langerin. J. Am. Chem. Soc. 138, 12176–12186. doi: 10.1021/jacs.6b05458
Hanske, J., Schulze, J., Aretz, J., McBride, R., Loll, B., Schmidt, H., et al. (2017). Bacterial polysaccharide specificity of the pattern recognition receptor langerin is highly species-dependent. J. Biol. Chem. 292, 862–871. doi: 10.1074/jbc.M116.751750
Harman, A. N., Bye, C. R., Nasr, N., Sandgren, K. J., Kim, M., Mercier, S. K., et al. (2013). Identification of lineage relationships and novel markers of blood and skin human dendritic cells. J. Immunol. 190, 66–79. doi: 10.4049/jimmunol.1200779
He, Y. X., Ye, C. L., Zhang, P., Li, Q., Park, C. G., Yang, K., et al. (2019). Yersinia pseudotuberculosis exploits CD209 receptors for promoting host dissemination and infection. Infect. Immun. 87:e00654–1. doi: 10.1128/IAI.00654-18
Heger, L., Balk, S., Luhr, J. J., Heidkamp, G. F., Lehmann, C. H. K., Hatscher, L., et al. (2018). CLEC10A Is a specific marker for human CD1c(+) dendritic cells and enhances their toll-like receptor 7/8-induced cytokine secretion. Front. Immunol. 9:744. doi: 10.3389/fimmu.2018.00744
Hossain, M. K., and Wall, K. A. (2019). Use of dendritic cell receptors as targets for enhancing anti-cancer immune responses. Cancers 11:418. doi: 10.3390/cancers11030418
Hoving, J. C., Wilson, G. J., and Brown, G. D. (2014). Signalling C-type lectin receptors, microbial recognition and immunity. Cell. Microbiol. 16, 185–194. doi: 10.1111/cmi.12249
Hsu, K. L., and Mahal, L. K. (2009). Sweet tasting chips: microarray-based analysis of glycans. Curr. Opin. Chem. Biol. 13, 427–432. doi: 10.1016/j.cbpa.2009.07.013
Hsu, K. L., Pilobello, K. T., and Mahal, L. K. (2006). Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2, 153–157. doi: 10.1038/nchembio767
Hsu, T. L., Cheng, S. C., Yang, W. B., Chin, S. W., Chen, B. H., Huang, M. T., et al. (2009). Profiling carbohydrate-receptor interaction with recombinant innate immunity receptor-Fc fusion proteins. J. Biol. Chem. 284, 34479–34489. doi: 10.1074/jbc.M109.065961
Iborra, S., Martinez-Lopez, M., Cueto, F. J., Conde-Garrosa, R., Del Fresno, C., Izquierdo, H. M., et al. (2016). Leishmania uses mincle to target an inhibitory ITAM signaling pathway in dendritic cells that dampens adaptive immunity to infection. Immunity 45, 788–801. doi: 10.1016/j.immuni.2016.09.012
Imai, T., Matsumura, T., Mayer-Lambertz, S., Wells, C. A., Ishikawa, E., Butcher, S. K., et al. (2018). Lipoteichoic acid anchor triggers Mincle to drive protective immunity against invasive group A Streptococcus infection. Proc. Natl. Acad. Sci. U.S.A. 115, E10662–E10671. doi: 10.1073/pnas.1809100115
Jahnsen, F. L., Strickland, D. H., Thomas, J. A., Tobagus, I. T., Napoli, S., Zosky, G. R., et al. (2006). Accelerated antigen sampling and transport by airway mucosal dendritic cells following inhalation of a bacterial stimulus. J. Immunol. 177, 5861–5867. doi: 10.4049/jimmunol.177.9.5861
Janela, B., Patel, A. A., Lau, M. C., Goh, C. C., Msallam, R., Kong, W. T., et al. (2019). A subset of type I conventional dendritic cells controls cutaneous bacterial infections through VEGFalpha-mediated recruitment of neutrophils. Immunity 50, 1069–1083.e1068. doi: 10.1016/j.immuni.2019.03.001
Jegouzo, S. A., Quintero-Martinez, A., Ouyang, X., dos Santos, A., Taylor, M. E., and Drickamer, K. (2013). Organization of the extracellular portion of the macrophage galactose receptor: a trimeric cluster of simple binding sites for N-acetylgalactosamine. Glycobiology 23, 853–864. doi: 10.1093/glycob/cwt022
Kawata, K., Illarionov, P., Yang, G. X., Kenny, T. P., Zhang, W., Tsuda, M., et al. (2012). Mincle and human B cell function. J. Autoimmun. 39, 315–322. doi: 10.1016/j.jaut.2012.04.004
Kostarnoy, A. V., Gancheva, P. G., Lepenies, B., Tukhvatulin, A. I., Dzharullaeva, A. S., Polyakov, N. B., et al. (2017). Receptor mincle promotes skin allergies and is capable of recognizing cholesterol sulfate. Proc. Natl. Acad. Sci. U.S.A. 114, E2758–E2765. doi: 10.1073/pnas.1611665114
Kubo, A., Nagao, K., Yokouchi, M., Sasaki, H., and Amagai, M. (2009). External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. J. Exp. Med. 206, 2937–2946. doi: 10.1084/jem.20091527
Lang, R., Schoenen, H., and Desel, C. (2011). Targeting Syk-Card9-activating C-type lectin receptors by vaccine adjuvants: findings, implications and open questions. Immunobiology 216, 1184–1191. doi: 10.1016/j.imbio.2011.06.005
Lepenies, B., Lee, J., and Sonkaria, S. (2013). Targeting C-type lectin receptors with multivalent carbohydrate ligands. Adv. Drug Deliv. Rev. 65, 1271–1281. doi: 10.1016/j.addr.2013.05.007
Lewis, A. L., Nizet, V., and Varki, A. (2004). Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc. Natl. Acad. Sci. U.S.A. 101, 11123–11128. doi: 10.1073/pnas.0403010101
Li, R. E., Hogervorst, T. P., Achilli, S., Bruijns, S. C., Arnoldus, T., Vives, C., et al. (2019). Systematic dual targeting of dendritic cell C-type lectin receptor DC-SIGN and TLR7 using a trifunctional mannosylated antigen. Front. Chem. 7:650. doi: 10.3389/fchem.2019.00650
Li, T. H., Liu, L., Hou, Y. Y., Shen, S. N., and Wang, T. T. (2019). C-type lectin receptor-mediated immune recognition and response of the microbiota in the gut. Gastroenterol. Rep. 7, 312–321. doi: 10.1093/gastro/goz028
Liu, C. H., Liu, H., and Ge, B. (2017). Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell. Mol. Immunol. 14, 963–975. doi: 10.1038/cmi.2017.88
Lobato-Pascual, A., Saether, P. C., Fossum, S., Dissen, E., and Daws, M. R. (2013). Mincle, the receptor for mycobacterial cord factor, forms a functional receptor complex with MCL and FcepsilonRI-gamma. Eur. J. Immunol. 43, 3167–3174. doi: 10.1002/eji.201343752
Lu, X., Nagata, M., and Yamasaki, S. (2018). Mincle: 20 years of a versatile sensor of insults. Int. Immunol. 30, 233–239. doi: 10.1093/intimm/dxy028
Lundberg, A. H., Fukatsu, K., Gaber, L., Callicutt, S., Kotb, M., Wilcox, H., et al. (2001). Blocking pulmonary ICAM-1 expression ameliorates lung injury in established diet-induced pancreatitis. Ann. Surg. 233, 213–220. doi: 10.1097/00000658-200102000-00010
Lundberg, K., Albrekt, A. S., Nelissen, I., Santegoets, S., de Gruijl, T. D., Gibbs, S., et al. (2013). Transcriptional profiling of human dendritic cell populations and models–unique profiles of in vitro dendritic cells and implications on functionality and applicability. PLoS ONE 8:e52875. doi: 10.1371/journal.pone.0052875
Maalej, M. M., Forgione, R. E., Marchetti, R., Bulteau, F. B., Thepaut, M. T., Lanzetta, R., et al. (2019). The human macrophage galactose-type lectin, MGL, recognizes the outer core of E. coli lipooligosaccharide. Chembiochem 20, 1778–1782. doi: 10.1002/cbic.201900087
Mader, U., Nicolas, P., Depke, M., Pane-Farre, J., Debarbouille, M., van der Kooi-Pol, M. M., et al. (2016). Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions. PLoS Genet. 12:e1005962. doi: 10.1371/journal.pgen.1005962
Maglinao, M., Eriksson, M., Schlegel, M. K., Zimmermann, S., Johannssen, T., Gotze, S., et al. (2014). A platform to screen for C-type lectin receptor-binding carbohydrates and their potential for cell-specific targeting and immune modulation. J. Control. Release 175, 36–42. doi: 10.1016/j.jconrel.2013.12.011
Mairpady Shambat, S., Chen, P., Nguyen Hoang, A. T., Bergsten, H., Vandenesch, F., Siemens, N., et al. (2015). Modelling staphylococcal pneumonia in a human 3D lung tissue model system delineates toxin-mediated pathology. Dis. Model. Mech. 8, 1413–1425. doi: 10.1242/dmm.021923
Marcelo, F., Garcia-Martin, F., Matsushita, T., Sardinha, J., Coelho, H., Oude-Vrielink, A., et al. (2014). Delineating binding modes of Gal/GalNAc and structural elements of the molecular recognition of tumor-associated mucin glycopeptides by the human macrophage galactose-type lectin. Chemistry 20, 16147–16155. doi: 10.1002/chem.201404566
Masterson, A. J., Sombroek, C. C., De Gruijl, T. D., Graus, Y. M., van der Vliet, H. J., Lougheed, S. M., et al. (2002). MUTZ-3, a human cell line model for the cytokine-induced differentiation of dendritic cells from CD34+ precursors. Blood 100, 701–703. doi: 10.1182/blood.V100.2.701
Matsumoto, M., Tanaka, T., Kaisho, T., Sanjo, H., Copeland, N. G., Gilbert, D. J., et al. (1999). A novel LPS-inducible C-type lectin is a transcriptional target of NF-IL6 in macrophages. J. Immunol. 163, 5039–5048.
Matsumura, T., Ikebe, T., Arikawa, K., Hosokawa, M., Aiko, M., Iguchi, A., et al. (2019). Sequential sensing by TLR2 and mincle directs immature myeloid cells to protect against invasive group A streptococcal infection in mice. Cell Rep. 27, 561–571.e566. doi: 10.1016/j.celrep.2019.03.056
Mayer, S., Moeller, R., Monteiro, J. T., Ellrott, K., Josenhans, C., and Lepenies, B. (2018). C-Type Lectin Receptor (CLR)-Fc fusion proteins as tools to screen for novel CLR/bacteria interactions: an exemplary study on preselected Campylobacter jejuni isolates. Front. Immunol. 9:213. doi: 10.3389/fimmu.2018.00213
Medzhitov, R., and Janeway, C. A. Jr. (2002). Decoding the patterns of self and nonself by the innate immune system. Science 296, 298–300. doi: 10.1126/science.1068883
Mitchell, D. A., Fadden, A. J., and Drickamer, K. (2001). A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276, 28939–28945. doi: 10.1074/jbc.M104565200
Mizoguchi, I., Ohashi, M., Chiba, Y., Hasegawa, H., Xu, M., Owaki, T., et al. (2017). Prediction of chemical respiratory and contact sensitizers by OX40L expression in dendritic cells using a novel 3D coculture system. Front. Immunol. 8:929. doi: 10.3389/fimmu.2017.00929
Mnich, M. E., van Dalen, R., Gerlach, D., Hendriks, A., Xia, G., Peschel, A., et al. (2019). The C-type lectin receptor MGL senses N-acetylgalactosamine on the unique Staphylococcus aureus ST395 wall teichoic acid. Cell. Microbiol. 21:e13072. doi: 10.1111/cmi.13072
Nakatsuji, T., Chiang, H. I., Jiang, S. B., Nagarajan, H., Zengler, K., and Gallo, R. L. (2013). The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 4:1431. doi: 10.1038/ncomms2441
Neuhaus, K., Wamhoff, E. C., Freichel, T., Grafmuller, A., Rademacher, C., and Hartmann, L. (2019). Asymmetrically branched precision glycooligomers targeting langerin. Biomacromolecules 20, 4088–4095. doi: 10.1021/acs.biomac.9b00906
Neumann, A. K., Thompson, N. L., and Jacobson, K. (2008). Distribution and lateral mobility of DC-SIGN on immature dendritic cells–implications for pathogen uptake. J. Cell Sci. 121, 634–643. doi: 10.1242/jcs.022418
Niess, J. H., Brand, S., Gu, X., Landsman, L., Jung, S., McCormick, B. A., et al. (2005). CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258. doi: 10.1126/science.1102901
Nordenfelt, P., and Bjorck, L. (2013). IgG-binding bacterial proteins and pathogenesis. Future Microbiol. 8, 299–301. doi: 10.2217/fmb.13.6
Olaniyi, R. O., Pancotto, L., Grimaldi, L., and Bagnoli, F. (2018). Deciphering the pathological role of staphylococcal alpha-toxin and panton-valentine leukocidin using a novel ex vivo human skin model. Front. Immunol. 9:951. doi: 10.3389/fimmu.2018.00951
Ouchi, T., Kubo, A., Yokouchi, M., Adachi, T., Kobayashi, T., Kitashima, D. Y., et al. (2011). Langerhans cell antigen capture through tight junctions confers preemptive immunity in experimental staphylococcal scalded skin syndrome. J. Exp. Med. 208, 2607–2613. doi: 10.1084/jem.20111718
Patel, V. I., and Metcalf, J. P. (2018). Airway macrophage and dendritic cell subsets in the resting human lung. Crit. Rev. Immunol. 38, 303–331. doi: 10.1615/CritRevImmunol.2018026459
Patterson, B. K., Landay, A., Siegel, J. N., Flener, Z., Pessis, D., Chaviano, A., et al. (2002). Susceptibility to human immunodeficiency virus-1 infection of human foreskin and cervical tissue grown in explant culture. Am. J. Pathol. 161, 867–873. doi: 10.1016/S0002-9440(10)64247-2
Porkolab, V., Chabrol, E., Varga, N., Ordanini, S., Sutkeviciu Te, I., Thepaut, M., et al. (2018). Rational-differential design of highly specific glycomimetic ligands: targeting DC-SIGN and excluding langerin recognition. ACS Chem. Biol. 13, 600–608. doi: 10.1021/acschembio.7b00958
Prado Acosta, M., and Lepenies, B. (2019). Bacterial glycans and their interactions with lectins in the innate immune system. Biochem. Soc. Trans. 47, 1569–1579. doi: 10.1042/BST20170410
Pupovac, A., Senturk, B., Griffoni, C., Maniura-Weber, K., Rottmar, M., and McArthur, S. L. (2018). Toward Immunocompetent 3D Skin Models. Adv. Healthc. Mater. 7:e1701405. doi: 10.1002/adhm.201701405
Quaresma, J. A. S. (2019). Organization of the skin immune system and compartmentalized immune responses in infectious diseases. Clin. Microbiol. Rev. 32:e00034–18. doi: 10.1128/CMR.00034-18
Raes, G., Brys, L., Dahal, B. K., Brandt, J., Grooten, J., Brombacher, F., et al. (2005). Macrophage galactose-type C-type lectins as novel markers for alternatively activated macrophages elicited by parasitic infections and allergic airway inflammation. J. Leukoc. Biol. 77, 321–327. doi: 10.1189/jlb.0304212
Rappuoli, R. (2018). Glycoconjugate vaccines: principles and mechanisms. Sci. Transl. Med. 10:eaat4615. doi: 10.1126/scitranslmed.aat4615
Rathinam, V. A., Appledorn, D. M., Hoag, K. A., Amalfitano, A., and Mansfield, L. S. (2009). Campylobacter jejuni-induced activation of dendritic cells involves cooperative signaling through toll-like receptor 4 (TLR4)-MyD88 and TLR4-TRIF axes. Infect. Immun. 77, 2499–2507. doi: 10.1128/IAI.01562-08
Reed, W. P., Metzler, C., and Albright, E. (1994). Streptococcal adherence to Langerhans cells: a possible step in the pathogenesis of streptococcal pharyngitis. Clin. Immunol. Immunopathol. 70, 28–31. doi: 10.1006/clin.1994.1006
Regnier, M., Patwardhan, A., Scheynius, A., and Schmidt, R. (1998). Reconstructed human epidermis composed of keratinocytes, melanocytes and langerhans cells. Med. Biol. Eng. Comput. 36, 821–824. doi: 10.1007/BF02518889
Rescigno, M., Urbano, M., Valzasina, B., Francolini, M., Rotta, G., Bonasio, R., et al. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2, 361–367. doi: 10.1038/86373
Rillahan, C. D., and Paulson, J. C. (2011). Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 80, 797–823. doi: 10.1146/annurev-biochem-061809-152236
Saijo, S., Ikeda, S., Yamabe, K., Kakuta, S., Ishigame, H., Akitsu, A., et al. (2010). Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 32, 681–691. doi: 10.1016/j.immuni.2010.05.001
Santegoets, S. J., Masterson, A. J., van der Sluis, P. C., Lougheed, S. M., Fluitsma, D. M., van den Eertwegh, A. J., et al. (2006). A CD34(+) human cell line model of myeloid dendritic cell differentiation: evidence for a CD14(+)CD11b(+) Langerhans cell precursor. J. Leukoc. Biol. 80, 1337–1344. doi: 10.1189/jlb.0206111
Sato, K., Yang, X. L., Yudate, T., Chung, J. S., Wu, J., Luby-Phelps, K., et al. (2006). Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J. Biol. Chem. 281, 38854–38866. doi: 10.1074/jbc.M606542200
Schaudinn, C., Dittmann, C., Jurisch, J., Laue, M., Gunday-Tureli, N., Blume-Peytavi, U., et al. (2017). Development, standardization and testing of a bacterial wound infection model based on ex vivo human skin. PLoS ONE 12:e0186946. doi: 10.1371/journal.pone.0186946
Schneider, H., Griffiss, J. M., Boslego, J. W., Hitchcock, P. J., Zahos, K. M., and Apicella, M. A. (1991). Expression of paragloboside-like lipooligosaccharides may be a necessary component of gonococcal pathogenesis in men. J. Exp. Med. 174, 1601–1605. doi: 10.1084/jem.174.6.1601
Schneider, T., and Sahl, H. G. (2010). An oldie but a goodie - cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 300, 161–169. doi: 10.1016/j.ijmm.2009.10.005
Schutgens, F., and Clevers, H. (2020). Human organoids: tools for understanding biology and treating diseases. Annu. Rev. Pathol. 15, 211–234. doi: 10.1146/annurev-pathmechdis-012419-032611
Sender, R., Fuchs, S., and Milo, R. (2016a). Are We really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340. doi: 10.1016/j.cell.2016.01.013
Sender, R., Fuchs, S., and Milo, R. (2016b). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Sharma, A., Simonson, T. J., Jondle, C. N., Mishra, B. B., and Sharma, J. (2017). Mincle-mediated neutrophil extracellular trap formation by regulation of autophagy. J. Infect. Dis. 215, 1040–1048. doi: 10.1093/infdis/jix072
Sharma, A., Steichen, A. L., Jondle, C. N., Mishra, B. B., and Sharma, J. (2014). Protective role of Mincle in bacterial pneumonia by regulation of neutrophil mediated phagocytosis and extracellular trap formation. J. Infect. Dis. 209, 1837–1846. doi: 10.1093/infdis/jit820
Shiokawa, M., Yamasaki, S., and Saijo, S. (2017). C-type lectin receptors in anti-fungal immunity. Curr. Opin. Microbiol. 40, 123–130. doi: 10.1016/j.mib.2017.11.004
Sidorin, E. V., and Solov'eva, T. F. (2011). IgG-binding proteins of bacteria. Biochemistry 76, 295–308. doi: 10.1134/S0006297911030023
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. doi: 10.1101/cshperspect.a000414
Soilleux, E. J., Morris, L. S., Leslie, G., Chehimi, J., Luo, Q., Levroney, E., et al. (2002). Constitutive and induced expression of DC-SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J. Leukoc. Biol. 71, 445–457. doi: 10.1189/jlb.71.3.445
Sparber, F., De Gregorio, C., Steckholzer, S., Ferreira, F. M., Dolowschiak, T., Ruchti, F., et al. (2019). The skin commensal yeast malassezia triggers a type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation. Cell Host Microbe. 25, 389–403.e386. doi: 10.1016/j.chom.2019.02.002
Stambach, N. S., and Taylor, M. E. (2003). Characterization of carbohydrate recognition by langerin, a C-type lectin of langerhans cells. Glycobiology 13, 401–410. doi: 10.1093/glycob/cwg045
Steeghs, L., van Vliet, S. J., Uronen-Hansson, H., van Mourik, A., Engering, A., Sanchez-Hernandez, M., et al. (2006). Neisseria meningitidis expressing lgtB lipopolysaccharide targets DC-SIGN and modulates dendritic cell function. Cell. Microbiol. 8, 316–325. doi: 10.1111/j.1462-5822.2005.00623.x
Sung, S. S., Fu, S. M., Rose, C. E. Jr., Gaskin, F., Ju, S. T., and Beaty, S. R. (2006). A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J. Immunol. 176, 2161–2172. doi: 10.4049/jimmunol.176.4.2161
Suzuki, N., Yamamoto, K., Toyoshima, S., Osawa, T., and Irimura, T. (1996). Molecular cloning and expression of cDNA encoding human macrophage C-type lectin. Its unique carbohydrate binding specificity for Tn antigen. J. Immunol. 156, 128–135.
Takeuchi, O., and Akira, S. (2010). Pattern recognition receptors and inflammation. Cell 140, 805–820. doi: 10.1016/j.cell.2010.01.022
Takeuchi, O., Hoshino, K., and Akira, S. (2000). Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J. Immunol. 165, 5392–5396. doi: 10.4049/jimmunol.165.10.5392
Tanaka, J., Gleinich, A. S., Zhang, Q., Whitfield, R., Kempe, K., Haddleton, D. M., et al. (2017). Specific and differential binding of n-acetylgalactosamine glycopolymers to the human macrophage galactose lectin and asialoglycoprotein receptor. Biomacromolecules. 18, 1624–1633. doi: 10.1021/acs.biomac.7b00228
Tateno, H., Ohnishi, K., Yabe, R., Hayatsu, N., Sato, T., Takeya, M., et al. (2010). Dual specificity of langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J. Biol. Chem. 285, 6390–6400. doi: 10.1074/jbc.M109.041863
Taylor, P. R., Reid, D. M., Heinsbroek, S. E., Brown, G. D., Gordon, S., and Wong, S. Y. (2005). Dectin-2 is predominantly myeloid restricted and exhibits unique activation-dependent expression on maturing inflammatory monocytes elicited in vivo. Eur. J. Immunol. 35, 2163–2174. doi: 10.1002/eji.200425785
Thépaut, M., Valladeau, J., Nurisso, A., Kahn, R., Arnou, B., Vivès, C., et al. (2009). Structural studies of langerin and Birbeck granule: a macromolecular organization model. Biochemistry 48, 2684–2698. doi: 10.1021/bi802151w
Thornton, E. E., Looney, M. R., Bose, O., Sen, D., Sheppard, D., Locksley, R., et al. (2012). Spatiotemporally separated antigen uptake by alveolar dendritic cells and airway presentation to T cells in the lung. J. Exp. Med. 209, 1183–1199. doi: 10.1084/jem.20112667
Tytgat, H. L., and Lebeer, S. (2014). The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol. Mol. Biol. Rev. 78, 372–417. doi: 10.1128/MMBR.00007-14
Valladeau, J., Ravel, O., Dezutter-Dambuyant, C., Moore, K., Kleijmeer, M., Liu, Y., et al. (2000). Langerin, a novel C-type lectin specific to Langerhans cells, is an endocytic receptor that induces the formation of Birbeck granules. Immunity 12, 71–81. doi: 10.1016/S1074-7613(00)80160-0
van Dalen, R., De La Cruz Diaz, J. S., Rumpret, M., Fuchsberger, F. F., van Teijlingen, N. H., Hanske, J., et al. (2019a). Langerhans cells sense staphylococcus aureus wall teichoic acid through langerin to induce inflammatory responses. MBio 10:e00330–19. doi: 10.1128/mBio.00330-19
van Dalen, R., Fuchsberger, F. F., Rademacher, C., van Strijp, J. A. G., and van Sorge, N. M. (2019b). A Common Genetic Variation in Langerin (CD207) Compromises cellular uptake of staphylococcus aureus. J. Innate Immun. 12, 191–200. doi: 10.1159/000500547
van der Aar, A. M., Sylva-Steenland, R. M., Bos, J. D., Kapsenberg, M. L., de Jong, E. C., and Teunissen, M. B. (2007). Loss of TLR2, TLR4, and TLR5 on Langerhans cells abolishes bacterial recognition. J. Immunol. 178, 1986–1990. doi: 10.4049/jimmunol.178.4.1986
van der Es, D., Hogendorf, W. F., Overkleeft, H. S., van der Marel, G. A., and Codee, J. D. (2017). Teichoic acids: synthesis and applications. Chem. Soc. Rev. 46, 1464–1482. doi: 10.1039/C6CS00270F
van der Vlist, M., de Witte, L., de Vries, R. D., Litjens, M., de Jong, M. A., Fluitsma, D., et al. (2011). Human langerhans cells capture measles virus through langerin and present viral antigens to CD4(+) T cells but are incapable of cross-presentation. Eur. J. Immunol. 41, 2619–2631. doi: 10.1002/eji.201041305
van Kooyk, Y. (2008). C-type lectins on dendritic cells: key modulators for the induction of immune responses. Biochem. Soc. Trans. 36, 1478–1481. doi: 10.1042/BST0361478
van Kooyk, Y., and Geijtenbeek, T. B. (2003). DC-SIGN: escape mechanism for pathogens. Nat. Rev. Immunol. 3, 697–709. doi: 10.1038/nri1182
van Sorge, N. M., Bleumink, N. M., van Vliet, S. J., Saeland, E., van der Pol, W. L., van Kooyk, Y., et al. (2009). N-glycosylated proteins and distinct lipooligosaccharide glycoforms of Campylobacter jejuni target the human C-type lectin receptor MGL. Cell. Microbiol. 11, 1768–1781. doi: 10.1111/j.1462-5822.2009.01370.x
van Vliet, S. J., Bay, S., Vuist, I. M., Kalay, H., Garcia-Vallejo, J. J., Leclerc, C., et al. (2013). MGL signaling augments TLR2-mediated responses for enhanced IL-10 and TNF-alpha secretion. J. Leukoc. Biol. 94, 315–323. doi: 10.1189/jlb.1012520
van Vliet, S. J., Paessens, L. C., Broks-van den Berg, V. C., Geijtenbeek, T. B., and van Kooyk, Y. (2008). The C-type lectin macrophage galactose-type lectin impedes migration of immature APCs. J. Immunol. 181, 3148–3155. doi: 10.4049/jimmunol.181.5.3148
van Vliet, S. J., Steeghs, L., Bruijns, S. C., Vaezirad, M. M., Snijders Blok, C., Arenas Busto, J. A., et al. (2009). Variation of Neisseria gonorrhoeae lipooligosaccharide directs dendritic cell-induced T helper responses. PLoS Pathog. 5:e1000625. doi: 10.1371/journal.ppat.1000625
van Vliet, S. J., van Liempt, E., Saeland, E., Aarnoudse, C. A., Appelmelk, B., Irimura, T., et al. (2005). Carbohydrate profiling reveals a distinctive role for the C-type lectin MGL in the recognition of helminth parasites and tumor antigens by dendritic cells. Int. Immunol. 17, 661–669. doi: 10.1093/intimm/dxh246
Wamhoff, E. C., Schulze, J., Bellmann, L., Rentzsch, M., Bachem, G., Fuchsberger, F. F., et al. (2019). A specific, glycomimetic langerin ligand for human langerhans cell targeting. ACS Cent Sci. 5, 808–820. doi: 10.1021/acscentsci.9b00093
Winstel, V., Liang, C., Sanchez-Carballo, P., Steglich, M., Munar, M., Broker, B. M., et al. (2013). Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat. Commun. 4:2345. doi: 10.1038/ncomms3345
Winstel, V., Sanchez-Carballo, P., Holst, O., Xia, G., and Peschel, A. (2014). Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. MBio 5:e00869. doi: 10.1128/mBio.00869-14
Wittmann, A., Lamprinaki, D., Bowles, K. M., Katzenellenbogen, E., Knirel, Y. A., Whitfield, C., et al. (2016). Dectin-2 recognizes mannosylated O-antigens of human opportunistic pathogens and augments lipopolysaccharide activation of myeloid cells. J. Biol. Chem. 291, 17629–17638. doi: 10.1074/jbc.M116.741256
Xia, G., Maier, L., Sanchez-Carballo, P., Li, M., Otto, M., Holst, O., et al. (2010). Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J. Biol. Chem. 285, 13405–13415. doi: 10.1074/jbc.M109.096172
Yamasaki, S., Ishikawa, E., Sakuma, M., Hara, H., Ogata, K., and Saito, T. (2008). Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat. Immunol. 9, 1179–1188. doi: 10.1038/ni.1651
Yan, H., Kamiya, T., Suabjakyong, P., and Tsuji, N. M. (2015). Targeting C-type lectin receptors for cancer immunity. Front. Immunol. 6:408. doi: 10.3389/fimmu.2015.00408
Yanagimachi, M. D., Niwa, A., Tanaka, T., Honda-Ozaki, F., Nishimoto, S., Murata, Y., et al. (2013). Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS ONE 8:e59243. doi: 10.1371/journal.pone.0059243
Yang, K., He, Y., Park, C. G., Kang, Y. S., Zhang, P., Han, Y., et al. (2019). Yersinia pestis interacts with SIGNR1 (CD209b) for promoting host dissemination and infection. Front. Immunol. 10:96. doi: 10.3389/fimmu.2019.00096
Yang, K., Park, C. G., Cheong, C., Bulgheresi, S., Zhang, S., Zhang, P., et al. (2015). Host Langerin (CD207) is a receptor for Yersinia pestis phagocytosis and promotes dissemination. Immunol. Cell Biol. 93, 815–824. doi: 10.1038/icb.2015.46
Yoshida, K., Kubo, A., Fujita, H., Yokouchi, M., Ishii, K., Kawasaki, H., et al. (2014). Distinct behavior of human langerhans cells and inflammatory dendritic epidermal cells at tight junctions in patients with atopic dermatitis. J. Allergy Clin. Immunol. 134, 856–864. doi: 10.1016/j.jaci.2014.08.001
Yu, Q. D., Oldring, A. P., Powlesland, A. S., Tso, C. K., Yang, C., Drickamer, K., et al. (2009). Autonomous tetramerization domains in the glycan-binding receptors DC-SIGN and DC-SIGNR. J. Mol. Biol. 387, 1075–1080. doi: 10.1016/j.jmb.2009.02.046
Zhang, G. L., and Ye, X. S. (2018). Synthetic glycans and glycomimetics: a promising alternative to natural polysaccharides. Chemistry 24, 6696–6704. doi: 10.1002/chem.201705469
Zhang, P., Schwartz, O., Pantelic, M., Li, G., Knazze, Q., Nobile, C., et al. (2006a). DC-SIGN (CD209) recognition of Neisseria gonorrhoeae is circumvented by lipooligosaccharide variation. J. Leukoc. Biol. 79, 731–738. doi: 10.1189/jlb.0405184
Zhang, P., Skurnik, M., Zhang, S. S., Schwartz, O., Kalyanasundaram, R., Bulgheresi, S., et al. (2008). Human dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin (CD209) is a receptor for Yersinia pestis that promotes phagocytosis by dendritic cells. Infect. Immun. 76, 2070–2079. doi: 10.1128/IAI.01246-07
Zhang, P., Snyder, S., Feng, P., Azadi, P., Zhang, S., Bulgheresi, S., et al. (2006b). Role of N-acetylglucosamine within core lipopolysaccharide of several species of gram-negative bacteria in targeting the DC-SIGN (CD209). J. Immunol. 177, 4002–4011. doi: 10.4049/jimmunol.177.6.4002
Keywords: bacteria, antigen-presenting cells, immunity, glycan, host-pathogen interaction, C-type lectin, pattern-recognition receptor
Citation: Mnich ME, van Dalen R and van Sorge NM (2020) C-Type Lectin Receptors in Host Defense Against Bacterial Pathogens. Front. Cell. Infect. Microbiol. 10:309. doi: 10.3389/fcimb.2020.00309
Received: 13 March 2020; Accepted: 22 May 2020;
Published: 07 July 2020.
Edited by:
Amanda S. MacLeod, Duke University, United StatesReviewed by:
Cornelis Hendrik Hokke, Leiden University Medical Center, NetherlandsWendy W. J. Unger, Sophia Children's Hospital, Netherlands
Copyright © 2020 Mnich, van Dalen and van Sorge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nina M. van Sorge, bi5tLnZhbnNvcmdlQGFtc3RlcmRhbXVtYy5ubA==
 Malgorzata E. Mnich
Malgorzata E. Mnich Rob van Dalen
Rob van Dalen Nina M. van Sorge
Nina M. van Sorge