
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol., 19 February 2020
Sec. Microbiome in Health and Disease
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00009
This article is part of the Research TopicThe Microbiota and InterventionsView all 6 articles
 Raphaël Enaud1,2,3*†
Raphaël Enaud1,2,3*† Renaud Prevel2,3,4†
Renaud Prevel2,3,4† Eleonora Ciarlo5
Eleonora Ciarlo5 Fabien Beaufils2,3,6
Fabien Beaufils2,3,6 Gregoire Wieërs7
Gregoire Wieërs7 Benoit Guery5
Benoit Guery5 Laurence Delhaes2,3,8
Laurence Delhaes2,3,8The gut and lungs are anatomically distinct, but potential anatomic communications and complex pathways involving their respective microbiota have reinforced the existence of a gut–lung axis (GLA). Compared to the better-studied gut microbiota, the lung microbiota, only considered in recent years, represents a more discreet part of the whole microbiota associated to human hosts. While the vast majority of studies focused on the bacterial component of the microbiota in healthy and pathological conditions, recent works have highlighted the contribution of fungal and viral kingdoms at both digestive and respiratory levels. Moreover, growing evidence indicates the key role of inter-kingdom crosstalks in maintaining host homeostasis and in disease evolution. In fact, the recently emerged GLA concept involves host–microbe as well as microbe–microbe interactions, based both on localized and long-reaching effects. GLA can shape immune responses and interfere with the course of respiratory diseases. In this review, we aim to analyze how the lung and gut microbiota influence each other and may impact on respiratory diseases. Due to the limited knowledge on the human virobiota, we focused on gut and lung bacteriobiota and mycobiota, with a specific attention on inter-kingdom microbial crosstalks which are able to shape local or long-reached host responses within the GLA.
Recent advances in microbiota explorations have led to an improved knowledge of the communities of commensal microorganisms within the human body. Human skin and mucosal surfaces are associated with rich and complex ecosystems (microbiota) composed of bacteria (bacteriobiota), fungi (mycobiota), viruses (virobiota), phages, archaea, protists, and helminths (Cho and Blaser, 2012). The role of the gut bacteriobiota in local health homeostasis and diseases is being increasingly investigated, but its long-distance impacts still need to be clarified (Chiu et al., 2017). Among the relevant inter-organ connections, the gut–lung axis (GLA) remains less studied than the gut–brain axis. So far, microbiota studies mainly focused on the bacterial component, neglecting other microbial kingdoms. However, the understanding of mycobiota involvement in human health and inter-organ connections should not be overlooked (Nguyen et al., 2015; Enaud et al., 2018). Viruses are also known to be key players in numerous respiratory diseases and to interact with the human immune system, but technical issues still limit the amount of data regarding virobiota (Mitchell and Glanville, 2018). Therefore, we will focus on bacterial and fungal components of the microbiota and their close interactions that are able to shape local or long-reached host responses within the GLA. While GLA mycobiota also influences chronic gut diseases such as IBD, we will not address this key role in the present review: we aimed at analyzing how lung and gut bacteriobiota and mycobiota influence each other, how they interact with the human immune system, and their role in respiratory diseases.
The gut microbiota has been the most extensively investigated. The majority of genes (99%) amplified in human stools are from bacteria, which are as numerous as human cells and comprise 150 distinct bacterial species, belonging mainly to Firmicutes and Bacteroidetes phyla. Proteobacteria, Actinobacteria, Cyanobacteria, and Fusobacteria are also represented in healthy people (Sekirov et al., 2010; Human Microbiome Project Consortium, 2012).
More recently, fungi have been recognized as an integral part of our commensal flora, and their role in health and diseases is increasingly considered (Huffnagle and Noverr, 2013; Huseyin et al., 2017). Fungi are about 100 times larger than bacteria, so even if fungal sequences are 100 to 1,000 times less frequent than bacterial sequences, fungi must not be neglected in the gastrointestinal ecosystem. In contrast with the bacteriobiota, the diversity of the gut mycobiota in healthy subjects is limited to few genera, with a high prevalence of Saccharomyces cerevisiae, Malassezia restricta, and Candida albicans (Nash et al., 2017).
Although often dichotomized due to technical and analysis sequencing issues, critical interactions exist between bacteriobiota and mycobiota (Peleg et al., 2010). The most appropriate approach to decipher the role of gut microbiota is therefore considering the gut as an ecosystem in which inter-kingdom interactions occur and have major implications as suggested by the significant correlations between the gut bacteriobiota and mycobiota profiles among healthy subjects (Hoffmann et al., 2013). Yeasts, e.g., Saccharomyces boulardii and C. albicans, or fungus wall components, e.g., β-glucans, are able to inhibit the growth of some intestinal pathogens (Zhou et al., 2013; Markey et al., 2018). S. boulardii also produces proteases or phosphatases that inactivate the toxins produced by intestinal bacteria such as Clostridium difficile and Escherichia coli (Castagliuolo et al., 1999; Buts et al., 2006). In addition, at physiological state and during gut microbiota disturbances (e.g., after a course of antibiotics), fungal species may take over the bacterial functions of immune modulation, preventing mucosal tissue damages (Jiang et al., 2017). Vice versa, bacteria can also modulate fungi: fatty acids locally produced by bacteria impact on the phenotype of C. albicans (Noverr and Huffnagle, 2004; Tso et al., 2018).
Beside the widely studied gut microbiota, microbiotas of other sites, including the lungs, are essential for host homeostasis and disease. The lung microbiota is now recognized as a cornerstone in the physiopathology of numerous respiratory diseases (Soret et al., 2019; Vandenborght et al., 2019).
The lung microbiota represents a significantly lower biomass than the gut microbiota: about 10 to 100 bacteria per 1,000 human cells (Sze et al., 2012). Its composition depends on the microbial colonization from the oropharynx and upper respiratory tract through salivary micro-inhalations, on the host elimination abilities (especially coughing and mucociliary clearance), on interactions with the host immune system, and on local conditions for microbial proliferation, such as pH or oxygen concentration (Gleeson et al., 1997; Wilson and Hamilos, 2014). The predominant bacterial phyla in lungs are the same as in gut, mainly Firmicutes and Bacteroidetes followed by Proteobacteria and Actinobacteria (Charlson et al., 2011). In healthy subjects, the main identified fungi are usually environmental: Ascomycota (Aspergillus, Cladosporium, Eremothecium, and Vanderwaltozyma) and Microsporidia (Systenostrema) (Nguyen et al., 2015; Vandenborght et al., 2019). In contrast to the intestinal or oral microbiota, data highlighting the interactions between bacteria and fungi in the human respiratory tract are more scattered (Delhaes et al., 2012; Soret et al., 2019). However, data from both in vitro and in vivo studies suggest relevant inter-kingdom crosstalk (Delhaes et al., 2012; Xu and Dongari-Bagtzoglou, 2015; Lof et al., 2017; Soret et al., 2019). This dialogue may involve several pathways as physical interaction, quorum-sensing molecules, production of antimicrobial agents, immune response modulation, and nutrient exchange (Peleg et al., 2010). Synergistic interactions have been documented between Candida and Streptococcus, such as stimulation of Streptococcus growth by Candida, increasing biofilm formation, or enhancement of the Candida pathogenicity by Streptococcus (Diaz et al., 2012; Xu et al., 2014). In vitro studies exhibited an increased growth of Aspergillus fumigatus in presence of Pseudomonas aeruginosa, due to the mold's ability in to assimilate P. aeruginosa-derived volatile sulfur compounds (Briard et al., 2019; Scott et al., 2019). However, the lung microbiota modulation is not limited to local inter-kingdom crosstalk and also depends on inter-compartment crosstalk between the gut and lungs.
From birth throughout the entire life span, a close correlation between the composition of the gut and lung microbiota exists, suggesting a host-wide network (Grier et al., 2018). For instance, modification of newborns' diet influences the composition of their lung microbiota, and fecal transplantation in rats induces changes in the lung microbiota (Madan et al., 2012; Liu et al., 2017).
The host's health condition can impact this gut–lung interaction too. In cystic fibrosis (CF) newborns, gut colonizations with Roseburia, Dorea, Coprococcus, Blautia, or Escherichia presaged their respiratory appearance, and their gut and lung abundances are highly correlated over time (Madan et al., 2012). Similarly, the lung microbiota is enriched with gut bacteria, such as Bacteroides spp., after sepsis (Dickson et al., 2016).
Conversely, lung microbiota may affect the gut microbiota composition. In a pre-clinical model, influenza infection triggers an increased proportion of Enterobacteriaceae and decreased abundances of Lactobacilli and Lactococci in the gut (Looft and Allen, 2012). Consistently, lipopolysaccharide (LPS) instillation in the lungs of mice is associated with gut microbiota disturbances (Sze et al., 2014).
Although gastroesophageal content inhalations and sputum swallowing partially explain this inter-organ connection, GLA also involves indirect communications such as host immune modulation.
Gut microbiota effects on the local immune system have been extensively reviewed (Elson and Alexander, 2015). Briefly, the gut microbiota closely interacts with the mucosal immune system using both pro-inflammatory and regulatory signals (Skelly et al., 2019). It also influences neutrophil responses, modulating their ability to extravasate from blood (Karmarkar and Rock, 2013). Toll-like receptor (TLR) signaling is essential for microbiota-driven myelopoiesis and exerts a neonatal selection shaping the gut microbiota with long-term consequences (Balmer et al., 2014; Fulde et al., 2018). Moreover, the gut microbiota communicates with and influences immune cells expressing TLR or GPR41/43 by means of microbial associated molecular patterns (MAMPs) or short-chain fatty acids (SCFAs) (Le Poul et al., 2003). Data focused on the gut mycobiota's impact on the immune system are sparser. Commensal fungi seem to reinforce bacterial protective benefits on both local and systemic immunity, with a specific role for mannans, a highly conserved fungal wall component. Moreover, fungi are able to produce SCFAs (Baltierra-Trejo et al., 2015; Xiros et al., 2019). Therefore, gut mycobiota perturbations could be as deleterious as bacteriobiota ones (Wheeler et al., 2016; Jiang et al., 2017).
A crucial role of lung microbiota in the maturation and homeostasis of lung immunity has emerged over the last few years (Dickson et al., 2018). Colonization of the respiratory tract provides essential signals for maturing local immune cells with long-term consequences (Gollwitzer et al., 2014). Pre-clinical studies confirm the causality between airway microbial colonization and the regulation and maturation of the airways' immune cells. Germ-free mice exhibit increased local Th2-associated cytokine and IgE production, promoting allergic airway inflammation (Herbst et al., 2011). Consistently, lung exposure to commensal bacteria reduces Th2-associated cytokine production after an allergen challenge and induces regulatory cells early in life (Russell et al., 2012; Gollwitzer et al., 2014). The establishment of resident memory B cells in lungs also requires encountering lung microbiota local antigens, especially regarding immunity against viruses such as influenza (Allie et al., 2019).
Interactions between lung microbiota and immunity are also a two-way process; a major inflammation in the lungs can morbidly transform the lung microbiota composition (Molyneaux et al., 2013).
Beyond the local immune regulation by the site-specific microbiota, the long-reaching immune impact of gut microbiota is now being recognized, especially on the pulmonary immune system (Chiu et al., 2017). The mesenteric lymphatic system is an essential pathway between the lungs and the intestine, through which intact bacteria, their fragments, or metabolites (e.g., SCFAs) may translocate across the intestinal barrier, reach the systemic circulation, and modulate the lung immune response (Trompette et al., 2014; Bingula et al., 2017; McAleer and Kolls, 2018). SCFAs, mainly produced by the bacterial dietary fibers' fermentation especially in case of a high-fiber diet (HFD), act in the lungs as signaling molecules on resident antigen-presenting cells to attenuate the inflammatory and allergic responses (Anand and Mande, 2018; Cait et al., 2018). SCFA receptor–deficient mice show increased inflammatory responses in experimental models of asthma (Trompette et al., 2014). Fungi, including A. fumigatus, can also produce SCFAs or create a biofilm enhancing the bacterial production of SCFAs, but on the other hand, bacterial SCFAs can dampen fungal growth (Hynes et al., 2008; Baltierra-Trejo et al., 2015; Xiros et al., 2019). The impact of fungal production of SCFAs on the host has not been assessed so far.
Other important players of this long-reaching immune effect are gut segmented filamentous bacteria (SFBs), a commensal bacteria colonizing the ileum of most animals, including humans, and involved in the modulation of the immune system's development (Yin et al., 2013). SFBs regulate CD4+ T-cell polarization into the Th17 pathway, which is implicated in the response to pulmonary fungal infections and lung autoimmune manifestations (McAleer et al., 2016; Bradley et al., 2017). Recently, innate lymphoid cells, involved in tissue repair, have been shown to be recruited from the gut to the lungs in response to inflammatory signals upon IL-25 (Huang et al., 2018). Finally, intestinal TLR activation, required for the NF-κB–dependent pathways of innate immunity and inflammation, is associated with an increased influenza-related lung response in mice (Ichinohe et al., 2011).
Other mechanisms may be involved in modulating the long-reaching immune response related to gut microbiota, exemplified by the increased number of mononuclear leukocytes and an increased phagocytic and lytic activity after treatment with Bifidobacterium lactis HN019 probiotics (Gill et al., 2001). Diet, especially fiber intake, which increases the systemic level of SCFAs, or probiotics influence the pulmonary immune response and thus impact the progression of respiratory disorders (King et al., 2007; Varraso et al., 2015; Anand and Mande, 2018).
The GLA immune dialogue remains a two-way process. For instance, Salmonella nasal inoculation promotes a Salmonella-specific gut immunization which depends on lung dendritic cells (Ruane et al., 2013). Respiratory influenza infection also modulates the composition of the gut microbiota as stated above. These intestinal microbial disruptions seem to be unrelated to an intestinal tropism of influenza virus but mediated by Th17 cells (Wang et al., 2014).
In summary, GLA results from complex interactions between the different microbial components of both the gut and lung microbiotas combined with local and long-reaching immune effects. All these interactions strongly suggest a major role for the GLA in respiratory diseases, as recently documented in a mice model (Skalski et al., 2018).
Regarding influenza infection and the impact of gut and lung microbiota, our knowledge is still fragmentary; human data are not yet available. However, antibiotic treatment causes significantly reduced immune responses against influenza virus in mice (Ichinohe et al., 2011). Conversely, influenza-infected HFD-fed mice exhibit increased survival rates compared to infected controls thanks to an enhanced generation of Ly6c-patrolling monocytes. These monocytes increase the numbers of macrophages that have a limited capacity to produce CXCL1 locally, reducing neutrophil recruitment to the airways and thus tissue damage. In parallel, diet-derived SCFAs boost CD8+ T-cell effector function in HFD-fed mice (Trompette et al., 2018).
Both lung and gut microbiota are essential against bacterial pneumonia. The lung microbiota is able to protect against respiratory infections with Streptococcus pneumoniae and Klebsiella pneumoniae by priming the pulmonary production of granulocyte-macrophage colony-stimulating factor (GM-CSF) via IL-17 and Nod2 stimulation (Brown et al., 2017). The gut microbiota also plays a crucial role in response to lung bacterial infections. Studies on germ-free mice showed an increased morbidity and mortality during K. pneumoniae, S. pneumoniae, or P. aeruginosa acute lung infection (Fagundes et al., 2012; Fox et al., 2012; Brown et al., 2017). The use of broad-spectrum antibiotic treatments, to disrupt mouse gut microbiota, results in worse outcome in lung infection mouse models (Schuijt et al., 2016; Robak et al., 2018). Mechanistically, alveolar macrophages from mice deprived of gut microbiota through antibiotic treatment are less responsive to stimulation and show reduced phagocytic capacity (Schuijt et al., 2016). Interestingly, priming of antibiotic-treated animals with TLR agonists restores resistance to pulmonary infections (Fagundes et al., 2012). SFBs appear to be an important gut microbiota component for lung defense against bacterial infection thanks to their capacity to induce the production of the Th17 cytokine, IL-22, and to increase neutrophil counts in the lungs during Staphylococcus aureus pneumonia (Gauguet et al., 2015).
Modulating chronic infectious diseases will similarly depend on gut and lung microbiotas. For instance, Mycobacterium tuberculosis infection severity is correlated with gut microbiota (Namasivayam et al., 2018).
Multiple studies have addressed the impact of gut and lung microbiota on chronic respiratory diseases such as chronic obstructive pulmonary disease (COPD), asthma, and CF (Table 1).
Decreased lung microbiota diversity and Proteobacteria expansion are associated with both COPD severity and exacerbations (Garcia-Nuñez et al., 2014; Wang et al., 2016, 2018; Mayhew et al., 2018). The fact that patients with genetic mannose binding lectin deficiency exhibit a more diverse pulmonary microbiota and a lower risk of exacerbation suggests not only association but also causality (Dicker et al., 2018). Besides the lung flora, the gut microbiota is involved in exacerbations, as suggested by the increased gastrointestinal permeability in patients admitted for COPD exacerbations (Sprooten et al., 2018). Whatever the permeability's origin (hypoxemia or pro-inflammatory status), the level of circulating gut microbiota–dependent trimethylamine-N-oxide has been associated with mortality in COPD patients (Ottiger et al., 2018). This association being explained by comorbidities and age, its impact per se is not guaranteed. Further studies are warranted to investigate the role of GLA in COPD and to assess causality.
Early-life perturbations in fungal and bacterial gut colonization, such as low gut microbial diversity, e.g., after neonatal antibiotic use, are critical to induce childhood asthma development (Abrahamsson et al., 2014; Metsälä et al., 2015; Arrieta et al., 2018). This microbial disruption is associated with modifications of fecal SCFA levels (Arrieta et al., 2018). Causality has been assessed in murine models. Inoculation of the bacteria absent in the microbiota of asthmatic patients decreases airways inflammation (Arrieta et al., 2015). Furthermore, Bacteroides fragilis seems to play a major role in immune homeostasis, balancing the host systemic Th1/Th2 ratio and therefore conferring protection against allergen-induced airway disorders (Mazmanian et al., 2005; Panzer and Lynch, 2015; Arrieta et al., 2018). Nevertheless, it is still not fully deciphered, as some studies conversely found that an early colonization with Bacteroides, including B. fragilis, could be an early indicator of asthma later in life (Vael et al., 2008). Regarding fungi, gut fungal overgrowth (after antibiotic administration or a gut colonization protocol with Candida or Wallemia mellicola) increases the occurrence of asthma via IL-13 without any fungal expansion in the lungs (Noverr et al., 2005; Wheeler et al., 2016; Skalski et al., 2018). The prostaglandin E2 produced in the gut by Candida can reach the lungs and promotes lung M2 macrophage polarization and allergic airway inflammation (Kim et al., 2014). In mice, a gut overrepresentation of W. mellicola associated with several intestinal microbiome disturbances appears to have long-reaching effects on the pulmonary immune response and severity of asthma, by involving the Th2 pathways, especially IL-13 and to a lesser degree IL-17, goblet cell differentiation, fibroblasts activation, and IgE production by B cells (Skalski et al., 2018). Taken together, these results indicate that the GLA, mainly through the gut microbiota, is likely to play a major role in asthma.
In CF patients, gut and lung microbiota are distinct from those of healthy subjects, and disease progression is associated with microbiota alterations (Madan et al., 2012; Stokell et al., 2015; Nielsen et al., 2016). Moreover, the bacterial abundances at both sites are highly correlated and have similar trends over time (Madan et al., 2012), especially regarding Streptococcus, which is found in higher proportion in CF stools, gastric contents, and sputa (Al-Momani et al., 2016; Nielsen et al., 2016). Moreover, CF patients with a documented intestinal inflammation exhibit a higher Streptococcus abundance in the gut (Enaud et al., 2019), suggesting the GLA's involvement in intestinal inflammation. Of note, gut but not lung microbiota alteration is associated with early-life exacerbations: some gut microbiota perturbations, such as a decrease of Parabacteroides, are predictive of airway colonization with P. aeruginosa (Hoen et al., 2015). Furthermore, oral administration of probiotics to CF patients leads to a decreased number of exacerbations (Anderson et al., 2016). While the mycobiota has been recently studied in CF (Nguyen et al., 2015; Soret et al., 2019), no data on the role of the fungal component of the GLA are currently available in CF, which deserves to be more widely studied.
The role of inter-compartment and inter-kingdom interactions within the GLA in those pulmonary diseases now has to be further confirmed and causality to be assessed. Diet, probiotics, or more specific modulations could be, in the near future, novel essential tools in therapeutic management of these respiratory diseases.
The gut–lung axis or GLA has emerged as a specific axis with intensive dialogues between the gut and lungs, involving each compartment in a two-way manner, with both microbial and immune interactions (Figure 1). Each kingdom and compartment plays a crucial role in this dialogue, and consequently in host health and diseases. The roles of fungal and viral kingdoms within the GLA still remain to be further investigated. Their manipulation, as for the bacterial component, could pave the way for new approaches in the management of several respiratory diseases such as acute infections, COPD, asthma, and/or CF.
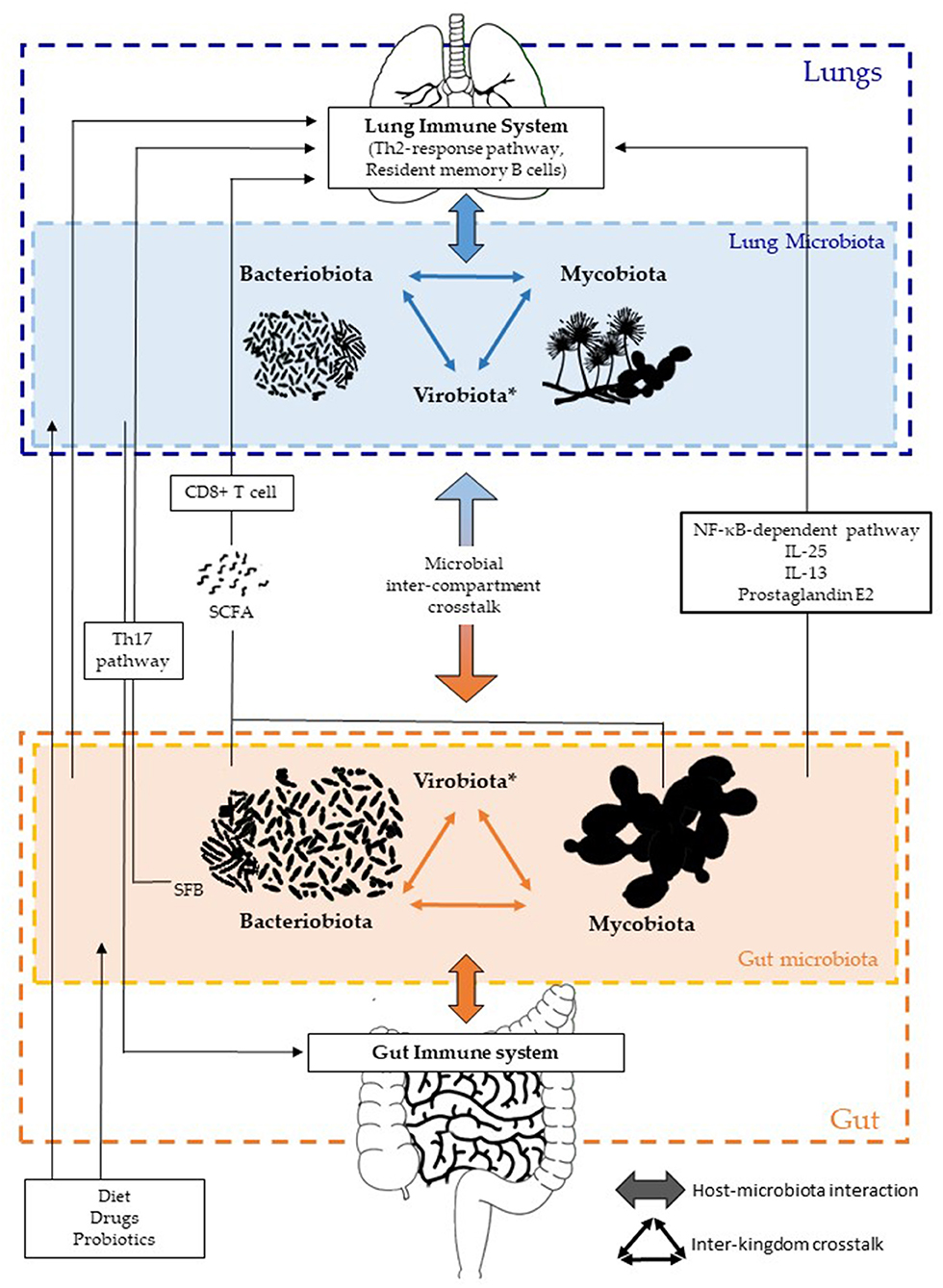
Figure 1. Inter-kingdom and inter-compartment crosstalks within the gut–lung axis. Bacteriobiota, mycobiota, and virobiota are closely interacting within each organ by either direct or indirect mechanisms shaping each other. Gut microbiota influences both the gut immune system and the lung immune system via local or long-reaching interactions, which involve either CD8+ T cell, Th17, IL-25, IL-13, prostaglandin E2, and/or NF-κB–dependent pathways. The lung microbiota impacts the mucosal immunity and contributes to immune tolerance, through neutrophil recruitment, production of pro-inflammatory cytokines mediated by receptor 2 (TLR2), and the release of antimicrobial peptides, such as β-defensin 2 stimulated by T helper 17 (Th17) cells. On the other hand, the lung microbiota also influences the gut immune system, but precise mechanisms remain to be deciphered, even if an intestinal microbial disruption has been associated with Th17 cell mediation after influenza virus lung infection. Several factors are well-known to influence the composition of the intestinal and/or lung microbiota, such as diet, drugs, and probiotics. *Of note, the virobiota is not covered in this review.
RE, RP, EC, FB, GW, BG, and LD conceptualized and wrote the manuscript.
LD has received research grants from Vaincre La Mucoviscidose (RF20160501626) and from the Fondation de l'Université de Bordeaux (Bronch-Asp project); she is member of ESGHAMI and NGS-MycA working groups of ESCMID and ECCM, respectively. RP and RE has received Ph.D. research grants from University Hospital of Bordeaux. All the funders had no role in article design, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Laure De Rosamel for her precious proofreading of the manuscript.
Abrahamsson, T. R., Jakobsson, H. E., Andersson, A. F., Björkstén, B., Engstrand, L., and Jenmalm, M. C. (2012). Low diversity of the gut microbiota in infants with atopic eczema. J. Allergy Clin. Immunol. 129, 434–440, 440.e1–2. doi: 10.1016/j.jaci.2011.10.025
Abrahamsson, T. R., Jakobsson, H. E., Andersson, A. F., Björkstén, B., Engstrand, L., and Jenmalm, M. C. (2014). Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 44, 842–850. doi: 10.1111/cea.12253
Allie, S. R., Bradley, J. E., Mudunuru, U., Schultz, M. D., Graf, B. A., Lund, F. E., et al. (2019). The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 20, 97–108. doi: 10.1038/s41590-018-0260-6
Al-Momani, H., Perry, A., Stewart, C. J., Jones, R., Krishnan, A., Robertson, A. G., et al. (2016). Microbiological profiles of sputum and gastric juice aspirates in cystic fibrosis patients. Sci. Rep. 6:26985. doi: 10.1038/srep26985
Anand, S., and Mande, S. S. (2018). Diet, microbiota and gut-lung connection. Front. Microbiol. 9:2147. doi: 10.3389/fmicb.2018.02147
Anderson, J. L., Miles, C., and Tierney, A. C. (2016). Effect of probiotics on respiratory, gastrointestinal and nutritional outcomes in patients with cystic fibrosis: a systematic review. J. Cyst. Fibros. 16, 186–197. doi: 10.1016/j.jcf.2016.09.004
Arrieta, M.-C., Arévalo, A., Stiemsma, L., Dimitriu, P., Chico, M. E., Loor, S., et al. (2018). Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 142, 424–434.e10. doi: 10.1016/j.jaci.2017.08.041
Arrieta, M.-C., Stiemsma, L. T., Dimitriu, P. A., Thorson, L., Russell, S., Yurist-Doutsch, S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152. doi: 10.1126/scitranslmed.aab2271
Balmer, M. L., Schürch, C. M., Saito, Y., Geuking, M. B., Li, H., Cuenca, M., et al. (2014). Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 193, 5273–5283. doi: 10.4049/jimmunol.1400762
Baltierra-Trejo, E., Sánchez-Yáñez, J. M., Buenrostro-Delgado, O., and Márquez-Benavides, L. (2015). Production of short-chain fatty acids from the biodegradation of wheat straw lignin by Aspergillus fumigatus. Bioresour. Technol. 196, 418–425. doi: 10.1016/j.biortech.2015.07.105
Bingula, R., Filaire, M., Radosevic-Robin, N., Bey, M., Berthon, J.-Y., Bernalier-Donadille, A., et al. (2017). Desired turbulence? Gut-lung axis, immunity, and lung cancer. J. Oncol. 2017:5035371. doi: 10.1155/2017/5035371
Björkstén, B., Sepp, E., Julge, K., Voor, T., and Mikelsaar, M. (2001). Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 108, 516–520. doi: 10.1067/mai.2001.118130
Bradley, C. P., Teng, F., Felix, K. M., Sano, T., Naskar, D., Block, K. E., et al. (2017). Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 22, 697–704.e4. doi: 10.1016/j.chom.2017.10.007
Briard, B., Mislin, G. L. A., Latgé, J.-P., and Beauvais, A. (2019). Interactions between Aspergillus fumigatus and pulmonary bacteria: current state of the field, new data, and future perspective. J Fungi (Basel) 5:E48. doi: 10.3390/jof5020048
Brown, R. L., Sequeira, R. P., and Clarke, T. B. (2017). The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 8:1512. doi: 10.1038/s41467-017-01803-x
Buts, J.-P., Dekeyser, N., Stilmant, C., Delem, E., Smets, F., and Sokal, E. (2006). Saccharomyces boulardii produces in rat small intestine a novel protein phosphatase that inhibits Escherichia coli endotoxin by dephosphorylation. Pediatr. Res. 60, 24–29. doi: 10.1203/01.pdr.0000220322.31940.29
Cait, A., Hughes, M. R., Antignano, F., Cait, J., Dimitriu, P. A., Maas, K. R., et al. (2018). Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal. Immunol. 11, 785–795. doi: 10.1038/mi.2017.75
Castagliuolo, I., Riegler, M. F., Valenick, L., LaMont, J. T., and Pothoulakis, C. (1999). Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect. Immun. 67, 302–307. doi: 10.1128/IAI.67.1.302-307.1999
Charlson, E. S., Bittinger, K., Haas, A. R., Fitzgerald, A. S., Frank, I., Yadav, A., et al. (2011). Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184, 957–963. doi: 10.1164/rccm.201104-0655OC
Chiu, L., Bazin, T., Truchetet, M.-E., Schaeverbeke, T., Delhaes, L., and Pradeu, T. (2017). Protective microbiota: from localized to long-reaching co-immunity. Front. Immunol. 8:1678. doi: 10.3389/fimmu.2017.01678
Cho, I., and Blaser, M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. doi: 10.1038/nrg3182
Coburn, B., Wang, P. W., Diaz Caballero, J., Clark, S. T., Brahma, V., Donaldson, S., et al. (2015). Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 5:10241. doi: 10.1038/srep10241
Cox, M. J., Allgaier, M., Taylor, B., Baek, M. S., Huang, Y. J., Daly, R. A., et al. (2010). Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS ONE 5:e11044. doi: 10.1371/journal.pone.0011044
Delhaes, L., Monchy, S., Fréalle, E., Hubans, C., Salleron, J., Leroy, S., et al. (2012). The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PLoS ONE 7:e36313. doi: 10.1371/journal.pone.0036313
Denner, D. R., Sangwan, N., Becker, J. B., Hogarth, D. K., Oldham, J., Castillo, J., et al. (2016). Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J. Allergy Clin. Immunol. 137, 1398–1405.e3. doi: 10.1016/j.jaci.2015.10.017
Diaz, P. I., Xie, Z., Sobue, T., Thompson, A., Biyikoglu, B., Ricker, A., et al. (2012). Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80, 620–632. doi: 10.1128/IAI.05896-11
Dicker, A. J., Crichton, M. L., Cassidy, A. J., Brady, G., Hapca, A., Tavendale, R., et al. (2018). Genetic mannose binding lectin deficiency is associated with airway microbiota diversity and reduced exacerbation frequency in COPD. Thorax 73, 510–518. doi: 10.1136/thoraxjnl-2016-209931
Dickson, R. P., Erb-Downward, J. R., Falkowski, N. R., Hunter, E. M., Ashley, S. L., and Huffnagle, G. B. (2018). The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am. J. Respir. Crit. Care Med. 198, 497–508. doi: 10.1164/rccm.201711-2180OC
Dickson, R. P., Singer, B. H., Newstead, M. W., Falkowski, N. R., Erb-Downward, J. R., Standiford, T. J., et al. (2016). Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 1:16113. doi: 10.1038/nmicrobiol.2016.113
Durack, J., Lynch, S. V., Nariya, S., Bhakta, N. R., Beigelman, A., Castro, M., et al. (2017). Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 140, 63–75. doi: 10.1016/j.jaci.2016.08.055
Elson, C. O., and Alexander, K. L. (2015). Host-microbiota interactions in the intestine. Dig. Dis. 33, 131–136. doi: 10.1159/000369534
Enaud, R., Hooks, K. B., Barre, A., Barnetche, T., Hubert, C., Massot, M., et al. (2019). Intestinal inflammation in children with cystic fibrosis is associated with crohn's-like microbiota disturbances. J. Clin. Med. 8:E645. doi: 10.3390/jcm8050645
Enaud, R., Vandenborght, L.-E., Coron, N., Bazin, T., Prevel, R., Schaeverbeke, T., et al. (2018). The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms 6:E22. doi: 10.3390/microorganisms6010022
Fagundes, C. T., Amaral, F. A., Vieira, A. T., Soares, A. C., Pinho, V., Nicoli, J. R., et al. (2012). Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J. Immunol. 188, 1411–1420. doi: 10.4049/jimmunol.1101682
Fox, A. C., McConnell, K. W., Yoseph, B. P., Breed, E., Liang, Z., Clark, A. T., et al. (2012). The endogenous bacteria alter gut epithelial apoptosis and decrease mortality following Pseudomonas aeruginosa pneumonia. Shock 38, 508–514. doi: 10.1097/SHK.0b013e31826e47e8
Fraczek, M. G., Chishimba, L., Niven, R. M., Bromley, M., Simpson, A., Smyth, L., et al. (2018). Corticosteroid treatment is associated with increased filamentous fungal burden in allergic fungal disease. J. Allergy Clin. Immunol. 142, 407–414. doi: 10.1016/j.jaci.2017.09.039
Frayman, K. B., Armstrong, D. S., Carzino, R., Ferkol, T. W., Grimwood, K., Storch, G. A., et al. (2017). The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax 72, 1104–1112. doi: 10.1136/thoraxjnl-2016-209279
Fulde, M., Sommer, F., Chassaing, B., van Vorst, K., Dupont, A., Hensel, M., et al. (2018). Neonatal selection by toll-like receptor 5 influences long-term gut microbiota composition. Nature 560, 489–493. doi: 10.1038/s41586-018-0395-5
Garcia-Nuñez, M., Millares, L., Pomares, X., Ferrari, R., Pérez-Brocal, V., Gallego, M., et al. (2014). Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 52, 4217–4223. doi: 10.1128/JCM.01967-14
Gauguet, S., D'Ortona, S., Ahnger-Pier, K., Duan, B., Surana, N. K., Lu, R., et al. (2015). Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect. Immun. 83, 4003–4014. doi: 10.1128/IAI.00037-15
Gill, H. S., Rutherfurd, K. J., Cross, M. L., and Gopal, P. K. (2001). Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am. J. Clin. Nutr. 74, 833–839. doi: 10.1093/ajcn/74.6.833
Gleeson, K., Eggli, D. F., and Maxwell, S. L. (1997). Quantitative aspiration during sleep in normal subjects. Chest 111, 1266–1272. doi: 10.1378/chest.111.5.1266
Gollwitzer, E. S., Saglani, S., Trompette, A., Yadava, K., Sherburn, R., McCoy, K. D., et al. (2014). Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 20, 642–647. doi: 10.1038/nm.3568
Green, B. J., Wiriyachaiporn, S., Grainge, C., Rogers, G. B., Kehagia, V., Lau, L., et al. (2014). Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS ONE 9:e100645. doi: 10.1371/journal.pone.0100645
Grier, A., McDavid, A., Wang, B., Qiu, X., Java, J., Bandyopadhyay, S., et al. (2018). Neonatal gut and respiratory microbiota: coordinated development through time and space. Microbiome 6:193. doi: 10.1186/s40168-018-0566-5
Hampton, T. H., Green, D. M., Cutting, G. R., Morrison, H. G., Sogin, M. L., Gifford, A. H., et al. (2014). The microbiome in pediatric cystic fibrosis patients: the role of shared environment suggests a window of intervention. Microbiome 2:14. doi: 10.1186/2049-2618-2-14
Herbst, T., Sichelstiel, A., Schär, C., Yadava, K., Bürki, K., Cahenzli, J., et al. (2011). Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 184, 198–205. doi: 10.1164/rccm.201010-1574OC
Hilty, M., Burke, C., Pedro, H., Cardenas, P., Bush, A., Bossley, C., et al. (2010). Disordered microbial communities in asthmatic airways. PLoS ONE 5:e8578. doi: 10.1371/journal.pone.0008578
Hoen, A. G., Li, J., Moulton, L. A., O'Toole, G. A., Housman, M. L., Koestler, D. C., et al. (2015). Associations between gut microbial colonization in early life and respiratory outcomes in cystic fibrosis. J. Pediatr. 167, 138–147.e3. doi: 10.1016/j.jpeds.2015.02.049
Hoffman, L. R., Pope, C. E., Hayden, H. S., Heltshe, S., Levy, R., McNamara, S., et al. (2014). Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin. Infect. Dis. 58, 396–399. doi: 10.1093/cid/cit715
Hoffmann, C., Dollive, S., Grunberg, S., Chen, J., Li, H., Wu, G. D., et al. (2013). Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE 8:e66019. doi: 10.1371/journal.pone.0066019
Huang, Y., Mao, K., Chen, X., Sun, M.-A., Kawabe, T., Li, W., et al. (2018). S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119. doi: 10.1126/science.aam5809
Huang, Y. J., Nariya, S., Harris, J. M., Lynch, S. V., Choy, D. F., Arron, J. R., et al. (2015). The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 136, 874–884. doi: 10.1016/j.jaci.2015.05.044
Huffnagle, G. B., and Noverr, M. C. (2013). The emerging world of the fungal microbiome. Trends Microbiol. 21, 334–341. doi: 10.1016/j.tim.2013.04.002
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Huseyin, C. E., O'Toole, P. W., Cotter, P. D., and Scanlan, P. D. (2017). Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 41, 479–511. doi: 10.1093/femsre/fuw047
Hynes, M. J., Murray, S. L., Khew, G. S., and Davis, M. A. (2008). Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics 178, 1355–1369. doi: 10.1534/genetics.107.085795
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza a virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. doi: 10.1073/pnas.1019378108
Jiang, T. T., Shao, T.-Y., Ang, W. X. G., Kinder, J. M., Turner, L. H., Pham, G., et al. (2017). Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe 22, 809–816.e4. doi: 10.1016/j.chom.2017.10.013
Karmarkar, D., and Rock, K. L. (2013). Microbiota signalling through MyD88 is necessary for a systemic neutrophilic inflammatory response. Immunology 140, 483–492. doi: 10.1111/imm.12159
Kim, S. H., Clark, S. T., Surendra, A., Copeland, J. K., Wang, P. W., Ammar, R., et al. (2015). Global analysis of the fungal microbiome in cystic fibrosis patients reveals loss of function of the transcriptional repressor Nrg1 as a mechanism of pathogen adaptation. PLoS Pathog. 11:e1005308. doi: 10.1371/journal.ppat.1005308
Kim, Y.-G., Udayanga, K. G. S., Totsuka, N., Weinberg, J. B., Núñez, G., and Shibuya, A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 15, 95–102. doi: 10.1016/j.chom.2013.12.010
King, D. E., Egan, B. M., Woolson, R. F., Mainous, A. G., Al-Solaiman, Y., and Jesri, A. (2007). Effect of a high-fiber diet vs a fiber-supplemented diet on C-reactive protein level. Arch. Intern. Med. 167, 502–506. doi: 10.1001/archinte.167.5.502
Le Poul, E., Loison, C., Struyf, S., Springael, J.-Y., Lannoy, V., Decobecq, M.-E., et al. (2003). Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 278, 25481–25489. doi: 10.1074/jbc.M301403200
Liu, T., Yang, Z., Zhang, X., Han, N., Yuan, J., and Cheng, Y. (2017). 16S rDNA analysis of the effect of fecal microbiota transplantation on pulmonary and intestinal flora. 3 Biotech 7:370. doi: 10.1007/s13205-017-0997-x
Lof, M., Janus, M. M., and Krom, B. P. (2017). Metabolic interactions between bacteria and fungi in commensal oral biofilms. J Fungi (Basel) 3:40. doi: 10.3390/jof3030040
Looft, T., and Allen, H. K. (2012). Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 3, 463–467. doi: 10.4161/gmic.21288
Madan, J. C., Koestler, D. C., Stanton, B. A., Davidson, L., Moulton, L. A., Housman, M. L., et al. (2012). Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio 3, e00251–12. doi: 10.1128/mBio.00251-12
Markey, L., Shaban, L., Green, E. R., Lemon, K. P., Mecsas, J., and Kumamoto, C. A. (2018). Pre-colonization with the commensal fungus Candida albicans reduces murine susceptibility to Clostridium difficile infection. Gut Microbes 9, 497–509. doi: 10.1080/19490976.2018.1465158
Marri, P. R., Stern, D. A., Wright, A. L., Billheimer, D., and Martinez, F. D. (2013). Asthma-associated differences in microbial composition of induced sputum. J. Allergy Clin. Immunol. 131, 346–352.e1–3. doi: 10.1016/j.jaci.2012.11.013
Mayhew, D., Devos, N., Lambert, C., Brown, J. R., Clarke, S. C., Kim, V. L., et al. (2018). Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 73, 422–430. doi: 10.1136/thoraxjnl-2017-210408
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
McAleer, J. P., and Kolls, J. K. (2018). Contributions of the intestinal microbiome in lung immunity. Eur. J. Immunol. 48, 39–49. doi: 10.1002/eji.201646721
McAleer, J. P., Nguyen, N. L. H., Chen, K., Kumar, P., Ricks, D. M., Binnie, M., et al. (2016). Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J. Immunol. 197, 97–107. doi: 10.4049/jimmunol.1502566
Metsälä, J., Lundqvist, A., Virta, L. J., Kaila, M., Gissler, M., and Virtanen, S. M. (2015). Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin. Exp. Allergy 45, 137–145. doi: 10.1111/cea.12356
Mitchell, A. B., and Glanville, A. R. (2018). Introduction to techniques and methodologies for characterizing the human respiratory virome. Methods Mol. Biol. 1838, 111–123. doi: 10.1007/978-1-4939-8682-8_9
Molyneaux, P. L., Mallia, P., Cox, M. J., Footitt, J., Willis-Owen, S. A. G., Homola, D., et al. (2013). Outgrowth of the bacterial airway microbiome after rhinovirus exacerbation of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 188, 1224–1231. doi: 10.1164/rccm.201302-0341OC
Namasivayam, S., Sher, A., Glickman, M. S., and Wipperman, M. F. (2018). The microbiome and tuberculosis: early evidence for cross talk. mBio 9, e01420–e01418. doi: 10.1128/mBio.01420-18
Nash, A. K., Auchtung, T. A., Wong, M. C., Smith, D. P., Gesell, J. R., Ross, M. C., et al. (2017). The gut mycobiome of the human microbiome project healthy cohort. Microbiome 5:153. doi: 10.1186/s40168-017-0373-4
Nguyen, L. D. N., Deschaght, P., Merlin, S., Loywick, A., Audebert, C., Van Daele, S., et al. (2016). Effects of Propidium Monoazide (PMA) treatment on mycobiome and bacteriome analysis of cystic fibrosis airways during exacerbation. PLoS ONE 11:e0168860. doi: 10.1371/journal.pone.0168860
Nguyen, L. D. N., Viscogliosi, E., and Delhaes, L. (2015). The lung mycobiome: an emerging field of the human respiratory microbiome. Front. Microbiol. 6:89. doi: 10.3389/fmicb.2015.00089
Nielsen, S., Needham, B., Leach, S. T., Day, A. S., Jaffe, A., Thomas, T., et al. (2016). Disrupted progression of the intestinal microbiota with age in children with cystic fibrosis. Sci. Rep. 6:24857. doi: 10.1038/srep24857
Noverr, M. C., Falkowski, N. R., McDonald, R. A., McKenzie, A. N., and Huffnagle, G. B. (2005). Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect. Immun. 73, 30–38. doi: 10.1128/IAI.73.1.30-38.2005
Noverr, M. C., and Huffnagle, G. B. (2004). Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 72, 6206–6210. doi: 10.1128/IAI.72.11.6206-6210.2004
Ottiger, M., Nickler, M., Steuer, C., Bernasconi, L., Huber, A., Christ-Crain, M., et al. (2018). Gut, microbiota-dependent trimethylamine-N-oxide is associated with long-term all-cause mortality in patients with exacerbated chronic obstructive pulmonary disease. Nutrition 45, 135–141.e1. doi: 10.1016/j.nut.2017.07.001
Panzer, A. R., and Lynch, S. V. (2015). Influence and effect of the human microbiome in allergy and asthma. Curr. Opin. Rheumatol. 27, 373–380. doi: 10.1097/BOR.0000000000000191
Peleg, A. Y., Hogan, D. A., and Mylonakis, E. (2010). Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8, 340–349. doi: 10.1038/nrmicro2313
Penders, J., Thijs, C., van den Brandt, P. A., Kummeling, I., Snijders, B., Stelma, F., et al. (2007). Gut microbiota composition and development of atopic manifestations in infancy: the KOALA birth cohort study. Gut 56, 661–667. doi: 10.1136/gut.2006.100164
Renwick, J., McNally, P., John, B., DeSantis, T., Linnane, B., Murphy, P., et al. (2014). The microbial community of the cystic fibrosis airway is disrupted in early life. PLoS ONE 9:e109798. doi: 10.1371/journal.pone.0109798
Robak, O. H., Heimesaat, M. M., Kruglov, A. A., Prepens, S., Ninnemann, J., Gutbier, B., et al. (2018). Antibiotic treatment-induced secondary IgA deficiency enhances susceptibility to Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 128, 3535–3545. doi: 10.1172/JCI97065
Ruane, D., Brane, L., Reis, B. S., Cheong, C., Poles, J., Do, Y., et al. (2013). Lung dendritic cells induce migration of protective T cells to the gastrointestinal tract. J. Exp. Med. 210, 1871–1888. doi: 10.1084/jem.20122762
Russell, S. L., Gold, M. J., Hartmann, M., Willing, B. P., Thorson, L., Wlodarska, M., et al. (2012). Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13, 440–447. doi: 10.1038/embor.2012.32
Schuijt, T. J., Lankelma, J. M., Scicluna, B. P., de Sousa e Melo, F., Roelofs, J. J., de Boer, J. D., et al. (2016). The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65, 575–583. doi: 10.1136/gutjnl-2015-309728
Scott, J., Sueiro-Olivares, M., Ahmed, W., Heddergott, C., Zhao, C., Thomas, R., et al. (2019). Pseudomonas aeruginosa-derived volatile sulfur compounds promote distal Aspergillus fumigatus growth and a synergistic pathogen-pathogen interaction that increases pathogenicity in co-infection. Front. Microbiol. 10:2311. doi: 10.3389/fmicb.2019.02311
Sekirov, I., Russell, S. L., Antunes, L. C. M., and Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Skalski, J. H., Limon, J. J., Sharma, P., Gargus, M. D., Nguyen, C., Tang, J., et al. (2018). Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 14:e1007260. doi: 10.1371/journal.ppat.1007260
Skelly, A. N., Sato, Y., Kearney, S., and Honda, K. (2019). Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 19, 305–323. doi: 10.1038/s41577-019-0144-5
Soret, P., Vandenborght, L.-E., Francis, F., Coron, N., Enaud, R., The Mucofong Investigation Group, et al. (2019). Respiratory mycobiome and suggestion of inter-kingdom network during acute pulmonary exacerbation in cystic fibrosis. Sci. Rep. doi: 10.1038/s41598-020-60015-4. [Epub ahead of print].
Sprooten, R. T. M., Lenaerts, K., Braeken, D. C. W., Grimbergen, I., Rutten, E. P., Wouters, E. F. M., et al. (2018). Increased small intestinal permeability during severe acute exacerbations of COPD. Respiration 95, 334–342. doi: 10.1159/000485935
Stiemsma, L. T., Arrieta, M.-C., Dimitriu, P. A., Cheng, J., Thorson, L., Lefebvre, D. L., et al. (2016). Shifts in lachnospira and clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin. Sci. 130, 2199–2207. doi: 10.1042/CS20160349
Stokell, J. R., Gharaibeh, R. Z., Hamp, T. J., Zapata, M. J., Fodor, A. A., and Steck, T. R. (2015). Analysis of changes in diversity and abundance of the microbial community in a cystic fibrosis patient over a multiyear period. J. Clin. Microbiol. 53, 237–247. doi: 10.1128/JCM.02555-14
Sverrild, A., Kiilerich, P., Brejnrod, A., Pedersen, R., Porsbjerg, C., Bergqvist, A., et al. (2017). Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J. Allergy Clin. Immunol. 140, 407–417.e11. doi: 10.1016/j.jaci.2016.10.046
Sze, M. A., Dimitriu, P. A., Hayashi, S., Elliott, W. M., McDonough, J. E., Gosselink, J. V., et al. (2012). The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care. Med. 185, 1073–1080. doi: 10.1164/rccm.201111-2075OC
Sze, M. A., Tsuruta, M., Yang, S.-W. J., Oh, Y., Man, S. F. P., Hogg, J. C., et al. (2014). Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS ONE 9:e111228. doi: 10.1371/journal.pone.0111228
Trompette, A., Gollwitzer, E. S., Pattaroni, C., Lopez-Mejia, I. C., Riva, E., Pernot, J., et al. (2018). Dietary fiber confers protection against flu by shaping Ly6c- patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity 48, 992–1005.e8. doi: 10.1016/j.immuni.2018.04.022
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Tso, G. H. W., Reales-Calderon, J. A., Tan, A. S. M., Sem, X., Le, G. T. T., Tan, T. G., et al. (2018). Experimental evolution of a fungal pathogen into a gut symbiont. Science 362, 589–595. doi: 10.1126/science.aat0537
Vael, C., Nelen, V., Verhulst, S. L., Goossens, H., and Desager, K. N. (2008). Early intestinal Bacteroides fragilis colonisation and development of asthma. BMC Pulm. Med. 8:19. doi: 10.1186/1471-2466-8-19
van Nimwegen, F. A., Penders, J., Stobberingh, E. E., Postma, D. S., Koppelman, G. H., Kerkhof, M., et al. (2011). Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 128, 948–955.e1–3. doi: 10.1016/j.jaci.2011.07.027
van Woerden, H. C., Gregory, C., Brown, R., Marchesi, J. R., Hoogendoorn, B., and Matthews, I. P. (2013). Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect. Dis. 13:69. doi: 10.1186/1471-2334-13-69
Vandenborght, L.-E., Enaud, R., Coron, N., Denning, D. W., and Delhaes, L. (2019). “From culturomics to metagenomics: the mycobiome in chronic respiratory diseases,” in The Lung Microbiome (Norwich: European Respiratory Society), 88–118. doi: 10.1183/2312508X.10015918
Varraso, R., Chiuve, S. E., Fung, T. T., Barr, R. G., Hu, F. B., Willett, W. C., et al. (2015). Alternate healthy eating index 2010 and risk of chronic obstructive pulmonary disease among US women and men: prospective study. BMJ 350:h286. doi: 10.1136/bmj.h286
Wang, J., Li, F., Wei, H., Lian, Z.-X., Sun, R., and Tian, Z. (2014). Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 211, 2397–2410. doi: 10.1084/jem.20140625
Wang, Z., Bafadhel, M., Haldar, K., Spivak, A., Mayhew, D., Miller, B. E., et al. (2016). Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J. 47, 1082–1092. doi: 10.1183/13993003.01406-2015
Wang, Z., Singh, R., Miller, B. E., Tal-Singer, R., Van Horn, S., Tomsho, L., et al. (2018). Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: an analysis of the COPDMAP study. Thorax 73, 331–338. doi: 10.1136/thoraxjnl-2017-210741
Wheeler, M. L., Limon, J. J., Bar, A. S., Leal, C. A., Gargus, M., Tang, J., et al. (2016). Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19, 865–873. doi: 10.1016/j.chom.2016.05.003
Willger, S. D., Grim, S. L., Dolben, E. L., Shipunova, A., Hampton, T. H., Morrison, H. G., et al. (2014). Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2:40. doi: 10.1186/2049-2618-2-40
Wilson, M. T., and Hamilos, D. L. (2014). The nasal and sinus microbiome in health and disease. Curr. Allergy Asthma Rep. 14:485. doi: 10.1007/s11882-014-0485-x
Worlitzsch, D., Rintelen, C., Böhm, K., Wollschläger, B., Merkel, N., Borneff-Lipp, M., et al. (2009). Antibiotic-resistant obligate anaerobes during exacerbations of cystic fibrosis patients. Clin. Microbiol. Infect. 15, 454–460. doi: 10.1111/j.1469-0691.2008.02659.x
Xiros, C., Shahab, R. L., and Studer, M. H.-P. (2019). A cellulolytic fungal biofilm enhances the consolidated bioconversion of cellulose to short chain fatty acids by the rumen microbiome. Appl. Microbiol. Biotechnol. 103, 3355–3365. doi: 10.1007/s00253-019-09706-1
Xu, H., and Dongari-Bagtzoglou, A. (2015). Shaping the oral mycobiota: interactions of opportunistic fungi with oral bacteria and the host. Curr. Opin. Microbiol. 26, 65–70. doi: 10.1016/j.mib.2015.06.002
Xu, H., Sobue, T., Thompson, A., Xie, Z., Poon, K., Ricker, A., et al. (2014). Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell. Microbiol. 16, 214–231. doi: 10.1111/cmi.12216
Yin, Y., Wang, Y., Zhu, L., Liu, W., Liao, N., Jiang, M., et al. (2013). Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 7, 615–621. doi: 10.1038/ismej.2012.128
Zhang, Q., Cox, M., Liang, Z., Brinkmann, F., Cardenas, P. A., Duff, R., et al. (2016). Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS ONE 11:e0152724. doi: 10.1371/journal.pone.0152724
Zhao, J., Schloss, P. D., Kalikin, L. M., Carmody, L. A., Foster, B. K., Petrosino, J. F., et al. (2012). Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. U.S.A. 109, 5809–5814. doi: 10.1073/pnas.1120577109
Keywords: Gut-Lung Axis, Mycobiota, Microbiome, Respiratory disease, Dysbiosis
Citation: Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B and Delhaes L (2020) The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell. Infect. Microbiol. 10:9. doi: 10.3389/fcimb.2020.00009
Received: 28 August 2019; Accepted: 10 January 2020;
Published: 19 February 2020.
Edited by:
Yongqun Oliver He, University of Michigan, United StatesReviewed by:
Xingmin Sun, University of South Florida, United StatesCopyright © 2020 Enaud, Prevel, Ciarlo, Beaufils, Wieërs, Guery and Delhaes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raphaël Enaud, cmFwaGFlbF9lbmF1ZEB5YWhvby5mcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.