- Department of Biotechnology, Indian Institute of Technology Hyderabad, Kandi, India
The emergence of multidrug-resistant strains of Gram-negative Klebsiella species is an urgent global threat. The World Health Organization has listed Klebsiella pneumoniae as one of the global priority pathogens in critical need of next-generation antibiotics. Compared to other Gram-negative pathogens, K. pneumoniae accumulates a greater diversity of antimicrobial-resistant genes at a higher frequency. The evolution of a hypervirulent phenotype of K. pneumoniae is yet another concern. It has a broad ecological distribution affecting humans, agricultural animals, plants, and aquatic animals. Extracellular polysaccharides of Klebsiella, such as lipopolysaccharides, capsular polysaccharides, and exopolysaccharides, play crucial roles in conferring resistance against the host immune response, as well as in colonization, surface adhesion, and for protection against antibiotics and bacteriophages. These extracellular polysaccharides are major virulent determinants and are highly divergent with respect to their antigenic properties. Wzx/Wzy-, ABC-, and synthase-dependent proteinaceous nano-machineries are involved in the biosynthesis, transport, and cell surface expression of these sugar molecules. Although the proteins involved in the biosynthesis and surface expression of these sugar molecules represent potential drug targets, variation in the amino acid sequences of some of these proteins, in combination with diversity in their sugar composition, poses a major challenge to the design of a universal drug for Klebsiella infections. This review discusses the challenges in universal Klebsiella vaccine and drug development from the perspective of antigen sugar compositions and the proteins involved in extracellular antigen transport.
Introduction
Klebsiella species (spp.) are rod-shaped and encapsulated Gram-negative bacteria in the Enterobacteriaceae family (Podschun and Ullmann, 1998; Kenneth and Ryan, 2003; Murray and Baron, 2007; Paczosa and Mecsas, 2016). Eleven species have been identified in the Klebsiella genus, namely, Klebsiella pneumoniae (K. pneumoniae) (subsp. pneumoniae, subsp. ozaenae, subsp. rhinoscleromatis), Klebsiella oxytoca (K. oxytoca), Klebsiella ornithinolytica (K. ornithinolytica), Klebsiella planticola (K. planticola), Klebsiella terrigena (K. terrigena) (Murray and Baron, 2007), Klebsiella variicola (K. variicola) (Rosenblueth et al., 2004) [subsp. tropicalensis (Rodrigues et al., 2019)], Klebsiella granulomatis (K. granulomatis) (Carter et al., 1999), Klebsiella aerogenes (K. aerogenes) (Tindall et al., 2017), Klebsiella africanensis (K. africanensis) (Rodrigues et al., 2019), Klebsiella grimontii (K. grimontii) (Passet and Brisse, 2018), and Klebsiella quasipneumoniae (K. quasipneumoniae) (subsp. quasipneumoniae and subsp. similipneumoniae) (Brisse et al., 2014). Klebsiella pneumoniae (K. pneumonia) (Kp) are further classified into classical (cKp) and hypervirulent (hvKp) strains based on their phenotype and nature of pathogenicity (Shon et al., 2013; Russo et al., 2018). Klebsiella spp. are generally found in animal and human gut microbiota (Selden et al., 1971; Taur and Pamer, 2013; Bilinski et al., 2016; Paczosa and Mecsas, 2016). They colonize a wide range of hosts including plants and mammals (Bagley, 1985; Podschun and Ullmann, 1998; Podschun et al., 2001; Wyres and Holt, 2018) and can grow ubiquitously in water and soil (Bagley, 1985; Podschun and Ullmann, 1998; Podschun et al., 2001; Rock et al., 2014).
Klebsiella spp. are generally opportunistic pathogens (Wyres and Holt, 2018) and do not usually affect healthy individuals (Bagley, 1985; Centers For Disease Control Prevention, 2010). Generally, it is immunocompromised individuals, such as patients undergoing chemotherapy, neonates, and the elderly, that are affected by cKp infections. In contrast, hvKp can infect healthy individuals of any age and can infect nearly every site of the body and spread metastatically (Liu et al., 1986; Fang et al., 2007; Russo et al., 2018). Klebsiella spp. utilize the following virulence traits to protect themselves from the host immune response (Davies, 2003; Lavender et al., 2004; Mishra et al., 2015; Paczosa and Mecsas, 2016; Hsieh et al., 2019): capsular polysaccharides (CPS), lipopolysaccharides (LPS), siderophores, fimbriae (alternatively, pili), a type VI secretion system, outer-membrane proteins, porins, efflux pumps, an iron transport system, biofilms, and allantoin metabolism. Among these, CPS, LPS, siderophores, and fimbriae are well-characterized virulence factors of Klebsiella spp. (Paczosa and Mecsas, 2016). These virulence factors assist Klebsiella spp. in evading the innate immune response of the host and to survive in different sites within the host, rather than actively suppressing host immune system components (Domenico et al., 1994; Hsieh et al., 2019). Notably, increased production of CPS and aerobactin (an iron-chelating siderophore) is specific to the hvKp pathotype (Cheng et al., 2010; Russo et al., 2018), as increased production of CPS results in a hypermucoviscous phenotype that has a viscous string length >5 mm (Cheng et al., 2010). Nevertheless, hypermucoviscosity is not specific to the hvKp pathotype, as cKp can also exhibit such a phenotype (Catalan-Najera et al., 2017; Russo et al., 2018). Furthermore, hvKp strains are not always hypermucoviscous (Catalan-Najera et al., 2017; Russo et al., 2018). Thus, the genes involved in the regulation of CPS (Cheng et al., 2010) and aerobactin production are used to distinguish the cKp and hvKp pathotypes (Russo et al., 2018). These are not elaborated here, as it is beyond the scope of this review.
Klebsiella spp. cause a variety of opportunistic nosocomial and community-acquired infections (Podschun and Ullmann, 1998; Tsai et al., 2008; Lin et al., 2010; Paczosa and Mecsas, 2016; Martin and Bachman, 2018; Vading et al., 2018; Juan et al., 2019), such as urinary tract infection (Goldstein et al., 1978; Sewify et al., 2016), soft tissue infection (Goldstein et al., 1978), pneumonia (Lee et al., 1996; Tan et al., 1998), septicemia (Arredondo-Garcia et al., 1992; Al-Anazi et al., 2008), bacteremia (Goldstein et al., 1978; Lin et al., 1997), meningitis (Price and Sleigh, 1972; Ku et al., 2017; Khaertynov et al., 2018), and pyogenic liver abscesses (Chowdhury and Stein, 1992; Youssef et al., 2012). As Klebsiella spp. have acquired resistance against various antimicrobials, they often become a challenge in treating these infections (Bengoechea and Sa Pessoa, 2019). For instance, Kp isolates have continuously accumulated resistance against four important classes of antibiotics, namely, the third-generation cephalosporins, aminoglycosides, fluoroquinolones, and carbapenems (Navon-Venezia et al., 2017; The European Antimicrobial Resistance Surveillance Network, 2018). Multiple drug resistance such as this eventually leads to extremely drug-resistant Klebsiella strains (XDR) (Magiorakos et al., 2012; Navon-Venezia et al., 2017).
In general, Kp is a hospital-associated pathogen that is subjected to continuous selective pressure due to continuous exposure to multiple antibiotics. K. pneumoniae inactivates a spectrum of beta-lactams through the action of carbapenemases and an extended spectrum of beta-lactamases (ESBL). As a consequence, Kp can become resistant to beta-lactams and thrive in healthcare settings (Hawkey and Jones, 2009; D'andrea et al., 2013; Andrade et al., 2014; Zhang et al., 2016; Feng et al., 2018; Fu et al., 2018). For example, a New Delhi metallo-β-lactamase 1 (NDM-1)-producing Kp strain originating from India has now disseminated across the globe (Yong et al., 2009; Khan et al., 2017). In 2016, a patient infected with NDM-1-producing Kp died due to a lack of treatment options in Nevada (Chen et al., 2017). Colistin, a drug of last resort that has been used against carbapenem-resistant Enterobacteriaceae, targets bacterial lipid A. K. pneumoniae has developed resistance against colistin through mutations in lipid A modification regulatory genes such as mgrB (Cannatelli et al., 2013; Jayol et al., 2014; Olaitan et al., 2014; Poirel et al., 2015; Wright et al., 2015). Although both cKp and hvKp are global pathogens, the former is predominantly found in Western countries, while the latter is observed in the Asia-Pacific Rim (Fazili et al., 2016; Rossi et al., 2018; Russo and Marr, 2019). However, the evolution of hvKp strains with multiple drug resistance (MDR) and extreme drug resistance (XDR) is due to either hvKp acquiring drug-resistant plasmids from cKp (Zhang et al., 2015, 2016; Wei et al., 2016; Feng et al., 2018; Fu et al., 2018; Yao et al., 2018) or cKp acquiring an hvKp virulence plasmid (Gu et al., 2018). Both pose a significant challenge with respect to the treatment of infection.
Klebsiella pneumoniae has evolved several mechanisms to resist antibiotics. In comparison to Escherichia coli (E. coli), Kp has acquired double the number (more than 400) of antimicrobial-resistant (AMR) genes (Wyres and Holt, 2018). Interestingly, ESBL-producing Kp exhibits carbapenem resistance as a result of alterations in permeability due to loss of porins (Bradford et al., 1997; Martinez-Martinez, 2008; Leavitt et al., 2009) and overexpression of efflux pumps (Van De Klundert et al., 1988). Klebsiella pneumoniae has also acquired AMR through horizontal gene transfer enabled by plasmids and a mobile genetic environment (Pendleton et al., 2013). The emergence of plasmids with ESBL genes in Kp is one such example (Wachino et al., 2004; Queenan and Bush, 2007; Woodford et al., 2011; Lee et al., 2016). The translocation of carbapenemase-encoding genes from Kp plasmids onto a chromosome makes infections almost impossible to control (Lee et al., 2016). Due to chromosomal mutations, Kp has also become resistant to the antimicrobial peptide colistin (Olaitan et al., 2014; Doorduijn et al., 2016; Liu et al., 2016), leaving very few therapeutic options for the treatment of patients infected with Kp. Thus, it has become increasingly challenging to treat Kp infections, as reflected by the increase in the number of severe infections and the scarcity of effective antimicrobials (Paczosa and Mecsas, 2016).
As Klebsiella spp. are reservoirs for antibiotic-resistant genes (Navon-Venezia et al., 2017; Bengoechea and Sa Pessoa, 2019), they can act as key traffickers of AMR genes to other environmentally and clinically important Gram-negative bacteria. One such example is spread of carbapenem resistance genes from Kp (Sidjabat et al., 2009) strains originated in the United States (Smith Moland et al., 2003) to other Gram-negative bacterial species such as Salmonella (Miriagou et al., 2003), Enterobacter spp. (Hossain et al., 2004), Escherichia coli (Bratu et al., 2007), and Proteus mirabilis (Tibbetts et al., 2008). Such examples of interspecies spread have been observed for quite some time. Similar to Kp, other Klebsiella species have also acquired resistance against antibiotics. Klebsiella grimontii (which is closely related to Klebsiella oxytoca) is a newly added species to the Klebsiella genus (Passet and Brisse, 2018) and has acquired resistance against carbapenem (Liu et al., 2018). These events prompted the World Health Organization (WHO) to call for a global effort to develop next-generation antibiotics against Klebsiella infections (World Health Organization, 2017, 2018).
The rise in multidrug-resistant Klebsiella spp. (as well as hvKp strains) and their periodic outbreak and global spread (Navon-Venezia et al., 2017) warrant a new treatment strategy, along with a new set of antibiotics and vaccines for Klebsiella infections. Targeting bacterial survival mechanisms (rather than destroying the bacteria) exerts less selective pressure on the bacteria. For example, targeting the sugary armor of Klebsiella spp., such as the LPS, CPS, and exopolysaccharide (EPS), would be an efficient alternative strategy. Though structural information and mechanical insights relating to the transport of CPS and LPS onto the bacterial surface through various proteinaceous nanomachines are available (Rahn et al., 1999, 2003; Kos et al., 2009; Ruiz et al., 2009; Shu et al., 2009; Freinkman et al., 2012; Sachdeva et al., 2017; Bi et al., 2018), only a fragmented picture of their utility as potential drug and vaccine targets exists. To this end, this review focuses on targeting the CPS, LPS, and EPS armors of Klebsiella spp.
Host Innate Immune Defenses Against Klebsiella Species
When a pathogen enters a host, it must contend with the mechanical, chemical, and cellular barriers exhibited by the host, and Klebsiella spp. is no exception (Zhang et al., 2000). Initially, it has to overcome mechanical barriers such as the epithelia of the skin, mucociliary clearance, the low-pH environment of the genitourinary tract or gastrointestinal tract, etc. Subsequent to this, the pathogen must circumvent the humoral and cellular innate defenses. Several humoral defenses (opsonic, bactericidal, and bacteriostatic) are used by the host for bacterial clearance (Kabha et al., 1997; Zhang et al., 2000; Ivin et al., 2017). One such humoral defense is the complement system, which is activated in three different pathways (namely, the classical, alternative, and mannose-binding lectin pathways) (Murphy et al., 2012) for the purpose of clearing bacteria. In addition, the pathogen has to deceive antimicrobial peptides, collectins, and cellular components (i.e., neutrophils, monocytes/macrophages, dendritic cells, and innate lymphoid cells) of the innate immune defense to survive and maintain its growth in the host (Murphy et al., 2012). The mechanisms of Klebsiella spp. defense against the host are covered in detail in recent reviews (Doorduijn et al., 2016; Paczosa and Mecsas, 2016; Bengoechea and Sa Pessoa, 2019).
Once Klebsiella spp. overcome the mechanical barriers of the host, a variety of host immune defense pathways are activated by pathogen recognition receptors (PRRs) such as “Toll-like” receptors (TLRs), nucleotide-binding oligomerization domain-like receptors (NLRs), etc. (Takeuchi and Akira, 2010) through the detection of pathogen-associated molecular patterns (PAMPs). As CPS and LPS are major pathogen surface components, many of the PRRs activate these immune response pathways primarily mediated by the detection of LPS and CPS. For instance, upon binding to TLR4, the CPS activates the NF-κB-mediated inflammatory and immune response pathways (Regueiro et al., 2006, 2009; Yang et al., 2011). Interaction of LPS with TLR4 and MD2 receptors on the host innate immune cells also induces the NF-κB-mediated inflammatory response (Kawai and Akira, 2010; Maeshima and Fernandez, 2013). The lung collectins SP-A and SP-D, which are soluble PRRs, bind to LPS and facilitate agglutination and phagocytosis by macrophages. The recruitment of the classical complement pathway (following LPS detection) and that of the lectin-mediated complement pathway (upon detection of CPS) are some of the major host strategies for bacterial clearance (Walport, 2001; Ricklin et al., 2010; Holers, 2014; Gomez-Simmonds and Uhlemann, 2017). On detection of LPS, NLR protein family members assemble to form inflammasome, which activates caspase 4/5 in humans and caspase-11 in the mouse. This triggers the activation of non-canonical inflammasome to produce IL1β and induce bacterial cell death (Hagar et al., 2013; Shi et al., 2014). Opsonophagocytosis mediated by neutrophils and macrophages is also a major bacterial clearance strategy (Domenico et al., 1994; Salo et al., 1995; Regueiro et al., 2006).
Host Immune Evasion Strategies of Klebsiella spp.
Klebsiella spp. make use of several sophisticated stealth immune evasion strategies to escape from the host innate immune response, rather than actively suppressing it. However, recent research indicates that Klebsiella spp. have also developed several anti-immune strategies that involve the attack of key regulators and effectors of the host immune system. This makes them formidable pathogens capable of disseminating and growing across a variety of sites in their hosts (Paczosa and Mecsas, 2016; Bengoechea and Sa Pessoa, 2019). To establish in the host, the pathogen has to counteract the host innate immune defenses (Zhang et al., 2000). The surface oligosaccharide molecules (CPS and LPS) are some of the major virulence factors that Klebsiella spp. use to protect themselves from the host immune response.
Capsular Polysaccharide
The surface of Klebsiella spp. is shielded by a thick layer of CPS fibers that protect the bacteria from the environment (Amako et al., 1988). The polysaccharide capsule assists the bacteria in surviving stressful environmental conditions such as desiccation and exposure to detergents. High-molecular-weight CPS, consisting of linear or branched oligosaccharides, form a shield around the Klebsiella spp. cell surface and represent a physical barrier against the complement system, as also seen in E. coli (Meri and Pangburn, 1994; Alvarez et al., 2000; Cortes et al., 2002b; Abreu and Barbosa, 2017). This shield plays a crucial role in protecting Kp against innate immune response mechanisms, evading complement deposition and opsonization, reducing recognition, and adhesion by epithelial cells and phagocytes, and abrogating lysis by antimicrobial peptides and complement cascades (Podschun and Ullmann, 1998; Fang et al., 2004; Lin et al., 2004; Pomakova et al., 2012; Paczosa and Mecsas, 2016; Martin and Bachman, 2018). Poorly encapsulated Kp strains are readily vulnerable to phagocytosis (Cortes et al., 2002a; De Astorza et al., 2004). As compared to capsular Kp strains, acapsular Kp strains are more easily phagocytosed by innate immune cells (Domenico et al., 1994; Yoshida et al., 2000; Cortes et al., 2002b; Lawlor et al., 2005, 2006). Deletion of the genes responsible for capsule formation in the clinical strains ideally leads to a non-pathogenic bacterium by drastically impairing the virulence of Kp (Cortes et al., 2002b; Lawlor et al., 2006). It has been shown that the thickness of CPS (rather than its chemical composition) determines the extent of protection it confers to Klebsiella spp. (De Astorza et al., 2004). Not surprisingly, hvKp exhibits enhanced resistance to a variety of humoral defenses such as complement killing, HBD-1 to HBD-3 [human beta-defensin (HBD)], and to antimicrobial peptides such as neutrophil protein 1 and lactoferrin (Fang et al., 2004).
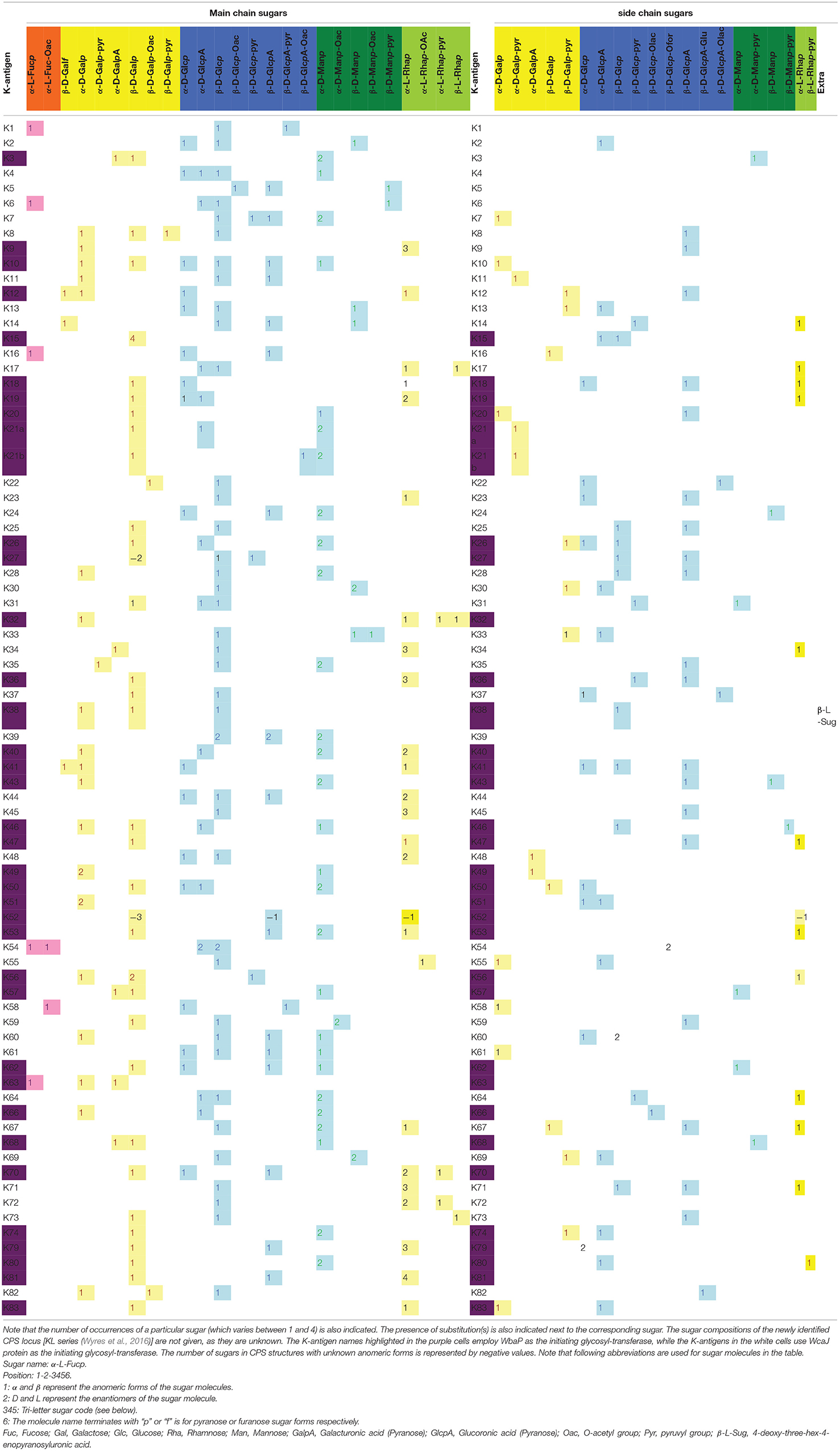
The capsule type, also known as the K-antigen or K-type, is Klebsiella species-specific and is widely used in the serotyping of Klebsiella spp. Traditionally, Klebsiella spp. are identified as having 77 K-antigens (viz., K1–K81, excluding K75–K78) based on the diversity in their sugar composition, type of glycosidic linkage, and the nature of enantiomeric and epimeric forms (https://iith.ac.in/K-PAM/, K-PAM unpublished) (Pan et al., 2015). Recently, additional K-types have been identified based on the CPS locus or K-locus (KL) arrangement. These are known as the KL series (KL1–KL81, KL101–KL149, KL151, KL153–KL155, and KL157–159) (Wyres et al., 2016). It is noteworthy that the KL1–KL81 locus types and the K1–K81 K-types are synonymously used. However, the sugar compositions of the remaining antigens in the KL series are as yet unknown. The variation in the repeating units of different K-antigens leads to varying degrees of detection of Klebsiella spp. by the innate immune system (Kabha et al., 1995; Doorduijn et al., 2016). Among the 134 K-types (including the KL series) identified so far (Wyres et al., 2016), only a few of them are frequently found in the strains isolated from clinical samples (Cryz et al., 1986). Due to the increased production of CPS, hypervirulent Kp strains produce a hypercapsule, which is a hypermucoviscous EPS bacterial coating that may significantly contribute to Kp pathogenicity (Shon et al., 2013). Klebsiella pneumoniae strains with a hypercapsule are less sensitive to complement detection and elimination (Pomakova et al., 2012) and also have increased resistance to phagocytosis (Fang et al., 2004; Lin et al., 2004; Pomakova et al., 2012) compared to the classical strains. However, some cKp strains are also found to have a hypermucoviscous coating (Catalan-Najera et al., 2017; Russo et al., 2018). Notably, the presence of fucose in the hypercapsule has been implicated in the evasion of the immune response for the K1 antigen (Wu et al., 2008; Yeh et al., 2010). Although Kp strains possessing K1 and K2 serotypes are often found to be hypervirulent (Fung et al., 2002; Struve et al., 2015), other capsule types such as K5, K20, K47, K54, K57, and K64 are also found in hvKp strains (Yu et al., 2008; Shon et al., 2013; Russo et al., 2018).
Klebsiella spp. K-antigens are negatively charged (as is the case for other Gram-negative bacteria) and consist of up to six monosaccharides in their main chain as well as in the branch: D-mannose, D-glucose, D-galactose, L-fucose, and L-rhamnose. Of particular note is a completely new monosaccharide, 4-deoxy-threo-hex-4-enopyranosyluronic acid, that is found in K38 but is absent in any other K-antigen structure (Jansson et al., 1994). Detailed analyses of Klebsiella spp. K-antigen sugar compositions (Table 1) reveal that as with E. coli (Kunduru et al., 2016), K-antigens are negatively charged due to the presence of uronic acid and or pyruvate substitutions (https://iith.ac.in/K-PAM/, unpublished work). Additionally, they also have O-acetyl, O-lactose, O-formyl, and glutamate substitutions. The evolution and variability in the sugar composition of CPS are one of the major advantages possessed by Klebsiella when evading the host immune response.
Lipopolysaccharide
Pathogenicity factor LPS, also known as endotoxin, is found on the bacterial outer leaflet of the outer membrane and plays an important role in offering protection against cationic antimicrobial peptides (Clements et al., 2007; Llobet et al., 2015) and the complement system in certain serotypes (Merino et al., 1992). Klebsiella pneumoniae exploits the versatility of both CPS and LPS to counteract the complement system (Ciurana and Tomas, 1987; Alvarez et al., 2000; Shankar-Sinha et al., 2004; Doorduijn et al., 2016; Adamo and Margarit, 2018). It has been shown that purified LPS from Kp inhibits serum-mediated clearance (Merino et al., 1992). The structure of LPS consists of lipid A, core oligosaccharides, and O-antigens, among which the O-antigen composition is highly variable across different strains of Klebsiella spp. (Table 2, http://iith.ac.in/K-PAM/o_antigen.html) (Lugo et al., 2007). Unlike K-antigens, Klebsiella spp. has only 11 O-antigens (Clarke et al., 2018). O-antigen is also used in the typing of Klebsiella spp. Among the 11 O-antigen types found in Klebsiella spp., O1, O2, O3, and O5 are found in clinically imported strains (Hansen et al., 1999; Follador et al., 2016). O-antigens of Klebsiella spp. consist of D-galactose, D-galactofuranose, D-mannose, D-ribofuranose, and N-acetyl-D-glucosamine sugars. Their composition varies between different O-antigens, leading to differences in their antigenicity. Like the K-antigen, the O-antigens differ from each other in terms of sugar composition, glycosidic linkage, number of repeating units, and epimeric and enantiomeric forms (https://iith.ac.in/K-PAM/, unpublished work) (Follador et al., 2016; Clarke et al., 2018). Unlike the K-antigens, only acetyl group substitution is observed in O-antigens (and only in one of the O-antigens). While Klebsiella spp. strains that have truncated O-antigen or lack O-antigen (termed as “rough LPS”) are susceptible to complement system-mediated killing, the full-length O antigen or smooth LPS-containing Klebsiella spp. strains are resistant to complement system-mediated killing (Ciurana and Tomas, 1987; Mccallum et al., 1989; Merino et al., 1992). Although the complement-resistant strains activate the complement cascade, they are not susceptible to killing, as O-antigen variability protects the Kp surface molecules (Merino et al., 1992; Alberti et al., 1996; Shankar-Sinha et al., 2004; Merle et al., 2015).
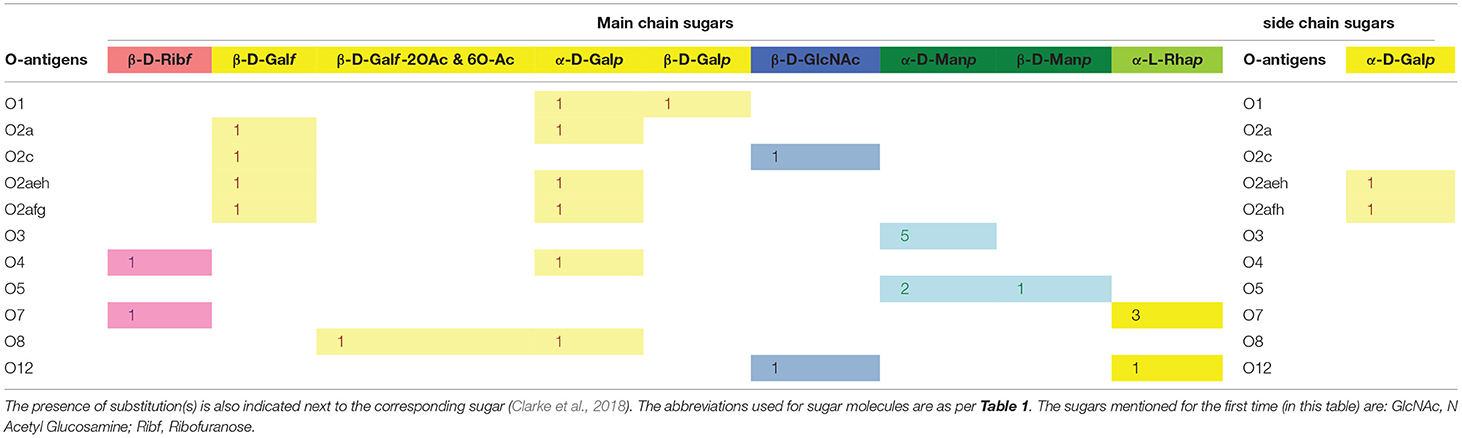
Table 2. Main-chain and side-chain sugar compositions of 11 O-antigens of Klebsiella spp. Note that the number of occurrences of a particular sugar (which varies between 1 and 5) is also indicated.
Exopolysaccharide
The extracellular matrix (which is a component of bacterial biofilm) of Klebsiella spp. is composed of proteinaceous adhesins, nucleic acids, and EPS (Sutherland, 2001; Branda et al., 2005; Vu et al., 2009). Compared to other surface-attached polysaccharides, little information is available on biofilm-associated EPS, which is yet another virulence factor of Kp (Cescutti et al., 2016). It has been shown that biofilm polysaccharides of Kp to some extent reduce antimicrobial peptide activity by preventing it from reaching the bacterial membrane or by impeding interaction with the membrane (Bellich et al., 2018). Genetic information regarding the biosynthesis of EPS is encoded in specific operons on the bacterial genome and 30 ORFs have been identified for the hetero-capsular EPS K40-type of Klebsiella spp. (Pan et al., 2015). In general, EPS contains rare sugars such as L-fucose, L-rhamnose, or uronic acids (Kumar et al., 2007), and Klebsiella is no exception. For example, hexasaccharide repeats of Klebsiella I-714 EPS have a high L-rhamnose content in addition to D-galactose and D-glucuronic acid (López-Santin, 1995; Roca et al., 2015). The primary structures of EPS extracted from K. pneumoniae strain KpTs113 have K24 CPS-repeating units and the K. pneumoniae strain KpTs101 is identical to the O1 antigen of LPS. This observation is supported by the finding that CPS and LPS are required for building the mature biofilm architecture (Balestrino et al., 2008; Benincasa et al., 2016; Cescutti et al., 2016). However, the KpMn7 strain has a rare sugar (rhamnose) in the repeating unit and is highly similar (but not identical) to the K24 CPS unit. Given this intriguing finding, further research is warranted on novel EPS structures found in Klebsiella spp. (Kubler-Kielb et al., 2013; Bellich et al., 2018).
Extracellular Polysaccharide Biosynthesis and Transportation Pathways
The aforementioned extracellular polysaccharide virulence factors are biosynthesized in the cytoplasm and transported through sophisticated proteinaceous nano-machines onto the bacterial surface. In general, bacteria use three different pathways for the transport of extracellular polysaccharides: (i) a Wzx/Wzy-dependent pathway (Rahn et al., 1999; Whitfield, 2006; Kalynych et al., 2014), (ii) an adenosine tri-phosphate (ATP)-binding cassette (ABC) transporter-dependent pathway (Cuthbertson et al., 2010; Greenfield and Whitfield, 2012; Kalynych et al., 2014), and (iii) a synthase-dependent pathway (Whitney and Howell, 2013). In addition to these, a fourth pathway (the dextrase/sucrase-dependent pathway) has also been identified for EPS secretion (Whitney and Howell, 2013; Schmid et al., 2015; Schmid, 2018).
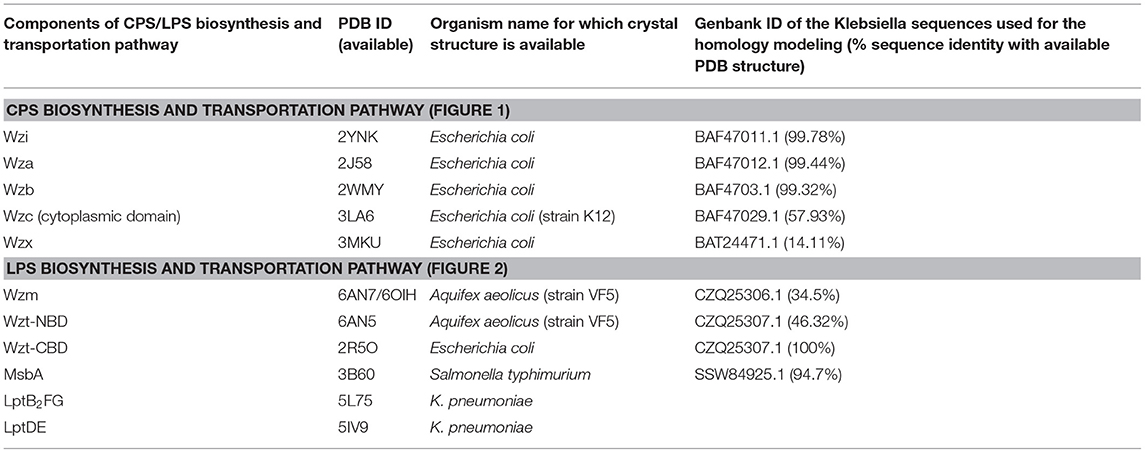
Klebsiella spp. use a Wzx/Wzy-dependent pathway for CPS secretion (Rahn et al., 1999) that is similar to Group 1 CPS surface export in E. coli (Rahn et al., 1999; Whitfield and Paiment, 2003; Sachdeva et al., 2017). Klebsiella spp. use three independent ABC transporter-dependent pathways for LPS secretion. Although it is known that E. coli uses a Wzx/Wzy-dependent pathway for EPS secretion (Reid and Whitfield, 2005), there is no information available regarding EPS secretion in Klebsiella spp. Understanding the mechanisms of transport and the structural features of the proteins involved in such transport is essential for the identification of potential antimicrobial targets and the development of novel antimicrobials. As information on the EPS-secretion pathway is not available for Klebsiella spp., the following sections are limited to a review of CPS and LPS transportation strategies used by Klebsiella. The structures of the proteins involved in Klebsiella CPS and LPS export (with the exceptions of LptDE and LptB2FG) have been obtained by homology modeling using known template structures from other organisms (see Table 3).
The Wzx/Wzy-Dependent Secretion Pathway
The chromosomal cps gene cluster harbors genes that are essential for the biosynthesis of sugar precursor molecules, assembly of the repeating unit, flipping of the repeating unit to the periplasmic side, polymerization of the repeating unit, transport of the nascent CPS, and anchorage of CPS onto the surface of Klebsiella (Pan et al., 2015). Klebsiella spp. utilize a Wzx/Wzy-dependent CPS secretion pathway, which is similar to that for Group 1 capsule production in E. coli (Whitfield and Paiment, 2003; Sachdeva et al., 2017). The process of CPS export in Klebsiella spp. begins with the biosynthesis of nucleotide sugar precursors corresponding to a particular K-type and the assembly of the repeat unit at the cytoplasmic face. This occurs with the help of sugar-specific glycosyl transferases encoded by genes such as wbaP, wcaN, manC, rmlA, wcaA, wcuD, wcuM, wckA, and wclH (Rahn et al., 1999; Shu et al., 2009; Pan et al., 2015). Subsequently, recognition of the specific CPS-repeating unit by the flippase Wzx occurs with the first sugar linked to undecaprenol-pyrophosphate (Und-PP), followed by flipping to the periplasmic side. Repeat unit polymerization is facilitated by Wzy copolymerase (Whitfield and Paiment, 2003; Li et al., 2016). Finally, Wza (an outer-membrane translocon), Wzc (a tyrosin autokinase), and Wzb (a phosphatase) synergistically transport CPS onto the bacterial surface and anchor the CPS onto the outer-membrane protein Wzi (Rahn et al., 2003; Whitfield, 2006; Woodward et al., 2010). This CPS export pathway is common to all Klebsiella spp. (as they all have cps locus genes). Gene sequences of the cps locus (specifically wzi and wzc) are used in the K-typing of Klebsiella spp., owing to limitations in conventional K-typing (Brisse et al., 2013; Pan et al., 2013; Wyres et al., 2016). Klebsiella spp. cps gene sequences (e.g., wzi, wza, wzb, wzc, wbap, wcaj, wzx, and wzy) vary according to their K-antigen composition and are used in genome-based surveillance of Klebsiella spp. (Pan et al., 2015; Wyres et al., 2016) (https://iith.ac.in/K-PAM/, unpublished work). Notably, a recent study has shown that the arrangement of the genes in the CPS locus is K-type-specific and this finding has been successfully applied to Klebsiella spp. K-typing (Pan et al., 2015; Wyres et al., 2016; Wick et al., 2018). It has been found that a Klebsiella spp. strain can either contain initialization glycosyl transferase WbaP or WcaJ, but not both (Shu et al., 2009). Sugar composition analysis indicates that the serotypes K1, K2, K4-K8, K11, K13, K14, K16, K17, K22–K25, K28, K30, K31, K33–K35, K37, K39, K44, K45, K48, K54, K55, K58–K61, K64, K67, K69, K71–K73, and K82 have WcaJ and use glucose-Und-PP as an initializing sugar. On the other hand, K3, K9, K10, K12, K15, K18–K21, K26, K27, K32, K36, K38, K40, K41–K43, K46, K47, K49–K53, K56, K57, K62, K63, K66, K68, K70, K74, and K79–K81 use galactose-Und-PP as the initializing sugar and have WbaP in their cps gene cluster.
LPS Biosynthesis in the Cytoplasm by ABC-Dependent Pathway
LPS are glycolipids that encompass three structural moieties, namely, lipid A, core oligosaccharides (core-OS), and the O-antigenic polysaccharide (O-PS) (Whitfield and Trent, 2014). The lipid A (the lipid moiety of LPS) is highly conserved and anchors the LPS on the outer leaflet of the outer bacterial membrane. The core-OS is conserved and acts as a linkage between the lipid A and O-PS. The O-PS is highly variable across different Klebsiella spp.
Such complexity in the LPS structure leads to a complex biosynthesis pathway that takes place at the cytosolic and periplasmic faces of the inner membrane: (i) biosynthesis of lipid A through the Raetz pathway, (ii) attachment of core-OS to the lipid A, (iii) flipping of the lipid A-core-OS to the periplasmic end, (iv) biosynthesis of O-PS at the cytoplasmic end, (v) flipping of O-PS to the periplasmic region, and (vi) ligation of O-PS to lipid A-core-OS in the periplasmic region (Raetz and Whitfield, 2002; Whitfield and Trent, 2014). Finally, the LPS molecule assembled in the periplasmic region is exported to the bacterial surface wherein lipid A acts as the anchorage point for the LPS (Okuda et al., 2016). The entire process of LPS biosynthesis and surface export involves four different gene clusters: lpx, waa, rfb, and lpt. The gene products of lpx, waa, and rfb are involved in the biosynthesis of lipid A, core-OS, and O-PS, respectively (Regue et al., 2005; Fresno et al., 2007; Okuda et al., 2016). The lpt gene products are involved in the transport of the LPS molecule to the extracellular side of Klebsiella. The other protein involved in this biosynthesis process is MsbA, which is part of a different gene cluster. Intriguingly, Klebsiella spp. LPS biosynthesis and transportation are driven by ATP hydrolysis at three different stages: flipping of the lipid A-core-OS, flipping of O-PS, and transport of LPS from the periplasmic end to the bacterial extracellular region. These steps are outlined below.
Biosynthesis of Kdo2-lipid A–core-OS
The biosynthesis of lipid A begins in the cytosolic region with the involvement of nine enzymes synthesized from the lpx gene cluster (Raetz et al., 2009). The first step is the substitution of an acyl chain to the 3-OH group of uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) (Anderson et al., 1985; Anderson and Raetz, 1987), followed by the release of an acetate group and the addition of the second acyl side chain. Two such monosaccharides are glycosylated, wherein one is phosphorylated (called lipid X) prior to the reaction, following which disaccharide-1-phosphate is again phosphorylated to synthesize Lipid IVA at the cytoplasmic face of the inner membrane. The matured lipid IVA is glycosylated with two 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) residues, which are incorporated by WaaA (a product of the waa gene cluster) to produce Kdo2-lipid IVA. Subsequently, Kdo2-lipid A is synthesized by the acylation of Kdo2-lipid IVA. This entire Raetz pathway takes place in the cytoplasmic end of the inner membrane and is mediated by several glycosyltransferases, along with other enzymes (Raetz and Whitfield, 2002).
In the next step, core-OS is synthesized by extending Kdo2-lipid A with the help of several glycosyltransferase enzymes (which vary as per the sugar components of different O-antigens). In general, the core-OS is conceptually divided into two regions, namely, the conserved inner core and the variable outer core. The inner core typically has Kdo2 and L-glycero-D-manno-heptopyranose (L, D-Hep). The outer core consists of three to six sugars, whose compositions are variable.
Export of Kdo2-lipid A–core-OS Across the Inner Membrane
The nascent Kdo2-lipid A–core-OS intermediate is subsequently flipped to the periplasmic end of the inner membrane through an ABC transporter, MsbA. MsbA is a “half” transporter as it contains two different polypeptide chains wherein each chain contains a nucleotide-binding domain (NBD) and a transmembrane domain (TMD). MsbA uses an “outward only” mechanism to flip Kdo2-lipid A–core across the inner bacterial membrane. In this mechanism, MsbA remains in a resting state with an (inward) open conformation at the cytoplasmic side when ATP is not bound. This inward open form allows Kdo2-lipid A–core-OS entry. Stable Kdo2-lipid A–core-OS binding aligns TMD for ATP binding and restricts the opening of TMD. Upon ATP binding, the NBD domain intertwines, and Kdo2-lipid A–core-OS moves toward the periplasmic side. This coordinated movement of MsbA conformational change and LPS translocation leads to ATP hydrolysis, thus restoring the ground state inward for open confirmation of MsbA. This “outward only” mechanism for Kdo2-lipid A–core-OS export across the inner membrane is established based on different conformations of MsbA, derived from different Gram-negative bacterial species (Doerrler et al., 2004; Arai et al., 2017; Mi et al., 2017; Ford and Beis, 2019). Upon transport to the periplasmic region, Kdo2-lipid A–core-OS can undergo environmentally regulated modifications.
O-PS Biosynthesis and Transportation Machinery
Klebsiella spp. O-PS biosynthesis takes place separately at the cytoplasmic end of the inner bacterial membrane. The O-PS has four conceptually different regions: primer, adaptor, repeating unit, and terminal modification domains (Raetz and Whitfield, 2002). In general, Klebsiella spp. have two to five sugars in the O-PS repeating unit that are highly variable for different O-antigens (Clarke et al., 2018). The O-PS repeating unit is assembled on a lipid carrier undecaprenyl phosphate (embedded in the inner membrane) with the help of several glycosyltransferases encoded by wecA, gmlABC, wbbMNO, wbmV, wbmW, and wbmX genes and is transported to the periplasm with the help of an ABC transporter (Clarke et al., 2018). O-PS biosynthesis requires a polyisoprenoid derivative, namely, C55-undecaprenol phosphate (Und-P), which serves as an acceptor for O-PS chain assembly. The reaction begins with the transfer of N-acetylglucosamine (GlcNAc)-1-phosphate onto Und-P. This reaction is facilitated by GlcNAc-1-phosphate transferase (WecA) and produces Und-PP-GlcNAc, which is the primer region of O-PS. The O-PS is extended on Und-PP-GlcNAc with the help of glycosyltransferases, depending on the sugar composition of the O-PS (Meier-Dieter et al., 1992; Rick et al., 1994; Clarke et al., 1995; Guan et al., 2001; Kos et al., 2009). Depending on the O-antigen, the O-PS biosynthesis rfb gene cluster has 6 to 13 genes that are required for O-PS synthesis, of which 6 are essential genes (Clarke and Whitfield, 1992; Clarke et al., 2018). The 5′ end of the gene cluster has genes that encode for ABC transporters, and the 3′ end of the cluster has genes that produce glycosyltransferases. The adaptor domain, which occurs only once in an O-PS chain and acts as the connection between Und-PP-GlcNAc and the repeat unit domain, is subsequently attached to the growing O-PS. The O-PS chain extension takes place by the addition of a repeat-unit domain. The growth of O-PS occurs at the non-reducing end of the polysaccharide chain. Finally, the O-antigen length is regulated either through a covalent modification at the terminal residue of the O-PS (terminal capping/modification) or as a result of the stoichiometry of the Wzm-Wzt ABC transporter that transfers the Und-PP linked O-PS to the periplasmic end (see below).
O-antigen Transport Through the Wzm/Wzt System
After polymerization, the O-antigens are transported to the periplasmic-leaflet of the inner membrane by an ABC transporter that has two transmembrane domains (TMDs) (named Wzm) and two nucleotide-binding domains (NBDs) (named Wzt) (Kos et al., 2009). This O-antigen ABC transporter system is common in most of the Gram-negative bacteria. Intriguingly, in some of the Klebsiella spp., the O-antigen ABC transporter has an additional carbohydrate-binding domain (CBD) that is fused to the C-terminus of the NBD (Cuthbertson et al., 2005, 2007; Liston et al., 2017). Chemical modifications, such as the addition of a phosphate or methyl group at the non-reducing end of some O-antigens, provide the biosynthesis completion signal, which is recognized by the CBD to accomplish the transport (Liston et al., 2017). O12 is one such antigen that has the CBD, while such a mechanism is absent in the uncapped O-antigen biosynthesis in Kp (Bi et al., 2018).
Although structural information pertaining to the Klebsiella spp. Wzm/Wzt ABC transporter is unavailable, its homologous structure from Aquifex aeolicus has provided insights into the mechanism of O-antigen transport. Wzm/Wzt structures determined from A. aeolicus in ATP-free (Bi et al., 2018) and ATP-bound (Caffalette et al., 2019) forms reveal that the formation of a continuous inner transmembrane (TM) channel is wide enough to accommodate an O-antigen chain in the nucleotide-unbound conformation. ATP is seen in the bound conformation at the conserved Walker A, Walker B, and H-loop signature motifs of NDB (Davidson et al., 2008; Locher, 2016). These motifs are conserved between Klebsiella spp. and A. aeolicus and are essential for the transport of O-antigens across the inner membrane. In the complex form, the NBD adopts a compact structure and interacts with the Wzm dimer. The O-antigen chain bound to the Wzm/Wzt transporter is passed through the TM channel to reach the periplasmic face of the inner membrane, following which the lipid portion of the Und-PP-N-acetamido sugar moiety is inserted into the inner-membrane periplasmic leaflet (onto which the O-antigen is anchored) (Figure 2, left).
LPS Maturation in the Periplasm
The LPS intermediates (Und-PP-linked O-PS and Kdo2-lipid A–core-OS) that are transported to the periplasm are ligated with the help of WaaL ligase (a product of the waa gene cluster) (Regue et al., 2005). The Und-PP-linked O-PS is transferred to Kdo2-lipid A–core-OS by the formation of a glycosidic bond between the first sugar of the O-PS and the sugar in the outer core.
LPS Transport to the Outer Membrane Through LptA-G
The LPS is transported to the outer membrane through a transport system comprising seven proteins, namely, LptABCDEFG (LptA–G) (Sperandeo et al., 2007; Ruiz et al., 2008; Freinkman et al., 2011, 2012; Villa et al., 2013). All seven of the protein structures of the LPS transport system have been fully characterized (Botos et al., 2016; Dong et al., 2017; Vetterli et al., 2018; Li et al., 2019; Owens et al., 2019). Among these proteins, the LptDE and LptB2FG complex structures are known for Klebsiella spp. (Table 3), while structural information for the remaining components is available for other Gram-negative bacterial species (Vetterli et al., 2018; Li et al., 2019; Owens et al., 2019). This structural information, combined with existing knowledge of the associated transport mechanisms, has been used here to explain LPS transport in Klebsiella spp. Strikingly, the portal for transport of LPS molecules is formed by LptD and LptE, which is connected to a pump-like system formed by the LptB2FG ABC-transporter through a bridge-like structure consisting of LptA and LptC (Bishop, 2019; Li et al., 2019; Owens et al., 2019). The individual sections of this integrated LPS transporter are discussed below. As the LptA–G transporter is distributed across the inner membrane, periplasmic region, and outer membrane, this nano-machine represents a promising antimicrobial target.
Insertion and Translocation of LPS Into LptB2FG
LPS is driven across the ABC transporter LptB2FG (Okuda et al., 2012; Sherman et al., 2014) in a continuous flow from the periplasmic leaflet of the inner membrane to the periplasmic domain of LptC and through the transmembrane helix of LptC (Sperandeo et al., 2008, 2011; Narita and Tokuda, 2009). This is accomplished by utilizing energy derived from the ATP-hydrolysis activity of LptB (Narita and Tokuda, 2009; Sherman et al., 2014). The LptB2FG complex contains two transmembrane domains (LptF and LptG) and two nucleotide-binding domains (LptB2) (Ruiz et al., 2008; Narita and Tokuda, 2009). Both LptF and LptG contain a periplasmic β-jelly roll domain that is unique to this ABC transporter (LptB2FG). LPS passes into LptFG through a lateral opening formed by transmembrane helix 1 (TM1) of LptF and TM5 of LptG through an electrostatic gating mechanism (Dong et al., 2017). The LPS subsequently travels to the periplasmic domain helix (locked in-between TM1 of LptG and TM5 of LptF) of LptC (Okuda et al., 2016) in a stepwise manner (Owens et al., 2019). The soluble periplasmic protein LptA bridges LptC and the N-terminal domain of outer-membrane protein LptD by forming a head-to-tail oligomer (Suits et al., 2008) with a continuous hydrophobic groove (Bowyer et al., 2011; Sperandeo et al., 2011; Grabowicz et al., 2013; Villa et al., 2013). LptA shares a β-jelly roll fold with the periplasmic domain of LptC (Tran et al., 2010) and the N-terminal domain of LptD (Qiao et al., 2014). Strikingly, a β-jelly roll fold arrangement with a similar hydrophobic groove has also been observed in the periplasmic domain of LptF (Dong et al., 2017; Li et al., 2019; Owens et al., 2019), which could explain the transport of LPS to the outer membrane of the bacteria (as mediated by the LptFG complex).
LPS Assembly Onto the Outer Leaflet of the Outer Membrane
The N-terminal domain of the outer-membrane LptD is thought to be very flexible in order to maintain the physical connection and integrity of the LptCAD scaffold (Botos et al., 2016). Soon after the N-terminal domain of LptD accepts the LPS from the periplasmic protein LptA, it undergoes a significant conformational change in such a way as to open up a luminal gate formed by two periplasmic loops of LptE with LptD. The opening of the LptDE lateral gate facilitates LPS transit through the periplasmic hydrophobic groove to the extracellular region (Botos et al., 2016). Subsequent to this, the lipid A section of LPS is inserted directly into the membrane and facilitates the transition of the polysaccharide fragment through the barrel lumen to the extracellular space (Gu et al., 2015; Botos et al., 2016; Dong et al., 2017).
A Therapeutic Perspective for Combating Klebsiella spp. Infections
Although antibiotics such as third-generation cephalosporins, aminoglycosides, fluoroquinolones, and carbapenems (Navon-Venezia et al., 2017; The European Antimicrobial Resistance Surveillance Network, 2018) have contributed dramatically to the reduction of morbidity and mortality associated with Klebsiella spp. infections, the continued emergence of cKp strains with extreme drug resistance and the newly emerged multidrug-resistant hypervirulent Klebsiella strains (Gu et al., 2018) limit current treatment options to eradicate infections (Brisse et al., 2009; Magiorakos et al., 2012; Doorduijn et al., 2016; Navon-Venezia et al., 2017; Martin and Bachman, 2018). Alarmingly, recent evidence suggests that Klebsiella has also evolved mechanisms to actively suppress innate immune responses (Bengoechea and Sa Pessoa, 2019), in addition to other well-known stealthy Klebsiella immune evasion strategies. Although many virulence factors are thought to be involved in the counteraction of host defenses by Klebsiella, only a few of these are well-studied, including CPS, LPS, fimbriae, and siderophores (Paczosa and Mecsas, 2016). As CPS and LPS actively participate in hijacking host defenses to establish infection, targeting these can prevent the growth of Klebsiella spp. (rather than killing the pathogen) by imposing less intense selective pressure. Ultimately, this may limit the evolution of resistant strains. Here, the biosynthesis and export of these surface-associated polysaccharides are discussed from the perspective of the treatment of Klebsiella infections.
CPS and LPS protect Klebsiella spp. from the action of complement cascade and antimicrobial peptides, as well as from engulfment and phagocytosis by host immune cells (Alvarez et al., 2000; Regueiro et al., 2006; Pan et al., 2011). In addition, CPS acts as a physical barrier to protect LPS (Merino et al., 1992; Alvarez et al., 2000). EPS (another surface-associated polysaccharide) is a component of biofilm and has been shown to interfere with the action of antimicrobial peptides of the host immune system (Bellich et al., 2018). Thus, inhibition of the LPS, CPS, and EPS biosynthesis and surface expression would be an effective approach to counteract Klebsiella anti-immune strategies. As the EPS secretion pathway and its structural composition in Klebsiella spp. is not well-understood, this review discusses the treatment strategies of Klebsiella infections from the perspective of CPS and LPS.
CPS and LPS are biosynthesized in cytoplasmic/periplasmic regions of the inner bacterial membrane and are transported to the bacterial surface with the help of sophisticated proteinaceous nano-machines. Thus, blocking the biosynthesis of CPS and LPS or disrupting the assembly of these nano-machineries can block the surface expression of these molecules that offer protection from the host immune response. For instance, LPS biosynthesis can be targeted in three different stages: lipid A, core-OS, and O-PS biosynthesis. Targeting the components of biosynthesis may prevent the formation of LPS and render Klebsiella vulnerable to host defenses. A possibility for novel antibiotic development could involve targeting the lipid A synthesizing enzymes (synthesized by the lpx locus), as there are no human homologs for them (Whitfield and Trent, 2014). Indeed, a recent study drawing on multi-omics data from sources including genomics, transcriptomics, structuromic, and metabolic information has listed LpxA, LpxB, LpxC, and LpxD as prioritized non-host homologous protein targets (Ramos et al., 2018). Targeting the LPS export pathway proteins represents yet another strategy. Specifically, the outer-membrane proteins [LptD and LptE (Figure 2)] involved in LPS export represent potential antibiotics targets, given that they are easily accessible (Srinivas et al., 2010; Robinson, 2019). Producing antibodies against these outer-membrane proteins is also of particular clinical interest (Storek et al., 2019). Similarly, Wzm, Wzt, and MsbA could also be targets for the development of novel antimicrobials (Alexander et al., 2018; Ho et al., 2018).
Targeting the proteins that participate in Wzx/Wzy-dependent CPS transport and the surface expression pathway (Figure 1) may interfere with CPS export to the bacterial surface (Sachdeva et al., 2017). For instance, manipulating the function of the aqua-lecto-porin Wzi (Bushell et al., 2013; Sachdeva et al., 2016), as well as capping the extracellular side of Wza (Dong et al., 2006) involved in CPS surface expression through a novel antibiotic, would be potential targets similar to that for E. coli. It is worth noting that a similar strategy has been successfully demonstrated in E. coli Wza (Kong et al., 2013; Sachdeva et al., 2017). However, the sequence diversity of the surface-exposed region of Wza across various K-types may present a challenge in designing a common antibiotic (boxed region in Figure 3A). In contrast, Wzi is highly conserved and is a potential target for all Klebsiella spp. (Figure 3B).
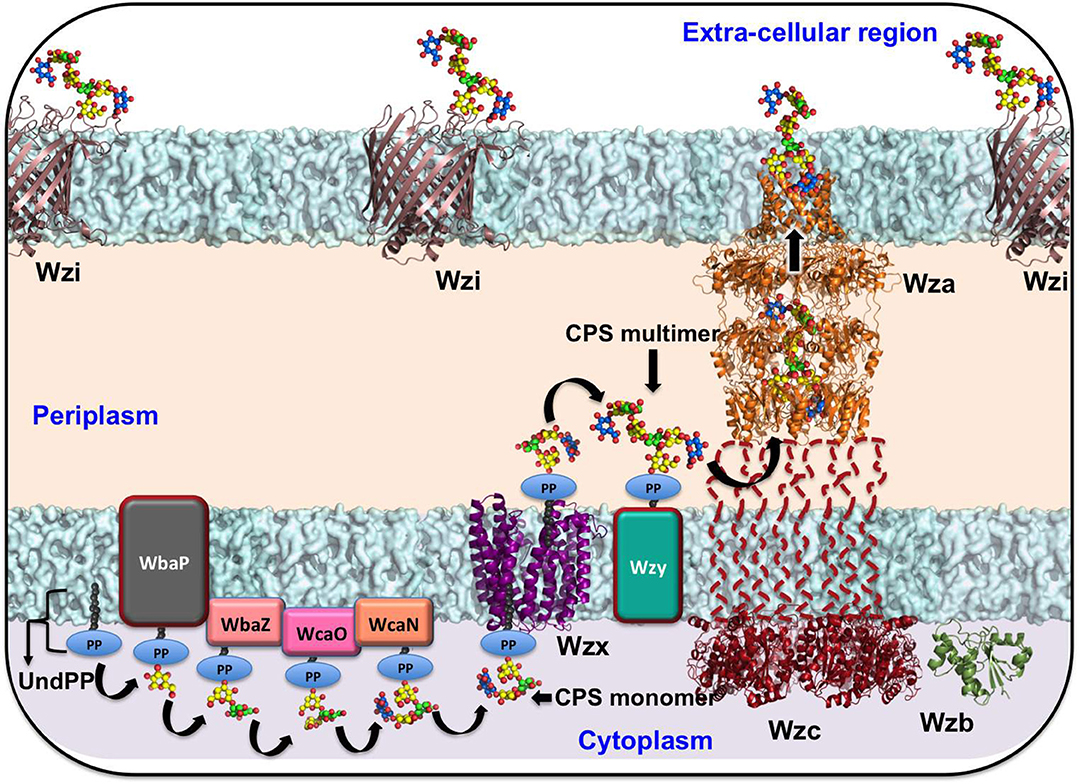
Figure 1. Schematic representation of Klebsiella spp. CPS biosynthesis and surface export machinery. The sugar precursors biosynthesized in the cytoplasm are subsequently assembled in the cytoplasmic face of the inner membrane to form the repeating unit with the help of sugar-specific glycosyl transferases WbaP (or WcaJ), followed by WbaZ, WcaN, WcaJ, and WcaO. The recognition of the CPS repeating unit by the first sugar linked to undecaprenol-pyrophosphate (Und-PP) by Wzx (a flippase) facilitates the flipping of the repeating unit to the periplasmic side. Subsequent to this event, Wzy (a copolymerase) polymerizes the repeating units. Finally, Wza (an outer-membrane translocon), Wzc (a tyrosin autokinase), and Wzb (a phosphatase) synergistically transport CPS onto the bacterial surface and anchor the CPS onto the outer-membrane protein Wzi (a lecto-aqua-porin). As structural information on the representative proteins from Kp is unknown, the structures of Wzi, Wza, Wzb, Wzc (cytoplasmic domain), and Wzx have been modeled from available reference structures through the SWISS-MODEL server (Schwede et al., 2003). Klebsiella pneumoniae (K20) accession numbers corresponding to Wzi, Wza, Wzb, Wzc, and Wzx are BAF47011.1, BAF47012.1, BAF4703.1, BAF47029.1, and BAT24471.1, respectively. The corresponding PDB IDs used as templates in the modeling are 2YNK (99.78%), 2J58 (99.44%), 2WMY (99.32%), 3LA6 (57.93%), and 3MKU (14.11%), respectively. The sequence identity between the query and template is indicated in the bracket.
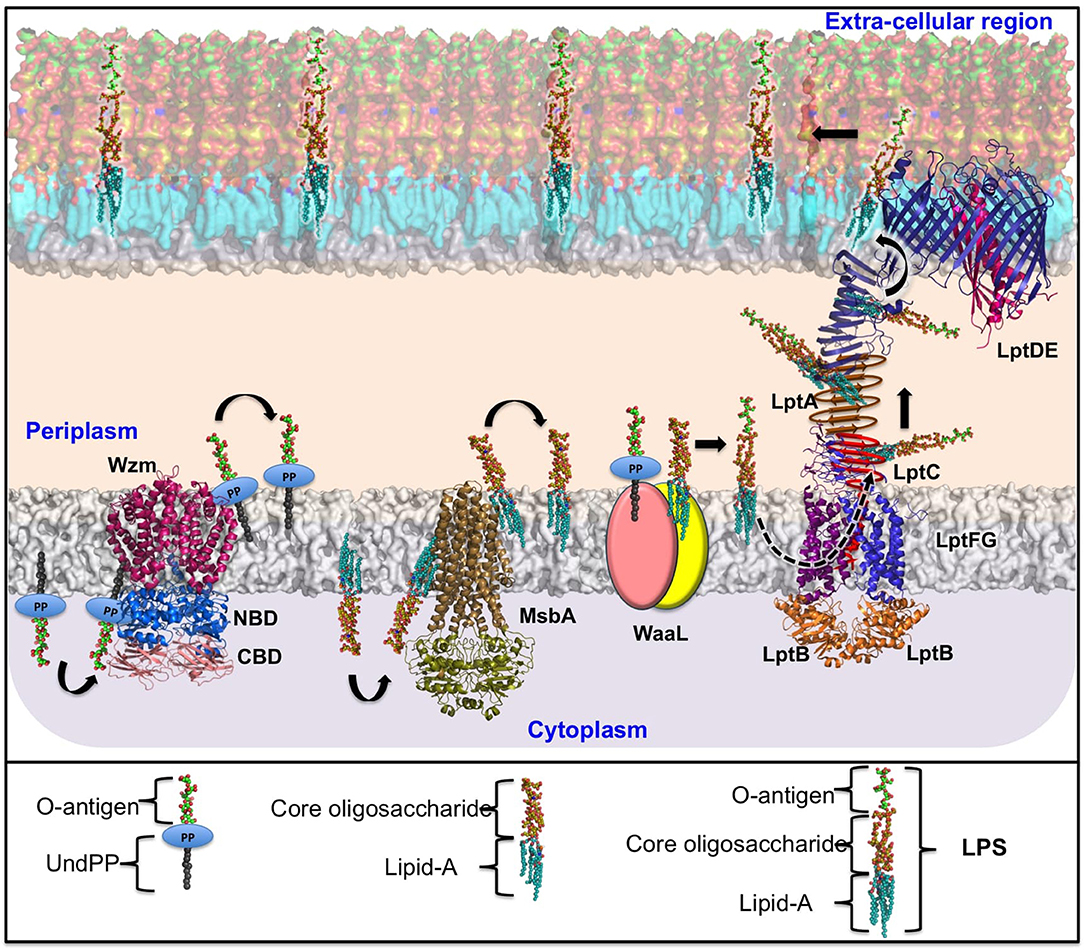
Figure 2. Schematic representation of Klebsiella spp. ABC transporter-dependent LPS assembly and transport. The O-antigen repeating unit is synthesized in the cytosol with the help of the corresponding glycosyltransferases and is subsequently polymerized by Wzy and transferred to the periplasm by an ABC transporter (Wzm/Wzt complex). In a similar fashion, the Kdo2-lipid A–core oligosaccharide biosynthesized in the cytoplasmic region is flipped to the periplasmic region through the ATP-driven MsbA. Following this, the matured O-polysaccharide and lipid A-core-oligosaccharide are ligated by WaaL ligase in the periplasmic region. The completely grown LPS is transported to the bacterial surface through LptA-G assembly as indicated. For the purpose of illustration, the LptB2FG (PDB ID: 5L75) and LptDE (PDB ID: 5IV9) structures are taken directly from Kp, while the Wzm/Wzt complex and MsbA proteins are homology-modeled using structures available in other organisms as templates. The reference PDB IDs for Wzm, Wzt-NBD, and Wzt-CBD are 6AN7 (34.5%), 6AN5 (46.32%), and 2R5O (100%), respectively. The sequence identity between the template and the Kp are given in brackets. The NCBI accession numbers corresponding to the Kp protein sequences are CZQ25306.1 (Wzm) and CZQ25307.1 (Wzt-NBD and Wzt-CBD). LptA and LptC are indicated by schematic representation. The helical and beta-jelly conformation of LptC is shown in red. The beta-jelly conformation of LptA is colored dark gold. As WaaL structural information for any Gram-negative organism is unavailable, the two domains of WaaL are represented in yellow and peach-colored ovals. Note that for the purpose of illustration, O3 has been considered as a case in point. Individual parts of the LPS and O-antigen are annotated separately at the bottom of the figure.
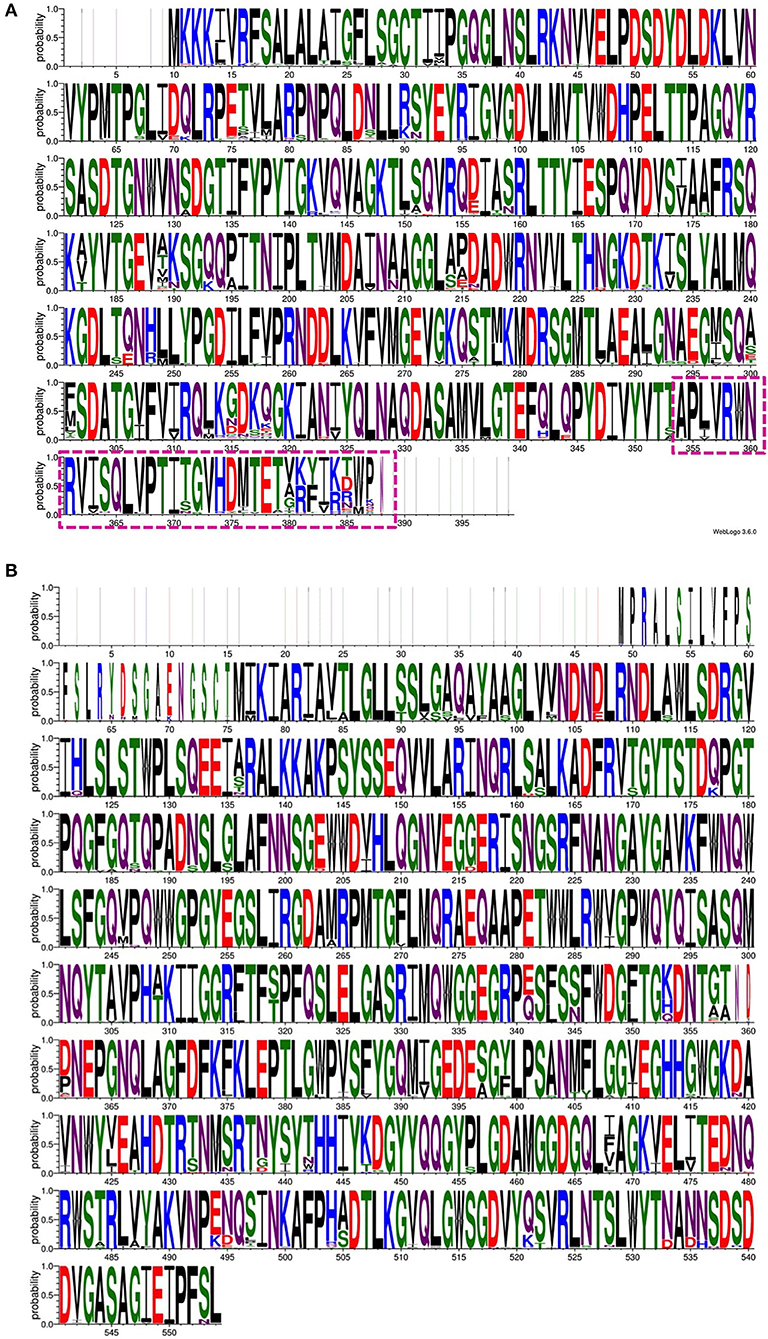
Figure 3. An amino acid sequence logo constructed using the multiple sequence alignment of 139 Wza protein sequences (A) and 138 Wzi protein sequences (B). Note the variation observed in the C-terminal region (transmembrane region) of Wza that faces the extracellular region of the bacterial cell (dash-box) (A). In contrast, Wzi sequences are highly conserved between different serotypes of Klebsiella spp. Note that non-redundant sequences that have a defined K-type are used to generate sequence logo.
Another approach for the treatment of Klebsiella infection involves the development of antibodies targeting CPS and LPS (Szijarto et al., 2017; Diago-Navarro et al., 2018; Kobayashi et al., 2018). Cell surface carbohydrate-based vaccines (Hutter and Lepenies, 2015) can be an effective choice for combating Klebsiella infections (Cryz et al., 1985; Cross, 2014; Seeberger et al., 2017; Adamo and Margarit, 2018; Hegerle et al., 2018; Micoli et al., 2018). Glycan epitopes, namely, the antibody-interacting and minimal antigenic determinant of O- or K- antigens, can be used in vaccine development. The heterogeneity and complexity of O- and K-antigens of different Klebsiella serotypes may pose a challenge to the development of a polyvalent vaccine against all Klebsiella infections. Fortunately, only a few O- and K-antigens are found in clinical isolates; thus, they can be used in the development of a novel immunogenic polyvalent glycoconjugate Klebsiella vaccine with the help of improved vaccine technology. Multiple interactions between protein and glycan is essential at different stages of the immune response. Identification of surface saccharide epitope patterns in clinical/hypervirulent strains and their use in the design of a unique synthetic glycan epitope conjugated with an immunogenic carrier protein may be useful in the development of an effective multivalent glycoconjugate Klebsiella vaccine.
Although Klebsiella spp. have 12 O-antigens, seroepidemiological investigations have revealed only four Klebsiella O serotypes found in clinical isolates (Edelman et al., 1994; Cryz et al., 1995; Trautmann et al., 2004). Thus, Klebsiella anti-endotoxin vaccines/antibodies can be developed based on the O-antigen structure of clinical isolates of Klebsiella spp. Protection against Kp through anti-LPS antibodies has been successfully demonstrated (Cohen et al., 2017; Pennini et al., 2017; Hegerle et al., 2018). Although thermostable LPS is a strong immune activator, Kp quite often uses modifications of lipid A of LPS in such a way that it is no longer recognized by certain immune receptors such as TLR4 (Llobet et al., 2015). This helps it evade the complement system and to survive within the host during colonization and infection (Llobet et al., 2011, 2015; Kidd et al., 2017; Mills et al., 2017). Modification of the polysaccharide composition of the O-antigen side chain (which is exposed to antibodies) and elongation of the O-antigen has also been documented (Doorduijn et al., 2016). Kp strains with a long O-antigen produce a high-molecular-weight (smooth phenotype) LPS that is less susceptible to serum killing, as compared to strains lacking an O-antigen side chain with a low-molecular-weight (rough phenotype) LPS (Ciurana and Tomas, 1987; Mccallum et al., 1989). For example, D-galactan I to D-galactan III structure modification of the O-antigen is found to improve Kp survival in human serum compared to strains expressing D-galactan I (Szijarto et al., 2016). Similarly, an epidemic multidrug-resistant Kp clone (Tzouvelekis et al., 2013) was found to have a modified O-antigen structure (Wyres et al., 2015; Szijarto et al., 2016). Modification of the glycan structures at the terminal end of the O-antigen has also been shown to alter complement activation in Kp (Tytgat and Lebeer, 2014; Adamo and Margarit, 2018).
CPS could also be exploited to counteract Klebsiella anti-immune strategies. Recognition of this possibility has led to the development of a 24-valent CPS-based vaccine for Klebsiella (Cryz et al., 1991; Edelman et al., 1994; Campbell et al., 1996; Donta et al., 1996). Although a phase 1 trial of the vaccine has shown it to be immunogenic and non-toxic (Edelman et al., 1994), no further developments have been reported in the last two decades. Similar to LPS, the capsule also undergoes modifications to resist the host complement system (Wyres et al., 2015; Szijarto et al., 2016). This may pose a challenge in developing a vaccine against Klebsiella spp. infections. Chemical modifications in K-antigen structures, such as acetylation and deacetylation (Hsu et al., 2016), may also bring about differential effects in CPS antigenicity, representing yet another challenge in the development of vaccines against Klebsiella spp. K2-antigen-lacking mannobiose or rhamnobiose produced by a Kp strain escapes host recognition during the host innate immune response (Sahly et al., 2009). It is worth noting that hvKp strains are frequently found to have K2 antigens.
Use of exogenous cholesterol and bacteriophage depolymerase against Klebsiella infections represents yet another promising approach. It has been shown that exogenous cholesterol increases macrophage-mediated phagocytosis by down-regulating the expression of genes responsible for LPS core oligosaccharides production, as well as reducing the anti-phagocytic properties of the Kp capsule (Ares et al., 2019). The discovery that bacteriophage capsule depolymerases can be used against Klebsiella capsule types KN1, KN3, KN4, and K56 represents a potential approach for the treatment of Kp infections (Pan et al., 2019).
Significant progress has been made in understanding Klebsiella immune evasion strategies. As the CPS and LPS of Klebsiella spp. play an important role in hijacking host defenses, targeting these virulence factors may be an efficient strategy against Klebsiella infections. The known structural components of Klebsiella CPS and LPS export machineries could be useful in the design of novel antibiotics. However, heterogeneity in sugar composition, glycosidic linkage, stereoisomeric forms, and the concomitant variation in the proteins involved in biosynthesis and transport may pose a challenge in the design of antibiotics and vaccines that can be used against diverse Klebsiella spp. In addition, the ability of Klebsiella spp. to modify components of the CPS and LPS may be another concern. Recent developments in gene sequencing techniques in combination with a metagenomic approach to the investigation of Kp clinical strains help in the design of polyvalent vaccines. A combinatorial therapy involving Klebsiella vaccines against surface polysaccharides and antibiotics inhibiting surface antigen assembly may represent the most promising approach.
Author Contributions
TR designed and supervised the entire project. LP and TR wrote the manuscript.
Funding
We greatly acknowledge the support from BIRAC-SRISTI (SAN No. BIRAC-SRISTI GYTI - PMU_2017_010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Mr. C. Sathyaseelan for his assistance with regard to homology modeling. The authors also thank Ms. Vinothini and Ms. Madhushree for their proofreading of the manuscript.
References
Abreu, A. G., and Barbosa, A. S. (2017). How Escherichia coli circumvent complement-mediated killing. Front. Immunol. 8:452. doi: 10.3389/fimmu.2017.00452
Adamo, R., and Margarit, I. (2018). Fighting antibiotic-resistant klebsiella pneumoniae with “sweet” immune targets. Mbio 9:e00874–18. doi: 10.1128/mBio.00874-18
Al-Anazi, K. A., Al-Jasser, A. M., Al-Zahrani, H. A., Chaudhri, N., and Al-Mohareb, F. I. (2008). Klebsiella oxytoca bacteremia causing septic shock in recipients of hematopoietic stem cell transplant: two case reports. Cases J. 1:160. doi: 10.1186/1757-1626-1-160
Alberti, S., Alvarez, D., Merino, S., Casado, M. T., Vivanco, F., Tomas, J. M., et al. (1996). Analysis of complement c3 deposition and degradation on Klebsiella pneumoniae. Infect. Immun. 64, 4726–4732.
Alexander, M. K., Miu, A., Oh, A., Reichelt, M., Ho, H., Chalouni, C., et al. (2018). Disrupting gram-negative bacterial outer membrane biosynthesis through inhibition of the lipopolysaccharide transporter Msba. Antimicrob. Agents Chemother. 62:e01142–18. doi: 10.1128/AAC.01142-18
Alvarez, D., Merino, S., Tomas, J. M., Benedi, V. J., and Alberti, S. (2000). Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide o side chain-deficient klebsiella pneumoniae clinical isolates. Infect. Immun. 68, 953–955. doi: 10.1128/IAI.68.2.953-955.2000
Amako, K., Meno, Y., and Takade, A. (1988). Fine structures of the capsules of Klebsiella pneumoniae and Escherichia coli K1. J. Bacteriol. 170, 4960–4962. doi: 10.1128/jb.170.10.4960-4962.1988
Anderson, M. S., Bulawa, C. E., and Raetz, C. R. (1985). The biosynthesis of gram-negative endotoxin. formation of lipid a precursors from udp-glcnac in extracts of Escherichia coli. J. Biol. Chem. 260, 15536–15541.
Anderson, M. S., and Raetz, C. R. (1987). Biosynthesis of lipid a precursors in Escherichia coli. A cytoplasmic acyltransferase that converts Udp-N-acetylglucosamine to Udp-3-O-(R-3-Hydroxymyristoyl)-N-acetylglucosamine. J. Biol. Chem. 262, 5159–5169.
Andrade, L. N., Vitali, L., Gaspar, G. G., Bellissimo-Rodrigues, F., Martinez, R., and Darini, A. L. (2014). Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), Qnrs1-, Ctx-M-2-, and Kpc-2-producing Klebsiella pneumoniae St11 international high-risk clone. J. Clin. Microbiol. 52, 2530–2535. doi: 10.1128/JCM.00088-14
Arai, N., Furuta, T., and Sakurai, M. (2017). Analysis of an atp-induced conformational transition of Abc transporter msba using a coarse-grained model. Biophys. Physicobiol. 14, 161–171. doi: 10.2142/biophysico.14.0_161
Ares, M. A., Sansabas, A., Rodriguez-Valverde, D., Siqueiros-Cendon, T., Rascon-Cruz, Q., Rosales-Reyes, R., et al. (2019). The interaction of Klebsiella pneumoniae with lipid rafts-associated cholesterol increases macrophage-mediated phagocytosis due to down regulation of the capsule polysaccharide. Front. Cell. Infect. Microbiol. 9:255. doi: 10.3389/fcimb.2019.00255
Arredondo-Garcia, J. L., Diaz-Ramos, R., Solorzano-Santos, F., Sosa-Gonzalez, I. E., and Beltran-Zuniga, M. (1992). Neonatal septicaemia due to K. Pneumoniae. Septicaemia due to Klebsiella pneumoniae in newborn infants. Nosocomial outbreak in an intensive care unit. Rev. Latinoam. Microbiol. 34, 11–16.
Bagley, S. T. (1985). Habitat association of Klebsiella species. Infect. Control 6, 52–58. doi: 10.1017/S0195941700062603
Balestrino, D., Ghigo, J. M., Charbonnel, N., Haagensen, J. A., and Forestier, C. (2008). The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 10, 685–701. doi: 10.1111/j.1462-2920.2007.01491.x
Bellich, B., Lagatolla, C., Tossi, A., Benincasa, M., Cescutti, P., and Rizzo, R. (2018). Influence of bacterial biofilm polysaccharide structure on interactions with antimicrobial peptides: a study on Klebsiella pneumoniae. Int. J. Mol. Sci. 19:E1685. doi: 10.3390/ijms19061685
Bengoechea, J. A., and Sa Pessoa, J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43, 123–144. doi: 10.1093/femsre/fuy043
Benincasa, M., Lagatolla, C., Dolzani, L., Milan, A., Pacor, S., Liut, G., et al. (2016). Biofilms from Klebsiella pneumoniae: matrix polysaccharide structure and interactions with antimicrobial peptides. Microorganisms 4:26. doi: 10.3390/microorganisms4030026
Bi, Y., Mann, E., Whitfield, C., and Zimmer, J. (2018). Architecture of a channel-forming o-antigen polysaccharide abc transporter. Nature 553, 361–365. doi: 10.1038/nature25190
Bilinski, J., Grzesiowski, P., Muszynski, J., Wroblewska, M., Madry, K., Robak, K., et al. (2016). Fecal microbiota transplantation inhibits multidrug-resistant gut pathogens: preliminary report performed in an immunocompromised host. Arch. Immunol. Ther. Exp. 64, 255–258. doi: 10.1007/s00005-016-0387-9
Bishop, R. E. (2019). Ratcheting Up lipopolysaccharide transport. Nature 567, 471–472. doi: 10.1038/d41586-019-00802-w
Botos, I., Majdalani, N., Mayclin, S. J., Mccarthy, J. G., Lundquist, K., Wojtowicz, D., et al. (2016). Structural and functional characterization of the lps transporter lptde from gram-negative pathogens. Structure 24, 965–976. doi: 10.1016/j.str.2016.03.026
Bowyer, A., Baardsnes, J., Ajamian, E., Zhang, L., and Cygler, M. (2011). Characterization of interactions between lps transport proteins of the Lpt system. Biochem. Biophys. Res. Commun. 404, 1093–1098. doi: 10.1016/j.bbrc.2010.12.121
Bradford, P. A., Urban, C., Mariano, N., Projan, S. J., Rahal, J. J., and Bush, K. (1997). Imipenem resistance in Klebsiella pneumoniae is associated with the combination of Act-1, a plasmid-mediated Ampc Beta-Lactamase, and the foss of an outer membrane protein. Antimicrob. Agents Chemother. 41, 563–569. doi: 10.1128/AAC.41.3.563
Branda, S. S., Vik, S., Friedman, L., and Kolter, R. (2005). Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26. doi: 10.1016/j.tim.2004.11.006
Bratu, S., Brooks, S., Burney, S., Kochar, S., Gupta, J., Landman, D., et al. (2007). Detection and spread of Escherichia coli possessing the plasmid-borne carbapenemase Kpc-2 in brooklyn, New York. Clin. Infect. Dis. 44, 972–975. doi: 10.1086/512370
Brisse, S., Fevre, C., Passet, V., Issenhuth-Jeanjean, S., Tournebize, R., Diancourt, L., et al. (2009). Virulent Clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS ONE 4:E4982. doi: 10.1371/journal.pone.0004982
Brisse, S., Passet, V., and Grimont, P. A. (2014). Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. 64, 3146–3152. doi: 10.1099/ijs.0.062737-0
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). Wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 51, 4073–4078. doi: 10.1128/JCM.01924-13
Bushell, S. R., Mainprize, I. L., Wear, M. A., Lou, H., Whitfield, C., and Naismith, J. H. (2013). Wzi is an outer membrane lectin that underpins group 1 capsule assembly in Escherichia coli. Structure 21, 844–853. doi: 10.1016/j.str.2013.03.010
Caffalette, C. A., Corey, R. A., Sansom, M. S. P., Stansfeld, P. J., and Zimmer, J. (2019). A lipid gating mechanism for the channel-forming O antigen Abc transporter. Nat. Commun. 10:824. doi: 10.1038/s41467-019-08646-8
Campbell, W. N., Hendrix, E., Cryz, S. Jr., and Cross, A. S. (1996). Immunogenicity Of A 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent pseudomonas O-polysaccharide conjugate vaccine administered to victims of acute trauma. Clin. Infect. Dis. 23, 179–181. doi: 10.1093/clinids/23.1.179
Cannatelli, A., D'andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in klebsiella pneumoniae producing Kpc-type carbapenemases mediated by insertional inactivation of the Phoq/Phop Mgrb regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Carter, J. S., Bowden, F. J., Bastian, I., Myers, G. M., Sriprakash, K. S., and Kemp, D. J. (1999). Phylogenetic evidence for reclassification of calymmatobacterium granulomatis as klebsiella granulomatis comb. Nov. Int. J. Syst. Bacteriol. 49 Pt 4, 1695–1700. doi: 10.1099/00207713-49-4-1695
Catalan-Najera, J. C., Garza-Ramos, U., and Barrios-Camacho, H. (2017). Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. Phenotypes? Virulence 8, 1111–1123. doi: 10.1080/21505594.2017.1317412
Centers For Disease Control Prevention (2010). Division Of Healthcare Quality Promotion (Dhqp). Klebsiella pneumoniae in healthcare settings [Online]. Centers For Disease Control and Prevention. Available online at: https://www.cdc.gov/hai/organisms/klebsiella/klebsiella.html [accessed August, 15 2019].
Cescutti, P., De Benedetto, G., and Rizzo, R. (2016). Structural determination of the polysaccharide isolated from biofilms produced by a clinical strain of Klebsiella pneumoniae. Carbohydr. Res. 430, 29–35. doi: 10.1016/j.carres.2016.05.001
Chen, L., Todd, R., Kiehlbauch, J., Walters, M., and Kallen, A. (2017). Notes from the field: pan-resistant new delhi metallo-beta-lactamase-producing Klebsiella pneumoniae- Washoe County, Nevada, (2016). MMWR Morb. Mortal. Wkly. Rep. 66:33. doi: 10.15585/mmwr.mm6601a7
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., and Peng, H. L. (2010). Rmpa regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae Cg43. J. Bacteriol. 192, 3144–3158. doi: 10.1128/JB.00031-10
Chowdhury, P., and Stein, D. S. (1992). Pyogenic hepatic abscess and septic pulmonary emboli associated with Klebsiella ozaenae bacteremia. South. Med. J. 85, 638–641. doi: 10.1097/00007611-199206000-00014
Ciurana, B., and Tomas, J. M. (1987). Role of lipopolysaccharide and complement in susceptibility of Klebsiella pneumoniae to nonimmune serum. Infect. Immun. 55, 2741–2746.
Clarke, B. R., Bronner, D., Keenleyside, W. J., Severn, W. B., Richards, J. C., and Whitfield, C. (1995). Role of Rfe and Rfbf in the initiation of biosynthesis of D-Galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae serotype O1. J. Bacteriol. 177, 5411–5418. doi: 10.1128/jb.177.19.5411-5418.1995
Clarke, B. R., Ovchinnikova, O. G., Kelly, S. D., Williamson, M. L., Butler, J. E., Liu, B., et al. (2018). Molecular basis for the structural diversity in serogroup O2-antigen polysaccharides in Klebsiella pneumoniae. J. Biol. Chem. 293, 4666–4679. doi: 10.1074/jbc.RA117.000646
Clarke, B. R., and Whitfield, C. (1992). Molecular cloning of the Rfb region of Klebsiella pneumoniae serotype O1:K20: the Rfb gene cluster is responsible for synthesis of the D-Galactan I O polysaccharide. J. Bacteriol. 174, 4614–4621. doi: 10.1128/jb.174.14.4614-4621.1992
Clements, A., Tull, D., Jenney, A. W., Farn, J. L., Kim, S. H., Bishop, R. E., et al. (2007). Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J. Biol. Chem. 282, 15569–15577. doi: 10.1074/jbc.M701454200
Cohen, T. S., Pelletier, M., Cheng, L., Pennini, M. E., Bonnell, J., Cvitkovic, R., et al. (2017). Anti-Lps antibodies protect against Klebsiella pneumoniae by empowering neutrophil-mediated clearance without neutralizing Tlr4. Jci Insight 2:e92774. doi: 10.1172/jci.insight.92774
Cortes, G., Alvarez, D., Saus, C., and Alberti, S. (2002a). Role of lung epithelial cells in defense against Klebsiella pneumoniae pneumonia. Infect. Immun. 70, 1075–1080. doi: 10.1128/IAI.70.3.1075-1080.2002
Cortes, G., Borrell, N., De Astorza, B., Gomez, C., Sauleda, J., and Alberti, S. (2002b). Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella Pneumoniae in a murine model of pneumonia. Infect. Immun. 70, 2583–2590. doi: 10.1128/IAI.70.5.2583-2590.2002
Cross, A. S. (2014). Anti-endotoxin vaccines: back to the future. Virulence 5, 219–225. doi: 10.4161/viru.25965
Cryz, S. J. Jr., Furer, E., and Germanier, R. (1985). Purification and vaccine potential of Klebsiella capsular polysaccharides. Infect. Immun. 50, 225–230.
Cryz, S. J. Jr., Furer, E., Sadoff, J. C., Fredeking, T., Que, J. U., and Cross, A. S. (1991). Production and characterization of a human hyperimmune intravenous immunoglobulin against pseudomonas aeruginosa and Klebsiella species. J. Infect. Dis. 163, 1055–1061. doi: 10.1093/infdis/163.5.1055
Cryz, S. J. Jr., Mortimer, P. M., Mansfield, V., and Germanier, R. (1986). Seroepidemiology of Klebsiella bacteremic isolates and implications for vaccine development. J. Clin. Microbiol. 23, 687–690.
Cryz, S. J. Jr., Que, J. O., Cross, A. S., and Furer, E. (1995). Synthesis and characterization of a polyvalent Escherichia coli O-polysaccharide-toxin a conjugate vaccine. Vaccine 13, 449–453. doi: 10.1016/0264-410X(94)00009-C
Cuthbertson, L., Kimber, M. S., and Whitfield, C. (2007). Substrate binding by a bacterial abc transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. U.S.A. 104, 19529–19534. doi: 10.1073/pnas.0705709104
Cuthbertson, L., Kos, V., and Whitfield, C. (2010). Abc transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74, 341–362. doi: 10.1128/MMBR.00009-10
Cuthbertson, L., Powers, J., and Whitfield, C. (2005). The C-terminal domain of the nucleotide-binding domain protein Wzt determines substrate specificity in the Atp-binding cassette transporter for the lipopolysaccharide O-antigens in Escherichia coli serotypes O8 and O9a. J. Biol. Chem. 280, 30310–30319. doi: 10.1074/jbc.M504371200
D'andrea, M. M., Arena, F., Pallecchi, L., and Rossolini, G. M. (2013). Ctx-M-type beta-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303, 305–317. doi: 10.1016/j.ijmm.2013.02.008
Davidson, A. L., Dassa, E., Orelle, C., and Chen, J. (2008). Structure, function, and evolution of bacterial Atp-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–64. doi: 10.1128/MMBR.00031-07
Davies, D. (2003). Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122. doi: 10.1038/nrd1008
De Astorza, B., Cortes, G., Crespi, C., Saus, C., Rojo, J. M., and Alberti, S. (2004). C3 promotes clearance of Klebsiella pneumoniae by A549 epithelial cells. Infect. Immun. 72, 1767–1774. doi: 10.1128/IAI.72.3.1767-1774.2004
Diago-Navarro, E., Motley, M. P., Ruiz-Perez, G., Yu, W., Austin, J., Seco, B. M. S., et al. (2018). Novel, broadly reactive anticapsular antibodies against carbapenem-resistant Klebsiella pneumoniae protect from infection. Mbio 9:e00091–18. doi: 10.1128/mBio.00091-18
Doerrler, W. T., Gibbons, H. S., and Raetz, C. R. (2004). Msba-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 279, 45102–45109. doi: 10.1074/jbc.M408106200
Domenico, P., Salo, R. J., Cross, A. S., and Cunha, B. A. (1994). Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 62, 4495–4499.
Dong, C., Beis, K., Nesper, J., Brunkan-Lamontagne, A. L., Clarke, B. R., Whitfield, C., et al. (2006). Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229. doi: 10.1038/nature05267
Dong, H., Zhang, Z., Tang, X., Paterson, N. G., and Dong, C. (2017). Structural and functional insights into the lipopolysaccharide Abc transporter Lptb2fg. Nat. Commun. 8:222. doi: 10.1038/s41467-017-00273-5
Donta, S. T., Peduzzi, P., Cross, A. S., Sadoff, J., Haakenson, C., Cryz, S. J. Jr., et al. (1996). Immunoprophylaxis against Klebsiella and Pseudomonas aeruginosa infections. The federal hyperimmune immunoglobulin trial study group. J. Infect. Dis. 174, 537–543. doi: 10.1093/infdis/174.3.537
Doorduijn, D. J., Rooijakkers, S. H., Van Schaik, W., and Bardoel, B. W. (2016). Complement resistance mechanisms of Klebsiella pneumoniae. Immunobiology 221, 1102–1109. doi: 10.1016/j.imbio.2016.06.014
Edelman, R., Taylor, D. N., Wasserman, S. S., Mcclain, J. B., Cross, A. S., Sadoff, J. C., et al. (1994). Phase 1 trial of A 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent pseudomonas o-polysaccharide conjugate vaccine administered simultaneously. Vaccine 12, 1288–1294. doi: 10.1016/S0264-410X(94)80054-4
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., and Wang, J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Fang, C. T., Lai, S. Y., Yi, W. C., Hsueh, P. R., Liu, K. L., and Chang, S. C. (2007). Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45, 284–293. doi: 10.1086/519262
Fazili, T., Sharngoe, C., Endy, T., Kiska, D., Javaid, W., and Polhemus, M. (2016). Klebsiella pneumoniae liver abscess: an emerging disease. Am. J. Med. Sci. 351, 297–304. doi: 10.1016/j.amjms.2015.12.018
Feng, Y., Lu, Y., Yao, Z., and Zong, Z. (2018). Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob. Agents Chemother. 62:e02644–17. doi: 10.1128/AAC.02644-17
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom. 2:E000073. doi: 10.1099/mgen.0.000073
Ford, R. C., and Beis, K. (2019). Learning the abcs one at a time: structure and mechanism of Abc transporters. Biochem. Soc. Trans. 47, 23–36. doi: 10.1042/BST20180147
Freinkman, E., Chng, S. S., and Kahne, D. (2011). The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 108, 2486–2491. doi: 10.1073/pnas.1015617108
Freinkman, E., Okuda, S., Ruiz, N., and Kahne, D. (2012). Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51, 4800–4806. doi: 10.1021/bi300592c
Fresno, S., Jimenez, N., Canals, R., Merino, S., Corsaro, M. M., Lanzetta, R., et al. (2007). A second galacturonic acid transferase is required for core lipopolysaccharide biosynthesis and complete capsule association with the cell surface in Klebsiella pneumoniae. J. Bacteriol. 189, 1128–1137. doi: 10.1128/JB.01489-06
Fu, L., Tang, L., Wang, S., Liu, Q., Liu, Y., Zhang, Z., et al. (2018). Co-location of the Blakpc-2, Blactx-M-65, Rmtb and virulence relevant factors in an incfii plasmid from a hypermucoviscous Klebsiella pneumoniae isolate. Microb. Pathog. 124, 301–304. doi: 10.1016/j.micpath.2018.08.055
Fung, C. P., Chang, F. Y., Lee, S. C., Hu, B. S., Kuo, B. I., Liu, C. Y., et al. (2002). A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50, 420–424. doi: 10.1136/gut.50.3.420
Goldstein, E. J., Lewis, R. P., Martin, W. J., and Edelstein, P. H. (1978). Infections caused by Klebsiella ozaenae: a changing disease spectrum. J. Clin. Microbiol. 8, 413–418.
Gomez-Simmonds, A., and Uhlemann, A. C. (2017). Clinical implications of genomic adaptation and evolution of carbapenem-resistant Klebsiella pneumoniae. J. Infect. Dis. 215, S18–S27. doi: 10.1093/infdis/jiw378
Grabowicz, M., Yeh, J., and Silhavy, T. J. (2013). Dominant negative lpte mutation that supports a role for Lpte as a plug in the Lptd barrel. J. Bacteriol. 195, 1327–1334. doi: 10.1128/JB.02142-12
Greenfield, L. K., and Whitfield, C. (2012). Synthesis of lipopolysaccharide O-antigens by Abc transporter-dependent pathways. Carbohydr. Res. 356, 12–24. doi: 10.1016/j.carres.2012.02.027
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of St11 carbapenem-resistant hypervirulent klebsiella pneumoniae in a chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Gu, Y., Stansfeld, P. J., Zeng, Y., Dong, H., Wang, W., and Dong, C. (2015). Lipopolysaccharide is inserted into the outer membrane through an intramembrane hole, a lumen gate, and the lateral opening of Lptd. Structure 23, 496–504. doi: 10.1016/j.str.2015.01.001
Guan, S., Clarke, A. J., and Whitfield, C. (2001). Functional analysis of the galactosyltransferases required for biosynthesis of D-Galactan I, A component of the lipopolysaccharide O1 antigen of Klebsiella pneumoniae. J. Bacteriol. 183, 3318–3327. doi: 10.1128/JB.183.11.3318-3327.2001
Hagar, J. A., Powell, D. A., Aachoui, Y., Ernst, R. K., and Miao, E. A. (2013). Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253.
Hansen, D. S., Mestre, F., Alberti, S., Hernandez-Alles, S., Alvarez, D., Domenech-Sanchez, A., et al. (1999). Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J. Clin. Microbiol. 37, 56–62.
Hawkey, P. M., and Jones, A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64 (Suppl 1), I3–10. doi: 10.1093/jac/dkp256
Hegerle, N., Choi, M., Sinclair, J., Amin, M. N., Ollivault-Shiflett, M., Curtis, B., et al. (2018). Development of a broad spectrum glycoconjugate vaccine to prevent wound and disseminated infections with Klebsiella pneumoniae and Pseudomonas aeruginosa. PLoS ONE 13:E0203143. doi: 10.1371/journal.pone.0203143
Ho, H., Miu, A., Alexander, M. K., Garcia, N. K., Oh, A., Zilberleyb, I., et al. (2018). Structural basis for dual-mode inhibition of the Abc transporter Msba. Nature 557, 196–201. doi: 10.1038/s41586-018-0083-5
Holers, V. M. (2014). Complement and its receptors: new insights into human disease. Annu. Rev. Immunol. 32, 433–459. doi: 10.1146/annurev-immunol-032713-120154
Hossain, A., Ferraro, M. J., Pino, R. M., Dew, R. B. III., Moland, E. S., Lockhart, T. J., et al. (2004). Plasmid-mediated carbapenem-hydrolyzing enzyme Kpc-2 in an Enterobacter Sp. Antimicrob. Agents Chemother. 48, 4438–4440. doi: 10.1128/AAC.48.11.4438-4440.2004
Hsieh, P. F., Lu, Y. R., Lin, T. L., Lai, L. Y., and Wang, J. T. (2019). Klebsiella pneumoniae type Vi secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 219, 637–647. doi: 10.1093/infdis/jiy534
Hsu, C. R., Liao, C. H., Lin, T. L., Yang, H. R., Yang, F. L., Hsieh, P. F., et al. (2016). Identification of a capsular variant and characterization of capsular acetylation in Klebsiella pneumoniae Pla-associated type K57. Sci. Rep. 6:31946. doi: 10.1038/srep31946
Hutter, J., and Lepenies, B. (2015). Carbohydrate-based vaccines: an overview. Methods Mol. Biol. 1331, 1–10. doi: 10.1007/978-1-4939-2874-3_1
Ivin, M., Dumigan, A., De Vasconcelos, F. N., Ebner, F., Borroni, M., Kavirayani, A., et al. (2017). Natural killer cell-intrinsic type i Ifn signaling controls Klebsiella pneumoniae growth during lung infection. PLoS Pathog. 13:E1006696. doi: 10.1371/journal.ppat.1006696
Jansson, P. E., Lindberg, B., Manca, M. C., Nimmich, W., and Widmalm, G. (1994). Structural studies of the capsular polysaccharide from Klebsiella type 38: a reinvestigation. Carbohydr. Res. 261, 111–118. doi: 10.1016/0008-6215(94)80010-3
Jayol, A., Poirel, L., Brink, A., Villegas, M. V., Yilmaz, M., and Nordmann, P. (2014). Resistance to colistin associated with a single amino acid change in protein pmrb among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58, 4762–4766. doi: 10.1128/AAC.00084-14
Juan, C. H., Chuang, C., Chen, C. H., Li, L., and Lin, Y. T. (2019). Clinical characteristics, antimicrobial resistance and capsular types of community-acquired, healthcare-associated, and nosocomial Klebsiella pneumoniae Bacteremia. Antimicrob. Resist. Infect. Control 8:1. doi: 10.1186/s13756-018-0426-x
Kabha, K., Nissimov, L., Athamna, A., Keisari, Y., Parolis, H., Parolis, L. A., et al. (1995). Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect. Immun. 63, 847–852.
Kabha, K., Schmegner, J., Keisari, Y., Parolis, H., Schlepper-Schaeffer, J., and Ofek, I. (1997). Sp-A enhances phagocytosis of Klebsiella by interaction with capsular polysaccharides and alveolar macrophages. Am. J. Physiol. 272, L344–L352. doi: 10.1152/ajplung.1997.272.2.L344
Kalynych, S., Morona, R., and Cygler, M. (2014). Progress in understanding the assembly process of bacterial O-antigen. FEMS Microbiol. Rev. 38, 1048–1065. doi: 10.1111/1574-6976.12070
Kawai, T., and Akira, S. (2010). The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11, 373–384. doi: 10.1038/ni.1863
Khaertynov, K. S., Anokhin, V. A., Rizvanov, A. A., Davidyuk, Y. N., Semyenova, D. R., Lubin, S. A., et al. (2018). Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front. Med. 5:225. doi: 10.3389/fmed.2018.00225
Khan, A. U., Maryam, L., and Zarrilli, R. (2017). Structure, genetics and worldwide spread of New Delhi Metallo-Beta-Lactamase (Ndm): a threat to public health. BMC Microbiol. 17:101. doi: 10.1186/s12866-017-1012-8
Kidd, T. J., Mills, G., Sa-Pessoa, J., Dumigan, A., Frank, C. G., Insua, J. L., et al. (2017). A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol. Med. 9, 430–447. doi: 10.15252/emmm.201607336
Kobayashi, S. D., Porter, A. R., Freedman, B., Pandey, R., Chen, L., Kreiswirth, B. N., et al. (2018). Antibody-mediated killing of carbapenem-resistant St258 Klebsiella pneumoniae by human neutrophils. Mbio 9:e00297–18. doi: 10.1128/mBio.00297-18
Kong, L., Harrington, L., Li, Q., Cheley, S., Davis, B. G., and Bayley, H. (2013). Single-molecule interrogation of a bacterial sugar transporter allows the discovery of an extracellular inhibitor. Nat. Chem. 5, 651–659. doi: 10.1038/nchem.1695
Kos, V., Cuthbertson, L., and Whitfield, C. (2009). The Klebsiella Pneumoniae O2a antigen defines a second mechanism for o antigen Atp-binding cassette transporters. J. Biol. Chem. 284, 2947–2956. doi: 10.1074/jbc.M807213200
Ku, Y. H., Chuang, Y. C., Chen, C. C., Lee, M. F., Yang, Y. C., Tang, H. J., et al. (2017). Klebsiella pneumoniae isolates from meningitis: epidemiology, virulence and antibiotic resistance. Sci. Rep. 7:6634. doi: 10.1038/s41598-017-06878-6
Kubler-Kielb, J., Vinogradov, E., Ng, W. I., Maczynska, B., Junka, A., Bartoszewicz, M., et al. (2013). The capsular polysaccharide and lipopolysaccharide structures of two carbapenem resistant Klebsiella pneumoniae outbreak isolates. Carbohydr. Res. 369, 6–9. doi: 10.1016/j.carres.2012.12.018
Kumar, A. S., Mody, K., and Jha, B. (2007). Bacterial exopolysaccharides–a perception. J. Basic Microbiol. 47, 103–117. doi: 10.1002/jobm.200610203
Kunduru, B. R., Nair, S. A., and Rathinavelan, T. (2016). Ek3d: an E. coli K antigen 3-dimensional structure database. Nucleic Acids Res. 44, D675–D681. doi: 10.1093/nar/gkv1313
Lavender, H. F., Jagnow, J. R., and Clegg, S. (2004). Biofilm formation in vitro and virulence in vivo of mutants of Klebsiella pneumoniae. Infect. Immun. 72, 4888–4890. doi: 10.1128/IAI.72.8.4888-4890.2004
Lawlor, M. S., Handley, S. A., and Miller, V. L. (2006). Comparison of the host responses to wild-type and cpsb mutant Klebsiella pneumoniae infections. Infect. Immun. 74, 5402–5407. doi: 10.1128/IAI.00244-06
Lawlor, M. S., Hsu, J., Rick, P. D., and Miller, V. L. (2005). Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol. Microbiol. 58, 1054–1073. doi: 10.1111/j.1365-2958.2005.04918.x
Leavitt, A., Chmelnitsky, I., Colodner, R., Ofek, I., Carmeli, Y., and Navon-Venezia, S. (2009). Ertapenem resistance among extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae isolates. J. Clin. Microbiol. 47, 969–974. doi: 10.1128/JCM.00651-08
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Lee, K. H., Hui, K. P., Tan, W. C., and Lim, T. K. (1996). Severe community-acquired pneumonia in Singapore. Singapore Med. J. 37, 374–377.
Li, L., Woodward, R. L., Han, W., Qu, J., Song, J., Ma, C., et al. (2016). Chemoenzymatic synthesis of the bacterial polysaccharide repeating unit undecaprenyl pyrophosphate and its analogs. Nat. Protoc. 11, 1280–1298. doi: 10.1038/nprot.2016.067
Li, Y., Orlando, B. J., and Liao, M. (2019). Structural basis of lipopolysaccharide extraction by the Lptb2fgc complex. Nature 567, 486–490. doi: 10.1038/s41586-019-1025-6
Lin, J. C., Chang, F. Y., Fung, C. P., Xu, J. Z., Cheng, H. P., Wang, J. J., et al. (2004). High prevalence of phagocytic-resistant capsular serotypes of Klebsiella pneumoniae in liver abscess. Microbes Infect. 6, 1191–1198. doi: 10.1016/j.micinf.2004.06.003
Lin, R. D., Hsueh, P. R., Chang, S. C., Chen, Y. C., Hsieh, W. C., and Luh, K. T. (1997). Bacteremia due to Klebsiella oxytoca: clinical features of patients and antimicrobial susceptibilities of the isolates. Clin. Infect. Dis 24, 1217–1222. doi: 10.1086/513637
Lin, W. H., Wang, M. C., Tseng, C. C., Ko, W. C., Wu, A. B., Zheng, P. X., et al. (2010). Clinical and microbiological characteristics of Klebsiella pneumoniae isolates causing community-acquired urinary tract infections. Infection 38, 459–464. doi: 10.1007/s15010-010-0049-5
Liston, S. D., Mann, E., and Whitfield, C. (2017). Glycolipid substrates for Abc transporters required for the assembly of bacterial cell-envelope and cell-surface glycoconjugates. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1394–1403. doi: 10.1016/j.bbalip.2016.10.008
Liu, L., Feng, Y., Hu, Y., Kang, M., Xie, Y., and Zong, Z. (2018). Klebsiella grimontii, a new species acquired carbapenem resistance. Front. Microbiol. 9:2170. doi: 10.3389/fmicb.2018.02170
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism Mcr-1 in animals and human beings in china: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Llobet, E., Campos, M. A., Gimenez, P., Moranta, D., and Bengoechea, J. A. (2011). Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun. 79, 3718–3732. doi: 10.1128/IAI.05226-11
Llobet, E., Martinez-Moliner, V., Moranta, D., Dahlstrom, K. M., Regueiro, V., Tomas, A., et al. (2015). Deciphering tissue-induced Klebsiella pneumoniae lipid a structure. Proc. Natl. Acad. Sci. U.S.A. 112, E6369–E6378. doi: 10.1073/pnas.1508820112
Locher, K. P. (2016). Mechanistic diversity in Atp-binding cassette (Abc) transporters. Nat. Struct. Mol. Biol. 23, 487–493. doi: 10.1038/nsmb.3216
López-Santin, J. M. S. C. G. S. (1995). Production and purification of rhamnose from microbial polysaccharide produced by Klebsiella sp. I-714. Bioproc. Eng. 12, 287–291. doi: 10.1007/BF00369505
Lugo, J. Z., Price, S., Miller, J. E., Ben-David, I., Merrill, V., Mancuso, P., et al. (2007). Lipopolysaccharide O-antigen promotes persistent murine bacteremia. Shock 27, 186–191. doi: 10.1097/01.shk.0000238058.23837.21
Maeshima, N., and Fernandez, R. C. (2013). Recognition of lipid a variants by the Tlr4-Md-2 receptor complex. Front. Cell. Infect. Microbiol. 3:3. doi: 10.3389/fcimb.2013.00003
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Martin, R. M., and Bachman, M. A. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8:4. doi: 10.3389/fcimb.2018.00004
Martinez-Martinez, L. (2008). Extended-spectrum beta-lactamases and the permeability barrier. Clin. Microbiol. Infect. 14(Suppl 1), 82–89. doi: 10.1111/j.1469-0691.2007.01860.x
Mccallum, K. L., Schoenhals, G., Laakso, D., Clarke, B., and Whitfield, C. (1989). A high-molecular-weight fraction of smooth lipopolysaccharide in klebsiella serotype O1:K20 contains a unique o-antigen epitope and determines resistance to nonspecific serum killing. Infect. Immun. 57, 3816–3822.
Meier-Dieter, U., Barr, K., Starman, R., Hatch, L., and Rick, P. D. (1992). Nucleotide sequence of the Escherichia coli Rfe gene involved in the synthesis of enterobacterial common antigen. molecular cloning of the Rfe-Rff gene cluster. J. Biol. Chem. 267, 746–753.
Meri, S., and Pangburn, M. K. (1994). Regulation of alternative pathway complement activation by glycosaminoglycans: specificity of the polyanion binding site on factor H. Biochem. Biophys. Res. Commun. 198, 52–59. doi: 10.1006/bbrc.1994.1008
Merino, S., Camprubi, S., Alberti, S., Benedi, V. J., and Tomas, J. M. (1992). Mechanisms Of Klebsiella pneumoniae resistance to complement-mediated killing. Infect. Immun. 60, 2529–2535.
Merle, N. S., Noe, R., Halbwachs-Mecarelli, L., Fremeaux-Bacchi, V., and Roumenina, L. T. (2015). Complement system part Ii: role in immunity. Front. Immunol. 6:257. doi: 10.3389/fimmu.2015.00257
Mi, W., Li, Y., Yoon, S. H., Ernst, R. K., Walz, T., and Liao, M. (2017). Structural basis of Msba-mediated lipopolysaccharide transport. Nature 549, 233–237. doi: 10.1038/nature23649
Micoli, F., Costantino, P., and Adamo, R. (2018). Potential targets for next generation antimicrobial glycoconjugate vaccines. FEMS Microbiol. Rev. 42, 388–423. doi: 10.1093/femsre/fuy011
Mills, G., Dumigan, A., Kidd, T., Hobley, L., and Bengoechea, J. A. (2017). Identification and characterization of two Klebsiella pneumoniae Lpxl lipid a late acyltransferases and their role in virulence. Infect. Immun. 85:e00068–17. doi: 10.1128/IAI.00068-17
Miriagou, V., Tzouvelekis, L. S., Rossiter, S., Tzelepi, E., Angulo, F. J., and Whichard, J. M. (2003). Imipenem resistance in a salmonella clinical strain due to plasmid-mediated class a carbapenemase Kpc-2. Antimicrob. Agents Chemother. 47, 1297–1300. doi: 10.1128/AAC.47.4.1297-1300.2003
Mishra, S. K., Basukala, P., Basukala, O., Parajuli, K., Pokhrel, B. M., and Rijal, B. P. (2015). Detection of biofilm production and antibiotic resistance pattern in clinical isolates from indwelling medical devices. Curr. Microbiol. 70, 128–134. doi: 10.1007/s00284-014-0694-5
Murphy, K., Travers, P., Walport, M., and Janeway, C. (2012). Janeway's Immunobiology. New York, NY: Garland Science.
Narita, S., and Tokuda, H. (2009). Biochemical characterization of an Abc transporter lptbfgc complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583, 2160–2164. doi: 10.1016/j.febslet.2009.05.051
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Okuda, S., Freinkman, E., and Kahne, D. (2012). Cytoplasmic Atp hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217. doi: 10.1126/science.1228984
Okuda, S., Sherman, D. J., Silhavy, T. J., Ruiz, N., and Kahne, D. (2016). Lipopolysaccharide transport and assembly at the outer membrane: the Pez model. Nat. Rev. Microbiol. 14, 337–345. doi: 10.1038/nrmicro.2016.25
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Owens, T. W., Taylor, R. J., Pahil, K. S., Bertani, B. R., Ruiz, N., Kruse, A. C., et al. (2019). Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553. doi: 10.1038/s41586-019-1039-0
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pan, Y. J., Lin, T. L., Chen, C. T., Chen, Y. Y., Hsieh, P. F., Hsu, C. R., et al. (2015). Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella Spp. Sci. Rep. 5:15573. doi: 10.1038/srep15573
Pan, Y. J., Lin, T. L., Chen, Y. H., Hsu, C. R., Hsieh, P. F., Wu, M. C., et al. (2013). Capsular types of Klebsiella pneumoniae revisited by Wzc sequencing. PLoS ONE 8:E80670. doi: 10.1371/journal.pone.0080670
Pan, Y. J., Lin, T. L., Chen, Y. Y., Lai, P. H., Tsai, Y. T., Hsu, C. R., et al. (2019). Identification of three podoviruses infecting Klebsiella encoding capsule depolymerases that digest specific capsular types. Microb. Biotechnol. 12, 472–486. doi: 10.1111/1751-7915.13370
Pan, Y. J., Lin, T. L., Hsu, C. R., and Wang, J. T. (2011). Use of a dictyostelium model for isolation of genetic loci associated with phagocytosis and virulence in Klebsiella pneumoniae. Infect. Immun. 79, 997–1006. doi: 10.1128/IAI.00906-10
Passet, V., and Brisse, S. (2018). Description of Klebsiella Grimontii Sp. Nov. Int. J. Syst. Evol. Microbiol. 68, 377–381. doi: 10.1099/ijsem.0.002517
Pendleton, J. N., Gorman, S. P., and Gilmore, B. F. (2013). Clinical relevance of the eskape pathogens. Exp. Rev. Anti Infect. Ther. 11, 297–308. doi: 10.1586/eri.13.12
Pennini, M. E., De Marco, A., Pelletier, M., Bonnell, J., Cvitkovic, R., Beltramello, M., et al. (2017). Immune stealth-driven O2 serotype prevalence and potential for therapeutic antibodies against multidrug resistant Klebsiella pneumoniae. Nat. Commun. 8:1991. doi: 10.1038/s41467-017-02223-7
Podschun, R., Pietsch, S., Holler, C., and Ullmann, U. (2001). Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 67, 3325–3327. doi: 10.1128/AEM.67.7.3325-3327.2001
Podschun, R., and Ullmann, U. (1998). Klebsiella Spp. As nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Poirel, L., Jayol, A., Bontron, S., Villegas, M. V., Ozdamar, M., Turkoglu, S., et al. (2015). The Mgrb gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Pomakova, D. K., Hsiao, C. B., Beanan, J. M., Olson, R., Macdonald, U., Keynan, Y., et al. (2012). Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 31, 981–989. doi: 10.1007/s10096-011-1396-6
Price, D. J., and Sleigh, J. D. (1972). Klebsiella meningitis–report of nine cases. J. Neurol. Neurosurg. Psychiatr. 35, 903–908. doi: 10.1136/jnnp.35.6.903
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C., and Huang, Y. (2014). Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111. doi: 10.1038/nature13484
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–58. doi: 10.1128/CMR.00001-07
Raetz, C. R., Guan, Z., Ingram, B. O., Six, D. A., Song, F., Wang, X., et al. (2009). Discovery of new biosynthetic pathways: the lipid a story. J. Lipid Res. 50 (Suppl), S103–S108. doi: 10.1194/jlr.R800060-JLR200
Raetz, C. R., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1146/annurev.biochem.71.110601.135414
Rahn, A., Beis, K., Naismith, J. H., and Whitfield, C. (2003). A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J. Bacteriol. 185, 5882–5890. doi: 10.1128/JB.185.19.5882-5890.2003
Rahn, A., Drummelsmith, J., and Whitfield, C. (1999). Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 181, 2307–2313.
Ramos, P. I. P., Fernandez Do Porto, D., Lanzarotti, E., Sosa, E. J., Burguener, G., Pardo, A. M., et al. (2018). An integrative, multi-omics approach towards the prioritization of klebsiella pneumoniae drug Targets. Sci. Rep. 8:10755. doi: 10.1038/s41598-018-28916-7
Regue, M., Izquierdo, L., Fresno, S., Pique, N., Corsaro, M. M., Naldi, T., et al. (2005). A second outer-core region in Klebsiella pneumoniae lipopolysaccharide. J. Bacteriol. 187, 4198–4206. doi: 10.1128/JB.187.12.4198-4206.2005
Regueiro, V., Campos, M. A., Pons, J., Alberti, S., and Bengoechea, J. A. (2006). The uptake of a Klebsiella pneumoniae capsule polysaccharide mutant triggers an inflammatory response by human airway epithelial cells. Microbiology 152, 555–566. doi: 10.1099/mic.0.28285-0
Regueiro, V., Moranta, D., Campos, M. A., Margareto, J., Garmendia, J., and Bengoechea, J. A. (2009). Klebsiella pneumoniae increases the levels of toll-like receptors 2 And 4 in human airway epithelial cells. Infect. Immun. 77, 714–724. doi: 10.1128/IAI.00852-08
Reid, A. N., and Whitfield, C. (2005). Functional analysis of conserved gene products involved in assembly of Escherichia coli capsules and exopolysaccharides: evidence for molecular recognition between Wza and Wzc for colanic acid biosynthesis. J. Bacteriol. 187, 5470–5481. doi: 10.1128/JB.187.15.5470-5481.2005
Rick, P. D., Hubbard, G. L., and Barr, K. (1994). Role of the Rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J. Bacteriol. 176, 2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994
Ricklin, D., Hajishengallis, G., Yang, K., and Lambris, J. D. (2010). Complement: a key system for immune surveillance and homeostasis. Nat. Immunol. 11, 785–797. doi: 10.1038/ni.1923
Robinson, J. A. (2019). Folded synthetic peptides and other molecules targeting outer membrane protein complexes in gram-negative bacteria. Front. Chem. 7:45. doi: 10.3389/fchem.2019.00045
Roca, C., Alves, V. D., Freitas, F., and Reis, M. A. (2015). Exopolysaccharides enriched in rare sugars: bacterial sources, production, and applications. Front. Microbiol. 6:288. doi: 10.3389/fmicb.2015.00288
Rock, C., Thom, K. A., Masnick, M., Johnson, J. K., Harris, A. D., and Morgan, D. J. (2014). Frequency of Klebsiella pneumoniae carbapenemase (Kpc)-producing and non-Kpc-producing Klebsiella species contamination of healthcare workers and the environment. Infect. Control Hosp. Epidemiol. 35, 426–429. doi: 10.1086/675598
Rodrigues, C., Passet, V., Rakotondrasoa, A., Diallo, T. A., Criscuolo, A., and Brisse, S. (2019). Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. 170, 165–170. doi: 10.1101/516278
Rosenblueth, M., Martinez, L., Silva, J., and Martinez-Romero, E. (2004). Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst. Appl. Microbiol. 27, 27–35. doi: 10.1078/0723-2020-00261
Rossi, B., Gasperini, M. L., Leflon-Guibout, V., Gioanni, A., De Lastours, V., Rossi, G., et al. (2018). Hypervirulent Klebsiella pneumoniae in cryptogenic liver abscesses, Paris, France. Emerg. Infect. Dis. 24, 221–229. doi: 10.3201/eid2402.170957
Ruiz, N., Gronenberg, L. S., Kahne, D., and Silhavy, T. J. (2008). Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 5537–5542. doi: 10.1073/pnas.0801196105
Ruiz, N., Kahne, D., and Silhavy, T. J. (2009). Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 7, 677–683. doi: 10.1038/nrmicro2184
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32:e00001–19. doi: 10.1128/CMR.00001-19
Russo, T. A., Olson, R., Fang, C. T., Stoesser, N., Miller, M., Macdonald, U., et al. (2018). Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. Pneumoniae. J. Clin. Microbiol. 56:e00776–18. doi: 10.1128/JCM.00776-18
Sachdeva, S., Kolimi, N., Nair, S. A., and Rathinavelan, T. (2016). Key diffusion mechanisms involved in regulating bidirectional water permeation across E. coli outer membrane lectin. Sci. Rep. 6:28157. doi: 10.1038/srep28157
Sachdeva, S., Palur, R. V., Sudhakar, K. U., and Rathinavelan, T. (2017). E. coli group 1 capsular polysaccharide exportation nanomachinary as a plausible antivirulence target in the perspective of emerging antimicrobial resistance. Front. Microbiol. 8:70. doi: 10.3389/fmicb.2017.00070
Sahly, H., Keisari, Y., and Ofek, I. (2009). Manno(Rhamno)Biose-containing capsular polysaccharides of Klebsiella pneumoniae enhance opsono-stimulation of human polymorphonuclear leukocytes. J. Innate Immun. 1, 136–144. doi: 10.1159/000154812
Salo, R. J., Domenico, P., Tomas, J. M., Straus, D. C., Merino, S., Benedi, V. J., et al. (1995). Salicylate-enhanced exposure of Klebsiella pneumoniae subcapsular components. Infection 23, 371–377. doi: 10.1007/BF01713568
Schmid, J. (2018). Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies. Curr. Opin. Biotechnol. 53, 130–136. doi: 10.1016/j.copbio.2018.01.005
Schmid, J., Sieber, V., and Rehm, B. (2015). Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol. 6:496. doi: 10.3389/fmicb.2015.00496
Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003). Swiss-model: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385. doi: 10.1093/nar/gkg520
Seeberger, P. H., Pereira, C. L., Khan, N., Xiao, G., Diago-Navarro, E., Reppe, K., et al. (2017). A semi-synthetic glycoconjugate vaccine candidate for carbapenem-resistant Klebsiella pneumoniae. Angew. Chem. Int. Ed. Engl. 56, 13973–13978. doi: 10.1002/anie.201700964
Selden, R., Lee, S., Wang, W. L., Bennett, J. V., and Eickhoff, T. C. (1971). Nosocomial Klebsiella infections: intestinal colonization as a reservoir. Ann. Intern. Med. 74, 657–664. doi: 10.7326/0003-4819-74-5-657
Sewify, M., Nair, S., Warsame, S., Murad, M., Alhubail, A., Behbehani, K., et al. (2016). Prevalence of urinary tract infection and antimicrobial susceptibility among diabetic patients with controlled and uncontrolled glycemia in Kuwait. J. Diabetes Res. 2016:6573215. doi: 10.1155/2016/6573215
Shankar-Sinha, S., Valencia, G. A., Janes, B. K., Rosenberg, J. K., Whitfield, C., Bender, R. A., et al. (2004). The Klebsiella pneumoniae O antigen contributes to bacteremia and lethality during murine Pneumonia. Infect. Immun. 72, 1423–1430. doi: 10.1128/IAI.72.3.1423-1430.2004
Sherman, D. J., Lazarus, M. B., Murphy, L., Liu, C., Walker, S., Ruiz, N., et al. (2014). Decoupling catalytic activity from biological function of the Atpase that powers lipopolysaccharide transport. Proc. Natl. Acad. Sci. U.S.A. 111, 4982–4987. doi: 10.1073/pnas.1323516111
Shi, J., Zhao, Y., Wang, Y., Gao, W., Ding, J., Li, P., et al. (2014). Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192.
Shon, A. S., Bajwa, R. P., and Russo, T. A. (2013). Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Shu, H. Y., Fung, C. P., Liu, Y. M., Wu, K. M., Chen, Y. T., Li, L. H., et al. (2009). Genetic diversity of capsular polysaccharide biosynthesis in Klebsiella pneumoniae clinical isolates. Microbiology 155, 4170–4183. doi: 10.1099/mic.0.029017-0
Sidjabat, H. E., Silveira, F. P., Potoski, B. A., Abu-Elmagd, K. M., Adams-Haduch, J. M., Paterson, D. L., et al. (2009). Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin. Infect. Dis. 49, 1736–1738. doi: 10.1086/648077
Smith Moland, E., Hanson, N. D., Herrera, V. L., Black, J. A., Lockhart, T. J., Hossain, A., et al. (2003). Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, Kpc-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51, 711–714. doi: 10.1093/jac/dkg124
Sperandeo, P., Cescutti, R., Villa, R., Di Benedetto, C., Candia, D., Deho, G., et al. (2007). Characterization of Lpta and Lptb, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 189, 244–253. doi: 10.1128/JB.01126-06
Sperandeo, P., Lau, F. K., Carpentieri, A., De Castro, C., Molinaro, A., Deho, G., et al. (2008). Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190, 4460–4469. doi: 10.1128/JB.00270-08
Sperandeo, P., Villa, R., Martorana, A. M., Samalikova, M., Grandori, R., Deho, G., et al. (2011). New insights into the Lpt machinery for lipopolysaccharide transport to the cell surface: Lpta-Lptc interaction and Lpta stability as sensors of a properly assembled transenvelope complex. J. Bacteriol. 193, 1042–1053. doi: 10.1128/JB.01037-10
Srinivas, N., Jetter, P., Ueberbacher, B. J., Werneburg, M., Zerbe, K., Steinmann, J., et al. (2010). Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas Aeruginosa. Science 327, 1010–1013. doi: 10.1126/science.1182749
Storek, K. M., Chan, J., Vij, R., Chiang, N., Lin, Z., Bevers, J. III, et al. (2019). Massive antibody discovery used to probe structure-function relationships of the essential outer membrane protein Lptd. Elife 8:e46258. doi: 10.7554/eLife.46258
Struve, C., Roe, C. C., Stegger, M., Stahlhut, S. G., Hansen, D. S., Engelthaler, D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella Pneumoniae. MBio 6:E00630. doi: 10.1128/mBio.00630-15
Suits, M. D., Sperandeo, P., Deho, G., Polissi, A., and Jia, Z. (2008). Novel structure of the conserved gram-negative lipopolysaccharide transport protein a and mutagenesis analysis. J. Mol. Biol. 380, 476–488. doi: 10.1016/j.jmb.2008.04.045
Sutherland, I. (2001). Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147, 3–9. doi: 10.1099/00221287-147-1-3
Szijarto, V., Guachalla, L. M., Hartl, K., Varga, C., Badarau, A., Mirkina, I., et al. (2017). Endotoxin neutralization by an O-antigen specific monoclonal antibody: a potential novel therapeutic approach against Klebsiella pneumoniae St258. Virulence 8, 1203–1215. doi: 10.1080/21505594.2017.1279778
Szijarto, V., Guachalla, L. M., Hartl, K., Varga, C., Banerjee, P., Stojkovic, K., et al. (2016). Both clades of the epidemic Kpc-producing Klebsiella pneumoniae clone St258 share a modified galactan O-antigen type. Int. J. Med. Microbiol. 306, 89–98. doi: 10.1016/j.ijmm.2015.12.002
Takeuchi, O., and Akira, S. (2010). Pattern recognition receptors and inflammation. Cell 140, 805–820. doi: 10.1016/j.cell.2010.01.022
Tan, Y. K., Khoo, K. L., Chin, S. P., and Ong, Y. Y. (1998). Aetiology and outcome of severe community-acquired pneumonia in Singapore. Eur. Respir. J. 12, 113–115. doi: 10.1183/09031936.98.12010113
Taur, Y., and Pamer, E. G. (2013). The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr. Opin. Infect. Dis. 26, 332–337. doi: 10.1097/QCO.0b013e3283630dd3
The European Antimicrobial Resistance Surveillance Network, E.-N. (2018). “Antimicrobial Resistance (Amr) Reporting Protocol 2018,” in Ecdc. Sweden: Tessy - The European Surveillance System.
Tibbetts, R., Frye, J. G., Marschall, J., Warren, D., and Dunne, W. (2008). Detection of Kpc-2 in a clinical isolate of proteus mirabilis and first reported description of carbapenemase resistance caused by a Kpc Beta-Lactamase In P. Mirabilis. J. Clin. Microbiol. 46, 3080–3083. doi: 10.1128/JCM.00979-08
Tindall, B. J., Sutton, G., and Garrity, G. M. (2017). Enterobacter aerogenes Hormaeche and Edwards 1960 (Approved Lists 1980) and Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980) share the same nomenclatural type (ATCC 13048) on the Approved Lists and are homotypic synonyms, with consequences for the name Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980). Int. J. Syst. Evol. Microbiol. 67, 502–504. doi: 10.1099/ijsem.0.001572
Tran, A. X., Dong, C., and Whitfield, C. (2010). Structure and functional analysis of Lptc, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539. doi: 10.1074/jbc.M110.144709
Trautmann, M., Held, T. K., and Cross, A. S. (2004). O antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Vaccine 22, 818–821. doi: 10.1016/j.vaccine.2003.11.026
Tsai, F. C., Huang, Y. T., Chang, L. Y., and Wang, J. T. (2008). Pyogenic liver abscess as endemic disease, Taiwan. Emerg. Infect. Dis. 14, 1592–1600. doi: 10.3201/eid1410.071254
Tytgat, H. L., and Lebeer, S. (2014). The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol. Mol. Biol. Rev. 78, 372–417. doi: 10.1128/MMBR.00007-14
Tzouvelekis, L. S., Miriagou, V., Kotsakis, S. D., Spyridopoulou, K., Athanasiou, E., Karagouni, E., et al. (2013). Kpc-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob. Agents Chemother. 57, 5144–5146. doi: 10.1128/AAC.01052-13
Vading, M., Naucler, P., Kalin, M., and Giske, C. G. (2018). Invasive infection caused by Klebsiella pneumoniae is a disease affecting patients with high comorbidity and associated with high long-term mortality. PLoS ONE 13:E0195258. doi: 10.1371/journal.pone.0195258
Van De Klundert, J. A., Van Gestel, M. H., Meerdink, G., and De Marie, S. (1988). Emergence of bacterial resistance to cefamandole in vivo due to outer membrane protein deficiency. Eur. J. Clin. Microbiol. Infect. Dis. 7, 776–778. doi: 10.1007/BF01975046
Vetterli, S. U., Zerbe, K., Muller, M., Urfer, M., Mondal, M., Wang, S. Y., et al. (2018). Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 4:Eaau2634. doi: 10.1126/sciadv.aau2634
Villa, R., Martorana, A. M., Okuda, S., Gourlay, L. J., Nardini, M., Sperandeo, P., et al. (2013). The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108. doi: 10.1128/JB.02057-12
Vu, B., Chen, M., Crawford, R. J., and Ivanova, E. P. (2009). Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554. doi: 10.3390/molecules14072535
Wachino, J., Doi, Y., Yamane, K., Shibata, N., Yagi, T., Kubota, T., et al. (2004). Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class a Beta-Lactamase, Ges-4, possessing a single G170s substitution in the omega-loop. Antimicrob. Agents Chemother. 48, 2905–2910. doi: 10.1128/AAC.48.8.2905-2910.2004
Walport, M. J. (2001). Complement. First of two parts. N. Engl. J. Med. 344, 1058–1066. doi: 10.1056/NEJM200104053441406
Wei, D. D., Wan, L. G., Deng, Q., and Liu, Y. (2016). Emergence of Kpc-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 In Mainland China. Diagn. Microbiol. Infect. Dis. 85, 192–194. doi: 10.1016/j.diagmicrobio.2015.03.012
Whitfield, C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68. doi: 10.1146/annurev.biochem.75.103004.142545
Whitfield, C., and Paiment, A. (2003). Biosynthesis and assembly of group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr. Res. 338, 2491–2502. doi: 10.1016/j.carres.2003.08.010
Whitfield, C., and Trent, M. S. (2014). Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128. doi: 10.1146/annurev-biochem-060713-035600
Whitney, J. C., and Howell, P. L. (2013). Synthase-dependent exopolysaccharide secretion in gram-negative bacteria. Trends Microbiol. 21, 63–72. doi: 10.1016/j.tim.2012.10.001
Wick, R. R., Heinz, E., Holt, K. E., and Wyres, K. L. (2018). Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 56:e00197–18. doi: 10.1128/JCM.00197-18
Woodford, N., Turton, J. F., and Livermore, D. M. (2011). Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755. doi: 10.1111/j.1574-6976.2011.00268.x
Woodward, R., Yi, W., Li, L., Zhao, G., Eguchi, H., Sridhar, P. R., et al. (2010). In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy And Wzz. Nat. Chem. Biol. 6, 418–423. doi: 10.1038/nchembio.351
World Health Organization, W. (2017). Who Publishes List Of Bacteria For Which New Antibiotics Are Urgently Needed.
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., Van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-14
Wu, J. H., Wu, A. M., Tsai, C. G., Chang, X. Y., Tsai, S. F., and Wu, T. S. (2008). Contribution of fucose-containing capsules in Klebsiella pneumoniae to bacterial virulence in mice. Exp. Biol. Med. 233, 64–70. doi: 10.3181/0706-RM-170
Wyres, K. L., Gorrie, C., Edwards, D. J., Wertheim, H. F., Hsu, L. Y., Van Kinh, N., et al. (2015). Extensive capsule locus variation and large-scale genomic recombination within the Klebsiella pneumoniae clonal group 258. Genome Biol. Evol. 7, 1267–1279. doi: 10.1093/gbe/evv062
Wyres, K. L., and Holt, K. E. (2018). Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 45, 131–139. doi: 10.1016/j.mib.2018.04.004
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2:E000102. doi: 10.1099/mgen.0.000102
Yang, F. L., Yang, Y. L., Liao, P. C., Chou, J. C., Tsai, K. C., Yang, A. S., et al. (2011). Structure and immunological characterization of the capsular polysaccharide of a pyrogenic liver abscess caused by Klebsiella pneumoniae: activation of macrophages through toll-like receptor 4. J. Biol. Chem. 286, 21041–21051. doi: 10.1074/jbc.M111.222091
Yao, H., Qin, S., Chen, S., Shen, J., and Du, X. D. (2018). Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 18:25. doi: 10.1016/S1473-3099(17)30628-X
Yeh, K. M., Lin, J. C., Yin, F. Y., Fung, C. P., Hung, H. C., Siu, L. K., et al. (2010). Revisiting the importance of virulence determinant maga and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J. Infect. Dis. 201, 1259–1267. doi: 10.1086/606010
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, Bla(Ndm-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Yoshida, K., Matsumoto, T., Tateda, K., Uchida, K., Tsujimoto, S., and Yamaguchi, K. (2000). Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J. Med. Microbiol. 49, 1003–1010. doi: 10.1099/0022-1317-49-11-1003
Youssef, D., Shams, W., Kareem Abu Malouh, A., and Al-Abbadi, M. A. (2012). Chronic organizing retroperitoneal abscess caused by Klebsiella oxytoca masquerading as sarcoma: recognition by diff-quik stain on FNA material. Diagn. Cytopathol. 40, 747–750. doi: 10.1002/dc.21701
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, C. C., Lai, C. C., and Chuang, Y. C. (2008). Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and Non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 62, 1–6. doi: 10.1016/j.diagmicrobio.2008.04.007
Zhang, P., Summer, W. R., Bagby, G. J., and Nelson, S. (2000). Innate immunity and pulmonary host defense. Immunol. Rev. 173, 39–51. doi: 10.1034/j.1600-065X.2000.917306.x
Zhang, R., Lin, D., Chan, E. W., Gu, D., Chen, G. X., and Chen, S. (2016). Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob. Agents Chemother. 60, 709–711. doi: 10.1128/AAC.02173-15
Keywords: Klebsiella species, multidrug resistance, lipopolysaccharide, capsular polysaccharide, exopolysaccharide, complement system, vaccine, antibiotics
Citation: Patro LPP and Rathinavelan T (2019) Targeting the Sugary Armor of Klebsiella Species. Front. Cell. Infect. Microbiol. 9:367. doi: 10.3389/fcimb.2019.00367
Received: 07 April 2019; Accepted: 09 October 2019;
Published: 08 November 2019.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Muhammad Ammar Zafar, Wake Forest School of Medicine, United StatesMichael Bachman, University of Michigan, United States
Copyright © 2019 Patro and Rathinavelan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thenmalarchelvi Rathinavelan, dHJAaWl0aC5hYy5pbg==
 L. Ponoop Prasad Patro
L. Ponoop Prasad Patro Thenmalarchelvi Rathinavelan
Thenmalarchelvi Rathinavelan