
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 16 April 2019
Sec. Parasite and Host
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00096
This article is part of the Research Topic CRISPR/Cas9 and Its Asset to Infectious Disease Research View all 5 articles
 Catherin Marin-Mogollon1
Catherin Marin-Mogollon1 Ahmed M. Salman2
Ahmed M. Salman2 Karin M. J. Koolen3
Karin M. J. Koolen3 Judith M. Bolscher3
Judith M. Bolscher3 Fiona J. A. van Pul1
Fiona J. A. van Pul1 Shinya Miyazaki1
Shinya Miyazaki1 Takashi Imai1,4
Takashi Imai1,4 Ahmad Syibli Othman1,5
Ahmad Syibli Othman1,5 Jai Ramesar1
Jai Ramesar1 Geert-Jan van Gemert6
Geert-Jan van Gemert6 Hans Kroeze1
Hans Kroeze1 Severine Chevalley-Maurel1
Severine Chevalley-Maurel1 Blandine Franke-Fayard1
Blandine Franke-Fayard1 Robert W. Sauerwein3,6
Robert W. Sauerwein3,6 Adrian V. S. Hill2
Adrian V. S. Hill2 Koen J. Dechering3
Koen J. Dechering3 Chris J. Janse1
Chris J. Janse1 Shahid M. Khan1*
Shahid M. Khan1*Transgenic malaria parasites expressing fluorescent and bioluminescent proteins are valuable tools to interrogate malaria-parasite biology and to evaluate drugs and vaccines. Using CRISPR/Cas9 methodology a transgenic Plasmodium falciparum (Pf) NF54 line was generated that expresses a fusion of mCherry and luciferase genes under the control of the Pf etramp10.3 gene promoter (line bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw==). Pf etramp10.3 is related to rodent Plasmodium uis4 and the uis4 promoter has been used to drive high transgene expression in rodent parasite sporozoites and liver-stages. We examined transgene expression throughout the complete life cycle and compared this expression to transgenic lines expressing mCherry-luciferase and GFP-luciferase under control of the constitutive gapdh and eef1a promoters. The bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== parasites express mCherry in gametocytes, sporozoites, and liver-stages. While no mCherry signal was detected in asexual blood-stage parasites above background levels, luciferase expression was detected in asexual blood-stages, as well as in gametocytes, sporozoites and liver-stages, with the highest levels of reporter expression detected in stage III-V gametocytes and in sporozoites. The expression of mCherry and luciferase in gametocytes and sporozoites makes this transgenic parasite line suitable to use in in vitro assays that examine the effect of transmission blocking inhibitors and to analyse gametocyte and sporozoite biology.
Transgenic rodent and human malaria parasites expressing fluorescent and bioluminescent proteins are used extensively to interrogate parasite biology and host-parasite interactions associated with malaria pathology and as tools to evaluate anti-parasite inhibitors and vaccines (Othman et al., 2017). In comparison to transgenic rodent malaria parasites (RMP) only a few Plasmodium falciparum (Pf) transgenic parasites expressing fluorescent or luminescent proteins are available. Transgenic Pf parasites have been used to quantify effects of inhibitors and antibodies either on blood-stage growth in vitro using standard growth inhibition assays (Wilson et al., 2010) or on transmission-blocking (TB) activity in the mosquito in high-throughput screens using standard membrane feeding assays (Vos et al., 2015; Lucantoni et al., 2016). For the TB assays transgenic Pf (NF54 strain) parasite lines expressing the GFP-luciferase fusion protein under control of either the strong constitutive Pf hsp70 (Vos et al., 2015) or the gametocyte-specific pfs16 promoters have been used (Lucantoni et al., 2016). In addition, a transgenic Pf NF54 has been described that expresses a GFP-luciferase fusion protein under control of the constitutive Pf eef1a promoter (Vaughan et al., 2012). This reporter line has been used to analyze liver infection in immune-deficient humanized mice engrafted with human liver tissue ((Sack et al., 2014; Flannery et al., 2018); (Foquet et al., 2018)).
In multiple RMP transgenic lines the uis4 gene promoter has been used to drive expression of different transgenes in sporozoites and liver-stages, such as genes encoding mCherry, ovalbumin or human Plasmodium proteins (Combe et al., 2009; Panchal et al., 2012; Montagna et al., 2014; Hopp et al., 2015; Longley et al., 2015, 2017; Singer et al., 2015) The uis4 gene, a member of the small Plasmodium etramp gene family, is highly transcribed in sporozoites and liver-stages and encodes a parasitophorous vacuole membrane (PVM) protein that surrounds the parasite in the infected hepatocyte. Although uis4 transcripts are translationally repressed in sporozoites (Silvie et al., 2014; Silva et al., 2016), transgenes expressed under control of uis4 regulatory sequences are expressed as protein in sporozoites since uis4 translational repression in sporozoites is dependent on DNA sequences present within the uis4 open reading frame (Silvie et al., 2014; Hopp et al., 2015). In this study, we generated a transgenic Pf parasite, bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw==, which encodes a fusion of the proteins mCherry and luciferase (mCherry-Luc) expressed under the control of the promoter of etramp10.3 (PF3D7_1016900). We selected this promoter since etramp10.3 is related to uis4 and it also belongs to the etramp gene family and both genes have the same syntenic genomic location. It has been previously reported that etramp10.3 is expressed in Pf sporozoites as well as in blood- and liver-stages, where the protein is located at the PVM, similar to the location of UIS4 in liver-stages of rodent malaria parasites (Mackellar et al., 2010). In this study, as a blood-stage reporter control line, we also generated a transgenic Pf parasite that expresses mCherry-Luc under the control of the promoter of gapdh (PF3D7_1462800), mCherry-luc@gapdh. The gapdh gene is constitutively expressed in blood-stages (Aurrecoechea et al., 2009) and we recently showed that the gapdh promoter drives high GFP expression in both asexual blood-stages and in gametocytes (Mogollon et al., 2016).
We chose to generate mCherry-expressing Pf reporter lines as such lines could be used to visualize interactions of Plasmodium parasites with host-cells (e.g., cells of the immune system or hepatocytes), which are often labeled with green fluorescent proteins. Moreover, we fused the mCherry gene to firefly luciferase, as luminescence can be used to quantify parasite numbers (e.g., sporozoites and liver-stages) using standardized and sensitive assays (Annoura et al., 2013; Le Bihan et al., 2016; Swann et al., 2016). The mCherry-luciferase expression cassettes were introduced using a CRISPR/Cas9 methodology into the p47 gene locus, a non-essential locus that has been previously used to introduce transgenes into the Pf genome (Talman et al., 2010; Vaughan et al., 2012; Vos et al., 2015; Lucantoni et al., 2016). Reporter gene expression of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== line was analyzed throughout the complete parasite life cycle and compared to transgenic lines expressing mCherry-luciferase or GFP-luciferase under control of the constitutive gapdh or eef1a promoters. We demonstrate mCherry and luciferase expression in bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== gametocytes, sporozoites and liver-stages.
P. falciparum (Pf) parasites from the NF54 strain were used (Mogollon et al., 2016). Parasites were cultured using standard culture conditions in a semi-automated culture system as described (Mogollon et al., 2016) Fresh human serum and human red blood cells (RBC) were obtained from the Dutch National Blood Bank (Sanquin Amsterdam, the Netherlands; permission granted from donors for the use of blood products for malaria research and microbiology tested for safety). RBC of different donors were pooled every 2 weeks, washed twice in serum free RPMI-1640 and suspended in complete culture medium to 50% hematocrit. Human serum of different donors were pooled every 4–6 months and stored at −20°C until required. Pf gametocytes cultures were generated using standard culture conditions with some modifications as described (Mogollon et al., 2016). Briefly, parasites from asexual stage cultures were diluted to a final parasitemia of 0.5% and cultures were followed during 14 days without refreshing RBC. At day 14 the cultures were analyzed for mature, stage V, gametocytes.
In addition, a transgenic P. falciparum (NF54) line (gfp-luc@eef1a) was used that contains a reporter cassette with a fusion gene of GFP and luciferase (GFP-Luc) under control of eukaryotic elongation factor 1 alpha (eef1a) promoter (Vaughan et al., 2012).
To create the mCherry-luc@etramp reporter line we used a previously described Cas9 construct (pLf0019), containing the Cas9 expression cassette with a blasticidin (BSD) drug-selectable marker cassette (Mogollon et al., 2016) in combination with a sgRNA donor-DNA plasmid (pLf0049). The sgRNA-donor DNA construct (pLf0049) contains a hdhfr-yfcu drug-selectable marker (SM) cassette for selection with the drug WR99210. To generate pLf0049, the intermediate plasmid pLf0039 (Supplementary Figure 1) was modified by introducing two homology regions targeting of p47 (PF3D7_1346800). Homology region 1 (HR1) was amplified using primers P1/P2 and homology region 2 (HR2) with P3/P4 from Pf NF54 genomic DNA (see Supplementary Table 1 for primer details). HR1 was cloned in pLf0039 using restriction sites StuII/SacII and HR2 using ApaI/HindIII. Subsequently, a guide RNA sgRNA targeting the p47 locus (gRNA019) was selected using the Protospacer software (alphaversion; https://sourceforge.net/projects/ protospacerwb/files/Release/), based on the best off targets hits score throughout the genome given by Protospacer and the total number of mismatches of the sgRNA with respect to the PAM site. A 20 bp guide sgRNA sequence (using the primers P5/P6), flanked on both sides by a 15 bp DNA sequence necessary for In Fusion cloning (HD Cloning Kit; Clontech), was annealed and used to replace the BtgZI adaptor in pLf0039 as previously described (Ghorbal et al., 2014), resulting in pLf0047. The plasmid pLf0047 was digested with BlnI and NruI to confirm successful cloning of the sgRNA and the correct sequence of the sgRNA (using primes P7/P8) confirmed by Sanger sequencing. An additional intermediate plasmid, pLf0130 (Supplementary Figure 1) was modified by replacing the existing promotor region of the gapdh gene (PF3D7_1462800) by the promoter region of etramp10.3 promoter (PF3D7_1016900) of a reporter cassette containing mCherry fused to luciferase with the 3'UTR region of histidine-rich protein II (PF3D7_0831800; 626 bp obtained by digestion of an intermediate plasmid pLf0053 Mogollon et al., 2016 with restriction sites ApaI/XbaI). The etramp10.3 promoter region (1.7kb) was amplified from Pf NF54 genomic DNA using the primers P9/P10 and cloned into pLf0130 using restriction sites SacIII/XhoI or KpnI. Next, to obtain the complete bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== expression cassette, pLf0130 was digested with ApaI/SacII and this cassette was cloned in pLf0047 using the same enzymes, resulting in the final construct pLf0049.
To create the mCherry-luc@gapdh reporter line we used the Cas9 construct (pLf0019), described above, in combination with a sgRNA donor-DNA plasmid (pLf0048). The sgRNA-donor DNA construct (pLf0048) contains a hdhfr-yfcu drug-selectable marker (SM) cassette for selection with the drug WR99210. To generate pLf0048, an intermediate plasmid CM269 (Supplementary Figure 1) was modified by inserting the promoter region (1.6 kb) of the gapdh gene (PF3D7_1462800) that was PCR-amplified from Pf NF54 genomic DNA using the primers P11/P12 (Supplementary Table 1). CM269 was digested with restriction enzymes NruI/XhoI and the gapdh promoter was cloned into CM269 using the same enzymes, resulting in construct pLf0130 (Supplementary Figure 1). Finally, to obtain the mcherry-luc@gapdh expression cassette pLf0130 was digested with ApaI/SacII and this cassette was cloned in pLf0047 (see above) using the same enzymes, resulting in the final construct pLf0048 (Supplementary Figure 1).
Isolation of plasmids for transfection and transfection of synchronized ring stage parasites was performed as described (Mogollon et al., 2016) and parasites were transfected with a mixture of ~50 μg of each circular plasmid (Cas9 and sgRNA/Donor DNA construct). After transfection, parasite cultures were maintained under standard culture conditions in the semi-automated culture system (see above). Selection of transformed parasites was performed by applying “double” positive selection 24 h after transfection using the drugs WR99210 (2.6 nM) and BSD (5 μg/ml) as described (Mogollon et al., 2016). Drug pressure in the cultures was maintained until thin blood-smears were parasite-positive (usually after 14 to 26 days). Positive selection will select for the parasites that were transfected successfully with both plasmids (Cas9 and sgRNA/Donor constructs). Subsequently, both drugs were removed from the cultures for 2–4 days, followed by applying negative selection by addition of 5-Fluorocytosine as described (Mogollon et al., 2016) in order to eliminate parasites that retained the crRNA/Donor construct as episomal plasmid and enriching for the transfected parasites containing the donor DNA integrated into the genome. Negative drug pressure in the cultures was maintained until thin blood-smears were parasite-positive (usually after 7 days). After negative selection infected RBC (iRBC) were harvested from cultures with a parasitemia of 4 to 10% for genotyping by diagnostic PCR and Southern analysis (see next sections). Subsequently, selected parasites were cloned by limiting dilution as has been described previously (Mogollon et al., 2016). Cloned parasites were transferred in 10 ml culture flasks at 5% haematocrit and cultured under standard culture conditions (see above) in the semi-automated culture system for collection of parasites for further genotype and phenotype analyses (see next section).
For genotyping diagnostic PCR and Southern analysis of digested DNA were performed using DNA isolated from iRBC obtained from 10 ml cultures (parasitemia 3–10%). DNA was isolated as described (Mogollon et al., 2016). Correct integration of the donors constructs was analyzed by PCR amplification of the mCherry-luciferase gene (primers P13/P14), the fragment for 3' integration (3'int; primers P15/P16) and the p47 gene (ORF; primers P17/P18) (see Supplementary Table 1 for primer sequences). The PCR fragments were amplified using Go-taq® DNA polymerase (Promega) following standard conditions with an annealing temperature of 56°C for 20 s and a elongation step of 72°C for 4 min. All other PCR settings were according to manufacturer's instructions.
Southern blot analysis of digested DNA was performed with HpaI digested genomic DNA (4 h at 37°C). Digested DNA was hybridized with probes targeting the p47 homology region 1 (HR1), amplified from Pf NF54 genomic DNA by PCR using the primers P1/P2 and a second probe targeting ampicillin (Amp) gene, obtained by digestion of the intermediate plasmid pLf0040 with AatII/PvuI (550bp).
The growth rate of asexual blood-stages (parasitemia) was monitored by determination of parasitemia in standard in vitro cultures for a period of 4 days with a starting parasitemia between 0.1 and 0.5%. Parasitemia was determined either by FACS analysis of iRBC or by counting parasitemia in Giemsa-stained thin blood films. For FACS analysis iRBC were stained with the DNA-specific dye Hoechst-33258 in 1 ml of PBS by adding 4 μl of a 500 μM stock-solution (final concentration 2 μM), as has been described previously (Mogollon et al., 2016). mCherry expression of asexual blood-stages was analyzed by standard fluorescence microscopy. In brief, 200 μl samples of iRBC were collected from 10 ml cultures with a parasitemia between 4 and 10% and stained with the DNA-specific dye Hoechst-33342 by adding 4 μl of a 500 μM stock-solution (final concentration 10 μM) for 20 min at 37°C. Subsequently, 5 μl was placed on a microscopic slide (mounted under a cover slip) and fluorescence of live iRBC analyzed using a Leica fluorescence MDR microscope (100x magnification). Pictures were recorded with a DC500 digital camera microscope using Leica LAS X software and with the following exposure times: mCherry 0.6 s; Hoechst-33342 0.136 s; bright field 0.62 s (1x gain). Luciferase expression in mCherry-luc@etramp asexual blood-stages was determined as follows. A serial dilution of asexual blood-stages was prepared from parasites samples that were collected (in triplicate) from asexual blood-stage cultures with a final number of 5 × 106 parasites per sample. These were diluted with RPMI-1640 culture medium containing uninfected RBC (5% haematocrit) to prepare (triplicate) samples with 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, 1 × 101 parasites, respectively. Samples of 40 μl containing only uninfected RBC (5% haematocrit) were used as controls. The cells were pelleted by centrifugation (800 g; 30 s) and were lysed with 40 μl of cell culture Lysis 5X reagent from Promega (1 in 5 dilution in miliQ water). The complete lysates were collected in black 96-well plate (flat bottom) and luciferase activity was measured after adding 50 μl of the Luciferase substrate (Luciferase Assay System Promega). Luciferase activity (in relative light units; RLU) was measured using the Glomax multi detection system Luminometer (Promega) and the Instinct software (Promega).
Gametocyte production (stage V male/female gametocytes) and exflagellation of male gametocytes were analyzed in gametocyte cultures established as described previously (van Dijk et al., 2010). For analysis of mCherry expression, stage II -V gametocytes were collected at days 7, 9, 11, and 14. Samples (200 μl) were collected, pelleted by centrifugation (800 g; 30 s) and stained with Hoechst-33342 and mCherry expression analysis using fluorescence microscopy as described for asexual blood-stages. Luciferase expression was determined in mCherry-luc@etramp gametocytes (stage III-IV) collected at day 11. A similar serial dilution of gametocytes was prepared as described for asexual blood-stages and luciferase activity in gametocytes was determined as described for the asexual blood-stages.
For analysis of mosquito stages (oocysts and sporozoites), Anopheles stephensi mosquitoes were infected with day 14 gametocytes cultures using the standard membrane feeding assay (SMFA) (Ponnudurai et al., 1987, 1989). Oocysts numbers and mCherry expression in oocysts was determined at day 8 and 10 after infection. mCherry expression was analyzed using a Leica fluorescence MDR microscope (100x magnification). Pictures were recorded with a DC500 digital camera microscope using Leica LAS X software and with the following exposure times: mCherry 0.6 s; Hoechst-33342 0.136 s; bright field 0.62 s (1x gain). Collection of salivary gland sporozoites for counting numbers and expression of mCherry was performed at day 14 and 21 after feeding. For counting sporozoites, salivary glands from 30 to 60 mosquitoes were dissected, collected in 100 μl of RPMI-1640 pH 7.2 and homogenized using a grinder. Sporozoites were counted using a Bürker cell counter using phase-contrast microscopy. For mCherry expression, the isolated sporozoite were pelleted by centrifugation (800 g; 5 min). The pellet was suspended in 40 μl 1X PBS and sporozoites stained with Hoechst-33342 (10 μM) for 30 min at 37°C. Of this solution, 5 μl was placed on a microscopic slide (mounted under a cover slip) and fluorescence of sporozoites in live was analyzed using a Leica fluorescence MDR microscope (100x magnification). Pictures were recorded with a DC500 digital camera microscope using Leica LAS X software and with the following exposure times: mCherry 0.6 s; Hoechst-33342 0.136 s; bright field 0.62 s (1x gain).
Luciferase activity in mCherry-luc@etramp sporozoites was determined in duplicate samples (total volume of 40 μl of RPMI) of a serial dilution of 0.31 × 104-5.0 × 104 salivary glands sporozoites. Fifty microliter of D-Luciferin (0.4 mg/ml; Perkin Elmer Life Sciences, Waltham, USA) was added to the 40 μl of diluted sporozoites in a black 96-well plate (flat bottom). The in vivo imaging system Lumina (Caliper Life Sciences, USA) was used to measure luciferase activity. Imaging data were analyzed using the Living Image® 4.5.5 software (Caliper Life Sciences, USA). Bioluminescence images were acquired with a 12.5 cm field of view (FOV), medium binning factor and an “auto-exposure” time of maximum 2 min.
Liver-stages of mCherry-luc@etramp and gfp-luc@eef1a were cultured in vitro using cryopreserved primary human hepatocytes obtained from Tebu-bio (Tebu-bio.com–Life science Research) and thawed according to the instructions of Sekisui/Xenotech (Sekisui XenoTech, LLC; Kansas City). Cells were cultured in Williams's E culture medium supplemented with 10% FCS, 1% penicillin-streptomycin, 1% fungizone, 0.1 IU/ml insulin and 70 μM hydrocortisone 21-hemisuccinate (Sigma). Hepatocytes were seeded in Greiner clear bottom white 96-well plates at a density of 5 × 104 cells per well, 2 days before infection with sporozoites as described previously (Boes et al., 2016). Sporozoites were collected from infected mosquitoes at day 21 after as described above and hepatocyte cultures (at 37°C) were infected with 5 × 104 sporozoites per well. Three hours (h) after the addition of sporozoites, the cultures were washed three times with 1X PBS to remove mosquito material as well as sporozoites and complete Williams's E medium was added and cultures which were incubated overnight at 37°C. The day after, the culture medium was replaced and then was changed every 48 h until day 7.
Cultured cryopreserved primary human hepatocytes were infected with 5 × 104 bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== and GFP-luc@eef1α sporozoites per well of a 96-wells plate. Development of liver-stages of these two transgenic lines at day 2, 4, and 6 after infection was compared with WT liver-stages by immunofluorescence using antibodies against the cytoplasmic protein Pf HSP70 (rabbit, anti-Pf HSP70; 1:75 dilution of 1 mg/ml stock solution StressMarqBiosciences) and secondary antibody (goat anti-rabbit IgG AF594; 1:200 dilution of 4 mg/ml stock solution Invitrogen). Hepatocyte and parasite nuclei were stained with 300nM DAPI (Invitrogen D1306). Fluorescence signals were visualized using a Cytation Imager (Biotek, Winooski, VT). The percentage of infected hepatocytes, the size of the liver-stages and the intensity of Pf HSP70 staining were analyzed using the FIJI image analysis software package (Schindelin et al., 2012).
For analysis of mCherry expression in liver-stages, infected hepatocytes were fixed (at day 3, 4, and 5) with 4% paraformaldehyde in 1X PBS during 1 h at room temperature. After fixation the wells were washed three times with 1X PBS and permeabilized with 20 μl of 0.5% triton in 1X PBS and then blocked with 10% of FCS in 1X PBS for 1 h. Fixed cells were washed with 1X PBS and incubated with monoclonal antibodies against PfHSP70 (rabbit, anti-Pfhsp70; 1:200 dilution of 100 μg/ml stock solution StressMarqBiosciences) and against mCherry (goat, anti-mCherry Mab Sicgen antibodies; 1:200 dilution of 3 mg/ml stock solution) for 1 h at room temperature. Subsequently, cells were rinsed 3 times with 1X PBS and incubated with the secondary antibodies Alexa Fluor®488/594-conjugated chicken anti-rabbit and anti-goat (Invitrogen Detection Technologies at 1:200). Finally, the cells were washed again three times with 1X PBS and stained with the DNA-specific dye Hoechst-33342 at a final concentration of 10μM. Fixed cells were covered with 1-2 drops of an anti-fading agent (Vectashield), and stained cells were analyzed for fluorescence using a Leica fluorescence MDR microscope (100x magnification). Pictures were recorded with a DC500 digital camera microscope using Leica LAS X software with the following exposure times: Alexa 488: 0.7s; Alexa 594: 0.6s; Hoechst 0.136s; bright field 0.62s (1x gain). Luciferase expression in liver-stages was monitored daily by adding 150 μl of Bright-Glo luciferase assay substrate (Promega, Madison, WI) to 150 μl of culture medium to each well and luminescence was quantified using a Synergy 2 multi-purpose plate reader (Biotek, Winooski, VT). Background was determined by measuring wells with uninfected hepatocytes.
The sensitivity of liver-stage development to atovaquone determined by measurement of luminescence of infected primary human hepatocytes maintained in 96-well plates and incubated with a serial dilution of atovaquone. Atovaquone was serially diluted in DMSO and then in Williams E culture medium to reach a final DMSO concentration of 0.1%. Atovaquone was added to the 96-well cultures 3hrs after adding 5x104 sporozoites to cultures of primary human hepatocytes. Medium containing drug was refreshed each day and the cultures were allowed to proceed for 4 days after which luminescence was determined as describe above.
Data were analyzed using GraphPad Prism software package 5.04 (GraphPad Software, Inc). Significance difference analyses between WT and the reporter lines mcherry-luc@etramp 10.3 and gfp-luc@eef1a was performed using the unpaired Student's t-test.
In order to create a P. falciparum parasite that has high reporter protein expression in sporozoites and/or liver-stages, we selected the promoter of the etramp10.3 gene (PF3D7_1016900) to drive gene expression. We selected the P. falciparum p47 gene (PF3D7_1346800) locus for introduction of the transgene expression cassette into the genome as it has been shown to be suitable for transgene expression without compromising parasite development in blood- and liver-stages as well as in A. stephensi mosquitoes (Talman et al., 2010; Vaughan et al., 2012; Vos et al., 2015). We had previously generated various transgenic reporter P. falciparum lines, where GFP-expression cassettes were introduced into the p230p gene locus (PF3D7_0208900), but disruption of this gene resulted in parasites that could not complete mosquito stage development (Marin-Mogollon et al., 2018). Using CRISPR/Cas9 methodology a transgenic P. falciparum (Pf) parasite line was created that encodes an mCherry-luciferase fusion gene under the control of 1.7 kb of 5' UTR of the etramp10.3 gene. This expression cassette was introduced into the neutral p47 gene locus (PF3D7_1346800) using a two plasmid CRISPR/Cas9 method; a previously described Cas9 construct (pLf0019) was used that contains the Cas9 expression cassette and a blasticidin (BSD) drug-selectable marker cassette (Mogollon et al., 2016) and a second plasmid (pLf0049) containing both the p47 sgRNA and the donor DNA sequences (Supplementary Figure 1). The pLf0049 plasmid contains the p47 targeting sequences, the bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== expression cassette and a hdhfr-yfcu drug-selectable marker cassette (see Figure 1A and Materials and Methods section for details of the generation of the constructs). This plasmid is designed to introduce the reporter bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== cassette into the p47 gene locus by double cross-over homologous recombination to repair the double strand break (DSB) introduced into the p47 gene locus (Figure 1A).
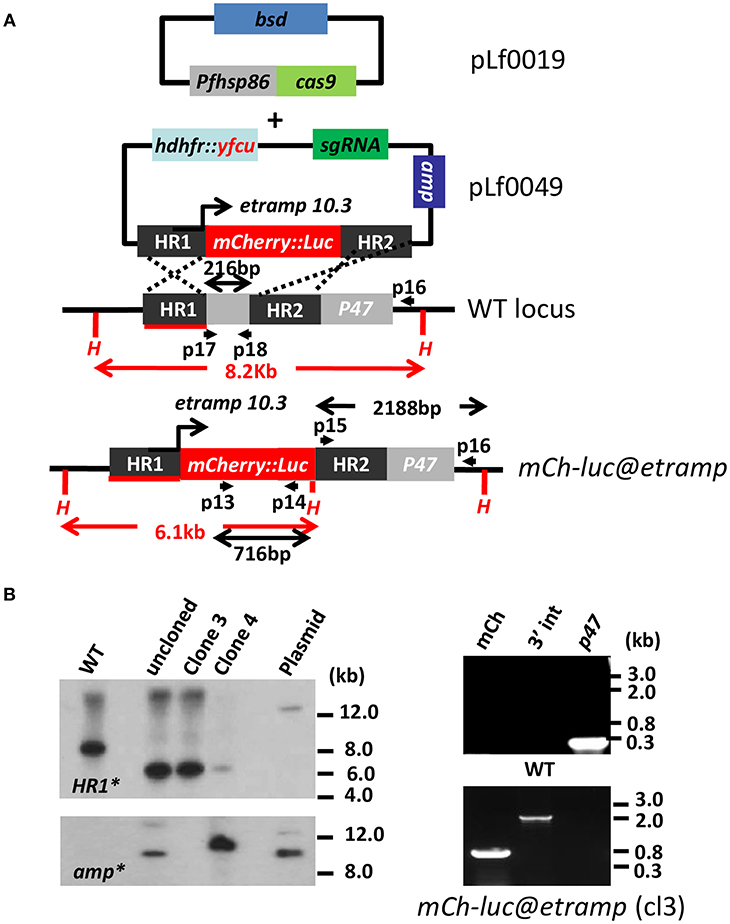
Figure 1. Generation and characterization of a P. falciparum reporter line expressing mCherry-luciferase under control of the etramp10.3 promoter. (A) Schematic representation of the Cas9 (pLf0019) and sgRNA/donor (pL0049) constructs used to introduce the mCherry-luciferase expression cassette into the P. falciparum p47 gene locus. The mCherry-luciferase fusion gene is under the control of the promoter of the etramp10.3 gene. The p47 homology regions (HR1, HR2) used to introduce the donor DNA (i.e., the mCherry-luciferase expression cassette), location of primers (p), sizes of restriction fragments (H: HpaI; in red), and PCR amplicons (in black) are indicated. Primer sequences (shown in black and bold) are shown in Supplementary Table 1. WT–wild type; bsd–blasticidin selectable marker (SM); hdhfr::yfcu–SM in donor plasmid. mCh-luc@etramp–the final reporter line bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw==. (B) Southern analysis of HpaI restricted DNA (left panel) and diagnostic PCR (right panel) to confirm correct integration of construct pLf0049 into the p47 locus [wild type (WT), transfected, uncloned (uncl) parasites, clones 3 and 4 of mCherry-luc@etramp parasites, and plasmid (PL)]. Digested DNA was hybridized with a probe targeting the homology region 1 of p47 [HR1; primers p3/p4; see (A)], showing the expected different-sized DNA fragments [shown in red in (A)] in WT (8.2 kb) and bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== parasites (6.1 kb). The absence of hybridization of digested DNA with a probe for ampicillin (amp) confirms the absence of donor-DNA plasmid and single cross-over event integration of construct pLf0049 in clone 3. Diagnostic PCR of mCherry-luc@etramp clone 3 confirms the presence of the mCherry gene (lane 1; primers p13/p14; 716 bp), correct 3′ integration of the construct (lane 2; primers p15/p16; 2,188 bp) and absence of the p47 gene (lane 3; primers p17/p18; 216 bp). Primer locations and product sizes are shown in (A) and primer sequences in Supplementary Table 1). Uncropped images of the PCR and Southern analyses are shown in Supplementary Figures 5, 6.
As a control line we created also a transgenic P. falciparum (Pf) parasite line that contains an mCherry-luciferase fusion gene under control of 1.6kb 5'UTR of the gene encoding the glycolytic enzyme, gapdh (PF3D7_1462800). Recently we have shown that this promoter drives high GFP expression in P. falciparum blood-stages (Mogollon et al., 2016). The same Cas9 construct, pLf0019, was used in combination with a plasmid (pLf0048) expressing the p47 sgRNA and p47 donor-DNA sequences (Supplementary Figure 1). This plasmid is designed to introduce the reporter mCherry-luc@gapdh and the hdhfr-yfcu drug-selectable marker cassettes into the p47 gene locus by double cross-over homologous recombination to repair the double strand break (DSB) introduced into the p47 gene locus (see Supplementary Figure 2A and Materials and Methods section for details of the generation of the pLf0048 construct).
Transfection of Pf wild type (WT) NF54 parasites was performed using synchronized ring-stage parasites that were transfected with ~50 μg of each circular plasmid (Cas9 and sgRNA/donor-DNA constructs; see Materials and Methods section). Selection of transfected parasites containing both plasmids was performed by applying “double” positive selection with the drugs BSD and WR99210 until parasites were detectable by thin blood-smear analysis (between day 14 to 26 post transfection). Subsequently, parasites were cultured for 2–4 days without drugs, followed by the application of negative (5-FC) selection to eliminate parasites that retain transfection constructs (i.e., donor-DNA) as episomal plasmids and to enrich for parasites in which the donor-DNA construct has integrated into the parasite genome. Subsequently, drug-selected parasites were cloned by limiting dilution. Genotyping of two clones of the bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== line (exp.0064 cl3 and cl4) and four clones of the mCherry-luc@gapdh line (exp.0055cl2, 3, 4 and 6) by Southern analysis and PCR revealed integration of the mCherry-luciferase cassettes into the p47 locus (Figure 1B; Supplementary Figure 2B). Whereas, in all clones of mCherry-luc@gapdh the donor DNA appears to be integrated by double cross-over integration, plasmid DNA is present in clone 4 of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== line (Figure 1B) indicating single cross-over integration of pLf0049 in this clone. For further phenotype characterization of the cloned transgenic lines we selected bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== clone 3 and mCherry-luc@gapdh clone 2. PCR analyses of these two clones confirmed correct integration of the expression cassettes into the p47 gene locus (Figure 1B; Supplementary Figure 2B). In vitro growth of asexual blood-stages of clones of both lines was comparable to the growth of asexual blood-stages of the parent wild type (WT) NF54 strain (Supplementary Figure 2C) and the two clones of both lines produced WT-comparable numbers of mature stage V male and female gametocytes in standardized gametocyte cultures (Supplementary Table 2).
We analyzed mCherry expression in the reporter parasites by fluorescence-microscopy of cultured asexual blood-stages. The bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== line did not exhibit clear mCherry expression in asexual blood-stages (ring-forms, trophozoites, and schizonts) and the fluorescent signals were indistinguishable from background of uninfected red blood cells (Figure 2A). In contrast, asexual blood-stages of the control mCherry-luc@gapdh line showed mCherry expression, in agreement with our previous analyses of GFP expression in blood-stages of the GFP@gapdh line, where the gene encoding GFP was under the control of the gapdh promoter (Mogollon et al., 2016) (Figure 2A; Supplementary Figure 3A).
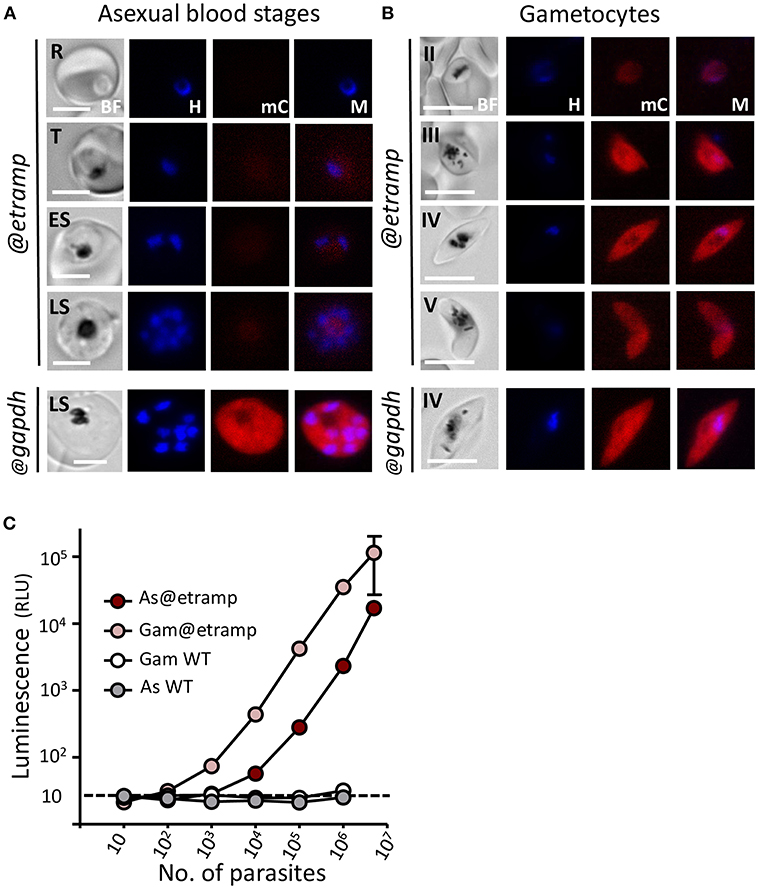
Figure 2. Expression of mCherry and luciferase in asexual blood-stages and gametocytes of mcherry-luc@etramp10.3 and mcherry-luc@gapdh parasites. (A) Fluorescence microscopy analysis of live mcherry-luc@etramp10.3 asexual blood-stages (@etramp). No mCherry fluorescence signal above background were detected in the different stages. R, rings; T, trophozoites; ES, early schizonts; LS, late schizonts. Lower panel, an mCherry-positive schizont of the control line mcherry-luc@gapdh (@gapdh). See Supplementary Figure 3A for other asexual stages of this control line. Nuclei were stained with Hoechst-33342. All pictures were recorded with standardized exposure/gain times to visualize differences in fluorescence intensity [mCherry 0.7 s; Hoechst 0.136 s; bright field 0.62 s (1x gain)]. Bright field (BF), Hoechst (H), mCherry (mC), Merge (M). Scale bar, 4 μm. (B) Fluorescence microscopy analysis of mCherry expression in live mcherry-luc@etramp10.3 gametocytes (@etramp). Gametocyte stage II, III, IV and V are shown. Lower panel, stage IV gametocyte of the control line mcherry-luc@gapdh (@gapdh). See Supplementary Figure 3B for other gametocyte stages of this control line. Nuclei were stained with Hoechst-33342. All pictures were recorded with standardized exposure/gain times to visualize differences in fluorescence intensity [mCherry 0.7 s; Hoechst 0.136 s; bright field 0.62 s (1x gain)]. Bright field (BF), Hoechst (H), mCherry (mC), Merge (M). Scale bar, 7 μm. (C) Correlation between luciferase expression (luminescence, RLU, photons/sec) and number of parasites in serial dilutions series of asexual blood-stages (AS) and gametocytes stage III/IV (Gam) of the mcherry-luc@etramp10.3 line (3 independent experiments). Wild type Pf NF54 parasites (WT) were used as a control. Black dotted line: luminescence value of uninfected cells. The mean luminescence value of triplicate samples is shown; error bars represent the standard deviation. Correlation coefficient r (two-tailed Spearman's test: 103-107 parasites): 1.00; p = 0.016 for AS and 1.0; p = 0.016 for Gam.
Gametocytes of mCherry-luc@gapdh were mCherry positive, again in agreement with our previous observations of the GFP@gapdh parasites (Marin-Mogollon et al., 2018) (Figure 2B; Supplementary Figure 3B). However, mCherry signals in gametocytes were relatively weak in all stages during their development (stage II-IV). The bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== parasites exhibit strong mCherry expression in stage III-V gametocytes (Figure 2B), with weak mCherry signals being detectable in at least 20% of stage II gametocytes, increasing to more than 95% of stage III-V gametocytes that were strongly mCherry positive. The high etramp10.3 promoter activity in gametocytes is in agreement with the high levels of etramp10.3 transcripts and ETRAMP10.3 protein previously reported in gametocytes in genome-wide transcriptomic and mass spectrometry analyses. Peak etramp10.3 transcript abundance was observed in stage III gametocytes (López-Barragán et al., 2011) and ETRAMP10.3 peptides were detected in proteomic analyses of mature male and female gametocytes (Lasonder et al., 2016; Miao et al., 2017), with higher protein expression in gametocytes than in asexual blood-stages.
The absence of clear mCherry signals in bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== asexual blood-stages was unexpected since expression of ETRAMP10.3 in asexual blood-stages had been detected by mass spectrometry-based proteomics (Florens et al., 2002; Silvestrini et al., 2010) and also been confirmed by immunofluorescence analysis using anti-ETRAMP10.3 antibodies (Mackellar et al., 2010). Moreover, unsuccessful attempts to delete the P. falciparum etramp10.3 gene indicates that it is essential during asexual blood-stage development (Mackellar et al., 2010). It is possible that fluorescence microscopy analysis is too insensitive to detect low level mCherry expression from the mCherry-luciferase fusion protein in asexual blood-stages. We therefore performed luminescence assays to analyse luciferase expression in bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== asexual blood-stages. Luminescence signals in parasites obtained from pure asexual blood-stage cultures were significantly higher than those of uninfected red blood cells (p < 0.0005 in culture wells with more than 104 parasites; Figure 2C), indicating that asexual blood-stages express the mCherry-luciferase protein. Stage IV/V gametocytes obtained from gametocyte cultures had on average 30-fold higher luminescence values (3 exp.; range 15–60 fold) compared to asexual blood-stages. The luminescence values obtained from bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== gametocyte (IV/V) or mixed asexual stage dilution series demonstrate a linear relationship between the number of parasites and signal intensity in the range of 1 × 103 to 1 × 107 parasites for gametocytes and 1 × 104 to 1 × 107 for asexual blood-stage parasites (Figure 2C). Combined our observations show that bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== parasites have high level expression of mCherry and luciferase in stage III-V gametocytes and low-level expression of luciferase in asexual blood-stages.
The essential role of ETRAMP10.3 during asexual blood-stage development and its expression in asexual blood-stages and gametocytes is in contrast to expression of the UIS4 protein of the rodent parasites, P. berghei and P. yoelii. The P. berghei and P. yoelii uis4 gene is dispensable for blood-stage development s (Kaiser et al., 2004; Mueller et al., 2005) and there is no evidence that uis4 is expressed in blood-stages either from proteomic or transcriptomic analyses [www.PlasmoDB.org; (Otto et al., 2014)]. In studies using transgenic parasites expressing fluorescent and/or luminescent proteins either under control of the uis4 promoter or fused to UIS4 we are unable able to detect these reporters in blood-stage parasites (Grützke et al., 2014; Hopp et al., 2015). In contrast UIS4 has been show to play a vital role during liver-stage development in both P. berghei and P. yoelii (Kaiser et al., 2004; Mueller et al., 2005). In a previous study, where the P. yoelii uii4 gene was replaced with the etramp10.3 gene, it was demonstrated that ETRAMP10.3 is unable to complement the function of UIS4 in P. yoelii liver-stages (Mackellar et al., 2010). Combined, these observations indicate that UIS4 and ETRAMP10.4 may have different, or only partially overlapping, roles in rodent and human malaria parasites.
We examined mCherry expression in oocysts and sporozoites, collected from Anopheles stephensi mosquitoes, which were fed with bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== gametocytes using the standard membrane feeding assay. This line produced oocysts and sporozoites in the same range as wild type (WT) NF54 parasites (Supplementary Table 2). Weak mCherry signals were detected in maturing bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== oocysts containing sporozoites (at day 8 and 11 after feeding; Figure 3A), but mCherry signals were not above the background mCherry signals observed with WT oocysts. In contrast, the oocysts (day 8) of the control line mCherry-luc@gapdh were clearly mCherry-positive (Figure 3A).
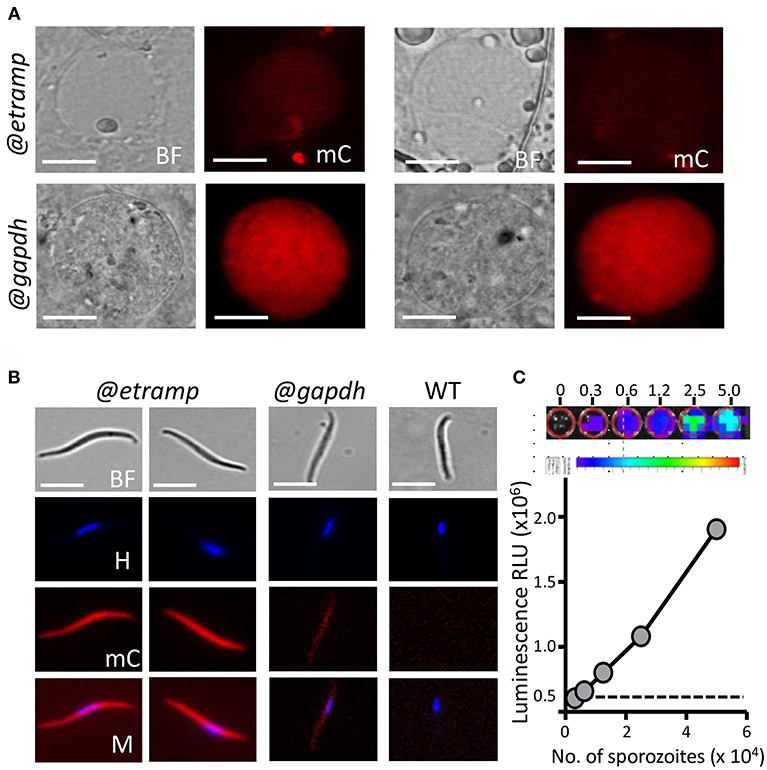
Figure 3. Expression of mCherry and luciferase in oocyst and sporozoites of bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== and mcherry-luc@gapdh parasites. (A) Upper panel: Representative mCherry-fluorescence microscopy image of a live bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== oocyst in A. stephensi mosquitoes at day 10 after infection (@etramp). No mCherry fluorescence signal above background was detected in oocysts. Lower panel: Representative mCherry-fluorescence microscopy image of a live oocysts of the control line mcherry-luc@gapdh (@gapdh). Bright field (BF), mCherry (mC). Scale bar, 20 μm. (B) Representative mCherry-fluorescence microscopy images of live salivary glands sporozoites collected at day 21 after infection of mosquitoes. Left panel: bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== (@etramp) sporozoites. Middle panel: sporozoites of the control line mcherry-luc@gapdh (@gapdh). Right panel: wild type (WT) Pf NF54 sporozoites. Nuclei were stained with Hoechst33342. Bright field (BF), Hoechst (H), mCherry (mC), Merge (M). Scale bar, 7 μm. (C) Correlation between luciferase expression (luminescence, RLU) and number of sporozoites in serial dilutions series (in duplicate) of live bUNoZXJyeUBldHJhbXAxMC4z salivary gland sporozoites. Upper panel: Representative luminescence measurements of live sporozoites determined in 96-well plates. From left to right, rainbow images of a serial dilution of sporozoites (from 0, 0.3, 0.6, 1.2, 2.5, and 5.0 × 104 sporozoites). Relative levels of luminescence range from low (blue), to medium (green), to high (yellow/red). Lower panel: Correlation between luciferase expression (luminescence, RLU, photons/sec) and number of sporozoites. Black dotted line: luminescence values of wells without sporozoites. The mean luminescence values of duplicate samples is shown; error bars represent the standard deviation. Correlation coefficient r (two-tailed Spearman's test): 0.99; p = 0.016.
In contrast to the mCherry negative bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== oocysts, bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== sporozoites collected from salivary glands exhibited strong mCherry expression, whereas lower mCherry signals were detected in mCherry-luc@gapdh sporozoites (Figure 3B; Supplementary Figure 4). The activity of the etramp10.3 promoter in salivary gland sporozoites is in agreement with detection of multiple ETRAMP10.3 peptides in P. falciparum sporozoite proteomes (Lindner et al., 2013; Swearingen et al., 2016). Expression of luciferase in bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== sporozoites was examined and luminescence signals from a dilution series of purified sporozoites exhibit a linear relationship between sporozoite number and luminescence intensity in the range of 1.25 × 104 to 5 × 104 sporozoites (Figure 3C). We compared the luminescence signals of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== sporozoites (5 × 104) with that those of a previously described transgenic sporozoite line that expresses GFP-luciferase fusion protein under control of the eef1α promoter (GFP-luc@eef1α) (Vaughan et al., 2012). The luminescence signal of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== sporozoites was 14-fold higher than the luminescence signal obtained from the same number of GFP-luc@eef1α sporozoites (Figure 4).
Figure 4. Liver stage development of bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== parasites and expression of mCherry and luciferase. (A) Development of liver-stages of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== in cultured cryopreserved primary human hepatocytes and comparison with wild type (WT) and GFP-luc@eef1α. Shown are the percentage of infected hepatocytes (left graph), the size of liver-stages (middle graph; mean surface area; arbitrary units - a.u.) and the fluorescence intensity of Pf HSP70 staining (right graph; arbitrary units a.u. × 106). Hepatocytes were infected with 5 × 104 sporozoites and liver-stage development was analyzed at day 2, 4, and 6 after infection. At least 20 parasites were assessed at each time point. nd: not determined. Significance values (unpaired two-tailed t-test): n.s.–not significant. (B) Representative mCherry-fluorescence microscopy images of a fixed bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== liver stages at day 3, 4, and 5 after infecting cryopreserved primary human hepatocytes with sporozoites. Fixed hepatocytes were stained with rabbit anti-Pf HSP70 and goat anti-mCherry antibodies. Secondary conjugated antibodies used: anti-IgG from rabbit Alexa Fluor® 488 (green) or anti-IgG from goat Alexa Fluor® 594 (red). Nuclei stained with Hoechst-33342. All pictures were recorded with standardized exposure/gain times; Alexa Fluor® 488 (green) 0.7 s; anti-IgG Alexa Fluor® 594 (red) 0.6 s; Hoechst (blue) 0.136 s; bright field 0.62 s (1x gain). Bright field (BF), Hoechst (H), anti-Pf HSP70 (αh), mCherry (mC), Merge (M). Scale bar, 10 μm. mCherry fluorescence was analyzed using bWNoZXJyeS1sdWNAZXRyYW1wMTAuMw== cl3 in two independent experiments. (C) luciferase expression (luminescence, RLU, photons/sec) in sporozoites (spz) and in liver-stages from mCherry-luc@etramp10.3 cl3 and the control line gfp-luc@eef1a. Cultured cryopreserved primary human hepatocytes were infected with 5 × 104 sporozoites and luminescence was measured during a 7 days period. Uninfected hepatocytes were used as a control (Black circle). The mean luminescence value of duplicate samples is shown; error bars represent the standard deviation. (D) Sensitivity of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== liver-stages to atovaquone ([ATQμM], concentration in micromolar). Inhibition of liver-stage development was determined in a single experiment by measurement of luminescence at day 4 after infection of cultured cryopreserved primary human hepatocytes with 5 × 104 sporozoites. The IC50 value (7.9 nM) was calculated by non-linear regression using GraphPad Prism software package 5.04.
We next analyzed reporter expression in liver-stages; however, since mCherry-luc@gapdh sporozoites showed low mCherry expression, we compared bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== liver-stages directly with GFP-luc@eef1α parasites as it has been reported that sporozoites and liver-stages of this line expresses both GFP and luciferase and this line has been used to analyse Pf liver stage development (Vaughan et al., 2012; Sack et al., 2014; Flannery et al., 2018; Foquet et al., 2018). Cultured cryopreserved primary human hepatocytes were infected with 5x104 bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== or GFP-luc@eef1α sporozoites per well of a 96-wells plate. Development of liver-stages at day 2, 4, and 6 after infection of the two transgenic lines and WT liver-stages was examined by immunofluorescence using a rabbit polyclonal antibody against the cytoplasmic P. falciparum protein PfHSP70. The percentage of infected hepatocytes, the size of the liver-stages and the intensity of HSP70 staining were comparable between the transgenic and WT parasites at day 4 after infection and liver-stages of both transgenic lines were comparable at day 6 (Figure 4A).
Expression of mCherry in bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== liver-stages was analyzed at day 3, 4, and 5 after hepatocyte infection by immunofluorescence assay using anti-mCherry antibodies; mCherry signals were detectable in every Hsp70 positive-liver stage parasite on all days (Figure 4B). Expression of luciferase in liver-stages was also examined by luminescence assays (Figure 4C; in duplicate) and we compared the luminescence signals of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== with GFP-luc@eef1α at the same point of development. The luminesce signals of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== liver-stages, while significantly higher than background levels with a peak of expression at day 5 (unpaired T-test:***p ≤ 0.0001), were between 2.2 to 7.6 fold lower than GFP-luc@eef1α from day 3 to 7 (Figure 4C).
To determine if bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== parasites could be used in a plate-based assay to test drug sensitivity of liver-stage parasites we performed a drug assay using atovaquone, which has potent activity against intra-hepatic parasites (Baragaña et al., 2015). The in vitro inhibition of P. falciparum liver-stage development was determined by measuring luminescence signals of bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw==-infected hepatocytes maintained in 96-well plates and incubated with different concentrations of atovaquone (0.001–100 nM). Atovaquone was added to the 96-well cultures of primary human hepatocytes 3 h after the addition of 5 × 104 sporozoites and the cultures were allowed to proceed for 4 days, after which luminescence was determined. In this assay we determined an IC50 value of atovaquone of 7.9 nM (Figure 4D), which is comparable to the previously established IC50 value of atovaquone against liver-stages (Delves et al., 2012).
Combined, our observation show high mCherry and luciferase expression in gametocytes and sporozoites of the bUNoZXJyeS1sdWNAZXRyYW1wMTAuMw== line. Although both mCherry and luciferase expression is detected in liver stages, luciferase expression driven by the etramp10.3 promoter is lower than luciferase expression under the eef1α promoter.
In summary: As with rodent malaria parasites the development and expansion of reporter lines in P. falciparum would increase the range of analyses that can more easily be performed, both to interrogate P. falciparum gene function at different points of development and to permit the miniaturization and rapid screening of compounds or immune sera that target the parasite at different points of development. Specifically, the expression of mCherry (red) instead of GFP (green) can be an critical advantage, especially when transgenic parasites/sporozoites will be used in assays using GFP-expressing host cells. In addition, parasites expressing mCherry in gametocytes will be valuable tools in cross-fertilization assays between GFP-expressing and mCherry expressing parasites (Ukegbu et al., 2015, 2017) for gaining a better understanding of parasite fertilization and mosquito-stage development.
The knowledge of expression of reporters under different promoters, such as the etramp10.3, can be useful for generation of additional (gene knock-out/mutated) mutants that require the expression of transgenes in specific life cycle stages. In contrast to the high expression in gametocytes and sporozoites our studies indicate that the etramp10.3 promoter is less suitable for high reporter expression in liver stages, which is unexpected based on the use of the uis4 promoter of rodent parasites. We demonstrate that the reporter expression in sporozoites under control of the etramp10.3 promoter is much higher than under control of the eef1a promoter and therefore this line can be a valuable tool for studies on the biology of P. falciparum sporozoites, for example to visualize sporozoite migration studies in human skin explants (performed in our laboratory) or to visualize and quantify P. falciparum sporozoite invasion of human hepatocytes.
CM-M, CJ, and SK came up with the study concept and design. CM-M, AS, KK, JB, FP, SM, TI, AO, JR, G-JG, SC-M, and BF-F acquired the data. CM-M, CJ, SK, KD, and BF-F conducted analysis and interpretation of the data. CM-M, CJ, and SK wrote the draft of the manuscript. CM-M, AS, BF-F, KD, JB, CJ, SK, RS, and AH critically revised the manuscript for important intellectual content. CM-M, AS, FP, JR, AO, G-JG, HK, SC-M, SM, and BF-F provided technical and/or material support. CJ and SK supervised the study. All authors reviewed the manuscript.
CM-M was supported by Colciencias Ph.D. fellowship (Call 568 from 2012 Resolution 01218 Bogotá, Colombia). AO is supported by a Skim Latihan Akademik IPTA—SLAI (Ministry of Higher Education, Malaysia). TI was, in part, supported by Uehara Memorial Foundation grant. KD and RS hold stock in TropIQ Health Sciences.
KD, RS, KK, and JB are employed by the company TropIQ Health Sciences.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with one of the authors SK.
The authors are grateful to Professor Stefan Kappe for providing the GFP-luc@eef1α parasite line.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00096/full#supplementary-material
Annoura, T., Chevalley, S., Janse, C. J., Franke-Fayard, B., and Khan, S. M. (2013). Quantitative analysis of Plasmodium berghei liver stages by bioluminescence imaging. Methods Mol. Biol. 923, 429–443. doi: 10.1007/978-1-62703-026-7_30
Aurrecoechea, C., Brestelli, J., Brunk, B. P., Dommer, J., Fischer, S., Gajria, B., et al. (2009). PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37:D539–543. doi: 10.1093/nar/gkn814
Baragaña, B., Hallyburton, I., Lee, M. C., Norcross, N. R., Grimaldi, R., Otto, T. D., et al. (2015). A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315–320. doi: 10.1038/nature14451
Boes, A., Spiegel, H., Kastilan, R., Bethke, S., Voepel, N., Chudobová, I., et al. (2016). Analysis of the dose-dependent stage-specific in vitro efficacy of a multi-stage malaria vaccine candidate cocktail. Malar. J. 15:279. doi: 10.1186/s12936-016-1328-0
Combe, A., Giovannini, D., Carvalho, T. G., Spath, S., Boisson, B., Loussert, C., et al. (2009). Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe 5, 386–396. doi: 10.1016/j.chom.2009.03.008
Delves, M., Plouffe, D., Scheurer, C., Meister, S., Wittlin, S., Winzeler, E. A., et al. (2012). The activities of current antimalarial drugs on the life cycle stages of Plasmodium: a comparative study with human and rodent parasites. PLoS Med. 9:e1001169. doi: 10.1371/journal.pmed.1001169
Flannery, E. L., Foquet, L., Chuenchob, V., Fishbaugher, M., Billman, Z., Navarro, M. J., et al. (2018). Assessing drug efficacy against Plasmodium falciparum liver stages in vivo. JCI Insight 3:1. doi: 10.1172/jci.insight.92587
Florens, L., Washburn, M. P., Raine, J. D., Anthony, R. M., Grainger, M., Haynes, J. D., et al. (2002). A proteomic view of the Plasmodium falciparum life cycle. Nature 419, 520–526. doi: 10.1038/nature01107
Foquet, L., Schafer, C., Minkah, N. K., Alanine, D. G. W., Flannery, E. L., Steel, R. W. J., et al. (2018). Plasmodium falciparum liver stage infection and transition to stable blood stage infection in liver-humanized and blood-humanized FRGN KO mice enables testing of blood stage inhibitory antibodies (reticulocyte-binding protein homolog 5) in vivo. Front. Immunol. 9:524. doi: 10.3389/fimmu.2018.00524
Ghorbal, M., Gorman, M., Macpherson, C. R., Martins, R. M., Scherf, A., and Lopez-Rubio, J. J. (2014). Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 32, 819–821. doi: 10.1038/nbt.2925
Grützke, J., Rindte, K., Goosmann, C., Silvie, O., Rauch, C., Heuer, D., et al. (2014). The spatiotemporal dynamics and membranous features of the Plasmodium liver stage tubovesicular network. Traffic 15, 362–382. doi: 10.1111/tra.12151
Hopp, C. S., Chiou, K., Ragheb, D. R., Salman, A. M., Khan, S. M., Liu, A. J., et al. (2015). Longitudinal analysis of Plasmodium sporozoite motility in the dermis reveals component of blood vessel recognition. Elife 4. doi: 10.7554/eLife.07789
Kaiser, K., Matuschewski, K., Camargo, N., Ross, J., and Kappe, S. H. (2004). Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol. Microbiol. 51, 1221–1232. doi: 10.1046/j.1365-2958.2003.03909.x
Lasonder, E., Rijpma, S. R., van Schaijk, B. C., Hoeijmakers, W. A., Kensche, P. R., Gresnigt, M. S., et al. (2016). Integrated transcriptomic and proteomic analyses of P. falciparum gametocytes: molecular insight into sex-specific processes and translational repression. Nucleic Acids Res. 44, 6087–6101. doi: 10.1093/nar/gkw536
Le Bihan, A., de Kanter, R., Angulo-Barturen, I., Binkert, C., Boss, C., Brun, R., et al. (2016). Characterization of novel antimalarial compound ACT-451840: preclinical assessment of activity and dose-efficacy modeling. PLoS Med. 13:e1002138. doi: 10.1371/journal.pmed.1002138
Lindner, S. E., Swearingen, K. E., Harupa, A., Vaughan, A. M., Sinnis, P., Moritz, R. L., et al. (2013). Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol. Cell. Proteomics 12, 1127–1143. doi: 10.1074/mcp.M112.024505
Longley, R. J., Halbroth, B. R., Salman, A. M., Ewer, K. J., Hodgson, S. H., Janse, C. J., et al. (2017). Assessment of the Plasmodium falciparum preerythrocytic antigen UIS3 as a potential candidate for a malaria vaccine. Infect. Immun. 85:3. doi: 10.1128/IAI.00641-16
Longley, R. J., Salman, A. M., Cottingham, M. G., Ewer, K., Janse, C. J., Khan, S. M., et al. (2015). Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates. Sci. Rep. 5:11820. doi: 10.1038/srep11820
López-Barragán, M. J., Lemieux, J., Quinones, M., Williamson, K. C., Molina-Cruz, A., Cui, K., et al. (2011). Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12:587. doi: 10.1186/1471-2164-12-587
Lucantoni, L., Fidock, D. A., and Avery, V. M. (2016). Luciferase-based, high-throughput assay for screening and profiling transmission-blocking Compounds against Plasmodium falciparum gametocytes. Antimicrob. Agents Chemother. 60, 2097–2107. doi: 10.1128/AAC.01949-15
Mackellar, D. C., O'Neill, M. T., Aly, A. S., Sacci, J. B. Jr., Cowman, A. F., and Kappe, S. H. (2010). Plasmodium falciparum PF10_0164 (ETRAMP10.3) is an essential parasitophorous vacuole and exported protein in blood stages. Eukaryot. Cell 9, 784–794. doi: 10.1128/EC.00336-09
Marin-Mogollon, C., van de Vegte-Bolmer, M., van Gemert, G. J., van Pul, F. J. A., Ramesar, J., Othman, A. S., et al. (2018). The Plasmodium falciparum male gametocyte protein P230p, a paralog of P230, is vital for ookinete formation and mosquito transmission. Sci. Rep. 8:14902. doi: 10.1038/s41598-018-33236-x
Miao, J., Chen, Z., Wang, Z., Shrestha, S., Li, X., Li, R., et al. (2017). Sex-specific biology of the human malaria parasite revealed from the proteomes of mature male and female gametocytes. Mol. Cell Proteomics 16, 537–551. doi: 10.1074/mcp.M116.061804
Mogollon, C. M., van Pul, F. J., Imai, T., Ramesar, J., Chevalley-Maurel, S., de Roo, G. M., et al. (2016). Rapid generation of marker-free P. falciparum fluorescent reporter lines using modified CRISPR/Cas9 constructs and selection protocol. PLoS ONE. 11:e0168362. doi: 10.1371/journal.pone.0168362
Montagna, G. N., Beigier-Bompadre, M., Becker, M., Kroczek, R. A., Kaufmann, S. H., and Matuschewski, K. (2014). Antigen export during liver infection of the malaria parasite augments protective immunity. MBio 5:e01321–e01314. doi: 10.1128/mBio.01321-14
Mueller, A. K., Camargo, N., Kaiser, K., Andorfer, C., Frevert, U., Matuschewski, K., et al. (2005). Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. U. S. A. 102, 3022–3027. doi: 10.1073/pnas.0408442102
Othman, A. S., Marin-Mogollon, C., Salman, A. M., Franke-Fayard, B. M., Janse, C. J., and Khan, S. M. (2017). The use of transgenic parasites in malaria vaccine research. Expert. Rev. Vaccines 16, 1–13. doi: 10.1080/14760584.2017.1333426
Otto, T. D., Böhme, U., Jackson, A. P., Hunt, M., Franke-Fayard, B., Hoeijmakers, W. A., et al. (2014). A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 12:86. doi: 10.1186/s12915-014-0086-0
Panchal, D., Govindasamy, K., Rana, A., and Bhanot, P. (2012). Improved Plasmodium berghei lines for conditional mutagenesis. Mol. Biochem. Parasitol. 184, 52–54. doi: 10.1016/j.molbiopara.2012.03.005
Ponnudurai, T., Lensen, A. H., Van Gemert, G. J., Bensink, M. P., Bolmer, M., and Meuwissen, J. H. (1989). Infectivity of cultured Plasmodium falciparum gametocytes to mosquitoes. Parasitology 98 (Pt 2), 165–173. doi: 10.1017/S0031182000062065
Ponnudurai, T., van Gemert, G. J., Bensink, T., Lensen, A. H., and Meuwissen, J. H. (1987). Transmission blockade of Plasmodium falciparum: its variability with gametocyte numbers and concentration of antibody. Trans. R. Soc. Trop. Med. Hyg. 81, 491–493. doi: 10.1016/0035-9203(87)90172-6
Sack, B. K., Miller, J. L., Vaughan, A. M., Douglass, A., Kaushansky, A., Mikolajczak, S., et al. (2014). Model for in vivo assessment of humoral protection against malaria sporozoite challenge by passive transfer of monoclonal antibodies and immune serum. Infect. Immun. 82, 808–817. doi: 10.1128/IAI.01249-13
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Silva, P. A., Guerreiro, A., Santos, J. M., Braks, J. A., Janse, C. J., and Mair, G. R. (2016). Translational control of UIS4 protein of the host-parasite interface is mediated by the RNA binding protein Puf2 in Plasmodium berghei sporozoites. PLoS ONE. 11:e0147940. doi: 10.1371/journal.pone.0147940
Silvestrini, F., Lasonder, E., Olivieri, A., Camarda, G., van Schaijk, B., Sanchez, M., et al. (2010). Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol. Cell Proteomics 9, 1437–1448. doi: 10.1074/mcp.M900479-MCP200
Silvie, O., Briquet, S., Müller, K., Manzoni, G., and Matuschewski, K. (2014). Post-transcriptional silencing of UIS4 in Plasmodium berghei sporozoites is important for host switch. Mol. Microbiol. 91, 1200–1213. doi: 10.1111/mmi.12528
Singer, M., Marshall, J., Heiss, K., Mair, G. R., Grimm, D., Mueller, A. K., et al. (2015). Zinc finger nuclease-based double-strand breaks attenuate malaria parasites and reveal rare microhomology-mediated end joining. Genome Biol. 16:249. doi: 10.1186/s13059-015-0811-1
Swann, J., Corey, V., Scherer, C. A., Kato, N., Comer, E., Maetani, M., et al. (2016). High-throughput luciferase-based assay for the discovery of therapeutics that prevent malaria. ACS Infect. Dis. 2, 281–293. doi: 10.1021/acsinfecdis.5b00143
Swearingen, K. E., Lindner, S. E., Shi, L., Shears, M. J., Harupa, A., Hopp, C. S., et al. (2016). Interrogating the plasmodium sporozoite surface: identification of surface-exposed proteins and demonstration of glycosylation on CSP and TRAP by mass spectrometry-based proteomics. PLoS Pathog. 12:e1005606. doi: 10.1371/journal.ppat.1005606
Talman, A. M., Blagborough, A. M., and Sinden, R. E. (2010). A Plasmodium falciparum strain expressing GFP throughout the parasite's life-cycle. PLoS ONE 5:e9156. doi: 10.1371/journal.pone.0009156
Ukegbu, C. V., Cho, J. S., Christophides, G. K., and Vlachou, D. (2015). Transcriptional silencing and activation of paternal DNA during Plasmodium berghei zygotic development and transformation to oocyst. Cell Microbiol. 17, 1230–1240. doi: 10.1111/cmi.12433
Ukegbu, C. V., Giorgalli, M., Yassine, H., Ramirez, J. L., Taxiarchi, C., Barillas-Mury, C., et al. (2017). Plasmodium berghei P47 is essential for ookinete protection from the Anopheles gambiae complement-like response. Sci. Rep. 7:6026. doi: 10.1038/s41598-017-05917-6
van Dijk, M. R., van Schaijk, B. C., Khan, S. M., van Dooren, M. W., Ramesar, J., Kaczanowski, S., et al. (2010). Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog. 6:e1000853. doi: 10.1371/journal.ppat.1000853
Vaughan, A. M., Mikolajczak, S. A., Camargo, N., Lakshmanan, V., Kennedy, M., Lindner, S. E., et al. (2012). A transgenic Plasmodium falciparum NF54 strain that expresses GFP-luciferase throughout the parasite life cycle. Mol. Biochem. Parasitol. 186, 143–147. doi: 10.1016/j.molbiopara.2012.10.004
Vos, M. W., Stone, W. J., Koolen, K. M., van Gemert, G. J., van Schaijk, B., Leroy, D., et al. (2015). A semi-automated luminescence based standard membrane feeding assay identifies novel small molecules that inhibit transmission of malaria parasites by mosquitoes. Sci. Rep. 5:18704. doi: 10.1038/srep18704
Keywords: Plasmodium falciparum, transgenes, reporters, mCherry, luciferase, CRISPR/Cas9
Citation: Marin-Mogollon C, Salman AM, Koolen KMJ, Bolscher JM, van Pul FJA, Miyazaki S, Imai T, Othman AS, Ramesar J, van Gemert G-J, Kroeze H, Chevalley-Maurel S, Franke-Fayard B, Sauerwein RW, Hill AVS, Dechering KJ, Janse CJ and Khan SM (2019) A P. falciparum NF54 Reporter Line Expressing mCherry-Luciferase in Gametocytes, Sporozoites, and Liver-Stages. Front. Cell. Infect. Microbiol. 9:96. doi: 10.3389/fcimb.2019.00096
Received: 14 December 2018; Accepted: 22 March 2019;
Published: 16 April 2019.
Edited by:
Justin Boddey, Walter and Eliza Hall Institute of Medical Research, AustraliaReviewed by:
Vasant Muralidharan, University of Georgia, United StatesCopyright © 2019 Marin-Mogollon, Salman, Koolen, Bolscher, van Pul, Miyazaki, Imai, Othman, Ramesar, van Gemert, Kroeze, Chevalley-Maurel, Franke-Fayard, Sauerwein, Hill, Dechering, Janse and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shahid M. Khan, cy5tLmtoYW5AbHVtYy5ubA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.