- 1Department of Immunology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand
- 2Microbiology Laboratory, Faculty of Veterinary Science, Veterinary Diagnostic Center, Mahidol University, Nakhon Pathom, Thailand
- 3The Monitoring Surveillance Center for Zoonotic Diseases in Wildlife and Exotic Animals, Faculty of Veterinary Science, Mahidol University, Nakhon Pathom, Thailand
- 4The Roslin Institute and Royal (Dick) School of Veterinary Studies, University of Edinburgh, Midlothian, United Kingdom
The intracellular pathogen Burkholderia pseudomallei, the etiological agent of melioidosis in humans and various animals, is capable of survival and movement within the cytoplasm of host cells by a process known as actin-based motility. The bacterial factor BimA is required for actin-based motility through its direct interaction with actin, and by mediating actin polymerization at a single pole of the bacterium to promote movement both within and between cells. However, little is known about the other bacterial proteins required for this process. Here, we have investigated the role of the bimC gene (bpss1491) which lies immediately upstream of the bimA gene (bpss1492) on the B. pseudomallei chromosome 2. Conserved amongst all B. pseudomallei, B. mallei and B. thailandensis strains sequenced to date, this gene encodes an iron-binding protein with homology to a group of proteins known as the bacterial autotransporter heptosyltransferase (BAHT) family. We have constructed a B. pseudomallei bimC deletion mutant and demonstrate that it is defective in intracellular survival in HeLa cells, but not in J774.1 macrophage-like cells. The bimC mutant is defective in cell to cell spread as demonstrated by ablation of plaque formation in HeLa cells, and by the inability to form multi-nucleated giant cells in J774.1 cells. These phenotypes in intracellular survival and cell to cell spread are not due to the loss of expression and polar localization of the BimA protein on the surface of intracellular bacteria, however they do correlate with an inability of the bacteria to recruit and polymerize actin. Furthermore, we also establish a role for bimC in virulence of B. pseudomallei using a Galleria mellonella larvae model of infection. Taken together, our findings indicate that B. pseudomallei BimC plays an important role in intracellular behavior and virulence of this emerging pathogen.
Introduction
Burkholderia pseudomallei, a Gram-negative environmental saprophyte, is the causative agent of melioidosis, a potentially life-threatening infectious disease affecting both humans and animals in certain areas of the tropics (Cheng and Currie, 2005). The pathogen is endemic in South East Asia (Limmathurotsakul et al., 2014) and Northern Australia where the bacterium has been detected in drinking water and soil (Currie et al., 2001; Kaestli et al., 2009; Draper et al., 2010). The infection is typically acquired through inhalation, skin penetration or possibly through ingestion; and the resulting disease can manifest with clinically diverse signs including skin abscesses, severe pneumonia and septic shock (Cheng et al., 2013). In North East Thailand, the case fatality rate of melioidosis is around 40% and the incidence of melioidosis continues to increase annually (Limmathurotsakul et al., 2010). Additionally, recent modeling data suggests that the incidence of melioidosis is around 165,000 cases per year worldwide, of which 89,000 are fatal (Limmathurotsakul et al., 2016). B. pseudomallei has gained much attention in recent years over concerns regarding its potential for use as a biological warfare agent. The aerosol infectivity, high mortality rate, and the absence of an effective human vaccine for the prevention of melioidosis has resulted in B. pseudomallei being listed as a Tier 1 select agent in the U.S. There is a continuing need to characterize virulence mechanisms of B. pseudomallei pathogenesis to inform novel strategies for disease prevention and control.
As a facultative intracellular bacterium, B. pseudomallei is capable of entering both phagocytic and non-phagocytic cells. Following phagocytosis, B. pseudomallei has evolved mechanisms to escape the phagosome into the host cell cytoplasm (Jones et al., 1996; Harley et al., 1998; Kespichayawattana et al., 2000), where it replicates and causes host cell death by induction of apoptosis (Kespichayawattana et al., 2000; Sun et al., 2005). Whilst in the cytoplasm, B. pseudomallei subverts host cellular actin dynamics to induce actin polymerization at one bacterial pole leading to its movement within and between infected cells by a process known as actin-based motility, as seen for several other intracellular pathogens (Stevens et al., 2002, 2005b; Gouin et al., 2005). Actin-based motility is thought to underlie the ability of B. pseudomallei to spread into adjacent cells in the absence of immune surveillance, thereby contributing to the formation of multi-nucleated giant cells (MNGCs) (Kespichayawattana et al., 2000) and virulence. This unique phenotype may be relevant to pathogenesis, since granuloma formation and generation of MNGCs are also found in tissues of melioidosis patients (Wong et al., 1995). Infection of J774.2 murine macrophage-like cells with a B. pseudomallei bimA mutant showed a complete lack of actin tail formation, demonstrating that this process is dependent on the BimA protein (Stevens et al., 2005b), a putative Type V autosecreted protein exhibiting carboxyl-terminal homology to the oligomeric Yersinia adhesion protein and bacterial autotransporter YadA (Stevens et al., 2005b; Lazar Adler et al., 2011). The BimA protein is encoded by all B. pseudomallei strains sequenced to date (Sitthidet et al., 2008). BimA is anchored to one pole of the bacterial outer membrane during actin tail formation in infected cells, interacts directly with cellular actin and is capable of polymerizing actin into filaments in the absence of any other cellular or bacterial proteins (Stevens et al., 2005b). Specific domains of the BimA protein with homology to eukaryotic proteins have been implicated in actin polymerization and cell to cell spread (Sitthidet et al., 2011).
Recently a proteomic approach has identified candidate host cell proteins involved in actin polymerization mediated by BimA (Jitprasutwit et al., 2016). For example the cellular scaffold protein IQGAP1 plays a role in regulating the actin density and tail length of B. pseudomallei in infected HeLa cells (Jitprasutwit et al., 2016). Interestingly, BimA proteins from the closely related virulent B. mallei and the avirulent B. thailandensis differ significantly in amino acid sequence at the N-terminus of the proteins, resulting in the use of distinctly different mechanisms of actin polymerization between B. pseudomallei and B. thailandensis (Stevens et al., 2005a; Sitthidet et al., 2010; Benanti et al., 2015).
The B. pseudomallei bimA gene (annotated as bpss1492 in the reference K96243 genome) is encoded on the second smaller chromosome within an operon of several co-regulated genes, including genes encoding the VirAG two component system required for regulation of the virulence-associated T6SS of B. pseudomallei (Burtnick and Brett, 2013). B. pseudomallei bimA lies downstream of a gene encoding a putative glycosyltransferase (bpss1491, bimC). The predicted BimC protein shows amino acid sequence homology with members of the bacterial autotransporter heptosyltransferase (BAHT) family (Lu et al., 2014) including the diarrheagenic Escherichia coli AAH protein (Autotransporter Adhesin Heptosyltransferase). AAH protein catalyzes the addition of heptose molecules to the Escherichia coli virulence factor AIDA-I (Adhesin Involved in Diffuse Adherence) to mediate bacterial autoaggregation and adhesion to the host cell (Sherlock et al., 2004; Benz and Schmidt, 2011). Interestingly, a Tn5-based transposon mutagenesis screen in B. thailandensis revealed a role for the bimC gene in actin tail formation and MNGC formation (Lu et al., 2015). In addition, the authors demonstrated that expression of an N-terminal tagged B. thailandensis BimC protein from a prokaryotic vector was required for polar localization of a similarly expressed N-terminal tagged B. thailandensis BimA protein, likely facilitated through a direct interaction between the two proteins (Lu et al., 2015).
In the present study, we aimed to characterize the role of native BimC protein in a virulent B. pseudomallei strain. We constructed and characterized a B. pseudomallei ΔbimC deletion mutant and determined its role in B. pseudomallei intracellular survival and virulence. Interestingly we showed that the mutant was defective in net intracellular replication in HeLa cells, a finding that was not reproduced in the macrophage-like cell line J774.1. Unlike the findings of Lu et al. (2015), who studied the B. thailandensis bimC gene, deletion of bimC in B. pseudomallei did not affect the expression or polar localization of native BimA protein on the surface of intracellular bacteria, although it did prevent actin recruitment and polymerization. The ability of the B. pseudomallei ΔbimC deletion mutant to induce plaques and MNGCs were also studied in vitro, and the role of bimC in pathogenesis was assessed using a Galleria mellonella larvae surrogate model of infection.
Materials and Methods
Bacterial Strains, Cell Lines, and Culture Conditions
B. pseudomallei strain 10276, a clinical isolate from a human melioidosis patient, was kindly provided by Prof. Ty Pitt (Health Protection Agency, Colindale, United Kingdom) (Maegraith and Leithead, 1964). Construction of the ΔbimA mutant is described in Jitprasutwit et al. (2016). All experiments with B. pseudomallei and its derivatives were conducted in a Biosafety Level-3 Laboratory at Mahidol University with Institutional Biosafety Committee approval. E. coli DH5α, λpir-116, and S17-1 λpir strains were used as bacterial host strains for molecular cloning or conjugation. Bacteria were routinely cultured at 37°C in Luria-Bertani (LB; Difco) agar or broth. Chloramphenicol (10 μg/ml for E. coli and 50 μg/ml for B. pseudomallei) was added to the medium when required. Yeast extract tryptone (YT) agar (Difco) lacking NaCl and supplemented with 15% sucrose was used for resolution of merodiploids during allelic exchange (Logue et al., 2009). Murine macrophage J774A.1 cells and human epithelial HeLa cells were obtained from the American Type Culture Collection (ATCC) and were cultured in Dulbecco's Modified Eagle medium (DMEM; Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone) at 37°C in the presence of 5% CO2. Unless otherwise stated, all reagents were purchased from Sigma-Aldrich.
Construction of an Isogenic ΔbimC Deletion Mutant and Complemented Strain
Mutagenesis of the B. pseudomallei bimC gene was performed using a homology recombination and sacB counter-selection approach using the suicide replicon pDM4, essentially as previously described (Logue et al., 2009). To produce the ΔbimC deletion mutant, ~400-bp DNA upstream and downstream of B. pseudomallei bimC (bpss1491) were amplified (BimC-P1/P2 and BimC-P3/P4; Table S1), kinase-treated and ligated before a second round of PCR using primers BimC-P1 and BimC-P4. The resulting PCR product was digested with SpeI and XbaI before ligating with similarly digested pDM4. The recombinant pDM4 ΔbimC plasmid was introduced into B. pseudomallei strain 10276 by conjugation. Merodiploids were selected on agar plates containing 50 μg/ml chloramphenicol, and merodiploid resolution was selected by plating on YT agar (without NaCl) supplemented with 15% sucrose at 30°C (Logue et al., 2009). Construction of the ΔbimC deletion mutant was confirmed by PCR (using the primers listed in Table S1) and DNA sequencing.
For complementation of the ΔbimC mutant, pBHR1-bimC encoding the complete bimC ORF was constructed by PCR amplification of the 10276 bimC gene using the primers listed in Table S1. Following verification by DNA sequencing, the plasmid was introduced into the ΔbimC mutant by electroporation (Choi and Schweizer, 2005) to produce the strain ΔbimC/pBHR1-bimC, which was verified by plasmid DNA extraction and further DNA sequencing.
Net Intracellular Replication Assay
The net intracellular replication of B. pseudomallei in macrophage-like and epithelial cells were assessed as described previously (Muangsombut et al., 2008) with some modifications. Essentially, J774A.1 or HeLa cells were seeded at a density of 2.5 x 105 cells per well of a 24-well tissue culture plate and infected ~24 h later with B. pseudomallei wild-type (10276) or its derivatives at a multiplicity of infection (MOI) of 0.5 or 50, respectively. At 2 h post infection, infected cells were overlaid with DMEM medium containing kanamycin (250 μg/ml) to kill extracellular bacteria. The infected cells were subsequently lysed at 2, 6, 12, 18, and 24 h post infection with PBS containing 0.1% Triton X-100. The intracellular bacteria were serially diluted and plated on tryptic soy agar. Colony forming units (CFU) were counted after 36–48 h incubation at 37°C.
Plaque and MNGC Formation Assays
Burkholderia-induced plaque assays were performed as described earlier (Pumirat et al., 2014) with some modifications. Cells were infected with B. pseudomallei wild-type or its derivatives at MOI of 50 for 2 h. Then, the infected cell monolayers were overlaid with fresh medium containing 250 μg/ml of kanamycin to kill extracellular bacteria and incubated statically for 24–48 h before crystal violet staining. Plaque-forming efficiency was calculated as the number of plaques divided by bacterial CFU added per well. Detection of B. pseudomallei-induced MNGC formation in J774A.1 and HeLa cells was assessed by confocal microscopy.
Immunostaining and Confocal Microscopy
J774A.1 or HeLa cells were infected with B. pseudomallei wild-type or its derivatives at a multiplicity of infection (MOI) of 0.5 or 50, respectively. At the experimental end-point, the infected cells were fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) overnight. Cells were permeabilized with PBS containing 0.1% (v/v) Triton X-100 for 30 min and incubated with PBS containing 1% (w/v) bovine serum albumin (BSA) for 30 min at room temperature. Bacteria were stained with rabbit polyclonal anti-B. pseudomallei lipopolysaccharide antibody (kindly provided by Prof. R.W. Titball, Exeter University, UK) at 37°C for 1 h, washed with PBS and then incubated with goat anti-rabbit antibody-Alexa Fluor488 (Molecular Probes). Nuclei were stained with DAPI (4', 6-Diamidino-2-Phenylindole, Dihydrochloride; Molecular Probes). F-actin was directly stained using phalloidin568 (Molecular Probes). BimA protein was detected with a panel of three previously described monoclonal antibodies (Stevens et al., 2005b). Bound anti-BimA was detected with goat anti-mouse antibody-AlexaFluor568 (Molecular Probes). Images were captured using an LSM 510 laser scanning confocal microscope (Carl Zeiss).
Galleria Mellonella Killing Assay
G. mellonella killing assays were performed according to Wand et al. (2011), with some modifications. Larvae between 2 and 2.5 cm and free of melanization or injury were used in the experiments. To prepare the bacterial culture for infection, the overnight culture of B. pseudomallei wild-type or its derivatives were adjusted to the concentration to 104 CFU per ml in PBS. A Hamilton syringe was used to inject 10 μl bacterial suspension into the G. mellonella larvae. Injections were performed directly into the larval body cavity and groups of 10 larvae were injected with each bacterial strain. Control larvae were injected with PBS. Following injection, larvae were incubated in the dark at 37°C and the number of dead larvae were recorded at a variety of times post injection. The virulence test was performed three times on separate occasions.
Statistical Analysis
For in vivo mutant characterization, a log-rank (Mantel-Cox) test was used to compare survival curves using Graph Pad Prism7 software. Data from the experiments for comparison between groups were collected from three independent experiments and analyzed using the student's unpaired t-test using Graph Pad Prism7 software. The results were considered significant if the P < 0.05.
Results
Generation of a B. pseudomallei BimC Deletion Mutant and a Complemented Strain
BimC is completely conserved amongst all B. pseudomallei, B. mallei, B. thailandensis and B. oklahomensis strains studied to date, with over 90% amino acid identity. Although it is notable that B. pseudomallei BimC proteins possess a 13 amino acid C-terminal extension missing from the BimC proteins of the other closely related Burkholderia species (Figure S1).
The bimC gene (bpss1491) is situated directly upstream of the bimA gene (bpss1492) within an operon of genes including the two-component regulator virAG (Figure S2A). To investigate the function of the bimC gene, we created a B. pseudomallei bimC deletion mutant (ΔbimC) using the suicide replicon pDM4 and unmarked allelic-exchange mutagenesis as previously described (Logue et al., 2009). The bimC merodiploid strain yielded DNA fragments consistent with both the presence of the wild-type allele and the deletion allele introduced on the pDM4 plasmid (Figure S2B). Following growth on sucrose plates, deletion of the bimC gene was confirmed by PCR using primers flanking the deletion region (Figure S2B). This strain was also verified across the deletion junction by DNA sequencing. A constitutive prokaryotic expression plasmid containing bimC (pBHR1-bimC) was introduced into the ΔbimC mutant to give rise to the complemented strain ΔbimC/pBHR1-bimC. Neither deletion of the bimC gene nor introduction of the plasmid-borne bimC gene resulted in any detectable difference in growth of the mutants in LB broth compared to the isogenic parent strain 10276 (data not shown).
BimC Is Essential for Actin-Based Motility of B. pseudomallei in Cultured Cells
B. pseudomallei expresses the BimA protein to recruit cellular actin at the pole of the bacterium where it catalyzes its polymerization to promote its movement within and between infected host cells (Stevens et al., 2005b; Sitthidet et al., 2011). To determine the effect of bimC deletion on B. pseudomallei induced-actin tail formation, HeLa cells were infected with the B. pseudomallei 10276 and mutant (ΔbimC, ΔbimA, and ΔbimC/pBHR1-bimC) strains, and the cells were analyzed by immunofluorescence staining and confocal microscopy. At 12 h post infection, wild-type B. pseudomallei exhibited many intracellular bacteria with intense filamentous actin staining proximal to one bacterial pole (Figure 1A). In contrast, no such membrane protrusions or F-actin filaments were detected in any of the cells infected with the ΔbimC or ΔbimA mutant (Figures 1B,C), despite demonstrating efficient escape of these strains into the cell cytoplasm that was indistinguishable from WT bacteria (see Figure S3 and Supplemental Methods). The deficiency of actin tail formation of the ΔbimC mutant could be partially restored by plasmid-borne bimC (ΔbimC/pBHR1-bimC) (Figure 1D).
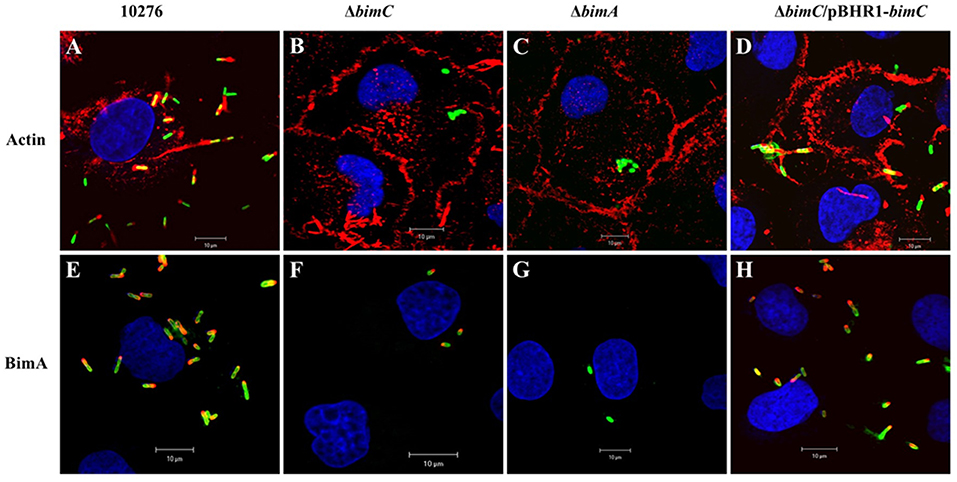
Figure 1. Actin-based motility of B. pseudomallei strains in HeLa cells. B. pseudomallei wild-type strain 10276 (A,E), ΔbimC mutant (B,F), ΔbimA mutant (C,G), and ΔbimC/pBHR1-bimC (D,H) strains were used to infect HeLa cells. At 12 h post infection, the infected epithelial cells and bacteria were stained for bacteria and actin-tails (A–D), or bacteria and BimA protein expression (E–H). Bacteria (green) were stained with anti-B. pseudomallei lipopolysaccharide antibody and detected with anti-rabbit antibody-AlexaFluor488. F-actin (red) was stained with phalloidin-AlexaFluor568 and nuclei (blue) were stained with DAPI. BimA protein (red) was stained with a panel of three monoclonal antibodies detected with anti-mouse antibody-AlexaFluor568. Scale bar = 10 μm.
Interestingly, at the same time point (12 h post infection), intracellular bacteria with polar expression of the BimA protein were visible in cells infected with the ΔbimC mutant, similar to the wild-type strain (Figures 1E,F,H). As expected, the ΔbimA mutant lacked any BimA expression (Figure 1G). Similar data was obtained when J774.1 cells were infected with the same B. pseudomallei strains (Figure S4). Notably, the ΔbimC mutant failed to display actin-based motility, even though intracellular bacteria with polar expression of the BimA protein were detected in infected J774A.1 cells (Figure S4). These results indicate that BimC is required for BimA-mediated B. pseudomallei actin-based motility in infected cells, and that unlike the studies on B. thailandensis bimC (Lu et al., 2015), B. pseudomallei bimC is not required for localization of B. pseudomallei BimA at the pole of the bacterium.
BimC Is Necessary for B. pseudomallei Survival in HeLa Cells
B. pseudomallei has the ability to multiply and survive within a wide range of both phagocytic and non-phagocytic cells. To assess the role of bimC in intracellular survival in non-phagocytic cells, the net intracellular replication of the B. pseudomallei ΔbimC mutant was assessed in comparison with wild-type, ΔbimC/pBHR1-bimC and ΔbimA mutant strains. At 6 h post infection, all B. pseudomallei strains showed a similar number of viable bacteria (CFU) within infected cells, without any statistical difference (Figure 2). In contrast, the number of viable ΔbimC mutant recovered was significantly lower than the wild-type strain at 12, 18, and 24 h post infection (P = 0.0054 at 24 h post infection) (Figure 2B). This defect in intracellular survival and replication could be partially restored by a plasmid-borne bimC gene (Figure 2), indicating that bimC is required for B. pseudomallei intracellular replication within HeLa cells. The intracellular replication defect of the ΔbimC mutant is indistinguishable from that of the ΔbimA mutant (this study and Sitthidet et al., 2011).
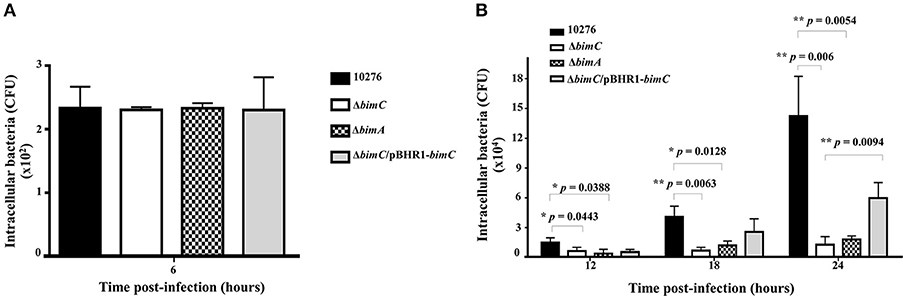
Figure 2. Net intracellular replication and survival of B. pseudomallei strains within HeLa cells. HeLa cells were infected with B. pseudomallei strains at an MOI of 50. At the indicated time point, the numbers of viable intracellular bacteria (CFU) were determined. Data from 6 h post-infection is shown in A and data from 12, 18, and 24 h post-infection in B. The graphs show data for the wild-type (black bars), ΔbimC mutant (white bars), ΔbimA mutant (dotted bars), and ΔbimC/pBHR1-bimC (gray bars) strains. Error bars represent standard errors of the means from three independent experiments (n = 3 biological replicates). Asterisks indicate significant differences (P < 0.01, t-test).
In addition to HeLa cells, intracellular replication within J774A.1 macrophage-like phagocytic cells was also investigated. Interestingly there was no significant difference in the intracellular bacterial load of the wild-type, ΔbimC, ΔbimC/pBHR1-bimC, and ΔbimA mutants in J774A.1 cells, across all time points studied (Figure S5). This may highlight a different role for actin-based motility in these two different cell types, or differences in the pathogen recognition and innate immunity pathways these cell types may deploy to control intracellular replication of B. pseudomallei.
BimC Facilitates B. pseudomallei Intercellular Spreading
We have previously demonstrated a role for bimA in the intercellular spreading of B. pseudomallei in HeLa cell monolayers using a plaque-forming efficiency assay (Sitthidet et al., 2011). Here, at 30 h post infection, HeLa cells infected with B. pseudomallei wild-type demonstrated numerous plaques within the cell monolayer. In contrast, no plaque formation was observed in cells infected with both ΔbimC and ΔbimA mutants. The ΔbimC/pBHR1-bimC complemented strain could partially restore this phenotype (Figure 3). Ablation of plaque formation is likely the result of the limited intracellular replication of the ΔbimC and ΔbimA mutants in HeLa cells, as shown in Figure 2B.
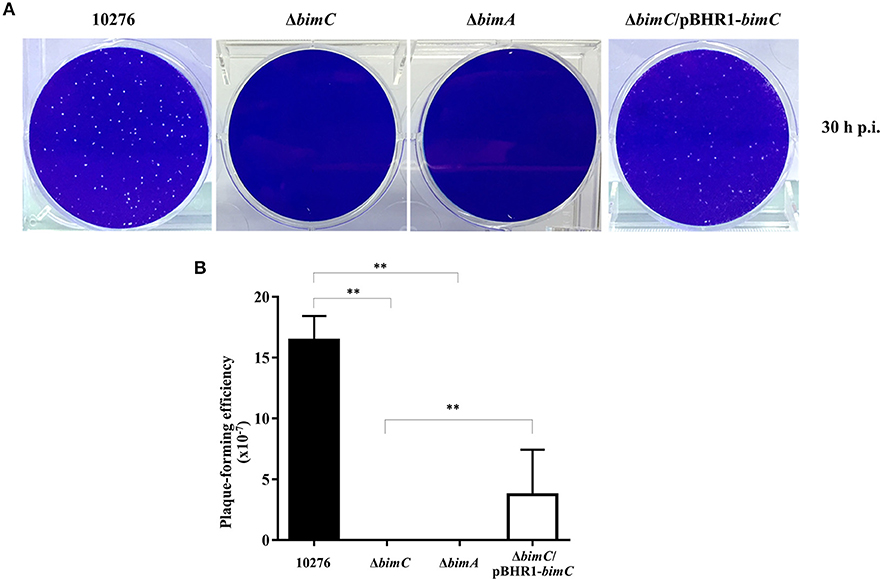
Figure 3. Intercellular spreading of B. pseudomallei strains in HeLa cells. (A) Representative micrographs of cell monolayers and (B) plaque-forming efficiency of HeLa cells infected with B. pseudomallei wild-type (10276), ΔbimC mutant, ΔbimA mutant, or ΔbimC/pBHR1-bimC strains. Plaque-forming efficiency was calculated as the number of plaques at 30 h post infection divided by bacterial CFU added per well. Error bars represent standard errors of the means from three independent experiments (n = 3 biological replicates). Asterisks indicate significant differences (P < 0.01, t-test).
BimC Is Required for B. pseudomallei–Induced MNGC Formation
Another remarkable feature of B. pseudomallei infected host cells is the formation of multi-nucleated giant cells (MNGCs) in both phagocytic and non-phagocytic cell lines (Kespichayawattana et al., 2000). To assess whether bimC had an effect on B. pseudomallei induced MNGC formation, J774A.1 cells were infected with B. pseudomallei, ΔbimC, ΔbimA, and ΔbimC/pBHR1-bimC mutants and subjected to immunofluorescence staining. At 24 h post infection, MNGCs with 10–15 nuclei/cell were readily observed in cells infected with the wild-type strain (Figure 4A). In contrast, no MNGC formation was observed in cells infected with the ΔbimC mutant (Figure 4B) or the ΔbimA mutant (Figure 4C), despite the presence of mutant bacteria within the cytoplasm of these cells. Moreover, cells infected with the ΔbimC/pBHR1-bimC complemented strain displayed MNGC formation, albeit with fewer nuclei (3–5 nuclei/cell) (Figure 4D). The finding that cells infected with the ΔbimC and ΔbimA mutants failed to fuse despite harboring viable bacteria, may indicate a role for these proteins in the fusion process. Until recently the only bacterial factors implicated in this process where LfpA (Boddey et al., 2007), the T6SS-5 (Burtnick et al., 2011) and its associated effector protein Vgr-5 (Schwarz et al., 2014; Toesca et al., 2014).
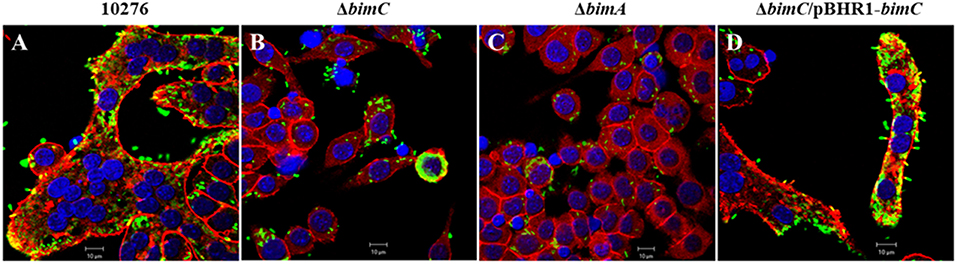
Figure 4. MNGC formation in J774A.1 cells. B. pseudomallei wild-type 10276 (A), ΔbimC mutant (B), ΔbimA mutant (C), and ΔbimC/pBHR1-bimC (D) strains were used to infect J774A.1 cells. At 24 h post infection, the bacteria were stained with anti-B. pseudomallei lipopolysaccharide antibody and anti-rabbit-AlexaFluor488. F-actin (red) was stained with phalloidin-AlexaFluor568 and nuclei (blue) were stained with DAPI. Scale bar = 10 μm.
BimC Contributes to Pathogenesis of B. pseudomallei in a Surrogate Galleria Mellonella Model of Virulence
Data from the in vitro cell-based assays described above indicated that the BimC protein plays roles in various stages of the intracellular life of B. pseudomallei in both phagocytic and non-phagocytic cells. We next investigated the effect of bimC mutation on B. pseudomallei virulence in a Galleria mellonella larvae model. This model has been successfully employed to measure virulence of a wide range of bacterial pathogens including Burkholderia species (Wand et al., 2011). On three separate occasions, G. mellonella larvae were challenged with 100 CFU of B. pseudomallei strains (wild-type, ΔbimC, ΔbimA, and ΔbimC/pBHR1-bimC) and monitored for signs of melanization and death over a 48 h time period. Figure 5 shows representative data from a single experiment. In all experiments, infection of Galleria with either the ΔbimC or ΔbimA mutant showed attenuation of virulence demonstrated by a statistically significant mean time to death (MTTD, p < 0.05). In this assay system we were able to restore the virulence of the ΔbimC strain to levels similar to wild-type B. pseudomallei by expressing bimC on a plasmid (ΔbimC/pBHR1-bimC). This is the first demonstration of a role for the bimC gene in B. pseudomallei virulence.
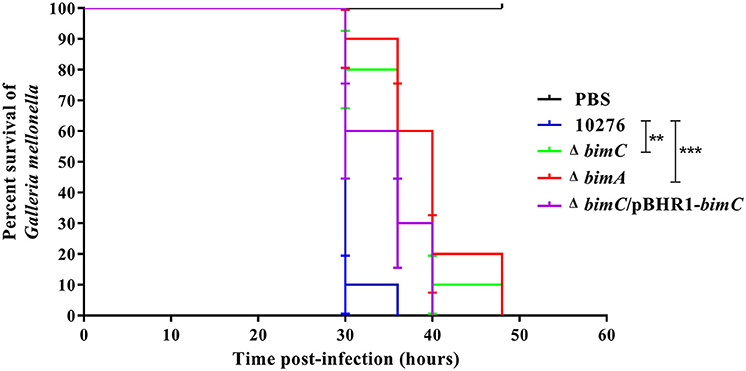
Figure 5. Virulence of B. pseudomallei strains in Galleria mellonella larvae. Representative data from an experiment where groups of 10 insect larvae were challenged with 100 CFU of either B. pseudomallei wild-type 10276, ΔbimC mutant, ΔbimA mutant, or the ΔbimC/pBHR1-bimC strain. The numbers of dead larvae were scored at 24, 30, 36, 40, and 48 h post infection. GraphPad Prism software was used to graph and analyze the data using a Log-rank (Mantel-Cox) test. Asterisks indicate significant differences (P < 0.05) in mean time to death (MTTD) between larvae infected with B. pseudomallei wild-type 10276 and the mutant strains. Data is representative of that obtained in three independent experiments (n = 3 biological replicates).
Discussion
Lu et al. (2014) have recently characterized the B. thailandensis BimC protein, demonstrating a role for this protein in actin-based motility and in the polar localization of BimA, using exogenously expressed tagged proteins. However, the role of B. thailandensis BimC in intracellular survival or pathogenesis was not assessed.
Here we have studied the importance of endogenously expressed B. pseudomallei BimC in actin-based motility, and confirm that it is absolutely required for this process. Furthermore, the B. pseudomallei BimC protein does not affect polar localization of native BimA, as demonstrated by the polar localization of BimA on the surface of ΔbimC bacteria in infected cells (Figure 1F and Figure S4F). This is contradictory to the findings of Lu et al. (2014), where exogenously expressed and tagged B. thailandensis BimC was required for the polar localization of the a similarly exogenously expressed and tagged B. thailandensis BimA protein. The presence of a 13 amino acid C-terminal extension on all of the available B. pseudomallei BimC protein sequences could account for this difference in function. Polar localization of the B. pseudomallei BimA protein may involve an alternative bacterial factor, or factors. Alternatively, it is possible that the study of an exogenously expressed B. thailandensis BimA protein containing a 3X HA tag between the extended signal sequence and the passenger domain, which are required for transport across the inner membrane via the Sec pathway and interaction with the Bam complex for transport across the outer membrane, respectively, may have abrogated its natural export route.
With regard to the role of BimC in intracellular life of B. pseudomallei, here we demonstrate a role for BimC in intracellular survival in HeLa cells. Ablation of the bimC gene resulted in a significant reduction in the net intracellular replication in HeLa cells (around 10-fold compared to wild-type bacteria at 24 h post infection) (Figure 2B), which also led to a complete lack of plaque formation in a cell monolayer at 30 h post infection (Figure 3). This data was mirrored by the ΔbimA mutant, which taken together suggests a key role for both of these proteins involved in actin-based motility in intracellular survival, cell to cell spread, and cell fusion events in this cell line. Perhaps of most interest is the finding that the requirement for both BimC and BimA in intracellular survival was not replicated in the murine macrophage-like J774.1 cells (Figure S5), although a defect in cell to cell spread was observed in this cell type (Figure 4). This may indicate a difference between the cell types in their innate immune responses to invading cytoplasmic bacteria. It is also possible that the macrophage-like cells require the presence of a priming agent such as LPS or IFN-γ for optimal bactericidal activity.
Finally, we utilized the surrogate Galleria mellonella larvae model of B. pseudomallei virulence to assess the role of both BimA and BimC in pathogenesis. A bimA insertion mutant has previously been shown to be attenuated following intraperitoneal injection of BALB/c mice (Lazar Adler et al., 2015) and intranasal infection of C57/Bl mice (M. Stevens and G. Bancroft, unpublished data), a finding that we reproduced in the G. mellonella model using a bimA deletion mutant. In addition, significant attenuation of the ΔbimC mutant was also observed, which was restored by expression of bimC in trans, confirming a role for BimC in virulence.
Whilst we have demonstrated a role for B. pseudomallei BimC in intracellular survival in epithelial cells, BimA-mediated actin-based motility and virulence, we do not know how this protein affects these processes directly. Using a Yeast two hybrid approach, Lu et al. (2014) showed that the B. thailandensis BimC protein is an iron-binding protein that directly interacts with the B. thailandensis BimA protein. The authors also showed that the B. thailandensis BimC protein lacks the predicted glycosyltransferase activity displayed by other members of the BAHT family of glycosyltransferases (Lu et al., 2014), and were unable to hypothesize an alternative biochemical function for the B. thailandensis BimC protein. Since the B. pseudomallei BimC protein clearly does not play a role in the polar localization of the cognate BimA protein, and is unlikely to be a functional glycosyltransferase; further molecular studies will be required to unravel the mechanism underlying the role of the B. pseudomallei BimC protein in pathogenesis of this fascinating micro-organism.
Conclusion
Here we have studied the importance of the BimC protein of B. pseudomallei, the causative agent of melioidosis, in the intracellular life of the pathogen. We have shown that the protein is required for actin-based motility within the host cell cytoplasm. This is associated with a role in intracellular survival and cell to cell spread of the bacterium in HeLa cells, and a significant role in virulence in a surrogate insect model system.
Author Contributions
VS carried out the experiments and was involved in the preparation of the manuscript. SC carried out the experiments. SK was involved in the study design and manuscript preparation. JS contributed to the study design, experiments, and manuscript writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
VS is supported by the Royal Golden Jubilee-Ph.D. program (PHD0177/2554) and Ph.D. scholarship from the Faculty of Medicine Siriraj Hospital, Mahidol University. We are grateful to M. Thongdee (The Monitoring Surveillance Center for Zoonotic Diseases in Wildlife and Exotic Animals, Faculty of Veterinary Science), V. Muangsombat (Department of Immunology, Siriraj Hospital), and P. Songprakhon (Department of Research and Development, Siriraj Hospital) for their kind technical assistance. JS is supported by a BBSRC Institute Strategic Programme Grant (BBS/E/D/20231761).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00063/full#supplementary-material
References
Benanti, E. L., Nguyen, C. M., and Welch, M. D. (2015). Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell 161, 348–360. doi: 10.1016/j.cell.2015.02.044
Benz, I., and Schmidt, M. A. (2011). Structures and functions of autotransporter proteins in microbial pathogens. Int. J. Med. Microbiol. 301, 461–468. doi: 10.1016/j.ijmm.2011.03.003
Boddey, J. A., Day, C. J., Flegg, C. P., Ulrich, R. L., Stephens, S. R., Beacham, I. R., et al. (2007). The bacteria‘l gene lfpA influences the potent induction of calcitonin receptor and osteoclast-related genes in Burkholderia pseudomallei-induced TRAP-positive multinucleated giant cells. Cell. Microbiol. 9, 514–531. doi: 10.1111/j.1462-5822.2006.00807.x
Burtnick, M. N., and Brett, P. J. (2013). Burkholderia mallei and Burkholderia pseudomallei cluster 1 type VI secretion system gene expression is negatively regulated by iron and zinc. PLoS ONE 8:e76767. doi: 10.1371/journal.pone.0076767
Burtnick, M. N., Brett, P. J., Harding, S. V., Ngugi, S. A., Ribot, W. J., Chantratita, N., et al. (2011). The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79, 1512–1525. doi: 10.1128/IAI.01218-10
Cheng, A. C., and Currie, B. J. (2005). Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18, 383–416. doi: 10.1128/CMR.18.2.383-416.2005
Cheng, A. C., Currie, B. J., Dance, D. A., Funnell, S. G., Limmathurotsakul, D., Simpson, A. J., et al. (2013). Clinical definitions of melioidosis. Am. J. Trop. Med. Hyg. 88, 411–413. doi: 10.4269/ajtmh.12-0555
Choi, K. H., and Schweizer, H. P. (2005). An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. doi: 10.1186/1471-2180-5-30
Currie, B. J., Mayo, M., Anstey, N. M., Donohoe, P., Haase, A., and Kemp, D. J. (2001). A cluster of melioidosis cases from an endemic region is clonal and is linked to the water supply using molecular typing of Burkholderia pseudomallei isolates. Am. J. Trop. Med. Hyg. 65, 177–179. doi: 10.4269/ajtmh.2001.65.177
Draper, A. D., Mayo, M., Harrington, G., Karp, D., Yinfoo, D., Ward, L., et al. (2010). Association of the melioidosis agent Burkholderia pseudomallei with water parameters in rural water supplies in Northern Australia. Appl. Environ. Microbiol. 76, 5305–5307. doi: 10.1128/AEM.00287-10
Gouin, E., Welch, M. D., and Cossart, P. (2005). Actin-based motility of intracellular pathogens. Curr. Opin. Microbiol. 8, 35–45. doi: 10.1016/j.mib.2004.12.013
Harley, V. S., Dance, D. A., Tovey, G., Mccrossan, M. V., and Drasar, B. S. (1998). An ultrastructural study of the phagocytosis of Burkholderia pseudomallei. Microbios 94, 35–45.
Jitprasutwit, N., Zainal-Abidin, N., Vander Broek, C., Kurian, D., Korbsrisate, S., Stevens, M. P., et al. (2016). Identification of candidate host cell factors required for actin-based motility of Burkholderia pseudomallei. J. Proteome Res. 15, 4675–4685. doi: 10.1021/acs.jproteome.6b00760
Jones, A. L., Beveridge, T. J., and Woods, D. E. (1996). Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64, 782–790.
Kaestli, M., Mayo, M., Harrington, G., Ward, L., Watt, F., Hill, J. V., et al. (2009). Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in northern Australia. PLoS Negl. Trop. Dis. 3:e364. doi: 10.1371/journal.pntd.0000364
Kespichayawattana, W., Rattanachetkul, S., Wanun, T., Utaisincharoen, P., and Sirisinha, S. (2000). Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 68, 5377–5384. doi: 10.1128/IAI.68.9.5377-5384.2000
Lazar Adler, N. R., Stevens, J. M., Stevens, M. P., and Galyov, E. E. (2011). Autotransporters and their role in the virulence of burkholderia pseudomallei and Burkholderia mallei. Front. Microbiol. 2:151. doi: 10.3389/fmicb.2011.00151
Lazar Adler, N. R., Stevens, M. P., Dean, R. E., Saint, R. J., Pankhania, D., Prior, J. L., et al. (2015). Systematic mutagenesis of genes encoding predicted autotransported proteins of Burkholderia pseudomallei identifies factors mediating virulence in mice, net intracellular replication and a novel protein conferring serum resistance. PLoS ONE 10:e0121271. doi: 10.1371/journal.pone.0121271
Limmathurotsakul, D., Golding, N., Dance, D. A., Messina, J. P., Pigott, D. M., Moyes, C. L., et al. (2016). Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 1:15008. doi: 10.1038/nmicrobiol.2015.8
Limmathurotsakul, D., Wongratanacheewin, S., Teerawattanasook, N., Wongsuvan, G., Chaisuksant, S., Chetchotisakd, P., et al. (2010). Increasing incidence of human melioidosis in Northeast Thailand. Am. J. Trop. Med. Hyg. 82, 1113–1117. doi: 10.4269/ajtmh.2010.10-0038
Limmathurotsakul, D., Wongsuvan, G., Aanensen, D., Ngamwilai, S., Saiprom, N., Rongkard, P., et al. (2014). Melioidosis caused by Burkholderia pseudomallei in drinking water, Thailand, 2012. Emerging Infect. Dis. 20, 265–268. doi: 10.3201/eid2002.121891
Logue, C. A., Peak, I. R., and Beacham, I. R. (2009). Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. J. Microbiol. Methods 76, 320–323. doi: 10.1016/j.mimet.2008.12.007
Lu, Q., Xu, Y., Yao, Q., Niu, M., and Shao, F. (2015). A polar-localized iron-binding protein determines the polar targeting of Burkholderia BimA autotransporter and actin tail formation. Cell. Microbiol. 17, 408–424. doi: 10.1111/cmi.12376
Lu, Q., Yao, Q., Xu, Y., Li, L., Li, S., Liu, Y., et al. (2014). An iron-containing dodecameric heptosyltransferase family modifies bacterial autotransporters in pathogenesis. Cell Host Microbe 16, 351–363. doi: 10.1016/j.chom.2014.08.008
Maegraith, B. G., and Leithead, C. S. (1964). Melioidosis: a case-report. Lancet 1, 862–863. doi: 10.1016/S0140-6736(64)91581-8
Muangsombut, V., Suparak, S., Pumirat, P., Damnin, S., Vattanaviboon, P., Thongboonkerd, V., et al. (2008). Inactivation of Burkholderia pseudomallei bsaQ results in decreased invasion efficiency and delayed escape of bacteria from endocytic vesicles. Arch. Microbiol. 190, 623–631. doi: 10.1007/s00203-008-0413-3
Pumirat, P., Broek, C. V., Juntawieng, N., Muangsombut, V., Kiratisin, P., Pattanapanyasat, K., et al. (2014). Analysis of the prevalence, secretion and function of a cell cycle-inhibiting factor in the melioidosis pathogen Burkholderia pseudomallei. PLoS ONE 9:e96298. doi: 10.1371/journal.pone.0096298
Schwarz, S., Singh, P., Robertson, J. D., Leroux, M., Skerrett, S. J., Goodlett, D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452. doi: 10.1128/IAI.01368-13
Sherlock, O., Schembri, M. A., Reisner, A., and Klemm, P. (2004). Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186, 8058–8065. doi: 10.1128/JB.186.23.8058-8065.2004
Sitthidet, C., Korbsrisate, S., Layton, A. N., Field, T. R., Stevens, M. P., and Stevens, J. M. (2011). Identification of motifs of Burkholderia pseudomallei BimA required for intracellular motility, actin binding, and actin polymerization. J. Bacteriol. 193, 1901–1910. doi: 10.1128/JB.01455-10
Sitthidet, C., Stevens, J. M., Chantratita, N., Currie, B. J., Peacock, S. J., Korbsrisate, S., et al. (2008). Prevalence and sequence diversity of a factor required for actin-based motility in natural populations of Burkholderia species. J. Clin. Microbiol. 46, 2418–2422. doi: 10.1128/JCM.00368-08
Sitthidet, C., Stevens, J. M., Field, T. R., Layton, A. N., Korbsrisate, S., and Stevens, M. P. (2010). Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J. Bacteriol. 192, 5249–5252. doi: 10.1128/JB.00608-10
Stevens, J. M., Ulrich, R. L., Taylor, L. A., Wood, M. W., Deshazer, D., Stevens, M. P., et al. (2005a). Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J. Bacteriol. 187, 7857–7862. doi: 10.1128/JB.187.22.7857-7862.2005
Stevens, M. P., Stevens, J. M., Jeng, R. L., Taylor, L. A., Wood, M. W., Hawes, P., et al. (2005b). Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 56, 40–53. doi: 10.1111/j.1365-2958.2004.04528.x
Stevens, M. P., Wood, M. W., Taylor, L. A., Monaghan, P., Hawes, P., Jones, P. W., et al. (2002). An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46, 649–659. doi: 10.1046/j.1365-2958.2002.03190.x
Sun, G. W., Lu, J., Pervaiz, S., Cao, W. P., and Gan, Y. H. (2005). Caspase-1 dependent macrophage death induced by Burkholderia pseudomallei. Cell. Microbiol. 7, 1447–1458. doi: 10.1111/j.1462-5822.2005.00569.x
Toesca, I. J., French, C. T., and Miller, J. F. (2014). The Type VI secretion system spike protein VgrG5 mediates membrane fusion during intercellular spread by pseudomallei group Burkholderia species. Infect. Immun. 82, 1436–1444. doi: 10.1128/IAI.01367-13
Wand, M. E., Muller, C. M., Titball, R. W., and Michell, S. L. (2011). Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis. BMC Microbiol. 11:11. doi: 10.1186/1471-2180-11-11
Keywords: Burkholderia pseudomallei, BimA, BimC, actin-based motility, intracellular survival, multi-nucleated giant cell, virulence
Citation: Srinon V, Chaiwattanarungruengpaisan S, Korbsrisate S and Stevens JM (2019) Burkholderia pseudomallei BimC Is Required for Actin-Based Motility, Intracellular Survival, and Virulence. Front. Cell. Infect. Microbiol. 9:63. doi: 10.3389/fcimb.2019.00063
Received: 03 September 2018; Accepted: 28 February 2019;
Published: 22 March 2019.
Edited by:
Stephanie M. Seveau, The Ohio State University, United StatesReviewed by:
Javier Pizarro-Cerda, Institut Pasteur, FranceJonathan Mark Warawa, University of Louisville, United States
Copyright © 2019 Srinon, Chaiwattanarungruengpaisan, Korbsrisate and Stevens. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanne M. Stevens, am8uc3RldmVuc0Byb3NsaW4uZWQuYWMudWs=
 Varintip Srinon
Varintip Srinon Somjit Chaiwattanarungruengpaisan
Somjit Chaiwattanarungruengpaisan Sunee Korbsrisate
Sunee Korbsrisate Joanne M. Stevens
Joanne M. Stevens