
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 15 February 2019
Sec. Clinical Microbiology
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00021
A correction has been applied to this article in:
Corrigendum: Comparison of 21-Plex PCR and API 20C AUX, MALDI-TOF MS, and rDNA Sequencing for a Wide Range of Clinically Isolated Yeast Species: Improved Identification by Combining 21-Plex PCR and API 20C AUX as an Alternative Strategy for Developing Countries
 Amir Arastehfar1†
Amir Arastehfar1† Farnaz Daneshnia1†
Farnaz Daneshnia1† Mohammad Kord2
Mohammad Kord2 Maryam Roudbary3
Maryam Roudbary3 Hossein Zarrinfar4
Hossein Zarrinfar4 Wenjie Fang5
Wenjie Fang5 Sayed Jamal Hashemi2,6
Sayed Jamal Hashemi2,6 Mohammad Javad Najafzadeh7
Mohammad Javad Najafzadeh7 Sadegh Khodavaisy2,8*
Sadegh Khodavaisy2,8* Weihua Pan5*
Weihua Pan5* Wanqing Liao5
Wanqing Liao5 Hamid Badali9
Hamid Badali9 Sassan Rezaie2
Sassan Rezaie2 Kamiar Zomorodian10
Kamiar Zomorodian10 Ferry Hagen1
Ferry Hagen1 Teun Boekhout1,5,11
Teun Boekhout1,5,11Occurrence of non-Candida albicans Candida (NCAC) species that are associated with elevated MIC values and therapeutic failures are increasing. As a result, timely and accurate means of identification to the species level is becoming an essential part of diagnostic practices in clinical settings. In this study, 301 clinically isolated yeast strains recovered from various anatomical sites [Blood (n = 145), other sites (n = 156)] were used to assess the accuracy and practicality of API 20C AUX and 21-plex PCR compared to MALDI-TOF MS and large subunit rDNA (LSU rDNA). MALDI-TOF MS correctly identified 98.33% of yeast isolates, 100% of top five Candida species, 95.7% of rare yeast species, while 1.3% of isolates were misidentified. API 20C AUX correctly identified 83.7% of yeast isolates, 97.2% of top five Candida species, 61.8% of rare yeast species, while 16.2% of yeast isolates were misidentified. The 21-plex PCR, accurately identified 87.3% of yeast isolates, 100% of top five Candida species, 72% of rare yeast species, but it misidentified 1.3% of rare yeast species while 9.9% of whole yeast isolates were not identified. The combination of rapidity of 21-plex PCR and comprehensiveness of API 20C AUX, led to correct identification of 92% of included yeast isolates. Due to expensiveness of MALDI-TOF MS and sequencing, this combination strategy could be the most accurate and inexpensive alternative identification strategy for developing countries. Moreover, by the advent and development of cost-effective, reliable, and rapid PCR machines that cost 130 US dollars, 21-plex could be integrated in routine laboratories of developing and resource-limited countries to specifically identify 95% causative agents of yeast-related infections in human. Databases of MALDI-TOF MS, API 20C AUX, and the number of target species identified by 21-plex require further improvement to keep up with the diverse spectrum of yeast species.
Increasing population of immunocompromised patients and administration of broad-spectrum antibiotics etc. (Pappas, 2006), led to a higher occurrence of fungal infections in clinical settings (Yapar, 2014). Among opportunistic yeast species, Candida albicans is continuously reported to be the most commonly encountered yeast species (Pappas et al., 2010). However, applying changes to the clinical practices and interventions resulted in epidemiological landscape and emergence of non-Candida albicans Candida (NCAC) species (Pham et al., 2014). For instance, since the introduction of echinocandins as a prophylactic antifungal, selective pressure has aided in emergence of NCAC species that are less susceptible to this class of antifungals (Pham et al., 2014). Moreover, more frequent isolation of yeast species exhibited inherent less susceptibility/acquired resistance to fluconazole and those with multi-drug resistant traits (MDR) highlight the importance of correct identification (Pfaller et al., 2008; Bizerra et al., 2014; Pham et al., 2014; Chowdhary et al., 2016). Due to the availability of trifle classes of antifungals, monitoring frequency, and epidemiology of yeast species would become an imperative practice in clinical routine laboratories.
Traditionally, phenotypic assays such as direct microscopy, biochemical characterization, and culture are among the most widely used technique to identify yeast species (Posteraro et al., 2015). API 20C AUX, Vitek2 YST ID Card, and AuxaColor are among the most widely exploited biochemical means of identification (Posteraro et al., 2015; Zhao et al., 2017). However, these techniques are time-consuming, labor-intensive, and expensive (Posteraro et al., 2013). Many studies showed that API kits cannot reliably identify rare yeast species, which are less susceptible to routinely used antifungals (Castanheira et al., 2013; Magobo et al., 2014). On the contrary, Sanger sequencing of common barcoding regions and MALDI-TOF MS proved to be the most accurate identification tools (Criseo et al., 2015). Although, these techniques are used in routine laboratories in developed countries, they are regarded as unaffordable devices in developed countries (Posteraro et al., 2013; Criseo et al., 2015; Aslani et al., 2018).
Advances in the machinery of polymerase chain reaction (PCR) has made this device as an affordable identification tool for developing and low-resourced countries. Moreover, due to showing a reasonable reproducibility, WHO recommended PCR as a reliable identification tool in developing countries (Ragheb and Jimenez, 2014). Although, commercial PCR machines still are considered to be expensive (2000–4000 US dollars), it has been shown that using the most basic off-the-shelf tools, it is possible to develop reliable, rapid, and cheap (130 US dollars) PCR machines that without continuous power supply and as efficient as commercial PCR machines can amplify PCR products larger than 1,509 bps (Wong et al., 2015). These kinds of PCR machines can be easily made and expanded as an in on-site diagnostic tools in resource-limited countries (Wong et al., 2015). Unfortunately, there are few PCR-based techniques that can target a comprehensive list of opportunistic yeast species. Automated rep-PCR proved to be a reliable, but expensive assay (Zhao et al., 2017). Recently, Arastehfar et al., have developed a 21-plex PCR assay that targets the most clinically important yeast species, which uses the basic chemistry and devices that are used in routine laboratories (Arastehfar et al., 2018). As 21-plex intended to be used in developing countries, we evaluated its practicality and accuracy compared to a time-consuming and widely used biochemical technique in developing countries, namely API 20C AUX, MALDI-TOF MS, and sequencing of large subunit D1/D2 domains of rDNA (LSU rDNA) as a gold standard.
Due to the diagnostic nature of this study, and the fact that we did not include any clinical samples (blood, serum, CSF, urine, etc…) or biopsies derived from patients, we did not have any consent forms from patients. Isolates investigated in this study were part of previous studies that had been approved by the local ethical committees of Mashhad University of Medical Sciences and Tehran University of Medical Sciences under the following ethical code numbers IR MUMS fm REC.1397.268, and IR. TUMS. .SPH.REC.1396.4195. As such, inclusion of clinical strains in our study did not require institutional ethical approval according to institutional and national guidelines.
Two hundred and ninety-eight clinical yeast strains encompassing a wide range of yeast species that were recovered from clinical sources [blood (n = 145), vagina (n = 71), sputum (n = 35), oral swabs (n = 21), Cerebrospinal fluid (CSF) (n = 8), urine (n = 7), nail (n = 6), tube aspirate (n = 2), penis (n = 1), and throat (n = 1)] were retrospectively collected from Iran and China (Table 1). Due to the importance of Candida auris as an emerging yeast species, and lack of this species in our clinical collection, three CBS reference strains were included. These 301 strains were serially numbered from 1 to 301, prepared as a blinded test set, and three centers were involved for their identification. In the Netherlands two technicians separately performed MALDI-TOF MS and API 20C AUX, In China sequencing of D1/D2 domains of rDNA was carried out, and in Iran, as an example of a developing country a multiplex PCR known as 21-plex was utilized. Strains comprised a diverse range of yeast species, including most and less prevalent Candida species and basidiomyceteous yeasts, including Trichosporon, Cryptococcus, and Rhodotorula. Strains were grown on GYPA and SDA media for 48 h at 25°C, single colonies were struck on SDA and GYPA media, incubated another 48 h at 25°C, and from those pure cultures identifications were performed.
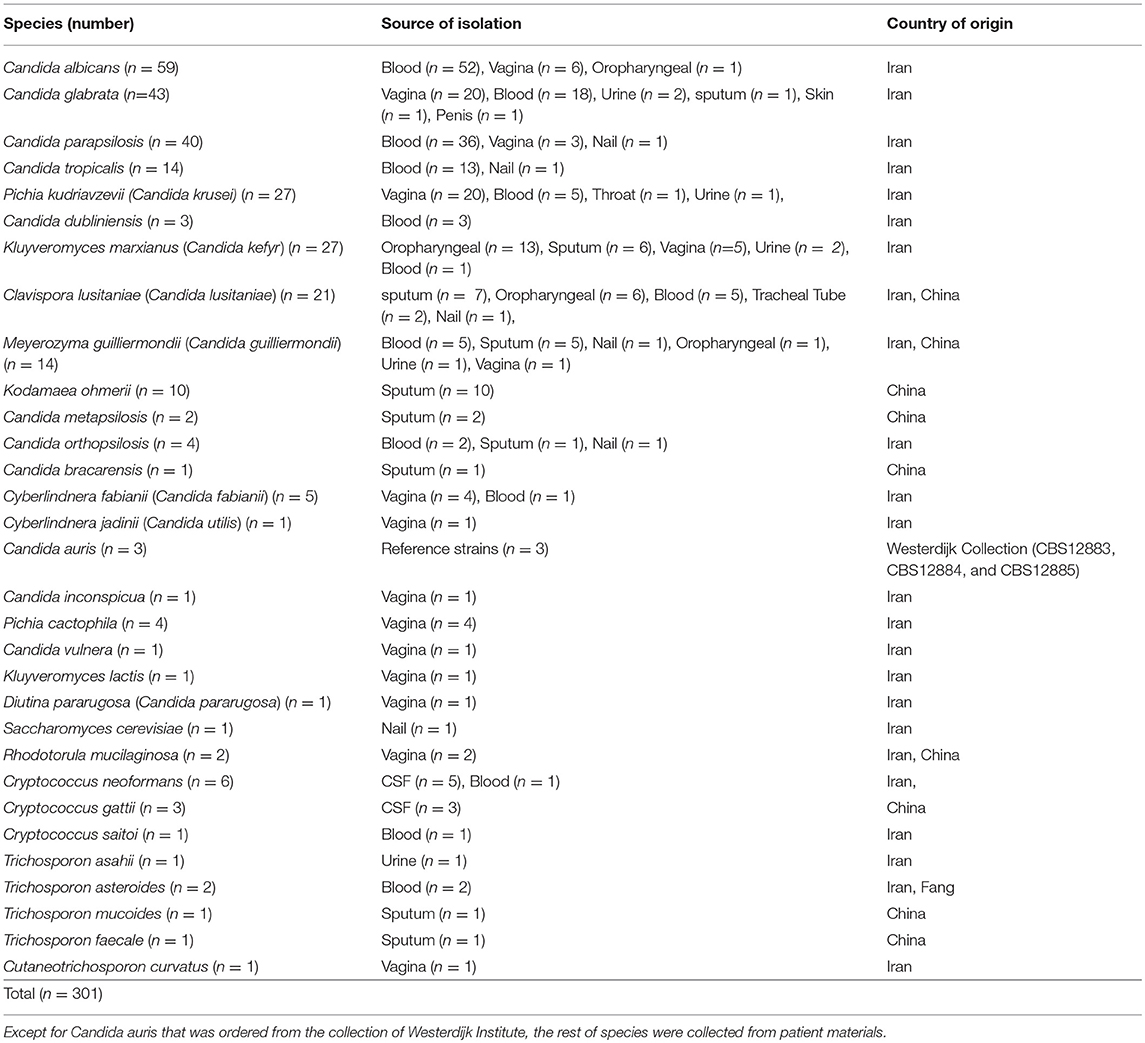
Table 1. Summary of species utilized in this along with their source of isolation and the country of origin.
One full loop of pure colonies (with the volume of 10 μl) was suspended in 100 μl of TaKaRa Lysis buffer (TaKaRa, Japan), vortexed thoroughly, and incubated at 95°C for 30. After 15 min incubation at 95°C, lysates were vortexed vigorously and incubation for another 15 min at 95°C was continued. In the last step, lysates were vortexed again and centrifuged at 14,000 rpm for 5 min. Two microliters of obtained supernatants were used as the PCR template.
One technician was responsible for performing sequencing of D1/D2 domains of rDNA (LSU rDNA), as described previously (Stielow et al., 2015). Bidirectional chain Terminated Sanger sequencing using referenced primers were performed. Obtained sequences were searched in BLAST database (https://blast.ncbi.nlm.nih.gov) and the identity of each strain was assigned accordingly. This experiment was carried out in China.
Identification of 301 using 21-plex PCR was performed in Iran. This technique contains three multiplex PCR reactions, with the first one identifying the most prevalent Candida species (Table 2), the second one targeting rare Candida species, and the third multiplex reaction identifying the most clinically important basidiomyceteous yeast species, i.e., Trichosporon, Cryptococcus, Geotrichum, and Rhodotorula (Arastehfar et al., 2018). Authors claimed that except for Candida zeylanoides, the rest of target species were correctly identified. PCR reaction and program were used same same as described, previously (Arastehfar et al., 2018). PCR products were run on 2% agarose gel (voltage of 135, 60 min), stained with Gel Red (BioTium Corporation, USA), and visualized by Gel Doc (Gel Doc XR+, BioRad, California, USA). This experiment was performed in Iran.
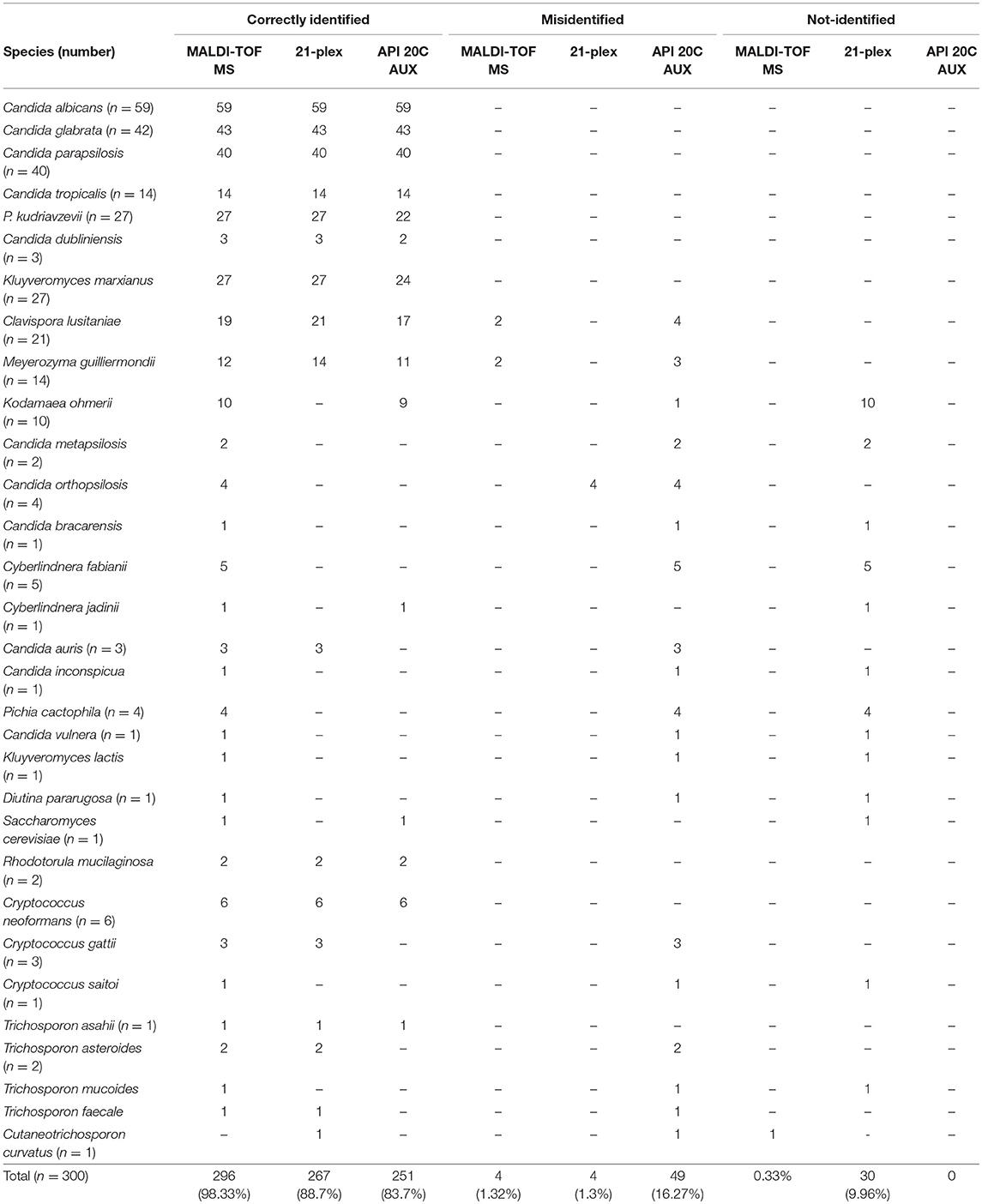
Table 2. Summary of species identification of wide range of clinically obtained yeast species using three approaches, MALDI-TOF MS, 21-plex, API 20C AUX, and their comparison with large subunit of rDNA domain sequencing.
Full-extraction method as it was utilized as suggested, previously (Marklein et al., 2009), and identification was carried out by using Microflex LT, MALDI-TOF MS device (Bruker Daltonics, Bremen Germany). Scores lower than 1.7, ≥1.7 <2, and above 2 were considered as not reliable identification, identification at the genus level, and identification at the species level, respectively. MALDI-TOF MS was performed in Netherlands.
API 20C AUX (BioMerieux, France), based on assimilation of 19 sugars and presence or absence of hyphal/pseudohyphal formation identifies clinically important yeast species. API strips were prepared as suggested by the manufacturer and incubated at 30°C for 48–72 h. Besides of results obtained from sugar assimilation profile, the possibility of hyphal/pseudohyphal formation was investigated as described previously (Keçeli et al., 2016). As 72 h incubation of API strips improved the accuracy of results (Willemsen et al., 1997), final sugar assimilation patterns were read after 72 h incubation at 25°C. Accurate identification was based on identity and T indices greater than 90% and 0.75, respectively. For hints lower than those values, the first proposed identity was assigned as the species name. API 20C AUX experiments were carried out in the Netherlands.
The strength of agreements between API 20C AUX and sequencing, 21-plex PCR and sequencing, MALDI-TOF MS and sequencing was assessed by Kappa coefficient value. Kappa coefficient value was calculated by SPSS v.23 software (Chicago, USA).
Two hundred and ninety-six of isolates (98.33%) were correctly identified, four isolates (1.3%) misidentified, and only one isolate was not identified using MALDI-TOF MS (Table 2). All of those yeast isolates were identified with the score of over two, indicating reliable identification at the species level. Surprisingly, two isolates of Meyerozyma guilliermondii and two isolates of Clavispora lusitaniae were misidentified as C. dendronema and Wickerhamiela pagnoccae (Table 3), respectively. Despite of repeated efforts and experiments using full-extraction method, the same results were obtained. All of top 5 Candida species [C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and Pichia kudriavzevii (Candida krusei)] and 95.7% of rare yeast species were identified, correctly. In total, MALDI-TOF MS identified 98.33% of yeast isolates. The Kappa coefficient value for MALDI-TOF MS and sequencing was 0.991.
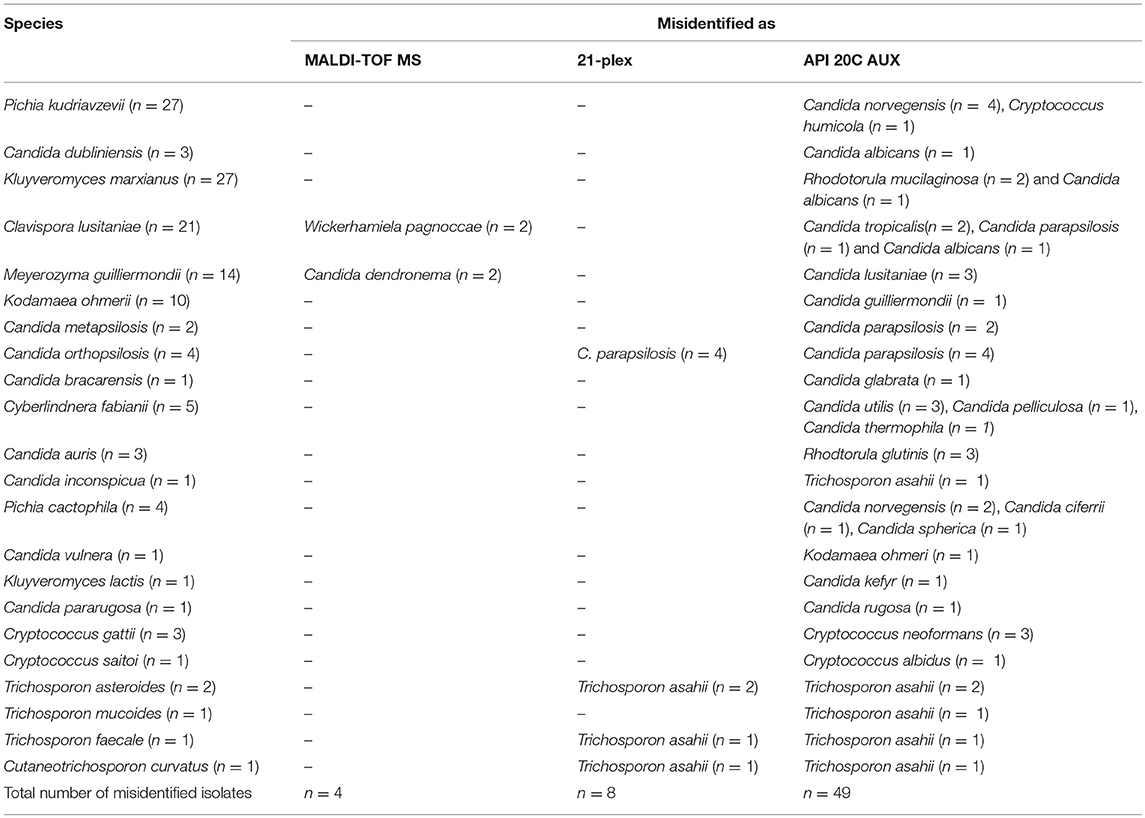
Table 3. Misidentified isolates using MALDI-TOF MS, 21-plex, and API 20C AUX compared to LSU rDNA sequencing as the gold standard method.
Two hundred and sixty-seven isolates (88.7%) were correctly identified, 4 isolates (1.3%) were misidentified, and 29 isolates (9.96%) were not identified by the 21-plex PCR technique (Table 2). Both misidentified and none-identified isolates were rare yeast species (Tables 2, 3), while all of top 5 Candida species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and P. kudriavzevii) were identified, correctly. The Kappa coefficient value for 21-plex PCR and sequencing was 0.943.
Two hundred and fifty-one isolates (83.7%) were correctly identified, 49 (16.2%) isolates were misidentified, and there was no species without identification using API 20C AUX (Table 2). The majority of misidentified yeast isolates were among rare species (n = 45), and only 4 strains of P. kudriavzevii were among top 5 Candida species (Table 3). API 20C AUX showed the lowest Kappa Coefficient value (0.918) when compared to sequencing.
Although, the 21-plex PCR showed higher accuracy, API 20C AUX more comprehensively identified yeast species. For instance, species such as Saccharomyces cerevisiae (1/1) and Kodamaea ohmerii (9/10) were correctly identified by API 20C AUX, while they were not identified by 21-plex PCR. By integrating the rapidity of PCR and comprehensiveness of API 20C AUX we could correctly identify 92% of yeast isolates included in our study. As 21-plex PCR represented a fast and reliable technique for the majority of the more prevalent yeast species and API 20C AUX requires 48–72 h for identification, we used 21-plex as the first line and rapid identification tool and in case of encountering with negative results, API 20C AUX could be used as the alternative technique.
Because the number of yeast species causing infection in human is increasing, fast, and accurate identification of clinically obtained isolates is highly important to initiate appropriate antifungal regimen (Pincus et al., 2007). Sequencing of commonly used phylogenetic markers, MALDI-TOF MS, PCR-based techniques, and biochemical and phenotypic assays are considered as the most popular identification systems. Herein, we have compared the accuracy of the API 20C AUX and 21-plex PCR methods in the light of MALDI-TOF MS and sequencing of D1/D2 domains of rDNA.
In our study, MALDI-TOF MS showed a good accuracy for identification of a diverse range of opportunistic yeast species (98.3%). All of five top Candida species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and P. kudriavzveii) and 95.7% of rare yeast species were identified successfully (Kappa value of 0.991). Despite the close genetic background of cryptic species complexes, MALDI-TOF MS identified them to the species level (note that we did not have C. africana). This is in agreement with other studies, where cryptic species complexes of C. albicans, C. glabrata, and C. parapsilosis were correctly identified (Santos et al., 2011). Moreover, FDA-approved spectra of wide-spreading multidrug-resistant yeast species, i.e., Candida auris, has been added to the clinical database of MALDI-TOF MS, leading to rapid and reliable identification of this organism (Bao et al., 2018). Although, cases of misidentification for Cryptococcus and Trichosporon species had been reported previously (Kolecka et al., 2013; Sendid et al., 2013; Ling et al., 2014; Zhao et al., 2017), except for Cutaneotrichosporon curvatus, all of our clinical isolates of aforementioned species were correctly identified (the reference spectra of this species is not included in the MALDI-TOF MS database). Consistent with the other studies and due to hardship of obtaining proper spectra for Meyerozyma guilliermondii (Ling et al., 2014; Zhao et al., 2017), using MALDI-TOF MS M. guilliemondii (n = 2/14) and Cl. lusitaniae (n = 2/21) were misidentified in our study. MALDI-TOF MS, despite of being fast, robust, and providing accurate strain identity is still considered as an economical burden, especially for developing countries, not only to purchase the device but also as it requires trained technicians and periodical maintenances (Posteraro et al., 2013; Criseo et al., 2015; Aslani et al., 2018). Due to occurrence of misidentification of some rare yeast species in our study and the other studies (Sendid et al., 2013; Zhao et al., 2017), improvement of the MALDI-TOF MS library can enhance the accuracy of this technique.
Although Vitek 2 YST ID Card reported amongst the most popular biochemical assays used in routine laboratories (Posteraro et al., 2015), API 20C AUX showed a higher agreement with sequencing of ITS and D1/D2 domains of rDNA (Zhao et al., 2017). As a result, herein API 20C AUX was used as the representative of biochemical assays. In our study, API 20C AUX correctly identified 83.7% of all included yeast isolates, the vast majority (97.26%) of the most prevalent Candida species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and P. kudriavzveii) and 61.8% of rare yeast species which is consistent with previous studies (Keçeli et al., 2016; Zhao et al., 2017) (Kappa value of 0.918). Despite the fact that, API 20C AUX misidentified the majority (4/5) of Pichia kudriavzevii strains as Pichia norvegensis, studies have revealed that both species are inherently resistant to fluconazole and genetically are close to each other (Sandven et al., 1997; Musso et al., 2014). Although, identification to the species level is an integral part of epidemiological studies, grouping of few species with the same antifungal susceptibility pattern in routine settings may hasten the timely administration of antifungals and as a consequence it may contribute to a lower the mortality rate. For instance, C. albicans, C. parapsilosis, C. tropicalis, and C. dubliniensis are all susceptible to fluconazole, and hence, using one probe they were identified as the fluconazole susceptible group (McMullan et al., 2008). On the other hand, in vitro studies have shown that C. dubliniensis compared to C. albicans more rapidly can acquire resistance to antifungals(Moran et al., 1997), underscoring the importance of specie level identification in such circumstances.In our study, 66.6% (2/3) of C. dubliniensis strains were correctly identified, whereas other cryptic complex species of C. metapsilosis, C. orthospsilosis, and C. bracarensis were all identified as C. parapsilosis and C. glabrata, which is in agreement with other reports (Keçeli et al., 2016; Zhao et al., 2017). For other clinically rare Candida species including M. guilliermondii, Cl. lusitaniae, and Kl. marxianus we observed few misidentification cases. All CBS reference strains of C. auris in concordance with other studies were misidentified as Rhodotorula glutinis (Magobo et al., 2014). In our setting, application of API 20C AUX for the most prevalent basidiomyceteous yeasts did not obtain satisfactory results for Cr. gattii, Trichosporon mucoides, and T. asteroides as seen with the previous studies (Guo et al., 2011). Biochemical assays in general and API 20C AUX in particular, despite of generating satisfactory results for rare yeast species, are labor-intensive, time-consuming, and interpretation of sugar assimilation profiles is sometimes subjective. Moreover, in order to generate accurate identity, API 20C AUX requires further testing of yeast isolates for hyphal/pseudohyphal formation (Guo et al., 2011). Numerous of reports have shown that biochemical assays can lead to underestimation of some rare yeast species and ignoring them as etiological agents of infection in human (Kathuria et al., 2015; Svobodova et al., 2016). As an example, all biochemical assays provide inaccurate identity for C. auris and it is mistaken for other yeast species such as C. parapslosis, C. famata, Rhodotorula glutinis etc (Kordalewska et al., 2017), leading to its persistence as a colonizer in hospital environment and source of future outbreaks. Given the rise in occurrence of rare yeast species that are less susceptible to fluconazole (Miceli et al., 2011), this could be of a great importance, as in developing countries due to limited economical support, this drug is administered as the drug of choice for the first line therapy (Kordalewska et al., 2018). Consequently, for a species like C. auris that exhibited resistance to all classes of antifungals, especially more than 90% resistance to fluconazole (Kathuria et al., 2015), along with high rate of mortality of 30–60% (Chowdhary et al., 2017), such misidentifications could be accompanied by adverse consequences.
Recently, we have developed a multiplex PCR that in a stepwise manner identifies the majority of yeast species regularly encountered in clinical settings as the cause of infection in human (Arastehfar et al., 2018). With the application of 21-plex PCR we correctly identified 87.3% of all included yeast species, 100% of most prevalent Candida species and 72% of rare yeast species (Kappa value of 0.943). As this assay originally was not intended for identification of other rare yeast species, such as S. cerevisiae, Cyberlindnera fabianii, Cyberlindnera jadinii, Kodamaea ohmerii, C. cactophila, C. norvegensis, Kl. lactis, Cryptococcus saitoi, and T. mucoides, they were not identified, accordingly. This assay showed a high degree of specificity (98.7%). Although, T. asteroides (n = 2), T. faecale (n = 1), and Cutaneotrichosporon curvatus (n = 1) were identified as T. asahii, 21-plex utilizes one universal primer to identify most clinically important Trichosporon species in the genus level, and hence, these cases were not considered as misidentification. Although, a slight difference in susceptibility pattern of triazoles (ravuconazole, itraconazole, and voriconazole) and AMB between T.asahii and non-T. asahii strains have been observed (Paphitou et al., 2002), MIC values are not always correlated with clinical outcomes (Paphitou et al., 2002). Accordingly, in order to prove the difference in clinical outcomes, in vivo testing with neutropenic and immunocompromised mice is still required. As a result, identification of species of non-T. asahii and T. asahii to the genus level (only as Trichosporon) will be clinically relevant, unless otherwise is proved. In concordance with sequencing of D1/D2 domains of rDNA and MALDI-TOF MS, all included strains of C. auris were correctly identified. Other PCR-based techniques such Rep-PCR shown to be a robust technique to identify a wide range of yeast species (Pincus et al., 2007; Zhao et al., 2017), but it requires tedious DNA extraction methods, capillary electrophoresis for separation of amplified PCR product, and highly trained personnel (Pincus et al., 2007; Zhao et al., 2017). Moreover, using rep-PCR technique, identification of a single isolate requires 90 USD, while in our setting using 21-plex costs 0.75–1 euros. Although 21-plex PCR exhibited a high degree of sensitivity (98.7%), there were some species (K. ohmerii, C. metapsilosis, and C. bracarensis, Cy. fabianii and Cr. satoi) that were not identified. As a result, including more multiplex PCR assays to identify other clinically important yeast species, can improve the sensitivity of 21-plex PCR.
Sole dependence on phenotypic assays could result in misidentification and subsequently oblivion of emerging and important yeast species, while combination of comprehensiveness of biochemical and phenotypic assays with the rapidity of PCR-based techniques (21-plex PCR) could increase the accuracy of identification, reduces required time and expenses, and circumvent the imperfection of either assays. For instance, despite of obtaining negative results using the 21-plex PCR for strains of K. ohmerii, Cy. jadinii, S. cerevisiae, they were correctly identified by API 20C AUX. Moreover, 21-plex PCR due to its rapidity and possessing high specificity and sensitivity, if accompanied by inexpensive and reliable PCR devices, it could be used as a reliable means of identification in developing and resource-limited countries. This could be relevant for routine laboratories, especially in developing countries, where robust and accurate means of identification such as MALDI-TOF MS and Sanger sequencing are lacking. In terms of required expenses, MALDI-TOF MS was the least expensive (less than 0.3 Euros), followed by 21-plex PCR (0.75–1 Euros/reaction), sequencing (3 Euros) and API 20C AUX (5.9 Euros/reaction). As a result, API 20C AUX was the most expensive and least accurate identification tool.
Despite the fact that we included various rare Candida and yeast species obtained from multiple healthcare facilities in Iran and China, our study still could benefit from addition of other rare yeast species, such Debaromyces hansinii, Diutina rugose, Yarrowia lipolytica, and the like. Moreover, we did not obtain clinical isolates of cryptic species complexes of C. nivariensis and C. africana and hence we could not observe how well they could be differentiated from C. glabrata and C. albicans using applied techniques.
AA, FD, SK, WP, MK, and TB have designed the study, did the experiments, and participated in draft preparation and revision. MR, HZ, WF, MN, WL, SR, HB, KZ, and FH have provided the isolates, participated in carrying out the experiments, and assisted in paper revision. SH provided strains and participated in performing 21-plex PCR.
This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 642095, National Health Department of China [2018ZX10101003], National Natural Science Foundation of China [31770161], Second Military Medical University [2017JZ47], and Shanghai Science and Technology Committee [14DZ2272900 and 14495800500]. This research has been financially supported by Tehran University of Medical Sciences & Health Services grant.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arastehfar, A., Fang, W., Pan, W., Lackner, M., Liao, W., Badiee, P., et al. (2018). YEAST PANEL multiplex PCR for identification of clinically important yeast species: stepwise diagnostic strategy, useful for developing countries. Diagn. Microbiol. Infect. Dis. 93, 112–119. doi: 10.1016/j.diagmicrobio.2018.09.007
Aslani, N., Janbabaei, G., Abastabar, M., Meis, J. F., Babaeian, M., Khodavaisy, S., et al. (2018). Identification of uncommon oral yeasts from cancer patients by MALDI-TOF mass spectrometry. BMC Infect. Dis. 18:24. doi: 10.1186/s12879-017-2916-5
Bao, J. R., Master, R. N., Azad, K. N., Schwab, D. A., Clark, R. B., Jones, R. S., et al. (2018). Rapid, accurate identification of Candida auris by using a novel matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) database (Library). J. Clin. Microbiol. 56:e01700–17. doi: 10.1128/JCM.01700-17
Bizerra, F. C., Jimenez-Ortigosa, C., Souza, A. C. R., Breda, G. L., Queiroz-Telles, F., Perlin, D. S., et al. (2014). Breakthrough candidemia due to multidrug-resistant Candida glabrata during prophylaxis with a low dose of micafungin. Antimicrob. Agents Chemother. 58, 2438–2440. doi: 10.1128/AAC.02189-13
Castanheira, M., Woosley, L. N., Diekema, D. J., Jones, R. N., and Pfaller, M. A. (2013). Candida guilliermondii and other species of Candida misidentified as Candida famata: assessment by Vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry in two global antifungal surveillance programs. J. Clin. Microbiol. 51, 117–124. doi: 10.1128/JCM.01686-12
Chowdhary, A., Sharma, C., and Meis, J. F. (2017). Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 13:e1006290. doi: 10.1371/journal.ppat.1006290
Chowdhary, A., Voss, A., and Meis, J. F. (2016). Multidrug-resistant Candida auris: ‘new kid on the block’ in hospital-associated infections? J. Hosp. Infect. 94, 209–212. doi: 10.1016/j.jhin.2016.08.004
Criseo, G., Scordino, F., and Romeo, O. (2015). Current methods for identifying clinically important cryptic Candida species. J. Microbiol. Methods 111, 50–56. doi: 10.1016/j.mimet.2015.02.004
Guo, L. N., Xiao, M., Kong, F., Chen, S. C. A., Wang, H., Sorrell, T. C., et al. (2011). Three-locus identification, genotyping, and antifungal susceptibilities of medically important Trichosporon species from China. J. Clin. Microbiol. 49, 3805–3811. doi: 10.1128/JCM.00937-11
Kathuria, S., Singh, P. K., Sharma, C., Prakash, A., Masih, A., Kumar, A., et al. (2015). Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by vitek 2, CL. J. Clin. Microbiol. 53, 1823–1830. doi: 10.1128/JCM.00367-15
Keçeli, S. A., Dündar, D., and Tamer, G. S. (2016). Comparison of vitek matrix-assisted laser desorption/ionization time-of-flight mass spectrometry versus conventional methods in Candida identification. Mycopathologia 181, 67–73. doi: 10.1007/s11046-015-9944-8
Kolecka, A., Khayhan, K., Groenewald, M., Theelen, B., Arabatzis, M., Velegraki, A., et al. (2013). Identification of medically relevant species of arthroconidial yeasts by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51, 2491–2500. doi: 10.1128/JCM.00470-13
Kordalewska, M., Lee, A., Park, S., Berrio, I., Chowdhary, A., Zhao, Y., et al. (2018). Understanding echinocandin resistance in the emerging pathogen Candida auris. Antimicrob. Agents Chemother. 62:e00238–18. doi: 10.1128/AAC.00238-18
Kordalewska, M., Zhao, Y., Lockhart, S. R., Chowdhary, A., Berrio, I., and Perlin, D. S. (2017). Rapid and accurate molecular identification of the emerging multidrug resistant pathogen Candida auris. J. Clin. Microbiol. 55, 2445–2452. doi: 10.1128/JCM.00630-17
Ling, H., Yuan, Z., Shen, J., Wang, Z., and Xu, Y. (2014). Accuracy of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinical pathogenic fungi: a meta-analysis. J. Clin. Microbiol. 52, 2573–2582. doi: 10.1128/JCM.00700-14
Magobo, R. E., Corcoran, C., Seetharam, S., and Govender, N. P. (2014). Candida auris –Associated Candidemia, South Africa. Emerg. Infect. Dis. 20, 1250–1251. doi: 10.3201/eid2007.131765
Marklein, G., Josten, M., Klanke, U., Müller, E., Horré, R., Maier, T., et al. (2009). Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47, 2912–2917. doi: 10.1128/JCM.00389-09
McMullan, R., Metwally, L., Coyle, P. V., Hedderwick, S., McCloskey, B., O'Neill, H. J., et al. (2008). A prospective clinical trial of a real-time polymerase chain reaction assay for the diagnosis of candidemia in nonneutropenic, critically Ill adults. Clin. Infect. Dis. 46, 890–896. doi: 10.1086/528690
Miceli, M. H., Díaz, J. A., and Lee, S. A. (2011). Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151. doi: 10.1016/S1473-3099(10)70218-8
Moran, G. P., Sullivan, D. J., Henman, M. C., Creary, C. E. M. C., Harrington, B. J., Shanley, D. B., et al. (1997). Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV) -infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. 41, 617–623.
Musso, M., Giannella, M., Antonini, M., Bordi, E., Ettorre, G. M., Tessitore, L., et al. (2014). Invasive candidiasis due to Candida norvegensis in a liver transplant patient: case report and literature review. Infect. Dis. Rep. 6, 1–4. doi: 10.4081/idr.2014.5374
Paphitou, N. I., Ostrosky-Zeichner, L., Paetznick, V. L., Rodriguez, J. R., Chen, E., and Rex, J. H. (2002). In vitro antifungal susceptibilities of Trichosporon species. Antimicrob. Agents Chemother. 46, 1144–1146. doi: 10.1128/AAC.46.4.1144-1146.2002
Pappas, P. G. (2006). Invasive candidiasis. Infect. Dis. Clin. North Am. 20, 485–506. doi: 10.1016/j.idc.2006.07.004
Pappas, P. G., Alexander, B. D., Andes, D. R., Hadley, S., Kauffman, C. A., Freifeld, A., et al. (2010). Invasive fungal infections among organ transplant recipients: results of the transplant-associated infection surveillance network (TRANSNET). Clin. Infect. Dis. 50, 1101–1111. doi: 10.1086/651262
Pfaller, M. A., Diekema, D. J., Gibbs, D. L., Newell, V. A., Nagy, E., Dobiasova, S., et al. (2008). Candida krusei, a multidrug-resistant opportunistic fungal pathogen: Geographic and temporal trends from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J. Clin. Microbiol. 46, 515–521. doi: 10.1128/JCM.01915-07
Pham, C. D., Iqbal, N., Bolden, C. B., Kuykendall, R. J., Harrison, L. H., Farley, M. M., et al. (2014). Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob. Agents Chemother. 58, 4690–4696. doi: 10.1128/AAC.03255-14
Pincus, D. H., Orenga, S., and Chatellier, S. (2007). Yeast identification - Past, present, and future methods. Med. Mycol. 45, 97–121. doi: 10.1080/13693780601059936
Posteraro, B., De Carolis, E., Vella, A., and Sanguinetti, M. (2013). MALDI-TOF mass spectrometry in the clinical mycology laboratory: identification of fungi and beyond. Expert Rev. Proteomics 10, 151–164. doi: 10.1586/epr.13.8
Posteraro, B., Efremov, L., Leoncini, E., Amore, R., Posteraro, P., Ricciardi, W., et al. (2015). Are the conventional commercial yeast identification methods still helpful in the era of new clinical microbiology diagnostics? A meta-analysis of their accuracy. J. Clin. Microbiol. 53, 2439–2450. doi: 10.1128/JCM.00802-15
Ragheb, S. M., and Jimenez, L. (2014). Polymerase chain reaction/rapid methods are gaining a foothold in developing countries. PDA J. Pharm. Sci. Technol. 68, 239–255. doi: 10.5731/pdajpst.2014.00979
Sandven, P., Nilsen, K., Digranes, A., Tjade, T., and Lassen, J. (1997). Candida norvegensis: a fluconazole-resistant species. Antimicrob. Agents Chemother. 41, 1375–1376.
Santos, C., Lima, N., Sampaio, P., and Pais, C. (2011). Matrix-assisted laser desorption/ionization time-of-flight intact cell mass spectrometry to detect emerging pathogenic Candida species. Diagn. Microbiol. Infect. Dis. 71, 304–308. doi: 10.1016/j.diagmicrobio.2011.07.002
Sendid, B., Ducoroy, P., François, N., Lucchi, G., Spinali, S., Vagner, O., et al. (2013). Evaluation of MALDI-TOF mass spectrometry for the identification of medically-important yeasts in the clinical laboratories of Dijon and Lille hospitals. Med. Mycol. 51, 25–32. doi: 10.3109/13693786.2012.693631
Stielow, J. B., Lévesque, C. A., Seifert, K. A., Meyer, W., Irinyi, L., Smits, D., et al. (2015). One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia Mol. Phylogeny Evol. Fungi 35, 242–263. doi: 10.3767/003158515X689135
Svobodova, L., Bednarova, D., Ruzicka, F., Chrenkova, V., Dobias, R., Mallatova, N., et al. (2016). High frequency of Candida fabianii among clinical isolates biochemically identified as Candida pelliculosa and Candida utilis. Mycoses 59, 241–246. doi: 10.1111/myc.12454
Willemsen, M., Breynaert, J., and Lauwers, S. (1997). Comparison of Auxacolor with API 20 C Aux in yeast identification. Clin. Microbiol. Infect. 3, 369–375.
Wong, G., Wong, I., Chan, K., Hsieh, Y., and Wong, S. (2015). A rapid and low-cost PCR thermal cycler for low resource settings. PLoS ONE 10:e0131701. doi: 10.1371/journal.pone.0131701
Yapar, N. (2014). Epidemiology and risk factors for invasive candidiasis. Ther. Clin. Risk Manag. 10, 95–105. doi: 10.2147/TCRM.S40160
Keywords: API 20C AUX, 21-plex PCR, MALDI-TOF MS, LSU rDNA sequencing, developing countries
Citation: Arastehfar A, Daneshnia F, Kord M, Roudbary M, Zarrinfar H, Fang W, Hashemi SJ, Najafzadeh MJ, Khodavaisy S, Pan W, Liao W, Badali H, Rezaie S, Zomorodian K, Hagen F and Boekhout T (2019) Comparison of 21-Plex PCR and API 20C AUX, MALDI-TOF MS, and rDNA Sequencing for a Wide Range of Clinically Isolated Yeast Species: Improved Identification by Combining 21-Plex PCR and API 20C AUX as an Alternative Strategy for Developing Countries. Front. Cell. Infect. Microbiol. 9:21. doi: 10.3389/fcimb.2019.00021
Received: 23 October 2018; Accepted: 22 January 2019;
Published: 15 February 2019.
Edited by:
Rong Fang, The University of Texas Medical Branch at Galveston, United StatesReviewed by:
Maurizio Sanguinetti, Catholic University of Sacred Heart, ItalyCopyright © 2019 Arastehfar, Daneshnia, Kord, Roudbary, Zarrinfar, Fang, Hashemi, Najafzadeh, Khodavaisy, Pan, Liao, Badali, Rezaie, Zomorodian, Hagen and Boekhout. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sadegh Khodavaisy, c2FkZWdoXzczOTIwMDhAeWFob28uY29t
Weihua Pan, cGFud2VpaHVhQHNtbXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.