- 1Department of Molecular Sciences, Faculty of Science and Engineering, Macquarie University, Sydney, NSW, Australia
- 2Department of Pharmaceutical Biosciences, Centre of Integrative Microbial Evolution, School of Pharmacy, University of Oslo, Oslo, Norway
- 3Microscopy Unit, Faculty of Science and Engineering, Macquarie University, Sydney, NSW, Australia
- 4Department of Biomedical Sciences, Faculty of Medicine and Health Sciences, Macquarie University, Sydney, NSW, Australia
- 5Department of Biosciences, The Faculty of Mathematic and Natural Sciences, University of Oslo, Oslo, Norway
Pseudomonas aeruginosa is a significant cause of mortality in patients with cystic fibrosis (CF). To explore the interaction of the CF isolate P. aeruginosa PASS1 with the innate immune response, we have used Danio rerio (zebrafish) as an infection model. Confocal laser scanning microscopy (CLSM) enabled visualization of direct interactions between zebrafish macrophages and P. aeruginosa PASS1. Dual RNA-sequencing of host-pathogen was undertaken to profile RNA expression simultaneously in the pathogen and the host during P. aeruginosa infection. Following establishment of infection in zebrafish embryos with PASS1, 3 days post infection (dpi), there were 6739 genes found to be significantly differentially expressed in zebrafish and 176 genes in PASS1. A range of virulence genes were upregulated in PASS1, including genes encoding pyoverdine biosynthesis, flagellin, non-hemolytic phospholipase C, proteases, superoxide dismutase and fimbrial subunits. Additionally, iron and phosphate acquisition genes were upregulated in PASS1 cells in the zebrafish. Transcriptional changes in the host immune response genes highlighted phagocytosis as a key response mechanism to PASS1 infection. Transcriptional regulators of neutrophil and macrophage phagocytosis were upregulated alongside transcriptional regulators governing response to tissue injury, infection, and inflammation. The zebrafish host showed significant downregulation of the ribosomal RNAs and other genes involved in translation, suggesting that protein translation in the host is affected by PASS1 infection.
Introduction
Cystic fibrosis (CF) is an autosomal recessive disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator gene (Freedman et al., 1999; Phennicie et al., 2010). It is most prevalent among the Caucasian population, affecting 1 in 2,500 new-borns (Freedman et al., 1999). CF lung disease is the major cause of morbidity and mortality among CF patients and is a result of colonization and infection of airways with bacteria, fungi, and viruses (Cantin et al., 2015). In early childhood, the CF lung is mainly colonized by Staphylococcus aureus and Haemophilus influenzae, although later in a CF patient's life, P. aeruginosa typically takes over as the dominant pathogen (Davies, 2002).
P. aeruginosa is a versatile Gram-negative microorganism found commonly in both terrestrial and aquatic environments (Whitehead et al., 2006; Sousa and Pereira, 2014). Its 6.3 Mb sized genome supports its metabolic versatility and, consequently, its adaptability to diverse environments (Blázquez et al., 2006). This opportunistic pathogen can cause acute and chronic infections in immunocompromised people, such as AIDS sufferers and neutropenic patients undergoing chemotherapy, and patients with injuries, catheters, burn wounds, and non-CF-associated pulmonary infections (Lyczak et al., 2000; Papaioannou et al., 2013). P. aeruginosa infection often becomes the major cause of morbidity and mortality in CF patients (Folkesson et al., 2012).
The respiratory pathogenesis of P. aeruginosa can be attributed to an array of key virulence factors, including flagella, type III secretion system, phenazines, the iron scavenging siderophores pyochelin and pyoverdine, lipopolysaccharide, elastase, alkaline proteases, hemolysins (phospholipase and lecithinase), cytotoxins (leukocidin), and exotoxin A (Sadikot et al., 2005; Gellatly and Hancock, 2013). P. aeruginosa chronic infections in the CF lung also provoke aggressive inflammatory reactions, such as the host neutrophilic response which releases oxidants and enzymes detrimental to the host tissue (Davies, 2002; Phennicie et al., 2010; Gellatly and Hancock, 2013).
The pathogenesis of P. aeruginosa has been studied using diverse model host organisms including Dictyostelium discoideum, Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Galleria mellonella, rodents, and zebrafish (Clatworthy et al., 2009). Zebrafish has been previously used to study aspects of pathogenesis of P. aeruginosa (Phennicie et al., 2010; Diaz-Pascual et al., 2017), as well as other bacterial pathogens including Salmonella typhimurium, S. aureus, Burkholderia cenocepacia, H. influenzae, Leptospira interrogans, and Listeria monocytogenes (Meijer and Spaink, 2011).
Zebrafish is a teleost fish with a total genome size of 1.412 gigabases (van der Sar et al., 2003; Howe et al., 2013). Genome comparison has revealed that ~70% of human genes have a clear zebrafish ortholog (Howe et al., 2013). Remarkable similarities have been observed with human transcriptional regulators, immune effectors, immune recognition systems, defense signaling pathways, and macrophage lineages (Cui et al., 2011; Meijer and Spaink, 2011; Hall et al., 2013) which make zebrafish a good model for studying host-pathogen interaction.
There are several other advantages in using zebrafish embryos as a model organism for study of human infections. These include ease in handling, low cost, rapid development and the ability of a single pair of adult zebrafish to produce hundreds of offspring every week (Lessman, 2011; Meijer and Spaink, 2011). It is optically transparent and the availability of transgenic lines with fluorescently marked immune cells allows for real-time visualization of in vivo microbe-phagocyte interactions at high resolution throughout the organism (Clatworthy et al., 2009; Lessman, 2011; Meijer and Spaink, 2011).
In zebrafish, the innate and adaptive immune systems develop sequentially. The innate immune response is developed at the embryonic and early larval stages with early lymphocytes making their first appearance in the 4-day-old larvae, and a full adaptive immune system developing at about 3 weeks of age (Torraca et al., 2014). The innate immune system is the host's first line of defense against infections and includes physical barriers, cellular, and humoral components such as complement and acute phase proteins (Cui et al., 2011; van Soest et al., 2011). It is responsible for early recognition of pathogens and triggering an appropriate pro-inflammatory response (Mogensen, 2009; van der Vaart et al., 2012; Torraca et al., 2014). The main phagocytic cell types of the innate immune system are macrophages and neutrophils (Sieger et al., 2009; Torraca et al., 2014). In zebrafish embryos, as early as one day post fertilization, functional macrophages are capable of sensing and responding to microbial infections (Meijer and Spaink, 2011). Both neutrophils and macrophages can lead to bacterial clearance by engulfing and killing bacterial pathogens. The bacterial pathogens shown to be engulfed by these phagocytic cells include P. aeruginosa and S. aureus (Brannon et al., 2009; Clatworthy et al., 2009; Sieger et al., 2009).
Global gene expression studies of wild-type zebrafish embryos using a zebrafish microarray have been conducted following static immersion with Edwardsiella tarda, Escherichia coli, P. aeruginosa strain PA14 and strain PAO1, whereas systemic infection has been studied with E. tarda and S. typhimurium (Ordas et al., 2011; van Soest et al., 2011). Infection in zebrafish embryos is often established via injection into the blood circulation, and response to infection with various bacterial pathogens has been subjected to microarray analysis (Stockhammer et al., 2009; van der Sar et al., 2009; van Soest et al., 2011; van der Vaart et al., 2013; Lima et al., 2014). Specifically, zebrafish embryos are usually microinjected with bacterial pathogens directly into the blood circulation at 1-3 day post fertilization, mostly using the posterior blood island or into the duct of Cuvier, a wide blood circulation valley on the yolk sac connecting the heart to the trunk vasculature (Meijer and Spaink, 2011).
RNA sequencing (RNA-Seq) has previously been used to study innate immune response of zebrafish embryo to systemic S. typhimurium infection (Ordas et al., 2011). Ordas et al. (2011) used a combination of Tag-Seq, RNA-Seq and microarray transcriptome data to compile an annotated reference set of infection-responsive genes in the zebrafish embryos. These included genes encoding transcription factors, signal transduction proteins, cytokines and chemokines, complement factors, proteins involved in apoptosis and proteolysis, proteins with antimicrobial activities, as well as many known or novel proteins not previously linked to the immune response.
Despite the importance of P. aeruginosa pathogenesis in CF lung infection, the use of zebrafish as an alternative model to understand host-pathogen interaction until recently had remained unexplored. Diaz-Pascual et al. (2017) have recently profiled the global proteome of both zebrafish and P. aeruginosa PAO1 following establishment of P. aeruginosa PAO1 infection via immersion and injection (Diaz-Pascual et al., 2017). However, the interaction of zebrafish and P. aeruginosa remains to be elucidated at a global transcriptome scale. Hence, the aim of this study was to investigate the interaction of zebrafish embryos with the virulent P. aeruginosa CF isolate PASS1 (Penesyan et al., 2015) by visualizing macrophage-PASS1 interaction and analyzing the simultaneous global gene expression profiles of both organisms via RNA sequencing (RNA-Seq). To our knowledge, this is the first study describing the dual transcriptome of P. aeruginosa-zebrafish interaction.
Methods
P. aeruginosa Strain and Growth Conditions
The bacterial strains used in this study were P. aeruginosa PASS1 (Penesyan et al., 2015) obtained from the sputum of a 40-year old female patient and yellow fluorescent protein (YFP)-tagged P. aeruginosa PASS1 (Kaur et al., 2015). The strains were maintained in a glycerol stock at −80°C, and prior to each experiment, were grown on Luria Bertani (LB) agar plates and incubated at 37°C till isolated colonies were obtained. The isolated colonies were then cultured in LB broth overnight at 37°C with constant shaking at 150 rpm. Cells from overnight cultures were washed, pelleted at 6,000 g for 10 min at 4°C and resuspended into sterile PBS. The cell concentration was estimated by measuring the optical density at 600 nm. Following estimation of cell concentration, the cells were diluted in PBS to an optical density OD600 of 2.0 which corresponds to 4.08 × 108 CFU/mL. For visualization of the bacterial suspension during injection of zebrafish embryos an aliquot of phenol red sodium salt stock solution was added to a final concentration of 0.01%.
Visualization of PASS1-YFP Infection in Zebrafish Embryos by Confocal Microscopy
The injection apparatus for the zebrafish embryos was set up as described by Brudal et al. (2014). Zebrafish embryos derived from adults of the Tg(mpeg1:Gal4, UAS;mCherry-CAAX) (Ellett et al., 2011) were manually dechorionated and maintained at 29°C prior to injection at 48 hours post fertilization (hpf). The embryos were anesthetized with 0.005 w/vol % ethyl 3-aminobenzoate methanesulfonate (Tricaine) for 1–2 min and placed on 2% agarose plates for injection. PASS1-YFP cells (in a volume of 0.7 to 1 nl) were microinjected into the duct of Cuvier, as visually ascertained under the stereomicroscope. Infected embryos were returned to a petri dish with fresh embryo medium and incubated for 6 h at 29°C prior to confocal microscopy. A mock-infection with only sterile PBS was also set-up for comparison with the PASS1-YFP infection. Prior to confocal microscopy zebrafish embryos were anesthetized with Tricaine as described above, and then transferred to a glass bottom petri dish with glass cover slip containing a mixture of embryo water (Zebrafish embryo medium, 2011) and Tricaine. The anesthetized embryos in the petri dish were then covered with 1.3% low-melting-point agarose. Confocal microscopy was performed with an Olympus Fluoview FV1000 IX81 inverted confocal microscope.
RNA Extraction and RNA-Seq Transcriptomics
The zebrafish embryos derived from adults of the AB wild-type line were infected with PASS1 or mock-infected with PBS with microinjection into the duct of Cuvier (48 hpf) according to the procedure described above. Infected embryos were returned to a petri dish with fresh embryo medium and incubated for 3 dpi at 29°C prior to RNA isolation. At 3 dpi, 9 randomly chosen zebrafish embryos from each group were euthanized by a prolonged immersion in an overdose of 50 mg/L Tricaine solution and transferred into 1.5 ml Eppendorf tubes. Three embryos were then pooled to represent one sample to allow for sufficient amount of starting material for RNA isolation. The embryo water was replaced by RNAlater (Ambion) immediately after transfer of embryos to fresh 1.5 ml Eppendorf tubes. The samples were kept at 4°C until RNA was isolated. For the extraction of total RNA, RNAlater was replaced with 600 μl of Qiazol, and the tissue was homogenized using a pestle motor followed by drawing of sample with a needle 5 times till tissue was completely homogenized. As a control, RNA was isolated from PASS1 cultures grown in LB to an OD600 = 1.0. For both the zebrafish and bacterial culture, RNA extractions were performed using the miRNeasy Mini kit (Qiagen) according to the manufacturer's protocols. An additional DNase treatment was performed with a TURBO DNA-free kit (Ambion) according to the manufacturer's protocols. The concentration of the extracted RNA was measured using a NanoDrop Spectrophotometer. The total RNA samples were subjected to ribosomal RNA (rRNA) depletion, zebrafish samples mock-infected with PBS were treated with Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) (Illumina), zebrafish infected with PASS1 were treated with Ribo-Zero rRNA Removal Kit (Gram-negative Bacteria) (Illumina) followed by Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) and the PASS1 sample grown in LB was treated with Ribo-Zero rRNA Removal Kit (Gram-negative Bacteria). The depletion steps and subsequent 125 bp paired-end RNA Sequencing on a HiSeq2500 (Illumina) were performed at the Australian Genome Research Facility (Melbourne, Australia).
Bioinformatic Analyses of Transcriptomic Data
Sequencing data were assessed for quality using FastQC software (Babraham Bioinformatics). The transcriptomes of zebrafish infected with PASS1 and PBS were mapped against the zebrafish genome (Ensembl). Bacterial transcriptomic data were mapped against the PAO1 (NCBI) genome. Transcriptome mapping was undertaken with TopHat2 and normalized based on FPKM differential expression calculation using Cuffdiff (Trapnell et al., 2012).
Significantly differentially expressed genes in PASS1 (p ≤ 0.01 and log2 fold-changes cut-off −1≥ to ≤1) were mapped to annotated pathways and to cluster of orthologous groups of P. aeruginosa strain PAO1, obtained from the Pseudomonas Database (Winsor et al., 2016).
Significantly differentially expressed genes in zebrafish (p ≤ 0.01 and log2 fold-changes cut-off −1≥ to ≤1) were functionally annotated using Ingenuity Pathway Analysis (Ingenuity Systems Inc., Redwood City, CA). A total of 3,238 differentially expressed genes were successfully mapped (Table S6). Functional annotation and gene ontology (GO) classification was conducted separately of the upregulated and downregulated genes in zebrafish-P. aeruginosa PASS1 infection using DAVID (The Database for Annotation, Visualization, and Integration Discovery) version 6.8 (Huang da et al., 2009a,b).
Results and Discussion
Confocal Laser Scanning Microscopy of Macrophage - P. aeruginosa PASS1 Interaction in Zebrafish
Macrophages are important effector cells of the innate immune response that can rapidly phagocytose bacteria and alert the immune system to danger (Kline et al., 2009). We have previously generated a (YFP)-labeled derivative of the CF isolate P. aeruginosa PASS1, PASS1-YFP (Kaur et al., 2015). The PASS1-YFP cells were injected into the Duct of Cuvier of transgenic zebrafish embryos Tg(mpeg1:Gal4, UAS;mCherry-CAAX) (Ellett et al., 2011) which produce mCherry-labeled macrophages. This enabled the analysis of macrophage behavior in zebrafish by confocal laser scanning microscopy (CLSM) visualization.
Strikingly, within 6 h post infection (hpi), P. aeruginosa PASS1-YFP cells were predominantly found to be associated or engulfed by macrophages (Figures 1A–C). Previous studies have shown that macrophages can kill both Gram-positive and Gram-negative bacteria, including P. aeruginosa, via phagocytosis (Brannon et al., 2009). Brannon et al. (2009) have shown the phagocytosis of P. aeruginosa strains PAO1 and PAK by macrophages to occur within 2 hpi (Brannon et al., 2009).
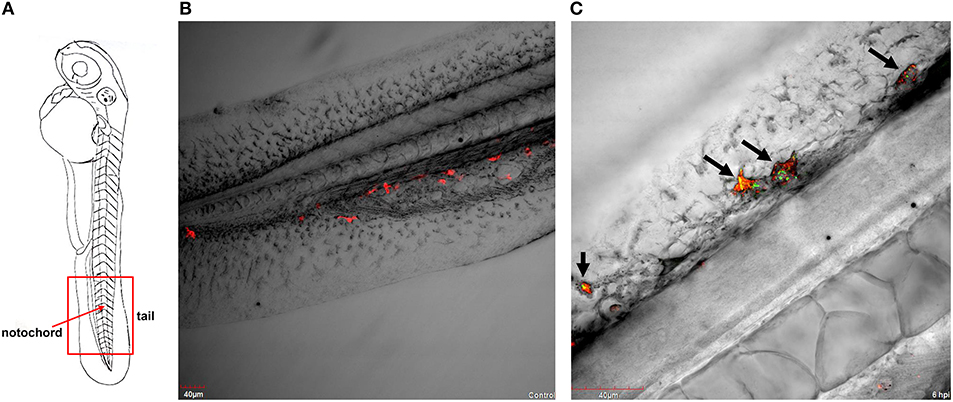
Figure 1. P. aeruginosa PASS1-macrophage interaction in zebrafish at 6 hpi. (A) A schematic view of a zebrafish embryo. The red box represents the tail region, and the red arrow indicates vertebrate notochord. Confocal laser scanning microscopy of transgenic embryos of Danio rerio Tg(mpeg1:Gal4, UAS;mCherry-CAAX) injected into the duct of Cuvier with (B) phosphate buffered saline and with (C) YFP-labelled P. aeruginosa PASS1 at 48 hpf. (B) Post-infection with phosphate buffered saline, macrophages are localized in the tail region of the vertebrate. (C) The black arrows on the vertebrate notochord in the tail region of zebrafish embryos indicate either an association or engulfment of P. aeruginosa PASS1-YFP cells (in green) by macrophages (in red).
A central advantage of the zebrafish embryo model is the ability to monitor infection at a detailed cellular level in real time. Our CLSM results corroborate previous observations that macrophages are capable of phagocytosing and killing P. aeruginosa (Tang et al., 1995; Brannon et al., 2009). Earlier studies have used the laboratory strains PAO1 and PAK, this is the first zebrafish study to use a CF isolate. Our previous work showed that PASS1 is non-mucoid and has a mutation in the lasR gene, and displays significant phenotypic differences compared with PAO1, including increased biofilm formation and production of virulence factors such as phenazines (Penesyan et al., 2015).
Generation of a Dual Host-Pathogen Transcriptome
The use of a zebrafish embryo model allows the possibility of global gene expression analysis of both host and microbe in parallel. This provides an opportunity to investigate the molecular mechanisms of the interaction between the host innate immune system and the pathogen. The survival of zebrafish embryo infected with PASS1 displayed increased mortality within 24 h of infection followed by gradual decrease in embryo survival rate till 3 days post infection (dpi) (Figure 2). To investigate host-pathogen interaction prior to increase in mortality, we isolated total RNA from PASS1-infected zebrafish (3 dpi), and RNA-Seq was used to examine the zebrafish and PASS1 transcriptomes in parallel. The infected zebrafish transcriptome was compared with phosphate buffered saline (PBS)-injected zebrafish as a negative control. The transcriptome of P. aeruginosa PASS1 in zebrafish was compared with PASS1 grown in Luria-Bertani (LB) culture medium.
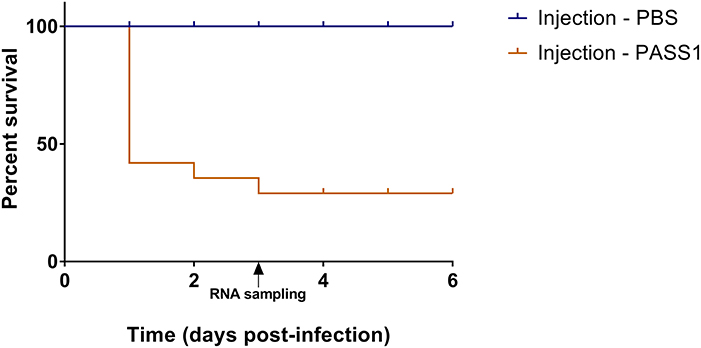
Figure 2. Percentage survival of zebrafish embryos infected with P. aeruginosa PASS1. Kaplan-Meier representation of the survival of zebrafish embryos infected with PASS1 and mock-infected with PBS via injection into the duct of Cuvier.
A total of 214,749,236 sequence reads were generated from the total RNA extracted from three independent biological replicates of PASS1-infected zebrafish. Around 53.2% of the reads aligned with the P. aeruginosa reference genome while 90% aligned with the zebrafish reference genome (Table 1).
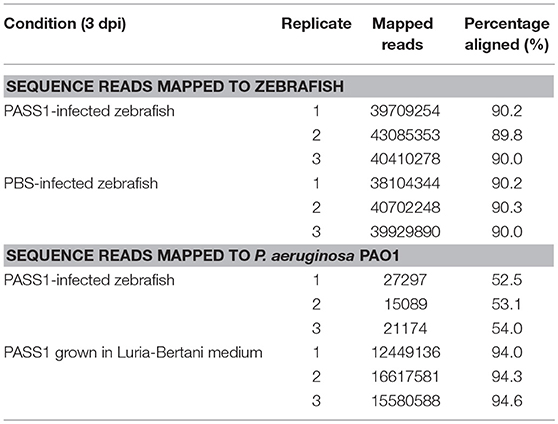
Table 1. Summary of P. aeruginosa PASS1 and zebrafish embryos mapped reads at 3 days post-infection.
Whole-Cell Transcriptome Analysis of P. aeruginosa Infected Into Zebrafish
Analysis of the P. aeruginosa transcriptome data revealed 176 genes to be differentially expressed in P. aeruginosa within the infected zebrafish (p ≤ 0.01 and log2 fold-changes cut-off −1≥ to ≤1) with 140 genes upregulated and 36 genes downregulated (Figure 3). Ribosomal RNA genes were the most upregulated transcripts in P. aeruginosa PASS1 during zebrafish infection (Figure 3). A number of studies have shown a correlation between growth rate and rRNA concentration (Bartlett and Gourse, 1994; Rang et al., 1999; Ramos et al., 2000; Schneider et al., 2003; Dennis et al., 2004; Benítez-Páez et al., 2012; Blazewicz et al., 2013). Additionally, translation initiation and elongation factor genes were also more highly expressed by PASS1 within the zebrafish (Table S1). This suggests that within the host there is a higher rate of protein synthesis and cell proliferation at 3 dpi compared to the late log (OD600 = 1.0) culture of PASS1 in LB medium.
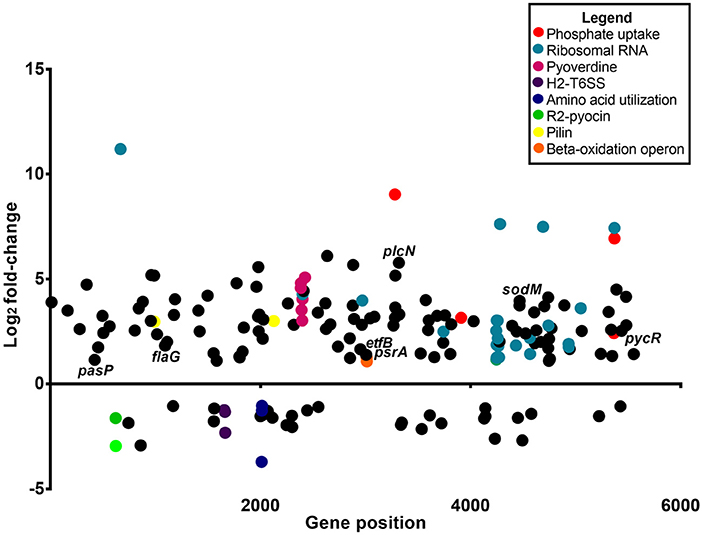
Figure 3. Differential gene expression of P. aeruginosa PASS1 in zebrafish compared to PASS1 cells grown in Luria-Bertani medium. Each dot represents a gene within the P. aeruginosa PASS1 genome (x-axis) and its fold-change (log2) expression in vivo, 3 dpi. Only significantly differentially expressed genes are shown (p ≤ 0.01 and log2 fold-change cut-off −1≥ to ≤1).
Expression of Virulence Genes in PASS1 Cells in a Zebrafish Model
Vertebrates are known to deplete both inorganic phosphate and iron in response to bacterial infection (Skaar, 2010). Consistent with this the P. aeruginosa phosphate transport and iron acquisition genes were highly expressed in zebrafish compared with PASS1 culture in LB (Figure 4). The pyrophosphate porin gene oprO and the phosphate-binding protein gene pstS were highly upregulated (9 log2 fold-change and 6.9 log2 fold-change, respectively). Phosphate regulation (phoU), and DNA degradation (eddA) genes were also upregulated. All four genes have been identified to be upregulated under phosphate limitation in vitro (Hancock and Brinkman, 2002; Bains et al., 2012). P. aeruginosa is able to obtain phosphate from the host cell membrane via hydrolysis of the phospholipids using phospholipases (Sadikot et al., 2005). The non-hemolytic phospholipase C gene was highly upregulated 5.8 log2 fold-change in P. aeruginosa PASS1 during zebrafish infection (Figure 4).
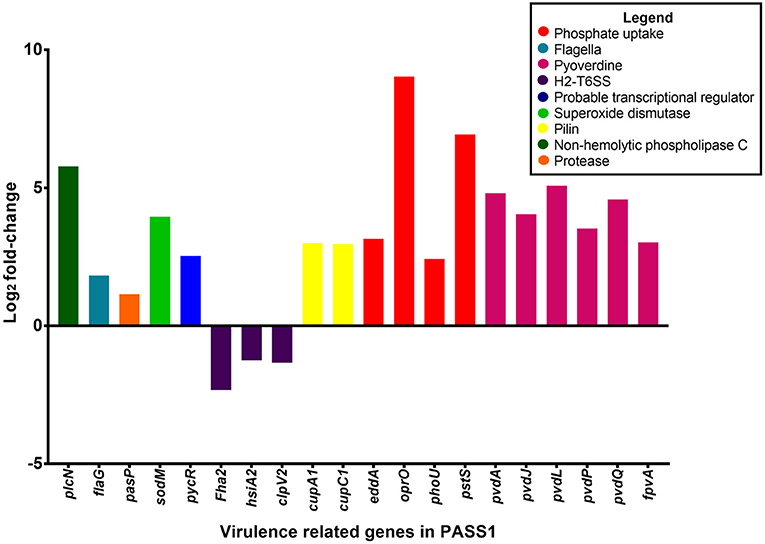
Figure 4. Differential gene expression of virulence-related genes of P. aeruginosa PASS1 during infection of zebrafish compared to PASS1 cells grown in Luria-Bertani medium. Log2 fold-change differential expression of known virulence genes (p ≤ 0.01 and log2 fold-changes cut-off −1≥ to ≤1).
Also upregulated were iron scavenging systems of PASS1. Pyoverdine biosynthesis genes and the ferri-pyoverdine receptor gene of the PASS1 strain were significantly upregulated in zebrafish. P. aeruginosa synthesizes and secretes the siderophore pyoverdine to scavenge ferric iron from host to overcome iron limitation during infection (Lamont et al., 2002; Konings et al., 2013; Nguyen et al., 2014).
In addition, a range of known P. aeruginosa virulence genes were differentially expressed in the zebrafish host compared with the culture in LB medium. These include genes encoding flagella biogenesis and the PasP protease (Figure 4). PasP is an extracellular protease (Pelzer et al., 2015) able to cleave collagen, contributing to the loss of epithelial cells (Tang et al., 2009, 2013).
The P. aeruginosa flaG flagellin gene was upregulated 1 log2 fold-change in the zebrafish host. The delivery of bacterial flagellin into the mice macrophage cytosol has been shown to trigger the NLRC4 inflammasome which mediates activation of the protease, caspase-1 (upregulated in our data set by 1 log2 fold-change (Mariathasan et al., 2004; Sutterwala et al., 2007). The activation of caspase-1 promotes the secretion of the proinflammatory cytokines IL-1β and IL-18 as well as pyroptosis, a form of cell death induced by bacterial pathogens (Franchi et al., 2007).
Expression of the cupA1 and cupC1 genes, encoding fimbrial subunits of chaperone-usher type fimbriae, were both increased in PASS1 in zebrafish. These class of fimbriae are important tissue-specific adhesins in many pathogens, and are presumably playing a role in adhesion to zebrafish cells.
The pycR gene encoding a LysR-type transcriptional regulator was upregulated by 2.5 log2 fold-change. This regulator modulates expression of virulence factors such as lipase/esterase and biofilm formation, as well as genes implicated in lipid metabolism and anaerobic respiration (Kukavica-Ibrulj and Levesque, 2008) (Figure S1).
The P. aeruginosa sodM superoxide dismutase gene was upregulated by 4 log2 fold-change in zebrafish. SodM protects P. aeruginosa against toxic effects of superoxides (Iiyama et al., 2007), so the increased expression level of sodM may be a defensive response against oxidative killing in the zebrafish macrophage phagolysosome. Iron limitation has been reported to lead to an increase in SodM activity in P. aeruginosa (Chang et al., 2005), which may be an alternate explanation for increased sodM expression. All of the P. aeruginosa type VI secretion system genes (orthologous to PA1656-PA1671 of PAO1) encoded within the Hcp secretion island-2 (H2-T6SS) showed lower expression levels in zebrafish compared with the culture in LB medium, although not all of them were below the significance threshold (p ≤ 0.01) (Table S1). This type VI secretion system in PAO1 has been shown to be important in virulence in a worm model and in mammalian cell cultures, and its expression has been shown to be induced by the Fur regulator during iron limitation and by quorum sensing (Sana et al., 2012). However, our transcriptomic data indicates that PASS1 is responding to iron limited conditions, thus the downregulation of the H2-T6SS genes in our infected zebrafish model suggests that there are other unknown regulatory pathways governing expression of this secretion system.
Other Differentially Expressed PASS1 Genes in a Zebrafish Model
The PASS1 genes faoA and foaB located in the fadBA5 β-oxidation operon were upregulated in the infected zebrafish. β-oxidative enzymes have been shown to be induced in vivo during lung infection in CF patients (Kang et al., 2008). In vitro studies have demonstrated that the fadBA5 operon is required for phosphatidylcholine (PC) and fatty acid (FA) degradation (Kang et al., 2008; Turner et al., 2014). The lung surfactant consists of ~10% surfactant proteins and ~90% lipids with phosphatidylcholine (PC) accounting for ~80% of the lipids (Griese, 1999; Son et al., 2007). The most abundant lipids in the zebrafish embryo are cholesterol, PC, and triglyceride (Fraher et al., 2016). These lipids are processed within the yolk prior to mobilization to the embryonic body (Hölttä-Vuori et al., 2010; Fraher et al., 2016). The PASS1 psrA gene encoding a TetR family transcriptional regulator required for regulation of the fadBA5 operon also showed increased expression in the zebrafish host (Figure S1). PsrA has been reported to also regulate the electron transfer flavoprotein B-subunit, etfB gene during stationary phase of bacterial growth (Kojic et al., 2005), and etfB also showed increased expression in PASS1 in zebrafish.
A variety of amino acid utilization genes, for example, liuA, liuB, and liuE encoding enzymes in the branched chain amino acid degradation pathway, showed decreased levels of expression in PASS1 cells in zebrafish. Conversely, a variety of PASS1 amino acid biosynthesis genes showed increased levels of expression in the zebrafish infection model. This most likely reflects the availability of amino acids in LB medium that was used for the in vitro control PASS1 culture.
The PA0622 and PA0623 genes in the bacteriocin R2 pyocin gene locus (Waite and Curtis, 2009; Purschke et al., 2012) were downregulated in PASS1 cells in the zebrafish model (Figure 3, Figure S2). This gene cluster has been implicated in the production of a cryptic prophage endolysin that mediates P. aeruginosa explosive cell lysis (Turnbull et al., 2016). This cell lysis results in the production of extracellular DNA that facilitates biofilm formation. Other biofilm-related genes, such as GacS/GacA and RetS/LadS two-component systems and quorum-sensing systems including las, rhl, and pqs (Rasamiravaka et al., 2015) were not significantly differentially expressed (Table S1).
Whole-Cell Transcriptome Analysis of Zebrafish Embryos Infected With P. aeruginosa PASS1
The transcriptome of zebrafish infected with PASS1 was compared with PBS-injected zebrafish to identify genes upregulated in response to P. aeruginosa infection. RNA-Seq analysis revealed 6,739 genes to be differentially expressed (p ≤ 0.01 and log2 fold-changes cut-off −1≥ to ≤1). This represents a quarter of the protein-encoding genes in the zebrafish genome, suggesting that there is a dramatic transcriptional response to infection. Of the differentially expressed genes, 2,510 were found to be upregulated and 4,229 were downregulated. The complete list of genes is provided in Table S2, and their log2 fold change in expression (p ≤ 0.01, log2 fold-change cut-off −1≥ to ≤1) are shown graphically in Figure 5.
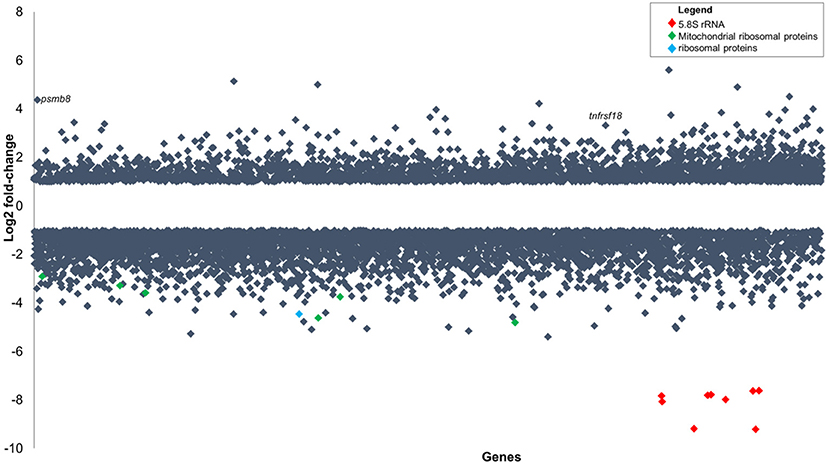
Figure 5. Gene expression changes in zebrafish embryos (log2 fold-change) infected with P. aeruginosa PASS1 compared to zebrafish embryos injected with phosphate buffered saline. Each dot represents a gene within the zebrafish genome (x-axis) and its fold-change (log2) expression 3 dpi (p ≤ 0.01 and log2 fold-changes cut-off −1≥ to ≤1).
Two of the most highly upregulated genes in the infected zebrafish are PSMB8 (4.4 log2 fold-change) and TNFRSF18 (3.3 log2 fold-change) (Figure 5). The PSMB8 gene has been linked to a number of auto-inflammatory diseases and found to be induced during the innate immune response of zebrafish to bacterial infection (Meijer and Spaink, 2011; Warnatsch et al., 2013). The TNFRSF18 gene is part of the TNF receptor signaling family and plays a role in anti-apoptotic signaling via TRAF2 (upregulated 0.4 log2 fold-change), which is thought to be involved in protection of lymphocytes against activation-induced cell death (Donaldson et al., 2005). Mycobacterial infection of zebrafish has suggested that TNF receptor signaling mediates resistance against mycobacteria (Meijer and Spaink, 2011). PSMB8 and TRAF2 genes may play a similar role in the zebrafish immune response against P. aeruginosa PASS1.
The most significantly downregulated genes in the zebrafish infected with P. aeruginosa PASS1 were the 5.8S rRNA genes (Figure 5). RNA isolated from both the infected and uninfected control cells underwent an rRNA depletion step, which makes it difficult to conclusively draw inferences about translation, as the differences in the 5S rRNA abundance could be due to artifacts introduced by the depletion process. Nevertheless, 22 mitochondrial ribosomal proteins, 9 ribosomal proteins, as well as several ribosomal proteins modifying enzymes all showed decreased expression in the infected zebrafish cells compared with the uninfected control (Table S2). This suggests that translation in the zebrafish is negatively impacted by the bacterial infection.
The downregulation of transcription of rRNA genes and ribosomal protein genes has been previously reported to occur due to intracellular and extracellular stressors (Xiao and Grove, 2009; Hayashi et al., 2014). Stress leads to the induction of processes such as cell cycle arrest, apoptosis or autophagy (Naora, 1999; Gupta et al., 2012; Hayashi et al., 2014). Consistent with this the PASS1-infected zebrafish transcriptome was significantly enriched in transcripts related to organismal injury and cell death (Figure S3).
PASS1-infected zebrafish displayed decreased expression of the prohibitin 2 (PHB2) mitochondrial protein, a coordinator/communication protein for cell division, metabolism, and cell death (Bavelloni et al., 2015). Proteins known to interact with PHB2, including transcription factors ATF2, MEF2A, TEAD3, DNA modifying proteins, SIRT2, HDAC5, RNF2, protease, AFG3L2, RNA binding/ processing proteins, AGO3, DDX20, cell cycle, KIF23, cytoskeleton/structural protein, NUP93, signal transduction, ADRB2, ATP5B, COX4I1, cellular respiration protein, COX6C, mitochondrial transport/translation TIMM50 were also significantly downregulated in PASS1-infected zebrafish, suggesting that PASS1 infection is impacting a swathe of activities linked to PHB2.
Cellular and Humoral Innate Immune Response in Zebrafish Infected With PASS1
Pathway analysis of the zebrafish transcriptome using the Ingenuity package revealed that several canonical pathways within the category of cellular and humoral innate immunity were highly enriched in zebrafish upon infection (Figure S4). These included phagosome maturation, leukocyte extravasation signaling, Fcɤ receptor-mediated phagocytosis in macrophages and monocytes, CXCR4 signaling, clathrin-mediated endocytosis signaling, IL-8 signaling, caveolar-mediated endocytosis signaling, fMLP signaling in neutrophils, production of nitric oxide and reactive oxygen species in macrophages, and macropinocytosis signaling.
In the leukocyte extravasation signaling pathway, the chemokines CXCR3 and CXCR4 was upregulated by 2 log2 fold-change and 1.1 log2 fold-change, respectively (Figure 6). The leukocyte extravasation signaling pathway involves the movement of leukocytes out of the circulatory system and toward the site of tissue damage and infection. Members of the CXC chemokine family have a role in inducing neutrophil recruitment (Gellatly and Hancock, 2013). Previously, the CXC chemokine family has been shown to be involved in the inflammatory response in mice to Pseudomonas lung infection (Tsai et al., 2000). These chemokines are presumably involved in enhancing migration of leukocytes to the site of bacterial infection.
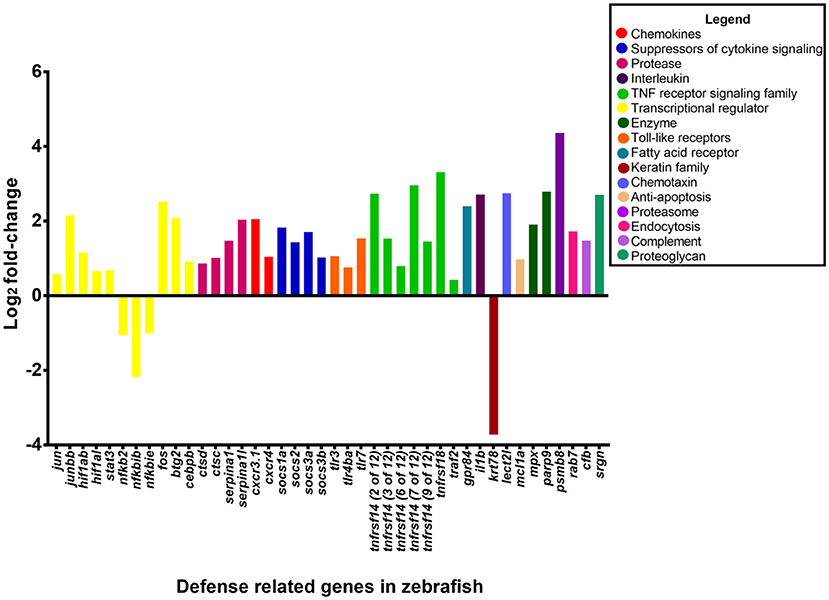
Figure 6. Expression of host defense-related genes in response to infection with P. aeruginosa PASS1. The upregulated genes in zebrafish infected with PASS1 (p ≤ 0.01 and log2 fold-change cut-off −1≥ to ≤1).
Toll like receptors (TLRs) which are expressed by neutrophils and macrophages play a key role in recognition of bacterial ligands (Lloyd et al., 2007; Mittal et al., 2014). TLR3, 4, and 7 were significantly upregulated in the zebrafish transcriptome following PASS1 infection (Table S2). In particular, TLR4, which can recognize bacterial lipolysaccharide, plays a significant role in the response to P. aeruginosa infections in mammalian lungs (Campodónico et al., 2008). TLRs also play important roles in regulating phagocytosis at multiple steps including internalization and enhancement of phagosome maturation (Blander and Medzhitov, 2004; Kagan and Iwasaki, 2012). RAB7 which is a mediator of late phagosome process (Flannagan et al., 2009) was upregulated by 1.7 log2 fold-change, suggestive of phagosomal activity against PASS1 inside the zebrafish host, which is consistent with interaction between PASS1 and phagosomes observed in our confocal microscopy (Figure 1).
Key transcriptional regulators in the acute phase response to tissue injury, infection, and inflammation, including FOS, JUN, and STAT3 were upregulated, while the NFKB2, NFKBIB, NFKBIE regulators were downregulated in response to PASS1 infection (Moshage, 1997). Previous studies using zebrafish embryos have shown upregulation of FOS and STAT3 in response to S. typhimurium and M. marinum (van der Vaart et al., 2012). The JUN and FOS transcriptional regulators together are known as activating protein 1 (AP-1), both are conserved between mammals and zebrafish (Meijer and Spaink, 2011; Ordas et al., 2011). AP-1 is involved in cellular expression, cell proliferation and differentiation, with its activation dependent on a variety of stress-related stimuli (Kim et al., 2003).
The intracellular suppressors of cytokine signaling (SOCS) genes SOCS1, SOCS2, and SOCS3 were upregulated in the acute phase signaling pathway by 1.8-, 1.4-, and 1 log2 fold-change, respectively, in response to PASS1 infection. The SOCS proteins are important regulators of acute phase response and cytokine signaling pathways as they regulate the balance between pro- and anti-inflammatory signals during infection (Wang et al., 2011; Brudal et al., 2014).
Identification of Genes Previously Linked to Cell Infection
Based on the Ingenuity Pathway Analysis there were 233 differentially expressed genes related to infection of cells (Figure S5). The 233 differentially expressed genes comprised genes encoding enzymes, G-protein coupled receptors, ion channels, growth factors, kinases, ligand–dependent nuclear receptors, peptidases, transcription regulators, translational regulators, transmembrane receptors, and transporters. The genes with the highest expression changes (> 2 log2 fold-change) were the transmembrane receptor (tumor necrosis factor receptor superfamily 14, TNFRS14), poly (ADP-ribose) polymerase PARP9, transcription regulator (BTG2) and the serine protease inhibitor SERPINA1 genes (Figure 6). This suggests a complex cascade of cellular events in response to P. aeruginosa infection.
Other Genes Differentially Expressed in Zebrafish in Response to Bacteria
Functional analysis of upregulated genes using DAVID (Huang da et al., 2009a,b) (Tables S3, S4) showed enrichment within the GO category “response to bacterium.” The genes upregulated by 2 log2 fold-change included G protein-coupled receptor 84 (GPR84), leukocyte cell-derived chemotaxin 2 like (LECT2L), and the tumor necrosis factor receptors TNFRSF18 and TNFRSF14. GPR84 is expressed in leukocytes, monocytes and macrophages, and is known to play a critical role in immune regulation (Cha et al., 2013; Figure 6). The acute phase response LECT2 protein attracts neutrophils (Škugor et al., 2009) and studies of mammalian LECT2 indicate that it plays a role in immune regulation (Chen et al., 2010). Infection studies with Aeromonas salmonicida and S. aureus have shown high induction of LECT2 in adult zebrafish (Lin et al., 2007). MPX, which is a zebrafish ortholog of the mammalian MPO gene was upregulated suggesting that there is presence of neutrophils at the site of infection which are undergoing apoptosis (Mathias et al., 2009).
Comparison of Zebrafish Infection With Various Pathogens
Previously Ordas et al. (2011) have compared Salmonella infection of zebrafish embryos with M. marinum infection of adult zebrafish (Hegedus et al., 2009). Transcriptomic datasets from zebrafish were compared to identify the overlap between the up- and downregulated transcripts of the Salmonella- and Mycobacterium-infected zebrafish. This revealed 288 and 3 commonly up- or downregulated transcripts. Comparison of our dataset to both these infection studies revealed 47 and 1 common up- or downregulated transcripts (Table S5). The one downregulated gene common to these two datasets, as well as our PASS1 zebrafish embryo infection, was KRT78, involved in translation.
The common set of 47 upregulated genes included 19 previously implicated in the vertebrate immune response. The MCL1A gene was upregulated by 1 log2 fold-change protects against apoptosis during initial steps of differentiation in human macrophages (Arslan et al., 2012). Complement factor B in macrophages was upregulated by 1.5 log2 fold-change, and its expression was proposed to be facilitated by TLR3, TLR4, and TRIF (Li et al., 2011). The transcriptional regulator CEBPβ was upregulated by 0.9 log2 fold-change, and it has been suggested to influence expression of the IL-1β gene (Didon et al., 2011), which, in turn, was upregulated by 2.7 log2 fold-change in our study, and is known to activate neutrophils and macrophages in bacterial phagocytosis (Didon et al., 2011). HIF-1α, a global regulator of macrophage and neutrophil inflammatory and innate immune functions that is stimulated by TLR4 (Zinkernagel et al., 2007), was upregulated by 0.7 log2 fold-change.
The gene encoding SRGN which interacts with inflammatory mediators such as IL-1β and TNF (Korpetinou et al., 2014) was upregulated by 1.4 log2 fold-change. The protease cathepsin C gene was upregulated by 1 log2 fold-change and is involved in the activation of granule serine peptidases in inflammatory cells (Turk et al., 2001). The cathepsin D protease gene was also upregulated (0.8 log2 fold-change) and has been implicated in macrophage apoptosis (Bewley et al., 2011).
Comparison of transcriptomic data from infection studies with different bacterial pathogens can thus be used to collectively define a common set of innate host genes expressed in response to infection. Comparison of zebrafish embryo infection studies with adult zebrafish infection studies provides an opportunity to dissect the innate immune response separate from the adaptive immune response. The generation of transcriptomics data investigating response to infection to various pathogens is valuable for future host-pathogen interaction studies as well as developing targeted therapeutics.
Conclusions
Previously, zebrafish was used as a model organism for P. aeruginosa infection by looking at the expression of specific immune related genes and in vivo interaction of the pathogen with phagocytes. In this study we report, for the first time, the simultaneous global gene expression of a zebrafish-P. aeruginosa systemic infection. RNA-Seq analysis has yielded a detailed view of both host and pathogen transcriptional responses. During infection, PASS1 displayed increased expression of an array of genes shown previously to be important in pathogenesis. We have also shown that phosphate and iron acquisition genes are significantly upregulated in PASS1, suggesting these are limiting nutrients within the zebrafish host. The response of zebrafish to PASS1 infection involved both humoral and cellular components of the innate immune system. Significant upregulation was observed for genes involved in bacterial recognition and clearance, inflammation and tissue injury. Based on the transcriptomic data, we present a schematic overview of the key response mechanisms in both host and pathogen during PASS1 infection of zebrafish embryos (Figure 7).
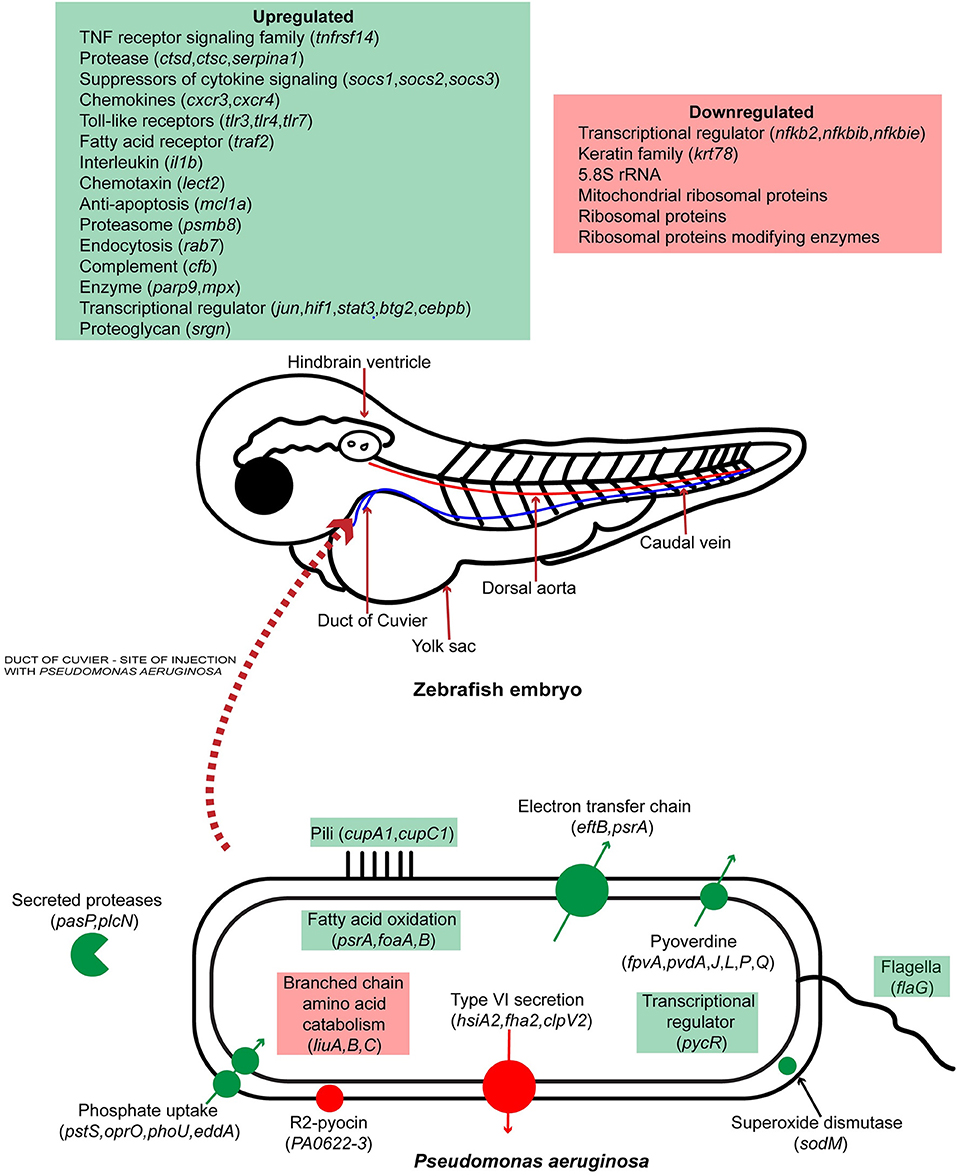
Figure 7. Schematic representation of host-pathogen interactions of zebrafish embryos infected with P. aeruginosa PASS1. Up and down regulated processes and genes are highlighted in green and red, respectively.
Data Availability Statement
We provide two supplementary tables with the gene expression profile of Pseudomonas aeruginosa PASS1 and Danio rerio following host-pathogen interaction. The P. aeruginosa PASS1 and Danio rerio transcriptomic data can be found in Dryad under the accession number doi:10.5061/dryad.3vk38 at http://datadryad.org/reviewdoi=doi:10.5061/dryad.3vk38.
Impact Statement
This study represents the first global expression view of the molecular interactions between Pseudomonas aeruginosa and zebrafish. P. aeruginosa is an opportunistic pathogen that is the major cause of mortality among cystic fibrosis patients. Our transcriptomic data suggests that key virulence mechanisms for P. aeruginosa PASS1 in zebrafish include adherence to host cells via Cup fimbriae, iron and phosphate scavenging, protease cleavage of host proteins such as collagen, and superoxide dismutase as a defense mechanism against oxidative killing. The host gene expression during P. aeruginosa PASS1 infection shows upregulation of an array of genes including transcriptional regulators, toll-like receptors and chemokines to be involved in the initiation, and process of phagocytosis. Decreased expression levels of ribosomal RNAs and translation proteins suggests that protein translation in the host is impacted by bacterial infection.
Ethics Statement
All zebrafish experiments were performed with the approval of University of Oslo (Animal ethic approval number: 6981) and Macquarie University (Animal ethic approval number: 5201500513) animal ethics committee and Macquarie University Internal Biosafety Committee (NLRD 5201500584). Zebrafish embryos were utilized 48 hpf for all infection experiments.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
The project was funded by the Australian Research Council (http://www.arc.gov.au). Super Science grant FS110200026. IP was supported by an Australian Research Council Laureate Fellowship (FL140100021). SK was supported by a Macquarie University Research Excellence Scholarship and the Australian Cystic Fibrosis Research Trust Postgraduate Studentship. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Rola Bazzi for assistance with zebrafish breeding at Macquarie University. Also, we would like to thank Anita Sulen at Norwegian University of Life Sciences, Norway for breeding of zebrafish. We are very grateful for the facilities provided for use during conducting these experiments by Gareth Griffith's laboratory and the School of Pharmacy laboratory at the University of Oslo. We would specifically like to thank the following members of these labs for their advice and assistance: Håkon Høgset, Leidy Lagos, Ewa Jaroszewicz, Anne Lise Rishovd, and Lilia Ulanova. We would like to thank Garry Myers, Taotao Huang, and Martin Ostrowski for valuable advice on bioinformatic analysis of dual RNA-seq data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00406/full#supplementary-material
Abbreviations
CF, cystic fibrosis; CLSM, confocal laser scanning microscopy; YFP, yellow fluorescent protein; HPI, hours post infection; DPI, days post infection; PBS, phosphate buffered saline; LB, Luria-Bertani; SOCS, suppressors of cytokine signaling; HPF, hours post fertilization.
References
Arslan, S. Y., Son, K.-N., and Lipton, H. L. (2012). The antiapoptotic protein Mcl-1 controls the type of cell death in theiler's virus-infected BHK-21 cells. J. Virol. 86, 1922–1929. doi: 10.1128/JVI.06516-11
Bains, M., Fernández, L., and Hancock, R. E. W. (2012). Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78, 6762–6768. doi: 10.1128/AEM.01015-12
Bartlett, M. S., and Gourse, R. L. (1994). Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J. Bacteriol. 176, 5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994
Bavelloni, A., Piazzi, M., Raffini, M., Faenza, I., and Blalock, W. L. (2015). Prohibitin 2, At a communications crossroads. Int. Union Biochem. Mol. Biol. Life 67, 239–254. doi: 10.1002/iub.1366
Benítez-Páez, A., Villarroya, M., and Armengod, M. E. (2012). Regulation of expression and catalytic activity of Escherichia coli RsmG methyltransferase. RNA 18, 795–806. doi: 10.1261/rna.029868.111
Bewley, M. A., Marriott, H. M., Tulone, C., Francis, S. E., Mitchell, T. J., Read, R. C., et al. (2011). A cardinal role for Cathepsin D in co-ordinating the host-mediated apoptosis of macrophages and killing of Pneumococci. PLoS Pathog. 7:e1001262. doi: 10.1371/journal.ppat.1001262
Blander, J. M., and Medzhitov, R. (2004). Regulation of phagosome maturation by signals from toll-like receptors. Science 304, 1014–1018. doi: 10.1126/science.1096158
Blazewicz, S. J., Barnard, R. L., Daly, R. A., and Firestone, M. K. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068. doi: 10.1038/ismej.2013.102
Blázquez, J., Gómez-Gómez, J. M., Oliver, A., Juan, C., Kapur, V., and Martín, S. (2006). PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol. Microbiol. 62, 84–99. doi: 10.1111/j.1365-2958.2006.05366.x
Brannon, M. K., Davis, J. M., Mathias, J. R., Hall, C. J., Emerson, J. C., Crosier, P. S., et al. (2009). Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell microbiology. 11, 755–768. doi: 10.1111/j.1462-5822.2009.01288.x
Brudal, E., Ulanova, L. S., O Lampe, E., Rishovd, A. L., Griffiths, G., and Winther-Larsen, H. C. (2014). Establishment of three Francisella infections in zebrafish embryos at different temperatures. Infect. Immun. 82, 2180–2194. doi: 10.1128/IAI.00077-14
Campodónico, V. L., Gadjeva, M., Paradis-Bleau, C., Uluer, A., and Pier, G. B. (2008). Airway epithelial control of Pseudomonas aeruginosa infection in cystic fibrosis. Trends Mol. Med. 14, 120–133. doi: 10.1016/j.molmed.2008.01.002
Cantin, A. M., Hartl, D., Konstan, M. W., and Chmiel, J. F. (2015). Inflammation in cystic fibrosis lung disease: pathogenesis and therapy. J. Cyst. Fibr. 14, 419–430. doi: 10.1016/j.jcf.2015.03.003
Cha, S. B., Lee, W. J., Shin, M. K., Jung, M. H., Shin, S. W., Yoo, A. N., et al. (2013). Early transcriptional responses of internalization defective Brucella abortus mutants in professional phagocytes, RAW 264.7. BMC Genomics 14:426. doi: 10.1186/1471-2164-14-426
Chang, W., Small, D. A., Toghrol, F., and Bentley, W. E. (2005). Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. doi: 10.1186/1471-2164-6-115
Chen, J., Lu, X. J., Yang, H. Y., and Shi, Y. H. (2010). An interaction between a C-type lectin receptor and leukocyte cell-derived chemotaxin 2 of ayu, Plecoglossus altivelis. Fish Shellf. Immunol. 28, 245–248. doi: 10.1016/j.fsi.2009.10.011
Clatworthy, A. E., Lee, J. S., Leibman, M., Kostun, Z., Davidson, A. J., and Hung, D. T. (2009). Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 77, 1293–1303. doi: 10.1128/IAI.01181-08
Cui, C., Benard, E. L., Kanwal, Z., Stockhammer, O. W., van der Vaart, M., Zakrzewska, A., et al. (2011). Infectious disease modeling and innate immune function in zebrafish embryos. Methods Cell Biol. 105, 273–308. doi: 10.1016/B978-0-12-381320-6.00012-6
Davies, J. C. (2002). Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr. Respir. Rev. 3, 128–134. doi: 10.1016/S1526-0550(02)00003-3
Dennis, P. P., Ehrenberg, M., and Bremer, H. (2004). Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68, 639–668. doi: 10.1128/MMBR.68.4.639-668.2004
Diaz-Pascual, F., Ortiz-Severin, J., Varas, M. A., Allende, M. L., and Chavez, F. P. (2017). In vivo Host-pathogen interaction as revealed by global proteomic profiling of zebrafish larvae. Front. Cell. Infect. Microbiol. 7:334. doi: 10.3389/fcimb.2017.00334
Didon, L., Barton, J. L., Roos, A. B., Gaschler, G. J., Bauer, C. M., Berg, T., et al. (2011). Lung epithelial CCAAT/enhancer-binding protein-beta is necessary for the integrity of inflammatory responses to cigarette smoke. Am. J. Respir. Crit. Care Med. 184, 233–242. doi: 10.1164/rccm.201007-1113OC
Donaldson, L., Vuocolo, T., Gray, C., Strandberg, Y., Reverter, A., McWilliam, S., et al. (2005). Construction and validation of a bovine innate immune microarray. BMC Genomics 6, 135–135. doi: 10.1186/1471-2164-6-135
Ellett, F., Pase, L., Hayman, J. W., Andrianopoulos, A., and Lieschke, G. J. (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117:e49–56. doi: 10.1182/blood-2010-10-314120
Flannagan, R. S., Cosio, G., and Grinstein, S. (2009). Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366. doi: 10.1038/nrmicro2128
Folkesson, A., Jelsbak, L., Yang, L., Johansen, H. K., Ciofu, O., Høiby, N., et al. (2012). Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat. Rev. Microbiol. 10, 841–851. doi: 10.1038/nrmicro2907
Fraher, D., Sanigorski, A., Mellett, N. A., Meikle, P. J., Sinclair, A. J., and Gibert, Y. (2016). Zebrafish embryonic lipidomic analysis reveals that the yolk cell is metabolically active in processing lipid. Cell Rep. 14, 1317–1329. doi: 10.1016/j.celrep.2016.01.016
Franchi, L., Stoolman, J., Kanneganti, T. D., Verma, A., Ramphal, R., and Núñez, G. (2007). Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur. J. Immunol. 37, 3030–3039. doi: 10.1002/eji.200737532
Freedman, S. D., Katz, M. H., Parker, E. M., Laposata, M., Urman, M. Y., and Alvarez, J. G. (1999). A membrane lipid imbalance plays a role in the phenotypic expression of cystic fibrosis in cftr(–/–) mice. Proc. Natl. Acad. Sci. U.S.A. 96, 13995–14000. doi: 10.1073/pnas.96.24.13995
Gellatly, S. L., and Hancock, R. E. (2013). Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173. doi: 10.1111/2049-632X.12033
Griese, M. (1999). Pulmonary surfactant in health and human lung diseases: state of the art. Eur. Respir. J. 13, 1455–1476. doi: 10.1183/09031936.99.13614779
Gupta, R., Kim, S., and Taylor, M. W. (2012). Suppression of ribosomal protein synthesis and protein translation factors by Peg-interferon alpha/ribavirin in HCV patients blood mononuclear cells (PBMC). J. Transl. Med. 10, 54–54. doi: 10.1186/1479-5876-10-54
Hall, C. J., Boyle, R. H., Astin, J. W., Flores, M. V., Oehlers, S. H., Sanderson, L. E., et al. (2013). Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating beta-oxidation-dependent mitochondrial ROS production. Cell Metab. 18, 265–278. doi: 10.1016/j.cmet.2013.06.018
Hancock, R. E., and Brinkman, F. S. (2002). Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56, 17–38. doi: 10.1146/annurev.micro.56.012302.160310
Hayashi, Y., Kuroda, T., Kishimoto, H., Wang, C., Iwama, A., and Kimura, K. (2014). Downregulation of rRNA transcription triggers cell differentiation. PLoS ONE. 9:e98586. doi: 10.1371/journal.pone.0098586
Hegedus, Z., Zakrzewska, A., Agoston, V. C., Ordas, A., Rácz, P., Mink, M., et al. (2009). Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol. Immunol. 46, 2918–2930. doi: 10.1016/j.molimm.2009.07.002
Hölttä-Vuori, M., Salo, V. T., Nyberg, L., Brackmann, C., Enejder, A., Panula, P., et al. (2010). Zebrafish: gaining popularity in lipid research. Biochem. J. 429, 235–242. doi: 10.1042/BJ20100293
Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. doi: 10.1038/nature12111
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Iiyama, K., Chieda, Y., Lee, J. M., Kusakabe, T., Yasunaga-Aoki, C., and Shimizu, S. (2007). Effect of superoxide dismutase gene inactivation on virulence of Pseudomonas aeruginosa PAO1 toward the silkworm, Bombyx mori. Appl. Environ. Microbiol. 73, 1569–1575. doi: 10.1128/AEM.00981-06
Kagan, J. C., and Iwasaki, A. (2012). Phagosome as the organelle linking innate and adaptive immunity. Traffic 13, 1053–1061. doi: 10.1111/j.1600-0854.2012.01377.x
Kang, Y., Nguyen, D. T., Son, M. S., and Hoang, T. T. (2008). The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 beta-oxidation operon. Microbiology 154(Pt 6), 1584–1598. doi: 10.1099/mic.0.2008/018135-0
Kaur, J., Pethani, B. P., Kumar, S., Kim, M., Sunna, A., Kautto, L., et al. (2015). Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front. Microbiol. 6:866. doi: 10.3389/fmicb.2015.00866
Kim, J., Woolridge, S., Biffi, R., Borghi, E., Lassak, A., Ferrante, P., et al. (2003). Members of the AP-1 family, c-Jun and c-Fos, functionally interact with jc virus early regulatory protein large t antigen. J. Virol. 77, 5241–5252. doi: 10.1128/JVI.77.9.5241-5252.2003
Kline, K. A., Fälker, S., Dahlberg, S., Normark, S., and Henriques-Normark, B. (2009). Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 5, 580–592. doi: 10.1016/j.chom.2009.05.011
Kojic, M., Jovcic, B., Vindigni, A., Odreman, F., and Venturi, V. (2005). Novel target genes of PsrA transcriptional regulator of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 246, 175–181. doi: 10.1016/j.femsle.2005.04.003
Konings, A. F., Martin, L. W., Sharples, K. J., Roddam, L. F., Latham, R., Reid, D. W., et al. (2013). Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect. Immun. 81, 2697–2704. doi: 10.1128/IAI.00418-13
Korpetinou, A., Skandalis, S. S., Labropoulou, V. T., Smirlaki, G., Noulas, A., Karamanos, N. K., et al. (2014). Serglycin: at the crossroad of inflammation and malignancy. Front. Oncol. 3:327. doi: 10.3389/fonc.2013.00327
Kukavica-Ibrulj, I., and Levesque, R. C. (2008). Animal models of chronic lung infection with Pseudomonas aeruginosa: useful tools for cystic fibrosis studies. Lab. Anim. 42, 389–412. doi: 10.1258/la.2007.06014e
Lamont, I. L., Beare, P. A., Ochsner, U., Vasil, A. I., and Vasil, M. L. (2002). Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 99, 7072–7077. doi: 10.1073/pnas.092016999
Lessman, C. A. (2011). The developing zebrafish (Danio rerio): a vertebrate model for high-throughput screening of chemical libraries. Birth Defects Res. Part C 93, 268–280. doi: 10.1002/bdrc.20212
Li, Q., Li, Y. X., Stahl, G. L., Thurman, J. M., He, Y., and Tong, H. H. (2011). Essential role of factor B of the alternative complement pathway in complement activation and opsonophagocytosis during acute pneumococcal otitis media in mice. Infect. Immun. 79, 2578–2585. doi: 10.1128/IAI.00168-11
Lima, A., Cha, B. J., Amin, J., Smith, L. K., and Anderson, B. (2014). Zebrafish embryo model of Bartonella henselae infection. Zebrafish 11, 434–446. doi: 10.1089/zeb.2014.1001
Lin, B., Chen, S., Cao, Z., Lin, Y., Mo, D., Zhang, H., et al. (2007). Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: Striking similarities and obvious differences with mammals. Mol. Immunol. 44:295–301. doi: 10.1016/j.molimm.2006.03.001
Lloyd, D. H., Viac, J., Werling, D., Rème, C. A., and Gatto, H. (2007). Role of sugars in surface microbe-host interactions and immune reaction modulation. Vet. Dermatol. 18, 197–204. doi: 10.1111/j.1365-3164.2007.00594.x
Lyczak, J. B., Cannon, C. L., and Pier, G. B. (2000). Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2, 1051–1060. doi: 10.1016/S1286-4579(00)01259-4
Mariathasan, S., Newton, K., Monack, D. M., Vucic, D., French, D. M., Lee, W. P., et al. (2004). Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218. doi: 10.1038/nature02664
Mathias, J. R., Dodd, M. E., Walters, K. B., Yoo, S. K., Ranheim, E. A., and Huttenlocher, A. (2009). Characterization of zebrafish larval inflammatory macrophages. Dev. Comp. Immunol. 33, 1212–1217. doi: 10.1016/j.dci.2009.07.003
Meijer, A. H., and Spaink, H. P. (2011). Host-pathogen interactions made transparent with the zebrafish model. Curr. Drug Targets. 12, 1000–1017. doi: 10.2174/138945011795677809
Mittal, M., Siddiqui, M. R., Tran, K., Reddy, S. P., and Malik, A. B. (2014). Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 20, 1126–1167. doi: 10.1089/ars.2012.5149
Mogensen, T. H. (2009). Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240–273. doi: 10.1128/CMR.00046-08
Naora, H. (1999). Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol. Cell Biol. 77, 197–205. doi: 10.1046/j.1440-1711.1999.00816.x
Nguyen, A. T., O'Neill, M. J., Watts, A. M., Robson, C. L., Lamont, I. L., Wilks, A., et al. (2014). Adaptation of iron homeostasis pathways by a Pseudomonas aeruginosa pyoverdine mutant in the cystic fibrosis lung. J. Bacteriol. 196, 2265–2276. doi: 10.1128/JB.01491-14
Ordas, A., Hegedus, Z., Henkel, C. V., Stockhammer, O. W., Butler, D., Jansen, H. J., et al. (2011). Deep sequencing of the innate immune transcriptomic response of zebrafish embryos to Salmonella infection. Fish Shellf. Immunol. 31, 716–724. doi: 10.1016/j.fsi.2010.08.022
Papaioannou, E., Utari, P. D., and Quax, W. J. (2013). Choosing an appropriate infection model to study quorum sensing inhibition in Pseudomonas infections. Int. J. Mol. Sci. 14, 19309–19340. doi: 10.3390/ijms140919309
Pelzer, A., Schwarz, C., Knapp, A., Wirtz, A., Wilhelm, S., Smits, S., et al. (2015). Functional expression, purification, and biochemical properties of subtilase SprP from Pseudomonas aeruginosa. Microbiol. Open 4, 743–752. doi: 10.1002/mbo3.275
Penesyan, A., Kumar, S. S., Kamath, K., Shathili, A. M., Venkatakrishnan, V., Krisp, C., et al. (2015). Genetically and phenotypically distinct Pseudomonas aeruginosa cystic fibrosis isolates share a core proteomic signature. PLoS ONE 10:e0138527. doi: 10.1371/journal.pone.0138527
Phennicie, R. T., Sullivan, M. J., Singer, J. T., Yoder, J. A., and Kim, C. H. (2010). Specific resistance to Pseudomonas aeruginosa infection in zebrafish is mediated by the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 78, 4542–4550. doi: 10.1128/IAI.00302-10
Purschke, F. G., Hiller, E., Trick, I., and Rupp, S. (2012). Flexible survival strategies of Pseudomonas aeruginosa in biofilms result in increased fitness compared with Candida albicans. Mol. Cell. Prot. 11, 1652–1669. doi: 10.1074/mcp.M112.017673
Ramos, C., Molbak, L., and Molin, S. (2000). Bacterial activity in the rhizosphere analyzed at the single-cell level by monitoring ribosome contents and synthesis rates. Appl. Environ. Microbiol. 66, 801–809. doi: 10.1128/AEM.66.2.801-809.2000
Rang, C. U., Licht, T. R., Midtvedt, T., Conway, P. L., Chao, L., Krogfelt, K. A., et al. (1999). Estimation of growth rates of Escherichia coli BJ4 in streptomycin-treated and previously germfree mice by in situ rRNA hybridization. Clin. Diagn. Lab. Immunol. 6, 434–436.
Rasamiravaka, T., Labtani, Q., Duez, P., and El Jaziri, M. (2015). The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res. Int. 2015:17. doi: 10.1155/2015/759348
Sadikot, R. T., Blackwell, T. S., Christman, J. W., and Prince, A. S. (2005). Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171, 1209–1223. doi: 10.1164/rccm.200408-1044SO
Sana, T. G., Hachani, A., Bucior, I., Soscia, C., Garvis, S., Termine, E., et al. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J. Biol. Chem. 287, 27095–27105. doi: 10.1074/jbc.M112.376368
Schneider, D. A., Ross, W., and Gourse, R. L. (2003). Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 6, 151–156. doi: 10.1016/S1369-5274(03)00038-9
Sieger, D., Stein, C., Neifer, D., van der Sar, A. M., and Leptin, M. (2009). The role of gamma interferon in innate immunity in the zebrafish embryo. Dis. Model. Mech. 2, 571–581. doi: 10.1242/dmm.003509
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. doi: 10.1371/journal.ppat.1000949
Škugor, S., Jørgensen, S. M., and Gjerde, B., Krasnov, A. (2009). Hepatic gene expression profiling reveals protective responses in Atlantic salmon vaccinated against furunculosis. BMC Genomics 10, 503–503. doi: 10.1186/1471-2164-10-503
Son, M. S., Matthews, W. J. Jr., Kang, Y., Nguyen, D. T., and Hoang, T. T. (2007). In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75, 5313–5324. doi: 10.1128/IAI.01807-06
Sousa, A. M., and Pereira, M. O. (2014). Pseudomonas aeruginosa Diversification during infection development in cystic fibrosis lungs—A review. Pathogens 3, 680–703. doi: 10.3390/pathogens3030680
Stockhammer, O. W., Zakrzewska, A., Hegedus, Z., Spaink, H. P., and Meijer, A. H. (2009). Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J. Immunol. 182, 5641–5653. doi: 10.4049/jimmunol.0900082
Sutterwala, F. S., Mijares, L. A., Li, L., Ogura, Y., Kazmierczak, B. I., and Flavell, R. A. (2007). Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204, 3235–3245. doi: 10.1084/jem.20071239
Tang, A., Caballero, A. R., Marquart, M. E., and O'Callaghan, R. J. (2013). Pseudomonas aeruginosa small protease (PASP), a keratitis virulence factor. Invest. Ophthalmol. Vis. Sci. 54, 2821–2828. doi: 10.1167/iovs.13-11788
Tang, A., Marquart, M. E., Fratkin, J. D., McCormick, C. C., Caballero, A. R., Gatlin, H. P., et al. (2009). Properties of PASP: a Pseudomonas protease capable of mediating corneal erosions. Invest. Ophthalmol. Vis. Sci. 50, 3794–3801. doi: 10.1167/iovs.08-3107
Tang, H., Kays, M., and Prince, A. (1995). Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect. Immun. 63, 1278–1285.
Torraca, V., Masud, S., Spaink, H. P., and Meijer, A. H. (2014). Macrophage-pathogen interactions in infectious diseases: new therapeutic insights from the zebrafish host model. Dis. Model. Mech. 7, 785–797. doi: 10.1242/dmm.015594
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Tsai, W. C., Strieter, R. M., Mehrad, B., Newstead, M. W., Zeng, X., and Standiford, T. J. (2000). CXC chemokine receptor CXCR2 is essential for protective innate host response in murine Pseudomonas aeruginosa pneumonia. Infect. Immun. 68, 4289–4296. doi: 10.1128/IAI.68.7.4289-4296.2000
Turk, D., Janjić, V., Stern, I., Podobnik, M., Lamba, D., Dahl, S. W., et al. (2001). Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 20, 6570–6582. doi: 10.1093/emboj/20.23.6570
Turnbull, L., Toyofuku, M., Hynen, A. L., Kurosawa, M., Pessi, G., Petty, N. K., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7:11220. doi: 10.1038/ncomms11220
Turner, K. H., Everett, J., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2014). Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound Infection. PLoS Genet. 10:e1004518. doi: 10.1371/journal.pgen.1004518
van der Sar, A. M., Musters, R. J., van Eeden, F. J., Appelmelk, B. J., Vandenbroucke-Grauls, C. M., and Bitter, W. (2003). Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell Microbiol. 5, 601–611. doi: 10.1046/j.1462-5822.2003.00303.x
van der Sar, A. M., Spaink, H. P., Zakrzewska, A., Bitter, W., and Meijer, A. H. (2009). Specificity of the zebrafish host transcriptome response to acute and chronic mycobacterial infection and the role of innate and adaptive immune components. Mol. Immunol. 46, 2317–2332. doi: 10.1016/j.molimm.2009.03.024
van der Vaart, M., Spaink, H. P., and Meijer, A. H. (2012). Pathogen recognition and activation of the innate immune response in zebrafish. Adv. Hematol. 2012:19. doi: 10.1155/2012/159807
van der Vaart, M., van Soest, J. J., Spaink, H. P., and Meijer, A. H. (2013). Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis. Model. Mech. 6, 841–854. doi: 10.1242/dmm.010843
van Soest, J. J., Stockhammer, O. W., Ordas, A., Bloemberg, G. V., Spaink, H. P., and Meijer, A. H. (2011). Comparison of static immersion and intravenous injection systems for exposure of zebrafish embryos to the natural pathogen Edwardsiella tarda. BMC Immunol. J. 12:58. doi: 10.1186/1471-2172-12-58
Waite, R. D., and Curtis, M. A. (2009). Pseudomonas aeruginosa PAO1 pyocin production affects population dynamics within mixed-culture biofilms. J. Bacteriol. 191, 1349–1354. doi: 10.1128/JB.01458-08
Wang, T., Gorgoglione, B., Maehr, T., Holland, J. W., Vecino, J. L., Wadsworth, S., et al. (2011). Fish suppressors of cytokine signaling (SOCS): Gene discovery, modulation of expression and function. J. Signal Transduct. 2011:20. doi: 10.1155/2011/905813
Warnatsch, A., Bergann, T., and Kruger, E. (2013). Oxidation matters: the ubiquitin proteasome system connects innate immune mechanisms with MHC class I antigen presentation. Mol. Immunol. 55, 106–109. doi: 10.1016/j.molimm.2012.10.007
Whitehead, K. A., Rogers, D., Colligon, J., Wright, C., and Verran, J. (2006). Use of the atomic force microscope to determine the effect of substratum surface topography on the ease of bacterial removal. Colloids Surf. B 51, 44–53. doi: 10.1016/j.colsurfb.2006.05.003
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., and Brinkman, F. S. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucl. Acids Res. 44(Database issue), D646–D653. doi: 10.1093/nar/gkv1227
Xiao, L., and Grove, A. (2009). Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr. Genomics 10, 198–205. doi: 10.2174/138920209788185261
Keywords: RNA-Seq, host-pathogen interactions, virulence, Pseudomonas aeruginosa, zebrafish, innate immunity
Citation: Kumar SS, Tandberg JI, Penesyan A, Elbourne LDH, Suarez-Bosche N, Don E, Skadberg E, Fenaroli F, Cole N, Winther-Larsen HC and Paulsen IT (2018) Dual Transcriptomics of Host-Pathogen Interaction of Cystic Fibrosis Isolate Pseudomonas aeruginosa PASS1 With Zebrafish. Front. Cell. Infect. Microbiol. 8:406. doi: 10.3389/fcimb.2018.00406
Received: 12 August 2018; Accepted: 29 October 2018;
Published: 22 November 2018.
Edited by:
Matthew C. Wolfgang, University of North Carolina at Chapel Hill, United StatesReviewed by:
Melody N. Neely, University of Maine, United StatesMurugesan V. S. Rajaram, The Ohio State University, United States
Copyright © 2018 Kumar, Tandberg, Penesyan, Elbourne, Suarez-Bosche, Don, Skadberg, Fenaroli, Cole, Winther-Larsen and Paulsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian T. Paulsen, aWFuLnBhdWxzZW5AbXEuZWR1LmF1
†Present Address: Nadia Suarez-Bosche, Illawarra Health and Medical Research Institute, University of Wollongong, Wollongong, NSW, Australia
 Sheemal S. Kumar
Sheemal S. Kumar Julia I. Tandberg
Julia I. Tandberg Anahit Penesyan
Anahit Penesyan Liam D. H. Elbourne
Liam D. H. Elbourne Nadia Suarez-Bosche
Nadia Suarez-Bosche Emily Don
Emily Don Eline Skadberg
Eline Skadberg Federico Fenaroli
Federico Fenaroli Nicholas Cole
Nicholas Cole Hanne Cecilie Winther-Larsen
Hanne Cecilie Winther-Larsen Ian T. Paulsen
Ian T. Paulsen