- 1Key Laboratory of Animal Epidemiology and Zoonosis, College of Veterinary Medicine, China Agricultural University, Beijing, China
- 2Institute of Animal Husbandry and Veterinary Medicine, Beijing Academy of Agricultural and Forestry Sciences, Beijing, China
- 3Veterinary Diagnostic Laboratory, China Animal Disease Control Center, Beijing, China
MGA_0676 has been characterized as a Mycoplasma gallisepticum nuclease that can induce apoptosis of chicken cells. However, the mechanism by which MGA_0676 induces apoptosis has remained unclear. In this study, we evaluated MGA_0676-induced apoptosis and internalization in immortalized chicken embryo fibroblasts (DF-1) and cancer cell lines. The internalization of MGA_0676 was proven through caveolin-mediated endocytosis by blocking the endocytosis with specific inhibitors or with siRNA. We identified the Thif domain of NEDD8-activating enzyme E1 regulatory subunit (NAE) in DF-1 as the target region interacting with the SNC domain of MGA_0676. The interaction between the Thif and SNC domains was observed co-located in the perinuclear and nuclear of DF-1. We found that the interaction between NAE and MGA_0676 increased the ability of apoptosis and accelerated the process of cullin neddylation in DF-1 cells, in turn activating NF-κB. This resulted in the observed aggregation of NF-κB in the nuclei of DF-1 cells. Moreover, the apoptosis induced by MGA_0676 decreased significantly when NF-κB was inhibited by siRNA or BAY 11-7082 or when NAE was silenced by siRNA. Overall, our results demonstrate that MGA_0676 is internalized through caveolin-mediated endocytosis, interacts with SNC-dependent Thif to accelerate the process of cullin neddylation and activates NF-κB in DF-1 cells, ultimately playing a key role in apoptosis in chicken cells. Our results indicate MGA_0676 constitutes a critical etiological virulence factor of the respiratory disease caused by M. gallisepticum. This study also opens a venue to investigate MGA_0676 as a potential candidate as pro-apoptotic drug in cancer studies.
Introduction
Microbes have been reported to mediate pathogenicity in host cells by utilizing their virulence proteins (Fitzgerald et al., 2006; Humphrys et al., 2013; Krachler and Orth, 2013). Many of these virulence-associated proteins have also been found to play a significant role in cytotoxicity through their interaction with host cell proteins (Casadevall and Pirofski, 2001; Sourjik and Berg, 2004; Chen et al., 2011). Previous reports have shown that proteins of microbes with nuclease activity are cytotoxic to host cells, and are directly related to their pathogenicity (Pediaditakis et al., 2012; Milner et al., 2014; Sacco et al., 2014). Nucleases are expressed by many different pathogenic microbes and have been shown to play a role in creating favorable nutrient-rich environments (Cuatrecasas et al., 1967; Ma and Goodridge, 1992; Suh and Benedik, 1997). Recently, it has been demonstrated that many pathogenic microbes possess diverse nucleases with biological functions related to the metabolism of nitrogen, phosphate and carbohydrates, as well as their survival and virulence (Suciu and Inouye, 1996; Dominski, 2007; Lennon, 2007). Other studies have indicated that microbial nucleases degrade neutrophil extracellular traps (NETs) and evade the host cell innate immune responses (Papayannopoulos and Zychlinsky, 2009; Thammavongsa et al., 2013; Toid, 2014). Furthermore, some bacteria possess more than one kind of nucleases, which are vital to virulence and contribute to a wide variety of cellular biological processes, such as inducing host cell death, repairing DNA damage, and evading immune clearance from host tissues (Rottem, 2003; Amundsen et al., 2008; Delaney et al., 2012; Derré-Bobillot et al., 2013; Shields et al., 2013). Due to their extremely limited biosynthetic capabilities, Mycoplasma adopts a parasitic lifestyle in order to obtain their nutritional needs from host cells (Chung et al., 2010; Fan et al., 2010; Großhennig et al., 2013). Without the ability to synthesize de novo purine and pyrimidine bases, Mycoplasma has to salvage nucleotide bases to produce nucleotide precursors (Wanga et al., 2014). However, these salvage pathways result in a series of pathological cellular processes, such as inflammation and apoptosis (Razin, 1999; Nakhyung, 2009). Numerous intracellular, extracellular and, particularly, membrane-associated nucleases have been reported in different Mycoplasma species, many of which are implicated in host pathogenicity and cytotoxicity through the degradation of nucleotides and induction of apoptosis-like cell death (Pollack and Hoffmann, 1982; Minion et al., 1993; Paddenberg et al., 1998). Some membrane-associated nucleases have been shown to have a SNC region and able to translocate into cells, a process followed by cytotoxic effects and induction of apoptosis, such as MPN133 in M. pneumoniae, mhp379 in M. hyopneumoniae, and MG186 in M. genitalium (Schmidt et al., 2007; Li et al., 2010; Somarajan et al., 2010). Therefore, it is worthwhile to examine the biological properties and mechanisms of mycoplasmal membrane-associated nucleases.
Previously, we found that M. gallisepticum MGA_0676 was a Ca2+-dependent cytotoxic nuclease containing a SNC region similar to other mycoplasmal nucleases, which could translocate into chicken cells and induce apoptosis in a SNC-dependent manner (Xu et al., 2015). However, the mechanism by which MGA_0676 induced apoptosis remained unclear. Nuclear factor-kappa B (NF-κB) is a very important molecule associated with many signaling pathways, but few studies have been made to investigate the relationship between NF-κB and apoptosis. To evaluate these mechanisms, in the present study we show that MGA_0676 internalizes through caveolin-mediated endocytosis, interacts with Thif-dependent SNC, accelerating the process of cullin neddylation and activating NF-κB in DF-1 cells, ultimately inducing apoptosis. In addition, we also show that MGA_0676 may be an important etiological virulence factor of the respiratory disease caused by M. gallisepticum, and that it may be involved in the immunosuppression of the infected birds.
Materials and Methods
Bacterial Strains, Plasmids, and DNA Manipulations
M. gallisepticum from the BJ44T strain (CVCC350, preserved in China Veterinary Culture Collection Center, Beijing, China) were grown in PPLO medium (BD, Franklin Lakes, NJ, USA) as described previously (Xu et al., 2015). Escherichia coli (E. coli) T1 (Invitrogen, Carlsbad, CA, USA) and E. coli BL21(DE3) pLysS competent E. coli (TransGen Biotech, Beijing, China) were grown in Luria–Bertani (LB) broth and used to clone and express M. gallisepticum nuclease (MGA_0676, AE015450.2). Vectors pGEX-6p-1, pET28a, pEGF-N1, pCMV-HA-tag plamid, and pCMV-Myc-tag plamid (Novagen, Darmstadt, Germany) were used for DNA manipulations.
Cell Lines, Proteins, Antibodies, and Reagents
Immortal chicken embryo fibroblasts (DF-1) and human embryonic kidney 293T cells (HEK293T) were obtained from ATCC (American Type Culture Collection, Manassas, VA, USA). All cells were cultured in Dulbecco's modified Eagle medium (DMEM, Invitrogen, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) in a 5% CO2 incubator. All restriction enzymes were purchased from New England Biolabs (Ipswich, MA, USA). Annexin V/PI apoptosis assay kits were purchased from BD (Franklin Lakes, NJ, USA). Anti-GST polyclonal antibody, anti-GFP polyclonal antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Rabbit polyclonal anti-clathrin-1, anti-cholera antibodies anti-transferrin antibody, and anti-cleaved caspase 3 antibodies were obtained from Abcam (Cambridge MA, USA). Anti-HA monoclonal antibodies, anti-Myc antibodies, anti-Rela antibodies, anti-IκBα antibodies, anti p-IκBα antibodies, and β-actin antibodies were obtained from Abclonal Inc. (Cambridge MA, USA). Mouse anti-NAE polyclonal antibody was prepared with purified recombinant NAE protein according to a standard molecular biology technique (Xu et al., 2015). Mouse anti-MGA_0676 monoclonal antibody was prepared according to a previously reported standard protocol (Fu et al., 2014). Alexa Fluor 555-conjugated phalloidin (red) and Lipofectamine® LTX DNA transfection reagents were purchased from Invitrogen. Human holo-transferrin (Tf), cholera toxin-FITC, monodansylcadaverine (MDC), and filipin were purchased from Sigma-Aldrich (Louis, MO, USA). All other chemicals reagents used in the study were of analytical grade.
Computer-Assisted Sequence Analysis
The M. gallisepticum MGA_0676 sequence (MGA_0676) was downloaded from the UniProtKB database at http://www.uniprot.org/uniprot/C0SKM0. The NEDD8-activating enzyme E1 regulatory subunit (NAE, NP_001006129.1) of DF-1 cells was downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/protein/57524906). Domains of MGA_0676 and NAE were analyzed using the PROSITE database (http://www.expasy.ch/tools/scanprosite/). Prediction of the signal peptide cleavage sites in MGA_0676 was performed using SignalP Server (http://www.cbs.dtu.dk/services/SignalP/). Sequence alignment analysis and three-dimensional (3D) structure modeling were performed using ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) and homology remodeling tools (http://swissmodel.expasy.org/workspace/index). Prediction of the interactions between proteins was done using the Hexserver tool (http://hexserver.loria.fr/). Software programs, such as DNAstar, DNAMAN, and Primer Premier 5, were also used in the analyses.
Preparation of Recombinant Proteins
Recombinant His-tag MGA_0676 (rMGA_0676) and recombinant His-tag MGA_0676ΔSNC (rMGA_0676ΔSNC) were prepared as described previously (Xu et al., 2015). Recombinant GST-tag MGA_0676 was prepared with vector pGEX-6p-1 and recombinant His-tag NAE was prepared with Vector pET28a. All proteins were purified using standard molecular biology techniques. Protein concentrations were determined by the Bradford protein assay using bovine serum albumin (BSA) as a standard. Nuclease activity of rMGA_0676 and rMGA_0676ΔSNC was detected as described previously (Xu et al., 2015).
Construction of Plasmids
The MGA_0676 gene of M. gallisepticum was PCR amplified from the M. gallisepticum strain BJ44T chromosomal DNA. After changing the TGA encoded tryptophan codons using specific primers, the M. gallisepticum MGA_0676 gene was subcloned in the pEGF-N1 and pCMV-HA expression vectors. The plasmid was then introduced into E. coli T1 and extracted using a Qiagen plasmid kit. MGA_0676ΔSNC (MGA_0676 without the SNC region) was amplified by OE-PCR using specific primers and construction of recombinant plasmid (pEGF-N1-MGA_0676ΔSNC and pCMV-HA-MGA_0676ΔSNC) was performed as described previously (Xu et al., 2015).
The NEDD8-activating enzyme E1 regulatory subunit (NAE, 57524906) was cloned from the cDNA of DF-1 cells using specific primers (Table S1). The pCMV-Myc-NAE and pCMV-Myc-NAEΔThif (NAE without Thif region) expression plasmids were constructed using standard molecular biology techniques. All the primers were synthesized by Sangon Biotech (Beijing, China) (Table S1).
Detection of rMGA_0676 Internalization in Cells
For immunofluorescence antibody assays (IFA), HepsG2, A549, Hela, and 293A cells were treated with rMGA_0676 or rMGA_0676ΔSNC (40 μg/ml) for 24 h at 37°C. DF-1 cells treated with recombinant proteins were used as positive controls. PBS-treated cells were used as negative controls. After incubation, cells were fixed with 4% paraformaldehyde, permeabilized with 0.25% TritonX-100 for 5–10 min, blocked with 3% bovine serum albumin, and incubated with mouse anti-His monoclonal antibody overnight at 4°C. Cells were then incubated with FITC-conjugated goat anti mouse IgG antibody (green) and cellular F-actin was stained with Alexa Fluor 555-conjugated phalloidin (red). Nuclei were counterstained with DAPI (blue). Cell samples were examined with a laser confocal scanning microscope (Leica, Saarbrücken, Germany).
DF-1 cells were treated with rMGA_0676 at concentrations of 20, 40, and 80 μg/ml for 24 h at 37°C to study the dose and time-dependence internalization of rMGA_0676. DF-1 cells were also treated with rMGA_0676 (40 μg/ml) at 37°C for 12, 24, and 36 h. All treated samples underwent IFA, as described above.
DF-1 cells were treated with filipin (0, 1.5, 3.75, 7.5 μM) and monodansylcadaverine (MDC) (0, 50, 100, or 200 μM) followed by rMGA_0676 for 24 h at 37°C to detect the inhibition of rMGA_0676 internalization. The samples underwent IFA and were examined with fluorescence microscopy. DF-1 cells were also pretreated with small interfering RNA to inhibit expression of caveolin or clathrin, and treated with rMGA_0676 for 24 h at 37°C, then analyzed by IFA, and observed using fluorescence microscopy.
RNA Interference (RNAi) Knockdown of NAE, Caveolin, Clathrin, and NF-κB
SiRNAs, designed by Genepharma (Shanghai, China), were used to knock down NAE, caveolin, clathrin, and NF-κB in DF-1 cells. The siRNA sequences for targeting proteins in DF-1 cells were shown in Table S1. Cells were transfected with siRNA using RNAiMAX reagent, according to manufacturer's instructions (Invitrogen). Analysis was performed with western blots. Knockdown of NAE in DF-1 cells for evaluation of impact on apoptosis and the activation of NF-κB under different conditions. Knockdown of caveolin and clathrin in DF-1 cells for evaluation of internalization of rMGA_0676 under rMGA_0676 treated. Knockdown of NF-κB in DF-1 cells for evaluation of impact on activation of NF-κB under treated conditions.
Apoptosis Assays
Cells were cultured in six-well flat-bottomed plates and exposed to rMGA_0676 or rMGA_0676ΔSNC (40 μg/ml) for 24 h to determine whether rMGA_0676 and/or rMGA_0676ΔSNC induced apoptosis in cells. PI positive cells were then analyzed using the Annexin V-FITC/PI apoptosis detection kit, according to manufacturer's instructions.
Cells were cultured in six-well flat-bottomed plates and exposed to rMGA_0676 (40 μg/ml), MGA_0676Δ92−221 (40 μg/ml), or M. gallisepticum (MOI = 10) for 24 h to determine whether rMGA_0676 or M. gallisepticum induced apoptosis after NAE knockdown or treatment with NF-κB siRNA. Apoptotic cells were analyzed using the Annexin V-FITC/PI apoptosis detection kit, according to manufacturer's instructions. Apoptotic DF-1 cells were analyzed by flow cytometry. The above experiments were carried out in triplicate and the data were analyzed using SPSS software.
Pull Down, Co-immunoprecipitation, and Western Blot Analysis
For the pull down, DF-1 cells were seeded on six-well plates, cultured for 24 h, and then treated for 24 h with rGST-MGA_0676 (40 μg/ml); rGST was used as a negative control. DF-1 cells were also transfected with pEGF-N1-MGA_0676 for 24 h; pEGF-N1 was used as a negative control. All cell lysate samples were prepared with a non-denaturing lysis buffer. The pull down technology was performed with a detection kit, according to manufacturer's instructions.
For co-immunoprecipitation, HEK293T cells or DF-1 cells were seeded on plates and cultured for 24 h before co-transfection with pCMV-HA-MGA_0676, pCMV-HA-MGA_0676ΔSNC, pCMV-Myc-NAE, and pCMV-Myc-NAEΔThif; empty vectors were used as controls. Twenty-four hours after transfection, cell lysates were prepared using a non-denaturing lysis buffer. Cell lysates were incubated with 5 μg of anti-HA antibody at 4°C for 2 h and then mixed with 25 μl of a 50% slurry of protein A/G plus agarose and incubated for another 2 h. Beads were washed three times with lysis buffer and boiled with 6×SDS loading buffer for 5 min. The samples were fractionated by electrophoresis on 12% SDS-polyacrylamide gels and resolved proteins were transferred onto polyvinylidene difluoride membranes. After blocking with 5% skim milk, the membranes were incubated with either anti-HA or anti-Myc antibodies, followed by an appropriate horseradish peroxidase-conjugated secondary antibody. For the endogenous NAE assay, DF-1 cells were transfected with pCMV-HA-MGA_0676 or with an empty vector. Twenty-four hours after transfection, cell lysates were subjected to immunoprecipitation with anti-HA antibody and immunoblotted with anti-NAE or anti-FLAG antibodies. For the M. gallisepticum MGA_0676 and endogenous NAE assays, DF-1 cells were infected with M. gallisepticum (MOI = 10) for 36 h and the cell lysates were subjected to immunoprecipitation with anti-MGA_0676 antibody and immunoblotting with anti-NAE.
Confocal Laser Scanning Microscopy Assays
HEK293T or DF-1 cells were seeded on cover slips in 24-well plates and cultured for 18–24 h, for overexpression of pCMV-HA-MGA_0676 and pCMV-Myc-NAE. The two plasmids were then co-transfected with LTX reagents. For overexpression of MGA_0676, DF-1 cells were treated only with rMGA_0676 (40 μg/ml) or transfected with pCMV-HA-MGA_0676. Twenty-four hours after transfection or treatment, cells were fixed with 1% paraformaldehyde, permeabilized with 0.1% TritonX-100 for 3–5 min and blocked with 3% bovine serum albumin. For co-localization of HA-MGA_0676 and Myc-NAE in DF-1 cells, the slides were incubated with mouse anti-HA antibody and rabbit anti-Myc antibody for 1.5 h at 37°C. For co-localization of HA-MGA_0676 and endogenous NAE in DF-1 cells, the slides were incubated with mouse anti-HA antibody and rabbit anti-NAE antibody for 1.5 h at 37°C. For co-localization of rMGA_0676 and endogenous NAE in DF-1 cells, the slides were incubated with mouse anti-His antibody and rabbit anti-NAE antibody for 1.5 h at 37°C. For co-localization of rMGA_0676 and caveolin, the sections were incubated with mouse anti-His antibody and rabbit anti-caveolin antibody for 1.5 h at 37°C. Sections were then incubated with FITC-conjugated goat anti-mouse IgG antibody (green) and rhodamine (tetramethyl rhodamine isocyanate [TRITC])-conjugated goat anti-rabbit antibody (red). Nuclei were counterstained with DAPI (blue).
Detection of NF-κB Activity
To detect NF-κB activity, DF-1 cells were seeded on six-well plates and cultured for 24 h, then treated with rMGA_0676 at concentrations of 0, 20, 40, or 80 μg/ml for 24 h. TNF-α treated cells were used as positive control. Cell lysates were prepared using a non-denaturing lysis buffer, and boiled with 6×SDS loading buffer for 10 min. The samples underwent immunoblotting (according to the manufacturer's instructions) for detection of Rela in the nucleus and IκBα and p-IκBα in the cytoplasm. PCNA was used as the nucleus internal reference and β-actin was used as the cytoplasm internal reference. Translocation of Rela was also detected in DF-1 cells. DF-1 cells were seeded on 24-well plates, cultured for 18–24 h and then treated with either rMGA_0676 (40 μg/ml) or M. gallisepticum (MOI = 10) for 24 h. Cells then underwent IFA, as previously described, after incubation with mouse anti-Rela antibody for 1.5 h at 37°C, followed by incubation with FITC-conjugated goat anti-mouse IgG antibody (green) and cellular F-actin staining with Alexa Fluor 555-conjugated phalloidin (red). Nuclei were counterstained with DAPI (blue). Cells were then observed with a laser confocal scanning microscope (Leica). DF-1 cells were treated with rMGA_0676 (40 μg/ml) or rMGA_0676ΔSNC (40 μg/ml) for 24 h as described above to determine whether NF-κB is activated via interaction of MGA_0676 and NAE. DF-1 cells with silenced NAE were also treated with rMGA_0676 (40 μg/ml), and NC (unrelated RNA oligo) was used as negative control. The samples were analyzed as described above.
Detection of Activation of Cullin Neddylation in DF-1 Cells
To detect activation of cullin neddylation, DF-1 cells were seeded on six-well plates and cultured for 24 h, then treated with rMGA_0676 at concentrations of 0, 20, 40, or 80 μg/ml for 24 h. Then cell lysates were prepared for western blot to detect the expression of cull-NEDD8 based on the anti-chicken cull antibody or anti-NEDD8 antibody.β-actin was used as internal reference. DF-1 cells were seeded on six-well plates and cultured for 24 h, then treated with MLN4924 (500 ng/l) for 12–18 h or MGA_0676 at concentrations of 40 μg/ml for 24 h. Then cell lysates were prepared for western blot to detect expression of cull-NEDD8 and NF-κB activity as described above. β-actin was used as internal reference.
Statistical Analysis
All the tests were triplicated, and the statistical differences between treated and control groups were determined and analyzed by analysis of variance (ANOVA) using SPSS software, version 18.0 (SPSS, Chicago, IL, USA), and the graphs were made using GraphPad Prism 5.0. “ns” represented no significant differences (P > 0.05). “*” represented statistically differences (P < 0.05), “**” represented statistically significant differences (P < 0.01), “***” represented statistically significant differences (P < 0.001).
Results
Caveolin-Mediated Endocytic Mechanisms Are Implicated in Internalization of rMGA_0676
Internalization of rMGA_0676 was determined by incubating chicken embryo fibroblasts (DF-1 cells) with different doses of recombinant nuclease at 37°C for 24 h. Initial determinations of the mechanism and internalization rates were shown in Figure S1. Internalization of rMGA_0676 by DF-1 cells increased with nuclease concentrations (Figure S1A). Internalization rates were also determined by incubating DF-1 cells with rMGA_0676 (40 μg/ml) at different times, and were found to increase with incubation times (Figure S1B).
Bacterial toxins are endocytosed through caveolin-mediated or clathrin-dependent mechanisms. For instance, filipin can prevent caveolae vesicle formation while monodansylcadaverine (MDC) can inhibit clathrin-dependent vacuole formations (Orlandi and Fishman, 1998; Rejman et al., 2004; Veiga and Cossart, 2006; Kamashev et al., 2011). Therefore, we pretreated DF-1 cells with both filipin and MDC, and found that the internalization of rMGA_0676 was suppressed in a dose-dependent manner in cells pretreated with filipin (Figure S2A, Figures 1A,B), but pretreatment with MDC did not change the internalization of rMGA_0676 (Figures 1C,D). SiRNA technology was also employed to verify the above results. Figure S2C shows a reduction of caveolin in DF-1 cells treated with siRNA and a significant inhibition of the internalization of rMGA_0676 (Figure S2B). Inhibition of internalization can also be observed by confocal laser scanning microscopy (Figures 1E,F), from which rMGA_0676 and caveolin were found to be co-localizing in DF-1 cells (Figure 1I). The above results indicate that caveolin-mediated endocytosis is a dominant pathway for rMGA_0676 internalization. Clathrin was also silenced with siRNA, but no obvious effect on the internalization of rMGA_0676 was observed (Figures 1G,H).
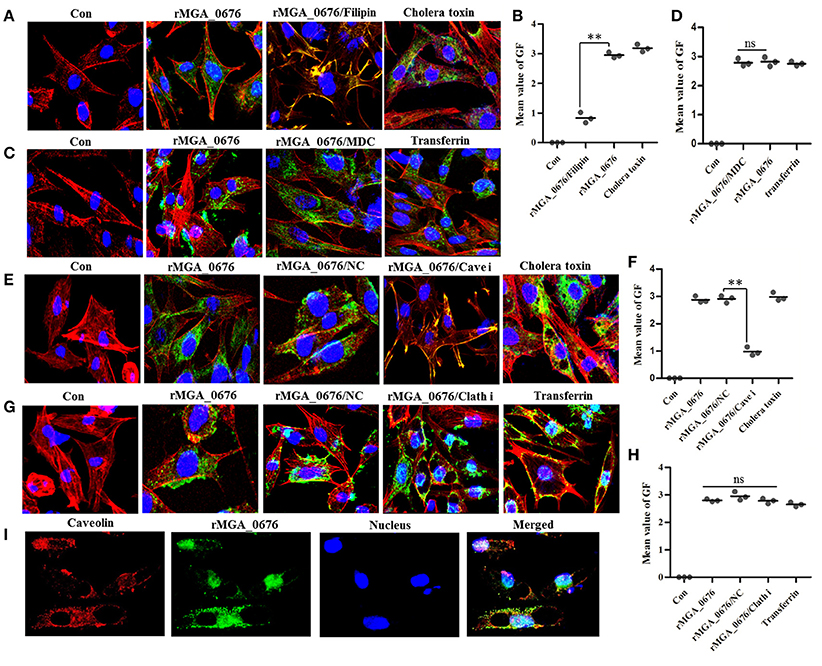
Figure 1. Caveolin-mediated endocytic mechanisms are implicated in internalization of rMGA_0676. (A) Internalization of rMGA_0676 was blocked by caveolin-mediated endocytic inhibitor filipin. DF-1 cells were treated with filipin followed by rMGA_0676 (40 μg/ml) for 24 h at 37°C to detect the inhibition of rMGA_0676 internalization. The samples underwent IFA and were examined with a laser confocal scanning microscope. Cellular F-actin was stained with Alexa Fluor 555-conjugated phalloidin. The nuclei were counterstained with DAPI (blue). The rMGA_0676 was hybridized with mouse anti-His monoclonal antibody and labeled with FITC-conjugated goat anti mouse IgG antibody green fluorescence (GF). The normal DF-1 cells as the negative control (Con), cells were treated with Cholera toxin-FITC as the caveolin-mediated endocytic positive control (Cholera toxin). (B) The mean value of GF was used to quantify the positive staining for rMGA_0676 in filipin treated DF-1 cells (A), the data were analyzed using SPSS software, and the graph was made using GraphPad Prism 5.0. “**”represented statistically significant differences (P < 0.01). (C) Internalization of rMGA_0676 was not blocked by clathrin-mediated endocytic inhibitor MDC. DF-1 cells were treated with MDC followed by rMGA_0676 (40 μg/ml) for 24 h at 37°C and detected the inhibition of rMGA_0676 internalization underwent IFA as described above. The normal DF-1 cells as the negative control (Con), cells were treated with transferrin as the clathrin-mediated endocytic positive control (transferrin). (D) The mean value of GF was used to quantify the positive staining for rMGA_0676 in MDC treated DF-1 cells (B), the data were analyzed using SPSS software, and the graph was made using GraphPad Prism 5.0. (E) Internalization of rMGA_0676 was reduced by knockdown of caveolin in DF-1 cells by siRNA. The caveolin siRNA sequence(si Cave) and its negative control siRNA sequence(NC) were transfected into DF-1 cells using RNAiMAX reagent followed by rMGA_0676 (40 μg/ml) for 24 h at 37°C and detected the inhibition of rMGA_0676 internalization underwent IFA as described above. The normal DF-1 cells as the negative control (Con), cells transfected with negative control siRNA sequence followed by rMGA_0676 for 24 h at 37°C as the siRNA negative control (NC), cells were treated with Cholera toxin-FITC as the caveolin-mediated endocytic positive control (Cholera toxin). (F) The mean value of GF was used to quantify the positive staining for rMGA_0676 in caveolin siRNA sequence (Cave i) transfected DF-1 cells (C), the data were analyzed using SPSS software, and the graph was made using GraphPad Prism 5.0. “**”represented statistically significant differences (P < 0.01), “ns” represented no significant differences. (G) Internalization of rMGA_0676 was not reduced by knockdown of clathrin in DF-1 cells by siRNA. The clathrin siRNA sequence(Clath i) and its negative control siRNA sequence(NC) were transfected into DF-1 cells using RNAiMAX reagent followed by rMGA_0676 (40 μg/ml) for 24 h at 37°C and detected the inhibition of rMGA_0676 internalization underwent IFA as described above. The normal DF-1 cells as the negative control (Con), cells transfected with negative control siRNA sequence followed by rMGA_0676 for 24 h at 37°C as the siRNA negative control (NC), cells were treated with transferrin as the clathrin-mediated endocytic positive control (transferrin). (H) The mean value of GF was used to quantify the positive staining for rMGA_0676 in clathrin siRNA sequence (Clath i) transfected DF-1 cells (D), the data were analyzed using SPSS software, and the graph was made using GraphPad Prism 5.0. “**”represented statistically significant differences (P < 0.01), “ns” represented no significant differences. (I) Co-localization of MGA_0676 and caveolin in DF-1 cells. The sections were treated with rMGA_0676 (40 μg/ml) for 24 h at 37°C followed by IFA as described above. The Sections were incubated with mouse anti-His antibody and rabbit anti-caveolin antibody for 1.5 h at 37°C. Sections were then incubated with FITC-conjugated goat anti mouse IgG antibody (green) and rhodamine (tetramethyl rhodamine isocyanate [TRITC])-conjugated goat antirabbit antibody (red). Nuclei were counterstained with DAPI (blue). All the cell samples were examined with a laser confocal scanning microscope. Scale bar = 10 μm.
MGA_0676 Interacts with NEDD8-Activating Enzyme (NAE)
In order to determine the target proteins of MGA_0676, GST fusion MGA_0676 protein was expressed and purified from recombinant E. coli (Figure S3A), and used as bait in the pull down system to screen proteins from DF-1 cells. Two proteins (a and b) were selected as candidate target proteins (Figure 2A). GFP-MGA_0676 expressed in DF-1 cells was also used to screen proteins from DF-1 cells by pull down, and three proteins (c, d, and e) were selected as candidate target proteins (Figure 2B). Mass spectrometry sequencing (MS) results of the above five proteins showed that NEDD8 activity enzyme E1 (NAE) chain B (structure Appbp1-Uba3-Nedd8-Mgatp-Ubc12) had the highest MS sequencing score, which is the most likely target protein interacting with MGA_0676 (Table 1).
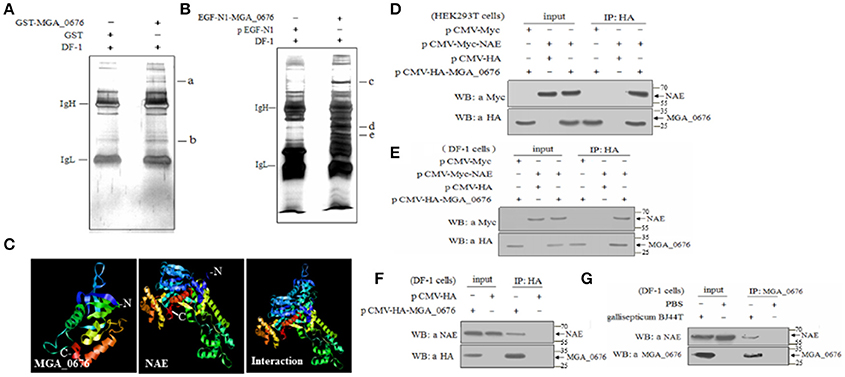
Figure 2. MGA_0676 interacts with NEDD8- activating enzyme (NAE). (A) Pull down assays based on GST. IgH and IgL indicate the heavy and light chains in the antibodies; a and b show the target proteins' interactions with MGA_0676. (B) Pull down assays based on GFP. IgH and IgL indicate the heavy and light chains in the antibodies; c, d, and e show the target proteins' interactions with MGA_0676. (C) A model of MGA_0676 binding to NAE. MGA_0676 can bind NAE through an interaction between the C-terminal of MGA_0676 and the N-terminal of NAE. (D) HEK293T cells were transfected with the indicated expression plasmids. (E) DF-1 cells were transfected with the indicated expression plasmids. Cell lysates were immunoprecipitated (IP) with anti-HA antibody and immunoblotted with anti-HA or anti-Myc antibodies. (F) Interaction of MGA_0676 with endogenous NAE. IP of cell lysates with anti-HA antibody, and immunoblotted with anti-HA or anti-NAE antibodies. (G) Interaction of M. gallisepticum MGA_0676 with endogenous NAE. DF-1 cells were infected with M. gallisepticum or PBS as a control. IP with anti-MGA_0676 antibody and immunoblotted with anti-MGA_0676 or anti-NAE antibodies.
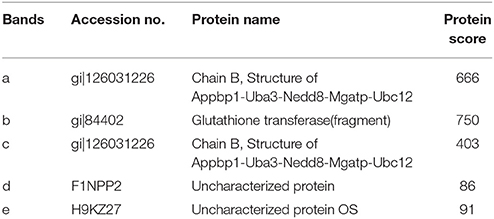
Table 1. Proteins present in two specific bands were observed in the GST and GFP lanes as identified by MS.
In order to test the potential interaction, a model of the MGA_0676-NAE complex was constructed, analysis of the structure prediction models suggested that MGA_0676 could perfectly interact with NAE, and that the interacting domain should be located at the C-terminal of MGA_0676 and the N-terminal of NAE (Figure 2C). Furthermore, plasmids expressing HA-MGA_0676 or Myc-NAE were constructed to analyze their interactions within mammalian and chicken cells. When lysates of HEK293 T cells expressing both HA-MGA_0676 and Myc-NAE were immunoprecipitated with HA monoclonal antibody, Myc-NAE was detected in the precipitate, which indicates that MGA_0676 interacts with NAE ectopically expressed in mammalian cells (Figure 2D). Similar results were obtained in chicken DF-1 cells (Figure 2E), indicating that the interaction between the two proteins is not cell type specific. To confirm the interaction between the protein and host cells, DF-1 cells were transfected with pCMV-HA-MGA_0676 expression plasmid and an immunoprecipitation assay was performed with anti-HA monoclonal antibody. NAE was detected in the lysate of HA-MGA_0676-expressing cells after immunoprecipitation with anti-HA antibody (Figure 2F), which indicates that MGA_0676 interacts with endogenous NAE. DF-1 cells were then treated with M. gallisepticum BJ44T, lysed and immunoprecipitated with anti-MGA_0676 monoclonal antibody, and the precipitate was blotted with anti-NAE antibody. Both MGA_0676 and NAE were found in the precipitate (Figure 2G), indicating that M. gallisepticum MGA_0676 also interacted with endogenous NAE in host cells.
Furthermore, the interaction between MGA_0676 and NAE was observed in HEK293T and DF-1 cells expressing HA-MGA_0676 and Myc-NAE by confocal microscopy, which shows HA-MGA_0676 and Myc-NAE co-localize in the perinuclear and nuclear regions of cells (Figures 3A–H). To determine whether endogenous NAE co-localizes in the same cellular compartment, DF-1 cells were transfected to express HA-MGA_0676 or treated with rMGA_0676, and a similar co-localization between HA-MGA_0676 and NAE was found (Figures 3I–P). DF-1 cells were also treated with M. gallisepticum and IFA was done using anti-MGA_0676 and anti-NAE antibodies. As expected, endogenous NAE also co-localizes with MGA_0676 in M. gallisepticum-infected cells (Figures 3Q–T).
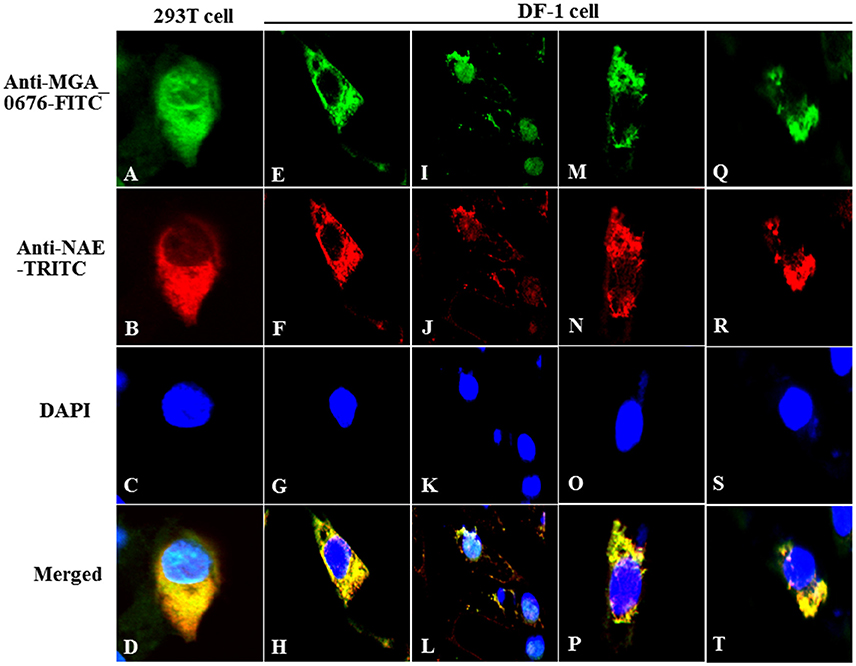
Figure 3. Co-localization of MGA_0676 with NAE in the perinuclear and nuclear regions of cells. HEK293T cells (A–D) or DF-1 cells (E–G) were co-transfected with pCMV-HA-MGA_0676 and pCMV-Myc-NAE. Twenty-four hours after transfection, cells underwent IFA based on anti-HA or anti-Myc antibodies. MGA_0676 was stained with FITC-conjugated goat anti mouse IgG antibody (green), NAE was stained with rhodamine (TRITC)-conjugated goat anti-rabbit antibody (red), and cell nuclei were counterstained with DAPI (blue). Cell samples were observed with a laser confocal scanning microscope. Co-localization of MGA_0676 with endogenous NA, DF-1 cells were transfected with pCMV-HA-MGA_0676 plasmid (I–L) or incubated with rMGA_0676 (M–P). Twenty-four hours after treatment, cells underwent IFA based on anti-HA or anti-NAE antibodies; cell samples were observed with a laser confocal scanning microscope. Co-localization of M. gallisepticum MGA_0676 with endogenous NAE in M. gallisepticum-infected cells (Q–T). DF-1 cells were infected with M. gallisepticum. Twenty-four hours after treatment, cells underwent IFA based on anti-MGA_0676 or anti-NAE antibodies; IFA and the following results were observed with a laser confocal scanning microscope. Scale bar = 10 μm.
The SNC Region of MGA_0676 Interacts With the Thif Region of NAE
To determine the region of MGA_0676 that interacts with NAE, a HA tag fused MGA_0676 mutant (MGA_0676ΔSNC) was constructed with a deleted SNC region that located at 92 to 221 amino acids of MGA_0676. The MGA_0676ΔSNC derivative was expressed in HEK293T cells, and immunoprecipitation results indicated that MGA_0676ΔSNC lost the ability to interact with NAE (Figure 4A). To determine the region of NAE that interacts with MGA_0676, a Myc tag fused NAE mutant (NAEΔThif) was constructed with a deleted Thif region that located at 29 to 166 amino acids of NAE. Immunoprecipitation results showed that the Thif region of NAE is necessary to interact with MGA_0676 (Figure 4B). The above results indicate that the SNC region of MGA_0676 interacts with the Thif region of NAE.
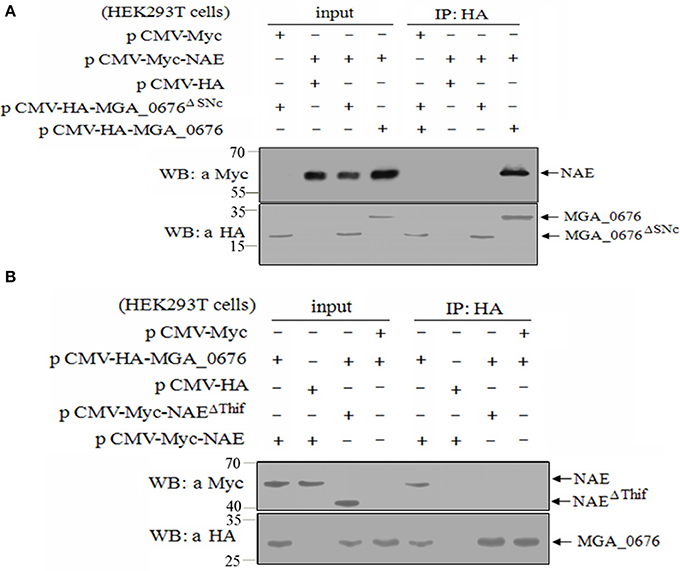
Figure 4. The SNC region of MGA_0676 interacts with the Thif region of NAE. (A) Endogenous NAE cannot interact with MGA_0676ΔSNC (The SNC portion of MGA_0676, from amino acids 92 to 221 was deleted). HEK293T cells were transfected with full-length HA-MGA_0676 and HA-MGA_0676ΔSNC molecules or empty vectors. Twenty-four hours after transfection, cell lysates were prepared and IP with anti-HA monoclonal antibody. The pellets were examined by western blotting using anti-Myc antibody. (B) Endogenous NAEΔThif cannot interact with MGA_0676. HEK293T cells were transfected with full-length Myc-NAE and Myc-NAEΔThif (The Thif region of NAE from amino acids 29 to 166 was deleted) molecules or empty vectors. Twenty-four hours after transfection, cell lysates were prepared and IP with anti-HA monoclonal antibody. The pellets were examined by western blotting using anti-Myc antibody.
MGA_0676 Interacts With NAE to Induce Apoptosis in DF-1 Cells
The binding of MGA_0676 to NAE suggests that NAE plays an important role in MGA_0676-induced apoptosis. When NAE of DF-1 cells was silenced via siRNA (Figure 5A), rMGA_0676 or M. gallisepticum did not induce obvious morphological changes (Figure 5B), rMGA_0676 did not induce observable apoptosis (Figure 5C), and the apoptosis of M. gallisepticum-infected cells decreased significantly (Figure 5D). The results of these experiments suggest that the interaction of NAE and MGA_0676 is related to the apoptosis of DF-1 cells induced by MGA_0676.
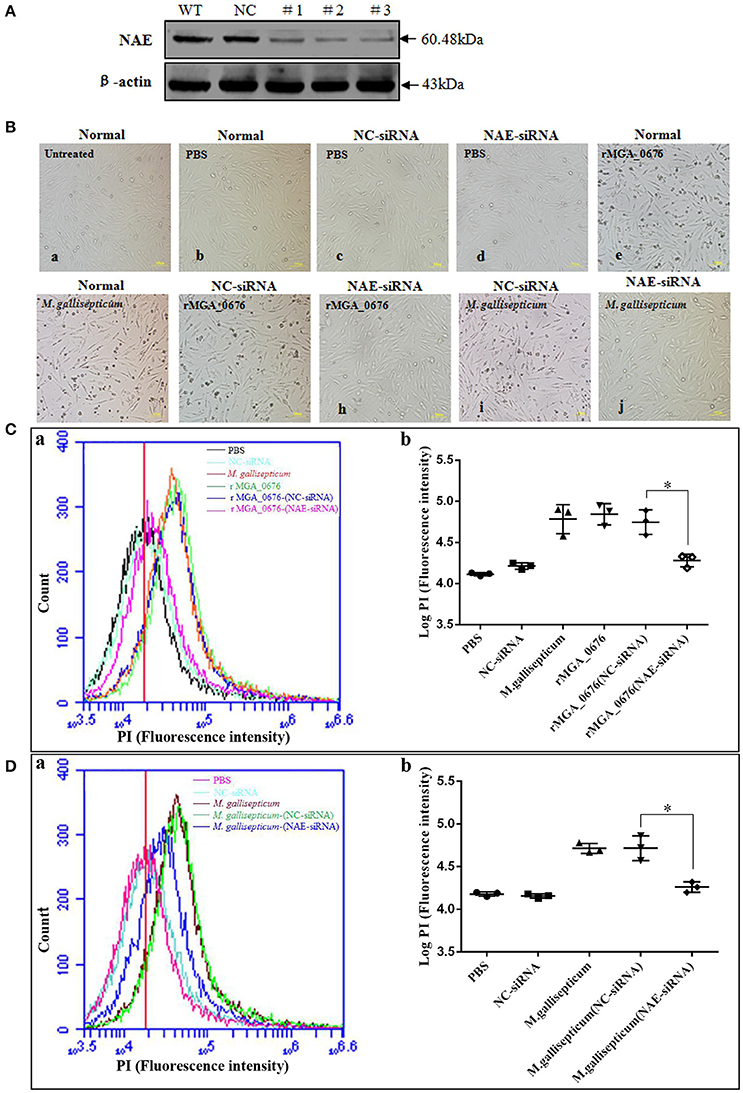
Figure 5. MGA_0676 interacts with NAE inducing apoptosis in DF-1 cells. (A) Effect of NAE RNAi on the expression of endogenous NAE. DF-1 cells were transfected with siRNA (#1 to #3) or controls (WT and NC), as described in Materials and Methods. Forty-eight hours after the second transfection, cell lysates were prepared and examined by western blotting with anti-NAE antibody. Endogenous β-actin expression was used as an internal control. (B) Pathological changes in NAE inhibited DF-1 cells after rMGA_0676 treatment. All cell groups were observed with a microscope after treatment as follows. a: normal cells; b: PBS-treated; c: siRNA NC; d: siRNA NAE; e: rMGA_0676 (40 μg/ml); f: M. gallisepticum (MOI = 10); g: siRNA NC pre-treatment, rMGA_0676 (40 μg/ml) treatment; h: siRNA NAE pre-treatment, rMGA_0676 (40 μg/ml) treatment; i: siRNA NC pre-treatment, M. gallisepticum (MOI = 10) treatment; j: siRNA NAE pre-treatment, M. gallisepticum (MOI = 10) treatment. Scale bar = 100 μm. (C) Apoptosis induction by rMGA_0676 in NAE arrested DF-1 cells. The curves (a) and the scatter plot (b) represent apoptotic cells in (B) treated groups. PBS (5Bb); NC-siRNA (5Bc); rMGA_0676 (5Be); M. gallisepticum (5Bf); rMGA_0676-(NC-siRNA) (5Bg); rMGA_0676-(NAE-siRNA) (5Bh). “*”represented statistically significant differences (P < 0.05). (D) Apoptosis induction by M. gallisepticum in NAE arrested DF-1 cells. The curves (a) and the scatter plot (b) also represent apoptotic cells in (B) treated groups. PBS (5Bb); NC-siRNA (5Bc); M. gallisepticum (5Bf); M. gallisepticum-(NC-siRNA) (5Bi); M. gallisepticum-(NAE-siRNA) (5Bj). “*”represented statistically significant differences (P < 0.05).
Nuclear Factor kappa B (NF-κB) Is Activated via Interaction of MGA_0676 With NAE
The activity of nuclear factor kappa B (NF-κB) was examined by immunoblotting and laser scanning confocal microscopy to explore the biological effects of the interactions between MGA_0676 and NAE. Levels of NF-κB in the nucleus and of p-IκBα in the cytoplasm increased while IκBα in the cytoplasm decreased significantly after DF-1 cells were treated with rMGA_0676 (Figure 6A). In addition, Rela (p65 in mammalian cells) translocated from the cytoplasm to the nucleus in DF-1 cells treated with rMGA_0676 or TNF-α, as shown in Figure 6B. Furthermore, NF-κB was not activated in DF-1 cells treated with MGA_0676ΔSNC (Figure 6C) or with MGA_0676 after knockdown of NAE (Figure 6D). Laser scanning confocal microscopy confirmed that NF-κB lost the ability to translocate to the nucleus in DF-1 cells treated with MGA_0676ΔSNC and in rMGA_0676-treated DF-1 cells that had undergone NAE-knockdown (Figure 6E). The above results indicate that MGA_0676 activates NF-κB via interaction with NAE.
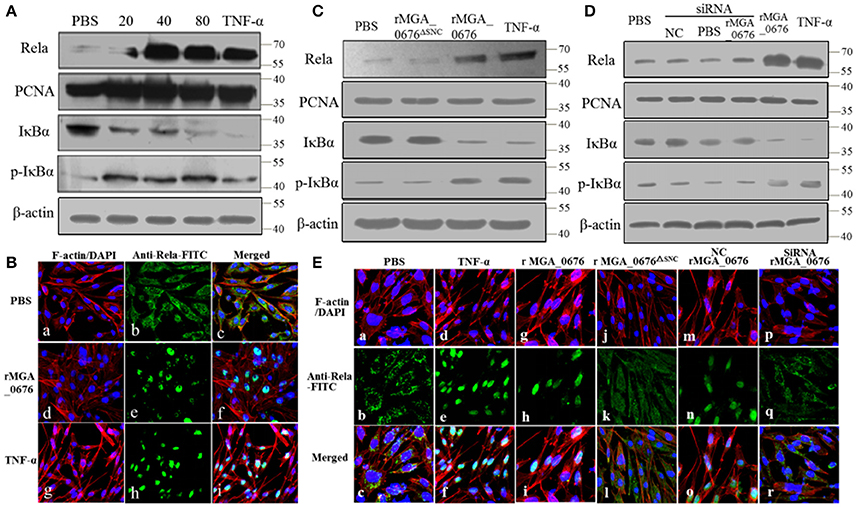
Figure 6. NF-κB is activated via interaction between MGA_0676 and NAE. (A) rMGA_0676 activated NF-κB in DF-1 cells were detected by western blot. The activity of NF-κB is rMGA_0676 dose dependent in DF-1 cells. DF-1 cells were treated with PBS or different doses of rMGA_0676 at 37°C. Twenty-four hours after transfection, cell lysates were prepared to detect the expression of Rela in the nucleus; PCNA was used as an internal control. Expression of IκBα and p-IκBα in the cytoplasm; β-actin expression was used as an internal control. (B) Translocation of Rela from the cytoplasm to the nucleus in DF-1 cells after rMGA_0676 treatment. (a–c) cells were treated with PBS as a negative control and then subjected to IFA based on anti-Rela antibody, followed by incubation with FITC-conjugated goat anti-mouse IgG antibody (green) and cellular F-actin staining with Alexa Fluor 555-conjugated phalloidin (red); nuclei were counterstained with DAPI (blue). The cell samples were observed with a laser confocal scanning microscope. (d–f) cells were treated with rMGA_0676 (40 μg/ml) for 24 h at 37°C; IFA and sample observation were done with a laser confocal scanning microscope. (j–i) cells were treated with TNF-α as a positive control. Scale bar = 50 μm. (C) NF-κB was not activated by MGA_0676ΔSNC. DF-1 cells were treated with PBS, rMGA_0676, or rMGA_0676ΔSNC; TNF-α was used as a positive control. After twenty-four hours, cell lysates were prepared to detect the expression levels of Rela, IKBα, and p-IkBα in the cells. PCNA was used as the nucleus internal control and β-actin was used as the cytoplasm internal control. (D) NF-κB was not activated by MGA_0676 in DF-1 cells after knockdown of NAE. DF-1 cells were pretreated with siRNA and then treated with rMGA_0676. Cells treated with PBS, NC, and siRNA were used as negative control; cells treated with rMGA_0676 or TNF-α were used as positive control. After 24 h, Rela, IκBα, and p-IκBα were detected in the cells as described above. (E) Rela was not translocated from the cytoplasm to the nucleus in DF-1 cells after disruption of the interaction between MGA_0676 and NAE. DF-1 cells were subjected to IFA as described in Materials and Methods. (a–c) cells treated with PBS were used as negative control; (d–f) cells treated with TNF-α were used as positive control; (g–i) cells were treated with rMGA_0676; (j–l) cells were treated with rMGA_0676ΔSNC: (m–o) cells were pretreated with siRNA (negative control) and then treated with rMGA_0676; (p–r) cells were pretreated with siRNA NAE and then treated with rMGA_0676.
MGA_0676 Activates NF-κB Through Acceleration of Cullin Neddylation of DF-1 Cells
To explore the influence of cullin neddylation of DF-1 after treatment with MGA_0676, the cullin-NEDD8 complex of DF-1 cells was detected by western blot. As seen in Figure 7A, compared with the PBS treated DF-1 cells, expression of cullin-NEDD8 clearly increased in MGA_0676 treated DF-1 cells, which indicated that cullin neddylation of DF-1 was activated by MGA_0676. However, we also have shown above that MGA_0676 could activate NF-κB in DF-1 cells (Figure 6). For this reason, to investigate the possible interaction between neddylation and NF-κB in DF-1 cells after treatment with MGA_0676, we inhibited neddylation in DF-1 cells using a specific inhibitor, MLN4924. We found that neddylation specific inhibitor MLN4924 blocked the activation of NF-κB induced by MGA_0676 (Figure 7B), for further confirming this results, we observed the translocation of Rela in DF-1 cells by confocal microscopy, from the Figure 7C, we found that MGA_0676 lost its ability of inducing NF-κB translocated into nucleus when the DF-1 was pretreated with MLN4924. All these results demonstrated that MGA_0676 activated NF-κB through acceleration of cullin neddylation in DF-1 cells.
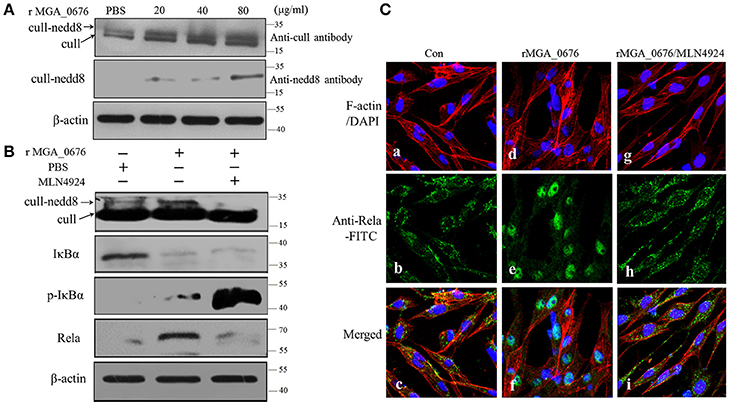
Figure 7. MGA_0676 activates NF-κB through accelerating the process of cullin neddylation in DF-1 cells (A) MGA_0676 accelerated the process of cullin neddylation in DF-1 cells. DF-1 cells were seeded on six-well plates and cultured for 24 h and then treated with rMGA_0676 at concentrations of 0, 20, 40, or 80 μg/ml for 24 h. Afterwards, cell lysates were prepared for western blot to detect the expression of cull-NEDD8 based on the anti-chicken cull antibody or anti-NEDD8 antibody. β-actin was used as internal reference. (B) Neddylation specific inhibitor MLN4924 blocked the activity of NF-kB induced by MGA_0676. DF-1 cells were seeded on six-well plates and cultured for 24 h and then treated with MLN4924 or MGA_0676 for 24 h. Afterwards, cell lysates were prepared to detect the expression of cull-NEDD8, IkBα, p-IkBα, and p65 (Rela) by western blot. β-actin was used as internal reference. (C) Translocation of Rela from the cytoplasm to the nucleus in DF-1 cells. (a–c) cells were treated with PBS as a negative control(Con); (d–f) cells were treated with rMGA_0676 (40 μg/ml) for 24 h at 37°C; (g–i) cells were treated with MLN4924 (500 ng/l) for 12–18 h at 37°C, then inclubated with rMGA_0676 (40 μg/ml) for 24 h at 37°C. All the cell samples were subjected to IFA based on anti-Rela antibody, followed by incubation with FITC-conjugated goat anti-mouse IgG antibody (green) and cellular F-actin staining with Alexa Fluor 555-conjugated phalloidin (red); nuclei were counterstained with DAPI (blue). The slices were observed with a laser confocal scanning microscope. Scale bar = 50 μm.
MGA_0676 Induces Apoptosis of Chicken Embryo Fibroblast by Activating NF-κB
Since we observed that the interaction between MGA_0676 and NAE activated NF-κB and was related to the apoptosis in DF-1 cells, we suspected NF-κB could play an important role during the apoptosis induced by MGA_0676. Therefore, in a subsequent experiment, we inhibited NF-κB with BAY 11-7082 (an inhibitor of NF-κB) and the apoptosis of DF-1 cells induced by MGA_0676 was examined. Figure 8AI and Figure S4 showed that the cell death phenotype treated with rMGA_0676 was significantly reduced by the addition of BAY 11-7082 (d and e). The key molecule of apoptosis cleaved caspase 3 was detected in apoptosis-induced group by western blot assays (Figure 8AII; d and f), and anti-cleaved caspase 3 antibodies and HRP-conjugated goat-anti-rabbit antibodies were used as primary and secondary antibodies, and the procedure was as described in “Material and Methods”. NF-κB was effectively silenced by siRNA (Figure 8B), and the apoptosis induced by rMGA_0676 decreased accordingly as shown by flow cytometry using PI staining (Figure 8CI, Figure S5), and cleaved caspase 3 was also detected in apoptosis-induced group (Figure 8CII; e, f, and h). These data strongly suggest that MGA_0676 induces apoptosis by activating NF-κB in DF-1 cells.
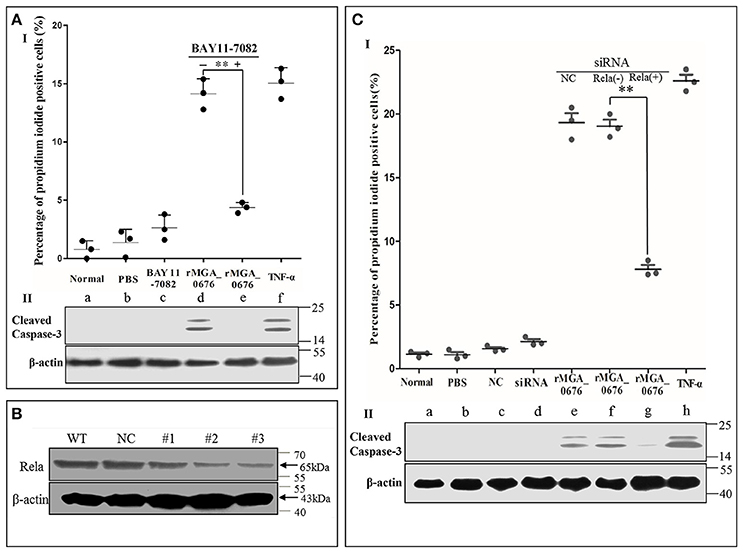
Figure 8. rMGA_0676 activates NF-κB inducing apoptosis in DF-1 cells. (AI) rMGA_0676-induced apoptosis in DF-1 cells was arrested by an NF-κB inhibitor. The cells were seeded on 12-well plates and cultured overnight. Apoptosis was assessed by the percentage of propidium iodide-positive cells following the treatment. a: normal cells; b: cells were treated with PBS; c: cells were treated with BAY 11-7082; d: cells were treated with rMGA_0676; e: cells were pretreated with BAY 11-7082 for 1 h at 37°C and then treated with rMGA_0676; f: cells were treated with TNF-α. The data were analyzed using SPSS software, and the graph was made using GraphPad Prism 5.0. “**”represented statistically significant differences (P < 0.01). (AII) Cleaved caspase 3 was detected in apoptosis-induced group (d and f). (B) Effect of Rela RNAi on the expression of endogenous Rela. DF-1 cells were transfected with siRNA (#1 to #3) or controls (WT and NC), as described in Materials and Methods. Forty-eight hours after the second transfection, cell lysates were prepared and examined by western blotting with anti-Rela antibody. Endogenous β-actin expression was used as an internal control. (CI) rMGA_0676-induced apoptosis in DF-1 cells was also arrested by knockdown of NF-κB. The cells were seeded on 12-well plates and cultured overnight, apoptosis was assessed by the percentage of propidium iodide positive cells following the treatment. a: normal cells; b: cells were treated with PBS; c: cells were treated with siRNA negative control (NC); d: cells were treated with siRNA NAE; e: cells were pretreated with siRNA NC for 48 h at 37°C and then treated with rMGA_0676; f: cells were treated with rMGA_0676; g: cells were pretreated with siRNA NAE for 48 h at 37°C and then treated with rMGA_0676; h: cells were treated with TNF-α. the data were analyzed using SPSS software, and the graph was made using GraphPad Prism 5.0. “**”represented statistically significant differences (P < 0.01). (CII) Cleaved caspase 3 was detected in apoptosis-induced group (e, f, and h).
Discussion
Endocytosis is involved in cell signaling generally mediated by clathrin (cytosolic protein) or caveolin (cholesterol-binding protein) (Pascual-Lucas et al., 2014; Haucke, 2015; Garcia et al., 2017). Mycoplasma nucleases have been found to be internalized host cells through endocytosis which are relevant to the process of infection (Schmidt et al., 2007; Li et al., 2010; Somarajan et al., 2010). Among of them, rMGA_0676 is a nuclease with a SNC region in M. gallisepticum, which can translocate into chicken cells and induce cell apoptosis, and MGA_0676 plays a important role in the process of infection of M. gallisepticum (Xu et al., 2015). In the present study, we show that endocytosis is the major process by which rMGA_0676 translocate into cells, and that it is mediated by caveolin (Figure S2, Figure 1). Unlike clathrin-dependent endocytosis, caveolar endocytosis has an important function in the cellular uptake of some bacterial toxins, viruses and circulating proteins, and it is usually mediated by the cellular receptors in caveolae of the host cells (Chaudhary et al., 2014; Schmidt et al., 2015). This implies there should be receptors of MGA_0676 on the membrane of chicken cells worthy of being identified.
Several nucleases (MPN133, MG_186, or mhp379, among others) have been discovered in Mycoplasma, and all of them are membrane-associated proteins that can induce apoptosis of host cells. These nucleases are thought to contribute to the pathological damage, immune evasion, and persistent infection of the host cells (Jones and Falkow, 1996; Schmidt et al., 2007; Li et al., 2010; Somarajan et al., 2010). NEDD8-activating enzyme (NAE) has been proposed to be an important member of the neddylation pathway in the cell cycle (Chen et al., 2000; Soucy et al., 2009). Furthermore, bacterial effector proteins can lead to ubiquitin/NEDD8 dysfunction via glutamine deamidation and induce macrophage-specific apoptosis by arresting the activity of NAE in host cells (Munro et al., 2007; Cui et al., 2010; Jubelin et al., 2010). In our study, we show that the apoptosis of DF-1 cells induced by rMGA_0676 decrease significantly after knockdown of endogenous NAE, suggesting that NAE is a crucial factor in MGA_0676-induced apoptosis. Moreover, MGA_0676 is found to interact with NAE by their co-location in the perinuclear and nuclear regions of DF-1 cells (Figures 2, 3), and the interaction happens between the SNC region of MGA_0676 and the Thif domain of NAE (Figure 4). The most important is the interaction of NAE and MGA_0676 is related to the apoptosis of DF-1 cells induced by MGA_0676 (Figure 5), which indicates the neddylation pathway maybe associated with apoptosis of DF-1 cells induced by MGA_0676.
NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) is one of the first responders to harmful cellular stimuli since it belongs to the category of “rapid-acting” primary transcription factors. The activation of NF-κB may induce or inhibit apoptosis depending on the cell type and context of the stimulus (Chen et al., 2003; Brasier, 2006; Huang et al., 2015). Several mycoplasmal lipoproteins have been proven to induce apoptotic and necrotic cell death in lymphocytes and monocytes/macrophages through TLR2-mediated signaling, which leads to activation of NF-κB (Into et al., 2004; Wu et al., 2008), M. gallisepticum lipid associated membrane proteins through an NF-κB dependent pathway up-regulate inflammatory genes in chicken tracheal epithelial cells (Majumder et al., 2014). Similar activation of NF-κB and induction of apoptosis are found in our studies, which implies that toll-like receptors that recognize pathogen-associated molecules may be involved in the pathogenic process of MGA_0676 (Lee et al., 2014; Okumura and Nizet, 2014). Additional results in our study indicated that the activation of NF-κB is dependent on the interaction between MGA_0676 and NAE in DF-1 cells (Figure 6), and for this reason the exact molecular mechanism is also explored.
MGA_0676 activated NF-κB by accelerating the process of cullin neddylation in DF-1 cells (Figure 7), while the activation of NF-κB induced apoptosis of DF-1 cells (Figure 8). These findings indicated a relevant link between neddylation and NF-κB in chicken embryo fibroblasts after treatment by MGA_0676. These results were also the first to show the apoptosis induction mechanism of mycoplasmal nuclease. In addition, our results indicated that MGA_0676 may be an important etiological virulence factor of the respiratory disease caused by M. gallisepticum, and that it may be involved in the immunosuppression of the infected birds. MGA_0676 may also participate in the evasion from the immune clearance by the host, due to its ability of inducing apoptosis, which was probably mediated through the activation of NF-κB (Adegboye, 1978; Papayannopoulos, 2014). Knock-out of MGA_0676 or SNC region may decrease the virulence of M. gallisepticum, and the attenuated strain could be used to prepare vaccines to prevent the infection caused by M. gallisepticum in the future. In conclusion, in our study we found that the mycoplasmal nuclease MGA_0676 interacted with NAE and accelerated cullin neddylation, which activated the NF-κB to induce apoptosis of chicken embryo fibroblasts, but how this mechanism plays its vital role in the M. gallisepticum growth and infections remain further explored.
Ethics Statement
All animal care procedures and experiments were approved by the Beijing Association for Science and Technology (approval ID SYXK (Beijing) 2007-0023) and were in compliance with the Beijing Laboratory Animal Welfare and Ethics Guidelines issued by the Beijing Administration Committee of Laboratory Animals. All animal studies were performed in accordance with the China Agricultural University Institutional Animal Care and Use Committee Guidelines (ID: SKLAB-B-2010-003) and were approved by the Animal Welfare Committee of China Agricultural University.
Author Contributions
WW and JX conceived and designed the experiments. PL, JX, HR, XL, and YZ performed the experiments. WW and JX analyzed the data. FJ contributed reagents, materials, analysis tools. PL, JX and WW wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 31472190). We thank Prof. Antonio Peramo for providing suggestions for this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00105/full#supplementary-material
References
Adegboye, D. S. (1978). A review of mycoplasma-induced immunosuppression. Br. Vet. J. 134, 556–560. doi: 10.1016/S0007-1935(17)33337-7
Amundsen, S. K., Fero, J., Hansen, L. M., Cromi, G. A., Solnick, J. V., Smith, G. R., et al. (2008). Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol. Microbiol. 69, 994–1007. doi: 10.1111/j.1365-2958.2008.06336.x
Brasier, A. R. (2006). The NF-κB regulatory network. Cardiovasc. Toxicol. 6, 111–130. doi: 10.1385/CT:6:2:111
Casadevall, A., and Pirofski, L. (2001). Host-pathogen interactions: the attributes of virulence. J. Infect. Dis. 184, 337–344. doi: 10.1086/322044
Chaudhary, N., Gomez, G. A., Howes, M. T., Lo, H. P., McMahon, K. A., Rae, J. A., et al. (2014). Endocytic crosstalk: cavins, caveolins, and caveolae regulate clathrin-independent endocytosis. PLoS Biol. 12:e1001832. doi: 10.1371/journal.pbio.1001832
Chen, B., Zhang, A., Xu, Z. M., Li, R., Chen, H. C., and Jin, M. L. (2011). Large-scale identification of bacteria-host crosstalk by affinity chromatography: capturing the interactions of streptococcus suis proteins with host cells. J. Proteome Res. 10, 5163–5174. doi: 10.1021/pr200758q
Chen, X., Kandasamy, K., and Srivastava, R. K. (2003). Differential roles of RelA (p65) and c-Rel subunits of nuclear factor kappa B in tumor necrosis factor-related apoptosis-inducing ligand signaling. Cancer Res. 63, 1059–1066.
Chen, Y., Mcphie, D. L., Hirschberg, J., and Neve, R. L. (2000). The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J. Biol. Chem. 275, 8929–8935. doi: 10.1074/jbc.275.12.8929
Chung, H. R., Dunkel, I., Heise, F., Linke, C., Krobitsch, S., Ehrenhofer-Murray, A. E., et al. (2010). The effect of micrococcal nuclease digestion on nucleosome positioning data. PLoS ONE 5:e15754. doi: 10.1371/journal.pone.0015754
Cuatrecasas, P., Fuchs, S., and Anfinsen, C. B. (1967). The interaction of nucleotides with the active site of staphylococcal nuclease. J. Biol. Chem. 242, 4759–4767.
Cui, J., Yao, Q., Li, S., Ding, X., Lu, Q., Mao, H., et al. (2010). Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science 329, 1215–1218. doi: 10.1126/science.1193844
Delaney, N. F., Balenger, S., Bonneaud, C., Marx, C. J., Hill, G. E., Ferguson-Noel, N., et al. (2012). Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 8:e1002511. doi: 10.1371/journal.pgen.1002511
Derré-Bobillot, A., Cortes-Perez, N. G., Yamamoto, Y. J., Kharrat, P., Couvé, E., Cunha, V. D., et al. (2013). Nuclease A (Gbs0661), an extracellular nuclease of Streptococcus agalactiae, attacks the neutrophil extracellular traps and is needed for full virulence. Mol. Microbiol. 89, 518–531. doi: 10.1111/mmi.12295
Dominski, Z. (2007). Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. 42, 67–93. doi: 10.1080/10409230701279118
Fan, X., Moqtaderi, Z., Jin, Y., Zhang, Y., Liu, X. S., and Struhl, K. (2010). Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3'-end formation. Proc. Natl. Acad. Sci. U.S.A. 107, 17945–17950. doi: 10.1073/pnas.1012674107
Fitzgerald, J. R., Foster, T. J., and Cox, D. (2006). The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 4, 445–457. doi: 10.1038/nrmicro1425
Fu, P., Sun, Z., Zhang, Y., Yu, Z., Zhang, H., Su, D., et al. (2014). Development of a direct competitive ELISA for the detection of Mycoplasma bovis infection based on a monoclonal antibody of P48 protein. BMC Vet. Res. 10:42. doi: 10.1186/1746-6148-10-42
Garcia, M. D., Formoso, K., Aparicio, G. I., Frasch, A. C. C., and Scorticati, C. (2017). The Membrane glycoprotein M6a endocytic/recycling pathway involves clathrin-mediated endocytosis and affects neuronal synapses. Front. Mol. Neurosci. 10:296. doi: 10.3389/fnmol.2017.00296
Großhennig, S., Schmidl, S. R., Schmeisky, G., Busse, J., and Stülke, J. (2013). Implication of glycerol and phospholipid transporters in Mycoplasma pneumoniae growth and virulence. Infect. Immun. 81, 896–904. doi: 10.1128/IAI.01212-12
Haucke, V. (2015). Cell biology: on the endocytosis rollercoaster. Nature 517, 446–447. doi: 10.1038/nature14081
Huang, J., Yu, S., Ji, C., and Li, J. (2015). Structural basis of cell apoptosis and necrosis in TNFR signaling. Apoptosis 20, 210–215. doi: 10.1007/s10495-014-1061-5
Humphrys, M. S., Creasy, T., Sun, Y., Shetty, A. C., Chibucos, M. C., Drabek, E. F., et al. (2013). Simultaneous transcriptional profiling of bacteria and their host cells. PLoS ONE 8:e80597. doi: 10.1371/journal.pone.0080597
Into, T., Kiura, K., Yasuda, M., Kataoka, H., Inoue, N., Hasebe, A., et al. (2004). Stimulation of human toll-like receptor (TLR) 2 and TLR6 with membrane lipoproteins of Mycoplasma fermentans induces apoptotic cell death after NF-κB activation. Cell. Microbiol. 6, 187–199. doi: 10.1046/j.1462-5822.2003.00356.x
Jones, B. D., and Falkow, S. (1996). Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14, 533–561. doi: 10.1146/annurev.immunol.14.1.533
Jubelin, G., Taieb, F., Duda, D. M., Hsu, Y., Samba-Louaka, A., Nobe, R., et al. (2010). Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 6:e1001128. doi: 10.1371/journal.ppat.1001128
Kamashev, D., Oberto, J., Serebryakova, M., Gorbachev, A., Zhukova, Y., Levitskii, S., et al. (2011). Mycoplasma gallisepticum produces a histone-like protein that recognizes base mismatches in DNA. Biochemistry 50, 8692–8702. doi: 10.1021/bi2009097
Krachler, A. M., and Orth, K. (2013). Targeting the bacteria -host interface strategies in anti-adhesion therapy. Virulence 4, 284–294. doi: 10.4161/viru.24606
Lee, S. M., Kok, K. H., Jaume, M., Cheung, T. K., Yipa, T. F., Laia, J. C., et al. (2014). Toll-like receptor 10 is involved in induction of innate immune responses to influenza virus infection. Proc. Natl. Acad. Sci. U.S.A. 111, 3793–3798. doi: 10.1073/pnas.1324266111
Lennon, J. T. (2007). Diversity and metabolism of marine bacteria cultivated on dissolved DNA. Appl. Environ. Microbiol. 73, 2799–2805. doi: 10.1128/AEM.02674-06
Li, L., Krishnan, M., Baseman, J. B., and Kannan, T. R. (2010). Molecular cloning, expression, and characterization of a Ca2+-dependent, membrane -associated nuclease of Mycoplasma genitalium. J. Bacteriol. 192, 4876–4884. doi: 10.1128/JB.00401-10
Ma, X. J., and Goodridge, A. G. (1992). Nutritional regulation of nucleosomal structure at the chicken malic enzyme promoter in liver. Nucleic Acids Res. 20, 4997–5002. doi: 10.1093/nar/20.19.4997
Majumder, S., Zappulla, F., and Silbart, L. K. (2014). Mycoplasma gallisepticum lipid associated membrane proteins up-regulate inflammatory genes in chicken tracheal epithelial cells via TLR-2 ligation through an NF-κB dependent pathway. PLoS ONE 9:e112796. doi: 10.1371/journal.pone.0112796
Milner, D. S., Till, R., Cadby, I., Lovering, A. L., Basford, S. M., Saxon, E. B., et al. (2014). Ras GTPase-like protein MglA, a controller of bacterial social-motility in Myxobacteria, has evolved to control bacterial predation by bdellovibrio. PLoS Genet. 10:e1004253. doi: 10.1371/journal.pgen.1004253
Minion, F. C., Jarvill-Taylor, K. J., Billings, D. E., and Tigges, E. (1993). Membrane-associated nuclease activities in Mycoplasmas. J. Bacteriol. 175, 7842–7847. doi: 10.1128/jb.175.24.7842-7847.1993
Munro, P., Flatau, G., and Lemichez, E. (2007). Bacteria and the ubiquitin pathway. Curr. Opin. Microbiol. 10, 39–46. doi: 10.1016/j.mib.2006.11.002
Nakhyung, L. (2009). Characterization of an ATP-binding Cassette (ABC) Transport System Involved in Nucleoside Uptake in Mycoplasma Bovis Strain M23, and discoveRy of its Pathogenicity geneS. Ph.D. thesis, Iowa State University, Ames, IA.
Okumura, C. Y., and Nizet, V. (2014). Subterfuge and sabotage: evasion of host innate defenses by invasive Gram-positive bacterial pathogens. Annu. Rev. Microbiol. 68, 439–458. doi: 10.1146/annurev-micro-092412-155711
Orlandi, P. A., and Fishman, P. H. (1998). Fishman filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J. Cell Biol. 141, 905–915. doi: 10.1083/jcb.141.4.905
Paddenberg, R., Weber, A., Wulf, S., and Mannherz, H. G. (1998). Mycoplasma nucleases able to induce internucleosomal DNA degradation in cultured cells possess many characteristics of eukaryotic apoptotic nucleases. Cell Death Differ. 5, 517–528. doi: 10.1038/sj.cdd.4400380
Papayannopoulos, V. (2014). Infection: microbial nucleases turn immune cells against each other. Curr. Biol. 24, 123–125. doi: 10.1016/j.cub.2013.12.027
Papayannopoulos, V., and Zychlinsky, A. (2009). NETs: a new strategy for using old weapons. Trends Immunol. 30, 513–521. doi: 10.1016/j.it.2009.07.011
Pascual-Lucas, M., Fernandez-Lizarbe, S., Montesinos, J., and Guerri, C. (2014). LPS or ethanol triggers clathrin- and rafts/caveolae-dependent endocytosis of TLR4 in cortical astrocytes. J. Neurochem. 129, 448–462. doi: 10.1111/jnc.12639
Pediaditakis, M., Kaufenstein, M., and Graumann, P. L. (2012). Bacillus subtilis hlpB encodes a conserved stand-alone HNH nuclease-like protein that is essential for viability unless the hlpB deletion is accompanied by the deletion of genes encoding the AddAB DNA repair complex. J. Bacteriol. 194, 6184–6194. doi: 10.1128/JB.05283-11
Pollack, J. D., and Hoffmann, P. J. (1982). Properties of the nucleases of mollicutes. J. Bacteriol. 152, 538–541.
Razin, S. (1999). Adherence of pathogenic mycoplasmas to host cells. Biosci. Rep. 19, 367–372. doi: 10.1023/A:1020204020545
Rejman, J., Oberle, V., Zuhorn, I. S., and Hoekstra, D. (2004). Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 377, 159–169. doi: 10.1042/bj20031253
Rottem, S. (2003). Interaction of Mycoplasmas with host cells. Physiol. Rev. 83, 417–432. doi: 10.1152/physrev.00030.2002
Sacco, E., Cortes, M., Josseaume, N., Rice, L. B., Mainardi, J. L., and Arthur, M. (2014). Serine/threonine protein phosphatase-mediated control of the peptidoglycan cross-linking L,D-transpeptidase pathway in Enterococcus faecium. MBio 5, e01446–e01414. doi: 10.1128/mBio.01446-14
Schmidt, G., Papatheodorou, P., and Aktories, K. (2015). Novel receptors for bacterial protein toxins. Curr. Opin. Microbiol. 23, 55–61. doi: 10.1016/j.mib.2014.11.003
Schmidt, J. A., Browning, G. F., and Markham, P. F. (2007). Mycoplasma hyopneumoniae mhp379 is a Ca2+-dependent, sugar-nonspecific exonuclease exposed on the cell surface. J. Bacteriol. 189, 3414–3424. doi: 10.1128/JB.01835-06
Shields, R. C., Mokhtar, N., Ford, M., Hall, M. J., Burgess, J. G., ElBadawey, M. R., et al. (2013). Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS ONE 8:e55339. doi: 10.1371/journal.pone.0055339
Somarajan, S. R., Kannan, T. R., and Baseman, J. B. (2010). Mycoplasma pneumoniae MPN133 is a cytotoxic nuclease with a glutamic acid-, lysine- and serine-rich region essential for binding and internalization but not enzymatic activity. Cell. Microbiol. 12, 1821–1831. doi: 10.1111/j.1462-5822.2010.01513.x
Soucy, T. A., Smith, P. G., Milhollen, M. A., Berger, A. J., Gavin, J. M., Adhikari, S., et al. (2009). An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736. doi: 10.1038/nature07884
Sourjik, V., and Berg, H. C. (2004). Functional interactions between receptors in bacterial chemotaxis. Nature 428, 437–441. doi: 10.1038/nature02406
Suciu, D., and Inouye, M. (1996). The 19-residue pro-peptide of staphylococcal nuclease has a profound secretion-enhancing ability in Escherichia coli. Mol. Microbiol. 21, 181–195. doi: 10.1046/j.1365-2958.1996.6211341.x
Suh, Y., and Benedik, M. J. (1997). Secretion of nuclease across the outer membrane of Serratia marcescens and its energy requirement. J. Bacteriol. 179, 677–683.
Thammavongsa, V., Missiakas, D. M., and Schneewind, O. (2013). Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342, 863–866. doi: 10.1126/science.1242255
Toid, A. D. (2014). Staphylococcus slips through the net. Nat. Rev. Microbiol. 12, 6–7. doi: 10.1038/nrmicro3179
Veiga, E., and Cossart, P. (2006). The role of clathrin-dependent endocytosis in bacterial internalization. Trends Cell Biol. 16, 499–504. doi: 10.1016/j.tcb.2006.08.005
Wanga, L., Schmidlb, S. R., and Stülkeb, J. (2014). Mycoplasma pneumoniae thymidine phosphorylase. Nucleos. Nucleot. Nucl. 33, 296–304. doi: 10.1080/15257770.2013.853783
Wu, Y., Qiu, H., Zeng, Y., You, X., Deng, Z., Yu, M., et al. (2008). Mycoplasma genitalium lipoproteins induce human monocytic cell expression of proinflammatory cytokines and apoptosis by activating nuclear factor B. Mediat. Inflamm. 2008:195427. doi: 10.1155/2008/195427
Xu, J., Teng, D., Jiang, F., Zhang, Y. W., El-Saeed, A., Wang, H., et al. (2015). Mycoplasma gallisepticum MGA_0676 is a membrane-associated cytotoxic nuclease with a staphylococcal nuclease region essential for nuclear translocation and apoptosis induction in chicken cells. Appl. Microbiol. Biotechnol. 99, 1859–1871. doi: 10.1007/s00253-014-6185-6
Keywords: mycoplasmal nuclease MGA_0676, apoptosis, endocytosis, NEDD8-activating enzyme E1 regulatory subunit, activation of NF-κB
Citation: Li P, Xu J, Rao H, Li X, Zhang Y, Jiang F and Wu W (2018) Mechanism of Apoptosis Induction by Mycoplasmal Nuclease MGA_0676 in Chicken Embryo Fibroblasts. Front. Cell. Infect. Microbiol. 8:105. doi: 10.3389/fcimb.2018.00105
Received: 29 January 2018; Accepted: 19 March 2018;
Published: 04 April 2018.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Xiaoming Liu, Ningxia Medical University, ChinaYujiong Wang, Ningxia University, China
Copyright © 2018 Li, Xu, Rao, Li, Zhang, Jiang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-xue Wu, bGFiYm9hcmRAMTI2LmNvbQ==
†These authors have contributed equally to this work.
 Peng Li
Peng Li Jian Xu
Jian Xu Hong-mei Rao
Hong-mei Rao Xia Li
Xia Li Yun-ke Zhang
Yun-ke Zhang Fei Jiang
Fei Jiang Wen-xue Wu
Wen-xue Wu