- 1State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2The Cooperative Innovation Center for Sustainable Pig Production, Huazhong Agricultural University, Wuhan, China
- 3Key Laboratory of Development of Veterinary Diagnostic Products of Ministry of Agriculture, Huazhong Agricultural University, Wuhan, China
- 4Hubei Key Laboratory of Animal Embryo and Molecular Breeding, Institute of Animal Husbandry and Veterinary Sciences, Hubei Academy of Agricultural Sciences, Wuhan, China
Gram-negative bacteria have evolved numerous two-component systems (TCSs) to cope with external environmental changes. The CpxA/CpxR TCS consisting of the kinase CpxA and the regulator CpxR, is known to be involved in the biofilm formation and virulence of Escherichia coli. However, the role of CpxA/CpxR remained unclear in Actinobacillus pleuropneumoniae, a bacterial pathogen that can cause porcine contagious pleuropneumonia (PCP). In this report, we show that CpxA/CpxR contributes to the biofilm formation ability of A. pleuropneumoniae. Furthermore, we demonstrate that CpxA/CpxR plays an important role in the expression of several biofilm-related genes in A. pleuropneumoniae, such as rpoE and pgaC. Furthermore, The results of electrophoretic mobility shift assays (EMSAs) and DNase I footprinting analysis demonstrate that CpxR-P can regulate the expression of the pgaABCD operon through rpoE. In an experimental infection of mice, the animals infected with a cpxA/cpxR mutant exhibited delayed mortality and lower bacterial loads in the lung than those infected with the wildtype bacteria. In conclusion, these results indicate that the CpxA/CpxR TCS plays a contributing role in the biofilm formation and virulence of A. pleuropneumoniae.
Introduction
Actinobacillus pleuropneumoniae is a species of Gram-negative bacteria that causes porcine contagious pleuropneumonia (PCP), which is characterized by fibrinous pleurisy and fibrinous pneumonia. Each year this pathogen causes substantial economic damages in the pork industry globally (Zimmerman et al., 2012). Previous studies have discovered many virulence factors of A. pleuropneumoniae (Fuller et al., 2000; Sheehan et al., 2003). These virulence factors, such as Apx toxins, capsule, adhesins, transferrin-binding proteins, and outer membrane proteins, play crucial roles in the pathogenicity of A. pleuropneumoniae (Chiers et al., 2010).
To increase their adaptability, bacteria generally utilize two-component signal transduction systems (TCSs) to perceive and respond to environmental influences. A representative TCS consists of a histidine kinase (HK) in the inner membrane and a response regulator (RR) in the cytoplasm (Buelow and Raivio, 2010). After perceiving an environmental signal, the HK subsequently autophosphorylates and transfers the same phosphate group to the RR, after which the RR can influence the transcription level of downstream genes by binding to their promoter regions (Vogt and Raivio, 2012). Whole genome sequencing of A. pleuropneumoniae serovar 3 strain JL03 revealed five putative TCSs: ArcA/ArcB, QseB/QseC, CpxR/CpxA, NarP/NarQ, and PhoB/PhoR (Xu et al., 2008).
When it was initially characterized, the TCS CpxA/CpxR was viewed as a newly discovered envelope stress response system. Experimental evidence in Escherichia coli then provided a model: when misfolded envelope proteins accumulate, CpxA autophosphorylates and then transfers the phosphate group to CpxR, which causes the upregulation of a series of chaperonins and proteases that can degrade or refold the misfolded proteins, thus alleviating the envelope stress (Vogt and Raivio, 2012). In recent years, the CpxA/CpxR system has been demonstrated to be involved in the virulence of uropathogenic Escherichia coli (Debnath et al., 2013), Salmonella enterica serotype Typhimurium (Humphreys et al., 2004), and Vibro cholerae (Acosta et al., 2015). The CpxA/CpxR system has also been shown to be involved in the biofilm formation of E. coli (Dorel et al., 1999; Ma and Wood, 2009; Dudin et al., 2014). In addition, the CpxA/CpxR system has been implicated in antibiotic resistance (Mahoney and Silhavy, 2013).
Biofilms are aggregated bacterial groups that are embedded in a matrix of extracellular polymeric substances (Donlan and Costerton, 2002). Biofilm formation of A. pleuropneumoniae in vitro (such as on polystyrene microtiter plates) depends on the lipopolysaccharides (LPS), the capsular polysaccharides (CPS) and the yields of β-1,6-N-acetyl-D-glucosamine (PGA) polymer (Kaplan et al., 2004; Hathroubi et al., 2015b). In A. pleuropneumoniae, the production of PGA depends on the expression of the pgaABCD operon, which is repressed by the protein H-NS and upregulated by the alternative sigma factor σE (Bosse et al., 2010). Kaplan et al. found that only two of the fifteen A. pleuropneumoniae reference strains, including 5b and 11, were able to form a pronounced biofilm on polystyrene microtiter plates (Kaplan and Mulks, 2005). However, Hathroubi and colleagues found that when cultured in the presence of sub-minimum inhibitory concentration of penicillin G, many A. pleuropneumoniae strains can form pronounced biofilms and this is likely the result of a cell envelope stress sensed by the CpxRA system resulting in an increased production of PGA and other matrix components (Hathroubi et al., 2015a). Several studies have also proposed that CpxA/CpxR may be involved in the mechanism of biofilm formation in A. pleuropneumoniae (Tremblay et al., 2013; Hathroubi et al., 2015b). However, these studies did not use a deletion mutant to confirm the phenotype directly.
Although previous work has demonstrated that CpxA/CpxR plays an important role in biofilm formation and virulence in E. coli and other bacteria, the role of CpxA/CpxR in A. pleuropneumoniae remained unclear. In this study, we found new growth conditions that induce biofilm formation in A. pleuropneumoniae (i.e., 42°C, TSB, static). Based on this new system, we investigated the role of CpxA/CpxR in biofilm formation, focusing on the rpoE gene and its relationship to the expression of the pga operon. We also assessed whether or not CpxA/CpxR contributes to the virulence of A. pleuropneumoniae serotype 1 strain S4074.
Materials and Methods
Bacterial Strains and Culture Conditions
The bacterial strains, primers, and plasmids used in this study are listed in Table 1. A. pleuropneumoniae strains were cultured at 37°C or 42°C in tryptic soy broth (TSB; Difco Laboratories, Detroit, MI, USA) or on tryptic soy agar (TSA; Difco Laboratories) supplemented with nicotinamide adenine dinucleotide (10 μg mL−1) and 10% (v/v) newborn calf serum. Additionally, A. pleuropneumoniae strains were also cultured in brain heart infusion broth (BHIB; Oxoid Ltd., Hampshire, United Kingdom) or on brain heart infusion agar (BHIA; Oxoid Ltd.) supplemented with 15 μg mL−1 NAD (Solarbio, Beijing, China) at 37°C. For the selection of A. pleuropneumoniae transformants, chloramphenicol (5 μg mL−1) was added. When culturing different E. coli strains, appropriate antibiotics were added in Luria-Bertani (LB) broth or agar (Haibo, Qingdao, China). For cultivation of E. coli β2155, diaminopimelic acid (dapA; 50 μg mL−1) (Sigma-Aldrich, St. Louis, USA) was added to LB medium (Yuan et al., 2014).
Construction of the A. pleuropneumoniae Mutant ΔcpxAR and Complement CΔcpxAR
To construct a cpxA/cpxR (cpxAR) gene deletion mutant, the upstream and downstream fragments of these two genes were amplified from the genomic DNA of A. pleuropneumoniae strain S4074 (wildtype, WT) using primers cpxAR-up F/R or cpxAR-down F/R. These PCR products were cloned sequentially into the suicide vector pEMOC2 to generate plasmid pEΔcpxAR. The mutant ΔcpxAR was then constructed using the plasmid pEΔcpxAR as previously described (Liu et al., 2015).
For complementation studies, the intact cpxAR genes were cloned with their promoters from S4074 using primers cpxAR-F and cpxAR-R. The entire fragment was then ligated into the T vector pMD18-T, generating plasmid pMD18-T-cpxAR. For constructing complementation plasmid pCΔcpxAR, the 2.3-kb fragment from pMD18-T-cpxAR containing the cpxAR genes was ligated into E. coli–A. pleuropneumoniae shuttle vector pJFF224-XN via PstI–NotI restriction sites (Yuan et al., 2014). Then, pCΔcpxAR was electroporated (2.5 KV, 25 μFD, 800 Ω) into the mutant strain ΔcpxAR. The correct CΔcpxAR was selected via its chloramphenicol resistance. When the CΔcpxAR strain was cultured, 2 μg mL−1 chloramphenicol was always added in the culture medium.
The resultant mutant strain ΔcpxAR and complement strain CΔcpxAR were verified by both PCR amplification with primers cpxAR-exterior F/R and cpxAR-interior F/R and RT-PCR amplification with primers cpxA-F/R and cpxR-F/R. The sequence accuracy of each strain was verified by DNA sequencing (data not shown).
Biofilm Assay
The microtiter plate biofilm assay is especially advantageous for examining early processes in biofilm formation (Merritt et al., 2005). Overnight inoculums were balanced to the same optical density (OD) with fresh TSB, and 100 μL of the 1:100-diluted balanced inoculums were transferred to each well of a 96-well microtiter plate (Corning, USA). Following an incubation of 36, 48, or 60 h at 37°C or 42°C, each well was washed three times with 200 μL of sterile PBS to remove slackly adherent cells. To fix the remaining attached bacteria, 100 μL of methanol was added to each well. After air-drying, the wells were then stained with 100 μL per well of crystal violet (0.1%) for 10 min at room temperature. After removing the crystal violet solution, the wells were washed under running tap water and dried in a 37°C incubator for 30 min. At this point, the plates were photographed. Last, 100 μL per well of glacial acetic acid (33%, v/v) was added, and the OD590nm of each well was monitored by a Multi-Detection Microplate Reader. Furthermore, we also used this method to detect the biofilm formation of A. pleuropneumoniae strains grown in BHIB, since this is the common method used inducing biofilm formation in A. pleuropneumoniae (Labrie et al., 2010). Both assays were performed in triplicate.
RNA Extraction, RT-PCR, and Real-Time RT-PCR
The WT (S4074), cpxAR mutant, and complement strains were grown in TSB in six-well plates for 4 h at 37°C or 42°C. Ice-cold methanol was used to prevent changes in transcript levels after the planktonic cells of each strain were collected (Subashchandrabose et al., 2013). A total of 1 mL of bacterial cells was blended with 1 mL of 100% ice-cold methanol and centrifuged at 10,000 × g for 3 min at 4°C. For real-time RT-PCR (qRT-PCR), total RNA from the WT, mutant, and complement strains was extracted using the Bacteria Total RNA Isolation Kit (Sangon Biotech, China) and was then reverse-transcribed using the HiScript II Q RT SuperMix for qRT-PCR (+gDNA wiper) (Vazyme, China) following the manufacturer's instructions. The real-time PCR experimental method was based on SYBR-Green dye, and all reactions were performed in triplicate. The 10-μL real-time PCR reaction mixtures contained 5 μL of 2× SYBR Green Master Mix (Vazyme, China), 0.2 μL of Rox Reference Dye 2, 0.2 μL each of the forward and reverse primers (10 μM), 1 μL of template cDNA, and 3.4 μL of ddH2O. The real-time PCR amplification conditions were as follows: 5 min at 50°C, then 5 min at 95°C, followed by 40 cycles of 10 s at 95°C and 35 s at 60°C. The ViiA™ 7 real-time PCR system was used for qRT-PCR analysis by the 2−ΔCt method (Pfaffl, 2001). For normalizing the relative expression of target genes, the 16S rRNA gene was used as an endogenous control (Hathroubi et al., 2015b). The cDNA of WT was also used for RT-PCR by primers cpxAR-RT-F/R.
Expression of His-CpxR Protein in E. coli
The cpxR gene was amplified via PCR from the genomic DNA of A. pleuropneumoniae S4074 using primers PcpxR-F and PcpxR-R (Table 2) and then cloned into the BamHI and HindIII sites of prokaryotic expression plasmid pET-28a, generating the recombinant plasmid pET-cpxR; the accuracy of the inserted sequence was confirmed by DNA sequencing. The plasmid pET-cpxR was transferred into BL21 (DE3) strain and grown at 37°C with energetic shaking in 1 L of LB broth containing 50 μg mL−1 kanamycin to an OD600 of 0.6–0.8. The E. coli BL21 (DE3) cells containing the plasmid pET-cpxR were then induced by isopropyl-β -D-thiogalactoside (0.25 mM) at 25°C for 5 h. A Low Temperature Ultra-high Pressure Continuous Flow Homogenizer (JNBIO, China) was used to disrupt cells, and the resulting cellular debris and membranes were removed by centrifugation at 12,000 × g for 20 min at 4°C. The recombinant protein was purified by Ni-NTA resin affinity chromatography, as described in the OIAexpress manual (Qiagen, Germany). The purified protein rCpxR was dissolved by elution buffer and was stored at −80°C until use. The expression of His-RpoE was also performed as above described.
Electrophoretic Mobility Shift Assays
The promoter regions of rpoE and pgaABCD genes were cloned into the T-vector pMD19-T to generate pMD19-T-rpoE and pMD19-T-pgaABCD. The promoter regions of rpoE and pgaABCD were then amplified via PCR from the plasmid using primers M13F-47 (FAM) and M13R-48, generating fluorescent FAM-labeled probes.
The recombinant protein CpxR was phosphorylated by acetyl phosphate (Sigma, USA) (Pogliano et al., 1997). Electrophoretic mobility shift assays (EMSAs) were carried out in 20 μL of reaction buffer (50 mM Tris-HCl (pH 8.0), 2.5 mM MgCl2, 100 mM KCl, 2 μg of salmon sperm DNA, 0.2 mM DTT, and 10% glycerol) that contained 40 ng of probe and varied quantities of CpxR or RpoE proteins. After incubation for 30 min at 30°C, the reaction liquid was loaded onto 4% non-denaturing PAGE gels in 0.5× TBE [5.4 g L−1 of Tris base, 2.75 g L−1 of boric acid, 2 mL L−1 of 0.5 M EDTA (pH 8.0)]. The resulting gel was photographed using an ImageQuant LAS 4000 mini system (GE Healthcare, USA).
DNase I Footprinting Assay
The DNase I footprinting assay was carried out as previously described (Wang et al., 2012). For each assay, 40 μL of the total reaction mixture contained 400 ng of the probe and different amounts of CpxR. After incubating for 30 min at room temperature, a 10-μL solution [0.015 units of DNase I (Promega, USA) and 100 nmol of freshly prepared CaCl2] was added to the mixture and incubated for 1 min at room temperature. To stop the reaction, 140 μL of DNase I stop solution, which contained 200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS, was added to the mixture. The processing method of samples and the DNA ladder, electrophoresis, and data analysis were all performed as previously described (Wang et al., 2012), except that the GeneScan-LIZ500 size standard (Applied Biosystems, USA) was used in our assay.
Bacterial Virulence in Vivo
Six-week-old BALB/c mice were purchased from the Wuhan Institute of Biological Products Co., Ltd. (Wuhan, China). All procedures and handling techniques were approved by the Laboratory Animal Monitoring Committee of Huazhong Agricultural University and were performed accordingly. We equally divided 30 female BALB/c mice into three groups: WT, ΔcpxAR, and CΔcpxAR. Briefly, all strains were cultured in TSB at 37°C, harvested in logarithmic phase (OD600 = 0.6–0.8), and washed three times with PBS. For each group of test mice, bacteria at 1 × 107 CFU mouse−1 were injected via the abdominal cavity, while an additional five control mice were injected with same volume of sterile PBS. The clinical signs, such as appetite, dyspnea, and lethargy, and mortality rates for each group were recorded twice daily.
To evaluate the colonization ability of ΔcpxAR in susceptible mouse tissue (lung), another 18 female BALB/c mice were divided into three equal groups (6 mice/group). Groups 1 and 3 were intraperitoneally administered 100 μL of PBS containing the WT strain (5 × 105 CFU mouse−1) or the CΔcpxAR strain (5 × 105 CFU mouse−1), respectively, while group 2 received a dose of ΔCpxAR (6.5 × 105 CFU mouse−1). After 72 h, the blood of each mouse was removed via cardiac perfusion, and the lung was aseptically removed. Each lung was homogenized (100 mg weight mL−1 of PBS) using a Tissuelyser (Jingxin, Shanghai, China), and 100 μL of the resulting homogenate were processed for determining the CFU counts.
Bioinformatic and Statistical Analyses
The prediction of bacterial promoters was carried out using the tool in http://fruitfly.org/seq_tools/promoter.html. All data are shown as means ± standard deviation. The statistical analyses of the results were performed using a two-tailed Student's t-test or a one- or two-way analysis of variance, and comparisons with a p < 0.05 were considered as statistically significant. The data and analyses were graphed using GraphPad Prism.
Results
Bioinformatics Analysis of the cpxAR Genes and RT-PCR Verification
To gain a thorough understanding of the role of CpxA/CpxR in A. pleuropneumoniae, we first analyzed the genetic organization of the cpxAR operon in silico. The results indicate that the cpxA and cpxR genes form an operon (Figures 1A,B). We also found that the CpxA and CpxR proteins of A. pleuropneumoniae each have a relatively high amino acid sequence identity with those of E. coli and Salmonella enterica serovar Typhimurium (Figures 1C,D). BlastP analysis revealed that the A. pleuropneumoniae CpxA has 41% amino acid sequence identity each with the CpxA from E. coli and Salmonella enterica serovar Typhimurium, while CpxR has 58 and 59% amino acid sequence identity, respectively, with the CpxR of these two species.
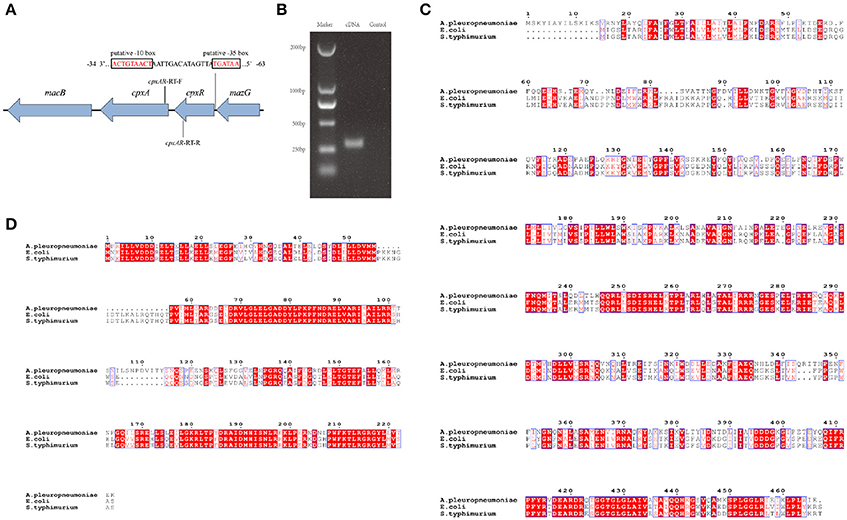
Figure 1. Schematic structure of the cpxAR genes, RT-PCR verification and Bioinformatics analysis. (A) Schematic structure of the cpxA and cpxR genes. There have 64 bp intergenic spacing between the cpxA and cpxR genes. The figure shows the genetic organization of genes up- and down-stream of cpxAR. There is a putative −10 box and a putative −35 box before the cpxR start codon. The promoter of the cpxR gene was predicted by http://fruitfly.org/seq_tools/promoter.html. The E. coli consensus for the −35 box is TTGACA and the −10 box is TATAAT (Shimada et al., 2014). The schematic is not to scale. (B) The primers cpxAR-RT-F/R were used to amplify the junction regions of cpxA–cpxR by RT-PCR. Multiple sequence alignments of CpxA (C) and CpxR (D) were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Construction of the ΔcpxAR Mutant and Complemented Strain CΔcpxAR
An unmarked, in-frame mutant strain ΔcpxAR was designed based on the genome sequence of A. pleuropneumoniae strain S4074, along with a complemented strain CΔcpxAR to confirm that there was no polarity effect in the ΔcpxAR using plasmid pJFF224-XN. The successful constructions of mutant strain ΔcpxAR and complemented strain CΔcpxAR were confirmed by PCR using primers cpxAR-exterior F/R and cpxAR-interior F/R (Supplementary Figure 1) and by RT-PCR using primers cpxA-F/R and cpxR-F/R (Supplementary Figure 2). Furthermore, DNA sequencing results confirmed the sequence accuracy of these two strains (data not shown).
CpxA/CpxR Is Required for Biofilm Formation
No biofilm growth was observed when the WT S4074 strain was grown in TSB at 37°C (Supplementary Figure 3), which confirms the results of previous studies (Kaplan and Mulks, 2005). Figure 2 illustrates the biofilm formation of WT, ΔcpxAR, and CΔcpxAR at 42°C. At this temperature, the WT and CΔcpxAR strains formed a pronounced biofilm that increased over time, whereas the mutant strain ΔcpxAR was unable to form any visible biofilm at 42°C at any time point (Figure 2A). A quantitative assay, which monitored the OD590, also revealed a significant difference between the ΔcpxAR mutant strain and the WT and CΔcpxAR strains in their abilities to form a biofilm (Figure 2B). Additionally, we also compared biofilm formation by the WT and cpxAR mutant strain when the bacteria were grown for 24 h in BHIB at 37°C, which is the most frequently-used culturing condition. The results similarly show that the biofilm formed by the ΔcpxAR mutant was significantly smaller than those formed by the WT and CΔcpxAR strains (Supplementary Figure 4).
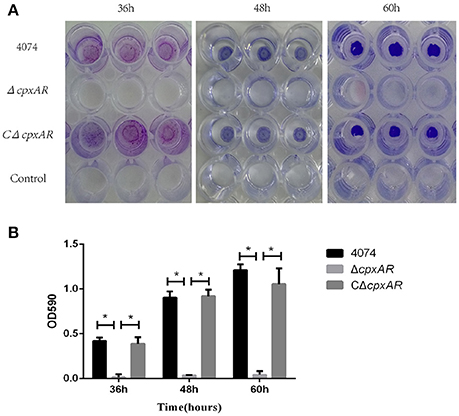
Figure 2. Polystyrene microtiter plate biofilm assay of A. pleuropneumoniae grown in TSB. (A) Biofilm formation of the S4074, ΔcpxAR, and CΔcpxAR strains in the wells of 96-well polystyrene microtiter plates at 42°C. The plates were stained with crystal violet. (B) Quantitative determination of biofilm formation. The S4074, ΔcpxAR, and CΔcpxAR strains were grown in TSB at 42°C. The optical density of the bacterial biofilm was monitored at OD590 after 36, 48, and 60 h of incubation. The data represents the mean ± S.D. of three independent experiments performed in duplicate. *p < 0.05.
Growth Analysis of the WT, ΔcpxAR, and CΔcpxAR Strains
The growth rates of the WT, ΔcpxAR, and CΔcpxAR strains in TSB were analyzed at 37°C and 42°C under shaking and static conditions (Figure 3). A lower growth rate was observed with mutant strain ΔcpxAR compared with the WT and complemented mutant strains at both temperatures. The lack of a growth defect in CΔcpxAR demonstrates that CpxA/CpxR slightly affects the growth rate and indicates that there likely were not any polar effects of the mutations or undetected mutations at other genetic loci.
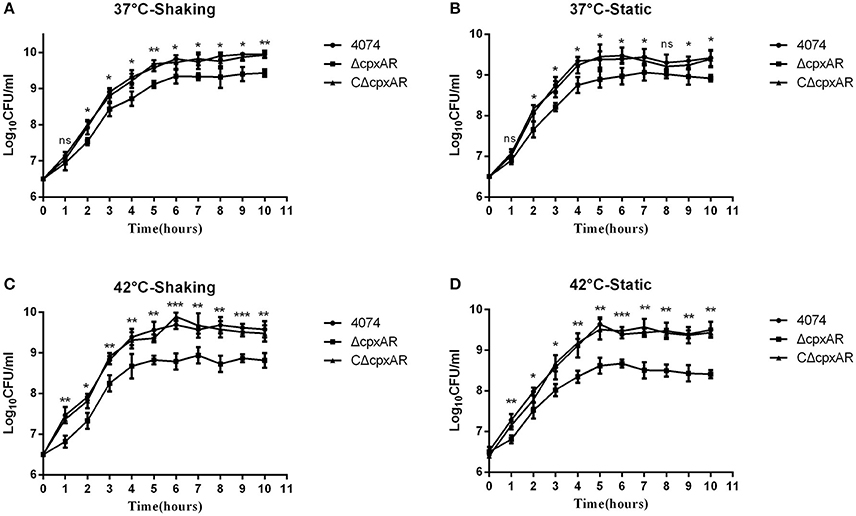
Figure 3. Growth curves of the WT, mutant, and complemented strains of A. pleuropneumoniae. Growth curves of the WT (S4074), mutant (ΔcpxAR), and complemented (CΔcpxAR) strains of A. pleuropneumoniae at 37°C under shaking (A) or static (B) conditions or at 42°C under shaking (C) or static (D) conditions are shown. Bacterial growth was monitored by measurement of viable cell counts expressed as LogCFU/mL. The data represents the mean ± S.D. of three independent experiments performed in duplicate. ***p < 0.001, **p < 0.01, *p < 0.05.
CpxA/CpxR Regulates the mRNA Levels of Some Important Biofilm-Related Genes
Many biofilm-related genes have been identified in recent years, and selected important genes, such as pgaC (encoded within the pgaABCD operon; Kaplan et al., 2004) and rpoE (Bosse et al., 2010) were subsequently studied further. Our results indicate that the pgaC and rpoE genes were each expressed at lower levels in the mutant strain ΔcpxAR when grown at 42°C (Figure 4). Interestingly, there were also remarkably similar fold-changes in these genes at 37°C, even though ΔcpxAR was not able to form a biofilm at this temperature. Additionally, these results show that the expression levels of the pgaC and rpoE genes were each upregulated (p < 0.05) in the parental strain S4074 grown at 42°C compared with their expression levels when the bacteria were grown at 37°C.
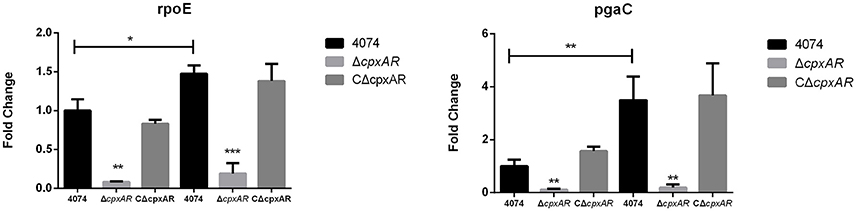
Figure 4. Effects of the cpxA/cpxR deletion on the transcription of biofilm-related genes. QRT-PCR analysis of the transcription of important biofilm-related genes (pgaC and rpoE) in the WT S4074, ΔcpxAR, and CΔcpxAR strains at different temperatures. The mRNA levels of biofilm-related genes were normalized to those of the 16S RNA gene for each strain. Data presented are the mean ± S.D. from three independent experiments performed in duplicate. ***p < 0.001, **p < 0.01, *p < 0.05.
CpxR Binds to the Promoter Region of the rpoE Gene in A. pleuropneumoniae
The recognition site of phosphorylated CpxR (CpxR-P) has a conserved sequence GTAAA-(N)4−8-GTAAA (Yamamoto and Ishihama, 2006; Srinivasan et al., 2012; Bernal-Cabas et al., 2015). SDS-PAGE and Western blot results show that the His-CpxR and His-RpoE were successfully expressed in supernatant (Supplementary Figure 5). EMSAs revealed that CpxR-P and RpoE clearly bound to the promoter region of the rpoE gene and the pga operon respectively in a dose-dependent manner, and these binding were abolished by adding excess unlabeled competitor DNA (Figure 5). Furthermore, a DNase I footprinting analysis also revealed that the promoter region of rpoE had a CpxR-P-binding site (5′-AGTATTTGTAAATAAT-3′), which was 25 bp upstream of the putative transcription start site (Figure 6A and Supplementary Figure 7). The CpxR-P-binding site partially coincided with the conserved sequence GTAAA-(N)4−8-GTAAA, and overlapped the promoter −35 region (Figure 6B). These data show that CpxR-P binds directly to the promoter region of rpoE. Taken together, these data support that under this biofilm formation condition (42°C, TSB, static), CpxR-P can regulate the expression of the pga operon through rpoE.
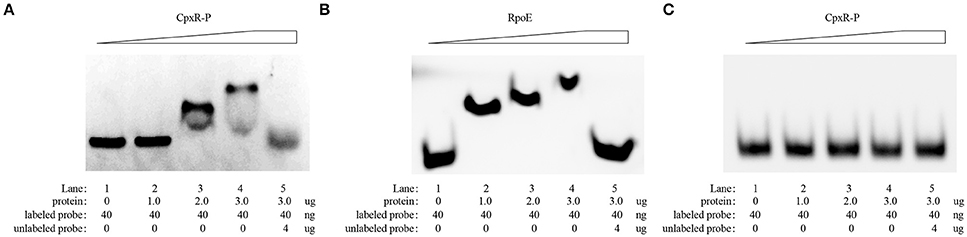
Figure 5. EMSA evaluation of the in vitro direct binding of CpxR-P and RpoE to the target promoter region of rpoE and pga respectively. An EMSA was performed to evaluate the in vitro direct binding of CpxR-P and RpoE to the target promoter region of rpoE (A) and pga (B). Protein (0, 1.0, 2.0, and 3.0 μg) and FAM-labeled DNA fragments (40 ng) were added to the binding reaction. For the competition control, an excess amount of unlabeled competitor DNA (4 μg) was added to the reaction mixture. (C) Binding of CpxR-P to the target promoter region of pga was negative control.
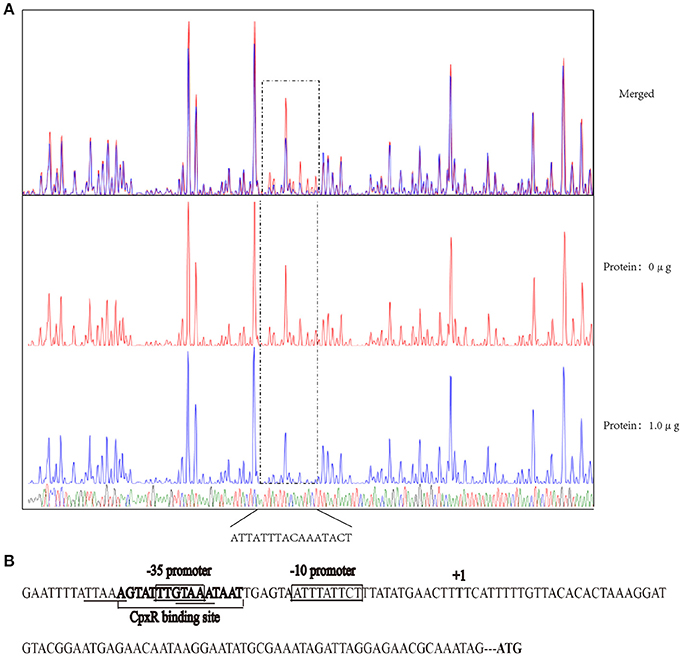
Figure 6. DNase I footprinting assay assessing the in vitro direct binding of CpxR-P to the promoter region of rpoE. (A) A DNase I footprinting assay of the rpoE promoter DNA fragment was performed in the absence or presence of CpxR-P. The FAM-labeled 273-bp DNA fragments (400 ng) that had been pre-incubated in the absence or presence of 1.0 μg of CpxR-P were subjected to DNase I digestion and a subsequent fragment length analysis. The fluorescent signals for the labeled DNA fragments are shown plotted against the sequences of the fragments. (B) Summary of the results from the DNase I footprinting assay described above. The sequence of the predicted motif is shown with the putative core regions underlined and the sequence determined by the DNase I footprinting assay in bold. The promoter region of rpoE gene is shown enclosed in a box.
Virulence of the cpxAR Mutant in Mice
The role of CpxA/CpxR in the virulence of A. pleuropneumoniae was evaluated in vivo in BALB/c mice, which serve as an appropriate model for A. pleuropneumoniae infection (Chiang et al., 2009; Seo et al., 2013; Yuan et al., 2014; Wang et al., 2015; Xie et al., 2016). As shown in Figure 7A, 90% of the mice in the WT and CΔcpxAR groups died within 7 days, but only two mice in the ΔcpxAR group died within this time frame. Overall, the survival rate of the ΔcpxAR group was significantly higher than those of the WT and CΔcpxAR groups (p < 0.05).
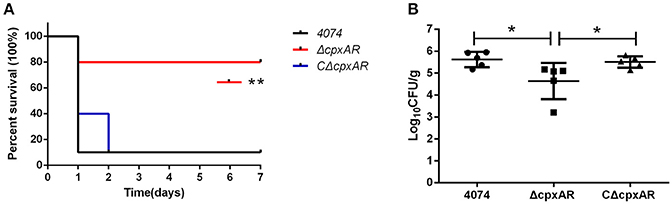
Figure 7. Survival curves and lung bacteria loads of mice infected with A. pleuropneumoniae. (A) Survival curves for Balb/c mice infected with the WT, ΔcpxAR, and CΔcpxAR strains. Six-week-old Balb/c mice were inoculated intraperitoneally with 1 × 107 CFU of bacteria, and their survival was monitored over a 7 day period. Data were analyzed using the log rank test. (B) The A. pleuropneumoniae load in the lungs of infected mice. The horizontal lines denote the median number of bacteria in each group of 6 mice. The data is representative of one of three independent experiments. **p < 0.01, *p < 0.05.
As shown in Figure 7B, the amount of colonization by the ΔcpxAR strain in the lungs was significantly lower compared with those by the WT and CΔcpxAR strains (p < 0.001). This result reveals that the CpxA/CpxR TCS is necessary for A. pleuropneumoniae colonization in vivo. Taken together, these results indicate that the CpxA/CpxR TCS contributes to the virulence of A. pleuropneumoniae.
Discussion
Although many studies regarding biofilm formation in A. pleuropneumoniae have cited cpxAR and consider cpxAR to be a biofilm-related factor, no reports have directly validated the function of cpxAR in A. pleuropneumoniae (Tremblay et al., 2013; Hathroubi et al., 2015a,b). A mutation of cpxAR in A. pleuropneumoniae resulted in a lower growth rate than observed for the corresponding WT strain.
Previous work showed that A. pleuropneumoniae strain S4074 can produce a pronounced biofilm in BHI broth that had lower than normal levels of zinc, but this condition is inconvenient for use in research (Labrie et al., 2010). In this study, our microtiter plate biofilm assays revealed that although S4074 cannot produce a detectable biofilm at 37°C, it does produce a pronounced biofilm at 42°C. Furthermore, the qRT-PCR results revealed that the expression levels of three biofilm-related genes (rpoE and pgaC) in S4074 were significantly upregulated at 42°C compared with their expression levels at 37°C. Thus, our qRT-PCR results were consistent with the results of the biofilm assays. In addition, the expression of cpxR was upregulated in S4074 at 42°C compared with at 37°C (Supplementary Figure 6). Together, these results indicate that high temperature can induce a pronounced biofilm through upregulating the cpxR gene in S4074.
CpxR was previously shown to bind to the promoter region of the regulator σE in E. coli (De Wulf et al., 2002). In this study, qRT-PCR revealed that CpxA/CpxR positively influences transcription of the rpoE gene in A. pleuropneumoniae. Additionally, the sequence GTAAA-(N)4−8-GTAAA is the CpxR-binding site in many other bacteria (Feldheim et al., 2016; Tian et al., 2016). Here, an in silico assessment identified a putative CpxR-binding sequence in the vicinity of the promoter region of sigma factor rpoE. We also demonstrated via EMSA that CpxR can directly bind to the promotor region of the rpoE gene. Furthermore, our DNase I footprinting analysis found that the CpxR-binding site was partially consistent with the putative sequence and was 25 bp away from the putative transcription start site. The CpxR-binding site is generally within 100 bp in the 5′ direction from the transcriptional start site, which is essential for CpxR-dependent activation of the target promoters in E. coli (De Wulf and Lin, 2000; Raffa and Raivio, 2002). Previous studies demonstrated that CpxR primarily functions as a class I factor binding upstream of the promoter −35 region in E. coli (Yamamoto and Ishihama, 2006). In this study, we observed that CpxR activates rpoE, as the CpxR binding site overlaps the −35 box sequence in A. pleuropneumoniae.
Notably, in TSB, the cpxAR mutant showed a remarkable fold-change in the expression of the pgaC gene, which was lower than that in the WT strain. Previous studies found that RpoE positively regulates the transcription of the pga operon, and the rpoE promoter sequence (GAACTT-n16-TCAAA) is 468 bp upstream from the pgaA translational start in A. pleuropneumoniae (Bosse et al., 2010). PgaC synthesizes the major biofilm matrix polysaccharide PGA, which is a component of A. pleuropneumoniae biofilm colonies and may prevent the access of antimicrobial agents to the bacterial cells within the biofilm (Costerton et al., 1999; Kaplan et al., 2004). In this study, we also demonstrated via EMSA that RpoE can directly bind to the promotor region of the pga operon. In light of these previous findings and those in the present study, we demonstrated that the CpxA/CpxR TCS in A. pleuropneumoniae is able to regulate the expression of the pga operon through rpoE to facilitate biofilm formation.
Our experimental results suggest that cpxAR could be a virulence-associated gene and, therefore, may be a potential therapeutic target (Wassenaar and Gaastra, 2001). Many major virulence factors reside in the cell envelope, so it is reasonable that the Cpx response is involved in the development of host infection caused by various bacterial pathogens (Vogt and Raivio, 2012). Additionally, RpoE plays an important role in the virulence of Salmonella Typhimurium, Burkholderia pseudomallei, Vibrio cholera, and Vibrio harveyi (Humphreys et al., 1999; Kovacikova and Skorupski, 2002; Thongboonkerd et al., 2007; Rattanama et al., 2012). Furthermore, it has been observed that the pgaBC genes were upregulated in A. pleuropneumoniae following contact with St. Jude porcine lung cells (Auger et al., 2009). In this study, the colonization assay and survival results demonstrate that CpxA/CpxR contributes to the virulence of A. pleuropneumoniae. Taken together, these results indicate that the regulation of biofilm formation by CpxA/CpxR may be involved in the pathogenesis of A. pleuropneumoniae.
Biofilm formation is reportedly involved in escaping from the host immune system, thereby playing an important role in the pathogenicity of A. pleuropneumoniae in vivo (Bosse et al., 2010; Hathroubi et al., 2017). The predominant sign of acute pleuropneumonia is high fever that body temperature can elevated up to 41.8°C (Menzel et al., 2014; Sassu et al., 2017). Here we demonstrate that the fever temperature contribute to biofilm formation in A. pleuropneumoniae. Therefore, the mechanism may promote the understanding of the pathogenesis of A. pleuropneumoniae-induced infection.
Author Contributions
WB, HL, FL conceived and designed the experiments. HL, KY, WP, HZ, HuiC, and TL performed the experiments. FL, HL, PC analyzed the data. FY, HuaC contributed reagents, materials and analysis tools.
Funding
This work was supported by grants from the Ministry of Agriculture Public Welfare Industry (Agriculture) Research (No. 201303034-10, No. 201303034-9), Competitive Planning Project from Hubei Academy of Agricultural Sciences (No. 2015jzxjh03), the Foundation of State Key Laboratory of Veterinary Biotechnology (No. SKLVBF 201603), and Natural Science Foundation of Hubei Province (No.2016CFA015).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Gerald-F. Gerlach (Institute for Microbiology, Department of Infectious Diseases, University of Veterinary Medicine Hannover, Germany) for the generous donation of E. coli β2155 strain and vector pEMOC2. They also thank Katie Oakley, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00072/full#supplementary-material
Supplementary Figure 1. Identification of the A. pleuropneumoniae mutant strain and the complemented strain by PCR. Identification of the ΔcpxAR mutant strain and the CΔcpxAR complemented strain by PCR, using PCR primer pairs cpxAR-exterior F/R (2,935 bp for WT, 573 bp for ΔcpxAR and CΔcpxAR) and cpxAR-interior F/R (506 bp for WT and CΔcpxAR, no fragment for ΔcpxAR).
Supplementary Figure 2. RT-PCR analysis of cpxAR transcripts. Total RNA was extracted from the WT, ΔcpxAR, and CΔcpxAR strains. cDNA generated from these RNA samples was subjected to RT-PCR analysis with primer pairs cpxA-F/R and cpxR-F/R. The products were analyzed by electrophoresis. The DL 2000 DNA Marker is shown on the left (lane M).
Supplementary Figure 3. Polystyrene microtiter plate biofilm assay of A. pleuropneumoniae grown in TSB at 37°C. The S4074, ΔcpxAR, and CΔcpxAR strains were grown in TSB at 37°C. The resulting bacterial biofilms were photographed (A) and measured (B) at OD590 after 36, 48, and 60 h of incubation. Each data point represents the mean ± S.D. from three independent experiments performed in duplicate.
Supplementary Figure 4. Polystyrene microtiter plate biofilm assay of A. pleuropneumoniae grown in BHI at 37°C for 24 h. The S4074, ΔcpxAR, and CΔcpxAR strains were grown in BHI at 37°C. The optical density of the bacterial biofilm was monitored at OD590 after 24 h of incubation. Each data point represents the mean ± S.D. from three independent experiments performed in duplicate. *p < 0.05.
Supplementary Figure 5. Identification of purified His-CpxR protein and His-RpoE protein. Sodium dodecyl sulfate polyacrylamide gel electrophoresis behavior (A,C) and Western blot analyses (B,D) of the purified His-CpxR protein and His-RpoE protein respectively.
Supplementary Figure 6. Effects of temperature on the transcription of cpxAR genes. A qRT-PCR analysis of cpxAR gene transcriptions in the WT S4074 strain at different temperatures is shown. The mRNA levels of cpxAR genes were normalized to those of the 16S RNA gene. Data presented are the mean ± S.D. from three independent experiments performed in duplicate. *p < 0.05.
Supplementary Figure 7. DNase I footprinting assay. The FAM-labeled 273-bp DNA fragments (400 ng) that had been pre-incubated with 1.0 μg (A) or 2.0 μg (B) of CpxR-P were subjected to DNase I digestion and a subsequent fragment length analysis.
References
Acosta, N., Pukatzki, S., and Raivio, T. L. (2015). The Cpx system regulates virulence gene expression in Vibrio cholerae. Infect. Immun. 83, 2396–2408. doi: 10.1128/IAI.03056-14
Auger, E., Deslandes, V., Ramjeet, M., Contreras, I., Nash, J. H., Harel, J., et al. (2009). Host-pathogen interactions of Actinobacillus pleuropneumoniae with porcine lung and tracheal epithelial cells. Infect. Immun. 77, 1426–1441. doi: 10.1128/IAI.00297-08
Bernal-Cabas, M., Ayala, J. A., and Raivio, T. L. (2015). The Cpx envelope stress response modifies peptidoglycan cross-linking via the L,D-transpeptidase LdtD and the novel protein YgaU. J. Bacteriol. 197, 603–614. doi: 10.1128/JB.02449-14
Bosse, J. T., Sinha, S., Li, M. S., O'Dwyer, C. A., Nash, J. H., Rycroft, A. N., et al. (2010). Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. J. Bacteriol. 192, 2414–2423. doi: 10.1128/JB.01513-09
Buelow, D. R., and Raivio, T. L. (2010). Three (and more) component regulatory systems - auxiliary regulators of bacterial histidine kinases. Mol. Microbiol. 75, 547–566. doi: 10.1111/j.1365-2958.2009.06982.x
Chiang, C. H., Huang, W. F., Huang, L. P., Lin, S. F., and Yang, W. J. (2009). Immunogenicity and protective efficacy of ApxIA and ApxIIA DNA vaccine against Actinobacillus pleuropneumoniae lethal challenge in murine model. Vaccine 27, 4565–4570. doi: 10.1016/j.vaccine.2009.05.058
Chiers, K., De Waele, T., Pasmans, F., Ducatelle, R., and Haesebrouck, F. (2010). Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet. Res. 41:65. doi: 10.1051/vetres/2010037
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Debnath, I., Norton, J. P., Barber, A. E., Ott, E. M., Dhakal, B. K., Kulesus, R. R., et al. (2013). The Cpx stress response system potentiates the fitness and virulence of uropathogenic Escherichia coli. Infect. Immun. 81, 1450–1459. doi: 10.1128/IAI.01213-12
De Wulf, P., and Lin, E. C. (2000). Cpx two-component signal transduction in Escherichia coli: excessive CpxR-P levels underlie CpxA* phenotypes. J. Bacteriol. 182, 1423–1426. doi: 10.1128/JB.182.5.1423-1426.2000
De Wulf, P., McGuire, A. M., Liu, X., and Lin, E. C. (2002). Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277, 26652–26661. doi: 10.1074/jbc.M203487200
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Dorel, C., Vidal, O., Prigent-Combaret, C., Vallet, I., and Lejeune, P. (1999). Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178, 169–175. doi: 10.1111/j.1574-6968.1999.tb13774.x
Dudin, O., Geiselmann, J., Ogasawara, H., Ishihama, A., and Lacour, S. (2014). Repression of flagellar genes in exponential phase by CsgD and CpxR, two crucial modulators of Escherichia coli biofilm formation. J. Bacteriol. 196, 707–715. doi: 10.1128/JB.00938-13
Feldheim, Y. S., Zusman, T., Speiser, Y., and Segal, G. (2016). The Legionella pneumophila CpxRA two-component regulatory system: new insights into CpxR's function as a dual regulator and its connection to the effectors regulatory network. Mol. Microbiol. 99, 1059–1079. doi: 10.1111/mmi.13290
Frey, J. (1992). Construction of a broad host range shuttle vector for gene cloning and expression in Actinobacillus pleuropneumoniae and other Pasteurellaceae. Res. Microbiol. 143, 263–269. doi: 10.1016/0923-2508(92)90018-J
Fuller, T. E., Martin, S., Teel, J. F., Alaniz, G. R., Kennedy, M. J., and Lowery, D. E. (2000). Identification of Actinobacillus pleuropneumoniae virulence genes using signature-tagged mutagenesis in a swine infection model. Microb. Pathog. 29, 39–51. doi: 10.1006/mpat.2000.0364
Hathroubi, S., Fontaine-Gosselin, S. E., Tremblay, Y. D., Labrie, J., and Jacques, M. (2015a). Sub-inhibitory concentrations of penicillin G induce biofilm formation by field isolates of Actinobacillus pleuropneumoniae. Vet. Microbiol. 179, 277–286. doi: 10.1016/j.vetmic.2015.06.011
Hathroubi, S., Hancock, M. A., Bosse, J. T., Langford, P. R., Tremblay, Y. D., Labrie, J., et al. (2015b). Surface polysaccharide mutants reveal that absence of O antigen reduces biofilm formation of Actinobacillus pleuropneumoniae. Infect. Immun. 84, 127–137. doi: 10.1128/IAI.00912-15
Hathroubi, S., Loera-Muro, A., Guerrero-Barrera, A. L., Tremblay, Y. D. N., and Jacques, M. (2017). Actinobacillus pleuropneumoniae biofilms: role in pathogenicity and potential impact for vaccination development. Anim. Health Res. Rev. doi: 10.1017/S146625231700010X. [Epub ahead of print].
Humphreys, S., Rowley, G., Stevenson, A., Anjum, M. F., Woodward, M. J., Gilbert, S., et al. (2004). Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 72, 4654–4661. doi: 10.1128/IAI.72.8.4654-4661.2004
Humphreys, S., Stevenson, A., Bacon, A., Weinhardt, A. B., and Roberts, M. (1999). The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67, 1560–1568.
Kaplan, J. B., and Mulks, M. H. (2005). Biofilm formation is prevalent among field isolates of Actinobacillus pleuropneumoniae. Vet. Microbiol. 108, 89–94. doi: 10.1016/j.vetmic.2005.02.011
Kaplan, J. B., Velliyagounder, K., Ragunath, C., Rohde, H., Mack, D., Knobloch, J. K., et al. (2004). Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186, 8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004
Kovacikova, G., and Skorupski, K. (2002). The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70, 5355–5362. doi: 10.1128/IAI.70.10.5355-5362.2002
Labrie, J., Pelletier-Jacques, G., Deslandes, V., Ramjeet, M., Auger, E., Nash, J. H., et al. (2010). Effects of growth conditions on biofilm formation by Actinobacillus pleuropneumoniae. Vet. Res. 41:3. doi: 10.1051/vetres/2009051
Liu, J., Hu, L., Xu, Z., Tan, C., Yuan, F., Fu, S., et al. (2015). Actinobacillus pleuropneumoniae two-component system QseB/QseC regulates the transcription of PilM, an important determinant of bacterial adherence and virulence. Vet. Microbiol. 177, 184–192. doi: 10.1016/j.vetmic.2015.02.033
Ma, Q., and Wood, T. K. (2009). OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ. Microbiol. 11, 2735–2746. doi: 10.1111/j.1462-2920.2009.02000.x
Mahoney, T. F., and Silhavy, T. J. (2013). The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J. Bacteriol. 195, 1869–1874. doi: 10.1128/JB.02197-12
Menzel, A., Beyerbach, M., Siewert, C., Gundlach, M., Hoeltig, D., Graage, R., et al. (2014). Actinobacillus pleuropneumoniae challenge in swine: diagnostic of lung alterations by infrared thermography. BMC Vet. Res. 10:199. doi: 10.1186/s12917-014-0199-2
Merritt, J. H., Kadouri, D. E., and O'Toole, G. A. (2005). Growing and analyzing static biofilms. Curr. Protoc. Microbiol. Chapter 1, Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pogliano, J., Lynch, A. S., Belin, D., Lin, E. C., and Beckwith, J. (1997). Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11, 1169–1182. doi: 10.1101/gad.11.9.1169
Raffa, R. G., and Raivio, T. L. (2002). A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45, 1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x
Rattanama, P., Thompson, J. R., Kongkerd, N., Srinitiwarawong, K., Vuddhakul, V., and Mekalanos, J. J. (2012). Sigma E regulators control hemolytic activity and virulence in a shrimp pathogenic Vibrio harveyi. PLoS ONE 7:e32523. doi: 10.1371/journal.pone.0032523
Sassu, E. L., Bosse, J. T., Tobias, T. J., Gottschalk, M., Langford, P. R., and Hennig-Pauka, I. (2017). Update on Actinobacillus pleuropneumoniae-knowledge, gaps and challenges. Transbound. Emerg. Dis. doi: 10.1111/tbed.12739. [Epub ahead of print].
Seo, K. W., Kim, S. H., Park, J., Son, Y., Yoo, H. S., Lee, K. Y., et al. (2013). Nasal immunization with major epitope-containing ApxIIA toxin fragment induces protective immunity against challenge infection with Actinobacillus pleuropneumoniae in a murine model. Vet. Immunol. Immunopathol. 151, 102–112. doi: 10.1016/j.vetimm.2012.10.011
Sheehan, B. J., Bosse, J. T., Beddek, A. J., Rycroft, A. N., Kroll, J. S., and Langford, P. R. (2003). Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect. Immun. 71, 3960–3970. doi: 10.1128/IAI.71.7.3960-3970.2003
Shimada, T., Yamazaki, Y., Tanaka, K., and Ishihama, A. (2014). The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS ONE 9:e90447. doi: 10.1371/journal.pone.0090447
Srinivasan, V. B., Vaidyanathan, V., Mondal, A., and Rajamohan, G. (2012). Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 7:e33777. doi: 10.1371/journal.pone.0033777
Subashchandrabose, S., Leveque, R. M., Kirkwood, R. N., Kiupel, M., and Mulks, M. H. (2013). The RNA chaperone Hfq promotes fitness of Actinobacillus pleuropneumoniae during porcine pleuropneumonia. Infect. Immun. 81, 2952–2961. doi: 10.1128/IAI.00392-13
Thongboonkerd, V., Vanaporn, M., Songtawee, N., Kanlaya, R., Sinchaikul, S., Chen, S. T., et al. (2007). Altered proteome in Burkholderia pseudomallei rpoE operon knockout mutant: insights into mechanisms of rpoE operon in stress tolerance, survival, and virulence. J. Proteome Res. 6, 1334–1341. doi: 10.1021/pr060457t
Tian, Z. X., Yi, X. X., Cho, A., O'Gara, F., and Wang, Y. P. (2016). CpxR activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa. PLoS Pathog. 12:e1005932. doi: 10.1371/journal.ppat.1005932
Tremblay, Y. D., Deslandes, V., and Jacques, M. (2013). Actinobacillus pleuropneumoniae genes expression in biofilms cultured under static conditions and in a drip-flow apparatus. BMC Genomics 14:364. doi: 10.1186/1471-2164-14-364
Vogt, S. L., and Raivio, T. L. (2012). Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326, 2–11. doi: 10.1111/j.1574-6968.2011.02406.x
Wang, L., Qin, W., Yang, S., Zhai, R., Zhou, L., Sun, C., et al. (2015). The Adh adhesin domain is required for trimeric autotransporter Apa1-mediated Actinobacillus pleuropneumoniae adhesion, autoaggregation, biofilm formation and pathogenicity. Vet. Microbiol. 177, 175–183. doi: 10.1016/j.vetmic.2015.02.026
Wang, Y., Cen, X. F., Zhao, G. P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-12
Wassenaar, T. M., and Gaastra, W. (2001). Bacterial virulence: can we draw the line? FEMS Microbiol. Lett. 201, 1–7. doi: 10.1111/j.1574-6968.2001.tb10724.x
Xie, F., Li, G., Zhang, Y., Zhou, L., Liu, S., Liu, S., et al. (2016). The Lon protease homologue LonA, not LonC, contributes to the stress tolerance and biofilm formation of Actinobacillus pleuropneumoniae. Microb. Pathog. 93, 38–43. doi: 10.1016/j.micpath.2016.01.009
Xu, Z., Zhou, Y., Li, L., Zhou, R., Xiao, S., Wan, Y., et al. (2008). Genome biology of Actinobacillus pleuropneumoniae JL03, an isolate of serotype 3 prevalent in China. PLoS ONE 3:e1450. doi: 10.1371/journal.pone.0001450
Yamamoto, K., and Ishihama, A. (2006). Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 70, 1688–1695. doi: 10.1271/bbb.60024
Keywords: Actinobacillus pleuropneumoniae, CpxA/CpxR, biofilm, rpoE, virulence
Citation: Li H, Liu F, Peng W, Yan K, Zhao H, Liu T, Cheng H, Chang P, Yuan F, Chen H and Bei W (2018) The CpxA/CpxR Two-Component System Affects Biofilm Formation and Virulence in Actinobacillus pleuropneumoniae. Front. Cell. Infect. Microbiol. 8:72. doi: 10.3389/fcimb.2018.00072
Received: 06 December 2017; Accepted: 27 February 2018;
Published: 20 March 2018.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Skander Hathroubi, University of California, Santa Cruz, United StatesAnn Karen Brassinga, University of Manitoba, Canada
Victor H. Bustamante, Universidad Nacional Autónoma de México, Mexico
Copyright © 2018 Li, Liu, Peng, Yan, Zhao, Liu, Cheng, Chang, Yuan, Chen and Bei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weicheng Bei, YmVpd2NAbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Huan Li1,2,3†
Huan Li1,2,3† Huanchun Chen
Huanchun Chen Weicheng Bei
Weicheng Bei