
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 12 October 2017
Sec. Molecular Bacterial Pathogenesis
Volume 7 - 2017 | https://doi.org/10.3389/fcimb.2017.00442
This article is part of the Research Topic The Twin Threats of Klebsiella pneumoniae Infections: Antimicrobial Drug Resistance and Hypervirulence View all 8 articles
The type VI secretion system (T6SS) is a class of sophisticated cell contact-dependent apparatus with anti-eukaryotic or anti-bacterial function. Klebsiella pneumoniae is one of the most common bacterial pathogens with resistance to the carbapenem antibiotics. However, little is known about the antibacterial T6SS in K. pneumoniae. Using core-component protein searches, we identified a putative T6SS gene cluster on the chromosome of the carbapenemase-producing K. pneumoniae (CRKP) strain HS11286. Intraspecies and interspecies competition assays revealed an antibacterial function of the HS11286 T6SS. The phospholipase Tle1KP was found to be an effector protein that is transferred by T6SS. The overexpression of this effector gene in the periplasm caused severe growth inhibition of Escherichia coli. A sub-inhibitory concentration of β-lactam antibiotics stimulated the expression and secretion of the HS11286 T6SS and enhanced T6SS-dependent killing. It suggested that the antibiotics might be an impact factor for the T6SS secretion and antibacterial activity.
The type VI secretion system (T6SS) is structurally related to the cell-puncturing device of tailed bacteriophages and functions as a contractile injection machinery that perforates eukaryotic and prokaryotic target membranes (Pukatzki et al., 2006; Kapitein and Mogk, 2013; Alteri and Mobley, 2016). T6SS gene cluster contains the core component genes and variable regions that encode effector and immunity proteins (Zhang et al., 2012; see the comprehensive review of Russell et al., 2014). The VipA and VipB proteins form a dynamic tubular sheath that switches between extended and contracted states within the bacterial cytosol (Basler and Mekalanos, 2012). Upon contraction of the cytoplasmic VipA/VipB tube, the hemolysin-coregulated protein (Hcp) capped with valine-glycine repeat protein G (VgrG) were ejected from the bacterium (Bönemann et al., 2009; Leiman et al., 2009; Basler and Mekalanos, 2012). ClpV (a class of ATPases) is required to disassemble the contracted VipA/VipB sheath (Bönemann et al., 2009; Basler and Mekalanos, 2012). Two core proteins, Hcp and VgrG, have frequently been used as markers to investigate the T6SS translocation of secreted effectors (Pukatzki et al., 2006, 2009; Li et al., 2016).
T6SS occur in many pathogenic bacteria and are implicated in virulence in important pathogens, including Pseudomonas aeruginosa (Mougous et al., 2006), Vibrio cholerae (Pukatzki et al., 2006), Edwardsiella tarda (Zheng and Leung, 2007), Burkholderia mallei (Schell et al., 2007), Burkholderia cenocepacia (Aubert et al., 2008), and Aeromonas hydrophila (Suarez et al., 2008). In several cases, the host-pathogen interaction of such “anti-eukaryotic” T6SSs resulted in disruption of the actin cytoskeleton (Pukatzki et al., 2007; Aubert et al., 2008; Suarez et al., 2010). Recently, T6SS has been reported as weaponry in interbacterial warfare (Hood et al., 2010; see the detailed review of Hood et al., 2017). It provides a fitness advantage by delivery of effector proteins to hydrolyze cell walls, cell membranes, and nucleic acids of opponent bacteria (Kapitein and Mogk, 2013; Jiang et al., 2014; Russell et al., 2014). Each of these effectors exhibited toxicity to bacteria and was located adjacent to the genes encoding proteins that conferred immunity to the toxin, thereby preventing self-intoxication (Russell et al., 2011; Zhang et al., 2012). Among these, membrane-targeting effectors were identified and found to have lipase activities (Dong et al., 2013; Russell et al., 2013). They are classified into five families, Tle1 to Tle5 (Russell et al., 2014).
The bacterial species Klebsiella pneumoniae is an increasingly important human pathogen. Dramatic increases in the levels of multidrug resistance associated with this species pose an emerging global problem (Broberg et al., 2014), particularly for carbapenemase-producing K. pneumoniae (CRKP; Tzouvelekis et al., 2012; Cubero et al., 2015). The in silico analysis showed that the putative T6SS gene clusters were present in the complete genome sequence of K. pneumoniae NTUH-K2044, MG78578, and Kp342 (Sarris et al., 2011). Furthermore, a T6SS in the hypervirulent K. pneumoniae strain Kp52.145 with the K2 capsular serotype transfers the effector phospholipase D1 (Tle5, PLD1), which has been reported as a novel virulence factor (Lery et al., 2014). However, little is known about the antibacterial function of T6SS in K. pneumoniae (Hood et al., 2017). A recent finding suggested that the rice pathogenic bacterium Acidovorax avenae strain RS-1 exposure to the β-lactam antibiotics enhanced the virulence of T6SS (Li et al., 2016). Thus, a better understanding of the role of T6SS in CRKP under antibiotic stress is warranted.
We have recently reported the complete genome of K. pneumoniae HS11286, an ST11, carbapenemase (KPC)-2-producing clinical isolate collected in 2011 from the sputum specimen of an inpatient in Shanghai, China (Liu et al., 2012). K. pneumoniae ST11 is a dominant KPC-producing clone in China and is closely related to the worldwide-dominant CRKP clone ST258. In this study, we annotated an entire T6SS gene cluster of K. pneumoniae HS11286. The anti-bacterial function of this T6SS was subsequently investigated with bacterial competition assays and overexpression of the effector gene tle1KP in E. coli.
In this study, sampling collection of patient sputum is a routine hospital procedure. As such, verbal informed consent was obtained from the volunteer. The bacterial sample and data sheet were anonymized. This study protocol, including the verbally informed consent procedure, was approved by the ethics committee of the School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, China.
The strains, plasmids, and primers used in this study are listed in Supplementary Tables 1–3, respectively. K. pneumoniae strain HS11286 was used as the sample strain in all experiments in this study unless otherwise noted in the figure legend. E. coli strain DH10B (streptomycin resistant) and E. coli strain BL21 were used for cloning and expressing the target gene, respectively. Unless stated otherwise, bacteria were grown in Luria-Bertani (LB) broth at 37°C with shaking motion (220 rpm). The antibiotic concentrations used were 100 μg/mL ampicillin, 25 μg/mL chloramphenicol, 100 μg/mL streptomycin, 50 μg/mL kanamycin, 200 μg/mL hygromycin, and 50 μg/mL apramycin. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at a final concentration of 0.5 mM to induce the expression of the T7 promoter.
The nucleotide sequence and annotation of the completely sequenced K. pneumoniae HS11286 chromosome were downloaded from the NCBI RefSeq Project (accession number NC_016845.1). The putative T6SS gene clusters were identified and aligned by using the web-based tool VRprofile (http://bioinfo-mml.sjtu.edu.cn/VRprofile/; Li et al., 2017). Briefly, it performs the Hidden Markov Model (HMM)-based detection of T6SS core components encoded by K. pneumoniae chromosome sequences. The annotated T6SS core components are searched against the VRprofile-specific HMM profiles. Significant hits within a defined gene distance are subsequently grouped to detect putative T6SS gene clusters. The minimum number of the colocalized T6SS core components is five to exclude false positives. Besides, VRprofile also employs BLAST searches to examine the rearranged architecture of a given T6SS gene cluster among multiple K. pneumoniae genomes under investigation. It aids identification of putative effectors and immunity protein genes inserted within or around the T6SS core components.
Bacterial strains were grown overnight to stationary phase and suspended in LB medium. We washed strains with 10 mM MgSO4 and diluted to an optical density at OD600 of 0.2 before mixing at a ratio of 1:1 (attacker: prey). Cells of each strain were spotted on a sterile 0.22 μm filter (Millipore) on LB solid medium (casein tryptone 10 g/L; yeast extract 5 g/L; NaCl 5 g/L; agar 3%). Competitions were incubated for 5 h at 37 °C. Each group of bacteria was harvested, and serial dilutions were grown in selective culture medium containing 200 μg/mL hygromycin. Cells of each strain were grown overnight. The CFU per milliliter of the surviving prey strain was measured by counting single clones. The prey strain was the Δtle1KPΔtli1KP::hph of K. pneumoniae HS11286 carrying the hygromycin-resistance gene (Supplementary Table 1). The fatality rate was calculated as follows:
where CFUcompetition is the difference between the CFU of the surviving prey strain (Δtle1KPΔtli1KP::hph) that was co-incubated with attacker strain ΔvipA and the CFU of the same prey strain when co-incubated with wild type attacker strain. The CFUcontrol is the CFU of the prey strain (Δtle1KPΔtli1KP::hph) without growth competition by any attacker strain.
For gene mutation, the gene was first replaced by an FRT site-flanking hph cassette (Supplementary Figure 1) via lambda red recombination, resulting in an intermediate strain. Then, the hph cassette was eliminated via Flp-FRT recombination to obtain a markerless in-frame indel mutant strain (Supplementary Figure 2). In-frame deletion of a gene was performed as described previously (Hoang et al., 1998; Chaveroche et al., 2000; Bi et al., 2015).
An Hcp antibody was produced by B&M Biotech (Beijing, China). Before immune injection into rabbits, we checked whether the rabbit serum gave an immune response to the whole protein extract of HS11286 at the target protein length to avoid a false-positive response. Rabbits were grown in specific-pathogen-free (SPF) conditions to ensure a non-specific immune response. Upon receiving the antibody, we first tested its specificity by blotting against Hcp protein. The specificity of antibody was shown in Supplementary Figure 3. For western blot, proteins were resolved on a precast 15% SDS/PAGE gel and transferred to a PVDF membrane (Millipore) by electrophoresis. The membrane was then blocked with 5% skimmed milk for 1 h at room temperature and incubated with primary antibodies (MBL Biotech) at 4°C overnight. The membrane was washed three times with TBST buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, pH: 7.6) and incubated with an HRP-conjugated secondary antibody (Pierce) for 1 h at room temperature. Signals were detected using DAB (diaminobenzidine, Dako Denmark A/S) solution.
The wild type and point mutants of tle1KP were sub-cloned into the pET-22b vector containing N-terminal PelB signal peptide sequences. The plasmid pET28a was used to express tle1KP in the cytoplasm. A single colony harboring the expression plasmid was grown in LB medium at 37°C. After overnight culture to the stationary phase and suspended in LB medium, we diluted strains to an optical density at OD600 of 0.2. Then the cells were serially diluted in 10-fold steps and grown in select culture medium with 0.05 mM IPTG and 50 μg/mL ampicillin (or 200 μg/mL kanamycin). The plates were prepared for imaging and plate counts after an additional 20 h incubation at 37°C. The CFU per milliliter of the surviving prey strain was measured by counting single clones (Hu et al., 2014).
Data are shown as mean ± SD. The t-test and one-way ANOVA were used to compare continuous variables. A P < 0.05 was considered statistically significant. Data were analyzed with the R package.
We detected an entire 23-gene T6SS cluster (KPHS_22970-23190) on the chromosome of K. pneumoniae strain HS11286 (Supplementary Table 4). This T6SS consisted of 12 core components (Figure 1). Interestingly, we found a 4.7 kb insertion region that contained KPHS_23060-23110, which is located between the core component genes icmF and vgrG. There is a sequencing mistake in the insertion region. According to Sanger sequencing of the PCR amplification, a homopolymer error within KPHS_23100 was made by 454 sequencing. This error resulted in a frame-shift mutation. After correcting the sequencing mistake, the region from KPHS_23100 to KPHS_23110 was combined into a new protein-coding gene that we named KPHS_23105 (Supplementary Figure 4). This gene coded for a putative effector of T6SS that we called Tle1KP. This effector contains a conserved alpha/beta hydrolase domain (DUF2235) like the reported T6SS effector Tle1 in P. aeruginosa (Hu et al., 2014). A tandem array of four genes (KPHS_23060-23090) were predicted to code for immunity proteins Tli1KP that were related to Tle1KP. These genes exhibited 94% nucleotide sequence identities to each other. In addition, VRprofile typed 254 putative T6SS gene clusters in the 107 completely sequenced K. pneumoniae genomes (including HS11286), of which, 42 of the gene clusters code for homologous effectors of Tle1KP (numbers 1–42 in Supplementary Figure 5).
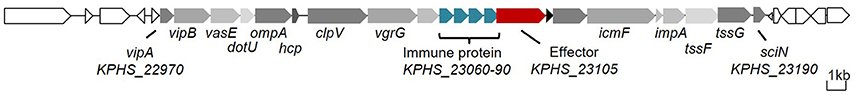
Figure 1. T6SS gene cluster of K. pneumoniae HS11286. The core component genes are marked in gray (Supplementary Table 4), the effector gene in red, and the immunity protein genes in blue.
To characterize the predicted T6SS, we first determined the expression of the T6SS core component and effectors in the logarithmic and stationary phases of K. pneumoniae grown in LB medium. The marker genes that we used were gapA, vipA, clpV, hcp, and tle1KP, which encode the housekeeping protein GapA, a VipA sheath, an ATPase, a hallmark effector and the Tle1KP effector, respectively. The semi-quantitative PCR assays showed that there was no detectable band of the four marker genes (besides the housekeeping gapA) in either the logarithmic or the stationary phase (Figure 2A), suggesting that T6SS was inactive when cultured in LB broth. We also observed that T6SS was inactive in the M9 medium culture conditions (Supplementary Figure 7). Previously, it was reported that T6SS could be induced under certain conditions, such as temperature, pH, and the presence of chitin or with antibiotics (Cerith et al., 2013; Borgeaud et al., 2015). Accordingly, we tested whether the expression of T6SS in strain HS11286 could be induced with a sub-inhibitory concentration of β-lactam antibiotics (meropenem and ceftazidime). As shown in Figure 2B, vipA, clpV, hcp, and tle1KP exhibited strong bright bands in the presence of a sub-inhibitory concentration of meropenem (4 mg/L) or ceftazidime (32 mg/L) compared to control conditions without antibiotics. The determination of sub-inhibitory concentrations of the antibiotics is shown in Supplementary Figure 8.
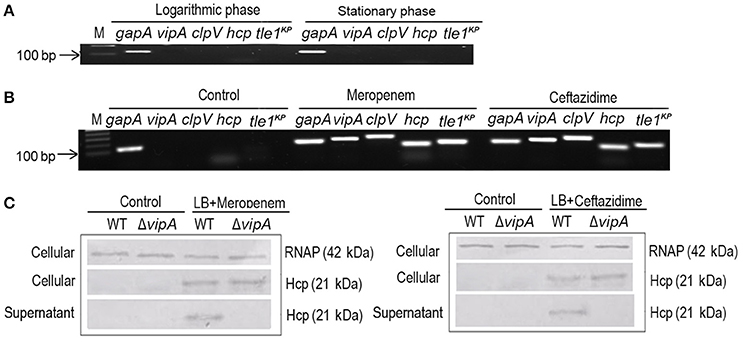
Figure 2. The expression and secretion of T6SS of K. pneumoniae HS11286. (A) Semi-quantitative PCR was used to detect the expression of T6SS, with gapA as the reference gene, and samples were cultured with LB medium. vipA, clpV, hcp, and tle1KP coded for tube sheath, ATPase, hallmark effector, and putative effector, respectively. (B) The expression of T6SS of K. pneumoniae HS11286 cultured with the addition of the sub-inhibitory concentration of 4 mg/L meropenem or 32 mg/L ceftazidime (Supplementary Figure 8). The gels were cropped from the original images available at the Supplementary Figure 6. (C) Immunoblots in the supernatant and cellular fractions of the HS11286 wide-type and ΔvipA mutant using specific antibodies against Hcp and RNAP (cellular control).
To further explore whether the T6SS effectors could be secreted in K. pneumoniae under sub-inhibitory antibiotic stress, we produced a ΔvipA mutant that cannot secrete T6SS effectors due to the lack of the VipA sheath element. Then, we examined Hcp, one of the reported T6SS effectors, in both the cellular and supernatant fractions by immunoblotting. As shown in Figure 2C, Hcp was absent in both the cellular and supernatant fractions of the wild type and ΔvipA mutant when cultured with LB broth. In contrast, Hcp was easily detected in the cellular fraction of both the wild type and ΔvipA mutant under β-lactam antibiotic stimulation, indicating that the T6SS of HS11286 could be produced upon exposure to a sub-inhibitory concentration of β-lactam antibiotics. We also observed that Hcp was present only in the supernatant of the wild type strain but not in ΔvipA mutant (Figure 2C), suggesting that the secretory function of T6SS occurred under antibiotic stimulation. Also, we identified the specificity of antibiotics by using apramycin (Supplementary Figure 9) and got the similar results. It was to say that not only β-lactam antibiotic can induce the secretion of T6SS.
Bioinformatics analysis predicted that Tle1KP in HS11286 is an effector of T6SS. To test this, we examined Tle1KP expression in the supernatant of both the wild type and the T6SS-apparatus-deletion mutant (ΔvipA) treated with ceftazidime. The results showed that Tle1KP was present in both the cytosol and supernatant fractions of the wild type strain with antibiotic treatment but was absent in the supernatant of the ΔvipA mutant strain, indicating that Tle1KP is a T6SS effector (Figure 3A).
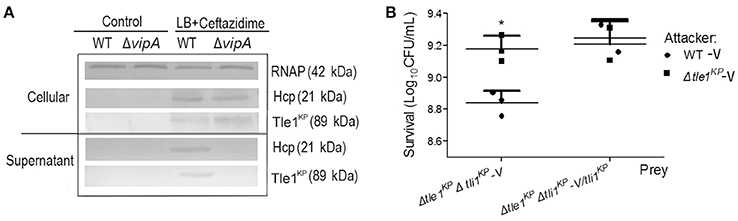
Figure 3. The identification of effector and immune protein pair. (A) Immunoblots in the supernatant and cellular fractions of the HS11286 wild-type and ΔvipA mutant using specific antibodies against Tle1KP, Hcp, and RNAP (cellular control). Samples acquired in liquid culture with 32 mg/L ceftazidime (Supplementary Figure 8). The westernblot membranes were cropped from the original images available at the Supplementary Figure 10. (B) Bacterial competition assays. Deletion mutant strain (Δtle1KPΔtli1KP-V) and complementation of Δtle1KPΔtli1KP mutant with tli1KP (Δtle1KPΔtli1KP-V/tli1KP) as the prey strains following co-culture with the attacking strains (the WT-V and Δtle1KP -V mutant, Supplementary Table 1). All strains carried the vector control plasmid pBAD33-Apra. Each group has three replications. *P < 0.05.
The toxicity of Tle1KP was predicted to be neutralized by the upstream cognate immune protein Tli1KP. T6SS-dependent killing (dueling) was performed to confirm this prediction, which uses co-incubation of attacker and prey bacterial strains to show whether a predicted immune protein will protect the prey from killing. The attacker and prey strains are co-cultured on agar, and the survival strains are quantified. The double deletion mutant lacking the effector and immune protein pair (strain Δtle1KPΔtli1KP-V) was used as the prey that could be susceptible to T6SS. The wild type and Δtle1KP-V mutant were the attackers. As shown in Figure 3B, the WT-V (Supplementary Table 1) caused a statistically significant impairment/killing of the Δtle1KPΔtli1KP-V mutant, but the Δtle1KP-V mutant did not. Additionally, when the prey (Δtle1KPΔtli1KP-V) was complemented with tli1KP, the inhibition by the attacker WT-V was same to the attacker Δtle1KP-V. These results show that the Tli1KP protein counteracts the toxicity of effector Tle1KP. Hence, Tle1KP and Tli1KP represent a cognate effector and immunity protein pair.
We performed the competition assays to investigate the antibacterial activity of the K. pneumoniae HS11286 T6SS. The wild type, the T6SS-apparatus-deletion mutant (ΔvipA), and the transferred effector deletion mutant (Δtle1KP) were employed as the attacker strains. The effector and immune protein gene double deletion mutant strain (Δtle1KPΔtli1KP) served as the prey strain. As shown in Figure 4A, the killing activity of the deletion mutant lacking the T6SS apparatus or the effector was lower than that of the wild type (The complementary of Tle1KP in Supplementary Figure 11). The survival of the prey was greater with the ΔvipA mutant than with Δtle1KP, which indicated the T6SS might contain another toxic effector. Also, the fatality rate of the antibiotics-free group (the control) is nearly two-fold less than that of the meropenem or ceftazidime group (Figure 4B). It indicated that the T6SS killing activated by cell-to-cell contact might become stronger under antibiotics stress.
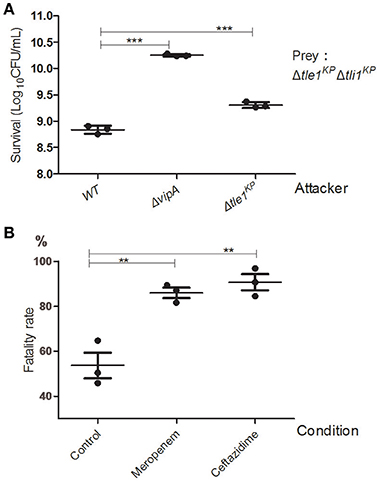
Figure 4. Anti-bacterial activity of the HS11286 T6SS. (A) Intraspecies competition assays. Different strains were individually mixed with Δtle1KPΔtli1KP on LB medium and then viability on selective medium was determined. Δtle1KPΔtli1KP was the prey. (B) T6SS-dependent killing activity under antibiotic stress. The attacker strains (WT and ΔvipA) were individually mixed with the prey strain (Δtle1KPΔtli1KP) on different antibiotic media and then titered for viable counts on selective media. The fatality rate was calculated based on viable cell count of the prey with or without competence. CFUcompetition/CFUcontrol. The sub-inhibitory concentrations of meropenem (4 mg/L) and ceftazidime (32 mg/L). Control was added (ddH2O). Each group in (A,B) has three replications. **P < 0.01; ***P < 0.001.
We also confirmed the T6SS-mediated interspecies competition between K. pneumoniae and E. coli. We utilized E. coli DH10B as the prey strain. The HS11286 wild type, the ΔvipA mutant, and the Δtle1KP mutant were employed as the attacker strains. The attacker strains were individually mixed with the prey and grown on LB agar, and the prey survival was quantified after co-incubation. As expected, the wild type exhibited the strongest killing activity compared to the ΔvipA mutant and the Δtle1KP mutant (Supplementary Figure 12).
The Tle1 family effector member from P. aeruginosa (Tle1PA) can hydrolyze cell membranes (Hu et al., 2014). To confirm the membrane-targeting activity of Tle1KP, this effector was produced in E. coli strain BL21 and designed to target to the periplasm (Hu et al., 2014). As expected, the presence of Tle1KP in the periplasm inhibited the growth of E. coli with significant difference (Figure 5A, P = 0.0004), suggesting that Tle1KP is an active membrane-targeting phospholipase effector in K. pneumoniae. Meanwhile, no decrease in viability was observed after overexpressing tle1KP in the cytoplasm (Figure 5A), suggesting that Tle1KP is not toxic in the cytoplasm.
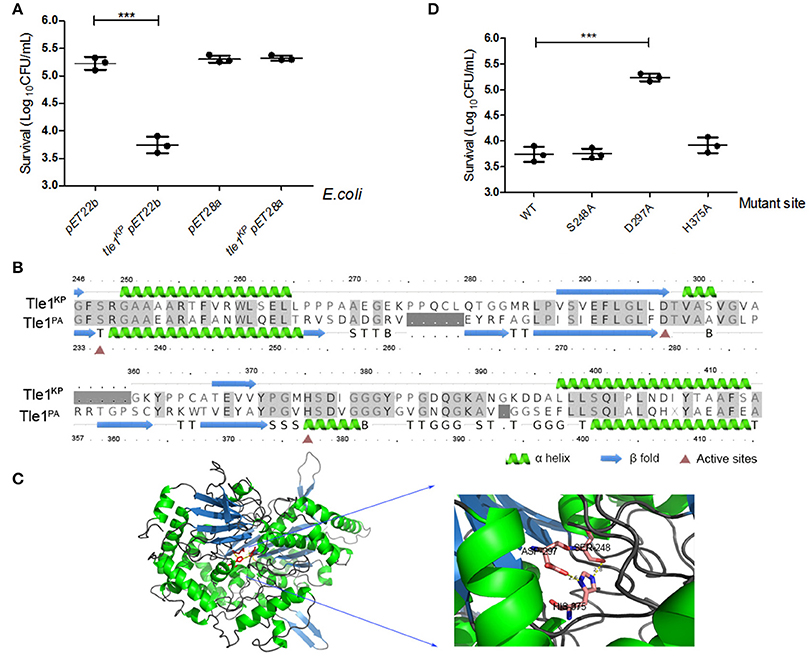
Figure 5. Characterization of the T6SS effector Tle1KP. (A) The growth of E. coli BL21 containing tle1KP pET22b and tle1KP pET28a on LB agar induced by IPTG. BL21 with an empty vector was included as a control. pET22b contains a periplasm-targeting signal. Ten-fold serial dilutions of overnight culture were grown on LA media plates with selective culture medium. (B) Amino acid sequence alignment for Tle1KP and the Tle1PA of P. aeruginosa (Hu et al., 2014). Red triangles highlight the characterized active sites of phospholipase Tle1PA. (C) The 3D structure and active site of Tle1KP predicted by Phrye2 based on the reported structure of Tle1PA (PDB entry 405p). (D) Identification of the active sites of Tle1KP. According to the active sites shown in (C), E. coli BL21 harboring pET22b expressing wild type and its point mutants in the periplasm were grown in agar plates. The cells were prepared with serial 10-fold dilutions on LA medium and induced by IPTG. Each group in (A,D) has three replications. The CFU per milliliter of the surviving prey strain was measured by counting single clones. ***P < 0.001.
In addition, the structure and active domain of Tle1PA have been reported recently (Hu et al., 2014). The protein sequence alignment between Tle1KP and Tle1PA exhibited 43% identities. According to homology modeling, Tle1KP also contains a catalytic triad (S248-D297-H375), like the one that has been characterized in Tle1PA (Figures 5B,C). Thus, we constructed three tle1KP mutant strains (S248A, D297A, and H375A) by site-directed mutagenesis for a toxicity assay, as described above. Only the D297A mutant showed an impaired antibacterial effect (Figure 5D, P = 0.0003), suggesting that D297 is indispensable in catalytic activity. In Tle1PA, site-directed mutations that alter each of these three amino acids lost activity, which implies that Tle1KP and Tle1PA have different active centers that may have different functions.
In this study, we identified and characterized a new T6SS with the antibacterial effector Tle1KP in K. pneumoniae HS11286. This T6SS is silent in basic medium without an induction signal. It follows that expression and assembly of this structure would be tightly regulated. In some cases, there is evidence for transcriptional regulation of the T6SS via quorum sensing (Gueguen et al., 2013; Salomon et al., 2013), biofilm formation (Aubert et al., 2008; Hood et al., 2010), iron limitation (Brunet et al., 2011; Chakraborty et al., 2011), and temperature variation (Salomon et al., 2013), which may react to stress responses (Gueguen et al., 2013). In P. aeruginosa, kanamycin stimulated T6SS expression but did not affect the cognate effector secretion (Cerith et al., 2013). But in K. pneumoniae HS11286, a sub-inhibitory concentration of the β-lactam antibiotics could induce both T6SS expression and secretion of the effectors Hcp and Tle1KP.
Furthermore, antibiotics also enhanced the antibacterial activity of the HS11286 T6SS. We propose that antibiotics induce the expression and secretion of T6SS and might make the attacker T6SS+ strains more aggressive in the competition for growth. A previous report (Li et al., 2016) has shown a similar phenomenon in Acidovorax avenae subsp. avenae (Aaa) strain RS-1. Exposure of RS-1 to ampicillin alters the virulence, colonization capacity, composition of extracellular polymeric substances and secretion of the T6SS effector Hcp. Thus, for CRKP HS11286 in the clinical practice, antibiotics may not inhibit its proliferation but instead may induce the activity of T6SS, making HS11286 more aggressive (Supplementary Figure 13). Under antibiotic stress, CRKP HS1186 thus dominates the growth superiority comparing to the T6SS−/multidrug resistant strain.
There are five known Tle family T6SS effectors (Tle1-5; Lu et al., 2014), and the newly identified effector encoded within the T6SS gene cluster of K. pneumoniae HS11286 belongs to Tle1. Tle1-Tle4 families contain a conserved G-X-S-X-G motif, and Tle5 features a conserved H-X-K-X-X-X-X-D motif (Durand et al., 2014; Russell et al., 2014). Members of Tle1, Tle2, and Tle5 had been experimentally confirmed to possess phospholipase A2, A1, and D activities, respectively (Durand et al., 2014). After being injected into their periplasmic space by the T6SS, Tle1 of Burkholderia thailandensis and Tle2 of V. cholerae can hydrolyze the membrane phospholipids of neighboring cells, causing an increase in cellular permeability (Russell et al., 2013; Hu et al., 2014). According to homologous model, Tle1KP of K. pneumoniae HS11286 contains a Tle1 family conserved motif G-X-S-X-G and exhibits periplasmic activity. But the active motif of Tle1KP may not be the same as Tle1PA, as shown in our point mutations results. And the amino acid sequence identities between reported Tle1PA and Tle1KP were just 43%, also indicating that they might have different active amino acids. Similarly, Tle4 of P. aeruginosa possesses an unusual pentapeptide motif T-X-S-X-G (Lu et al., 2014), different from the canonical hydrolases with the (G-X-S-X-G) motif (Durand et al., 2014; Russell et al., 2014).
In conclusion, the anti-bacterial function of the T6SS of K. pneumoniae HS11286 was confirmed with the intraspecies and interspecies competition assays. Overexpression of the effector gene tle1KP in the periplasm caused severe growth retardation of E. coli. The results also indicated that the antibiotics could be an important factor for the T6SS secretion and antibacterial activity. To our knowledge, this is the first report about the antibacterial function of T6SS in the K. pneumoniae. This information might deepen our understanding of the T6SS-carrying CRKP under antibiotic treatments.
HO and YY conceived and designed the experiments. LL and XL performed the experiments. LL, XL, MY, JL, HO, and YY analyzed the data. MY, JL, YY, HO, and ZD contributed reagents/materials/analysis tools. LL, YY and HO wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by grants from the National Natural Science Foundation of China (31670074 and 31270173), and the 973 program, Ministry of Science and Technology, China (2015CB554200).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2017.00442/full#supplementary-material
Alteri, C. J., and Mobley, H. L. T. (2016). The versatile type VI secretion system. Microbiol. Spectrum. 2:15. doi: 10.1128/microbiolspec.VMBF-0026-2015
Aubert, D. F., Flannagan, R. S., and Valvano, M. A. (2008). A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 76, 1979–1991. doi: 10.1128/IAI.01338-07
Basler, M., and Mekalanos, J. J. (2012). Type 6 secretion dynamics within and between bacterial cells. Science 337:815. doi: 10.1126/science.1222901
Bi, D. X., Jiang, X. F., Sheng, Z. K., Ngmenterebo, D., Tai, C., Wang, M., et al. (2015). Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J. Antimicrob. Chemother. 70, 2770–2774. doi: 10.1093/jac/dkv204
Bönemann, G., Pietrosiuk, A., Diemand, A., Zentgraf, H., and Mogk, A. (2009). Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325. doi: 10.1038/emboj.2008.269
Borgeaud, S., Metzger, L. C., Scrignari, T., and Blokesch, M. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:4. doi: 10.1126/science.1260064
Broberg, C. A., Palacios, M., and Miller, V. L. (2014). Klebsiella: a long way to go towards understanding this enigmatic jet-setter. F1000Prime Rep. 6:64. doi: 10.12703/P6-64
Brunet, Y. R., Bernard, C. S., Gavioli, M., Lloubes, R., and Cascales, E. (2011). An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 7:e1002205. doi: 10.1371/journal.pgen.1002205
Cerith, J., Luke, A., Jack, H., Hemantha, K., and Alain, F. (2013). Subinhibitory concentration of kanamycin induces the Pseudomonas aeruginosa type VI secretion system. PLoS ONE 8:e81132. doi: 10.1371/journal.pone.0081132
Chakraborty, S., Sivaraman, J., Leung, K. Y., and Mok, Y. K. (2011). Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J. Biol. Chem. 286, 39417–39430. doi: 10.1074/jbc.M111.295188
Chaveroche, M.-K., Ghigo, J.-M., and d'Enfert, C. (2000). A rapidmethod for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:e97. doi: 10.1093/nar/28.22.e97
Cubero, M., Cuervo, G., Dominguez, M. A., Tubau, F., Marti, S., Sevillano, E., et al. (2015). Carbapenem-resistant and carbapenem-susceptible isogenic isolates of Klebsiella pneumoniae ST101 causing infection in a tertiary hospital. BMC Microbiol. 15:177. doi: 10.1186/s12866-015-0510-9
Dong, T. G., Ho, B. T., Yoder-Himes, D. R., and Mekalanos, J. J. (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628. doi: 10.1073/pnas.1222783110
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Gueguen, E., Durand, E., Zhang, X. Y., d'Amalric, Q., Journet, L., and Cascales, E. (2013). Expression of a Yersinia pseudotuberculosis type VI secretion system is responsive to envelope stresses through the OmpR transcriptional activator. PLoS ONE 8:e66615. doi: 10.1371/journal.pone.0066615
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. T., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Hood, R. D., Peterson, S. B., and Mougous, J. D. (2017). From striking out to striking gold: discovering that Type VI secretion targets bacteria. Cell Host Microbe 21, 286–289. doi: 10.1016/j.chom.2017.02.001
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Hu, H., Zhang, H., Gao, Z., Wang, D., Liu, G., Xu, J., et al. (2014). Structure of the type VI secretion phospholipase effector Tle1 provides insight into its hydrolysis and membrane targeting. Acta Crystallogr. D Biol. Crystallogr. 70(Pt 8), 2175–2185. doi: 10.1107/S1399004714012899
Jiang, F., Waterfield, N. R., Yang, J., Yang, G., and Jin, Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15, 600–610. doi: 10.1016/j.chom.2014.04.010
Kapitein, N., and Mogk, A. (2013). Deadly syringes: type VI secretion system activities in pathogenicity and interbacterial competition. Curr. Opin. Microbiol. 16, 52–58. doi: 10.1016/j.mib.2012.11.009
Leiman, P. G., Basler, M., Ramagopal, U. A., Bonanno, J. B., Sauder, J. M., Pukatzki, S., et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159. doi: 10.1073/pnas.0813360106
Lery, L. M., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., et al. (2014). Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 12:41. doi: 10.1186/1741-7007-12-41
Li, B., Ge, M., Zhang, Y., Wang, L., Ibrahim, M., Wang, Y., et al. (2016). New insights into virulence mechanisms of rice pathogen Acidovorax avenae subsp. avenae strain RS-1 following exposure to ss-lactam antibiotics. Sci. Rep. 6:22241. doi: 10.1038/srep22241
Li, J., Tai, C., Deng, Z., Zhong, W., He, Y., and Ou, H.-Y. (2017). VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief. Bioinformatics. doi: 10.1093/bib/bbw141. [Epub ahead of print].
Liu, P., Li, P., Jiang, X., Bi, D., Xie, Y., Tai, C., et al. (2012). Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 194, 1841–1842. doi: 10.1128/JB.00043-12
Lu, D., Zheng, Y., Liao, N., Wei, L., Xu, B., Liu, X., et al. (2014). The structural basis of the Tle4-Tli4 complex reveals the self-protection mechanism of H2-T6SS in Pseudomonas aeruginosa. Acta Crystallogr. D Biol. Crystallogr. 70(Pt 12), 3233–3243. doi: 10.1107/S1399004714023967
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513. doi: 10.1073/pnas.0706532104
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Pukatzki, S., McAuley, S. B., and Miyata, S. T. (2009). The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17. doi: 10.1016/j.mib.2008.11.010
Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W., and Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244
Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Salomon, D., Gonzalez, H., Updegraff, B. L., and Orth, K. (2013). Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 8:e61086. doi: 10.1371/journal.pone.0061086
Sarris, P. F., Zoumadakis, C., Panopoulos, N. J., and Scoulica, E. V. (2011). Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect. Genet. Evol. 11, 157–166. doi: 10.1016/j.meegid.2010.09.006
Schell, M. A., Ulrich, R. L., Ribot, W. J., Brueggemann, E. E., Hines, H. B., Chen, D., et al. (2007). Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64, 1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x
Suarez, G., Sierra, J. C., Erova, T. E., Sha, J., Horneman, A. J., and Chopra, A. K. (2010). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168. doi: 10.1128/JB.01260-09
Suarez, G., Sierra, J. C., Sha, J., Wang, S., Erova, T. E., Fadl, A. A., et al. (2008). Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44, 344–361. doi: 10.1016/j.micpath.2007.10.005
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Zhang, D., de Souza, R. F., Anantharaman, V., Iyer, L. M., and Aravind, L. (2012). Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7:18. doi: 10.1186/1745-6150-7-18
Keywords: type VI secretion system, Klebsiella pneumoniae, antibacterial, phospholipase, antibiotic stress
Citation: Liu L, Ye M, Li X, Li J, Deng Z, Yao Y-F and Ou H-Y (2017) Identification and Characterization of an Antibacterial Type VI Secretion System in the Carbapenem-Resistant Strain Klebsiella pneumoniae HS11286. Front. Cell. Infect. Microbiol. 7:442. doi: 10.3389/fcimb.2017.00442
Received: 21 June 2017; Accepted: 28 September 2017;
Published: 12 October 2017.
Edited by:
Yunn Hwen Gan, National University of Singapore, SingaporeReviewed by:
Andrés Esteban Marcoleta, University of Chile, ChileCopyright © 2017 Liu, Ye, Li, Li, Deng, Yao and Ou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Yu Ou, aHlvdUBzanR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.