
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 06 September 2017
Sec. Molecular Bacterial Pathogenesis
Volume 7 - 2017 | https://doi.org/10.3389/fcimb.2017.00393
The enterobacterium, Klebsiella pneumoniae invades the intestinal epithelium of humans by interfering with multiple host cell response. To uncover a system-level overview of host response during infection, we analyzed the global dynamics of protein profiling in Caenorhabditis elegans using quantitative proteomics approach. Comparison of protein samples of nematodes exposed to K. pneumoniae for 12, 24, and 36 h by 2DE revealed several changes in host proteome. A total of 266 host-encoded proteins were identified by 2DE MALDI-MS/MS and LC-MS/MS and the interacting partners of the identified proteins were predicted by STRING 10.0 analysis. In order to understand the interacting partners of regulatory proteins with similar or close pI ranges, a liquid IEF was performed and the isolated fractions containing proteins were identified by LC-MS/MS. Functional bioinformatics analysis on identified proteins deciphered that they were mostly related to the metabolism, dauer formation, apoptosis, endocytosis, signal transduction, translation, developmental, and reproduction process. Gene enrichment analysis suggested that the metabolic process as the most overrepresented pathway regulated against K. pneumoniae infection. The dauer-like formation in infected C. elegans along with intestinal atrophy and ROS during the physiological analysis indicated that the regulation of metabolic pathway is probably through the involvement of mTOR. Immunoblot analysis supported the above notion that the K. pneumoniae infection induced protein mis-folding in host by involving PI3Kinase/AKT-1/mTOR mediated pathway. Furthermore, the susceptibility of pdi-2, akt-1, and mTOR C. elegans mutants confirmed the role and involvement of PI3K/AKT/mTOR pathway in mediating protein mis-folding which appear to be translating the vulnerability of host defense toward K. pneumoniae infection.
Infection by Klebsiella pneumoniae is one of the most significant problems facing human health. Although several necessary measures have been taken to control the infections caused by K. pneumoniae, the emergence of new drug-resistance superbugs poses a great level of threat. In spite of the risk associated with K. pneumoniae to human health, it has been investigated to a minimal extent. Therefore, insightful knowledge on how pathogen alters the host cellular immune defense using a eukaryotic model organism may potentially open a new avenue for therapeutic interventions. An appropriate in vivo model system is supposed to not only satisfy the scientific community, but also the research ethics (Zak and O'Reilly, 1993). Moreover, it has also been considered that not all outcomes of pre-clinical studies using model animals could be effectively extrapolated to humans (Fuchs et al., 2009; Evans et al., 2010). Hitherto, there are several mammalian models with diversified physiological, anatomical, molecular, and genetic characteristics have comprehensively been utilized to understand host-pathogen interactions as well as in pre-clinical assessment of drugs (Means and Aballay, 2011). Of late, numerous reports have argued the extensive use of several alternative models such as insects, small vertebrates, and nematodes for studying bacterial infections (Kurz and Ewbank, 2007; Lopez Hernandez et al., 2015).
In the midst, Caenorhabditis elegans, a nematode model provides an excellent model to explore the cellular and molecular impacts in host by K. pneumoniae, particularly since we established it as a better surrogate host for K. pneumoniae infection (Kamaladevi and Balamurugan, 2015). Several studies on host-pathogen interaction using C. elegans against various bacterial pathogens provided insight knowledge on cellular and molecular immune signaling pathways that are conserved to mammals during infections (Kim, 2008; Marsh and May, 2012). Albeit, C. elegans has been extensively employed in forward and reverse genetic approaches to derive wide knowledge at the transcriptome level, the prominence of proteomic changes is still remains at its preliminary stage. Hence, exploring C. elegans with different pathogens complement abundant information to the existing transcriptomics data that will increases our understanding on host response to infections. For the past few decades, several key findings were discovered in C. elegans with high relevance to mammals (Grompone et al., 2012). Also, the shared common mechanisms between nematodes and mammals have been validated by using relevant in vivo model organism like C. elegans complemented with advanced tools in proteomics required addressing the host-pathogen interaction mediated complications.
The innate immune system is considered as a first line of defense against invading pathogens. Generally, it is an evolutionarily ancient and phylogenetically conserved from nematodes to mammals. Stimulation of innate immune system orchestrates several signaling pathways involved in defending the invading pathogens and protects the host system against infections (Akira, 2009). This response required an organized coordination between genomic and proteomic machineries. In C. elegans, host response against infection has been resulted in activation of complex network of pathways involved in metabolism, immunity, stress pathway, and aging (Singh and Aballay, 2006). This indicated that host defense is not only a single process but it is a process of complicated networks. Hence, analyzing the global level response either at the transcriptional or translational level will advance our understanding in host-pathogen interactions.
Hitherto several researches applied the combination of both transcriptional and translational tools to understand the host-pathogen interaction using C. elegans. Bogaerts et al., utilized C. elegans to decipher the defense mechanism of host at their protein level against Staphylococcus aureus (Bogaerts et al., 2010a) and Aeromonas hydrophila (Bogaerts et al., 2010b). The kinetic analysis of C. elegans proteome by using 2D-DIGE at different time-points revealed involvement of citric acid cycle and chaperon molecules in host defense against Gram positive bacteria S. aureus. Whereas, these regulatory proteins were regulated in an opposite manner in C. elegans infected with Gram negative bacteria A. hydrophila. Comparing the outcome of both studies they concluded that the host response is more specific to pathogens. Though data on proteomics advanced our understanding to next level, the connection between the regulation of proteins and genes against any infection process was not well-understood. Later in 2015, the connections between the transcriptional and translational regulations of C-type lectins and AMP-activated protein kinase were deciphered through genomics and proteomics analysis in C. elegans against Bacillus thuringiensis infection (Yang et al., 2015). Since characterizing the transcriptional and translational relationship is critical in understanding the host defense during host-pathogen interaction, we analyze the regulation of host regulatory proteins at their transcriptional and translational levels during K. pneumoniae infection. Previously, our group reported the role of few regulatory proteins and signaling pathway(s) involved in host defense of C. elegans against Vibrio alginolyticus (Durai et al., 2014), Pseudomonas aeruginosa (Balasubramanian et al., 2016), and Protease mirabilis (Jebamercy et al., 2016) using both proteomics and transcriptional tools.
With this background, the present study aimed to investigate the system-level overview of changes in host proteome using C. elegans against K. pneumoniae infection. Here we employed a conventional 2DE, liquid phase IEF, MALDI-MS/MS, nano LC-MS/MS, bioinformatics analysis along with molecular analysis (Western blot and real-time) to decipher the host defense in C. elegans against K. pneumoniae infection.
K. pneumoniae was obtained from American Type Culture Condition (ATCC). Escherichia coli OP50 was provided by Caenorhabditis Genetic Center (CGC), MN, USA. Both the cultures were inoculated in Luria Bertani medium and incubated at 37°C.
The C. elegans wild type and mutant strains were originally obtained from the CGC, MN, USA and grown on a solid nematode growth medium (NGM) plates containing E. coli OP50 as a standard food source. The C. elegans strains used in the present study were Bristol N2 (wild-type), VC2312 (let-363) mTOR mutant, BQ1 (akt-1), and VC858 (pdi-2). The age-synchronized L4 C. elegans were used in all bioassays. To obtain L4 staged C. elegans, the gravid hermaphrodites containing eggs inside the uterus were treated with commercial bleach solution containing 5 M potassium hydroxide in a ratio of 1:1 and the resulting solution containing eggs were transferred to the fresh NGM plates seeded with E. coli OP50. The plates containing eggs were incubated at 20°C for favoring the hatching. Thus, obtained stage-synchronized C. elegans were allowed to reach the L4 stage and used in all bioassays.
The age-synchronized L4 stage C. elegans were infected with K. pneumoniae for different time points (12, 24, and 36 h). Here, the nematodes fed on E. coli OP50 at respective time points were served as controls. After infection, the nematodes were washed thoroughly with sterile M9 medium to remove the bacterial contaminants. The samples were then flash frozen in lysis buffer [7 M urea, 2 M thiourea, 4% CHAPS, and 30 mM Tris-HCl, pH 8.5, and protease inhibitor cocktail (Sigma)] and stored at −86°C. When required, the samples were homogenized on ice for 1 min with 5 s pulses for every 5 s by employing sonicator. The insoluble cellular debris was separated by centrifugation at 12,000 rpm for 15 min at 4°C. The protein concentrations were determined with a Bradford assay Kit (Bio-Rad) using bovine serum albumin as a standard.
The 2D-gel electrophoresis was performed according to Balasubramanian et al. (2016) with minor modifications. A concentration of 1 mg of protein was taken in the volume range of (50–75 μl). The samples were processed to remove the non-specific biological contaminants such as lipids, nucleic acids, and salts using 2D cleanup kit (GE healthcare). The protein samples were dissolved in sample buffer containing urea and thiourea. Prior to 2DE, the sample containing total protein was loaded onto an immobilized pH gradient (IPG) strips (24 cm, pH 3–10 NL, GE Healthcare) and rehydrated for 12 h at room temperature. The rehydrated strips were subjected to isoelectric focusing (IEF) using Ettan™ IPGphor 3 isoelectric focusing system and accessories purchased from GE Healthcare. Then the strips were equilibrated in SDS equilibration buffer containing DTT (10 mg/ml) and iodoacetamide (25 mg/ml) for first and second equilibration respectively for 20 min each. The second dimensional separation of proteins was performed using 12% denaturant SDS polyacrylamide gels. After electrophoresis, the gels were stained with Coomassie Brilliant Blue (CBB) G-250.
The protein separated in SDS gels were digitized using Gel scanner-III and the scanned gel images were analyzed with the ImageMaster 2D platinum version 7 software (GE Healthcare). Briefly, after automatic spot detection, the manual editing of images like images like adding, splitting and removing of spots were done to increase the recognition of majority of spots. To match the spots across gels in each replicates, a class and match set were obtained with the available gel images. Among them, based on the gel and spot quality one gel was selected as the master gel for each time points, against which all other respective gels were matched. Spots, found in a match set member but absent in the master gel were added manually to the master gel. Then the automatic matching of spots on each gel was performed and matching across gels was manually verified. For each, match set, each gel was normalized to minimize the variability due to slight difference in protein load per gel, staining efficiency and image capture. This normalization was obtained by dividing the raw intensity of each spot in a gel by the total intensity of all spots in that gel that was included in the master gel. And all the analyses were performed on normalized quantities. The normalized values of all proteins were exported to in-built statistical tools for statistical analysis. The protein spots which had significant changes of >1.5-fold and p < 0.05 were considered as differentially regulated proteins.
The differentially regulated proteins were identified by following the procedure of Durai et al. (2014) with minor modifications. In brief, the protein spots of interest were manually exercised from the SDS PAGE gel with end-remove pipet tips to accommodate the various diameters of spots. The gel pieces in the microtube was destained and dehydrated with 30% acetonitrile (ACN) and 100 mM ammonium bicarbonate in 1:1 ratio. After washing, the wash solution was removed and the gel pieces were lyophilized. The lyophilized gel pieces were further dehydrated thrice with 50 μl of ACN and subsequently reswollen with 50 μl of 50 mM ammonium bicarbonate for 10 min. Again the protein spots were rehydrated with Acetonitrile and lyophilized. The protein containing gel spots were subjected to reduction and alkylation by adding 10 mM DTT and 50 mM iodoacetamide, respectively. Subsequently, the traces of DTT and iodoacetamide were removed from the gel spots by dehydrating and lyophilization. Then the dried proteins spots were rehydrated in a protease solution containing 2.5 ng/μl of trypsin (Sigma) solution and incubated in ice for 45 min. The mixture was incubated at 37°C for trypsinization. After 16 h of enzymatic digestion and vortexing for 2 min at high speed, the peptides were extracted by adding extraction buffer containing 50 mM ammonium bicarbonate, 50% can, and 0.1% formic acid. The step was repeated twice to increase the yield. The extracts were pooled together and concentrated by using speed Vac. The peptides were dissolved in MS grade water and were desalted using ZipTipC 18 pipet tips (Millipore). About 0.5 μl of α-Cyano-4-hydroxycinnamic acid was mixed with equal volume of desalted samples and allowed to dry for 4–6 h. The MALDI TOF/TOF analysis was performed using MALDI-TOF/TOF analyzer (AXIMA Performance, SHIMADZU BIOTECH) in a positive reflectron ion mode. With peptide mass fingerprint (PMF) and MALDI TOF/TOF, the proteins were identified against the all entries of C. elegans database using the MASCOT (Matrix Science Version 2.2) as a search engine. During search, the carbamidomethylation and oxidation was set as fixed and variable modifications, respectively. In the MASCOT search, the mass tolerance of 1.2 Da with one missed cleavage per peptide was allowed in all searches. Proteins with constant hit against different mass tolerance and significant scores were considered. All the proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al., 2016) partner database with the dataset identifier PXD007151.
The protein-protein interaction among the differentially regulated proteins identified at different time points were explored and the interacting-network map was created by using STRING 10.0 with a high confidence score of 0.7. The interacting partners were identified for their functions and annotated manually (Schmutz et al., 2013). The Gene ontology (GO) and the functional enrichment was performed using DAVID and KEGG pathway analysis. In order to eliminate the redundancy of proteins and to reduce the number of terms to a smaller meaningful set, the list of identified proteins were analyzed by ReviGO tool (Supek et al., 2013; Larance et al., 2015).
The proteins isolated from C. elegans exposed to K. pneumoniae were separated by liquid-phase isoelectric focusing (IEF) with the MicroRotofor cell (Bio-Rad Laborotaries), according to their isoelectric points (pI). Approximately 2 mg of protein samples were mixed with IEF buffer [7 M urea, 2 M thiourea, 4% CHAPS and 0.24% Triton X100, 5% glycerol, and 1.6% ampholytes (pH 3–10)]. The focusing chamber of MicroRotofor cell was loaded with 2.5 ml of samples and the electrode cells were filled with appropriate electrolytes. The IEF was performed by applying a constant power of 1 W at room temperature. After 3 h of focusing, protein fractions were separated into different compartments (~200 μl) and were collected immediately to avoid the diffusion of separated fractions. For identification purpose, the collected fractions were trypsinized and analyzed using nano-RPLC coupled with an Orbitrap Elite Mass spectrometer (Thermo Scientific, USA). The digested peptides were dissolved in 2% ACN containing 0.1% formic acid and loaded to a C18 guard column (5 μm, 100 μm × 2 cm) (Thermo Scientific). The injected piptides were allowed to pass through a linear solvent system of 5–100% ACN for 80 min with a flow-rate of 300 nl/min and given voltage of 1.9 kV. The final MS data acquired were over the mass range of m/z 350–4,000 Da and visualized using Xcalibur software (version 2.2.SP1.48) (Thermo Scientific USA) (Ananthi et al., 2011). The data analysis was done using Proteome Discover software v. 1.4 based on sequest algorithm using a database downloaded from Uniprot. During search, the carbamidomethylation of cysteine was considered as fixed modification and methionine oxidation, N-terminal acetylation and phosphorylation (S, T, Y) as variable modifications. Here, the precursor mass tolerance of 10 ppm, fragment mass tolerance of 0.5 Da with 2 missed cleavages was allowed in protein identification.
For blotting, nematodes exposed to K. pneumoniae for different time-points (12, 24, and 36 h) were washed thoroughly with M9 buffer and homogenized in ice cold Phosphate buffered saline containing a protease inhibitor cocktail. The soluble fraction of lysates was separated from the mixture by centrifugation at 12, 000 rpm for 5 min at 4°C. The final protein concentration of the samples was determined with Bradford solution (Bio-Rad). For each experimental run, equal concentration of proteins were taken and mixed with a SDS-sample loading buffer. The mixture was boiled at 100°C for 2–3 min. After heat-denaturation, the samples were subjected to SDS-polyacrylamide gel electrophoresis, followed by electrophoretic transfer onto the polyvinylidene fluoride (PVDF) membranes (Amersham Biosciences). Then the membranes were transferred to the primary antibodies solution (1:2,000) and incubated for 6 h at 4°C. After required time of incubation, the membranes were washed thrice with tris-buffered saline containing 0.1% tween-20 and transferred to the secondary antibodies solution (1:1,000) conjugated with alkaline phosphatase for 4 h at 4°C. Finally, the membranes were transferred to the developing solution containing nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP). The membranes were allowed to develop until the intense bands were observed (Kamaldevi and Balamurugan, 2016). The antibodies used in the current study are, AKT, pAKT, mTOR, pmTOR (Cell signaling), actin (sigma), PI3-kinase, pPI3-kinase, PDI-2, anti-rabbit and anti-mouse conjugated with alkaline phosphatase (Santa Cruz Biotechnology).
Approximately 10 worms were transferred to the sterile M9 buffer containing K. pneumoniae (20%, 3 × 105 cells/ml), in a sterile 24-well culture plate and incubated at 20°C. The survival of the nematodes was monitored at an interval of an hour till it attained complete mortality. The nematodes that did not show any response to touch and lack of pharyngeal pumping were considered as dead. Here, the nematodes fed on E. coli OP50 were considered as control. Each experiment was performed atleast three times independently under the similar conditions.
The defect caused by the K. pneumoniae in the nematodes' intestine was monitored under microscope. The infected nematodes were washed thrice with M9 buffer containing 1 mM sodium azide (anesthetizing agent) and placed on a 2% agarose pad. The physiological changes with the exposed C. elegans were examined under inverted fluorescence microscope (Nikon, Japan).
The reactive oxygen species (ROS) production in nematodes during K. pneumoniae infection was quantitatively analyzed by confocal laser scanning microscopy (CLSM). The nematodes exposed to pathogen were washed thrice to remove the surface bacteria and transferred to the liquid M9 medium containing 5 μg/ml of 2′, 7′-dichlorofluoresceine-diacetate (DCFH-DA). The content was incubated in dark for 10 min at room temperature. Then the nematodes were washed thoroughly with PBS to remove the excess DCFH-DA and placed the worms on an agar pad (2%) containing an anesthetizer (30 mM sodium azide). Finally, the slides were subjected to microscopic observations (Carl zeiss, Germany). The relative fluorescent intensity of 2′, 7′-dichlorofluorescein (DCF) in the nematodes represents the amount of ROS production in nematodes (Kamaladevi et al., 2013).
The quantitative measurement of reactive oxygen species was performed by (DCFH-DA) method (Scherz-Shouval et al., 2007). The nematodes exposed to K. pneumoniae for different time-points (12, 24, 36 h) were washed thoroughly with M9 buffer. The infected nematodes were homogenized and the concentration of proteins was determined by Bradford's method. An equal concentration of protein was mixed with 5 μM DCFH-DA in a final volume of 2 ml of Tris-HCl. The mixture was incubated at room temperature for 45 min in dark condition. The level of DCF produced by ROS mediated oxidation of DCFH-DA was measured spectroscopically at 530 nm, following excitation at 485 nm and the level of DCF was quantified against DCF standard. Results were expressed as nmol of DCF/mg of protein.
The total RNA was isolated from N2 wild-type and mutants exposed to K. pneumoniae for different time-points (12, 24, and 36 h) by TRIzol method. For RT-PCR, the oligo-dT method was followed to synthesis a cDNA strands. According to instruction of SuperScript III kit (Invitrogen Inc., USA), the CDNA template was generated from 100 ng of RNA isolated from nematodes. PCR was carried out to amplify from the cDNA by using forward and reverse primers. Quantitative real time (qRT-PCR) was performed by using the Applied Biosystem SYBR Green fluorescence dye. The expression of the candidate genes were normalized by act-2. The relative gene expression was determined by calculating 2−ΔΔct.
The 2D-gel electrophoresis was performed at least three times from the biologically different samples for consistency of differentially regulated proteins and statistical significance of fold-changes. Real-time analysis and quantification of ROS were conducted in triplicates and the statistical analysis was performed by one-way ANOVA using SPAA software 10.0. Significant differences between the groups were calculated by Dunnet's multiple range test (P < 0.05).
In our previous study Kamaladevi and Balamurugan (2015), we have reported that K. pneumoniae required 48 h for complete killing of nematodes. Hence, to understand the host response against K. pneumoniae pathogenesis at their proteome level, 12, 24, and 36 h were considered. Using conventional proteomic approach, infection induced host response against K. pneumoniae was monitored kinetically. The CBB stained gel images of respective time-points were compared and approximately > 300 protein spots were detected in each time-points. Figure 1 showed the CBB stained gel images of 12 (Figure 1A), 24 (Figure 1B), and 36 h (Figure 1C). The protein spots matched between the OP50 control and K. pneumoniae infected at 12, 24, and 36 h were 381, 630, and 742, respectively. We performed three biologically independent experiments and used the stringent criteria of a 1.2 Da mass tolerance and more than 1.5-fold regulation. This resulted in an identification of a total of 53, 82, and 43 regulatory proteins at 12 Supplementary Table 1, 24 Supplementary Table 2, and 36 h Supplementary Table 3, respectively.
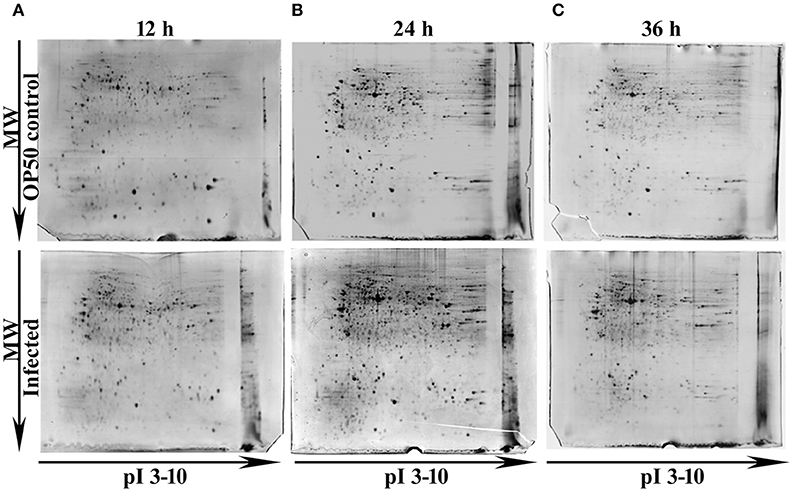
Figure 1. Representative gel images showing the detected protein spots at different time points (A) 12 h, (B) 24 h and (C) 36 h. The Upper panel representing the proteome of control nematodes fed on E. coli OP50 and the Lower panel representing the proteome of K. pneumoniae infected nematodes.
To decipher the interaction between the identified regulatory proteins and their role in biological activities during host defense, STRING 10.0 analysis was performed. The interactions between the identified proteins were analyzed based on the literature with a high confidence score of 0.7. The interaction between the proteins identified at 12 Figure 2A https://string-db.org/cgi/network.pl?taskId=2ewoymMJQSou), 24 Figure 2B https://string-db.org/cgi/network.pl?taskId=KS9tVO9Zw468), and 36 h (Figure 2C https://string-db.org/cgi/network.pl?taskId=iBw1EvsLlGVD) were analyzed separately and their biological functions were determined by Gene ontology (GO) database. The GO results revealed that the major group of identified proteins at different time points were predominantly encodes for metabolism, development, translation, reproduction, protein unfolding, and apoptosis (Figures 2D–F). Concomitantly, in order to investigate the interactions between the proteins identified at 12, 24, and 36 h and their biological significance in elucidating the host defense against K. pneumoniae infection, STRING analysis was performed with the total list of identified proteins and they were manually annotated (Figure 3A https://string-db.org/cgi/network.pl?taskId=I10REy8oLgGn). Based on their biological functions, the identified proteins and their interacting partners were classified into six major classes such as metabolism, translation, dauer formation, signal transduction, endocytosis, apoptosis, and protein folding (Figures 3A,B).
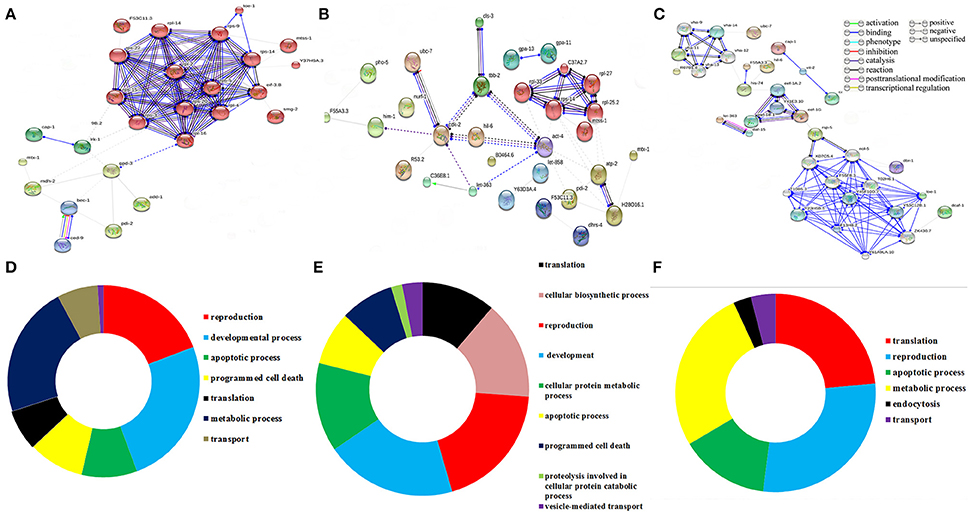
Figure 2. Protein-protein interaction of the proteins identified at (A) 12 h, (B) 24 h, and (C) 36 h using STRING 10.0 software and based on their function the identified proteins were classified using DAVID (D–F). The interactive modes of STRING analysis results were available at their website 12 h (https://string-db.org/cgi/network.pl?taskId=2ewoymMJQSou), 24 h (https://string-db.org/cgi/network.pl?taskId=KS9tVO9Zw468), and 36 h (https://string-db.org/cgi/network.pl?taskId=iBw1EvsLlGVD).
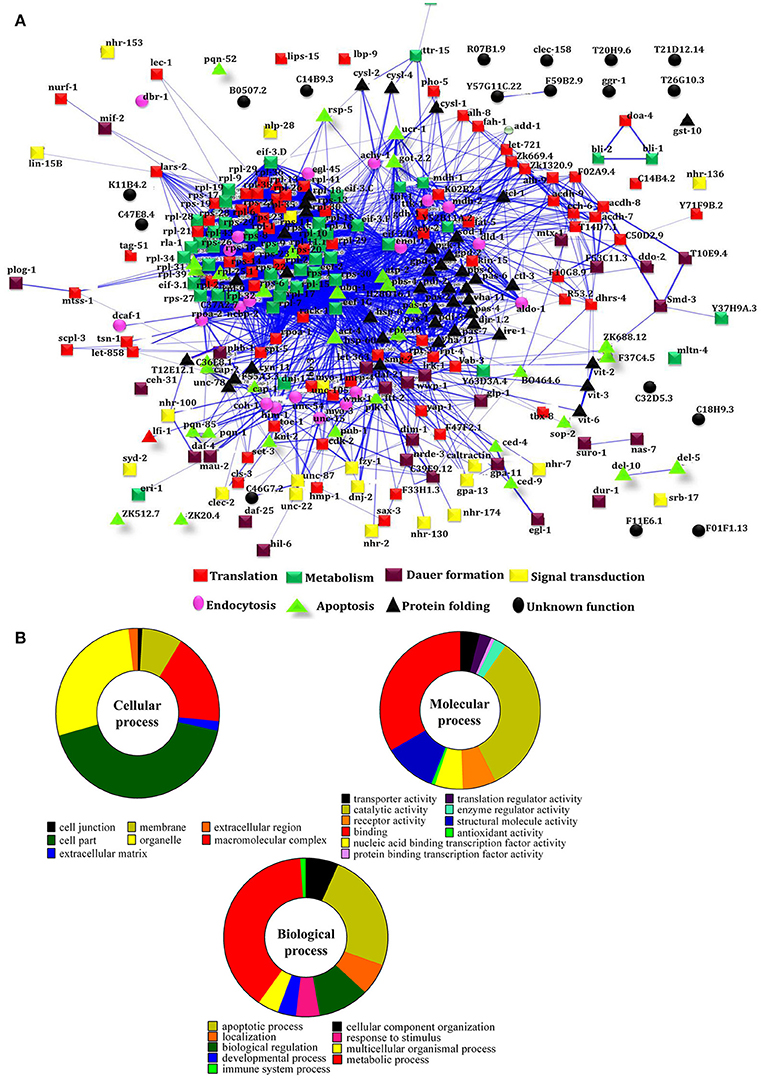
Figure 3. (A) Protein-protein interaction of all the identified proteins were manually annotated based on their function. The interactive mode of STRING analysis results was available at their website https://string-db.org/cgi/network.pl?taskId=I10REy8oLgGn (B) Gene ontology analysis for the identified proteins based on their molecular, biological and cellular functions
In order to separate the interacting partners of regulatory proteins, a liquid IEF was performed. Proteins isolated from the control and experimental C. elegans homogenate were separated on the basis of their pI (pH 3–10) in solution. After the IEF separation, SDS-PAGE was performed to determine the separation of the proteins in all the 10 fractions Figure 4. Since most of the interacting proteins were identified to be in the pI ranges 5–9, the fraction numbers 5–8 were pooled together and enzymatically digested with trypsin. The peptides were analyzed by nano LC-MS/MS. A total of 88 proteins were identified in the IEF fractionations. The list of identified proteins was given in Supplementary Table 4.
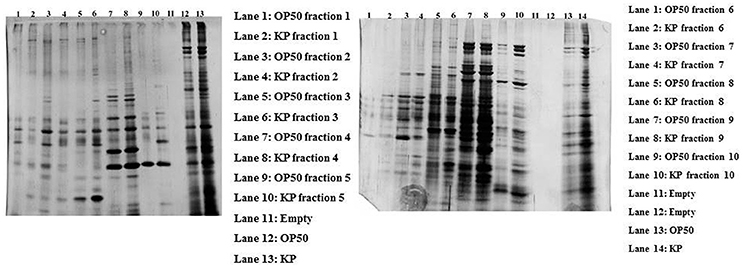
Figure 4. The gel representing the profile of proteins separated using liquid phase IEF. The control and experimental proteins were separated based on their pI using liquid IEF and the collected fractions were subjected to SDS-PAGE. The left gel shows the proteins present in the fractions 1–5 with an approximate pI of 3–6.8. The right gel shows the profile of proteins found in the fractions 6–10 with an approximate pI of 7–10.
To identify the protein that have major role in activating the host response against K. pneumoniae infection, several bioinformatics analysis were performed. The total of 266 proteins were enriched using the MetaCore software (DAVID) (Figure 5A) following enrichment analysis as described in the methods (Bessarabova et al., 2015). In proportion with the large number of regulated proteins in response to K. pneumoniae infection, more enriched GO categories were identified. In order to reduce the number of GO terms, enriched GO categories with false discovery rate (FDR) < 0.05 from DAVID analysis were submitted to the REVIGO tool (Supek et al., 2013). This tool uses the Uniprot as background database and has default semantic similarity measure (simrel). Investigation by this tool clearly showed that the biological process associated with metabolism, translation, protein metabolism, cellular homeostasis, protein folding, reproduction etc. were significantly over-represented among the proteins regulated in C. elegans during K. pneumoniae infection (Figure 5B). The lists of top 10 GO processes for these proteins in the host functions are provided in the Figures 2D–F. In addition, KEGG pathway analysis revealed that most of the identified proteins were regulated for metabolic pathway (Figure 5C). Concerning the GO process, the most regulated proteins were involved in metabolism, translation, apoptosis, development, immunity, and response to stimuli. Further, to identify the contribution of each protein in the different biological functions like metabolic process, translation, protein folding or oxidative stress, apoptosis and dauer formation, a Venn diagram were drawn with the list of proteins contributing to each function. The results revealed several overlapping proteins among the identified functions. Interestingly, a protein mTOR (mammalian target of Rapamycin) was found to have a contribution in all the biological functions (Figure 5D) and hence, was selected for further characterization studies.
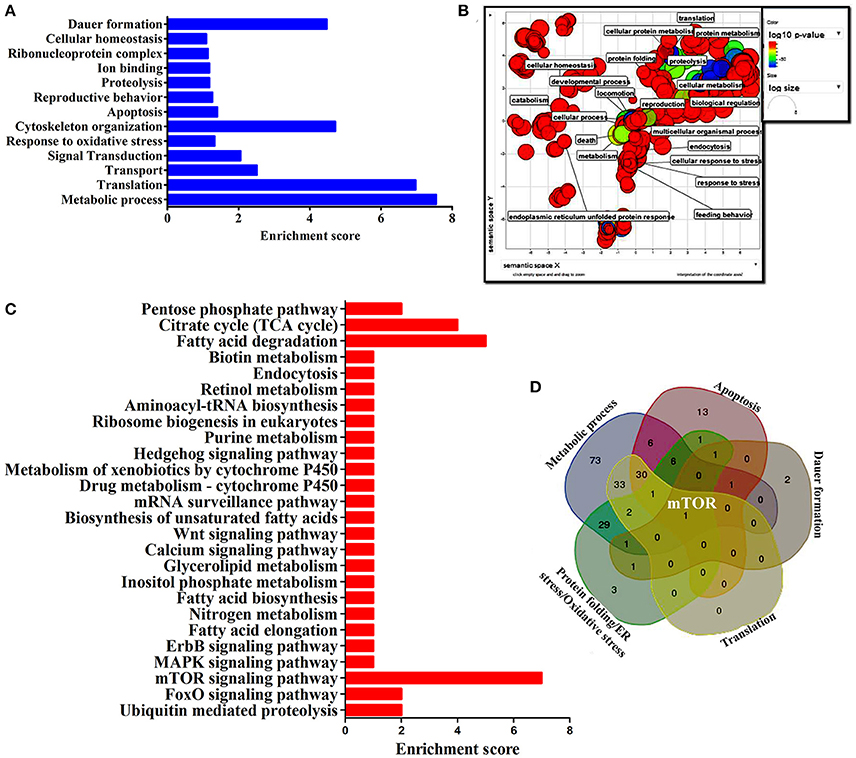
Figure 5. (A) The histogram representing the functional enrichment analysis for identified proteins based on their functions. (B) The enriched protein functional categories were defined by GO terms and the highly redundant functions were removed and the significant functions were plotted. (C) The histogram representing the enriched pathway based on the KEGG pathway analysis. (D) The Venn diagram represents the regulatory proteins which appears to be commonly involved in major biological functions.
mTOR, a mammalian Target of Rapamycin regulates several cellular process in normal system. However, the impact of bacterial infection on mTOR signaling remains unclear. In the present study, the proteomics approach coupled with bioinformatics analysis revealed mTOR has appeared to be involving in regulating the different biological functions. So we examined the regulation of mTOR in host during infection with K. pneumoniae in 2D gel analysis. Interestingly, we found that mTOR was significantly (P < 0.05) down regulated in 12 h when compared to the control. The downregulation of mTOR was showed in the 3D images Figure 6. The down regulation of mTOR was believed to induce a metabolic stress in host (Jia et al., 2004). mTOR is a major component for regulating the metabolic process (Tattoli et al., 2012). In C. elegans, metabolic stress acts as a double edged-sword (Kenific et al., 2010; Tattoli et al., 2012).
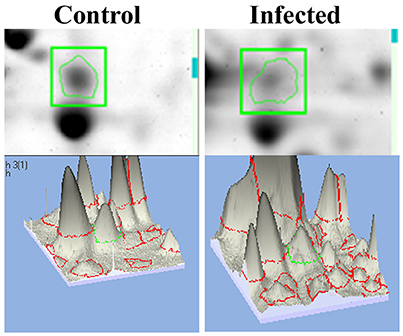
Figure 6. The representative spot showing the downregulation of mTOR and the 3D images of the control and infected samples confirmed their inhibition against K. pneumoniae.
Intestinal atrophy in a biological system was usually determined by the level of autophagy. It is well-established that autophagy is induced following the inhibition of mTOR in multicellular systems (Mortimore and Schworer, 1977; Chakrabarti et al., 2012; Kapuy et al., 2014). To examine the mTOR mediated autophagy, we monitored the physiological alteration in C. elegans intestine and marker for autophagy. The microscopic analysis of C. elegans exposed to K. pneumoniae displayed a distended intestine Figure 7. Due to the defective intestine, the infected nematodes could not absorb the nutrients from the ingested food. This promoted a dauer formation in infected nematodes. The results of short-time exposure assay indicated the presence of dauer nematodes during the course of infection (Table 1). This explicates that the infection with K. pneumoniae induced an abnormal dauer formation in nematodes. Interestingly, the dauer formation due to the deficient mTOR shares few unique and common features of normally formed dauer due to food starvation where it is not having severe intestinal atrophy (Long et al., 2002). Thus, the dauer formation along with intestinal atrophy in infected nematodes suggested that might be due to the downregulation of mTOR during infection. Furthermore, the increased autophagy was also confirmed at the molecular level by measuring the level of bec-1, a gene responsible for autophagy (Figure 8A). The upregulation of bec-1 during infection suggested the activation of autophagic process during K. pneumoniae infection.
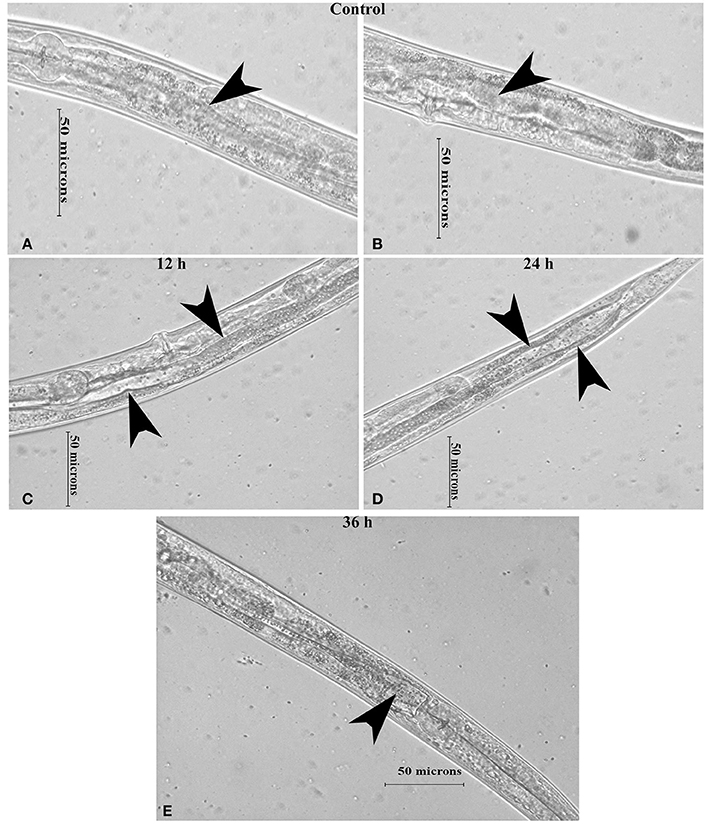
Figure 7. The micrographs representation the intestinal atrophy in nematodes exposed to K. pneumoniae at 12 (C), 24 (D), and 36 h (E) whereas the control (A,B) showed the normal intact intestine. The arrow head in the control panels indicates the normal intestine and the infected panels indicate the distended pharynx
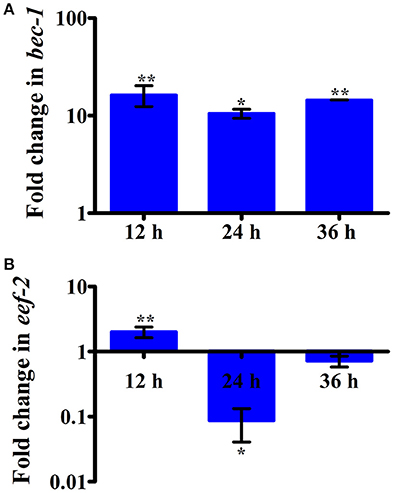
Figure 8. The histograms representing the upregulation of (A) bec-1 which indicating the increased apoptosis in nematodes and downregulation of (B) eef-2 indicating the inhibition of translation elongation factor in C. elegans against K. pneumoniae infection. Data are presented as mean ± SD of three biological replicates and the level of significance was analyzed by one-way ANOVA followed by Duncan's post-hoc analysis (*P < 0.05; **P < 0.005).
An inhibition of translation machinery is the other negative consequence of the inhibition of mTOR. The results of 2D gel electrophoresis also suggested that the infection with K. pneumoniae in C. elegans regulated several proteins that are encoded for translation. Hence, to investigate the status of translational regulatory gene in the C. elegans during infection, the mRNA level of eef-2 was examined. The eef-2 is the downstream regulator of mTOR, which is also an essential component for translation. The regulation of eef-2 in the nematodes infected with K. pneumoniae was significantly downregulated during course of infection (Figure 8B). This result indicated that the downregulation of mTOR influences the regulation of eef-2.
To elucidate the role of mTOR signaling in host defense against K. pneumoniae infection, the upstream regulators of mTOR were also examined for their regulation at their protein level using western blot analysis. The PI3-kinase and AKT were found to be the upstream regulators of mTOR. Hence, the level of expression of PI3-kinase, AKT, and mTOR were investigated. The PI3-Kinase activates the AKT and induced the phosphorylation of mTOR in normal conditions (Schmitz et al., 2008; Rafii et al., 2015). Our results showed that the level of PI3-kinase and AKT was inhibited in infected nematodes Figures 9A,B. This in turn inhibited the phosphorylation of mTOR Figures 9A,B. The inhibition of PI3-kinase/AKT/mTOR signaling pathway appeared to be increased the risk of infection (Figure 9C) (Rafii et al., 2015). Additionally, PDI-2, the protein encoded for protein folding was also examined for their regulation. The PDI is a protein-di-isomerase, which catalyzes the protein folding. The downregulation of PDI-2 (Figures 9A,B) indicated that the K. pneumoniae infection not favors the protein not to fold properly at 24 h in the host system. The inhibition of PDI-2 in the K. pneumoniae infected samples in western blot analysis suggested the possibilities of important regulatory proteins were not in fully or properly folded state in C. elegans. The result of the present study goes well with the previous study wherein the similar kind of regulatory event was observed in C. elegans during infection with V. alginolyticus (Durai et al., 2014).
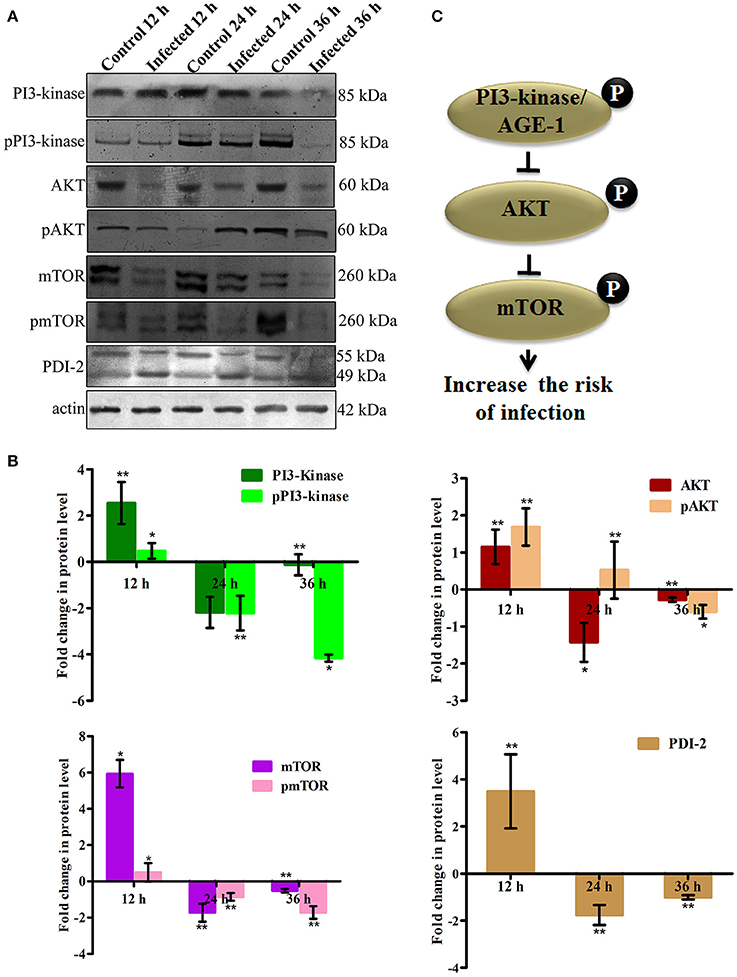
Figure 9. (A) The western blot analysis showing the regulation of upstream regulators of mTOR in C. elegans at different time points (12, 24, and 36 h). The downregulation of PI3-kinase, pPI3-kinase, AKT, pAKT, mTOR, pmTOR, and PDI-2 were observed during the K. pneumoniae infection in nematodes. The membrane containing proteins were transferred to the developing solution containing nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) and allowed to develop until the intense bands were observed. (B) The histograms representing the fold change in the regulation of total and phospho PI3-kinase, AKT, mTOR and PDI-2 in C. elegans during K. pneumoniae infection. (C) The schematic diagram representing the PI3-kinase/AKT/mTOR pathway in increasing the risk of host susceptibility towards infection.
To further confirm the role of PI3-kinase/AKT/mTOR signaling pathway and PDI-2 in host defense against K. pneumoniae infection, the C. elegans with specific mutation in AKT, mTOR and PDI-2 were examined for their survival during K. pneumoniae infection. The mutants of akt (Figure 10A), mTOR (Figure 10B), and pdi-2 (Figure 10C) exposed with K. pneumoniae exhibited a significant (P < 0.05) shorter life span (mean lifespan of 27 ± 5, 20 ± 2, and 12 ± 7 h for AKT, mTOR, and PDI-2 mutants, respectively) than the controls (N2 mean lifespan of 48 ± 5 h) (Figure 10). The susceptibility of AKT, mTOR, and PDI-2 mutants to K. pneumoniae suggested the role and importance of PI3-kinase/AKT/mTOR pathway and unresponsive protein folding in C. elegans during host defense.
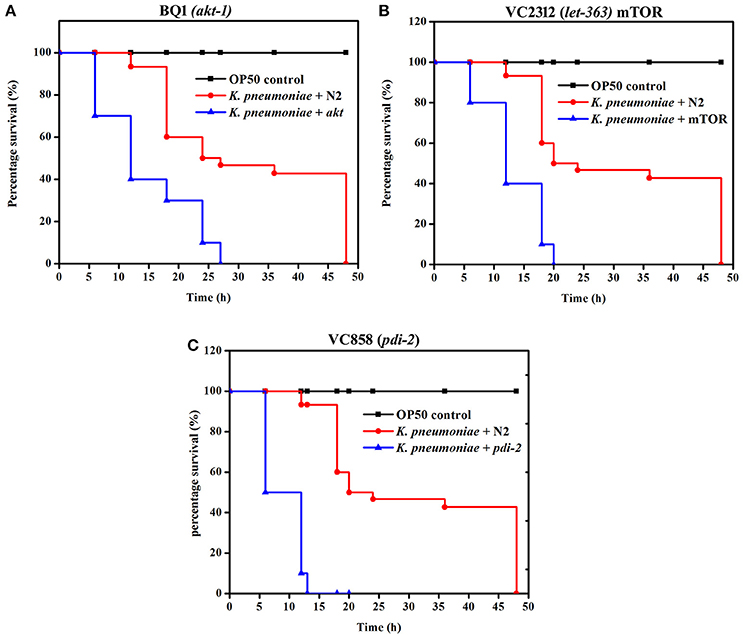
Figure 10. The role of PI3-kinase/ AKT/mTOR pathway and PDI-2 in protein misfolding was analyzed using the survival of (A) BQ1 (akt), (B) VC2312 (let-363) mTOR, and (C) VC858 (pdi-2) mutants.
In our proteomic analysis, we identified several stress responsive, detoxification, and protein mis-folding genes strongly induced after K. pneumoniae infection. The process of increased autophagy and translation blockage is also another source of oxidative burst (Chavez et al., 2007; Schmitz et al., 2008; Chakrabarti et al., 2012). The increased ROS is an alternate part of innate immune defense mechanism (Chavez et al., 2007). A significant increase in the level of intracellular ROS in the C. elegans exposed to K. pneumoniae than the control revealed the oxidative burst in host during infection (Figure 11A). Concomitantly, a high level of DCF fluorescence in the C. elegans with the increase in an infection time corroborated the generation of ROS due to infection mediated oxidative stress in the host (Figure 11B). Furthermore, in our 2D analysis, several proteins that encoded for antioxidants like gst-8, sod-3, catalase (cat-1, cat-2, and cat-3) were also found to be regulated. To validate these regulations, the mRNA levels of the identified proteins were determined by qPCR using specific primers. The mRNA levels of all the selected antioxidant enzymes were significantly (P < 0.05) upregulated during course of infection except cat-2 (Figure 12). It appears that the levels of antioxidant enzymes increased to combat the elevated ROS during the K. pneumoniae infection in the host system.
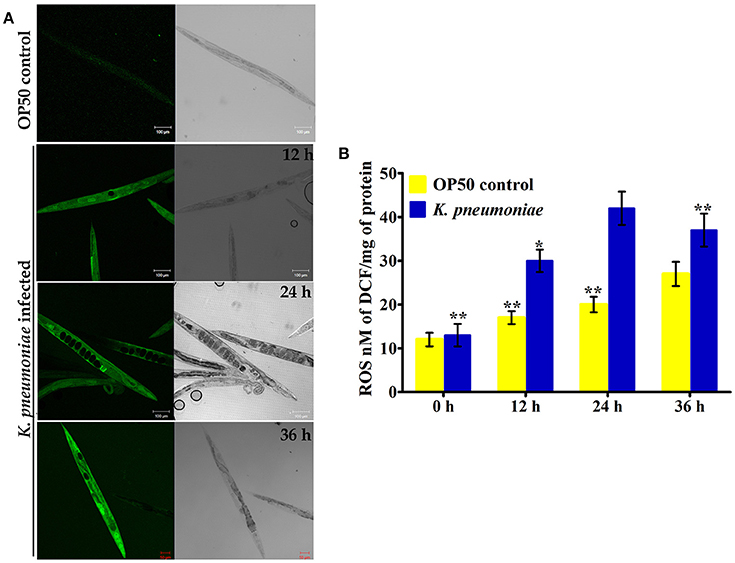
Figure 11. (A) The confocal laser scanning micrographs showing the ROS production in nematodes exposed to K. pneumoniae at different time-points (12, 24, and 36 h) by DCFH-DA staining method. (B) The histogram indicates the quantitative amount of intracellular ROS produced in host against K. pneumoniae infection. The values are expressed as mean ± SD of triplicates. Statistical analysis was performed by one-way ANOVA followed by Duncan's post-hoc analysis (*P < 0.05;**P < 0.005).
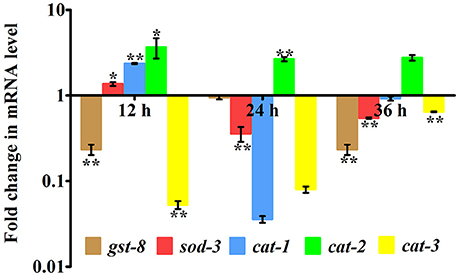
Figure 12. Quantitative real-time PCR showing the relative expression of the candidate antioxidant genes (gst-8, sod-3, cat-1, cat-2, cat-3, and B-actin) normalized over the relative expression of housekeeping gene, β-actin of respective time-points. Data are presented as mean ± SD of three biological replicates and the level of significance was analyzed by one-way ANOVA followed by Duncan's post-hoc analysis (*P < 0.05; **P < 0.005).
K. pneumoniae, an opportunistic pathogen has been well-documented for its severity in causing community and nosocomial-acquired infections. Though it has been tagged as a most virulent pathogen, the lack of knowledge on host response against infection failed to eradicate their infections with available tools. Interestingly, C. elegans shares similarity to mammalian immune system, mainly signaling cascades in innate immunity toward bacterial infection (Alper et al., 2008). An inevitable characteristics of consuming microorganisms as a food has evident the selection pressure to evolve and maintain immune defense mechanism (Kim et al., 2002; Alper et al., 2010; Shivers et al., 2010). Although human lungs and nematodes' intestine vary in several aspects, it could be possible to characterize the virulence traits essential for infecting lung as well as respiratory tract, and also to understand the host innate immune response. Hence, the C. elegans, which is devoid of lungs system, has globally been accepted as a suitable model (Singh and Aballay, 2006; Akira, 2009) to study host response against lung and respiratory tract infecting microorganisms such as P. aeruginosa (Tan et al., 1999; Kirienko et al., 2013; Balasubramanian et al., 2016), Streptococcus pneumoniae (Bolm et al., 2004), Streptococcus pyogens (Jansen et al., 2002), Microbacterium Spp. (Hodgkin et al., 2000), Mycoplasma iowae (Pritchard et al., 2014), S. aureus (Bogaerts et al., 2010a; JebaMercy et al., 2011), Legionella pneumoniae (Brassinga et al., 2010; Komura et al., 2010; Hellinga et al., 2015), Coxiella burnetti (Battisti et al., 2017), and Mycobacterium tuberculosis (Galbadage et al., 2016). In addition, a study by Green et al. (2009) utilized C. elegans to characterize the impact of cigarette smoke on innate immune response of host fortifies the idea of using nematode as lung and respiratory disease model. In this scenario, the current study is aimed to demonstrate for the first time a detailed characterization of the host response or defense against K. pneumoniae infection using C. elegans. Here, the conventional proteomic method coupled with MALDI-MS/MS, LC-MS/MS, bioinformatics, and molecular approaches were utilized to decipher the involvement of regulatory signaling pathway(s) and other consequences in host during K. pneumoniae infection.
Earlier, the proteomic studies in C. elegans against different pathogens including S. aureus (Bogaerts et al., 2010a), A. hydrophila (Bogaerts et al., 2010b), V. alginolyticus (Durai et al., 2014), P. aeruginosa (Balasubramanian et al., 2016), and P. mirabilis (Jebamercy et al., 2016) exhibited the strong regulation of specific regulatory protein(s) at particular time-point. Such regulation depicted the specific role of particular protein in host defense. With this understanding, our previous knowledge on host-pathogen interaction between the C. elegans and K. pneumoniae at physiological levels, an early, mid, and late time-point of infections were considered for the present proteomics based investigations. In our previous study, we reported that K. pneumoniae required 48 ± 5 h for the complete killing of nematode (Kamaladevi and Balamurugan, 2015). Therefore, we opted to look at the changes in the proteomes at early (12 h), mid (24 h), and late (36 h) hours of infection.
A total of 266 differentially regulated proteins were identified at 12, 24, and 36 h respectively using conventional 2D method coupled with MALDI-MS/MS and liquid IEF coupled with LC-MS/MS analysis. The STRING analysis provided a network or interaction of the proteins identified at the different time-points. These identified proteins were subdivided into categories based on their biological functions which demonstrated that how the infection is anticipated at the proteome levels of whole organism at different time-points. The proteins which were identified, encodes for the biological functions like metabolism, translation, apoptosis, dauer formation, protein folding, signal transduction, and endocytosis. The functional enrichment results revealed that metabolic processes were enriched among the annotated functions. Furthermore, the mTOR was found to be involved in all identified functions. The mTOR is a key regulatory kinase that acts at the nexus of diverse nutrient-sensitive signals to regulate cellular metabolism and thus it controls the immune activation (Schmitz et al., 2008; Russell et al., 2011). The reduced feeding is the one of the prime factors that affects the nutrient signals. Previously we reported that infection with K. pneumoniae damaged the pharyngeal region of the C. elegans (Kamaladevi and Balamurugan, 2015). The affected pharynx attributed to the inhibited feeding and subsequently deprives nutrient signal during infection. The present study indicated that in 12 and 24 h of infection, the mTOR was downregulated during K. pneumoniae infection which clearly corroborated with earlier findings on deprived nutrient signals. The infection with Shigella, Salmonella, P. entomophila, and P. aeruginosa also reported to inhibit the level of expression of mTOR (Mohr and Sonenberg, 2012).
Nutrient starvation is known to inhibit mTOR, increase autophagy and inhibit translation (Jia et al., 2004; Watanabe et al., 2011; Chakrabarti et al., 2012). In C. elegans there has been an advantage of detecting the mTOR inhibition by physiological read-outs. The intestinal atrophy and dauer formation were the chief information to confirm the inhibition of mTOR. An intestinal atrophy is defined by the increased autophagy in the intestinal region of nematodes (Long et al., 2002). The distended intestine with an increased intestinal width corroborated the intestinal atrophy or increased autophagy in C. elegans by K. pneumoniae. Furthermore, the significant (P < 0.05) upregulation of bec-1, a marker for autophagy (Jia et al., 2009; Zou et al., 2014) in C. elegans confirmed the activation of autophagy in host during K. pneumoniae infection. From our 2D data, we showed that infection with K. pneumoniae regulated several proteins that codes for translation machinery. Among them, eef-2 the gene which codes for elongation was studied previously to confirm the inhibition of translation machinery against P. aeruginosa infection (Balasubramanian et al., 2016). Similarly, the inhibited level of eef-2 mRNA in C. elegans suggested that the K. pneumoniae infection affected the translational events in the host. Inhibition of protein translation reduced the synthesis of several immune genes which makes the host most vulnerable to an infection (Chakrabarti et al., 2012; Tattoli et al., 2012; Balasubramanian et al., 2016).
mTOR signaling, induces a Endoplasmic reticulum (ER) stress in host. The ER stress triggers the protein mis-folding and thus elevated the ROS level in host (Jia et al., 2004; Tattoli et al., 2012). An inhibited levels of mTOR and PDI-2 in western blot analysis suggested that the regulatory proteins are not folded properly in C. elegans during infection. Moreover, the elevated levels of intracellular and extracellular ROS in nematodes exposed to K. pneumoniae suggested the generation of oxidative stress in host against infection. Additionally, the significant upregulation of antioxidant genes corroborated the oxidative stress in host against K. pneumoniae infection. The elevation of oxidative stress is necessary to block the translation machinery during infection (Chakrabarti et al., 2012). Concurrently, the KEGG pathway analysis revealed that PI3K/AKT/mTOR pathway is responsible for host defense against K. pneumoniae infection. To confirm the role of this pathway against infection, the western blot analysis and survival assays using respective gene specific mutant C. elegans were performed. The inhibited activation of PI3-Kinase, AKT, and mTOR confirmed that K. pneumoniae arrested this pathway and the susceptibility of C. elegans having mutation in pdi-2, akt-1, and mTOR confirmed the above results. Recently, Rafii et al. (2015), through their clinical trials confirmed that inhibition of PI3K/AKT/mTOR increased the risk of infections in host. Therefore, present study deciphered that the inhibition of PI3K/AKT/mTOR pathway increased the susceptibility of host to K. pneumoniae infection.
In summary, the data of the current study uncovered the critical role of PI3/AKT/mTOR pathway in the host response against K. pneumoniae infection in C. elegans Figure 13. The system-level over-view of host proteome during K. pneumoniae differentially regulated the proteins that encodes for metabolism, translation, signal transduction, apoptosis, protein misfolding, and dauer formation. Inhibition of mTOR, followed by subsequent changes in host cellular activities and hyper-susceptibility of PI3 kinase, akt-1, mTOR, pdi-2, and eef-2 mutants unveiled the role of PI3K/AKT/mTOR pathway, protein folding, and translation machinery in host defense against infection. Moreover, the importance of host metabolic perturbations in infected host in addition to severe defects like autophagy, dauer formation, protein mis-folding, and oxidative stress advance our understanding in the multiple aspects of the host response against K. pneumoniae. These results will pave a way to the advancement of new therapeutic strategies against K. pneumoniae infection.
AK and KB conceived and designed the experiments. AK performed the experiments, analyzed the data, and prepared the manuscript.
Research was supported by the DBT (Grant No. BT/PR14932/MED/29/233/2010), ICMR (Sanction No: 5/3/3/13/2010-ECD-I), DST-SERB (No.SR/SO/AS-80/2010), UGC Major Research Project [No. 42-222/2013 (SR)], CSIR [No. 37(1460)/11/EMR-II], AU-ITC, FIST (Grant No. SR-FST/LSI-087/2008), and UGC SAP-DRS-I [Grant No. F.3-28/2011 (SAP-II)], New Delhi, India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors have gratefully thanked the Caenorhabditis Genetics Center, MN which is funded by the National Institutes of Health, National Center for Research Resources for providing C. elegans strains. We also thankfully acknowledged Dr. K. Emmanuvel Rajan, Department of Animal Science, School of Life Sciences, Bharathidasan University, Trichirappalli, India for providing phosphoAKT antibody. Authors have also gratefully acknowledge the computational and bioinformatics facility provided by the Alagappa University Bioinformatics Infrastructure Facility (funded by DBT, GOI; Grant No. BT/BI/25/001/2006). Proteomics instrumentation Facilities provided by DST-PURSE [Grant No. SR/S9Z-23/2010/42(G)] is gratefully acknowledged.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00393/full#supplementary-material
Supplementary Table 1. List of proteins identified in nematodes exposed to K. pneumoniae for 12 h by MALDI-TOF/TOF analysis.
Supplementary Table 2. List of proteins identified in nematodes exposed to K. pneumoniae for 24 h by MALDI-TOF/TOF analysis.
Supplementary Table 3. List of proteins identified in nematodes exposed to K. pneumoniae for 36 h by MALDI-TOF/TOF analysis.
Supplementary Table 4. List of proteins separated by liquid phase IEF and identified by nano LC-MS/MS analysis.
Akira, S. (2009). Pathogen recognition by innate immunity and its signaling. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 143–156. doi: 10.2183/pjab.85.143
Alper, S., Laws, R., Lackford, B., Boyd, W. A., Dunlap, P., Freedman, J. H., et al. (2008). Identification of innate immunity genes and pathways using a comparative genomics approach. Proc. Natl. Acad. Sci. USA 105, 7016–7021. doi: 10.1073/pnas.0802405105
Alper, S., Mcelwee, M. K., Apfeld, J., Lackford, B., Freedman, J. H., and Schwartz, D. A. (2010). The Caenorhabditis elegans germ line regulates distinct signaling pathways to control lifespan and innate immunity. J. Biol. Chem. 285, 1822–1828. doi: 10.1074/jbc.M109.057323
Ananthi, S., Santhosh, R. S., Nila, M. V., Prajna, N. V., Lalitha, P., and Dharmalingam, K. (2011). Comparative proteomics of human male and female tears by two-dimensional electrophoresis. Exp. Eye Res. 92, 454–463. doi: 10.1016/j.exer.2011.03.002
Balasubramanian, V., Sellegounder, D., Suman, K., and Krishnaswamy, B. (2016). Proteome analysis reveals translational inhibition of Caenorhabditis elegans enhances susceptibility to Pseudomonas aeruginosa PAO1 pathogenesis. J. Proteomics 16, 30146–30154 doi: 10.1016/j.jprot.2016.04.025
Battisti, J. M., Watson, L. A., Naung, M. T., Drobish, A. M., Voronina, E., and Minnick, M. F. (2017). Analysis of the Caenorhabditis elegans innate immune response to Coxiella burnetii. Innate Immun. 23, 111–127. doi: 10.1177/1753425916679255
Bessarabova, M., Ishkin, A., Jebailey, L., Nikolskaya, T., and Nikolsky, Y. (2015). Knowledge-based analysis of proteomics data. BMC Bioinformatics 13(Suppl. 16):S13. doi: 10.1186/1471-2105-13-S16-S13
Bogaerts, A., Beets, I., Temmerman, L., Schoofs, L., and Verleyen, P. (2010a). Proteome changes of Caenorhabditis elegans upon a Staphylococcus aureus infection. Biol. Direct 5:11. doi: 10.1186/1745-6150-5-11
Bogaerts, A., Temmerman, L., Boerjan, B., Husson, S. J., Schoofs, L., and Verleyen, P. (2010b). A differential proteomics study of Caenorhabditis elegans infected with Aeromonas hydrophila. Dev. Comp. Immunol. 34, 690–698. doi: 10.1016/j.dci.2010.02.003
Bolm, M., Jansen, W. T., Schnabel, R., and Chhatwal, G. S. (2004). Hydrogen peroxide-mediated killing of Caenorhabditis elegans: a common feature of different streptococcal species. Infect. Immun. 72, 1192–1194. doi: 10.1128/IAI.72.2.1192-1194.2004
Brassinga, A. K., Kinchen, J. M., Cupp, M. E., Day, S. R., Hoffman, P. S., and Sifri, C. D. (2010). Caenorhabditis is a metazoan host for Legionella. Cell. Microbiol. 12, 343–361 doi: 10.1111/j.1462-5822.2009.01398.x
Chakrabarti, S., Liehl, P., Buchon, N., and Lemaitre, B. (2012). Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12, 60–70. doi: 10.1016/j.chom.2012.06.001
Chavez, V., Mohri-Shiomi, A., Maadani, A., Vega, L. A., and Garsin, D. A. (2007). Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics 176, 1567–1577. doi: 10.1534/genetics.107.072587
Durai, S., Singh, N., Kundu, S., and Balamurugan, K. (2014). Proteomic investigation of Vibrio alginolyticus challenged Caenorhabditis elegans revealed regulation of cellular homeostasis proteins and their role in supporting innate immune system. Proteomics 14, 1820–1832. doi: 10.1002/pmic.201300374
Evans, S. E., Tuvim, M. J., Zhang, J., Larson, D. T., García, C. D., Pro, S. M., et al. (2010). Host lung gene expression patterns predict infectious etiology in a mouse model of pneumonia. Respir. Res. 11:101. doi: 10.1186/1465-9921-11-101
Fuchs, H., Gailus-Durner, V., Adler, T., Pimentel, J. A., Becker, L., Bolle, I., et al. (2009). The german mouse clinic: a platform for systemic phenotype analysis of mouse models. Curr. Pharm. Biotechnol. 10, 236–243. doi: 10.2174/138920109787315051
Galbadage, T., Shepherd, T. F., Cirillo, S. L., Gumienny, T. L., and Cirillo, J. D. (2016). The Caenorhabditis elegans p38 MAPK Gene plays a key role in protection from mycobacteria. Microbiologyopen 5, 436–452. doi: 10.1002/mbo3.341
Green, R. M., Gally, F., Keeney, J. G., Alper, S., Gao, B., Han, M., et al. (2009). Impact of cigarette smoke exposure on innate immunity: a Caenorhabditis elegans model. PLoS ONE 4:e6860. doi: 10.1371/journal.pone.0006860
Grompone, G., Martorell, P., Llopis, S., Gonzalez, N., Genoves, S., Mulet, A. P., et al. (2012). Anti-inflammatory Lactobacillus rhamnosus CNCM I-3690 strain protects against oxidative stress and increases lifespan in Caenorhabditis elegans. PLoS ONE 7:e52493. doi: 10.1371/journal.pone.0052493
Hellinga, J. R., Garduno, R. A., Kormish, J. D., Tanner, J. R., Khan, D., Buchko, K., et al. (2015). Identification of vacuoles containing extraintestinal differentiated forms of Legionella pneumophila in colonized Caenorhabditis elegans soil nematodes. Microbiologyopen 4, 660–681. doi: 10.1002/mbo3.271
Hodgkin, J., Kuwabara, P. E., and Corneliussen, B. (2000). A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 10, 1615–1618. doi: 10.1016/S0960-9822(00)00867-8
Jansen, W. T., Bolm, M., Balling, R., Chhatwal, G. S., and Schnabel, R. (2002). Hydrogen peroxide-mediated killing of Caenorhabditis elegans by Streptococcus pyogenes. Infect. Immun. 70, 5202–5207. doi: 10.1128/IAI.70.9.5202-5207.2002
Jebamercy, G., Durai, S., Prithika, U., Marudhupandiyan, S., Dasauni, P., Kundu, S., et al. (2016). Role of DAF-21protein in Caenorhabditis elegans immunity against Proteus mirabilis infection. J. Proteomics 16, 30114–30122. doi: 10.1016/j.jprot.2016.03.047
JebaMercy, G., Pandian, S. K., and Balamurugan, K. (2011). Changes in Caenorhabditis elegans life span and selective innate immune genes during Staphylococcus aureus infection. Folia Microbiol. 56, 373–380. doi: 10.1007/s12223-011-0060-y
Jia, K., Chen, D., and Riddle, D. L. (2004). The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131, 3897–3906. doi: 10.1242/dev.01255
Jia, K., Thomas, C., Akbar, M., Sun, Q., Adams-Huet, B., Gilpin, C., et al. (2009). Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl. Acad. Sci. U.S.A. 106, 14564–14569. doi: 10.1073/pnas.0813319106
Kamaladevi, A., and Balamurugan, K. (2015). Role of PMK-1/p38 MAPK defense in Caenorhabditis elegans against Klebsiella pneumoniae infection during host-pathogen interaction. Pathog. Dis. 73, 1–9. doi: 10.1093/femspd/ftv021
Kamaladevi, A., Ganguli, A., Kmar, M., and Balamurugan, K. (2013). Lactobacillus Casei protects malathion induced oxidative stress and macromolecular changes in Caenorhabditis elegans. Pest Biochem. Physiol. 105, 213–223. doi: 10.1016/j.pestbp.2013.02.005
Kamaldevi, A., and Balamurugan, K. (2016). Lipopolysaccharide of Klebsiella pneumoniae attenuates immunity of Caenorhabditis elegans and evades by altering its supramolecular structure. RSC Adv. 6, 30070–30080. doi: 10.1039/C5RA18206A
Kapuy, O., Vinod, P. K., and Banhegyi, G. (2014). mTOR inhibition increases cell viability via autophagy induction during endoplasmic reticulum stress - an experimental and modeling study. FEBS Open Biol. 4, 704–713. doi: 10.1016/j.fob.2014.07.006
Kenific, C. M., Thorburn, A., and Debnath, J. (2010). Autophagy and metastasis: another double-edged sword. Curr. Opin. Cell Biol. 22, 241–245. doi: 10.1016/j.ceb.2009.10.008
Kim, D. (2008). Studying host-pathogen interactions and innate immunity in Caenorhabditis elegans. Dis. Model. Mech. 1, 205–208. doi: 10.1242/dmm.000265
Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., et al. (2002). A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626. doi: 10.1126/science.1073759
Kirienko, N. V., Kirienko, D. R., Larkins-Ford, J., Wahlby, C., Ruvkun, G., and Ausubel, F. M. (2013). Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 13, 406–416. doi: 10.1016/j.chom.2013.03.003
Komura, T., Yasui, C., Miyamoto, H., and Nishikawa, Y. (2010). Caenorhabditis elegans as an alternative model host for Legionella pneumophila, and protective effects of Bifidobacterium infantis. Appl. Environ. Microbiol. 76, 4105–4108. doi: 10.1128/AEM.03021-09
Kurz, C. L., and Ewbank, J. J. (2007). Infection in a dish: high-throughput analyses of bacterial pathogenesis. Curr. Opin. Microbiol. 10, 10–16. doi: 10.1016/j.mib.2006.12.001
Larance, M., Pourkarimi, E., Wang, B., Brenes Murillo, A., Kent, R., Lamond, A. I., et al. (2015). Global proteomics analysis of the response to starvation in C. elegans. Mol. Cell Proteomics 14, 1989–2001. doi: 10.1074/mcp.M114.044289
Long, X., Spycher, C., Han, Z. S., Rose, A. M., Muller, F., and Avruch, J. (2002). TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mrna translation. Curr. Biol. 12, 1448–1461. doi: 10.1016/S0960-9822(02)01091-6
Lopez Hernandez, Y., Yero, D., Pinos-Rodriguez, J. M., and Gibert, I. (2015). Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Front. Microbiol. 6:38. doi: 10.3389/fmicb.2015.00038
Marsh, E. K., and May, R. C. (2012). Caenorhabditis elegans, a model organism for investigating immunity. Appl. Environ. Microbiol. 78, 2075–2081. doi: 10.1128/AEM.07486-11
Means, T. K., and Aballay, A. (2011). Models to study ancient host-pathogen interactions: lessons from Crete. EMBO Rep. 12, 5–7. doi: 10.1038/embor.2010.205
Mohr, I., and Sonenberg, N. (2012). Host translation at the nexus of infection and immunity. Cell Host Microbe 12, 470–483. doi: 10.1016/j.chom.2012.09.006
Mortimore, G. E., and Schworer, C. M. (1977). Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 270, 174–176. doi: 10.1038/270174a0
Pritchard, R. E., Prassinos, A. J., Osborne, J. D., Raviv, Z., and Balish, M. F. (2014). Reduction of hydrogen peroxide accumulation and toxicity by a catalase from Mycoplasma iowae. PLoS ONE 9:e105188. doi: 10.1371/journal.pone.0105188
Rafii, S., Roda, D., Geuna, E., Jimenez, B., Rihawi, K., Capelan, M., et al. (2015). Higher risk of infections with PI3K-AKT-mTOR pathway inhibitors in patients with advanced solid tumors on phase I clinical trials. Clin. Cancer Res. 21, 1869–1876. doi: 10.1158/1078-0432.CCR-14-2424
Russell, R. C., Fang, C., and Guan, K. L. (2011). An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development 138, 3343–3356. doi: 10.1242/dev.058230
Scherz-Shouval, R., Shvets, E., Fass, E., Shorer, H., Gil, L., and Elazar, Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760. doi: 10.1038/sj.emboj.7601623
Schmitz, F., Heit, A., Dreher, S., Eisenacher, K., Mages, J., Haas, T., et al. (2008). Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur. J. Immunol. 38, 2981–2992. doi: 10.1002/eji.200838761
Schmutz, C., Ahrne, E., Kasper, C. A., Tschon, T., Sorg, I., Dreier, R. F., et al. (2013). Systems-level overview of host protein phosphorylation during Shigella flexneri infection revealed by phosphoproteomics. Mol. Cell. Proteomics 12, 2952–2968. doi: 10.1074/mcp.M113.029918
Shivers, R. P., Pagano, D. J., Kooistra, T., Richardson, C. E., Reddy, K. C., Whitney, J. K., et al. (2010). Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet 6:e1000892. doi: 10.1371/journal.pgen.1000892
Singh, V., and Aballay, A. (2006). Heat shock and genetic activation of HSF-1 enhance immunity to bacteria. Cell Cycle 5, 2443–2446. doi: 10.4161/cc.5.21.3434
Supek, F., Bosnjak, M., Skunca, N., and Smuc, T. (2013). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6:e21800. doi: 10.1371/journal.pone.0021800
Tan, M. W., Mahajan-Miklos, S., and Ausubel, F. M. (1999). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 96, 715–720. doi: 10.1073/pnas.96.2.715
Tattoli, I., Sorbara, M. T., Vuckovic, D., Ling, A., Soares, F., Carneiro, L. A., et al. (2012). Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575. doi: 10.1016/j.chom.2012.04.012
Vizcaino, J. A., Csordas, A., del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., et al. (2016). Update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456. doi: 10.1093/nar/gkv1145
Watanabe, R., Wei, L., and Huang, J. (2011). mTOR signaling, function, novel inhibitors, and therapeutic targets. J. Nucl. Med. 52, 497–500. doi: 10.2967/jnumed.111.089623
Yang, W., Dierking, K., Esser, D., Tholey, A., Leippe, M., Rosenstiel, P., et al. (2015). Overlapping and unique signatures in the proteomic and transcriptomic responses of the nematode Caenorhabditis elegans toward pathogenic Bacillus thuringiensis. Dev. Comp. Immunol. 51, 1–9. doi: 10.1016/j.dci.2015.02.010
Zak, O., and O'Reilly, T. (1993). Animal infection models and ethics – the perfect infection model. J. Antimicrob. Chemother. 31, 193–205. doi: 10.1093/jac/31.suppl_D.193
Keywords: Caenorhabditis elegans, Klebsiella pneumoniae, proteomics, host-pathogen interaction, liquid isoelectric focusing, PI3K/AKT/mTOR pathway
Citation: Kamaladevi A and Balamurugan K (2017) Global Proteomics Revealed Klebsiella pneumoniae Induced Autophagy and Oxidative Stress in Caenorhabditis elegans by Inhibiting PI3K/AKT/mTOR Pathway during Infection. Front. Cell. Infect. Microbiol. 7:393. doi: 10.3389/fcimb.2017.00393
Received: 27 March 2017; Accepted: 22 August 2017;
Published: 06 September 2017.
Edited by:
Amal O. Amer, The Ohio State University, Department of Internal Medicine, United StatesReviewed by:
Janakiram Seshu, University of Texas at San Antonio, United StatesCopyright © 2017 Kamaladevi and Balamurugan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krishnaswamy Balamurugan, YnN1cnlhckB5YWhvby5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.