- 1The State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
- 2College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 3Department of Biosciences, COMSATS Institute of Information Technology, Sahiwal, Pakistan
- 4Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
- 5Key Laboratory of Development of Veterinary Diagnostic Products, Ministry of Agriculture, Huazhong Agricultural University, Wuhan, China
- 6Hubei International Scientific and Technological Cooperation Base of Veterinary Epidemiology, Huazhong Agricultural University, Wuhan, China
Mycoplasma bovis is an important cause of bovine respiratory disease worldwide. To understand its virulence mechanisms, we sequenced three attenuated M. bovis strains, P115, P150, and P180, which were passaged in vitro 115, 150, and 180 times, respectively, and exhibited progressively decreasing virulence. Comparative genomics was performed among the wild-type M. bovis HB0801 (P1) strain and the P115, P150, and P180 strains, and one 14.2-kb deleted region covering 14 genes was detected in the passaged strains. Additionally, 46 non-sense single-nucleotide polymorphisms and indels were detected, which confirmed that more passages result in more mutations. A subsequent collective bioinformatics analysis of paralogs, metabolic pathways, protein-protein interactions, secretory proteins, functionally conserved domains, and virulence-related factors identified 11 genes that likely contributed to the increased attenuation in the passaged strains. These genes encode ascorbate-specific phosphotransferase system enzyme IIB and IIA components, enolase, L-lactate dehydrogenase, pyruvate kinase, glycerol, and multiple sugar ATP-binding cassette transporters, ATP binding proteins, NADH dehydrogenase, phosphate acetyltransferase, transketolase, and a variable surface protein. Fifteen genes were shown to be enriched in 15 metabolic pathways, and they included the aforementioned genes encoding pyruvate kinase, transketolase, enolase, and L-lactate dehydrogenase. Hydrogen peroxide (H2O2) production in M. bovis strains representing seven passages from P1 to P180 decreased progressively with increasing numbers of passages and increased attenuation. However, eight mutants specific to eight individual genes within the 14.2-kb deleted region did not exhibit altered H2O2 production. These results enrich the M. bovis genomics database, and they increase our understanding of the mechanisms underlying M. bovis virulence.
Introduction
Mycoplasma bovis is a member of the Mycoplasmataceae family in the class of Mollicutes. It was first identified as a causative agent of bovine mastitis in 1961, and it was later recognized as an important pathogen of bovine respiratory disease in 1976 (Caswell and Archambault, 2007). Although it has been 56 years since M. bovis has been identified, there is limited understanding regarding its pathogenesis and virulence.
Compared with other bacteria, pathogenic Mycoplasma species have not been found to produce conventional toxins. Although ADP-ribosyl-transferase was preliminarily described as a possible toxin in a Mycoplasma pneumoniae strain that exhibits ADP-ribosyltransferase activity and elicits a distinct pattern of cytopathology in mammalian cells (Kannan and Baseman, 2006), it is difficult to distinguish pathogenic and non-pathogenic Mollicutes based on such virulence-related factors. Liproproteins and secretory proteins might contribute to bacterial virulence. Membrane lipoproteins, such as variable surface proteins (Vsps), enolase, and Vpmax, play significant roles in the adhesion of M. bovis to host cells (Burki et al., 2015). Subsequent invasion of host cells may be beneficial for in vivo survival and the dissemination of M. bovis to different sites in its hosts (Kleinschmidt et al., 2013). Regarding secretory proteins, only a few, including one M. bovis secretory protein, have been discovered (Zhang et al., 2016). Secondary metabolites such as hydrogen peroxide (H2O2) are considered to play a significant role in the pathogenesis of some Mycoplasma species, including M. pneumoniae (Hames et al., 2009) and Mycoplasma mycoides subsp. mycoides small colony (MmmSC) (Pilo et al., 2005). However, variations in H2O2 production might not correlate with M. bovis virulence (Schott et al., 2014). Instead, M. bovis might modulate the host immune response by suppressing interferon-γ and tumor necrosis factor-α production by invading immune cells to support its persistence and systemic dissemination (Mulongo et al., 2014). Genome sequences might provide more evidence that explains Mycoplasma pathogenesis at the genetic level. Currently, the genomes of 28 M. bovis strains, including the wild-type strain HB0801 and the three attenuated strains in the present study, have been sequenced and published (Li et al., 2011; Wise et al., 2011; Qi et al., 2012). Pathogenicity islands (PAIs) play a significant role in genome evolution and pathogenesis because many virulence-related factors are shared and acquired by PAIs. However, no PAIs and secretory systems have been detected in any Mycoplasma species (Guo and Wei, 2012). Using the virulence factors database (VFDB), some virulence genes were identified in the M. bovis genome (Parker et al., 2016), but their impact on M. bovis virulence remains to be investigated.
M. bovis was first isolated from the milk of a cow with mastitis in 1983 (Chen et al., 1983) and subsequently from lesioned lung tissue of a calf with pneumonia in 2008 in China (Qi et al., 2012). To develop candidate live vaccines against M. bovis, one strain, HB0801, which was isolated from lesioned lung tissue, was passaged continuously in vitro, and three strains that were passaged 115, 150, and 180 times, designated as M. bovis HB0801-P115, HB0801-P150, and HB0801-P180, respectively, were tested individually in cattle. The resulting clinical signs and pathological changes demonstrated that their virulence decreased with increasing numbers of passages (Zhang et al., 2014). Thus, a comparative genomics analysis of the virulent, wild-type strain HB0801 and these three attenuated strains might reveal some novel clues regarding the pathogenesis and virulence mechanisms of M. bovis.
Hence, in this study, we sequenced the complete genomes of these three attenuated strains and performed a comprehensive genomic analysis between the wild-type and the three attenuated strains. Based on the results, we hypothesize that a 14.2-kb deleted DNA fragment, single-nucleotide polymorphisms (SNPs), and indels probably affect the expression of some potential virulence-related proteins in the attenuated strains. In addition, the decreased capability to produce H2O2 in the attenuated strains was confirmed.
Materials and Methods
Mycoplasma Strains and Culture Conditions
M. bovis strain HB0801 (GenBank accession no. NC_018077.1) was isolated from the lung of infected beef cattle in Hubei Province, China, and its genome was fully sequenced by our laboratory (Qi et al., 2012). The M. bovis HB0801 attenuated strains HB0801-P115, HB0801-P150, and HB0801-P180, abbreviated as P115, P150, and P180, respectively, which exhibit progressively decreasing virulence, were used (Zhang et al., 2014). All the strains were propagated in pleuropneumonia-like organism (PPLO) medium supplemented with 10% horse serum (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C for 48–72 h as described previously (Zhang et al., 2014).
Library Construction, DNA Sequencing, and Assembly
The DNA of strains P115, P150, and P180 was extracted using bacterial genomic DNA extraction kits (Tiangen, Beijing, China). The 454 pyrosequencing method was used to determine the whole genome sequences of strains P115, P150, and P180. Three paired-end sequencing libraries with 8-kb inserts were constructed at the China Tianjin Biochip Corporation (Tianjin, China). For each sample, one-fourth of a PicoTiterPlate was run on a Roche/454 GS FLX sequencer (454 Life Sciences, Branford, CT, USA) using titanium chemistry according to the manufacturer's recommendations. Finally, 83,750,973 bases with 286,408 reads were obtained for strain P115, while 80,961,239 bases with 277,020 reads were obtained for strain P150, and 83,589,289 bases with 260,056 reads were obtained for strain P180, resulting in 85.7-fold (P115), 82.8-fold (P150), and 85.5-fold (P180) depths of sequencing. All the reads for each genome were assembled de novo by the GS De Novo Assembler (version 2.6). Approximately 95% of reads were assembled for each genome, resulting in 11 scaffolds with 52 non-redundant contigs for strain P115, three scaffolds with 49 non-redundant contigs for strain P150, and one scaffold with 53 non-redundant contigs for strain P180. All the scaffolds were ordered and oriented according to the genome architecture of strain HB0801 (Qi et al., 2012). The N50 contig lengths of the large contigs (>1 kb) of each strain were 31,437 bp (P115), 31,456 bp (P150), and 31,042 bp (P180). The total numbers of base pairs of the non-redundant contigs were 916,922 bp (P115), 919,584 bp (P150), and 918,470 bp (P180), which agrees with the previously reported genome size of 991,702 bp for strain HB0801. To fill gaps within the scaffolds, polymerase chain reactions (PCRs) and capillary electrophoresis sequencing were performed with primers designed near the gaps. After gap filling, all the generated reads were respectively mapped to the corresponding full genome sequence using Burrows–Wheeler Aligner software (Li and Durbin, 2010). With the help of deep sequencing coverage, possible homopolymer errors resulting from the 454 sequencing method were rectified manually.
Genome Annotation and Analysis
Open reading frames of the three passaged genomes (P115, P150, and P180) were predicted initially using Glimmer 3.02 (http://www.cbcb.umd.edu/software/glimmer/) and then modified using the translated nucleotide Basic Local Alignment Search Tool (BLAST) algorithm (http://blast.ncbi.nlm.nih.gov/) and compared to the genome of the primary strain HB0801 (GenBank accession no. CP002058). The functions of the coding sequences in the passaged strains were defined by referencing strain HB0801 (Qi et al., 2012). A comparative analysis between these four strains (P115, P150, P180, and HB0801) was conducted using MEGA 5.0 (Tamura et al., 2011) and Mauve 2.3.1 (Darling et al., 2004) genome alignment software. In addition, we detected differences in SNPs and indel sites among these strains.
Confirmation of the 14.2-kb Deleted Region by PCR
To confirm the presence of the 14.2-kb deleted region during continuous passage, a putative lipoprotein-encoding gene (Mbov_0732) present in the deleted region was selected for PCR with forward (5′–AGCGACCAAAATACTAGAC –3′) and reverse (5′–TCGTTGCCACTGTATTCA–3′) primers using the following program: 95°C 3 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C 2 min, followed by 72°C for 15 min and 16°C for 5 min.
Confirmation of SNPs and Indels
SNPs and indels might affect the expression of essential genes. Sixty-seven pairs of primers specific to the flanking sequences of the SNPs and indel sites were designed (Table S1) and Sanger DNA sequencing was performed with an ABI 3730 sequencer by China Tianjin Biochip Corporation (Tianjin, China).
Bioinformatics Analysis of Predicted Proteins
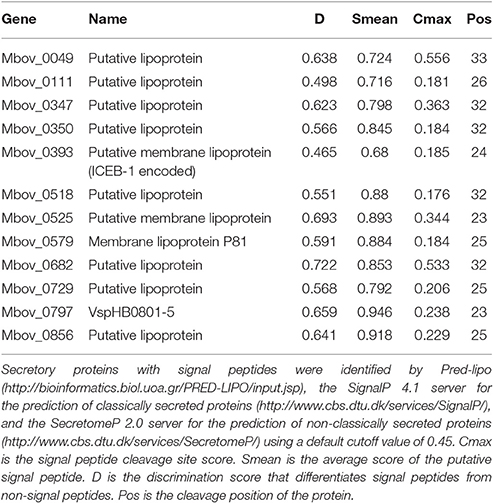
Differentially expressed proteins predicted by genome comparisons of the virulent HB0801 strain to its attenuated P115, P150, and P180 derivatives were classified into two categories: (i) proteins deleted in P115, P150, or P180 (Table 1) and (ii) proteins displaying non-sense SNPs and indels (Table 2). Protein functions and related metabolic pathways were assigned according to the Uniprot database (http://www.uniprot.org/) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=mbi), respectively. To determine whether proteins are secreted and contain a signal peptide, we used Pred-lipo (http://bioinformatics.biol.uoa.gr/PRED-LIPO/input.jsp), the SignalP 4.1 server for the prediction of classical secreted proteins (http://www.cbs.dtu.dk/services/SignalP/), and the SecretomeP 2.0 server for the prediction of non-classical secreted proteins (http://www.cbs.dtu.dk/services/SecretomeP/). Moreover, to assess whether the SNPs were present at active sites or domains of the proteins and to obtain the conserved domains in the secretory proteins, we used the National Center for Biotechnology Information (NCBI) conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Paralogs of the genes and percentages of identity between paralogs were determined with the NCBI BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Protein–Protein interactions were obtained using the STRING database (www.string-db.org).
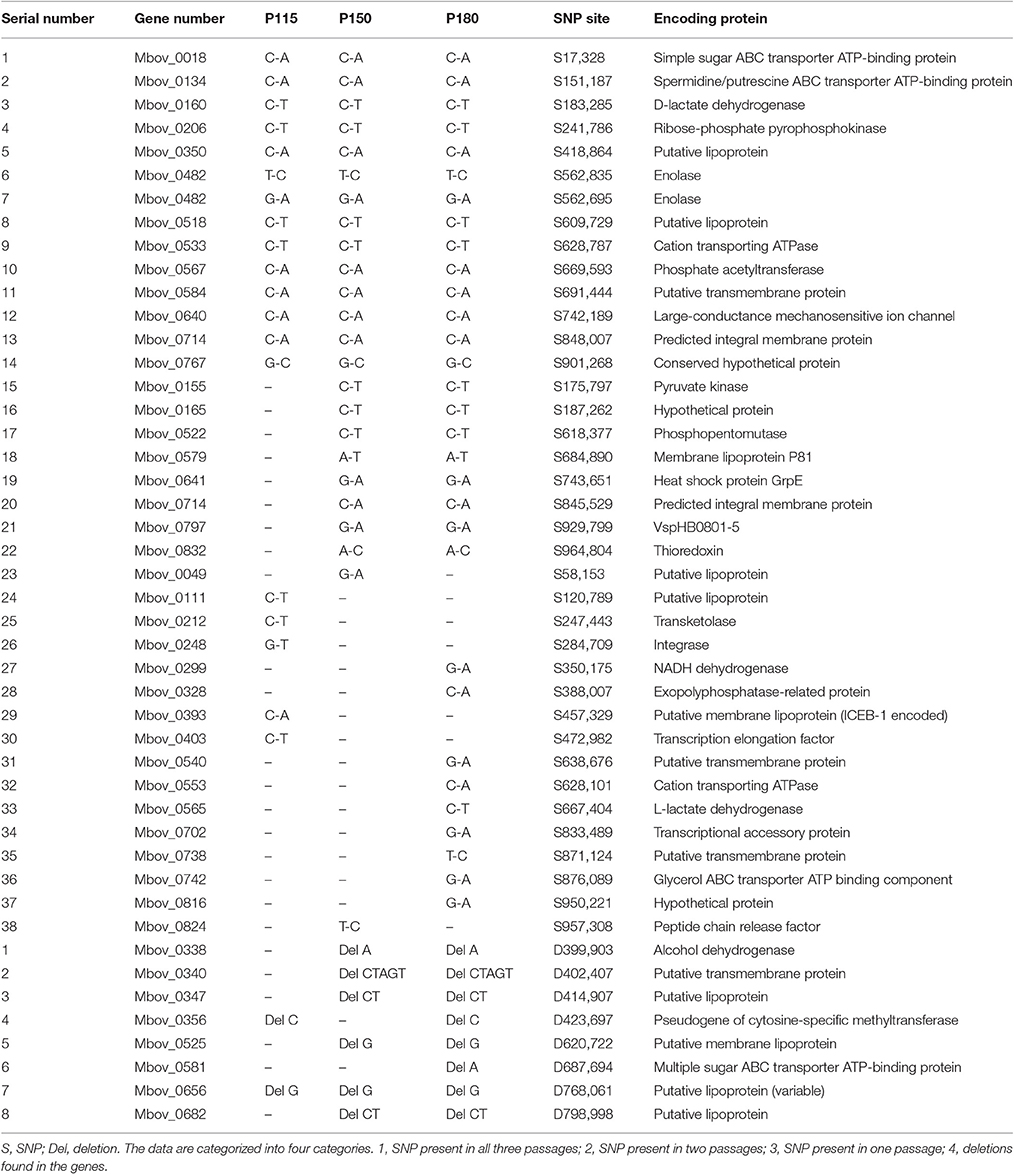
Table 2. SNPs and indels in the three attenuated strains, compared with strain HB0801, after resequencing and PCR confirmation.
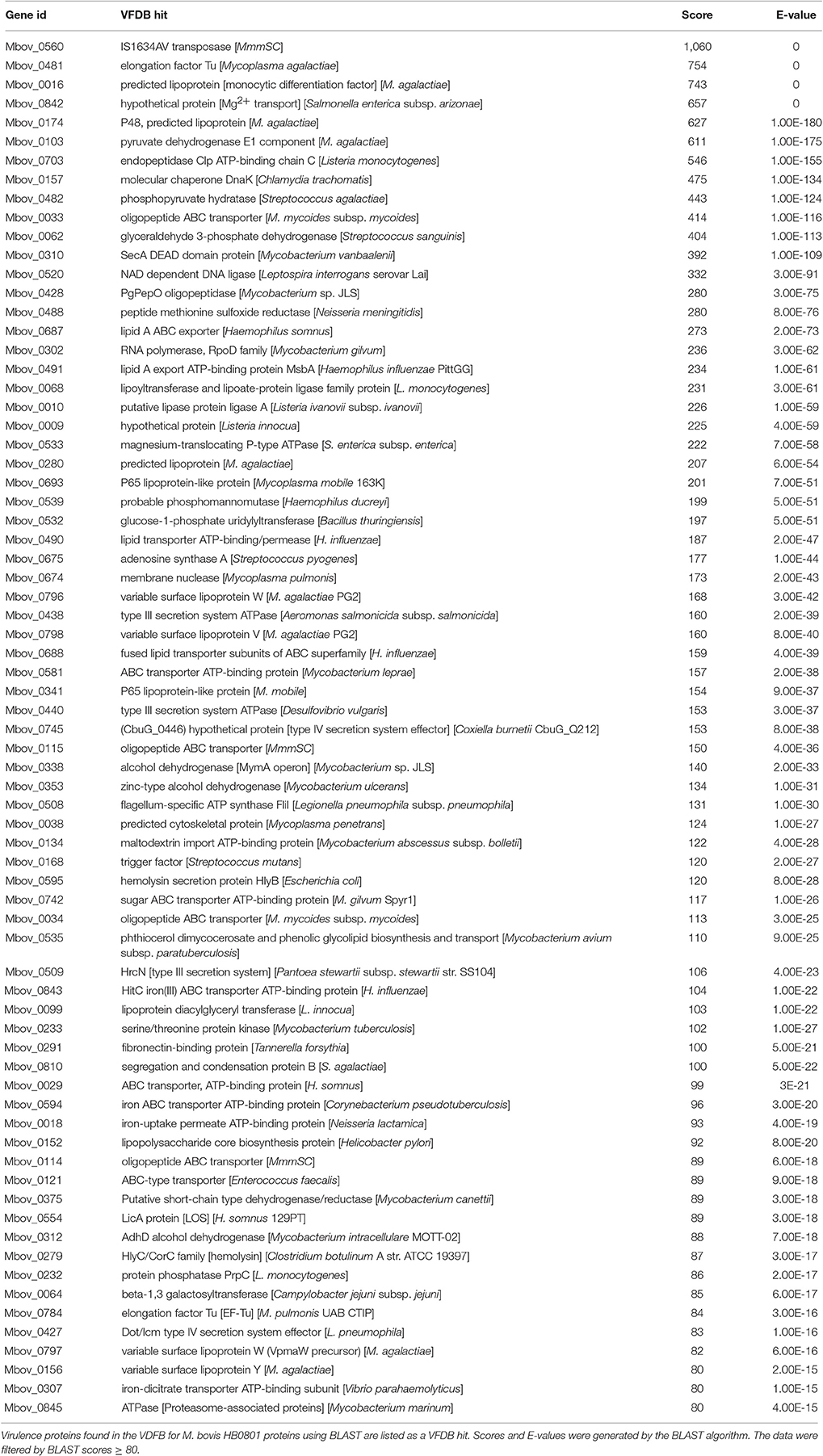
The involvement of these proteins in virulence was further confirmed based on the whole genome of M. bovis HB0801 using the VFDB (http://www.mgc.ac.cn/VFs/) (Chen et al., 2016). The amino acid sequences of all the proteins were obtained from the NCBI. Each protein was aligned individually against the VFDB full dataset by the BLAST algorithm. A matrix was created by VFDB output, including VFDB hits against each protein in M. bovis, a related BLAST score, and an E-value. The matrix was filtered based on a BLAST score ≥80.
H2O2 Production Assay for Representative Passaged Strains and Mutants
H2O2 production was measured as described previously in Mycoplasma mycoides subsp. capri (Allam et al., 2012). Briefly, eight transposon-disrupted mutants in the deleted region of the attenuated strains, including Mbov_0723, Mbov_0724, Mbov_0725, Mbov_0727, Mbov_0730, Mbov_0732, Mbov_0734, and Mbov_0735, as well as different passages of M. bovis HB0801, including P1, P25, P50, P75, P100, P115, P150, and P180, were grown to mid-logarithmic phase. The transposon-disrupted mutants were prepared using the wild-type HB0801 strain and a Tn4001 transposon. The transposon disrupted the abovementioned genes that are present in the deleted region mentioned in Table 1. Moreover, the Mbov_0723, Mbov_0724, Mbov_0725, Mbov_0727, Mbov_0730, Mbov_0732, Mbov_0734, and Mbov_0735 genes were transposon-disrupted at positions 85,6071, 85,7345, 858,521, 860,287, 863,914, 864,069, 866,042, and 866,786, respectively, in the genome. To assess H2O2 production, bacteria were harvested by centrifugation at 15,400 g for 4 min at 4°C. The pellets were washed three times in incubation buffer (67.7 mM HEPES, pH 7.3; 140 mM NaCl, and 7 mM MgCl2). After the final wash, the M. bovis pellets were resuspended in incubation buffer to a density of 108 cells/ml and incubated for 20 min at 37°C. After the incubation, 10 μl of glycerol (10 mM) was added, and the samples were kept at 37°C for 30 min. H2O2 production was determined using commercial H2O2 assay kits (Cayman Chemical, Ann Arbor, MI, USA). Standard curve determination and specificity quality controls were performed according to the instructional manual. The experiments were performed in three independent replicates.
Statistical Analysis
To determine the statistical significance among the differences in SNPs after comparing all the genes of M. bovis, as well as among different SNP and indel types, a z-test for proportions (z = pˆ-p0/√p0(1 − p0)/n) was applied to generate a p-value. Moreover, to determine the statistical significance of the genes involved in different KEGG pathways, a KEGG enrichment analysis was performed using the KOBAS 2.0 web server, and Fisher's exact test was used to analyze the data (Xie et al., 2011). To analyze differences in H2O2 production among the different strains, two-way analysis of variance was performed using GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA). p < 0.05 were considered to be significantly different and are marked with an asterisk in the figures, while p < 0.01 were considered to very significantly different and are marked with two asterisks in the figures.
Nucleotide Sequence Accession Numbers
The complete genome sequences of the three attenuated strains were deposited in the GenBank database with the accession numbers CP007589 for P115, CP007590 for P150, and CP007591 for P180.
Results and Discussion
A 14.2-kb Deleted Fragment is Present in the Genomes of the Three Attenuated Strains
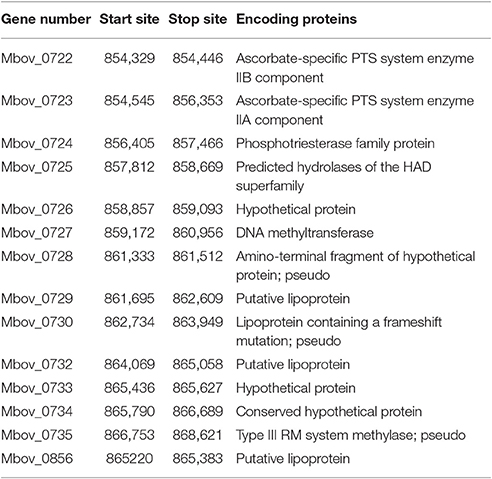
Because of a lack of genetic tools and limited research techniques, there is limited understanding of the pathogenesis and virulence-related factors of M. bovis. Hence, the whole genomes of strains P115, P150, and P180 were sequenced and assembled, and a comparative study was performed to identify significant virulence-related factors. The genome structures of the virulent M. bovis HB0801 strain and the three attenuated strains are shown in Figure 1A. An analysis of the three attenuated strains and the wild-type HB0801 strain revealed that all the genomes of strains P115, P150, and P180 lost a 14.2-kb fragment compared with strain HB0801 (Figure 1B and Table 1). Further analysis of the deleted fragment showed that it consists of 14 putative genes encoding ascorbate-specific phosphotransferase system (PTS) system enzyme IIB and IIA components, a phosphotriesterase family protein, predicted hydrolases of the haloacid dehalogenase (HAD) superfamily, DNA methyltransferase, and a type III restriction-modification (RM) system methylase, as shown in Table 1. In addition, 38 strains that were passage between one and 115 times were shown by PCR to have a deletion in the Mbov_0732 gene. The results demonstrated that the 14.2-kb deletion occurred in passage 115 and onward as shown in Figure S1, indicating that the deletion was stably maintained in the passaged strains after its occurrence.
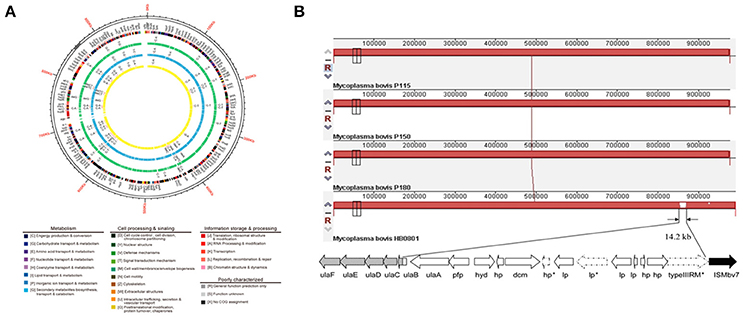
Figure 1. Genomic comparison of M. bovis strains HB0801 (P1), P115, P150, and P180. (A) Genome structures of the M. bovis HB0801 strain and the three attenuated strains; (B) Serial propagation of HB0801 resulted in the deletion of a 14.2-kb fragment in the genome of the three attenuated strains. The red lines show the lengths of the genomes of respective strains. The lengths of the P115, P150, and P180 strains are reduced and shown in dashes at the ends of the lines.
SNPs and Indels
In addition to the 14.2-kb deleted region, a genome comparison between strain HB0801 and the attenuated strains revealed many SNPs and indels. Sixty-seven non-sense SNP sites were tested with PCR and DNA sequencing, and 38 SNPs were confirmed (Table 2). In agreement with the increasing attenuation tendency resulting from continuous passage, strains that were subjected to more passages had more SNPs, and the SNPs in the strains that were subjected to fewer passages were shared by the higher passaged strains. The numbers of SNPs were 31, 24, and 19 for the P180, P150, and P115 strains, respectively. In addition, eight nucleotide deletions were found, and their frequencies of occurrence followed a pattern that was similar to that of the SNPs (Table 2). These changes in the genomes were analyzed statistically with a z-test of proportions, and the results showed that there were significant differences in the numbers of SNPs, indels, and SNPs+indels between the wild-type strain and each of the attenuated strains, and between any two attenuated strains (normalized p < 0.00001).
Detection of Paralogs in the M. bovis Genome
Theoretically, the functions of some genes containing SNPs or deletions could be compensated by their paralogs in the genome. Hence, the FASTA sequences of such paralogs were aligned with a reference protein by the BLAST algorithm to determine the percentage of identity between them (Table S2); a reference protein is a protein that is found in our genomics data (Tables 1, 2). The results showed that the proteins that have paralogs in our genomics data included four of the 14 genes in the 14.2-kb deleted region (Mbov_0727, Mbov_0729, Mbov_0732, and Mbov_0856) and 14 of the 46 genes containing SNPs or indels (Mbov_0018, Mbov_0049, Mbov_0111, Mbov_0134, Mbov_0338, Mbov_0347, Mbov_0350, Mbov_0393, Mbov_0518, Mbov_0525, Mbov_0581, Mbov_0682, Mbov_0742, and Mbov_0832). Hence, the genes responsible for virulence attenuation would likely include the other 10 genes in the 14.2-kb deleted region and the 32 genes with SNPs and indels, but which lacked paralogs.
Prediction of Protein–Protein Interactions
The proteins listed in Tables 1, 2 were input into the STRING database to predict potential protein-protein interactions using the type strain M. bovis PG45 as the database default reference. Protein–protein interaction maps are given for 13 of the 60 input proteins (Figure 2). Different colored lines show different types of interactions. Hence, more lines show more interactions and an increased probability of a protein-protein interaction. The interactions among the proteins are highlighted in evidence view (Figure 2A). In evidence view, all possible interactions are shown. Different colored lines show different types of interactions e.g., gene fusion, co-occurrence, co-expression, experiments, databases, text mining etc. While in interactive view, the interactive proteins are clustered together based on interaction and co-occurrence. The evidence (Figure 2A) and interactive views (Figure 2B) showed that the most significant proteins that are likely to interact include enolase (Mbov_0482), pyruvate kinase (Mbov_0155), transketolase (Mbov_0212), L-lactate dehydrogenases (Mbov_0565), and D-lactate dehydrogenases (Mbov_0160), suggesting that these proteins might function together, and thus, that they are the strongest candidates for explaining the attenuated virulence of highly passage M. bovis strains.
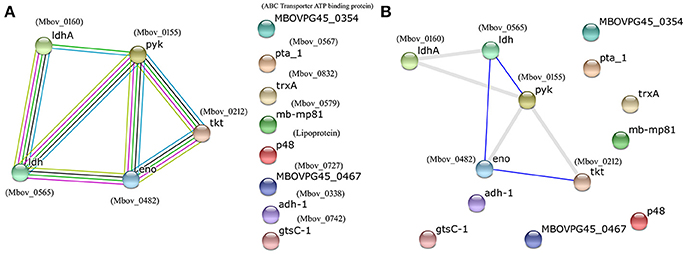
Figure 2. Protein–protein interaction map of the available proteins from the list of analyzed proteins from Tables 1, 2. Different colored lines show different types of interactions. Hence, more lines show more interactions and an increased probability of a protein–protein interaction. The image and related details were retrieved from the STRING database (http://www.string-db.org/). (A) Evidence view; (B) Interactive view. In evidence view, all possible interactions are shown. Different colored lines show different types of interactions e.g., gene fusion, co-occurrence, co-expression, experiments, databases, text mining etc. While in interactive view, the interactive proteins are clustered together based on interaction and co-occurrence.
Pathway Enrichment Assay
A KEGG enrichment analysis was performed to determine whether the mutated genes in Tables 1, 2 are enriched in some important metabolic pathways. Fifteen genes were shown to be enriched in 15 metabolic pathways. Furthermore, 10 of the 15 genes were involved in more than one pathway (Figure 3 and Table S3). The 10 genes and the involved numbers of pathways numbers are Mbov_0155 and Mbov_0482 (seven), Mbov_0206 and Mbov_0565 (six), Mbov_0212 (five), Mbov_0567 (four), Mbov_0338 (three), and Mbov_0522, Mbov_0722, and Mbov_0723 (two). Among them, Mbov_0155, Mbov_0212, Mbov_0482, and Mbov_0565 were also identified in the protein-protein interaction analysis. Theoretically, proteins involved in more pathways should be more likely to play a significant role in M. bovis virulence. However, among the pathways, biosynthesis of secondary metabolites, biosynthesis of antibiotics, carbon metabolism, pyruvate metabolism, biosynthesis of amino acids, glycolysis/gluconeogenesis, pentose phosphate pathway, purine metabolism, and ATP-binding cassette (ABC) transporters were enriched in more pathways (six, six, five, four, four, four, three, three, and three, respectively).
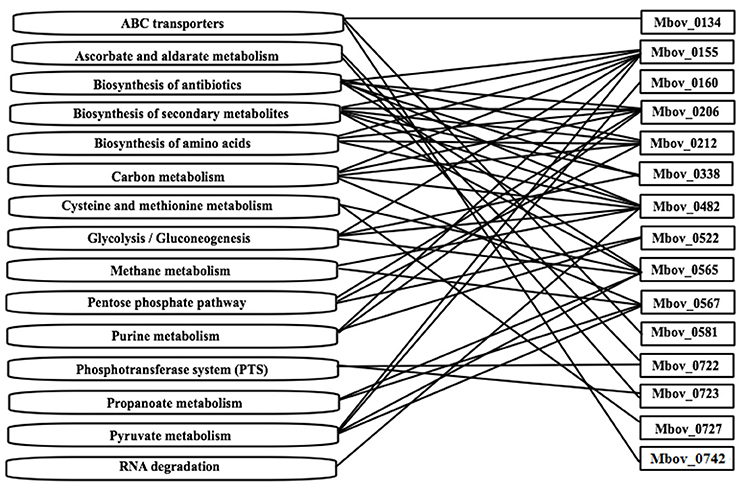
Figure 3. The mutated genes from Tables 1, 2 that are enriched in some metabolic pathways. Fifteen genes were shown to be enriched in 15 metabolic pathways. Ten of the fifteen genes are involved in more than one pathway.
Conserved Domain Analysis
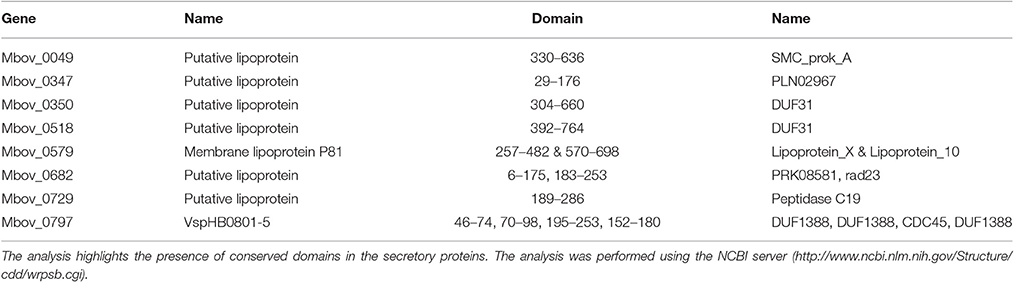
Twelve predicted secretory proteins with signal peptides, which includes those encoded by 10 genes (Mbov_0049, Mbov_0111, Mbov_0347, Mbov_0350, Mbov_0393, Mbov_0518, Mbov_0525, Mbov_0579, Mbov_0682, and Mbov_0797) that have SNPs (Table 2) and the Mbov_0729 and Mbov_0856 genes, which are deleted, were identified (Table 1). A conserved domain analysis (Table 4) identified the presence of conserved domains in the eight secretory proteins mentioned in Table 3, but not in Mbov_0111, Mbov_0393, Mbov_0525, and Mbov_0856. Liproproteins and secretory proteins might contribute to bacterial virulence and pathogenesis because they are involved in many phenomena, ranging from cellular physiology through immune responses to virulence. Some secretory proteins were discovered previously in M. bovis (Khan et al., 2016; Zhang et al., 2016). Many other genes in the M. bovis genome are predicted to encode putative lipoproteins and secretory proteins, but whether these proteins exist in a functional form and how they are related to virulence need to be determined. The most significant conserved domains with available functions include CDC45 (Mbov_0797), DUF1388 (Mbov_0797), structural maintenance of chromosomes (SMC) (Mbov_0049), Rad23 (Mbov_0682), and peptidase C19 (Mbov_0729) (Table 4). CDC45 is required for the initiation of DNA replication (Saha et al., 1998). Members of the DUF1388 family function as the main targets for neurofilament-directed protein kinases in vivo. SMC proteins are essential for successful chromosome transmission during replication, and segregation of the genome, including chromosome condensation, recombination, DNA repair, and epigenetic silencing of gene expression (Haering et al., 2002). Rad23 family proteins are used for targeting nucleotide excision repair to specific parts of the genome. Peptidase C19 is a deubiquitinating enzyme that can deconjugate ubiquitin or ubiquitin-like proteins from ubiquitin-conjugated proteins (De Jong et al., 2006). The functions of the other domains are currently unknown.
The Virulence-Related Factors Identified by the VFDB
To further investigate the virulence-related factors that contribute to the attenuation of highly passaged M. bovis strains, all the proteins of M. bovis were analyzed using the VFDB. In the VFDB full dataset, all proteins related to known and predicted virulence-related factors are present. Seventy-two genes in the M. bovis genome were shown to encode virulence-related factors based on a BLAST score ≥80 (Table 5). Although no virulence-related factor-encoding genes were found in the 14.2-kb deleted region, eight genes overlapped with those containing SNPs and indels (Table 2). In decreasing order of BLAST scores, they are Mbov_0482, Mbov_0533, Mbov_0581, Mbov_0338, Mbov_0134, Mbov_0742, Mbov_0018, and Mbov_0797.
Analysis of Critical Deleted Genes and Genes with SNPs and Indels
By combining the above results for the deleted genes and the genes with SNPs and indels, 11 critical genes that likely contribute to the attenuation of highly passaged M. bovis strains were identified, and they include (in order of decreasing importance) Mbov_0722, Mbov_0723, Mbov_0482, Mbov_0565, Mbov_0155, Mbov_0581, Mbov_0742, Mbov_0299, Mbov_0212, Mbov_0797, and Mbov_0567.
Among the deleted genes, Mbov_0722 and Mbov_0723 are highlighted. They encode ascorbate-specific PTS enzyme IIB and IIA components, respectively, which function in ascorbate and aldarate metabolism pathways by involving the PTS, the major carbohydrate transport system in bacteria, and they are responsible for the conversion of L-ascorbate into L-ascorbate-6 phosphate, as shown in Figure S2 (Postma et al., 1993). The final product of this pathway is D-xylulose-5 phosphate, which is an intermediate in the pentose phosphate pathway. As is known, the primary purpose of this pathway is to generate a reducing equivalent of NADPH that can be used in reductive biosynthesis reactions, while the production of the pentose phosphate pathway intermediates ribose 5-phosphate and erythrose 4-phosphate are used to synthesize nucleotides and nucleic acids, and aromatic amino acids, respectively. In addition, this sugar has a role in gene expression, mainly by promoting the ChREBP transcription factor in the well-fed state (Iizuka and Horikawa, 2008). Hence, because of the deficiencies of both proteins in the attenuated strains, these related metabolic functions would be less efficient. Because the occurrence of the 14.2-kb deleted region began at passage 115 and was maintained from passages 115 through 180, these metabolic defects might contribute to the increasing attenuation of highly passaged M. bovis strains.
The enolase encoded by Mbov_0482 is considered to be a virulence-related factor, and it participates in seven metabolic pathways and interacts with other proteins as described previously. It catalyzes the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate. This reaction is present in the glycolysis/gluconeogenesis pathways of M. bovis, as shown in Figure S3. Prokaryotic α-enolase may contribute to pathophysiological processes (Pancholi, 2001). A surface-associated enolase is an adhesion-related factor of M. bovis that contributes to adherence by binding plasminogen (Song et al., 2012). The immunogenicity of enolase has also been observed in Mycoplasma synoviae (Bercic et al., 2008) and Mycoplasma capricolum subsp. capripneumoniae (Zhao et al., 2012). Hence, enolase might be a significant protein that contributes to the virulence of M. bovis. However, two SNPs were not detected inside the enolase-encoding gene, but were located approximately 300 bp upstream of the gene, suggesting that both SNPs might cause some change in the promoter region.
Like enolase, L-lactate dehydrogenase encoded by Mbov_0565 is another protein that is important for metabolism and as a virulence-related factor. It interconverts L-lactate to pyruvate. Additionally, this enzyme works in many metabolic pathways, including glycolysis/gluconeogenesis, cysteine and methionine metabolism, pyruvate metabolism, and propanoate metabolism. Hence, this lactate dehydrogenase plays significant roles in a variety of metabolic processes. Moreover, L-lactate dehydrogenase was shown to be surface expressed and to interact with plasminogen (Grundel et al., 2015). Furthermore, because of the SNP in the L-lactate dehydrogenase-encoding gene, a polar amino acid (threonine) was converted to a non-polar amino acid (methionine).
Pyruvate kinase (Mbov_0155) is also a very active protein in metabolism and energy production. It catalyzes the conversion of phosphoenolpyruvate to pyruvate (Figure S4). Then, mycoplasmas generate ATP via a proton-translocating ATP synthase by oxidizing organic acids (pyruvate and lactate) to acetate and CO2. Moreover, the pyruvate kinase present in the genome of Mycoplasma suis is proposed to be required for the conversion of all NDPs and dNDPs to NTPs and dNTPs, respectively (Pollack et al., 2002). Hence, this protein is very important for energy production in Mycoplasma species, and the SNP in the pyruvate kinase-encoding gene may lead to the attenuation of M. bovis.
The different molecular functions of transketolase (Mbov_0212) include metal ion binding and transferase activity, and it is a key enzyme in the non-oxidative branch of the pentose phosphate pathway that transfers a two-carbon glycolaldehyde unit from a ketose donor to aldose-acceptor sugars (Jores et al., 2009). Moreover, transketolase is an immunodominant membrane protein that may be used as a biomarker for the serological diagnosis of contagious agalactia caused by M. mycoides subsp. capri (Corona et al., 2013). Hence, this protein may be involved in the immunogenicity and virulence of M. bovis.
ABC transporter proteins have different functions, including ATP binding, ATPase activity, and catalyzing the transmembrane movement of substances, such as importing sugars, amino acids, peptides, metal ions, and phosphates, and effluxing toxins, drugs, and proteins (Higgins et al., 1986). The Mbov_0742 gene, which contained a SNP only in strain P180, encodes a glycerol ABC transporter protein. The Mbov_0581 gene, which had an indel only in strain P180, encodes a multiple sugar ABC transporter protein. Both genes are considered to encode virulence-related factors in the VFDB. In addition, the Mbov_0134 and Mbov_0018 genes encoding ABC proteins responsible for the uptake of simple sugar and spermidine/putrescine were found to have SNPs in all three attenuated strains.
NADH dehydrogenase (Mbov_0299) is considered to be a virulence-related factor. It generates energy by transferring electrons from NADH (oxidation) to quinine. NADH and NADPH are possibly essential for the growth of M. suis (Guimaraes et al., 2011). In Mycobacterium tuberculosis, a mutant lacking NuoG, a subunit of the type I NADH dehydrogenase complex, exhibited attenuated growth in vivo (Blomgran et al., 2012). Hence, this gene mutation might be significantly related to the further attenuation of M. bovis strain P180.
Phosphate acetyltransferase (PTA) (Mbov_0567) catalyzes the formation of acetyl phosphate and acetyl CoA, which are used in many metabolic pathways, e.g., in taurine and hypotaurine metabolism, pyruvate metabolism, propanoate metabolism, and methane metabolism (Figure S5). Acetyl-phosphate regulates various cellular processes, including cell division, outer membrane protein expression, osmoregulation, and biofilm development. Moreover, acetyl phosphate is required for the activation of the Rrp2-RpoN-RpoS pathway, which serves as a global signal in bacterial pathogenesis by activating virulent genes (Xu et al., 2010). Moreover, Salmonella enterica serovar Typhimurium (Kim et al., 2006) and Vibrio cholerae (Chiang and Mekalanos, 1998) PTA mutants were shown to exhibit impaired growth and attenuated virulence. In addition, because of a SNP in Mbov_0567, a proline residue was converted to threonine, resulting in polarity change that might be significant in virulence attenuation.
In addition, membrane proteins influence cell shape, cell division, motility, and adhesion to host cells, and they are thought to be integrally involved in the pathogenesis of mycoplasmas. As is known, adhesion and invasion are generally considered to be virulence-associated processes. M. bovis can adhere to and invade epithelial cells and immune cells. Membrane lipoproteins, such as Vsps, enolase, and Vpmax, play significant roles in the adhesion of M. bovis to host cells (Burki et al., 2015). Five genes encoding membrane proteins were shown to have SNPs, and four of them were predicted to encode secretory proteins with signal peptides, including Mbov_0393, Mbov_0525, Mbov_0579, and Mbov_0797. Among them, Mbov_0797 encodes a Vsp, and it was predicted to be a virulence-related factor, Mbov_0579 was predicted to encode the ADP-ribosyltransferase CDTa, which contains functional domains of the community-acquired respiratory distress syndrome toxin of M. pneumoniae (Kannan et al., 2014).
Alternation of H2O2 Production in the Attenuated Strains
H2O2 is thought to be a significant virulence related factor in M. pneumoniae, M. mycoides, and Mycoplasma ovipneumoniae. Secondary metabolites are considered to play significant roles in the pathogenesis of some Mycoplasma species (Pilo et al., 2005; Hames et al., 2009). For example, H2O2 was demonstrated to be a major virulence-related factor that leads to cell death and lipid peroxidation in M. pneumoniae (Hames et al., 2009) and MmmSC (Pilo et al., 2005). Moreover, activation of glycerol utilization and overproduction of H2O2 occurred during intracellular infection with Mycoplasma gallisepticum (Matyushkina et al., 2016). Although an in vitro H2O2 assay for M. bovis field strains showed that variations in H2O2 production did not correlate with M. bovis virulence (Schott et al., 2014), in vitro passaging an M. bovis strain resulted in decreased levels of H2O2 production (Khan et al., 2005).
To confirm that H2O2 production was affected by the mutations in the attenuated strains, H2O2 production was tested in the wild-type strain HB0801 (P1), strains of various passages (25, 50, 75, 100, 115, 150, and 180), and eight mutants specific to the genes in the 14.2-kb deleted region (Mbov_0723, Mbov_0724, Mbov_0725, Mbov_0727, Mbov_0730, Mbov_0732, Mbov_0734, and Mbov_0735). The results showed a decreasing tendency of H2O2 production following increasing numbers of passages in M. bovis. The differences in H2O2 production between passage 1 and the serially passaged strains began to be statistically significant from passage 115 and onward (p < 0.01) (Figure 4A). Interestingly, other researchers measured the production of H2O2 by in vitro passaged strains including the 50th, 100th, and 200th passages of M. bovis, and they showed a similar decreasing tendency of H2O2 production with increasing numbers of passages (Khan et al., 2005). Random transposon mutagenesis was used recently to generate M. bovis mutants (Sharma et al., 2014). With the availability and further improvement of these techniques, it should be possible to obtain detailed information about the interactions of M. bovis with its host in the near future. To differentiate the effects resulting from the 14.2-kb deleted region and the SNPs in related genes on H2O2 production, transposon-disrupted mutants were further characterized. However, there was no significant difference in H2O2 production by the mutants compared with the P1 strain (Figure 4B). Therefore, these genes in the 14.2-kb deleted region might not affect H2O2 production, or a single gene could only contribute slightly to H2O2 production. In fact, other genes outside the 14.2-kb deleted region that have SNPs and indels might be associated with altered H2O2 production via the impairment of various pathways (Figure 5), such as ABC transporters (Figure S6), carbon metabolism (Figure S7), biosynthesis of amino acids (Figure S8), glycolysis/gluconeogenesis (Figure S9), and pyruvate metabolism (Figure S10).
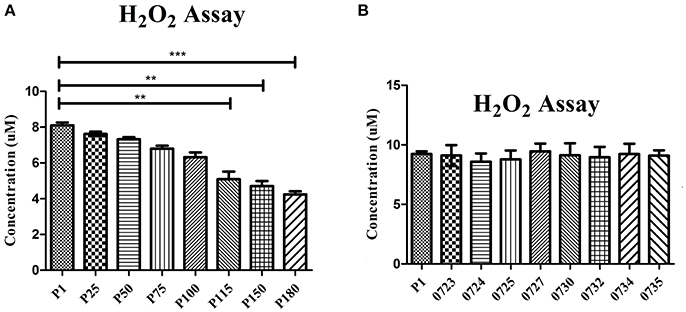
Figure 4. Production of H2O2 by 108 cells/ml determined after a 30-min incubation with 10 mM glycerol. Significant differences between groups are highlighted with asterisks. (A) Comparison of the wild-type M. bovis HB0801 strain and strains that were subjected to different numbers of passages during in vitro growth. (B) Comparison of the wild-type M. bovis HB0801 with different transposon-disrupted mutants during in vitro growth. These transposon-disrupted mutants were produced using the virulent M. bovis HB0801 and a Tn4001 transposon. Gene numbers of M. bovis given are as follows. 0723 = Mbov_0723, 0724 = Mbov_0724, 0725 = Mbov_0725, 0727 = Mbov_0727, 0730 = Mbov_0730, 0732 = Mbov_0732, 0734 = Mbov_0734, and 0735 = Mbov_0735.
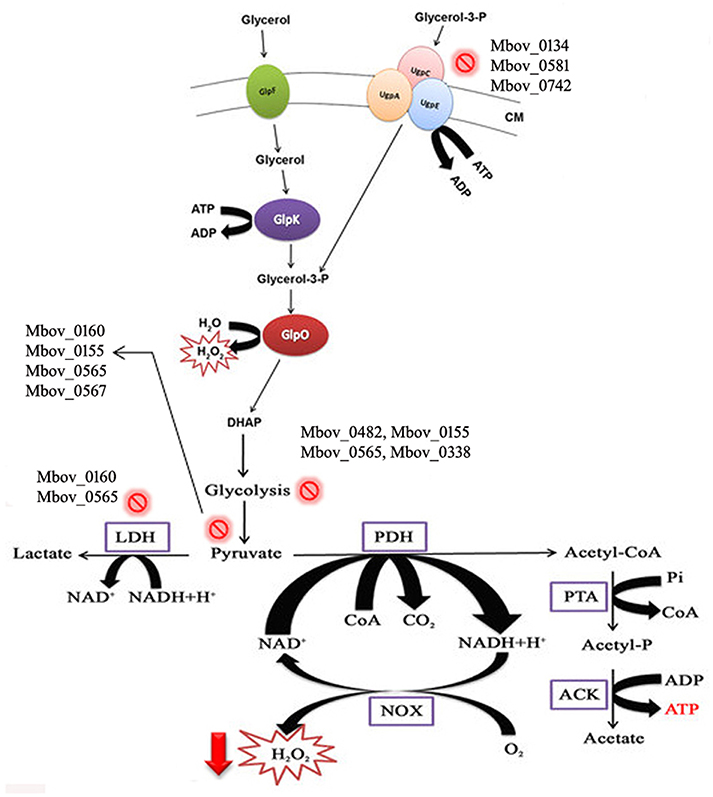
Figure 5. Schematic illustration of H2O2 production in mycoplasmas affected by multiple pathways, including glycerol metabolism. Glycerol is transported into the cell by the GlpF membrane transporter. Glycerol is phosphorylated to glycerol-3-P by GlpK (glycerol kinase). Moreover, glycerol-3-phosphate is also recycled through ABC transporters (UgpA, C, and E). Glycerol-3-phosphate is converted to dihydroxyacetone phosphate (DHAP) by oxidoreductase GlpO. H2O2 is secreted during and after the whole process. Mutations in the genes of mycoplasmas related to these metabolic pathways lead to decreased H2O2 production. CM, cell membrane. The  sign shows the mutations in the pathways, and mutated genes are listed with this sign.
sign shows the mutations in the pathways, and mutated genes are listed with this sign.
Conclusions
We first sequenced the genomes of three attenuated M. bovis strains, HB0801-P115, HB0801-P150, and HB0801-P180, with various levels of virulence, and we performed a comprehensive comparative genomics analysis of these strains. The results not only enrich the basic genomic data for further research of M. bovis, but they also provide guidance for exploring the molecular mechanisms of M. bovis virulence and pathogenesis.
Author Contributions
MR performed experiments, analyses, and wrote the manuscript. JQ, XZ, and HC performed experiments. HM, FK, GZ, and MZ performed analyses. AG, HCC, YC, and CH designed experiments and revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant Nos. 31661143015 and 31302111), the Special Fund for the Chinese Agricultural Research System (Beef/yaks) (grant No. CARS-38), the National Key Research and Development Program of China (grant No.2016YFD0500906) and the Special Fund for National Distinguished Scholars in Agricultural Research and the Technical Innovative Team.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00177/full#supplementary-material
References
Allam, A. B., Brown, M. B., and Reyes, L. (2012). Disruption of the S41 peptidase gene in Mycoplasma mycoides capri impacts proteome profile, H2O2 production, and sensitivity to heat shock. PLoS ONE 7:e51345. doi: 10.1371/journal.pone.0051345
Bercic, R. L., Slavec, B., Lavric, M., Narat, M., Bidovec, A., Dovc, P., et al. (2008). Identification of major immunogenic proteins of Mycoplasma synoviae isolates. Vet. Microbiol. 127, 147–154. doi: 10.1016/j.vetmic.2007.07.020
Blomgran, R., Desvignes, L., Briken, V., and Ernst, J. D. (2012). Mycobacterium tuberculosis inhibits neutrophil apoptosis, leading to delayed activation of naive CD4 T cells. Cell Host Microbe 11, 81–90. doi: 10.1016/j.chom.2011.11.012
Burki, S., Frey, J., and Pilo, P. (2015). Virulence, persistence and dissemination of Mycoplasma bovis. Vet. Microbiol. 179, 15–22. doi: 10.1016/j.vetmic.2015.02.024
Caswell, J. L., and Archambault, M. (2007). Mycoplasma bovis pneumonia in cattle. Anim. Health Res. Rev. 8, 161–186. doi: 10.1017/S1466252307001351
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Chen, Z. F., Zhao, H., and Xu, R. S. Z. (1983). Isolation and identification of Mycoplasma gallisepticum Shanghai dairy cows. J. Anim. Husbandry Vet. Med. 4, 60–66.
Chiang, S. L., and Mekalanos, J. J. (1998). Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27, 797–805. doi: 10.1046/j.1365-2958.1998.00726.x
Corona, L., Cillara, G., and Tola, S. (2013). Proteomic approach for identification of immunogenic proteins of Mycoplasma mycoides subsp. capri. Vet. Microbiol. 167, 434–439. doi: 10.1016/j.vetmic.2013.08.024
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
De Jong, R. N., Ab, E., Diercks, T., Truffault, V., Daniels, M., Kaptein, R., et al. (2006). Solution structure of the human ubiquitin-specific protease 15 DUSP domain. J. Biol. Chem. 281, 5026–5031. doi: 10.1074/jbc.M510993200
Grundel, A., Pfeiffer, M., Jacobs, E., and Dumke, R. (2015). Network of surface-displayed glycolytic enzymes in Mycoplasma pneumoniae and their interactions with human plasminogen. Infect. Immun. 84, 666–676. doi: 10.1128/IAI.01071-15
Guimaraes, A. M., Santos, A. P., Sanmiguel, P., Walter, T., Timenetsky, J., and Messick, J. B. (2011). Complete genome sequence of Mycoplasma suis and insights into its biology and adaption to an erythrocyte niche. PLoS ONE 6:e19574. doi: 10.1371/journal.pone.0019574
Guo, F. B., and Wei, W. (2012). Prediction of genomic islands in three bacterial pathogens of pneumonia. Int. J. Mol. Sci. 13, 3134–3144. doi: 10.3390/ijms13033134
Haering, C. H., Lowe, J., Hochwagen, A., and Nasmyth, K. (2002). Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788. doi: 10.1016/S1097-2765(02)00515-4
Hames, C., Halbedel, S., Hoppert, M., Frey, J., and Stulke, J. (2009). Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 191, 747–753. doi: 10.1128/JB.01103-08
Higgins, C. F., Hiles, I. D., Salmond, G. P., Gill, D. R., Downie, J. A., Evans, I. J., et al. (1986). A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323, 448–450. doi: 10.1038/323448a0
Iizuka, K., and Horikawa, Y. (2008). ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr. J. 55, 617–624. doi: 10.1507/endocrj.K07E-110
Jores, J., Meens, J., Buettner, F. F., Linz, B., Naessens, J., and Gerlach, G. F. (2009). Analysis of the immunoproteome of Mycoplasma mycoides subsp. mycoides small colony type reveals immunogenic homologues to other known virulence traits in related Mycoplasma species. Vet. Immunol. Immunopathol. 131, 238–245. doi: 10.1016/j.vetimm.2009.04.016
Kannan, T. R., and Baseman, J. B. (2006). ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 103, 6724–6729. doi: 10.1073/pnas.0510644103
Kannan, T. R., Krishnan, M., Ramasamy, K., Becker, A., Pakhomova, O. N., Hart, P. J., et al. (2014). Functional mapping of community-acquired respiratory distress syndrome (CARDS) toxin of Mycoplasma pneumoniae defines regions with ADP-ribosyltransferase, vacuolating and receptor-binding activities. Mol. Microbiol. 93, 568–581. doi: 10.1111/mmi.12680
Khan, F. A., Faisal, M., Chao, J., Liu, K., Chen, X., Zhao, G., et al. (2016). Immunoproteomic identification of MbovP579, a promising diagnostic biomarker for serological detection of Mycoplasma bovis infection. Oncotarget 7, 39376–39395. doi: 10.18632/oncotarget.9799
Khan, L. A., Miles, R. J., and Nicholas, R. A. (2005). Hydrogen peroxide production by Mycoplasma bovis and Mycoplasma agalactiae and effect of in vitro passage on a Mycoplasma bovis strain producing high levels of H2O2. Vet. Res. Commun. 29, 181–188. doi: 10.1023/B:VERC.0000047506.04096.06
Kim, Y. R., Brinsmade, S. R., Yang, Z., Escalante-Semerena, J., and Fierer, J. (2006). Mutation of phosphotransacetylase but not isocitrate lyase reduces the virulence of Salmonella enterica serovar Typhimurium in mice. Infect. Immun. 74, 2498–2502. doi: 10.1128/IAI.74.4.2498-2502.2006
Kleinschmidt, S., Spergser, J., Rosengarten, R., and Hewicker-Trautwein, M. (2013). Long-term survival of Mycoplasma bovis in necrotic lesions and in phagocytic cells as demonstrated by transmission and immunogold electron microscopy in lung tissue from experimentally infected calves. Vet. Microbiol. 162, 949–953. doi: 10.1016/j.vetmic.2012.11.039
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Li, Y., Zheng, H., Liu, Y., Jiang, Y., Xin, J., Chen, W., et al. (2011). The complete genome sequence of Mycoplasma bovis strain Hubei-1. PLoS ONE 6:e20999. doi: 10.1371/journal.pone.0020999
Matyushkina, D., Pobeguts, O., Butenko, I., Vanyushkina, A., Anikanov, N., Bukato, O., et al. (2016). Phase transition of the bacterium upon invasion of a host cell as a mechanism of adaptation: a Mycoplasma gallisepticum model. Sci. Rep. 6:35959. doi: 10.1038/srep35959
Mulongo, M., Prysliak, T., Scruten, E., Napper, S., and Perez-Casal, J. (2014). In vitro infection of bovine monocytes with Mycoplasma bovis delays apoptosis and suppresses production of gamma interferon and tumor necrosis factor alpha but not interleukin-10. Infect. Immun. 82, 62–71. doi: 10.1128/IAI.00961-13
Pancholi, V. (2001). Multifunctional alpha-enolase: its role in diseases. Cell. Mol. Life Sci. 58, 902–920. doi: 10.1007/PL00000910
Parker, A. M., Shukla, A., House, J. K., Hazelton, M. S., Bosward, K. L., Kokotovic, B., et al. (2016). Genetic characterization of Australian Mycoplasma bovis isolates through whole genome sequencing analysis. Vet. Microbiol. 196, 118–125. doi: 10.1016/j.vetmic.2016.10.010
Pilo, P., Vilei, E. M., Peterhans, E., Bonvin-Klotz, L., Stoffel, M. H., Dobbelaere, D., et al. (2005). A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. J. Bacteriol. 187, 6824–6831. doi: 10.1128/JB.187.19.6824-6831.2005
Pollack, J. D., Myers, M. A., Dandekar, T., and Herrmann, R. (2002). Suspected utility of enzymes with multiple activities in the small genome Mycoplasma species: the replacement of the missing “household” nucleoside diphosphate kinase gene and activity by glycolytic kinases. OMICS 6, 247–258. doi: 10.1089/15362310260256909
Postma, P. W., Lengeler, J. W., and Jacobson, G. R. (1993). Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594.
Qi, J., Guo, A., Cui, P., Chen, Y., Mustafa, R., Ba, X., et al. (2012). Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS ONE 7:e38239. doi: 10.1371/journal.pone.0038239
Saha, P., Thome, K. C., Yamaguchi, R., Hou, Z., Weremowicz, S., and Dutta, A. (1998). The human homolog of Saccharomyces cerevisiae CDC45. J. Biol. Chem. 273, 18205–18209. doi: 10.1074/jbc.273.29.18205
Schott, C., Cai, H., Parker, L., Bateman, K. G., and Caswell, J. L. (2014). Hydrogen peroxide production and free radical-mediated cell stress in Mycoplasma bovis pneumonia. J. Comp. Pathol. 150, 127–137. doi: 10.1016/j.jcpa.2013.07.008
Sharma, S., Markham, P. F., and Browning, G. F. (2014). Genes found essential in other mycoplasmas are dispensable in Mycoplasma bovis. PLoS ONE 9:e97100. doi: 10.1371/journal.pone.0097100
Song, Z., Li, Y., Liu, Y., Xin, J., Zou, X., and Sun, W. (2012). α-Enolase, an adhesion-related factor of Mycoplasma bovis. PLoS ONE 7:e38836. doi: 10.1371/journal.pone.0038836
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Wise, K. S., Calcutt, M. J., Foecking, M. F., Roske, K., Madupu, R., and Methe, B. A. (2011). Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523). Infect. Immun. 79, 982–983. doi: 10.1128/IAI.00726-10
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, H., Caimano, M. J., Lin, T., He, M., Radolf, J. D., Norris, S. J., et al. (2010). Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog. 6:e1001104. doi: 10.1371/annotation/88293342-3534-4b8b-8800-c63a8c86bf6d
Zhang, H., Zhao, G., Guo, Y., Menghwar, H., Chen, Y., Chen, H., et al. (2016). Mycoplasma bovis MBOV_RS02825 encodes a secretory nuclease associated with cytotoxicity. Int. J. Mol. Sci. 17:628. doi: 10.3390/ijms17050628
Zhang, R., Han, X., Chen, Y., Mustafa, R., Qi, J., Chen, X., et al. (2014). Attenuated Mycoplasma bovis strains provide protection against virulent infection in calves. Vaccine 32, 3107–3114. doi: 10.1016/j.vaccine.2013.12.004
Keywords: attenuation, bioinformatics, genome, Mycoplasma bovis, virulence
Citation: Rasheed MA, Qi J, Zhu X, Chenfei H, Menghwar H, Khan FA, Zhao G, Zubair M, Hu C, Chen Y, Chen H and Guo A (2017) Comparative Genomics of Mycoplasma bovis Strains Reveals That Decreased Virulence with Increasing Passages Might Correlate with Potential Virulence-Related Factors. Front. Cell. Infect. Microbiol. 7:177. doi: 10.3389/fcimb.2017.00177
Received: 23 February 2017; Accepted: 24 April 2017;
Published: 11 May 2017.
Edited by:
Yongqun “Oliver” He, University of Michigan Health System, USAReviewed by:
Weihuan Fang, Zhejiang University, ChinaHongjie Fan, Nanjing Agricultural University, China
Copyright © 2017 Rasheed, Qi, Zhu, Chenfei, Menghwar, Khan, Zhao, Zubair, Hu, Chen, Chen and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aizhen Guo, YWl6aGVuQG1haWwuaHphdS5lZHUuY24=
†These authors have contributed equally to this work.
 Muhammad A. Rasheed
Muhammad A. Rasheed Jingjing Qi1,4†
Jingjing Qi1,4† Harish Menghwar
Harish Menghwar Farhan A. Khan
Farhan A. Khan Huanchun Chen
Huanchun Chen Aizhen Guo
Aizhen Guo