- 1Laboratory of Medical Microbiology, Vaccine and Infectious Disease Institute, University of Antwerp, Wilrijk, Belgium
- 2Molecular Pathology Group, Cell Biology and Histology, University of Antwerp, Wilrijk, Belgium
The significance of polymicrobial infections is increasingly being recognized especially in a biofilm context wherein multiple bacterial species—including both potential pathogens and members of the commensal flora—communicate, cooperate, and compete with each other. Two important bacterial pathogens that have developed a complex network of evasion, counter-inhibition, and subjugation in their battle for space and nutrients are Pseudomonas aeruginosa and Staphylococcus aureus. Their strain- and environment-specific interactions, for instance in the cystic fibrosis lung or in wound infections, show severe competition that is generally linked to worse patient outcomes. For instance, the extracellular factors secreted by P. aeruginosa have been shown to subjugate S. aureus to persist as small colony variants (SCVs). On the other hand, data also exist where S. aureus inhibits biofilm formation by P. aeruginosa but also protects the pathogen by inhibiting its phagocytosis. Interestingly, such interspecies interactions differ between the planktonic and biofilm phenotype, with the extracellular matrix components of the latter likely being a key, and largely underexplored, influence. This review attempts to understand the complex relationship between P. aeruginosa and Staphylococcus spp., focusing on S. aureus, that not only is interesting from the bacterial evolution point of view, but also has important consequences for our understanding of the disease pathogenesis for better patient management.
Introduction
Over the past decade there is a growing appreciation that the biofilm mode of growth is the most common lifestyle adopted by bacteria (Hall-Stoodley et al., 2004; Burmolle et al., 2014). Biofilms can be defined as surface-associated, structured bacterial communities embedded in an extracellular matrix (Hall-Stoodley et al., 2004). Living in a biofilm provides protection in a stressful environment where mechanical stress, desiccation, and biocides are common threats (Donlan and Costerton, 2002; Flemming and Wingender, 2010). Multiple species frequently exist together in a single biofilm, where they either improve the fitness of one another or compete for space and nutrients (Jefferson, 2004; Billings et al., 2013; Burmolle et al., 2014; DeLeon et al., 2014). Most bacteria have developed interaction strategies to communicate within and between species in a cell density-dependent manner, for example, by using small diffusible molecules in a process called quorum-sensing (Federle and Bassler, 2003; Li and Tian, 2012). Furthermore, many bacteria excrete antimicrobial components, also often regulated by quorum-sensing, to eliminate competitors (Federle and Bassler, 2003; Li and Tian, 2012). Indeed, these multispecies interactions within the biofilm are important for the inhabiting bacteria and, given the increasing evidence of the link between biofilm-associated pathogens and disease, also from a clinical point of view (Donlan and Costerton, 2002; Li and Tian, 2012).
S. aureus and P. aeruginosa are important pathogens causing a wide variety of infections, including pneumonia in cystic fibrosis (CF) patients, healthcare associated pneumonia and chronic wounds (Harrison, 2007; Fazli et al., 2009; Cystic Fibrosis Foundation Patient Registry, 2012). Initially, only an antagonistic relationship between both organisms was described as the presence of one is associated with the absence of the other in CF and both are rarely found in close association in chronic wounds. S. aureus mostly resides on the wound surface whereas P. aeruginosa is found in the deep layers (Kirketerp-Moller et al., 2008; Fazli et al., 2009). We also recently showed a negative correlation between presence of P. aeruginosa and the total species diversity in in vivo endotracheal tube biofilms and a low co-occurrence of P. aeruginosa with Staphylococcus epidermidis (Hotterbeekx et al., 2016). Nonetheless, recent studies have also co-isolated P. aeruginosa and Gram-positive bacteria, including S. aureus, from the same infection site where increased virulence and/or antibiotic resistance is described (Duan et al., 2003; Kirketerp-Moller et al., 2008; Fazli et al., 2009; Dalton et al., 2011; Korgaonkar et al., 2013). After describing first in vivo observations occurring in human diseases, we will discuss and summarize in vitro data from the current literature on potential mechanisms of interactions between P. aeruginosa and Staphylococcus spp., primarily S. aureus.
Co-Occurrence of P. aeruginosa and S. aureus in vivo is Linked to Worse Disease Outcomes
CF is a typical example of a biofilm-related infection wherein P. aeruginosa and S. aureus are frequently isolated from the lungs of these patients (Harrison, 2007; Hauser et al., 2011; Cystic Fibrosis Foundation, 2014). While, S. aureus is mostly acquired during childhood, the presence of P. aeruginosa is associated with increasing age and worsening patient prognosis (Sagel et al., 2009; Hauser et al., 2011; Cystic Fibrosis Foundation, 2014). An increasing incidence of P. aeruginosa with age has been shown to coincide with a decreasing S. aureus incidence in CF patients (Harrison, 2007; Cystic Fibrosis Foundation, 2014), data that primarily indicates an antagonistic relationship between the two pathogens. However, in cases where P. aeruginosa and S. aureus have been co-isolated, both pathogens seem to contribute independently and additively to the disease severity (Sagel et al., 2009; Hauser et al., 2011), presenting as increased lung inflammation and consequently increased lung damage compared to infection with a single pathogen (Sagel et al., 2009). Furthermore, due to repeated antibiotic therapy, CF patients also carry higher levels of methicillin-resistant S. aureus (MRSA) that is associated with a worse lung function compared to methicillin- sensitive S. aureus (MSSA) but only in combination with P. aeruginosa (Hubert et al., 2013).
Chronic wounds are another example of biofilm-related infections wherein co-presence of P. aeruginosa and S. aureus has been shown to result in delayed wound healing compared to single species infections (Dalton et al., 2011; Seth et al., 2012; Pastar et al., 2013). In a pig wound model, infections initiated by in vitro preformed dual species biofilm caused a significant suppression of keratinocyte growth factor 1 (KGF1), which is responsible for re-epithelialization and wound closure (Pastar et al., 2013). In a rabbit ear-wound model, mixed species infection of S. aureus and P. aeruginosa caused an increased expression of the pro-inflammatory cytokines IL-1β and TNF-α, indicating a higher inflammatory response compared to single species infection (Seth et al., 2012). Moreover, S. aureus and P. aeruginosa reached an equilibrium after 12 days of infection, with P. aeruginosa being the dominant pathogen (Seth et al., 2012). In a mouse chronic wound model infected with in vitro preformed four-species biofilm and monitored up to 12 days, presence of multiple species was found to significantly delay wound healing only at 8 days post-infection (Dalton et al., 2011). However, polymicrobial infections showed increased antimicrobial tolerance compared to single species infection with P. aeruginosa in this study (Dalton et al., 2011). These studies suggest that, despite the constraints of different host backgrounds, multispecies infections can lead to delayed wound healing, increased inflammation and increased antibiotic tolerance, which all add to a worse patient outcome. P. aeruginosa is often the dominant pathogen due to its wide array of mechanisms to adapt to changing hostile environments, which allows colonization in a variety of niches. When P. aeruginosa encounters other bacteria like S. aureus, it can co-exist or take over the biofilm through production of various quorum-sensing regulated factors. Section Extracellular products of Staphylococcus spp. impact P. aeruginosa virulence in vivo discusses in vivo animal studies exploring production of P. aeruginosa virulence factors in the presence of S. aureus.
Extracellular Products of Staphylococcus spp. Impact P. aeruginosa Virulence in vivo
P. aeruginosa possesses a wide range of extracellular factors to survive and invade human tissues, often by modulating the immune system. The complex interplay between biofilms and the host immune response are reviewed in detail by Watters et al. (2016). Here, we discuss four molecules in particular which are upregulated in the presence of Gram-positive bacteria: LasB elastase, rhamnolipids, exotoxins, and phenazines (Figure 1). LasB elastase is an extracellular protease capable of digesting the lung surfactant, the pulmonary antimicrobial enzyme lysozyme, and transferrin, as well as slowing down the ciliary movement (Hauser, 2009). In addition, LasB impairs uptake of P. aeruginosa by macrophages and its protease activity leads to lung tissue damage, thereby decreasing pulmonary function and facilitating dissemination into the bloodstream (Strateva and Mitov, 2011). Similar to LasB, rhamnolipids are glycolipidic biosurfactants that interfere with the lung surfactant activity by solubilizing the phospholipids and with airway immune response by disrupting the polymorphonuclear leucocyte chemotaxis and macrophage function, and also inhibit ciliary beating (Soberon-Chavez et al., 2005; Jensen et al., 2007). Furthermore, rhamnolipids increase inflammation by stimulating the release of the pro-inflammatory cytokines IL-6 and IL-8 by the airway epithelium (Soberon-Chavez et al., 2005). An excessive inflammatory response and associated tissue damage is also induced by the release of exotoxins ExoT, ExoS, and ExoY by the type III secretion system. The type III secretion system is a needle-like structure directly injecting exotoxins into other bacteria, macrophages and epithelial cells, thereby killing them (Hauser, 2009). The fourth type of molecules, phenazines, are pigments produced by a large number of Pseudomonas spp., and have been shown to be involved in mediating microbial interactions as well as in CF disease progression. Pyocyanin is the most important phenazine and its production in the CF lung was shown to lead to goblet cell hyperplasia, airway fibrosis and alveolar airspace destruction (Caldwell et al., 2009; Strateva and Mitov, 2011). Pyocyanin causes an imbalance between the T helper type 1 (Th1) and type 2 (Th2) cytokines, leading to overproduction of Th2 cytokines IL-4 and IL-13 and increased macrophage infiltration (Caldwell et al., 2009). Essentially, release of these extracellular molecules by P. aeruginosa, partly in response to the presence of S. aureus, leads to increased tissue damage due to their cytotoxic and immune-modulatory effects, which also helps P. aeruginosa survival. Interestingly, not only S. aureus but Gram-positive commensals (coagulase negative staphylococci and viridans streptococci) can also alter the virulence of P. aeruginosa in a similar fashion (Duan et al., 2003). Such immune modulation and evasion by collective bacterial species might underlie the worsened patient prognosis observed in multispecies infections (Figure 1). Sections AI-2 in the CF Lung Increases P. aeruginosa Virulence and Might Be an Important Therapeutic Target and N-Acetyl Glucosamine Sensing Enhances the Production of P. aeruginosa Extracellular Virulence Factors discuss the known mechanisms of how extracellular products of staphylococci modulate the four major virulence factors P. aeruginosa that were discussed above.
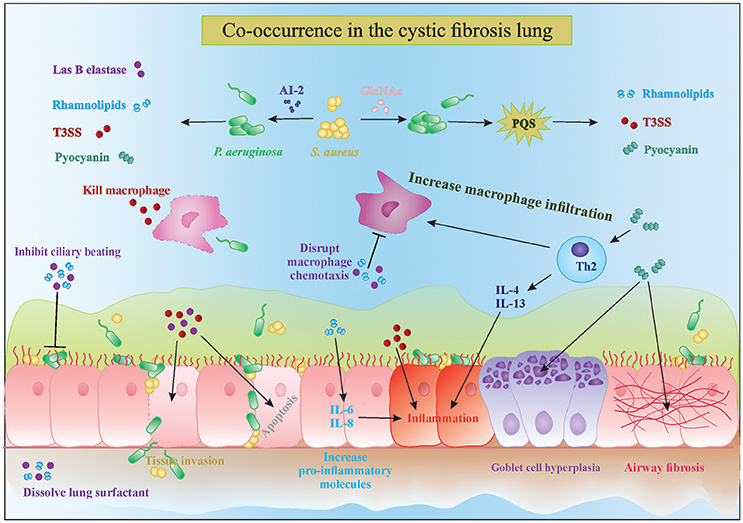
Figure 1. Interactions in the cystic fibrosis lung. The complex interplay between microbial competition and the human immune system results in increased secretion of pro-inflammatory cytokines, microbial virulence factors and consequently tissue damage and bacterial invasion of the epithelial barrier. Mixed species infections lead to a stronger decrease in ciliary beating and increased goblet cell hyperplasia and tissue fibrosis, which are characteristic of cystic fibrosis disease progression. PQS, Pseudomonas quinolone signal; T3SS, type 3 secretion system; AI-2, autoinducer 2.
AI-2 in the CF Lung Increases P. aeruginosa Virulence and Might Be an Important Therapeutic Target
Autoinducer-2 (AI-2) is a small diffusible quorum-sensing molecule produced by several bacteria, including staphylococci, and has been shown to cause upregulation of several major virulence genes of P. aeruginosa discussed above, including extracellular protease (lasB), rhamnosyltransferase involved in rhamnolipid synthesis (rhlA), exotoxins (exoT, exoS, exoY) and phenazines (phzA1 and phzA2; Li et al., 2015). Induction of P. aeruginosa virulence by AI-2 was shown both in vitro after screening of a random lux reporter-based promotor library and in vivo in rat lung infection and Drosophila chronic infection models (Duan et al., 2003; Sibley et al., 2008; Li et al., 2015). AI-2 mediated quorum-sensing is now recognized as a universal language of interspecies communication regulating a wide variety of genes involved in virulence and biofilm formation in a cell-density dependent manner in a number of micro-organisms, including non-producers like P. aeruginosa (Rezzonico et al., 2012). Furthermore, AI-2 has been detected in substantial amounts in the sputum of CF patients and in infected rats (Duan et al., 2003), raising the possibility of interruption of AI-2 signaling to either slow down disease progression or hasten the healing process.
A promising approach is the use of AI-2 analogs like D-ribose that block the AI-2 pathway and inhibit P. aeruginosa virulence. Wang et al showed in a rat model of mechanical ventilation that co-inoculation of P. aeruginosa and Streptococcus mitis resulted in increased biomass, lung damage, and rat mortality compared to infection with only P. aeruginosa (Wang et al., 2016). Treatment with D-ribose of both the single and dual species infections showed a significant decrease in biomass and lowering of rat mortality in the latter group due to interference with AI-2 signaling (Wang et al., 2016). Inhibition of P. aeruginosa virulence is not only beneficial because the P. aeruginosa-mediated damage is reduced but also because the immune system is less stimulated (Figure 1). Further studies showing the benefits of non-toxic biofilm inhibitors such as D-ribose in patient populations are awaited.
N-Acetyl Glucosamine Sensing Enhances the Production of P. aeruginosa Extracellular Virulence Factors
Another molecule that increases the virulence of P. aeruginosa and is commonly found in the CF lung is N-acetyl glucosamine (GlcNAc). GlcNAc is part of the Gram-positive cell wall polymer peptidoglycan and induces the virulence of P. aeruginosa by enhancing the Pseudomonas quinolone signal (PQS), which controls the production of extracellular virulence factors like pyocyanin, elastase, rhamnolipids and HQNO (discussed in Section Pseudomonas Quinolone Signal Regulates the Production of Anti-Staphylococcal 4-Hydroxy-2-Heptylquinoline N-Oxide (HQNO); Deziel et al., 2004; Williams and Camara, 2009; Jimenez et al., 2012; Korgaonkar et al., 2013; Figures 1, 2). The PQS, with 2-heptyl-3-hydroxy-4-quilonone as the main effector molecule, is one of the three quorum-sensing systems present in P. aeruginosa. PQS is positively regulated by LasR and negatively regulated by RhlR, the two other quorum-sensing systems of P. aeruginosa with N-acylhomoserine lactone as main effector molecule (Jimenez et al., 2012). P. aeruginosa has the ability to sense the peptidoglycan shed by the Gram positive commensal flora and in response increase the production of antimicrobials. The enhanced virulence in the presence of GlcNAc from Gram-positive bacteria was demonstrated in vivo in a Drosophila and Galleria mellonella infection model (Korgaonkar et al., 2013; Whiley et al., 2014). Both GlcNAc and AI-2 sensing are examples where P. aeruginosa can sense its environment and generate the appropriate response to eliminate competitors by producing several virulence factors, which also has a negative impact on the host.
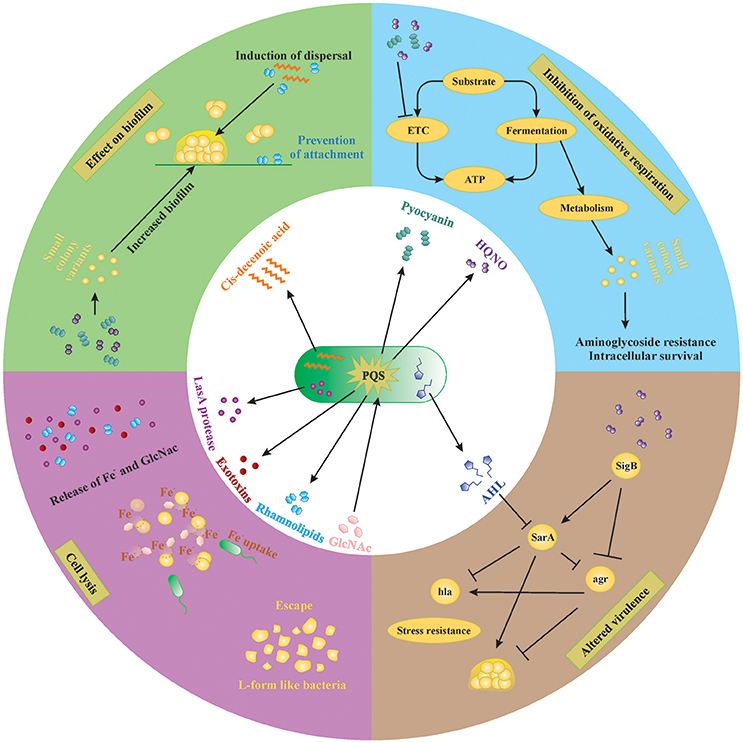
Figure 2. Small molecules secreted by P. aeruginosa and S. aureus. Extracellular factors produced by P. aeruginosa affect biofilm formation, oxidative respiration, cell lysis and virulence of S. aureus. Lysis of S. aureus leads to increased extracellular iron and N-acetyl glucosamine (GlcNac), which are sensed by P. aeruginosa. AHL, N-acyl homoserine lactone; HQNO, 4-hydroxy-2-heptylquinoline N-oxide; PQS, Pseudomonas quinolone signal; GlcNac, N-acetyl glucosamine.
P. aeruginosa Produces a Wide Variety of Molecules that Inhibit S. aureus in vitro
P. aeruginosa produces many molecules to compete with other microorganisms for space and nutrients. The number of molecules, quantities produced and even the structure of these molecules vary between different strains of P. aeruginosa as well as between different growth conditions (planktonic vs. biofilm; the presence of host factors, antibiotics etc.). In order to achieve a better understanding of complex interplay of the different compounds in in vivo biofilm-related infections, it is necessary to dissect this complex system into individual subsystems and investigate each compound individually. In the following sections, we discuss the effect of different molecules produced by P. aeruginosa on S. aureus in vitro. An overview of the extracellular molecules produced by P. aeruginosa and their effect on S. aureus is shown in Figure 2.
Pseudomonas Quinolone Signal Regulates the Production of Anti-Staphylococcal 4-Hydroxy-2-Heptylquinoline N-Oxide (HQNO)
P. aeruginosa strongly reduces or completely outcompetes S. aureus during co-culture in many in vitro model systems, both planktonic and biofilm (Palmer et al., 2005; Baldan et al., 2014; DeLeon et al., 2014). This anti-staphylococcal activity of P. aeruginosa was first described by Lightbown and Jackson (1956), who identified 4-hydroxy-2-heptylquinoline N-oxide (HQNO) as a major compound produced by P. aeruginosa that inhibited the cytochrome systems of some bacteria, including S. aureus (Lightbown and Jackson, 1956). The same phenomenon was again described by Machan et al. in 1991 by testing the culture supernatant of fifty P. aeruginosa clinical isolates on 261 staphylococci (Machan et al., 1991). The growth of all staphylococci was reduced by each of the P. aeruginosa strains, although the extent of inhibition was strain-dependent. The factor responsible for this phenomenon was again identified as HQNO (Machan et al., 1992). HQNO is the major compound produced by the pqsABCDE operon, which is regulated by the quorum-sensing system PQS. Although HQNO is described as an antistaphylococcal compound, it has no lytic activity against S. aureus itself but rather slows down the growth by inhibiting oxidative respiration (Figure 2, right upper panel; Williams and Camara, 2009). Exposure to an HQNO source suppresses the growth of S. aureus, resulting in small colonies which are easily missed in diagnostic cultures (Hoffman et al., 2006). These so-called SCVs represent a different phenotype with specific characteristics and will be discussed later in this review. Furthermore, HQNO can be detected at active concentrations in the sputum of CF patients infected with P. aeruginosa, suggesting that HQNO has the same effect in the lungs of CF patients as it has in vitro (Hoffman et al., 2006). Although HQNO is one of the most important and well-studied antistaphylococcal compounds, it is not the only factor slowing down the growth and inhibiting oxidative respiration in staphylococci.
Pyocyanin Inhibits Oxidative Respiration in S. aureus
Pyocyanin is one of the numerous pigmented phenazines produced by P. aeruginosa and an important virulence factor. Pyocyanin is produced during Pseudomonas biofilm formation, has a role in acute and chronic airway infections, enables anaerobic survival and serves as a redox-active antimicrobial compound (Biswas et al., 2009; Caldwell et al., 2009). Furthermore, by its inter-and intracellular signaling, pyocyanin enables P. aeruginosa to successfully compete with other bacteria and even fungi (Gibson et al., 2009; Toyofuku et al., 2010; Tashiro et al., 2013). Like HQNO, pyocyanin also blocks the oxidative respiration and inhibits growth of S. aureus, also selecting for the SCV phenotype (Biswas et al., 2009; Figure 2, right upper panel). The production of pyocyanin can be observed after 8 h of culture, around the same time when a strong reduction of S. aureus cells occurs during co-culture (Biswas et al., 2009; Tashiro et al., 2013). Furthermore, the presence of Gram-positive organisms, including some Staphylococcus spp., can induce pyocyanin production in P. aeruginosa by stimulating the PQS system (Korgaonkar and Whiteley, 2011; Whiley et al., 2014; Figure 2, middle panel). In addition, exposure to pyocyanin in the airways leads to pulmonary damage and contributes to CF pathogenesis (Caldwell et al., 2009). Therefore, pyocyanin seems to be an antagonistic compound secreted to provide a competitive advantage to P. aeruginosa by harming S. aureus, other Gram-positive bacteria as well as the host.
LasA Protease or Staphylolysin Effectively Lyses S. aureus Cells
P. aeruginosa secretes a staphylolytic endopeptidase called LasA protease or staphylolysin, which degrading pentaglycine in the cell wall of S. aureus causing cell lysis (Kessler et al., 1993). P. aeruginosa might use LasA protease to compete with staphylococci but also to acquire iron from S. aureus (Mashburn et al., 2005; Figure 2, left lower panel). Because freely available iron is often limited, P. aeruginosa has developed several strategies to scavenge iron, like the synthesis of iron chelating siderophores, pyoverdin, and pyochelin (Diggle et al., 2007). Transcription patterns of iron-regulated genes of P. aeruginosa in the presence of S. aureus in vivo are the same as in high-iron conditions in vitro, suggesting that S. aureus might be an iron source for P. aeruginosa (Mashburn et al., 2005). This type of interaction is, however, only important when both species are located close together, like in multi-species biofilms (Mashburn et al., 2005). The specificity of the LasA protease for staphylococci makes it a potential therapeutic candidate against staphylococcal infections especially those cause by antibiotic resistant MRSA strains as shown in a rat model of endophthalmitis (Barequet et al., 2009). However, S. aureus can survive LasA by the emergence of L-form-like colonies, which lack a cell wall (Falcon et al., 1989), although the role of L-form like colonies in disease remains rather vague.
Cis-2-Decenoic Acid Induces Biofilm Dispersal in a Broad Range of Organisms Including P. aeruginosa
Interspecies competition in biofilms not only occurs by inhibiting or killing the other species but also by inducing its dispersal. Biofilm dispersal is mainly induced when the environment becomes less favorable, like in case of nutrient depletion, and is extensively reviewed by Petrova and Sauer (2016). The exact mechanisms that induce biofilm dispersal are currently unknown, although several factors have been investigated (Hall-Stoodley and Stoodley, 2005; Davies and Marques, 2009). Since most bacteria reside in a biofilm consisting of multiple species in vivo, the dispersal signal must be recognized by a wide range of species (Davies and Marques, 2009). One class of such molecules are the cis-monosaturated fatty acids, which are small extracellular messenger molecules with broad inter-phylum and even inter-kingdom activities (Davies and Marques, 2009). P. aeruginosa produces cis-2-decenoic acid, which induces a dispersion response in biofilms formed by a range of Gram-negative and Gram-positive bacteria, including S. aureus (Figure 2, left upper panel), yeast as well as in P. aeruginosa (Davies and Marques, 2009). Interestingly, Davies et al showed that dispersion was only induced when the microcolonies reached a minimum of 40 μm diameter and 10 μm of thickness, indicating that a certain threshold concentration is needed for cis-2-decenoic acid to become active (Davies and Marques, 2009). This molecule could possibly be employed to disrupt biofilms on surfaces, followed by disinfectants that can successfully clear planktonic bacteria.
Rhamnolipids Promote Biofilm Dispersal and Inhibit Adhesion
Most P. aeruginosa strains produce rhamnolipids, biosurfactants consisting of one or two rhamnose molecules linked to one or two fatty acids (Soberon-Chavez et al., 2005). While cis-2-decenoic acid is mainly used as a common signal for dispersion at the final biofilm stages, rhamnolipids are used to dislodge competing bacteria from the biofilm. Many different rhamnolipid homologs are produced, depending on the Pseudomonas strain and carbon source, and their synthesis is quorum-sensing regulated (Soberon-Chavez et al., 2005). Rhamnolipids were shown to reduce the surface tension and to have an anti-adhesive and antimicrobial effect on many micro-organisms (Haba et al., 2003; Rodrigues et al., 2006; Zezzi do Valle Gomes and Nitschke, 2012). The amphiphilic nature of rhamnolipids enables them to intercalate into the cell membranes of different microorganisms and form complexes, thereby permeabilizing the membranes and causing leakage of intracellular material (Sotirova et al., 2008). Gram-positive organisms seem more susceptible to rhamnolipid permeabilization than Gram-negative because the presence of lipopolysaccharides protects the cell membranes of the latter against the effect of surfactants (Soberon-Chavez et al., 2005). Furthermore, rhamnolipids were shown to promote biofilm dispersal in many different microorganisms, including P. aeruginosa itself, although this effect is strain dependent (Rodrigues et al., 2006). For S. aureus and S. epidermidis, rhamnolipids were shown to induce biofilm dispersal and to inhibit adhesion in a dose-dependent manner (Rodrigues et al., 2006; Zezzi do Valle Gomes and Nitschke, 2012; Pihl et al., 2013; Figure 2, left upper panel).
Long-Chain AHLs Reduce Growth and Virulence of S. aureus
The N-acylhomoserine lactone (AHL) system is the most important and most extensively studied quorum-sensing system in P. aeruginosa. Many diverse AHLs are produced by various Gram-negative bacteria, all consisting of a homoserine lactone ring that is N-acylated with a fatty acyl group (Jimenez et al., 2012). The length of the acyl chains may vary from 4 to 18 carbons and P. aeruginosa mainly produces a short-chain C4-HSL and a long-chain 3-oxo-C12-HSL, although other lengths might also occur (Jimenez et al., 2012). The production of many virulence factors is regulated by AHL, including that of pyocyanin and rhamnolipids (Jimenez et al., 2012). Although it is currently not described that Gram-positive bacteria produce AHLs, they might still be influenced by them (Qazi et al., 2006). For example, growth of S. aureus is inhibited by several long chain 3-oxo-AHLs (including C8, C10, C12, and C14 chains) in a concentration dependent manner, with C12 and C14 being the most effective (Qazi et al., 2006). At concentrations below growth inhibition, the function of staphylococcal accessory regulator sarA and accessory gene regulator agr are strongly reduced (see Section SCV Induction by P. aeruginosa is Sigma B Dependent), and consequently their dependent virulence factors like hemolysins, TSST-1, protein A, and fibronectin-binding proteins (Figure 2, right lower panel; Qazi et al., 2006). Moreover, inhibition of agr might lead to more biofilm formation in S. aureus due to reduced detachment, although this study only tested planktonic conditions and requires further research (Qazi et al., 2006). The short chain AHL produced by P. aeruginosa seems to have no effect on growth and agr expression of S. aureus (Qazi et al., 2006). Furthermore, long-chain AHLs produced by other Gram-negative bacteria are likely to have similar effects in S. aureus, although, again, more studies are required here.
P. aeruginosa Might also Cause Increased Expression of S. aureus Virulence Factors
As described before, wounds infected with both S. aureus and P. aeruginosa generally show delayed closure compared to the single species infected wounds (Dalton et al., 2011; Seth et al., 2012). In addition to host related factors, one of the reasons for this phenomenon might be the upregulation of S. aureus virulence factors during co-infection, as was demonstrated for the MRSA strain USA300 (Pastar et al., 2013). Interestingly, although the growth of USA300 was strongly inhibited by P. aeruginosa in vitro, this effect was much weaker in vivo (Pastar et al., 2013). Furthermore, co-infection of wounds in a pig model induced S. aureus virulence factors hla and pvl, encoding α-hemolysin and Panton-Valentine leucocidin (Figure 2, right lower panel; Pastar et al., 2013). Another example of increased virulence of S. aureus in the presence of P. aeruginosa is the induction of staphyloxanthin production observed in a white S. aureus variant isolated from a soft tissue wound (Antonic et al., 2013). This strain possessed an intact and functional crtOPQMN operon, which is essential for production of the staphyloxanthin pigment, but was unable to induce pigment production on its own. Interestingly, staphyloxanthin production was induced by a P. aeruginosa co-isolate (Antonic et al., 2013). Furthermore, the pigment production in a characteristically golden-yellow S. aureus strain, which was co-isolated with the Pseudomonas strain and the white S. aureus variant, remained unaffected. However, this contradicts other studies that report an inhibition of S. aureus pigment production by pyocyanin and pyoverdin produced by P. aeruginosa (Biswas et al., 2009). Another result of this study (Antonic et al., 2013) that is in contradiction with other studies is that there was an unchanged expression of sigB that encodes the alternative transcription factor sigma B and which is previously reported to be upregulated in the presence of P. aeruginosa (Mitchell et al., 2010). The discrepancies in results between different studies indicate the importance of co-evolution and adaptation of the different isolates to each other and their environment. Adaptation of S. aureus to P. aeruginosa might lead to an expression pattern that is similar to a stress-resistant phenotype.
S. aureus Survives in The Presence of P. aeruginosa as The Small Colony Variant Phenotype
As a defense mechanism, S. aureus has also devised strategies to survive in the presence of P. aeruginosa. One of these is the switch to the SCV, a well-characterized phenotype detected in various diseases, including CF and device-related infections (Proctor et al., 2006). SCVs appear as small, smooth colonies on a culture plate and grow significantly slower compared to wild type colonies. The SCV phenotype might appear naturally and is caused by a defective or inhibited electron transport pathway that switches S. aureus to a fermentative growth state. In addition to a decreased growth rate, SCVs also demonstrate decreased ATP yield, decreased pigmentation and often hemin or menadione auxotrophy (Proctor et al., 2006; Biswas et al., 2009). Remarkably, the switch to a SCV phenotype increases survival of S. aureus in unfavorable conditions as it exhibits an increased aminoglycoside resistance, biofilm formation, and intracellular survival (Hoffman et al., 2006; Proctor et al., 2006; Biswas et al., 2009; Atalla et al., 2011). Prolonged co-culture with P. aeruginosa or exposure to pure HQNO leads to a high proportion of stable S. aureus SCVs, an effect that is increased by the presence of aminoglycosides (Hoffman et al., 2006). It has also been proposed that the reason why S. aureus and P. aeruginosa are not frequently detected together in diagnostic cultures of sputum of CF patients is because of the existence of S. aureus as SCVs that are more difficult to detect due to their small size and fastidious growth requirements (Proctor et al., 2006; Atalla et al., 2011).
SCV Induction by P. aeruginosa Is Sigma B Dependent
After the induction of the SCV phenotype during exposure to HQNO, the expression of three main regulatory mechanisms of virulence and biofilm formation is altered. First, the alternative transcription factor sigma B (SigB) is upregulated (Mitchell et al., 2010). SigB regulates the general stress response of Gram-positive bacteria, repressing the expression of most exoenzymes and toxins, stimulating the expression of adhesins and promoting the persistence of S. aureus in host cells (van Schaik and Abee, 2005; Atalla et al., 2011). Second, stimulation of SigB was shown to stimulate the expression of the Staphylococcal accessory regulator SarA, which modulates the expression of the pore-forming toxin α-hemolysin (hla) and increases biofilm formation (Valle et al., 2003; Oscarsson et al., 2006; Mitchell et al., 2010). In addition, SigB represses a third important regulator of S. aureus biofilm formation, the accessory gene regulator (agr) system, which induces biofilm dispersal thereby decreasing the total biomass and increases the expression of hla (Boles and Horswill, 2008; Mitchell et al., 2010; Atalla et al., 2011). In conclusion, HQNO reduces the production of the toxin hla by increasing the expression of SigB, leading to an increased expression of SarA and a decreased expression of agr. Reduced expression of toxins helps S. aureus to remain intracellular and thus increase its chances of survival in the human host. The stimulation of sarA by upregulation of SigB might be counteracted by long chain AHLs, which inhibit sarA (Figure 2, right lower panel). The net effect is probably dependent on the Pseudomonas strain involved since the production of both factors might be variable between isolates (Qazi et al., 2006; Fugère et al., 2014).
P. aeruginosa and S. aureus in Dual Species Biofilms
During dual species biofilm formation, the balance of attacking, evading and counter-attacking is even more important and the properties of each strain as well as some environmental factors will determine if a dual species biofilm will be formed. For example, the presence of environmental selection pressure, like antibiotics or the host immune system, stimulates a more synergistic relationship and biofilm formation as the tolerance of S. aureus to antibiotics is significantly higher during co-culture with P. aeruginosa (DeLeon et al., 2014; Kumar and Ting, 2015). In the following paragraphs we describe how certain extracellular factors influence the structure, characteristics and composition of the dual species biofilms (Figure 3).
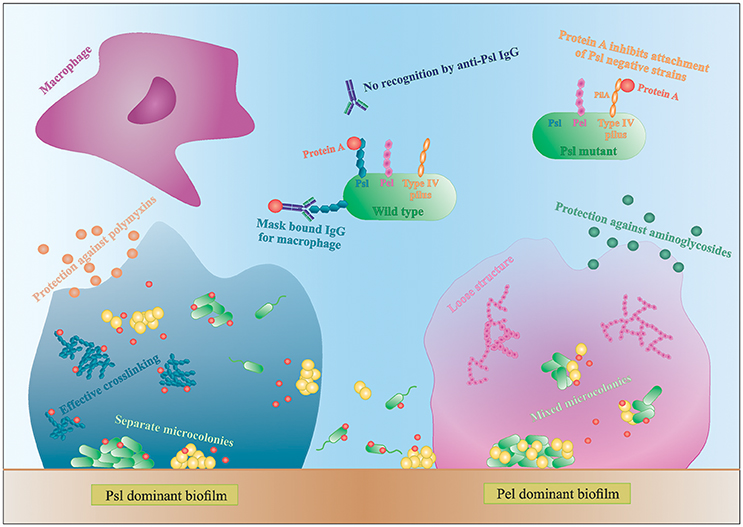
Figure 3. Interactions in mixed species biofilms. Differences in biofilm formation by S. aureus with a Psl- or Pel-dominant P. aeruginosa strain. IgG, immunoglobulin G.
P. aeruginosa Extracellular Polysaccharides Are Important in the Formation of Multi-Species Biofilms
P. aeruginosa produces three main exopolysaccharides (EPS): alginate, Pel, and Psl, which form the extracellular matrix in the biofilm and have a structural and protective function (Leid et al., 2005; Ryder et al., 2007; Colvin et al., 2011, 2012). Pel and Psl are the main EPS in non-mucoid strains (Colvin et al., 2012). The Psl polysaccharide is recently identified as repeating units of glucose-, mannose-, and rhamnose-sugars and is mainly produced during the attachment phase of the biofilm (Colvin et al., 2012). Psl-positive strains have an elastic matrix with highly effective cross-linking of the matrix components (Chew et al., 2014). Pel is glucose-rich and is mainly involved in pellicle formation and later stages of biofilm formation (Colvin et al., 2012). Contrary to Psl, Pel dominant strains form loose biofilm structures since Pel reduces the effective cross-linking in the matrix network (Figure 3; Chew et al., 2014). Consequently, Pel-mediated loosening of the P. aeruginosa biofilm allows S. aureus to infiltrate into the biofilm and form multi-species biofilms (Chew et al., 2014). In contrast, the role of Psl in multi-species biofilm formation is not very clear. Chew et al showed that co-culture of a Psl-positive strain (PAO1) and S. aureus resulted in separated microcolonies without much association between both species (Chew et al., 2014). Billings et al., on the other hand, showed that S. aureus was incorporated in the air-liquid interface of a Psl producing P. aeruginosa biofilm. Both studies use the same biofilm assay and the same P. aeruginosa strains, PAO1 and mutants derived from PAO1, but different S. aureus strains. Because the biofilm structure and dual species interactions are dependent on both P. aeruginosa and the Staphylococcus strains, this might explain the discrepancy between the studies. Nonetheless, both studies concluded that the EPS provides protection against antibiotics to all inhabitants of the biofilm, even the non-producers, although the biofilm as a whole is weakened (Billings et al., 2013). More specifically, Psl functions as a protective barrier against the antibiotics colistin and polymyxin B, whereas Pel offers a protective barrier against aminoglycosides (Figure 3; Colvin et al., 2011; Billings et al., 2013). These findings indicate that a minimum amount of EPS per cell present in the biofilm is needed for optimal protection against antibiotics (Billings et al., 2013). These data suggests, if S. aureus is able to survive killing by P. aeruginosa and to co-exist in a multi-species biofilm, it benefits from the antimicrobial barrier formed by the P. aeruginosa matrix components. However, the third EPS, alginate, was not shown to have an effect on S. aureus and S. epidermidis biofilm formation. Alginate is mainly associated with chronic infections as its overproduction leads to the mucoid phenotype that frequently arises during long-term CF lung infection (Ryder et al., 2007). The switch to a mucoid phenotype contributes to the establishment of chronic colonization since alginate offers structural protection against uptake by macrophages and antimicrobials by forming a barrier limiting the penetration of antimicrobials, macrophages and macrophage-derived products, such as the pro-phagocytic cytokine IFN-γ (Leid et al., 2005; Ryder et al., 2007). Moreover, Leid et al. suggest that alginate might cause the transition from acute to chronic infection by limiting IFN-mediated clearance by macrophages, which is the main mechanism of bacterial clearance during acute infection (Leid et al., 2005).
S. aureus Protein a Binds to Psl and Type IV Pili of P. aeruginosa
In addition to EPS, other extracellular factors are important in dual species biofilm formation. Yang et al showed that only P. aeruginosa strains producing type IV pili co-aggregate with S. aureus in microcolonies (Yang et al., 2011). Type IV pili probably facilitate biofilm formation by binding to extracellular DNA (eDNA), which is derived from dead bacteria and part of the biofilm matrix (Yang et al., 2011). P. aeruginosa strains defective for the production of type IV pili or even treating the biofilm with DNAse I was shown to reduce the growth of mixed-species microcolonies (Yang et al., 2011). Furthermore, using a single S. aureus laboratory strain, Armbruster et al. showed inhibition of surface attachment of some P. aeruginosa clinical isolates due to the secretion of protein A (SpA) by S. aureus. SpA is a cell-wall associated extracellular adhesive protein of S. aureus that mediates biofilm formation and disrupts phagocytosis (by binding to the Fc portion of IgG antibodies) and its secretion was shown to be increased in artificial sputum (Armbruster et al., 2016). Secreted SpA was shown to specifically bind both Psl and type IV pili of P. aeruginosa, stressing the importance of these two molecules in multispecies interactions (Figure 3; Armbruster et al., 2016). In a Psl producing P. aeruginosa, all SpA seem to bind to the Psl, leaving the type IV pili free to mediate biofilm formation. In absence of Psl, SpA binds to the PilA component of type IV pili and inhibits adhesion of P. aeruginosa (Armbruster et al., 2016). Furthermore, SpA seems to protect P. aeruginosa from phagocytosis, as the Psl-SpA complex is no longer recognized by anti-Psl IgG antibodies. SpA can also bind to the Fc domain of anti-Psl IgG antibodies and prevent recognition by neutrophils (Figure 3; Armbruster et al., 2016).
P. aeruginosa and Other Staphylococci
P. aeruginosa Induces Biofilm Dispersal in S. epidermidis
The antistaphylococcal molecules produced by P. aeruginosa are also active against other staphylococci, including S. epidermidis, although some are more resistant to killing compared to S. aureus. P. aeruginosa was shown to effectively inhibit and disrupt established S. epidermidis biofilms and induce detachment without killing during dual species biofilm formation (Qin et al., 2009; Pihl et al., 2010a,b). After co-inoculation in equal proportions, P. aeruginosa and S. epidermidis could coexist for up to 18 h. After this time point, the S. epidermidis cells in the biofilm are lysed by P. aeruginosa (Pihl et al., 2010a). These data suggest that there are two stages in interactions between P. aeruginosa and S. epidermidis, the first includes the induction of detachment of viable S. epidermidis cells from the biofilm, while in the second stage cell lysis causes the total detachment (Pihl et al., 2010a). Similar to S. aureus, the effect of P. aeruginosa on S. epidermidis is strain dependent as some P. aeruginosa strains have a more pronounced effect on some S. epidermidis strains while others are more resistant to P. aeruginosa (Pihl et al., 2010b). Nevertheless, extracellular products that prevent initial attachment of some S. epidermidis strains to surfaces might be an interesting option for the development of coatings for indwelling medical devices, like peritoneal dialysis catheters (Pihl et al., 2013). Moreover, in this model, P. aeruginosa supernatant components replaced serum proteins on the catheter surface and reduced S. epidermidis attachment (Pihl et al., 2013). In addition, exposure of a S. epidermidis biofilm on a catheter to P. aeruginosa supernatant also caused dispersal of S. epidermidis (Pihl et al., 2013). The dispersed cells are, however, not killed making it less suitable as a treatment option and only interesting as a prevention strategy.
Yayurea A and B from the S. intermedius Group Are Quorum-Quenching Molecules Which Provide Protection against Gram Negative Bacteria
P. aeruginosa is originally an environmental bacterium and shares a niche with many other, non-pathogenic staphylococci like the Staphylococcus intermedius group consisting of S. delphini, S. intermedius, S. lutrae, S. pseudointermedius and S. schleiferi. All are common colonizers of various animals and rarely occur in humans (Simou et al., 2005; Ruscher et al., 2009). This group of staphylococci produces two low molecular weight compounds, yayurea A and B, that inhibit the production of quorum-sensing regulated products in Gram negative bacteria and provide protection against extracellular compounds produced by P. aeruginosa (Chu et al., 2013). For example, the growth of S. delphini is not suppressed by respiratory toxins during co-culture with P. aeruginosa. Moreover, S. delphini is able to completely inhibit the production of pyocyanin (Chu et al., 2013). The quorum-quenching effect of yayurea A and B covers a broad spectrum of Gram negative bacteria, including P. aeruginosa, Serratia marcescens, Vibrio harveyi, and Chromobacterium subtsugae (Chu et al., 2013). Quenching of the quorum-sensing system of these Gram negative bacteria does not kill them but rather maintains their physiological state as if the cell density is low, even though density is in fact high. This increases the chances of survival of the staphylococci since toxin production usually begins at high cell density (Chu et al., 2013). Surprisingly, other staphylococci seem to be resistant to both molecules, even though they are not producers (Chu et al., 2013). Interestingly, S. aureus is protected from killing by P. aeruginosa when yayurea A and B are added to the medium without the former having to undergo physiological changes (SCV formation), amd represent promising candidates for inhibition studies of P. aeruginosa virulence and biofilm formation.
Variations in CydAB from S. carnosus Provides Protection Against Killing by P. aeruginosa
In addition to the S. intermedius group, several other non-pathogenic staphylococci (S. carnosus, S. piscifermentans, and S. simulans) seem to be resistant to respiratory toxins secreted by P. aeruginosa due to alterations in the cydAB genes. These genes encode the two subunits of cytochrome bd quinol oxidase, of which homologs are also present in the genomes of S. aureus and S. epidermidis (Voggu et al., 2006). However, only the cytochrome bd quinol oxidase of the first group is resistant to the respiratory toxin, pyocyanin (Voggu et al., 2006). Furthermore, cloning of the S. carnosus cydAB cluster into S. aureus confers resistance to respiratory inhibitors produced by P. aeruginosa (Voggu et al., 2006). Further research showed that, whereas the CydA subunit is more conserved in staphylococci, CydB underwent a microevolution with relatively higher identity within than between the groups of pathogenic and non-pathogenic staphylococci (Voggu et al., 2006). This asymmetric evolution of CydB could be explained by the fact that the non-pathogenic staphylococci frequently inhabit the same environment as Pseudomonas spp. and were therefore selected for a higher resistance to respiratory toxins.
Concluding Remarks
Both P. aeruginosa and staphylococci are highly versatile organisms, which readily adapt to a wide variety of environments and stress factors. In the first glance, these bacteria seem to have an antagonistic relationship as P. aeruginosa produces a wide variety of molecules inhibiting staphylococci and frequently outcompetes S. aureus and S. epidermidis during co-culture. This antagonistic behavior is mainly shown during planktonic growth and under traditional culture conditions, where no host factors or antibiotics are present. However, under some in vitro and in vivo circumstances, both bacteria are able to co-exist and form dual species biofilms. These circumstances are dependent on a combination of strain-dependent properties of both species and the presence or absence of certain environmental factors like antibiotics or host factors, the sum of which might tip the balance toward either killing or co-existence. The presence of some sort of selection pressure or presence of a preformed matrix seems to favor dual species biofilm formation whereas planktonic co-culture without selection pressure leads to domination of P. aeruginosa. Interspecies competition often leads to an increased production of virulence factors in both P. aeruginosa and S. aureus, which are also harmful to the human host. In addition, escaping the antistaphylococcal compounds results in a more stress-resistant phenotype of S. aureus, which is more difficult to be cleared by the immune system, to be eradicated by antibiotics and to be detected in diagnostic cultures. Furthermore, the presence of an extracellular matrix was shown to be beneficial for all biofilm inhabitants, providing protection against classical antibiotics and the host immune system, although the exact composition might be variable depending on the species present and the environment. The role of these matrix components (exopolysaccharides, eDNA, matrix proteins, host-derived factors etc.) in interspecies interactions and their role in disease pathogenesis provides an exciting opportunity for future research toward better patient care. When strains are co-existing for a longer time, they might evolve to a phenotype that is better adapted to the presence of the other. For example, non-pathogenic staphylococci that are frequently encountering P. aeruginosa have developed strategies to continue growing in the presence of P. aeruginosa antistaphylcoccal compounds, indicating parallel evolution. Moreover, S. aureus and P. aeruginosa strains isolated from the same chronic CF lung infection are less sensitive to, and produce less, HQNO, respectively. This strain adaptation and the underestimation of the co-existence of P. aeruginosa and S. aureus might still have a large impact on the clinical outcome of a patient and therefore should be a subject of continuing investigation.
Author Contributions
AH collected literature. All authors contributed in drafting the review.
Funding
AH was supported by funding from the Innovative Medicines Initiative project COMBACTE-MAGNET (Combatting Bacterial Resistance in Europe—Molecules Against Gram Negative Infections) (IMI Grant Agreement No 115737-2) and funding from Research Foundation Flanders (FWO-F, G.0513.12).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Antonic, V., Stojadinovic, A., Zhang, B., Izadjoo, M. J., and Alavi, M. (2013). Pseudomonas aeruginosa induces pigment production and enhances virulence in a white phenotypic variant of Staphylococcus aureus. Infect. Drug Resist. 6, 175–186. doi: 10.2147/IDR.S49039
Armbruster, C. R., Wolter, D. J., Mishra, M., Hayden, H. S., Radey, M. C., Merrihew, G., et al. (2016). Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. MBio 7:e00538–16. doi: 10.1128/mBio.00538-16
Atalla, H., Gyles, C., and Mallard, B. (2011). Staphylococcus aureus small colony variants (SCVs) and their role in disease. Anim. Health Res. Rev. 12, 33–45. doi: 10.1017/S1466252311000065
Baldan, R., Cigana, C., Testa, F., Bianconi, I., De Simone, M., Pellin, D., et al. (2014). Adaptation of Pseudomonas aeruginosa in Cystic Fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS ONE 9:89614. doi: 10.1371/journal.pone.0089614
Barequet, I. S., Habot-Wilner, Z., Mann, O., Safrin, M., Ohman, D. E., Kessler, E., et al. (2009). Evaluation of Pseudomonas aeruginosa staphylolysin (LasA protease) in the treatment of methicillin-resistant Staphylococcus aureus endophthalmitis in a rat model. Graefes Arch. Clin. Exp. Ophthalmol. 247, 913–917. doi: 10.1007/s00417-009-1061-2
Billings, N., Millan, M., Caldara, M., Rusconi, R., Tarasova, Y., Stocker, R., et al. (2013). The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 9:e1003526. doi: 10.1371/journal.ppat.1003526
Biswas, L., Biswas, R., Schlag, M., Bertram, R., and Gotz, F. (2009). Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75, 6910–6912. doi: 10.1128/aem.01211-09
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Burmolle, M., Ren, D., Bjarnsholt, T., and Sorensen, S. J. (2014). Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 22, 84–91. doi: 10.1016/j.tim.2013.12.004
Cystic Fibrosis Foundation (2014). 2013 patient registry annual data report. Bathesda, MD: Cystic Fibrosis Foundation.
Cystic Fibrosis Foundation Patient Registry (2012). 2011 annual data report. Bathesda, MD: Cystic Fibrosis Foundation.
Caldwell, C. C., Chen, Y., Goetzmann, H. S., Hao, Y., Borchers, M. T., Hassett, D. J., et al. (2009). Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am. J. Pathol. 175, 2473–2488. doi: 10.2353/ajpath.2009.090166
Chew, S. C., Kundukad, B., Seviour, T., van der Maarel, J. R., Yang, L., Rice, S. A., et al. (2014). Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. MBio 5, e01536–e01514. doi: 10.1128/mBio.01536-14
Chu, Y. Y., Nega, M., Wolfle, M., Plener, L., Grond, S., Jung, K., et al. (2013). A new class of quorum quenching molecules from Staphylococcus species affects communication and growth of gram-negative bacteria. PLoS Pathog. 9:e1003654. doi: 10.1371/journal.ppat.1003654
Colvin, K. M., Gordon, V. D., Murakami, K., Borlee, B. R., Wozniak, D. J., Wong, G. C., et al. (2011). The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264. doi: 10.1371/journal.ppat.1001264
Colvin, K. M., Irie, Y., Tart, C. S., Urbano, R., Whitney, J. C., Ryder, C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14, 1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x
Dalton, T., Dowd, S. E., Wolcott, R. D., Sun, Y., Watters, C., Griswold, J. A., et al. (2011). An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS ONE 6:e27317. doi: 10.1371/journal.pone.0027317
Davies, D. G., and Marques, C. N. (2009). A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403. doi: 10.1128/jb.01214-08
DeLeon, S., Clinton, A., Fowler, H., Everett, J., Horswill, A. R., and Rumbaugh, K. P. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728. doi: 10.1128/iai.02198-14
Deziel, E., Lepine, F., Milot, S., He, J., Mindrinos, M. N., Tompkins, R. G., et al. (2004). Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. U.S.A. 101, 1339–1344. doi: 10.1073/pnas.0307694100
Diggle, S. P., Matthijs, S., Wright, V. J., Fletcher, M. P., Chhabra, S. R., Lamont, I. L., et al. (2007). The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 14, 87–96. doi: 10.1016/j.chembiol.2006.11.014
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Duan, K., Dammel, C., Stein, J., Rabin, H., and Surette, M. G. (2003). Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x
Falcon, M. A., Mansito, T. B., Carnicero, A., and Gutierrez-Navarro, A. M. (1989). L-form-like colonies of Staphylococcus aureus induced by an extracellular lytic enzyme from Pseudomonas aeruginosa. J. Clin. Microbiol. 27, 1650–1654.
Fazli, M., Bjarnsholt, T., Kirketerp-MØller, K., JØrgensen, B., Andersen, A. S., Krogfelt, K. A., et al. (2009). Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47, 4084–4089. doi: 10.1128/jcm.01395-09
Federle, M. J., and Bassler, B. L. (2003). Interspecies communication in bacteria. J. Clin. Invest. 112, 1291–1299. doi: 10.1172/jci20195
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Fugére, A., Lalonde Séguin, D., Mitchell, G., Déziel, E., Dekimpe, V., Cantin, A. M., et al. (2014). Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS ONE 9:e86705. doi: 10.1371/journal.pone.0086705
Gibson, J., Sood, A., and Hogan, D. A. (2009). Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 75, 504–513. doi: 10.1128/AEM.01037-08
Haba, E., Pinazo, A., Jauregui, O., Espuny, M. J., Infante, M. R., and Manresa, A. (2003). Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 81, 316–322. doi: 10.1002/bit.10474
Hall-Stoodley, L., and Stoodley, P. (2005). Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13, 7–10. doi: 10.1016/j.tim.2004.11.004
Hall-Stoodley, L., Costerton, J. W., and Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Harrison, F. (2007). Microbial ecology of the cystic fibrosis lung. Microbiology 153, 917–923. doi: 10.1099/mic.0.2006/004077-0
Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665. doi: 10.1038/nrmicro2199
Hauser, A. R., Jain, M., Bar-Meir, M., and McColley, S. A. (2011). Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 24, 29–70. doi: 10.1128/CMR.00036-10
Hoffman, L. R., Deziel, E., D'Argenio, D. A., Lepine, F., Emerson, J., McNamara, S., et al. (2006). Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 103, 19890–19895. doi: 10.1073/pnas.0606756104
Hotterbeekx, A., Xavier, B. B., Bielen, K., Lammens, C., Moons, P., Schepens, T., et al. (2016). The endotracheal tube microbiome associated with Pseudomonas aeruginosa or Staphylococcus epidermidis. Sci. Rep. 6:36507. doi: 10.1038/srep36507
Hubert, D., Réglier-Poupet, H., Sermet-Gaudelus, I., Ferroni, A., Le Bourgeois, M., Burgel, P. R., et al. (2013). Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. J. Cyst. Fibros. 12, 497–503. doi: 10.1016/j.jcf.2012.12.003
Jefferson, K. K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x
Jensen, P. Ø., Bjarnsholt, T., Phipps, R., Rasmussen, T. B., Calum, H., Christoffersen, L., et al. (2007). Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153, 1329–1338. doi: 10.1099/mic.0.2006/003863-0
Jimenez, P. N., Koch, G., Thompson, J. A., Xavier, K. B., Cool, R. H., and Quax, W. J. (2012). The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65. doi: 10.1128/MMBR.05007-11
Kessler, E., Safrin, M., Olson, J. C., and Ohman, D. E. (1993). Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 268, 7503–7508.
Kirketerp-MØller, K., Jensen, P. Ø., Fazli, M., Madsen, K. G., Pedersen, J., Moser, C., et al. (2008). Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46, 2717–2722. doi: 10.1128/jcm.00501-08
Korgaonkar, A. K., and Whiteley, M. (2011). Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J. Bacteriol. 193, 909–917. doi: 10.1128/JB.01175-10
Korgaonkar, A., Trivedi, U., Rumbaugh, K. P., and Whiteley, M. (2013). Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. U.S.A. 110, 1059–1064. doi: 10.1073/pnas.1214550110
Kumar, A., and Ting, Y. P. (2015). Presence of Pseudomonas aeruginosa influences biofilm formation and surface protein expression of Staphylococcus aureus. Environ. Microbiol. 17, 4459–4468. doi: 10.1111/1462-2920.12890
Leid, J. G., Willson, C. J., Shirtliff, M. E., Hassett, D. J., Parsek, M. R., and Jeffers, A. K. (2005). The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J. Immunol. 175, 7512–7518. doi: 10.4049/jimmunol.175.11.7512
Li, H., Li, X., Wang, Z., Fu, Y., Ai, Q., Dong, Y., et al. (2015). Autoinducer-2 regulates Pseudomonas aeruginosa PAO1 biofilm formation and virulence production in a dose-dependent manner. BMC Microbiol. 15:192. doi: 10.1186/s12866-015-0529-y
Li, Y. H., and Tian, X. (2012). Quorum sensing and bacterial social interactions in biofilms. Sensors 12, 2519–2538. doi: 10.3390/s120302519
Lightbown, J. W., and Jackson, F. L. (1956). Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem. J. 63, 130–137. doi: 10.1042/bj0630130
Machan, Z. A., Pitt, T. L., White, W., Watson, D., Taylor, G. W., Cole, P. J., et al. (1991). Interaction between Pseudomonas aeruginosa and Staphylococcus aureus: description of an anti-staphylococcal substance. J. Med. Microbiol. 34, 213–217. doi: 10.1099/00222615-34-4-213
Machan, Z. A., Taylor, G. W., Pitt, T. L., Cole, P. J., and Wilson, R. (1992). 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30, 615–623. doi: 10.1093/jac/30.5.615
Mashburn, L. M., Jett, A. M., Akins, D. R., and Whiteley, M. (2005). Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187, 554–566. doi: 10.1128/JB.187.2.554-566.2005
Mitchell, G., Séguin, D. L., Asselin, A. E., Deziel, E., Cantin, A. M., Frost, E. H., et al. (2010). Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 10:33. doi: 10.1186/1471-2180-10-33
Oscarsson, J., Kanth, A., Tegmark-Wisell, K., and Arvidson, S. (2006). SarA is a repressor of hla (α-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J. Bacteriol. 188, 8526–8533. doi: 10.1128/JB.00866-06
Palmer, K. L., Mashburn, L. M., Singh, P. K., and Whiteley, M. (2005). Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187, 5267–5277. doi: 10.1128/jb.187.15.5267-5277.2005
Pastar, I., Nusbaum, A. G., Gil, J., Patel, S. B., Chen, J., Valdes, J., et al. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 8:e56846. doi: 10.1371/journal.pone.0056846
Petrova, O. E., and Sauer, K. (2016). Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microbiol. 30, 67–78. doi: 10.1016/j.mib.2016.01.004
Pihl, M., Arvidsson, A., Skepö, M., Nilsson, M., Givskov, M., Tolker-Nielsen, T., et al. (2013). Biofilm formation by Staphylococcus epidermidis on peritoneal dialysis catheters and the effects of extracellular products from Pseudomonas aeruginosa. Pathog. Dis. 67, 192–198. doi: 10.1111/2049-632X.12035
Pihl, M., Chavez de Paz, L. E., Schmidtchen, A., Svensater, G., and Davies, J. R. (2010b). Effects of clinical isolates of Pseudomonas aeruginosa on Staphylococcus epidermidis biofilm formation. FEMS Immunol. Med. Microbiol. 59, 504–512. doi: 10.1111/j.1574-695X.2010.00707.x
Pihl, M., Davies, J. R., Chavez de Paz, L. E., and Svensater, G. (2010a). Differential effects of Pseudomonas aeruginosa on biofilm formation by different strains of Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 59, 439–446. doi: 10.1111/j.1574-695X.2010.00697.x
Proctor, R. A., von Eiff, C., Kahl, B. C., Becker, K., McNamara, P., Herrmann, M., et al. (2006). Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305. doi: 10.1038/nrmicro1384
Qazi, S., Middleton, B., Muharram, S. H., Cockayne, A., Hill, P., O'Shea, P., et al. (2006). N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 74, 910–919. doi: 10.1128/IAI.74.2.910-919.2006
Qin, Z., Yang, L., Qu, D., Molin, S., and Tolker-Nielsen, T. (2009). Pseudomonas aeruginosa extracellular products inhibit staphylococcal growth, and disrupt established biofilms produced by Staphylococcus epidermidis. Microbiology 155, 2148–2156. doi: 10.1099/mic.0.028001-0
Rezzonico, F., Smits, T. H., and Duffy, B. (2012). Detection of AI-2 receptors in genomes of Enterobacteriaceae suggests a role of type-2 quorum sensing in closed ecosystems. Sensors 12, 6645–6665. doi: 10.3390/s120506645
Rodrigues, L. R., Banat, I. M., van der Mei, H. C., Teixeira, J. A., and Oliveira, R. (2006). Interference in adhesion of bacteria and yeasts isolated from explanted voice prostheses to silicone rubber by rhamnolipid biosurfactants. J. Appl. Microbiol. 100, 470–480. doi: 10.1111/j.1365-2672.2005.02826.x
Ruscher, C., Lubke-Becker, A., Wleklinski, C. G., Soba, A., Wieler, L. H., and Walther, B. (2009). Prevalence of Methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet. Microbiol. 136, 197–201. doi: 10.1016/j.vetmic.2008.10.023
Ryder, C., Byrd, M., and Wozniak, D. J. (2007). Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10, 644–648. doi: 10.1016/j.mib.2007.09.010
Sagel, S. D., Gibson, R. L., Emerson, J., McNamara, S., Burns, J. L., Wagener, J. S., et al. (2009). Impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J. Pediatr. 154, 183–188. doi: 10.1016/j.jpeds.2008.08.001
Seth, A. K., Geringer, M. R., Galiano, R. D., Leung, K. P., Mustoe, T. A., and Hong, S. J. (2012). Quantitative comparison and analysis of species-specific wound biofilm virulence using an in vivo, rabbit-ear model. J. Am. Coll. Surg. 215, 388–399. doi: 10.1016/j.jamcollsurg.2012.05.028
Sibley, C. D., Duan, K., Fischer, C., Parkins, M. D., Storey, D. G., Rabin, H. R., et al. (2008). Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4:e1000184. doi: 10.1371/journal.ppat.1000184
Simou, C., Hill, P. B., Forsythe, P. J., and Thoday, K. L. (2005). Species specificity in the adherence of staphylococci to canine and human corneocytes: a preliminary study. Vet. Dermatol. 16, 156–161. doi: 10.1111/j.1365-3164.2005.00452.x
Soberón-Chávez, G., Lépine, F., and Déziel, E. (2005). Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 68, 718–725. doi: 10.1007/s00253-005-0150-3
Sotirova, A. V., Spasova, D. I., Galabova, D. N., Karpenko, E., and Shulga, A. (2008). Rhamnolipid-biosurfactant permeabilizing effects on gram-positive and gram-negative bacterial strains. Curr. Microbiol. 56, 639–644. doi: 10.1007/s00284-008-9139-3
Strateva, T., and Mitov, I. (2011). Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann. Microbiol. 61, 717–732. doi: 10.1007/s13213-011-0273-y
Tashiro, Y., Yawata, Y., Toyofuku, M., Uchiyama, H., and Nomura, N. (2013). Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 28, 13–24. doi: 10.1264/jsme2.ME12167
Toyofuku, M., Nakajima-Kambe, T., Uchiyama, H., and Nomura, N. (2010). The effect of a cell-to-cell communication molecule, Pseudomonas quinolone signal (PQS), produced by P. aeruginosa on other bacterial species. Microbes Environ. 25, 1–7. doi: 10.1264/jsme2.ME09156
Valle, J., Toledo-Arana, A., Berasain, C., Ghigo, J. M., Amorena, B., Penades, J. R., et al. (2003). SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x
van Schaik, W., and Abee, T. (2005). The role of sigmaB in the stress response of Gram-positive bacteria – targets for food preservation and safety. Curr. Opin. Biotechnol. 16, 218–224. doi: 10.1016/j.copbio.2005.01.008
Voggu, L., Schlag, S., Biswas, R., Rosenstein, R., Rausch, C., and Gotz, F. (2006). Microevolution of cytochrome bd oxidase in Staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J. Bacteriol. 188, 8079–8086. doi: 10.1128/jb.00858-06
Wang, Z., Xiang, Q., Yang, T., Li, L., Yang, J., Li, H., et al. (2016). Autoinducer-2 of Streptococcus mitis as a target molecule to inhibit pathogenic multi-species biofilm formation in vitro and in an endotracheal intubation rat model. Front. Microbiol. 7:88. doi: 10.3389/fmicb.2016.00088
Watters, C., Fleming, D., Bishop, D., and Rumbaugh, K. P. (2016). Host responses to biofilm. Prog. Mol. Biol. Transl. Sci. 142, 193–239. doi: 10.1016/bs.pmbts.2016.05.007
Whiley, R. A., Sheikh, N. P., Mushtaq, N., Hagi-Pavli, E., Personne, Y., Javaid, D., et al. (2014). Differential potentiation of the virulence of the Pseudomonas aeruginosa cystic fibrosis liverpool epidemic strain by oral commensal Streptococci. J. Infect. Dis. 209, 769–780. doi: 10.1093/infdis/jit568
Williams, P., and Camara, M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12, 182–191. doi: 10.1016/j.mib.2009.01.005
Yang, L., Liu, Y., Markussen, T., HØiby, N., Tolker-Nielsen, T., and Molin, S. (2011). Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. 62, 339–347. doi: 10.1111/j.1574-695X.2011.00849.x
Keywords: microbial interactions, S. aureus, quorum-sensing, cystic fibrosis, biofilm
Citation: Hotterbeekx A, Kumar-Singh S, Goossens H and Malhotra-Kumar S (2017) In vivo and In vitro Interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 7:106. doi: 10.3389/fcimb.2017.00106
Received: 30 January 2017; Accepted: 16 March 2017;
Published: 03 April 2017.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Robert J. C. McLean, Texas State University, USASarah Maddocks, Cardiff Metropolitan University, UK
Copyright © 2017 Hotterbeekx, Kumar-Singh, Goossens and Malhotra-Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Surbhi Malhotra-Kumar, c3VyYmhpLm1hbGhvdHJhQHVhbnR3ZXJwZW4uYmU=
 An Hotterbeekx
An Hotterbeekx Samir Kumar-Singh
Samir Kumar-Singh Herman Goossens1
Herman Goossens1 Surbhi Malhotra-Kumar
Surbhi Malhotra-Kumar