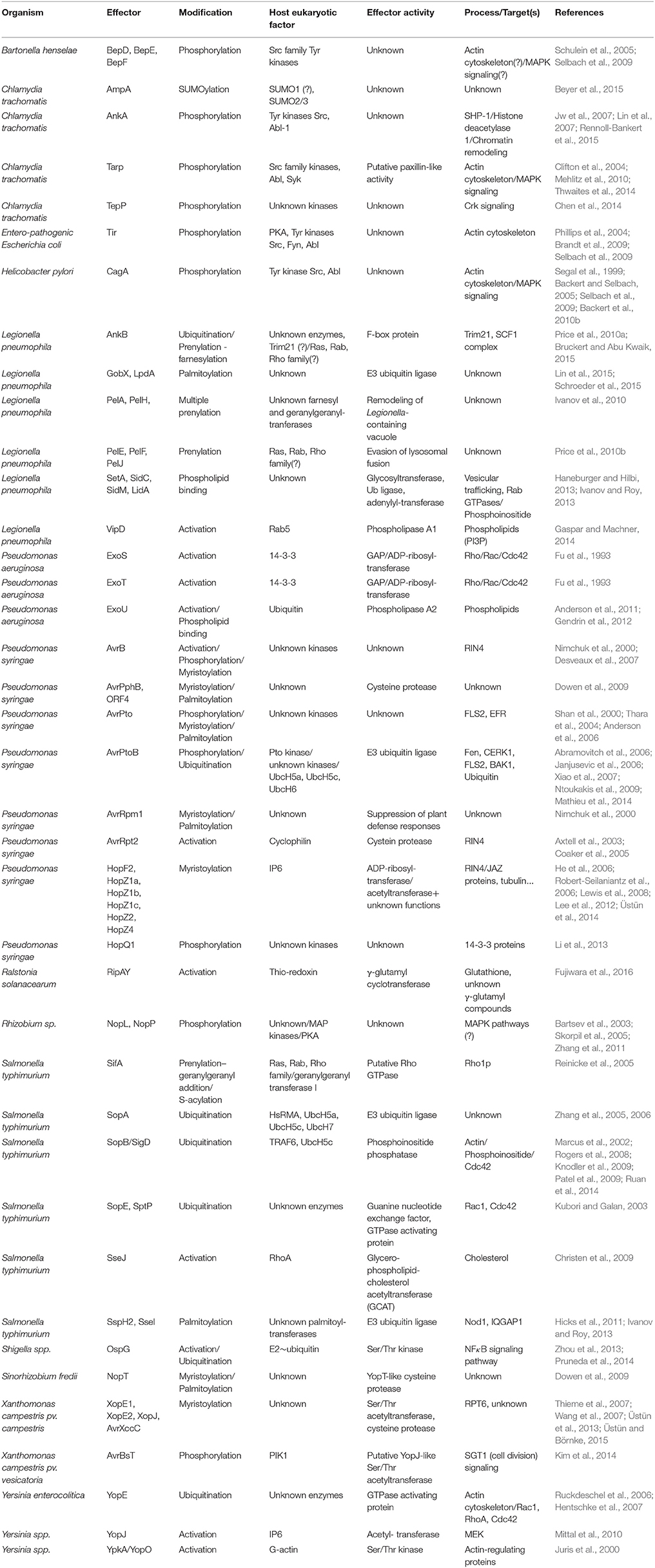
- 1Department of Genetics, Centre for Research in Agricultural Genomics (CSIC-IRTA-UAB), Universitat de Barcelona, Barcelona, Spain
- 2Department of Applied Biological Science, Faculty of Agriculture, Kagawa University, Kagawa, Japan
Pathogenic bacteria manipulate their hosts by delivering a number of virulence proteins -called effectors- directly into the plant or animal cells. Recent findings have shown that such effectors can suffer covalent modifications inside the eukaryotic cells. Here, we summarize the recent reports where effector modifications by the eukaryotic machinery have been described. We restrict our focus on proteins secreted by the type III or type IV systems, excluding other bacterial toxins. We describe the known examples of effectors whose enzymatic activity is triggered by interaction with plant and animal cell factors, including GTPases, E2-Ubiquitin conjugates, cyclophilin and thioredoxins. We focus on the structural interactions with these factors and their influence on effector function. We also review the described examples of host-mediated post-translational effector modifications which are required for proper subcellular location and function. These host-specific covalent modifications include phosphorylation, ubiquitination, SUMOylation, and lipidations such as prenylation, fatty acylation and phospholipid binding.
Activation of Bacterial Effectors through Interaction with Host Factors
Many effectors from bacterial pathogens of both animals and plants contain catalytic domains on their primary sequences with predicted enzymatic activities, such as phospholipase, protease, protein kinase, transferase, etc. and some of them have been found to be highly active enzymes that can outcompete their eukaryotic counterparts (Levin et al., 2010). However, not all effectors exhibit the enzyme activity when expressed in bacterial systems, but rather require interaction with additional eukaryotic factors for activation. Recent findings provide remarkable examples of spatiotemporal regulation of bacterial effectors by coupling the catalytic activity to the arrival into a host cell cytoplasm (Table 1; Anderson et al., 2015). Here, we will describe the available examples of the structure-based activation of effectors from animal and plant bacterial pathogens through interaction with host cell factors.
Allosteric Activation of Legionella Effector VipD by Host GTPase Rab5
Upon uptake by macrophages, Legionella pneumophila, the causative agent of legionnaires' disease (Horwitz and Silverstein, 1980), injects more than 250 effector proteins through the Dot/Icm type IV secretion system (T4SS) into the host cell. These effectors allow escape from the phagosomal maturation and establishment of a Legionella-containing vacuole (LCV) that supports bacterial proliferation (Ensminger and Isberg, 2009). The effector VipD can remove the endosomal specific phospholipid, phosphaptidyinositol-3 phosphate, PI(3)P by its robust phospholipase A1 (PLA1) activity, which is stimulated by the host GTPase Rab5, a key regulator of endosomes (Gaspar and Machner, 2014). Depletion of PI(3)P by VipD causes membrane disassociation of the endosomal fusion proteins including the tethering protein early endosomal antigen (EEA)1, resulting in inhibition of phagosomal maturation and allowing endosomal avoidance by LCVs. The N-terminal half of VipD possesses high homology to patatin, a lipid acyl hydrolase present in the potato tuber, whereas the C-terminal half of VipD is required for binding to the Rab5 to trigger PLA1 activity within the N-terminal domain (Gaspar and Machner, 2014). The crystal structure of VipD confirmed the predicted bimodular organization and in addition, revealed a surface loop called “lid,” that obstructs a PLA1 active site (Figure 1A, Closed Lid), explaining why recombinant VipD alone exhibits little or no PLA1 activity in vitro (Ku et al., 2012). The crystal structure of VipD in complex with constitutively active Rab5 provides evidence for a heterotropic allosteric activation mechanism in which locally induced structural changes through Rab5-binding are transmitted from the C-terminal domain of VipD to the N-terminal PLA1 domain, causing the reposition of the lid and exposure of the catalytic pocket (Figure 1A, Open Lid; Lucas et al., 2014).
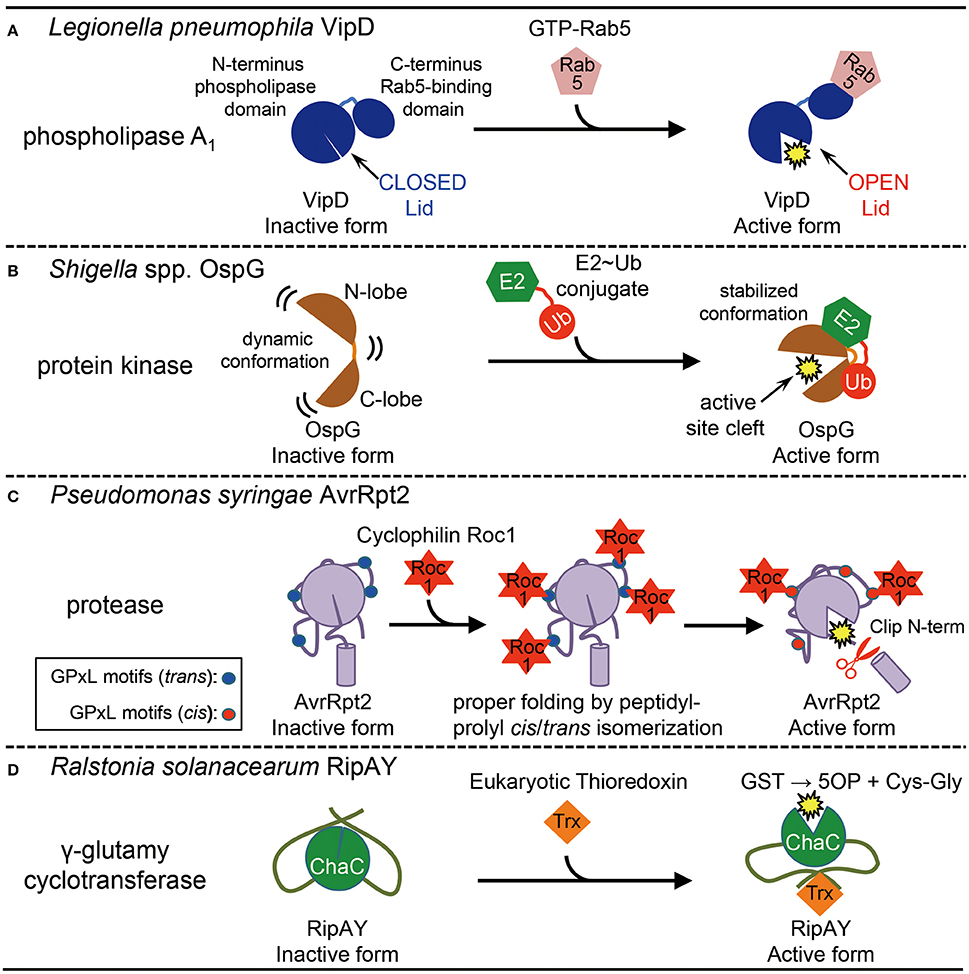
Figure 1. Activation of bacterial effector proteins by host eukaryotic factors. (A) Activation of Legionella pneumophila effector VipD by host small GTPase Rab5. Rab5-binding induces conformational change to open the lid, which results in the activation of its phospholipase A1 activity. (B) Activation of Shigella spp. effector OspG by host E2-ubiquitin (Ub) conjugate. E2~Ub binding stabilizes an active site cleft of the kinase, which stimulates enzymatic activity. (C) Activation of Pseudomonas syringae effector AvrRpt2 by host cyclophilin Roc1. Roc1 binds AvrRpt2 at the four potential cyclophilin-binding sites (GPxL) and properly folds the protein by cis/trans peptidyl-prolyl isomerization, which stimulates the protease activity. (D) Activation of Ralstonia solanacearum effector RipAY by host eukaryotic thioredoxins. Binding of eukaryotic thioredoxins stimulates the γ-glutamyl cyclotransferase activity of ChaC domain in RipAY by unknown mechanisms. GST, glutathione; 5OP, 5-oxoproline; Cys-Gly, cysteinylglycine.
E2-Ub Conjugates Stabilize an Active Conformation of Shigella Effector OspG
Shigella spp. are human pathogens that cause shigellosis and utilize a type III secretion system (T3SS) to deliver over 20 effector proteins to hijack cellular processes of the host and promote bacterial invasion, survival, and proliferation (Buchrieser et al., 2000). The Shigella effector OspG represents a minimal kinase domain that retains key catalytic elements, but lacks additional structural features typically found in eukaryotic kinases (Kim et al., 2005). Using a yeast two-hybrid analysis and pull-down experiments, Kim et al. (2005) identified that OspG binds to the ubiquitin (Ub)-conjugating enzymes (E2s) covalently linked with Ub (E2~Ub) and exhibits weak kinase activity in vitro (Kim et al., 2005). The crystal structure of OspG in complex with E2~Ub conjugate uncovered how E2~Ub conjugate-binding stimulates the kinase activity of OspG (Grishin et al., 2014; Pruneda et al., 2014). While OspG alone appears to be highly dynamic and weakly active, complex formation stabilizes a highly active conformation via simultaneous interaction with both subunits of the E2~Ub conjugate (Figure 1B). In vitro kinase assay revealed that OspG alone exhibits very weak kinase activity, while OspG in complex with E2~Ub conjugate exhibits substantially greater activity. Consistent with a model in which the OspG kinase domain is stabilized by binding to an intact E2~Ub conjugate, the covalent linkage between E2 enzyme and Ub is required for maximal activation of the kinase in vitro. It was recently showen that OspG also binds ubiquitin and polyubiqutin chains and this binding stimulates its kinase activity (see below).
Eukaryotic Cyclophilin-Dependent Conformational Change Activates P. syringae Effector AvrRpt2
The plant bacterial pathogen, Pseudomonas syringae injects between 20 and 30 effector proteins into host plant cells via the T3SS (Chang et al., 2005). Delivery of the effector AvrRpt2 to Arabidopsis thaliana plants expressing the plant resistance protein RPS2 specifically induces a hypersensitive response leading to disease resistance (Day et al., 2005). AvrRpt2 possesses cysteine protease activity and cleaves the Arabidopsis protein RIN4, which negatively regulates resistance interacting with RPS2 (Axtell et al., 2003). RPS2 is activated following RIN4 cleavage, thereby indirectly detecting AvrRpt2's enzymatic activity (Axtell and Staskawicz, 2003; Mackey et al., 2003). Interestingly, AvrRpt2 is delivered into plant cells as an inactive protease that is activated in planta and autoprocessed to trigger RIN4 degradation and subsequent activation of RPS2. Coaker et al. (2005), demonstrated that plant cyclophilin triggers both self-cleavage of AvrRpt2 and limited degradation of RIN4 (Coaker et al., 2005). Cyclophilin possesses peptidyl-prolyl cis/trans isomerase activity, which facilitates protein folding catalyzing the trans to cis isomerization of peptide bonds at proline residues (Kiefhaber et al., 1990). Enzymes from Arabidopsis plant deficient in peptidyl-prolyl cis/trans isomerization were unable to activate AvrRpt2 in vitro, indicating that this activity is key for AvrRpt2 activation (Coaker et al., 2006). Interestingly, AvrRpt2 possesses four consensus cyclophilin-binding motifs, GPxLs which are located in close proximity to ArRpt2's catalytic triad and are required for enzymatic activity both in vitro and in planta. Nuclear magnetic resonance spectra and gel filtration chromatography suggest that AvrRpt2 may only be structured and active when ROC1 or another cyclophilins is bound in order to maintain one or more proline residues in the appropriate isomerization state (Figure 1C; Coaker et al., 2006), which is also supported by analysis of protease activity using synthetic protease substrate (Aumüller et al., 2010).
Activation of R. solanacearum Effector RipAY by Host Thioredoxins
R. solanacearum is a widely distributed soil borne phytopathogen that possesses an exceptionally large (60–75) T3SS effector repertoire, for which only a few members have been assigned a molecular function (Coll and Valls, 2013). RipAY was identified as one of the few Ralstonia effectors causing growth inhibition in yeast (Fujiwara et al., 2016). RipAY contains a ChaC domain, which is conserved in all phyla and recently shown to encode γ-glutamyl cyclotransferase (GGCT) activity specifically to degrade glutathione (Kumar et al., 2012, 2015). RipAY has N- and C-terminal extension sequences outside of its ChaC domain, so that is much larger than other ChaC proteins (416 amino acids vs ~200 amino). In spite of the limited identity of its ChaC domain to the consensus sequence, RipAY was found to exhibit a robust GGCT activity in yeast cells. In addition, intracellular glutathione levels were significantly decreased following ripAY expression in yeast or inoculation of R. solanacearum wild-type, but not a RipAY-deficient strain, into plant leaves. Recombinant RipAY protein purified from a bacterial expression system showed undetectable GGCT activity, but addition of eukaryotic thioredoxins (Trxs) stimulated this activity in vitro (Figure 1D; Fujiwara et al., 2016). Yeast two hybrid analysis revealed that RipAY bound to plant cytoplasmic thioredoxins in an isoform-specific manner. RipAY preferentially bond to the plant cytoplasmic thioredoxin Trx-h5, whose expression is specifically induced during pathogen infection and the GGCT activity of RipAY was most efficiently stimulated by Trx-h5. Unlike the requirement of cyclophilin enzymatic activity for AvrRpt2 activation, the Trx-h5 oxide reductase activity is not indispensable, although a Trx-h5 redox inactive mutant showed decreased binding and activation of RipAY. The crystal structure of RipAY in complex with thioredoxin will uncover the mechanisms underlying recognition and activation of RipAY at the molecular level.
Post-Translational Modification of Pathogen Effectors Inside the Host Cells
Besides interacting with the host components, bacterial effectors can suffer a number of different post-translational modifications in the eukaryotic environment. These host-specific covalent modifications include phosphorylation, ubiquitination, SUMOylation and lipidation—mainly prenylation and fatty acylation. The main function of these modifications seems to be effector targeting to a precise subcellular compartment or regulation of its biological activity
Serine and Threonine Phosphorylation of Plant-Associated Type III Effectors
Specific kinases attach negatively charged phosphate groups to the phosphorylatable aminoacid residues (histidine, serine, threonine, and tyrosine) of their substrates, which regulates their function and changes their biochemical properties (Backert and Selbach, 2005; Korkuc and Walther, 2016). Such post-translational modifications by the host are essential for some plant-associated virulence proteins to manipulate host defense signaling. It is the case of Pseudomonas syringae type III effectors AvrPto and AvrPtoB, known to elicit plant resistance in the form of a hypersensitive cell death (Coll et al., 2011) after interaction with the tomato immunity-associated kinase Pto (Kim et al., 2002). Phosphorylation on two serine residues of AvrPto C-terminal domain was shown to contribute to both its virulence and avirulence activity inside the host cell (Anderson et al., 2006; Yeam et al., 2010). P. syringae strains carrying mutations in AvrPto S149, a phosphorylation site confirmed in vivo, and in S147 caused less severe disease symptoms in susceptible tomato plants lacking Pto kinase (Anderson et al., 2006). By contrast, these alterations still elicited cell death in resistant tomato cultivars, as a result of unaffected Pto-mediated recognition of AvrPto (Yeam et al., 2010). Still, phosphorylation on S147 and S149 was required for AvrPto recognition by a putative resistance protein in Nicotiana sylvestris and Nicotiana tabacum (Yeam et al., 2010). The Pto-independent kinase activity responsible for AvrPto phosphorylation was observed in various plant species (Anderson et al., 2006), however it seems that not all hosts use similar recognition mechanisms for the same effector.
Similarly, AvrPtoB was shown to be phosphorylated on two amino acid residues by host kinases (Xiao et al., 2007; Ntoukakis et al., 2009). Whereas phosphorylation on serine-258 contributed to AvrPtoB full activity (Xiao et al., 2007), phosphorylation on threonine-450 by Pto kinase itself had controversial outcomes on AvrPtoB-mediated recognition in resistant tomatoes. Ntoukakis and coworkers demonstrated that Pto kinase phosphorylates AvrPtoB on a threonine residue in its E3 ubiquitin ligase C-terminal domain, leading to effector inactivation and its inability to degrade the tomato kinase and suppress the immune response (Ntoukakis et al., 2009). On the contrary, another study described that Pto binding to the N-terminal domain of AvrPtoB (which includes residue serine-258), and not T450 phosphorylation, allows this kinase to evade degradation and activate immunity in response to the effector protein (Mathieu et al., 2014).
Other effectors from phytobacteria are substrates for host immunity kinases. Recent findings highlight phosphorylation as a key event during the resistance response of the host plant to the action of Xanthomonas campestris pv vesicatoria AvrBsT effector (Kim et al., 2014). Phosphorylation of AvrBsT by PIK1 (Pathogen-Induced Protein Kinase1), dependent on the presence of SGT1 (part of a protein complex with AvrBsT and PIK1), was shown to play a role in effector recognition and cell death-associated phenotype in N. benthamiana leaves (Kim et al., 2014). In a likewise manner, P. syringae HopQ1 phosphorylation on serine-51 residue strongly promoted bacterial virulence and modulated effector interaction with multiple tomato 14-3-3 proteins (Li et al., 2013). This is in accordance to a recent study, confirming a regulatory role of phosphorylation in compound binding (Korkuc and Walther, 2016).
Finally, two effectors from the symbiotic Rhizobium strain NGR234, NopL and NopP, are phosphorylated in vitro by different plant kinases, including MAPKs (Bartsev et al., 2003, 2004; Skorpil et al., 2005). Further experiments confirmed four phosphorylated serines of NopL and revealed this effector interference with MAPK pathways in yeast and tobacco, but it is yet unknown whether phosphorylation is involved in NopL and NopP function (Zhang et al., 2011).
Tyrosine Phosphorylation of Effector Proteins from Animal-Associated Bacteria
Phosphoproteomic studies have demonstrated that phosphorylation of proteins on tyrosine residues occurs at a minor ratio compared to serine and threonine phosphorylation (Olsen et al., 2006). Nevertheless, tyrosine phosphorylation is crucial for the regulation of processes like growth, division and differentiation in all eukaryotes, and it has recently emerged as a key circuit controlling many cellular functions in bacteria (Hunter, 2009; Whitmore and Lamont, 2012). In the last decades, many effector proteins from animal-associated bacteria such as enteropathogenic Escherichia coli, Helicobacter pylori, Chlamydia trachomatis, Bartonella henselae, and Anaplasma phagocytophilum have been shown to target and perturb host tyrosine (Tyr) phosphorylation mechanisms. H. pylori effector CagA and Tir from E. coli possess Tyr phosphorylation sites within conserved Glu-Pro-Ile-Tyr-Ala or related sequence motifs, described to be modified by host cytosolic kinases involved in signal transduction, including Src, Abl, and Fyn (Phillips et al., 2004; Tegtmeyer and Backert, 2011). Time-dependent regulation of CagA tyrosine phosphorylation process by host Src and Abl family kinases (Mueller et al., 2012) plays a direct role in effector activation and triggering of host cell morphological changes related to cytoskeletal rearrangements and induction of cell elongation (Backert et al., 2010b; Sougleri et al., 2016). Similarly, manipulation of actin signaling by E. coli Tir requires phosphorylated tyrosine-454 and tyrosine-474 residues, suggesting these phosphorylation events are triggered by infection with the bacteria and are crucial for the effector activity inside host (Campellone and Leong, 2003; Bommarius et al., 2007). Furthermore, E. coli and wild-type Tir specifically activate host protein kinase A (PKA), which phosphorylates Tir at serine-434 and serine-463, modifications that, in contrast to CagA-induced phenotype, inhibit cell elongation (Brandt et al., 2009; Backert et al., 2010a). Recent studies concluded multiple phosphorylation sites of type III or type IV effectors like CagA (H. pylori), Tir (EPEC E. coli), BepD-F (Bartonella henselae), Tarp (Chlamydia trachomatis) and AnkA (Anaplasma phagocytolium) allow these bacterial proteins to interfere with host cellular signaling at different levels, by recruiting a rich repertoire of interacting partners (Selbach et al., 2009; Hayashi et al., 2013).
Chlamydia trachomatis Tarp is another effector shown to undergo tyrosine phosphorylation, immediately after its translocation inside the host cell (Clifton et al., 2004). In this case, Tarp phosphorylation was required and led to an increase in the number of effector interactions with host partners, such as the human adaptor protein SHC1, involved in activation of growth and MAPK signaling (Mehlitz et al., 2010). Recent findings identified a novel Chlamydia type III effector, TepP, whose interaction with Crk, another host adaptor protein, depends on effector phosphorylation at tyrosine and serine residues (Chen et al., 2014). Interestingly, tyrosine phosphorylation of TepP was shown to occur later than Tarp phosphorylation, suggesting that C. trachomatis together with a type III secretion chaperone Slc1 are able to regulate translocation of the effector repertoire to the pathogen's benefit (Chen et al., 2014).
Ubiquitin-Dependent Function of Pseudomonas syringae, Salmonella, Yersinia and Legionella Effector Proteins
Ubiquitination is a post-translational protein modification involving the addition of the small (8.5 kDa) ubiquitin molecule on lysine residues (rarely on cysteine and serine) in the substrate N-terminus (Behrends and Harper, 2011). Attachment of a single ubiquitin moiety is called monoubiquitination, and it can affect the localization or the activity of the target protein (Ramanathan and Ye, 2012). The ubiquitin (Ub) subunit is itself modified on one or more of its seven lysine residues, leading to the formation of a poly-Ub chain (polyubiquitination), which can constitute a signal for target protein degradation or play a role in modulating substrate function (Behrends and Harper, 2011). Because ubiquitination regulates and participates in many cellular functions such as protein degradation, cell cycle, vesicle trafficking or immune responses, some bacterial effectors have evolved to exploit this system by binding to or modifying host ubiquitin components, while other effectors are themselves subjected to ubiquitination (Angot et al., 2007; Zhou and Zhu, 2015). Yersinia YopE and Salmonella thyphimurium SopA, SopB/SigD, SopE, and SptP type III effectors are all ubiquitinated after their translocation inside the host cell (Ruckdeschel et al., 2006; Narayanan and Edelmann, 2014). Time-dependent ubiquitination of SopE and SptP leaded to their degradation by the host ubiquitin-proteasome system (Kubori and Galan, 2003). Similarly, YopE belonging to Y. enterocolitica serotype O8, but not its homologs from serogroups O3 and O9, was shown to be polyubiquitinated at lysine-62 and lysine-72, suggesting that some effectors have evolved to escape ubiquitination and subsequent degradation by the host (Hentschke et al., 2007). This is also the case of type III effector SopB, whose host-mediated ubiquitination serves as a non-proteolytic signal and contributes to effector function and intracellular localization (Thomas and Holden, 2009; Narayanan and Edelmann, 2014). For example, a SopB mutant that cannot be ubiquitinated demonstrated that effector modification on any of its nine lysine residues is required for redistribution from the plasma membrane to the Salmonella-containing vacuole (SCV) and recruitment of the small GTPase Rab5 (Patel et al., 2009). Importantly, SopB forms -regardless of their ubiquitination status- are still present at the plasma membrane and function to stimulate bacterial internalization and actin remodeling. Effector delivery to SCV by the host-ubiquitin machinery concentrated SopB activities, such as alteration of phosphoinositide metabolism, at this site, which allows Salmonella to escape degradation by the lysosomes (Knodler et al., 2009; Patel et al., 2009). Regarding the host enzyme(s) that modify SopB, it was recently described that its ubiquitination is mediated by the E2 ubiquitin-conjugating UbcH5c enzyme and the TRAF6 member of E3 ubiquitin ligases (Ruan et al., 2014), enzymes regulating substrate specificity in the ubiquitination process (Berndsen and Wolberger, 2014). SopA is itself an ubiquitin E3 ligase, polyubiquitinated by the host HsRMA1, with the same E3 ligase activity (Zhang et al., 2005, 2006). Although SopA ubiquitination by HsRMA1 finally leads to effector “sacrifice” and its proteasomal degradation, it also serves as a signal regulating Salmonella escape into the cytosol, where it can rapidly multiply (Zhang et al., 2005). In a likewise manner, ubiquitination of P. syringae AvrPtoB by host enzymes, together with the effector intrinsic E3 ligase activity, play a role in AvrPtoB interaction with ubiquitin itself and suppression of plant immunity (Abramovitch et al., 2006; Janjusevic et al., 2006).
Finally, Shigella spp. OspG and L. pneumophila AnkB are also ubiquitinated. OspG binds ubiquitin and polyubiqutin chains, which stimulates its kinase activity (Zhou et al., 2013) and AnkB is polyubiquitinated through Lys11 (Bruckert and Abu Kwaik, 2015). Lys11-linked polyubiquitinated AnkB is not degraded by the proteasome, suggesting this post-translational effector modification might lead to other cellular outcomes, distinct from the established function of Lys11-linked chains as proteasomal targeting signals (Behrends and Harper, 2011; Bruckert and Abu Kwaik, 2015). Additional data would be needed to thoroughly understand the biological significance of the host-mediated polyubiquitination of this Legionella effector protein.
AmpA, a Bacterial SUMOylated Effector
Functionally distinct from the ubiquitin pathway, protein SUMOylation involves target substrate modification of one or more lysine residues by covalent attachment of a member of the small ubiquitin-like modifier (SUMO) family of proteins (Guo and Henley, 2014). By altering interactions of the modified substrate, or changing its localization, stability and activity, SUMO conjugation controls a broad network of cellular processes, including nuclear processes, metabolic pathways, endocytic trafficking of receptors and resistance to pathogens (Wilson, 2012).
Many pathogenic bacteria were described to exploit and negatively regulate host SUMOylation system (Wilson, 2012; Verma et al., 2015), however there are few cases when pathogens utilize this essential pathway to “adorn” their own effectors. Effector protein AmpA (Anaplasma phagocytophilum) was shown to be poly-SUMOylated by conjugation to SUMO2/3 and this modification promoted bacterial survival inside the host (Beyer et al., 2015). Although the molecular consequences of AmpA host-mediated SUMOylation are yet unknown, Beyer and coworkers insinuate this modification would offer the possibility to manipulate a wide range of host activities to a bacterium with a limited number of effector proteins (Beyer et al., 2015).
Prenylation of the Effectors AnkB and SifA
S-prenylation covalently adds isoprene groups, usually farnesyl (15-carbon) and geranylgeranyl (20-carbon), to specific cysteine residues within 5 amino acids from a protein C-terminus via thioether linkages. The CaaX (Cys—aliphatic—aliphatic—X) motif is the most common prenylation site in proteins, a reaction carried out by farnesyl transferase, Caax protease and geranylgeranyl transferase I (Casey and Seabra, 1996). Prenyl moieties can play an important role in increasing molecular hydrophobicity, so that they serve as mediators of membrane association or determine specific protein-protein interaction (Ivanov and Roy, 2013). Unlike S-palmitoylation (see below), S-prenylation is an irreversible process.
PelH and AnkB from Legionella pneumophila are known examples of farnesylated bacterial effectors (Price et al., 2010a,b). Specific inhibitors and mutant cell lines showed that host-dependent farnesylation -but not geranylgeranylation- of AnkB was shown to be indispensable for its anchoring to the cytosolic face of the membrane surrounding the LCV. This modification was also shown to be essential for biological function, as bacteria bearing a mutation in the farnesylated cysteine showed a reduced capacity to proliferate in mice lungs (Price et al., 2010a). The same experiments performed with PelH demonstrated that its farnesylation is essential for proper membrane location (Price et al., 2010b).
Prenylation by geranylgeranyl addition has been indirectly proven for the Salmonella typhimurium effector protein SifA. SifA is required for maintenance of the membrane that surrounds replicating bacteria in the so-called SCV. It was shown that for SifA targeting and association to membranes a C-terminal cysteine in a conserved CAAX and Rab geranylgeranyl transferase prenylation motif was required (Reinicke et al., 2005). This cysteine residue within the CAAX was shown to be modified by isoprenoid addition through the action of protein geranylgeranyl transferase I (Reinicke et al., 2005).
Prenylation may be a conserved mechanism for effector modification in animal pathogens, as in silico analyses show that most bacterial species contain effectors with the conserved prenylation motif. An exhaustive in silico screen of microbial genomes for C-terminal CXXX-motif-containing proteins identified 56 proteins (Al-Quadan et al., 2011), 10 of them corresponding to Legionella pneumophila type IV effectors (Ivanov et al., 2010). Mutation of this motif in the Legionella effectors or inhibition of isoprenoid biosynthesis in the host cell confirmed lipidation of AnkB, and PelH, and showed altered membrane localization of PelA, PelE, PelJ, and PelF. Treatment with specific enzyme inhibitors showed that AnkB and PelJ are farnesylated, whereas PelE and PelF are modified by a geranylgeranyltransferase (Ivanov et al., 2010). Prenylated eukaryotic proteins include Ras and members of the Rab and Rho families. It will be interesting to check whether effectors mimicking these eukaryotic activities (Popa et al., 2016) are also prenylated.
Effector Fatty Acylation: Myristoylation in Plant Cells and Palmitoylation in Animal Cells
Acylation involves the covalent attachment of fatty acids at certain amino acid residues. The saturated myristic (14-carbon) acid and palmitic acid (16-carbon) are the most common fatty acids covalently attached to proteins, providing different biochemical characteristics to the protein. Myristoylation is a common acylation through which a myristic acid is attached to the α-amino group of an N-terminal glycine residue through an amide linkage. This irreversible protein modification typically occurs co-translationally (Martin et al., 2011). Addition to a myristoyl group provides proteins sufficient hydrophobicity and affinity for membranes, but it is insufficient to maintain permanent association with them (Resh, 2006). For this reason, myristoylation is often combined with S-acylation on proximal cysteine residues (see below). S-palmitoylation is another acylation in which a palmitic acid is attached to the thiolate side chain of a cysteine residue via thioester linkage. In contrast to N-myristoylation, S-acylation is a post-translational and reversible modification and no consensus sequence for protein palmitoylation has been identified so far. Because of its long hydrophobic group, S-palmitoylation can permanently anchor the protein to the membrane, sometimes concentrated at lipid rafts and thioesterases can release the protein by cleaving the linkage to the lipid (Resh, 2006).
Myristoylation has been described in a number of effectors from bacterial plant pathogens. The Pseudomonas syringae effectors AvrRpm1 and AvrB were the first shown to require a consensus Glycine2 fatty acid acylation site for full functionality and to be myristoylated in the plant host cell (Nimchuk et al., 2000). This seems a common strategy for P. syringae effectors, as myristoylation sites in HopF2, AvrPphB, AvrPto, and four of the five HopZ family effectors (HopZ1a, HopZ1b, HopZ1c, and HopZ2) are also required for targeting these effectors to the plant plasma membrane (He et al., 2006; Robert-Seilaniantz et al., 2006; Lewis et al., 2008). The putative myristoylation site was essential for HopZ2 and HopF2 virulence functions and for HopZ1a and AvrPto recognition by the plant immune system. In the case of AvrPphB, the eukaryotic N-myristoylation site was only exposed after protein autoprocessing in the plant cell and direct binding of the lipid was proven (Nimchuk et al., 2000). In a recent report, HopZ4 was also shown to the plasma membrane and this location was required for its activity as a proteasome inhibitor (Üstün et al., 2014). The Xanthomonas effectors XopE1, XopE2, XopJ, and AvrXccC are also anchored to the plant plasma membrane via myristoylation, as point mutations in their putative myristoylated G2 glycine residues resulted in cytoplasmic localization (Thieme et al., 2007; Wang et al., 2007). In the case of XopJ -a cysteine protease that degrades its target RPT6 (Üstün and Börnke, 2015)- a G2A exchange in the N-terminal myristoylation motif also abolished its proteasome inhibitor activity inside host cells (Üstün et al., 2013). Although these results suggest that acylation plays an important role in effector function, direct myristoylation in plant host cells has not been demonstrated so far for any of them.
Contrary to mysristoylation, palmitoylation is more common in effectors from animal bacterial pathogens. For instance, the Salmonella effector proteins SspH2 and SseI are localized to the plasma membrane of host cells through S-palmitoylation of a conserved cysteine residue within their N-terminal domains. In these cases, lipidation is mediated by specific palmitoyltransferases from the host cell and is critical for effector function (Hicks et al., 2011; Ivanov and Roy, 2013). Interestingly, palmitoylated SspH2, and SseI are targeted to different domains of the plasma membrane, suggesting that this modification is not sufficient for proper localisation. Legionella effectors GobX and LpdA were also shown to be post-translationally modified by palmitoylation, which targets them to the Golgi or the Rab4- and Rab14-containing endosomes, respectively (Lin et al., 2015; Schroeder et al., 2015). AvrPto and AvrPphB from the plant pathogen P. syringae is an exception to the described specific acylation in animal vs plant cells, as they can be palmitoylated. For AvrPto disruption of a putative myristoylation motif abolished membrane association and its avirulence activity in tomato and tobacco. Regarding AvrPphB was shown be palmitoylated -as well as myristoylated- inside the plant cells (see below).
Phospholipid Binding to Effectors in Animal Cells
Phosphoinositideipids are phosphorylated derivatives of phosphatidylinositol and control key cellular processes, such as vesicular trafficking. Different phosphoinositide species target different intracellular membranes, so that they can play an essential role as anchor moieties to target proteins to precise locations. L. pneumophila type IV effectors SetA, SidC, SidM, and LidA have been shown to bind different phosphoinositides to target the LCV (Haneburger and Hilbi, 2013). This lipid association and the ensuing location is essential to carry out their function promoting interaction of the LCV with the host organelles (Ivanov and Roy, 2013). The type III-secreted effector ExoU from Pseudomonas aeruginosa is also modified by phosphoinositide binding. ExoU acts as a phospholipase that is localized to the plasma membrane. Effector binding to precise phosphatidylinositol species that are abundant at the cytoplasmic side of the plasma membrane was shown to be required for its location and activity as a necrotic factor to promote bacterial multiplication (Gendrin et al., 2012).
Multiple Lipidation of Effectors
Protein modification with palmitate can stably target to the plasma membrane proteins previously modified by other types of lipidation, such as myristoylation or farnesylation (Resh, 2006). It has been proposed that multiple protein lipidation would start with N-myristoylation, which would target the protein to the endomembrane system, followed by S-acylation—normally S-palmitoylation of nearby cysteines-, which enhances membrane association of lipidated proteins (Resh, 1999). This process also takes place for some effectors once in their eukaryotic target cells. The first report of multiple lipidation was Pseudomonas syringae effector AvrRpm1, in which a G2A mutation in the myristoylation site eliminated membrane localization, while a C3A exchange in the putative palmitoylation site reduced membrane association (Nimchuk et al., 2000). This is logical given the requirement for myristoylation to occur before palmitoylation. In this same work, palmitoylation consensus sequences were identified in AvrRpm1, AvrB, AvrC, AvrPto, and AvrPphB. Some years later, multiple lipidation by both myristoylation and palmitoylation were demonstrated by heterologous expression in yeast for the Pseudomonas syringae effectors AvrPphB and ORF4 and their related effector NopT from Sinorhizobium fredii (Dowen et al., 2009). As described for AvrPphB, these effectors are auto-processed inside the plant cell, exposing the previously hidden acylation motifs. Lipidation targeted the effectors to the plasma membrane, which was required, at least for AvrPphB, to exert its functions. Interestingly, myristoylation-deficient variants of these effectors were also not palmitoylated, indicating that the former modification is required for subsequent acylation (Dowen et al., 2009). Although not proven biochemically, conservation of the predicted dual acylation motif containing G2 and a proximal cysteine suggests that at least 10 more Pseudomonas and Xanthomonas effectors may be modified in plant cells by myristoylation and S-acylation (Hicks and Galán, 2013).
The S. typhimurium effector protein SifA was shown to be modified both by the animal host cell prenylation (see above) and the S-acylation machineries (Reinicke et al., 2005). Interestingly, mutation of the S-acylation motif in SifA did not affect bacterial survival in the host, whereas disruption of the prenylated residue attenuated bacterial growth in the rat liver, suggesting the latter modification plays a more prominent role in bacterial virulence (Reinicke et al., 2005).
Finally, various forms of prenylation have also been proposed to coexist in a single effector. For instance, location of the L. pneumophila type IV effectors PelA and PelH is dependent on prenylation but neither farnesyltransferase nor geranylgeranyltransferase inhibitors perturbed their localization, suggesting that both enzymes can modify them (Ivanov et al., 2010).
Author Contributions
CP wrote the manuscript. MT wrote the manuscript. MV coordinated manuscript preparation and wrote the manuscript.
Funding
This work was funded by projects AGL2013-46898-R (MINECO, Spain) to MV and COST Action FA1208 Sustain funded by the European Union H2020. This work was also supported by Japan Society of the Promotion of Science KAKENHI Grant 16K07668.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abramovitch, R. B., Janjusevic, R., Stebbins, C. E., and Martin, G. B. (2006). Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 2851–2856. doi: 10.1073/pnas.0507892103
Al-Quadan, T., Price, C. T., London, N., Schueler-Furman, O., and AbuKwaik, Y. (2011). Anchoring of bacterial effectors to host membranes through host-mediated lipidation by prenylation: a common paradigm. Trends Microbiol. 19, 573–579. doi: 10.1016/j.tim.2011.08.003
Anderson, D. M., Feix, J. B., and Frank, D. W. (2015). Cross Kingdom activators of five classes of bacterial effectors. PLoS Pathog 11:e1004944. doi: 10.1371/journal.ppat.1004944
Anderson, D. M., Schmalzer, K. M., Sato, H., Casey, M., Terhune, S. S., Haas, A. L., et al. (2011). Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol. Microbiol. 82, 1454–1467. doi: 10.1111/j.1365-2958.2011.07904.x
Anderson, J. C., Pascuzzi, P. E., Xiao, F., Sessa, G., and Martin, G. B. (2006). Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 18, 502–514. doi: 10.1105/tpc.105.036590
Angot, A., Vergunst, A., Genin, S., and Peeters, N. (2007). Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog 3:e3. doi: 10.1371/journal.ppat.0030003
Aumüller, T., Jahreis, G., Fischer, G., and Schiene-Fischer, C. (2010). Role of prolyl cis/trans isomers in cyclophilin-assisted Pseudomonas syringae AvrRpt2 protease activation. Biochemistry 49, 1042–1052. doi: 10.1021/bi901813e
Axtell, M. J., Chisholm, S. T., Dahlbeck, D., and Staskawicz, B. J. (2003). Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 49, 1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x
Axtell, M. J., and Staskawicz, B. J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. doi: 10.1016/S0092-8674(03)00036-9
Backert, S., Kenny, B., Gerhard, R., Tegtmeyer, N., and Brandt, S. (2010a). PKA-mediated phosphorylation of EPEC-Tir at serine residues 434 and 463: a novel pathway in regulating Rac1 GTPase function. Gut Microbes 1, 94–99. doi: 10.4161/gmic.1.2.11437
Backert, S., and Selbach, M. (2005). Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol. 13, 476–484. doi: 10.1016/j.tim.2005.08.002
Backert, S., Tegtmeyer, N., and Selbach, M. (2010b). The versatility of Helicobacter pylori CagA effector protein functions: the master key hypothesis. Helicobacter 15, 163–176. doi: 10.1111/j.1523-5378.2010.00759.x
Bartsev, A. V., Boukli, N. M., Deakin, W. J., Staehelin, C., and Broughton, W. J. (2003). Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 554, 271–274. doi: 10.1016/S0014-5793(03)01145-1
Bartsev, A. V., Deakin, W. J., Boukli, N. M., McAlvin, C. B., Stacey, G., Malnoë, P., et al. (2004). NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 134, 871–879. doi: 10.1104/pp.103.031740
Behrends, C., and Harper, J. W. (2011). Constructing and decoding unconventional ubiquitin chains. Nat. Struct. Mol. Biol. 18, 520–528. doi: 10.1038/nsmb.2066
Berndsen, C. E., and Wolberger, C. (2014). New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307. doi: 10.1038/nsmb.2780
Beyer, A. R., Truchan, H. K., May, L. J., Walker, N. J., Borjesson, D. L., and Carlyon, J. A. (2015). The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell. Microbiol. 17, 504–519. doi: 10.1111/cmi.12380
Bommarius, B., Maxwell, D., Swimm, A., Leung, S., Corbett, A., Bornmann, W., et al. (2007). Enteropathogenic Escherichia coli Tir is an SH2/3 ligand that recruits and activates tyrosine kinases required for pedestal formation. Mol. Microbiol. 63, 1748–1768. doi: 10.1111/j.1365-2958.2007.05626.x
Brandt, S., Kenny, B., Rohde, M., Martinez-Quiles, N., and Backert, S. (2009). Dual infection system identifies a crucial role for PKA-mediated serine phosphorylation of the EPEC-Tir-injected effector protein in regulating Rac1 function. Cell Microbiol. 11, 1254–1271. doi: 10.1111/j.1462-5822.2009.01330.x
Bruckert, W. M., and Abu Kwaik, Y. (2015). Lysine11-Linked Polyubiquitination of the AnkB F-Box Effector of Legionella pneumophila. Infect. Immun. 84, 99–107. doi: 10.1128/IAI.01165-15
Buchrieser, C., Glaser, P., Rusniok, C., Nedjari, H., D'Hauteville, H., Kunst, F., et al. (2000). The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38, 760–771. doi: 10.1046/j.1365-2958.2000.02179.x
Campellone, K. G., and Leong, J. M. (2003). Tails of two Tirs: actin pedestal formation by enteropathogenic E. coli and enterohemorrhagic E. coli O157:H7. Curr. Opin. Microbiol. 6, 82–90. doi: 10.1016/S1369-5274(03)00005-5
Chang, J. H., Urbach, J. M., Law, T. F., Arnold, L. W., Hu, A., Gombar, S., et al. (2005). A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. U.S.A. 102, 2549–2554. doi: 10.1073/pnas.0409660102
Chen, Y. S., Bastidas, R. J., Saka, H. A., Carpenter, V. K., Richards, K. L., Plano, G. V., et al. (2014). The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog. 10:e1003954. doi: 10.1371/journal.ppat.1003954
Christen, M., Coye, L. H., Hontz, J. S., LaRock, D. L., Pfuetzner, R. A., Megha, et al. (2009). Activation of a bacterial virulence protein by the GTPase RhoA. Sci. Signal 2, ra71. doi: 10.1126/scisignal.2000430
Clifton, D. R., Fields, K. A., Grieshaber, S. S., Dooley, C. A., Fischer, E. R., Mead, D. J., et al. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U.S.A. 101, 10166–10171. doi: 10.1073/pnas.0402829101
Coaker, G., Falick, A., and Staskawicz, B. (2005). Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308, 548–550. doi: 10.1126/science.1108633
Coaker, G., Zhu, G., Ding, Z., Van Doren, S. R., and Staskawicz, B. (2006). Eukaryotic cyclophilin as a molecular switch for effector activation. Mol. Microbiol. 61, 1485–1496. doi: 10.1111/j.1365-2958.2006.05335.x
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Coll, N. S., and Valls, M. (2013). Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. 6, 614–620. doi: 10.1111/1751-7915.12056
Day, B., Dahlbeck, D., Huang, J., Chisholm, S. T., Li, D., and Staskawicz, B. J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17, 1292–1305. doi: 10.1105/tpc.104.030163
Desveaux, D., Singer, A. U., Wu, A. J., McNulty, B. C., Musselwhite, L., Nimchuk, Z., et al. (2007). Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 3:e48. doi: 10.1371/journal.ppat.0030048
Dowen, R. H., Engel, J. L., Shao, F., Ecker, J. R., and Dixon, J. E. (2009). A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J. Biol. Chem. 284, 15867–15879. doi: 10.1074/jbc.M900519200
Ensminger, A. W., and Isberg, R. R. (2009). Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 12, 67–73. doi: 10.1016/j.mib.2008.12.004
Fu, H., Coburn, J., and Collier, R. J. (1993). The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 90, 2320–2324.
Fujiwara, S., Kawazoe, T., Ohnishi, K., Kitagawa, T., Popa, C., Valls, M., et al. (2016). RipAY, a plant pathogen effector protein exhibits robust gamma-glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 291, 6813–6830. doi: 10.1074/jbc.M115.678953
Gaspar, A. H., and Machner, M. P. (2014). VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc. Natl. Acad. Sci. U.S.A. 111, 4560–4565. doi: 10.1073/pnas.1316376111
Gendrin, C., Contreras-Martel, C., Bouillot, S., Elsen, S., Lemaire, D., Skoufias, D. A., et al. (2012). Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog. 8:e1002637. doi: 10.1371/journal.ppat.1002637
Grishin, A. M., Condos, T. E., Barber, K. R., Campbell-Valois, F. X., Parsot, C., Shaw, G. S., et al. (2014). Structural basis for the inhibition of host protein ubiquitination by Shigella effector kinase OspG. Structure 22, 878–888. doi: 10.1016/j.str.2014.04.010
Guo, C., and Henley, J. M. (2014). Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life 66, 71–77. doi: 10.1002/iub.1244
Haneburger, I., and Hilbi, H. (2013). Phosphoinositide lipids and the Legionella pathogen vacuole. Curr. Top. Microbiol. Immunol. 376, 155–173. doi: 10.1007/82_2013_341
Hayashi, T., Morohashi, H., and Hatakeyama, M. (2013). Bacterial EPIYA effectors–where do they come from? What are they? Where are they going? Cell Microbiol. 15, 377–385. doi: 10.1111/cmi.12040
He, P., Shan, L., Lin, N. C., Martin, G. B., Kemmerling, B., Nurnberger, T., et al. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575. doi: 10.1016/j.cell.2006.02.047
Hentschke, M., Trulzsch, K., Heesemann, J., Aepfelbacher, M., and Ruckdeschel, K. (2007). Serogroup-related escape of Yersinia enterocolitica YopE from degradation by the ubiquitin-proteasome pathway. Infect. Immun. 75, 4423–4431. doi: 10.1128/IAI.00528-07
Hicks, S. W., Charron, G., Hang, H. C., and Galan, J. E. (2011). Subcellular targeting of Salmonella virulence proteins by host-mediated S-palmitoylation. Cell Host Microbe 10, 9–20. doi: 10.1016/j.chom.2011.06.003
Hicks, S. W., and Galán, J. E. (2013). Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat. Rev. Microbiol. 11, 316–326. doi: 10.1038/nrmicro3009
Horwitz, M. A., and Silverstein, S. C. (1980). Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Invest. 66, 441–450. doi: 10.1172/JCI109874
Hunter, T. (2009). Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21, 140–146. doi: 10.1016/j.ceb.2009.01.028
Ivanov, S. S., Charron, G., Hang, H. C., and Roy, C. R. (2010). Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J. Biol. Chem. 285, 34686–34698. doi: 10.1074/jbc.M110.170746
Ivanov, S. S., and Roy, C. (2013). Host lipidation: a mechanism for spatial regulation of Legionella effectors. Curr. Top. Microbiol. Immunol. 376, 135–154. doi: 10.1007/82_2013_344
Janjusevic, R., Abramovitch, R. B., Martin, G. B., and Stebbins, C. E. (2006). A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311, 222–226. doi: 10.1126/science.1120131
Juris, S. J., Rudolph, A. E., Huddler, D., Orth, K., and Dixon, J. E. (2000). A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc. Natl. Acad. Sci. U.S.A. 97, 9431–9436. doi: 10.1073/pnas.170281997
Jw, I. J., Carlson, A. C., and Kennedy, E. L. (2007). Anaplasma phagocytophilum AnkA is tyrosine-phosphorylated at EPIYA motifs and recruits SHP-1 during early infection. Cell. Microbiol. 9, 1284–1296. doi: 10.1111/j.1462-5822.2006.00871.x
Kiefhaber, T., Grunert, H. P., Hahn, U., and Schmid, F. X. (1990). Replacement of a cis proline simplifies the mechanism of ribonuclease T1 folding. Biochemistry 29, 6475–6480.
Kim, D. W., Lenzen, G., Page, A. L., Legrain, P., Sansonetti, P. J., and Parsot, C. (2005). The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U.S.A. 102, 14046–14051. doi: 10.1073/pnas.0504466102
Kim, N. H., Kim, D. S., Chung, E. H., and Hwang, B. K. (2014). Pepper suppressor of the G2 allele of skp1 interacts with the receptor-like cytoplasmic kinase1 and type III effector AvrBsT and promotes the hypersensitive cell death response in a phosphorylation-dependent manner. Plant Physiol. 165, 76–91. doi: 10.1104/pp.114.238840
Kim, Y. J., Lin, N. C., and Martin, G. B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109, 589–598. doi: 10.1016/S0092-8674(02)00743-2
Knodler, L. A., Winfree, S., Drecktrah, D., Ireland, R., and Steele-Mortimer, O. (2009). Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell. Microbiol. 11, 1652–1670. doi: 10.1111/j.1462-5822.2009.01356.x
Korkuc, P., and Walther, D. (2016). Spatial proximity statistics suggest a regulatory role of protein phosphorylation on compound binding. Proteins 84, 565–579. doi: 10.1002/prot.25001
Ku, B., Lee, K. H., Park, W. S., Yang, C. S., Ge, J., Lee, S. G., et al. (2012). VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog. 8:e1003082. doi: 10.1371/journal.ppat.1003082
Kubori, T., and Galan, J. E. (2003). Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115, 333–342. doi: 10.1016/S0092-8674(03)00849-3
Kumar, A., Tikoo, S., Maity, S., Sengupta, S., Sengupta, S., Kaur, A., et al. (2012). Mammalian proapoptotic factor ChaC1 and its homologues function as gamma-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 13, 1095–1101. doi: 10.1038/embor.2012.156
Kumar, S., Kaur, A., Chattopadhyay, B., and Bachhawat, A. K. (2015). Defining the Cytosolic Pathway of Glutathione Degradation in Arabidopsis thaliana: role of the ChaC/GCG Family of gamma-glutamyl cyclotransferases as Glutathione Degrading Enzymes and AtLAP1 as the Cys-Gly Peptidase. Biochem. J. 468, 73–85. doi: 10.1042/BJ20141154
Lee, A. H., Hurley, B., Felsensteiner, C., Yea, C., Ckurshumova, W., Bartetzko, V., et al. (2012). A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 8:e1002523. doi: 10.1371/journal.ppat.1002523
Levin, I., Eakin, C., Blanc, M. P., Klevit, R. E., Miller, S. I., and Brzovic, P. S. (2010). Identification of an unconventional E3 binding surface on the UbcH5 ~ Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc. Natl. Acad. Sci. U.S.A. 107, 2848–2853. doi: 10.1073/pnas.0914821107
Lewis, J. D., Abada, W., Ma, W., Guttman, D. S., and Desveaux, D. (2008). The HopZ family of Pseudomonas syringae type III effectors require myristoylation for virulence and avirulence functions in Arabidopsis thaliana. J. Bacteriol. 190, 2880–2891. doi: 10.1128/JB.01702-07
Li, W., Yadeta, K. A., Elmore, J. M., and Coaker, G. (2013). The Pseudomonas syringae effector HopQ1 promotes bacterial virulence and interacts with tomato 14-3-3 proteins in a phosphorylation-dependent manner. Plant Physiol. 161, 2062–2074. doi: 10.1104/pp.112.211748
Lin, M., den Dulk-Ras, A., Hooykaas, P. J., and Rikihisa, Y. (2007). Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 9, 2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x
Lin, Y. H., Doms, A. G., Cheng, E., Kim, B., Evans, T. R., and Machner, M. P. (2015). Host Cell-catalyzed S-Palmitoylation Mediates Golgi Targeting of the Legionella Ubiquitin Ligase GobX. J. Biol. Chem. 290, 25766–25781. doi: 10.1074/jbc.M115.637397
Lucas, M., Gaspar, A. H., Pallara, C., Rojas, A. L., Fernández-Recio, J., Machner, M. P., et al. (2014). Structural basis for the recruitment and activation of the Legionella phospholipase VipD by the host GTPase Rab5. Proc. Natl. Acad. Sci. U.S.A. 111, E3514– E3523. doi: 10.1073/pnas.1405391111
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R., and Dangl, J. L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. doi: 10.1016/S0092-8674(03)00040-0
Marcus, S. L., Knodler, L. A., and Finlay, B. B. (2002). Salmonella enterica serovar Typhimurium effector SigD/SopB is membrane-associated and ubiquitinated inside host cells. Cell. Microbiol. 4, 435–446. doi: 10.1046/j.1462-5822.2002.00202.x
Martin, D. D., Beauchamp, E., and Berthiaume, L. G. (2011). Post-translational myristoylation: fat matters in cellular life and death. Biochimie 93, 18–31. doi: 10.1016/j.biochi.2010.10.018
Mathieu, J., Schwizer, S., and Martin, G. B. (2014). Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog. 10:e1004227. doi: 10.1371/journal.ppat.1004227
Mehlitz, A., Banhart, S., Mäurer, A. P., Kaushansky, A., Gordus, A. G., Zielecki, J., et al. (2010). Tarp regulates early Chlamydia-induced host cell survival through interactions with the human adaptor protein SHC1. J. Cell Biol. 190, 143–157. doi: 10.1083/jcb.200909095
Mittal, R., Peak-Chew, S. Y., Sade, R. S., Vallis, Y., and McMahon, H. T. (2010). The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J. Biol. Chem. 285, 19927–19934. doi: 10.1074/jbc.M110.126581
Mueller, D., Tegtmeyer, N., Brandt, S., Yamaoka, Y., De Poire, E., Sgouras, D., et al. (2012). c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J. Clin. Invest. 122, 1553–1566. doi: 10.1172/JCI61143
Narayanan, L. A., and Edelmann, M. J. (2014). Ubiquitination as an efficient molecular strategy employed in salmonella infection. Front. Immunol. 5:558. doi: 10.3389/fimmu.2014.00558
Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R. T., Katagiri, F., and Dangl, J. L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363. doi: 10.1016/S0092-8674(00)80846-6
Ntoukakis, V., Mucyn, T. S., Gimenez-Ibanez, S., Chapman, H. C., Gutierrez, J. R., Balmuth, A. L., et al. (2009). Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324, 784–787. doi: 10.1126/science.1169430
Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., et al. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648. doi: 10.1016/j.cell.2006.09.026
Patel, J. C., Hueffer, K., Lam, T. T., and Galán, J. E. (2009). Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137, 283–294. doi: 10.1016/j.cell.2009.01.056
Phillips, N., Hayward, R. D., and Koronakis, V. (2004). Phosphorylation of the Enteropathogenic E. coli receptor by the Src-family kinase c-Fyn triggers actin pedestal formation. Nat. Cell Biol 6, 618–625. doi: 10.1038/ncb1148
Popa, C., Coll, N. S., Valls, M., and Sessa, G. (2016). Yeast as a Heterologous Model System to Uncover Type III Effector Function. PLoS Pathog. 12:e1005360. doi: 10.1371/journal.ppat.1005360
Price, C. T., Al-Quadan, T., Santic, M., Jones, S. C., and Abu Kwaik, Y. (2010a). Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J. Exp. Med. 207, 1713–1726. doi: 10.1084/jem.20100771
Price, C. T., Jones, S. C., Amundson, K. E., and Kwaik, Y. A. (2010b). Host-mediated post-translational prenylation of novel dot/icm-translocated effectors of Legionella pneumophila. Front. Microbiol. 1:131. doi: 10.3389/fmicb.2010.00131
Pruneda, J. N., Smith, F. D., Daurie, A., Swaney, D. L., Villén, J., Scott, J. D., et al. (2014). E2~Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. EMBO J. 33, 437–449. doi: 10.1002/embj.201386386
Ramanathan, H. N., and Ye, Y. (2012). Cellular strategies for making monoubiquitin signals. Crit. Rev. Biochem. Mol. Biol. 47, 17–28. doi: 10.3109/10409238.2011.620943
Reinicke, A. T., Hutchinson, J. L., Magee, A. I., Mastroeni, P., Trowsdale, J., and Kelly, A. P. (2005). A Salmonella typhimurium effector protein SifA is modified by host cell prenylation and S-acylation machinery. J. Biol. Chem. 280, 14620–14627. doi: 10.1074/jbc.M500076200
Rennoll-Bankert, K. E., Garcia-Garcia, J. C., Sinclair, S. H., and Dumler, J. S. (2015). Chromatin-bound bacterial effector ankyrin A recruits histone deacetylase 1 and modifies host gene expression. Cell. Microbiol. 17, 1640–1652. doi: 10.1111/cmi.12461
Resh, M. D. (1999). Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16.
Resh, M. D. (2006). Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590. doi: 10.1038/nchembio834
Robert-Seilaniantz, A., Shan, L., Zhou, J. M., and Tang, X. (2006). The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant Microbe Interact. 19, 130–138. doi: 10.1094/MPMI-19-0130
Rogers, L. D., Kristensen, A. R., Boyle, E. C., Robinson, D. P., Ly, R. T., Finlay, B. B., et al. (2008). Identification of cognate host targets and specific ubiquitylation sites on the Salmonella SPI-1 effector SopB/SigD. J. Proteomics 71, 97–108. doi: 10.1016/j.jprot.2008.01.011
Ruan, H. H., Li, Y., Zhang, X. X., Liu, Q., Ren, H., Zhang, K. S., et al. (2014). Identification of TRAF6 as a ubiquitin ligase engaged in the ubiquitination of SopB, a virulence effector protein secreted by Salmonella typhimurium. Biochem. Biophys. Res. Commun. 447, 172–177. doi: 10.1016/j.bbrc.2014.03.126
Ruckdeschel, K., Pfaffinger, G., Trülzsch, K., Zenner, G., Richter, K., Heesemann, J., et al. (2006). The proteasome pathway destabilizes Yersinia outer protein E and represses its antihost cell activities. J. Immunol. 176, 6093–6102. doi: 10.4049/jimmunol.176.10.6093
Schroeder, G. N., Aurass, P., Oates, C. V., Tate, E. W., Hartland, E. L., Flieger, A., et al. (2015). Legionella pneumophila Effector LpdA Is a Palmitoylated Phospholipase D Virulence Factor. Infect. Immun. 83, 3989–4002. doi: 10.1128/IAI.00785-15
Schulein, R., Guye, P., Rhomberg, T. A., Schmid, M. C., Schröder, G., Vergunst, A. C., et al. (2005). A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. U.S.A. 102, 856–861. doi: 10.1073/pnas.0406796102
Segal, E. D., Cha, J., Lo, J., Falkow, S., and Tompkins, L. S. (1999). Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 96, 14559–14564.
Selbach, M., Paul, F. E., Brandt, S., Guye, P., Daumke, O., Backert, S., et al. (2009). Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe 5, 397–403. doi: 10.1016/j.chom.2009.03.004
Shan, L., Thara, V. K., Martin, G. B., Zhou, J. M., and Tang, X. (2000). The pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell 12, 2323–2338. doi: 10.1105/tpc.12.12.2323
Skorpil, P., Saad, M. M., Boukli, N. M., Kobayashi, H., Ares-Orpel, F., Broughton, W. J., et al. (2005). NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 57, 1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x
Sougleri, I. S., Papadakos, K. S., Zadik, M. P., Mavri-Vavagianni, M., Mentis, A. F., and Sgouras, D. N. (2016). Helicobacter pylori CagA protein induces factors involved in the epithelial to mesenchymal transition (EMT) in infected gastric epithelial cells in an EPIYA- phosphorylation-dependent manner. FEBS J. 283, 206–220. doi: 10.1111/febs.13592
Tegtmeyer, N., and Backert, S. (2011). Role of Abl and Src family kinases in actin-cytoskeletal rearrangements induced by the Helicobacter pylori CagA protein. Eur. J. Cell Biol. 90, 880–890. doi: 10.1016/j.ejcb.2010.11.006
Thara, V. K., Seilaniantz, A. R., Deng, Y., Dong, Y., Yang, Y., Tang, X., et al. (2004). Tobacco genes induced by the bacterial effector protein AvrPto. Mol. Plant Microbe Interact. 17, 1139–1145. doi: 10.1094/MPMI.2004.17.10.1139
Thieme, F., Szczesny, R., Urban, A., Kirchner, O., Hause, G., and Bonas, U. (2007). New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol. Plant Microbe Interact. 20, 1250–1261. doi: 10.1094/MPMI-20-10-1250
Thomas, M., and Holden, D. W. (2009). Ubiquitination - a bacterial effector's ticket to ride. Cell Host Microbe 5, 309–311. doi: 10.1016/j.chom.2009.03.010
Thwaites, T., Nogueira, A. T., Campeotto, I., Silva, A. P., Grieshaber, S. S., and Carabeo, R. A. (2014). The Chlamydia effector TarP mimics the mammalian leucine-aspartic acid motif of paxillin to subvert the focal adhesion kinase during invasion. J. Biol. Chem. 289, 30426–30442. doi: 10.1074/jbc.M114.604876
Üstün, S., Bartetzko, V., and Bornke, F. (2013). The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic-acid mediated plant defence. PLoS Pathog. 9, e1003427. doi: 10.1371/journal.ppat.1003427
Üstün, S., and Börnke, F. (2015). The Xanthomonas campestris type III effector XopJ proteolytically degrades proteasome subunit RPT6. Plant Physiol. 168, 107–119. doi: 10.1104/pp.15.00132
Üstün, S., König, P., Guttman, D. S., and Börnke, F. (2014). HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Mol. Plant Microbe Interact. 27, 611–623. doi: 10.1094/MPMI-12-13-0363-R
Verma, S., Mohapatra, G., Ahmad, S. M., Rana, S., Jain, S., Khalsa, J. K., et al. (2015). Salmonella Engages Host MicroRNAs To Modulate SUMOylation: a New Arsenal for Intracellular Survival. Mol. Cell. Biol. 35, 2932–2946. doi: 10.1128/MCB.00397-15
Wang, L., Tang, X., and He, C. (2007). The bifunctional effector AvrXccC of Xanthomonas campestris pv. campestris requires plasma membrane-anchoring for host recognition. Mol. Plant Pathol. 8, 491–501. doi: 10.1111/j.1364-3703.2007.00409.x
Whitmore, S. E., and Lamont, R. J. (2012). Tyrosine phosphorylation and bacterial virulence. Int. J. Oral Sci. 4, 1–6. doi: 10.1038/ijos.2012.6
Wilson, V. G. (2012). Sumoylation at the host-pathogen interface. Biomolecules 2, 203–227. doi: 10.3390/biom2020203
Xiao, F., Giavalisco, P., and Martin, G. B. (2007). Pseudomonas syringae type III effector AvrPtoB is phosphorylated in plant cells on serine 258, promoting its virulence activity. J. Biol. Chem. 282, 30737–30744. doi: 10.1074/jbc.M705565200
Yeam, I., Nguyen, H. P., and Martin, G. B. (2010). Phosphorylation of the Pseudomonas syringae effector AvrPto is required for FLS2/BAK1-independent virulence activity and recognition by tobacco. Plant J. 61, 16–24. doi: 10.1111/j.1365-313X.2009.04028.x
Zhang, L., Chen, X. J., Lu, H. B., Xie, Z. P., and Staehelin, C. (2011). Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 286, 32178–32187. doi: 10.1074/jbc.M111.265942
Zhang, Y., Higashide, W., Dai, S., Sherman, D. M., and Zhou, D. (2005). Recognition and ubiquitination of Salmonella type III effector SopA by a ubiquitin E3 ligase, HsRMA1. J. Biol. Chem. 280, 38682–38688. doi: 10.1074/jbc.M506309200
Zhang, Y., Higashide, W. M., McCormick, B. A., Chen, J., and Zhou, D. (2006). The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 62, 786–793. doi: 10.1111/j.1365-2958.2006.05407.x
Zhou, Y., Dong, N., Hu, L., and Shao, F. (2013). The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. PLoS ONE 8:e57558. doi: 10.1371/journal.pone.0057558
Keywords: type III secretion system, type IV secretion system, bacterial effector, bacterial virulence, eukaryotic host, animal pathogens, plant pathogens
Citation: Popa CM, Tabuchi M and Valls M (2016) Modification of Bacterial Effector Proteins Inside Eukaryotic Host Cells. Front. Cell. Infect. Microbiol. 6:73. doi: 10.3389/fcimb.2016.00073
Received: 31 May 2016; Accepted: 27 June 2016;
Published: 20 July 2016.
Edited by:
Damien F. Meyer, CIRAD, FranceReviewed by:
Robert B. Abramovitch, Michigan State University, USASuayib Üstün, Swedish University of Agricultural Sciences, Sweden
Copyright © 2016 Popa, Tabuchi and Valls. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mitsuaki Tabuchi, bXRhYnVjaGlAYWcua2FnYXdhLXUuYWMuanA=
Marc Valls, bWFyY3ZhbGxzQHViLmVkdQ==
 Crina M. Popa
Crina M. Popa Mitsuaki Tabuchi
Mitsuaki Tabuchi Marc Valls
Marc Valls