Abstract
Brucella spp. are intracellular bacterial pathogens that cause infection in domestic and wild animals. They are often used as model organisms to study intracellular bacterial infections. Brucella VirB T4SS is a key virulence factor that plays important roles in mediating intracellular survival and manipulating host immune response to infection. In this review, we discuss the roles of Brucella VirB T4SS and 15 effectors that are proposed to be crucial for Brucella pathogenesis. VirB T4SS regulates the inflammation response and manipulates vesicle trafficking inside host cells. VirB T4SS also plays crucial roles in the inhibition of the host immune response and intracellular survival during infection. Here, we list the key molecular events in the intracellular life cycle of Brucella that are potentially targeted by the VirB T4SS effectors. Elucidating the functions of these effectors will help clarify the molecular role of T4SS during infection. Furthermore, studying the effectors secreted by Brucella spp. might provide insights into the mechanisms used by the bacteria to hijack the host signaling pathways and aid in the development of better vaccines and therapies against brucellosis.
Introduction
Brucella spp. cause brucellosis in domestic and wild animals. Their primary hosts include sheep, cattle, swine, dogs, camels, and desert woodrats (Martirosyan et al., 2011; Bargen et al., 2012). Humans are secondary or accidental hosts and usually contract brucellosis through contact with infected animals or animal products (Atluri et al., 2011; Bargen et al., 2012). Brucellosis is a chronic disease and may last several weeks or months. If it is not treated effectively, the disease can lead to pathologies in the liver, spleen, lymph nodes, bone marrow, reproductive tract, and skeletal system (Atluri et al., 2011). Some complications, including arthritis, liver abscess, epididymo-orchitis, neurobrucellosis, and endocarditis, often tend to relapse, resulting in long-term disability and suffering (Colmenero et al., 1996; Franco et al., 2007).
In natural hosts, Brucella can penetrate the mucous membranes of the reproductive, respiratory, and digestive tracts and the conjunctiva; in humans, the most common sites of entry are the respiratory and digestive tracts (Atluri et al., 2011; Bargen et al., 2012). Once the bacteria break the mucosal barrier, they enter the regional lymph nodes and disseminate through the body (Atluri et al., 2011). Inside the host, the bacteria reside in the macrophages for several days and replicate to high numbers asymptomatically (Martirosyan et al., 2011). Therefore, elucidating the mechanisms involved in their intracellular survival and their ability to evade host immunity is crucial for understanding the pathogenesis of Brucella spp., which are used as model organisms to study intracellular bacterial infections.
On entering the host cells, the bacteria interact with the early and late endosomes and acquire several markers, including Rab5, early endosome antigen (EEA) 1, and Rab7, resulting in the formation of a “Brucella-containing vacuole” (BCV) (Pizarro-Cerdá et al., 1998; Chaves-Olarte et al., 2002; Celli et al., 2003; Starr et al., 2008; Lee et al., 2013). The BCVs then fuse rapidly with the lysosome in a controlled manner, as suggested by the presence of the lysosomal markers, lysosomal-associated membrane protein (LAMP), and CD63, but not the luminal lysosome enzyme, cathepsin D, on the surface of bacteria (Pizarro-Cerdá et al., 1998; Celli et al., 2003; Starr et al., 2008). In this transient stage, most of the contents of the BCVs are subjected to phagolysosomal degradation, and 90% of internalized Brucella are killed by the action of hydrolyzing enzymes (Celli et al., 2003). However, the remaining 10% evade the host killing mechanisms through an unknown mechanism that probably involves the acidification of the BCVs, subsequent triggering of the virB operon [which encodes the type IV secretion system (T4SS)], and release of a large variety of effectors into the host cells' cytosol (Boschiroli et al., 2002). The bacteria then traffic and arrive at the endoplasmic reticulum (ER). Within the ER, the bacteria survive and establish their replicative niche, and multiply to large numbers (Celli et al., 2003). However, the ER is not the final site of this intracellular journey; recent studies have shown that autophagy-like vacuoles provide a replication-permissive compartment for Brucella following the ER stage (Starr et al., 2012). Because the “Brucella-replicating organelle” possesses autophagic features, it is also called the autophagic BCV (aBCV). The formation of the aBCV is essential for the completion of the intracellular lifecycle of Brucella and for cell-to-cell spreading (Starr et al., 2012).
Although several studies have attempted to elucidate the pathogenesis of Brucella, the components that are responsible for their virulence and their ability to establish systemic infection have not been identified. For a long time, it was thought that Brucella spp. lacked the characteristic virulence factors that are present in other bacteria (Fugier et al., 2007). However, in recent years, various virulence factors that are essential for infection, including lipopolysaccharide (LPS) (Lapaque et al., 2005), β-cyclic glucan (Arellano-Reynoso et al., 2005; Martirosyan et al., 2012), BvrS/BvrR (Guzman-Verri et al., 2002; Martín-Martín et al., 2012), outer membrane proteins (Omps) (Lim et al., 2012; Vizcaíno and Cloeckaert, 2012), BmaC (Posadas et al., 2012), MucR (Mirabella et al., 2013), SagA (Del Giudice et al., 2013), BtaE (Ruiz-Ranwez et al., 2013), BacA (Martín-Martín et al., 2012), and BetB (Lee et al., 2014), have been identified in Brucella VirB. T4SS is another key virulence factor that plays important roles in mediating intracellular survival and manipulating host immune response to infection. Many studies have elucidated various aspects of VirB T4SS from Brucella spp., such as its genetic organization, structure, functions, and transcriptional regulation. Some studies have used T4SS as targets for small molecular inhibitors (Paschos et al., 2011; Smith et al., 2012). T4SS functions in the translocation of effector proteins across bacterial and host membranes into the host cytosol. Therefore, to understand the functions of T4SS, it is important to identify these effectors. Recent studies have identified 15 effectors that are secreted by Brucella in a T4SS-dependent manner. In this review, we discuss the roles of Brucella T4SS and its 15 effectors, as well as the associations between bacterial phenotype and effector functions.
Brucella type IV secretion system
Organization, structure, and regulation
In Brucella, T4SS is encoded by the virB operon, which consists of 12 genes (virB1–12) located on chromosome II. Transcription of the virB operon is controlled by the promoter upstream of virB1 (O'Callaghan et al., 1999; Sieira et al., 2000). The virB operon was first identified in B. suis. Subsequently, it was found to be highly conserved in all Brucella spp. for which genomic sequences were available, indicating a potentially critical role for this operon (O'Callaghan et al., 1999). Although the structure of T4SS from Brucella is unknown, the architecture of two closely related T4SSs, one from Agrobacterium tumefaciens (VirB/D4 system) and the other from Escherichia coli (encoded on the R388 conjugative plasmid), has been examined. The T4SS apparatus is a large macromolecular complex comprising 12 subunits. The complex can be divided into five parts: the stretching needle complex (composed of VirB2), the core/outer membrane complex (composed of VirB7, VirB9, and VirB10), the linking stalk (probably composed of fragments from VirB5 or VirB10), the inner membrane complex (composed of VirB3, VirB4, VirB6, VirB8, and the N-terminus of VirB10), and the ATPases/energy center (consisting of VirB4 andVirB11) (Fronzes et al., 2009; Low et al., 2014; Trokter et al., 2014). Except for VirB1, VirB7, and VirB12, all the other subunits play crucial roles in the virulence of Brucella (as inferred from studies involving mice), which may reflect their status in the whole machinery (Comerci et al., 2001; den Hartigh et al., 2004, 2008; Sun et al., 2005).
The underlying molecular mechanisms involved in the initiation of T4SS assembly are still unknown; however, some intracellular signals are thought to be critical for inducing the assembly of T4SS. Several transcriptional regulators may be involved in this process. The low pH condition that occurs within the BCV (as a result of acidification when fusing with the lysosome) probably serves as the key signal that induces the upregulation of the virB operon in the bacteria (Porte et al., 1999; Boschiroli et al., 2002; Köhler et al., 2002). Regulators that regulate the expression of the virB operon include vacuolar hijacking Brucella regulator VjbR (Arocena et al., 2010), Brucella luxR-like regulator (BlxR) (Rambow-Larsen et al., 2008), histidine utilization regulator (HutC) (Sieira et al., 2010), the two-component regulatory system BvrR/BvrS (López-Goñi et al., 2002; Martínez-Núñez et al., 2010), RelA/SpoT homolog (Rsh) (Dozot et al., 2006), MarR-like sodium deoxycholate-responsive activator (MdrA) (Sieira et al., 2012), Brucella quorum-sensing regulator (BabR) (Caswell et al., 2012), and integration host factor (IHF) (Sieira et al., 2004). However, their exact roles in the induction and assembly of T4SS during infection remain to be evaluated.
Role of T4SS during infection
Since its identification, T4SS from Brucella has been studied extensively. Several in vitro and in vivo studies have improved our understanding of the phenotypes, including long-term survival within the host, immune inhibition, and the underlying mechanism of action of T4SS. Most of these studies have concentrated on four critical aspects of Brucella infection: phenotypes of the infected animals, surviving ability in cultured cells, utilization of the host vesicle trafficking pathway, and regulation of the host inflammatory response. The results indicate that T4SS plays crucial roles in the inhibition of the host innate immune response and in intracellular survival during infection.
In animal models, mainly mice and goats, screening of genes involved in Brucella virulence by signature-tagged mutagenesis (STM) led to the identification of T4SS as the crucial factor required to establish successful infection in vivo (Hong et al., 2000; Lestrate et al., 2000, 2003; Kahl-McDonagh et al., 2006; Zygmunt et al., 2006). In these studies, screening for genes essential for the acute phase of infection did not identify T4SS; however, screening for genes required for chronic persistence identified T4SS as the virulence determinant (Hong et al., 2000; Lestrate et al., 2003). Consistent with these results, subsequent analysis of the kinetics of infection in mice and calves revealed that T4SS is not needed for initial systematic colonization within the spleen, liver, kidney, and lungs, but is required for chronic persistence (Rajashekara et al., 2005; Rolán and Tsolis, 2007; Rossetti et al., 2013). In goats, a similar screening experiment suggested that T4SS is involved in bacterial survival in vivo. Another study reported that, unlike wild-type B. melitensis 16M, attenuated B. melitensis lacking virB2 does not cause abortions or colonize fetal tissues in pregnant goats (Kahl-McDonagh et al., 2006; Zygmunt et al., 2006). Recent studies on T4SS in two less virulent strains, B. ovis and B. microti, revealed that T4SS-deficient strains are significantly attenuated in both the early and late stages of infection (Hanna et al., 2011; Sá et al., 2012).
In cultured cells, including THP1, J774, dendritic cells, murine bone marrow-derived macrophages (BMMs), primary human monocytes, and HeLa cells, T4SS has been shown to be essential for the intracellular growth of Brucella during infection (O'Callaghan et al., 1999; Foulongne et al., 2000; Pizarro-Cerdá et al., 2000; Sieira et al., 2000; Comerci et al., 2001; Delrue et al., 2001; Celli et al., 2003; Salcedo et al., 2008; den Hartigh et al., 2008). Screening for the intracellular growth ability of Brucella mutants in HeLa and THP1 cells revealed that the disruption of T4SS leads to impaired intracellular survival, thereby demonstrating the importance of this secretion machinery in vitro (Foulongne et al., 2000; Delrue et al., 2001). Among the proteins that comprise the T4SS apparatus, VirB2–6 and VirB8-11, were found to be essential for the survival and replication of Brucella within host cells (Guzman-Verri et al., 2002; Celli et al., 2003; Fronzes et al., 2009; Lee et al., 2013; Trokter et al., 2014), indicating the importance of the integrity of this secretion apparatus and the differing roles of its various components. T4SS-deficient mutants exhibited complete loss of the ability to grow within cells, in vitro; however, they exhibited no attenuation in the initial systematic spreading stage, albeit a diminished replicating ability during the later stage was observed in vivo. This suggests that, in the initial invasion stage of an in vivo infection, bacterial survival does not rely on T4SS. This may be attributed to the extracellular localization of the bacteria and the favorable microenvironment. These aspects need to be studied in more detail in the future.
To elucidate the mechanism by which T4SS mediates the intracellular survival ability of Brucella, VirB10 polar mutants, which lacked the virB10 gene and were therefore, deficient in T4SS, were used. The results revealed that T4SS-deficient Brucella still invaded host cells and interacted with the early and late endosomes in a manner similar to that by wild-type bacteria, as suggested by the acquisition of the early and late endosome markers (EEA1 and LAMP1, respectively) by the BCVs (Delrue et al., 2001; Celli et al., 2003). However, unlike wild-type Brucella, BCVs of the mutant bacteria did not exclude LAMP1 over time, and remained LAMP1-positive. Therefore, these mutants were engulfed by lysosomes and eventually targeted for degradation (Comerci et al., 2001; Delrue et al., 2001; Celli et al., 2003). Thus, unlike wild-type Brucella, the T4SS-deficient mutants did not complete their intracellular lifecycle, which involves escaping from the late endosome, reaching the ER and establishing the replicating niche, and escaping from the ER to utilize autophagic vacuoles for survival (Starr et al., 2012). Therefore, what happens to the BCVs within the late endosome/lysosome is very critical, and determines the fate of intracellular Brucella. This divergence may depend on the induction of the virB operon and subsequent delivery of the effectors into the host cytosol. The signals that trigger the induction may originate from the environment around Brucella, such as acidification during BCV fusion with the late endosome/lysosome (Pizarro-Cerdá et al., 1998; Starr et al., 2008).
Brucella use the stealth strategy to avoid immune recognition by the host, and elicit a much lower immune response compared to other bacterial infections, such as those caused by Salmonella (Barquero-Calvo et al., 2007). Although T4SS may play a role in regulating the host immune response, the molecular mechanism underlying this regulation is unclear. Some studies have indicated that T4SS is required for Brucella to elicit relatively low levels of immune responses (Rolán and Tsolis, 2007, 2008; Rolán et al., 2009). Conversely, experiments performed on other bacteria have indicated roles for T4SS in inhibiting host immune responses, as evidenced by the fact that T4SS-deficient mutants generally induce a stronger immune response in the hosts than wild-type strains (Luo, 2012; van Schaik et al., 2013). The phenotype of the infected cells suggested that T4SS-deficient Brucella are quickly degraded in the lysosomes, before they can ignite an immune response. On the other hand, wild-type Brucella activate the host immune response at a later stage in their intracellular lifecycle. Another possibility is that the effectors secreted by T4SS have inflammation-inhibitory activities within host cells. Therefore, we believe that the loss of a functional T4SS may lead to impaired effector secretion and upregulated immune responses.
Like for many other bacteria, T4SS is essential for the virulence of Brucella infections, in both animal models and cultured cells. The cellular and molecular mechanisms underlying this virulence may include hijacking the vesicle trafficking pathway and manipulating the innate and adaptive immune responses of the host. T4SS secretes effectors into host cells. These effectors then target various host mechanisms and perpetuate the infection. Therefore, identifying the effectors secreted by Brucella through the T4SS machinery and determining their target host pathways are critical for understanding their pathogenesis.
Molecular events that are potentially targeted by Brucella T4SS effectors
To date, 15 T4SS effectors have been identified in Brucella. The phenotypes presented by T4SS-deficient mutants and studies on some well-known T4SS effectors from other bacteria suggest that, in Brucella, T4SS effectors might function in the following molecular events associated with infection: (1) excluding markers of late endosome or lysosome, (2) acquiring ER markers, (3) interacting with secretory pathways, (4) acquiring markers for autophagosomes, (5) resisting the harsh intracellular environment, and (6) regulating the activation of vital immune pathways (Figure 1). However, their exact mechanisms of action have not been clarified. Every event may be targeted by one or more effectors, and an effector may act on one or more processes. In addition, the effectors may exhibit overlapping functions, thereby complicating the elucidation of their individual functions.
Figure 1
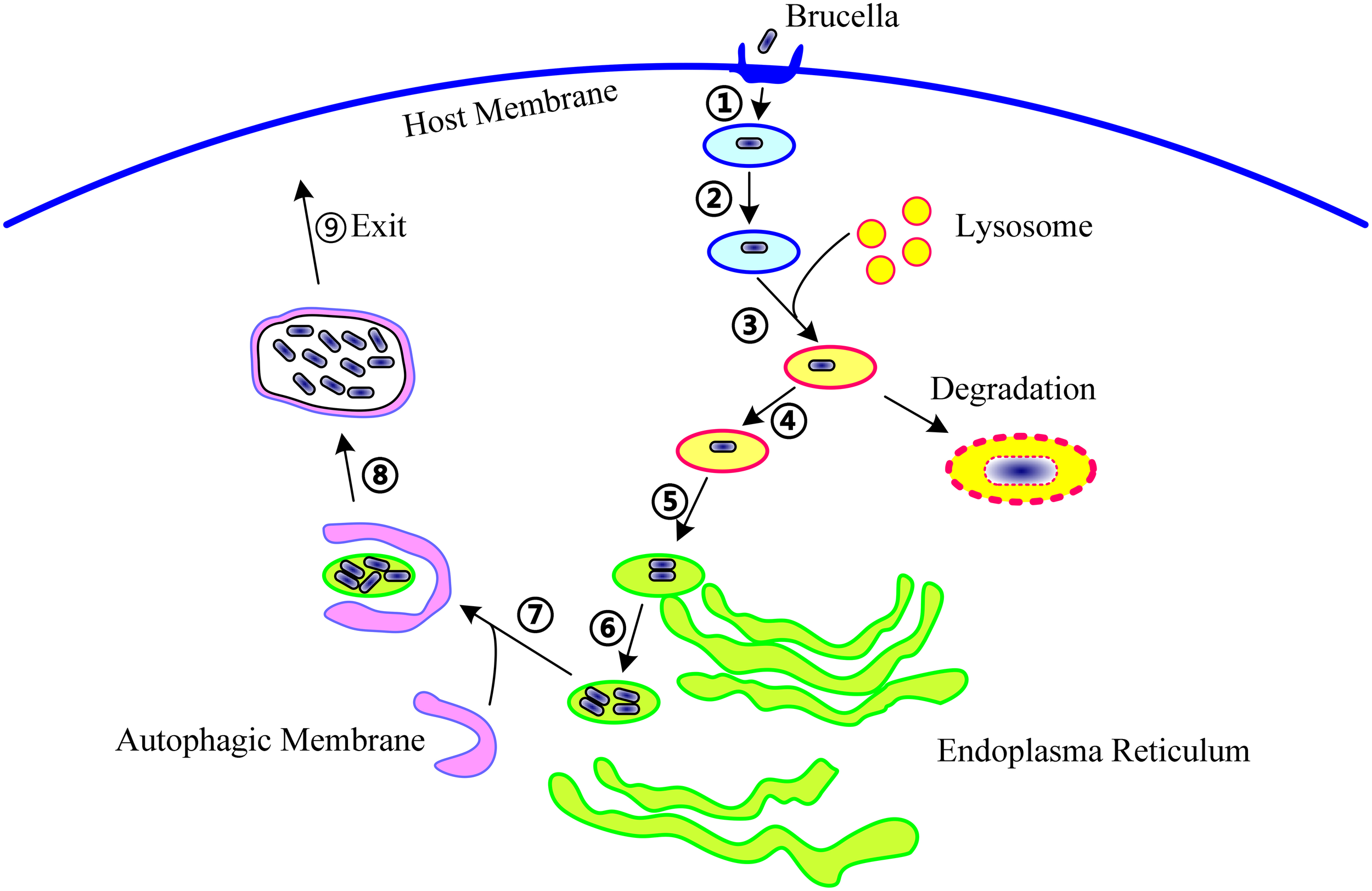
The life cycle of Brucella within host cells. During infection, Brucella first invade the host cells (①), form Brucella-containing vacuoles (BCVs) (②), and undergo fusion with the lysosome in a controlled manner (③). In this step, about 90% of the Brucella are degraded, and the remaining 10% survive (④). Then, the BCV traffic to and reach the endoplasmic reticulum (ER) (⑤), and establish the replicative site (⑥). After ER replication, the Brucella traffic toward the autophagy-like vacuoles (⑦), survive within these compartments (⑧), and finally, leave the host cells to promote cell-to-cell spreading (⑨). The following molecular events involved in these steps are potentially targeted by T4SS: excluding markers of the late endosome or lysosome, acquiring ER markers, interacting with secretory pathways, acquiring markers for autophagosomes, resisting the harsh intracellular environment, and regulating the activation of vital immune pathways.
Known effectors secreted by Brucella through T4SS
Identification of effectors
Although T4SS was identified in Brucella spp. approximately 15 years ago, the effectors of T4SS remained uncharacterized until recently. The first two effectors, VirB-co-regulated effector (Vec) A and VecC, were identified while screening for genes whose transcription was co-regulated by the virB operon regulator, VjbR (de Jong et al., 2008). Screening of the interactions between human proteins and predicted Brucella proteins using a yeast two hybrid (Y2H) system led to the identification of the Rab2 interacting conserved protein A (RicA)-Rab2 interaction pair. Subsequently, RicA was identified as a T4SS-dependent effector (de Barsy et al., 2011). Genome-wide bioinformatics screening for putative T4SS effector proteins using distinct filtering criteria led to the identification of nine T4SS-dependent effector proteins (Marchesini et al., 2011; Myeni et al., 2013). Bacterial toll-Interleukin receptor (TIR) domain-containing proteins are thought to be involved in the virulence of Brucella; screening for TIR-containing proteins in Brucella led to the identification of two proteins, Brucella TIR protein (Btp) A and BtpB, as effectors that are translocated by T4SS into host cells (Salcedo et al., 2013). Recently, a secretory protein, secreted effector protein A (SepA), which is encoded within a horizontally transmitted region, was confirmed as a novel T4SS substrate (Döhmer et al., 2014). Together, these 15 effectors constitute the repertoire of T4SS substrates in Brucella spp. identified to date (Table 1).
Table 1
| Name | ORF in B. abortus | Amino acids | Conserved domains | Targets within hosts | Functions | Methods | References |
|---|---|---|---|---|---|---|---|
| RicA | BAB1_1279 | 175 | Carbonic anhydrase | Rab2 | Regulating vesicle trafficking | TEM1 | de Barsy et al., 2011; Nkengfac et al., 2012; Herrou and Crosson, 2013 |
| VceA | BAB1_1652 | 105 | Non | Unknown | Unknown | CyaA, TEM1 | de Jong et al., 2008 |
| VceC | BAB1_1058 | 418 | The Proline-rich domain | Bip/Grp78 | Activating Unfolded Protein Response | CyaA, TEM1 | de Jong et al., 2008, 2013 |
| BPE005 | BAB1_2005 | 153 | The Coiled-coil domain | Unknown | Unknown | CyaA | Marchesini et al., 2011 |
| BPE043 | BAB1_1043 | 1553 | The Cyclic nucleotide binding domain | Unknown | Unknown | CyaA | Marchesini et al., 2011 |
| BPE275 | BAB1_1275 | 253 | The Rhomboid-like domain | Unknown | Unknown | CyaA | Marchesini et al., 2011 |
| BPE123 | BAB2_0123 | 153 | The Apolipoprotein domain | Unknown | Unknown | CyaA | Marchesini et al., 2011 |
| BtpA/TcpB/Btp1 | BAB1_0279 | 250 | TIR domain | MAL | Inhibiting TLR pathways | CyaA | Cirl et al., 2008; Radhakrishnan et al., 2009; Radhakrishnan and Splitter, 2010; Sengupta et al., 2010; Salcedo et al., 2013 |
| BtpB | BAB1_0756 | 292 | TIR domain | Unknown | Inhibiting TLR pathways | CyaA, TEM1 | Salcedo et al., 2013 |
| BspA | BAB1_0678 | 191 | The Pfam domain DUF2062 | Unknown | Inhibiting the secretory pathway | CyaA, TEM1 | Myeni et al., 2013 |
| BspB | BAB1_0712 | 187 | The SCOP structural domain (d2gsaa) | Unknown | Inhibiting the secretory pathway | CyaA, TEM1 | Myeni et al., 2013 |
| BspC | BAB1_0847 | 137 | The predicted Sec-dependent signal peptide | Unknown | Unknown | CyaA, TEM1 | Myeni et al., 2013 |
| BspE | BAB1_1671 | 117 | Coiled-coil (CC) and TM domains | Unknown | Unknown | CyaA, TEM1 | Myeni et al., 2013 |
| BspF | BAB1_1948 | 428 | the GNAT-family acetyltransferase domain | Unknown | Inhibiting the secretory pathway | CyaA, TEM1 | Myeni et al., 2013 |
| SepA | BAB1_1492 | 130 | Non | Unknown | Inhibiting BCV fusion with the lysosome | TEM1 | Döhmer et al., 2014 |
The 15 effectors secreted by Brucella through T4SS.
A candidate protein may be identified as a T4SS effector protein if it fulfills two criteria: the protein must be secreted into host cells and the secretion must be through the T4SS machinery. The former may be validated by TEM-1 lactamase or calmodulin-dependent adenylate cyclase (CyaA) assays and the latter, by constructing a T4SS-deficient mutant. All strategies used to identify effectors in Brucella are confined by the limited number of potential proteins; similar problems were encountered during the identification of effectors in other bacteria as well. By extending the screening of potential targets to the whole genome, the Dot/Icm T4SS machinery of Legionella pneumophila and Coxiella burnetii were identified to secrete approximately 300 and 100 proteins, respectively (Chen et al., 2010; Voth et al., 2011; Zhu et al., 2011; Lifshitz et al., 2013). Similarly, extending the screening method described above to the whole Brucella genome may lead to the identification of many more effectors.
VceA and VceC
Both VceA and VceC, which are composed of 105 and 418 amino acids, respectively, are highly conserved in all Brucella spp. They share an 18-base pair (bp) conserved promoter box, similar to the virB promoter, and their transcription is regulated by the same transcriptional activator, VjbR (de Jong et al., 2008). Like in the case of the virB promoter, VjbR expressed under in vitro conditions could bind to DNA fragments containing the VceC promoter in electrophoretic mobility shift assays (EMSA). Expression of vceC and virB was downregulated in B. abortus vjbR mutants, as determined by lacZ fusion and β-galactosidase activity assays (de Jong et al., 2008), suggesting that both genes are co-regulated by the same transcriptional factor.
Analysis of the various domains of VceC revealed that 20 amino acids present at its C-terminus are necessary for its translocation into host cells, and 38 amino acids present at its N-terminus, which form a hydrophobic transmembrane domain, are required for its localization to and perturbation of the ER (de Jong et al., 2008, 2013). Immunoprecipitation-mass spectrometry (IP-MS) studies revealed that the ER chaperone protein, Bip/Grp78, binds to the central proline-rich region of VceC. This proline-rich region is the only characterized domain of VceC (de Jong et al., 2013). Consistent with the localization and identity of its interacting partners inside host cells, VceC was shown to activate the unfolded protein response (UPS), cause ER stress, and induce pro-inflammatory responses during infection. The N-terminal ER-targeting domain of VceC was found to be involved in the activation of these processes (de Jong et al., 2013). However, whether the activation of UPS by VceC contributes to its cytotoxity toward macrophages is not clear (de Jong et al., 2008). Further studies are required to elucidate the nature and consequences of the interaction between VceC and Bip. In addition, the co-regulation of vceC and virB by VjbR suggests that the expression of virB and its effectors might be temporally regulated in response to the microenvironment around the BCVs.
RicA
RicA is a mono-domain protein composed of 175 amino acids (de Barsy et al., 2011). Extensive mutagenesis studies have revealed that there are two key elements in RicA, a β-sheet and an isoleucine-glycine-phenylalanine-proline (IGFP) loop, both of which are thought to be involved in protein folding and interaction with Rab2 (Nkengfac et al., 2012). The crystal structure of RicA revealed that the structure of this protein is highly similar to those of the γ-carbonic anhydrase (CA) family of proteins, all of which contain a bound zinc ion. However, RicA does not possess carbonic anhydrase activity, suggesting that it may have evolved a unique function that remains to be elucidated (Herrou and Crosson, 2013).
RicA interacts both in vivo and in vitro with guanosine diphosphate (GDP)-free and GDP-bound forms of Rab2, but not with the GTP-bound form, suggesting that RicA is not a GTPase-activating protein (GAP) (de Barsy et al., 2011; Herrou and Crosson, 2013). Further assays revealed that this protein does not function as a guanine nucleotide exchange factor for Rab2. Whether RicA functions as a GDP dissociation inhibitor or a Rab escort protein remains to be clarified. A ricA deletion mutant did not exhibit attenuated virulence in either mice or HeLa cells. In addition, the internalization and intracellular survival abilities of the mutant remained unaltered (de Barsy et al., 2011). Surprisingly, ΔricA mutants showed faster loss of LAMP1 inside host cells compared with wild-type Brucella (de Barsy et al., 2011), suggesting that ΔricA mutants escape earlier from late lysosomes, arrive at the ER, and establish a replicative niche faster than wild-type bacteria.
BtpA and BtpB
BtpA (also known as Btp1/TcpB) and BtpB share a conserved TIR-containing domain, which is widely present in mammals and is responsible for mediating the signaling cascades of innate immune recognition (Salcedo et al., 2013). Both these proteins were confirmed to be translocated into host cells, as verified by the CyaA assay; however, in the widely used TEM-1 lactamase assay, BtpB showed markedly lower translocating efficiency than did other effectors or BtpA. However, one must note that the TEM-1 lactamase assay is less sensitive than the CyaA assay (Salcedo et al., 2013). Both proteins inhibit the maturation of dendritic cells and are essential for virulence; however, they show opposite effects on the activation of NF-κB, with BtpA inhibiting and BtpB inducing the activation of NF-κB (Salcedo et al., 2008, 2013; Sengupta et al., 2010). The function and mechanism of action of BtpA has been studied extensively (Cirl et al., 2008; Radhakrishnan et al., 2009; Radhakrishnan and Splitter, 2010; Sengupta et al., 2010), and a recent crystal structure revealed that BtpA specifically targets the host signaling adapter protein, MyD88 adapter-like (MAL)/TIRAP, through a unique BB loop region within the TIR domain (Alaidarous et al., 2014; Snyder et al., 2014).
In order to mediate the inhibition of the host TLR signaling pathway, BtpA must be translocated into the host cell from the bacteria; for Brucella, the T4SS machinery is probably the most efficient system for exerting this translocation. TIR domain-containing proteins are present in many other bacteria, such as TlpA in Salmonella enterica, TcpC in Escherichia coli strain CFT073, TirS in Staphylococcus aureus, YpTdp in Yersinia pestis, and PdTLP in Paracoccus denitrificans. These proteins also have the ability to inhibit the toll-like receptor (TLR) pathway (Newman et al., 2006; Chan et al., 2009; Spear et al., 2009; Rana et al., 2011, 2013; Snyder et al., 2013; Askarian et al., 2014); whether or how they are delivered into host cells during infection remains to be elucidated.
BspA, BspB, and BspF
BspA, BspB, and BspF, together with BspC and BspE, were first predicted by bioinformatics approaches and then identified by the TEM-1-fusing protein method (Myeni et al., 2013). BspA, BspB, and BspF are composed of 191, 187, and 428 amino acids, and contain the DUF2062 domain (Domain of Unknown Function 2062, Pfam database), the structural classification of proteins (SCOP) structural domain (flanked by 2 transmembrane domains), and the Gcn5-related N-acetyltransferases (GNAT)-family acetyltransferase domain, respectively (Myeni et al., 2013). Ectopically expressed BspA and BspB appear to localize at the ER, while BspF localizes throughout the cytosol and plasma membrane (Myeni et al., 2013). These three effectors inhibit the host cells' protein secretory pathway when overexpressed in transfected cells, and inhibit cellular secretion during infection (Myeni et al., 2013). Although deletion of one of these effectors alone did not impair the replicating abilities of the mutants in infected BMMs, as determined by enumerating colony forming units (CFUs), the triple ΔbspABF mutant showed decreased intracellular growth (Myeni et al., 2013). Nevertheless, both ΔbspB and ΔbspABF mutants displayed a similar replication deficiency when the percentage of infected cells supporting bacterial replication and containing at least 10 bacteria were scored (Myeni et al., 2013). Consistent with these results, in mice models, the triple ΔbspABF mutant partially lost its ability to survive in the liver, in contrast to the wild-type strain and the single deletion mutants (Myeni et al., 2013). However, neither single nor triple deletions of these three effectors affected the ability to establish systematic infection (Myeni et al., 2013), suggesting that these effectors exert their effects only in the later stages of infection and have complementary functions. Further research on these effectors needs to focus on the identification of their interacting partners, elucidation of their biochemical mechanisms, and resolution of their crystal structures.
SepA
SepA is a 130-amino acid protein that does not contain any previously characterized domain. Secretion of SepA occurs in the very early stages of infection (as early as 30 min post-infection), in a T4SS-dependent manner (Döhmer et al., 2014). However, strikingly, SepA is not directly delivered into host cells. Instead it goes through a periplasm-aggregating intermediate stage before leaving the bacterium, unlike other effectors secreted by bacterial T4SS or T3SS (Döhmer et al., 2014). A ΔsepA mutant exhibited slightly diminished replicating ability 48 h post-infection in infected macrophages, and dramatically decreased replicating ability in the first 4–24 h post-infection, suggesting that SepA is involved in the initial stages of intracellular survival, consistent with its early secretion (Döhmer et al., 2014). The absence of attenuation in the ΔsepA mutant might be because other effectors have similar functions. As the intracellular cycle of Brucella proceeds, the effects of SepA deficiency may be rescued by these effectors, resulting in attenuation at the initial stages and recovery at later stages. Interestingly, although the ΔsepA mutant was as virulent as the wild-type strain, a SepA-overexpressing mutant was found to be attenuated, indicating that the expression of SepA is tightly regulated (Döhmer et al., 2014). Because the other effectors exert their effects in the later stages of the intracellular lifecycle of Brucella, early secretion is a unique feature of SepA. Therefore, sepA mutants may be used to study the early stages of infection by Brucella.
Other effectors
BspC, BspE, BPE123, BPE005, BPE275, and BPE043 were identified as effectors after experimental validation of potential proteins as described above. Although they are highly conserved among the Brucella, little is known about their roles and mechanisms of action. When expressed ectopically, both BspC and BspE localize around the cell nucleus, but with some differences; the former possibly co-localizes with the Golgi apparatus but the latter forms discrete vesicles (Myeni et al., 2013). BspC contains an N-terminal Sec-dependent signal peptide and can induce an ER stress response without affecting the protein secretion pathway (Myeni et al., 2013). BPE005, BPE275, and BPE043 are widely found in other kinds of bacteria, and BPE123 is present in several species, including Bartonella bacilliformis, Ochrobactrum anthropi, and O. intermedium (Marchesini et al., 2011). BPE123 is positioned at the surface of the BCV during infection. Twenty-five amino acids located at the N-terminus of the protein might function as a signal sequence for its translocation into the host cell (Marchesini et al., 2011). The multiplication abilities of BPE123-deficient and wild-type strains were found to be indistinguishable in several cell types and mouse models (Marchesini et al., 2011).
Future perspective
The identification of the effectors secreted by Brucella will enable the elucidation of the functions and mechanism of action of T4SS. In addition, it facilitates the comparison of the various roles of T4SS among gram-negative bacteria. Effectors secreted by T4SS from different bacteria, albeit with similar sequences, may display distinct structures, functions, and mechanisms, contributing to diverse bacterial pathogenesis. Because Brucella is used as a model organism to study intracellular infections, studying the effectors from this species might provide insights into the functioning of T4SS in intracellular bacteria, which may, in turn, aid in understanding the pathogenesis of intracellular bacterial pathogen.
Compared to other bacteria, studies on Brucella T4SS are limited in number, partly because the effectors were identified only recently. On the basis of the information available on the pathogenesis of Brucella, we believe that the following aspects should be considered in future studies: (1) characterization of the biochemistry of these effectors, including identification of the binding targets within host cells, elucidation of the biochemical mechanisms underlying their action on targets, investigations on whether they contain featured domains or novel enzymatic activities such as kinase or phosphatase activities etc., and their intracellular co-localization with binding partners when ectopically expressed or injected during infection; (2) characterization of the cell biology of these effectors, including their effects on intracellular persistence, intracellular apparatus or cytoplasmic membrane docking, signaling pathways targeted by them, and regulation of BCV trafficking [including evading fusion with lysosomes, arriving at ERs, moving to autophagic compartments, and spreading into neighboring cells (see Figure 1)]; (3) elucidation of the contribution of these effectors to virulence in vivo, including their necessity, roles in establishing systematic infection, processing during chronic infection, or eliciting inflammatory response, and whether these effectors can explain the typical intracellular survival performance of Brucella species; (4) investigation of effector microevolution, including similarities and diversities among closely associated species, distribution of homologs in other intracellular bacterial pathogens, horizontal transfer among bacteria, and conservation of function or structure with proteins in other bacteria.
In past decades, our knowledge of vacuolar trafficking of eukaryotic cells has significantly advanced; as a result, cues and tools to explore the intracellular life style of Brucella spp. are now available. Here, on the basis of the information available on the pathogenesis of Brucella and the roles of its T4SS outlined above and elsewhere (Atluri et al., 2011; Martirosyan et al., 2011; Bargen et al., 2012), we suggest that the following events are targeted by the 15 effectors of Brucella during infection (Figure 1): acquiring or losing markers for late endosomes, ERs, or autophagosomes; resisting and surviving the harsh microenvironment; hijacking and utilizing the secretory pathways of the ER and the Golgi; and usurping or regulating the innate immune response. Analyzing the roles of all 15 effectors in these cascades will further our understanding of Brucella pathogenesis.
Effectors that are translocated into host cells by bacteria provide an interface between the bacterium and the host, and lie in the frontline of the battle between the two. Studying the effectors is not only important for understanding bacterial pathogenesis but also provides insights into the immune response of eukaryotic hosts. The lack of an adequate immune response is one of the primary reasons for the inability of the host to overcome the Brucella infection (Martirosyan et al., 2011; Bargen et al., 2012). Therefore, studying the effectors secreted by Brucella might provide insights into the mechanisms by which this intracellular bacterium specifically hijacks the host immune signaling pathways. Knowledge of these mechanisms will help develop better vaccines and therapies against Brucella.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Statements
Acknowledgments
This work was supported by the National Basic Research Program of China (Grant No. 2009CB522602), the Beijing Novo Program (Z131102000413062), the National Key Program for Infectious Diseases of China (2013ZX10004-203, 2013ZX10004-217-002, and 2013ZX10004805-006), and the National Natural Science Foundation of China (81071399, 81071320, 31000041, 81301405, and 30672133).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Alaidarous M. Ve T. Casey L. W. Valkov E. Ericsson D. J. Ullah M. O. et al . (2014). Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin-1 receptor domain-containing protein TcpB. J. Biol. Chem.289, 654–668. 10.1074/jbc.M113.523274
2
Arellano-Reynoso B. Lapaque N. Salcedo S. Briones G. Ciocchini A. E. Ugalde R. et al . (2005). Cyclic β-1, 2-glucan is a Brucella virulence factor required for intracellular survival. Nat. Immunol.6, 618–625. 10.1038/ni1202
3
Arocena G. M. Sieira R. Comerci D. J. Ugalde R. A. (2010). Identification of the quorum-sensing target DNA sequence and N-Acyl homoserine lactone responsiveness of the Brucella abortus virB promoter. J. Bacteriol.192, 3434–3440. 10.1128/JB.00232-10
4
Askarian F. van Sorge N. M. Sangvik M. Beasley F. C. Henriksen J. R. Sollid J. U. et al . (2014). A Staphylococcus aureus TIR domain protein virulence factor blocks TLR2-mediated NF-kappaB signaling. J. Innate Immun.6, 485–498. 10.1159/000357618
5
Atluri V. L. Xavier M. N. de Jong M. F. den Hartigh A. B. Tsolis R. M. (2011). Interactions of the human pathogenic Brucella species with their hosts. Annu. Rev. Microbiol.65, 523–541. 10.1146/annurev-micro-090110-102905
6
Bargen K. Gorvel J. P. Salcedo S. P. (2012). Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol. Rev.36, 533–562. 10.1111/j.1574-6976.2012.00334.x
7
Barquero-Calvo E. Chaves-Olarte E. Weiss D. S. Guzmán-Verri C. Chacón-Diaz C. Rucavado A. et al . (2007). Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS ONE2:e631. 10.1371/journal.pone.0000631
8
Boschiroli M. L. Ouahrani-Bettache S. Foulongne V. Michaux-Charachon S. Bourg G. Allardet-Servent A. et al . (2002). The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U.S.A.99, 1544–1549. 10.1073/pnas.032514299
9
Caswell C. C. Gaines J. M. Roop R. M. II. (2012). The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J. Bacteriol.194, 3–14. 10.1128/JB.05623-11
10
Celli J. de Chastellier C. Franchini D.-M. Pizarro-Cerda J. Moreno E. Gorvel J.-P. (2003). Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med.198, 545–556. 10.1084/jem.20030088
11
Chan S. L. Low L. Y. Hsu S. Li S. Liu T. Santelli E. et al . (2009). Molecular mimicry in innate immunity: crystal structure of a bacterial TIR domain. J. Biol. Chem.284, 21386–21392. 10.1074/jbc.C109.007591
12
Chaves-Olarte E. Guzmán-Verri C. Méresse S. Desjardins M. Pizarro-Cerdá J. Badilla J. et al . (2002). Activation of Rho and Rab GTPases dissociates Brucella abortus internalization from intracellular trafficking. Cell. Microbiol.4, 663–676. 10.1046/j.1462-5822.2002.00221.x
13
Chen C. Banga S. Mertens K. Weber M. M. Gorbaslieva I. Tan Y. et al . (2010). Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc. Natl. Acad. Sci. U.S.A.107, 21755–21760. 10.1073/pnas.1010485107
14
Cirl C. Wieser A. Yadav M. Duerr S. Schubert S. Fischer H. et al . (2008). Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med.14, 399–406. 10.1038/nm1734
15
Colmenero J. D. Reguera J. M. Martos F. Sánchez-De-Mora D. Delgado M. Causse M. et al . (1996). Complications associated with Brucella melitensis infection: a study of 530 cases. Medicine75, 195–211. 10.1097/00005792-199607000-00003
16
Comerci D. J. Martínez-Lorenzo M. J. Sieira R. Gorvel J. P. Ugalde R. A. (2001). Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol.3, 159–168. 10.1046/j.1462-5822.2001.00102.x
17
de Barsy M. Jamet A. Filopon D. Nicolas C. Laloux G. Rual J. F. et al . (2011). Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell. Microbiol.13, 1044–1058. 10.1111/j.1462-5822.2011.01601.x
18
de Jong M. F. Starr T. Winter M. G. den Hartigh A. B. Child R. Knodler L. A. et al . (2013). Sensing of bacterial type IV secretion via the unfolded protein response. MBio4, e00418–e00412. 10.1128/mBio.00418-12
19
de Jong M. F. Sun Y. H. den Hartigh A. B. van Dijl J. M. Tsolis R. M. (2008). Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol. Microbiol.70, 1378–1396. 10.1111/j.1365-2958.2008.06487.x
20
Del Giudice M. G. Ugalde J. E. Czibener C. (2013). A lysozyme-like protein in Brucella abortus is involved in the early stages of intracellular replication. Infect. Immun.81, 956–964. 10.1128/IAI.01158-12
21
Delrue R. M. Martinez-Lorenzo M. Lestrate P. Danese I. Bielarz V. Mertens P. et al . (2001). Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol.3, 487–497. 10.1046/j.1462-5822.2001.00131.x
22
den Hartigh A. B. Rolan H. G. de Jong M. F. Tsolis R. M. (2008). VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J. Bacteriol.190, 4427–4436. 10.1128/JB.00406-08
23
den Hartigh A. B. Sun Y. H. Sondervan D. Heuvelmans N. Reinders M. O. Ficht T. A. et al . (2004). Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect. Immun.72, 5143–5149. 10.1128/IAI.72.9.5143-5149.2004
24
Döhmer P. H. Valguarnera E. Czibener C. Ugalde J. E. (2014). Identification of a type IV secretion substrate of Brucella abortus that participates in the early stages of intracellular survival. Cell. Microbiol.16, 396–410. 10.1111/cmi.12224
25
Dozot M. Boigegrain R. A. Delrue R. M. Hallez R. Ouahrani-Bettache S. Danese I. et al . (2006). The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell. Microbiol.8, 1791–1802. 10.1111/j.1462-5822.2006.00749.x
26
Foulongne V. Bourg G. Cazevieille C. Michaux-Charachon S. O'Callaghan D. (2000). Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun.68, 1297–1303. 10.1128/IAI.68.3.1297-1303.2000
27
Franco M. P. Mulder M. Smits H. L. (2007). Persistence and relapse in brucellosis and need for improved treatment. Trans. R. Soc. Trop. Med. Hyg.101, 854–855. 10.1016/j.trstmh.2007.05.016
28
Fronzes R. Schäfer E. Wang L. Saibil H. R. Orlova E. V. Waksman G. (2009). Structure of a type IV secretion system core complex. Science323, 266–268. 10.1126/science.1166101
29
Fugier E. Pappas G. Gorvel J. P. (2007). Virulence factors in brucellosis: implications for aetiopathogenesis and treatment. Expert Rev. Mol. Med.9, 1–10. 10.1017/s1462399407000543
30
Guzman-Verri C. Manterola L. Sola-Landa A. Parra A. Cloeckaert A. Garin J. et al . (2002). The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U.S.A.99, 12375–12380. 10.1073/pnas.192439399
31
Hanna N. Jiménez de Bagüés M. P. Ouahrani-Bettache S. El Yakhlifi Z. Köhler S. Occhialini A. (2011). The virB operon is essential for lethality of Brucella microti in the Balb/c murine model of infection. J. Infect. Dis.203, 1129–1135. 10.1093/infdis/jiq163
32
Herrou J. Crosson S. (2013). Molecular structure of the Brucella abortus metalloprotein RicA, a Rab2-binding virulence effector. Biochemistry52, 9020–9028. 10.1021/bi401373r
33
Hong P. C. Tsolis R. M. Ficht T. A. (2000). Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun.68, 4102–4107. 10.1128/IAI.68.7.4102-4107.2000
34
Kahl-McDonagh M. M. Elzer P. H. Hagius S. D. Walker J. V. Perry Q. L. Seabury C. M. et al . (2006). Evaluation of novel Brucella melitensis unmarked deletion mutants for safety and efficacy in the goat model of brucellosis. Vaccine24, 5169–5177. 10.1016/j.vaccine.2006.04.005
35
Köhler S. Porte F. Jubier-Maurin V. Ouahrani-Bettache S. Teyssier J. Liautard J. P. (2002). The intramacrophagic environment of Brucella suis and bacterial response. Vet. Microbiol.90, 299–309. 10.1016/S0378-1135(02)00215-8
36
Lapaque N. Moriyon I. Moreno E. Gorvel J.-P. (2005). Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol.8, 60–66. 10.1016/j.mib.2004.12.003
37
Lee J. J. Kim D. G. Kim D. H. Simborio H. L. Min W. Lee H. J. et al . (2013). Interplay between clathrin and Rab5 controls the early phagocytic trafficking and intracellular survival of Brucella abortus within HeLa cells. J. Biol. Chem.288, 28049–28057. 10.1074/jbc.M113.491555
38
Lee J. J. Kim J. H. Kim D. G. Kim D. H. Simborio H. L. Min W. G. et al . (2014). Characterization of betaine aldehyde dehydrogenase (BetB) as an essential virulence factor of Brucella abortus. Vet. Microbiol.168, 131–140. 10.1016/j.vetmic.2013.10.007
39
Lestrate P. Delrue R. M. Danese I. Didembourg C. Taminiau B. Mertens P. et al . (2000). Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol.38, 543–551. 10.1046/j.1365-2958.2000.02150.x
40
Lestrate P. Dricot A. Delrue R.-M. Lambert C. Martinelli V. De Bolle X. et al . (2003). Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect. Immun.71, 7053–7060. 10.1128/IAI.71.12.7053-7060.2003
41
Lifshitz Z. Burstein D. Peeri M. Zusman T. Schwartz K. Shuman H. A. et al . (2013). Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. U.S.A.110, E707–E715. 10.1073/pnas.1215278110
42
Lim J. J. Kim D. H. Lee J. J. Kim D. G. Min W. Lee H. J. et al . (2012). Protective effects of recombinant Brucella abortus Omp28 against infection with a virulent strain of Brucella abortus 544 in mice. J. Vet. Sci.13, 287–292. 10.4142/jvs.2012.13.3.287
43
López-Goñi I. Guzmán-Verri C. Manterola L. Sola-Landa A. Moriyon I. Moreno E. (2002). Regulation of Brucella virulence by the two-component system BvrR/BvrS. Vet. Microbiol.90, 329–339. 10.1016/S0378-1135(02)00218-3
44
Low H. H. Gubellini F. Rivera-Calzada A. Braun N. Connery S. Dujeancourt A. et al . (2014). Structure of a type IV secretion system. Nature508, 550–553. 10.1038/nature13081
45
Luo Z. Q. (2012). Legionella secreted effectors and innate immune responses. Cell. Microbiol.14, 19–27. 10.1111/j.1462-5822.2011.01713.x
46
Marchesini M. I. Herrmann C. K. Salcedo S. P. Gorvel J. P. Comerci D. J. (2011). In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell. Microbiol.13, 1261–1274. 10.1111/j.1462-5822.2011.01618.x
47
Martínez-Núñez C. Altamirano-Silva P. Alvarado-Guillén F. Moreno E. Guzmán-Verri C. Chaves-Olarte E. (2010). The two-component system BvrR/BvrS regulates the expression of the type IV secretion system VirB in Brucella abortus. J. Bacteriol.192, 5603–5608. 10.1128/JB.00567-10
48
Martín-Martín A. I. Sancho P. de Miguel M. J. Fernández-Lago L. Vizcaíno N. (2012). Quorum-sensing and BvrR/BvrS regulation, the Type IV secretion system, cyclic glucans, and BacA in the virulence of Brucella ovis: similarities to and differences from smooth Brucellae. Infect. Immun.80, 1783–1793. 10.1128/IAI.06257-11
49
Martirosyan A. Moreno E. Gorvel J. P. (2011). An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol. Rev.240, 211–234. 10.1111/j.1600-065X.2010.00982.x
50
Martirosyan A. Pérez-Gutierrez C. Banchereau R. Dutartre H. Lecine P. Dullaers M. et al . (2012). Brucella β 1, 2 cyclic glucan is an activator of human and mouse dendritic cells. PLoS Pathog.8:e1002983. 10.1371/journal.ppat.1002983
51
Mirabella A. Terwagne M. Zygmunt M. S. Cloeckaert A. De Bolle X. Letesson J. (2013). Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J. Bacteriol.195, 453–465. 10.1128/JB.01336-12
52
Myeni S. Child R. Ng T. W. Kupko J. J. III Wehrly T. D. Porcella S. F. et al . (2013). Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog.9:e1003556. 10.1371/journal.ppat.1003556
53
Newman R. M. Salunkhe P. Godzik A. Reed J. C. (2006). Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun.74, 594–601. 10.1128/IAI.74.1.594-601.2006
54
Nkengfac B. Pouyez J. Bauwens E. Vandenhaute J. Letesson J. J. Wouters J. et al . (2012). Structural analysis of Brucella abortus RicA substitutions that do not impair interaction with human Rab2 GTPase. BMC Biochem.13:16. 10.1186/1471-2091-13-16
55
O'Callaghan D. Cazevieille C. Allardet-Servent A. Boschiroli M. L. Bourg G. Foulongne V. et al . (1999). A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol.33, 1210–1220. 10.1046/j.1365-2958.1999.01569.x
56
Paschos A. den Hartigh A. Smith M. A. Atluri V. L. Sivanesan D. Tsolis R. M. et al . (2011). An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect. Immun.79, 1033–1043. 10.1128/IAI.00993-10
57
Pizarro-Cerdá J. Méresse S. Parton R. G. van der Goot G. Sola-Landa A. Lopez-Goñi I. et al . (1998). Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun.66, 5711–5724.
58
Pizarro-Cerdá J. Moreno E. Gorvel J. P. (2000). Invasion and intracellular trafficking of Brucella abortus in nonphagocytic cells. Microbes Infect.2, 829–835. 10.1016/S1286-4579(00)90368-X
59
Porte F. Liautard J.-P. Köhler S. (1999). Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect. Immun.67, 4041–4047.
60
Posadas D. M. Ruiz-Ranwez V. Bonomi H. R. Martín F. A. Zorreguieta A. (2012). BmaC, a novel autotransporter of Brucella suis, is involved in bacterial adhesion to host cells. Cell. Microbiol.14, 965–982. 10.1111/j.1462-5822.2012.01771.x
61
Radhakrishnan G. K. Splitter G. A. (2010). Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem. Biophys. Res. Commun.397, 59–63. 10.1016/j.bbrc.2010.05.056
62
Radhakrishnan G. K. Yu Q. Harms J. S. Splitter G. A. (2009). Brucella TIR domain-containing protein mimics properties of the toll-like receptor adaptor protein TIRAP. J. Biol. Chem.284, 9892–9898. 10.1074/jbc.M805458200
63
Rajashekara G. Glover D. A. Krepps M. Splitter G. A. (2005). Temporal analysis of pathogenic events in virulent and avirulent Brucella melitensis infections. Cell. Microbiol.7, 1459–1473. 10.1111/j.1462-5822.2005.00570.x
64
Rambow-Larsen A. A. Rajashekara G. Petersen E. Splitter G. (2008). Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J. Bacteriol.190, 3274–3282. 10.1128/JB.01915-07
65
Rana R. R. Simpson P. Zhang M. Jennions M. Ukegbu C. Spear A. M. et al . (2011). Yersinia pestis TIR-domain protein forms dimers that interact with the human adaptor protein MyD88. Microb. Pathog.51, 89–95. 10.1016/j.micpath.2011.05.004
66
Rana R. R. Zhang M. Spear A. M. Atkins H. S. Byrne B. (2013). Bacterial TIR-containing proteins and host innate immune system evasion. Med. Microbiol. Immunol.202, 1–10. 10.1007/s00430-012-0253-2
67
Rolán H. G. Tsolis R. M. (2007). Mice lacking components of adaptive immunity show increased Brucella abortus virB mutant colonization. Infect. Immun.75, 2965–2973. 10.1128/IAI.01896-06
68
Rolán H. G. Tsolis R. M. (2008). Inactivation of the type IV secretion system reduces the Th1 polarization of the immune response to Brucella abortus infection. Infect. Immun.76, 3207–3213. 10.1128/IAI.00203-08
69
Rolán H. G. Xavier M. N. Santos R. L. Tsolis R. M. (2009). Natural antibody contributes to host defense against an attenuated Brucella abortus virB mutant. Infect. Immun.77, 3004–3013. 10.1128/IAI.01114-08
70
Rossetti C. A. Drake K. L. Siddavatam P. Lawhon S. D. Nunes J. E. Gull T. et al . (2013). Systems biology analysis of Brucella infected Peyer's patch reveals rapid invasion with modest transient perturbations of the host transcriptome. PLoS ONE8:e81719. 10.1371/journal.pone.0081719
71
Ruiz-Ranwez V. Posadas D. M. Van der Henst C. Estein S. M. Arocena G. M. Abdian P. L. et al . (2013). BtaE, an adhesin that belongs to the trimeric autotransporter family, is required for full virulence and defines a specific adhesive pole of Brucella suis. Infect. Immun.81, 996–1007. 10.1128/IAI.01241-12
72
Sá J. C. Silva T. M. Costa E. A. Silva A. P. Tsolis R. M. Paixão T. A. et al . (2012). The virB-encoded type IV secretion system is critical for establishment of infection and persistence of Brucella ovis infection in mice. Vet. Microbiol.159, 130–140. 10.1016/j.vetmic.2012.03.029
73
Salcedo S. P. Marchesini M. I. Degos C. Terwagne M. Von Bargen K. Lepidi H. et al . (2013). BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front. Cell. Infect. Microbiol.3:28. 10.3389/fcimb.2013.00028
74
Salcedo S. P. Marchesini M. I. Lelouard H. Fugier E. Jolly G. Balor S. et al . (2008). Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog.4:e21. 10.1371/journal.ppat.0040021
75
Sengupta D. Koblansky A. Gaines J. Brown T. West A. P. Zhang D. et al . (2010). Subversion of innate immune responses by Brucella through the targeted degradation of the TLR signaling adapter, MAL. J. Immunol.184, 956–964. 10.4049/jimmunol.0902008
76
Sieira R. Arocena G. M. Bukata L. Comerci D. J. Ugalde R. A. (2010). Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters. J. Bacteriol.192, 217–224. 10.1128/JB.01124-09
77
Sieira R. Arocena G. M. Zorreguieta A. Comerci D. J. Ugalde R. A. (2012). A MarR-Type regulator directly activates transcription from the Brucella abortus virB promoter by sharing a redundant role with HutC. J. Bacteriol.194, 6431–6440. 10.1128/JB.01007-12
78
Sieira R. Comerci D. J. Pietrasanta L. I. Ugalde R. A. (2004). Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol.54, 808–822. 10.1111/j.1365-2958.2004.04316.x
79
Sieira R. Comerci D. J. Sánchez D. O. Ugalde R. A. (2000). A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol.182, 4849–4855. 10.1128/JB.182.17.4849-4855.2000
80
Smith M. A. Coincon M. Paschos A. Jolicoeur B. Lavallée P. Sygusch J. et al . (2012). Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem. Biol.19, 1041–1048. 10.1016/j.chembiol.2012.07.007
81
Snyder G. A. Cirl C. Jiang J. Chen K. Waldhuber A. Smith P. et al . (2013). Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A.110, 6985–6990. 10.1073/pnas.1215770110
82
Snyder G. A. Deredge D. Waldhuber A. Fresquez T. Wilkins D. Z. Smith P. T. et al . (2014). Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry. J. Biol. Chem.289, 669–679. 10.1074/jbc.M113.523407
83
Spear A. M. Loman N. J. Atkins H. S. Pallen M. J. (2009). Microbial TIR domains: not necessarily agents of subversion?Trends Microbiol.17, 393–398. 10.1016/j.tim.2009.06.005
84
Starr T. Child R. Wehrly T. D. Hansen B. Hwang S. López-Otin C. et al . (2012). Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe11, 33–45. 10.1016/j.chom.2011.12.002
85
Starr T. Ng T. W. Wehrly T. D. Knodler L. A. Celli J. (2008). Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic9, 678–694. 10.1111/j.1600-0854.2008.00718.x
86
Sun Y.-H. Rolán H. G. den Hartigh A. B. Sondervan D. Tsolis R. M. (2005). Brucella abortus virB12 is expressed during infection but is not an essential component of the type IV secretion system. Infect. Immun.73, 6048–6054. 10.1128/IAI.73.9.6048-6054.2005
87
Trokter M. Felisberto-Rodrigues C. Christie P. J. Waksman G. (2014). Recent advances in the structural and molecular biology of type IV secretion systems. Curr. Opin. Struct. Biol.27C, 16–23. 10.1016/j.sbi.2014.02.006
88
van Schaik E. J. Chen C. Mertens K. Weber M. M. Samuel J. E. (2013). Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol.11, 561–573. 10.1038/nrmicro3049
89
Vizcaíno N. Cloeckaert A. (2012). Biology and genetics of the Brucella outer membrane, in Brucella Molecular Microbiology and Genomics, eds Lopez-GoniI.O'CallaghanD. (Norfolk, UK: Caister Academic Press), 133–161.
90
Voth D. E. Beare P. A. Howe D. Sharma U. M. Samoilis G. Cockrell D. C. et al . (2011). The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J. Bacteriol.193, 1493–1503. 10.1128/JB.01359-10
91
Zhu W. Banga S. Tan Y. Zheng C. Stephenson R. Gately J. et al . (2011). Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE6:e17638. 10.1371/journal.pone.0017638
92
Zygmunt M. S. Hagius S. D. Walker J. V. Elzer P. H. (2006). Identification of Brucella melitensis 16M genes required for bacterial survival in the caprine host. Microbes Infect.8, 2849–2854. 10.1016/j.micinf.2006.09.002
Summary
Keywords
Brucella , type IV secretion, intracellular survival, effector, signaling pathway
Citation
Ke Y, Wang Y, Li W and Chen Z (2015) Type IV secretion system of Brucella spp. and its effectors. Front. Cell. Infect. Microbiol. 5:72. doi: 10.3389/fcimb.2015.00072
Received
23 May 2015
Accepted
28 September 2015
Published
13 October 2015
Volume
5 - 2015
Edited by
Matthew S. Francis, Umeå University, Sweden
Reviewed by
Jianwu Pei, American Animal Health, Inc., USA; Juan Esteban Ugalde, Universidad Nacional de San Martin, Argentina; Carlos Rossetti, Instituto Nacional de Tecnología Agropecuaria, Argentina
Updates
Copyright
© 2015 Ke, Wang, Li and Chen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wengfeng Li lwf304@163.com;
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.