- Microbial Biology, Ruhr University Bochum, Bochum, Germany
Temperature is one of the decisive signals that a mammalian pathogen has entered its warm-blooded host. Among the many ways to register temperature changes, bacteria often use temperature-modulated structures in the untranslated region of mRNAs. In this article, we describe how such RNA thermometers (RNATs) have been discovered one by one upstream of heat shock and virulence genes in the past, and how next-generation sequencing approaches are able to reveal novel temperature-responsive RNA structures on a global scale.
How RNA Thermometers Work
RNA molecules are not linear, but fold into complex three-dimensional structures. Base pairing of proximal nucleotides generates secondary structures, like stem-loops. Long distance interactions allow formation of tertiary structures, like pseudoknots or kissing loops. It is well established that the structure is fundamental for the biological function of non-coding RNA such as tRNA and rRNA. What is now emerging is that the three-dimensional architecture of mRNA influences its entire life cycle: transcription, maturation, translation and degradation. RNA-mediated gene regulation is relevant as it is fast and energy-saving because it bypasses the expression of transcription factors. Structured RNA elements are known to respond to different stimuli, for example metabolite-sensing riboswitches (Serganov and Nudler, 2013). RNA thermometers (RNATs) modulate translation efficiency of an mRNA according to the ambient temperature (Kortmann and Narberhaus, 2012). They are usually located in the 5′-untranslated region (5′-UTR) of an mRNA and form a base-paired structure that involves the ribosome binding site (RBS) and/or the translation initiation codon (Figure 1). Increasing or decreasing temperatures alter the conformation of that structure, allowing or preventing ribosome access and thus translation. RNAT have been localized not only at the 5′-end of a transcript but also in intercistronic regions, where they differentially control gene expression (Krajewski and Narberhaus, 2014). The instantaneous response makes RNAT suitable for regulation of heat shock and virulence genes. The reversibility of the melting process permits simple bidirectional control of translation because the structure melts open and allows translation while the temperature increases, but refolds and blocks translation when the temperature drops again (Chowdhury et al., 2003; Kortmann et al., 2011).
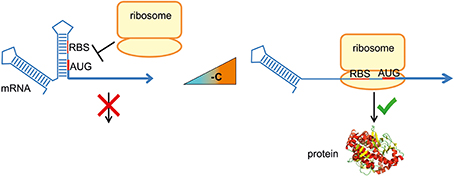
Figure 1. RNA thermometer-mediated translational regulation. RNA thermometers trap the ribosome binding site (RBS) and/or the translation initiation codon (AUG) of an mRNA by base pairing within a secondary structure. An increase in temperature to 37°C (virulence genes) or 40–42°C (heat shock genes) destabilizes the structure in a reversible, zipper-like manner. Liberation of the RBS permits formation of the translation initiation complex and translation occurs.
In contrast to fairly conserved ligand-binding riboswitches, the structural elements found in RNATs show little sequence conservation, if any (Kortmann and Narberhaus, 2012). Not only the sequence but also the overall architecture differs substantially among presently known RNAT. A single short stem-loop structure can be sufficient to confer thermoregulation, as shown for the 46-nt long hsp17 RNAT from Synechocystis sp. PCC 6803 (Kortmann et al., 2011). Other RNAT, like the ROSE (Repression Of heat-Shock gene Expression) element, have a very complex structure ranging from two to four stem-loops (Nocker et al., 2001a,b; Chowdhury et al., 2003; Waldminghaus et al., 2005; Krajewski et al., 2013). Temperature sensing is based on several non-canonical, heat-labile base pairs (Chowdhury et al., 2006). Another poorly conserved RNAT class is characterized by four consecutive uridines that base-pair with the RBS. In the best characterized representative, the Salmonella enterica FourU thermometer of the small heat shock gene agsA, Mg2+ ions play a crucial role in stabilization of the closed conformation at low temperature (Waldminghaus et al., 2007b; Rinnenthal et al., 2010, 2011).
Long and complex structures involving regions in both the untranslated and coding regions are used to permit translation at low temperature. The E. coli cspA transcript adopts two mutually exclusive conformations at 37 and 10°C. The conformational switch affects the translatability and stability of the mRNA resulting in massive induction of the cold shock protein CspA at low temperatures (Yamanaka et al., 1999; Giuliodori et al., 2010).
In this article, we briefly recapitulate how RNATs have been discovered in the past, before we go on to discuss the potential of recently established next-generation sequencing techniques for genome-wide identification of new regulatory RNA elements.
Serendipitous Discovery of RNA Thermometers
The first reported RNAT are unique and rather complex. Their discovery has been preceded by decade-long research on phage λ and the Escherichia coli heat shock response. The 5′-UTR of the λ cIII transcript forms two mutually exclusive conformations whose equilibrium strictly depends on the temperature (Altuvia et al., 1989). At temperatures below 37°C, the RNA adopts a conformation, in which the RBS is accessible and translation occurs. At a higher temperature (45°C), the equilibrium shifts toward the conformation that partially occludes the RBS. Low levels of cIII in turn initiate the lytic cycle of phage λ. An RNAT that liberates the SD sequence with increasing temperature was first found in the E. coli rpoH gene encoding the alternative sigma factor σ32, the master regulator of the heat shock response. The temperature-sensing structure is very complex and involves the 5′-UTR and up to 229 nucleotides of the coding region (Morita et al., 1999a,b).
These seminal findings along with quantitative studies on the role of secondary structures in translation initiation (De Smit and Van Duin, 1990) established the concept that temperature-dependent modulation of RNA structures can regulate translation efficiency. Since then numerous RNAT-controlled heat shock and virulence genes have been discovered. Probably, the most abundant class of RNAT is the ROSE family, always associated with bacterial small heat shock genes. The first ROSE element was found upstream of the Bradyrhizobium japonicum heat shock protein A gene (hspA) (Nocker et al., 2001a,b). Conserved nucleotides upstream of the open reading frame were first believed to serve as binding site for a temperature-responsive transcription factor (Narberhaus et al., 1998) but later shown to be involved in RNA structure formation (Nocker et al., 2001a). In silico prediction of the RNA structure and reporter gene fusions (Nocker et al., 2001a; Chowdhury et al., 2003), CD spectroscopy (Chowdhury et al., 2003), and NMR (Chowdhury et al., 2006) helped to understand the underlying RNA-based control mechanism.
Almost simultaneously, the first RNAT-regulated virulence gene was described in Listeria monocytogenes (Johansson et al., 2002). The prfA gene encodes a transcription factor that controls the synthesis of a number of important virulence factors. A hairpin structure in its 5′-UTR partially masks the ribosome binding region and permits efficient translation only at host body temperature. The prfA thermosensor is peculiar as it integrates not only the temperature signal by structural changes but also metabolic information via a riboswitch-derived small regulatory RNA (Loh et al., 2009).
Only recently, three new RNATs helping to escape the human immune system at increasing temperature were identified in Neisseria meningitidis (Loh et al., 2013). Several N. meningitidis strains resistant to complement-mediated killing were found to contain an 8-nucleotide deletion upstream of the css operon causing increased production of CssA involved in capsule production. After establishing that 38 transcription factors did not control cssA expression, it emerged that an RNA structure is responsible for this effect. At low temperature, the 5′-UTR assumes a hairpin structure that prevents ribosome binding. This structure is much weaker in the mutated RNA. Two other Neisseria genes involved in immune escape are regulated by RNATs. They encode the factor H binding protein and Lst involved in lipopolysaccharide modification.
Guilty by Association: Heat Shock and Virulence Genes
The growing awareness of temperature-responsive RNA structures triggered systematic searches for RNATs upstream of heat shock and virulence genes. Computational predictions by mfold (Zuker, 2003) or other programs (Seetin and Mathews, 2012) can be used to locate RNA structures in the 5′-UTR of selected genes or in entire genomes (Waldminghaus et al., 2007a). Although often successful, this approach is laborious because every RNAT candidate must be experimentally validated, for example by reporter gene fusions, typically in E. coli (Klinkert et al., 2012), and by site-directed mutagenesis, structure probing and toeprinting analysis.
The most common genes subjected to RNAT-mediated regulation code for small heat shock proteins (sHSPs), ATP-independent chaperones that maintain client proteins in a folding-competent state. The systematic search for potential RNATs upstream of sHSP genes has led to the identification of more than 40 ROSE-type RNATs in diverse alpha- and gamma-proteobacteria (Waldminghaus et al., 2005). Structurally very diverse RNATs control sHSP genes in cyanobacteria. A short single-hairpin structure is sufficient to confer thermoregulation to hsp17 of Synechocystis sp. PCC 6803 (Kortmann et al., 2011). Anabaena variabilis encodes two sHSPs. One is controlled by a short RNAT that primarily blocks the start codon, the other controls access to the SD sequence by a ROSE-like sequence and contains an additional extended hairpin structure that might be involved in tertiary RNA-RNA interactions (Cimdins et al., 2014). This study also provided evidence for a complex RNA structure controlling translation of the hspA gene of Thermosynechococcus elongatus, a thermophilic cyanobacterium with an optimal growth temperature at 57°C.
The founding member of the fourU class of RNATs was found to control the Salmonella sHSP gene agsA (Waldminghaus et al., 2007b). These RNATs are characterized by a stretch of four uridines that pair with the AGGA sequence of the SD region. Other members of this class control synthesis of the periplasmic protease HtrA in Salmonella (Klinkert et al., 2012), of the virulence regulator LcrF (VirF) in Yersinia pestis (Hoe and Goguen, 1993) and Yersinia pseudotuberculosis (Böhme et al., 2012), and of the heme receptor proteins ChuA or ShuA in pathogenic E. coli strains and in Shigella dysenteriae, respectively (Kouse et al., 2013).
There is accumulating evidence that various other heat shock and virulence genes are under control of gene-specific RNATs. Differential regulation of individual genes in poly-cistronic heat shock operons is achieved by an RNAT upstream of groES in the Salmonella groESL operon (Cimdins et al., 2013) and two unrelated RNATs upstream of hspX and hspY in the Pseudomonas putida hspXYZ operon (Krajewski et al., 2014). Two identical RNATs were found upstream of the Leptospira interrogans virulence genes ligA and ligB coding for putative lipoproteins important for adhesion and complement resistance (Matsunaga et al., 2013).
Restricting searches for novel RNATs to regions upstream of annotated heat shock and virulence genes is unlikely to reveal RNATs in unexpected places. New bioinformatic tools, such as RNAtips (temperature-induced perturbation of structure) (Chursov et al., 2013), RNAthermsw (Churkin et al., 2014) or other programs able to predict transient RNA structures (Zhu et al., 2013) may help reveal temperature-responsive RNA structures on a genome-wide scale. However, while computational methods are advanced enough to accurately predict short and stable secondary structures, their reliability decreases substantially with increasing length of the RNA molecule or when complex structures, such as pseudoknots and other tertiary interactions, come into play. Therefore, unbiased experimental high-throughput approaches are desirable for the identification of regulatory RNA structures on a global scale.
The RNA Structurome Revealed by Next-Generation Sequencing
The genome-wide assessment of RNA structures relies on structure-probing techniques able to distinguish single- and double-stranded regions. Traditional structure-probing experiments are based on the in vitro treatment of a single RNA species with a variety of chemical or enzymatic probes, capable to modify or cut selectively paired or unpaired nucleotides (Ehresmann et al., 1987; Weeks, 2010). Several chemical reagents modify or cleave unpaired or flexible bases, such as Pb2+ (Gornicki et al., 1989), dimethyl sulfide (DMS), N-methylisatoic anhydride (NMIA), kethoxal, and 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulphonate (CMCT). Enzymatic single-strand specific probes include nucleases S1, A, T1, I. The RNase V1 enzyme cleaves at double-stranded nucleotides. Hydroxyl radicals cleave at RNA bases that are solvent-exposed. Upon treatment, modified or cut positions are mapped by polyacrylamide gel electrophoresis, if necessary after reverse transcription. These approaches are well established and were successfully applied in the structural analysis of a wide range of RNA molecules.
RNA secondary structure can also be sampled in vivo. Chemicals that quickly penetrate the cell membrane, such as Pb2+ (Lindell et al., 2002, 2005) and DMS (Wells et al., 2000; Liebeg and Waldsich, 2009), can be employed to probe the intracellular RNA structure. The in vitro conformation of some RNAs can differ from its functional conformation in vivo, due to the different solvent conditions and the presence of ligands, proteins or other RNAs that can bind to the RNA and alter its architecture (Zemora and Waldsich, 2010). Another important aspect is that the folding process of a nascent bacterial RNA is coupled to its transcription and translation. Cotranscriptional folding and the progressive binding of proteins and ribosomes guide the folding into secondary structures that can differ from the structure of an RNA molecule folded and probed in vitro. In vivo structure probing averages the structural state of each nucleotide from all conformations the RNA molecule adopts during its life cycle. Together, in vivo and in vitro data provide valuable complementary information to unveil biologically relevant structures and their dynamics.
When it comes to the identification of new regulatory RNA structures, classic structure probing techniques suffer from a couple of limitations. First, only a single species of RNA can be tested per experiment, making this technique suitable for the validation of individual structures but not for global screening purposes. Second, only a relatively short region of several 100 nucleotides of an in vitro synthesized and labeled RNA can be investigated. The selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) approach takes advantage of capillary electrophoresis of cDNA obtained from RNA treated with NMIA, or its derivatives (Merino et al., 2005). NMIA attacks flexible (unpaired) bases and forms 2-O adducts that terminate reverse transcription. SHAPE could successfully probe the structure of the 9-kb HIV RNA genome leading to the identification of structural regions that interact with nucleocapsid proteins and elements important for the regulation of viral gene expression (Watts et al., 2009). The development of new chemical probes allowed the application of the SHAPE protocol to the analysis of RNA structure within living cells (Spitale et al., 2013).
Only recently high-throughput sequencing technologies have been successfully applied to RNA structure probing in order to obtain experimentally-derived genome-wide insights into RNA folding. The global landscape of the structural organization of a whole transcriptome of an organism has been referred as the “RNA structurome” (Wan et al., 2011). In this case, a complex RNA population is cleaved or modified with structure-specific probes prior to cDNA synthesis and sequencing. The sequencing reads are mapped to the reference genome or transcriptome and the position of each read along the transcript provides information on single- and double-stranded nucleotides. Structural data can be used to constrain RNA structure prediction algorithms in order to obtain more accurate experimentally-derived secondary structure models of all the sequenced transcripts. The bioinformatic analysis of the raw data is a challenge and considerable effort has been put on software development in order to facilitate data interpretation (Aviran et al., 2011; Ouyang et al., 2013; Zhong and Zhang, 2014).
The first pioneering studies took advantage of structure-specific enzymatic probes. Two examples are the PARS (Parallel Analysis of RNA Structure) (Kertesz et al., 2010) and the Frag-seq (Fragmentation Sequencing) (Underwood et al., 2010) approaches that were first applied to the analysis of the yeast Saccharomyces cerevisiae poly(A) transcriptome and mice nuclear transcriptome, respectively (Figure 2A). In a typical PARS experiment, poly(A) enriched total RNA is isolated, refolded in vitro and partially digested either with the single-strand specific nuclease S1 or the double-strand specific RNaseV1. Paired and unpaired regions of the whole transcriptome are deduced from a comparison of the two sequenced libraries. This approach unveiled the secondary structure profile of more than 3000 yeast transcripts and revealed interesting structural features, such as a higher average secondary structure occurrence in coding regions compared to untranslated regions, a three-nucleotide periodicity of secondary structure across coding regions and correlation between translation efficiency and the structure around the translation start site (Kertesz et al., 2010). Most recently, the PARS approach was applied to the human transcriptome (Wan et al., 2014). Beside the structural features that demarcate coding regions, splicing junctions and microRNA binding site, the authors could identify over 1900 single nucleotide variants that alter the local RNA structure. These variations can affect gene expression and microRNA and protein binding to the RNA molecule.
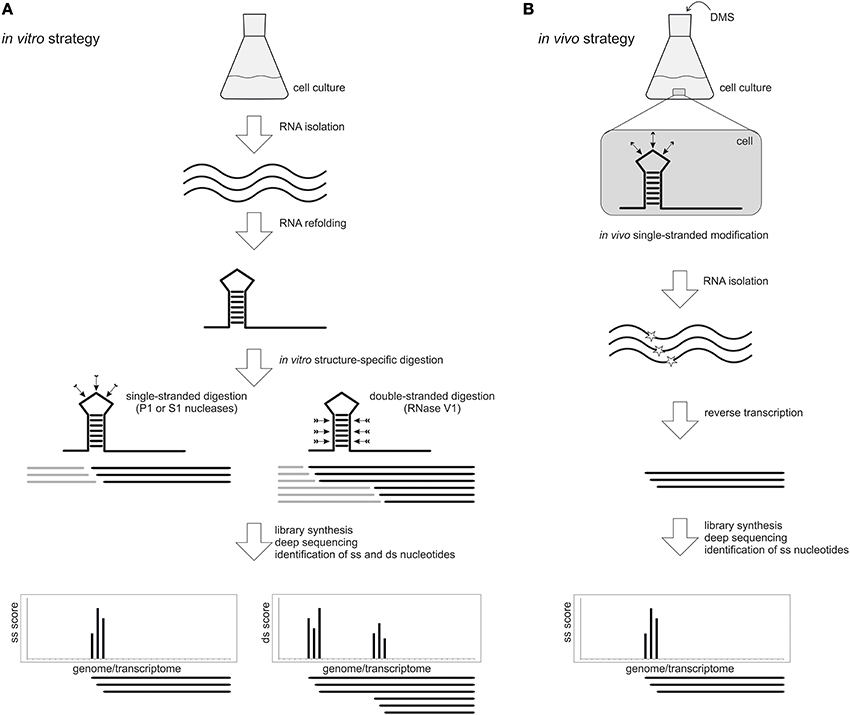
Figure 2. High-throughput genome-wide RNA structure probing. (A) In an in vitro approach, RNA is isolated from a cell culture and re-folded prior to treatment with single-stranded (ss) or double-stranded (ds) specific probes like nucleases P1 and S1 or RNase V1, respectively. After library preparation and deep sequencing, the resulting reads are mapped to the reference genome or transcriptome. The score at each nucleotide indicates whether it is in a single-stranded (ss) or double-stranded (ds) conformation. (B) The in vivo approach allows to probe native RNA structures directly inside the cell using chemical probes that penetrate the membranes and modify nucleotides in a ss conformation. A widely used probe is DMS, which methylates unpaired adenine and cytosine bases. After DMS treatment, the RNA is isolated and the modification position is detected by reverse transcription and deep sequencing. This approach permits the identification of ss regions only.
Purified nuclear RNA from mice, which was refolded in vitro and partially digested with the single-strand specific nuclease P1, has been analyzed by Frag-seq. This approach could successfully confirm single-strand regions of non-coding RNA with known structure and unveiled the secondary structure of non-coding RNA with previously unknown structure (Underwood et al., 2010). In a SHAPE-seq pipeline, the SHAPE method was coupled to high-throughput sequencing to simultaneously read out the structures of a mixture of 7 different RNA molecules (Lucks et al., 2011). Recently, the whole RNA structuromes of Arabidopsis thaliana (Zheng et al., 2010; Li et al., 2012b), Drosophila melanogaster and Caenorhabditis elegans (Li et al., 2012a) have been analyzed by coupling nuclease digestion and high-throughput sequencing. Such global RNA folding profiles allow the identification of structural features involved in RNA-related processes, such as translation regulation, splicing and microRNA-mediated regulation.
It is easily conceivable that temperature-responsive structures such as RNATs can be discovered by this technology and a first step in this direction has been undertaken by probing the whole yeast transcriptome at different temperatures (Wan et al., 2012). The melting temperature of each sequenced transcript was measured at a single nucleotide resolution, which led to the identification of RNA regions that undergo conformational changes in a physiological range of temperature. These regions might have a role in temperature-mediated post-transcriptional gene regulation.
In vivo global structure probing strategies have been attempted only very recently and applied to A. thaliana seedlings (Ding et al., 2014), S. cerevisiae and mammalian cells (Rouskin et al., 2014) and S. cerevisiae (Talkish et al., 2014) (Figure 2B). All these approaches were based on the treatment of living cells with DMS, a chemical probe that can quickly cross the membranes and modify preferentially unpaired adenine and cytosine residues. Modified bases block reverse transcription and sequencing of the resulting fragments permits the identification of the reactive sites and the mapping of single-stranded nucleotides. Comparison between in vitro and in vivo data will ultimately provide a detailed picture of the RNA structurome in its physiological context.
Conclusion
RNA structure discovery has entered a new era by coupling structure probing to next-generation sequencing. Quite surprisingly, no prokaryotic RNA structurome has been reported so far. Given that the principle has been established in yeast (Wan et al., 2012), it is likely that it will be used to identify temperature-sensitive RNA elements in bacteria in the near future. It will be particularly interesting to learn whether RNATs exist in other places than heat shock and virulence gene transcripts.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by the Cluster of Excellence RESOLV (EXC 1069) and grant NA 240/10-1 funded by the Deutsche Forschungsgemeinschaft (DFG).
References
Altuvia, S., Kornitzer, D., Teff, D., and Oppenheim, A. B. (1989). Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J. Mol. Biol. 210, 265–280. doi: 10.1016/0022-2836(89)90329-X
Aviran, S., Trapnell, C., Lucks, J. B., Mortimer, S. A., Luo, S., Schroth, G. P., et al. (2011). Modeling and automation of sequencing-based characterization of RNA structure. Proc. Natl. Acad. Sci. U.S.A. 108, 11069–11074. doi: 10.1073/pnas.1106541108
Böhme, K., Steinmann, R., Kortmann, J., Seekircher, S., Heroven, A. K., Berger, E., et al. (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. doi: 10.1371/journal.ppat.1002518
Chowdhury, S., Maris, C., Allain, F. H., and Narberhaus, F. (2006). Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 25, 2487–2497. doi: 10.1038/sj.emboj.7601128
Chowdhury, S., Ragaz, C., Kreuger, E., and Narberhaus, F. (2003). Temperature-controlled structural alterations of an RNA thermometer. J. Biol. Chem. 278, 47915–47921. doi: 10.1074/jbc.M306874200
Churkin, A., Avihoo, A., Shapira, M., and Barash, D. (2014). RNAthermsw: direct temperature simulations for predicting the location of RNA thermometers. PLoS ONE 9:e94340. doi: 10.1371/journal.pone.0094340
Chursov, A., Kopetzky, S. J., Bocharov, G., Frishman, D., and Shneider, A. (2013). RNAtips: analysis of temperature-induced changes of RNA secondary structure. Nucleic Acids Res. 41, W486–W491. doi: 10.1093/nar/gkt486
Cimdins, A., Klinkert, B., Aschke-Sonnenborn, U., Kaiser, F. M., Kortmann, J., and Narberhaus, F. (2014). Translational control of small heat shock genes in mesophilic and thermophilic cyanobacteria by RNA thermometers. RNA Biol. 11, 594–608. doi: 10.4161/rna.28648
Cimdins, A., Rossmanith, J., Langklotz, S., Bandow, J. E., and Narberhaus, F. (2013). Differential control of Salmonella heat shock operons by structured mRNAs. Mol. Microbiol. 89, 715–731. doi: 10.1111/mmi.12308
De Smit, M. H., and Van Duin, J. (1990). Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. U.S.A. 87, 7668–7672. doi: 10.1073/pnas.87.19.7668
Ding, Y., Tang, Y., Kwok, C. K., Zhang, Y., Bevilacqua, P. C., and Assmann, S. M. (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700. doi: 10.1038/nature12756
Ehresmann, C., Baudin, F., Mougel, M., Romby, P., Ebel, J. P., and Ehresmann, B. (1987). Probing the structure of RNAs in solution. Nucleic Acids Res. 15, 9109–9128. doi: 10.1093/nar/15.22.9109
Giuliodori, A. M., Di Pietro, F., Marzi, S., Masquida, B., Wagner, R., Romby, P., et al. (2010). The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37, 21–33. doi: 10.1016/j.molcel.2009.11.033
Gornicki, P., Baudin, F., Romby, P., Wiewiorowski, M., Kryzosiak, W., Ebel, J. P., et al. (1989). Use of lead(II) to probe the structure of large RNA's. Conformation of the 3′ terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biomol. Struct. Dyn. 6, 971–984. doi: 10.1080/07391102.1989.10506525
Hoe, N. P., and Goguen, J. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909.
Johansson, J., Mandin, P., Renzoni, A., Chiaruttini, C., Springer, M., and Cossart, P. (2002). An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561. doi: 10.1016/S0092-8674(02)00905-4
Kertesz, M., Wan, Y., Mazor, E., Rinn, J. L., Nutter, R. C., Chang, H. Y., et al. (2010). Genome-wide measurement of RNA secondary structure in yeast. Nature 467, 103–107. doi: 10.1038/nature09322
Klinkert, B., Cimdins, A., Gaubig, L. C., Rossmanith, J., Aschke-Sonnenborn, U., and Narberhaus, F. (2012). Thermogenetic tools to monitor temperature-dependent gene expression in bacteria. J. Biotechnol. 160, 55–63. doi: 10.1016/j.jbiotec.2012.01.007
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi: 10.1038/nrmicro2730
Kortmann, J., Sczodrok, S., Rinnenthal, J., Schwalbe, H., and Narberhaus, F. (2011). Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res. 39, 2855–2868. doi: 10.1093/nar/gkq1252
Kouse, A. B., Righetti, F., Kortmann, J., Narberhaus, F., and Murphy, E. R. (2013). RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS ONE 8:e63781. doi: 10.1371/journal.pone.0063781
Krajewski, S. S., Joswig, M., Nagel, M., and Narberhaus, F. (2014). A tricistronic heat shock operon is important for stress tolerance of Pseudomonas putida and conserved in many environmental bacteria. Environ. Microbiol. 16, 1835–1853. doi: 10.1111/1462-2920.12432
Krajewski, S. S., Nagel, M., and Narberhaus, F. (2013). Short ROSE-like RNA thermometers control IbpA synthesis in Pseudomonas species. PLoS ONE 8:e65168. doi: 10.1371/journal.pone.0065168
Krajewski, S. S., and Narberhaus, F. (2014). Temperature-driven differential gene expression by RNA thermosensors. Biochim. Biophys. Acta. doi: 10.1016/j.bbagrm.2014.03.006. [Epub ahead of print].
Li, F., Zheng, Q., Ryvkin, P., Dragomir, I., Desai, Y., Aiyer, S., et al. (2012a). Global analysis of RNA secondary structure in two metazoans. Cell Rep. 1, 69–82. doi: 10.1016/j.celrep.2011.10.002
Li, F., Zheng, Q., Vandivier, L. E., Willmann, M. R., Chen, Y., and Gregory, B. D. (2012b). Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 24, 4346–4359. doi: 10.1105/tpc.112.104232
Liebeg, A., and Waldsich, C. (2009). Probing RNA structure within living cells. Methods Enzymol. 468, 219–238. doi: 10.1016/S0076-6879(09)68011-3
Lindell, M., Brannvall, M., Wagner, E. G., and Kirsebom, L. A. (2005). Lead(II) cleavage analysis of RNase P RNA in vivo. RNA 11, 1348–1354. doi: 10.1261/rna.2590605
Lindell, M., Romby, P., and Wagner, E. G. (2002). Lead(II) as a probe for investigating RNA structure in vivo. RNA 8, 534–541. doi: 10.1017/S1355838201020416
Loh, E., Dussurget, O., Gripenland, J., Vaitkevicius, K., Tiensuu, T., Mandin, P., et al. (2009). A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139, 770–779. doi: 10.1016/j.cell.2009.08.046
Loh, E., Kugelberg, E., Tracy, A., Zhang, Q., Gollan, B., Ewles, H., et al. (2013). Temperature triggers immune evasion by Neisseria meningitidis. Nature 502, 237–240. doi: 10.1038/nature12616
Lucks, J. B., Mortimer, S. A., Trapnell, C., Luo, S., Aviran, S., Schroth, G. P., et al. (2011). Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. U.S.A. 108, 11063–11068. doi: 10.1073/pnas.1106501108
Matsunaga, J., Schlax, P. J., and Haake, D. A. (2013). Role for cis-acting RNA sequences in the temperature-dependent expression of the multiadhesive lig proteins in Leptospira interrogans. J. Bacteriol. 195, 5092–5101. doi: 10.1128/JB.00663-13
Merino, E. J., Wilkinson, K. A., Coughlan, J. L., and Weeks, K. M. (2005). RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231. doi: 10.1021/ja043822v
Morita, M., Kanemori, M., Yanagi, H., and Yura, T. (1999a). Heat-induced synthesis of sigma32 in Escherichia coli: structural and functional dissection of rpoH mRNA secondary structure. J. Bacteriol. 181, 401–410.
Morita, M. T., Tanaka, Y., Kodama, T. S., Kyogoku, Y., Yanagi, H., and Yura, T. (1999b). Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev. 13, 655–665. doi: 10.1101/gad.13.6.655
Narberhaus, F., Käser, R., Nocker, A., and Hennecke, H. (1998). A novel DNA element that controls bacterial heat shock gene expression. Mol. Microbiol. 28, 315–323. doi: 10.1046/j.1365-2958.1998.00794.x
Nocker, A., Hausherr, T., Balsiger, S., Krstulovic, N. P., Hennecke, H., and Narberhaus, F. (2001a). A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 29, 4800–4807. doi: 10.1093/nar/29.23.4800
Nocker, A., Krstulovic, N.-P., Perret, X., and Narberhaus, F. (2001b). ROSE elements occur in disparate rhizobia and are functionally interchangeable between species. Arch. Microbiol. 176, 44–51. doi: 10.1007/s002030100294
Ouyang, Z., Snyder, M. P., and Chang, H. Y. (2013). SeqFold: genome-scale reconstruction of RNA secondary structure integrating high-throughput sequencing data. Genome Res. 23, 377–387. doi: 10.1101/gr.138545.112
Rinnenthal, J., Klinkert, B., Narberhaus, F., and Schwalbe, H. (2010). Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 38, 3834–3847. doi: 10.1093/nar/gkq124
Rinnenthal, J., Klinkert, B., Narberhaus, F., and Schwalbe, H. (2011). Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res. 39, 8258–8270. doi: 10.1093/nar/gkr314
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M., and Weissman, J. S. (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705. doi: 10.1038/nature12894
Seetin, M. G., and Mathews, D. H. (2012). RNA structure prediction: an overview of methods. Methods Mol. Biol. 905, 99–122. doi: 10.1007/978-1-61779-949-5_8
Serganov, A., and Nudler, E. (2013). A decade of riboswitches. Cell 152, 17–24. doi: 10.1016/j.cell.2012.12.024
Spitale, R. C., Crisalli, P., Flynn, R. A., Torre, E. A., Kool, E. T., and Chang, H. Y. (2013). RNA SHAPE analysis in living cells. Nat. Chem. Biol. 9, 18–20. doi: 10.1038/nchembio.1131
Talkish, J., May, G., Lin, Y., Woolford, J. L. Jr., and McManus, C. J. (2014). Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA 20, 713–720. doi: 10.1261/rna.042218.113
Underwood, J. G., Uzilov, A. V., Katzman, S., Onodera, C. S., Mainzer, J. E., Mathews, D. H., et al. (2010). FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods 7, 995–1001. doi: 10.1038/nmeth.1529
Waldminghaus, T., Fippinger, A., Alfsmann, J., and Narberhaus, F. (2005). RNA thermometers are common in alpha- and gamma-proteobacteria. Biol. Chem. 386, 1279–1286. doi: 10.1515/BC.2005.145
Waldminghaus, T., Gaubig, L. C., and Narberhaus, F. (2007a). Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers. Mol. Genet. Genomics 278, 555–564. doi: 10.1007/s00438-007-0272-7
Waldminghaus, T., Heidrich, N., Brantl, S., and Narberhaus, F. (2007b). FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65, 413–424. doi: 10.1111/j.1365-2958.2007.05794.x
Wan, Y., Kertesz, M., Spitale, R. C., Segal, E., and Chang, H. Y. (2011). Understanding the transcriptome through RNA structure. Nat. Rev. Genet. 12, 641–655. doi: 10.1038/nrg3049
Wan, Y., Qu, K., Ouyang, Z., Kertesz, M., Li, J., Tibshirani, R., et al. (2012). Genome-wide measurement of RNA folding energies. Mol. Cell 48, 169–181. doi: 10.1016/j.molcel.2012.08.008
Wan, Y., Qu, K., Zhang, Q. C., Flynn, R. A., Manor, O., Ouyang, Z., et al. (2014). Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709. doi: 10.1038/nature12946
Watts, J. M., Dang, K. K., Gorelick, R. J., Leonard, C. W., Bess, J. W. Jr., Swanstrom, R., et al. (2009). Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716. doi: 10.1038/nature08237
Weeks, K. M. (2010). Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 20, 295–304. doi: 10.1016/j.sbi.2010.04.001
Wells, S. E., Hughes, J. M., Igel, A. H., and Ares, M. Jr. (2000). Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 318, 479–493. doi: 10.1016/S0076-6879(00)18071-1
Yamanaka, K., Mitta, M., and Inouye, M. (1999). Mutation analysis of the 5′ untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 181, 6284–6291.
Zemora, G., and Waldsich, C. (2010). RNA folding in living cells. RNA Biol. 7, 634–641. doi: 10.4161/rna.7.6.13554
Zheng, Q., Ryvkin, P., Li, F., Dragomir, I., Valladares, O., Yang, J., et al. (2010). Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 6:e1001141. doi: 10.1371/journal.pgen.1001141
Zhong, C., and Zhang, S. (2014). Simultaneous folding of alternative RNA structures with mutual constraints: an application to next-generation sequencing-based RNA structure probing. J. Comput. Biol. 21, 609–621. doi: 10.1089/cmb.2013.0044
Zhu, J. Y., Steif, A., Proctor, J. R., and Meyer, I. M. (2013). Transient RNA structure features are evolutionarily conserved and can be computationally predicted. Nucleic Acids Res. 41, 6273–6285. doi: 10.1093/nar/gkt319
Keywords: regulatory RNA, RNA structure, heat shock, virulence, next-generation sequencing
Citation: Righetti F and Narberhaus F (2014) How to find RNA thermometers. Front. Cell. Infect. Microbiol. 4:132. doi: 10.3389/fcimb.2014.00132
Received: 30 June 2014; Paper pending published: 21 July 2014;
Accepted: 02 September 2014; Published online: 18 September 2014.
Edited by:
Wyndham W. Lathem, Northwestern University Feinberg School of Medicine, USAReviewed by:
Jon Audia, University of South Alabama School of Medicine, USAJörgen Johansson, Umeå University, Sweden
Copyright © 2014 Righetti and Narberhaus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Franz Narberhaus, Lehrstuhl für Biologie der Mikroorganismen, Ruhr-Universität Bochum, Universitätsstrasse 150, NDEF 06/783, Bochum, D-44801, Germany e-mail:ZnJhbnoubmFyYmVyaGF1c0BydWIuZGU=
 Francesco Righetti
Francesco Righetti Franz Narberhaus
Franz Narberhaus