- Department of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven, CT, USA
Tsetse flies are the primary vectors of African trypanosomes, which cause Human and Animal African trypanosomiasis in 36 countries in sub-Saharan Africa. These flies have also established symbiotic associations with bacterial and viral microorganisms. Laboratory-reared tsetse flies harbor up to four vertically transmitted organisms—obligate Wigglesworthia, commensal Sodalis, parasitic Wolbachia and Salivary Gland Hypertrophy Virus (SGHV). Field-captured tsetse can harbor these symbionts as well as environmentally acquired commensal bacteria. This microbial community influences several aspects of tsetse's physiology, including nutrition, fecundity and vector competence. This review provides a detailed description of tsetse's microbiome, and describes the physiology underlying host-microbe, and microbe-microbe, interactions that occur in this fly.
Introduction
Tsetse flies (Glossina sp.) serve as hosts to numerous microorganisms. This insect is the primary vector of Trypanosoma brucei parasites that cause a chronic wasting disease in humans (human African trypanosomiasis, or HAT) and domesticated animals (animal African trypanosomiasis, or AAT) in 36 countries throughout sub-Saharan Africa. Both HAT and AAT are pernicious diseases that inflict untold hardships upon their mammalian hosts. In fact, African trypanosomes are regarded as one of the greatest constraints to Africa's economic development (Simarro et al., 2008; Welburn and Maudlin, 2012).
Tsetse also has a relationship with multiple bacterial species and at least one virus. Tsetse's bacterial partners can include 3 maternally-transmitted endosymbionts as well as a taxonomically diverse collection of commensals that the fly acquires from it's environment. Additionally, many tsetse flies also harbor a salivary gland-associated DNA virus. These microbes are intimately associated with many important aspects of their host's biology. Thus, increasing our fundamental knowledge regarding how tsetse interacts with it's microbiota will allow us to develop novel and innovative control strategies aimed at reducing tsetse populations and/or tsetse vector competence.
Exploiting Tsetse's Unusual Reproductive Physiology for Population Control
Tsetse flies lead a relatively sterile existence when compared to the vast majority of other insects. Adult males and females feed exclusively on sterile vertebrate blood. Also, unlike other oviparous insects, female tsetse produce only one egg per gonotrophic cycle (Tobe, 1978). Tsetse offspring develop in their mother's uterus. 3rd instar larvae are deposited and immediately pupate. Adult flies emerge 30 days later. This mode of reproduction results in deposition of only 8–10 progeny per female over her lifespan.
During larvagenesis, tsetse progeny receive nourishment via highly modified maternal accessory gland (referred to as the milk gland) secretions that contain proteins, lipids and amino acids (Attardo et al., 2008, 2012; Benoit et al., 2012; Wang and Aksoy, 2012). Several milk associated protein-encoding genes, including milk gland proteins (mgp) 1–10, transferrin and acid sphingomyelinase 1, are differentially expressed during pregnancy in the milk gland organ. All 10 milk proteins are up-regulated during lactation to support larval growth, and then down-regulated drastically following parturition. Interference with the expression of milk proteins through a double-stranded RNA interference approach has negative effects on fecundity by extending larvagenesis and/or inducing premature abortions (Attardo et al., 2010; Yang et al., 2010; S. Aksoy and J. Benoit, personal communication). Thus, compounds that interfere with milk production could be used as novel vector control tools to reduce vector populations. Understanding the mechanism(s) that regulates the coordinated transcription of milk proteins may lead to the discovery of key regulatory factors that could be exploited to reduce fecundity.
The Biology of Tsetse Symbiosis
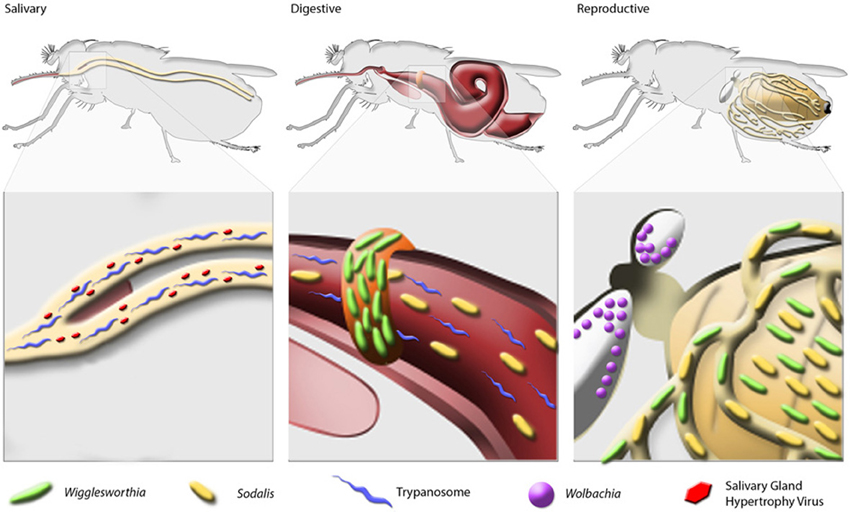
Bacterial endosymbionts play vital roles in insect metabolic processes, the maintenance of fecundity and immune system development (Douglas, 2011). Tsetse flies house 3 endogenous symbionts, Wigglesworthia, Sodalis and Wolbachia, which also impact the physiology of their host (Figure 1).
Wigglesworthia
Maternal milk is more than a source of nourishment for developing tsetse offspring. This substance also serves as a conduit through which vertically-transmitted symbiotic bacteria colonize developing larvae. All laboratory reared tsetse flies examined to date house up to 3 maternally transmitted symbiotic bacteria. The first of these symbionts is the obligate mutualist Wigglesworthia. Tsetse's association with this bacterium began 50-80 million years ago, and since this time Wigglesworthia has been subjected to strict vertical transmission within its species-specific tsetse host (Chen et al., 1999). This contiguous association accounts for Wigglesworthia's extraordinarily streamlined chromosome, which is about 700 kb in size (Akman et al., 2002; Rio et al., 2012). Comparison of the structural organization and gene content of two distant Wigglesworthia species analyzed from Glossina brevipalpis and G. morsitans revealed overall high synteny between the two genomes, similarly high AT biases (over 80%) and highly conserved coding capacity including the absence of dnaA-based DNA replication mechanisms. However, despite extensive conservation, unique genes were identified between the two symbiont genomes that may result in divergent metabolomes impacting host physiology (Rio et al., 2012). Analysis of the Wigglesworthia morsitans specific gene set reveals the presence of a complete shikimate biosynthetic pathway, in which 3-deoxyD-arabino-heptulosonate-7-phosphate (DAHP) can be converted into chorismate. This pathway is degraded in the Wigglesworthia brevipalpis genome. Downstream of the chorismate pathway, W. morsitans' chromosome encodes pabA, pabB (aminodeoxychorismate synthase II and I, respectively) and pabC (4-amino-4-deoxychorismate lyase). These enzymes catalyze the formation of p-aminobenzoate from chorismate–an essential component in folate biosynthesis. The W. morsitans genome also contains an aspC homolog that can be used following chorismate biosynthesis to produce phenylalanine. Interestingly, African trypanosomes are unable to synthesize phenylalanine and folate, yet their genome encodes transporters capable of salvaging both metabolites from their host environment (Berriman et al., 2005). It remains to be seen whether genomic variations noted in the two Wigglesworthia species lead to different biosynthetic capabilities. Additionally, further investigations are required to determine if these metabolic distinctions contribute to the differential trypanosome vector competences exhibited by G. morsitans and G. brevipalpis flies (Harley and Wilson, 1968; Moloo and Kutuza, 1988).
Wigglesworthia provides two well-documented functional benefits to its tsetse host. The first benefit is nutritional, as in the absence of this bacterium, intrauterine larval development is stunted and progeny are aborted (Schlein, 1977; Nogge, 1978; Nogge and Gerresheim, 1982; Pais et al., 2008). Interestingly, Wigglesworthia's contracted genome encodes an unusually high number of putative vitamin biosynthesis pathways (Akman et al., 2002; Rio et al., 2012). More than 10% of the retained CDSs are involved in the biosynthesis of cofactors, prosthetic groups and carriers, supporting Wigglesworthia's genetic contributions to de novo metabolism of biotin, thiazole, lipoic acid, FAD (riboflavin, B2), folate, pantothenate, thiamine (B1), pyridoxine (B6), and protoheme. This genotypic feature supports the theory that Wigglesworthia supplements its tsetse host with nutritious metabolites that are naturally present at low titers in vertebrate blood. Similarly, in mosquitoes, some bacteria, including Serratia, Enterobacter, and Asaia, regulate blood digestion and supplement their host with vitamins (Minard et al., 2013). These observations indicate that hematophagy depends on the presence of gut-associated microbes that facilitate blood meal metabolism.
Wigglesworthia's second function in tsetse is immunological. More specifically, when flies undergo intrauterine larval development in the absence of this bacterium they present a severely compromised immune system during adulthood. Under these conditions, Wigglesworthia-free tsetse are unusually susceptible to infection with normally non-pathogenic E. coli K12 and trypanosomes (Wang et al., 2009; Weiss et al., 2011, 2012, 2013).
Tsetse houses two distinct populations of Wigglesworthia. The first resides intracellularly within bacteriocytes, which collectively form an organ called the bacteriome that is found immediately adjacent to tsetse's anterior midgut (Aksoy, 1995, 2000). Tsetse's second population of Wigglesworthia is found extracellularly in milk gland secretions (Attardo et al., 2008). Interestingly, Wigglesworthia's genome contains genes that encode a complete flagellar structure (Akman et al., 2002). Flagella-associated genes (fliC, motA) are highly expressed in Wigglesworthia that reside extracellularly in tsetse's milk gland, but not in the intracellular bacteriome population (Rio et al., 2012). Expression of a functional flagellum in milk-associated Wigglesworthia may facilitate vertical transmission of this bacterium from the pregnant female to the developing intrauterine larvae, where the symbiont colonizes juvenile tissues.
Sodalis
Tsetse's second endosymbiont is the commensal Sodalis, which is a gram-negative organism closely related to free-living microbes within the Enterobacteriacaea. In addition to tsetse, Sodalis-allied bacteria are present in several other insects, including stinkbugs and weevils (Kaiwa et al., 2010; Toju et al., 2010). Unlike Wigglesworthia, Sodalis exhibits a wide tissue tropism and can be found both intra and extra-cellularly in various tissues including midgut, fat body, milk gland, salivary glands and hemocoel (Cheng and Aksoy, 1999; Balmand et al., 2013). Sodalis' 4.2 Mb chromosome is similar in size to those of its free living ancestors. However, Sodalis' genome exhibits a low coding capacity and an unusually high number of pseudogenes (over 600 genes) in pathways that it likely no longer requires as a resident tsetse endosymbiont (Toh et al., 2006). Interestingly, Sodalis' genome contains features typically associated with pathogenic lifestyles, including 3 type three secretion systems (TTSS). The Sodalis TTSSs, which function during tsetse's juvenile developmental stages, share high homology with those from Salmonella and Yersinia. Interestingly, while structural genes associated with these systems are retained, effector protein genes, the products of which are pathogenic to host cells, are lacking (Toh et al., 2006). Sodalis also has retained the capacity to synthesize a complete flagellar structure, which may be important for it to colonize intrauterine tsetse offspring. This bacterium can be cultured in cell-free medium, which further indicates its recent association with tsetse and its intermediate status between free living and obligate intracellular bacteria.
Sodalis lacks a clearly defined functional role within its tsetse host. More so, several natural tsetse populations lack this bacterium, suggesting that it presents a truly commensal phenotype within its tsetse host. However, several studies indicate that Sodalis may play a role in tsetse's ability to vector pathogenic trypanosomes. Sodalis has been reported to increase tsetse's susceptibility to trypanosomes by obstructing the trypanocidal activity of host midgut lectins (Welburn et al., 1993). Specifically, a G. morsitans line that harbored a high Sodalis infection prevalence exhibited greater susceptibility to trypanosomes than did a G. austeni line that harbored a low Sodalis infection prevalence (Welburn et al., 1993). Additionally, treatment of tsetse with the antibiotic streptozotocin results in the elimination of Sodalis exclusively. While the fecundity of these flies does not change, they become more resistant to trypanosome infections (Dale and Welburn, 2001). In G. palpalis populations collected in Cameroon, parasite infection prevalence positively correlated with the presence of Sodalis in examined flies (Farikou et al., 2010; Soumana et al., 2013). These results suggest that in contrast to Wigglesworthia, which increases tsetse refractoriness to trypanosomes, Sodalis appears to favor the establishment of trypanosome infections in tsetse.
Wolbachia
Some tsetse populations also harbor a parasitic bacterium from the genus Wolbachia. Wolbachia is a wide spread alpha-proteobacteria endosymbiont, infecting approximately 70% insects (Hilgenboecker et al., 2008). This bacterium manipulates the reproductive biology of its host through a variety of mechanisms, including cytoplasmic incompatibility (CI), male killing, feminization and parthenogenesis (Werren et al., 2008). Expression of CI occurs when a Wolbachia infected male mates with an uninfected female, causing developmental arrest during embryogenesis. In contrast, Wolbachia infected females can mate with uninfected males, or with a male infected with the same Wolbachia strain, and produce viable offspring. In tsetse, Wolbachia is localized exclusively intracellularly in germ line tissues and can be detected in early oocyte, embryo and larvae (Cheng et al., 2000; Balmand et al., 2013). Unlike Sodalis and Wigglesworthia, which are transmitted via milk gland secretions, Wolbachia is transmitted transovarially via germ line cells. In G. morsitans laboratory colonies, Wolbachia induces CI when Wolbachia-cured females mate with wild-type Wolbachia-infected males. Offspring from these crosses perish during early embryogenesis while the reciprocal cross survives and produces viable progeny (Alam et al., 2011).
Salivary Gland Hypertrophy Virus (SGHV)
Both colony-reared and natural tsetse populations harbor a rod-shaped, enveloped DNA virus called Salivary Gland Hypertrophy Virus (SGHV). In tsetse, SGHV can cause hypertrophy of the salivary glands and gonadal lesions (Jaenson, 1978). SGHV can be vertically transmitted via maternal milk gland secretions or horizontally during the feeding process (Abd-Alla et al., 2011). While the majority of SGHV-infected flies are asymptomatic with no apparent loss of host fitness, flies infected with high virus titers exhibit reduced fecundity and lifespan, and display hypertrophied salivary glands (Sang et al., 1999; Abd-Alla et al., 2011). In field populations, infection prevalence of SGHV varies in different species and locations (Alam et al., 2012; Malele et al., 2013).
Metabolic Interdependence of Tsetse's Microbiota
Selective elimination of tsetse's microbial partners via treatment with antibiotics revealed that these microorganisms may be metabolically dependent on one another for their survival in the tsetse host (Belda et al., 2010; Snyder et al., 2010; Snyder and Rio, 2013; Wang et al., 2013). For example, offspring from female tsetse that are fed a diet supplemented with ampicillin lack obligate Wigglesworthia but still retain Sodalis and Wolbachia (Pais et al., 2008). In the absence of Wigglesworthia, Sodalis densities decline and are eventually eliminated from subsequent generations. Wigglesworthia was shown to provide thiamine to Sodalis, which lacks the ability to synthesize this metabolite but has retained a transporter to scavenge it from the environment (Snyder et al., 2010; Wang et al., 2013). The fate of SGHV in tsetse also depends upon the presence of the fly's endogenous bacterial symbionts. Specifically, virus proliferation and transmission decreased in tsetse that lacked either Wigglesworthia or all 3 maternally transmitted microbes (Boucias et al., 2013; Wang et al., 2013). Taken together these findings indicate that nutritional dependencies between tsetse's endogenous symbionts not only influence host fitness, but also regulate microbial density and transmission dynamics of the individual partners.
Establishment of Symbiont Infections in the Gut
A complete understanding of the mechanisms that allow commensal bacteria to evade their host's gut immune mechanisms and enable them to colonize their host's gut is vital for successful paratransgenic applications. Recent work using the tsetse model system indicates that bacterial surface coat modifications may be intimately involved in determining symbiont infection outcomes. One such molecule, outer membrane protein A (OmpA), regulates Sodalis' ability to form biofilms, which are essential for this bacterium to colonize tsetse's gut. A mutant Sodalis (SodalisΔompA) line that does not express ompA was unable to form biofilms in vitro. In contrast, wild-type Sodalis, and biofilm-defective E. coli (E. coliΔompA) genetically modified to express Sodalis ompA were able to produce these structures (Maltz et al., 2012). OmpA also influences Sodalis' in vivo colonization competency. Following per os inoculation into tsetse that lack their endogenous microbiome (which serves as an alternative source of OmpA), SodalisΔompA are unable to colonize the fly gut. In contrast, wild-type cells persist in this niche (Maltz et al., 2012). This finding suggests that biofilm formation is essential for Sodalis to be able to colonize the host gut, as these structures may protect the bacteria from the hostile effects of the tsetse's immune system. The identification and functional characterization of additional commensal symbiont colonization factors will be useful for enhancing the efficacy of paratransgenesis in insect disease vectors.
Tsetse-Microbe Associations in Natural Populations
To date, all field-collected tsetse flies examined harbor obligate Wigglesworthia. In contrast, Sodalis infection prevalence in tsetse field populations varies from 0 to 85% (Maudlin et al., 1990; Farikou et al., 2010; Lindh and Lehane, 2011; Alam et al., 2012). Additionally, multiple Sodalis genotypes are present in some field-captured tsetse, suggesting that beyond prevalence Sodalis is genetically distant between different tsetse species (Geiger et al., 2005; Farikou et al., 2011). Like Sodalis, Wolbachia infection prevalence in field-captured tsetse differed significantly between different host species and different populations within the same species (Alam et al., 2012; Doudoumis et al., 2012). Based on standard PCR assays, Wolbachia can be detected in G. m. morsitans, G. m. centralis and G. austeni populations, but not G. tachinoides. Additionally, low density Wolbachia infections are reported in several species and populations, including G. fuscipes and G. morsitans subspecies (Alam et al., 2012; Schneider et al., 2013). In field G. m. morsitans, Wolbachia infection prevalence ranges from 10 to 100% depending on their resident habitats (Doudoumis et al., 2012). Some G. fuscipes individuals also harbor infections with multiple Wolbachia strains (Symula et al., 2013). For example, Wolbachia strains detected in Ugandan G. fuscipes belong to two different lineages. Furthermore, unusually high groEL sequence diversity was present both within and between individuals. Thus, there appears to be high diversity in infection prevalence, density and the circulating strains associated with different tsetse natural populations. Interestingly, three Wolbachia genes, 16S rRNA, fbpA and wsp, have inserted into the G. m. morsitans host chromosome and the ongoing G. m morsitans genome project has revealed that insertions from Wolbachia into host chromosome may be more extensive (Doudoumis et al., 2012; S. Aksoy and K. Bourtzis, personal communication).
Tsetse's viviparous mode of reproduction serves as a rigorous barrier between immature developmental stages and the microbe-rich external environment. However, several environmentally acquired bacterial commensals were recently found in the guts of wild tsetse (Geiger et al., 2013). Bacteria belonging to 3 phyla (23 species), the Firmicutes, Proteobacteria and Actinobacteria, were characterized from east African G. fuscipes (Lindh and Lehane, 2011). G. p. palpalis, G. pallicera, G. nigrofusca, and G. caliginea collected in Cameroon and Angola were found to harbor cultured bacteria from 9 distinct genera (Geiger et al., 2009, 2011). The relative densities of these infections and their persistent association with the tsetse remains to be elucidated. Taxonomically diverse commensal bacteria housed in wild tsetse flies may also influence their host's physiology and vector competence. However, further studies are required to verify this hypothesis.
Conclusions and Future Directions
Tsetse flies house several microorganisms, all of which interact with one another and their host to manipulate the physiology of the system as a whole. With the aid of the Wigglesworthia, Sodalis, Wolbachia, SGHV and T. brucei sequenced and annotated genomes (the annotation of G. morsitans is almost complete), more in depth studies on the interactions between tsetse's multiple partners can be performed. Work to date has focused primarily on host-microbe interactions in laboratory-reared tsetse. However, a growing number of studies using tsetse, and other insect disease vectors, indicate greater spatial and temporal diversity of symbiotic composition, density and prevalence in natural populations. Future work with natural populations will be important to understand the functional role of this diversity on host fitness and pathogen infection outcomes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to many members of our laboratory who contributed to the data we describe in this short review. We especially thank Geoffrey Attardo for editorial assistance. This study received support from NIH AI051584 and Li Foundation and Ambrose Monell Foundation awards to S.A. We are also grateful to the CRPs organized through the Joint FAO/International Atomic Energy Agency, Seibersdorf Austria for discussion and to Slovak Academy of Sciences, Bratislava, Slovakia for providing tsetse puparia for colony maintenance.
References
Abd-Alla, A. M., Parker, A. G., Vreysen, M. J., and Bergoin, M. (2011). Tsetse salivary gland hypertrophy virus: hope or hindrance for tsetse control. PLoS Negl. Trop. Dis. 5:e1220. doi: 10.1371/journal.pntd.0001220
Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., et al. (2002). Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32, 402–407. doi: 10.1038/ng986
Aksoy, S. (1995). Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int. J. Syst. Bacteriol. 45, 848–851. doi: 10.1099/00207713-45-4-848
Aksoy, S. (2000). Tsetse—a haven for microorganisms. Parasitol. Today 16, 114–118. doi: 10.1016/S0169-4758(99)01606-3
Alam, U., Hyseni, C., Symula, R. E., Brelsfoard, C., Wu, Y., Kruglov, O., et al. (2012). Implications of microfauna-host interactions for trypanosome transmission dynamics in Glossina fuscipes fuscipes in Uganda. Appl. Environ. Microbiol. 78, 4627–4637. doi: 10.1128/AEM.00806-12
Alam, U., Medlock, J., Brelsfoard, C., Pais, R., Lohs, C., Balmand, S., et al. (2011). Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 7:e1002415. doi: 10.1371/journal.ppat.1002415
Attardo, G. M., Benoit, J. B., Michalkova, V., Yang, G., Roller, L., Bohova, J., et al. (2012). Analysis of lipolysis underlying lactation in the tsetse fly, Glossina morsitans. Insect Biochem. Mol. Biol. 42, 360–370. doi: 10.1016/j.ibmb.2012.01.007
Attardo, G. M., Lohs, C., Heddi, A., Alam, U. H., Yildirim, S., and Aksoy, S. (2008). Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J. Insect Physiol. 54, 1236–1242. doi: 10.1016/j.jinsphys.2008.06.008
Attardo, G. M., Ribeiro, J. M., Wu, Y., Berriman, M., and Aksoy, S. (2010). Transcriptome analysis of reproductive tissue and intrauterine developmental stages of the tsetse fly (Glossina morsitans morsitans). BMC Genomics 11, 160–169. doi: 10.1186/1471-2164-11-160
Balmand, S., Lohs, C., Aksoy, S., and Heddi, A. (2013). Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 112(Suppl.) S116–S122. doi: 10.1016/j.jip.2012.04.002
Belda, E., Moya, A., Bentley, S., and Silva, F. J. (2010). Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 22, 449–465. doi: 10.1186/1471-2164-11-449
Benoit, J. B., Attardo, G. M., Michalkova, V., Takac, P., Bohova, J., and Aksoy, S. (2012). Sphinomyelinase activity in the mother's milk is essential for juvenile development: a case from lactating tsetse flies. Biol. Reprod. 87, 1–10. doi: 10.1095/biolreprod.112.100008
Berriman, M., Ghedin, E., Hertz-Fowler, C., Blandin, G., Renauld, H., Bartholomeu, D. C. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422. doi: 10.1126/science.1112642
Boucias, D. G., Kariithi, H. M., Bourtzis, K., Schneider, D. I., Kelley, K., Miller, W. J., et al. (2013). Transgenerational transmission of the Glossina pallidipes hytrosavirus depends on the presence of a functional symbiome. PLoS ONE 22:e61150. doi: 10.1371/journal.pone.0061150
Chen, X., Li, S., and Aksoy, S. (1999). Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J. Mol. Evol. 48, 49–58. doi: 10.1007/PL00006444
Cheng, Q., and Aksoy, S. (1999). Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 8, 125–132. doi: 10.1046/j.1365-2583.1999.810125.x
Cheng, Q., Ruel, T. D., Zhou, W., Moloo, S. K., Majiwa, P., O'Neill, S. L., et al. (2000). Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med. Vet. Entomol. 14, 44–50. doi: 10.1046/j.1365-2915.2000.00202.x
Dale, C., and Welburn, S. C. (2001). The endosymbionts of tsetse flies: manipulating host-parasite interactions. Int. J. Parasitol. 1, 628–631. doi: 10.1016/S0020-7519(01)00151-5
Doudoumis, V., Tsiamis, G., Wamwiri, F., Brelsfoard, C., Alam, U., Aksoy, E., et al. (2012). Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus Glossina). BMC Microbiology 12(Suppl.) S1–S3. doi: 10.1186/1471-2180-12-S1-S3
Douglas, A. E. (2011). Lessons from studying insect symbioses. Cell Host Microbe 20, 359–367. doi: 10.1016/j.chom.2011.09.001
Farikou, O., Njiokou, F., Mbida Mbida, J. A., Njitchouang, G. R., Djeunga, H. N., Asonganyi, T., et al. (2010). Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes–an epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect. Genet. Evolut. 10, 115–121. doi: 10.1016/j.meegid.2009.10.008
Farikou, O., Thevenon, S., Njiokou, F., Allal, F., Cuny, G., and Geiger, A. (2011). Genetic diversity and population structure of the secondary symbiont of tsetse flies, Sodalis glossinidius, in sleeping sickness foci in Cameroon. PLoS Negl. Trop. Dis. 5:e1281. doi: 10.1371/journal.pntd.0001281
Geiger, A., Cuny, G., and Frutos, R. (2005). Two Tsetse fly species, Glossina palpalis gambiensis and Glossina morsitans morsitans, carry genetically distinct populations of the secondary symbiont Sodalis glossinidius. Appl. Environ. Microbiol. 71, 8941–8943. doi: 10.1128/AEM.71.12.8941-8943.2005
Geiger, A., Fardeau, M. L., Grebaut, P., Vatunga, G., Josénando, T., Herder, S., et al. (2009). First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infect. Genet. Evol. 9, 1364–1370. doi: 10.1016/j.meegid.2009.09.013
Geiger, A., Fardeau, M. L., Njiokou, F., Joseph, M., Asonganyi, T., Ollivier, B., et al. (2011). Bacterial diversity associated with populations of Glossina spp. from Cameroon and distribution within the Campo sleeping sickness focus. Microb. Ecol. 62, 632–643. doi: 10.1007/s00248-011-9830-y
Geiger, A., Fardeau, M. L., Njiokou, F., and Ollivier, B. (2013). Glossina spp. Gut bacterial flora and their putative role fly-hosted trypanosome development. Cell. Infect. Microbiol. 3, 1–8. doi: 10.3389/fcimb.2013.00034
Harley, J. M. B., and Wilson, A. J. (1968). Comparison between Glossina morsitans, G. pallidipes and G. fuscipes as vectors of trypanosomes of the Trypanosoma congolense group: the proportions infected experimentally and the numbers of infective organisms extruded during feeding. Ann. Trop. Med. Parasitol. 62, 178–187.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia?–A statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220. doi: 10.1111/j.1574-6968.2008.01110.x
Jaenson, T. G. (1978). Virus-like rods associated with salivary gland hyperplasia in tsetse, Glossina pallidipes. Trans. R. Soc. Trop. Med. Hyg. 978, 234–238. doi: 10.1016/0035-9203(78)90200-6
Kaiwa, N., Hosokawa, T., Kikuchi, Y., Nikoh, N., Meng, X. Y., Kimura, N., et al. (2010). Primary gut symbiont and secondary, Sodalisallied symbiont of the Scutellerid stinkbug Cantao ocellatus. Appl. Environ. Microbiol. 76, 3486–3494. doi: 10.1128/AEM.00421-10
Lindh, J. M., and Lehane, M. J. (2011). The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek 99, 711–720. doi: 10.1007/s10482-010-9546-x
Malele, I. I., Manangwa, O., Nyingilili, H. H., Kitwika, W. A., Lyaruu, E. A., Msangi, A. R., et al. (2013). Prevalence of SGHV among tsetse species of economic importance in Tanzania and their implication for SIT application. J. Invertebr. Pathol. 112(Suppl.) S133–S137. doi: 10.1016/j.jip.2012.07.018
Maltz, M. A., Weiss, B. L., O'Neill, M., Wu, Y., and Aksoy, S. (2012). OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl. Environ. Microbiol. 78, 7760–7768. doi: 10.1128/AEM.01858-12
Maudlin, I., Welburn, S. C., and Mehlitz, D. (1990). The relationship between rickettsia-like-organisms and trypanosome infections in natural populations of tsetse in Liberia. Trop. Med. Parasitol. 41, 265–267.
Minard, G., Mavingui, P., and Moro, C. V. (2013). Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit. Vectors 6, 146–157. doi: 10.1186/1756-3305-6-146
Moloo, S. K., and Kutuza, S. B. (1988). Comparative study on the susceptibility of different Glossina species to Trypanosoma brucei brucei infection. Trop. Med. Parasitol. 39, 211–213.
Nogge, G. (1978). Aposymbiotic tsetse flies, Glossina morsitans morsitans obtained by feeding on rabbits immunized specifically with symbionts. J. Invertebr. Pathol. 24, 299–304. doi: 10.1016/0022-1910(78)90026-4
Nogge, G., and Gerresheim, A. (1982). Experiments on the elimination of symbionts from the Tsetse-Fly, Glossina-Morsitans-Morsitans (Diptera, Glossinidae), by antibiotics and lysozyme. J. Invertebr. Pathol. 40, 166–179. doi: 10.1016/0022-2011(82)90112-4
Pais, R., Lohs, C., Wu, Y., Wang, J., and Aksoy, S. (2008). The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74, 5965–5974. doi: 10.1128/AEM.00741-08
Rio, R. V., Symula, R. E., Wang, J., Lohs, C., Wu, Y., Snyder, A. K., et al. (2012). Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. MBio 14:e00240. doi: 10.1128/mBio.00240-11
Sang, R. C., Jura, W. G., Otieno, L. H., Mwangi, R. W., and Ogaja, P. (1999). The effects of a tsetse DNA virus infection on the functions of the male accessory reproductive gland in the host fly Glossina morsitans centralis (Diptera; Glossinidae). Curr. Microbiol. 38, 349–354. doi: 10.1007/PL00006815
Schlein, Y. (1977). Lethal effect of tetracycline on tsetse flies following damage to bacterioid symbionts. Experientia 33, 450–451. doi: 10.1007/BF01922204
Schneider, D. I., Garschall, K. I., Parker, A. G., Abd-Alla, A. M., and Miller, W. J. (2013). Global Wolbachia prevalence, titer fluctuations and their potential of causing cytoplasmic incompatibilities in tsetse flies and hybrids of Glossina morsitans subgroup species. J. Invertebr. Pathol. 112(Suppl.) S104–S115. doi: 10.1016/j.jip.2012.03.024
Simarro, P. P., Jannin, J., and Cattand, P. (2008). Eliminating human African trypanosomiasis: where do we stand and what comes next. PLoS Med. 5:e55. doi: 10.1371/journal.pmed.0050055
Snyder, A. K., and Rio, R. V. (2013). The interwoven biology of the tsetse holobiont. J. Bacteriol. 195, 4322–4330. doi: 10.1128/JB.00487-13
Snyder, A. K., Deberry, J. W., Runyen-Janecky, L., and Rio, R. V. (2010). Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc. R. Soc. 7, 2389–2397. doi: 10.1098/rspb.2010.0364
Soumana, I. H., Simo, G., Njiokou, F., Tchicaya, B., Abd-Alla, A. M., Cuny, G., et al. (2013). The bacterial flora of tsetse fly midgut and its effect on trypanosome transmission. J. Invertebr. Pathol. 112 (Suppl.) S89–S93. doi: 10.1016/j.jip.2012.03.029
Symula, R. E., Alam, U., Brelsfoard, C., Wu, Y., Echodu, R., Okedi, L. M., et al. (2013). Wolbachia association with the tsetse fly, Glossina fuscipes fuscipes, reveals high levels of genetic diversity and complex evolutionary dynamics. BMC Evol. Biol. 13, 31–42. doi: 10.1186/1471-2148-13-31
Tobe, S. S. (1978). Reproductive physiology of Glossina. Annu. Rev. Entomol. 23, 283–307. doi: 10.1146/annurev.en.23.010178.001435
Toh, H., Weiss, B. L., Perkin, S. A., Yamashita, A., Oshima, K., Hattori, M., et al. (2006). Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16, 149–156. doi: 10.1101/gr.4106106
Toju, H., Hosokawa, T., Koga, R., Nikoh, N., Meng, X. Y., Kimura, N., et al. (2010). “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl. Environ. Microbiol. 76, 275–282. doi: 10.1128/AEM.02154-09
Wang, J., and Aksoy, S. (2012). PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse's offspring. Proc. Natl. Acad. Sci. U.S.A. 109, 10552–10557. doi: 10.1073/pnas.1116431109
Wang, J., Brelsfoard, C., Wu, Y., and Aksoy, S. (2013). Intercommunity effects on microbiome and GpSGHV density regulation in tsetse flies. J. Invertebr. Pathol. 112(Suppl.) S32–S39. doi: 10.1016/j.jip.2012.03.028
Wang, J., Wu, Y., Yang, G., and Aksoy, S. (2009). Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. U.S.A. 106, 12133–12138. doi: 10.1073/pnas.0901226106
Weiss, B. L., Maltz, M., and Aksoy, S. (2012). Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188, 3395–3403. doi: 10.4049/jimmunol.1103691
Weiss, B. L., Wang, J., and Aksoy, S. (2011). Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9:e1000619. doi: 10.1371/journal.pbio.1000619
Weiss, B. L., Wang, J., Maltz, M. A., Wu, Y., and Aksoy, S. (2013). Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog. 9:e1003318. doi: 10.1371/journal.ppat.1003318
Welburn, S. C., and Maudlin, I. (2012). Priorities for the elimination of sleeping sickness. Adv. Parasitol. 79, 299–337. doi: 10.1016/B978-0-12-398457-9.00004-4
Welburn, S. C., Arnold, K., Maudlin, I., and Gooday, G. W. (1993). Rickettsia-like organisms and chitinase production in relation to transmission of trypanosomes by tsetse flies. Parasitology 107, 141–145. doi: 10.1017/S003118200006724X
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751. doi: 10.1038/nrmicro1969
Keywords: tsetse fly, symbiont, wigglesworthia, Sodalis, wolbachia
Citation: Wang J, Weiss BL and Aksoy S (2013) Tsetse fly microbiota: form and function. Front. Cell. Infect. Microbiol. 3:69. doi:10.3389/fcimb.2013.00069
Received: 13 August 2013; Accepted: 11 October 2013;
Published online: 29 October 2013.
Edited by:
Brice Rotureau, Institut Pasteur, FranceReviewed by:
Jan Van Den Abbeele, Institute of Tropical Medicine, BelgiumAnne Geiger, Institut de Recherche pour le Développement (IRD), France
Steve Perlman, University of Victoria, Canada
Copyright © 2013 Wang, Weiss and Aksoy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingwen Wang, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, 60 College Street, LEPH 624, New Haven, CT 06510-3201, USA e-mail:amluZ3dlbi53YW5nQHlhbGUuZWR1
 Jingwen Wang
Jingwen Wang Brian L. Weiss
Brian L. Weiss Serap Aksoy
Serap Aksoy