
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 28 September 2011
Sec. Virus and Host
Volume 1 - 2011 | https://doi.org/10.3389/fcimb.2011.00005
This article is part of the Research Topic Haemophilus influenzae View all 9 articles
 Nicole A. Spahich1,2 and Joseph W. St. Geme III1,2*
Nicole A. Spahich1,2 and Joseph W. St. Geme III1,2*Autotransporters are a large class of proteins that are found in the outer membrane of Gram-negative bacteria and are almost universally implicated in virulence. These proteins consist of a C-terminal β-domain that is embedded in the outer membrane and an N-terminal domain that is exposed on the bacterial surface and is endowed with effector function. In this article, we review and compare the structural and functional characteristics of the Haemophilus influenzae IgA1 protease and Hap monomeric autotransporters and the H. influenzae Hia and Hsf trimeric autotransporters. All of these proteins play a role in colonization of the upper respiratory tract and in the pathogenesis of H. influenzae disease.
Autotransporters are a large family of proteins that are inserted into the outer membrane in Gram-negative bacteria and have a variety of effector functions mediated by an extracellular domain, including adherence, invasion, protease activity, and cytotoxicity, among others (Henderson and Nataro, 2001; Pallen et al., 2003). All autotransporters share a common domain structure consisting of an N-terminal signal sequence, a C-terminal β-barrel (translocator) domain, and an internal passenger domain (Desvaux et al., 2004). In the first step of secretion, the cleavable signal sequence directs the nascent protein to the Sec machinery, which facilitates translocation across the inner membrane. Subsequently, the β-barrel domain targets the protein to the outer membrane, where it inserts and facilitates presentation of the internal passenger domain on the bacterial surface.
There are two forms of autotransporter proteins, namely monomeric and trimeric. Monomeric autotransporters consist of a single polypeptide that contains a C-terminal β-barrel domain that is approximately 300 residues in length and folds into 12 β-strands (Klauser et al., 1993; Maurer et al., 1999; Oomen et al., 2004). In contrast, trimeric autotransporters have a short β-barrel domain that is about 70 residues in length and forms four β-strands. Because of the shortened β-barrel domain in trimeric autotransporters, trimerization must occur to form a functional, full-size β-barrel with 12 β-strands (Roggenkamp et al., 2003; Surana et al., 2004).
Initially, it was believed that all the components necessary for successful folding and insertion of autotransporters into the outer membrane were contained in a single polypeptide and that translocation of the passenger domain across the outer membrane occurred without the aid of accessory proteins. However, recent experimental evidence has established that the term “autotransporter” is a misnomer, as outer membrane localization and passenger domain surface presentation require the BamA integral outer membrane protein (Voulhoux et al., 2003; Jain and Goldberg, 2007; Lehr et al., 2010). Understanding the mechanism of outer membrane insertion and passenger domain surface presentation via the Bam complex is still limited, and a number of models exist to explain this process.
Folding of the β-barrel domain occurs as a result of interactions with the Bam complex, with at least partial folding of some translocator domains occurring in the periplasm (Ieva et al., 2008). Regarding transport of the passenger domain across the outer membrane, multiple mechanisms have been proposed. One possibility is that threading of the passenger domain occurs in an N- to C-terminal direction. However, deletions in the N-terminal region of autotransporters do not inhibit translocation, making this scenario unlikely (Bernstein, 2007). Another possibility is that the C-terminal end of the passenger domain forms a hairpin within the translocator pore as an initial step. According to this model, movement through the pore occurs in a C-terminal to N-terminal direction, with folding of the passenger domain providing the energy needed for secretion. In support of this model, there is evidence that the C-terminal end passes through the β-barrel pore as an initial step in passenger domain secretion and that folding of this region facilitates translocation of the rest of the protein (Junker et al., 2009; Peterson et al., 2010). The passenger domain may be partially folded while in the periplasm, requiring addition space in the β-barrel pore during secretion (Skillman et al., 2005). To accommodate a monomeric passenger domain with some degree of tertiary structure, the β-barrel pore may be held in an open conformation by the Bam complex until the passenger domain is fully translocated. In support of this model, several members of the Bam complex have been shown to interact with autotransporter β-barrel and passenger domains at multiple locations along the protein, presumably aiding in protein folding and translocation (Bernstein, 2007; Peterson et al., 2010; Ieva et al., 2011). In the case of trimeric autotransporters, three passenger domains must be secreted and trimerized. It is difficult to envision how three passenger domains could fit through a single β-barrel pore. Translocation of one monomer at a time through the pore would require folding and trimerization to occur as a last step, which would not supply energy for the translocation process. Secretion of a trimeric passenger domain through the small β-barrel pore could be explained if the Bam complex holds the trimeric β-barrel pore in an open configuration, as in the proposed monomeric secretion model, creating enough space for the secretion of all three subunits.
IgA1 protease was initially identified in several Neisseria and Streptococcus species as a secreted bacterial protein able to cleave human immunoglobulin A1 (IgA1), the IgA subclass that predominates in the nasopharyngeal cavity (Mehta et al., 1973; Plaut et al., 1975, 1977). IgA antibody protects mucus membranes by agglutinating invading bacteria, thus inhibiting bacterial interaction with the epithelial surface, and by binding to and neutralizing microbial enzymes and toxins (Kett et al., 1986). IgA cleavage eliminates these protective mechanisms, presumably enhancing bacterial colonization. Based on the observation that only pathogenic bacteria produce IgA1 proteases, it seems likely that these proteins are virulence factors; however, because the IgA1 proteases are specific for human, chimpanzee, and gorilla IgA1, there is no animal model to directly test their pathogenic role (Mulks and Plaut, 1978; Kilian et al., 1979). Haemophilus influenzae IgA1 protease shares significant homology with the Neisseria IgA1 proteases and has the characteristic autotransporter extended N-terminal signal sequence, a C-terminal β-barrel domain called Igaβ, and an internal passenger domain called Igap (Figure 1; Pohlner et al., 1987). The iga gene from H. influenzae type b strain HK368 spans 4,646 bp and encodes a protein with a predicted molecular mass of 169 kDa, while iga in the prototypic laboratory strain Rd is slightly larger, spanning 5,082 bp and encoding a protein with a predicted molecular mass greater than 180 kDa. H. influenzae IgA1 protease undergoes autoproteolytic cleavage, resulting in release of the passenger domain from the membrane-embedded β-barrel domain (Grundy et al., 1987; Poulsen et al., 1989; Plaut et al., 2000). Most H. influenzae strains are capable of cleaving serum IgA1 and, to a lesser extent, the dimeric secretory form of IgA1, including encapsulated meningitis isolates, non-typeable otitis media isolates, and nasopharyngeal isolates from healthy individuals (Kilian et al., 1979). As a reflection of the clonal population structure of encapsulated isolates of H. influenzae, typeable strains have similar IgA1 proteases with similar cleavage capabilities. In contrast, non-typeable H. influenzae (NTHi) IgA1 proteases are highly variable antigenically, presumably due to horizontal gene transfer and recombination between multiple colonizing strains to avoid immune detection (Musser et al., 1986; Lomholt et al., 1993). This variability leads to strain to strain differences in protease activity, with disease isolates from NTHi having a higher level of activity than colonizing isolates of NTHi (Vitovski et al., 2002).
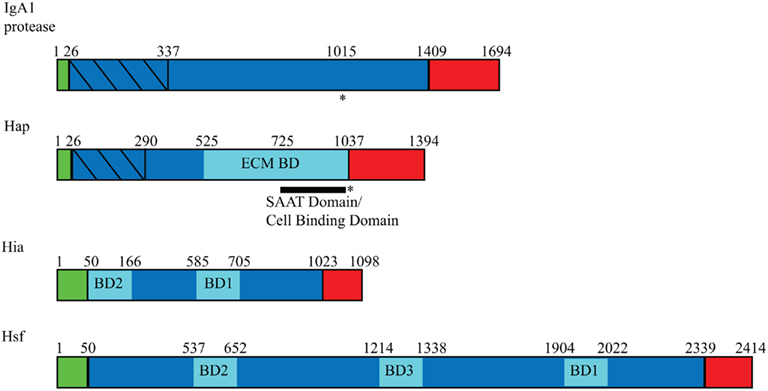
Figure 1. Domain arrangement of Haemophilus influenzae autotransporters. Autotransporters contain three general domains: a signal peptide (green), a passenger domain (blue), and an outer membrane β-barrel domain (red). Within the passenger domains, binding domains are represented in pale blue, and protease domains are represented with diagonal lines. Asterisks (*) indicate regions of autoproteolysis, and SAAT refers to self-associating autotransporter.
The crystal structure of the IgA1 protease passenger domain reveals an N-terminal trypsin/chymotrypsin-like protease domain and a β-helical spine as the major structural components (Figure 2A; Johnson et al., 2009). The spine has a core of hydrophobic residues and an exterior of stacked serine, threonine, and asparagine residues and serves to extend the N-terminal domain away from the bacterial membrane. The N-terminal domain has a globular structure with unique loops in the chymotrypsin fold that are important for substrate selection and a catalytic triad that is responsible for protease activity (Perona and Craik, 1995). The active site of the protease domain is shallow, creating a perfect environment for proline residues typically found at IgA1 protease cleavage sites. Of note, a small domain (Domain 2) protrudes from the stalk of the β-helix, giving the protein a Y-shaped structure. Computational docking studies of the crystal structure of human IgA1 with IgA1 protease have shown that the IgA1 Fc domain binds in a valley formed by Domain 2 and the protease domain, implicating this small region in substrate recognition (Johnson et al., 2009).
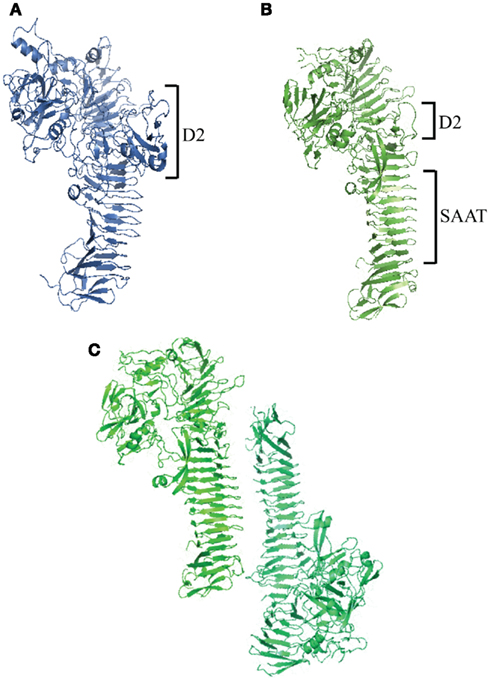
Figure 2. Haemophilus influenzae autotransporter structures. (A) Crystal structure of IgA1 protease passenger domain (IgAp) with an N-terminal globular protease domain and a larger Domain 2 (D2) compared to Haps. (B) Crystal structure of Hap passenger domain (Haps) with an N-terminal globular protease domain and a C-terminal SAAT β-helical domain. (C) Hap dimer highlighting Hap–Hap interactions via the SAAT domain.
The H. influenzae IgA1 protease is a serine protease that mediates autoproteolytic cleavage near the junction of the N-terminal passenger domain and the C-terminal β-barrel domain and cleavage of the hinge region of human IgA1. The N-terminal protease domain contains the sequence GDSGSPLF, where S is the active-site serine characteristic of serine proteases (Bachovchin et al., 1990). Autoproteolytic cleavage occurs at a proline-rich sequence in the C-terminal region of the passenger domain, releasing the extracellular domain from the bacterial surface. Within this sequence, there are four sites that have the potential for cleavage (Poulsen et al., 1989). A similar proline-rich sequence exists in the IgA1 hinge region. Cleavage of IgA1 at the hinge region separates the Fab regions from the Fc region. The IgA1 Fab regions bind antigens, while the IgA1 Fc region interacts with receptors on mucins and various immune cells (Mestecky and McGhee, 1987; Phalipon et al., 2002). When cleaved, Fab fragments can still bind surface antigens, masking epitopes, and preventing recognition of these epitopes by uncleaved antibodies. In this way, cleavage by IgA1 protease may facilitate bacterial survival on mucosal surfaces (Mansa and Kilian, 1986; Kilian et al., 1988).
IgA1 proteases are classified as type 1 or type 2 and are distinguished by the bond that they cleave in the IgA1 hinge region. Type 1 proteases cleave at the proline–serine bond between residues 231 and 232, and type 2 proteases cleave at the proline–threonine bond between residues 235 and 236 (Kilian et al., 1983). In encapsulated H. influenzae strains, the enzyme type is correlated with the polysaccharide capsule serotype. Type 1 IgA1 proteases are produced by serotypes a, b, d, and f, while type 2 IgA1 proteases are produced by serotype c and e strains. Non-typeable strains can have either or both enzyme types (Mulks et al., 1982).
Hap is a 155-kDa monomeric autotransporter that was identified when a genomic library of the clinical H. influenzae isolate N187 was screened for factors that promote interaction with respiratory epithelial cells (Figure 1; St. Geme et al., 1994). A clone expressing a secreted high-molecular mass protein demonstrated adherence and low-level invasion in assays with Chang epithelial cells. This transformant contained a 4182-bp gene that was named hap for Haemophilus adherence and penetration and encoded a predicted protein with homology to serine-type IgA1 proteases from H. influenzae strains and N. gonorrhoeae strains. The Hap passenger domain harbors an adhesive region that promotes adherence to human epithelial cell, extracellular matrix (ECM) proteins, and neighboring Hap-expressing bacteria and a serine protease domain that mediates autoproteolytic cleavage.
Hap is a unique protein that has characteristics of both self-associating autotransporters (SAATs) that promote bacterial aggregation and serine protease autotransporters and may be an evolutionary link between the two autotransporter types Hendrixson and St.Geme, 1998; Klemm et al., 2006). In light of this observation, efforts were made to understand the structure of the Hap passenger domain (Haps) in detail. Initially, electron microscopy of purified Haps revealed an arrowhead shape similar to the crystal structure of E. coli hemoglobin protease (Hbp), another serine protease autotransporter (Otto et al., 2005; Unzai et al., 2005). Crystallization of Haps revealed a structure similar to the structures of Hbp and IgA1 protease, with an N-terminal globular serine protease domain and a C-terminal β-spine (Figure 2B; Meng et al., 2011). Haps also has four smaller domains that may play structural roles. Domain 2 (D2) is similar to IgA1 protease D2 and forms a loose hairpin structure that protrudes away from the molecule. Compared to Hap D2, IgA1 protease D2 is much larger and has a branched, Y-shaped morphology that could inhibit self-association of the C-terminal β-spine by hindering close association of nearby IgA1 protease molecules.
As in IgA1 protease, the Haps globular domain has a V-shaped substrate-binding pocket with a catalytic triad located in the center that mediates protease activity. As predicted by mutational analysis, the shallow depth of the binding pocket suggests that a leucine at the P1 position of the cleavage site is the preferred amino acid for efficient substrate recognition; however, a nearby disordered loop could explain the ability of Haps to cleave peptides with larger side chains such as the phenylalanine side chain at the P1 position (Kenjale et al., 2009).
The Haps crystal structure revealed a multimer, reflecting the activity of the SAAT domain, which lies C-terminal to the serine protease domain. The SAAT domain consists of a 7-turn β-helix with a triangular prism morphology. The core of this structure consists of hydrophobic and aromatic residues, while the exterior is made up of hydrophilic residues at the edges and stacked asparagine and aspartic acid ladders. The β-helix region is straighter and more ordered compared to other autotransporters (e.g., IgA1 protease), allowing for the close association of Hap proteins on neighboring bacteria that is necessary for oligomerization. The shape of this region taken together with the Y-structure and the short β-spine of IgA1 protease explains the differences in self-association between Hap and IgA1 protease, which are structurally similar otherwise. Haps molecules pack against each other along the SAAT domain in a tail-to-tail configuration (Figure 2C). Higher order Hap multimers could form via associations at this interface, overcoming repulsive forces between adjacent cells to form microcolonies. Disruptions in this region truncate the edges of interaction and lead to a decrease in Hap-mediated bacterial aggregation, implicating this prism structure in microcolony formation (Meng et al., 2011).
A region of the Hap passenger domain with homology to IgA1 protease was found to have a similar catalytic site comprising the sequence GDSGSPLF, where S is the active-site serine (Brenner, 1988; St. Geme et al., 1994). In spite of the same active site, Hap is unable to cleave human IgA1 and instead mediates autoproteolytic cleavage at four C-terminal sites (St. Geme et al., 1994; Hendrixson et al., 1997). This cleavage releases Haps from Hapβ and the bacterial cell surface. The catalytic region responsible for proteolysis is consistent with that of typical SA (chymotrypsin) clan serine proteases and consists of amino acids H98, N140, and S243 (Fink et al., 2001). Mutation of these amino acids individually to alanine abolishes protease activity and results in accumulation of full-length Hap in the bacterial outer membrane. Mutations in the proposed substrate-binding groove have identified amino acids N274, L263, R264, and E265 as important for mediating interactions between the catalytic site and its substrate (Kenjale et al., 2009). The catalytic site is responsible for cleavage of the passenger domain predominantly between residues L1036 and N1037 (P1 and P1′); however, when this bond is unavailable, other cleavage sites are recognized at positions L1046-T1047, F1077-A1078, and F1067-S1068, the secondary, tertiary, and quaternary sites, respectively (Hendrixson et al., 1997; Kenjale et al., 2009). The consensus sequence for cleavage at these sites is (Q/R)(A/S)X(L/F) in the P4–P1 positions. Further analysis of the cleavage sites revealed more efficient cleavage at the primary and secondary sites compared to the tertiary and quaternary sites, due to a preference for leucine rather than phenylalanine at the P1 position.
In general, H. influenzae preferentially binds to non-ciliated cells, areas of damaged epithelium, and mucus in the respiratory tract. The C-terminal 311 amino acids of Haps mediate bacterial binding to cultured epithelial cells by interacting with an unknown receptor (Fink et al., 2003). Additionally, the C-terminal 511 amino acids of Haps facilitate H. influenzae binding to several ECM proteins, including fibronectin, laminin, and collagen IV (Fink et al., 2002). Inhibition of Hap serine protease activity results in an increase in Hap-mediated adherence to epithelial cells and ECM proteins, reflecting retention of the Hap passenger domain on the bacterial surface (Fink et al., 2002, 2003).
Haemophilus influenzae interactions with epithelial cells and the ECM are enhanced by bacterial autoaggregation and microcolony formation, which are mediated by a region within Haps. Aggregation occurs due to intercellular multimerization of neighboring Hap molecules at the SAAT domain. This interaction generates van der Waals forces that mediate stable cell–cell interactions (Meng et al., 2011). Like the E. coli autotransporter Antigen 43, this aggregation can be appreciated in liquid culture and may have a role in biofilm formation, potentially facilitating H. influenzae persistence in middle ear effusions and in the lower respiratory tract in patients with cystic fibrosis or chronic obstructive pulmonary disease (COPD; Kjaergaard et al., 2000a,b; Fink et al., 2003).
Hap autoproteolytic activity is inhibited by secretory leukocyte protease inhibitor (SLPI), a small protein found in human upper respiratory secretions that is upregulated by TNFα and IL-1β Hendrixson and St.Geme, 1998. SLPI is believed to protect the respiratory epithelium from injury due to neutrophil elastase and other proteases involved in acute inflammation (Gauthier et al., 1982). Similar to mutations in the Hap catalytic triad, inhibition of Hap autoproteolysis by SLPI results in increased Hap-mediated bacterial adherence to respiratory epithelial cells and ECM proteins, bacterial autoaggregation, and bacterial microcolony formation because of the retention of Haps on the bacterial surface Hendrixson and St.Geme, 1998. During natural infection, inhibition of Hap autoproteolysis presumably facilitates H. influenzae colonization of the respiratory mucosa, while release of Haps may result in dispersal and migration from the site of infection.
Haemophilus influenzae has been observed inside cells in adenoidal tissue obtained from patients with otitis media and in bronchial cells from individuals with COPD, potentially representing a mechanism to evade local immune mechanisms (Forsgren et al., 1994; Moller et al., 1998). Invasion has also been shown in vitro and is mediated at least in part by Hap (St. Geme et al., 1994). H. influenzae invasion into epithelial cells is dependent on actin and microtubules and begins with the formation of lamellipodia around the invading bacterium (St.Geme and Falkow, 1990; Ketterer et al., 1999). While invasion occurs at low levels, it is possible that an intracellular population of organisms accounts for the recurrence of H. influenzae infection in patients with recurrent otitis media or COPD.
Hia and Hsf are homologous trimeric autotransporters found in non-typeable and encapsulated strains of H. influenzae, respectively (Figure 1; St.Geme and Cutter, 2000; Cotter et al., 2005). Hia was first identified in strain 11, a strain lacking HMW1/HMW2-like proteins but still able to adhere efficiently to epithelial cells (Barenkamp and St. Geme et al., 1996). A recombinant phage library of strain 11 was constructed and was screened for reactivity with convalescent serum from a child naturally infected with strain 11, resulting in recovery of a clone expressing a high-molecular mass protein important for adherence to epithelial cells. The 3294-bp open reading frame encodes a protein that was named Hia for H. influenzae adhesin. The predicted molecular mass of Hia from strain 11 is 114.1 kDa, but the protein trimerizes, causing it to migrate on SDS-PAGE gels at a size greater than 300 kDa. The hia gene is found in about 25% of non-typeable strains, corresponding to nearly all strains that lack HMW1/HMW2-like proteins (Barenkamp and St. Geme et al., 1996). Shortly after the discovery of hia, a similar locus ubiquitous in encapsulated H. influenzae strains was characterized and was designated hsf, for Haemophilus surface fibrils, reflecting the fact that this gene encodes short thin surface fibrils. The hsf gene from a prototypic type b strain encodes a protein with a predicted molecular mass of greater than 230 kDa and promotes adherence to human respiratory epithelial cells (St.Geme and Cutter, 1995). Hia and Hsf are 81% similar and 72% identical, with the regions of greatest homology at their N- and C-terminal ends.
The Hia passenger domain consists of repeating domains that are structurally similar, including five Trp-ring domains connected by two IsNeck domains and two KG domains (Figure, 3; Meng et al., 2008). Hsf has a similar architecture, but is extended with 14 Trp-ring domains, three IsNeck domains, and four KG domains, among others. This extension may be needed for Hsf to extend past the capsule in encapsulated strains. The crystal structures of three different fragments of the Hia passenger domain reveal a trimeric architecture in all cases. The trimers are held together by a hydrophobic core that consists primarily of phenylalanine and tryptophan residues. Both Hia and Hsf have two binding domains located at opposite ends of the passenger domain that mediate adherence to epithelial cells (Laarmann et al., 2002; Cotter et al., 2005). These binding domains are always found in regions of the passenger domain where an IsNeck domain is immediately followed by a Trp-ring domain. The binding domains contain an acidic pocket that is necessary for adherence. A single trimeric binding domain contains three identical binding pockets, possibly allowing the protein to interact with three separate receptor molecules or with related domains on one receptor molecule (Yeo et al., 2004).
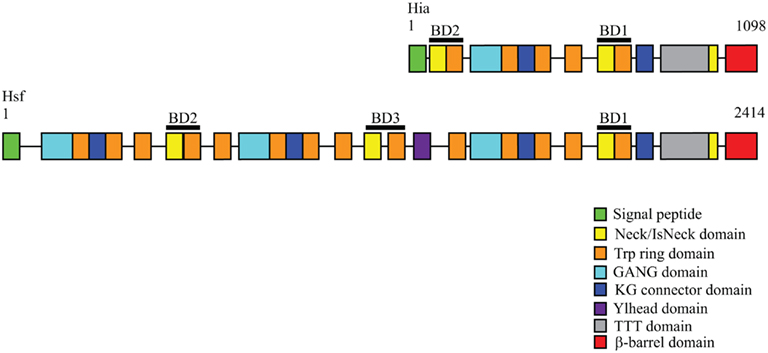
Figure 3. Domain arrangements of Hia and Hsf trimeric autotransporters. The Hia passenger domain is characterized by a repetitive architecture consisting of multiple domain types. Hsf consists of a similar but extended domain arrangement compared to Hia. Domain arrangements were obtained from the daTAA server (http://toolkit.tuebingen.mpg.de/dataa).
The Hia passenger domain is connected to the C-terminal β-barrel domain by a Neck domain (Meng et al., 2008). Like other trimeric autotransporters, Hia contains a relatively short β-barrel translocator domain (Surana et al., 2004). Analysis of the Hia β-barrel domain has shown that it is a trimer that consists of 12 transmembrane β-strands, including four strands from each monomer (Figure 3). The outer surface of the barrel is hydrophobic, facilitating its insertion into the outer membrane. The translocator domain has a central channel that is 1.8 nm and is filled with three N-terminal α-helices that are essential for stability of the trimer. Interestingly, this region contains a ring of basic residues that may coordinate the trimerization of the N-terminal passenger domain (Meng et al., 2006). The Hia β-barrel trimer has a structure that is similar to the C-terminal domain of monomeric autotransporters, with the same number of β-strands, the same angle of the strands relative to the axis of the barrel, and similar pore sizes. These structural similarities suggest a common mechanism for translocation among all autotransporters.
The Hia and Hsf proteins contribute to H. influenzae adherence to epithelial cells. With both Hia and Hsf, the passenger domain contains highly homologous binding domains that mediate the adhesive activity (Figure 1; St. Geme et al., 1996; Laarmann et al., 2002; Cotter et al., 2005). The Hia passenger domain harbors two binding domains that must trimerize for full adhesive activity, resulting in three identical binding pockets (Yeo et al., 2004; Cotter et al., 2006). Both binding domains interact with the same host receptor, but with differing affinities. The primary binding domain of Hia (HiaBD1) is located adjacent to the C-terminal translocator domain. HiaBD1 lies closer to the bacterial surface than the secondary binding domain (HiaBD2) and has greater affinity for the yet unknown host receptor (Laarmann et al. 2002). The core of the HiaBD1 binding pocket is comprised of an aspartic acid, an alanine, and a valine, while the HiaBD2 pocket is comprised of a glutamine, an alanine, and a valine. Therefore, the different binding affinities of HiaBD1 and HiaBD2 appear to be due to differences in the aspartic acid and glutamine side chains (Yeo et al. 2004). One model of Hia binding proposes that the distal HiaBD2 is responsible for the initial interaction with the receptor, bringing the host cell within reach of the bacteria. The higher affinity HiaBD1 may then competitively displace HiaBD2 from the cellular receptor, resulting in more intimate bacterial attachment to the epithelial cell layer. Unlike Hap, the passenger domains of Hia and Hsf are not cleaved from the C-terminus and remain cell associated, enhancing adherence to host cells (St.Geme and Cutter, 2000).
The Hsf passenger domain contains three regions homologous to the Hia binding domains; however, only two have acidic binding pockets that mediate adhesive activity. Similar to Hia, both binding domains recognize the same receptor, with the proximal Hsf binding domain (HsfBD1) having a higher affinity for the host cell receptor than does the distal Hsf binding domain (HsfBD2; Cotter et al. 2005; Radin et al. 2009). The key contact residues in the HsfBD1 and HsfBD2 binding pockets are identical except for an aspartic acid in HsfBD1 and a glutamic acid in HsfBD2, accounting for the different binding affinities Radin et al. 2009). The Hia binding model can be applied to Hsf, where HsfBD2 initiates binding to the cellular receptor and HsfBD1 later displaces HsfBD2, bringing the bacteria into closer contact with the host cell. For Hsf to interact with the host, the passenger domain must extend past the polysaccharide capsule. The intervening region between HsfBD1 and HsfBD2 is three times longer than the sequence between HiaBD1 and HiaBD2 and may be necessary for this extension. The third region of homology to the Hia binding domains is called HsfBD3 and is located in this intervening region. While HsfBD3 is structurally similar to the other binding domains, it lacks an acidic binding pocket and does not mediate adherence to epithelial cells Cotter et al. 2005). Instead, HsfBD3 may play a role in stabilizing the passenger domain or may mediate adherence to yet untested cell types or other host molecules.
The H. influenzae IgA1 protease, Hap, Hia, and Hsf autotransporters are virulence factors that promote colonization and persistence in the human host. Adherence to the respiratory epithelium, microcolony formation leading to biofilm formation, and protease activity facilitating bacterial spread and immune evasion are important pathogenic mechanisms that are mediated by these proteins. The mechanisms by which these virulence factors may work together to establish a H. influenzae infection are outlined in Figure 4. Hia-mediated adherence to epithelial cells and Hap-mediated adherence to both epithelial cells and the ECM may be responsible for initial contact with the host, while IgA1 protease cleaves IgA1, protecting the bacteria from the innate immune response. As infection progresses, Hap-Hap interactions result in microcolony formation and may eventually lead to biofilm formation, another mechanism of immune evasion. Because of Hap autoproteolytic activity, some bacteria may be released from the biofilm to colonize elsewhere. Finally, bacterial invasion mediated by Hap can lead to an intracellular bacterial reservoir that may be responsible for the recurrent infections seen in COPD and otitis media. Further examination of these proteins will aid in understanding H. influenzae disease progression and may facilitate development of novel antimicrobials.
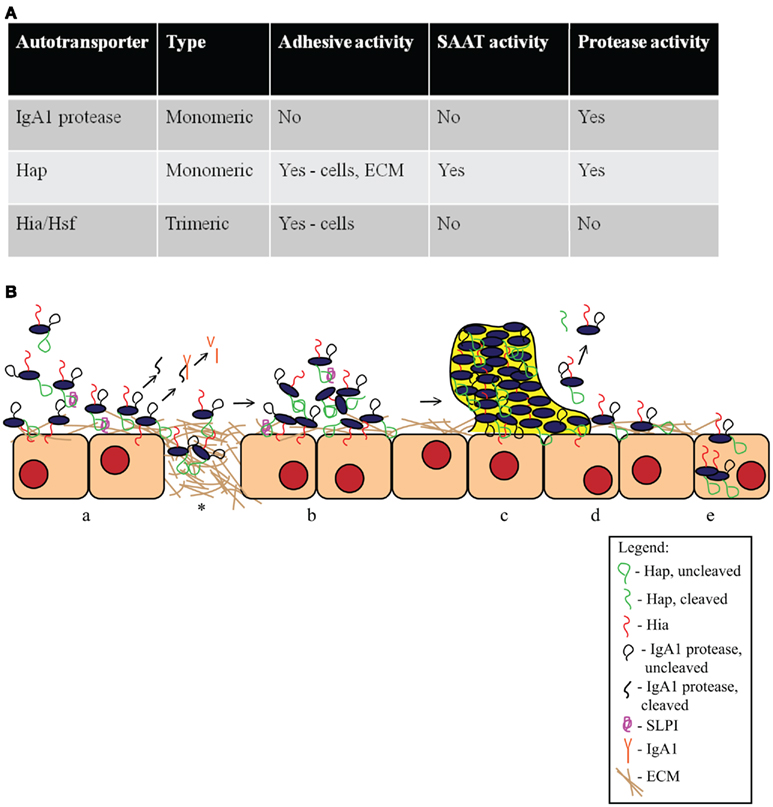
Figure 4. Haemophilus influenzae autotransporter proteins. (A) Summary of key features of H. influenzae autotransporter proteins. (B) Schematic of the role of H. influenzae autotransporter proteins in the pathogenesis of disease. (a) Hap and Hia facilitate H. influenzae attachment to epithelial cells. Hap also promotes adherence to the ECM on the cell layer and at areas of damaged epithelium (*). Hap autoproteolysis is inhibited by SLPI, resulting in the retention of Haps on the bacterial surface. IgA1 protease is released from the bacterial surface and cleaves human IgA1. (b) Once bacteria have adhered, Hap–Hap interactions lead to microcolony formation. (c) Microcolony formation leads to the establishment of a biofilm that is resistant to host immune factors. (d) In the absence of SLPI, Hap autoproteolytic activity allows some bacteria to disperse, presumably to new sites of infection. (e) Hap-mediated attachment promotes bacterial invasion into epithelial cells, potentially providing a protected niche, and allowing bacterial evasion of local immune mechanisms.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Bachovchin, W. W., Plaut, A. G., Flentke, G. R., Lynch, M., and Kettner, C. A. (1990). Inhibition of IgA1 proteinases from Neisseria gonorrhoeae and Haemophilus influenzae by peptide prolyl boronic acids. J. Biol. Chem. 265, 3738–3743.
Barenkamp, S. J., and St. Geme, J. W. III. (1996). Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19, 1215–1223.
Bernstein, H. D. (2007). Are bacterial ‘autotransporters’ really transporters? Trends Microbiol. 15, 441–447.
Brenner, S. (1988). The molecular evolution of genes and proteins: a tale of two serines. Nature 334, 528–530.
Cotter, S. E., Surana, N. K., Grass, S., and St. Geme, J. W. III. (2006). Trimeric autotransporters require trimerization of the passenger domain for stability and adhesive activity. J. Bacteriol. 188, 5400–5407.
Cotter, S. E., Yeo, H. J., Juehne, T., and St. Geme, J. W. III. (2005). Architecture and adhesive activity of the Haemophilus influenzae Hsf adhesin. J. Bacteriol. 187, 4656–4664.
Desvaux, M., Parham, N. J., and Henderson, I. R. (2004). Type V protein secretion: simplicity gone awry? Curr. Issues Mol. Biol. 6, 111–124.
Fink, D. L., Buscher, A. Z., Green, B., Fernsten, P., and St. Geme, J. W. III. (2003). The Haemophilus influenzae Hap autotransporter mediates microcolony formation and adherence to epithelial cells and extracellular matrix via binding regions in the C-terminal end of the passenger domain. Cell. Microbiol. 5, 175–186.
Fink, D. L., Cope, L. D., Hansen, E. J., and St. Geme, J. W. III. (2001). The Hemophilus influenzae Hap autotransporter is a chymotrypsin clan serine protease and undergoes autoproteolysis via an intermolecular mechanism. J. Biol. Chem. 276, 39492–39500.
Fink, D. L., Green, B. A., and St. Geme, J. W. III. (2002). The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, and collagen IV. Infect. Immun. 70, 4902–4907.
Forsgren, J., Samuelson, A., Ahlin, A., Jonasson, J., Rynnel-Dagoo, B., and Lindberg, A. (1994). Haemophilus influenzae resides and multiplies intracellularly in human adenoid tissue as demonstrated by in situ hybridization and bacterial viability assay. Infect. Immun. 62, 673–679.
Gauthier, F., Fryksmark, U., Ohlsson, K., and Bieth, J. G. (1982). Kinetics of the inhibition of leukocyte elastase by the bronchial inhibitor. Biochim. Biophys. Acta 700, 178–183.
Grundy, F. J., Plaut, A. G., and Wright, A. (1987). Haemophilus influenzae IgA1 proteases: determinants of cleavage site specificity and secretion. Adv. Exp. Med. Biol. 216B, 1251–1260.
Henderson, I. R., and Nataro, J. P. (2001). Virulence functions of autotransporter proteins. Infect. Immun. 69, 1231–1243.
Hendrixson, D. R., De La Morena, M. L., Stathopoulos, C., and St. Geme, J. W. III. (1997). Structural determinants of processing and secretion of the Haemophilus influenzae hap protein. Mol. Microbiol. 26, 505–518.
Hendrixson, D. R., and St. Geme, J. W. III. (1998). The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell 2, 841–850.
Ieva, R., Skillman, K. M., and Bernstein, H. D. (2008). Incorporation of a polypeptide segment into the beta-domain pore during the assembly of a bacterial autotransporter. Mol. Microbiol. 67, 188–201.
Ieva, R., Tian, P., Peterson, J. H., and Bernstein, H. D. (2011). Sequential and spatially restricted interactions of assembly factors with an autotransporter {beta} domain. Proc. Natl. Acad. Sci. U.S.A. 108, E383–E391.
Jain, S., and Goldberg, M. B. (2007). Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J. Bacteriol. 189, 5393–5398.
Johnson, T. A., Qiu, J., Plaut, A. G., and Holyoak, T. (2009). Active-site gating regulates substrate selectivity in a chymotrypsin-like serine protease the structure of Haemophilus influenzae immunoglobulin A1 protease. J. Mol. Biol. 389, 559–574.
Junker, M., Besingi, R. N., and Clark, P. L. (2009). Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol. Microbiol. 71, 1323–1332.
Kenjale, R., Meng, G., Fink, D. L., Juehne, T., Ohashi, T., Erickson, H. P., Waksman, G., and St. Geme, J. W. III. (2009). Structural determinants of autoproteolysis of the Haemophilus influenzae Hap autotransporter. Infect. Immun. 77, 4704–4713.
Kett, K., Brandtzaeg, P., Radl, J., and Haaijman, J. J. (1986). Different subclass distribution of IgA-producing cells in human lymphoid organs and various secretory tissues. J. Immunol. 136, 3631–3635.
Ketterer, M. R., Shao, J. Q., Hornick, D. B., Buscher, B., Bandi, V. K., and Apicella, M. A. (1999). Infection of primary human bronchial epithelial cells by Haemophilus influenzae: macropinocytosis as a mechanism of airway epithelial cell entry. Infect. Immun. 67, 4161–4170.
Kilian, M., Mestecky, J., and Russell, M. W. (1988). Defense mechanisms involving Fc-dependent functions of immunoglobulin A and their subversion by bacterial immunoglobulin A proteases. Microbiol. Rev. 52, 296–303.
Kilian, M., Mestecky, J., and Schrohenloher, R. E. (1979). Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect. Immun. 26, 143–149.
Kilian, M., Thomsen, B., Petersen, T. E., and Bleeg, H. (1983). Molecular biology of Haemophilus influenzae IgA1 proteases. Mol. Immunol. 20, 1051–1058.
Kjaergaard, K., Schembri, M. A., Hasman, H., and Klemm, P. (2000a). Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182, 4789–4796.
Kjaergaard, K., Schembri, M. A., Ramos, C., Molin, S., and Klemm, P. (2000b). Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2, 695–702.
Klauser, T., Kramer, J., Otzelberger, K., Pohlner, J., and Meyer, T. F. (1993). Characterization of the Neisseria Iga beta-core. The essential unit for outer membrane targeting and extracellular protein secretion. J. Mol. Biol. 234, 579–593.
Klemm, P., Vejborg, R. M., and Sherlock, O. (2006). Self-associating autotransporters, SAATs: functional and structural similarities. Int. J. Med. Microbiol. 296, 187–195.
Laarmann, S., Cutter, D., Juehne, T., Barenkamp, S. J., and St. Geme, J. W. (2002). The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Mol. Microbiol. 46, 731–743.
Lehr, U., Schutz, M., Oberhettinger, P., Ruiz-Perez, F., Donald, J. W., Palmer, T., Linke, D., Henderson, I. R., and Autenrieth, I. B. (2010). C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA. Mol. Microbiol. 78, 932–946.
Lomholt, H., Van Alphen, L., and Kilian, M. (1993). Antigenic variation of immunoglobulin A1 proteases among sequential isolates of Haemophilus influenzae from healthy children and patients with chronic obstructive pulmonary disease. Infect. Immun. 61, 4575–4581.
Mansa, B., and Kilian, M. (1986). Retained antigen-binding activity of Fab alpha fragments of human monoclonal immunoglobulin A1 (IgA1) cleaved by IgA1 protease. Infect. Immun. 52, 171–174.
Maurer, J., Jose, J., and Meyer, T. F. (1999). Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181, 7014–7020.
Mehta, S. K., Plaut, A. G., Calvanico, N. J., and Tomasi, T. B. Jr. (1973). Human immunoglobulin A: production of an Fc fragment by an enteric microbial proteolytic enzyme. J. Immunol. 111, 1274–1276.
Meng, G., Spahich, N. A., Kenjale, R., Waksman, G., and St. Geme, J. W. III. (2011). Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 30, 3864–3874.
Meng, G., St. Geme, J. W. III, and Waksman, G. (2008). Repetitive architecture of the Haemophilus influenzae Hia trimeric autotransporter. J. Mol. Biol. 384, 824–836.
Meng, G., Surana, N. K., St. Geme, J. W. III, and Waksman, G. (2006). Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 25, 2297–2304.
Mestecky, J., and McGhee, J. R. (1987). Immunoglobulin A (IgA): molecular and cellular interactions involved in IgA biosynthesis and immune response. Adv. Immunol. 40, 153–245.
Moller, L. V., Timens, W., Van Der Bij, W., Kooi, K., De Wever, B., Dankert, J., and Van Alphen, L. (1998). Haemophilus influenzae in lung explants of patients with end-stage pulmonary disease. Am. J. Respir. Crit. Care Med. 157, 950–956.
Mulks, M. H., Kornfeld, S. J., Frangione, B., and Plaut, A. G. (1982). Relationship between the specificity of IgA proteases and serotypes in Haemophilus influenzae. J. Infect. Dis. 146, 266–274.
Mulks, M. H., and Plaut, A. G. (1978). IgA protease production as a characteristic distinguishing pathogenic from harmless neisseriaceae. N. Engl. J. Med. 299, 973–976.
Musser, J. M., Barenkamp, S. J., Granoff, D. M., and Selander, R. K. (1986). Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52, 183–191.
Oomen, C. J., Van Ulsen, P., Van Gelder, P., Feijen, M., Tommassen, J., and Gros, P. (2004). Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23, 1257–1266.
Otto, B. R., Sijbrandi, R., Luirink, J., Oudega, B., Heddle, J. G., Mizutani, K., Park, S. Y., and Tame, J. R. (2005). Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J. Biol. Chem. 280, 17339–17345.
Pallen, M. J., Chaudhuri, R. R., and Henderson, I. R. (2003). Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6, 519–527.
Perona, J. J., and Craik, C. S. (1995). Structural basis of substrate specificity in the serine proteases. Protein Sci. 4, 337–360.
Peterson, J. H., Tian, P., Ieva, R., Dautin, N., and Bernstein, H. D. (2010). Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc. Natl. Acad. Sci. U.S.A. 107, 17739–17744.
Phalipon, A., Cardona, A., Kraehenbuhl, J. P., Edelman, L., Sansonetti, P. J., and Corthesy, B. (2002). Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity 17, 107–115.
Plaut, A. G., Gilbert, J. V., Artenstein, M. S., and Capra, J. D. (1975). Neisseria gonorrhoeae and Neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science 190, 1103–1105.
Plaut, A. G., Gilbert, J. V., and Wistar, R. Jr. (1977). Loss of antibody activity in human immunoglobulin A exposed extracellular immunoglobulin A proteases of Neisseria gonorrhoeae and Streptococcus sanguinis. Infect. Immun. 17, 130–135.
Plaut, A. G., Qiu, J., and St. Geme, J. W. III. (2000). Human lactoferrin proteolytic activity: analysis of the cleaved region in the IgA protease of Haemophilus influenzae. Vaccine 19(Suppl. 1), S148–S152.
Pohlner, J., Halter, R., Beyreuther, K., and Meyer, T. F. (1987). Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325, 458–462.
Poulsen, K., Brandt, J., Hjorth, J. P., Thogersen, H. C., and Kilian, M. (1989). Cloning and sequencing of the immunoglobulin A1 protease gene (iga) of Haemophilus influenzae serotype b. Infect. Immun. 57, 3097–3105.
Radin, J. N., Grass, S. A., Meng, G., Cotter, S. E., Waksman, G., and St. Geme, J. W. III. (2009). Structural basis for the differential binding affinities of the HsfBD1 and HsfBD2 domains in the Haemophilus influenzae Hsf adhesin. J. Bacteriol. 191, 5068–5075.
Roggenkamp, A., Ackermann, N., Jacobi, C. A., Truelzsch, K., Hoffmann, H., and Heesemann, J. (2003). Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185, 3735–3744.
Skillman, K. M., Barnard, T. J., Peterson, J. H., Ghirlando, R., and Bernstein, H. D. (2005). Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol. Microbiol. 58, 945–958.
St. Geme, J. W. III, and Cutter, D. (1995). Evidence that surface fibrils expressed by Haemophilus influenzae type b promote attachment to human epithelial cells. Mol. Microbiol. 15, 77–85.
St. Geme, J. W. III, and Cutter, D. (2000). The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182, 6005–6013.
St. Geme, J. W. III, Cutter, D., and Barenkamp, S. J. (1996). Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 178, 6281–6287.
St. Geme, J. W. III, De La Morena, M. L., and Falkow, S. (1994). A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14, 217–233.
St. Geme, J. W. III, and Falkow, S. (1990). Haemophilus influenzae adheres to and enters cultured human epithelial cells. Infect. Immun. 58, 4036–4044.
Surana, N. K., Cutter, D., Barenkamp, S. J., and St. Geme, J. W. III. (2004). The Haemophilus influenzae Hia autotransporter contains an unusually short trimeric translocator domain. J. Biol. Chem. 279, 14679–14685.
Unzai, S., Park, S. Y., and Tame, J. R. (2005). Crystal structure of heme binding protein, an autotransporter hemoglobin protease from pathogenic Escherichia coli. Tanpakushitsu Kakusan Koso 50, 1322–1327.
Vitovski, S., Dunkin, K. T., Howard, A. J., and Sayers, J. R. (2002). Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA 287, 1699–1705.
Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M., and Tommassen, J. (2003). Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265.
Keywords: Haemophilus influenzae, autotransporter, adhesin, serine protease
Citation: Spahich NA and St. Geme JW III (2011) Structure and function of the Haemophilus influenzae autotransporters. Front. Cell. Inf. Microbio. 1:5. doi: 10.3389/fcimb.2011.00005
Received: 07 July 2011; Accepted: 03 September 2011;
Published online: 28 September 2011.
Edited by:
W. Edward Swords, Wake Forest University Health Sciences, USAReviewed by:
David Hendrixson, University of Texas Southwestern Medical Center, USACopyright: © 2011 Spahich and St. Geme. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Joseph W. St. Geme III, Departments of Pediatrics and Molecular Genetics and Microbiology, Children’s Health Center, Duke University Medical Center, Room T901, DUMC Box 3352, Durham, NC 27710, USA. e-mail:ai5zdGdlbWVAZHVrZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.