- Department of Genetics, Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo, Japan
Caspases are cysteine aspartic acid proteases conserved in animals that not only execute apoptosis, but also regulate diverse cellular processes independent of apoptosis, which are termed caspase-dependent non-lethal cellular processes (CDPs). Owing to its strong genetics to detect and manipulate caspase activity in cells of interest in vivo, Drosophila melanogaster serves as an excellent model organism for analyzing CDPs. This is further supported by the fact that apoptotic signaling, as well as CDPs and their mechanisms, are, in part, conserved in other animals. Here, we present a review to guide researchers studying CDPs using Drosophila. In this review, we provide an overview of the current understanding of apoptotic signaling, which regulates caspase activation in Drosophila as well as available genetic tools and their characteristics for detecting and manipulating caspase activity so that researchers can choose appropriate tools for their own experimental settings. We also introduce the CDPs identified in Drosophila, including a brief description of their discovery and characterization as non-lethal processes. We further describe the underlying molecular mechanisms of several well-characterized CDPs, including the regulatory mechanisms that enable non-lethal caspase activation. Finally, we introduce the use of proximity labeling techniques, especially TurboID, for studying CDPs, which facilitates the analysis of underlying molecular mechanisms. Because caspases regulate various non-lethal cellular functions, their activation is no longer considered a point of no return in cell death. Understanding CDPs will advance our understanding of the states of living and dying cells, along with the intermediate states.
1 Introduction
Caspases are cysteine aspartic acid proteases that are conserved in multicellular organisms, and primarily execute apoptosis by cleaving hundreds of substrates (Newton et al., 2024). For examples, caspases cleave inhibitor of caspase-activated DNase (ICAD) to activate caspase-activated DNase (CAD) for DNA fragmentation during apoptosis (Enari et al., 1998; Sakahira et al., 1998). Caspases also cleave phospholipid scramblase Xkr8 and phospholipid flippases ATP11A/C to externalize phosphatidylserine which acts as an “eat-me” signal of apoptotic cells (Suzuki et al., 2013; Segawa et al., 2014; Segawa et al., 2016). In addition, caspases also regulate a wide range of cellular processes, from cell differentiation to organismal aging, without inducing cell death during development or beyond. In 2017, these processes were termed caspase-dependent non-lethal cellular processes (CDPs) (Aram et al., 2017). New CDPs are continually being discovered in diverse animals, although the underlying mechanisms that enable non-lethal caspase activation without causing cell death or downstream signaling of caspase activation are lagging.
Drosophila melanogaster is a widely used model organism for analyzing diverse biological processes, from development to aging. Caspases and their upstream apoptotic signaling in Drosophila are, in part, conserved among other animals (Kuranaga and Miura, 2007; Nakajima and Kuranaga, 2017). Thus, along with strong Drosophila genetics to detect and manipulate caspase activity in cells of interest in vivo, Drosophila serves as one of the best model organisms not only for describing CDPs but also for characterizing their underlying molecular mechanisms. A wide variety of CDPs have been discovered in Drosophila (Nakajima and Kuranaga, 2017). Notably, some CDPs and their mechanisms are conserved in other animals, further supporting the idea that Drosophila is an excellent model organism for studying CDPs.
In this review, we provide an overview of the current understanding of apoptotic signaling that regulates caspase activation in Drosophila. In the following section, we summarize the currently available genetic tools and their characteristics for detecting and manipulating caspase activity so that researchers can choose appropriate tools for their own experimental settings. We also introduce the currently identified CDPs in Drosophila, including a brief background on their discovery and characterization as non-lethal processes. We further describe the underlying molecular mechanisms of some intensively studied CDPs, including the regulatory mechanisms of non-lethal caspase activation. Finally, we introduce the use of proximity labeling techniques, especially TurboID, to study CDPs as a recently emerging method for analyzing caspase-proximal proteins, which facilitates the analysis of the molecular mechanisms of CDPs. We hope that researchers will find ways to study CDPs after reviewing this review.
2 Caspases and apoptotic signaling in Drosophila
2.1 Caspases and their classification in Drosophila
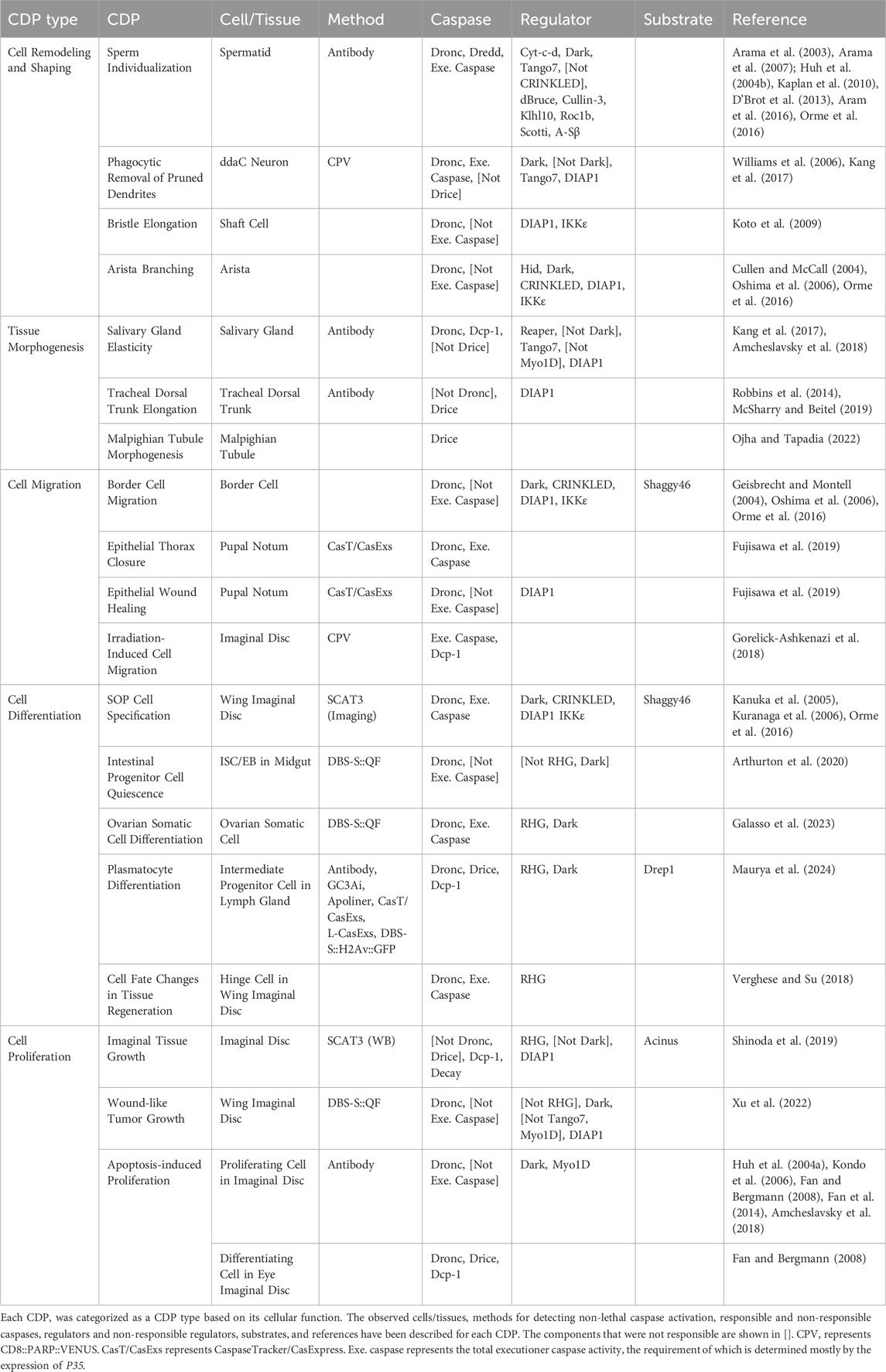
Caspases are cysteine aspartic acid proteases which execute apoptosis in multicellular organisms (Newton et al., 2024). Caspases primarily cleave their substrates at the C-terminal of aspartic acid at P1 position. Preference for P1’ has been observed in smaller amino acids (glycine, serine, alanine) and amino acids with large aromatic rings (phenylalanine, tyrosine) (Timmer and Salvesen, 2007). Structurally, caspase, which is first synthesized as an inactive zymogen, consists of a pro-domain and large and small subunits. For full activation, caspases need to be cleaved by themselves or by other caspases, resulting in a heterotetramer composed of two large and two small subunits (Julien and Wells, 2017) (Figure 1A). In Drosophila, there are seven caspases which are divided into two groups (Kumar and Doumanis, 2000) (Figure 1B): initiator caspases (class I caspases) which have a large pro-domain, including Dronc (Dorstyn et al., 1999a), Strica (Doumanis et al., 2001), and Dredd (Chen et al., 1998), and executioner caspases (class II caspases) which have a short pro-domain, including Drice (Fraser and Evan, 1997), Dcp-1 (Song et al., 1997), Decay (Dorstyn et al., 1999b), and Damm (Harvey et al., 2001). Among the seven caspases, the initiator caspase Dronc (Chew et al., 2004; Daish et al., 2004; Xu et al., 2005) and the executioner caspases Drice and Dcp-1 (Xu et al., 2006; Kondo et al., 2006; Florentin and Arama, 2012) are well described in their requirements for executing apoptosis, where Drice is more effective in inducing apoptosis than Dcp-1 (Florentin and Arama, 2012).
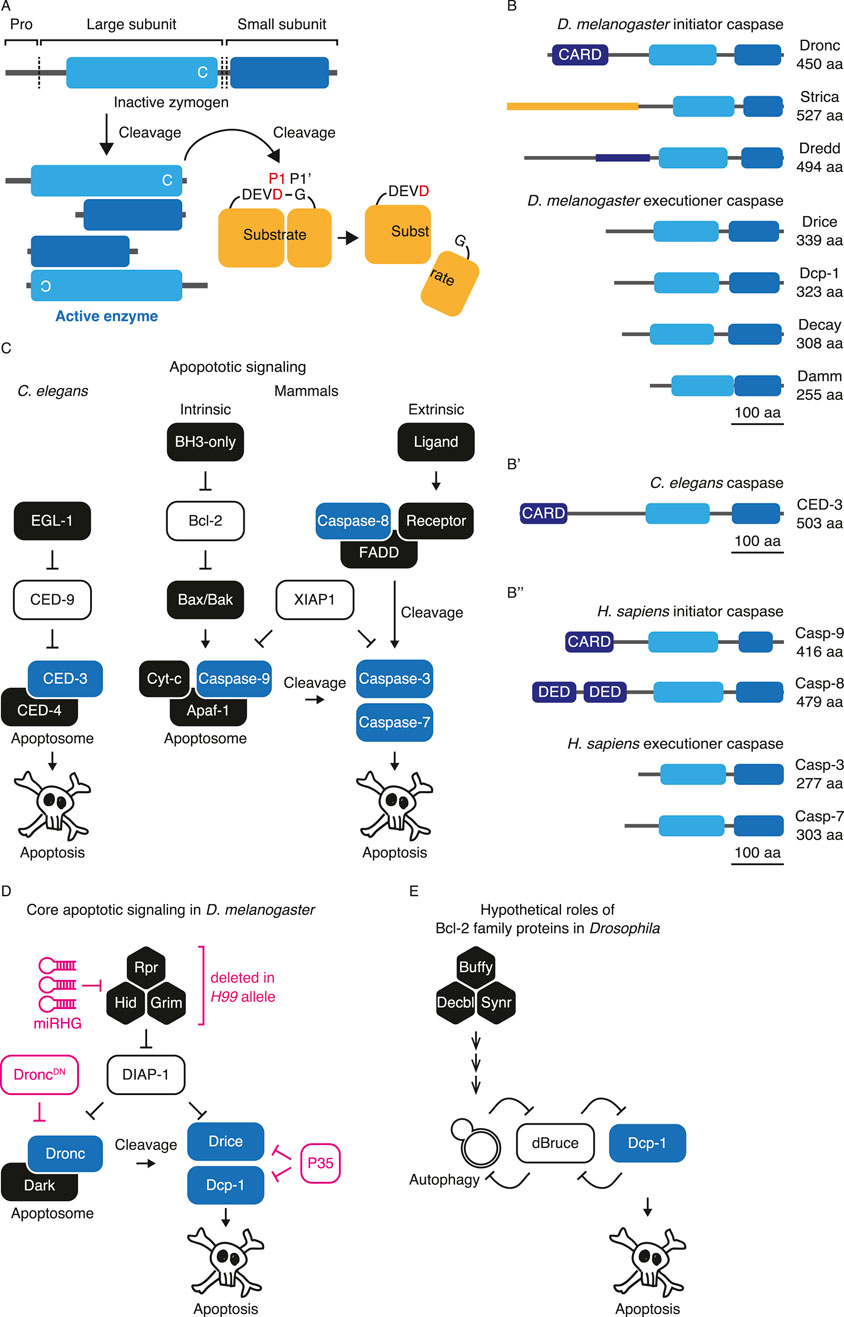
Figure 1. Caspases and apoptotic signaling in Drosophila. (A) Schematic structures and activation machinery of caspase. Inactive zymogen is activated by cleavages (dotted lines) followed by hetero-tetramerization which consists of two large subunits (light blue) and two small subunits (blue). Active enzyme typically cleaves C-terminus of aspartic acid at P1 position of substrates. (B) Schematic structures of seven Drosophila caspases (B), C. elegans CED-3 (B’), and four representative mammalian caspases (B”). Three Drosophila caspases (Dronc, Strica, Dredd) which have long pro-domains are categorized as initiator caspases while four Drosophila caspases (Drice, Dcp-1, Decay, Damm) which have small pro-domains are categorized as executioner caspases. Purple boxes show CARD and DED domains. Light blue shows large subunits and blue shows small subunits. Domain information is from Pfam database [http://pfam.xfam.org/]. Although not in Pfam databse, Strica has serine-threonine rich region indicated by the orange line and Dredd has DID domain indicated by the purple line at their N termini, respectively. (C) Apoptotic signaling in C. elegans and mammals. Caspases are shown in blue and their activators are shown in black while inhibiting components are shown in white. (D) Core apoptotic signaling in Drosophila. Caspases are shown in blue and their activators are shown in black while an inhibiting component is shown in white. Widely used genetic methods for manipulating the signaling are shown in magenta. (E) Hypothetical roles of Bcl-2 family proteins in Drosophila. Caspase is shown in blue and their activators are shown in black while an inhibiting component is shown in white.
While we primarily reviewed the role of non-lethal caspase functions of apoptotic components, it should be mentioned that few caspases are not involved in apoptosis. Dredd (Chen et al., 1998) is not involved in apoptosis but rather regulates innate immune pathways in Drosophila. The Toll (Valanne et al., 2011) and immune deficiency (IMD) pathways (Myllymäki et al., 2014) are the two major innate immune pathways in Drosophila. IMD pathway is activated by infection with gram-negative bacteria and eventually activate Relish, an ortholog of mammalian NF-κB, to induce antimicrobial peptide (AMP) expression. IMD protein binds to the bacterial peptidoglycan (PGN) receptor PGRP-LC, and the adaptor proteins FADD and Dredd. FADD-Dredd interaction is mediated through death-inducing domain (DID) (Hu and Yang, 2000). DIAP2 binds to Dredd and promotes K63 ubiquitination and activation. Activated Dredd cleaves IMD and Relish, and activates the IMD pathway (Meinander et al., 2012; Kleino and Silverman, 2014). C-terminal ankyrin repeat region of Relish is cleaved by Dredd and N-terminal NF-κB portion translocates to the nucleus to induce AMP expression. Dredd is activated by PGN in S2 cells. Dredd is not cleaved for activation and its activity cannot be inhibited by a baculovirus-derived protein, P35 (Kim et al., 2014).
2.2 Core apoptotic signaling in Drosophila
Apoptotic signaling for caspase activation in Drosophila is partially conserved among multicellular organisms (Kuranaga and Miura, 2007; Nakajima and Kuranaga, 2017) (Figures 1C, D). In Caenorhabditis elegans, caspase, CED-3 (Figure 1B’), is activated through the formation of apoptosome complex with CED-4. The apoptosome complex formation is inhibited by CED-9, which is further antagonized by EGL-1 upon apoptotic stimuli (Horowitz and Shaham, 2024) (Figure 1C). In contrast, mammals have two apoptotic signaling pathways called intrinsic and extrinsic apoptotic signaling pathways. Similar to Drosophila caspases, mammalian caspases are also categorized into initiator caspases (Caspase-8 and Caspase-9) and executioner caspases (Caspase-3 and Caspase-7) (Figure 1B”). In both pathways, executioner caspases are activated by the initiator caspase. In intrinsic apoptotic signaling pathway, Caspase-9 is activated through the formation of apoptosome complex with apoptotic peptidase activating factor 1 (Apaf-1), an ortholog of CED-4, and mitochondria-derived cytochrome c, the release of which is mediated by mitochondrial outer membrane permeabilization (MOMP) by Bax and Bak. Bax and Bak are negatively regulated by B-cell lymphoma-2 (Bcl-2), an ortholog of CED-9, which is further antagonized by BH3-only proteins, orthologs of EGL-1, upon apoptotic stimuli. In extrinsic apoptotic signaling pathway, Caspase-8 is activated by extracellular death ligands-stimulated death receptors and its adaptor, FADD. In mammals, caspases are also negatively regulated by inhibitor of apoptosis proteins (IAPs), E3 ubiquitin ligases, including X-linked inhibitor of apoptosis protein (XIAP) (Newton et al., 2024) (Figure 1C).
Core apoptotic signaling in Drosophila is most similar to mammalian intrinsic apoptotic signaling pathway. Upon apoptotic stimuli, Drosophila Apaf-1-related killer (Dark)/homolog of Apaf-1 (Hac-1)/Dapaf-1 (Kanuka et al., 1999; Rodriguez et al., 1999; Zhou et al., 1999) forms apoptosome complex with the initiator caspase, Dronc, for activation via autocatalytic cleavage (Tian et al., 2024). Unlike mammals, apoptosome formation does not require mitochondria-derived cytochrome c (Dorstyn et al., 2002; Dorstyn et al., 2004; Yu et al., 2006). Activated Dronc cleaves the downstream executioner caspases Drice (Hawkins et al., 2000) and Dcp-1 to induce apoptosis. In living cells, caspase activation is negatively regulated by the Drosophila inhibitor of apoptosis protein 1 (DIAP1), an E3 ubiquitin ligase (Hay et al., 1995). DIAP1 ubiquitinates Dronc (Wilson et al., 2002), Drice, and Dcp-1 to inactivate them in a nondegradative (independent of proteasomal degradation) manner (Ditzel et al., 2008). Upon apoptotic stimuli, the pro-apoptotic IAP antagonists reaper (Rpr), head involution defective (Hid), and Grim, together called RHG proteins, the genes of which are deleted in H99 allele (White et al., 1994), are synthesized. RHG proteins bind to DIAP1 to facilitate DIAP1 degradation through auto-ubiquitination (Ryoo et al., 2002). Developmental and stress-induced cell death is mostly suppressed in mutants of RHG (White et al., 1994), Dronc (Chew et al., 2004; Daish et al., 2004; Xu et al., 2005) and double mutants of Drice and Dcp-1 (Kondo et al., 2006; Xu et al., 2006; Florentin and Arama, 2012), which are regarded as components of the core apoptotic signaling pathway in Drosophila (Figure 1D).
2.3 Bcl-2 family proteins in Drosophila
In addition to the well-characterized core apoptotic signaling pathway, Drosophila has two Bcl-2-like proteins, Death executioner Bcl-2 (Debcl)/Drob-1 (Colussi et al., 2000; Igaki et al., 2000) and Buffy (Quinn et al., 2003). In mammals, the Bcl-2 family proteins regulate apoptosis through the regulation of MOMP mediated by the pore-forming proteins, Bax and Bak (Figure 1C) (Singh et al., 2019). Since apoptosome formation in Drosophila does not require mitochondria-derived cytochrome c, the actions of Debcl/Drob-1 and Buffy seem to differ from those in mammals (Igaki and Miura, 2004). Recently, a BH3-only-like protein, Sayonara (Synr), whose overexpression induces caspase activation through autophagy, was described (Ikegawa et al., 2023). Synr-induced caspase activation is antagonized by Debcl/Drob-1, Buffy or Dcp-1 knockdown. In addition, Debcl/Drob-1, Buffy, and Dcp-1 are all required for starvation-induced autophagy in Drosophila cell lines (Hou et al., 2008). Thus, it seems likely that the Bcl-2 family proteins in Drosophila regulate apoptosis through autophagy and the executioner caspase Dcp-1. Since autophagy degrades another giant IAP, dBruce, for caspase activation in nurse cells during oogenesis (Nezis et al., 2010), it is possible that autophagy activates caspase via dBruce degradation (Figure 1E). Importantly, both Debcl/Drob-1, Buffy double mutant (Sevrioukov et al., 2007) and Synr mutant (Ikegawa et al., 2023) are developmentally viable, suggesting that Bcl-2 family proteins in Drosophila act not as core apoptotic machinery, but rather only in a specific biological context such as starvation.
3 Detecting and manipulating caspase activation in Drosophila
3.1 Genetic manipulations of core apoptotic signaling in Drosophila
Using Drosophila strong genetics, we can detect and manipulate caspase activation in cells of interest to analyze functions and mechanisms of caspase activation in vivo. In addition to the mutant alleles and the Gal4/UAS system (Brand and Perrimon, 1993) -mediated knockdown of each apoptotic component, several other manipulations have been well established (Figure 1D). One such manipulation is the expression of the baculovirus-derived protein P35 which inhibits whole executioner caspase activation (Hay et al., 1994), however not initiator caspase activation (Meier et al., 2000). P35 expression suppresses both Drice and Dcp-1, thereby avoiding the redundancy of each caspase to execute apoptosis. To inhibit all executioner caspase activities, combined RNAi against Drice, Dcp-1, Decay, and Damm is sometimes used to completely deplete their activity (Arthurton et al., 2020). The expression of Dronc dominant-negative (DroncDN), which harbors a mutation in the catalytic cysteine, is frequently used to suppress apoptosis (Meier et al., 2000). Furthermore, inhibition of upstream RHG genes by simultaneous knockdown using microRNAs (miRHG) strongly suppresses caspase activation (Siegrist et al., 2010). Genetic manipulations of caspase activity in vivo not only identify the role of apoptotic cells during development and homeostasis but also lead to the identification of numerous non-lethal caspase functions (see Section 4).
3.2 Antibody staining for detecting cleaved (activated) executioner caspases
One of the strong advantages of Drosophila in studying caspase function is the variety of biochemical and genetic tools for detecting caspase activation. Because caspase activation relies on its own cleavage, anti-cleaved caspase antibodies, including anti-cleaved (activated) Caspase-3 (Asp175) antibody (#9661, Cell Signaling Technology), anti-cleaved (activated) Dcp-1 (Asp215) antibody (#9578, Cell Signaling Technology) and anti-active Drice antibody (Dorstyn et al., 2002), are frequently used to detect activated caspases and are thus sometimes used as markers for apoptosis. However, when using these antibodies, researchers must be careful while interpreting of the results. Because anti-cleaved Caspase-3 (Asp175) antibody immunoreactivity persists in Dcp-1 and Drice (Drosophila orthologs of Caspase-3) double mutants and immunoreactivity is lost in both Dronc and Dark (Drosophila orthologs of Caspase-9 and Apaf-1, respectively) mutants, this antibody is considered a marker of Dronc activity in Drosophila (Fan and Bergmann, 2010). Anti-cleaved Dcp-1 (Asp215) antibody immunoreactivity is lost in Drice and Dcp-1 double mutants, but not in Dcp-1 mutant, and thus serves as a specific marker for activated executioner caspases of Drice and Dcp-1 however is not specific to activated Dcp-1 in Drosophila (Li et al., 2019). Because immunohistochemistry using antibodies preserves the spatial information of caspase activation, sometimes even inside the cell, it is useful not only for detecting apoptotic cells within tissues, but also for examining the spatial patterns of caspase activation in non-lethal scenarios (Kang et al., 2017; Amcheslavsky et al., 2018; McSharry and Beitel, 2019). However, since antibody staining only captures a snapshot of caspase activation at a single point in time, it is unclear whether cells ultimately die. Thus, live cell monitoring of caspase activity using genetically-encoded caspase activity probes and reporters helps trace the fate of cells to characterize the processes as CDPs.
3.3 Genetically-encoded caspase activity probes and reporters
A more direct method for measuring caspase activity is the use of caspase activity probes and reporters. Since caspases recognize specific amino acid sequences with a high preference for aspartic acid at the P1 position (canonically, DEVD for executioner caspases) (Talanian et al., 1997), probes and reporters utilizing these properties are often developed in Drosophila. These include sensor for activated caspase-3 based on FRET (SCAT3) (Takemoto et al., 2003; Takemoto et al., 2007), VENUS-based variant of caspase-3-like proteases activity indicator (VC3Ai) (Zhang et al., 2013; Schott et al., 2017), infrared fluorescent executioner-caspase reporter (iCasper) (To et al., 2015), CD8::PARP::VENUS (Williams et al., 2006), Apoliner (Bardet et al., 2008), SR4VH (Pop et al., 2020), CaspaseTracker/CasExpress (Tang et al., 2015; Ding et al., 2016) and their variants (Baena-Lopez et al., 2018; Sun et al., 2020; Muramoto et al., 2024). As these probes are based on different rationales and have distinct characteristics, each probe has its own advantages and disadvantages in detecting caspase activity.
3.3.1 A FRET-based probe for detecting caspase activity
SCAT3 is a FRET-based caspase activity probe that detects caspase activation with high temporal resolution during live cell imaging (Figure 2A). SCAT3 is composed of ECFP and VENUS linked by 18-amino acids including the caspase recognition sequence DEVD. Caspase activity was detected by a reduction in the VENUS/ECFP FRET ratio due to the cleavage-mediated release of VENUS (Takemoto et al., 2003). SCAT3 is useful for detecting caspase activity in live cell/tissue imaging, such as in Drosophila pupal salivary gland, abdomen, male genitalia, and notum, with high temporal resolution (Takemoto et al., 2007; Koto et al., 2011; Kuranaga et al., 2011; Nakajima et al., 2011; Levayer et al., 2016; Fujisawa et al., 2019; Fujisawa et al., 2020). A nuclear-localized signal-tagged variant of SCAT3 (NLS::SCAT3) is also available (Takemoto et al., 2003; Koto et al., 2009), and is useful for the identification of individual cells in complex tissues. The drawback of SCAT3 is that FRET imaging requires an ECFP-exciting laser such as 448 nm diode laser, and post-acquisition image processing to calculate the FRET ratio, which needs to be prepared before experiments by researchers.
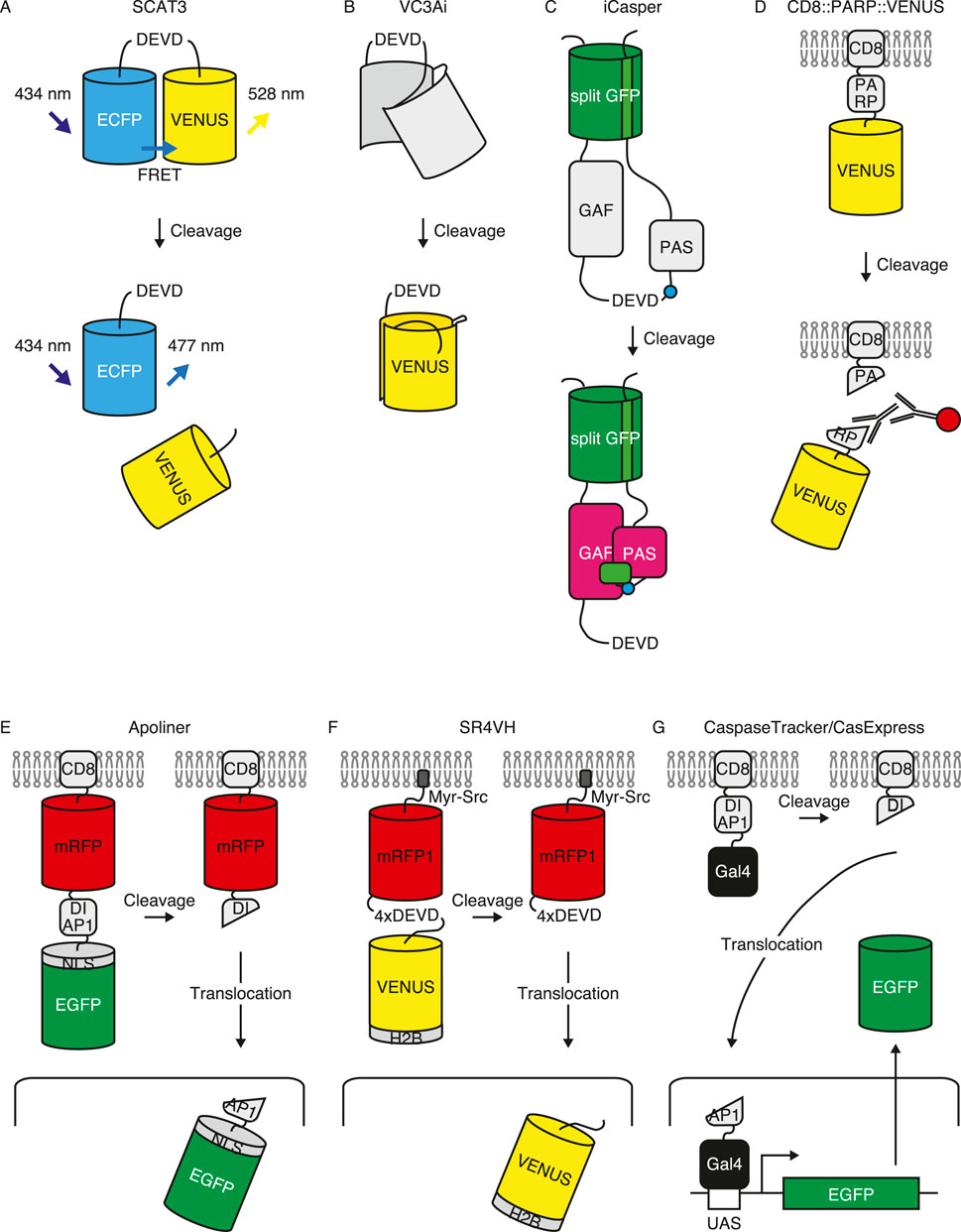
Figure 2. Genetically-encoded probes for detecting caspase activity in Drosophila. (A) Schematic structures of FRET-based probe, SCAT3. Caspase-mediated cleavage diminishes FRET between ECFP and VENUS. Caspase activity is measured by the reduction in VENUS/ECFP FRET ratio. The peak wavelengths for excitation/emission of fluorescent proteins are shown. (B) Schematic structures of fluorescent switch-on probe, VC3Ai. Caspase-mediated cleavage makes the probe fluorescent. Caspase activity is measured by the intensity of the probe. (C) Schematic structures of fluorescent switch-on probe, iCasper, which is tagged with a split GFP at new N′- and C′-terminal. A blue circle shows catalytic cysteine, while a green box shows biliverdin. Caspase-mediated cleavage makes the probe fluorescent. Caspase activity is measured by the intensity of the probe. Cells expressing iCasper are detected by split-GFP fluorescence. (D) Schematic structures of CD8::PARP::VENUS. Caspase-mediated cleavage generates neo-epitope of anti-cleaved PARP antibody. Caspase activity is measured by the intensity of the immunohistochemistry. (E) Schematic structures of fluorescent translocation probe, Apoliner. Caspase-mediated cleavage releases NLS::EGFP translocating to the nucleus. Caspase activity is measured by the intensity of NLS::EGFP in the nucleus. (F) Schematic structures of fluorescent translocation probe, SR4VH. Caspase-mediated cleavage releases VENUS::H2B translocating to the nucleus. Caspase activity is measured by the intensity of VENUS::H2B in the nucleus. (G) Schematic structures of transcription factors activating probes, CaspaseTracker/CasExpress. Caspase-mediated cleavage releases Gal4 translocating to the nucleus to drive fluorescent protein expressions (e.g., EGFP) downstream of the UAS sequence. Caspase activity is measured by the intensity of the synthesized EGFP or other fluorescent proteins.
3.3.2 Fluorescent switch-on probes for detecting caspase activity
VC3Ai is a circularly permuted VENUS (cpVENUS) caspase activity probe that detects caspase activation by switching on VENUS fluorescence (Figure 2B). VC3Ai is composed of cpVENUS with its N- and C-terminal fused with caspase recognition sequence, DEVD, with a split Npu DnaE intein at new N′- and C′-terminal to enhance the efficiency of complementation upon cleavage. VC3Ai was originally synthesized as a non-fluorescent indicator that becomes fluorescent upon cleavage. In addition to the VENUS fluorescent protein, multi-fluorescent color variants of CC3Ai based on Cerulean, GC3Ai based on superfolder GFP, and RC3Ai based on mCherry are available (Zhang et al., 2013). VC3Ai is useful in detecting caspase activity with a standard fluorescent microscope system in live and fixed cell/tissue such as Drosophila imaginal discs (Schott et al., 2017; Matamoro-Vidal et al., 2024) and pupal notum (Fujisawa et al., 2019; Fujisawa et al., 2020; Villars et al., 2022). VC3Ai is good for detecting apoptosis mediated by strong caspase activation. However, it may lack the sensitivity to detect non-lethal caspase activation (Schott et al., 2017; Fujisawa et al., 2019).
iCasper is a cpIFP-based caspase activity probe that detects caspase activation using switched-on IFP fluorescence (Figure 2C). iCasper is composed of cpIFP with its N- and C-terminal fused with the caspase recognition sequence, DEVD, with a split GFP at the new N′- and C′-terminal to ensure that following caspase-mediated cleavage, the cleaved probes remain intact. iCasper was originally synthesized as a non-IFP fluorescent indicator that becomes fluorescent upon cleavage, with split GFP fluorescence. Since IFP requires biliverdin at its chromophore, iCasper also requires it to be fluorescent (To et al., 2015). iCasper is useful for detecting caspase activity, at least in apoptotic cells in live and fixed cells/tissues, such as embryos, larval brains (To et al., 2015), and pupal nota (Canales Coutiño et al., 2020). Because the number of reports using iCasper is still limited, its characteristics, including sensitivity compared to other probes as well as the potential of this probe to be applicable for analyzing non-lethal caspase activation, should be described in the future.
3.3.3 Antibody that recognizes cleaved neoepitope of the probe for detecting caspase activity
CD8::PARP::VENUS is a probe that was developed to detect localized caspase activity in neurons. CD8::PARP::VENUS is composed of the extracellular and transmembrane domains of mouse CD8 fused to 40 amino acids from human PARP containing a caspase cleavage site and VENUS. Caspase activity was detected by immunohistochemistry using an anti-cleaved PARP antibody that recognizes cleaved fragments of the probe (Williams et al., 2006) (Figure 2D). Probe overexpression enhances the sensitivity of the immunoreaction to antibodies in the respective cells. CD8::PARP::VENUS is useful not only for detecting caspase activation in neurons (Williams et al., 2006; Chihara et al., 2014), but also in other tissues, including wing imaginal discs (Florentin and Arama, 2012; Shinoda et al., 2016; Shinoda et al., 2019; Gorelick-Ashkenazi et al., 2018). It is also used to detect local caspase activation in the pruned dendrites of neurons (Williams et al., 2006). However, because CD8::PARP::VENUS requires immunohistochemistry, it is not applicable to live cell/tissue imaging.
3.3.4 Fluorescent protein translocation probes for detecting caspase activity
Apoliner is an mRFP- and EGFP-based probe that detects caspase activation via translocation of EGFP into the nucleus (Figure 2E). Apoliner is composed of the transmembrane domain of mouse CD8 fused with mRFP, a caspase cleavage site from DIAP1, and EGFP with a nuclear localization signal of the large T antigen (NLS::EGFP). Apoliner resides on the membrane. Following caspase activation, NLS::EGFP is released via caspase-mediated cleavage and accumulates in the nucleus (Bardet et al., 2008). Apoliner is useful for detecting caspase activity in live and fixed cells of embryo (Bardet et al., 2008; Kolahgar et al., 2011).
SR4VH is an RFP- and VENUS-based probe that detects caspase activation with the translocation of VENUS into the nucleus, the rationale for which is similar to that of Apoliner. SR4VH is composed of a membrane-bound RFP (Src::RFP) fused with four tandem repeats of the amino acid sequences DEVD and VENUS tagged with Histone H2B (VENUS::H2B) (Figure 2F). Following caspase activation, VENUS::H2B is released by caspase-mediated cleavage, translocated, and accumulated in the nucleus. While the design was similar to that of Apoliner, the authors discussed that the myristoylation signal from Src differentiates its subcellular localization from that of Apoliner. They also reported that the nuclear localization signal from H2B allow for efficient sequestration, even in the late stages of apoptosis (Pop et al., 2020). SR4VH is useful for detecting caspase activity in live and fixed cells/tissues, such as larval brains (Pop et al., 2020), wing imaginal discs, and dendritic arborization neurons (Mukherjee et al., 2021). Both Apoliner and SR4VH have sufficient sensitivity to detect apoptosis. However, whether these probes can be used to analyze non-lethal caspase activation needs to be further tested.
3.3.5 Transcription factors activating probes for detecting caspase activity
CaspaseTracker/CasExpress is a caspase-activated transcription factor-based reporter that drives the expression of downstream fluorescent proteins for visualization. The caspase-activatable transcription factor is composed of a transmembrane protein of mouse CD8 fused with a caspase-binding and cleavage domain from DIAP1 and the transcription factor Gal4. In the systems, caspase activatable-transcription factor is ubiquitously expressed using ubiquitin promoter. Following caspase activation, the cell membrane-sequestered Gal4 is released by caspase-mediated cleavage and Gal4 accumulates in the nucleus to drive gene expression downstream of UAS sequence (Tang et al., 2015; Ding et al., 2016) (Figure 2G). Because of its Gal4/UAS-mediated binary gene expression system, CaspaseTracker/CasExpress is highly useful for detecting both lethal and non-lethal caspase activation throughout the body with high sensitivity. Moreover, CaspaseTracker/CasExpress is more sensitive than VC3Ai and SCAT3 (Schott et al., 2017; Fujisawa et al., 2019). It may also be more sensitive than all other probes. By combining the probe with the lineage-tracing system G-TRACE, the authors identified a wide range of developmentally caspase-activated but surviving cells at the adult stage, suggesting diverse non-lethal caspase functions in Drosophila (Tang et al., 2015; Ding et al., 2016). However, because CaspaseTracker/CasExpress relies on the Gal4/UAS-mediated binary gene expression system, the temporal resolution should not be as high as that of SCAT3 and other probes. In addition, the reliance on Gal4 to report caspase activation in CaspaseTracker/CasExpress presents a challenge in reconciling Gal4/UAS-dependent gene manipulations, such as RNAi or overexpression. Thus, L-CasExpress, in which Gal4 is replaced with LexA, has been developed (Sun et al., 2020). Furthermore, to restrict the expression of the probe within the cells of interest, a Gal4-Manipulated Area Specific CaspaseTracker/CasExpress (MASCaT) has been developed, where Gal4 was replaced by QF2 and probe expression was regulated by FLP/FRT-mediated excision of the STOP cassette downstream of Gal4/UAS expression system (Muramoto et al., 2024). Both systems further improve the usability of CaspaseTracker/CasExpress. Similar to CaspaseTracker/CasExpress, which detects executioner caspase activity, a Drice-based sensor (DBS), specifically CD8::DriceC211A-short (DBS-S), was developed to detect the activity of initiator caspase, Dronc. DBS-S is composed of a transmembrane domain of CD8 fused with an enzymatically inactive Drice, DriceC211A, which retains only 16 amino acids downstream of the Dronc cleavage site and Histone2Av::GFP or transcription factor QF. Following caspase activation, cell membrane sequestered Histone2Av::GFP or QF is released by caspase-mediated cleavage and accumulates in the nucleus (Baena-Lopez et al., 2018). DBS-S::Histone2Av::GFP is useful for detecting Dronc activity in live and fixed cells or tissues, such as wing imaginal discs (Baena-Lopez et al., 2018). In combination with the QF/QUAS-mediated binary gene expression system, DBS-S::QF is useful for detecting caspase activation throughout the body with high sensitivity. DBS-S::QF is useful for a variety of tissues, including imaginal discs (Baena-Lopez et al., 2018; Xu et al., 2022), midguts (Baena-Lopez et al., 2018; Arthurton et al., 2020), and ovaries (Galasso et al., 2023). CaspaseTracker/CasExpress and their variants have the best sensitivity for detecting caspase activation. Thus, they should be listed as the first-choice reporters for analyzing CDPs.
3.4 Monitoring DIAP1 protein status
In addition to directly detecting caspase activity, there are several ways to monitor the upstream components of apoptotic signaling. Increased or decreased DIAP1 expression is crucial for caspase activation and function. DIAP1 is an E3 ubiquitin ligase that is ubiquitinated and degraded by the proteasome when RHG expression is induced by apoptotic stimulation. A pre-apoptosis signal-detecting probe based on diap1 degradation (PRAP) was developed to monitor DIAP1 status in vivo (Koto et al., 2009). In this probe, VENUS is linked to mutant forms of DIAP1 that are unable to inhibit caspases because of mutations in the caspase-binding sites of the baculovirus IAP repeat (BIR) domains. VENUS fluorescence intensity monitors DIAP1 status. In larval epidermal cells, during metamorphosis, VENUS expression is rapidly reduced before nuclear condensation during apoptosis (Koto et al., 2009). PRAP is also useful for detecting DIAP1 status in live cell/tissue imaging, such as Drosophila pupal notum (Koto et al., 2009). Although direct examination of caspase activity is necessary for subsequent analyses because DIAP1 protein status does not always correlate with caspase activity, it is informative for analyzing the upstream signaling that regulates caspase activation.
3.5 Monitoring RHG genes status
DIAP1 protein levels are regulated by RHG proteins whose expression is transcriptionally induced by apoptotic stimulation. Thus, monitoring RHG genes expression using enhancer trap lines is useful for indirect detection of caspase activation patterns. Frequently used reporters include Rpr-11-LacZ (Nordstrom et al., 1996), hid05014-LacZ (Grether et al., 1995), and hid20-10-LacZ (Fan et al., 2010). In addition to each single RHG gene reporter, the irradiation-responsive enhancer region (IRER) mediates the expression of surrounding pro-apoptotic genes, including Rpr. The epigenetic status of the IRER correlates pro-apoptotic gene expression propensity (Zhang et al., 2008). Thus, IRER {ubi-DsRed} reporter, where DsRed expression is driven by the 2.2 kb upstream regulatory sequence of the ubiquitin-63E gene, is inserted into IRER, and can be used for monitoring pro-apoptotic genes expression propensity (Zhang et al., 2015). Similar to DIAP1, although a direct examination of caspase activity is necessary for subsequent analysis, it is informative for analyzing the upstream signaling that regulates caspase activation.
3.6 Summary
A wide variety of methods for detecting caspase activation and its upstream status in vivo have not only uncovered when and where cells undergo apoptosis within tissues, but also identified cells undergoing caspase activation without cell death. This led to the identification of diverse non-lethal caspase functions in Drosophila (see Section 4).
4 Caspase-dependent non-lethal cellular processes in Drosophila
In addition to their roles in apoptosis, caspases exhibit diverse non-lethal cellular functions in many organisms (Kuranaga and Miura, 2007; Nakajima and Kuranaga, 2017). CDPs identified in Drosophila include cell remodeling and shaping (Arama et al., 2003; Huh et al., 2004b; Oshima et al., 2006; Williams et al., 2006; Koto et al., 2009), tissue morphogenesis (Kang et al., 2017; McSharry and Beitel, 2019; Ojha and Tapadia, 2022), cell migration (Geisbrecht and Montell, 2004; Gorelick-Ashkenazi et al., 2018; Fujisawa et al., 2019), cell differentiation (Kanuka et al., 2005; Verghese and Su, 2018; Arthurton et al., 2020; Galasso et al., 2023; Maurya et al., 2024), cell proliferation (Huh et al., 2004a; Pérez-Garijo et al., 2004; Ryoo et al., 2004; Kondo et al., 2006; Shinoda et al., 2019; Xu et al., 2022) (Table 1). A single caspase family regulates various cellular processes. The regulatory mechanisms of CDPs are not unique, but rather diversified for different outcomes. In this section, we introduce the CDPs identified in Drosophila with a background of their discovery and characterization as non-lethal processes. We also introduce the functions and mechanisms of the CDPs to understand the diversity of non-lethal caspase functions. As observed with respect to several CDPs, we introduce similar examples observed in animals apart from Drosophila, to understand that CDPs are not limited to Drosophila.
4.1 Cell remodeling and shaping
4.1.1 Sperm individualization
One representative CDP in Drosophila is sperm individualization (Arama et al., 2003; Huh et al., 2004b). Spermatogenesis occurs within individual units known as cysts, which contain 64 spermatids that remain connected via cytoplasmic bridges after meiosis. During terminal differentiation, round spermatids transform into thin spermatozoa. In the final stage of spermatogenesis, termed individualization, the cytoplasmic bridges are disconnected, and most of the cytoplasm is expelled, leading to individual sperms, events that are reminiscent of apoptosis. Using anti-active mouse caspase-3 antibody (also known as CM1 (IDUN Pharmaceuticals) which is no longer available (Fan and Bergmann, 2010)) and anti-active Drice antibody (Dorstyn et al., 2002), the executioner caspase was found to be activated at the onset of individualization. In addition, they showed that the inhibition of caspase activity by zVAD-fmk, a broad-spectrum caspase inhibitor, or P35 expression results in defects in individualization, suggesting that caspase activity non-lethally regulates sperm individualization. Furthermore, the authors showed that a specific cytochrome c, cyt-c-d, is required for caspase activation, and that dBruce is required to protect spermatids during individualization (Arama et al., 2003). The molecular mechanisms that enable caspase activation without causing cell death are intensively studied by Eli Arama and others, which is described in the following section (see Section 5).
4.1.2 Phagocytic removal of pruned dendrites
Another CDP for cell remodeling and shaping is the phagocytic removal of pruned dendrites from neurons (Williams et al., 2006). During metamorphosis, the dendritic arborizing (da) sensory neuron ddaC undergoes dendrite pruning while the soma remains intact. The authors first showed that inhibition of the apoptotic machinery using DroncDN, P35, and DIAP1 expression, as well as using Dronc and Dark mutant alleles, suppressed the removal of pruned dendrites however did not suppress dendrite severing by itself. Subsequently, using the CD8::PARP::VENUS probe, they showed that caspase activation was locally observed in severed branches, but not in branches attached to the soma (Williams et al., 2006). These results suggest that non-lethal local caspase activation at pruned dendrites facilitates ddaC neuronal remodeling through phagocytic removal of dendrites.
4.1.3 Bristle elongation
CDPs also participate in cellular shaping during bristle-length regulation (Koto et al., 2009). During Drosophila sensory organ development, the socket and shaft cells are generated by asymmetric division from the same precursor cell, pIIa. Live imaging analysis using PRAP, an indicator of DIAP1 status, revealed that DIAP1 levels decreased during asymmetric division and then increased in both socket and shaft cells. While high DIAP1 level is maintained in the socket cell, DIAP1 is degraded by IKKε, which promotes DIAP1 degradation through phosphorylation (Kuranaga et al., 2006) during elongation in the shaft cell. IKKε knockdown reduces the elongation, which is further suppressed by simultaneous expression of DroncDN, however not by P35, suggesting that non-lethal Dronc activation promotes shaft cell elongation under IKKε knockdown, thus regulating bristle length (Koto et al., 2009). These results suggest that the dynamics of DIAP1 protein status control the timing of non-apoptotic caspase activation and function.
4.1.4 Arista branching
DIAP1, IKKε, and Dronc also regulate cellular morphogenesis in branching of arista, a feather-like structure at the tip of the antenna (Cullen and McCall, 2004; Oshima et al., 2006). It was first reported that arista branching reduced in DIAP1 mutant, but increased in Hid mutant (Cullen and McCall, 2004). The following study shows that IKKε negatively regulates F-actin assembly. Moreover, when IKKε is suppressed, arista showed increased lateral branching. Apoptotic component including Dronc, Dark, and DIAP1 genetically interact with the phenotype under the inhibition of IKKε activity. Specifically, reducing Dronc activity, but not inhibiting executioner caspase activity by P35 overexpression, resulted in a further increase in lateral branching (Oshima et al., 2006). These results suggest that caspase, specifically Dronc, non-lethally suppressed ectopic branching of arista under IKKε knockdown.
4.1.5 CDPs for cell remodeling and shaping in other animals
Caspase also regulates cell remodeling and shaping in other animals. During rat lens fiber differentiation, the authors first found that differentiating lens fibers were TUNEL-positive and PARP was cleaved for denucleation, suggesting that caspase was activated. In addition, they found that zVAD-fmk, a broad-spectrum caspase inhibitor, blocked the denucleation process during lens fiber differentiation in cultured explants, suggesting that caspase non-lethally regulates the denucleation process in mammals (Ishizaki et al., 1998). During retinal ganglion cell axon pathfinding in zebrafish embryos, caspases are locally activated at the branch tip of dynamic arbors, which can be detected using SCAT3. Genetic inhibition of caspase using an antisense morpholino results in a reduction in axon arbor dynamics, suggesting that caspase non-lethally promote axon arbor dynamics during development (Campbell and Okamoto, 2013). As shown, CDPs for cell remodeling and shaping are not specific to Drosophila but are rather conserved among animals.
4.2 Tissue morphogenesis
4.2.1 Salivary gland elasticity
As caspases can shape cell morphology, it is not surprising that caspases can also regulates tissue morphogenesis. In the larval salivary glands, caspases regulate tissue elasticity (Kang et al., 2017). Just before pupation, when no cell death occurs in the salivary gland, a weak pulse of ecdysone induces weak pro-apoptotic Rpr expression in the larval salivary gland. This leads to the activation of caspase, specifically in the cell cortex, which is detected by the anti-cleaved Dcp-1 antibody. Using Dronc and Dcp-1 mutants, the authors showed that caspase activation in the cortex is mediated by Dronc and Dcp-1. Moreover, using Dronc and Dcp-1 mutants, both caspases were required for the dismantling of cortical F-actin. However, Drice was not required for these processes. The dismantling of cortical F-actin was also suppressed by Rpr mutant and knockdown, Dronc knockdown, and Diap1 and P35 overexpression. Cortical caspase activation requires Tango7, which physically interacts with Dronc (D’Brot et al., 2013), but does not require Dark, suggesting that Tango7 regulates cortical caspase activation. Furthermore, using Dcp-1 mutant, the authors showed that caspase activation is required for luminal expansion of the tissue, which allows the secretion of mucin-like glue protein into the lumen (Kang et al., 2017). These results suggest that caspase activity in the cortex regulates tissue morphogenesis by regulating tissue elasticity.
4.2.2 Tracheal dorsal trunk elongation
Caspases also regulate the tracheal dorsal trunk elongation (Robbins et al., 2014; McSharry and Beitel, 2019). The tracheal system delivers oxygen to the tissues through a network of epithelial tubes. The tracheal dorsal trunk tubes are elongated by changing the cell shape and junctions without changing the cell number during the late stages of embryonic development. The authors first showed that Drice mutant, as well as P35 and DIAP1 overexpression, however not Dronc mutant, reduced tracheal dorsal trunk length. The overall tracheal cell number was not influenced by these conditions, suggesting that caspases, especially Drice, non-lethally elongate the tracheal dorsal trunk, potentially downstream of the Hippo signaling pathway (Robbins et al., 2014). In a subsequent study, this was further supported by the fact that Drice overexpression in the tracheal system elongated the dorsal tank without inducing apoptosis, suggesting that Drice promotes elongation non-lethally. Using an anti-cleaved Drice antibody, they showed that numerous cleaved Drice puncta were enriched at the apical surface of the tracheal cells, which partially colocalize with Clathrin. Furthermore, the abundance and trafficking of several cargo proteins required for tracheal size control are altered in Drice mutant (McSharry and Beitel, 2019). These results suggest that caspases, particularly Drice, non-lethally regulate tracheal dorsal trunk elongation by regulating endocytic trafficking.
4.2.3 Malpighian tubule morphogenesis
In addition, Drice regulates tissue morphogenesis in malpighian tubules (Ojha and Tapadia, 2022). Malpighian tubule, a tissue responsible for excretion and osmoregulation, shows no apoptosis at the larval and pupal stages of development, although it does express apoptotic proteins (Shukla and Tapadia, 2011). The authors showed that Drice mutant exhibited deformed malpighian tubule morphology with reduced tubule length and a disorganized cytoskeleton in the third-instar larvae, suggesting that Drice non-lethally regulates malpighian tubule morphogenesis. This mutant also showed deposition of uric acid crystals suggesting the functional breakdown of malpighian tubule (Ojha and Tapadia, 2022). Since salivary glands, tracheal system, and malpighian tubules are all tubular tissues, caspases may regulate tubular tissue morphogenesis in other animals.
4.3 Cell migration
4.3.1 Border cell migration
As caspases regulate the actin cytoskeleton, caspases can also regulate cell migration in a non-lethal manner. During oogenesis in the Drosophila egg chamber which consists of 15 nurse cells and one oocyte, 6–10 cells emerge from the surrounding follicle cells to form border cell clusters. The cluster migrates through the center of the egg chamber of the nurse cells to the anterior border of the oocyte. The migration process, called border cell migration, starts during stage 9 and is completed by stage 10 (Montell, 2003). Migration is inhibited when dominant-negative form of a small GTPase, Rac (RacN17), is ectopically expressed in the border cells. Through genetic screening, the authors found that DIAP1 overexpression rescued the migration defects caused by RacN17 overexpression. Conversely, DIAP1 hypomorphic mutant shows border cell migration defects without excess cell death. Since IKKε overexpression causes border cell migration defects, this also supports the involvement of DIAP1 for the regulation of border cell migration (Oshima et al., 2006). Furthermore, the authors showed that inhibiting Dronc activity, however not executioner caspase activity by P35 overexpression, also rescued the migration defect mediated by RacN17 overexpression. Thus, caspases, specifically Dronc, suppresses border cell migration (Geisbrecht and Montell, 2004).
4.3.2 Epithelial thorax closure and wound healing
Caspases also regulate cell migration during epithelial thorax closure and wound healing (Fujisawa et al., 2019). During thorax closure in the pupal stage, where a pair of wing imaginal disc joints form the adult thorax, caspase activation is specifically observed in the leading-edge cells using CaspaseTracker/CasExpress, mostly not undergoing delamination, as revealed by live imaging. Inhibition of Dronc or executioner caspase activity results in an increase in the speed of thorax closure, suggesting that caspases non-lethally attenuate thorax closing speeds. In addition, the authors found that numerous caspase-activating cells, mostly without delamination as revealed by live imaging, emerged around the wound site ablated by the laser at the developing pupal notum. Genetic inhibition of Dronc activity and DIAP1 overexpression, but not P35 overexpression, promotes wound closure, which is different from thorax closure in executioner caspase-dependency (Fujisawa et al., 2019). Therefore, the suppression of cell migration by caspases, specifically Dronc, seems rather general in Drosophila, whereas its dependency on executioner caspases differs depending on the situation.
4.3.3 Irradiation-induced cell migration
In addition to the initiator caspase, Dronc, executioner caspases also suppress cell migration under other conditions (Gorelick-Ashkenazi et al., 2018). In imaginal discs, which are composed of a monolayer of columnar epithelial cells, ionizing irradiation induces caspase activation, resulting in apoptosis (Florentin and Arama, 2012). However, in Drice mutant, which only contains Dcp-1 executioner caspase, ionizing irradiation did not induce apoptosis, as detected by TUNEL staining, even though caspase activity was still detected using the CD8::PARP::VENUS probe. Under these conditions, irradiation-induced cell migration involving basal delamination was observed. Because cell migration is further enhanced by additional Dcp-1 mutations, the authors suggested that non-lethal executioner caspase activity, especially that of Dcp-1, inhibits irradiation-induced cell migration (Gorelick-Ashkenazi et al., 2018). Since border cell migration and epithelial thorax wound healing are not dependent on executioner caspase activity, the underlying mechanisms seem different from those of irradiation-induced cell migration.
4.3.4 CDPs for cell migration in other animals
Caspases also regulate mammalian cell migration. For example, caspase-11-deficient leukocytes show defective migration, suggesting that caspase-11 promotes cell migration (Li et al., 2007). Caspase-8-deficient mouse embryonic fibroblasts exhibit reduced cell motility, suggesting that caspase-8 promotes cell motility (Helfer et al., 2006). Because caspases also negatively regulate cell migration in Drosophila as shown above (Geisbrecht and Montell, 2004; Gorelick-Ashkenazi et al., 2018; Fujisawa et al., 2019), caspases do not always regulate cell migration in a single direction, i.e., caspases can promote and suppress cell migration. Studying the downstream molecular mechanisms helps to understand their bidirectional ability to regulate cell motility, depending on the context and evolutionary conservation.
4.4 Cell differentiation
4.4.1 Sensory organ precursor cell specification
Caspases regulate cell differentiation in a non-lethal manner. Drosophila constantly produces 4 mechanosensory bristles called macrochaetae in their scutellum, which emerge from sensory organ precursor (SOP) cells selected within the proneural cluster of wing imaginal discs. Using Dark mutant as well as proneural cluster specific P35 overexpression, the authors first noticed that ectopic bristles emerged in the adult scutellum. While no apoptosis was detected by TUNEL staining in the proneural cluster, caspase activity was detected using SCAT3 FRET imaging. Genetic screening revealed that Shaggy, especially the alternative isoform Shaggy46, which is a substrate of caspase whose cleavage activates Shaggy46 kinase, is responsible for the suppression of ectopic bristles. Thus, the authors concluded that caspase activity suppresses the emergence of ectopic bristles in the scutellum through non-lethal substrate cleavage of Shaggy46 for kinase activation (Kanuka et al., 2005). The ectopic bristle also emerges upon DIAP1 overexpression (Kanuka et al., 2005) and IKKε knockdown (Kuranaga et al., 2006), suggesting that this phenomenon is regulated by IKKε-mediated DIAP1 degradation.
4.4.2 Intestinal progenitor cell quiescence
Caspases also preserve cells in quiescence (Arthurton et al., 2020). In intestinal progenitor cells, which consist of intestinal stem cells and intermediate progenitor cells termed enteroblasts, Dronc is non-lethally activated under normal conditions, as detected using DBS-S::QF. Using the conditional Dronc knockout allele, they found that Dronc activity is required to prevent an increase in the number of intestinal precursor cells as well as to prevent their unintended differentiation. This phenotype was not dependent on RHG, Dark, or executioner caspase activity, but was solely dependent on Dronc. Although the exact substrate has not been determined, Dronc activity limits Notch signaling to regulate cellular quiescence (Arthurton et al., 2020).
4.4.3 Ovarian somatic cell differentiation
Dronc also regulates ovarian somatic cell proliferation and differentiation (Galasso et al., 2023). In germarium, a tissue composed of germline cells and supporting somatic cells of Drosophila ovary, the authors first found Dronc activation in surviving somatic cells by using DBS-S::QF with lineage tracing. Using the conditional Dronc knockout allele, they found that Dronc was required for the proliferation and differentiation of ovarian somatic cells. In contrast to the role of Dronc in preserving intestinal progenitor cell quiescence (Arthurton et al., 2020), the entire apoptotic pathway, including RHG, DIAP1, Dark, and executioner caspase activity, is involved in these processes. Subsequently, Dronc deficiency reduced Hedgehog signaling through excess autophagy (Galasso et al., 2023), suggesting that caspases non-lethally prevent excess autophagy to sustain Hedgehog signaling for proliferation and differentiation.
4.4.4 Plasmatocyte differentiation
Caspases also participate in the differentiation of plasmatocyte, a macrophage-like cell in Drosophila (Maurya et al., 2024). During the larval stage, plasmatocytes differentiate from the lymph gland, the larval hematopoietic organ, which includes multipotent progenitors, intermediate progenitors, and differentiating cells. Using a wide variety of caspase activity probes, including GC3Ai, Apoliner, CaspaseTracker/CasExpress, L-CasExpress, DBS-S::Histone2Av::GFP as well as anti-cleaved Dcp-1 antibody staining, it was found that caspase is activated during differentiation, especially in intermediate progenitor cells. Moreover, using L-CasExpress combined with lineage tracing (L-Trace), the authors found that caspase-activated cells survived in the differentiation zone, suggesting that transiently activated caspases do not induce apoptosis. Subsequent analysis showed that caspase activation relies on the entire core apoptotic signaling pathway, including RHG, Dronc, Drice, and Dcp-1, and induces DNA damage through Drep4, a Drosophila ortholog of CAD, and Drep1, a Drosophila ortholog of ICAD, most likely through its cleavage. DNA damage is required for plasmatocyte differentiation, potentially by altering gene expression. Thus, developmentally programmed non-lethal caspase activation is required for plasmatocyte differentiation by inducing programmed DNA breaks (Maurya et al., 2024).
4.4.5 Cell fate changes in tissue regeneration
Caspases regulate cell fate changes during tissue regeneration (Verghese and Su, 2018). Imaginal discs are epithelial tissues capable of regeneration after damage (Hariharan and Serras, 2017). During the ionizing irradiation of wing imaginal discs, it was found that dorsal hinge cells were resistant to apoptosis (Verghese and Su, 2016). Moreover, hinge cells changed their fate and translocated to the wing pouch region during ionizing radiation-induced regeneration. This process was inhibited by the inhibition of caspase activity in the H99 allele, in P35 and DroncDN overexpression (Verghese and Su, 2018), suggesting that caspase activation is non-lethally required for cell fate changes from hinge cells to wing pouch cells during regeneration.
4.4.6 CDPs for cell differentiation in other animals
Regulation of cell differentiation by non-lethal caspase activation is also common in other animals. Similar to plasmatocyte differentiation in Drosophila, caspase activation-mediated DNA damage is a conserved mechanism of cellular differentiation. These include skeletal muscle differentiation, which requires caspase activation and subsequent CAD activation (Fernando et al., 2002; Larsen et al., 2010) and Myc-induced oncogenic transformation of human mammary epithelial cells, which requires caspase activation and subsequent ENDOG-mediated DNA damage (Cartwright et al., 2017). In addition to cell differentiation, iPSC induction from human fibroblasts requires caspase activation, wherein retinoblastoma is cleaved for reprogramming (Li et al., 2010).
4.5 Cell proliferation
4.5.1 Imaginal tissue growth
While caspases can induce apoptosis to reduce the cell number in tissues, they increase the tissue size of growing epithelial tissues in a non-lethal manner (Shinoda et al., 2019). During imaginal disc growth, the inhibition of executioner caspase activity by overexpression of P35, but not by overexpression of DroncDN or knockdown of Dark, reduces adult wing size. This effect is specific to the executioner caspases Dcp-1 and Decay, but not to Drice. Even in the absence of Dronc, basal caspase activity was detected by western blotting using a SCAT3 probe, suggesting that Dronc-independent basal executioner caspase activity exists. As Dcp-1 or Decay knockdown did not result in any reduction in the apoptotic cell number detected by TUNEL staining, it was shown that basal executioner caspase activity, independent of Dronc activity, sustained imaginal tissue growth. Mechanistically, uncleavable Acinus, a Dcp-1 substrate (Nandi et al., 2014), showed reduced wing size (Shinoda et al., 2019). These results suggest that at least in part, Dcp-1-mediated basal Acinus cleavage sustains imaginal tissue growth.
4.5.2 Wound-like tumor growth
Caspases can limit tumor growth in a non-lethal manner, even in the same tissue (Xu et al., 2022). In wound-like tumors induced by simultaneous activation of the EGFR and JAK/STAT pathways in wing imaginal discs, most of the cells that survived underwent Dronc activation, which was detected using DBS-S::QF with lineage tracing. The authors showed that the tumor size increased upon genetic inhibition of Dronc activity, suggesting that Dronc limits tumor growth in a non-lethal manner. While this effect was observed with Dark knockdown and DIAP1 overexpression, it was not observed with RHG knockdown or P35 overexpression. Dronc activity limits JNK signaling, independent of Eiger and reactive oxygen species, which are required for tumor overgrowth (Xu et al., 2022).
4.5.3 Apoptosis-induced proliferation
Apoptosis-induced proliferation (AiP), which does not necessarily require apoptosis to induce nascent cell proliferation, is also categorized as a non-lethal process in which undead cells secrete mitogens into surrounding tissues, resulting in the over-proliferation of surrounding cells (Huh et al., 2004a; Pérez-Garijo et al., 2004; Ryoo et al., 2004; Kondo et al., 2006). Typically, AiP is induced by apoptosis induction, such as overexpression of pro-apoptotic genes, including Rpr, and simultaneous inhibition of apoptosis by expressing P35 in wing imaginal discs. In proliferating cells, AiP requires Dronc, but not executioner caspase activity (Kondo et al., 2006). Using eye imaginal discs, a study showed that AiP in proliferating cells of eye imaginal discs is dependent on Dronc, but not on executioner caspase activity, while AiP in differentiating cells is dependent on executioner caspases Drice and Dcp-1, suggesting distinct mechanisms of AiP depending on their cell status (Fan and Bergmann, 2008). With respect to AiP in proliferating cells, Dark is required (Fan et al., 2014). However, the catalytic activity of Dronc may not be required because the substitution of the catalytic cysteine with alanine does not interfere with AiP if it simultaneously carries a substitution from lysine 78 to arginine, where inhibitory mono-ubiquitylation occurs (Kamber Kaya et al., 2017). With respect to Dronc-dependent AiP in proliferating cells, undead cells secrete mitogens, including Wingless and Decapentaplegic to stimulate tissue growth by inducing the proliferation of surrounding cells (Pérez-Garijo et al., 2009). The detailed molecular mechanism that enables the non-lethal function of caspase in AiP has long been pursued by Andreas Bergmann and Yun Fan, and is further described in the next section.
4.5.4 CDPs for tissue size regulation in other animals
Caspases non-lethally promote organ growth in the sebaceous glands of mice (Yosefzon et al., 2018). Caspase-3 cleaves α-Catenin which leads to the nuclear translocation of yes-associated protein (YAP), a vital regulator of organ size (Yosefzon et al., 2018). Conversely, examining the role of apoptosis in tissue size regulation by caspase inhibition should be considered, as caspase has non-lethal functions in regulating tissue size.
4.6 Summary
As mentioned above, studies in Drosophila lead to the identification of wide variety of non-lethal caspase functions, mainly in developmental processes. Because several CDPs are also observed in other animals, and the mechanisms of which are conserved in some cases, Drosophila studies will serve as a guide for CDPs studies in other animals. From the pathological perspective, because CDPs include cell proliferation and cell migration, it is conceivable that caspases can non-lethally regulate cancer development. Indeed, it is reported that minority MOMP, where only a minority of mitochondria undergoes MOMP, induces sub-lethal caspase activation that causes DNA damage and genomic instability, promoting transformation and tumorigenesis of primary mouse embryonic fibroblasts (Ichim et al., 2015). Thus, manipulation of caspase activity in both directions, suppressing the activity for reducing non-lethal effect and enhancing the activity for inducing cell death, can open therapeutic possibilities combined with anti-cancer regimes.
Important processes of CDPs studies include the detection of caspase activation without cell death (often analyzed by TUNEL staining), manipulation of the core apoptotic machinery at each level, and description of the cellular phenotype. Substrates are not often identified, which benefits the analysis of downstream events. Compared to the physiological functions of non-lethal caspase activation, the underlying mechanism that enables non-lethal caspase activation without cell death and regulates downstream events is rather lagging due to technical shortages in identifying substrates. As described above, the core apoptotic components, including caspases, are not always the same; some require both Dronc and executioner caspases, whereas others require only Dronc or executioner caspases. Thus, researchers must be aware that the underlying mechanisms differ depending on the analyzed CDPs. Yet, overlapping components are reproducibly used in some CDPs: IKKε-DIAP1-Dronc axis regulates wide range of phenotypes with respect to cytoskeleton dynamics. Thus, testing similarities will benefit the CDPs research. In the following section, we summarize several well-studied mechanisms that enable non-lethal caspase activation.
5 Mechanisms of controlling caspase activation without inducing cell death
The growing body of CDPs clearly show that their underlying molecular mechanisms are diverse. However, extensive analyses of several CDPs have revealed the underlying mechanisms and similarities between other CDPs. Specifically, evidence indicates the importance of restricting caspase activation to a specific compartment of cells shaped by caspase-interacting and proximal proteins. In this section, we introduce the underlying mechanisms that enable non-lethal caspase activation, specifically focusing on interacting proteins of caspase. In particular, analyses of sperm individualization, AiP, and SOP specification revealed subcellular restriction of caspase activation as a key regulator of non-lethal activation.
5.1 Restriction of caspase activation to the vicinity of mitochondrial outer membrane
Continuous analysis of non-lethal caspase functions in sperm individualization by Eli Arama and colleagues first identified the Cullin-3-dependent ubiquitin ligase complex from the genetic screening of a large collection of sterile male fly mutants. Mutations in the testis-specific isoform of Cullin-3, the small RING protein Roc1b, or a Drosophila ortholog of the mammalian BTB-Kelch protein, Klhl10, result in no caspase activation during sperm individualization (Arama et al., 2007). Subsequently, they conducted a yeast two-hybrid screen to identify the binding partners of Klhl10, an E3 substrate-recruitment subunit. They found that Scotti (Soti) is a binding partner of Klhl10. Soti mutant exhibited elevated caspase activation, suggesting that Soti antagonized this process. Moreover, caspase activation is not uniform in individualizing spermatids but is rather high in the head and low in the tail. Soti expression was inversely correlated with activation patterns. A giant IAP called dBruce, which competes with Soti, is a target of Cullin-3. This results in the degeneration of dBruce and graded activation patterns of caspase, showing that the distal part is protected from excessive caspase activation and cell death (Kaplan et al., 2010). Furthermore, using yeast two hybrid screening against Cullin-3, they found a Krebs cycle component, the ATP-specific form of the succinyl-CoA synthetase β subunit (A-Sβ) as an interacting partner. A-Sβ localizes at the mitochondrial outer membrane and restricts Cullin-3 to the proximity of the membrane, thereby limiting caspase activation to close proximity of the mitochondrial outer membrane (Aram et al., 2016). Thus, a series of studies by Eli Arama and colleagues provided a model for restricting caspase activation to a specific subcellular compartment; head-to-tail descending activation as well as proximal to the mitochondrial outer membrane activation are the keys to escape cell death caused by excessive caspase activation (Figure 3A, left).
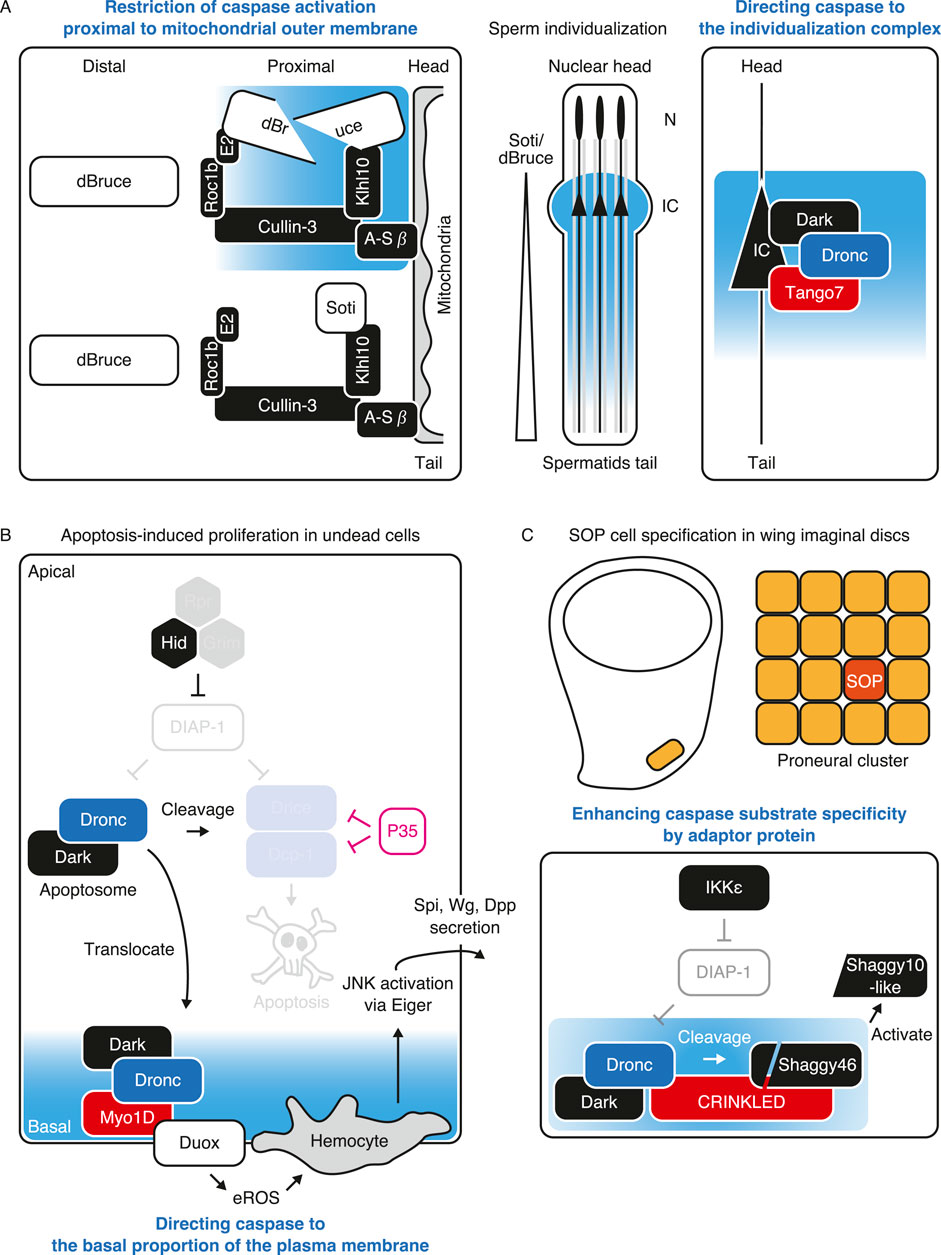
Figure 3. Mechanisms of controlling caspase activation without inducing cell death. (A) Regulation of non-lethal caspase activation during sperm individualization. During sperm individualization, caspase activation, which peaks in individualization complex (IC), forms descending gradient to the tail. This gradient activation is regulated by inverse gradient expression of Soti and dBruce which antagonize caspase activation. In addition, caspase activation is restricted to the outer membrane of mitochondria where Cullin-3 complex is tethered by A-Sβ (left). Alongside, Dronc is directed to the individualization complex by Tango7 (right). Caspase activating regions are shown in light blue. (B) Regulation of non-lethal caspase activation during apoptosis-induced proliferation in undead cells. Undead cells are induced by simultaneous activation of apoptotic signaling and inhibition of executioner caspase activity by P35 expression. In undead cells, Dronc translocates to the basal side of the plasma membrane with help of Myo1D where activation is restricted. Dronc promotes activation of Duox to produce extracellular reactive oxygen species (eROS), which accumulates hemocyte for JNK activation resulting in mitogen production in the undead cell for inducing nascent cell proliferation. Caspase activating region is shown in light blue. (C) Regulation of non-lethal caspase activation during sensory organ precursor (SOP) cell specification. During SOP cell (orange) specification from proneural cluster cells (yellow) (top), IKKε-mediated DIAP1 degradation induced caspase activation. CRINKLED, an adaptor protein of Dronc, proximates Dronc and Shaggy46, the cleavage of which by executioner caspases generates active Shaggy10-like kinase (bottom) and negatively regulates SOP cell specification, thus inhibiting the emergence of ectopic SOP cells. Caspase activating region is shown in light blue.
5.2 Directing caspase activation at the individualization complex
Dronc-interacting protein Tango7, the Drosophila translation initiation factor eIF3 subunit m, is required for activation during sperm individualization. Tango7 physically interacts with Dronc and localizes to the individualization complex where caspase activity peaks (D’Brot et al., 2013). Therefore, by caspase-interacting proteins and not by IAP-mediated activity regulation, its direct localization to the site of interest is similarly important (Figure 3A, right). Tango7 is also required for regulating salivary gland cell elasticity, as well as phagocytic removal of pruned dendrites in ddaC neurons. Both phenotypes appear to be independent of Dark, suggesting that Dark is required for cytosolic Dronc activation, whereas Tango7 regulates cortical non-lethal Dronc activation (Kang et al., 2017).
5.3 Directing caspase activation at the basal side of the plasma membrane
As suggested, controlling the caspase activation site within cells appears to be the key to regulating non-lethal caspase activation. This has also been observed with respect to AiP. Andreas Bergmann and colleagues investigated the mechanism of AiP through genetic screening. A series of studies have identified an unconventional Myosin1D (Myo1D) that is required for AiP. Myo1D interacts physically with Dronc. They further found that endogenous Dronc proteins localized to the basal side of disc proper cells in undead conditions with Myo1D. Myo1D is required for the translocation of Dronc to the basal side of the plasma membrane in imaginal discs, where it activates Duox and induces AiP in surrounding cells. These results suggest that Myo1D localizes Dronc to the basal side of the plasma membrane for non-lethal activation (Amcheslavsky et al., 2018) (Figure 3B). Moreover, Myo1D works not only in imaginal discs of the AiP model, but also in midgut enterocytes, where they undergo transient non-lethal activation, suggesting that translocation of the caspase molecules to the restricted cellular compartment is key for non-lethal (transiently undead) activation (Amcheslavsky et al., 2020). Although Myo1D is expressed and localized on the basal side of the salivary gland and colocalizes with Dronc, it is not required for Dronc localization in the salivary gland, potentially because Dronc localization is regulated by Tango7 in this tissue under the normal condition. Alternatively, under undead condition in the salivary gland, the localization of Dronc to the basal side of the plasma membrane increases, which is dependent on Myo1D (Amcheslavsky et al., 2018). This suggests that Dronc mediates distinct non-lethal cellular processes by changing adaptor proteins to direct distinct subcellular compartments, depending on the conditions.
5.4 Enhancing substrate specificity by an adaptor protein
CRINKLED (CK), a Drosophila unconventional myosin, was identified in an IP-MS experiment using Dronc. Genetic analysis revealed that CK is required for efficient Dronc-dependent induction of apoptosis in response to high-level expression of IAP antagonists. In addition, further genetic analysis revealed that CK is involved in arista branching, SOP specification and border cell migration, a process which involves non-lethal caspase activation of Dronc, mediated by DIAP1 and IKKε (Geisbrecht and Montell, 2004; Kanuka et al., 2005; Kuranaga et al., 2006; Oshima et al., 2006). CK physically interacts with Shaggy46, a caspase substrate responsible for SOP cell specification (Kanuka et al., 2005). They showed that Shaggy46 is also involved in border cell migration. Thus, they proposed a model where CK interacts with both Dronc and Shaggy46 to induce specific substrate cleavage, providing that caspase-interacting proteins not only restrict their activity to a specific cellular compartment but also engage in substrate cleavage (Orme et al., 2016). IKKε also regulates SOP cell specification in a non-apoptotic scenario (Kuranaga et al., 2006). Thus, limiting activation level by IKKε-DIAP1 axis is necessary for regulating Dronc-CK-Shaggy46 mediated non-lethal functions (Figure 3C). CK is not involved in sperm individualization (Orme et al., 2016), further supporting the idea that Dronc mediates distinct non-lethal cellular processes by altering adaptor proteins.
5.5 Summary
Regulating caspase activity in restricted regions, as well as limiting the activation level or timing, is key to regulating non-lethal caspase activity. While their functions are different, caspases often exploit the same set of mechanisms for regulating caspase activation and function. Thus, it is possible that the molecular mechanisms of CDPs involve a pair of caspases and their adaptor proteins. It is noteworthy that although limiting tumor growth is mediated by Dronc, this CDP is not mediated by Tango7 or Myo1D (Xu et al., 2022), suggesting that there is another adaptor protein that mediates other types of CDPs. As in most cases, non-lethal caspase activity is regulated by its interacting and proximal proteins, identifying these proteins in cells of interest within living animals is critical. In the following section, we introduce a new approach to identify such components.
6 Analyzing caspase-proximal proteins for CDPs using TurboID
Proximity labeling techniques are emerging as means of detecting proximal proteins of interest inside cells or organisms (Qin et al., 2021). One such technique is APEX2, which is hydrogen peroxide-dependent proximity labeling with high resolution in a span of 1 min (Lam et al., 2015; Hung et al., 2016). However, the use of a toxic reagent, makes it difficult for its application in animals, including Drosophila. TurboID is another proximity labeling technique that uses ATP to convert biotin into the reactive intermediate biotin-AMP, which covalently labels the lysine residues of proximal proteins within 10 min. Because TurboID labeling is hydrogen peroxide-free, it also enables its application in Drosophila (Branon et al., 2018) (Figure 4A). In Drosophila, biotin is supplied through feeding. The labelled proteins were purified using streptavidin beads for further analysis, including mass spectrometry for proteomics.
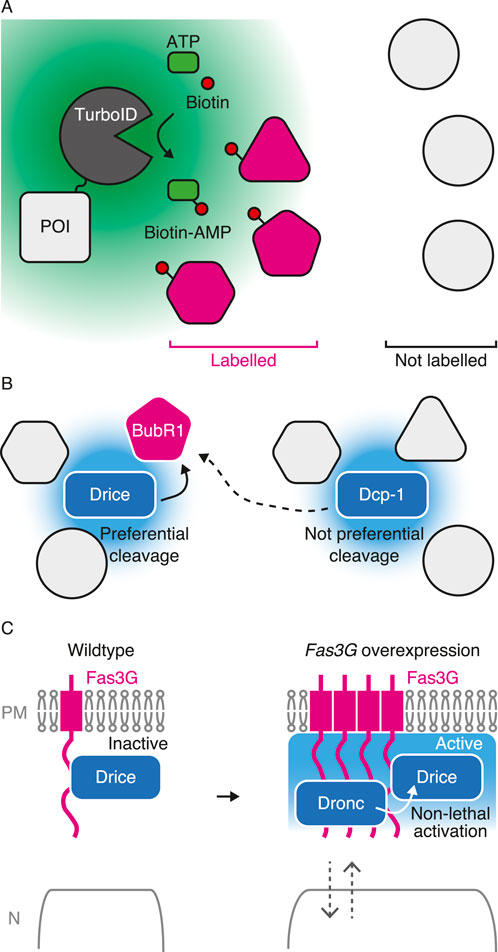
Figure 4. Analyzing caspase-proximal proteins in non-lethal scenario using TurboID. (A) Schematic diagram of TurboID-mediated biotinylation of proximal proteins. TurboID generates reactive biotin-AMP from ATP (green) and biotin (magenta), which conjugates to lysine residue of proximal proteins (magenta) within approximately 10 nm (green region) within 10 min. (B) Proximal proteins can be preferentially cleaved by proximal caspase. BubR1 (magenta), a substrate of caspase, is proximal to Drice rather than Dcp-1 which is preferentially cleaved by Drice rather than Dcp-1. Hypothetical caspase activating region of each caspase is shown in light blue. (C) Proximal proteins can regulate non-lethal caspase activation. Fasciclin 3 isoform G (Fas3G; magenta) is one of proximal proteins to Drice. Overexpression of Fas3G promotes Dronc expression which also comes close to Fas3G for facilitating non-lethal activation. Caspase activating region is shown in light blue.
Recently, we established C-terminal TurboID knocked-in strains in Drosophila with three major apoptotic caspases: Dronc, Drice, and Dcp-1 (Shinoda et al., 2019). As TurboID-tagged single caspase biotinylates numerous nearby proteins, we can examine the expression patterns of each caspase using streptavidin with high sensitivity (Shinoda et al., 2019; Arthurton et al., 2020; Muramoto et al., 2024). Executioner caspases are the “executioner” of cell death and other cellular processes as executioner caspases cleave wide ranges of substrate compared to initiator caspases. Because of the lack of protein-interacting motifs at their N-termini, the identification of their interacting proteins is rather limited. Moreover, caspase-proximal proteins are not identical between Drice and Dcp-1 in wing imaginal discs (Shinoda et al., 2019), suggesting that executioner caspases sometimes have distinct functions in CDPs even though they prefer similar primary sequences for cleavages. We showed that BubR1, one of the spindle assembly checkpoint components, is a substrate of caspase, the cleavage of which regulates organismal lifespan. Importantly, using the TurboID technique, we showed that BubR1 is proximal to Drice but not to Dcp-1 in the ovaries, even though both are almost equally expressed. Accordingly, BubR1 is preferentially cleaved by Drice rather than Dcp-1 in S2 cells during non-lethal activation (Shinoda et al., 2023). These results indicate that caspase-proximal proteins include substrates of caspases that are preferentially cleaved by nearby proteins (Figure 4B). Because the cleavage of limited substrates is the key for non-lethal functions and as identifying responsible substrates is always challenging (Table 1), TurboID-mediated identification of CDP-responsible substrates helps in understanding the effectors of CDPs. In addition to CDPs, TurboID can also be applied for analyzing proteins related to cell death. Using BioID, a less efficient proximity labeling enzyme, to human Caspase-1, p62/sequestosome-1 is identified as a substrate which only interacts with Caspase-1 upon inflammasome activation (Jamilloux et al., 2018). Thus, TurboID is useful for identifying context-dependent substrates of caspases both in lethal and non-lethal scenarios.
We recently analyzed the proximal proteins of Drice in the adult brain. Although Drice is primarily a cytosolic protein, its proximity lies within the membrane fraction. Drice is only specifically proximal to the specific isoform of Fasciclin 3, isoform G, and not to other isoforms even though they are overexpressed. Thus, Drice might be compartmentalized to a specific microdomain of the membrane compartment. In addition, overexpression of one of the Drice proximal proteins, Fasciclin 3 isoform G, activates caspase in the olfactory receptor neurons without inducing cell death, suggesting that proximal proteins regulate non-lethal caspase activation. Fasciclin 3 isoform G overexpression induces Dronc expression, which also comes close to Fasciclin 3 isoform G. Dronc is required for caspase activation induced by Fasciclin 3 isoform G overexpression (Muramoto et al., 2024) (Figure 4C). In addition to the translocation of initiator caspase Dronc to the membrane for non-lethal activation, these results suggest that executioner caspase is pre-compartmentalized in proximity to a specific set of proteins prior to activation which defines the “hotspot” for non-lethal caspase activation. TurboID-mediated identification of proximal proteins, including substrates and regulators, in living animals will enhance our understanding of the regulation of caspase activation without inducing cell death in a wide range of CDPs in a variety of tissues.
7 Discussion
Diverse CDPs have been identified using advanced Drosophila genetic tools to detect and manipulate caspase activity in specific cells in vivo. Continuous analyses have revealed that non-lethal caspase activation can occur through limited activation in specific subcellular compartments, specifically mediated by caspase-interacting proteins. However, we still do not fully understand how the intensity and duration of caspase activation for non-lethal processes are regulated. To enhance our understanding, it is necessary to develop genetic tools capable of detecting and manipulating caspase activity with a high spatial resolution, potentially at the subcellular level, in a highly quantitative manner. Development of such tools will significantly advance our understanding of non-lethal functions mediated by caspase activity. Identifying critical substrates for non-lethal functions is equally important. TurboID will serve as a vital methodology for identifying critical substrates within cells of interest in vivo. In addition, it is critical to analyze the kinetics of substrates cleavage within cells of interest in vivo. Given that enzymes with multiple substrates, such as caspase, are multifunctional, understanding the action of caspase will broaden our understanding of the actions of other multifunctional enzymes, both in cells and in vivo.
Because caspase regulates various non-lethal functions, its activation is no longer considered a point of no return in cell death. Understanding the non-lethal functions of caspases has also led to a deeper understanding of the molecular mechanisms of cell death, including the identification of critical substrates for the process of cell death. In the Drosophila pupal notum, caspase-mediated microtubule disassembly is the key rate-limiting step in cell extrusion, the process of expelling dying epithelial cells (Villars et al., 2022). Studying CDPs will advance our understanding of cell death. This will clarify the states of the living and dead cells, along with their intermediate states.
Author contributions
NS: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing. MM: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (KAKENHI Grant Numbers 19K16137, 21K15080, 22H05586, and 23K05747 to NS and 21H04774, 23H04766, and 24H00567 to MM) and grants from the Takeda Science Foundation and Sumitomo Foundation to NS.
Acknowledgments
We apologize for omitting several relevant publications because of space limitations. We thank Editage [www.editage.jp] for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amcheslavsky, A., Lindblad, J. L., and Bergmann, A. (2020). Transiently “undead” enterocytes mediate homeostatic tissue turnover in the adult Drosophila midgut. Cell Rep. 33, 108408. doi:10.1016/j.celrep.2020.108408
Amcheslavsky, A., Wang, S., Fogarty, C. E., Lindblad, J. L., Fan, Y., and Bergmann, A. (2018). Plasma membrane localization of apoptotic caspases for non-apoptotic functions. Dev. Cell 45, 450–464. doi:10.1016/j.devcel.2018.04.020
Aram, L., Braun, T., Braverman, C., Kaplan, Y., Ravid, L., Levin-Zaidman, S., et al. (2016). A Krebs cycle component limits caspase activation rate through mitochondrial surface restriction of CRL activation. Dev. Cell 37, 15–33. doi:10.1016/j.devcel.2016.02.025
Aram, L., Yacobi-Sharon, K., and Arama, E. (2017). CDPs: caspase-dependent non-lethal cellular processes. Cell Death Differ. 24, 1307–1310. doi:10.1038/cdd.2017.111
Arama, E., Agapite, J., and Steller, H. (2003). Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4, 687–697. doi:10.1016/s1534-5807(03)00120-5
Arama, E., Bader, M., Rieckhof, G. E., and Steller, H. (2007). A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 5, e251. doi:10.1371/journal.pbio.0050251
Arthurton, L., Nahotko, D. A., Alonso, J., Wendler, F., and Baena-Lopez, L. A. (2020). Non-apoptotic caspase activation preserves Drosophila intestinal progenitor cells in quiescence. EMBO Rep. 21, e48892. doi:10.15252/embr.201948892
Baena-Lopez, L. A., Arthurton, L., Bischoff, M., Vincent, J.-P., Alexandre, C., and McGregor, R. (2018). Novel initiator caspase reporters uncover previously unknown features of caspase-activating cells. Development 145, dev170811. doi:10.1242/dev.170811
Bardet, P.-L., Kolahgar, G., Mynett, A., Miguel-Aliaga, I., Briscoe, J., Meier, P., et al. (2008). A fluorescent reporter of caspase activity for live imaging. Proc. Natl. Acad. Sci. U. S. A. 105, 13901–13905. doi:10.1073/pnas.0806983105
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. doi:10.1242/dev.118.2.401
Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., et al. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887. doi:10.1038/nbt.4201
Campbell, D. S., and Okamoto, H. (2013). Local caspase activation interacts with Slit-Robo signaling to restrict axonal arborization. J. Cell Biol. 203, 657–672. doi:10.1083/jcb.201303072
Canales Coutiño, B., Cornhill, Z. E., Couto, A., Mack, N. A., Rusu, A. D., Nagarajan, U., et al. (2020). A genetic analysis of tumor progression in Drosophila identifies the cohesin complex as a suppressor of individual and collective cell invasion. iScience 23, 101237. doi:10.1016/j.isci.2020.101237
Cartwright, I. M., Liu, X., Zhou, M., Li, F., and Li, C.-Y. (2017). Essential roles of Caspase-3 in facilitating Myc-induced genetic instability and carcinogenesis. Elife 6, e26371. doi:10.7554/elife.26371
Chen, P., Rodriguez, A., Erskine, R., Thach, T., and Abrams, J. M. (1998). Dredd, a novel effector of the apoptosis activators reaper, grim, and hid in Drosophila. Dev. Biol. 201, 202–216. doi:10.1006/dbio.1998.9000
Chew, S. K., Akdemir, F., Chen, P., Lu, W.-J., Mills, K., Daish, T., et al. (2004). The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev. Cell 7, 897–907. doi:10.1016/j.devcel.2004.09.016
Chihara, T., Kitabayashi, A., Morimoto, M., Takeuchi, K.-I., Masuyama, K., Tonoki, A., et al. (2014). Caspase inhibition in select olfactory neurons restores innate attraction behavior in aged Drosophila. PLoS Genet. 10, e1004437. doi:10.1371/journal.pgen.1004437
Colussi, P. A., Quinn, L. M., Huang, D. C., Coombe, M., Read, S. H., Richardson, H., et al. (2000). Debcl, a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J. Cell Biol. 148, 703–714. doi:10.1083/jcb.148.4.703
Cullen, K., and McCall, K. (2004). Role of programmed cell death in patterning the Drosophila antennal arista. Dev. Biol. 275, 82–92. doi:10.1016/j.ydbio.2004.07.028
Daish, T. J., Mills, K., and Kumar, S. (2004). Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev. Cell 7, 909–915. doi:10.1016/j.devcel.2004.09.018
D’Brot, A., Chen, P., Vaishnav, M., Yuan, S., Akey, C. W., and Abrams, J. M. (2013). Tango7 directs cellular remodeling by the Drosophila apoptosome. Genes Dev. 27, 1650–1655. doi:10.1101/gad.219287.113
Ding, A. X., Sun, G., Argaw, Y. G., Wong, J. O., Easwaran, S., and Montell, D. J. (2016). CasExpress reveals widespread and diverse patterns of cell survival of caspase-3 activation during development in vivo. Elife 5, e10936. doi:10.7554/eLife.10936
Ditzel, M., Broemer, M., Tenev, T., Bolduc, C., Lee, T. V., Rigbolt, K. T. G., et al. (2008). Inactivation of effector caspases through nondegradative polyubiquitylation. Mol. Cell 32, 540–553. doi:10.1016/j.molcel.2008.09.025
Dorstyn, L., Colussi, P. A., Quinn, L. M., Richardson, H., and Kumar, S. (1999a). DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. U. S. A. 96, 4307–4312. doi:10.1073/pnas.96.8.4307
Dorstyn, L., Mills, K., Lazebnik, Y., and Kumar, S. (2004). The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J. Cell Biol. 167, 405–410. doi:10.1083/jcb.200408054
Dorstyn, L., Read, S., Cakouros, D., Huh, J. R., Hay, B. A., and Kumar, S. (2002). The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 156, 1089–1098. doi:10.1083/jcb.200111107
Dorstyn, L., Read, S. H., Quinn, L. M., Richardson, H., and Kumar, S. (1999b). DECAY, a novel Drosophila caspase related to mammalian caspase-3 and caspase-7. J. Biol. Chem. 274, 30778–30783. doi:10.1074/jbc.274.43.30778
Doumanis, J., Quinn, L., Richardson, H., and Kumar, S. (2001). STRICA, a novel Drosophila melanogaster caspase with an unusual serine/threonine-rich prodomain, interacts with DIAP1 and DIAP2. Cell Death Differ. 8, 387–394. doi:10.1038/sj.cdd.4400864
Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A., and Nagata, S. (1998). A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391, 43–50. doi:10.1038/34112
Fan, Y., and Bergmann, A. (2008). Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev. Cell 14, 399–410. doi:10.1016/j.devcel.2008.01.003
Fan, Y., and Bergmann, A. (2010). The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 17, 534–539. doi:10.1038/cdd.2009.185
Fan, Y., Lee, T. V., Xu, D., Chen, Z., Lamblin, A.-F., Steller, H., et al. (2010). Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ. 17, 912–921. doi:10.1038/cdd.2009.182
Fan, Y., Wang, S., Hernandez, J., Yenigun, V. B., Hertlein, G., Fogarty, C. E., et al. (2014). Genetic models of apoptosis-induced proliferation decipher activation of JNK and identify a requirement of EGFR signaling for tissue regenerative responses in Drosophila. PLoS Genet. 10, e1004131. doi:10.1371/journal.pgen.1004131
Fernando, P., Kelly, J. F., Balazsi, K., Slack, R. S., and Megeney, L. A. (2002). Caspase 3 activity is required for skeletal muscle differentiation. Proc. Natl. Acad. Sci. U. S. A. 99, 11025–11030. doi:10.1073/pnas.162172899
Florentin, A., and Arama, E. (2012). Caspase levels and execution efficiencies determine the apoptotic potential of the cell. J. Cell Biol. 196, 513–527. doi:10.1083/jcb.201107133
Fraser, A. G., and Evan, G. I. (1997). Identification of a Drosophila melanogaster ICE/CED-3-related protease, drICE. EMBO J. 16, 2805–2813. doi:10.1093/emboj/16.10.2805
Fujisawa, Y., Kosakamoto, H., Chihara, T., and Miura, M. (2019). Non-apoptotic function of Drosophila caspase activation in epithelial thorax closure and wound healing. Development 146, dev169037. doi:10.1242/dev.169037
Fujisawa, Y., Shinoda, N., Chihara, T., and Miura, M. (2020). ROS regulate caspase-dependent cell delamination without apoptosis in the Drosophila pupal notum. iScience 23, 101413. doi:10.1016/j.isci.2020.101413
Galasso, A., Xu, D. C., Hill, C., Iakovleva, D., Stefana, M. I., and Baena-Lopez, L. A. (2023). Non-apoptotic caspase activation ensures the homeostasis of ovarian somatic stem cells. EMBO Rep. 24, e51716. doi:10.15252/embr.202051716
Geisbrecht, E. R., and Montell, D. J. (2004). A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell 118, 111–125. doi:10.1016/j.cell.2004.06.020
Gorelick-Ashkenazi, A., Weiss, R., Sapozhnikov, L., Florentin, A., Tarayrah-Ibraheim, L., Dweik, D., et al. (2018). Caspases maintain tissue integrity by an apoptosis-independent inhibition of cell migration and invasion. Nat. Commun. 9, 2806. doi:10.1038/s41467-018-05204-6
Grether, M. E., Abrams, J. M., Agapite, J., White, K., and Steller, H. (1995). The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9, 1694–1708. doi:10.1101/gad.9.14.1694
Hariharan, I. K., and Serras, F. (2017). Imaginal disc regeneration takes flight. Curr. Opin. Cell Biol. 48, 10–16. doi:10.1016/j.ceb.2017.03.005
Harvey, N. L., Daish, T., Mills, K., Dorstyn, L., Quinn, L. M., Read, S. H., et al. (2001). Characterization of the DrosophilaCaspase, DAMM. J. Biol. Chem. 276, 25342–25350. doi:10.1074/jbc.M009444200
Hawkins, C. J., Yoo, S. J., Peterson, E. P., Wang, S. L., Vernooy, S. Y., and Hay, B. A. (2000). The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J. Biol. Chem. 275, 27084–27093. doi:10.1074/jbc.M000869200
Hay, B. A., Wassarman, D. A., and Rubin, G. M. (1995). Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 83, 1253–1262. doi:10.1016/0092-8674(95)90150-7
Hay, B. A., Wolff, T., and Rubin, G. M. (1994). Expression of baculovirus P35 prevents cell death in Drosophila. Development 120, 2121–2129. doi:10.1242/dev.120.8.2121
Helfer, B., Boswell, B. C., Finlay, D., Cipres, A., Vuori, K., Bong Kang, T., et al. (2006). Caspase-8 promotes cell motility and calpain activity under nonapoptotic conditions. Cancer Res. 66, 4273–4278. doi:10.1158/0008-5472.CAN-05-4183
Horowitz, L. B., and Shaham, S. (2024). Apoptotic and nonapoptotic cell death in Caenorhabditis elegans development. Annu. Rev. Genet. doi:10.1146/annurev-genet-111523-102051
Hou, Y.-C. C., Chittaranjan, S., Barbosa, S. G., McCall, K., and Gorski, S. M. (2008). Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182, 1127–1139. doi:10.1083/jcb.200712091
Hu, S., and Yang, X. (2000). dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J. Biol. Chem. 275, 30761–30764. doi:10.1074/jbc.C000341200
Huh, J. R., Guo, M., and Hay, B. A. (2004a). Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 14, 1262–1266. doi:10.1016/j.cub.2004.06.015
Huh, J. R., Vernooy, S. Y., Yu, H., Yan, N., Shi, Y., Guo, M., et al. (2004b). Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2, E15. doi:10.1371/journal.pbio.0020015
Hung, V., Udeshi, N. D., Lam, S. S., Loh, K. H., Cox, K. J., Pedram, K., et al. (2016). Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475. doi:10.1038/nprot.2016.018
Ichim, G., Lopez, J., Ahmed, S. U., Muthalagu, N., Giampazolias, E., Delgado, M. E., et al. (2015). Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol. Cell 57, 860–872. doi:10.1016/j.molcel.2015.01.018
Igaki, T., Kanuka, H., Inohara, N., Sawamoto, K., Núñez, G., Okano, H., et al. (2000). Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc. Natl. Acad. Sci. U. S. A. 97, 662–667. doi:10.1073/pnas.97.2.662
Igaki, T., and Miura, M. (2004). Role of Bcl-2 family members in invertebrates. Biochim. Biophys. Acta Mol. Cell Res. 1644, 73–81. doi:10.1016/j.bbamcr.2003.09.007
Ikegawa, Y., Combet, C., Groussin, M., Navratil, V., Safar-Remali, S., Shiota, T., et al. (2023). Evidence for existence of an apoptosis-inducing BH3-only protein, sayonara, in Drosophila. EMBO J. 42, e110454. doi:10.15252/embj.2021110454
Ishizaki, Y., Jacobson, M. D., and Raff, M. C. (1998). A role for caspases in lens fiber differentiation. J. Cell Biol. 140, 153–158. doi:10.1083/jcb.140.1.153
Jamilloux, Y., Lagrange, B., Di Micco, A., Bourdonnay, E., Provost, A., Tallant, R., et al. (2018). A proximity-dependent biotinylation (BioID) approach flags the p62/sequestosome-1 protein as a caspase-1 substrate. J. Biol. Chem. 293, 12563–12575. doi:10.1074/jbc.RA117.000435
Julien, O., and Wells, J. A. (2017). Caspases and their substrates. Cell Death Differ. 24, 1380–1389. doi:10.1038/cdd.2017.44
Kamber Kaya, H. E., Ditzel, M., Meier, P., and Bergmann, A. (2017). An inhibitory mono-ubiquitylation of the Drosophila initiator caspase Dronc functions in both apoptotic and non-apoptotic pathways. PLoS Genet. 13, e1006438. doi:10.1371/journal.pgen.1006438
Kang, Y., Neuman, S. D., and Bashirullah, A. (2017). Tango7 regulates cortical activity of caspases during reaper-triggered changes in tissue elasticity. Nat. Commun. 8, 603. doi:10.1038/s41467-017-00693-3
Kanuka, H., Kuranaga, E., Takemoto, K., Hiratou, T., Okano, H., and Miura, M. (2005). Drosophila caspase transduces Shaggy/GSK-3beta kinase activity in neural precursor development. EMBO J. 24, 3793–3806. doi:10.1038/sj.emboj.7600822
Kanuka, H., Sawamoto, K., Inohara, N., Matsuno, K., Okano, H., and Miura, M. (1999). Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol. Cell 4, 757–769. doi:10.1016/s1097-2765(00)80386-x
Kaplan, Y., Gibbs-Bar, L., Kalifa, Y., Feinstein-Rotkopf, Y., and Arama, E. (2010). Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev. Cell 19, 160–173. doi:10.1016/j.devcel.2010.06.009
Kim, C.-H., Paik, D., Rus, F., and Silverman, N. (2014). The caspase-8 homolog Dredd cleaves Imd and Relish but is not inhibited by p35. J. Biol. Chem. 289, 20092–20101. doi:10.1074/jbc.M113.544841
Kleino, A., and Silverman, N. (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev. Comp. Immunol. 42, 25–35. doi:10.1016/j.dci.2013.05.014
Kolahgar, G., Bardet, P.-L., Langton, P. F., Alexandre, C., and Vincent, J.-P. (2011). Apical deficiency triggers JNK-dependent apoptosis in the embryonic epidermis of Drosophila. Development 138, 3021–3031. doi:10.1242/dev.059980
Kondo, S., Senoo-Matsuda, N., Hiromi, Y., and Miura, M. (2006). DRONC coordinates cell death and compensatory proliferation. Mol. Cell. Biol. 26, 7258–7268. doi:10.1128/MCB.00183-06
Koto, A., Kuranaga, E., and Miura, M. (2009). Temporal regulation of Drosophila IAP1 determines caspase functions in sensory organ development. J. Cell Biol. 187, 219–231. doi:10.1083/jcb.200905110
Koto, A., Kuranaga, E., and Miura, M. (2011). Apoptosis ensures spacing pattern formation of Drosophila sensory organs. Curr. Biol. 21, 278–287. doi:10.1016/j.cub.2011.01.015
Kumar, S., and Doumanis, J. (2000). The fly caspases. Cell Death Differ. 7, 1039–1044. doi:10.1038/sj.cdd.4400756
Kuranaga, E., Kanuka, H., Tonoki, A., Takemoto, K., Tomioka, T., Kobayashi, M., et al. (2006). Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell 126, 583–596. doi:10.1016/j.cell.2006.05.048
Kuranaga, E., Matsunuma, T., Kanuka, H., Takemoto, K., Koto, A., Kimura, K.-I., et al. (2011). Apoptosis controls the speed of looping morphogenesis in Drosophila male terminalia. Development 138, 1493–1499. doi:10.1242/dev.058958
Kuranaga, E., and Miura, M. (2007). Nonapoptotic functions of caspases: caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 17, 135–144. doi:10.1016/j.tcb.2007.01.001
Lam, S. S., Martell, J. D., Kamer, K. J., Deerinck, T. J., Ellisman, M. H., Mootha, V. K., et al. (2015). Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54. doi:10.1038/nmeth.3179
Larsen, B. D., Rampalli, S., Burns, L. E., Brunette, S., Dilworth, F. J., and Megeney, L. A. (2010). Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc. Natl. Acad. Sci. U. S. A. 107, 4230–4235. doi:10.1073/pnas.0913089107
Levayer, R., Dupont, C., and Moreno, E. (2016). Tissue crowding induces caspase-dependent competition for space. Curr. Biol. 26, 670–677. doi:10.1016/j.cub.2015.12.072
Li, F., He, Z., Shen, J., Huang, Q., Li, W., Liu, X., et al. (2010). Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell 7, 508–520. doi:10.1016/j.stem.2010.09.003
Li, J., Brieher, W. M., Scimone, M. L., Kang, S. J., Zhu, H., Yin, H., et al. (2007). Caspase-11 regulates cell migration by promoting Aip1-Cofilin-mediated actin depolymerization. Nat. Cell Biol. 9, 276–286. doi:10.1038/ncb1541
Li, M., Sun, S., Priest, J., Bi, X., and Fan, Y. (2019). Characterization of TNF-induced cell death in Drosophila reveals caspase- and JNK-dependent necrosis and its role in tumor suppression. Cell Death Dis. 10, 613. doi:10.1038/s41419-019-1862-0
Matamoro-Vidal, A., Cumming, T., Davidović, A., Levillayer, F., and Levayer, R. (2024). Patterned apoptosis has an instructive role for local growth and tissue shape regulation in a fast-growing epithelium. Curr. Biol. 34, 376–388.e7. doi:10.1016/j.cub.2023.12.031
Maurya, D., Rai, G., Mandal, D., and Mondal, B. C. (2024). Transient caspase-mediated activation of caspase-activated DNase causes DNA damage required for phagocytic macrophage differentiation. Cell Rep. 43, 114251. doi:10.1016/j.celrep.2024.114251
McSharry, S. S., and Beitel, G. J. (2019). The Caspase-3 homolog DrICE regulates endocytic trafficking during Drosophila tracheal morphogenesis. Nat. Commun. 10, 1031. doi:10.1038/s41467-019-09009-z
Meier, P., Silke, J., Leevers, S. J., and Evan, G. I. (2000). The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 19, 598–611. doi:10.1093/emboj/19.4.598
Meinander, A., Runchel, C., Tenev, T., Chen, L., Kim, C.-H., Ribeiro, P. S., et al. (2012). Ubiquitylation of the initiator caspase DREDD is required for innate immune signalling. EMBO J. 31, 2770–2783. doi:10.1038/emboj.2012.121
Montell, D. J. (2003). Border-cell migration: the race is on. Nat. Rev. Mol. Cell Biol. 4, 13–24. doi:10.1038/nrm1006
Mukherjee, A., Pop, S., Kondo, S., and Williams, D. W. (2021). Proapoptotic RHG genes and mitochondria play a key non-apoptotic role in remodelling theDrosophilasensory system. bioRxiv. doi:10.1101/2021.01.15.426850
Muramoto, M., Hanawa, N., Okumura, M., Chihara, T., Miura, M., and Shinoda, N. (2024). Executioner caspase is proximal to Fasciclin 3 which facilitates non-lethal activation in Drosophila olfactory receptor neurons. eLife 13, RP99650. doi:10.7554/eLife.99650.1
Myllymäki, H., Valanne, S., and Rämet, M. (2014). The Drosophila imd signaling pathway. J. Immunol. 192, 3455–3462. doi:10.4049/jimmunol.1303309
Nakajima, Y.-I., and Kuranaga, E. (2017). Caspase-dependent non-apoptotic processes in development. Cell Death Differ. 24, 1422–1430. doi:10.1038/cdd.2017.36
Nakajima, Y.-I., Kuranaga, E., Sugimura, K., Miyawaki, A., and Miura, M. (2011). Nonautonomous apoptosis is triggered by local cell cycle progression during epithelial replacement in Drosophila. Mol. Cell. Biol. 31, 2499–2512. doi:10.1128/MCB.01046-10
Nandi, N., Tyra, L. K., Stenesen, D., and Krämer, H. (2014). Acinus integrates AKT1 and subapoptotic caspase activities to regulate basal autophagy. J. Cell Biol. 207, 253–268. doi:10.1083/jcb.201404028
Newton, K., Strasser, A., Kayagaki, N., and Dixit, V. M. (2024). Cell death. Cell 187, 235–256. doi:10.1016/j.cell.2023.11.044
Nezis, I. P., Shravage, B. V., Sagona, A. P., Lamark, T., Bjørkøy, G., Johansen, T., et al. (2010). Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J. Cell Biol. 190, 523–531. doi:10.1083/jcb.201002035
Nordstrom, W., Chen, P., Steller, H., and Abrams, J. M. (1996). Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 180, 213–226. doi:10.1006/dbio.1996.0296
Ojha, S., and Tapadia, M. G. (2022). Nonapoptotic role of caspase-3 in regulating Rho1GTPase-mediated morphogenesis of epithelial tubes of Drosophila renal system. Dev. Dyn. 251, 777–794. doi:10.1002/dvdy.437
Orme, M. H., Liccardi, G., Moderau, N., Feltham, R., Wicky-John, S., Tenev, T., et al. (2016). The unconventional myosin CRINKLED and its mammalian orthologue MYO7A regulate caspases in their signalling roles. Nat. Commun. 7, 10972. doi:10.1038/ncomms10972
Oshima, K., Takeda, M., Kuranaga, E., Ueda, R., Aigaki, T., Miura, M., et al. (2006). IKKɛ regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr. Biol. 16, 1531–1537. doi:10.1016/j.cub.2006.06.032
Pérez-Garijo, A., Martín, F. A., and Morata, G. (2004). Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development 131, 5591–5598. doi:10.1242/dev.01432
Pérez-Garijo, A., Shlevkov, E., and Morata, G. (2009). The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 136, 1169–1177. doi:10.1242/dev.034017
Pop, S., Chen, C.-L., Sproston, C. J., Kondo, S., Ramdya, P., and Williams, D. W. (2020). Extensive and diverse patterns of cell death sculpt neural networks in insects. Elife 9, e59566. doi:10.7554/eLife.59566
Qin, W., Cho, K. F., Cavanagh, P. E., and Ting, A. Y. (2021). Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143. doi:10.1038/s41592-020-01010-5
Quinn, L., Coombe, M., Mills, K., Daish, T., Colussi, P., Kumar, S., et al. (2003). Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 22, 3568–3579. doi:10.1093/emboj/cdg355
Robbins, R. M., Gbur, S. C., and Beitel, G. J. (2014). Non-canonical roles for Yorkie and Drosophila Inhibitor of Apoptosis 1 in epithelial tube size control. PLoS One 9, e101609. doi:10.1371/journal.pone.0101609
Rodriguez, A., Oliver, H., Zou, H., Chen, P., Wang, X., and Abrams, J. M. (1999). Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat. Cell Biol. 1, 272–279. doi:10.1038/12984
Ryoo, H. D., Bergmann, A., Gonen, H., Ciechanover, A., and Steller, H. (2002). Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 4, 432–438. doi:10.1038/ncb795
Ryoo, H. D., Gorenc, T., and Steller, H. (2004). Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell 7, 491–501. doi:10.1016/j.devcel.2004.08.019
Sakahira, H., Enari, M., and Nagata, S. (1998). Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391, 96–99. doi:10.1038/34214
Schott, S., Ambrosini, A., Barbaste, A., Benassayag, C., Gracia, M., Proag, A., et al. (2017). A fluorescent toolkit for spatiotemporal tracking of apoptotic cells in living Drosophila tissues. Development 144, 3840–3846. doi:10.1242/dev.149807
Segawa, K., Kurata, S., and Nagata, S. (2016). Human type IV P-type ATPases that work as plasma membrane phospholipid flippases and their regulation by caspase and calcium. J. Biol. Chem. 291, 762–772. doi:10.1074/jbc.M115.690727
Segawa, K., Kurata, S., Yanagihashi, Y., Brummelkamp, T. R., Matsuda, F., and Nagata, S. (2014). Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168. doi:10.1126/science.1252809
Sevrioukov, E. A., Burr, J., Huang, E. W., Assi, H. H., Monserrate, J. P., Purves, D. C., et al. (2007). Drosophila Bcl-2 proteins participate in stress-induced apoptosis, but are not required for normal development. Genesis 45, 184–193. doi:10.1002/dvg.20279
Shinoda, N., Hanawa, N., Chihara, T., Koto, A., and Miura, M. (2019). Dronc-independent basal executioner caspase activity sustains Drosophila imaginal tissue growth. Proc. Natl. Acad. Sci. U. S. A. 116, 20539–20544. doi:10.1073/pnas.1904647116
Shinoda, N., Horikoshi, M., Taira, Y., Muramoto, M., Hirayama, S., Murata, S., et al. (2023). Caspase cleaves Drosophila BubR1 to modulate spindle assembly checkpoint function and lifespan of the organism. FEBS J. 290, 4200–4223. doi:10.1111/febs.16811
Shinoda, N., Obata, F., Zhang, L., and Miura, M. (2016). Drosophila SETDB1 and caspase cooperatively fine-tune cell fate determination of sensory organ precursor. Genes cells. 21, 378–386. doi:10.1111/gtc.12348
Shukla, A., and Tapadia, M. G. (2011). Differential localization and processing of apoptotic proteins in Malpighian tubules of Drosophila during metamorphosis. Eur. J. Cell Biol. 90, 72–80. doi:10.1016/j.ejcb.2010.08.010
Siegrist, S. E., Haque, N. S., Chen, C.-H., Hay, B. A., and Hariharan, I. K. (2010). Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Curr. Biol. 20, 643–648. doi:10.1016/j.cub.2010.01.060
Singh, R., Letai, A., and Sarosiek, K. (2019). Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20, 175–193. doi:10.1038/s41580-018-0089-8
Song, Z., McCall, K., and Steller, H. (1997). DCP-1, a Drosophila cell death protease essential for development. Science 275, 536–540. doi:10.1126/science.275.5299.536
Sun, G., Ding, X. A., Argaw, Y., Guo, X., and Montell, D. J. (2020). Akt1 and dCIZ1 promote cell survival from apoptotic caspase activation during regeneration and oncogenic overgrowth. Nat. Commun. 11, 5726. doi:10.1038/s41467-020-19068-2
Suzuki, J., Denning, D. P., Imanishi, E., Horvitz, H. R., and Nagata, S. (2013). Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406. doi:10.1126/science.1236758
Takemoto, K., Kuranaga, E., Tonoki, A., Nagai, T., Miyawaki, A., and Miura, M. (2007). Local initiation of caspase activation in Drosophila salivary gland programmed cell death in vivo. Proc. Natl. Acad. Sci. U. S. A. 104, 13367–13372. doi:10.1073/pnas.0702733104
Takemoto, K., Nagai, T., Miyawaki, A., and Miura, M. (2003). Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J. Cell Biol. 160, 235–243. doi:10.1083/jcb.200207111
Talanian, R. V., Quinlan, C., Trautz, S., Hackett, M. C., Mankovich, J. A., Banach, D., et al. (1997). Substrate specificities of caspase family proteases. J. Biol. Chem. 272, 9677–9682. doi:10.1074/jbc.272.15.9677
Tang, H. L., Tang, H. M., Fung, M. C., and Hardwick, J. M. (2015). In vivo CaspaseTracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Sci. Rep. 5, 9015. doi:10.1038/srep09015
Tian, L., Li, Y., and Shi, Y. (2024). Dark and dronc activation in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 121, e2312784121. doi:10.1073/pnas.2312784121
Timmer, J. C., and Salvesen, G. S. (2007). Caspase substrates. Cell Death Differ. 14, 66–72. doi:10.1038/sj.cdd.4402059
To, T.-L., Piggott, B. J., Makhijani, K., Yu, D., Jan, Y. N., and Shu, X. (2015). Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc. Natl. Acad. Sci. U. S. A. 112, 3338–3343. doi:10.1073/pnas.1502857112
Valanne, S., Wang, J.-H., and Rämet, M. (2011). The Drosophila Toll signaling pathway. J. Immunol. 186, 649–656. doi:10.4049/jimmunol.1002302
Verghese, S., and Su, T. T. (2016). Drosophila Wnt and STAT define apoptosis-resistant epithelial cells for tissue regeneration after irradiation. PLoS Biol. 14, e1002536. doi:10.1371/journal.pbio.1002536
Verghese, S., and Su, T. T. (2018). Ionizing radiation induces stem cell-like properties in a caspase-dependent manner in Drosophila. PLoS Genet. 14, e1007659. doi:10.1371/journal.pgen.1007659
Villars, A., Matamoro-Vidal, A., Levillayer, F., and Levayer, R. (2022). Microtubule disassembly by caspases is an important rate-limiting step of cell extrusion. Nat. Commun. 13, 3632. doi:10.1038/s41467-022-31266-8
White, K., Grether, M. E., Abrams, J. M., Young, L., Farrell, K., and Steller, H. (1994). Genetic control of programmed cell death in Drosophila. Science 264, 677–683. doi:10.1126/science.8171319
Williams, D. W., Kondo, S., Krzyzanowska, A., Hiromi, Y., and Truman, J. W. (2006). Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9, 1234–1236. doi:10.1038/nn1774
Wilson, R., Goyal, L., Ditzel, M., Zachariou, A., Baker, D. A., Agapite, J., et al. (2002). The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 4, 445–450. doi:10.1038/ncb799
Xu, D., Li, Y., Arcaro, M., Lackey, M., and Bergmann, A. (2005). The CARD-carrying caspase Dronc is essential for most, but not all,developmental cell death inDrosophila. Development 132, 2125–2134. doi:10.1242/dev.01790
Xu, D., Wang, Y., Willecke, R., Chen, Z., Ding, T., and Bergmann, A. (2006). The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ. 13, 1697–1706. doi:10.1038/sj.cdd.4401920
Xu, D. C., Wang, L., Yamada, K. M., and Baena-Lopez, L. A. (2022). Non-apoptotic activation of Drosophila caspase-2/9 modulates JNK signaling, the tumor microenvironment, and growth of wound-like tumors. Cell Rep. 39, 110718. doi:10.1016/j.celrep.2022.110718
Yosefzon, Y., Soteriou, D., Feldman, A., Kostic, L., Koren, E., Brown, S., et al. (2018). Caspase-3 regulates YAP-dependent cell proliferation and organ size. Mol. Cell 70, 573–587. doi:10.1016/j.molcel.2018.04.019
Yu, X., Wang, L., Acehan, D., Wang, X., and Akey, C. W. (2006). Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 355, 577–589. doi:10.1016/j.jmb.2005.10.040
Zhang, C., Casas-Tintó, S., Li, G., Lin, N., Chung, M., Moreno, E., et al. (2015). An intergenic regulatory region mediates Drosophila Myc-induced apoptosis and blocks tissue hyperplasia. Oncogene 34, 2385–2397. doi:10.1038/onc.2014.160
Zhang, J., Wang, X., Cui, W., Wang, W., Zhang, H., Liu, L., et al. (2013). Visualization of caspase-3-like activity in cells using a genetically encoded fluorescent biosensor activated by protein cleavage. Nat. Commun. 4, 2157. doi:10.1038/ncomms3157
Zhang, Y., Lin, N., Carroll, P. M., Chan, G., Guan, B., Xiao, H., et al. (2008). Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev. Cell 14, 481–493. doi:10.1016/j.devcel.2008.01.018
Keywords: caspase, caspase-dependent non-lethal cellular processes, Drosophila, caspase activity probes, TurboID, mass spectrometry
Citation: Shinoda N and Miura M (2024) Exploring caspase-dependent non-lethal cellular processes using Drosophila. Front. Cell. Death 3:1472108. doi: 10.3389/fceld.2024.1472108
Received: 28 July 2024; Accepted: 04 October 2024;
Published: 21 October 2024.
Edited by:
Yun Fan, University of Birmingham, United KingdomReviewed by:
Hui Xiao, Shaanxi Normal University, ChinaOlivier Meurette, Université Claude Bernard Lyon 1, France
Copyright © 2024 Shinoda and Miura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natsuki Shinoda, Zi1zaGlub2RhLm5hdHN1a2lAZy5lY2MudS10b2t5by5hYy5qcA==; Masayuki Miura, bWl1cmFAbW9sLmYudS10b2t5by5hYy5qcA==
 Natsuki Shinoda
Natsuki Shinoda Masayuki Miura
Masayuki Miura