
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 31 March 2025
Sec. Stem Cell Research
Volume 13 - 2025 | https://doi.org/10.3389/fcell.2025.1564889
With the progression of cardiovascular disease (CVD) treatment technologies, conventional animal models face limitations in clinical translation due to interspecies variations. Recently, human cardiac organoids (hCOs) have emerged as an innovative platform for CVD research. This review provides a comprehensive overview of the definition, characteristics, classifications, application and development of hCOs. Furthermore, this review examines the application of hCOs in models of myocardial infarction, heart failure, arrhythmias, and congenital heart diseases, highlighting their significance in replicating disease mechanisms and pathophysiological processes. It also explores their potential utility in drug screening and the development of therapeutic strategies. Although challenges persist regarding technical complexity and the standardization of models, the integration of multi-omics and artificial intelligence (AI) technologies offers a promising avenue for the clinical translation of hCOs.
Cardiovascular disease (CVD) encompasses a range of disorders affecting the circulatory system and constitutes a significant contributor to the global disease burden, with a reported prevalence of 523 million cases (Roth et al., 2020; Tsao et al., 2022). According to data published by the WHO, approximately 17 million individuals succumb to cardiovascular-related diseases each year, representing nearly 31% of global deaths. Projections indicate that by 2030, the annual mortality from CVD will rise to an estimated 23.3 million individuals (Chen et al., 2023). The prevalence and incidence of CVD vary markedly in different countries and regions, but the overall trend suggests that the burden of these diseases is increasing (Yeh et al., 2010; Dyrbuś et al., 2019; Mosterd and Hoes, 2007; Kusumoto et al., 2019; Heeringa et al., 2006). In recent years, the global prevalence of CVD has shown an expansion towards younger age groups (Andersson and Vasan, 2018), which is closely related to the increase in unhealthy lifestyle habits and environmental factors. Despite significant advances in CVD therapies, the emergence of novel agents such as proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors (LDL-cholesterol-lowering) (O'Donoghue et al., 2019) and sodium-glucose cotransporter 2 (SGLT2) inhibitors (endothelial function-improving) (Vallon and Verma, 2021) has redefined cardioprotection. By targeting distinct pathological pathways such as atherosclerosis via LDL-C reduction and endothelial dysfunction via glucuretic/anti-inflammatory effects, these drugs synergistically mitigate atherosclerotic progression and cardiovascular mortality. Contemporary animal models utilized in CVD research are typically genetically or surgically altered to manifest a spectrum of symptoms pertinent to specific diseases. While these models are instrumental in advancing research, the intrinsic species differences between humans and animals, coupled with the disparities at the organ and cellular levels, pose significant challenges in extrapolating animal study results to human clinical trials, often resulting in a clinical translational bottleneck (Wang et al., 2017). In contrast, the cultivation of human cardiac organoids (hCOs) in laboratory settings offers a promising avenue to bridge the gap between animal studies and clinical research (Keung et al., 2019). This approach provides an innovative method for investigating the physiological functions and pathological mechanisms of human heart diseases. As a next-generation model, hCOs largely preserve the biological properties and functions of in vivo cells, thereby significantly advancing CVD research.
The origin of organoid technology can be traced back to cell culture technology in the 1990s (Folkman and Moscona, 1978), and early cardiac research relied on two-dimensional (2D) cell culture models, which, although playing an important role in basic research, often failed to realistically reproduce the complex physiological environment and function of the heart (Schaaf et al., 2011). In contrast, this field has developed significantly in recent years with advances in microfluidics and biomaterials science (Nahak et al., 2022).The application of organoid technology in cardiac research has covered a wide range of aspects, including the establishment of cardiac disease models, drug screening and toxicity assessment, cardiac development and regeneration studies. For example, by combining oxygen diffusion gradients and norepinephrine stimulation, hCOs can accurately mimic myocardial infarction and hypoxia-enhanced doxorubicin cardiotoxicity in humans, which provides a valuable tool for drug screening and development (Richards et al., 2020). In addition, Plansky Hoang et al. identified a new combination of biomaterial-based cell patterning and stem cell organoid engineering technology to create 3D cardiac microchambers from 2D induced pluripotent stem cells (iPSCs), which can mimic early cardiac development and be used as an embryotoxicity screening assay (Hoang et al., 2018). Lee et al. (2020) developed a hCOs model derived from mouse embryonic stem cells to study the spatiotemporal regulation mechanisms of the heart, which was able to self-organize to form tissue structures similar to the real heart under specific conditions, with cardiac tissue structure of a real heart with myocardial contraction and action potential properties, providing a promising research tool for cardiac development and drug testing. In addition, organoid technology has been widely used for screening and toxicity testing of new drugs, providing more accurate drug response data than traditional cell culture models. Kenji Kosuke et al. cultured miniature colonic organoids in 96-well plates and successfully screened compounds for 2000 potential drugs (Kozuka et al., 2017). In conclusion, organoid technology has gained increasing importance in biomedical research as an important tool for exploring organ function, disease mechanisms and drug screening and development (Hofbauer et al., 2021; Ma et al., 2021).
Organoids are three dimensional (3D) cellular aggregates generated in vitro from stem cells or human induced pluripotent stem cells (hiPSCs). hCOs, which currently lack a standardized definition, are typically 3D microstructures that spontaneously arise from hiPSCs or other cardiac-related cells, partially mimicking the structure or function of cardiac tissues. These organoids encompass major cardiac cell types. These structures mimic the development, architecture, and function of the heart in vitro (Kim et al., 2020; Voges et al., 2017). Cardiac organs have many features: (1) 3D structure: Compared with traditional 2D cell culture models, 3D models can better reproduce the natural structure of cardiac tissues, including the arrangement of myocardial layers and the formation of vascular networks (Zhang and Radisic, 2017; Thavandiran et al., 2013), This is primarily due to the ability of 3D models to provide mechanical and biochemical cues that regulate cardiomyocyte alignment, extracellular matrix (ECM) deposition, and vascular development (Beauchamp et al., 2015). Furthermore, 3D hCOs create physiologically relevant cell-cell and cell-matrix interactions, supporting the maturation of cardiomyocytes and the development of functional cardiac tissue. These features enable hCOs to better mimic the dynamic microenvironment of the heart; (2) Cellular heterogeneity: A notable feature of hCOs is their high cellular heterogeneity, which allows them to mimic the complex cellular diversity of native cardiac tissue. hCOs typically contain various cell types, including cardiomyocytes, endothelial cells, fibroblasts, and cardiac progenitor cells (Ma et al., 2021; Ronaldson-Bouchard et al., 2018). This multicellular composition not only supports the structural integrity of cardiac tissue but also promotes the formation of a functional microenvironment (Ergir et al., 2022); (3) Functional analysis: weave-engineered cardiac patches promoted the functional maturation of human embryonic stem cell (hESC) derived cardiomyocytes (Zhang et al., 2013), utilized electrophysiological tests and other techniques to assess the functional properties of hCOs, including the excitability and conductivity of cardiomyocytes; (4) Disease modelling function: cardiac class organs can be used to simulate a variety of cardiac disorders. For example, Richards et al. (2020) demonstrated that hCOs can replicate the structural features of the heart after myocardial infarction, including the infarcted, marginal, and distal zones, making them a promising model for studying heart repair. Tiburcy et al. (2017) established a systematic approach for generating hCOs that closely mimic the structural and functional properties of the postnatal myocardium. Together, these studies highlight the potential of hCOs to serve as accurate models for both myocardial infarction and postnatal heart function, providing valuable insights for therapeutic development.
HCOs can be categorized into several types, each with unique methods and applications:
• Embryoid Bodies (EBs)-derived Organoids: Burridge and Paul W. utilized embryoid bodies’ cardiac differentiation potential to generate 3D cardiac tissues under specific culture conditions, which has provided valuable insights into early cardiac development and cell differentiation (Burridge et al., 2014). These models offer a foundational approach to studying cardiogenesis.
•Cardiac Microtissues: Through bioprinting technology, the precise arrangement of cardiac cells and biomaterials has been used to create tissues with complex structures and functions. Lind et al. (2017) advanced this field by developing instrumented cardiac microphysiological devices, allowing for high-throughput drug screening, toxicity assessments, and investigations into cardiac contraction and development. This approach enhances the fidelity and versatility of cardiac models for therapeutic testing.
•Engineered Cardiac Tissue Blocks: These models involve the 3D culturing of cardiomyocytes or stem cells on biological scaffolds to create functional cardiac tissues. Tao et al. (2018) demonstrated the creation of cardiac structures by seeding adult rat cardiomyocytes onto fibrin gel matrices, providing a useful tool for disease modeling and drug screening.
Together, these diverse types of hCOs contribute to a more comprehensive understanding of cardiac function and disease, with specific applications in drug discovery, disease modeling, and regenerative medicine.
Currently, organoids are classified as scaffold-free self-organization structures and co-cultures incorporating active materials (as shown in Figure 1).
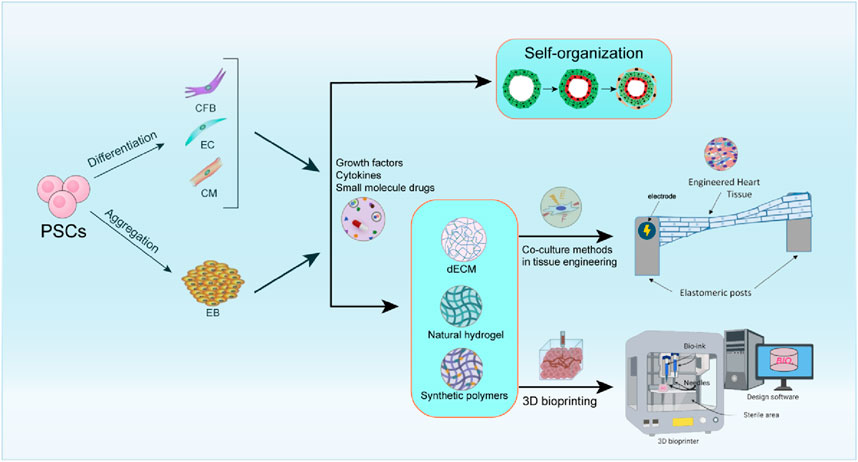
Figure 1. Overview of methods for the preparation of cardiac organoids. This includes scaffold-free self-organization methods, where cells assemble themselves to form cardiac tissue (Filippo Buono et al., 2020); tissue engineering co-culture methods, which combine different cell types to simulate the cardiac microenvironment; and 3D printing methods, which utilize precise bioprinting techniques to construct complex cardiac structures. Each of these approaches has unique advantages and contributes to advancements in cardiac regenerative medicine and disease modeling.
Self-organizing methodologies utilize stem cells with high differentiation potential alongside specific developmental signaling molecules to generate hCOs. These approaches elucidate cardiac development by directing stem cells toward the cardiac mesodermal lineage, promoting self-organization that generates diverse cell populations and enhances physiologically relevant cellular interactions critical for cardiac morphology and function (Yang et al., 2023). For example, Keung et al. (2019) was able to more realistically mimic the physiological and pharmacological properties of the human heart by using human hiPSCs to self-organize to generate organoid and tissue strips with ventricular features, providing a reliable model for drug screening and heart disease research, and this technological platform is particularly suited to the evaluation of drugs that affect myocardial contractility (i.e., positive and negative inotropic drugs). Recently, Hofbauer et al. (2021) developed a temporally regulated differentiation protocol utilizing key cardiogenic signaling pathway, encompassing both hESCs and hiPSCs. This method was facilitated through the incorporation of laminin 521/511, hPSCs demonstrated a self-assembly efficiency exceeding 90% into mesoderm, cardiac mesoderm, and beating cardiac progenitors within 2D cultures. Furthermore, these cells rapidly self-organized into beating luminal structures, termed “cardiac shapes,” in 3D non-adherent high-throughput cultures without the addition of exogenous extracellular matrix, also achieving 90% efficiency. These structures exhibited tissue characteristics reminiscent of myocardial nodules and intercalated discs. Electrophysiological assessments and calcium imaging were employed to evaluate the myocardial contractile function and electrical activity of the organoids. Lewis-Israeli et al. (2021) have successfully developed a time-regulated differentiation methodology that leverages key cardiogenic signaling pathways to facilitate the self-assembly of physiologically functional 3D hCOs. These organoids express a range of myocardial markers. This hCO model not only exemplifies the self-organization principles inherent in cardiac development but also offers a novel platform for the investigation of congenital heart disease and the screening of pharmaceutical compounds. Recently, Branco et al. (2022) successfully developed a self-organized multispectral embryonic organoid exhibiting early embryonic structures, such as primitive endoderm (PE), septal mesenchymal stromal cells, and liver buds, through the modulation of the WNT signaling pathway in induced hiPSCs. Richards et al. (2017) utilized hiPSCs in conjunction with cardiac fibroblasts to engineer biohybrid human heart-like structures. Firstly, scaffold-free self-organization techniques are constrained by inadequate nutrient and oxygen delivery, rendering them unsuitable for extensive clinical application. Secondly, these methods lack uniformity and do not permit precise regulation of cell proportions. Thirdly, self-assembled tissues exhibit greater fragility compared to those produced through conventional tissue engineering techniques, primarily due to a lack of adequate support materials and 3D structural integrity. Furthermore, self-assembled hCOs typically display immaturity. Therefore, additional research is needed to promote tissue and cell maturation in self-assembled hCOs.
Tissue engineering co-culture methodologies comprise three fundamental components: seed cells, scaffolds that provide structural support, and bioactive factors that modulate cellular behavior, all of which are integral to the development of 3D tissue and organ models (Borovjagin et al., 2017). The co-culture system significantly enhances the physiological relevance of heart organoids by simulating in vivo multicellular interactions. hiPSCs, with their robust proliferative capacity and patient-specific origin, have become the ideal seed cells in co-culture systems (Clevers, 2016). iPSC-derived hCOs can recapitulate the complete genomic features of the donor, providing an important tool for personalized drug testing and disease modeling (Yang et al., 2023).
Co-culture methodologies capitalize on the synergistic interactions between cardiomyocytes (CMs), endothelial cells (ECs), fibroblasts, and other cardiac cell types to recapitulate the complex cellular architecture of the heart.
In 2017, Polonchuk et al. (2017) have developed 3D in vitro models of the human heart, known as “cardiac spheroids,” by co-culturing human primary or iPSC-derived cardiomyocytes, endothelial cells, and fibroblasts in ratios that approximate those found in vivo. This approach successfully replicates the human cardiac microenvironment. Similarly, Mills et al. (2019) developed a 96-well high-throughput screening plate, the Heart-Dyno, for high-throughput screening of human pluripotent stem cell-derived heart organoids and optimization of their maturation conditions. This device promotes the formation of compact myofibrils and is capable of automatically analyzing the contraction force of heart organoids, significantly reducing the use of cells and reagents. The resulting hCOs exhibit structural similarities to native cardiac tissue, including highly organized cardiomyocytes, stromal cells, endothelial microvascular networks, and epicardial layers. Under culture conditions that simulate the postnatal metabolic environment, the maturity of hCOs is further enhanced, providing an efficient tool for drug screening and disease modeling. More recently, Drakhlis et al. (2021) successfully generated complex, highly structured 3D heart-forming organoids (HFOs) by embedding hPSCs aggregates in Matrigel and directing cardiac differentiation through biphasic WNT pathway modulation using small molecules. HFOs consist of a myocardial layer lined with endocardial-like cells and surrounded by septum-transversum-like tissue. Additionally, they contain spatially and molecularly distinct anterior and posterior foregut endoderm tissues, along with a developing vascular network. The architecture of HFOs closely resembles aspects of early native heart anlagen, particularly before heart tube formation. This model provides a new perspective for studying human heart development mechanisms and congenital heart diseases.
Biological active factors regulate cell signaling pathways in co-culture systems, driving the self-organization and functional maturation of hCOs (Unal and West, 2020). As Hofbauer et al. (2021) demonstrated, WNT and ACTIVIN dosages during the early stages of mesoderm differentiation influence the self-organization of cardiac-like organs. This regulation affects the patterning and morphogenesis of CMs and EC profiles. Additionally, the specification and patterning of the EC layer within the cardiac mesoderm are directed by VEGF levels and Other growth factors (Iyer et al., 2012; Montessuit et al., 2006; Yang et al., 2014; Parikh et al., 2017; Lee et al., 2017).
In co-culture systems, scaffold materials for cardiac organoids serve a pivotal role, primarily by mimicking the natural extracellular matrix (ECM) to offer structural support, biochemical signals, and mechanical cues (Capulli et al., 2016) (Xia and Chen, 2022). Scaffold materials can be classified into two primary categories based on their origin and properties: natural biomaterials (e.g., hydrogels and decellularized ECM) and synthetic polymers (Sharma et al., 2019; Tian et al., 2015; Cho et al., 2021; Pok et al., 2013). 1)Natural biomaterials, including natural hydrogels and decellularized extracellular matrix (dECM), offer significant advantages such as excellent biocompatibility and the ability to mimic natural ECM components. Hydrogels, which are 3D network structures constructed from natural or synthetic polymers, are particularly notable for their tunable mechanical properties and degradation rates (Radhakrishnan et al., 2014). However, their relatively low mechanical strength limits their application in high-load environments such as cardiac and other mechanically demanding organoids (Li et al., 2024). Decellularized extracellular matrix (dECM) retains the structural and biochemical components of the natural ECM through decellularization techniques, making it a popular scaffold material in cardiac bioengineering (Yang et al., 2023; Zhang et al., 2022; Jang et al., 2017). Jang et al. (2017) used dECM derived from cardiac tissues containing stem cells to enable improved intercellular interactions and differentiation capacity of stem cells, to provide a cardiac-like microenvironment for pre-vascularized constructs, and to have a beneficial impact on cardiac repair. 2) synthetic polymers are frequently employed in the construction of engineered cardiac tissues (Unal and West, 2020; Coenen et al., 2018; Saludas et al., 2017; Reis et al., 2016). Due to their customizability and reproducibility, they play a pivotal role in cardiac organoid scaffolds. Pok et al. (2013) engineered a polycaprolactone scaffold integrated with a gelatin-chitosan hydrogel by amalgamating diverse scaffolding materials. This innovation facilitated the seeding and migration of cardiomyocytes within a biomimetic environment while ensuring adequate tensile strength. Gelmi et al. (2016) developed an electroactive composite scaffold composed of PLGA fibers and a polypyrrole coating. This material integrates electrical conductivity, mechanical driving capability, and a biomimetic topological structure, providing a dynamic physico-chemical environment for hiPSCs. The scaffold can provide both electrical and mechanical stimulation without exhibiting cytotoxicity, significantly enhancing the expression of cardiac biomarkers.
Although co-culture systems have made significant progress in cardiac organoid development, they still face a series of challenges that limit their widespread application. The main issues include insufficient cell maturation, vascularization defects, and the difficulty of standardizing co-culture systems. Currently, some solutions have been proposed. For example, external stimuli such as mechanical stretching (Chiou et al., 2016; Lu et al., 2021; Li et al., 2020) and electrical pulses (Heidi Au et al., 2009) have been shown to promote the maturation of cardiomyocytes. In addition, the introduction of pro-angiogenic factors, such as VEGF, effectively promotes the proliferation of EC, thereby forming functional vascular networks within the cardiac organoids (Li et al., 2024).
The process of 3D bioprinting involves layer-by-layer positioning of biological agents within 3D layers of tissue, thereby accurately replicating complex geometries of the heart such as cardiac spheroids (Beauchamp et al., 2020; Giacomelli et al., 2020; Filippo Buono et al., 2020), cardiac patches (Zhang et al., 2013; Jackman et al., 2018; Gao et al., 2018), cardiac specimens (Feric et al., 2019), and cardiac rings (Goldfracht et al., 2020) for a various applications. 3D bioprinting technology has the capability to fabricate perfused vascular networks and diverse cardiac tissue structures (Borovjagin et al., 2017). Basara et al. (2021) prepared a bioink was formulated by combining human induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CMs) with hCFs. This bioink was utilized to print the infarct border region, effectively replicating the infarcted area surrounded by healthy tissue, but it was not able to achieve physiologically accurate stiffness of myocardium. Lee et al. (2019) employed collagen as a bioink to fabricate the outer shell of a human ventricular model. This model demonstrated synchronized contraction and directional propagation of action potentials, with the ventricular wall exhibiting a 14% increase in thickness during contraction after 7 days. Additionally, the researchers produced three 28 mm collagen heart valves and improved their mechanical properties using a decellularized porcine heart valve fixation protocol. At the adult scale, the collagen valves had well-separated leaflets, were well-suited to handling in air, and demonstrated mechanical integrity and functionality. Gaetani and colleagues (Gaetani et al., 2012) demonstrated that microstructural tissue printing using a combination of alginate scaffolds with human fetal cardiomyocyte progenitors (hCMPCs) could be used to fabricate cardiac-derived patches with defined pore sizes and improved viability. A novel cardiogenic scaffold consisting of hCMPCs and hyaluronic acid/gelatin (HA/gel)-based biomaterials was created to advance the attachment and survival of the hCMPCs without compromising their growth and differentiation potential. Kawai et al. (2021) utilized hiPSCs to develop engineered heart tissue (EHT), which holds promise as a potential cardiac tissue replacement for heart failure treatment. To fabricate pulsatile catheters for patients with congenital heart disease, they employed hiPSC-COs and bio-3D printing to construct stentless tubular EHT (T-EHT) and transplante into the abdominal aorta and around the inferior vena cava (IVC) of NOG mice. Post-transplantation, the T-EHT was enveloped by the omentum, and the abdominal cavity was sutured closed upon completion of the procedure. Furthermore, to evaluate the functional performance of hiPSC-COs in comparison to T-EHTs, the hiPSC-COs were transplanted in proximity to the aorta and IVC, as well as injected into the subcutaneous tissues. The beating of T-EHTs in mice was observed 1 month after the transplantation procedure. In histological analyses, T-EHTs showed distinct myocardial striations and angiogenesis compared with hiPSC-COs. T-EHTs bio-3D printed showed better maturation in vivo than hiPSC-COs. These beating T-EHTs may serve as a conduit for patients with congenital heart disease, making T-EHT transplantation a therapeutic option. By integrating hiPSC-CMs, ECs, and ECM, Noor et al. (2019) created cardiac parenchymal tissue, blood vessels, and functional cardiac patches. Kupfer et al. (2020) fabricate vascular inlet and outflow constructs using optimized ECM protein bio-inks. Nonetheless, several challenges persist in this field.
Although promising, the field of 3D bioprinting for cardiac applications continues to face significant challenges, including achieving precise tissue mechanics, scalability, and long-term tissue functionality post-transplantation (Basara et al., 2021; Hwang et al., 2024; Mehrotra et al., 2020). Further advancements are needed to enhance the viability, maturity, and integration of printed tissues in clinical settings.
Significant advancements have been achieved thus far in the development of platforms for cardiovascular modeling. hCOs provide extensive advantages for investigating cardiac function under pathological condition. In addition to myocardial ischemic injury, contractile dysfunction, and aberrant electrophysiological activity, these advancements may stimulate research in various fields of cardiac physiology (as shown in Figure 2).
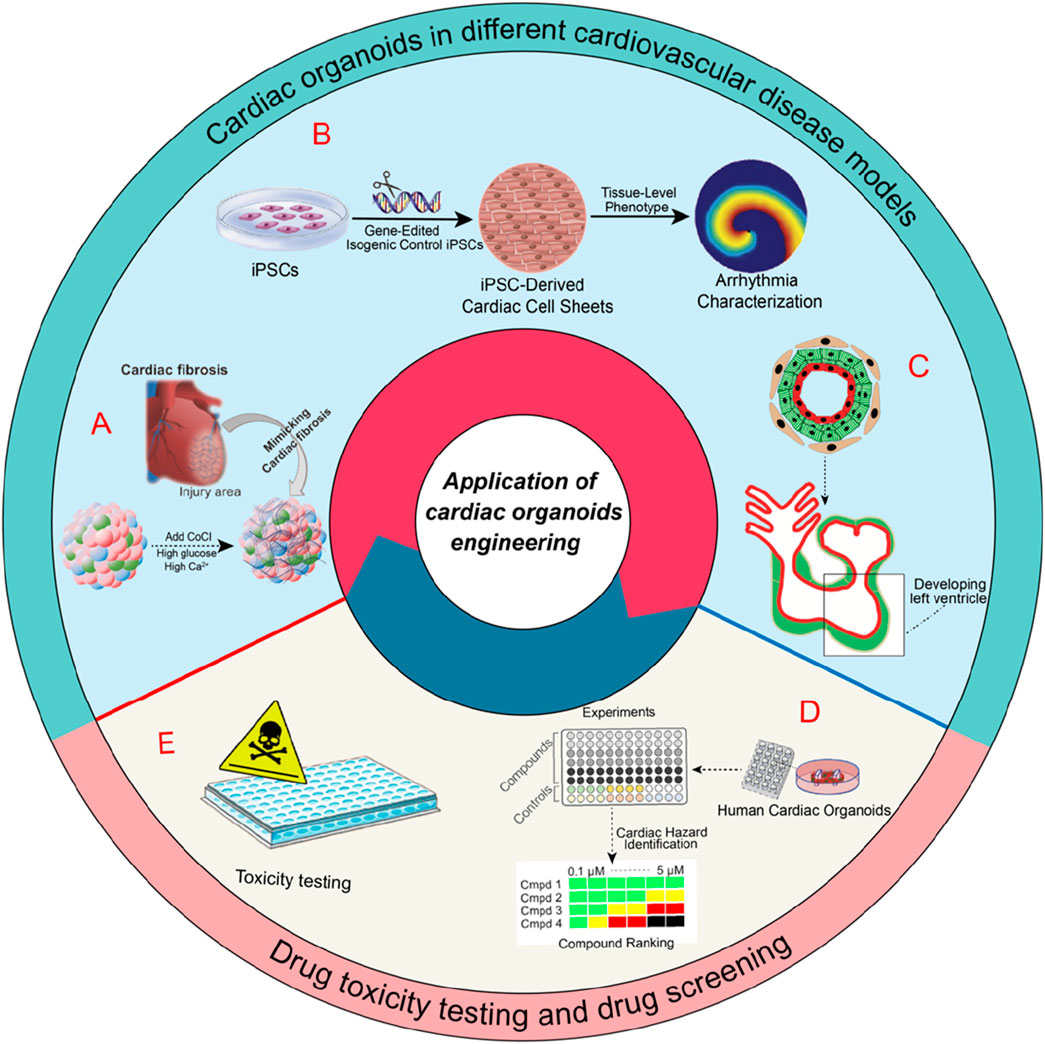
Figure 2. Applications of cardiac organoids in various cardiovascular disease models and their roles in drug toxicity testing and drug screening. A) schematic of an experiment mimicking heart disease in organoids via the IR injury mechanism occurring in the human adult heart, followed by the study of fibrogenesis (Song et al., 2024). B) cardiac organoids in arrhythmia research (Shinnawi et al., 2019). C) modeling heart chamber development and disease (Hofbauer et al., 2021). D) Illustrates the role of hCOs in drug screening for cardiovascular diseases. E) their application in drug toxicity testing.
Organoid models of myocardial infarction or injury represent some of the earliest established hCO models for studying cardiovascular disease. Voges and his colleagues (Voges et al., 2017) developed hCOs by employing collagen-gel cultures of hPSC-CMs within circular molds. They successfully simulated acute myocardial infarction by subjecting the hCOs to cryoinjury, resulting in localized tissue damage. Richards et al. (2020) employed non-adhesive agarose hydrogel molds to construct a composite of hiPSC-CMs and non-cardiomyocytes, which were utilized to generate hCOs. Using transcriptomic, structural, and functional characteristics of myocardial infarction, the researchers successfully developed a non-genetic in vitro model of hCOs. This approach facilitated the replication of infarcts, border zones, and remote regions of the post-infarction heart.
Nevertheless, the aforementioned hCOs lack the ability to incorporate the full spectrum of heart cell types found in vivo. After myocardial infarction, immune and inflammatory cells are crucial to the initial response and the subsequent fibrotic response (Forte et al., 2018). Song et al. (2024) employed hiPSCs to generate self-organizing heart-like organoids (HOs). These organoids comprised cardiomyocytes, fibroblasts, and endothelial cells, effectively replicating the cellular composition of the human heart and the multicellular architecture of HCOs. The organoids were then exposed to hypoxia-induced ischemia and ischemia-reperfusion (IR) injury under controlled conditions, which resulted in a phenotype analogous to acute myocardial infarction. Furthermore, IR-exposed HOs showed significant cardiac fibrosis, collagen deposition, and electrophysiological abnormalities similar to heart disease. The results are significant for the advancement of 3D cardiac models and myocardial infarction models in vivo.
The key pathophysiological features of Heart failure (HF)include systolic and diastolic dysfunction, cardiomyocyte hypertrophy (Du et al., 2024), apoptosis, and neurohumoral activation (e.g., catecholamine overstimulation) (Ren et al., 2024). Despite advancements in therapeutic strategies, HF remains a leading cause of morbidity and mortality worldwide (Najjar et al., 2024), underscoring the urgent need for innovative preclinical models to study its mechanisms and develop targeted therapies. Similarly, Tiburcy et al. (2017) presented a method for developing hCOs derived from postnatal myocardium utilizing hiPSC-CMs and fibroblasts. To simulate human heart failure, they implemented a neurohumoral overstimulation protocol. hCOs models showed responses to chronic catecholamine toxicity, including systolic dysfunction, a diminution in positive force-frequency response, cardiomyocyte hypertrophy, cardiomyocyte apoptosis, and desensitization to adrenergic signaling, and the release of cardiac biomarkers. These responses collectively represent hallmark characteristics of heart failure. Significantly, the inhibition of β1-adrenergic and α1-adrenergic receptors by metoprolol and phenoxybenzamine partially or entirely mitigates the pathological phenotype associated with heart failure. However, immune cells are crucial in the pathophysiology of myocardial damage (Swirski and Nahrendorf, 2018). Nonetheless, using cardiac-like organoids are deficient in immune cells and immature cardiomyocytes. This deficiency presents a significant challenge in accurately replicating the functional roles of immune cells and mature CMs in these conditions within hCOs.
hCOs surpass 2D models in arrhythmia research by leveraging their enlarged surface area and 3D geometry to model reentrant circuits through spatially heterogeneous electrical propagation (Drakhlis et al., 2021). Therefore, hCOs are an excellent disease model for studying arrhythmias. Shinnawi et al. (2019) advanced the application of hCOs in conjunction with distinctive hiPSC-CM sheets. A patient-specific hiPSC-CM approach combines CRISPR/Cas9 genome editing technologies with organoids to replicate arrhythmic events. The study demonstrated that the increased susceptibility of reentrant arrhythmias in thin slices of cardiomyocytes derived from short QT syndrome (SQTS) hiPSCs was consistent with the heightened inducibility of ventricular fibrillation. The authors employed the hCOs model to analyze the role of novel antiarrhythmic drug candidates. The findings indicated that quinidine and dipyridamole resulted in the prolongation of action potentials and action potential duration, as well as the suppression of arrhythmias. This study offers evidence that hCOs can effectively generalize the phenotype of SQTS. Gold and colleagues (Goldfracht et al., 2020) engineered hCOs comprising chamber-specific hPSC-CMs, and the study suggests that atrial hCOs are promising models for studying recurrent arrhythmias and shows that flecainide and vernakalant can effectively terminate reentrant activity in these models.
Laksman et al. (2017) generated atrial-specific tissue models for pharmacological testing using hESC. Compared with ventricular-like cardiomyocytes, they generated atrial-like cardiomyocytes that expressed atrial-specific genes and had shorter action potential (AP) durations. Using optical labeling techniques, they generated atrial-like CM slices and observed fast-folding rotor patterns in these slices. Using hESC-derived atrial CM preparations, they then demonstrated that flecainide and dovetailed modulate the foldback arrhythmogenic rotor activation pattern, which explains their efficacy in treating and preventing atrial fibrillation. It is noteworthy, however, that these hCOs lack relative chamber-specific structures and the expression of ion channel-related genes differs from that of adults. hCOs represent a significant advancement in the modeling of arrhythmias, offering a platform to study disease-specific mechanisms, test novel therapeutics, and explore the electrophysiological properties of human cardiac tissues. However, to fully realize their potential, further efforts are needed to enhance their maturity, ion channel expression, and structural complexity, ensuring that these models can accurately replicate the full range of arrhythmic events observed in humans.
For several decades, the genetic underpinnings of familial cardiomyopathies, including hypertrophic and dilated cardiomyopathy, have been recognized, and congenital heart defects are identified as the most prevalent human birth anomalies. Nonetheless, the comprehension of the pathogenesis of these conditions is constrained by the current limitations in accurately replicating the human heart in vitro. hiPSC-derived cellular models exhibit numerous characteristics emblematic of human diseases, such as disease-specific gene expression profiles, altered cellular morphology, dysfunctional ion channel activity, and abnormal contractile behavior (Pré et al., 2022), making them valuable tools for investigating the genetic and congenital underpinnings of these conditions. Researchers are increasingly using multicellular hCOs to examine cell-cell interactions in disease pathogenesis, advancing disease research. Marini et al. (2022) used hiPSCs from patients with Duchenne Muscular Dystrophy (DMD) to self-organize into hCOs (DMD-hCOs) in long-term cultures, but the DMD-hCOs were not proliferative at the beginning. A novel mutation in myosin heavy chain 7 (E848G) in familial cardiomyopathies was investigated by Yang et al. (2018), in which hCOs were interconnected to a force transducer and a length controller to assess tissue tectonic forces. Importantly, hCOs derived from familial cardiomyopathy exhibited significantly less alignment compared to the broader hCO cohort. Additionally, hCOs carrying the E848G mutation demonstrated a marked reduction in systolic function, while diastolic function remained largely unaffected. These results hold greater physiological relevance as they successfully replicate the contraction of aligned myogenic fibers within a 3D system. Lewis-Israeli et al. (2021) reported a method for generating developmentally relevant human cardiac-like organs by self-assembly using hiPSCs. At the transcriptomic, structural, and cellular levels, cardiac-like organs are comparable to age-matched fetal heart tissue. They can effectively model features of congenital heart disease induced by pregestational diabetes, offering valuable insights into the molecular pathology of these conditions.
Hereditary and congenital heart diseases remain challenging to model accurately due to limitations in hCO maturation and structural complexity. Future research should focus on improving maturation, vascularization, and chamber-specific models to better replicate conditions like familial cardiomyopathies and congenital defects such as septal defects (Fang et al., 2023; Ching et al., 2022).
hCOs can also be used to study the developmental biology of the embryo. Rossi and his colleagues (Rossi et al., 2021) successfully modeled the early stages of cardiac development, including the formation of a vascular-like network, the production of FHF/SHF, and the emergence of crescentic structures using axial-mode mouse gastrula-like proteins. A study by Wu et al. (2022) explored the effects of cadmium (Cd) on development of the heart in 2D and 3D using an in vitro embryonic model as well as cardiomyocytes in 2D and 3D, alongside a hCOs formation model that simulates early cardiac development. Their study conclusively demonstrated that exposure to Cd at a concentration of 0.6 mM resulted in a transient upregulation of the mesoderm-related transcription factors MESP1 and EOMES during the initial phase of mesoderm induction, followed by a downregulation during the later stages of heart induction. This effect was attributed to the inhibition of the Wnt/β-catenin signaling pathway. Branco et al. (2022) employed hiPSCs cultured in a 3D environment to promote the differentiation of proepicardial (PE) and epicardial cells through the modulation of WNT, BMP, and RA signaling pathways, thereby facilitating the formation of PE/STM/PFH-like organoids. These organoids were subsequently co-cultured with cardiomyocyte aggregates to produce cardiac-like organoids, termed engineered myocardial organoids. The findings from this study underscore the pivotal role of WNT and RA signaling in the canonical development of organoids, while also highlighting the active involvement of BMP4 signaling in the canonical development of liver bud-like cells. Mills et al. (2017) demonstrated that a shift in fatty acid metabolism plays a pivotal role in cardiac maturation by suppressing key proliferative pathways, particularly those involving β-catenin and YAP1. Their findings revealed that the proliferation impairment induced by fatty acid metabolism could be rescued through overexpression of β-catenin and YAP1 or pharmacological activation of these pathways using a small molecule (Compound 6.28). These results provide critical insights into the metabolic mechanisms underlying fatty acid metabolism-induced cell cycle arrest in cardiomyocytes, highlighting potential therapeutic strategies to modulate cardiac proliferation and maturation. Lewis-Israeli et al. (2021) investigated the impact of preconception diabetes on cardiac development by culturing hCOs in conditions of elevated glucose and insulin. These methodologies facilitate the rapid in vitro modeling of certain aspects of cardiac development. Nevertheless, the current applicability of these models is restricted to the initial stages of embryonic development. Consequently, there is a pressing need to advance the design of more precise cardiac organoids to establish highly accurate and reproducible culture models.
To date, the modeling of hCOs has proven effective in simulating pathological processes and cardiac development, serving as a complementary tool to preclinical models. However, hCOs have not yet fully supplanted characteristic parameters of animal models.
There is a high likelihood of adverse drug reactions in the heart and liver because they are particularly sensitive organs. Cardiotoxicity is responsible for 31% of adverse drug reactions (Wilke et al., 2007). There can be structural damage to cardiac tissue and impairment of cardiac function as a result of drug-induced cardiovascular toxicity, manifested as arrhythmias and altered cardiac contractility (Bowes et al., 2012). hiPSC-CM tissues have been shown to be excellent predictors of drug-induced arrhythmias and contractility by incorporating them (Peters et al., 2015). Beck et al. (2022) employed hCOs to evaluate the direct cardiotoxic effects of trametinib, observing a reduction in both the diameter and contractility of trametinib-treated hCOs in comparison to untreated controls, findings that align with existing in vivo data. Furthermore, the Cardiac Arrhythmia Suppression Test (CAST) has been developed to demonstrate the effectiveness of antiarrhythmic drugs in preventing ventricular arrhythmias associated with ischemic cardiomyopathy. However, some outcomes were attributed to an unforeseen increase in mortality due to arrhythmic events and shock. Kopljar et al. (2018) employed a calcium transient screening assay in cardiac cells derived from hiPSC-CMs to establish a hazard scoring system for assessing cardiac electrical liability. This methodology facilitated the integration of diverse pharmacological effects observed in hiPSC-CMs into a unified hazard label, thereby enabling the evaluation of antiarrhythmic drugs for potential drug toxicity. Sharma et al. (2017) employed hiPSC-CMs to assess cardiomyocyte viability, contractility, and changes in electrophysiology, calcium handling, and signaling pathways for screening the cardiotoxicity of tyrosine kinase inhibitors (TKIs). Based on these assessments, they developed a “cardiac safety index” to quantify the cardiotoxic potential of existing TKIs. They found that TKIs inhibited vascular endothelial growth factor receptor 2 (VEGFR2) and platelet-derived growth factor receptor (PDGFR) to cause cardiotoxicity in hiPSC-CMs, hiPSC-ECs, and hiPSC-CFs. Through phosphoprotein analysis, the researchers ascertained that the inhibition of VEGFR2/PDGFR by TKIs led to a compensatory enhancement in cardioprotective signaling pathways mediated by insulin and IGF in hiPSC-CMs. The upregulation of these cardioprotective pathways through the administration of exogenous insulin or IGF1 was found to enhance the viability of hiPSC-CMs during concurrent treatment with cardiotoxic TKIs targeting VEGFR2/PDGFR. Consequently, hiPSC-CMs serve as a viable model for detecting cardiovascular toxicity associated with anticancer TKIs, with the experimental findings demonstrating a correlation with clinical phenotypes.
Because of their accessibility, scalability, and genetic stability, organoids are extensively used in high-throughput drug screening. hCOs can replicate the microenvironment of the native heart, potentially making them superior models for preclinical drug screening. For instance, hiPSC-CMs have been employed to detect adverse reactions to targeted pharmaceuticals utilizing next-generation sequencing and proteomic analyses. A recent investigation utilized hydrogen peroxide to induce ischemic injury in hiPSC-CMs in vitro, leading to the identification of a novel small molecule compound that enhances the survival of hiPSC-CMs post-ischemic insult. This study underscores the potential for tissue-specific high-throughput screening of drug compounds derived from hiPSCs (Fiedler et al., 2020). Mills et al. (2019) employed the hCOs platform to evaluate 105 small molecules with regenerative potential. Their study revealed significant disparities between the hCO system and traditional 2D assays. Furthermore, hCO platforms are applicable for screening drugs that facilitate organoid growth, maturation, and function. The advancement of complex organoids to simulate organ-organ interactions has emerged as a critical objective in the screening of drug candidates. Skardal et al. (2020) developed a sophisticated multi-organ-on-a-chip platform, utilizing 3D organoid technology derived from primary stem cells. This platform not only sustains in vitro survival for a minimum of 28 days but also preserves long-term viability and functionality. A multi-organ-on-a-chip system was employed to screen drugs withdrawn by the Food and Drug Administration (FDA).
While hCOs serve as valuable models for drug testing, they remain significantly different from a natural heart. Enhancing the structural and functional resemblance of hCOs to the natural heart, alongside investigating the impact of the cardiac microenvironment and other organ systems on cardiac drug toxicity, could improve the reliability of drug screening and toxicity assessments (as shown in Figure 2).
Known as miniature and simplified in vitro models, organoids simulate the structural and functional attributes of organs and have garnered significant interest for possible applications in disease modeling, drug screening, and personalized medicine. However, they face many challenges, particularly when it comes to model complexity, data analysis, and standardization. Your choice of matrix material is crucial for your model construction (Jose et al., 2020; Xu et al., 2021; Kloxin et al., 2009). The advent of genome editing techniques has significantly advanced the development of homologous hPSCs, which are extensively utilized in two-dimensional disease models. In parallel, the integration of genome editing technologies enables precise modification of human cerebral organoids to correct mutations, thereby facilitating the creation of innovative and personalized therapeutic platforms for disease modeling. The CRISPR/Cas9 system represents a cutting-edge genome editing technology for the correction and mitigation of disease-associated mutations (Yang et al., 2018). Multidimensional interactions among diverse cell types are typically involved in the growth and development of organoids. The analysis and processing of multi-omics sequencing data presents substantial challenges to conventional analytical methodologies in organoid research (Zhou et al., 2023; Tang et al., 2023). In the broader field of data analysis, the absence of standardized protocols and real-time monitoring techniques exacerbates this issue, introducing considerable variability into the system. This variability is further intensified by manual data processing and subjective interpretations (Peng et al., 2018; Lukonin et al., 2021). As organoids increase in complexity, there is a pressing demand for the development of novel technologies to address the challenges of complexity and standardization inherent in organoid research. Recently, the advent of artificially intelligent organoids and artificial intelligence (AI) interfaces has the potential to expedite the development and clinical application of organoids by offering innovative insights and methodologies that could transform the field (Bai et al., 2024). Machine learning has emerged as a potent methodology for analyzing cellular electrochemical impedance spectroscopy measurements, demonstrating its capacity to distinguish between proliferative and differentiation behaviors. This capability holds significant potential for diverse applications in stem cell implantation therapies (Cunha et al., 2019). Kegeles et al. (2020) have developed a deep learning-based computational algorithm for the identification and prediction of retinal structures in stem cell-derived organoids using bright-field imaging. The differentiation of pluripotent stem cells into 3D organoids, representing the retina and other neural tissues, has emerged as a prominent in vitro strategy for modeling developmental processes. Meanwhile, Park et al. (2023) employed AI algorithms to predict the differentiation status of stem cells into kidney organoids, leading to the formation of more complex organoids. The integration of AI in organoid development holds significant potential for enhancing our understanding of the intricate biological systems inherent in various organoids. Furthermore, the emergence of multi-omics analyses, integrating genomic, transcriptomic, proteomic, and metabolomic data, has allowed researchers to model the development of human cerebral organoids with greater precision, while also shedding light on the intricate mechanisms underlying disease. According to Gu et al. (2022), several molecular events underlie the maturation and development of the developing heart based on an integrated multi-omics analysis. Their findings provide novel insights into the multilevel regulatory mechanisms governing developmental transitions.
In particular, AI technologies can be applied to optimize the design of matrix gels with desirable physicochemical properties (Suwardi et al., 2022; Verheyen et al., 2023). Moreover, the refinement of cell culture conditions through machine learning techniques holds the potential to diminish variability and minimize errors by automating the cell culture process. This automation facilitates the achievement of more consistent and reproducible results, while also supporting higher throughput (Kanda et al., 2022). Using extensive datasets of organoid-scale images, we can train machine learning algorithms to recognize and quantify diverse organoid features (Gritti et al., 2021). AI plays a pivotal role in the preclinical evaluation and application stages of organoid research. Through the utilization of predictive models and optimization algorithms, AI facilitates the assessment of mechanisms underlying organoid intervention development, the screening of potential pharmacological agents, and the construction of in vitro disease models. These applications are crucial for bridging the gap between fundamental research and clinical implementation (He et al., 2023; Kong et al., 2020; Monzel et al., 2020). The application of AI in multi-omics data analysis facilitates the integration of diverse functional information and the establishment of construction parameters. This approach enhances the efficiency and quality of organoid development, thereby expediting the progression from laboratory research to clinical applications.
The heart is an essential organ with critical functions, yet the complexities of cardiogenesis are not fully understood. Consequently, there is a pressing need to develop in vitro models of cardiac development and cardiovascular disease. hCOs have thus far demonstrated their value as complementary tools to in vivo studies. However, some challenges remain for the organoids. Firstly, the large-scale differentiation of organoid cells continues to present substantial challenges in terms of efficiency, quality, and orientation. Secondly, there is a pressing need to amalgamate advanced expertise in biology, materials science, and bioengineering to facilitate the construction of more precise and complex organ systems. Thirdly, the spontaneous formation of organoids relies on endogenous physiological mechanisms. The precise coordination of these signaling pathways during the developmental process is of paramount importance. It is essential to integrate these processes with validated methodologies, utilizing a range of established technologies, to advance hCOs technology from basic scientific applications to translational research with extensive and practical implications.
Facts:
• hCOs as an innovative model for CVD research, offering human-relevant insights.
•Applications in modeling myocardial infarction, heart failure, arrhythmias, and congenital heart diseases.
•Potential for drug screening and developing personalized therapeutic strategies.
•Integration of multi-omics and AI for improving model accuracy and clinical translation.
•Challenges in large-scale differentiation and model standardization.
Open questions:
•How to improve large-scale differentiation and reproducibility of hCOs?
•What are the key signaling pathways involved in hCO development?
•How to integrate bioengineering to enhance hCO complexity?
•Can hCOs be used for personalized medicine in CVD?
• What are the long-term implications of hCOs in drug testing and clinical research?
HZ: Writing – original draft, Writing – review and editing. PQ: Writing – original draft. JL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. PC: Writing – review and editing. QL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review and editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by National Natural Science Foundation of China (82470290), Sichuan Provincial Science and Technology Program (2023YFS0036) and Natural Science Foundation of Sichuan Province (No. 2023NSFSC0585).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Andersson, C., and Vasan, R. S. (2018). Epidemiology of cardiovascular disease in young individuals. Nat. Rev. Cardiol. 15 (4), 230–240. doi:10.1038/nrcardio.2017.154
Bai, L., Wu, Y., Li, G., Zhang, W., Zhang, H., and Su, J. (2024). AI-enabled organoids: construction, analysis, and application. Bioact. Mater 31, 525–548. doi:10.1016/j.bioactmat.2023.09.005
Basara, G., Ozcebe, S. G., Ellis, B. W., and Zorlutuna, P. (2021). Tunable human myocardium derived decellularized extracellular matrix for 3D bioprinting and cardiac tissue engineering. Gels 7 (2), 70. doi:10.3390/gels7020070
Beauchamp, P., Jackson, C. B., Ozhathil, L. C., Agarkova, I., Galindo, C. L., Sawyer, D. B., et al. (2020). 3D Co-culture of hiPSC-derived cardiomyocytes with cardiac fibroblasts improves tissue-like features of cardiac spheroids. Front. Mol. Biosci. 7, 14. doi:10.3389/fmolb.2020.00014
Beauchamp, P., Moritz, W., Kelm, J. M., Ullrich, N. D., Agarkova, I., Anson, B. D., et al. (2015). Development and characterization of a scaffold-free 3D spheroid model of induced pluripotent stem cell-derived human cardiomyocytes. Tissue Eng. Part C Methods 21 (8), 852–861. doi:10.1089/ten.TEC.2014.0376
Beck, T. C., Arhontoulis, D. C., Morningstar, J. E., Hyams, N., Stoddard, A., Springs, K., et al. (2022). Cellular and molecular mechanisms of MEK1 inhibitor-induced cardiotoxicity. JACC CardioOncol 4 (4), 535–548. doi:10.1016/j.jaccao.2022.07.009
Borovjagin, A. V., Ogle, B. M., Berry, J. L., and Zhang, J. (2017). From microscale devices to 3D printing: advances in fabrication of 3D cardiovascular tissues. Circ. Res. 120 (1), 150–165. doi:10.1161/CIRCRESAHA.116.308538
Bowes, J., Brown, A. J., Hamon, J., Jarolimek, W., Sridhar, A., Waldron, G., et al. (2012). Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat. Rev. Drug Discov. 11 (12), 909–922. doi:10.1038/nrd3845
Branco, M. A., Dias, T. P., Cabral, J. M. S., Pinto-do-Ó, P., and Diogo, M. M. (2022). Human multilineage pro-epicardium/foregut organoids support the development of an epicardium/myocardium organoid. Nat. Commun. 13 (1), 6981. doi:10.1038/s41467-022-34730-7
Burridge, P. W., Matsa, E., Shukla, P., Lin, Z. C., Churko, J. M., Ebert, A. D., et al. (2014). Chemically defined generation of human cardiomyocytes. Nat. Methods 11 (8), 855–860. doi:10.1038/nmeth.2999
Capulli, A. K., MacQueen, L. A., Sheehy, S. P., and Parker, K. K. (2016). Fibrous scaffolds for building hearts and heart parts. Adv. Drug Deliv. Rev. 96, 83–102. doi:10.1016/j.addr.2015.11.020
Chen, M. X., Deng, B. Y., Liu, S. T., Wang, Z. B., and Wang, S. Z. (2023). Salusins: advance in cardiovascular disease research. J. Pharm. Pharmacol. 75 (3), 363–369. doi:10.1093/jpp/rgac087
Ching, T., Vasudevan, J., Chang, S. Y., Tan, H. Y., Sargur Ranganath, A., Lim, C. T., et al. (2022). Biomimetic vasculatures by 3D-printed porous molds. Small 18 (39), e2203426. doi:10.1002/smll.202203426
Chiou, K. K., Rocks, J. W., Chen, C. Y., Cho, S., Merkus, K. E., Rajaratnam, A., et al. (2016). Mechanical signaling coordinates the embryonic heartbeat. Proc. Natl. Acad. Sci. U. S. A. 113 (32), 8939–8944. doi:10.1073/pnas.1520428113
Cho, S., Lee, C., Skylar-Scott, M. A., Heilshorn, S. C., and Wu, J. C. (2021). Reconstructing the heart using iPSCs: engineering strategies and applications. J. Mol. Cell Cardiol. 157, 56–65. doi:10.1016/j.yjmcc.2021.04.006
Clevers, H. (2016). Modeling development and disease with organoids. Cell 165 (7), 1586–1597. doi:10.1016/j.cell.2016.05.082
Coenen, A. M. J., Bernaerts, K. V., Harings, J. A. W., Jockenhoevel, S., and Ghazanfari, S. (2018). Elastic materials for tissue engineering applications: natural, synthetic, and hybrid polymers. Acta Biomater. 79, 60–82. doi:10.1016/j.actbio.2018.08.027
Cunha, A. B., Hou, J., and Schuelke, C. (2019). Machine learning for stem cell differentiation and proliferation classification on electrical impedance spectroscopy. J. Electr. Bioimpedance 10 (1), 124–132. doi:10.2478/joeb-2019-0018
Drakhlis, L., Biswanath, S., Farr, C. M., Lupanow, V., Teske, J., Ritzenhoff, K., et al. (2021). Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 39 (6), 737–746. doi:10.1038/s41587-021-00815-9
Du, B. B., Shi, H. T., Xiao, L. L., Li, Y. P., Yao, R., Liang, C., et al. (2024). Melanoma differentiation-associated protein 5 prevents cardiac hypertrophy via apoptosis signal-regulating kinase 1-c-Jun N-terminal kinase/p38 signaling. Int. J. Biol. Macromol. 264 (Pt 1), 130542. doi:10.1016/j.ijbiomac.2024.130542
Dyrbuś, K., Gąsior, M., Desperak, P., Osadnik, T., Nowak, J., and Banach, M. (2019). The prevalence and management of familial hypercholesterolemia in patients with acute coronary syndrome in the Polish tertiary centre: results from the TERCET registry with 19,781 individuals. Atherosclerosis 288, 33–41. doi:10.1016/j.atherosclerosis.2019.06.899
Ergir, E., Oliver-De La Cruz, J., Fernandes, S., Cassani, M., Niro, F., Pereira-Sousa, D., et al. (2022). Generation and maturation of human iPSC-derived 3D organotypic cardiac microtissues in long-term culture. Sci. Rep. 12 (1), 17409. doi:10.1038/s41598-022-22225-w
Fang, Y., Guo, Y., Wu, B., Liu, Z., Ye, M., Xu, Y., et al. (2023). Expanding embedded 3D bioprinting capability for engineering complex organs with freeform vascular networks. Adv. Mater 35 (22), e2205082. doi:10.1002/adma.202205082
Feric, N. T., Pallotta, I., Singh, R., Bogdanowicz, D. R., Gustilo, M. M., Chaudhary, K. W., et al. (2019). Engineered cardiac tissues generated in the biowire II: a platform for human-based drug discovery. Toxicol. Sci. 172 (1), 89–97. doi:10.1093/toxsci/kfz168
Fiedler, L. R., Chapman, K., Xie, M., Maifoshie, E., Jenkins, M., Golforoush, P. A., et al. (2020). MAP4K4 inhibition promotes survival of human stem cell-derived cardiomyocytes and reduces infarct size in vivo. Cell Stem Cell 26 (3), 458. doi:10.1016/j.stem.2020.01.015
Filippo Buono, M., von Boehmer, L., Strang, J., Hoerstrup, S. P., Emmert, M. Y., and Nugraha, B. (2020). Human cardiac organoids for modeling genetic cardiomyopathy. Cells 9 (7), 1733. doi:10.3390/cells9071733
Folkman, J., and Moscona, A. (1978). Role of cell shape in growth control. Nature 273 (5661), 345–349. doi:10.1038/273345a0
Forte, E., Furtado, M. B., and Rosenthal, N. (2018). The interstitium in cardiac repair: role of the immune-stromal cell interplay. Nat. Rev. Cardiol. 15 (10), 601–616. doi:10.1038/s41569-018-0077-x
Gaetani, R., Doevendans, P. A., Metz, C. H. G., Alblas, J., Messina, E., Giacomello, A., et al. (2012). Cardiac tissue engineering using tissue printing technology and human cardiac progenitor cells. Biomaterials 33 (6), 1782–1790. doi:10.1016/j.biomaterials.2011.11.003
Gao, L., Gregorich, Z. R., Zhu, W., Mattapally, S., Oduk, Y., Lou, X., et al. (2018). Large cardiac muscle patches engineered from human induced-pluripotent stem cell-derived cardiac cells improve recovery from myocardial infarction in swine. Circulation 137 (16), 1712–1730. doi:10.1161/CIRCULATIONAHA.117.030785
Gelmi, A., Cieslar-Pobuda, A., de Muinck, E., Los, M., Rafat, M., and Jager, E. W. H. (2016). Direct mechanical stimulation of stem cells: a beating electromechanically active scaffold for cardiac tissue engineering. Adv. Healthc. Mater 5 (12), 1471–1480. doi:10.1002/adhm.201600307
Giacomelli, E., Meraviglia, V., Campostrini, G., Cochrane, A., Cao, X., van Helden, R. W. J., et al. (2020). Human-iPSC-Derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell 26 (6), 862–879.e11. doi:10.1016/j.stem.2020.05.004
Goldfracht, I., Protze, S., Shiti, A., Setter, N., Gruber, A., Shaheen, N., et al. (2020). Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 11 (1), 75. doi:10.1038/s41467-019-13868-x
Gritti, N., Lim, J. L., Anlaş, K., Pandya, M., Aalderink, G., Martínez-Ara, G., et al. (2021). MOrgAna: accessible quantitative analysis of organoids with machine learning. Development 148 (18), dev199611. doi:10.1242/dev.199611
Gu, Y., Zhou, Y., Ju, S., Liu, X., Zhang, Z., Guo, J., et al. (2022). Multi-omics profiling visualizes dynamics of cardiac development and functions. Cell Rep. 41 (13), 111891. doi:10.1016/j.celrep.2022.111891
He, C., Kalafut, N. C., Sandoval, S. O., Risgaard, R., Sirois, C. L., Yang, C., et al. (2023). BOMA, a machine-learning framework for comparative gene expression analysis across brains and organoids. Cell Rep. Methods 3 (2), 100409. doi:10.1016/j.crmeth.2023.100409
Heeringa, J., van der Kuip, D. A. M., Hofman, A., Kors, J. A., van Herpen, G., Stricker, B. H. C., et al. (2006). Prevalence, incidence and lifetime risk of atrial fibrillation: the Rotterdam study. Eur. Heart J. 27 (8), 949–953. doi:10.1093/eurheartj/ehi825
Heidi Au, H. T., Cui, B., Chu, Z. E., Veres, T., and Radisic, M. (2009). Cell culture chips for simultaneous application of topographical and electrical cues enhance phenotype of cardiomyocytes. Lab. Chip 9 (4), 564–575. doi:10.1039/b810034a
Hoang, P., Wang, J., Conklin, B. R., Healy, K. E., and Ma, Z. (2018). Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat. Protoc. 13 (4), 723–737. doi:10.1038/nprot.2018.006
Hofbauer, P., Jahnel, S. M., Papai, N., Giesshammer, M., Deyett, A., Schmidt, C., et al. (2021). Cardioids reveal self-organizing principles of human cardiogenesis. Cell 184 (12), 3299–3317.e22. doi:10.1016/j.cell.2021.04.034
Hwang, D. G., Choi, H., Yong, U., Kim, D., Kang, W., Park, S. M., et al. (2024). Bioprinting-Assisted tissue assembly for structural and functional modulation of engineered heart tissue mimicking left ventricular myocardial fiber orientation. Adv. Mater 36 (34), e2400364. doi:10.1002/adma.202400364
Iyer, R. K., Odedra, D., Chiu, L. L. Y., Vunjak-Novakovic, G., and Radisic, M. (2012). Vascular endothelial growth factor secretion by nonmyocytes modulates Connexin-43 levels in cardiac organoids. Tissue Eng. Part A 18 (17-18), 1771–1783. doi:10.1089/ten.TEA.2011.0468
Jackman, C. P., Ganapathi, A. M., Asfour, H., Qian, Y., Allen, B. W., Li, Y., et al. (2018). Engineered cardiac tissue patch maintains structural and electrical properties after epicardial implantation. Biomaterials 159, 48–58. doi:10.1016/j.biomaterials.2018.01.002
Jang, J., Park, H. J., Kim, S. W., Kim, H., Park, J. Y., Na, S. J., et al. (2017). 3D printed complex tissue construct using stem cell-laden decellularized extracellular matrix bioinks for cardiac repair. Biomaterials 112, 264–274. doi:10.1016/j.biomaterials.2016.10.026
Jose, G., Shalumon, K. T., and Chen, J. P. (2020). Natural polymers based hydrogels for cell culture applications. Curr. Med. Chem. 27 (16), 2734–2776. doi:10.2174/0929867326666190903113004
Kanda, G. N., Tsuzuki, T., Terada, M., Sakai, N., Motozawa, N., Masuda, T., et al. (2022). Robotic search for optimal cell culture in regenerative medicine. Elife 11, e77007. doi:10.7554/eLife.77007
Kawai, Y., Tohyama, S., Arai, K., Tamura, T., Soma, Y., Fukuda, K., et al. (2021). Scaffold-free tubular engineered heart tissue from human induced pluripotent stem cells using bio-3D printing technology in vivo. Front. Cardiovasc Med. 8, 806215. doi:10.3389/fcvm.2021.806215
Kegeles, E., Naumov, A., Karpulevich, E. A., Volchkov, P., and Baranov, P. (2020). Convolutional neural networks can predict retinal differentiation in retinal organoids. Front. Cell Neurosci. 14, 171. doi:10.3389/fncel.2020.00171
Keung, W., Chan, P. K. W., Backeris, P. C., Lee, E. K., Wong, N., Wong, A. O. T., et al. (2019). Human cardiac ventricular-like organoid chambers and tissue strips from pluripotent stem cells as a two-tiered assay for inotropic responses. Clin. Pharmacol. Ther. 106 (2), 402–414. doi:10.1002/cpt.1385
Kim, J., Koo, B. K., and Knoblich, J. A. (2020). Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21 (10), 571–584. doi:10.1038/s41580-020-0259-3
Kloxin, A. M., Kasko, A. M., Salinas, C. N., and Anseth, K. S. (2009). Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324 (5923), 59–63. doi:10.1126/science.1169494
Kong, J., Lee, H., Kim, D., Han, S. K., Ha, D., Shin, K., et al. (2020). Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nat. Commun. 11 (1), 5485. doi:10.1038/s41467-020-19313-8
Kopljar, I., Lu, H. R., Van Ammel, K., Otava, M., Tekle, F., Teisman, A., et al. (2018). Development of a human iPSC cardiomyocyte-based scoring system for cardiac hazard identification in early drug safety de-risking. Stem Cell Rep. 11 (6), 1365–1377. doi:10.1016/j.stemcr.2018.11.007
Kozuka, K., He, Y., Koo-McCoy, S., Kumaraswamy, P., Nie, B., Shaw, K., et al. (2017). Development and characterization of a human and mouse intestinal epithelial cell monolayer platform. Stem Cell Rep. 9 (6), 1976–1990. doi:10.1016/j.stemcr.2017.10.013
Kupfer, M. E., Lin, W. H., Ravikumar, V., Qiu, K., Wang, L., Gao, L., et al. (2020). In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3D bioprinted, chambered organoid. Circ. Res. 127 (2), 207–224. doi:10.1161/CIRCRESAHA.119.316155
Kusumoto, F. M., Schoenfeld, M. H., Barrett, C., Edgerton, J. R., Ellenbogen, K. A., Gold, M. R., et al. (2019). 2018 ACC/AHA/HRS guideline on the evaluation and management of patients with bradycardia and cardiac conduction delay: a report of the American college of cardiology/American heart association task force on clinical practice guidelines and the heart rhythm society. Circulation 140 (8), e382–e482. doi:10.1161/CIR.0000000000000628
Laksman, Z., Wauchop, M., Lin, E., Protze, S., Lee, J., Yang, W., et al. (2017). Modeling atrial fibrillation using human embryonic stem cell-derived atrial tissue. Sci. Rep. 7 (1), 5268. doi:10.1038/s41598-017-05652-y
Lee, A., Hudson, A. R., Shiwarski, D. J., Tashman, J. W., Hinton, T. J., Yerneni, S., et al. (2019). 3D bioprinting of collagen to rebuild components of the human heart. Science 365 (6452), 482–487. doi:10.1126/science.aav9051
Lee, J., Sutani, A., Kaneko, R., Takeuchi, J., Sasano, T., Kohda, T., et al. (2020). In vitro generation of functional murine heart organoids via FGF4 and extracellular matrix. Nat. Commun. 11 (1), 4283. doi:10.1038/s41467-020-18031-5
Lee, J. H., Protze, S. I., Laksman, Z., Backx, P. H., and Keller, G. M. (2017). Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21 (2), 179–194.e4. doi:10.1016/j.stem.2017.07.003
Lewis-Israeli, Y. R., Wasserman, A. H., Gabalski, M. A., Volmert, B. D., Ming, Y., Ball, K. A., et al. (2021). Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nat. Commun. 12 (1), 5142. doi:10.1038/s41467-021-25329-5
Li, J., Zhang, L., Yu, L., Minami, I., Miyagawa, S., Hörning, M., et al. (2020). Circulating re-entrant waves promote maturation of hiPSC-derived cardiomyocytes in self-organized tissue ring. Commun. Biol. 3 (1), 122. doi:10.1038/s42003-020-0853-0
Li, M., Liu, Z., Shen, Z., Han, L., Wang, J., and Sang, S. (2024). A heparin-functionalized bioink with sustained delivery of vascular endothelial growth factor for 3D bioprinting of prevascularized dermal constructs. Int. J. Biol. Macromol. 262 (Pt 1), 130075. doi:10.1016/j.ijbiomac.2024.130075
Lind, J. U., Busbee, T. A., Valentine, A. D., Pasqualini, F. S., Yuan, H., Yadid, M., et al. (2017). Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater 16 (3), 303–308. doi:10.1038/nmat4782
Lu, K., Seidel, T., Cao-Ehlker, X., Dorn, T., Batcha, A. M. N., Schneider, C. M., et al. (2021). Progressive stretch enhances growth and maturation of 3D stem-cell-derived myocardium. Theranostics 11 (13), 6138–6153. doi:10.7150/thno.54999
Lukonin, I., Zinner, M., and Liberali, P. (2021). Organoids in image-based phenotypic chemical screens. Exp. Mol. Med. 53 (10), 1495–1502. doi:10.1038/s12276-021-00641-8
Ma, C., Peng, Y., Li, H., and Chen, W. (2021). Organ-on-a-Chip: a new paradigm for drug development. Trends Pharmacol. Sci. 42 (2), 119–133. doi:10.1016/j.tips.2020.11.009
Marini, V., Marino, F., Aliberti, F., Giarratana, N., Pozzo, E., Duelen, R., et al. (2022). Long-term culture of patient-derived cardiac organoids recapitulated Duchenne muscular dystrophy cardiomyopathy and disease progression. Front. Cell Dev. Biol. 10, 878311. doi:10.3389/fcell.2022.878311
Mehrotra, S., de Melo, B. A. G., Hirano, M., Keung, W., Li, R. A., Mandal, B. B., et al. (2020). Nonmulberry silk based ink for fabricating mechanically robust cardiac patches and endothelialized myocardium-on-a-chip application. Adv. Funct. Mater 30 (12), 1907436. doi:10.1002/adfm.201907436
Mills, R. J., Parker, B. L., Quaife-Ryan, G. A., Voges, H. K., Needham, E. J., Bornot, A., et al. (2019). Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell 24 (6), 895–907.e6. doi:10.1016/j.stem.2019.03.009
Mills, R. J., Titmarsh, D. M., Koenig, X., Parker, B. L., Ryall, J. G., Quaife-Ryan, G. A., et al. (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 114 (40), E8372–e8381. doi:10.1073/pnas.1707316114
Montessuit, C., Palma, T., Viglino, C., Pellieux, C., and Lerch, R. (2006). Effects of insulin-like growth factor-I on the maturation of metabolism in neonatal rat cardiomyocytes. Pflugers Arch. 452 (4), 380–386. doi:10.1007/s00424-006-0059-4
Monzel, A. S., Hemmer, K., Kaoma, T., Smits, L. M., Bolognin, S., Lucarelli, P., et al. (2020). Machine learning-assisted neurotoxicity prediction in human midbrain organoids. Park. Relat. Disord. 75, 105–109. doi:10.1016/j.parkreldis.2020.05.011
Mosterd, A., and Hoes, A. W. (2007). Clinical epidemiology of heart failure. Heart 93 (9), 1137–1146. doi:10.1136/hrt.2003.025270
Nahak, B. K., Mishra, A., Preetam, S., and Tiwari, A. (2022). Advances in organ-on-a-chip materials and devices. ACS Appl. Bio Mater 5 (8), 3576–3607. doi:10.1021/acsabm.2c00041
Najjar, R. S., Roy, R. K., Stern, J. E., and Feresin, R. G. (2024). Raspberry polyphenols target molecular pathways of heart failure. J. Nutr. Biochem. 124, 109535. doi:10.1016/j.jnutbio.2023.109535
Noor, N., Shapira, A., Edri, R., Gal, I., Wertheim, L., and Dvir, T. (2019). 3D printing of personalized thick and perfusable cardiac patches and hearts. Adv. Sci. (Weinh) 6 (11), 1900344. doi:10.1002/advs.201900344
O'Donoghue, M. L., Fazio, S., Giugliano, R. P., Stroes, E. S. G., Kanevsky, E., Gouni-Berthold, I., et al. (2019). Lipoprotein(a), PCSK9 inhibition, and cardiovascular risk. Circulation 139 (12), 1483–1492. doi:10.1161/CIRCULATIONAHA.118.037184
Parikh, S. S., Blackwell, D. J., Gomez-Hurtado, N., Frisk, M., Wang, L., Kim, K., et al. (2017). Thyroid and glucocorticoid hormones promote functional T-tubule development in human-induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 121 (12), 1323–1330. doi:10.1161/CIRCRESAHA.117.311920
Park, K., Lee, J. Y., Lee, S. Y., Jeong, I., Park, S. Y., Kim, J. W., et al. (2023). Deep learning predicts the differentiation of kidney organoids derived from human induced pluripotent stem cells. Kidney Res. Clin. Pract. 42 (1), 75–85. doi:10.23876/j.krcp.22.017
Peng, W., Datta, P., Wu, Y., Dey, M., Ayan, B., Dababneh, A., et al. (2018). Challenges in bio-fabrication of organoid cultures. Adv. Exp. Med. Biol. 1107, 53–71. doi:10.1007/5584_2018_216
Peters, M. F., Lamore, S. D., Guo, L., Scott, C. W., and Kolaja, K. L. (2015). Human stem cell-derived cardiomyocytes in cellular impedance assays: bringing cardiotoxicity screening to the front line. Cardiovasc Toxicol. 15 (2), 127–139. doi:10.1007/s12012-014-9268-9
Pok, S., Myers, J. D., Madihally, S. V., and Jacot, J. G. (2013). A multilayered scaffold of a chitosan and gelatin hydrogel supported by a PCL core for cardiac tissue engineering. Acta Biomater. 9 (3), 5630–5642. doi:10.1016/j.actbio.2012.10.032
Polonchuk, L., Chabria, M., Badi, L., Hoflack, J. C., Figtree, G., Davies, M. J., et al. (2017). Cardiac spheroids as promising in vitro models to study the human heart microenvironment. Sci. Rep. 7 (1), 7005. doi:10.1038/s41598-017-06385-8
Pré, D., Wooten, A. T., Biesmans, S., Hinckley, S., Zhou, H., Sherman, S. P., et al. (2022). Development of a platform to investigate long-term potentiation in human iPSC-derived neuronal networks. Stem Cell Rep. 17 (9), 2141–2155. doi:10.1016/j.stemcr.2022.07.012
Radhakrishnan, J., Krishnan, U. M., and Sethuraman, S. (2014). Hydrogel based injectable scaffolds for cardiac tissue regeneration. Biotechnol. Adv. 32 (2), 449–461. doi:10.1016/j.biotechadv.2013.12.010
Reis, L. A., Chiu, L. L. Y., Feric, N., Fu, L., and Radisic, M. (2016). Biomaterials in myocardial tissue engineering. J. Tissue Eng. Regen. Med. 10 (1), 11–28. doi:10.1002/term.1944
Ren, F. F., Zhao, L., Jiang, X. Y., Zhang, J. J., Gou, J. M., Yu, X. Y., et al. (2024). Sphingosylphosphorylcholine alleviates pressure overload-induced myocardial remodeling in mice via inhibiting CaM-JNK/p38 signaling pathway. Acta Pharmacol. Sin. 45 (2), 312–326. doi:10.1038/s41401-023-01168-6
Richards, D. J., Coyle, R. C., Tan, Y., Jia, J., Wong, K., Toomer, K., et al. (2017). Inspiration from heart development: biomimetic development of functional human cardiac organoids. Biomaterials 142, 112–123. doi:10.1016/j.biomaterials.2017.07.021
Richards, D. J., Li, Y., Kerr, C. M., Yao, J., Beeson, G. C., Coyle, R. C., et al. (2020). Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 4 (4), 446–462. doi:10.1038/s41551-020-0539-4
Ronaldson-Bouchard, K., Ma, S. P., Yeager, K., Chen, T., Song, L., Sirabella, D., et al. (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556 (7700), 239–243. doi:10.1038/s41586-018-0016-3
Rossi, G., Broguiere, N., Miyamoto, M., Boni, A., Guiet, R., Girgin, M., et al. (2021). Capturing cardiogenesis in gastruloids. Cell Stem Cell 28 (2), 230–240.e6. doi:10.1016/j.stem.2020.10.013
Roth, G. A., Mensah, G. A., Johnson, C. O., Addolorato, G., Ammirati, E., Baddour, L. M., et al. (2020). Global burden of cardiovascular diseases and risk factors, 1990-2019: update from the GBD 2019 study. J. Am. Coll. Cardiol. 76 (25), 2982–3021. doi:10.1016/j.jacc.2020.11.010
Saludas, L., Pascual-Gil, S., Prósper, F., Garbayo, E., and Blanco-Prieto, M. (2017). Hydrogel based approaches for cardiac tissue engineering. Int. J. Pharm. 523 (2), 454–475. doi:10.1016/j.ijpharm.2016.10.061
Schaaf, S., Shibamiya, A., Mewe, M., Eder, A., Stöhr, A., Hirt, M. N., et al. (2011). Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS One 6 (10), e26397. doi:10.1371/journal.pone.0026397
Sharma, A., Burridge, P. W., McKeithan, W. L., Serrano, R., Shukla, P., Sayed, N., et al. (2017). High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 9 (377), eaaf2584. doi:10.1126/scitranslmed.aaf2584
Sharma, D., Ferguson, M., Kamp, T. J., and Zhao, F. (2019). Constructing biomimetic cardiac tissues: a review of scaffold materials for engineering cardiac patches. Emergent Mater 2, 181–191. doi:10.1007/s42247-019-00046-4
Shinnawi, R., Shaheen, N., Huber, I., Shiti, A., Arbel, G., Gepstein, A., et al. (2019). Modeling reentry in the short QT syndrome with human-induced pluripotent stem cell-derived cardiac cell sheets. J. Am. Coll. Cardiol. 73 (18), 2310–2324. doi:10.1016/j.jacc.2019.02.055
Skardal, A., Aleman, J., Forsythe, S., Rajan, S., Murphy, S., Devarasetty, M., et al. (2020). Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication 12 (2), 025017. doi:10.1088/1758-5090/ab6d36
Song, M., Choi, D. B., Im, J. S., Song, Y. N., Kim, J. H., Lee, H., et al. (2024). Modeling acute myocardial infarction and cardiac fibrosis using human induced pluripotent stem cell-derived multi-cellular heart organoids. Cell Death Dis. 15 (5), 308. doi:10.1038/s41419-024-06703-9
Suwardi, A., Wang, F., Xue, K., Han, M. Y., Teo, P., Wang, P., et al. (2022). Machine learning-driven biomaterials evolution. Adv. Mater 34 (1), e2102703. doi:10.1002/adma.202102703
Swirski, F. K., and Nahrendorf, M. (2018). Cardioimmunology: the immune system in cardiac homeostasis and disease. Nat. Rev. Immunol. 18 (12), 733–744. doi:10.1038/s41577-018-0065-8
Tang, X., Zhang, J., He, Y., Zhang, X., Lin, Z., Partarrieu, S., et al. (2023). Explainable multi-task learning for multi-modality biological data analysis. Nat. Commun. 14 (1), 2546. doi:10.1038/s41467-023-37477-x
Tao, Z. W., Mohamed, M., Jacot, J. G., and Birla, R. K. (2018). Bioengineering cardiac tissue constructs with adult rat cardiomyocytes. Asaio J. 64 (5), e105–e114. doi:10.1097/MAT.0000000000000765
Thavandiran, N., Dubois, N., Mikryukov, A., Massé, S., Beca, B., Simmons, C. A., et al. (2013). Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proc. Natl. Acad. Sci. U. S. A. 110 (49), E4698–E4707. doi:10.1073/pnas.1311120110
Tian, S., Liu, Q., Gnatovskiy, L., Ma, P. X., and Wang, Z. (2015). Heart regeneration with embryonic cardiac progenitor cells and cardiac tissue engineering. J. Stem Cell Transpl. Biol. 1 (1), 104. doi:10.19104/jstb.2015.104
Tiburcy, M., Hudson, J. E., Balfanz, P., Schlick, S., Meyer, T., Chang Liao, M. L., et al. (2017). Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation 135 (19), 1832–1847. doi:10.1161/CIRCULATIONAHA.116.024145
Tsao, C. W., Aday, A. W., Almarzooq, Z. I., Alonso, A., Beaton, A. Z., Bittencourt, M. S., et al. (2022). Heart disease and stroke statistics-2022 update: a report from the American heart association. Circulation 145 (8), e153–e639. doi:10.1161/CIR.0000000000001052
Unal, A. Z., and West, J. L. (2020). Synthetic ECM: bioactive synthetic hydrogels for 3D tissue engineering. Bioconjug Chem. 31 (10), 2253–2271. doi:10.1021/acs.bioconjchem.0c00270
Vallon, V., and Verma, S. (2021). Effects of SGLT2 inhibitors on kidney and cardiovascular function. Annu. Rev. Physiol. 83, 503–528. doi:10.1146/annurev-physiol-031620-095920
Verheyen, C. A., Uzel, S. G., Kurum, A., Roche, E. T., and Lewis, J. A. (2023). Integrated data-driven modeling and experimental optimization of granular hydrogel matrices. Matter 6 (3), 1015–1036. doi:10.1016/j.matt.2023.01.011
Voges, H. K., Mills, R. J., Elliott, D. A., Parton, R. G., Porrello, E. R., and Hudson, J. E. (2017). Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 144 (6), 1118–1127. doi:10.1242/dev.143966
Wang, Z., Wang, S. N., Xu, T. Y., Miao, Z. W., Su, D. F., and Miao, C. Y. (2017). Organoid technology for brain and therapeutics research. CNS Neurosci. Ther. 23 (10), 771–778. doi:10.1111/cns.12754
Wilke, R. A., Lin, D. W., Roden, D. M., Watkins, P. B., Flockhart, D., Zineh, I., et al. (2007). Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat. Rev. Drug Discov. 6 (11), 904–916. doi:10.1038/nrd2423
Wu, X., Chen, Y., Luz, A., and Tokar, E. J. (2022). Cardiac development in the presence of cadmium: an in vitro study using human embryonic stem cells and cardiac organoids. Environ. Health Perspect. 130 (11), 117002. doi:10.1289/EHP11208
Xia, B., and Chen, G. (2022). Research progress of natural tissue-derived hydrogels for tissue repair and reconstruction. Int. J. Biol. Macromol. 214, 480–491. doi:10.1016/j.ijbiomac.2022.06.137
Xu, J., Shamul, J. G., Staten, N. A., White, A. M., Jiang, B., and He, X. (2021). Bioinspired 3D culture in nanoliter hyaluronic acid-rich core-shell hydrogel microcapsules isolates highly pluripotent human iPSCs. Small 17 (33), e2102219. doi:10.1002/smll.202102219
Yang, K. C., Breitbart, A., De Lange, W. J., Hofsteen, P., Futakuchi-Tsuchida, A., Xu, J., et al. (2018). Novel adult-onset systolic cardiomyopathy due to MYH7 E848G mutation in patient-derived induced pluripotent stem cells. JACC Basic Transl. Sci. 3 (6), 728–740. doi:10.1016/j.jacbts.2018.08.008
Yang, X., Rodriguez, M., Pabon, L., Fischer, K. A., Reinecke, H., Regnier, M., et al. (2014). Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell Cardiol. 72, 296–304. doi:10.1016/j.yjmcc.2014.04.005
Yang, Z., Zhang, Y., Wang, J., Yin, J., Wang, Z., and Pei, R. (2023). Cardiac organoid: multiple construction approaches and potential applications. J. Mater Chem. B 11 (32), 7567–7581. doi:10.1039/d3tb00783a
Yeh, R. W., Sidney, S., Chandra, M., Sorel, M., Selby, J. V., and Go, A. S. (2010). Population trends in the incidence and outcomes of acute myocardial infarction. N. Engl. J. Med. 362 (23), 2155–2165. doi:10.1056/NEJMoa0908610
Zhang, B., and Radisic, M. (2017). Organ-on-a-chip devices advance to market. Lab. Chip 17 (14), 2395–2420. doi:10.1039/c6lc01554a
Zhang, D., Shadrin, I. Y., Lam, J., Xian, H. Q., Snodgrass, H. R., and Bursac, N. (2013). Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials 34 (23), 5813–5820. doi:10.1016/j.biomaterials.2013.04.026
Zhang, X., Chen, X., Hong, H., Hu, R., Liu, J., and Liu, C. (2022). Decellularized extracellular matrix scaffolds: recent trends and emerging strategies in tissue engineering. Bioact. Mater 10, 15–31. doi:10.1016/j.bioactmat.2021.09.014
Keywords: human cardiac organoids, cardiovascular disease, clinical translation, disease models, drug screening
Citation: Zhang H, Qu P, Liu J, Cheng P and Lei Q (2025) Application of human cardiac organoids in cardiovascular disease research. Front. Cell Dev. Biol. 13:1564889. doi: 10.3389/fcell.2025.1564889
Received: 22 January 2025; Accepted: 19 March 2025;
Published: 31 March 2025.
Edited by:
Daniel Hesselson, Garvan Institute of Medical Research, AustraliaReviewed by:
Pratima Kumari, Georgia State University, United StatesCopyright © 2025 Zhang, Qu, Liu, Cheng and Lei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Lei, bGVpcWlhbmdnaEAxNjMuY29t; Panke Cheng, Y2hlbmdwazEwMDJAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.