- Departamento de Genética, Facultad de Biología, Universidad de Sevilla, Sevilla, Spain
In this mini review, we discussed the functional roles of PIWI proteins and their associated small RNAs, piRNAs, in regulating gene expression within stem cell biology. Guided by piRNAs, these proteins transcriptionally and post-transcriptionally repress transposons using mechanisms such as the ping-pong amplification cycle and phasing to protect germline genomes. Initially identified in Drosophila melanogaster, the piRNA pathway regulate germline stem cell self-renewal and differentiation via cell-autonomous and non-cell-autonomous mechanisms. Precisely, in GSCs, PIWI proteins and piRNAs regulate gene expression by modulating chromatin states and directly influencing mRNA translation. For instance, the PIWI protein Aubergine loaded with piRNAs promotes and represses translation of certain mRNAs to balance self-renewal and differentiation. Thus, the piRNA pathway exhibits dual regulatory roles in mRNA stability and translation, highlighting its context-dependent functions. Moreover, PIWI proteins are essential in somatic stem cells to support the regenerative capacity of highly regenerative species, such as planarians. Similarly, in Drosophila intestinal stem cells, the PIWI protein Piwi regulates metabolic pathways and genome integrity, impacting longevity and gut homeostasis. In this case, piRNAs appear absent in the gut, suggesting piRNA-independent regulatory mechanisms. Together, PIWI proteins and piRNAs demonstrate evolutionary conservation in stem cell regulation, integrating TE silencing and gene expression regulation at chromatin and mRNA levels in somatic and germline lineages. Beyond their canonical roles, emerging evidence reveal their broader significance in maintaining stem cell properties and organismal health under physiological and pathological conditions.
Introduction
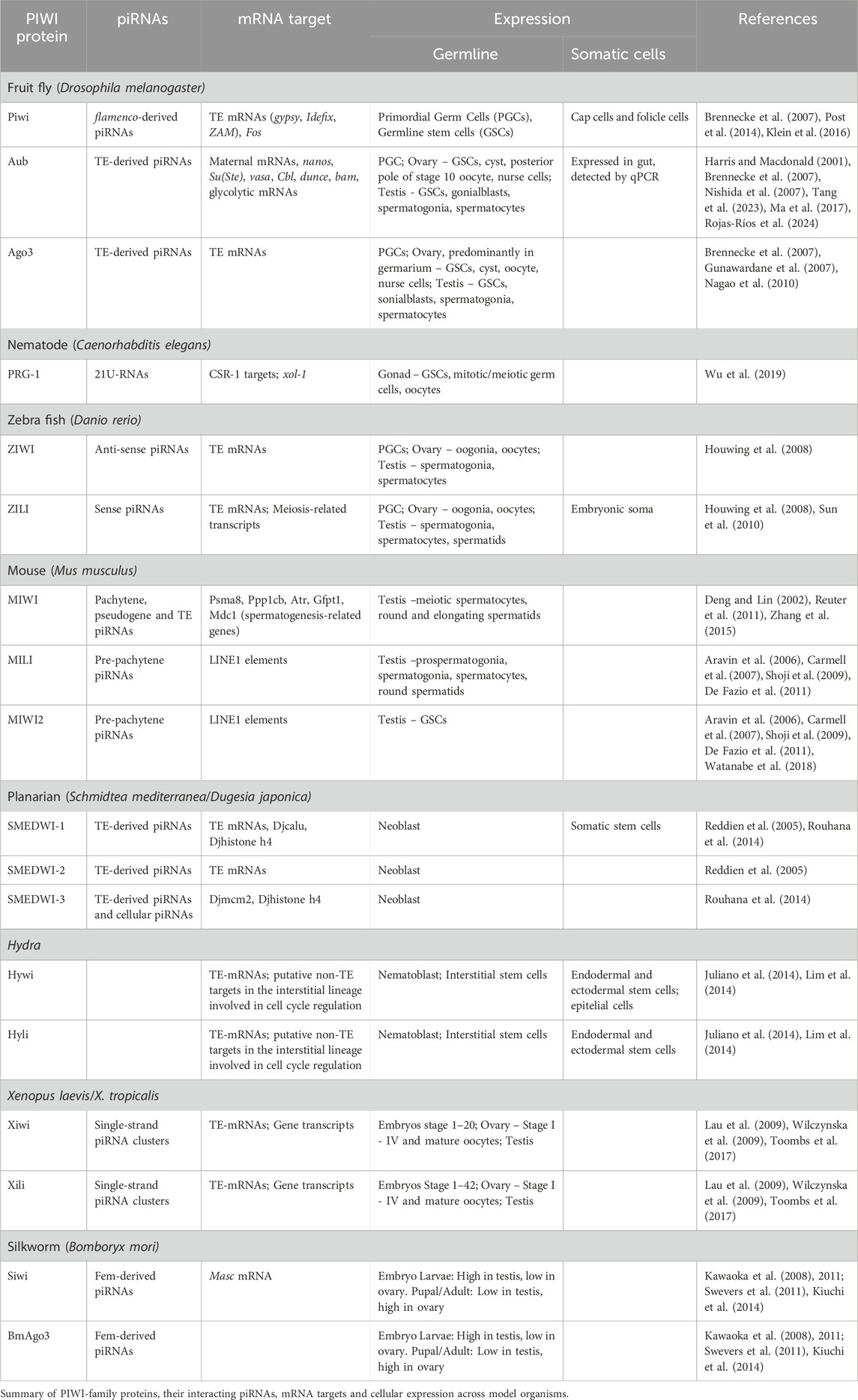
The piwi (for P-element induced wimpy testis) gene was initially identified in a genetic screen of single P-element mutants as a key regulator of Germline Stem Cell (GSC) asymmetric division in Drosophila melanogaster (Lin and Spradling, 1997). Several years later, a novel class of small non-coding RNAs, termed PIWI-interacting RNAs (piRNAs), was discovered in the germline of various animal species (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Lau et al., 2006; Vagin et al., 2006). These RNAs were named piRNAs because they associate with the PIWI-clade subfamily of the Argonaute protein family. PIWI proteins are highly expressed in animal gonads and exhibit RNA-endonucleolytic activity guided by piRNAs. piRNAs are typically 23–31 nucleotides long. Unlike the PIWI proteins, which are highly conserved throughout evolution, piRNAs exhibit low sequence conservation across species (Özata et al., 2020). Despite this variability, the presence and biological roles of piRNAs are well conserved in the germline of many animals, where they are crucial for repressing transposable elements (TEs). TEs are parasitic and highly abundant DNA sequences capable of replicative transposition, enabling them to move to new genomic regions. Thus, the movement of TEs compromises genome integrity, and piRNAs play an essential role in protecting the genome from such damage. The mode of action of the piRNA pathway in TE silencing is either at the transcriptional or at post-transcriptional levels depending on the PIWI proteins. However, these mechanisms tolerate a low level of TE transposition which serves as both a driver of evolutionary processes and a source for basal piRNA biogenesis. Notably, piRNAs are mainly encoded by TE sequences localized in specific genomic regions forming arrays known as piRNA clusters (Brennecke et al., 2007; Gunawardane et al., 2007). These clusters, sometimes referred to as a genomic immune system, are classified as either uni-strand or dual-strand clusters, depending on whether they transcribe from one or both DNA strands.
Even though their differences, all piRNA clusters generate long precursor transcripts, which are transported to electro-dense cytoplasmic perinuclear foci. These foci are called Yb-bodies in gonadal somatic cells and nuage in germline cells in Drosophila (Huang et al., 2017). Within the nuage, piRNAs are amplified through a mechanism called the “ping-pong” cycle. Briefly, the piRNA precursors are cleaved to produce a first round of piRNAs. These primary piRNAs, guided by sequence complementarity, target to TE mRNAs and, through the endonucleolytic activity of PIWI proteins, cut the TE mRNAs precisely 10 nucleotides upstream of the 5′-end of the guide piRNA. This cleavage generates the 5′-end of a secondary piRNA on the opposite strand. A distinctive hallmark of ping-pong piRNAs is the presence of a 10-nucleotide overlap at their 5′-ends, with Aub-loaded piRNAs typically starting with an Uracil and Ago3-loaded piRNAs displaying an Adenine at position 10 from the 5′-end (Brennecke et al., 2007; Gunawardane et al., 2007; Nishida et al., 2007). In addition to the ping-pong cycle, an alternative piRNA biogenesis mechanism occurs at the outer mitochondrial membrane in both gonadal somatic and germline cells. This process, known as “phasing,” involves the RNA helicase Armitage (Armi) and the endonuclease protein Zucchini (Zuc) (Han et al., 2015; Mohn et al., 2015). Briefly, Armi facilitates the transport of Aub-bound pre-piRNA to the outer mitochondrial membrane, where it is processed by Zuc to generate an initial piRNA. Zuc cleaves the piRNA precursor to define its 3′-end, while the Piwi protein processes the 5′-end. This produces phased piRNAs that are loaded onto Piwi protein and subsequently translocated to the nucleus, where it transcriptionally silences TEs (Figure 1A). Thus, although Piwi protein is primarily localized in the nucleus, where it transcriptionally silences TEs, a small portion may transiently reside in the cytoplasm and contribute to the phasing process, unlike Aub and Ago3, which are predominantly cytoplasmic. Through these complementary mechanisms, the piRNA pathway effectively safeguards genome integrity while maintaining a controlled level of transpositional activity to support evolutionary dynamics and piRNA production. The choice between phasing and ping-pong processing for Aub depends on piRNA-guided slicing and Armi availability. Phasing processing relies on cleavages from the ping-pong cycle, which Armi facilitates by shuttling precursors from the nuage to mitochondria. Armi’s role is crucial, as its disruption halts phased piRNA production (Ge et al., 2019). Also, depletion of Aub or Ago3 significantly reduces Piwi-bound piRNAs. The frequency of ping-pong cleavages further regulates substrate supply, linking both pathways despite their spatial separation (Senti et al., 2015; Wang et al., 2015; Chary and Hayashi, 2023).
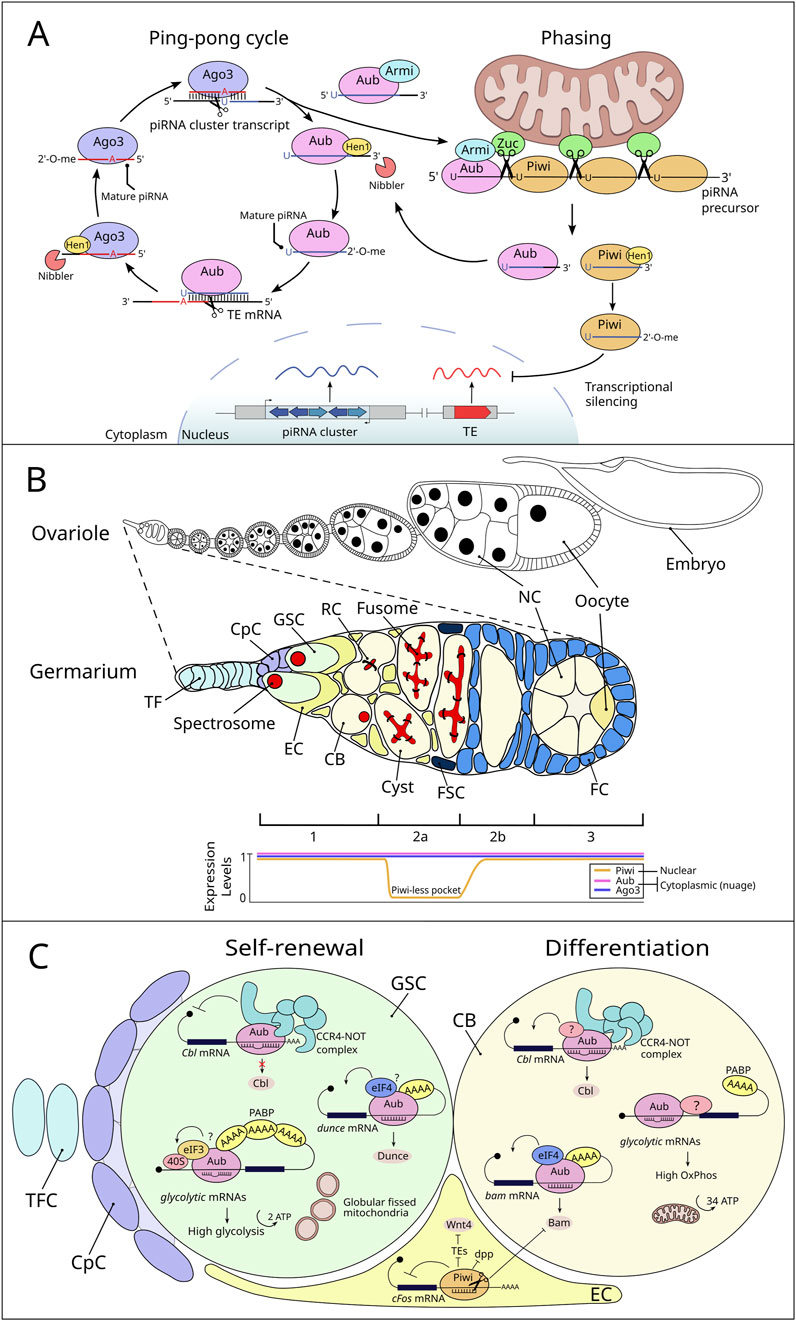
Figure 1. piRNA biogenesis in Drosophila germline and mRNA regulation by PIWI proteins and piRNAs in the GSC niche. (A) piRNA biogenesis in Drosophila germline. The ping-pong cycle occurs in the nuage and involves reciprocal cleavage of transposon mRNAs and piRNA cluster transcripts by two PIWI proteins, Aubergine (Aub) and Argonaute 3 (Ago3). Aub, guided by antisense primary piRNAs, cleaves transposon mRNAs to generate the 5′ ends of sense secondary piRNAs. Ago3-bound secondary piRNAs then cleave piRNA cluster precursors, producing 5′ ends of new primary piRNAs. The 3′ ends of pre-piRNAs are processed by the exonuclease Nibbler and modified with 2′-O-methylation by the methyltransferase Hen1, protecting them from degradation and resulting in mature piRNAs. In phased biogenesis, long piRNA precursors bound to Aub are transported by the RNA helicase Armitage (Armi) to the mitochondrial outer membrane. There, the endonuclease Zucchini (Zuc) cleaves the precursor, generating intermediate piRNAs with a characteristic 5′ uridine bias. The first piRNA returns to the nuage, while subsequent cleavages by Zuc create a series of Piwi-bound piRNAs. These are methylated by Hen1 and transported into the nucleus for transposon silencing. The choice between phasing and ping-pong processing is governed by the availability of substrates produced through secondary piRNA-guided cleavages and the regulatory role of Armi, which couples the two pathways despite their physical separation. (B) Schematic representation of a Drosophila ovariole and germarium with associated PIWI protein expression patterns. The top section depicts a Drosophila ovariole, showcasing the sequential stages of oogenesis. A zoomed-in view of the germarium is depicted below, detailing its cellular components (TF: terminal filament, CpC: cap cell, GSC: germline stem cell, EC: escort cell, CB: cystoblast, FC: follicle cell, FSC: follicle stem cell, NC: nurse cell). The bottom panel shows the expression pattern of the Drosophila PIWI protein family members (Piwi, Aub, and Ago3) in the germarium. These patterns highlight the “Piwi-less pocket” in region 2a, where Piwi expression is absent (Dufourt et al., 2014), while Aub and Ago3 are expressed across regions 1, 2a, 2b, and 3 and latter oogenesis. (C) Roles of PIWI/piRNAs in GSC maintenance and differentiation by gene expression regulation. Aub/piRNA complex regulates GSC self-renewal and differentiation by repressing Cbl mRNA through the CCR4-NOT complex, a repression reduced in cystoblasts (CBs), leading to higher Cbl levels. Additionally, Aub positively controls dunce and bam mRNAs in GSCs and CBs, respectively. Aub also influences the metabolic state of GSCs. Directed by piRNAs, Aub associates with glycolytic mRNAs to promote their translation in GSCs, resulting in increased glycolytic enzyme production. This mechanism likely involves Aub interacting with translation initiation factors such as PABP and eIF3. During differentiation, mitochondria undergo maturation, becoming more fused and structured with developed cristae, facilitating a metabolic shift towards oxidative phosphorylation. Other unidentified components (indicated by a question mark) may work alongside Aub to restrict glycolytic enhancement specifically to GSCs. In escort cells, Piwi plays a crucial role in regulating key signaling pathways and maintaining cellular functions. It suppresses Dpp signaling through an-as-yet-unknown mechanism. Piwi also targets cFos mRNA in the cytoplasm of cap and escort cells by binding to its 3′UTR, which leads to its cleavage into piRNAs and subsequent degradation of the cFos transcript. Furthermore, Piwi influences the expression of Wnt4 in escort cells by silencing transposable elements (TEs) at the transcriptional level.
Beyond their canonical role in silencing TEs in animal gonads, piRNAs and PIWI proteins have been increasingly recognized for their functional roles in regulating gene expression at both transcriptional and post-transcriptional levels across diverse biological systems (Rojas-Ríos and Simonelig, 2018; Wang et al., 2023) (Table 1). The first direct evidence of piRNA involvement in mRNA regulation was observed in the regulation of the maternal gene nanos (nos) during early Drosophila embryogenesis (Rouget et al., 2010). This study demonstrated that piRNAs derived from the TEs roo and 412 guide the PIWI proteins Aubergine (Aub) and Argonaute 3 (Ago3) to the 3′UTR of nos mRNA, leading to its degradation via CCR4-NOT-mediated deadenylation. Subsequently, this novel role of piRNAs in mRNA destabilization has been extended to hundreds of mRNAs, particularly during two critical developmental processes: the maternal-to-zygotic transition (when the zygotic genome becomes transcriptionally active) and mouse spermiogenesis (Gou et al., 2014; Barckmann et al., 2015). During these processes, the piRNA pathway plays an essential role in orchestrating large-scale mRNA decay. Interestingly, PIWI proteins loaded with piRNAs have also been implicated in stabilizing cellular mRNAs by promoting poly(A) tail elongation and enhancing translational initiation. They activate mRNA translation through imperfect base-pairing interactions between piRNAs and their target mRNAs (Vourekas et al., 2016; Dufourt et al., 2017; Dai et al., 2019; Ramat et al., 2020). Thus, the piRNA pathway demonstrates versatile regulatory functions in mRNA processing and stability, which are critical not only for the aforementioned developmental processes but also for others, such as sex determination and fertility (Gou et al., 2014; Kiuchi et al., 2014). In addition to these roles, piRNAs and PIWI proteins are crucial for gene expression regulation at both chromatin and mRNA levels in stem cell biology. This regulation influences key processes such as chromatin remodeling, transcriptional silencing, and post-transcriptional control of mRNAs. This mini review highlights the emerging roles of PIWI proteins and piRNAs in regulating gene expression at both transcriptional and post-transcriptional levels, with a particular focus on their contributions to stem cell biology in highly regenerative species and D. melanogaster.
PIWI proteins and piRNAs in somatic stem cells of highly regenerative species
A diverse range of studies indicates that PIWI proteins are specifically expressed and required in somatic stem cells to support the regenerative capacity of several species, including sponges, acoels, cnidarians and planaria (Reddien et al., 2005; Palakodeti et al., 2008; Krishna et al., 2013; Rinkevich et al., 2013; Juliano et al., 2014; Lim et al., 2014; Ross et al., 2014; Bradshaw et al., 2015). In addition, piRNAs are expressed in the soma of Hydra, Nematostella, and the jellyfish Sanderia malayensis (Juliano et al., 2014; Praher et al., 2017; Nong et al., 2020). In planarian, PIWI proteins are expressed in somatic stem cells (known as neoblasts), and the loss of function of specific piwi genes compromises the ability of these animals to regenerate body parts due to defects in neoblast maintenance. Moreover, small RNAs produced by PIWI proteins are present in planarian soma cells, although only a small portion are complementary to TEs (Resch and Palakodeti, 2012; Shibata et al., 2016).
The functional role of PIWI and piRNAs in planarians involves silencing TEs as well as protein-coding genes. Notably, the nuclear PIWI protein regulates the expression of essential functional genes, such as Djmcm2, Djhistone h4, and Djcalu, which are involved in neoblast self-renewal and differentiation (Kashima et al., 2018). Interestingly, another study on planarian PIWI proteins, specifically the cytoplasmic SMEDWI-1 and SMEDWI-3, demonstrated their role in the localization of histone H4 mRNA to chromatoid bodies in stem cells (Rouhana et al., 2014) suggesting distinct regulatory mechanisms employed by PIWI proteins for specific functional genes. Additionally, the nuclear PIWI plays a key role in TE silencing and regulates neoblast differentiation during cell specialization in the planarian Dugesia japonica (Shibata et al., 2016). An independent study revealed that SMEDWI-3 has a dual role in mRNA turnover in planarian neoblasts (Kim et al., 2019). It degrades certain mRNAs through a homotypic ping-pong cycle while binding to others, guided by antisense piRNAs, without causing degradation. These distinct functions are determined by the level of complementarity between the target mRNAs and antisense piRNAs, highlighting the critical regulation of neoblast mRNA turnover in planarians by piRNAs. Furthermore, a recent study showed that the planarian PIWI protein SMEDWI-2 is crucial for guiding stem cells through chromatin transitions during differentiation in the planarian Schmidtea mediterranea (Li et al., 2021). Overall, PIWI proteins and their associated piRNAs are integral to somatic stem cell function and regenerative processes suggesting that this mechanism is a conserved feature through evolution.
Non-cell-autonomous function of PIWI and piRNAs in GSC self-renewal and differentiation
Like its role in silencing TEs, the regulation of gene expression by the piRNA pathway has been extensively studied in the GSCs of the Drosophila female. The Drosophila ovary provides an exceptional model for investigating stem cell regulation in vivo (Rojas-Ríos and González-Reyes, 2014; Rosales-Nieves and González-Reyes, 2014). Each ovary consists of 16–20 ovarioles, each composed of an anterior germarium that transitions into progressively maturing follicles (Figure 1B). Within the germarium, two to three GSCs reside in a somatic cellular niche of three distinct somatic cell types: terminal filament cells (TFCs), cap cells (CpCs), and escort cells (ECs) (Losick et al., 2011; Jin and Zhao, 2023). The GSCs divide asymmetrically inside the niche to give rise to a cystoblast (CB) that undergoes four rounds of synchronous divisions to generate a 16-cell germline cyst that will ultimately produce a mature oocyte. Within the GSCs themselves, spectrosomes-dynamic membranous structures- undergo shape changes throughout the cell cycle and are critical for proper mitotic spindle orientation (Villa-Fombuena et al., 2021; Sánchez-Gómez et al., 2024). This ensures the asymmetric division necessary to maintain the stem cell pool while producing differentiating daughter cells. GSCs are anchored to the niche via E-cadherin and adherens junctions, effectively maintaining their self-renewal capacity and preventing their migration outside the niche (Dansereau and Lasko, 2008). The main signaling system from niche cells is Decapentaplegic (Dpp), a bone morphogenetic protein (BMP) ligand that promotes GSC self-renewal within a short range (Harris and Ashe, 2011). Dpp signaling represses bam expression specifically within GSCs, essential for GSC differentiation since Bam is required and sufficient for germline differentiation (Xie and Spradling, 1998). This repression is relieved once a GSC daughter leaves niche since the movement and stability of Dpp is restricted in the niche by Collagen IV and Glypican Dally, a protein whose expression is controlled by the EGFR-MAPK signaling pathway (Wang X. et al., 2008; Guo and Wang, 2009; Hayashi et al., 2009; Liu et al., 2010). The expression of Dpp in the GSC niche is highly controlled by different mechanisms including the JAK-STAT, Hedgehog and Piwi pathways (López-Onieva et al., 2008; Wang L. et al., 2008; Rojas-Ríos et al., 2012; Jin et al., 2013).
Early studies using piwi mutants demonstrated that piwi regulates GSC self-renewal and differentiation (Cox et al., 1998; 2000; Szakmary et al., 2005). Further studies of the role of piwi in GSC biology revealed that its cell-autonomous function regulates GSC self-renewal and asymmetric division, while its non-cell-autonomous function in the niche is essential for early germline differentiation. Specifically, piwi acts in ECs to promote GSC differentiation by regulating several signaling pathways (Figure 1C). Genetic analyses indicate that piwi represses dpp expression in ECs to limit its diffusion, thereby promoting germline differentiation (Jin et al., 2013). However, dpp is not the only factor regulated by piwi in ECs to maintain GSCs and support differentiation. A noteworthy study demonstrated that piwi represses the c-Fos proto-oncogene at the mRNA level in ECs. Importantly, this post-transcriptional repression occurs through the 3′UTR of c-Fos mRNA, leading to its cleavage and the production of piRNAs. This finding indicates that cellular c-Fos mRNA serves as a source for piRNA biogenesis in somatic cells of the gonad (Klein et al., 2016). Additionally, Piwi control of TEs in ECs plays a critical role in repressing Wnt4 expression, a key signal involved in germline differentiation and cystoblast encapsulation (Upadhyay et al., 2016). Specifically, mutations in soma piRNA pathway components, such as piwi and flamenco (but not aubergine), were shown to reduce Wnt4 expression, as demonstrated by qRT-PCR and in situ hybridization. These mutants also exhibited germline differentiation defects, further underscoring the importance of Wnt4 regulation. These findings suggest that Piwi promotes GSC differentiation by repressing Wnt4 expression in ECs through its TE control function (Figure 1C). By maintaining TE silencing, Piwi ensures proper signaling in the somatic niche, allowing for the differentiation of GSCs and the proper encapsulation of CBs, highlighting its pivotal role in regulating the balance between stem cell self-renewal and differentiation. A novel identified role of Piwi in the GSC niche involves maintaining GSC adhesion to CpCs. Like its function in ECs, Piwi silences TEs to prevent the activation of Toll-GSK3 signaling, which would otherwise lead to the degradation of β-catenin (a critical component of the Cadherin-Catenin-Actin complex that mediates cell adhesion). Importantly, this recent study demonstrated that aging CpCs express reduced levels of Piwi. This decline results in TE-dependent activation of the Toll receptor through an unknown mechanism, disrupting β-catenin stability. Consequently, reduced Piwi levels in aged niches lead to GSC detachment from CpCs, impairing GSC self-renewal and contributing to age-associated niche deterioration. This highlights the essential role of Piwi in preserving the structural integrity and functionality of the GSC niche during adulthood (Lin et al., 2020).
Cell-autonomous functions of PIWI and piRNAs in GSC self-renewal
An epigenetic mechanism involving Piwi has been identified as essential for GSC maintenance in female Drosophila (Peng et al., 2016). A genome-wide screen designed to identify suppressors of piwi uncovered a partner protein associated with Polycomb group (PcG) proteins (Smulders-Srinivasan and Lin, 2003). PcG proteins are crucial epigenetic regulators that modulate chromatin through histone methylation, particularly by adding H3K27me3 marks. These marks are generally associated with transcriptional repression, while reduced H3K27me3 levels correlate with active transcription mediated by RNA polymerase II. Piwi interacts with the Polycomb Repressive Complex 2 (PRC2) in the nucleoplasm, playing a key role in regulating chromatin state. This interaction inhibits PRC2 binding to genomic regions that do not directly interact with Piwi, resulting in reduced H3K27me3 levels and altered transcriptional activity, which are critical for oogenesis and GSC maintenance. Piwi appears to modulate RNA polymerase II function by sequestering PRC2 in the nucleoplasm, thereby restricting its access to genomic targets and preventing excessive transcriptional repression. These findings suggest a dual role for Piwi: modulating chromatin states in both niche cells and GSCs to create a favorable transcriptional environment for self-renewal and differentiation.
Aub, another Drosophila PIWI protein, plays a crucial role in gene expression regulation through direct binding to specific mRNAs in GSCs. Recently, it has been described that Aub binds mRNAs encoding glycolytic enzymes like Enolase (Eno), to promote their translational activation. This process is guided by piRNAs, which enable Aub to interact with regions of target mRNAs, such as untranslated regions (UTRs). Mutations in the piRNA-binding sites within the Eno 5′UTR result in reduced Eno expression and subsequent GSC loss, underscoring the importance of precise mRNA regulation in maintaining glycolytic flux and GSC self-renewal (Rojas-Ríos et al., 2024). High glycolytic activity is essential for GSC maintenance, while disruptions in Aub function led to a metabolic shift towards oxidative phosphorylation (oxphos), characterized by reduced glycolytic enzyme levels, increased ATP synthase expression, and premature mitochondrial maturation in GSCs. These metabolic changes are incompatible with the maintenance of GSCs, as increasing glycolysis via Phosphofructokinase overexpression partially rescues GSC loss in aub mutants. In addition to glycolytic mRNAs, Aub regulates a broader spectrum of transcripts critical for GSC fate transitions. For instance, Aub represses Cbl mRNA to support GSC self-renewal by recruiting the CCR4-NOT deadenylation complex, which is also required for maintaining GSC self-renewal (Joly et al., 2013). Notably, this repression occurs without poly(A) tail shortening, suggesting an alternative mechanism of translational inhibition (Rojas-Ríos et al., 2017). Conversely, Aub also activates dunce mRNA translation, further illustrating its capacity for dual regulatory roles depending on the specific target and cellular context (Ma et al., 2017). Additionally, PIWI proteins are capable of positively regulate target mRNAs under certain conditions. For example, Aub promotes the translation of mRNAs like bam and dunce at precise stages of the GSC lineage, ensuring precise temporal controls over developmental processes. These findings illustrate that the piRNA-PIWI pathway employs a context-dependent mechanism to either repress or activate mRNA targets, thereby maintaining GSC properties such as self-renewal and differentiation.
Piwi in intestinal stem cells of Drosophila
Recent research has explored the role of Piwi in regulating intestinal homeostasis in Drosophila (Tang et al., 2023). Piwi expression has been detected in the adult gut at both mRNA and protein levels, as confirmed by RT-PCR and Western blot analyses. Additionally, Piwi-Gal4-driven GFP expression reveals that intestinal stem cells (ISCs) and gut progenitors express Piwi specifically. Immunostaining analysis further shows that Piwi protein is localized in the cytoplasm of ISCs, suggesting a potential role in post-transcriptional regulation. As Piwi protein is primarily nuclear in the gonads, and the analysis lacks an antibody specificity control for the gut, the claim regarding Piwi’s cytoplasmic localization should be interpreted with caution. Nevertheless, to identify Piwi’s target genes in the gut, mRNA sequencing of piwi mutant guts reveals hundreds of dysregulated protein-coding genes (Tang et al., 2023). Gene ontology analysis indicates that several metabolic processes, including carbohydrate metabolism and reactive oxygen species (ROS) response pathways, are affected. Given that ROS levels impact various stem cell populations, Piwi may play a critical role in maintaining stem cell homeostasis through ROS regulation. Notably, Piwi’s involvement in ISC maintenance appears to influence adult longevity. Other Argonaute family members, such as Aub, Ago3, and Ago2, also impact lifespan, suggesting a broader role for silencing pathways in adult survival. Surprisingly, small RNA sequencing fails to detect piRNAs in the gut, a finding confirmed by an independent study (Siudeja et al., 2021), suggesting a piRNA-independent role in the gut for PIWI proteins. Additionally, this work identified TE insertions in the tumor suppressor gene Notch within Drosophila ISCs, which may contribute to the development of gut neoplasia (Siudeja et al., 2021). Moreover, Piwi function has been examined under acute proliferative conditions, such as cancer development or enteropathogenic infection (Sousa-Victor et al., 2017). In these contexts, Jak/STAT-dependent Piwi activation in ISCs is essential for the proliferative response, TE silencing, genome integrity, and apoptosis suppression. Together, these studies underscore the essential role of Piwi in somatic stem cell regulation under both physiological and pathological conditions, highlighting its involvement in broader molecular mechanisms such as TE repression and cellular metabolism.
Discussion
Stem cells, characterized by their capacity for self-renewal and differentiation into specialized cell types, play a fundamental role in development and tissue maintenance throughout life. A notable feature of many stem cells populations is their reliance on glycolysis over mitochondrial oxphos. This preference is reminiscent of the Warburg effect in cancer cells, where high glycolytic activity supports rapid proliferation (Liberti and Locasale, 2016; Liu and Chen, 2021). However, the exact functional significance of elevated glycolysis in stem cells remains incompletely understood.
Emerging evidence suggests that glycolytic enzymes may play roles beyond metabolism, functioning as RNA-binding proteins (RBPs) that regulate post-transcriptional gene expression (Castello et al., 2012). Various metabolic enzymes, including glycolytic enzymes such as Aldolase, Enolase, Hexokinase and Pyruvate Kinase, have been identified as RBPs (Baltz et al., 2012). Recent studies underscore the multifunctionality of these enzymes. For example, Enolase was shown to regulate embryonic stem cell differentiation through riboregulation of specific mRNAs (Huppertz et al., 2022), while Hexokinase demonstrated a nuclear role in hematopoietic stem cell maintenance by modulating chromatin accessibility and DNA integrity (Thomas et al., 2022). Moreover, in Drosophila, glycolytic enzymes have been implicated in piRNA biogenesis by binding Tudor proteins (Gao et al., 2015). Additionally, research has revealed that glycolytic enzymes are essential for the maintenance and function of GSCs in Drosophila (Rojas-Ríos et al., 2024). Specifically, Ald, Eno, and Pyk are expressed at higher levels in GSCs compared to differentiated germline cells and are required for both GSC maintenance and piRNA biogenesis (Gao et al., 2015; Rojas-Ríos et al., 2024). Importantly, Aub regulation of glycolytic mRNAs establishes a direct connection between translational control, metabolic programming, and piRNAs in stem cell biology. Furthermore, the mechanisms by which PIWI proteins regulate mRNAs appear to be evolutionarily conserved across species. For instance, in mammalian systems, homologs such as Miwi similarly activate translation by interacting with poly(A)-binding proteins and eIF3 subunits (Dai et al., 2019; Ramat et al., 2020). Overall, these parallels suggest that the PIWI-mediated regulation of cellular mRNA translation is a fundamental feature of developmental biology. Since the expression of PIWI proteins and piRNAs, along with elevated glycolysis, are hallmark features of both stem cells and cancer cells, it would be highly interesting to investigate whether the piRNA pathway regulates energy metabolism in cancer cells and whether glycolytic enzymes perform moonlighting functions as RBPs in these cell types.
Author contributions
FC-L: Writing–original draft, Writing–review and editing. PR-R: Funding acquisition, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. PR-R is supported by Contrato de Acceso de I+D+i from VI PPIT—Universidad de Sevilla and Programa EMERGIA 2023 from Junta de Andalucía (DGP_EMEC_2023_00239). FC-L was supported by “Beca de Iniciación a la Investigación” from VII PPIT-Seville University. Work in R-RP’s laboratory is supported by Programa EMERGIA 2023 from Junta de Andalucía (DGP_EMEC_2023_00239), VI and VII PPIT-Seville University (2021/00001269 and 2024/00000593).
Acknowledgments
The authors apologize that space constraints have prevented the inclusion of some valuable contributions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. doi:10.1038/nature04916
Baltz, A. G., Munschauer, M., Schwanhäusser, B., Vasile, A., Murakawa, Y., Schueler, M., et al. (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690. doi:10.1016/j.molcel.2012.05.021
Barckmann, B., Pierson, S., Dufourt, J., Papin, C., Armenise, C., Port, F., et al. (2015). Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 12, 1205–1216. doi:10.1016/j.celrep.2015.07.030
Bradshaw, B., Thompson, K., and Frank, U. (2015). Distinct mechanisms underlie oral vs aboral regeneration in the cnidarian Hydractinia echinata. eLife 4, e05506. doi:10.7554/eLife.05506
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. doi:10.1016/j.cell.2007.01.043
Carmell, M. A., Girard, A., Kant, H. J. G., Bourc’his, D., Bestor, T. H., Rooij, D. G., et al. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514. doi:10.1016/j.devcel.2007.03.001
Castello, A., Fischer, B., Eichelbaum, K., Horos, R., Beckmann, B. M., Strein, C., et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406. doi:10.1016/j.cell.2012.04.031
Chary, S., and Hayashi, R. (2023). The absence of core piRNA biogenesis factors does not impact efficient transposon silencing in Drosophila. PLoS Biol. 21, e3002099. doi:10.1371/journal.pbio.3002099
Cox, D. N., Chao, A., Baker, J., Chang, L., Qiao, D., and Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727. doi:10.1101/gad.12.23.3715
Cox, D. N., Chao, A., and Lin, H. (2000). Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514. doi:10.1242/dev.127.3.503
Dai, P., Wang, X., Gou, L.-T., Li, Z.-T., Wen, Z., Chen, Z.-G., et al. (2019). A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell 179, 1566–1581. doi:10.1016/j.cell.2019.11.022
Dansereau, D. A., and Lasko, P. (2008). The development of germline stem cells in Drosophila. Methods Mol. Biol. 450, 3–26. doi:10.1007/978-1-60327-214-8_1
De Fazio, S., Bartonicek, N., Di Giacomo, M., Abreu-Goodger, C., Sankar, A., Funaya, C., et al. (2011). The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263. doi:10.1038/nature10547
Deng, W., and Lin, H. (2002). Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830. doi:10.1016/S1534-5807(0200165-X)
Dufourt, J., Bontonou, G., Chartier, A., Jahan, C., Meunier, A.-C., Pierson, S., et al. (2017). piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nat. Commun. 8, 1305. doi:10.1038/s41467-017-01431-5
Dufourt, J., Dennis, C., Boivin, A., Gueguen, N., Théron, E., Goriaux, C., et al. (2014). Spatio-temporal requirements for transposable element piRNA-mediated silencing during Drosophila oogenesis. Nucleic Acids Res. 42, 2512–2524. doi:10.1093/nar/gkt1184
Gao, M., Thomson, T. C., Creed, T. M., Tu, S., Loganathan, S. N., Jackson, C. A., et al. (2015). Glycolytic enzymes localize to ribonucleoprotein granules in Drosophila germ cells, bind Tudor and protect from transposable elements. EMBO Rep. 16, 379–386. doi:10.15252/embr.201439694
Ge, D. T., Wang, W., Tipping, C., Gainetdinov, I., Weng, Z., and Zamore, P. D. (2019). The RNA-binding ATPase, armitage, couples piRNA amplification in nuage to phased piRNA production on mitochondria. Mol. Cell 74, 982–995. doi:10.1016/j.molcel.2019.04.006
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A. (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202. doi:10.1038/nature04917
Gou, L.-T., Dai, P., Yang, J.-H., Xue, Y., Hu, Y.-P., Zhou, Y., et al. (2014). Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24, 680–700. doi:10.1038/cr.2014.41
Grivna, S. T., Beyret, E., Wang, Z., and Lin, H. (2006). A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714. doi:10.1101/gad.1434406
Gunawardane, L. S., Saito, K., Nishida, K. M., Miyoshi, K., Kawamura, Y., Nagami, T., et al. (2007). A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science 315, 1587–1590. doi:10.1126/science.1140494
Guo, Z., and Wang, Z. (2009). The glypican Dally is required in the niche for the maintenance of germline stem cells and short-range BMP signaling in the Drosophilaovary. Development 136, 3627–3635. doi:10.1242/dev.036939
Han, B. W., Wang, W., Li, C., Weng, Z., and Zamore, P. D. (2015). Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821. doi:10.1126/science.aaa1264
Harris, A. N., and Macdonald, P. M. (2001). Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128, 2823–2832. doi:10.1242/dev.128.14.2823
Harris, R. E., and Ashe, H. L. (2011). Cease and desist: modulating short-range Dpp signalling in the stem-cell niche. EMBO Rep. 12, 519–526. doi:10.1038/embor.2011.80
Hayashi, Y., Kobayashi, S., and Nakato, H. (2009). Drosophila glypicans regulate the germline stem cell niche. J. Cell Biol. 187, 473–480. doi:10.1083/jcb.200904118
Houwing, S., Berezikov, E., and Ketting, R. F. (2008). Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27, 2702–2711. doi:10.1038/emboj.2008.204
Huang, X., Fejes Tóth, K., and Aravin, A. A. (2017). piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 33, 882–894. doi:10.1016/j.tig.2017.09.002
Huppertz, I., Perez-Perri, J. I., Mantas, P., Sekaran, T., Schwarzl, T., Russo, F., et al. (2022). Riboregulation of Enolase 1 activity controls glycolysis and embryonic stem cell differentiation. Mol. Cell 82, 2666–2680.e11. doi:10.1016/j.molcel.2022.05.019
Jin, J., and Zhao, T. (2023). Niche formation and function in developing tissue: studies from the Drosophila ovary. Cell Commun. Signal. 21, 23. doi:10.1186/s12964-022-01035-7
Jin, Z., Flynt, A. S., and Lai, E. C. (2013). Drosophila piwi mutants exhibit germline stem cell tumors that are sustained by elevated dpp signaling. Curr. Biol. 23, 1442–1448. doi:10.1016/j.cub.2013.06.021
Joly, W., Chartier, A., Rojas-Rios, P., Busseau, I., and Simonelig, M. (2013). The CCR4 deadenylase acts with nanos and pumilio in the fine-tuning of mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep. 1, 411–424. doi:10.1016/j.stemcr.2013.09.007
Juliano, C. E., Reich, A., Liu, N., Götzfried, J., Zhong, M., Uman, S., et al. (2014). PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 337–342. doi:10.1073/pnas.1320965111
Kashima, M., Agata, K., and Shibata, N. (2018). Searching for non-transposable targets of planarian nuclear PIWI in pluripotent stem cells and differentiated cells. Dev. Growth Differ. 60, 260–277. doi:10.1111/dgd.12536
Kawaoka, S., Arai, Y., Kadota, K., Suzuki, Y., Hara, K., Sugano, S., et al. (2011). Zygotic amplification of secondary piRNAs during silkworm embryogenesis. RNA 17, 1401–1407. doi:10.1261/rna.2709411
Kawaoka, S., Minami, K., Katsuma, S., Mita, K., and Shimada, T. (2008). Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem. Biophysical Res. Commun. 367, 755–760. doi:10.1016/j.bbrc.2008.01.013
Kim, I. V., Duncan, E. M., Ross, E. J., Gorbovytska, V., Nowotarski, S. H., Elliott, S. A., et al. (2019). Planarians recruit piRNAs for mRNA turnover in adult stem cells. Genes Dev. 33, 1575–1590. doi:10.1101/gad.322776.118
Kiuchi, T., Koga, H., Kawamoto, M., Shoji, K., Sakai, H., Arai, Y., et al. (2014). A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636. doi:10.1038/nature13315
Klein, J. D., Qu, C., Yang, X., Fan, Y., Tang, C., and Peng, J. C. (2016). c-Fos repression by piwi regulates Drosophila ovarian germline formation and tissue morphogenesis. PLoS Genet. 12, e1006281. doi:10.1371/journal.pgen.1006281
Krishna, S., Nair, A., Cheedipudi, S., Poduval, D., Dhawan, J., Palakodeti, D., et al. (2013). Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata. Nucleic Acids Res. 41, 599–616. doi:10.1093/nar/gks1020
Lau, N. C., Ohsumi, T., Borowsky, M., Kingston, R. E., and Blower, M. D. (2009). Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 28, 2945–2958. doi:10.1038/emboj.2009.237
Lau, N. C., Seto, A. G., Kim, J., Kuramochi-Miyagawa, S., Nakano, T., Bartel, D. P., et al. (2006). Characterization of the piRNA complex from rat testes. Science 313, 363–367. doi:10.1126/science.1130164
Li, D., Taylor, D. H., and Van Wolfswinkel, J. C. (2021). PIWI-mediated control of tissue-specific transposons is essential for somatic cell differentiation. Cell Rep. 37, 109776. doi:10.1016/j.celrep.2021.109776
Liberti, M. V., and Locasale, J. W. (2016). The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 41, 211–218. doi:10.1016/j.tibs.2015.12.001
Lim, R. S. M., Anand, A., Nishimiya-Fujisawa, C., Kobayashi, S., and Kai, T. (2014). Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev. Biol. 386, 237–251. doi:10.1016/j.ydbio.2013.12.007
Lin, H., and Spradling, A. C. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463–2476. doi:10.1242/dev.124.12.2463
Lin, K.-Y., Wang, W.-D., Lin, C.-H., Rastegari, E., Su, Y.-H., Chang, Y.-T., et al. (2020). Piwi reduction in the aged niche eliminates germline stem cells via Toll-GSK3 signaling. Nat. Commun. 11, 3147. doi:10.1038/s41467-020-16858-6
Liu, M., Lim, T. M., and Cai, Y. (2010). The Drosophila female germline stem cell lineage acts to spatially restrict DPP function within the niche. Sci. Signal 3, ra57. doi:10.1126/scisignal.2000740
Liu, W., and Chen, G. (2021). Regulation of energy metabolism in human pluripotent stem cells. Cell. Mol. Life Sci. 78, 8097–8108. doi:10.1007/s00018-021-04016-0
López-Onieva, L., Fernández-Miñán, A., and González-Reyes, A. (2008). Jak/Stat signalling in niche support cells regulates dpptranscription to control germline stem cell maintenance in the Drosophila ovary. Development 135, 533–540. doi:10.1242/dev.016121
Losick, V. P., Morris, L. X., Fox, D. T., and Spradling, A. (2011). Drosophila Stem Cell Niches: a decade of discovery suggests a unified view of stem cell regulation. Dev. Cell 21, 159–171. doi:10.1016/j.devcel.2011.06.018
Ma, X., Zhu, X., Han, Y., Story, B., Do, T., Song, X., et al. (2017). Aubergine controls germline stem cell self-renewal and progeny differentiation via distinct mechanisms. Dev. Cell 41, 157–169. doi:10.1016/j.devcel.2017.03.023
Mohn, F., Handler, D., and Brennecke, J. (2015). Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817. doi:10.1126/science.aaa1039
Nagao, A., Mituyama, T., Huang, H., Chen, D., Siomi, M. C., and Siomi, H. (2010). Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16, 2503–2515. doi:10.1261/rna.2270710
Nishida, K. M., Saito, K., Mori, T., Kawamura, Y., Nagami-Okada, T., Inagaki, S., et al. (2007). Gene silencing mechanisms mediated by Aubergine–piRNA complexes in Drosophila male gonad. RNA 13, 1911–1922. doi:10.1261/rna.744307
Nong, W., Cao, J., Li, Y., Qu, Z., Sun, J., Swale, T., et al. (2020). Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat. Commun. 11, 3051. doi:10.1038/s41467-020-16801-9
Özata, D. M., Yu, T., Mou, H., Gainetdinov, I., Colpan, C., Cecchini, K., et al. (2020). Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 4, 156–168. doi:10.1038/s41559-019-1065-1
Palakodeti, D., Smielewska, M., Lu, Y.-C., Yeo, G. W., and Graveley, B. R. (2008). The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14, 1174–1186. doi:10.1261/rna.1085008
Peng, J. C., Valouev, A., Liu, N., and Lin, H. (2016). Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Polycomb group proteins. Nat. Genet. 48, 283–291. doi:10.1038/ng.3486
Post, C., Clark, J. P., Sytnikova, Y. A., Chirn, G.-W., and Lau, N. C. (2014). The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 20, 1977–1986. doi:10.1261/rna.046300.114
Praher, D., Zimmermann, B., Genikhovich, G., Columbus-Shenkar, Y., Modepalli, V., Aharoni, R., et al. (2017). Characterization of the piRNA pathway during development of the sea anemone Nematostella vectensis. RNA Biol. 14, 1727–1741. doi:10.1080/15476286.2017.1349048
Ramat, A., Garcia-Silva, M.-R., Jahan, C., Naït-Saïdi, R., Dufourt, J., Garret, C., et al. (2020). The PIWI protein Aubergine recruits eIF3 to activate translation in the germ plasm. Cell Res. 30, 421–435. doi:10.1038/s41422-020-0294-9
Reddien, P. W., Oviedo, N. J., Jennings, J. R., Jenkin, J. C., and Alvarado, A. S. (2005). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330. doi:10.1126/science.1116110
Resch, A. M., and Palakodeti, D. (2012). Small RNA pathways in Schmidtea mediterranea. Int. J. Dev. Biol. 56, 67–74. doi:10.1387/ijdb.113436ar
Reuter, M., Berninger, P., Chuma, S., Shah, H., Hosokawa, M., Funaya, C., et al. (2011). Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267. doi:10.1038/nature10672
Rinkevich, Y., Voskoboynik, A., Rosner, A., Rabinowitz, C., Paz, G., Oren, M., et al. (2013). Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Dev. Cell 24, 76–88. doi:10.1016/j.devcel.2012.11.010
Rojas-Ríos, P., Chartier, A., Enjolras, C., Cremaschi, J., Garret, C., Boughlita, A., et al. (2024). piRNAs are regulators of metabolic reprogramming in stem cells. Nat. Commun. 15, 8405. doi:10.1038/s41467-024-52709-4
Rojas-Ríos, P., Chartier, A., Pierson, S., and Simonelig, M. (2017). Aubergine and pi RNA s promote germline stem cell self-renewal by repressing the proto-oncogene Cbl. EMBO J. 36, 3194–3211. doi:10.15252/embj.201797259
Rojas-Ríos, P., and González-Reyes, A. (2014). Concise review: the plasticity of stem cell niches: a general property behind tissue homeostasis and repair. Stem Cells 32, 852–859. doi:10.1002/stem.1621
Rojas-Ríos, P., Guerrero, I., and González-Reyes, A. (2012). Cytoneme-mediated delivery of Hedgehog regulates the expression of bone morphogenetic proteins to maintain germline stem cells in Drosophila. PLOS Biol. 10, e1001298. doi:10.1371/journal.pbio.1001298
Rojas-Ríos, P., and Simonelig, M. (2018). piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 145, dev161786. doi:10.1242/dev.161786
Rosales-Nieves, A. E., and González-Reyes, A. (2014). Genetics and mechanisms of ovarian cancer: parallels between Drosophila and humans. Semin. Cell Dev. Biol. 28, 104–109. doi:10.1016/j.semcdb.2014.03.031
Ross, R. J., Weiner, M. M., and Lin, H. (2014). PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359. doi:10.1038/nature12987
Rouget, C., Papin, C., Boureux, A., Meunier, A.-C., Franco, B., Robine, N., et al. (2010). Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467, 1128–1132. doi:10.1038/nature09465
Rouhana, L., Weiss, J., King, R., and Newmark, P. (2014). PIWI homologs mediate histone H4 mRNA localization to planarian chromatoid bodies. Development 141, 2592–2601. doi:10.1242/dev.101618
Sánchez-Gómez, H., Garrido-Maraver, J., and Rojas-Ríos, P. (2024). Extended live imaging of female Drosophila melanogaster germline stem cell niches. J. Vis. Exp. doi:10.3791/67389
Senti, K.-A., Jurczak, D., Sachidanandam, R., and Brennecke, J. (2015). piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 29, 1747–1762. doi:10.1101/gad.267252.115
Shibata, N., Kashima, M., Ishiko, T., Nishimura, O., Rouhana, L., Misaki, K., et al. (2016). Inheritance of a nuclear PIWI from pluripotent stem cells by somatic descendants ensures differentiation by silencing transposons in planarian. Dev. Cell 37, 226–237. doi:10.1016/j.devcel.2016.04.009
Shoji, M., Tanaka, T., Hosokawa, M., Reuter, M., Stark, A., Kato, Y., et al. (2009). The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell 17, 775–787. doi:10.1016/j.devcel.2009.10.012
Siudeja, K., Van Den Beek, M., Riddiford, N., Boumard, B., Wurmser, A., Stefanutti, M., et al. (2021). Unraveling the features of somatic transposition in the Drosophila intestine. EMBO J. 40, e106388. doi:10.15252/embj.2020106388
Smulders-Srinivasan, T. K., and Lin, H. (2003). Screens for piwi suppressors in Drosophila identify dosage-dependent regulators of germline stem cell division. Genetics 165, 1971–1991. doi:10.1093/genetics/165.4.1971
Sousa-Victor, P., Ayyaz, A., Hayashi, R., Qi, Y., Madden, D. T., Lunyak, V. V., et al. (2017). Piwi is required to limit exhaustion of aging somatic stem cells. Cell Rep. 20, 2527–2537. doi:10.1016/j.celrep.2017.08.059
Sun, H., Li, D., Chen, S., Liu, Y., Liao, X., Deng, W., et al. (2010). Zili inhibits transforming growth factor-beta signaling by interacting with Smad4. J. Biol. Chem. 285, 4243–4250. doi:10.1074/jbc.M109.079533
Swevers, L., Liu, J., Huvenne, H., and Smagghe, G. (2011). Search for limiting factors in the RNAi pathway in silkmoth tissues and the Bm5 cell line: the RNA-binding proteins R2D2 and translin. PLOS ONE 6, e20250. doi:10.1371/journal.pone.0020250
Szakmary, A., Cox, D. N., Wang, Z., and Lin, H. (2005). Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 15, 171–178. doi:10.1016/j.cub.2005.01.005
Tang, X., Liu, N., Qi, H., and Lin, H. (2023). Piwi maintains homeostasis in the Drosophila adult intestine. Stem Cell Rep. 18, 503–518. doi:10.1016/j.stemcr.2023.01.001
Thomas, G. E., Egan, G., García-Prat, L., Botham, A., Voisin, V., Patel, P. S., et al. (2022). The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness. Nat. Cell Biol. 24, 872–884. doi:10.1038/s41556-022-00925-9
Toombs, J. A., Sytnikova, Y. A., Chirn, G., Ang, I., Lau, N. C., and Blower, M. D. (2017). Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome. RNA 23, 504–520. doi:10.1261/rna.058859.116
Upadhyay, M., Martino Cortez, Y., Wong-Deyrup, S., Tavares, L., Schowalter, S., Flora, P., et al. (2016). Transposon dysregulation modulates dWnt4 signaling to control germline stem cell differentiation in Drosophila. PLoS Genet. 12, e1005918. doi:10.1371/journal.pgen.1005918
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. doi:10.1126/science.1129333
Villa-Fombuena, G., Lobo-Pecellín, M., Marín-Menguiano, M., Rojas-Ríos, P., and González-Reyes, A. (2021). Live imaging of the Drosophila ovarian niche shows spectrosome and centrosome dynamics during asymmetric germline stem cell division. Development 148, dev199716. doi:10.1242/dev.199716
Vourekas, A., Alexiou, P., Vrettos, N., Maragkakis, M., and Mourelatos, Z. (2016). Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 531, 390–394. doi:10.1038/nature17150
Wang, L., Li, Z., and Cai, Y. (2008a). The JAK/STAT pathway positively regulates DPP signaling in the Drosophila germline stem cell niche. J. Cell Biol. 180, 721–728. doi:10.1083/jcb.200711022
Wang, W., Han, B. W., Tipping, C., Ge, D. T., Zhang, Z., Weng, Z., et al. (2015). Slicing and binding by Ago3 or aub trigger piwi-bound piRNA production by distinct mechanisms. Mol. Cell 59, 819–830. doi:10.1016/j.molcel.2015.08.007
Wang, X., Harris, R. E., Bayston, L. J., and Ashe, H. L. (2008b). Type IV collagens regulate BMP signalling in Drosophila. Nature 455, 72–77. doi:10.1038/nature07214
Wang, X., Ramat, A., Simonelig, M., and Liu, M.-F. (2023). Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat. Rev. Mol. Cell Biol. 24, 123–141. doi:10.1038/s41580-022-00528-0
Watanabe, T., Cui, X., Yuan, Z., Qi, H., and Lin, H. (2018). MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. EMBO J. 37, e95329. doi:10.15252/embj.201695329
Wilczynska, A., Minshall, N., Armisen, J., Miska, E. A., and Standart, N. (2009). Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA 15, 337–345. doi:10.1261/rna.1422509
Wu, W.-S., Brown, J. S., Chen, T.-T., Chu, Y.-H., Huang, W.-C., Tu, S., et al. (2019). piRTarBase: a database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 47, D181–D187. doi:10.1093/nar/gky956
Xie, T., and Spradling, A. C. (1998). Decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94, 251–260. doi:10.1016/s0092-8674(00)81424-5
Keywords: PIWI proteins, piRNAs, germline stem cells, Drosophila, mRNA regulation
Citation: Claro-Linares F and Rojas-Ríos P (2025) PIWI proteins and piRNAs: key regulators of stem cell biology. Front. Cell Dev. Biol. 13:1540313. doi: 10.3389/fcell.2025.1540313
Received: 05 December 2024; Accepted: 20 January 2025;
Published: 06 February 2025.
Edited by:
Myon Hee Lee, East Carolina University, United StatesReviewed by:
Kyung Won Kim, Hallym University, Republic of KoreaCopyright © 2025 Claro-Linares and Rojas-Ríos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patricia Rojas-Ríos, cHJvamFzQHVzLmVz
 Fernando Claro-Linares
Fernando Claro-Linares Patricia Rojas-Ríos
Patricia Rojas-Ríos