
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 06 March 2025
Sec. Epigenomics and Epigenetics
Volume 13 - 2025 | https://doi.org/10.3389/fcell.2025.1533148
5-Methylcytosine (m5C) is a widespread RNA methylation modification, wherein a methyl group is enzymatically transferred to specific RNA sites by methyltransferases, such as the NSUN family and DNMT2. The m5C modification not only impacts RNA structure and stability but also governs post-transcriptional regulation by influencing RNA transport, translation, and protein interactions. Recently, the functional importance of m5C in complex diseases, including cancer, has gained substantial attention. Increasing evidence highlights the critical roles of m5C in digestive system malignancies, where it contributes to tumor progression by modulating oncogene expression and regulating processes such as tumor cell proliferation, migration, invasion, and resistance to chemotherapy. Furthermore, m5C’s involvement in non-coding RNAs reveals additional dimensions in elucidating their roles in cancer. This review summarizes recent advances in m5C RNA methylation research within digestive system tumors, focusing on its functional mechanisms, clinical significance, and potential applications. Specifically, it aims to explore m5C’s role in tumor diagnosis, prognosis, and treatment, while proposing future directions to address current challenges and broaden its clinical utility.
RNA modifications have emerged as a pivotal aspect of epigenetics, drawing increasing attention in recent years (Barbieri and Kouzarides, 2020; Li G. et al., 2024; Orsolic et al., 2023). Various chemical alterations, including N6-methyladenosine (m6A), 5-methylcytosine (m5C), and N1-methyladenosine (m1A), play critical roles in regulating gene expression, RNA processing, stability, nuclear export, and translation (Orsolic et al., 2023; Roundtree et al., 2017; Shahrajabian and Sun, 2023; Zhang et al., 2023c). Aberrant RNA modifications are frequently implicated in the onset and progression of numerous diseases, particularly cancer, where they are key contributors to tumorigenesis, progression, and drug resistance (Han et al., 2023; Huang et al., 2020; Yang B. et al., 2021; Zhuang et al., 2023).
Among these modifications, m5C is a prominent methylation mark found in diverse RNA species (Sun et al., 2023; Wiener and Schwartz, 2021; Zheng et al., 2023). It is predominantly catalyzed by RNA methyltransferases, such as the NSUN family and DNMT2, and is commonly present in tRNA, rRNA, mRNA, as well as long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) (Cusenza et al., 2023; He et al., 2020; Nombela et al., 2021; Wang Y. et al., 2023). m5C serves multiple regulatory functions by modulating RNA structure, stability, nuclear export, and translation. It enhances RNA stability by preventing degradation and influences post-transcriptional processes, including splicing, transport, and translation (Squires et al., 2012). Recent advances in high-throughput sequencing technologies have gradually illuminated the role of m5C in cancer. Research suggests that m5C modification regulates oncogene expression and plays a role in controlling cancer cell proliferation, migration, invasion, and chemoresistance (Yang et al., 2017). Specifically, NSUN2-mediated m5C modification stabilizes oncogene mRNA, facilitating tumor progression (Chen S. J. et al., 2024). Furthermore, m5C’s involvement in non-coding RNAs, such as lncRNAs and circRNAs, has gained increasing recognition, as it modulates their function and protein interactions, thereby impacting cancer development and progression (Zheng et al., 2022).
In digestive system malignancies, dysregulated m5C expression is closely linked to tumorigenesis, progression, and patient prognosis (Lin and Kuang, 2024). Studies have revealed abnormal expression patterns of m5C methyltransferases, including NSUN2 and NSUN6, as well as m5C-binding proteins like YBX1 and ALYREF, in cancers such as esophageal, gastric, hepatocellular, colorectal, and pancreatic cancers. These alterations significantly affect tumor progression by regulating oncogene expression, tumor cell proliferation, migration, and responsiveness to chemotherapy. Consequently, investigating the role of m5C modifications in digestive system cancers is essential for deciphering the molecular mechanisms underlying tumor development and for devising novel diagnostic and therapeutic approaches. This review aims to comprehensively summarize the current advancements in m5C RNA methylation research within digestive system cancers, with a focus on its functional and molecular mechanisms across various tumor types, while exploring its potential as a biomarker for diagnosis, prognosis, and as a therapeutic target.
The modification of RNA by m5C was first identified in the 1950s, when it was detected in tRNA and rRNA (Amos and Korn, 1958). However, only with the advent of high-throughput sequencing technologies did comprehensive investigations confirm the presence of m5C across a broader range of RNA species, including mRNA, miRNA, lncRNA, and circRNA (Amos and Korn, 1958; Li and Huang, 2024). In recent years, m5C has emerged as a pivotal epigenetic regulatory mechanism, garnering significant attention in cancer biology. Compared to other RNA modifications, m5C exhibits distinct and intricate patterns of distribution and function across various RNA types. Research has shown that m5C affects RNA structure and stability while playing a critical role in regulating gene expression and translation, thus contributing to cancer initiation and progression (Cusenza et al., 2023; He et al., 2020; Nombela et al., 2021; Squires et al., 2012; Wang Y. et al., 2023). Notably, the dynamic and adaptable nature of m5C modification underscores its significant role within the tumor microenvironment, potentially influencing cell fate decisions and cancer heterogeneity (Han et al., 2023; Yang B. et al., 2021; Zhang et al., 2021).
With ongoing technological advancements, m5C RNA methylation detection methods have evolved significantly, moving from early quantitative techniques to modern, high-throughput approaches that allow precise localization (Motorin et al., 2010). One of the earliest methods, high-performance liquid chromatography (HPLC), facilitated the quantification of m5C content by separating and analyzing RNA fragments. While effective, HPLC’s limitations in identifying the exact location of m5C modifications have led to the development of more precise tools (Schaefer et al., 2009). Mass spectrometry (MS), recognized for its high sensitivity and accuracy, offers detailed insights into RNA molecules. By integrating efficient sample preparation with advanced analytical processes, MS generates refined maps of m5C modifications (Dominissini et al., 2012; Edelheit et al., 2013). As next-generation sequencing (NGS) technologies matured, chemical labeling approaches enabled more comprehensive detection. For example, bisulfite conversion combined with RNA-seq (BS-seq) selectively converts cytosine into uracil while preserving m5C, facilitating its identification in sequencing data (Cui et al., 2017; Frommer et al., 1992). Additionally, methylated RNA immunoprecipitation sequencing (MeRIP-seq) employs specific m5C antibodies to enrich m5C-modified RNA, significantly improving sensitivity and specificity (Zhang et al., 2024). At the forefront of technological innovation, single-molecule real-time sequencing (SMRT) has begun to reveal unique advantages (Ardui et al., 2018; Fang et al., 2012). SMRT enables direct RNA sequencing without requiring chemical labeling or immunoprecipitation, allowing the detection of various RNA modifications, including m5C, at the single-molecule level. Its ability to identify multiple RNA modifications simultaneously positions it as a promising tool in RNA modification research. These advanced detection techniques, particularly when employed in combination, provide higher resolution and sensitivity in m5C modification analysis, propelling further exploration in this rapidly advancing field.
m5C modifications play an essential role in regulating various RNA molecules, with their functions intricately linked to specific regulatory factors (Chen et al., 2021). The primary m5C methyltransferases are members of the NSUN protein family (NSUN1-7) and DNMT2 (Figure 1) (Chen T. et al., 2023; He et al., 2020; Li et al., 2022; Sun et al., 2022) These enzymes catalyze m5C modifications at designated RNA sites, impacting a broad spectrum of biological functions. Each NSUN family member exhibits distinct functional specificity for different RNA types. For instance, NSUN2 is the predominant methyltransferase responsible for mRNA and tRNA methylation, whereas NSUN6 primarily modifies rRNA. DNMT2, initially identified as a DNA methyltransferase, was later recognized for its role in catalyzing m5C modifications in tRNA as well (Cheng et al., 2018; Li H. et al., 2024; Zhang et al., 2018).
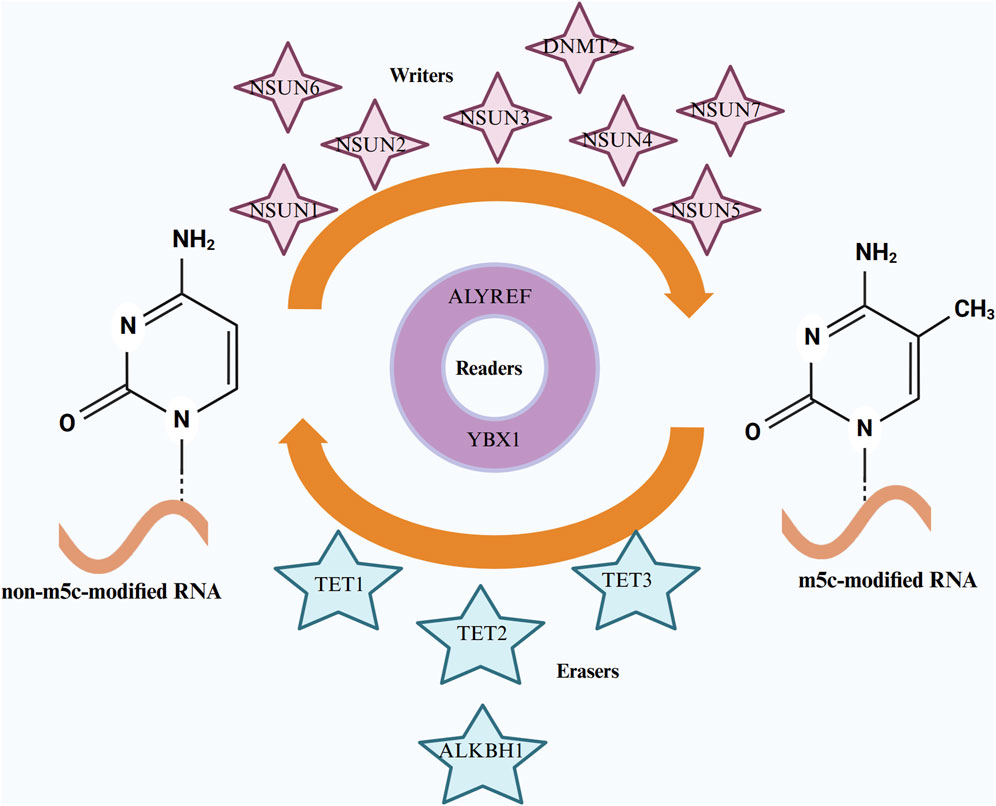
Figure 1. The regulatory mechanisms of RNA m5C methylation involving “Writers,” “Erasers,” and “Readers.” “Writers” are methyltransferases, such as the NSUN family members and DNMT2, responsible for adding m5C methyl groups to RNA. “Erasers,” including demethylases like the TET family and ALKBH1, remove these m5C modifications. “Readers,” such as ALYREF and YBX1, recognize and bind to m5C-modified RNA. Additionally, chemical structures of both unmodified RNA and m5C-modified RNA are depicted for comparison.
The dynamic regulation of m5C modifications is not solely governed by methyltransferases but also relies on demethylases, which enable the reversibility and precise modulation of these modifications (Chen et al., 2021). The TET family of proteins (TET1, TET2, and TET3), originally characterized for their role in DNA demethylation, has been suggested to function in RNA demethylation as well, providing new perspectives on the plasticity of m5C modifications (Dusadeemeelap et al., 2022; Hu et al., 2013; Ito et al., 2011; Li et al., 2023b; Shen et al., 2021). In addition to the TET proteins, ALKBH1 (AlkB homolog 1), a key demethylase, has been shown to remove m5C modifications from RNA (Arguello et al., 2022). ALKBH1, a Fe(II)/α-ketoglutarate-dependent dioxygenase, was initially known for its role in demethylating N1-methyladenine and N3-methylcytosine in DNA (Zhong et al., 2024). More recent research has demonstrated that ALKBH1 can also demethylate m5C in RNA, particularly in tRNA and rRNA, where its activity is crucial for maintaining RNA stability and function (Arguello et al., 2022).
Beyond the methyltransferases and demethylases, the biological effects of m5C modifications are further modulated by m5C-binding proteins (Wang N. et al., 2023). ALYREF (Aly/REF export factor) is a key m5C-binding protein that recognizes and binds to m5C-modified RNA, regulating processes such as RNA transport, nuclear export, and translation (Yang et al., 2017; Zhao et al., 2024). The interaction between ALYREF and m5C-modified RNA enhances both the stability and translational efficiency of the RNA, a function particularly critical in tumor cells (Klec et al., 2022; Nagy et al., 2021; Yang et al., 2023). Other m5C-binding proteins, including YBX1 and FMRP, also play vital roles in downstream regulation. YBX1 binds to m5C-modified mRNA, influencing its stability, splicing, and translation efficiency. In the context of cancer, YBX1 promotes tumor growth and metastasis by upregulating oncogene expression, and its overexpression is frequently linked to poor prognosis (Chen et al., 2019; Zou et al., 2024). Under stress conditions, YBX1 stabilizes mRNA, aiding tumor cells in surviving adverse environments (Liu X. et al., 2024; Meng H. et al., 2024; Wang et al., 2022). FMRP, primarily active in the nervous system, binds to m5C-modified RNA to regulate nuclear export and the localized translation of synaptic mRNAs (Chen Y. S. et al., 2023). The loss or dysfunction of FMRP is closely associated with neurodevelopmental disorders such as Fragile X syndrome, where it plays a pivotal role in maintaining neuronal function by regulating synaptic mRNA dynamics. These m5C-binding proteins perform multifaceted regulatory roles by interacting with m5C-modified RNA across various cellular contexts, contributing to disease development and progression (Chen Y. S. et al., 2023; Yang et al., 2022). Through the coordinated actions of methyltransferases, demethylases, and RNA-binding proteins (readers), the status of m5C modifications is finely tuned to meet specific physiological demands. This dynamic and reversible regulation ensures appropriate RNA stability, translational efficiency, transport, and RNA-protein interactions. The ability to tightly control m5C modifications is not only critical for normal cellular processes but also plays a significant role in cancer development and progression.
The intricate regulatory networks outlined above set the stage for understanding how m5C modifications impact specific RNA classes and their associated biological functions. The role of m5C modifications in mRNA has been extensively explored, revealing their critical impact on mRNA stability, nuclear export, and translation efficiency (Boo and Kim, 2020; Sun et al., 2023; Zhao et al., 2017). Research indicates that m5C modifications enhance mRNA stability by protecting it from ribonuclease-mediated degradation, thereby prolonging mRNA expression within cells (Selmi et al., 2021; Squires et al., 2012; Yang et al., 2022; Zhang et al., 2023b). NSUN2 introduces m5C modifications in the 5′UTR or 3′UTR regions of mRNA, which contributes to increased mRNA stability and augments translation efficiency (Chen S. J. et al., 2024; Yang et al., 2017). Furthermore, m5C modifications are intricately linked to mRNA nuclear export. Proteins such as ALYREF, which specifically bind to m5C-modified mRNA, facilitate its transport from the nucleus to the cytoplasm, thereby influencing gene expression regulation (Fan et al., 2019). These processes underscore the multifaceted regulatory functions of m5C modifications at the mRNA level, highlighting their pivotal role in the precise control of gene expression.
In the realm of non-coding RNAs, m5C modifications also exert significant influence (Fabian and Sonenberg, 2012; Ferragut Cardoso et al., 2021; Xue et al., 2022). miRNAs, which are short non-coding RNAs involved in post-transcriptional gene regulation, are subject to regulation by m5C modifications. These modifications can affect miRNA precursor processing, thereby modulating the levels and activity of mature miRNAs, which in turn impacts critical cellular processes such as proliferation, differentiation, and apoptosis (Carissimi et al., 2021; Tang et al., 2023). Similarly, m5C modifications play a vital role in the regulation of lncRNAs (Ali and Grote, 2020; Herman et al., 2022). By influencing the stability and secondary structure of lncRNAs, m5C modifications enable lncRNAs to interact with proteins or DNA, thereby exerting control over downstream gene expression (Huang et al., 2023; Jiang et al., 2024; Pan et al., 2022). circRNAs, a unique class of non-coding RNAs characterized by their covalently closed circular structure, are also influenced by m5C modifications (Xue et al., 2021; Zhou et al., 2020). These modifications have been shown to affect both the biogenesis and functional roles of circRNAs. In the context of cancer, m5C-modified circRNAs are associated with promoting cancer cell proliferation, metastasis, and resistance to chemotherapy (Hou et al., 2024).
The elevated RNA m5C methylation observed in ESCC tumors stems from the overexpression of the m5C methyltransferase NSUN2 and the m5C “reader” Y-box-binding protein 1 (YBX1) (Liu L. et al., 2024; Niu et al., 2022; Su et al., 2021). Both NSUN2 and YBX1 are markedly upregulated in esophageal cancer tissues compared to adjacent normal tissues (Table 1). Higher NSUN2 expression is linked to more advanced cancer stages and heightened drug resistance, while elevated YBX1 expression correlates with poorer patient survival (Liu L. et al., 2024; Niu et al., 2022; Su et al., 2021). Functionally, NSUN2 overexpression significantly promotes cell proliferation, migration, and invasion (Table 2) (Liu L. et al., 2024; Niu et al., 2022; Su et al., 2021) In vivo experiments reveal that tumor growth and lung metastasis are markedly suppressed in NSUN2 knockout mice. NSUN2 also enhances resistance to irradiation in vivo (Niu et al., 2022). Moreover, YBX1 facilitates proliferation, invasion, and pluripotency maintenance in ESCC cells in vitro, increasing the sphere-forming ability of TE1 cells and regulating the expression of EMT and stem cell-associated proteins, including MMP1, MMP2, and β-catenin (Liu L. et al., 2024). YBX1 overexpression further promotes the growth and metastasis of esophageal cancer. Mechanistically, NSUN2 overexpression is positively regulated by E2F1, and NSUN2 induces m5C modification of growth factor receptor-bound protein 2 (GRB2), stabilizing its mRNA. LIN28B recognizes this modification, further stabilizing GRB2 mRNA (Su et al., 2021). Additionally, increased NSUN2 activity upregulates numerous oncogenes via m5C methylation, driving ESCC progression and the emergence of chemoradiotherapy resistance (Niu et al., 2022). YBX1, in an NSUN2- and m5C-dependent manner, binds to and stabilizes SMOX mRNA (Liu L. et al., 2024). The YBX1/m5C-SMOX axis accelerates ESCC progression by activating mTORC1 signaling.
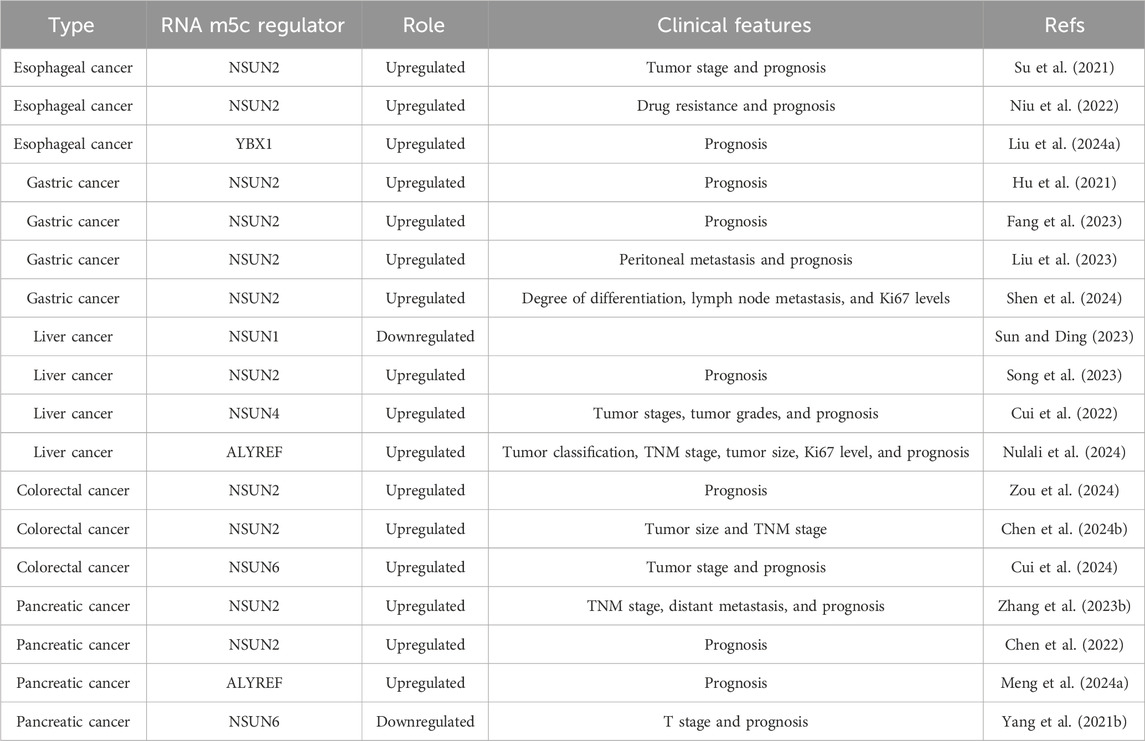
Table 1. Expression profiles and associated clinical features of various RNA m5C modification regulators in digestive system cancers.
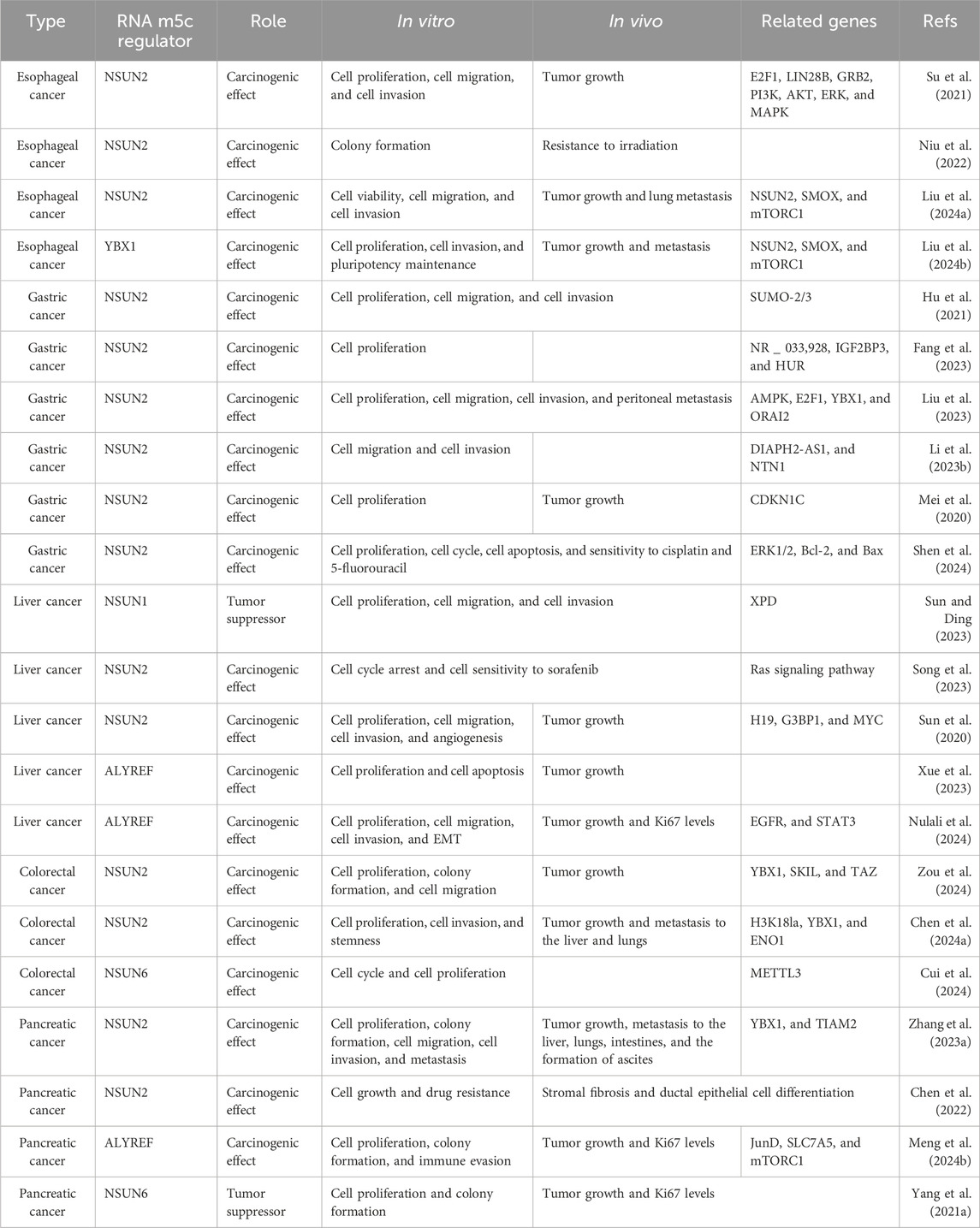
Table 2. Roles of various RNA m5C regulators and their associated genes in multiple digestive system cancers.
In gastric cancer (GC) tissues, NSUN2 is significantly upregulated compared to adjacent normal tissues, with its expression positively correlated to factors such as tumor differentiation, lymph node metastasis, elevated Ki67 levels, and peritoneal metastasis (Table 1) (Fang et al., 2023; Hu et al., 2021; Liu et al., 2023; Mei et al., 2020; Shen et al., 2024) Higher NSUN2 expression is associated with reduced overall survival (OS) in patients with GC, and univariate analysis identifies it as an independent prognostic risk factor for OS (Fang et al., 2023; Hu et al., 2021; Liu et al., 2023). Functionally, NSUN2 inhibition suppresses GC cell proliferation, migration, invasion, and peritoneal metastasis in vitro, while inducing cell cycle arrest and promoting apoptosis (Hu et al., 2021; Li et al., 2023a; Liu et al., 2023; Mei et al., 2020). Conversely, NSUN2 overexpression accelerates in vivo tumor growth, resulting in increased tumor volume and weight (Mei et al., 2020).
Mechanistically, the small ubiquitin-like modifier (SUMO)-2/3 directly interacts with and stabilizes NSUN2 and facilitates its nuclear translocation, enhancing its oncogenic function (Figure 2) (Hu et al., 2021) Additionally, the lncRNA NR_033928, methylated and upregulated by NSUN2 in an m5C-dependent manner, plays a critical role in promoting GC progression by enhancing its stability and expression (Fang et al., 2023). The transcription factor E2F1 further stimulates NSUN2 expression by binding to specific cis-regulatory elements (Liu et al., 2023). NSUN2-mediated m5C modification increases ORAI2 expression via YBX1-dependent stabilization of ORAI2 mRNA. Similarly, DIAPH2-AS1 binds to NSUN2, enhancing its stability by preventing ubiquitin-proteasome-mediated degradation (Li et al., 2023a). NSUN2 also promotes the upregulation of NTN1 through m5C modification and may facilitate GC progression by inhibiting CDKN1C (p57Kip2), a downstream target, in an m5C-dependent manner (Mei et al., 2020). Furthermore, NSUN2 inhibition reduces the phosphorylation of ERK1/2, leading to decreased levels of the anti-apoptotic protein Bcl-2 and increased levels of the pro-apoptotic protein Bax (Figure 2), thereby sensitizing GC cells to 5-FU/CDDP by enhancing apoptosis (Shen et al., 2024).
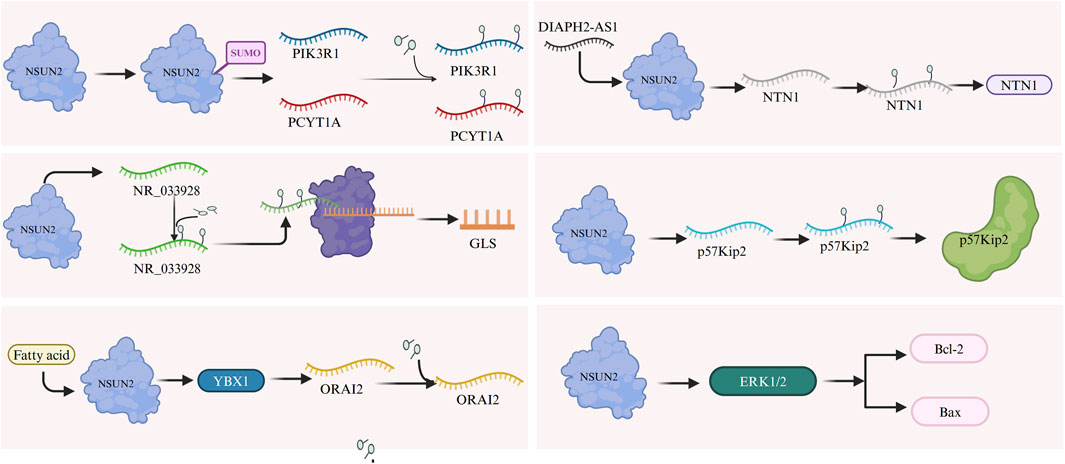
Figure 2. The m5C RNA methylation regulator NSUN2 promotes gastric cancer progression through several mechanisms. SUMO-2/3 interacts with and stabilizes NSUN2, facilitating its nuclear translocation and enhancing its oncogenic activity. NSUN2 methylates and stabilizes lncRNA NR_033928, driving cancer progression. E2F1 stimulates NSUN2 expression, while NSUN2 stabilizes ORAI2 mRNA via YBX1, increasing ORAI2 expression. DIAPH2-AS1 binds NSUN2, preventing its degradation through the ubiquitin-proteasome pathway, which enhances NSUN2 stability and upregulates NTN1 expression. NSUN2 may also inhibit CDKN1C (p57Kip2) through m5C modification, contributing to tumor progression. Furthermore, NSUN2 inhibition decreases ERK1/2 phosphorylation, reducing anti-apoptotic Bcl-2 levels and increasing pro-apoptotic Bax levels.
In hepatocellular carcinoma (HCC) tissues and cells, NSUN2, NSUN4, and ALYREF are significantly upregulated, while NSUN1 is notably downregulated (Table 1) (Cui et al., 2022; Nulali et al., 2024; Song et al., 2023; Sun and Ding, 2023; Xue et al., 2023) ALYREF expression correlates positively with tumor classification, TNM stage, tumor size, and Ki67 levels, and elevated NSUN4 levels are indicative of more advanced tumor stages and grades (Cui et al., 2022; Nulali et al., 2024). Patients with higher expression levels of NSUN2, NSUN4, and ALYREF typically exhibit worse prognoses (Cui et al., 2022; Nulali et al., 2024; Song et al., 2023). ALYREF shows high diagnostic accuracy for HCC, with an AUC of 0.88, while NSUN4 expression serves as an independent prognostic risk factor (Cui et al., 2022; Nulali et al., 2024). Functionally, NSUN2 deficiency inhibits HCC cell proliferation, migration, invasion, and angiogenesis, while also increasing sensitivity to sorafenib (Song et al., 2023; Sun et al., 2020). Notably, chronic hepatitis B virus (HBV) infection is a key contributor to HCC. NSUN2 deficiency downregulates HBV expression, reducing HBV replication, whereas TET2 deficiency upregulates HBV expression (Feng et al., 2023). ALYREF further promotes proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) in HCC cells while suppressing apoptosis (Nulali et al., 2024; Xue et al., 2023). Silencing ALYREF significantly decreases tumor growth in vivo by reducing HCC cell proliferation.
Mechanistically, NSUN1 modulates XPD levels via m5C methylation, thereby inhibiting HCC progression (Sun and Ding, 2023). NSUN2 influences sorafenib resistance through the regulation of Ras pathway activity (Song et al., 2023). NSUN2-mediated RNA methylation promotes H19 lncRNA expression, with methylated H19 interacting with the oncoprotein G3BP1 to delay MYC mRNA decay, thereby driving tumor progression (Sun et al., 2020). Additionally, ALYREF recognizes the m5C modification of EGFR and regulates its levels, activating the STAT3 signaling pathway and further promoting HCC progression (Nulali et al., 2024).
In colorectal cancer (CRC) tissues and cells, NSUN2 and NSUN6 are significantly upregulated (Chen B. et al., 2024; Cui et al., 2024; Zou et al., 2024). Specifically, NSUN2 expression is positively correlated with tumor size, TNM stage, and overall tumor stage, while NSUN6 expression is closely associated with ethnicity and tumor stage (Table 1) (Chen B. et al., 2024; Cui et al., 2024; Zou et al., 2024) Patients with CRC exhibiting elevated NSUN2 expression have poorer OS and disease-free survival (DFS) rates compared to those with lower NSUN2 levels (Chen B. et al., 2024; Zou et al., 2024). ROC curve analysis has highlighted NSUN6’s considerable diagnostic value across different cohorts (Tables 1, 2) (Cui et al., 2024) Silencing NSUN2 significantly reduces the stemness of CRC cells, and its knockdown effectively impairs tumor growth and metastasis to the liver and lungs in vivo. Furthermore, the NSUN2 inhibitor, Nsun2-i4, has demonstrated efficacy in significantly curbing tumor growth and reducing tumor burden in CRC (Zou et al., 2024). When combined with PD-1 therapy, Nsun2-i4 further amplifies tumor growth inhibition compared to monotherapy.
Mechanistically, NSUN2 positively regulates the expression of the SKI-like oncogene (SKIL) through RNA m5C modification in a YBX1-dependent manner, thereby upregulating SKIL mRNA (Zou et al., 2024). Elevated SKIL expression promotes tumor progression by activating the transcriptional coactivator with PDZ-binding motif (TAZ) (Zou et al., 2024). Additionally, NSUN2 and YBX1 jointly target ENO1 in an m5C-dependent fashion, facilitating glucose metabolism reprogramming (Chen B. et al., 2024). Lactate produced by CRC cells enhances NSUN2 expression and its RNA-binding affinity through histone H3K18 lactylation (H3K18la), promoting m5C-mediated CRC progression and metastasis (Chen B. et al., 2024). Moreover, NSUN6 knockdown decreases m5C levels on METTL3, leading to METTL3 upregulation, which can partially offset the cell cycle arrest and proliferation inhibition triggered by NSUN6 knockdown (Figure 3) (Cui et al., 2024).
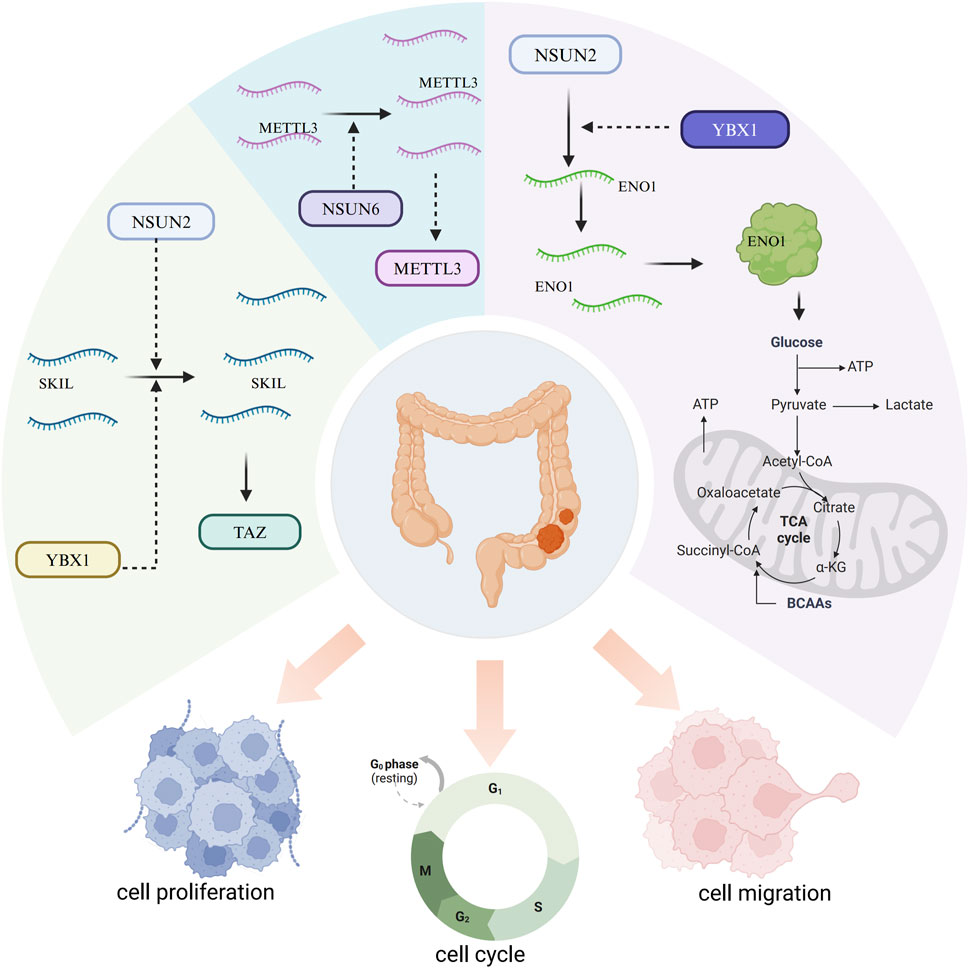
Figure 3. Mechanisms of m5C RNA methylation regulators in colorectal cancer. NSUN2 positively regulates the SKI-like oncogene (SKIL) via m5C modification in a YBX1-dependent manner, promoting tumor progression by activating TAZ. NSUN2 and YBX1 also collaboratively target Eno1 in an m5C-dependent manner, facilitating the reprogramming of glucose metabolism. NSUN6 knockdown reduces m5C modification on METTL3, resulting in METTL3 upregulation, which partially compensates for the cell cycle arrest and proliferation inhibition caused by NSUN6 knockdown.
In pancreatic cancer tissues, NSUN2 and ALYREF are significantly upregulated, with NSUN2 expression positively correlating with TNM stage and distant metastasis (Table 1) (Chen et al., 2022; Meng Q. et al., 2024; Zhang G. et al., 2023) Elevated levels of NSUN2 and ALYREF are associated with shorter OS in patients with pancreatic cancer (Chen et al., 2022; Meng Q. et al., 2024; Zhang G. et al., 2023). In contrast, NSUN6 expression is reduced, and its levels are significantly correlated with the T stage and Ki67+ cell rate, showing a negative correlation with both OS and DFS (Yang R. et al., 2021). Regression analysis indicates that ALYREF and NSUN6 function as independent prognostic biomarkers for predicting pancreatic cancer outcomes (Meng Q. et al., 2024; Yang R. et al., 2021). Functionally, NSUN2 knockdown exerts a modest impact on pancreatic cancer cell growth and drug sensitivity, with effects becoming more pronounced over time (Chen et al., 2022). In vivo, NSUN2 knockdown leads to reduced stromal fibrosis and the restoration of ductal epithelial differentiation (Zhang G. et al., 2023). NSUN2 also promotes cell migration, invasion, and metastasis, and in vivo studies show that NSUN2 enhances tumor growth and metastasis to the liver, lungs, intestines, and ascites formation. Additionally, ALYREF has been shown to promote tumor growth and increase Ki-67 expression in tumor tissues, while NSUN6 inhibits pancreatic cancer cell proliferation and in vivo tumor growth (Meng Q. et al., 2024; Yang R. et al., 2021). Mechanistically, NSUN2-mediated m5C modification suppresses TIAM2 expression through YBX1, and disruption of the NSUN2/TIAM2 axis impairs the EMT, thereby slowing pancreatic cancer progression (Zhang G. et al., 2023). ALYREF, by enhancing CD8+ T cell functionality, contributes to the delay of pancreatic cancer development (Meng Q. et al., 2024). Furthermore, ALYREF directly regulates JunD in an m5C-dependent manner, leading to the transcriptional activation of SLC7A5. This activation of the JunD-SLC7A5-mTORC1 signaling pathway drives the proliferation of pancreatic ductal adenocarcinoma (PDAC) cells and facilitates tumor immune evasion (Meng Q. et al., 2024).
m5C modifications have emerged as promising biomarkers for the diagnosis and prognosis of digestive system cancers. Research has established that the overexpression of m5C methyltransferases, including NSUN2, NSUN4, and NSUN6, as well as m5C “readers” such as YBX1 and ALYREF, is strongly linked to tumor progression, poor prognosis, and treatment resistance across cancers like esophageal squamous cell carcinoma (ESCC), GC, HCC, CRC, and PDAC. For instance, in ESCC, elevated NSUN2 levels are associated with advanced tumor stages and reduced patient survival, emphasizing its potential as a prognostic marker (Niu et al., 2022; Su et al., 2021). Similarly, in GC, NSUN2 overexpression is correlated with decreased overall survival and increased metastasis risk (Liu et al., 2023). In HCC, NSUN4 and ALYREF are independent prognostic indicators with high diagnostic accuracy (Cui et al., 2022; Nulali et al., 2024; Xue et al., 2023). NSUN6 plays a critical role in CRC and PDAC; in CRC, higher NSUN6 expression is linked to advanced tumor stages and lower survival rates, while in PDAC, reduced NSUN6 expression negatively correlates with prognostic markers like T stage and Ki67 positivity (Cui et al., 2024; Yang R. et al., 2021). ROC curve analysis further underscores NSUN6’s diagnostic significance across diverse populations, highlighting its potential as a biomarker for digestive system cancers (Yang R. et al., 2021). Despite its central importance in cancer biology, research on m5C faces technical challenges, such as the limited sensitivity and specificity of current detection methods and obstacles in translating these findings into clinical applications.
Targeting m5C modification pathways presents a promising therapeutic strategy for digestive system cancers (Wang C. et al., 2023). Inhibitors of m5C methyltransferases, such as NSUN2 inhibitors, have been shown to significantly reduce tumor growth and metastasis while enhancing the sensitivity of cancer cells to chemotherapeutic agents like cisplatin and 5-fluorouracil (Liu L. et al., 2024; Shen et al., 2024). In gastric and colorectal cancers, NSUN2 inhibition effectively suppresses cancer cell proliferation and migration, while improving the response to chemotherapy (Shen et al., 2024). NSUN6’s role in digestive tumors also suggests its potential as a therapeutic target. In PDAC, low NSUN6 expression is associated with enhanced cancer cell proliferation and decreased sensitivity to chemotherapy, whereas restoring NSUN6 activity inhibits tumor cell proliferation and reduces invasiveness, improving patient outcomes (Yang R. et al., 2021). In CRC, NSUN6 knockdown decreases m5C levels and upregulates METTL3 expression, which partially counteracts the cell cycle arrest and proliferation inhibition induced by NSUN6 knockdown (Cui et al., 2024). Additionally, targeting the interactions between m5C “readers” like YBX1 and ALYREF with m5C-modified RNA opens new avenues for disrupting key oncogenic pathways (Liu L. et al., 2024; Meng Q. et al., 2024). For example, inhibiting NSUN2 in PDAC reduces stromal fibrosis and restores ductal epithelial differentiation, thus slowing tumor progression and reinforcing NSUN2’s potential as a therapeutic target (Chen et al., 2022). Overall, targeting m5C modifications and their regulatory proteins offers substantial potential for the treatment of digestive system cancers and represents a critical avenue for advancing precision medicine strategies.
As a vital epigenetic modification, m5C RNA methylation plays a pivotal role in the initiation, progression, and treatment of digestive system cancers. Studies have demonstrated that aberrant expression of m5C methyltransferases, such as NSUN2, NSUN4, and NSUN6, and m5C-binding proteins like YBX1 and ALYREF, is closely linked to the progression of various digestive tumors, including esophageal, gastric, hepatocellular, colorectal, and pancreatic cancers. Elevated levels of m5C modifications are frequently associated with advanced tumor stages, poor prognoses, and resistance to standard therapies, positioning m5C modifications as valuable biomarkers for cancer diagnosis and prognosis. On the therapeutic front, targeting m5C modifications and their regulatory proteins presents substantial promise. Inhibiting the activity or expression of m5C methyltransferases, such as NSUN2, can significantly suppress tumor cell proliferation, migration, and invasion while enhancing sensitivity to chemotherapy. Furthermore, disrupting the interaction between m5C-binding proteins, such as YBX1 and ALYREF, and m5C-modified RNA could interfere with key signaling pathways that drive tumor progression, offering new opportunities for the development of Therapeutic strategies targeting m5C modifications. Despite progress in understanding the role of m5C modifications in digestive system cancers, many aspects remain unexplored. Future research could focus on several critical areas: first, elucidating the dynamic regulatory mechanisms governing m5C modifications, especially the role of demethylases; second, investigating the tumor-specific effects of m5C modifications and their influence on tumor heterogeneity; and finally, developing more precise and effective m5C-targeted therapies aimed at improving clinical outcomes while minimizing adverse effects. These efforts will be crucial for unlocking the full potential of m5C modifications in cancer treatment.
XH: Conceptualization, Formal Analysis, Investigation, Visualization, Writing–original draft, Writing–review and editing. YL: Conceptualization, Investigation, Methodology, Writing–review and editing. SZ: Conceptualization, Formal Analysis, Methodology, Writing–review and editing. KL: Formal Analysis, Investigation, Methodology, Writing–review and editing. XG: Conceptualization, Funding acquisition, Supervision, Validation, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 81600512) and Science and technology Research program of Henan Province (No. 242102311156).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, T., and Grote, P. (2020). Beyond the rna-dependent function of lncrna genes. Elife 9, e60583. doi:10.7554/eLife.60583
Amos, H., and Korn, M. (1958). 5-methyl cytosine in the rna of escherichia coli. Biochim. Biophys. Acta 29, 444–445. doi:10.1016/0006-3002(58)90214-2
Ardui, S., Ameur, A., Vermeesch, J. R., and Hestand, M. S. (2018). Single molecule real-time (smrt) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 46, 2159–2168. doi:10.1093/nar/gky066
Arguello, A. E., Li, A., Sun, X., Eggert, T. W., Mairhofer, E., and Kleiner, R. E. (2022). Reactivity-dependent profiling of rna 5-methylcytidine dioxygenases. Nat. Commun. 13, 4176. doi:10.1038/s41467-022-31876-2
Barbieri, I., and Kouzarides, T. (2020). Role of rna modifications in cancer. Nat. Rev. Cancer 20, 303–322. doi:10.1038/s41568-020-0253-2
Boo, S. H., and Kim, Y. K. (2020). The emerging role of rna modifications in the regulation of mrna stability. Exp. Mol. Med. 52, 400–408. doi:10.1038/s12276-020-0407-z
Carissimi, C., Laudadio, I., Lorefice, E., Azzalin, G., De Paolis, V., and Fulci, V. (2021). Bisulphite mirna-seq reveals widespread cpg and non-cpg 5-(hydroxy)methyl-cytosine in human micrornas. RNA Biol. 18, 2226–2235. doi:10.1080/15476286.2021.1927423
Chen, B., Deng, Y., Hong, Y., Fan, L., Zhai, X., Hu, H., et al. (2024a). Metabolic recoding of nsun2-mediated m(5)c modification promotes the progression of colorectal cancer via the nsun2/ybx1/m(5)c-eno1 positive feedback loop. Adv. Sci. (Weinh) 11, e2309840. doi:10.1002/advs.202309840
Chen, S. J., Zhang, J., Zhou, T., Rao, S. S., Li, Q., Xiao, L. Y., et al. (2024b). Epigenetically upregulated nsun2 confers ferroptosis resistance in endometrial cancer via m(5)c modification of slc7a11 mrna. Redox Biol. 69, 102975. doi:10.1016/j.redox.2023.102975
Chen, S. Y., Chen, K. L., Ding, L. Y., Yu, C. H., Wu, H. Y., Chou, Y. Y., et al. (2022). Rna bisulfite sequencing reveals nsun2-mediated suppression of epithelial differentiation in pancreatic cancer. Oncogene 41, 3162–3176. doi:10.1038/s41388-022-02325-7
Chen, T., Xu, Z. G., Luo, J., Manne, R. K., Wang, Z., Hsu, C. C., et al. (2023a). Nsun2 is a glucose sensor suppressing cgas/sting to maintain tumorigenesis and immunotherapy resistance. Cell Metab. 35, 1782–1798.e8. doi:10.1016/j.cmet.2023.07.009
Chen, X., Li, A., Sun, B. F., Yang, Y., Han, Y. N., Yuan, X., et al. (2019). 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mrnas. Nat. Cell Biol. 21, 978–990. doi:10.1038/s41556-019-0361-y
Chen, Y. S., Dong, J., Tan, W., Liu, H., Zhang, S. M., Zou, J., et al. (2023b). The potential role of ribonucleic acid methylation in the pathological mechanisms of fragile x syndrome. Behav. Brain Res. 452, 114586. doi:10.1016/j.bbr.2023.114586
Chen, Y. S., Yang, W. L., Zhao, Y. L., and Yang, Y. G. (2021). Dynamic transcriptomic m(5) c and its regulatory role in rna processing. Wiley Interdiscip. Rev. RNA 12, e1639. doi:10.1002/wrna.1639
Cheng, J. X., Chen, L., Li, Y., Cloe, A., Yue, M., Wei, J., et al. (2018). Rna cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat. Commun. 9, 1163. doi:10.1038/s41467-018-03513-4
Cui, M., Qu, F., Wang, L., Liu, X., Yu, J., Tang, Z., et al. (2022). M5c rna methyltransferase-related gene nsun4 stimulates malignant progression of hepatocellular carcinoma and can be a prognostic marker. Cancer Biomark. 33, 389–400. doi:10.3233/cbm-210154
Cui, X., Liang, Z., Shen, L., Zhang, Q., Bao, S., Geng, Y., et al. (2017). 5-methylcytosine rna methylation in arabidopsis thaliana. Mol. Plant 10, 1387–1399. doi:10.1016/j.molp.2017.09.013
Cui, Y., Lv, P., and Zhang, C. (2024). Nsun6 mediates 5-methylcytosine modification of mettl3 and promotes colon adenocarcinoma progression. J. Biochem. Mol. Toxicol. 38, e23749. doi:10.1002/jbt.23749
Cusenza, V. Y., Tameni, A., Neri, A., and Frazzi, R. (2023). The lncrna epigenetics: the significance of m6a and m5c lncrna modifications in cancer. Front. Oncol. 13, 1063636. doi:10.3389/fonc.2023.1063636
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6a rna methylomes revealed by m6a-seq. Nature 485, 201–206. doi:10.1038/nature11112
Dusadeemeelap, C., Rojasawasthien, T., Matsubara, T., Kokabu, S., and Addison, W. N. (2022). Inhibition of tet-mediated DNA demethylation suppresses osteoblast differentiation. Faseb J. 36, e22153. doi:10.1096/fj.202101402R
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O., and Sorek, R. (2013). Transcriptome-wide mapping of 5-methylcytidine rna modifications in bacteria, archaea, and yeast reveals m5c within archaeal mrnas. PLoS Genet. 9, e1003602. doi:10.1371/journal.pgen.1003602
Fabian, M. R., and Sonenberg, N. (2012). The mechanics of mirna-mediated gene silencing: a look under the hood of mirisc. Nat. Struct. Mol. Biol. 19, 586–593. doi:10.1038/nsmb.2296
Fan, J., Wang, K., Du, X., Wang, J., Chen, S., Wang, Y., et al. (2019). Alyref links 3'-end processing to nuclear export of non-polyadenylated mrnas. Embo J. 38, e99910. doi:10.15252/embj.201899910
Fang, G., Munera, D., Friedman, D. I., Mandlik, A., Chao, M. C., Banerjee, O., et al. (2012). Genome-wide mapping of methylated adenine residues in pathogenic escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239. doi:10.1038/nbt.2432
Fang, L., Huang, H., Lv, J., Chen, Z., Lu, C., Jiang, T., et al. (2023). M5c-methylated lncrna nr_033928 promotes gastric cancer proliferation by stabilizing gls mrna to promote glutamine metabolism reprogramming. Cell Death Dis. 14, 520. doi:10.1038/s41419-023-06049-8
Feng, J., Xu, T., He, M., Li, J., Yao, P., Ma, C., et al. (2023). Nsun2-mediated m5c modification of hbv rna positively regulates hbv replication. PLoS Pathog. 19, e1011808. doi:10.1371/journal.ppat.1011808
Ferragut Cardoso, A. P., Banerjee, M., Nail, A. N., Lykoudi, A., and States, J. C. (2021). Mirna dysregulation is an emerging modulator of genomic instability. Semin. Cancer Biol. 76, 120–131. doi:10.1016/j.semcancer.2021.05.004
Frommer, M., McDonald, L. E., Millar, D. S., Collis, C. M., Watt, F., Grigg, G. W., et al. (1992). A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A. 89, 1827–1831. doi:10.1073/pnas.89.5.1827
Han, M., Sun, H., Zhou, Q., Liu, J., Hu, J., Yuan, W., et al. (2023). Effects of rna methylation on tumor angiogenesis and cancer progression. Mol. Cancer 22, 198. doi:10.1186/s12943-023-01879-8
He, Y., Zhang, Q., Zheng, Q., Yu, X., and Guo, W. (2020). Distinct 5-methylcytosine profiles of circular rna in human hepatocellular carcinoma. Am. J. Transl. Res., 12, 5719–5729. doi:10.7554/eLife.56205
Herman, A. B., Tsitsipatis, D., and Gorospe, M. (2022). Integrated lncrna function upon genomic and epigenomic regulation. Mol. Cell 82, 2252–2266. doi:10.1016/j.molcel.2022.05.027
Hou, C., Liu, J., Liu, J., Yao, D., Liang, F., Qin, C., et al. (2024). 5-methylcytosine-mediated upregulation of circular rna 0102913 augments malignant properties of colorectal cancer cells through a microrna-571/rac family small gtpase 2 axis. Gene 901, 148162. doi:10.1016/j.gene.2024.148162
Hu, L., Li, Z., Cheng, J., Rao, Q., Gong, W., Liu, M., et al. (2013). Crystal structure of tet2-DNA complex: insight into tet-mediated 5mc oxidation. Cell 155, 1545–1555. doi:10.1016/j.cell.2013.11.020
Hu, Y., Chen, C., Tong, X., Chen, S., Hu, X., Pan, B., et al. (2021). Nsun2 modified by sumo-2/3 promotes gastric cancer progression and regulates mrna m5c methylation. Cell Death Dis. 12, 842. doi:10.1038/s41419-021-04127-3
Huang, F., Wang, X., Zhong, J., Chen, H., Song, D., Xu, T., et al. (2023). Using integrated analysis from multicentre studies to identify rna methylation-related lncrna risk stratification systems for glioma. Cancer Cell Int. 23, 156. doi:10.1186/s12935-023-03001-w
Huang, H., Weng, H., and Chen, J. (2020). M(6)a modification in coding and non-coding rnas: roles and therapeutic implications in cancer. Cancer Cell 37, 270–288. doi:10.1016/j.ccell.2020.02.004
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. doi:10.1126/science.1210597
Jiang, J., Duan, M., Wang, Z., Lai, Y., Zhang, C., and Duan, C. (2024). Rna epigenetics in pulmonary diseases: insights into methylation modification of lncrnas in lung cancer. Biomed. Pharmacother. 175, 116704. doi:10.1016/j.biopha.2024.116704
Klec, C., Knutsen, E., Schwarzenbacher, D., Jonas, K., Pasculli, B., Heitzer, E., et al. (2022). Alyref, a novel factor involved in breast carcinogenesis, acts through transcriptional and post-transcriptional mechanisms selectively regulating the short neat1 isoform. Cell Mol. Life Sci. 79, 391. doi:10.1007/s00018-022-04402-2
Li, G., Yao, Q., Liu, P., Zhang, H., Liu, Y., Li, S., et al. (2024a). Critical roles and clinical perspectives of rna methylation in cancer. MedComm 5 (2020), e559. doi:10.1002/mco2.559
Li, H., Liu, H., Zhu, D., Dou, C., Gang, B., Zhang, M., et al. (2024b). Biological function molecular pathways and druggability of dnmt2/trdmt1. Pharmacol. Res. 205, 107222. doi:10.1016/j.phrs.2024.107222
Li, M., Tao, Z., Zhao, Y., Li, L., Zheng, J., Li, Z., et al. (2022). 5-methylcytosine rna methyltransferases and their potential roles in cancer. J. Transl. Med. 20, 214. doi:10.1186/s12967-022-03427-2
Li, P., and Huang, D. (2024). Nsun2-mediated rna methylation: molecular mechanisms and clinical relevance in cancer. Cell Signal 123, 111375. doi:10.1016/j.cellsig.2024.111375
Li, Y., Xia, Y., Jiang, T., Chen, Z., Shen, Y., Lin, J., et al. (2023a). Long noncoding rna diaph2-as1 promotes neural invasion of gastric cancer via stabilizing nsun2 to enhance the m5c modification of ntn1. Cell Death Dis. 14, 260. doi:10.1038/s41419-023-05781-5
Li, Y., Xue, M., Deng, X., Dong, L., Nguyen, L. X. T., Ren, L., et al. (2023b). Tet2-mediated mrna demethylation regulates leukemia stem cell homing and self-renewal. Cell Stem Cell 30, 1072–1090.e10. doi:10.1016/j.stem.2023.07.001
Lin, S., and Kuang, M. (2024). Rna modification-mediated mrna translation regulation in liver cancer: mechanisms and clinical perspectives. Nat. Rev. Gastroenterol. Hepatol. 21, 267–281. doi:10.1038/s41575-023-00884-y
Liu, K., Xu, P., Lv, J., Ge, H., Yan, Z., Huang, S., et al. (2023). Peritoneal high-fat environment promotes peritoneal metastasis of gastric cancer cells through activation of nsun2-mediated orai2 m5c modification. Oncogene 42, 1980–1993. doi:10.1038/s41388-023-02707-5
Liu, L., Chen, Y., Zhang, T., Cui, G., Wang, W., Zhang, G., et al. (2024a). YBX1 promotes esophageal squamous cell carcinoma progression via m5C-dependent SMOX mRNA stabilization. Adv. Sci. (Weinh) 11, e2302379. doi:10.1002/advs.202302379
Liu, X., Wei, Q., Yang, C., Zhao, H., Xu, J., Mobet, Y., et al. (2024b). Rna m(5)c modification upregulates e2f1 expression in a manner dependent on ybx1 phase separation and promotes tumor progression in ovarian cancer. Exp. Mol. Med. 56, 600–615. doi:10.1038/s12276-024-01184-4
Mei, L., Shen, C., Miao, R., Wang, J. Z., Cao, M. D., Zhang, Y. S., et al. (2020). Rna methyltransferase nsun2 promotes gastric cancer cell proliferation by repressing p57(kip2) by an m(5)c-dependent manner. Cell Death Dis. 11, 270. doi:10.1038/s41419-020-2487-z
Meng, H., Miao, H., Zhang, Y., Chen, T., Yuan, L., Wan, Y., et al. (2024a). Ybx1 promotes homologous recombination and resistance to platinum-induced stress in ovarian cancer by recognizing m5c modification. Cancer Lett. 597, 217064. doi:10.1016/j.canlet.2024.217064
Meng, Q., Xie, Y., Sun, K., He, L., Wu, H., Zhang, Q., et al. (2024b). Alyref-jund-slc7a5 axis promotes pancreatic ductal adenocarcinoma progression through epitranscriptome-metabolism reprogramming and immune evasion. Cell Death Discov. 10, 97. doi:10.1038/s41420-024-01862-2
Motorin, Y., Lyko, F., and Helm, M. (2010). 5-methylcytosine in rna: detection, enzymatic formation and biological functions. Nucleic Acids Res. 38, 1415–1430. doi:10.1093/nar/gkp1117
Nagy, Z., Seneviratne, J. A., Kanikevich, M., Chang, W., Mayoh, C., Venkat, P., et al. (2021). An alyref-mycn coactivator complex drives neuroblastoma tumorigenesis through effects on usp3 and mycn stability. Nat. Commun. 12, 1881. doi:10.1038/s41467-021-22143-x
Niu, X., Peng, L., Liu, W., Miao, C., Chen, X., Chu, J., et al. (2022). A cis-eqtl in nsun2 promotes esophageal squamous-cell carcinoma progression and radiochemotherapy resistance by mrna-m(5)c methylation. Signal Transduct. Target Ther. 7, 267. doi:10.1038/s41392-022-01063-2
Nombela, P., Miguel-López, B., and Blanco, S. (2021). The role of m(6)a, m(5)c and ψ rna modifications in cancer: novel therapeutic opportunities. Mol. Cancer 20, 18. doi:10.1186/s12943-020-01263-w
Nulali, J., Zhang, K., Long, M., Wan, Y., Liu, Y., Zhang, Q., et al. (2024). Alyref-mediated rna 5-methylcytosine modification promotes hepatocellular carcinoma progression via stabilizing egfr mrna and pstat3 activation. Int. J. Biol. Sci. 20, 331–346. doi:10.7150/ijbs.82316
Orsolic, I., Carrier, A., and Esteller, M. (2023). Genetic and epigenetic defects of the rna modification machinery in cancer. Trends Genet. 39, 74–88. doi:10.1016/j.tig.2022.10.004
Pan, Q., Yi, C., and Zhang, Y. (2022). Overall survival signature of 5-methylcytosine regulators related long non-coding rna in hepatocellular carcinoma. Front. Oncol. 12, 884377. doi:10.3389/fonc.2022.884377
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic rna modifications in gene expression regulation. Cell 169, 1187–1200. doi:10.1016/j.cell.2017.05.045
Schaefer, M., Pollex, T., Hanna, K., and Lyko, F. (2009). Rna cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12. doi:10.1093/nar/gkn954
Selmi, T., Hussain, S., Dietmann, S., Heiß, M., Borland, K., Flad, S., et al. (2021). Sequence- and structure-specific cytosine-5 mrna methylation by nsun6. Nucleic Acids Res. 49, 1006–1022. doi:10.1093/nar/gkaa1193
Shahrajabian, M. H., and Sun, W. (2023). Survey on multi-omics, and multi-omics data analysis, integration andApplication application. Curr. Pharm. Anal., 19, 267–281. doi:10.2174/1573412919666230406100948
Shen, H., Ontiveros, R. J., Owens, M. C., Liu, M. Y., Ghanty, U., Kohli, R. M., et al. (2021). Tet-mediated 5-methylcytosine oxidation in trna promotes translation. J. Biol. Chem. 296, 100087. doi:10.1074/jbc.RA120.014226
Shen, X., Sun, H., Shu, S., Tang, W., Yuan, Y., Su, H., et al. (2024). Suppression of nsun2 enhances the sensitivity to chemosensitivity and inhibits proliferation by mediating cell apoptosis in gastric cancer. Pathol. Res. Pract. 253, 154986. doi:10.1016/j.prp.2023.154986
Song, D., An, K., Zhai, W., Feng, L., Xu, Y., Sun, R., et al. (2023). Nsun2-mediated mrna m(5)c modification regulates the progression of hepatocellular carcinoma. Genomics Proteomics Bioinforma. 21, 823–833. doi:10.1016/j.gpb.2022.09.007
Squires, J. E., Patel, H. R., Nousch, M., Sibbritt, T., Humphreys, D. T., Parker, B. J., et al. (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding rna. Nucleic Acids Res. 40, 5023–5033. doi:10.1093/nar/gks144
Su, J., Wu, G., Ye, Y., Zhang, J., Zeng, L., Huang, X., et al. (2021). Nsun2-mediated rna 5-methylcytosine promotes esophageal squamous cell carcinoma progression via lin28b-dependent grb2 mrna stabilization. Oncogene 40, 5814–5828. doi:10.1038/s41388-021-01978-0
Sun, G., Ma, S., Zheng, Z., Wang, X., Chen, S., Chang, T., et al. (2022). Multi-omics analysis of expression and prognostic value of nsun members in prostate cancer. Front. Oncol. 12, 965571. doi:10.3389/fonc.2022.965571
Sun, G. F., and Ding, H. (2023). Nop2-mediated m5c methylation of xpd is associated with hepatocellular carcinoma progression. Neoplasma 70, 340–349. doi:10.4149/neo_2023_230110N17
Sun, H., Li, K., Liu, C., and Yi, C. (2023). Regulation and functions of non-m(6)a mrna modifications. Nat. Rev. Mol. Cell Biol. 24, 714–731. doi:10.1038/s41580-023-00622-x
Sun, Z., Xue, S., Zhang, M., Xu, H., Hu, X., Chen, S., et al. (2020). Aberrant nsun2-mediated m(5)c modification of h19 lncrna is associated with poor differentiation of hepatocellular carcinoma. Oncogene 39, 6906–6919. doi:10.1038/s41388-020-01475-w
Tang, F., Liu, Y., Sun, Y., Xiong, Y., Gu, Y., Zhou, J., et al. (2023). Construction of a serum diagnostic signature based on m5c-related mirnas for cancer detection. Front. Endocrinol. (Lausanne) 14, 1099703. doi:10.3389/fendo.2023.1099703
Wang, C., Hou, X., Guan, Q., Zhou, H., Zhou, L., Liu, L., et al. (2023a). Rna modification in cardiovascular disease: implications for therapeutic interventions. Signal Transduct. Target Ther. 8, 412. doi:10.1038/s41392-023-01638-7
Wang, N., Chen, R. X., Deng, M. H., Wei, W. S., Zhou, Z. H., Ning, K., et al. (2023b). M(5)c-dependent cross-regulation between nuclear reader alyref and writer nsun2 promotes urothelial bladder cancer malignancy through facilitating rabl6/tk1 mrnas splicing and stabilization. Cell Death Dis. 14, 139. doi:10.1038/s41419-023-05661-y
Wang, Y., Wei, J., Feng, L., Li, O., Huang, L., Zhou, S., et al. (2023c). Aberrant m5c hypermethylation mediates intrinsic resistance to gefitinib through nsun2/ybx1/qsox1 axis in egfr-mutant non-small-cell lung cancer. Mol. Cancer 22, 81. doi:10.1186/s12943-023-01780-4
Wang, Z. Z., Meng, T., Yang, M. Y., Wang, W., Zhang, Y., Liu, Y., et al. (2022). Alyref associated with immune infiltration is a prognostic biomarker in hepatocellular carcinoma. Transl. Oncol. 21, 101441. doi:10.1016/j.tranon.2022.101441
Wiener, D., and Schwartz, S. (2021). The epitranscriptome beyond m(6)a. Nat. Rev. Genet. 22, 119–131. doi:10.1038/s41576-020-00295-8
Xue, C., Gu, X., Bao, Z., Su, Y., Lu, J., and Li, L. (2022). The mechanism underlying the ncrna dysregulation pattern in hepatocellular carcinoma and its tumor microenvironment. Front. Immunol. 13, 847728. doi:10.3389/fimmu.2022.847728
Xue, C., Gu, X., Zheng, Q., Shi, Q., Yuan, X., Su, Y., et al. (2023). Alyref mediates rna m(5)c modification to promote hepatocellular carcinoma progression. Signal Transduct. Target Ther. 8, 130. doi:10.1038/s41392-023-01395-7
Xue, C., Li, G., Lu, J., and Li, L. (2021). Crosstalk between circrnas and the pi3k/akt signaling pathway in cancer progression. Signal Transduct. Target Ther. 6, 400. doi:10.1038/s41392-021-00788-w
Yang, B., Wang, J. Q., Tan, Y., Yuan, R., Chen, Z. S., and Zou, C. (2021a). Rna methylation and cancer treatment. Pharmacol. Res. 174, 105937. doi:10.1016/j.phrs.2021.105937
Yang, H., Wang, Y., Xiang, Y., Yadav, T., Ouyang, J., Phoon, L., et al. (2022). Fmrp promotes transcription-coupled homologous recombination via facilitating tet1-mediated m5c rna modification demethylation. Proc. Natl. Acad. Sci. U. S. A. 119, e2116251119. doi:10.1073/pnas.2116251119
Yang, Q., Wang, M., Xu, J., Yu, D., Li, Y., Chen, Y., et al. (2023). Linc02159 promotes non-small cell lung cancer progression via alyref/yap1 signaling. Mol. Cancer 22, 122. doi:10.1186/s12943-023-01814-x
Yang, R., Liang, X., Wang, H., Guo, M., Shen, H., Shi, Y., et al. (2021b). The rna methyltransferase nsun6 suppresses pancreatic cancer development by regulating cell proliferation. EBioMedicine 63, 103195. doi:10.1016/j.ebiom.2020.103195
Yang, X., Yang, Y., Sun, B. F., Chen, Y. S., Xu, J. W., Lai, W. Y., et al. (2017). 5-methylcytosine promotes mrna export - nsun2 as the methyltransferase and alyref as an m(5)c reader. Cell Res. 27, 606–625. doi:10.1038/cr.2017.55
Zhang, G., Liu, L., Li, J., Chen, Y., Wang, Y., Zhang, Y., et al. (2023a). Nsun2 stimulates tumor progression via enhancing tiam2 mrna stability in pancreatic cancer. Cell Death Discov. 9, 219. doi:10.1038/s41420-023-01521-y
Zhang, L. S., Dai, Q., and He, C. (2024). Base-resolution sequencing methods for whole-transcriptome quantification of mrna modifications. Acc. Chem. Res. 57, 47–58. doi:10.1021/acs.accounts.3c00532
Zhang, M., Song, J., Yuan, W., Zhang, W., and Sun, Z. (2021). Roles of rna methylation on tumor immunity and clinical implications. Front. Immunol. 12, 641507. doi:10.3389/fimmu.2021.641507
Zhang, Y., Chen, X. N., Zhang, H., Wen, J. K., Gao, H. T., Shi, B., et al. (2023b). Cdk13 promotes lipid deposition and prostate cancer progression by stimulating nsun5-mediated m5c modification of acc1 mrna. Cell Death Differ. 30, 2462–2476. doi:10.1038/s41418-023-01223-z
Zhang, Y., Hu, W., and Li, H. B. (2023c). Rna modification-mediated translational control in immune cells. RNA Biol. 20, 603–613. doi:10.1080/15476286.2023.2246256
Zhang, Y., Zhang, X., Shi, J., Tuorto, F., Li, X., Liu, Y., et al. (2018). Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding rnas. Nat. Cell Biol. 20, 535–540. doi:10.1038/s41556-018-0087-2
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mrna modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42. doi:10.1038/nrm.2016.132
Zhao, Y., Xing, C., and Peng, H. (2024). Alyref (aly/ref export factor): a potential biomarker for predicting cancer occurrence and therapeutic efficacy. Life Sci. 338, 122372. doi:10.1016/j.lfs.2023.122372
Zheng, H., Zhu, M., Li, W., Zhou, Z., and Wan, X. (2022). M(5) c and m(6) a modification of long noncoding nkila accelerates cholangiocarcinoma progression via the mir-582-3p-yap1 axis. Liver Int. 42, 1144–1157. doi:10.1111/liv.15240
Zheng, L., Duan, Y., Li, M., Wei, J., Xue, C., Chen, S., et al. (2023). Deciphering the vital roles and mechanism of m5c modification on rna in cancers. Am. J. Cancer Res., 13, 334899-e33546.
Zhong, J., Xu, Z., Ding, N., Wang, Y., and Chen, W. (2024). The biological function of demethylase alkbh1 and its role in human diseases. Heliyon, 10, e33489. doi:10.1016/j.heliyon.2024.e33489
Zhou, W. Y., Cai, Z. R., Liu, J., Wang, D. S., Ju, H. Q., and Xu, R. H. (2020). Circular rna: metabolism, functions and interactions with proteins. Mol. Cancer 19, 172. doi:10.1186/s12943-020-01286-3
Zhuang, H., Yu, B., Tao, D., Xu, X., Xu, Y., Wang, J., et al. (2023). The role of m6a methylation in therapy resistance in cancer. Mol. Cancer 22, 91. doi:10.1186/s12943-023-01782-2
Keywords: RNA methylation, m5C modification, post-transcriptional regulation, biomarker, digestive system tumors
Citation: Hu X, Liu Y, Zhang S, Liu K and Gu X (2025) The multifaceted role of m5C RNA methylation in digestive system tumorigenesis. Front. Cell Dev. Biol. 13:1533148. doi: 10.3389/fcell.2025.1533148
Received: 23 November 2024; Accepted: 05 February 2025;
Published: 06 March 2025.
Edited by:
Sujay Paul, Monterrey Institute of Technology and Higher Education (ITESM), MexicoReviewed by:
David Valle-Garcia, National Institute of Neurology and Neurosurgery “Manuel Velasco Suárez”, MexicoCopyright © 2025 Hu, Liu, Zhang, Liu and Gu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinyu Gu, aGtkZ3V4eUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.