
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 04 February 2025
Sec. Cellular Biochemistry
Volume 13 - 2025 | https://doi.org/10.3389/fcell.2025.1523958
Gastric cancer (GC) is one of the most common and highly lethal malignant tumors worldwide, and its occurrence and development are regulated by multiple molecular mechanisms. Post-translational modifications (PTM) common forms include ubiquitylation, phosphorylation, acetylation and methylation. Emerging research has highlighted lactylation and glycosylation. The diverse realm of PTM and PTM crosstalk is linked to many critical signaling events involved in neoplastic transformation, carcinogenesis and metastasis. This review provides a comprehensive overview of the impact of PTM on the occurrence and progression of GC. Specifically, aberrant PTM have been shown to alter the proliferation, migration, and invasion capabilities of GC cells. Moreover, PTM are closely associated with resistance to chemotherapeutic agents in GC. Notably, this review also discusses the phenomenon of PTM crosstalk, highlighting the interactions among PTM and their roles in regulating signaling pathways and protein functions. Therefore, in-depth investigation into the mechanisms of PTM and the development of targeted therapeutic strategies hold promise for advancing early diagnosis, treatment, and prognostic evaluation of GC, offering novel insights and future research directions.
Gastric cancer (GC) is a malignant tumor originating from the gastric mucosa, usually developed from glandular cells in the stomach (Smyth et al., 2020). GC is a public health problem worldwide. Exposition to Helicobacter pylori infection and dietary risk factors for GC shape the epidemiology of this disease (Tirado-Hurtado et al., 2019; Parsonnet et al., 1991; Wang et al., 2014). The incidence rate of GC varies significantly worldwide, especially in East Asia (such as China, Japan and South Korea) (Lopez et al., 2023; Davis and Sano, 2001; Bray et al., 2015). GC is the fifth most common cancer and the fifth most common cause of cancer death globally (Bray et al., 2024).
Post translational modifications (PTM) refer to a series of chemical modifications that occur after protein synthesis is completed (Deribe et al., 2010). Common PTM include phosphorylation, acetylation, glycosylation, ubiquitination, methylation, lactylation, etc., (Khoury et al., 2011; Pan and Chen, 2022). PTM can affect cell proliferation, apoptosis, invasion and metastasis by regulation protein activity, stability, localization and interactions with other molecules (Lee et al., 2023; Deribe et al., 2010; Vu et al., 2018; Pienkowski et al., 2023). Different types of PTM together form a complex network for protein functional regulation (Figure 1). In summary, PTM of proteins play a crucial role in biological processes. Therefore, studying PTM is crucial for understanding cell biology and developing new therapeutic strategies.
The recent research showed that the occurrence and development of GC are closely related to protein PTM (Paska and Hudler, 2015; Tan et al., 2007; Ramesh et al., 2023). Understanding the role of PTM of proteins in the occurrence and development of GC plays an important role in the treatment and prognosis of GC. Nowadays, there are many FDA approved targeted drugs on PTM (Table 1).
Several PTM-targeted therapies have already been approved by the FDA. These therapies are characterized by their high specificity, enabling precise modulation of critical signaling pathways while minimizing off-target effects. Additionally, their dynamic and reversible nature provides greater flexibility and adaptability in therapeutic applications. However, PTM-targeted drugs also face certain limitations. The intricate biological mechanisms underlying PTM complicate target identification and drug design. Furthermore, the high spatial and temporal specificity of certain PTM may restrict the applicability of these drugs across different tissues or diseases.
The purpose of this review is to outline the role of common protein PTM in GC.
In recent years, important results have been achieved regarding the role of ubiquitination. Ubiquitin is a highly conserved small molecule protein that exists in all eukaryotic cells (Popovic et al., 2014). It is composed of 76 amino acids and has a molecular weight of approximately 8.5 kDa (Popovic et al., 2014; Cockram et al., 2021). Ubiquitination regulation is a dynamic process regulated by both ubiquitinases and deubiquitinases (Figure 2). It is a PTM process in which ubiquitin is covalently attached to target proteins through three main steps: activation, conjugation, and ligation (Dikic and Schulman, 2023). First, the E1 ubiquitin-activating enzyme activates ubiquitin via ATP hydrolysis, forming an E1-ubiquitin thioester intermediate (Dikic and Schulman, 2023). Subsequently, the activated ubiquitin is transferred to the E2 ubiquitin-conjugating enzyme (Dikic and Schulman, 2023). Finally, the E3 ubiquitin ligase recognizes specific target proteins and catalyzes the transfer of ubiquitin from the E2-ubiquitin complex to a lysine residue on the target protein, resulting in ubiquitinated proteins (Dikic and Schulman, 2023). Through repeated cycles, polyubiquitin chains can be formed, which regulate various biological functions such as protein degradation, signal transduction, and subcellular localization (Dang et al., 2021; Li et al., 2021; Sampson et al., 2023; Qiu et al., 2023).
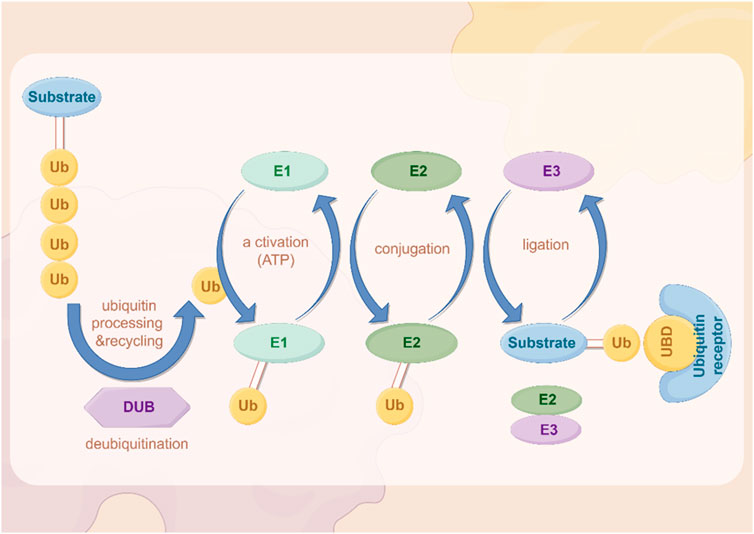
Figure 2. The process of ubiquitination. The figure was drawled by Figdraw (www.figdraw.com/#).
Ubiquitin complexes can be degraded by ubiquitinases, and this process is reversible, with deubiquitinases (DUBs) removing ubiquitin molecules from target proteins (Harrigan et al., 2018; Dewson et al., 2023). DUBs recover ubiquitin by hydrolyzing the heteropeptide bond between ubiquitin and target proteins, regulating protein degradation and cellular function (Mevissen and Komander, 2017). This step plays an important role in maintaining cellular homeostasis and regulating protein degradation balance.
In the occurrence and progression of GC, abnormalities in the ubiquitination system can lead to the degradation of tumor suppressor genes and excessive activation of oncogenes, thereby promoting the occurrence and development of tumors (Wang D. et al., 2022; Liu et al., 2020; Sokolova and Naumann, 2021; Li K. Q. et al., 2024; Hou and Deng, 2015; Sun et al., 2020b).
The ubiquitination system can also promote tumor growth by regulating the stability of certain oncogenes (Popovic et al., 2014). While the roles of DUBs in GC have been recently reviewed (An et al., 2022), here, some E3 ubiquitin ligases may enhance the function of oncogenes by protecting them from degradation, thereby promoting the growth and metastasis of GC. For example, studies have shown that the ubiquitination system can promote the proliferation and survival of GC cells by regulating cellular signaling pathways such as the NF - κB pathway (Yang W. et al., 2023).The documented roles of these proteins in GC are summarized in Table 2.
The ubiquitination system plays a critical role in cell cycle regulation and DNA repair processes (Louie and Kurzrock, 2020; Dagar et al., 2023). Abnormal ubiquitination can lead to uncontrolled cell cycle and obstacles to DNA damage repair, thereby increasing the risk of GC. Research has shown that E3 ubiquitin ligase SKP2 can promote the degradation of cyclin inhibitor p27, leading to uncontrolled cell cycle, which is related to the development of GC (Wen et al., 2016; Ge et al., 2023).
Abnormalities in the ubiquitination system are also closely related to the resistance of GC patients to chemotherapy drugs (Gonzalez et al., 2023; Narayanan et al., 2020). GC cells promote the stability of anti-apoptotic proteins by upregulating specific ubiquitinases, thereby evading the effects of chemotherapy drugs (Niu et al., 2021; Xu et al., 2014).
In the tumor microenvironment of GC, ubiquitination regulates the expression and function of oncogenic genes, influencing the interactions between tumor cells and their surrounding microenvironment (Aichem and Groettrup, 2016). Additionally, ubiquitination modifications modulate immune evasion mechanisms, enabling cancer cells to evade recognition and attack by the host immune system, thereby promoting tumor progression and recurrence (Zhang C. et al., 2023). Furthermore, the association between ubiquitination and cancer treatment has become increasingly significant, particularly in chemotherapy and targeted therapies. Abnormal ubiquitination may affect the efficacy of therapeutic agents and contribute to the development of drug resistance in cancer cells, driving the advancement of personalized treatment strategies (Sun W. et al., 2022).
Due to the important role of the ubiquitination system in GC, targeted therapy targeting the ubiquitination process may become a new approach for treating GC. In summary, abnormalities in the ubiquitination system play a key role in the occurrence, progression, and drug resistance of GC. In depth research on the mechanism of ubiquitination and its specific regulatory pathways in GC can help discover new therapeutic targets and improve the prognosis of GC patients.
Protein phosphorylation is the most common and important in PTM (Zhang W. J. et al., 2023). Approximately 30% of the human proteome is phosphorylated, which is involved in almost all cellular life processes such as cell division, protein breakdown, signal transduction, gene expression regulation, and protein interactions (Li Y. et al., 2023; Singh et al., 2017). Many phosphorylation pathways, including MAPK, PI3K/Akt, tyrosine kinase, cadherin catenin complex, cyclin dependent kinase, NF -κB, TGF -β signaling, etc., which pathway play important roles in cancer development (Yuan et al., 2020; Cargnello and Roux, 2011; Fresno Vara et al., 2004; Koromilas and Mounir, 2013; Du and Lovly, 2018; Hubbard and Till, 2000; Le et al., 2019; Singh et al., 2017; Fischer et al., 2022; Karin and Ben-Neriah, 2000; Wang et al., 2023c; Zhang Q. et al., 2019).
Phosphorylation regulates many key molecules and signaling pathways associated with GC, and abnormal phosphorylation levels may promote the occurrence, progression, and metastasis of GC (Mun et al., 2019; Miao et al., 2023; Jiang et al., 2021). Cytoplasmic adapter proteins that become phosphorylated and activated downstream of many kinases are a link between kinases and other events of signaling cascades (Figure 3). Research has shown that phosphorylation of EGFR receptors activates downstream pathways (Cardoso et al., 2014; Zhang G. et al., 2023). In GC, p53 gene mutations often lead to ineffective phosphorylation regulation, further promoting the development of cancer (Yuan et al., 2022). The PI3K/AKT/mTOR signaling pathway is a key pathway that promotes cell proliferation, survival, and metabolism (Glaviano et al., 2023). In GC patients, key components of this pathway are often abnormally activated by phosphorylation, especially the excessive phosphorylation of AKT, which is associated with tumor proliferation and metastasis (Shen et al., 2023). The increase of AKT phosphorylation can not only inhibit cell apoptosis, but also promote protein synthesis and cell growth by affecting mTOR, further promoting the progression of GC (Wang C. et al., 2021; Zhong et al., 2023). Phosphorylation also plays an important role in regulating the activity of cell cycle proteins and apoptosis related proteins. In GC cells, abnormal phosphorylation levels can inhibit cell apoptosis and promote tumor cell survival (Rong et al., 2020). The invasion and metastasis of GC are one of the main reasons for poor prognosis in patients (Matsuoka and Yashiro, 2023). The signaling pathway regulated by phosphorylation plays a crucial role in cell movement, matrix degradation, and invasion processes. Abnormal phosphorylation of ERK can activate downstream molecules and enhance the migration and invasion ability of GC cells (Kim et al., 2024; Wu et al., 2010). Phosphorylation abnormalities are closely related to the resistance of GC to chemotherapy and targeted therapy (Wu et al., 2023c). Research has shown that GC cells can evade chemotherapy induced apoptosis by activating phosphorylation of key proteins on the PI3K/AKT pathway (Rong et al., 2020). The efficacy of drugs targeting EGFR in GC is also reduced due to resistance caused by phosphorylation activation (Cao et al., 2022). The documented roles of these kinases in GC are summarized in Table 3.
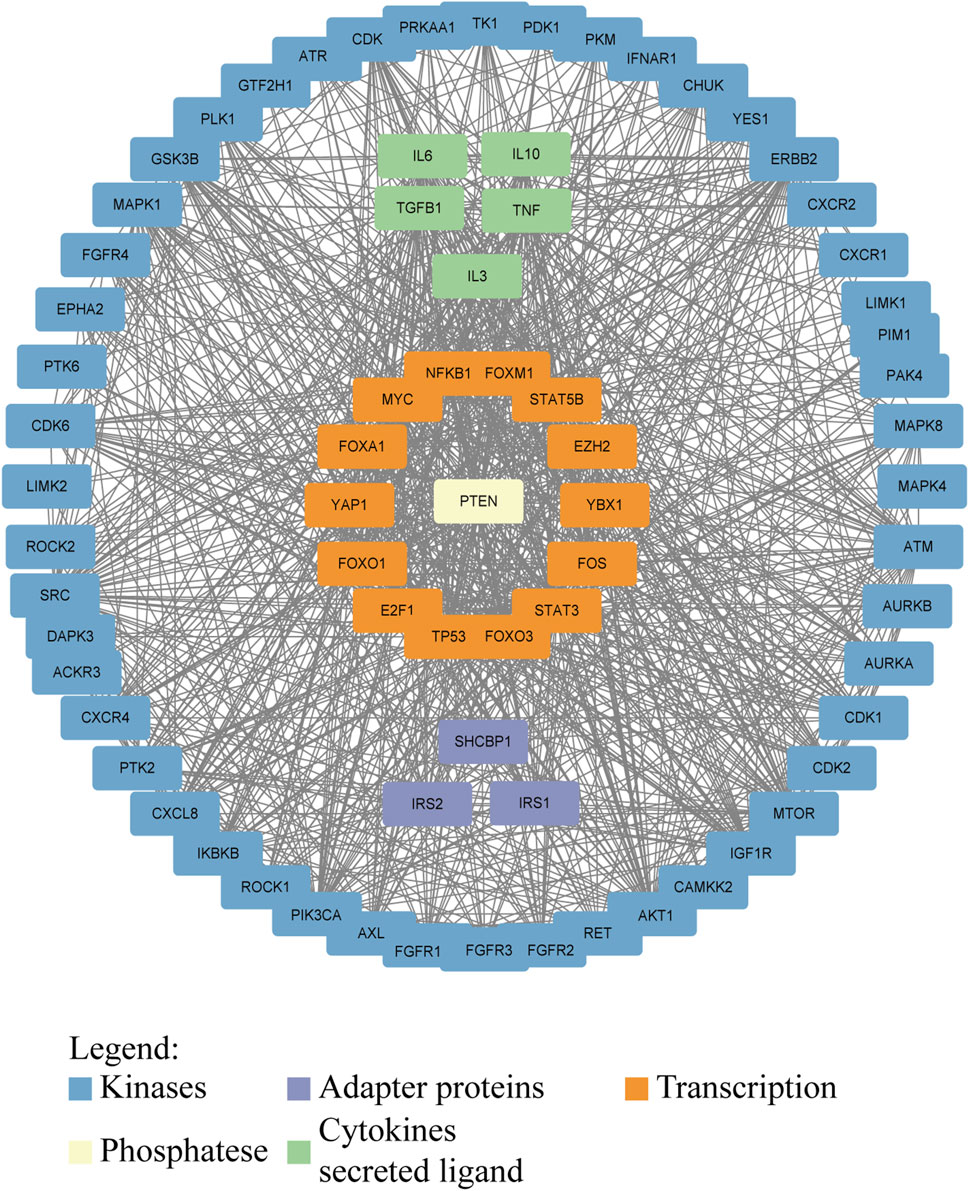
Figure 3. Interaction network of proteins involved in or affected by phosphorylation in GC. Kinases, adapter proteins, and transcription factors are shown to visualize the network that drives GC progression. Protein–protein interactions were downloaded from the STRING database (https://cn.string-db.org/) and visualized in Cytoscape.
Phosphorylation plays an important role in the occurrence, development, invasion, and drug resistance of GC. Dysregulation of phosphorylation of many oncogenes and tumor suppressor genes is one of the key mechanisms underlying the progression of GC. Studying the abnormal phosphorylation phenomenon in GC can help deepen our understanding of its pathological process and provide new ideas for developing targeted treatment plans.
Acetylation is one of the important forms of PTM of proteins, which refers to the addition of acetyl groups (CH3CO) to amino acid residues in proteins, especially lysine residues (Shvedunova and Akhtar, 2022) (Figure 4). Zinc (Zn2+)-dependent histone deacetylases (HDACs) are classified into four major classes: class I (HDACs 1, 2, 3, and 8), class II (HDACs 4, 5, 6, 7, 9, and 10), and class IV, which includes only HDAC11 (Shvedunova and Akhtar, 2022). The class III deacetylases cover the NAD-dependent deacetylases SIRT1–7 (Shvedunova and Akhtar, 2022). Acetylation not only regulates the structure and function of proteins, but also extensively participates in important biological processes such as gene expression, chromatin remodeling, cell cycle regulation, and metabolism (Dang and Wei, 2022; Li and Seto, 2016). Acetylation abnormalities play a crucial role in the occurrence and development of GC (Badie et al., 2022). Histone acetylation is the most common form of acetylation that regulates gene expression. Histones are the core components of chromatin, and by regulating their acetylation levels, the structure of chromatin can be altered, thereby affecting gene expression (Geffen et al., 2023). Acetylation of histones is usually associated with gene activation, which enhances chromatin openness and makes transcription factors more likely to bind to DNA, initiating gene transcription (Zaib et al., 2022). Acetylation not only acts on histones, but also affects the function of various non histone proteins, altering their stability, subcellular localization, interactions, and activity (Narita et al., 2019).
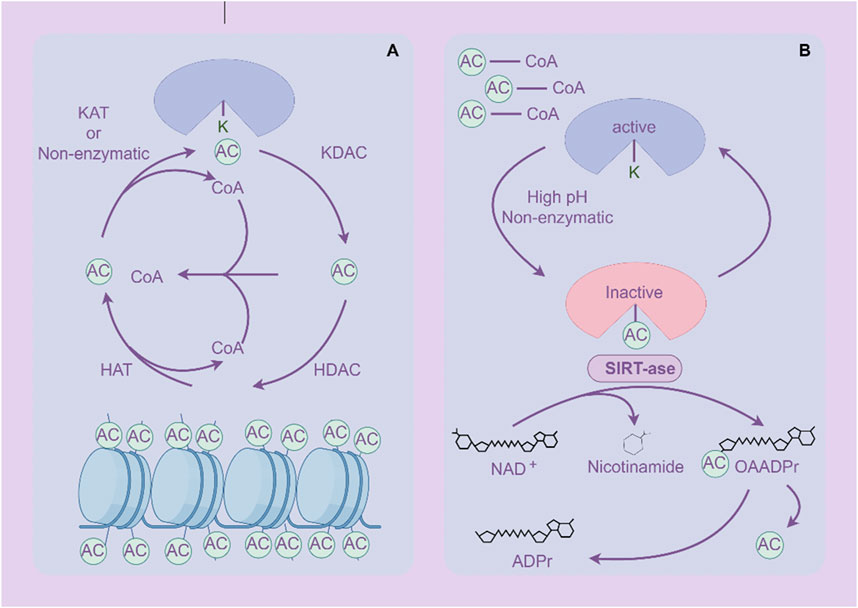
Figure 4. Acetylation and deacetylation processes of proteins. (A) Histone protein de/acetylation Process (HDACs family). (B) Protein de/acetylation Process (SIRTs family). The figure was drawled by Figdraw (www.figdraw.com/#).
Histone acetyltransferases (HATs) are key regulatory factors in acetylation modification, which can enhance their function by adding acetyl groups to proteins (White et al., 2024). In GC, overexpression of HAT promotes acetylation of histones and non-histones, activating the expression of tumor related genes (Jie et al., 2020; Guo et al., 2022).
Histone deacetylases (HDACs) are important inverse regulators of acetylation modification, inhibiting their function by removing acetyl groups from proteins (Li and Seto, 2016). HDACs are highly expressed in GC, leading to deacetylation of histones and non-histones, and inhibiting the expression and function of tumor suppressor genes (Lin et al., 2023; Jenke et al., 2024). HDAC inhibitors, as a potential anti-cancer treatment, have been applied in the treatment of GC (Jenke et al., 2024). By inhibiting HDACs, the expression of tumor suppressor genes can be restored, inducing apoptosis and differentiation of cancer cells (McClure et al., 2018).
The Sirtuins family is a homolog of yeast chromatin silencing signal regulator 2, which is an NAD+- dependent three class histone deacetylase widely distributed in the body (Nassir, 2022). This family influences the occurrence and development of tumor cells through various pathways, such as regulating gene stability, inflammatory response, cellular stress, apoptosis, energy metabolism of GC cells, and altering the tumor microenvironment (Lagunas-Rangel, 2024; Poniewierska-Baran et al., 2022; Yu L. et al., 2024).
Acetylation is associated with the invasion and metastasis ability of GC (Li et al., 2018). Research has shown that E-cadherin is an important molecule that inhibits cell invasion and metastasis, and its expression and function can be regulated by (Zhao et al., 2019; Tanaka et al., 2002). In GC, HDACs inhibit the expression of E-cadherin through deacetylation, leading to reduced intercellular adhesion and enhancing the invasion and metastasis ability of cancer cells (Decourtye-Espiard et al., 2021). The abnormality of acetylation is closely related to the resistance of GC cells to chemotherapy and targeted therapy. The abnormal expression of HDACs may help GC cells evade chemotherapy induced apoptosis by altering the expression of apoptosis related genes (Regel et al., 2012). In addition, changes in acetylation levels of certain transcription factors may also affect the sensitivity of cells to anticancer drugs (Kokate et al., 2018). The documented roles of these proteins in GC are summarized in Table 4.
Due to the important role of acetylation in the occurrence and progression of GC, targeted acetylation therapy strategies are becoming a promising anti-cancer pathway. HDAC inhibitors have shown certain anti GC effects by inhibiting HDAC activity, restoring the expression and function of tumor suppressor genes. In addition, other molecules that target acetylation regulation (histone acetyltransferases, HATs) are also expected to become new therapeutic targets. By regulating acetylation levels, cancer cell proliferation can be effectively inhibited, apoptosis can be promoted, and drug resistance can be reduced (Wu et al., 2020; Marmorstein and Zhou, 2014).
Glycosylation is a process in which a protein or lipid is attached to a carbohydrate under the control of an enzyme, aiming to regulate the structure and function of proteins (Eichler, 2019). Glycosylation is one of the important processes in protein PTM. As a common and complex modification, glycosylation plays a crucial role in biological processes such as protein folding, stability, intercellular recognition, and signal transduction (Eichler, 2019). Abnormal glycosylation is closely related to the occurrence and progression of cancer (Figure 5).
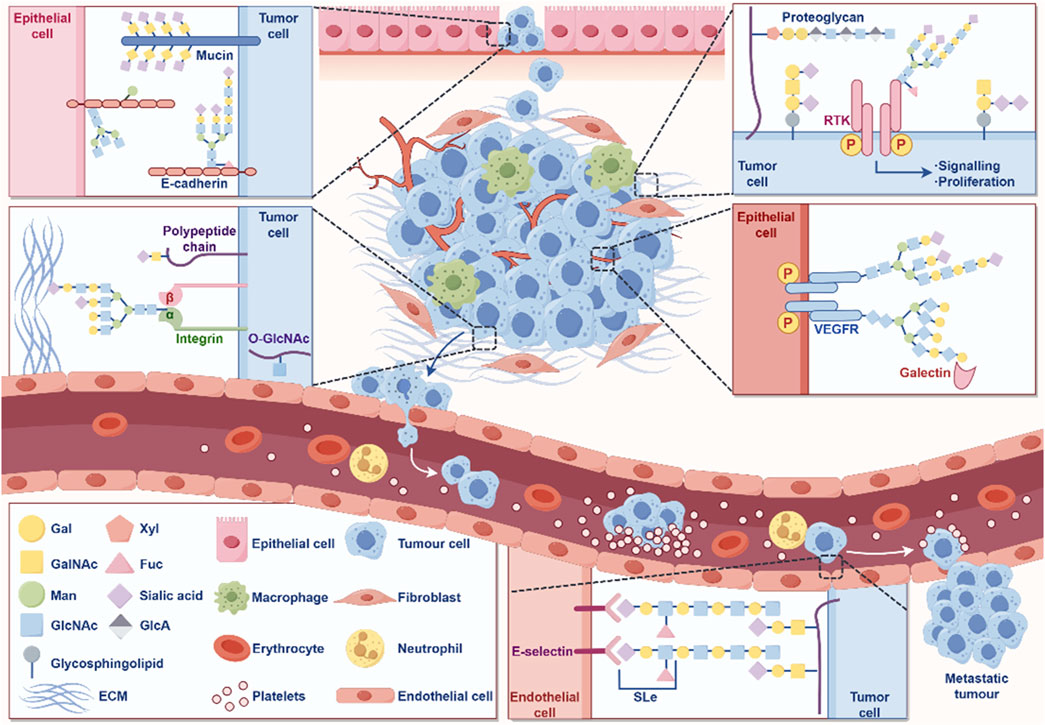
Figure 5. The role of glycosylation in the occurrence and development of cancer. The figure was drawled by Figdraw (www.figdraw.com/#).
In GC, glycosylation abnormalities are manifested in changes in the sugar chain structure and modification patterns of various proteins, which affect the behavioral characteristics of cells and promote the occurrence, progression, and malignant transformation of tumors (Arai et al., 2024; Ferreira et al., 2017). Cancer cells often exhibit abnormally glycosylated sugar chain structures on their surface, including high mannose type and hyper branched structures (Pinho and Reis, 2015; Stowell et al., 2015). These abnormal sugar chains can alter the function of cell membrane receptors, thereby enhancing the activity of signaling pathways, promoting cell proliferation and anti-apoptotic ability (Pinho and Reis, 2015; Stowell et al., 2015). In GC cells, glycosylation modification of EGFR increases its stability on the cell membrane, further activating signaling pathways related to cell proliferation and survival, accelerating tumor growth and malignant progression (Hu et al., 2018). E-cadherin is a key protein that inhibits cell migration, and changes in its glycosylation can affect intercellular adhesion. The abnormal glycosylation of E-cadherin can weaken the adhesion ability between cells and enhance the invasion and metastasis potential of GC cells (Carvalho et al., 2016).
Glycosylation abnormalities are closely related to the expression and activity of multidrug resistance related proteins. The glycosylation of P-gp can enhance its ability to pump chemotherapy drugs, leading to resistance of GC cells to chemotherapy drugs (Liang et al., 2009). Meanwhile, glycosylation modification can alter the expression of surface antigens and affect the recognition of the immune system. GC cells reduce the probability of immune system recognition through abnormal glycosylation, thereby helping them evade immune surveillance, promoting tumor survival and chemotherapy resistance (Sun et al., 2021; Stanczak et al., 2022).The glycosylation process is catalyzed by glycosyltransferases, and the expression and activity of glycosyltransferases in GC often undergo abnormal changes (Pinho and Reis, 2015). GnT-V (N-acetylglucosyltransferase V) is a glycosyltransferase upregulated in GC, which can catalyze the formation of complex sugar chains and is associated with the malignant progression of GC (Huang et al., 2023; Huang et al., 2014). Upregulation of GnT-V can promote the proliferation, invasion, and migration of GC cells, making it a potential therapeutic target (Huang et al., 2014). In summary, glycosylation is crucial for the occurrence and development of GC (Table 5).
Glycosylation plays an important role in the occurrence, progression, metastasis, and drug resistance of GC. We have summarized the specific mechanisms by which various types of glycosylation modifications contribute to the onset and progression of GC (Table 6). Abnormal glycosylation not only alters the proliferation and invasion behavior of GC cells, but is also closely related to the tumor’s resistance to chemotherapy and immunotherapy. By conducting in-depth research on the regulatory mechanisms of glycosylation and developing targeted glycosylation treatment methods, it is expected to provide new ideas and means for the diagnosis, prognosis, and personalized treatment of GC.
Methylation is a form of PTM of proteins, particularly DNA and histone methylation, which plays a crucial role in gene expression regulation (Dai et al., 2021; Mattei et al., 2022; Yang B. et al., 2021). Methylation affects the transcriptional activity of genes, the structure of DNA, and the state of chromatin by adding methyl groups (-CH3) at specific base positions (Moore et al., 2013). Methylation remodeling of DNA, RNA, histone, and nonhistone proteins contributes to tumor initiation and progression (Figure 6). In GC, abnormal methylation patterns are closely related to the occurrence, development, invasion, and drug resistance of tumors (Qu et al., 2013; Zeng et al., 2017).
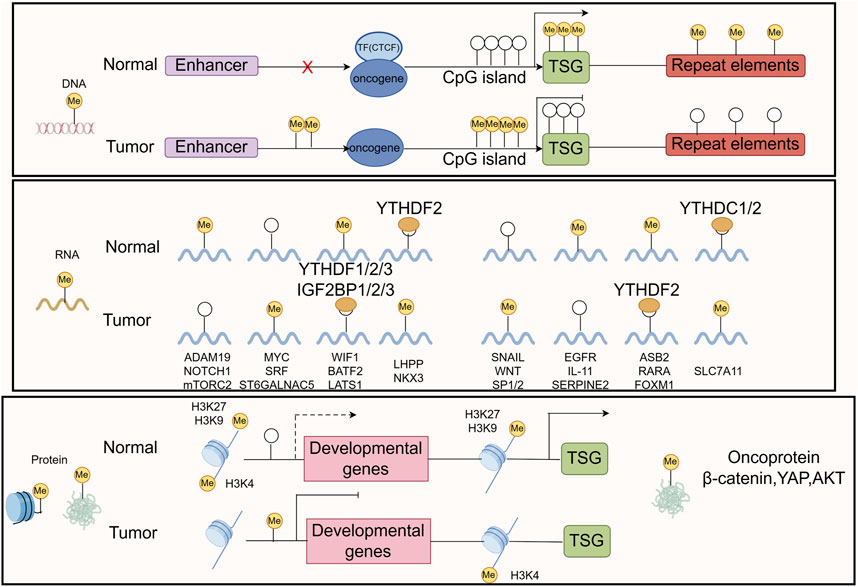
Figure 6. The common mechanisms that cause oncogene/TSG disturbance by methylation remodeling at DNA, RNA, and protein levels are recapitulated in the boxes. The figure was drawled by Figdraw (www.figdraw.com/#).
This abnormal methylation leads to the inactivation of tumor suppressor genes, inhibiting functions such as cell cycle regulation, DNA repair, and apoptosis, thereby promoting the proliferation and survival of tumor cells (Wang F. et al., 2022; Zhang N. et al., 2023; Mo et al., 2024). Research has found that common DNA methylation changes in GC tissue are associated with patient prognosis (Usui et al., 2021; Sogutlu et al., 2022), therefore, targeted DNA methylation therapy strategies are considered to have potential clinical application value. Histone methylation plays an important role in regulating gene transcription, chromatin structure, and gene expression. The methylation status of histones H3 and H4 can affect the biological behavior of tumor cells (Audia and Campbell, 2016; Michalak et al., 2019; Liu et al., 2023). Abnormal histone methylation patterns may lead to the inactivation or abnormal expression of tumor related genes, thereby promoting the occurrence and progression of GC (Michalak et al., 2019). The abnormal expression of histone demethylase may be related to the malignant characteristics of GC (Li et al., 2023a; Dong et al., 2023).
In the microenvironment of GC, abnormal methylation can regulate the function of tumor associated macrophages (TAMs) and other immune cells, thereby affecting the tumor’s immune escape ability (Mittelstaedt et al., 2021; Li Y. et al., 2024). Tumor cells evade immune system surveillance and promote cancer progression by altering the phenotype and function of immune cells. The methylation status of drug metabolism related genes in GC cells may affect the tumor’s sensitivity to chemotherapy drugs. Abnormal methylation of some genes can lead to tumor cells developing resistance to chemotherapy drugs, affecting treatment efficacy (Wu Q. et al., 2021; Nagaraju et al., 2021).
Methylation plays an important role in the occurrence, development, invasion, and drug resistance of GC. Abnormal methylation of DNA and histones leads to the inactivation of tumor suppressor genes, promoting the proliferation and survival of cancer cells. Meanwhile, methylation changes are closely related to the tumor microenvironment and drug resistance. By conducting in-depth research on the regulatory mechanisms of methylation and developing targeted methylation therapy methods, it is expected to provide new ideas for early diagnosis, prognosis evaluation, and personalized treatment of GC.
Lactation is a newly discovered PTM of proteins in recent years, which refers to the covalent addition of lactate molecules (-C3H6O3) to lysine residues in proteins (Fan et al., 2023) (Figure 7). This modification plays an important role in cellular metabolism, signal transduction, and gene expression regulation (Zhang D. et al., 2019). In tumor cells, due to the increased metabolic demand, there is usually a phenomenon of enhanced glycolysis, known as the Warburg effect, which leads to an increase in lactate production (Zhang D. et al., 2019). Tumor cells regulate the functions of various proteins through lactylation, thereby adapting to changes in the tumor microenvironment and promoting cell growth and proliferation (Xie et al., 2023; Qu et al., 2023). Lactation may affect the energy metabolism of tumor cells by regulating the activity or stability of metabolism related enzymes. This modification can increase the flexibility of metabolic pathways and help tumor cells survive under low oxygen and nutrient deficient conditions (Yang H. et al., 2023; Dai et al., 2024; Yang W. et al., 2021).
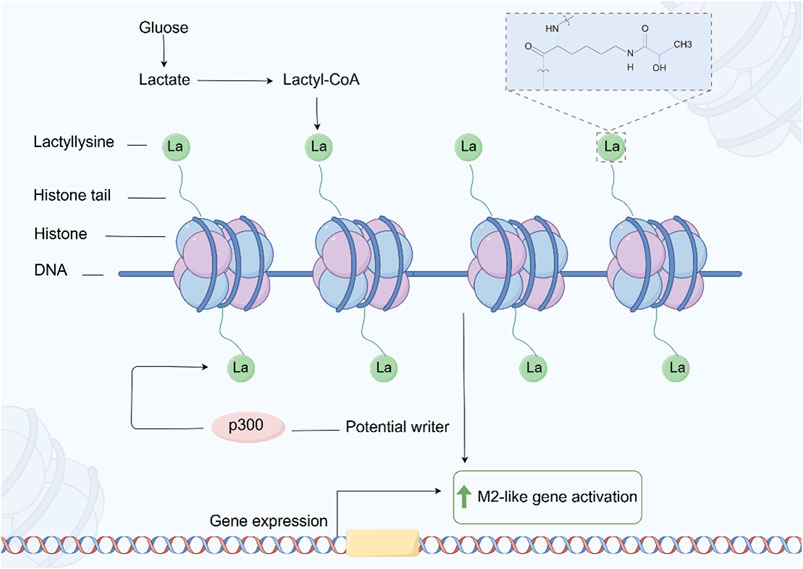
Figure 7. The process of protein Lactylation. The figure was drawled by Figdraw (www.figdraw.com/#).
In GC, the increase in lactate may enhance the migration ability of cancer cells by regulating the reorganization of the cytoskeleton and the expression of intercellular adhesion molecules (Zhao et al., 2024; Li Z. et al., 2024). Studies have shown that lactylation may affect signaling pathways related to cell adhesion and migration (Wang J. et al., 2022). The drug resistance of GC cells in chemotherapy and targeted therapy is often related to metabolic reprogramming and changes in intracellular signaling pathways (Bin et al., 2021). Lactic acid may promote cancer cell tolerance to treatment by regulating signaling pathways related to drug resistance. Lactic acid modification of certain key proteins may affect drug targeting, leading to increased excretion of chemotherapy drugs in cancer cells or loss of target function (Yu X. et al., 2024; Zha et al., 2024; Chen H. et al., 2024).
Lactic acid plays an important role in the metabolic regulation, gene expression, tumor microenvironment, and drug resistance of GC. Lactic acid promotes the development and malignant progression of GC by regulating protein functions related to metabolism, cell proliferation, and immune escape. In depth research on the mechanism of lactylation and the development of treatment strategies targeting lactylation are expected to provide new ideas for the early diagnosis, treatment, and prognosis evaluation of GC.
SUMOylation (Small Ubiquitin like Modifier) refers to a PTM that covalently attaches SUMO proteins to lysine residues of target proteins (Han et al., 2018). Similar to ubiquitination, SUMOylation regulates various cellular processes by altering protein stability, activity, subcellular localization, or interactions with other proteins (Wei et al., 2023; Hu et al., 2021; Wu et al., 2023b). In recent years, the role of SUMOylation in tumor biology has gradually received attention, especially in GC, where abnormal SUMOylation is closely related to the occurrence, development, invasion, and drug resistance of tumors (Xie et al., 2020; Seeler and Dejean, 2017).
The increase or decrease of SUMOylation can promote the occurrence and development of GC by inhibiting or enhancing the expression of specific genes (Zhao Y. Q. et al., 2023; Wang T. et al., 2023). SUMOylation can also regulate gene expression by binding to transcription factors (Tian et al., 2024). In GC cells, abnormal SUMOylation may lead to uncontrolled cell cycle and promote abnormal proliferation of cancer cells (Fang et al., 2017; Gu et al., 2024).
SUMOylation also plays an important role in the process of DNA damage repair. By regulating the SUMOylation status of proteins involved in DNA repair, it can affect the efficiency of DNA repair and genomic stability (Zhang F. L. et al., 2023). In GC, DNA repair defects are closely related to tumor development, and abnormal SUMOylation may lead to the accumulation of DNA damage, promoting the occurrence of cancer (Zhang M. et al., 2019). SUMOylation can affect the migration and invasion ability of GC cells by regulating the functions of cytoskeleton related proteins and cell adhesion molecules (Wang Q. et al., 2021; Liu et al., 2021). The SUMOylation of intercellular adhesion molecules and integrins may alter their functions, promoting cell detachment from the primary tumor and migration to distant organs. SUMOylation may also affect the progression of GC by regulating the interaction between tumor cells and the surrounding microenvironment (Gu et al., 2023). The low oxygen state in the tumor microenvironment can regulate the stability of hypoxia inducible factors (HIFs) through SUMOylation, promoting GC angiogenesis and tumor cell survival under low oxygen conditions (Filippopoulou et al., 2020; Zhou et al., 2021). In addition, SUMOylation plays an important role in the drug resistance of GC. SUMOylation may affect the efficacy of chemotherapy drugs by regulating proteins involved in drug metabolism, leading to drug resistance in GC cells (Gu et al., 2024; Huang et al., 2022a).
SUMOylation, as a key protein PTM, plays multiple roles in the occurrence, development, invasion, and drug resistance of GC. By regulating the SUMOylation status of transcription factors, cell cycle proteins, DNA repair related proteins, and cell migration related factors, GC cells can acquire the ability to proliferate, invade, and resist treatment. Therefore, in-depth research on the specific mechanism of SUMOylation in GC and the development of targeted SUMOylation treatment methods will provide new ideas for the treatment of GC.
PTM crosstalk refers to the phenomenon of mutual influence between different types of PTM, which plays an important role in regulating protein function, stability, and interaction networks (Huang et al., 2019; Geffen et al., 2023; Cutler et al., 2021). PTM crosstalk can occur in both intraprotein and interprotein contexts, involving the same or different types of modifications. Regardless of the specific mechanisms, PTM crosstalk can orchestrate complex interactions among various PTM, influencing protein functions, signaling pathways, and the regulation of protein networks in tumorigenesis. This interplay plays a crucial role in the development and progression of tumors, highlighting the profound impact of PTM on cellular fate and pathological processes (Wang W. et al., 2024; Li et al., 2023b; Wu et al., 2019; Hernandez-Valladares et al., 2019).
In GC, common PTM include phosphorylation, acetylation, methylation, and ubiquitination, and the interactions between these modifications may significantly affect protein activity. PTM crosstalk also plays an important role in cellular signaling pathways. Taking the NF - κB signaling pathway as an example, this pathway plays a crucial role in the development of various tumors. The activity of NF - κB is regulated by various PTM such as phosphorylation, acetylation, and ubiquitination. Research has shown that acetylation modification of NF - κB can enhance its transcriptional activity, while phosphorylation may affect its transcriptional activity in the nucleus by altering its affinity for binding proteins. In addition, ubiquitination modification of NF - κB can promote its degradation, thereby regulating its stability in cells. These complex PTM interactions enable NF - κB to flexibly regulate its function in different cellular environments (Ito, 2007). In the RAS/MAPK pathway, KRAS and other signaling mediators are influenced by various PTM, including phosphorylation, ubiquitination, farnesylation, proteolysis, methylation, and palmitoylation (Ahearn et al., 2011; Laude and Prior, 2008). Many signaling mediators in the TGF - β pathway are widely influenced by PTM, including phosphorylation and ubiquitination, which are crucial for initiating and regulating signal transduction to the nucleus (Xu et al., 2016). The activation/inactivation of tumor suppressor gene p53 function is regulated by various PTM, including phosphorylation, ubiquitination, acetylation, and methylation (Bode and Dong, 2004; Dai and Gu, 2010).
As an emerging field of PTM research, the study of PTM crosstalk in cancer is still somewhat blank. Therefore, understanding the mechanism of PTM crosstalk is particularly important for developing new therapeutic strategies, especially when targeting specific signaling pathways or regulating protein functions, which can provide new ideas and methods for precision medicine.
Although PTM play a crucial role in cell biology, there are still significant limitations to current research on their use in GC. PTM such as ubiquitination, phosphorylation, acetylation, glycosylation, methylation, lactylation, and SUMOylation regulate protein stability, activity, and interactions, but how these modifications alter tumor behavior in GC has not been fully elucidated. Most of the research has focused on genomic and epigenetic regulation, while there is relatively little research on the detailed role and crosstalk of PTM in GC. he complexity of PTM mechanisms makes target selection and drug design challenging, especially in cases where significant differences exist between cancer subtypes and individuals, limiting the broad applicability of PTM-targeted therapies. Additionally, the high cost and complexity of research technologies restrict the widespread clinical application of these methods. The challenge of individualized treatment is another critical issue, as variations in PTM across different patients may lead to differential drug responses, making precise treatment difficult. PTM-targeted therapies may influence off-target genes, potentially inducing side effects or affecting normal cell functions. Furthermore, the prolonged use of PTM-targeted drugs may lead to drug resistance, impacting the long-term effectiveness of treatment. These limitations necessitate further scientific research and technological advancements to overcome these challenges and enhance the clinical utility of PTM-targeted therapies. Filling this gap is expected to reveal new biological mechanisms and potential therapeutic targets.
In GC, PTM (ubiquitination, phosphorylation, acetylation, glycosylation, methylation, lactylation and SUMOylation, etc.) affect biological processes by regulating protein stability, activity, and interactions. For example, ubiquitination regulates protein degradation (Sun T. et al., 2020), phosphorylation participates in the activation of key signaling pathways (Agashe et al., 2022; Luo et al., 2020; Ebert et al., 2022), acetylation and methylation affect gene expression, while glycosylation plays a role in intercellular signaling (Xu and Wan, 2023; Bao and Wong, 2021; Ramaiah et al., 2021; Jarrold and Davies, 2019; Li et al., 2020; Locke et al., 2019). Lactylation is associated with metabolic reprogramming (Sun L. et al., 2022; Lv et al., 2023), while SUMO modification is associated with tumor drug resistance and progression (Chang and Yeh, 2020). In addition, the crosstalk between different modifications makes the regulatory mechanism more complex, which affects protein function and tumor cell behavior, especially playing an important role in the invasion and metastasis of GC.
The complexity of PTM is reflected in the interplay and crosstalk between different types of PTM. Various modifications such as ubiquitination, phosphorylation, and acetylation play a critical role in regulating tumor cell processes, including growth, migration, invasion, and immune evasion. For instance, the interplay between phosphorylation and ubiquitination can enhance kinase activity, promoting tumor cell survival and dissemination (Cutler et al., 2021; Barbour et al., 2023). Additionally, acetylation and SUMOylation contribute to the regulation of protein stability and function. PTM crosstalk not only affects the individual roles of specific PTM but also integrates multiple signaling pathways to control the complex behaviors of tumor cells (Barbour et al., 2023). These mechanisms play a pivotal role in the progression and drug resistance observed in GC, where tumor cells exploit the PTM network to evade therapeutic inhibition. Therefore, a deeper understanding of PTM crosstalk mechanisms is essential for the development of more precise and effective targeted therapies for GC.
As research progresses, PTM-targeted therapies are increasingly being recognized as a crucial strategy in the treatment of GC, aiming to disrupt abnormal signaling pathways in tumor cells through targeted modifications. For instance, drugs targeting phosphorylation kinases or ubiquitination-regulated proteins can interfere with these modifications to inhibit tumor cell proliferation and migration (Wang et al., 2020; Su et al., 2022).
In GC research, PTM and their crosstalk mechanisms play critical roles in regulating various biological processes in tumor cells. Despite significant advances, there remain substantial challenges and limitations. Current studies primarily focus on certain PTM types, such as phosphorylation and ubiquitination, while the functional mechanisms of less-studied PTM, such as glycosylation and lactylation, are still underexplored. With ongoing research, more novel PTM are being identified, yet studies on these modifications remain at the preliminary stages of screening and validation, with limited clinical applicability. Moreover, the dynamic nature of PTM and their intricate networks within the tumor microenvironment add layers of complexity to the selection of therapeutic targets and the development of effective treatment strategies. Many PTM-targeted drugs face challenges related to target generalization, lacking precise interventions for specific PTM or PTM networks.
Future research should delve deeper into several key areas. First, leveraging high-throughput omics technologies, such as mass spectrometry and single-cell RNA sequencing, to comprehensively characterize the dynamic changes in PTM networks and identify critical modification sites with functional significance in various cellular states (Gillette et al., 2024; Li and Zhan, 2020; Kirsch et al., 2020; Qin et al., 2020). Second, integrating bioinformatics and machine learning approaches to predict and screen effective drugs targeting PTM while optimizing the selectivity and efficacy of existing PTM-targeted therapies. Additionally, research should address the variability of PTM responses among individuals, tumor subtypes, and their microenvironments to design more personalized and adaptable therapeutic strategies (Wang et al., 2023d; Hegde et al., 2020).
Another significant challenge lies in addressing the long-term safety and resistance associated with PTM-targeted drugs. Prolonged use of such therapies may prompt tumor cells to remodel PTM networks, enabling them to evade drug inhibition and develop resistance (Wang Y. et al., 2023; Onglao et al., 2022). Therefore, future efforts should prioritize exploring combination targeting strategies, integrating multiple PTM and diverse biological pathways to enhance therapeutic efficacy and mitigate resistance risks. By adopting these comprehensive strategies, PTM-targeted therapies could more precisely and effectively disrupt the complex biological mechanisms of GC, ultimately improving clinical outcomes for patients.
Multiple protein PTM mechanisms are closely involved in the occurrence, progression, and treatment tolerance of GC. Ubiquitination affects the proliferation and apoptosis of cancer cells by regulating the degradation of key proteins; Acetylation modification regulates gene expression, especially at the epigenetic level, by affecting the activity of oncogenes and tumor suppressor genes through histone acetylation and deacetylation; Abnormal glycosylation alters the invasiveness and immune escape ability of cancer cells; Methylation is involved in gene silencing and oncogene activation, and is a common epigenetic change in GC; Lactic acid modification, as an emerging research field, may be related to metabolic reprogramming in the tumor microenvironment; Phosphorylation is the core of signal pathway regulation, affecting cell proliferation and survival; SUMOylation plays an important role in cancer drug resistance by regulating protein stability and DNA repair. These modifications together form a complex network for the development of GC and provide multiple potential targets for diagnosis and therapeutic interventions. Overall, these PTM participate in the multifaceted regulation of GC through synergistic or independent pathways, and provide rich potential targets for the development of diagnostic biomarkers and targeted therapy strategies.
HS: Writing–original draft, Writing–review and editing, Formal Analysis. MZ: Writing–original draft, Writing–review and editing. CG: Data curation, Writing–review and editing. XG: Data curation, Writing–review and editing. YqM: Data curation, Writing–review and editing. YnM: Funding acquisition, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by Gansu Provincial Science and Technology Plan (Joint Scientific Research Fund) Project (24JRRA885); Natural Science Foundation of Gansu Province funding project (22JR5RA663); Research Project of Gansu Provincial People’s Hospital (2024KYQDJ-A-14). The APC was funded by Natural Science Foundation of Gansu Province funding project (22JR5RA663).
The authors thank all the members of Department of General Surgery of Gansu Provincial People’s Hospital for the discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agashe, R. P., Lippman, S. M., and Kurzrock, R. (2022). JAK: not just another kinase. Mol. Cancer Ther. 21, 1757–1764. doi:10.1158/1535-7163.MCT-22-0323
Ahearn, I. M., Haigis, K., Bar-Sagi, D., and Philips, M. R. (2011). Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13, 39–51. doi:10.1038/nrm3255
Aichem, A., and Groettrup, M. (2016). The ubiquitin-like modifier FAT10 in cancer development. Int. J. Biochem. Cell Biol. 79, 451–461. doi:10.1016/j.biocel.2016.07.001
Aldaz, P., Otaegi-Ugartemendia, M., Saenz-Antonanzas, A., Garcia-Puga, M., Moreno-Valladares, M., Flores, J. M., et al. (2020). SOX9 promotes tumor progression through the axis BMI1-p21(CIP). Sci. Rep. 10, 357. doi:10.1038/s41598-019-57047-w
Alexander, K. L., Serrano, C. A., Chakraborty, A., Nearing, M., Council, L. N., Riquelme, A., et al. (2020). Modulation of glycosyltransferase ST6Gal-I in gastric cancer-derived organoids disrupts homeostatic epithelial cell turnover. J. Biol. Chem. 295, 14153–14163. doi:10.1074/jbc.RA120.014887
An, T., Lu, Y., Gong, Z., Wang, Y., Su, C., Tang, G., et al. (2022). Research progress for targeting deubiquitinases in gastric cancers. Cancers (Basel) 14, 5831. doi:10.3390/cancers14235831
Arai, J., Hayakawa, Y., Tateno, H., Murakami, K., Hayashi, T., Hata, M., et al. (2024). Impaired glycosylation of gastric mucins drives gastric tumorigenesis and serves as a novel therapeutic target. Gastroenterology 167, 505–521 e19. doi:10.1053/j.gastro.2024.03.037
Argnani, L., Broccoli, A., and Zinzani, P. L. (2017). Cutaneous T-cell lymphomas: focusing on novel agents in relapsed and refractory disease. Cancer Treat. Rev. 61, 61–69. doi:10.1016/j.ctrv.2017.10.007
Audia, J. E., and Campbell, R. M. (2016). Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 8, a019521. doi:10.1101/cshperspect.a019521
Aziz, F., Khan, I., Shukla, S., Dey, D. K., Yan, Q., Chakraborty, A., et al. (2022). Partners in crime: the Lewis Y antigen and fucosyltransferase IV in Helicobacter pylori-induced gastric cancer. Pharmacol. Ther. 232, 107994. doi:10.1016/j.pharmthera.2021.107994
Badie, A., Gaiddon, C., and Mellitzer, G. (2022). Histone deacetylase functions in gastric cancer: therapeutic target? Cancers (Basel) 14, 5472. doi:10.3390/cancers14215472
Bao, M. H., and Wong, C. C. (2021). Hypoxia, metabolic reprogramming, and drug resistance in liver cancer. Cells 10, 1715. doi:10.3390/cells10071715
Barbour, H., Nkwe, N. S., Estavoyer, B., Messmer, C., Gushul-Leclaire, M., Villot, R., et al. (2023). An inventory of crosstalk between ubiquitination and other post-translational modifications in orchestrating cellular processes. iScience 26, 106276. doi:10.1016/j.isci.2023.106276
Bartlett, J. B., Dredge, K., and Dalgleish, A. G. (2004). The evolution of thalidomide and its IMiD derivatives as anticancer agents. Nat. Rev. Cancer 4, 314–322. doi:10.1038/nrc1323
Bin, Y. L., Hu, H. S., Tian, F., Wen, Z. H., Yang, M. F., Wu, B. H., et al. (2021). Metabolic reprogramming in gastric cancer: trojan horse effect. Front. Oncol. 11, 745209. doi:10.3389/fonc.2021.745209
Bode, A. M., and Dong, Z. (2004). Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4, 793–805. doi:10.1038/nrc1455
Bokemeyer, C., Ciardiello, F., Dubreuil, O., Guigay, J., Kasper, S., Pfeiffer, P., et al. (2024). Cetuximab every 2 weeks versus standard weekly dosing administration schedule. Future Oncol. 20, 393–407. doi:10.2217/fon-2023-0282
Bray, F., Ferlay, J., Laversanne, M., Brewster, D. H., Gombe Mbalawa, C., Kohler, B., et al. (2015). Cancer Incidence in Five Continents: inclusion criteria, highlights from Volume X and the global status of cancer registration. Int. J. Cancer 137, 2060–2071. doi:10.1002/ijc.29670
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., et al. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263. doi:10.3322/caac.21834
Bykov, V. J. N., Issaeva, N., Zache, N., Shilov, A., Hultcrantz, M., Bergman, J., et al. (2017). Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J. Biol. Chem. 292, 19607. doi:10.1074/jbc.AAC117.000815
Cao, T., Lu, Y., Wang, Q., Qin, H., Li, H., Guo, H., et al. (2022). A CGA/EGFR/GATA2 positive feedback circuit confers chemoresistance in gastric cancer. J. Clin. Invest 132, e154074. doi:10.1172/JCI154074
Cardoso, A. P., Pinto, M. L., Pinto, A. T., Oliveira, M. I., Pinto, M. T., Goncalves, R., et al. (2014). Macrophages stimulate gastric and colorectal cancer invasion through EGFR Y(1086), c-Src, Erk1/2 and Akt phosphorylation and smallGTPase activity. Oncogene 33, 2123–2133. doi:10.1038/onc.2013.154
Cargnello, M., and Roux, P. P. (2011). Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83. doi:10.1128/MMBR.00031-10
Carvalho, S., Catarino, T. A., Dias, A. M., Kato, M., Almeida, A., Hessling, B., et al. (2016). Preventing E-cadherin aberrant N-glycosylation at Asn-554 improves its critical function in gastric cancer. Oncogene 35, 1619–1631. doi:10.1038/onc.2015.225
Chai, A. W. Y., Tan, Y. H., Ooi, S., Yee, P. S., Yee, S. M., and Cheong, S. C. (2024). TNO155 is a selective SHP2 inhibitor to target PTPN11-dependent oral squamous cell carcinoma. Heliyon 10, e39677. doi:10.1016/j.heliyon.2024.e39677
Chang, H. M., and Yeh, E. T. H. (2020). SUMO: from bench to bedside. Physiol. Rev. 100, 1599–1619. doi:10.1152/physrev.00025.2019
Chen, H., Li, Y., Li, H., Chen, X., Fu, H., Mao, D., et al. (2024a). NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature 631, 663–669. doi:10.1038/s41586-024-07620-9
Chen, J. J., Ren, Y. L., Shu, C. J., Zhang, Y., Chen, M. J., Xu, J., et al. (2020). JP3, an antiangiogenic peptide, inhibits growth and metastasis of gastric cancer through TRIM25/SP1/MMP2 axis. J. Exp. Clin. Cancer Res. 39, 118. doi:10.1186/s13046-020-01617-8
Chen, T., Jinlin, D., Wang, F., Yuan, Z., Xue, J., Lu, T., et al. (2022). GSTM3 deficiency impedes DNA mismatch repair to promote gastric tumorigenesis via CAND1/NRF2-KEAP1 signaling. Cancer Lett. 538, 215692. doi:10.1016/j.canlet.2022.215692
Chen, Y., Li, Q., Yu, X., Lu, L., Zhou, Z., Li, M., et al. (2024b). The microprotein HDSP promotes gastric cancer progression through activating the MECOM-SPINK1-EGFR signaling axis. Nat. Commun. 15, 8381. doi:10.1038/s41467-024-50986-7
Cheng, L., Li, X., Dong, W., Yang, J., Li, P., Qiang, X., et al. (2024). LAMC2 regulates the proliferation, invasion, and metastasis of gastric cancer via PI3K/Akt signaling pathway. J. Cancer Res. Clin. Oncol. 150, 230. doi:10.1007/s00432-024-05720-7
Cockram, P. E., Kist, M., Prakash, S., Chen, S. H., Wertz, I. E., and Vucic, D. (2021). Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 28, 591–605. doi:10.1038/s41418-020-00708-5
Cutler, J. A., Perner, F., and Armstrong, S. A. (2021). Histone PTM crosstalk stimulates Dot1 methyltransferase activity. Trends Biochem. Sci. 46, 522–524. doi:10.1016/j.tibs.2021.04.001
Dagar, G., Kumar, R., Yadav, K. K., Singh, M., and Pandita, T. K. (2023). Ubiquitination and deubiquitination: implications on cancer therapy. Biochim. Biophys. Acta Gene Regul. Mech. 1866, 194979. doi:10.1016/j.bbagrm.2023.194979
Dai, C., and Gu, W. (2010). p53 post-translational modification: deregulated in tumorigenesis. Trends Mol. Med. 16, 528–536. doi:10.1016/j.molmed.2010.09.002
Dai, E., Wang, W., Li, Y., Ye, D., and Li, Y. (2024). Lactate and lactylation: behind the development of tumors. Cancer Lett. 591, 216896. doi:10.1016/j.canlet.2024.216896
Dai, X., Ren, T., Zhang, Y., and Nan, N. (2021). Methylation multiplicity and its clinical values in cancer. Expert Rev. Mol. Med. 23, e2. doi:10.1017/erm.2021.4
Dang, F., Nie, L., and Wei, W. (2021). Ubiquitin signaling in cell cycle control and tumorigenesis. Cell Death Differ. 28, 427–438. doi:10.1038/s41418-020-00648-0
Dang, F., and Wei, W. (2022). Targeting the acetylation signaling pathway in cancer therapy. Semin. Cancer Biol. 85, 209–218. doi:10.1016/j.semcancer.2021.03.001
Davis, P. A., and Sano, T. (2001). The difference in gastric cancer between Japan, USA and Europe: what are the facts? what are the suggestions? Crit. Rev. Oncol. Hematol. 40, 77–94. doi:10.1016/s1040-8428(00)00131-1
Debsharma, S., Pramanik, S., Bindu, S., Mazumder, S., Das, T., Pal, U., et al. (2024). NSAID targets SIRT3 to trigger mitochondrial dysfunction and gastric cancer cell death. iScience 27, 109384. doi:10.1016/j.isci.2024.109384
Decourtye-espiard, L., Bougen-Zhukov, N., Godwin, T., Brew, T., Schulpen, E., Black, M. A., et al. (2021). E-Cadherin-Deficient epithelial cells are sensitive to HDAC inhibitors. Cancers (Basel) 14, 175. doi:10.3390/cancers14010175
Deng, M., Liu, B., Song, H., Yu, R., Zou, D., Chen, Y., et al. (2020). β-Elemene inhibits the metastasis of multidrug-resistant gastric cancer cells through miR-1323/Cbl-b/EGFR pathway. Phytomedicine 69, 153184. doi:10.1016/j.phymed.2020.153184
Deribe, Y. L., Pawson, T., and Dikic, I. (2010). Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672. doi:10.1038/nsmb.1842
Deshpande, A., and Munoz, J. (2022). Targeted and cellular therapies in lymphoma: mechanisms of escape and innovative strategies. Front. Oncol. 12, 948513. doi:10.3389/fonc.2022.948513
Dewson, G., Eichhorn, P. J. A., and Komander, D. (2023). Deubiquitinases in cancer. Nat. Rev. Cancer 23, 842–862. doi:10.1038/s41568-023-00633-y
Dhillon, S. (2020). Decitabine/cedazuridine: first approval. Drugs 80, 1373–1378. doi:10.1007/s40265-020-01389-7
Dikic, I., and Schulman, B. A. (2023). An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 24, 273–287. doi:10.1038/s41580-022-00543-1
Diniz, F., Lamas, S., Osorio, H., Aguiar, P., Freitas, D., Gartner, F., et al. (2023). Nanoparticles targeting Sialyl-Tn for efficient tyrosine kinase inhibitor delivery in gastric cancer. Acta Biomater. 170, 142–154. doi:10.1016/j.actbio.2023.08.014
Dong, L., Zhu, J., Deng, A., Wei, J., Li, J., Mao, X., et al. (2023). Relationship between histone demethylase LSD family and development and prognosis of gastric cancer. Front. Immunol. 14, 1170773. doi:10.3389/fimmu.2023.1170773
Dowell, J., Minna, J. D., and Kirkpatrick, P. (2005). Erlotinib hydrochloride. Nat. Rev. Drug Discov. 4, 13–14. doi:10.1038/nrd1612
Du, Z., and Lovly, C. M. (2018). Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 17, 58. doi:10.1186/s12943-018-0782-4
Duarte, H. O., Rodrigues, J. G., Gomes, C., Hensbergen, P. J., Ederveen, A. L. H., DE Ru, A. H., et al. (2021). ST6Gal1 targets the ectodomain of ErbB2 in a site-specific manner and regulates gastric cancer cell sensitivity to trastuzumab. Oncogene 40, 3719–3733. doi:10.1038/s41388-021-01801-w
Ebert, T., Tran, N., Schurgers, L., Stenvinkel, P., and Shiels, P. G. (2022). Ageing - oxidative stress, PTMs and disease. Mol. Asp. Med. 86, 101099. doi:10.1016/j.mam.2022.101099
Fan, H., Yang, F., Xiao, Z., Luo, H., Chen, H., Chen, Z., et al. (2023). Lactylation: novel epigenetic regulatory and therapeutic opportunities. Am. J. Physiol. Endocrinol. Metab. 324, E330–E338. doi:10.1152/ajpendo.00159.2022
Fang, H., Zhou, Y., Bai, X., Che, W., Zhang, W., Zhang, D., et al. (2024). The VEGFA-induced MAPK-AKT/PTEN/TGFβ signal pathway enhances progression and MDR in gastric cancer. Genes (Basel) 15, 1266. doi:10.3390/genes15101266
Fang, S., Hong, H., Li, L., He, D., Xu, Z., Zuo, S., et al. (2017). Plasminogen kringle 5 suppresses gastric cancer via regulating HIF-1α and GRP78. Cell Death Dis. 8, e3144. doi:10.1038/cddis.2017.528
Feng, W., Ma, C., Rao, H., Zhang, W., Liu, C., Xu, Y., et al. (2023). Setd2 deficiency promotes gastric tumorigenesis through inhibiting the SIRT1/FOXO pathway. Cancer Lett. 579, 216470. doi:10.1016/j.canlet.2023.216470
Ferreira, J. A., Magalhaes, A., Gomes, J., Peixoto, A., Gaiteiro, C., Fernandes, E., et al. (2017). Protein glycosylation in gastric and colorectal cancers: toward cancer detection and targeted therapeutics. Cancer Lett. 387, 32–45. doi:10.1016/j.canlet.2016.01.044
Filippopoulou, C., Simos, G., and Chachami, G. (2020). The role of sumoylation in the response to hypoxia: an overview. Cells 9, 2359. doi:10.3390/cells9112359
Fischer, M., Schade, A. E., Branigan, T. B., Muller, G. A., and Decaprio, J. A. (2022). Coordinating gene expression during the cell cycle. Trends Biochem. Sci. 47, 1009–1022. doi:10.1016/j.tibs.2022.06.007
Fresno Vara, J. A., Casado, E., DE Castro, J., Cejas, P., Belda-Iniesta, C., and Gonzalez-Baron, M. (2004). PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 30, 193–204. doi:10.1016/j.ctrv.2003.07.007
Fukuda, A., and Okuma, Y. (2024). From rarity to reality: osimertinib's promising horizon in treating uncommon EGFR mutations in non-small cell lung cancer. Clin. Cancer Res. 30, 3128–3136. doi:10.1158/1078-0432.CCR-23-4035
Ge, L., Zhao, G., Lan, C., Song, H., Qi, D., Huang, P., et al. (2023). MESP2 binds competitively to TCF4 to suppress gastric cancer progression by regulating the SKP2/p27 axis. Cell Death Discov. 9, 79. doi:10.1038/s41420-023-01367-4
Geffen, Y., Anand, S., Akiyama, Y., Yaron, T. M., Song, Y., Johnson, J. L., et al. (2023). Pan-cancer analysis of post-translational modifications reveals shared patterns of protein regulation. Cell 186, 3945–3967 e26. doi:10.1016/j.cell.2023.07.013
Gillette, M. A., Jimenez, C. R., and Carr, S. A. (2024). Clinical proteomics: a promise becoming reality. Mol. Cell Proteomics 23, 100688. doi:10.1016/j.mcpro.2023.100688
Glaviano, A., Foo, A. S. C., Lam, H. Y., Yap, K. C. H., Jacot, W., Jones, R. H., et al. (2023). PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 22, 138. doi:10.1186/s12943-023-01827-6
Gonzalez, A., Covarrubias-Pinto, A., Bhaskara, R. M., Glogger, M., Kuncha, S. K., Xavier, A., et al. (2023). Ubiquitination regulates ER-phagy and remodelling of endoplasmic reticulum. Nature 618, 394–401. doi:10.1038/s41586-023-06089-2
Gu, Y., Fang, Y., Wu, X., Xu, T., Hu, T., Xu, Y., et al. (2023). The emerging roles of SUMOylation in the tumor microenvironment and therapeutic implications. Exp. Hematol. Oncol. 12, 58. doi:10.1186/s40164-023-00420-3
Gu, Y., Xu, T., Fang, Y., Shao, J., Hu, T., Wu, X., et al. (2024). CBX4 counteracts cellular senescence to desensitize gastric cancer cells to chemotherapy by inducing YAP1 SUMOylation. Drug Resist Updat 77, 101136. doi:10.1016/j.drup.2024.101136
Guo, X., Li, Y., Wan, B., Lv, Y., Wang, X., Liu, G., et al. (2022). KAT7 promoted gastric cancer progression through promoting YAP1 activation. Pathol. Res. Pract. 237, 154020. doi:10.1016/j.prp.2022.154020
Hajek, R., Bryce, R., Ro, S., Klencke, B., and Ludwig, H. (2012). Design and rationale of FOCUS (PX-171-011): a randomized, open-label, phase 3 study of carfilzomib versus best supportive care regimen in patients with relapsed and refractory multiple myeloma (R/R MM). BMC Cancer 12, 415. doi:10.1186/1471-2407-12-415
Hameed, H., and Cassidy, J. (2011). Use of capecitabine in management of early colon cancer. Cancer Manag. Res. 3, 295–299. doi:10.2147/CMR.S12704
Han, Z. J., Feng, Y. H., Gu, B. H., Li, Y. M., and Chen, H. (2018). The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 52, 1081–1094. doi:10.3892/ijo.2018.4280
Harrigan, J. A., Jacq, X., Martin, N. M., and Jackson, S. P. (2018). Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78. doi:10.1038/nrd.2017.152
He, J., Yi, J., Ji, L., Dai, L., Chen, Y., and Xue, W. (2024). ECHDC2 inhibits the proliferation of gastric cancer cells by binding with NEDD4 to degrade MCCC2 and reduce aerobic glycolysis. Mol. Med. 30, 69. doi:10.1186/s10020-024-00832-9
He, Y., Zhou, J., and Wan, Q. (2021). The E3 ligase HUWE1 mediates TGFBR2 ubiquitination and promotes gastric cancer cell proliferation, migration, and invasion. Invest New Drugs 39, 713–723. doi:10.1007/s10637-020-01041-x
Hegde, S., Soory, A., Kaduskar, B., and Ratnaparkhi, G. S. (2020). SUMO conjugation regulates immune signalling. Fly. (Austin) 14, 62–79. doi:10.1080/19336934.2020.1808402
Hernandez-Valladares, M., Wangen, R., Berven, F. S., and Guldbrandsen, A. (2019). Protein post-translational modification crosstalk in acute myeloid leukemia calls for action. Curr. Med. Chem. 26, 5317–5337. doi:10.2174/0929867326666190503164004
Hideshima, T., Richardson, P. G., and Anderson, K. C. (2011). Mechanism of action of proteasome inhibitors and deacetylase inhibitors and the biological basis of synergy in multiple myeloma. Mol. Cancer Ther. 10, 2034–2042. doi:10.1158/1535-7163.MCT-11-0433
Hong, R., Zhang, X., Zhang, Y., Bei, L., Yang, J., Xia, J., et al. (2024). The serine protease CORIN promotes progression of gastric cancer by mediating the ERK1/2 MAPK pathway. Mol. Carcinog. 63, 1500–1514. doi:10.1002/mc.23739
Hosamani, K. R., K, H., Pal, R., Matada, G. S. P., B, K., I, A., et al. (2024). Pyrazole, pyrazoline, and fused pyrazole derivatives: new horizons in EGFR-targeted anticancer agents. Chem. Biodivers. 21, e202400880. doi:10.1002/cbdv.202400880
Hou, J., Huang, P., Lan, C., Geng, S., Xu, M., Liu, Y., et al. (2022). ZC3H15 promotes gastric cancer progression by targeting the FBXW7/c-Myc pathway. Cell Death Discov. 8, 32. doi:10.1038/s41420-022-00815-x
Hou, Y. C., and Deng, J. Y. (2015). Role of E3 ubiquitin ligases in gastric cancer. World J. Gastroenterol. 21, 786–793. doi:10.3748/wjg.v21.i3.786
Hu, W. T., Yeh, C. C., Liu, S. Y., Huang, M. C., and Lai, I. R. (2018). The O-glycosylating enzyme GALNT2 suppresses the malignancy of gastric adenocarcinoma by reducing EGFR activities. Am. J. Cancer Res. 8, 1739–1751.
Hu, Y., Chen, C., Tong, X., Chen, S., Hu, X., Pan, B., et al. (2021). NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 12, 842. doi:10.1038/s41419-021-04127-3
Huang, B., Wu, Q., Ge, Y., Zhang, J., Sun, L., Zhang, Y., et al. (2014). Expression of N-acetylglucosaminyltransferase V in gastric cancer correlates with metastasis and prognosis. Int. J. Oncol. 44, 849–857. doi:10.3892/ijo.2014.2248
Huang, G., Cai, G., Hu, D., Li, J., Xu, Q., Chen, Z., et al. (2022a). Low SP1 SUMOylation-dependent SNHG17 upregulation promotes drug resistance of gastric cancer through impairing hsa-miR-23b-3p-induced Notch2 inhibition. Cell Oncol. (Dordr) 45, 1329–1346. doi:10.1007/s13402-022-00722-4
Huang, G., Xiang, Z., Wu, H., He, Q., Dou, R., Lin, Z., et al. (2022b). The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int. J. Biol. Sci. 18, 1415–1433. doi:10.7150/ijbs.69454
Huang, K. Y., Lee, T. Y., Kao, H. J., Ma, C. T., Lee, C. C., Lin, T. H., et al. (2019). dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 47, D298–D308. doi:10.1093/nar/gky1074
Huang, N., He, H. W., He, Y. Y., Gu, W., Xu, M. J., and Liu, L. (2023). Xiaotan Sanjie recipe, a compound Chinese herbal medicine, inhibits gastric cancer metastasis by regulating GnT-V-mediated E-cadherin glycosylation. J. Integr. Med. 21, 561–574. doi:10.1016/j.joim.2023.11.001
Huang, N., Xu, C., Deng, L., Li, X., Bian, Z., Zhang, Y., et al. (2020). PAICS contributes to gastric carcinogenesis and participates in DNA damage response by interacting with histone deacetylase 1/2. Cell Death Dis. 11, 507. doi:10.1038/s41419-020-2708-5
Huang, Y., Yang, X., Wei, M., Yang, X., Yuan, Z., Huang, J., et al. (2024). FUT11 expression in gastric cancer: its prognostic significance and role in immune regulation. Discov. Oncol. 15, 250. doi:10.1007/s12672-024-01120-y
Huangfu, L., He, Q., Han, J., Shi, J., Li, X., Cheng, X., et al. (2021). MicroRNA-135b/CAMK2D Axis contribute to malignant progression of gastric cancer through EMT process remodeling. Int. J. Biol. Sci. 17, 1940–1952. doi:10.7150/ijbs.58062
Hubbard, S. R., and Till, J. H. (2000). Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 69, 373–398. doi:10.1146/annurev.biochem.69.1.373
Ito, K. (2007). Impact of post-translational modifications of proteins on the inflammatory process. Biochem. Soc. Trans. 35, 281–283. doi:10.1042/BST0350281
Ito, T., Ando, H., Suzuki, T., Ogura, T., Hotta, K., Imamura, Y., et al. (2010). Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350. doi:10.1126/science.1177319
Jaffry, U., and Wells, G. (2023). Small molecule and peptide inhibitors of βTrCP and the βTrCP-NRF2 protein-protein interaction. Biochem. Soc. Trans. 51, 925–936. doi:10.1042/BST20220352
Jang, T. J., and Kim, U. J. (2016). O-GlcNAcylation is associated with the development and progression of gastric carcinoma. Pathol. Res. Pract. 212, 622–630. doi:10.1016/j.prp.2016.04.002
Jarrold, J., and Davies, C. C. (2019). PRMTs and arginine methylation: cancer's best-kept secret? Trends Mol. Med. 25, 993–1009. doi:10.1016/j.molmed.2019.05.007
Jenke, R., Oliinyk, D., Zenz, T., Korfer, J., Schaker-Hubner, L., Hansen, F. K., et al. (2024). HDAC inhibitors activate lipid peroxidation and ferroptosis in gastric cancer. Biochem. Pharmacol. 225, 116257. doi:10.1016/j.bcp.2024.116257
Jiang, H. Y., Chen, Y. L., Xu, X. X., Li, C. Y., Chen, Y., Li, D. P., et al. (2022). Ubiquitylation of cyclin C by HACE1 regulates cisplatin-associated sensitivity in gastric cancer. Clin. Transl. Med. 12, e770. doi:10.1002/ctm2.770
Jiang, Q., Miao, J., Wu, F., Li, F., Zhang, J., Jing, X., et al. (2023). RNF6 promotes gastric cancer progression by regulating CCNA1/CREBBP transcription. Cell Cycle 22, 2018–2037. doi:10.1080/15384101.2023.2275899
Jiang, T., Xia, Y., Lv, J., Li, B., Li, Y., Wang, S., et al. (2021). A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 20, 66. doi:10.1186/s12943-021-01358-y
Jie, M., Wu, Y., Gao, M., Li, X., Liu, C., Ouyang, Q., et al. (2020). CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. Cancer 19, 56. doi:10.1186/s12943-020-01160-2
Karin, M., and Ben-Neriah, Y. (2000). Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18, 621–663. doi:10.1146/annurev.immunol.18.1.621
Khoury, G. A., Baliban, R. C., and Floudas, C. A. (2011). Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 90. doi:10.1038/srep00090
Kim, H., Jang, B., Zhang, C., Caldwell, B., Park, D. J., Kong, S. H., et al. (2024). Targeting stem cells and dysplastic features with dual MEK/ERK and STAT3 suppression in gastric carcinogenesis. Gastroenterology 166, 117–131. doi:10.1053/j.gastro.2023.09.040
Kim, S., Kim, W., Kim, D. H., Jang, J. H., Kim, S. J., Park, S. A., et al. (2020). Resveratrol suppresses gastric cancer cell proliferation and survival through inhibition of PIM-1 kinase activity. Arch. Biochem. Biophys. 689, 108413. doi:10.1016/j.abb.2020.108413
Kirsch, R., Jensen, O. N., and Schwammle, V. (2020). Visualization of the dynamics of histone modifications and their crosstalk using PTM-CrossTalkMapper. Methods 184, 78–85. doi:10.1016/j.ymeth.2020.01.012
Kokate, S. B., Dixit, P., Das, L., Rath, S., Roy, A. D., Poirah, I., et al. (2018). Acetylation-mediated Siah2 stabilization enhances PHD3 degradation in Helicobacter pylori-infected gastric epithelial cancer cells. FASEB J. 32, 5378–5389. doi:10.1096/fj.201701344RRR
Koromilas, A. E., and Mounir, Z. (2013). Control of oncogenesis by eIF2α phosphorylation: implications in PTEN and PI3K-Akt signaling and tumor treatment. Future Oncol. 9, 1005–1015. doi:10.2217/fon.13.49
Lagunas-Rangel, F. A. (2024). The dark side of SIRT7. Mol. Cell Biochem. 479, 2843–2861. doi:10.1007/s11010-023-04869-y
Lakshmi Ch, N. P., Sivagnanam, A., Raja, S., and Mahalingam, S. (2021). Molecular basis for RASSF10/NPM/RNF2 feedback cascade-mediated regulation of gastric cancer cell proliferation. J. Biol. Chem. 297, 100935. doi:10.1016/j.jbc.2021.100935
Laude, A. J., and Prior, I. A. (2008). Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J. Cell Sci. 121, 421–427. doi:10.1242/jcs.020107
Le, S., Yu, M., and Yan, J. (2019). Phosphorylation reduces the mechanical stability of the α-catenin/β-catenin complex. Angew. Chem. Int. Ed. Engl. 58, 18663–18669. doi:10.1002/anie.201911383
Lee, J. M., Hammaren, H. M., Savitski, M. M., and Baek, S. H. (2023). Control of protein stability by post-translational modifications. Nat. Commun. 14, 201. doi:10.1038/s41467-023-35795-8
Li, G., Yao, Q., Liu, P., Zhang, H., Liu, Y., Li, S., et al. (2020). Critical roles and clinical perspectives of RNA methylation in cancer. MedComm 5, e559. doi:10.1002/mco2.559
Li, J., Lan, Z., Liao, W., Horner, J. W., Xu, X., Liu, J., et al. (2023a). Histone demethylase KDM5D upregulation drives sex differences in colon cancer. Nature 619, 632–639. doi:10.1038/s41586-023-06254-7
Li, J., Liu, X., Peng, B., Feng, T., Zhou, W., Meng, L., et al. (2023b). O-GlcNAc has crosstalk with ADP-ribosylation via PARG. J. Biol. Chem. 299, 105354. doi:10.1016/j.jbc.2023.105354
Li, K. Q., Bai, X., Ke, A. T., Ding, S. Q., Zhang, C. D., and Dai, D. Q. (2024). Ubiquitin-specific proteases: from biological functions to potential therapeutic applications in gastric cancer. Biomed. Pharmacother. 173, 116323. doi:10.1016/j.biopha.2024.116323
Li, N., and Zhan, X. (2020). Mass spectrometry-based mitochondrial proteomics in human ovarian cancers. Mass Spectrom. Rev. 39, 471–498. doi:10.1002/mas.21618
Li, S., Xiao, B., Zhan, Y., Wu, Z., Zhang, W., Pan, H., et al. (2024). Rps3 attenuates gastric precancerous lesions by promoting dendritic cells maturation via AKT/β-Catenin pathway. J. Proteome Res. 23, 4579–4588. doi:10.1021/acs.jproteome.4c00472
Li, X., Yang, K. B., Chen, W., Mai, J., Wu, X. Q., Sun, T., et al. (2021). CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy 17, 4323–4340. doi:10.1080/15548627.2021.1912270
Li, Y., Jin, H., Li, Q., Shi, L., Mao, Y., and Zhao, L. (2024). The role of RNA methylation in tumor immunity and its potential in immunotherapy. Mol. Cancer 23, 130. doi:10.1186/s12943-024-02041-8
Li, Y., and Seto, E. (2016). HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 6, a026831. doi:10.1101/cshperspect.a026831
Li, Y., Tian, Z., Tan, Y., Lian, G., Chen, S., Chen, S., et al. (2020). Bmi-1-induced miR-27a and miR-155 promote tumor metastasis and chemoresistance by targeting RKIP in gastric cancer. Mol. Cancer 19, 109. doi:10.1186/s12943-020-01229-y
Li, Y., Wang, F., Chen, X., Wang, J., Zhao, Y., Li, Y., et al. (2019). Zinc-dependent deacetylase (HDAC) inhibitors with different zinc binding groups. Curr. Top. Med. Chem. 19, 223–241. doi:10.2174/1568026619666190122144949
Li, Y., Zhang, M., Dorfman, R. G., Pan, Y., Tang, D., Xu, L., et al. (2018). SIRT2 promotes the migration and invasion of gastric cancer through RAS/ERK/JNK/MMP-9 pathway by increasing PEPCK1-related metabolism. Neoplasia 20, 745–756. doi:10.1016/j.neo.2018.03.008
Li, Y., Zhang, R., and Hei, H. (2023c). Advances in post-translational modifications of proteins and cancer immunotherapy. Front. Immunol. 14, 1229397. doi:10.3389/fimmu.2023.1229397
Li, Z., Liang, P., Chen, Z., Chen, Z., Jin, T., He, F., et al. (2024). CAF-secreted LOX promotes PD-L1 expression via histone Lactylation and regulates tumor EMT through TGFβ/IGF1 signaling in gastric Cancer. Cell Signal 124, 111462. doi:10.1016/j.cellsig.2024.111462
Liang, J., Ge, F., Guo, C., Luo, G., Wang, X., Han, G., et al. (2009). Inhibition of PI3K/Akt partially leads to the inhibition of PrP(C)-induced drug resistance in gastric cancer cells. FEBS J. 276, 685–694. doi:10.1111/j.1742-4658.2008.06816.x
Lin, D. C., Zheng, S. Y., Zhang, Z. G., Luo, J. H., Zhu, Z. L., Li, L., et al. (2021). TRPC3 promotes tumorigenesis of gastric cancer via the CNB2/GSK3β/NFATc2 signaling pathway. Cancer Lett. 519, 211–225. doi:10.1016/j.canlet.2021.07.038
Lin, L., Zheng, X., Wu, M., Chen, Y., Nian, Q., Lin, Y., et al. (2024). A real-world disproportionality analysis of FDA Adverse Event Reporting System (FAERS) events for ramucirumab. Expert Opin. Drug Saf., 1–10. doi:10.1080/14740338.2024.2441286
Lin, Y., Jing, X., Chen, Z., Pan, X., Xu, D., Yu, X., et al. (2023). Histone deacetylase-mediated tumor microenvironment characteristics and synergistic immunotherapy in gastric cancer. Theranostics 13, 4574–4600. doi:10.7150/thno.86928
Liu, C., Hao, D., Ai, M., Zhang, Y., Li, J., and Xu, C. (2022). The long non-coding RNA UPAT promotes gastric cancer cell progression via UHRF1. Genes Genomics 44, 1283–1300. doi:10.1007/s13258-022-01235-y
Liu, D., Li, Z., Yang, Z., Ma, J., and Mai, S. (2021). Ginkgoic acid impedes gastric cancer cell proliferation, migration and EMT through inhibiting the SUMOylation of IGF-1R. Chem. Biol. Interact. 337, 109394. doi:10.1016/j.cbi.2021.109394
Liu, J. Z., Hu, Y. L., Feng, Y., Jiang, Y., Guo, Y. B., Liu, Y. F., et al. (2020). BDH2 triggers ROS-induced cell death and autophagy by promoting Nrf2 ubiquitination in gastric cancer. J. Exp. Clin. Cancer Res. 39, 123. doi:10.1186/s13046-020-01620-z
Liu, L., Liu, L., Wang, Y., Fang, Z., Bian, Y., Zhang, W., et al. (2024a). Robust glycoproteomics platform reveals a tetra-antennary site-specific glycan capping with sialyl-lewis antigen for early detection of gastric cancer. Adv. Sci. (Weinh) 11, e2306955. doi:10.1002/advs.202306955
Liu, R., Zhao, E., Yu, H., Yuan, C., Abbas, M. N., and Cui, H. (2023). Methylation across the central dogma in health and diseases: new therapeutic strategies. Signal Transduct. Target Ther. 8, 310. doi:10.1038/s41392-023-01528-y
Liu, S., Wang, Z., Hu, L., Ye, C., Zhang, X., Zhu, Z., et al. (2024b). Pan-cancer analysis of super-enhancer-induced LINC00862 and validation as a SIRT1-promoting factor in cervical cancer and gastric cancer. Transl. Oncol. 45, 101982. doi:10.1016/j.tranon.2024.101982
Locke, W. J., Guanzon, D., Ma, C., Liew, Y. J., Duesing, K. R., Fung, K. Y. C., et al. (2019). DNA methylation cancer biomarkers: translation to the clinic. Front. Genet. 10, 1150. doi:10.3389/fgene.2019.01150
Lopez, M. J., Carbajal, J., Alfaro, A. L., Saravia, L. G., Zanabria, D., Araujo, J. M., et al. (2023). Characteristics of gastric cancer around the world. Crit. Rev. Oncol. Hematol. 181, 103841. doi:10.1016/j.critrevonc.2022.103841
Lopez-Girona, A., Mendy, D., Ito, T., Miller, K., Gandhi, A. K., Kang, J., et al. (2012). Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 26, 2326–2335. doi:10.1038/leu.2012.119
Louie, B. H., and Kurzrock, R. (2020). BAP1: not just a BRCA1-associated protein. Cancer Treat. Rev. 90, 102091. doi:10.1016/j.ctrv.2020.102091
Lu, J., Li, D., Jiang, H., Li, Y., Lu, C., Chen, T., et al. (2023). The aryl sulfonamide indisulam inhibits gastric cancer cell migration by promoting the ubiquitination and degradation of the transcription factor ZEB1. J. Biol. Chem. 299, 103025. doi:10.1016/j.jbc.2023.103025
Lu, S., Dong, X., Jian, H., Chen, J., Chen, G., Sun, Y., et al. (2022). AENEAS: a randomized phase III trial of aumolertinib versus gefitinib as first-line therapy for locally advanced or MetastaticNon-small-cell lung cancer with EGFR exon 19 deletion or L858R mutations. J. Clin. Oncol. 40, 3162–3171. doi:10.1200/JCO.21.02641
Luo, Y., Ma, J., and Lu, W. (2020). The significance of mitochondrial dysfunction in cancer. Int. J. Mol. Sci. 21, 5598. doi:10.3390/ijms21165598
Lv, X., Lv, Y., and Dai, X. (2023). Lactate, histone lactylation and cancer hallmarks. Expert Rev. Mol. Med. 25, e7. doi:10.1017/erm.2022.42
Ma, J., Shi, Y., Lu, Q., and Huang, D. (2024). Inflammation-related gene ADH1A regulates the polarization of macrophage M1 and influences the malignant progression of gastric cancer. J. Inflamm. Res. 17, 4647–4665. doi:10.2147/JIR.S452670
Ma, J., Zhao, G., DU, J., Li, J., Lin, G., and Zhang, J. (2021). LncRNA FENDRR inhibits gastric cancer cell proliferation and invasion via the miR-421/SIRT3/notch-1 Axis. Cancer Manag. Res. 13, 9175–9187. doi:10.2147/CMAR.S329419
Marmorstein, R., and Zhou, M. M. (2014). Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762. doi:10.1101/cshperspect.a018762
Martin Moyano, P., Nemec, V., and Paruch, K. (2020). Cdc-like kinases (CLKs): biology, chemical probes, and therapeutic potential. Int. J. Mol. Sci. 21, 7549. doi:10.3390/ijms21207549
Matsuoka, T., and Yashiro, M. (2023). Molecular insight into gastric cancer invasion-current status and future directions. Cancers (Basel) 16, 54. doi:10.3390/cancers16010054
Mattei, A. L., Bailly, N., and Meissner, A. (2022). DNA methylation: a historical perspective. Trends Genet. 38, 676–707. doi:10.1016/j.tig.2022.03.010
Mcclure, J. J., Li, X., and Chou, C. J. (2018). Advances and challenges of HDAC inhibitors in cancer therapeutics. Adv. Cancer Res. 138, 183–211. doi:10.1016/bs.acr.2018.02.006
Mevissen, T. E. T., and Komander, D. (2017). Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192. doi:10.1146/annurev-biochem-061516-044916
Miao, Z., Li, J., Wang, Y., Shi, M., Gu, X., Zhang, X., et al. (2023). Hsa_circ_0136666 stimulates gastric cancer progression and tumor immune escape by regulating the miR-375/PRKDC Axis and PD-L1 phosphorylation. Mol. Cancer 22, 205. doi:10.1186/s12943-023-01883-y
Michalak, E. M., Burr, M. L., Bannister, A. J., and Dawson, M. A. (2019). The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 20, 573–589. doi:10.1038/s41580-019-0143-1
Mittelstaedt, N. N., Becker, A. L., DE Freitas, D. N., Zanin, R. F., Stein, R. T., and Duarte DE Souza, A. P. (2021). DNA methylation and immune memory response. Cells 10, 2943. doi:10.3390/cells10112943
Mo, W., Deng, L., Cheng, Y., Ge, S., and Wang, J. (2024). IGFBP7 regulates cell proliferation and migration through JAK/STAT pathway in gastric cancer and is regulated by DNA and RNA methylation. J. Cell Mol. Med. 28, e70080. doi:10.1111/jcmm.70080
Moore, L. D., LE, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi:10.1038/npp.2012.112
Morgan, E. L., Toni, T., Viswanathan, R., Robbins, Y., Yang, X., Cheng, H., et al. (2023). Inhibition of USP14 promotes TNFα-induced cell death in head and neck squamous cell carcinoma (HNSCC). Cell Death Differ. 30, 1382–1396. doi:10.1038/s41418-023-01144-x
Mun, D. G., Bhin, J., Kim, S., Kim, H., Jung, J. H., Jung, Y., et al. (2019). Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell 35, 111–124 e10. doi:10.1016/j.ccell.2018.12.003
Nagaraju, G. P., Kasa, P., Dariya, B., Surepalli, N., Peela, S., and Ahmad, S. (2021). Epigenetics and therapeutic targets in gastrointestinal malignancies. Drug Discov. Today 26, 2303–2314. doi:10.1016/j.drudis.2021.04.013
Narayanan, S., Cai, C. Y., Assaraf, Y. G., Guo, H. Q., Cui, Q., Wei, L., et al. (2020). Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist Updat 48, 100663. doi:10.1016/j.drup.2019.100663
Narita, T., Weinert, B. T., and Choudhary, C. (2019). Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174. doi:10.1038/s41580-018-0081-3
Nassir, F. (2022). NAFLD: mechanisms, treatments, and biomarkers. Biomolecules 12, 824. doi:10.3390/biom12060824
Ni, T., Chu, Z., Tao, L., Zhao, Y., Zhu, M., Luo, Y., et al. (2023). PTBP1 drives c-Myc-dependent gastric cancer progression and stemness. Br. J. Cancer 128, 1005–1018. doi:10.1038/s41416-022-02118-5
Niu, Z., Li, X., Dong, S., Gao, J., Huang, Q., Yang, H., et al. (2021). The E3 ubiquitin ligase HOIP inhibits cancer cell apoptosis via modulating PTEN stability. J. Cancer 12, 6553–6562. doi:10.7150/jca.61996
O'Connor, O. A., Ma, H., Chan, J. Y. S., Kim, S. J., Yoon, S. E., and Kim, W. S. (2024). Peripheral T-cell lymphoma: from biology to practice to the future. Cancer Treat. Rev. 129, 102793. doi:10.1016/j.ctrv.2024.102793
Onglao, W., Khew-Goodall, Y., Belle, L., and Lonic, A. (2022). Aberrant post-translational modifications in endosomal trafficking are potential therapeutic targets to avert therapy resistance in solid cancers: dysregulation of PTM-regulated endosomal interactions presents an opportunity to block oncogenic signalling from multiple receptors by targeting common trafficking pathways: dysregulation of PTM-regulated endosomal interactions presents an opportunity to block oncogenic signalling from multiple receptors by targeting common trafficking pathways. Bioessays 44, e2100192. doi:10.1002/bies.202100192
Pan, S., and Chen, R. (2022). Pathological implication of protein post-translational modifications in cancer. Mol. Asp. Med. 86, 101097. doi:10.1016/j.mam.2022.101097
Pang, J., Li, G., Qian, H., Wu, Y., and Chen, Y. (2022). Secretory type II cGMP-dependent protein kinase blocks activation of PDGFRβ via Ser254 in gastric cancer cells. Cell Biol. Int. 46, 747–754. doi:10.1002/cbin.11766
Parsonnet, J., Friedman, G. D., Vandersteen, D. P., Chang, Y., Vogelman, J. H., Orentreich, N., et al. (1991). Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325, 1127–1131. doi:10.1056/NEJM199110173251603
Paska, A. V., and Hudler, P. (2015). Aberrant methylation patterns in cancer: a clinical view. Biochem. Med. Zagreb. 25, 161–176. doi:10.11613/BM.2015.017
Peng, W., Zhang, J., Wang, S., Wang, F., Wang, K., Geng, R., et al. (2023). LncRNA MIR31HG controls the proliferation and metastasis of gastric cancer by c-CBL-mediated degradation of β-catenin. Genes Dis. 10, 712–715. doi:10.1016/j.gendis.2022.12.022
Pienkowski, T., Kowalczyk, T., Cysewski, D., Kretowski, A., and Ciborowski, M. (2023). Glioma and post-translational modifications: a complex relationship. Biochim. Biophys. Acta Rev. Cancer 1878, 189009. doi:10.1016/j.bbcan.2023.189009
Pinho, S. S., and Reis, C. A. (2015). Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555. doi:10.1038/nrc3982
Poniewierska-Baran, A., Warias, P., and Zgutka, K. (2022). Sirtuins (SIRTs) as a novel target in gastric cancer. Int. J. Mol. Sci. 23, 15119. doi:10.3390/ijms232315119
Popovic, D., Vucic, D., and Dikic, I. (2014). Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253. doi:10.1038/nm.3739
Qi, M., Jiao, M., Li, X., Hu, J., Wang, L., Zou, Y., et al. (2021). Correction: CUL4B promotes gastric cancer invasion and metastasis-involvement of upregulation of HER2. Oncogene 40, 6140–6141. doi:10.1038/s41388-021-01995-z
Qin, X., Sufi, J., Vlckova, P., Kyriakidou, P., Acton, S. E., Li, V. S. W., et al. (2020). Cell-type-specific signaling networks in heterocellular organoids. Nat. Methods 17, 335–342. doi:10.1038/s41592-020-0737-8
Qiu, L., Xu, W., Lu, X., Chen, F., Chen, Y., Tian, Y., et al. (2023). The HDAC6-RNF168 axis regulates H2A/H2A.X ubiquitination to enable double-strand break repair. Nucleic Acids Res. 51, 9166–9182. doi:10.1093/nar/gkad631
Qu, J., Li, P., and Sun, Z. (2023). Histone lactylation regulates cancer progression by reshaping the tumor microenvironment. Front. Immunol. 14, 1284344. doi:10.3389/fimmu.2023.1284344
Qu, Y., Dang, S., and Hou, P. (2013). Gene methylation in gastric cancer. Clin. Chim. Acta 424, 53–65. doi:10.1016/j.cca.2013.05.002
Radziejewska, I., Supruniuk, K., and Bielawska, A. (2021). Anti-cancer effect of combined action of anti-MUC1 and rosmarinic acid in AGS gastric cancer cells. Eur. J. Pharmacol. 902, 174119. doi:10.1016/j.ejphar.2021.174119
Ramaiah, M. J., Tangutur, A. D., and Manyam, R. R. (2021). Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 277, 119504. doi:10.1016/j.lfs.2021.119504
Ramesh, P., Behera, S. K., Kotimoole, C. N., Mohanty, V., Raju, R., Prasad, T. S. K., et al. (2023). Mining proteomics data to extract post-translational modifications associated with gastric cancer. Amino Acids 55, 993–1001. doi:10.1007/s00726-023-03287-0
Regel, I., Merkl, L., Friedrich, T., Burgermeister, E., Zimmermann, W., Einwachter, H., et al. (2012). Pan-histone deacetylase inhibitor panobinostat sensitizes gastric cancer cells to anthracyclines via induction of CITED2. Gastroenterology 143, 99–109 e10. doi:10.1053/j.gastro.2012.03.035
Ren, H., and Tang, L. (2021). HDAC3-mediated lncRNA-LOC101928316 contributes to cisplatin resistance in gastric cancer via activating the PI3K-Akt-mTOR pathway. Neoplasma 68, 1043–1051. doi:10.4149/neo_2021_210317N356
Rong, L., Li, Z., Leng, X., Li, H., Ma, Y., Chen, Y., et al. (2020). Salidroside induces apoptosis and protective autophagy in human gastric cancer AGS cells through the PI3K/Akt/mTOR pathway. Biomed. Pharmacother. 122, 109726. doi:10.1016/j.biopha.2019.109726
Sampson, C., Wang, Q., Otkur, W., Zhao, H., Lu, Y., Liu, X., et al. (2023). The roles of E3 ubiquitin ligases in cancer progression and targeted therapy. Clin. Transl. Med. 13, e1204. doi:10.1002/ctm2.1204
Seeler, J. S., and Dejean, A. (2017). SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197. doi:10.1038/nrc.2016.143
Shen, Q., Yang, L., Li, C., Wang, T., Lv, J., Liu, W., et al. (2023). Metformin promotes cGAS/STING signaling pathway activation by blocking AKT phosphorylation in gastric cancer. Heliyon 9, e18954. doi:10.1016/j.heliyon.2023.e18954
Shi, L., Wang, X., Guo, S., Gou, H., Shang, H., Jiang, X., et al. (2024). TMEM65 promotes gastric tumorigenesis by targeting YWHAZ to activate PI3K-Akt-mTOR pathway and is a therapeutic target. Oncogene 43, 931–943. doi:10.1038/s41388-024-02959-9
Shvedunova, M., and Akhtar, A. (2022). Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349. doi:10.1038/s41580-021-00441-y
Singh, V., Ram, M., Kumar, R., Prasad, R., Roy, B. K., and Singh, K. K. (2017). Phosphorylation: implications in cancer. Protein J. 36, 1–6. doi:10.1007/s10930-017-9696-z
Sivaraj, D., Green, M. M., and Gasparetto, C. (2017). Panobinostat for the management of multiple myeloma. Future Oncol. 13, 477–488. doi:10.2217/fon-2016-0329
Smyth, E. C., Nilsson, M., Grabsch, H. I., VAN Grieken, N. C., and Lordick, F. (2020). Gastric cancer. Lancet 396, 635–648. doi:10.1016/S0140-6736(20)31288-5
Sogutlu, F., Pekerbas, M., and Biray Avci, C. (2022). Epigenetic signatures in gastric cancer: current knowledge and future perspectives. Expert Rev. Mol. Diagn 22, 1063–1075. doi:10.1080/14737159.2022.2159381
Sokolova, O., and Naumann, M. (2021). Manifold role of ubiquitin in Helicobacter pylori infection and gastric cancer. Cell Mol. Life Sci. 78, 4765–4783. doi:10.1007/s00018-021-03816-8
Spaety, M. E., Gries, A., Badie, A., Venkatasamy, A., Romain, B., Orvain, C., et al. (2019). HDAC4 levels control sensibility toward cisplatin in gastric cancer via the p53-p73/BIK pathway. Cancers (Basel) 11, 1747. doi:10.3390/cancers11111747
Stanczak, M. A., Rodrigues Mantuano, N., Kirchhammer, N., Sanin, D. E., Jacob, F., Coelho, R., et al. (2022). Targeting cancer glycosylation repolarizes tumor-associated macrophages allowing effective immune checkpoint blockade. Sci. Transl. Med. 14, eabj1270. doi:10.1126/scitranslmed.abj1270
Stowell, s. R., Ju, T., and Cummings, R. D. (2015). Protein glycosylation in cancer. Annu. Rev. Pathol. 10, 473–510. doi:10.1146/annurev-pathol-012414-040438
Su, T., Wang, T., Zhang, N., Shen, Y., Li, W., Xing, H., et al. (2022). Long non-coding RNAs in gastrointestinal cancers: implications for protein phosphorylation. Biochem. Pharmacol. 197, 114907. doi:10.1016/j.bcp.2022.114907
Sun, J., Piao, J., Li, N., Yang, Y., Kim, K. Y., and Lin, Z. (2020a). Valproic acid targets HDAC1/2 and HDAC1/PTEN/Akt signalling to inhibit cell proliferation via the induction of autophagy in gastric cancer. FEBS J. 287, 2118–2133. doi:10.1111/febs.15122
Sun, J., Shi, X., Mamun, M. A. A., and Gao, Y. (2020b). The role of deubiquitinating enzymes in gastric cancer. Oncol. Lett. 19, 30–44. doi:10.3892/ol.2019.11062
Sun, L., Zhang, H., and Gao, P. (2022a). Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 13, 877–919. doi:10.1007/s13238-021-00846-7
Sun, R., Kim, A. M. J., and Lim, S. O. (2021). Glycosylation of immune receptors in cancer. Cells 10, 1100. doi:10.3390/cells10051100
Sun, T., Liu, Z., and Yang, Q. (2020c). The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer 19, 146. doi:10.1186/s12943-020-01262-x
Sun, W., Wang, X., Wang, D., Lu, L., Lin, H., Zhang, Z., et al. (2022b). CD40×HER2 bispecific antibody overcomes the CCL2-induced trastuzumab resistance in HER2-positive gastric cancer. J. Immunother. Cancer 10, e005063. doi:10.1136/jitc-2022-005063
Syed, Y. Y. (2017). Lenalidomide: a review in newly diagnosed multiple myeloma as maintenance therapy after asct. Drugs 77, 1473–1480. doi:10.1007/s40265-017-0795-0
Tam, B. Y., Chiu, K., Chung, H., Bossard, C., Nguyen, J. D., Creger, E., et al. (2020). The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett. 473, 186–197. doi:10.1016/j.canlet.2019.09.009
Tan, S., Liang, C. R., Yeoh, K. G., So, J., Hew, C. L., and Chung, M. C. (2007). Gastrointestinal fluids proteomics. Proteomics Clin. Appl. 1, 820–833. doi:10.1002/prca.200700169
Tanaka, M., Kitajima, Y., Edakuni, G., Sato, S., and Miyazaki, K. (2002). Abnormal expression of E-cadherin and beta-catenin may be a molecular marker of submucosal invasion and lymph node metastasis in early gastric cancer. Br. J. Surg. 89, 236–244. doi:10.1046/j.0007-1323.2001.01985.x
Tian, P., Xu, Z., Guo, J., Zhao, J., Chen, W., Huang, W., et al. (2024). Hypoxia causes trophoblast cell ferroptosis to induce miscarriage through lnc-HZ06/HIF1α-SUMO/NCOA4 axis. Redox Biol. 70, 103073. doi:10.1016/j.redox.2024.103073
Tirado-Hurtado, I., Carlos, C., Lancho, L., Alfaro, A., Ponce, R., Schwarz, L. J., et al. (2019). Helicobacter pylori: history and facts in Peru. Crit. Rev. Oncol. Hematol. 134, 22–30. doi:10.1016/j.critrevonc.2018.12.005
Usui, G., Matsusaka, K., Mano, Y., Urabe, M., Funata, S., Fukayama, M., et al. (2021). DNA methylation and genetic aberrations in gastric cancer. Digestion 102, 25–32. doi:10.1159/000511243
Vu, L. D., Gevaert, K., and DE Smet, I. (2018). Protein Language: post-translational modifications talking to each other. Trends Plant Sci. 23, 1068–1080. doi:10.1016/j.tplants.2018.09.004
Wan, M., Gan, A., Dai, J., Lin, F., Wang, R., Wu, B., et al. (2024). Rhein induces apoptosis of AGS and MGC803 cells by regulating the Ras/PI3K/AKT and p38/MAPK signaling pathway. J. Pharm. Pharmacol., rgae115. doi:10.1093/jpp/rgae115
Wang, C., Yang, Z., Xu, E., Shen, X., Wang, X., Li, Z., et al. (2021a). Apolipoprotein C-II induces EMT to promote gastric cancer peritoneal metastasis via PI3K/AKT/mTOR pathway. Clin. Transl. Med. 11, e522. doi:10.1002/ctm2.522
Wang, D., Xu, C., Yang, W., Chen, J., Ou, Y., Guan, Y., et al. (2022a). E3 ligase RNF167 and deubiquitinase STAMBPL1 modulate mTOR and cancer progression. Mol. Cell 82, 770–784 e9. doi:10.1016/j.molcel.2022.01.002
Wang, F., Meng, W., Wang, B., and Qiao, L. (2014). Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 345, 196–202. doi:10.1016/j.canlet.2013.08.016
Wang, F., Zhang, J., Tang, H., Pang, Y., Ke, X., Peng, W., et al. (2022b). Nup54-induced CARM1 nuclear importation promotes gastric cancer cell proliferation and tumorigenesis through transcriptional activation and methylation of Notch2. Oncogene 41, 246–259. doi:10.1038/s41388-021-02078-9
Wang, F. L., Chang, X., Shi, Y., Yang, T., Li, J., Dong, H., et al. (2024a). β-Ionone enhances the inhibitory effects of 5-fluorouracil on the proliferation of gastric adenocarcinoma cells by the GSK-3β signaling pathway. PLoS One 19, e0309014. doi:10.1371/journal.pone.0309014
Wang, G., Xu, Z., Sun, J., Liu, B., Ruan, Y., Gu, J., et al. (2023a). O-GlcNAcylation enhances Reticulon 2 protein stability and its promotive effects on gastric cancer progression. Cell Signal 108, 110718. doi:10.1016/j.cellsig.2023.110718
Wang, H., Breadner, D. A., Deng, K., and Niu, J. (2023b). CircRHOT1 restricts gastric cancer cell ferroptosis by epigenetically regulating GPX4. J. Gastrointest. Oncol. 14, 1715–1725. doi:10.21037/jgo-23-550
Wang, J., Jia, Q., Jiang, S., Lu, W., and Ning, H. (2024b). POU6F1 promotes ferroptosis by increasing lncRNA-CASC2 transcription to regulate SOCS2/SLC7A11 signaling in gastric cancer. Cell Biol. Toxicol. 40, 3. doi:10.1007/s10565-024-09843-y
Wang, J., Liu, Z., Xu, Y., Wang, Y., Wang, F., Zhang, Q., et al. (2022c). Enterobacterial LPS-inducible LINC00152 is regulated by histone lactylation and promotes cancer cells invasion and migration. Front. Cell Infect. Microbiol. 12, 913815. doi:10.3389/fcimb.2022.913815
Wang, J., Wu, J., Wang, L., Min, X., and Chen, Z. (2021b). The linc00152/miR-138 Axis facilitates gastric cancer progression by mediating SIRT2. J. Oncol. 2021, 1173869. doi:10.1155/2021/1173869
Wang, L., Gu, S., Chen, F., Yu, Y., Cao, J., Li, X., et al. (2023c). Imatinib blocks tyrosine phosphorylation of Smad4 and restores TGF-β growth-suppressive signaling in BCR-ABL1-positive leukemia. Signal Transduct. Target Ther. 8, 120. doi:10.1038/s41392-023-01327-5
Wang, L., Zhang, L., Li, S., Cao, L., Li, K., and Zhao, W. (2023d). A novel acetylation-immune subtyping for the identification of a BET inhibitor-sensitive subgroup in melanoma. Pharm. (Basel) 16, 1037. doi:10.3390/ph16071037
Wang, M., Dai, W., Ke, Z., and Li, Y. (2020). Functional roles of E3 ubiquitin ligases in gastric cancer. Oncol. Lett. 20, 22. doi:10.3892/ol.2020.11883
Wang, M., Zhao, H., Chen, W., Bie, C., Yang, J., Cai, W., et al. (2023e). The SMAD2/miR-4256/HDAC5/p16(INK4a) signaling axis contributes to gastric cancer progression. Oncol. Res. 31, 515–541. doi:10.32604/or.2023.029101
Wang, Q., Xu, C., Fan, Q., Yuan, H., Zhang, X., Chen, B., et al. (2021c). Positive feedback between ROS and cis-axis of PIASxα/p38α-SUMOylation/MK2 facilitates gastric cancer metastasis. Cell Death Dis. 12, 986. doi:10.1038/s41419-021-04302-6
Wang, T., Min, L., Gao, Y., Zhao, M., Feng, S., Wang, H., et al. (2023f). SUMOylation of TUFT1 is essential for gastric cancer progression through AKT/mTOR signaling pathway activation. Cancer Sci. 114, 533–545. doi:10.1111/cas.15618
Wang, W., Li, Y., Tang, L., Shi, Y., Li, W., Zou, L., et al. (2024d). Cross-talk between BCKDK-mediated phosphorylation and STUB1-dependent ubiquitination degradation of BCAT1 promotes GBM progression. Cancer Lett. 591, 216849. doi:10.1016/j.canlet.2024.216849
Wang, Y., Hu, J., Wu, S., Fleishman, J. S., Li, Y., Xu, Y., et al. (2023g). Targeting epigenetic and posttranslational modifications regulating ferroptosis for the treatment of diseases. Signal Transduct. Target Ther. 8, 449. doi:10.1038/s41392-023-01720-0
Wang, K., He, Z., Jin, G., Jin, S., DU, Y., Yuan, S., et al. (2024c). Targeting DNA methyltransferases for cancer therapy. Bioorg Chem. 151, 107652. doi:10.1016/j.bioorg.2024.107652
Wawruszak, A., Borkiewicz, L., Okon, E., Kukula-Koch, W., Afshan, S., and Halasa, M. (2021). Vorinostat (SAHA) and breast cancer: an overview. Cancers (Basel) 13, 4700. doi:10.3390/cancers13184700
Wei, M., Huang, X., Liao, L., Tian, Y., and Zheng, X. (2023). SENP1 decreases RNF168 phase separation to promote DNA damage repair and drug resistance in colon cancer. Cancer Res. 83, 2908–2923. doi:10.1158/0008-5472.CAN-22-4017
Wei, X., Liu, J., Cheng, J., Cai, W., Xie, W., Wang, K., et al. (2024). Super-enhancer-driven ZFP36L1 promotes PD-L1 expression in infiltrative gastric cancer. Elife 13. doi:10.7554/eLife.96445
Wen, Y., Wang, K., and Yang, K. (2016). Inhibiting the role of Skp2 suppresses cell proliferation and tumorigenesis of human gastric cancer cells via the upregulation of p27kip1. Mol. Med. Rep. 14, 3917–3924. doi:10.3892/mmr.2016.5676
White, J., Derheimer, F. A., Jensen-Pergakes, K., O'Connell, S., Sharma, S., Spiegel, N., et al. (2024). Histone lysine acetyltransferase inhibitors: an emerging class of drugs for cancer therapy. Trends Pharmacol. Sci. 45, 243–254. doi:10.1016/j.tips.2024.01.010
Wu, D., Qiu, Y., Jiao, Y., Qiu, Z., and Liu, D. (2020). Small molecules targeting HATs, HDACs, and BRDs in cancer therapy. Front. Oncol. 10, 560487. doi:10.3389/fonc.2020.560487
Wu, F., Cao, L., Zhang, J., Cai, S., Wu, H., Miao, J., et al. (2024). FUT3 promotes gastric cancer cell migration by synthesizing Lea on ITGA6 and GLG1, affecting adhesion and vesicle distribution. Life Sci. 359, 123193. doi:10.1016/j.lfs.2024.123193
Wu, J., Ye, F., and Xu, T. (2023a). Celastrol impairs tumor growth by modulating the CIP2A-GSK3β-MCL-1 axis in gastric cancer cells. Aging (Albany NY) 15, 6894–6904. doi:10.18632/aging.204879
Wu, Q., Schapira, M., Arrowsmith, C. H., and Barsyte-Lovejoy, D. (2021a). Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 20, 509–530. doi:10.1038/s41573-021-00159-8
Wu, W. K., Cho, C. H., Lee, C. W., Wu, Y. C., Yu, L., Li, Z. J., et al. (2010). Macroautophagy and ERK phosphorylation counteract the antiproliferative effect of proteasome inhibitor in gastric cancer cells. Autophagy 6, 228–238. doi:10.4161/auto.6.2.11042
Wu, X., Li, J. H., Xu, L., Li, Y. X., Zhu, X. X., Wang, X. Y., et al. (2023b). SUMO specific peptidase 3 halts pancreatic ductal adenocarcinoma metastasis via deSUMOylating DKC1. Cell Death Differ. 30, 1742–1756. doi:10.1038/s41418-023-01175-4
Wu, X., Xu, M., Geng, M., Chen, S., Little, P. J., Xu, S., et al. (2023c). Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies. Signal Transduct. Target Ther. 8, 220. doi:10.1038/s41392-023-01439-y
Wu, Z., Ding, Z., Cheng, B., and Cui, Z. (2021b). The inhibitory effect of human DEFA5 in growth of gastric cancer by targeting BMI1. Cancer Sci. 112, 1075–1083. doi:10.1111/cas.14827
Wu, Z., Huang, R., and Yuan, L. (2019). Crosstalk of intracellular post-translational modifications in cancer. Arch. Biochem. Biophys. 676, 108138. doi:10.1016/j.abb.2019.108138
Xiang, T., Yuan, C., Guo, X., Wang, H., Cai, Q., Xiang, Y., et al. (2021). The novel ZEB1-upregulated protein PRTG induced by Helicobacter pylori infection promotes gastric carcinogenesis through the cGMP/PKG signaling pathway. Cell Death Dis. 12, 150. doi:10.1038/s41419-021-03440-1
Xiao, L., Zhang, Y., Luo, Q., Guo, C., Chen, Z., and Lai, C. (2023). DHRS4-AS1 regulate gastric cancer apoptosis and cell proliferation by destabilizing DHX9 and inhibited the association between DHX9 and ILF3. Cancer Cell Int. 23, 304. doi:10.1186/s12935-023-03151-x
Xie, B., Lin, J., Chen, X., Zhou, X., Zhang, Y., Fan, M., et al. (2023). CircXRN2 suppresses tumor progression driven by histone lactylation through activating the Hippo pathway in human bladder cancer. Mol. Cancer 22, 151. doi:10.1186/s12943-023-01856-1
Xie, M., Yu, J., Ge, S., Huang, J., and Fan, X. (2020). SUMOylation homeostasis in tumorigenesis. Cancer Lett. 469, 301–309. doi:10.1016/j.canlet.2019.11.004
Xiong, J. X., Li, Y. T., Tan, X. Y., Chen, T., Liu, B. H., and Fu, L. (2024). Targeting PRSS23 with tipranavir induces gastric cancer stem cell apoptosis and inhibits growth of gastric cancer via the MKK3/p38 MAPK-IL24 pathway. Acta Pharmacol. Sin. 45, 405–421. doi:10.1038/s41401-023-01165-9
Xu, L., Xiang, W., Yang, J., Gao, J., Wang, X., Meng, L., et al. (2024). PHB2 promotes SHIP2 ubiquitination via the E3 ligase NEDD4 to regulate AKT signaling in gastric cancer. J. Exp. Clin. Cancer Res. 43, 17. doi:10.1186/s13046-023-02937-1
Xu, P., Lin, X., and Feng, X. H. (2016). Posttranslational regulation of smads. Cold Spring Harb. Perspect. Biol. 8, a022087. doi:10.1101/cshperspect.a022087
Xu, W., Wang, S., Chen, Q., Zhang, Y., Ni, P., Wu, X., et al. (2014). TXNL1-XRCC1 pathway regulates cisplatin-induced cell death and contributes to resistance in human gastric cancer. Cell Death Dis. 5, e1055. doi:10.1038/cddis.2014.27
Xu, Y., and Wan, W. (2023). Acetylation in the regulation of autophagy. Autophagy 19, 379–387. doi:10.1080/15548627.2022.2062112
Yang, B., Wang, J. Q., Tan, Y., Yuan, R., Chen, Z. S., and Zou, C. (2021a). RNA methylation and cancer treatment. Pharmacol. Res. 174, 105937. doi:10.1016/j.phrs.2021.105937
Yang, H., Zou, X., Yang, S., Zhang, A., Li, N., and Ma, Z. (2023a). Identification of lactylation related model to predict prognostic, tumor infiltrating immunocytes and response of immunotherapy in gastric cancer. Front. Immunol. 14, 1149989. doi:10.3389/fimmu.2023.1149989
Yang, W., Cui, X., Sun, D., Sun, G., Yan, Z., Wei, M., et al. (2023b). POU5F1 promotes the proliferation, migration, and invasion of gastric cancer cells by reducing the ubiquitination level of TRAF6. Cell Death Dis. 14, 802. doi:10.1038/s41419-023-06332-8
Yang, W., Wang, P., Cao, P., Wang, S., Yang, Y., Su, H., et al. (2021b). Hypoxic in vitro culture reduces histone lactylation and impairs pre-implantation embryonic development in mice. Epigenetics Chromatin 14, 57. doi:10.1186/s13072-021-00431-6
Yang, Z., Jiang, X., Zhang, Z., Zhao, Z., Xing, W., Liu, Y., et al. (2021c). HDAC3-dependent transcriptional repression of FOXA2 regulates FTO/m6A/MYC signaling to contribute to the development of gastric cancer. Cancer Gene Ther. 28, 141–155. doi:10.1038/s41417-020-0193-8
Yu, G., Chen, W., Li, X., Yu, L., Xu, Y., Ruan, Q., et al. (2022). TWIST1-EP300 expedites gastric cancer cell resistance to apatinib by activating the expression of COL1A2. Anal. Cell Pathol. (Amst) 2022, 5374262. doi:10.1155/2022/5374262
Yu, L., Li, Y., Song, S., Zhang, Y., Wang, Y., Wang, H., et al. (2024a). The dual role of sirtuins in cancer: biological functions and implications. Front. Oncol. 14, 1384928. doi:10.3389/fonc.2024.1384928
Yu, X., Yang, J., Xu, J., Pan, H., Wang, W., Yu, X., et al. (2024b). Histone lactylation: from tumor lactate metabolism to epigenetic regulation. Int. J. Biol. Sci. 20, 1833–1854. doi:10.7150/ijbs.91492
Yuan, J., Dong, X., Yap, J., and Hu, J. (2020). The MAPK and AMPK signalings: interplay and implication in targeted cancer therapy. J. Hematol. Oncol. 13, 113. doi:10.1186/s13045-020-00949-4
Yuan, Y., Zhang, X., DU, K., Zhu, X., Chang, S., Chen, Y., et al. (2022). Circ_CEA promotes the interaction between the p53 and cyclin-dependent kinases 1 as a scaffold to inhibit the apoptosis of gastric cancer. Cell Death Dis. 13, 827. doi:10.1038/s41419-022-05254-1
Yue, F., Peng, K., Zhang, L., and Zhang, J. (2021). Circ_0004104 accelerates the progression of gastric cancer by regulating the miR-539-3p/RNF2 Axis. Dig. Dis. Sci. 66, 4290–4301. doi:10.1007/s10620-020-06802-5
Yun, R., Hong, E., Kim, J., Park, B., Kim, S. J., Lee, B., et al. (2023). N-linked glycosylation is essential for anti-tumor activities of KIAA1324 in gastric cancer. Cell Death Dis. 14, 546. doi:10.1038/s41419-023-06083-6
Zaib, S., Rana, N., and Khan, I. (2022). Histone modifications and their role in epigenetics of cancer. Curr. Med. Chem. 29, 2399–2411. doi:10.2174/0929867328666211108105214
Zang, W. J., Hu, Y. L., Qian, C. Y., Feng, Y., Liu, J. Z., Yang, J. L., et al. (2022). HDAC4 promotes the growth and metastasis of gastric cancer via autophagic degradation of MEKK3. Br. J. Cancer 127, 237–248. doi:10.1038/s41416-022-01805-7
Zeng, X. Q., Wang, J., and Chen, S. Y. (2017). Methylation modification in gastric cancer and approaches to targeted epigenetic therapy (Review). Int. J. Oncol. 50, 1921–1933. doi:10.3892/ijo.2017.3981
Zha, J., Zhang, J., Lu, J., Zhang, G., Hua, M., Guo, W., et al. (2024). A review of lactate-lactylation in malignancy: its potential in immunotherapy. Front. Immunol. 15, 1384948. doi:10.3389/fimmu.2024.1384948
Zhang, C., and Huang, Z. (2024). KAT2A promotes the succinylation of PKM2 to inhibit its activity and accelerate glycolysis of gastric cancer. Mol. Biotechnol. 66, 1446–1457. doi:10.1007/s12033-023-00778-z
Zhang, C., Wei, S., Dai, S., Li, X., Wang, H., Zhang, H., et al. (2023a). The NR_109/FUBP1/c-Myc axis regulates TAM polarization and remodels the tumor microenvironment to promote cancer development. J. Immunother. Cancer 11, e006230. doi:10.1136/jitc-2022-006230
Zhang, D., Tang, Z., Huang, H., Zhou, G., Cui, C., Weng, Y., et al. (2019a). Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580. doi:10.1038/s41586-019-1678-1
Zhang, F. L., Yang, S. Y., Liao, L., Zhang, T. M., Zhang, Y. L., Hu, S. Y., et al. (2023b). Dynamic SUMOylation of MORC2 orchestrates chromatin remodelling and DNA repair in response to DNA damage and drives chemoresistance in breast cancer. Theranostics 13, 973–990. doi:10.7150/thno.79688
Zhang, G., Dong, J., Lu, L., Liu, Y., Hu, D., Wu, Y., et al. (2023c). Acacetin exerts antitumor effects on gastric cancer by targeting EGFR. Front. Pharmacol. 14, 1121643. doi:10.3389/fphar.2023.1121643
Zhang, G., Yang, R., Wang, B., Yan, Q., Zhao, P., Zhang, J., et al. (2024). TRIP13 regulates progression of gastric cancer through stabilising the expression of DDX21. Cell Death Dis. 15, 622. doi:10.1038/s41419-024-07012-x
Zhang, H., Li, L., Yuan, C., Wang, C., Gao, T., and Zheng, Z. (2020). MiR-489 inhibited the development of gastric cancer via regulating HDAC7 and PI3K/AKT pathway. World J. Surg. Oncol. 18, 73. doi:10.1186/s12957-020-01846-3
Zhang, J., Liu, X., Chen, J., and Xia, S. (2021a). HDAC3-Mediated repression of LncRNA-LET regulates gastric cancer cell growth proliferation, invasion, migration, and apoptosis via MiR-548k. J. Environ. Pathol. Toxicol. Oncol. 40, 21–32. doi:10.1615/JEnvironPatholToxicolOncol.2021039050
Zhang, L., Liu, F., Meng, Z., Luo, Q., Pan, D., and Qian, Y. (2021b). Inhibited HDAC3 promotes microRNA-376c-3p to suppress malignant phenotypes of gastric cancer cells by reducing WNT2b. Genomics 113, 3512–3522. doi:10.1016/j.ygeno.2021.07.018
Zhang, L., Wang, N., Chen, M., Wu, S., Zeng, J., Zhou, F., et al. (2022). HDAC6/FOXP3/HNF4α axis promotes bile acids induced gastric intestinal metaplasia. Am. J. Cancer Res. 12, 1409–1422.
Zhang, M., Jiang, D., Xie, X., He, Y., Lv, M., and Jiang, X. (2019b). miR-129-3p inhibits NHEJ pathway by targeting SAE1 and represses gastric cancer progression. Int. J. Clin. Exp. Pathol. 12, 1539–1547.
Zhang, N., Gao, X., Yuan, Q., Fu, X., Wang, P., Cai, F., et al. (2023d). E3 ubiquitin ligase RNF180 prevents excessive PCDH10 methylation to suppress the proliferation and metastasis of gastric cancer cells by promoting ubiquitination of DNMT1. Clin. Epigenetics 15, 77. doi:10.1186/s13148-023-01492-y
Zhang, Q., Xiao, M., Gu, S., Xu, Y., Liu, T., Li, H., et al. (2019c). ALK phosphorylates SMAD4 on tyrosine to disable TGF-β tumour suppressor functions. Nat. Cell Biol. 21, 179–189. doi:10.1038/s41556-018-0264-3
Zhang, W. J., Zhou, Y., Zhang, Y., Su, Y. H., and Xu, T. (2023e). Protein phosphorylation: a molecular switch in plant signaling. Cell Rep. 42, 112729. doi:10.1016/j.celrep.2023.112729
Zhang, X. W., Sheng, Y. P., Li, Q., Qin, W., Lu, Y. W., Cheng, Y. F., et al. (2010). BMI1 and Mel-18 oppositely regulate carcinogenesis and progression of gastric cancer. Mol. Cancer 9, 40. doi:10.1186/1476-4598-9-40
Zhang, Z., Peng, L., Yang, W., Li, B., Hua, Y., and Luo, S. (2023f). PHF5A facilitates the development and progression of gastric cancer through SKP2-mediated stabilization of FOS. J. Transl. Med. 21, 5. doi:10.1186/s12967-022-03821-w
Zhao, B., Wu, J., Cha, X., Mao, G., Shi, H., Fei, S., et al. (2023a). Effect of COP1 in promoting the tumorigenesis of gastric cancer by down-regulation of CDH18 via PI3K/AKT signal pathway. Anal. Cell Pathol. (Amst) 2023, 5617875. doi:10.1155/2023/5617875
Zhao, C., Miao, J., Sun, R., Liang, R., Chen, W., Gao, Y., et al. (2022). MBD1/HDAC3-miR-5701-FGFR2 axis promotes the development of gastric cancer. Aging (Albany NY) 14, 5878–5894. doi:10.18632/aging.204190
Zhao, H., Ding, Y., and Zhang, L. (2023b). SIRT1/APE1 promotes the viability of gastric cancer cells by inhibiting p53 to suppress ferroptosis. Open Med. (Wars) 18, 20220620. doi:10.1515/med-2022-0620
Zhao, Y., Jiang, J., Zhou, P., Deng, K., Liu, Z., Yang, M., et al. (2024). H3K18 lactylation-mediated VCAM1 expression promotes gastric cancer progression and metastasis via AKT-mTOR-CXCL1 axis. Biochem. Pharmacol. 222, 116120. doi:10.1016/j.bcp.2024.116120
Zhao, Y., Nakagawa, T., Itoh, S., Inamori, K., Isaji, T., Kariya, Y., et al. (2006). N-acetylglucosaminyltransferase III antagonizes the effect of N-acetylglucosaminyltransferase V on alpha3beta1 integrin-mediated cell migration. J. Biol. Chem. 281, 32122–32130. doi:10.1074/jbc.M607274200
Zhao, Y., Yu, T., Zhang, N., Chen, J., Zhang, P., Li, S., et al. (2019). Nuclear E-cadherin acetylation promotes colorectal tumorigenesis via enhancing β-catenin activity. Mol. Cancer Res. 17, 655–665. doi:10.1158/1541-7786.MCR-18-0637
Zhao, Y. Q., Jin, H. R., Kim, D., Jung, S. H., Liu, S., Wan, J., et al. (2023c). SUMO1 degrader induces ER stress and ROS accumulation through deSUMOylation of TCF4 and inhibition of its transcription of StarD7 in colon cancer. Mol. Carcinog. 62, 1249–1262. doi:10.1002/mc.23560
Zhong, Q., Wang, H. G., Yang, J. H., Tu, R. H., Li, A. Y., Zeng, G. R., et al. (2023). Loss of ATOH1 in pit cell drives stemness and progression of gastric adenocarcinoma by activating AKT/mTOR signaling through GAS1. Adv Sci. (Weinh) 10, e2301977. doi:10.1002/advs.202301977
Zhou, B., Zhu, Y., Xu, W., Zhou, Q., Tan, L., Zhu, L., et al. (2021). Hypoxia stimulates SUMOylation-dependent stabilization of KDM5B. Front. Cell Dev. Biol. 9, 741736. doi:10.3389/fcell.2021.741736
Zhou, L., Wang, H., Zhong, M., Fang, Z., LE, Y., Nie, F., et al. (2022). The E3 ubiquitin ligase TRIM11 facilitates gastric cancer progression by activating the wnt/β-catenin pathway via destabilizing Axin1 protein. J. Oncol. 2022, 8264059. doi:10.1155/2022/8264059
Zhu, Y., Zhou, B., Hu, X., Ying, S., Zhou, Q., Xu, W., et al. (2022). LncRNA LINC00942 promotes chemoresistance in gastric cancer by suppressing MSI2 degradation to enhance c-Myc mRNA stability. Clin. Transl. Med. 12, e703. doi:10.1002/ctm2.703
Zinzani, P. L., Izutsu, K., Mehta-Shah, N., Barta, S. K., Ishitsuka, K., Cordoba, R., et al. (2024). Valemetostat for patients with relapsed or refractory peripheral T-cell lymphoma (VALENTINE-PTCL01): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 25, 1602–1613. doi:10.1016/S1470-2045(24)00503-5
Keywords: gastric cancer, ubiquitination, phosphorylation, acetylation, glycosylation, methylation, lactylation, SUMOylation
Citation: Song H, Zhang M, Guo C, Guo X, Ma Y and Ma Y (2025) Implication of protein post translational modifications in gastric cancer. Front. Cell Dev. Biol. 13:1523958. doi: 10.3389/fcell.2025.1523958
Received: 06 November 2024; Accepted: 10 January 2025;
Published: 04 February 2025.
Edited by:
Francesco Esposito, National Research Council (CNR), ItalyReviewed by:
Zaid Altaany, Yarmouk University, JordanCopyright © 2025 Song, Zhang, Guo, Guo, Ma and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuntao Ma, MzU3NTUxNTY2NUBxcS5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.