- 1College of Fisheries, Huazhong Agricultural University, Wuhan, Hubei, China
- 2Engineering Research Center of Green development for Conventional Aquatic Biological Industry in the Yangtze River Economic Belt, Ministry of Education, Wuhan, China
- 3Hubei Hongshan Laboratory, Fishery College, Huazhong Agricultural University, Wuhan, China
Introduction: Learning and memory allow individuals to adapt to their environmental needs and survive. Fish have the ability to solve complex learning tasks, associative learning, and flexible spatial memory. The proto-oncogene fos (c-fos) has been reported to be involved in brain development, learning and memory in mammals. However, whether the c-fos plays a vital role in learning and memory in fish is unclear.
Methods: Almost all fish have two paralogues of c-fos named fosaa and fosab. We used CRISPR/Cas9 technology to generate fosaa and fosab knockout zebrafish models.
Results: In this study, we discovered the brain weight marked reduction in fosaa−/− and fosab−/− zebrafish compared with the wild-type (WT) (AB strain) zebrafish. In the T-maze behavioral assay, the fosab−/− zebrafish took significantly more than the average time to complete the assigned trial as the increase in the days compared to WT zebrafish, while the fosaa−/− zebrafish did not show a notable variance. The average time to complete the trial in fosab−/− zebrafish was significantly higher than in WT zebrafish. The relative mRNA expression level of c-jun in fosab−/− zebrafish was significantly higher than that in WT zebrafish, while the fosaa−/− zebrafish has no discernible trend. Additionally, the phylogenetic and multiple amino acid alignment results indicated that fish fosab has a higher identity with mammals Fos.
Discussion: By integrating the above results, we found that fosab, but not fosaa, may possess a learning and memory function in fish. For the first time, we illustrated the role of fosaa and fosab in learning and memory via c-fos knockout in fish, which can provide new insights into environmental adaptation.
1 Introduction
Learning and memory are essential to organism survival and are conserved across various species, especially vertebrates (Tan et al., 2022). Mammals are able to continually learn and acquire skills to respond to the demands of a changing environment (Barron, 2021). Proto-oncogene fos (c-fos), an immediate early response gene, provides an essential function in the regulatory component of the nuclear response to mitogens and other extracellular stimuli and is a candidate gene for the role of an adaptive regulator (Curran and Morgan, 1987; Randall et al., 1992). Adaptive regulators are proteins that modulate the expression of genes involved in the long-term response, while learning is one of the most representative examples of long-term alteration (Curran and Morgan, 1987). The c-fos gene is widely used as a functional marker of brain activity in terms of neuroscience (Ariyasiri et al., 2021; Cai et al., 2022; Kubra et al., 2022; Morris et al., 2000). Emerging evidence suggested c-fos mutant could impair neuronal excitability, brain development and long-term learning and memory in mice (Fleischmann et al., 2003; Kim et al., 2003; Paylor et al., 1994; Velazquez et al., 2015; Zhang et al., 2002). Previous studies have demonstrated that the dorsolateral telencephalon of fish could be homologous to the mammalian hippocampus sharing functional similarities (Gómez et al., 2020; Naderi et al., 2016). Fish also have the ability to solve complex learning tasks. However, there is a paucity of information on whether c-fos plays a vital role in learning and memory in fish.
Zebrafish are a valuable, useful and promising vertebrate model organism to study a wide range of biological disciplines, including neuroscience on account of their short reproductive cycle, rapid external development, high genetic homology to humans, and various tools available to investigate (de Abreu et al., 2019; Soussi-Yanicostas, 2022). The National Center for Biotechnology Information (NCBI)’s research and literature showed that almost all fish have two paralogues of c-fos, named fosaa and fosab, individually. The fosaa and fosab genes are highly expressed in the brain of zebrafish (Kubra et al., 2022; Maili et al., 2023). Thus, we identified and performed an evolutionary analysis of the c-fos gene from different vertebrates and used zebrafish to explore if the function of learning and memory is regulated by the c-fos (fosaa and fosab) gene in fish as it is in mammals.
Behavioral assays are broadly applied to evaluate the associative learning and memory abilities of mice and zebrafish, with the T-maze test being one of the most commonly employed methods (Bicakci et al., 2021; Kim J. et al., 2019; Kim Y.-H. et al., 2019; Song et al., 2022; Yoo et al., 2021b). Zebrafish also showed a color preference that could be targeted when designing color-based learning and memory experiments involving fear, anxiety, or reward in the zebrafish (Avdesh et al., 2012). Based on this, we assessed the function of the fosaa and fosab genes in learning and memory by combining color and food rewards to evaluate associative learning abilities in zebrafish between different groups.
The cholinergic system plays an important role in regulating learning and memory in mammals (Haam and Yakel, 2017). Cholinergic transmission is involved in memory formation and consolidation, even over long periods (Oda, 2010; Phillis, 2005). Acetylcholinesterase (AChE) participates at the end of cholinergic transmission based on the breakdown of acetylcholine in choline and acetic acid, and acetylcholine transferase (AChT) is involved in the production of acetylcholine (Ichikawa et al., 1997). Inhibition of AChE activity and upregulation of AChT activity will enhance cholinergic neurotransmission, which can improve impaired memory in mice (He et al., 2015; Nogami-Hara et al., 2018). The hyperglycemia promoted memory deficit in adult zebrafish, which results from increased AChE activity (Katiucia et al., 2014). Fos-immunoreactive cells were identified as neurons, predominantly clustered in areas corresponding to the regions exhibiting high acetylcholinesterase (AChE) activity (Liu et al., 1991). Whether the fosaa and fosab affect learning and memory through the cholinergic system in zebrafish is unclear.
Brain-derived neurotrophic factor (BDNF), a neurotrophin, promotes neuronal survival, differentiation, and growth (Ismail et al., 2021). BDNF is one of the upstream signals for c-fos expression and positively related to long-term memory formation and memory in rats (Bekinschtein et al., 2014; Yoo et al., 2021a). C-JUN, a component of the AP-1 (Activator Protein 1) transcription factor, regulates cell proliferation, differentiation, survival, and responses to stress and injury (Ganguly et al., 2013). Investigating how the bdnf and c-jun expression is altered in the fosaa and fosab knockout zebrafish offers valuable insights into the molecular mechanisms underlying learning and memory.
In the present study, we identified the c-fos genes and predicted phylogenetic and evolutionary relationships in several vertebrates. To investigate whether c-fos has the function of learning and memory, we generated fosaa and fosab knockout zebrafish models using CRISPR/Cas9 technology. Behavioral experiments, morphology of brain observation, and cholinergic system detection were utilized to evaluate the function. In addition, the mRNA expression of brain derived neurotrophic factor (bdnf) and jun proto-oncogene (c-jun) was measured.
2 Materials and methods
2.1 Sequence alignment and phylogenetic analysis of c-fos genes
c-fos gene sequences from various vertebrate species were retrieved from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/). Species included were: human (Homo sapiens), mouse (Mus musculus), chimpanzee (Pan troglodytes), chicken (Gallus gallus), pig (Sus scrofa), common frog (Rana temporaria), tropical clawed frog (Xenopus tropicalis), zebra finch (Taeniopygia guttata), rabbit (Oryctolagus cuniculus), vaquita (Phocoena sinus), West African lungfish (Protopterus annectens), gray bichir (Polypterus senegalus), coelacanth (Latimeria chalumnae), zebrafish (Danio rerio), medaka (Oryzias latipes), mummichog (Fundulus heteroclitus), Klamath speckled dace (Rhinichthys klamathensis goyatoka), large yellow croaker (Larimichthys crocea), oriental weatherfish (Misgurnus anguillicaudatus), minute mudskipper (Periophthalmus magnuspinnatus), channel catfish (Ictalurus punctatus), chum salmon (Oncorhynchus keta), Wuchang bream (Megalobrama amblycephala), snailfish (Pseudoliparis swirei), yellowtail (Seriola aureovittata), smalleye Pacific opah (Lampris incognitus), ringed pipefish (Dunckerocampus dactyliophorus), North African catfish (Clarias gariepinus), mandarinfish (Synchiropus splendidus), striped catfish (Pangasianodon hypophthalmus), European seabass (Dicentrarchus labrax), Asian seabass (Lates calcarifer), Gulf pipefish (Syngnathus scovelli), silver crucian carp (Carassius gibelio), dwarf seahorse (Hippocampus zosterae), grass carp (Ctenopharyngodon idella), southern bluefin tuna (Thunnus maccoyii), and swordfish (Xiphias gladius). Accession numbers of sequences are listed in Supplementary Table S1. Amino acid sequences of Fos were aligned using Clustal X, and phylogenetic analysis was performed with MEGA X. A neighbor-joining tree was constructed using the JTT model and evaluated through 1,000 bootstrap replicates.
2.2 Zebrafish maintenance and treatment
Adult wild-type (AB strain) zebrafish were purchased from the National Zebrafish Resource Center, Institute of Hydrobiology, Chinese Academy of Sciences (http://www.zfish.cn/). Wild-type and knockout zebrafish were maintained in the Chinses perch Center of Huazhong University with a recirculating water system (Meifengbishui Environmental Technology, China, Wuhan), the water at 26°C–28°C, the pH at 6.5–7.5, the dissolved oxygen at least 6.0 mg/L, and conductivity was 400–600 μs/cm. The fish were also given a 14-hour light/10-hour dark cycle. The recirculating water system was equipped with a sediment filter, post-carbon filter, fluorescent UV light, and sterilizing filter. Artemia was used to feed both the Larvae following near depletion of the yolk sac and adult zebrafish twice daily. The adult zebrafish were kept in tanks measuring 10 cm × 26 cm × 15 cm at a density 6 fish/L. All experimental procedures of zebrafish were approved by the Institutional Animal Care and Use Ethics Committee of Huazhong Agricultural University (Approval Number: HZAUFI-2020-0032).
2.3 Construction of zebrafish fosaa and fosab mutants by CRISPR/Cas9 technology
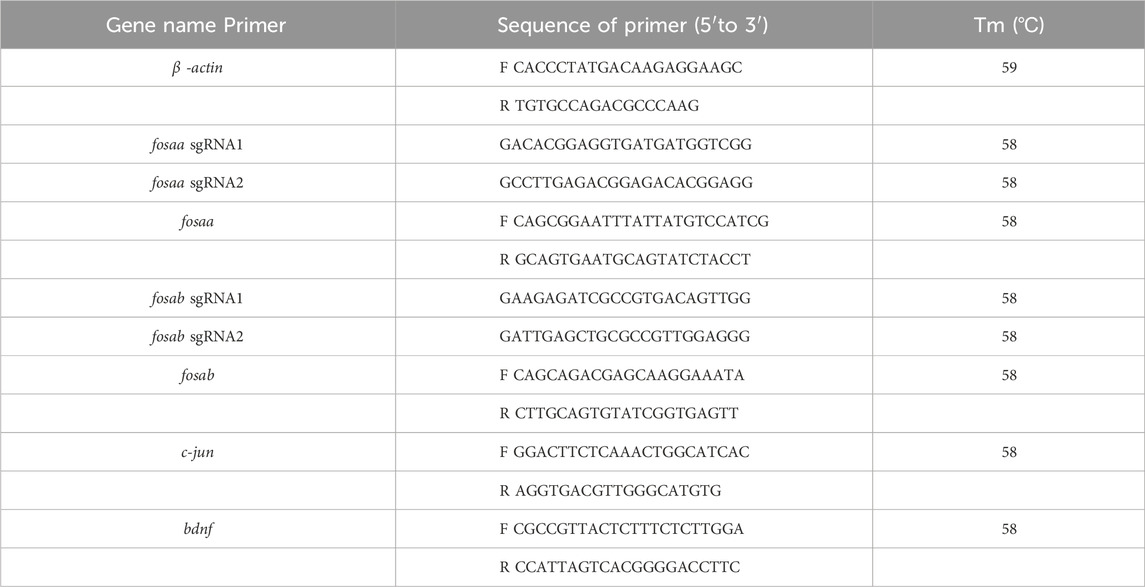
The wild-type (WT) zebrafish of AB strain was used to produce mutant lines. CRISPR/Cas9 technology was employed to generate fosaa and fosab zebrafish mutants. Single-guide RNAs (sgRNAs) were designed using the CCTOP tool (https://cctop.cos.uni-heidelberg.de/), and the sequences of the sgRNAs and PCR primers are listed in Table 1. sgRNAs were cloned into the pMD-18T vector and synthesized using the TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific, Waltham, MA, United States). The night before microinjection, the male and female of the zebrafish were paired to produce 3-5 parents with well development and stable spawning. Female and male fish were separated by a partition, the partition was removed before microinjection on the second day, and the embryos were collected for 15 min after the parents had finished laying eggs. A mixture of sgRNA (50 ng/μL) and Cas9 protein (New England Biolabs, Ipswich, MA, United States) was co-injected into wild-type embryos at the 1–2 cell stage. The F0 generation was crossed with wild-type fish to produce F1 offspring. F1 heterozygotes zebrafish of fosaa and fosab were named fos+/− and fosab+/−. F1 heterozygotes were then crossed to generate F2 homozygous mutants, and all experiments were conducted using F3 homozygous zebrafish. The same WT zebrafish stock was bred in parallel to generate the +/+ animals. fosaa and fosab mutants of zebrafish were named fosaa−/− and fosab−/− zebrafish. All of the experiments were conducted on the basis of known zebrafish genotypes in the study.
2.4 Growth performance and brain weight assay
Wild-type (WT), fosaa−/−, and fosab−/− zebrafish were maintained at identical stocking densities and culture conditions. Length and weight were measured at 273 dpf (day post-fertilization) for each group. Six zebrafish of each genotype and theirs corresponding WT zebrafish were anesthetized with 80 mg/L MS-222 prior to measurement, placed on clean gauze to absorb surface water to measure length and weight. Then, the same fish were executed by decapitation, and the brains were carefully dissected, weighed, and preserved in formaldehyde fixative.
2.5 T-maze test
The T-mazed assay was conducted based on the protocol by (Colwill et al., 2005; Pilehvar et al., 2020), with modifications in apparatus size. The test tank consisted of a T-shaped transparent maze equipped with two removable color sleeves (green and red). A start box, measuring 10 cm × 10 cm × 10 cm, was located at the base of the maze and could be sealed off. Fish were fed only during the daily training sessions. The test spanned 25 days and comprised three phases: (ⅰ) Pre-training (days 1–2): a period for zebrafish to adapt with conditioning of the T-maze without the colour stimulus. They were given 2 min in the start box to acclimate. Once the fish exited, the start box door was closed, and Artemia was manually added as a reward upon entering the open arm. The delay interval prior to reward delivery following correct performance was approximately 3–5 s. The T-maze was rinsed and refilled after each trial to remove residual odors. The total time to and complete the trial were recorded using timer manually for each trial. (ⅱ) Discrimination (days 1–16): a period of 16 days for zebrafish taking choice between the two coloured arms (red and green) of T-maze. Each consisting of four trials with 30-second intervals. The position of colored sleeves alternated (RGGRGRRG). Correct choices were rewarded with food, while incorrect choices triggered a correction procedure where the incorrect arm was blocked, forcing the fish to choose the correct arm. Half of the fish (n = 6) were rewarded for choosing red, the other half for green. Time to leave the start box and enter an arm, as well as the colour of the arm chosen was recorded using timer manually in discrimination trials. (ⅲ) Extinction (days 1–7): a period of 7 days for zebrafish performing the same procedure as the discrimination training with two exceptions: no food reward and no correction rounds were given. The T-maze test was carried out at 280 dpf of each zebrafish group, and the behavioral experiments performed in same room as they were housed at 9:00 a.m. each day. The experimenters maintained a distance of 0.5 m from the fish during the experiment. A total of 48 zebrafish, including fosaa−/− zebrafish (n = 12), fosab−/− zebrafish (n = 12), and their corresponding WT zebrafish (n = 12), were tested, with food provided only during training sessions. The recorded time in three training phases was analyzed by SPSS 19.0 software Levene’s test or Kruskal-Wallis test.
2.6 Estimation of acetylcholinesterase (AChE) acetylcholine and acetylcholine transferase (AChT) activity in the brain of zebrafish
Three zebrafish of each genotype, and along with their corresponding WT zebrafish, were anesthetized with 80 mg/L MS-222 prior to decapitation. Subsequently, the brains were excised and placed in 1.5 mL tubes. The tissues were weighed, and saline was added at a ratio of 9:1 (volume: weight). Brains were mechanically homogenized in an ice-water bath at 2,500 r/min, followed by centrifugation for 10 min. The supernatant was collected to measure AChE and AChT activity using specific reagent kits (AChE, catalog no. A024-1-1; AChT, catalog no. A079-1) from Jian-cheng Institute of Biotechnology (Nanjing, China), following the manufacturer’s instructions. AChE activity was measured by quantifying the yellow product, TNB (Symmetric Trinitrobenzene), formed when AChE hydrolyzes acetylcholine. The absorbance was measured spectrophotometrically at 420 nm. AChT activity was determined using acetyl coenzyme A and choline as substrates, and the absorbance of the reaction products was measured at 324 nm.
2.7 RNA isolation and RT-PCR
Six zebrafish of each genotype, along with their corresponding WT zebrafish, were anesthetized with 80 mg/L MS-222 prior to decapitation. The brains were excised and placed in 1.5 mL tubes, and then stored at −80°C refrigerator for subsequent processing. Total RNA was isolated from zebrafish brain using the Trizol Reagent (TaKaRa, Tokyo, Japan) as per the manufacturer’s instructions and stored at −80°C. RNA purity was assessed using a BioTek Synergy™2 Multi-detection Microplate Reader (BioTek Instruments, Winooski, VT, United States) and agarose gel electrophoresis. cDNA synthesis was carried out using the HiScript II Q RT SuperMix reverse transcriptase (Vazyme, Piscataway, NJ, United States). Gene-specific primers for mandarin fish were designed with Primer 5.0 software and synthesized by Sangon (Shanghai, China). The sequences are listed in Table 1. β-actin was used as the housekeeping gene due to its stability (Zhang et al., 2016). Real-time qPCR was performed using a quantitative thermal cycler (BIO-RAD, United States) with a 20 μL reaction volume consisting of 10 μL AceQ qPCR SYBR Green Master Mix (Vazyme, United States), 8.2 μL double-distilled water, 1 μL template (cDNA), and 0.4 μL each of forward and reverse primers. PCR conditions were 95°C for 5 min, followed by 40 cycles at 95°C for 10 s, annealing at Tm for 30 s, and a melt curve step with acquisition every 6 s. Gene expression was calculated relative to β-actin using the comparative Ct (2−ΔΔCt) method. Results were expressed as mean ± SEM (n = 6).
2.8 Data analysis
Statistical analyses were performed using SPSS 19.0 software. Data were expressed as mean ± SEM. Homogeneity of variance was assessed using Levene’s test. When normality and equal variance between sample groups was achieved, the data were assessed using one-way ANOVA, and differences between group means were compared by Duncan’s post-hoc multiple comparison. When normality failed, Kruskal-Wallis test was performed, and followed by Dunn’s multiple comparisons test. Statistical significance was set at P < 0.05.
3 Results
3.1 Synteny analysis, sequence alignment and phylogenetic analysis of c-fos gens
We found that c-fos has no paralogues among the mammals, birds, reptiles and amphibians through searching in NCBI. However, almost all of the fish have two paralogues named fosaa and fosab except individual original fish such as the West African lungfish (P. annectens), Gray bichir (P. senegalus) and coelacanth (L. chalumnae). The flanking genes of c-fos were conserved in mammals, birds, reptiles, and amphibians, while the flanking genes of fosaa and fosab exist with deletions or other gene additions in the majority of fish (Figure 1). The multiple sequence alignments of vertebrate Fos indicated that amino acids are not completely conserved, and some relatively conserved amino acid sites altered mainly in fish (Figure 2). To investigate the evolutionary relationships of Fos among the vertebrates, we constructed the phylogenetic trees using amino sequences. The results showed that the Fos protein diverged into two groups, which the mammals, birds, reptiles and amphibians were grouped together with the fosab protein of fish, and the fosaa protein of fish stands alone as a group. The Fos protein of mammals is increasingly distant from that of birds, reptiles, amphibians, and fish in kinship ordering, which could be used as identity genes in evolution. Besides, the fish species with one paralogue named fosab have a higher amino acid sequence similarity to Fos in mammals and amphibians (Figure 3).
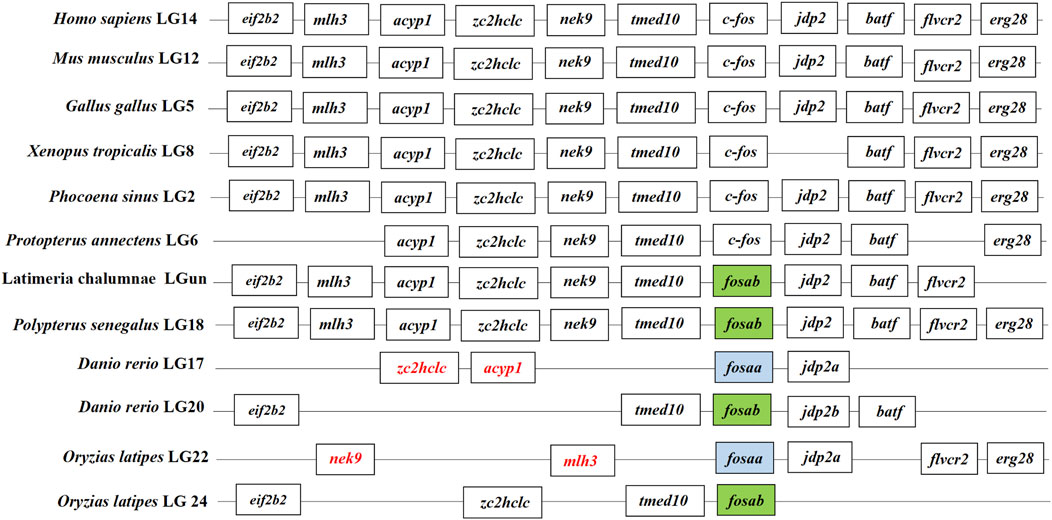
Figure 1. Synteny analysis of c-fos genes. The synteny analysis was performed by searching genes flanking c-fos in genomes of human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), tropical clawed frog (Xenopus tropicalis), Vaquita (Phocoena sinus), West African lungfish (Protopterus annectens), Gray bichir (Polypterus senegalus), coelacanth (Latimeria chalumnae), zebrafish (Danio rerio), medaka (Oryzias latipes). The color of the font in the box was marked in red indicated that the change in gene location.
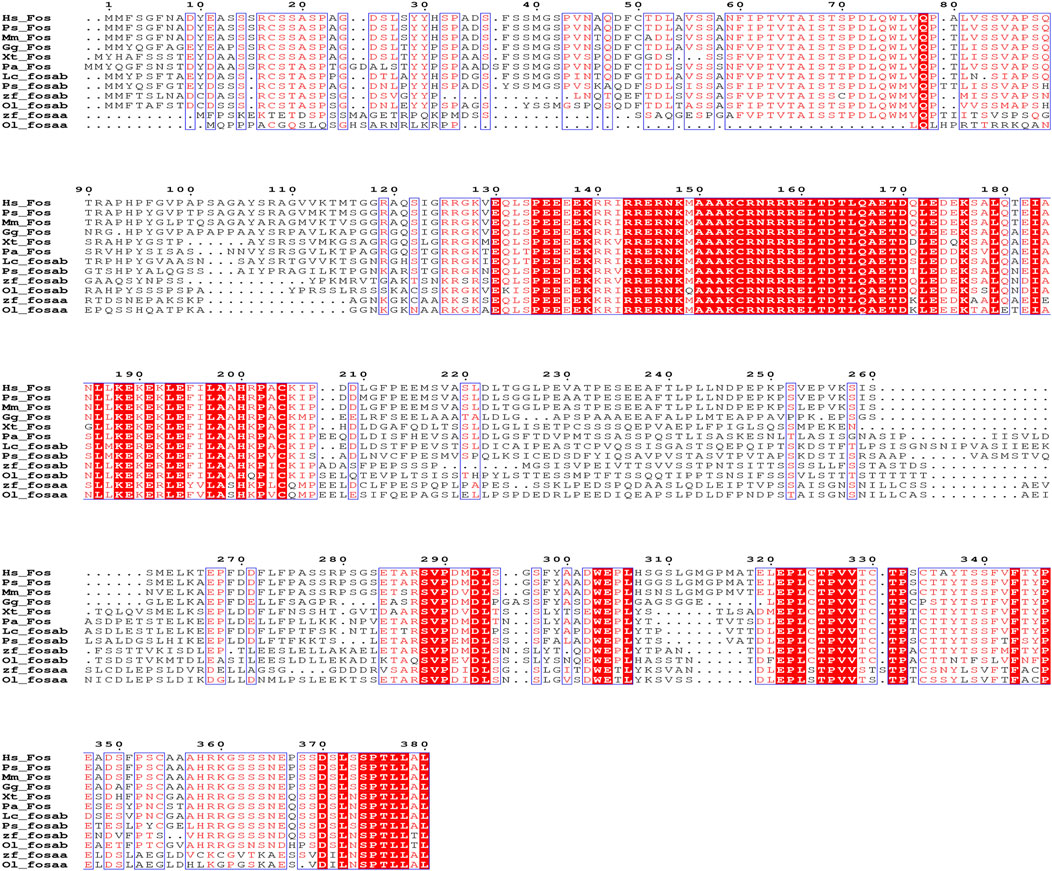
Figure 2. Sequence alignment of human (Homo sapiens, Hs), Vaquita (Phocoena sinus, Ps), mouse (Mus musculus, Mm), chicken (Gallus gallus, Gg), tropical clawed frog (Xenopus tropicalis, Xt), West African lungfish (Protopterus annectens, Pa), coelacanth (Latimeria chalumnae, Lc), Gray bichir (Polypterus senegalus, Ps), zebrafish (Danio rerio, Dr) and medaka (Oryzias latipes, Ol). Red with white letters represents fully conserved residues.
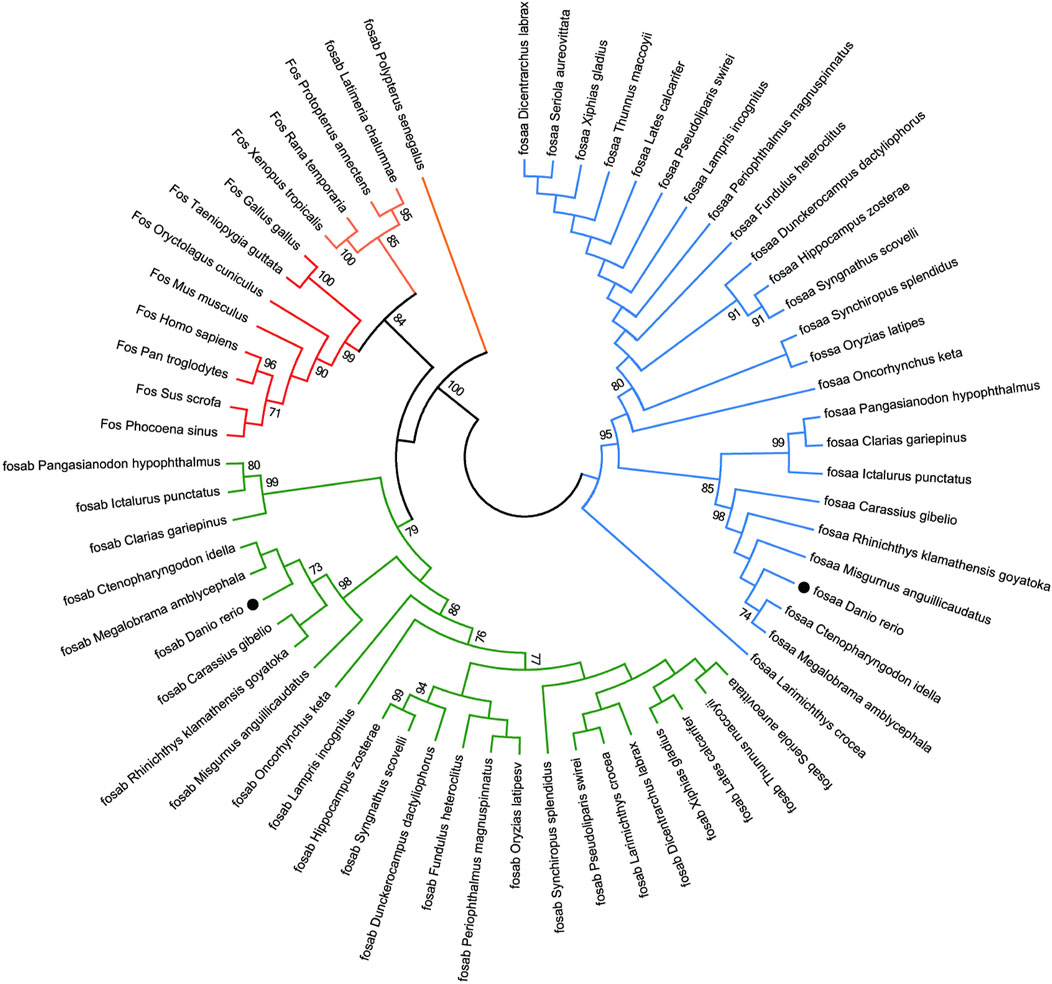
Figure 3. The Neighbor-joining tree based on the Fos proteins. The tree was built by the JTT model bootstrap method with 1,000 replications. The lines with the same color (red, blue or green) belong to one branch. The zebrafish fosaa and fosab were marked with a black dot.
3.2 Generation of fosaa and fosab knockout zebrafish
We constructed the fosaa and fosab mutants using the CRISPR/Cas9 technique in zebrafish, and sgRNA is located at the second exon adjacent motif (APM) individually (Figure 4A). The fosaa and fosab homozygous mutants (fosaa−/− and fosab−/−) resulted in a frameshift and premature truncation of proteins with 73 bp deletion and 76 bp insert, respectively (Figures 4B, C). F1 heterozygous were crossed to produce F2 homozygous and generated F3 stable genetic mutant lines (Figure 4D). The knockout zebrafish are viable and fertile. Further RT-qPCR analysis showed that fosaa−/−and fosab−/− mRNA expression was significantly decreased compared with wild-type (WT) (Figure 4E).
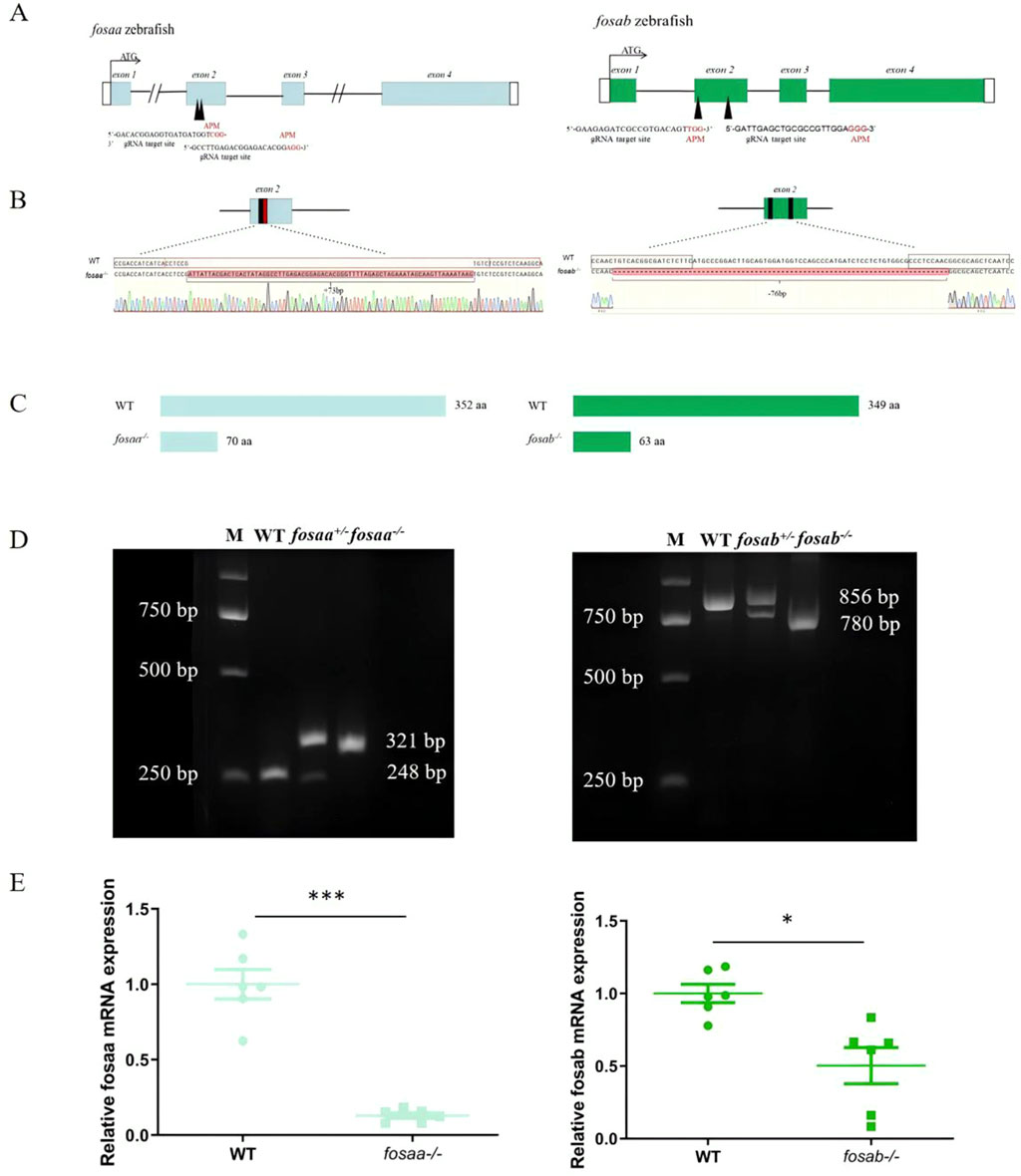
Figure 4. Generation of fosaa−/− and fosab−/− zebrafish using CRISPR/Cas9. (A): Schematic representation of gene structure for fosaa and fosab. The open reading frame (ORF) of fosaa and fosab are respectively indicated by blue and green boxes. The black lines show untranslated introns. The write boxes show regions. The translation start site (ATG) is indicated by a broken line. Both fosaa gene and fosab gene have 4 exons and the target sites are located in exon 2 by black arrowhead. The black and red letters respectively represent the target region and protospacer adjacent motif (PAM). (B): Mutation pattern of fosaa and fosab knockout. The black and red boxes represent sgRNA target binding site and PAM respectively. The red letters and hyphens represent nucleotide insertion and deletion. (C): The fosaa and fosab deletions and insertions caused frame-shift early termination of translation, resulting in fosaa−/− and fosab−/− proteins with only 70 and 63 amino acids (aa) individually. (D): Left and right panels show PCR genotyping of fosaa and fosab knockout zebrafish. WT, +/− and −/− indicate wild-type, heterozygous and homozygous zebrafish, respectively. (E): Relative mRNA expression of fosaa and fosab in WT zebrafish, fosaa−/− and fosab−/− zebrafish. Data represent mean ± SEM (n = 6). Values marked with * is significantly different with the WT group (P < 0.05) and *** is significantly different with the WT group (P < 0.001).
3.3 Brain weight is reduced in adult fosaa−/− and fosab−/− zebrafish
The growth performance was observed in adults fosaa−/−, fosab−/− and WT zebrafish, and the results indicated that there was no difference with body length and weight (P > 0.05) (Figure 5A). The brain weight showed a marked reduction in fosaa−/− (P < 0.05) and fosab−/− (P < 0.001) groups compared with the WT group (Figure 5B).
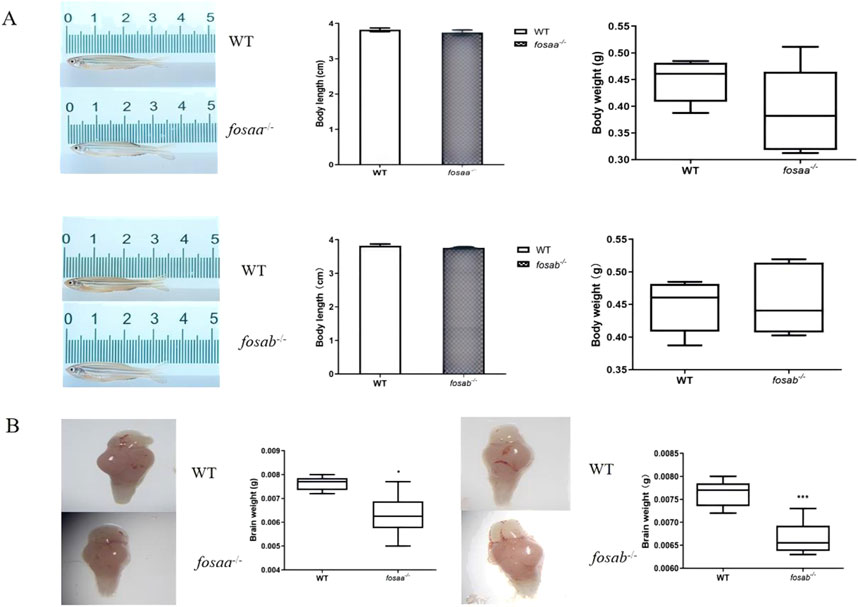
Figure 5. Growth performance and brain weight were observed in fosaa−/−, fosab−/− and WT groups. (A): Body length and body weight of fosaa−/−, fosab−/− and WT groups. (B): Brain weight of fosaa−/−, fosab−/− and WT groups. The data in the figure were expressed as mean ± S.E.M. (n = 6), and values marked with * is significantly different with the WT group (P < 0.05) and *** is significantly different with the WT group (P < 0.001).
3.4 T-maze test
The average time taken for fosaa−/−, fosab−/− and corresponding WT zebrafish to accomplish the trial for each session were shown in Figure 4. There was no significant difference in the average time taken for fosaa−/−, fosab−/− zebrafish compared with the corresponding WT zebrafish in the 2 pretraining sessions (P > 0.05) except for the first day of fosaa−/− (P < 0.05). In the discrimination sessions, the WT zebrafish had a marked decrease in the average time taken to complete the trial compared with fosab−/− zebrafish as the increase in the days, while did not show a notable variance with the fosaa−/− zebrafish although it also has a decreasing trend. The average time taken to complete the trial in fosab−/− zebrafish was significantly higher than the WT zebrafish (P < 0.05, P < 0.01, P < 0.001) (Figure 6). However, the average time taken to complete the trial had no difference between fosaa−/− and WT zebrafish in the discrimination sessions (P > 0.05) (Figure 6). In the extinction sessions, the average time taken to complete the trial in fosab−/− zebrafish was significantly higher than the WT zebrafish, while the fosab−/− zebrafish had no obvious difference compared with WT zebrafish (Figure 6).
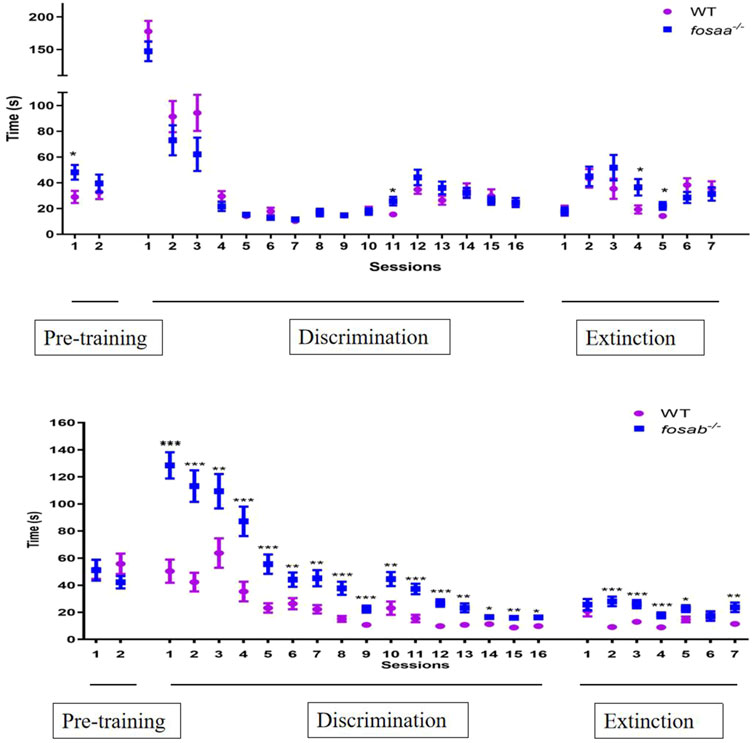
Figure 6. The average time required by zebrafish to complete trials in pre-training (2 days), discrimination learning training (16 days) and decision test (7 days). The data represented by mean ± SEM (n = 6). *, **, *** indicated significant differences (P < 0.05) (P < 0.01) (P < 0.001) compared with the WT group at the same session respectively.
3.5 The acetylcholinesterase (AChE) and acetylcholine transferase (AChT) activity in the brain of fosaa−/− and fosab−/− zebrafish
The acetylcholinesterase activity of the fosaa−/− and fosab−/− zebrafish were not significantly different from that of the WT (P > 0.05) (Figure 7A). By detecting the activity of acetylcholine transferase, it was found that there was no significant difference in the activity of acetylcholinesterase between the fosaa−/− and fosab−/− zebrafish compared with the WT zebrafish (P > 0.05) (Figure 7B).
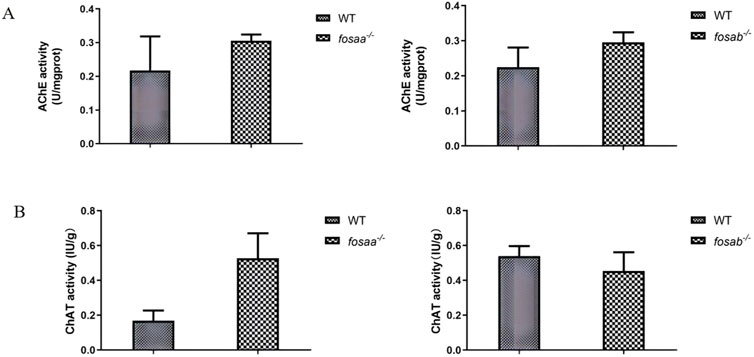
Figure 7. The acetylcholinesterase (AChE) acetylcholine and transferase (AChT) activity in the brain of WT, fosaa−/− and fosab−/− zebrafish. (A): The acetylcholinesterase (AChE) acetylcholine in the brain of WT, fosaa−/− and fosab−/− zebrafish. (B): The transferase (AChT) activity in the brain of WT, fosaa−/− and fosab−/− zebrafish. The data in the figure were expressed as mean ± S.E.M. (n = 3).
3.6 The relative mRNA expression of bdnf and c-jun in the brain of fosaa−/− and fosab−/− zebrafish
To evaluate how the bdnf and c-jun expression was altered in the fosaa and fosab knockout zebrafish, the relative mRNA expression of bdnf and c-jun was detected. The results indicated that the relative mRNA expression of bdnf and c-jun was not significant in fosaa−/− zebrafish (P > 0.05), while the c-jun in fosab−/− zebrafish was notably higher than the WT zebrafish (P < 0.05) (Figure 8).
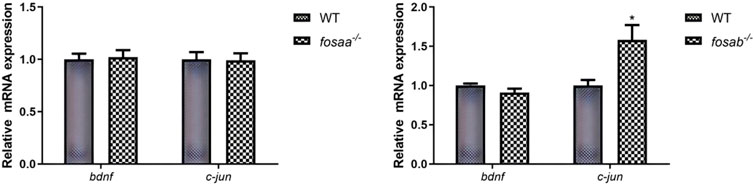
Figure 8. Relative mRNA expression of t of bdnf and c-jun in the brain of fosaa−/− and fosab−/− zebrafish. Data represent mean ± SEM (n = 6). Values marked with * are significantly different with the WT group (P < 0.05).
4 Discussion
Learning and memory are prevalent in animals, from the tiny nematode worm Caenorhabditis elegans to humans (Johnson and Hasher, 1987; Morrison et al., 1999). Variation in learning and memory results from an adaptation to ecological conditions, which allows an individual to adapt to environmental changes (Mery, 2013). Previous studies have demonstrated that the dorsolateral telencephalon of fish could be homologous to the mammalian hippocampus, sharing functional similarities, and fish have the ability to solve complex learning tasks, while its regulatory mechanisms have not been characterized (Gómez et al., 2020; Naderi et al., 2016). The c-fos gene is involved in regulating learning and memory in mammals (Cai et al., 2022; Velazquez et al., 2015). However, studies on fish are limited. The c-fos only has one paralogue in mammals, birds, reptiles, and amphibians, while most of the fish have two paralogues, excluding individual original fish like West African lungfish (P. annectens), Gray bichir (P. senegalus), and coelacanth (L. chalumnae). The results indicated that there is a gene duplication of c-fos in the fish, which might be attributed to homologous recombination errors or large-scale duplication of chromosomal segments with whole genomes in the evolutionary process (Tian et al., 2021; Zhang et al., 2003).
To investigate whether the c-fos gene in fish regulates learning and memory like mammals, we utilized the zebrafish to generate fosaa and fosab zebrafish mutant lines. The mutant zebrafish did not exhibit significant difference in the length and weight compared to the WT zebrafish, which is consistent with homozygous mutant mice (Randall et al., 1992). While the brain weight of knockout zebrafish showed a significant reduction compared with WT zebrafish, and the fosab−/− zebrafish had a more significant decrease than the fosaa−/− zebrafish might be due to the c-fos gene assuming a functional role in the brain. Maili et al. (2023) reported that fos knockout in zebrafish resulted in an abnormal craniofacial phenotype characterized by a hypoplastic oral cavity and significant changes in midface dimensions. The reduced brain weight observed in fosaa−/− and fosab−/ zebrafish may be attributed to these craniofacial abnormalities. The high brain weight of mice had a more rapid brain growth and behavioral maturation compared with medium and low brain, which suggests that there is a positive correlation between brain weight and brain development and learning and memory (Fuller and Herman, 1974; Paylor et al., 1994). The brain weight was reduced in the DPP6 knockout mice with slower learning and reduced memory performance in adults (Lin et al., 2018). The c-fos−/− mice showed a marked decrease in the brain weight resulting from a decrease in the number of cells in their organs and tissues (Velazquez et al., 2015). Thus, we figured that the fosaa and fosab knockout zebrafish could lead to impeded brain development via abnormal craniofacial and alleviating brain weight. Meanwhile, the function of learning and memory might be disrupted in the fosaa−/− and fosab−/− zebrafish.
The mice lacking c-fos in the Central Nervous System (CNS) showed impaired spatial and associative learning tasks (Fleischmann et al., 2003; Guzowski, 2002). Some behaviors that interfere with the evaluation of learning on the Morris water task were affected in the c-fos mutant mouse (Paylor et al., 1994). To further investigate whether the learning and memory ability of fosaa−/− and fosab−/− zebrafish is impaired, we performed a 24-day behavioral experiment utilizing the T-maze based on associative learning, where a visual stimulus (colour) was linked with a natural stimulus (food reward). The T-maze test can be used to perform associative learning tasks to assess the learning capabilities of zebrafish (Hieu et al., 2020; Jouandot et al., 2012; Yoo et al., 2021a). Our results found that the time both wild and knockout zebrafish took to complete a given colour choice decreased with increasing learning days in the T-maze test. It is worth noting that there is a marked decrease in the average time taken to complete the trial as the increase in the days between fosab−/− and WT zebrafish, while the average time taken to complete the trial had no difference between fosaa−/− and WT zebrafish in the discrimination and extinction sessions. The results suggested that zebrafish did have learning and memory, which may be regulated by fosab rather than fosaa. In addition to memory and learning, the c-fos gene is also involved in DNA damage repair, immunity, and craniofacial development (Maili et al., 2023; Yao et al., 2024; Zhang et al., 2022). fosaa may play an important role in other functions in fish, which deserves to be studied in depth further.
The synteny and phylogenetic analysis of c-fos could act as evidence for the functional differentiation of fosaa and fosab. The flanking genes of c-fos were conserved in mammals, birds, reptiles, amphibians, and several original fish, while the flanking genes of fosaa and fosab exist deletions or other gene additions in the majority of fish. The phylogenetic trees showed that the Fos diverge into two groups, which Fos of the mammals, birds, reptiles and amphibians were grouped together with fosab of fish, and fosaa stands alone as a group. Protein BLAST analysis showed that human c-FOS is 48% identical to Fosaa and 55% to Fosab, and the phosphorylation sites in mammalian FOS proteins are highly conserved in zebrafish Fosab but not in Fosaa (Kubra et al., 2022). Besides, (Kubra et al., 2022), observed that the pattern of fosaa and fosab expression were similar in early embryos but persisted until 4 dpf in multiple regions of the embryos via wholemount in situ hybridisation (WISH) analysis, which provided key insights into the tissue-specific functions of fosaa and fosab.
Fos-immunoreactive cells were identified as neurons, predominantly clustered in areas corresponding to the regions exhibiting high acetylcholinesterase (AChE) activity (Liu et al., 1991). The hyperglycemia promoted memory deficit in adult zebrafish, which results from increased AChE activity (Katiucia et al., 2014). We determined the activity of AChE and AChT in the brain of fosaa−/− and fosab−/− zebrafish, and both of them had no significant difference in fosaa−/− and fosab−/− zebrafish compared with WT zebrafish individually. Acute exposure to copper induces significant changes in the behavior of zebrafish, and the AChE activity decreased significantly in zebrafish muscle, but there were no significant changes in cerebral AChE activity (Haverroth et al., 2015). Cholinergic blocker scopolamine treatment in zebrafish has been confirmed to induce cognitive impairment, and the induced memory deficits were solely due to the drug effect on the cholinergic system (Cognato et al., 2012; Richetti et al., 2011). Gac fruit extract exhibits protective activity against scopolamine-induced memory impairment in zebrafish, while the lack of observed significant differences in AChE activity may be attributed to insufficient analysis of the adult zebrafish central nervous system (Singsai et al., 2024). Our data is in accordance with literature, indicating that the lack of differences in the enzyme activity of AChE and AChT may be attributed to the presence of several isoforms of AChE in the central nervous system and the limited number of examined samples. Therefore, whether the cholinergic system is involved in the disruption of learning and memory in the fosaa−/− and fosab−/− zebrafish requires future studies with diverse testing methods and larger sample sizes to determine.
The relative mRNA expression level of c-jun in fosab−/− zebrafish was significantly higher than in WT zebrafish, while the fosaa−/− zebrafish has no discernible trend. The result is consistent with our experimental results using the T-maze to assess learning and memory ability in fosaa−/− and fosab−/− zebrafish. Jun-Fos heterodimers have stronger DNA-binding activity and serve primarily as a vehicle for short-term stimulation to link synapses to long-term changes (Kuzniewska et al., 2013; Messaoudi et al., 2002). The reason for fosab−/− zebrafish impaired learning and memory may be via the reduction of Jun-Fos heterodimer synthesis (Cynthia and Medina, 2015). showed that BDNF is one of the upstream signals for c-Fos expression. Blocking BDNF synthesis prevents inhibitory avoidance (IA)-induced increase in c-Fos levels and affects memory persistence (Cynthia and Medina, 2015). Our results showed that the mRNA expression of bdnf has no significant change in fosaa−/− and fosab−/− zebrafish compared with wild zebrafish (Lucon-Xiccato et al., 2022). illustrated that zebrafish’s learning abilities were positively correlated with bdnf expression, and bdnf−/− zebrafish had a remarkable learning deficit. However, the researchers did not investigate the relationship between bdnf and foaa and fosab further by using the bdnf−/− zebrafish. In the brain of Triclocarban (TCC)-exposed zebrafish could reduce the cognitive performance of zebrafish and upregulation of c-fos and downregulation of bdnf (Lucon-Xiccato et al., 2023). Based on the available results, we hypothesized that bdnf might be a system that regulates learning and memory independently of fosaa and fosab in zebrafish.
In conclusion, the present study revealed that fosab, but not fosaa, may possess the function of learning and memory via synthesis heterodimers with c-jun in fish. Questions remain regarding the fosaa play which kind of role in fish needs to be further investigated. For the first time, we illustrated the role of fosaa and fosab in learning and memory via c-fos knockout in fish, which can provide new insights into environmental adaptation.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by HZAUFI-2020-0032, the Tab of Animal Experimental Ethical Inspection of Laboratory Animal Centre, Huazhong Agriculture University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QW: Conceptualization, Data curation, Formal Analysis, Validation, Writing–original draft, Writing–review and editing. LZ: Conceptualization, Data curation, Formal Analysis, Validation, Writing–original draft, Writing–review and editing. CZ: Data curation, Formal Analysis, Writing–review and editing. KL: Data curation, Formal Analysis, Writing–review and editing. JW: Data curation, Formal Analysis, Writing–review and editing. X-FL: Conceptualization, Data curation, Funding acquisition, Supervision, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by Fishery Seed Industry Joint Breeding Project of Jiangxi Province (2023yyzygg-02), the Key Research and Development Program of Jiangxi Province (20232BBF6001) and The National Natural Science Foundation of China (31972809).
Acknowledgments
We thank members of Chinses Perch Research Center for technical support and discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1503066/full#supplementary-material
References
Ariyasiri, K., Choi, T.-I., Gerlai, R., and Kim, C. H. (2021). Acute ethanol induces behavioral changes and alters c-fos expression in specific brain regions, including the mammillary body, in zebrafish. Prog. Neuro-Psychopharmacology Biol. Psychiatry 109, 110264. doi:10.1016/j.pnpbp.2021.110264
Avdesh, A., Martin-Iverson, M. T., Mondal, A., Chen, M., Askraba, S., Morgan, N., et al. (2012). Evaluation of color preference in zebrafish for learning and memory. J. Alzheimer's Dis. 28 (2), 459–469. doi:10.3233/jad-2011-110704
Barron, H. C. (2021). Neural inhibition for continual learning and memory. Curr. Opin. Neurobiol. 67, 85–94. doi:10.1016/j.conb.2020.09.007
Bekinschtein, P., Cammarota, M., and Medina, J. H. (2014). BDNF and memory processing. Neuropharmacol. 76, 677–683. doi:10.1016/j.neuropharm.2013.04.024
Bicakci, A. O., Tsai, P. L., Kahl, E., Mayer, D., and Fendt, M. (2021). Dissociative effects of neuropeptide S receptor deficiency and nasal neuropeptide S administration on T-maze discrimination and reversal learning. Pharmaceuticals 14 (7), 643. doi:10.3390/ph14070643
Cai, G., Lu, Y., Chen, J., Yang, D., Yan, R., Ren, M., et al. (2022). Brain-wide mapping of c-Fos expression with fluorescence micro-optical sectioning tomography in a chronic sleep deprivation mouse model. Neurobiol. Stress 20, 100478. doi:10.1016/j.ynstr.2022.100478
Cognato, G. d. P., Bortolotto, J. W., Blazina, A. R., Christoff, R. R., Lara, D. R., Vianna, M. R., et al. (2012). Y-Maze memory task in zebrafish (Danio rerio): the role of glutamatergic and cholinergic systems on the acquisition and consolidation periods. Neurobiol. Learn. Mem. 98 (4), 321–328. doi:10.1016/j.nlm.2012.09.008
Colwill, R. M., Raymond, M. P., Ferreira, L., and Escudero, H. (2005). Visual discrimination learning in zebrafish (Danio rerio). Behav. Process. 70 (1), 19–31. doi:10.1016/j.beproc.2005.03.001
Curran, T., and Morgan, J. I. (1987). Memories of fos. BioEssays 7 (6), 255–258. doi:10.1002/bies.950070606
Cynthia, K. J., and Medina, J. H. (2015). Requirement of an early activation of BDNF/c-Fos cascade in the retrosplenial cortex for the persistence of a long-lasting aversive memory. Cereb. Cortex 27 (2), 1060–1067. doi:10.1093/cercor/bhv284
de Abreu, M. S., Giacomini, A. C. V. V., Sysoev, M., Demin, K. A., Alekseeva, P. A., Spagnoli, S. T., et al. (2019). Modeling gut-brain interactions in zebrafish. Brain Res. Bull. 148, 55–62. doi:10.1016/j.brainresbull.2019.03.003
Fleischmann, A., Hvalby, O., Jensen, V., Strekalova, T., Zacher, C., Layer, L. E., et al. (2003). Impaired long-term memory and nr2a-type NMDA receptor-dependent synaptic plasticity in mice lacking c-fos in the CNS. J. Neuroence 23 (27), 9116–9122. doi:10.1523/JNEUROSCI.23-27-09116.2003
Fuller, J. L., and Herman, B. H. (1974). Effect of genotype and practice upon behavioral development in mice. Dev. Psychobiol. 7, 21–30. doi:10.1002/dev.420070105
Ganguly, K., Rejmak, E., Mikosz, M., Nikolaev, E., Knapska, E., and Kaczmarek, L. (2013). Matrix metalloproteinase (MMP) 9 transcription in mouse brain induced by fear learning. J. Biol. Chem. 288 (29), 20978–20991. doi:10.1074/jbc.M113.457903
Gómez, Y., Vargas, J. P., López, J. C., and Portavella, M. (2020). Inhibition of brain NOS activity impair spatial learning acquisition in fish. Brain Res. Bull. 164, 29–36. doi:10.1016/j.brainresbull.2020.08.007
Guzowski, J. F. (2002). Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus 12 (1), 86–104. doi:10.1002/hipo.10010
Haam, J., and Yakel, J. L. (2017). Cholinergic modulation of the hippocampal region and memory function. J. Neurochem. 142, 111–121. doi:10.1111/jnc.14052
Haverroth, G. M. B., Welang, C., Mocelin, R. N., Postay, D., Bertoncello, K. T., Franscescon, F., et al. (2015). Copper acutely impairs behavioral function and muscle acetylcholinesterase activity in zebrafish (Danio rerio). Ecotoxicol. Environ. Saf. 122 (DEC.), 440–447. doi:10.1016/j.ecoenv.2015.09.012
He, D., Wu, H., Wei, Y., Liu, W., Huang, F., Shi, H., et al. (2015). Effects of harmine, an acetylcholinesterase inhibitor, on spatial learning and memory of APP/PS1 transgenic mice and scopolamine-induced memory impairment mice. Eur. J. Pharmacol. 768, 96–107. doi:10.1016/j.ejphar.2015.10.037
Hieu, B. T. N., Anh, N. T. N., Audira, G., Juniardi, S., Liman, R. A. D., Villaflores, O. B., et al. (2020). Development of a modified three-day T-maze protocol for evaluating learning and memory capacity of adult zebrafish. Int. J. Mol. Sci. 21 (4), 1464. doi:10.3390/ijms21041464
Ichikawa, T., Ajiki, K., Matsuura, J., and Misawa, H. (1997). Localization of two cholinergic makers, choline acetyltransferase and vesicular acetylcholine transporter in the central nervous system of the rat : in situ hybridization histochemistry and immunohistochemistry. J. Chem. Neuroanat. 13 (1), 39. doi:10.1016/s0891-0618(97)00021-5
Ismail, N. A., Abdullah, M. F. I. L., Hami, R., and Yusof, H. A. (2021). A narrative review of brain-derived neurotrophic factor (BDNF) on cognitive performance in Alzheimer's disease. Growth factors., 1–17. doi:10.1080/08977194.2020.1864347
Johnson, M. K., and Hasher, L. (1987). Human learning and memory. Annu. Rev. Psychol. 38, 631–668. doi:10.1146/annurev.ps.38.020187.003215
Jouandot, D. J., Echevarria, D. J., and Lamb, E. A. (2012). The utility of the T-maze in assessing learning, memory, and models of neurological disorders in the zebrafish. Behaviour 149 (10-12), 1081–1097. doi:10.1163/1568539x-00003027
Katiucia, M. C., De Moraes, D. A., Menezes, F. P., Kist, L. W., Bogo, M. R., and Da Silva, R. S. (2014). Hyperglycemia induces memory impairment linked to increased acetylcholinesterase activity in zebrafish (Danio rerio). Behav. Brain Res. 274, 319–325. doi:10.1016/j.bbr.2014.08.033
Kim, J., Lee, S., and Cho, E. (2019a). Acer okamotoanum and isoquercitrin improve cognitive function via attenuation of oxidative stress in high fat diet- and amyloid beta-induced mice. Food and Funct. 10 (10), 6803–6814. doi:10.1039/c9fo01694e
Kim, M. S., Kim, J. H., Kry, D., Ae Choi, M., Ok Choi, D., Gon Cho, B., et al. (2003). Effects of acute hypotension on expression of cFos-like protein in the vestibular nuclei of rats. Brain Res. 962 (1-2), 111–121. doi:10.1016/S0006-8993(02)03977-X
Kim, Y.-H., Lee, K.-S., Kim, Y.-S., and Kim, J. H. (2019b). Effects of hypoxic preconditioning on memory evaluated using the T-maze behavior test. Anim. Cells Syst. 23, 10–17. doi:10.1080/19768354.2018.1557743
Kubra, K., Gaddu, G. K., Liongue, C., Heidary, S., Ward, A. C., Dhillon, A. S., et al. (2022). Phylogenetic and expression analysis of fos transcription factors in zebrafish. Int. J. Mol. Sci. 23 (17), 10098. doi:10.3390/ijms231710098
Kuzniewska, B., Rejmak, E., Malik, A. R., Jaworski, J., Kaczmarek, L., and Kalita, K. (2013). Brain-derived neurotrophic factor induces matrix metalloproteinase 9 expression in neurons via the serum response factor/c-Fos pathway. Mol. Cell Biol. 33 (11), 2149–2162. doi:10.1128/MCB.00008-13
Lin, L., Murphy, J. G., Karlsson, R.-M., Petralia, R. S., Gutzmann, J. J., Abebe, D., et al. (2018). DPP6 loss impacts hippocampal synaptic development and induces behavioral impairments in recognition, learning and memory. Front. Cell. Neurosci. 12, 84. doi:10.3389/fncel.2018.00084
Liu, F. C., Dunnett, S. B., Robertson, H. A., and Graybiel, A. M. (1991). Intrastriatal grafts derived from fetal striatal primordia. III. Induction of modular patterns of fos-like immunoreactivity by cocaine. Exp. Brain Res. 85, 501–506. doi:10.1007/BF00231733
Lucon-Xiccato, T., Montalbano, G., Gatto, E., Frigato, E., D’Aniello, S., and Bertolucci, C. (2022). Individual differences and knockout in zebrafish reveal similar cognitive effects of BDNF between teleosts and mammals. Proc. R. Soc. B, 289. doi:10.1098/rspb.2022.2036
Lucon-Xiccato, T., Sava, B. B., Merola, C., Benedetti, E., Caioni, G., Aliko, V., et al. (2023). Environmentally relevant concentrations of triclocarban affect behaviour, learning, and brain gene expression in fish. Sci. Total Environ. 903, 166717. doi:10.1016/j.scitotenv.2023.166717
Maili, L., Tandon, B., Yuan, Q., Menezes, S., Chiu, F., Hashmi, S. S., et al. (2023). Disruption of fos causes craniofacial anomalies in developing zebrafish. Front. Cell Dev. Biol. 11, 1141893. doi:10.3389/fcell.2023.1141893
Mery, F. (2013). Natural variation in learning and memory. Curr. Opin. Neurobiol. 23 (1), 52–56. doi:10.1016/j.conb.2012.09.001
Messaoudi, E., Ying, S. W., Kanhema, T., Croll, S. D., and Bramham, C. R. (2002). Brain-derived neurotrophic factor triggers transcription-dependent, late phase long-term potentiation In Vivo. J. Neurosci.: The Journal of neuroscience 22 (17), 7453–7461. doi:10.1523/JNEUROSCI.22-17-07453.2002
Morris, B. J., Newman-Tancredi, A., Audinot, V., Simpson, C. S., and Millan, M. J. (2000). Activation of dopamine D3 receptors induces c-fos expression in primary cultures of rat striatal neurons. J. Neurosci. Res. 59 (6), 740–749. doi:10.1002/(sici)1097-4547(20000315)59:6<740::Aid-jnr6>3.0.Co;2-f
Morrison, G. E., Wen, J. Y., Runciman, S., and van der Kooy, D. (1999). Olfactory associative learning in Caenorhabditis elegans is impaired in lrn-1 and lrn-2 mutants. Behav. Neurosci. 113, 358–367. doi:10.1037//0735-7044.113.2.358
Naderi, M., Jamwal, A., Ferrari, M. C. O., Niyogi, S., and Chivers, D. P. (2016). Dopamine receptors participate in acquisition and consolidation of latent learning of spatial information in zebrafish (Danio rerio). Prog. Neuro-Psychopharmacology Biol. Psychiatry 67, 21–30. doi:10.1016/j.pnpbp.2016.01.002
Nogami-Hara, A., Nagao, M., Takasaki, K., Egashira, N., Fujikawa, R., Kubota, K., et al. (2018). The Japanese Angelica acutiloba root and yokukansan increase hippocampal acetylcholine level, prevent apoptosis and improve memory in a rat model of repeated cerebral ischemia. J. Ethnopharmacol. 214, 190–196. doi:10.1016/j.jep.2017.12.025
Oda, Y. (2010). Choline acetyltransferase: The structure, distribution and pathologic changes in the central nervous system. Pathol. Int. 49 (11), 921–937. doi:10.1046/j.1440-1827.1999.00977.x
Paylor, R., Johnson, R. S., Papaioannou, V., Spiegelman, B. M., and Wehner, J. M. (1994). Behavioral assessment of c-fos mutant mice. Brain Res. 651 (1–2), 275–282. doi:10.1016/0006-8993(94)90707-2
Phillis, J. W. (2005). Acetylcholine release from the central nervous system: a 50-year retrospective. Crit. Rev. Neurobiol. 17 (3–4), 161–217. doi:10.1615/CritRevNeurobiol.v17.i3-4.30
Pilehvar, A., Town, R. M., and Blust, R. (2020). The effect of copper on behaviour, memory, and associative learning ability of zebrafish (Danio rerio). Ecotoxicol. Environ. Saf. 188, 109900. doi:10.1016/j.ecoenv.2019.109900
Randall, S., Johnson, , and Papaioannou, V. (1992). Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell 71, 577–586. doi:10.1016/0092-8674(92)90592-Z
Richetti, S. K., Blank, M., Capiotti, K. M., Piato, A. L., Bogo, M. R., Vianna, M. R., et al. (2011). Quercetin and rutin prevent scopolamine-induced memory impairment in zebrafish. Behav. Brain Res. 217 (1), 10–15. doi:10.1016/j.bbr.2010.09.027
Singsai, K., Saksit, N., and Chaikhumwang, P. (2024). Brain acetylcholinesterase activity and the protective effect of Gac fruit on scopolamine-induced memory impairment in adult zebrafish. IBRO Neurosci. Rep. 16, 368–372. doi:10.1016/j.ibneur.2024.02.004
Song, Y., Liu, S., Jiang, X., Ren, Q., Deng, H., Paudel, Y. N., et al. (2022). Benzoresorcinol induces developmental neurotoxicity and injures exploratory, learning and memorizing abilities in zebrafish. Sci. Total Environ. 834 (834-), 155268. doi:10.1016/j.scitotenv.2022.155268
Soussi-Yanicostas, N. (2022). Zebrafish as a model for neurological disorders. Int. J. Mol. Sci. 23 (8), 4321. doi:10.3390/ijms23084321
Tan, J. K., Nazar, F. H., Makpol, S., and Teoh, S. L. (2022). Zebrafish: a pharmacological model for learning and memory research. Molecules 27 (21), 7374. doi:10.3390/molecules27217374
Tian, Y., Jiang, C., Pan, Y., Guo, Z., Wang, W., Luo, X., et al. (2021). Bombyx neuropeptide G protein-coupled receptor A14 and A15 are two functional G protein-coupled receptors for CCHamide neuropeptides. Insect Biochem. Mol. Biol. 131, 103553. doi:10.1016/j.ibmb.2021.103553
Velazquez, F. N., Prucca, C. G., Etienne, O., D'Astolfo, D. S., Silvestre, D. C., Boussin, F. D., et al. (2015). Brain development is impaired in c-fos -/- mice. Oncotarget 6 (19), 16883–16901. doi:10.18632/oncotarget.4527
Yao, H., Wu, Y., Zhong, Y., Huang, C., Guo, Z., Jin, Y., et al. (2024). Role of c-Fos in DNA damage repair. J. Cell Physiol. 239 (5), e31216. doi:10.1002/jcp.31216
Yoo, J. W., Sohn, S.-H., Kim, Y.-H., and Min, T. J. (2021a). The effect of ulinastatin to the learning and memory in zebrafish. NeuroMolecular Med. 23 (4), 511–520. doi:10.1007/s12017-021-08653-3
Yoo, J. W., Sohn, S. H., Kim, Y. H., and Min, T. J. (2021b). The effect of ulinastatin to the learning and memory in zebrafish. NeuroMolecular Med. 23, 511–520. doi:10.1007/s12017-021-08653-3
Zhang, H., Xie, G., Jin, Z., Zhang, C., and Huang, Y. (2022). The transcription factor c-Fos plays a negative role in prawn immunity against WSSV infection. Aquaculture 553, 738080. doi:10.1016/j.aquaculture.2022.738080
Zhang, J., Zhang, D., McQuade, J. S., Behbehani, M., Tsien, J. Z., and Xu, M. (2002). c-fos regulates neuronal excitability and survival. Nat. Genet. 30 (4), 416–420. doi:10.1038/ng859
Zhang, P., Gu, Z., and Li, W. H. (2003). Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 4 (9), R56. doi:10.1186/gb-2003-4-9-r56
Keywords: fosaa, fosab, learning and memory, brain development, zebrafish
Citation: Wang Q, Zhang L, Zhu C, Lu K, Wu J and Liang X-F (2025) Fosab, but not fosaa, plays important role in learning and memory in fish—insights from zebrafish gene knockout study. Front. Cell Dev. Biol. 13:1503066. doi: 10.3389/fcell.2025.1503066
Received: 28 September 2024; Accepted: 07 January 2025;
Published: 27 January 2025.
Edited by:
James Alan Marrs, Purdue University Indianapolis, United StatesReviewed by:
Petronella Kettunen, University of Gothenburg, SwedenJosé Miguel Cerda-Reverter, Spanish National Research Council (CSIC), Spain
Darlan Gusso, Pontifical Catholic University of Rio Grande do Sul, Brazil
Copyright © 2025 Wang, Zhang, Zhu, Lu, Wu and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu-Fang Liang, eHVmYW5nX2xpYW5nQGhvdG1haWwuY29t
†These authors have contributed equally to this work
 Qiuling Wang1,2,3†
Qiuling Wang1,2,3† Xu-Fang Liang
Xu-Fang Liang