- 1Departments of Pathology and Cell Biology, Emory University School of Medicine, Atlanta, GA, United States
- 2Winship Cancer Institute, Emory University School of Medicine, Atlanta, GA, United States
Lifeact is a short peptide that is widely utilized as a probe for actin filaments in live imaging. However, high concentrations of Lifeact can alter actin filament dynamics and cause artificial modifications to the actin cytoskeleton. Here, I evaluated Caenorhabditis elegans strains expressing Lifeact fused to fluorescent proteins in the body wall muscle. I found that, while low-level expression of Lifeact from a single-copy transgene was appropriate for labeling sarcomeric actin filaments, overexpression of Lifeact from an extrachromosomal array causes severe disorganization of muscle sarcomeres and lethality at an embryonic or larval stage. Therefore, for imaging studies in C. elegans, Lifeact needs to be kept at a low level by proper management of the expression system.
Introduction
Actin is one of the major cytoskeletal proteins and it plays important roles in many cell biological events (Pollard, 2016). The actin cytoskeleton is constantly reorganized, and its dynamic behaviors are often critical for its functions. The development of genetically encodable actin probes in combination with fluorescent proteins as fusion partners has dramatically advanced the field of analyzing the dynamics of actin filaments in living cells (Melak et al., 2017). Lifeact, a 17-amino-acid peptide from yeast Abp140, is one of the most utilized F-actin probes for live imaging (Riedl et al., 2008). Lifeact labels actin filaments in cells from many different species but has some limitations, such as not detecting stress-induced actin-cofilin rods (Munsie et al., 2009) and poorly labeling certain filopodia and lamellar actin networks (Belin et al., 2014).
However, the major disadvantage is the detrimental effects of overexpressed Lifeact on the actin cytoskeleton. Overexpression of Lifeact causes F-actin disorganization and sterility in the Drosophila ovary (Spracklen et al., 2014), cardiac dysfunction in the zebrafish heart (Xu and Du, 2021), F-actin stabilization in the Arabidopsis root (van der Honing et al., 2011), prolonged endocytosis and cytokinesis in fission yeast (Courtemanche et al., 2016), and alterations in nuclear and cytoplasmic actin filaments in cultured mammalian cells (Du et al., 2015; Flores et al., 2019). In in vitro experiments, Lifeact alters rates of nucleation and elongation of actin filaments and inhibits filament severing by cofilin (Courtemanche et al., 2016), indicating that excessive Lifeact can artificially modify actin filament dynamics. Nonetheless, when Lifeact expression is managed at low levels, it is still a very useful F-actin probe for live imaging (Courtemanche et al., 2016; Alvarez et al., 2024; Wirshing and Goode, 2024).
In this study, I evaluated Lifeact as a probe for actin filaments in the body wall muscle of the nematode Caenorhabditis elegans, in which actin and myosin are organized in a sarcomeric pattern (Benian and Epstein, 2011; Ono, 2014). Since many structural and regulatory proteins for assembly and maintenance of sarcomeric actin filaments are conserved among vertebrates and invertebrates (Ono, 2010), the C. elegans body wall muscle is an excellent model for studies on the structure and function of the striated muscle. One of the major advantages of C. elegans is its transparent body, allowing live fluorescence imaging of the body wall muscle in intact animals (Ghosh and Hope, 2010). Previously, we have reported that green fluorescent protein (GFP)-tagged actin (GFP::ACT-4) could be incorporated into sarcomeres in the C. elegans body wall muscle (Hayashi et al., 2019). However, several studies have indicated that GFP-actin does not behave in the same manner as untagged actin (Doyle and Botstein, 1996; Aizawa et al., 1997; Chen et al., 2012). Therefore, it would be desirable to assess other less invasive methods for in vivo actin labeling than direct actin tagging with a fluorescent protein. By comparing previously published strains, I found that while stable low expression of Lifeact causes no detectable effects on the sarcomeres, overexpression of Lifeact in the body wall muscle causes severe sarcomere disorganization and embryonic or larval lethality. Accordingly, Lifeact, at low concentrations, is a useful F-actin probe in C. elegans muscle, but artificial detrimental effects of Lifeact need to be carefully evaluated.
Results
Overexpression of Lifeact in the body wall muscle causes embryonic or larval lethality
During the previous studies to localize CLIK-1, a calponin-related protein, in the sarcomeres of the C. elegans body wall muscle (Ono and Ono, 2020; Ono, 2024), we utilized a strain, KAG190, containing a transgene to express mCherry::Lifeact in the body wall muscle driven by the myo-3 promoter (Brouilly et al., 2015). The myo-3 gene encodes a myosin heavy chain that is predominantly expressed in the body wall muscle (Miller et al., 1983; Waterston, 1989; Okkema et al., 1993). This transgene was maintained as an extrachromosomal array that can contain a high copy number of the transgene and is often inherited in a mosaic manner within a transgenic animal (Mello et al., 1991; Mello and Fire, 1995). These characteristics can cause overexpression of a transgene and/or cell-to-cell variability in the expression levels. When mCherry::Lifeact was expressed at low levels in KAG190, it properly labeled sarcomeric actin filaments in the muscle cells (Brouilly et al., 2015; Ono, 2024). However, I also noticed a high occurrence of arrested embryos and L1 larvae when the expression of mCherry::Lifeact was high. Quantitation of lethality indicated that 43% ± 3.1% of mCherry::Lifeact-positive animals were arrested either as late embryos or L1 larvae (Figure 1). Most of the arrested embryos were elongated and immobile, suggesting that they failed to hatch. In contrast, wild-type (N2) animals with no transgene rarely produced arrested embryos or larvae (Figure 1). A strain that carries a single-copy transgene encoding Lifeact::mRuby with the myo-3 promoter (RHS41) expressed uniformly low levels of Lifeact::mRuby throughout all body wall muscle cells (Higuchi-Sanabria et al., 2018) and rarely produced arrested embryos or larvae (Figure 1). As a control to examine the effect of mCherry, I examined ABR14 carrying a transgene in an extrachromosomal array to express mCherry alone by the myo-3 promoter (Han et al., 2017). The lethality of this strain was slightly higher than that of N2 wild-type or RHS41 (single-copy Lifeact::mRuby) but much lower than that of KAG190 (mCherry::Lifeact overexpression) (Figure 1), indicating that overexpression of Lifeact caused lethality. These results indicate that overexpression of Lifeact in the body wall muscle is sufficient to cause embryonic or larval lethality.
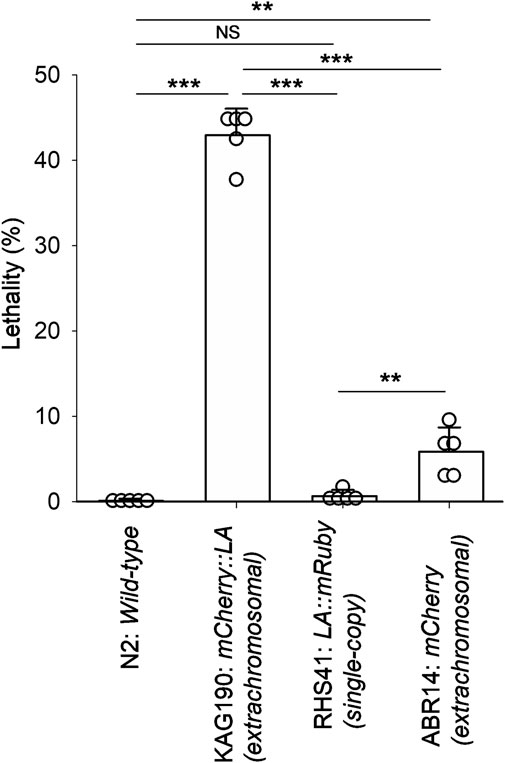
Figure 1. Expression of mCherry::Lifeact in the C. elegans body wall muscle from an extrachromosomal array causes high lethality. Lethality of embryos and larvae was quantified for N2 (wild-type), KAG190 [mCherry::Lifeact (abbreviated to LA) expressed from an extrachromosomal array], RHS41 (Lifeact::mRuby expressed from a single-copy transgene), and ABR14 (mCherry alone expressed from an extrachromosomal array). For transgenic strains, only animals with the expression of the fluorescent proteins were counted. Data are presented as average ± standard deviation (n = 5). **, 0.01 < P < 0.001. ***, P < 0.001. NS, not significant.
Overexpression of Lifeact in the body wall muscle causes sarcomere disorganization
Examination of the actin filament organization showed that Lifeact overexpression caused severe sarcomere disorganization in the body wall muscle (Figures 2, 3). Although arrested embryos with mCherry::Lifeact overexpression showed highly disorganized actin filaments in the body wall muscle, these embryos were elongated and folded within confined eggshells, which made microscopic imaging of subcellular structures difficult. For clear comparison among the strains, I examined actin organization in the body wall muscle cells of L1 larvae (Figure 2) and adults (Figure 3) after staining with Alexa 488-labeled phalloidin. In wild-type L1 larvae, F-actin was normally accumulated into linear structures representing contractile apparatuses (Figure 2A, arrow). When the expression of mCherry::Lifeact was low, these F-actin structures were labeled, although some non-uniform distribution of F-actin was also detected (Figure 2B, arrows), suggesting minor disorganization. However, when the expression of mCherry::Lifeact was high, abnormal wavy F-actin bundles, which were heavily decorated by mCherry::Lifeact, were formed (Figure 2C, arrows). Lifeact::mRuby from a single-copy transgene was uniformly expressed in all body wall muscle cells at low levels and properly labeled F-actin structures without a detrimental effect (Figure 2D, arrows). When mCherry alone was overexpressed, aggregates of mCherry were formed, but sarcomeric actin organization was not visibly affected (Figure 2E, arrows).
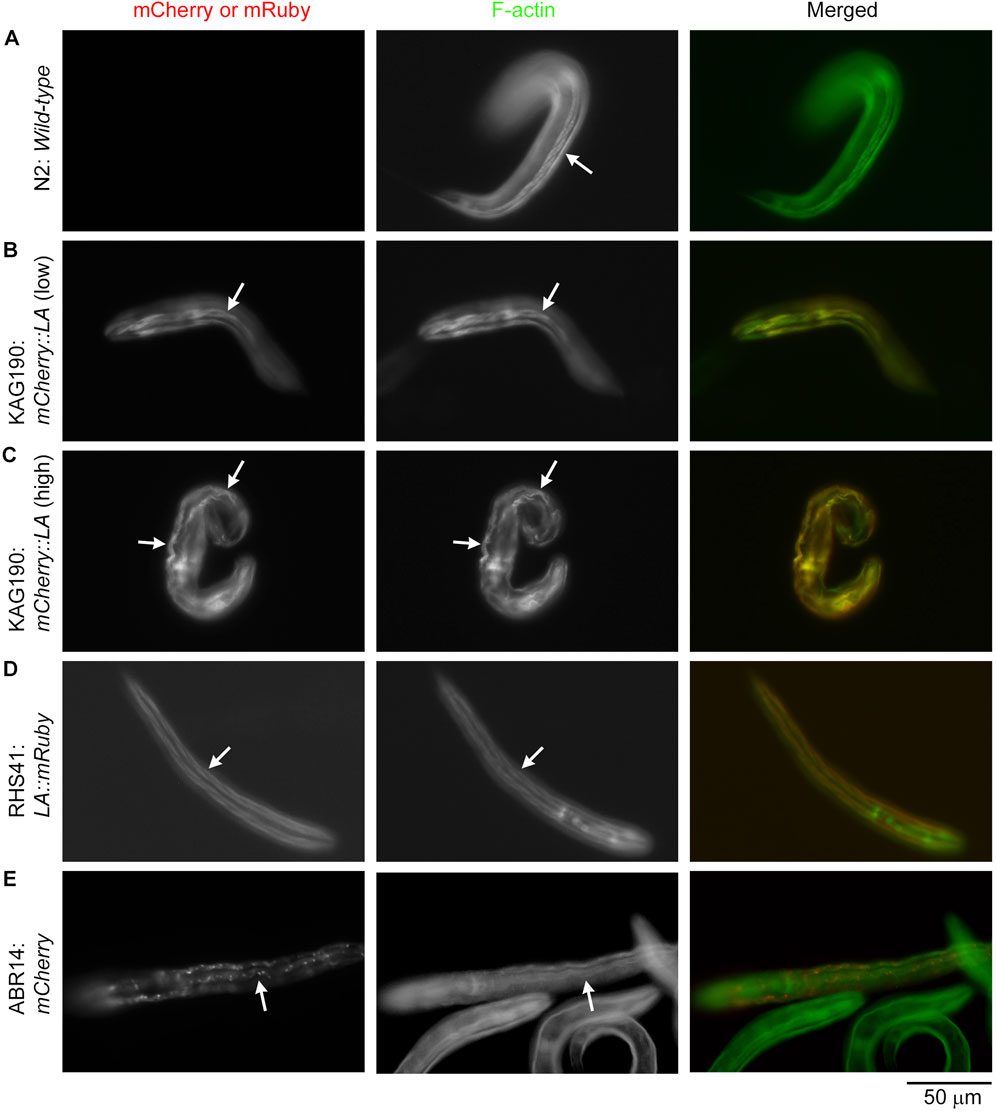
Figure 2. Overexpression of mCherry::Lifeact in the C. elegans body wall muscle causes severe disorganization of actin filaments in L1 larvae. L1 larvae of the indicated strains (Lifeact is abbreviated to LA) were stained with Alexa 488-phalloidin to visualize F-actin. Strains were N2 (A), KAG190 [(B): low expression; (C): high expression], RHS41 (D), and ABR14 (E). Images shown are mCherry or mRuby (left), F-actin (middle), and merged (right: mCherry or mRuby in red; F-actin in green). Arrows indicate representative regions of the body wall muscle. Bar, 50 μm.
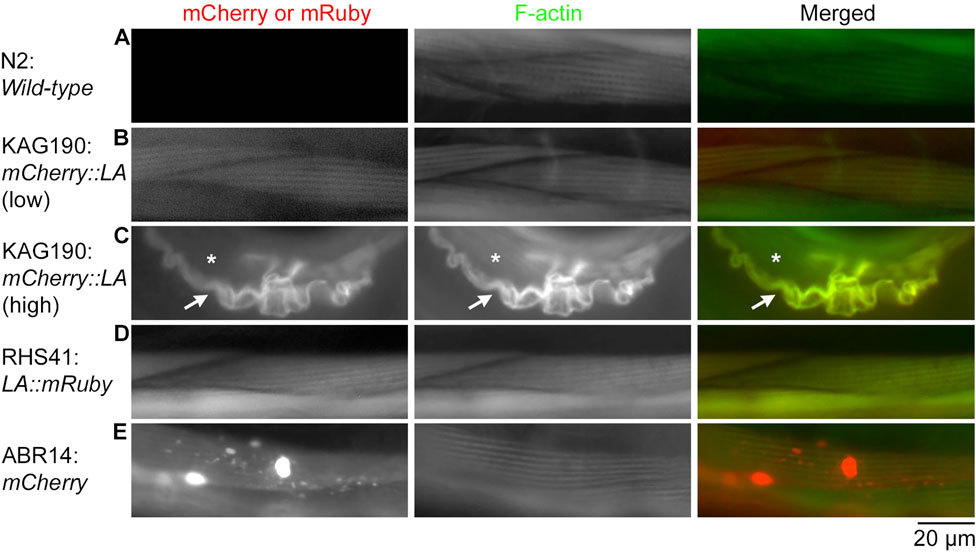
Figure 3. Overexpression of mCherry::Lifeact in the C. elegans body wall muscle causes severe disorganization of actin filaments in adult worms. Adult worms of the indicated strains (Lifeact is abbreviated to LA) were stained with Alexa 488-phalloidin to visualize F-actin, and their body wall muscle cells were examined. Strains were N2 (A), KAG190 [(B): low expression; (C): high expression], RHS41 (D), and ABR14 (E). Images shown are mCherry or mRuby (left), F-actin (middle), and merged (right: mCherry or mRuby in red; F-actin in green). In panel C, arrows indicate a muscle cell that overexpressed mCherry::Lifeact, and asterisks indicate a neighboring muscle cell that did not express mCherry::Lifeact and had intact sarcomeric F-actin. Bar, 20 μm.
Similarly, in adult worms, low-level expression of mCherry::Lifeact from the extrachromosomal array in KAG190 (Figure 3B) and Lifeact::mRuby from a single-copy transgene in RHS41 (Figure 3D) labeled F-actin in a striated sarcomeric pattern without disturbing the structure (compare with wild-type in Figure 3A). However, when mCherry::Lifeact was expressed at high levels, abnormally thick F-actin bundles containing mCherry::Lifeact were formed with no detectable sarcomere formation (Figure 3C). Because C. elegans body wall muscle cells are mononucleated and do not fuse, F-actin bundles are present only in the cell overexpressing mCherry::Lifeact (Figure 3C, arrow) but not in the neighboring cell without the expression of mCherry::Lifeact (Figure 3C, asterisk). When mCherry alone was overexpressed, aggregates of mCherry were formed with no detectable alterations in sarcomeric actin organization (Figure 3E). Therefore, overexpression of Lifeact in striated muscle cells is highly detrimental to the sarcomeric actin assembly, suggesting strongly that the muscle defects are the major cause of lethality.
Discussion
Overexpression of mCherry::Lifeact in C. elegans body wall muscle caused severe disorganization of sarcomeric actin filaments and embryonic/larval lethality. This is consistent with the previous reports that severe defects in the sarcomeres of body wall muscle cause lethality at a late embryonic or L1 larval stage with defective body elongation (Waterston, 1989; Barstead and Waterston, 1991; Williams and Waterston, 1994; Ono et al., 2020). Since contraction of the body wall muscle is required for the body-axis elongation of C. elegans embryos (Zhang et al., 2011; Lardennois et al., 2019), severe sarcomere disorganization by Lifeact overexpression may prevent muscle contraction and affect embryonic and larval development. Although effects of overexpressed Lifeact on sarcomere components other than F-actin were not analyzed in this study, our previous studies indicate that disorganization of sarcomeric actin filaments causes relatively weak structural alterations in dense bodies and myosin thick filaments (Ono et al., 2003; Ono et al., 2020). Within the muscle cells, an antagonistic effect of Lifeact on cofilin in actin filament severing (Courtemanche et al., 2016) is expected to be impactful to sarcomere disorganization because UNC-60B, a muscle-specific actin depolymerizing factor/cofilin isoform, is essential for the assembly of sarcomeric actin filaments in the C. elegans body wall muscle (Ono et al., 1999; Ono et al., 2003). A recent structural study also showed that the Lifeact-binding site on F-actin overlaps with binding sites for cofilin and myosin (Belyy et al., 2020), suggesting that actomyosin contractility may be affected by Lifeact.
Aggregation of overexpressed mCherry may contribute to enhancing the artificial effects of Lifeact. Aggregate formation of mCherry has also been reported in cultured mammalian neurons (Ning et al., 2022). When multiple mCherry::Lifeact molecules associate, actin filaments can be bundled together and then recruit additional mCherry::Lifeact. Whether mRuby is also prone to aggregation remains unknown. In addition to the stabilizing effects of Lifeact on F-actin (Courtemanche et al., 2016), multimerization or aggregation of the tagged fluorescent protein is a factor that needs to be considered to avoid unfavorable effects on the actin cytoskeleton. Nonetheless, despite the fact that overexpressed Lifeact causes disorganization of the actin cytoskeleton, properly managed Lifeact is still considered one of the most useful probes for actin filaments, given that artificial effects of Lifeact and/or a fluorescent protein are carefully assessed.
Materials and methods
C. elegans strains and culture
The worms were cultured at 20°C, following standard methods using Escherichia coli OP50 for feeding (Stiernagle, 2006). Wild-type N2, ABR14 shEx34 [myo-3p::mCherry] (Han et al., 2017), and RHS41 uthSi7 [myo-3p::Lifeact::mRuby] (Higuchi-Sanabria et al., 2018) were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). KAG190 Ex[myo-3p::mCherry::Lifeact; myo-2p::mCherry] (Brouilly et al., 2015) was obtained from Kathrin Gieseler (University of Lyon, Villeurbanne, France) and maintained by picking mCherry-positive worms.
Quantification of embryonic and larval lethality
Three to five gravid adult hermaphrodites were placed in an E. coli OP50-seeded 60-mm Nematode Growth Medium plate for 24 h and were let to lay eggs. After removing the adult worms, the remaining embryos and larvae on the plate were cultured for an additional 24 h and scored for dead or arrested embryos and larvae and healthy larvae (83–219 animals per plate, n = 5). For transgenic strains, only animals with the expression of the fluorescent proteins were counted. Since L1 larvae normally hatch ∼9 h after eggs are laid on the plate (Sulston et al., 1983), healthy worms are normally developed into the L2, L3, or L4 stage at the time of quantification. Data were analyzed by one-way analysis of variance with the Holm–Sidak method for pairwise comparison using SigmaPlot 15.0 (Grafiti LLC).
Fluorescence microscopy
Staining of whole worms with Alexa 488-phalloidin (Thermo Fisher Scientific, catalog # A12379) was performed, as described previously (Ono, 2001; Ono, 2022). Samples were observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope (Nikon Instruments, Tokyo, Japan) with a CFI Plan Fluor ELWD 40× (dry; NA 0.60) objective. Images were captured using a Hamamatsu ORCA Flash 4.0 LT sCMOS camera (Hamamatsu Photonics, Shizuoka, Japan) and processed by NIS-Elements (Nikon Instruments) and Adobe Photoshop CS3 (Adobe).
Data availability statement
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding author/s.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SO: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, validation, visualization, writing–original draft, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by a grant from the National Institutes of Health (R01-GM144563) to SO.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aizawa, H., Sameshima, M., and Yahara, I. (1997). A green fluorescent protein-actin fusion protein dominantly inhibits cytokinesis, cell spreading, and locomotion in Dictyostelium. Cell. Struct. Funct. 22 (3), 335–345. doi:10.1247/csf.22.335
Alvarez, Y. D., van der Spuy, M., Wang, J. X., Noordstra, I., Tan, S. Z., Carroll, M., et al. (2024). A Lifeact-EGFP quail for studying actin dynamics in vivo. J. Cell. Biol. 223 (9), e202404066. doi:10.1083/jcb.202404066
Barstead, R. J., and Waterston, R. H. (1991). Vinculin is essential for muscle function in the nematode. J. Cell. Biol. 114(4), 715–724. doi:10.1083/jcb.114.4.715
Belin, B. J., Goins, L. M., and Mullins, R. D. (2014). Comparative analysis of tools for live cell imaging of actin network architecture. Bioarchitecture 4 (6), 189–202. doi:10.1080/19490992.2014.1047714
Belyy, A., Merino, F., Sitsel, O., and Raunser, S. (2020). Structure of the Lifeact-F-actin complex. PLoS Biol. 18 (11), e3000925. doi:10.1371/journal.pbio.3000925
Benian, G. M., and Epstein, H. F. (2011). Caenorhabditis elegans muscle: a genetic and molecular model for protein interactions in the heart. Circ. Res. 109 (9), 1082–1095. doi:10.1161/CIRCRESAHA.110.237685
Brouilly, N., Lecroisey, C., Martin, E., Pierson, L., Mariol, M. C., Qadota, H., et al. (2015). Ultra-structural time-course study in the C. elegans model for Duchenne muscular dystrophy highlights a crucial role for sarcomere-anchoring structures and sarcolemma integrity in the earliest steps of the muscle degeneration process. Hum. Mol. Genet. 24 (22), 6428–6445. doi:10.1093/hmg/ddv353
Chen, Q., Nag, S., and Pollard, T. D. (2012). Formins filter modified actin subunits during processive elongation. J. Struct. Biol. 177 (1), 32–39. doi:10.1016/j.jsb.2011.10.005
Courtemanche, N., Pollard, T. D., and Chen, Q. (2016). Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat. Cell. Biol. 18 (6), 676–683. doi:10.1038/ncb3351
Doyle, T., and Botstein, D. (1996). Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. U. S. A. 93 (9), 3886–3891. doi:10.1073/pnas.93.9.3886
Du, J., Fan, Y. L., Chen, T. L., and Feng, X. Q. (2015). Lifeact and Utr230 induce distinct actin assemblies in cell nuclei. Cytoskelet. Hob. 72 (11), 570–575. doi:10.1002/cm.21262
Flores, L. R., Keeling, M. C., Zhang, X., Sliogeryte, K., and Gavara, N. (2019). Lifeact-GFP alters F-actin organization, cellular morphology and biophysical behaviour. Sci. Rep. 9 (1), 3241. doi:10.1038/s41598-019-40092-w
Ghosh, S. R., and Hope, I. A. (2010). Determination of the mobility of novel and established Caenorhabditis elegans sarcomeric proteins in vivo. Eur. J. Cell. Biol. 89 (6), 437–448. doi:10.1016/j.ejcb.2009.11.027
Han, S., Schroeder, E. A., Silva-Garcia, C. G., Hebestreit, K., Mair, W. B., and Brunet, A. (2017). Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544 (7649), 185–190. doi:10.1038/nature21686
Hayashi, Y., Ono, K., and Ono, S. (2019). Mutations in Caenorhabditis elegans actin, which are equivalent to human cardiomyopathy mutations, cause abnormal actin aggregation in nematode striated muscle. F1000Res 8, 279. doi:10.12688/f1000research.18476.1
Higuchi-Sanabria, R., Paul, J. W., Durieux, J., Benitez, C., Frankino, P. A., Tronnes, S. U., et al. (2018). Spatial regulation of the actin cytoskeleton by HSF-1 during aging. Mol. Biol. Cell. 29 (21), 2522–2527. doi:10.1091/mbc.E18-06-0362
Lardennois, A., Pasti, G., Ferraro, T., Llense, F., Mahou, P., Pontabry, J., et al. (2019). An actin-based viscoplastic lock ensures progressive body-axis elongation. Nature 573 (7773), 266–270. doi:10.1038/s41586-019-1509-4
Melak, M., Plessner, M., and Grosse, R. (2017). Actin visualization at a glance. J. Cell. Sci. 130 (3), 525–530. doi:10.1242/jcs.189068
Mello, C., and Fire, A. (1995). Chapter 19 DNA transformation. Methods Cell. Biol. 48, 451–482. doi:10.1016/S0091-679X(08)61399-0
Mello, C. C., Kramer, J. M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10 (12), 3959–3970. doi:10.1002/j.1460-2075.1991.tb04966.x
Miller, D. M., Ortiz, I., Berliner, G. C., and Epstein, H. F. (1983). Differential localization of two myosins within nematode thick filaments. Cell. 34 (2), 477–490. doi:10.1016/0092-8674(83)90381-1
Munsie, L. N., Caron, N., Desmond, C. R., and Truant, R. (2009). Lifeact cannot visualize some forms of stress-induced twisted F-actin. Nat. Methods 6 (5), 317. doi:10.1038/nmeth0509-317
Ning, L., Geng, Y., Lovett-Barron, M., Niu, X., Deng, M., Wang, L., et al. (2022). A bright, nontoxic, and non-aggregating red fluorescent protein for long-term labeling of fine structures in neurons. Front. Cell. Dev. Biol. 10, 893468. doi:10.3389/fcell.2022.893468
Okkema, P. G., Harrison, S. W., Plunger, V., Aryana, A., and Fire, A. (1993). Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135 (2), 385–404. doi:10.1093/genetics/135.2.385
Ono, K., Parast, M., Alberico, C., Benian, G. M., and Ono, S. (2003). Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell. Sci. 116 (10), 2073–2085. doi:10.1242/jcs.00421
Ono, K., Qin, Z., Johnsen, R. C., Baillie, D. L., and Ono, S. (2020). Kettin, the large actin-binding protein with multiple immunoglobulin domains, is essential for sarcomeric actin assembly and larval development in Caenorhabditis elegans. FEBS J. 287 (4), 659–670. doi:10.1111/febs.15039
Ono, S. (2001). The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell. Biol. 152 (6), 1313–1319. doi:10.1083/jcb.152.6.1313
Ono, S. (2010). Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskelet. Hob. 67 (11), 677–692. doi:10.1002/cm.20476
Ono, S. (2014). Regulation of structure and function of sarcomeric actin filaments in striated muscle of the nematode Caenorhabditis elegans. Anat. Rec. Hob. 297 (9), 1548–1559. doi:10.1002/ar.22965
Ono, S. (2022). Imaging of actin cytoskeleton in the nematode Caenorhabditis elegans. Methods Mol. Biol. 2364, 149–158. doi:10.1007/978-1-0716-1661-1_7
Ono, S. (2024). Segregated localization of two calponin-related proteins within sarcomeric thin filaments in Caenorhabditis elegans striated muscle. Cytoskelet. Hob. 81 (2-3), 127–140. doi:10.1002/cm.21794
Ono, S., Baillie, D. L., and Benian, G. M. (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell. Biol. 145 (3), 491–502. doi:10.1083/jcb.145.3.491
Ono, S., and Ono, K. (2020). Two Caenorhabditis elegans calponin-related proteins have overlapping functions that maintain cytoskeletal integrity and are essential for reproduction. J. Biol. Chem. 295 (34), 12014–12027. doi:10.1074/jbc.RA120.014133
Pollard, T. D. (2016). Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8 (8), a018226. doi:10.1101/cshperspect.a018226
Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5 (7), 605–607. doi:10.1038/nmeth.1220
Spracklen, A. J., Fagan, T. N., Lovander, K. E., and Tootle, T. L. (2014). The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev. Biol. 393 (2), 209–226. doi:10.1016/j.ydbio.2014.06.022
Sulston, J. E., Schierenberg, E., White, J. G., and Thomson, J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100 (1), 64–119. doi:10.1016/0012-1606(83)90201-4
van der Honing, H. S., van Bezouwen, L. S., Emons, A. M., and Ketelaar, T. (2011). High expression of Lifeact in Arabidopsis thaliana reduces dynamic reorganization of actin filaments but does not affect plant development. Cytoskelet. Hob. 68 (10), 578–587. doi:10.1002/cm.20534
Waterston, R. H. (1989). The minor myosin heavy chain, mhcA, of Caenorhabditis elegans is necessary for the initiation of thick filament assembly. EMBO J. 8 (11), 3429–3436. doi:10.1002/j.1460-2075.1989.tb08507.x
Williams, B. D., and Waterston, R. H. (1994). Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell. Biol. 124(4), 475–490. doi:10.1083/jcb.124.4.475
Wirshing, A. C. E., and Goode, B. L. (2024). Improved tools for live imaging of F-actin structures in yeast. Mol. Biol. Cell. 35 (9), mr7. doi:10.1091/mbc.E24-05-0212-T
Xu, R., and Du, S. (2021). Overexpression of Lifeact-GFP disrupts F-actin organization in cardiomyocytes and impairs cardiac function. Front. Cell Dev. Biol. 9, 746818. doi:10.3389/fcell.2021.746818
Keywords: actin, muscle, sarcomere, thin filaments, Caenorhabditis elegans
Citation: Ono S (2024) Overexpression of Lifeact in the C. elegans body wall muscle causes sarcomere disorganization and embryonic or larval lethality. Front. Cell Dev. Biol. 12:1504980. doi: 10.3389/fcell.2024.1504980
Received: 01 October 2024; Accepted: 25 October 2024;
Published: 13 November 2024.
Edited by:
Lei-Miao Yin, Shanghai University of Traditional Chinese Medicine, ChinaReviewed by:
Jörg Großhans, Philipps University, GermanyWorawit Suphamungmee, Mahidol University, Thailand
Copyright © 2024 Ono. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shoichiro Ono, c29ub0BlbW9yeS5lZHU=
 Shoichiro Ono
Shoichiro Ono