- Department of Cell and Cancer Biology, College of Medicine and Life Sciences, The University of Toledo, Toledo, OH, United States
Heat Shock Factor 1 (HSF1) is a major transcriptional factor regulating the heat shock response and has become a potential target for overcoming cancer chemoresistance. This review comprehensively examines HSF1’s role in chemoresistance and its potential as a therapeutic target in cancer. We explore the complex, intricate mechanism that regulates the activation of HSF1, HSF1’s function in promoting resistance to chemotherapy, and the strategies used to manipulate HSF1 for therapeutic benefit. In addition, we discuss emerging research implicating HSF1’s roles in autophagy, apoptosis, DNA damage repair, drug efflux, and thus chemoresistance. This article highlights the significance of HSF1 in cancer chemoresistance and its potential as a target for enhancing cancer treatment efficacy.
1 Introduction
In 2024, roughly two million people are estimated to be diagnosed with cancer, with 611,720 cancer-related deaths in the United States (Siegel et al., 2024). Approximately 80%–90% of cancer-related deaths are attributed to the development of chemoresistance in responders (Ramos et al., 2021). Chemoresistance is responsible for most relapses, contributing to metastasis and poor rate of survival in patients (Ramos et al., 2021). Chemoresistance often leads to the need for more intensive chemotherapy regimens in second and later lines of cancer treatment as the initial therapies become less effective. The action of chemotherapies typically involves cellular uptake, intracellular activation, acting on the target site, and ultimately inducing cell death (Florea and Busselberg, 2011; Tilsed et al., 2022). Chemoresistance can occur at any of these stages, leading to failure of drug response. Cancers have been shown to develop resistance to various chemotherapies rapidly (Vasan et al., 2019; Ramos et al., 2021; Khan et al., 2024). For example, the resistance to the paclitaxel and 5-fluorouracil combination treatment was observed within 2 years since it being introduced to clinical use (Harris et al., 2006; Szakacs et al., 2006; Murray et al., 2012; Vulsteke et al., 2014). With the advancement in new targeted therapies and drug development, novel drugs show efficacy in many cancer types; however, these treated tumors often develop resistance to these drugs over time, making resistance a significant barrier to successful cancer treatment (Vasan et al., 2019; Khan et al., 2024). Hence, it is no surprise that chemoresistance contributes to about 80%–90% of cancer-related mortality.
Cancer cells utilize complex interplays between innate, intrinsic, and acquired factors to develop resistance to an administered drug during tumor development (Dzobo et al., 2018). In general, intrinsic chemoresistance is due to cell heterogeneity, whereas acquired resistance develops from chemotherapy-induced changes. The intrinsic chemoresistance of a tumor is evident when there is a lack of an initial response to the administered drug. The existence of intrinsic chemoresistance is largely due to the heterogeneity of the cancer cell population, especially the presence of cancer stem cells and mutations in key genes involved in cellular homeostasis and metabolism. These mutations can confer resistance by altering metabolic pathways, enhancing survival signaling, and impairing apoptotic response, thereby allowing cancer cells to survive and proliferate despite chemotherapy (Cancer Genome Atlas Research, 2014; Giordano, 2014; Hasan et al., 2018). Intrinsic chemoresistance is influenced by various factors, including the activation of signaling pathways like phosphoinositide 3-kinase (PI3K)/Akt, hedgehog, nuclear factor-κB (NFkB), and mitogen-activated protein kinase (MAPK), which have been shown to confer resistance to chemotherapeutic agents such as gemcitabine in pancreatic cancer (Rajabpour et al., 2017; He et al., 2021; Guo et al., 2024). The acquired chemoresistance only develops as a response to chemotherapy treatment and involves drug target mutations, tumor microenvironment modifications, and epigenetic changes such as methylation, acetylation, and microRNA (miRNA) expression survival (Mansoori et al., 2017; Emran et al., 2022). These alterations modulate upstream/downstream intracellular cell growth signaling such as MAPK and mammalian target of rapamycin (mTOR), modify the cell cycle checkpoints, inhibit apoptosis, and alter DNA replication, thereby enhancing cancer cell growth.
In addition to the mechanisms mentioned above that contribute to chemoresistance, accumulating evidence shows that stress response pathways are exploited by cancer cells to further support the chemoresistance processes (Vasan et al., 2019; Ramos et al., 2021; Khan et al., 2024). Stress responses, such as those triggered by oxidative stress, hypoxia, and DNA damage, initiate the survival and adaptive responses of cancer cells (Fulda et al., 2010; Dai et al., 2012; Chen and Xie, 2018). Stress responses also play crucial roles in the development and enhancement of resistance to chemotherapy. By inducting cytoprotective responses such as autophagy (Suh et al., 2012), the expression of heat shock proteins (HSPs) (Chatterjee and Burns, 2017), and the unfolded protein response, cancer cells can mitigate the effects of therapeutic stress, thereby promoting their continued growth and survival despite the treatment.
A better understanding of these chemoresistance mechanisms is essential for developing strategies to overcome drug resistance and improve the efficacy of cancer treatments. Recently, heat shock factor 1 (HSF1) has emerged as an intriguing player in tumorigenesis and chemoresistance (Alasady and Mendillo, 2020; Prince et al., 2020; Zhang et al., 2021). HSF1 is originally characterized as a transcription factor for the expression of HSPs responsible for the initiation of the proteotoxic stress response (Alasady and Mendillo, 2020; Zhang et al., 2021). This cellular mechanism safeguards cells from protein misfolding and aggregation under stress conditions (Alasady and Mendillo, 2020; Zhang et al., 2021; Ghai et al., 2024). HSF1 has since also been recognized for its pro-oncogenic properties, contributing to cancer initiation, progression, and chemoresistance (Kijima et al., 2019; Lee et al., 2021; Cyran and Zhitkovich, 2022; Gumilar et al., 2023).
The role of HSF1 in cancer has been extensively studied. HSF1 is downstream of the Kirsten rat sarcoma viral oncogene homolog (KRAS) signaling (Tang et al., 2015; Dai and Sampson, 2016). The KRAS gene is one of the most frequently mutated oncogenes in human cancers, with mutations commonly identified in approximately 90%–95% of pancreatic ductal adenocarcinoma (PDAC), 40% of colorectal cancers (CRC), and 25%–30% of non-small cell lung cancers (NSCLC) (Bailey et al., 2016; Zhu et al., 2021; Reita et al., 2022; Nusrat et al., 2024). Cancers with KRAS mutations are particularly lethal due to their role in promoting rapid cell proliferation and resulting in highly aggressive tumor phenotype (Huang et al., 2021). Recent research suggests that by regulating autophagy, apoptosis, DNA damage repair, or drug efflux mechanism, cancer cells can develop resistance to chemotherapy, thereby promoting tumor survival and progression (Dai and Sampson, 2016; Cyran and Zhitkovich, 2022; Gumilar et al., 2023). In this review, we will discuss the role of HSF1 in chemoresistance and its potential as a therapeutic target in tumorigenesis.
2 HSF1 in tumorigenesis
2.1 HSF1 is a key transcription factor in cancer development
When cells undergo proteotoxic stress from exposure to environmental factors such as heat shock, an adaptive cytoprotective mechanism, known as the heat shock response (HSR) or proteotoxic stress response (PSR), is activated. The HSR maintains the protein quality and prevents protein aggregation in the cells, ensuring the proteome homeostasis (proteostasis) in the cells (Lindquist, 1986). Immediately after the accumulation of unfolded proteins, HSF1, a master regulator of proteotoxic stress, is activated and, in turn, activates the transcription of genes encoding HSPs (Ghai et al., 2023). HSPs are molecular chaperones of the cells that enable the proper folding of the unfolded proteins and the degrading of the unrequired proteins through ubiquitination (Bukau et al., 2006; Gidalevitz et al., 2011; Dai and Sampson, 2016) (Figure 1).
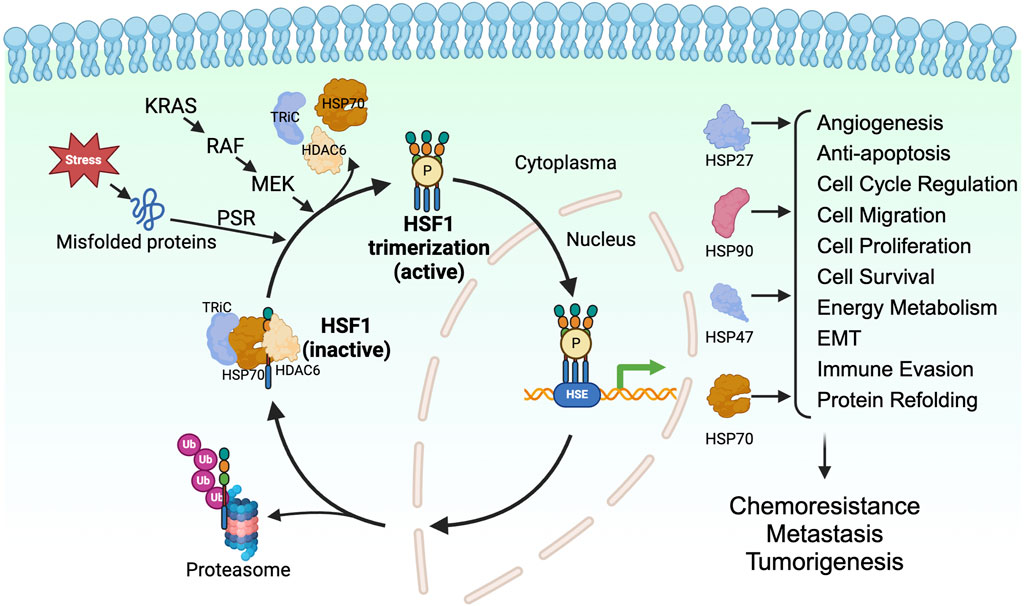
Figure 1. HSF1 is a transcriptional factor for HSPs in tumorigenesis. HSF1 is a downstream effector of KRAS-RAF-MEK signaling. HSF1 is activated upon proteotoxic stress in the cells, which allows its trimerization, phosphorylation, and translocation to the nucleus. In the nucleus, HSF1 binds to heat shock elements and allows expression of HSP27, HSP47, HSP70, and HSP90, which directly and indirectly affects tumor progression. EMT, epithelial-mesenchymal transition; HDAC6, histone deacetylase 6; HSE, heat shock element; HSF1, heat shock factor 1; HSP, heat shock protein; KRAS, Kirsten rat sarcoma viral oncogene homolog; MEK, mitogen-activated protein kinase kinase; P, phosphorylation; PSR, proteotoxic stress response; RAF, rapidly accelerated fibrosarcoma; TRiC, T-complex protein ring complex; Ub, ubiquitin.
HSF1 is a transcription factor and is an evolutionarily conserved member of the HSF family proteins (Wu, 1995; Anckar and Sistonen, 2011). HSF1 comprises the N-terminal helix-turn-helix DNA-binding domain, the oligomerization domain, and the C-terminal transactivation domain. The N-terminal DNA-binding domain of HSF1 binds to the inverted repeats of the heat shock elements (HSE) (Buchwalter et al., 2004). The oligomerization domain of HSF1 comprises four leucine zipper repeats, allowing HSF1 to trimerize and become an active transcription factor (Neudegger et al., 2016; Gomez-Pastor et al., 2018). The C-terminal transactivation domain of HSF1 is essential for the elongation process of transcription (Vihervaara et al., 2017). In cells under normal physiological conditions, HSF1 is present as a monomer and is sequestered by the HSP70, T-complex protein ring complex (TRiC), and histone deacetylase 6 (HDAC6) complex in the cytoplasm (Neef et al., 2014). Misfolded proteins resulting from proteotoxic stresses induced by various stimuli compete with HSF1 for binding to the chaperones, allowing the release of HSF1 from the HSP70/HDAC6 complex (Shi et al., 1998; Gomez-Pastor et al., 2018; Kijima et al., 2018). Along with the proteotoxic stress, post-translational modifications such as phosphorylation, SUMOylation, and acetylation are known to regulate the activation of HSF1. Phosphorylation of HSF1 at Ser326 is a commonly used marker for the activation of HSF1. In addition, phosphorylation of HSF1 on Ser230 and Ser320 positively regulates the HSF1 activity (Holmberg et al., 2001; Guettouche et al., 2005; Zhang et al., 2011; Chou et al., 2012a) whereas phosphorylation on Ser121 negatively regulates the activity of HSF1 (Dai et al., 2015; Swan and Sistonen, 2015). The acetylation of HSF1 on K298 prevents the proteosome-dependent degradation of the HSF1, thus increases the stability of HSF1 (Wu, 1995; Anckar and Sistonen, 2011). In contrast, the SUMOylation of HSF1 at K298 inhibits the activity of HSF1 (Wu, 1995; Hietakangas et al., 2003; Anckar and Sistonen, 2011). The activated HSF1 can trans-localize to the nucleus and regulate the transcription of genes encoding chaperone proteins. After the restoration of proteostasis, the HSP90 binds to the HSF1, leading to the inactivation of HSF1 (Gomez-Pastor et al., 2018; Kijima et al., 2018) (Figure 1).
Several studies using different murine models have shown HSF1 as a pro-oncogenic factor. A pioneering work performed in skin carcinogenesis mouse models shows reduced oncogenic Ras-induced tumor formation upon the loss of Hsf1 (Dai et al., 2007). Hsf1 deletion also leads to reduced tumor burden in other mouse models, including chemical carcinogenesis-driven hepatocellular carcinoma, mammary tumorigenesis resulting from p53 and neurofibromatosis type 1 (Nf1) loss, and lymphomas caused by p53-deficiency (Min et al., 2007; Jin et al., 2011; Xi et al., 2012). Along with that, increased HSF1 expression has been found in a wide range of human cancers, including cervix, colon, breast, lung, liver, pancreatic, and prostate carcinomas (Dudeja et al., 2011; Santagata et al., 2011; Fang et al., 2012; Mendillo et al., 2012). These reports indicate that HSF1 plays a crucial role in mediating tumorigenesis.
The increased rate of protein synthesis in cells allows for a high number of unfolded or misfolded proteins. Similarly, inflammation and stresses, such as hypoxia, activate HSF1 in the cells (Luo et al., 2009; Oromendia et al., 2012; Santagata et al., 2013). Besides this general activation of HSF1, cancer cells can constitutively activate HSF1, thus allowing its cytoprotective effect through various mechanisms. The mammalian target of rapamycin complex 1 (mTORC1) activated by PI3K/AKT phosphorylates HSF1 at Ser326 and accentuates the HSF1 transcriptional activity (Chou et al., 2012b). The HSF1 activation promotes cell proliferation while circumventing the cells' apoptosis and senescence (Meng et al., 2010; Carpenter et al., 2015). Besides, the Ras-MEK signaling phosphorylates HSF1 at Ser326 to activate HSF1’s transcriptional activity (Tang et al., 2015). In addition, the metabolic sensor AMP-activated protein kinase (AMPK) is reported to suppress the activity of HSF1 through phosphorylating the HSF1 Ser121 (Dai et al., 2015; Swan and Sistonen, 2015). In PDAC, the loss of AMPK allows the activation of HSF1, promoting the invasion and migration of PDAC (Chen et al., 2017). In summary, the activity of HSF1 is regulated by multiple upstream kinases and regulatory factors.
HSF1 activation in cancers thus is not only in response to different stresses but also through a wide array of other mechanisms, including those driven by oncogenic signaling and those that elevate HSF1 expression levels. This diverse range of HSF1 activation pathways collectively accounts for the extensive activation and functional roles of HSF1 in tumorigenesis.
2.2 HSF1-mediated upregulation of HSPs
Because HSF1 is a major transcriptional factor for HSPs, we will discuss the role of HSF1 in the chemoresistance mechanism. HSPs, including HSP70, HSP27, HSP90, HSP40, and HSP60, are crucial players in cancer chemoresistance (Zhang et al., 2021). These molecular chaperones facilitate protein folding, maintain protein stability, and, in some conditions, promote the degradation of misfolded or aggregated proteins, contributing to the maintenance of proteostasis within cancer cells. For example, HSP27 regulates the Salvador–Warts–Hippo pathway (Hippo pathway) known to control tumor progression and cancer stem cell reprogramming (Vahid et al., 2016). HSP27 forms multimeric complexes to stabilize denatured and aggregated proteins, making them functional (Vahid et al., 2016). HSP40 aids proper protein folding, translation, translocation, and degradation (Takashima et al., 2018). Several HSP40 family members are highly expressed in various types of human cancer, including colorectal, gastric, and KRAS-mutated lung cancers. Concerning colorectal cancer, HSP40 has demonstrated metastatic promoting behavior (Sterrenberg et al., 2011; He et al., 2015; Yamashita et al., 2016; Yang et al., 2020). Elevated levels of HSPs can promote chemoresistance by aiding the proper folding of oncoproteins, thereby sustaining malignant processes. HSP90 is overexpressed in multiple cancer types and regulates the stability, activity, maturation, and proteolytic degradation of various oncogenic kinases. HSP90 interacts with its substrates through its N-terminal ATPase domain, which is enhanced by the binding of co-chaperones such as HSP70 (Chiosis et al., 2004; Workman et al., 2007; Chatterjee et al., 2016).
HSPs also directly regulate kinases. For example, overexpressed HSP90AA1 upon chemotherapy dissociates phosphorylated AKT and c-Jun N-terminal kinase (JNK), inducing protective autophagy while inhibiting apoptosis and hence contributing to chemoresistance (Xiao et al., 2018). This highlights the important role of HSPs in chemoresistance and underscores how highly regulated cellular pathways are hijacked by cancer cells to survive stress and chemotherapy. Furthermore, cancer cells exploit external signaling by binding extracellular HSP90α to lipoprotein receptor-related protein 1 (LRP1), which has been shown to promote PDAC metastasis via AKT activation. LRP1 is associated with poor PDAC patient survival and LRP1 silencing increases the susceptibility of PDAC cells to doxorubicin and gemcitabine (Xue et al., 2022). Disruption of HSP47 in PDAC cells increases intracellular reactive oxygen species (ROS) and subsequent Ca2+ levels, resulting in the activation of the caspase-12/caspase-9/caspase-3 axis, which may sensitize cells to chemotherapy (Yoneda et al., 2021). Inhibition of HSP27 in human colon cancer cells reduces their acquired resistance to 5-fluorouracil (Asada et al., 2021). This suggests that HSPs play a role in protecting cells from ROS-induced damage and cellular stress. There is still a significant gap in our understanding of how HSPs modulate oxidative stress and provide protection against ROS, further investigation into these mechanisms could shed light on their contributions to chemoresistance.
In summary, HSF1-mediated regulation of HSPs provides diverse mechanisms of chemoresistance by preserving proteomic integrity and modulating cell death pathways. The upregulation of HSPs through HSF1 thus promotes tumor growth and contributes to chemoresistance (Figure 1). Given this understanding, targeting HSF1 in various cancers appears to be a promising therapeutic strategy. We will further discuss the role of HSF1 in chemotherapy and its effect on chemoresistance. The combination of HSF1 inhibition with existing chemotherapeutic agents may represent a potential avenue for enhancing treatment efficacy and warrants further investigation. We have summarized recent studies and the current mechanisms on the role of HSF1 in driving chemoresistance, which may provide new avenues for targeting HSF1 in various cancers.
3 HSF1 as a regulator of chemoresistance
3.1 HSF1 and autophagy
PSR regulates both the ubiquitin-proteasome system (UPS) and autophagy for the degradation of dysfunctional and toxic proteins to maintain proteostasis in response to stress. HSF1 induces autophagy by acting as a transcription factor of multiple autophagy-related genes (ATG), such as ATG5, ATG7, and ATG12. HSF1 also induces the autophagy marker sequestosome 1 (SQSTM1)/p62 activity through the regulation of its upstream kinases (Watanabe et al., 2017). However, the autophagy flux and the p62 level are increased in HSF1-deficient mice (Dayalan Naidu et al., 2017). This suggests that the relationship between HSF1 and autophagy may be context dependent.
In the past decade, various studies have demonstrated that autophagy plays an important role in conferring chemoresistance. ATGs, such as ATG3, ATG5, ATG6 (Beclin-1), ATG7, and ATG12, and autophagy markers, such as microtubule-associated protein 1A/1B-light chain 3 (LC3) and p62, are the major regulators of autophagy and chemoresistance (Li et al., 2019). ATG3 facilitates the conversion of LC3-I to LC3-II and promotes endoplasmic reticulum (ER) stress. However, the downregulation of ATG3 increases the sensitivity to erlotinib in the erlotinib-resistant lung cancer cell lines (Lee and Wu, 2012). ATG5 normally participates in the elongation of the autophagosome membrane. However, the interaction of the long non-coding RNA (lncRNA) gallbladder cancer drug resistance-associated lncRNA1 with phosphoglycerate kinase 1 in gallbladder cancer cells prevents ATG5 degradation and induces the ATG5-ATG12 complex formation, promoting autophagy and doxorubicin resistance (Cai et al., 2019). Additionally, suppression of ATG6, a crucial protein in the ATG6 and Vps-34 complex that promotes autophagosome formation, downregulates HER2 expression and enhances tamoxifen sensitivity in estrogen receptor-positive breast cancer cells in vitro (Gu et al., 2017). ATG7, an activator of ATG8 involved in the expansion of phagophore, confers chemoresistance in the acute myeloid leukemia cell lines against cytarabine and idarubicin treatment, knockdown of ATG7 increased apoptosis and DNA damage (Piya et al., 2016). Hence, autophagy is critical in chemoresistance, involving multiple autophagy-related genes and proteins (Debnath et al., 2023; Mohammed et al., 2024). This observation supports the well-established phenomenon of autophagy functioning in an oncogenic manner, contingent upon the stage of tumorigenesis. Autophagy is cytoprotective under stress conditions, such as chemotherapy, and helps maintaining the cellular homeostasis in the surviving cancer cells, such as colorectal and hepatocellular carcinoma cells (Xu et al., 2013; Lan et al., 2024). Therefore, monitoring the stages of tumorigenesis and autophagy and employing autophagy inhibitors may open new therapeutic avenues to combat chemoresistance.
HSF1 is correlated with various ATGs and their activity. Higher levels of nuclear HSF1 result in a higher H3 acetylation level and enhanced activity of the ATG7 promoter. The absence of HSF1 prevents the ATG7 promoter activity and increases the chemosensitivity to carboplatin in the MDA-MB-231 breast cancer cell line (Desai et al., 2013). Thus, inhibiting cytoprotective autophagy induced by HSF1 knockdown could provide a better therapeutic efficacy against drug resistance. In a study showing that BAG3 contributes to chemoresistance, 5-Fluorouracil and Doxorubicin-resistant triple negative breast cancer cell lines show higher expression of ATG5, LC3-II, and Beclin-1, suggesting a correlation between drug resistance and autophagy (Das et al., 2018). Importantly, inhibiting the transcriptional activity of HSF1 significantly increases the sensitivity of these resistant cell lines to the treatment. Bcl2-associated athanogene 3 (BAG3) also works together with the molecular chaperones HSP70 and HSPB8 along with the ubiquitin receptor SQSTM1/p62 to selectively direct aggregation-prone proteins for degradation via autophagy (Minoia et al., 2014; Sturner and Behl, 2017). Moreover, transmission electron microscopy revealed a distinct accumulation of small vacuoles in the cytoplasm of cells expressing BAG3, indicating enhanced autophagic flux, in HepG2 and MCF7 cells. The enhanced autophagic flux is further supported by a significant increase in LC3-II and p62 levels (Zhao et al., 2019). These studies highlight the novel HSF1-BAG3 axis, targeting which may pave the way to increase chemosensitivity to current therapies in cancer (Figure 2).
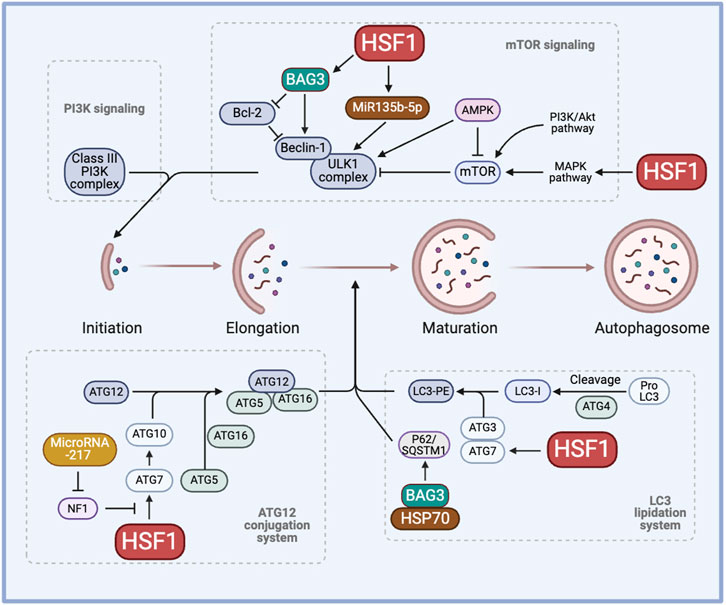
Figure 2. The role of HSF1 in cancer autophagy. HSF1 is involved in cancer autophagy by regulating the initiation process through the PI3K-mTOR signaling, the elongation process through the ATG12 conjunction system, and the maturation of autophagosome formation through the LC3 lipidation system. This regulation induces cytoprotective autophagy, hence regulating chemoresistance. AMPK, AMP-activated protein kinase; ATG, autophagy-related genes/proteins; BAG3, Bcl2-associated athanogene 3; HSP, heat shock proteins; HSF1, heat shock factor 1; LC3, microtubule-associated protein 1 light chain 3; LC3-PE, LC3-phosphatidylethanolamine conjugate; MAPK, mitogen-activated protein kinase; miR, micro RNA, mTOR, mammalian target of rapamycin; NF1, neurofibromin 1; PI3K, phosphoinositide 3-kinase; SQSTM1, sequestosome 1/p62; ULK1, Unc-51 like autophagy activating kinase 1.
HSF1 has been shown to induce miR-135b-5p overexpression, which induces protective autophagy, in colorectal cancer following oxaliplatin treatment. MiR135b-5p stabilizes Unc-51 like autophagy activating kinase 1 (ULK1) by inhibiting mitochondrial E3 ubiquitin protein ligase 1 and its E3 ubiquitin ligase activity on ULK1, thereby inducing protective autophagy and resistance against oxaliplatin (Wang et al., 2021). MiR-217 is known to regulate the HSF1-ATG7 axis by inhibiting the NF1 activity and enhancing chemoresistance (Li et al., 2023a). Other microRNAs, such as miR-107, have been shown to inhibit autophagy and decrease breast cancer progression by targeting HMGB1. Therefore, overexpressing miR-107 can serve as a strategy to hinder cancer progression whereas inhibiting miR-217 or miR-135b-5p may aid in regulating autophagy to combat drug resistance (Ai et al., 2019). Given the diverse roles of microRNAs across various cancers and stress factors, it is crucial to adopt a more mechanistic perspective to comprehend the clinical significance of microRNAs in chemoresistance (Figure 2).
The role of HSF1 and HSPs in tumor autophagy remains a topic of ongoing debate. HSF1 promotes autophagy by upregulating ATG10 through binding to the Atg10 promoter, thereby enhancing the lipidation of LC3-II. In contrast, HSF1 depletion reduces ATG10 expression and increases the production of inflammatory cytokines in lipopolysaccharide-treated peritoneal macrophages (Tan et al., 2023). Manganese exposure induces hepatic mitochondrial oxidative stress, leading to HSF1 phosphorylation at Ser326 and activation of autophagy. Knockdown of HSF1 prevents manganese-induced autophagosome formation in hepatocytes of yellow catfish (Zhao et al., 2024). Conversely, HSF1 regulates JNK1-mediated mTORC1 activation, suggesting an inhibitory role in autophagy (Su et al., 2016; Su and Dai, 2016; Su and Dai, 2017). Furthermore, HSF1 knockdown activates AMPK and promotes mitophagy, leading to reduced mitochondrial mass (Su et al., 2019). Moreover, recent studies show that HSP70 negatively regulates autophagy and that HSP70 inhibition, along with autophagy blockade, promotes cell death in NSCLC cells (Alhasan et al., 2024). These findings demonstrate the complexity of HSF1 in autophagy, highlighting the need for further investigation to clarify its functions in autophagy regulation and its contribution to chemotherapy resistance (Figure 2).
Future research should focus on clarifying the dual role of HSF1 in autophagy regulation, investigating the specific conditions and signaling pathways that determine whether HSF1 acts as a promoter or inhibitor of autophagy. Additionally, exploring the impact of HSF1-mediated autophagy on chemoresistance in cancer could reveal new strategies to enhance the therapeutic sensitivity in autophagy-dependent tumors. Finally, studying the interactions between HSF1, HSP70, and key pathways, such as kinase activities and ATG protein expression, may provide insights into combination therapies targeting autophagy and chemoresistance in cancer treatment.
3.2 HSF1 and apoptosis
Both intrinsic and extrinsic pathways of apoptosis contribute to chemoresistance. The involvement of the intrinsic pathway is indicated by the upregulation of anti-apoptotic proteins, such as BCL-2 family proteins, and the downregulation of proapoptotic proteins, such as BAX and BAK (Singh et al., 2019). Decreased expression and increased endocytosis of molecules involved in the extrinsic pathway, such as tumor necrosis factor superfamily proteins, FAS ligands, and death receptor (DR) 4 and DR5, potentially confer drug resistance (Ashkenazi, 2015).
HSF1 primarily functions as a transcription factor of HSP70 and HSP90, which are well-known for their roles in inhibiting apoptosis. For instance, HSP70 plays a protective role in heat-induced apoptosis by stabilizing the anti-apoptotic protein MCL-1, which prevents BAX activation and cytochrome-c release (Stankiewicz et al., 2009). Similarly, HSP90 prevents the release of cytochrome c by inhibiting the activity of the proapoptotic protein FKBP38. Increased HSP90 expression is correlated with elevated levels of antiapoptotic proteins BCL-2 and BCL-xL (Edlich et al., 2007). On the other hand, inhibiting HSP27 triggers the release of SMAC protein, a key regulator of the mitochondrial apoptotic pathway, in dexamethasone-resistant myeloma cell lines and promotes the activation of caspase-9 and caspase-3, suggesting that HSP27 contributes to resistance against dexamethasone (Chauhan et al., 2003).
Moreover, genetic knockdown of HSF1 has been shown to directly link to enhancing apoptosis through the regulation of the mitochondrial apoptosis pathway. In breast cancer, HSF1 knockdown enhances BAX expression and cisplatin-induced apoptosis, whereas restoring HSF1 expression significantly reduces cisplatin-induced apoptosis (Liu and Ma, 2021). In a study on pancreatic tumorigenesis, HSF1 silencing led to the upregulation of pro-apoptotic proteins, including SMAC, cytochrome c, Apaf1, and cleaved caspase-3 and -9, suggesting that HSF1 inhibits the mitochondrial apoptosis pathway to promote tumor growth (Liang et al., 2017). Inhibiting HSP70 prevents HSP70 from stabilizing anti-apoptotic proteins and blocking apoptosis, thus reducing chemoresistance, in bladder cancer (Wei et al., 2024). Inhibiting HSF1 by small molecules enhances the effectiveness of the aurora kinase inhibitor efficacy in NSCLC by promoting apoptosis, potentially overcoming chemoresistance through PI3K/AKT pathway downregulation and ROS activation (Zhang et al., 2024). This suggests that inhibiting HSF1 enhances the effectiveness of chemotherapy by overcoming HSF1- or HSP70-mediated anti-apoptosis mechanisms.
BAG3, a member of the co-chaperone family, is a well-known non-HSP substrate of HSF1 and contributes to chemoresistance in cancer (Jacobs and Marnett, 2009; Antonietti et al., 2017; Guo et al., 2022). Generally, HSP70’s function depends on its interactions with other chaperones like HSP90 and the co-chaperone BAG3. In an interesting study, the HSF1/HSP70/BAG3 pathway is investigated and confirmed to contribute to chemoresistance, particularly through its role in the overexpression of pro-survival BCL-2 family proteins and the subsequent resistance to cell death in gliomas (Das et al., 2018). In another study, the overexpression of Bag-1, another protein from the BAG family, modulates HSP levels by phosphorylating HSF1 at Ser326 via the PI3K/AKT/mTOR pathway in HER2-positive and HER2-negative breast cancer cells. BAG1 explicitly enhances the expression of HSP70 and HSP27, contributing to breast cancer cell survival and, potentially, drug resistance (Kizilboga et al., 2024). HSF1 influences apoptosis and chemoresistance through its interactions with co-chaperones like BAG3 and BAG1 and it promotes cell survival by upregulating pro-survival BCL-2 family proteins and HSPs across various cancer types (Figure 3).
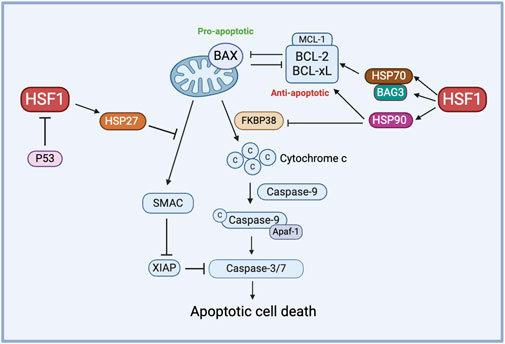
Figure 3. The role of HSF1 in cancer apoptosis. HSF1 mediates pro-apoptotic and anti-apoptotic pathways in the chemoresistance by regulating the expression of downstream HSPs. HSP70 controls the function of anti-apoptotic proteins via the HSP70-BAG3 axis, HSP90 regulates anti-apoptotic proteins and inhibits FKBP38, and HSP27 prevents the release of SMAC. Additionally, P53 inhibits HSF1 to avert this process of apoptosis. Apaf-1, apoptotic protease activating factor 1; BAG3, Bcl2-associated athanogene 3; BAX, Bcl-2-associated X protein; BCL-xL, B-cell lymphoma-extra large; c, Cytochrome c; FKBP38, FK506-binding protein 38; HSF1, heat shock factor 1; HSP, heat shock protein; MCL-1, myeloid cell leukemia-1; P53, tumor protein p53; SMAC, second mitochondria-derived activator of caspases; XIAP, X-linked inhibitor of apoptosis protein.
Other stress response mechanisms also contribute to the HSF1-mediated chemoresistance. For example, ER stress-induced HSF1 promotes chemoresistance to ubiquitin-specific protease 7 (USP7) inhibitors via the protein kinase R-like ER kinase (PERK) pathway, suggesting that targeting HSF1 or PERK could improve USP7 inhibitor-based chemotherapy (Lim et al., 2024). Besides, The Munc18-1 interacting protein 3- (Mint3)-activated hypoxia-indued factor 1α (HIF-1α) signaling promotes chemoresistance in triple-negative breast cancer (TNBC) by increasing the HSP70 expression. Mint3 depletion induces energy stress, which inactivates HSF1 via the AMPK/mTOR pathway, reducing HSP70 levels and enhancing the effectiveness of doxorubicin in TNBC (Tanaka et al., 2023). These findings suggest crosstalk among stress response signaling pathways, with HSF1 driving chemoresistance through mechanisms like ER stress via PERK and HIF-1α signaling, ultimately enhancing cancer cell survival during chemotherapy.
In addition, the interaction of HSF1 with multiple oncogenic transcription factors is crucial for tumorigenesis and apoptosis. The wildtype p53 is a tumor suppressor, inducing cell cycle arrest and apoptosis when DNA damage cannot be repaired. Surprisingly, mutant p53 directly interacts with HSF1, aiding the proper binding of HSF1 to the target HSPs and regulating the activity of HSPs, thereby contributing to cell survival. Moreover, mutant p53 is also known to activate mTOR, MAPK, and PI3K pathways via erythroblastic oncogene B (ErbB) family, including epidermal growth factor receptor (EGFR) and HER2, inducing HSF1 activation and contributing to apoptosis and chemoresistance (Toma-Jonik et al., 2019; Zhang et al., 2021). Furthermore, SUMOylation of HSF1 at the K298 enhances its stability, nuclear localization, and mitochondrial unfolded protein response, which promotes glioblastoma cell proliferation, migration, and resistance to apoptosis (Li et al., 2024). This SUMOylation-modified HSF1 activity contributes to chemoresistance by supporting mitochondrial function and increasing the expression of mitochondrial chaperones, which may help cancer cells to evade chemotherapy-induced stress. Future research should explore how targeting HSF1 and its downstream pathways can be used to overcome chemoresistance, focusing on both intrinsic and extrinsic apoptotic mechanisms and the HSF1’s effect on other programmed cell death signaling.
3.3 HSF1 and DNA damage repair
The DNA damage response (DDR) is a complex network of pathways that maintain the genome integrity. A key mechanism of DDR is base excision repair, which is mediated by Poly (ADP-ribose) polymerase (PARP) and AP endonuclease1 (APE1) as the responsible proteins. Other pathways contributing to DDR include non-homologous end joining (NHEJ) and homologous recombination (HR). DNA–dependent protein kinase (DNA-PK) plays an important role in NHEJ for double-strand breaks (DSBs) and contributes to chemo-radiotherapy resistance. DSBs are critical DNA lesions repair mainly by HR or NHEJ. Ataxia-telangiectasia mutated and ataxia telangiectasia and Rad3-related (ATR) kinases detect DSBs, activating p53 for cell cycle arrest or apoptosis and Breast cancer genes (BRCA) 1/2 for accurate repair through HR (Jackson and Bartek, 2009). This coordinated response maintains genomic stability, preventing mutations that can lead to cancer. Cell cycle checkpoint kinases are activated during DDR, of which checkpoint kinase 1 (CHK1) and CHK2 are downstream substrates of ataxia-telangiectasia mutated (ATM)/ATR. ATR and ATM are activated during single-strand and double-strand breaks, respectively. The enhanced DNA damage repair capacity (DRC) in tumor cells contributes significantly to their drug resistance, including targeted and immune therapy For example, cisplatin-resistant tumor cells often demonstrate higher expression of DNA damage repair-related genes and higher DRC (Fujimoto et al., 2017). Furthermore, inhibiting the nucleotide excision repair (NER) pathway further increased the sensitivity of tumor cells to another chemotherapy drug, cisplatin, complementing the cytotoxic effects (Oliver et al., 2010; Wang et al., 2011). HSF1 plays a crucial role in DNA damage repair-mediated chemoresistance, particularly through its involvement in the NHEJ pathway. HSF1 inhibits NHEJ by interacting with Lupus Ku autoantigen protein p70 (Ku70) and Ku86 to disrupt their heterodimeric interaction, leading to defective DNA repair and genomic instability. This is a potential mechanism for HSF1-mediated carcinogenesis (Kang et al., 2015). This inhibition of NHEJ by HSF1 contributes to chemoresistance by promoting genomic instability, which can drive cancer progression and resistance to therapy. Notably, HSF1’s function in NHEJ appears to be independent of its traditional role as a transcription factor, it instead operates through direct protein-protein interactions within the DNA damage repair machinery.
Single-strand DNA breaks are repaired through the base excision repair pathway, where PARP detects the damage sites and recruits repair factors to them while proliferating cell nuclear antigen (PCNA) acts as a sliding clamp to facilitate the recruitment of DNA polymerases, ensuring efficient repair and genome stability. HSF1 also contributes to DNA damage response by forming a complex with PARP1 and PARP13 and redistributing PARP1 to DNA lesions (Fujimoto et al., 2017). PARP1 contains a C-terminal catalytic domain that helps the synthesis of PAR, and autoPARylation of PARP1 regulates chromatin remodeling and DNA damage repair. Deficiency of HSF1 also reduces the expression of DNA damage repair factors such as RAD51 and 53BP1 and reduces DNA damage repair efficiency. BRCA1 and BRCA2 are DNA damage repair genes; many mutations in these genes result in a dysfunctional DNA damage response. HSF1 deficiency has been shown to reduce the proliferation of mammary tumors having dysfunctional HR due to BRCA1 mutations (Fujimoto et al., 2017). This correlated with HSF1’s crucial role in maintaining the genome integrity as a part of the PSR, contributing to BRCA-mutated tumor cells' addiction to HSF1 (Figure 4). This addiction can be leveraged as a targeted therapy approach, increasing the susceptibility of BRCA-mutated cells to chemotherapy.
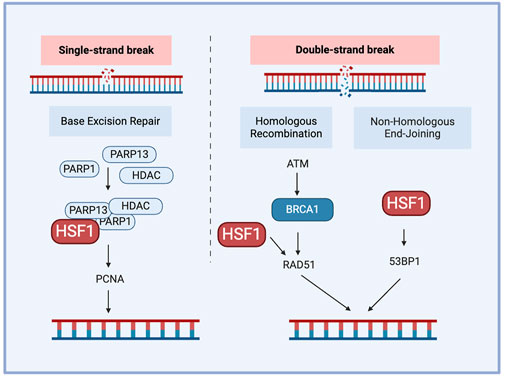
Figure 4. The role of HSF1 in cancer DNA damage repair. HSF1 participates in DNA damage repair by forming interactions with PARP1 and PARP13, facilitating the localization of PARP1 to DNA single-strain break sites and regulating RAD51 or 53BP1 during double-strain break repair. 53BP1. tumor protein p53 binding protein 1; ATM, ataxia-telangiectasia mutated; BRCA1, breast cancer type 1 susceptibility protein; HDAC, histone deacetylase; HSF1, heat shock factor 1; PARP, poly (ADP-ribose) polymerase; PCNA, proliferating cell nuclear antigen; RAD51, RAD51 recombinase.
Besides, HSF1 can form a complex with DNA damage kinases ATR and CHK1, facilitating p53 phosphorylation in response to DNA damage (Logan et al., 2009). HSF1 activation increases DNA damage repair, contributing to radiation resistance and promoting cancer cell survival under treatment stress in colorectal cancer (Li et al., 2023b). MDM2-mediated ubiquitination of HSF1 under stress conditions affects HSF1 stability, facilitating DNA damage repair processes and potentially contributing to resistance against DNA-damaging treatments (Xiang et al., 2023). Besides, inhibiting HSF1 with KRIBB11 disrupts these pathways, reduces MDM2 and other survival proteins, induces apoptosis, and enhances sensitivity to HSP90 inhibitors, making HSF1 a promising target to overcome DNA damage repair-mediated chemoresistance in adult T-cell leukemia (Ishikawa and Mori, 2023). These findings highlight the potential for targeting HSF1 as a therapeutic approach to overcome DNA damage repair-mediated chemoresistance.
Although it is promising to target HSF1 in DNA damage repair chemoresistance, currently there are gaps in our understanding of HSF1’s role in DNA damage repair involving its dual function in promoting or inhibiting repair pathways, its specific interactions with DNA repair proteins, and its context-specific effects across different cancer types. Addressing these questions could improve our understanding of HSF1 as a potential therapeutic target for developing targeted therapies to overcome chemoresistance.
3.4 HSF1 and drug efflux transporters
ATP-binding cassette (ABC) transporters contribute to drug resistance. At the basal level, these transporters export hydrophobic molecules to the outside of cells. However, in cancer cells, the upregulated efflux of drugs through ABC transporters contributes to another potential drug resistance mechanism (Gumilar et al., 2023). ATP-binding cassette sub-family G member 2 (ABCG2), commonly known as breast cancer resistance protein (BCRP), is an important factor in the development of chemoresistance in many malignancies. ABCG2 actively remove a variety of chemotherapeutic drugs out of cancer cells, lowering intracellular drug accumulation and effectiveness (Doyle and Ross, 2003; Noguchi et al., 2014; Mao and Unadkat, 2015). ABCG2 is extensively expressed in some cancer stem cells and its expression is often increased in response to chemotherapy, which helps these cells surviving treatments. Overexpression of ABCG2 has been associated with resistance to many chemotherapeutic treatments, including mitoxantrone, topotecan, and doxorubicin. Targeting ABCG2-mediated drug efflux therefore has emerged as a promising strategy for combating chemoresistance and improving treatment results in cancer patients (Zhang, 2007; Peng et al., 2010; Mo and Zhang, 2012). ABCG1, another member of the ABCG subfamily of ABC transporter, regulates and maintains cellular cholesterol homeostasis and is critical for the survival and function of normal cells. HSF1 overexpression has been seen in melanoma cell lines, contributing to greater drug efflux (Vydra et al., 2013). HSF1 regulates drug resistance through multiple mechanisms. As the mere binding of HSF1 to the HSE element on the ABCG1 gene is insufficient to activate the ABCG1 expression, the detailed mechanism of HSF1 in regulating ABGC1 protein is not understood. However, there are reasonable hypotheses that the ABCG1 activity is post-transcriptionally regulated by the overexpression of HSF1 (Vydra et al., 2013). Interestingly, ABCG1 and ABCG2 are co-expressed in metastatic colon cancer cells, with ABCG1 influencing ABCG2 expression through the modulation of HIF-1α (Namba et al., 2018). This interaction suggests a crosstalk between ABCG1 and ABCG2, implicating HSF1 in regulating drug resistance and tumorigenesis by affecting ABCG2 through ABCG1.
Another key mechanism is that HSF1 acts as the major transcriptional regulator of the multidrug-resistant 1 gene (MDR1), which encodes the ABC transporter P-glycoprotein responsible for driving chemoresistance (Krishnamurthy et al., 2012; Gumilar et al., 2023). Overexpression of activated HSF1 increases the MDR1 mRNA level along with enhanced P-glycoprotein cell surface expression (Vilaboa et al., 2000). Interestingly, this function of HSF1 persists even in HSF1 mutants that are unable to increase the transcription of HSP genes, implying that HSF1 may also activate MDR1 through a non-transcriptional, stress-independent pathway (Tchenio et al., 2006). This finding expands the understanding of HSF1’s role in chemoresistance, which may involve additional, pathways that do not rely solely on stress-induced HSP gene activation. HSF1, through its regulation of HSPs and interaction with SIRT1, contributes to 17-allylamino-17-demethoxygeldanamycin (17-AAG)-mediated chemoresistance in cancer stem-like cells by stabilizing key proteins and enhancing MDR1 drug efflux mechanisms (Kim et al., 2015). F-Box And WD Repeat Domain Containing 7 (FBXW7) has been reported to regulate the stability of nuclear HSF1 by binding to phosphorylated HSF1 at Ser303/307 and leading to HSF1 degradation (Kourtis et al., 2015; Yu et al., 2024). In drug-resistant cells, decreased FBXW7 expression driven by ERK1/2 activation stabilizes phosphorylated HSF1, which in turn enhances the MDR1 transcription by directly binding to the MDR1 promoter (Mun et al., 2020). This reveals the post-translational regulation of HSF1 in MDR1-mediated drug resistance (Figure 5).
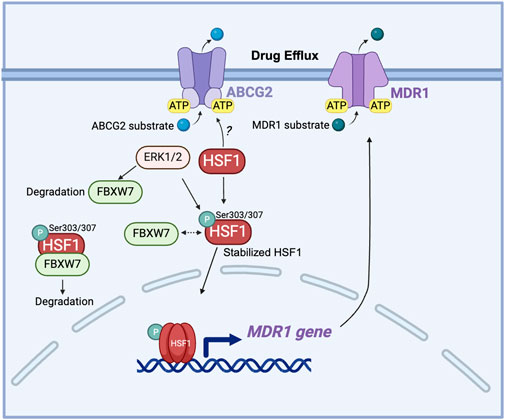
Figure 5. The role of HSF1 in cancer drug efflux. HSF1 upregulates the MDR1 gene expression in drug-resistant cells and is associated with the expression of ABCG2. ERK1/2 phosphorylates FBXW7 at the Thr205 residue, leading to the degradation of FBXW7. This degradation of FBXW7 reduces its ability to ubiquitinate and target HSF1 for proteasomal degradation, resulting in the stabilization of HSF1. The stabilized HSF1 then enhances the transcription of the MDR1 gene, increasing MDR1 protein expression. Elevated levels of MDR1 contribute to drug resistance by actively pumping chemotherapeutic drugs out of cancer cells, thereby reducing their efficacy. Dashed arrow: dissociation. ABCG2, ATP-binding cassette sub-family G member 2; ATP, adenosine triphosphate; ERK: extracellular signal-regulated kinase; FBXW7, F-box and WD repeat domain-containing 7; HSF1, heat shock factor 1; MDR1, multidrug resistance protein 1 (also known as P-glycoprotein); P, phosphorylation; Ser, serine.
Targeting HSF1 in conjunction with ABC transporters, particularly ABCG2 and ABCG1, represents a promising strategy for overcoming chemoresistance in various cancers. Inhibiting HSP90 with geldanamycin (GDN) or 17-AAG induces significant apoptotic cell death in glioma cell lines, where ABCG2 shows minimal and ineffective efflux of GDN and 17-AAG (Pastvova et al., 2021). HSP90 inhibitors have been known to induce HSF1 activation (Kijima et al., 2018); however, whether HSF1 regulates ABCG2 stability and expression is still unknown. Furthermore, HSF1 has both transcriptional and non-transcriptional roles in regulating drug efflux pathways, adding complexity to resistance mechanisms but reinforcing its potential as a valuable target for overcoming transporters-mediated resistance, including through MDR1. Further research is needed to elucidate the mechanisms of interaction between HSF1 and ABC transporters to optimize therapeutic strategies in cancer chemoresistance.
3.5 HSF1 inhibition in KRAS-mutated cancer
Cancers driven by KRAS mutants often show significant resistance to conventional and targeted chemotherapies, making them particularly challenging to treat (Negri et al., 2022; Singhal et al., 2024). For example, KRAS mutations are known to confer primary and secondary resistance to EGFR-targeted therapies in CRC and NSCLC. This is mainly because that KRAS functions downstream of receptor tyrosine kinases (RTKs), thus, constitutive KRAS activation will activate downstream signalings independently of the upstream RTK activity (Siddiqui and Piperdi, 2010; Zhao et al., 2017; Zhu et al., 2021). To effectively treat KRAS-driven tumors and to overcome chemoresistance conferred by KRAS mutants, effective inhibition of KRAS mutants would be ideal. Despite four decades of intensive research, targeting KRAS mutations directly remains challenging due to the protein’s structural characteristics, which lack deep binding pockets, limiting the efficacy of traditional small-molecule inhibitors (Huang et al., 2021; Wadood et al., 2022; Zhu et al., 2022). Additionally, the mechanisms by which KRAS mutations confer chemoresistance, including interactions with downstream signaling pathways such as HSF1-mediated stress responses, are not fully understood.
An exciting recent advance in inhibiting KRAS mutants is the approval of KRAS(G12C)-mutant inhibitors for the clinical use. Both adagrasib and sotorasib are developed to target the KRAS(G12C)-mutant by covalently binding to the mutant cysteine residue in KRAS(G12C), leading to irreversible inhibition of the KRAS activity and blocking KRAS downstream signaling pathways essential for cancer cell proliferation. Both adagrasib and sotorasib have been approved explicitly for treating KRAS(G12C)-mutant NSCLC (Huang et al., 2021; Janne et al., 2022). However, G12C is only one of many mutations that having been found in KRAS in tumors and acquired resistance to KRAS(G12C) inhibitors has been observed. KRAS mutant tumors treated with KRAS inhibitors often develop resistance to these inhibitors through acquired KRAS alternations, MET amplification or oncogenic BRAF gene fusions and mutations. These tumors may also undergo phenotypic changes, such as epithelial-mesenchymal transition (EMT), or adopt survival mechanisms like metabolic rewiring and autophagy (Awad et al., 2021; Yaeger et al., 2023). For example, although adagrasib can reverse multidrug resistance mediated by the MDR1 transporter (Zhang et al., 2022), the resistance to adagrasib has emerged through heterogeneous subclonal mutations in RAS-MAPK pathway components (e.g., NRAS, BRAF, and MAP2K1) that enable NSCLC tumor cells to reactivate MAPK signaling and bypass KRAS(G12C) inhibition (Tanaka et al., 2021). In order to obtain more durable responses to KRAS inhibitors, researchers are exploring combination therapies, such as pairing KRAS inhibitors with MEK or SHP2 inhibitors or with immune checkpoint inhibitors (Khan and O'Bryan, 2021; Hegazi et al., 2024). Thus, targeting KRAS for cancer treatment remains challenging, particularly in overcoming chemoresistance.
HSF1 is a downstream effector of the RAS-MEK pathway (Tang et al., 2015; Dai and Sampson, 2016) and KRAS-mutant cancers, due to their elevated levels of cellular stress resulting from rapid proliferation and metabolic demands, have a higher dependence on HSF1. Therefore, HSF1 represents a promising target for KRAS-mutant cancers, potentially addressing some resistance mechanisms by disrupting cancer cell stress responses. By targeting HSF1, it may be possible to exploit this stress vulnerability, making KRAS-mutant cancer cells more susceptible to therapeutic intervention. Targeting HSF1 in KRAS-mutant cancers, such as PDAC and NSCLC, where chemoresistance is common and therapeutic options are limited could open new avenues for treatment. By specifically disrupting the HSF1-mediated stress response in these tumors, HSF1 inhibitors may provide a novel approach to sensitize these cancers to existing therapies and improve clinical outcomes.
3.6 Challenges and opportunities in targeting HSF1 for cancer therapy
Several small molecules targeting HSF1 have been synthesized and their effects have been tested, primarily in in vitro and in vivo preclinical models. However, because HSF1 is a transcription factor and lacks clearly targetable sites, developing drugs that specifically target HSF1 remains highly challenging (Gumilar et al., 2023). Several HSF1 inhibitors have been discussed broadly (Dong et al., 2019; Cyran and Zhitkovich, 2022). Most current inhibitors target HSF1 indirectly and suffer from limited specificity and potency. Additionally, HSF1’s role in tumorigenesis is complex, involving multiple signaling pathways that vary across different cancer types. Although several potential HSF1 inhibitors have been identified, we will mainly discuss current small molecules that directly interact with HSF1 in this review.
KRIBB11 is a small molecule known to directly associate with HSF1, disrupting its functional activity by preventing the recruitment of pTEFB on the promoter region of HSP70, as seen in a colorectal carcinoma cell line (Yoon et al., 2011). This blocks the transcription of HSP70 and HSP27 and the downstream stress response while inducing growth arrest and triggering caspase-dependent apoptosis. For example, in lung cancer, KRIBB11 induces apoptosis by reducing the level of Mcl-1, an anti-apoptotic protein (Kang et al., 2017). KRIBB11 induces apoptosis and cell cycle arrest in NSCLC, especially in combination therapies, by inhibiting the PI3K/AKT pathway, increasing ROS, and activating DNA damage responses (Zhang et al., 2024). In another study of NSCLC, KRIBB11 is shown to reduce drug resistance potentially associated with EMT by downregulating EMT-associated proteins, such as N-cadherin and vimentin, and EGFR, along with other key signaling molecules (Shibue and Weinberg, 2017; Lee et al., 2021). Although KRIBB11 directly associates with HSF1, it may also interact with other proteins or pathways, leading to off-target effects that lead to toxicities and narrowing the therapeutic window, thus complicating its clinical application.
DTHIB binds HSF1 directly and degrades HSF1 in the nucleus, thereby decreasing its nuclear activity and ultimately lowering its transcriptional activity in prostate cancer (Dong et al., 2020). DTHIB induces the degradation of HSF1 through the proteasome and the E3 ligase component FBXW7. Additionally, DTHIB effectively attenuated tumor growth in four therapy-resistant prostate cancer animal models, including a neuroendocrine prostate cancer model, where it caused profound tumor regression (Dong et al., 2019; Dong et al., 2020). However, further development and optimization are necessary to assess its efficacy in KRAS mutant cancers before advancing to clinical trials.
NXP800 (CCT361814) is currently the only inhibitor targeting the HSF1 pathway that has advanced to clinical trials, it is now in phase Ib clinical trial in platinum-resistant, ARID1a-mutated ovarian cancer (NCT05226507) (Cheeseman et al., 2017; clinicaltrials.gov 2021; Pasqua et al., 2023). This marks a significant milestone in HSF1-targeted cancer therapy. NXP800 has been shown to inhibit HSF1 transcriptional activity (Menezes et al., 2017; Workman et al., 2022). NXP800 inhibits HSF1 indirectly by activating the integrated stress response (ISR) through the general control nonderepressible 2 (GCN2), Activation of GCN2 leads to phosphorylation of eukaryotic translation initiation factor 2 α (eIF2α), which reduces global protein synthesis and selectively increases the translation of stress-responsive genes like activating transcription factor 4 (ATF4). Elevated ATF4 levels subsequently inhibit HSF1 activation, thereby diminishing the expression of HSF1-regulated genes This mechanism has been observed in human carcinoma cell lines and tumor xenograft models, where NXP800-induced ISR activation resulted in decreased HSF1 activity and reduced tumor cell proliferation (Cheeseman et al., 2017; Pasqua et al., 2023). NXP800-mediated inhibition of HSF1 induces sustained cellular stress, ultimately triggering programmed cell death. This therapeutic mechanism is particularly effective in cancers that heavily rely on HSF1 for protection against stress-induced damage, as it weakens the cells’ defenses against therapeutic intervention. However, further extensive research is necessary to optimize this therapeutic approach, including investigations into optimal dosing strategies, potential synergistic effects with various chemotherapy agents, and a deeper understanding of the molecular pathways by which HSF1 inhibition impacts cancer cell survival and treatment resistance.
Technological advancements are revolutionizing the creation of HSF1 inhibitors to address chemoresistance. Proteolysis-targeting chimeras, or PROTACs, offer a strong alternative by completely degrading HSF1 rather than just inhibiting it, which has the potential to reduce the possibility of resistance. A proof-of-concept study has demonstrated the feasibility of HSF1 degradation using a bifunctional PROTAC molecular that leverages KRIBB11-mediated binding and E3 ligase-mediated proteolysis (Sharma et al., 2022). Although this study has demonstrated the utility of the PROTAC approach for HSF1 inhibition, the effect of this approach on chemoresistance remains to be evaluated. Besides, an RNA aptamer that binds specifically and tightly to the DNA-binding domain of HSF1 has been developed and demonstrated to be able to block HSF1 from binding to DNA when delivered using a synthetic gene and strong promoter (Salamanca et al., 2011). Overall, the study highlights aptamers’ potential for precise inhibition of HSF1 in cancer while minimizing off-target effects, providing a novel strategy to suppress cancer cell growth and survival by blocking HSF1’s DNA-binding activity (Salamanca et al., 2014). Recently, nanoparticle drug delivery systems have demonstrated effective in enhancing the potency of HSF1 inhibitors by targeting them directly to tumor tissues. This approach reduces off-target side effects and maintains reliable treatment drug concentration. For example, one study shows that functionalized nanomaterials can efficiently transport small-molecule HSP inhibitors to tumor locations, boosting the effectiveness of both photothermal and photodynamic therapies (Premji et al., 2024). In addition, studies on hybrid nanoparticles enhanced with hyaluronic acid for delivering an HSP90 inhibitor emphasize their potential in targeted cancer therapy by enhancing drug delivery efficiency and specificity (Pan et al., 2021). Another study has developed aqueous bovine serum albumin nanoparticles for controlled delivery of the Hsp90 inhibitor luminespib. Through in vitro characterization and evaluations, this approach has demonstrated its potential as a nanoformulation for pancreatic and breast cancer therapy (K Rochani et al., 2020). Using advanced patient-derived models like organoids and xenografts allows for the assessment of HSF1 inhibitors in settings that closely resemble human tumors. This method results in more reliable predictions about their effectiveness and possible resistance (Yang et al., 2018). In summary, these advancements are driving the development of more effective HSF1-targeted therapies to tackle the issue of chemoresistance in cancer treatment specifically.
4 Conclusion and perspective
This review explores mechanisms of the HSF1’s role in the proteotoxic stress response and its function during chemotherapy exposure. HSF1 regulates multiple pathways that contribute to chemoresistance throughout tumorigenesis. Because HSF1 is active in normal and cancerous cells, complete inhibition of HSF1 as a cancer treatment approach remains challenging. Among autophagy, apoptosis resistance, DNA damage repair, and drug efflux mechanisms, autophagy is often considered the key downstream pathway regulated by HSF1 in the context of chemotherapy resistance. HSF1-mediated autophagy enables cancer cells to manage cellular stress by degrading and recycling damaged proteins and organelles, supporting cell survival under chemotherapy-induced stress. This autophagy-driven survival mechanism is crucial, allowing cancer cells to resist apoptosis and maintain functionality despite therapeutic interventions. Therefore, while drug efflux, apoptosis resistance, and DNA damage repair are important, autophagy is likely the primary downstream pathway through which HSF1 exerts its protective effects against chemotherapy. Given the high expression of HSF1 in various cancer types and its role in chemoresistance, inhibiting HSF1 may offer a promising therapeutic strategy to counteract chemoresistance. This strategy could potentially disrupt the mechanisms that enable cancer cells to resist chemotherapy, enhancing the efficacy of treatment.
Author contributions
SG: Writing–original draft, Writing–review and editing. RS: Writing–original draft, Writing–review and editing. K-HS: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the University of Toledo Fund Number #110917 and the National Cancer Institute of the National Institutes of Health under Award Number K22CA248616. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgments
The images were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ai, H., Zhou, W., Wang, Z., Qiong, G., Chen, Z., and Deng, S. (2019). microRNAs-107 inhibited autophagy, proliferation, and migration of breast cancer cells by targeting HMGB1. J. Cell Biochem. 120 (5), 8696–8705. doi:10.1002/jcb.28157
Alasady, M. J., and Mendillo, M. L. (2020). The multifaceted role of HSF1 in tumorigenesis. Adv. Exp. Med. Biol. 1243, 69–85. doi:10.1007/978-3-030-40204-4_5
Alhasan, B., Gladova, Y. A., Sverchinsky, D. V., Aksenov, N. D., Margulis, B. A., and Guzhova, I. V. (2024). Hsp70 negatively regulates autophagy via governing AMPK activation, and dual hsp70-autophagy inhibition induces synergetic cell death in NSCLC cells. Int. J. Mol. Sci. 25 (16), 9090. doi:10.3390/ijms25169090
Anckar, J., and Sistonen, L. (2011). Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80, 1089–1115. doi:10.1146/annurev-biochem-060809-095203
Antonietti, P., Linder, B., Hehlgans, S., Mildenberger, I. C., Burger, M. C., Fulda, S., et al. (2017). Interference with the HSF1/HSP70/BAG3 pathway primes glioma cells to matrix detachment and BH3 mimetic-induced apoptosis. Mol. Cancer Ther. 16 (1), 156–168. doi:10.1158/1535-7163.MCT-16-0262
Asada, Y., Tsuruta, M., Okabayashi, K., Shigeta, K., Ishida, T., Shimada, T., et al. (2021). Inhibition of heat-shock protein 27 reduces 5-Fluorouracil-acquired resistance in human colon cancer cells. Anticancer Res. 41 (3), 1283–1290. doi:10.21873/anticanres.14885
Ashkenazi, A. (2015). Targeting the extrinsic apoptotic pathway in cancer: lessons learned and future directions. J. Clin. Invest 125 (2), 487–489. doi:10.1172/JCI80420
Awad, M. M., Liu, S., Rybkin, I. I., Arbour, K. C., Dilly, J., Zhu, V. W., et al. (2021). Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 384 (25), 2382–2393. doi:10.1056/NEJMoa2105281
Bailey, P., Chang, D. K., Nones, K., Johns, A. L., Patch, A. M., Gingras, M. C., et al. (2016). Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531 (7592), 47–52. doi:10.1038/nature16965
Buchwalter, G., Gross, C., and Wasylyk, B. (2004). Ets ternary complex transcription factors. Gene 324, 1–14. doi:10.1016/j.gene.2003.09.028
Bukau, B., Weissman, J., and Horwich, A. (2006). Molecular chaperones and protein quality control. Cell 125 (3), 443–451. doi:10.1016/j.cell.2006.04.014
Cai, Q., Wang, S., Jin, L., Weng, M., Zhou, D., Wang, J., et al. (2019). Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 18 (1), 82. doi:10.1186/s12943-019-1016-0
Cancer Genome Atlas Research, N. (2014). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511 (7511), 543–550. doi:10.1038/nature13385
Carpenter, R. L., Paw, I., Dewhirst, M. W., and Lo, H. W. (2015). Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene 34 (5), 546–557. doi:10.1038/onc.2013.582
Chatterjee, S., Bhattacharya, S., Socinski, M. A., and Burns, T. F. (2016). HSP90 inhibitors in lung cancer: promise still unfulfilled. Clin. Adv. Hematol. Oncol. 14 (5), 346–356.
Chatterjee, S., and Burns, T. F. (2017). Targeting heat shock proteins in cancer: a promising therapeutic approach. Int. J. Mol. Sci. 18 (9), 1978. doi:10.3390/ijms18091978
Chauhan, D., Li, G., Hideshima, T., Podar, K., Mitsiades, C., Mitsiades, N., et al. (2003). Hsp27 inhibits release of mitochondrial protein Smac in multiple myeloma cells and confers dexamethasone resistance. Blood 102 (9), 3379–3386. doi:10.1182/blood-2003-05-1417
Cheeseman, M. D., Chessum, N. E., Rye, C. S., Pasqua, A. E., Tucker, M. J., Wilding, B., et al. (2017). Discovery of a chemical probe bisamide (CCT251236): an orally bioavailable efficacious pirin ligand from a heat shock transcription factor 1 (HSF1) phenotypic screen. J. Med. Chem. 60 (1), 180–201. doi:10.1021/acs.jmedchem.6b01055
Chen, K., Qian, W., Li, J., Jiang, Z., Cheng, L., Yan, B., et al. (2017). Loss of AMPK activation promotes the invasion and metastasis of pancreatic cancer through an HSF1-dependent pathway. Mol. Oncol. 11 (10), 1475–1492. doi:10.1002/1878-0261.12116
Chen, M., and Xie, S. (2018). Therapeutic targeting of cellular stress responses in cancer. Thorac. Cancer 9 (12), 1575–1582. doi:10.1111/1759-7714.12890
Chiosis, G., Vilenchik, M., Kim, J., and Solit, D. (2004). Hsp90: the vulnerable chaperone. Drug Discov. Today 9 (20), 881–888. doi:10.1016/S1359-6446(04)03245-3
Chou, C. H., Hwang, C. L., and Wu, Y. T. (2012a). Effect of exercise on physical function, daily living activities, and quality of life in the frail older adults: a meta-analysis. Arch. Phys. Med. Rehabil. 93 (2), 237–244. doi:10.1016/j.apmr.2011.08.042
Chou, S. D., Prince, T., Gong, J., and Calderwood, S. K. (2012b). mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One 7 (6), e39679. doi:10.1371/journal.pone.0039679
clinicaltrials.gov (2021). A phase 1 clinical study of NXP800 in subjects with advanced cancers and expansion in subjects with ovarian cancer.
Cyran, A. M., and Zhitkovich, A. (2022). Heat shock proteins and HSF1 in cancer. Front. Oncol. 12, 860320. doi:10.3389/fonc.2022.860320
Dai, C., Dai, S., and Cao, J. (2012). Proteotoxic stress of cancer: implication of the heat-shock response in oncogenesis. J. Cell Physiol. 227 (8), 2982–2987. doi:10.1002/jcp.24017
Dai, C., and Sampson, S. B. (2016). HSF1: guardian of proteostasis in cancer. Trends Cell Biol. 26 (1), 17–28. doi:10.1016/j.tcb.2015.10.011
Dai, C., Whitesell, L., Rogers, A. B., and Lindquist, S. (2007). Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130 (6), 1005–1018. doi:10.1016/j.cell.2007.07.020
Dai, S., Tang, Z., Cao, J., Zhou, W., Li, H., Sampson, S., et al. (2015). Suppression of the HSF1-mediated proteotoxic stress response by the metabolic stress sensor AMPK. EMBO J. 34 (3), 275–293. doi:10.15252/embj.201489062
Das, C. K., Linder, B., Bonn, F., Rothweiler, F., Dikic, I., Michaelis, M., et al. (2018). BAG3 overexpression and cytoprotective autophagy mediate apoptosis resistance in chemoresistant breast cancer cells. Neoplasia 20 (3), 263–279. doi:10.1016/j.neo.2018.01.001
Dayalan Naidu, S., Dikovskaya, D., Gaurilcikaite, E., Knatko, E. V., Healy, Z. R., Mohan, H., et al. (2017). Transcription factors NRF2 and HSF1 have opposing functions in autophagy. Sci. Rep. 7 (1), 11023. doi:10.1038/s41598-017-11262-5
Debnath, J., Gammoh, N., and Ryan, K. M. (2023). Autophagy and autophagy-related pathways in cancer. Nat. Rev. Mol. Cell Biol. 24 (8), 560–575. doi:10.1038/s41580-023-00585-z
Desai, S., Liu, Z., Yao, J., Patel, N., Chen, J., Wu, Y., et al. (2013). Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J. Biol. Chem. 288 (13), 9165–9176. doi:10.1074/jbc.M112.422071
Dong, B., Jaeger, A. M., Hughes, P. F., Loiselle, D. R., Hauck, J. S., Fu, Y., et al. (2020). Targeting therapy-resistant prostate cancer via a direct inhibitor of the human heat shock transcription factor 1. Sci. Transl. Med. 12 (574), eabb5647. doi:10.1126/scitranslmed.abb5647
Dong, B., Jaeger, A. M., and Thiele, D. J. (2019). Inhibiting heat shock factor 1 in cancer: a unique therapeutic opportunity. Trends Pharmacol. Sci. 40 (12), 986–1005. doi:10.1016/j.tips.2019.10.008
Doyle, L., and Ross, D. D. (2003). Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2). Oncogene 22 (47), 7340–7358. doi:10.1038/sj.onc.1206938
Dudeja, V., Chugh, R. K., Sangwan, V., Skube, S. J., Mujumdar, N. R., Antonoff, M. B., et al. (2011). Prosurvival role of heat shock factor 1 in the pathogenesis of pancreatobiliary tumors. Am. J. Physiol. Gastrointest. Liver Physiol. 300 (6), G948–G955. doi:10.1152/ajpgi.00346.2010
Dzobo, K., Senthebane, D. A., Thomford, N. E., Rowe, A., Dandara, C., and Parker, M. I. (2018). Not everyone fits the mold: intratumor and intertumor heterogeneity and innovative cancer drug design and development. OMICS 22 (1), 17–34. doi:10.1089/omi.2017.0174
Edlich, F., Erdmann, F., Jarczowski, F., Moutty, M. C., Weiwad, M., and Fischer, G. (2007). The Bcl-2 regulator FKBP38-calmodulin-Ca2+ is inhibited by Hsp90. J. Biol. Chem. 282 (21), 15341–15348. doi:10.1074/jbc.M611594200
Emran, T. B., Shahriar, A., Mahmud, A. R., Rahman, T., Abir, M. H., Siddiquee, M. F., et al. (2022). Multidrug resistance in cancer: understanding molecular mechanisms, immunoprevention and therapeutic approaches. Front. Oncol. 12, 891652. doi:10.3389/fonc.2022.891652
Fang, F., Chang, R., and Yang, L. (2012). Heat shock factor 1 promotes invasion and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer 118 (7), 1782–1794. doi:10.1002/cncr.26482
Florea, A. M., and Busselberg, D. (2011). Cisplatin as an anti-tumor drug: cellular mechanisms of activity, drug resistance and induced side effects. Cancers (Basel) 3 (1), 1351–1371. doi:10.3390/cancers3011351
Fujimoto, M., Takii, R., Takaki, E., Katiyar, A., Nakato, R., Shirahige, K., et al. (2017). The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat. Commun. 8 (1), 1638. doi:10.1038/s41467-017-01807-7
Fulda, S., Gorman, A. M., Hori, O., and Samali, A. (2010). Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010, 214074. doi:10.1155/2010/214074
Ghai, S., Young, A., and Su, K. H. (2023). Proteotoxic stress response in atherosclerotic cardiovascular disease: emerging role of heat shock factor 1. Front. Cardiovasc Med. 10, 1155444. doi:10.3389/fcvm.2023.1155444
Ghai, S., Shrestha, R., Hegazi, A., Boualoy, V., Liu, S. H., and Su, K. H. (2024). The role of heat shock factor 1 in preserving proteomic integrity during copper-induced cellular toxicity. Int. J. Mol. Sci. 25 (21), 11657. doi:10.3390/ijms252111657
Gidalevitz, T., Prahlad, V., and Morimoto, R. I. (2011). The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb. Perspect. Biol. 3 (6), a009704. doi:10.1101/cshperspect.a009704
Giordano, T. J. (2014). The cancer genome atlas research network: a sight to behold. Endocr. Pathol. 25 (4), 362–365. doi:10.1007/s12022-014-9345-4
Gomez-Pastor, R., Burchfiel, E. T., and Thiele, D. J. (2018). Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19 (1), 4–19. doi:10.1038/nrm.2017.73
Gu, Y., Chen, T., Li, G., Xu, C., Xu, Z., Zhang, J., et al. (2017). Lower Beclin 1 downregulates HER2 expression to enhance tamoxifen sensitivity and predicts a favorable outcome for ER positive breast cancer. Oncotarget 8 (32), 52156–52177. doi:10.18632/oncotarget.11044
Guettouche, T., Boellmann, F., Lane, W. S., and Voellmy, R. (2005). Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 6, 4. doi:10.1186/1471-2091-6-4
Gumilar, K. E., Chin, Y., Ibrahim, I. H., Tjokroprawiro, B. A., Yang, J. Y., Zhou, M., et al. (2023). Heat shock factor 1 inhibition: a novel anti-cancer strategy with promise for precision oncology. Cancers (Basel) 15 (21), 5167. doi:10.3390/cancers15215167
Guo, J., Du, X., and Li, C. (2022). BAG family proteins contributes to autophagy-mediated multidrug resistance of tumor. Clin. Transl. Oncol. 24 (8), 1492–1500. doi:10.1007/s12094-022-02819-6
Guo, Q., Jin, Y., Chen, X., Ye, X., Shen, X., Lin, M., et al. (2024). NF-κB in biology and targeted therapy: new insights and translational implications. Signal Transduct. Target Ther. 9 (1), 53. doi:10.1038/s41392-024-01757-9
Harris, L. N., Broadwater, G., Lin, N. U., Miron, A., Schnitt, S. J., Cowan, D., et al. (2006). Molecular subtypes of breast cancer in relation to paclitaxel response and outcomes in women with metastatic disease: results from CALGB 9342. Breast Cancer Res. 8 (6), R66. doi:10.1186/bcr1622
Hasan, S., Taha, R., and Omri, H. E. (2018). Current opinions on chemoresistance: an overview. Bioinformation 14 (2), 80–85. doi:10.6026/97320630014080
He, H. L., Lee, Y. E., Chen, H. P., Hsing, C. H., Chang, I. W., Shiue, Y. L., et al. (2015). Overexpression of DNAJC12 predicts poor response to neoadjuvant concurrent chemoradiotherapy in patients with rectal cancer. Exp. Mol. Pathol. 98 (3), 338–345. doi:10.1016/j.yexmp.2015.03.029
He, Y., Sun, M. M., Zhang, G. G., Yang, J., Chen, K. S., Xu, W. W., et al. (2021). Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target Ther. 6 (1), 425. doi:10.1038/s41392-021-00828-5
Hegazi, A., Rager, L. E., Watkins, D. E., and Su, K. H. (2024). Advancing immunotherapy in pancreatic cancer. Int. J. Mol. Sci. 25 (21), 11560. doi:10.3390/ijms252111560
Hietakangas, V., Ahlskog, J. K., Jakobsson, A. M., Hellesuo, M., Sahlberg, N. M., Holmberg, C. I., et al. (2003). Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell Biol. 23 (8), 2953–2968. doi:10.1128/MCB.23.8.2953-2968.2003
Holmberg, E. B., Hillman, R. E., Hammarberg, B., Sodersten, M., and Doyle, P. (2001). Efficacy of a behaviorally based voice therapy protocol for vocal nodules. J. Voice 15 (3), 395–412. doi:10.1016/S0892-1997(01)00041-8
Huang, L., Guo, Z., Wang, F., and Fu, L. (2021). KRAS mutation: from undruggable to druggable in cancer. Signal Transduct. Target Ther. 6 (1), 386. doi:10.1038/s41392-021-00780-4
Ishikawa, C., and Mori, N. (2023). Heat shock factor 1 is a promising therapeutic target against adult T-cell leukemia. Med. Oncol. 40 (6), 172. doi:10.1007/s12032-023-02042-5
Jackson, S. P., and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461 (7267), 1071–1078. doi:10.1038/nature08467
Jacobs, A. T., and Marnett, L. J. (2009). HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J. Biol. Chem. 284 (14), 9176–9183. doi:10.1074/jbc.M808656200
Janne, P. A., Riely, G. J., Gadgeel, S. M., Heist, R. S., Ou, S. I., Pacheco, J. M., et al. (2022). Adagrasib in non-small-cell lung cancer harboring a G12C mutation. N. Engl. J. Med. 387 (2), 120–131. doi:10.1056/NEJMoa2204619
Jin, X., Moskophidis, D., and Mivechi, N. F. (2011). Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab. 14 (1), 91–103. doi:10.1016/j.cmet.2011.03.025
Kang, G. Y., Kim, E. H., Lee, H. J., Gil, N. Y., Cha, H. J., and Lee, Y. S. (2015). Heat shock factor 1, an inhibitor of non-homologous end joining repair. Oncotarget 6 (30), 29712–29724. doi:10.18632/oncotarget.5073
Kang, M. J., Yun, H. H., and Lee, J. H. (2017). KRIBB11 accelerates Mcl-1 degradation through an HSF1-independent, Mule-dependent pathway in A549 non-small cell lung cancer cells. Biochem. Biophys. Res. Commun. 492 (3), 304–309. doi:10.1016/j.bbrc.2017.08.118
Khan, I., and O'Bryan, J. P. (2021). Probing RAS function with monobodies. Methods Mol. Biol. 2262, 281–302. doi:10.1007/978-1-0716-1190-6_17
Khan, S. U., Fatima, K., Aisha, S., and Malik, F. (2024). Unveiling the mechanisms and challenges of cancer drug resistance. Cell Commun. Signal 22 (1), 109. doi:10.1186/s12964-023-01302-1
Kijima, T., Prince, T., Neckers, L., Koga, F., and Fujii, Y. (2019). Heat shock factor 1 (HSF1)-targeted anticancer therapeutics: overview of current preclinical progress. Expert Opin. Ther. Targets 23 (5), 369–377. doi:10.1080/14728222.2019.1602119
Kijima, T., Prince, T. L., Tigue, M. L., Yim, K. H., Schwartz, H., Beebe, K., et al. (2018). HSP90 inhibitors disrupt a transient HSP90-HSF1 interaction and identify a noncanonical model of HSP90-mediated HSF1 regulation. Sci. Rep. 8 (1), 6976. doi:10.1038/s41598-018-25404-w
Kim, H. B., Lee, S. H., Um, J. H., Kim, M. J., Hyun, S. K., Gong, E. J., et al. (2015). Sensitization of chemo-resistant human chronic myeloid leukemia stem-like cells to Hsp90 inhibitor by SIRT1 inhibition. Int. J. Biol. Sci. 11 (8), 923–934. doi:10.7150/ijbs.10896
Kizilboga, T., Ozden, C., Can, N. D., Onay Ucar, E., and Dinler Doganay, G. (2024). Bag-1-mediated HSF1 phosphorylation regulates expression of heat shock proteins in breast cancer cells. FEBS Open Bio 14 (9), 1559–1569. doi:10.1002/2211-5463.13843
Kourtis, N., Moubarak, R. S., Aranda-Orgilles, B., Lui, K., Aydin, I. T., Trimarchi, T., et al. (2015). FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat. Cell Biol. 17 (3), 322–332. doi:10.1038/ncb3121
Krishnamurthy, K., Vedam, K., Kanagasabai, R., Druhan, L. J., and Ilangovan, G. (2012). Heat shock factor-1 knockout induces multidrug resistance gene, MDR1b, and enhances P-glycoprotein (ABCB1)-based drug extrusion in the heart. Proc. Natl. Acad. Sci. U. S. A. 109 (23), 9023–9028. doi:10.1073/pnas.1200731109
K Rochani, A., Balasubramanian, S., Ravindran Girija, A., Maekawa, T., Kaushal, G., and Kumar, D. S. (2020). Heat shock protein 90 (Hsp90)-Inhibitor-Luminespib-Loaded-Protein-Based nanoformulation for cancer therapy. Polym. (Basel) 12 (8), 1798. doi:10.3390/polym12081798
Lan, N., Su, Y., Zeng, Q., Zhou, P., Hu, Y., Zhang, Z., et al. (2024). JD-02, a novel Hsp90 inhibitor, induces ROS/SRC axis-dependent cytoprotective autophagy in colorectal cancer cells. Mol. Carcinog. 63 (6), 1038–1050. doi:10.1002/mc.23706
Lee, J. G., and Wu, R. (2012). Combination erlotinib-cisplatin and Atg3-mediated autophagy in erlotinib resistant lung cancer. PLoS One 7 (10), e48532. doi:10.1371/journal.pone.0048532
Lee, S., Jung, J., Lee, Y. J., Kim, S. K., Kim, J. A., Kim, B. K., et al. (2021). Targeting HSF1 as a therapeutic strategy for multiple mechanisms of EGFR inhibitor resistance in EGFR mutant non-small-cell lung cancer. Cancers (Basel) 13 (12), 2987. doi:10.3390/cancers13122987
Li, W., Yang, C., Li, J., Li, X., and Zhou, P. (2023a). MicroRNA-217 aggravates breast cancer through activation of NF1-mediated HSF1/ATG7 axis and c-Jun/ATF3/MMP13 axis. Hum. Cell 36 (1), 377–392. doi:10.1007/s13577-022-00817-y
Li, X., Wang, Z., Gao, B., Dai, K., Wu, J., Shen, K., et al. (2024). Unveiling the impact of SUMOylation at K298 site of heat shock factor 1 on glioblastoma malignant progression. Neoplasia 57, 101055. doi:10.1016/j.neo.2024.101055
Li, X., Zhou, Y., Li, Y., Yang, L., Ma, Y., Peng, X., et al. (2019). Autophagy: a novel mechanism of chemoresistance in cancers. Biomed. Pharmacother. 119, 109415. doi:10.1016/j.biopha.2019.109415
Li, Y., Wang, B., Ma, F., Jiang, D., Wang, Y., Li, K., et al. (2023b). Proteomic characterization of the colorectal cancer response to chemoradiation and targeted therapies reveals potential therapeutic strategies. Cell Rep. Med. 4 (12), 101311. doi:10.1016/j.xcrm.2023.101311
Liang, W., Liao, Y., Zhang, J., Huang, Q., Luo, W., Yu, J., et al. (2017). Heat shock factor 1 inhibits the mitochondrial apoptosis pathway by regulating second mitochondria-derived activator of caspase to promote pancreatic tumorigenesis. J. Exp. Clin. Cancer Res. 36 (1), 64. doi:10.1186/s13046-017-0537-x
Lim, C. H., Fang, X. Q., Kang, H., Oh, T., Lee, S., Kim, Y. S., et al. (2024). ER stress-activated HSF1 governs cancer cell resistance to USP7 inhibitor-based chemotherapy through the PERK pathway. Int. J. Mol. Sci. 25 (5), 2768. doi:10.3390/ijms25052768
Lindquist, S. (1986). The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191. doi:10.1146/annurev.bi.55.070186.005443
Liu, K., and Ma, R. (2021). MicroRNA-615-5p regulates the proliferation and apoptosis of breast cancer cells by targeting HSF1. Exp. Ther. Med. 21 (3), 192. doi:10.3892/etm.2021.9624
Logan, I. R., McNeill, H. V., Cook, S., Lu, X., Meek, D. W., Fuller-Pace, F. V., et al. (2009). Heat shock factor-1 modulates p53 activity in the transcriptional response to DNA damage. Nucleic Acids Res. 37 (9), 2962–2973. doi:10.1093/nar/gkp180
Luo, J., Solimini, N. L., and Elledge, S. J. (2009). Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136 (5), 823–837. doi:10.1016/j.cell.2009.02.024
Mansoori, B., Mohammadi, A., Davudian, S., Shirjang, S., and Baradaran, B. (2017). The different mechanisms of cancer drug resistance: a brief review. Adv. Pharm. Bull. 7 (3), 339–348. doi:10.15171/apb.2017.041
Mao, Q., and Unadkat, J. D. (2015). Role of the breast cancer resistance protein (BCRP/ABCG2) in drug transport--an update. AAPS J. 17 (1), 65–82. doi:10.1208/s12248-014-9668-6
Mendillo, M. L., Santagata, S., Koeva, M., Bell, G. W., Hu, R., Tamimi, R. M., et al. (2012). HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150 (3), 549–562. doi:10.1016/j.cell.2012.06.031
Menezes, K., Aram, G., Mirabella, F., Johnson, D. C., Sherborne, A. L., Houlston, R. S., et al. (2017). The novel protein HSF1 stress pathway inhibitor bisamide CCT361814 demonstrates pre-clinical anti-tumor activity in myeloma. Blood 130, 3072. doi:10.1182/blood.V130.Suppl_1.3072.3072
Meng, L., Gabai, V. L., and Sherman, M. Y. (2010). Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene 29 (37), 5204–5213. doi:10.1038/onc.2010.277
Min, J. N., Huang, L., Zimonjic, D. B., Moskophidis, D., and Mivechi, N. F. (2007). Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 26 (35), 5086–5097. doi:10.1038/sj.onc.1210317
Minoia, M., Boncoraglio, A., Vinet, J., Morelli, F. F., Brunsting, J. F., Poletti, A., et al. (2014). BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: implications for a proteasome-to-autophagy switch. Autophagy 10 (9), 1603–1621. doi:10.4161/auto.29409
Mo, W., and Zhang, J. T. (2012). Human ABCG2: structure, function, and its role in multidrug resistance. Int. J. Biochem. Mol. Biol. 3 (1), 1–27.
Mohammed, W. H., Sulaiman, G. M., Abomughaid, M. M., Klionsky, D. J., and Abu-Alghayth, M. H. (2024). The dual role of autophagy in suppressing and promoting hepatocellular carcinoma. Front. Cell Dev. Biol. 12, 1472574. doi:10.3389/fcell.2024.1472574
Mun, G. I., Choi, E., Lee, Y., and Lee, Y. S. (2020). Decreased expression of FBXW7 by ERK1/2 activation in drug-resistant cancer cells confers transcriptional activation of MDR1 by suppression of ubiquitin degradation of HSF1. Cell Death Dis. 11 (5), 395. doi:10.1038/s41419-020-2600-3
Murray, S., Briasoulis, E., Linardou, H., Bafaloukos, D., and Papadimitriou, C. (2012). Taxane resistance in breast cancer: mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat. Rev. 38 (7), 890–903. doi:10.1016/j.ctrv.2012.02.011
Namba, Y., Sogawa, C., Okusha, Y., Kawai, H., Itagaki, M., Ono, K., et al. (2018). Depletion of lipid efflux pump ABCG1 triggers the intracellular accumulation of extracellular vesicles and reduces aggregation and tumorigenesis of metastatic cancer cells. Front. Oncol. 8, 376. doi:10.3389/fonc.2018.00376
Neef, D. W., Jaeger, A. M., Gomez-Pastor, R., Willmund, F., Frydman, J., and Thiele, D. J. (2014). A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 9 (3), 955–966. doi:10.1016/j.celrep.2014.09.056
Negri, F., Bottarelli, L., de'Angelis, G. L., and Gnetti, L. (2022). KRAS: a druggable target in colon cancer patients. Int. J. Mol. Sci. 23 (8), 4120. doi:10.3390/ijms23084120
Neudegger, T., Verghese, J., Hayer-Hartl, M., Hartl, F. U., and Bracher, A. (2016). Structure of human heat-shock transcription factor 1 in complex with DNA. Nat. Struct. Mol. Biol. 23 (2), 140–146. doi:10.1038/nsmb.3149
Noguchi, K., Katayama, K., and Sugimoto, Y. (2014). Human ABC transporter ABCG2/BCRP expression in chemoresistance: basic and clinical perspectives for molecular cancer therapeutics. Pharmgenomics Pers. Med. 7, 53–64. doi:10.2147/PGPM.S38295
Nusrat, F., Khanna, A., Jain, A., Jiang, W., Lavu, H., Yeo, C. J., et al. (2024). The clinical implications of KRAS mutations and variant allele frequencies in pancreatic ductal adenocarcinoma. J. Clin. Med. 13 (7), 2103. doi:10.3390/jcm13072103
Oliver, T. G., Mercer, K. L., Sayles, L. C., Burke, J. R., Mendus, D., Lovejoy, K. S., et al. (2010). Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 24 (8), 837–852. doi:10.1101/gad.1897010
Oromendia, A. B., Dodgson, S. E., and Amon, A. (2012). Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 26 (24), 2696–2708. doi:10.1101/gad.207407.112
Pan, C., Zhang, T., Li, S., Xu, Z., Pan, B., Xu, S., et al. (2021). Hybrid nanoparticles modified by hyaluronic acid loading an HSP90 inhibitor as a novel delivery system for subcutaneous and orthotopic colon cancer therapy. Int. J. Nanomedicine 16, 1743–1755. doi:10.2147/IJN.S275805
Pasqua, A. E., Sharp, S. Y., Chessum, N. E. A., Hayes, A., Pellegrino, L., Tucker, M. J., et al. (2023). HSF1 pathway inhibitor clinical candidate (CCT361814/NXP800) developed from a phenotypic screen as a potential treatment for refractory ovarian cancer and other malignancies. J. Med. Chem. 66 (8), 5907–5936. doi:10.1021/acs.jmedchem.3c00156
Pastvova, N., Dolezel, P., and Mlejnek, P. (2021). Heat shock protein inhibitor 17-allyamino-17-demethoxygeldanamycin, a potent inductor of apoptosis in human glioma tumor cell lines, is a weak substrate for ABCB1 and ABCG2 transporters. Pharm. (Basel) 14 (2), 107. doi:10.3390/ph14020107
Peng, H., Qi, J., Dong, Z., and Zhang, J. T. (2010). Dynamic vs static ABCG2 inhibitors to sensitize drug resistant cancer cells. PLoS One 5 (12), e15276. doi:10.1371/journal.pone.0015276
Piya, S., Kornblau, S. M., Ruvolo, V. R., Mu, H., Ruvolo, P. P., McQueen, T., et al. (2016). Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood 128 (9), 1260–1269. doi:10.1182/blood-2016-01-692244
Premji, T. P., Dash, B. S., Das, S., and Chen, J. P. (2024). Functionalized nanomaterials for inhibiting ATP-dependent heat shock proteins in cancer photothermal/photodynamic therapy and combination therapy. Nanomater. (Basel). 14 (1), 112. doi:10.3390/nano14010112
Prince, T. L., Lang, B. J., Guerrero-Gimenez, M. E., Fernandez-Munoz, J. M., Ackerman, A., and Calderwood, S. K. (2020). HSF1: primary factor in molecular chaperone expression and a major contributor to cancer morbidity. Cells 9 (4), 1046. doi:10.3390/cells9041046
Rajabpour, A., Rajaei, F., and Teimoori-Toolabi, L. (2017). Molecular alterations contributing to pancreatic cancer chemoresistance. Pancreatology 17 (2), 310–320. doi:10.1016/j.pan.2016.12.013
Ramos, A., Sadeghi, S., and Tabatabaeian, H. (2021). Battling chemoresistance in cancer: root causes and strategies to uproot them. Int. J. Mol. Sci. 22 (17), 9451. doi:10.3390/ijms22179451
Reita, D., Pabst, L., Pencreach, E., Guerin, E., Dano, L., Rimelen, V., et al. (2022). Direct targeting KRAS mutation in non-small cell lung cancer: focus on resistance. Cancers (Basel) 14 (5), 1321. doi:10.3390/cancers14051321
Salamanca, H. H., Antonyak, M. A., Cerione, R. A., Shi, H., and Lis, J. T. (2014). Inhibiting heat shock factor 1 in human cancer cells with a potent RNA aptamer. PLoS One 9 (5), e96330. doi:10.1371/journal.pone.0096330
Salamanca, H. H., Fuda, N., Shi, H., and Lis, J. T. (2011). An RNA aptamer perturbs heat shock transcription factor activity in Drosophila melanogaster. Nucleic Acids Res. 39 (15), 6729–6740. doi:10.1093/nar/gkr206
Santagata, S., Hu, R., Lin, N. U., Mendillo, M. L., Collins, L. C., Hankinson, S. E., et al. (2011). High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 108 (45), 18378–18383. doi:10.1073/pnas.1115031108
Santagata, S., Mendillo, M. L., Tang, Y. C., Subramanian, A., Perley, C. C., Roche, S. P., et al. (2013). Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science 341 (6143), 1238303. doi:10.1126/science.1238303
Sharma, C., Choi, M. A., Song, Y., and Seo, Y. H. (2022). Rational design and synthesis of HSF1-PROTACs for anticancer drug development. Molecules 27 (5), 1655. doi:10.3390/molecules27051655
Shi, Y., Mosser, D. D., and Morimoto, R. I. (1998). Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12 (5), 654–666. doi:10.1101/gad.12.5.654
Shibue, T., and Weinberg, R. A. (2017). EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14 (10), 611–629. doi:10.1038/nrclinonc.2017.44
Siddiqui, A. D., and Piperdi, B. (2010). KRAS mutation in colon cancer: a marker of resistance to EGFR-I therapy. Ann. Surg. Oncol. 17 (4), 1168–1176. doi:10.1245/s10434-009-0811-z
Siegel, R. L., Giaquinto, A. N., and Jemal, A. (2024). Cancer statistics, 2024. CA Cancer J. Clin. 74 (1), 12–49. doi:10.3322/caac.21820
Singh, R., Letai, A., and Sarosiek, K. (2019). Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 20 (3), 175–193. doi:10.1038/s41580-018-0089-8
Singhal, A., Li, B. T., and O'Reilly, E. M. (2024). Targeting KRAS in cancer. Nat. Med. 30 (4), 969–983. doi:10.1038/s41591-024-02903-0
Stankiewicz, A. R., Livingstone, A. M., Mohseni, N., and Mosser, D. D. (2009). Regulation of heat-induced apoptosis by Mcl-1 degradation and its inhibition by Hsp70. Cell Death Differ. 16 (4), 638–647. doi:10.1038/cdd.2008.189
Sterrenberg, J. N., Blatch, G. L., and Edkins, A. L. (2011). Human DNAJ in cancer and stem cells. Cancer Lett. 312 (2), 129–142. doi:10.1016/j.canlet.2011.08.019
Sturner, E., and Behl, C. (2017). The role of the multifunctional BAG3 protein in cellular protein quality control and in disease. Front. Mol. Neurosci. 10, 177. doi:10.3389/fnmol.2017.00177
Su, K. H., Cao, J., Tang, Z., Dai, S., He, Y., Sampson, S. B., et al. (2016). HSF1 critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promote growth. Nat. Cell Biol. 18 (5), 527–539. doi:10.1038/ncb3335
Su, K. H., and Dai, C. (2016). Protein quantity-quality balance licenses growth. Cell Cycle 15 (23), 3155–3156. doi:10.1080/15384101.2016.1220714
Su, K. H., and Dai, C. (2017). mTORC1 senses stresses: coupling stress to proteostasis. Bioessays 39 (5). doi:10.1002/bies.201600268
Su, K. H., Dai, S., Tang, Z., Xu, M., and Dai, C. (2019). Heat shock factor 1 is a direct antagonist of AMP-activated protein kinase. Mol. Cell 76 (4), 546–561. doi:10.1016/j.molcel.2019.08.021
Suh, D. H., Kim, M. K., Kim, H. S., Chung, H. H., and Song, Y. S. (2012). Unfolded protein response to autophagy as a promising druggable target for anticancer therapy. Ann. N. Y. Acad. Sci. 1271 (1), 20–32. doi:10.1111/j.1749-6632.2012.06739.x
Swan, C. L., and Sistonen, L. (2015). Cellular stress response cross talk maintains protein and energy homeostasis. EMBO J. 34 (3), 267–269. doi:10.15252/embj.201490757
Szakacs, G., Paterson, J. K., Ludwig, J. A., Booth-Genthe, C., and Gottesman, M. M. (2006). Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 5 (3), 219–234. doi:10.1038/nrd1984
Takashima, K., Oshiumi, H., Matsumoto, M., and Seya, T. (2018). DNAJB1/HSP40 suppresses melanoma differentiation-associated gene 5-mitochondrial antiviral signaling protein function in conjunction with HSP70. J. Innate Immun. 10 (1), 44–55. doi:10.1159/000480740
Tan, H., Huang, F., Huang, M., Wu, X., and Tong, Z. (2023). HSF1 attenuates the release of inflammatory cytokines induced by lipopolysaccharide through transcriptional regulation of Atg10. Microbiol. Spectr. 11 (1), e0305922. doi:10.1128/spectrum.03059-22
Tanaka, N., Lin, J. J., Li, C., Ryan, M. B., Zhang, J., Kiedrowski, L. A., et al. (2021). Clinical acquired resistance to KRAS(G12C) inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 11 (8), 1913–1922. doi:10.1158/2159-8290.CD-21-0365
Tanaka, N., Okada, H., Yamaguchi, K., Seki, M., Matsubara, D., Gotoh, N., et al. (2023). Mint3-depletion-induced energy stress sensitizes triple-negative breast cancer to chemotherapy via HSF1 inactivation. Cell Death Dis. 14 (12), 815. doi:10.1038/s41419-023-06352-4
Tang, Z., Dai, S., He, Y., Doty, R. A., Shultz, L. D., Sampson, S. B., et al. (2015). MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell 160 (4), 729–744. doi:10.1016/j.cell.2015.01.028
Tchenio, T., Havard, M., Martinez, L. A., and Dautry, F. (2006). Heat shock-independent induction of multidrug resistance by heat shock factor 1. Mol. Cell Biol. 26 (2), 580–591. doi:10.1128/MCB.26.2.580-591.2006
Tilsed, C. M., Fisher, S. A., Nowak, A. K., Lake, R. A., and Lesterhuis, W. J. (2022). Cancer chemotherapy: insights into cellular and tumor microenvironmental mechanisms of action. Front. Oncol. 12, 960317. doi:10.3389/fonc.2022.960317
Toma-Jonik, A., Vydra, N., Janus, P., and Widlak, W. (2019). Interplay between HSF1 and p53 signaling pathways in cancer initiation and progression: non-oncogene and oncogene addiction. Cell Oncol. (Dordr) 42 (5), 579–589. doi:10.1007/s13402-019-00452-0
Vahid, S., Thaper, D., Gibson, K. F., Bishop, J. L., and Zoubeidi, A. (2016). Molecular chaperone Hsp27 regulates the Hippo tumor suppressor pathway in cancer. Sci. Rep. 6, 31842. doi:10.1038/srep31842
Vasan, N., Baselga, J., and Hyman, D. M. (2019). A view on drug resistance in cancer. Nature 575 (7782), 299–309. doi:10.1038/s41586-019-1730-1
Vihervaara, A., Mahat, D. B., Guertin, M. J., Chu, T., Danko, C. G., Lis, J. T., et al. (2017). Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat. Commun. 8 (1), 255. doi:10.1038/s41467-017-00151-0
Vilaboa, N. E., Galan, A., Troyano, A., de Blas, E., and Aller, P. (2000). Regulation of multidrug resistance 1 (MDR1)/P-glycoprotein gene expression and activity by heat-shock transcription factor 1 (HSF1). J. Biol. Chem. 275 (32), 24970–24976. doi:10.1074/jbc.M909136199
Vulsteke, C., Pfeil, A. M., Schwenkglenks, M., Pettengell, R., Szucs, T. D., Lambrechts, D., et al. (2014). Impact of genetic variability and treatment-related factors on outcome in early breast cancer patients receiving (neo-) adjuvant chemotherapy with 5-fluorouracil, epirubicin and cyclophosphamide, and docetaxel. Breast Cancer Res. Treat. 147 (3), 557–570. doi:10.1007/s10549-014-3105-5
Vydra, N., Toma, A., Glowala-Kosinska, M., Gogler-Piglowska, A., and Widlak, W. (2013). Overexpression of Heat Shock Transcription Factor 1 enhances the resistance of melanoma cells to doxorubicin and paclitaxel. BMC Cancer 13, 504. doi:10.1186/1471-2407-13-504
Wadood, A., Ajmal, A., and Rehman, A. U. (2022). Strategies for targeting KRAS: a challenging drug target. Curr. Pharm. Des. 28 (23), 1897–1901. doi:10.2174/1381612828666220506144046
Wang, H., Wang, X., Zhang, H., Deng, T., Liu, R., Liu, Y., et al. (2021). The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene 40 (28), 4695–4708. doi:10.1038/s41388-021-01898-z
Wang, L. E., Yin, M., Dong, Q., Stewart, D. J., Merriman, K. W., Amos, C. I., et al. (2011). DNA repair capacity in peripheral lymphocytes predicts survival of patients with non-small-cell lung cancer treated with first-line platinum-based chemotherapy. J. Clin. Oncol. 29 (31), 4121–4128. doi:10.1200/JCO.2010.34.3616
Watanabe, Y., Tsujimura, A., Taguchi, K., and Tanaka, M. (2017). HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy 13 (1), 133–148. doi:10.1080/15548627.2016.1248018
Wei, Y., Zhuang, Y., Zhang, Y., Luo, L., Yu, B., and Zeng, J. (2024). Role of heat shock protein 70 in silibinin-induced apoptosis in bladder cancer. J. Cancer 15 (1), 79–89. doi:10.7150/jca.88668
Workman, P., Burrows, F., Neckers, L., and Rosen, N. (2007). Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann. N. Y. Acad. Sci. 1113, 202–216. doi:10.1196/annals.1391.012
Workman, P., Clarke, P. A., Te Poele, R., Powers, M., Box, G., De Billy, E., et al. (2022). Discovery and validation of biomarkers to support clinical development of NXP800: a first-in-class orally active, small-molecule HSF1 pathway inhibitor. Eur. J. Cancer 174, S35. doi:10.1016/s0959-8049(22)00893-0
Wu, C. (1995). Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11, 441–469. doi:10.1146/annurev.cb.11.110195.002301
Xi, J., Liu, Y., Liu, H., Chen, H., Emborg, M. E., and Zhang, S. C. (2012). Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells 30 (8), 1655–1663. doi:10.1002/stem.1152
Xiang, W., Yang, Y., Weng, L., Ye, Z., Ding, P., Li, H., et al. (2023). Hyperhomocysteinemia activates NLRP3 inflammasome to cause hepatic steatosis and insulin resistance via MDM2-mediated ubiquitination of HSF1. Int. Immunopharmacol. 118, 110085. doi:10.1016/j.intimp.2023.110085
Xiao, X., Wang, W., Li, Y., Yang, D., Li, X., Shen, C., et al. (2018). HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J. Exp. Clin. Cancer Res. 37 (1), 201. doi:10.1186/s13046-018-0880-6
Xu, Y., An, Y., Wang, Y., Zhang, C., Zhang, H., Huang, C., et al. (2013). miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep. 29 (5), 2019–2024. doi:10.3892/or.2013.2338
Xue, N., Du, T., Lai, F., Jin, J., Ji, M., and Chen, X. (2022). Secreted HSP90α-LRP1 signaling promotes tumor metastasis and chemoresistance in pancreatic cancer. Int. J. Mol. Sci. 23 (10), 5532. doi:10.3390/ijms23105532
Yaeger, R., Mezzadra, R., Sinopoli, J., Bian, Y., Marasco, M., Kaplun, E., et al. (2023). Molecular characterization of acquired resistance to KRASG12C-EGFR inhibition in colorectal cancer. Cancer Discov. 13 (1), 41–55. doi:10.1158/2159-8290.CD-22-0405
Yamashita, M., Hirohashi, Y., Torigoe, T., Kusumoto, H., Murai, A., Imagawa, T., et al. (2016). Dnajb8, a member of the heat shock protein 40 family has a role in the tumor initiation and resistance to docetaxel but is dispensable for stress response. PLoS One 11 (1), e0146501. doi:10.1371/journal.pone.0146501
Yang, H., Sun, L., Liu, M., and Mao, Y. (2018). Patient-derived organoids: a promising model for personalized cancer treatment. Gastroenterol. Rep. (Oxf). 6 (4), 243–245. doi:10.1093/gastro/goy040
Yang, S., Ren, X., Liang, Y., Yan, Y., Zhou, Y., Hu, J., et al. (2020). KNK437 restricts the growth and metastasis of colorectal cancer via targeting DNAJA1/CDC45 axis. Oncogene 39 (2), 249–261. doi:10.1038/s41388-019-0978-0
Yoneda, A., Minomi, K., and Tamura, Y. (2021). Heat shock protein 47 confers chemoresistance on pancreatic cancer cells by interacting with calreticulin and IRE1α. Cancer Sci. 112 (7), 2803–2820. doi:10.1111/cas.14976
Yoon, Y. J., Kim, J. A., Shin, K. D., Shin, D. S., Han, Y. M., Lee, Y. J., et al. (2011). KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J. Biol. Chem. 286 (3), 1737–1747. doi:10.1074/jbc.M110.179440
Yu, J., Zhao, Y., and Xie, Y. (2024). Advances of E3 ligases in lung cancer. Biochem. Biophys. Rep. 38, 101740. doi:10.1016/j.bbrep.2024.101740
Zhang, B., Fan, Y., Cao, P., and Tan, K. (2021). Multifaceted roles of HSF1 in cell death: a state-of-the-art review. Biochim. Biophys. Acta Rev. Cancer 1876 (2), 188591. doi:10.1016/j.bbcan.2021.188591
Zhang, J. T. (2007). Biochemistry and pharmacology of the human multidrug resistance gene product, ABCG2. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 32 (4), 531–541.
Zhang, X., Lei, Y., Chen, X., He, J., Liu, Z., Zhu, W., et al. (2024). Suppression of NSCLC progression via the co-administration of Danusertib, an AURK inhibitor, and KRIBB11, an HSF1 inhibitor. Biochem. Pharmacol. 223, 116155. doi:10.1016/j.bcp.2024.116155
Zhang, Y., Li, C., Xia, C., Wah To, K. K., Guo, Z., Ren, C., et al. (2022). Adagrasib, a KRAS G12C inhibitor, reverses the multidrug resistance mediated by ABCB1 in vitro and in vivo. Cell Commun. Signal 20 (1), 142. doi:10.1186/s12964-022-00955-8
Zhang, Y., Murshid, A., Prince, T., and Calderwood, S. K. (2011). Protein kinase A regulates molecular chaperone transcription and protein aggregation. PLoS One 6 (12), e28950. doi:10.1371/journal.pone.0028950
Zhao, B., Wang, L., Qiu, H., Zhang, M., Sun, L., Peng, P., et al. (2017). Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget 8 (3), 3980–4000. doi:10.18632/oncotarget.14012
Zhao, S., Wang, J. M., Yan, J., Zhang, D. L., Liu, B. Q., Jiang, J. Y., et al. (2019). BAG3 promotes autophagy and glutaminolysis via stabilizing glutaminase. Cell Death Dis. 10 (4), 284. doi:10.1038/s41419-019-1504-6
Zhao, T., Zheng, H., Xu, J. J., Pantopoulos, K., Xu, Y. C., Liu, L. L., et al. (2024). MnO(2) nanoparticles trigger hepatic lipotoxicity and mitophagy via mtROS-dependent Hsf1(Ser326) phosphorylation. Free Radic. Biol. Med. 210, 390–405. doi:10.1016/j.freeradbiomed.2023.11.037
Zhu, C., Guan, X., Zhang, X., Luan, X., Song, Z., Cheng, X., et al. (2022). Targeting KRAS mutant cancers: from druggable therapy to drug resistance. Mol. Cancer 21 (1), 159. doi:10.1186/s12943-022-01629-2
Keywords: heat shock factor 1, chemoresistance, proteotoxic stress response, autophagy, apoptosis, drug efflux
Citation: Ghai S, Shrestha R and Su K-H (2025) HSF1 at the crossroads of chemoresistance: from current insights to future horizons in cell death mechanisms. Front. Cell Dev. Biol. 12:1500880. doi: 10.3389/fcell.2024.1500880
Received: 24 September 2024; Accepted: 18 December 2024;
Published: 09 January 2025.
Edited by:
Jing Pu, University of New Mexico, United StatesReviewed by:
Tejeshwar Rao, University of Houston, United StatesChunyan Wang, Shenzhen Maternity and Child Healthcare Hospital, China
Copyright © 2025 Ghai, Shrestha and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuo-Hui Su, a3VvLWh1aS5zdUB1dG9sZWRvLmVkdQ==
 Shruti Ghai
Shruti Ghai Rejina Shrestha
Rejina Shrestha Kuo-Hui Su
Kuo-Hui Su