- 1Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, Russia
- 2Department of Regenerative Medicine, Pirogov Russian National Research Medical University, Moscow, Russia
Whole-mount in situ hybridization (WISH) is a widely used method that supports the concept of “seeing is believing” by enabling the visualization of gene expression patterns in whole-mount multicellular samples or sections. This technique is essential in the study of epimorphic regeneration in cold-blooded vertebrates, where complex three-dimensional organs such as tails, limbs, and eyes are completely restored after loss. The tadpoles of the frog X. laevis serve as a convenient model for studying regeneration, as they can regenerate their tails within a week after amputation. Modern high-throughput sequencing methods have identified various cell populations involved in the regeneration process and determined the repertoire of genes activated during this time. Specifically, a population of reparative myeloid cells expressing mmp9 as a marker gene has been shown to be crucial for the initial stages of tail regeneration in X. laevis tadpoles. The validation of these data and further examination using WISH offers the advantage of providing detailed information on the spatial and temporal dynamics of target gene expression levels. However, detecting mRNA by WISH can be challenging when mRNA levels are very low, transcripts are localized in hard-to-access areas, or tissue samples are prone to background staining, as is the case with X. laevis regenerating tail samples. Here, we describe additional treatments for regenerating tail samples that minimize background staining and enhance the visualization of cells containing target RNA through in situ hybridization. Using an optimized WISH protocol on X. laevis tadpole tail regenerates, we obtained novel data on the mmp9 expression pattern during the first day post-amputation at the regeneration-competent stage 40 and the regeneration-incompetent stage 47 (refractory period). The significant differences in the expression patterns indicate that mmp9 activity is positively correlated with regeneration competence.
1 Introduction
One of the most frequently used methods in the concept of “seeing is believing” is in situ hybridization, which allows the visualization of the spatio-temporal expression pattern of a gene of interest in the cells of whole organisms or tissues (Herrmann and Neidhardt, 2006). This method involves the hybridization of a labeled antisense RNA probe with the corresponding endogenous mRNA in the sample, followed by a label-determined staining step. The importance of this method among developmental molecular biologists cannot be overstated.
Despite the emergence of high-throughput methods such as single-cell or bulk RNA sequencing, spatial transcriptomics, and proteomics, the results obtained by in situ hybridization remain a staple in many publications, providing crucial validating data. However, detecting mRNA by WISH can be challenging when mRNA expression levels are very low, the transcripts are localized in hard-to-reach or hard-to-view areas, or tissue samples are prone to background staining, which decreases the signal-to-noise ratio.
One of such case is WISH in regenerating tail of Xenopus frog tadpoles, which can regenerate an amputated tail in a week and a hind limb in a month (Beck C.W. 2012). This model along with other models of fins, tails and limbs regeneration in fishes and amphibians is widely used to study mechanisms of regeneration in amniotic species, which are distinguishing by their amazing ability to restore these body appendages (Murawala et al., 2012). The study of genes associated with regeneration in model anamniotic species is of great interest not only for basic science, but also for the development of approaches that could enhance regeneration in amniotes, especially in human. The point is that amniotes (reptiles, birds and mammals) cannot regenerate body appendages because such important stages of regeneration as the formation of the regenerative epithelia and blastema cannot be realized in them. In particular, this is attributed by the destruction of gene network and loss of many genes responsible for these processes occurred during amniotes evolution (Zaraisky et al., 2024).
Here we describe additional treatments for Xenopus laevis tadpoles' regenerating tail samples, which allow us to obtain clear, high-contrast images of target RNA-containing cells by WISH. The most challenging aspect of this method is minimizing background staining in the sample to achieve high-sensitivity detection of cells expressing the target gene. For wild-type X. laevis tadpole tails at different stages of regeneration, this presented a double challenge.
Firstly, melanosomes (pigment granules) actively migrate with cells to the amputation site and can therefore interfere with the BM Purple stain signal. Additionally, due to the numerous melanophores, visualization and photodetection of the staining signal are very difficult (El Mir et al., 2024). If melanophores are not the focus of the investigation, we suggest adding a bleaching step for the tadpoles, which has proven effective for decoloring both melanosomes and melanophores after fixation and before the pre-hybridization stages.
Secondly, tail fins are very loose tissues, and the main problem during in situ hybridization is strong background staining, especially when the target RNA is not highly expressed and requires long staining incubation. We have observed that tadpole samples fixed immediately after amputation (0 hpa, hours post amputation) exhibited the lowest background staining of fins. To address the background problem, we recommend making fin incisions in a fringe-like pattern at some distance from the area of interest in the regenerating tail. This tail fin notching procedure improved the washing out of all solutions, preventing BM Purple from getting trapped in the loose fin tissues and causing non-specific autocromogenic reactions. Even after 3–4 days of staining, no background staining was detected. The combination of these procedures before hybridizing the regenerating tail samples allowed us to obtain high-contrast images of gene expression patterns without background interference.
To test various WISH protocols, we chose the gene encoding Zn2+-dependent extracellular matrix metalloproteinase 9 (mmp9), whose expression pattern was firstly detected in X. laevis albino embryos and tadpoles during normal development and hindlimb bud and lens regeneration (Carinato et al., 2000). Recently mmp9 was identified as one of the genes specific to the population of reparative myeloid cells that plays a key role in tail regeneration in X. laevis tadpoles (Aztekin et al., 2019; Aztekin et al., 2020). This lineage of cells is essential for the early steps of regeneration, as it quickly replaces the inflammatory myeloid lineage and induces subsequent processes necessary for regeneration progression: apoptosis and tissue remodeling, which ultimately lead to the relocalization of regeneration-organizing cells responsible for progenitor proliferation (Aztekin et al., 2019; Aztekin et al., 2020). According to yet unpublished data, mmp9, along with mpepa1, junb, mmp1 and mmp8, are unique markers of regeneration-inducing cells (RICs), which are formed transiently from basal epidermal cells. These markers are critical for modifying the surrounding extracellular matrix to facilitate the migration of other cell types, such as regeneration-organizing cells that further promote regeneration. These markers were identified through bulk, single-cell, and spatial RNA-seq (Radek et al., 2023).
As a result, we demonstrate, for the first time to our knowledge, the detailed expression pattern of mmp9 during the early stages (0, 3, 6, and 24 h post-amputation) of tail regeneration in tadpoles at stage 40. Additionally, we show that the activity of mmp9 is closely linked to regeneration competency. Its expression pattern significantly differs in tails amputated during the refractory period (stages 45–47), when regeneration is temporarily blocked as part of the normal developmental program (Beck et al., 2003). The high-quality images of regenerating tails, stained for mmp9-expressing cells using our optimized WISH protocol, allowed us to observe the behavior of these cells during the early stages of regeneration and to clarify and supplement data obtained by high-throughput methods, such as bulk- and sc-RNAseq.
2 Design of experiments
To optimize the in situ hybridization staining in the regenerating tails of X. laevis tadpoles, we tested several additional treatments, as shown in Figure 1. We used tadpoles at stage 40 with tails regenerating for 0 or 6 hpa to evaluate different protocol variants. Samples at 0 and 6 hpa (12–15 tadpoles) for each protocol variant were collected in at least three independent experiments.
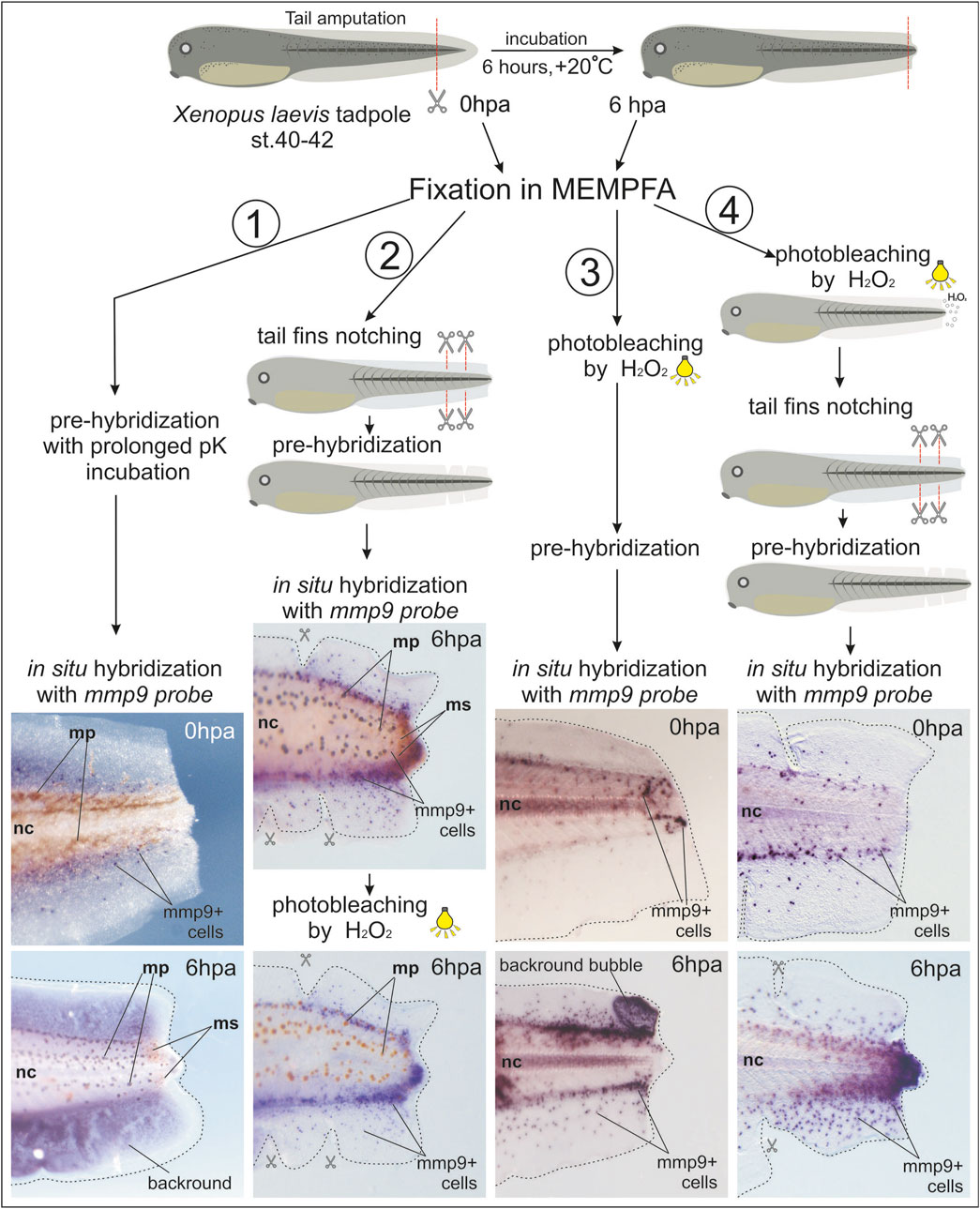
Figure 1. Comparison of different treatments for whole-mount samples of regenerating tails of Xenopus laevis tadpoles to obtain a clearer in situ hybridization signal from mmp9+ cells. 1st variant: Prolonged proteinase K (pK) incubation during the pre-hybridization step, which did not improve staining signal clarity or reduce background staining. 2nd variant: Combination of tail fin notching before WISH and photo-bleaching after BM Purple staining. 3rd variant: Addition of a photo-bleaching step before WISH. 4th variant: Early photo-bleaching after fixation in MEMPFA and rehydration, followed by tail fin notching. This variant provided the clearest image results. Mp–melanophores, ms–melanosomes, nc–notochord.
Treating samples with proteinase K helps to remove nucleases and makes the tissue more permeable to reagents. Lengthening the incubation time with proteinase K solution for samples at later developmental stages can increase the sensitivity of in situ hybridization and reduce non-specific staining.
In the first variant, we extended the proteinase K incubation time for tadpole tail samples to 30 min, but the staining results were unimpressive, with mmp9+ cells overlapping with strong background staining.
In the second variant, we partially notched the edges of the fin to help wash out reagents causing background staining from the loose tissues. This approach allowed us to observe many more mmp9+ cells. To reduce overlapping of the BM Purple stain with melanosomes and melanophores, we added a post-staining photo-bleaching step, as recommended by Harland (1991). The result was improved imaging, but melanophores only faded to brown.
In the third variant, we moved the photo-bleaching step to the very beginning of the protocol, immediately after fixation in MEMPFA and the dehydration step. This resulted in perfectly albino tails, but some samples developed large bubbles in the tail fin area filled with non-specific BM Purple staining after in situ hybridization.
Finally, in the fourth variant, we combined the early photo-bleaching step with caudal fin cutting before sample hybridization, resulting in very clear images of the specific staining of mmp9+ cells. The step-by-step procedures for this optimized protocol are described below.
2.1 Materials
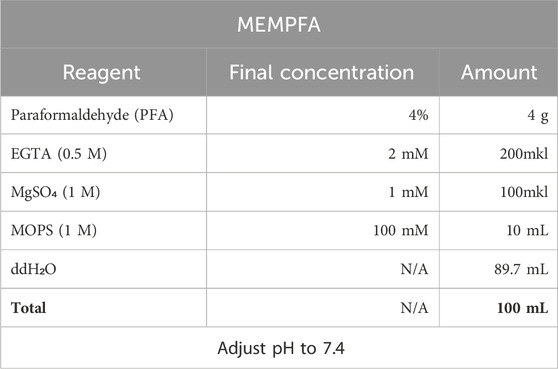
1. MEMPFA solution for samples fixation.
Procedure for preparing MEMPFA:
First add PFA powder to half the total volume of water, add NaOH (approximately 100 µL per 100 mL) and heat to 60°C for better dissolution. Once the PFA is completely dissolved, cool the solution to room temperature, add the remaining reagents and adjust the pH to 7.4.
Note: MEMPFA solution stored at +4°C can be used to fix samples for 2 weeks. It can subsequently be used for postfixation and storage of samples after in situ hybridization.
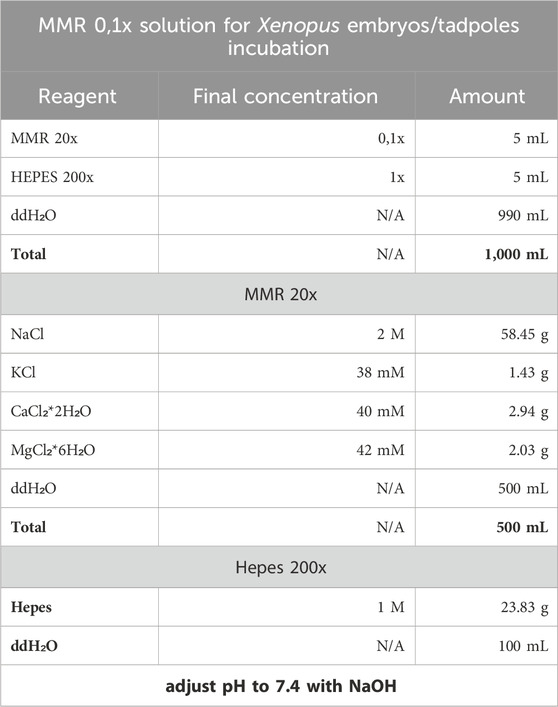
2. The Xenopus embryos and tadpoles are incubated in MMR 0,1x with Hepes 1x.
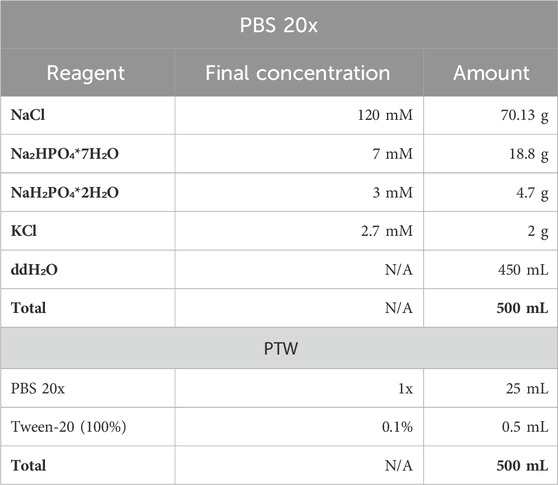
3. PBS 1x solution for washes was prepared from PBS 20x stock. PTW solution was prepared from PBS 1x by addition of Tween20 detergent till 0.1% final concentration.
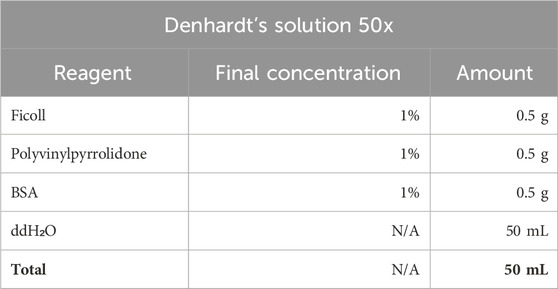
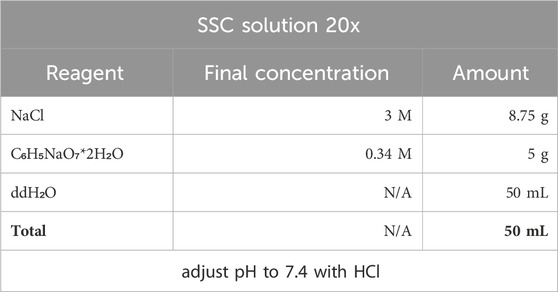
4. Pre-Hybridization Buffer and its ingridients.
Note: When preparing the PH buffer use RNase-free tubes, pippets, tips and wear gloves. RNase contamination can lead to dig-RNA degradation in the hybridization probe and in situ hybridization failure. The PH-buffer can be stored at −20oC in tube with parafilme-wrapped lid.
Stock Denhardt’s solution (50x) can be stored at −20oC.
Stock solution can be stored at −20oC. SSC 2x and 0,2x solutions for washing off dig-probe are prepared from stock solution by dilution 10 or 100 times with double distilled water.
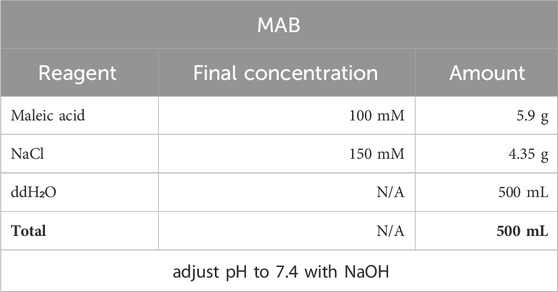
5. MAB solution for incubation with anti-bodies and further washing off.
Prepare a fresh MAB solution and use it for one WISH cycle, storing at +4C between incubation and antibody washout.
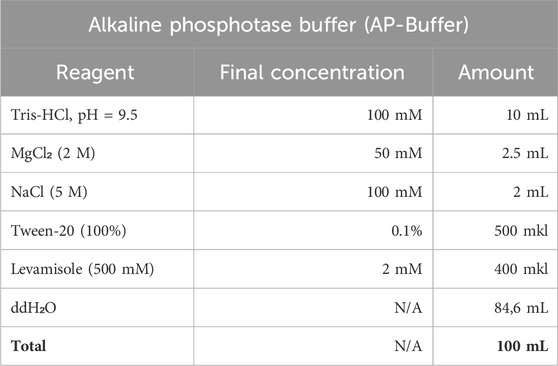
6. Alkaline phosphatase buffer is prepared fresh with an endogenous AP inhibitor (levamisole) for preparation for staining and without it for washing.
7. The BM Purple substrate (Roche, MERCK) is used with addition of levamisole till 1 mM final concentration.
2.2 Equipment
The following equipment was used in the current work:
Applied biosystems ProFlex PCR system (Thermo Fisher Scientific), thermostate “GNOM” (DNA-technologies), Eppendorf MiniSpin centrifuge, Leica KL300 LED binocular, thermostate incubator (Thermo Fisher Scientific), Vannas scissors, orbital shaker OS-20 (Biosan), hot shaker (Bellco Biotechnology), Leica binocular M205 with camera Leica DC 400F, fluorescent lamp light (OSRAM L 18W/77), cryotome MICROM HM 525 (Thermo Scientific).
3 Step-by-step procedures
Our laboratory has historically used the in situ hybridization protocol described by Richard Harland (Harland et al., 2004) as it is optimized for X. laevis embryos. However, in regenerating tail samples, this protocol resulted in strong background staining of the tail fins. Below, we describe our additional sample treatments and modifications to the in situ protocol for regenerating tadpole tail samples that significantly improved the signal-to-noise ratio.
The first step in preparing for an in situ hybridization experiment is to clone the gene of interest and synthesize the corresponding antisense RNA labeled with dig-/fluorescein-/dnp-uridine residues and prepare a hybridization probe (Long and Rebagliati, 2002). Hybridization with labeled sense RNA, which does not bind to the RNA of interest and allows detection of the presence of basic background staining, was used as a control.
3.1 Mmp9 cloning, dig-RNA synthesis and dig-probe preparation
1) The fragment of mmp9. S cDNA (GenBank accession number NM_001086503) was amplified using PCR method. The DNA matrix was presented by the cDNA reversely transcribed by MMLV-RT (Evrogen) from mRNA of 1dpa (days post amputation) regenerating tails of X. laevis tadpoles extracted by RNAextract reagent (Evrogen). The following primers were used: mmp9. S F 5’ – TTCACTCGTATATACAGC; mmp9. S FTest 5’ - TATCTTCGACGGAGTGTCAT and mmp9. S R 5’ – TGCACATCACTGTGATCCA. After purificatioin the PCR-fragment was cloned into pAL2-T vector (Evrogen) and then several clones were sequenced. On the base of correct clone the PCR-fragment of mmp9 with SP6 promoter in the reverse orientation was obtained and purified with elution in RNAse-free water (Encyclo plus PCR kit, CleanUp Standard, Evrogen).
2) The Dig-labeled RNA antisense probe for the whole-mount in situ hybridization was synthesized by SP6 polymerase from SP6 transcription kit (mMessage mMachine, Thermo Fisher Scientific) with DIG RNA Labeling Mix (Roche) and purified mmp9 PCR-product obtained with mmp9. S F and M13 reverse primers as a matrix. The dig-labeled sense-RNA for mmp9 was synthesized by T7 polymerase from T7 transcription kit (mMessage mMachine, Thermo Fisher Scientific) with DIG RNA Labeling Mix (Roche) and purified mmp9 PCR-product obtained with M13 forward and mmp9. S R primers as a matrix. The obtained dig-RNAs were purified by CleanRNA Standard kit (Evrogen) and stored at −20oC.
3) To prepare mmp9 dig-RNA probe 1000 ng of antisense or sense dig-RNA was diluted in 1 mL of pre-hybridization buffer (PHB). The probe can be stored at −20oC.
Note: The probe can be re-used 2–3 times, on the 4th time we usually add 500 ng of dig-RNA to 1 mL of old probe and use it 2-3 more times.
3.2 Tadpoles tail amputation, fixation and dehydration
4) Tadpoles were incubated in 0,1xMMR till the 40–41 stage or 45–46 according to normal tables of Xenopus development (Zahn et al., 2022). Before amputation tadpoles were placed into Petri dish covered with 2% agarose and filled with anesthetic solution. The quarter of tail was amputated by Vannas scissors and 0 hpa tadpoles were replaced into fresh 0,1MMR solution and have been incubated for 6 h at room temperature.
5) The anesthetized regenerating tadpoles were placed in 11 mL-glass vials containing 5–7 mL of cold, freshly prepared MEMPFA solution and incubated overnight at 4oC. All incubations are hold on the slightly rotating (<60 rpm) platform of orbital shaker at room temperature or in incubated orbital shaker.
6) Afterwards the MEMPFA solution was replaced by 1xPBS (5–7 mL) and washed three times for 5–10 min at RT (room temperature).
7) The graded dehydratation of samples was made by washing with 5–7 mL of the following solutions at RT on the rotating platform:
a) 25% EtOH/75% PBS – 5–10 min
b) 50% EtOH/50% PBS - 5–10 min
c) 75% EtOH/25% PBS -5–10 min
d) 96% EtOH - 5–10 min
e) 96% EtOH -storage
II Pause point. Dehydrated samples can be stored at −20°C for several months until the inspiration arises to perform in situ hybridization.
3.3 Tadpoles photobleaching and post-fixation
In the case of albino tadpoles, the bleaching step can be skipped. However, if only wild-type tadpoles are available, the problem of accumulating near the amputation line melanosomes (granules of embryonic pigment) and melanophores (black pigment cells) complicating the visualization of in situ hybridization staining arises. Here we describe the optimized bleaching protocol for regenerating tails samples.
Note: Wear gloves and lab coat.
8) Replace 96% EtOH with bleaching solution – 3%H₂O₂ in 96%EtOH.
Note: the bleaching solution must be freshly prepared.
9) Lie down glass vials with samples in bleaching solution on a tray lined with foil. Place the tray on the orbital shaker under the fluorescent lamp light (OSRAM L 18W/77) and bleach for 12–24 h.
10) Wash samples twice by 5 mL of 96% EtOH.
11) Rehydrate samples by sequential washes
a) 5 mL 75% EtOH/25% PBS 5–10 min
b) 5 mL 50% EtOH/50% PBS 5–10 min
c) 5 mL 25% EtOH/75% PBS 5–10 min
d) 5 mL 1xPBS 5–10 min
e) 5 mL 1xPBS 5–10 min
12) Post-fixate samples in 5 mL MEMPFA for 1 h at room temperature.
Note: We found that melanophores are discolored best of all if bleached before the WISH pre-hybridization, but the melanosomes can be bleached before as well as after WISH procedures (see images on Figure 1).
3.4 Tail fin notching
The background staining of tail fins makes it difficult to distinguish weak target staining. We hypothesized that the background staining in long-lasting colorings is due to the loose structure of fin tissues, which obstructs effective washing out of non-specific hybrids, unbound probes, and staining solutions, leading to non-specific BM Purple staining. To minimize this issue, we attempted to increase the time of proteinase K treatment, but this had no effect. Previous results have shown that mmp9 is expressed primarily in the peri-trunk area at stage 40 (Carinato et al., 2000). Therefore, we made 3-4 cuts on the upper and 2-3 cuts on the lower tail fins using Vannas scissors or an ophthalmic scalpel. The distance between cuts was 600–800 μm, and the depth of the cut corresponded to the width of the fin from the outer edge to the trunk muscles (in the range of 200–300 μm for the upper and 400–600 μm for the lower fin). We do not recommend cutting the muscle tissue, as it leads to a stronger deformation of the sample after hybridization procedures and sometimes causes background staining of the edges of the cut muscle tissue, which makes it difficult to visualize the true-positive signal.
13) Wash samples with 5 mL of PBS three times for 5–7 min each. Place tadpoles in an agarose-coated Petri dish with PBS. Make several cuts in the tail fins under a binocular microscope. Return the tadpoles to vials and process according to the WISH protocol optimized in our laboratory for X. laevis.
3.5 Whole-mount in situ hybridization
3.5.1 Pre-hybridization
All washes/incubations are hold on the slightly rotating (<60 rpm) platform of orbital shaker by default at room temperature unless otherwise specified.
14) Remove PBS and add 5 mL of pK-solution (recommended working concentration of Proteinase K is 1 mkg/mL), incubate 10–15 min
15) Change solution on Glycin solution (working concentration is 2 mg/mL) incubate for 10 min
16) Wash twice with 5 mL PTW for 5–7 min
17) Remove PTW and add 3 mL of MEMPFA for post-fixation during 20 min
18) Do two washes by 5 mL PTW for 5–7 min each
19) Remove PTW and add 4 mL of PTW and 1 mL of Pre-hybridization buffer (PHB), incubate for 15 min
20) Set change solution on 1 mL of PHB and incubate at +60oC.
Note: X. laevis embryos at early developmental stages (till st.25) can be moved to +60°C after room temperature without any harm, but the tadpoles may curl up, making imaging awkward. To minimize distortion, we gradually increase the temperature in the incubated orbital shaker, starting from +35°C to +60°C with two 10°C intervals and one 5°C interval. We maintain incubation at each temperature level for 20–30 min. Finally, the samples are incubated in PHB at +60°C for at least 30 min.
21) Change PHB on fresh pre-heated at +60oC 0.5 mL PHB and incubate vials at +60oC for 4–16 h.
3.5.2 Dig-RNA-hybridization
22) Replace the PHB with a 0.5 mL dig-RNA probe preheated at +60°C and incubate for 12–15 h in an orbital shaker at +60°C.
3.5.3 Dig-probe washing out and incubation with antibodies
23) Carefully remove dig-probe with a pipette into eppendorf, wrap in parafilm and store at −20 for re-use. Add pre-heated at +60oC 0.5 mL PHB. Incubate 30 min at +60oC.
24) Remove PHB and add pre-heated at +60oC 0.5 mL PHB, incubate 30 min at +60oC.
25) Wash twice with 5 mL of pre-heated at +60oC 2x SSC, incubate 1 h at +60°C.
26) Remove 2xSSC, add 5 mL of pre-heated at +60oC 0,2xSSC and transfer vials on the orbital shaker at room temperature, incubate 30 min. All further incubations are hold at room temperature.
27) Wash with 5 mL 0,2xSSC for 30 min.
28) Change 0,2xSSC on 5 mL MAB and incubate 10 min.
29) Wash with 5 mL MAB for 10 min.
30) Remove MAB, add 1 mL of pre-Blocking Solution (without calf serum) (final concentration of blocking reagent is 2%) and incubate 20 min.
31) Remove pre-Blocking solution, add 1 mL Blocking solution (+20% Heat inactivated calf serum) and incubate 1–2 h.
32) Change solution on 0.5 mL Blocking solution +0.1% anti-dig-AP antibodies and incubate at +4°C overnight on the orbital shaker.
3.5.4 Anti-bodies wash, chromogenic staining and post-fixation
33) Remove solution with antibodies, add 5 mL MAB and incubate 1 h. Make in sum 5 washes with MAB.
II Pause point: The final rinse can be left in the refrigerator overnight.
34) Prepair fresh AP-Buffer with levamisole, inhibitor of endogenous alkaline phosphatase. Change MAB to 3 mL of AP Buffer (lev+), incubate 15–20 min.
35) Repeat AP buffer (lev+) rinse for 15–20 min.
36) Add 1mkl levamisole into 0.5 mL of BM Purple, change AP buffer to BM Purple (lev+) and place the vials into dark room/box/wrap into foil. Incubate at RT till appropriate staining. If necessary the vial(s) may be placed into refrigerator and left overnight, the rate of staining at +4oC is very low. It is better to change the BM Purple (lev+) if it starts to turn blue.
Note: BM Purple is sensitive to the light and must be kept in darkness, otherwise it starts autochromogenic reaction with multiple colored sediments, sticking on the samples.
37) Upon reaching the required color intensity, replace BM Purple with AP buffer (lev-), rinse twice with 2–3 mL for 15–20 min.
38) Change AP buffer to MEMPFA for staining fixation.
Note: Of course, like all methods, this one has its own sensitivity limits. According to our experience, to detect genes that are revealed in transcriptome sequencing (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE88975 and our unpublished data, platform Illumina 2000) in the amount of 70–170 normalized by DESeq counts, staining step should be continued for 3–4 days, putting the samples in the refrigerator overnight and changing the BM Purple solution to a fresh one as it turns blue. The background staining can grow in the notochord, but at the same time the pattern of the gene of interest gradually appears. If it is not diffuse, but concentrated in certain cells or their clusters, then the pattern will be clearly visible. Staining of genes with over 500 counts requires 1–2 days of development (for mmp9 at 24hpa 890, our unpublished data), over 3,000–4–5 h.
3.5.5 Samples imaging and storage
39) To photograph the samples wash them with PBS three times and make images on agarose-coated Petri dish with PBS under the stereomicroscope (in our case it was Leica M205) with camera (Leica DC 400F).
40) For storage change PBS to MEMPFA and store at +4oC or perform graded dehydration as in step 4) and store in EtOH 96% at −20oC.
41) For more detailed pattern describing the samples after in situ hybridization can be cryosectioned, treated with dapi and embedded into mowiol for imaging.
4 Results and discussion
Although the expression activity of mmp9 during early Xenopus development, wounding, and hindlimb regeneration was demonstrated as early as 2000 (Carinato et al., 2000), the first data on its spatiotemporal expression pattern in the regenerating tadpole tail tip appeared relatively recently. However, these data are quite contradictory and not complete. For the first time, mmp9 expression during Xenopus tadpole tail regeneration was reported at 1, 2, and 3 dpa in reparative myeloid cells using scRNA-seq (Aztekin et al., 2019). As the authors of this study showed, after amputation at stage 40 (0 dpa), the number of mmp9 transcript counts decreased up to 1 dpa with a subsequent increase to 3 dpa. However, in the work of other authors, an increase in mmp9 expression over background level was detected using bulk HiSeq already at 0.5-1 hpa, followed by further rise in expression up to 6 hpa and a subsequent drop down to 3 dpa (Radek et al., 2023). These authors also validated mmp9 expression by in situ hybridization and spatial transcriptomics only at 1 dpa, and both methods showed rather blurred images of expression regions along the trunk and on the amputated tail tip.
To clarify the data on the temporal-spatial distribution of cells with mmp9 expression, we tested mmp9 gene activity in tissues of the regenerating tail of X. laevis tadpoles using our new in situ hybridization protocol optimized for these samples. Using the fourth variant of treatments (see Figure 1), we performed in situ hybridization on tadpole tails amputated at stage 40 (regeneration-competent) or at refractory stages 46–47 (regeneration-incompetent) and fixed at 0, 3, 6, and 24 hpa (Figure 2). Samples for each time point (3, 6, 24 hpa, 12–15 tadpoles) were collected in three independent experiments. Images in Figure 2 show one tadpole out of ≥75% of tadpoles from each group at a given time point that have the same expression pattern of mmp9 mRNA in cells of regenerating tails.
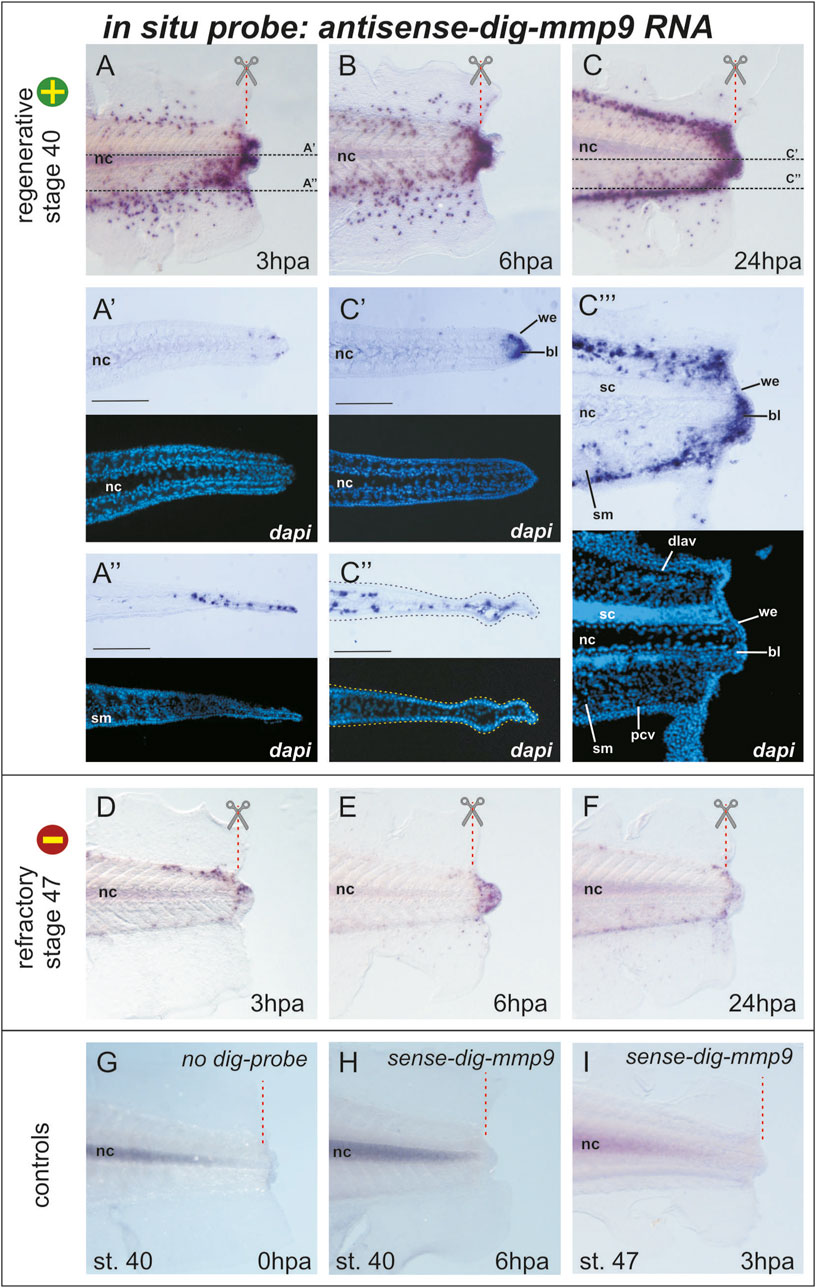
Figure 2. Mmp9 expression pattern in the tail of Xenopus laevis tadpoles after amputation at stage 40 (regeneration competent) and stage 47 (regeneration incompetent) revealed by an optimized whole-mount in situ hybridization protocol followed by cryo-sectioning (A–C) Images of the distal part of the tail 3, 6, or 24 h post-amputation stained (blue-violet dots) for cells expressing mmp9. The red dashed line with scissors indicates the amputation line, and black dashed lines indicate the position of the frontal cryosections of (A’, A”) and (C’, C”). Light microscopy images of sections are combined with DAPI fluorescence. Scale bar 200 µm (C’’’) The light microscopy image of the sagittal cryo-section of the tail at 24 hpa after in situ hybridization for mmp9 expression with the corresponding DAPI fluorescence image. Bl–blastema, dlav–dorsal longitudinal anastomosing vessel, nc–notochord, pcv–posterior cardinal vein, sc–spinal cord, sm–somatic muscles, we–wound epithelium (D–F) Mmp9 expression pattern in tadpole tails 3, 6, or 24 h after amputation during the refractory period (stage 47, regeneration incompetent) (G–I) Images of control in situ hybridization performed without dig-labeled RNA probe (G) or with sense-dig-labeled mmp9 RNA probe show similar background staining of the notochord. The intensity of notochord staining was positively correlated with the duration of the chromogenic reaction. No positive signals were detected in other tissues of regenerating tail.
As shown in Figure 1, taken immediately after tail amputation, mmp9+ cells are predominantly scattered in the area of the dorsal and ventral tail fins. When comparing mmp9 expression patterns in the regenerating tail at 0, 3, 6, and 24 hpa, a sharp increase in mmp9+ cells can be observed in the distal region of the caudal fins, as well as in the notochord and neural tube below the amputation line (Figures 1, 2A). The staining of the notochord in the proximal region of the tail is background, as it appears during hybridization both without RNA-probe and with the sense-RNA probe (compare Figures 2B, H). Considering that in early development, mmp7, mmp9, and mmp18 are active in macrophages and are required for their migration (Tomlinson et al., 2008) new data on the mmp9 expression pattern during regeneration are likely the result of active migration of myeloid cells towards the injury. At the same time, we cannot exclude the possibility that, in addition to the migration of mmp9+ cells, there is also an induction of mmp9 expression in the cells of the tail stump. Furthermore, by 24 h post-amputation, the orderliness of migrating mmp9+ cells increases greatly, resulting in a concentration of these cells along the main vascular pathways. Notably, the temporal mmp9 expression profile revealed here using WISH is consistent with the mmp9 temporal profile obtained by other authors using bulk RNA-seq of the regenerating tail in the more rapidly developing species Xenopus tropicalis, in which the expression peak of this gene was observed as early as 15 hpa (Chang et al., 2017).
To determine in which tissues mmp9+ cells are localized, we made frontal and sagittal cryo-sections (20 µm thick) of tails stained by WISH. Figure 2 shows that at the level of the notochord, cells expressing mmp9 appear near its distal tip, and their number increases significantly at 24 hpa, filling the space between the tip of the notochord and the wound epithelium. This area is usually defined as a blastema (Figures 2A’, C’). At first glance, it appears that the mmp9+ cells in the fin region lie in the outer epithelial layer. However, a frontal section at the level of the ventral caudal fin shows that mmp9+ cells are distributed underneath it in the stroma or vessels of the fin (Figures 2C’, C’’). Additionally, the sagittal section at 24 hpa demonstrates stained cells concentrated above and below the notochord and spinal cord. According to the literature, these cells correspond to the dorsolateral anastomosing vessel and posterior cardinal vein (Levine et al., 2003; Senevirathne et al., 2020). It is also clear that the closer the myeloid cells are to the distal end, the more actively they spread to the area of muscle tissue and the spinal cord (Figure 2C’’’).
In vivo tracing of labeled myeloid lineages, including granulocytes and macrophages, during wounding and infection in Xenopus tadpoles has shown that myeloid cells near the injury site start migrating as early as 20 min post-injury. The myeloid cells roll along the vascular endothelium or extravasate from vessels, migrating toward the wound area (Paredes et al., 2015). Mmp9 (gelatinase B) is a secreted matrix metalloproteinase that degrades proteins of the extracellular matrix, particularly collagen IV, VII, and X (Fu et al., 2009). The distribution of type IV collagen was detected at stage 56 in the basement membrane within the skin, between muscle cells, and around the spinal cord and notochord lamella (Nakajima and Yaoita, 2003). The detected pattern of the reparative myeloid marker mmp9 during the first day after tail amputation can be explained by combining all these data. Myeloid cells, likely attracted by chemokines or other agents, move toward the damage (amputation line) along vessels and also between cells of tissues containing collagen IV, VII, or X, cutting a path through the ECM using mmp9 as a machete. Such active ECM disruption near the amputation line could induce the release of ECM-linked factors, alter the cellular connectome and signaling repertoire, stimulate cellular dedifferentiation for blastema formation, and provide spatial freedom for further proliferation.
The next question we addressed was whether and how the expression pattern of mmp9 changes if the tail is amputated during stages when it is incompetent for regeneration, i.e., during the so-called refractory period (stages 45–47). Indeed, WISH staining of mmp9 transcripts at 3, 6, and 24 hpa at refractory stages differed significantly from that at the regeneration-competent stage 40 (compare Figures 2A–F). As shown in Figures 2D–F, the number of mmp9+ cells in refractory tadpoles is catastrophically low beneath the amputation line as well as in the tail fin tissues. By 6 hpa, mmp9+ cells are concentrated in the tail tip, and by 24 hpa, the number of cells decreases. At 2 and 3 dpa, mmp9 expression drops to zero levels, similar to 0 dpa (data not shown). These results suggest an interconnection between myeloid cell activity and regeneration progression.
Comparing the obtained results with scRNA-seq data, some inconsistencies in expression levels can be seen. The mmp9 expression, as determined by the scRNA-seq transcriptomic atlas, is downregulated by 1 dpa (Aztekin et al., 2019), in contrast to the apparent upregulation detected using our optimized in situ hybridization protocol. The expression level of mmp9 in the refractory period according to scRNA-seq data is higher than at stage 40, while we demonstrate the opposite results. Undoubtedly, the large volume of new omics data is indispensable and very useful for guiding research. However, validating the data using proven old-fashioned methods is still a necessary step. The obtained high-quality imaging data of regenerating tails stained for mmp9-expressing cells by the optimized WISH protocol made it possible to observe the life of these cells at the early stages of regeneration and to clarify and supplement the data obtained by high-throughput methods. It is likely that the proposed protocol will be suitable for addressing signal-to-noise ratio problems in similar samples, such as the regenerating fin of Danio rerio, the tail of X. laevis during normal development or the regenerating hindlimb, regenerating tail, or limb of Ambystoma mexicanum. In addition, we believe it is worth testing whether our additional sample treatments are applicable to variations of in situ hybridization, such as two-color WISH or fluorescent multichannel WISH (Long and Rebagliati, 2002; Vize et al., 2009).
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Review Board (or Ethics Committee) of the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry (Moscow, Russia, protocol code IACUC 229 and 1 February 2018). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AS: Data curation, Formal Analysis, Investigation, Validation, Visualization, Writing–original draft. EP: Methodology, Resources, Writing–review and editing. AZ: Data curation, Funding acquisition, Writing–review and editing. MT: Conceptualization, Methodology, Project administration, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by RSF grant 23-74-30005 (AZ).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aztekin, C., Hiscock, T. W., Butler, R., De Jesús Andino, F., Roberts, J., Gurdon, J. B., et al. (2020). The myeloid lineage is required for the emergence of a regeneration permissive environment following Xenopus tail amputation. Dev. Dev. 147, 185496. doi:10.1242/dev.185496
Aztekin, C., Hiscock, T. W., Marioni, J. C., Gurdon, J. B., Simons, B. D., and Jullien, J. (2019). Identification of a regeneration-organizing cell in the Xenopus tail. Science 364, 653–658. doi:10.1126/science.aav9996
Beck, C. W. (2012). “Studying regeneration in Xenopus,” in Xenopus protocols. Editors S. Hoppler,, and P. D. Vize (Totowa, NJ: Humana Press), 525–539. doi:10.1007/978-1-61779-992-1_30
Beck, C. W., Christen, B., and Slack, J. M. (2003). Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev. Cell 5 (3), 429–439. doi:10.1016/s1534-5807(03)00233-8
Carinato, M. E., Walter, B. E., and Henry, J. J. (2000). Xenopus laevis gelatinase B (Xmmp-9): development, regeneration, and wound healing. Dev. Dyn. 217, 377–387. doi:10.1002/(SICI)1097-0177(200004)217:4<377::AID-DVDY5>3.0.CO;2-U
Chang, J., Baker, J., and Wills, A. (2017). T ranscriptional dynamics of tail regeneration in Xenopus tropicalis. Genesis 55, e23015. doi:10.1002/dvg.23015
El Mir, J., Nasrallah, A., Thézé, N., Cario, M., Fayyad-Kazan, H., Thiébaud, P., et al. (2024). Xenopus as a model system for studying pigmentation and pigmentary disorders. Pigment. Cell Melanoma Res. Pcmr., 13178. doi:10.1111/pcmr.13178
Fu, L., Das, B., Mathew, S., and Shi, Y.-B. (2009). Genome-wide identification of Xenopus matrix metalloproteinases: conservation and unique duplications in amphibians. BMC Genomics 10, 81. doi:10.1186/1471-2164-10-81
Harland, R. M. (1991). “Appendix G: in situ hybridization: an improved Whole-Mount method for Xenopus embryos,” in Methods in cell biology (Elsevier), 685–695. doi:10.1016/S0091-679X(08)60307-6
Harrison, M., Abu-Elmagd, M., Grocott, T., Yates, C., Gavrilovic, J., and Wheeler, G. N. (2004). Matrix metalloproteinase genes in Xenopus development. Dev. Dyn. 231, 214–220. doi:10.1002/dvdy.20113
Herrmann, B. G., and Neidhardt, L. (2006). “Whole Mount ISH,” in Encyclopedic reference of genomics and proteomics in molecular medicine (Springer Berlin Heidelberg), 1994–1998. doi:10.1007/3-540-29623-9_1540
Levine, A. J., Munoz-Sanjuan, I., Bell, E., North, A. J., and Brivanlou, A. H. (2003). Fluorescent labeling of endothelial cells allows in vivo, continuous characterization of the vascular development of Xenopus laevis. Dev. Biol. 254, 50–67. doi:10.1016/S0012-1606(02)00029-5
Long, S., and Rebagliati, M. (2002). Sensitive two-color Whole-Mount in situ hybridizations using digoxygenin- and dinitrophenol-labeled RNA probes. BioTechniques 32, 494–500. doi:10.2144/02323bm04
Murawala, P., Tanaka, E. M., and Currie, J. D. (2012). Regeneration: the ultimate example of wound healing. Seminars Cell and Dev. Biol. 23, 954–962. doi:10.1016/j.semcdb.2012.09.013
Nakajima, K., and Yaoita, Y. (2003). Dual mechanisms governing muscle cell death in tadpole tail during amphibian metamorphosis. Dev. Dyn. 227, 246–255. doi:10.1002/dvdy.10300
Paredes, R., Ishibashi, S., Borrill, R., Robert, J., and Amaya, E. (2015). Xenopus: an in vivo model for imaging the inflammatory response following injury and bacterial infection. Dev. Biol. 408, 213–228. doi:10.1016/j.ydbio.2015.03.008
Radek, S., Pavel, A., Daniel, Z., Ravindra, N., Daniel, K., Jiri, N., et al. (2023). Characterization of regeneration initiating cells during Xenopus laevis tail regeneration. doi:10.1101/2023.03.30.534908
Senevirathne, G., Baumgart, S., Shubin, N., Hanken, J., and Shubin, N. H. (2020). Ontogeny of the anuran urostyle and the developmental context of evolutionary novelty. Proc. Natl. Acad. Sci. U.S.A. 117, 3034–3044. doi:10.1073/pnas.1917506117
Tomlinson, M. L., Garcia-Morales, C., Abu-Elmagd, M., and Wheeler, G. N. (2008). Three matrix metalloproteinases are required in vivo for macrophage migration during embryonic development. Mech. Dev. 125, 1059–1070. doi:10.1016/j.mod.2008.07.005
Vize, P. D., McCoy, K. E., and Zhou, X. (2009). Multichannel wholemount fluorescent and fluorescent/chromogenic in situ hybridization in Xenopus embryos. Nat. Protoc. 4, 975–983. doi:10.1038/nprot.2009.69
Zahn, N., James-Zorn, C., Ponferrada, V. G., Adams, D. S., Grzymkowski, J., Buchholz, D. R., et al. (2022). Normal Table of Xenopus development: a new graphical resource. Development 149, dev200356. doi:10.1242/dev.200356
Keywords: whole-mount in situ hybridization, photobleaching, tail regeneration, refractory period, Xenopus laevis, myeloid cells, MMP9
Citation: Shitikov AD, Parshina EA, Zaraisky AG and Tereshina MB (2024) An improved method for whole-mount in situ hybridization in regenerating tails of Xenopus laevis tadpoles. Front. Cell Dev. Biol. 12:1487644. doi: 10.3389/fcell.2024.1487644
Received: 28 August 2024; Accepted: 14 November 2024;
Published: 09 December 2024.
Edited by:
Imadeldin Yahya, University of Khartoum, SudanReviewed by:
Sylvain Marcellini, University of Concepcion, ChileJian Sun, University of Delaware, United States
Caroline Beck, University of Otago, New Zealand
Copyright © 2024 Shitikov, Parshina, Zaraisky and Tereshina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. B. Tereshina, bWEtdHJlc2hrYUB5YW5kZXgucnU=
 A. D. Shitikov
A. D. Shitikov E. A. Parshina
E. A. Parshina A. G. Zaraisky
A. G. Zaraisky M. B. Tereshina
M. B. Tereshina