
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 31 October 2024
Sec. Cell Growth and Division
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1485773
The liver possesses an impressive capability to regenerate following various injuries. Given its profound implications for the treatment of liver diseases, which afflict millions globally, liver regeneration stands as a pivotal area of digestive organ research. Zebrafish (Danio rerio) has emerged as an ideal model organism in regenerative medicine, attributed to their remarkable ability to regenerate tissues and organs, including the liver. Many fantastic studies have been performed to explore the process of liver regeneration using zebrafish, especially the extreme hepatocyte injury model. Biliary-mediated liver regeneration was first discovered in the zebrafish model and then validated in mammalian models and human patients. Considering the notable expansion of biliary epithelial cells in many end-stage liver diseases, the promotion of biliary-mediated liver regeneration might be another way to treat these refractory liver diseases. To date, a comprehensive review discussing the current advancements in zebrafish liver regeneration models is lacking. Therefore, this review aims to investigate the utility of different zebrafish models in exploring liver regeneration, highlighting the genetic and cellular insights gained and discussing the potential translational impact on human health.
Liver diseases encompass a range of conditions affecting the liver, including hepatitis, cirrhosis, liver cancer, and fatty liver disease. These illnesses can significantly impair liver function, often leading to severe health complications and, in advanced stages, liver failure (Wang et al., 2014; Xiao et al., 2019). Given the central roles of the liver in detoxifying the blood, aiding digestion, and regulating metabolism (Miyajima et al., 2014; Michalopoulos and Bhushan, 2021), maintaining its health is critical.
Liver regeneration is a remarkable natural process where the liver regenerates lost or damaged tissue, a vital capability for survival after injury or surgical removal of the liver. Unlike most organs, the liver can regenerate to its full size and function from as little as 30% of its original mass (Gilgenkrantz and de l’Hortet, 2018; Yagi et al., 2020). This regeneration involves complex interactions among various types of cells and signaling pathways (Kiseleva et al., 2021; Michalopoulos and Bhushan, 2021). Understanding these mechanisms is crucial for developing treatments for liver diseases, as enhancing the liver’s natural regenerative abilities could potentially reverse or mitigate the effects of liver conditions. Researchers often study model organisms, such as mice and zebrafish, to gain insights into the cellular and molecular foundations of liver regeneration, aiming to apply these findings to improve human health (Forbes and Newsome, 2016; Van Haele et al., 2019).
Zebrafish are increasingly used as a model organism in regenerative medicine research, particularly for studying organ regeneration (Wang et al., 2017; Marques et al., 2019). This small tropical fish possesses remarkable regenerative abilities, capable of repairing and regrowing several organs and tissues, including the heart, spinal cord, retina, and liver, even as adults (Marques et al., 2019). The high regenerative capacity of zebrafish offers valuable insights into the biological mechanisms that could be harnessed to improve regenerative therapies in humans. Moreover, genes are highly conserved between humans and zebrafish, making it a valuable system for studying the basic mechanisms of liver disease (Goessling and Sadler, 2015; Wrighton et al., 2019; Wang et al., 2021).
Except for the Kupffer cell, the zebrafish liver contains all other cell types of the mammalian liver (Goessling and Sadler, 2015; Cai et al., 2021). Zebrafish are similar to mammals in hepatic cellular composition, function, signaling, and response to injury, as well as the cellular processes that mediate liver diseases. Importantly, all the liver functions, including bile secretion, glycogen and lipid storage, insulin responsiveness, xenobiotic and ammonia metabolism, and secretion of serum proteins, are fulfilled in zebrafish liver as early as 5 days post-fertilization (dpf) (Goessling and Sadler, 2015). Compared to mammalian models such as mice and monkeys, zebrafish presents several advantages for studying liver regeneration, including embryonic transparency, rapid growth rate, genetic manipulability, and conserved regulatory mechanisms (MacRae and Peterson, 2015; Huang et al., 2016; van der Helm et al., 2018). Zebrafish have become a widely used model for studying liver regeneration, primarily due to the development of several liver-specific transgenic lines (Figure 1). Research on zebrafish liver regeneration focuses on three models: partial hepatectomy, drug-induced liver injury, and genetic ablation. This review will discuss the current findings of these three frequently used and three other uncommon zebrafish models, especially the genetic ablation model that first uncovered the process of biliary-to-hepatocyte transition, and summarize the regulatory factors involved in these regenerative processes (Table 1).
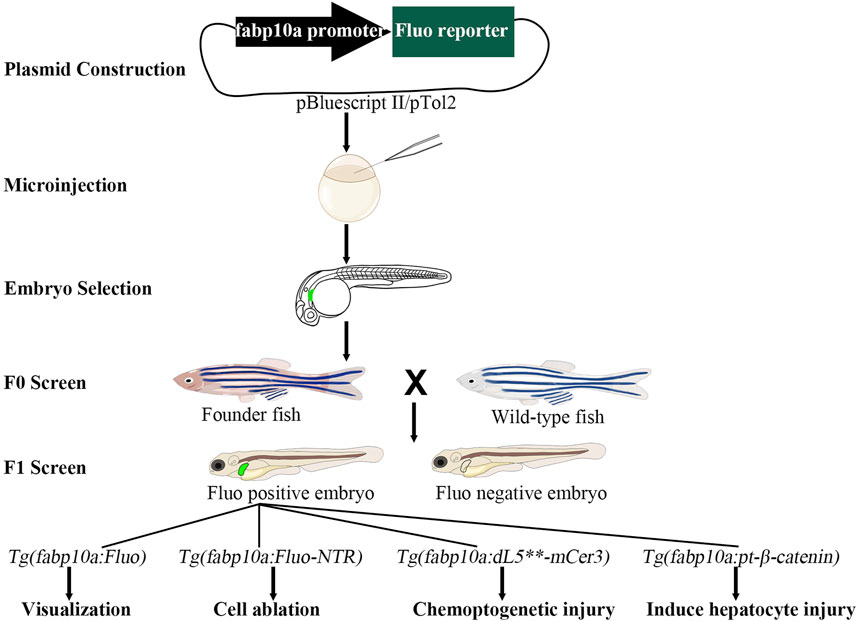
Figure 1. The construction of transgenic models in zebrafish. The fluorescent reporter genes are initially linked to the fabp10a promoter in the pBluescript II or pTol2 vector. The resulting plasmid is then microinjected into one-cell stage embryos. At 3–5 days post-fertilization, embryos expressing fluorescent proteins are selected for further development. At the adult stage, founder (F0) fish are crossed with wild-type fish to produce F1 embryos. Further crossing of F1 fish with wild-type fish results in the establishment of a stable transgenic line. These transgenic lines can be utilized for applications such as visualization, cell ablation, chemoptogenetic injury, and induced hepatocyte injury studies. Fluo, fluorescent.
Despite the high conservation of cell types within the liver, zebrafish possess a unique hepatic anatomy and cellular architecture compared to mammals. The adult zebrafish liver is composed of three contiguous lobes (two lateral and one ventral), which lack the pedicle that separates distinct lobes in mammalian livers. Instead of a portal architecture, fish livers feature hepatocytes arranged in tubules, with bile ductules running between two rows of hepatocytes. The apical membranes face the interior of the tubule, while sinusoids follow the basal side of hepatocytes (Goessling and Sadler, 2015). From the ScRNA-seq data, several cell types were identified in zebrafish livers, including hepatocytes, biliary epithelial cells (BECs), vascular endothelial cells, blood cells, and hepatic stellate cells (Cai et al., 2021). The only difference between the cell types of zebrafish and mammalian livers is the existence of hepatic immune cells (such as Kuffer cells), which could not detected in normal zebrafish livers (Goessling and Sadler, 2015). Fish usually possess two types of BECs: small preductal BECs, which form intracellular lumens to transport bile from hepatocytes, and larger columnar cholangiocytes, which construct the complete intrahepatic biliary system (Weis, 1972; Hampton et al., 1988; Hampton et al., 1989). The formation of small preductal BECs was revealed by time-lapse imaging of living zebrafish larvae, in which bile duct lumen occurs typically through the fusion of cytoplasmic vesicles between two adjacent BECs (Lorent et al., 2010). Even the fact that zebrafish owns unique liver structure, the high conservation of genes between humans and zebrafish makes zebrafish a valuable model for studying the fundamental mechanisms of liver injury and regeneration.
Since the liver weight varies between individuals, the standard parameter used to study zebrafish liver regrowth is the liver/body weight ratio (L/B ratio), which is constant within other species (Weglarz and Sandgren, 2000). Likewise, despite the fact that a difference in L/B ratio between males and females had been observed in adult zebrafish, the L/B ratio in the same gender is constant and does not depend on age (Kan et al., 2009), making it easy to evaluate the efficiency of liver regeneration.
The adult zebrafish liver is structurally different from the mammalian liver. It contains three liver lobes, two dorsal lobes, and a single ventral lobe flattened along the intestine (Unterweger et al., 2023). Unlike the 2/3 PH in mammals, the resection of any two liver lobes led to intense bleeding, disrupted blood circulation, and subsequent mortality (Kan et al., 2009). Thus, the zebrafish PH model is characterized as a 1/3 partial hepatectomy in which the entire ventral lobe is surgically removed (Oderberg and Goessling, 2021).
After the PH surgery, the L/B ratio would be immediately reduced to approximately 65% of uninjured fish, indicating that the injury model is a 1/3 hepatectomy (Figure 2). The liver mass would be recovered to the average level 7 days post-surgery, as reflected by the L/B ratio. Over the next 2–3 days, an additional increase of L/B ratio (10% higher than normal fish) would be observed. After this, the L/B ratio decreased to 90% at 14 days post-surgery and then normalized at 28 days post-surgery (Kan et al., 2009). This pattern of liver regeneration in zebrafish is similar to mammalian liver regeneration, marked by an initial increase in liver mass, followed by a slow return to the original liver mass (Michalopoulos and DeFrances, 1997). Despite the L/B ratio presenting gender differences, males and females show similar regenerative dynamics after surgery (Kan et al., 2009).
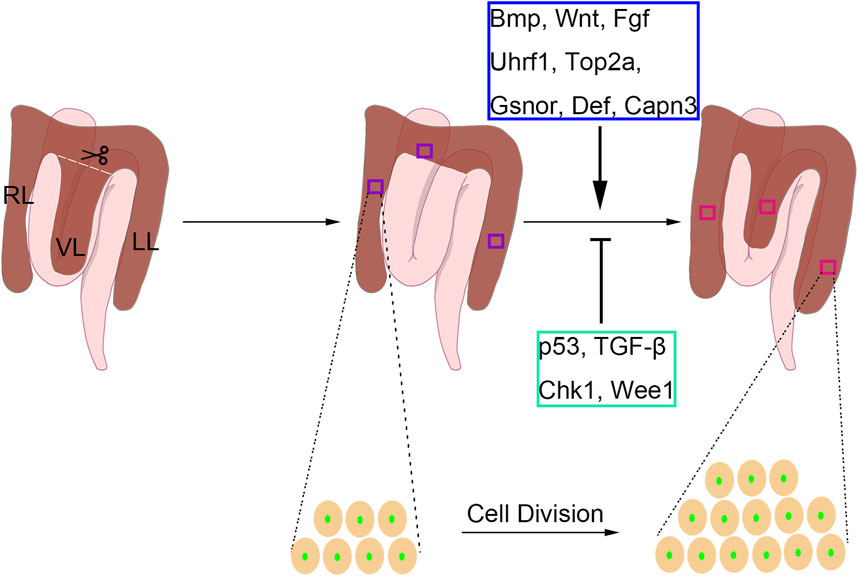
Figure 2. The partial hepatectomy (PH) model in adult zebrafish. In the zebrafish one-third PH model, the entire ventral lobe (VL) is removed, and the left lobe (LL) and right lobe (RL) are retained. The local regeneration of VL and compensatory regeneration of LL and RL are both present upon PH-induced liver injury in adult zebrafish. The liver regenerative types differ from different PH protocols. The liver regeneration is achieved by intrahepatic cell division. During the regenerative process, Bmp, Wnt, Fgf, Uhrf1, Gsnor, Def, and Capn3 play positive roles, while p53, TGF-β, Chk1, and Wee1 have negative roles.
Like the mammalian PH model (Michalopoulos and Bhushan, 2021), liver regeneration in the zebrafish PH model is achieved by the contribution of uninjured hepatocyte proliferation. Lineage tracing data reveal that BECs fail to contribute to hepatocyte regeneration after PH in zebrafish (Zhang et al., 2021), indicating the conserved regenerative process between zebrafish and mammals. However, zebrafish liver regeneration after PH could be divided into two ways: local regeneration and compensatory regeneration.
After 1/3 PH surgery in zebrafish liver, the regeneration of the missing ventral lobe could result in compensatory regeneration in the dorsal lobes other than the ventral lobe and ultimately lead to the recovery of liver mass within a week (Kan et al., 2009; Oderberg and Goessling, 2021). The compensatory growth of the liver after hepatectomy of the ventral lobe occurs in zebrafish, and this process, similar to those in rodents and humans, is closely associated with the activation and proliferation of hepatocytes (Kan et al., 2009). Transcriptomic profiling of the dorsal lobes following resection of the ventral lobe revealed significant changes related to compensatory regeneration (Feng et al., 2015). Meanwhile, another research group reported that the ventral lobe could mostly regenerate over 36 days post-surgery (Sadler et al., 2007), suggesting that the zebrafish owns the capacity for epimorphic liver regeneration, which differs in mammals (Goessling and Sadler, 2015). Considering the liver regeneration models vary with the extent of the injury, the technical variation in the PH protocol between research groups may lead to discrepancies in results.
In mammals, several signaling pathways have been shown to regulate PH-induced liver regeneration, including BMP, FGF, and Wnt signaling (Forbes and Newsome, 2016), which are also involved in regulating liver development (Zong and Stanger, 2012). Similar results were carried out using the zebrafish PH model. By using the zebrafish transgenic lines Tg(hsp:dnBPMR-GFP) and Tg(hsp:dnFGFR1-GFP), BMP and FGF were proved to be essential for hepatocyte proliferation after PH in both males and females (Kan et al., 2009). The Wnt/β-catenin pathway was shown to be activated after PH (Kan et al., 2009), implying that Wnt signaling also regulates liver regeneration in zebrafish. Indeed, Wnt activation by Wnt8a overexpression would promote PH-induced liver regeneration in zebrafish, while Wnt inhibition by dnTCF expression would reduce the regenerated liver mass (Goessling et al., 2008). Because PH-induced liver regeneration in zebrafish is achieved by hepatocyte proliferation, the modulation of cell cycle regulators could affect the regenerative process. Uhrf1 regulates the outgrowth of the developmental liver by inducing the genes involved in the cell cycle. Besides, uhrf1 heterozygous mutants exhibited defective regeneration after PH, indicating the essential roles of Uhrf1 in regulating the cell cycle upon acute liver injury (Sadler et al., 2007). Top2a has traditionally served as a marker for proliferation in both normal and cancer tissues, and Uhrf1 is recognized as a known positive regulator of Top2a activity (Wang et al., 1997; Hopfner et al., 2000; Zhu et al., 2002). Another study reported that top2a heterozygosity also caused the deficiency of liver regeneration in adult zebrafish (Dovey et al., 2009), suggesting that the promotion of Uhrf1-Top2a axis in cell proliferation is essential for liver regeneration in the PH model. The nucleolus complex Def-Capn3 also participates in PH-induced liver regeneration in zebrafish. Haploinsufficiency of def activates p53-dependent TGF-β signaling and causes scar formation after PH (Zhu et al., 2014), and Def-Capn3 complex the cell cycle reentry of hepatocytes by inhibiting Chk1 and Wee1 during liver regeneration (Chen et al., 2020). In addition to these factors, s-nitrosoglutathione reductase (GSNOR) plays opposing roles in PH-induced zebrafish liver regeneration, which was confirmed by the promoted phenotype upon genetic mutation or pharmacological inhibition of GSNOR (Cox et al., 2014).
The regulatory factors revealed in the zebrafish PH model deepen our understanding of hepatocyte proliferation upon hepatectomy (Figure 2). However, most of these findings have been previously reported in mammals, thus limiting the novelty of these studies. Additionally, due to the circulating breed system, the zebrafish PH model could hardly be used for drug treatment and screening. Hence, zebrafish is not a popular model to study PH-induced liver regeneration.
Since nitroreductase (NTR) could convert the nontoxic prodrug metronidazole (Mtz) to the cytotoxic form, Mtz treatment specifically ablates cells that express NTR. The Mtz/NTR system is cell-cycle independent and applicable to any target cell population (Bridgewater et al., 1995). The NTR enzyme is initially reduced by NADH or NADPH. Following this reduction, NTR binds to Mtz, reducing it to a powerful DNA interstrand cross-linking agent, leading to cell death (Curado et al., 2007). Until now, Mtz/NTR system has been used to study several organ or tissue regeneration in zebrafish, such as cerebrovascular, retina, heart, liver, pancreas, and fin (Gemberling et al., 2013; Chen et al., 2019; Mi et al., 2024).
Based on the Mtz/NTR system, the transgenic lines Tg(fabp10a:Dendra2-NTR), Tg(fabp10a:mCherry-NTR), and Tg(fabp10a:CFP-NTR) were generated by two research groups (Choi et al., 2014; He et al., 2014; Choi et al., 2015). The hepatocytes of these three lines express fluorescent proteins that could be observed by fluorescent microscopes, and the hepatocytes can also be ablated by Mtz treatment (Figure 3). After 10 mM Mtz treatment for 24 h, nearly all hepatocytes would suffer from apoptosis, leading to extreme liver injury in zebrafish. In larval zebrafish, the injured livers could be functionally recovered at 48 h post Mtz treatment, making the larval zebrafish an ideal model to study liver regeneration (He et al., 2014). The Mtz/NTR system also works in adult zebrafish, and only 10 h of Mtz treatment could cause extreme hepatocyte ablation and functional regeneration can be achieved 5 days post-injury (He et al., 2014).
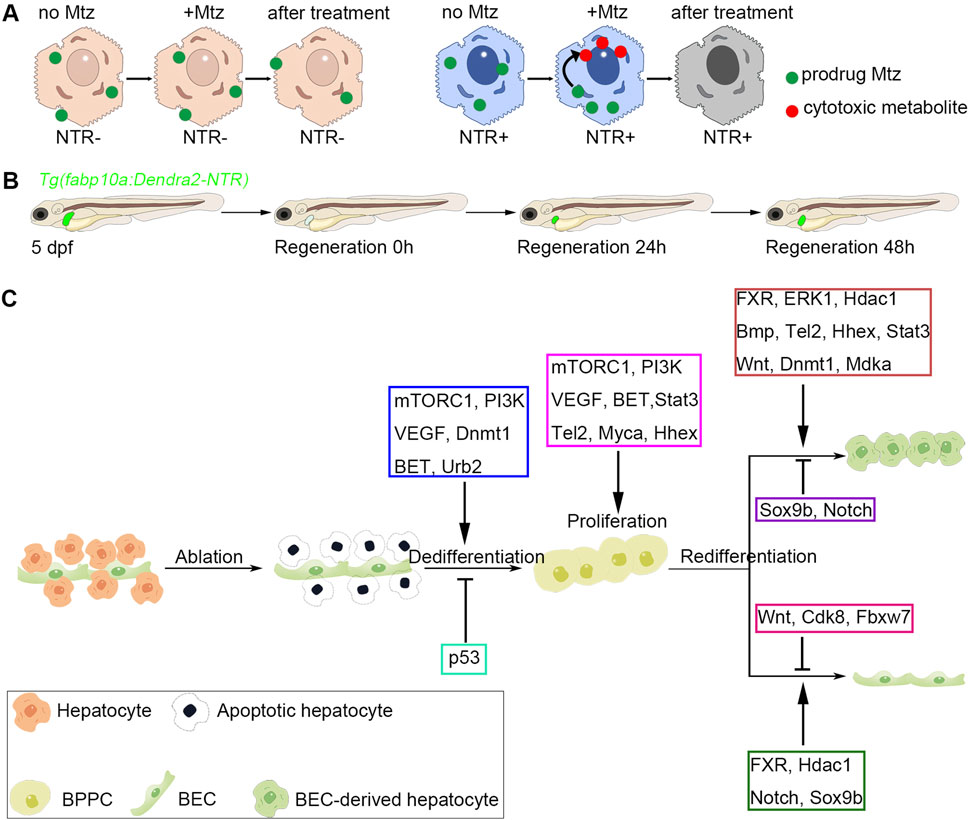
Figure 3. Genetic ablation of hepatocytes and biliary-mediated liver regeneration in zebrafish. (A) The work model of Mtz/NTR system in zebrafish liver. Nitroreductase (NTR) converts the nontoxic prodrug metronidazole (Mtz) to the cytotoxic metabolite, which could lead to the cell death of hepatocytes. (B) The schematic diagram of zebrafish biliary-mediated liver regeneration model. The fluorescent images of regenerating livers could be obtained by a confocal microscope. (C) Regulatory factors involved in different regenerative stages. In the zebrafish extreme hepatocyte injury model, nitroreductase (NTR)-expressed hepatocytes are ablated upon metronidazole treatment. Then, the biliary epithelial cells (BECs) would dedifferentiate into bipotential progenitor cells (BPPCs). mTORC1, PI3K, VEGF, Dnmt1, BET, and Urb2 are essential for BEC-to-BPPC dedifferentiation, while p53 activation inhibits the dedifferentiation process. BPPCs proliferate rapidly after BEC dedifferentiation, and mTORC1, PI3K, VEGF, BET, Stat3, Tel2, Myca, and Hhex positively regulate BPPC proliferation. Lastly, BPPCs re-differentiated into hepatocytes and BECs. During the BPPC-to-hepatocyte redifferentiation process, FXR, ERK1, Hdac1, Bmp, Tel2, Hhex, Stat3, Wnt, Dnmt1, and Mdka exert positive roles, while Sox9b and Notch show negative roles. During the BPPC-to-BEC redifferentiation process, FXR, Hdac1, Notch, and Sox9b play positive roles, while Wnt, Cdk8, and Fbxw7 exhibit negative roles.
In addition to the above three commonly used transgenic lines that could induce extreme hepatocyte injury, another research group reported a new Mtz/NTR-based double transgenic model that could trigger moderate liver injury. This model contains a driver line Tg(fabp10a:GAL4-VP16,myl7:Cerulean) and a effector line: Tg(UAS:NTR-mcherry) (Jagtap et al., 2020). By crossing these two single lines with each other, hepatocyte-specific expressed GAL4 would activate the expression of UAS downstream genes (Halpern et al., 2008), leading to the hepatocyte-specific expression of NTR. In this model, even 2.5 mM Mtz treatment for 3 h would cause lethality of adult zebrafish. Thus, 1.5 mM Mtz treatment for 3 h was performed to induce hepatocyte injury. A large number of pyknotic nuclei and enucleated cells were detected at 3 h post-injury, suggesting that the hepatocytes were indeed suffering the injury. Unlike the extreme hepatocyte injury model, this model should be categorized as moderate to intermediate. The expression of hepatocyte marker fabp10a could always be detected at high levels during the regeneration process (Jagtap et al., 2020), indicating the moderate damage induced by Mtz/NTR system in this model.
Unlike hepatocyte proliferation-mediated liver regeneration, hepatocyte regeneration upon extreme liver injury is contributed by BEC transdifferentiation, which is validated by the Cre/Loxp-based lineage tracing system (Choi et al., 2014; He et al., 2014). The BEC-mediated liver regeneration consists of three steps: BEC dedifferentiation to bipotential progenitor cells (BPPCs), BPPC proliferation, and BPPC redifferentiation to hepatocytes and BECs. Firstly, extreme loss of hepatocytes leads to alterations of BEC morphologies and induction of the hepatoblast markers such as hhex and foxa3, indicating the dedifferentiation of BECs to BPPCs. Then, the BPPCs rapidly proliferate, which could be detected by EdU and PCNA staining. Lastly, BPPCs re-differentiate into nascent hepatocytes, which express the mature hepatocyte markers such as gc, bhmt, and tfa (He et al., 2019; Cai et al., 2021; Cai et al., 2023). In adult zebrafish, hepatocyte regeneration is also achieved by BEC transdifferentiation (Choi et al., 2014; He et al., 2014; Oderberg and Goessling, 2023), indicating the conserved regenerative process between larval and adult zebrafish.
In the moderate hepatocyte injury model Tg(fabp10a:GAL4-VP16,myl7:Cerulean; UAS:NTR-mcherry), hepatocytes but not BECs may play the major roles in liver regeneration. Because lineage tracing data were lacking in the model, all the conclusions made by the authors were based on the RNA-seq data (Jagtap et al., 2020). Even though some BEC markers showed induction at 12 h after injury, the changes were not significant, indicating that BECs would not rapidly proliferate upon hepatocyte injury. On the other hand, the continuous high levels of fabp10a also suggested that not all hepatocytes were injured, and the proliferation of uninjured hepatocytes could contribute to normal liver regeneration (Jagtap et al., 2020). However, the contribution of BEC transdifferentiation to liver regeneration could not be absolutely excluded, and the application of lineage tracing experiments is the only way to determine the exact sources of hepatocyte regeneration in this model.
The BEC dedifferentiation initiates the BEC-mediated liver regeneration (He et al., 2019; Cai et al., 2021). The mammalian target of rapamycin complex 1 (mTORC1) signaling is the first pathway that was reported to govern BEC dedifferentiation (He et al., 2019). In the mouse/rat PH-induced liver injury model, PI3K/AKT activates mTOR to regulate the cell cycle and cell proliferation (Chen et al., 2009), and mTOR-dependent phosphorylation and activation of ribosomal S6 protein kinase 1 (S6K) play dominant roles in regulating the cell cycle during liver regeneration (Espeillac et al., 2011). Upon Mtz-induced hepatocyte injury in zebrafish, mTORC1 signaling would be activated in BECs. Chemical inhibition or genetic inactivation of mTORC1 signaling would disrupt the morphological changes of BECs and reduce the expression of BPPC markers. Mechanistically, mTORC1 regulates BEC-mediated liver regeneration through transcriptionally controlling the ribosome biogenesis protein Urb2 (He et al., 2019). The essential roles of mTORC1-Urb2 axis may imply that protein synthesis is the primary force to drive the cell fate conversion of BECs to liver progenitor cells. Indeed, another study also proved the requirement of mTORC1 signaling in zebrafish liver regeneration, and promoting mTORC1 activation may facilitate the BEC-to-hepatocyte transition (Chaturantabut et al., 2019).
DNA methylation, an essential epigenetic mechanism, plays crucial roles in regulating gene expression and organ regeneration (Albogami, 2019). After extreme hepatocyte injury in zebrafish, dnmt1, but not dnmt3a or dnmt3b, was strongly induced in the liver region, and DNA methylation was continuously maintained in the BECs during the regenerative process. Both pharmacological and genetic inhibitions of DNA methylation would disrupt the dedifferentiation process, thus inhibiting the BEC transdifferentiation to hepatocytes (He et al., 2022). Inhibition of DNA methylation would disrupt the maintenance of DNA methylation at the p53 locus, thus upregulating the transcription of p53. The mutation of p53 could correct the regenerative defects upon DNA methylation inhibition, while p53 overexpression in BECs would trigger defective liver regeneration. Besides, the activation of mTORC1 signaling was reduced upon the inhibition of DNA methylation (He et al., 2022), thus linking the DNA methylation to mTOR activation in the process of BEC dedifferentiation.
The well-known role of VEGF signaling is its regulation on angiogenesis, even in normal or pathological conditions (Hu et al., 2014; Simons et al., 2016). In the zebrafish extreme liver injury model, VEGF ligands were mainly secreted by active hepatic stellate cells upon injury, and then the downstream signaling was activated through the receptor VEGFR2 in BECs. VEGF signaling controls the activation of PI3K-mTORC1 axis, which is essential for BEC dedifferentiation (Cai et al., 2023). Another lab also reported the involvement of VEGF signaling in regulating BEC-mediated liver regeneration (Rizvi et al., 2023). Moreover, VEGFA supplement in mice strongly promoted the dedifferentiation of BECs to BPPCs and therefore increased the number of BEC-derived hepatocytes upon liver injury (Rizvi et al., 2023). These two studies prove that VEGF signaling as an important regulator of BEC dedifferentiation and provides VEGF as a potential modulator to treat end-stage liver diseases.
Through chemical screening, the inhibitors of bromodomain and extraterminal (BET) proteins were found to suppress BEC dedifferentiation (Ko et al., 2016). The BET protein family members brd2a, brd3a, brd3b and brd4 would be upregulated in the regenerating livers. BET inhibition could reduce the expression of BPPC markers and the subsequent hepatocyte regeneration. However, once the BET inhibitors were removed, BEC dedifferentiation would be resumed, indicating that temporal BET inhibition does not permanently impair BEC-mediated liver regeneration. Besides, BET proteins are also involved in hepatocyte proliferation in the mouse PH model (Russell et al., 2017), indicating that the BEC transdifferentiation-mediated and hepatocyte proliferation-mediated liver regeneration share some critical regulatory factors.
After the dedifferentiation of BECs into BPPCs, the proliferation of BPPCs occurs. Because the progenitor cells have higher proliferating capacity (Liu et al., 2019; Long and Huttner, 2022), those genes involved in BEC dedifferentiation often control BPPC proliferation simultaneously. Indeed, previous studies in zebrafish have shown that the deficiency of mTORC1, VEGF, or BET disrupted BPPC formation and proliferation and thereby led to compromised liver regeneration (He et al., 2019; He et al., 2022; Cai et al., 2023). Myca is reported to regulate BPPC proliferation downstream of BET proteins, and myca overexpression could partially rescue the defects caused by BET inhibition (Ko et al., 2016). Unlike those three factors, telomere maintenance 2 (Tel2) does not control the BEC dedifferentiation but regulates BPPC proliferation via hematopoietically expressed homeobox (Hhex) in BPPCS. However, the prominent roles of tel2 are involved in the BPPC redifferentiation (Zhang et al., 2022), which will be discussed in the following section.
The final step of BEC-mediated liver regeneration is BPPC redifferentiation, which contains two directions: hepatocytes and BECs. Farnesoid X receptor (FXR) is a central factor that controls the redifferentiation of BPPCs to both directions. The regulation of FXR on cell proliferation during liver regeneration has been reported in the CCl4- and PH-induced liver injury model (Huang et al., 2006; Meng et al., 2010). In zebrafish, FXR was induced in the BECs upon extreme hepatocyte injury and then reduced to normal levels when regeneration is completed. FXR inhibition would disrupt BPPC redifferentiation, leading to the accumulation of BPPCs and the defects in hepatocyte and BEC regeneration. However, FXR is dispensable for BEC dedifferentiation or BPPC proliferation. FXR controls BPPC redifferentiation to hepatocytes and BECs through transcriptional regulating erk1 and notch3, respectively (Cai et al., 2021). Despite these findings, another group reported that FXR activation may impair BEC-mediated liver regeneration in zebrafish (Jung et al., 2021). However, differing from the prior study that contains both pharmacological and genetic data, the findings in the latter study are mainly based on chemical treatments. Indeed, high doses of FXR agonists caused the apoptosis of BECs and BEC-derived cells, which may be attributed to the toxicological effects beyond FXR. Besides, the lower and appropriate dose of FXR agonist treatments could promote hepatocyte regeneration (Cai et al., 2021), suggesting the requirements of FXR in BEC-mediated liver regeneration.
Through chemical screening, the inhibitors of HDAC1/2 and KDM1A were found to impair the redifferentiation of BPPCs into hepatocytes. Because zebrafish only have Hdac1 and Hdac1 protein was highly induced in the BPPCs (Noel et al., 2008; Ko et al., 2019), Hdac1 is thought to be the essential factor of BEC-mediated liver regeneration. Even though the hdac1 homozygous mutants could not survive to 5 days and the heterozygous mutants exhibited no obvious regenerative phenotype, the authors found that the reduced dosage of HDAC inhibitor would lead to regenerative defects in heterozygous mutants but not in wild-type. During the redifferentiation process, the hyperacetylation of the sox9b genomic locus depends on Hdac1 activation, and sox9b mutation could rescue the defects of hepatocyte differentiation in hdac1 heterozygous mutants. On the other hand, Hdac1 controls BPPC-to-BEC redifferentiation by enhancing Notch activity, which is achieved by repressing the Cdk8/Fbxw7-mediated degradation pathway of NICD (Ko et al., 2019). However, whether Hdac1 directly regulates the acetylation of Cdk8/Fbxw7 genomic locus remains unknown.
The essential roles of VEGF signaling imply the requirements of activated hepatic stellate cells, which could secrete the VEGF and other signaling ligands (Cai et al., 2023). A recently published paper further proved this hypothesis. Midkine (MDK), a heparin-binding growth factor, interacts with a range of cell surface receptors, thereby modulating processes like cell proliferation, migration, and differentiation (Muramatsu, 2010). The expression of mdka significantly increased in the stellate cells upon injury, and its receptor ncl upregulated in the BECs. Genetic mutation of mdka and ncl both blocked the redifferentiation of BPPCs into hepatocytes (Zhang et al., 2024). Compared to the transcriptional control of ERK1 by FXR (Cai et al., 2021), Mdka-Ncl signaling regulates BPPC redifferentiation by activating ERK1/2 (Zhang et al., 2024).
Bone morphogenetic protein (Bmp) signaling seems to be a regulatory factor that balances the redifferentiation to hepatocyte and BEC directions (Choi et al., 2017). Upon extreme hepatocyte loss, those genes involved in Bmp signaling were upregulated at the redifferentiation stages. Bmp inhibition maintained the BPPCs as undifferentiated, thus disrupting the direction of hepatocyte differentiation. However, differing from Hdac1 or FXR inhibition, Bmp inhibition would increase the number of newly regenerative BECs, implying that Bmp signaling may control the hepatocyte differentiation direction to antagonize the BEC direction. The authors claimed that this phenomenon was caused by the abnormal proliferation of BECs. However, we thought that the promotion of BPPC-to-BEC redifferentiation may be one reason to explain this phenotype.
Tel2 also regulates BPPC redifferentiation in addition to its role in promoting BPPC proliferation. The roles of Tel2 in regulating telomere length and localizing telomeric DNA were first identified in yeast and nematodes (Kota and Runge, 1998; Lim et al., 2001). The tel2 mutants showed redundant BPPCs and defective hepatocyte regeneration. Interestingly, Tel2 regulates the transcription of hhex in BECs and BPPC-to-hepatocyte redifferentiation independent of telomere-related function, and the hhex heterozygous mutants showed defective BPPC-to-hepatocyte redifferentiation (Zhang et al., 2022).
By combining the Mtz/NTR system with alcohol exposure, BECs are proved to be the central resource of hepatocyte regeneration in zebrafish fibrotic livers (Huang et al., 2014). Consistent with its roles in liver development, Wnt signaling is found to be required for BPPC-to-hepatocyte redifferentiation and antagonizes the Notch signaling to ensure the hepatocyte direction in BEC-mediated liver regeneration (Huang et al., 2014; Russell et al., 2019a). Notch signaling is critical for both BEC development and regeneration (Zong et al., 2009; Ko et al., 2019; Cai et al., 2021). The mutation of notch3 completely blocks the redifferentiation of BPPCs into BECs (Ko et al., 2019). Moreover, Notch inhibition would promote the BPPC-to-hepatocyte redifferentiation through repressing sox9b (Russell et al., 2019a), thus confirming the roles of Notch and Sox9b in BPPC-to-BEC differentiation direction (Ko et al., 2019). Hence, Wnt and Notch signaling may be the key effectors that control hepatocyte direction and BEC direction, respectively.
DNA methylation was continuously maintained in the BECs during BEC-mediated liver regeneration, which may be the reason why Dnmt1 is also involved in BPPC redifferentiation (He et al., 2022). DNA methylation inhibition at later regenerative stages also released the DNA methylation in the p53 genomic locus and increased the transcription of p53, leading to the BPPC accumulation and BPPC redifferentiation defects. Besides, the activation of Bmp signaling would be blocked upon late DNA methylation inhibition. p53 has been reported to be a regulator of BMP signaling (Liu et al., 2013; Balboni et al., 2015). Thus, the authors thought that p53 activation contributes to the Bmp repression after DNA methylation inhibition.
In addition to these factors, the MRN complex is reported to prevent the BEC-derived hepatocytes from apoptosis through the ATR-Chk1 pathway. Either rad50 or nbn mutation would activate DNA damage response and thus trigger apoptosis in BEC-derived hepatocytes (Song et al., 2023). Signal transducer and activator 3(Stat3) has been reported to be involved in hepatocyte proliferation-mediated liver regeneration (Moh et al., 2007) and BEC-derived oval cell proliferation (Sánchez et al., 2004). Like the roles of Tel2, Stat3 is crucial for the BPPC proliferation and redifferentiation. However, despite the BPPC-to-hepatocyte direction was blocked, the BPPC-to-BEC direction seemed to be unaffected upon Stat3 inhibition (Khaliq et al., 2018). Epithelial cell adhesion molecule (EpCAM) is dispensable for BPPC formation, proliferation, and redifferentiation but regulates the maturation and reconstruction of biliary network (Lee et al., 2024). Those factors found in zebrafish BEC-mediated liver regeneration provide many insights into mammal liver recovery (Figure 3). The roles of Wnt and Notch were validated in mouse models (Pu et al., 2023), and VEGF overexpression was reported to promote the BEC-to-hepatocyte transdifferentiation and thus accelerate liver recovery upon chronic liver injury in mice (Rizvi et al., 2023). Even though these studies reported in recent years have deepened our understanding of BEC-mediated liver regeneration, the regulatory mechanisms remain largely unknown, especially how BECs sense the deficiency of hepatocyte proliferation and then start to trans-differentiate upon acute and chronic liver injury.
Utilizing hepatotoxic over-the-counter medications such as acetaminophen (APAP), tetracycline, erythromycin, aspirin, amiodarone, and cyclosporine A, it has been established that drug-induced liver injury manifests similarly in both zebrafish and humans (Shimizu et al., 2023). Unlike those drugs frequently used to assess hepatotoxicity in zebrafish, APAP could also be used to study liver regeneration after its acute injury to the liver (Driessen et al., 2015; Yoon et al., 2016).
APAP produced similar effects on embryonic and larval hepatocytes, and APAP diminished liver size in a dose- and time-dependent fashion in larval zebrafish (North et al., 2010). The larval zebrafish was often used to assess liver regeneration upon APAP-induced liver injury. 10 mM APAP was used to treat zebrafish larvae from 3.5 to 5 dpf, and the regenerative effects were checked at 2 days after APAP exposure. Hepatocyte proliferation, but not BEC transdifferentiation, contributes to liver regeneration after APAP-induced liver injury (Russell et al., 2017). Using the APAP-induced liver injury model, the chemicals PGE2 and NAC were found to reduce the toxicological effects of APAP and promote liver regeneration after APAP exposure (North et al., 2010). Similar to BEC-mediated liver regeneration, BET inhibition would reduce hepatocyte proliferation in the zebrafish APAP-induced injury model (Russell et al., 2017), indicating the conserved roles of BET proteins in both hepatocyte- and BEC-mediated liver regeneration. By performing SLAM-ITseq, the nascent transcriptome was investigated during the initiation of liver injury and regeneration after APAP exposure. A swift metabolic shift from the postprandial to the fasting state was observed, leading to the induction of the nuclear erythroid 2-related factor (Nrf2) antioxidant program. Activation of Nrf2 in hepatocytes is crucial for the initiation of the pentose phosphate pathway (PPP), thereby enhancing liver regeneration and survival in cases of APAP-induced liver injury (Tan et al., 2024). Additionally, pharmacological activation of FXR could promote liver regeneration after APAP exposure, confirming the roles of FXR in hepatocyte proliferation-mediated liver regeneration cross species (Huang et al., 2006; Jung et al., 2021). Although APAP could be used to study zebrafish liver regeneration, this model is not commonly used due to the vast researches in mouse models.
Optogenetic approaches offer unprecedented precision in spatially and temporally regulating tissue manipulation, enabling minute control over biological processes (Portugues et al., 2013). To create a chemoptogenetic hepatocyte ablation tool for extended live imaging, the transgenic line Tg(fabp10a:dL5**-mCer3), abbreviated Tg(LiverZap), was generated. In this model, the exposure of 12 min NIR light would induce cell death of hepatocytes by producing ROS in larval zebrafish. Eight hours after the injury, apoptotic activity was observed in the liver region. Hepatocyte ablation was categorized into two distinct types: mild (30%) and severe (70%). Mildly ablated livers regenerated within 3 days post-injury, while severely ablated livers recovered within 7 days. Notably, the mechanisms of liver regeneration differed between the two conditions. Mildly ablated livers regenerated primarily through hepatocyte proliferation, whereas severely ablated livers were repaired via transdifferentiation of BECs. Additionally, using the LiverZap tool, the researchers demonstrated that targeted ablation of hepatocytes in a discrete region of interest is unexpectedly effective in triggering BEC-mediated regeneration, thereby challenging current perspectives on liver progenitor cell activation (So et al., 2020). Dynamic rearrangement of the biliary network and E-cadherin re-localization could both be observed in this model, indicating that cell adhesion modulation may be a pivotal step in BEC-mediated liver regeneration (Ambrosio et al., 2024). This model expands the current regeneration toolkit and enables detailed analysis of critical cellular dynamics, which are essential for understanding how the liver’s complex architecture is deconstructed during injury and reconstructed during repair. However, due to the variability in regeneration speed and dependency on injury severity, this model is limited in its ability to investigate the key factors involved in hepatocyte proliferation or BEC-mediated liver regeneration.
To identify small molecules that can facilitate liver progenitor cell (LPC)-to-hepatocyte differentiation, the Tg(fabp10a:pt-β-catenin) zebrafish transgenic line was constructed for LPC-mediated liver regeneration in which a mutated, stable form of Xenopus β-catenin is overexpressed in hepatocytes (So et al., 2021). The pt-β-catenin variant includes four point mutations (S33A, S37A, T41A, and S45A) at putative phosphorylation sites, and these mutations activate β-catenin by preventing its phosphorylation and subsequent degradation (Evason et al., 2015). Zebrafish hepatocyte-specific overexpression of pt-β-catenin would cause hepatocellular carcinoma (HCC) and recapitulate the pathologic features of human HCC (Evason et al., 2015). As early as 7 dpf, the hepatocytes of Tg(fabp10a:pt-β-catenin) larvae exhibited DNA damage, apoptosis, and senescence, which would be partially recovered at 30 dpf. The lineage tracing data showed that liver regeneration was achieved by both hepatocyte and BEC contributions. Thus, this model differs from the Mtz/NTR model. Using this model, the EGFR-ERK-Sox9 axis was found to play opposing roles during LPC-mediated liver regeneration, and the treatment of EGFR and ERK inhibitors would accelerate the differentiation of hepatocytes from LPCs (So et al., 2021). The inhibitory role of Sox9 in hepatocyte differentiation has also been observed in BEC-mediated liver regeneration, suggesting a conserved function across different models. Notably, EGFR and ERK signaling pathways exhibit opposing roles in BEC- and LPC-mediated liver regeneration (Cai et al., 2021; So et al., 2021; Oderberg and Goessling, 2023), highlighting the distinct regenerative mechanisms employed in different liver injury models. Additionally, PPARα activation is found to augment the differentiation of LPCs to hepatocytes by suppressing YAP signaling in this model (Kim et al., 2023).
A recent paper published in Development reports a new liver injury model in which liver cryoinjury is induced by adapting the CUBIC tissue-clearing approach in adult zebrafish. The ventral lobe was selected for cryoinjury due to its surgical accessibility. To ensure a reproducible and consistent injury, all procedures were performed at the level of the anterior fins and towards the midline. By 14 days post-cryoinjury, the injured area was nearly fully repaired. The cryoinjury would induce a localized necrotic and apoptotic lesion characterized by inflammation and infiltration of innate immune cells. Following the initial phase, the liver would suffer from fibrosis, which then be resolved by liver regeneration within 30 days. Cryoinjury would induce both localized and distal compensatory hyperplasia through trigger cell proliferation. The transcriptional landscape following cryoinjury has also been discovered (Sande-Melon et al., 2024). However, despite this, more information about this model needs further investigation, especially the functional validation of the regulatory factors.
After discovering BEC-mediated liver regeneration in zebrafish, this type of liver regeneration was also found in several mouse liver injury models. Three years after the zebrafish studies were published, by performing hepatocyte-specific knock-out of β1-integrin or overexpression of p21 combined with DDC/CDE/MCD-induced liver injury in mice, 15%–25% of the newly regenerated hepatocytes were found to be derived from BECs. However, the total experimental periods of this model reach about 2 months, and the contribution of BECs to hepatocytes is relatively low (Raven et al., 2017). Using the TAA/DDC-induced chronic liver injury model in mice, another group reported that chronic liver injury would also generate BEC-derived hepatocytes. Upon chronic injury for 6 months, about 10% of new hepatocytes were derived from BECs. These BEC-derived hepatocytes own a high proliferating capacity, and the ratio of these cells would reach 55% upon liver injury for 13 months (Deng et al., 2018). Besides, hepatocyte-specific deletion of β-catenin in mice could also trigger the BEC-to-hepatocyte transdifferentiation upon CDE-induced liver injury for 2 weeks. Statistically, about 20% of hepatocytes come from BEC transdifferentiation after liver recovery for 2 weeks, and this ratio reaches 70% after 6 months of recovery, confirming the proliferative capacity of BEC-derived hepatocytes (Russell et al., 2019b). Long-term CCl4 treatment could also trigger BEC-derived hepatocyte regeneration in mice. However, like all the other models above, the contribution ratio of BECs is low, only up to 13% after 4 weeks of recovery upon CCl4-induced injury for 16 weeks (Manco et al., 2019). Thus, these mouse models have similar disadvantages, including long experimental periods, operating difficulty, and low contribution ratio.
Compared to the mouse models, almost all newly regenerated hepatocytes are attributed to the BEC transdifferentiation in the zebrafish model (Choi et al., 2014; He et al., 2014). The Mtz/NTR system ensures extreme hepatocyte loss in zebrafish, thus providing a more specific model to study BEC-mediated liver regeneration. Additionally, the zebrafish larvae at 5 dpf could be used to induce liver injury, and the regeneration process can be completed as quickly as 48 h after Mtz treatment (He et al., 2014; Cai et al., 2023). Therefore, the experimental period of the zebrafish model is much shorter than that of the mouse models. This advantage provides the zebrafish BEC-mediated liver regeneration model as ideal for drug screening and regulatory factor exploration.
The process of liver regeneration via hepatocyte proliferation seems to be conserved, as the majority of hepatocytes in the resected lobe of zebrafish begin to proliferate within 2–3 days following partial hepatectomy (Kan et al., 2009). Research in zebrafish has identified multiple mechanisms that control liver regeneration, which is similar to mammals. However, the difference exists between zebrafish and mammals. In mammals, the hepatocytes have been proven to be heterogeneous by several genetic lineage tracing and single-cell sequencing studies (Aizarani et al., 2019). By using lineage tracing from the telomerase reverse transcriptase (Tert) locus in mice, rare hepatocytes with high telomerase expression are found to be distributed throughout the liver lobule. Upon injury, the repopulating activity of these Terthigh hepatocytes accelerates. When Terthigh hepatocytes are genetically ablated in combination with chemical-induced liver injury, there is a marked increase in stellate cell activation and fibrosis (Lin et al., 2018), suggesting the contribution of these cells in liver regeneration. Additionally, Axin2+ pericentral hepatocytes may contribute to liver homeostasis and repair (Wang et al., 2015) despite the controversial conclusion (Sun et al., 2020). These studies adequately prove the heterogeneity of hepatocytes in mammalian livers. However, no evidence has been reported supporting the hepatocyte heterogeneity in zebrafish. Besides, a systematic analysis of zebrafish liver is lacking to compare the difference between zebrafish and mammals.
Compared to hepatocytes, BECs exhibit similar plasticity in zebrafish, mice, and humans. Upon extreme or chronic hepatocyte injury, BECs similarly have the capacity to transdifferentiate into functional hepatocytes (He et al., 2014; Raven et al., 2017; Gribben et al., 2024). The regulatory factors involved in BEC-mediated liver regeneration seem to be conserved cross species. Wnt signaling controls the redifferentiation of BPPCs into hepatocytes in both mice and zebrafish (Huang et al., 2014; Pu et al., 2023), while Notch signaling also governs the redifferentiation of BPPCs into BECs in mice and zebrafish (Ko et al., 2019; Cai et al., 2021; Pu et al., 2023). Similar roles of FXR, Hdac1, VEGF, Mdka, and BET have been proved in mouse and zebrafish BEC-mediated liver regeneration models (Ko et al., 2016; Ko et al., 2019; Cai et al., 2021; Cai et al., 2023; Rizvi et al., 2023; Zhang et al., 2024). Besides, the PI3K-AKT-mTORC1 pathway regulates the BEC-to-hepatocyte plasticity in both zebrafish and humans (He et al., 2019; Cai et al., 2023; Gribben et al., 2024).
Structural and cellular differences in the liver between mammals and zebrafish may lead to distinct regenerative responses. In zebrafish, the bile duct structure is defined by a single layer of cuboidal cells, which is specific to hilar bile ducts and not present in peripheral bile ducts (Cai et al., 2021). The lumen formation of peripheral bile ducts typically occurs through the fusion of cytoplasmic vesicles between adjacent biliary cells (Lorent et al., 2010), a process distinct from that in mammals. This unique structure of the zebrafish liver may account for the rapid response of BECs to hepatocyte injury, potentially explaining the high frequency of BEC-mediated liver regeneration observed in zebrafish injury models (He et al., 2014; Ambrosio et al., 2024). Furthermore, zebrafish lack the liver-resident macrophage Kupffer cell, which is found in mammals (Goessling and Sadler, 2015). In mammals, Kupffer cells secrete pro-inflammatory cytokines upon liver injury, which are crucial for hepatocyte proliferation and liver regeneration (Wang et al., 2024). The absence of Kupffer cells in zebrafish may explain the lack of inflammatory factors involved in BEC-mediated liver regeneration and could influence the preferred pathway for liver regeneration.
Liver transplantation is the only way to treat end-stage liver diseases, which have high mortality (Fisher, 2017). However, the shortage of organ donors and graft rejection limit the application of liver transplantation to many patients (Fisher, 2017). In patients with end-stage liver diseases, hepatocyte proliferation is compromised due to liver steatosis, inflammation, fibrosis, and cirrhosis (Stanger, 2015). Thus, the facilitation of hepatocyte proliferation-mediated liver regeneration seems to be infeasible for liver repair in these cases. Ductular reaction, reflected by BEC proliferation and progenitor activation, is the hallmark of almost all chronic and acute liver diseases (Sato et al., 2019). After the findings of BEC-mediated liver regeneration, promoting the BEC-to-hepatocyte transdifferentiation was postulated to be the alternative way to alleviate severe hepatocyte injury in end-stage liver diseases.
Using the zebrafish extreme liver injury model, VEGF signaling was found to be essential for BEC-to-hepatocyte transdifferentiation (Cai et al., 2023; Rizvi et al., 2023). Based on this phenomenon, another group explored the clinical benefit of VEGF activation in promoting liver repair and restoring liver function in several mammal liver injury models. Using the mRNA-lipid nanoparticles (LNP) delivery system, the controllable transient VEGFA expression in the liver could be archived (Maestro et al., 2021). VEGF mRNA-LNP robustly induced the BEC-to-hepatocyte transdifferentiation and promoted the recovery of liver function in both chronic and acute mouse liver injury models (Rizvi et al., 2023), suggesting the potential clinical benefits of VEGFA mRNA-LNP to alleviate liver diseases. In the livers of patients with end-stage liver diseases, intermediate hepatocyte-like cells exist and express both hepatocyte and BEC markers. Moreover, the presence of these cells is associated with the expression of VEGF receptor KDR (Rizvi et al., 2023), implying that VEGF activation may also stimulate BEC-mediated liver regeneration for human liver disease intervention. Additionally, even though there is a lack of data from mammals, the supplement of either 17 β-estradiol (Chaturantabut et al., 2019), FGF21 protein (Qiang et al., 2021), or PPARα agonist (Kim et al., 2023) could promote the transdifferentiation of BECs to hepatocytes in zebrafish. These findings suggest that using zebrafish as the model of BEC-mediated liver regeneration could indeed provide a potential translational impact on human health.
Although significant progress has been made in studying liver regeneration using zebrafish models, a comprehensive understanding of hepatic biology and disease pathology requires the integration of diverse research tools. The development of liver organoids, a self-organizing and self-renewing three-dimensional cell culture model, has significantly advanced liver research. Liver organoids offer a physiologically relevant platform for drug screening and development, personalized medicine, disease modeling, and the study of liver regeneration (Chawla and Das, 2023). Recently, liver organoids have been used to alleviate liver damage and promote liver repair in mice (Aloia et al., 2019; Yuan et al., 2024), indicating the huge therapeutic potential for liver diseases. However, the emergence of organoid technology is accompanied by several limitations that must be addressed, including the high cost of these models, limited availability of source tissues, and the need for multilineage liver organoids to accurately replicate the cellular heterogeneity of the liver (Chawla and Das, 2023). In contrast, zebrafish offer several advantages, including low cost, high reproductive capacity, and the ability to replicate an in vivo microenvironment. Therefore, considering the distinct benefits of zebrafish models and organoids, integrating these tools is recommended for studying liver regeneration, particularly in pre-clinical research.
In conclusion, the utilization of the zebrafish as an animal model in liver regeneration studies has significantly contributed to the progression and advancement of this research field. The advantages of zebrafish make it suitable for small molecule and drug screening, providing new insights for treating human liver diseases. The scope of hepatobiliary diseases being investigated in zebrafish models is rapidly expanding. Alongside this, novel tools and methodologies are being formulated, encompassing precision gene editing techniques to introduce mutations in specific genes, sophisticated imaging techniques to monitor hepatic cells, and new sequencing methods (such as scRNA-seq and stRNA-seq) to assess the intracellular transcriptional changes upon injury. Studies of liver regeneration using zebrafish models not only enhance our comprehension of the underlying mechanisms of liver diseases but also aid in identifying novel therapeutic targets and potential candidate compounds for treating liver diseases.
DM: Conceptualization, Funding acquisition, Investigation, Writing–original draft. ML: Funding acquisition, Investigation, Writing–original draft. XM: Conceptualization, Investigation, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32260177), the Basic Research Program of Guizhou Provincial Department of Science and Technology Agency [Qiankehe Basic-(2024) Youth 235], and the Basic Research Program of Guizhou Provincial Department of Science and Technology Agency [ZK (2024)-143].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aizarani, N., Saviano, A., Sagar, , Mailly, L., Durand, S., Herman, J. S., et al. (2019). A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572 (7768), 199–204. doi:10.1038/s41586-019-1373-2
Albogami, S. (2019). Epigenetic regulator signatures in regenerative capacity. Curr. Stem Cell Res. and Ther. 14 (7), 598–606. doi:10.2174/1574888x14666190618125111
Aloia, L., McKie, M. A., Vernaz, G., Cordero-Espinoza, L., Aleksieva, N., van den Ameele, J., et al. (2019). Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol. 21 (11), 1321–1333. doi:10.1038/s41556-019-0402-6
Ambrosio, E. M. G., Bailey, C. S. L., Unterweger, I. A., Christensen, J. B., Bruchez, M. P., Lundegaard, P. R., et al. (2024). LiverZap: a chemoptogenetic tool for global and locally restricted hepatocyte ablation to study cellular behaviours in liver regeneration. Development 151 (4), dev202217. doi:10.1242/dev.202217
Balboni, A. L., Cherukuri, P., Ung, M., DeCastro, A. J., Cheng, C., and DiRenzo, J. (2015). p53 and ΔNp63α coregulate the transcriptional and cellular response to TGFβ and BMP signals. Mol. Cancer Res. 13 (4), 732–742. doi:10.1158/1541-7786.Mcr-14-0152-t
Bridgewater, J. A., Springer, C. J., Knox, R. J., Minton, N. P., Michael, N. P., and Collins, M. K. (1995). Expression of the bacterial nitroreductase enzyme in mammalian cells renders them selectively sensitive to killing by the prodrug CB1954. Eur. J. Cancer 31A (13-14), 2362–2370. doi:10.1016/0959-8049(95)00436-x
Cai, P., Mao, X., Zhao, J., Nie, L., Jiang, Y., Yang, Q., et al. (2021). Farnesoid X receptor is required for the redifferentiation of bipotential progenitor cells during biliary-mediated zebrafish liver regeneration. Hepatology 74 (6), 3345–3361. doi:10.1002/hep.32076
Cai, P., Ni, R., Lv, M., Liu, H., Zhao, J., He, J., et al. (2023). VEGF signaling governs the initiation of biliary-mediated liver regeneration through the PI3K-mTORC1 axis. Cell Rep. 42 (9), 113028. doi:10.1016/j.celrep.2023.113028
Chaturantabut, S., Shwartz, A., Evason, K. J., Cox, A. G., Labella, K., Schepers, A. G., et al. (2019). Estrogen activation of G-protein-coupled estrogen receptor 1 regulates phosphoinositide 3-kinase and mTOR signaling to promote liver growth in zebrafish and proliferation of human hepatocytes. Gastroenterology 156 (6), 1788–1804. doi:10.1053/j.gastro.2019.01.010
Chawla, S., and Das, A. (2023). Preclinical-to-clinical innovations in stem cell therapies for liver regeneration. Curr. Res. Transl. Med. 71 (1), 103365. doi:10.1016/j.retram.2022.103365
Chen, F., Huang, D., Shi, H., Gao, C., Wang, Y., and Peng, J. (2020). Capn3 depletion causes Chk1 and Wee1 accumulation and disrupts synchronization of cell cycle reentry during liver regeneration after partial hepatectomy. Cell Regen. Lond. Engl. 9 (1), 8. doi:10.1186/s13619-020-00049-1
Chen, J., He, J., Ni, R., Yang, Q., Zhang, Y., and Luo, L. (2019). Cerebrovascular injuries induce lymphatic invasion into brain parenchyma to guide vascular regeneration in zebrafish. Dev. Cell 49 (5), 697–710. doi:10.1016/j.devcel.2019.03.022
Chen, P., Yan, H., Chen, Y., and He, Z. (2009). The variation of AkT/TSC1-TSC1/mTOR signal pathway in hepatocytes after partial hepatectomy in rats. Exp. Mol. Pathology 86 (2), 101–107. doi:10.1016/j.yexmp.2009.01.013
Choi, T.-Y., Khaliq, M., Ko, S., So, J., and Shin, D. (2015). Hepatocyte-specific ablation in zebrafish to study biliary-driven liver regeneration. Jove-Journal Vis. Exp. 99, e52785. doi:10.3791/52785
Choi, T.-Y., Khaliq, M., Tsurusaki, S., Ninov, N., Stainier, D. Y. R., Tanaka, M., et al. (2017). Bone morphogenetic protein signaling governs biliary-driven liver regeneration in zebrafish through Tbx2b and Id2a. Hepatology 66 (5), 1616–1630. doi:10.1002/hep.29309
Choi, T.-Y., Ninov, N., Stainier, D. Y. R., and Shin, D. (2014). Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146 (3), 776–788. doi:10.1053/j.gastro.2013.10.019
Cox, A. G., Saunders, D. C., Kelsey, P. B., Conway, A. A., Tesmenitsky, Y., Marchini, J. F., et al. (2014). S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell Rep. 6 (1), 56–69. doi:10.1016/j.celrep.2013.12.007
Curado, S., Anderson, R. M., Jungblut, B., Mumm, J., Schroeter, E., and Stainier, D. Y. R. (2007). Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn. 236 (4), 1025–1035. doi:10.1002/dvdy.21100
Deng, X., Zhang, X., Li, W., Feng, R.-X., Li, L., Yi, G.-R., et al. (2018). Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell 23 (1), 114–122. doi:10.1016/j.stem.2018.05.022
Dovey, M., Patton, E. E., Bowman, T., North, T., Goessling, W., Zhou, Y., et al. (2009). Topoisomerase II alpha is required for embryonic development and liver regeneration in zebrafish. Mol. Cell. Biol. 29 (13), 3746–3753. doi:10.1128/mcb.01684-08
Driessen, M., Vitins, A. P., Pennings, J. L. A., Kienhuis, A. S., van de Water, B., and van der Ven, L. T. M. (2015). A transcriptomics-based hepatotoxicity comparison between the zebrafish embryo and established human and rodent in vitro and in vivo models using cyclosporine A, amiodarone and acetaminophen. Toxicol. Lett. 232 (2), 403–412. doi:10.1016/j.toxlet.2014.11.020
Espeillac, C., Mitchell, C., Celton-Morizur, S., Chauvin, C., Koka, V., Gillet, C., et al. (2011). S6 kinase 1 is required for rapamycin-sensitive liver proliferation after mouse hepatectomy. J. Clin. Investigation 121 (7), 2821–2832. doi:10.1172/jci44203
Evason, K. J., Francisco, M. T., Juric, V., Balakrishnan, S., Pazmino, M. D. P. L., Gordan, J. D., et al. (2015). Identification of chemical inhibitors of β-catenin-driven liver tumorigenesis in zebrafish. Plos Genet. 11 (7), e1005305. doi:10.1371/journal.pgen.1005305
Feng, G., Long, Y., Peng, J., Li, Q., and Cui, Z. (2015). Transcriptomic characterization of the dorsal lobes after hepatectomy of the ventral lobe in zebrafish. Bmc Genomics 16, 979. doi:10.1186/s12864-015-2145-5
Fisher, R. A. (2017). Living donor liver transplantation: eliminating the wait for death in end-stage liver disease? Nat. Rev. Gastroenterology and Hepatology 14 (6), 373–382. doi:10.1038/nrgastro.2017.2
Forbes, S. J., and Newsome, P. N. (2016). Liver regeneration - mechanisms and models to clinical application. Nat. Rev. Gastroenterology and Hepatology 13 (8), 473–485. doi:10.1038/nrgastro.2016.97
Gemberling, M., Bailey, T. J., Hyde, D. R., and Poss, K. D. (2013). The zebrafish as a model for complex tissue regeneration. Trends Genet. 29 (11), 611–620. doi:10.1016/j.tig.2013.07.003
Gilgenkrantz, H., and de l’Hortet, A. C. (2018). Understanding liver regeneration: from mechanisms to regenerative medicine. Am. J. Pathology 188 (6), 1316–1327. doi:10.1016/j.ajpath.2018.03.008
Goessling, W., North, T. E., Lord, A. M., Ceol, C., Lee, S., Weidinger, G., et al. (2008). APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev. Biol. 320 (1), 161–174. doi:10.1016/j.ydbio.2008.05.526
Goessling, W., and Sadler, K. C. (2015). Zebrafish: an important tool for liver disease research. Gastroenterology 149 (6), 1361–1377. doi:10.1053/j.gastro.2015.08.034
Gribben, C., Galanakis, V., Calderwood, A., Williams, E. C., Chazarra-Gil, R., Larraz, M., et al. (2024). Acquisition of epithelial plasticity in human chronic liver disease. Nature 630, 166–173. doi:10.1038/s41586-024-07465-2
Halpern, M. E., Rhee, J., Goll, M. G., Akitake, C. M., Parsons, M., and Leach, S. D. (2008). Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5 (2), 97–110. doi:10.1089/zeb.2008.0530
Hampton, J. A., Lantz, R. C., Goldblatt, P. J., Lauren, D. J., and Hinton, D. E. (1988). Functional units in rainbow trout (Salmo gairdneri, Richardson) liver: II. The biliary system. Anat. Rec. 221 (2), 619–634. doi:10.1002/ar.1092210208
Hampton, J. A., Lantz, R. C., and Hinton, D. E. (1989). Functional units in rainbow trout (Salmo gairdneri, Richardson) liver: III. Morphometric analysis of parenchyma, stroma, and component cell types. Am. J. Anat. 185 (1), 58–73. doi:10.1002/aja.1001850107
He, J., Chen, J., Wei, X., Leng, H., Mu, H., Cai, P., et al. (2019). Mammalian target of rapamycin complex 1 signaling is required for the dedifferentiation from biliary cell to bipotential progenitor cell in zebrafish liver regeneration. Hepatology 70 (6), 2092–2106. doi:10.1002/hep.30790
He, J., Lu, H., Zou, Q., and Luo, L. (2014). Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology 146 (3), 789–800. doi:10.1053/j.gastro.2013.11.045
He, J., Zhou, Y., Qian, C., Wang, D., Yang, Z., Huang, Z., et al. (2022). DNA methylation maintenance at the p53 locus initiates biliary-mediated liver regeneration. Npj Regen. Med. 7 (1), 21. doi:10.1038/s41536-022-00217-8
Hopfner, R., Mousli, M., Jeltsch, J. M., Voulgaris, A., Lutz, Y., Marin, C., et al. (2000). ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIalpha expression. Cancer Res. 60 (1), 121–128.
Hu, J., Srivastava, K., Wieland, M., Runge, A., Mogler, C., Besemfelder, E., et al. (2014). Endothelial cell-derived angiopoietin-2 controls liver regeneration as a spatiotemporal rheostat. Science 343 (6169), 416–419. doi:10.1126/science.1244880
Huang, M., Chang, A., Choi, M., Zhou, D., Anania, F. A., and Shin, C. H. (2014). Antagonistic interaction between wnt and notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology 60 (5), 1753–1766. doi:10.1002/hep.27285
Huang, M., Xu, J., and Shin, C. H. (2016). Development of an ethanol-induced fibrotic liver model in zebrafish to study progenitor cell-mediated hepatocyte regeneration. Jove-Journal Vis. Exp. 111, 54002. doi:10.3791/54002
Huang, W. D., Ma, K., Zhang, J., Qatanani, M., Cuvillier, J., Liu, J., et al. (2006). Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 312 (5771), 233–236. doi:10.1126/science.1121435
Jagtap, U., Sivadas, A., Basu, S., Verma, A., Sivasubbu, S., Scaria, V., et al. (2020). A temporal map of gene expression pattern during zebrafish liver regeneration. Zebrafish 17 (1), 1–10. doi:10.1089/zeb.2019.1790
Jung, K., Kim, M., So, J., Lee, S.-H., Ko, S., and Shin, D. (2021). Farnesoid X receptor activation impairs liver progenitor cell-mediated liver regeneration via the PTEN-PI3K-AKT-mTOR Axis in zebrafish. Hepatology 74 (1), 397–410. doi:10.1002/hep.31679
Kan, N. G., Junghans, D., and Izpisua Belmonte, J. C. (2009). Compensatory growth mechanisms regulated by BMP and FGF signaling mediate liver regeneration in zebrafish after partial hepatectomy. Faseb J. 23 (10), 3516–3525. doi:10.1096/fj.09-131730
Khaliq, M., Ko, S., Liu, Y., Wang, H., Sun, Y., Solnica-Krezel, L., et al. (2018). Stat3 regulates liver progenitor cell-driven liver regeneration in zebrafish. Gene Expr. 18 (3), 157–170. doi:10.3727/105221618x15242506133273
Kim, M., So, J., and Shin, D. (2023). PPARα activation promotes liver progenitor cell-mediated liver regeneration by suppressing YAP signaling in zebrafish. Sci. Rep. 13 (1), 18312. doi:10.1038/s41598-023-44935-5
Kiseleva, Y. V., Antonyan, S. Z., Zharikova, T. S., Tupikin, K. A., Kalinin, D. V., and Zharikov, Y. O. (2021). Molecular pathways of liver regeneration: a comprehensive review. World J. Hepatology 13 (3), 270–290. doi:10.4254/wjh.v13.i3.270
Ko, S., Choi, T.-Y., Russell, J. O., So, J., Monga, S. P. S., and Shin, D. (2016). Bromodomain and extraterminal (BET) proteins regulate biliary-driven liver regeneration. J. Hepatology 64 (2), 316–325. doi:10.1016/j.jhep.2015.10.017
Ko, S., Russell, J. O., Tian, J., Gao, C., Kobayashi, M., Feng, R., et al. (2019). Hdac1 regulates differentiation of bipotent liver progenitor cells during regeneration via Sox9b and Cdk8. Gastroenterology 156 (1), 187–202. doi:10.1053/j.gastro.2018.09.039
Kota, R. S., and Runge, K. W. (1998). The yeast telomere length regulator TEL2 encodes a protein that binds to telomeric DNA. Nucleic Acids Res. 26 (6), 1528–1535. doi:10.1093/nar/26.6.1528
Lee, S., Memon, A., Chae, S.-C., Shin, D., and Choi, T.-Y. (2024). Epcam regulates intrahepatic bile duct reconstruction in zebrafish, providing a potential model for primary cholangitis model. Biochem. biophysical Res. Commun. 696, 149512. doi:10.1016/j.bbrc.2024.149512
Lim, C. S., Mian, I. S., Dernburg, A. F., and Campisi, J. (2001). C. elegans clk-2, a gene that limits life span, encodes a telomere length regulator similar to yeast telomere binding protein Tel2p. Curr. Biol. 11 (21), 1706–1710. doi:10.1016/s0960-9822(01)00526-7
Lin, S., Nascimento, E. M., Gajera, C. R., Chen, L., Neuhofer, P., Garbuzov, A., et al. (2018). Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556 (7700), 244–248. doi:10.1038/s41586-018-0004-7
Liu, H., Jia, D., Li, A., Chau, J., He, D., Ruan, X., et al. (2013). p53 regulates neural stem cell proliferation and differentiation via BMP-smad1 signaling and Id1. Stem Cells Dev. 22 (6), 913–927. doi:10.1089/scd.2012.0370
Liu, L., Michowski, W., Kolodziejczyk, A., and Sicinski, P. (2019). The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 21 (9), 1060–1067. doi:10.1038/s41556-019-0384-4
Long, K. R., and Huttner, W. B. (2022). The role of the extracellular matrix in neural progenitor cell proliferation and cortical folding during human neocortex development. Front. Cell. Neurosci. 15, 804649. doi:10.3389/fncel.2021.804649
Lorent, K., Moore, J. C., Siekmann, A. F., Lawson, N., and Pack, M. (2010). Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev. Dyn. 239 (3), 855–864. doi:10.1002/dvdy.22220
MacRae, C. A., and Peterson, R. T. (2015). Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14 (10), 721–731. doi:10.1038/nrd4627
Maestro, S., Weber, N. D., Zabaleta, N., Aldabe, R., and Gonzalez-Aseguinolaza, G. (2021). Novel vectors and approaches for gene therapy in liver diseases. Jhep Rep. 3 (4), 100300. doi:10.1016/j.jhepr.2021.100300
Manco, R., Clerbaux, L.-A., Verhulst, S., Nader, M. B., Sempoux, C., Ambroise, J., et al. (2019). Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury. J. Hepatology 70 (6), 1180–1191. doi:10.1016/j.jhep.2019.02.003
Marques, I. J., Lupi, E., and Mercader, N. (2019). Model systems for regeneration: zebrafish. Development 146 (18), dev167692. doi:10.1242/dev.167692
Meng, Z., Wang, Y., Wang, L., Jin, W., Liu, N., Pan, H., et al. (2010). FXR regulates liver repair after CCl4-induced toxic injury. Mol. Endocrinol. 24 (5), 886–897. doi:10.1210/me.2009-0286
Mi, J., Ren, L., and Andersson, O. (2024). Leveraging zebrafish to investigate pancreatic development, regeneration, and diabetes. Trends Mol. Med. 30, 932–949. doi:10.1016/j.molmed.2024.05.002
Michalopoulos, G. K., and Bhushan, B. (2021). Liver regeneration: biological and pathological mechanisms and implications. Nat. Rev. Gastroenterology and Hepatology 18 (1), 40–55. doi:10.1038/s41575-020-0342-4
Michalopoulos, G. K., and DeFrances, M. C. (1997). Liver regeneration. Science 276 (5309), 60–66. doi:10.1126/science.276.5309.60
Miyajima, A., Tanaka, M., and Itoh, T. (2014). Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14 (5), 561–574. doi:10.1016/j.stem.2014.04.010
Moh, A., Iwamoto, Y., Chai, G.-X., Zhang, S.S.-M., Kano, A., Yang, D. D., et al. (2007). Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab. Investig. 87 (10), 1018–1028. doi:10.1038/labinvest.3700630
Muramatsu, T. (2010). Midkine, a heparin-binding cytokine with multiple roles in development, repair and diseases. Proc. Jpn. Acad. Ser. B-Physical Biol. Sci. 86 (4), 410–425. doi:10.2183/pjab.86.410
Noel, E. S., Casal-Sueiro, A., Busch-Nentwich, E., Verkade, H., Dong, P. D. S., Stemple, D. L., et al. (2008). Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev. Biol. 322 (2), 237–250. doi:10.1016/j.ydbio.2008.06.040
North, T. E., Babu, I. R., Vedder, L. M., Lord, A. M., Wishnok, J. S., Tannenbaum, S. R., et al. (2010). PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc. Natl. Acad. Sci. U. S. A. 107 (40), 17315–17320. doi:10.1073/pnas.1008209107
Oderberg, I. M., and Goessling, W. (2021). Partial hepatectomy in adult zebrafish. Jove-Journal Vis. Exp. 170. doi:10.3791/62349
Oderberg, I. M., and Goessling, W. (2023). Biliary epithelial cells are facultative liver stem cells during liver regeneration in adult zebrafish. Jci Insight 8 (1), e163929. doi:10.1172/jci.insight.163929
Portugues, R., Severi, K. E., Wyart, C., and Ahrens, M. B. (2013). Optogenetics in a transparent animal: circuit function in the larval zebrafish. Curr. Opin. Neurobiol. 23 (1), 119–126. doi:10.1016/j.conb.2012.11.001
Pu, W., Zhu, H., Zhang, M., Pikiolek, M., Ercan, C., Li, J., et al. (2023). Bipotent transitional liver progenitor cells contribute to liver regeneration. Nat. Genet. 55 (4), 651–664. doi:10.1038/s41588-023-01335-9
Qiang, W., Shen, T., Noman, M., Guo, J., Jin, Z., Lin, D., et al. (2021). Fibroblast growth factor 21 augments autophagy and reduces apoptosis in damaged liver to improve tissue regeneration in zebrafish. Front. Cell Dev. Biol. 9, 756743. doi:10.3389/fcell.2021.756743
Raven, A., Lu, W.-Y., Man, T. Y., Ferreira-Gonzalez, S., O'Duibhir, E., Dwyer, B. J., et al. (2017). Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547 (7663), 350–354. doi:10.1038/nature23015
Rizvi, F., Lee, Y.-R., Diaz-Aragon, R., Bawa, P. S., So, J., Florentino, R. M., et al. (2023). VEGFA mRNA-LNP promotes biliary epithelial cell-to-hepatocyte conversion in acute and chronic liver diseases and reverses steatosis and fibrosis. Cell Stem Cell 30 (12), 1640–1657.e8. doi:10.1016/j.stem.2023.10.008
Russell, J. O., Ko, S., Monga, S. P., and Shin, D. (2019a). Notch inhibition promotes differentiation of liver progenitor cells into hepatocytes via sox9b repression in zebrafish. Stem Cells Int. 2019, 8451282. doi:10.1155/2019/8451282
Russell, J. O., Ko, S., Shin, D., and Monga, S. P. (2017). Bromodomain and extraterminal (BET) proteins regulate hepatocyte proliferation in hepatocyte-driven liver regeneration. Faseb J. 31. doi:10.1096/fasebj.31.1_supplement.531.7
Russell, J. O., Lu, W.-Y., Okabe, H., Abrams, M., Oertel, M., Poddar, M., et al. (2019b). Hepatocyte-specific β-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology 69 (2), 742–759. doi:10.1002/hep.30270
Sadler, K. C., Krahn, K. N., Gaur, N. A., and Ukomadu, C. (2007). Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc. Natl. Acad. Sci. U. S. A. 104 (5), 1570–1575. doi:10.1073/pnas.0610774104
Sánchez, A., Factor, V. M., Schroeder, I. S., Nagy, P., and Thorgeirsson, S. S. (2004). Activation of NF-kappaB and STAT3 in rat oval cells during 2-acetylaminofluorene/partial hepatectomy-induced liver regeneration. Hepatology 39 (2), 376–385. doi:10.1002/hep.20040
Sande-Melon, M., Bergemann, D., Fernandez-Lajarin, M., Gonzalez-Rosa, J. M., and Cox, A. G. (2024). Development of a hepatic cryoinjury model to study liver regeneration. Development 151 (5), dev203124. doi:10.1242/dev.203124
Sato, K., Marzioni, M., Meng, F., Francis, H., Glaser, S., and Alpini, G. (2019). Ductular reaction in liver diseases: pathological mechanisms and translational significances. Hepatology 69 (1), 420–430. doi:10.1002/hep.30150
Shimizu, N., Shiraishi, H., and Hanada, T. (2023). Zebrafish as a useful model system for human liver disease. Cells 12 (18), 2246. doi:10.3390/cells12182246
Simons, M., Gordon, E., and Claesson-Welsh, L. (2016). Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 17 (10), 611–625. doi:10.1038/nrm.2016.87
So, J., Kim, A., Lee, S.-H., and Shin, D. (2020). Liver progenitor cell-driven liver regeneration. Exp. Mol. Med. 52 (8), 1230–1238. doi:10.1038/s12276-020-0483-0
So, J., Kim, M., Lee, S.-H., Ko, S., Lee, D. A., Park, H., et al. (2021). Attenuating the epidermal growth factor receptor-extracellular signal-regulated kinase-sex-determining region Y-box 9 Axis promotes liver progenitor cell-mediated liver regeneration in zebrafish. Hepatology 73 (4), 1494–1508. doi:10.1002/hep.31437
Song, J., Ma, J., Liu, X., Huang, Z., Li, L., Li, L., et al. (2023). The MRN complex maintains the biliary-derived hepatocytes in liver regeneration through ATR-Chk1 pathway. Npj Regen. Med. 8 (1), 20. doi:10.1038/s41536-023-00294-3
Stanger, B. Z. (2015). “Cellular homeostasis and repair in the mammalian liver,”in Annual review of physiology. Editor D. Julius, 77, 179–200. doi:10.1146/annurev-physiol-021113-170255
Sun, T., Pikiolek, M., Orsini, V., Bergling, S., Holwerda, S., Morelli, L., et al. (2020). AXIN2+ pericentral hepatocytes have limited contributions to liver homeostasis and regeneration. Cell Stem Cell 26 (1), 97–107. doi:10.1016/j.stem.2019.10.011
Tan, V. W. T., Salmi, T. M., Karamalakis, A. P., Gillespie, A., Ong, A. J. S., Balic, J. J., et al. (2024). SLAM-ITseq identifies that Nrf2 induces liver regeneration through the pentose phosphate pathway. Dev. Cell 59 (7), 898–910.e6. doi:10.1016/j.devcel.2024.01.024
Unterweger, I. A., Klepstad, J., Hannezo, E., Lundegaard, P. R., Trusina, A., and Ober, E. A. (2023). Lineage tracing identifies heterogeneous hepatoblast contribution to cell lineages and postembryonic organ growth dynamics. PLoS Biol. 21 (10), e3002315. doi:10.1371/journal.pbio.3002315
van der Helm, D., Groenewoud, A., de Jonge-Muller, E. S. M., Barnhoorn, M. C., Schoonderwoerd, M. J. A., Coenraad, M. J., et al. (2018). Mesenchymal stromal cells prevent progression of liver fibrosis in a novel zebrafish embryo model. Sci. Rep. 8, 16005. doi:10.1038/s41598-018-34351-5
Van Haele, M., Snoeck, J., and Roskams, T. (2019). Human liver regeneration: an etiology dependent process. Int. J. Mol. Sci. 20 (9), 2332. doi:10.3390/ijms20092332
Wang, B., Zhao, L., Fish, M., Logan, C. Y., and Nusse, R. (2015). Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature 524 (7564), 180–185. doi:10.1038/nature14863
Wang, F.-S., Fan, J.-G., Zhang, Z., Gao, B., and Wang, H.-Y. (2014). The global burden of liver disease: the major impact of China. Hepatology 60 (6), 2099–2108. doi:10.1002/hep.27406
Wang, Q. J., Zambetti, G. P., and Suttle, D. P. (1997). Inhibition of DNA topoisomerase II alpha gene expression by the p53 tumor suppressor. Mol. Cell. Biol. 17 (1), 389–397. doi:10.1128/mcb.17.1.389
Wang, S., Miller, S. R., Ober, E. A., and Sadler, K. C. (2017). “Making it new again: insight into liver development, regeneration, and disease from zebrafish research,” in Zebrafish at the interface of development and disease research. Editor K. C. Sadler, 161–195.
Wang, X., Copmans, D., and de Witte, P. A. M. (2021). Using zebrafish as a disease model to study fibrotic disease. Int. J. Mol. Sci. 22 (12), 6404. doi:10.3390/ijms22126404
Wang, Y., Rodrigues, R. M., Chen, C., Feng, D., Maccioni, L., and Gao, B. (2024). Macrophages in necrotic liver lesion repair: opportunities for therapeutical applications. Am. J. Physiology-Cell Physiology 326 (5), C1556–C1562. doi:10.1152/ajpcell.00053.2024
Weglarz, T. C., and Sandgren, E. P. (2000). Timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous. Proc. Natl. Acad. Sci. U. S. A. 97 (23), 12595–12600. doi:10.1073/pnas.220430497
Weis, P. (1972). Hepatic ultrastructure in 2 species of normal, fasted and gravid teleost fishes. Am. J. Anat. 133 (3), 317–and. doi:10.1002/aja.1001330306
Wrighton, P. J., Oderberg, I. M., and Goessling, W. (2019). There is something fishy about liver cancer: zebrafish models of hepatocellular carcinoma. Cell. Mol. Gastroenterology Hepatology 8 (3), 347–363. doi:10.1016/j.jcmgh.2019.05.002
Xiao, J., Wang, F., Wong, N.-K., He, J., Zhang, R., Sun, R., et al. (2019). Global liver disease burdens and research trends: analysis from a Chinese perspective. J. Hepatology 71 (1), 212–221. doi:10.1016/j.jhep.2019.03.004
Yagi, S., Hirata, M., Miyachi, Y., and Uemoto, S. (2020). Liver regeneration after hepatectomy and partial liver transplantation. Int. J. Mol. Sci. 21 (21), 8414. doi:10.3390/ijms21218414
Yoon, E., Babar, A., Choudhary, M., Kutner, M., and Pyrsopoulos, N. (2016). Acetaminophen-induced hepatotoxicity: a comprehensive update. J. Clin. Transl. hepatology 4 (2), 131–142. doi:10.14218/jcth.2015.00052
Yuan, X., Wu, J., Sun, Z., Cen, J., Shu, Y., Wang, C., et al. (2024). Preclinical efficacy and safety of encapsulated proliferating human hepatocyte organoids in treating liver failure. Cell Stem Cell 31 (4), 484–498.e5. doi:10.1016/j.stem.2024.02.005
Zhang, J., Zhou, Y., Li, S., Mo, D., Ma, J., Ni, R., et al. (2022). Tel2 regulates redifferentiation of bipotential progenitor cells via Hhex during zebrafish liver regeneration. Cell Rep. 39 (1), 110596. doi:10.1016/j.celrep.2022.110596
Zhang, W., Chen, J., Ni, R., Yang, Q., Luo, L., and He, J. (2021). Contributions of biliary epithelial cells to hepatocyte homeostasis and regeneration in zebrafish. Iscience 24 (3), 102142. doi:10.1016/j.isci.2021.102142
Zhang, X., Liu, H., Cai, P., Huang, Z., Ma, J., and Luo, L. (2024). Mdka produced by the activated hepatic stellate cells drives bipotential progenitor cell redifferentiation during zebrafish biliary-mediated liver regeneration. Hepatology. doi:10.1097/HEP.0000000000001031
Zhu, H., Chang, B.-D., Uchiumi, T., and Roninson, I. B. (2002). Identification of promoter elements responsible for transcriptional inhibition of polo-like kinase 1 and topoisomerase IIα genes by p21WAF1/CIP1/SDI1. Cell Cycle 1 (1), 55–62. doi:10.4161/cc.1.1.101
Zhu, Z., Chen, J., Xiong, J.-W., and Peng, J. (2014). Haploinsufficiency of Def activates p53-dependent TGFβ signalling and causes scar formation after partial hepatectomy. Plos One 9 (5), e96576. doi:10.1371/journal.pone.0096576
Zong, Y., Panikkar, A., Xu, J., Antoniou, A., Raynaud, P., Lemaigre, F., et al. (2009). Notch signaling controls liver development by regulating biliary differentiation. Development 136 (10), 1727–1739. doi:10.1242/dev.029140
Keywords: zebrafish, liver injury, biliary epithelial cell, hepatocyte, regeneration
Citation: Mo D, Lv M and Mao X (2024) Using different zebrafish models to explore liver regeneration. Front. Cell Dev. Biol. 12:1485773. doi: 10.3389/fcell.2024.1485773
Received: 24 August 2024; Accepted: 22 October 2024;
Published: 31 October 2024.
Edited by:
Nai Yang Fu, Duke-NUS Medical School, SingaporeReviewed by:
Alberto Dinarello, University of Copenhagen, DenmarkCopyright © 2024 Mo, Lv and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dashuang Mo, d3VtYW5zaHVhbmdAaG90bWFpbC5jb20=; Xiaoyu Mao, am9keTA0MTdAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.