- Cell Biology Unit, Children’s Medical Research Institute, Faculty of Medicine and Health, University of Sydney, Westmead, NSW, Australia
Telomeres are the protective caps at the ends of linear chromosomes of eukaryotic organisms. Telomere binding proteins, including the six components of the complex known as shelterin, mediate the protective function of telomeres. They do this by suppressing many arms of the canonical DNA damage response, thereby preventing inappropriate fusion, resection and recombination of telomeres. One way this is achieved is by facilitation of DNA replication through telomeres, thus protecting against a “replication stress” response and activation of the master kinase ATR. On the other hand, DNA damage responses, including replication stress and ATR, serve a positive role at telomeres, acting as a trigger for recruitment of the telomere-elongating enzyme telomerase to counteract telomere loss. We postulate that repression of telomeric replication stress is a shared mechanism of control of telomerase recruitment and telomere length, common to several core telomere binding proteins including TRF1, POT1 and CTC1. The mechanisms by which replication stress and ATR cause recruitment of telomerase are not fully elucidated, but involve formation of nuclear actin filaments that serve as anchors for stressed telomeres. Perturbed control of telomeric replication stress by mutations in core telomere binding proteins can therefore cause the deregulation of telomere length control characteristic of diseases such as cancer and telomere biology disorders.
1 Introduction
Telomeres are nucleoprotein complexes located at the ends of linear chromosomes which serve to maintain genomic integrity and ensure cellular survival. Telomeres were first identified in fruit flies and corn (McClintock, 1941; Muller, 1938) and have since been characterized in a range of eukaryotes. Human telomeres are comprised of tandem TTAGGG repeats which extend for 3–18 kb (Moyzis et al., 1988), ending in a single-stranded G-rich overhang 12–400 nucleotides long (Makarov et al., 1997; McElligott and Wellinger, 1997; Zhao et al., 2008). This overhang, which if exposed would resemble DNA damage, can strand invade the double-stranded region of the telomere to form a telomere-loop (t-loop; Figure 1A), which serves to protect the telomere from being misrecognized as a double-strand break (DSB) and activating a DNA damage response (DDR) (Doksani et al., 2013; Griffith et al., 1999; Van Ly et al., 2018).
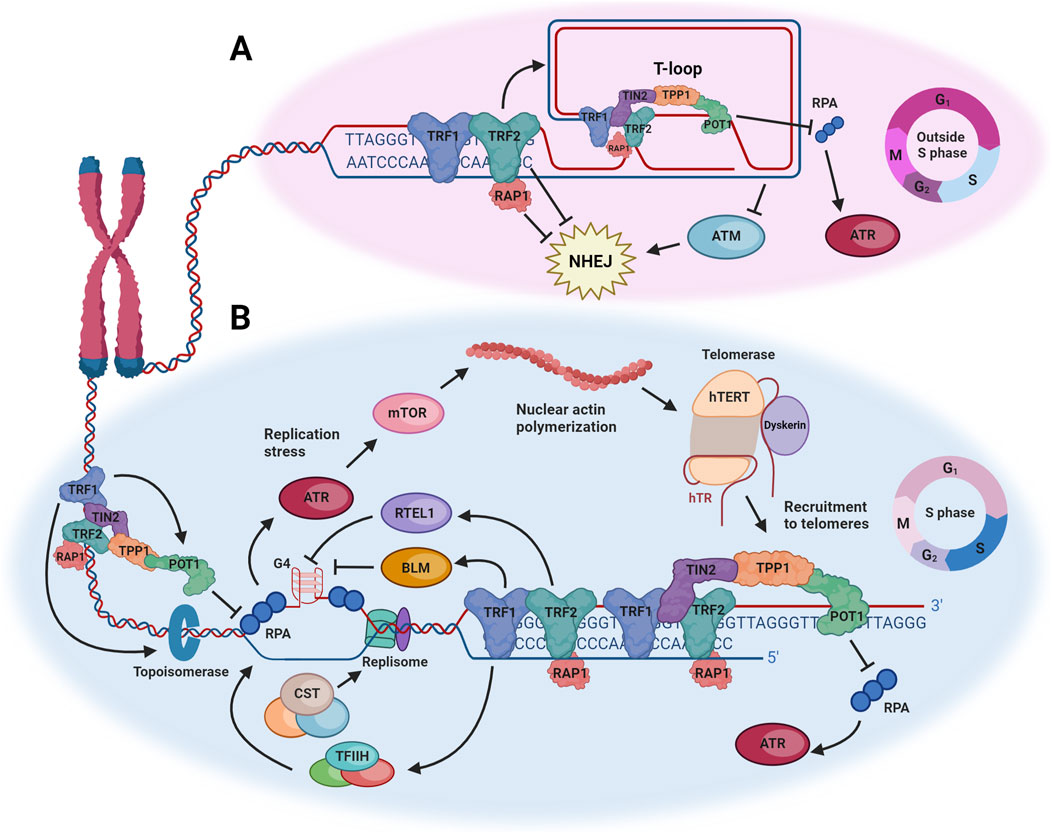
Figure 1. Summary of mechanisms by which the DNA damage response is repressed at telomeres or harnessed to facilitate telomerase-mediated telomere maintenance. (A) Shelterin protects telomeres from inappropriately activating a DNA damage response in phases of the cell cycle when the telomere can fold into a t-loop. TRF2 promotes t-loop formation, which prevents activation of ATM, and TRF2 also directly suppresses NHEJ. POT1 protects against activation of ATR. (B) During S phase, the t-loop is unwound to allow replication of the telomere by the canonical replication machinery, which is impeded by the repetitive sequence of telomeres, their occupation by shelterin, and propensity to form G-quadruplexes (G4). Multiple components of shelterin counteract the resulting replication stress, but any remaining stress activates ATR, which facilitates the polymerization of nuclear actin and ultimately the recruitment of telomerase to telomeres. See text for more details. Figure created with Biorender.com.
Telomeric DNA is bound by a six protein complex named shelterin, which helps maintain telomere structure and function in many different ways (de Lange, 2005; de Lange, 2018; Figure 1). Shelterin binds to telomeres through TRF1 and TRF2 (telomere-repeat binding factor 1 and 2), which both recognize and bind to the double-stranded portion of the telomere (Bilaud et al., 1997; Broccoli et al., 1997; Chong et al., 1995). The telomere is also bound by POT1 (protection of telomere 1) which forms a heterodimer with TPP1 (TINT1/PTOP/PIP1) (Houghtaling et al., 2004; Liu et al., 2004; Ye et al., 2004b) and binds to the single-stranded overhang specifically through its oligonucleotide/oligosaccharide binding (OB) folds (Baumann and Cech, 2001; Loayza and De Lange, 2003). Rap1 is a conserved shelterin subunit and is recruited to telomeres by forming a complex with TRF2 (Li et al., 2000; Sfeir et al., 2010). The final shelterin component is TIN2 (TRF1-interacting factor 2) which bridges TRF1/2 and the TPP1-POT1 complex and helps to stabilize the latter in addition to the TRF2-Rap1 complex (Houghtaling et al., 2004; Kim et al., 1999; Liu et al., 2004; Ye and de Lange, 2004; Ye et al., 2004a). Shelterin is crucial for telomere protection, as loss of shelterin binding leads to deprotection of telomeres, unwinding of the t-loop and activation of at least seven different DDR pathways (de Lange, 2018; Sfeir and de Lange, 2012), culminating in genomic instability, cell cycle arrest and cell death. Conversely, the shelterin complex can also harness several DDR pathways to assist in maintenance of telomere length and avoidance of the cell cycle arrest and cell death that result from critically short telomeres. This article will discuss the complicated and paradoxical relationship between telomeres, the DDR, and telomere maintenance by the ribonucleoprotein enzyme telomerase, with a focus on events in human cells.
2 DNA damage and its suppression at the telomere
Given that the structure of a telomere resembles damaged DNA, it is imperative that the DDR is suppressed to prevent unnecessary repair of the telomere which could result in chromosome end-to-end fusions. Global DNA damage is detected largely by three phosphatidylinositol 3-kinase-related protein kinases (PIKKs): ATR (ataxia telangiectasia and Rad3-related), ATM (ataxia telangiectasia mutated) and DNA-PK (DNA-dependent protein kinase) (Blackford and Jackson, 2017). These kinases are responsible for the activation of various signaling pathways which trigger cell cycle arrest and the promotion of DNA repair mechanisms to resolve the DNA damage (Harrison and Haber, 2006). Should the extent of damage be irreparable then the kinases can instead signal the cell to undergo apoptosis or senescence (Nowsheen and Yang, 2012). ATR is activated in response to single-stranded DNA, which can occur at replication forks that are slowed or stalled in situations where DNA replication is impeded (a state known as “replication stress”) (Saldivar et al., 2017; Yang et al., 2004). Conversely, both ATM and DNA-PK can be activated in response to DSBs, although they each employ different pathways to repair the damage, through homology-directed repair or non-homologous end joining (NHEJ) respectively (Davis et al., 2014; Shiloh and Ziv, 2013).
The components of shelterin both directly and indirectly suppress an unwanted telomeric DDR. Indirect suppression relies on t-loop formation, as telomeres in this closed state are unlikely to trigger a DDR (Figure 1A). This conformation would prevent the telomere end from being detected by factors which sense exposed DNA ends and activate the DDR, such as MRN (Mre11, Rad50, Nbs1) or the Ku70/80 complex (Gottlieb and Jackson, 1993; Lee and Paull, 2005). The key regulator of t-loop formation is TRF2, as its depletion from cells results in reduced t-loop formation (Doksani et al., 2013; Van Ly et al., 2018). Furthermore, TRF2 can promote strand invasion in vitro, as it binds to DNA and promotes formation of structures which resemble t-loops (Amiard et al., 2007; Stansel et al., 2001). As telomeres shorten, or following moderate TRF2 depletion, t-loops unfold and telomere ends become accessible to the DNA damage sensing machinery. These are termed “intermediate-state” telomeres (Cesare and Karlseder, 2012; Cesare et al., 2009), which trigger a DDR predominately through ATM (Karlseder et al., 2004; Van Ly et al., 2018), leading to cell cycle arrest and senescence. However, critically short telomeres or extreme loss of TRF2 instead results in deprotected telomeres (an “uncapped-state”) which undergo ATM-dependent end-to-end chromosomal fusions, resulting in genome wide instability (Celli and de Lange, 2005; Denchi and de Lange, 2007). This is a consequence of TRF2 also directly repressing the downstream consequences of ATM activation at telomeres, via a short region named the iDDR (inhibitor of DDR) within the hinge domain of TRF2 (Okamoto et al., 2013). This motif inhibits telomeric localization of the E3 ubiquitin ligase RNF168, in turn preventing 53BP1 accumulation at telomeres and chromosomal fusion via NHEJ (Okamoto et al., 2013).
On the other hand, ATR activation resulting from an exposed single-stranded telomeric overhang is thought to be primarily suppressed by POT1, since POT1 depletion activates ATR and results in ATR-dependent DNA damage signaling at telomeres (Denchi and de Lange, 2007; Figures 1A, B). This is dependent on the ability of POT1 to bind to telomeres, as preventing its recruitment via TPP1 or TIN2 inhibition also results in ATR activation (Hockemeyer et al., 2007; Takai et al., 2011). POT1 binds to single-stranded telomeric repeats, including the 3′telomeric overhang, preventing binding of RPA (replication protein A) (Gong and de Lange, 2010), which coats single-stranded DNA and is a key factor in the activation of ATR (Zou and Elledge, 2003).
3 Shelterin suppression of the DDR resulting from telomere replication
While the structural similarity of exposed telomeres and damaged DNA suggests that telomere-associated proteins would strive to keep DDR proteins away, paradoxically this does not always appear to occur. In fact, many DDR proteins are recruited to telomeres, not just in cancer cells, but also in normal somatic cells. For example, ATR localizes to telomeres during S phase in normal human fibroblasts (Verdun and Karlseder, 2006), and is necessary to protect telomere fidelity in both human and mouse cells (McNees et al., 2010; Pennarun et al., 2010). One possible explanation for this phenomenon is that telomeres are difficult regions for the DNA replication machinery to efficiently and correctly traverse, as repetitive DNA sequences (termed “fragile sites”) are highly prone to stalling replication machinery at the replication fork. Replication fork pausing or stalling has been observed in telomeres from yeast to humans (Fouche et al., 2006; Ivessa et al., 2002; Makovets et al., 2004; Sfeir et al., 2009), and may be further compounded at telomeres by shelterin binding also impeding DNA replication (Ohki and Ishikawa, 2004). Additionally, given their G-rich composition, telomeres readily form G-quadruplexes (G4s), DNA secondary structures which can also impede replisome progression (Bochman et al., 2012; Bryan, 2020). The inhibition of replication fork progression requires activation of processes to stabilize, repair and ultimately restart the replication fork. This response is regulated by ATR, which is activated by RPA-coated single-stranded DNA at the stalled replication fork (Shechter et al., 2004). Excessive replication stress at telomeres which cannot be resolved results in a “fragile telomere” phenotype: telomeres with breaks which appear as elongated or multiple signals at the ends of metaphase chromosomes (Sfeir et al., 2009). Given this, it is imperative that replication of the telomere is regulated to ensure its completion to prevent significant loss of telomeric DNA.
To prevent the generation of replication stress, shelterin helps to promote efficient telomere replication [reviewed in Bonnell et al. (2021); Higa et al. (2017a); Figure 1B]. The promotion of telomere replication by shelterin proteins is conserved across evolution, since Taz1, the fission yeast orthologue of TRF1, also has this capability (Miller et al., 2006). Mammalian TRF1 specifically mitigates lagging strand replication stress by promoting the recruitment of BLM (Bloom syndrome) (Martinez et al., 2009; Sfeir et al., 2009; Zimmermann et al., 2014), a RecQ family helicase which is capable of unwinding G4s which would impede replisome progression (Drosopoulos et al., 2015; Sun et al., 1998). A similar RecQ helicase, WRN (Werner syndrome), is also important for telomere replication as its depletion results in large telomeric deletions (Crabbe et al., 2004). WRN is capable of unwinding G4s (Drosopoulos et al., 2015; Mohaghegh et al., 2001), co-localizes at stalled replication forks with ATR and PCNA (proliferating cell nuclear antigen), a DNA clamp component of the replisome (Constantinou et al., 2000; Rodrı́guez-López et al., 2003), and is capable of maintaining telomeric overhangs in vitro in a DNA-PK-dependent manner (Kusumoto-Matsuo et al., 2010). Furthermore, both BLM and WRN have been shown to interact with TRF1, TRF2 and POT1 in vitro, which in most cases stimulates their helicase activity (Lillard-Wetherell et al., 2004; Opresko et al., 2005; Opresko et al. 2004; Opresko et al. 2002). The specific ability of TRF1 to recruit helicases to telomeres during S phase is modulated by post-translational modification of TRF1, including phosphorylation and poly-ADP-ribosylation (Li et al., 2018; Maresca et al., 2023).
RTEL1 (regulator of telomere elongation helicase 1) is another important regulator of telomeric replication, as it is capable of unwinding G4s and t-loops to promote replisome progression (Vannier et al., 2012; Vannier et al., 2013). RTEL1 is recruited to telomeres in S phase by TRF2 and interacts with PCNA (Sarek et al., 2015; Vannier et al., 2013). Loss of RTEL1, or inhibition of its interaction with PCNA or TRF2, results in replication fork stalling, fragile telomeres and t-loop cleavage (Sarek et al., 2015; Vannier et al., 2012; Vannier et al., 2013). These observations suggest that RTEL1 is capable of unwinding t-loops to facilitate complete replication of telomeres, which is further supported by the ability of RTEL1 to unwind t-loop-like structures in vitro (Youds et al., 2010).
Another way in which TRF1 and TRF2 help telomeres avoid replication stress is by the recruitment of topoisomerase IIα and the nuclease Apollo to relieve topological stress at telomeres (d'Alcontres et al., 2014; Ye et al., 2010). Furthermore, TRF1-mediated recruitment of the transcription factor and nucleotide excision repair complex TFIIH is required to suppress the fragile telomere phenotype (Yang et al., 2022). If replication fork stalling does occur, TRF1 also helps mitigate the resulting ATR-mediated DDR by virtue of its tethering TIN2/POT1/TPP1 to the telomere, which may be able to displace RPA from exposed single-stranded DNA at replication forks (Zimmermann et al., 2014). The shelterin complex thus employs a full armory of defenses against the many ways in which telomeres could provoke unwanted DDRs, particularly during DNA replication.
4 Replication stress is a trigger for telomere maintenance by telomerase
Shelterin proteins also ensure complete telomere maintenance through their role in recruiting the telomere lengthening enzyme telomerase. Telomerase is an evolutionarily conserved ribonucleoprotein complex (Greider and Blackburn, 1985; Morin, 1989), containing two major components: the catalytic protein subunit telomerase reverse transcriptase (hTERT in humans; Nakamura et al., 1997), and the telomerase RNA subunit (hTR in humans; Feng et al., 1995), which provides the template for de novo telomere synthesis. Most normal human somatic cells lack telomerase expression (Nakamura et al., 1997), and hence experience telomere shortening over successive population doublings (Harley et al., 1990), due to the inability of the conventional DNA replication machinery to copy the extreme ends of linear DNA molecules (Lingner et al., 1995; Takai et al., 2024). Approximately 85% of cancer cells overcome telomere shortening via activation of telomerase; however, telomerase is also active within highly proliferative cells, including stem and germline cells [reviewed in Bryan and Cohen (2023); Roake and Artandi (2020)]. Following synthesis of a telomeric repeat, telomerase translocates along the newly synthesized repeat to continue extending the 3′overhang of the same telomere (Greider, 1991; Wu et al., 2017). Following this, the complementary C-rich telomeric strand is filled in by the CST-Polα/primase complex [reviewed in Cai and de Lange (2023)].
Telomerase activity relies upon its successful recruitment to telomeres; this process is very tightly regulated and typically only occurs during S phase (Diede and Gottschling, 1999; Jady et al., 2006; Marcand et al., 2000; Tomlinson et al., 2006). The timing of recruitment appears to be regulated over the cell cycle, at least partially, by human TRF1 or its fission yeast orthologue Taz1, as their depletion results in accumulation of telomerase at telomeres outside S phase (Dehe et al., 2012; Tong et al., 2015). Stable recruitment of human telomerase is dependent on its interaction with TPP1, specifically an N-terminal patch termed the TEL patch within the TPP1 OB fold (Grill et al., 2018; Nandakumar et al., 2012; Zhong et al., 2012). The interaction between POT1 and TPP1 is also essential for telomerase processivity (Latrick and Cech, 2010; Wang et al., 2007), while the depletion of TPP1 and TIN2 (which bridges TPP1 and TRF1/2) results in reduced telomerase presence at telomeres (Abreu et al., 2010). The interaction of hTERT with TPP1 relies upon the TEN domain within hTERT (Schmidt et al., 2014; Stern et al., 2012), and a region within the reverse transcriptase domain (Padmanaban et al., 2023).
It has become clear that in addition to their role in ensuring accurate telomere replication, DDR proteins are also important regulators of telomerase recruitment to the telomere. In particular, the telomeric replication stress response can be considered an evolutionarily conserved trigger for bringing telomerase to those telomeres experiencing stress. The requirement for an ATR-mediated DDR for telomerase recruitment to telomeres was initially demonstrated in the fission yeast Schizosaccharomyces pombe, as depletion of Rad3 (the fission yeast homolog of ATR) negatively impacts telomerase recruitment in this organism (Moser et al., 2011; Moser et al., 2009; Yamazaki et al., 2012). In humans, patients with ATR mutations have poor telomere maintenance, resulting in a range of telomere abnormalities (Pennarun et al., 2010); depletion or chemical inhibition of ATR also results in reduced telomerase recruitment to telomeres in human cells (Tong et al., 2015). This may involve the 9-1-1 (Rad9-Rad1-Hus1) checkpoint complex, which is needed for full catalytic activation of ATR at single-stranded DNA (Majka et al., 2006), and also shows an association with active human telomerase (Francia et al., 2006). Inversely, induction of replication stress can promote the recruitment of telomerase and therefore its ability to lengthen telomeres. Chemical induction of replication stress using the DNA polymerase inhibitor aphidicolin results in telomere lengthening (Sfeir et al., 2009) and an ATR-dependent increased accumulation of telomerase at telomeres (Tong et al., 2015). This regulation of telomerase recruitment to telomeres by ATR appears to be highly specific to natural chromosome ends, as contrastingly it has recently been demonstrated that ATR inhibits de novo telomere formation by telomerase at resected DSBs (Kinzig et al., 2024). Induction of interstitial DSBs, using either Cas9 or I-SceI, resulted in telomerase-mediated de novo telomere formation which was significantly upregulated after inhibition of ATR, but not ATM (Kinzig et al., 2024).
As described briefly above, components of shelterin have long been known to play important roles in regulating telomere length by modulation of the access of telomerase to telomeres [reviewed in Smogorzewska and de Lange (2004)]. There have been various models proposed to provide conceptual frameworks for how this achieved; for example, the “protein counting model” postulates that the overall abundance of shelterin proteins at telomeres provides a negative regulatory signal for telomerase recruitment and/or extension (Marcand et al., 1997; Smogorzewska and de Lange, 2004). An alternative model proposes that telomerase travels along the telomere with the replication fork, and the chances of it reaching the end of the telomere are impacted by its encounters with shelterin proteins and the distance from subtelomeric origins of replication (Greider, 2016). Here, we would like to instead argue that the role of shelterin proteins in controlling telomere replication stress, discussed above, is a major component of their ability to also regulate telomerase recruitment to telomeres (Figure 1B).
As discussed above, a major function of TRF1 at telomeres is to recruit and coordinate other proteins to reduce telomeric replication stress, and TRF1 is also a major negative regulator of telomerase-mediated telomere lengthening (Smogorzewska et al., 2000). TRF1 levels at human telomeres decrease during S phase (Maresca et al., 2023; Verdun et al., 2005); this decrease is partially regulated by ATM, since experimental depletion of ATM results in accumulation of TRF1 at telomeres (Wu et al., 2007). Phosphorylation of S367 on TRF1 by ATM results in telomere elongation, due to removal of TRF1 from the telomere (McKerlie et al., 2012). This phosphorylation event also promotes telomerase recruitment to the telomere, as a phospho-null mutation of TRF1 S367 diminishes telomerase recruitment (Tong et al., 2015). Concordantly, experimental depletion or inhibition of ATM results in telomere shortening (Wu et al., 2007), and reduced recruitment of telomerase (Lee et al., 2015; Tong et al., 2015), and cells from patients with mutations in ATM have short telomeres (Metcalfe et al., 1996). Together, these data support a model in which partial depletion of TRF1 during S phase triggers telomere replication stress that results in increased telomerase recruitment. This model is supported by data from S. pombe, showing that deletion of the TRF1 orthologue Taz1 results in deregulated arrival of replicative polymerases at telomeres, resulting in an extended period in which both RPA and TERT are simultaneously and aberrantly recruited to telomeres, implicating a replication stress-induced ATR response in telomerase recruitment (Chang et al., 2013).
This may also be an explanation for the telomere lengthening induced by mutant versions of POT1. The first hints that POT1 is also involved in facilitating telomere replication came from the identification of familial or somatic mutations in patients with cancer; these mutations induced fragile telomeres (Calvete et al., 2015; Ramsay et al., 2013; Shi et al., 2014), and were directly shown to result in increased fork stalling in a DNA combing replication assay (Pinzaru et al., 2016). Expression of POT1 lacking its OB fold (POT1-ΔOB), rendering it incapable of binding the telomeric overhang, also results in telomeric replication stress (Pinzaru et al., 2020), which ultimately leads to increased telomerase recruitment (Laprade et al., 2020) and telomere lengthening (Loayza and De Lange, 2003; Tong et al., 2015). Consistently, the cancers that carry mutations in the POT1 OB fold also have long telomeres (Calvete et al., 2015; Ramsay et al., 2013; Robles-Espinoza et al., 2014; Shi et al., 2014). This has also been observed in mice, which possess two POT1 homologs with separation of function; the major role of POT1a is to suppress the DDR at telomeres, while POT1b also regulates telomeric overhang length (He et al., 2006; Hockemeyer et al., 2006; Wu et al., 2006). When POT1b is depleted from mouse cells, their telomeres initially shorten, triggering a DDR and an increase in ATR-mediated telomerase recruitment to telomeres (Gu et al., 2021; Takasugi et al., 2023). These cells ultimately develop ultralong telomeres as a result of the increased telomerase presence at telomeres (Takasugi et al., 2023). While physical sequestration of the 3′ telomeric overhang may contribute to the negative regulation of telomerase by POT1, it is likely that its role in preventing replication stress also contributes to the same outcome.
Another protein complex that both helps to overcome replication stress at telomeres and participates in telomere length control is the CST complex (consisting of proteins CTC1, STN1 and TEN1 in humans) (Olson and Wuttke, 2024). Human CST promotes replication restart after fork stalling; it does this through promotion of latent origin firing (Stewart et al., 2012), recruitment of the replication stress-response protein RAD51 (Chastain et al., 2016) and activation of the ATR pathway [Ackerson et al. (2020); reviewed in Olson and Wuttke (2024)]. In addition, human CST depletion causes a telomerase-dependent increase in telomere length (Chen et al., 2012). A mutation in CTC1 that leads to telomere elongation also causes an increase in the recruitment of telomerase to telomeres (Gu et al., 2018). While this may be at least partially explained by the inhibitory effect of wild-type CST on telomerase binding to its DNA substrate (Chen et al., 2012; Zaug et al., 2021), resulting in dissociation of telomerase from the telomere, it is also possible that some CTC1 mutations increase telomerase recruitment by increasing telomeric replication stress. Given that mutations in CST components cause the telomere biology disorder Coats Plus, and that these patients do not always show the telomere shortening typical of telomere biology disorders (Revy et al., 2023), further understanding of the interplay between CST, replication stress and telomerase recruitment may shed light on the telomere dysfunction underlying their disease.
5 Mechanisms and outcomes of replication stress-induced telomerase recruitment
Although the ways in which ATR and the replication stress response result in telomerase recruitment are incompletely understood, recent evidence implicates nuclear actin as one downstream element of this pathway. A large body of work in recent years has identified that nuclear filamentous actin (F-actin) is a key regulator of the DDR. Actin rapidly polymerizes within the nucleus in response to replication stress and DSBs; under replication stress conditions this polymerization is dependent on ATR, whose activity is required for downstream phosphorylation of another PIKK family kinase, mTOR (Lamm et al., 2020), in turn regulating F-actin through the Wiskott-Aldrich syndrome protein (WASP) family (Mok et al., 2015). F-actin facilitates the DDR by re-localization of damaged DNA to the nuclear periphery for fork restart or DNA repair (Belin et al., 2015; Caridi et al., 2018; Lamm et al., 2020; Palumbieri et al., 2023; Ryu et al., 2015). Stressed telomeres are processed by the same pathway; they have also been shown to move toward the periphery under conditions of replication stress in an F-actin-dependent manner (Lamm et al., 2020; Pinzaru et al., 2020). It has also been previously demonstrated that telomeres located closer to the nuclear periphery are more likely to be later replicating (Arnoult et al., 2010); however, it is unclear whether this timing is solely due to their position within the nucleus, or if these are stressed telomeres.
Telomerase recruitment to telomeres also requires the function of nuclear F-actin; inhibition of actin polymerization, or regulators of its polymerization or function, results in decreased telomerase recruitment under endogenous conditions in human cell lines, as well as abrogating replication stress-mediated recruitment (Harman et al., 2024). Furthermore, nuclear F-actin serves as a direct site for telomerase recruitment to stressed telomeres, which reside along these actin fibers (Harman et al., 2024). The requirement for nuclear F-actin in this process, as well as its role in telomere replication, provides a further link which connects telomerase activity to telomere replication and the DDR.
Another mechanism by which the replication stress response may increase telomerase recruitment involves the known role of ATR in promoting firing of “dormant replication origins” (Ge and Blow, 2010) within a region under replicative stress, which allows replication in problematic regions to be efficiently completed. Replication of telomeres in human cells occurs across S phase and largely originates from origins within subtelomeric regions, proceeding unidirectionally through the telomere (Drosopoulos et al., 2015; Drosopoulos et al., 2012; Higa et al., 2017a). While this appears to account for most telomeric replication, replication origins have been observed within telomeric regions (Drosopoulos et al., 2012). These origins appear to be dormant, as they are fired following replication stress in a TRF2-dependent manner (Drosopoulos et al., 2020). This likely occurs via the recruitment of ORC proteins by TRF2 (Deng et al., 2007; Higa et al., 2017b; Tatsumi et al., 2008).
There is evidence that recruitment of telomerase to telomeres needs the passage of the replication fork. In budding yeast, telomerase cannot extend an artificial minichromosome unless it contains an origin of replication (Marcand et al., 2000), and generation of a 3′ telomeric overhang (the substrate of telomerase) does not happen without passage of a replication fork (Dionne and Wellinger, 1998). In human cells, telomerase acts after the conventional replication machinery has finished replicating the rest of the telomere (Hirai et al., 2012; Zhao et al., 2009). Therefore, it is possible that increased firing of dormant replication origins within telomeres during replication stress, mediated by ATR, increases the likelihood of the replication machinery reaching the end of the telomere, triggering telomerase recruitment.
The conservation of the link between replication stress and telomerase recruitment to telomeres implies that telomerase serves a vital cellular role after such stress. Indeed, there is an increasing amount of evidence that yeast need telomerase to survive replication stress, even when global telomere length has not decreased (Chang et al., 2009; Jay et al., 2016; Noel and Wellinger, 2011; Xie et al., 2015). Replication stress can cause sudden loss of telomeres (Crabbe et al., 2004; Zhang et al., 2017), so it is likely that telomerase is needed to counteract this potentially lethal event. If the requirement for telomerase to survive replication stress extends to human cells, this could open up ways to rapidly target cancer cells by combining replication stress-inducing chemotherapeutic agents with telomerase inhibition.
6 Conclusion
Telomeres impose a difficult balancing act that cells must perform to maintain their proliferative capabilities and genome integrity while also avoiding aberrant telomere overextension. Cells possess a tightly controlled regulatory system which impedes the DDR from inappropriately recognizing telomeres and causing telomere loss. Concurrently, the DDR is utilized to ensure that telomeres are correctly replicated and that their length is not shortened as a byproduct of replication stress, whether exogenous or endogenous. The interactions between telomere binding proteins and the telomeric replication stress response are emerging as major factors in the maintenance of telomere lengths within defined limits, which can be perturbed in diseases such as cancer and telomere biology disorders. Further elucidation of this interplay is therefore likely to increase understanding of the mechanisms underlying these diseases.
Data availability statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding author.
Author contributions
AH: Visualization, Writing–original draft, Writing–review and editing. TB: Conceptualization, Funding acquisition, Visualization, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. AH was supported by the Neil and Norma Hill Foundation. TB was supported by the Arcus Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abreu, E., Aritonovska, E., Reichenbach, P., Cristofari, G., Culp, B., Terns, R. M., et al. (2010). TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell Biol. 30 (12), 2971–2982. doi:10.1128/MCB.00240-10
Ackerson, S. M., Gable, C. I., and Stewart, J. A. (2020). Human CTC1 promotes TopBP1 stability and CHK1 phosphorylation in response to telomere dysfunction and global replication stress. Cell Cycle 19 (24), 3491–3507. doi:10.1080/15384101.2020.1849979
Amiard, S., Doudeau, M., Pinte, S., Poulet, A., Lenain, C., Faivre-Moskalenko, C., et al. (2007). A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 14 (2), 147–154. doi:10.1038/nsmb1192
Arnoult, N., Schluth-Bolard, C., Letessier, A., Drascovic, I., Bouarich-Bourimi, R., Campisi, J., et al. (2010). Replication timing of human telomeres is chromosome arm–specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 6 (4), e1000920. doi:10.1371/journal.pgen.1000920
Baumann, P., and Cech, T. R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292 (5519), 1171–1175. doi:10.1126/science.1060036
Belin, B. J., Lee, T., and Mullins, R. D. (2015). DNA damage induces nuclear actin filament assembly by Formin -2 and Spire-½ that promotes efficient DNA repair. Elife 4, e07735. doi:10.7554/eLife.07735
Bilaud, T., Brun, C., Ancelin, K., Koering, C. E., Laroche, T., and Gilson, E. (1997). Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17 (2), 236–239. doi:10.1038/ng1097-236
Blackford, A. N., and Jackson, S. P. (2017). ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66 (6), 801–817. doi:10.1016/j.molcel.2017.05.015
Bochman, M. L., Paeschke, K., and Zakian, V. A. (2012). DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13 (11), 770–780. doi:10.1038/nrg3296
Bonnell, E., Pasquier, E., and Wellinger, R. J. (2021). Telomere replication: solving multiple end replication problems. Front. Cell Dev. Biol. 9, 668171. doi:10.3389/fcell.2021.668171
Broccoli, D., Smogorzewska, A., Chong, L., and de Lange, T. (1997). Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17 (2), 231–235. doi:10.1038/ng1097-231
Bryan, T. M. (2020). G-quadruplexes at telomeres: friend or foe? Molecules 25 (16), 3686. doi:10.3390/molecules25163686
Bryan, T. M., and Cohen, S. B. (2023). “Telomerase,” in Handbook of chemical biology of nucleic acids (Springer), 1–26. doi:10.1007/978-981-19-9776-1_47
Cai, S. W., and de Lange, T. (2023). CST–Polα/Primase: the second telomere maintenance machine. Genes and Dev. 37 (13-14), 555–569. doi:10.1101/gad.350479.123
Calvete, O., Martinez, P., Garcia-Pavia, P., Benitez-Buelga, C., Paumard-Hernandez, B., Fernandez, V., et al. (2015). A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat. Commun. 6, 8383. doi:10.1038/ncomms9383
Caridi, C. P., D’Agostino, C., Ryu, T., Zapotoczny, G., Delabaere, L., Li, X., et al. (2018). Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559 (7712), 54–60. doi:10.1038/s41586-018-0242-8
Celli, G. B., and de Lange, T. (2005). DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7 (7), 712–718. doi:10.1038/ncb1275
Cesare, A. J., and Karlseder, J. (2012). A three-state model of telomere control over human proliferative boundaries. Curr. Opin. Cell Biol. 24 (6), 731–738. doi:10.1016/j.ceb.2012.08.007
Cesare, A. J., Kaul, Z., Cohen, S. B., Napier, C. E., Pickett, H. A., Neumann, A. A., et al. (2009). Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat. Struct. Mol. Biol. 16 (12), 1244–1251. doi:10.1038/nsmb.1725
Chang, M., Luke, B., Kraft, C., Li, Z., Peter, M., Lingner, J., et al. (2009). Telomerase is essential to alleviate pif1-induced replication stress at telomeres. Genetics 183 (3), 779–791. doi:10.1534/genetics.109.107631
Chang, Y.-T., Moser, B. A., and Nakamura, T. M. (2013). Fission yeast shelterin regulates DNA polymerases and Rad3 ATR kinase to limit telomere extension. PLoS Genet. 9 (11), e1003936. doi:10.1371/journal.pgen.1003936
Chastain, M., Zhou, Q., Shiva, O., Whitmore, L., Jia, P., Dai, X., et al. (2016). Human CST facilitates genome-wide RAD51 recruitment to GC-rich repetitive sequences in response to replication stress. Cell Rep. 16 (5), 2048–2114. doi:10.1016/j.celrep.2016.08.008
Chen, L. Y., Redon, S., and Lingner, J. (2012). The human CST complex is a terminator of telomerase activity. Nature 488, 540–544. doi:10.1038/nature11269
Chong, L., van Steensel, B., Broccoli, D., Erdjument-Bromage, H., Hanish, J., Tempst, P., et al. (1995). A human telomeric protein. Science 270 (5242), 1663–1667. doi:10.1126/science.270.5242.1663
Constantinou, A., Tarsounas, M., Karow, J. K., Brosh, R. M., Bohr, V. A., Hickson, I. D., et al. (2000). Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1 (1), 80–84. doi:10.1093/embo-reports/kvd004
Crabbe, L., Verdun, R. E., Haggblom, C. I., and Karlseder, J. (2004). Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306 (5703), 1951–1953. doi:10.1126/science.1103619
d'Alcontres, M. S., Palacios, J. A., Mejias, D., and Blasco, M. A. (2014). TopoIIα prevents telomere fragility and formation of ultra thin DNA bridges during mitosis through TRF1-dependent binding to telomeres. Cell Cycle 13 (9), 1463–1481. doi:10.4161/cc.28419
Davis, A. J., Chen, B. P., and Chen, D. J. (2014). DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA repair 17, 21–29. doi:10.1016/j.dnarep.2014.02.020
Dehe, P. M., Rog, O., Ferreira, M. G., Greenwood, J., and Cooper, J. P. (2012). Taz1 enforces cell-cycle regulation of telomere synthesis. Mol. Cell 46 (6), 797–808. doi:10.1016/j.molcel.2012.04.022
de Lange, T. (2005). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19 (18), 2100–2110. doi:10.1101/gad.1346005
de Lange, T. (2018). Shelterin-mediated telomere protection. Annu. Rev. Genet. 52, 223–247. doi:10.1146/annurev-genet-032918-021921
Denchi, E. L., and de Lange, T. (2007). Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448 (7157), 1068–1071. doi:10.1038/nature06065
Deng, Z., Dheekollu, J., Broccoli, D., Dutta, A., and Lieberman, P. M. (2007). The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr. Biol. 17, 1989–1995. doi:10.1016/j.cub.2007.10.054
Diede, S. J., and Gottschling, D. E. (1999). Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases a and d. Cell 99, 723–733. doi:10.1016/s0092-8674(00)81670-0
Dionne, I., and Wellinger, R. J. (1998). Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res. 26, 5365–5371. doi:10.1093/nar/26.23.5365
Doksani, Y., Wu, J. Y., de Lange, T., and Zhuang, X. (2013). Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155 (2), 345–356. doi:10.1016/j.cell.2013.09.048
Drosopoulos, W. C., Deng, Z., Twayana, S., Kosiyatrakul, S. T., Vladimirova, O., Lieberman, P. M., et al. (2020). TRF2 mediates replication initiation within human telomeres to prevent telomere dysfunction. Cell Rep. 33 (6), 108379. doi:10.1016/j.celrep.2020.108379
Drosopoulos, W. C., Kosiyatrakul, S. T., and Schildkraut, C. L. (2015). BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J. Cell Biol. 210 (2), 191–208. doi:10.1083/jcb.201410061
Drosopoulos, W. C., Kosiyatrakul, S. T., Yan, Z., Calderano, S. G., and Schildkraut, C. L. (2012). Human telomeres replicate using chromosome-specific, rather than universal, replication programs. J. Cell Biol. 197 (2), 253–266. doi:10.1083/jcb.201112083
Feng, J., Funk, W. D., Wang, S. S., Weinrich, S. L., Avilion, A. A., Chiu, C. P., et al. (1995). The RNA component of human telomerase. Science 269 (5228), 1236–1241. doi:10.1126/science.7544491
Fouche, N., Ozgur, S., Roy, D., and Griffith, J. D. (2006). Replication fork regression in repetitive DNAs. Nucleic Acids Res. 34 (20), 6044–6050. doi:10.1093/nar/gkl757
Francia, S., Weiss, R. S., Hande, M. P., Freire, R., and di Fagagna, F. d. A. (2006). Telomere and telomerase modulation by the mammalian Rad9/Rad1/Hus1 DNA-damage-checkpoint complex. Curr. Biol. 16 (15), 1551–1558. doi:10.1016/j.cub.2006.06.066
Ge, X. Q., and Blow, J. J. (2010). Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J. Cell Biol. 191 (7), 1285–1297. doi:10.1083/jcb.201007074
Gong, Y., and de Lange, T. (2010). A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol. Cell 40 (3), 377–387. doi:10.1016/j.molcel.2010.10.016
Gottlieb, T. M., and Jackson, S. P. (1993). The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72 (1), 131–142. doi:10.1016/0092-8674(93)90057-w
Greider, C. W. (1991). Telomerase is processive. Mol. Cell. Biol. 11 (9), 4572–4580. doi:10.1128/mcb.11.9.4572
Greider, C. W. (2016). Regulating telomere length from the inside out: the replication fork model. Genes Dev. 30 (13), 1483–1491. doi:10.1101/gad.280578.116
Greider, C. W., and Blackburn, E. H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43 (2 Pt 1), 405–413. doi:10.1016/0092-8674(85)90170-9
Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., et al. (1999). Mammalian telomeres end in a large duplex loop. Cell 97 (4), 503–514. doi:10.1016/s0092-8674(00)80760-6
Grill, S., Tesmer, V. M., and Nandakumar, J. (2018). The N terminus of the OB domain of telomere protein TPP1 is critical for telomerase action. Cell Rep. 22 (5), 1132–1140. doi:10.1016/j.celrep.2018.01.012
Gu, P., Jia, S., Takasugi, T., Smith, E., Nandakumar, J., Hendrickson, E., et al. (2018). CTC1-STN1 coordinates G- and C-strand synthesis to regulate telomere length. Aging Cell 17, e12783. doi:10.1111/acel.12783
Gu, P., Jia, S., Takasugi, T., Tesmer, V. M., Nandakumar, J., Chen, Y., et al. (2021). Distinct functions of POT1 proteins contribute to the regulation of telomerase recruitment to telomeres. Nat. Commun. 12 (1), 5514. doi:10.1038/s41467-021-25799-7
Harley, C. B., Futcher, A. B., and Greider, C. W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345 (6274), 458–460. doi:10.1038/345458a0
Harman, A., Kartawinata, M., Maroun, N. M., Nguyen, D. R., Hughes, W. E., Winardi, K., et al. (2024). Nuclear actin and DNA replication stress regulate the recruitment of human telomerase to telomeres. bioRxiv. 2024.03. 25.586711. doi:10.1101/2024.03.25.586711
Harrison, J. C., and Haber, J. E. (2006). Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40, 209–235. doi:10.1146/annurev.genet.40.051206.105231
He, H., Multani, A. S., Cosme-Blanco, W., Tahara, H., Ma, J., Pathak, S., et al. (2006). POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 25, 5180–5190. doi:10.1038/sj.emboj.7601294
Higa, M., Fujita, M., and Yoshida, K. (2017a). DNA replication origins and fork progression at mammalian telomeres. Genes 8 (4), 112. doi:10.3390/genes8040112
Higa, M., Kushiyama, T., Kurashige, S., Kohmon, D., Enokitani, K., Iwahori, S., et al. (2017b). TRF2 recruits ORC through TRFH domain dimerization. Biochim. Biophys. Acta Mol. Cell Res. 1864 (1), 191–201. doi:10.1016/j.bbamcr.2016.11.004
Hirai, Y., Masutomi, K., and Ishikawa, F. (2012). Kinetics of DNA replication and telomerase reaction at a single-seeded telomere in human cells. Genes cells. 17, 186–204. doi:10.1111/j.1365-2443.2012.01581.x
Hockemeyer, D., Daniels, J. P., Takai, H., and de Lange, T. (2006). Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126 (1), 63–77. doi:10.1016/j.cell.2006.04.044
Hockemeyer, D., Palm, W., Else, T., Daniels, J. P., Takai, K. K., Ye, J. Z., et al. (2007). Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat. Struct. Mol. Biol. 14 (8), 754–761. doi:10.1038/nsmb1270
Houghtaling, B. R., Cuttonaro, L., Chang, W., and Smith, S. (2004). A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14 (18), 1621–1631. doi:10.1016/j.cub.2004.08.052
Ivessa, A. S., Zhou, J.-Q., Schulz, V. P., Monson, E. K., and Zakian, V. A. (2002). Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes and Dev. 16 (11), 1383–1396. doi:10.1101/gad.982902
Jady, B. E., Richard, P., Bertrand, E., and Kiss, T. (2006). Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell 17 (2), 944–954. doi:10.1091/mbc.e05-09-0904
Jay, K. A., Smith, D. L., and Blackburn, E. H. (2016). Early loss of telomerase action in yeast creates a dependence on the DNA damage response adaptor proteins. Mol. Cell Biol. 36 (14), 1908–1919. doi:10.1128/MCB.00943-15
Karlseder, J., Hoke, K., Mirzoeva, O. K., Bakkenist, C., Kastan, M. B., Petrini, J. H., et al. (2004). The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2 (8), E240. doi:10.1371/journal.pbio.0020240
Kim, S. H., Kaminker, P., and Campisi, J. (1999). TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23 (4), 405–412. doi:10.1038/70508
Kinzig, C. G., Zakusilo, G., Takai, K. K., Myler, L. R., and de Lange, T. (2024). ATR blocks telomerase from converting DNA breaks into telomeres. Science 383 (6684), 763–770. doi:10.1126/science.adg3224
Kusumoto-Matsuo, R., Opresko, P. L., Ramsden, D., Tahara, H., and Bohr, V. A. (2010). Cooperation of DNA-PKcs and WRN helicase in the maintenance of telomeric D-loops. Aging (Albany NY) 2 (5), 274–284. doi:10.18632/aging.100141
Lamm, N., Read, M. N., Nobis, M., Van Ly, D., Page, S. G., Masamsetti, V. P., et al. (2020). Nuclear F-actin counteracts nuclear deformation and promotes fork repair during replication stress. Nat. Cell Biol. 22 (12), 1460–1470. doi:10.1038/s41556-020-00605-6
Laprade, H., Querido, E., Smith, M. J., Guerit, D., Crimmins, H., Conomos, D., et al. (2020). Single-molecule imaging of telomerase RNA reveals a recruitment-retention model for telomere elongation. Mol. Cell 79 (1), 115–126. doi:10.1016/j.molcel.2020.05.005
Latrick, C. M., and Cech, T. R. (2010). POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 29 (5), 924–933. doi:10.1038/emboj.2009.409
Lee, J.-H., and Paull, T. T. (2005). ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308 (5721), 551–554. doi:10.1126/science.1108297
Lee, S. S., Bohrson, C., Pike, A. M., Wheelan, S. J., and Greider, C. W. (2015). ATM kinase is required for telomere elongation in mouse and human cells. Cell Rep. 13 (8), 1623–1632. doi:10.1016/j.celrep.2015.10.035
Li, B., Oestreich, S., and de Lange, T. (2000). Identification of human Rap1: implications for telomere evolution. Cell 101 (5), 471–483. doi:10.1016/s0092-8674(00)80858-2
Li, F., Kim, H., Ji, Z., Zhang, T., Chen, B., Ge, Y., et al. (2018). The BUB3-BUB1 complex promotes telomere DNA replication. Mol. Cell 70 (3), 395–407. doi:10.1016/j.molcel.2018.03.032
Lillard-Wetherell, K., Machwe, A., Langland, G. T., Combs, K. A., Behbehani, G. K., Schonberg, S. A., et al. (2004). Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 13 (17), 1919–1932. doi:10.1093/hmg/ddh193
Lingner, J., Cooper, J. P., and Cech, T. R. (1995). Telomerase and DNA end replication: no longer a lagging strand problem? Science 269 (5230), 1533–1534. doi:10.1126/science.7545310
Liu, D., Safari, A., O'Connor, M. S., Chan, D. W., Laegeler, A., Qin, J., et al. (2004). PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6 (7), 673–680. doi:10.1038/ncb1142
Loayza, D., and De Lange, T. (2003). POT1 as a terminal transducer of TRF1 telomere length control. Nature 423 (6943), 1013–1018. doi:10.1038/nature01688
Majka, J., Niedziela-Majka, A., and Burgers, P. M. (2006). The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol. Cell 24 (6), 891–901. doi:10.1016/j.molcel.2006.11.027
Makarov, V. L., Hirose, Y., and Langmore, J. P. (1997). Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88 (5), 657–666. doi:10.1016/s0092-8674(00)81908-x
Makovets, S., Herskowitz, I., and Blackburn, E. H. (2004). Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell Biol. 24 (9), 4019–4031. doi:10.1128/MCB.24.9.4019-4031.2004
Marcand, S., Brevet, V., Mann, C., and Gilson, E. (2000). Cell cycle restriction of telomere elongation. Curr. Biol. 10, 487–490. doi:10.1016/s0960-9822(00)00450-4
Marcand, S., Gilson, E., and Shore, D. (1997). A protein-counting mechanism for telomere length regulation in yeast. Science 275 (5302), 986–990. doi:10.1126/science.275.5302.986
Maresca, C., Dello Stritto, A., D’Angelo, C., Petti, E., Rizzo, A., Vertecchi, E., et al. (2023). PARP1 allows proper telomere replication through TRF1 poly (ADP-ribosyl) ation and helicase recruitment. Commun. Biol. 6 (1), 234. doi:10.1038/s42003-023-04596-6
Martinez, P., Thanasoula, M., Munoz, P., Liao, C., Tejera, A., McNees, C., et al. (2009). Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 23 (17), 2060–2075. doi:10.1101/gad.543509
McClintock, B. (1941). The stability of broken ends of chromosomes in zea mays. Genetics 26 (2), 234–282. doi:10.1093/genetics/26.2.234
McElligott, R., and Wellinger, R. J. (1997). The terminal DNA structure of mammalian chromosomes. EMBO J. 16 (12), 3705–3714. doi:10.1093/emboj/16.12.3705
McKerlie, M., Lin, S., and Zhu, X.-D. (2012). ATM regulates proteasome-dependent subnuclear localization of TRF1, which is important for telomere maintenance. Nucleic acids Res. 40 (9), 3975–3989. doi:10.1093/nar/gks035
McNees, C. J., Tejera, A. M., Martínez, P., Murga, M., Mulero, F., Fernandez-Capetillo, O., et al. (2010). ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J. Cell Biol. 188 (5), 639–652. doi:10.1083/jcb.200908136
Metcalfe, J. A., Parkhill, J., Campbell, L., Stacey, M., Biggs, P., Byrd, P. J., et al. (1996). Accelerated telomere shortening in ataxia telangiectasia. Nat. Genet. 13 (3), 350–353. doi:10.1038/ng0796-350
Miller, K. M., Rog, O., and Cooper, J. P. (2006). Semi-conservative DNA replication through telomeres requires Taz1. Nature 440 (7085), 824–828. doi:10.1038/nature04638
Mohaghegh, P., Karow, J. K., Brosh Jr, R. M., Bohr, V. A., and Hickson, I. D. (2001). The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic acids Res. 29 (13), 2843–2849. doi:10.1093/nar/29.13.2843
Mok, K.-W., Chen, H., Lee, W. M., and Cheng, C. Y. (2015). rpS6 regulates blood-testis barrier dynamics through Arp3-mediated actin microfilament organization in rat Sertoli cells. An in vitro study. Endocrinology 156 (5), 1900–1913. doi:10.1210/en.2014-1791
Morin, G. B. (1989). The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59 (3), 521–529. doi:10.1016/0092-8674(89)90035-4
Moser, B. A., Chang, Y. T., Kosti, J., and Nakamura, T. M. (2011). Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat. Struct. Mol. Biol. 18 (12), 1408–1413. doi:10.1038/nsmb.2187
Moser, B. A., Subramanian, L., Khair, L., Chang, Y. T., and Nakamura, T. M. (2009). Fission yeast Tel1(ATM) and Rad3(ATR) promote telomere protection and telomerase recruitment. PLoS Genet. 5 (8), e1000622. doi:10.1371/journal.pgen.1000622
Moyzis, R. K., Buckingham, J. M., Cram, L. S., Dani, M., Deaven, L. L., Jones, M. D., et al. (1988). A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. U. S. A. 85, 6622–6626. doi:10.1073/pnas.85.18.6622
Nakamura, T. M., Morin, G. B., Chapman, K. B., Weinrich, S. L., Andrews, W. H., Lingner, J., et al. (1997). Telomerase catalytic subunit homologs from fission yeast and human. Science 277 (5328), 955–959. doi:10.1126/science.277.5328.955
Nandakumar, J., Bell, C. F., Weidenfeld, I., Zaug, A. J., Leinwand, L. A., and Cech, T. R. (2012). The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492 (7428), 285–289. doi:10.1038/nature11648
Noel, J. F., and Wellinger, R. J. (2011). Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase. DNA Repair (Amst. ) 10, 271–282. doi:10.1016/j.dnarep.2010.11.010
Nowsheen, S., and Yang, E. (2012). The intersection between DNA damage response and cell death pathways. Exp. Oncol. 34 (3), 243–254.
Ohki, R., and Ishikawa, F. (2004). Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 32 (5), 1627–1637. doi:10.1093/nar/gkh309
Okamoto, K., Bartocci, C., Ouzounov, I., Diedrich, J. K., Yates, J. R., and Denchi, E. L. (2013). A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 494 (7438), 502–505. doi:10.1038/nature11873
Olson, C. L., and Wuttke, D. S. (2024). Guardians of the genome: how the single-stranded DNA-binding proteins RPA and CST facilitate telomere replication. Biomolecules 14 (3), 263. doi:10.3390/biom14030263
Opresko, P. L., Mason, P. A., Podell, E. R., Lei, M., Hickson, I. D., Cech, T. R., et al. (2005). POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem. 280 (37), 32069–32080. doi:10.1074/jbc.M505211200
Opresko, P. L., Otterlei, M., Graakjaer, J., Bruheim, P., Dawut, L., Kolvraa, S., et al. (2004). The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 14 (6), 763–774. doi:10.1016/j.molcel.2004.05.023
Opresko, P. L., von Kobbe, C., Laine, J. P., Harrigan, J., Hickson, I. D., and Bohr, V. A. (2002). Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277 (43), 41110–41119. doi:10.1074/jbc.M205396200
Padmanaban, S., Tesmer, V. M., and Nandakumar, J. (2023). Interaction hub critical for telomerase recruitment and primer-template handling for catalysis. Life Sci. Alliance 6 (6), e202201727. doi:10.26508/lsa.202201727
Palumbieri, M. D., Merigliano, C., González-Acosta, D., Kuster, D., Krietsch, J., Stoy, H., et al. (2023). Nuclear actin polymerization rapidly mediates replication fork remodeling upon stress by limiting PrimPol activity. Nat. Commun. 14 (1), 7819. doi:10.1038/s41467-023-43183-5
Pennarun, G., Hoffschir, F., Revaud, D., Granotier, C., Gauthier, L. R., Mailliet, P., et al. (2010). ATR contributes to telomere maintenance in human cells. Nucleic acids Res. 38 (9), 2955–2963. doi:10.1093/nar/gkp1248
Pinzaru, A. M., Hom, R. A., Beal, A., Phillips, A. F., Ni, E., Cardozo, T., et al. (2016). Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. 15 (10), 2170–2184. doi:10.1016/j.celrep.2016.05.008
Pinzaru, A. M., Kareh, M., Lamm, N., Lazzerini-Denchi, E., Cesare, A. J., and Sfeir, A. (2020). Replication stress conferred by POT1 dysfunction promotes telomere relocalization to the nuclear pore. Genes Dev. 34 (23-24), 1619–1636. doi:10.1101/gad.337287.120
Ramsay, A. J., Quesada, V., Foronda, M., Conde, L., Martinez-Trillos, A., Villamor, N., et al. (2013). POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 45 (5), 526–530. doi:10.1038/ng.2584
Revy, P., Kannengiesser, C., and Bertuch, A. A. (2023). Genetics of human telomere biology disorders. Nat. Rev. Genet. 24 (2), 86–108. doi:10.1038/s41576-022-00527-z
Roake, C. M., and Artandi, S. E. (2020). Regulation of human telomerase in homeostasis and disease. Nat. Rev. Mol. Cell Biol. 21 (7), 384–397. doi:10.1038/s41580-020-0234-z
Robles-Espinoza, C. D., Harland, M., Ramsay, A. J., Aoude, L. G., Quesada, V., Ding, Z., et al. (2014). POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 46, 478–481. doi:10.1038/ng.2947
Rodrı́guez-López, A. M., Jackson, D. A., Nehlin, J. O., Iborra, F., Warren, A. V., and Cox, L. S. (2003). Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA. Mech. ageing Dev. 124 (2), 167–174. doi:10.1016/s0047-6374(02)00131-8
Ryu, T., Spatola, B., Delabaere, L., Bowlin, K., Hopp, H., Kunitake, R., et al. (2015). Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 17 (11), 1401–1411. doi:10.1038/ncb3258
Saldivar, J. C., Cortez, D., and Cimprich, K. A. (2017). The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18 (10), 622–636. doi:10.1038/nrm.2017.67
Sarek, G., Vannier, J. B., Panier, S., Petrini, J. H. J., and Boulton, S. J. (2015). TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol. Cell 57 (4), 622–635. doi:10.1016/j.molcel.2014.12.024
Schmidt, J. C., Dalby, A. B., and Cech, T. R. (2014). Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife 3, e03563. doi:10.7554/eLife.03563
Sfeir, A., and de Lange, T. (2012). Removal of shelterin reveals the telomere end-protection problem. Science 336 (6081), 593–597. doi:10.1126/science.1218498
Sfeir, A., Kabir, S., van Overbeek, M., Celli, G. B., and de Lange, T. (2010). Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327 (5973), 1657–1661. doi:10.1126/science.1185100
Sfeir, A., Kosiyatrakul, S. T., Hockemeyer, D., MacRae, S. L., Karlseder, J., Schildkraut, C. L., et al. (2009). Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138 (1), 90–103. doi:10.1016/j.cell.2009.06.021
Shechter, D., Costanzo, V., and Gautier, J. (2004). Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 3 (8-9), 901–908. doi:10.1016/j.dnarep.2004.03.020
Shi, J., Yang, X. R., Ballew, B., Rotunno, M., Calista, D., Fargnoli, M. C., et al. (2014). Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 46, 482–486. doi:10.1038/ng.2941
Shiloh, Y., and Ziv, Y. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14 (4), 197–210. doi:10.1038/nrm3546
Smogorzewska, A., and de Lange, T. (2004). Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73, 177–208. doi:10.1146/annurev.biochem.73.071403.160049
Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M. R., Schnapp, G., et al. (2000). Control of human telomere length by TRF1 and TRF2. Mol. Cell Biol. 20 (5), 1659–1668. doi:10.1128/MCB.20.5.1659-1668.2000
Stansel, R. M., de Lange, T., and Griffith, J. D. (2001). T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. EMBO J. 20 (19), 5532–5540. doi:10.1093/emboj/20.19.5532
Stern, J. L., Zyner, K. G., Pickett, H. A., Cohen, S. B., and Bryan, T. M. (2012). Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol. Cell Biol. 32 (13), 2384–2395. doi:10.1128/MCB.00379-12
Stewart, J. A., Wang, F., Chaiken, M. F., Kasbek, C., Chastain, P. D., Wright, W. E., et al. (2012). Human CST promotes telomere duplex replication and general replication restart after fork stalling. Embo J. 31 (17), 3537–3549. doi:10.1038/emboj.2012.215
Sun, H., Karow, J. K., Hickson, I. D., and Maizels, N. (1998). The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273 (42), 27587–27592. doi:10.1074/jbc.273.42.27587
Takai, H., Aria, V., Borges, P., Yeeles, J. T. P., and de Lange, T. (2024). CST–polymerase α-primase solves a second telomere end-replication problem. Nature 627 (8004), 664–670. doi:10.1038/s41586-024-07137-1
Takai, K. K., Kibe, T., Donigian, J. R., Frescas, D., and de Lange, T. (2011). Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 44 (4), 647–659. doi:10.1016/j.molcel.2011.08.043
Takasugi, T., Gu, P., Liang, F., Staco, I., and Chang, S. (2023). Pot1b−/− tumors activate G-quadruplex-induced DNA damage to promote telomere hyper-elongation. Nucleic Acids Res. 51 (17), 9227–9247. doi:10.1093/nar/gkad648
Tatsumi, Y., Ezura, K., Yoshida, K., Yugawa, T., Narisawa-Saito, M., Kiyono, T., et al. (2008). Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes cells. 13 (10), 1045–1059. doi:10.1111/j.1365-2443.2008.01224.x
Tomlinson, R. L., Ziegler, T. D., Supakorndej, T., Terns, R. M., and Terns, M. P. (2006). Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17 (2), 955–965. doi:10.1091/mbc.e05-09-0903
Tong, A. S., Stern, J. L., Sfeir, A., Kartawinata, M., de Lange, T., Zhu, X. D., et al. (2015). ATM and ATR signaling regulate the recruitment of human telomerase to telomeres. Cell Rep. 13 (8), 1633–1646. doi:10.1016/j.celrep.2015.10.041
Van Ly, D., Low, R. R. J., Frolich, S., Bartolec, T. K., Kafer, G. R., Pickett, H. A., et al. (2018). Telomere loop dynamics in chromosome end protection. Mol. Cell 71 (4), 510–525. doi:10.1016/j.molcel.2018.06.025
Vannier, J.-B., Pavicic-Kaltenbrunner, V., Petalcorin, M. I., Ding, H., and Boulton, S. J. (2012). RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149 (4), 795–806. doi:10.1016/j.cell.2012.03.030
Vannier, J.-B., Sandhu, S., Petalcorin, M. I., Wu, X., Nabi, Z., Ding, H., et al. (2013). RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 342 (6155), 239–242. doi:10.1126/science.1241779
Verdun, R. E., Crabbe, L., Haggblom, C., and Karlseder, J. (2005). Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell 20 (4), 551–561. doi:10.1016/j.molcel.2005.09.024
Verdun, R. E., and Karlseder, J. (2006). The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127 (4), 709–720. doi:10.1016/j.cell.2006.09.034
Wang, F., Podell, E. R., Zaug, A. J., Yang, Y., Baciu, P., Cech, T. R., et al. (2007). The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445 (7127), 506–510. doi:10.1038/nature05454
Wu, L., Multani, A. S., He, H., Cosme-Blanco, W., Deng, Y., Deng, J. M., et al. (2006). Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126, 49–62. doi:10.1016/j.cell.2006.05.037
Wu, R. A., Upton, H. E., Vogan, J. M., and Collins, K. (2017). Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 86 (1), 439–460. doi:10.1146/annurev-biochem-061516-045019
Wu, Y., Xiao, S., and Zhu, X. D. (2007). MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 14 (9), 832–840. doi:10.1038/nsmb1286
Xie, Z., Jay, K. A., Smith, D. L., Zhang, Y., Liu, Z., Zheng, J., et al. (2015). Early telomerase inactivation accelerates aging independently of telomere length. Cell 160 (5), 928–939. doi:10.1016/j.cell.2015.02.002
Yamazaki, H., Tarumoto, Y., and Ishikawa, F. (2012). Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 26 (3), 241–246. doi:10.1101/gad.177873.111
Yang, J., Xu, Z. P., Huang, Y., Hamrick, H. E., Duerksen-Hughes, P. J., and Yu, Y. N. (2004). ATM and ATR: sensing DNA damage. World J. Gastroenterol. 10 (2), 155–160. doi:10.3748/wjg.v10.i2.155
Yang, Z., Sharma, K., and de Lange, T. (2022). TRF1 uses a noncanonical function of TFIIH to promote telomere replication. Genes Dev. 36 (17-18), 956–969. doi:10.1101/gad.349975.122
Ye, J., Lenain, C., Bauwens, S., Rizzo, A., Saint-Leger, A., Poulet, A., et al. (2010). TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142 (2), 230–242. doi:10.1016/j.cell.2010.05.032
Ye, J. Z., and de Lange, T. (2004). TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 36 (6), 618–623. doi:10.1038/ng1360
Ye, J. Z., Donigian, J. R., van Overbeek, M., Loayza, D., Luo, Y., Krutchinsky, A. N., et al. (2004a). TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279 (45), 47264–47271. doi:10.1074/jbc.M409047200
Ye, J. Z., Hockemeyer, D., Krutchinsky, A. N., Loayza, D., Hooper, S. M., Chait, B. T., et al. (2004b). POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18 (14), 1649–1654. doi:10.1101/gad.1215404
Youds, J. L., Mets, D. G., McIlwraith, M. J., Martin, J. S., Ward, J. D., Nj, O. N., et al. (2010). RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327 (5970), 1254–1258. doi:10.1126/science.1183112
Zaug, A. J., Lim, C. J., Olson, C. L., Carilli, M. T., Goodrich, K. J., Wuttke, D. S., et al. (2021). CST does not evict elongating telomerase but prevents initiation by ssDNA binding. Nucleic Acids Res. 49 (20), 11653–11665. doi:10.1093/nar/gkab942
Zhang, T., Zhang, Z., Li, F., Hu, Q., Liu, H., Tang, M., et al. (2017). Looping-out mechanism for resolution of replicative stress at telomeres. EMBO Rep. 18 (8), 1412–1428. doi:10.15252/embr.201643866
Zhao, Y., Hoshiyama, H., Shay, J. W., and Wright, W. E. (2008). Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 36, e14. doi:10.1093/nar/gkm1063
Zhao, Y., Sfeir, A. J., Zou, Y., Buseman, C. M., Chow, T. T., Shay, J. W., et al. (2009). Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138 (3), 463–475. doi:10.1016/j.cell.2009.05.026
Zhong, F. L., Batista, L. F., Freund, A., Pech, M. F., Venteicher, A. S., and Artandi, S. E. (2012). TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150 (3), 481–494. doi:10.1016/j.cell.2012.07.012
Zimmermann, M., Kibe, T., Kabir, S., and de Lange, T. (2014). TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 28 (22), 2477–2491. doi:10.1101/gad.251611.114
Keywords: telomere maintenance, telomerase, shelterin, DNA damage response, replication stress, telomere replication, nuclear actin
Citation: Harman A and Bryan TM (2024) Telomere maintenance and the DNA damage response: a paradoxical alliance. Front. Cell Dev. Biol. 12:1472906. doi: 10.3389/fcell.2024.1472906
Received: 30 July 2024; Accepted: 07 October 2024;
Published: 17 October 2024.
Edited by:
Elaine Sanij, Peter MacCallum Cancer Centre, AustraliaReviewed by:
Kazunori Tomita, Brunel University London, United KingdomRanjodh Singh Sandhu, National Institutes of Health (NIH), United States
Copyright © 2024 Harman and Bryan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tracy M. Bryan, dGJyeWFuQGNtcmkub3JnLmF1
 Ashley Harman
Ashley Harman Tracy M. Bryan
Tracy M. Bryan