- 1Department of Biotechnology, College of Applied Sciences, University of Technology, Baghdad, Iraq
- 2Department of Medical Laboratory Sciences, College of Applied Medical Sciences, University of Bisha, Bisha, Saudi Arabia
- 3Life Sciences Institute, University of Michigan, Ann Arbor, MI, United States
The 5-year survival rate for hepatocellular carcinoma (HCC), a deadly form of liver cancer, is quite low. Although drug therapy is successful, patients with advanced liver cancer frequently develop resistance because of the significant phenotypic and genetic heterogeneity of these cells. The overexpression of drug efflux transporters, downstream adaptive responses, malfunctioning DNA damage repair, epigenetic modification, the tumor microenvironment, and the extracellular matrix can all be linked to drug resistance. The evolutionary process of autophagy, which is in charge of intracellular breakdown, is intimately linked to medication resistance in HCC. Autophagy is involved in both the promotion and suppression of cancer by influencing treatment resistance, metastasis, carcinogenesis, and the viability of stem cells. Certain autophagy regulators are employed in anticancer treatment; however, because of the dual functions of autophagy, their use is restricted, and therapeutic failure is increased. By focusing on autophagy, it is possible to reduce HCC expansion and metastasis, and enhance tumor cell reactivity to treatment. Macroautophagy, the best-characterized type of autophagy, involves the formation of a sequestering compartment termed a phagophore, which surrounds and encloses aberrant or superfluous components. The phagophore matures into a double-membrane autophagosome that delivers the cargo to the lysosome; lysosomes and autophagosomes fuse to degrade and recycle the cargo. Macroautophagy plays dual functions in both promoting and suppressing cancer in a variety of cancer types.
Introduction
The predominant liver cancer subtype, which ranks second in terms of approximate mortality after pancreatic cancer, is hepatocellular carcinoma (HCC) (Siegel et al., 2020). The survival rate after 5 years for HCC is 18%. Treatments for HCC include surgical resection, liver transplantation, chemotherapy, radiofrequency ablation, targeted therapy, transarterial chemoembolization, and immunotherapy (Villanueva, 2019). Nevertheless, the majority of individuals with advanced or intermediate stages of HCC do not effectively react to anticancer medications, and only a small percentage of individuals are susceptible to these treatments. In HCC patients where medication resistance has already developed, the total survival rate is decreased (Llovet et al., 2008). To save HCC patients, it is crucial to understand HCC’s mechanism of drug resistance, pinpoint the targets of drug resistance, and optimize the therapy regimen.
HCC is a naturally drug-resistant tumor. Drug resistance is caused by the transmembrane ATP-binding cassette (ABC) transporter protein, which prevents anticancer medications from penetrating cells. Drugs are delivered to cancer cells by inhibition of the solute carrier (SLC) family, which also leads to multidrug resistance (MDR) development in HCC cells (Lei et al., 2024), chemotherapeutic medications typically have little effect on HCC patients. During treatment, the associated tumors are likely to develop MDR, which lowers survival and worsens the prognosis (Wang et al., 2014; Koltai, 2022). The restricted accumulation of anticancer medicines within tumors is one of the factors behind resistance to these medications. In this sense, pharmaceuticals are transported across the cellular membrane via membrane transporters, which are primarily responsible for the influx (mostly by members of the SLC transporter family) and efflux (primarily by members of the ABC transporter family). Thus, transporters control the concentrations of drug at the target site in both healthy tissues and cancer cells, which has an impact on therapeutic results. One of the main challenges in anticancer drug delivery and a major factor in the failure of cancer medication therapy is transporter-mediated drug resistance. Because SLC transporters are essential for the drug’s intracellular absorption drug therapy may be rendered ineffective if cancer cells exhibit downregulated expression and function of SLC transporters, which restricts the absorption of medications into the tumor cells. The phenomenon known as multidrug resistance occurs when a high percentage of patients become non-responsive to numerous functionally and structurally varied anticancer medications as a result of developing drug resistance to chemotherapy and molecularly targeted therapies (Puris et al., 2023). Exogenous and intrinsic factors are the two main causes of medication failure in cancer patients. For example, the result of genetic changes that were already present in tumor cells prior to the initiation of therapy is considered intrinsic resistance, sometimes referred to as primary resistance. Included in this are treatment resistance linked to cancer stem cells/CSCs (Chan et al., 2015), as well as elevated expression of drug efflux transporters (Alisi et al., 2013), which bind and eliminate chemotherapeutic agents. Exogenous resistance, sometimes referred to as developed resistance, occurs when cancer cells that are originally responsive to drugs later become resistant to them following a course of chemotherapy (Chan et al., 2015).
Autophagy is a system that has been maintained over evolution. It plays a role in the turnover of organelles and proteins, as well as in the regulation of metabolism and cell quality control (Kumariya et al., 2021; Raudenska et al., 2021). Lysosomes are necessary for the process of autophagy, which has as its primary objectives maintaining cellular quality control and the production of energy and nutrition through the breakdown and release of cytoplasmic substituents. To maintain cellular health and balance, the autophagy-lysosomal system—a combination of autophagy and the lysosomal system—eradicates waste products from cells, including protein aggregates, damaged organelles, and invasive microbes. Consequently, dysfunction of the autophagy-lysosomal system contributes to several pathophysiological states, including neurological disorders, cancer, inflammatory and immunological diseases, and metabolic abnormalities (Kumar et al., 2021).
Additionally, autophagy is regulated to protect cells from various types of stress. Including malnutrition, hypoxia, DNA damage, chemotherapeutic exposure, and meeting cells’ metabolic demands to keep organelles and cellular signaling pathways intact (Glick et al., 2010; Parzych and Klionsky, 2014). The human body uses autophagy frequently in response to different stimuli (Huang F. et al., 2018). The primary routes of autophagy can be broadly categorized into five stages: initiation, expansion, closure, maturation, and degradation; the final step involves the release of breakdown products back into the cytoplasm (see below under Autophagy basic mechanism). Cellular homeostasis is maintained through autophagy by the elimination of improperly folded and aggregated proteins, and damaged organelles (He and Klionsky, 2009). Autophagy also plays a crucial role in the immune response, for example, by regulating the activation of inflammasomes by primary macrophages generated from bone marrow (Jabir et al., 2014; Jabir et al., 2021).
Hepatocellular carcinoma
In terms of mortality, hepatocellular carcinoma, a type of primary liver cancer, is ranked second and according to the frequency of occurrence is the seventh globally (Bray et al., 2018; Katherine et al., 2021). HCC is the most prevalent kind of liver cancer, comprising up to 75% of cases. The top rates of liver cancer occurrence are discovered in Asia and Africa (Zhang et al., 2023). Up to a half million new cases of HCC are identified each year, contributing to a high mortality rate (Tsochatzis et al., 2014; Petrick et al., 2020). Five percent of patients with HCC survive longer than 5 years after the initial diagnosis, which is a relatively poor survival rate. This poor prognosis is connected to the fact that only 15% of these patients are eligible for or capable of undergoing surgery and liver transplantation due to the late identification of HCC. Thirty-five percent or more of HCC patients receive the greatest care at the time of diagnosis, and fifty percent have non-surgical therapy (El-Serag, 2011; Yun et al., 2020). In hepatotoxicity, usually associated with different liver injuries (Hashemi et al., 2023), the oxidative process, which is thought to be the primary factor causing a shift toward inflammation, fibrosis, and hepatocellular damage, is triggered by free radical damage and many other factors, which lead to HCC (Mohamed et al., 2020a; Mohamed et al., 2020b). Race, gender and age are factors that affect the chance of getting HCC. Men are two to four times more likely than women to develop HCC. Up to 75 years of age, HCC exhibits a positive relationship with age (Petrick et al., 2020). Hepatitis virus infection, alcohol consumption, nonalcoholic fatty liver disease and cirrhosis are among the risk factors for this disease (Mohamed et al., 2022).
HCC progression is influenced by various molecular pathways and processes (Deldar Abad Paskeh et al., 2021; Yu et al., 2021). For example, FIS1 (fission, mitochondrial 1) provides one example of how the cellular machinery for mitochondrial fission intersects with HCC. This protein was thought to have a critical role in mitochondrial fission; however, mammalian cells lacking FIS1 do not exhibit an obvious defect in this process (Yamano et al., 2014; Wong S. W. et al., 2018). Nonetheless, FIS1 phosphorylation via MET (MET proto-oncogene, receptor tyrosine kinase) at Y38 facilitates mitochondrial fission by recruiting the mitochondrial fission GTPase DNM1L/Drp1 (dynamin 1 like) to mitochondria (Yu et al., 2021; Zhang et al., 2021); however, it has not been established that FIS1, an outer mitochondrial membrane protein, is a DNM1L receptor (Otera et al., 2010). HCC cells can metastasize both in vitro and in vivo by the development of lamellipodia or invadopodia, which is facilitated by fragmented mitochondria that drive actin filament remodeling. By activating MET kinase, which directly affects mitochondrial fission as described above, HGF (hepatocyte growth factor) plays a crucial role in the migration and invasion of cancer cells in the HCC microenvironment. Based on this, MET-targeted inhibitors have been used in HCC clinical trials, and FIS1 has been identified as a novel, significant downstream target that is regulated by MET kinase (Deldar Abad Paskeh et al., 2021).
The inflammatory state of the HCC microenvironment makes HCC immunotherapy modalities such as immune checkpoint inhibitors, anti-CTLA4 (cytotoxic T-lymphocyte associated protein 4), and anti-PDCD1/PD-1 (programmed cell death 1) potentially helpful. These alterations may result in changes in the number of regulatory T cells, the induction of dendritic cells, and the release of immune-modulating factors (Shokoohian et al., 2021; Minaei et al., 2023). However, the effectiveness of chemotherapy and radiation therapy in extending the life expectancy of HCC patients is hindered by the formation of several resistance mechanisms during treatment (Niu et al., 2020; Xia et al., 2021; Seydi et al., 2023).
The plasma membrane protein CD44 (CD44 molecule (IN blood group)), which functions as a cell-cell and cell-stromal adhesion protein and is frequently overexpressed in tumor cells, is a marker of poor survival in the majority of solid tumors (Wan et al., 2014). CD44 is linked to tumor malignancy and denotes a poor prognosis for a number of cancers, including liver cancer (Dhar et al., 2018). One of the main factors contributing to a worse prognosis is extrahepatic metastasis (EHM), which can happen when a patient has advanced HCC at the time of presentation or at the time of recurrence (Becker et al., 2014; Yoon et al., 2020). One major obstacle to increasing HCC patients’ overall survival (OS) is the existence of EHM. For individuals with HCC, CD44 is a strong predictor of both EHM and OS. In HCC cells and patient specimens with a high risk for malignancy, CD44 is abundantly expressed. Patients with HCC who overexpress CD44 have a lower OS and a greater cumulative recurrence rate compared to those with low levels of CD44 expression. Experiments conducted in vitro and in vivo demonstrate that CD44 downregulation inhibits the formation, migration, invasion, growth, and metastasis of HCC cells. Additionally, the pro-metastasis effect of CD44 is mediated through the MAPK/ERK (mitogen-activated protein kinase)-AKT/protein kinase B (AKT serine/threonine kinase)-CXCR4 (C-X-C chemokine receptor 4) axis. The aggressive clinical characteristics of HCCs are compatible with CD44’s documented ability to stimulate CXCR4 expression and increase the tendency of tumors to infiltrate and metastasize to distant organs (Xie et al., 2022). Finally, in HCC, the overexpression of CD44 facilitates the growth and migration of HCC cells through the action of oncogenic YAP1 (Yes1 associated transcriptional regulator), a crucial downstream regulator in the Hippo pathway. These results imply that CD44-YAP1 is most likely a significant axis in the pathophysiology of HCC, offering insights into the etiology of HCC and possible targets for HCC treatments (Yu et al., 2021). Obesity, PIK3CG/PI3Kγ (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma) ablation, ZRANB1 (zinc finger RANBP2-type containing 1), and ADORA2A-AS1 (ADORA2A antisense RNA 1) all contribute to HCC development. There are genetic factors that contribute to the development of HCC. PIK3CG/PI3Kγ, a type of phosphatidylinositol 3-kinase, is often overexpressed in HCC and promotes tumorigenesis. ZRANB1, a zinc finger protein, is downregulated in several types of cancer, including HCC. ADORA2A-AS1, an antisense RNA, is an oncogenic factor that regulates gene expression and is associated with tumorigenesis. Abnormalities in these genes may play a crucial role in the development of HCC. To fully understand their roles, further research is needed (Becattini et al., 2021; Zhang et al., 2021).
Autophagy basic mechanism
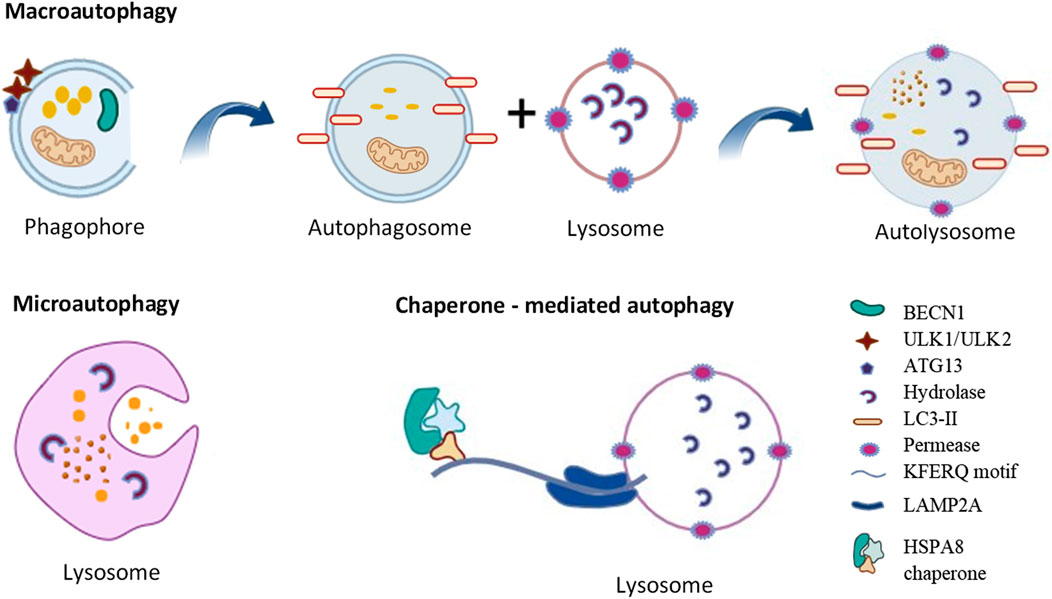
Autophagy is a self-degradation and internal recycling mechanism that is highly evolutionarily conserved, carrying out metabolic requirements and upholding homeostasis (Li Q. et al., 2021). As a homeostatic mechanism, autophagy facilitates the proteolytic breakdown of large cytosolic cellular constituents and aggregates in lysosomes, particularly those that are not susceptible to ubiquitin-proteasome pathway-mediated degradation (Pu et al., 2021). Different kinds of cellular stressors, such as cellular injury, the synthesis of defective proteins, and hunger, and the presence of excess or damaged organelles, pathogens and some microorganisms can trigger the autophagic process (White et al., 2015). Chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy are the three primary forms of autophagy (Figure 1). Microautophagy is a particular kind of direct autophagy in which cargo is captured and cytosolic components are invaginated directly at the lysosomal membrane (Jabir et al., 2018). Lysosomal membrane receptors, that include LAMP2A (lysosomal associated membrane protein 2A), detect and translocate cargo in the form of individual proteins that are complexed with chaperone proteins (such as HSPA8/HSC70 [heat shock protein family A (Hsp70) member 8]); these substrates are unfolded as they cross the membrane into the lysosomal lumen during CMA, a type of selective autophagy that involves the recognition of a KFERQ amino acid motif within the cargo (Yun and Lee, 2018).
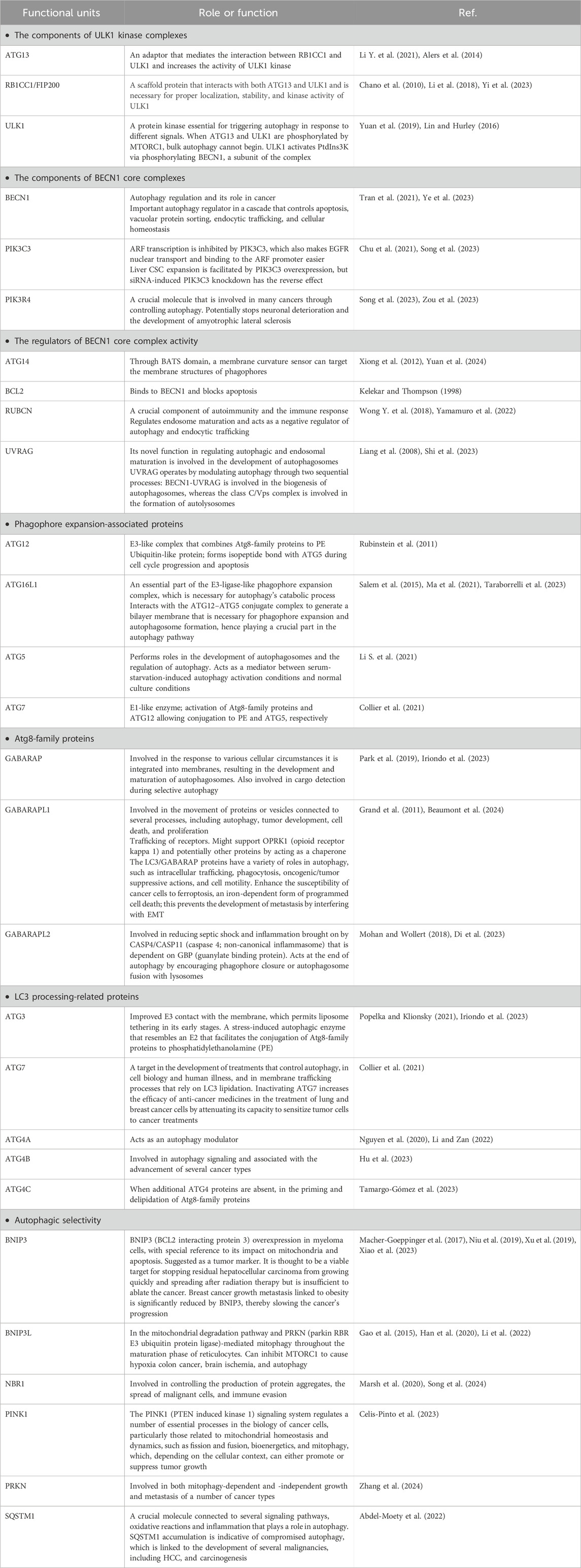
The best characterized of these autophagic processes is macroautophagy (referred to as autophagy hereafter). Once cytoplasmic cargo is isolated and incorporated into phagophores, the latter mature into double-membrane autophagosomes. Phagophore nucleation, which is the first step in autophagy, is brought on by the stimulation of the ULK1 (unc-51 like autophagy activating kinase 1) kinase complex, which includes ATG13 (autophagy related 13), ATG101 and RB1CC1/FIP200 (RB1 inducible coiled-coil 1). Expansion of the phagophore involves additional ATG (autophagy related) proteins including the ubiquitin-like proteins ATG12 and yeast Atg8 homologs (composed of the MAP1LC3/LC3 [microtubule associated protein 1 light chain 3] and GABARAP [GABA type A receptor-associated protein] subfamilies) and associated components that are required for their conjugation. The ATG2 (autophagy related 2)-ATG9 (autophagy related 9) complex also plays a critical role in delivering lipids for membrane expansion. Upon completion of sequestration, autophagosomes form. The autophagosomes fuse with lysosomes to create autolysosomes, which perform the breakdown and recycling of the cargo (Yorimitsu and Klionsky, 2005). It has been proposed that ATG2, a rod-shaped protein, is necessary for phagophore expansion because it tethers phosphatidylinositol-3-phosphate-enriched phagophores to the endoplasmic reticulum (ER) (Maeda et al., 2019). Because ATG2 mediates lipid transfer and re-equilibrates membranes in conjunction with ATG9 to facilitate autophagosome formation, it plays a crucial function in autophagy (Wang et al., 2024).
Small vesicles containing the membrane protein ATG9 seed the creation of the phagophore (Maeda et al., 2020; Guardia et al., 2020). ATG9A scramblase moves phospholipids delivered by ATG2 across both sides of the phagophore membrane. Furthermore, it appears from molecular dynamics simulation that lipids can flip across the bilayers because a central pore in Atg9 widens laterally to make room for lipid headgroups. Based on cryo-EM, fungal Atg9 forms a homotrimer containing two linked pores that create a pathway between the two membrane leaflets: one pore at the protomer opens laterally to the cytoplasmic leaflet, while the second pore at the trimer center travels vertically through the membrane (Matoba et al., 2020). At the growing margin of the phagophore, where Atg2 obtains phospholipids from the endoplasmic reticulum, Atg2 and Atg9 colocalize. Lipid scrambling by ATG9A is necessary for membrane growth, as evidenced by the fact that mutations in the pore decrease scrambling activity and result in noticeably smaller autophagosomes.
Although autophagy is primarily a cytoprotective mechanism, cell death may eventually ensue from an overabundance of autophagy and cellular self-consumption brought on by cellular damage—autophagy is considered as one type of programmed cell death (Oku and Sakai, 2018).
A number of autophagy-related proteins, including BECN1 (beclin 1), MAP1LC3/LC3 (microtubule associated protein 1 light chain 3), and ULK1/ULK2 (Table 1) play a critical role in this process. The development of the autophagosome requires ATG7, the ATG12–ATG5 conjugate, Atg8-family proteins such as LC3, and SQSTM1/p62 (sequestosome 1). RABs, SNAREs, and tethers work together to cause lysosomes to fuse with autophagosomes so they can break down cargo and return nutrients to the cytoplasm (Huang F. et al., 2018). Autophagy breaks down the multifunctional protein SQSTM1, which is important in cell development, survival, and death. Tumor development has been linked to SQSTM1 gene amplification as well as abnormal SQSTM1 accumulation and phosphorylation. By activating the transcription factor NFE2L2/Nrf2 (NFE2 like bZIP transcription factor 2), phosphorylation of SQSTM1 at Ser349 routes glutamine into glutathione production and glucose toward the glucuronate pathway. These alterations confer proliferative potency and resistance to anti-cancer medications on HCC cells. An inhibitor of phosphorylated SQSTM1-dependent NFE2L2 activation blocks HCC growth and resistance to anticancer drugs. In individuals with HCV-positive HCC, an NFE2L2 inhibitor may be able to reduce cancer cell resistance to anticancer medications (Saito et al., 2016).
Linkage of the KEAP1 (kelch like ECH associated protein 1)-NFE2L2 system to autophagy is implied by the assembly of SQSTM1 on specific autophagic cargos, such as ubiquitinated organelles, and its subsequent phosphorylation in an MTORC1-dependent manner. The development of HCCs is facilitated by the continuous activation of NFE2L2 caused by the buildup of phosphorylated SQSTM1. Therefore, inhibitors of the connection between KEAP1 and phosphorylated SQSTM1 show potential as therapeutic drugs against human HCC. The KEAP1-NFE2L2 pathway and selective autophagy are interdependent (Ichimura et al., 2013; Inami et al., 2011).
The role of autophagy in the development of tumors
Autophagy suppresses tumorigenesis by preserving physiological homeostasis and preventing cell conversion to malignancy in part by lowering the quantity of damaged mitochondria; the elimination of the latter helps prevent the formation of reactive oxygen species that can further cause damage of cellular components including DNA (Nazio et al., 2019; Jawad et al., 2023). Accordingly, inhibition of autophagy promotes oncogenesis and malignant transformation, and a high carcinogenic incidence (Takahashi et al., 2007; Jabir et al., 2015; Yun et al., 2020). BECN1 has been linked to the start of autophagy as well as a number of other cellular functions, including cell death, development, aging, and stress adaptation. Furthermore, BECN1 controls autophagic activity via interacting with other ATG proteins, the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, and TP53/p53 (tumor protein p53), among other autophagy mediators, which in turn affects the initiation and progression of cancer (Yun et al., 2020). A reduction in BECN1 expression leads to both cancer growth and tumorigenesis (Vara-Perez et al., 2019), whereas BECN1 regulates autophagic activity to prevent the growth of tumors. BECN1 promotes autophagy and inhibits the growth of malignancies that are mediated by ERBB2/HER2 (erb-b2 receptor tyrosine kinase 2). Mitophagy is a crucial type of selective autophagy of mitochondria, removing damaged or abnormal organelles to maintain mitochondrial homeostasis (Chang et al., 2017). Defects in mitophagy can lead to mitochondrial damage, tumorigenesis, and tumor growth in different types of cancer (Vara-Perez et al., 2019). Mitophagy function varies depending on tumor stage, with mutations or functional changes causing an accumulation of impaired mitochondria and tumorigenesis (Yun et al., 2020). Mitophagy inhibits cancer during the early stages of carcinogenesis while maintaining the metabolic needs of healthy cells. Conversely, mitophagy promotes cell tolerance and accelerates tumor growth during the later stages of tumor development (Wang et al., 2020).
Autophagy’s dual role in cancer
As discussed above, autophagy breaks down and recycles long-lived, misfolded or damaged proteins, and aberrant or damaged organelles to preserve cellular homeostasis (Huang T. et al., 2018; Onorati et al., 2018). Moreover, autophagy regulation protects against various types of cellular stressors, including starvation, hypoxia, DNA damage, and chemotherapeutic exposure, as well as fulfilling the metabolic needs of cells to maintain the functionality of organelles and cellular communication channels. As an integral process of cell physiology, both the maintenance of health and the development of diseases can be linked to autophagy or defects in the autophagy pathway. Numerous illnesses, such as cardio-related diseases (Zhang et al., 2023), neurological disease (Chu, 2019), gastrointestinal diseases (Zhang et al., 2023), lung diseases (Yun et al., 2020), cancer (Raudenska et al., 2021), and type II diabetes (Hashemi et al., 2023), are linked to abnormal autophagy. Both the initiation and spread of malignant tumors are influenced by tumor-suppressive autophagy. In contrast, the removal of aberrant cells and organelles, as well as the limitation of cell division and genetic instability in cancer, due to normal autophagy produce tumor inhibitory effects (Dong et al., 2019).
Numerous studies have suggested that autophagy has a dual role in the onset and progression of cancer (Li et al., 2020). However, there is an ongoing debate as to whether autophagy functions primarily as a pro- or anti-tumor mechanism (Zhang et al., 2023). Autophagy is involved in the quality control of proteins and organelles during the early stages of tumorigenesis (Barnard et al., 2016). It does this by preserving genomic stability, guarding against tissue damage over time, and preventing the accumulation of oncogenic proteins linked to inflammation. These actions prevent the initiation, proliferation, invasion, and metastasis of tumors (Guo et al., 2013). Research has shown that artificially limiting autophagy (for example, by atg5 deletion in mice) enhances the early development of liver cancers, suggesting that tumor suppression is a key function of autophagy in hepatocytes (Takamura et al., 2011).
In contrast, when a tumor reaches an advanced stage, autophagy becomes a shield for the tumor cells, protecting against DNA damage and increasing the survival of cancer cells by causing resistance to drugs (Wu et al., 2012; Zhang et al., 2023). Thus, autophagy promotes cancer cells by triggering chemoresistance and satisfying the growing metabolic requirements of cancer cells (Anderson and Macleod, 2019; Xiong et al., 2019). According to Liu et al. (2018), autophagy increases the expression of the transcription factor NANOG (Nanog homeobox) and suppresses TP53, encouraging hepatocarcinogenesis in benign liver tumors in a process involving hepatoma stem cells (Liu et al., 2018). The dual roles of autophagy in cancer, both supporting and suppressing tumor growth, are illustrated in Figure 2 (Yun et al., 2020).
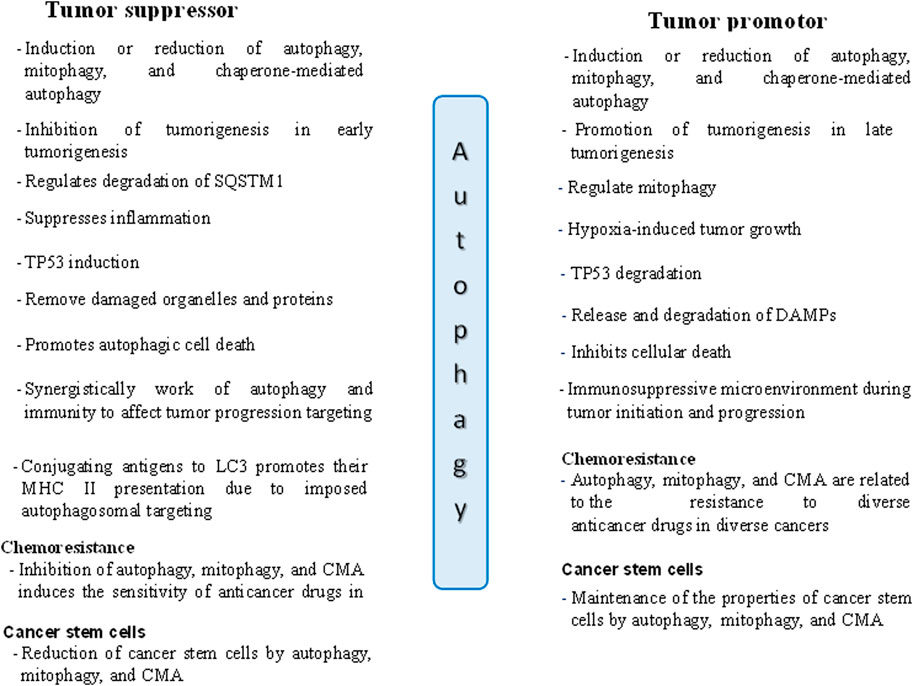
Figure 2. The dual roles of autophagy in tumor suppression and promotion in cancer cells. DAMPs: Danger/damage associated molecular patterns.
Roles of autophagy in HCC
In HCC, autophagy has a conflicting function, both in preventing early-stage carcinogenesis and in accelerating the growth of tumors in later stages (Ni et al., 2014). This dual role demonstrates how difficult it is to target autophagy in the treatment of HCC. Autophagy and the control of the development and progression of HCC are mediated by autophagy-associated genes, non-coding RNAs, and associated signaling pathways (Figure 3) (Yang et al., 2019). Autophagy has two roles in the onset and progression of hepatocellular cancer and many factors trigger the activation of hepatic autophagy. First, autophagy can function as a tumor suppressor during the initiation stage of hepatoneogenesis by reducing inflammation, SQSTM1 accumulation, the oxidative stress response, and ultimately genomic instability and through autophagic cell death. Second, however, at other stages of hepatoneogenesis, autophagy can promote cancer through its cytoprotective functions (Lee and Jang, 2015). Autophagy’s precise role in HCC is debatable and remains incompletely understood. Extensive investigation is necessary to comprehend the function of autophagy in the progression of HCC.
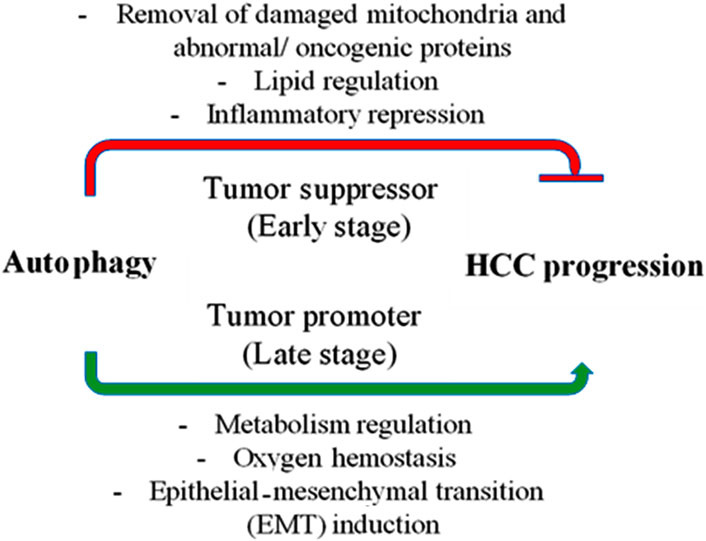
Figure 3. Autophagy’s role in hepatocellular carcinoma (HCC). In the early stages of cancer, autophagy suppresses tumors by removing damaged mitochondria and abnormal proteins, controlling hepatic lipid metabolism, and reducing inflammation (represented by the red line). However, once cancer has taken hold, autophagy becomes a tumor promoter by controlling metabolism and preserving the oxygen equilibrium, which helps cancer cells survive. Additionally, autophagy promotes the progression of the disease by inducing the epithelial-mesenchymal transition (EMT).
Autophagy and proteins associated with autophagy in HCC
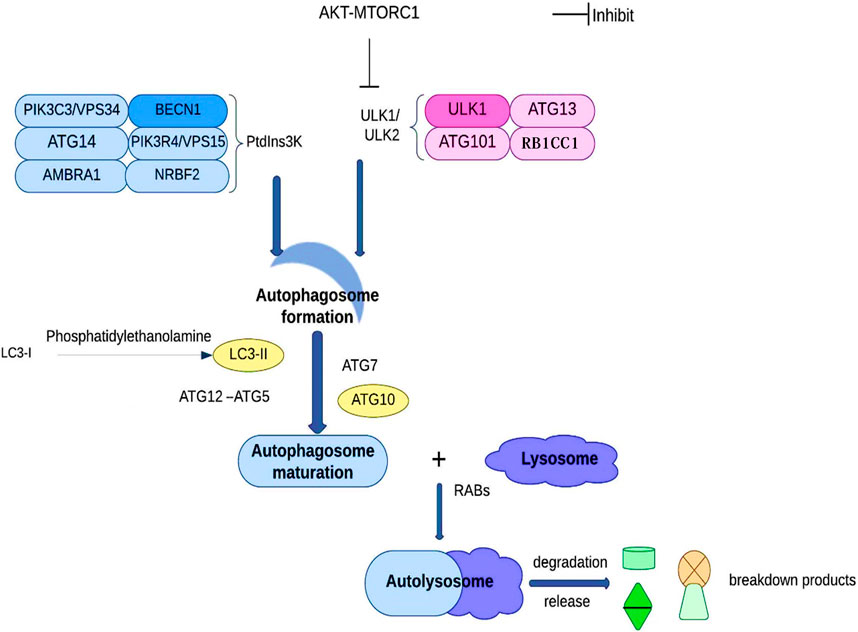
In order to maintain the equilibrium of cell component production and breakdown, autophagy involves the following steps: phagophore formation, autophagosome maturation, autolysosome formation, and cargo degradation (Levy et al., 2017). Autophagy contributes to the preservation of the nitrogen balance and the equilibrium of the cell environment by using the lysosomal route to break down macromolecules when the cell is malnourished (Kim and Lee, 2014; Wu et al., 2021). Based on the transport pathways and on their capacity to break down specific cargo, they can be categorized as reticulophagy (endoplasmic reticulum), pexophagy (peroxisomes), ribophagy (ribosomes), xenophagy (invasive microbes), mitophagy (mitochondria), etc. (Chu, 2019; Kirkin, 2020). Cargo sequestration, autophagosome development and autolysosome-dependent degradation are the three main components of autophagy. ATG101, ATG13, RB1CC1, and ULK1/ULK2 make up the ULK1/ULK2 complex during the early stages of autophagy development. The PtdIns3K complex, which has the lipid kinase PIK3C3/VPS34 at its core, plays a critical role by synthesizing phosphatidylinositol-3-phosphate on the phagophore, allowing the recruitment of other proteins leading to the autophagosome’s development. Subsequently, LC3-I is converted into LC3-II through conjugation to phosphatidylethanolamine, along with ATG12–ATG5 conjugation, and the action of specific receptors such as SQSTM1 to promote autophagosome maturation. Ultimately, the autophagosome fuses with a lysosome under the direction of RAB proteins to carry out the cargo breakdown and release (Figure 4) (Wu et al., 2021).
Autophagy and HCC drug resistance
Chemoresistance in HCC has proven to be a difficult problem in recent years, but it can be overcome by stimulating oxidative stress and inhibiting mitochondrial respiration. In addition, the interplay between different factors can influence autophagy levels, affecting cancer development and progression. For example, HCC is associated with ICMT (isoprenylcysteine carboxyl methyltransferase), NFE2L2, and USP7 (ubiquitin specific peptidase 7) via a number of pathways, including control over cell survival, proliferation, and metabolism. In HCC, overexpression of ICMT results in doxorubicin resistance and inhibits apoptosis (Kiruthiga et al., 2020). Conversely, cancer cells become more drug sensitive when NFE2L2/Nrf2 is overexpressed because it causes chemoresistance in HCC (Cai et al., 2020), whereas medication sensitivity is caused by downregulation of USP7, which also inhibits HCC cell proliferation and metastasis (Zhang et al., 2020). However, depending on the particular signaling pathways involved and the setting, the precise interaction among these components in HCC may differ. Thus, to fully understand the connections between ICMT, NFE2L2, and USP7 in HCC, more research is required (Ying et al., 2023). One major pathogenic role for NFE2L2 deregulation in HCC is hypothesized. When defective autophagy occurs under specific pathophysiological conditions, such as oxidative stress, NFE2L2 activation follows, which has negative effects that promote HCC survival and proliferation. Through autophagic pathways, NFE2L2 is involved in the migration, invasion, and proliferation of HCC. For example, NFE2L2 is negatively controlled by KEAP1 (kelch like ECH associated protein 1), which contributes to HCC carcinogenesis by increasing the production of ROS; autophagy may help HCC cells undergo an oxidative metabolic reprogramming (Moon et al., 2012).
Comprehensive evidence substantiates the association between autophagy and drug resistance, development, migration, and cancer (Russo and Russo, 2018). However, it is still unknown how, particularly in cases of treatment resistance, autophagy flux and tumor cell state control autophagy to either become a tumor guardian or a tumor killer (Sun et al., 2013). Medication therapy stimulates autophagy and increases its flux; nevertheless, elevated autophagy flux also stimulates drug resistance or, by modifications to autophagy-related proteins, causes tumor cell death (Sheng et al., 2018).
Because autophagy helps tumor cells survive under therapeutic stress, it is also thought to be a significant factor of drug resistance (Lei et al., 2024). Therefore, HCC cells may become more sensitive to chemotherapeutic treatments if autophagy is suppressed (Wang et al., 2017).
To put it briefly, learning more about how autophagy influences drug resistance in HCC, such as the initially prescribed drug, traditional chemotherapeutic medications, and cutting-edge anticancer agents, is extremely important and needs further research.
Conclusion
The main treatment for HCC, a common malignant tumor and a major subtype of liver cancer, is surgical excision. 65%–70% of patients are at a middle or advanced stage and need chemotherapy. Treatment failure, however, might result from drug resistance to chemotherapeutic agents. Drug efflux pump transport, DNA repair ability, hereditary variables, and adaptive responses are some of the mechanisms underlying drug resistance. In both healthy and malignant cells, autophagy is an essential biochemical process that has many applications based on the situation. While autophagy helps normal cells break down toxic substances and ageing organelles, cancer cells can either stimulate or block autophagy in order to increase their chances of surviving. Drug resistance is a complex process involving autophagy, with dysregulation of autophagy activation being one of the contributing factors. Dysregulation of the expression of BECN1 or LC3, or SQSTM1-induced activation of autophagy contribute to medication resistance. Targeting proteins related to autophagy, signaling pathways, and exosomes can help reverse drug resistance. Autophagy affects both survival and death in HCC; pro-survival autophagy increases cell viability, while pro-death autophagy hinders tumor growth. Increasing sensitivity to medications and radiation can be achieved by targeting autophagy, which may benefit HCC patients’ prognosis, survival, and course of treatment. Treating advanced and metastatic stages of HCC presents obstacles, including early diagnosis and therapeutic resistance.
However, the intricacy of autophagy, the heterogeneity of the disease, and the need for customized treatment make it difficult to combine autophagy inhibition with currently available HCC medications. There are targeted therapies that rely on the role of autophagy in HCC resistance using drugs such as sorafenib, cisplatin, 5-fluorouracil, oxaliplatin, and Pirarubicin (THP-adriamycin) and doxorubicin. To improve the sensitivity of HCC to anticancer medications, a better knowledge of autophagy’s role in drug resistance is necessary. Additionally, further multicenter medical studies are needed for the therapy of anti-HCC in conjunction with the suppression of autophagy.
Author contributions
WHM: Conceptualization, Writing–original draft, Writing–review and editing. GMS: Conceptualization, Writing–original draft, Writing–review and editing. MMA: Funding acquisition, Software, Writing–review and editing. DJK: Writing–original draft, Writing–review and editing. MHA-A: Funding acquisition, Software, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors are thankful to the Deanship of Graduate Studies and Scientific Research at the University of Bisha for supporting this work through the Fast-Track Research Support Program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Moety, A., Baddour, N., Salem, P., El-Tobgy, H., and El-Shendidi, A. (2022). SQSTM1 expression in hepatocellular carcinoma and relation to tumor recurrence after radiofrequency ablation. J. Clin. Exp. Hepatology 12, 774–784. doi:10.1016/j.jceh.2021.12.001
Alers, S., Wesselborg, S., and Stork, B. (2014). ATG13: just a companion, or an executor of the autophagic program?. Autophagy. 10, 944–956. doi:10.4161/auto.28987
Alisi, A., Cho, W. C., Locatelli, F., and Fruci, D. (2013). Multidrug resistance and cancer stem cells in neuroblastoma and hepatoblastoma. Int. J. Mol. Sci. 14, 24706–24725. doi:10.3390/ijms141224706
Anderson, C. M., and Macleod, K. F. (2019). Autophagy and cancer cell metabolism. Int. Rev. Cell Mol. Biol. 347, 145–190. doi:10.1016/bs.ircmb.2019.06.002
Barnard, R. A., Regan, D. P., Hansen, R. J., Maycotte, P., Thorburn, A., and Gustafson, D. L. (2016). Autophagy inhibition delays early but not late-stage metastatic disease. J. Pharmacol. Exp. Ther. 358, 282–293. doi:10.1124/jpet.116.233908
Beaumont, J. E. J., Ju, J., Barbeau, L. M. O., Demers, I., Savelkouls, K. G., Derks, K., et al. (2024). GABARAPL1 is essential in extracellular vesicle cargo loading and metastasis development. Radiotherapy Oncol. 190, 109968. doi:10.1016/j.radonc.2023.109968
Becattini, B., Breasson, L., Sardi, C., Zani, F., and Solinas, G. (2021). PI3Kγ promotes obesity-associated hepatocellular carcinoma by regulating metabolism and inflammation. Jhep Rep. 3, 100359. doi:10.1016/j.jhepr.2021.100359
Becker, A. K., Tso, D. K., Harris, A. C., Malfair, D., and Chang, S. D. (2014). Extrahepatic metastases of hepatocellular carcinoma: a spectrum of imaging findings. Can. Assoc. Radiologists J. 65, 60–66. doi:10.1016/j.carj.2013.05.004
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Cai, L., Jin, X., Zhang, J., Li, L., and Zhao, J. (2020). Metformin suppresses Nrf2-mediated chemoresistance in hepatocellular carcinoma cells by increasing glycolysis. Aging 12, 17582–17600. doi:10.18632/aging.103777
Celis-Pinto, J. C., Fernández-Velasco, A. A., Corte-Torres, M. D., Santos-Juanes, J., Blanco-Agudín, N., Piña Batista, K. M., et al. (2023). PINK1 immunoexpression predicts survival in patients undergoing hepatic resection for colorectal liver metastases. Int. J. Mol. Sci. 24, 6506–6513. doi:10.3390/ijms24076506
Chan, L. H., Luk, S. T., and Ma, S. (2015). Turning hepatic cancer stem cells inside out - a deeper understanding through multiple perspectives. Mol. Cells 38, 202–209. doi:10.14348/molcells.2015.2356
Chang, J. Y., Yi, H. S., Kim, H. W., and Shong, M. (2017). Dysregulation of mitophagy in carcinogenesis and tumor progression. Biochimica Biophysica Acta - Bioenergetics 1858, 633–640. doi:10.1016/j.bbabio.2016.12.008
Chano, T., Ikebuchi, K., Ochi, Y., Tameno, H., Tomita, Y., Jin, Y., et al. (2010). RB1CC1 activates RB1 pathway and inhibits proliferation and cologenic survival in human cancer. Plos One 5, e11404. doi:10.1371/journal.pone.0011404
Chu, C.-A., Wang, Y.-W., Chen, Y.-L., Chen, H.-W., Chuang, J.-J., Chang, H.-Y., et al. (2021). The role of phosphatidylinositol 3-kinase catalytic subunit type 3 in the pathogenesis of human cancer. Int. J. Mol. Sci. 22, 10964. doi:10.3390/ijms222010964
Chu, C. T. (2019). Mechanisms of selective autophagy and mitophagy: implications for neurodegenerative diseases. Neurobiol. Dis. 122, 23–34. doi:10.1016/j.nbd.2018.07.015
Collier, J. J., Suomi, F., Oláhová, M., McWilliams, T. G., and Taylor, R. W. (2021). Emerging roles of ATG7 in human health and disease. Embo Mol. Med. 13, 148244–e14920. doi:10.15252/emmm.202114824
Deldar Abad Paskeh, M., Mirzaei, S., Ashrafizadeh, M., Zarrabi, A., and Sethi, G. (2021). Wnt/β-Catenin signaling as a driver of hepatocellular carcinoma progression: an emphasis on molecular pathways. J. Hepatocell. Carcinoma 8, 1415–1444. doi:10.2147/JHC.S336858
Dhar, D., Antonucci, L., Nakagawa, H., Kim, J. Y., Glitzner, E., Caruso, S., et al. (2018). Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell 33, 1061–1077. doi:10.1016/j.ccell.2018.05.003
Di, C., Du, Y., Zhang, R., Zhang, L., and Wang, S. (2023). Identification of autophagy-related genes and immune cell infiltration characteristics in sepsis via bioinformatic analysis. J. Thorac. Dis. 15, 1770–1784. doi:10.21037/jtd-23-312
Dong, Y., Wu, Y., Zhao, G. L., Ye, Z. Y., Xing, C. G., and Yang, X. D. (2019). Inhibition of autophagy by 3-MA promotes hypoxia-induced apoptosis in human colorectal cancer cells. Eur. Rev. Med. Pharmacol. Sci. 23, 1047–1054. doi:10.26355/eurrev_201902_16992
El-Serag, H. B. (2011). Hepatocellular carcinoma. N. Engl. J. Med. 365, 1118–1127. doi:10.1056/NEJMra1001683
Gao, F., Chen, D., Si, J., Hu, Q., Qin, Z., Fang, M., et al. (2015). The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum. Mol. Genet. 24, 2528–2538. doi:10.1093/hmg/ddv017
Glick, D., Barth, S., and Macleod, K. F. (2010). Autophagy: cellular and molecular mechanisms. J. Pathology 221, 3–12. doi:10.1002/path.2697
Grand, J. N.Le, Chakrama, F. Z., Seguin-Py, S., Fraichard, A., Delage-Mourroux, R., Jouvenot, M., et al. (2011). GABARAPL1 (GEC1): original or copycat?. Autophagy. 7, 1098–1107. doi:10.4161/auto.7.10.15904
Guardia, C. M., Tan, X., Lian, T., Rana, M. S., Zhou, W., Christenson, E. T., et al. (2020). Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep. 31, 107837. doi:10.1016/j.celrep.2020.107837
Guo, J. Y., Xia, B., and White, E. (2013). Autophagy-mediated tumor promotion. Cell. 155, 1216–1219. doi:10.1016/j.cell.2013.11.019
Han, S., Jeong, Y. Y., Sheshadri, P., Su, X., and Cai, Q. (2020). Mitophagy regulates integrity of mitochondria at synapses and is critical for synaptic maintenance. Embo Rep. 21, e49801–e49819. doi:10.15252/embr.201949801
Hashemi, M., Nadafzadeh, N., Imani, M. H., Rajabi, R., Ziaolhagh, S., Bayanzadeh, S. D., et al. (2023). Targeting and regulation of autophagy in hepatocellular carcinoma: revisiting the molecular interactions and mechanisms for new therapy approaches. Cell Commun. Signal. 21, 32–22. doi:10.1186/s12964-023-01053-z
He, C., and Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. doi:10.1146/annurev-genet-102808-114910
Hu, W. H., Liu, T. T., Liu, P. F., Morgan, P., Lin, I. L., Tsai, W. L., et al. (2023). ATG4B and pS383/392-ATG4B serve as potential biomarkers and therapeutic targets of colorectal cancer. Cancer Cell Int. 23, 63–17. doi:10.1186/s12935-023-02909-7
Huang, F., Wang, B. R., and Wang, Y. G. (2018). Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J. Gastroenterology. 24, 4643–4651. doi:10.3748/wjg.v24.i41.4643
Huang, T., Song, X., Yang, Y., Wan, X., Alvarez, A. A., Sastry, N., et al. (2018). Autophagy and hallmarks of cancer. Crit. Rev. Oncog. 23, 247–267. doi:10.1615/CritRevOncog.2018027913
Ichimura, Y., Waguri, S., Sou, Y., Kageyama, S., Hasegawa, J., Ishimura, R., et al. (2013). Phosphorylation of p62 activates the keap1-nrf2 pathway during selective autophagy. Mol. Cell 51, 618–631. doi:10.1016/j.molcel.2013.08.003
Inami, Y., Waguri, S., Sakamoto, A., Kouno, T., Nakada, K., Hino, O., et al. (2011). Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 193 (2), 275–284. doi:10.1083/jcb.201102031
Iriondo, M. N., Etxaniz, A., Varela, Y. R., Ballesteros, U., Lázaro, M., Valle, M., et al. (2023). Effect of ATG12–ATG5-ATG16L1 autophagy E3-like complex on the ability of LC3/GABARAP proteins to induce vesicle tethering and fusion. Cell Mol. Life Sci. 80, 56. doi:10.1007/s00018-023-04704-z
Jabir, M. S., Hopkins, L., Ritchie, N. D., Ullah, I., Bayes, H. K., Li, D., et al. (2015). Mitochondrial damage contributes to pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy. 11, 166–182. doi:10.4161/15548627.2014.981915
Jabir, M. S., Ritchie, N. D., Li, D., Bayes, H. K., Tourlomousis, P., Puleston, D., et al. (2014). Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and β-interferon production during Pseudomonas aeruginosa infection. Cell Host Microbe. 15, 214–227. doi:10.1016/j.chom.2014.01.010
Jabir, M. S., Saleh, Y. M., Sulaiman, G. M., Yaseen, N. Y., Sahib, U. I., Dewir, Y. H., et al. (2021). Green synthesis of silver nanoparticles using Annona muricata extract as an inducer of apoptosis in cancer cells and inhibitor for NLRP3 inflammasome via enhanced autophagy. Nanomaterials. 11, 384–422. doi:10.3390/nano11020384
Jabir, M. S., Sulaiman, G. M., Taqi, Z. J., and Li, D. (2018). Iraqi propolis increases degradation of IL-1β and NLRC4 by autophagy following Pseudomonas aeruginosa infection. Microbes Infect. 20, 89–100. doi:10.1016/j.micinf.2017.10.007
Jawad, M. H., Jabir, M. S., Ozturk, K., Sulaiman, G. M., Abomughaid, M. M., Albukhaty, S., et al. (2023). Induction of apoptosis and autophagy via regulation of AKT and JNK mitogen-activated protein kinase pathways in breast cancer cell lines exposed to gold nanoparticles loaded with TNF-α and combined with doxorubicin. Nanotechnol. Rev. 12. doi:10.1515/ntrev-2023-0148
Katherine, A. M., Jessica, L., and Petrick, H. B. (2021). Epidemiology of hepatocellular carcinoma. Hepatology 73, 4–13. doi:10.1002/hep.31288
Kelekar, A., and Thompson, C. B. (1998). Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 8, 324–330. doi:10.1016/s0962-8924(98)01321-x
Kim, K. H., and Lee, M. S. (2014). Autophagy - a key player in cellular and body metabolism. Nat. Rev. Endocrinol. 10, 322–337. doi:10.1038/nrendo.2014.35
Kirkin, V. (2020). History of the selective autophagy research: how did it begin and where does it stand today? J. Mol. Biol. 432, 3–27. doi:10.1016/j.jmb.2019.05.010
Kiruthiga, C., Devi, K. P., Nabavi, S. M., and Bishayee, A. (2020). Autophagy: a potential therapeutic target of polyphenols in hepatocellular carcinoma. Cancers. 12, 562–631. doi:10.3390/cancers12030562
Koltai, T. (2022). The complex relationship between multiple drug resistance and the tumor pH gradient: a review. Cancer Drug Resist. 5, 277–303. doi:10.20517/cdr.2021.134
Kumar, S., Sánchez-Álvarez, M., Lolo, F.-N., Trionfetti, F., Strippoli, R., and Cordani, M. (2021). Autophagy and the lysosomal system in cancer. Cells. 10, 2752. doi:10.3390/cells10102752
Kumariya, S., Ubba, V., Jha, R. K., and Gayen, J. R. (2021). Autophagy in ovary and polycystic ovary syndrome: role, dispute and future perspective. Autophagy. 17, 2706–2733. doi:10.1080/15548627.2021.1938914
Lee, Y. J., and Jang, B. K. (2015). The role of autophagy in hepatocellular carcinoma. Int. J. Mol. Sci. 16, 26629–26643. doi:10.3390/ijms161125984
Lei, Y. R., He, X. L., Li, J., and Mo, C. F. (2024). Drug resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Front. Biosci. - Landmark 29, 52–17. doi:10.31083/j.fbl2902052
Levy, J. M. M., Towers, C. G., and Thorburn, A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer. 17, 528–542. doi:10.1038/nrc.2017.53
Li, L., Wang, G., Hu, J. S., Zhang, G. Q., Chen, H. Z., Yuan, Y., et al. (2018). RB1CC1-enhanced autophagy facilitates PSCs activation and pancreatic fibrogenesis in chronic pancreatitis. Cell Death Dis. 9, 952. doi:10.1038/s41419-018-0980-4
Li, Q., Chao, Q., Liu, Y., Fang, J., Xie, J., Zhen, J., et al. (2021). Loss of TARBP2 drives the progression of hepatocellular carcinoma via miR-145-SERPINE1 Axis. Am. J. cancer Res. 11, 4807–4825. doi:10.3389/fonc.2021.620912
Li, Q.Gf. G., and Zan, L. G. G. (2022). Knockdown of ATG4A inhibits breast cancer progression and promotes tamoxifen chemosensitivity by suppressing autophagy. Mol. Med. Rep. 25, 101–109. doi:10.3892/mmr.2022.12617
Li, S., Zhang, L., Zhang, G., Shangguan, G., Hou, X., Duan, W., et al. (2021). A nonautophagic role of ATG5 in regulating cell growth by targeting c-Myc for proteasome-mediated degradation. iScience. 24, 103296. doi:10.1016/j.isci.2021.103296
Li, X., He, S., and Ma, B. (2020). Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 19, 12–16. doi:10.1186/s12943-020-1138-4
Li, Y., Wang, R., Wang, H., Pu, F., Feng, X., Jin, L., et al. (2021). Codon usage bias in autophagy-related gene 13 in eukaryotes: uncovering the genetic divergence by the interplay between nucleotides and codon usages. Front. Cell. Infect. Microbiol. 11, 771010–771015. doi:10.3389/fcimb.2021.771010
Li, Y., Zheng, W., Lu, Y., Zheng, Y., Pan, L., Wu, X., et al. (2022). BNIP3L/NIX-mediated mitophagy: molecular mechanisms and implications for human disease. Cell Death Dis. 13, 14. doi:10.1038/s41419-021-04469-y
Liang, C., Sir, D., Lee, S., Ou, J. H. J., and Jung, J. U. (2008). Beyond autophagy: the role of UVRAG in membrane trafficking. Autophagy. 4, 817–820. doi:10.4161/auto.6496
Lin, M. G., and Hurley, J. H. (2016). Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 39, 61–68. doi:10.1016/j.ceb.2016.02.010
Liu, K., Lee, J., and Ou, J. H. J. (2018). Autophagy and mitophagy in hepatocarcinogenesis. Mol. Cell. Oncol. 5, e1405142–2. doi:10.1080/23723556.2017.1405142
Llovet, J. M., Ricci, S., Mazzaferro, V., Hilgard, P., Gane, E., Blanc, J.-F., et al. (2008). Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390. doi:10.1056/nejmoa0708857
Ma, C., Storer, C. E., Chandran, U., LaFramboise, W. A., Petrosko, P., Frank, M., et al. (2021). Crohn’s disease-associated ATG16L1 T300A genotype is associated with improved survival in gastric cancer. EBioMedicine. 67, 103347. doi:10.1016/j.ebiom.2021.103347
Macher-Goeppinger, S., Keith, M., Hatiboglu, G., Hohenfellner, M., Schirmacher, P., Roth, W., et al. (2017). Expression and functional characterization of the BNIP3 protein in renal cell carcinomas. Transl. Oncol. 10, 869–875. doi:10.1016/j.tranon.2017.08.008
Maeda, S., Otomo, C., and Otomo, T. (2019). The autophagic membrane tether ATG2A transfers lipids between membranes. eLife. 8, e45777. doi:10.7554/eLife.45777
Maeda, S., Yamamoto, H., Kinch, L. N., Garza, C. M., Takahashi, S., Otomo, C., et al. (2020). Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 27, 1194–1201. doi:10.1038/s41594-020-00520-2
Marsh, T., Kenific, C. M., Suresh, D., Gonzalez, H., Shamir, E. R., Mei, W., et al. (2020). Autophagic degradation of NBR1 restricts metastatic outgrowth during mammary tumor progression. Dev. Cell 52, 591–604. doi:10.1016/j.devcel.2020.01.025
Matoba, K., Kotani, T., Tsutsumi, A., Tsuji, T., Mori, T., Noshiro, D., et al. (2020). Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 27, 1185–1193. doi:10.1038/s41594-020-00518-w
Minaei, N., Ramezankhani, R., Tamimi, A., Piryaei, A., Zarrabi, A., Aref, A. R., et al. (2023). Immunotherapeutic approaches in Hepatocellular carcinoma: building blocks of hope in near future. Eur. J. Cell Biol. 102, 151284. doi:10.1016/j.ejcb.2022.151284
Mohamed, E. A., Bordean, D. M., Radulov, I., Moruzi, R. F., Hulea, C. I., Orǎan, S. A., et al. (2020a). Sea buckthorn and grape antioxidant effects in hyperlipidemic rats: relationship with the atorvastatin therapy. Evidence-based Complementary Altern. Med. 2020, 1736803. doi:10.1155/2020/1736803
Mohamed, E. A., Dumitrescu, E., Muselin, F., Moruzi, R. F., Or, S. A., and Cristina, R. T. (2022). The phytotherapy effect on liver’s cytoarchitecture of high-fat diet rats treated with statins.
Mohamed, E. A., Tulcan, C., Alexa, E., Morar, D., Dumitrescu, E., Muselin, F., et al. (2020b). Sea buckthorn and grape extract might be helpful and sustainable phyto-resources as associated hypolipidemic agents—preliminary study. Sustain. Switz. 12, 9297–9312. doi:10.3390/su12219297
Mohan, J., and Wollert, T. (2018). Human ubiquitin-like proteins as central coordinators in autophagy. Interface Focus 8, 20180025. doi:10.1098/rsfs.2018.0025
Moon, M. S., McDevitt, E. I., Zhu, J., Stanley, B., Krzeminski, J., Amin, S., et al. (2012). Elevated hepatic iron activates NF-E2–Related factor 2–regulated pathway in a dietary iron overload mouse model. Toxicol. Sci. 129, 74–85. doi:10.1093/toxsci/kfs193
Nazio, F., Bordi, M., Cianfanelli, V., Locatelli, F., and Cecconi, F. (2019). Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death Differ. 26, 690–702. doi:10.1038/s41418-019-0292-y
Nguyen, N., Olivas, T. J., Mires, A., Jin, J., Yu, S., Luan, L., et al. (2020). The insufficiency of ATG4A in macroautophagy. J. Biol. Chem. 295, 13584–13600. doi:10.1074/jbc.ra120.013897
Ni, H.-M., Woolbright, B. L., Williams, J., Copple, B., Cui, W., Luyendyk, J. P., et al. (2014). Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J. Hepatology. 61, 617–625. doi:10.1016/j.jhep.2014.04.043
Niu, Y., Lin, Z., Wan, A., Chen, H., Liang, H., Sun, L., et al. (2019). RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer. 18, 46–16. doi:10.1186/s12943-019-1004-4
Niu, Z. S., Wang, W. H., Dong, X. N., and Tian, L. M. L. (2020). Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J. Gastroenterology. 26, 4240–4260. doi:10.3748/WJG.V26.I29.4240
Oku, M., and Sakai, Y. (2018). Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays. 40, 18000088–e1800016. doi:10.1002/bies.201800008
Onorati, A. V., Dyczynski, M., Ojha, R., and Amaravadi, R. K. (2018). Targeting autophagy in cancer. Cancer. 124, 3307–3318. doi:10.1002/cncr.31335
Otera, H., Wang, C., Cleland, M. M., Setoguchi, K., Yokota, S., Youle, R. J., et al. (2010). Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191 (6), 1141–1158. doi:10.1083/jcb.201007152
Park, S. W., Jeon, P., Jun, Y. W., Park, J. H., Lee, S. H., Lee, S., et al. (2019). Monitoring LC3- or GABARAP-positive autophagic membranes using modified RavZ-based probes. Sci. Rep. 9, 16593–16611. doi:10.1038/s41598-019-53372-2
Parzych, K. R., and Klionsky, D. J. (2014). An overview of autophagy: morphology, mechanism, and regulation. Antioxidants Redox Signal. 20, 460–473. doi:10.1089/ars.2013.5371
Petrick, J. L., Florio, A. A., Znaor, A., Ruggieri, D., Laversanne, M., Alvarez, C. S., et al. (2020). International trends in hepatocellular carcinoma incidence, 1978–2012. Int. J. Cancer. 147, 317–330. doi:10.1002/ijc.32723
Popelka, H., and Klionsky, D. J. (2021). Multiple structural rearrangements mediated by high-plasticity regions in Atg3 are key for efficient conjugation of Atg8 to PE during autophagy. Autophagy. 17, 1805–1808. doi:10.1080/15548627.2021.1954457
Pu, J., Zhang, Y., Wang, A., Qin, Z., Zhuo, C., Li, W., et al. (2021). ADORA2A-AS1 restricts hepatocellular carcinoma progression via binding HuR and repressing FSCN1/AKT Axis. Front. Oncol. 11, 754835–754917. doi:10.3389/fonc.2021.754835
Puris, E., Fricker, G., and Gynther, M. (2023). The role of solute carrier transporters in efficient anticancer drug delivery and therapy. Pharmaceutics. 15, 364. doi:10.3390/pharmaceutics15020364
Raudenska, M., Balvan, J., and Masarik, M. (2021). Crosstalk between autophagy inhibitors and endosome-related secretory pathways: a challenge for autophagy-based treatment of solid cancers. Mol. Cancer. 20, 140–227. doi:10.1186/s12943-021-01423-6
Rubinstein, A. D., Eisenstein, M., Ber, Y., Bialik, S., and Kimchi, A. (2011). The autophagy protein atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell. 44, 698–709. doi:10.1016/j.molcel.2011.10.014
Russo, M., and Russo, G. L. (2018). Autophagy inducers in cancer. Biochem. Pharmacol. 153, 51–61. doi:10.1016/j.bcp.2018.02.007
Saito, T., Ichimura, Y., Taguchi, K., Suzuki, T., Mizushima, T., Takagi, K., et al. (2016). p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 7, 12030. doi:10.1038/ncomms12030
Salem, M., Ammitzboell, M., Nys, K., Seidelin, J. B., and Nielsen, O. H. (2015). ATG16L1: a multifunctional susceptibility factor in crohn disease. Autophagy. 11, 585–594. doi:10.1080/15548627.2015.1017187
Seydi, H., Nouri, K., Rezaei, N., Tamimi, A., Hassan, M., Mirzaei, H., et al. (2023). Autophagy orchestrates resistance in hepatocellular carcinoma cells. Biomed. Pharmacother. 161, 114487. doi:10.1016/j.biopha.2023.114487
Sheng, J., Qin, H., Zhang, K., Li, B., and Zhang, X. (2018). Targeting autophagy in chemotherapy-resistant of hepatocellular carcinoma. Am. J. cancer Res. 8, 354–365.
Shi, M., An, G., Chen, N., Jia, J., Cui, X., Zhan, T., et al. (2023). UVRAG promotes tumor progression through regulating SP1 in colorectal cancer. Cancers. 15, 2502. doi:10.3390/cancers15092502
Shokoohian, B., Negahdari, B., Aboulkheyr Es, H., Abedi-Valugerdi, M., Baghaei, K., Agarwal, T., et al. (2021). Advanced therapeutic modalities in hepatocellular carcinoma: novel insights. J. Cell. Mol. Med. 25, 8602–8614. doi:10.1111/jcmm.16875
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. CA A Cancer J. Clin. 70, 7–30. doi:10.3322/caac.21590
Song, H., Zhu, Y., Hu, C., Liu, Q., Jin, Y., Tang, P., et al. (2024). Selective autophagy receptor NBR1 retards nucleus pulposus cell senescence by directing the clearance of SRBD1. Int. J. Biol. Sci. 20, 701–717. doi:10.7150/ijbs.90186
Song, W., Postoak, J. L., Yang, G., Guo, X., Pua, H. H., Bader, J., et al. (2023). Lipid kinase PIK3C3 maintains healthy brown and white adipose tissues to prevent metabolic diseases. Proc. Natl. Acad. Sci. U.S.A. 120, e2214874120. doi:10.1073/pnas.2214874120
Sun, K., Guo, X. L., Zhao, Q. D., Jing, Y. Y., Kou, X. R., Xie, X. Q., et al. (2013). Paradoxical role of autophagy in the dysplastic and tumor-forming stages of hepatocarcinoma development in rats. Cell Death Dis. 4, e501–e511. doi:10.1038/cddis.2013.35
Takahashi, Y., Coppola, D., Matsushita, N., Cualing, H. D., Sato, Y., Liang, C., et al. (2007). Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 9, 1142–1151. doi:10.1038/ncb1634
Takamura, A., Komatsu, M., Hara, T., Sakamoto, A., Kishi, C., Waguri, S., et al. (2011). Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800. doi:10.1101/gad.2016211
Tamargo-Gómez, I., Martínez-García, G. G., Suárez, M. F., Mayoral, P., Bretones, G., Astudillo, A., et al. (2023). Analysis of ATG4C function in vivo. Autophagy. 19, 2912–2933. doi:10.1080/15548627.2023.2234799
Taraborrelli, L., Şenbabaoğlu, Y., Wang, L., Lim, J., Blake, K., Kljavin, N., et al. (2023). Tumor-intrinsic expression of the autophagy gene Atg16l1 suppresses anti-tumor immunity in colorectal cancer. Nat. Commun. 14, 5945. doi:10.1038/s41467-023-41618-7
Tran, S., Fairlie, W. D., and Lee, E. F. (2021). Beclin1: protein structure, function and regulation. Cells. 10, 1522. doi:10.3390/cells10061522
Tsochatzis, E. A., Fatourou, E., O’Beirne, J., Meyer, T., and Burroughs, A. K. (2014). Transarterial chemoembolization and bland embolization for hepatocellular carcinoma. World J. Gastroenterology. 20, 3069–3077. doi:10.3748/wjg.v20.i12.3069
Vara-Perez, M., Felipe-Abrio, B., and Agostinis, P. (2019). Mitophagy in cancer: a tale of adaptation. Cells 8, 493–538. doi:10.3390/cells8050493
Villanueva, A. (2019). Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462. doi:10.1056/nejmra1713263
Wan, S., Zhao, E., Kryczek, I., Vatan, L., Sadovskaya, A., Ludema, G., et al. (2014). Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 147, 1393–1404. doi:10.1053/j.gastro.2014.08.039
Wang, M., Huang, C., Su, Y., Yang, C., Xia, Q., and Xu, D. J. (2017). Astragaloside II sensitizes human hepatocellular carcinoma cells to 5-fluorouracil via suppression of autophagy. J. Pharm. Pharmacol. 69, 743–752. doi:10.1111/jphp.12706
Wang, P. P., Xu, D. J., Huang, C., Wang, W. P., and Xu, W. K. (2014). Astragaloside Ⅳ reduces the expression level of P-glycoprotein in multidrug-resistant human hepatic cancer cell lines. Mol. Med. Rep. 9, 2131–2137. doi:10.3892/mmr.2014.2074
Wang, Y., Dahmane, S., Ti, R., Mai, X., Zhu, L., Carlson, L., et al. (2024). Structural basis for lipid transfer by the ATG2A–ATG9A complex. Nat. Struct. Mol. Biol. doi:10.1038/s41594-024-01376-6
Wang, Y., Liu, H. H., Cao, Y. T., Zhang, L. L., Huang, F., and Yi, C. (2020). The role of mitochondrial dynamics and mitophagy in carcinogenesis, metastasis and therapy. Front. Cell Dev. Biol. 8, 413–512. doi:10.3389/fcell.2020.00413
White, E., Mehnert, J. M., and Chan, C. S. (2015). Autophagy, metabolism, and cancer. Clin. Cancer Res. 21, 5037–5046. doi:10.1158/1078-0432.CCR-15-0490
Wong, S. W., Sil, P., and Martinez, J. (2018). Rubicon: LC3-associated phagocytosis and beyond. Febs J. 285, 1379–1388. doi:10.1111/febs.14354
Wong Y., Y., Ysselstein, D., and Krainc, D. (2018). Mitochondria–lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 554, 382–386. doi:10.1038/nature25486
Wu, W. K. K., Coffelt, S. B., Cho, C. H., Wang, X. J., Lee, C. W., Chan, F. K. L., et al. (2012). The autophagic paradox in cancer therapy. Oncogene. 31, 939–953. doi:10.1038/onc.2011.295
Wu, Y., Zhang, J., and Li, Q. (2021). Autophagy, an accomplice or antagonist of drug resistance in HCC? Cell Death Dis. 12, 266. doi:10.1038/s41419-021-03553-7
Xia, H., Green, D. R., Zou, W., Arbor, A., Arbor, A., Arbor, A., et al. (2021). Autophagy in tumour immunity and therapy. Autophagy. 21, 281–297. doi:10.1038/s41568-021-00344-2
Xiao, P., Chen, X., Dong, Z., Fan, W., Chen, Y., Su, J., et al. (2023). BNIP3 overexpression may promote myeloma cell apoptosis by enhancing sensitivity to bortezomib via the p38 MAPK pathway. Hematol. (United Kingdom) 28, 2231739. doi:10.1080/16078454.2023.2231739
Xie, P., Yan, J., Wu, M., Li, H., Chen, Z., Yu, M., et al. (2022). CD44 potentiates hepatocellular carcinoma migration and extrahepatic metastases via the AKT/ERK signaling CXCR4 axis. Ann. Transl. Med. 10, 689. doi:10.21037/atm-22-2482
Xiong, X., Lu, B., Tian, Q., Zhang, H., Wu, M., Guo, H., et al. (2019). Inhibition of autophagy enhances cinobufagin-induced apoptosis in gastric cancer. Oncol. Rep. 41, 492–500. doi:10.3892/or.2018.6837
Xiong, X., Tao, R., DePinho, R. A., and Dong, X. C. (2012). The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J. Biol. Chem. 287, 39107–39114. doi:10.1074/jbc.M112.412569
Xu, W. L., Wang, S. H., Sun, W. B., Gao, J., Ding, X. M., Kong, J., et al. (2019). Insufficient radiofrequency ablation-induced autophagy contributes to the rapid progression of residual hepatocellular carcinoma through the HIF-1α/BNIP3 signaling pathway. Bmb Rep. 52, 277–282. doi:10.5483/BMBRep.2019.52.4.263
Yamamuro, T., Nakamura, S., Yanagawa, K., Tokumura, A., Kawabata, T., Fukuhara, A., et al. (2022). Loss of RUBCN/rubicon in adipocytes mediates the upregulation of autophagy to promote the fasting response. Autophagy. 18, 2686–2696. doi:10.1080/15548627.2022.2047341
Yamano, K., Fogel, A. I., Wang, C., Bliek, A. M., and Youle, R. J. (2014). Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife. 3, e01612. doi:10.7554/eLife.01612
Yang, S., Yang, L., Li, X., Li, B., Li, Y., Zhang, X., et al. (2019). New insights into autophagy in hepatocellular carcinoma: mechanisms and therapeutic strategies. Am. J. cancer Res. 9, 1329–1353.
Ye, J., Zhang, J., Zhu, Y., Wang, L., Jiang, X., Liu, B., et al. (2023). Targeting autophagy and beyond: deconvoluting the complexity of Beclin-1 from biological function to cancer therapy. Acta Pharm. Sin. B 13, 4688–4714. doi:10.1016/j.apsb.2023.08.008
Yi, F., Cai, C., Ruan, B., Hao, M., Yeo, S. K., Haas, M., et al. (2023). Regulation of RB1CC1/FIP200 stability and autophagy function by CREBBP-mediated acetylation in an intrinsically disordered region. Autophagy. 19, 1662–1677. doi:10.1080/15548627.2022.2148432
Ying, H., Zhang, B., Cao, G., Wang, Y., and Zhang, X. (2023). Role for ubiquitin-specific protease 7 (USP7) in the treatment and the immune response to hepatocellular carcinoma: potential mechanisms. Transl. Cancer Res. 12, 3016–3033. doi:10.21037/tcr-23-153
Yoon, J. H., Goo, Y. J., Lim, C. J., Choi, S. K., Cho, S. B., Shin, S. S., et al. (2020). Features of extrahepatic metastasis after radiofrequency ablation for hepatocellular carcinoma. World J. Gastroenterology. 26, 4833–4845. doi:10.3748/WJG.V26.I32.4833
Yorimitsu, T., and Klionsky, D. J. (2005). Autophagy: molecular machinery for self-eating. Cell Death Differ. 12, 1542–1552. doi:10.1038/sj.cdd.4401765
Yu, Y., Peng, X. D., Qian, X. J., Zhang, K. M., Huang, X., Chen, Y. H., et al. (2021). Fis1 phosphorylation by Met promotes mitochondrial fission and hepatocellular carcinoma metastasis. Signal Transduct. Target. Ther. 6, 401–415. doi:10.1038/s41392-021-00790-2
Yuan, F., Jin, X., Li, D., Song, Y., Zhang, N., Yang, X., et al. (2019). ULK1 phosphorylates Mad1 to regulate spindle assembly checkpoint. Nucleic Acids Res. 47, 8096–8110. doi:10.1093/nar/gkz602
Yuan, Z., Cai, K., Li, J., Chen, R., Zhang, F., Tan, X., et al. (2024). ATG14 targets lipid droplets and acts as an autophagic receptor for syntaxin18-regulated lipid droplet turnover. Nat. Commun. 15, 631. doi:10.1038/s41467-024-44978-w
Yun, C. W., Jeon, J., Go, G., Lee, J. H., and Lee, S. H. (2020). The dual role of autophagy in cancer development and a therapeutic strategy for cancer by targeting autophagy. Int. J. Mol. Sci. 22, 179. doi:10.3390/ijms22010179
Yun, C. W., and Lee, S. H. (2018). The roles of autophagy in cancer. Int. J. Mol. Sci. 19, 3466–3518. doi:10.3390/ijms19113466
Zhang, J., He, X., Wan, Y., Zhang, H., Tang, T., Zhang, M., et al. (2021). CD44 promotes hepatocellular carcinoma progression via upregulation of YAP. Exp. Hematol. Oncol. 10, 54–4. doi:10.1186/s40164-021-00247-w
Zhang, K., Zhang, Q., Jia, R., Xiang, S., and Xu, L. (2023). A comprehensive review of the relationship between autophagy and sorafenib-resistance in hepatocellular carcinoma: ferroptosis is noteworthy. Front. Cell Dev. Biol. 11, 1156383–1156412. doi:10.3389/fcell.2023.1156383
Zhang, R., Jiang, W., Wang, G., Zhang, Y., Liu, W., Li, M., et al. (2024). Parkin inhibits proliferation and migration of bladder cancer via ubiquitinating Catalase. Commun. Biol. 7, 245. doi:10.1038/s42003-024-05935-x
Zhang, W., Zhang, J., Xu, C., Zhang, S., Bian, S., Jiang, F., et al. (2020). Ubiquitin-specific protease 7 is a drug-able target that promotes hepatocellular carcinoma and chemoresistance. Cancer Cell Int. 20, 28–12. doi:10.1186/s12935-020-1109-2
Keywords: autophagy, autophagy dual role, cancer, drug resistance, hepatocecllular carcimoa
Citation: Mohammed WH, Sulaiman GM, Abomughaid MM, Klionsky DJ and Abu-Alghayth MH (2024) The dual role of autophagy in suppressing and promoting hepatocellular carcinoma. Front. Cell Dev. Biol. 12:1472574. doi: 10.3389/fcell.2024.1472574
Received: 01 August 2024; Accepted: 25 September 2024;
Published: 11 October 2024.
Edited by:
Shigeomi Shimizu, Tokyo Medical and Dental University, JapanReviewed by:
Ghita Ghislat, Imperial College London, United KingdomKoji Yamano, Tokyo Medical and Dental University, Japan
Copyright © 2024 Mohammed, Sulaiman, Abomughaid, Klionsky and Abu-Alghayth. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ghassan M. Sulaiman, Z2hhc3Nhbi5tLnN1bGFpbWFuQHVvdGVjaG5vbG9neS5lZHUuaXE=; Daniel J. Klionsky, a2xpb25za3lAdW1pY2guZWR1
 Wasnaa H. Mohammed
Wasnaa H. Mohammed Ghassan M. Sulaiman
Ghassan M. Sulaiman Mosleh M. Abomughaid
Mosleh M. Abomughaid Daniel J. Klionsky
Daniel J. Klionsky Mohammed H. Abu-Alghayth
Mohammed H. Abu-Alghayth