
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 23 September 2024
Sec. Epigenomics and Epigenetics
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1460705
Ticks are important blood feeding ectoparasites that transmit pathogens to wildlife, domestic animals, and humans. Hard ticks can feed for several days to weeks, nevertheless they often go undetected. This phenomenon can be explained by a tick’s ability to release analgesics, immunosuppressives, anticoagulants, and vasodilators within their saliva. Several studies have identified extracellular vesicles (EVs) as carriers of some of these effector molecules. Further, EVs, and their contents, enhance pathogen transmission, modulate immune responses, and delay wound healing. EVs are double lipid-membrane vesicles that transport intracellular cargo, including microRNAs (miRNAs) to recipient cells. miRNAs are involved in regulating gene expression post-transcriptionally. Interestingly, tick-derived miRNAs have been shown to enhance pathogen transmission and affect vital biological processes such as oviposition, blood digestion, and molting. miRNAs have been found within tick salivary EVs. This review focuses on current knowledge of miRNA loading into EVs and homologies reported in ticks. We also describe findings in tick miRNA profiles, including miRNAs packed within tick salivary EVs. Although no functional studies have been done to investigate the role of EV-derived miRNAs in tick feeding, we discuss the functional characterization of miRNAs in tick biology and pathogen transmission. Lastly, we propose the possible uses of tick miRNAs to develop management tools for tick control and to prevent pathogen transmission. The identification and functional characterization of conserved and tick-specific salivary miRNAs targeting important molecular and immunological pathways within the host could lead to the discovery of new therapeutics for the treatment of tick-borne and non-tick-borne human diseases.
Ticks are important vectors of a wide range of disease-causing microorganisms that affect humans and other mammals (Anderson and Magnarelli, 2008; Nicholson et al., 2019). The incidence of tick-borne diseases has increased in recent years, raising public awareness about the importance of ticks. (Vayssier-Taussat et al., 2015; Lippi et al., 2021). In 2022, it was estimated that around 71 thousand people were infected with tick-borne pathogens, with reports of Babesiosis, Lyme Disease, Rocky Mountain Spotted Fever, and Anaplasmosis/Ehrlichiosis all rising in numbers (CDC, 2024b). The outcomes of these diseases range from self-resolving, in the case of Human Anaplasmosis, to fatality rates of 20%–30% for Rocky Mountain Spotted fever (Bakken and Dumler, 2015; Drexler et al., 2017; Bradshaw et al., 2020). Tick bites can also lead to the development of a delayed allergic hypersensitivity known as Alpha-Gal syndrome (AGS), which has also increased in incidence from 13,000 cases in 2017 to over 18,000 in 2021 (Thompson et al., 2023). According to the CDC, 110,000 cases of AGS have been reported between 2010 and 2022 (CDC, 2024a). In addition to their public health importance, ticks pose a global economic threat to livestock producers with $13.9–18.7 billion dollars loss annually (de Castro et al., 1997). Direct effects caused by tick feeding include cowhide damage, decreases in milk and meat yield, and anemia in the case of heavy infestations. Indirect effects from the transmission of tick-borne pathogens can result in animal mortality and abortions (Rajput et al., 2006).
Tick feeding and tick-borne pathogen transmission are facilitated by effector molecules within tick saliva that modulate immune responses at the bite site (Pham et al., 2021; Nuttall, 2023). Saliva assisted transmission has been reported during the inoculation of flaviviruses and bacteria in in vivo and in vitro experiments (Marchal et al., 2009; Hermance and Thangamani, 2015; Reynolds et al., 2023). Moreover, tick saliva can affect the clinical outcome of disease. In a recent study, Reynolds et al. (2023), showed that inoculation of the heartland virus along with salivary gland extracts (SGE) from Amblyomma americanum, the natural vector of this virus, enhanced virus persistence in the blood, had a significant effect on the reduction of lymphocytes, and led to higher liver inflammation in a murine model. Similar modulation of disease intensity and pathogen colonization has been observed using tick-salivary extracellular vesicles (EVs) (Oliva Chávez et al., 2021). However, large knowledge gaps concerning molecular factors involved in pathogen transmission and feeding hinder the development of efficient tick control tools and anti-tick transmission therapeutics and vaccines. Thus, understanding the molecular factors involved in pathogen transmission and feeding is essential to identify potential targets that can be used against ticks and tick-borne pathogens.
Although several groups have studied the properties of tick saliva, most studies have focused on profiling the sialome through proteomics, transcriptomics, or in the functional characterization of immunomodulatory or structural proteins (i.e., cement proteins), with little attention given to small RNAs (Madden et al., 2004; Perner et al., 2018; Martins et al., 2021). Small RNAs can be categorized as piwi-interacting RNA (piRNA), small interfering RNA (siRNA), microRNAs (miRNAs), and non-canonical small noncoding RNAs (sncRNAs), such as transfer RNAs (tRNAs), Y small RNAs, and others (Shi et al., 2022). From these small RNAs, miRNAs are the best studied due to their role in immune regulation and disease pathology. miRNAs are involved in post-transcriptional gene regulation to modulate homeostasis and respond to infection or environmental changes (Filipowicz et al., 2008; Hobert, 2008). In ticks, the role and characterization of miRNAs has just begun and there is limited understanding of their contribution in regulating tick biological processes. Even though it has not been shown experimentally, in silico analyses suggest that saliva-derived miRNAs may target transcripts of proteins involved in immune regulation in the host, potentially affecting their expression (Hackenberg et al., 2017). Recently, miRNAs were detected within tick salivary extracellular vesicles (EVs) secreted in the saliva of Haemaphysalis longicornis (Nawaz et al., 2020). The presence of miRNAs within the tick salivary EVs suggests that salivary miRNAs may act in the manipulation of host immune responses. Yet, most studies focused on salivary miRNAs, and EV-derived tick miRNAs, have been limited to profiling and putative function characterization. Therefore, the actual function of most of these miRNAS remains to be investigated. Similarly, how miRNAs are synthesized and the molecular mechanisms behind miRNAs loading into tick EVs are also largely unknown. This review discusses what is known about miRNA transport and packing into EVs in human cells as well as the tick miRNA profiles described to date. Lastly, we will finish with a discussion of potential avenues to exploit tick miRNAs.
EVs have been studied in the context of cancer research and infectious diseases for their role in cell-cell communication (van Niel et al., 2018). EVs are categorized into three classes: exomeres (0–50 nm), exosomes (50–150 nm), and microvesicles (150 nm-1 µm) (Zhang et al., 2018). Exosomes are formed via the inward budding of multivesicular bodies (MVB), whereas microvesicles are produced by the outward budding and fission of the plasma membrane (Sedgwick and D'Souza-Schorey, 2018). How exomeres are formed is poorly understood (Zijlstra et al., 2018). Because the focus of this review is about the transport of miRNAs and their function, we will not describe the biogenesis of EVs further. For a comprehensive review of EV biogenesis and their cargo, refer to van Niel et al. (2018); Sedgwick and D'Souza-Schorey (2018).
EVs have the capability of transferring intracellular cargo from a donor cell to a recipient cell and are involved in interspecies interactions, such as plant-fungal, vector-pathogens, and host-parasite (Mu et al., 2014; Jurkoshek et al., 2016; Oliva Chávez et al., 2019; Freitas et al., 2020). In ticks, recent studies have shown that salivary derived EVs can manipulate skin host immunity and wound healing responses to promote successful feeding (Zhou et al., 2020; Oliva Chávez et al., 2021; Butler et al., 2023). Oliva Chávez et al. (2021) demonstrated that tick-derived EVs interact with immune cells and affect murine resident γδ T-cell migration and proliferation within the epidermis of mice. These cells belong to the dendritic epidermal T cells (DETCs). The knock-down of the EV biogenesis resulted in deficient tick feeding, which was rescued when the ticks fed on γδ T-cell deficient mice. Interestingly, exosomes isolated from the salivary glands and saliva of Amblyomma maculatum and Ixodes scapularis delayed wound healing during in vitro experiments with the human epidermal keratinocyte (HaCaT) cell line (Zhou et al., 2020). Upon injury, keratinocytes and γδ T-cells play critical roles in inflammation and wound healing responses. Moreover, γδ T-cells are essential for maintaining keratinocyte’s homeostasis in the epidermis (Chen et al., 2021). Even though it is currently unknown what specific molecules within EVs are involved in the suppression of inflammatory responses and the alteration of cellular homeostasis in the host, in vivo and in vitro experiments have shown that tick salivary EVs prevent wound healing and allow attached ticks to feed unabated (Zhou et al., 2020; Oliva Chávez et al., 2021). Further, the inoculation of tick salivary EVs along with the intracellular pathogen Anaplasma phagocytophilum enhanced its colonization and establishment in the skin (Oliva Chávez et al., 2021) and tick cell EVs assist in the translocation of Langat virus in in vitro models (Zhou et al., 2018). Thus, defining EV derived molecules that act in the regulation of host responses and identifying the molecular mechanisms for their packing within salivary EVs has important public health implications.
miRNAs are small non-coding RNAs (snRNAs), ∼18–22 nucleotides in length, that are critical for regulating gene expression post-transcriptionally. miRNAs can inhibit protein translation or degrade target messenger RNA (mRNA) by binding to complementary sequences within the 3′ untranslated region (UTR) of mRNAs (Lu and Rothenberg, 2018). It is reported that miRNAs may regulate ∼60% of genes in humans and other mammals (Catalanotto et al., 2016; Shu et al., 2017). The biogenesis of miRNAs begins in the nucleus when a miRNA coding gene is transcribed into primary miRNAs (pri-miRNA, >1 kilobases) by RNA polymerase II (Lu and Rothenberg, 2018). Drosha and DiGeorge syndrome critical region 8 (DGCR or pasha), RNase III endoribonucleases, will cleave the pri-miRNAs into a miRNA precursor (pre-miRNA, ∼60–70 nt). The cleaving of pre-miRNAs by Drosha and DGCR leaves extended 5′phosphate and 3′ nucleotide overhangs (Kim et al., 2016; Sheng et al., 2018). The pre-miRNA is then transported to the cytoplasm by the Exportin 5 (XP05)/Ras-related nuclear protein GTPase (Ran-GTP) complex. The precursor extensions and loop structure is recognized and cleaved by DICER1, leaving a mature miRNA duplex known as the miRNA:miRNA* complex (miRNA = mature strand; miRNA* = guide strand (Yi et al., 2003; Kim et al., 2016). The mature duplex is transported into the RNA induced silencing complex (RISC) by the family of Argonaut (Ago) proteins, where the guide strand is degraded and only the mature strand is left (∼18–22 nucleotides). The Argonaut-miRNA duo is redirected to either repress or degrade the target mRNA (Sarshad et al., 2018; Iwakawa and Tomari, 2022). Conversely, how miRNAs are synthesized in ticks has yet to be defined. Studies assessing antiviral activity of the RNA interference (RNAi) pathway in ticks has shown that it is functional (Schnettler et al., 2014). Putative tick homologs of Drosophila melanogaster Ago-1 and Ago-2 have been identified in Rhipicephalus (Boophilus) microplus and I. scapularis (de la Fuente et al., 2007; Schnettler et al., 2014). Additionally, Dicer-1 and Dicer-2 homologs have been identified in I. scapularis (Schnettler et al., 2014). However, the involvement of these proteins in miRNA mediated interference has not been tested, but this canonical pathway is at least partially conserved in ticks as compared to Drosophila and humans (de la Fuente et al., 2007) (Figure 1).
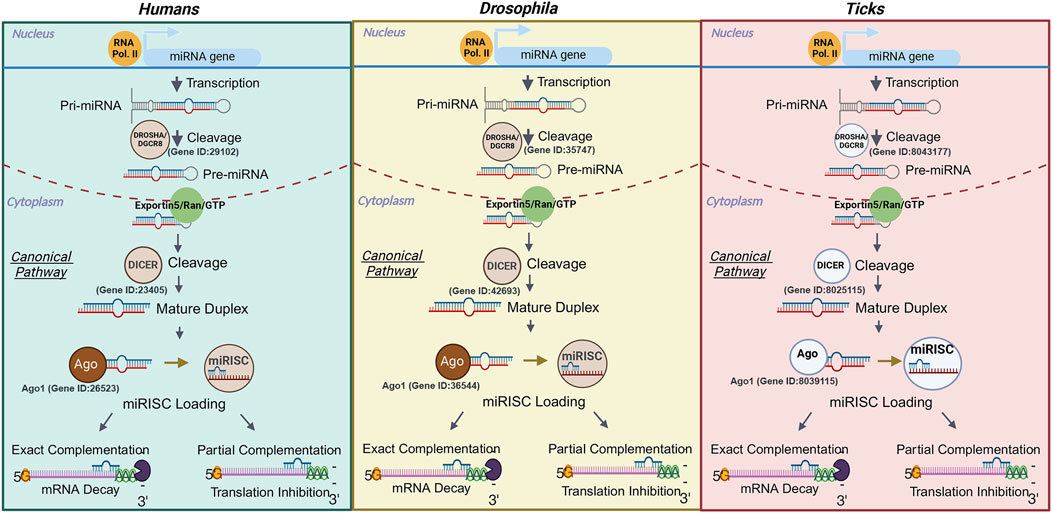
Figure 1. Comparison of the miRNA biogenesis canonical pathway in humans, Drosophila, and ticks. The transcription of a miRNA gene is completed by RNA polymerase II, which leads to the production of a miRNA. The primary miRNA is cleaved into a miRNA precursor by Drosha/DGCR8 and then exported out of the nucleus via Exportin 5/Ran/GTP. Next the precursor is cleaved by Dicer and forms a mature duplex that will be loaded into the miRNA Induced Silencing Complex by an Argonaut protein. The guide strand is then degraded, and the mature miRNA sequence will be translocated to its mRNA target. The exact complementation between the mRNA and miRNA leads to the degradation of the transcript, while the partial match represses translation. Ribonuclease III enzymes represented in dark or light brown have been functionally characterized in the species whereas their representation in white signifies that their role in silencing remains to be tested in members of this group. The ID of the gene encoding each of the ribonucleases has been added to each organism. In the case of ticks, the gene IDs from the Ixodes scapularis genome were used based on the results from Schnettler et al. (2014). Only Dicer 1 and Ago 1 IDs are provided as these proteins are involved in the miRNA silencing pathway. The IDs for Dicer 2 and Ago 2 in I. scapularis have been described in Schnettler et al. (2014). Images were created with BioRender.
Likewise, how miRNAs are packed within tick salivary exosomes for secretion is currently unknown. In human cells, the localization of miRNAs is determined by specific motifs that are recognized by transport proteins, which aid in their translocation into EVs (Lee et al., 2019; Groot and Lee, 2020; de Abreu et al., 2021). Villarroya-Beltri et al. (2013) demonstrated that when heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) undergoes SUMOylation, it facilitates the sorting of exosomal miRNAs through a GGAG/UGCA motif. Another member of the hnRNP family, Synaptotagmin-binding cytoplasmic RNA-interaction protein (SYNCRIP), also selectively sorted miRNAs containing a hEXOmotif GGCU (Santangelo et al., 2016; Hobor et al., 2018). Interestingly, these proteins do not appear to work in parallel, where SYNCRIP would not bind to miRNAs with the GGAG EXOmotif and hnRNPa2b1 would not bind with miRNAs containing the hEXOmotif GGCU (Santangelo et al., 2016). These findings suggest that RNA-binding proteins have a high level of specificity and each protein binds to unique EXOmotifs. A detailed review of the pathways involved in the miRNA sorting into exosomes in humans can be found in Xin et al. (2022). Interestingly, SYNCRIP is highly conserved between humans and arthropods, such as D. melanogaster, and domains important for RNA binding are present with high degree of similarity in these homologs (Hobor et al., 2018). This protein was recently shown to regulate the translation of mRNAs involved in synaptic function; however, this effect is due to direct binding of the protein to mRNAs (McDermott et al., 2014) rather than by the translocation of miRNAs into EVs. Likewise, a homolog of hnRNPa2b1, Hrb98DE, has been identified in Drosophila (Li et al., 2016). Nonetheless, the involvement of these proteins in the transport of miRNAs into Drosophila exosomes remains to be determined. Similarly, RNA -binding protein Musashi (B7Q6Y7), a homolog of human hnRNPa2b1, has been detected by proteomic analysis of EVs from I. scapularis salivary gland ex vivo cultures (Oliva Chávez et al., 2021). The presence of B7Q6Y7 within salivary vesicles could indicate an involvement of this protein in the translocation of exosomal tick miRNAs, warranting further investigation (Figure 2).
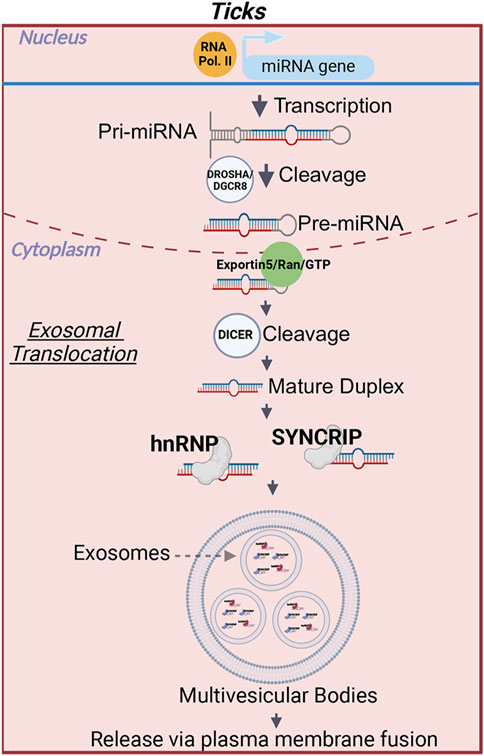
Figure 2. miRNA translocation into exosomes in ticks. After the cleavage of a precursor miRNA into a mature miRNA a motif in the seed sequence is recognized by RNA binding proteins, such as heterogenous nuclear ribonuclear protein A2B1 (hnRNPa2b1) and Synaptotagmin Binding Cytoplasmic RNA Interacting Protein (SYNCRIP). Bound miRNAs are loaded into exosomes. The exosomes will then be released when the multivesicular bodies fuse with the plasma membrane. The RNA binding proteins depicted in this figure have been shown to participate in miRNA translocation into exosomes in humans. Whether these proteins complete the same function in ticks remains to be determined. Image was created with BioRender.
In mammalian macrophages, Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) protein Syntaxin 5 also binds to miRNAs during the biosynthesis of exosomes and facilitates their export outside of the cell (Hom Choudhury et al., 2024). Human antigen R protein transfers miRNAs into a disordered domain in Syntaxin 5 that binds to miRNAs, such as let-7a and miR-122. Syntaxin 5 is then packed into exosomes along with these miRNAs, leading to their secretion outside the cell (Hom Choudhury et al., 2024). In ticks, SNARE proteins have been shown to assist in the secretion of tick salivary proteins (Karim et al., 2002; Karim et al., 2005) and tick derived exosomes (Oliva Chávez et al., 2021). Several SNARE proteins have been identified, including synaptobrevin-2, syntaxin-1A, syntaxin-2, SNAP-25, and Vamp33 (Karim et al., 2002; Oliva Chávez et al., 2021). By comparison, transport of miRNAs into microvesicles is less well defined, but experiments involving tumor cell derived microvesicles suggests that miRNA and pre-miRNA translocation is mediated by the interaction of the small GTPase ADP-ribosylation factor 6 (ARF6) and Exportin-5 (Muralidharan-Chari et al., 2009). Nonetheless, whether any of these mechanisms are involved in the trafficking of miRNAs into tick salivary vesicles remains to be determined. Corrado et al. (2021) and Groot and Lee (2020) expound on the mechanisms and details of miRNA transport and sorting.
Because few studies have functionally characterized miRNAs in ticks, profiling can be used as a baseline to provide insight into their putative roles. A miRNA profile includes both novel, or organism specific, and conserved miRNAs. In the case of conserved miRNAs, their potential function can be inferred from their known role in the biology of other organisms or by looking at their expression profiles in specific organs, life-stages, tick species, during infection with tick-borne pathogens or entomopathogenic microbes, and in response to environmental stressors. For example, miRNA expression profiles of Dermacentor silvarum females exposed to four different temperatures, 8°C, 4°C, 0°C, −4°C, were compared during a 10-day treatment (Agwunobi et al., 2022). MiR-2a expression peaked at day 6 at 4°C and 8°C, whereas miR-279 was lowest at day 3 at all four temperatures. To understand their potential role in cold tolerance, these miRNAs were inhibited, altering the expression of its target genes, like glycogen phosphorylase (GPase) (Agwunobi et al., 2022). The inhibition of miR-2a and miR-279 significantly increased the mortality rates of ticks exposed to −22°C for 2 h in a thermostatic bath and then placed in a normal incubator to recover for 24 h, confirming that these miRNAs contribute to cold tolerance in D. silvarum (Agwunobi et al., 2022). A list of studies done to characterize the miRNA profiles within different ticks can be found in Table 1.
Even though miRNAs have been identified in various tick species under different conditions (Table 1), only one study has profiled miRNAs-EVs in H. longicornis saliva. Nawaz et al. (2020) identified 36 conserved and 34 novel miRNAs-EVs after 4 days of feeding. Among miRNAs of interest, miR-315 was the most abundant miRNA in salivary EVs. This miRNA has previously been reported in the saliva of several arthropod species, including Aedes aegypti, Ae. albopictus, and Anopheles coluzzi (Maharaj et al., 2015; Feng et al., 2018; Liu et al., 2020; Nawaz et al., 2020). Curiously, miR-315 was among the ten most abundant miRNAs in H. longicornis saliva yet was not found in high abundance in the salivary glands of fed H. longicornis adults (Malik et al., 2019a). This suggests that miR-315 is potentially bound to transport proteins or EVs and released into the saliva from the salivary glands during active feeding. Cellular depletion of EV secreted miRNAs is observed in macrophages (Hom Choudhury et al., 2024). Even though the function of miR-315 has not been characterized in any of the vector species listed above, it has been shown that miR-315 regulates the expression of Notum and Axin, two proteins involved in the repression of the Wnt/Wg signaling pathway in Drosophila. This repression of Wnt/Wg signaling leads to the development of wings from the notum in Drosophila (Silver et al., 2007). In mammals, Wnt signaling affects the proliferation phase during skin wound repair by regulating keratinocyte migration, proliferation, and differentiation (Fathke et al., 2006; Zhang et al., 2015; Takaya et al., 2022). Activation of Wnt results in the phosphorylation of Dishevelled (Dsh) that binds Axin, leading to the accumulation of β-catenin in the cytoplasm (Kan et al., 2020). β-catenin is a protein involved in cell adhesion; however, it can act as a transcriptional regulator (Kim et al., 2017). The accumulation of β-catenin leads to its translocation to the nucleus where it activates keratinocyte proliferation (Bai et al., 2023). Yet, increased β-catenin in the nucleus of keratinocytes decreases wound healing by reducing keratinocyte migration (Stojadinovic et al., 2005).
Tick salivary exosomes from I. scapularis and A. maculatum delayed wound closure during in vitro tests using HaCaT cells, a human keratinocyte cell line. The molecular mechanism appears to be connected to the downregulation of CXCL12, which affects keratinocyte migration (Zhou et al., 2020). Activation of the canonical Wnt signaling pathway in stromal bone marrow cells reduces the expression and secretion of CXCL12 and decreases cell migration (Tamura et al., 2011). Given that miR-315 is found within the saliva of two different hematophagous arthropods and it is highly abundant in tick saliva and EVs (Malik et al., 2019a; Nawaz et al., 2020), it is highly likely this miRNA could target Axin in keratinocytes upregulating the Wnt signaling pathway and decreasing CXCL12 expression. Interestingly, target prediction analysis of genes potentially affected by mir-1_38, mir-252_6, and mir-252_2, miRNAs found within the saliva of Ornithodoros moubata and O. erraticus, showed an enrichment of genes involved in the Wnt signaling pathway and angiogenesis. Gene targets of interest included Chondromodulin (CNMD), TRAF interacting protein with forkhead associated domain (TIF), and Growth hormone secretagogue receptor (GHSR), which are involved in angiogenesis and immune signaling (Cano-Argüelles et al., 2023). Still, whether tick miRNAs and EV-miRNAs are involved in the repression of wound healing remains to be determined.
The miRNA seed region, the first 6-8 nucleotides of the mature sequence, or the flanking sequences of the precursor can be highly conserved across various arthropod taxa (Etebari et al., 2015; He et al., 2017; Qu et al., 2017). In some cases, these conserved miRNAs might maintain similar functions or play multiple roles, even within the same organism (Table 2). For instance, Let-7 is a highly conserved miRNA, that clusters with miR-100/-125, and plays a critical part in development, feeding, reproduction and pathogen replication in Ae. Albopictus, Hyalomma asiaticum, and Drosophila (Sokol et al., 2008; Gu et al., 2013; Wu et al., 2019). In H. anatolicum females, miR-1 was consistently expressed throughout each life stage and targeted the gene encoding Heat shock protein 60 (Hsp60) (Luo et al., 2021). When miR-1 was inhibited, the ticks displayed obvious deformities during the later developmental stages and slower engorgement time. Nonetheless, a compensatory effect by close miRNA family members is sometimes observed when another member is inhibited. This compensatory effect is observed by the upregulation of a miRNA that targets the same mRNA as the family member that has been knocked down. These miRNAs are often transcribed within the same miRNA encoding gene but may differ by one nucleotide (Pidíkova et al., 2020; Donnelly et al., 2022). This is a common phenomenon reported in invertebrates and vertebrates (Abbott et al., 2005; Trümbach and Prakash, 2015). This compensatory effect might explain the upregulation of miR-5, a miRNA closely related to miR-1 that also recognizes the same target site in the Hsp60 mRNA (Luo et al., 2021).
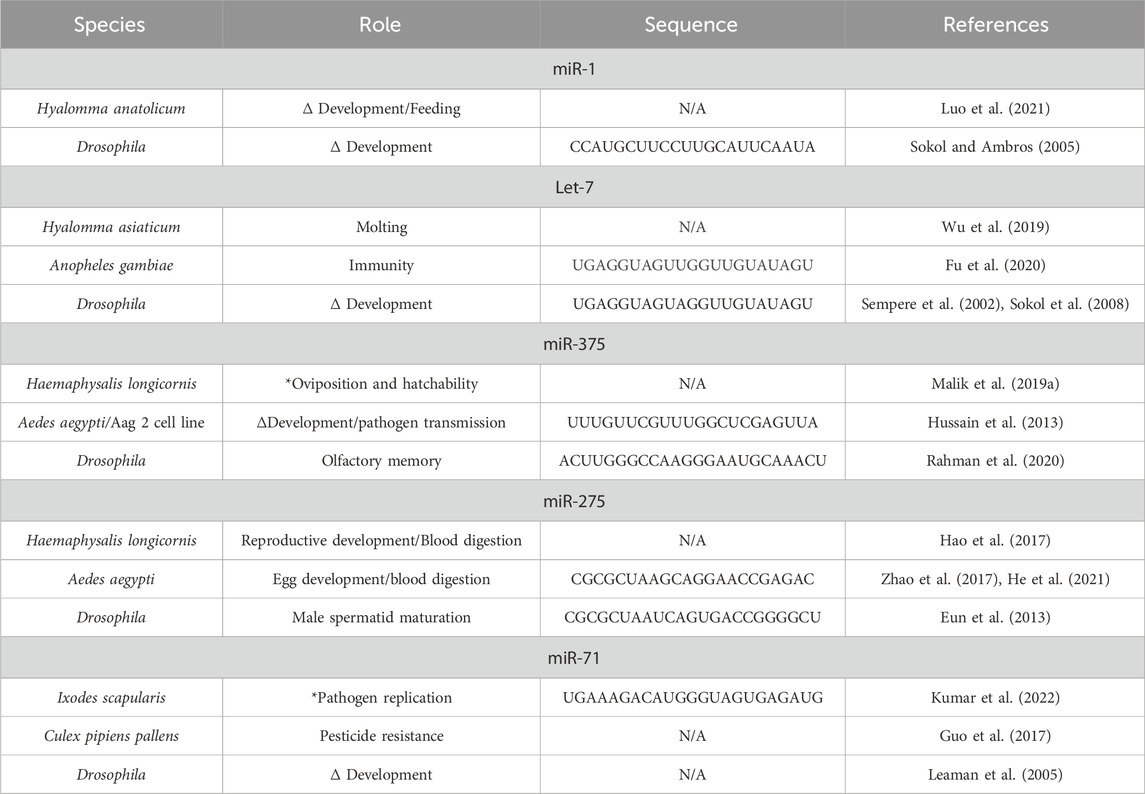
Table 2. Conserved microRNA and their putative or validated roles in hard ticks, mosquitoes, and Drosophila.
Some conserved miRNAs are regulators of biological processes, such as feeding or oviposition among hard ticks. A conserved miRNA, miR-375, is a highly expressed miRNA in the salivary glands, saliva and salivary EVs of H. longicornis and in the saliva of I. ricinus (Hackenberg et al., 2017; Malik et al., 2019a; Nawaz et al., 2020). Oddly, despite lower expressions of this miRNA in the ovaries and midgut, when miR-375 was inhibited in H. longicornis, feeding was not affected, instead resulting in a decrease of egg laying and eclosion (Malik et al., 2019a) (Figure 3). This may indicate miR-375 could directly or indirectly affect oviposition. MiR-375 is a prime example of a miRNA that is conserved across different taxa and has a diverse range of targets. For example, studies have shown that miR-375 can be involved in immunity, insulin secretion, and development in humans, A. aegypti, and Culex pipiens (Hussain et al., 2013; Jafarian et al., 2015; Meuti et al., 2018). A recent study using Ixodes ricinus showed that silencing an insulin receptor and two signaling proteins involved in the insulin signaling pathway, AKT and TOR, resulted in reduced weight during feeding and lower oviposition (Kozelková et al., 2021). Thus, miR-375 may affect several biological processes within ticks by regulating the insulin signaling pathway.
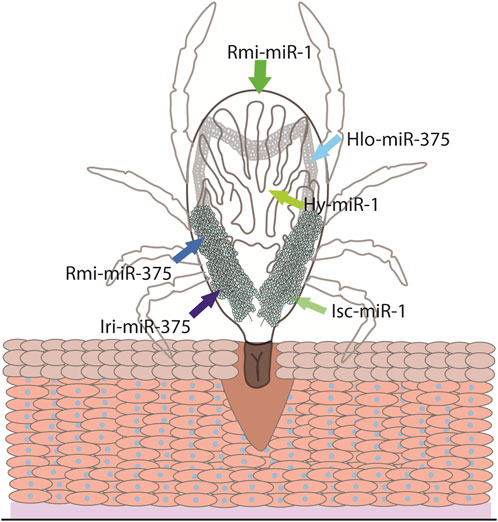
Figure 3. The conserved miRNA-1 and miRNA-375 and their site of expression. The site of expression of miRNA-1 (green arrows) and miRNA-375 (blue arrows) have been investigated in Ixodes scapularis (Isc), Rhipicephalus microplus (Rmi), Hyalomma Anatolicum (Hy), Haemaphysalis longicornis (Hlo), and Ixodes ricinus (Iri).
Unlike conserved miRNAs, novel miRNAs are species specific and share no homology with miRNAs identified in other organisms. In the case of tick specific miRNAs, these miRNAs might regulate biological processes unique to ticks. For instance, in H. asiaticum females, tick-specific miRNA-17 (nov-miR-17) was shown to affect blood-feeding by repressing the expression of the gene coding transforming growth factor-β (TGF-β)-activated kinase 1 binding protein (TAB2) and TGF-beta receptor type I (TβR-I) (Luo et al., 2022a). Injection of ticks with nov-miR-17 agomirs, synthetic mimics of endogenous miRNAs, led to the downregulation of TAB2 and TβR-I, resulting in reduced weight after engorgement. By contrast, antagomir treatment, chemically synthesized oligonucleotides complementary to miRNAs that leads to their silencing (Krützfeldt et al., 2005), led to enhanced expression of TAB2 and TβR-I and increased bloodmeal weight (Luo et al., 2022a). Although the exact molecular mechanism behind TAB2 and TβR-I regulation of tick feeding remains to be further described, TAB2 is a binding partner of the transforming growth factor-β activated kinase 1 (TAK1), part of the Immune Deficiency (IMD) signaling pathway in ticks (Oliva Chávez et al., 2017). Interestingly, the IMD pathway activates the shedding of epithelial cells in Drosophila midguts upon gram-negative bacteria infection (Zhai et al., 2018). Tick digestion occurs intracellularly within midgut epithelial cells that detach from the base membrane during gut cell remodeling (Sojka et al., 2013) and it is well documented that the microbiota of tick midguts is altered upon blood-feeding (Boulanger et al., 2023). Changes in microbiota during blood feeding could lead to the activation of the IMD pathway that may be further regulated by miRNA-17, hence affecting midgut epithelium shedding.
A tick-specific miRNA that appears to be involved in the regulation of development is novel miR-2 (Hlo-miR-2). This miRNA was found to be highly abundant in unfed ticks during all life stages, except in the eggs (Liu et al., 2020). This miRNA is highly expressed in epidermal and midgut tissues. Its target is the cuticular protein 1 (CPR1), which belongs to the most abundant family of insect cuticular proteins that bind to chitin and is present in most arthropods (Cornman et al., 2008). When Hlo-miR-2 was overexpressed, the expression of CPR1 was significantly reduced, resulting in delayed molting and obvious morphological defects in molted nymphs. Blocking miR-2 with antagomirs, on the other hand, reduced molting time in nymphs (Liu et al., 2020), therefore, demonstrating the importance of tick-specific miRNAs in regulating tick biological processes.
During active feeding, ticks and other parasitic arthropods release salivary immunomodulatory compounds into the skin bite site to counteract the host immune system (Šimo et al., 2017; Perner et al., 2018). There is no experimental demonstration of hematophagous arthropods utilizing their salivary miRNAs to manipulate host responses at the bite site. However, in vitro experiments transfecting VeroE6 cells, an epithelial cell line derived from African green monkey kidney, with inhibitors of 9 I. scapularis miRNAs upregulated during Powassan virus (POWV) infection, affected the replication of the virus in the mammalian cells. Transfection of VeroE6 cells with isc-miR-124 and isc-miR-184 inhibitors resulted in decreased viral loads compared to mock transfected cells. By contrast, 6 isc-miRNA inhibitors, including inhibitors against the tick-specific isc-nDS630914_20990, isc-nDS625977_65388, and isc-nDS752087_3745 inhibitors, led to increased viral titers (Hermance et al., 2019). Although, not tested by the authors, it is possible that the isc-miRNA inhibitors interacted with miRNAs in the mammalian host cells, mimicking the potential effect of the tick miRNAs. isc-miRNAs might target the same mRNAs as their mammalian homologs, leading to the repression of mammalian proteins. This phenomenon has not been reported during feeding of hematophagous arthropods in mammalian hosts. However, arthropods infesting plants can regulate plant genes by secreting miRNAs in their saliva. The rice planthopper (Nilaparvata lugens) secretes miR-7–5p, which represses the bZIP transcription factor 43 and plant defenses. This miRNA is injected into rice plants during feeding and its inhibition reduced phloem sap ingestion (Zhang et al., 2024). Thus, arthropod salivary miRNAs are capable of cross-kingdom RNAi interference; yet, whether hematophagous arthropods inject miRNAs that can reduce the expression of host genes remains to be determined.
In silico studies on hematophagous arthropods suggest that tick salivary miRNAs target host genes. Putative target analysis of miRNAs overexpressed in I. ricinus saliva, when compared to salivary glands, showed that saliva overexpressed miRNAs target host physiological responses. Four miRNAs overexpressed in I. ricinus saliva, miR-8–3p, miR-279a–3p, miR-bantam-3, and miR-317-3p potentially regulate mitogen-activated protein kinases (MAPK), like MAPK10 (Hackenberg et al., 2017), a JNK pathway signaling protein previously shown to be regulated by miRNAs and modulate wound healing in mice (Liu et al., 2023). Surprisingly, other mRNAs potentially targeted by these miRNAs included transcripts encoding proteins involved in the mTOR signaling pathway (Hackenberg et al., 2017). A potential similar effect has been predicted in other hematophagous arthropods. miRNAs released within the saliva of A. coluzzi can mimic human endogenous miRNAs to potentially manipulate their host (Arcà et al., 2019). Target prediction analysis of the eight most abundant miRNAs in A. coluzzi saliva identified potential mammalian targets like rapamycin (mTOR), Phosphatidylinositol −4,5-bisphosate 3-kinase catalytic subunit delta (PIK3CD), Fc fragment of IgG receptor IIIb (FCGR3B), and several other proteins involved in T cell regulation and inflammation. Other potential targets for A. coluzzi salivary miRNAs included the transcriptional factors NF-κB, NFAT1, and IRF4, chemokines (such as CCL2 and CCL8), the signaling factor MyD88, and other immune related genes. Interestingly, certain miRNAs identified in mosquito saliva have also been detected in the saliva of I. ricinus, including miR-276-3p, miR-263a–5p, miR-100-5p, and several others (Arcà et al., 2019). Given that salivary miRNAs and their targets are conserved among different arthropod species, it is likely that distinct tick species target similar pathways in different hosts, although the miRNA that is utilized for the regulation of a pathway may differ depending on the tick species. Even though no experimental data has been provided to validate this hypothesis; nonetheless, these studies provide insight into how ticks might potentially use miRNA to regulate host responses and facilitate their uninterrupted feeding.
Due to the importance of miRNAs in gene regulation, development, and host-arthropod interactions, miRNAs are being explored as novel pest management tools against herbivorous insects. Insect feeding or development can be disturbed by delivering miRNAs to disrupt important biological processes in a strategy known as trans-kingdom RNA interference (TK-RNAi) or through the delivery of endogenous miRNAs via viral-vectors, nanoparticles, or exosomes (Bordoloi and Agarwala, 2021; Zhang et al., 2021). In TK-RNAi, artificial miRNAs (amiRNAs) can be introduced into target pests when insects ingest bacteria modified to express endogenous miRNA precursors (Zhang et al., 2021), a technology that could be exploited to reduce blood feeding in livestock. Ticks are pool-feeders that intake blood accumulated at the bite site after cutting small blood vessels in the skin with their chelicerae. Therefore, bacteria from the skin are likely to be ingested during the bloodmeal and modify the microbiome present in ticks. In fact, Boulanger et al. (2023) showed that at least three different genera of bacteria found in the skin of mice, originally detected in low proportions in unfed ticks, increased in prevalence after feeding. Bacteria present in the skin microbiome of animals, like livestock, and shown to be ingested by ticks during feeding, could be genetically modified to express precursors from tick miRNAs that disrupt crucial biological processes, like oviposition or molting. These genetically modified bacteria could then be inoculated into the skin of these animals, potentially reducing tick populations by decreasing egg numbers or lowering ability to molt to their next life stage when ingested with the bloodmeal. Likewise, engineered exosomes could be used to distribute antagomirs targeting miRNAs involved in the immune modulation of the host to reduce tick feeding and pathogen transmission. These vesicles carrying antagomirs could also be used in conjunction with vaccination programs in high infestation areas to increase immune responses in livestock.
On the other hand, tick derived miRNAs could be explored as potential therapeutics. Ticks have evolved salivary compounds to modulate host processes. These compounds can be exploited in the discovery of novel therapeutics for humans and other vertebrates that can be used systemically for the treatment of non-tick related illnesses (Aounallah et al., 2020). One example is Amblyomin-X, a 13.5 kDa protein encoded within the Amblyomma cajennense genome that acts as a Factor Xa (FXa) inhibitor (Batista et al., 2010). This protein reduced viability of tumor cells by inducing ER stress (Morais et al., 2016) and can decrease melanoma size after intra-tumor injection in horses (Lichtenstein et al., 2020). Amblyomin-X is listed as an anti-tumor drug on the NIH National Cancer Institute website (NIH, 2024). Though experimental evidence displaying the effect of miRNAs in host gene regulation is still lacking, the conservation of miRNAs in blood-feeding arthropods (Arcà et al., 2019) and their potential targeting and manipulating host immune, inflammatory, and wound related pathways (Hackenberg et al., 2017; Cano-Argüelles et al., 2023) makes them interesting targets for the development of novel therapeutics for tick-borne and other human diseases. In fact, delivery of miRNAs within exosomes is being explored as an avenue to speed wound closure (Liu et al., 2023), increase the susceptibility of cancer cells to treatment (Lou et al., 2020), reduce aorta inflammation to increase survival during aneurysm (Hu et al., 2022), and other disorders affecting humans. Remarkably, some of the proteins and pathways being focused on for the development of exosome mediated therapeutics in humans are putative targets of tick miRNAs, including mTOR and MAPK10 (Lou et al., 2020; Liu et al., 2023). Thus, profiling of salivary tick miRNAs and EV-derived miRNAs will not only shed light on the biology of tick feeding and their interaction with their host but may also help identify unexplored avenues for the development of human and animal treatments against cancer and other illnesses.
Similarly, the study of other small RNAs within tick saliva and EVs may reveal important information about tick biology, modulation of immune responses in the host, and disease pathogenesis. Paradigm shifting studies have shown that glycosylated (sialylated and fucosylated) small RNAs are found exposed on the outside of the plasma membrane of mammalian cells, particularly Y RNAs, small nuclear RNAs (snuRNAs), small nucleolar RNAs (snoRNAs), and tRNAs (Flynn et al., 2021); populations of small RNAs known to be present within EVs (Dellar et al., 2022). These glycoRNAs play a role in neutrophil recruitment and migration through the endothelium (Zhang et al., 2024) and could potentially enter the secretory system displayed in EV membranes (Nachtergaele and Krishnan, 2021; Dellar et al., 2022). Although Y RNAs have not been reported within tick salivary EVs, tRNAs and snoRNAs have been identified in H. longicornis salivary EVs (Nawaz et al., 2020) and small RNAs between 20–150 nucleotides have been observed I. scapularis salivary EVs (Leal-Galvan et al., 2022). Glycosylation, sialylation, and fucosylation are associated with the development of AGS (Karim et al., 2023) and the entry of A. phagocytophilum, the causative agent of Human Granulocytic Anaplasmosis (HGA), into human, murine, and tick cells (Carlyon et al., 2003; Pedra et al., 2010; Ojogun et al., 2012). Curiously, A. phagocytophilum hijacks the secretory vacuoles where EVs are formed (Read et al., 2022), and it is known to increase EV secretion in tick cells (Oliva Chávez et al., 2021). Whether small RNAs from ticks are modified remains unknown, but its potential implication in AGS and other tick-borne disease pathologies warrants further exploration.
Because of their role in post-transcriptional gene regulation, miRNAs have been explored in various prospective applications ranging from disease diagnosis in humans (Ghamlouche et al., 2023) and in domestic animals (Winter et al., 2021) to the development of novel pest control in plants (Bordoloi and Agarwala, 2021; Zhang et al., 2021). Comparatively, our grasp of the capabilities of small non-coding RNA in regulating tick innate immunity, viral replication, and other biological processes has only begun. Nevertheless, progress in investigating the role of miRNAs in tick biology is held back by the difficulties of working with non-model organisms. The challenges when investigating miRNAs in hard ticks include the lack of well-annotated genomes (Xiong et al., 2020), the compensatory role of miRNA family members that may mask the results from functional analysis (Donnelly et al., 2022), non-coding RNA cross-talk (Tan et al., 2015), shortage of bioinformatic software tailored to non-model organisms (Bortolomeazzi et al., 2019), and presence of unidentified vectored pathogens that may alter miRNA profiles or results.
Several current efforts are trying to increase the number and quality of tick genomes available for bioinformatic analysis (Jia et al., 2020; Yu et al., 2022; De et al., 2023; Nuss et al., 2023). For example, initiatives within the USDA Agricultural Research Services, like the AgPest 100 and Veterinary Pest Genetics Research Unit, are developing high quality genome assemblies via PacBio HiFi reads and HiC contact mapping (I5K, 2021). However, investigators must still be mindful of the BUSCO score that signifies the completeness of the assembly and whether it is assembled at the chromosome or scaffold level (Yandell and Ence, 2012; Seppey et al., 2019). Currently, only the I. scapularis genome (GCA_031841145.1), Rhipicephalus sanguineus (GCA_013339695.2), R. B. microplus (GCA_013339725.1), D. silvarum (GCA_013339745.2) and H. longicornis (GCA_013339765.2) are annotated to the chromosome level with some unplaced scaffolds (Jia et al., 2020; Nuss et al., 2023). However, some tick genomes may not include the annotation of miRNAs, thus, software used to identify miRNAs would only provide the chromosome or genomic coordinates. Tools like miRDeep2 (Friedländer et al., 2008; Friedländer et al., 2012) or sRNAtoolbox (Aparicio-Puerta et al., 2022) identify sequences based on the miRNA biogenesis pathway. Still, these tools might detect false positives. Therefore, stringent filtering protocols to remove the false positives or low-quality miRNAs are needed; especially when reporting novel miRNAs. These improvements in analysis during profiling, in silico identification of miRNA targets, and in vitro and in vivo validation of miRNA function particularly in salivary glands and tick saliva will lead us to identify tick specific miRNAs. This information could potentially be used for the development of novel therapeutics for treatment in human related illnesses and the identification of targets that decrease the immunomodulation caused by ticks. Likewise, the recent finding that tick salivary EVs transport miRNAs (Nawaz et al., 2020) has opened a new array of possible miRNA delivery mechanisms utilized for tick control.
BL-G: Conceptualization, Writing–original draft, Writing–review and editing. DK: Writing–review and editing. SK: Writing–review and editing. PS: Writing–review and editing. DT: Writing–review and editing. AOC: Conceptualization, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was funded by the USDA National Institute of Food and Agriculture (NIFA) Hatch-MultiState Project #TEX0-1-7714, USDA NIFA award #2022-67015-42166, cooperative agreement #58-3094-1-003 by the USDA Agriculture Research Service (ARS), and University of Wisconsin, Madison start-up funds to AOC. BL-G is supported by an ORISE fellowship from the USDA and was a recipient of the Knippling-Bushland-Swahrf fellowship from the Department of Entomology at Texas A&M University.
We thank Dr. Zachary Adelman for the administration of the cooperative agreement #58-3094-1-003 at Texas A&M University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbott, A. L., Alvarez-Saavedra, E., Miska, E. A., Lau, N. C., Bartel, D. P., Horvitz, H. R., et al. (2005). The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9 (3), 403–414. doi:10.1016/j.devcel.2005.07.009
Agwunobi, D. O., Pei, T., Bai, R., Wang, Z., Shi, X., Zhang, M., et al. (2022). miR-2a and miR-279 are functionally associated with cold tolerance in Dermacentor silvarum (Acari: ixodidae). Comp. Biochem. Physiol. Part D. Genomics Proteomics 41, 100946. doi:10.1016/j.cbd.2021.100946
Anderson, J. F., and Magnarelli, L. A. (2008). Biology of ticks. Infect. Dis. Clin. North Am. 22 (2), 195–215. doi:10.1016/j.idc.2007.12.006
Aounallah, H., Bensaoud, C., M'Ghirbi, Y., Faria, F., Chmelar, J. I., and Kotsyfakis, M. (2020). Tick salivary compounds for targeted immunomodulatory therapy. Front. Immunol. 11, 583845. doi:10.3389/fimmu.2020.583845
Aparicio-Puerta, E., Gómez-Martín, C., Giannoukakos, S., Medina, J. M., Scheepbouwer, C., García-Moreno, A., et al. (2022). sRNAbench and sRNAtoolbox 2022 update: accurate miRNA and sncRNA profiling for model and non-model organisms. Nucleic Acids Res. 50 (W1), W710–w717. doi:10.1093/nar/gkac363
Arcà, B., Colantoni, A., Fiorillo, C., Severini, F., Benes, V., Di Luca, M., et al. (2019). MicroRNAs from saliva of anopheline mosquitoes mimic human endogenous miRNAs and may contribute to vector-host-pathogen interactions. Sci. Rep. 9 (1), 2955. doi:10.1038/s41598-019-39880-1
Bai, R., Guo, Y., Liu, W., Song, Y., Yu, Z., and Ma, X. (2023). The roles of WNT signaling pathways in skin development and mechanical-stretch-induced skin regeneration. Biomolecules 13 (12), 1702. doi:10.3390/biom13121702
Bakken, J. S., and Dumler, J. S. (2015). Human granulocytic anaplasmosis. Infect. Dis. Clin. North Am. 29 (2), 341–355. doi:10.1016/j.idc.2015.02.007
Barrero, R. A., Keeble-Gagnère, G., Zhang, B., Moolhuijzen, P., Ikeo, K., Tateno, Y., et al. (2011). Evolutionary conserved microRNAs are ubiquitously expressed compared to tick-specific miRNAs in the cattle tick Rhipicephalus (Boophilus) microplus. BMC Genomics 12, 328. doi:10.1186/1471-2164-12-328
Batista, I. F., Ramos, O. H., Ventura, J. S., Junqueira-de-Azevedo, I. L., Ho, P. L., and Chudzinski-Tavassi, A. M. (2010). A new Factor Xa inhibitor from Amblyomma cajennense with a unique domain composition. Arch. Biochem. Biophys. 493 (2), 151–156. doi:10.1016/j.abb.2009.10.009
Bordoloi, K. S., and Agarwala, N. (2021). MicroRNAs in plant insect interaction and insect pest control. Plant gene. 26, 100271. doi:10.1016/j.plgene.2021.100271
Bortolomeazzi, M., Gaffo, E., and Bortoluzzi, S. (2019). A survey of software tools for microRNA discovery and characterization using RNA-seq. Brief. Bioinform 20 (3), 918–930. doi:10.1093/bib/bbx148
Boulanger, N., Insonere, J. L., Van Blerk, S., Barthel, C., Serres, C., Rais, O., et al. (2023). Cross-alteration of murine skin and tick microbiome concomitant with pathogen transmission after Ixodes ricinus bite. Microbiome 11 (1), 250. doi:10.1186/s40168-023-01696-7
Bradshaw, M. J., Byrge, K. C., Ivey, K. S., Pruthi, S., and Bloch, K. C. (2020). Meningoencephalitis due to spotted fever rickettsioses, including Rocky Mountain Spotted fever. Clin. Infect. Dis. 71 (1), 188–195. doi:10.1093/cid/ciz776
Butler, L. R., Gonzalez, J., Pedra, J. H. F., and Oliva Chavez, A. S. (2023). Tick extracellular vesicles in host skin immunity and pathogen transmission. Trends Parasitol. 39 (10), 873–885. doi:10.1016/j.pt.2023.07.009
Cano-Argüelles, A. L., Pérez-Sánchez, R., and Oleaga, A. (2023). A microRNA profile of the saliva in the argasid ticks Ornithodoros erraticus and Ornithodoros moubata and prediction of specific target genes. Ticks Tick. Borne Dis. 14 (6), 102249. doi:10.1016/j.ttbdis.2023.102249
Carlyon, J. A., Akkoyunlu, M., Xia, L., Yago, T., Wang, T., Cummings, R. D., et al. (2003). Murine neutrophils require alpha1,3-fucosylation but not PSGL-1 for productive infection with Anaplasma phagocytophilum. Blood 102 (9), 3387–3395. doi:10.1182/blood-2003-02-0621
Catalanotto, C., Cogoni, C., and Zardo, G. (2016). MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 17 (10), 1712. doi:10.3390/ijms17101712
CDC (2024a). Alpha-gal syndrome. Available at: https://www.cdc.gov/alpha-gal-syndrome/about/index.html (Accessed August 26, 2024).
CDC (2024b). Tickborne disease surveillance data summary. Available at: https://www.cdc.gov/ticks/data-research/facts-stats/tickborne-disease-surveillance-data-summary.html (Accessed July 9, 2024).
Chen, C., Meng, Z., Ren, H., Zhao, N., Shang, R., He, W., et al. (2021). The molecular mechanisms supporting the homeostasis and activation of dendritic epidermal T cell and its role in promoting wound healing. Burns Trauma 9, tkab009. doi:10.1093/burnst/tkab009
Cornman, R. S., Togawa, T., Dunn, W. A., He, N., Emmons, A. C., and Willis, J. H. (2008). Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles gambiae. BMC Genomics 9, 22. doi:10.1186/1471-2164-9-22
Corrado, C., Barreca, M. M., Zichittella, C., Alessandro, R., and Conigliaro, A. (2021). Molecular mediators of RNA loading into extracellular vesicles. Cells 10 (12), 3355. doi:10.3390/cells10123355
De, S., Kingan, S. B., Kitsou, C., Portik, D. M., Foor, S. D., Frederick, J. C., et al. (2023). A high-quality Ixodes scapularis genome advances tick science. Nat. Genet. 55 (2), 301–311. doi:10.1038/s41588-022-01275-w
de Abreu, R. C., Ramos, C. V., Becher, C., Lino, M., Jesus, C., da Costa Martins, P. A., et al. (2021). Exogenous loading of miRNAs into small extracellular vesicles. J. Extracell. Vesicles 10 (10), e12111. doi:10.1002/jev2.12111
de Castro, J. J., James, A. D., Minjauw, B., Di Giulio, G. U., Permin, A., Pegram, R. G., et al. (1997). Long-term studies on the economic impact of ticks on Sanga cattle in Zambia. Exp. Appl. Acarol. 21 (1), 3–19. doi:10.1023/a:1018450824854
de la Fuente, J., Kocan, K. M., Almazán, C., and Blouin, E. F. (2007). RNA interference for the study and genetic manipulation of ticks. Trends Parasitol. 23 (9), 427–433. doi:10.1016/j.pt.2007.07.002
Dellar, E. R., Hill, C., Melling, G. E., Carter, D. R. F., and Baena-Lopez, L. A. (2022). Unpacking extracellular vesicles: RNA cargo loading and function. J. Extracell. Biol. 1 (5), e40. doi:10.1002/jex2.40
Donnelly, B. F., Yang, B., Grimme, A. L., Vieux, K. F., Liu, C. Y., Zhou, L., et al. (2022). The developmentally timed decay of an essential microRNA family is seed-sequence dependent. Cell Rep. 40 (6), 111154. doi:10.1016/j.celrep.2022.111154
Drexler, N. A., Yaglom, H., Casal, M., Fierro, M., Kriner, P., Murphy, B., et al. (2017). Fatal Rocky Mountain Spotted fever along the United States-Mexico border, 2013-2016. Emerg. Infect. Dis. 23 (10), 1621–1626. doi:10.3201/eid2310.170309
Etebari, K., Osei-Amo, S., Blomberg, S. P., and Asgari, S. (2015). Dengue virus infection alters post-transcriptional modification of microRNAs in the mosquito vector Aedes aegypti. Sci. Rep. 5, 15968. doi:10.1038/srep15968
Eun, S. H., Stoiber, P. M., Wright, H. J., McMurdie, K. E., Choi, C. H., Gan, Q., et al. (2013). MicroRNAs downregulate Bag of marbles to ensure proper terminal differentiation in the Drosophila male germline. Development 140 (1), 23–30. doi:10.1242/dev.086397
Fathke, C., Wilson, L., Shah, K., Kim, B., Hocking, A., Moon, R., et al. (2006). Wnt signaling induces epithelial differentiation during cutaneous wound healing. BMC Cell Biol. 7, 4. doi:10.1186/1471-2121-7-4
Feng, X., Zhou, S., Wang, J., and Hu, W. (2018). microRNA profiles and functions in mosquitoes. PLoS Negl. Trop. Dis. 12 (5), e0006463. doi:10.1371/journal.pntd.0006463
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9 (2), 102–114. doi:10.1038/nrg2290
Flynn, R. A., Pedram, K., Malaker, S. A., Batista, P. J., Smith, B. A. H., Johnson, A. G., et al. (2021). Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184 (12), 3109–3124.e22. doi:10.1016/j.cell.2021.04.023
Freitas, M. N., Marten, A. D., Moore, G. A., Tree, M. O., McBrayer, S. P., and Conway, M. J. (2020). Extracellular vesicles restrict dengue virus fusion in Aedes aegypti cells. Virology 541, 141–149. doi:10.1016/j.virol.2019.12.010
Friedländer, M. R., Chen, W., Adamidi, C., Maaskola, J., Einspanier, R., Knespel, S., et al. (2008). Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26 (4), 407–415. doi:10.1038/nbt1394
Friedländer, M. R., Mackowiak, S. D., Li, N., Chen, W., and Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40 (1), 37–52. doi:10.1093/nar/gkr688
Fu, X., Liu, P., Dimopoulos, G., and Zhu, J. (2020). Dynamic miRNA-mRNA interactions coordinate gene expression in adult Anopheles gambiae. PLoS Genet. 16 (4), e1008765. doi:10.1371/journal.pgen.1008765
Ghamlouche, F., Yehya, A., Zeid, Y., Fakhereddine, H., Fawaz, J., Liu, Y. N., et al. (2023). MicroRNAs as clinical tools for diagnosis, prognosis, and therapy in prostate cancer. Transl. Oncol. 28, 101613. doi:10.1016/j.tranon.2022.101613
Groot, M., and Lee, H. (2020). Sorting mechanisms for MicroRNAs into extracellular vesicles and their associated diseases. Cells 9 (4), 1044. doi:10.3390/cells9041044
Gu, J., Hu, W., Wu, J., Zheng, P., Chen, M., James, A. A., et al. (2013). miRNA genes of an invasive vector mosquito, Aedes albopictus. PLoS One 8 (7), e67638. doi:10.1371/journal.pone.0067638
Guo, Q., Huang, Y., Zou, F., Liu, B., Tian, M., Ye, W., et al. (2017). The role of miR-2∼13∼71 cluster in resistance to deltamethrin in Culex pipiens pallens. Insect Biochem. Mol. Biol. 84, 15–22. doi:10.1016/j.ibmb.2017.03.006
Hackenberg, M., Langenberger, D., Schwarz, A., Erhart, J., and Kotsyfakis, M. (2017). In silico target network analysis of de novo-discovered, tick saliva-specific microRNAs reveals important combinatorial effects in their interference with vertebrate host physiology. Rna 23 (8), 1259–1269. doi:10.1261/rna.061168.117
Hao, J., Luo, J., Chen, Z., Ren, Q., Guo, J., Liu, X., et al. (2017). MicroRNA-275 and its target Vitellogenin-2 are crucial in ovary development and blood digestion of Haemaphysalis longicornis. Parasit. Vectors 10 (1), 253. doi:10.1186/s13071-017-2153-1
He, R., Gu, X., Lai, W., Peng, X., and Yang, G. (2017). Transcriptome-microRNA analysis of Sarcoptes scabiei and host immune response. PLoS One 12 (5), e0177733. doi:10.1371/journal.pone.0177733
He, Y. Z., Aksoy, E., Ding, Y., and Raikhel, A. S. (2021). Hormone-dependent activation and repression of microRNAs by the ecdysone receptor in the dengue vector mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 118 (26), e2102417118. doi:10.1073/pnas.2102417118
Hermance, M. E., and Thangamani, S. (2015). Tick saliva enhances powassan virus transmission to the host, influencing its dissemination and the course of disease. J. Virol. 89 (15), 7852–7860. doi:10.1128/jvi.01056-15
Hermance, M. E., Widen, S. G., Wood, T. G., and Thangamani, S. (2019). Ixodes scapularis salivary gland microRNAs are differentially expressed during Powassan virus transmission. Sci. Rep. 9 (1), 13110. doi:10.1038/s41598-019-49572-5
Hobert, O. (2008). Gene regulation by transcription factors and microRNAs. Science 319 (5871), 1785–1786. doi:10.1126/science.1151651
Hobor, F., Dallmann, A., Ball, N. J., Cicchini, C., Battistelli, C., Ogrodowicz, R. W., et al. (2018). A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets. Nat. Commun. 9 (1), 831. doi:10.1038/s41467-018-03182-3
Hom Choudhury, S., Bhattacharjee, S., Mukherjee, K., and Bhattacharyya, S. N. (2024). Human antigen R transfers miRNA to Syntaxin 5 to synergize miRNA export from activated macrophages. J. Biol. Chem. 300 (4), 107170. doi:10.1016/j.jbc.2024.107170
Hu, J., Jiang, Y., Wu, X., Wu, Z., Qin, J., Zhao, Z., et al. (2022). Exosomal miR-17-5p from adipose-derived mesenchymal stem cells inhibits abdominal aortic aneurysm by suppressing TXNIP-NLRP3 inflammasome. Stem Cell Res. Ther. 13 (1), 349. doi:10.1186/s13287-022-03037-1
Hussain, M., Walker, T., O'Neill, S. L., and Asgari, S. (2013). Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 43 (2), 146–152. doi:10.1016/j.ibmb.2012.11.005
I5K (2021). Ag100Pest initiative. San Francisco, California: github. Available at: https://i5k.github.io/ag100pest (Accessed July 5, 2024).
Iwakawa, H. O., and Tomari, Y. (2022). Life of RISC: formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 82 (1), 30–43. doi:10.1016/j.molcel.2021.11.026
Jafarian, A., Taghikani, M., Abroun, S., Allahverdi, A., Lamei, M., Lakpour, N., et al. (2015). The generation of insulin producing cells from human mesenchymal stem cells by MiR-375 and anti-MiR-9. PLoS One 10 (6), e0128650. doi:10.1371/journal.pone.0128650
Jia, N., Wang, J., Shi, W., Du, L., Sun, Y., Zhan, W., et al. (2020). Large-scale comparative analyses of tick genomes elucidate their genetic diversity and vector capacities. Cell 182 (5), 1328–1340.e13. doi:10.1016/j.cell.2020.07.023
Jurkoshek, K. S., Wang, Y., Athman, J. J., Barton, M. R., and Wearsch, P. A. (2016). Interspecies communication between pathogens and immune cells via bacterial membrane vesicles. Front. Cell Dev. Biol. 4, 125. doi:10.3389/fcell.2016.00125
Kan, W., Enos, M. D., Korkmazhan, E., Muennich, S., Chen, D. H., Gammons, M. V., et al. (2020). Limited dishevelled/Axin oligomerization determines efficiency of Wnt/β-catenin signal transduction. Elife 9, e55015. doi:10.7554/eLife.55015
Karim, S., Essenberg, R. C., Dillwith, J. W., Tucker, J. S., Bowman, A. S., and Sauer, J. R. (2002). Identification of SNARE and cell trafficking regulatory proteins in the salivary glands of the lone star tick, Amblyomma americanum (L.). Insect Biochem. Mol. Biol. 32 (12), 1711–1721. doi:10.1016/s0965-1748(02)00111-x
Karim, S., Leyva-Castillo, J. M., and Narasimhan, S. (2023). Tick salivary glycans - a sugar-coated tick bite. Trends Parasitol. 39 (12), 1100–1113. doi:10.1016/j.pt.2023.09.012
Karim, S., Miller, N. J., Valenzuela, J., Sauer, J. R., and Mather, T. N. (2005). RNAi-mediated gene silencing to assess the role of synaptobrevin and cystatin in tick blood feeding. Biochem. Biophys. Res. Commun. 334 (4), 1336–1342. doi:10.1016/j.bbrc.2005.07.036
Kim, K., Cho, J., Hilzinger, T. S., Nunns, H., Liu, A., Ryba, B. E., et al. (2017). Two-element transcriptional regulation in the canonical Wnt pathway. Curr. Biol. 27 (15), 2357–2364. doi:10.1016/j.cub.2017.06.037
Kim, Y. K., Kim, B., and Kim, V. N. (2016). Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 113 (13), E1881–E1889. doi:10.1073/pnas.1602532113
Kozelková, T., Doležel, D., Grunclová, L., Kučera, M., Perner, J., and Kopáček, P. (2021). Functional characterization of the insulin signaling pathway in the hard tick Ixodes ricinus. Ticks Tick. Borne Dis. 12 (4), 101694. doi:10.1016/j.ttbdis.2021.101694
Krützfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs in vivo with 'antagomirs. Nature 438 (7068), 685–689. doi:10.1038/nature04303
Kumar, D., Downs, L. P., Embers, M., Flynt, A. S., and Karim, S. (2022). Identification of microRNAs in the Lyme disease vector Ixodes scapularis. Int. J. Mol. Sci. 23 (10), 5565. doi:10.3390/ijms23105565
Leal-Galvan, B., Harvey, C., Thomas, D., Saelao, P., and Oliva Chavez, A. S. (2022). Isolation of microRNAs from tick ex vivo salivary gland cultures and extracellular vesicles. J. Vis. Exp. 182. doi:10.3791/63618
Leaman, D., Chen, P. Y., Fak, J., Yalcin, A., Pearce, M., Unnerstall, U., et al. (2005). Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121 (7), 1097–1108. doi:10.1016/j.cell.2005.04.016
Lee, H., Li, C., Zhang, Y., Zhang, D., Otterbein, L. E., and Jin, Y. (2019). Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 216 (9), 2202–2220. doi:10.1084/jem.20182313
Li, S., Zhang, P., Freibaum, B. D., Kim, N. C., Kolaitis, R. M., Molliex, A., et al. (2016). Genetic interaction of hnRNPA2B1 and DNAJB6 in a Drosophila model of multisystem proteinopathy. Hum. Mol. Genet. 25 (5), 936–950. doi:10.1093/hmg/ddv627
Lichtenstein, F., Iqbal, A., de Lima Will, S. E. A., Bosch, R. V., DeOcesano-Pereira, C., Goldfeder, M. B., et al. (2020). Modulation of stress and immune response by Amblyomin-X results in tumor cell death in a horse melanoma model. Sci. Rep. 10 (1), 6388. doi:10.1038/s41598-020-63275-2
Lippi, C. A., Ryan, S. J., White, A. L., Gaff, H. D., and Carlson, C. J. (2021). Trends and opportunities in tick-borne disease geography. J. Med. Entomol. 58 (6), 2021–2029. doi:10.1093/jme/tjab086
Liu, W., Guo, J., Luo, J., Ren, Q., Chen, Z., Qu, Z., et al. (2020a). Analysis of microRNA expression profiles dynamic in different life stages of Haemaphysalis longicornis ticks by deep sequencing of small RNA libraries. Ticks Tick. Borne Dis. 11 (4), 101427. doi:10.1016/j.ttbdis.2020.101427
Liu, W. G., Luo, J., Ren, Q. Y., Qu, Z. Q., Lin, H. L., Xu, X. F., et al. (2020b). A novel miRNA-hlo-miR-2-Serves as a regulatory factor that controls molting events by targeting CPR1 in Haemaphysalis longicornis nymphs. Front. Microbiol. 11, 1098. doi:10.3389/fmicb.2020.01098
Liu, X., Jin, S., Liu, J., and Xu, X. (2023). MiR-223-3p overexpressed adipose mesenchymal stem cell-derived exosomes promote wound healing via targeting MAPK10. Acta histochem. 125 (8), 152102. doi:10.1016/j.acthis.2023.152102
Lou, G., Chen, L., Xia, C., Wang, W., Qi, J., Li, A., et al. (2020). MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J. Exp. Clin. Cancer Res. 39 (1), 4. doi:10.1186/s13046-019-1512-5
Lu, T. X., and Rothenberg, M. E. (2018). MicroRNA. J. Allergy Clin. Immunol. 141 (4), 1202–1207. doi:10.1016/j.jaci.2017.08.034
Luo, J., Liu, G. Y., Chen, Z., Ren, Q. Y., Yin, H., Luo, J. X., et al. (2015). Identification and characterization of microRNAs by deep-sequencing in Hyalomma anatolicum anatolicum (Acari: ixodidae) ticks. Gene 564 (2), 125–133. doi:10.1016/j.gene.2015.01.019
Luo, J., Ren, Q., Chen, Z., Liu, W., Qu, Z., Xiao, R., et al. (2019). Comparative analysis of microRNA profiles between wild and cultured Haemaphysalis longicornis (Acari, Ixodidae) ticks. Parasite 26, 18. doi:10.1051/parasite/2019018
Luo, J., Ren, Q., Liu, W., Qiu, X., Zhang, G., Tan, Y., et al. (2021). MicroRNA-1 expression and function in Hyalomma anatolicum anatolicum (Acari: ixodidae) ticks. Front. Physiol. 12, 596289. doi:10.3389/fphys.2021.596289
Luo, J., Wu, F., Liu, W., Ren, Q., Diao, P., Guan, G., et al. (2022a). A novel MicroRNA and the target gene TAB2 can regulate the process of sucking blood in and the spawn rate of Hyalomma asiaticum (Acari: ixodidae) ticks. Front. Immunol. 13, 930532. doi:10.3389/fimmu.2022.930532
Luo, J., Zhao, S., Ren, Q., Wang, Q., Chen, Z., Cui, J., et al. (2022b). Dynamic analysis of microRNAs from different life stages of Rhipicephalus microplus (Acari: ixodidae) by high-throughput sequencing. Pathogens 11 (10), 1148. doi:10.3390/pathogens11101148
Madden, R. D., Sauer, J. R., and Dillwith, J. W. (2004). A proteomics approach to characterizing tick salivary secretions. Exp. Appl. Acarol. 32 (1-2), 131–141. doi:10.1023/b:appa.0000018241.81636.91
Maharaj, P. D., Widen, S. G., Huang, J., Wood, T. G., and Thangamani, S. (2015). Discovery of mosquito saliva microRNAs during CHIKV infection. PLoS Negl. Trop. Dis. 9 (1), e0003386. doi:10.1371/journal.pntd.0003386
Malik, M. I., Nawaz, M., Hassan, I. A., Zhang, H., Gong, H., Cao, J., et al. (2019a). A microRNA profile of saliva and role of miR-375 in Haemaphysalis longicornis (Ixodida: ixodidae). Parasit. Vectors 12 (1), 68. doi:10.1186/s13071-019-3318-x
Malik, M. I., Nawaz, M., Wang, Y., Zhang, H., Cao, J., Zhou, Y., et al. (2019b). Localized expression and inhibition effect of miR-184 on blood digestion and oviposition in Haemaphysalis longicornis (Acari: ixodidae). Parasit. Vectors 12 (1), 500. doi:10.1186/s13071-019-3754-7
Marchal, C. M., Luft, B. J., Yang, X., Sibilia, J., Jaulhac, B., and Boulanger, N. M. (2009). Defensin is suppressed by tick salivary gland extract during the in vitro interaction of resident skin cells with Borrelia burgdorferi. J. Invest Dermatol 129 (10), 2515–2517. doi:10.1038/jid.2009.73
Martins, L. A., Bensaoud, C., Kotál, J., Chmelař, J., and Kotsyfakis, M. (2021). Tick salivary gland transcriptomics and proteomics. Parasite Immunol. 43 (5), e12807. doi:10.1111/pim.12807
McDermott, S. M., Yang, L., Halstead, J. M., Hamilton, R. S., Meignin, C., and Davis, I. (2014). Drosophila Syncrip modulates the expression of mRNAs encoding key synaptic proteins required for morphology at the neuromuscular junction. Rna 20 (10), 1593–1606. doi:10.1261/rna.045849.114
Meuti, M. E., Bautista-Jimenez, R., and Reynolds, J. A. (2018). Evidence that microRNAs are part of the molecular toolkit regulating adult reproductive diapause in the mosquito, Culex pipiens. PLoS One 13 (11), e0203015. doi:10.1371/journal.pone.0203015
Morais, K. L., Pacheco, M. T., Berra, C. M., Bosch, R. V., Sciani, J. M., Chammas, R., et al. (2016). Amblyomin-X induces ER stress, mitochondrial dysfunction, and caspase activation in human melanoma and pancreatic tumor cell. Mol. Cell Biochem. 415 (1-2), 119–131. doi:10.1007/s11010-016-2683-4
Mu, J., Zhuang, X., Wang, Q., Jiang, H., Deng, Z. B., Wang, B., et al. (2014). Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 58 (7), 1561–1573. doi:10.1002/mnfr.201300729
Muralidharan-Chari, V., Clancy, J., Plou, C., Romao, M., Chavrier, P., Raposo, G., et al. (2009). ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. 19 (22), 1875–1885. doi:10.1016/j.cub.2009.09.059
Nachtergaele, S., and Krishnan, Y. (2021). New vistas for cell-surface GlycoRNAs. N. Engl. J. Med. 385 (7), 658–660. doi:10.1056/NEJMcibr2108679
Nawaz, M., Malik, M. I., Zhang, H., Gebremedhin, M. B., Cao, J., Zhou, Y., et al. (2020). miRNA profile of extracellular vesicles isolated from saliva of Haemaphysalis longicornis tick. Acta Trop. 212, 105718. doi:10.1016/j.actatropica.2020.105718
Nicholson, W. L., Sonenshine, D. E., Noden, B. H., and Brown, R. N. (2019). “Chapter 27 - ticks (ixodida),” in Medical and veterinary Entomology. Editors G. R. Mullen,, and L. A. Durden Third Edition (Academic Press), 603–672.
NIH (2024). NIH amblyomin-X. Available at: https://www.cancer.gov/publications/dictionaries/cancer-drug/def/amblyomin-x (Accessed June 17, 2024).
Nuss, A. B., Lomas, J. S., Reyes, J. B., Garcia-Cruz, O., Lei, W., Sharma, A., et al. (2023). The highly improved genome of Ixodes scapularis with X and Y pseudochromosomes. Life Sci. Alliance 6 (12), e202302109. doi:10.26508/lsa.202302109
Nuttall, P. A. (2023). Tick saliva and its role in pathogen transmission. Wien Klin. Wochenschr 135 (7-8), 165–176. doi:10.1007/s00508-019-1500-y
Ojogun, N., Kahlon, A., Ragland, S. A., Troese, M. J., Mastronunzio, J. E., Walker, N. J., et al. (2012). Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect. Immun. 80 (11), 3748–3760. doi:10.1128/iai.00654-12
Oliva Chávez, A. S., O'Neal, A. J., Santambrogio, L., Kotsyfakis, M., and Pedra, J. H. F. (2019). Message in a vesicle - trans-kingdom intercommunication at the vector-host interface. J. Cell Sci. 132 (6), jcs224212. doi:10.1242/jcs.224212
Oliva Chávez, A. S., Shaw, D. K., Munderloh, U. G., and Pedra, J. H. (2017). Tick humoral responses: marching to the beat of a different drummer. Front. Microbiol. 8, 223. doi:10.3389/fmicb.2017.00223
Oliva Chávez, A. S., Wang, X., Marnin, L., Archer, N. K., Hammond, H. L., Carroll, E. E. M., et al. (2021). Tick extracellular vesicles enable arthropod feeding and promote distinct outcomes of bacterial infection. Nat. Commun. 12 (1), 3696. doi:10.1038/s41467-021-23900-8
Pedra, J. H., Narasimhan, S., Rendić, D., DePonte, K., Bell-Sakyi, L., Wilson, I. B., et al. (2010). Fucosylation enhances colonization of ticks by Anaplasma phagocytophilum. Cell Microbiol. 12 (9), 1222–1234. doi:10.1111/j.1462-5822.2010.01464.x
Perner, J., Kropáčková, S., Kopáček, P., and Ribeiro, J. M. C. (2018). Sialome diversity of ticks revealed by RNAseq of single tick salivary glands. PLoS Negl. Trop. Dis. 12 (4), e0006410. doi:10.1371/journal.pntd.0006410
Pham, M., Underwood, J., and Oliva Chávez, A. S. (2021). Changing the recipe: pathogen directed changes in tick saliva components. Int. J. Environ. Res. Public Health 18 (4), 1806. doi:10.3390/ijerph18041806
Pidíkova, P., Reis, R., and Herichova, I. (2020). miRNA clusters with down-regulated expression in human colorectal cancer and their regulation. Int. J. Mol. Sci. 21 (13), 4633. doi:10.3390/ijms21134633
Qu, Z., Bendena, W. G., Nong, W., Siggens, K. W., Noriega, F. G., Kai, Z. P., et al. (2017). MicroRNAs regulate the sesquiterpenoid hormonal pathway in Drosophila and other arthropods. Proc. Biol. Sci. 284 (1869), 20171827. doi:10.1098/rspb.2017.1827
Rahman, S., Modak, C., Akter, M., and Alam, M. S. (2020). Role of MicroRNA genes miR-1000 and miR-375 in forming olfactory conditional memory in Drosophila melanogaster. Microrna 9 (4), 283–288. doi:10.2174/2211536609666200204113403
Rajput, Z. I., Hu, S. H., Chen, W. J., Arijo, A. G., and Xiao, C. W. (2006). Importance of ticks and their chemical and immunological control in livestock. J. Zhejiang Univ. Sci. B 7 (11), 912–921. doi:10.1631/jzus.2006.B0912
Read, C. B., Lind, M. C. H., Chiarelli, T. J., Izac, J. R., Adcox, H. E., Marconi, R. T., et al. (2022). The obligate intracellular bacterial pathogen Anaplasma phagocytophilum exploits host cell multivesicular body biogenesis for proliferation and dissemination. mBio 13 (6), e0296122. doi:10.1128/mbio.02961-22
Reynolds, E. S., Wooldridge, J. T., Stevenson, H. L., and Thangamani, S. (2023). The Lone Star tick, Amblyomma americanum, salivary factors exacerbate the clinical outcome of Heartland virus disease in a small animal model. Sci. Rep. 13 (1), 13304. doi:10.1038/s41598-023-40397-x
Santangelo, L., Giurato, G., Cicchini, C., Montaldo, C., Mancone, C., Tarallo, R., et al. (2016). The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling MicroRNA sorting. Cell Rep. 17 (3), 799–808. doi:10.1016/j.celrep.2016.09.031
Sarshad, A. A., Juan, A. H., Muler, A. I. C., Anastasakis, D. G., Wang, X., Genzor, P., et al. (2018). Argonaute-miRNA complexes silence target mRNAs in the nucleus of mammalian stem cells. Mol. Cell 71 (6), 1040–1050. doi:10.1016/j.molcel.2018.07.020
Schnettler, E., Tykalová, H., Watson, M., Sharma, M., Sterken, M. G., Obbard, D. J., et al. (2014). Induction and suppression of tick cell antiviral RNAi responses by tick-borne flaviviruses. Nucleic Acids Res. 42 (14), 9436–9446. doi:10.1093/nar/gku657
Sedgwick, A. E., and D'Souza-Schorey, C. (2018). The biology of extracellular microvesicles. Traffic 19 (5), 319–327. doi:10.1111/tra.12558
Sempere, L. F., Dubrovsky, E. B., Dubrovskaya, V. A., Berger, E. M., and Ambros, V. (2002). The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev. Biol. 244 (1), 170–179. doi:10.1006/dbio.2002.0594
Seppey, M., Manni, M., and Zdobnov, E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962, 227–245. doi:10.1007/978-1-4939-9173-0_14
Shao, C. C., Xu, M. J., Chen, Y. Z., Tao, J. P., and Zhu, X. Q. (2015). Comparative profiling of MicroRNAs in male and female Rhipicephalus sanguineus. Appl. Biochem. Biotechnol. 176 (7), 1928–1936. doi:10.1007/s12010-015-1688-x
Sheng, P., Fields, C., Aadland, K., Wei, T., Kolaczkowski, O., Gu, T., et al. (2018). Dicer cleaves 5'-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res. 46 (11), 5737–5752. doi:10.1093/nar/gky306
Shi, J., Zhou, T., and Chen, Q. (2022). Exploring the expanding universe of small RNAs. Nat. Cell Biol. 24 (4), 415–423. doi:10.1038/s41556-022-00880-5
Shu, J., Silva, B., Gao, T., Xu, Z., and Cui, J. (2017). Dynamic and modularized MicroRNA regulation and its implication in human cancers. Sci. Rep. 7 (1), 13356. doi:10.1038/s41598-017-13470-5
Silver, S. J., Hagen, J. W., Okamura, K., Perrimon, N., and Lai, E. C. (2007). Functional screening identifies miR-315 as a potent activator of Wingless signaling. Proc. Natl. Acad. Sci. U. S. A. 104 (46), 18151–18156. doi:10.1073/pnas.0706673104
Šimo, L., Kazimirova, M., Richardson, J., and Bonnet, S. I. (2017). The essential role of tick salivary glands and saliva in tick feeding and pathogen transmission. Front. Cell Infect. Microbiol. 7, 281. doi:10.3389/fcimb.2017.00281
Sojka, D., Franta, Z., Horn, M., Caffrey, C. R., Mareš, M., and Kopáček, P. (2013). New insights into the machinery of blood digestion by ticks. Trends Parasitol. 29 (6), 276–285. doi:10.1016/j.pt.2013.04.002
Sokol, N. S., and Ambros, V. (2005). Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 19 (19), 2343–2354. doi:10.1101/gad.1356105
Sokol, N. S., Xu, P., Jan, Y. N., and Ambros, V. (2008). Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22 (12), 1591–1596. doi:10.1101/gad.1671708
Stojadinovic, O., Brem, H., Vouthounis, C., Lee, B., Fallon, J., Stallcup, M., et al. (2005). Molecular pathogenesis of chronic wounds: the role of beta-catenin and c-myc in the inhibition of epithelialization and wound healing. Am. J. Pathol. 167 (1), 59–69. doi:10.1016/s0002-9440(10)62953-7
Takaya, K., Sunohara, A., Aramaki-Hattori, N., Sakai, S., Okabe, K., Kanazawa, H., et al. (2022). Role of Wnt signaling in mouse fetal skin wound healing. Biomedicines 10 (7), 1536. doi:10.3390/biomedicines10071536
Tamura, M., Sato, M. M., and Nashimoto, M. (2011). Regulation of CXCL12 expression by canonical Wnt signaling in bone marrow stromal cells. Int. J. Biochem. Cell Biol. 43 (5), 760–767. doi:10.1016/j.biocel.2011.01.021
Tan, J. Y., Sirey, T., Honti, F., Graham, B., Piovesan, A., Merkenschlager, M., et al. (2015). Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in mouse embryonic stem cells. Genome Res. 25 (5), 655–666. doi:10.1101/gr.181974.114
Thompson, J. M., Carpenter, A., Kersh, G. J., Wachs, T., Commins, S. P., and Salzer, J. S. (2023). Geographic distribution of suspected alpha-gal syndrome cases - United States, january 2017-december 2022. MMWR Morb. Mortal. Wkly. Rep. 72 (30), 815–820. doi:10.15585/mmwr.mm7230a2
Trümbach, D., and Prakash, N. (2015). The conserved miR-8/miR-200 microRNA family and their role in invertebrate and vertebrate neurogenesis. Cell Tissue Res. 359 (1), 161–177. doi:10.1007/s00441-014-1911-z
van Niel, G., D'Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19 (4), 213–228. doi:10.1038/nrm.2017.125
Vayssier-Taussat, M., Cosson, J. F., Degeilh, B., Eloit, M., Fontanet, A., Moutailler, S., et al. (2015). How a multidisciplinary 'One Health' approach can combat the tick-borne pathogen threat in Europe. Future Microbiol. 10 (5), 809–818. doi:10.2217/fmb.15.15
Villarroya-Beltri, C., Gutiérrez-Vázquez, C., Sánchez-Cabo, F., Pérez-Hernández, D., Vázquez, J., Martin-Cofreces, N., et al. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4, 2980. doi:10.1038/ncomms3980
Wang, F., Gong, H., Zhang, H., Zhou, Y., Cao, J., and Zhou, J. (2015). Lipopolysaccharide-induced differential expression of miRNAs in male and female Rhipicephalus haemaphysaloides ticks. PLoS One 10 (10), e0139241. doi:10.1371/journal.pone.0139241
Winter, E., Cisilotto, J., Silva, A. H., Rosolen, D., Fabichak, A. P., Rode, M. P., et al. (2021). MicroRNAs: potential biomarkers for reproduction, diagnosis, prognosis, and therapeutic in domestic animals. Res. Vet. Sci. 142, 117–132. doi:10.1016/j.rvsc.2021.12.004
Wu, F., Luo, J., Chen, Z., Ren, Q., Xiao, R., Liu, W., et al. (2019). MicroRNA let-7 regulates the expression of ecdysteroid receptor (ECR) in Hyalomma asiaticum (Acari: ixodidae) ticks. Parasit. Vectors 12 (1), 235. doi:10.1186/s13071-019-3488-6
Xin, W., Qin, Y., Lei, P., Zhang, J., Yang, X., and Wang, Z. (2022). From cerebral ischemia towards myocardial, renal, and hepatic ischemia: exosomal miRNAs as a general concept of intercellular communication in ischemia-reperfusion injury. Mol. Ther. Nucleic Acids 29, 900–922. doi:10.1016/j.omtn.2022.08.032
Xiong, Q., Wan, A. T. Y., and Tsui, S. K. (2020). A mini-review of the genomes and allergens of mites and ticks. Curr. Protein Pept. Sci. 21 (2), 114–123. doi:10.2174/1389203720666190719150432
Yandell, M., and Ence, D. (2012). A beginner's guide to eukaryotic genome annotation. Nat. Rev. Genet. 13 (5), 329–342. doi:10.1038/nrg3174
Yi, R., Qin, Y., Macara, I. G., and Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17 (24), 3011–3016. doi:10.1101/gad.1158803
Yu, Z., He, B., Gong, Z., Liu, Y., Wang, Q., Yan, X., et al. (2022). The new Haemaphysalis longicornis genome provides insights into its requisite biological traits. Genomics 114 (2), 110317. doi:10.1016/j.ygeno.2022.110317
Zhai, Z., Boquete, J. P., and Lemaitre, B. (2018). Cell-specific imd-NF-κb responses enable simultaneous antibacterial immunity and intestinal epithelial cell shedding upon bacterial infection. Immunity 48 (5), 897–910. doi:10.1016/j.immuni.2018.04.010
Zhang, H., Freitas, D., Kim, H. S., Fabijanic, K., Li, Z., Chen, H., et al. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 20 (3), 332–343. doi:10.1038/s41556-018-0040-4
Zhang, N., Tang, W., Torres, L., Wang, X., Ajaj, Y., Zhu, L., et al. (2024a). Cell surface RNAs control neutrophil recruitment. Cell 187 (4), 846–860.e17. doi:10.1016/j.cell.2023.12.033
Zhang, Q., Dou, W., Taning, C. N. T., Smagghe, G., and Wang, J. J. (2021). Regulatory roles of microRNAs in insect pests: prospective targets for insect pest control. Curr. Opin. Biotechnol. 70, 158–166. doi:10.1016/j.copbio.2021.05.002
Zhang, Y., Tu, C., Zhang, D., Zheng, Y., Peng, Z., Feng, Y., et al. (2015). Wnt/β-Catenin and Wnt5a/Ca pathways regulate proliferation and apoptosis of keratinocytes in psoriasis lesions. Cell Physiol. Biochem. 36 (5), 1890–1902. doi:10.1159/000430158
Zhang, Z. L., Wang, X. J., Lu, J. B., Lu, H. B., Ye, Z. X., Xu, Z. T., et al. (2024b). Cross-kingdom RNA interference mediated by insect salivary microRNAs may suppress plant immunity. Proc. Natl. Acad. Sci. U. S. A. 121 (16), e2318783121. doi:10.1073/pnas.2318783121
Zhao, B., Lucas, K. J., Saha, T. T., Ha, J., Ling, L., Kokoza, V. A., et al. (2017). MicroRNA-275 targets sarco/endoplasmic reticulum Ca2+ adenosine triphosphatase (SERCA) to control key functions in the mosquito gut. PLoS Genet. 13 (8), e1006943. doi:10.1371/journal.pgen.1006943
Zhou, J., Zhou, Y., Cao, J., Zhang, H., and Yu, Y. (2013). Distinctive microRNA profiles in the salivary glands of Haemaphysalis longicornis related to tick blood-feeding. Exp. Appl. Acarol. 59 (3), 339–349. doi:10.1007/s10493-012-9604-3
Zhou, W., Tahir, F., Wang, J. C., Woodson, M., Sherman, M. B., Karim, S., et al. (2020). Discovery of exosomes from tick saliva and salivary glands reveals therapeutic roles for CXCL12 and IL-8 in wound healing at the tick-human skin interface. Front. Cell Dev. Biol. 8, 554. doi:10.3389/fcell.2020.00554
Zhou, W., Woodson, M., Neupane, B., Bai, F., Sherman, M. B., Choi, K. H., et al. (2018). Exosomes serve as novel modes of tick-borne flavivirus transmission from arthropod to human cells and facilitates dissemination of viral RNA and proteins to the vertebrate neuronal cells. PLoS Pathog. 14 (1), e1006764. doi:10.1371/journal.ppat.1006764
Keywords: extracellular vesicles, small RNA, tick feeding, RNA transport proteins, miRNA transport
Citation: Leal-Galvan B, Kumar D, Karim S, Saelao P, Thomas DB and Oliva Chavez A (2024) A glimpse into the world of microRNAs and their putative roles in hard ticks. Front. Cell Dev. Biol. 12:1460705. doi: 10.3389/fcell.2024.1460705
Received: 06 July 2024; Accepted: 10 September 2024;
Published: 23 September 2024.
Edited by:
Sujay Paul, Monterrey Institute of Technology and Higher Education (ITESM), MexicoReviewed by:
Vipin Rana, University of Maryland, United StatesCopyright © 2024 Leal-Galvan, Kumar, Karim, Saelao, Thomas and Oliva Chavez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adela Oliva Chavez, b2xpdmFjaHZlekB3aXNjLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.