- 1Laboratory of Functional Genomics, Koltzov Institute of Developmental Biology, Russian Academy of Sciences, Moscow, Russia
- 2Department of Molecular Mechanisms for Realization of Genetic Information, National Research Centre Kurchatov Institute, Moscow, Russia
DEAD-box RNA helicase Vasa is required for gonad development and fertility in multiple animals. Vasa is implicated in many crucial aspects of Drosophila oogenesis, including translation regulation, primordial germ cell specification, piRNA silencing of transposable elements, and maintenance of germline stem cells (GSCs). However, data about Vasa functions in Drosophila spermatogenesis remain controversial. Here we showed that loss-of-function vasa mutations led to failures of GSC maintenance in the testes, a severe loss of total germ cell content, and a cessation of male fertility over time. Defects in GSC maintenance in vasa mutant testes were not associated with an increasing frequency of programmed cell death, indicating that a premature loss of GSCs occurred via entering differentiation. We found that Vasa is implicated in the positive regulation of rhino expression both in the testes and ovaries. The introduction of a transgene copy of rhino, encoding a nuclear component of piRNA pathway machinery, in vasa mutant background allowed us to restore premeiotic stages of spermatogenesis, including the maintenance of GSCs and the development of spermatogonia and spermatocytes. However, piRNA-guided repression of Stellate genes in spermatocytes of vasa mutant testes with additional rhino copy was not restored, and male fertility was disrupted. Our study uncovered a novel mechanistic link involving Vasa and Rhino in a regulatory network that mediates GSC maintenance but is dispensable for the perfect biogenesis of Su(Ste) piRNAs in testes. Thus, we have shown that Vasa functions in spermatogenesis are essential at two distinct developmental stages: in GSCs for their maintenance and in spermatocytes for piRNA-mediated silencing of Stellate genes.
Introduction
DEAD-box RNA helicase Vasa (also known as DDX4) is a conserved protein involved in the germline specification and gametogenesis of multiple Metazoa species (Roussell and Bennett, 1993; Lasko, 2013; Hartung et al., 2014; Adashev et al., 2023). As all DEAD-box family proteins, Vasa possesses a highly conserved core helicase region that contains at least 12 conservative motifs distributed between two RecA-like domains, called the N-terminal DEAD-like helicase domain (DEXDc) and the C-terminal helicase superfamily domain (HELICc). These core domains bind both ATP and RNA molecules and ensure RNA helicase and ATPase activity (Linder and Jankowsky, 2011; Wang et al., 2015). The biochemical activities of DEAD-box helicases include the local RNA duplex unwinding, the remodeling of ribonucleoprotein complexes, the facilitation of RNA renaturation, and RNA clamping (Linder and Jankowsky, 2011; Kotov et al., 2014). DEAD-box helicases are known to contribute to all aspects of intracellular RNA metabolism.
Initially, gene vasa has been discovered in fruit flies from the screens for maternal-effect recessive lethal mutations (Schüpbach and Wieschaus, 1986; Schüpbach and Wieschaus, 1989). Functions of the RNA helicase Vasa in Drosophila are essential for primordial germ cell (PCG) specification and embryonic development, as well as for the progression of oogenesis (Lasko and Ashburner, 1990; Styhler et al., 1998; Durdevic et al., 2018; Durdevic and Ephrussi, 2019; Hansen and Pelegri, 2021). A deficiency of vasa contributes to multiple defects of oogenesis and leads to female sterility in Drosophila (Carrera et al., 2000; Johnstone and Lasko, 2004; Liu et al., 2009; Durdevic et al., 2018; Durdevic and Ephrussi, 2019). Only a small number of oocytes in vasa mutant females reach maturation, while the bulk of them undergo premature degeneration in the early oogenesis stages. In the ovaries of vasa mutants, the atrophy of germarium and a loss of early germ cells, including germline stem cells (GSCs) and cystoblasts, are often detected (Lasko and Ashburner, 1990; Styhler et al., 1998). However, the molecular functions of Vasa in oogenesis remain poorly understood.
In the Drosophila germline, Vasa is enriched in nuage, the perinuclear granules of ovarian nurse cells and premeiotic germ cells in the testes (Snee and Macdonald, 2004; Lim and Kai, 2007; Malone et al., 2009; Kibanov et al., 2011). Granules nuage are intracellular membraneless organelles that are assembled as liquid biomolecular condensates (Nott et al., 2015; So et al., 2021). They are known as the sites of posttranscriptional piRNA biogenesis and posttranscriptional piRNA-mediated silencing of selfish genomic elements, including transposons, genome repeats, and protein-coding genes (Lim and Kai, 2007; Malone et al., 2009; Kibanov et al., 2011; Webster et al., 2015). Vasa is a basic hierarchical structural component of nuage that recruits ARGONAUTE/PIWI family endonucleases Aubergine (Aub) and AGO3 in nuage (Lim and Kai, 2007; Kibanov et al., 2011; Yamazaki et al., 2023). Vasa appears to participate in the transfer of long piRNA precursors from the nucleus to nuage granules (Zhang et al., 2012) and contribute to the piRNA amplification ping-pong cycle carried out by Aub and AGO3 proteins for the tuning of target silencing process (Brennecke et al., 2007; Gunawardane et al., 2007; Malone et al., 2009; Xiol et al., 2014; Webster et al., 2015). Vasa helicase activity is required for the piRNA-mediated silencing of target transcripts (Durdevic and Ephrussi, 2019). Loss-of-function vasa mutations lead to a failure of nuage assembly. It causes the cytoplasmic distribution of Aub, AGO3, and other components of the piRNA machinery and the disruption of piRNA-mediated silencing in the germline (Vagin et al., 2006; Lim and Kai, 2007; Malone et al., 2009; Kibanov et al., 2011; Kotov et al., 2014). In vasa mutant females, a lack of piRNA production in ovarian nurse cells leads to the mobilization of multiple transposable elements (TEs) (Vagin et al., 2004; Malone et al., 2009; Czech et al., 2013). A transient loss of Vasa expression only between oogenesis stages 2 and 6 causes the accumulation of TE transcripts in ovarian nurse cells (Durdevic et al., 2018). High-level TE activation leads to the transportation of their transcripts in the developing oocyte and their integration into the oocyte genome (Wang et al., 2018). Insertions of TEs cause DNA double-strand breaks in various loci of the genome, causing a loss of chromatin integrity and replication stress. It can also trigger oogenesis arrest through activation of the checkpoint kinase 2 (Chk2) signaling cascade (Klattenhoff et al., 2007; Durdevic et al., 2018). Elevated level of TE activity can also result in vertical TE transfer from mothers to progeny, causing chromatin damage and mitotic arrest in early embryogenesis (Durdevic et al., 2018; Wang et al., 2018). Vasa is expressed in the germline throughout the whole fly oogenesis and is also enriched on the posterior pole of the developing oocyte in so-called pole granules or pole plasm that is essential for primordial germ cell specification in embryos (Hay et al., 1988; Lasko and Ashburner, 1990; Trcek and Lehmann, 2019).
Despite the fact that Vasa is expressed in both the ovarian and testis germline, data about its functions in spermatogenesis remain controversial. Fertility of males carrying vasa mutations leads to the proposal that vasa have no essential functions in spermatogenesis (Lasko and Ashburner, 1988; Lasko, 2013). In the testes of vasa mutant males, TE mobilization is less conspicuous, but protein-coding tandem Stellates genes become strongly derepressed in spermatocytes (Aravin et al., 2001; Vagin et al., 2006; Kibanov et al., 2011; Adashev et al., 2021), which leads to subsequent meiotic disorders and male fertility disturbances (Palumbo et al., 1994; Bozzetti et al., 1995). Thus, data about the involvement of Vasa in the piRNA pathway and Stellate gene silencing in testes clearly conflict with the above-mentioned point of view (Vagin et al., 2006; Kibanov et al., 2011). Since Vasa contributes to various intracellular processes in the germline, studying its functions is difficult. It remains unclear whether its functions in the piRNA pathway and in other developmental processes are mutually linked or relatively independent.
Adult males of Drosophila contain a pair of testes; each of them is represented by an elongated and twisted tube. In wild-type testes, Vasa-positive germline stem cells (GSCs) reside at the apical tip of the testis around a cohort of terminally differentiated somatic cells, representing a niche structure termed the hub (Figure 1A). The hub supports two adjacent stem cell populations with the aid of the secretion of signaling molecules: 9–12 germline stem cells (GSCs) and twice as many somatic cyst stem cells (CySCs). The self-renewal division of a GSC provides a new GSC and a goniablast (spermatogonium cell). The division of CySC generates a self-renewing CySC and a cyst cell. A pair of cyst cells overlap a spermatogonium, forming the cyst, a functional unit of spermatogenesis. Spermatogonial cells inside the cyst undergo four rapid mitotic divisions with incomplete cytokinesis, finally providing a 16-cell syncytium of primary spermatocytes surrounded by two undivided flattened CyCs. Primary spermatocytes enter a long premeiotic interphase accompanied by extensive cell growth. After that, mature spermatocytes undergo synchronous meiosis with the appearance of 64 haploid round spermatids. Spermatids elongate and undergo the individualization process, move to the basal end of the testis, enter the seminal vesicle, and are stored there as mature sperm before copulation (Spradling et al., 2011; Greenspan et al., 2015) (Figure 1A).
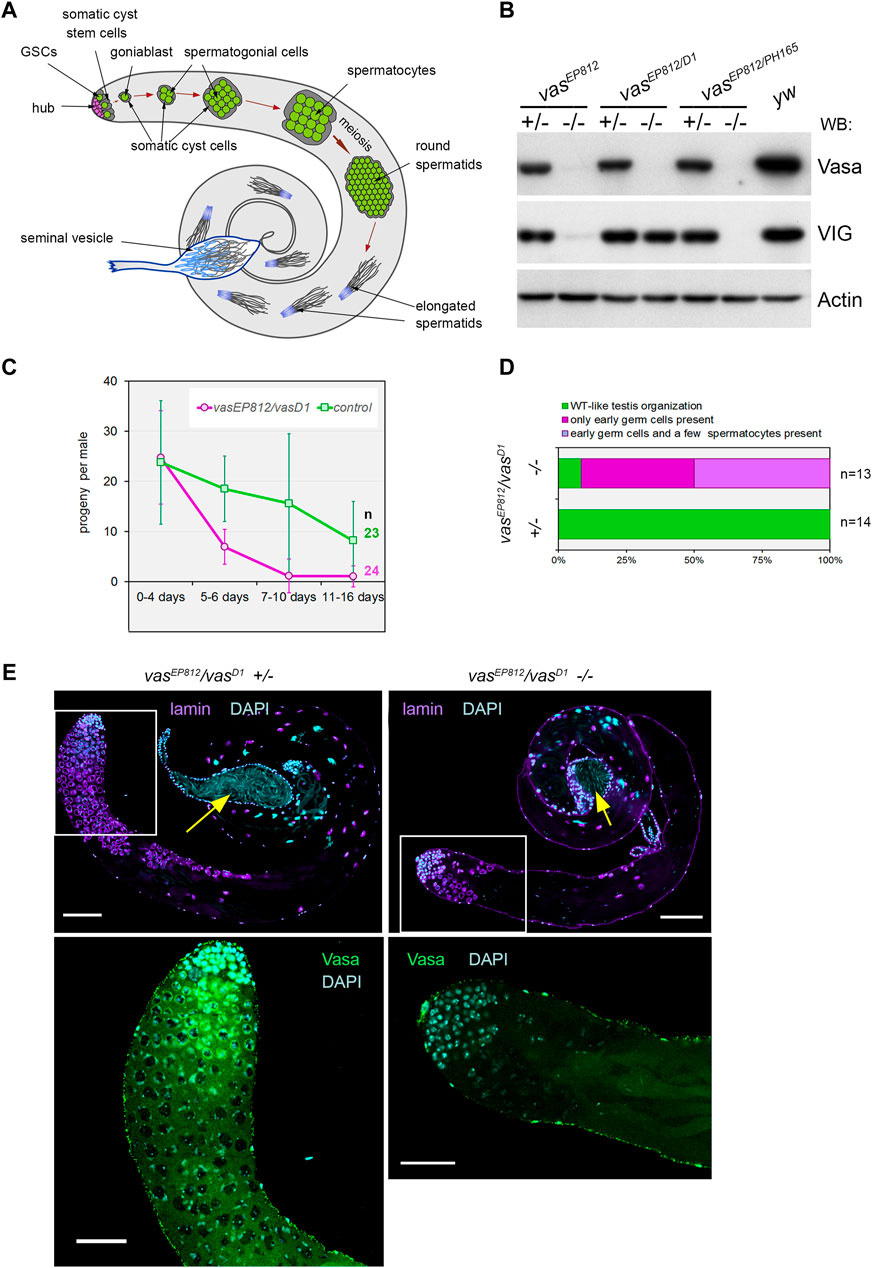
Figure 1. Vasa is required for male fertility and germ cell maintenance in the testes. (A) A scheme of Drosophila spermatogenesis. Germline stem cells (GSCs, green) are located adjacent to the hub (violet) at the apical testis tip and are encapsulated by two somatic cyst stem cells (CySCs, grey). The daughter of the GSC, the goniablast, undergoes four mitotic divisions to create cysts of 16 spermatogonia, overlapping two undivided somatic cyst cells. After that, the germ cells within the cyst switch to the spermatocyte program. Spermatocytes undergo extensive cell growth and then enter synchronous meiosis with the appearance of 64 haploid round spermatids. Spermatids elongate and move to the basal end of the testis. Mature individual spermatozoa enter the seminal vesicle and are stored there as mature sperm. (B) Western blot analysis of the testis lysates of vasa mutants. Note that the testes of vasEP812/vasD1 heteroallelic combination expressed VIG but not Vasa, whereas vasEP812/vasPH165 and vasEP812/vasEP812 mutations caused a loss of expression of both Vasa and VIG proteins. Anti-Actin antibodies were used as a loading control. (C) Fertility test of vasa mutant males with vasEP812/vasD1 heteroallelic combinations (violet lines) in comparison with their heterozygous siblings (green lines). The average offspring number of male per day with standard errors is presented for indicated time intervals after parent male eclosion. The number of examined males is indicated. (D) The bar diagram depicts the distribution of testis phenotypes of vasEP812/vasD1 mutants and control heterozygous males (3 days after eclosion): wild-type-like phenotype (green), only early germ cells (violet), early germ cells and a few spermatocytes (blue). The numbers of examined testes are indicated. (E) A loss of germ cells in the testes of vasEP812/vasD1 heteroallelic mutants. Left: testes of heterozygous control males (3 days after eclosion) exhibited a wild-type phenotype. Right: testes of vasEP812/vasD1 heteroallelic mutants (3 days after eclosion) rapidly lost germline content. Whole-mount fixed testes were immunostained with Vasa (green) and lamin (violet) antibodies, and chromatin was stained by DAPI (blue). Confocal slices with the apical tips of the testes oriented leftward are shown. Fragments in the white boxes of the top images are present in higher magnifications in the corresponding bottom images with Vasa and DAPI staining. Scale bars are 100 µm for the top images and 50 µm for the bottom ones. Yellow arrows indicate seminal vesicles containing mature sperm.
Our study explored the molecular functions of Vasa in testes of D. melanogaster. Here we found that loss-of-function vasa mutations led to a rapid loss of GSC number in testes and a decrease in male fertility with aging. Defects in GSC maintenance in vasa mutant testes were not associated with an elevated frequency of premature cell death, indicating that a loss of GSCs occurred through entering differentiation without a self-renewal. According to our data, two main functions of RNA helicase Vasa in the testes of Drosophila melanogaster are carried out in distinct cohorts of germ cells: the maintenance of GSCs in early spermatogenesis and piRNA silencing of Stellate genes, ensuring correct passage through meiosis, in spermatocytes. Both of these functions are necessary for male fertility, underscoring the essential role of Vasa in fly spermatogenesis.
Results
Vasa is required for male fertility in Drosopila
We revealed that vasEP812/vasPH165 and vasEP812/vasEP812 mutations caused a loss of expression of both Vasa and VIG (a product of the vasa intronic gene also encoding the vasa locus) proteins in the testes, whereas vasEP812/vasD1 heteroallelic combination only disrupted Vasa expression but did not disrupt half-dose VIG expression (Figure 1B). The vasD1 loss-of-function allele has been obtained using ethyl methanesulfonate mutagen with no changes in the coding region (Lasko and Ashburner, 1990). vasD1 is not a null allele that appears to be associated with a disturbance of vasa transcriptional regulation (Lasko and Ashburner, 1990; Liang et al., 1994). Using males carrying loss-of-function vasa mutations, we performed male fertility tests over a long period of time (described in Materials and Methods). We found that young vasEP812/vasD1 mutant males (0–4 days after eclosion) give rise to adult offspring in crossing with yw females, providing a progeny number comparable to that of control heterozygous siblings (Figure 1C). However, already from the fifth day onwards, a drastic fertility reduction of mutant males compared to control heterozygous siblings was observed. In subsequent time intervals (7–10 days and 11–16 days), most vasEP812/vasD1 males produce no progeny (Figure 1C). Testing males with heteroallelic combinations vasEP812/vasPH165, we found that even though mutant males were fertile at a low level up to the twentieth day after eclosion, in subsequent time intervals their fertility declined to a very low level (Supplementary Figure S1A). Taken together, these results demonstrate that male reproductive capacity is substantially lowered in the absence of Vasa expression, indicating the indispensable role of Vasa in male fertility maintenance.
Further, we examined the morphology of the testes of vasEP812/vasD1 mutant males (3 days after eclosion). We found that whereas the testes of heterozygous siblings exhibited a wild-type distribution of premeiotic Vasa-positive germ cells, the vasEP812/vasD1 testes exhibited a significant loss of the total pool of germ cells in the testes (Figures 1D, E). However, seminal vesicles in both control and mutant young males were filled with mature sperm (Figure 1E, yellow arrows), supporting that the first waves of spermatogenesis in vasa mutant males proceeded correctly. We also observed a similar loss of germline content for other vasa mutant males, vasEP812/vasPH165 (Supplementary Figures S1B, C). In sum, in vasa mutant males, a rapid decline in germ cell number in the testes is accompanied by reduced fertility.
Defects of germline stem cells maintenance in the testes of vasa mutant males
Then, we examined the effects of lacking vasa expression on the maintenance of testis GSCs. In wild-type males, Vasa-positive GSCs are located at the apical tip of the testis around the hub (Figure 1A). We identified the hub as a cluster of closely fitting small cells at the apical testis tip brightly expressing the marker protein Fasciclin III (FasIII) on their surface. We scored GSCs as Vasa-positive cells that directly contacted the hub and contained the α-spectrin-positive dot-like organelle, the spectrosomes, in control heterozygous testes. In the testes of vasa mutant males, we defined GSCs as cells in direct contact with the hub and carrying the spectrosomes (Figure 2A). We found a significant decline of GSCs in the testes of vasa heteroallelic mutant combinations, vasEP812/vasD1 and vasEP812/vasPH165, compared with heterozygous controls in two age groups (0 days after eclosion, p < 4.3E-8, and 6 days old, p < 2.3E-10, Wilcoxon/Mann-Whitney test, Figures 2A, B; Supplementary Figure S1B). These data indicate that Vasa is required intrinsically for the maintenance of testis GSCs.
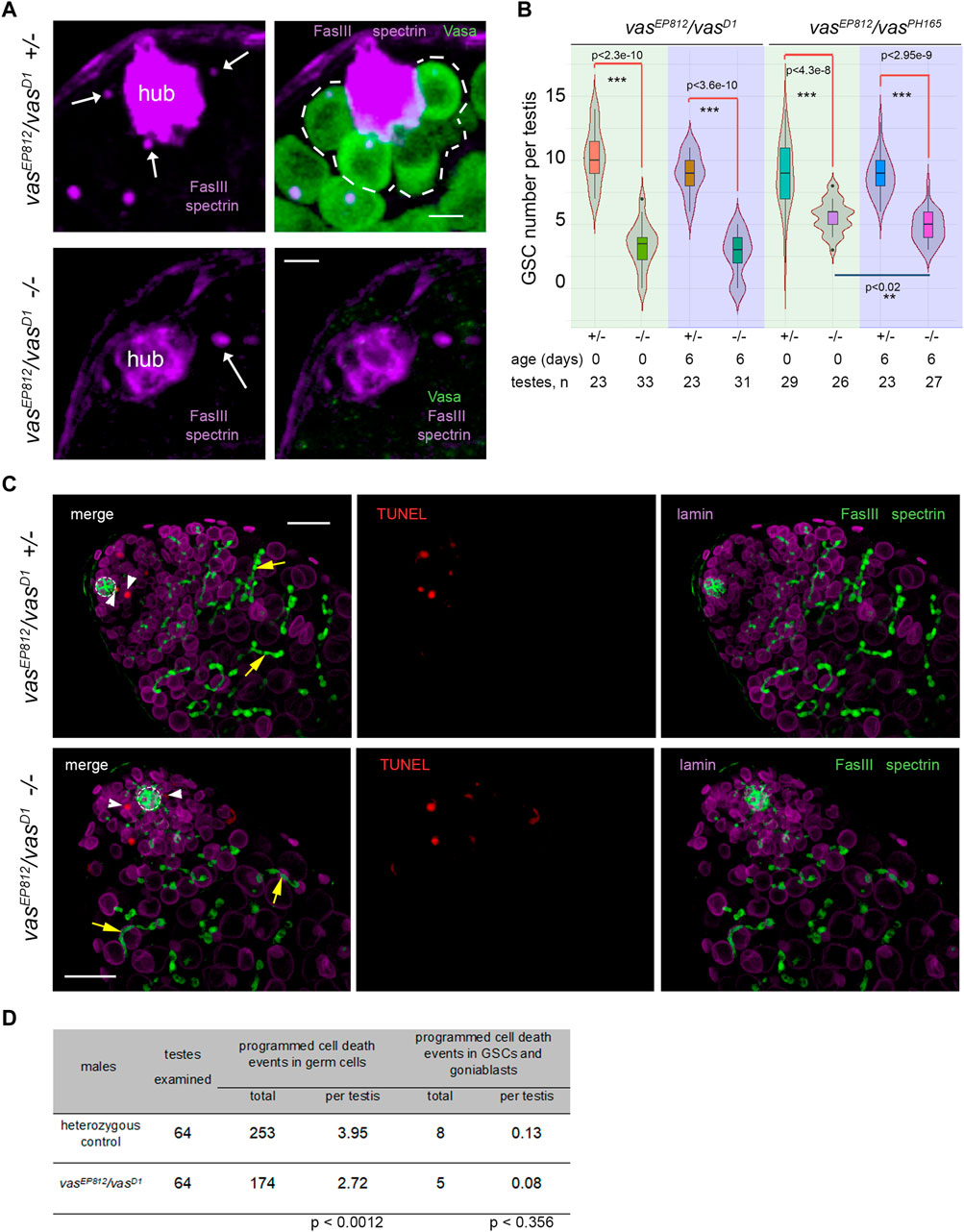
Figure 2. Analysis of the testes of vasa mutants. (A) GSCs in the testes of vasa mutants and control heterozygous siblings. Testes of mutant and heterozygous vasEP812/vasD1 control males (0 days) were immunostained with Vasa (green), Fasciclin III (violet, marker of hub), α-spectrin (violet) antibodies, and DAPI (blue) staining. White arrows indicate spectrosomes, dot-like GSC-specific organelles located near the hub. Single confocal slices of testis tip images are shown. Scale bars are 5 µm. (B) Loss of GSCs in the testes of vasEP812/vasD1and vasEP812/vasPH165 mutants. Violin plots present GSC number per testis in vasa mutant and heterozygous control testes of 0-day-old and 6-day-old males. The medians and quartiles of GSC number sets are marked. The number of testes counted for each genotype and the age of the analyzed males are shown at the bottom. *** Significant differences (from p < 4.3e-8 to p < 2.3e-10) are found by pairwise comparison of GSC numbers in the mutant and control testes of the same age (Wilcoxon/Mann-Whitney (U) test). ** Between the testes of 0- and 6-day vasEP812/vasPH165 mutant males, significant differences in GSC number were also found (p < 0.02). (C) TUNEL assay (red signals) with simultaneous immunostaining with antibodies to Fasciclin III (FasIII, hub marker, green), α-spectrin (marker spectrosomes and fusomes, green), and lamin (marker nuclear envelope, violet). Confocal slices with the apical tips of testes oriented leftward here and after are shown. The hubs are marked by white outlines. TUNEL signals in GSCs and goniablasts are indicated by white arrowheads. The fusomes are indicated by yellow arrows. Scale bars are 20 µm. (D) Analysis of programmed cell death events in mutant and control testes. Data are presented as counted TUNEL-positive signals and as the average number per testis for early germ cells and separately for GSCs and goniablasts. The Wilcoxon/Mann-Whitney (U) test was used for pairwise comparison of mutant and control testes, as indicated at the bottom of the table. See also Supplementary Figure S2 for additional TUNEL assay data.
We could expect a premature loss of GSCs according to one of two general models: a loss through programmed cell death or premature differentiation of germ cells followed by normal development of newly generated germline cysts. To estimate programmed cell death events during early spermatogenesis, we used the TUNEL assay, which detects massive DNA fragmentation associated with necrotic or apoptotic dying cells (Arama and Steller, 2006; Kibanov et al., 2013). Taking into account a rapid loss of total germline content in the testes of vasa mutants with aging, we performed TUNEL assay using the testes of very young males (0–1 day after eclosion) (Figure 2C; Supplementary Figure S2). We found that the average number of programmed cell death events per whole testis was 2.72 for mutants and 3.95 for heterozygous control (significant differences are found by Wilcoxon/Mann–Whitney test, p < 0.0012) (Figure 2D; Supplementary Figure S2). We believe that the observed difference in the average number of programmed cell death events per mutant and control testis is not biologically significant. According to our data, the total number of cases of programmed cell death was rather higher in control testes than in mutant testes. But in both cases, there are few apoptotic events per testis, and they do not have a noticeable effect on the process of spermatogenesis. Note that in the wild-type testes, apoptosis of single cysts occurs with low frequency, but it does not affect the development of adjacent cysts, and the lost cysts are rapidly replaced during the following waves of spermatogenesis (Arama and Steller, 2006; Kibanov et al., 2013). Thus, our results indicate that there is no increased frequency of total cell death events in mutant versus control testes. At the same time, programmed cell death events in GSCs and goniablasts consisted of 0.08 and 0.13 per testis in average for vasEP812/vasD1 mutants and heterozygous control, respectively (insignificant differences according to Wilcoxon/Mann-Whitney test, p < 0.356) (Figures 2C, D). We did not observe any morphological abnormalities of early germ cells, including giant nuclear and cellular sizes, in the mutant testes. Instead, we detected the presence of differentiating cysts marked with branched fusomes in the testes of vasa mutant males similar to the control (Figure 2C, yellow arrows). We also performed staining of the testes for the phosphorylated H2A variant (γ-H2Av), a marker for DNA double-strand breaks (DSBs). However, we reveal an accumulation of DSBs at the apical tips in the mutant testes with a frequency that is equal to that of heterozygous controls (Supplementary Figure S3), indicating that a loss of GSCs was not caused by unrepaired DNA breaks and subsequent cell death. Thus, the obtained results indicate that defects in the maintenance of GSCs in the testes of vasa mutant males are generally not associated with premature cell death and rather support a loss of GSCs by entering differentiation without a self-renewal.
Rhino mutants phenocopy disorders of testis morphology of vasa mutants
Recently published data demonstrate that loss-of-function mutations in the gene encoding nuclear piRNA pathway component, rhino (rhi), lead to morphological and functional abnormalities in the testes of Drosophila, including a premature loss of GSCs, sharply decreased total germ cell content, and reduced fertility by aging (Chen et al., 2021). Thus, rhi mutation defects resemble those of vasa mutant testes. It is also known that loss-of-function mutations of another piRNA pathway component, aubergine (aub), cause disturbances of GSC maintenance in the ovaries (Ma et al., 2017; Rojas-Ríos et al., 2017). Aub is autonomously required in GSCs for their self-renewal, and Aub loading by piRNAs is required for that (Rojas-Ríos et al., 2017).
Analyzing GSC maintenance in the testes of rhi2/KG and aubHN2/QC42 males, we found that GSC number decreased with aging compared to controls in both of the cases (Figure 3A). Nevertheless, only for rhi mutant testes we observed defects in GSC maintenance similar to those in the vasa testes a week later of fly eclosion. We employed the combination of aubHN2 and aubQC42 mutations harboring strong and null alleles (Schüpbach and Wieschaus, 1991) to examine the function of Aub in testis GSC maintenance. However, our analysis of aubHN2/QC42 testes revealed that a decline in GSC number was delayed in time compared with vasa or rhi testes, and similar effects of GSC loss were observed to the 20th day after male eclosion (Figure 3A).
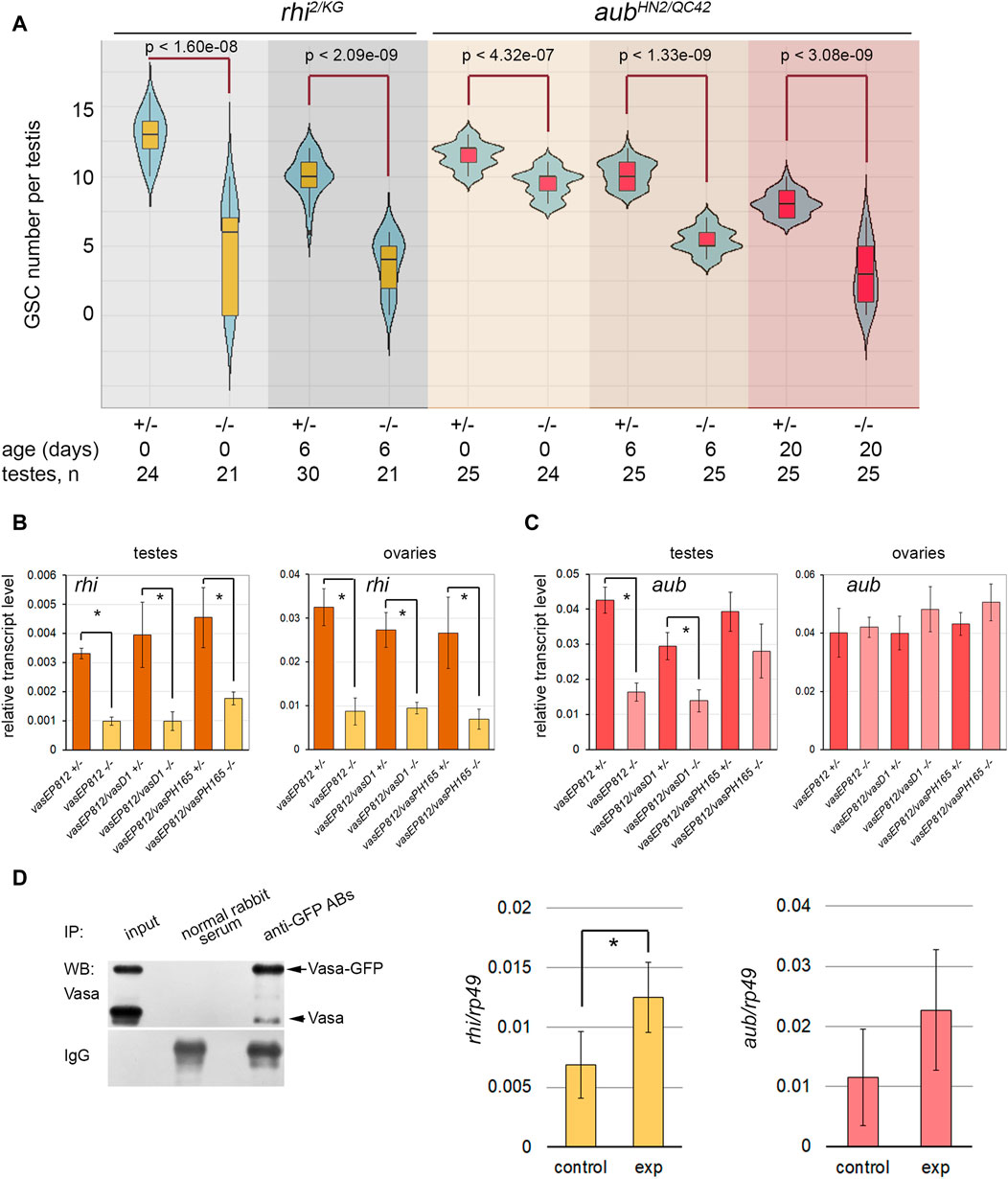
Figure 3. Identification of Vasa mRNA target. (A) Violin plots present GSC number per testis in the testes of rhi and aub mutants and heterozygous control. The medians and quartiles of GSC number sets are marked. The number of testes counted for each genotype and the age of the analyzed males are shown at the bottom. * Significant differences are found by the indicated pairwise comparison of GSC numbers in the mutant and control males of the same age (Wilcoxon/Mann-Whitney (U) test). (B, C) RT-qPCR analysis of transcription levels of rhi (B) and aub (C) in the testes and ovaries of vasa mutants. The expression levels of mRNAs were normalized to rp49 transcripts. Error bars represent standard errors of the mean. Differences between the mutant and control heterozygous samples siblings were analyzed by Student’s t-test. The asterisks indicate significant differences between conditions; *p < 0.05. (D) RIP-qPCR assay for Vasa target mRNA identification. Left: Western blot analysis of immunoprecipitation of vasa-GFP ovarian lysate using antibodies to GFP and normal rabbit serum for control. Middle and right: RT-qPCR analysis of experimental and control RNA-immunoprecipitates with transcript-specific primers for rhi and aub with normalization to rp49 transcripts. * The asterisks indicate that significant differences between the experiment and the control (Student’s t-test, p < 0.05) were found only for rhi transcripts.
To identify the relationship between Vasa and these genes, we performed RT-qPCR analysis using the gonads of vasa mutants. The relative level of rhi transcripts decreased more than three times in the testes of vasa mutant males (0-day-old after eclosion) compared to heterozygous control (Figure 3B). Significant differences were found for all three mutant combinations with p < 0.05. The level of rhi transcripts in the ovaries of vasa mutant females (3–5 days after eclosion) was also significantly reduced, p < 0.05 (Figure 3B). Thus, our results indicate the existence of genetic interaction between vasa and rhi. According to our data, the level of aub transcripts decreased in the mutant testes compared to the control, but only in the testes of vasEP812/vasEP812 and vasEP812/vasD1 males, differences were found to be significant with p < 0.05 (Figure 3C). The analysis of vasa mutant ovaries did not reveal significant differences for aub transcript abundance compared to control (Figure 3C). Using Western blot analysis, we defined that in the testes of 0-day-old vasa mutants, the level of Aub protein was sharply decreased (Supplementary Figure S4A). We observed that the level of Aub was also reduced in the ovaries of vasa mutant females (3–5 days after eclosion) (Supplementary Figure S4B). It should be proposed that Vasa is able to facilitate translation of aub mRNA or provide stabilization of Aub protein in nuage granules.
Since RNA helicase Vasa is known to be located only in the cytoplasmic compartment of germ cells (Snee and Macdonald, 2004; Lim and Kai, 2007; Malone et al., 2009; Kibanov et al., 2011), it cannot directly regulate gene transcription. We assumed that Vasa may specifically bind to rhi mRNA in gonads, promoting translation initiation and/or transcript stabilization, as previously shown for mei-P26 mRNA in the ovaries (Liu et al., 2009). To test this hypothesis, RNA immunoprecipitation (RIP) experiments were carried out using vasa-GFP D. melanogaster flies expressing in germ cells the functional vasa copy fused with the GFP tag (Kugler et al., 2010). Since rhi expression in the testes is restricted to a small cohort of early germ cells from GSCs to spermatogonia (Chen et al., 2021), we performed RNA immunoprecipitation (RIP) experiments using formaldehyde cross-linked lysates of vasa-GFP ovaries and antibodies to GFP with a high affinity. The recovery of Vasa-GFP protein in the immunoprecipitation procedure was monitored by Western blot analysis (Figure 3D, left). RIP-qPCR analysis showed significant enrichment (p < 0.05) (Figure 3D, middle) of rhi transcripts in complex with Vasa-GFP, according to the results of three independent experiments. Thus, our data in sum suggest that RNA helicase Vasa is a positive regulator of rhi expression in the gonads of D. melanogaster.
The enrichment of aub mRNA in RIP-qPCR experiments turned out to be insignificant (p < 0.1) (Figure 3D, right). Based on previously published data, Aub colocalizes with Vasa in nuage granules and directly interacts with Vasa during posttranscriptional piRNA biogenesis (Kibanov et al., 2011; Webster et al., 2015; Zheng et al., 2016; Durdevic and Ephrussi, 2019). A loss of Vasa, the top hierarchical nuage component, causes the mislocalization of Aub in the cytoplasm in the ovaries and testes (Lim and Kai, 2007; Malone et al., 2009; Kibanov et al., 2011). Taking this into account, our results allow us to propose that Vasa contributes to the stability of Aub protein within nuage granules.
Expression of transgene rhi copy partially suppresses premeiotic germ cell loss in the testes of vasa mutants
Based on our findings of genetic and molecular interactions between rhi and vasa, we investigated the potential effects of an additional rhi dose on spermatogenesis in the absence of vasa expression. We introduced the transgenic construct rhi-GFP (Mohn et al., 2014) in the background of the vasa mutation (vasEP812−/−) through successive crosses. We detected a high level of rhi transcripts in the testes of vasEP812;rhi-GFP males and a presence of Rhi-GFP fluorescent signal in early germ cells at the apical testis tip, corresponding to the Rhi expression pattern in the testes of wild-type males (Supplementary Figure S5). The testes of vasEP812;rhi-GFP males (6-day-old after eclosion) exhibited normal morphology and size and contained multiple germ cells, including spermatogonia and spermatocytes with branched fusomes. While the testes of vasEP812 mutant males of the same age were reduced in size and contained a small number of germ cells (Figures 4A, B). According to Western blot analysis (Figure 4C), in the testes of 0-day-old males from the experimental and control lines, we observed an equal intensity signal of βNACtes, a marker of spermatocytes (Kogan et al., 2022), which indicates a similar filling of the testes with premeiotic germ cells. However, in the testes of 6-day-old males, βNACtes signal of similar intensity was observed only for yw (wild type control) and vasEP812;rhi-GFP males, whereas for vasEP812 testes it was more than 10-fold weaker. This indicates that the amount of germ cells is sharply reduced in the testes of 6-day-old vasEP812 males but remains at a level comparable to the wild type in the testes of vasEP812;rhi-GFP males.
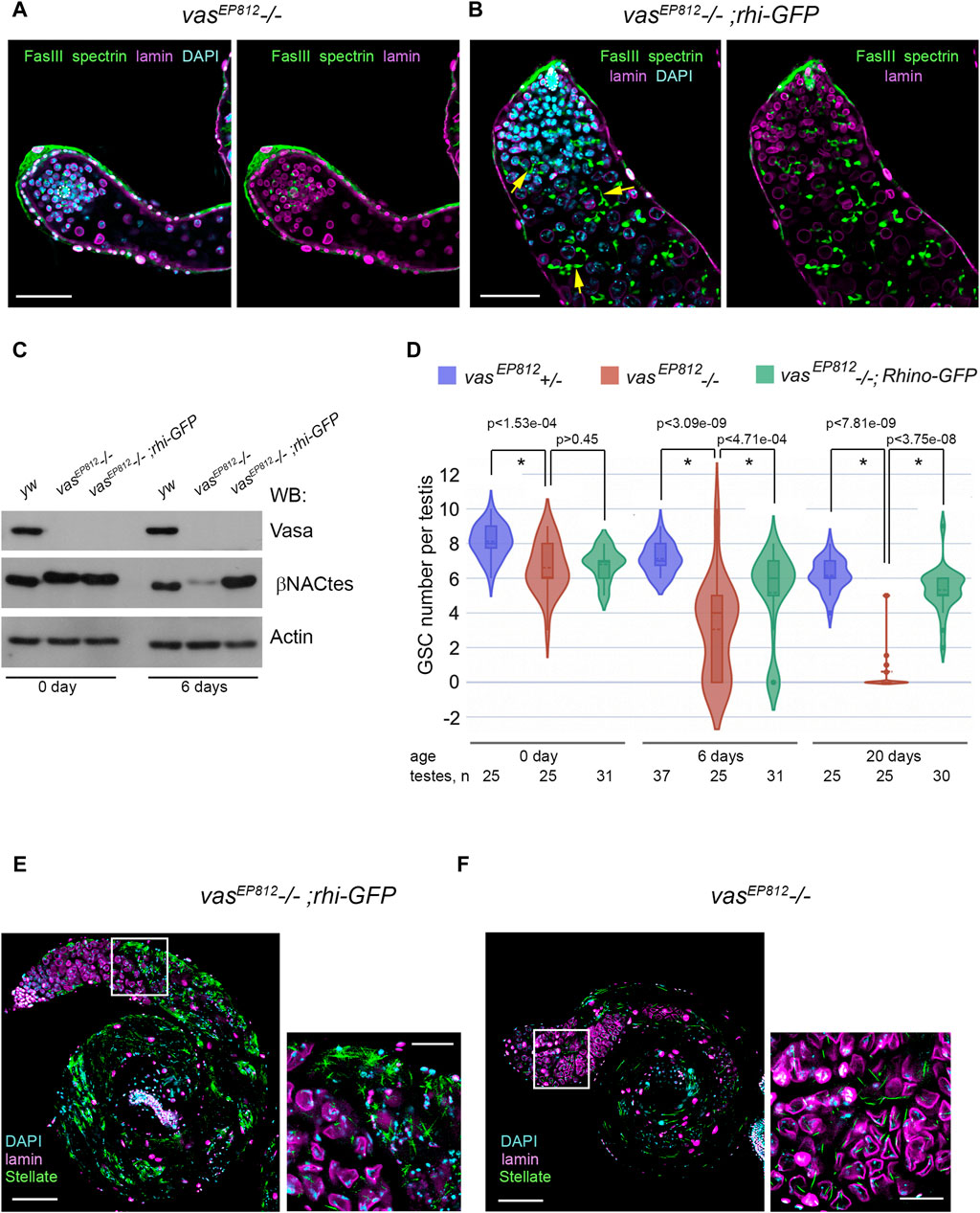
Figure 4. Effects of additional rhi expression in the testes of vasa mutants. (A, B) Immunofluorescence analysis of vasEP812−/− (A) and vasEP812;rhi-GFP (B) testes. Testes of 6-day-old males were stained with antibodies to Fasciclin III (FasIII, green), α-spectrin (green), and lamin (violet). Internal confocal slices of the immunostained whole-mount fixed testis preparations are shown. The hubs are marked by white outlines. The fusomes are indicated by yellow arrows (B). Scale bars are 50 µm. (C) Western blot analysis of testis lysates of 0- and 6-day-old yw, vasEP812−/−, and vasEP812;rhi-GFP males using antibodies to Vasa, βNACtes, and Actin. Actin was used as a loading control. βNACtes is a marker of spermatocytes, which in normal circumstances constitute the most significant part of the germinal content of testes. (D) Violin plots present GSC number per testis in vasEP812+/−, vasEP812−/−, and vasEP812;rhi-GFP males. The number of testes counted for each genotype and the age of males are shown at the bottom. * Significant differences are found by indicated pairwise comparisons (Wilcoxon/Mann-Whitney (U) test). (E, F) Stellate expression in the vasEP812;rhi-GFP (E) and vasEP812−/− (F) testes. Testes of 0-day-old males were stained with antibodies to Stellate (green) and lamin (violet). Chromatin was stained with DAPI (blue). Images of whole testes are presented on the left. Scale bars are 100 µm. Fragments in the white boxes of the left images are present in higher magnifications on the right. Scale bars are 30 µm.
To examine whether the GSC number is also restored in the testes of the vasEP812;rhi-GFP line, we counted them using the method described above (Figure 4D). Testes from vasEP812−/− males showed a rapid loss of GSCs, in accordance with data for other vasa mutant combinations as shown above (Figure 2B). However, for vasEP812;rhi-GFP males, the amount of GSCs per testis decreased more slowly and exhibited significant differences with vasEP812 mutants from the 6-day age and after (Figure 4D). In sum, our data demonstrate that the expression of additional rhi copy ameliorates the phenotype of vasa mutant testes, partially restoring not only premeiotic germ cell content in the testes of vasa mutants but also the maintenance of GSCs. We asked whether the expression of the additional rhi copy in the background of vasa mutation affected the late stages of spermatogenesis. It is known that tandemly repeated Stellate genes are the main targets of the piRNA pathway in D. melanogaster spermatocytes (Aravin et al., 2001; Vagin et al., 2006; Kotov et al., 2019). RNA helicase Vasa is required for Stellate silencing, being a basic architectural and functional component of nuage granules where posttranscriptional Su(Ste) piRNA biogenesis and piRNA silencing of Stellates proceed (Lim and Kai, 2007; Kibanov et al., 2011). We found that in the testes of vasEP812;rhi-GFP males, a strong derepression of Stellate genes occurred with the formation of crystalline aggregates of Stellate protein (Figure 4E). It has been shown previously that the presence of more than 50–60 Stellate gene copies in the genome in cases of their derepression leads to complete male sterility due to strong meiotic disorders (Palumbo et al., 1994). We showed that the derepression of Stellate genes occurs in the testes of all three vasa mutant combinations; however, the expression of Stellate protein appears to be relatively low (Figure 4F; Supplementary Figure S6A), presumably owing to the premature loss of premeiotic germ cells with aging and the small number of Stellate copies in the genome. Compared with that in the testes of vasEP812;rhi-GFP males, extremely high expression of Stellate protein was observed (Figure 4E; Supplementary Figure S6B). We developed a method for the estimation of Stellate gene copies in the genome (see Materials and Methods for details). For vasEP812 males, we determined about 19 Stellate copies; that is not enough for complete sterility (Figure 6C). We determined more than 160 Stellate copies for our “rescue” vasEP812;rhi-GFP line (Supplementary Figure S6C), obtained through several crossing rounds. We assume that both the rescue effect on germ cell maintenance and the large number of Stellate copies in the genome contribute to the observed testis phenotype. In accordance with these data, vasEP812;rhi-GFP testes demonstrated complete sterility with defects of the elongated spermatid individualization process and lacking of mature sperm in the seminal vesicles compared to vasEP812+/− control ones (Supplementary Figures S6D, E).
Thus, the expression of the additional rhi dose in the background of vasa mutation leads to a significant restoration of GSC number and total content of premeiotic germ cells in the testes but does not restore piRNA silencing of Stellate genes. This experimental model allowed us to clearly separate two distinct functions of Vasa protein in spermatogenesis of D. melanogaster.
Vasa is essential for ping-pong biogenesis of piRNAs directed to stellate silencing
To study the contribution of Vasa in Stellate silencing in more detail, we prepared and sequenced on the Illumina platform whole-transcriptome libraries from the testes of vasEP812−/−, vasEP812+/−, and vasEP812;rhi-GFP males. To evaluate the expression level of Stellate and Su(Ste) repeats, we mapped RNA-seq library data to their consensus sequences (Figure 5A). We evidently observed a strong increase in Sellate transcript abundance in the testes of vasa mutants (up to 17.7- and 418.9-fold higher than in heterozygous control for vasEP812−/− and vasEP812;rhi-GFP males, respectively). We found that in vasa mutant and vasEP812;rhi-GFP males, the level of antisense Su(Ste) transcripts was 2.8- and 2.4-fold higher than in the steady-state conditions in heterozygous males (Figure 5A). Thus, in the background of vasa mutation, the accumulation of unprocessed Su(Ste) transcripts occurred due to failures of their processing into mature piRNAs. At the same time, the additional rhi dose in the testes of vasEP812;rhi-GFP males did not significantly affect the level of Su(Ste) transcripts (Figure 5A), according to previously published data (Chen et al., 2021).
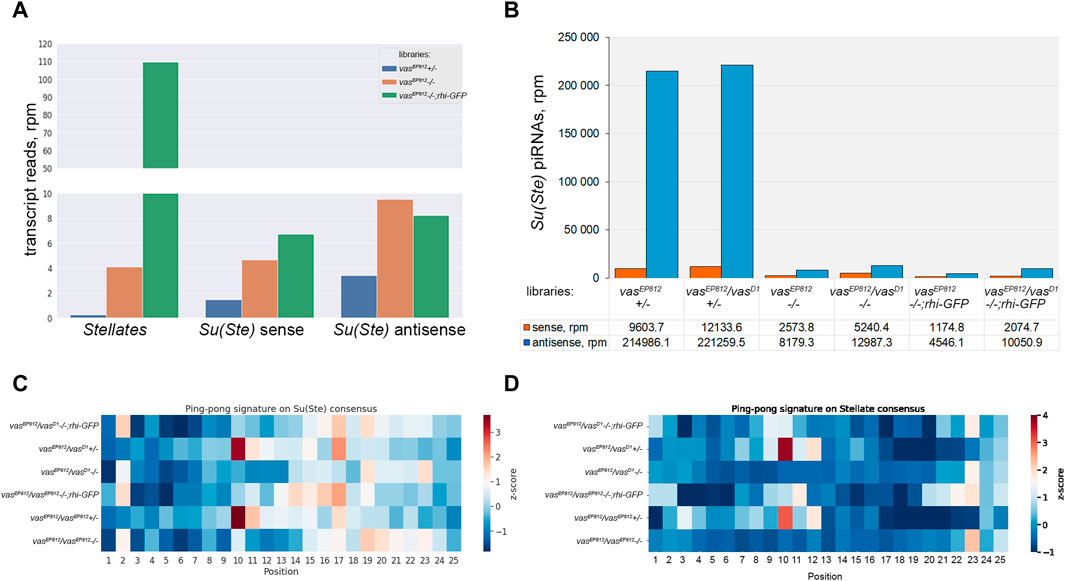
Figure 5. Analysis of the biogenesis of Su(Ste) piRNAs. (A) Evaluation of the expression level of Stellate and Su(Ste) repeats using RNA-seq library data. A strong increase in Sellate transcript abundance was found in vasEP812−/− and vasEP812;rhi-GFP testis libraries. In vasa mutant and vasEP812;rhi-GFP testes, the level of antisense Su(Ste) transcripts was 2.8- and 2.4-fold higher than in the testes of heterozygous males. Thus, it indicates that in vasa mutants, the accumulation of unprocessed antisense Su(Ste) transcripts occurred. (B) Comparative analysis of the biogenesis of Su(Ste) piRNAs in vasa mutants. In the testes of vasa mutant males, a strong decline in antisense Su(Ste) piRNA production, more than 25-fold compared to heterozygous controls, was found. Antisense Su(Ste) piRNAs detected in the testes of vasa mutants presumably are produced by the phasing mechanism without the ping-pong amplification process. (C) Heat map presentation of the ping-pong signatures (z10 score) for piRNA pairs mapped on the Su(Ste) consensus sequence with 0–2 mm. (D) Heat map presentation of the ping-pong signatures (z10 score) for piRNA pairs mapped on the Stellate consensus sequence with 0–2 mm. Enrichment of ping-pong pairs mapped on Stellate and Su(Ste) sequences was found in the testes of heterozygous males but not in vasa mutant testes. See also Supplementary Table S1 for z score data.
It is known that Stellate genes are transcriptionally silent in early germ cells, and they start to express in spermatocytes (Aravin et al., 2004; Egorova et al., 2009; Venkei et al., 2023; Shao et al., 2024). However, antisense transcription of Su(Ste) repeats was observed at an earlier stage in GSCs and spermatogonia (Aravin et al., 2004; Venkei et al., 2023; Shao et al., 2024). According to a recent study, antisense Su(Ste) transcripts undergo processing into piRNAs in early germ cells via Vasa-independent phased piRNA biogenesis triggered by maternally inherited transposon hoppel-derived piRNAs (Venkei et al., 2023). To analyze a generation pattern of Su(Ste) piRNAs, we prepared and sequenced libraries of 18–29 nt small RNAs isolated from the testes of vasa mutants, control heterozygous males, and males with the transgenic construct rhi-GFP in the background of vasa mutations.
We selected piRNA fractions (23–29 nt) of small RNA libraries and mapped them on the Su(Ste) consensus. Su(Ste)-mapped reads (with 0–2 mismatches) are present in the control heterozygous testis libraries in large amounts (Figure 5B). The ratios of antisense and sense Su(Ste) piRNAs were found in the range 18.2–22.4 (Figure 5B), which is in accordance with our data published earlier (Kotov et al., 2019). In the testes of vasa mutant males, we found a strong decline of antisense Su(Ste) piRNA production more than 25-fold compared to controls (Figure 5B). Similar results were obtained by analysis of the libraries from the testes of vasa mutants carrying rhi-GFP transgene, where antisense Su(Ste) piRNA amounts were found to be decreased from 22- to 47.3-fold relative to heterozygous controls (Figure 5B). Thus, antisense Su(Ste) piRNAs detected in the testes of vasa mutants appear to be those produced by the phasing mechanism. Despite presence, their amount appears to be insufficient to repress Stellate genes. The ping-pong amplification process is known as an adaptive mechanism of the piRNA biogenesis in the germline, ensuring effective repression of targets (Brennecke et al., 2007; Gunawardane et al., 2007). Accordingly, our analysis revealed ping-pong signatures (the prevalence of 10 nt overlapping sense and antisense piRNA pairs) for piRNAs mapped on both Su(Ste) and Stellate in the testes of control heterozygous males vasEP812+/−, vasEP812/vasD1+/−, with z10 score values are 3.41 and 3.69 for Su(Ste), 3.77 and 3.04 for Stellate, respectively (Figures 5C, D; Supplementary Table S1). In the case of vasa mutants, including those with transgene rhi-GFP copy, generation of piRNAs via the ping-pong process does not occur (Figures 5C, D).
Taken together, our data suggest that the presence of maternally inherited transposon-derived piRNAs for the initiation of Su(Ste) piRNA generation by the phasing process itself is not enough for perfect repression of Stellates. Vasa-dependent amplification ping-pong process is strongly required for effective Su(Ste) piRNA biogenesis and Stellate silencing.
Discussion
A large body of evidence indicates that Vasa plays an essential role in Drosophila oogenesis during the early and late stages (Lasko and Ashburner, 1990; Czech et al., 2013; Durdevic et al., 2018; Durdevic and Ephrussi, 2019). Females with vasa null alleles lay a few embryos; they do not form germ cells, and adult offspring possess abdominal defects and defects of gonad development (Lasko and Ashburner, 1988; Hay et al., 1988). The functions of Vasa in early oogenesis are also associated with the maintenance and self-renewal divisions of GSCs (Dehghani and Lasko, 2015). At the same time, Vasa functions in fly spermatogenesis have remained unclear to date. Moreover, it is supposed that Vasa is not required for spermatogenesis since young males, mutants for vasa, have been found to be fertile (Lasko and Ashburner, 1988; Dehghani and Lasko, 2017). This circumstance for a long time has supported the hypothesis about sexual dimorphism of Vasa functions in D. melanogaster, similar to that showing for mouse Vasa homologue MVH in Mus musculus (Tanaka et al., 2000; Kuramochi-Miyagawa et al., 2010).
Here we first demonstrated that RNA helicase Vasa is an essential protein for GSC maintenance in Drosophila testes. In loss-of-function vasa mutant males, the number of GSCs was significantly reduced in comparison with heterozygous control males of the same age (Figure 2B). The premature loss of GSCs caused a reduction in the total population of differentiating germ cells, suppression of spermatogenesis, and a sharp fertility decline with aging (Figures 1C–E). The loss of GSCs was not associated with increased incidences of programmed cell death (Figures 2C, D) and was rather a consequence of their poor maintenance in the niche. The fertility of young vasa males in a short time after eclosion indicates that age-dependent loss of GSCs appears to be a cell-autonomous process at the level of a single GSC, which loses contact with the niche and enters differentiation. In oogenesis, Vasa is known as a translational activator of specific transcripts, such as nanos, gurken, and mei-P26 mRNAs (Gavis et al., 1996; Styhler et al., 1998; Tomancak et al., 1998; Johnstone and Lasko, 2004; Liu et al., 2009). Other protein components of the piRNA pathway, Aub and Piwi in Drosophila and MIWI and MILI in mice, have been shown to involve translational regulation of protein-coding genes. Aub and MIWI physically interact with the components of translation machinery such as the cap-binding complex through their interactions with subunits of the eIF3 complex, eIF4E, PABP, and Aub is also able to recruit poly(A) polymerase Wispy for the stabilization of specific transcripts in the embryonic germ plasm (Unhavaithaya et al., 2009; Dufourt et al., 2017; Dai et al., 2019; Ramat et al., 2020; Ramat and Simonelig, 2021). Vasa presumably is a component of a protein-protein interaction network, including Aub, Rm62, PABP, and eIF3 proteins, that have been shown to control germ cell development and maintenance (Ma et al., 2017; Ramat et al., 2020). A set of factors of translation initiation involved in the formation of the pre-initiation complex, eIF2, eIF3, and the eIF4E-4G cap-binding complex has been shown to be coimmunoprecipitated with Vasa from Drosophila ovaries (Durdevic and Ephrussi, 2019; Bansal et al., 2020). However, the exact mechanism and direct mRNA targets of translational regulation with the aid of Vasa remain obscure to date, and nothing is known about Vasa targets in Drosophila spermatogenesis.
Here we uncovered a direct relationship between Vasa and rhi (Figures 3B, D). Significantly decreasing rhi transcript levels in vasa mutant testes and the association of Vasa with rhi transcripts indicate its involvement in the positive regulation of rhi expression at the posttranscriptional level. Rhi is an important piRNA pathway protein, a key component of the RDC complex, responsible for non-canonical transcription from double-stranded piRNA clusters (Klattenhoff et al., 2009; Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014). Recently published data demonstrate that mutations rhi exhibit morphological and functional abnormalities in the testes (Chen et al., 2021) that resemble those observed in vasa mutant testes in our study, including premature loss of GSCs and rapid decline of male fertility with age. Taking into account the effect of Vasa on rhi expression, we generated flies carrying the construct with the additional copy of rhi in the absence of vasa expression. This results in the restoration of normal testes morphology and the maintenance of GSCs for a long time, but not the restoration of male fertility (Figure 4; Supplementary Figure S6E).
The experiment for “rescuing” testis morphology with the aid of an additional rhi dose allowed us to identify two most critical developmental points of necessity in Vasa function for spermatogenesis. First is the maintenance of GSCs associated with positive regulation of rhi expression. It remains unclear how Rhi functions in maintaining GSCs in the testes. We assume that this function is not directly related to piRNA repression of TEs. In the recent study, it has been shown that, apart from piRNA clusters, Rhi protein is recruited to hundreds of chromatin sites in the ovaries with the aid of ovary-specific partner protein Kipferl (Baumgartner et al., 2022). The genetic and functional regulatory relationship established between Vasa and Rhi, components of the piRNA system, supports the assumption that the piRNA pathway could be involved in gene regulatory process directed to GSC maintenance in the testes of D. melanogaster. However, further studies are needed to explore the molecular mechanism of this process.
The second critical point for ensuring male fertility in D. melanogaster is Vasa-dependent Stellate silencing in premeiotic spermatocytes. According to the current view, antisense transcription of Y-linked Su(Ste) repeats begins in early germ cells of testes (Aravin et al., 2004; Venkei et al., 2023; Shao et al., 2024). Note that antisense transcription from the Su(Ste) cluster does not require Rhi, as has been shown previously (Chen et al., 2021). Su(Ste) is not a Rhi-dependent dual-stranded piRNA cluster with a non-canonical transcription mode described in Drosophila ovaries (Le Thomas et al., 2014; Mohn et al., 2014; Zhang et al., 2014). Transcription of sense Su(Ste) RNAs is carried out from the classical promoter region in a later developmental period than antisense RNAs (Aravin et al., 2004; Olenkina et al., 2012; Shao et al., 2024). Phasing-based production of antisense Su(Ste) piRNAs in early germ cells appears to initiate with the aid of maternally inherited piRNAs derived from transposon hoppel inserted in the promotor region of Su(Ste) repeats (Venkei et al., 2023). The authors of this paper emphasize that abundant antisense Su(Ste) piRNAs are produced by the phasing before starting transcription of their target Stellate genes. However, other sources demonstrate a significant overlap between the transcription windows of Su(Ste) and Stellate rather than their spatiotemporal separation (Shao et al., 2024). Taking advantage of small RNA library data analysis, we revealed that only a small amount of antisense Su(Ste) piRNAs were accumulated in vasa mutant testes, with a more than 25-fold reduction compared to heterozygous control (Figure 5B). Indeed, these piRNAs have no characteristic ping-pong signatures, indicating in favor of their processing by the phasing mechanism (Figure 5C). Because Stellate derepression is clearly observed in vasa mutant testes (Figure 4F; Supplementary Figure S6A), the amount of these Su(Ste) piRNAs is not enough for Stellate silencing. Note that in the testes of males carrying the additional copy of rhi in the absence of vasa expression, we revealed the similar disruption of antisense Su(Ste) piRNA biogenesis and the absence of the ping-pong process (Figures 5B–D). Thus, using comparative analysis of small RNA libraries, we confirmed that Vasa-dependent processing of abundant Su(Ste) piRNAs via the ping-pong mechanism is required for Stellate silencing (Figure 5C; Supplementary Table S1). This processing is Rhi-independent in accordance with previous findings (Chen et al., 2021).
Here we found a strong correlation between vasa deficiencies in the testes and ovaries and the decreased level of Aub protein, but not its transcripts (Figure 3C; Supplementary Figure S4). As indicated above in the testes of vasa mutant males, a strong, more than 25-fold reduction of antisense Su(Ste) piRNA production compared to heterozygous controls occurs (Figure 5B). A loss of Vasa, the top hierarchical nuage component, causes the mislocalization of Aub and AGO3 in the cytoplasm in the ovaries and testes (Lim and Kai, 2007; Malone et al., 2009; Kibanov et al., 2011; Kotov et al., 2014). Impaired nuage granule formation leads to a decline of posttranscriptional piRNA biogenesis (Lim and Kai, 2007; Kibanov et al., 2011). It is also shown that unloaded Aub with defective PAZ domain is unable to bind mRNA targets and did not localize to the germ plasm (Barckmann et al., 2015). We assume that empty Aub that is distributed in the cytoplasm undergoes a premature elimination. Note that several studies reported that unloaded ARGONAUTE family proteins undergo selective degradation in various species (Derrien et al., 2012; Martinez and Gregory, 2013; Kobayashi et al., 2019a; b). E3 ubiquitin-protein ligase Iruka recognizes miRNA-empty AGO1 in Drosophila and triggers its degradation via polyubiquitination of lysine residues across practically all domains, especially K514 in the linker two domain (Kobayashi et al., 2019a). Elimination of dysfunctional AGO1 that is not able to bind miRNA by proteasomal or autophagic mechanisms ensures a quality control of miRNA-mediated silencing (Kobayashi et al., 2019a; b). For unloaded PIWI subfamily of ARGONAUTE proteins, such mechanisms of elimination have not been shown yet, and this needs further studies. However, murine PIWI (MIWI) in late spermatids is ubiquitinated and degraded through the proteasome system, indicating a feedforward mechanism for coordinated elimination of the MIWI/piRNA machinery at the specific developmental stage (Zhao et al., 2013). Taking that into account, we propose that Vasa contributes to the stability of Aub protein by recruiting it into the nuage granules for assembling functional piRNA-Aub complexes and providing piRNA silencing of mRNA targets. Here we showed that RNA helicase Vasa essentially contributes to male fertility in D. melanogaster at least on two distinct stages: GSC maintenance in early spermatogenesis and piRNA-dependent Stellate silencing need for correct passage through meiosis in spermatocytes. Our study uncovers a novel mechanistic link involving Vasa and Rhi in a regulatory network that mediates GSC maintenance. The open questions include whether observed relationships indicate a possible role of piRNA silencing in this important developmental process.
Materials and methods
Fly stocks and genetics
Germinal tissues of adult D. melanogaster males and females generally raised at 23°C were used. We used following Drosophila stocks: vasEP812 (vasLYG2) mutant strain is originated as a result of P-element insertion into the last vig (vasa intronic gene) exon, adjacent to the 3′-splice site (Ashburner et al., 1999); vasD1 (b1vas3/CyO) loss-of-function allele was obtained using ethyl methanesulfonate mutagen with no changes in coding vasa sequence (Lasko and Ashburner, 1990); vasPH165 allele was obtained by imprecise excision of P-element, resulting in a 7343bp deletion removing the entire vasa coding region (Styhler et al., 1998). Flies with heteroallelic mutation combinations vasEP812/vasPH165, and vasEP812/vasD1 as well as homozygotes vasEP812 males were chosen for our experiments. yw flies (Df(1)w67c23(2)y), was used as wild-type controls. For RIP-PCR experiments we used yw strain carrying transgene construct vas-GFP (vascKa.GFP) under the control of endogenous vasa promoter kindly provided by P. Lasko (Kugler et al., 2010). The following fly stocks were also used: aubQC42 (BDSC4968), aubHN2 (BDSC8517), were obtained from the Bloomington Drosophila Stock Center (Indiana University, IN, USA); rhi2 (rhi02086) and rhiKG (rhiKG00910) lines were kindly provided from W. Theurkauf Lab. Rhi-GFP line (VDRC313340) were obtained from Vienna Drosophila Resource Center (Vienna, Austria); GFP-tagged rhi is a previously described transgene constructed by inserting N-terminal GFP into genomic BACs by recombineering (Mohn et al., 2014). cry1 line, ywf/c(1)DX, yf/cry1BS, Yy+, carries a deletion of the bulk of the Su(Ste) locus on the Y chromosome.
Immunofluorescence staining, TUNEL assay and confocal microscopy
Testes of adult males were dissected in phosphate-buffered saline (PBS) at 4°C, washed with PBT (1×PBS, 0.1% Tween 20) and fixed in 3.7% formaldehyde and PBT for 30 min at room temperature. All the following procedures were carried out as described previously (Kibanov et al., 2013). Staining was detected by laser scanning confocal microscopy using a Zeiss Axio Observer LSM 900 machine (Carl Zeiss). All images were taken with a z-resolution of 1 µm. The obtained pictures were imported into Imaris 5.0.1 (Bitplane AG) for subsequent processing. Counting of spectrosomes and the number of testis GSCs on the confocal images was carried out using Imaris software. The Click-iT TUNEL Alexa Fluor imaging assay kit (Invitrogen) was used for detection of DNA breaks that associated with necrotic or apoptotic cell death according to the published protocol (Kibanov et al., 2013). TUNEL signal of whole cyst of germ cells was estimated as a single event. p-values for pairwise comparison in control and experimental testes were calculated using Wilcoxon/Mann–Whitney (U) test.
Antibodies
The following antibodies were used for immunofluorescence staining: a mix of murine monoclonal anti-Lamin Dm0 ADL67.10 and ADL84.12 antibodies (Developmental Studies Hybridoma Bank, University of Iowa (DSHB)), 1:500; rabbit polyclonal anti-Lamin antibodies (Osouda et al., 2005), 1:500; rat monoclonal anti-Vasa antibody (DSHB), 1:100; murine monoclonal anti-α-spectrin 3A9 antibody (DSHB), 1:200; murine monoclonal anti-Fasciclin III 7G10 antibody (DSHB), 1:25; rabbit polyclonal anti-γH2Av pS137 antibodies (Rockland), 1:100. Alexa Fluor-labeled secondary goat anti-rat IgG, goat anti-rabbit IgG, and goat anti-mouse IgG (Invitrogen) were used as secondary reagents at a dilution of 1:500. DAPI (4′,6-diamidino-2-phenylindole) (Sigma) was used for chromatin staining.
For RNA immunoprecipitation (RIP) experiments, rabbit polyclonal anti-GFP antibody ab6556 (Abcam) was used.
For Western blot analysis, the following antibodies were used: murine monoclonal anti-β-Actin antibody ab8224 (Abcam), 1:2000; rat monoclonal anti-Vasa antibody (DSHB), 1:2000; rabbit polyclonal anti-VIG 1803 antibodies (Gracheva et al., 2009), 1:500; murine monoclonal anti-Aub 4D10 antibody (Nishida et al., 2007), 1:1500; rabbit polyclonal βNACtes antibodies (Kogan et al., 2022), 1:1000; murine polyclonal anti-Stellate antibodies, 1:500, (Egorova et al., 2009). Samples were resolved by SDS-PAGE and blotted onto PVDF membrane Immobilon-P (Sigma). Alkaline phosphatase-conjugated anti-mouse, anti-rabbit, anti-rat and anti-goat antibodies (Sigma) were used as secondary reagents at a dilution of 1:20,000. Blots were developed using the Immun-Star AP detection system (Bio-Rad Laboratories). All experiments were performed at least in triplicate with independent preparations of testis or ovarian lysates.
RNA extraction, reverse transcription and quantitative PCR
Total RNA was isolated from sets of 50–100 pairs of dissected testes or 30–50 pairs of ovaries, using TRIzol Reagent (Invitrogen) according to the manufacturer’s recommendations. cDNA was synthesized using random hexamers and SuperScript II reverse transcriptase (Invitrogen). cDNA samples were analyzed by real-time quantitative PCR using the incorporation of SYTO-13 (Invitrogen). Thermal cycling consisted of 5 min at 95°C, followed by 45 cycles of denaturation (94°C, 20 s), annealing (64°C, 20 s), extension (72°C, 20 s), and a final extension of 5 min at 72°C. We used rp49 (rpL32) as a loading control. All experiments were performed with at least three independent RNA samples; each sample was analyzed in duplicate. The following primers were used for RT-qPCR and RIP-RT-qPCR: rp49 fw 5′-ATGACCATCCGCCCAGCATAC-3′, rev 5′-GCTTAGCATATCGATCCGACTGG-3′; aub fw 5′-CATGAGTGAACATACCAGGCTGAA-3′, rew 5′-GCGGAGTCCAGCTCGATGTT-3′; rhi fw 5′-CGGTTTTCCGAACGAGAAC-3′, rew 5′-CGGCCTTCCGATGCA-3′.
Copy number of Stellate genes estimation
The number of X-linked Stellate genes in the genome was assessed by quantitative PCR with genomic DNA using highly specific Stellate primers, the efficiency of which was previously tested and compared with the efficiency of rp49 primers (normalization control). In these experiments, genomic DNA of both males and females of D. melanogaster was used as an additional control; the obtained value of the relative number of Stellate genes for males was 2-fold lower compared to females. Stellate primers: fw 5′-GATTGGTTCCTCGGGATCAA-3′, rev 5′-CCGTACAACAAGCCAGAGGAACT-3′.
Fertility tests
Sets of 25–30 experimental males and their heterozygous siblings as a control were analyzed for their fertility. Individual adult male (0 days after eclosion) was placed with three virgin yw females 4- to 6-day-old for 5 days in separate vials at 25°C. After that the parent flies were removed from the vial. The males were translocated in other vials with new three virgin females for the next 5 days up to six times. The adult progeny in each vial was counted within 13–18-day interval after mating. The tests were carried out at 25°C. The flies were subjected to a light–dark cycle of 12:12 h. At the end of the experiments, the offspring from one male were counted per female for each time period.
RNA immunoprecipitation
Ovaries of vas-GFP flies 3–5 days after eclosion were dissected in ice-cold phosphate-buffered saline (PBS). Freshly dissected gonads were cross-linked in PBS supplemented with 0.25% formaldehyde for 30 min, were washed by three times for 5 min and were stored at −70°C until further use. Total cellular lysates were obtained from 500 pair of cross-linked ovaries using pre-chilled Dounce homogenizer on ice in a cold lysis buffer (50 mM Tris-HCl pH 8.0; 100 mM KCl; 2 mM MgCl2; 1 mM DTT; 0.5% Nonidet P-40 (NP-40); in the presence of 1% protease and phosphatase inhibitors (Sigma) with the addition of 2% Ribolock (Thermo Fisher Scientific) by two series of 100 strokes of pestle A with a 5 min incubation between the series. The lysates were transferred to 1.7-mL polypropylene microcentrifuge tubes and were precipitated at 5000 g for 10 min at 4°C. The supernatant fractions (crude extracts) were cleared by subsequent centrifugation at 16,000 g for 20 min at 4°C. The clarified lysates were transferred to new tubes and diluted to protein concentration of 7–10 mg/mL with lysis buffer. We used 50 µL of 50%-slurry of Protein A beads (Invitrogen) for each tube for immunoprecipitation. The beads were previously washed two times with 500 µL cold PBS supplied by Tween 20 (1 × PBS; 0.1% Tween 20 (PBST)) and incubated with anti GFP-antibodies (Abcam) or normal rabbit serum for negative control in 200 µL PBST 20 min at room temperature on the rotator. The antibody-coated beads were washed and pre-equilibrated with lysis buffer. Then the equal volumes of clarified lysates were added to the experimental and control tubes for immunoprecipitation. After incubation 40 min at RT on the rotator, washing was performed three times with 1 mL washing buffer NT2 (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.5 mM DTT; 0.3% NP-40), three times with 1 mL washing buffer NT3-Urea (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1M urea; 0.5 mM DTT; 0.3% NP-40) and one time NT2 buffer. Small portions of immunoprecipitated material from each tube were put aside for Western blot analysis. For that the bound material was eluted from the beads by boiling in 30 μL 2×sample buffer containing 0.2 M DTT. Samples were subsequently resolved by % SDS-PAGE and blotted onto PVDF membrane Immobilon-P (Sigma). The rest parts of the material were subjected to proteolysis to remove a bulk of proteins performed on-bead in solution of 4 mg/ml Proteinase K (Promega) in PK2 buffer (10 mM Tris-HCl pH 7.5; 50 mM NaCl; 5 mM EDTA; 0.5% SDS; 1 mM DTT; in the presence of 2% Ribolock) for 2 hours at 55°C with extensive agitation. After that the tubes were subjected by incubation for 10 min at 65°C for removing of cross-linking between RNA and peptide material. Soluble fractions were separated from beads and were processed with TRIzol LS reagent (Thermo Fisher Scientific) followed by RNA isolation. RT-qPCR analysis was performed as mentioned above.
Library preparation
Total RNA for RNA-seq libraries were isolated from vasEP812−/−, vasEP812+/−, and vasEP812−/−; rhi-GFP D. melanogaster gonads with ExtractRNA (Evrogen). rRNA was depleted with Dynabeads MyOne Streptavidin C1 (Thermo Scientific Fisher) conjugated with synthesized anti-rDNA oligos. RNA-seq libraries were generated using the NEBNext Ultra™ II Directional RNA Library Prep Kit for Illumina (#E7760, NEB) according to the manufacturer’s instructions. Rhibo-depleted RNA-seq libraries were sequenced in 100 bp single-end mode on NovaSeq 6000 platform. Small RNAs of 18–29 nt in size from gonads of D. melanogaster vasEP812−/−, vasEP812+/−, vasEP812−/−; rhi-GFP, vasEP812/vasD1, mix of vasEP812/+ with vasD1/+, and vasEP812/vasD1; rhi-GFP were isolated with TRIzol Reagent (Invitrogen). Small RNA fraction (19–29 nt) was purified by 15% polyacrylamide gel and libraries were prepared using NEBNext Multiplex Small RNA Sample Prep Set for Illumina (E7300S) and sequenced on NovaSeq 6000 in 50 bp single-end mode.
Bioinformatics analysis
Quality control of the sequenced libraries was carried out using the FastQC tool. For subsequent analysis, only reads with fastq score >30 were used. To evaluate the expression level of the Stellate and Su(Ste) loci RNA-seq libraries were mapped to consensus sequences using bowtie version 1.1.0 (-l 150). Mapped reads were counted and normalized to the library depth, rpm. To evaluate the expression level of rhi RNA-seq libraries were pseudo-aligned to D. melanogaster transcriptome (GCF_000001215.4) by kallisto (v.0.50.0) with following parameters--single -l 100 -s 0.1. Quantified abundances of transcripts (transcript per million, tpm) for protein-coding genes were imported with tximport (v.1.24.0). Adapter sequences from small RNA libraries were removed by cutadapt 2.8. After quality control with FastQC, rRNA, snRNA, snoRNA, microRNA and tRNA were filtered from the libraries and piRNA were selected by size (23–30 nt). To evaluate the piRNA level and analyze ping-pong signature of piRNAs for Stellate and Su(Ste) filtered piRNA fraction were mapped to consensus sequences using bowtie with o-2 mm (-l 30). Mapped reads were normalized to the microRNA level. Ping-pong signatures were calculated using a signature. py script (Antoniewski, 2014).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE269987 and GSE269988.
Author contributions
VA: Conceptualization, Formal Analysis, Investigation, Visualization, Writing–original draft. AK: Conceptualization, Formal Analysis, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing. SB: Formal Analysis, Investigation, Writing–review and editing. IK: Formal Analysis, Investigation, Writing–review and editing. OM: Formal Analysis, Investigation, Writing–review and editing. AS: Formal Analysis, Investigation, Methodology, Writing–review and editing. LO: Conceptualization, Data curation, Project administration, Supervision, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was conducted under the Institute of Developmental Biology Russian Academy of Sciences Government basic research program in 2024, No. 0088–2024-0017. The funder had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Acknowledgments
We thank the Developmental Studies Hybridoma Bank and the Bloomington Stock Center for providing fly strains and antibodies used in this study. We thank A. Aravin, M. Siomi, P. Fisher, E. Gracheva, and G. Kogan for antibodies and flies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1450227/full#supplementary-material
References
Adashev, V. E., Kotov, A. A., Bazylev, S. S., Shatskikh, A. S., Aravin, A. A., and Olenina, L. V. (2021). Stellate genes and the piRNA pathway in speciation and reproductive isolation of Drosophila melanogaster. Front. Genet. 11, 610665. doi:10.3389/fgene.2020.610665
Adashev, V. E., Kotov, A. A., and Olenina, L. V. (2023). RNA helicase vasa as a multifunctional conservative regulator of gametogenesis in eukaryotes. Curr. Issues Mol. Biol. 45 (7), 5677–5705. doi:10.3390/cimb45070358
Antoniewski, C. (2014). Computing siRNA and piRNA overlap signatures. Methods Mol. Biol. 1173, 135–146. doi:10.1007/978-1-4939-0931-5_12
Arama, E., and Steller, H. (2006). Detection of apoptosis by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling and acridine orange in Drosophila embryos and adult male gonads. Nat. Protoc. 1 (4), 1725–1731. doi:10.1038/nprot.2006.235
Aravin, A. A., Klenov, M. S., Vagin, V. V., Bantignies, F., Cavalli, G., and Gvozdev, V. A. (2004). Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 24 (15), 6742–6750. doi:10.1128/MCB.24.15.6742-6750.2004
Aravin, A. A., Naumova, N. M., Tulin, A. V., Vagin, V. V., Rozovsky, Y. M., and Gvozdev, V. A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11 (13), 1017–1027. doi:10.1016/s0960-9822(01)00299-8
Ashburner, M., Misra, S., Roote, J., Lewis, S. E., Blazej, R., Davis, T., et al. (1999). An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: the Adh region. Genetics 153 (1), 179–219. doi:10.1093/genetics/153.1.179
Bansal, P., Madlung, J., Schaaf, K., Macek, B., and Bono, F. (2020). An interaction network of RNA-binding proteins involved in Drosophila oogenesis. Mol. Cell. Proteomics 19 (9), 1485–1502. doi:10.1074/mcp.RA119.001912
Barckmann, B., Pierson, S., Dufourt, J., Papin, C., Armenise, C., Port, F., et al. (2015). Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 12 (7), 1205–1216. doi:10.1016/j.celrep.2015.07.030
Baumgartner, L., Handler, D., Platzer, S. W., Yu, C., Duchek, P., and Brennecke, J. (2022). The Drosophila ZAD zinc finger protein Kipferl guides Rhino to piRNA clusters. Elife 11, e80067. doi:10.7554/eLife.80067
Bozzetti, M. P., Massari, S., Finelli, P., Meggio, F., Pinna, L. A., Boldyreff, B., et al. (1995). The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc. Natl. Acad. Sci. U. S. A. 92 (13), 6067–6071. doi:10.1073/pnas.92.13.6067
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell , 128 (6), 1089–1103. doi:10.1016/j.cell.2007.01.043
Carrera, P., Johnstone, O., Nakamura, A., Casanova, J., Jäckle, H., and Lasko, P. (2000). VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell. 5 (1), 181–187. doi:10.1016/s1097-2765(00)80414-1
Chen, P., Luo, Y., and Aravin, A. A. (2021). RDC complex executes a dynamic piRNA program during Drosophila spermatogenesis to safeguard male fertility. PLoS Genet. 17 (9), e1009591. doi:10.1371/journal.pgen.1009591
Czech, B., Preall, J. B., McGinn, J., and Hannon, G. J. (2013). A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell. 50 (5), 749–761. doi:10.1016/j.molcel.2013.04.007
Dai, P., Wang, X., Gou, L. T., Li, Z. T., Wen, Z., Chen, Z. G., et al. (2019). A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell 179 (7), 1566–1581.e16. doi:10.1016/j.cell.2019.11.022
Dehghani, M., and Lasko, P. (2015). In vivo mapping of the functional regions of the DEAD-box helicase Vasa. Biol. Open 4 (4), 450–462. doi:10.1242/bio.201410579
Dehghani, M., and Lasko, P. (2017). Multiple functions of the DEAD-box helicase vasa in Drosophila oogenesis. Results Probl. Cell. Differ. 63, 127–147. doi:10.1007/978-3-319-60855-6_6
Derrien, B., Baumberger, N., Schepetilnikov, M., Viotti, C., De Cillia, J., Ziegler-Graff, V., et al. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U. S. A. 109 (39), 15942–15946. doi:10.1073/pnas.1209487109
Dufourt, J., Bontonou, G., Chartier, A., Jahan, C., Meunier, A. C., Pierson, S., et al. (2017). piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nat. Commun. 8 (1), 1305. doi:10.1038/s41467-017-01431-5
Durdevic, Z., and Ephrussi, A. (2019). Germ cell lineage homeostasis in Drosophila requires the vasa RNA helicase. Genetics 213 (3), 911–922. doi:10.1534/genetics.119.302558
Durdevic, Z., Pillai, R. S., and Ephrussi, A. (2018). Transposon silencing in the Drosophila female germline is essential for genome stability in progeny embryos. Life Sci. Alliance 1, e201800179. doi:10.26508/lsa.201800179
Egorova, K. S., Olenkina, O. M., Kibanov, M. V., Kalmykova, A. I., Gvozdev, V. A., and Olenina, L. V. (2009). Genetically Derepressed Nucleoplasmic Stellate Protein in Spermatocytes of D. melanogaster interacts with the catalytic subunit of protein kinase 2 and carries histone-like lysine-methylated mark. J. Mol. Biol. 389 (5), 895–906. doi:10.1016/j.jmb.2009.04.064
Gavis, E. R., Lunsford, L., Bergsten, S. E., and Lehmann, R. (1996). A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122 (9), 2791–2800. doi:10.1242/dev.122.9.2791
Gracheva, E., Dus, M., and Elgin, S. C. (2009). Drosophila RISC component VIG and its homolog Vig2 impact heterochromatin formation. PLoS One 4, e6182. doi:10.1371/journal.pone.0006182
Greenspan, L. J., de Cuevas, M., and Matunis, E. (2015). Genetics of gonadal stem cell renewal. Annu. Rev. Cell. Dev. Biol. 31, 291–315. doi:10.1146/annurev-cellbio-100913-013344
Gunawardane, L. S., Saito, K., Nishida, K. M., Miyoshi, K., Kawamura, Y., Nagami, T., et al. (2007). A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science 315 (5818), 1587–1590. doi:10.1126/science.1140494
Hansen, C. L., and Pelegri, F. (2021). Primordial germ cell specification in vertebrate embryos: phylogenetic distribution and conserved molecular features of preformation and induction. Front. Cell Dev. Biol. 9, 730332. doi:10.3389/fcell.2021.730332
Hartung, O., Forbes, M. M., and Marlow, F. L. (2014). Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 81 (10), 946–961. doi:10.1002/mrd.22414
Hay, B., Jan, L. Y., and Jan, Y. N. (1988). A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55 (4), 577–587. doi:10.1016/0092-8674(88)90216-4
Johnstone, O., and Lasko, P. (2004). Interaction with eIF5B is essential for Vasa function during development. Development 131 (17), 4167–4178. doi:10.1242/dev.01286
Kibanov, M. V., Egorova, K. S., Ryazansky, S. S., Sokolova, O. A., Kotov, A. A., Olenkina, O. M., et al. (2011). A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing. Mol. Biol. Cell 22 (18), 3410–3419. doi:10.1091/mbc.E11-02-0168
Kibanov, M. V., Kotov, A. A., and Olenina, L. V. (2013). Multicolor fluorescence imaging of whole-mount Drosophila testes for studying spermatogenesis. Anal. Biochem. 436, 55–64. doi:10.1016/j.ab.2013.01.009
Klattenhoff, C., Bratu, D. P., McGinnis-Schultz, N., Koppetsch, B. S., Cook, H. A., and Theurkauf, W. E. (2007). Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 12 (1), 45–55. doi:10.1016/j.devcel.2006.12.001
Klattenhoff, C., Xi, H., Li, C., Lee, S., Xu, J., Khurana, J. S., et al. (2009). The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138 (6), 1137–1149. doi:10.1016/j.cell.2009.07.014
Kobayashi, H., Shoji, K., Kiyokawa, K., Negishi, L., and Tomari, Y. (2019a). Iruka eliminates dysfunctional argonaute by selective ubiquitination of its empty state. Mol. Cell 73 (1), 119–129.e5. doi:10.1016/j.molcel.2018.10.033
Kobayashi, H., Shoji, K., Kiyokawa, K., Negishi, L., and Tomari, Y. (2019b). VCP machinery mediates autophagic degradation of empty argonaute. Cell Rep. 28 (5), 1144–1153.e4. doi:10.1016/j.celrep.2019.07.003
Kogan, G. L., Mikhaleva, E. A., Olenkina, O. M., Ryazansky, S. S., Galzitskaya, O. V., Abramov, Y. A., et al. (2022). Extended disordered regions of ribosome-associated NAC proteins paralogs belong only to the germline in Drosophila melanogaster. Sci. Rep. 12 (1), 11191. doi:10.1038/s41598-022-15233-3
Kotov, A. A., Adashev, V. E., Godneeva, B. K., Ninova, M., Shatskikh, A. S., Bazylev, S. S., et al. (2019). piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster. Nucleic Acids Res. 47 (8), 4255–4271. doi:10.1093/nar/gkz130
Kotov, A. A., Akulenko, N. V., Kibanov, M. V., and Olenina, L. V. (2014). Dead-box RNA helicases in animal gametogenesis. Mol. Biol. (Mosk.) 48, 16–28. doi:10.1134/S0026893314010063
Kugler, J. M., Woo, J. S., Oh, B. H., and Lasko, P. (2010). Regulation of Drosophila vasa in vivo through paralogous cullin-RING E3 ligase specificity receptors. Mol. Cell. Biol. 30 (7), 1769–1782. doi:10.1128/MCB.01100-09
Kuramochi-Miyagawa, S., Watanabe, T., Gotoh, K., Takamatsu, K., Chuma, S., Kojima-Kita, K., et al. (2010). MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24 (9), 887–892. doi:10.1101/gad.1902110
Lasko, P. (2013). The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta 1829 (8), 810–816. doi:10.1016/j.bbagrm.2013.04.005
Lasko, P. F., and Ashburner, M. (1988). The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 335 (6191), 611–617. doi:10.1038/335611a0
Lasko, P. F., and Ashburner, M. (1990). Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 4 (6), 905–921. doi:10.1101/gad.4.6.905
Le Thomas, A., Stuwe, E., Li, S., Du, J., Marinov, G., Rozhkov, N., et al. (2014). Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 28 (15), 1667–1680. doi:10.1101/gad.245514.114
Liang, L., Diehl-Jones, W., and Lasko, P. (1994). Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120 (5), 1201–1211. doi:10.1242/dev.120.5.1201
Lim, A. K., and Kai, T. (2007). Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 104 (16), 6714–6719. doi:10.1073/pnas.0701920104
Linder, P., and Jankowsky, E. (2011). From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell. Biol. 12 (8), 505–516. doi:10.1038/nrm3154
Liu, N., Han, H., and Lasko, P. (2009). Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3´ UTR. Genes Dev. 23 (23), 2742–2752. doi:10.1101/gad.1820709
Ma, X., Zhu, X., Han, Y., Story, B., Do, T., Song, X., et al. (2017). Aubergine controls germline stem cell self-renewal and progeny differentiation via distinct mechanisms. Dev. Cell 41 (2), 157–169.e5. doi:10.1016/j.devcel.2017.03.023
Malone, C. D., Brennecke, J., Dus, M., Stark, A., McCombie, W. R., Sachidanandam, R., et al. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137 (3), 522–535. doi:10.1016/j.cell.2009.03.040
Martinez, N. J., and Gregory, R. I. (2013). Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA 19 (5), 605–612. doi:10.1261/rna.036434.112
Mohn, F., Sienski, G., Handler, D., and Brennecke, J. (2014). The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell , 157 (6), 1364–1379. doi:10.1016/j.cell.2014.04.031
Nishida, K. M., Saito, K., Mori, T., Kawamura, Y., Nagami-Okada, T., Inagaki, S., et al. (2007). Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13 (11), 1911–1922. doi:10.1261/rna.744307
Nott, T. J., Petsalaki, E., Farber, P., Jervis, D., Fussner, E., Plochowietz, A., et al. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57 (5), 936–947. doi:10.1016/j.molcel.2015.01.013
Olenkina, O. M., Egorova, K. S., Kibanov, M. V., Gervaziev, Y. V., Gvozdev, V. A., and Olenina, L. V. (2012). Promoter contribution to the testis-specific expression of Stellate gene family in Drosophila melanogaster. Gene 499 (1), 143–153. doi:10.1016/j.gene.2012.03.023
Osouda, S., Nakamura, Y., de Saint Phalle, B., McConnell, M., Horigome, T., Sugiyama, S., et al. (2005). Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev. Biol. 284 (1), 219–232. doi:10.1016/j.ydbio.2005.05.022
Palumbo, G., Bonaccorsi, S., Robbins, L. G., and Pimpinelli, S. (1994). Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics 138 (4), 1181–1197. doi:10.1093/genetics/138.4.1181
Ramat, A., Garcia-Silva, M. R., Jahan, C., Naït-Saïdi, R., Dufourt, J., Garret, C., et al. (2020). The PIWI protein Aubergine recruits eIF3 to activate translation in the germ plasm. Cell Res. 30 (5), 421–435. doi:10.1038/s41422-020-0294-9
Ramat, A., and Simonelig, M. (2021). Functions of PIWI proteins in gene regulation: new arrows added to the piRNA quiver. Trends Genet. 37 (2), 188–200. doi:10.1016/j.tig.2020.08.011
Rojas-Ríos, P., Chartier, A., Pierson, S., and Simonelig, M. (2017). Aubergine and piRNAs promote germline stem cell self-renewal by repressing the proto-oncogene Cbl. EMBO J. 36 (21), 3194–3211. doi:10.15252/embj.201797259
Roussell, D. L., and Bennett, K. L. (1993). glh-1, a germ-line putative RNA helicase from Caenorhabditis, has four zinc fingers. Proc. Natl. Acad. Sci. U. S. A. 90 (20), 9300–9304. doi:10.1073/pnas.90.20.9300
Schüpbach, T., and Wieschaus, E. (1986). Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev. Biol. 113 (2), 443–448. doi:10.1016/0012-1606(86)90179-x
Schüpbach, T., and Wieschaus, E. (1989). Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics 121 (1), 101–117. doi:10.1093/genetics/121.1.101
Schüpbach, T., and Wieschaus, E. (1991). Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129 (4), 1119–1136. doi:10.1093/genetics/129.4.1119
Shao, Z., Hu, J., Jandura, A., Wilk, R., Jachimowicz, M., Ma, L., et al. (2024). Spatially revealed roles for lncRNAs in Drosophila spermatogenesis, Y chromosome function and evolution. Nat. Commun. 15 (1), 3806. doi:10.1038/s41467-024-47346-w
Snee, M. J., and Macdonald, P. M. (2004). Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 117 (Pt 10), 2109–2120. doi:10.1242/jcs.01059
So, C., Cheng, S., and Schuh, M. (2021). Phase separation during germline development. Trends Cell. Biol. 31 (4), 254–268. doi:10.1016/j.tcb.2020.12.004
Spradling, A., Fuller, M. T., Braun, R. E., and Yoshida, S. (2011). Germline stem cells. Cold Spring Harb. Perspect. Biol. 3 (11), a002642. doi:10.1101/cshperspect.a002642
Styhler, S., Nakamura, A., Swan, A., Suter, B., and Lasko, P. (1998). Vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125 (9), 1569–1578. doi:10.1242/dev.125.9.1569
Tanaka, S. S., Toyooka, Y., Akasu, R., Katoh-Fukui, Y., Nakahara, Y., Suzuki, R., et al. (2000). The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14 (7), 841–853. doi:10.1101/gad.14.7.841
Tomancak, P., Guichet, A., Zavorszky, P., and Ephrussi, A. (1998). Oocyte polarity depends on regulation of gurken by Vasa. Development 125 (9), 1723–1732. doi:10.1242/dev.125.9.1723
Trcek, T., and Lehmann, R. (2019). Germ granules in Drosophila. Traffic 20 (9), 650–660. doi:10.1111/tra.12674
Unhavaithaya, Y., Hao, Y., Beyret, E., Yin, H., Kuramochi-Miyagawa, S., Nakano, T., et al. (2009). MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 284 (10), 6507–6519. doi:10.1074/jbc.M809104200
Vagin, V. V., Klenov, M. S., Kalmykova, A. I., Stolyarenko, A. D., Kotelnikov, R. N., and Gvozdev, V. A. (2004). The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 1 (1), 53–57. doi:10.4161/rna.1.1.943
Vagin, V. V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., and Zamore, P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 (5785), 320–324. doi:10.1126/science.1129333
Venkei, Z. G., Gainetdinov, I., Bagci, A., Starostik, M. R., Choi, C. P., Fingerhut, J. M., et al. (2023). A maternally programmed intergenerational mechanism enables male offspring to make piRNAs from Y-linked precursor RNAs in Drosophila. Nat. Cell. Biol. 25 (10), 1495–1505. doi:10.1038/s41556-023-01227-4
Wang, L., Dou, K., Moon, S., Tan, F. J., and Zhang, Z. Z. (2018). Hijacking oogenesis enables massive propagation of LINE and retroviral transposons. Cell 174 (5), 1082–1094.e12. doi:10.1016/j.cell.2018.06.040
Wang, S. C., Hsu, H. J., Lin, G. W., Wang, T. F., Chang, C. C., and Lin, M. D. (2015). Germ plasm localisation of the HELICc of Vasa in Drosophila: analysis of domain sufficiency and amino acids critical for localisation. Sci. Rep. 5, 14703. doi:10.1038/srep14703
Webster, A., Li, S., Hur, J. K., Wachsmuth, M., Bois, J. S., Perkins, E. M., et al. (2015). Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by krimper. Mol. Cell. 59 (4), 564–575. doi:10.1016/j.molcel.2015.07.017
Xiol, J., Spinelli, P., Laussmann, M. A., Homolka, D., Yang, Z., Cora, E., et al. (2014). RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157 (7), 1698–1711. doi:10.1016/j.cell.2014.05.018
Yamazaki, H., Namba, Y., Kuriyama, S., Nishida, K. M., Kajiya, A., and Siomi, M. C. (2023). Bombyx Vasa sequesters transposon mRNAs in nuage via phase separation requiring RNA binding and self-association. Nat. Commun. 14 (1), 1942. doi:10.1038/s41467-023-37634-2
Zhang, F., Wang, J., Xu, J., Zhang, Z., Koppetsch, B. S., Schultz, N., et al. (2012). UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151 (4), 871–884. doi:10.1016/j.cell.2012.09.040
Zhang, Z., Wang, J., Schultz, N., Zhang, F., Parhad, S. S., Tu, S., et al. (2014). The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157 (6), 1353–1363. doi:10.1016/j.cell.2014.04.030
Zhao, S., Gou, L. T., Zhang, M., Zu, L. D., Hua, M. M., Hua, Y., et al. (2013). piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev. Cell 24 (1), 13–25. doi:10.1016/j.devcel.2012.12.006
Keywords: piRNA pathway, germline stem cells, DEAD-box RNA helicase, spermatogenesis, male fertility
Citation: Adashev VE, Kotov AA, Bazylev SS, Kombarov IA, Olenkina OM, Shatskikh AS and Olenina LV (2024) Essential functions of RNA helicase Vasa in maintaining germline stem cells and piRNA-guided Stellate silencing in Drosophila spermatogenesis. Front. Cell Dev. Biol. 12:1450227. doi: 10.3389/fcell.2024.1450227
Received: 17 June 2024; Accepted: 29 July 2024;
Published: 09 August 2024.
Edited by:
Patricia Rojas Ríos, University of Seville, SpainReviewed by:
Xiaolei Li, University of Pennsylvania, United StatesJeremy Dufourt, UMR9004 Institut de Recherche en Infectiologie de Montpellier (IRIM), France
Copyright © 2024 Adashev, Kotov, Bazylev, Kombarov, Olenkina, Shatskikh and Olenina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ludmila V. Olenina, b2xlbmluYV9sdWRtaWxhQG1haWwucnU=
†These authors have contributed equally to this work and share first authorship
 Vladimir E. Adashev1†
Vladimir E. Adashev1† Alexei A. Kotov
Alexei A. Kotov Ilia A. Kombarov
Ilia A. Kombarov Aleksei S. Shatskikh
Aleksei S. Shatskikh Ludmila V. Olenina
Ludmila V. Olenina