- 1Department of Gastrointestinal surgery, The First Affiliated Hospital, College of Clinical Medicine, Henan University of Science and Technology, Luoyang, Henan, China
- 2Department of thyroid surgery, The First Affiliated Hospital, College of Clinical Medicine, Henan University of Science and Technology, Luoyang, Henan, China
Recent advances in high-resolution mass spectrometry-based proteomics have improved our understanding of lysine acetylation in proteins, including histones and non-histone proteins. Lysine acetylation, a reversible post-translational modification, is catalyzed by lysine acetyltransferases (KATs) and lysine deacetylases (KDACs). Proteins comprising evolutionarily conserved bromodomains (BRDs) recognize these acetylated lysine residues and consequently activate transcription. Lysine acetylation regulates almost all cellular processes, including transcription, cell cycle progression, and metabolic functions. Studies have reported the aberrant expression, translocation, and mutation of genes encoding lysine acetylation regulators in various cancers, including digestive tract cancers. These dysregulated lysine acetylation regulators contribute to the pathogenesis of digestive system cancers by modulating the expression and activity of cancer-related genes or pathways. Several inhibitors targeting KATs, KDACs, and BRDs are currently in preclinical trials and have demonstrated anti-cancer effects. Digestive tract cancers, including encompass esophageal, gastric, colorectal, liver, and pancreatic cancers, represent a group of heterogeneous malignancies. However, these cancers are typically diagnosed at an advanced stage owing to the lack of early symptoms and are consequently associated with poor 5-year survival rates. Thus, there is an urgent need to identify novel biomarkers for early detection, as well as to accurately predict the clinical outcomes and identify effective therapeutic targets for these malignancies. Although the role of lysine acetylation in digestive tract cancers remains unclear, further analysis could improve our understanding of its role in the pathogenesis of digestive tract cancers. This review aims to summarize the implications and pathogenic mechanisms of lysine acetylation dysregulation in digestive tract cancers, as well as its potential clinical applications.
Background
Protein acetylation, a post-translational modification (PTM), involves the covalent attachment of an acetyl group to proteins. Generally, the acetyl group is added to the lysine residues or the N-terminus through acetyltransferase-mediated mechanisms or non-enzymatic mechanisms (Christensen et al., 2019; Ree et al., 2018; Bloch and Borek, 1946). Previous studies have demonstrated that protein acetylation mediates diverse physiological and pathological functions by regulating various proteins and signaling pathways (Shen et al., 2015; Kong et al., 2022; Shvedunova and Akhtar, 2022; Liu et al., 2021). Lysine acetylation, which is the most extensively studied PTM, was first identified to be a histone modification by Vincent Allfrey et al., in 1964 (Phillips, 1963). In addition to histones, lysine acetylation has been subsequently identified in several eukaryotic non-histone proteins (Narita et al., 2019; Saxholm et al., 1982). Since the advent of antibody enrichment and mass spectrometry techniques in 2006, various acetylation-related enzymes and their functions have been identified (Verdin and Ott, 2015; Xue et al., 2022; VanDrisse and Escalante-Semerena, 2019; Morales-Tarré et al., 2021). Lysine acetylation is predominantly mediated through the transfer of an acetyl group from acetyl-CoA to the ε-amino side chain of lysine. This process is reversible, which is called lysine deacetylation, a process catalyzed by deacetylases (Marmorstein and Zhou, 2014). The enzymatic mechanisms are primarily mediated by the following two groups of enzymes: lysine acetyltransferases [KATs; previously called histone acetyltransferases (HATs)] and lysine deacetylases [KDACs; also called histone deacetylases (HDACs)] (Vo et al., 2018; Wu et al., 2019). Bromodomain-containing proteins (BRDs) specifically recognize and bind to acetylated lysine via an evolutionarily conserved asparagine residue within the bromodomain (Figure 1) (Del et al., 2019). Recent studies have reported that lysine can also be acetylated through non-enzymatic mechanisms in the mitochondrial matrix.
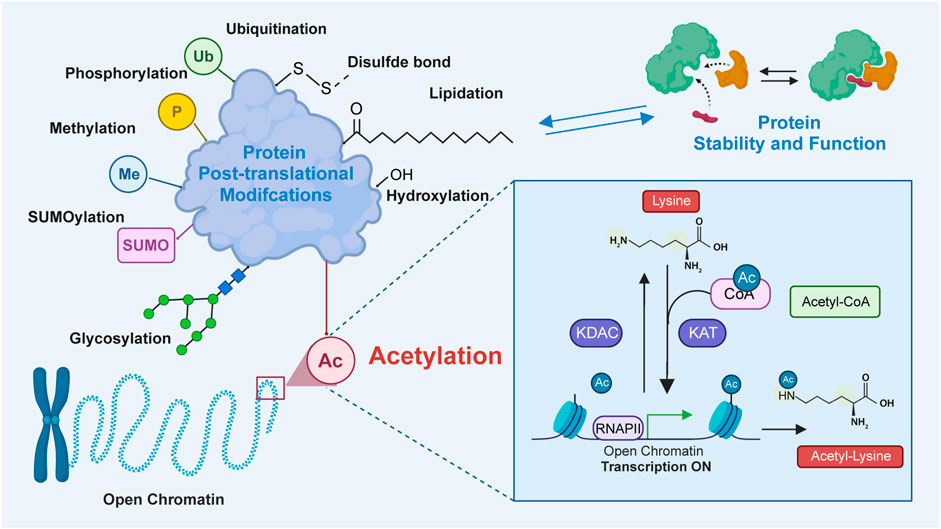
Figure 1. The process of lysine acetylation modification on proteins. In this process, acetyltransferase catalyzes the transfer of acetyl groups to specific lysine residues. The source of this acetyl group is mainly from acetyl-CoA. Acetyltransferase transfers acetyl groups to target proteins by recognizing lysine residues on them. Bromodomains recognize and bind to the acetylated substrates to activate transcription. Additionally, deacetylase, which reverses lysine acetylation, catalyzes the removal of acetyl groups and reduces the acetylation of lysine residues in proteins. Lysine acetylation plays an important role in the regulation of diverse protein functions by modulating protein localization, interaction, stability, and transcriptional activity.
Lysine acetylation affects various properties and functions of proteins through several regulatory mechanisms. In particular, lysine acetylation affects the stability, subcellular localization, and enzymatic activity of proteins, as well as their interactions with other proteins or DNA (Allfrey et al., 1964; Hebbes et al., 1988; Hebbes et al., 1992; Li et al., 2020). This modification can alter protein function and regulate cellular processes. Thus, lysine acetylation is the most crucial regulatory mechanism in various biological processes, such as DNA repair, chromatin remodeling, cell cycle progression, and cell apoptosis, which are essential for the development and progression of cancer (Kumar et al., 2016; Ding et al., 2022; You et al., 2012). Lysine acetylation is dysregulated in diverse cancer types, contributing to aberrant protein function and disruption of physiological cellular processes (Ramaiah et al., 2021; Kaypee et al., 2016; Jain and Barton, 2017). For example, the upregulated levels of lysine acetylation in some proteins involved in cell cycle regulation can result in uncontrolled cell proliferation, a hallmark of cancer. An understanding of the regulatory mechanisms of lysine acetylation will provide valuable insights into the development and progression of cancers.
Digestive tract cancers encompass a broad spectrum of malignancies that affect various organs within the gastrointestinal tract. These cancers can manifest as diverse types of cancer, such as esophageal cancer (EC), gastric cancer (GC), colorectal cancer (CRC), liver cancer, and pancreatic cancer (PC) (Sung et al., 2021; Ferlay et al., 2018; Smyth et al., 2020). The symptoms of digestive system tumors may vary depending on the affected organ. However, most digestive system tumors are not easily detectable in the early stages (Issa and Noureddine, 2017; Ladabaum and Mannalithara, 2016). Currently, some success has been achieved in the treatment of digestive tract cancers (Kaźmierczak-Siedlecka et al., 2022; Kronig et al., 2023; Rao et al., 2022). In particular, targeted therapy and immunotherapy have emerged as promising therapeutic modalities for advanced and refractory digestive tract cancers (Chen et al., 2021; Zhao et al., 2023; Patel and Cecchini, 2020; Xue et al., 2023; Ai et al., 2023). However, the efficacy of treatments for advanced metastatic tumors is suboptimal (Galun et al., 2017; Wang et al., 2019). Additionally, early diagnosis of some digestive tract cancers, such as PC, is challenging. Thus, most patients are diagnosed with advanced-stage cancer and exhibit poor prognosis (Klein, 2021; Stoffel et al., 2023; Cai et al., 2021). As there is a continuous demand for comprehensive treatment modalities and personalized treatment strategies, the pathogenesis of digestive tract cancers must be elucidated.
Previous studies have elucidated the correlation of the dysregulated KATs, KDACs, and BRDs (involved in lysine acetylation) with the progression of various digestive tract cancers. Thus, lysine acetylation is a potential therapeutic target for digestive tract cancers (Table 1) (Li and Seto, 2016; Zhao et al., 2022; Jin et al., 2021; Carbajo-Pescador et al., 2013; Cougot et al., 2007; Yang et al., 2023). This review provides an overview of the components and reversible modification process of lysine acetylation. Next, the regulatory mechanisms of lysine acetylation that contribute to the development of digestive tract cancers have been discussed. Additionally, advances in drug development and identification of prognosis biomarkers targeting lysine acetylation in digestive tract cancers have been summarized. Finally, potential future research directions and challenges associated with targeting lysine acetylation in digestive tract cancers have been discussed.
Components and dynamic processes of lysine acetylation
Acetylation of lysine residues is a reversible PTM process with critical roles in modulating protein functions and cellular processes. The dynamic balance between lysine acetylation and deacetylation involves multiple components, including enzymes (KATs and KDACs) and KAT substrate recognition proteins (BRDs). The functions of lysine acetylation in regulating various cellular processes are diverse and context-dependent. The dynamic nature of lysine acetylation is a key feature that allows organisms to respond to both intracellular and extracellular signals, leading to rapid and reversible modifications of proteins. This dynamic process enables cells to fine-tune protein functions and gene expression to adapt to constantly changing environmental conditions. In this section, a detailed overview of KATs and KDACs involved in lysine acetylation, as well as the BRDs that specifically recognize acetylated lysine residues, has been provided.
KATs
Based on the amino acid sequence homology, KATs are currently classified into the following three major families: the MYST (MOZ, Ybf2/Sas3, Sas2, and TIP60) family, the p300/cyclic AMP-response element (CRE)-binding protein (CREB)-binding protein (CBP) family, and the GCN5/PCAF (general control nonderepressible 5 and its human ortholog PCAF; also known as KAT2A/KAT2B) family (Roth et al., 2001; Yang et al., 2020; Simon et al., 2016; Yang, 2004; Wapenaar and Dekker, 2016). Among these families, the p300/CBP family members without the conserved motif found in other acetyltransferases comprise p300 and CBP (also known as KAT3B/KAT3A) and are mainly localized to the nucleus and cytoplasm. Additionally, the p300/CBP family members are involved in the targeted acetylation of specific chromatin domains (McIntyre et al., 2019; Ogryzko et al., 1996; Dutta et al., 2016).
KDACs
In mammals, the KDAC superfamily comprises 11 zinc-dependent HDACs and 7 nicotinamide adenine dinucleotide (NAD+)-dependent sirtuins (Shvedunova and Akhtar, 2022; Menzies et al., 2016; Ellmeier and Seiser, 2018). These two subgroups of KDACs regulate transcription. The function of HDACs is dependent on zinc ion (Zn2+) as a coenzyme, while that of sirtuins is dependent on NAD+ as a cofactor for the removal of acetyl groups (Penney and Tsai, 2014). Based on sequence similarity, KDACs can be divided into 4 different classes. Class I KDACs, which comprise four members (HDAC1, HDAC2, HDAC3, and HDAC8), are highly similar to the yeast protein RPD3 (Moreno-Yruela et al., 2022). Class II KDACs comprise Class IIa (HDAC4, HDAC5, HDAC7, and HDAC9) and class IIb (HDAC6 and HDAC10) enzymes. HDAC11 is a unique member of class IV KDACs. Sirtuins belong to class III KDACs with seven members (SIRT1 to SIRT7) and exhibit homology to yeast protein Sir2.
BRDs
In addition to KATs and KDACs, lysine acetylation is also regulated by KAT substrate recognition proteins (Gong et al., 2016; Zaware and Zhou, 2019). BRDs with approximately 110 amino acid structural domains recognize specific acetylated lysine residues on target proteins and recruit KATs or KDACs to regulate their acetylation status. Furthermore, the human bromodomain and extra-terminal (BET) family, which functions as a well-conserved class of transcriptional regulators, has piqued the interest of the scientific community (Filippakopoulos et al., 2010; Ferri et al., 2016; Taniguchi, 2016). The BET family in humans comprises four members (BRD2, BRD3, BRD4, and BRDT), which comprise two N-terminal bromodomains (BD1 and BD2).
These components function together in the process of lysine acetylation and regulate various cellular biological functions, including gene expression, cell cycle regulation, and cell signaling. The interplay between KATs, KDACs, and BRDs has a critical role in regulating diverse epigenetic processes and can be targeted to treat various diseases, including digestive tract cancers (Alleboina et al., 2021; Ackloo et al., 2017).
Lysine acetylation in digestive tract cancers
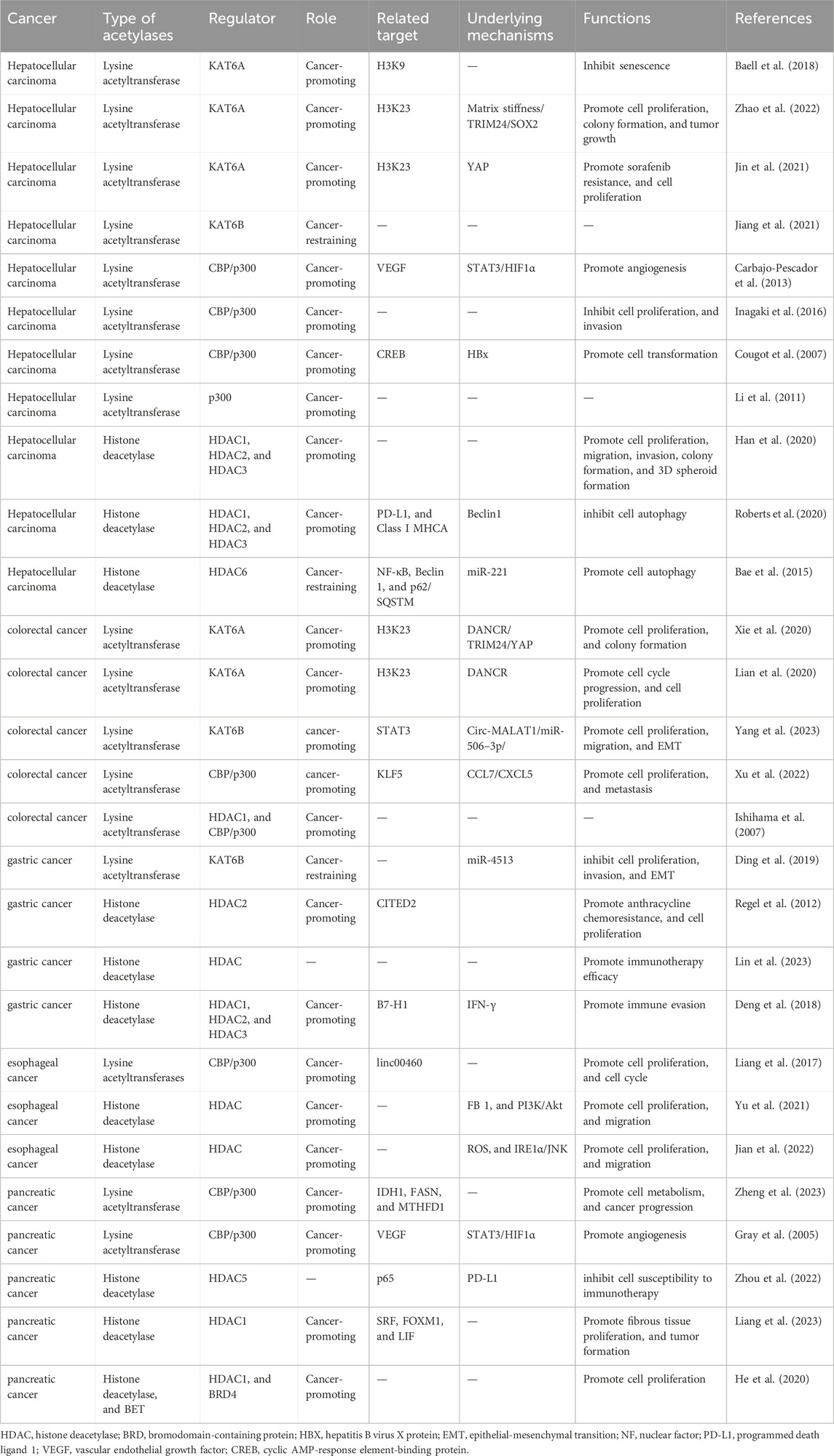
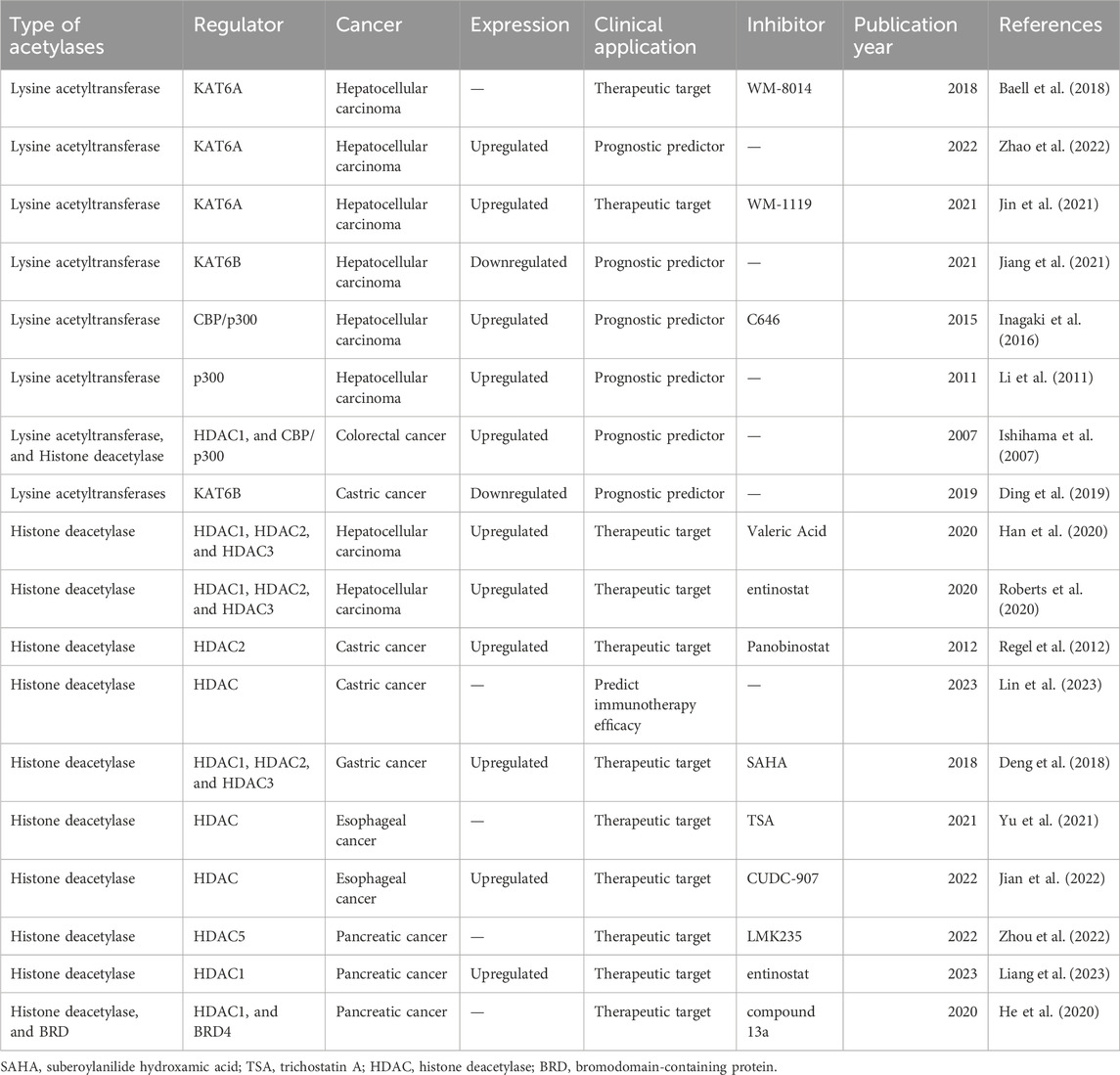
Several studies have demonstrated that lysine acetylation is dysregulated in multiple digestive tract cancers, including EC, GC, CRC, liver cancer, and PC. The dysregulation of lysine acetylation and deacetylation actively contributes to the malignant behaviors of digestive tract cancers, such as cell proliferation, migration, and metabolism, stem cell properties, and drug resistance through the regulation of cancer-associated genes and pathways (Figure 2) (Narita et al., 2019; Xu et al., 2014). Additionally, the expression profiles of various lysine acetylation regulators are closely associated with the clinicopathological indices and outcomes of patients with digestive tract cancers, suggesting their potential prognostic values (Figure 3). The regulatory effects of lysine acetylation on cancer-associated genes and pathways provide useful insights into the pathogenesis of digestive tract cancers from the PTM perspective. Thus, emerging inhibitors have been developed and tested in several digestive tract cancers (Table 2). In this section, the aberrant expression and functions of lysine acetylation regulators in diverse digestive tract cancers have been highlighted. Additionally, this section discusses the regulatory mechanisms of these regulators in the development of digestive tract cancers and their clinical applications.
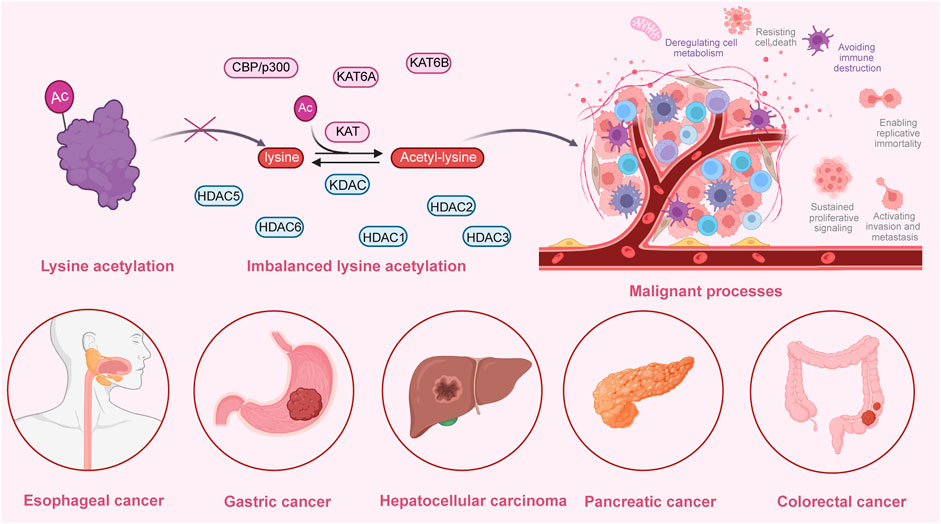
Figure 2. An overview of the dysregulated lysine acetylation in multiple digestive tract cancers. Various lysine acetyltransferases and deacetylases, such as KAT6A, KAT6B, CBP/p300, HDAC1, HDAC2, HDAC3, HDAC5, and HDAC6 are frequently dysregulated in several digestive tract cancers, including hepatocellular carcinoma, colorectal cancer, gastric cancer, esophageal cancer, and pancreatic cancer. Moreover, these dysregulated enzymes exert regulatory effects on the expression and functions of specific target molecules, modulating multiple malignant biological processes during the progression of cancer. These processes include cell proliferation, colony formation, cell metabolism, invasion, migration, autophagy, drug resistance, immune evasion, and angiogenesis.
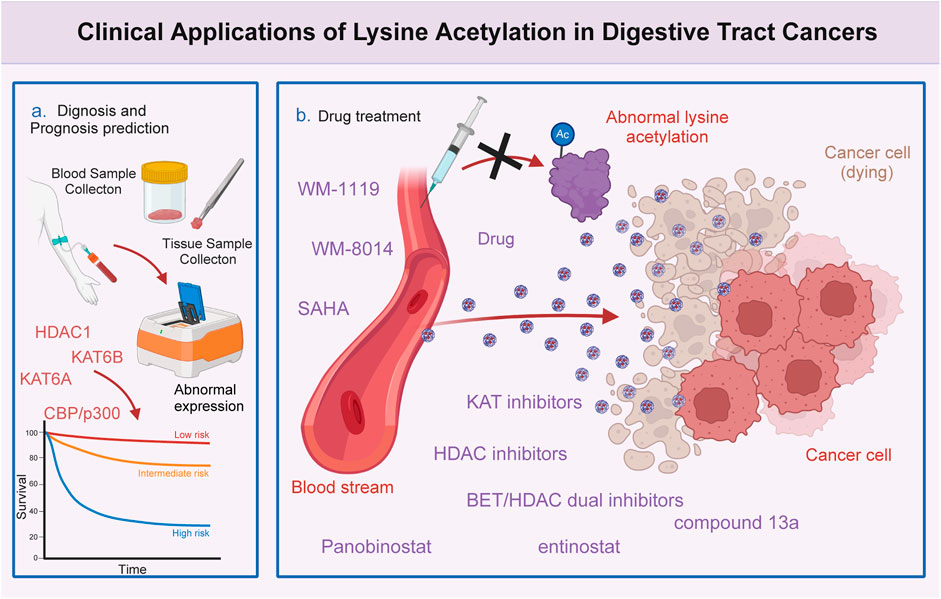
Figure 3. Clinical significance of lysine acetylation in digestive tract cancers. The dysregulation of multiple lysine acetyltransferases and deacetylases, including KAT6A, KAT6B, CBP/p300, and HDAC1, is closely associated with the clinical characteristics and prognosis of patients with digestive tract cancers. This association enables precise and sensitive prediction of patient prognosis by assessing the expression levels of these enzymes in patients with digestive tract cancers. Additionally, several lysine acetylation-targeting agents have been developed. Several promising candidates have shown encouraging efficacy in treating digestive tract cancers. Currently, various KAT and HDAC inhibitors, such as WM-8014, WM-1119, panobinostat, suberoylanilide hydroxamic acid (SAHA), and entinostat, exert effective anti-cancer effects in various models of digestive tract cancer. Furthermore, several bromodomain and extra-terminal domain (BET)/HDAC dual inhibitors are under development, and some drugs, such as compound 13a have exhibited excellent synergistic effects.
KATs and digestive tract cancers
According to The Cancer Genome Atlas data analysis, most KATs, including KAT8, KAT5, KAT7, KAT6A, and KAT6B, are frequently mutated in diverse digestive tract cancers, especially in CRC, GC, and EC (Chen et al., 2016; Wang et al., 2011; Ahn et al., 2014; Ng et al., 2022). The levels of lysine acetylation and lysine acetylation regulators are reported to be dysregulated in various digestive tract cancers, suggesting the correlation between lysine acetylation and the pathogenesis of digestive tract cancers (Ding et al., 2019).
Several studies have suggested that KAT6A is upregulated in hepatocellular carcinoma (HCC) tissues and cell lines, suggesting that KAT6A is correlated with the pathogenesis of HCC (Zhao et al., 2022). The upregulation of KAT6A induced by matrix stiffness promotes tumorigenesis during HCC progression, contributing to increased cell proliferation, colony formation, and tumor growth. In Huh7 cells, KAT6A upregulation promotes the acetylation of histone H3 at lysine 23 (H3K23) and its interaction with tripartite motif containing 24 (TRIM24). This modification subsequently activates the expression of SRY-box transcription factor 2 (SOX2), driving HCC tumorigenesis (Zhao et al., 2022). Furthermore, CBP/p300 is upregulated in HCC tissues. The transcriptional complex, which comprises phosphorylated signal transducer and activator of transcription 3 (STAT3), hypoxia-inducible factor 1 alpha (HIF1α), and CBP/p300 under hypoxic conditions, is actively recruited to the promoter region of vascular endothelial growth factor (VEGF)-encoding gene. This complex plays a crucial role in upregulating the transcription of VEGF in HepG2 cells. The upregulation of VEGF promotes tube formation in human umbilical vein endothelial cells (Carbajo-Pescador et al., 2013). The recruitment of hepatitis B virus (HBV) X protein (HBx) to CBP/p300 activates CREB-dependent gene transcription in human HCC cells, promoting HBV-mediated oncogenesis (Cougot et al., 2007). Additionally, KAT6A exerts oncogenic effects in CRC by interacting with long non-coding RNA (lncRNA) DANCR to form functional complexes and upregulating the expression of TRIM24 and cell cycle-related proteins (Lian et al., 2020; Xie et al., 2020). Subsequently, TRIM24 interacts with acetylated H3K23 and further activates the transcription of Yes-associated protein (YAP), promoting cell proliferation, cell cycle progression, and colony formation in LOVO cells (Xie et al., 2020). KAT6B is also upregulated in CRC tissues and cells, facilitating the proliferation, migration, and epithelial-mesenchymal transition (EMT) of CRC cells. Previous studies have revealed that circ-MALAT1 interacts with miR-506–3p, resulting in the upregulation of KAT6B expression. Subsequently, KAT6B enhances the acetylation of STAT3 in the CRC cell lines (LS513 and SW837 cells). The aberrant upregulation of STAT3 promotes the proliferation, migration, and EMT of CRC cells, highlighting a complex regulatory mechanism underlying the pathological role of lysine acetylation in the progression of CRC (Yang et al., 2023). C-C motif chemokine ligand 7 (CCL7), which is secreted by tumor-associated mesenchymal stromal cells (T-MSCs), stimulates the acetylation of Kruppel-like factor 5 (KLF5) through CBP/P300. This process leads to the upregulation of C-X-C motif chemokine ligand 5 (CXCL5). CXCL5 upregulation in the CRC microenvironment enhances the ability of CRC cells to proliferate and metastasize (Xu et al., 2022). KAT6B, which exerts tumor-suppressive effects in GC, is downregulated in GC cells. KAT6B knockdown in HGC27 and MGC803 cells promotes cell proliferation, invasion, and EMT caused by its upstream target miR-4513 (Ding et al., 2019). In PC, STAT3, HIF1α, and CBP/p300 also form a promoter complex to bind to the VEGF-encoding gene and activate its expression under hypoxic conditions. This process promotes PC angiogenesis, growth, and metastasis (Gray et al., 2005). Additionally, in response to the alterations in PC extracellular nutrient availability, CBP/p300, HDAC1, and HDAC3 modulate the dynamic crotonylation levels of the metabolic enzymes isocitrate dehydrogenase 1 (IDH1), fatty acid synthase (FASN), and methylenetetrahydrofolate dehydrogenase 1 (MTHFD1), enhancing PC cell metabolism and tumor progression (Zheng et al., 2023). CBP/p300 is reported to function as a co-activator to drive linc00460 transcription. The upregulated linc00460 promotes cell proliferation and inhibits apoptosis in EC cells (KYSE150 and KYSE450 cells) (Liang et al., 2017).
The expression of lysine acetylation regulators is dysregulated in various types of digestive tract cancers. Several studies have reported a significant correlation between the levels of lysine acetylation regulators and poor prognosis in patients with HCC, GC, EC, PC, and CRC (Ishihama et al., 2007). For example, clinical data suggest that KAT6A upregulation is associated with short survival and aggressive clinical features in HCC, including large tumor sizes, advanced tumor-node-metastasis stages, and advanced Edmondson-Steiner grade (Zhao et al., 2022). Furthermore, p300 upregulation is associated with aggressive characteristics of HCC, such as increased alpha-fetoprotein (AFP) level, large tumor size, multiplicity, poor differentiation, and advanced stage. Thus, p300 expression functions as a strong and independent predictor of HCC survival (Li et al., 2011; Inagaki et al., 2016). Conversely, KAT6B downregulation is associated with worse prognosis, as well as with large tumor size and elevated AFP level, in HCC (Jiang et al., 2021). Kaplan-Meier analysis revealed that KAT6B downregulation independently predicts decreased overall survival in patients with GC (Ding et al., 2019). These findings indicate the potential of lysine acetylation regulators to serve as prognostic factors for digestive tract cancers (Zhang et al., 2022). The elucidation of tumorigenesis mechanisms of lysine acetylation in digestive tract cancers will provide a theoretical foundation for the development of novel therapeutic strategies that directly target lysine acetylation. The identification of specific enzymes involved in lysine acetylation and their associated pathways will provide novel avenues for developing precision medicine strategies targeting lysine acetylation. For example, previous studies have reported that KAT6A is upregulated in sorafenib-resistant HCC clinical samples. Silencing KAT6A in sorafenib-resistant HCC cells decreases the enrichment of H3K23 acetylation and RNA polymerase II at the promoter region of the YAP-encoding gene, effectively suppressing cell proliferation and resistance to sorafenib. Furthermore, the combination of the KAT6A inhibitor WM-1119 and sorafenib markedly decreases the viability of sorafenib-resistant HCC cells, delaying HCC progression (Jin et al., 2021). Additionally, studies on the zebrafish model of HCC revealed that WM-8014 suppresses KAT6A-induced specific acetylation of H3K9 and cellular senescence. Treatment with WM-8014 significantly decreased tumor volume and promoted cell cycle exit and cellular senescence without adversely affecting the growth of healthy hepatocytes (Baell et al., 2018). The inhibition of CBP/p300 using a specific inhibitor (C646) impairs the proliferation, apoptotic sensitivity, and invasion of HCC cells in a dose-dependent manner (Inagaki et al., 2016). These results suggest the therapeutic potential of targeting lysine acetylation and contribute to the rapid development of treatments targeting lysine acetylation (Leaver et al., 2019).
KDACs and digestive tract cancers
Additionally, KDAC expression is dysregulated in several types of digestive tract cancers, contributing to cancer development and progression. Several KDAC inhibitors have been extensively studied for applications in tumor immunotherapy. Various small molecule HDAC inhibitors have been identified and their potential therapeutic efficacy as a cancer immunotherapeutic has been demonstrated (Cheng et al., 2024). HDAC2 is upregulated in human GC tissues, GC cells, and CEA424/SV40 T-antigen (CEA/Tag) transgenic mice, suggesting that HDAC2 is correlated with GC development (Regel et al., 2012). Previous studies have reported that the HDAC2 inhibitor LBH589 increases the acetylation levels and suppresses the expression of a cluster of anthracycline resistance-conferring genes (e.g. FOXM1, HMGB2, and TYMS) in GC cells (AGS and MKN-45 cells). Further studies on transgenic CEA/Tag mice revealed that the combination of LBH589 and anthracycline drugs upregulates the expression of CBP/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 2 (CITED2), enhancing apoptosis and decreasing the viability of AGS cells (Regel et al., 2012). Additionally, the expression levels of HDACs are correlated with the characteristics of the tumor microenvironment (TME) and the efficacy of immunotherapy in GC. Analysis of HDAC score (HDS), which is based on the expression of HDACs in GC, revealed that patients with a high HDS exhibit increased immune infiltration and improved prognosis (Lin et al., 2023). One study reported that HDACs increase the IFN-γ-induced histone acetylation of the B7 homolog 1 (B7-H1)-encoding gene promoter, enhancing B7-H1 expression and promoting tumor growth in GC. In a mouse model of subcutaneously grafted GC, pretreatment with the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) suppresses tumor growth by downregulating IFN-γ-induced B7-H1 expression (Deng et al., 2018). Recent studies have demonstrated that valeric acid functions as a novel HDAC inhibitor, exerting broad growth-inhibitory effects against liver cancer by inhibiting cell proliferation, colony formation, cell invasion, and spheroid formation (Han et al., 2020). Additionally, the interaction of the HDAC inhibitor entinostat with lenvatinib results in the inactivation of mammalian target of rapamycin complex 1 (mTORC1) and mTORC2, inducing autophagy-mediated liver cancer cell clearance. The enhanced autophagy leads to the upregulation of Beclin 1 (a stress response gene), which subsequently suppresses the levels of HDAC1, HDAC2, and HDAC3. This leads to the downregulation of programmed death ligand 1 (PD-L1) and human major histocompatibility complex class I A (MHCA) (Roberts et al., 2020). However, the downregulation of HDAC6 in liver cancer promotes autophagy-induced Hep3B cell death, exerting a tumor-suppressive effect. During hepatocarcinogenesis, miR-221 serves as a potent inhibitor of HDAC6, suppressing the levels of the autophagy-related molecules Beclin 1 and p62/SQSTM (p62), a process mediated by activated c-Met signaling in HCC cells. HDAC6 also reversely regulates miR-221 expression by suppressing nuclear factor kappa B (NF-κB) activation (Bae et al., 2015). In PC, HDAC5 upregulation is positively correlated with favorable clinical outcomes. Additionally, HDAC5 induces the phosphorylation of p65 at S311, leading to the inhibition of NF-κB-mediated PD-L1 expression. Silencing HDAC5 in Panc02 cells and treating mouse models with HDAC5 inhibitor LMK235 upregulated PD-L1 expression and increased the susceptibility of cancer cells to anti-PD1 antibody therapy in PC (Zhou et al., 2022). Recent studies have demonstrated that class I HDACs stimulate the transcription of pro-desmoplastic and pro-tumorigenic processes in pancreatic stellate cells (PSCs) and cancer-associated fibroblasts (CAFs) through the regulation of serum response factor (SRF), forkhead box M1 (FOXM1), and leukemia inhibitory factor (LIF) expression. In particular, HDACs play a central role in promoting pro-desmoplastic programs and activating PSCs by upregulating FOXM1 and SRF levels. Additionally, HDACs facilitate LIF expression to enhance the pro-tumorigenic characteristics of CAFs. Studies using genetically engineered mouse models treated with the HDAC inhibitor entinostat have further confirmed that HDAC downregulation inhibits the myofibroblastic transcriptional program, leading to the reversal of stromal activation and tumor progression in PC (Liang et al., 2023). In EC, the environmental contaminant fumonisin B1 (FB1) upregulated HDAC expression and activated the phosphoinositide 3 kinase (PI3K)/protein kinase B (Akt) signaling pathway, promoting cell proliferation and migration during esophageal carcinogenesis. The HDAC inhibitor trichostatin A (TSA) and myriocin (ISP-1) repress FB1-induced HDAC expression and inhibit carcinogenesis in human esophageal epithelial cells (Yu et al., 2021). Additionally, dual PI3K-HDAC inhibitor CUDC-907 has been developed to target the elevated activation of the PI3K-Akt pathway and the expression of HDACs in various types of EC cells. Studies using KYSE-450 xenograft nude mouse models have indicated that CUDC-907 inhibits the proliferation, invasion, and migration of EC cells by promoting reactive oxygen species (ROS)-inositol requiring enzyme 1 α (IRE1α)-Jun N-terminal kinase (JNK)-mediated cytotoxic autophagy (Jian et al., 2022). These studies indicate the therapeutic potential of HDAC inhibitors in digestive tract cancers. HDAC inhibitors potentially exert therapeutic effects on digestive tract cancers by modulating cellular signaling pathways and regulating the TME.
BRDs and digestive tract cancers
BRD dysregulation is reported to be associated with the tumorigenesis and progression of digestive tract cancers. Multiple BRDs recognize and bind to acetylated lysine residues of proteins, regulating the fundamental transcription mechanism. Studies on various disease models have revealed that targeting BET is a potent epigenetic therapeutic strategy (Schwalm and Knapp, 2022). The inhibition of the BET family members using small molecules or specific inhibitors can disrupt the interaction between BET proteins and acetylated histones, suppressing the expression of oncogenes and promoting the activation of tumor suppressor genes. This targeted epigenetic therapy has shown encouraging results in preclinical studies and early-phase clinical trials for digestive tract cancers, including CRC, GC, and EC. For example, the combination of the pan-BET inhibitor JQ1 and HDAC inhibitors (such as SAHA and CI994) exhibits a strong synergistic therapeutic effect on advanced PC, inducing cell apoptosis, reducing tumor volume, and decreasing relapse rate (He et al., 2020; Mazur et al., 2015; Zhang et al., 2020; Seyfinejad and Jouyban, 2022; Shahrajabian and Sun, 2023). Based on the excellent synergistic effect of BET and HDAC inhibitors on PC, several novel BET/HDAC dual inhibitors are being developed. Among these inhibitors, compound 13a exerts balanced inhibitory effects against both BRD4 and HDAC1 in Capan-1 PC xenograft models (He and Tang, 2020). Further studies are needed to determine the efficacy of this approach. However, these findings suggest that the anti-cancer efficacy of the combination of BET and HDAC inhibitors is higher than that of monotherapy (Akhtar et al., 2023; Li et al., 2024).
Conclusion
Lysine acetylation has been demonstrated to play a major role in the development and progression of various diseases, especially digestive tract cancers. This dynamic and reversible process regulates protein functions and further contributes to the modulation of complex cellular processes involved in tumorigenesis. However, several challenges must be addressed in the analysis of lysine acetylation in digestive tract cancers. The functions and mechanisms of lysine acetylation have been extensively studied. However, the precise role of lysine acetylation in digestive tract cancers has not been elucidated. For example, the exact preferred lysine sites and protein types of enzymes related to lysine acetylation have not been identified. Additionally, the mechanisms underlying the different activities and influences of the same acetylation enzyme in the same cancer type are unclear. A major challenge in the clinical setting is the development of sensitive and specific assays for the detection and quantification of lysine acetylation in cancer tissues. The identification of aberrant acetylation sites is crucial for cancer diagnosis and prognosis prediction. Improving the sensitivity and specificity of these acetylation-related biomarkers for clinical applications is challenging. Furthermore, the effectiveness and safety of KDAC inhibitors are major hurdles to their widespread application in the treatment of digestive tract cancers. Thus, future studies must focus on the precise mechanisms of lysine acetylation in the development and progression of digestive tract tumors. Additionally, large-scale clinical trials are required for the development of epigenetic drugs targeting lysine acetylation and the validation of their efficacy and safety. Continued investigation into the functions and mechanisms of lysine acetylation will provide valuable insights into the complex regulatory processes of lysine acetylation involved in tumorigenesis and may lead to the development of novel diagnostic and therapeutic strategies for digestive tract cancers.
Author contributions
PL: Conceptualization, Supervision, Writing–original draft, Writing–review and editing. YX: Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ackloo, S., Brown, P. J., and Müller, S. (2017). Chemical probes targeting epigenetic proteins: applications beyond oncology. Epigenetics 12 (5), 378–400. doi:10.1080/15592294.2017.1279371
Ahn, S. M., Jang, S. J., Shim, J. H., Kim, D., Hong, S. M., Sung, C. O., et al. (2014). Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 60 (6), 1972–1982. doi:10.1002/hep.27198
Ai, H., Yang, H., Li, L., Ma, J., Liu, K., and Li, Z. (2023). Cancer/testis antigens: promising immunotherapy targets for digestive tract cancers. Front. Immunol. 14, 1190883. doi:10.3389/fimmu.2023.1190883
Akhtar, M. J., Kumar, B., Paul, J., Singh, K., Pannu, S., Pal, R., et al. (2023). An update on recently developed analytical and bio-analytical methods for some anticancer drugs. Curr. Pharm. Anal. 19 (2), 117–135. doi:10.2174/1573412919666221123110420
Alleboina, S., Aljouda, N., Miller, M., and Freeman, K. W. (2021). Therapeutically targeting oncogenic CRCs facilitates induced differentiation of NB by RA and the BET bromodomain inhibitor. Mol. Ther. Oncolytics 23, 181–191. doi:10.1016/j.omto.2021.09.004
Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of rna synthesis. Proc. Natl. Acad. Sci. U. S. A. 51 (5), 786–794. doi:10.1073/pnas.51.5.786
Bae, H. J., Jung, K. H., Eun, J. W., Shen, Q., Kim, H. S., Park, S. J., et al. (2015). MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J. Hepatol. 63 (2), 408–419. doi:10.1016/j.jhep.2015.03.019
Baell, J. B., Leaver, D. J., Hermans, S. J., Kelly, G. L., Brennan, M. S., Downer, N. L., et al. (2018). Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature 560 (7717), 253–257. doi:10.1038/s41586-018-0387-5
Bloch, K., and Borek, E. (1946). Biological acetylation of natural amino acids. J. Biol. Chem. 164, 483–484. doi:10.1016/s0021-9258(18)43087-6
Cai, J., Chen, H., Lu, M., Zhang, Y., Lu, B., You, L., et al. (2021). Advances in the epidemiology of pancreatic cancer: trends, risk factors, screening, and prognosis. Cancer Lett. 520, 1–11. doi:10.1016/j.canlet.2021.06.027
Carbajo-Pescador, S., Ordoñez, R., Benet, M., Jover, R., García-Palomo, A., Mauriz, J. L., et al. (2013). Inhibition of VEGF expression through blockade of Hif1α and STAT3 signalling mediates the anti-angiogenic effect of melatonin in HepG2 liver cancer cells. Br. J. Cancer 109 (1), 83–91. doi:10.1038/bjc.2013.285
Chen, H., Zhuo, Q., Ye, Z., Xu, X., and Ji, S. (2021). Organoid model: a new hope for pancreatic cancer treatment? Biochim. Biophys. Acta Rev. Cancer 1875 (1), 188466. doi:10.1016/j.bbcan.2020.188466
Chen, J., Herlong, F. H., Stroehlein, J. R., and Mishra, L. (2016). Mutations of chromatin structure regulating genes in human malignancies. Curr. Protein Pept. Sci. 17 (5), 411–437. doi:10.2174/1389203717666160122120008
Cheng, B., Pan, W., Xiao, Y., Ding, Z., Zhou, Y., Fei, X., et al. (2024). HDAC-targeting epigenetic modulators for cancer immunotherapy. Eur. J. Med. Chem. 265, 116129. doi:10.1016/j.ejmech.2024.116129
Christensen, D. G., Baumgartner, J. T., Xie, X., Jew, K. M., Basisty, N., Schilling, B., et al. (2019). Mechanisms, detection, and relevance of protein acetylation in prokaryotes. mBio 10 (2), e02708. doi:10.1128/mBio.02708-18
Cougot, D., Wu, Y., Cairo, S., Caramel, J., Renard, C. A., Lévy, L., et al. (2007). The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J. Biol. Chem. 282 (7), 4277–4287. doi:10.1074/jbc.M606774200
Del, G. N., Di Costanzo, A., Liu, N. Q., Conte, L., Migliaccio, A., Vermeulen, M., et al. (2019). BRD9 binds cell type-specific chromatin regions regulating leukemic cell survival via STAT5 inhibition. Cell Death Dis. 10 (5), 338. doi:10.1038/s41419-019-1570-9
Deng, R., Zhang, P., Liu, W., Zeng, X., Ma, X., Shi, L., et al. (2018). HDAC is indispensable for IFN-γ-induced B7-H1 expression in gastric cancer. Clin. Epigenetics 10 (1), 153. doi:10.1186/s13148-018-0589-6
Ding, H., Shi, Y., Liu, X., and Qiu, A. (2019). MicroRNA-4513 promotes gastric cancer cell proliferation and epithelial-mesenchymal transition through targeting KAT6B. Hum. Gene Ther. Clin. Dev. 30 (3), 142–148. doi:10.1089/humc.2019.094
Ding, P., Ma, Z., Liu, D., Pan, M., Li, H., Feng, Y., et al. (2022). Lysine acetylation/deacetylation modification of immune-related molecules in cancer immunotherapy. Front. Immunol. 13, 865975. doi:10.3389/fimmu.2022.865975
Dutta, R., Tiu, B., and Sakamoto, K. M. (2016). CBP/p300 acetyltransferase activity in hematologic malignancies. Mol. Genet. Metab. 119 (1-2), 37–43. doi:10.1016/j.ymgme.2016.06.013
Ellmeier, W., and Seiser, C. (2018). Histone deacetylase function in CD4(+) T cells. Nat. Rev. Immunol. 18 (10), 617–634. doi:10.1038/s41577-018-0037-z
Ferlay, J., Colombet, M., Soerjomataram, I., Dyba, T., Randi, G., Bettio, M., et al. (2018). Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 103, 356–387. doi:10.1016/j.ejca.2018.07.005
Ferri, E., Petosa, C., and McKenna, C. E. (2016). Bromodomains: structure, function and pharmacology of inhibition. Biochem. Pharmacol. 106, 1–18. doi:10.1016/j.bcp.2015.12.005
Filippakopoulos, P., Qi, J., Picaud, S., Shen, Y., Smith, W. B., Fedorov, O., et al. (2010). Selective inhibition of BET bromodomains. Nature 468 (7327), 1067–1073. doi:10.1038/nature09504
Galun, D., Srdic-Rajic, T., Bogdanovic, A., Loncar, Z., and Zuvela, M. (2017). Targeted therapy and personalized medicine in hepatocellular carcinoma: drug resistance, mechanisms, and treatment strategies. J. Hepatocell. Carcinoma 4, 93–103. doi:10.2147/JHC.S106529
Gong, F., Chiu, L. Y., and Miller, K. M. (2016). Acetylation reader proteins: linking acetylation signaling to Genome maintenance and cancer. PLoS Genet. 12 (9), e1006272. doi:10.1371/journal.pgen.1006272
Gray, M. J., Zhang, J., Ellis, L. M., Semenza, G. L., Evans, D. B., Watowich, S. S., et al. (2005). HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene 24 (19), 3110–3120. doi:10.1038/sj.onc.1208513
Han, R., Nusbaum, O., Chen, X., and Zhu, Y. (2020). Valeric acid suppresses liver cancer development by acting as a novel HDAC inhibitor. Mol. Ther. Oncolytics 19, 8–18. doi:10.1016/j.omto.2020.08.017
He, S., Dong, G., Li, Y., Wu, S., Wang, W., and Sheng, C. (2020). Potent dual BET/HDAC inhibitors for efficient treatment of pancreatic cancer. Angew. Chem. Int. Ed. Engl. 59 (8), 3028–3032. doi:10.1002/anie.201915896
He, S., and Tang, S. (2020). WNT/β-catenin signaling in the development of liver cancers. Biomed. Pharmacother. 132, 110851. doi:10.1016/j.biopha.2020.110851
Hebbes, T. R., Thorne, A. W., Clayton, A. L., and Crane-Robinson, C. (1992). Histone acetylation and globin gene switching. Nucleic Acids Res. 20 (5), 1017–1022. doi:10.1093/nar/20.5.1017
Hebbes, T. R., Thorne, A. W., and Crane-Robinson, C. (1988). A direct link between core histone acetylation and transcriptionally active chromatin. Embo J. 7 (5), 1395–1402. doi:10.1002/j.1460-2075.1988.tb02956.x
Inagaki, Y., Shiraki, K., Sugimoto, K., Yada, T., Tameda, M., Ogura, S., et al. (2016). Epigenetic regulation of proliferation and invasion in hepatocellular carcinoma cells by CBP/p300 histone acetyltransferase activity. Int. J. Oncol. 48 (2), 533–540. doi:10.3892/ijo.2015.3288
Ishihama, K., Yamakawa, M., Semba, S., Takeda, H., Kawata, S., Kimura, S., et al. (2007). Expression of HDAC1 and CBP/p300 in human colorectal carcinomas. J. Clin. Pathol. 60 (11), 1205–1210. doi:10.1136/jcp.2005.029165
Issa, I. A., and Noureddine, M. (2017). Colorectal cancer screening: an updated review of the available options. World J. Gastroenterol. 23 (28), 5086–5096. doi:10.3748/wjg.v23.i28.5086
Jain, A. K., and Barton, M. C. (2017). Bromodomain histone readers and cancer. J. Mol. Biol. 429 (13), 2003–2010. doi:10.1016/j.jmb.2016.11.020
Jian, Z., Han, Y., Zhang, W., Li, C., Guo, W., Feng, X., et al. (2022). Anti-tumor effects of dual PI3K-HDAC inhibitor CUDC-907 on activation of ROS-IRE1α-JNK-mediated cytotoxic autophagy in esophageal cancer. Cell Biosci. 12 (1), 135. doi:10.1186/s13578-022-00855-x
Jiang, J., Wang, H. J., Mou, X. Z., Zhang, H., Chen, Y., and Hu, Z. M. (2021). Low expression of KAT6B may affect prognosis in hepatocellular carcinoma. Technol. Cancer Res. Treat. 20, 15330338211033063. doi:10.1177/15330338211033063
Jin, Y., Yang, R., Ding, J., Zhu, F., Zhu, C., Xu, Q., et al. (2021). KAT6A is associated with sorafenib resistance and contributes to progression of hepatocellular carcinoma by targeting YAP. Biochem. Biophys. Res. Commun. 585, 185–190. doi:10.1016/j.bbrc.2021.09.009
Kaypee, S., Sudarshan, D., Shanmugam, M. K., Mukherjee, D., Sethi, G., and Kundu, T. K. (2016). Aberrant lysine acetylation in tumorigenesis: implications in the development of therapeutics. Pharmacol. Ther. 162, 98–119. doi:10.1016/j.pharmthera.2016.01.011
Kaźmierczak-Siedlecka, K., Skonieczna-Żydecka, K., Hupp, T., Duchnowska, R., Marek-Trzonkowska, N., and Połom, K. (2022). Next-generation probiotics - do they open new therapeutic strategies for cancer patients? Gut Microbes 14 (1), 2035659. doi:10.1080/19490976.2022.2035659
Klein, A. P. (2021). Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 18 (7), 493–502. doi:10.1038/s41575-021-00457-x
Kong, F., Ma, L., Wang, X., You, H., Zheng, K., and Tang, R. (2022). Regulation of epithelial-mesenchymal transition by protein lysine acetylation. Cell Commun. Signal 20 (1), 57. doi:10.1186/s12964-022-00870-y
Kronig, M. N., Wehrli, M., Salas-Benito, D., and Maus, M. V. (2023). Hurdles race for CAR T-cell therapy in digestive tract cancer. Immunol. Rev. 320 (1), 100–119. doi:10.1111/imr.13273
Kumar, R., Li, D. Q., Müller, S., and Knapp, S. (2016). Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene 35 (34), 4423–4436. doi:10.1038/onc.2015.513
Ladabaum, U., and Mannalithara, A. (2016). Comparative effectiveness and cost effectiveness of a multitarget stool DNA test to screen for colorectal neoplasia. Gastroenterology 151 (3), 427–439. doi:10.1053/j.gastro.2016.06.003
Leaver, D. J., Cleary, B., Nguyen, N., Priebbenow, D. L., Lagiakos, H. R., Sanchez, J., et al. (2019). Discovery of benzoylsulfonohydrazides as potent inhibitors of the histone acetyltransferase KAT6A. J. Med. Chem. 62 (15), 7146–7159. doi:10.1021/acs.jmedchem.9b00665
Li, G., Yao, Q., Liu, P., Zhang, H., Liu, Y., Li, S., et al. (2024). Critical roles and clinical perspectives of RNA methylation in cancer. MedComm 5 (5), e559. doi:10.1002/mco2.559
Li, M., Luo, R. Z., Chen, J. W., Cao, Y., Lu, J. B., He, J. H., et al. (2011). High expression of transcriptional coactivator p300 correlates with aggressive features and poor prognosis of hepatocellular carcinoma. J. Transl. Med. 9 (5), 5. doi:10.1186/1479-5876-9-5
Li, P., Ge, J., and Li, H. (2020). Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat. Rev. Cardiol. 17 (2), 96–115. doi:10.1038/s41569-019-0235-9
Li, Y., and Seto, E. (2016). HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 6 (10), a026831. doi:10.1101/cshperspect.a026831
Lian, J., Zhang, H., Wei, F., Li, Q., Lu, Y., Yu, B., et al. (2020). Long non-coding RNA DANCR promotes colorectal tumor growth by binding to lysine acetyltransferase 6A. Cell Signal 67, 109502. doi:10.1016/j.cellsig.2019.109502
Liang, G., Oh, T. G., Hah, N., Tiriac, H., Shi, Y., Truitt, M. L., et al. (2023). Inhibiting stromal Class I HDACs curbs pancreatic cancer progression. Nat. Commun. 14 (1), 7791. doi:10.1038/s41467-023-42178-6
Liang, Y., Wu, Y., Chen, X., Zhang, S., Wang, K., Guan, X., et al. (2017). A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci. Rep. 37 (5). doi:10.1042/BSR20171019
Lin, Y., Jing, X., Chen, Z., Pan, X., Xu, D., Yu, X., et al. (2023). Histone deacetylase-mediated tumor microenvironment characteristics and synergistic immunotherapy in gastric cancer. Theranostics 13 (13), 4574–4600. doi:10.7150/thno.86928
Liu, M., Guo, L., Fu, Y., Huo, M., Qi, Q., and Zhao, G. (2021). Bacterial protein acetylation and its role in cellular physiology and metabolic regulation. Biotechnol. Adv. 53, 107842. doi:10.1016/j.biotechadv.2021.107842
Marmorstein, R., and Zhou, M. M. (2014). Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6 (7), a018762. doi:10.1101/cshperspect.a018762
Mazur, P. K., Herner, A., Mello, S. S., Wirth, M., Hausmann, S., Sánchez-Rivera, F. J., et al. (2015). Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat. Med. 21 (10), 1163–1171. doi:10.1038/nm.3952
McIntyre, J., Sobolewska, A., Fedorowicz, M., McLenigan, M. P., Macias, M., Woodgate, R., et al. (2019). DNA polymerase ι is acetylated in response to S(N)2 alkylating agents. Sci. Rep. 9 (1), 4789. doi:10.1038/s41598-019-41249-3
Menzies, K. J., Zhang, H., Katsyuba, E., and Auwerx, J. (2016). Protein acetylation in metabolism - metabolites and cofactors. Nat. Rev. Endocrinol. 12 (1), 43–60. doi:10.1038/nrendo.2015.181
Morales-Tarré, O., Alonso-Bastida, R., Arcos-Encarnación, B., Pérez-Martínez, L., and Encarnación-Guevara, S. (2021). Protein lysine acetylation and its role in different human pathologies: a proteomic approach. Expert Rev. Proteomics 18 (11), 949–975. doi:10.1080/14789450.2021.2007766
Moreno-Yruela, C., Zhang, D., Wei, W., Bæk, M., Liu, W., Gao, J., et al. (2022). Class I histone deacetylases (HDAC1-3) are histone lysine delactylases. Sci. Adv. 8 (3), eabi6696. doi:10.1126/sciadv.abi6696
Narita, T., Weinert, B. T., and Choudhary, C. (2019). Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20 (3), 156–174. doi:10.1038/s41580-018-0081-3
Ng, A. W. T., Contino, G., Killcoyne, S., Devonshire, G., Hsu, R., Abbas, S., et al. (2022). Rearrangement processes and structural variations show evidence of selection in oesophageal adenocarcinomas. Commun. Biol. 5 (1), 335. doi:10.1038/s42003-022-03238-7
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., and Nakatani, Y. (1996). The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87 (5), 953–959. doi:10.1016/s0092-8674(00)82001-2
Patel, T. H., and Cecchini, M. (2020). Targeted therapies in advanced gastric cancer. Curr. Treat. Options Oncol. 21 (9), 70. doi:10.1007/s11864-020-00774-4
Penney, J., and Tsai, L. H. (2014). Histone deacetylases in memory and cognition. Sci. Signal 7 (355), re12. doi:10.1126/scisignal.aaa0069
Phillips, D. M. (1963). The presence of acetyl groups of histones. Biochem. J. 87 (2), 258–263. doi:10.1042/bj0870258
Ramaiah, M. J., Tangutur, A. D., and Manyam, R. R. (2021). Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 277, 119504. doi:10.1016/j.lfs.2021.119504
Rao, X., Zhang, C., Luo, H., Zhang, J., Zhuang, Z., Liang, Z., et al. (2022). Targeting gastric cancer stem cells to enhance treatment response. Cells 11 (18), 2828. doi:10.3390/cells11182828
Ree, R., Varland, S., and Arnesen, T. (2018). Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 50 (7), 1–13. doi:10.1038/s12276-018-0116-z
Regel, I., Merkl, L., Friedrich, T., Burgermeister, E., Zimmermann, W., Einwächter, H., et al. (2012). Pan-histone deacetylase inhibitor panobinostat sensitizes gastric cancer cells to anthracyclines via induction of CITED2. Gastroenterology 143 (1), 99–109. doi:10.1053/j.gastro.2012.03.035
Roberts, J. L., Poklepovic, A., Booth, L., and Dent, P. (2020). The multi-kinase inhibitor lenvatinib interacts with the HDAC inhibitor entinostat to kill liver cancer cells. Cell Signal 70, 109573. doi:10.1016/j.cellsig.2020.109573
Roth, S. Y., Denu, J. M., and Allis, C. D. (2001). Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120. doi:10.1146/annurev.biochem.70.1.81
Saxholm, H. J., Pestana, A., O'Connor, L., Sattler, C. A., and Pitot, H. C. (1982). Protein acetylation. Mol. Cell Biochem. 46 (3), 129–153. doi:10.1007/BF00239663
Schwalm, M. P., and Knapp, S. (2022). BET bromodomain inhibitors. Curr. Opin. Chem. Biol. 68, 102148. doi:10.1016/j.cbpa.2022.102148
Seyfinejad, B., and Jouyban, A. (2022). Importance of method validation in the analysis of biomarker. Curr. Pharm. Anal. 18 (6), 567–569. doi:10.2174/1573412918666211213142638
Shahrajabian, M. H., and Sun, W. (2023). Survey on multi-omics, and multi-omics data analysis, integration and application. Curr. Pharm. Anal. 19 (4), 267–281. doi:10.2174/1573412919666230406100948
Shen, Y., Wei, W., and Zhou, D. X. (2015). Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 20 (10), 614–621. doi:10.1016/j.tplants.2015.07.005
Shvedunova, M., and Akhtar, A. (2022). Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23 (5), 329–349. doi:10.1038/s41580-021-00441-y
Simon, R. P., Robaa, D., Alhalabi, Z., Sippl, W., and Jung, M. (2016). KATching-Up on small molecule modulators of lysine acetyltransferases. J. Med. Chem. 59 (4), 1249–1270. doi:10.1021/acs.jmedchem.5b01502
Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C., and Lordick, F. (2020). Gastric cancer. Lancet 396 (10251), 635–648. doi:10.1016/S0140-6736(20)31288-5
Stoffel, E. M., Brand, R. E., and Goggins, M. (2023). Pancreatic cancer: changing epidemiology and new approaches to risk assessment, early detection, and prevention. Gastroenterology 164 (5), 752–765. doi:10.1053/j.gastro.2023.02.012
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Taniguchi, Y. (2016). The bromodomain and extra-terminal domain (BET) family: functional anatomy of BET paralogous proteins. Int. J. Mol. Sci. 17 (11), 1849. doi:10.3390/ijms17111849
VanDrisse, C. M., and Escalante-Semerena, J. C. (2019). Protein acetylation in bacteria. Annu. Rev. Microbiol. 73, 111–132. doi:10.1146/annurev-micro-020518-115526
Verdin, E., and Ott, M. (2015). 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16 (4), 258–264. doi:10.1038/nrm3931
Vo, T. T. L., Jeong, C. H., Lee, S., Kim, K. W., Ha, E., and Seo, J. H. (2018). Versatility of ARD1/NAA10-mediated protein lysine acetylation. Exp. Mol. Med. 50 (7), 1–13. doi:10.1038/s12276-018-0100-7
Wang, K., Kan, J., Yuen, S. T., Shi, S. T., Chu, K. M., Law, S., et al. (2011). Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 43 (12), 1219–1223. doi:10.1038/ng.982
Wang, W., Kandimalla, R., Huang, H., Zhu, L., Li, Y., Gao, F., et al. (2019). Molecular subtyping of colorectal cancer: recent progress, new challenges and emerging opportunities. Semin. Cancer Biol. 55, 37–52. doi:10.1016/j.semcancer.2018.05.002
Wapenaar, H., and Dekker, F. J. (2016). Histone acetyltransferases: challenges in targeting bi-substrate enzymes. Clin. Epigenetics 8 (59), 59. doi:10.1186/s13148-016-0225-2
Wu, H., Yang, K., Zhang, Z., Leisten, E. D., Li, Z., Xie, H., et al. (2019). Development of multifunctional histone deacetylase 6 degraders with potent antimyeloma activity. J. Med. Chem. 62 (15), 7042–7057. doi:10.1021/acs.jmedchem.9b00516
Xie, W., Zhang, Y., Wang, B., Hu, Y., Zhan, B., Wei, F., et al. (2020). Tripartite motif containing 24 regulates cell proliferation in colorectal cancer through YAP signaling. Cancer Med. 9 (17), 6367–6376. doi:10.1002/cam4.3310
Xu, W., Li, Y., Liu, C., and Zhao, S. (2014). Protein lysine acetylation guards metabolic homeostasis to fight against cancer. Oncogene 33 (18), 2279–2285. doi:10.1038/onc.2013.163
Xu, Z., Gao, H., Zhang, Y., Feng, W., Miao, Y., Xu, Z., et al. (2022). CCL7 and TGF-β secreted by MSCs play opposite roles in regulating CRC metastasis in a KLF5/CXCL5-dependent manner. Mol. Ther. 30 (6), 2327–2341. doi:10.1016/j.ymthe.2022.03.005
Xue, C., Yao, Q., Gu, X., Shi, Q., Yuan, X., Chu, Q., et al. (2023). Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders and cancer. Signal Transduct. Target Ther. 8 (1), 204. doi:10.1038/s41392-023-01468-7
Xue, M., Feng, T., Chen, Z., Yan, Y., Chen, Z., and Dai, J. (2022). Protein acetylation going viral: implications in antiviral immunity and viral infection. Int. J. Mol. Sci. 23 (19), 11308. doi:10.3390/ijms231911308
Yang, F. S., Gong, S. X., and Qiu, D. D. (2023). Circ-MALAT1 accelerates cell proliferation and epithelial mesenchymal transformation of colorectal cancer through regulating miR-506-3p/KAT6B axis. Kaohsiung J. Med. Sci. 39 (9), 862–872. doi:10.1002/kjm2.12698
Yang, M., Zhang, Y., and Ren, J. (2020). Acetylation in cardiovascular diseases: molecular mechanisms and clinical implications. Biochim. Biophys. Acta Mol. Basis Dis. 1866 (10), 165836. doi:10.1016/j.bbadis.2020.165836
Yang, X. J. (2004). The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32 (3), 959–976. doi:10.1093/nar/gkh252
You, L., Nie, J., Sun, W. J., Zheng, Z. Q., and Yang, X. J. (2012). Lysine acetylation: enzymes, bromodomains and links to different diseases. Essays Biochem. 52 (1-12), 1–12. doi:10.1042/bse0520001
Yu, S., Jia, B., Liu, N., Yu, D., Zhang, S., and Wu, A. (2021). Fumonisin B1 triggers carcinogenesis via HDAC/PI3K/Akt signalling pathway in human esophageal epithelial cells. Sci. Total Environ. 787, 147405. doi:10.1016/j.scitotenv.2021.147405
Zaware, N., and Zhou, M. M. (2019). Bromodomain biology and drug discovery. Nat. Struct. Mol. Biol. 26 (10), 870–879. doi:10.1038/s41594-019-0309-8
Zhang, T., Wang, B., Gu, B., Su, F., Xiang, L., Liu, L., et al. (2022). Genetic and molecular characterization revealed the prognosis efficiency of histone acetylation in pan-digestive cancers. J. Oncol. 2022, 3938652. doi:10.1155/2022/3938652
Zhang, X., Zegar, T., Weiser, T., Hamdan, F. H., Berger, B. T., Lucas, R., et al. (2020). Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma. Int. J. Cancer 147 (10), 2847–2861. doi:10.1002/ijc.33137
Zhao, W., Mo, H., Liu, R., Chen, T., Yang, N., and Liu, Z. (2022). Matrix stiffness-induced upregulation of histone acetyltransferase KAT6A promotes hepatocellular carcinoma progression through regulating SOX2 expression. Br. J. Cancer 127 (2), 202–210. doi:10.1038/s41416-022-01784-9
Zhao, Y., Tang, J., Jiang, K., Liu, S. Y., Aicher, A., and Heeschen, C. (2023). Liquid biopsy in pancreatic cancer - current perspective and future outlook. Biochim. Biophys. Acta Rev. Cancer 1878 (3), 188868. doi:10.1016/j.bbcan.2023.188868
Zheng, Y., Zhu, L., Qin, Z. Y., Guo, Y., Wang, S., Xue, M., et al. (2023). Modulation of cellular metabolism by protein crotonylation regulates pancreatic cancer progression. Cell Rep. 42 (7), 112666. doi:10.1016/j.celrep.2023.112666
Keywords: lysine acetylation, digestive tract cancers, expression, pathogenic mechanisms, clinical application
Citation: Li P and Xue Y (2024) Dysregulation of lysine acetylation in the pathogenesis of digestive tract cancers and its clinical applications. Front. Cell Dev. Biol. 12:1447939. doi: 10.3389/fcell.2024.1447939
Received: 04 July 2024; Accepted: 20 September 2024;
Published: 26 September 2024.
Edited by:
Shashi Anand, University of Mississippi Medical Center, United StatesReviewed by:
Sirin Saranyutanon, Burapha University, ThailandMary Patton, University of South Alabama, United States
Copyright © 2024 Li and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Penghui Li, bHBoMDgxOUAxNjMuY29t
 Penghui Li
Penghui Li Yuan Xue2
Yuan Xue2