- 1Integrative Chemical Biology (ICB), Institute for Stem Cell Science and Regenerative Medicine, Bangalore, India
- 2Manipal Academy of Higher Education, Manipal, India
- 3Stowers Institute for Medical Research, Kansas City, MO, United States
- 4Department of Life Science, Shiv Nadar Institution of Eminence, Greater Noida, Uttar Pradesh, India
Post-transcriptional regulation has emerged as a key mechanism for regulating stem cell renewal and differentiation, which is essential for understanding tissue regeneration and homeostasis. Poly(A)-binding proteins are a family of RNA-binding proteins that play a vital role in post-transcriptional regulation by controlling mRNA stability and protein synthesis. The involvement of poly(A) binding proteins in a wide range of cellular functions is increasingly being investigated. In this study, we used the regenerative model planarian organism Schmidtea mediterranea to demonstrate the critical role of poly(A)-binding protein 2 (PABP2) in regulating neoblast maintenance and differentiation. A deficit in PABP2 blocks the transition of neoblasts toward immediate early progenitors, leading to an enhanced pool of non-committed neoblasts and a decreased progenitor population. This is reflected in variations in the transcriptome profile, providing evidence of downregulation in multiple lineages. Thus, an insufficiency of PABP2 resulted in defective formation and organization of tissue, leading to abnormal regeneration. Our study reveals the essential role of PABP2 in regulating genes that mediate stem cell commitment to early progenitors during tissue regeneration.
Introduction
Highly regenerative planarians are an exceptional model system to elucidate in vivo stem cell functions. Their regenerative capability is due to their adult pluripotent stem cell population, called clonogenic neoblasts, that can differentiate into multiple cell lineages (Wagner et al., 2011). Neoblasts respond to injury with a systemic increase in their proliferation rate. In response to tissue loss, a second phase of proliferation occurs locally at the wounded region, which is critical for the formation of undifferentiated tissue known as a “blastema” (Wenemoser and Reddien, 2010). Several regenerative clues such as signals from injury, positional information, and the extracellular matrix are crucial for the expression of cell-type specific differentiation marks (Wenemoser and Reddien, 2010; Scimone et al., 2017; Cote et al., 2019). In planarians, muscle fibers express positional controlling genes in a spatiotemporal manner to provide positional information, helping in stem cell fate determination (Witchley et al., 2013). It has been shown that neoblasts at the S, G2, and M phases of the cell cycle express fate specific transcription factors (FSTF) followed by asymmetric division to form early progenitors (specialized neoblasts) and noncommitted neoblasts (Raz et al., 2021a). Some of the specialized neoblasts can undergo asymmetric division to give rise to daughter neoblasts with non-identical fate-specifying factors (Raz et al., 2021a; Lei et al., 2016). The perturbation of these processes including proliferation and differentiation mediated by asymmetric division or external cues can lead to defective regeneration and tissue turnover.
Studies show that neoblasts undergo massive changes in gene expression during the process of proliferation and differentiation. RNA binding proteins (RBPs) are critical for determining the functionality of transcripts, thereby regulating gene expression. Several genes encoding RNA binding proteins are enriched in the neoblast population, suggesting a prominent role for post-transcriptional regulation in stem cells during planarian regeneration (Krishna et al., 2019; Rouhana et al., 2010; Brais et al., 1999). Post-transcriptional processes such as alternative polyadenylation and poly A tail length modulation govern mRNA stability through intrinsic sequence alterations. Our previous study identified a homolog for CPSF, CsTF, CFI, CFII, and the cis sequence binding sites in planarians, critical for polyadenylation and their regulation (Lakshmanan et al., 2016). Polyadenylation at the 3′ UTR of mRNAs has a high affinity for highly conserved RBPs called “poly A binding proteins nuclear” (PABPN) in vertebrates, also known as “poly A binding protein 2” (PABP2) in invertebrates. In mouse models and human cell lines, PABPN mRNA and protein expression levels are lower in skeletal muscles at homeostasis. Interestingly, an increase in PABPN is observed during skeletal muscle regeneration, which suggests a role for this protein during tissue regeneration and repair (Apponi et al., 2013). However, the role of PABP2 in modulating stem cell function during regenerative processes is unknown. PABP2 is a multi-functional protein involved in several molecular processes. For example, it has been shown to interact with RNA polymerase II, suggesting a possible role in transcriptional regulation (Bear et al., 2003). It was also identified as a molecule involved in alternative polyadenylation (De Klerk et al., 2012; Jenal et al., 2012; Martine, 2012), RNA hyperadenylation (Bresson and Conrad, 2013), and the regulation of lncRNAs (Beaulieu et al., 2012). Although the molecular mechanisms of PABPN/PABP2 on RNA have been extensively studied, its impact on stem cell function remains to be defined.
In Schmidtea mediterranea, we identified pabp2 enriched in neoblasts. Here, we report a novel role for PABP2 in stem cell differentiation during tissue regeneration. PABP2 knockdown leads to altered phenotypes in both regenerative and homeostatic conditions. Further analysis showed that PABP2 is critical for stem cell regulation, and its downregulation restricts fate commitment during regenerative processes.
Materials and methods
Planarian culture
Schmidtea mediterranea (sexual strain- S2F2) were maintained at 18–20°C in an incubator (SANYO MIR 554). The culture was maintained in 1 X Montjuich salt media (1.6 mM NaCl, 1 mM CaCl2, 0.1 mM MgCl2, 1 mM MgSO4, 0.1 mM KCl, and 1.2 mM NaHCO3; pH - 7) in milli Q water. The media was filtered using 0.45 μm vacuum filters (membrane filter Millipore 654). Stock animals were fed twice per week with liver paste, and the experimental animals were starved for 7 days prior to the experiment. The experimental animals were maintained in Petri dishes (120 mm) and were cleaned every alternative day.
RNA extraction
TRIzol (Invitrogen, 15,596,026) reagent was used for RNA extraction. The tissue for RNA extraction was collected in Eppendorf tubes and incubated with 300μL of TRIzol at 80°C for 1 h; one-fifth of the total volume of chloroform was added, mixed by inversion, and kept on ice for 10 min. Subsequently the sample with the reagents was centrifuged at 21,000 g for 20 min at 4°C. The uppermost aqueous layer was collected into a fresh Eppendorf tube without disturbing the interface. An equal amount of pre-chilled isopropanol was added and mixed with the previously collected solution and incubated at −20°C for 30 min. Following incubation, RNA was pelleted at 18,000 g for 20 min at 4°C. The supernatant was discarded, and the pellet was washed twice with 70% ethanol (prepared in nuclease-free water from Invitrogen; code 10,793,837) by centrifuging at 18,000 g for 10 min at 4°C. During RNA extraction from the sorted cells, 1 μL of glycoblue (Invitrogen, AM9515) was added to visualize the RNA pellet. The washed RNA pellet was air dried and reconstituted with nuclease-free water (Invitrogen_10793837) and stored at −80°C.
cDNA preparation
cDNA was prepared using 1 μg of extracted RNA using a SuperScript II Reverse Transcriptase kit (Invitrogen 18,064,022), oligo (dT) 12–18 Primer (Invitrogen 18,418,012), and RNase OUT Recombinant Ribonuclease Inhibitor (Invitrogen 10,777,019). The reaction was set up as per the protocol manual. The RNA in the reaction mix was digested using RNase H treatment (Invitrogen 18,021,014).
Gene cloning
Gene specific primers were used to amplify specific genes (pabp2: 5′-TATGGTCGATAGCCTAAATGACA-3′ and 5′-ATTACTTTTATCTGACGACTGCG-3′). LA Taq DNA Polymerase (TaKaRa RR002C) was utilized for PCR amplification of targeted genes. Purified PCR products were inserted into the vector using a TA cloning kit as per their protocol manual (Invitrogen K207040). The ligated products were transformed into competent DH5α cells (Escherichia coli) and screened for blue and white colonies under a kanamycin resistant background. Plasmids were extracted from the selected colonies and the sequence confirmed with M13 primers (Invitrogen; N52002 and N53002).
RNA interference (RNAi)
pabp2 and gfp (control) dsRNA were prepared from gel eluted (Promega, A9282) PCR product using T7 RNA polymerase (NEB, M0251L) as per Buchholz et al. (2006). The product was then treated with DNase-1 (Sigma, 1,014,159,001), and RNA was precipitated with 4 M lithium chloride and 100% ice-cold ethanol. dsRNA synthesis was confirmed by running 1 μL of the purified product on 1.5% agarose gel with 0.5 μg/μL ethidium bromide (Amresco, X328) in TBE buffer (pH - 8.0). The concentration of RNA was assessed by comparing the known nucleic acid concentrations of the DNA ladder bands (NEB, N3232L). The microinjection protocol was followed as mentioned previously (Shibata and Agata, 2018) using Nanoject II injector (Drummond Scientific Company, Broomall, PA, USA, 3–000–204) for prepharyngeal injection of dsRNA. Three shots of 69 nL (stock concentration of dsRNA-1 μg/μL) were administered for three consecutive days followed by a recovery time of 4 days. This regimen was followed, and animals were cut into two (at the prepharyngeal region) on the 11th day and checked for phenotypic defects in a regeneration experiment. In homeostasis, the experiment’s injection regimen was followed for 28–30 days.
RNAi efficacy assessment
pabp2 was amplified from 1 µL of cDNA synthesized using 1 µg RNA extracted from control and pabp2 RNAi samples. Following 25 cycles of PCR amplification, 3 µL of PCR product was mixed with loading dye (NEB, B7021S) and subjected to agarose gel (1.5% agarose prepared in TBE buffer with 0.5 μg/mL EtBr) electrophoresis. Actin was used as a loading control.
Whole mount immunostaining
Animals treated with 2% HCl and fixed using Carnoy’s fixative (60% ethanol, 30% chloroform, 10% glacial acetic acid); they were stored at −20°C for at least an hour and then rehydrated before being bleached in hydrogen peroxide (20% H2O2 in methanol). The animals were incubated in blocking solution (10% horse serum in PBSTx) followed by primary antibody (anti arrestin/tmus/anti phospho-histone H3 ser10 (Abcam 47,297)/anti-acetylated tubulin (Sigma) incubation overnight at room temperature or 2–4 h at room temperature at dilutions of 1:5,000 (anti arrestin), 1:100 (tmus), 1:100 (anti phospho-histone 3 ser10), and 1:1,000 (anti-acetylated tubulin). Anti PIWI1 antibody was raised against NEPEGPTETDQSLS antigen in rabbits as described by Guo et al. (2006). The animals were washed with PBS-Tx and stained with secondary antibody (1:400 anti-mouse and rabbit conjugated with both Alexa Fluor 488 and Alexa Fluor 546, molecular probes) before being incubated overnight at 4°C or 2–4 h at room temperature. The animals were then washed and stained with Hoechst (33,342). They were washed, mounted on slides in Mowiol (Sigma, 81,381) containing Dabco (Sigma D2780), and stored in the dark at 4°C.
RNA probe preparation and fluorescence in-situ hybridization
Plasmid with the requisite gene sequence was linearized using NotⅠ or HindⅢ (New England Biolabs) restriction enzyme and was used as template. Digoxigenin-UTP RNA (Sigma, 11277073910) or dinitrophenol-UTP RNA labeling mix (Perkin Elmer NEL555001EA), SP6/T7 (Roche; 10,810,274,001/Invitrogen; AM2718) polymerase along with template was used for riboprobe synthesis, and RNase free DNase (NEB M0303S) was used for template digestion. RNA was purified using Bio-Rad spin mini-column (7,326,830) as per the manufacturer’s instruction. Whole-mount fluorescence in-situ hybridization and double fluorescence in-situ hybridization was performed as per Pearson et al. (2009) and King and Newmark (2013).
The following primers were used: agat-1, 5′-GGATTTCCACCGGTTTTCTGTG-3′ and 5′-AATTGA ACACGATGTAAGCAGTG-3’; wi-1, 5′-CTCGTTGGCAAGATTCATCG-3′ and 5′-TGACA CCAAATACAAAGAGACA-3’; h2b, 5′- TCTGTTAAGAAGATTTCAAAGG-3′ and 5′- TCCTGTGTATTTTGTAACAGC-3’; myoD, 5′ TCAACAATACCGATCCAGCCC-3′ and 5′ TCGGG CTTAGCGTCCATTG-3’; nkx1.1, 5′ ATTCCAAGTCAAACGATAAGCCT-3′ and 5′ TTCCGTTG GTATTTCTTTAACGG-3’; pabp2, 5′-AATCAATTGCATTTTTTATATCT-3′ and 5′-ATCTATC CAATTATTACTCATAA-3’.
Image acquisition and quantification
An Olympus SX-16 stereomicroscope was used to obtain darkfield images. Confocal images were acquired with an FV 3000 laser scanning microscope (Olympus) with Olympus Flow View software. Image processing and quantification were done using ImageJ software (https://imagej.nih.gov/ij).
Quantification of wi1 and pabp2 colocalization was done manually by quantifying the number of wi1+ cells and the number of wi1+ cells that co-expressed pabp2. Among the total wi1+ population (899 cells), the percentage of cells that were double-positive for both wi1 and pabp2 (244 cells) were calculated. wi1 and pabp2 colocalization were quantified from fluorescent in-situ hybridization images captured randomly across the whole animal (n = 7) using Fiji software. Blastemal size quantification was done by comparing the blastemal area to the total surface area of the worm (n = 6). Muscle fiber thickness was calculated as an average diameter of three random locations per muscle fiber (total number of fibers measured from a single worm = 30) (n = 5). Protonephridial images were collected dorsally from control and pabp2 RNAi animals, and the imaging parameters were kept constant. The protonephridial intensity captured per frame was quantified through ImageJ analysis (n = 6). Single fluorescent in-situ hybridization was quantified manually by comparing the number of positive cells across total dapi+ cells per frame imaged at the blastema (myoD (n = 6), nkx1.1 (n = 6) and agat 1 (n = 6) respectively). H3P analysis was done by comparing the total number of positive cells to the whole worm area (i.e., number of H3P+ cells/unit area (mm2)); (2 dpa, n = 10; 4 dpa, n = 5; 7 dpa, n = 3). For wi1/WI1 colocalization analysis, the stem cell population (wi1+/WI1+) and the immediate mitotic progeny population (WI1+) were manually counted using ImageJ at the blastema. The ratio of immediate early progeny to stem cells was calculated among each worm (n = 6). The ratios were compared across the control and pabp2 RNAi worms.
Statistical analysis
Statistical analysis was performed used Student’s t-test. An unpaired two-tailed t-test was used to check for the p-values. All experiments were conducted in biological duplicates, whereas the FACS analysis for mitochondrial potential was conducted in biological triplicates. GraphPad Prism software was used for data analysis.
Single-cell transcriptome analysis
We used a single-cell transcriptome dataset published from Peter Reddien’s laboratory (Fincher et al., 2018), GEO accession number GSE111764, to extract the cells that express pabp2. We used the data matrix submitted in the sequence read archive (SRA) to extract only the cells that express pabp2. We reanalyzed the single-cell data as described by Ross and colleagues (BioProject accession number PRJNA432445). We used Seurat (https://satijalab.org/seurat/) to analyze the single-cell transcriptome for the cells that express pabp2 mRNA (Butler et al., 2018; Stuart et al., 2019). Based on the markers from the single-cell transcriptome dataset (Fincher et al., 2018), we classified the uniform manifold approximation and projection (UMAP) clusters as cell types. We used the “log normalize” method of Seurat on the dataset, which was further scaled (linear transformation) using Seurat. This scaled value was further log-transformed and plotted as a heatmap for genes of interest. We used R ggplot2 GMD and heatmap.2 to derive all the plots.
Phylogenetic analysis
We aligned the planarian PABP2 sequence with known PABP sequences from other species (obtained from NCBI and GenBank) using MAFFT (version 7.310), and the multiple sequence alignment was visualized using Jalview. The phylogenetic tree was constructed using the IQTree webserver (IQ-TREE 1.6.12). In brief, the ModelFinder functionality implemented within IQTree was used to find the best substitution model for maximum likelihood estimation based on the Bayesian inference criterion (JTT + I + G4). A maximum likelihood tree was then generated using this model, along with bootstrap support (n = 1,000) in iqtree and was visualized using iTOL (iTOL: Interactive Tree of Life (embl.de)). The domain structure of the PABP2 sequence was identified using NCBI CDD.
Transcriptome analysis
RNA was extracted from the anterior and posterior blastema of the control and pabp2 RNAi animals at 0–3 dpa. The experiment was conducted in biological duplicates. NEB Next Poly(A) mRNA Magnetic Isolation Module (Catalog number - E7490L) was used for poly A selection, and NEBNext® Ultra™ II Directional RNA Library Preparation with Sample Purification Beads (Catalog number - E7765L) was used for transcriptome library preparation. Sequencing was done using the NovaSeq 6,000 platform using SP flowcell with 2 × 50 bp sequencing read length. All the samples were sequenced in biological replicates. Post sequencing, 11–16 million paired-end (2 * 50 bp) reads were obtained. Adapters were trimmed from the reads using cutadapt v2.10 (-a AGATCGGAAGAGCACACGTCTGAAC TCCAGTCA -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT -u 2 -U 2). The trimmed reads were mapped to the S. mediterranea transcriptome (Rozanski et al., 2019) using hisat2 v2.1.0 (--rna-strandness R). They (76%–80% mapping percentage) were counted using feature Counts v2.0.0. DESeq2 v1.40.1 was used to perform the read count normalization and differential expression analysis. The plots were generated in R v4.3.0. We have considered a log2FC threshold of 0.58 and an adjusted p-value (q) of less than 0.05 for classifying a gene as differentially expressed. Different planarian cell-type markers were obtained from the available single-cell transcriptome data (Fincher et al., 2018). The sequencing data reported in this manuscript have been deposited at NCBI- Sequence Read Archive (SRA), project ID: PRJNA1031933.
Fluorescence activated cell sorting
Cell suspension for FACS sorting was prepared as per Mohamed et al. (2021). The worms (n = 30) were macerated in calcium and magnesium-free buffer (CMFB) and mechanically sheared using a micropipette and strained using 70 μm cell strainers. The cells were centrifuged at 300 × g for 10 min, and the resulting single-cell suspension was stained with Hoechst 33,342 (40 μg/mL) for 40 min. Each subgroup among the control and pabp2 RNAi was FCCP treated (10 μM for 10 min). All samples were stained with rhodamine 123 (2 μM for 10 min) and analyzed using a BD Fortessa flow cytometer. The mitochondrial potential was assessed in control and pabp2 RNAi separately by calculating the decrease in potential in comparison with the FCCP-treated subgroups.
Results
pabp2 is enriched in the neoblast and epidermal cell populations
Previously, E2FB12/PABPC1 and PABPC2 were identified in S. mediterranea (Wang et al., 2010; Bansal et al., 2017). Here, the newly identified PABP2 sequence (dd_Smed_v6_21160_0_1) is clustered into a branch of PABP2 sequences from other species (Supplementary Figure S1A); it is evolutionarily distinct from the previously known PABPC. Domain analysis revealed the presence of a single RNA recognition motif (RRM) which is a feature of PABPN (Supplementary Figure S2B). Furthermore, the PABP2 sequence shows similarity with the PABPN sequences of various organisms in multiple sequence alignment (Supplementary Figure S1C). Thus, the identified pabp2 is presumed to be a pabp2 homologous gene in S. mediterranea.
We investigated the expression of pabp2 across different cell types using single-cell transcriptome data (https://radiant.wi.mit.edu/app/) and noticed the predominant expression of pabp2 in neoblasts (35%), followed by the major epidermal cluster (21%), cathepsin+ cells (11%), and the intestinal cell population (8%) (Figures 1A–C). The in-vivo expression of pabp2 in neoblasts was confirmed through double fluorescent in-situ hybridization using wi1 (pan neoblast marker) and pabp2 (Figure 1E). This analysis revealed that 27% of the neoblast population co-expressed pabp2, which is consistent with the single-cell data revealing 35% of neoblast co-expressing pabp2. From single cell RNA sequencing on neoblast enriched for both high wi1 transcript and protein expression, neoblasts have been classified into 12 different classes (Nb1-Nb12) (Zeng et al., 2018). Among these classes, the tspan1+ population (Nb2) was shown to exhibit clonogenic properties (Zeng et al., 2018). Some 35% of tspan1+ neoblasts, which co-express tgs1, was shown to be neuronal specialized neoblasts (Fincher et al., 2018). Expression of pabp2 was notably high in tspan+ neoblasts, suggesting a crucial role for PABP2 at stem cell regulation (Figure 1D). The second-highest enrichment of pabp2 was found in the epidermal cluster. The maintenance of epidermal integrity is imperative for wound healing and regeneration in planarians (Bansal et al., 2017). Altogether, pabp2 was predominantly expressed in neoblasts and the epidermis, suggesting a role for the protein in stem cell function and regeneration.
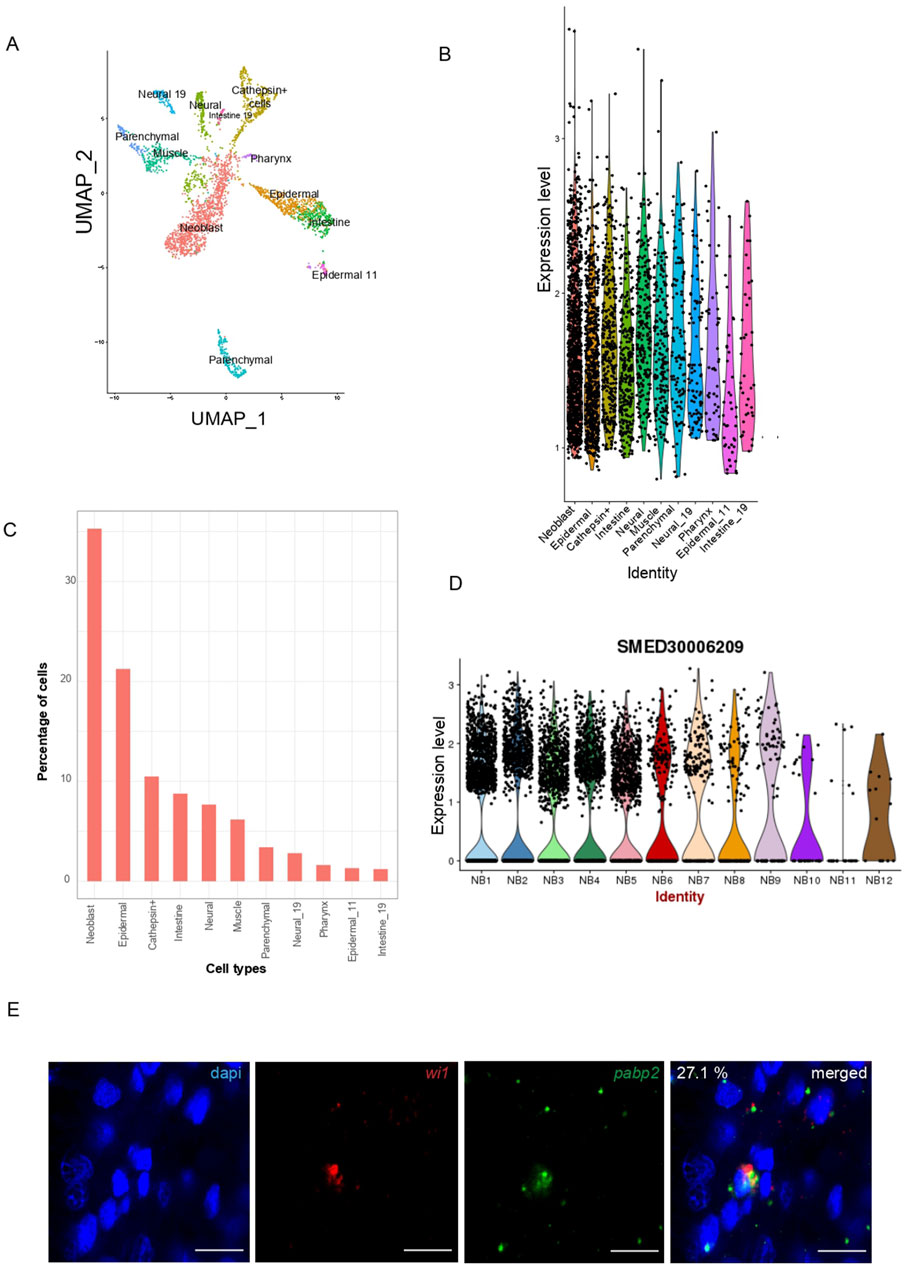
Figure 1. Characterization of pabp2. (A) UMAP plot obtained from single-cell transcriptome data (Fincher et al., 2018) showing pabp2 distribution across different cell types. (B) Violin plot depicting the expression level of pabp2 across different cell types. (C) Percentage of cells expressing pabp2 among different cell types. (D) Violin plots showing the distribution of pabp2 across 12 neoblast clusters. (E) Fluorescent in-situ hybridization to study the expression pattern of pabp2. Double fluorescent in-situ hybridization showing colocalization of pabp2 with wi1. 27.1% of wi1+ population co-expressed pabp2, i.e., among 899 wi1 expressing cells, 244 co-expressed pabp2. Probes are indicated (wi1-red, pabp2 - green), scale bars, 10 μm (n = 7).
pabp2 is an essential regulator of planarian regeneration and homeostasis
Previously, pabpc1 and pabpc2 were shown to have a role in meiotic progression and epidermal integrity, respectively (Wang et al., 2010; Bansal et al., 2017). To discern whether PABP2 has any functional significance during planarian regeneration and homeostasis, we knocked down pabp2 expression. RNAi based knockdown was done through microinjecting pabp2 or gfp (control) dsRNA for three consecutive days, followed by a 4-day recovery period. Following two rounds of injections, the animals were amputated for the regeneration experiment, and we observed that the pabp2 RNAi animals exhibited body lesions and underdeveloped photoreceptors (Figures 2A,B); they were subsequently lysed by day 20 of the injection regimen. The efficacy of pabp2 RNAi was assessed by PCR amplification (Supplementary Figure S2B). Phenotypes were observed in 90% of animals by 8 dpa (days post amputation). Blastema size quantification at various time points during regeneration revealed a significant (∼37.5%) reduction by 7 dpa at both anterior and posterior regenerating fragments (Supplementary Figure S2A). Under homeostatic conditions, pabp2 RNAi animals displayed head regression and had lesions on their body (Supplementary Figure S2C, D). Of the animals, 71% exhibited these phenotypes and underwent lysis within 30 days of the injection regimen. In summary, PABP2 is necessary for tissue regeneration as well as homeostasis.
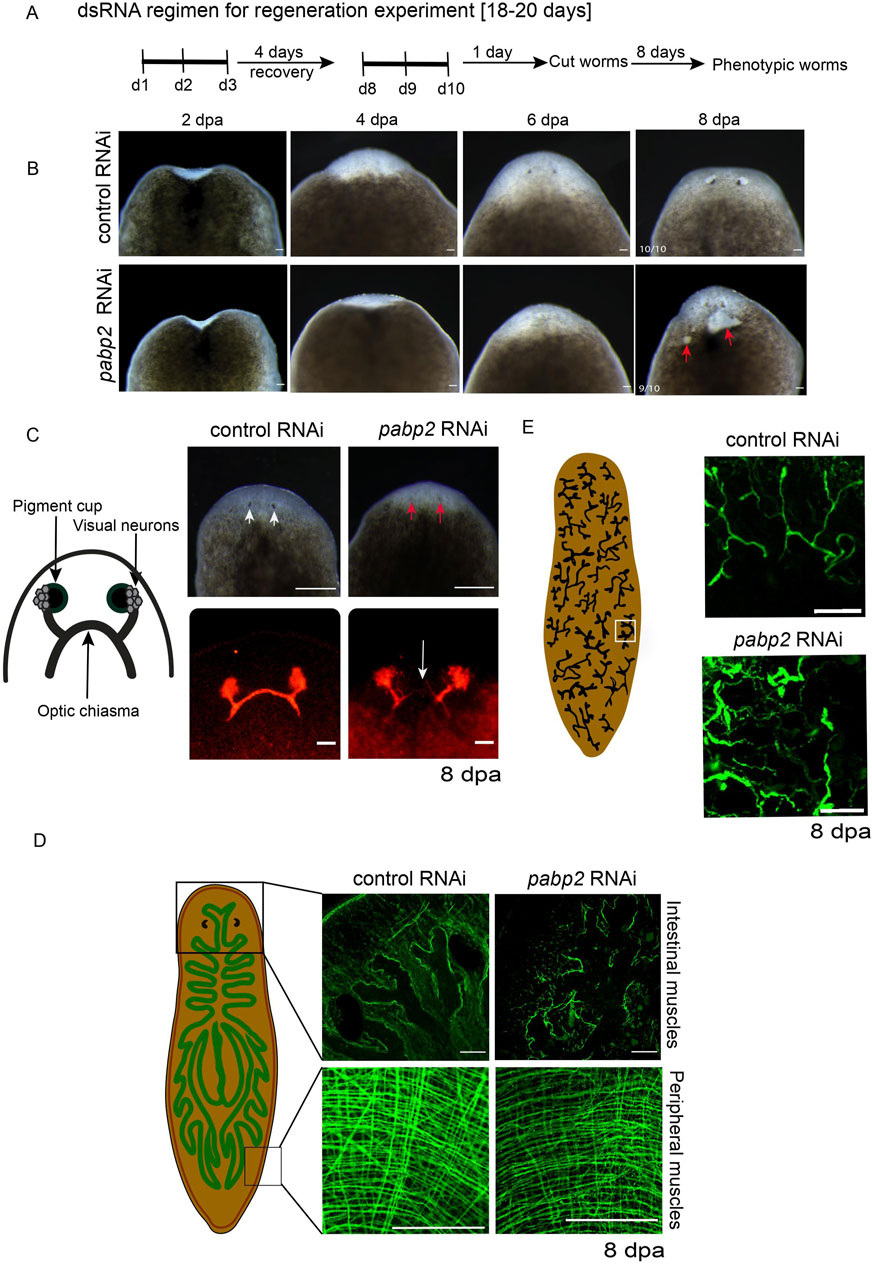
Figure 2. (A) Schematic representation showing the timeline of dsRNA administration for the regenerative RNAi experiment. (B) Representative images captured on 2, 4, 6, and 8 dpa. The red arrow points toward the lesions developed on the pabp2 RNAi animals during the course of regeneration (n = 10), scale bar, 200 μm. (C) Schematic representation representing the organization of photoreceptors, visual neurons, and optic chiasma in the planarian photoreceptor. Image of animals with photoreceptor where phenotypic animals exhibit underdeveloped eyes. White arrows indicate the photoreceptor pigment in control, and the red arrow represents the reduction in the photoreceptor pigment in pabp2 RNAi, scale bar, 200 μm. Confocal image of anti-arrestin immunostaining staining for photoreceptor neurons at 8 dpa. The white arrow represents lack of optic chiasma formation in pabp2 RNAi (n = 5), scale bar, 50 μm. (D) Schematic representation of peripheral and intestinal muscle fiber organization in planarian. Confocal image of whole mount tmus immunostaining to visualize the organization of anterior intestinal and peripheral muscle fibers at 8 dpa (n = 5), scale bar, 50 μm. (E) Schematic representation of the arrangement of protonephridia embedded in the epithelial tissue. Marked in the white box is a single protonephridia unit. Confocal image of anti-acetylated tubulin immunostaining showing organization of protonephridia in control RNAi and pabp2 RNAi at 8 dpa (n = 6), scale bar, 50 μm.
pabp2 knockdown impairs terminally differentiated tissues
The gross phenotypes in pabp2 RNAi are the stunted blastema growth and lesions in the regenerating animals, suggesting a defect in the blastema and the epidermal tissue. We further focused on delineating the effect of pabp2 RNAi on various tissues during the regenerative processes. At 8 dpa, animals were collected and experimentally assessed for regenerative defects. The planarian photoreceptor consists of pigment cups, photoreceptor neurons (PRN) and the axonal bundle arising from the dorsally located cell body which joins together to form the optic chiasma (Macrae, 1964; Agata et al., 1998; Shettigar et al., 2017). Phenotypic animals were observed with reduced photoreceptor pigmentation, suggesting a possible defect in eye regeneration (Figure 2C). We investigated the organization of photoreceptors through VC1 immunostaining and observed a lack of optic chiasma formation (Figure 2C). Previous studies reported mutations in PABP2 leading to oculopharyngeal muscular dystrophy patients (Brais et al., 1999; Brais et al., 1998). In planarians, tissues are surrounded by muscle fibers which provide support and also express positional control genes (PCG) critical for regeneration. Upon injury, the subepidermal muscle fibers provide positional cues essential for tissue regeneration (Witchley et al., 2013). We thus examined the possibility that an impaired muscle fiber organization may be responsible for the defective optic chiasma formation. Using tmus immunostaining, we observed that pabp2 RNAi perturbs the organization of both peripheral and intestinal muscles (Figure 2D). Planarian subepidermal myofibers are arranged in four different layers: outer circular, longitudinal, diagonal, and inner longitudinal (Cebrià et al., 1997). We found a 33% reduction in myofiber thickness upon quantification of circular muscle fiber thickness at 8 dpa (Supplementary Figure S2E). Since pabp2 RNAi leads to defective muscle organization, we also examined the organization of other tissues such as protonephridia, which is embedded in epithelial tissue and consists of multiple cell types that are organized to form a branched pattern (Rink et al., 2011) at 8 dpa. Anti-acetylated tubulin immunostaining revealed a disorganized protonephridial branching pattern (Figure 2E). Protonephridial intensity increased by 33%, indicating a spread of protonephridial organization (Supplementary Figure S2F). Taken together, we observed defects in terminally differentiated tissue, resulting either from an intrinsic neoblast defect failing to replace cells during regeneration or defective muscle fibers, which could lead to disorganized organ systems.
Knockdown of pabp2 leads to increased neoblasts at the blastema
Since pabp2 is enriched in the neoblast population, its depletion could either lead to a defect in proliferation, differentiation, or both. One of the possibilities is depletion of the neoblast population subsequently resulting in decreased progenitors. A second possibility could be an intrinsically defective neoblast population in pabp2 RNAi animals, affecting the later stages of differentiation. Thus, we checked the stem cell population by probing for h2b+ and wi1+ cells (neoblast markers) using whole mount fluorescent in-situ hybridization at 3 dpa (Figure 3A; Supplementary Figure S3A). We found a significant (p < 0.05) increase in h2b+ and wi1+ cells in pabp2 RNAi animals, suggesting an overall increase in the stem cell pool. Furthermore, we checked for mitotic neoblast (cells at G2 to M phase) using antibody against H3PS10 on 2, 4, and 7 dpa. We observed a slight increase (1.3 fold, p < 0.05) in the H3P+ population at 4 dpa, but no change in the numbers was observed at 2 dpa. To ensure that no major changes occurred in the mitotic cell numbers at later time points of regeneration, we left a few knockdown animals until 7 dpa and measured the number of H3P+ cells. We did not notice any observable changes in the H3P+ cells, suggesting that the pabp2 RNAi has no effect on the mitotic phase of the neoblast population (Figures 3C,D). Together, our data suggest that neoblast maintenance was not affected in the pabp2 RNAi animals. It is possible that the increased neoblast could impair differentiation, thereby leading to accumulation of unspecified neoblasts. However, the function of PABP2 in neoblast differentiation requires further investigation.
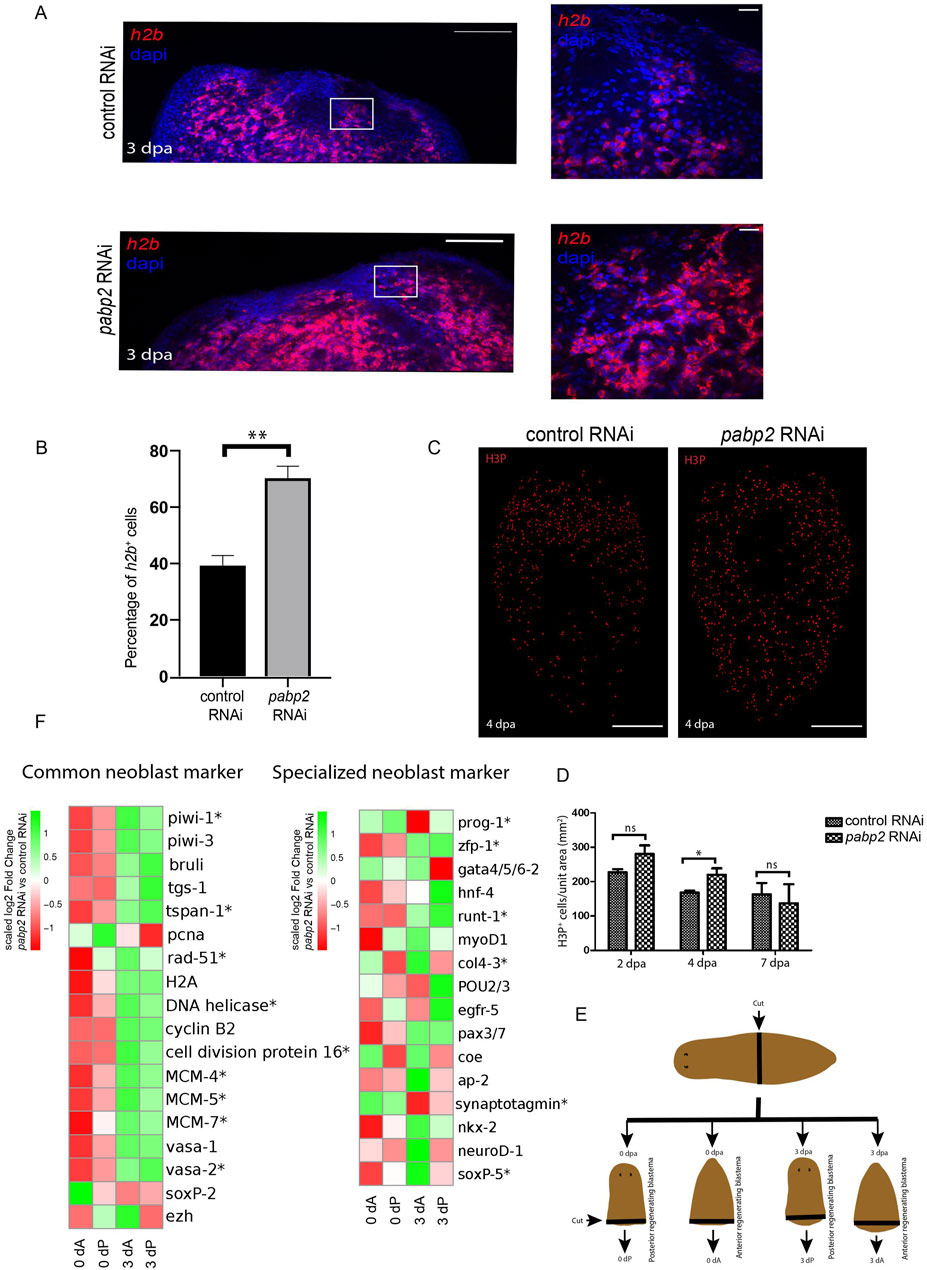
Figure 3. (A) Confocal images of fluorescent in-situ hybridization at the regenerating surface for h2b among control RNAi and pabp2 RNAi (scale bar, 100 μm) and the zoomed images (scale bar, 20 μm). White boxes indicate the magnified area in the zoomed images in the columns to the right. (B) Quantification of the percentage of h2b+ cells (n = 6) from the zoomed images, **p < 0.01. (C) Maximum intensity projections of confocal images showing H3PS10+ cells at 4 dpa among control RNAi and pabp2 RNAi (scale bar, 50 μm). (D) Quantification of the total number of mitotic cells (H3PS10+) at 2 dpa (n = 8), 4 dpa (n = 5, and 7 dpa (n = 3), *p < 0.05; n.s. p > 0.05. Number of H3PS10+ cells counted at 2 dpa (control RNAi- 4,188, pabp2 RNAi- 8,052), 4 dpa (control RNAi- 2,283, pabp2 RNAi- 4,475), and 7 dpa (control RNAi- 502, pabp2 RNAi- 572). (E) Schematic representation showing the methodology followed during sample collection for transcriptome analysis across control and pabp2 RNAi. The RNAi animals were cut at the prepharyngeal region into anterior regenerating and posterior regenerating animals. Blastema was collected at 0 and 3 dpa from anterior (0 dA, 3 dA) and posterior (0 dP, 3 dP) regenerating animals, respectively. (F) Heatmap depicting the expression of stem cell-specific markers at 0 and 3 dpa across anterior (0 dA, 3 dA) and posterior (0 dP, 3 dP) regenerating blastema, respectively. The asterisk indicates that at least one of the four comparisons included in the heatmap is statistically significance (q < 0.05) for that gene. Details about the fold change and the normalized read counts are included in Supplementary Tables S1, S2.
To further study the effect of pabp2 RNAi on neoblast maintenance, we conducted transcriptome profiling of 0 dpa and 3 dpa blastema from control and pabp2 RNAi animals. At 3 dpa, we observed a significant upregulation of neoblast-specific transcripts including wi1, tspan, vasa, and cell cycle proteins like DNA helicase and pcna (Figure 3E; Supplementary Table S1), which correlates with fluorescent in-situ hybridization experiments, suggesting an increased neoblast pool. Planarian neoblasts are a heterogeneous group consisting of distinct subpopulations known as “specialized neoblasts”. An extensive single-cell transcriptome analysis from the X1 population of neoblasts (dividing neoblasts) identified 14 different clusters of neoblasts, with neoblast cluster 2 revealing clonogenic properties. Further analysis also showed TSPAN as a surface marker expressed on the clonogenic neoblast. Another study showed that tspan+ neoblast co-expresses tgs-1, which also specifies neuronal specialized neoblasts (Fincher et al., 2018). Our transcriptome analysis showed increased expression of tspan in the pabp2 RNAi animals, suggesting the enrichment of clonogenic neoblasts. However, based on Fincher et al. (2018), we cannot also rule out that the neoblasts express neuronal lineages. It has been shown that the expression of fate-specific transcription factor (FSTF) regulates neoblast fate, and a neoblast can asymmetrically divide to give rise to fate-specified progeny (FSTF+) and a pluripotent neoblast (FSTF−) (Raz et al., 2021a). zfp-1, a FSTF expressed by epidermal specialized neoblasts, is essential to maintaining the epidermal lineage. Low levels of zfp1 expression in the intestinal neoblast coincide with endocyte-specific markers (King et al., 2024). zfp-1 was upregulated in pabp2 RNAi animals (Figure 3E; Supplementary Tables S2, S3), suggesting a possible defect in the epidermal or intestinal lineages. In addition, we also analyzed the transcriptome data for the expression of the early lineage markers. Most of the early lineage markers, such as ap2, coe, ston2 (neuronal marker), hnf4, and gata4/5/6 (intestinal marker), did not show any significant difference in expression (p > 0.05) or downregulation (p > 0.05) in the pabp2 RNAi animals (Figure 3E; Supplementary Table S2). In summary, our transcriptome data from the pabp2 RNAi animals showed upregulation of the global stem cell specific markers.
PABP2 is critical for cellular differentiation
Overall, an increase in neoblast number accompanied by defects in terminally differentiated tissue could result from defective commitment of neoblasts toward specific lineages (Cebrià et al., 2018). In order to determine if there is a differentiation defect in the pabp2 RNAi scenario, a colocalization experiment probing for wi1 (fluorescent in-situ hybridization for mRNA) and WI 1 (immunostaining for protein) was conducted. wi1 transcript is expressed in the neoblast whereas the corresponding protein is enriched both in the neoblast and the progenitors (Guo et al., 2006) (Supplementary Figure S4A). Stem cells express both wi1 mRNA and protein, and the WI1 protein endures for 72 h in post-mitotic cells (Guo et al., 2006). wi1 (−)/WI1 (+) cells represent the transition state from stem cell to postmitotic progeny, meaning that they mark the immediate early progeny. A change in the differentiation process can be measured by calculating the ratio of immediate early progeny with the stem cell population. An increase or decrease in this ratio will denote an increased or decreased differentiation process. Interestingly, the ratio at 3 dpa was 1.33 in control animals and decreased to 0.45 in pabp2 RNAi animals, thereby indicating a significant decrease in the immediate early progeny (Figures 4B,C). Taken together, these data demonstrate that a decrease in differentiation is reflected by an increase in the stem cell population.
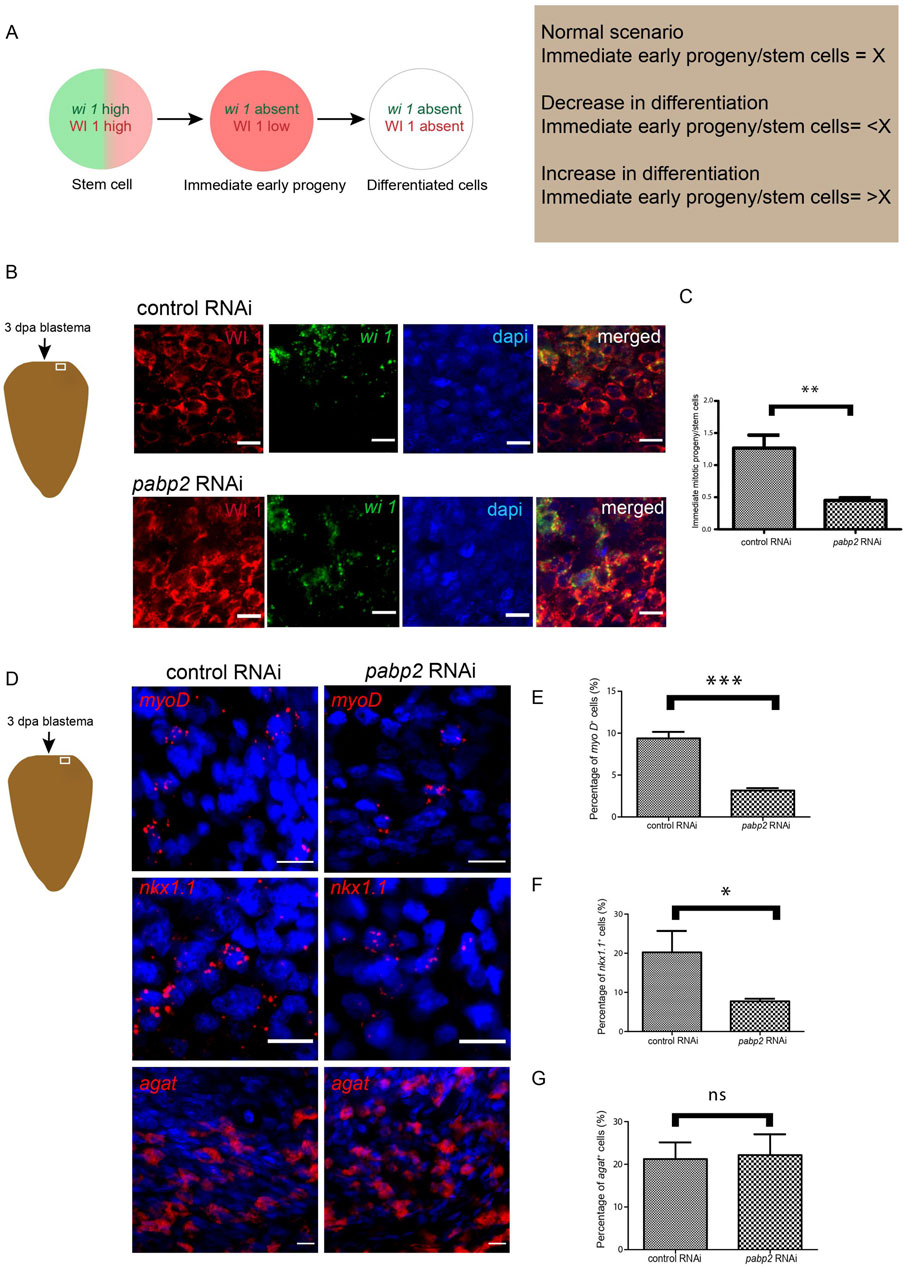
Figure 4. (A) Schematic representation showing the procedure adapted for quantifying differentiation defects. Green and red denote the presence of wi1 transcript and protein, respectively. Ratio of immediate early progeny to stem cell reflecting the variations in differentiation. (B) Schematic representation indicates the area of imaging at 3 dpa blastema. Confocal imaging of colocalization of wi1 (green) and WI1 (red) using fluorescent in-situ hybridization and immunofluorescence, respectively, scale bar, 10 μm. (C) Graphical representation of decrease in differentiation. Graph plotted from the quantified ratio of immediate early progeny to stem cell population (n = 6), **p < 0.01. (D) Fluorescent in-situ hybridization probing for various progenitor populations and its quantifications (myoD - longitudinal muscle fiber, nkx1.1 - circular muscle fiber, agat 1 - epidermal cells) across control RNAi and pabp2 RNAi, scale bar, 10 μm. (E–G) Representation of percentage of progenitor-expressing cells compared across control and pabp2 RNAi for myoD (n = 5), nkx1.1 (n = 6) and agat 1 (n = 6), respectively. *p < 0.05, ***p < 0.001, n.s. p > 0.05.
It has been shown that neoblasts express fate-specifying transcription factors required for their transition toward respective progenitors (Scimone et al., 2014). To decipher how the progenitor populations are affected, we performed fluorescent in-situ hybridization by probing for markers such as myoD and nkx1.1 which mark the longitudinal and circular muscle fibers respectively, and found a 60% decrease in myoD- and 50% decrease in nkx1.1-expressed cells (Figures 4E,F), corroborating the observed muscle fiber defect. Lineage specification during the epidermal differentiation process is very well studied in planarians. It is also known that the zeta neoblast lineage gives rise to epidermal tissue and that agat 1 marks the latter epidermal progenitors (Wolfswinkel et al., 2014; Tu et al., 2015). Since we found an enrichment of pabp2 in the epidermis (Figure 1), we quantified the epidermal progenitor population. However, there is no change in the agat 1+ epidermal progeny in pabp2 RNAi animals (Figure 4G). This also corroborates our previous finding that the pabp2 RNAi animals showed increased expression of zfp1—a zeta neoblast marker. The lesions observed could be due to the faulty body wall muscle organization, which was evident from muscle staining in the pabp2 RNAi animals (Figure 2D). Altogether, we found a decrease of myoD- and nkx 1.1-expressing cells, whereas agat 1+ cells remain unchanged, suggesting a possibility of muscle lineages being downregulated. To conclusively understand the effect of pabp2 RNAi on other lineages, it is essential to study the expression of a wide variety of lineage-specific markers.
PABP2 regulates transcripts essential for major epidermal population and intestinal lineages
Transcriptome analysis was performed to understand the effect of pabp2 RNAi on commitment toward various cell lineages. It has been reported that most lineage specification marks are expressed by 3 dpa (Wenemoser and Reddien, 2010). Transcriptome sequencing was conducted at 0 dpa and 3 dpa blastema in control and pabp2 RNAi animals to investigate the expression of different cell-type markers (Fincher et al., 2018). Among the various cell types, transcripts representing major epidermal clusters were drastically downregulated, such as egr 5, zpuf 6, and vim 3 by 3 dpa (Figure 5A; Supplementary Table S3), which are critical for later stages of epidermal differentiation (Tu et al., 2015). At 3 dpa, there was a minor change in prog expression (log2FC −0.83 and −0.38, respectively, in the anterior and posterior regenerating blastema). In the anterior and posterior regenerating blastema, we did not observe a reduction in agat 1 expression at 3 dpa (log2FC −0.05 and −0.01), and our fluorescent in-situ hybridization showed no change in agat 1+ cell population (Figure 4D). Together, our results suggest that epidermal lineage defects in pabp2 RNAi are caused by defects in later stages of differentiation among epidermal cell types rather than early progenitors.
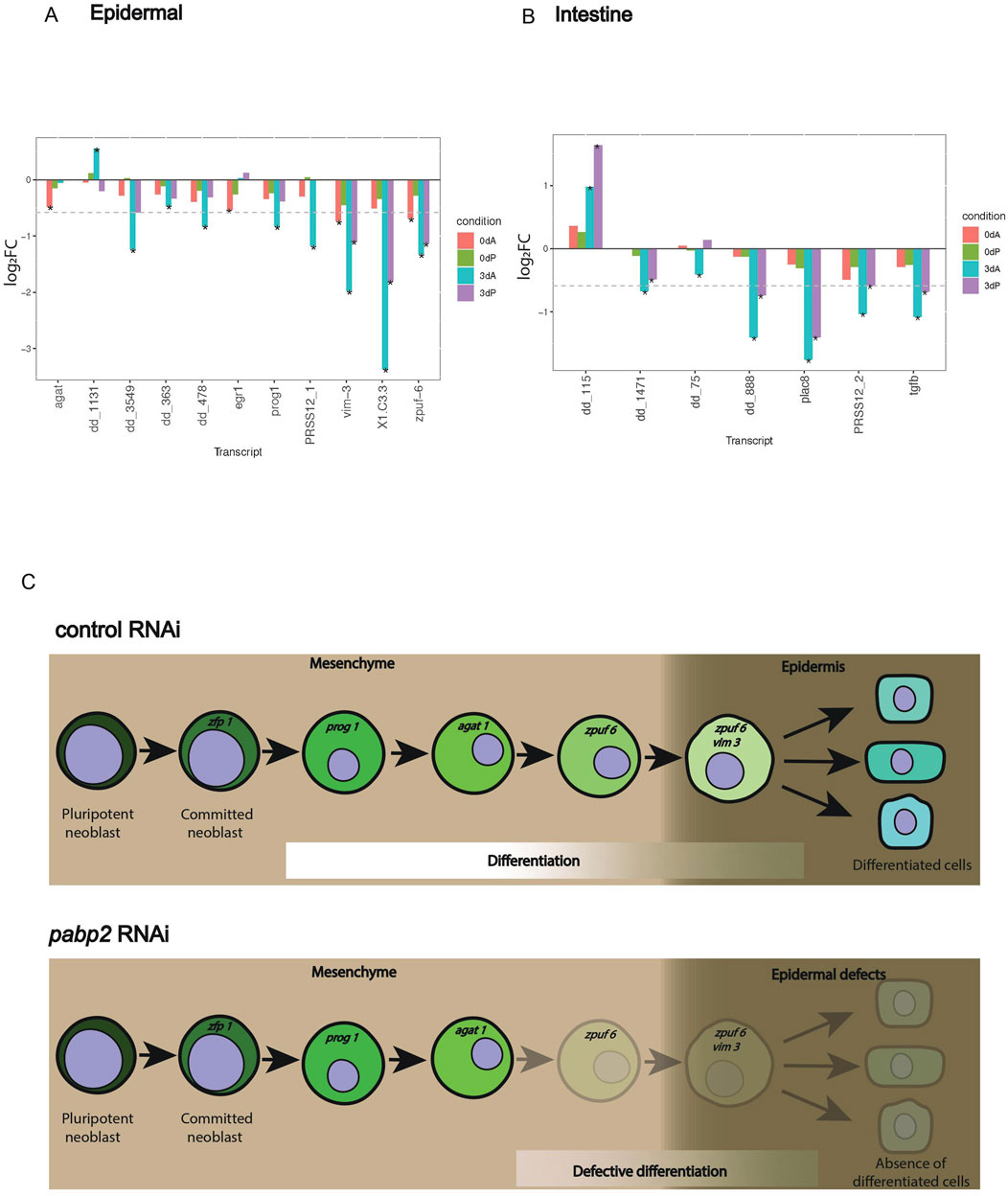
Figure 5. (A, B) Bar plot showing gene expression variation among specific lineages. Transcripts with variations among (A) epidermal and (B) intestinal markers compared between pabp2 RNAi and control RNAi at various time points (as denoted by color bars within plots) among anterior and posterior regenerating blastema. The comparison is between pabp2 RNAi versus control RNAi in each of the condition denoted by the bar color within the plots. Transcript PRSS12_1 (dd_Smed_v6_351_0_1) is specific to epidermis, and PRSS12_2 (dd_Smed_v6_790_0_1) is specific to intestine. The asterisk indicates statistical significance (q < 0.05), and the dotted line on y-axis represents log2FC −0.58. All cell-type specific transcripts with q < 0.05 in at least one of the four comparisons are included in the graph. Details about fold change and normalized read counts are provided in Supplementary Table S3. (C) Working model depicting the critical role of PABP2 in epidermal differentiation. The epidermal markers from the lineage progression during its transition from mesenchyme to epidermis across control and pabp2 RNAi. pabp2 RNAi leads to defective differentiation post-agat 1 stage.
Our analysis revealed a drastic decrease in intestinal-specific transcripts across anterior and posterior developing blastema (Figures 3E, 5B). The cathepsin+ cluster consists of a heterogeneous group of cells which includes pigment cells, glial cells, and several other unknown cell types (Lucila et al., 2018). Cela1 and jag1 specific for cathepsin+ cells showed downregulation at 3 dpa (Supplementary Figure S5A, Supplementary Table S3), suggesting that among the mixed population of cathepsin+ cells, a few subtypes were affected upon pabp2 RNAi. In the posterior developing blastema, we also observed few transcripts being downregulated among the pharyngeal cluster (Supplementary Figure S5B, Supplementary Table S3). Furthermore, we also observed few muscle-specific transcripts being downregulated which could possibly result from an indirect effect of suboptimal levels of pabp2 (Supplementary Figure S5C, Supplementary Table S3). The cell types which did not show significant variation include the parapharyngeal, neuronal, ciliated neuronal, and non-ciliated neuronal lineages upon pabp2 RNAi (Supplementary Figure S5D, S5E, S5F, S5G, Supplementary Table S3). In summary, our bulk transcriptome data from the control and pabp2 RNAi animals overlaid on the existing single cell data showed defects predominantly in the major epidermal population, a subset of cathepsin+ cell types and intestinal lineage.
Discussion
Our study revealed a role for PABP2 in the transition of stem cells to differentiated cell types. PABP2 insufficiency leads to enhanced self-renewal of intrinsically defective neoblasts and dysregulated differentiation. Hence, a defect in PABP2 functioning affects the process of differentiation, thereby leading to an insufficient progenitor population resulting in improper tissue regeneration and organization.
Cellular differentiation is the process by which committed cells undergo drastic gene expression changes to become specialized cells. In humans, defective differentiation can lead to several disease conditions affecting specific cell lineages, such as muscle (Storey et al., 2020) and B cells (Tangye et al., 2023). Here, reduction of pabp2 causes an increase in the neoblast population along with a decrease in their differentiation, resulting in a defect in lineage progression toward single or multiple cell types. We tackled the effect of PABP2 deficiency in regulating differentiation using epidermal lineages as a proxy. Planarian fate diversity occurs at various stages among different lineages. A recent study demonstrated the direct specification for epidermal, intestinal, muscle, and parenchymal lineages—fate specification occurs at the neoblast stage for these lineages (King et al., 2024). Planarian epidermal lineage commitment has been extensively studied, and many markers of epidermal lineage progression have been identified (Tu et al., 2015). The upregulation of zfp1 indicates the commitment of neoblasts to the epidermal lineage. Simultaneously, this is also evident from the maintenance of agat1+ cells, an epidermal lineage marker. Together, these provide evidence that the stem cell self-renewal and commitment to epidermal lineages are unaffected. However, our transcriptome data revealed that the consequence of pabp2 RNAi was observed in the latter stages of differentiation, which is evident from the drastic decrease in zpuf 6, and vim 3 expression (late epidermal progeny markers) (Figure 5C). This late-stage differentiation defect could either be an effect of an intrinsically defective early progenitor or could result from the depletion of the late progenitor-specific transcript which restricted the epidermal lineage progression. These observations point to a critical role for PABP2 in regulating cellular differentiation.
In addition to the significant reduction in transcripts specific to major epidermal population, a subset of the cathepsin+ cell type and intestinal populations is observed; we also observed a significant decrease in the nkx1.1+ and myoD+ cells in the pabp2 RNAi animals. This suggests that PABP2 is crucial for the regeneration and maintenance of the muscle fibers. Here, it is likely that lesions observed in the pabp2 RNAi animals could be due to the defective turnover of the epidermis and muscle. Embedded in the epidermis and surrounded by the muscle are the excretory units or protonephridia that regulate osmotic pressure. We did not see a depletion of protonephridial transcripts, although an abrupted protonephridial branching pattern was observed. Here, the defect at protonephridia seems like a defect caused either by an epidermal irregularity or due to an indirect effect of defective muscle cells. Several intestinal specialized neoblast markers were downregulated among pabp2 RNAi animals. Our transcriptome shows an enrichment of zfp1 (Figure 3E; Supplementary Tables S2, S3). zfp1 expression is observed among intestinal neoblast expressing markers for endocytes (King et al., 2024). This could be explained as an accumulation of intrinsically defective intestinal specialized neoblasts or a defective differentiation process. Together, our study demonstrates that PABP2 is a critical regulator for stem cell differentiation toward multiple cell lineages. A detailed analysis of the impact of pabp2 RNAi on other cell types is required, prioritizing intestinal lineages and cathepsin+ subtypes expressing cela 1 and jag 1.
The current study provides insights into the role of PABP2 in regulating stem cell differentiation in planaria. However, it is necessary to delineate the molecular mechanism through which multifunctional PABP2 elicits its effects during regeneration. Developing PABP2 antibodies specific to S. meditteranea would be necessary to understand the direct interacting partners. This would enable the delineation of the mechanistic role of PABP2 in stem cell functioning. Studies have shown that mitochondrial dynamics regulate stem cell self-renewal and differentiation (Khacho and Slack, 2017; Khacho et al., 2016; Seo et al., 2018). We observed the downregulation of mitochondrial potential among pabp2 RNAi animals (Supplementary Figure S6), suggesting a possibility that PABP2 modulates mitochondrial transcripts specific for stem cell fate determination. It is possible that PABP2 could play an inevitable role in regulating the mitochondrial transcripts necessary for stem cell differentiation. Considering the phenotype, which include lesions and subsequent lysis of animals, another aspect that requires attention is the investigation of the effect of pabp2 RNAi on apoptosis.
Conventionally, PABP2 is believed to be ubiquitously involved in active transcription. A number of studies have analyzed its multifaceted functions, including transcriptional regulation (Bear et al., 2003), alternative polyadenylation (De Klerk et al., 2012; Jenal et al., 2012; Martine, 2012), and the hyperadenylation and decay of RNA (Bresson et al., 2013). Research on xenopus, mouse, and MEL (mouse erythroleukemia) cells have reported the role of poly A binding proteins in the regulation of maternal transcript processing and erythroid differentiation, thereby extrapolating its function to stem cell regulation (Cragle et al., 2019; Dai et al., 2022; Kini et al., 2014). Several molecules requisite for neoblast differentiation have been identified among S. meditteranea: mex-3, CHD-4, apob-1, apob-2, egfr-1, nrg-1, p-66, and cbp-3 (Scimone et al., 2010; Wong et al., 2022; Zhu et al., 2015; Vásquez-Doorman and Petersen, 2016; Barberán et al., 2016; Fraguas et al., 2021). Our study demonstrates the requirement for PABP2 during stem cell differentiation in planaria.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The manuscript presents research on animals that did not require ethical approval for their study.
Author contributions
NM: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing–original draft, and writing–review and editing. NH: data curation, formal analysis, and writing–review and editing. VS: methodology and writing–review and editing. VL: data curation, formal analysis, and writing–review and editing. DP: conceptualization, funding acquisition, investigation, project administration, resources, supervision, and writing–review and editing. CJ: funding acquisition, investigation, supervision, writing–original draft, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. NM thanks the Institute for Stem Cell Science and Regenerative Medicine (inStem) for PhD fellowship funding. NH was supported by inStem core grant. VL was supported by the Council of Scientific and Industrial Research (CSIR)- SRF. VS was funded by the Stowers Institute for Medical Research. This project was funded by an inStem core grant and Department of Science and Technology (DST) -Swarnajayanti fellowship (DST/SJE/LSA-02/2015–16) to DP. Work in the CJ lab is supported by core funds from inStem and grants from the Government of Biotechnology of the Government of India (BT/PR8738/AGR/36/770/2013, BT/PR32539/BRB/10/1814/2019).
Acknowledgments
The authors are grateful for the immense technical support from the Central Imaging and Flow cytometry Facility (CIFF) and Sequencing Facility (special thanks to Awadhesh Pandit) at the BLiSc Bio-Cluster. They thank the Phil Newmark laboratory for their kind gift of Schmidtea mediterranea (S2F2-sexual strain). They thank Sanchez’s and Agata’s laboratories for their generous gifts of tmus and VC1 antibodies. They also acknowledge the support from Thermo Scientific for developing WI1 antibody. They thank Mohamed Haroon for help with flow cytometry analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1433142/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Related to Figure 1: (A) Phylogenetic tree showing the relatedness of PABP2 to other PABPs across different species. The PABP2 gene cluster with its homologs. Each node metadata represents bootstrap support (%). (B) Domain organization of PABP2 protein predicted using the conserved domain database (CDD). PABP2 protein has a single RNA recognition motif (RRM). (C) Multiple sequence alignment of PABP2 with PABPs of other model organisms.
SUPPLEMENTARY FIGURE S2 | Related to Figure 2: (A) Blastema-size quantification from days 1–7 post-amputation (n = 6). (B) Agarose gel electrophoresis demonstrating pabp2 RNAi efficacy analyzed by PCR amplification. Actin was used as a loading control. (C) Timeline showing the injection regime where dsRNA was injected during the homeostasis experiment. (D) Homeostasis knockdown animals with lesions at the anterior region (n = 7), scale bar, 200 μm. Images were taken on the day 28 from the beginning of the dsRNA regimen. (E) Quantification of peripheral muscle fiber thickness (three frames from a single worm where 10 fibers were measured from each of the frames. Total fibers measured from a single worm = 30, (n = 5), ***P < 0.001. (F) Quantification of intensity of anti-acetylated tubulin staining for protonephridia, (n = 6), ***P < 0.001.
SUPPLEMENTARY FIGURE S3 | Related to Figure 3: (A) Confocal images of fluorescent in-situ hybridization at the regenerating surface for wi1 (scale bar, 100 μm) and the zoomed images (scale bar, 20 μm). (B) Quantification of the percentage of wi1+ cells, (n = 5), ***P < 0.001. (C) Maximum intensity projections of confocal images showing H3PS10+ cells at 2 dpa and 7 dpa among control RNAi and pabp2 RNAi, scale bar, 50 μm.
SUPPLEMENTARY FIGURE S4 | Related to Figure 4: (A) Confocal images of entire animal showing distribution of the stem cell population (wi1-green) differentiating cell population (WI1- red) and immediate early progenitors (merged-yellow) (n = 6), scale bar, 100 μm.
SUPPLEMENTARY FIGURE S5 | Related to Figure 5: Bar plot depicting gene expression variations. Transcripts with variations among (A) cathepsin+ cells, (B) pharynx, (C) muscle, (D) parapharyngeal, (E) neural, (F) ciliated neurons, and (G) non-ciliated neurons compared with pabp2 RNAi and control RNAi animals during time points (as denoted by color bars within the plots) among anterior and posterior regenerating blastema. The asterisk indicates statistical significance (q < 0.05), and the dotted line on y-axis represents log2FC −0.58. Comparison is between pabp2 RNAi versus control RNAi in each of the conditions denoted by the bar color within the plots. All cell-type specific transcripts with q < 0.05 in at least one of the four comparisons are included in the graph. Details about fold change and normalized read counts are provided in Supplementary Table S3.
SUPPLEMENTARY FIGURE S6 | FACS analysis demonstrating reduced mitochondrial potential in pabp2 RNAi. (A, B) Scatter plot from FACS analysis of control RNAi macerates stained with R123 (A) and control RNAi stained with R123, followed by FCCP treatment (B) (n = 30). (C, D) Scatter plot from FACS analysis of pabp2 RNAi macerates stained with R123 (C) and pabp2 RNAi macerates stained with R123, followed by FCCP treatment (D) (n = 30). (E, F, G) Graphs represent difference in mitochondrial potential among X1 (E), X2 (F), and Xins populations (G) compared with various cell populations across control and pabp2 RNAi. The error bar is drawn from biological triplicates. *P < 0.05.
SUPPLEMENTARY TABLE S1 | Related to Figure 3: Expression values of common neoblast markers from RNA sequencing analysis at 0 dpa and 3 dpa anterior and posterior regenerating blastema.
SUPPLEMENTARY TABLE S2 | Related to Figure 3: Expression values of specialized neoblast markers from RNA sequencing analysis.
SUPPLEMENTARY TABLE S3 | Related to Figure 5: Expression values of differentiation markers from RNA sequencing analysis.
References
Agata, K., Soejima, Y., Kato, K., Kobayashi, C., Umesono, Y., and Watanabe, K. (1998). Structure of the planarian central nervous system (CNS) revealed by neuronal cell markers. Zoological Sci. 15, 433–440. doi:10.2108/zsj.15.433
Apponi, L. H., Corbett, A. H., and Pavlath, G. K. (2013). Control of mRNA stability contributes to low levels of nuclear poly(A) binding protein 1 (PABPN1) in skeletal muscle. Skelet. Muscle 3, 23. doi:10.1186/2044-5040-3-23
Bansal, D., Kulkarni, J., Nadahalli, K., Lakshmanan, V., Krishna, S., Sasidharan, V., et al. (2017). Cytoplasmic poly (A)-binding protein critically regulates epidermal maintenance and turnover in the planarian Schmidtea mediterranea. Development 144, 3066–3079. doi:10.1242/dev.152942
Barberán, S., Fraguas, S., and Cebrià, F. (2016). The EGFR signaling pathway controls gut progenitor differentiation during planarian regeneration and homeostasis. Development 143, 2089–2102. doi:10.1242/dev.131995
Bear, D. G., Fomproix, N., Soop, T., Bjorkroth, B., Masich, S., and Daneholt, B. (2003). Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp. Cell Res. 286, 332–344. doi:10.1016/S0014-4827(03)00123-X
Beaulieu, Y. B., Kleinman, C. L., Landry-Voyer, A. M., Majewski, J., and Bachand, F. (2012). Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-Binding protein nuclear 1. PLoS Genet. 8, e1003078. doi:10.1371/journal.pgen.1003078
Brais, B., Bouchard, J., Xie, Y., Rochefort, D., Chretien, N., Tome, F., et al. (1998). Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 18 (2), 164–167. doi:10.1038/ng0298-164
Brais, B., Rouleau, G. A., Bouchard, J. P., Fardeau, M., and Tome, F. M. S. (1999). Oculopharyngeal muscular dystrophy. Seminars neurology 19, 59–66. doi:10.1055/s-2008-1040826
Bresson, S. M., and Conrad, N. K. (2013). The human nuclear poly(A)-Binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 9, e1003893. doi:10.1371/journal.pgen.1003893
Buchholz, F., Kittler, R., Slabicki, M., and Theis, M. (2006). Enzymatically prepared RNAi libraries. Nat. Methods 3, 696–700. doi:10.1038/NMETH912
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. doi:10.1038/nbt.4096
Cebrià, F., Adell, T., and Saló, E. (2018). Rebuilding a planarian: from early signaling to final shape. Int. J. Dev. Biol. 62, 537–550. doi:10.1387/ijdb.180042es
Cebrià, F., Vispo, M., Newmark, P., Bueno, D., and Romero, R. (1997). Myocyte differentiation and body wall muscle regeneration in the planarian Girardia tigrina. Dev. Genes Evol. 207, 306–316. doi:10.1007/s004270050118
Cote, L. E., Simental, E., and Reddien, P. W. (2019). Muscle functions as a connective tissue and source of extracellular matrix in planarians. Nat. Commun. 10, 1592. doi:10.1038/s41467-019-09539-6
Cragle, C. E., MacNicol, M., Byrum, S., Hardy, L., Mackintosh, S., Richardson, W., et al. (2019). Musashi interaction with poly(A)-binding protein is required for activation of target mRNA translation. J. Biol. Chem. 294, 10969–10986. doi:10.1074/jbc.RA119.007220
Dai, X., Pi, S., Zhao, L., Wu, Y., Shen, J., Zhang, S., et al. (2022). PABPN1 functions as a hub in the assembly of nuclear poly(A) domains that are essential for mouse oocyte development. Sci. Adv. 8 (43), eabn9016. doi:10.1126/sciadv.abn9016
De Klerk, E., Venema, A., Anvar, S., Goeman, J., Hu, O., Trollet, C., et al. (2012). Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 40, 9089–9101. doi:10.1093/nar/gks655
Fincher, C. T., Wurtzel, O., de Hoog, T., Kravarik, K. M., and Reddien, P. W. (2018). Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, eaaq1736. doi:10.1126/science.aaq1736
Fraguas, S., Carcel, S., Vivancos, C., Molina, M., Gines, J., Mazariegos, J., et al. (2021). CREB-binding protein (CBP) gene family regulates planarian survival and stem cell differentiation. Dev. Biol. 476, 53–67. doi:10.1016/j.ydbio.2021.02.008
Guo, T., Peters, AHFM, and Newmark, P. A. (2006). A bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159–169. doi:10.1016/j.devcel.2006.06.004
Jenal, M., Elkon, R., Loayza-Puch, F., Haaften, V., Kuhn, U., Menzies, F., et al. (2012). The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553. doi:10.1016/j.cell.2012.03.022
Khacho, M., Clark, A., Svoboda, D., Azzi, J., MacLaurin, J., Meghaizel, C., et al. (2016). Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell 19, 232–247. doi:10.1016/j.stem.2016.04.015
Khacho, M., and Slack, R. S. (2017). Mitochondrial activity in the regulation of stem cell self-renewal and differentiation. Curr. Opin. Cell Biol. 49, 1–8. doi:10.1016/j.ceb.2017.11.003
King, H. O., Owusu-Boaitey, K. E., Fincher, C. T., and Reddien, P. W. (2024). A transcription factor atlas of stem cell fate in planarians. Cell Rep. 43 (3), 113843. doi:10.1016/j.celrep.2024.113843
King, R. S., and Newmark, P. A. (2013). In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol. 13, 8. doi:10.1186/1471-213X-13-8
Kini, H. K., Kong, J., and Liebhaber, S. A. (2014). Cytoplasmic poly(A) binding protein C4 serves a critical role in erythroid differentiation. Mol. Cell Biol. 34, 1300–1309. doi:10.1128/mcb.01683-13
Krishna, S., Palakodeti, D., and Solana, J. (2019). Post-transcriptional regulation in planarian stem cells. Semin. Cell Dev. Biol. 87, 69–78. doi:10.1016/j.semcdb.2018.05.013
Lakshmanan, V., Bansal, D., Kulkarni, J., Poduval, D., Krishna, S., Sasidharan, V., et al. (2016). Genome-wide analysis of polyadenylation events in Schmidtea mediterranea. G3 Genes, Genomes, Genet. 6, 3035–3048. doi:10.1534/g3.116.031120
Lei, K., Thi-Kim, Vu H., Mohan, R. D., McKinney, S. A., Seidel, C. W., Alexander, R., et al. (2016). Egf signaling directs neoblast repopulation by regulating asymmetric cell division in planarians. Dev. Cell 176, 413–429. doi:10.1016/j.devcel.2016.07.012
Lucila, S. M., Wurtzel, O., Malecek, K., Fincher, C., Oderberg, I., Kravarik, K., et al. (2018). foxF-1 controls specification of non-body wall muscle and phagocytic cells in planarians. Curr. Biol. 28, 3787–3801. doi:10.1016/j.cub.2018.10.030
Macrae, E. K. (1964). Observations on the fine structure of photoreceptor cells in the planarian dugesia tigrina. J. Ultrastruct. Res. 10, 334–349. doi:10.1016/s0022-5320(64)80013-7
Martine, S. (2012). PABPN1 shuts down alternative poly(A) sites. Cell Res. 22, 1419–1421. doi:10.1038/cr.2012.86
Mohamed, H. M., Lakshmanan, V., Sarkar, S., Lei, K., Vemula, P., and Palakodeti, D. (2021). Mitochondrial state determines functionally divergent stem cell population in planaria. Stem Cell Rep. 16, 1302–1316. doi:10.1016/j.stemcr.2021.03.022
Pearson, B. J., Eisenhoffer, G., Gurley, K., Rink, J., Miller, D., and Alvarado, A. S. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443–450. doi:10.1002/dvdy.21849
Raz, A. A., Wurtzel, O., and Reddien, P. W. (2021a). Planarian stem cells specify fate yet retain potency during the cell cycle. Cell Stem Cell 28, 1307–1322.e5. doi:10.1016/j.stem.2021.03.021
Rink, J. C., Vu, H. T. K., and Alvarado, A. S. (2011). The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769–3780. doi:10.1242/dev.066852
Rouhana, L., Shibata, N., Nishimura, O., and Agata, K. (2010). Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 341, 429–443. doi:10.1016/j.ydbio.2010.02.037
Rozanski, A., Moon, H., Brandl, H., Martin-Duran, J., Grohme, M., Huttner, K., et al. (2019). PlanMine 3.0 - improvements to a mineable resource of flatworm biology and biodiversity. Nucleic Acids Res. 47, D812-D820–D820. doi:10.1093/nar/gky1070
Scimone, M. L., Cote, L. E., and Reddien, P. W. (2017). Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 551 (551), 623–628. doi:10.1038/nature24660
Scimone, M. L., Kravarik, K. M., Lapan, S. W., and Reddien, P. W. (2014). Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep. 3, 339–352. doi:10.1016/j.stemcr.2014.06.001
Scimone, M. L., Meisel, J., and Reddien, P. W. (2010). The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development 137, 1231–1241. doi:10.1242/dev.042051
Seo, B. J., Yoon, S. H., and Do, J. T. (2018). Mitochondrial dynamics in stem cells and differentiation. Int. J. Mol. Sci. 19 (12), 3893. doi:10.3390/ijms19123893
Shettigar, N., Joshi, A., Dalmeida, R., Gopalkrishna, R., Chakravarthy, A., Patnaik, S., et al. (2017). Hierarchies in light sensing and dynamic interactions between ocular and extraocular sensory networks in a flatworm. Sci. Adv. 3 (7), e1603025. doi:10.1126/sciadv.1603025
Shibata, N., and Agata, K. (2018) Chapter 18 RNA interference in planarians: feeding and injection of synthetic dsRNA. 1774, 455–466. doi:10.1007/978-1-4939-7802-1
Storey, E. C., Holt, I., Morris, G. E., and Fuller, H. R. (2020). Muscle cell differentiation and development pathway defects in Emery-Dreifuss muscular dystrophy. Neuromuscul. Disord. 30, 443–456. doi:10.1016/j.nmd.2020.04.002
Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W., et al. (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902. doi:10.1016/j.cell.2019.05.031
Tangye, S. G., Nguyen, T., Deenick, E. K., Bryant, V. L., and Ma, C. S. (2023). Inborn errors of human B cell development, differentiation, and function. J. Exp. Med. 220, e20221105–e20221107. doi:10.1084/jem.20221105
Tu, K. C., Cheng, L., Tk Vu, H., Lange, J., Mckinney, S., Seidel, C., et al. (2015). Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife 4, 10501. doi:10.7554/elife.10501
Vásquez-Doorman, C., and Petersen, C. P. (2016). The NuRD complex component p66 suppresses photoreceptor neuron regeneration in planarians. Regeneration 3, 168–178. doi:10.1002/reg2.58
Wagner, D. E., Wang, I. E., and Reddien, P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 332, 811–816. doi:10.1126/science.1203983
Wang, Y., Stary, J. M., Wilhelm, J. E., and Newmark, P. A. (2010). A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes & Dev. 24, 2081–2092. doi:10.1101/gad.1951010
Wenemoser, D., and Reddien, P. W. (2010). Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 344, 979–991. doi:10.1016/j.ydbio.2010.06.017
Witchley, J. N., Mayer, M., Wagner, D. E., Owen, J. H., and Reddien, P. W. (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633–641. doi:10.1016/j.celrep.2013.07.022
Wolfswinkel, V., Wagner, P. R., and Reddien, P. (2014). Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 23, 1–7. doi:10.1016/j.stem.2014.06.007
Wong, L. L., Bruxvoort, C., Cejda, N., Delaney, M., Otero, J., and Forsthoefel, D. (2022). Intestine-enriched apolipoprotein b orthologs are required for stem cell progeny differentiation and regeneration in planarians. Nat. Commun. 13 (1), 3803. doi:10.1038/s41467-022-31385-2
Zeng, A., Li, H., Guo, L., Gao, X., McKinney, S., Wang, Y., et al. (2018). Prospectively isolated tetraspanin + neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173, 1593–1608. doi:10.1016/j.cell.2018.05.006
Keywords: stem cell, differentiation, poly A binding protein, regeneration, post transcriptional regulation, planaria
Citation: Mukundan N, Hariharan N, Sasidharan V, Lakshmanan V, Palakodeti D and Jamora C (2024) Poly (A) binding protein 2 is critical for stem cell differentiation during regeneration in the planarian Schmidtea mediterranea. Front. Cell Dev. Biol. 12:1433142. doi: 10.3389/fcell.2024.1433142
Received: 15 May 2024; Accepted: 21 August 2024;
Published: 23 September 2024.
Edited by:
Prasad Pethe, Symbiosis International University, IndiaReviewed by:
Kshitiz Raj Shrestha, Independent Researcher, NepalRicardo M. Zayas, San Diego State University, United States
Chen Guangwen, Henan Normal University, China
Copyright © 2024 Mukundan, Hariharan, Sasidharan, Lakshmanan, Palakodeti and Jamora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Colin Jamora, Y29saW4uamFtb3JhQHNudS5lZHUuaW4=; Namita Mukundan, bmFtaXRhbUBpbnN0ZW0ucmVzLmlu
 Namita Mukundan
Namita Mukundan Nivedita Hariharan1
Nivedita Hariharan1 Vairavan Lakshmanan
Vairavan Lakshmanan Dasaradhi Palakodeti
Dasaradhi Palakodeti