
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 26 July 2024
Sec. Cell Adhesion and Migration
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1431690
This article is part of the Research Topic Tumor Cell Mechanosensitivity: Molecular Basis View all 9 articles
 Yukari Fujimoto1
Yukari Fujimoto1 Naotaka Nakazawa2*
Naotaka Nakazawa2*The cell has multiple mechanisms for sensing and responding to dynamic changes in the mechanical environment. In the process, intracellular signaling is activated to modulate gene expression. Recent studies have shown that multifunctional signaling molecules that link intracellular force and gene expression are important for understanding cellular functions in the mechanical environment. This review discusses recent studies on one of the mechanotransducers, Four-and-a-half LIM domains 2 (FHL2), which localizes to focal adhesions (FAs), actin cytoskeleton, and nucleus. FHL2 localizes to FAs and the actin cytoskeleton in the cell on stiff substrate. In this situation, intracellular tension of F-actin by Myosin II is critical for FHL2 localization to FAs and actin stress fibers. In the case, a conserved phenylalanine in each LIM domain is responsible for its localization to F-actin. On the other hand, lower tension of F-actin in the cell on a soft substrate causes FHL2 to be released into the cytoplasm, resulting in its localization in the nucleus. At the molecular level, phosphorylation of specific tyrosine in FHL2 by FAK, non-receptor tyrosine kinase, is critical to nuclear localization. Finally, by binding to transcription factors, FHL2 modulates gene expression for cell proliferation as a transcriptional co-factor. Thus, FHL2 is involved in mechano-sensing and -transduction in the cell in a mechanical environment.
Mechanical environments, such as substrate rigidity, substrate topography, and spatial confinement, are signals for the intracellular pathways in cell growth and differentiation (Vogel and Sheetz, 2006; Vining and Mooney, 2017; Sheetz, 2019; Chaudhuri et al., 2020). Intracellular signaling pathways activate the expression of downstream genes, which modulate cellular responses. Thus, intracellular signaling activated by the mechanical environment is critical to understanding how the mechanical environment leads to gene expression through mechanosensing mechanisms.
Four-and-a-half-LIM domain 2 (FHL2) is a LIM domain (domain discovered in the proteins, Lin11, Isl-1, and Mec-3) family protein that contains a specific structure with a tandem zinc-finger motif (Kadrmas and Beckerle, 2004; Anderson et al., 2021). Although a general zinc-finger motif binds directly to DNA, the LIM domain mediates protein-protein interaction. Previous studies have suggested that FHL2 mediates intracellular signaling under the control of cytoskeletal regulators (Müller et al., 2002; Schiller et al., 2011). Importantly, FHL2 shuttles between the cytoplasm and the nucleus in this process. Previous studies have reported that a transcriptional co-factor, such as YAP/TAZ and MRTF/MAL, works as a mechanotransducer by shuttling between the cytoplasm and the nucleus (Miralles et al., 2003; Dupont et al., 2011; Er et al., 2022). FHL2 is the first molecule reported as an adhesion protein that translocates from the adhesion site to the nucleus in response to changes in extracellular matrix stiffness and intracellular contractility (Nakazawa et al., 2016). Regarding the relationship between FHL2 and cancer phenotypes, the expression level of the FHL2 gene is upregulated in metastatic cell lines (Kleiber et al., 2007; Zhang et al., 2023). In addition, previous studies have reported that ectopic expression of FHL2 gene correlates with a pathological phenotype of tumor progression and growth in human patients, mice, and rats (Ding et al., 2009; Hua et al., 2016; Jin et al., 2016; Cai et al., 2018). This suggests that understanding the function of FHL2 may be potentially important for cancer therapy. Taken together, the function of FHL2 as a mechanotransducer appears to be important for understanding the links between mechanical effects on adhesion complexes and gene expression in the cancer cell, which may contribute to new insights for cancer therapy. Here, we discuss recent work on FHL2 functions as a mechanotransducer for cell proliferation as it relates to cancer cell dynamics in a mechanical environment.
FHL2 at FAs binds directly to focal adhesion components, including several integrins and FA kinase (FAK) (Wixler et al., 2000; Gabriel et al., 2004; Samson et al., 2004). This suggests that FHL2 is a potential scaffolding protein at FAs. A previous study indicates that the cell without FHL2 shows impairment of FA maturation (Park et al., 2008). However, this effect is due to the lack of ECM proteins through the modulation of gene expression (Park et al., 2008). Since knocking out of the FHL2 gene affects the expression of downstream genes, it is still unclear whether FHL2 itself contributes to adhesion assembly.
To distinguish the functions of FHL2 as a scaffolding component and a transcriptional co-factor, it might be necessary to elucidate the function of each FHL2 domain on FA and to perform rescue experiments with each domain in FHL2 in a knockout condition. Another way to identify the functions of FHL2 in FA may be to combine perturbation for FHL2 function and optogenetic tools such as chromophore-assisted light inactivation (CALI) (Liao et al., 1994; Ryu et al., 2021; Takemoto, 2021). Thus, it remains unclear how FHL2 contributes to FA maturation, but recruitment of FHL2 to FAs is Myosin II activity-dependent (Kuo et al., 2011; Wolfenson et al., 2016). Since conserved mechanisms of the LIM domains facilitate its localization to the actin filament, FHL2 recruitment may occur at the end of FA maturation (Sun et al., 2020; Winkelman et al., 2020). Although FHL2 silencing reduces cancer cell migration and invasion, there is no direct evidence that FHL2 contributes to cell migration and invasion as an adhesion component (Brun et al., 2013; Hua et al., 2016; Wang et al., 2020; Jiao et al., 2022). However, distinguishing the function of FHL2 may be important when considering FHL2-targeted cancer therapy, as a strategy of cancer therapies generally depends on the stage of cancer, in which case FHL2 may have different functions at different cancer stages (Umar et al., 2012; Klein, 2020).
As mentioned above, FHL2 localization at FAs is dependent on Myosin II activity (Kuo et al., 2011). This suggests that intracellular tension of F-actin is critical for FHL2 localization to FAs. A rigid substrate generally facilitates FA maturation through tense actin stress fibers. Our previous study demonstrated that FHL2 accumulates at FAs in the cell on the rigid substrate (Figure 1) (Nakazawa et al., 2016). Conversely, soft substrate facilitates the release of FHL2 into the cytoplasm, resulting in its localization to the nucleus (Figure 1). Since cells with perturbation of Myosin II activity of actin polymerization show nuclear localization of FHL2, cell contraction is critical for FHL2 localization. Our study found that cyclic stretch can rescue FHL2 nuclear localization by the soft substrate (Cui et al., 2015; Nakazawa et al., 2016). In this case, intracellular tension is important for FHL2 localization in the nucleus rather than the disassembly of FA.
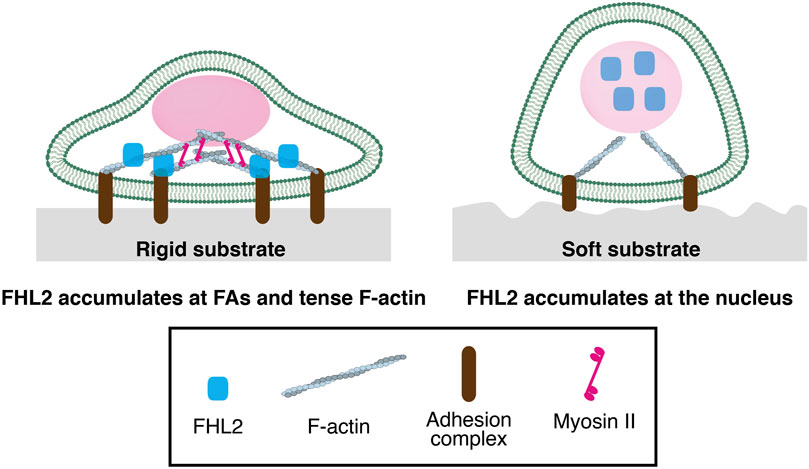
Figure 1. The functions of FHL2 as a mechanotransducer in the cell on a rigid or soft substrate (Left) FHL2 localizes at Focal adhesion or tense F-actin in the cell on rigid substrate (Right) FHL2 localizes to the nucleus on soft substrate, resulting in its binding to a specific promoter of the gene with transcriptional factor for cell proliferation.
FA kinase (FAK) is a non-receptor tyrosine kinase that plays critical role in the maturation of FA (Dawson et al., 2021; Le Coq et al., 2022). Previous studies have confirmed the biochemical interaction between FAK and FHL2 (Gabriel et al., 2004; Nakazawa et al., 2016). FAK phosphorylates Y93 in FHL2, which facilitates the nuclear localization of FHL2 dependent on substrate rigidity or intracellular tension. Other tyrosines, with the exception of Y93, are phosphorylated by another non-receptor tyrosine kinase, c-Abl. Therefore, phosphorylation of Y93 in FHL2 by FAK appears to be specifically dependent on intracellular force and substrate rigidity (Wang G.-F. et al., 2021).
Following its shuttling to the nucleus, FHL2 co-localizes with the active RNA polymerase (Pol) II on the promoter sequence of p21/CDKN1A gene. Since p21/CDKN1A is a negative regulator of cell proliferation, thus FHL2 modulates cell proliferation through p21/CDKN1A gene expression in a force- or substrate-rigidity-dependent manner (Nakazawa et al., 2016). However, the molecular mechanisms of FHL2 nuclear transport remain unclear so far. Since FHL2 is a small protein (32.2 kDa), active transport to the nucleus might not be required. One possibility is that phosphorylation of FHL2 by FAK might enhance its concentration at the active RNA pol II site. Recent studies on phase separation of the molecules suggest that protein modification, including phosphorylation, impacts transcriptional machinery (Hnisz et al., 2017; Boija et al., 2018; Sabari et al., 2018). It is notable that LIMD1, another LIM domain protein, has been reported to contribute to FA matulation through phase separation (Wang Y. et al., 2021). Further research is required to elucidate the effect of chemical modification in FHL2 on its accumulation at the specific region in the nucleus.
Previous studies have reported that FHL2 localizes to F-actin. A recent study observed that some LIM domain family proteins, including FHL2, HIC5, and Zyxin, are recruited to tense actin fibers (Sun et al., 2020). Surprisingly, direct binding to tense actin fibers depends on a conserved phenylalanine among these LIM domain proteins (Sun et al., 2020; Sun and Alushin, 2023). Furthermore, FHL2 with mutations at specific amino acid residues (F80A, F141A, F200A, F263A) accumulates at the nucleus in the cell on a rigid substrate (Sun et al., 2020). These findings indicate that FHL2 functions as a mechanosensor of actin fibers, not only as a mechanotransducer.
In this review, we summarized the functions of FHL2 as a mechanotransducer and a mechanosensor in the cell in a mechanical environment. FHL2 localizes to FA in the cell on a rigid substrate dependent on intracellular tension through F-actin. At the same time, FHL2 recruitment is facilitated to tense F-actin. FHL2 is released from FAs and F-actin when intracellular tension becomes low on a soft substrate, which leads to shuttling to the nucleus. Thus, FHL2 works as a transcriptional co-factor in the nucleus to modulate gene expression for cell proliferation in a tension-dependent manner.
To understand how FHL2 mediates mechanical signaling, the overall picture of proteins interacting with FHL2 still needs to be identified (Tran et al., 2016). Since binding partners of FHL2 may be different at several locations in the cell on soft/rigid substrates, proteome analysis focusing on the binding partners of FHL2 in the cell on different substrate rigidity might be important to understand how FHL2 functions as a multifunctional protein in a force-dependent manner. In this sense, a technique to check a history of protein-protein association, such as BioID, TurboID, and AirID, might be a useful method to identify the binding partner of FHL2 in different situations (Choi-Rhee et al., 2004; Roux et al., 2012; Branon et al., 2018; Kido et al., 2020). Previous studies using BioID focusing on components of FA have provided a new insight into the interaction between scaffolding proteins (Dong et al., 2016; Chastney et al., 2020; He et al., 2023). However, screening of proteins that interact with FHL2 in different mechanical environments has yet to be performed.
As mentioned, FHL2 in the nucleus modulates p21/CDKN1A gene expression for cell proliferation in a force- or substrate rigidity-dependent manner (Nakazawa et al., 2016). Although some FHL2 target genes are reported, other downstream genes that are regulated by the mechanical environment in FHL2 dependent manner have not been explored (Morlon and Sassone-Corsi, 2003; Martin et al., 2007; Labalette et al., 2008; Wong et al., 2012; Ng et al., 2014; Dahan et al., 2017). Comparison of transcriptome analysis with and without FHL2 in different mechanical environments will offer further understanding of FHL2 functions for cancer metastasis from a mechanobiological viewpoint.
YF: Writing–original draft, Writing–review and editing. NN: Conceptualization, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by KAKENHI from JSPS (Grant numbers 18K14819, 18KK0212, 21H05126 to NN) and FOREST from JST (Grant number JPMJFR225J).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anderson, C. A., Kovar, D. R., Gardel, M. L., and Winkelman, J. D. (2021). LIM domain proteins in cell mechanobiology. Cytoskeleton 78, 303–311. doi:10.1002/cm.21677
Boija, A., Klein, I. A., Sabari, B. R., Dall’Agnese, A., Coffey, E. L., Zamudio, A. V., et al. (2018). Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855. doi:10.1016/j.cell.2018.10.042
Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., et al. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887. doi:10.1038/nbt.4201
Brun, J., Dieudonné, F.-X., Marty, C., Müller, J., Schüle, R., Patiño-García, A., et al. (2013). FHL2 silencing reduces Wnt signaling and osteosarcoma tumorigenesis in vitro and in vivo. PLoS One 8, e55034. doi:10.1371/journal.pone.0055034
Cai, T., Sun, D., Duan, Y., Qiu, Y., Dai, C., Yang, J., et al. (2018). FHL2 promotes tubular epithelial-to-mesenchymal transition through modulating β-catenin signalling. J. Cell. Mol. Med. 22, 1684–1695. doi:10.1111/jcmm.13446
Chastney, M. R., Lawless, C., Humphries, J. D., Warwood, S., Jones, M. C., Knight, D., et al. (2020). Topological features of integrin adhesion complexes revealed by multiplexed proximity biotinylation. J. Cell Biol. 219, e202003038. doi:10.1083/jcb.202003038
Chaudhuri, O., Cooper-White, J., Janmey, P. A., Mooney, D. J., and Shenoy, V. B. (2020). Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 584, 535–546. doi:10.1038/s41586-020-2612-2
Choi-Rhee, E., Schulman, H., and Cronan, J. E. (2004). Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 13, 3043–3050. doi:10.1110/ps.04911804
Cui, Y., Hameed, F. M., Yang, B., Lee, K., Pan, C. Q., Park, S., et al. (2015). Cyclic stretching of soft substrates induces spreading and growth. Nat. Commun. 6, 6333. doi:10.1038/ncomms7333
Dahan, J., Levillayer, F., Xia, T., Nouët, Y., Werts, C., Fanton d’Andon, M., et al. (2017). LIM-only protein FHL2 is a negative regulator of transforming growth factor β1 expression. Mol. Cell. Biol. 37, e00636–16. doi:10.1128/MCB.00636-16
Dawson, J. C., Serrels, A., Stupack, D. G., Schlaepfer, D. D., and Frame, M. C. (2021). Targeting FAK in anticancer combination therapies. Nat. Rev. Cancer 21, 313–324. doi:10.1038/s41568-021-00340-6
Ding, L., Wang, Z., Yan, J., Yang, X., Liu, A., Qiu, W., et al. (2009). Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-beta-like signaling pathway. J. Clin. Invest. 119, 349–361. doi:10.1172/JCI35930
Dong, J.-M., Tay, F. P.-L., Swa, H. L.-F., Gunaratne, J., Leung, T., Burke, B., et al. (2016). Proximity biotinylation provides insight into the molecular composition of focal adhesions at the nanometer scale. Sci. Signal. 9, rs4. doi:10.1126/scisignal.aaf3572
Dupont, S., Morsut, L., Aragona, M., Enzo, E., Giulitti, S., Cordenonsi, M., et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183. doi:10.1038/nature10137
Er, E. E., Tello-Lafoz, M., and Huse, M. (2022). Mechanoregulation of metastasis beyond the matrix. Cancer Res. 82, 3409–3419. doi:10.1158/0008-5472.CAN-22-0419
Gabriel, B., Mildenberger, S., Weisser, C. W., Metzger, E., Gitsch, G., Schüle, R., et al. (2004). Focal adhesion kinase interacts with the transcriptional coactivator FHL2 and both are overexpressed in epithelial ovarian cancer. Anticancer Res. 24, 921–927.
He, Q., Sze, S. K., Ng, K. S., and Koh, C.-G. (2023). Paxillin interactome identified by SILAC and label-free approaches coupled to TurboID sheds light on the compositions of focal adhesions in mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 680, 73–85. doi:10.1016/j.bbrc.2023.09.017
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K., and Sharp, P. A. (2017). A phase separation model for transcriptional control. Cell 169, 13–23. doi:10.1016/j.cell.2017.02.007
Hua, G., He, C., Lv, X., Fan, L., Wang, C., Remmenga, S. W., et al. (2016). The four and a half LIM domains 2 (FHL2) regulates ovarian granulosa cell tumor progression via controlling AKT1 transcription. Cell Death Dis. 7, e2297. doi:10.1038/cddis.2016.207
Jiao, Y., Wei, J., Li, Z., Zhou, J., and Liu, Y. (2022). High FHL2 mRNA expression and its prognostic value in lung cancer. Aging 14, 7986–8000. doi:10.18632/aging.204328
Jin, H., Lee, K., Kim, Y.-H., Oh, H. K., Maeng, Y.-I., Kim, T.-H., et al. (2016). Scaffold protein FHL2 facilitates MDM2-mediated degradation of IER3 to regulate proliferation of cervical cancer cells. Oncogene 35, 5106–5118. doi:10.1038/onc.2016.54
Kadrmas, J. L., and Beckerle, M. C. (2004). The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5, 920–931. doi:10.1038/nrm1499
Kido, K., Yamanaka, S., Nakano, S., Motani, K., Shinohara, S., Nozawa, A., et al. (2020). AirID, a novel proximity biotinylation enzyme, for analysis of protein-protein interactions. Elife 9, e54983. doi:10.7554/eLife.54983
Kleiber, K., Strebhardt, K., and Martin, B. T. (2007). The biological relevance of FHL2 in tumour cells and its role as a putative cancer target. Anticancer Res. 27, 55–61.
Klein, C. A. (2020). Cancer progression and the invisible phase of metastatic colonization. Nat. Rev. Cancer 20, 681–694. doi:10.1038/s41568-020-00300-6
Kuo, J.-C., Han, X., Hsiao, C.-T., Yates, J. R., and Waterman, C. M. (2011). Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13, 383–393. doi:10.1038/ncb2216
Labalette, C., Nouët, Y., Levillayer, F., Armengol, C., Renard, C.-A., Soubigou, G., et al. (2008). The LIM-only protein FHL2 mediates ras-induced transformation through cyclin D1 and p53 pathways. PLoS One 3, e3761. doi:10.1371/journal.pone.0003761
Le Coq, J., Acebrón, I., Rodrigo Martin, B., López Navajas, P., and Lietha, D. (2022). New insights into FAK structure and function in focal adhesions. J. Cell Sci. 135, jcs259089. doi:10.1242/jcs.259089
Liao, J. C., Roider, J., and Jay, D. G. (1994). Chromophore-assisted laser inactivation of proteins is mediated by the photogeneration of free radicals. Proc. Natl. Acad. Sci. U. S. A. 91, 2659–2663. doi:10.1073/pnas.91.7.2659
Martin, B. T., Kleiber, K., Wixler, V., Raab, M., Zimmer, B., Kaufmann, M., et al. (2007). FHL2 regulates cell cycle-dependent and doxorubicin-induced p21Cip1/Waf1 expression in breast cancer cells. Cell Cycle 6, 1779–1788. doi:10.4161/cc.6.14.4448
Miralles, F., Posern, G., Zaromytidou, A.-I., and Treisman, R. (2003). Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113, 329–342. doi:10.1016/s0092-8674(03)00278-2
Morlon, A., and Sassone-Corsi, P. (2003). The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc. Natl. Acad. Sci. U. S. A. 100, 3977–3982. doi:10.1073/pnas.0735923100
Müller, J. M., Metzger, E., Greschik, H., Bosserhoff, A.-K., Mercep, L., Buettner, R., et al. (2002). The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J. 21, 736–748. doi:10.1093/emboj/21.4.736
Nakazawa, N., Sathe, A. R., Shivashankar, G. V., and Sheetz, M. P. (2016). Matrix mechanics controls FHL2 movement to the nucleus to activate p21 expression. Proc. Natl. Acad. Sci. U. S. A. 113, E6813–E6822. doi:10.1073/pnas.1608210113
Ng, C.-F., Xu, J.-Y., Li, M.-S., and Tsui, S. K.-W. (2014). Identification of FHL2-regulated genes in liver by microarray and bioinformatics analysis. J. Cell. Biochem. 115, 744–753. doi:10.1002/jcb.24714
Park, J., Will, C., Martin, B., Gullotti, L., Friedrichs, N., Buettner, R., et al. (2008). Deficiency in the LIM-only protein FHL2 impairs assembly of extracellular matrix proteins. FASEB J. 22, 2508–2520. doi:10.1096/fj.07-095521
Roux, K. J., Kim, D. I., Raida, M., and Burke, B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810. doi:10.1083/jcb.201112098
Ryu, K. A., Kaszuba, C. M., Bissonnette, N. B., Oslund, R. C., and Fadeyi, O. O. (2021). Interrogating biological systems using visible-light-powered catalysis. Nat. Rev. Chem. 5, 322–337. doi:10.1038/s41570-021-00265-6
Sabari, B. R., Dall’Agnese, A., Boija, A., Klein, I. A., Coffey, E. L., Shrinivas, K., et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. doi:10.1126/science.aar3958
Samson, T., Smyth, N., Janetzky, S., Wendler, O., Müller, J. M., Schüle, R., et al. (2004). The LIM-only proteins FHL2 and FHL3 interact with alpha- and beta-subunits of the muscle alpha7beta1 integrin receptor. J. Biol. Chem. 279, 28641–28652. doi:10.1074/jbc.M312894200
Schiller, H. B., Friedel, C. C., Boulegue, C., and Fässler, R. (2011). Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12, 259–266. doi:10.1038/embor.2011.5
Sheetz, M. (2019). A tale of two States: normal and transformed, with and without rigidity sensing. Annu. Rev. Cell Dev. Biol. 35, 169–190. doi:10.1146/annurev-cellbio-100818-125227
Sun, X., and Alushin, G. M. (2023). Cellular force-sensing through actin filaments. FEBS J. 290, 2576–2589. doi:10.1111/febs.16568
Sun, X., Phua, D. Y. Z., Axiotakis, L., Smith, M. A., Blankman, E., Gong, R., et al. (2020). Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev. Cell 55, 468–482. doi:10.1016/j.devcel.2020.09.022
Takemoto, K. (2021). Optical manipulation of molecular function by chromophore-assisted light inactivation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 97, 197–209. doi:10.2183/pjab.97.011
Tran, M. K., Kurakula, K., Koenis, D. S., and de Vries, C. J. M. (2016). Protein-protein interactions of the LIM-only protein FHL2 and functional implication of the interactions relevant in cardiovascular disease. Biochim. Biophys. Acta 1863, 219–228. doi:10.1016/j.bbamcr.2015.11.002
Umar, A., Dunn, B. K., and Greenwald, P. (2012). Future directions in cancer prevention. Nat. Rev. Cancer 12, 835–848. doi:10.1038/nrc3397
Vining, K. H., and Mooney, D. J. (2017). Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 18, 728–742. doi:10.1038/nrm.2017.108
Vogel, V., and Sheetz, M. (2006). Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275. doi:10.1038/nrm1890
Wang, C., Lv, X., He, C., Davis, J. S., Wang, C., and Hua, G. (2020). Four and a half LIM domains 2 (FHL2) contribute to the epithelial ovarian cancer carcinogenesis. Int. J. Mol. Sci. 21, 7751. doi:10.3390/ijms21207751
Wang, G.-F., Niu, X., Liu, H., Dong, Q., Yao, Y., Wang, D., et al. (2021). c-Abl kinase regulates cell proliferation and ionizing radiation-induced G2/M arrest via phosphorylation of FHL2. FEBS Open Bio 11, 1731–1738. doi:10.1002/2211-5463.13177
Wang, Y., Zhang, C., Yang, W., Shao, S., Xu, X., Sun, Y., et al. (2021). LIMD1 phase separation contributes to cellular mechanics and durotaxis by regulating focal adhesion dynamics in response to force. Dev. Cell 56, 1313–1325.e7. doi:10.1016/j.devcel.2021.04.002
Winkelman, J. D., Anderson, C. A., Suarez, C., Kovar, D. R., and Gardel, M. L. (2020). Evolutionarily diverse LIM domain-containing proteins bind stressed actin filaments through a conserved mechanism. Proc. Natl. Acad. Sci. U. S. A. 117, 25532–25542. doi:10.1073/pnas.2004656117
Wixler, V., Geerts, D., Laplantine, E., Westhoff, D., Smyth, N., Aumailley, M., et al. (2000). The LIM-only protein DRAL/FHL2 binds to the cytoplasmic domain of several alpha and beta integrin chains and is recruited to adhesion complexes. J. Biol. Chem. 275, 33669–33678. doi:10.1074/jbc.M002519200
Wolfenson, H., Meacci, G., Liu, S., Stachowiak, M. R., Iskratsch, T., Ghassemi, S., et al. (2016). Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat. Cell Biol. 18, 33–42. doi:10.1038/ncb3277
Wong, C.-H., Mak, G. W.-Y., Li, M.-S., and Tsui, S. K.-W. (2012). The LIM-only protein FHL2 regulates interleukin-6 expression through p38 MAPK mediated NF-κB pathway in muscle cells. Cytokine 59, 286–293. doi:10.1016/j.cyto.2012.04.044
Keywords: FHL2, substrate rigidity, LIM domain protein, gene expression, mechanotransduction
Citation: Fujimoto Y and Nakazawa N (2024) The roles of FHL2 as a mechanotransducer for cellular functions in the mechanical environment. Front. Cell Dev. Biol. 12:1431690. doi: 10.3389/fcell.2024.1431690
Received: 12 May 2024; Accepted: 03 July 2024;
Published: 26 July 2024.
Edited by:
Xian Hu, University of Oslo, NorwayReviewed by:
Mahesh Agarwal, University of California, Los Angeles, United StatesCopyright © 2024 Fujimoto and Nakazawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naotaka Nakazawa, bmFrYXphd2EubmFvdGFrYUBlbWF0LmtpbmRhaS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.