- 1Animal Genomics Laboratory, Animal Biotechnology Division, National Dairy Research Institute, Karnal, India
- 2ICAR- Central Institute of Research on Goat, Mathura, Uttar Pradesh, India
- 3Central Institute for Research on Buffaloes, Hisar, Haryana, India
Buffalo bulls are backbone of Indian dairy industry, and the quality of semen donating bulls determine the overall production efficiency of dairy farms. Seminal plasma harbor millions of lipid bilayer nanovesicles known as extracellular vesicles (EVs). These EVs carry a heterogenous cargo of essential biomolecules including fertility-associated proteins which contribute to fertilizing potential of spermatozoa. In this study, we explored size, concentration, and complete proteome profiles of SP EVs from two distinct fertility groups to uncover proteins influencing bull fertility. Through Dynamic Light Scattering (DLS) it was found that purified EVs were present in 7–14 size exclusion chromatographic (SEC) fractions with sizes ranging from 146.5 to 258.7 nm in high fertile (HF) and low fertile (LF) bulls. Nanoparticle Tracking Analysis (NTA) confirmed the size of seminal EVs up to 200 nm, and concentrations varying from 2.84 to 6.82 × 1011 and 3.57 to 7.74 × 1011 particles per ml in HF and LF bulls, respectively. No significant difference was observed in size and concentration of seminal EVs between two groups. We identified a total of 1,862 and 1,807 proteins in seminal EVs of HF and LF bulls, respectively using high throughput LC-MS/MS approach. Out of these total proteins, 1,754 proteins were common in both groups and about 87 proteins were highly abundant in HF group while 1,292 were less abundant as compared to LF bulls. Gene ontology (GO) analysis, revealed that highly abundant proteins in HF group were mainly part of the nucleus and involved in nucleosome assembly along with DNA binding. Additionally, highly abundant proteins in EVs of HF group were found to be involved in spermatogenesis, motility, acrosome reaction, capacitation, gamete fusion, and cryotolerance. Two highly abundant proteins, protein disulfide-isomerase A4 and gelsolin, are associated with sperm-oocyte fusion and acrosome reaction, respectively, and their immunolocalization on spermatozoa may indicate that these proteins are transferred through EVs. Our evidences support that proteins in EVs and subsequently their presence on sperm, are strongly associated with sperm functions. Altogether, our investigation indicates that SPEVs possess crucial protein repertoires that are essential for enhancing sperm fertilizing capacity.
1 Introduction
Spermatozoa are exquisitely crafted, motile gametes possessing the male’s genetic information which is transferred to the next-generation upon fertilization. In mammals, spermatozoa are suspended in seminal plasma (SP), which is a complex fluid consisting of bioactive molecules produced by secretions from the male reproductive tract, mainly from the epididymis and accessory sex glands (Juyena and Stelletta, 2012). SP accompanies spermatozoa from its maturation up to the time of its deposition in the female reproductive tract (FRT). It directly influences the sperms’ essential functions such as motility, capacitation, protection from oxidative stress, and modulation of the uterine immune response (Potts et al., 2000; Boccia et al., 2013; Rodríguez et al., 2013; Pereira et al., 2017). In addition to free bioactive molecules, some biomolecules including proteins, lipids, and nucleic acids in SP are encased in 30–350 nm lipid bilayer extracellular vesicles (EVs) which are delivered to the various target cells and hence, are essential for cell-to-cell communication (Zaborowski et al., 2015; Keerthikumar et al., 2016; Jeppesen et al., 2019).
Spermatozoa are hypothesized to be transcriptionally and translationally inactive and as a result, they carry pre-synthesized proteins as final products (Rahman et al., 2013; Sun et al., 2021). Consequently, EVs play a pivotal role in delivering the necessary protein cargo to meet the requirements of spermatozoa (Leahy et al., 2020). Most seminal EVs are secreted by the epididymis and prostate gland which are referred to as epididymosomes and prostasomes, respectively (Aalberts et al., 2012; Barrachina et al., 2022). Epididymosomes show different proteome profiles along the length of epididymis which lead to the maturation of spermatozoa, acquisition of motility, and the ability to bind with zona pellucida (Girouard et al., 2011; Rowlison et al., 2020). The ZP family proteins ZP3R and ZPBP2 in mouse epididymosomes have been shown to play a crucial role in zona-pellucida binding (Nixon et al., 2019). Seminal EV proteins in sheep have been found to correlate with vesicle biogenesis, metabolism, and membrane adhesion proteins which are essential for sperm fertilizing capacity (Leahy et al., 2020). In a study of seminal EVs from roosters of contrasting fertility, smaller EVs appeared to be more abundant in high fertile when compared to sub-fertile roosters (Cordeiro et al., 2021).
Seminal EVs are also known to enhance essential spermatozoal functions. In a previous report, exosomes from normal spermatozoa when co-incubated with spermatozoa from astenozoospermic men were able to enhance their progressive motility, capacitation, and acrosome reaction (Murdica et al., 2019). EVs with different densities are also known to differentially improve progressive motility and capacitation, among which the EVs with the highest density exhibited maximum improvement while the medium-density EVs were found to be enriched with antioxidant enzymes like GSTM2 (Wang et al., 2022). In a study on porcine semen, seminal EVs promoted acrosome reaction when they were co-incubated with spermatozoa (Siciliano et al., 2008). Prostasomes have also been reported to respond to progesterone and deliver Ca2+ under capacitating conditions (Aalberts et al., 2013). Additionally, EVs interact with endometrial stromal cells, promoting decidualization and prolactin secretion for implantation (Rodriguez-Caro et al., 2019). Additionally, seminal EVs from high fertile (HF) and low fertile (LF) animals display key differences in the type of cargo they carry. For example, cysteine-rich secretory protein 1 (CRISP1) was significantly less abundant in exosomes from astenozoospermic samples (Murdica et al., 2019). CRISP1 has been reported to regulate CatSper, the principal calcium channel in sperm that modulates sperm orientation and hyperactivation (Ernesto et al., 2015). Another protein Clusterin that plays a key role in capacitation and motility is less abundant in seminal EVs of non-normozoospermic men (Vickram et al., 2022). In another study, epididymal EVs from Teratospermic cats showed a significant deficiency in zona pellucida binding protein 1 which helps in species-specific sperm binding with oocyte when compared to normozoospermic cats (Rowlison et al., 2020). The sEVs not only transfer proteins but are also major carriers of miRNA, mRNA, piRNA, and snRNA. In a comprehensive study focusing on boar seminal EVs, the analysis revealed the presence of 288 known miRNAs, 37 novel miRNAs, and 19,749 piRNAs. Some of these mRNAs are known to modulate male reproductive physiology (Xu et al., 2020). Despite being one of the bodily fluids in which EVs were first identified nearly 50 years ago (Metz et al., 1968), SP has received less research focus in the context of EVs when compared to other bodily fluids (Royo et al., 2020). Research on the proteomic content of seminal extracellular vesicles (EVs) and its connection to buffalo bull fertility is a relatively unexplored field. To bridge this gap using a high throughput LC-MS/MS approach, the current study aims to identify differentially abundant proteins in the seminal EVs of Murrah buffalo bulls with distinct fertility and to elucidate their potential roles in fertilizing capacity.
2 Materials and methods
2.1 Chemicals and plastic wares
All chemicals and reagents used in this study were obtained from Sigma-Aldrich, India, ThermoFisher Scientific, and Invitrogen, United States, unless stated otherwise. Plasticwares were procured from Tarsons, India, and ThermoFisher Scientific, United States.
2.2 Selection of bulls and semen collection
Murrah bulls (Bubalus bubalis) were selected based on their conception rate (CR) data obtained from ABRC, NDRI, and the Central Institute for Research on Buffaloes (CIRB), Hisar. The buffalo bulls (n = 6) used in the study were categorized into two groups: high-fertile (HF, n = 3) with CR ranging from 38% to 48%, and low-fertile (LF, n = 3) with CR ranging from 6% to 34% (Supplementary Table S1) (Verma et al., 2014; Batra et al., 2020; Karanwal et al., 2023). All the bulls were maintained in the Artificial Breeding Research Centre (ABRC), NDRI. The semen samples were collected as per the minimum standard for production of bovine frozen semen, at ABRC. The semen ejaculate for each bull was collected weekly and number of samples per animal collected were approximately 24. After semen collection the physical parameters of semen were determine such as volume, colour, sperm concentration using a coulter counter and total motility (% of the total sperm population both motile and nonmotile). Any ejaculates failing to meet minimum standards for production of bovine frozen semen were not included in the experimentation.
2.3 Isolation of seminal plasma
The collected semen ejaculates were transported to the laboratory at 37°C without any delay (within half an hour) and centrifuged at 1,520 g for 15 min. The supernatant was collected in another tube and was centrifuged at 850 g for 5 min at 37°C. The final supernatant obtained was used for isolation of EVs or kept at −80°C until further use.
2.4 Isolation of seminal EVs using size exclusion chromatography (SEC)
For size exclusion chromatography, the column packaging and EV isolation was performed as previously described with minimal changes (Böing et al., 2014). A 10 mL of Sepharose CL-2b (Sigma-Aldrich, CL2B300) was packed in 20 mL column and washed with PBS (pH- 7.4). For EV isolation, 1 mL SP (thawed/fresh) was loaded on the column with 1 mL elution buffer, and a total of 26 fractions of 0.5 mL each were collected and were named in numerical order. After elution, the column was washed with 1 SEC volume (1 SEC volume = 12 mL) of 0.5 M NaOH followed by 2X washing with 0.1% Triton X. The column was then washed with distilled water and 0.05% sodium azide (2x SEC volumes). The SEC column filled with sodium azide was kept at 4°C for later use.
2.5 Dynamic light scattering (DLS) analysis of EVs
The size distribution of EVs was measured by Dynamic light scattering using a Zetasizer Nano ZS ZEN3600 (Malvern Instruments, Malvern, United Kingdom). After the isolation of seminal EVs, EVs were analyzed by pooling two consecutive fractions (fraction 1–2, fraction 3–4, and so on). The DLS sample comprised of seminal EVs and PBS in a 1:1 ratio. To prevent aggregation, these samples were sonicated using an ultrasonic water bath for 2 min and the size of EVs was measured. Three independent measurements were performed for each sample and the result was the average of three readings. Data analysis was performed using Zetasizer software version 7.03.
2.6 Nanoparticle tracking analysis (NTA) of EVs
The concentration and size distribution measurements were carried out through NTA in Malvern NS300 system. To perform NTA, fractions 7–9, 10–12, and 13–15 were diluted with PBS in a ratio of 1:1,000 to reduce the number of valid tracks below 200 per image. Each sample was measured five times, and 30–60 s videos were collected with less than 200 valid tracks recorded per video, and then the particles tracked were calculated and plotted.
2.7 Transmission electron microscopy (TEM) of EVs
For TEM, the grid was prepared as described previously (Pal et al., 2023). For sample preparation, EVs from fraction 7–14 were pooled and concentrated to 500 µL and incubated for 2 h in 4% PFA. About 4 μL EV sample was loaded on glow-discharged Formvar film-coated 300-mesh transmission electron microscopy grids. The sample was air-dried for 15 min followed by 3X washing with PBS. The EVs were then fixed by incubating in 1% glutaraldehyde for 15 min. After fixation, the grid was washed thrice with MilliQ water and stained with 1% phosphotungstenic acid for 45 s. Grid imaging was performed using a JEM1400FLASH transmission electron microscope (JEOL USA Inc., Peabody, MA, United States) at 120 kV and viewed under ×250,000 magnification.
2.8 Characterization of EVs through Western blot
To isolate proteins, the 7–14 fractions were first concentrated from 4 mL to 300 µL. The concentrated EVs were then incubated with Radioimmunoprecipitation assay buffer (RIPA buffer, Sigma-Aldrich, United States), in a ratio of 1:1.25 and 2 µL protease inhibitor (ThermoFisher USA) for 1 h, followed by syringe sonication for 24 times. A 100 μL EV lysate was then added into 900 µL chilled acetone and incubated for 22 h at −80°C. After incubation, the lysate was then centrifuged at 16,000 to 17,000 g for 30 min at 4°C, and the protein was redissolved in 80 µL of 0.5 M Tris HCl (pH- 6.8). For quantification of the protein, a standard Bradford graph was prepared using “Quick Start Bradford assay kit” according to the manufacturer’s instructions. SDS-PAGE of the protein was performed to evaluate its quality by utilizing the protocol described earlier (Karanwal et al., 2023).
To detect the EV biomarkers TSG101 and CD63, a Western blot assay was performed as described earlier (Pal et al., 2023). In brief, protein from HF and LF was separately pooled, then electrophoresed through SDS-PAGE was transferred from the gel to Immobilon-FL polyvinylidene difluoride membranes (PVDF, Millipore, Billerica, MA, United States). Before the transfer, membrane was first charged by immersing it for 1 min each in methanol and water. Subsequently, the membrane and the gel were immersed in the transfer buffer. The proteins on the gel were transferred onto PVDF membrane by using a power blotter station semi-dry blot (Invitrogen, United States) using voltage and current of 25 V and 2.5 A respectively at RT for 20 min. The blot was blocked by 5% BSA in Tris buffer saline containing Tween-20 (TBS-T) and incubated overnight at 4°C with gentle agitation. The membrane was incubated with primary mouse polyclonal anti-TSG101 (dilution-1:10,000, SC-7964, Santa Cruz Biotechnology, United States) and anti-CD63 (dilution-1:30,000, STJI40029, St. John’s laboratory, London, United Kingdom). Subsequently, the membrane was washed 3X with TBS-T for 15 min. The membrane was then incubated with HRP-conjugated secondary antibodies such as an anti-mouse secondary antibody for TSG101 (dilution-1:25,000, SC-516102, Santa Cruz Biotechnology) and an anti-rabbit secondary antibody for CD63 (dilution–1:140,000, STJ99512, St. Johns laboratory) for 2 h with constant shaking. The membrane was then washed 5X with TBS-T and then the immunocomplexes were detected using the ECL-HRP substrate (WBULS0100, Sigma). Finally, the signal was captured on an X-ray film and its image was taken using an Epson-perfection v600 film scanner, United States.
2.9 Preparation of protein samples for LC-MS/MS
For LC-MS/MS sample preparation, 50 µg seminal EV protein from each biological replicate was pooled to produce 150 µg protein from each fertility group. Out of 150 µg stock solution, 50 µg of protein sample was digested and reduced with 5 mM tris(2-carboxyethyl) phosphine (TCEP). The protein sample was further alkylated with 50 mM iodoacetamide and digested with trypsin (1:50, Trypsin/lysate ratio), and then incubated for 16 h at 37°C. The dried pellet was resuspended in buffer A (2% acetonitrile, 0.1% formic acid) for downstream LC-MS/MS analysis.
2.10 LC-MS/MS analysis of peptide mixtures and data processing
LC-MS/MS analysis was performed by utilizing an Easy-nlc-1000 system coupled with an Orbitrap Exploris mass spectrometer (Thermo Fisher Scientific, United States). To generate peptide spectra, 1 µg of peptide sample was injected in C18 column 15 cm, 3.0 μm Acclaim PepMap and separated with a 0%–40% gradient of buffer B (80% acetonitrile, 0.1% formic acid) at a flow rate of 300 μL/min. MS1 spectra were acquired in the Orbitrap (Max IT = 25 ms, AGQ target = 300%; RF Lens = 70%; R = 60K, mass range = 375–1,500).
The raw data from six technical replicates, three each for HF and LF groups were processed by retrieving the raw files. The data was analyzed with Proteome Discoverer (v2.5) against the UniProt Bovine database. Briefly, the precursor and fragment mass tolerances were set at 10 ppm and 0.02 Da respectively for Sequest and Amanda search. Both peptide spectrum match and false discovery rate (FDR) was set to <0.01 FDR to increase the confidence and remove the false positives. Proteins with Unique peptide ≥1 were selected and Keratin proteins were removed. Proteins with log2 fold change ≥2 were considered high abundant in HF and proteins with log2 fold change ≤ 0.5 were considered low abundant in HF bulls. Further, these proteins were divided into four categories namely, Known, predicted, uncharacterized, and LOC (i.e., when a published reference is not available and orthologs have not yet been determined, gene will provide a symbol that is known as “LOC” + gene ID) (Karanwal et al., 2023). The composition of unique proteins in HF and LF, and common proteins between HF and LF were visualized through Venn diagrams that were created using an online tool (https://bioinformatics.psb.ugent.be/webtools/Venn/).
2.11 Gene ontology and KEGG pathway analysis
To understand the functional roles of the identified proteins, differentially abundant proteins (DAPs) and unique proteins from both fertility groups were subjected to GO (Gene Ontology) analysis and KEGG (Kyoto Encyclopedia of Gene and Genomes) pathway analysis. The functional profiling of DAPs and unique proteins was carried out using UniProt and the Database for Annotation, Visualization, and Integrated Discovery (DAVID) gene enrichment tool v6.8. The proteins were categorized based on GO terms including cellular component (CC), molecular function (MF), and biological function (BP). Further KEGG pathway analysis was executed to understand the functional roles of these shortlisted proteins and their involvement in different pathways.
2.12 Immuno-localization of selected proteins on spermatozoa
To immunolocalize the selected proteins on spermatozoa, 100 µL of fresh semen was washed by adding 8 mL of sp-TALP (Sperm Tyrode’s albumin lactate pyruvate −50 mM NaCl, 5 mM HEPES, 1.55 mM KCl, 0.2 mM EDTA, 0.2 mM MgCl2.6H2O, 0.15 mM NaH2PO4.2H2O, 1 mM 60% Na-lactate, and 0.98 Mm Na-Pyruvate) followed by centrifugation at 800 g at 37°C for 7 min followed by incubation for 15 min in the incubator (New Brunswick, United Kingdom) with 5% CO2 maintained at 37°C. The washing step was repeated thrice. About 10 µL spermatozoa pellet was added on top of a poly-L lysine-coated glass slide (Poly-prep slides, Sigma-Aldrich) and were fixed using 4% Paraformaldehyde. Thereafter, the slide was washed thrice in 1X PBS for 2 min each. After washing, the slides were incubated in a permeabilization solution for 15 min (0.1% Triton X-100 in PBS) followed by three washing in PBS for 2 min each. Blocking was done for 1 h in 3% BSA prepared in PBST (0.1% Tween 20 in 1X PBS). Subsequently, the slides were washed thrice in PBST followed by overnight incubation with primary antibodies namely, mouse anti-PDIA4 at a dilution of 1:500 (Catalog no - RPD774Mu01, Cloud-Clone Corp.) and mouse Monoclonal Anti-Gelsolin antibody (Catalog no - G4896, Sigma-Aldrich) at a dilution of 1:500. Thereafter, the slides were washed 5 times in PBST. FITC labelled secondary goat anti-mouse IgG antibody (dilution 1:500, Catalog no–F4018, Sigma Aldrich) incubation was done for 1 h at RT. Following this, the slides were washed 5 times in PBST and counter-stained with 0.1 μg/mL DAPI (DNA stain Catalog no. D9542, Sigma Aldrich) for 1 min. The stained slides were washed 5 times in PBST for 2 min each. Thereafter, the slides were dried and a coverslip was mounted using DABCO to visualize the cells under the IX73 Olympus fluorescence microscope, United Kingdom at ×60 magnification. Along with immuno-localization of PDIA4 and GSN, secondary antibody control experiment was also performed (dilution 1:500) (Supplementary Figure S3).
3 Results
3.1 Isolation and characterization of seminal EVs
Seminal EVs were isolated using SEC, and all fractions were analyzed for the presence of seminal EVs using DLS. Out of 26 fractions, seminal EVs were mainly detected in SEC fractions of 7–8, 9–10, 11–12, and 13–14 as described earlier (Böing et al., 2014). Any differences in the concentration and the size of the EVs between the Murrah buffalo bulls of contrasting fertility were also assessed. The size of seminal EVs in the HF (high fertile) and LF (low fertile) bulls ranged from 146.5 nm to 200.3 nm and 160.1–258.7 nm, respectively (Supplementary Tables S2, S3). A p-value of 0.1161 indicated no significant difference in the size of the particles between the HF and the LF groups (Figure 1). The concentration of EVs was analyzed through NTA, and the results showed that in all the fractions (7–15), the concentration ranged from 2.84 × 1011 to 6.82 × 1011 particles per mL in the HF bulls, whereas in the case of LF bulls, the concentration of EVs ranged from 3.57 × 1011 to 7.74 × 1011 particles per mL (Supplementary Tables S4, S5). No significant difference was observed in the concentration of EVs between the HF and LF groups (p-value 0.4588) (Figure 2). Transmission electron microscopy (TEM) was employed to examine the morphology of SP EVs, which revealed their intact cup-shaped structure and membrane integrity in both, the HF and LF groups. The TEM results were consistent with both DLS and NTA findings and confirmed that the diameter of seminal EVs ranged from 50 to 150 nm (Figure 3). For biochemical characterization, the two EV biomarkers CD63 and TSG101 were confirmed for their presence in fractions 7–14 with molecular weight of 30 kDa and 45 kDa respectively (Supplementary Figure S2; Figure 3). The data collectively demonstrated the successful isolation of SP EVs by SEC.
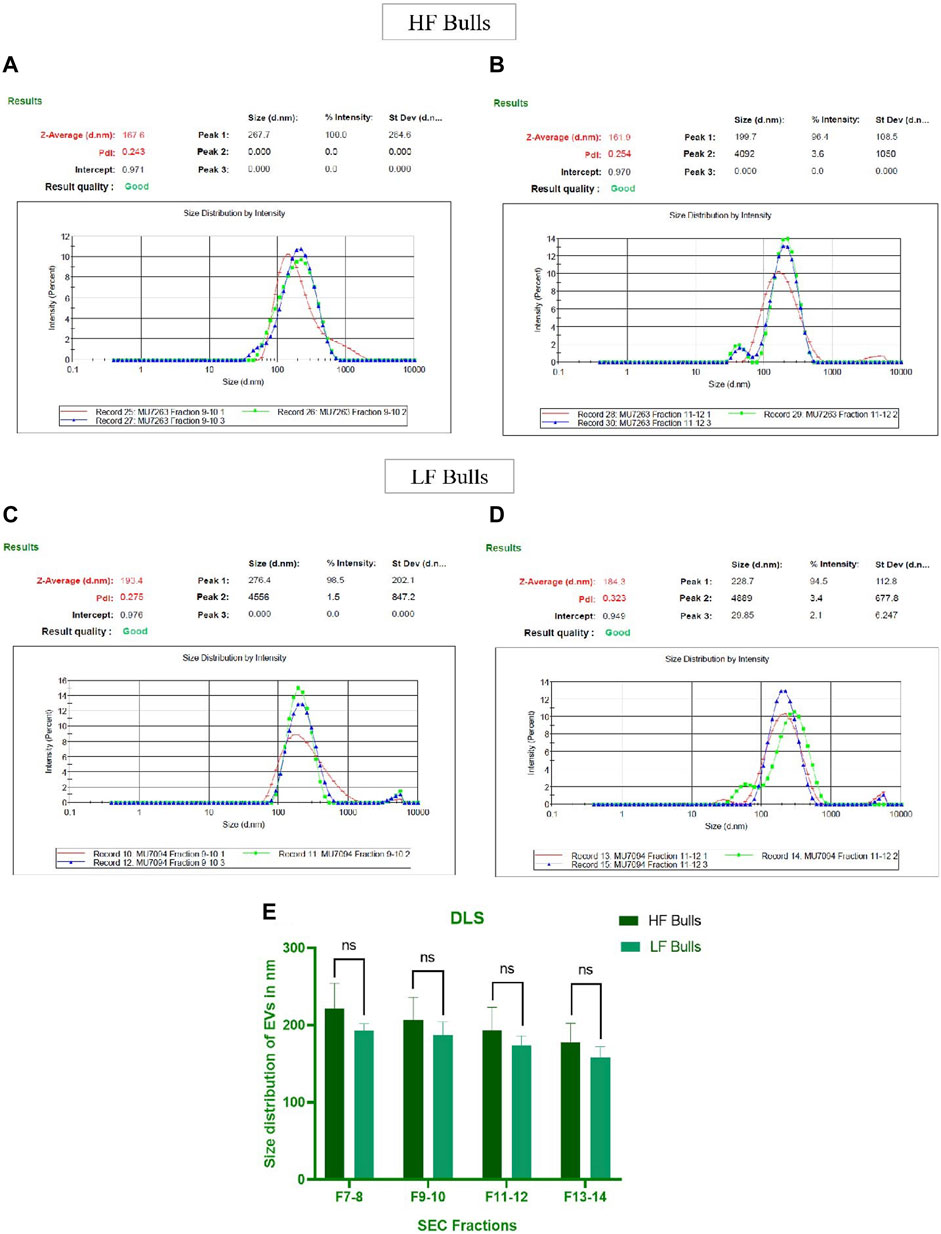
Figure 1. Characterization of seminal EVs. Intensity-based size distribution of seminal EVs analyzed by Zetasizer nano zs particle sizer. Each graph shows mean ± SD (n = 3), PDI, and particle size distribution for (A) Fractions 9–10 (B) fractions 11–12 of seminal EVs of HF bulls, and (C) fractions 9–10 (D) fractions 11–12 of seminal EVs from LF bulls. (E) Comparison of seminal EV size between the HF and LF bulls in which the x-axis represents the SEC fractions, and the y-axis represents the size of EVs in “nm.” The obtained p-value (0.1161) indicated that there was no statistically significant difference between the size of seminal EVs.
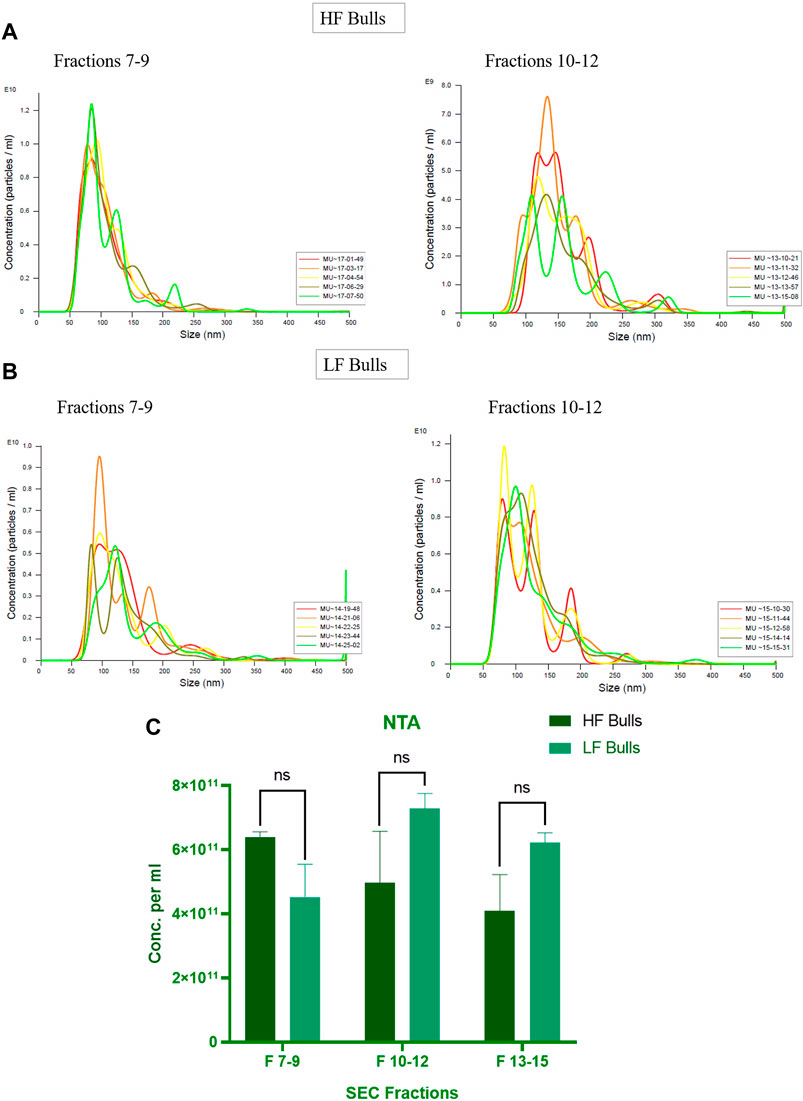
Figure 2. Characterization of seminal EVs Through Nanoparticle Tracking Analysis (NTA). The EVs were diluted 100X for NTA. The concentration vs. size graphs were plotted for (A) fractions 7–9, 10–12 of HF bulls and (B) for fractions 7–9, 10–12 of LF bulls. (C) Comparison of seminal EV concentration between the HF and LF bulls in which the x-axis represents the SEC fractions, and the y-axis represents the concentration of EVs in “particles per mL.” The obtained p-value (0.4588) indicated that there was no significant difference between the concentration of the seminal EVs between the HF and the LF groups.
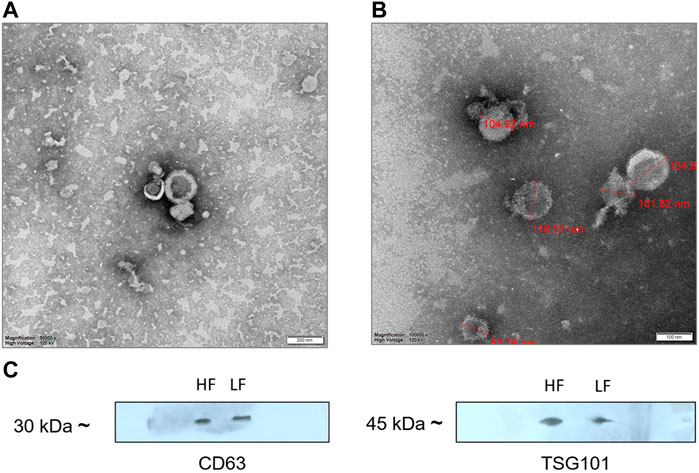
Figure 3. TEM images of (A) HF bulls (B) LF bulls seminal EVs that were negatively stained with phosphotungstic acid stain captured at ×250,000 magnification; scale bar–200 nm and 100 nm at 120 kV. (C) Identification of the CD63 and TSG101 EVs specific protein biomarkers by Western blot analysis. Protein extract loaded in each lane was 15 µg.
3.2 Proteome profiling of seminal EV proteins of HF and LF bulls
The seminal EV proteome profiling revealed that EVs from HF and LF bulls possessed 1862 and 1807 proteins, respectively. Out of the total proteins, 1,754 were common in both HF and LF bulls, while 108 and 53 proteins were unique to HF and LF bulls, respectively (FDR < 0.01, ≥1 peptide, p < 0.05). These common proteins were described as “Differentially Abundant Proteins” (DAPs). Among the DAPs, 87 proteins were highly abundant in HF bull EVs (log2 fold change ≥ 2), and 1,292 were low abundant in HF bull EVs (Log2 fold change ≤ 0.5) as compared to LF bull EVs. Further, these proteins were categorized into four groups: Known, Predicted, Characterized, and LOC. The number of HF unique, LF unique, and common proteins was represented in the Venn diagram (Figure 4). The DAPs were also visualized using a heatmap and volcano plot (Figures 5, 6).
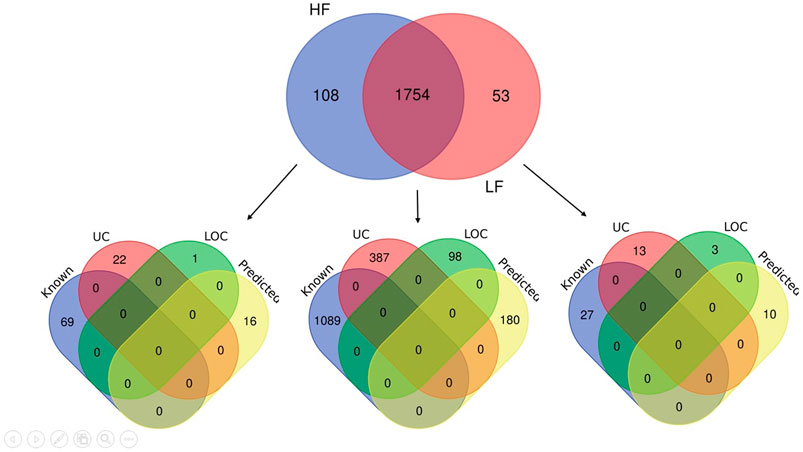
Figure 4. Venn diagram showing the number of identified proteins in seminal EVs of HF and LF bulls. A total of 1,862 and 1,807 proteins were present in seminal EVs of HF and LF bulls respectively. Out of these, 1,754 proteins were common (p < 0.05, FDR < 0.01). Some of the proteins were known in nature, followed by predicted, uncharacterized (UC), and LOC proteins.
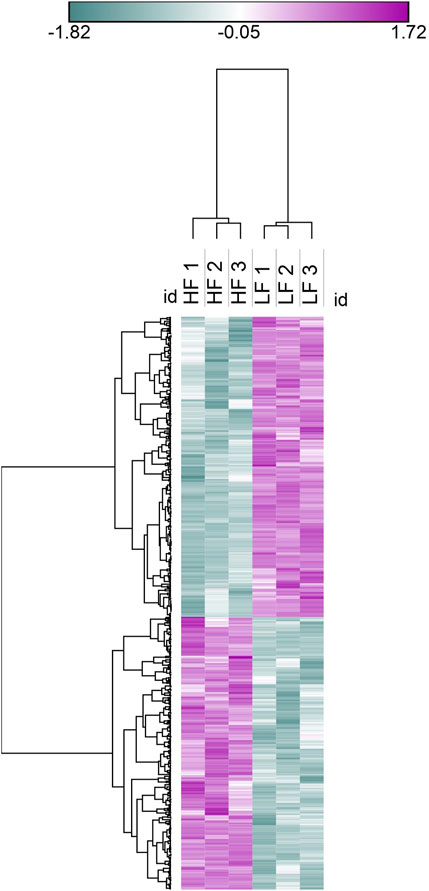
Figure 5. Heat map representation of DAPs in seminal EVs, Blue colour represents low abundant proteins in HF bulls and purple colour represents highly abundant proteins in HF bulls.
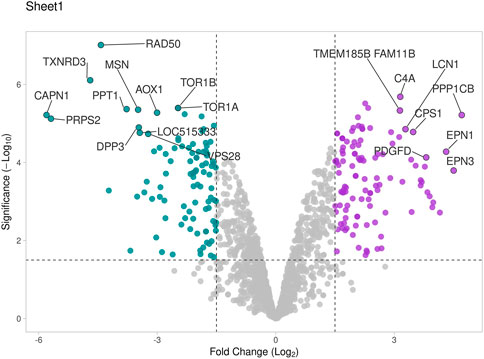
Figure 6. Volcano plot representing total DAPs identified in proteome profile of seminal EVs that were determined by log2 fold change vs. −log10 (p-value). Purple dots represent high abundant proteins in HF bulls, blue dots represent low abundant proteins in HF bulls, and grey dots represent neutral abundance of proteins.
3.3 GO and KEGG pathways enrichment of high abundant proteins in seminal EVs of HF bulls
GO of high abundant proteins in HF bulls revealed that these were associated with biological functions like nucleosome assembly (A7MAZ5, P62803, etc.; p-value = 1.3E-17), DNA templated transcription and initiation (P62803, G3X807, etc.; p-value = 9.4 E-13), and heterochromatin assembly (P0C0S9, F2Z4G5, etc.; p-value = 4.8E-20). In cellular components, many proteins were part of the nucleus (A7MAZ5, P0C0S9, etc.; p-value = 3.7E-6), nucleosome (F2Z416, Q3ZBX9, F2Z4J1, etc.; p-value = 1.3E-40), clathrin-coated vesicles (P04973, F1N579, and F1N4F8. p-value = 5.9E-4), and podosome (Q2KJA1, A4FUG5 and F1N1I6; p-value = 4.0E-3). In the case of molecular function, most proteins were involved in protein heteromerization activity (F2Z4I6, F2Z416, etc.; p-value = 5.9E-29) and DNA binding (F2Z4G6, F2Z4J1, etc.; p-value = 4.4E-19). KEGG pathway analysis revealed that the maximum number of proteins were involved in neutrophil extracellular trap formation (F1MLQ1, E1B7N2, G3N2B8, etc.; p-value = 4.4E-25) and necroptosis (P0C0S9, F2Z4G5, etc.; p-value = 4.7E-11) (Supplementary Table S6; Figure 7). Further, the list of high abundant proteins associated with fertility is given in Table 1 along with their log2 fold changes and p-values.
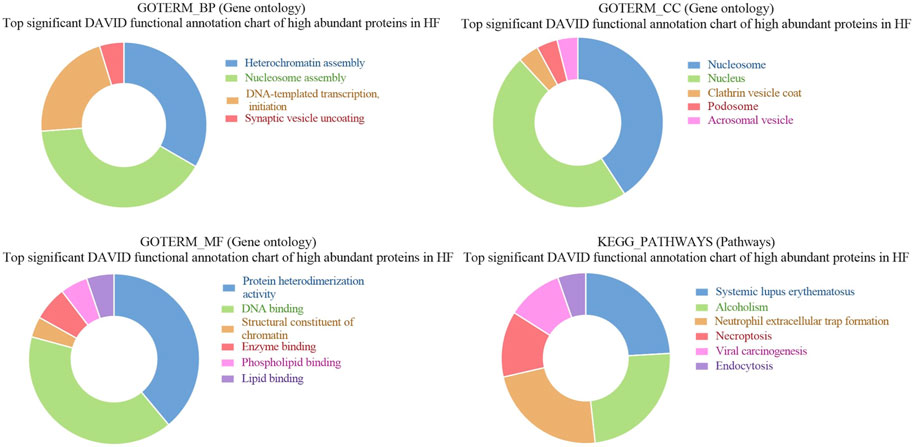
Figure 7. Pie chart representation of Gene ontology (GO) terms biological process (BP), Cellular component (CC), Molecular function (MF), and KEGG pathway enrichment of high abundant proteins in HF bulls.
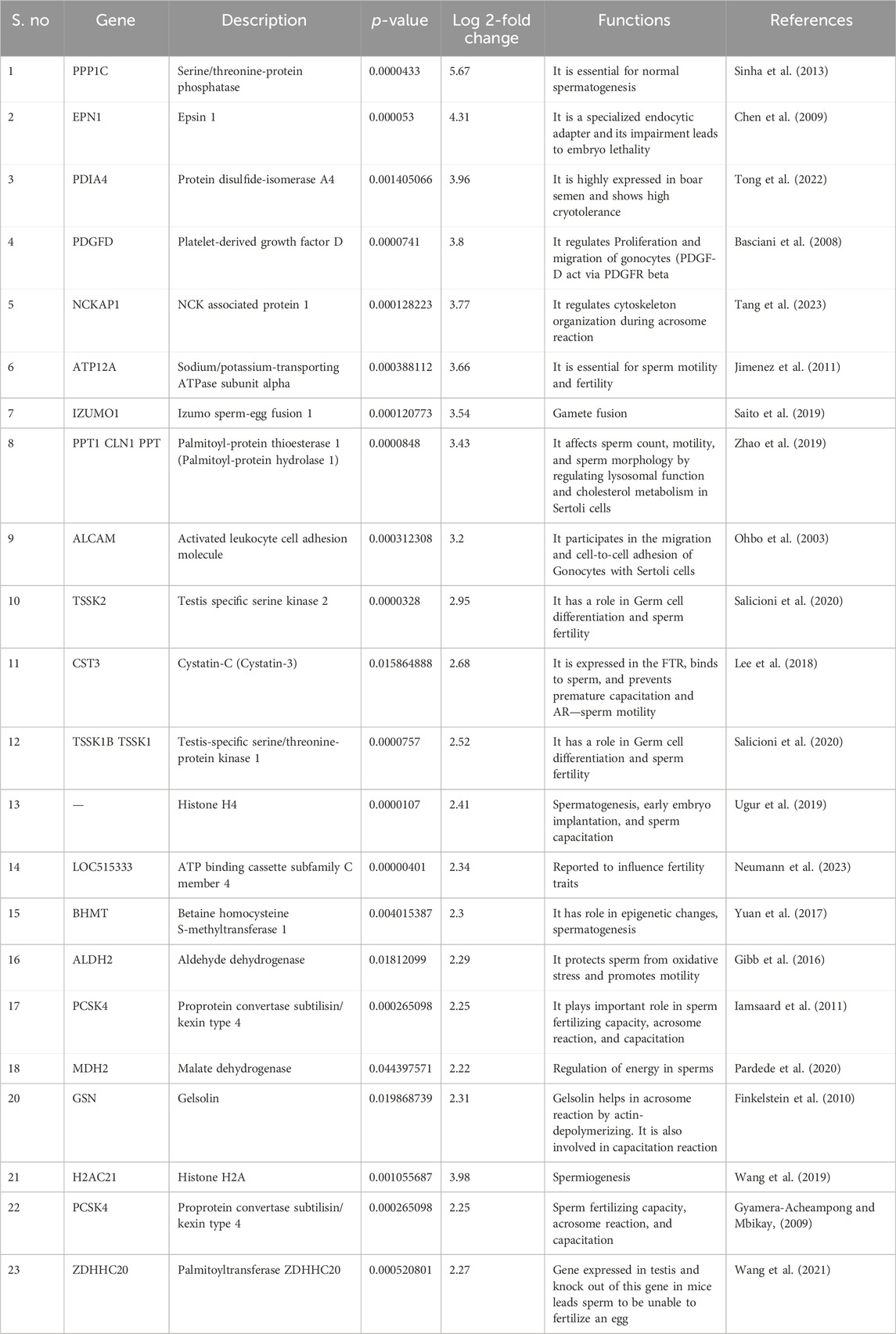
Table 1. List of top 20 fertility-associated proteins that were highly abundant in HF bulls along with their functions and log fold change.
3.4 GO and KEGG pathways enrichment of low abundant proteins in seminal EVs of HF bulls
Gene ontology analysis of low abundant proteins in HF bulls revealed that the majority of proteins were found to be associated with biological functions like protein transport (E1BP90, Q9XSK2, P2856, etc.; p-value = 1.5E-9), Glycolytic processes (E1B959, A6QLL8, F1MX69, etc.; p-value = 1.4E-12), proteolysis (Q3T0X8, Q5E946, G3NOF4, etc.; p-value = 1.9E-10), binding of sperm to zona pellucida (E1BG77, F1MFC4, Q32LB5, etc.; p-value = 7.3E-9). In cellular components, proteins were associated with cytosol (Q05927, Q0VCK0, P84080, etc.; p-value = 1.2E-36), Cytoplasm (F1MZP0, E1BDS9, P06623, etc.; p-value = 1.2E-34), apical plasma membrane (Q4GZT4, P31404, F1MXW4, etc.; p-value = 1.7E-22), melanosome (P06623, P31408, F1M132, etc.; p-value = 2.3E-22). In the case of molecular function, the proteins were associated with GTPase activity (Q5E196, G3N3N1, F1MN18, etc.; p-value = 2.4E-26), GTP binding (P84080, Q3SZF2, etc.; p-value = 3.4E-23), ATP binding (G3MXV4, E1BCR1, Q32PF2, etc.; p-value = 3.8E-19), protein binding (Q10741, P84080, etc.; p-value = 6.0E-16). In KEGG pathway analysis, the proteins were found to be associated with the proteasome (Q3T0X5, Q3T0Y5, Q3ZCK9, etc.; p-value = 1.9E-25), endocytosis (P884081, E1BP90, F1N371, etc.; p-value = 6.4E-21), metabolic pathways (F1MZP0, Q3ZCK3, Q05927, etc.; p-value = 9.8E-16) (Supplementary Table S6; Figure 8). The list of fertility-associated proteins that were low abundant in seminal EVs of HF bulls along with their function, Log2 fold change and the p-values is given in Table 2.
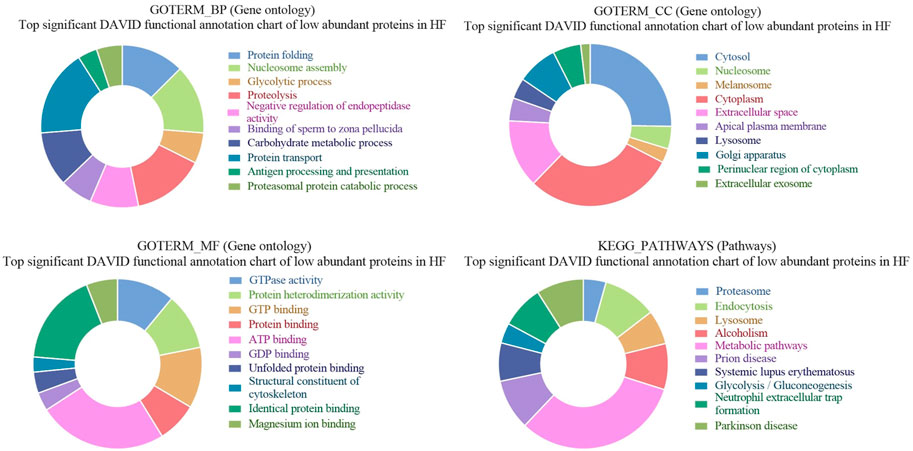
Figure 8. Pie chart representation of GO terms biological process (BP), Cellular component (CC), Molecular function (MF), and KEGG pathway enrichment of low abundant proteins in HF bulls.
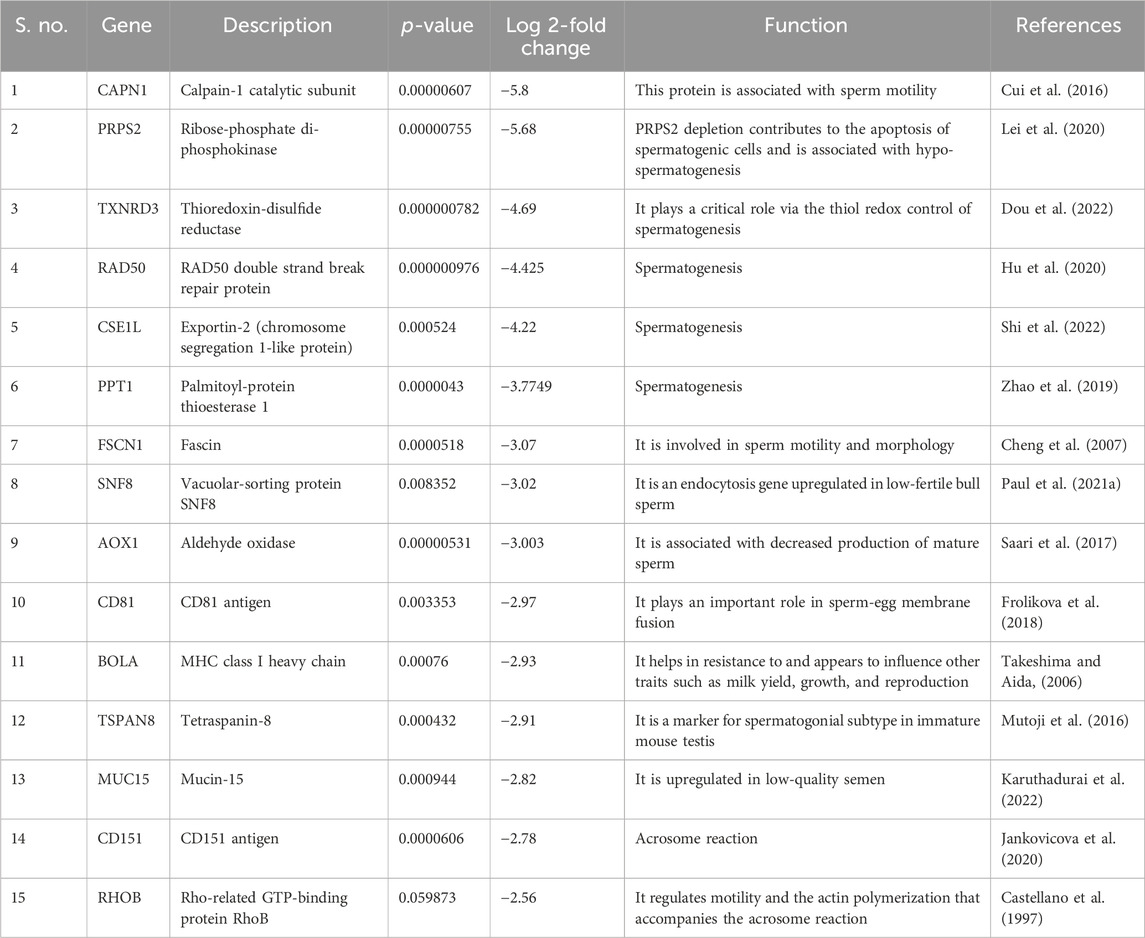
Table 2. List of fertility-associated proteins that were low abundant in HF bull EVs along with their functions and log fold changes.
3.5 GO and KEGG pathways enrichment of unique proteins in seminal EVs of HF bulls
GO of unique proteins in HF bulls revealed that these were associated with biological functions like a cellular response to calcium ion (A5D784, E1BID8, F1MLJ3; etc., p-value = 1.70E-06), malonyl-CoA biosynthetic process (E1BGH6, F1MSC3, Q9TTS3; p-value = 7.39E-05), acetyl-CoA metabolic process (E1BGH6, F1MSC3, Q9TTS3; p-value = 3.66E-04). In cellular components number of proteins were associated with organelles like cytosol (F6QS88, Q2KHU0, F1N189, etc.; p-value = 6.73E-05), phosphopyruvate hydratase complex (A6QR19, Q3ZC09; p-value = 0.016945), myelin sheath abaxonal region (A6QLD1, P53712; p-value = 0.021137), neuromuscular junction (Q2KJA7, F1MC13, P53712; p-value = 0.025453). In case of molecular function, many proteins were associated with GO terms like calcium-dependent phospholipid binding (A5D784, E1BID8, F1MLJ3, etc.; p-value = 1.71E-07), acetyl-CoA carboxylase activity (E1BGH6, F1MSC3, Q9TTS3; p-value = 5.80E-05), magnesium ion binding (F6QS88, G1K192, F1MWJ3, etc.; p-value = 2.98E-04), integrin binding (F1MDH3, Q2KJA7, F1MC13, etc.; p-value = 0.001399). In KEGG pathway analysis, the proteins were found to be associated with Carbon metabolism (Q148D3, A1A4J1, A6QR19, etc.; p-value = 2.13E-06), Biosynthesis of amino acids (A1A4J1, A6QR19, Q2KHU0, etc.; p-value = 4.39E-05), Metabolic pathways (F6QS88, Q3ZC09, Q2KHU0, etc.; p-value = 1.39E-04), Glycolysis/Gluconeogenesis (A1A4J1, A6QR19, Q3ZC09, etc.; p-value = 3.86E-04) (Figure 9).
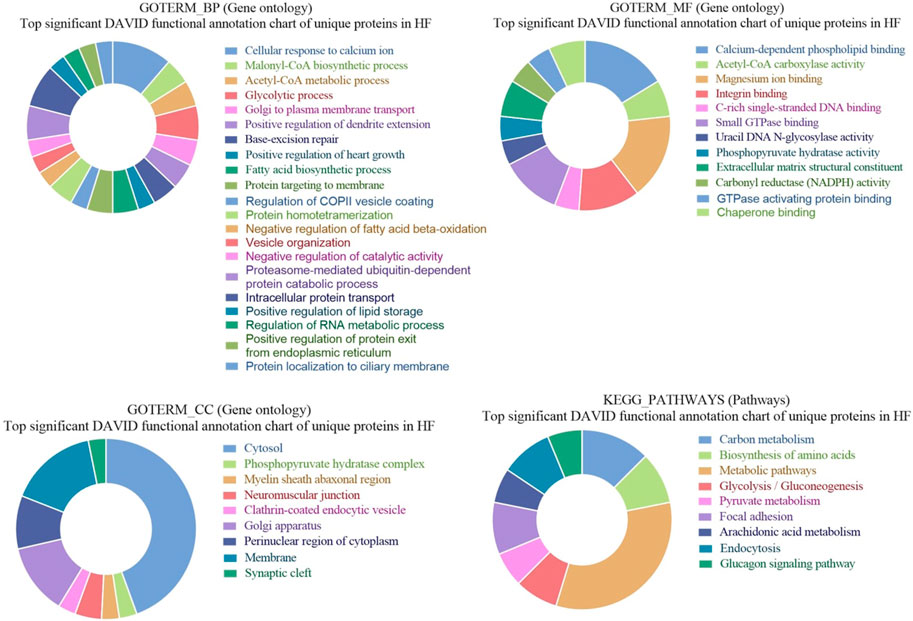
Figure 9. Pie chart representation of GO terms biological process (BP), Cellular component (CC), Molecular function (MF), and KEGG pathway enrichment of unique proteins in HF bulls.
3.6 GO and KEGG pathways enrichment of unique proteins in seminal EVs of LF bulls
GO of unique proteins in LF bulls revealed that these were associated with biological functions like carbohydrate metabolic process (Q5EAB4, G3MXK6, F1MDX6, A7MBC0; p-value = 0.027632), regulation of long-term neuronal synaptic (Q0IIG7, Q3SZ33; p-value = 0.029595), nucleosome assembly (A6H767, F1N7X3, Q2TA40, G3X7M5; p-value = 0.040927), negative regulation of intrinsic apoptotic signaling (G3MYD5, P68002; p-value = 0.058338). In cellular components, proteins were associated with extracellular space (Q8SPU5, Q5EAB4, E1B9Y3, etc.; 0.001648), membrane (F1N5M4, Q2TBQ5, P00171, etc.; p-value = 0.006135), chromatin (A6H767, F1N7X3, A6QLY7, etc.; p-value = 0.018316), chromaffin granule (F1MWI1, A6QPQ2; p-value = 0.026138). In the case of molecular function, the proteins were associated with histone binding (A6H767, F1N7X3, Q2TA40, G3X7M5; p-value = 0.031620912). In KEGG pathway analysis, the proteins were found to be associated with Choline metabolism in cancer (F1N5M4, Q2KJ15, A2VDK6; p-value = 0.009219) (Figure 10).
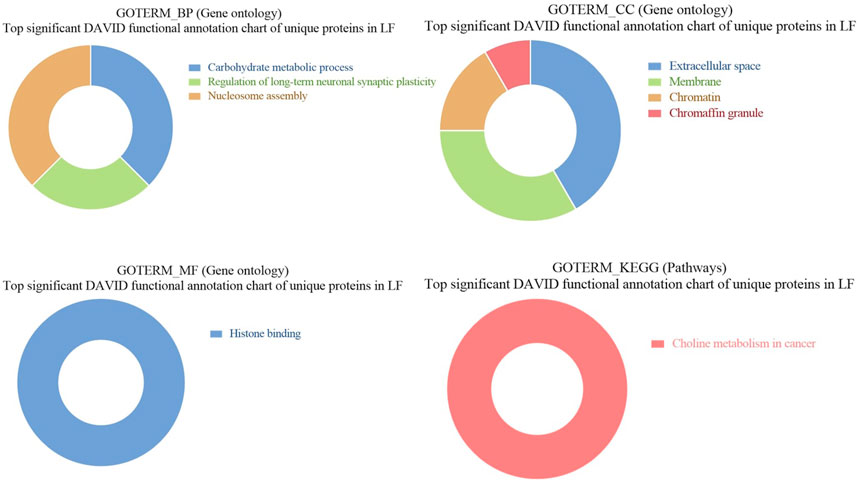
Figure 10. Pie chart representation of GO terms biological process (BP), Cellular component (CC), Molecular function (MF), and KEGG pathway enrichment of unique proteins in LF bulls.
3.7 Immuno-localization of gelsolin and PDIA4 on the spermatozoa
Two proteins PDIA4 and GSN that were highly abundant in HF bull EVs were subjected to immunolocalization on spermatozoa. It was observed that PDIA4 was present on the acrosome region and mid-piece of spermatozoa while Gelsolin was present on the acrosome region of the spermatozoa of bulls (Figure 11). In this study we also performed the secondary antibody control that does not shows any binding of secondary antibody on the spermatozoa (Supplementary Figure S3).
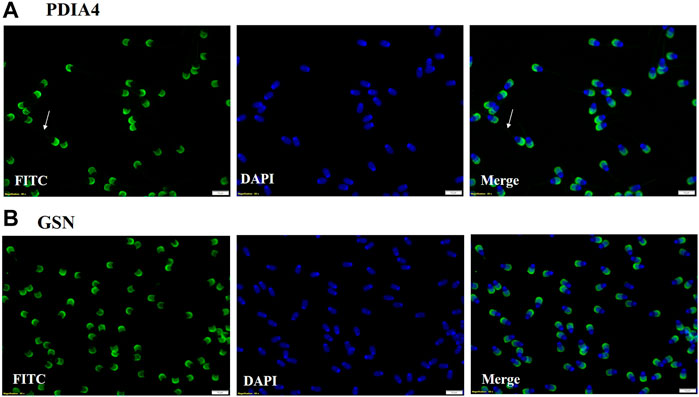
Figure 11. (A) Immuno-localization of PDIA4 was done on spermatozoa. ICC results show that PDIA4 is present on the acrosome region and mid-piece of spermatozoa. (B) Immuno-localization of Gelsolin on spermatozoa. ICC results showed that Gelsolin is present on the acrosome region of spermatozoa.
4 Discussion
This research was conducted to investigate whether there are differences in the size, concentration, and proteomic cargo of EVs based on the fertility status of buffalo bulls.
Seminal EVs were isolated using SEC and further characterized by DLS, NTA, TEM, and Western blotting. Fractions 7–14 carried the EVs with mean size of 200 nm in both the fertility groups as reported in previous studies (Luo et al., 2022; Barranco et al., 2023). The particles in the sample were homogenous as indicated by polydispersity index (PDI) ranging from 0.2 to 0.35 in the DLS data from EVs of both the fertility groups. In addition to the DLS method, the NTA assay re-established not only the size of the seminal EVs but also confirmed the abundance of EVs in buffalo SP. TEM also illustrated round cup-shaped characteristic morphology of seminal EVs in both HF and LF bulls falling in the desired size range (Xu et al., 2020; Yu et al., 2023).
The seminal EV proteomic profile of HF and LF bulls was deciphered with the help of LC-MS/MS and computational biology. From the data, 1,862 and 1,807 proteins were identified in HF and LF seminal EVs respectively. The highly abundant proteins present in HF bulls were further examined to determine their roles in the fertilizing potential of spermatozoa and based on previous literature, numerous proteins were found to be associated with the fertilizing capacity as mentioned in Tables 1, 2.
EVs are released by most internal organs of the male reproductive system like epithelial cells of vesicular glands (anatomically referred to as seminal vesicles) and the ampulla of the ductus deferens (Agrawal and Vanha-Perttula, 1987; Renneberg et al., 1995). Furthermore, Sertoli cells have also been reported to release EVs (Mancuso et al., 2018). All the EVs released by the organs constitute the diverse population of EVs present in SP.
Spermatogenesis is a complex series of events that includes the mitotic proliferation of spermatogonial cells, the meiotic division of spermatocytes, and their further maturation into spermatozoa. Various proteins involved in different aspects of sperm development were found to be highly abundant in HF seminal EVs. Activated leukocyte cell adhesion molecule (ALCAM) (log2 fold change - 3.2057; p-value = 0.00031) is expressed transiently in gonocytes and facilitates gonocyte-sertoli cell adhesion and migration of gonoyctes toward the basement membrane (Ohbo et al., 2003). Platelet-derived growth factor D (PDGFD) (log2 fold change - 3.8084; p-value = 7.41E-05) is localized in Sertoli cells in prenatal and early postnatal testis and its interaction with PDGFR-β is crucial for gonocyte development (Basciani et al., 2008). Testis-specific serine kinase 2 (TSSK 1,2) (TSSK1–log2 fold change-2.5187; p-value = 7.57E-05, TSSK2 – log2 fold-2.9543; p-value = 3.28E-05) are testis-specific kinases localized in spermatids and spermatozoa and deletion of Tssk1 and 2 causes male infertility due to haploinsufficiency (Xu et al., 2008), TSSKs have a crucial role in germ cell differentiation and sperm function (Salicioni et al., 2020). Betaine--homocysteine S-methyltransferase 1 (BHMT) (log2 fold change - 2.3045; p-value = 0.00401) is an important methyl donor and aberrant expression of BHMT leads to global DNA hypomethylation and modulation of spermiogenesis (Zhang et al., 2015; Yuan et al., 2017). Palmitoyl-protein thioesterase 1 (PPT1) (log2 fold change–3.4280; p-value = 8.48E-05) is a lysosomal depalmitoylating enzyme and its absence in Sertoli cells leads to disruption in adhesion of developing germ cells to Sertoli cells that causes decline in motility and number of sperms along with increase in number of sperms with deformed heads (Zhao et al., 2019). Nudix hydrolase 5 (NUDT15) (log2 fold change = 2.3856; p-value = 0.00130) is a gene of the KRAB transcription family and it might be involved in early embryonic maintenance or methylation of genomic regions (Lalancette et al., 2009). Histone 4 was previously reported to be localized in head region of sperm in Holstein bulls. It showed high expression in HF bulls compared to LF (Ugur et al., 2019).
Motility is a substantial trait of spermatozoa and numerous proteins were found highly abundant in HF bulls that are known to play essential roles in motility. Protein phosphatase (log2 fold change = 2.5762; p-value = 0.000511) is predominantly expressed in the testis and distributed over the flagellum and post-acrosomal region of the sperm head in dog (Tash et al., 1988). PPP1CA, PPP1CB, and PPP1CC1 are ubiquitously present, and PPP1CC2 is present at high levels in adult testis and spermatozoa. Deletion of the Ppp1cc gene causes loss of both isoforms resulting in oligo-terato-asthenozoospermia and thus, causing male infertility due to impaired spermiogenesis (Sinha et al., 2013). Another protein Sodium/potassium-transporting ATPase subunit alpha (ATP12A) (log2 fold change = 3.6687; p-value = 0.00038) also showed high abundance in HF bulls EVs. Inhibition of ATP12A has been reported to cause impairment of sperm motility and capacitation in vitro (Escoffier et al., 2020). ATP12A is localized on the acrosome region of spermatozoa in B. bubalis, Bos taurus, and Ovis aries and its expression increases progressively in bovine sperm acrosome region while moving from epididymis to deferent ducts (Favia et al., 2022). Sodium/potassium-transporting ATPase subunit alpha has 3 isoforms out of which, alpha4 is expressed in male germ cells and localized on the flagellum of spermatozoa which plays a pivotal role in motility (Woo et al., 2000). Aldehyde dehydrogenase (ALDH2) (log2 fold change = 2.2975; p-value = 0.01812) was also highly abundant in HF bull EVs which also plays an essential role in sperm motility. A recent study on stallion sperm showed a positive correlation between ALDH expression and progressive as well as rapid motility. ALDH was expressed all over the spermatozoa and the strongest expression was observed in the head region and mid-piece (Gibb et al., 2016). It is also highly expressed in spermatozoa of HF bulls (Karanwal et al., 2023). Malate dehydrogenase MDH2 (log2 fold change = 2.2166; p-value = 0.04439) is associated with energy regulation of sperm (Pardede et al., 2020), and comparative proteomics in a previous report revealed MDH2 as a fertility biomarker in spermatozoa in crossbred bulls (Muhammad Aslam et al., 2018). MDH has also been found to be involved in the formation of acrosomal membrane in Eriocheir sinensis (Li et al., 2023).
Capacitation and acrosome reaction are two important processes that occur before the fusion of sperm and oocyte. Capacitation involves many orchestrated processes including activation of cAMP-dependent phosphorylation pathways, removal of cholesterol, hyperpolarization, and change in ion permeability of sperm plasma membrane (Stival et al., 2016). Proteomic analysis of seminal EVs revealed many proteins that have been reported to play crucial roles in capacitation and acrosome reaction. Cystatin-C (CST3) (log2 fold change = 2.6770; p-value = 0.01586) is highly expressed in the female reproductive tract and it can bind to the post-acrosomal head region, midpiece and tail of spermatozoa. It enhances sperm motility and inhibits premature capacitation (Lee et al., 2018). It is reported to be associated with prostasomes (Carlsson et al., 2011). Another protein Gelsolin (GSN) (log2 fold change = 2.3103; p-value = 0.01986) was also found to be highly abundant in HF bulls EVs. Gelsolin is inactive during capacitation and the release of bound gelsolin causes F-actin depolymerization and it also enhances acrosome reaction (Finkelstein et al., 2010). Proprotein convertase subtilisin/kexin type 4 (PCSK4) (log2 fold change = 2.2510; p-value = 0.00026) is essential in sperm fertilizing ability localized along the acrosomal plasma membrane of mammalian spermatozoa (Gyamera-Acheampong and Mbikay, 2009). Inhibition of PCSK4 in rat spermatozoa has been shown to interfere with oocyte penetration (Dahril et al., 2019). Izumo sperm-egg fusion 1 (IZUMO1) (log2 fold change = 3.5414; p-value = 0.00012), a sperm adhesion protein, was also found highly abundant in HF bull EVs which aids in sperm—egg fusion by interacting with oocyte-specific GPI-anchored receptor JUNO. The mouse lines with disrupted Izumo gene produced healthy mice with spermatozoa that penetrated zona pellucida but were unable to fuse with eggs (Saito et al., 2019). Palmitoyl transferase (ZDHHC20) (log2 fold = 2.2691; p-value = 0.00052) is a testis-enriched gene and it was also found to be highly abundant in HF bull sperm EVs. Its deletion results in an abnormality in sperm head and tail, decreased motility, and disturbed acrosome reaction leading to male sterility (Wang et al., 2021).
Approximately 26 percent of the detected DAPs were identified as histone proteins and modified histone proteins. These proteins are mostly comprised of H2A and H4 histones. In a recent report, H4 and acetylated H4 were more abundant in spermatozoa of HF bulls (Ugur et al., 2019). Histones and their modification play a significant role in DNA packaging (Miller et al., 2010). Previous studies demonstrated that H2B, H3.3, and H4 were detectable in bull spermatozoa, and the condensation status of sperm chromatin, influenced by histone retention, is associated with the fertility of bulls (De Oliveira et al., 2013). Two H2A variants H2AFV and H2AFZ that are known to influence global DNA methylation and spermatogenesis were also highly abundant in HF Murrah bull spermatozoa (Zhao et al., 2013; Madakashira et al., 2017; Karanwal et al., 2023).
In case of low-abundant proteins in HF bulls, three proteins were negatively correlated with semen quality. These proteins are Vacuolar-sorting protein SNF8, aldehyde oxidase, and Mucin 15. Vacuolar-sorting protein SNF8 (log2 fold = −3.2285; p-value = 1.85E-05), an endocytosis gene, was previously found to be upregulated in low-fertile bull sperm (Paul et al., 2021a), while aldehyde oxidase (AOX1) (log2 fold change = −3.0038; p-value = 5.31E-06) has been associated with decreased production of mature sperm (Saari et al., 2017). Additionally, Mucin 15 (MUC15) (log2 fold change = −2.8176; p-value = 0.00094) was found to be upregulated in low-quality semen (Karuthadurai et al., 2022). Along with our findings, the available reports substantiate that EVs not only carry proteins responsible for enhancing fertilizing potential but also those proteins that harm sperm quality. The data generated from this study show high abundance of these three proteins in LF bull seminal EVs, suggesting that they may collectively contribute to the low quality of semen in LF bulls.
Two of the significantly abundant proteins in HF seminal EVs were also immunolocalized on the spermatozoa. Protein disulfide isomerase A4 (PDIA4) (log2 fold change = 3.9638; p-value = 0.00140) is a protein localized in the endoplasmic reticulum (ER) lumen that catalyzes the formation, isomerization, and reduction of disulfide bonds (Turano et al., 2002). These PDI proteins are found in the equatorial segment of mature spermatozoa (Ellerman et al., 2006) that play a crucial role during sperm-oocyte fusion by forming multimolecular complexes that are critical for the fusion process (Yanagimachi, 1994). In the context of predicting the freezability of Erhualian boars, PDIA4 has been identified as a potential biomarker. Immunolocalization of PDIA4 on spermatozoa showed its presence on the acrosome region and mid-piece of spermatozoa. Another protein that was localized was Gelsolin (GSN) (log2 fold change = 2.3103; p-value = 0.01986) which plays an important role in the sperm acrosome reaction. The cortical cytoskeleton is disassembled during the exocytotic acrosome reaction, which fuses the plasma membrane with the outer cytoskeleton membrane. This barrier can be broken by proteins of the gelsolin family (Finkelstein et al., 2010). Gelsolin also showed high expression in spermatozoa of HF Murrah bulls (Karanwal et al., 2023). Immunolocalization of gelsolin on spermatozoa revealed that it is present in the acrosome region of spermatozoa. Our findings demonstrate that PDIA and gelsolin remained differentially abundant in seminal EVs and its detection on sperm indicates that these proteins might be getting transferred to the spermatozoa through EVs as spermatozoa are translationally inactive. However, further studies need to be carried out to robustly validate this claim. Therefore, the overall data collectively indicate that the fertility status of the Murrah bulls can be predicted based on the proteomic profile of their seminal EVs while their size and concentration do not exhibit significant differences with respect to fertility status. However, there are some limitations in the study. For example, the assessment of the field fertility in buffalo bulls faced limitations due to the small sample size, and evaluating their fertility takes a considerable amount of time. Samples from both groups were pooled for LC-MS/MS and Western blotting due to technical and financial constraints. However, the pooling of sample increases homogeneity within the group detecting a maximum number of proteins while maintaining a high degree of confidence (Park et al., 2012; Somashekar et al., 2017; Muhammad Aslam et al., 2019). Conducting additional experiments involving a larger sample size within the same group will undoubtedly contribute to a more comprehensive understanding of the findings presented in the current study. Additionally, Western blot experiments also need to be performed to establish the differential abundance of the seminal EV proteins that were detected through LC-MS/MS. Experiments involving coincubation of seminal EVs and spermatozoa will further aid in establishing the transfer of the protein cargo from EVs to spermatozoa along with their roles in enhancing or reducing sperm functions.
5 Conclusion
In conclusion, our study shows a discrete proteome profile of seminal EVs of high vis a vis low fertile buffalo bulls using label-free LC-MS/MS. In addition, our study also indicates that the fertility status of the buffalo bulls is not dependent on the size and concentration of the seminal plasma EVs. However, based on comparative proteome data of seminal EVs of two distinct fertility groups, we identified many differentially abundant proteins and further explained the most prominent proteins associated with sperm function. The highly abundant proteins identified in the seminal EVs of HF bulls were found to be associated with key sperm functions such as sperm motility, capacitation, acrosome reaction, and pellucida binding, reflecting their crucial and supportive roles in various sperm functions. LF bull EVs exhibited the presence of certain proteins with detrimental effects on semen quality, nevertheless, a few were associated with spermatogenesis and motility. The presence of PDIA4 and GSN on spermatozoa may indicate that these proteins are being transferred on the spermatozoa through EVs although, further studies are required to verify this claim. Altogether, this study provides a unique insight into the dynamics of protein repertoire in seminal EVs in relation to buffalo bull fertility. Our current findings need further validation using additional protein candidates and co-incubation experiments of EVs with spermatozoa and subsequently, monitoring the changes in the fertilizing potential of spermatozoa.
Data availability statement
The data presented in the study are deposited in the PRIDE repository, accession number; PXD050211.
Ethics statement
The animal study was approved by Institutional Animal Ethics Committee (IAEC), National Dairy Research Institute, Karnal. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SB: Writing–original draft, Writing–review and editing. SK: Formal Analysis, Writing–original draft, Writing–review and editing. AP: Supervision, Writing–original draft. JC: Supervision, Writing–original draft, Writing–review and editing. VC: Methodology, Formal Analysis, Supervision, Writing–review and editing. AP: Supervision, Writing–original draft. MB: Resources, Writing–original draft. TD: Funding acquisition, Resources, Writing–original draft. RK: Conceptualization, Funding acquisition, Resources, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Department of Biotechnology, New Delhi, India- Funded project (Grant No. BT/PR34220/AAQ/1/771/2020) and Bill and Melinda Gates Foundation (Grant number OPP1154401) provided the funds for performing the experiments under this research work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1400323/full#supplementary-material
References
Aalberts, M., Sostaric, E., Wubbolts, R., Wauben, M. W. M., Nolte-’T Hoen, E. N. M., Gadella, B. M., et al. (2013). Spermatozoa recruit prostasomes in response to capacitation induction. Biochim. Biophys. Acta Proteins Proteom 1834 (11), 2326–2335. doi:10.1016/j.bbapap.2012.08.008
Aalberts, M., van Dissel-Emiliani, F. M. F., van Adrichem, N. P. H., van Wijnen, M., Wauben, M. H. M., Stout, T. A. E., et al. (2012). Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol. Reprod. 86 (3), 82. doi:10.1095/biolreprod.111.095760
Agrawal, Y., and Vanha-Perttula, T. (1987). Effect of secretory particles in bovine seminal vesicle secretion on sperm motility and acrosome reaction. J. reproduction Fertil. 79 (2), 409–419. doi:10.1530/jrf.0.0790409
Barrachina, F., Battistone, M. A., Castillo, J., Mallofré, C., Jodar, M., Breton, S., et al. (2022). Sperm acquire epididymis-derived proteins through epididymosomes. Hum. Reprod. 37, 651–668. doi:10.1093/humrep/deac015
Barranco, I., Sanchez-López, C. M., Bucci, D., Alvarez-Barrientos, A., Rodriguez-Martinez, H., Marcilla, A., et al. (2023). The proteome of large or small extracellular vesicles in pig seminal plasma differs, defining sources and biological functions. Mol. Cell. Proteomics 22 (4), 100514. doi:10.1016/j.mcpro.2023.100514
Basciani, S., De Luca, G., Dolci, S., Brama, M., Arizzi, M., Mariani, S., et al. (2008). Platelet-derived growth factor receptor beta-subtype regulates proliferation and migration of gonocytes. Endocrinology 149, 6226–6235. doi:10.1210/en.2008-0349
Batra, V., Dagar, K., Nayak, S., Kumaresan, A., Kumar, R., and Datta, T. K. (2020). A higher abundance of O-linked glycans confers a selective advantage to high fertile Buffalo spermatozoa for immune-evasion from neutrophils. Front. Immunol. 11, 1928. doi:10.3389/fimmu.2020.01928
Boccia, L., Di Francesco, S., Neglia, G., De Blasi, M., Longobardi, V., Campanile, G., et al. (2013). Osteopontin improves sperm capacitation and invitro fertilization efficiency in buffalo (Bubalus bubalis). Theriogenology 80, 212–217. doi:10.1016/j.theriogenology.2013.04.017
Böing, A. N., van der Pol, E., Grootemaat, A. E., Coumans, F. A. W., Sturk, A., and Nieuwland, R. (2014). Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 3. doi:10.3402/jev.v3.23430
Carlsson, L., Ronquist, G., Ronquist, G., Eliasson, R., Egberg, N., and Larsson, A. (2011). Association of cystatin c with prostasomes in human seminal plasma. Int. J. Androl. 34, 363–368. doi:10.1111/j.1365-2605.2010.01090.x
Castellano, L. E., Martínez-Cadena, G., López-Godínez, J., Obregón, A., and García-Soto, J. (1997). Subcellular localization of the GTP-binding protein Rho in the sea urchin sperm. Eur. J. Cell Biol. 74 (4), 329–335.
Chen, H., Ko, G., Zatti, A., Di Giacomo, G., Liu, L., Raiteri, E., et al. (2009). Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both epsin 1 and epsin 2 in mice. Proc. Natl. Acad. Sci. U. S. A. 106 (33), 13838–13843. doi:10.1073/pnas.0907008106
Cheng, Y., Buffone, M. G., Kouadio, M., Goodheart, M., Page, D. C., Gerton, G. L., et al. (2007). Abnormal sperm in mice lacking the Taf7l gene. Mol. Cell Biol. 27, 2582–2589. doi:10.1128/mcb.01722-06
Cordeiro, L., Lin, H. L. H., Carvalho, A. V., Grasseau, I., Uzbekov, R., and Blesbois, E. (2021). First insights on seminal extracellular vesicles in chickens of contrasted fertility. Reproduction 161 (5), 489–498. doi:10.1530/REP-20-0462
Cui, X., Sun, Y., Wang, X., Yang, C., Ju, Z., Jiang, Q., et al. (2016). A g.-1256 A > C in the promoter region of CAPN1 is associated with semen quality traits in Chinese Holstein bulls. Reproduction 152 (1), 101–109. doi:10.1530/REP-15-0535
Dahril, A., Keumala, V., and Mustafa, A. (2019). Human spermatozoa anti-proprotein convertase subtilisin/kexin type 4 synthesis using New Zealand rabbit for novel immunocontraception in males. Investig. Clin. Urol. 60, 303–311. doi:10.4111/icu.2019.60.4.303
De Oliveira, R. V., Dogan, S., Belser, L. E., Kaya, A., Topper, E., Moura, A., et al. (2013). Molecular morphology and function of bull spermatozoa linked to histones and associated with fertility. Reproduction 146, 263–272. doi:10.1530/REP-12-0399
Dou, Q., Turanov, A. A., Mariotti, M., Hwang, J. Y., Wang, H., Lee, S. G., et al. (2022). Selenoprotein TXNRD3 supports male fertility via the redox regulation of spermatogenesis. J. Biol. Chem. 298 (8), 102183. doi:10.1016/j.jbc.2022.102183
Ellerman, D. A., Myles, D. G., and Primakoff, P. (2006). A role for sperm surface protein disulfide isomerase activity in gamete fusion: evidence for the participation of ERp57. Dev. Cell 10 (6), 831–837. doi:10.1016/j.devcel.2006.03.011
Ernesto, J. I., Weigel Muñoz, M., Battistone, M. A., Vasen, G., Martínez-López, P., Orta, G., et al. (2015). CRISP1 as a novel CatSper regulator that modulates sperm motility and orientation during fertilization. J. Cell Biol. 210 (7), 1213–1224. doi:10.1083/jcb.201412041
Escoffier, J., Arnaud, B., Kaba, M., Hograindleur, J. P., Le Blévec, E., Martinez, G., et al. (2020). Pantoprazole, a proton-pump inhibitor, impairs human sperm motility and capacitation in vitro. Andrology 8, 1795–1804. doi:10.1111/andr.12855
Favia, M., Gerbino, A., Notario, E., Tragni, V., Sgobba, M. N., Dell’aquila, M. E., et al. (2022). The non-gastric H+/K+ ATPase (ATP12A) is expressed in mammalian spermatozoa. Int. J. Mol. Sci. 23 (3), 1048. doi:10.3390/ijms23031048
Finkelstein, M., Etkovitz, N., and Breitbart, H. (2010). Role and regulation of sperm gelsolin prior to fertilization. J. Biol. Chem. 285, 39702–39709. doi:10.1074/jbc.M110.170951
Frolikova, M., Manaskova-Postlerova, P., Cerny, J., Jankovicova, J., Simonik, O., Pohlova, A., et al. (2018). CD9 and CD81 interactions and their structural modelling in sperm prior to fertilization. Int. J. Mol. Sci. 19 (4), 1236. doi:10.3390/ijms19041236
Gibb, Z., Lambourne, S. R., Curry, B. J., Hall, S. E., and Aitken, R. J. (2016). Aldehyde dehydrogenase plays a pivotal role in the maintenance of stallion sperm motility. Biol. Reprod. 94 (6), 133. doi:10.1095/biolreprod.116.140509
Girouard, J., Frenette, G., and Sullivan, R. (2011). Comparative proteome and lipid profiles of bovine epididymosomes collected in the intraluminal compartment of the caput and cauda epididymidis. Int. J. Androl. 34 (2), e475–e486. doi:10.1111/j.1365-2605.2011.01203.x
Gyamera-Acheampong, C., and Mbikay, M. (2009). Proprotein convertase subtilisin/kexin type 4 in mammalian fertility: a review. Hum. Reprod. Update 15, 237–247. doi:10.1093/humupd/dmn060
Hu, M., Li, L., Liu, S., Lou, Y., Wang, L., Le, F., et al. (2020). Decreased expression of MRE11 and RAD50 in testes from humans with spermatogenic failure. J. Assist. Reprod. Genet. 37 (2), 331–340. doi:10.1007/s10815-019-01686-5
Iamsaard, S., Vanichviriyakit, R., Hommalai, G., Saewu, A., Srakaew, N., Withyachumnarnkul, B., et al. (2011). Enzymatic activity of sperm proprotein convertase is important for mammalian fertilization. J. Cell Physiol. 226 (11), 2817–2826. doi:10.1002/jcp.22626
Jankovicova, J., Frolikova, M., Palenikova, V., Valaskova, E., Cerny, J., Secova, P., et al. (2020). Expression and distribution of CD151 as a partner of alpha6 integrin in male germ cells. Sci. Rep. 10 (1), 4374. doi:10.1038/s41598-020-61334-2
Jeppesen, D. K., Fenix, A. M., Franklin, J. L., Higginbotham, J. N., Zhang, Q., Zimmerman, L. J., et al. (2019). Reassessment of exosome composition. Cell 177, 428–445. doi:10.1016/j.cell.2019.02.029
Jimenez, T., McDermott, J. P., Sánchez, G., and Blanco, G. (2011). Na,K-ATPase alpha4 isoform is essential for sperm fertility. Proc. Natl. Acad. Sci. U. S. A. 108, 644–649. doi:10.1073/pnas.1016902108
Juyena, N. S., and Stelletta, C. (2012). Seminal plasma: an essential attribute to spermatozoa. J. Androl. 33, 536–551. doi:10.2164/jandrol.110.012583
Karanwal, S., Pal, A., Chera, J. S., Batra, V., Kumaresan, A., Datta, T. K., et al. (2023). Identification of protein candidates in spermatozoa of water buffalo (Bubalus bubalis) bulls helps in predicting their fertility status. Front. Cell Dev. Biol. 11, 1119220. doi:10.3389/fcell.2023.1119220
Karuthadurai, T., Das, D. N., Kumaresan, A., Sinha, M. K., Kamaraj, E., Nag, P., et al. (2022). Sperm transcripts associated with odorant binding and olfactory transduction pathways are altered in breeding bulls producing poor-quality semen. Front. Vet. Sci. 9, 799386. doi:10.3389/fvets.2022.799386
Keerthikumar, S., Chisanga, D., Ariyaratne, D., Al Saffar, H., Anand, S., Zhao, K., et al. (2016). ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 428, 688–692. doi:10.1016/j.jmb.2015.09.019
Lalancette, C., Platts, A. E., Johnson, G. D., Emery, B. R., Carrell, D. T., and Krawetz, S. A. (2009). Identification of human sperm transcripts as candidate markers of male fertility. J. Mol. Med. 87, 735–748. doi:10.1007/s00109-009-0485-9
Leahy, T., Rickard, J. P., Pini, T., Gadella, B. M., and de Graaf, S. P. (2020). Quantitative proteomic analysis of seminal plasma, sperm membrane proteins, and seminal extracellular vesicles suggests vesicular mechanisms aid in the removal and addition of proteins to the ram sperm membrane. Proteomics 20 (12), e1900289. doi:10.1002/pmic.201900289
Lee, R. K. K., Tseng, H. C., Hwu, Y. M., Fan, C. C., Lin, M. H., Yu, J. J., et al. (2018). Expression of cystatin C in the female reproductive tract and its effect on human sperm capacitation. Reproductive Biol. Endocrinol. 16 (1), 8. doi:10.1186/s12958-018-0327-0
Lei, B., Xie, L. X., Zhang, S. B., Wan, B., Zhong, L. R., Zhou, X. M., et al. (2020). Phosphoribosyl-pyrophosphate synthetase 2 (PRPS2) depletion regulates spermatogenic cell apoptosis and is correlated with hypospermatogenesis. Asian J. Androl. 22 (5), 493–499. doi:10.4103/aja.aja_122_19
Li, C., Yu, R., Liu, H., Qiao, J., Zhang, F., Mu, S., et al. (2023). Sperm acrosomal released proteome reveals MDH and VDAC3 from mitochondria are involved in acrosome formation during spermatogenesis in Eriocheir sinensis. Gene 887, 147784. doi:10.1016/J.GENE.2023.147784
Luo, X., Guo, Y., Huang, Y., Cheng, M., Wu, X., and Gong, Y. (2022). Characterization and proteomics of chicken seminal plasma extracellular vesicles. Reproduction Domest. Animals 57 (1), 98–110. doi:10.1111/rda.14033
Madakashira, B., Corbett, L., Zhang, C., Paoli, P., Casement, J. W., Mann, J., et al. (2017). Variant Histone H2afv reprograms DNA methylation during early zebrafish development. Epigenetics 12, 811–824. doi:10.1080/15592294.2017.1359382
Mancuso, F., Calvitti, M., Milardi, D., Grande, G., Falabella, G., Arato, I., et al. (2018). Testosterone and FSH modulate Sertoli cell extracellular secretion: proteomic analysis. Mol. Cell Endocrinol. 476, 1–7. doi:10.1016/J.MCE.2018.04.001
Metz, C. B., Hinsch, G. W., and Anika, J. L. (1968). Ultrastructure and antigens of particles from rabbit semen. J. reproduction Fertil. 17 (1), 195–198. doi:10.1530/jrf.0.0170195
Miller, D., Brinkworth, M., and Iles, D. (2010). Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 139, 287–301. doi:10.1530/REP-09-0281
Muhammad Aslam, M. K., Kumaresan, A., Yadav, S., Mohanty, T. K., and Datta, T. K. (2019). Comparative proteomic analysis of high- and low-fertile buffalo bull spermatozoa for identification of fertility-associated proteins. Reproduction Domest. Animals 54, 786–794. doi:10.1111/rda.13426
Muhammad Aslam, M. K., Sharma, V. K., Pandey, S., Kumaresan, A., Srinivasan, A., Datta, T. K., et al. (2018). Identification of biomarker candidates for fertility in spermatozoa of crossbred bulls through comparative proteomics. Theriogenology 119, 43–51. doi:10.1016/J.THERIOGENOLOGY.2018.06.021
Murdica, V., Giacomini, E., Alteri, A., Bartolacci, A., Cermisoni, G. C., Zarovni, N., et al. (2019). Seminal plasma of men with severe asthenozoospermia contain exosomes that affect spermatozoa motility and capacitation. Fertil. Steril. 111, 897–908. doi:10.1016/j.fertnstert.2019.01.030
Mutoji, K., Singh, A., Nguyen, T., Gildersleeve, H., Kaucher, A. V., Oatley, M. J., et al. (2016). TSPAN8 expression distinguishes spermatogonial stem cells in the prepubertal mouse testis. Biol. Reprod. 95, 117. doi:10.1095/biolreprod.116.144220
Neumann, G. B., Korkuć, P., Arends, D., Wolf, M. J., May, K., König, S., et al. (2023). Genomic diversity and relationship analyses of endangered German Black Pied cattle (DSN) to 68 other taurine breeds based on whole-genome sequencing. Front. Genet. 13, 993959. doi:10.3389/fgene.2022.993959
Nixon, B., De Iuliis, G. N., Hart, H. M., Zhou, W., Mathe, A., Bernstein, I. R., et al. (2019). Proteomic profiling of mouse epididymosomes reveals their contributions to post-testicular sperm maturation. Mol. Cell. Proteomics 18 (Suppl. 1), S91–S108. doi:10.1074/mcp.RA118.000946
Ohbo, K., Yoshida, S., Ohmura, M., Ohneda, O., Ogawa, T., Tsuchiya, H., et al. (2003). Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Dev. Biol. 258, 209–225. doi:10.1016/S0012-1606(03)00111-8
Pal, A., Karanwal, S., Chera, J. S., Batra, V., Kumaresan, A., Sarwalia, P., et al. (2023). Circulatory extracellular vesicle derived miR-195-5p promotes cellular apoptosis and suppresses cell proliferation in the buffalo endometrial primary cell culture. Sci. Rep. 13 (1), 16703. doi:10.1038/s41598-023-43530-y
Pardede, B. P., Agil, M., and Supriatna, I. (2020). Protamine and other proteins in sperm and seminal plasma as molecular markers of bull fertility. Vet. World 13 (3), 556–562. doi:10.14202/vetworld.2020.556-562
Park, Y. J., Kwon, W. S., Oh, S. A., and Pang, M. G. (2012). Fertility-related proteomic profiling bull spermatozoa separated by percoll. J. Proteome Res. 11, 4162–4168. doi:10.1021/pr300248s
Paul, N., Kumaresan, A., Das Gupta, M., Nag, P., Guvvala, P. R., Kuntareddi, C., et al. (2021a). Transcriptomic profiling of Buffalo spermatozoa reveals dysregulation of functionally relevant mRNAs in low-fertile bulls. Front. Vet. Sci. 7, 609518. doi:10.3389/fvets.2020.609518
Pereira, R., Sá, R., Barros, A., and Sousa, M. (2017). Major regulatory mechanisms involved in sperm motility. Asian J. Androl. 19, 5–14. doi:10.4103/1008-682X.167716
Potts, R. J., Notarianni, L. J., and Jefferies, T. M. (2000). Seminal plasma reduces exogenous oxidative damage to human sperm, determined by the measurement of DNA strand breaks and lipid peroxidation. Mutat. Res. 447 (2), 249–256. doi:10.1016/s0027-5107(99)00215-8
Rahman, M. S., Lee, J. S., Kwon, W. S., and Pang, M. G. (2013). Sperm proteomics: road to male fertility and contraception. Int. J. Endocrinol. 2013, 360986. doi:10.1155/2013/360986
Renneberg, H., Konrad, L., and Aumüller, G. (1995). Immunohistochemistry of secretory particles (“vesiculosomes”) from the epithelium of bovine seminal vesicles and ampulla of the vas deferens. Acta anat. 153 (4), 273–281. doi:10.1159/000147728
Rodríguez, A. L., Rijsselaere, T., Beek, J., Vyt, P., Van Soom, A., and Maes, D. (2013). Boar seminal plasma components and their relation with semen quality. Syst. Biol. Reprod. Med. 59 (1), 5–12. doi:10.3109/19396368.2012.725120
Rodriguez-Caro, H., Dragovic, R., Shen, M., Dombi, E., Mounce, G., Field, K., et al. (2019). In vitro decidualisation of human endometrial stromal cells is enhanced by seminal fluid extracellular vesicles. J. Extracell. Vesicles 8 (1), 1565262. doi:10.1080/20013078.2019.1565262
Rowlison, T., Cleland, T. P., Ottinger, M. A., and Comizzoli, P. (2020). Novel proteomic profiling of epididymal extracellular vesicles in the domestic cat reveals proteins related to sequential sperm maturation with differences observed between normospermic and teratospermic individuals. Mol. Cell. Proteomics 19 (12), 2090–2103. doi:10.1074/mcp.RA120.002251
Royo, F., Théry, C., Falcón-Pérez, J. M., Nieuwland, R., and Witwer, K. W. (2020). Methods for separation and characterization of extracellular vesicles: results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells 9 (9), 1955. doi:10.3390/cells9091955
Saari, S., Andjelković, A., Garcia, G. S., Jacobs, H. T., and Oliveira, M. T. (2017). Expression of Ciona intestinalis AOX causes male reproductive defects in Drosophila melanogaster. BMC Dev. Biol. 17 (1), 9. doi:10.1186/s12861-017-0151-3
Saito, T., Wada, I., and Inoue, N. (2019). Sperm IZUMO1-dependent gamete fusion influences male fertility in mice. Int. J. Mol. Sci. 20 (19), 4809. doi:10.3390/ijms20194809
Salicioni, A. M., Gervasi, M. G., Sosnik, J., Tourzani, D. A., Nayyab, S., Caraballo, D. A., et al. (2020). Testis-specific serine kinase protein family in male fertility and as targets for non-hormonal male contraception. Biol. Reproduction 103 (2), 264–274. doi:10.1093/biolre/ioaa064
Shi, J., Qiao, F., Ye, M., Jiang, T., Liu, J., Zhang, M., et al. (2022). CSE1L/CAS regulates cell proliferation through CDK signalling in mouse spermatogenesis. Cell Prolif. 55 (11), e13334. doi:10.1111/cpr.13334
Siciliano, L., Marcianò, V., and Carpino, A. (2008). Prostasome-like vesicles stimulate acrosome reaction of pig spermatozoa. Reproductive Biol. Endocrinol. 6, 5. doi:10.1186/1477-7827-6-5
Sinha, N., Puri, P., Nairn, A. C., and Vijayaraghavan, S. (2013). Selective ablation of Ppp1cc gene in testicular germ cells causes oligo-teratozoospermia and infertility in mice. Biol. Reprod. 89 (5), 128. doi:10.1095/biolreprod.113.110239
Somashekar, L., Selvaraju, S., Parthipan, S., Patil, S. K., Binsila, B. K., Venkataswamy, M. M., et al. (2017). Comparative sperm protein profiling in bulls differing in fertility and identification of phosphatidylethanolamine-binding protein 4, a potential fertility marker. Andrology 5, 1032–1051. doi:10.1111/andr.12404
Stival, C., Puga Molina, L. D. C., Paudel, B., Buffone, M. G., Visconti, P. E., and Krapf, D. (2016). “Sperm capacitation and acrosome reaction in mammalian sperm,” in Advances in anatomy embryology and cell Biology (Springer Verlag), 93–106. doi:10.1007/978-3-319-30567-7_5
Sun, Y. H., Wang, A., Song, C., Shankar, G., Srivastava, R. K., Au, K. F., et al. (2021). Single-molecule long-read sequencing reveals a conserved intact long RNA profile in sperm. Nat. Commun. 12 (1), 1361. doi:10.1038/s41467-021-21524-6
Takeshima, S. N., and Aida, Y. (2006). Structure, function and disease susceptibility of the bovine major histocompatibility complex. Animal Sci. J. 77, 138–150. doi:10.1111/j.1740-0929.2006.00332.x
Tang, Y., Sun, L., Li, S., Liu, H., Luo, L., Chen, Z., et al. (2023). Role of cytoskeleton-related proteins in the acrosome reaction of Eriocheir sinensis spermatozoa. BMC Genom Data 24 (1), 4. doi:10.1186/s12863-023-01112-x
Tash, J. S., Krinks, M., Patel, J., Means, R. L., Klee, C. B., and Means, A. R. (1988). Identification, characterization, and functional correlation of calmodulin-dependent protein phosphatase in sperm. J. Cell Biol. 106 (5), 1625–1633. doi:10.1083/jcb.106.5.1625
Tong, S., Yin, C., Ge, Y., Ren, Z., Tao, J., and Liu, Y. (2022). Albumin (ALB) and protein disulfide isomerase family A member 4 (PDIA4) are novel markers to predict sperm freezability of Erhualian boar. Cryobiology 109, 37–43. doi:10.1016/j.cryobiol.2022.09.006
Turano, C., Coppari, S., Altieri, F., and Ferraro, A. (2002). Proteins of the PDI family: unpredicted non-ER locations and functions. J. Cell Physiol. 193 (2), 154–163. doi:10.1002/jcp.10172
Ugur, M. R., Kutchy, N. A., Menezes, E. B., Ul-Husna, A., Haynes, B. P., Uzun, A., et al. (2019). Retained acetylated histone four in bull sperm associated with fertility. Front. Vet. Sci. 6, 223. doi:10.3389/fvets.2019.00223
Verma, A., Rajput, S., De, S., Kumar, R., Chakravarty, A. K., and Datta, T. K. (2014). Genome-wide profiling of sperm DNA methylation in relation to buffalo (Bubalus bubalis) bull fertility. Theriogenology 82, 750–759. doi:10.1016/j.theriogenology.2014.06.012
Vickram, A. S., Anbarasu, K., Gulothungan, G., Thanigaivel, S., Nanmaran, R., and Palanivelu, J. (2022). Characterization of human prostasomes protein Clusterin (macromolecule)–a novel biomarker for male infertility diagnosis and prognosis. J. Biomol. Struct. Dyn. 40, 3979–3988. doi:10.1080/07391102.2020.1852960
Wang, H., Zhu, Y., Tang, C., Zhou, Z., Wang, Z., Li, Z., et al. (2022). Reassessment of the proteomic composition and function of extracellular vesicles in the seminal plasma. Endocrinol. (United States) 163 (1), bqab214. doi:10.1210/endocr/bqab214
Wang, S., Qiao, H., Wang, P., Wang, Y., and Qin, D. (2021). ZDHHC19 is dispensable for spermatogenesis, but is essential for sperm functions in mice. Int. J. Mol. Sci. 22 (16), 8894. doi:10.3390/ijms22168894
Wang, X., Kang, J. Y., Wei, L., Yang, X., Sun, H., Yang, S., et al. (2019). PHF7 is a novel histone H2A E3 ligase prior to histone-toprotamine exchange during spermiogenesis. Dev. Camb. 146 (13), dev175547. doi:10.1242/dev.175547
Woo, A. L., James, P. F., and Lingrel, J. B. (2000). Sperm motility is dependent on a unique isoform of the Na,K-ATPase. J. Biol. Chem. 275, 20693–20699. doi:10.1074/jbc.M002323200
Xu, B., Hao, Z., Jha, K. N., Zhang, Z., Urekar, C., Digilio, L., et al. (2008). Targeted deletion of Tssk1 and 2 causes male infertility due to haploinsufficiency. Dev. Biol. 319 (2), 211–222. doi:10.1016/j.ydbio.2008.03.047
Xu, Z., Xie, Y., Zhou, C., Hu, Q., Gu, T., Yang, J., et al. (2020). Expression pattern of seminal plasma extracellular vesicle small RNAs in boar semen. Front. Vet. Sci. 7, 585276. doi:10.3389/fvets.2020.585276
Yanagimachi, R. (1994). Fertility of mammalian spermatozoa: its development and relativity. Zygote 2 (4), 371–372. doi:10.1017/s0967199400002240
Yu, K., Xiao, K., Sun, Q. Q., Liu, R. F., Huang, L. F., Zhang, P. F., et al. (2023). Comparative proteomic analysis of seminal plasma exosomes in buffalo with high and low sperm motility. BMC Genomics 24 (1), 8. doi:10.1186/s12864-022-09106-2
Yuan, B., Wu, W., Chen, M., Gu, H., Tang, Q., Guo, D., et al. (2017). From the cover: metabolomics reveals a role of betaine in prenatal DBP exposure-induced epigenetic transgenerational failure of spermatogenesis in rats. Toxicol. Sci. 158, 356–366. doi:10.1093/toxsci/kfx092
Zaborowski, M. P., Balaj, L., Breakefield, X. O., and Lai, C. P. (2015). Extracellular vesicles: composition, biological relevance, and methods of study. Bioscience 65, 783–797. doi:10.1093/biosci/biv084
Zhang, B., Denomme, M. M., White, C. R., Leung, K. Y., Lee, M. B., Greene, N. D. E., et al. (2015). Both the folate cycle and betaine-homocysteine methyltransferase contribute methyl groups for DNA methylation in mouse blastocysts. FASEB J. 29, 1069–1079. doi:10.1096/fj.14-261131
Zhao, W., Su, J., Wang, Y., Qian, T., and Liu, Y. (2019). Functional importance of palmitoyl protein thioesterase 1 (PPT1) expression by Sertoli cells in mediating cholesterol metabolism and maintenance of sperm quality. Mol. Reprod. Dev. 86 (8), 984–998. doi:10.1002/mrd.23173
Keywords: buffalo, bull fertility, seminal plasma extracellular vesicles, high-throughput proteomics, immunolocalization
Citation: Badrhan S, Karanwal S, Pal A, Chera JS, Chauhan V, Patel A, Bhakat M, Datta TK and Kumar R (2024) Differential protein repertoires related to sperm function identified in extracellular vesicles (EVs) in seminal plasma of distinct fertility buffalo (Bubalus bubalis) bulls. Front. Cell Dev. Biol. 12:1400323. doi: 10.3389/fcell.2024.1400323
Received: 13 March 2024; Accepted: 03 July 2024;
Published: 29 July 2024.
Edited by:
Pascale Lybaert, Université libre de Bruxelles, BelgiumReviewed by:
Clara Isabel Marin-Briggiler, CONICET Institute of Biology and Experimental Medicine (IBYME), ArgentinaSouvik Dey, Manipal Institute of Technology, India
Copyright © 2024 Badrhan, Karanwal, Pal, Chera, Chauhan, Patel, Bhakat, Datta and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakesh Kumar, cmFrZXNoY2lmdEBnbWFpbC5jb20=
 Shiva Badrhan1
Shiva Badrhan1 Seema Karanwal
Seema Karanwal Ankit Pal
Ankit Pal Jatinder Singh Chera
Jatinder Singh Chera Tirtha K. Datta
Tirtha K. Datta Rakesh Kumar
Rakesh Kumar