- Department of Endodontics, Stomatological Hospital, School of Stomatology, Southern Medical University, Guangzhou, Guangdong, China
Epigenetics refers to the mechanisms such as DNA methylation and histone modification that influence gene expression without altering the DNA sequence. These epigenetic modifications can regulate gene transcription, splicing, and stability, thereby impacting cell differentiation, development, and disease occurrence. The formation of dentin is intrinsically linked to the odontogenic differentiation of dental pulp stem cells (DPSCs), which are recognized as the optimal cell source for dentin-pulp regeneration due to their varied odontogenic potential, strong proliferative and angiogenic characteristics, and ready accessibility Numerous studies have demonstrated the critical role of epigenetic regulation in DPSCs differentiation into specific cell types. This review thus provides a comprehensive review of the mechanisms by which epigenetic regulation controls the odontogenesis fate of DPSCs.
1 Introduction
Pulpitis refers to the inflammation of the dental pulp, typically caused by bacterial infection, resulting in severe pain (Kakehashi et al., 1965; Brännström and Nyborg, 1973) Over time, the pulp tissue may degenerate, leading to apical periodontitis, osteomyelitis, and eventually necessitating tooth extraction (Shi et al., 2005). However, effective management of pulp infections can stimulate the formation of reparative dentin and protect the pulp from further damage (Tronstad and Mjör, 1972). Among the treatment options, vital pulp therapy (VPT) presents a less invasive and more conservative approach compared to root canal therapy (RCT) (Ferracane et al., 2010), although the inability to accurately assess the severity of pulpitis clinically limits its potential for restorative success (Barthel et al., 2000; Ricucci et al., 2014). As regenerative medicine advances, modulating the expression of specific molecules can effectively suppress inflammation, promote the growth of reparative dentin, and enhance the overall regenerative capacity of the pulp (Gangolli et al., 2019).
Human dental pulp-derived stem cells (hDPSCs) are a type of mesenchymal stem cell developed from neural crest cells, which originate from ectodermal tissue (Nuti et al., 2016). In 2000, Gronthos et al. (2000) were the first to successfully culture colony-like cells from human third molars in 2000. These cells were found capable of differentiating into odontoblast-like cells and forming dentin-like structures, indicating the presence of stem cells in pulp tissue and leading to the initial concept of DPSCs. Subsequently, Miura et al. (2003a) isolated stem cells from the pulp of human exfoliated deciduous teeth (SHEDs), which are similar to mesenchymal stem cells (MSCs) in their undifferentiated or hypodifferentiated state. DPSCs are highly valued for pulp regeneration due to their unique differentiation advantages. Under specific induction conditions, DPSCs can differentiate into various functional cells such as osteoblasts, adipocytes, chondrocytes, odontoblast-like cells, and neuronal cells (Anitua et al., 2018). The odontogenic differentiation of dental pulp stem cells is presently a field that lacks comprehensive study, and the active investigation of the molecular pathways of dentin differentiation, along with the creation of reliable biomarkers, is imperative for examining the formation of the pulp-dentin complex. It has been discovered that chromatin state, transcriptional regulation, and epigenetic modifications collectively perform a part during the self-renewal and differentiation process of DPSCs (Bayarsaihan, 2016). Epigenetics has emerged as a prominent research focus due to its ability to modulate functional genes without altering DNA sequences (Wu and Morris, 2001). Thus, this paper offers both theoretical and empirical support for clinical endodontic practices, especially VPT, by exploring the immunophenotypes and related epigenetic processes that guide DPSCs potential for odontogenic differentiation.
2 Epigenetic
Epigenetics is a branch of genetics that gradually took shape in the 1980s to investigate how changes in non-genetic sequences impact gene expression levels (Goldberg et al., 2007). The term was initially introduced by Conrad Waddington in 1942 to illustrate the intricate developmental journey from genotype to phenotype (Waddington, 2012). Research has demonstrated that epigenetic modifications play a critical role in the regulation of embryonic stem cells (ESCs) and DPSCs, particularly in governing their self-renewal and differentiation capabilities (Anitua et al., 2018). The most extensively studied epigenetic mechanisms encompass DNA methylation, histone modifications, and non-coding RNA-mediated regulation of gene transcription (Figure 1). Given the breadth of research on classical modifications in contemporary stem cell studies, we aim to revisit and update the existing research on the epigenetic mechanisms of odontogenic differentiation.
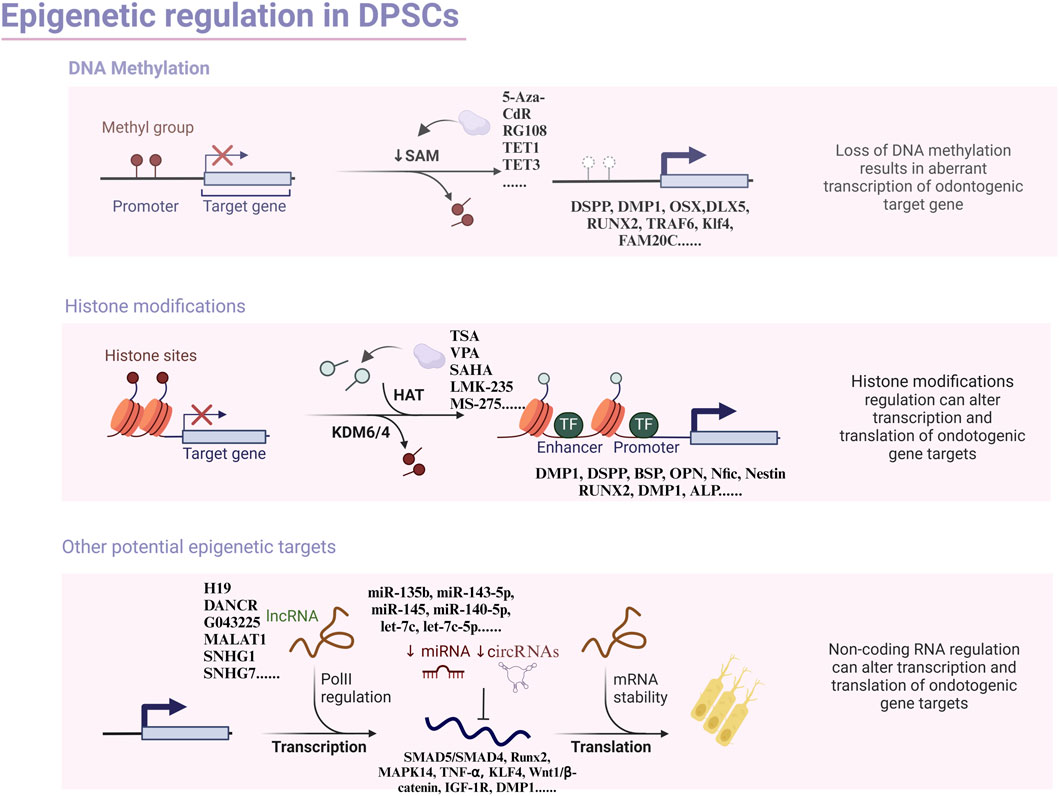
Figure 1. Overview of the principal types of epigenetic modification with focus on odontogenic differentiation in dental pulp stem cells.
2.1 DNA methylation
DNA methylation is a ubiquitous process in eukaryotic cells that serves as an established and inheritable epigenetic marker, influencing chromatin organization and gene transcription. This process entails attaching a methyl group to the 5′carbon atom of cytosine (C) within the cytosine-phosphothio-nyl-guanine (CpG) dinucleotide, creating 5-methylcytosine (5 mC). The methyl donor, S-adenosylmethionine (SAM), provides the necessary methyl group, and DNA methyltransferase (DNMT) enzymes catalyze the reaction. (Radhakrishnan et al., 2011). The primary target of DNA methylation is the CpG island, a genomic region rich in cytosine and guanine (CpG) sequences, primarily located near gene promoters and exon 1. CpG islands are typically unmethylated, but high levels of methylation result in stable gene silencing (Bird, 2002). The regulation of CpG methylation levels is dynamic and controlled by DNA methyltransferase (DNMT) and DNA demethylase (TET) enzymes (Li et al., 2015a; Zhang et al., 2018). The DNMT family comprises DNMT1, DNMT3A, DNMT3B, and DNMT3L, with only DNMTs (−1, -3A, and -3B) involved in DNA methylation, while DNMT-3L lacks enzyme activity (Kareta et al., 2006). The TET family, including TET1, TET2, and TET3, and activation-induced deaminases (AID) are required to demethylate DNA by lowering methylation marks in the course of DNA replication (Bochtler et al., 2017). The expression of CpG genes is regulated through two main mechanisms. First, methyl CpG-binding proteins recognize and bind specifically to methylated CpG regions, attracting transcriptional repression cofactors that suppress DNA transcription (Gao et al., 2014), Second, methylation at CpG sites can physically block the binding of DNA-binding proteins, such as transcription factors, to their target sites, thereby inhibiting transcription (Hlady et al., 2014).
2.2 Histone modifications
Histones, which encompass H1, H2A, H2B, H3 as well as H4, are incorporated with DNA to constitute nucleosomes, the fundamental units of chromatin. It has been demonstrated that histones play a vital role in promoting transcription in promoter regions (Lee et al., 2004). Moreover, histones undergo various post-translational protein modifications (PTPM), such as methylation, acetylation, ubiquitination, phosphorylation, succination, and ADP-ribosylation (Jenuwein and Allis, 2001), that change their interactions with DNA and with each other (Venkatesh and Workman, 2015). These modifications are crucial for sustaining chromatin structure, nucleosome stability, and gene expression (Zheng et al., 2014; Lawrence et al., 2016; Bascom and Schlick, 2018; Yi and Kim, 2018). Among all the histone post-translational modifications (HPTMs), methylation and acetylation are the most extensively investigated.
2.2.1 Histone methylation
Histone methylation, a covalent modification, occurs on arginine and lysine residues. This process involves transferring a methyl group from SAM to a lysine or arginine site in the presence of histone methyltransferase (Strahl and Allis, 2000). Typically, histone methylation occurs at the N-terminal end of histone H3 or H4. Arginine can undergo mono- or dimethylation, while lysine can be mono-, di-, or trimethylated (Pedersen and Helin, 2010). Stem cell differentiation processes produce enzymes that modify methylation levels at specific genes, influencing gene expression. Depending on the gene site, methylation can either repress or promote transcription.
2.2.2 Histone acetylation
Histone acetylation is predominantly facilitated by histone acetyltransferases (HAT) and histone deacetylases (HDAC) and typically occurs on lysine residues. This is a reversible chemical reaction where HAT catalyzes the acetylation of histone N-terminal lysine residues, neutralizing the positive charge on histones. This, in turn, reduces the affinity of histones for negatively charged DNA, loosening the chromatin structure and ultimately promoting the binding of transcription factors to DNA for the activation of specific genes (Galvani and Thiriet, 2015). Conversely, HDAC-mediated histone deacetylation can lead to gene silencing, as DNA neutralization by histone binding, condensing the chromatin structure, and inhibiting transcription factor binding (Meier and Brehm, 2014). Our comprehension of the mechanisms underlying alternative forms of histone modifications and the implicated signaling pathways remains constrained, warranting further investigations.
2.3 ncRNAs
Non-coding RNAs are synthesized from the reverse transcription of protein-encoding genes and lack an open reading frame. This group includes ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and microRNA (miRNA) (Kaikkonen et al., 2011). Non-coding RNA transcripts outnumber mRNA transcripts, yet they do not translate into proteins; instead, they play a role in epigenetically regulating gene expression (Palazzo and Lee, 2015). Among these, miRNA, a highly conserved non-coding single-stranded RNA consisting of 20–23 nucleotides, is found in eukaryotes and has been a central focus of epigenetics research. MiRNA is transcribed from intergenic DNA sequences as an initial transcript known as pri-mRNA, which is then processed into pre-mRNA by the nuclease Drosha and further refined into mature miRNA by Dicer. (Lee et al., 2003; Liu et al., 2004; Cipriani et al., 2022). The mature miRNA is subsequently incorporated into the AGO family of proteins to form the RNA-induced silencing complex (RISC). miRNAs can bind to the 3′-untranslated region (UTR) of mRNAs, resulting in the degradation or post-transcriptional repression of target mRNAs, thereby modulating gene expression (Ha and Kim, 2014; Seong et al., 2014). Additionally, competing endogenous RNAs (ceRNAs) indirectly regulate downstream targets by competing with miRNAs for binding. Among ceRNAs, circular RNA (circRNA), a family of ncRNAs with a ring-like covalent structure, resists RNA enzyme digestion, making it more stable and less susceptible to degradation (Qu et al., 2015; Guo et al., 2020; Zhang et al., 2020). Furthermore, circRNA is widely distributed in living organisms and exhibits expression specificity and high evolutionary conservation (Jeck et al., 2013). In some instances, the richness of circRNA is more than 10-fold that of the relevant linear mRNA, making it an ideal disease marker and treatment target with important clinical and research implications (Li et al., 2015b; Wang et al., 2016). Long non-coding RNAs (LncRNAs) are a new heterogeneous linear ncRNA with more than 200 nucleotides, and their functions are mainly divided into three aspects: modifying chromosomes for apparent regulation, participation in transcriptional regulation through interaction with transcription factors, and post-transcriptional or translation regulation by affecting mRNA processing. LncRNAs may serve as a biomarker in multiple biological processes (Carpenter et al., 2013; Wang et al., 2013; Legnini et al., 2014; Zhu et al., 2014), and increasing evidence suggests that lncRNAs have a major contribution to the regulation of odontogenic differentiation in hDPSCs.
3 Dental pulp stem cells
3.1 Discovery and characterization of DPSCs
Stem cells possess the remarkable ability to replicate, renew, and give rise to specific cell types, significantly influencing tissue development, repair, and homeostasis (McNeely et al., 2020) Dental pulp stem cells, categorized as DPSC and SHED depending on their origin, were successfully isolated and identified in 2000 and 2003 (Gronthos et al., 2000; Miura et al., 2003b). Both types exhibit the typical characteristics of MSC. DPSCs were the first MSCs of oral and maxillofacial origin to be isolated, paving the way for subsequent discoveries of similar cells, such as SHED, periodontal stem cells, and apical papilla stem cells (Esposito et al., 2003; Innes, 2010; Guven et al., 2011; Jeong et al., 2014). These stem cells express MSC markers but lack haematopoietic markers (Liu et al., 2015a) However, DPSCs are heterogeneous, with varying marker expressions. Specific cell surface markers enable the isolation of DPSC subpopulations for targeted differentiation in clinical settings. Researchers suggest that CD271+ DPSCs possess greater potential for odontogenic differentiation (Alvarez et al., 2015), while the expression level of CD146 correlates with the functionality of dental pulp stem cells, marking it as a key functional indicator of their efficacy (Ma et al., 2021).
3.2 Differentiation and application of DPSCs
DPSCs share similarities with bone marrow MSCs and express markers related to mineralization and osteogenesis (Chen et al., 2005; Ching et al., 2017). With proper induction, dental pulp stem cells can differentiate into osteoblasts, chondrocytes, adipocytes, and neural-like cells (Zhu et al., 2018) (Figure 2). In a study conducted by Alge et al. (2010), DPSCs demonstrated a higher proliferation rate and greater clonal potential compared to bone marrow MSCs. Lineage tracing in 2014 revealed that DPSCs originate from peripheral nerve-associated glia (Kaukua et al., 2014), which can differentiate into pulp cells in adult mice incisors. Furthermore, the perivascular niche, a distinct environment for various MSCs (Crisan et al., 2008), houses pulp stem cells within the perivascular region of pulp tissue (Shi and Gronthos, 2003). In vivo experiments have shown the involvement of dental pulp stem cells in angiogenesis (Iohara et al., 2009; Iohara et al., 2013), demonstrating their strong capacity for promoting pulp regeneration (Yang et al., 2015). In a groundbreaking study conducted in 2000 (Gronthos et al., 2000), Gronthos et al. transplanted DPSC-complexed hydroxyapatite/triple calcium phosphate scaffolds into the dermis of immunodeficient mice, resulting in the formation of pulpal and dentin-like tissues with organized collagen matrices, albeit without haematopoietic or adipocyte formation. This indicates that odontogenic differentiation is a major default phenotype of DPSCs. Furthermore, DPSCs derived from human mammary dental stem cells were integrated with recombinant collagen and injected into the pulp cavity. This integration enabled them to maintain their viability, rebuild vascularized pulp tissues, and differentiate into dentin cells expressing dentin sialophosphoproteins and dentin matrix proteins (Itoh et al., 2018). The diverse and complementary unity of cell populations found in pulp stem cells plays a vital role in repairing and maintaining the local homeostasis of pulp tissues. Achieving functional pulp regeneration through pulp stem cells may rely on the dynamic structure and unity of such diverse cell populations.
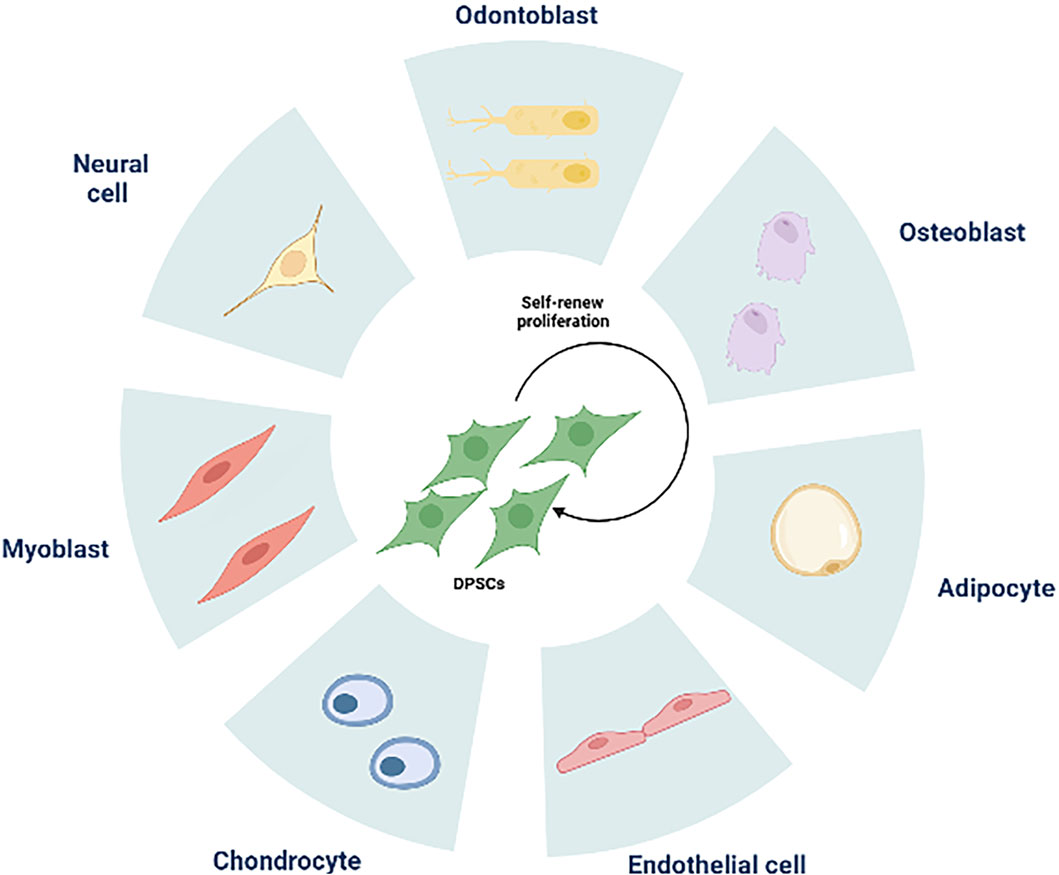
Figure 2. The multilineage differentiation potential of DPSCs. DPSCs can differentiate into odontoblasts, osteoblasts, adipocytes, endothelial cells, chondrocytes, myoblasts, neural cells.
4 Epigenetic mechanisms
4.1 DNA methylation associated with odontogenic differentiation in DPSCs
DNA demethylation activates gene transcription, while DNA methylation typically leads to gene repression. DNA methylation patterns vary among different cell types and contribute to the diverse differentiation potential of stem cells. Through the reprogramming of stem cells, pluripotent stem cells (iPSCs) can be generated for clinical therapy, with DNA methylation playing a pivotal role in this process (Godini et al., 2018) (Table 1). Genome-wide DNA methylation analysis has revealed that the methylation profile of DPSCs closely resembles that of embryonic stem (ES) and induced pluripotent stem (iPS) cells. Specifically, the downregulation of HERV-FRD and upregulation of PAX9 increase the efficacy of DPSCs in differentiating into iPS cells (Dunaway et al., 2017). DNA methylation patterns influence the expression of DPSC genes, with DNA demethylases and DNA methyltransferase (DNMT) activity affecting the differentiation potential of DPSCs. Treatment with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-Aza-CdR) reduced the proliferative capacity of DPSCs but upregulated odontogenic markers (DMP1 and DSPP) and transcription factors (OSX, RUNX2, and DLX5), increasing ALP activity and accelerating the formation of calcified nodules, thereby enhancing odontogenic differentiation capacity (Zhang et al., 2015). The novel DNA methyltransferase inhibitor RG108 also regulates DSPP and DMP1, promoting odontogenic differentiation of DPSCs (Sun et al., 2019). Evidence suggests that DNA methylation affects the transactivation of transcription factors (TFs). Kruppel-like factor (KLF4), a critical mediator of cell differentiation and proliferation, undergoes promoter region demethylation during odontoblast differentiation, enabling efficient SP1 binding and transcriptional regulation that upregulates Klf4 expression (Sun et al., 2019). This process promotes odontoblast differentiation and inhibits the growth of primary DPSCs (Lin et al., 2011).
The fate of DPSCs is also influenced by DNA demethylases, which regulate DNA methylation dynamics alongside DNA methyltransferases. The TET family of demethylases, including TET1, TET2, and TET3, plays a key role in this regulation. TET1 promotes odontoblast differentiation in DPSCs while inhibiting adipogenic and osteogenic differentiation. (Wiehle et al., 2016; Cakouros et al., 2019; Qian et al., 2021). As a DNA dioxygenase, TET1 is present in both the nucleus and cytoplasm of DPSCs, and its expression increases during odontogenic induction. However, TET1 expression decreases during early cell passage (less than six generations) (Li et al., 2014). Knockdown of TET1 inhibits both proliferation and odontogenic differentiation (Rao et al., 2016). Additionally, TET1 can promote odontogenic differentiation by regulating FAM20C demethylation, thereby upregulating FAM20C expression (Li et al., 2018a). Interestingly, TET3 is expressed gradually inPLSCs during adipogenic induction, suggesting a tissue-specific role in regulating PLSC cell fate by enhancing adipocyte differentiation. While TET3 may also play a part in the odontogenic differentiation of DPSCs, further research is needed to validate this hypothesis through extensive experiments (Argaez-Sosa et al., 2022).
Reparative dentinogenesis is a delicate balance between odontogenic differentiation and inflammation. DNA methylation plays a role in the inflammatory responses that occur in human dental pulp. During lipopolysaccharide (LPS)-induced inflammation, DNMT1 mRNA and protein levels decrease in DPSCs. DNMT1 influences the promoter methylation of the MyD88 gene and suppresses the expression of miR-146a-5p. Reduction in DNMT1 can activate the NF-κB pathway, thereby intensifying the inflammatory response (Meng et al., 2019; Mo et al., 2019). LPS-treated DPSCs exhibit elevated expression of pro-inflammatory cytokines such as GM-CSF, MCP-2, RANTES, IL-8, and IL-6. Additionally, 5-Aza-CdR activates the NF-κB and MAPK signaling pathways by decreasing the methylation level of the TRAF6 promoter in DPSCs. These findings suggest that 5-Aza-CdR promotes inflammation by activating TRAF6 (Feng et al., 2019). TET2 also modulates inflammation in dental pulp tissue by converting 5-methylcytosine to 5-hydroxymethylcytosine, which promotes DNA demethylation. Evidence suggests that TET2 enhances LPS-induced inflammation by regulating MyD88 hydroxymethylation in an in vitro culture model (Wang et al., 2018a). These results imply that DNA methylation plays a crucial role in promoting DPSCs’ odontogenic differentiation and the formation of reparative dentin both in vitro and in vivo.
In conclusion, further investigation is necessary to uncover the intricate mechanisms governing the methylation-dependent regulation of DPSCs. Such efforts will pave the way for using DPSCs in regenerative tissue bioengineering and vital pulp therapy with optimal safety and efficacy.
4.2 Histone modifications associated with odontogenic differentiation in DPSCs
4.2.1 Histone methylation
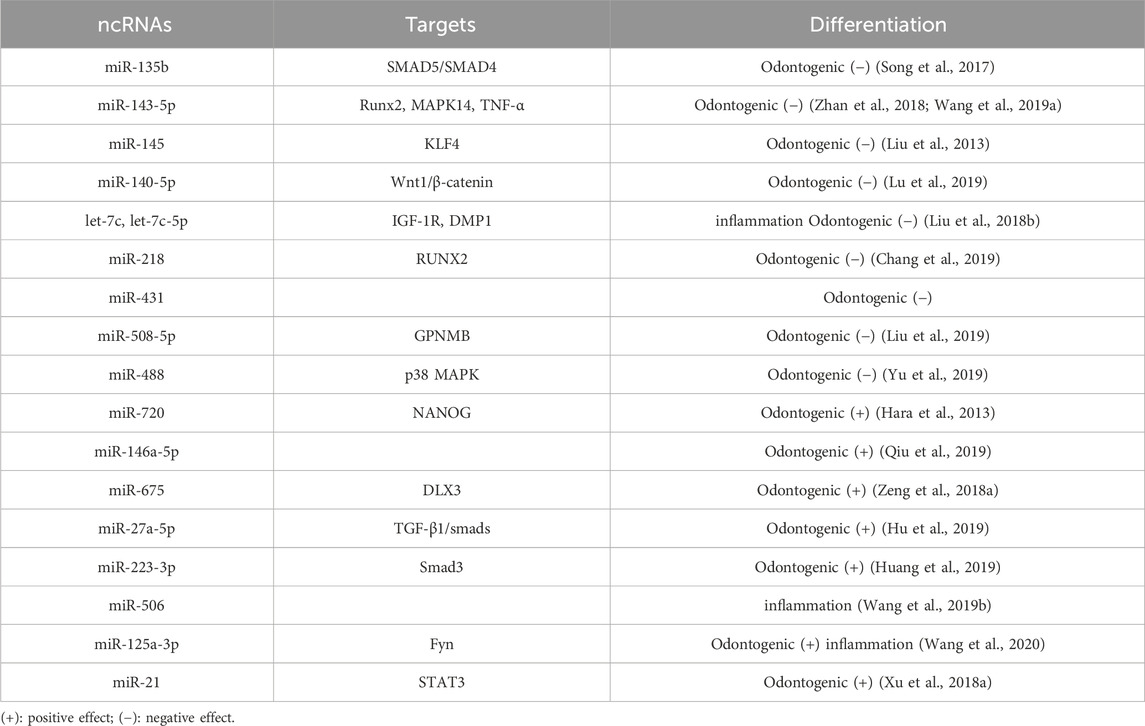
Histone methylation, a well-researched form of histone modification, involves the methylation of lysine or arginine residues in histone tails and is dynamically regulated by enzymes that control histone methylation and demethylation (Shen et al., 2017) (Table 2). A comparison of the epigenetic status between dental follicle cells (DFCs) and DPSCs showed that H3K27me3-mediated suppression of dental-derived genes DMP1 and DSPP in DFCs was not observed in DPSCs. It has been demonstrated that DPSCs exhibit higher expression of DMP1 and DSPP under osteoinductive conditions (Gopinathan et al., 2013), which is partially attributed to differences in histone methylation profiles. The histone methyltransferase EZH2, which methylates H3K27me3, decreases in activity during odontogenic differentiation, leading to lower levels of H3K27me3. Although a lack of methyltransferase activity in EZH2 was initially not expected to impact DPSCs’ differentiation into odontogenic cells, it has been shown to negatively affect this process. siEZH2 transfection of β-TCP/DPSCs implanted subcutaneously in nude mice has been observed to promote mineralized tissue formation. Additionally, chromatin immunoprecipitation (ChIP) analysis revealed that EZH2 downregulated β-catenin expression by increasing H3K27me3 methylation levels in the β-catenin promoter region, thereby inhibiting the Wnt/β-catenin signaling pathway and suppressing odontogenic differentiation (Li et al., 2018b). Histone methylation of H3K9 plays a role in the osteogenesis of DPSCs. Euchromatin Histone Methyltransferase-1 (EHMT1), an enzyme that represses transcription, demethylates H3K9, thereby promoting cellular differentiation (Kramer, 2016). During osteogenesis stimulated by BMP-2, the co-repressor CBFA22T reduces H3K9me methylation at the Runx promoter, which plays a key role in cellular mineralization and can inhibit H3K9me2 normally mediated by Euchromatin Histone Methyltransferase 1 (EHMT1) at the RUNX2 promoter. Downregulation of CBFA2T2 increases EHMT1 expression, leading to an increase in H3K9me2 methylation and subsequent inhibition of osteogenic differentiation (Huang et al., 2018).
Histone methylation is a reversible process regulated by demethylases. KDM2A, a histone demethylase, inhibits the osteogenic and odontogenic differentiation of dental DPSCs by demethylating H3K4 or H3K36, which suppresses the expression of genes such as BSP, OPN, and OCN (Du et al., 2013). Du et al. (2013) found that KDM2A initiates its demethylation activity by binding to the BCL6 co-repressor and targeting the promoter of the epiregulin (EREG) gene, reducing the methylation of H3K4 and H3K36 and thus inhibiting EREG expression, further suppressing osteogenic and odontogenic differentiation in dental-derived mesenchymal stem cells. Dong et al. (2013) observed that using shRNA to interfere with KDM2A expression in dental papilla stem cells resulted in significant increases in stemness-related genes such as SOX2 and H3K4me3 in the promoter region of NANOG. This led to an increase in SOX2 and NANOG gene expression, promoting lipogenic and chondrogenic differentiation. The Jumonji domain-containing protein D3 (JMJD3), also known as lysine-specific demethylase 6B (KDM6B), is a protein that specifically removes H3K27me2/3 methyl marks, thereby regulating gene expression and odontogenic differentiation through multiple mechanisms. Increased expression of JMJD3 enhances odontogenic differentiation, whereas its suppression through the addition of alcohol to the mineralization induction medium inhibits odontogenesis (Hoang et al., 2016). JMJD3 removes the H3K27me3 methylation mark by binding to the BMP2 promoter during odontogenic induction, thus activating the expression of transcriptional proteins associated with BMP2 and odontogenic differentiation (Xu et al., 2013). The promoter region of the Wnt5a gene contains a “bivalent domain” with both repressive H3K27me3 and active H3K4me3 markers. These modifications maintain the Wnt5a gene in a state of balance, allowing its transcription to be either repressed or activated depending on various stimuli (Bernstein et al., 2006). JMJD3 erases H3K27me3 on the Wnt5a promoter and activates Wnt5a in DPSCs under odontogenic stimulation. Conversely, JMJD3 deficiency leads to an increase in H3K27me3, which silences Wnt5a and impedes odontogenesis. Additionally, JMJD3 inhibits odontogenesis through interaction with the mixed-lineage leukemia complex (H3K4me3 methyltransferase) (Zhou et al., 2018).
An additional histone demethylase, known as lysine-specific demethylase 5A (KDM5A), is highly specific for the activity marker H3K4me3 (Shi, 2007). Depletion of KDM5A results in increased levels of H3K4me3 at the promoters of odontogenic differentiation genes, such as DMP1, OSX, OCN, and DSPP, suggesting a link between H3K4me3 and odontogenesis (Li et al., 2020). Another histone demethylase, JHDM1D, also referred to as KDM7A, is used to remove dimethyl tags at lysine nine and lysine 27 on histone H3 of target gene promoters (Huang et al., 2010; Yokoyama et al., 2010). According to a report by Yang et al. (2019), JHDM1D may inhibit osteogenesis in mouse bone marrow cells (ST2) by deactivating the Wnt signaling pathway. JHDM1D knockdown leads to increased mineralized nodule formation in human dental pulp-derived stem cells (hDPSCs) and upregulation of β-catenin expression, which is achieved by downregulating DKK1. Above results suggest that inhibiting JHDM1D expression could activate the Wnt/β-catenin signaling pathway to regulate odontogenesis in hDPSCs.
Furthermore, it has been discovered that modifications to histone methylation are implicated in the pulpal inflammatory response (Hui et al., 2017). Moreover, EZH2 has been shown to serve a key role in the inflammation and regeneration of dental pulp tissue (Hui et al., 2018). Upon TNF-α stimulation, DPSCs differentiate into dentin-producing odontoblasts and migrate to the site of infection to generate reparative dentin. Studies using immunohistochemistry and immunofluorescence have shown that infected cells exhibit significant reductions in EZH2 and H3K27me3 levels. Inhibiting EZH2 downregulates inflammatory factors such as IL-1β, IL-6, and IL-8, which suppresses DPSC proliferation but enhances osteogenesis. This suggests that EZH2 enhances inflammation and cell proliferation while suppressing osteogenic differentiation (Hui et al., 2014). Given that histone methylation is a reversible epigenetic modification, understanding its regulatory mechanism in DPSCs offers the potential to use histone methylation inhibitors in vital pulp therapy to modulate the inflammatory process.
4.2.2 Histone acetylation
The process of histone acetylation is under the control of HAT and HDAC enzymes (Marmorstein and Zhou, 2014), and it has profound consequences on the growth and specialization of various cells (Clayton et al., 2006; Fukuda et al., 2006; Cameron et al., 2016) (Table 2). HAT enzymes can enhance the odontogenesis of DPSCs by increasing histone H3 acetylation on the DSPP gene (Gu et al., 2013). For instance, the HAT enzyme p300, which is a member of the lysine acetyltransferase three family, transfers acetyl groups to lysine residues on histones. p300 predominantly targets the promoter regions of the OCN and DSPP genes, acetylating histone H3K9 and thereby promoting gene transcription and enhancing mineralization and differentiation in dental pulp cells (Wang et al., 2014; Liu et al., 2015b). The overexpression of p300 increases H3K9ac labeling on the OCN and DSPP genes, and it also regulates the promoter regions of NANOG and SOX2, ensuring sufficient expression of stemness-related genes and maintaining stemness (Liu et al., 2015b). Knocking down p300 in DPSCs inhibits proliferation and odontogenic differentiation, consistent with a previous report (Haigis and Guarente, 2006). HDAC enzymes also play a role in histone acetylation by removing acetyl groups from histones and facilitating the densification of chromatin structure (Fukuda et al., 2006). There are 18 known human HDACs, categorized into four classes based on homology to yeast HDACs. Classes I (HDAC1, HDAC2, HDAC3, HDAC8), II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, HDAC10), and IV (HDAC11) are zinc-dependent enzymes with high sequence conservation (Wang et al., 2018b). Also, Class III HDAC (usually known as sirtuins) is a nicotinamide adenine dinucleotide-reliant enzyme (Haigis and Guarente, 2006). HDAC6 was found to promote the DPSCs odontogenic differentiation, and the mineralization and ALP activity capability in pDPSCs were increased when HDAC6 was knocked down (Wang et al., 2018b). In mouse tooth papillary mesenchymal cells subjected to odontoblast induction, elevated p300 expression and decreased HDAC3 expression were observed, leading to an increase in H3K9ac and H3K27ac levels. This suggests that HATs and HDACs regulate odontogenic specialization in a coordinated manner (Tao et al., 2020). For further exploration, the transcription factor KLF4 is found to have a trans-activation domain that directly attaches to target gene promoters, recruiting either co-repressors like HDAC3 or coactivators like p300 (Tao et al., 2019). Chromatin immunoprecipitation (ChIP) assays have shown that HDAC3 and p300 regulate odontogenic differentiation in a time-sensitive manner through interactions with KLF4. When dental pulp cells are induced into odontoblasts, HDAC3 primarily interacts with KLF4 at the Sp7 and Dmp1 promoters on the first day of induction, keeping DMP1 and OSX expression at moderate levels. However, on day 7, HDAC3 relocates to the cytoplasm, allowing KLF4 to link with p300 and transactivate Dmp1 and Osx, thereby enhancing odontoblast differentiation. These findings reveal that KLF4 can influence histone acetylation in the promoter regions of both Sp7 and Dmp1, regulating odontoblast differentiation through interactions with HDAC3 and p300 (Tao et al., 2019).
It has been documented that HDAC inhibitors (HDACi) have the potential to modulate gene expression through the regulation of histone acetylation levels. These inhibitors have been studied for their therapeutic potential in cancer treatment (Lakshmaiah et al., 2014), inflammatory diseases (Hull et al., 2016), and neurodegenerative disorders (Didonna and Opal, 2015). These inhibitors also influence the differentiation and proliferation of DPSCs, showing promising prospects for pulp regeneration. Pan-HDAC inhibitors such as Trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), and valproic acid (VPA) hold potential for use in dental restorations (Duncan et al., 2011). Recent research demonstrates that TSA, an isohydroxamic acid, can inhibit the activity of all HDACs except class IIA, enhancing the expression of proliferating cell nuclear antigen (PCNA), cell cycle-related protein CCDN1, DSPP, DMP1, and BSP, thus promoting mineralization and differentiation of DPSCs (Jin et al., 2013). The Smad2 and Smad3 signaling pathways play a key role in mediating mineralization and differentiation of DPSCs stimulated by TSA. Moreover, recent in vivo studies have explored the developmental effects of HDAC inhibitors on dental pulp by injecting pregnant mice with TSA. Compared to control groups, an increase of 1.64-fold in the dentin matrix and 1.74-fold in the number of odontoblasts was observed, suggesting that histone acetylation plays a critical role in mouse tooth development (Jin et al., 2013). VPA, a short-chain fatty acid, can inhibit class I HDACs. Research suggests that low concentrations of VPA promote DPSC mineralization and correlate with increased expression of osteopontin (OSP) and bone sialoprotein (BSP) (Paino et al., 2014). VPA heightens the expression of OPN and BMP through HDAC2 while simultaneously reducing OCN, a belated marker of osteogenic differentiation. This suggests that VPA enhances early osteogenic differentiation, but not the terminal differentiation phase. Gene expression levels were accompanied by the suppression of another class I HDAC, HDAC-2 (Paino et al., 2014). SAHA, a type of pan-inhibitor of HDACs, activates Nfic, increasing DSPP expression and thus enhancing the DPSCs odontogenesis (Kwon et al., 2012). From a mechanistic perspective, SAHA was recently discovered to encourage matrix metalloproteinase 13 (MMP-13) expression in rat DPSCs to promote mineralization and migration. Despite this, low concentrations of SAHA did not affect cell proliferation (Duncan et al., 2016). As a means of reducing off-target effects and improving specificity over conventional pan-HDAC inhibitors, the development of subtype-specific HDACs has been underway for about a decade (Balasubramanian et al., 2009). A recent study demonstrated that LMK-235, a selective inhibitor of HDAC-4 and HDAC-5, enhances the differentiation of human DPSCs into odontoblast-like cells while having no effect on cell proliferation (Liu et al., 2018a). Similarly, MS-275 is a specific HDACi that targets both HDAC1 and HDAC3. Administration of MS-275 in normal cultures of pDPSCs can upregulate the expression of ALP, RUNX2, DMP1, and DSPP without being cytotoxic at a concentration of 20 nmol/L. Remarkably, the MAPK signaling system remains largely inactive upon MS-275 stimulation, indicating that it is unlikely that MAPK signaling pathway transduction is involved in MS-275-induced odontogenesis (Lee et al., 2020). The odontogenic potential of MS-275 has also been observed in MDPC-23 cells, a murine odontoblast-like cell line, where MS-275 alone can enhance the expression of Runx2, Bmp2, Bmp4, Ocn, Dspp, Dmp1, Klf5, Col1α1, and Msx1, and increase calcified nodule formation and ALP activity without mineralized medium induction (Sultana et al., 2021). These findings support the use of target-specific HDACis to promote odontoblast differentiation, thereby creating a promising avenue for potential therapeutic applications (Manaspon et al., 2021). Histone acetylation regulates a variety of physiological processes in DPSCs, which can impact their fate. While there is still much to be explored, As a potential mineralized regeneration tool, HDACis may hold promise.
4.3 ncRNAs associated with odontogenic differentiation in DPSCs
4.3.1 MiRNAs
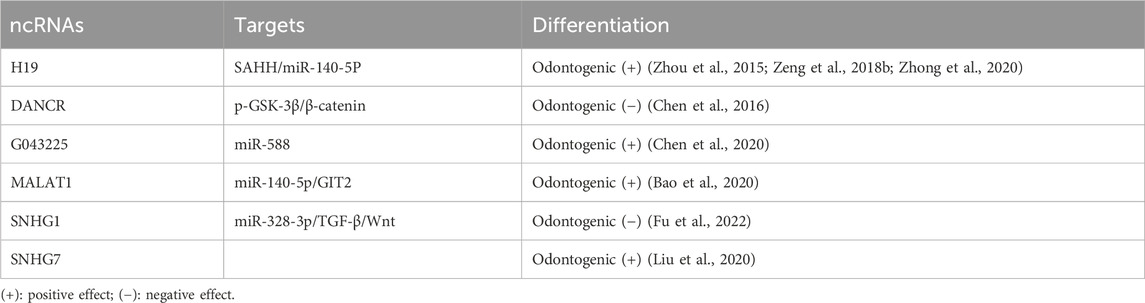
The field of epigenetics has seen a surge of interest in miRNAs, with numerous studies on their role in the odontogenic differentiation of DPSCs(Table 3). Using PCR analysis of 104 known miRNAs, 48 were differentially expressed between DPSCs and BM-MSCs, with 17 upregulated and 29 downregulated (Vasanthan et al., 2015). High-throughput microarray analysis revealed that 22 miRNAs were differentially expressed in odontogenic differentiated DPSCs, with 12 upregulated and 10 downregulated. It is of particular interest to note that miR-135b was significantly downregulated in the course of pulp cell mineralization. In light of the similarity between odontogenesis and osteogenesis and miR-135b’s significance in osteogenesis, it is postulated that miR-135b may serve a key part in regulating differentiated DPSCs mineralization (Schaap-Oziemlak et al., 2010). A recent study targeting miR-135b revealed its ability to inhibit odontogenic differentiation of DPSCs by modulating both SMAD5 and SMAD4 (Song et al., 2017). The miR-143 family exerts a negative regulatory effect on the odontogenic and osteogenic differentiation of DPSCs. Specifically, miR-143-5p is known to target RUNX2 to impair odontogenic differentiation through the OPG/RANKL pathway (Zhan et al., 2018). A number of studies have shown that miR-143-5p interacts with MAPK14, thereby reducing its expression. This leads to an induction of MAPK14 expression upon miR-143-5p knockdown, activating the p38 MAPK signaling pathway and enhancing odontogenesis (Wang et al., 2019a). Additionally, microarray analyses have revealed that miR-143 and miR-145 are significantly downregulated in mouse DPSCs and can form a negative regulatory feedback loop with KLF4 to inhibit the expression of Dspp and Dmp1 mRNA and protein, consequently having a negative impact on odontogenic differentiation (Liu et al., 2013). Importantly, this represents the first report of a feedback loop regulation involving miRNA during odontogenic differentiation.
Various other miRNAs have been affiliated with the DPSCs osteogenic and odontogenic differentiation. Upon induction of DPSCs into odontoblasts, miR-140-5p is decreased, and miR-140-5p mimics can impede odontogenic differentiation by targeting Wnt1 to repress the Wnt1/β-catenin signaling pathway (Lu et al., 2019). During odontogenesis induction, a gradual decline in miR-508-5p and a concomitant increase in glycoprotein non-metastatic melanoma protein B (GPNMB) can also be observed. Subsequent investigations have demonstrated that the knockdown of miR508-5p, which releases GPNMB expression, bolsters odontogenesis in DPSCs (Liu et al., 2019). In DPSCs, according to Chang et al. (2019), miR-218 expression is differentially downregulated, and miR-218 mimics considerably decrease DPSC proliferation and differentiation. It has also been established that insulin-like growth factor 1 induces DPSCs proliferation and osteogenic/odontogenic differentiation via activating the JNK and P38MAPK pathways. Increasing let-7c inhibits the insulin-like factor-1 receptor (IGF-1R) by reversing the process as well as inhibiting activation of the JNK/P38 MAPK pathway (Liu et al., 2018b). This process, including miR-488, may also be adversely involved (Yu et al., 2019).
In addition to their inhibitory effects, miRNAs can promote odontogenic differentiation in DPSCs (Sun et al., 2015). Studies have shown that miR-720 can promote odontogenic differentiation while concurrently reducing the proliferative capacity of DPSCs. As such, miR-720 represents a novel miRNA involved in the process of DPSCs differentiation (Hara et al., 2013). Kato (2018) demonstrated that lentiviral transfection of hDPSCs with miR-146a-5p resulted in its overexpression, which led to a significant upregulation in the expression of odontogenic differentiation markers (Dspp and Dmp1), suggesting a positive regulatory role of miR-146a-5p in odontogenic differentiation of hDPSCs. Through the inhibition of DNMT3B, miR-675 promotes the odontogenic differentiation of DPSCs by mediating DLX3 methylation (Qiu et al., 2019). Another miRNA, miR-27a-5p, positively regulates odontogenic differentiation by suppressing the LTBP1 molecule and activating the TGF-β1/Smad pathway (Hu et al., 2019).
Interestingly, this miRNA is found in the exosomes secreted by DPSCs during odontogenic induction. There is a conserved motif in miRNA, which can be recognized by RNA-binding proteins and transported in a cell-specific manner. When miRNA is present in the EXOmotif, it is recognized by specific RNA-binding proteins, loaded into vesicles, transported to other cells, and recognized by other cells for cell interaction; miRNA in vesicles can also inhibit gene expression in the receiving cells (Garcia-Martin et al., 2022). In the future, RNA editing technology can be used to specifically change the motif of specific miRNA molecules and solve the problem caused by disordered cell interaction, opening up new ideas for future treatment in the oral field.
The modulation of miRNA is an essential aspect during the development of dental pulp inflammation, and studies have identified its distinct expression profiles in healthy and inflamed dental pulp (Zhong et al., 2012). The odontoblast differentiation of pDPSCs can be stimulated by LPS or TNF-α (He et al., 2015). In most cases, the protective effect of miRNA is achieved by mitigating the inflammatory response or promoting odontogenic differentiation. A significantly increased miRNA in inflamed pulp tissue is miR-223-3p, which can be detected in pulp tissue. Overexpression of miR-223-3p has been found to promote DPSCs odontoblast differentiation in vitro (Huang et al., 2019). Both let-7c-5p and miR-506 can safeguard against LPS-induced pDPSC inflammation by reducing the pro-inflammatory cytokines performance (Wang et al., 2019b; Huang et al., 2019). In vivo experiments have confirmed that let-7c-5p reduces the ability of LPS to induce pulpitis in rats (Yuan et al., 2018). Furthermore, let-7c-5p has additional potential for promoting bone growth in inflamed pDPSCs(Yuan et al., 2019). Knocking down miR-140-5p has been found to enhance odontoblast differentiation, and Toll-like receptor-4 is involved in the miR-140-5p-mediated effects on pDPSCs, inhibiting pDPSC proliferation upon LPS stimulation (Sun et al., 2017). Fyn is a member of the Src kinase family, which has been reported to be upregulated in the microenvironment of deep caries. The miR-125a-3p has been identified as an upstream factor of Fyn and a positive factor in regulating pDPSC odontoblast differentiation upon TNF-αstimulation (Wang et al., 2020). At low concentrations (1–10 ng/mL), TNF-αenhances odontogenic differentiation, while at high concentrations, it suppresses it. As shown above, miR-21 is detected even at low levels of TNF-α, along with an increase in the expression of signal transduction and activator of transcription3 (STAT3). When high concentrations are present, the opposite results are observed. STAT and miR-21 are both involved in positive feedback loops regulating odontogenic differentiation (Xu et al., 2018a).
MiRNA, as a subject of epigenetic research, has attracted much attention, with many theories constantly being updated and improved. One such theory is target-directed miRNA degradation (TDMD), where target genes can degrade miRNA by binding to it (Figure 3). The form of miRNA and target gene binding influences the stability of miRNA. When the 3′end of miRNA does not fully bind to target genes, miRNA remains relatively stable. However, miRNA is easily degraded when the 3′end of miRNA can fully bind to target genes (Kato, 2018). For instance, research has shown that the 3′end of miR-382-3P can be completely complementary to the upstream gene GAS5, leading to the autodegradation of miR-382-3p. This process relieves the inhibitory effect of the downstream target gene TAF1, promoting the osteogenic differentiation of hBMSCs (Song et al., 2022). Another emerging concept is the nuclear activating miRNA (NamiRNA), which investigates the epigenetic regulatory mechanisms of miRNAs themselves. A systematic analysis of 1,594 miRNA precursors across seven different tissue types revealed more than 300 miRNA precursors with genomic positions overlapping enhancer histone modification markers H3K4me1 or H3K27ac. This association between tissue-specific miRNA and enhancers led to the discovery that the activation of these nuclear miRNAs relies on the integrity of the enhancer region. Knocking down specific sequences of the enhancer can prevent miRNA upregulation, affecting tumor cells’ proliferation and migration capabilities. Some scholars suggest that miRNAs function as bifunctional molecules. In the cytoplasm, miRNAs act as negative regulators, blocking mRNA translation and suppressing gene expression. Conversely, in the nucleus, miRNAs serve as activators, modulating the chromatin state of enhancers to stimulate gene transcription (Xiao et al., 2017).
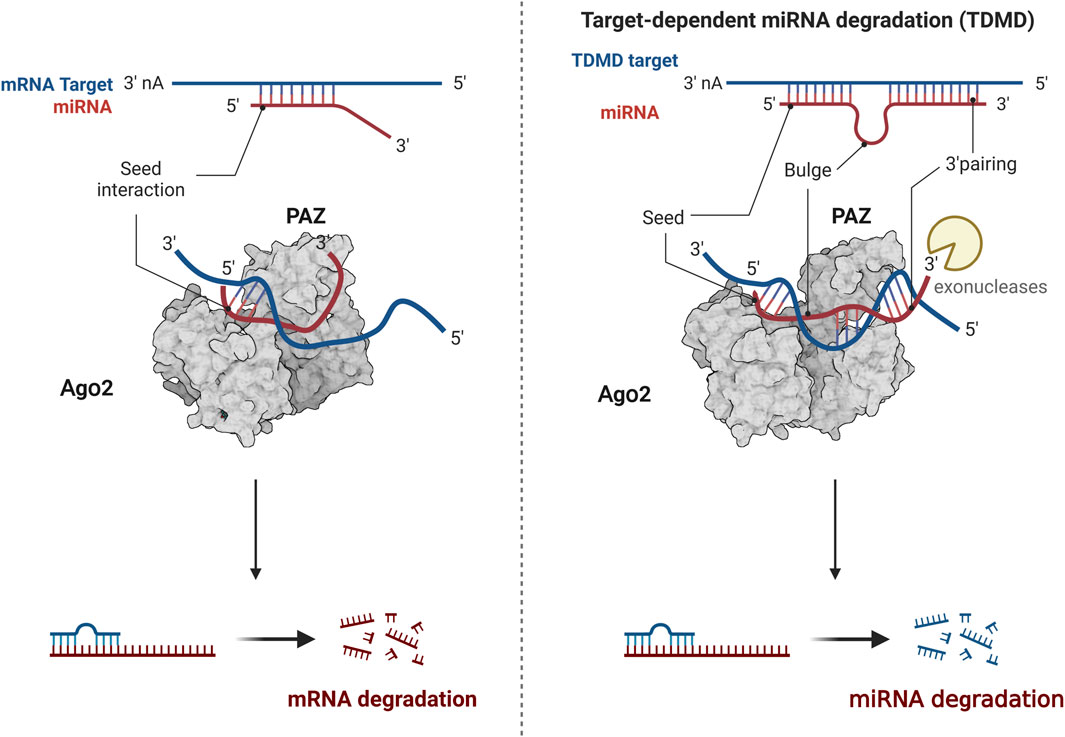
Figure 3. The modulation of TDMD mechanism. The 3′end of miRNA does not fully bind to target genes, miRNA remains relatively stable. The 3′end of miRNA can fully bind to target genes, miRNA is easily degraded. Created with BioRender.com.
While these theories have yet to be applied to the odontogenic differentiation of DPSCs, they present innovative concepts and perspectives that could stimulate scientific inquiry and inspire further research into restorative dentin and pulp regeneration. These ideas offer potential pathways for more in-depth exploration of these fields, potentially leading to breakthroughs in regenerative dental therapies.
4.3.2 LncRNAs
A mounting body of research underscores the significant regulatory role of lncRNA in the differentiation process of hDPSCs(Table 4). One such lncRNA, H19, is an imprinted paternal gene that encodes a 2.3-kb noncoding RNA and exhibits multiple biological effects, including promoting the ability of hDPSCs proliferation, migration, and differentiation (Huang et al., 2016). Research indicates that H19 inhibits the expression of LATS1 by increasing zeste homolog two-induced trimethylation levels of histone three at lysine 27. Conversely, LATS1 suppresses the differentiation, proliferation, and migration of hDPSCs by decreasing YAP and TAZ levels, thereby activating the Hippo-Yes-related protein (YAP) signaling pathway. Transplanting hDPSCs overexpressing lncRNA H19 into nude mice showed that lncRNA H19 inhibits LATS1 and promotes the production of odontoblast cells in vivo (Du et al., 2021). H19 also upregulates S-adenosyl homocysteine (SAH), an inhibitor of S-adenosylmethionine-dependent methyltransferase (Zhou et al., 2015). It enhances the odontogenic differentiation of DPSCs by downregulating the methylation level of the distal-free homology cassette 3 (DLX3) gene to upregulate the expression of DLX3 (Zeng et al., 2018b). Furthermore, H19 can bind to miR-140-5P via the sponge mechanism, which derepresses miR-140-5P on the downstream edentulous target genes (BMP-2, FGF9) to promote odontogenic differentiation (Zhong et al., 2020). Nonetheless, research has shown that lncRNA H19 expression was not significantly detected during odontogenesis. This discrepancy may arise from the fact that differentiated and undifferentiated cells used in the studies were derived from different individuals, potentially leading to varying results. Additionally, phenotypic differences and variations in sequencing methods may introduce errors. Therefore, large-scale studies are required to confirm these findings and provide more definitive conclusions (Zhong et al., 2020). In addition to chromosomal modifications that participate in epigenetic regulation, long non-coding RNAs (LncRNAs) may also engage with the key molecules of related signaling pathways to regulate the odontogenic differentiation of DPSCs. The LncRNA anti-differentiation non-coding RNA (ANCR), also known as differentiation antagonistic non-protein coding RNA (DANCR), was initially discovered in 2012 to inhibit differentiation and promote undifferentiation of somatic progenitor cells (Kretz et al., 2012). Furthermore, studies have revealed that inhibition of ANCR leads to neurogenic, osteogenic, and lipogenic differentiation in DPSCs without impeding their proliferation (Jia et al., 2016). ANCR impedes the odontogenic differentiation of DPSCs by targeting the Wnt/β-catenin signaling pathway (Chen et al., 2016).
The ability of dental pulp stem cells (DPSCs) to undergo odontogenic differentiation has been associated with the activity of lncRNAs. RNA-sequencing analysis has revealed a correlation between the loss of odontogenic differentiation potential and the downregulation of 108 lncRNAs, as well as the upregulation of 36 lncRNAs (Zheng and Jia, 2016). Moreover, inducing odontoblastic differentiation in DPSCs differentially alters the expression of 132 lncRNAs and 114 miRNAs. Bioinformatic analysis has identified lncRNAs linked to odontogenic proteins such as retinoid and fibronectin 1. Their associated competing endogenous RNA (ceRNA) networks play a role in the complex differentiation of DPSCs. Further studies have shown that lncRNA G043225 inhibits the expression of FBN1 by acting as an endogenous miRNA sponge, competitively inhibiting miR-588, thereby promoting odontogenic differentiation (Chen et al., 2020).
Prominent researchers have meticulously investigated the novel regulatory circuit governing the odontogenic differentiation of hDPSCs. This circuit involves an interplay between the long non-coding RNA metastasis-related lung adenocarcinoma transcript 1 (MALAT1), microRNA-140-5p (miR-140-5p), and the G protein-coupled receptor kinase two interaction protein 2 (GIT2). During the odontogenic differentiation of hDPSCs, miR-140-5p is significantly downregulated. Dual-luciferase reporter gene assays have demonstrated that miR-140-5p directly targets GIT2. RNA pull-down assays have further confirmed that MALAT1 can bind to miR-140-5p, thereby positively regulating GIT2 expression. Notably, increased miR-140-5p levels or GIT2 knockdown in hDPSCs (via exogenous transfection or lentiviral infection) inhibits the expression of key markers of odontogenic differentiation, such as ALP activity, DSPP, DMP-1, and DLX3. Collectively, these findings suggest that MALAT1 may play a crucial role in regulating odontogenic differentiation in hDPSCs (Bao et al., 2020). Analogously, the lncRNA colon cancer-associated transcript 1 (CCAT1) was initially found to participate in various aspects of metabolism, migration, and proliferative processes in select cancers (Li et al., 2018c). Subsequently, researchers have established that CCAT1 can also promote the DPSCs proliferation of odontogenic differentiation and proliferation. Remarkably, luciferase assays have evinced that CCAT1 is bound to miR-218, leading to negative modulation of miR-218 expression (Zhong et al., 2019). The most recent investigation has unveiled that the impact of small nucleolar RNA host gene 1 (SNHG1) can influence the progression of osteosarcoma by controlling miR-101-3p, miR-326, and miR-577 (Wang et al., 2018c; Jiang et al., 2018; Deng et al., 2019). Furthermore, SNHG1 can also impede the proliferation of colorectal cancer cells through miR-154-5p (Xu et al., 2018b). The overexpression of SNHG1 is observed during the differentiation of hDPSCs into odontoblast-like cells. SNHG1 expression increases progressively on days 0, 3, and seven of hDPSCs’ odontogenic differentiation. The overexpression of SNHG1 boosts the mRNA and protein expression of DSPP, DMP-1, and ALP. Mechanistically, SNHG1 can bind to miR-328-3p, lifting its inhibitory effect on target genes to activate the Wnt/β-catenin pathway, thereby promoting odontogenic differentiation (Fu et al., 2022). Recent research has highlighted substantial alterations in SNHG7 expression during the odontogenic differentiation of DPSCs. By culturing human DPSCs in an osteogenic/odontogenic differentiation medium for 14 days and analyzing the cells’ RNA sequences, scientists identified 89 differentially expressed lncRNAs, 1,636 mRNAs, and 113 miRNAs. Silencing SNHG7 was found to inhibit the odontogenic/osteogenic differentiation of DPSCs, suggesting it as a promising target for dentin-pulp complex regeneration and tissue engineering (Liu et al., 2020).
Despite progress, significant knowledge gaps remain in understanding the role of lncRNAs in odontogenic differentiation. Further research is needed to confirm the functions of various molecules involved. The current understanding centers around competing endogenous RNA (ceRNA) competition, transcription factor regulation, and DNA methylation interactions. However, more intricate and nuanced epigenetic mechanisms should be explored. It is clear that lncRNAs play a critical role in the differentiation potential of hDPSCs and represent a promising target for new treatments in VPT.
4.3.3 CircRNAs
CircRNA, with its closed-loop system, is highly conserved and extremely resistant to RNA enzymes, making it a noteworthy molecular entity. Moreover, it is recognized as a potent miRNA sponge due to its ability to competitively bind miRNAs. First discovered in viruses in 1976, circRNA has since been identified as a pivotal post-transcriptional regulator of cell proliferation, differentiation, and apoptosis (Wang et al., 2019c).
Numerous studies have verified the significant role of circRNAs in regulating the osteogenic differentiation of dental stem cells. However, there have been few studies on the effects of circRNAs on the odontogenic differentiation of DPSCs. Among them, Chen et al. (2020) have utilized microarray analysis to determine that 187 circRNAs have been differentially expressed more than 1.5-fold during odontogenic differentiation in the induced group compared to the non-mineralized induced group, of which 44 circRNAs have been upregulated, and 143 have been downregulated. In the induced group, hsa_circRNA_005044, hsa_circRNA_005044, hsa_circRNA_406763, and hsa_circRNA_104101 have been upregulated 3.78, 8.29, 15.95, 19.03-fold, respectively, compared to the uninduced group. Furthermore, hsa_circRNA_079813 has undergone a 3.85-fold downregulation and hsa_circRNA_008336 has undergone a 3.74-fold downregulation. Among the 187 circRNAs that have significantly different expressions prior to mineralization induction, these six circRNAs exhibit the most pronounced changes. Knockdown of circRNA_104101 has resulted in varying degrees of reduction in odontogenic markers, such as ALP, DSPP, DMP1, and OCN, after the 7th and 14th days of induction. Furthermore, based on bioinformatics analysis, it is indicated that the Wnt/TGF-β signaling pathway may be involved; however, the precise mechanism still requires further exploration. Li and Jiang (2019) have identified 1314 upregulated and 1780 downregulated circular RNAs, respectively, during odontogenic induction using high-throughput sequencing. The expression trends obtained by high-throughput sequencing have been consistent with the results of RT-qPCR screening and validation of two circular RNAs with large fold changes (hsa_circ_0015260 and hsa_circ_0006984). Additionally, the MAPK signaling pathway has been predicted to be part of odontogenic differentiation, but further investigation is necessary to determine specific results and mechanisms.
The bulk of research on the odontogenic differentiation of DPSCs has primarily centered on miRNAs, with circRNAs receiving relatively little attention. Given their inherent stability and significant abundance in physiological systems, circRNAs represent a promising avenue for biomarker research related to disease diagnosis (Han et al., 2018). However, numerous molecules and mechanisms remain to be explored in greater depth, with circRNAs presenting as potentially powerful targets for VPT. This paper serves as a concise summary intended to provide a reference point for the continued investigation into the regulatory mechanisms of circRNAs in the odontogenic differentiation of DPSCs.
5 Conclusion
This comprehensive review examines the odontogenic differentiation of DPSCs with a focus on the critical role of epigenetic modifications, including DNA methylation, histone modifications, and non-coding RNA regulation. Although most current research emphasizes in vitro studies, further in vivo research and animal disease models are essential for advancing our understanding. Additionally, the field shows great potential for progress in circRNA research. It is important to highlight two complementary concepts, TDMD and NamiRNA, which enhance our understanding of the unique impacts of miRNA molecules across different cellular locations. Given the wide range of epigenetic modifications and underlying mechanisms, our comprehension of epigenetic regulatory influences remains in its early stages, with a focus on classical epigenetic modifications and their respective locations.
Currently, clinical practice mainly employs biomaterials embedded with HDACi and integrates inhibitors into biologically derived dental restorative biomaterials for pulp restoration in exposed areas. However, advances in biotechnology are leading to the discovery of new epigenetic modifications, such as DNA 6 mA modifications, tRNA-enriched epigenetic modifications, and molecular variant shearing, all of which play roles in regulating odontogenic differentiation in DPSCs.The complex interplay of these various modifications presents a challenge in unraveling the epigenetic regulatory mechanisms that govern the odontogenic differentiation of dental pulp stem cells.
6 Future direction
Current trends in dentistry emphasize minimally invasive and biologically oriented restorative strategies, focusing on preserving the vital pulp and harnessing its natural regenerative abilities to facilitate self-renewal. The use of calcium silicate-based materials has increased the success of pulp regeneration therapies. However, limitations persist with existing dental materials, such as significant cytotoxicity, reduced tissue repair capacity, and a lack of target specificity. A comprehensive understanding of the biological mechanisms of pulp repair and regeneration is crucial for advancing treatment options. While DPSCs have been employed in vitro and in animal models for tissue engineering, their clinical applications remain limited. The expansion and cultivation of DPSCs involve complex procedures that require specialized culture media and conditions to preserve their pluripotency and multipotency. These challenges demand urgent solutions, particularly for large-scale clinical use. Given the variation in the differentiation potential of DPSCs across individuals and sources, patient outcomes may differ.
Looking forward, there are several key areas of focus: 1. Enhancing research on the growth and culture conditions of DPSCs is vital to optimize their expansion and stability while retaining their totipotency and pluripotency; 2. Developing a deeper understanding of the mechanisms and regulatory networks that govern DPSC differentiation is essential. Exploring additional epigenetic processes, such as lactation and succination modifications, can promote targeted differentiation of DPSCs into specific cell lines and improve the efficiency of regenerative therapies; 3. Improving regenerative therapy efficiency through the integration of biomaterials and bioengineering technologies is crucial. By combining these approaches to design suitable scaffolds and carriers, optimal growth environments can be created for DPSCs, promoting their directed differentiation and facilitating dental pulp tissue regeneration and repair; 4. Advances in pharmacology offer opportunities to target essential pulp-preserving therapies against epigenetic or other cellular markers, opening the door to clinical applications of novel endodontic restorative materials. However, the use of epigenetic agents presents several unresolved challenges that warrant attention, including: i. The absence of robust clinical trial data regarding the induction of dentition, raising significant ethical and regulatory concerns; ii. Limited specificity, as inhibitors targeting particular HDAC classes have been developed but lack total specificity, potentially leading to associated side effects; iii. Heavy reliance on the Zn2+ binding motif, which may strongly bind to other vital metalloenzymes, inducing cytotoxicity and restricting the clinical applicability of the inhibitors. Consequently, the future may call for the creation of highly precise inhibitors or the examination of combination therapies to mitigate dose dependency and enhance drug efficacy.
Author contributions
LH: Conceptualization, Data curation, Formal Analysis, Validation, Visualization, Writing–original draft. XC: Formal Analysis, Methodology, Supervision, Validation, Writing–review and editing. XXY: Supervision, Writing–review and editing. YCZ: Writing–review and editing. YYL: Writing–review and editing. XLQ: Funding acquisition, Resources, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Clinical Research Initiation Plan Project of Stomatological Hospital, Southern Medical University (grant number: KQIIT2021005), New Technology New Projects of Stomatological Hospital, Southern Medical University (grant number: NTP20220110703), and the Research and Cultivation Project of Stomatological Hospital, Southern Medical University (grant number: PY2020029).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor FF declared a shared parent affiliation with the authors at the time of the review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alge, D. L., Zhou, D., Adams, L. L., Wyss, B. K., Shadday, M. D., Woods, E. J., et al. (2010). Donor-matched comparison of dental pulp stem cells and bone marrow-derived mesenchymal stem cells in a rat model. J. tissue Eng. Regen. Med. 4, 73–81. doi:10.1002/term.220
Alvarez, R., Lee, H. L., Hong, C., and Wang, C. Y. (2015). Single CD271 marker isolates mesenchymal stem cells from human dental pulp. Int. J. oral Sci. 7, 205–212. doi:10.1038/ijos.2015.29
Anitua, E., Troya, M., and Zalduendo, M. (2018). Progress in the use of dental pulp stem cells in regenerative medicine. Cytotherapy 20, 479–498. doi:10.1016/j.jcyt.2017.12.011
Argaez-Sosa, A. A., Rodas-Junco, B. A., Carrillo-Cocom, L. M., Rojas-Herrera, R. A., Coral-Sosa, A., Aguilar-Ayala, F. J., et al. (2022). Higher Expression of DNA (de)methylation-Related Genes Reduces Adipogenicity in Dental Pulp Stem Cells. Front. Cell. Dev. Biol. 10, 791667. doi:10.3389/fcell.2022.791667
Balasubramanian, S., Verner, E., and Buggy, J. J. (2009). Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 280, 211–221. doi:10.1016/j.canlet.2009.02.013
Bao, M., Liu, G., Song, J., and Gao, Y. (2020). Long non-coding RNA MALAT1 promotes odontogenic differentiation of human dental pulp stem cells by impairing microRNA-140-5p-dependent downregulation of GIT2. Cell. tissue Res. 382, 487–498. doi:10.1007/s00441-020-03246-1
Barthel, C. R., Rosenkranz, B., Leuenberg, A., and Roulet, J. F. (2000). Pulp capping of carious exposures: treatment outcome after 5 and 10 years: a retrospective study. J. Endod. 26, 525–528. doi:10.1097/00004770-200009000-00010
Bascom, G. D., and Schlick, T. (2018). Chromatin fiber folding directed by cooperative histone tail acetylation and linker histone binding. Biophysical J. 114, 2376–2385. doi:10.1016/j.bpj.2018.03.008
Bayarsaihan, D. (2016). Deciphering the epigenetic code in embryonic and dental pulp stem cells. Yale J. Biol. Med. 89, 539–563.
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125, 315–326. doi:10.1016/j.cell.2006.02.041
Bird, A. (2002). DNA methylation patterns and epigenetic memory. Genes. and Dev. 16, 6–21. doi:10.1101/gad.947102
Bochtler, M., Kolano, A., and Xu, G. L. (2017). DNA demethylation pathways: additional players and regulators. BioEssays news Rev. Mol. Cell. Dev. Biol. 39, 1–13. doi:10.1002/bies.201600178
Brännström, M., and Nyborg, H. (1973). Cavity treatment with a microbicidal fluoride solution: growth of bacteria and effect on the pulp. J. Prosthet. Dent. 30, 303–310. doi:10.1016/0022-3913(73)90187-x
Cakouros, D., Hemming, S., Gronthos, K., Liu, R., Zannettino, A., Shi, S., et al. (2019). Specific functions of TET1 and TET2 in regulating mesenchymal cell lineage determination. Epigenetics chromatin 12, 3. doi:10.1186/s13072-018-0247-4
Cameron, A. M., Lawless, S. J., and Pearce, E. J. (2016). Metabolism and acetylation in innate immune cell function and fate. Seminars Immunol. 28, 408–416. doi:10.1016/j.smim.2016.10.003
Carpenter, S., Aiello, D., Atianand, M. K., Ricci, E. P., Gandhi, P., Hall, L. L., et al. (2013). A long noncoding RNA mediates both activation and repression of immune response genes. Sci. (New York, N.Y.) 341, 789–792. doi:10.1126/science.1240925
Chang, K., Chen, R. S., Chang, F. H., and Chen, M. H. (2019). Promoting dentinogenesis of DPSCs through inhibiting microRNA-218 by using magnetic nanocarrier delivery. J. Formos. Med. Assoc. = Taiwan yi zhi 118, 1005–1013. doi:10.1016/j.jfma.2018.10.018
Chen, L., Song, Z., Huang, S., Wang, R., Qin, W., Guo, J., et al. (2016). lncRNA DANCR suppresses odontoblast-like differentiation of human dental pulp cells by inhibiting wnt/β-catenin pathway. Cell. tissue Res. 364, 309–318. doi:10.1007/s00441-015-2333-2
Chen, S., Rani, S., Wu, Y., Unterbrink, A., Gu, T. T., Gluhak-Heinrich, J., et al. (2005). Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J. Biol. Chem. 280, 29717–29727. doi:10.1074/jbc.M502929200
Chen, Z., Zhang, K., Qiu, W., Luo, Y., Pan, Y., Li, J., et al. (2020). Genome-wide identification of long noncoding RNAs and their competing endogenous RNA networks involved in the odontogenic differentiation of human dental pulp stem cells. Stem Cell. Res. Ther. 11, 114. doi:10.1186/s13287-020-01622-w
Ching, H. S., Luddin, N., Rahman, I. A., and Ponnuraj, K. T. (2017). Expression of odontogenic and osteogenic markers in DPSCs and SHED: a review. Curr. stem Cell. Res. Ther. 12, 71–79. doi:10.2174/1574888x11666160815095733
Cipriani, N. A., Johnson, D. N., Sarne, D. H., Angelos, P., Reeves, W., and Antic, T. (2022). The significance of RAS-like mutations and MicroRNA profiling in predicting malignancy in thyroid biopsy specimens. Endocr. Pathol. 33, 446–456. doi:10.1007/s12022-022-09734-0
Clayton, A. L., Hazzalin, C. A., and Mahadevan, L. C. (2006). Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 23, 289–296. doi:10.1016/j.molcel.2006.06.017
Crisan, M., Yap, S., Casteilla, L., Chen, C. W., Corselli, M., Park, T. S., et al. (2008). A perivascular origin for mesenchymal stem cells in multiple human organs. Cell. stem Cell. 3, 301–313. doi:10.1016/j.stem.2008.07.003
Deng, R., Zhang, J., and Chen, J. (2019). lncRNA SNHG1 negatively regulates miRNA-101-3p to enhance the expression of ROCK1 and promote cell proliferation, migration and invasion in osteosarcoma. Int. J. Mol. Med. 43, 1157–1166. doi:10.3892/ijmm.2018.4039
Didonna, A., and Opal, P. (2015). The promise and perils of HDAC inhibitors in neurodegeneration. Ann. Clin. Transl. neurology 2, 79–101. doi:10.1002/acn3.147
Dong, R., Yao, R., Du, J., Wang, S., and Fan, Z. (2013). Depletion of histone demethylase KDM2A enhanced the adipogenic and chondrogenic differentiation potentials of stem cells from apical papilla. Exp. Cell. Res. 319, 2874–2882. doi:10.1016/j.yexcr.2013.07.008
Du, J., Ma, Y., Ma, P., Wang, S., and Fan, Z. (2013). Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem cells Dayt. Ohio 31, 126–136. doi:10.1002/stem.1255
Du, Z., Shi, X., and Guan, A. (2021). lncRNA H19 facilitates the proliferation and differentiation of human dental pulp stem cells via EZH2-dependent LATS1 methylation. Mol. Ther. Nucleic acids 25, 116–126. doi:10.1016/j.omtn.2021.04.017
Dunaway, K., Goorha, S., Matelski, L., Urraca, N., Lein, P. J., Korf, I., et al. (2017). Dental pulp stem cells model early life and imprinted DNA methylation patterns. Stem cells Dayt. Ohio 35, 981–988. doi:10.1002/stem.2563
Duncan, H. F., Smith, A. J., Fleming, G. J., and Cooper, P. R. (2011). HDACi: cellular effects, opportunities for restorative dentistry. J. Dent. Res. 90, 1377–1388. doi:10.1177/0022034511406919
Duncan, H. F., Smith, A. J., Fleming, G. J., Partridge, N. C., Shimizu, E., Moran, G. P., et al. (2016). The histone-deacetylase-inhibitor suberoylanilide hydroxamic acid promotes dental pulp repair mechanisms through modulation of matrix metalloproteinase-13 activity. J. Cell. physiology 231, 798–816. doi:10.1002/jcp.25128
Esposito, M., Coulthard, P., and Worthington, H. V. (2003). Enamel matrix derivative (Emdogain) for periodontal tissue regeneration in intrabony defects. Cochrane database Syst. Rev., Cd003875. doi:10.1002/14651858.Cd003875
Feng, Z., Zhan, M., Meng, R., Wang, X., and Xu, Q. (2019). 5-Aza-2'-deoxycytidine enhances lipopolysaccharide-induced inflammatory cytokine expression in human dental pulp cells by regulating TRAF6 methylation. Bioengineered 10, 197–206. doi:10.1080/21655979.2019.1621135
Ferracane, J. L., Cooper, P. R., and Smith, A. J. (2010). Can interaction of materials with the dentin-pulp complex contribute to dentin regeneration? Odontology 98, 2–14. doi:10.1007/s10266-009-0116-5
Fu, T., Liu, Y., Huang, X., Guo, Y., Shen, J., and Shen, H. (2022). lncRNA SNHG1 regulates odontogenic differentiation of human dental pulp stem cells via miR-328-3p/Wnt/β-catenin pathway. Stem Cell. Res. Ther. 13, 311. doi:10.1186/s13287-022-02979-w
Fukuda, H., Sano, N., Muto, S., and Horikoshi, M. (2006). Simple histone acetylation plays a complex role in the regulation of gene expression. Briefings Funct. genomics and proteomics 5, 190–208. doi:10.1093/bfgp/ell032
Galvani, A., and Thiriet, C. (2015). Nucleosome dancing at the tempo of histone tail acetylation. Genes. 6, 607–621. doi:10.3390/genes6030607
Gangolli, R. A., Devlin, S. M., Gerstenhaber, J. A., Lelkes, P. I., and Yang, M. (2019). A bilayered poly (lactic-Co-glycolic acid) scaffold provides differential cues for the differentiation of dental pulp stem cells. Tissue Eng. Part A 25, 224–233. doi:10.1089/ten.TEA.2018.0041
Gao, X. D., Qu, J. H., Chang, X. J., Lu, Y. Y., Bai, W. L., Wang, H., et al. (2014). Hypomethylation of long interspersed nuclear element-1 promoter is associated with poor outcomes for curative resected hepatocellular carcinoma. Liver Int. official J. Int. Assoc. Study Liver 34, 136–146. doi:10.1111/liv.12264
Garcia-Martin, R., Wang, G., Brandão, B. B., Zanotto, T. M., Shah, S., Kumar Patel, S., et al. (2022). MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 601, 446–451. doi:10.1038/s41586-021-04234-3
Godini, R., Lafta, H. Y., and Fallahi, H. (2018). Epigenetic modifications in the embryonic and induced pluripotent stem cells. Gene Expr. patterns GEP 29, 1–9. doi:10.1016/j.gep.2018.04.001
Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007). Epigenetics: a landscape takes shape. Cell. 128, 635–638. doi:10.1016/j.cell.2007.02.006
Gopinathan, G., Kolokythas, A., Luan, X., and Diekwisch, T. G. (2013). Epigenetic marks define the lineage and differentiation potential of two distinct neural crest-derived intermediate odontogenic progenitor populations. Stem cells Dev. 22, 1763–1778. doi:10.1089/scd.2012.0711
Gronthos, S., Mankani, M., Brahim, J., Robey, P. G., and Shi, S. (2000). Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 97, 13625–13630. doi:10.1073/pnas.240309797
Gu, S., Liang, J., Wang, J., and Liu, B. (2013). Histone acetylation regulates osteodifferentiation of human dental pulp stem cells via DSPP. Front. Biosci. Landmark Ed. 18, 1072–1079. doi:10.2741/4163
Guo, M., Qiu, J., Shen, F., Wang, S., Yu, J., Zuo, H., et al. (2020). Comprehensive analysis of circular RNA profiles in skeletal muscles of aging mice and after aerobic exercise intervention. Aging 12, 5071–5090. doi:10.18632/aging.102932
Guven, E. P., Yalvac, M. E., Sahin, F., Yazici, M. M., Rizvanov, A. A., and Bayirli, G. (2011). Effect of dental materials calcium hydroxide-containing cement, mineral trioxide aggregate, and enamel matrix derivative on proliferation and differentiation of human tooth germ stem cells. J. Endod. 37, 650–656. doi:10.1016/j.joen.2011.02.008
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 15, 509–524. doi:10.1038/nrm3838
Haigis, M. C., and Guarente, L. P. (2006). Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes. and Dev. 20, 2913–2921. doi:10.1101/gad.1467506
Han, B., Chao, J., and Yao, H. (2018). Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol. Ther. 187, 31–44. doi:10.1016/j.pharmthera.2018.01.010
Hara, E. S., Ono, M., Eguchi, T., Kubota, S., Pham, H. T., Sonoyama, W., et al. (2013). miRNA-720 controls stem cell phenotype, proliferation and differentiation of human dental pulp cells. PloS one 8, e83545. doi:10.1371/journal.pone.0083545
He, W., Wang, Z., Luo, Z., Yu, Q., Jiang, Y., Zhang, Y., et al. (2015). LPS promote the odontoblastic differentiation of human dental pulp stem cells via MAPK signaling pathway. J. Cell. physiology 230, 554–561. doi:10.1002/jcp.24732
Hlady, R. A., Tiedemann, R. L., Puszyk, W., Zendejas, I., Roberts, L. R., Choi, J. H., et al. (2014). Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget 5, 9425–9443. doi:10.18632/oncotarget.2444
Hoang, M., Kim, J. J., Kim, Y., Tong, E., Trammell, B., Liu, Y., et al. (2016). Alcohol-induced suppression of KDM6B dysregulates the mineralization potential in dental pulp stem cells. Stem Cell. Res. 17, 111–121. doi:10.1016/j.scr.2016.05.021
Hu, X., Zhong, Y., Kong, Y., Chen, Y., Feng, J., and Zheng, J. (2019). Lineage-specific exosomes promote the odontogenic differentiation of human dental pulp stem cells (DPSCs) through TGFβ1/smads signaling pathway via transfer of microRNAs. Stem Cell. Res. Ther. 10, 170. doi:10.1186/s13287-019-1278-x
Huang, C., Xiang, Y., Wang, Y., Li, X., Xu, L., Zhu, Z., et al. (2010). Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell. Res. 20, 154–165. doi:10.1038/cr.2010.5
Huang, H., Dou, L., Song, J., and Luo, J. (2018). CBFA2T2 is required for BMP-2-induced osteogenic differentiation of mesenchymal stem cells. Biochem. biophysical Res. Commun. 496, 1095–1101. doi:10.1016/j.bbrc.2018.01.144
Huang, X., Liu, F., Hou, J., and Chen, K. (2019). Inflammation-induced overexpression of microRNA-223-3p regulates odontoblastic differentiation of human dental pulp stem cells by targeting SMAD3. Int. Endod. J. 52, 491–503. doi:10.1111/iej.13032
Huang, Y., Zheng, Y., Jin, C., Li, X., Jia, L., and Li, W. (2016). Long non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci. Rep. 6, 28897. doi:10.1038/srep28897
Hui, T., Wang, C., Chen, D., Zheng, L., Huang, D., and Ye, L. (2017). Epigenetic regulation in dental pulp inflammation. Oral Dis. 23, 22–28. doi:10.1111/odi.12464
Hui, T., A, P., Zhao, Y., Wang, C., Gao, B., Zhang, P., et al. (2014). EZH2, a potential regulator of dental pulp inflammation and regeneration. J. Endod. 40, 1132–1138. doi:10.1016/j.joen.2014.01.031
Hui, T., A, P., Zhao, Y., Yang, J., Ye, L., and Wang, C. (2018). EZH2 regulates dental pulp inflammation by direct effect on inflammatory factors. Archives oral Biol. 85, 16–22. doi:10.1016/j.archoralbio.2017.10.004
Hull, E. E., Montgomery, M. R., and Leyva, K. J. (2016). HDAC inhibitors as epigenetic regulators of the immune system: impacts on cancer therapy and inflammatory diseases. BioMed Res. Int. 2016, 8797206. doi:10.1155/2016/8797206
Innes, N. (2010). Enamel matrix derivative for direct pulp capping. Evidence-based Dent. 11, 45–46. doi:10.1038/sj.ebd.6400719
Iohara, K., Murakami, M., Takeuchi, N., Osako, Y., Ito, M., Ishizaka, R., et al. (2013). A novel combinatorial therapy with pulp stem cells and granulocyte colony-stimulating factor for total pulp regeneration. Stem cells Transl. Med. 2, 818. doi:10.5966/sctm.2012-0132erratum
Iohara, K., Zheng, L., Ito, M., Ishizaka, R., Nakamura, H., Into, T., et al. (2009). Regeneration of dental pulp after pulpotomy by transplantation of CD31(-)/CD146(-) side population cells from a canine tooth. Regen. Med. 4, 377–385. doi:10.2217/rme.09.5
Itoh, Y., Sasaki, J. I., Hashimoto, M., Katata, C., Hayashi, M., and Imazato, S. (2018). Pulp regeneration by 3-dimensional dental pulp stem cell constructs. J. Dent. Res. 97, 1137–1143. doi:10.1177/0022034518772260
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, N.Y.) 19, 141–157. doi:10.1261/rna.035667.112
Jenuwein, T., and Allis, C. D. (2001). Translating the histone code. Sci. (New York, N.Y.) 293, 1074–1080. doi:10.1126/science.1063127
Jeong, Y., Yang, W., Ko, H., and Kim, M. (2014). The effects of bone morphogenetic protein-2 and enamel matrix derivative on the bioactivity of mineral trioxide aggregate in MC3T3-E1cells. Restor. Dent. Endod. 39, 187–194. doi:10.5395/rde.2014.39.3.187
Jia, Q., Chen, X., Jiang, W., Wang, W., Guo, B., and Ni, L. (2016). The regulatory effects of long noncoding RNA-ANCR on dental tissue-derived stem cells. Stem cells Int. 2016, 3146805. doi:10.1155/2016/3146805
Jiang, Z., Jiang, C., and Fang, J. (2018). Up-regulated lnc-SNHG1 contributes to osteosarcoma progression through sequestration of miR-577 and activation of WNT2B/Wnt/β-catenin pathway. Biochem. biophysical Res. Commun. 495, 238–245. doi:10.1016/j.bbrc.2017.11.012
Jin, H., Park, J. Y., Choi, H., and Choung, P. H. (2013). HDAC inhibitor trichostatin A promotes proliferation and odontoblast differentiation of human dental pulp stem cells. Tissue Eng. Part A 19, 613–624. doi:10.1089/ten.TEA.2012.0163
Kaikkonen, M. U., Lam, M. T., and Glass, C. K. (2011). Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 90, 430–440. doi:10.1093/cvr/cvr097
Kakehashi, S., Stanley, H. R., and Fitzgerald, R. J. (1965). THE EFFECTS OF SURGICAL EXPOSURES OF DENTAL PULPS IN GERM-FREE AND CONVENTIONAL LABORATORY RATS. Oral Surg. oral Med. oral pathology 20, 340–349. doi:10.1016/0030-4220(65)90166-0
Kareta, M. S., Botello, Z. M., Ennis, J. J., Chou, C., and Chédin, F. (2006). Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 281, 25893–25902. doi:10.1074/jbc.M603140200
Kato, M. (2018). Target RNA-directed microRNA degradation; which controls which? Non-coding RNA Investig. 2, 62. doi:10.21037/ncri.2018.11.01
Kaukua, N., Shahidi, M. K., Konstantinidou, C., Dyachuk, V., Kaucka, M., Furlan, A., et al. (2014). Glial origin of mesenchymal stem cells in a tooth model system. Nature 513, 551–554. doi:10.1038/nature13536
Kramer, J. M. (2016). Regulation of cell differentiation and function by the euchromatin histone methyltranserfases G9a and GLP. Biochem. Cell. Biol. = Biochimie Biol. Cell. 94, 26–32. doi:10.1139/bcb-2015-0017
Kretz, M., Webster, D. E., Flockhart, R. J., Lee, C. S., Zehnder, A., Lopez-Pajares, V., et al. (2012). Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes. and Dev. 26, 338–343. doi:10.1101/gad.182121.111
Kwon, A., Park, H. J., Baek, K., Lee, H. L., Park, J. C., Woo, K. M., et al. (2012). Suberoylanilide hydroxamic acid enhances odontoblast differentiation. J. Dent. Res. 91, 506–512. doi:10.1177/0022034512443367
Lakshmaiah, K. C., Jacob, L. A., Aparna, S., Lokanatha, D., and Saldanha, S. C. (2014). Epigenetic therapy of cancer with histone deacetylase inhibitors. J. cancer Res. Ther. 10, 469–478. doi:10.4103/0973-1482.137937
Lawrence, M., Daujat, S., and Schneider, R. (2016). Lateral thinking: how histone modifications regulate gene expression. Trends Genet. TIG 32, 42–56. doi:10.1016/j.tig.2015.10.007
Lee, C. K., Shibata, Y., Rao, B., Strahl, B. D., and Lieb, J. D. (2004). Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36, 900–905. doi:10.1038/ng1400
Lee, E. C., Kim, Y. M., Lim, H. M., Ki, G. E., and Seo, Y. K. (2020). The histone deacetylase inhibitor (MS-275) promotes differentiation of human dental pulp stem cells into odontoblast-like cells independent of the MAPK signaling system. Int. J. Mol. Sci. 21, 5771. doi:10.3390/ijms21165771
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419. doi:10.1038/nature01957
Legnini, I., Morlando, M., Mangiavacchi, A., Fatica, A., and Bozzoni, I. (2014). A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell. 53, 506–514. doi:10.1016/j.molcel.2013.12.012
Li, B., Yu, F., Wu, F., Hui, T., A, P., Liao, X., et al. (2018b). EZH2 impairs human dental pulp cell mineralization via the wnt/β-catenin pathway. J. Dent. Res. 97, 571–579. doi:10.1177/0022034517746987
Li, C., and Jiang, H. (2019). Altered expression of circular RNA in human dental pulp cells during odontogenic differentiation. Mol. Med. Rep. 20, 871–878. doi:10.3892/mmr.2019.10359
Li, D., Guo, B., Wu, H., Tan, L., and Lu, Q. (2015a). TET family of dioxygenases: crucial roles and underlying mechanisms. Cytogenet. genome Res. 146, 171–180. doi:10.1159/000438853
Li, J., Yang, J., Zhou, P., Le, Y., Zhou, C., Wang, S., et al. (2015b). Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am. J. cancer Res. 5, 472–480.
Li, N., Jiang, K., Fang, L. P., Yao, L. L., and Yu, Z. (2018c). Knockdown of long noncoding RNA CCAT1 inhibits cell growth, invasion and peritoneal metastasis via downregulation of Bmi-1 in gastric cancer. Neoplasma 65, 736–744. doi:10.4149/neo_2018_171206N801
Li, Q., Rao, L., Zhang, D., and Xu, Q. (2014). Expression features of DNA methylcytosine dioxygenase ten-eleven translocation 1 in human dental pulp cells. J. Endod. 40, 1791–1795. doi:10.1016/j.joen.2014.07.003
Li, Q., Yi, B., Feng, Z., Meng, R., Tian, C., and Xu, Q. (2018a). FAM20C could be targeted by TET1 to promote odontoblastic differentiation potential of human dental pulp cells. Cell. Prolif. 51, e12426. doi:10.1111/cpr.12426
Li, Q. M., Li, J. L., Feng, Z. H., Lin, H. C., and Xu, Q. (2020). Effect of histone demethylase KDM5A on the odontogenic differentiation of human dental pulp cells. Bioengineered 11, 449–462. doi:10.1080/21655979.2020.1743536
Lin, H., Xu, L., Liu, H., Sun, Q., Chen, Z., Yuan, G., et al. (2011). KLF4 promotes the odontoblastic differentiation of human dental pulp cells. J. Endod. 37, 948–954. doi:10.1016/j.joen.2011.03.030
Liu, F., Wang, X., Yang, Y., Hu, R., Wang, W., and Wang, Y. (2019). The suppressive effects of miR-508-5p on the odontogenic differentiation of human dental pulp stem cells by targeting glycoprotein non-metastatic melanomal protein B. Stem Cell. Res. Ther. 10, 35. doi:10.1186/s13287-019-1146-8
Liu, G. X., Ma, S., Li, Y., Yu, Y., Zhou, Y. X., Lu, Y. D., et al. (2018b). Hsa-let-7c controls the committed differentiation of IGF-1-treated mesenchymal stem cells derived from dental pulps by targeting IGF-1R via the MAPK pathways. Exp. Mol. Med. 50, 25–14. doi:10.1038/s12276-018-0048-7
Liu, H., Lin, H., Zhang, L., Sun, Q., Yuan, G., Zhang, L., et al. (2013). miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J. Biol. Chem. 288, 9261–9271. doi:10.1074/jbc.M112.433730
Liu, H. J., Wang, T., Li, Q. M., Guan, X. Y., and Xu, Q. (2015b). Knock-down of p300 decreases the proliferation and odontogenic differentiation potentiality of HDPCs. Int. Endod. J. 48, 976–985. doi:10.1111/iej.12392
Liu, J., Carmell, M. A., Rivas, F. V., Marsden, C. G., Thomson, J. M., Song, J. J., et al. (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Sci. (New York, N.Y.) 305, 1437–1441. doi:10.1126/science.1102513
Liu, J., Yu, F., Sun, Y., Jiang, B., Zhang, W., Yang, J., et al. (2015a). Concise reviews: characteristics and potential applications of human dental tissue-derived mesenchymal stem cells. Stem cells Dayt. Ohio 33, 627–638. doi:10.1002/stem.1909
Liu, Z., Chen, T., Han, Q., Chen, M., You, J., Fang, F., et al. (2018a). HDAC inhibitor LMK-235 promotes the odontoblast differentiation of dental pulp cells. Mol. Med. Rep. 17, 1445–1452. doi:10.3892/mmr.2017.8055
Liu, Z., Xu, S., Dao, J., Gan, Z., and Zeng, X. (2020). Differential expression of lncRNA/miRNA/mRNA and their related functional networks during the osteogenic/odontogenic differentiation of dental pulp stem cells. J. Cell. physiology 235, 3350–3361. doi:10.1002/jcp.29223
Lu, X., Chen, X., Xing, J., Lian, M., Huang, D., Lu, Y., et al. (2019). miR-140-5p regulates the odontoblastic differentiation of dental pulp stem cells via the Wnt1/β-catenin signaling pathway. Stem Cell. Res. Ther. 10, 226. doi:10.1186/s13287-019-1344-4
Ma, L., Huang, Z., Wu, D., Kou, X., Mao, X., and Shi, S. (2021). CD146 controls the quality of clinical grade mesenchymal stem cells from human dental pulp. Stem Cell. Res. Ther. 12, 488. doi:10.1186/s13287-021-02559-4
Manaspon, C., Jongwannasiri, C., Chumprasert, S., Sa-Ard-Iam, N., Mahanonda, R., Pavasant, P., et al. (2021). Human dental pulp stem cell responses to different dental pulp capping materials. BMC oral health 21, 209. doi:10.1186/s12903-021-01544-w
Marmorstein, R., and Zhou, M. M. (2014). Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762. doi:10.1101/cshperspect.a018762
McNeely, T., Leone, M., Yanai, H., and Beerman, I. (2020). DNA damage in aging, the stem cell perspective. Hum. Genet. 139, 309–331. doi:10.1007/s00439-019-02047-z
Meier, K., and Brehm, A. (2014). Chromatin regulation: how complex does it get? Epigenetics 9, 1485–1495. doi:10.4161/15592294.2014.971580
Meng, R., Li, D., Feng, Z., and Xu, Q. (2019). MyD88 hypermethylation mediated by DNMT1 is associated with LTA-induced inflammatory response in human odontoblast-like cells. Cell. tissue Res. 376, 413–423. doi:10.1007/s00441-019-02993-0
Miura, M., Gronthos, S., Zhao, M., Lu, B., Fisher, L. W., Robey, P. G., et al. (2003a). SHED: stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. U. S. A. 100, 5807–5812. doi:10.1073/pnas.0937635100
Miura, M., Gronthos, S., Zhao, M., Lu, B., Fisher, L. W., Robey, P. G., et al. (2003b). SHED: stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. U. S. A., 100, 5807–5812. doi:10.1073/pnas.0937635100
Mo, Z., Li, Q., Cai, L., Zhan, M., and Xu, Q. (2019). The effect of DNA methylation on the miRNA expression pattern in lipopolysaccharide-induced inflammatory responses in human dental pulp cells. Mol. Immunol. 111, 11–18. doi:10.1016/j.molimm.2019.03.012
Nuti, N., Corallo, C., Chan, B. M., Ferrari, M., and Gerami-Naini, B. (2016). Multipotent differentiation of human dental pulp stem cells: a literature review. Stem Cell. Rev. Rep. 12, 511–523. doi:10.1007/s12015-016-9661-9
Paino, F., La Noce, M., Tirino, V., Naddeo, P., Desiderio, V., Pirozzi, G., et al. (2014). Histone deacetylase inhibition with valproic acid downregulates osteocalcin gene expression in human dental pulp stem cells and osteoblasts: evidence for HDAC2 involvement. Stem cells Dayt. Ohio 32, 279–289. doi:10.1002/stem.1544
Palazzo, A. F., and Lee, E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6, 2. doi:10.3389/fgene.2015.00002
Pedersen, M. T., and Helin, K. (2010). Histone demethylases in development and disease. Trends Cell. Biol. 20, 662–671. doi:10.1016/j.tcb.2010.08.011
Qian, H., Zhao, J., Yang, X., Wu, S., An, Y., Qu, Y., et al. (2021). TET1 promotes RXRα expression and adipogenesis through DNA demethylation. Biochimica biophysica acta. Mol. Cell. Biol. lipids 1866, 158919. doi:10.1016/j.bbalip.2021.158919
Qiu, Z., Lin, S., Hu, X., Zeng, J., Xiao, T., Ke, Z., et al. (2019). Involvement of miR-146a-5p/neurogenic locus notch homolog protein 1 in the proliferation and differentiation of STRO-1(+) human dental pulp stem cells. Eur. J. oral Sci. 127, 294–303. doi:10.1111/eos.12624
Qu, S., Yang, X., Li, X., Wang, J., Gao, Y., Shang, R., et al. (2015). Circular RNA: a new star of noncoding RNAs. Cancer Lett. 365, 141–148. doi:10.1016/j.canlet.2015.06.003
Radhakrishnan, R., Kabekkodu, S., and Satyamoorthy, K. (2011). DNA hypermethylation as an epigenetic mark for oral cancer diagnosis. J. oral pathology Med. official Publ. Int. Assoc. Oral Pathologists Am. Acad. Oral Pathology 40, 665–676. doi:10.1111/j.1600-0714.2011.01055.x
Rao, L. J., Yi, B. C., Li, Q. M., and Xu, Q. (2016). TET1 knockdown inhibits the odontogenic differentiation potential of human dental pulp cells. Int. J. oral Sci. 8, 110–116. doi:10.1038/ijos.2016.4
Ricucci, D., Loghin, S., and Siqueira, J. F. (2014). Correlation between clinical and histologic pulp diagnoses. J. Endod. 40, 1932–1939. doi:10.1016/j.joen.2014.08.010
Schaap-Oziemlak, A. M., Raymakers, R. A., Bergevoet, S. M., Gilissen, C., Jansen, B. J., Adema, G. J., et al. (2010). MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem cells Dev. 19, 877–885. doi:10.1089/scd.2009.0112
Seong, Y., Lim, D. H., Kim, A., Seo, J. H., Lee, Y. S., Song, H., et al. (2014). Global identification of target recognition and cleavage by the Microprocessor in human ES cells. Nucleic acids Res. 42, 12806–12821. doi:10.1093/nar/gku957
Shen, H., Xu, W., and Lan, F. (2017). Histone lysine demethylases in mammalian embryonic development. Exp. Mol. Med. 49, e325. doi:10.1038/emm.2017.57
Sheng, R., Wang, Y., Wu, Y., Wang, J., Zhang, S., Li, Q., et al. (2021). METTL3-Mediated m(6) A mRNA methylation modulates tooth root formation by affecting NFIC translation. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 36, 412–423. doi:10.1002/jbmr.4180
Shi, S., Bartold, P. M., Miura, M., Seo, B. M., Robey, P. G., and Gronthos, S. (2005). The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod. craniofacial Res. 8, 191–199. doi:10.1111/j.1601-6343.2005.00331.x
Shi, S., and Gronthos, S. (2003). Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 18, 696–704. doi:10.1359/jbmr.2003.18.4.696
Shi, Y. (2007). Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 8, 829–833. doi:10.1038/nrg2218
Song, Y., Zhang, H., Song, Z., Yang, Y., Zhang, S., Wang, W., et al. (2022). Long noncoding RNA GAS5 inhibits osteogenic differentiation through MicroRNA 382-3p/TAF1 signaling. Mol. Cell. Biol. 42, e0054120. doi:10.1128/mcb.00541-20
Song, Z., Chen, L. L., Wang, R. F., Qin, W., Huang, S. H., Guo, J., et al. (2017). MicroRNA-135b inhibits odontoblast-like differentiation of human dental pulp cells by regulating Smad5 and Smad4. Int. Endod. J. 50, 685–693. doi:10.1111/iej.12678
Strahl, B. D., and Allis, C. D. (2000). The language of covalent histone modifications. Nature 403, 41–45. doi:10.1038/47412
Sultana, S., Uehara, O., Yoshida, K., Saito, T., and Abiko, Y. (2021). The histone deacetylase inhibitor, entinostat (MS-275), induces the odontogenic differentiation of an odontoblast-like cell line in the absence of an osteoblast mineralization medium. Odontology 109, 661–671. doi:10.1007/s10266-020-00588-8
Sun, D. G., Xin, B. C., Wu, D., Zhou, L., Wu, H. B., Gong, W., et al. (2017). miR-140-5p-mediated regulation of the proliferation and differentiation of human dental pulp stem cells occurs through the lipopolysaccharide/toll-like receptor 4 signaling pathway. Eur. J. oral Sci. 125, 419–425. doi:10.1111/eos.12384
Sun, Q., Liu, H., and Chen, Z. (2015). The fine tuning role of microRNA-RNA interaction in odontoblast differentiation and disease. Oral Dis. 21, 142–148. doi:10.1111/odi.12237
Sun, Z., Yu, S., Chen, S., Liu, H., and Chen, Z. (2019). SP1 regulates KLF4 via SP1 binding motif governed by DNA methylation during odontoblastic differentiation of human dental pulp cells. J. Cell. Biochem. 120, 14688–14699. doi:10.1002/jcb.28730
Tao, H., Li, Q., Lin, Y., Zuo, H., Cui, Y., Chen, S., et al. (2020). Coordinated expression of p300 and HDAC3 upregulates histone acetylation during dentinogenesis. J. Cell. Biochem. 121, 2478–2488. doi:10.1002/jcb.29470
Tao, H., Lin, H., Sun, Z., Pei, F., Zhang, J., Chen, S., et al. (2019). Klf4 promotes dentinogenesis and odontoblastic differentiation via modulation of TGF-β signaling pathway and interaction with histone acetylation. J. bone mineral Res. official J. Am. Soc. Bone Mineral Res. 34, 1502–1516. doi:10.1002/jbmr.3716
Tronstad, L., and Mjör, I. A. (1972). Capping of the inflamed pulp. Oral Surg. oral Med. oral pathology 34, 477–485. doi:10.1016/0030-4220(72)90327-1
Vasanthan, P., Govindasamy, V., Gnanasegaran, N., Kunasekaran, W., Musa, S., and Abu Kasim, N. H. (2015). Differential expression of basal microRNAs' patterns in human dental pulp stem cells. J. Cell. Mol. Med. 19, 566–580. doi:10.1111/jcmm.12381
Venkatesh, S., and Workman, J. L. (2015). Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell. Biol. 16, 178–189. doi:10.1038/nrm3941
Waddington, C. H. (2012). The epigenotype. 1942. Int. J. Epidemiol. 41, 10–13. doi:10.1093/ije/dyr184
Wang, B. L., Wang, Z., Nan, X., Zhang, Q. C., and Liu, W. (2019a). Downregulation of microRNA-143-5p is required for the promotion of odontoblasts differentiation of human dental pulp stem cells through the activation of the mitogen-activated protein kinases 14-dependent p38 mitogen-activated protein kinases signaling pathway. J. Cell. physiology 234, 4840–4850. doi:10.1002/jcp.27282
Wang, F., Nazarali, A. J., and Ji, S. (2016). Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. cancer Res. 6, 1167–1176.
Wang, H., Feng, C., Jin, Y., Tan, W., and Wei, F. (2019c). Identification and characterization of circular RNAs involved in mechanical force-induced periodontal ligament stem cells. J. Cell. physiology 234, 10166–10177. doi:10.1002/jcp.27686
Wang, J., Cao, L., Wu, J., and Wang, Q. (2018c). Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int. J. Oncol. 52, 77–88. doi:10.3892/ijo.2017.4187
Wang, J., Du, Y., Deng, J., Wang, X., Long, F., and He, J. (2019b). MicroRNA-506 is involved in regulation of the occurrence of lipopolysaccharides (LPS)-Induced pulpitis by sirtuin 1 (SIRT1). Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 25, 10008–10015. doi:10.12659/msm.918172
Wang, J., Zheng, Y., Bai, B., Song, Y., Zheng, K., Xiao, J., et al. (2020). MicroRNA-125a-3p participates in odontoblastic differentiation of dental pulp stem cells by targeting Fyn. Cytotechnology 72, 69–79. doi:10.1007/s10616-019-00358-7
Wang, T., Liu, H., Ning, Y., and Xu, Q. (2014). The histone acetyltransferase p300 regulates the expression of pluripotency factors and odontogenic differentiation of human dental pulp cells. PloS one 9, e102117. doi:10.1371/journal.pone.0102117
Wang, X., Feng, Z., Li, Q., Yi, B., and Xu, Q. (2018a). DNA methylcytosine dioxygenase ten-eleven translocation 2 enhances lipopolysaccharide-induced cytokine expression in human dental pulp cells by regulating MyD88 hydroxymethylation. Cell. tissue Res. 373, 477–485. doi:10.1007/s00441-018-2826-x
Wang, Y., Shi, Z. Y., Feng, J., and Cao, J. K. (2018b). HDAC6 regulates dental mesenchymal stem cells and osteoclast differentiation. BMC oral health 18, 190. doi:10.1186/s12903-018-0624-1
Wang, Y., Xu, Z., Jiang, J., Xu, C., Kang, J., Xiao, L., et al. (2013). Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 25, 69–80. doi:10.1016/j.devcel.2013.03.002
Wiehle, L., Raddatz, G., Musch, T., Dawlaty, M. M., Jaenisch, R., Lyko, F., et al. (2016). Tet1 and Tet2 protect DNA methylation canyons against hypermethylation. Mol. Cell. Biol. 36, 452–461. doi:10.1128/mcb.00587-15
Wu, C., and Morris, J. R. (2001). Genes, genetics, and epigenetics: a correspondence. Sci. (New York, N.Y.) 293, 1103–1105. doi:10.1126/science.293.5532.1103
Xiao, M., Li, J., Li, W., Wang, Y., Wu, F., Xi, Y., et al. (2017). MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 14, 1326–1334. doi:10.1080/15476286.2015.1112487
Xu, J., Yu, B., Hong, C., and Wang, C. Y. (2013). KDM6B epigenetically regulates odontogenic differentiation of dental mesenchymal stem cells. Int. J. oral Sci. 5, 200–205. doi:10.1038/ijos.2013.77
Xu, K., Xiao, J., Zheng, K., Feng, X., Zhang, J., Song, D., et al. (2018a). MiR-21/STAT3 signal is involved in odontoblast differentiation of human dental pulp stem cells mediated by TNF-α. Cell. Reprogr. 20, 107–116. doi:10.1089/cell.2017.0042
Xu, M., Chen, X., Lin, K., Zeng, K., Liu, X., Pan, B., et al. (2018b). The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol. cancer 17, 141. doi:10.1186/s12943-018-0894-x
Yang, J. W., Zhang, Y. F., Wan, C. Y., Sun, Z. Y., Nie, S., Jian, S. J., et al. (2015). Autophagy in SDF-1α-mediated DPSC migration and pulp regeneration. Biomaterials 44, 11–23. doi:10.1016/j.biomaterials.2014.12.006
Yang, X., Wang, G., Wang, Y., Zhou, J., Yuan, H., Li, X., et al. (2019). Histone demethylase KDM7A reciprocally regulates adipogenic and osteogenic differentiation via regulation of C/EBPα and canonical Wnt signalling. J. Cell. Mol. Med. 23, 2149–2162. doi:10.1111/jcmm.14126
Yi, S. J., and Kim, K. (2018). Histone tail cleavage as a novel epigenetic regulatory mechanism for gene expression. BMB Rep. 51, 211–218. doi:10.5483/bmbrep.2018.51.5.053
Yokoyama, A., Okuno, Y., Chikanishi, T., Hashiba, W., Sekine, H., Fujiki, R., et al. (2010). KIAA1718 is a histone demethylase that erases repressive histone methyl marks. Genes. cells devoted Mol. Cell. Mech. 15, 867–873. doi:10.1111/j.1365-2443.2010.01424.x
Yu, D., Zhao, X., Cheng, J. Z., Wang, D., Zhang, H. H., and Han, G. H. (2019). Downregulated microRNA-488 enhances odontoblast differentiation of human dental pulp stem cells via activation of the p38 MAPK signaling pathway. J. Cell. physiology 234, 1442–1451. doi:10.1002/jcp.26950
Yuan, H., Zhang, H., Hong, L., Zhao, H., Wang, J., Li, H., et al. (2018). MicroRNA let-7c-5p suppressed lipopolysaccharide-induced dental pulp inflammation by inhibiting dentin matrix protein-1-mediated nuclear factor kappa B (NF-κB) pathway in vitro and in vivo. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 24, 6656–6665. doi:10.12659/msm.909093
Yuan, H., Zhao, H., Wang, J., Zhang, H., Hong, L., Li, H., et al. (2019). MicroRNA let-7c-5p promotes osteogenic differentiation of dental pulp stem cells by inhibiting lipopolysaccharide-induced inflammation via HMGA2/PI3K/Akt signal blockade. Clin. Exp. Pharmacol. physiology 46, 389–397. doi:10.1111/1440-1681.13059
Zeng, L., Sun, S., Han, D., Liu, Y., Liu, H., Feng, H., et al. (2018b). Long non-coding RNA H19/SAHH axis epigenetically regulates odontogenic differentiation of human dental pulp stem cells. Cell. Signal. 52, 65–73. doi:10.1016/j.cellsig.2018.08.015
Zeng, L., Zhao, N., Li, F., Han, D., Liu, Y., Liu, H., et al. (2018a). miR-675 promotes odontogenic differentiation of human dental pulp cells by epigenetic regulation of DLX3. Exp. Cell. Res. 367, 104–111. doi:10.1016/j.yexcr.2018.03.035
Zhan, F. L., Liu, X. Y., and Wang, X. B. (2018). The role of MicroRNA-143-5p in the differentiation of dental pulp stem cells into odontoblasts by targeting Runx2 via the OPG/RANKL signaling pathway. J. Cell. Biochem. 119, 536–546. doi:10.1002/jcb.26212
Zhang, C., Zhang, B., Yuan, B., Chen, C., Zhou, Y., Zhang, Y., et al. (2020). RNA-Seq profiling of circular RNAs in human small cell lung cancer. Epigenomics 12, 685–700. doi:10.2217/epi-2019-0382
Zhang, D., Li, Q., Rao, L., Yi, B., and Xu, Q. (2015). Effect of 5-Aza-2'-deoxycytidine on odontogenic differentiation of human dental pulp cells. J. Endod. 41, 640–645. doi:10.1016/j.joen.2014.12.006
Zhang, Z. M., Lu, R., Wang, P., Yu, Y., Chen, D., Gao, L., et al. (2018). Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554, 387–391. doi:10.1038/nature25477
Zheng, L. W., Zhang, B. P., Xu, R. S., Xu, X., Ye, L., and Zhou, X. D. (2014). Bivalent histone modifications during tooth development. Int. J. oral Sci. 6, 205–211. doi:10.1038/ijos.2014.60
Zheng, Y., and Jia, L. (2016). Long noncoding RNAs related to the odontogenic potential of dental mesenchymal cells in mice. Archives oral Biol. 67, 1–8. doi:10.1016/j.archoralbio.2016.03.001
Zhong, J., Tu, X., Kong, Y., Guo, L., Li, B., Zhong, W., et al. (2020). LncRNA H19 promotes odontoblastic differentiation of human dental pulp stem cells by regulating miR-140-5p and BMP-2/FGF9. Stem Cell. Res. Ther. 11, 202. doi:10.1186/s13287-020-01698-4
Zhong, S., Zhang, S., Bair, E., Nares, S., and Khan, A. A. (2012). Differential expression of microRNAs in normal and inflamed human pulps. J. Endod. 38, 746–752. doi:10.1016/j.joen.2012.02.020
Zhong, Y. X., Li, W. S., Liao, L. S., and Liang, L. (2019). LncRNA CCAT1 promotes cell proliferation and differentiation via negative modulation of miRNA-218 in human DPSCs. Eur. Rev. Med. Pharmacol. Sci. 23, 3575–3583. doi:10.26355/eurrev_201905_17779
Zhou, J., Yang, L., Zhong, T., Mueller, M., Men, Y., Zhang, N., et al. (2015). H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 6, 10221. doi:10.1038/ncomms10221
Zhou, Y., Zheng, L., Li, F., Wan, M., Fan, Y., Zhou, X., et al. (2018). Bivalent histone codes on WNT5A during odontogenic differentiation. J. Dent. Res. 97, 99–107. doi:10.1177/0022034517728910
Zhu, J. G., Shen, Y. H., Liu, H. L., Liu, M., Shen, Y. Q., Kong, X. Q., et al. (2014). Long noncoding RNAs expression profile of the developing mouse heart. J. Cell. Biochem. 115, 910–918. doi:10.1002/jcb.24733
Keywords: epigenetic regulation, odontogenic differentiation, pulp, DPSC, LncRNA, long noncoding RNA
Citation: Huang L, Chen X, Yang X, Zhang Y, Liang Y and Qiu X (2024) Elucidating epigenetic mechanisms governing odontogenic differentiation in dental pulp stem cells: an in-depth exploration. Front. Cell Dev. Biol. 12:1394582. doi: 10.3389/fcell.2024.1394582
Received: 01 March 2024; Accepted: 13 May 2024;
Published: 28 May 2024.
Edited by:
Fuchun Fang, Southern Medical University, ChinaReviewed by:
Chao Liu, Dalian Medical University, ChinaHaoqing Yang, Capital Medical University, China
Copyright © 2024 Huang, Chen, Yang, Zhang, Liang and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoling Qiu, bGluZ2xpbmdqb2pvQDE2My5jb20=
 Lei Huang
Lei Huang Xuan Chen
Xuan Chen