- 1Hunan International Joint Laboratory of Animal Intestinal Ecology and Health, Laboratory of Animal Nutrition and Human Health, College of Life Sciences, Hunan Normal University, Changsha, Hunan, China
- 2Hunan Provincial Key Laboratory of Animal Intestinal Function and Regulation, Changsha, Hunan, China
Background: The COBLL1 gene has been implicated in human central obesity, fasting insulin levels, type 2 diabetes, and blood lipid profiles. However, its molecular mechanisms remain largely unexplored.
Methods: In this study, we established cobll1a mutant lines using the CRISPR/Cas9-mediated gene knockout technique. To further dissect the molecular underpinnings of cobll1a during early development, transcriptome sequencing and bioinformatics analysis was employed.
Results: Our study showed that compared to the control, cobll1a−/− zebrafish embryos exhibited impaired development of digestive organs, including the liver, intestine, and pancreas, at 4 days post-fertilization (dpf). Transcriptome sequencing and bioinformatics analysis results showed that in cobll1a knockout group, the expression level of genes in the Retinoic Acid (RA) signaling pathway was affected, and the expression level of lipid metabolism-related genes (fasn, scd, elovl2, elovl6, dgat1a, srebf1 and srebf2) were significantly changed (p < 0.01), leading to increased lipid synthesis and decreased lipid catabolism. The expression level of apolipoprotein genes (apoa1a, apoa1b, apoa2, apoa4a, apoa4b, and apoea) genes were downregulated.
Conclusion: Our study suggest that the loss of cobll1a resulted in disrupted RA metabolism, reduced lipoprotein expression, and abnormal lipid transport, therefore contributing to lipid accumulation and deleterious effects on early liver development.
1 Introduction
Vitamin A, known as retinol, is essential for normal physiological functions in living organisms. Retinol’s metabolite, retinoic acid (RA), is pivotal in cellular proliferation and differentiation (Gudas and Wagner, 2011), embryonic growth (Zile, 2001), organogenesis (Stafford et al., 2006), visual transduction (Zhong et al., 2012), immune regulation (Bono et al., 2016), and glucose and lipid metabolism (Tanumihardjo et al., 2016). Retinol Ester (RE) in food is hydrolyzed to retinol by retinol ester hydrolase (REH) (Boily et al., 2003). Afterward, retinol forms a complex with retinol binding protein 4 (RBP4) and plasma transthyretin (TTR) secreted by the liver before being transported to peripheral tissues (Ronne et al., 1983). This RBP4-TTR-retinol complex binds to the signaling receptor and transporter of retinol, STRA6 (STRA6), and undergoes two dehydrogenation reactions to produce RA (Theodosiou et al., 2010; Sun and Kawaguchi, 2011). RA is subsequently rapidly degraded by P450 family enzymes (Pennimpede et al., 2010). Vitamin A deficiency (VAD) is associated with metabolic syndrome (MetS) with liver issues like steatosis seen in rats lacking Vitamin A (Baybutt et al., 2000). Approximately 80% of retinol is stored as RE in the lipid droplets of quiescent hepatic stellate cells (HSCs) (Haaker et al., 2020). Liver injury leads to fibrosis of hematopoietic stem cells, which ultimately leads to retinol deficiency (Saeed et al., 2017). It is reported that decreased vitamin A intake result in nonalcoholic fatty liver disease (NAFLD) (Musso et al., 2007), and obese patients with NAFLD display reduced serum retinol (Villaça Chaves et al., 2008). NAFLD is a hepatic manifestation of MetS characterized by hepatic steatosis, fibrogenesis, and inflammation (Friedman et al., 2018).
The COBLL1 gene, originally isolated from a human brain cDNA library, is highly expressed in a variety of tissues including the liver, lung, kidney, pancreas, ovary, spinal cord, and brain (Nagase et al., 1999). It is a negative regulator of tumor cell apoptosis and is implicated in the progress of various cancers (Han et al., 2017; Takayama et al., 2018), as well as in insulin resistance-related metabolic diseases (Mancina et al., 2013). Genome-wide association studies have revealed the localization of the human COBLL1 gene in genomic regions associated with metabolism, including high-density cholesterol, triglyceride metabolism, insulin levels, type 2 diabetes, and other metabolic disorders (Manning et al., 2012; Albrechtsen et al., 2013; Desmarchelier et al., 2014). Furthermore, the SNP rs7607980 in the COBLL1 gene affects insulin resistance (Kraja et al., 2014), and its polymorphic form has been identified in patients with MetS (Albrechtsen et al., 2013). Loss of COBLL1 results in excessive lipid accumulation and increased lipolysis in preadipocytes (Simpson-Golabi-Behmel syndrome), suggesting that COBLL1 may have a crucial role in lipid metabolism (Chen et al., 2020). In zebrafish, two COBLL1 homologs have been identified: cobll1a on chromosome 9 and cobll1b on chromosome 6. Both homologs possessed a pair of KRAP domains, which are pivotal for protein-protein interactions and subcellular localization (Schwintzer et al., 2011). It is reported that cobll1b plays an important role in hematopoiesis during early embryonic development (Kim et al., 2017). Utilizing CRISPR/Cas9 gene editing technology, we constructed zebrafish cobll1a gene knockout lines to ascertain the role of this gene in embryonic development. Our study found that the digestive organs, such as the liver, intestine, and pancreas developed abnormally in 4 dpf cobll1a−/− embryos, compared to the control. To investigate the molecular mechanism underlying cobll1a gene regulation of digestive organ development, transcriptome sequencing and bioinformatics analysis were conducted. We found a significant alteration in the expression levels of genes related to lipid metabolism in the cobll1a gene knockout. Loss of cobll1a led to abnormal RA metabolism, increased lipid synthesis, decreased lipolysis metabolism, disrupted lipoprotein metabolism, abnormal lipid transport, and subsequent lipid accumulation. In conclusion, the cobll1a gene is critical for the RA signaling pathway and lipid metabolism in zebrafish.
2 Materials and methods
2.1 Zebrafish breeding
Tuebingen (TU) zebrafish were bred and maintained in our laboratory under the following conditions: a water temperature of 28°C, pH 6.5–7.5, salinity of 450–500 μs/cm, and a 14 h/10 h light and dark cycle. Fish were crossed weekly, the embryos were cultivated at 28.5°C in E3 solution.
2.2 Establishment of zebrafish cobll1a mutant lines
Mutant lines of zebrafish cobll1a were established using CRISPR/Cas9 gene editing technology. The complete gene and amino acid sequence of zebrafish cobll1a were obtained from the NCBI database, and the corresponding genome sequence was accessed from UCSC. The target sequence of cobll1a was identified via the CRISPOR website, and the target sites were localized in exon 4. Two target sequences were selected: sequence 1 (sgRNA-F1) is 5′-ACCAGTTATGGATGTTC-3′, and sequence 2 (sgRNA-F2) is 5′-TGATCGGCTCTCTCGAAT-3’. The core sequence of T7 RNA polymerase promoter was added to the 5′ end of the target sequence to generate forward primers. The PCR amplification, using forward primers (sgRNA-F1/2) and reverse primers (sgRNA-R), the purified PCR product as a template, sgRNAs were synthesized in vitro with the T7 in vitro transcription kit (ThermoFisher). Purified sgRNAs were recovered with the RNA purification kit (Qiagen) and mixed with Cas9 protein (ThermoFisher) for injection. Zebrafish embryos were injected at the one-cell stage and then incubated at 28.5°C. Genomic DNA was extracted from the F0 embryos and tested for cobll1a mutation using primers 5′-TGCATATACTGTATGTGGGACA-3′ and 5′-TCTTGTTGTCGTCACTTCCT-3’. DNA showing the deletions or insertions was sequenced and the genomic DNA of F1 and F2 generations were amplified using the following primers (F) 5′-TGCATATACTGTATGTGGGACA-3′ and (R) 5′-TGACAAAACTGACCACT-3’.
At 36 hours post-fertilization (hpf), the effciency of the sgRNAs was validated by randomly selecting injected and wild-type embryos. The PCR product of the control is 775 bp, with the two target sites being 134 bp apart. If both target sites work, a deletion or an insertion sequence would be generated, in comparison to the control. After validating the two target sites successfully work, the remaining embryos were raised to 45 dpf. Caudal fin clipping was performed individually, and genomic DNA was extracted for genotyping. Fish with missing or inserting DNA sequences were raised to adulthood (F0) and crossbred with wild-type to obtain F1 generation. Genotypes of F1 mutants were determined, and DNA fragments less than 775 bp were recovered and sequenced. Mutant lines resulting in protein-coding frame shift were selected for further studies.
2.3 Whole-mount in situ hybridization (WISH)
Zebrafish embryos at the desired stages were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. They were then rinsed twice with PBST, treated with a 10 mg/mL protease K solution, refixed with 4% PFA at room temperature for 20 min, and rinsed twice again with PBST. They were then incubated with DIG-labeled antisense RNA probe overnight at 68°C. On the following day, the embryos were washed with 50% formamide/2xSSCT, 2xSSCT and 0.2xSSCT solutions and incubated with anti-DIG antibodies overnight. After washing three times with MABT solution, with a 25 min interval, the staining reaction was performed with BCIP-NBT solution and imaged using a Leica stereo microscope. Antisense RNA probes labeled with digoxin-UTP for cobll1a, fabp10a, insulin, trypsin, fabp2, rdh10, radh1a2, cyp26a1, rbp4, and fasn genes were synthesized using the T7 in vitro transcription kit (Thermo Fisher). The primer sequences for these genes were listed in Supplementary Table S1.
2.4 Quantitative real-time polymerase chain reaction (qRT-PCR)
Fifty 52 hpf control and cobll1a−/− embryos were collected in triplicate, quickly frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted from the embryos using TRIZOL (Takara) reagent under manufacturer’s instructions. 1μg of RNA was reverse transcribed into cDNA using reverse transcriptase (TaKaRa) and oligonucleotide primers. Expression level of related genes was measured by quantitative PCR using 2×SYBR Green Master qPCR Mix (Vazyme). The qPCR primers used for this study are listed in Supplementary Table S2. Relative expression level of the tested mRNAs were determined using 18s as an internal reference and the comparative Ct (2−△△Ct) method (p < 0.05).
2.5 RNA sequencing and differentially expressed genes analysis
Fifty 52 hpf control and cobll1a−/− embryos were collected in triplicate and were subjected to RNA-seq and data analysis by Shanghai Ouyi Biotechnology Co, LTD. (Shanghai, China). Total RNA was extracted using the mirVana miRNA Separation Kit (Ambion), and the integrity of RNA was evaluated using the Agilent 2,100 bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Following manufacturer’s instructions, cDNA libraries were prepared and sequenced on the Illumina platform (HiSeqTM 2,500 or Illumina HiSeq X Ten) to produce paired-end reads of 125 bp/150 bp. The raw data were processed by Trimmomatic, and the cleaned read segments were mapped to the GRCz11 reference genome using Hisat2. FPKM was calculated using the output file from feature Counts. Genes with FPKM>1 were considered as expressed, and the DESeq2 (v 1.40.2) algorithm was employed for differential gene expression analysis. Differentially expressed genes (DEGs) were filtered with p < 0.05 and |log2FoldChange|>1 as thresholds. Volcano plots and heat maps were generated to display the distribution of overlapping DEGs between the knockout and the control groups. The R cluster Profiler package (v 4.8.3) was used for GO and KEGG enrichment analysis of DEGs. The enrichment results were visualized using the R package ggplot2 (v 3.4.3) and the online analysis website Sangerbox3.0 (http://www.sangerbox.com/). Gene Set Enrichment Analysis (GSEA) was also performed on the list of genes whose fold changes were detected in the experiment. The enrichment of up-regulated or down-regulated gene sets in the KEGG pathway database was calculated, excluding gene sets with fewer than 10 genes or more than 500 genes. The T-statistical average of genes in each KEGG pathway was calculated using 1,000 permutation tests. Pathways with a normalized enrichment score (NES) > 0 were considered up-regulated, and those with NES<0 were considered down-regulated. Key genes in each pathway were annotated using the biomaRt software package and the GSEA enrichment results were visualized through the function implementation package “GseaVis”.
2.6 Oil red O staining
Fixed zebrafish embryos were initially rinsed with phosphate-buffered saline (PBS) for 5 min, followed by permeabilization using sequentially 60% and 100% isopropyl alcohol for 30 min. Subsequently, the embryos were stained with 0.5% Oil Red O (Sigma, USA) for 3 h at room temperature in dark. After staining, the embryos were washed again with PBS for 5 min.
2.7 Statistical analysis
All experiments were repeated at least three times. Significant differences between different types of embryos were calculated using a t-test. *, **, *** denote p < 0.05, <0.01, and <0.001, respectively.
3 Results
3.1 Cobll1a expression pattern during embryonic development in zebrafish
In order to investigate the role of cobll1a in early zebrafish development, we employed Whole-Mount In Situ Hybridization (WISH) to observe the spatial-temporal expression pattern of cobll1a. WISH results demonstrated that cobll1a was expressed in the one-cell stage and eight-cell stage embryos (Figures 1A–C), ubiquitously expressed at 12 hpf (Figure 1D) and localized in the head at 18 somite stage (ss) (Figure 1E). Expression of cobll1a was detected in the spinal cord at 24 hpf and 36 hpf (Figures 1F, G), and notably enriched in the eye, brain, liver, and digestive organs at 72 hpf (Figure 1H). Additionally, RT-qPCR was utilized to measure cobll1a mRNA expression level at different developmental stages (Figure 1I). The RT-qPCR data revealed that cobll1a mRNA was highly expressed at the one-cell stage and gradually decreased, which was consistent with the WISH results (Figures 1A–H). These findings suggest that cobll1a may be involved in early development in zebrafish.
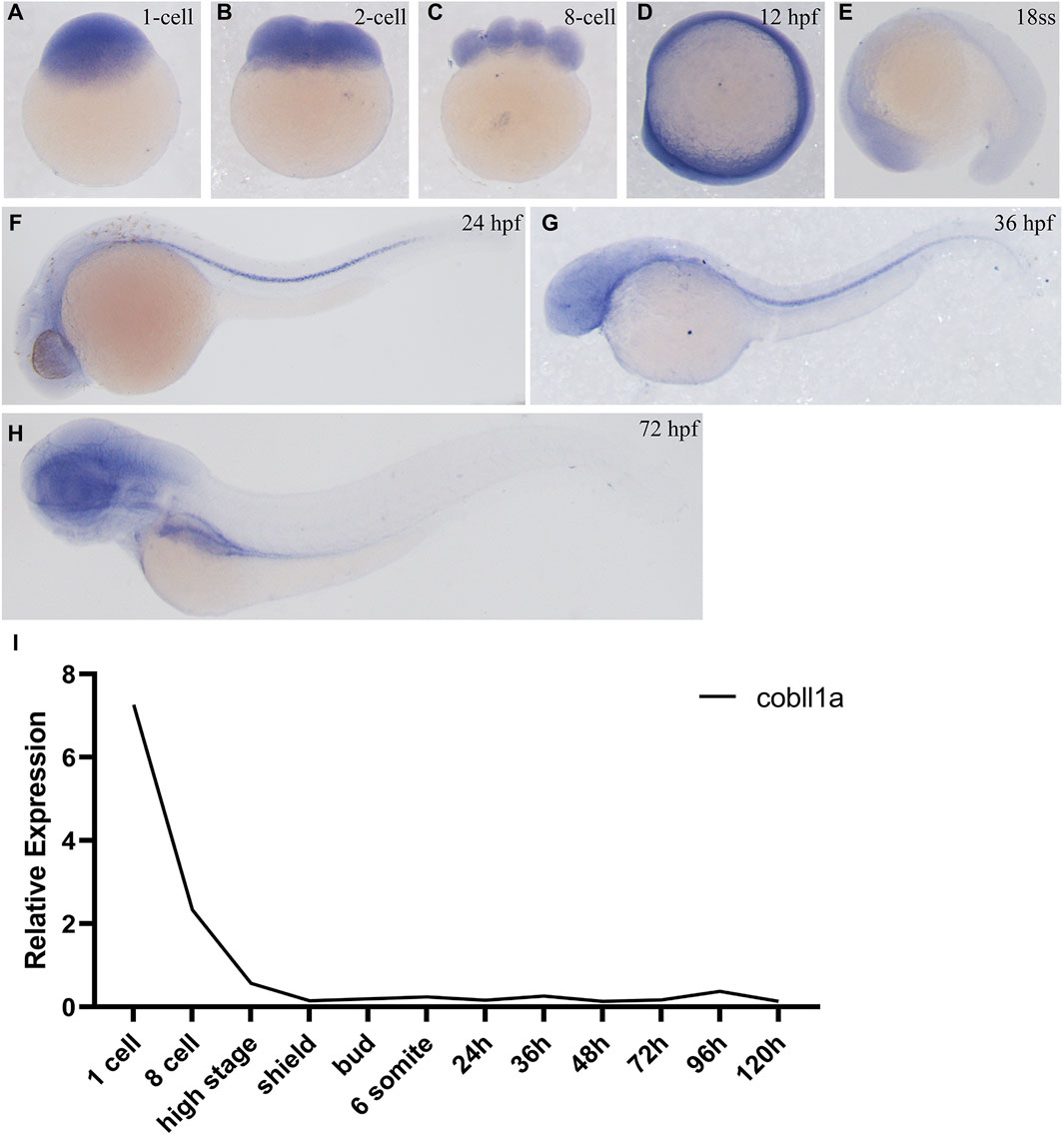
Figure 1. Expression pattern of cobll1a. (A–H) Spatio-temporal expression profile of the cobll1a gene detected by in situ hybridization in embryos. (A–C) cobll1a is expressed at the one to eight cell stages. (D) cobll1a is ubiquitously expressed at 12 hpf. (E) cobll1a is expressed in the head. (F, G) cobll1a was expressed in the notochord at 24 hpf and 36 hpf. (H) cobll1a is expressed in the eye, brain, liver and digestive system in 72 hpf embryos. (I) Using 18S as the reference gene, RT-qPCR results demonstrated the relative expression of the cobll1a at different stages.
3.2 Abnormal embryonic development due to cobll1a gene knockout
To elucidate the role of the cobll1a gene, CRISPR/Cas9 gene editing technology was employed to generate cobll1a mutant line. Initially, we conducted a bioinformatics analysis of the gene, selected a pair of knockout targets separated by 134 bp on exon4 (Supplementary Figure S2A), and established three independent mutant lines from different F0 mutants. Cobll1a line1 harbored a 2 bp insertion at target site 1 and a 77 bp deletion at target site 2 (Supplementary Figure S2B), cobll1a line2 had a a 4 bp deletion at target site 1 and a 100 bp deletion at target site 2 (Supplementary Figure S2C), while cobll1a line3 displayed a seven bp deletion at target site one and a 77 bp deletion at target site 2 (Supplementary Figure S2D). These alterations resulted in a frame shift in the open reading frames. In cobll1a line1, an early stop codon resulted in a truncated COBLL1A protein consisting of 104 amino acids (Supplementary Figure S2B). In cobll1a line2, the COBLL1A protein was altered at the 35th amino acid and translation was terminated after an additional 34 amino acids (Supplementary Figure S2C); whilst in cobll1a line3, the COBLL1A protein was altered at the 70th amino acid and ceased translation after an additional 31 amino acids (Supplementary Figure S2D). These nonsense mutations resulted in a loss of function of the cobll1a gene in all three mutant lines (Supplementary Figures S2B-S2D). Heterozygotes from these three lines exhibited no obvious phenotype and were able to mature into adult. The impact of the cobll1a gene’s impact on zebrafish development was studied by incrosing the cobll1a−/− and observed for 5 days. All the embryos of the three mutant lines developed normally, no significant difference was observed in the overall development of the 5 dpf cobll1a−/− homozygous mutant embryos compared to the control (Figure 2A). Given that each line resulted in a truncated COBLL1A protein, Line2 was chosen for subsequent experiments. To further confirm if cobll1a was knocked out successfully, the mRNA expression level of cobll1a was examined by RT-qPCR experiment. The results showed that a significant reduction in cobll1a gene expression was observed in cobll1a−/− embryos compared to the control group p < 0.001 (Figure 2B). Moreover, we used WISH to detect the expression level of cobll1a in WT and cobll1a−/− embryos, and the results showed that the expression of cobll1a in 24 hpf cobll1a−/− embryos decreased, compared to the control (Figure 2C). To validate the genotype, genomic DNA was extracted, followed by PCR and agarose gel electrophoresis. Results from the agarose gel electrophoresis showed that in lanes one to six, a band of 671 bp was observed, indicating that all of them are cobll1a−/− (Figure 2E). The 8-month-old cobll1a−/− adult fish exhibited an enlarged abdomen, more diminutive eyes, and smaller head phenotype, in contrast to the control (Figure 2D).
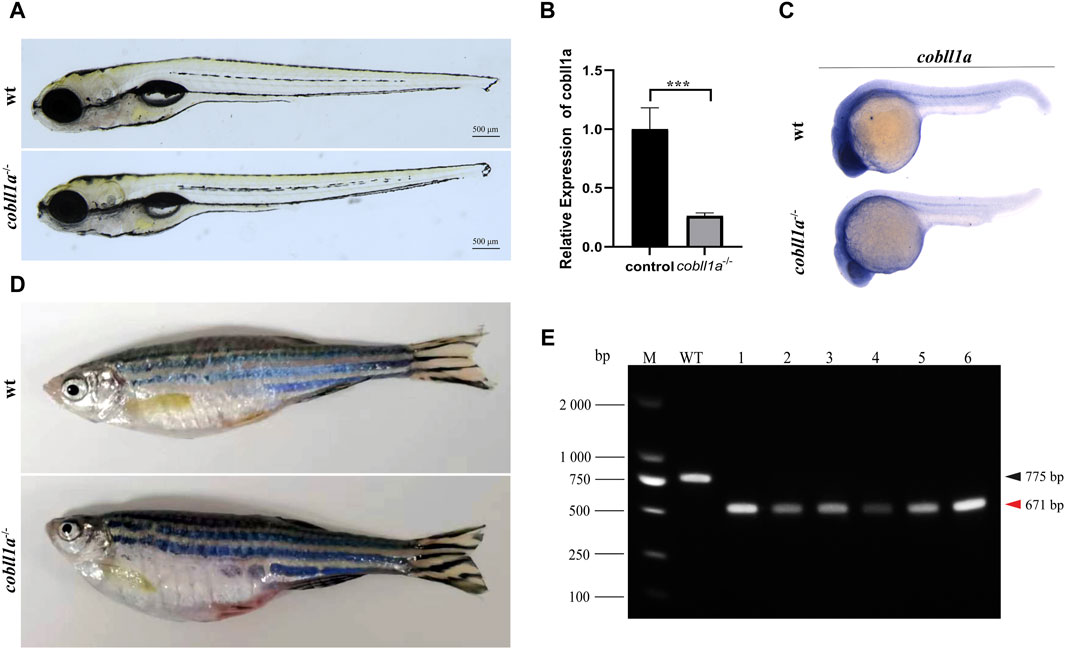
Figure 2. Loss of cobll1a results in a microcephalic phenotype. (A) cobll1a−/− larvae developed normally at 5 dpf. (B) Transcriptional expression level of cobll1a in cobll1a−/− embryos at 52 hpf were scrutinized by RT-qPCR, compared with the control. (C) An RNA probe of cobll1a was utilized in situ hybridization of WT and cobll1a−/− embryos at 24 hpf. (D) cobll1a−/− juveniles exhibited an enlarged abdomen and smaller eyes and head at 8 months. (E) Genotyping results of cobll1a−/− as shown in Figure 2A.
3.3 Loss of cobll1a impacts liver development
In situ hybridization results showed that cobll1a was expressed in the liver and other digestive organs at 72 hpf. Previous studies have shown that abnormalities in the human cobll1 gene lead to defects in glucose and lipid metabolism (Desmarchelier et al., 2014). Since the liver plays a crucial role in metabolism, we proposed that the cobll1a mutation in zebrafish may influence liver development and metabolism. To test this hypothesis, RT-qPCR was performed to measure the expression levels of liver development-related genes in 52 hpf cobll1a−/− embryos. The results indicated a decreased expression of hepatic progenitor cell markers hhex and cp in cobll1a mutants, compared to the control (Figure 3E). This suggests that the loss of cobll1a may led to abnormal liver development in zebrafish. It is reported that zebrafish’s foregut endoderm cells develop into progenitor cells of the liver, pancreas, and intestine, forming organ buds at specific locations before maturing into organs (Field et al., 2003). Whether the cobll1a mutation also affects the development of other digestive organs remains unclear. The liver cell-specific expression gene fabp10a, the early intestinal development marker fabp2, the exocrine pancreatic development marker trypsin, and the endocrine pancreatic marker insulin were examined. RT-qPCR results showed that the expression level of marker genes in digestive organs was reduced compared to the control (Figure 3E). To further verify the results that cobll1a mutation led to abnormal hepato-pancreatic intestine development in zebrafish, we used antisense digoxin-labeled RNA probes for WISH. The WISH results showed that the expression of fabp10a, fabp2, insulin, and trypsin in cobll1a−/− embryos were decreased (Figures 3A–D), which is consistent with the RT-qPCR results.
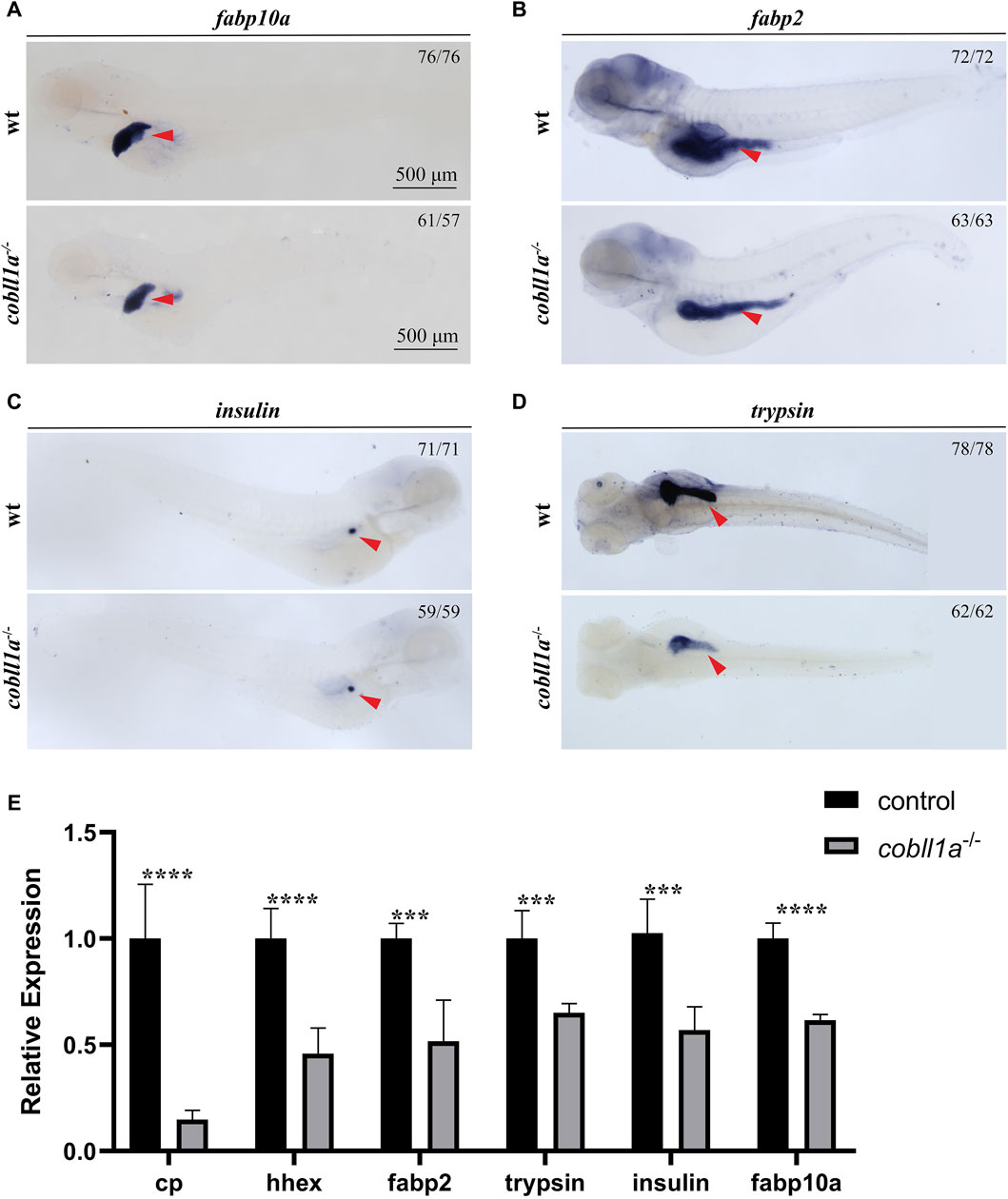
Figure 3. Depicts the abnormal liver development observed in the absence of cobll1a. (A) An RNA probe of fabp10a was utilized in situ hybridization of WT and cobll1a−/− embryos at 4 dpf. (B–D) WT and cobll1a−/− embryos at 4 dpf were detected with anti-sense RNA probes: intestinal marker fabp2 (B), endocrine pancreatic marker insulin (C), and excretory pancreatic marker trypsin (D) through WISH. (E) The mRNA expression level of hepatopancreatic-intestinal development-related genes in WT and cobll1a−/− embryos were analyzed using RT-qPCR at the 52 hpf stage.
3.4 The role of cobll1a in zebrafish earlydevelopment using a transcriptome profile
To uncover the molecular mechanism of cobll1a in liver development, three groups of 52 hpf wild type and cobll1a−/− zebrafish embryos were collected, with 50 embryos in each group. The specimens were sent to Shanghai Ouyi Biomedical Technology Co, Ltd. For high-throuphput transcriptome sequencing.
Transcriptome sequencing and subsequent bioinformatics analysis yielded an average of approximately 500,000 reads per sample, all mapped to the reference genome. Principal Component Analysis (PCA) results visualized the relationship between experimental and control samples, depicting a pronounced separation between the two groups (Figure 4A). Heat maps of DEGs (Differentially Expressed Genes) were established using gplot analysis, distinctly segregating experimental and control samples into two separate groups (Figure 4B). The screening threshold was set as |log2Foldchange| ≥ 1 and p-value≤0.05. A total of 832 genes were identified, of which 423 were down-regulated and 409 were up-regulated (Figure 4C). Volcano maps were used to illustrate the distribution of up-regulated and down-regulated genes (Figure 4D). Functional enrichment analysis of DEGs revealed significant enrichment in KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways such as “retinol metabolism”, “steroid hormone biosynthesis”, “drug metabolism-cytochrome P450”, and “PPAR signaling pathway” (Figure 4E). This suggests that the cobll1a knockout results in abnormal retinol and lipid metabolism. Additionally, 235 significantly enriched GO terms were identified, notably associated with the liver: “Lipoprotein complex”, “lipid metabolic process”, “cholesterol transport”, “lipid transport”, “triglyceride homeostasis”, “cholesterol effluence”, “cholesterol homeostasis”, “lipoprotein metabolic process”, “acylglycerol homeostasis”, “sterol transport”, “fatty acid binding” and other biological processes (Figure 4F). These results suggest that the deletion of the cobll1a gene disrupts lipid metabolism, lipid homeostasis, and lipid transport function, primarily affecting down-regulated genes enrichment. To further verify liver-related genes, DEGs such as apoa1a, apoa1b, apoa4a, and apoa4b.1 were selected from GO and KEGG for validation.
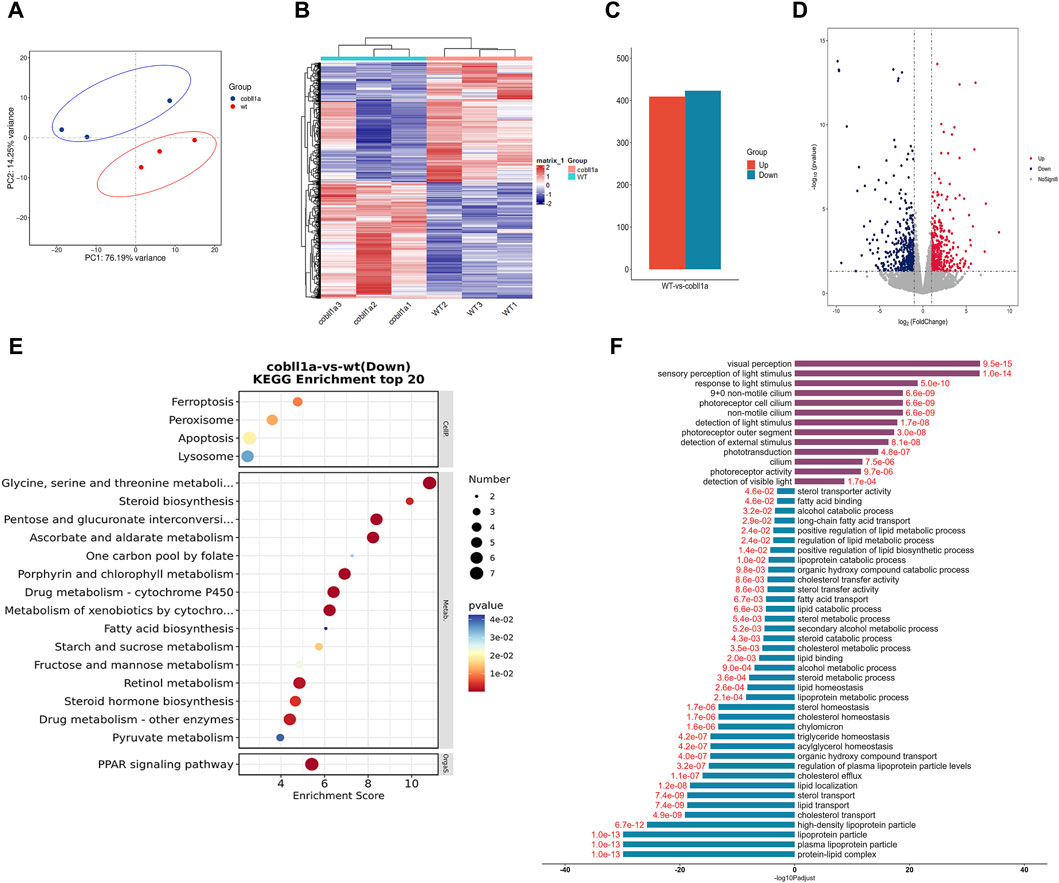
Figure 4. The DEGs analysis of WT and cobll1a−/− zebrafish embryo transcriptomes. (A) PCA cluster analysis of WT and cobll1a mutant zebrafish embryos demonstrated a significant correlation among three biological replicates in both the WT and cobll1a mutant groups. (B) Clustering heat maps of WT and cobll1a−/− were divided into two distinct groups. (C) The total number of differential genes in WT and cobll1a mutant zebrafish embryo samples was represented, with red denoting 409 up-regulated genes and blue indicating 423 down-regulated genes. (D) The volcano map displays DEGs between the two groups, with red representing up-regulated genes, blue signifying down-regulated genes, and gray denoting non-DEGs. Each dot represents a gene. (E) KEGG analysis of down-regulated DEGs revealed a significantly enriched pathway. (F) GO enrichment analysis of DEGs provides a description of their functions. Purple indicates up-regulated genes, blue represents down-regulated genes, and red fonts depict corrected p-values. Black font is used for the GO feature comment.
3.5 Cobll1a absence triggers disturbances in retinoic acid metabolism
Analysis of RNA-seq results revealed an abnormality in the RA metabolism following cobll1a mutation, including the PPAR signaling pathway and cytochrome p450 signaling pathway (Figure 5A). Consequently, focus shifted to the RA metabolic pathway. Retinol, oxidized to retinal by retinol dehydrogenase 10 (rdh10) (Metzler and Sandell, 2016), undergoes further dehydrogenation by retinal dehydrogenase to yield RA (Kedishvili, 2016). Humans and mice possess three variants of retinal dehydrogenases: aldh1a1, aldh1a2, and aldh1a3(Cunningham and Duester, 2015). However, zebrafish lack the aldh1a1 gene, and RA is primarily synthesized by aldh1a2(Grandel et al., 2002). Humans and mice encompass three RA receptors (RARα, β, γ), while zebrafish only have two homologues of RARα (raraa and rarab), two of RARγ (rarga and rargb), lacking the RARβ gene (Kastner et al., 1995). Genes associated with RA metabolism, such as aldh1a2 and rdh10, were detected among the DEGs. Consequently, we selected significantly altered RA metabolism-related genes for cluster analysis (Figure 5B) and RT-qPCR.Figure 5C illustrates significant changes in the expression of RA metabolism-related genes compared to the control. RT-qPCR results revealed a down-regulation in the expressions level of RA anabolic genes rdh10 and aldh1a2, and an up-regulation in the expression level of the RA catabolism gene cyp26a1. Additionally, the expression level of RARs genes raraa, rarab, rarga, and rargb were markedly down-regulated (p < 0.05) (Figure 5C). The expression level of retinol-binding protein rbp4 and retinoid-inducing protein stra6, responsible for retinol circulation, were significantly down-regulated (Figure 5C).
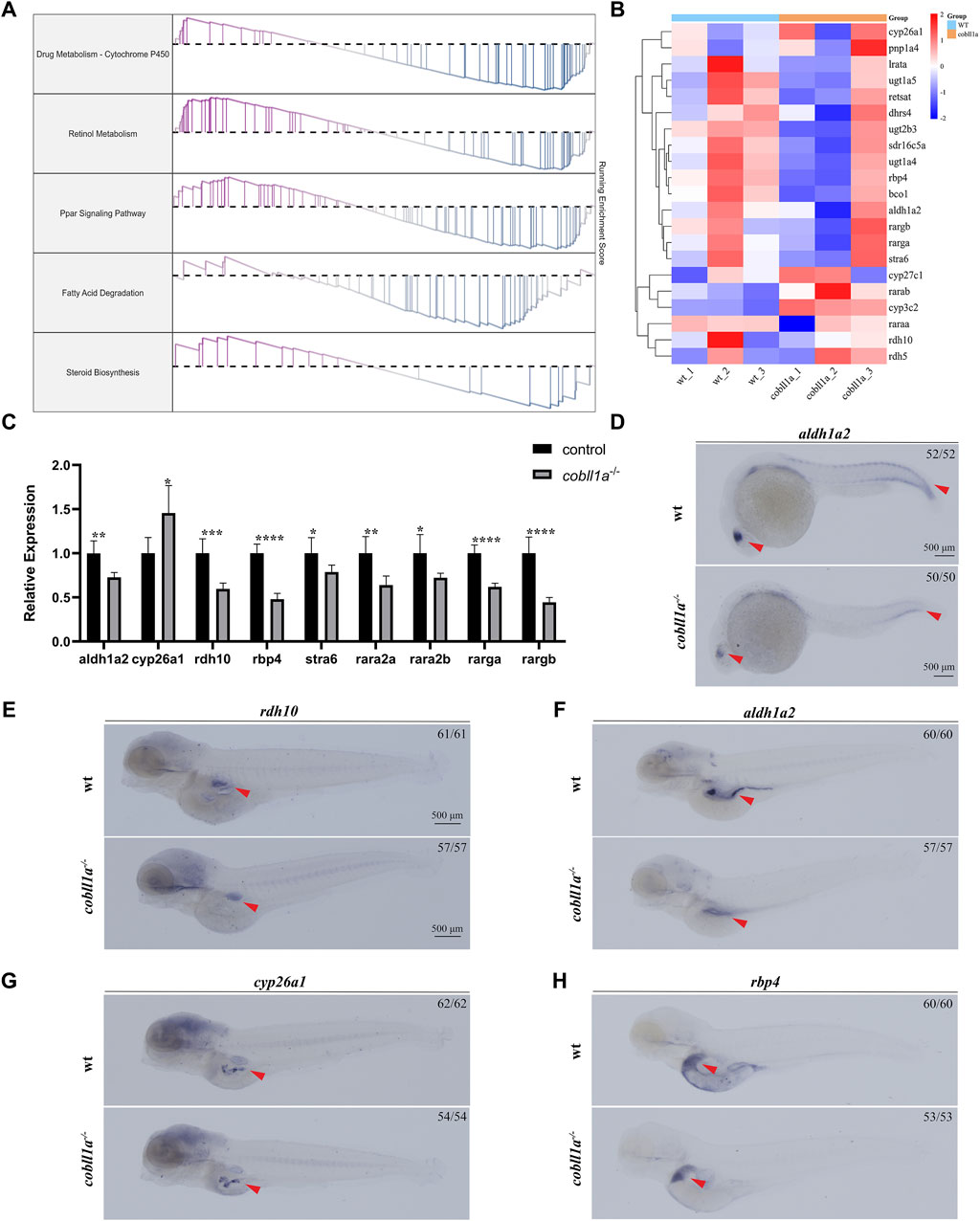
Figure 5. Loss of cobll1a leads to abnormal RA metabolism in zebrafish. (A) Line chart of GSEA results with the right ordinate representing the enrichment score. (B) Cluster heat maps of RA metabolism-related genes in DEGs, displaying significant differences between WT and cobll1a mutant. (C) RT-qPCR analysis of the transcriptional expression level of RA metabolism-related genes at 52 hpf cobll1a−/− compared with WT. (D, F) The expression of aldh1a2 was decreased in cobll1a−/−embryos, observed in eye and somatic cells at 24 hpf, in liver and intestinal tissues at four dpf. (E) At 4 dpf, the expression of rdh10 was decreased in cobll1a−/− embryos. (G) At 4 dpf, the expression of cyp26a1 was increased in cobll1a−/− embryos. (H) At 4dpf, the expression of rbp4 was decreased in cobll1a−/− embryos.
The expression pattern of rdh10, aldh1a2, cyp26a1, and rbp4 were detected using WISH, the results showed that they were expressed in intestine or liver (Figures 5E–H). Furthermore, the expression of aldh1a2 was reduced in 24 hpf cobll1a−/− embryos compared to control (Figure 5D). The expressions of rdh10, aldh1a2, and rbp4 in 4 dpf cobll1a−/− embryos were down-regulated (Figures 5E, F, H), and cyp26a1 was up-regulated (Figure 5G), corroborating the RT-qPCR results.
3.6 Cobll1a deficiency leads to hepatic lipid accumulation
Steatosis, a defining feature of NAFLD, progressively evolves into steatohepatitis and fibrosis (Angulo, 2002). It is chiefly characterized by lipid accumulation, including non-esterified fatty acids (NEFA), triglycerides, and non-esterified cholesterol. Studies suggest that exogenous RA treatment in wild-type mice results in decreased lipogenesis and enhanced catabolism (Amengual et al., 2010). Some researchers posit a potential correlation between liver triglyceride levels and reduced liver retinol in NAFLD (Chaves et al., 2014). Prior studies reveal that cobll1a mutation impacts RA metabolism and inhibits the RA signaling pathway. So we hypothesized that cobll1a mutation may result in an increase in hepatic fat anabolism, leading to steatosis and subsequent NAFLD progression. To verify whether cobll1a mutations contribute to enhanced hepatic fat anabolic metabolism, we analyzed fatty acid synthase (fasn), stearoyl-coA desaturase (scd), and elongation of very long-chain fatty acids protein two and six (elovl2, elovl6) using RT-qPCR. We also assessed the expression level of diacylglycerol oacyltransferase 1a (dgat1a) and sterol regulatory elements-binding transcription factor 1 and 2 (srebf1, srebf2) genes, crucial nuclear transcription factors regulating liver lipid metabolism. The cluster heat map and RT-qPCR results revealed significant up-regulation of fasn, scd, elovl2, elovl6, dgat1a, srebf1, and srebf2 genes in cobll1a−/− embryos compared to the control group, suggesting increased fat anabolism in cobll1a−/− embryos (Figures 6A, B). Moreover, the expression level of lipoprotein lipase (lpl) and liver lipase (lipc) was significantly down-regulated, indicating reduced lipid catabolism (Figure 6C). Subsequently, we utilized WISH to detect fasn expression. WISH results displayed upregulation of fasn expression in cobll1a−/− embryos (Figure 6D). Next, we used oil red O staining to detect the distribution and content of fat in cobll1a mutant and the control embryos, and the results showed that the oil red O positive region in cobll1a mutant was enlarged and there was obvious lipid accumulation compared with the control (Figure 6E).
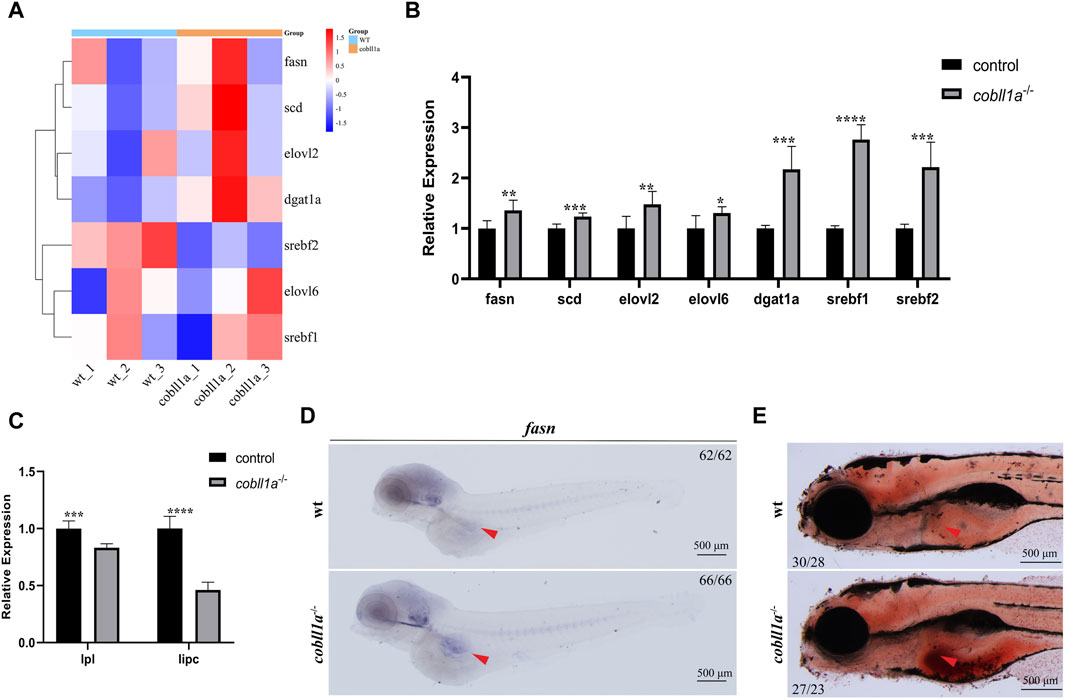
Figure 6. Loss of Cobll1a resulted in Abnormal Lipid Metabolism in Zebrafish. (A) Clustering heat maps of lipid anabolism-related genes in DEGs. WT and cobll1a mutated zebrafish embryo samples form two distinct groups. (B) RT-qPCR used to assess transcriptional expression level of lipid anabolism-related genes in cobll1a−/− embryos at 52 hpf, compared with WT. (C) RT-qPCR analysis of transcriptional expression level of lipid catabolism-related genes in cobll1a−/− embryos at 52 hpf, compared to the control. (D) At 4 dpf, fasn is expressed in adipocytes and the expression of fasn is up-regulated in cobll1a−/− embryos. (E) At 5 dpf, Oil Red O staining was used to assess the lipid distribution and content of WT and cobll1a−/− embryos.
3.7 Cobll1a deletion impairs lipid transport
The transport of lipids such as cholesterol, cholesterol esters, and triglycerides, which are fundamentally insoluble in aqueous environments, is dependent on their association with the lipoprotein complex (Illingworth, 1993). Plasma lipoprotein, a composite of proteins and lipids, is responsible for transporting dietary lipids from the small intestine to the liver, muscle, and adipose tissue. It also plays an instrumental role in lipid transportation from the liver to other tissues and in cholesterol reverse transport from peripheral tissues to the liver and intestine (Feingold, 2022). There are four primary types of plasma lipoproteins, namely, chylomicrons (CMs), very low-density lipoproteins (VLDL), low-density lipoproteins (LDL), and high-density lipoproteins (HDL). Apolipoproteins, the protein constituents of lipoprotein, include apoE, apoB, apoA-I, apoA-II, apoA-IV, apoC-I, etc. They contribute to the transport and redistribution of lipids between various types of cells and tissues, either as co-factors of lipid metabolic enzymes or by maintaining the structural stability of lipoprotein particles (Otis et al., 2015).
Our RNA-seq data identified a number of apolipoprotein-related genes in DEGs, with apolipoprotein expression in cobll1a−/− embryos significantly down-regulated compared to the control. GO gene enrichment analysis related to lipoproteins revealed that the primary biological processes affected were cholesterol transport, lipid transport, lipid localization, and triglyceride homeostasis, among others (Figure 7A). Abnormalities were observed in cellular components such as the lipoprotein complex, plasma lipoprotein particles, and lipoproteins (Figure 7B). Molecular functions like lipid binding, sterol transport activity, cholesterol transport activity, and fatty acid binding were also affected (Figure 7C). These GO analysis results indicated disturbances in lipoprotein metabolism and abnormalities in lipid transport, cholesterol efflux, and reverse transport upon cobll1a deletion. Cluster heat map analysis of apolipoproteins in DEGs showed significant differences in the expression of apolipoproteins between WT and cobll1a−/− embryos (Figure 7D). RT-qPCR was subsequently utilized to validate the expression level of apoa1a, apoa1b, apoa2, apoa4a, apoa4b, apoea, apoeb, apoba, apob.1, as depicted in Figure 7E, demonstrating a significant reduction in the expression of the related apolipoprotein genes.
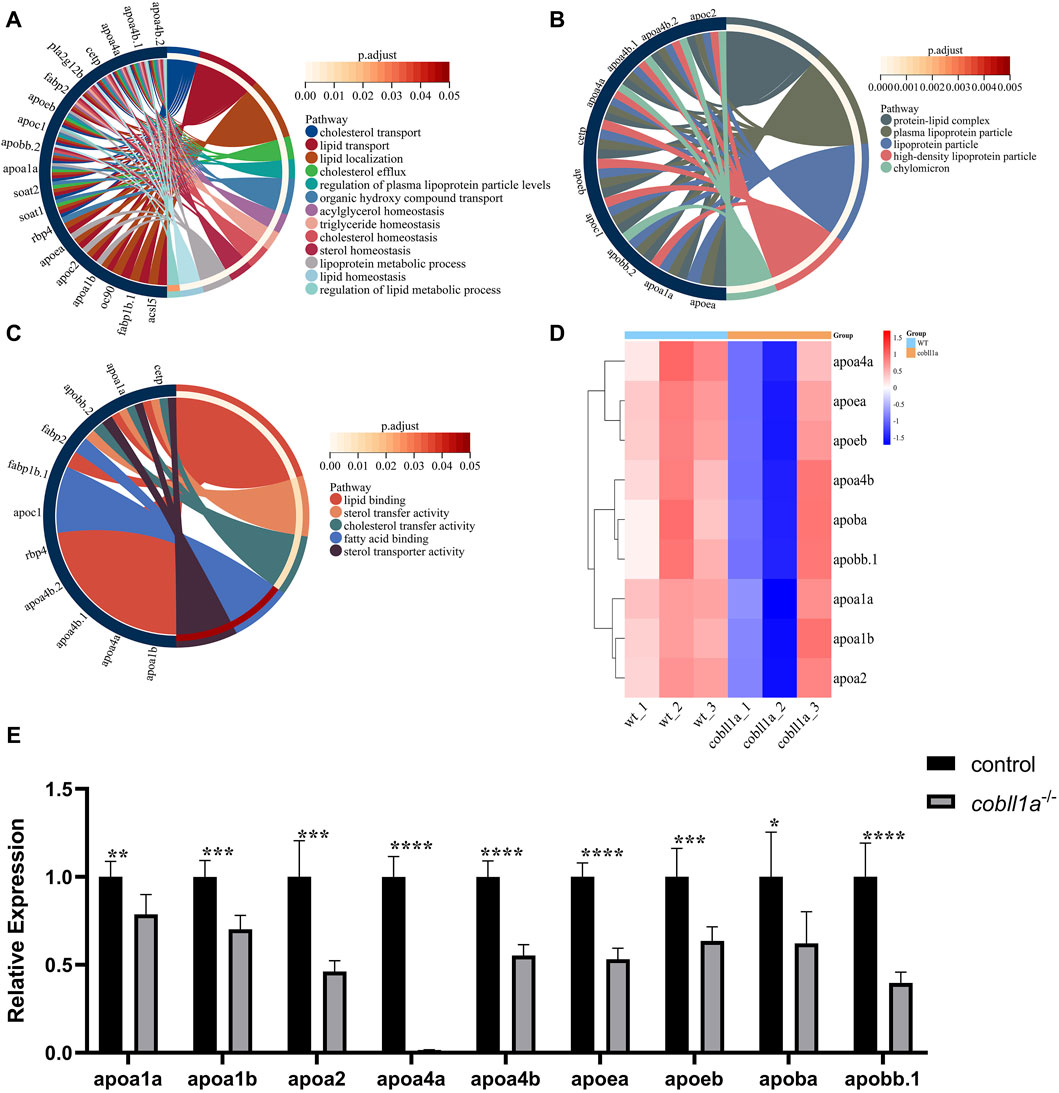
Figure 7. Zebrafish embryos exhibit abnormal lipoprotein metabolism in cobll1a mutant. (A–C) Enrichment analysis of GO molecular function (MF), biological process (BP), and cell component (CC) of biological processes in cobll1a−/− embryos. (D) Cluster heat maps of genes associated with lipoprotein metabolism, where samples from WT and cobll1a mutant zebrafish embryos are segregated into two distinct groups. (E) Transcriptional expression level of apolipoprotein metabolism-related genes in 52 hpf cobll1a−/− embryos were analyzed by RT-qPCR compared with WT.
4 Discussion
In human, COBLL1 is predominantly expressed in several organs including islets, kidneys, skeletal muscle, liver, mast cells, adipocytes, placenta, and esophagus (Plešingerová et al., 2018). The polymorphic sites of COBLL1, rs10195252 and rs3923113, are implicated in the regulation of fasting circulating levels of triglycerides and high-density lipoprotein cholesterol (Teslovich et al., 2010). This study elucidates the critical role of cobll1a in liver and RA metabolism in zebrafish. Zebrafish hhex is expressed in liver buds between 22 and 50 hpf, and is associated with a smaller liver in mutants (Wallace et al., 2001). The gene cp, expressed in hepatocytes at 32 hpf, serves as a specific molecular marker for early hepatocytes and the liver in zebrafish (Korzh et al., 2001). In cobll1a−/− mutant embryos, the expression of hhex and cp was significantly down-regulated compared to the control, indicating a reduction in liver progenitor cells, resulting in a small liver. In fact, the expression of the hepatocyte-specific gene fabp10a was significantly down-regulated at 72hpf, and there was a decrease in the size of both the intestine and pancreas. RNA-seq data revealed abnormal RA metabolism in cobll1a−/− mutant embryos, characterized by reduced RA synthesis. Prior studies have affirmed that RA is crucial for pancreatic and liver development in zebrafish, and its deficiency results in abnormal pancreas and liver development (Stafford and Prince, 2002). A deficiency in RA also induces microphthalmia and forebrain reduction in zebrafish embryos (Le et al., 2012), which is also observed in cobll1a−/− embryos.
To elucidate the molecular mechanism underpinning the abnormal liver development in cobll1a−/− mutants, RNA-seq was conducted. Results showed that down-regulated DEGs were predominantly concentrated in PPAR signaling pathway, cytochrome P450 signaling pathway, and other pathways involved in RA metabolism, cholesterol metabolism, and lipid metabolism. Peroxisome proliferator-activated receptors (PPARs) signaling pathway, a class of nuclear receptors, plays a crucial role in regulating lipid homeostasis during both development and adulthood, and disruption of PPAR signaling has been associated with diseases such as obesity and glucose intolerance (Rees et al., 2008). PPARs heterodimerize with the retinoid X receptor (RXR) to bind to the peroxisome proliferator response element (PPRE) promoter sequence, enabling the transcription of genes associated with fatty acid metabolism (Monsalve et al., 2013). RXR, a potent drug target for the treatment of metabolic syndrome and cancer, is a key member of nuclear receptors (Zhang et al., 2011). RXR forms homo-dimers and hetero-dimers with a range of other nuclear receptors, including fatty acid receptors (PPARs), bile acid receptors (farnesoid x receptors, FXR, pregnane x receptor (PXR), constitutive androstane receptor (CAR), vitamin D receptor (VDR), and RA receptor (RAR), liver x receptor (LXR), allowing these dimers to regulate the transcription of target genes by binding to their promoter regions (Dawson and Xia, 2012). Most RXR partners have roles in regulating lipid homeostasis (He et al., 2013). As most of these receptors are abundantly expressed in the liver, endogenous RA may regulate many hepatic nuclear receptor-mediated pathways. Cytochrome P450 (CYPs), important heme-containing proteins, plays key roles in the metabolism of exogenous substances and endogenous compounds (Tompkins and Wallace, 2007).
The regulation of retinol homeostasis is facilitated by a complex network of enzymes and proteins involved in retinol transportation, synthesis, and catabolism (Moise et al., 2007). Its physiological function is primarily mediated by its metabolites, retinal and RA (Napoli, 2012). The balance of RA is managed by enzyme expression responsible for its synthesis and oxidative degradation (Ross, 2003). Notably, the liver RA level in rdh10+/− mice was significantly reduced, with increased liver triglycerides and decreased gene expression involved in fatty acid β-oxidation (Yang et al., 2018). Mice expressing liver-specific RARα dominant negative protein exhibited microalveolar steatosis at 4 months, decreased mitochondrial β-oxidized fatty acids, and developed hepatocellular carcinoma and hepatic adenoma at 1 month (Yanagitani et al., 2004). Clinically, retinol analogs have been found to inhibit the occurrence of secondary primary tumors in patients diagnosed with hepatocellular carcinoma (HCC) (Muto et al., 1996). Previous studies have revealed a significant loss of liver retinol levels in NAFLD (Zhong et al., 2019), with the liver retinol level inversely correlated with the severity of liver damage and liver fat (Chaves et al., 2014; Trasino et al., 2015). In both mice and human NAFLD, liver retinol levels are inversely associated with liver steatosis (Friedman et al., 2018).
Fasn plays a pivotal role in lipid synthesis, responsible for combining palmitic acid from acetyl-CoA and malonyl-CoA into long-chain saturated fatty acids (Angeles and Hudkins, 2016). The elovls family of ultralong chain fatty acid elongase are crucial enzyme that manage fatty acid metabolism in animals (Morcillo et al., 2011). Elovl2 has been implicated in the biosynthesis of long-chain polyunsaturated fatty acids in mammals (Leonard et al., 2002), while elovl6 primarily catalyzes palmitate and palmitoleate into stearate and oleate (Moon et al., 2014). Dgat1a facilitates the conversion of diacylglycerol and fatty acyl-CoA into triacylglycerol (Barbosa et al., 2019). Srebf1 and srebf2 are significant nuclear transcription factors that regulate liver lipid metabolism and are crucial regulatory factors of fatty acid metabolism (Rong et al., 2017). Overexpression of srebf1 activates the synthesis of fatty acids and triglycerides (Horton et al., 2002), while overexpression of srebf2 activates cholesterol biosynthesis and uptake, and slightly triggers fatty acid biosynthesis (Rong et al., 2017). After the cobll1a mutation in this study, the expression of fasn, elovl2, elovl6, dgat1a, srebf1, and srebf2 genes was upregulated, thus promoting the production of new fat and eventually inducing steatosis (Anderson and Borlak, 2008).
Following absorption by intestinal cells, REs are carried and transported into circulation by CMs, HDL, LDL, and VLDL (Levi et al., 2012). Impairment in VLDL synthesis can result in the accumulation of triglycerides in the liver (Tanoli et al., 2004). Apolipoprotein B (ApoB) is a major component of CMs particles, VLDL, and LDL (Olofsson and Borèn, 2005), and defects in ApoB synthesis or secretion can induce high adipose degeneration in the liver (Di Filippo et al., 2014). Zebrafish embryonic hepatocytes exhibited lipid accumulation after the double mutation of apoBa and apoBb.1 of apoB genes (Templehof et al., 2021). Apolipoprotein A4 (ApoA4) is primarily expressed in the intestine and liver, reacts to fat absorption, and plays a role in regulating glucose homeostasis and lipid metabolism, thus reducing susceptibility to atherosclerosis (Li et al., 2021). ApoA4 knockout rats and mice demonstrated significant hepatic steatosis (Wang et al., 2019). Apolipoprotein AI (ApoAI), the main component of HDL, plays a crucial role in lipid transport and metabolism (Chistiakov et al., 2016). APOA1 promotes cholesterol efflux by interacting with the ATP-binding box (ABC) transporter (Navab et al., 2011). APOE, an exchangeable amphipathic apolipoprotein, binds to LDL receptors, thereby regulating lipid uptake (Mahley, 1988). Apoe-deficient mice developed severe atherosclerosis due to an increase in circulating LDL cholesterol (Nakashima et al., 1994). In this study, the expression of apoa1a, apoa1b, apoa2, apoa4a, apoa4b, apoea, apoeb, apoba, and apobb.1 genes was significantly down-regulated after cobll1a mutation, indicating that the loss of cobll1a resulted in lipid transport disorders in the liver of juvenile zebrafish (Xu et al., 2021).
In summary, cobll1a plays a crucial role in the liver lipid metabolism of zebrafish, and its mutation severely impairs RA metabolism and normal lipid homeostasis.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive under accession SRA: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1069604?reviewer=8sirsbvs21hj0a2cpo3fu6ea6j, accession number PRJNA1069604.
Ethics statement
The animal study was approved by the Biomedical Research Ethics Committee of Hunan Normal University (protocol code 2022/545). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
TZ: Data curation, Investigation, Project administration, Visualization, Writing–original draft. JL: Data curation, Investigation, Visualization, Writing–review and editing. JL: Formal Analysis, Writing–original draft. BX: Formal Analysis, Writing–original draft. LL: Data curation, Visualization, Writing–review and editing. YT: Data curation, Formal Analysis, Writing–original draft. JZ: Data curation, Formal Analysis, Writing–original draft. JJ: Data curation, Formal Analysis, Writing–original draft. HX: Conceptualization, Funding acquisition, Methodology, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (82170308), Xiaoxiang Scholar Distinguished Professor Startup Funding for H-pX (053112-3826).
Acknowledgments
We would like to express our appreciation to all the members of the Laboratory of Animal Nutrition and Human Health at Hunan Normal University for their assistance and encouragement.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1381362/full#supplementary-material
References
Albrechtsen, A., Grarup, N., Li, Y., Sparsø, T., Tian, G., Cao, H., et al. (2013). Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia 56 (2), 298–310. doi:10.1007/s00125-012-2756-1
Amengual, J., Ribot, J., Bonet, M. L., and Palou, A. (2010). Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell. Physiology Biochem. Int. J. Exp. Cell. Physiology, Biochem. Pharmacol. 25 (6), 657–666. doi:10.1159/000315085
Anderson, N., and Borlak, J. (2008). Molecular mechanisms and therapeutic targets in steatosis and steatohepatitis. Pharmacol. Rev. 60 (3), 311–357. doi:10.1124/pr.108.00001
Angeles, T. S., and Hudkins, R. L. (2016). Recent advances in targeting the fatty acid biosynthetic pathway using fatty acid synthase inhibitors. Expert Opin. Drug Discov. 11 (12), 1187–1199. doi:10.1080/17460441.2016.1245286
Angulo, P. (2002). Nonalcoholic fatty liver disease. N. Engl. J. Med. 346 (16), 1221–1231. doi:10.1056/NEJMra011775
Barbosa, M. A. G., Capela, R., Rodolfo, J., Fonseca, E., Montes, R., André, A., et al. (2019). Linking chemical exposure to lipid homeostasis: a municipal waste water treatment plant influent is obesogenic for zebrafish larvae. Ecotoxicol. Environ. Saf. 182, 109406. doi:10.1016/j.ecoenv.2019.109406
Baybutt, R. C., Hu, L., and Molteni, A. (2000). Vitamin A deficiency injures lung and liver parenchyma and impairs function of rat type II pneumocytes. J. Nutr. 130 (5), 1159–1165. doi:10.1093/jn/130.5.1159
Boily, M. H., Ndayibagira, A., and Spear, P. A. (2003). Retinoid metabolism (LRAT, REH) in the yolk-sac membrane of Japanese quail eggs and effects of mono-ortho-PCBs. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 134 (1), 11–23. doi:10.1016/s1532-0456(02)00146-1
Bono, M. R., Tejon, G., Flores-Santibañez, F., Fernandez, D., Rosemblatt, M., and Sauma, D. (2016). Retinoic acid as a m.odulator of T cell immunity. Nutrients 8 (6), 349. doi:10.3390/nu8060349
Chaves, G. V., Pereira, S. E., Saboya, C. J., Spitz, D., Rodrigues, C. S., and Ramalho, A. (2014). Association between liver vitamin A reserves and severity of nonalcoholic fatty liver disease in the class III obese following bariatric surgery. Obes. Surg. 24 (2), 219–224. doi:10.1007/s11695-013-1087-8
Chen, Z., Yu, H., Shi, X., Warren, C. R., Lotta, L. A., Friesen, M., et al. (2020). Functional screening of candidate causal genes for insulin resistance in human preadipocytes and adipocytes. Circulation Res. 126 (3), 330–346. doi:10.1161/CIRCRESAHA.119.315246
Chistiakov, D. A., Orekhov, A. N., and Bobryshev, Y. V. (2016). ApoA1 and ApoA1-specific self-antibodies in cardiovascular disease. Laboratory Investigation; a J. Tech. Methods Pathology 96 (7), 708–718. doi:10.1038/labinvest.2016.56
Cunningham, T. J., and Duester, G. (2015). Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 16 (2), 110–123. doi:10.1038/nrm3932
Dawson, M. I., and Xia, Z. (2012). The retinoid X receptors and their ligands. Biochimica Biophysica Acta 1821 (1), 21–56. doi:10.1016/j.bbalip.2011.09.014
Desmarchelier, C., Martin, J.-C., Planells, R., Gastaldi, M., Nowicki, M., Goncalves, A., et al. (2014). The postprandial chylomicron triacylglycerol response to dietary fat in healthy male adults is significantly explained by a combination of single nucleotide polymorphisms in genes involved in triacylglycerol metabolism. J. Clin. Endocrinol. Metabolism 99 (3), E484–E488. doi:10.1210/jc.2013-3962
Di Filippo, M., Moulin, P., Roy, P., Samson-Bouma, M. E., Collardeau-Frachon, S., Chebel-Dumont, S., et al. (2014). Homozygous MTTP and APOB mutations may lead to hepatic steatosis and fibrosis despite metabolic differences in congenital hypocholesterolemia. J. Hepatology 61 (4), 891–902. doi:10.1016/j.jhep.2014.05.023
Feingold, K. R. (2022). Lipid and lipoprotein metabolism. Endocrinol. Metabolism Clin. N. Am. 51 (3), 437–458. doi:10.1016/j.ecl.2022.02.008
Field, H. A., Ober, E. A., Roeser, T., and Stainier, D. Y. (2003). Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253 (2), 279–290. doi:10.1016/s0012-1606(02)00017-9
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M., and Sanyal, A. J. (2018). Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24 (7), 908–922. doi:10.1038/s41591-018-0104-9
Grandel, H., Lun, K., Rauch, G. J., Rhinn, M., Piotrowski, T., Houart, C., et al. (2002). Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development 129 (12), 2851–2865. doi:10.1242/dev.129.12.2851
Gudas, L. J., and Wagner, J. A. (2011). Retinoids regulate stem cell differentiation. J. Cell. Physiology 226 (2), 322–330. doi:10.1002/jcp.22417
Haaker, M. W., Vaandrager, A. B., and Helms, J. B. (2020). Retinoids in health and disease: a role for hepatic stellate cells in affecting retinoid levels. Biochimica Biophysica Acta. Mol. Cell Biol. Lipids 1865 (6), 158674. doi:10.1016/j.bbalip.2020.158674
Han, S. H., Kim, S. H., Kim, H. J., Lee, Y., Choi, S. Y., Park, G., et al. (2017). Cobll1 is linked to drug resistance and blastic transformation in chronic myeloid leukemia. Leukemia 31 (7), 1532–1539. doi:10.1038/leu.2017.72
He, Y., Gong, L., Fang, Y., Zhan, Q., Liu, H.-X., Lu, Y., et al. (2013). The role of retinoic acid in hepatic lipid homeostasis defined by genomic binding and transcriptome profiling. BMC Genomics 14, 575. doi:10.1186/1471-2164-14-575
Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest 109 (9), 1125–1131. doi:10.1172/jci15593
Illingworth, D. R. (1993). Lipoprotein metabolism. Am. J. Kidney Dis. 22 (1), 90–97. doi:10.1016/s0272-6386(12)70173-7
Kastner, P., Mark, M., and Chambon, P. (1995). Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell 83 (6), 859–869. doi:10.1016/0092-8674(95)90202-3
Kedishvili, N. Y. (2016). Retinoic acid synthesis and degradation. Subcell. Biochem. 81, 127–161. doi:10.1007/978-94-024-0945-1_5
Kim, H., Kim, D.-W., and Myung, K. (2017). Cobll1: a new player in CML. Oncotarget 8 (53), 90626–90627. doi:10.18632/oncotarget.21705
Korzh, S., Emelyanov, A., and Korzh, V. (2001). Developmental analysis of ceruloplasmin gene and liver formation in zebrafish. Mech. Dev. 103 (1-2), 137–139. doi:10.1016/s0925-4773(01)00330-6
Kraja, A. T., Chasman, D. I., North, K. E., Reiner, A. P., Yanek, L. R., Kilpeläinen, T. O., et al. (2014). Pleiotropic genes for metabolic syndrome and inflammation. Mol. Genet. Metabolism 112 (4), 317–338. doi:10.1016/j.ymgme.2014.04.007
Le, H. G., Dowling, J. E., and Cameron, D. J. (2012). Early retinoic acid deprivation in developing zebrafish results in microphthalmia. Vis. Neurosci. 29 (4-5), 219–228. doi:10.1017/s0952523812000296
Leonard, A. E., Kelder, B., Bobik, E. G., Chuang, L. T., Lewis, C. J., Kopchick, J. J., et al. (2002). Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 37 (8), 733–740. doi:10.1007/s11745-002-0955-6
Levi, L., Ziv, T., Admon, A., Levavi-Sivan, B., and Lubzens, E. (2012). Insight into molecular pathways of retinal metabolism, associated with vitellogenesis in zebrafish. Am. J. Physiology. Endocrinol. Metabolism 302 (6), E626–E644. doi:10.1152/ajpendo.00310.2011
Li, X., Liu, X., Zhang, Y., Cheng, C., Fan, J., Zhou, J., et al. (2021). Hepatoprotective effect of apolipoprotein A4 against carbon tetrachloride induced acute liver injury through mediating hepatic antioxidant and inflammation response in mice. Biochem. Biophysical Res. Commun. 534, 659–665. doi:10.1016/j.bbrc.2020.11.024
Mahley, R. W. (1988). Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science 240 (4852), 622–630. doi:10.1126/science.3283935
Mancina, R. M., Burza, M. A., Maglio, C., Pirazzi, C., Sentinelli, F., Incani, M., et al. (2013). The COBLL1 C allele is associated with lower serum insulin levels and lower insulin resistance in overweight and obese children. Diabetes/metabolism Res. Rev. 29 (5), 413–416. doi:10.1002/dmrr.2408
Manning, A. K., Hivert, M.-F., Scott, R. A., Grimsby, J. L., Bouatia-Naji, N., Chen, H., et al. (2012). A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44 (6), 659–669. doi:10.1038/ng.2274
Metzler, M. A., and Sandell, L. L. (2016). Enzymatic metabolism of vitamin A in developing vertebrate embryos. Nutrients 8 (12), 812. doi:10.3390/nu8120812
Moise, A. R., Noy, N., Palczewski, K., and Blaner, W. S. (2007). Delivery of retinoid-based therapies to target tissues. Biochemistry 46 (15), 4449–4458. doi:10.1021/bi7003069
Monsalve, F. A., Pyarasani, R. D., Delgado-Lopez, F., and Moore-Carrasco, R. (2013). Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediat. Inflamm. 2013, 549627. doi:10.1155/2013/549627
Moon, Y.-A., Ochoa, C. R., Mitsche, M. A., Hammer, R. E., and Horton, J. D. (2014). Deletion of ELOVL6 blocks the synthesis of oleic acid but does not prevent the development of fatty liver or insulin resistance. J. Lipid Res. 55 (12), 2597–2605. doi:10.1194/jlr.M054353
Morcillo, S., Martín-Núñez, G. M., Rojo-Martínez, G., Almaraz, M. C., García-Escobar, E., Mansego, M. L., et al. (2011). ELOVL6 genetic variation is related to insulin sensitivity: a new candidate gene in energy metabolism. PloS One 6 (6), e21198. doi:10.1371/journal.pone.0021198
Musso, G., Gambino, R., De Michieli, F., Biroli, G., Premoli, A., Pagano, G., et al. (2007). Nitrosative stress predicts the presence and severity of nonalcoholic fatty liver at different stages of the development of insulin resistance and metabolic syndrome: possible role of vitamin A intake. Am. J. Clin. Nutr. 86 (3), 661–671. doi:10.1093/ajcn/86.3.661
Muto, Y., Moriwaki, H., Ninomiya, M., Adachi, S., Saito, A., Takasaki, K. T., et al. (1996). Prevention of second primary tumors by an acyclic retinoid, polyprenoic acid, in patients with hepatocellular carcinoma. Hepatoma Prevention Study Group. N. Engl. J. Med. 334 (24), 1561–1567. doi:10.1056/nejm199606133342402
Nagase, T., Ishikawa, K., Suyama, M., Kikuno, R., Hirosawa, M., Miyajima, N., et al. (1999). Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 6 (1), 63–70. doi:10.1093/dnares/6.1.63
Nakashima, Y., Plump, A. S., Raines, E. W., Breslow, J. L., and Ross, R. (1994). ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler. Thromb. 14 (1), 133–140. doi:10.1161/01.atv.14.1.133
Napoli, J. L. (2012). Physiological insights into all-trans-retinoic acid biosynthesis. Biochimica Biophysica Acta 1821 (1), 152–167. doi:10.1016/j.bbalip.2011.05.004
Navab, M., Reddy, S. T., Van Lenten, B. J., and Fogelman, A. M. (2011). HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat. Rev. Cardiol. 8 (4), 222–232. doi:10.1038/nrcardio.2010.222
Olofsson, S. O., and Borèn, J. (2005). Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J. Intern Med. 258 (5), 395–410. doi:10.1111/j.1365-2796.2005.01556.x
Otis, J. P., Zeituni, E. M., Thierer, J. H., Anderson, J. L., Brown, A. C., Boehm, E. D., et al. (2015). Zebrafish as a model for apolipoprotein biology: comprehensive expression analysis and a role for ApoA-IV in regulating food intake. Dis. Models Mech. 8 (3), 295–309. doi:10.1242/dmm.018754
Pennimpede, T., Cameron, D. A., MacLean, G. A., Li, H., Abu-Abed, S., and Petkovich, M. (2010). The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res. Part A, Clin. Mol. Teratol. 88 (10), 883–894. doi:10.1002/bdra.20709
Plešingerová, H., Janovská, P., Mishra, A., Smyčková, L., Poppová, L., Libra, A., et al. (2018). Expression of COBLL1 encoding novel ROR1 binding partner is robust predictor of survival in chronic lymphocytic leukemia. Haematologica 103 (2), 313–324. doi:10.3324/haematol.2017.178699
Rees, W. D., McNeil, C. J., and Maloney, C. A. (2008). The roles of PPARs in the fetal origins of metabolic health and disease. PPAR Res. 2008, 459030. doi:10.1155/2008/459030
Rong, S., Cortés, V. A., Rashid, S., Anderson, N. N., McDonald, J. G., Liang, G., et al. (2017). Expression of SREBP-1c requires SREBP-2-mediated generation of a sterol ligand for LXR in livers of mice. ELife 6, e25015. doi:10.7554/eLife.25015
Ronne, H., Ocklind, C., Wiman, K., Rask, L., Obrink, B., and Peterson, P. A. (1983). Ligand-dependent regulation of intracellular protein transport: effect of vitamin a on the secretion of the retinol-binding protein. J. Cell Biol. 96 (3), 907–910. doi:10.1083/jcb.96.3.907
Ross, A. C. (2003). Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic Acid oxidation. J. Nutr. 133 (1), 291S–296S. doi:10.1093/jn/133.1.291S
Saeed, A., Dullaart, R. P. F., Schreuder, T. C. M. A., Blokzijl, H., and Faber, K. N. (2017). Disturbed vitamin A metabolism in non-alcoholic fatty liver disease (NAFLD). Nutrients 10 (1), 29. doi:10.3390/nu10010029
Schwintzer, L., Koch, N., Ahuja, R., Grimm, J., Kessels, M. M., and Qualmann, B. (2011). The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 30 (15), 3147–3159. doi:10.1038/emboj.2011.207
Stafford, D., and Prince, V. E. (2002). Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr. Biol. 12 (14), 1215–1220. doi:10.1016/s0960-9822(02)00929-6
Stafford, D., White, R. J., Kinkel, M. D., Linville, A., Schilling, T. F., and Prince, V. E. (2006). Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development 133 (5), 949–956. doi:10.1242/dev.02263
Sun, H., and Kawaguchi, R. (2011). The membrane receptor for plasma retinol-binding protein, a new type of cell-surface receptor. Int. Rev. Cell Mol. Biol. 288, 1–41. doi:10.1016/B978-0-12-386041-5.00001-7
Takayama, K.-I., Suzuki, T., Fujimura, T., Takahashi, S., and Inoue, S. (2018). COBLL1 modulates cell morphology and facilitates androgen receptor genomic binding in advanced prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 115 (19), 4975–4980. doi:10.1073/pnas.1721957115
Tanoli, T., Yue, P., Yablonskiy, D., and Schonfeld, G. (2004). Fatty liver in familial hypobetalipoproteinemia: roles of the APOB defects, intra-abdominal adipose tissue, and insulin sensitivity. J. Lipid Res. 45 (5), 941–947. doi:10.1194/jlr.M300508-JLR200
Tanumihardjo, S. A., Russell, R. M., Stephensen, C. B., Gannon, B. M., Craft, N. E., Haskell, M. J., et al. (2016). Biomarkers of nutrition for development (BOND)-Vitamin A review. J. Nutr. 146 (9), 1816S–1848S. doi:10.3945/jn.115.229708
Templehof, H., Moshe, N., Avraham-Davidi, I., and Yaniv, K. (2021). Zebrafish mutants provide insights into Apolipoprotein B functions during embryonic development and pathological conditions. JCI Insight 6 (13), e130399. doi:10.1172/jci.insight.130399
Teslovich, T. M., Musunuru, K., Smith, A. V., Edmondson, A. C., Stylianou, I. M., Koseki, M., et al. (2010). Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 (7307), 707–713. doi:10.1038/nature09270
Theodosiou, M., Laudet, V., and Schubert, M. (2010). From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell. Mol. Life Sci. CMLS 67 (9), 1423–1445. doi:10.1007/s00018-010-0268-z
Tompkins, L. M., and Wallace, A. D. (2007). Mechanisms of cytochrome P450 induction. J. Biochem. Mol. Toxicol. 21 (4), 176–181. doi:10.1002/jbt.20180
Trasino, S. E., Tang, X. H., Jessurun, J., and Gudas, L. J. (2015). Obesity leads to tissue, but not serum vitamin A deficiency. Sci. Rep. 5, 15893. doi:10.1038/srep15893
Villaça Chaves, G., Pereira, S. E., Saboya, C. J., and Ramalho, A. (2008). Non-alcoholic fatty liver disease and its relationship with the nutritional status of vitamin A in individuals with class III obesity. Obes. Surg. 18 (4), 378–385. doi:10.1007/s11695-007-9361-2
Wallace, K. N., Yusuff, S., Sonntag, J. M., Chin, A. J., and Pack, M. (2001). Zebrafish hhex regulates liver development and digestive organ chirality. Genesis 30 (3), 141–143. doi:10.1002/gene.1050
Wang, Z., Wang, L., Zhang, Z., Feng, L., Song, X., and Wu, J. (2019). Apolipoprotein A-IV involves in glucose and lipid metabolism of rat. Nutr. Metabolism 16, 41. doi:10.1186/s12986-019-0367-2
Xu, H., Jiang, Y., Miao, X.-M., Tao, Y.-X., Xie, L., and Li, Y. (2021). A model construction of starvation induces hepatic steatosis and transcriptome analysis in zebrafish larvae. Biology 10 (2), 92. doi:10.3390/biology10020092
Yanagitani, A., Yamada, S., Yasui, S., Shimomura, T., Murai, R., Murawaki, Y., et al. (2004). Retinoic acid receptor alpha dominant negative form causes steatohepatitis and liver tumors in transgenic mice. Hepatology 40 (2), 366–375. doi:10.1002/hep.20335
Yang, D., Vuckovic, M. G., Smullin, C. P., Kim, M., Lo, C.P.-S., Devericks, E., et al. (2018). Modest decreases in endogenous all-trans-retinoic acid produced by a mouse Rdh10 heterozygote provoke major abnormalities in adipogenesis and lipid metabolism. Diabetes 67 (4), 662–673. doi:10.2337/db17-0946
Zhang, H., Zhou, R., Li, L., Chen, J., Chen, L., Li, C., et al. (2011). Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor. J. Biol. Chem. 286 (3), 1868–1875. doi:10.1074/jbc.M110.166215
Zhong, G., Kirkwood, J., Won, K.-J., Tjota, N., Jeong, H., and Isoherranen, N. (2019). Characterization of vitamin A metabolome in human livers with and without nonalcoholic fatty liver disease. J. Pharmacol. Exp. Ther. 370 (1), 92–103. doi:10.1124/jpet.119.258517
Zhong, M., Kawaguchi, R., Kassai, M., and Sun, H. (2012). Retina, retinol, retinal and the natural history of vitamin A as a light sensor. Nutrients 4 (12), 2069–2096. doi:10.3390/nu4122069
Keywords: Zebrafish, liver, retinol, retinoic acid signaling pathway, NAFLD, apolipoprotein
Citation: Zeng T, Lv J, Liang J, Xie B, Liu L, Tan Y, Zhu J, Jiang J and Xie H (2024) Zebrafish cobll1a regulates lipid homeostasis via the RA signaling pathway. Front. Cell Dev. Biol. 12:1381362. doi: 10.3389/fcell.2024.1381362
Received: 03 February 2024; Accepted: 04 April 2024;
Published: 18 April 2024.
Edited by:
Natascia Tiso, University of Padua, ItalyCopyright © 2024 Zeng, Lv, Liang, Xie, Liu, Tan, Zhu, Jiang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaping Xie, aHB4aWVAaHVubnUuZWR1LmNu
†These authors share first authorship
 Ting Zeng1,2†
Ting Zeng1,2† Huaping Xie
Huaping Xie