
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 05 March 2024
Sec. Developmental Epigenetics
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1369751
This article is part of the Research TopicIn Celebration of Women in Developmental EpigeneticsView all 11 articles
DNA methylation (DNAme) has long been recognized as a host defense mechanism, both in the restriction modification systems of prokaryotes as well as in the transcriptional silencing of repetitive elements in mammals. When DNAme was shown to be implicated as a key epigenetic mechanism in the regulation of imprinted genes in mammals, a parallel with host defense mechanisms was drawn, suggesting perhaps a common evolutionary origin. Here we review recent work related to this hypothesis on two different aspects of the developmental imprinting cycle in mammals that has revealed unexpected roles for long terminal repeat (LTR) retroelements in imprinting, both canonical and noncanonical. These two different forms of genomic imprinting depend on different epigenetic marks inherited from the mature gametes, DNAme and histone H3 lysine 27 trimethylation (H3K27me3), respectively. DNAme establishment in the maternal germline is guided by transcription during oocyte growth. Specific families of LTRs, evading silencing mechanisms, have been implicated in this process for specific imprinted genes. In noncanonical imprinting, maternally inherited histone marks play transient roles in transcriptional silencing during preimplantation development. These marks are ultimately translated into DNAme, notably over LTR elements, for the maintenance of silencing of the maternal alleles in the extraembryonic trophoblast lineage. Therefore, LTR retroelements play important roles in both establishment and maintenance of different epigenetic pathways leading to imprinted expression during development. Because such elements are mobile and highly polymorphic among different species, they can be coopted for the evolution of new species-specific imprinted genes.
The first mouse imprinted genes, H19, Igf2, and Igf2r, were identified in 1991 (Barlow et al., 1991; Bartolomei et al., 1991; DeChiara et al., 1991). The imprinting of Snrpn was demonstrated the following year (Cattanach et al., 1992; Leff et al., 1992), and in 1993 the first reports presenting evidence supporting a role for DNAme in the imprinting mechanism were published (Bartolomei et al., 1993; Brandeis et al., 1993; Ferguson-Smith et al., 1993; Li et al., 1993). It was already recognized at the time that the DNAme machinery exploited in mammals was derived from bacterial immune systems that had been adapted for the transcriptional repression of repetitive sequences (Bestor, 1990). This led the late Denise Barlow to propose that genomic imprinting had evolved from host defense mechanisms, by co-opting DNAme-based functions in the parent-of-origin-specific silencing of imprinted genes (Barlow, 1993). Several different predictions of the model proposed have been confirmed, as recently reviewed (Ondičová et al., 2020). A related aspect of this model, which is the focus of this review, addresses the roles played by endogenous repetitive elements themselves in the regulation of imprinted gene expression. Here, we review recent evidence suggesting that LTR elements have been co-opted for both germline establishment and somatic maintenance of imprinted gene expression in early development.
Early studies on diploid biparental gynogenetic and androgenetic embryos suggested that genomic imprinting is established during gametogenesis (McGrath and Solter, 1984; Surani et al., 1984). Although the epigenetic mechanisms involved were not known at the time, DNAme was later shown to represent an important epigenetic mark, directly inherited from the mature gametes, and regulating imprinted gene expression. The monoallelic expression of canonical imprinted genes in somatic cells is maintained by differential DNAme marks (Tucci et al., 2019) established de novo during male or female gametogenesis by the sex-specific action of the DNA methyltransferase DNMT3A (Kaneda et al., 2004), and its co-factor DNMT3L (Bourc’his et al., 2001; Hata et al., 2002; Arima et al., 2006). Large fractions of the genome are differentially methylated between eggs and sperm, but unlike most of the differences, the gametic DNAme marks at imprinted genes survive the wave of demethylation occurring during pre-implantation stages. This survival of imprints requires the maintenance DNA methyltransferase DNMT1 (Li et al., 1993; Hirasawa et al., 2008), its partner UHRF1 (Sharif et al., 2007), and the DNAme-dependent DNA-binding factors ZFP57 and ZFP445. These KRAB zinc-finger proteins specifically bind the methylated allele of imprinted genes and protect it from demethylation during preimplantation stages via their recruitment of KAP1/TRIM28 and SETDB1. This histone methyltransferase establishes a H3K9me3 mark over the DNAme-marked region for preferential recruitment of DNMT1 via UHRF1, which recognizes H3K9me3 via its tandem Tudor domain and plant homeodomain (Li et al., 2008; Strogantsev and Ferguson-Smith, 2012; Takahashi et al., 2019; Janssen and Lorincz, 2021). In somatic cells, sequences carrying these DNAme imprints are detected as Differentially Methylated Regions (methylated on a single allele) of gametic origin (gDMR). Imprinted gDMRs are thought to be responsible for all canonical imprinted gene expression observed in embryonic and adult cells. Only 24 gDMRs have been identified in the mouse, 21 methylated in the oocyte and 3 in sperm (Proudhon et al., 2012; Bogutz et al., 2019). Both oocyte and sperm DNAme play essential roles in imprinting, but whereas most of the paternal DNAme is lost after fertilization, a portion of oocyte-derived 5-methylcytosine (5mC) survives the passive demethylation occurring during preimplantation (Smallwood et al., 2011). Although the function of most of this inherited maternal DNAme is still unknown, some of these maternal marks were shown to be required for silencing genes detrimental for placental development, the first demonstration of a role for maternal DNAme unrelated to genomic imprinting (Branco et al., 2016). Recent surveys suggest that the human genome contains more gDMRs (Zink et al., 2018; Akbari et al., 2022), with several maternally-inherited marks maintained only in the placenta (Court et al., 2014; Hamada et al., 2016; Hanna et al., 2016; Sanchez-Delgado et al., 2016).
The analysis of the DNAme profile of gametes at single-base resolution using whole-genome bisulphite sequencing (WGBS) revealed that there is nothing fundamentally unique about de novo establishment of imprinted gDMR. Rather, these sequences acquire DNAme as part of global mechanisms methylating the mouse sperm and oocyte genomes at >80% and ∼40% levels, respectively (Kobayashi et al., 2012). Mature gametes are also methylated at very different levels in human, with average levels of DNAme of ∼75% and ∼54% for sperm and egg, respectively (Okae et al., 2014a). In the mouse, whereas paternal gDMRs are DNA methylated in prospermatogonia from E14.5 to birth (Davis et al., 2000; Li et al., 2004), elegant embryological experiments showed that the establishment of functional maternal imprints occurs during the phase of oocyte growth taking place in postnatal ovaries (Kono et al., 1996; Obata et al., 1998; Obata and Kono, 2002). Accordingly, the genome of primary non-growing oocytes (NGO, from P1-P5 females), and of fully-grown, germinal vesicle stage oocytes (FGO or GVO, from mature females), show a drastic difference in average genomic DNAme levels, from 2% to 40%, including at several CpG islands (CGIs) (Shirane et al., 2013). The process of de novo DNAme therefore occurs postnatally in females, in non-dividing oocytes, and was shown to require DNMT3A and its cofactor DNMT3L, but not DNMT3B or the maintenance DNMT1 enzyme (Smallwood et al., 2011; Kobayashi et al., 2012; Shirane et al., 2013).
By comparing the DNA methylome and transcriptome of oocytes, as determined by RNA-seq, a direct correlation was observed between gene transcription and gene body DNAme (Smallwood et al., 2011; Kobayashi et al., 2012; Veselovska et al., 2015). Whereas promoter regions of active genes are hypomethylated (<15% 5 mC), their transcribed regions acquire 60%–90% DNAme, starting ∼2 kb downstream of their oocyte-specific transcription start site (TSS). Strikingly, transcribed regions account for 85%–90% of the methylome of FGOs, including DNAme at all imprinted maternal gDMRs (Veselovska et al., 2015). Pioneering work from the group of Gavin Kelsey showed that oocyte transcription across the gDMR region was required for DNAme establishment at the maternal Gnasxl/Nespas gDMR and that most imprinted maternal gDMRs are indeed covered by an oocyte transcript initiating at an upstream promoter (Chotalia et al., 2009). Following this work, a role for oocyte transcription in de novo DNAme at the gDMRs of the paternally expressed genes Snrpn, Plagl1, and Kcnq1ot1 was demonstrated directly in mouse mutants in which inserted transcription termination sequences prevent oocyte transcripts from extending across the DMR region (Smith et al., 2011; Veselovska et al., 2015; Singh et al., 2017). Although DNAme blocks are not as well defined in human oocytes, a strong correlation was also noted between methylated and transcribed regions, suggesting that the link between de novo methylation and transcription is conserved (Okae et al., 2014a). Interestingly, other examples of transcription-coupled acquisition of DNAme at imprinted promoters emerged from the analysis of retrogenes, inserted within a host gene expressed in oocytes (Cowley and Oakey, 2010). Because of their location, the promoter of these inserted retrogenes is covered by a transcript in oocytes and acquires a maternal gDMR, leading to silencing of the maternal allele and expression from the paternal allele of the retrogene in the progeny (Wood et al., 2007).
The mechanism whereby transcribed regions acquire DNAme in oocytes was shown to be guided by both negative and positive cross-talks with specific histone post-translational modifications. Unmethylated promoter CGIs are usually marked by H3K4me3 (Mikkelsen et al., 2007) and this mark has an inhibitory effect on the action of the DNMT3A-DNMT3L complex (Guo et al., 2015), protecting these CpG-rich sequences from de novo DNA methylation (Ooi et al., 2007). This implies that intragenic CpG-rich regions covered by an oocyte transcript would be refractory to de novo DNA methylation unless methylation marks at H3K4 are previously removed from those regions. Consistent with this prediction, CGIs acquiring DNAme during oocyte growth are devoid of or lose H3K4me2/3 marks in preparation for de novo DNA methylation, and the H3K4 lysine demethylase KDM1B plays a dominant role in the removal of these refractory marks (Ciccone et al., 2009; Stewart et al., 2015). Interestingly, a mutant version of DNMT3A carrying two point mutations within the ADD domain, which interfere with the binding of DNMT3A to H3K4me0 in vitro, was recently shown to lead to dwarfism and female infertility. The global DNAme of oocytes from homozygous females is severely affected (at only 17.6%, compared to 35.9% for wild-type oocytes), leading to stochastic loss of maternal imprinted methylation, abnormal expression of different imprinted genes in the progeny, with variations between individual embryos, several of which die before mid-gestation (Uehara et al., 2023).
The recruitment of the de novo DNA methyltransferases DNMT3A and 3B to transcribed regions is mediated by their PWWP domain, a reader for H3K36me2/3 marks (Dhayalan et al., 2010). As originally demonstrated in somatic cells, transcribed regions acquire an H3K36me3 domain via the recruitment of the histone methyltransferase SETD2, which is in a complex with the elongating RNA polymerase II (Yoh et al., 2008). The SETD2-deposited H3K36me3 marks then recruit DNMT3B to those regions via its PWWP domain, leading to the establishment of a DNAme block over transcribed regions in ESCs (Baubec et al., 2015; Neri et al., 2017). A similar recruitment mechanism is conserved during de novo DNA methylation in the germline, although different approaches are exploited to establish the H3K36me-marked domains in male and female gametes. In oocytes, both SETD2-deposited H3K36me3 marks over transcribed regions as well as H3K36me2, presumably deposited by the nuclear receptor-binding SET-domain proteins NSD1 or NSD2, are implicated in establishing the maternal methylome via recruitment of the DNMT3A-DNMT3L complex (Xu et al., 2019; Yano et al., 2022). In the male germline, where the genome is more than 80% methylated (Kobayashi et al., 2012), the recruitment of DNMT3A is mediated by NSD1-deposited H3K36me2 marks, which cover broad regions of the genome (Shirane et al., 2020).
Despite this simple model implicating direct DNMT3A-H3K36me2/3 interactions, some results on mouse mutants carrying specific mutations in the DNMT3A PWWP reader domain may suggest that additional mechanisms are also at play during de novo DNA methylation. Two point mutations within the PWWP domain abrogating the binding of DNMT3A to H3K36me2/3 in vitro have been modeled in mouse. Both of these alleles, D239A and W236R, lead to dominant growth retardation phenotypes characterized by abnormal gain of DNAme at H3K27me3-marked regions. Surprisingly, gene bodies where H3K36me3 is deposited were unaffected (Heyn et al., 2019; Sendžikaitė et al., 2019). Similar observations were also made in mutant oocytes expressing only the D239A variant, in which H3K36me2/3-marked regions still acquired DNAme (Kibe et al., 2021). Although those results may suggest the existence of an alternative recruitment mechanism for the DNMT3A/3L complex, the authors also raise the possibility of residual binding of the D239A mutant PWWP domain to H3K36me2/3 in vivo, or a compensation via interactions between DNMT3A and DNMT3B, which also features an H3K36me2/3-binding PWWP domain (Kibe et al., 2021). The resolution of these alternative scenarios will require the direct analysis of DNMT3A D239A binding specificity in vivo by ChIP-seq and studies involving the simultaneous deletion of Dnmt3b in oocytes.
Long-terminal-repeat retrotransposons (LTRs), also known as endogenous retroviruses (ERVs), are highly variable in mammalian genomes and constitute ∼10% and ∼9% of the mouse and human genomes, respectively (Chinwalla et al., 2002). Several families of transposable elements, mostly young LTRs, can promote transcription initiation and act as TSS during oocyte growth in both mouse and human (Peaston et al., 2004; Veselovska et al., 2015; Franke et al., 2017; Hendrickson et al., 2017). Therefore, although LTRs are usually silenced by epigenetic mechanisms implicating DNAme or repressive histone marks such as H3K9me3 (Liu et al., 2014), some of these elements, notably younger LTRs, evade these mechanisms and are active as promoter elements in growing oocytes. Some of these LTR-initiated transcripts are intergenic or antisense to known genes, but others act as oocyte-specific alternative promoters for annotated genes, forming chimeric transcripts with annotated downstream exons. This enormous potential of LTR elements to shape the oocyte transcriptome is conserved in mammals and has been documented by oocyte RNA-seq in several species, such as mouse, rat, hamster, human and cow (Franke et al., 2017; Hendrickson et al., 2017; Brind’Amour et al., 2018). Interestingly, oocytes utilize a paralogue of the general transcription factor TATA binding protein (TBP), called TBPL2 (also known as TRF3 or TBP2), for transcription initiation during oocyte growth (Gazdag et al., 2009). TBPL2 was shown to play an important role in oocyte transcription, including at LTR promoters, notably at those featuring a TATA-like motif (Yu et al., 2020). It will be interesting to document how the DNA methylome and imprinting are affected in Tbpl2−/− oocytes.
Given the high level of expression from specific LTR promoters in oocytes and the observation that transcribed regions are de novo methylated, a significant fraction of the oocyte methylome originates from transcription initiating in active LTR promoters (Brind’Amour et al., 2018). A comparative description of such an impact of LTR-initiated transcripts on the DNA methylome of mouse, rat, and human oocytes showed that transcriptionally active LTRs are responsible for wide differences in DNAme patterns in oocytes of different species (Brind’Amour et al., 2018). Note that as for non-repetitive oocyte promoters, the active LTRs themselves are not DNA methylated in oocytes and overlap with a peak of H3K4me3 active promoter mark. As for single-copy promoters, they also lead to the deposition of an H3K36me3 domain over the transcribed region and the subsequent formation of a block of DNAme starting ∼2 kb downstream of the TSS provided by the LTR element (Brind’Amour et al., 2018).
Since DNAme blocks acquired in oocytes and maintained during preimplantation stages play a critical role in imprinting, the work on LTR-driven transcription and DNAme in oocytes raised the following questions: Do some of the DNAme marks acquired in oocytes as a consequence of transcription from LTRs act as imprinted gDMRs allowing only paternal allele-specific expression of the downstream gene in the progeny? Has this mechanism contributed to the evolution of species-specific imprinted genes?
By analyzing known imprinted gDMRs established in mouse (21 gDMRs) and human (125 gDMRs) oocytes, 21 examples of methylated regions covered by oocyte transcripts initiated within an LTR element were identified, 4 in the mouse, and 17 in human (Bogutz et al., 2019). Based on a 2018 survey, the mouse and human genomes were found to contain approximately 260 and 228 imprinted genes, respectively, with 63 shared in both species (Tucci et al., 2019). From these figures, it follows that ∼1.5% and ∼7.5% of imprinted genes are regulated by oocyte promoters in mouse and human, respectively. Data from mouse oocytes show that transcription initiation from these LTRs, marked by H3K4me3, lead to downstream blocks of H3K36me3 and DNAme deposition over the transcribed region, covering the site of the associated gDMR (Figure 1). Interestingly, none of these are the 15 maternal gDMRs shared between those two species. Moreover, for the 4 mouse gDMRs, the oocyte transcripts all initiate within LTR families specific to rodents, and 12 of the 17 human gDMRs are covered by transcripts initiating from LTRs of primate-specific families, 9 of which appear conserved in chimpanzee. Whereas most of this data is correlational, CRISPR-Cas9 mediated deletions of the LTR elements acting as upstream oocyte promoters at the mouse Impact and Slc38a4 imprinted genes confirmed the importance of these elements in species-specific maternal imprints. For both LTR knockouts, DNAme is lost at the gDMR of those genes in oocytes of homozygous knockout females and imprinting is lost in the progeny, with biallelic transcription of each gene (Bogutz et al., 2019). Together, the analysis presented in this study highlights a previously unappreciated role for LTR elements of endogenous retroviruses: by acting as promoters in oocytes, some of these elements can induce DNAme at a downstream CpG-rich promoter that is otherwise kept unmethylated in sperm, and can therefore lead to the formation of a new paternally-expressed imprinted gene, assuming maintenance of this maternal DNAme mark post-fertilization (Figure 2). As mentioned above, the survival of DNAme marks at gDMRs during preimplantation development relies on the binding of ZFP57 and ZFP445 to methylated TGCCGC motifs. Most of the canonical imprinted genes regulated by oocyte-specific LTR promoters contain at least one such binding site (Table 1). For the 17 human genes, ZFP57 binding has been observed by ChIP-seq at the HTR5A and CLDN23 CGI promoters, which maintain their imprinted DNAme mark in many epiblast-derived tissues (Bogutz et al., 2019). For 14 of these genes, imprinted DNAme at the CGI has only been observed in the placenta, so ZFP57/445 binding would be expected to only be observed during preimplantation development and in extra-embryonic cells.
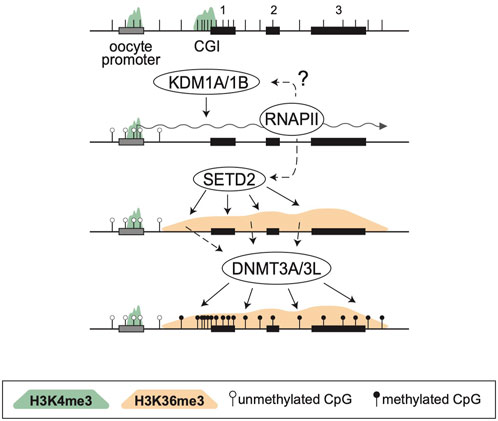
FIGURE 1. De novo DNA methylation during oocyte growth. Structure of a 3-exon gene is presented at the top, showing exons (black rectangles), a CGI promoter, the positions of CpG dinucleotides (vertical bars) and an upstream oocyte promoter (grey). In oocytes both promoters are marked with H3K4me3 (green shade) but KDM1A/1B, perhaps in association with RNAPII, remove this mark at the somatic CGI promoter. Simultaneously, SETD2 deposits H3K36me3 (orange shade) over the entire transcribed region. This mark is read by the PWWP domain of DNMT3A, which together with DNMT3L, methylates the transcribed region, including the CGI promoter.
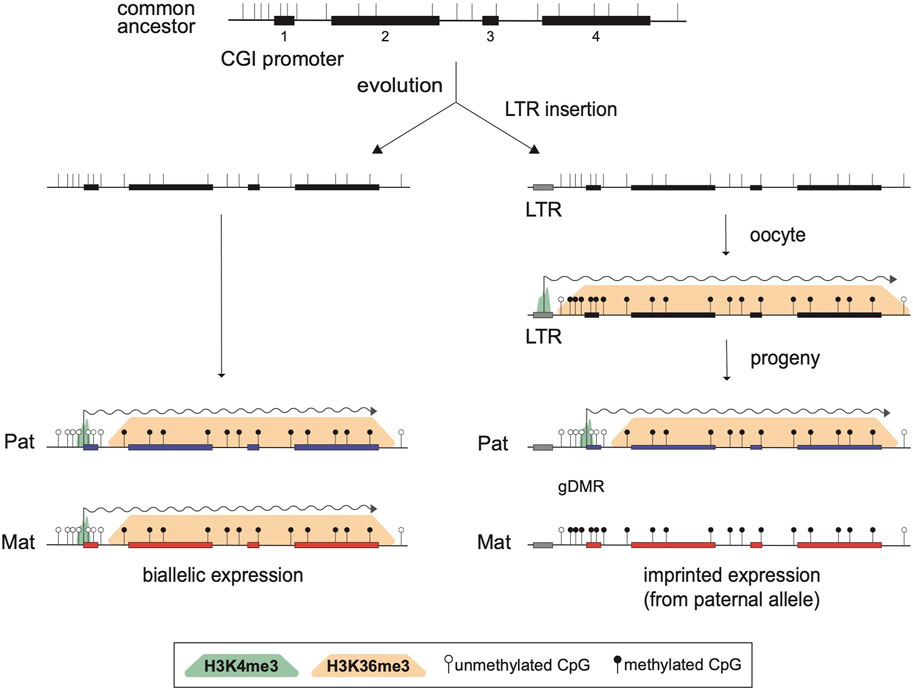
FIGURE 2. Model for the acquisition of imprinted expression via the insertion of an LTR element. Structure of a biallelically expressed ancestral gene is presented at the top, showing four exons (black rectangles), the positions of CpG dinucleotides (vertical bars), and a CpG island (CGI) promoter overlapping exon 1. Following evolution, two scenarios are considered. On the left, the locus is unchanged and the gene is expressed from both alleles in the progeny, as shown by biallelic active H3K4me3 marks (green shade) at the promoter and active transcription (wiggly arrow). One the right, a de novo retrotransposition event leading to the insertion of a solo LTR upstream of exon 1 is represented. The inserted LTR remains transcriptionally active in oocytes and induces the formation of blocks of H3K36me3 (orange shade) and DNAme (black lollipops) over the transcribed region. Consequently, the CGI promoter remains DNA methylated on the maternal allele in the progeny and the gene becomes a paternally expressed imprinted gene.
Although DNAme-based canonical imprinting provided an elegant mechanism to explain most imprinting effects, some observations suggested the existence of a parallel epigenetic pathway leading to parent-of-origin effects of gametic origin. For instance, a few cases of isolated imprinted genes lacking a gDMR were reported, such as the paternally expressed genes Sfmbt2 and Gab1 (Wang et al., 2011; Okae et al., 2012). In the placentae of cloned mice obtained by somatic cell nuclear transfer, using cumulus or Sertoli cells as nuclear donors, those same two genes, together with Slc38a4, were also shown to be consistently expressed from both alleles (Okae et al., 2014b). Furthermore, these three imprinted genes maintain at least some imprinted expression in the embryonic progeny of Dnmt3l or conditional Dnmt3a/3b null females, which fail to de novo methylate their oocyte genome (Okae et al., 2012; 2014b). Since the imprinted expression of canonical gDMR-regulated genes was faithfully maintained in most cloned mice, the authors concluded their study of Sfmbt2, Gab1, and Slc38a4 with this insightful prediction: “It is likely that an imprinting mark[s] other than DNA methylation may be required for the establishment of imprinting of these genes” (Okae et al., 2014b).
Similarly, research in the field of imprinted X chromosome inactivation (XCI) has hinted at a DNAme-independent mechanism responsible for the preferential inactivation of the paternal X in the extra-embryonic lineages of female mouse embryos (Takagi and Sasaki, 1975). Although earlier studies suggested that the maternally inherited allele of Xist, the lncRNA required for the initiation of XCI, is kept silent by DNAme directly inherited from oocytes (Ariel et al., 1995; Zuccotti and Monk, 1995), subsequent work with targeted or genome-wide bisulfite sequencing failed to confirm those results or reveal such a preemptive DNAme mark on the Xist promoter in eggs (McDonald et al., 1998; Shirane et al., 2013). Although the epigenetic imprint preventing silencing of the maternal X was shown to be established during oocyte growth, when DNAme marks are laid down (Tada et al., 2000), imprinted XCI was not perturbed in the progeny of Dnmt3a/3b mutant oocytes (Chiba et al., 2008), which fail to acquire DNAme (Hata et al., 2002; Kaneda et al., 2010; Shirane et al., 2013).
Together, these lines of evidence suggested that DNAme might not be the only epigenetic mark directly inherited from gametes that can lead to imprinted expression in the progeny. Note that all the evidence summarized above (for Xist and autosomal paternally expressed genes) pointed to a silencing mark inherited from the oocyte. The discovery of such a DNAme-independent mechanism, which has been called “noncanonical imprinting” (Inoue et al., 2017a), heralded new avenues of studies in genomic imprinting research in mammals. Features unique to noncanonical imprinting have been covered extensively by excellent recent reviews (Chen and Zhang, 2020; Hanna and Kelsey, 2021; Kobayashi, 2021; Albert and Greenberg, 2023; Inoue, 2023).
The discovery of noncanonical imprinting emerged from elegant studies mapping allele-specific DNase I hypersensitive sites (DHSs) in zygotes and morulae. For these experiments, the group of Yi Zhang first established a low-input protocol for the genome-wide mapping of DHSs, liDNase-seq, suitable for preimplantation work (Lu et al., 2016). By individually analyzing the profiles of DHSs in the paternal and maternal pronuclei, they identified parental allele-specific DHSs priming allele-specific expression at the 2-cell stage (Inoue et al., 2017a). Allele-specific regions of open chromatin in early mouse embryos have also been independently mapped by ATAC-seq (Wu et al., 2016). Most of these open chromatin regions were of paternal origin and since the protection of the maternal allele at 48% of these sites did not overlap with DNA methylated regions in oocytes, the results provided support for a DNAme-independent mechanism silencing the maternal alleles (Inoue et al., 2017a). By mining ChIP-seq data for the Polycomb Repressive Complex 2 (PRC2)-mediated H3K27me3 marks in oocytes and by injection of the mRNA for the H3K27me3-specific demethylase KDM6B, Inoue et al. further showed that maternally inherited H3K27me3 was responsible for the observed protection of the maternal allele and for imprinted expression of those genes from the paternal allele in morulae (Inoue et al., 2017a). Subsequent similar studies from this group showed that the imprinted expression of the lncRNA Xist, responsible for paternal X chromosome inactivation in extraembryonic tissues, is also controlled via a similar noncanonical imprinting mechanism via maternal H3K27me3 marks (Inoue et al., 2017b). The genetic requirement for a functional PRC2 in the establishment of oocyte H3K27me3 imprints was shown in two independent studies documenting the loss of non-canonical imprinting at autosomal genes and Xist in the progeny of embryonic ectoderm development (Eed)- deficient oocytes (Inoue et al., 2018; Harris et al., 2019). The observation that imprinted expression is maintained at DNAme-dependent canonical imprinted genes in those Eed maternal KO progeny highlights the functional independence of both imprinting mechanisms (Inoue et al., 2018). This conclusion is also supported by the maintenance of noncanonical imprinted expression in the progeny of Dnmt3l-deficient females, confirming that noncanonical imprinting is independent of oocyte DNAme (Chen et al., 2019). In addition to the protection of maternal alleles from assuming an open chromatin state, the maternal H3K27me3 imprints also prevent the acquisition of activating H3K4me3 marks on the maternal allele in preimplantation embryos (Chen et al., 2019). Although the available data are consistent with H3K27me3 being the epigenetic mark directly inherited from oocyte, an interplay with the Polycomb Repressive Complex 1 (PRC1)-mediated H2AK119ub1 mark has also been described: whereas H2AK119ub1 coexists with and might precede H3K27me3 establishment during oocyte growth, its depletion in zygotes does not disrupts noncanonical imprinting, unlike what was seen for the H3K27me3 marks (Chen et al., 2021; Mei et al., 2021). Nevertheless, deletions of the PRC1.6 subunits PCGF1/6 in oocytes lead to partial loss of noncanonical imprinting genes in morulae (at 9/16 genes) (Mei et al., 2021).
The work summarised above revealed a DNAme-independent mechanism of imprinting, called noncanonical imprinting, however important differences with DNAme-dependent canonical imprinting were noted. Although more than 70 genes have been detected as noncanonically imprinted and paternally expressed in preimplantation embryos, all of these genes lose their imprinted expression in epiblast-derived post-implantation tissues (Inoue et al., 2017a; Santini et al., 2021). Nevertheless, maintenance of noncanonical imprinted expression has been observed in extra-embryonic tissues, including visceral endoderm at E6.5, extra-embryonic ectoderm (EXE) at E6.5 and E7.5, ectoplacental cone at 6 somite stage (∼E8.5), as well as in E9.5 and E12.5 placentae (Inoue et al., 2017a; Hanna et al., 2019; Andergassen et al., 2021; Zeng et al., 2021). Similar tissue-specific maintenance of noncanonical imprinting only in extra-embryonic lineages was observed via the mapping of allelic H3K4me3 promoter marks (Hanna et al., 2019). One exception is Slc38a4, which shows imprinted expression from the paternal allele in E13 fetus as well as tissue-specifically imprinted in adult adrenals, heart, and skeletal muscle (Smith et al., 2003). This is explained by the fact that Slc38a4 is at least partially regulated by a gDMR, suggesting that both canonical and noncanonical mechanisms may regulate the expression of this gene in different tissues, perhaps via different isoforms (Smith et al., 2003; Inoue et al., 2017a; Bogutz et al., 2019; Chen and Zhang, 2020).
The expression data therefore suggest that noncanonical imprinting is mostly a transient mechanism, leading to paternal allele-specific imprinted expression of several genes (>70) in preimplantation embryos, but only maintained at some of these loci in extra-embryonic lineages (notably 7 genes: Gab1, Phf17, Platr20, Sall1, Sfmbt2, Slc38a4, and Smoc1). This would represent approximately 2.7% of mouse imprinted genes. The transient nature of this imprinting mechanism is consistent with the observation that the broad H3K27me3-marked regions inherited from the oocyte and required for noncanonical imprinting are largely maintained to the blastocyst stage, but are erased in E6.5 epiblast (Zheng et al., 2016), and are absent in embryonic stem cells, mouse embryonic fibroblasts, and adult somatic cells (Matoba et al., 2018). These observations provide an explanation for the biallelic expression seen for noncanonical imprinted genes in embryos generated by somatic cell nuclear transfer (cloning), since the maternal H3K27me3 marks responsible for noncanonical imprinting are absent in the somatic donor cells (Okae et al., 2014b; Matoba et al., 2018; Xie et al., 2022).
Although noncanonical imprinting has been detected in post-implantation extra-embryonic lineages via expression and H3K4me3 data, the maternal H3K27me3 marks do not survive past the blastocyst stage (Chen et al., 2019; 2021). This raises the question of what are the mechanisms guiding and maintaining this maternal allele-specific silencing in extra-embryonic lineages of post-implantation embryos. By comparing the genomic localisations of paternal H3K4me3 peaks associated with imprinted expression, a key difference was noted between the two families of imprinted genes: at canonical imprinted genes, those H3K4me3 peaks are mostly associated with promoter CpG islands, while at noncanonical genes, the active promoter marks map to endogenous retroviral elements, notably of the ERVK family (Hanna et al., 2019). While these noncanonical imprinted ERVK promoters are not marked by H3K27me3 in E6.5 EXE, they are in fact marked by DNAme on the silent maternal allele. Since these DNAme marks at imprinted ERVKs are not present in preimplantation embryos, they constitute classical somatic DMRs (sDMRs) (John and Lefebvre, 2011), acquired in post-implantation embryos (Chen et al., 2019; Hanna et al., 2019). An essential role for both DNMT3A and DNMT3B in this postimplantation de novo methylation pathway was confirmed by ablating both genes in zygotes using CRISPR-Cas9. However, Sfmbt2 appears to be an exception here, with its imprinted expression being maintained despite loss of DNAme, at least at E6.5 (Chen et al., 2019). Surprisingly, this effect at Sfmbt2 was not observed in zygotic euchromatic histone lysine N-methyltransferase 2 (Ehmt2)-null embryos (also known as G9a), in which the establishment of the sDMRs at noncanonical imprinted genes does not occur and biallelic expression is observed (Auclair et al., 2016; Zeng et al., 2021).
As expected, the sDMRs at imprinted ERVKs are also lost in the progeny of Eed-null oocytes, confirming the importance of the maternal H3K27me3 imprints in the initiation of this imprinting process. The observations that these ERVK elements become biallelically DNA methylated in the epiblast is consistent with the maintenance of noncanonical imprinting only in extra-embryonic lineages. Although some of these ERVKs, which are mostly solo LTR elements, were shown to act as alternative promoters for noncanonical imprinted genes, it remains to be seen whether some of these elements act as extra-embryonic enhancer elements, as has been previously reported for some LTR families (Chuong et al., 2013; Hanna et al., 2019; Figure 3). How the sDMRs are established specifically in extra-embryonic lineages but not in the epiblast-derived tissues is also currently unknown.
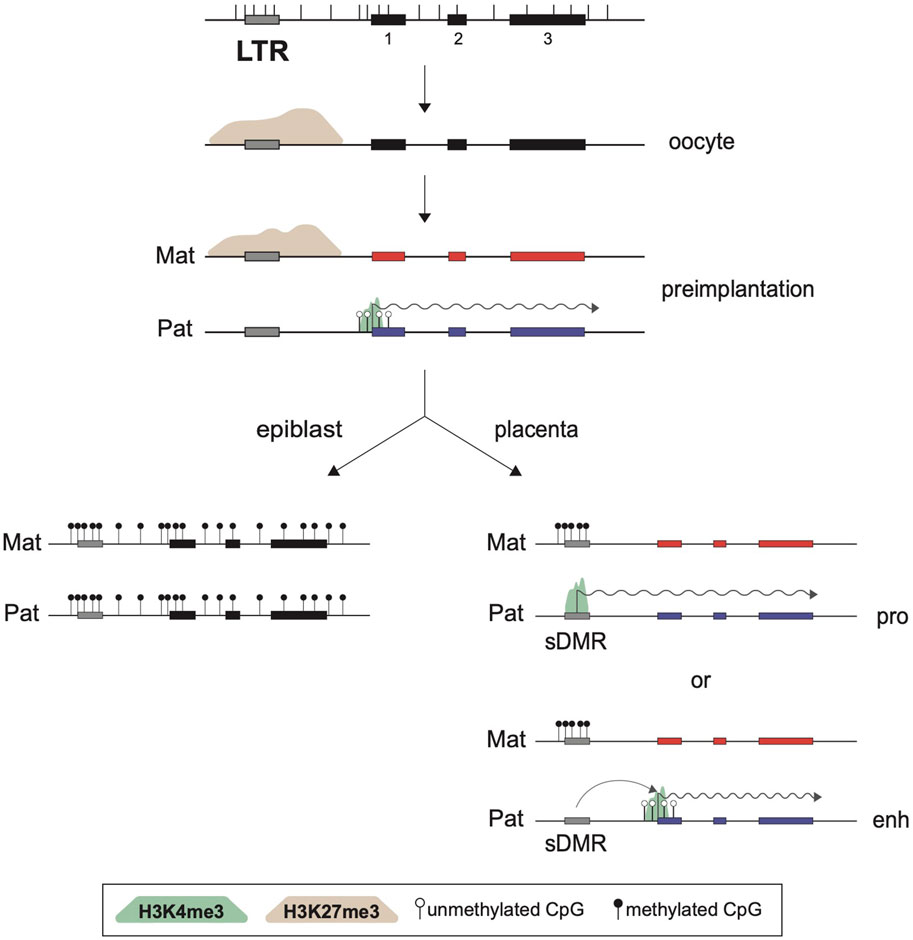
FIGURE 3. Role of LTRs in noncanonical imprinting maintenance. Structure of a noncanonical imprinted gene is presented at the top, showing three exons (black rectangles), the positions of CpG dinucleotides (vertical bars), and an upstream LTR element. In oocytes, part of the region is marked by a broad PRC2-deposited H3K27me3 domain (brown shade). This silencing epigenetic mark is inherited on the maternal allele such that only the paternal allele of the affected gene can be transcribed in preimplantation stages, as shown by the active H3K4me3 promoter mark (green shade) and transcription elongation (wiggly arrow). However, this histone imprint is only transient and is lost in all postimplantation cell lineages. In the epiblast, de novo DNA methylation leads to biallelic silencing, while in the extra-embryonic lineages a somatic DMR (sDMR) is generated over a nearby ERVK LTR element, with DNAme acquired exclusively on the previously H3K27me3-marked maternal allele. The LTR can then act as an alternative promoter (pro) or an enhancer (enh) to guide imprinted expression of the paternal allele. Not shown are the roles of PRC1 and its associated H2AK119ub mark in the establishment of the H3K27me3 domain in oocytes, or the implications of DNMT3A/3B, SMC hinge domain containing 1 (SMCHD1) and G9A/EHMT2 in formation of the extra-embryonic sDMRs themselves.
Together, this body of work has revealed that noncanonical imprinted genes, which so far have only been observed to be paternally expressed in extra-embryonic tissues, may rely on alternating allelic epigenetic marks for their allelic expression. The oocyte-derived H3K27me3 imprint, itself dependent on H2AK119ub1 at certain genes, must be converted into a DNAme somatic mark only on the maternal allele to achieve noncanonical imprinting in extra-embryonic tissues. This switch may be guided by the paternal H3K4me3 promoter marks over ERVK elements in preimplantation embryos, which would protect the paternal alleles from the action of the DNMT3A/3B de novo enzymes (Zhang et al., 2010). The observation that both the paternal H3K4me3 peaks and the sDMRs implicated in noncanonical imprinting map to endogenous retroviral promoters, notably of the ERVK family, suggests that these elements play critical roles in the maintenance of this unique tissue-specific imprinting pathway. In the cases where the imprinted ERVK element act as an alternative promoter for paternal allele-specific expression, a parallel can be drawn with the role of LTRs in the establishment of canonical imprinting, for which their activity as an oocyte promoter is critical.
Canonical imprints are essential for embryonic development, as shown by the early midgestational lethality of offspring obtained from oocytes deficient in de novo DNAme (Bourc’his et al., 2001; Hata et al., 2002; Kaneda et al., 2004). Although much remains to be determined regarding the biological functions of noncanonical imprinted genes in extraembryonic lineages, the global loss of maternal H3K27me3 marks in conditional Eed or Ezh2 mutant oocytes is compatible with development to term in the progeny, although with embryonic growth defects (Erhardt et al., 2003; Prokopuk et al., 2018). The shared placental overgrowth phenotypes observed in cloned mice and in Eed maternal KO conceptuses have been linked to the abnormal expression of noncanonical imprinted genes such as Slc38a4, Gm32885 (a transcript upstream of Slc38a4 on Chr 15), as well as a cluster of microRNAs coded within an intron of Sfmbt2, C2MC (Matoba et al., 2018; 2019; 2022). Together, these observations support key developmental roles for imprinted genes, both canonical and noncanonical, with an emphasis on the regulation of extra-embryonic lineages.
The fact that LTR elements, which are highly polymorphic in different mammalian species, are implicated in different aspects of both imprinting pathways suggests that they may play important roles in the emergence of new imprinted genes in different species. In their function as oocyte promoters for the establishment of maternal DNAme marks, LTRs were shown to be involved in the imprinting of non-overlapping sets of canonical imprinting genes in mouse and human (Bogutz et al., 2019). However, the three protein-coding genes imprinted by an oocyte LTR promoter in mouse (Slc38a4, Impact, and Chd15) are also imprinted in rat (Albert et al., 2023). A different picture emerges for noncanonical imprinted genes: although several noncanonical imprinted genes identified in mouse appear conserved in rat, profiling of allelic usage in this species also identified 8 rat-specific putative noncanonical imprinted genes, consistent with a rapid evolution of this imprinting mechanism in rodents (Albert et al., 2023). Nevertheless, whether the noncanonical pathway also operates in human embryos is unclear, since human XIST expression is not imprinted in preimplantation embryos nor in extra-embryonic membranes (Migeon and Do, 1979; Petropoulos et al., 2016), and most H3K27me3 marks are rapidly erased in human preimplantation embryo (Zheng et al., 2016; Xia et al., 2019; Lu et al., 2021). On the other hand, other studies have reported maternal-biased H3K27me3 marks and associated paternal allele-specific expression in human morulae (Zhang et al., 2019), as well as the presence of placental sDMRs corresponding to regions marked by H3K27me3 and hypomethylated in eggs, which are consistent with putative noncanonical imprinting (Hanna and Kelsey, 2021). Future work on other mammalian species will be important to establish the conservation and importance of LTR-based mechanisms of imprinting in the evolution of new imprinted genes.
SF: Visualization, Writing–original draft, Writing–review and editing. K-WC: Writing–original draft, Writing–review and editing. LL: Conceptualization, Funding acquisition, Project administration, Supervision, Visualization, Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by a Project Grant (#PJT-165992) from the Canadian Institutes of Health Research to LL.
The authors thank Aaron B. Bogutz for critical input.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akbari, V., Garant, J.-M., O’Neill, K., Pandoh, P., Moore, R., Marra, M. A., et al. (2022). Genome-wide detection of imprinted differentially methylated regions using nanopore sequencing. Elife 11, e77898. doi:10.7554/elife.77898
Albert, J. R., and Greenberg, M. V. C. (2023). Non-canonical imprinting in the spotlight. Development 150, dev201087. doi:10.1242/dev.201087
Albert, J. R., Kobayashi, T., Inoue, A., Monteagudo-Sánchez, A., Kumamoto, S., Takashima, T., et al. (2023). Conservation and divergence of canonical and non-canonical imprinting in murids. Genome Biol. 24, 48. doi:10.1186/s13059-023-02869-1
Andergassen, D., Smith, Z. D., Kretzmer, H., Rinn, J. L., and Meissner, A. (2021). Diverse epigenetic mechanisms maintain parental imprints within the embryonic and extraembryonic lineages. Dev. Cell 56, 2995–3005. doi:10.1016/j.devcel.2021.10.010
Anvar, Z., Cammisa, M., Riso, V., Baglivo, I., Kukreja, H., Sparago, A., et al. (2016). ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res. 44, 1118–1132. doi:10.1093/nar/gkv1059
Ariel, M., Robinson, E., McCarrey, J. R., and Cedar, H. (1995). Gamete-specific methylation correlates with imprinting of the murine Xist gene. Nat. Genet. 9, 312–315. doi:10.1038/ng0395-312
Arima, T., Hata, K., Tanaka, S., Kusumi, M., Li, E., Kato, K., et al. (2006). Loss of the maternal imprint in Dnmt3Lmat-/- mice leads to a differentiation defect in the extraembryonic tissue. Dev. Biol. 297, 361–373. doi:10.1016/j.ydbio.2006.05.003
Auclair, G., Borgel, J., Sanz, L. A., Vallet, J., Guibert, S., Dumas, M., et al. (2016). EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res. 26, 192–202. doi:10.1101/gr.198291.115
Barlow, D. P. (1993). Methylation and imprinting: from host defense to gene regulation? Science 260, 309–310. doi:10.1126/science.8469984
Barlow, D. P., Stöger, R., Herrmann, B. G., Saito, K., and Schweifer, N. (1991). The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349, 84–87. doi:10.1038/349084a0
Bartolomei, M. S., Webber, A. L., Brunkow, M. E., and Tilghman, S. M. (1993). Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7, 1663–1673. doi:10.1101/gad.7.9.1663
Bartolomei, M. S., Zemel, S., and Tilghman, S. M. (1991). Parental imprinting of the mouse H19 gene. Nature 351, 153–155. doi:10.1038/351153a0
Baubec, T., Colombo, D. F., Wirbelauer, C., Schmidt, J., Burger, L., Krebs, A. R., et al. (2015). Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247. doi:10.1038/nature14176
Bestor, T. H. (1990). DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos. Trans. R. Soc. Lond B Biol. Sci. 326, 179–187. doi:10.1098/rstb.1990.0002
Bogutz, A. B., Brind’Amour, J., Kobayashi, H., Jensen, K. N., Nakabayashi, K., Imai, H., et al. (2019). Evolution of imprinting via lineage-specific insertion of retroviral promoters. Nat. Commun. 10, 5674. doi:10.1038/s41467-019-13662-9
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B., and Bestor, T. H. (2001). Dnmt3L and the establishment of maternal genomic imprints. Sci. (New York, N.Y.) 294, 2536–2539. doi:10.1126/science.1065848
Branco, M. R., King, M., Perez-Garcia, V., Bogutz, A. B., Caley, M., Fineberg, E., et al. (2016). Maternal DNA methylation regulates early trophoblast development. Dev. Cell 36, 152–163. doi:10.1016/j.devcel.2015.12.027
Brandeis, M., Kafri, T., Ariel, M., Chaillet, J. R., McCarrey, J., Razin, A., et al. (1993). The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. Embo J. 12, 3669–3677. doi:10.1002/j.1460-2075.1993.tb06041.x
Brind’Amour, J., Kobayashi, H., Albert, J. R., Shirane, K., Sakashita, A., Kamio, A., et al. (2018). LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat. Commun. 9, 3331–3344. doi:10.1038/s41467-018-05841-x
Cattanach, B. M., Barr, J. A., Evans, E. P., Burtenshaw, M., Beechey, C. V., Leff, S. E., et al. (1992). A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat. Genet. 2, 270–274. doi:10.1038/ng1292-270
Chen, Z., Djekidel, M. N., and Zhang, Y. (2021). Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat. Genet. 53, 551–563. doi:10.1038/s41588-021-00821-2
Chen, Z., Yin, Q., Inoue, A., Zhang, C., and Zhang, Y. (2019). Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci. Adv. 5, eaay7246. doi:10.1126/sciadv.aay7246
Chen, Z., and Zhang, Y. (2020). Maternal H3K27me3-dependent autosomal and X chromosome imprinting. Nat. Rev. Genet. 21, 555–571. doi:10.1038/s41576-020-0245-9
Chiba, H., Hirasawa, R., Kaneda, M., Amakawa, Y., Li, E., Sado, T., et al. (2008). De novo DNA methylation independent establishment of maternal imprint on X chromosome in mouse oocytes. genesis 46, 768–774. doi:10.1002/dvg.20438
Chinwalla, A. T., Cook, L. L., Delehaunty, K. D., Fewell, G. A., Fulton, L. A., Fulton, R. S., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. doi:10.1038/nature01262
Chotalia, M., Smallwood, S. A., Ruf, N., Dawson, C., Lucifero, D., Frontera, M., et al. (2009). Transcription is required for establishment of germline methylation marks at imprinted genes. Genes and Dev. 23, 105–117. doi:10.1101/gad.495809
Chuong, E. B., Rumi, M. A. K., Soares, M. J., and Baker, J. C. (2013). Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329. doi:10.1038/ng.2553
Ciccone, D., Su, H., Hevi, S., Gay, F., Lei, H., Bajko, J., et al. (2009). KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418. doi:10.1038/nature08315
Court, F., Tayama, C., Romanelli, V., Martin-Trujillo, A., Iglesias-Platas, I., Okamura, K., et al. (2014). Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 24, 554–569. doi:10.1101/gr.164913.113
Cowley, M., and Oakey, R. J. (2010). Retrotransposition and genomic imprinting. Briefings Funct. Genomics 9, 340–346. doi:10.1093/bfgp/elq015
Davis, T. L., Yang, G. J., McCarrey, J. R., and Bartolomei, M. S. (2000). The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 9, 2885–2894. doi:10.1093/hmg/9.19.2885
DeChiara, T. M., Robertson, E. J., and Efstratiadis, A. (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859. doi:10.1016/0092-8674(91)90513-x
Dhayalan, A., Rajavelu, A., Rathert, P., Tamas, R., Jurkowska, R. Z., Ragozin, S., et al. (2010). The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114–26120. doi:10.1074/jbc.m109.089433
Erhardt, S., Su, I., Schneider, R., Barton, S., Bannister, A. J., Perez-Burgos, L., et al. (2003). Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130, 4235–4248. doi:10.1242/dev.00625
Ferguson-Smith, A. C., Sasaki, H., Cattanach, B. M., and Surani, M. A. (1993). Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362, 751–755. doi:10.1038/362751a0
Franke, V., Ganesh, S., Karlic, R., Malik, R., Pasulka, J., Horvat, F., et al. (2017). Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res. 27, 1384–1394. doi:10.1101/gr.216150.116
Gazdag, E., Santenard, A., Ziegler-Birling, C., Altobelli, G., Poch, O., Tora, L., et al. (2009). TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev. 23, 2210–2223. doi:10.1101/gad.535209
Guo, X., Wang, L., Li, J., Ding, Z., Xiao, J., Yin, X., et al. (2015). Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 517, 640–644. doi:10.1038/nature13899
Hamada, H., Okae, H., Toh, H., Chiba, H., Hiura, H., Shirane, K., et al. (2016). Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am. J. Hum. Genet. 99, 1045–1058. doi:10.1016/j.ajhg.2016.08.021
Hanna, C. W., and Kelsey, G. (2021). Features and mechanisms of canonical and noncanonical genomic imprinting. Genes and Dev. 35, 821–834. doi:10.1101/gad.348422.121
Hanna, C. W., Peñaherrera, M. S., Saadeh, H., Andrews, S., Mcfadden, D. E., Kelsey, G., et al. (2016). Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 26, 756–767. doi:10.1101/gr.196139.115
Hanna, C. W., Pérez-Palacios, R., Gahurova, L., Schubert, M., Krueger, F., Biggins, L., et al. (2019). Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol. 20, 225. doi:10.1186/s13059-019-1833-x
Harris, C., Cloutier, M., Trotter, M., Hinten, M., Gayen, S., Du, Z., et al. (2019). Conversion of random X-inactivation to imprinted X-inactivation by maternal PRC2. eLife 8, e44258. doi:10.7554/elife.44258
Hata, K., Okano, M., Lei, H., and Li, E. (2002). Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993. doi:10.1242/dev.129.8.1983
Hendrickson, P. G., Doráis, J. A., Grow, E. J., Whiddon, J. L., Lim, J.-W., Wike, C. L., et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 49, 925–934. doi:10.1038/ng.3844
Heyn, P., Logan, C. V., Fluteau, A., Challis, R. C., Auchynnikava, T., Martin, C.-A., et al. (2019). Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet. 51, 96–105. doi:10.1038/s41588-018-0274-x
Hirasawa, R., Chiba, H., Kaneda, M., Tajima, S., Li, E., Jaenisch, R., et al. (2008). Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes and Dev. 22, 1607–1616. doi:10.1101/gad.1667008
Inoue, A. (2023). Noncanonical imprinting: intergenerational epigenetic inheritance mediated by Polycomb complexes. Curr. Opin. Genet. Dev. 78, 102015. doi:10.1016/j.gde.2022.102015
Inoue, A., Chen, Z., Yin, Q., and Zhang, Y. (2018). Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Gene Dev. 32, 1525–1536. doi:10.1101/gad.318675.118
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017a). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424. doi:10.1038/nature23262
Inoue, A., Jiang, L., Lu, F., and Zhang, Y. (2017b). Genomic imprinting of Xist by maternal H3K27me3. Gene Dev. 31, 1927–1932. doi:10.1101/gad.304113.117
Janssen, S. M., and Lorincz, M. C. (2021). Interplay between chromatin marks in development and disease. Nat. Rev. Genet. 23, 137–153. doi:10.1038/s41576-021-00416-x
John, R. M., and Lefebvre, L. (2011). Developmental regulation of somatic imprints. Differ. Res. Biol. Divers. 81, 270–280. doi:10.1016/j.diff.2011.01.007
Kaneda, M., Hirasawa, R., Chiba, H., Okano, M., Li, E., and Sasaki, H. (2010). Genetic evidence for Dnmt3a-dependent imprinting during oocyte growth obtained by conditional knockout with Zp3-Cre and complete exclusion of Dnmt3b by chimera formation. Genes Cells 15, 169–179. doi:10.1111/j.1365-2443.2009.01374.x
Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E., et al. (2004). Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903. doi:10.1038/nature02633
Kibe, K., Shirane, K., Ohishi, H., Uemura, S., Toh, H., and Sasaki, H. (2021). The DNMT3A PWWP domain is essential for the normal DNA methylation landscape in mouse somatic cells and oocytes. PLoS Genet. 17, e1009570. doi:10.1371/journal.pgen.1009570
Kobayashi, H. (2021). Canonical and non-canonical genomic imprinting in rodents. Front. Cell Dev. Biol. 9, 713878. doi:10.3389/fcell.2021.713878
Kobayashi, H., Sakurai, T., Imai, M., Takahashi, N., Fukuda, A., Yayoi, O., et al. (2012). Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8, e1002440. doi:10.1371/journal.pgen.1002440
Kono, T., Obata, Y., Yoshimzu, T., Nakahara, T., and Carroll, J. (1996). Epigenetic modifications during oocyte growth correlates with extended parthenogenetic development in the mouse. Nat. Genet. 13, 91–94. doi:10.1038/ng0596-91
Leff, S. E., Brannan, C. I., Reed, M. L., Özçelik, T., Francke, U., Copeland, N. G., et al. (1992). Maternal imprinting of the mouse Snrpn gene and conserved linkage homology with the human Prader–Willi syndrome region. Nat. Genet. 2, 259–264. doi:10.1038/ng1292-259
Li, E., Beard, C., and Jaenisch, R. (1993). Role for DNA methylation in genomic imprinting. Nature 366, 362–365. doi:10.1038/366362a0
Li, J.-Y., Lees-Murdock, D. J., Xu, G.-L., and Walsh, C. P. (2004). Timing of establishment of paternal methylation imprints in the mouse. Genomics 84, 952–960. doi:10.1016/j.ygeno.2004.08.012
Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., et al. (2008). A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557. doi:10.1016/j.devcel.2008.08.014
Liu, S., Brind’Amour, J., Karimi, M. M., Shirane, K., Bogutz, A., Lefebvre, L., et al. (2014). Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes and Dev. 28, 2041–2055. doi:10.1101/gad.244848.114
Lu, F., Liu, Y., Inoue, A., Suzuki, T., Zhao, K., and Zhang, Y. (2016). Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165, 1375–1388. doi:10.1016/j.cell.2016.05.050
Lu, X., Zhang, Y., Wang, L., Wang, L., Wang, H., Xu, Q., et al. (2021). Evolutionary epigenomic analyses in mammalian early embryos reveal species-specific innovations and conserved principles of imprinting. Sci. Adv. 7, eabi6178. doi:10.1126/sciadv.abi6178
Matoba, S., Kozuka, C., Miura, K., Inoue, K., Kumon, M., Hayashi, R., et al. (2022). Noncanonical imprinting sustains embryonic development and restrains placental overgrowth. Gene Dev. 36, 483–494. doi:10.1101/gad.349390.122
Matoba, S., Nakamuta, S., Miura, K., Hirose, M., Shiura, H., Kohda, T., et al. (2019). Paternal knockout of Slc38a4/SNAT4 causes placental hypoplasia associated with intrauterine growth restriction in mice. Proc. Natl. Acad. Sci. 116, 21047–21053. doi:10.1073/pnas.1907884116
Matoba, S., Wang, H., Jiang, L., Lu, F., Iwabuchi, K. A., Wu, X., et al. (2018). Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23, 343–354. doi:10.1016/j.stem.2018.06.008
McDonald, L. E., Paterson, C. A., and Kay, G. F. (1998). Bisulfite genomic sequencing-derived methylation profile of the xist gene throughout early mouse development. Genomics 54, 379–386. doi:10.1006/geno.1998.5570
McGrath, J., and Solter, D. (1984). Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183. doi:10.1016/0092-8674(84)90313-1
Mei, H., Kozuka, C., Hayashi, R., Kumon, M., Koseki, H., and Inoue, A. (2021). H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat. Genet. 53, 539–550. doi:10.1038/s41588-021-00820-3
Migeon, B. R., and Do, T. T. (1979). In search of non-random X inactivation: studies of fetal membranes heterozygous for glucose-6-phosphate dehydrogenase. Am. J. Hum. Genet. 31, 581–585.
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. doi:10.1038/nature06008
Neri, F., Rapelli, S., Krepelova, A., Incarnato, D., Parlato, C., Basile, G., et al. (2017). Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77. doi:10.1038/nature21373
Obata, Y., Kaneko-Ishino, T., Koide, T., Takai, Y., Ueda, T., Domeki, I., et al. (1998). Disruption of primary imprinting during oocyte growth leads to the modified expression of imprinted genes during embryogenesis. Development 125, 1553–1560. doi:10.1242/dev.125.8.1553
Obata, Y., and Kono, T. (2002). Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 277, 5285–5289. doi:10.1074/jbc.m108586200
Okae, H., Chiba, H., Hiura, H., Hamada, H., Sato, A., Utsunomiya, T., et al. (2014a). Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 10, e1004868. doi:10.1371/journal.pgen.1004868
Okae, H., Hiura, H., Nishida, Y., Funayama, R., Tanaka, S., Chiba, H., et al. (2012). Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum. Mol. Genet. 21, 548–558. doi:10.1093/hmg/ddr488
Okae, H., Matoba, S., Nagashima, T., Mizutani, E., Inoue, K., Ogonuki, N., et al. (2014b). RNA sequencing-based identification of aberrant imprinting in cloned mice. Hum. Mol. Genet. 23, 992–1001. doi:10.1093/hmg/ddt495
Ondičová, M., Oakey, R. J., and Walsh, C. P. (2020). Is imprinting the result of “friendly fire” by the host defense system? Plos Genet. 16, e1008599. doi:10.1371/journal.pgen.1008599
Ooi, S. K. T., Qiu, C., Bernstein, E., Li, K., Jia, D., Yang, Z., et al. (2007). DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717. doi:10.1038/nature05987
Peaston, A. E., Evsikov, A. V., Graber, J. H., Vries, W. N. de, Holbrook, A. E., Solter, D., et al. (2004). Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606. doi:10.1016/j.devcel.2004.09.004
Petropoulos, S., Edsgärd, D., Reinius, B., Deng, Q., Panula, S. P., Codeluppi, S., et al. (2016). Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165, 1012–1026. doi:10.1016/j.cell.2016.03.023
Prokopuk, L., Stringer, J. M., White, C. R., Vossen, R. H. A. M., White, S. J., Cohen, A. S. A., et al. (2018). Loss of maternal EED results in postnatal overgrowth. Clin. Epigenetics 10, 95. doi:10.1186/s13148-018-0526-8
Proudhon, C., Duffié, R., Ajjan, S., Cowley, M., Iranzo, J., Carbajosa, G., et al. (2012). Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol. Cell 47, 909–920. doi:10.1016/j.molcel.2012.07.010
Sanchez-Delgado, M., Court, F., Vidal, E., Medrano, J., Monteagudo-Sánchez, A., Martin-Trujillo, A., et al. (2016). Human oocyte-derived methylation differences persist in the placenta revealing widespread transient imprinting. Plos Genet. 12, e1006427. doi:10.1371/journal.pgen.1006427
Santini, L., Halbritter, F., Titz-Teixeira, F., Suzuki, T., Asami, M., Ma, X., et al. (2021). Genomic imprinting in mouse blastocysts is predominantly associated with H3K27me3. Nat. Commun. 12, 3804. doi:10.1038/s41467-021-23510-4
Sendžikaitė, G., Hanna, C. W., Stewart-Morgan, K. R., Ivanova, E., and Kelsey, G. (2019). A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 10, 1884. doi:10.1038/s41467-019-09713-w
Sharif, J., Muto, M., Takebayashi, S.-I., Suetake, I., Iwamatsu, A., Endo, T. A., et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. doi:10.1038/nature06397
Shirane, K., Miura, F., Ito, T., and Lorincz, M. C. (2020). NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat. Genet. 52, 1088–1098. doi:10.1038/s41588-020-0689-z
Shirane, K., Toh, H., Kobayashi, H., Miura, F., Chiba, H., Ito, T., et al. (2013). Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 9, e1003439. doi:10.1371/journal.pgen.1003439
Singh, V. B., Sribenja, S., Wilson, K. E., Attwood, K. M., Hillman, J. C., Pathak, S., et al. (2017). Blocked transcription through KvDMR1 results in absence of methylation and gene silencing resembling Beckwith-Wiedemann syndrome. Development 144, 1820–1830. doi:10.1242/dev.145136
Smallwood, S. A., Tomizawa, S.-I., Krueger, F., Ruf, N., Carli, N., Segonds-Pichon, A., et al. (2011). Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814. doi:10.1038/ng.864
Smith, E. Y., Futtner, C. R., Chamberlain, S. J., Johnstone, K. A., and Resnick, J. L. (2011). Transcription is required to establish maternal imprinting at the Prader-Willi syndrome and Angelman syndrome locus. PLoS Genet. 7, e1002422. doi:10.1371/journal.pgen.1002422
Smith, R. J., Dean, W., Konfortova, G., and Kelsey, G. (2003). Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 13, 558–569. doi:10.1101/gr.781503
Stewart, K. R., Veselovska, L., Kim, J., Huang, J., Saadeh, H., Tomizawa, S.-I., et al. (2015). Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes and Dev. 29, 2449–2462. doi:10.1101/gad.271353.115
Strogantsev, R., and Ferguson-Smith, A. C. (2012). Proteins involved in establishment and maintenance of imprinted methylation marks. Briefings Funct. Genomics 11, 227–239. doi:10.1093/bfgp/els018
Strogantsev, R., Krueger, F., Yamazawa, K., Shi, H., Gould, P., Goldman-Roberts, M., et al. (2015). Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 16, 112. doi:10.1186/s13059-015-0672-7
Surani, M. A. H., Barton, S. C., and Norris, M. L. (1984). Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550. doi:10.1038/308548a0
Tada, T., Obata, Y., Tada, M., Goto, Y., Nakatsuji, N., Tan, S., et al. (2000). Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development 127, 3101–3105. doi:10.1242/dev.127.14.3101
Takagi, N., and Sasaki, M. (1975). Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642. doi:10.1038/256640a0
Takahashi, N., Coluccio, A., Thorball, C. W., Planet, E., Shi, H., Offner, S., et al. (2019). ZNF445 is a primary regulator of genomic imprinting. Genes and Dev. 33, 49–54. doi:10.1101/gad.320069.118
Tucci, V., Isles, A. R., Kelsey, G., Ferguson-Smith, A. C., Group, E. I., Tucci, V., et al. (2019). Genomic imprinting and physiological processes in mammals. Cell 176, 952–965. doi:10.1016/j.cell.2019.01.043
Uehara, R., Yeung, W. K. A., Toriyama, K., Ohishi, H., Kubo, N., Toh, H., et al. (2023). The DNMT3A ADD domain is required for efficient de novo DNA methylation and maternal imprinting in mouse oocytes. PLOS Genet. 19, e1010855. doi:10.1371/journal.pgen.1010855
Veselovska, L., Smallwood, S. A., Saadeh, H., Stewart, K. R., Krueger, F., Maupetit-Méhouas, S., et al. (2015). Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 16, 209. doi:10.1186/s13059-015-0769-z
Wang, Q., Chow, J., Hong, J., Smith, A. F., Moreno, C., Seaby, P., et al. (2011). Recent acquisition of imprinting at the rodent Sfmbt2 locus correlates with insertion of a large block of miRNAs. BMC Genom 12, 204. doi:10.1186/1471-2164-12-204
Wood, A. J., Roberts, R. G., Monk, D., Moore, G. E., Schulz, R., and Oakey, R. J. (2007). A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 3, e20. doi:10.1371/journal.pgen.0030020
Wu, J., Huang, B., Chen, H., Yin, Q., Liu, Y., Xiang, Y., et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657. doi:10.1038/nature18606
Xia, W., Xu, J., Yu, G., Yao, G., Xu, K., Ma, X., et al. (2019). Resetting histone modifications during human parental-to-zygotic transition. Science 365, 353–360. doi:10.1126/science.aaw5118
Xie, Z., Zhang, W., and Zhang, Y. (2022). Loss of Slc38a4 imprinting is a major cause of mouse placenta hyperplasia in somatic cell nuclear transferred embryos at late gestation. Cell Rep. 38, 110407. doi:10.1016/j.celrep.2022.110407
Xu, Q., Xiang, Y., Wang, Q., Wang, L., Brind’Amour, J., Bogutz, A. B., et al. (2019). SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat. Genet. 51, 844–856. doi:10.1038/s41588-019-0398-7
Yano, S., Ishiuchi, T., Abe, S., Namekawa, S. H., Huang, G., Ogawa, Y., et al. (2022). Histone H3K36me2 and H3K36me3 form a chromatin platform essential for DNMT3A-dependent DNA methylation in mouse oocytes. Nat. Commun. 13, 4440. doi:10.1038/s41467-022-32141-2
Yoh, S. M., Lucas, J. S., and Jones, K. A. (2008). The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes and Dev. 22, 3422–3434. doi:10.1101/gad.1720008
Yu, C., Cvetesic, N., Hisler, V., Gupta, K., Ye, T., Gazdag, E., et al. (2020). TBPL2/TFIIA complex establishes the maternal transcriptome through oocyte-specific promoter usage. Nat. Commun. 11, 6439. doi:10.1038/s41467-020-20239-4
Zeng, T.-B., Pierce, N., Liao, J., and Szabó, P. E. (2021). H3K9 methyltransferase EHMT2/G9a controls ERVK-driven noncanonical imprinted genes. Epigenomics-uk 13, 1299–1314. doi:10.2217/epi-2021-0168
Zhang, W., Chen, Z., Yin, Q., Zhang, D., Racowsky, C., and Zhang, Y. (2019). Maternal-biased H3K27me3 correlates with paternal-specific gene expression in the human morula. Gene Dev. 33, 382–387. doi:10.1101/gad.323105.118
Zhang, Y., Jurkowska, R., Soeroes, S., Rajavelu, A., Dhayalan, A., Bock, I., et al. (2010). Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 38, 4246–4253. doi:10.1093/nar/gkq147
Zheng, H., Huang, B., Zhang, B., Xiang, Y., Du, Z., Xu, Q., et al. (2016). Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079. doi:10.1016/j.molcel.2016.08.032
Zink, F., Magnusdottir, D. N., Magnusson, O. T., Walker, N. J., Morris, T. J., Sigurdsson, A., et al. (2018). Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet. 50, 1542–1552. doi:10.1038/s41588-018-0232-7
Keywords: genomic imprinting, DNA methylation, H3K27me3, endogenous retroviral elements, developmental epigenetics
Citation: Fang S, Chang K-W and Lefebvre L (2024) Roles of endogenous retroviral elements in the establishment and maintenance of imprinted gene expression. Front. Cell Dev. Biol. 12:1369751. doi: 10.3389/fcell.2024.1369751
Received: 12 January 2024; Accepted: 26 February 2024;
Published: 05 March 2024.
Edited by:
Mellissa Mann, University of Pittsburgh, United StatesReviewed by:
Courtney W. Hanna, University of Cambridge, United KingdomCopyright © 2024 Fang, Chang and Lefebvre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Louis Lefebvre, bG91aXMubGVmZWJ2cmVAdWJjLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.