
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 12 June 2024
Sec. Cancer Cell Biology
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1348894
This article is part of the Research TopiclncRNAs: Application in Immunotherapy, Radiotherapy, and ChemotherapyView all 9 articles
Long non-coding RNAs (lncRNAs) are a sort of transcripts that are more than 200 nucleotides in length. In recent years, many studies have revealed the modulatory role of lncRNAs in cancer. Typically, lncRNAs are linked to a variety of essential events, such as apoptosis, cellular proliferation, and the invasion of malignant cells. Simultaneously, autophagy, an essential intracellular degradation mechanism in eukaryotic cells, is activated to respond to multiple stressful circumstances, for example, nutrient scarcity, accumulation of abnormal proteins, and organelle damage. Autophagy plays both suppressive and promoting roles in cancer. Increasingly, studies have unveiled how dysregulated lncRNAs expression can disrupt autophagic balance, thereby contributing to cancer progression. Consequently, exploring the interplay between lncRNAs and autophagy holds promising implications for clinical research. In this manuscript, we methodically compiled the advances in the molecular mechanisms of lncRNAs and autophagy and briefly summarized the implications of the lncRNA-mediated autophagy axis.
As a heterogeneous group of non-protein coding transcripts, lncRNAs are not only poorly conserved but they can also controlling gene expression at different levels, such as the chromatin, transcriptional, and posttranscriptional levels (Liu et al., 2021). Historically, lncRNAs were dismissed as genomic “junk” and not accorded serious consideration (Statello et al., 2021). However, accumulating evidence now underscores that lncRNAs, in modulating transcription and translation, take a significant operational role. They act as sponges for microRNAs (miRNAs), bound to RNA-binding proteins (RBPs), function as scaffold for proteins, regulate transcription, and even serve as translation templates for peptides (Ransohoff et al., 2018; Ouyang et al., 2022). With increasing evidence pointing to their role in many diseases, especially cancer, lncRNAs are attracting more and more attention.
Autophagy is observed in nearly every type of eukaryotic cells and is a pervasive and highly conserved catabolic process (Zhou et al., 2022). This degradative process is central to cellular regulation, maintaining homeostasis by lysosomal breakdown of injured organelles and various proteins (Chen et al., 2021; Wang et al., 2021). To date, three forms of autophagy have been described: macroautophagy, microautophagy and chaperone-mediated autophagy, the latter of which is only found in mammalian cells (Ktistakis and Tooze, 2016; Xu et al., 2020). Under hypoxic, stressed and deprived conditions, phagophore begins to form and gradually matures into an autophagosome. It then fuses with a lysosome, resulting in the degradation of internal contents within the autolysosome and triggering macroautophagy (Jiang et al., 2021). Microautophagy involves the direct engulfment of small pieces of cytoplasm by the invagination of the lysosomal membrane, which is followed by lysis and subsequent degradation. Unlike the other types, chaperone-mediated autophagy does not entail membrane reorganization. It hinges on the recognition of substrate proteins containing KFERQ motifs, which then bind with cytosolic Hsc70 and cochaperones. This combination is translocated directly across the lysosomal membrane after binding to lysosomal Lamp-2A (Feng et al., 2014; Galluzzi and Green, 2019). Macroautophagy holds the utmost significance among autophagic pathways. For the purposes of the review, our focus will be on macroautophagy, hereafter referred to simply as autophagy.
Cancer is a major burden on socio-economic development and one of the world’s public health problems. In the US, there are projected to be 1,958,310 new cases of cancer and over 609,820 cancer-related deaths in 2023 (Siegel et al., 2023). While recent decades have witnessed remarkable progress to diagnose and treat cancer at an early stage, which has led to a significant reduction in both the number of new cases and the number of deaths, the decline in cancer mortality rates has plateaued worldwide since the 1990s. This stagnation is primarily attributed to the absence of groundbreaking therapies that promise improved prognoses for cancer patients in recent years (Sung et al., 2021). Consequently, there is an urgent requirement to discover novel mechanisms illustrating the etiology of cancer. Both lncRNAs and autophagy play integral roles in a wide array of biological activities and intricate signaling pathways. The existence of potential links between these two key regulatory mechanisms has already been established by mounting explorations (Bermúdez et al., 2019; Islam Khan et al., 2019). Therefore, summarizing the current knowledge on lncRNAs and autophagy is the aim of this review. We will discuss how lncRNAs induce and modulate autophagy in cancer and discuss the implications for clinical applications.
The truth is that less than 2 per cent of the human genome is made up of genes that code for proteins, with about 98 per cent of the rest being transcribed into RNA that does not code for proteins (Djebali et al., 2012; Lin et al., 2020). LncRNAs are mostly transcribed by RNA pol II, and sometimes by RNA pol III, and within the confines of some plant cells, by RNA pol IV and RNA pol V (Smolarz et al., 2021). Many lncRNAs, in addition to those derived from larger precursors, exhibit a conservative and stable protein secondary structure due to a 5′-end cap and 3′-end polyadenylation (e.g., intronic lncRNAs) (Nair et al., 2020; Smolarz et al., 2021). LncRNAs can be classified into five different groups according to the structure of the gene and the position of the gene relative to the protein-coding gene (Figure 1A): (1) sense lncRNAs or (2) antisense lncRNAs, which overlap with neighboring transcripts either in the same direction or in the opposite direction; (3) bidirectional lncRNAs, having their transcript start positions in common with the gene that codes for the protein on the opposite strand; (4) intronic lncRNAs, where the entire lncRNA transcript resides within a coding gene’s intron, and (5) intergenic lncRNAs, which are located in the interval of the genome between two genes (Lin, 2020; McCabe and Rasmussen, 2021).
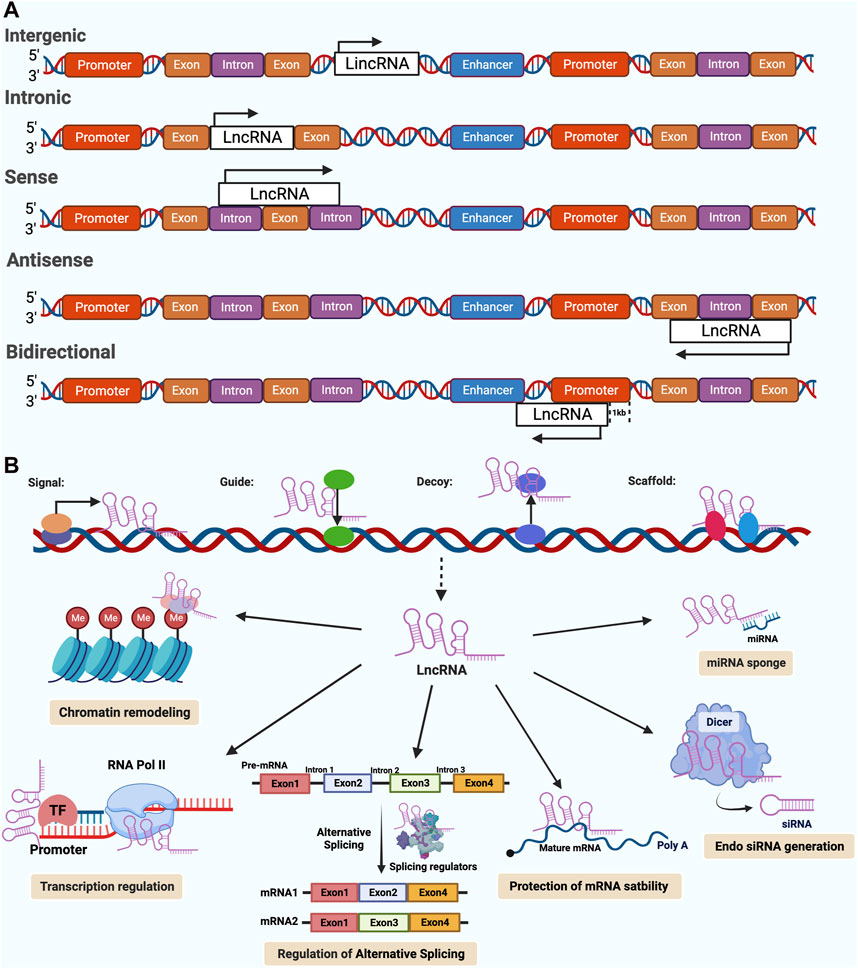
Figure 1. Classifications and functions of Long noncoding RNAs (lncRNAs) in cancer. [modified image from Abdelrahman M. Elsayed et al. (Elsayed et al., 2020)] (A) A schematic diagram showing the classification of lncRNAs according to their orientation and position, including intergenic, Intronic, sense, antisense, bidirectional lncRNAs. The arrow indicates the direction of transcription. (B) The biological functions of lncRNAs are generally categorized into four main archetypes of molecular mechanisms, including signals, guides, decoys, and scaffolds. Furthermore, lncRNAs have a variety of regulatory functions that are derived from these archetypes, such as; chromatin remodeling, regulation of transcription, alternative splicing and other post-transcriptional modifications, generation of endo siRNA, d protection of mRNA stability, miRNA sponges.
Since lncRNAs can be detected in the nucleus, cytoplasm or both, their functions are strongly dependent on their subcellular localization (Ransohoff et al., 2018). LncRNAs in the nucleus have a wide range of functions, including interacting with chromatin, regulating transcription and processing RNA. Conversely, lncRNAs in the cytoplasm may control the stability and translation of mRNAs and Impacts on cell signaling pathways (Chen et al., 2018). While the biological functions of lncRNAs are diverse, they tend to fall into four major categories according to their specific mechanisms of action (Figure 1B): signals, guides, decoys and scaffolds (Wang and Chang, 2011; Barangi et al., 2019). The signal lncRNAs are the ones that can be used as markers of notable biological events because they respond to certain stimulus. LncRNAs with guiding functions regulate target gene expression by binding to specific regulators, such as transcription factors and modifiers of chromatin, precise gene targeting for regulation in specific genomic regions (Kashi et al., 2016; Lin, 2020). In contrast to the guidance function, decoys are a class of lncRNAs that bind to or sequester miRNAs and negatively modulate the levels of specific genes, RNA-binding proteins and transcription factors (Wang and Chang, 2011). The most complex molecular archetype of lncRNAs is the scaffolds. They serve as operating workbenches upon which distinct effector molecules interact with each other, influencing their abilities to interact with various partners, ultimately resulting in either transcriptional repression or activation (Huang and Yu, 2015; Schmitz et al., 2016). Interestingly, lncRNAs are known to function in many different ways, and a single lncRNA may perform more than one archetypal function (Elsayed et al., 2020).
Inflammation is thought to play a major role in causing Cancer. Infection and chronic inflammation are responsible for about 25% of all cancers (Murata, 2018). In the 19th century, the German physician Rudolph Virchow first discovered and described the phenomenon of inflammatory cells infiltrating tumors (Rani et al., 2019). In the course of time, scientists discovered the existence of an inflammation-associated microenvironment consisting of cancer cells, immune cells, and several cytokines. These immune cell and cytokine abnormalities complicate the tumor microenvironment (TME), allowing inflammation to play a role in promoting or inhibiting tumor development (Hussain and Harris, 2007; Elinav et al., 2013). Tumor-induced inflammation is a dynamic process in which immune cells are infiltrated and activated. Accumulating evidence suggests that lncRNAs play an important role in this process. They are potent factors in the recruitment and activation of immune cells to regulate tumor development (Xu and Gewirtz, 2022).
In the past few years, with the continuous development of high-throughput sequencing technology, especially the groundbreaking lncRNA microarray and transcript sequencing (RNA-seq), scientists have been able to make enormous progress in the analysis of biomolecules. The latest statistics show that the number of identified human lncRNAs now exceeds 173,000 (Kopp and Mendell, 2018; Zhao et al., 2021). In the complex regulatory network of cancer, lncRNAs have been shown to play a critical role (Yan and Bu, 2021). Similar to other non-coding RNAs, lncRNAs can regulate gene expression through a variety of mechanisms, acting as either cancer-promoting or cancer-suppressing factors, which in turn can regulate the development and progression of cancer. lncRNAs can act as decoys for miRNAs, functioning as competitive endogenous RNAs (ceRNAs) that bind directly to miRNAs, preventing miRNAs from affecting downstream target mRNAs and thus maintaining their functional integrity (Wang et al., 2019). They are also able to influence the activation or repression status of target genes by fine-tuning the interaction between transcription factors and promoters. Furthermore, lncRNAs can act as scaffolding and RNA-binding proteins (RBPs) to directly participate post-transcriptional regulation, modulate protein-protein interactions and several related downstream signaling pathways (Bhat et al., 2016).
There has been evidence that autophagy plays a dual role in cancer, both promoting and inhibiting cancer growth and progression, since the earliest studies of cancer and autophagy (Amaravadi et al., 2019). Autophagy is one of the most important homeostatic mechanisms within the cell. In addition to responding to and alleviating various forms of cellular stress, such as starvation, organelle damage, and redox disturbances, it also contributes to cellular nutrient utilization and promotes metabolism (Debnath et al., 2023). For example, autophagy inhibits tumor growth through the removal of damaged mitochondria and the reduction of reactive oxygen species to inhibit glycolysis. Therefore, it is generally accepted that autophagy is degraded and recycled to inhibit tumor development (Russell and Guan, 2022). However, as cancer develops, the process of autophagy has been shown to be necessary to support uncontrolled growth and progressively increased metabolic activity of tumor cells, leading to tumor dependence on autophagy (Miller and Thorburn, 2021; Qiu et al., 2023). At this stage, aberrant autophagy may contribute to tumor cell proliferation and ongoing tumor progression by promoting cancer stem cell spreading (Ariosa et al., 2021a).
Studies of the BECN1 gene, which encodes beclin-1, provided the first evidence that autophagy plays a tumor suppressor role (Debnath et al., 2023). In the 1990s, Levine’s group identified BECN1 as a tumor suppressor, providing the first insight into autophagy’s role in cancer. Since then, a great deal of research has been undertaken on the role of autophagy in cancer, and it has been found that autophagy plays a cytoprotective role by maintaining cellular and genomic integrity during the early stages of tumor progression, thereby inhibiting tumor development (Ariosa et al., 2021a). Furthermore, ATG proteins have been reported as potential tumor suppressors. In experiments by Marsh et al. knockdown of Atg5 or Atg12 resulted in increased metastasis of cancer cells in a mouse model of breast cancer. This demonstrates a specific role for autophagy in suppressing cancer cell metastasis (Marsh et al., 2020). Eliminating the autophagy protein ATG4C, which is responsible for forming autophagosomes, also plays a role in suppressing tumors (Fu et al., 2019).
For autophagy, the process is complex and consists of several consecutive steps: (1) Initiation: the induction of autophagy; (2) Nucleation: the formation of the nucleus of the phagophore; (3) Elongation: during this stage, the phagophore expands, seals, and evolves into the autophagosome; (3) Fusion: autophagosome fusion with a lysosome; (4) Degradation: internal degradation of the material and recycling (Cao et al., 2021; Bai et al., 2022). Figure 2 shows the process and basic mechanism of autophagy.
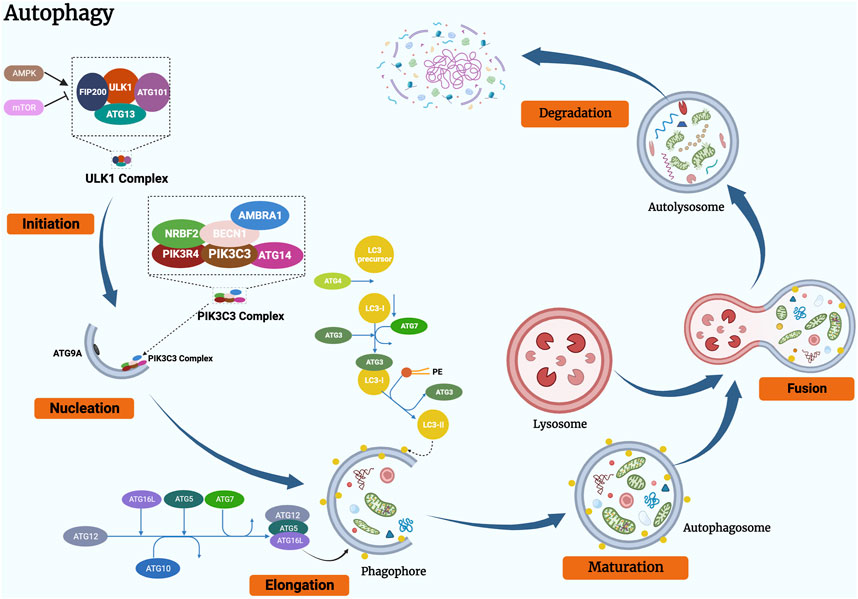
Figure 2. Schematic representation of the basic mechanism of autophagy. The process of autophagy can be divided into the following stages: (1) initiation, (2) vesicle nucleation, (3) vesicle elongation and maturation, (4) vesicle fusion, and (5) cargo degradation.
Autophagy is initiated under a variety of stress conditions (e.g., starvation, hypoxia and oxidative stress) by the release of signals that lead to the activation of the ULK1 complex, which consists of the central kinase protein ULK1, ATG13, FIP200 and ATG101 (Klionsky et al., 2021; C. W; Yun and Lee, 2018). AMPK and mTORC1 regulate autophagy induction (Ktistakis and Tooze, 2016; Han et al., 2022). Under nutrient-rich conditions, mTORC1 is a repressor of autophagy, which can bind ULK1 and avoid autophagy initiation. While AMPK acts as autophagy promoter that persists the balance between the production and consumption of adenosine triphosphate (ATP) (Ashrafizadeh et al., 2021).
Following the induction of autophagy, the ULK1 complex is phosphorylated and translocated into a specific region of the ER, where it facilitates the formation of the PtdIns3K complex, which consists of PIK3C3/VPS34, PIK3R4/VPS15, BECN1 (beclin 1), AMBRA1, ATG14, and NRBF2 (Wang et al., 2018; Kocaturk et al., 2019). Afterwards, the PtdIns3K complex cooperates with ATG9 (a multimembrane-spanning protein) containing vesicles, triggering nucleation of the phagophore and resulting in the phosphoinositide lipids bound to the membrane being phosphorylated, activating the local production of phosphatidylinositol-3-phosphate (PI3P) at the surface of the characteristic ER (called the omegasome) (Chao et al., 2020; Jing et al., 2020). On the omegasome, PI3P recognizes and attracts PI3P-binding proteins which mark the phagophore assembly sites (PAS), such as DFCP1 and WIPIs (here is WIPI2) (Glick et al., 2010; Smith and Macleod, 2019).
After autophagy is initiated, the phagophore continues to rely on two distinct conjugation systems for phagosomal expansion (Chen et al., 2019). The first is the ATG5 - ATG12 - ATG16L1 conjugative system. It consists of ATG12 (acts as a ubiquitin-like factor), ATG5 (acts as an ATG12 background), ATG16L1, ATG7 (acts as an E1 similar enzyme) and ATG10 (acts as an E2 similar enzyme) (Ariosa et al., 2021b). The second is the ATG8 family of proteins (ATG8s), which includes the MAP1LC3/LC3 subfamily and GABARAP subfamily (act as ubiquitin-like proteins), phosphatidylethanolamine (PE, act as substrate), ATG3 (act as a E2 similar carrier protein), ATG7 (act as a E1 similar enzyme), ATG12-ATG5-ATG16L1 complex (act as a E3 ligase), ATG4 (cysteine protease) (Liang et al., 1999; Zhao et al., 2021). At this stage, ATG12 is in the process of activation by ATG7, transferred by ATG10 and ultimately binds to ATG5 (Das et al., 2021). ATG12 and ATG5 subsequently interact with ATG16L1 to form a multimeric protein complex consisting of ATG12, ATG5 and ATG16 (Zhu et al., 2022). Recent studies indicate that ATG16L1 can directly bind WIPI2 on the omegasome, providing a membrane-binding platform for the ATG12-ATG5 conjugate and enhancing ATG3-mediated ATG8 family protein conjugation (J. Liang et al., 2021). LC3 is expressed as a full-length cytoplasmic protein in the majority of cell types. In such a conjugation, precursor LC3 is proteolytically cleaved by ATG4 (cysteine protease) to release a C-terminal glycine residue and to produce a cytoplasmically soluble free form (this form is termed LC3-I) (Xie et al., 2015; Li et al., 2021). Similar to the activation of ATG7 by ATG12, LC3-I was then activated in an ATP-dependent manner (Ohsumi, 2014). Activated LC3-I is delivered to ATG3 prior to covalent conjugation of the C-terminal glycine of LC3-I to PE lipid and formation of LC3-II (a membrane-bound, lipidated form of LC3), in which the ATG12, ATG5 and ATG16 protein complex functions as an E3 ligase to promote the process (Ashrafizadeh et al., 2021; C; Yun and Lee, 2018). The phagophore progressively elongates and becomes curved into a bowl-like structure which engulfs the cytoplasmic components of the cell, eventually closing to form a spherical autophagosome (Cocco et al., 2020). A key signature of autophagy is LC3-II, which marks the formation of autophagosomes (Ajoolabady et al., 2021). Another associated member of the ATG8 protein family, GABARAP, undergoes an analogous process and GABARAP-II localizes to autophagosomes together with LC3-II (Parzych and Klionsky, 2014). Research shows that LC3 proteins participate in vesicle elongation, while GABARAP is engaged in late autophagosome maturation (Weidberg et al., 2010).
ATG8s are not only critical for phagophore membrane expansion and closure, but also have a capacity that can integrate with components of LC3 interaction region (LIR) as well (Koustas et al., 2022). Furthermore, in selected autophagy, LC3 is mainly participate in sequestering specifically labelled cargo into autophagosomes through cargo receptors that contain the LIR (Parzych and Klionsky, 2014). The autophagosome membrane is sealed into a double-layered vesicle once the specifically labelled cargo has been chosen and bound to the membrane (de la Cruz-Ojeda et al., 2022).
The next stage of autophagy is the fusion of autophagosomes with lysosomes to form autolysosomes, after the phagophore membrane has been sealed and the autophagosome has matured (Glick et al., 2010; Parzych and Klionsky, 2014). Accumulating evidence suggests that the microtubule system and associated motor proteins contribute to the movement and traffic of autophagosomes to lysosomes. Autophagosomes and lysosomes are transported to the perinuclear region along the intracellular microtubule system by dynein-dependent mechanisms (C. Yun and Lee, 2018). In the perinuclear domain, SNARE proteins, the membrane-binding proteins and other fusion-associated protein families, such as LAMP-2 and RABs (RAB five and RAB 7), are involved in autolysosomal formation (Zhu et al., 2022). Specifically, the fusion requires the lysosome and autophagosome to be tethered to each other, which is normally controlled by the RAB7, RAB5 and LAMP-2 proteins (Rakesh, 1868; Jogalekar et al., 2021). SNARE proteins include STX17, SNAP29 and VAMP8. The HOPS complex, the best studied tethering protein, is also involved in this process (Rakesh et al., 2022). The HOPS have been identified to play a critical role in the fusion of autophagosomes with lysosomes by capturing Rab7-containing autophagosomes and binding with other adaptor proteins (Faruk et al., 2021; Rakesh et al., 2022).
Eventually, as the autolysosome forms, the lysosomal proteases break down the internal cargo and return nutrients (e.g., amino acids, fatty acids) to the cytosol for further use in different metabolic processes.
p62 was the first autophagy adaptor protein to be discovered in a mammalian organism. Shin, who discovered p62, named it Sequestosome 1 (SQSTM 1) because of its ability to form aggregates (Shin, 1998; Liu et al., 2016). However, it was not until Komatsu et al. reported a link between p62 and LC3, and found that p62 mediates the formation of protein aggregates for autophagic turnover, that the functional importance of p62 was appreciated. p62 is made up of 440 amino acids and has a multitude of structural domains with different functions. From the N-terminal to the C-segment, they are Phox-BEM1 structural domain (PB1), ZZ-type zinc finger structural domain, nuclear localization signal 1 (NLS1), tumor necrosis factor receptor-associated factor 6-binding (TRAF6-binding) TB domain, nuclear localization signal 1 (NLS2), export motif (NES), and LC3 interaction region (LIR), Keap1 interaction region (KIR) and ubiquitin-associated structural domain (UBA) (Liu et al., 2016; Cerda-Troncoso et al., 2020).
One of p62’s major roles in autophagy is to bind different types of ubiquitinated cargo, which are delivered to autophagosomes via its UBA domain, then to the lysosome via its LIR domain, and finally to the lysosome via its PB1 domain, leading to degradation (Shin et al., 2020; Kumar et al., 2022). In particular, the PB1 domain is able to polymerise with itself to transform into a dimeric form known as homo-oligomerisation. The dimeric form of p62, however, is essentially inactive with regard to autophagy. Despite this, the PB1 domain can also interact with the autophagy receptor NBR1 or other PB1-containing proteins in a process called hetero-oligomerisation. This promotes the polymerisation of the filamentous form of p62, giving the protein function. This structure is important for targeted delivery of ubiquitinated cargo to the autophagosome (Emanuele et al., 2020). There are also other structural domains in the p62 that play an important role in autophagy. For example, the ZZ-type zinc finger domain binds to hydrolysed protein cargo containing amino terminal arginine residues (Nt-Arg), interacting to promote autophagic degradation of the cargo (Cerda-Troncoso et al., 2020; Hennig et al., 2021).
The level of p62 expression in the cell is influenced by several factors. On the one hand it depends on transcriptional regulation, e.g., products of oxidative stress activation (Nrf2), the Ras/MAPK pathway and the JNK/c-Jun pathway all affect p62 transcription. Starvation and proteasome inhibitors also increase p62 transcription. On the other hand, because p62 is a substrate for autophagy activation, the expression level of p62 decreases as the degree of autophagic response increases (Moscat et al., 2016; Qian and Ding, 2023).
The involvement of lncRNAs in the regulation of p62-dependent autophagy has been demonstrated in many studies. New research has shown that the lncRNA CASC9 is upregulated in oral squamous cell carcinoma (OSCC), and knocking down CASC9 in OSCC cells significantly increases autophagy. There was also a significant decrease in the expression of P62 and other valuable biomarkers (Yang et al., 2019). In hepatocellular carcinoma (HCC) cells, lncRNA RP11-295G20.2 was significantly overexpressed and inhibited autophagy by targeting PTEN. Further studies demonstrated that the lncRNA RP11-295G20.2 binds directly to PTEN and promotes its interaction with the specific adaptor protein p62, which induces PTEN degradation via the autophagosome-lysosome pathway. This means that the lncRNA RP11-295G20.2, which mediates p62-dependent autophagy, plays a central role in the degradation of PTEN via the autophagosome-lysosome pathway (L. Liang et al., 2021). In a recent study, Hu and his team found that the lncRNA MITA1 induces autophagy in HCC827GR cells, promoting resistance to gefitinib. By overexpressing the lncRNA MITA1 in HCC827GR cells, they found that p62 levels decreased and the viability of HCC827GR cells improved subsequently. When HCC827-GR cells were treated with an autophagy inhibitor, lncRNA MITA1-mediated regulation of p62 expression was markedly abolished and the effects of lncRNA MITA1 on cell viability were ameliorated. Thus, it can be shown that the lncRNA MITA1 is able to promote resistance to gefitinib in HCC827GR cells through the induction of p62-dependent autophagy (Hu et al., 2021).
Many cancers have been reported to have abnormal expression of lncRNAs, and the association between lncRNAs and autophagy has been of particular interest in a variety of cancer types, such as lung cancer, gastric cancer, breast cancer and prostate cancer (Zhang et al., 2021; Zhang L. et al., 2022; Ma et al., 2022). In most of the studies presented, lncRNA regulation of autophagy was mainly mediated by miRNA sponging (ceRNA), RNA to RNA interaction, RNA to protein regulation or some other mechanism (de la Cruz-Ojeda et al., 2022). And these studies show that lncRNAs are involved in several stages of autophagy, from its initiation to its maturation. They mediate the initiation of autophagic phagocytosis by regulating ULK1, mTOR and Beclin-1, and the elongation of autophagic phagocytosis by regulating ATG3, ATG5, ATG4, ATG12 and ATG7. Hence, we have compiled the previous research on the relationships between lncRNA-mediated autophagy and associated phenotypes of cancer cells in Table 1 and Figure 3.
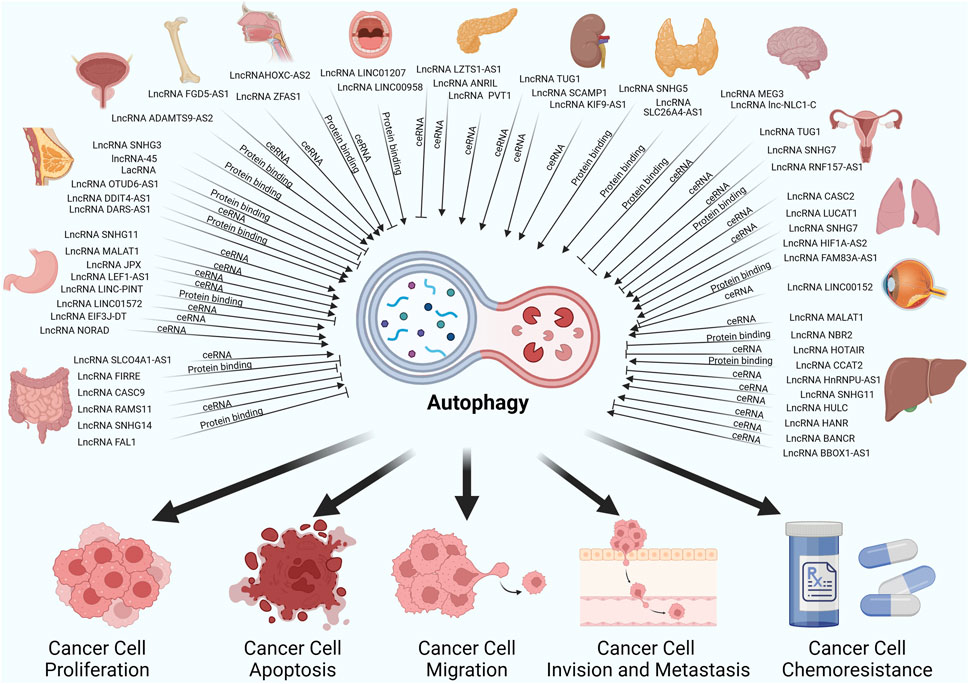
Figure 3. LncRNAs regulate autophagy through different mechanisms thereby affecting cancer cell-associated phenotypes in different cancers.
Firstly, proliferation is one of the most important malignant phenotypes of cancers, and apoptosis holds the same status in neoplastic development. While their functions are opposite and their interactions synergistically control the development of cancers (Evan and Vousden, 2001).
In digestive system cancers, the lncRNA SNHG11 is upregulated in gastric cancer (GC) and is associated with a poor prognosis of the patients, which post-transcriptionally upregulates ATG12 through miR-1276 to enhance autophagy and proliferation and further activate the Wnt/β-catenin signaling pathway (Wu et al., 2021). In a separate study, the lncRNA MALAT1 was shown to increase the autophagic capacity of GC cells by directly targeting miRNA-204 to increase the expression of LC3-II and mediate GC autophagy through the MALAT1/MiR-183/SIRT1 axis (Xu et al., 2021). LncRNA MALAT1 also exhibits different functions in hepatocellular carcinoma (HCC). The expression of the lncRNA MALAT1 was elevated in HCC tissues than in normal tissues. Silencing of MALAT1 promoted HCC autophagy by increasing LC3-II transformation and processing and suppressed HCC cell multiplication (Peng et al., 2020). In contrast, the lncRNA NBR2 acts as a tumor inhibitor in HCC, suppressing Beclin1 and autophagy via ERK/JNK pathways to limit HCC cell proliferation (Sheng et al., 2021). In colorectal cancer (CRC), lncRNA SLCO4A1-AS1 is an oncogenic factor that has a positive correlation with PARD3 and sponges miR-508-3p. SLCO4A1-AS1 promotes CRC cell proliferation and triggers autophagy through the miR-508-3p/PARD3 axis. However, this regulation was disrupted by treatment with 3-methyladenine (3-MA) (Wang and Jin, 2019). Another lncRNA among CRC called firre intergenic repeating RNA element (FIRRE), interacts directly with PTBP1 and enhances BECN1 mRNA stability, thus inducing the reduction of autophagy and promoting CRC cell proliferation (Wang et al., 2022). Significantly, among oral squamous cell carcinoma (OSCC) and cholangiocarcinoma (CCA), upregulated lncRNAs were found to suppress autophagy. In OSCC, high expression of LINC01207 was found in OSCC, and overexpression of LINC01207 promoted the proliferation of OSCC cells, but inhibited apoptosis and autophagy via the miR-1301-3p/LDHA axis (Lu et al., 2021). On the contrary, LINC00958 overexpression could reduce apoptosis and promote autophagy by upregulating the autophagy-related proteins Beclin-1 and Atg5, together with LC3-II/LC3-I ratio, via p53 mediated by SIRT1 (Jiang et al., 2021). Intriguingly, LINC00958 has another pathway to encourage OSCC cells in apoptosis, that is via LINC00958/miR-4306/GSDMD axis. In CCA, lncRNA HOTAIR significantly inhibits apoptotic and autophagic processes and promotes proliferation of CCA cells by targeting the miR-204-5p/HMGB1 axis (Lu et al., 2020). Pancreatic cancer (PANC) is one of the worst-prognostic malignancies, and the molecular mechanisms underlying how it progresses have not been fully elucidated. Wu’s team identified LZTS1-AS1, a highly expressed lncRNA, in PANC cells and tissues, and found that LZTS1-AS1 promotes PANC cell proliferation and inhibits apoptosis and autophagy through the miR-532/TWIST1 axis (Wu et al., 2023).
Among urinary system cancers, nearly 80% of all renal cell carcinomas (RCC) are diagnosed on pathology as clear cell renal cell carcinoma (ccRCC), and which account for 25% of cases, are at risk of developing metastases at an early stage. The group of Dong Lv learned that the high levels of the lncRNA TUG1 associated with ccRCC and confirmed that silencing the lncRNA TUG1 dramatically suppressed cell proliferation and promoted apoptosis, autophagy of ccRCC cells. This was thought to be mediated by the miR-31-5p/FLOT1 axis (Lv et al., 2020). LncRNA SCAMP1 is upregulated in RCC cells and tumor, regulating ZEB1/JUN and autophagy to promote oxidative stress-induced RCC in children through miR-429 (Shao et al., 2019). In bladder cancer (BLCA), the lncRNA autophagy network plays a critical role in BLCA progression. LncRNA ADAMTS9-AS2 is identified downregulated in BLCA and related to ADAMTS9, Zhang et al. showed that the lncRNA ADAMTS9-AS2 inhibits proliferation and elevates autophagy and apoptosis through the PI3K/AKT/mTOR pathway (Zhang et al., 2020).
Downregulation of the lncRNA CASC2 exacerbates NSCLC apoptosis and reduces ATG5-mediated autophagy via regulation of the miR-214/TRIM16 axis in A549 or H1299 NSCLC cells (Li et al., 2018). A reverse autophagy statue was observed in thyroid carcinoma (PTC), Overexpression of lncRNA SLC26A4-AS1 suppressed PTC cells proliferation and promoted autophagy through recruiting transcription factor ETS1 and increasing ITPR1 expression (Peng et al., 2021). Additionally, Qin et al. illuminated that lncRNA SNHG5 was stabilized by RBM47 and targeted to FOXO3, thereby inhibiting proliferation and activating autophagy in PTC cells via the RBM47/SNHG5/FOXO3 axis (Qin et al., 2022).
There is no doubt that the migration, invasion and metastasis of tumor cells are the key factors that lead to the deterioration and eventual death of patients suffering from solid tumors. Although researchers have thoroughly investigated various aspects of tumor growth, we still have little understanding of how lncRNAs and autophagy interact to influence tumor cell migration, invasion and metastasis, and further experimental studies are urgently needed to reveal their exact functions (Guo et al., 2023). In breast cancer (BC), Yu et al. explained how knocking down the lncRNA SNHG3 promotes autophagy by increasing autophagic vacuolization, which inhibits BC cells from migrating and invading (Yu et al., 2023). Analogously, lncRNA-45, a newly identified lncRNA that is transcribed by an internal region within the mTOR complex one gene, is the most upregulated lncRNA. It makes a contribution to BC cells invade and metastasize through activation of mTOR and inhibition of autophagy (Qiu et al., 2023). In addition, LacRNA (a novel LINC00478-associated cytoplasmic RNA) obviously blocked BC cell invasion and metastasis in vitro and in vivo via inhibiting the degradation of autophagy (Guo et al., 2023).
Hypopharyngeal carcinoma is the most aggressive form of squamous cell carcinoma of the head and neck. Xiang and his team found that the expression of lncRNA HOXC-AS2 and P62 protein in hypopharyngeal carcinoma tissues was significantly higher than that in normal hypopharyngeal tissues. Overexpressing lncRNA HOXC-AS2 could activate the NF-κB signaling pathway through binding to p62 protein, which suppressed the expression of Hmox1 protein, thereby inhibiting the autophagy of hypopharyngeal cancer cells and promoting their migration and invasion (Xiang et al., 2023). Among neurologic tumors, in neuroblastoma (NB), lncRNA MEG3 functions as an anti-tumor agent, and overexpressing MEG3 in NB cells decreased epithelial-mesenchymal transition invasion and metastasis via mTOR signaling and inhibited FOXO1-mediated autophagy (Ye et al., 2020). In retinoblastoma, the investigators observed high levels of LINC00152 expression, and silencing LINC00152 significantly reduced proliferation, invasion and autophagy, while reinforced apoptosis of retinoblastoma cells. Mechanistically, there was evidence that LINC00152 binds directly to miR-613 through ceRNA mechanism and targets YAP1 (Wang et al., 2020). In glioma, lnc-NLC1-C (narcolepsy candidate region one gene C) promotes glioma cell proliferation, migration and invasion and inhibits autophagy via lnc-NLC1-C/miR-383/PRDX-3 axis (Xu et al., 2021).
In lung cancer, the latest scientific findings indicate that the upregulation of lncRNA FAM83A-AS1 expression is not only closely related to the malignancy degree of the tumor, but also directly linked to the poorer quality of survival of patients. Specifically, the knockdown of FAM83A-AS1 gene showed a remarkable effect: it can effectively reduce the proliferation, migration and invasion ability of lung cancer cells. On a deeper level of analysis, the increase in phosphorylated AMPKα and ULK1 that was induced by the knockdown of FAM83A-AS1 revealed a possible molecular signaling pathway. This pathway is likely to involve the MET-AMPKα signaling pathway, which is thought to be an important switch in the control of cellular autophagy. Thus, to some extent, FAM83A-AS1 may contribute to cancer development by inhibiting autophagy through activation of the MET-AMPKα pathway by down-regulating the phosphorylation levels of AMPKα and ULK1. (Zhao et al., 2022). In HCC, Shi et al. revealed that lncRNA CCAT2 functions as an oncogene was upregulated in HCC tissues and cells. Further experiments showed that CCAT-2 was able to modulate miR-4496 and ELAVL-1, thereby inducing the autophagic process, which had the effect of increasing migration and invasion in vitro and in vivo (Shi et al., 2021). The lncRNA SNHG11/miR-184/AGO2 regulation axis is essential for the promotion of HCC cell proliferating, migrating, invading and autophagy (Huang et al., 2020). In contrast, the group of Li demonstrated that autophagy was shown to prevent invasion and migration of HCC cells. They found that low levels of lncRNA HnRNPU-AS1 positively correlated with bad prediction for HCC patients. Overexpressing HnRNPU-AS1 may constrain the proliferation, migration, and invasion while promoting autophagy in HCC cells via targeting the miR-556-3p and miR-580-3p/SOXS6 axis (Zhang K. et al., 2022). In GC, high lncRNA JPX expression in patients indicates bad prognosis, moreover, knocking down JPX inhibits GC cell activity, invasion and migration by sponging off miR-197, which modulates the downstream CXCR6 protein and promotes autophagy (Han and Liu, 2021). In addition, the lncRNA LEF1-AS1/miR-5100/DEK axis controls GC cell proliferation, invasion and metastasis by promoting autophagy and inhibiting apoptosis by way of the AMPK-mTOR pathway, according to the Zhang et al. study (Zhang et al., 2022).
Epithelial-mesenchymal transition (EMT) pathway takes an elemental role in cancer invasion and migration (Wang et al., 2021; Zhang et al., 2021). LncRNA CASC9 is significantly overexpressed in both the cell lines and tissues of the CRC. CASC9 silencing attenuates cell migration and induces autophagy through AKT/mTOR and EMT signaling pathways. In addition, onco-lncRNA RAMS11 promotes EMT and malignant phenotype of CRC cells by suppression of autophagy and apoptosis in an mTOR-dependent manner (Islam Khan and Law, 2021). A newly identified pseudogene, lnc-CTSLP8, in ovarian cancer which is obviously upregulated in metastatic tumor tissue in comparison to primary ovarian tumors. Mechanistically, lnc-CTSLP8 upregulates CTSL1 and sponges for miR-199a-5p, thereby increasing autophagy and EMT (Xu et al., 2021). In nasopharyngeal carcinoma (NPC), the lncRNA ZFAS1, whose RNA stability is enhanced by the m6A methyltransferase METTL3, promotes NPC cell proliferation, migration and tumor growth and regulates autophagy levels by modulating the miR-100-3p/ATG10 axis and through the PI3K/AKT pathway (Xu et al., 2023).
There have been multitudinous studies demonstrating the contribution of lncRNAs in the resistance of cancer to drugs and autophagy is increasingly recognized as a critical factor in tumor chemoresistance. LncRNAs can influence treatment-resistant phenotypes through regulation of autophagy, according to recent findings (Xu et al., 2020; Zhang and Lu, 2020; Entezari et al., 2022).
Since 1978, cisplatin (DDP) has been widely used as a first-choice chemotherapy drug to treat approximately half of all solid tumors, such as gastric, breast and lung cancer. The mechanisms that mediate the anti-tumor effects of DDP have been studied for decades, and the most important anti-tumor mechanism is DNA damage through the interaction with the purine bases of DNA (Ranasinghe et al., 2022; Xu and Gewirtz, 2022). However, DDP resistance limits the survival of patients, and a variety of researches reveal that lncRNA-mediated autophagy is one cause of DDP resistance (Xu and Gewirtz, 2022). The LINCRNA PINT/EZH2/ATG5 regulation axis in GC suppresses resistance to DDP by inhibiting the activation of autophagy (Zhang et al., 2022b). Additionally, silencing of LINC01572 inhibits autophagy and resistance to DDP through the miR-497-5p/ATG14 axis in GC cells (Song et al., 2020). In CRC, Han et al. performed that the lncRNA SNHG14/miR-186/ATG14 axis have an important impact on increasing autophagy and facilitating DDP resistance (Han et al., 2020). In NSCLC, the lncRNA LUCAT1 improves resistance to DDP chemotherapy and stimulates autophagy and metastasis of NSCLC cells via targeting the miR-514a-3p/ULK1 axis (Shen et al., 2020). In addition, the upregulated lncRNA SNHG7 promotes NSCLC progression and resistance to DDP by the induction of autophagy activity through the modulation of LC3B and BECN1 (She et al., 2023). In ovarian cancer, elimination of autophagy mediated by DIRAS3 and ULK1 via the lncRNA RNF157-AS1 reduced the resistance of ovarian cancer cell resistance to DDP (Yang et al., 2022). Oxaliplatin is also one of the platinum-based drugs that is commonly treated for cancer, and its resistance is also a major concern (Zhang et al., 2022a). LncRNA EIF3J-DT activates autophagy and contributes to the chemoresistance of oxaliplatin- and 5-Fu-treated cells via miR-188-3p/ATG14 axis among GC (Luo et al., 2021). Coincidentally, lnc-NORAD, which is triggered through both H3K27ac and CREBBP, increased the flux of autophagy in GC cells to repress oxidative stress-induced oxaliplatin resistance by the miR-433-3p/ATG5-ATG12 complex axis (W. J et al., 2021). The lncRNA FAL1, mainly derived from exosomal secretion by CAF, significantly inhibits autophagy induced by oxaliplatin and promotes oxaliplatin chemoresistance in CRC by acting as a scaffold for Beclin1 and TRIM3 to promote the polyubiquitylation of Beclin1 and its degradation (Zhu et al., 2023). In HCC, lncRNA HULC/miR-383-5p/VAMP2 axis promoted the protective autophagy and malignant progression of HCC cells and inhibited the chemosensitivity of oxaliplatin (Li et al., 2021).
5-Fluorouracil (5-FU) is one of the most commonly used chemotherapeutic drugs in the treatment of a wide range of malignancies, including gastrointestinal tumors. 5-FU is an anti-metabolic drug where the hydrogen at the C5 position is replaced by fluorine (Vodenkova et al., 2020). 5-FU interferes with DNA replication by inhibiting the intracellular activity of thymine nucleotide synthetase (TS), and also has some inhibitory effects on RNA synthesis (Ghafouri-Fard et al., 2021; Sethy and Kundu, 2021). LINC01871 was found to be expressed at low levels in CRC tissues and cell lines, and patients with low LINC01871 levels had significantly worse survival. LINC01871 can sponge miR-142-3p and regulate ZYG11B expression to induce autophagy, which increases CRC cell sensitivity to 5-FU and promotes CRC cell chemotherapy resistance (Duan et al., 2023). Similarly, knockdown of lncRNA NEAT1 significantly inhibited CRC cell proliferation, autophagy and enhanced 5-FU sensitivity via targeting miR-34a (Liu et al., 2020). In GC, lncRNA FEZF1-AS1 is upregulated in the tissues of chemotherapy-resistant gastric cancer. Downregulation of FEZF1-AS1 can directly modulate autophagy via ATG5, thereby enhancing multi-drug resistant and improving 5-FU sensitivity in GC cells (Gui et al., 2021).
Paclitaxel (PTX) is also a first-line chemotherapeutic agent for cancer that bonds to and stabilizes microtubules, leading to a disruption of the metaphase-to-anaphase junction during the mitosis process (Zhu and Chen, 2019). Recent evidence reveals a variety of mechanisms for PTX resistance, one of which is autophagic responses mediated by lncRNAs (Gu et al., 2020; Yu et al., 2022). In ovarian cancer, the lnc-TUG1/miRNA-29b-3p/Ago2 axis triggers autophagy and, as a result, leads to PTX resistance (Gu et al., 2020). In a similar investigation, researchers found that metformin could induce the sensitivity of paclitaxel in ovarian cancer by the regulation of lncRNA SNHG7/miRNA-3127-5p-mediated autophagy (Yu et al., 2022). In BC, oncogenic lnc-OTUD6B-AS1 drives resistance to paclitaxel and induces autophagy as well as DNA damage by regulating the miRNA-26a-5p/MTDH signaling pathway (Li et al., 2021). Intriguingly, the lnc-DDIT4-AS1 enhances autophagy and makes BC cells more sensitive to PTX by promoting the interacting DDIT4 mRNA with the AUF1 protein, resulting in inhibition of the mTOR pathway (Jiang et al., 2023).
Sorafenib, a novel multi-kinase inhibitor, is able to selectively disable the relevant kinases and abnormal signaling pathways. It is widely used in the treatment of RCC and HCC. In sorafenib-resistant HCC cells, Shi’s group discovered that the lnc-HANR was significantly hyper-expressed, which increases resistance to sorafenib via promoting autophagy through the axis of miRNA-29b/ATG9A (Shi et al., 2020). I In addition, the lnc-BANCR/miR-590-5P/OLR1 axis inhibited autophagy and promoted sorafenib responsiveness in HCC under the regulation of rutin, the major constituent of Potentilla discolor bunge (Zhou et al., 2021). A comparable study has been executed in HCC cells and proved that the lnc-BBOX1-AS1 enhances PHF8-driven autophagy and the resistance of HCC cells to the drug sorafenib by modulating the miRNA-361-3p/PHF8 (Tao et al., 2022). In RCC, lnc-KIF9-AS1 recruits and fixes miRNA -497-5p, regulates SMAD3, TGF-β and ATG9A-mediated autophagy and thus promotes the resistance to sorafenib (Jin et al., 2020).
Gemcitabine is a cytosine nucleotide analogue that is widely used as an anti-cancer drug to treat a number of conditions, especially PANC (Binenbaum et al., 2015). Wang’s research revealed that lnc-ANRIL is elevated in PANC tissue and that knockdown of ANRIL decreases chemoresistance to gemcitabine through the targeting of miRNA-181a/HMGB1-driven autophagy (Wang et al., 2021). Another oncogenic lnc-PVT1 increases chemoresistance to gemcitabine by sponging off miRNA-619-5p and through activation of the Pygo2/Wnt/β-catenin pathway and ATG14 mediated-autophagy pathway (Zhou et al., 2020). Interestingly, through the axis of miRNA-143/HIF-1α/VMP1, PVT1 knockdown reduces autophagic activity and increases gemcitabine sensitivity in PANC (Liu et al., 2021).
Doxorubicin (DOX) is an antibiotic chemotherapeutic agent which inhibits proliferation and induces apoptosis by blocking topoisomerase II activity and causing DNA breaks (Ashrafizaveh et al., 2021). lncRNA-mediated autophagy is closely in connection with DOX resistance in cancer cells, according to a growing number of studies (Guçlu et al., 2021; Fei et al., 2022). Among triple-negative breast cancer (TNBC), a new type of nanodrug delivery system based on CL4-modified exosomes was able to deliver the siRNA of lnc-DARS-AS1 to TNBC cells, and silencing of DARS-AS1 by this delivery system increased DOX sensitivity of BC cells by suppressing autophagy induced by the TGF-β/Smad3 axis (Liu et al., 2023). In lung cancer, knocking down the lncRNA HIF1A-AS2 made lung cancer cells more sensitive to DOX and reduced autophagy, but the detailed mechanism remained unclear (Guçlu et al., 2021). In osteosarcoma, the lnc-FGD5-AS1 is upregulated in osteosarcoma cells resistant to doxorubicin, and knockdown of its expression attenuates the chemoresistance to DOX by WNT5A-driven autophagy through miRNA-154-5p sponging (Fei et al., 2022).
Therefore, lncRNA-mediated autophagy can be expected to be a major contributor to overcoming chemoresistance in cancer cells. And, focusing on the lncRNA-autophagy axis has promise for accelerating the clinical translation of novel drugs.
To date, lncRNAs have emerged as a novel approach for investigating various facets of cancer, including detecting, diagnosing, responding to treatment and prognostic values. And the functional analysis of is especially important for understanding chemoresistance. Simultaneously, autophagy ubiquitously present in almost all eukaryotes, playing essential roles in the material homeostasis in cancer cells (Chang et al., 2021).
In this manuscript, we systematically explored the classifications and functions of lncRNAs, the essential cellular processes of autophagy, and the significance of lncRNA-mediated autophagy in the progression of various malignancies and chemotherapy resistance. We conclude that, lncRNAs regulate autophagy mechanisms primarily through two modes: ceRNAs and RBP interactions, as evidenced in the majority of studies. On one hand, lncRNAs modulate autophagy genes expression by binding to specific miRNAs, including proteins from the mTOR, ULK1 and ATG families. . . . . . . These genes are crucial for the activation of related pathways that influence the autophagy process. On the other hand, lncRNAs can bind directly to key proteins involved in autophagy initiation.
Notably, in a previous study, the findings of Liang et al. were quite remarkable. When they investigated the different mechanisms by which lncRNAs regulate autophagy, they found an interesting phenomenon: when lncRNAs work through non-ceRNA mechanisms, they tend to reduce the level of autophagy, but the opposite is often the case when lncRNAs work through ceRNA mechanisms (Yu et al., 2022). However, on the basis of recent studies, we found that lncRNAs also inhibit the level of autophagy through the ceRNA mechanism, such as HOTAIR in CCA, LZTS1 AS1 in PANC. Therefore, researchers urgently need to conduct further studies to better understand the complex and delicate mechanism by which lncRNAs and autophagy interact. These studies should focus on the identification and validation of specific lncRNAs that activate or inhibit the ceRNA machinery, as well as the exploration of the specific molecular details of how they affect autophagy. In addition, the deeper secrets behind the role of lncRNAs in regulating autophagy will be unraveled by combining multidisciplinary approaches such as molecular biology, biochemistry and genetics.
Furthermore, in our opinion, the lncRNA-mediated autophagy axis holds significant value for clinical applications. Primarily, given its deep involvement in the chemoresistance of cancers, targeting this axis could significantly improve chemotherapy efficacy. Secondly, considering the widespread participation of the lncRNA-mediated autophagy axis in anti-neoplastic activity of newly developing drugs, combining therapies that target this axis with these drugs may amplify therapeutic effects. Thus, extensive investigation is required to unravel the complicated interplay between lncRNAs and the complex regulatory autophagy process in order to identify both novel diagnostic biomarkers and potential therapeutic interventions.
Z-yL: Writing–original draft, Writing–review and editing. J-mT: Writing–review and editing. M-qY: Writing–review and editing. Z-hY: Writing–review and editing. J-zX: Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Key Project of Scientific Research of Jiangsu Commission of Health (ZDB2020026); Wuxi Taihu Lake Talent Plan, Team in Medical and Health Profession; Wuxi Medical Key Discipline Construction Project, Medical Development Discipline.
For their contributions to the publications cited in this review article, we thank the current and former members of our laboratories and collaborators. We apologize for not being able to cite all recent publications due to space limitations, as the fields of lncRNA and autophagy research are growing rapidly.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ajoolabady, A., Wang, S., Kroemer, G., Penninger, J. M., Uversky, V. N., Pratico, D., et al. (2021). Targeting autophagy in ischemic stroke: from molecular mechanisms to clinical therapeutics. Pharmacol. Ther. 225, 107848. doi:10.1016/j.pharmthera.2021.107848
Amaravadi, R. K., Kimmelman, A. C., and Debnath, J. (2019). Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 9, 1167–1181. doi:10.1158/2159-8290.CD-19-0292
Ariosa, A. R., Lahiri, V., Lei, Y., Yang, Y., Yin, Z., Zhang, Z., et al. (2021a). A perspective on the role of autophagy in cancer. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1867, 166262. doi:10.1016/j.bbadis.2021.166262
Ariosa, A. R., Lahiri, V., Lei, Y., Yang, Y., Yin, Z., Zhang, Z., et al. (2021b). A perspective on the role of autophagy in cancer. Biochim. Biophys. Acta Mol. Basis Dis. 1867, 166262. doi:10.1016/j.bbadis.2021.166262
Ashrafizadeh, M., Zarrabi, A., Orouei, S., Kiavash, H., Hakimi, A., Amirhossein, Z., et al. (2021). MicroRNA-mediated autophagy regulation in cancer therapy: the role in chemoresistance/chemosensitivity. Eur. J. Pharmacol. 892, 173660. doi:10.1016/j.ejphar.2020.173660
Ashrafizaveh, S., Ashrafizadeh, M., Zarrabi, A., Husmandi, K., Zabolian, A., Shahinozzaman, M., et al. (2021). Long non-coding RNAs in the doxorubicin resistance of cancer cells. Cancer Lett. 508, 104–114. doi:10.1016/j.canlet.2021.03.018
Bai, Z., Peng, Y., Ye, X., Liu, Z., Li, Y., and Ma, L. (2022). Autophagy and cancer treatment: four functional forms of autophagy and their therapeutic applications. J. Zhejiang Univ. Sci. B 23, 89–101. doi:10.1631/jzus.B2100804
Barangi, S., Hayes, A. W., Reiter, R., and Karimi, G. (2019). The therapeutic role of long non-coding RNAs in human diseases: a focus on the recent insights into autophagy. Pharmacol. Res. 142, 22–29. doi:10.1016/j.phrs.2019.02.010
Bermúdez, M., Aguilar-Medina, M., Lizárraga-Verdugo, E., Avendaño-Félix, M., Silva-Benítez, E., López-Camarillo, C., et al. (2019). LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front. Oncol. 9, 1008. doi:10.3389/fonc.2019.01008
Bhat, S. A., Ahmad, S. M., Mumtaz, P. T., Malik, A. A., Dar, M. A., Urwat, U., et al. (2016). Long non-coding RNAs: mechanism of action and functional utility. Non-coding RNA Res. 1, 43–50. doi:10.1016/j.ncrna.2016.11.002
Binenbaum, Y., Na'ara, S., and Gil, Z. (2015). Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist. Updat. Rev. Comment. Antimicrob. anticancer Chemother. 23, 55–68. doi:10.1016/j.drup.2015.10.002
Cao, W., Li, J., Yang, K., and Cao, D. (2021). An overview of autophagy: mechanism, regulation and research progress. Bull. Du. Cancer 108, 304–322. doi:10.1016/j.bulcan.2020.11.004
Cerda-Troncoso, C., Varas-Godoy, M., and Burgos, P. V. (2020). Pro-tumoral functions of autophagy receptors in the modulation of cancer progression. Front. Oncol. 10, 619727. doi:10.3389/fonc.2020.619727
Chang, C., Jensen, L. E., and Hurley, J. H. (2021). Autophagosome biogenesis comes out of the black box. Nat. Cell Biol. 23, 450–456. doi:10.1038/s41556-021-00669-y
Chao, X., Qian, H., Wang, S., Fulte, S., and Ding, W.-X. (2020). Autophagy and liver cancer. Clin. Mol. Hepatol. 26, 606–617. doi:10.3350/cmh.2020.0169
Chen, H.-T., Liu, H., Mao, M.-J., Tan, Y., Mo, X.-Q., Meng, X.-J., et al. (2019). Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Cancer 18, 101. doi:10.1186/s12943-019-1030-2
Chen, J.-F., Wu, P., Xia, R., Yang, J., Huo, X.-Y., Gu, D.-Y., et al. (2018). STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol. Cancer 17, 6. doi:10.1186/s12943-017-0756-y
Chen, L., He, M., Zhang, M., Sun, Q., Zeng, S., Zhao, H., et al. (2021). The Role of non-coding RNAs in colorectal cancer, with a focus on its autophagy. Pharmacol. Ther. 226, 107868. doi:10.1016/j.pharmthera.2021.107868
Cocco, S., Leone, A., Piezzo, M., Caputo, R., Di Lauro, V., Di Rella, F., et al. (2020). Targeting autophagy in breast cancer. IJMS 21, 7836. doi:10.3390/ijms21217836
Das, S., Shukla, N., Singh, S. S., Kushwaha, S., and Shrivastava, R. (2021). Mechanism of interaction between autophagy and apoptosis in cancer. Apoptosis 26, 512–533. doi:10.1007/s10495-021-01687-9
Debnath, J., Gammoh, N., and Ryan, K. M. (2023). Autophagy and autophagy-related pathways in cancer. Nat. Rev. Mol. Cell Biol. 24, 560–575. doi:10.1038/s41580-023-00585-z
de la Cruz-Ojeda, P., Flores-Campos, R., Navarro-Villarán, E., and Muntané, J. (2022). The role of non-coding RNAs in autophagy during carcinogenesis. Front. Cell Dev. Biol. 10, 799392. doi:10.3389/fcell.2022.799392
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi:10.1038/nature11233
Duan, B., Zhang, H., Zhu, Z., Yan, X., Ji, Z., and Li, J. (2023). LncRNA LINC01871 sponging miR-142-3p to modulate ZYG11B promotes the chemoresistance of colorectal cancer cells by inducing autophagy. Anti-cancer drugs 34, 827–836. doi:10.1097/CAD.0000000000001478
Elinav, E., Nowarski, R., Thaiss, C. A., Hu, B., Jin, C., and Flavell, R. A. (2013). Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer 13, 759–771. doi:10.1038/nrc3611
Elsayed, A. M., Amero, P., Salama, S. A., Abdelaziz, A. H., Lopez-Berestein, G., and Rodriguez-Aguayo, C. (2020). Back to the future: rethinking the great potential of lncRNAS for optimizing chemotherapeutic response in ovarian cancer. Cancers 12, 2406. doi:10.3390/cancers12092406
Emanuele, S., Lauricella, M., D’Anneo, A., Carlisi, D., De Blasio, A., Di Liberto, D., et al. (2020). p62: friend or foe? Evidences for OncoJanus and NeuroJanus roles. Int. J. Mol. Sci. 21, 5029. doi:10.3390/ijms21145029
Entezari, M., Taheriazam, A., Orouei, S., Fallah, S., Sanaei, A., Hejazi, E. S., et al. (2022). LncRNA-miRNA axis in tumor progression and therapy response: an emphasis on molecular interactions and therapeutic interventions. Biomed. Pharmacother. = Biomedecine Pharmacother. 154, 113609. doi:10.1016/j.biopha.2022.113609
Evan, G. I., and Vousden, K. H. (2001). Proliferation, cell cycle and apoptosis in cancer. Nature 411, 342–348. doi:10.1038/35077213
Faruk, M. O., Ichimura, Y., and Komatsu, M. (2021). Selective autophagy. Cancer Sci. 112, 3972–3978. doi:10.1111/cas.15112
Fei, D., Yuan, H., Zhao, M., and Zhao, D. (2022). LncRNA FGD5-AS1 potentiates autophagy-associated doxorubicin resistance by regulating the miR-154-5p/WNT5A axis in osteosarcoma. Cell Biol. Int. 46, 1937–1946. doi:10.1002/cbin.11889
Feng, Y., He, D., Yao, Z., and Klionsky, D. J. (2014). The machinery of macroautophagy. Cell Res. 24, 24–41. doi:10.1038/cr.2013.168
Fu, Y., Huang, Z., Hong, L., Lu, J. H., Feng, D., Yin, X. M., et al. (2019). Targeting ATG4 in cancer therapy. Cancers 11, 649. doi:10.3390/cancers11050649
Galluzzi, L., and Green, D. R. (2019). Autophagy-independent functions of the autophagy machinery. Cell 177, 1682–1699. doi:10.1016/j.cell.2019.05.026
Ghafouri-Fard, S., Abak, A., Tondro Anamag, F., Shoorei, H., Fattahi, F., Javadinia, S. A., et al. (2021). 5-Fluorouracil: a narrative review on the role of regulatory mechanisms in driving resistance to this chemotherapeutic agent. Front. Oncol. 11, 658636. doi:10.3389/fonc.2021.658636
Glick, D., Barth, S., and Macleod, K. F. (2010). Autophagy: cellular and molecular mechanisms. J. Pathol. 221, 3–12. doi:10.1002/path.2697
Gu, L., Li, Q., Liu, H., Lu, X., and Zhu, M. (2020). Long noncoding RNA TUG1 promotes autophagy-associated paclitaxel resistance by sponging miR-29b-3p in ovarian cancer cells. OncoTargets Ther. 13, 2007–2019. doi:10.2147/OTT.S240434
Guçlu, E., Eroğlu Güneş, C., Kurar, E., and Vural, H. (2021). Knockdown of lncRNA HIF1A-AS2 increases drug sensitivity of SCLC cells in association with autophagy. Med. Oncol. N. Lond. Engl. 38, 113. doi:10.1007/s12032-021-01562-2
Gui, Z., Zhao, Z., Sun, Q., Shao, G., Huang, J., Zhao, W., et al. (2021). LncRNA FEZF1-AS1 promotes multi-drug resistance of gastric cancer cells via upregulating ATG5. Front. Cell Dev. Biol. 9, 749129. doi:10.3389/fcell.2021.749129
Guo, R., Su, Y., Zhang, Q., Xiu, B., Huang, S., Chi, W., et al. (2023). LINC00478-derived novel cytoplasmic lncRNA LacRNA stabilizes PHB2 and suppresses breast cancer metastasis via repressing MYC targets. J. Transl. Med. 21, 120. doi:10.1186/s12967-023-03967-1
Han, X., and Liu, Z. (2021). Long non-coding RNA JPX promotes gastric cancer progression by regulating CXCR6 and autophagy via inhibiting miR-197. Mol. Med. Rep. 23, 60. doi:10.3892/mmr.2020.11698
Han, X., Mo, J., Yang, Y., Wang, Y., and Lu, H. (2022). Crucial roles of LncRNAs-mediated autophagy in breast cancer. Int. J. Med. Sci. 19, 1082–1092. doi:10.7150/ijms.72621
Han, Y., Zhou, S., Wang, X., Mao, E., and Huang, L. (2020). SNHG14 stimulates cell autophagy to facilitate cisplatin resistance of colorectal cancer by regulating miR-186/ATG14 axis. Biomed. Pharmacother. = Biomedecine Pharmacother. 121, 109580. doi:10.1016/j.biopha.2019.109580
Hennig, P., Fenini, G., Di Filippo, M., Karakaya, T., and Beer, H.-D. (2021). The pathways underlying the multiple roles of p62 in inflammation and cancer. Biomedicines 9, 707. doi:10.3390/biomedicines9070707
Hu, J., Dong, S.-W., Pei, Y., Wang, J., Zhang, J., and Wei, X.-P. (2021). LncRNA MITA1 promotes gefitinib resistance by inducing autophagy in lung cancer cells. Biochem. Biophys. Res. Commun. 551, 21–26. doi:10.1016/j.bbrc.2021.02.130
Huang, W., Huang, F., Lei, Z., and Luo, H. (2020). LncRNA SNHG11 promotes proliferation, migration, apoptosis, and autophagy by regulating hsa-miR-184/AGO2 in HCC. OncoTargets Ther. 13, 413–421. doi:10.2147/OTT.S237161
Huang, Y.-K., and Yu, J.-C. (2015). Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: an update and review. World J. Gastroenterol. 21, 9863–9886. doi:10.3748/wjg.v21.i34.9863
Hussain, S. P., and Harris, C. C. (2007). Inflammation and cancer: an ancient link with novel potentials. Int. J. cancer 121, 2373–2380. doi:10.1002/ijc.23173
Islam Khan, M. Z., and Law, H. K. W. (2021). RAMS11 promotes CRC through mTOR-dependent inhibition of autophagy, suppression of apoptosis, and promotion of epithelial-mesenchymal transition. Cancer Cell Int. 21, 321. doi:10.1186/s12935-021-02023-6
Islam Khan, M. Z., Tam, S. Y., and Law, H. K. W. (2019). Autophagy-modulating long non-coding RNAs (LncRNAs) and their molecular events in cancer. Front. Genet. 9, 750. doi:10.3389/fgene.2018.00750
Jiang, L., Ge, W., Cui, Y., and Wang, X. (2021). The regulation of long non-coding RNA 00958 (LINC00958) for oral squamous cell carcinoma (OSCC) cells death through absent in melanoma 2 (AIM2) depending on microRNA-4306 and Sirtuin1 (SIRT1) in vitro. Bioengineered 12, 5085–5098. doi:10.1080/21655979.2021.1955561
Jiang, N., Zhang, X., Gu, X., Li, X., and Shang, L. (2021). Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 7, 30. doi:10.1038/s41420-021-00407-1
Jiang, T., Zhu, J., Jiang, S., Chen, Z., Xu, P., Gong, R., et al. (2023). Targeting lncRNA DDIT4-AS1 sensitizes triple negative breast cancer to chemotherapy via suppressing of autophagy. Adv. Sci. (Weinheim, Baden-Wurttemberg, Ger.) 10, e2207257. doi:10.1002/advs.202207257
Jin, Y., Huang, R., Xia, Y., Huang, C., Qiu, F., Pu, J., et al. (2020). Long noncoding RNA KIF9-AS1 regulates transforming growth factor-β and autophagy signaling to enhance renal cell carcinoma chemoresistance via microRNA-497-5p. DNA Cell Biol. 39, 1096–1103. doi:10.1089/dna.2020.5453
Jing, Y., Liang, W., Liu, J., Zhang, L., Wei, J., Yang, J., et al. (2020). Autophagy-mediating microRNAs in cancer chemoresistance. Cell Biol. Toxicol. 36, 517–536. doi:10.1007/s10565-020-09553-1
Jogalekar, M. P., Veerabathini, A., and Gangadaran, P. (2021). Recent developments in autophagy-targeted therapies in cancer. Exp. Biol. Med. (Maywood) 246, 207–212. doi:10.1177/1535370220966545
Kashi, K., Henderson, L., Bonetti, A., and Carninci, P. (2016). Discovery and functional analysis of lncRNAs: methodologies to investigate an uncharacterized transcriptome. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1859, 3–15. doi:10.1016/j.bbagrm.2015.10.010
Klionsky, D. J., Petroni, G., Amaravadi, R. K., Baehrecke, E. H., Ballabio, A., Boya, P., et al. (2021). Autophagy in major human diseases. EMBO J. 40, e108863. doi:10.15252/embj.2021108863
Kocaturk, N. M., Akkoc, Y., Kig, C., Bayraktar, O., Gozuacik, D., and Kutlu, O. (2019). Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 134, 116–137. doi:10.1016/j.ejps.2019.04.011
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi:10.1016/j.cell.2018.01.011
Koustas, E., Trifylli, E.-M., Sarantis, P., Papadopoulos, N., Papanikolopoulos, K., Aloizos, G., et al. (2022). The emerging role of MicroRNAs and autophagy mechanism in pancreatic cancer progression: future therapeutic approaches. Genes 13, 1868. doi:10.3390/genes13101868
Ktistakis, N. T., and Tooze, S. A. (2016). Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 26, 624–635. doi:10.1016/j.tcb.2016.03.006
Kumar, A. V., Mills, J., and Lapierre, L. R. (2022). Selective autophagy receptor p62/SQSTM1, a pivotal player in stress and aging. Front. Cell Dev. Biol. 10, 793328. doi:10.3389/fcell.2022.793328
Li, J., Chen, X., Kang, R., Zeh, H., Klionsky, D. J., and Tang, D. (2021). Regulation and function of autophagy in pancreatic cancer. Autophagy 17, 3275–3296. doi:10.1080/15548627.2020.1847462
Li, P., Li, Y., and Ma, L. (2021). Long noncoding RNA highly upregulated in liver cancer promotes the progression of hepatocellular carcinoma and attenuates the chemosensitivity of oxaliplatin by regulating miR-383-5p/vesicle-associated membrane protein-2 axis. Pharmacol. Res. Perspect. 9, e00815. doi:10.1002/prp2.815
Li, P.-P., Li, R.-G., Huang, Y.-Q., Lu, J.-P., Zhang, W.-J., and Wang, Z.-Y. (2021). LncRNA OTUD6B-AS1 promotes paclitaxel resistance in triple negative breast cancer by regulation of miR-26a-5p/MTDH pathway-mediated autophagy and genomic instability. Aging 13, 24171–24191. doi:10.18632/aging.203672
Li, Q., Chen, K., Dong, R., and Lu, H. (2018). LncRNA CASC2 inhibits autophagy and promotes apoptosis in non-small cell lung cancer cells via regulating the miR-214/TRIM16 axis. RSC Adv. 8, 40846–40855. doi:10.1039/c8ra09573f
Liang, J., Zhang, L., and Cheng, W. (2021). Non-coding RNA-mediated autophagy in cancer: a protumor or antitumor factor? Biochimica Biophysica Acta (BBA) - Rev. Cancer 1876, 188642. doi:10.1016/j.bbcan.2021.188642
Liang, L., Huan, L., Wang, J., Wu, Y., Huang, S., and He, X. (2021). LncRNA RP11-295G20.2 regulates hepatocellular carcinoma cell growth and autophagy by targeting PTEN to lysosomal degradation. Cell Discov. 7, 118. doi:10.1038/s41421-021-00339-1
Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., et al. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676. doi:10.1038/45257
Lin, W., Zhou, Q., Wang, C.-Q., Zhu, L., Bi, C., Zhang, S., et al. (2020). LncRNAs regulate metabolism in cancer. Int. J. Biol. Sci. 16, 1194–1206. doi:10.7150/ijbs.40769
Lin, Y.-H. (2020). Crosstalk of lncRNA and cellular metabolism and their regulatory mechanism in cancer. Int. J. Mol. Sci. 21, 2947. doi:10.3390/ijms21082947
Liu, F., Ai, F. Y., Zhang, D. C., Tian, L., Yang, Z. Y., and Liu, S. J. (2020). LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 9, 1079–1091. doi:10.1002/cam4.2746
Liu, S. J., Dang, H. X., Lim, D. A., Feng, F. Y., and Maher, C. A. (2021). Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 21, 446–460. doi:10.1038/s41568-021-00353-1
Liu, W. J., Ye, L., Huang, W. F., Guo, L. J., Xu, Z. G., Wu, H. L., et al. (2016). p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 21, 29. doi:10.1186/s11658-016-0031-z
Liu, X., Zhang, G., Yu, T., Liu, J., Chai, X., Yin, D., et al. (2023). CL4-modified exosomes deliver lncRNA DARS-AS1 siRNA to suppress triple-negative breast cancer progression and attenuate doxorubicin resistance by inhibiting autophagy. Int. J. Biol. Macromol. 250, 126147. doi:10.1016/j.ijbiomac.2023.126147
Liu, Y. F., Luo, D., Li, X., Li, Z. Q., Yu, X., and Zhu, H. W. (2021). PVT1 knockdown inhibits autophagy and improves gemcitabine sensitivity by regulating the MiR-143/HIF-1α/VMP1 Axis in pancreatic cancer. Pancreas 50, 227–234. doi:10.1097/MPA.0000000000001747
Lu, M., Qin, X., Zhou, Y., Li, G., Liu, Z., Yue, H., et al. (2020). LncRNA HOTAIR suppresses cell apoptosis, autophagy and induces cell proliferation in cholangiocarcinoma by modulating the miR-204-5p/HMGB1 axis. Biomed. Pharmacother. 130, 110566. doi:10.1016/j.biopha.2020.110566
Lu, X., Chen, L., Li, Y., Huang, R., Meng, X., and Sun, F. (2021). Long non-coding RNA LINC01207 promotes cell proliferation and migration but suppresses apoptosis and autophagy in oral squamous cell carcinoma by the microRNA-1301-3p/lactate dehydrogenase isoform A axis. Bioengineered 12, 7780–7793. doi:10.1080/21655979.2021.1972784
Luo, Y., Zheng, S., Wu, Q., Wu, J., Zhou, R., Wang, C., et al. (2021). Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 17, 4083–4101. doi:10.1080/15548627.2021.1901204
Lv, D., Xiang, Y., Yang, Q., Yao, J., and Dong, Q. (2020). Long non-coding RNA TUG1 promotes cell proliferation and inhibits cell apoptosis, autophagy in clear cell renal cell carcinoma via MiR-31-5p/FLOT1 Axis. OncoTargets Ther. 13, 5857–5868. doi:10.2147/OTT.S254634
Ma, X., Ren, H., Zhang, Y., Wang, B., and Ma, H. (2022). LncRNA RHPN1-AS1 inhibition induces autophagy and apoptosis in prostate cancer cells via the miR-7-5p/EGFR/PI3K/AKT/mTOR signaling pathway. Environ. Toxicol. 37, 3013–3027. doi:10.1002/tox.23656
Marsh, T., Kenific, C. M., Suresh, D., Gonzalez, H., Shamir, E. R., Mei, W., et al. (2020). Autophagic degradation of NBR1 restricts metastatic outgrowth during mammary tumor progression. Dev. Cell 52, 591–604. doi:10.1016/j.devcel.2020.01.025
McCabe, E. M., and Rasmussen, T. P. (2021). lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Seminars Cancer Biol. 75, 38–48. doi:10.1016/j.semcancer.2020.12.012
Miller, D. R., and Thorburn, A. (2021). Autophagy and organelle homeostasis in cancer. Dev. Cell 56, 906–918. doi:10.1016/j.devcel.2021.02.010
Moscat, J., Karin, M., and Diaz-Meco, M. T. (2016). p62 in cancer: signaling adaptor beyond autophagy. Cell 167, 606–609. doi:10.1016/j.cell.2016.09.030
Murata, M. (2018). Inflammation and cancer. Environ. health Prev. Med. 23, 50. doi:10.1186/s12199-018-0740-1
Nair, L., Chung, H., and Basu, U. (2020). Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat. Rev. Mol. Cell Biol. 21, 123–136. doi:10.1038/s41580-019-0209-0
Ohsumi, Y. (2014). Historical landmarks of autophagy research. Cell Res. 24, 9–23. doi:10.1038/cr.2013.169
Ouyang, J., Zhong, Y., Zhang, Y., Yang, L., Wu, P., Hou, X., et al. (2022). Long non-coding RNAs are involved in alternative splicing and promote cancer progression. Br. J. Cancer 126, 1113–1124. doi:10.1038/s41416-021-01600-w
Parzych, K. R., and Klionsky, D. J. (2014). An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal 20, 460–473. doi:10.1089/ars.2013.5371
Peng, D., Li, W., Zhang, B., and Liu, X. (2021). Overexpression of lncRNA SLC26A4-AS1 inhibits papillary thyroid carcinoma progression through recruiting ETS1 to promote ITPR1-mediated autophagy. J. Cell. Mol. Med. 25, 8148–8158. doi:10.1111/jcmm.16545
Peng, N., He, J., Li, J., Huang, H., Huang, W., Liao, Y., et al. (2020). Long noncoding RNA MALAT1 inhibits the apoptosis and autophagy of hepatocellular carcinoma cell by targeting the microRNA-146a/PI3K/Akt/mTOR axis. Cancer Cell Int. 20, 165. doi:10.1186/s12935-020-01231-w
Qian, H., and Ding, W.-X. (2023). SQSTM1/p62 and hepatic mallory-denk body formation in alcohol-associated liver disease. Am. J. Pathol. 193, 1415–1426. doi:10.1016/j.ajpath.2023.02.015
Qin, Y., Sun, W., Wang, Z., Dong, W., He, L., Zhang, T., et al. (2022). RBM47/SNHG5/FOXO3 axis activates autophagy and inhibits cell proliferation in papillary thyroid carcinoma. Cell death Dis. 13, 270. doi:10.1038/s41419-022-04728-6
Qiu, J., Guo, Y., Wang, S., Ren, Q., Dong, Z., Gao, M., et al. (2023). Newly identified lncRNA-45 promotes breast cancer metastasis through activating the mTOR signaling pathway. Biochem. biophysical Res. Commun. 640, 40–49. doi:10.1016/j.bbrc.2022.11.099
Rakesh, R., PriyaDharshini, L. C., Sakthivel, K. M., and Rasmi, R. R. (2022). Role and regulation of autophagy in cancer. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1868, 166400. doi:10.1016/j.bbadis.2022.166400
Ranasinghe, R., Mathai, M. L., and Zulli, A. (2022). Cisplatin for cancer therapy and overcoming chemoresistance. Heliyon 8, e10608. doi:10.1016/j.heliyon.2022.e10608
Rani, A., Dasgupta, P., and Murphy, J. J. (2019). Prostate cancer: the role of inflammation and chemokines. Am. J. pathology 189, 2119–2137. doi:10.1016/j.ajpath.2019.07.007
Ransohoff, J. D., Wei, Y., and Khavari, P. A. (2018). The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157. doi:10.1038/nrm.2017.104
Russell, R. C., and Guan, K. L. (2022). The multifaceted role of autophagy in cancer. EMBO J. 41, e110031. doi:10.15252/embj.2021110031
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 73, 2491–2509. doi:10.1007/s00018-016-2174-5
Sethy, C., and Kundu, C. N. (2021). 5-Fluorouracil (5-FU) resistance and the new strategy to enhance the sensitivity against cancer: implication of DNA repair inhibition. Biomed. Pharmacother. = Biomedecine Pharmacother. 137, 111285. doi:10.1016/j.biopha.2021.111285
Shao, Q., Wang, Q., and Wang, J. (2019). LncRNA SCAMP1 regulates ZEB1/JUN and autophagy to promote pediatric renal cell carcinoma under oxidative stress via miR-429. Biomed. Pharmacother. = Biomedecine Pharmacother. 120, 109460. doi:10.1016/j.biopha.2019.109460
She, K., He, S., Lu, X., Yu, S., Li, M., Xiong, W., et al. (2023). LncRNA SNHG7 promotes non-small cell lung cancer progression and cisplatin resistance by inducing autophagic activity. J. Thorac. Dis. 15, 155–167. doi:10.21037/jtd-22-1826
Shen, Q., Xu, Z., and Xu, S. (2020). Long non-coding RNA LUCAT1 contributes to cisplatin resistance by regulating the miR-514a-3p/ULK1 axis in human non-small cell lung cancer. Int. J. Oncol. 57, 967–979. doi:10.3892/ijo.2020.5106
Sheng, J.-Q., Wang, M.-R., Fang, D., Liu, L., Huang, W.-J., Tian, D.-A., et al. (2021). LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. Biomed. Pharmacother. 133, 111023. doi:10.1016/j.biopha.2020.111023
Shi, J., Guo, C., and Ma, J. (2021). CCAT2 enhances autophagy-related invasion and metastasis via regulating miR-4496 and ELAVL1 in hepatocellular carcinoma. J. Cell. Mol. Med. 25, 8985–8996. doi:10.1111/jcmm.16859
Shi, Y., Yang, X., Xue, X., Sun, D., Cai, P., Song, Q., et al. (2020). HANR enhances autophagy-associated sorafenib resistance through miR-29b/atg9a Axis in hepatocellular carcinoma. OncoTargets Ther. 13, 2127–2137. doi:10.2147/OTT.S229913
Shin, J. (1998). P62 and the sequestosome, a novel mechanism for protein metabolism. Archives pharmacal Res. 21, 629–633. doi:10.1007/BF02976748
Shin, W. H., Park, J. H., and Chung, K. C. (2020). The central regulator p62 between ubiquitin proteasome system and autophagy and its role in the mitophagy and Parkinson’s disease. BMB Rep. 53, 56–63. doi:10.5483/BMBRep.2020.53.1.283
Siegel, R. L., Miller, K. D., Wagle, N. S., and Jemal, A. (2023). Cancer statistics, 2023. Cancer J. Clin. 73, 17–48. doi:10.3322/caac.21763
Smith, A. G., and Macleod, K. F. (2019). Autophagy, cancer stem cells and drug resistance. J. Pathol. 247, 708–718. doi:10.1002/path.5222
Smolarz, B., Zadrożna-Nowak, A., and Romanowicz, H. (2021). The role of lncRNA in the development of tumors, including breast cancer. IJMS 22, 8427. doi:10.3390/ijms22168427
Song, Z., Jia, N., Li, W., and Zhang, X. Y. (2020). LINC01572 regulates cisplatin resistance in gastric cancer cells by mediating miR-497-5p. OncoTargets Ther. 13, 10877–10887. doi:10.2147/OTT.S267915
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tao, H., Zhang, Y., Li, J., Liu, J., Yuan, T., Wang, W., et al. (2022). Oncogenic lncRNA BBOX1-AS1 promotes PHF8-mediated autophagy and elicits sorafenib resistance in hepatocellular carcinoma. Mol. Ther. oncolytics 28, 88–103. doi:10.1016/j.omto.2022.12.005
Vodenkova, S., Buchler, T., Cervena, K., Veskrnova, V., Vodicka, P., and Vymetalkova, V. (2020). 5-fluorouracil and other fluoropyrimidines in colorectal cancer: past, present and future. Pharmacol. Ther. 206, 107447. doi:10.1016/j.pharmthera.2019.107447
Wang, J., Sun, Y., Zhang, X., Cai, H., Zhang, C., Qu, H., et al. (2021). Oxidative stress activates NORAD expression by H3K27ac and promotes oxaliplatin resistance in gastric cancer by enhancing autophagy flux via targeting the miR-433-3p. Cell death Dis. 12, 90. doi:10.1038/s41419-020-03368-y
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi:10.1016/j.molcel.2011.08.018
Wang, L., Bi, R., Li, L., Zhou, K., and Yin, H. (2021). lncRNA ANRIL aggravates the chemoresistance of pancreatic cancer cells to gemcitabine by targeting inhibition of miR-181a and targeting HMGB1-induced autophagy. Aging 13, 19272–19281. doi:10.18632/aging.203251
Wang, L., Cho, K. B., Li, Y., Tao, G., Xie, Z., and Guo, B. (2019). Long noncoding RNA (lncRNA)-Mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 20, 5758. doi:10.3390/ijms20225758
Wang, P., Shao, B.-Z., Deng, Z., Chen, S., Yue, Z., and Miao, C.-Y. (2018). Autophagy in ischemic stroke. Prog. Neurobiol. 163–164, 98–117. doi:10.1016/j.pneurobio.2018.01.001
Wang, Y., Li, Z., Xu, S., Li, W., Chen, M., Jiang, M., et al. (2022). LncRNA FIRRE functions as a tumor promoter by interaction with PTBP1 to stabilize BECN1 mRNA and facilitate autophagy. Cell Death Dis. 13, 98. doi:10.1038/s41419-022-04509-1
Wang, Y., Qin, C., Yang, G., Zhao, B., and Wang, W. (2021). The role of autophagy in pancreatic cancer progression. Biochimica Biophysica Acta (BBA) - Rev. Cancer 1876, 188592. doi:10.1016/j.bbcan.2021.188592
Wang, Y., Xin, D., and Zhou, L. (2020). LncRNA LINC00152 increases the aggressiveness of human retinoblastoma and enhances carboplatin and adriamycin resistance by regulating MiR-613/yes-associated protein 1 (YAP1) Axis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 26, e920886. doi:10.12659/MSM.920886
Wang, Z., and Jin, J. (2019). LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging 11, 4876–4889. doi:10.18632/aging.102081
Weidberg, H., Shvets, E., Shpilka, T., Shimron, F., Shinder, V., and Elazar, Z. (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802. doi:10.1038/emboj.2010.74
Wu, H., Li, A., Zheng, Q., Gu, J., and Zhou, W. (2023). LncRNA LZTS1-AS1 induces proliferation, metastasis and inhibits autophagy of pancreatic cancer cells through the miR-532/TWIST1 signaling pathway. Cancer Cell Int. 23, 130. doi:10.1186/s12935-023-02979-7
Wu, Q., Ma, J., Wei, J., Meng, W., Wang, Y., and Shi, M. (2021). lncRNA SNHG11 promotes gastric cancer progression by activating the wnt/β-catenin pathway and oncogenic autophagy. Mol. Ther. 29, 1258–1278. doi:10.1016/j.ymthe.2020.10.011
Xiang, Y., Huang, G., Wang, J., and Hua, Q. (2023). lncRNA HOXC-AS2 promotes the progression of hypopharyngeal cancer by binding to the P62 protein mediating the autophagy process. Aging 15, 12476–12496. doi:10.18632/aging.205192
Xie, Y., Kang, R., Sun, X., Zhong, M., Huang, J., Klionsky, D. J., et al. (2015). Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11, 28–45. doi:10.4161/15548627.2014.984267
Xu, J., and Gewirtz, D. A. (2022). Is autophagy always a barrier to cisplatin therapy? Biomolecules 12, 463. doi:10.3390/biom12030463
Xu, J. L., Yuan, L., Tang, Y. C., Xu, Z. Y., Xu, H. D., Cheng, X. D., et al. (2020). The role of autophagy in gastric cancer chemoresistance: friend or foe? Front. Cell Dev. Biol. 8, 621428. doi:10.3389/fcell.2020.621428
Xu, J.-L., Yuan, L., Tang, Y.-C., Xu, Z.-Y., Xu, H.-D., Cheng, X.-D., et al. (2020). The role of autophagy in gastric cancer chemoresistance: friend or foe? Front. Cell Dev. Biol. 8, 621428. doi:10.3389/fcell.2020.621428
Xu, P., Xu, S., Pan, H., Dai, C., Xu, Y., Wang, L., et al. (2023). Differential effects of the LncRNA RNF157-AS1 on epithelial ovarian cancer cells through suppression of DIRAS3- and ULK1-mediated autophagy. Cell death Dis. 14, 140. doi:10.1038/s41419-023-05668-5
Xu, W., Ding, M., Wang, B., Cai, Y., Guo, C., and Yuan, C. (2021). Molecular mechanism of the canonical oncogenic lncRNA MALAT1 in gastric cancer. Curr. Med. Chem. 28, 8800–8809. doi:10.2174/0929867328666210521213352
Xu, Z., Chen, Q., Zeng, X., Li, M., and Liao, J. (2021). lnc-NLC1-C inhibits migration, invasion and autophagy of glioma cells by targeting miR-383 and regulating PRDX-3 expression. Oncol. Lett. 22, 640. doi:10.3892/ol.2021.12901
Yan, H., and Bu, P. (2021). Non-coding RNA in cancer. Essays Biochem. 65, 625–639. doi:10.1042/EBC20200032
Yang, P., Ding, J., Bian, Y., Ma, Z., Wang, K., and Li, J. (2022). Long non-coding RNAs and cancer mechanisms: immune cells and inflammatory cytokines in the tumor microenvironment. Med. Oncol. N. Lond. Engl. 39, 108. doi:10.1007/s12032-022-01680-5
Yang, Y., Chen, D., Liu, H., and Yang, K. (2019). Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 10, 41. doi:10.1038/s41419-018-1280-8
Ye, M., Lu, H., Tang, W., Jing, T., Chen, S., Wei, M., et al. (2020). Downregulation of MEG3 promotes neuroblastoma development through FOXO1-mediated autophagy and mTOR-mediated epithelial-mesenchymal transition. Int. J. Biol. Sci. 16, 3050–3061. doi:10.7150/ijbs.48126
Yu, H., Chen, Y., Lang, L., Liao, D., Liu, S., Yu, T., et al. (2023). BMP9 promotes autophagy and inhibits migration and invasion in breast cancer cells through the c-Myc/SNHG3/mTOR signaling axis. Tissue and Cell 82, 102073. doi:10.1016/j.tice.2023.102073
Yu, Z., Wang, Y., Wang, B., and Zhai, J. (2022). Metformin affects paclitaxel sensitivity of ovarian cancer cells through autophagy mediated by long noncoding RNASNHG7/miR-3127-5p Axis. Cancer biotherapy Radiopharm. 37, 792–801. doi:10.1089/cbr.2019.3390
Yun, C. W., and Lee, S. H. (2018). The roles of autophagy in cancer. Int. J. Mol. Sci. 19, 3466. doi:10.3390/ijms19113466
Zhang, C., Kang, T., Wang, X., Wang, J., Liu, L., Zhang, J., et al. (2022b). LINC-PINT suppresses cisplatin resistance in gastric cancer by inhibiting autophagy activation via epigenetic silencing of ATG5 by EZH2. Front. Pharmacol. 13, 968223. doi:10.3389/fphar.2022.968223
Zhang, C., Xu, C., Gao, X., and Yao, Q. (2022a). Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 12, 2115–2132. doi:10.7150/thno.69424
Zhang, F., Wang, H., Yu, J., Yao, X., Yang, S., Li, W., et al. (2021). LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol. Cancer 20, 6. doi:10.1186/s12943-020-01299-y
Zhang, H., and Lu, B. (2020). The roles of ceRNAs-mediated autophagy in cancer chemoresistance and metastasis. Cancers 12, 2926. doi:10.3390/cancers12102926
Zhang, H., Wang, J., Wang, Y., Li, J., Zhao, L., Zhang, T., et al. (2022). Long non-coding LEF1-AS1 sponge miR-5100 regulates apoptosis and autophagy in gastric cancer cells via the miR-5100/DEK/AMPK-mTOR Axis. Int. J. Mol. Sci. 23, 4787. doi:10.3390/ijms23094787
Zhang, H. G., Wang, F. J., Wang, Y., Zhao, Z. X., and Qiao, P. F. (2021). lncRNA GAS5 inhibits malignant progression by regulating macroautophagy and forms a negative feedback regulatory loop with the miR-34a/mTOR/SIRT1 pathway in colorectal cancer. Oncol. Rep. 45, 202–216. doi:10.3892/or.2020.7825
Zhang, K., Chen, J., Li, C., Yuan, Y., Fang, S., Liu, W., et al. (2022d). Exosome-mediated transfer of SNHG7 enhances docetaxel resistance in lung adenocarcinoma. Cancer Lett. 526, 142–154. doi:10.1016/j.canlet.2021.10.029
Zhang, L., Zhao, Y., Guan, H., and Zhang, D. (2022c). HnRNPU-AS1 inhibits the proliferation, migration and invasion of HCC cells and induces autophagy through miR-556-3p/miR-580-3p/SOCS6 axis. Cancer biomarkers Sect. A Dis. markers 34, 443–457. doi:10.3233/CBM-210261
Zhang, Z., Jia, J. P., Zhang, Y. J., Liu, G., Zhou, F., and Zhang, B. C. (2020). Long noncoding RNA ADAMTS9-AS2 inhibits the proliferation, migration, and invasion in bladder tumor cells. OncoTargets Ther. 13, 7089–7100. doi:10.2147/OTT.S245826
Zhao, H., Wang, Y., Wu, X., Zeng, X., Lin, B., Hu, S., et al. (2022). FAM83A antisense RNA 1 (FAM83A-AS1) silencing impairs cell proliferation and induces autophagy via MET-AMPKɑ signaling in lung adenocarcinoma. Bioengineered 13, 13312–13327. doi:10.1080/21655979.2022.2081457
Zhao, L., Wang, J., Li, Y., Song, T., Wu, Y., Fang, S., et al. (2021). NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic acids Res. 49, D165–D171. doi:10.1093/nar/gkaa1046
Zhao, Y. G., Codogno, P., and Zhang, H. (2021). Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 22, 733–750. doi:10.1038/s41580-021-00392-4
Zhou, C., Yi, C., Yi, Y., Qin, W., Yan, Y., Dong, X., et al. (2020). LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. Cancer 19, 118. doi:10.1186/s12943-020-01237-y
Zhou, M., Zhang, G., Hu, J., Zhu, Y., Lan, H., Shen, X., et al. (2021). Rutin attenuates sorafenib-induced chemoresistance and autophagy in hepatocellular carcinoma by regulating BANCR/miRNA-590-5P/OLR1 Axis. Int. J. Biol. Sci. 17, 3595–3607. doi:10.7150/ijbs.62471
Zhou, X., Lin, J., Wang, F., Chen, X., Zhang, Y., Hu, Z., et al. (2022). Circular RNA-regulated autophagy is involved in cancer progression. Front. Cell Dev. Biol. 10, 961983. doi:10.3389/fcell.2022.961983
Zhu, H., Wang, W., and Li, Y. (2022). Molecular mechanism and regulation of autophagy and its potential role in epilepsy. Cells 11, 2621. doi:10.3390/cells11172621
Zhu, L., and Chen, L. (2019). Progress in research on paclitaxel and tumor immunotherapy. Cell. Mol. Biol. Lett. 24, 40. doi:10.1186/s11658-019-0164-y
Keywords: lncRNA, cancer, autophagy, tumor progression, chemoresistance
Citation: Liu Z-y, Tang J-m, Yang M-q, Yang Z-h and Xia J-z (2024) The role of LncRNA-mediated autophagy in cancer progression. Front. Cell Dev. Biol. 12:1348894. doi: 10.3389/fcell.2024.1348894
Received: 03 December 2023; Accepted: 24 May 2024;
Published: 12 June 2024.
Edited by:
Wenjie Shi, Otto von Guericke University Magdeburg, GermanyReviewed by:
Suchandrima Saha, Stony Brook Medicine, United StatesCopyright © 2024 Liu, Tang, Yang, Yang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia-zeng Xia, eGp6X3d1eGlAYWx1bW5pLnNqdHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.