- 1Department of Biochemistry, School of Medicine, University of Washington, Seattle, WA, United States
- 2Institute for Stem Cell and Regenerative Medicine, School of Medicine, University of Washington, Seattle, WA, United States
- 3Department of Bioengineering, College of Engineering, University of Washington, Seattle, WA, United States
- 4Institute for Protein Design, University of Washington, Seattle, WA, United States
- 5Department of Comparative Medicine, School of Medicine, University of Washington, Seattle, WA, United States
Tissue repair is significantly compromised in the aging human body resulting in critical disease conditions (such as myocardial infarction or Alzheimer’s disease) and imposing a tremendous burden on global health. Reprogramming approaches (partial or direct reprogramming) are considered fruitful in addressing this unmet medical need. However, the efficacy, cellular maturity and specific targeting are still major challenges of direct reprogramming. Here we describe novel approaches in direct reprogramming that address these challenges. Extracellular signaling pathways (Receptor tyrosine kinases, RTK and Receptor Serine/Theronine Kinase, RSTK) and epigenetic marks remain central in rewiring the cellular program to determine the cell fate. We propose that modern protein design technologies (AI-designed minibinders regulating RTKs/RSTK, epigenetic enzymes, or pioneer factors) have potential to solve the aforementioned challenges. An efficient transdifferentiation/direct reprogramming may in the future provide molecular strategies to collectively reduce aging, fibrosis, and degenerative diseases.
1 Introduction
While the concept that mature cell states are stable holds the key for homeostasis of an organism, the long-held believe was that this state cannot ever be reversed. This fallacy has gradually broken down, most notably by 2012 Nobel-winning studies by John Gurdon and Shinya Yamanaka. In 1962 Prof. Gurdon showed that in specific conditions, the nucleus of a differentiated cell is capable of de-differentiating into the pluripotent stage. This revolutionary finding changed modern biology and marks the initiation of reprogramming field. Later studies revealed the reversion of epigenetic marks as the key mechanism of the process. Gurdon’s nuclear transfer work led to animal cloning from tadpoles to Dolly the sheep, to genetically modified pigs and cows, etc. Yamanaka’s work then identified the molecules capable of reprogramming adult cells into a pluripotent stage (induced pluripotent stem cells, iPSC) (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Takahashi and Yamanaka, 2016). The Yamanaka factors (OCT3/4, SOX2, KLF4, c-MyC) are now widely used not only for reprogramming but also for partial reprogramming that leads to rejuvenation of tissues (Ocampo et al., 2016; Browder et al., 2022).
Yet another kind of reprogramming was emerging from the basic science field, now dubbed direct reprogramming, or transdifferentiation (we use the terms interchangeably from here on). During transdifferentiation a differentiated cell changes its fate to another, more desired differentiated cell type, without entering a pluripotent stage. The first identified transcription factor capable of directly reprogramming fibroblasts to skeletal muscles was MyoD (Davis et al., 1987; Weintraub et al., 1989). Many other lineage-specific transcription factors capable of transdifferentiating a target cell have since been identified (Wang et al., 2021).
While transdifferentiation can be induced on demand using transcription factor overexpression, some endogenous transdifferentiation have been observed in vivo as well; from jellyfish to mammals, transdifferentiation has shown to play an important role in normal animal adaptation. Some of the naturally occurring transdifferentiation processes are: immune cell transdifferentiation in vertebrates and Drosophila melanogaster (Csordás et al., 2021), alpha to beta-cell transdifferentiation in pancreatic islets (Thorel et al., 2010) and the white adipocyte transdifferentiation to brown adipocytes due to cold induced transcriptional and epigenetic modifications (Roh et al., 2018).
Whether induced or endogenous process, in general, a pioneer factors (PF) act as the first responders in direct reprogramming by binding and opening closed chromatin. Transdifferentiation epigenetic factors are then recruited, a necessary step for maintaining the open chromatin state to activate transcription of key lineage-specifying genes. In general, pioneer factors have the capacity to bind a multitude of chromatin locations and thereby in a regulated manner affect an entire suite of genes destined to activate a particular cell fate in a regulated manner. Recent data suggest that pioneer factors may be more general than we have previously appreciated (Wang et al., 2022a). Hence today, it is not clear if each transdifferentiation lineage is regulated by a specific pioneer factor, or if a universal PF for transdifferentiation (capable of initiating multitude of direct lineage reversions) is still to be identified.
In general, transdifferentiation studies have unveiled the opportunities and offer applications in regenerative therapies, such as cell replacement therapy or immunotherapy (Wang et al., 2021). The key question, and the topic of this review is to identify new, feasible methods to induce specific, high efficiency and targeted transdifferentiation. These newest technologies may lead to the next-generation direct reprogramming that due to increased accuracy and efficiency may become invaluable for disease modeling, and for future therapeutics.
2 Pioneer factors in transdifferentiation
An exogenous expression of master regulators, lineage specific transcription factors (TF) has been one of the most reliable methods of transdifferentiation. For example, direct reprogramming of mouse fibroblast to myoblast was originally achieved by the exogenous expression of MyoD (Davis et al., 1987; Tapscott et al., 1988; Weintraub et al., 1991). Furthermore, a fusion of transcriptional activation domain VP-64 to N-terminus of wild-type MyoD increased the transdifferentiation efficiency in human fibroblasts (Kabadi et al., 2015). A combination of three transcription factors Gata4, Mef2c, and Tbx5 (GMT) induced transdifferentiation of mouse fibroblasts into functional cardiomyocytes in vitro and in vivo upon transplantation (Ieda et al., 2010; Qian et al., 2012; Song et al., 2012). For human fibroblasts, addition of MESP1 and MYOCD to the GMT cocktail was required for the cardiomyocyte conversion (Wada et al., 2013). The first direct neuronal reprogramming (mouse astrocytes to neurons, iN) was induced with the expression of either Pax6, Neurog2 or Ascl1 (Heins et al., 2002; Berninger et al., 2007). To convert both mouse and human fibroblasts into iNs required the use of three TFs: Brn2, Ascl1 and Mytl1 (BAM) (Vierbuchen et al., 2010; Pfisterer et al., 2011a; Pfisterer et al., 2011b). In all of the examples above, the key transcription factors acted by binding and opening the chromatin of the critical genes and were coined pioneer factors (PF) due to this capacity. Among the neural transdifferentiation, Ascl1 was shown to act as a pioneer factor, binding and opening chromatin of neural genes, allowing their expression (Wapinski et al., 2017; Wang et al., 2022a), while Brn2 and Mtl1 were used as secondary TFs binding to the newly opened regions (Vierbuchen et al., 2010; Ambasudhan et al., 2011; Mall and Wernig, 2017; Wapinski et al., 2017). Presently, a multitude of pioneer factors with their companioning TFs have been identified in transdifferentiation processes (Figure 1). However, despite of the master transcription factor overexpression the reprogramming efficiency has not been sufficient for efficient chronic disease modeling, or clinical trials. Eventually, epigenetic abnormalities, metabolic maturation, aging and inflammatory response are some of the major roadblocks of direct reprogramming today that need to be addressed.
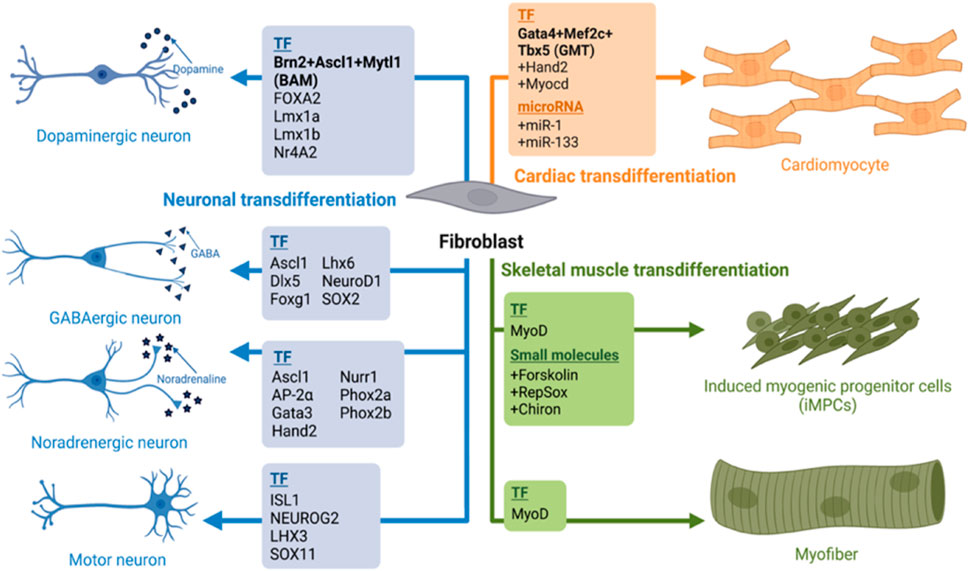
FIGURE 1. A summary of transdifferentiation fibroblast into skeletal, cardiac muscles and neurons. We summarize here various cocktails of transcription factors and their combination with small molecules or micoRNAs their lineage specific trans differentiation such as Dopaminergic neurons (Vierbuchen et al., 2010; Pfisterer et al., 2011a; Pfisterer et al., 2011b), GABAminergic (Lee et al., 2020; Bruzelius et al., 2021); Noradregenic neuron (Ainsworth et al., 2018); Myofibre (Davis et al., 1987; Tapscott et al., 1988; Weintraub et al., 1991); iMPCs (Qabrati et al., 2023) and Cardiomyocyte (Ieda et al., 2010; Qian et al., 2012; Song et al., 2012; Wada et al., 2013). Image created by Biorender.
3 Targeting signaling pathways augments transdifferentiation
Modulating signaling pathways has been another strategy for enhancing efficiency during transdifferentiation. Co-culturing mouse primary fibroblasts from various tissue origins with the C2C12 myoblasts in differentiation media, induced myogenic reprogramming in the fibroblasts (Iberite et al., 2022). This emphasized the importance of extrinsic signaling in cellular reprogramming. Inhibition of pathways such as TGF-β/activin pathway, BMP4 pathway, TNF-α and activation of canonical WNT signaling, IGF signalling, Follistatin pathway, GDF11 signaling, hGH pathway have shown to improve skeletal muscles trans differentiation (Boularaoui et al., 2018). Specifically, MyoD-induced myogenic transdifferentiation can be enhanced by IGF activation or BMP4 inhibition (Cacchiarelli et al., 2018). MyoD-E47 heterodimers can be inhibited by Id proteins that are downstream of BMP4 activation (Lassar et al., 1991). It is therefore plausible that BMP4 inhibition is beneficial for skeletal transdifferentiation via induced MyoD activity. Further, activation or inhibition of pathways such as FGF, WNT, NOTCH and IGF have increased the efficiency of cardiac transdifferentiation, as well as the maturity. BMPR/TGFbR inhibition has shown to be critical for guiding fibroblasts to the iN fate (Ladewig et al., 2012; Lu et al., 2013; Mertens et al., 2015; Pfisterer et al., 2016; Kim et al., 2018; Yang et al., 2019; Liu and Wang, 2020; Mall and Wernig, 2017; Bruzelius et al., 2021). Increasing activity of Wnt is also one of the most common strategies to improve the direct conversion to iNs. Indeed, Wnt activation, through the inhibition of GSK3β (CHIR99021) promotes iN generation (Dai et al., 2015; Mertens et al., 2015; Mall and Wernig, 2017; Kim et al., 2018; Bruzelius et al., 2021), presumably through activating neuron specific transcription factors (such as Ngn1, NeuroD and Brn3a). In addition to BMPR/TGFbR inhibition and Wnt activation, activity of the sonic hedgehog (SHH) pathway is also critical in neuron specification both from iPSC and from fibroblasts through direct conversion (Kriks et al., 2011; Arenas et al., 2015; Qin et al., 2020). Small molecules targeting specific signaling pathways can promote successful cardiac transdifferentiation through FGFR and VEGFR activation (Muraoka et al., 2019), or inhibition of TGF -
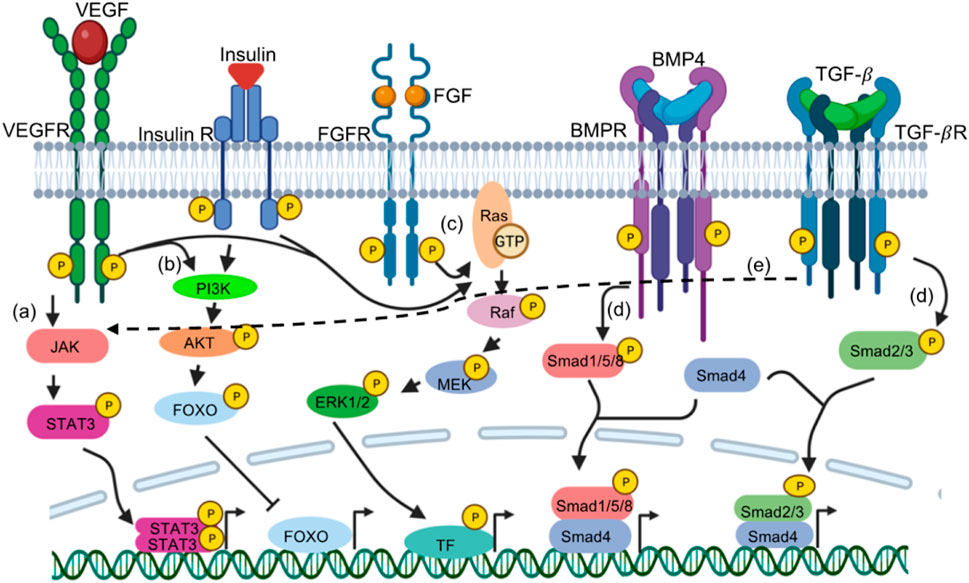
FIGURE 2. Signaling cascades augments transdifferentiation through gene expression. Various receptor tyrosine kinases (RTKs) or receptor Serine/Threonine Kinase (RSTKs) have been targeted in various direct reprogramming. RTKs such as VEGFR, FGFR, Insulin receptors acts through (A) JAK-STAT signalling, activated Janus Kinase downstream of the receptor phosphorylates STAT3. STAT3 dimers dependent gene expression is initiated. (C) MAPK pathway- RasGTP results in further phosphorylation of Raf, MEK and ERK1/2; phosphorylated ERK1/2 phosphorylate transcription factors (TF such as JUN,FOS, MYC, etc.) that activates their target genes. (B) PI3K pathway Activated PI3K downstream of the receptor phphorylates AKT. pAKT phosphorylation repress transcription factors such as FOXO (shown as example), BAD, TSC2, p27 while activates other transcription factors such as MDM2 and eNOS. RSTKs such as BMPR and TGF-
4 Correct epigenetic modifications are critical in transdifferentiation
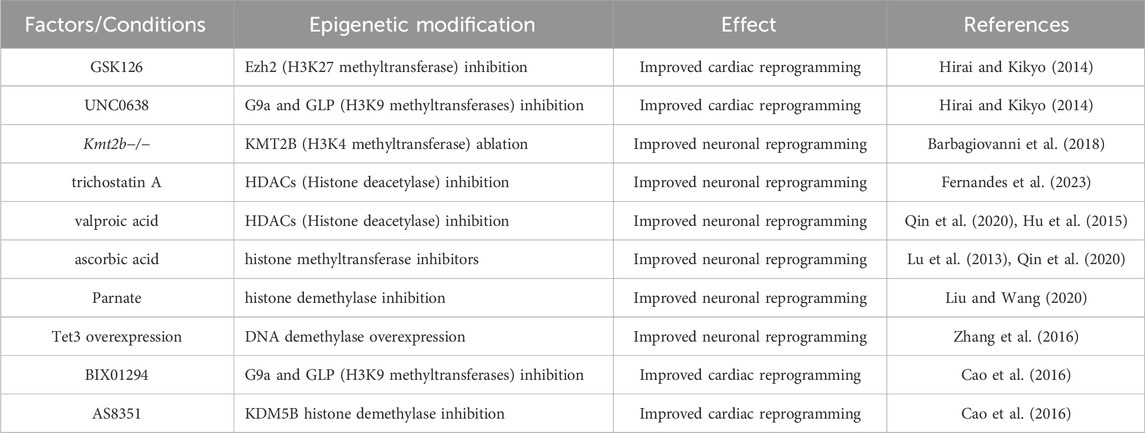
Direct reprogramming requires massive, but precise epigenomic alteration to ensure a proper cell fate change. Repatterning of H3K27me3, H3K4me3 and DNA methylation marks during fibroblast to cardiomyocyte direct reprogramming is one such example (Liu et al., 2016). The pioneer transcription factors such as MyoD, Ascl1 or GATA4 regulate the chromatin landscape through interaction with epigenetic modifiers (Molkentin et al., 1995). For example, MyoD as a pioneer factor, through epigenetic alterations allows access of chromatin to other transcription cofactors such as MEF2 proteins, Sp1, Pbx and Six proteins, that bind to flanking sequences of the E-motifs for synergistic transactivation, presumably through epigenetic alterations (Yun and Wold, 1996; Biesiada et al., 1999; Guo et al., 2003; Berkes et al., 2004; Liu et al., 2010; Taylor and Hughes, 2017). Studies have shown that modulation of epigenetic proteins can significantly improve the efficiency of reprogramming (Dal-Pra et al., 2017). For example, inhibition of Ezh2, the histone methyltransferase of PRC2, improves cardiac reprogramming (Hirai and Kikyo, 2014) and ablation of KMT2B, a histone methyltransferase, drastically reduces neuron reprogramming (Barbagiovanni et al., 2018).
The acquired DNA methylation pattern also correlates to the age of the donor (epigenetic aging-clock), representing an advantage of transdifferentiated neurons over iPSC-derived neurons for the study of age-dependent pathologies (Mertens et al., 2021). While this is an advantage in this case, it is also a barrier to efficient conversion as old fibroblasts conserve the hypermethylation pattern (Zhang et al., 2016). Various epigenetic modifiers have been utilized to improve transdiffererntiation (Table 1). For example, small molecules such as HDACs (Histone deacetylase) inhibitor trichostatin A (Fernandes et al., 2023), valproic acid (Hu et al., 2015; Qin et al., 2020) or the histone demethylase inhibitor parnate (Liu and Wang, 2020) have been used in the neuron conversion cocktails. Interestingly, the addition of ascorbic acid to the conversion media is also critical as it can activate both TET and Jumonji-C domain-containing histone demethylases leading to a more efficient epigenome rewiring (Lu et al., 2013; Lee Chong et al., 2019; Qin et al., 2020).
Further, non-coding RNAs have been utilized to enhance transdifferentiation efficiency. A cocktail of microRNAs including miR-1, miR-133, miR-208 and miR-499 was found to transdifferentiate mouse embryonic fibroblasts into cardiomyocyte-like cells (Jayawardena et al., 2012; Paoletti et al., 2020). Since then, studies have shown that different combinations of microRNAs and transcription factors can reprogram human fibroblasts with variable efficiencies (Nam et al., 2013; Muraoka et al., 2014). Some long noncoding RNAs such LncMyoD or Linc-RNA activator of myogenesis (Linc-RAM) and steroid receptor RNA activator (SRA) are known to promote myogenic transdifferentiation through modulating chromatin accessibility of several transcription factors (Yu et al., 2017; Dong et al., 2020).
5 Metabolic changes in transdifferentiation
While metabolic maturation has been a bottleneck in iPSC-based differentiation in general, the trans-differentiation process is shown (at least in the case of cardiomyocytes) to maintain the cellular metabolism observed in adult cells (Mathieu and Ruohola-Baker, 2017; Zhou et al., 2017; Miklas et al., 2019; Jahng et al., 2022). However, while both cell types (fibroblasts and transdifferentiated cells) are cells with mature metabolism, hence capable of utilizing a multitude of substrates for cellular energy, the demand for energy production in muscle is notably larger than in fibroblasts. Hence the number of mitochondria and their respiration capacity is expected to increase during direct reprogramming from fibroblasts to muscle cells. Accordingly, MyoD is shown to regulate skeletal muscle oxidative metabolism (Shintaku et al., 2016). Similarly single-cell RNA sequencing of GMT-induced transdifferentiation of fibroblasts to cardiomyocyte revealed dramatic metabolic alteration, and inhibition of oxidative metabolism impaired cardiomyocyte transdifferentiation (Zhou et al., 2017; Zhou et al., 2019; Jahng et al., 2022). A metabolic remodeling is also observed in neuronal transdifferentiation (Zheng et al., 2016). A mitochondrial mass and activity increase is driven by PGC-1α and ERRγ, two master regulators (Zheng et al., 2016). In accordance, an overexpression of the neuron-enriched mitochondrial proteins further boosted the conversion efficiency (Russo et al., 2021).
Interestingly metabolic changes due to aging or pathological conditions may transmit to the direct-reprogrammed cell types. This presents an opportunity to utilize transdifferentiated cells for disease modeling and search for therapies. For example, in Parkinson disorder, an impaired mitophagy (with parkin mutations) leading to metabolic stress is correlated to loss of neurons (Schwartzentruber et al., 2020). In Huntington’s disease, the increased ROS (reactive oxygen species) levels lead to DNA damage and neuronal loss (Victor et al., 2018). Fibroblast derived from an aged donor showed reduced mitochondrial activity and increased ROS levels (Huh et al., 2016; Kim et al., 2018), and neurons transdifferentiated from aged fibroblasts display reduced mitochondrial activity as well as reduced transdifferentiation efficiency (Kim et al., 2018). The efficiency of aged fibroblast conversion to neurons can be partially rescued by co-expression of either of an anti-apoptotic or antioxidant genes like BCL2 or SOD1 (Gascón et al., 2016; Russo et al., 2021). Furthermore, transdifferentiated adult cells may allow further dissection of the interdependence of metabolic, epigenetic and morphological maturation. For example, one could in the future test if the proteostasis index in these cells affects their health, regardless of their aging state.
6 Next-generation approaches in transdifferentiation
While above-described advances in Pioneer factor-, Signaling pathway-, Epigenetic- and Metabolic-regulation of the process have pushed the direct reprogramming approaches forward, major challenges still exist. Aging, inflammatory responses, fibrosis, metabolism and epigenetic barriers are the real, remaining challenges of direct reprogramming. Utilizing modern cutting edge technologies (including Artificial Intelligence (AI) based protein design) we can overcome some of these barriers. We discuss below some of the top-notch existing methods that have been less explored, and propose some novel methods of direct reprogramming that may revolutionize the personalized regenerative medicine.
6.1 Modulating signaling pathways using novel designed minibinders
Targeting signaling pathways with stable, ultra-specific and high affinity agonists and antagonists can be a monumental step forward in transdifferentiation. Recent advances in protein design now allow development of such powerful tools. Novel AI-based protein prediction and protein design (AlphaFold 2, RoseTTAFold, RFDiffusion) approaches are promising towards understanding disease, with a broad range of applications (Baek et al., 2021; Jumper et al., 2021; Bordin et al., 2023; Cheng et al., 2023; Watson et al., 2023). Since the cellular fate in direct reprogramming can be controlled by outside-in signaling (see Section 3), designed, highly specific and potent minibinder antagonists or agonists can potentially modulate the cellular fate to desired state. Synthetic agonists for TRKA receptor, EGFR, Tie2 receptor, FGFR1/2c, TGFbR or WNT (Janda et al., 2017; Zhao et al., 2021; Cao et al., 2022) allow precise analysis of specific splice variants and isomers of the receptors in multiple context, including transdifferentiation. These minibinders are precisely designed to target specific receptors and can also distinguish the receptor isoforms with minimal side effects which is not achievable with the natural ligands (Ocampo et al., 2016; Janda et al., 2017; Edman et al., 2023). An additional advantage of designed proteins is their greater stability (better half-lives) to ensure a sustained signaling response (Edman et al., 2023). One such example is usage of designed protein FGFR1/2c-isoform antagonist or agonist during pluripotent stem cell to endothelial cell differentiation. In the presence of FGFR1/2c designed miniprotein antagonist the pluripotent cells shift the fate completely to pericytes state instead of endothelial and in the presence of agonist iENDOs (iPSC-derived endothelial cells) shift to arterial vascular fate (Edman et al., 2023). These designed RTK agonists or antagonists can be utilized either in combination with transcription factors/pioneer factors, or plausibly in the future individually to reprogram the fibroblasts into desired tissues.
Designed minibinders may also be used to alleviate the problem of inflammation and fibrosis. Both inflammation and fibrosis have shown to be problematic for reprogramming. For instance, adult or neonatal fibroblast with increased inflammatory and fibrotic response show low efficiency of direct cardiac reprogramming (Chen et al., 2012; Zhao et al., 2015). Inhibiting the inflammatory chemokines or cytokines might improve the efficiency (Muraoka et al., 2019). Further, myocardial infarction triggers an inflammatory response in the myocardium that results in cardiac fibroblast activation. Recent advances in designing minibinders for IL (Interleukin)-receptors (Cao et al., 2022) may give a path for a new approach for reducing inflammation towards more robust direct reprogramming.
6.2 Direct reprogramming using mRNA
DNA delivery or viral delivery of transcription factors or microRNA faces clinical obstacles due to potential tumorigenic potential and immunogenic response. Injecting the mRNA or synthetic modified mRNA (modRNA; RNA with pseudoUridine, promises to offer a safer approach with potentially lower inflammation and immunogenicity and reduced risk of genomic integration (Karikó et al., 2008; Chien et al., 2014). For example, injecting synthetic VEGF-A modified mRNA (modRNA) can lead to efficient expansion and differentiation of cardiac progenitor cells (Zangi et al., 2013). For direct reprogramming of non-cardiomyocytes into cardiomyocytes modRNA has been used to express multiple transcription factors (Gata4, Mef2c, Tbx5, Hand2) and helper proteins [dominant-negative (DN)-TGFβ, DN-Wnt8a, and acid ceramidase] (Kaur et al., 2021). Further, transfection of MyoD-mRNA can reprogram MEFs into induced myogenic progenitor cell fate (iMPC) (Qabrati et al., 2023). These transgene free derived iMPCs successfully engrafted and restored dystrophin expression in DMD (Duchenne muscular dystrophy) mice (Qabrati et al., 2023). While exciting, further advances in efficiency and specificity of mRNA or modRNA delivery is required for therapeutic purposes.
6.3 CRISPR-mediated direct reprogramming
Transdifferentiation not only requires altering the expression of lineage-specific genes but also inheritable epigenetic changes. Mere overexpression of the lineage specific transcription factors is not enough to bring about efficient transdifferentiation. To overcome the epigenetic barriers microRNAs or small molecules have been used for more efficient transdifferentiation. There has been a considerable interest in utilizing targeted epigenetic tools to induce expression of lineage-specific transcription factors. Lineage-specific genes are mostly silenced in the fibroblast, and therefore require an exogenous expression of transcription factors for the reprogramming. Recently, alternatives to the overexpression of the TF transgenes have been developed, by opening the chromatin at precise loci to allow the endogenous target gene expression. Targeted epigenetic tools such as TALEN (Transcription activator-like effector nuclease) or CRISPR-based transcriptional activators such as CRISPRa-SAM (synergistic activation mediator) have been utilized to activate key genes in cellular reprogramming. One such example is activation of silenced chromatin with CRISPRa-SAM to reprogram fibroblasts into cardiac progenitor cells. Opening the silenced promoters of Gata4, Nkx2.5, and Tbx5 enabled reprogrammed fibroblasts into cardiac progenitors (Jiang et al., 2022). A simultaneous expression of endogenous lineage TF instead of overexpression of same set exogenous TF allows better reprogramming with a temporal and dosage control. Similarly, in another study, CRISPR mediated activation of neuron specific transcription factors such as Ascl1 or Ngn2 in mouse embryonic fibroblasts induced more efficient fate transition to neuronal lineage (Liu et al., 2018). Further, a dCas9 linked to the activating domain of VP64, was targeted to the promoter regions of neuronal inducers such as the BAM factors in fibroblasts to initiate the transcription (Black et al., 2016). This strategy was then further used for refining the TF involved in direct neuronal conversion (Hilton et al., 2015; Liu et al., 2018; Zhou et al., 2018). These epigenetic tools have great potential to make trans differentiation efficient and can be used in manipulating a gene of interest in case of genetic diseases.
6.4 Modulating epigenetic landscape using novel designed EpiBinders
A very recent epigenetic editing approach is to fuse catalytically dead Cas9 (RNA-guided endonuclease) to the core active domains of epigenetic modifier proteins. The dCas9 domain binds to the cognate sequences from guide RNA and the catalytic domain completes its function. Fusion proteins such dCas9-p300 (a Histone acetyltransferase) (Hilton et al., 2015), dCas9-DNMT3a (a DNA methyl transferase) (Stepper et al., 2017), [148] and others have been successful in altering global methylation levels. While exciting, their specificity has been a challenge since their loci-specific editing is less stringent, and it is unclear exactly why. One hypothesis is that these constructs are inevitably overexpressed in target cells, leading to an excess of catalytically active protein in the cell that then modifies DNA and chromatin (or other proteins) indiscriminately (Galonska et al., 2018). As an alternative, AI-designed small protein epigenetic binders, or EpiBinders, can adjust epigenetic Histone or DNA modifications at specific loci without increasing the functional expression level of editor proteins. The previously reported EBdCas9 (a de novo designed EED (a subunit of PRC2 complex)-binder, EB linked to dCas9) competes over EZH2 binding to EED and thereby inhibits histone3 lysine 27 methylation (H3K27Me3) on the specific promoter targeted by guide RNA, leading to upregulation of the specific gene expression (Levy et al., 2022). This EBdCas9/guide RNA directed tool sufficiently induced cellular reprogramming without off-target effects (Levy et al., 2022; Moody et al., 2017). One such example is EBdCas9/guide directed upregulation of a tumor suppressor p16 resulted in suppression of DMG (diffuse Midline glioma) cells (Levy et al., 2022). Similarly, designed EpiBinders to target other chromatin modifiers (DNMT, TET, histone methyltransferases, histone acetyltransferases, histone demethylases and histone deacetylases) can be utilized to activate or repress the gene of interest. After the recent development of an AI-based protein design algorithm, RFDiffusion designing small protein binders based on target structure alone has become significantly easier, enabling fast production of selective binding proteins (Watson et al., 2023). This approach now makes such efficient EpiBinder design to other epigenetic regulators highly feasible. Thus, the new generation of epigenetic editors will be smaller and modular, with the EpiBinder domains easily swapped out to match the desired modification.
6.5 Modulating the chromatin landscape by pioneer transcription factors
Among these various paths of cell transdifferentiation, the role of pioneer TFs is critical in rewiring the transcriptomic landscape and changing cellular identity. These factors are defined by their ability to bind closed nucleosomes and unwrap them, allowing the restructuring of chromatin for accessibility to other factors (transcription factors and epigenetic modifiers). This process of opening the chromatin is used most notably by Oct4, Sox2 and Klf4 during the reprogramming of somatic cells to the iPSC stage (Soufi et al., 2015; Chronis et al., 2017) (see Section 2 above). During the specific conversion from fibroblasts to other terminally differentiated cell types, the role of lineage related pioneer factors during the conversion process, with ASCL1 in iN conversion, MYOD in skeletal muscle and GATA4 in cardiomyocytes have been defined. Intriguingly, these factors have recently shown to have the ability to regulate the conversion toward other unrelated lineages as well; such as ASCL1, in combination with MEF2C, is shown to direct fibroblasts to a cardiomyocyte lineage (Wang et al., 2022a). To reach the full maturity however, addition of a cardiogenic specific factor, MEF2C, was required. Indeed, ASLC1 is able to bind chromatin regions corresponding to both neuronal and muscle genes, inducing chromatin opening of muscle genes to the same extent as the GMT cocktail (Wapinski et al., 2017; Wang et al., 2022a). This cross-lineage potential has been observed by MYOD which is known to maintain the muscle genome architecture but can also activate the neuronal transcriptomic landscape (Lee et al., 2020; Wang et al., 2022b). MYOD and ASCL1, both possess a very similar chromatin-binding capacity but with differences in quantitative binding (Lee et al., 2020). Blocking the muscle-specific lineage through the expression of Myt1l forced MYOD to induce the fibroblast to neuron conversion (Lee et al., 2020). This indicates the cross-lineage ability of pioneer factors drives lineage conversion through chromatin accessibility as the minimal requirement. An AI- designed transcription factor that can allow chromatin accessibility together with activation of the lineage specific genes will be one of the promising next-generation tools to regulate cell fate decision in near future. The reprogramming factor Sox2 is also able to enhance the neuronal conversion toward dopaminergic or GABA producing neurons when used in addition to other factors (Liu et al., 2012; Colasante et al., 2015; Tian et al., 2015). Recently, a triple mutant of SOX17 L111F/V118N/E122V has been reported to be more efficient than SOX2 in transactivation as well in inducing neuronal reprogramming (Weng et al., 2023). Interestingly, using the reprogramming and pioneer factors Oct4, Sox2 and Klf4 for a short time before launching the direct conversion of fibroblasts to cardiomyocytes greatly increased the transdifferentiation efficiency, by opening and priming the chromatin (Efe et al., 2011). These findings open the possibility that pioneer factors may have more general transdifferentiation potential than previously realized, and leave the door open to identifying, or AI-designing universal pioneer factors.
7 Conclusion
In this review, we discussed the classical methods of transdifferentiation that include transcription factors overexpression (Figure 1). Transdifferentiation has not been yet utilized at the clinical level, presumable due to low efficiency and targeting. In the future, modern tools may improve the transdifferentiation efficiency (Figure 3). Such modern tools discussed in this review are: AI-designed proteins that regulate signaling pathways or neutralize the inflammatory chemokines or cytokines, and pioneer factors or EpiBinders that open the silenced lineage genes. Further, predicted signaling pathways can also be utilized to induce a controlled transdifferentiation (Ainsworth et al., 2018; Tercan et al., 2022). These next-generation trans-differentiation approaches will come with better efficiency and plausibly with potential to treat diseases like Alzheimer’s disease, muscle injury, diabetes or myocardial infarction, resulting in elimination of the unsurmountable treatment issues at the moment (for example, finding a right donor or graft rejections). These novel approaches will enhance the efficacy and safety of direct reprogramming, allowing the ultimate decoding of the process towards plausibly resulting in 21st century personalized regenerative medicine.
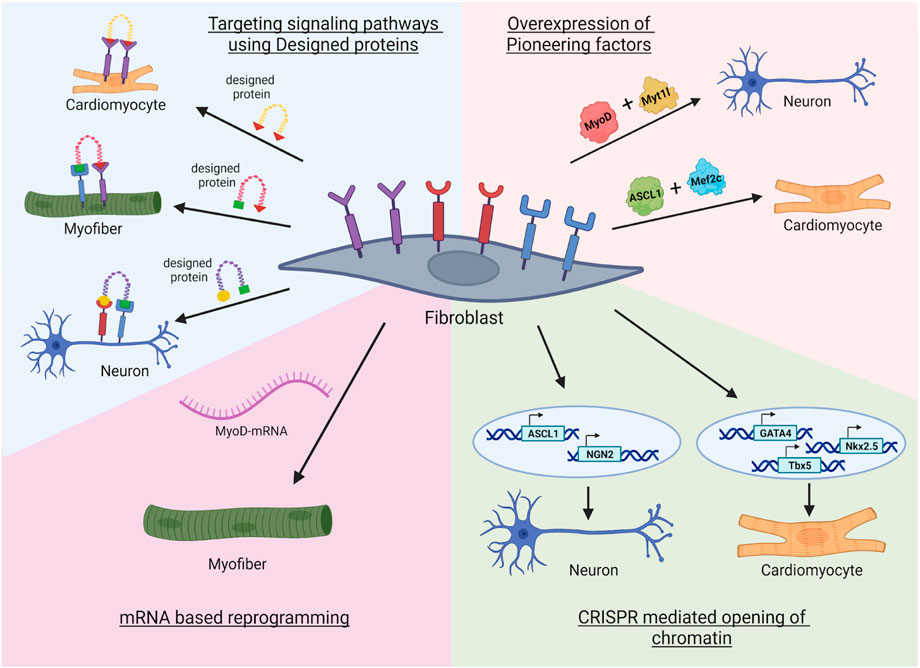
FIGURE 3. Novel approaches in transdifferentiation. First, the fibroblast receptors can be targeted with designed miniproteins that can enhance the reprogramming. Second, modified mRNA based reprogramming where transgene free expression of lineage specific transcription factors can reprogram the fibroblast to specific lineage (Qabrati et al., 2023). Third, the usage of epigenetic tools such as CRISPR mediated opening of promoters of specific pioneer transcription factor genes that are silenced in the fibroblast (Liu et al., 2018; Jiang et al., 2022). Fourth, combination of a pioneer factor with lineage specific transcription factor can open chromatin landscape allowing efficient reprogramming to multiple lineages. Image created by Biorender.
Author contributions
RK: Conceptualization, Formal Analysis, Investigation, Software, Writing–original draft, Writing–review and editing. DD: Conceptualization, Data curation, Investigation, Writing–original draft, Writing–review and editing. AP: Conceptualization, Data curation, Investigation, Writing–original draft, Writing–review and editing. CM: Writing–review and editing. SJ: Data curation, Software, Writing–original draft. TC: Data curation, Software, Writing–original draft. JM: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing. HR-B: Conceptualization, Funding acquisition, Investigation, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was partially supported by R01DE033016 for JM and HR-B.
Acknowledgments
We acknowledge rest of the HR-B lab’s members for helpful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abad, M., Hashimoto, H., Zhou, H., Morales, M. G., Chen, B., Bassel-Duby, R., et al. (2017). Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Rep. 8 (3), 548–560. doi:10.1016/j.stemcr.2017.01.025
Ainsworth, R. I., Ai, R., Ding, B., Li, N., Zhang, K., and Wang, W. (2018). Bayesian networks predict neuronal transdifferentiation. G3 Bethesda 8 (7), 2501–2511. doi:10.1534/g3.118.200401
Ambasudhan, R., Talantova, M., Coleman, R., Yuan, X., Zhu, S., Lipton, S. A., et al. (2011). Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9 (2), 113–118. doi:10.1016/j.stem.2011.07.002
Arenas, E., Denham, M., and Villaescusa, J. C. (2015). How to make a midbrain dopaminergic neuron. Dev. Camb. Engl. 142 (11), 1918–1936. doi:10.1242/dev.097394
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373 (6557), 871–876. doi:10.1126/science.abj8754
Barbagiovanni, G., Germain, P.-L., Zech, M., Atashpaz, S., Lo Riso, P., D’Antonio-Chronowska, A., et al. (2018). KMT2B is selectively required for neuronal transdifferentiation, and its loss exposes dystonia candidate genes. Cell Rep. 25 (4), 988–1001. doi:10.1016/j.celrep.2018.09.067
Berkes, C. A., Bergstrom, D. A., Penn, B. H., Seaver, K. J., Knoepfler, P. S., and Tapscott, S. J. (2004). Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14 (4), 465–477. doi:10.1016/s1097-2765(04)00260-6
Berninger, B., Guillemot, F., and Götz, M. (2007). Directing neurotransmitter identity of neurones derived from expanded adult neural stem cells. Eur. J. Neurosci. 25 (9), 2581–2590. doi:10.1111/j.1460-9568.2007.05509.x
Biesiada, E., Hamamori, Y., Kedes, L., and Sartorelli, V. (1999). Myogenic basic helix-loop-helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac alpha-actin promoter. Mol. Cell. Biol. 19 (4), 2577–2584. doi:10.1128/MCB.19.4.2577
Black, J. B., Adler, A. F., Wang, H.-G., D’Ippolito, A. M., Hutchinson, H. A., Reddy, T. E., et al. (2016). Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-Based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell 19 (3), 406–414. doi:10.1016/j.stem.2016.07.001
Bordin, N., Sillitoe, I., Nallapareddy, V., Rauer, C., Lam, S. D., Waman, V. P., et al. (2023). AlphaFold2 reveals commonalities and novelties in protein structure space for 21 model organisms. Commun. Biol. 6 (1), 160. doi:10.1038/s42003-023-04488-9
Boularaoui, S. M., Abdel-Raouf, K. M. A., Alwahab, N. S. A., Kondash, M. E., Truskey, G. A., Teo, J. C. M., et al. (2018). Efficient transdifferentiation of human dermal fibroblasts into skeletal muscle. J. Tissue Eng. Regen. Med. 12 (2), e918–e936. doi:10.1002/term.2415
Browder, K. C., Reddy, P., Yamamoto, M., Haghani, A., Guillen, I. G., Sahu, S., et al. (2022). In vivo partial reprogramming alters age-associated molecular changes during physiological aging in mice. Nat. Aging 2 (3), 243–253. doi:10.1038/s43587-022-00183-2
Bruzelius, A., Kidnapillai, S., Drouin-Ouellet, J., Stoker, T., Barker, R. A., and Rylander Ottosson, D. (2021). Reprogramming human adult fibroblasts into GABAergic interneurons. Cells 10 (12), 3450. doi:10.3390/cells10123450
Cacchiarelli, D., Qiu, X., Srivatsan, S., Manfredi, A., Ziller, M., Overbey, E., et al. (2018). Aligning single-cell developmental and reprogramming trajectories identifies molecular determinants of myogenic reprogramming outcome. Cell Syst. 7 (3), 258–268. doi:10.1016/j.cels.2018.07.006
Cao, L., Coventry, B., Goreshnik, I., Huang, B., Sheffler, W., Park, J. S., et al. (2022). Design of protein-binding proteins from the target structure alone. Nature 605 (7910), 551–560. doi:10.1038/s41586-022-04654-9
Cao, N., Huang, Y., Zheng, J., Spencer, C. I., Zhang, Y., Fu, J. D., et al. (2016). Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352 (6290), 1216–1220. doi:10.1126/science.aaf1502
Chen, J. X., Krane, M., Deutsch, M.-A., Wang, L., Rav-Acha, M., Gregoire, S., et al. (2012). Inefficient reprogramming of fibroblasts into cardiomyocytes using Gata4, Mef2c, and Tbx5. Mef2c, Tbx5. Circ. Res. 111 (1), 50–55. doi:10.1161/CIRCRESAHA.112.270264
Cheng, J., Novati, G., Pan, J., Bycroft, C., Žemgulytė, A., Applebaum, T., et al. (2023). Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381 (6664), eadg7492. doi:10.1126/science.adg7492
Chien, K. R., Zangi, L., and Lui, K. O. (2014). Synthetic chemically modified mRNA (modRNA): toward a new technology platform for cardiovascular biology and medicine. Cold Spring Harb. Perspect. Med. 5 (1), a014035. doi:10.1101/cshperspect.a014035
Chronis, C., Fiziev, P., Papp, B., Butz, S., Bonora, G., Sabri, S., et al. (2017). Cooperative binding of transcription factors orchestrates reprogramming. Cell 168 (3), 442–459. doi:10.1016/j.cell.2016.12.016
Colasante, G., Lignani, G., Rubio, A., Medrihan, L., Yekhlef, L., Sessa, A., et al. (2015). Rapid conversion of fibroblasts into functional forebrain GABAergic interneurons by direct genetic reprogramming. Cell Stem Cell 17 (6), 719–734. doi:10.1016/j.stem.2015.09.002
Csordás, G., Gábor, E., and Honti, V. (2021). There and back again: the mechanisms of differentiation and transdifferentiation in Drosophila blood cells. Dev. Biol. 469, 135–143. doi:10.1016/j.ydbio.2020.10.006
Dai, P., Harada, Y., and Takamatsu, T. (2015). Highly efficient direct conversion of human fibroblasts to neuronal cells by chemical compounds. J. Clin. Biochem. Nutr. 56 (3), 166–170. doi:10.3164/jcbn.15-39
Dal-Pra, S., Hodgkinson, C. P., Mirotsou, M., Kirste, I., and Dzau, V. J. (2017). Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by MiR combo. Circ. Res. 120 (9), 1403–1413. doi:10.1161/CIRCRESAHA.116.308741
Davis, R. L., Weintraub, H., and Lassar, A. B. (1987). Expression of a single transfected CDNA converts fibroblasts to myoblasts. Cell 51 (6), 987–1000. doi:10.1016/0092-8674(87)90585-x
Dong, A., Preusch, C. B., So, W.-K., Lin, K., Luan, S., Yi, R., et al. (2020). A long noncoding RNA, LncMyoD, modulates chromatin accessibility to regulate muscle stem cell myogenic lineage progression. Proc. Natl. Acad. Sci. U. S. A. 117 (51), 32464–32475. doi:10.1073/pnas.2005868117
Edman, N. I., Redler, R. L., Phal, A., Schlichthaerle, T., Srivatsan, S. R., Etemadi, A., et al. (2023). Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies. BioRxiv Prepr. Serv. Biol. 2023, 2023.03.14.532666. doi:10.1101/2023.03.14.532666
Efe, J. A., Hilcove, S., Kim, J., Zhou, H., Ouyang, K., Wang, G., et al. (2011). Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 13 (3), 215–222. doi:10.1038/ncb2164
Fernandes, G. S., Singh, R. D., De, D., and Kim, K. K. (2023). Strategic application of epigenetic regulators for efficient neuronal reprogramming of human fibroblasts. Int. J. Stem Cells 16 (2), 156–167. doi:10.15283/ijsc22183
Galonska, C., Charlton, J., Mattei, A. L., Donaghey, J., Clement, K., Gu, H., et al. (2018). Genome-wide tracking of DCas9-methyltransferase footprints. Nat. Commun. 9 (1), 597. doi:10.1038/s41467-017-02708-5
Gascón, S., Murenu, E., Masserdotti, G., Ortega, F., Russo, G. L., Petrik, D., et al. (2016). Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell 18 (3), 396–409. doi:10.1016/j.stem.2015.12.003
Guo, C. S., Degnin, C., Fiddler, T. A., Stauffer, D., and Thayer, M. J. (2003). Regulation of MyoD activity and muscle cell differentiation by MDM2, PRb, and Sp1. J. Biol. Chem. 278 (25), 22615–22622. doi:10.1074/jbc.M301943200
Heins, N., Malatesta, P., Cecconi, F., Nakafuku, M., Tucker, K. L., Hack, M. A., et al. (2002). Glial cells generate neurons: the role of the transcription factor Pax6. Nat. Neurosci. 5 (4), 308–315. doi:10.1038/nn828
Hilton, I. B., D’Ippolito, A. M., Vockley, C. M., Thakore, P. I., Crawford, G. E., Reddy, T. E., et al. (2015). Epigenome editing by a CRISPR-cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33 (5), 510–517. doi:10.1038/nbt.3199
Hirai, H., and Kikyo, N. (2014). Inhibitors of suppressive histone modification promote direct reprogramming of fibroblasts to cardiomyocyte-like cells. Cardiovasc. Res. 102 (1), 188–190. doi:10.1093/cvr/cvu023
Hu, W., Qiu, B., Guan, W., Wang, Q., Wang, M., Li, W., et al. (2015). Direct conversion of normal and Alzheimer’s disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17 (2), 204–212. doi:10.1016/j.stem.2015.07.006
Huh, C. J., Zhang, B., Victor, M. B., Dahiya, S., Batista, L. F., Horvath, S., et al. (2016). Maintenance of age in human neurons generated by MicroRNA-based neuronal conversion of fibroblasts. eLife 5, e18648. doi:10.7554/eLife.18648
Iberite, F., Gruppioni, E., and Ricotti, L. (2022). Skeletal muscle differentiation of human IPSCs meets bioengineering strategies: perspectives and challenges. NPJ Regen. Med. 7 (1), 23. doi:10.1038/s41536-022-00216-9
Ieda, M., Fu, J.-D., Delgado-Olguin, P., Vedantham, V., Hayashi, Y., Bruneau, B. G., et al. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142 (3), 375–386. doi:10.1016/j.cell.2010.07.002
Ifkovits, J. L., Addis, R. C., Epstein, J. A., and Gearhart, J. D. (2014). Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PloS One 9 (2), e89678. doi:10.1371/journal.pone.0089678
Jahng, J. W. S., Zhang, M., and Wu, J. C. (2022). The role of metabolism in directed differentiation versus trans-differentiation of cardiomyocytes. Semin. Cell Dev. Biol. 122, 56–65. doi:10.1016/j.semcdb.2021.05.018
Janda, C. Y., Dang, L. T., You, C., Chang, J., de Lau, W., Zhong, Z. A., et al. (2017). Surrogate Wnt agonists that phenocopy canonical Wnt and β-catenin signalling. Nature 545 (7653), 234–237. doi:10.1038/nature22306
Jayawardena, T. M., Egemnazarov, B., Finch, E. A., Zhang, L., Payne, J. A., Pandya, K., et al. (2012). MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 110 (11), 1465–1473. doi:10.1161/CIRCRESAHA.112.269035
Jiang, L., Liang, J., Huang, W., Ma, J., Park, K. H., Wu, Z., et al. (2022). CRISPR activation of endogenous genes reprograms fibroblasts into cardiovascular progenitor cells for myocardial infarction therapy. Mol. Ther. J. Am. Soc. Gene Ther. 30 (1), 54–74. doi:10.1016/j.ymthe.2021.10.015
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kabadi, A. M., Thakore, P. I., Vockley, C. M., Ousterout, D. G., Gibson, T. M., Guilak, F., et al. (2015). Enhanced MyoD-induced transdifferentiation to a myogenic lineage by fusion to a potent transactivation domain. ACS Synth. Biol. 4 (6), 689–699. doi:10.1021/sb500322u
Karikó, K., Muramatsu, H., Welsh, F. A., Ludwig, J., Kato, H., Akira, S., et al. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16 (11), 1833–1840. doi:10.1038/mt.2008.200
Kaur, K., Hadas, Y., Kurian, A. A., Żak, M. M., Yoo, J., Mahmood, A., et al. (2021). Direct reprogramming induces vascular regeneration post muscle ischemic injury. Mol. Ther. J. Am. Soc. Gene Ther. 29 (10), 3042–3058. doi:10.1016/j.ymthe.2021.07.014
Kim, Y., Zheng, X., Ansari, Z., Bunnell, M. C., Herdy, J. R., Traxler, L., et al. (2018). Mitochondrial aging defects emerge in directly reprogrammed human neurons due to their metabolic profile. Cell Rep. 23 (9), 2550–2558. doi:10.1016/j.celrep.2018.04.105
Kriks, S., Shim, J.-W., Piao, J., Ganat, Y. M., Wakeman, D. R., Xie, Z., et al. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480 (7378), 547–551. doi:10.1038/nature10648
Ladewig, J., Mertens, J., Kesavan, J., Doerr, J., Poppe, D., Glaue, F., et al. (2012). Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 9 (6), 575–578. doi:10.1038/nmeth.1972
Lassar, A. B., Davis, R. L., Wright, W. E., Kadesch, T., Murre, C., Voronova, A., et al. (1991). Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66 (2), 305–315. doi:10.1016/0092-8674(91)90620-e
Lee, Q. Y., Mall, M., Chanda, S., Zhou, B., Sharma, K. S., Schaukowitch, K., et al. (2020). Pro-neuronal activity of Myod1 due to promiscuous binding to neuronal genes. Nat. Cell Biol. 22 (4), 401–411. doi:10.1038/s41556-020-0490-3
Lee Chong, T., Ahearn, E. L., and Cimmino, L. (2019). Reprogramming the epigenome with vitamin C. Front. Cell Dev. Biol. 7, 128. doi:10.3389/fcell.2019.00128
Levy, S., Somasundaram, L., Raj, I. X., Ic-Mex, D., Phal, A., Schmidt, S., et al. (2022). DCas9 fusion to computer-designed PRC2 inhibitor reveals functional TATA box in distal promoter region. Cell Rep. 38 (9), 110457. doi:10.1016/j.celrep.2022.110457
Liu, X., Li, F., Stubblefield, E. A., Blanchard, B., Richards, T. L., Larson, G. A., et al. (2012). Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 22 (2), 321–332. doi:10.1038/cr.2011.181
Liu, Y., Chu, A., Chakroun, I., Islam, U., and Blais, A. (2010). Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 38 (20), 6857–6871. doi:10.1093/nar/gkq585
Liu, Y., and Wang, H. (2020). Modeling sporadic Alzheimer’s disease by efficient direct reprogramming of the elderly derived disease dermal fibroblasts into neural stem cells. J. Alzheimers Dis. Jad. 73 (3), 919–933. doi:10.3233/JAD-190614
Liu, Y., Yu, C., Daley, T. P., Wang, F., Cao, W. S., Bhate, S., et al. (2018). CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 23 (5), 758–771. doi:10.1016/j.stem.2018.09.003
Liu, Z., Chen, O., Zheng, M., Wang, L., Zhou, Y., Yin, C., et al. (2016). Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 16 (2), 507–518. doi:10.1016/j.scr.2016.02.037
Lu, J., Liu, H., Huang, C. T.-L., Chen, H., Du, Z., Liu, Y., et al. (2013). Generation of integration-free and region-specific neural progenitors from primate fibroblasts. Cell Rep. 3 (5), 1580–1591. doi:10.1016/j.celrep.2013.04.004
Mall, M., and Wernig, M. (2017). The novel tool of cell reprogramming for applications in molecular medicine. J. Mol. Med. Berl. Ger. 95 (7), 695–703. doi:10.1007/s00109-017-1550-4
Mathieu, J., and Ruohola-Baker, H. (2017). Metabolic remodeling during the loss and acquisition of pluripotency. Development 144 (4), 541–551. doi:10.1242/dev.128389
Mertens, J., Herdy, J. R., Traxler, L., Schafer, S. T., Schlachetzki, J. C. M., Böhnke, L., et al. (2021). Age-dependent instability of mature neuronal fate in induced neurons from Alzheimer’s patients. Cell Stem Cell 28 (9), 1533–1548.e6. doi:10.1016/j.stem.2021.04.004
Mertens, J., Paquola, A. C. M., Ku, M., Hatch, E., Böhnke, L., Ladjevardi, S., et al. (2015). Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell 17 (6), 705–718. doi:10.1016/j.stem.2015.09.001
Miklas, J. W., Clark, E., Levy, S., Detraux, D., Leonard, A., Beussman, K., et al. (2019). TFPa/HADHA is required for fatty acid beta-oxidation and cardiolipin Re-modeling in human cardiomyocytes. Nat. Commun. 10 (1), 4671. doi:10.1038/s41467-019-12482-1
Mohamed, T. M. A., Stone, N. R., Berry, E. C., Radzinsky, E., Huang, Y., Pratt, K., et al. (2017). Chemical enhancement of in vitro and in vivo direct cardiac reprogramming. Circulation 135 (10), 978–995. doi:10.1161/CIRCULATIONAHA.116.024692
Molkentin, J. D., Black, B. L., Martin, J. F., and Olson, E. N. (1995). Cooperative activation of muscle gene expression by MEF2 and myogenic BHLH proteins. Cell 83 (7), 1125–1136. doi:10.1016/0092-8674(95)90139-6
Moody, J. D., Levy, S., Mathieu, J., Xing, Y., Kim, W., Dong, C., et al. (2017). First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor. Proc. Natl. Acad. Sci. 114 (38), 10125–10130.
Muraoka, N., Nara, K., Tamura, F., Kojima, H., Yamakawa, H., Sadahiro, T., et al. (2019). Role of cyclooxygenase-2-mediated prostaglandin E2-prostaglandin E receptor 4 signaling in cardiac reprogramming. Nat. Commun. 10 (1), 674. doi:10.1038/s41467-019-08626-y
Muraoka, N., Yamakawa, H., Miyamoto, K., Sadahiro, T., Umei, T., Isomi, M., et al. (2014). MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 33 (14), 1565–1581. doi:10.15252/embj.201387605
Nam, Y.-J., Song, K., Luo, X., Daniel, E., Lambeth, K., West, K., et al. (2013). Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. U. S. A. 110 (14), 5588–5593. doi:10.1073/pnas.1301019110
Ocampo, A., Reddy, P., Martinez-Redondo, P., Platero-Luengo, A., Hatanaka, F., Hishida, T., et al. (2016). In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167 (7), 1719–1733. doi:10.1016/j.cell.2016.11.052
Paoletti, C., Divieto, C., Tarricone, G., Di Meglio, F., Nurzynska, D., and Chiono, V. (2020). MicroRNA-mediated direct reprogramming of human adult fibroblasts toward cardiac phenotype. Front. Bioeng. Biotechnol. 8, 529. doi:10.3389/fbioe.2020.00529
Pfisterer, U., Ek, F., Lang, S., Soneji, S., Olsson, R., and Parmar, M. (2016). Small molecules increase direct neural conversion of human fibroblasts. Sci. Rep. 6, 38290. doi:10.1038/srep38290
Pfisterer, U., Kirkeby, A., Torper, O., Wood, J., Nelander, J., Dufour, A., et al. (2011a). Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U. S. A. 108 (25), 10343–10348. doi:10.1073/pnas.1105135108
Pfisterer, U., Wood, J., Nihlberg, K., Hallgren, O., Bjermer, L., Westergren-Thorsson, G., et al. (2011b). Efficient induction of functional neurons from adult human fibroblasts. Tex 10 (19), 3311–3316. doi:10.4161/cc.10.19.17584
Qabrati, X., Kim, I., Ghosh, A., Bundschuh, N., Noé, F., Palmer, A. S., et al. (2023). Transgene-free direct conversion of murine fibroblasts into functional muscle stem cells. NPJ Regen. Med. 8 (1), 43. doi:10.1038/s41536-023-00317-z
Qian, L., Huang, Y., Spencer, C. I., Foley, A., Vedantham, V., Liu, L., et al. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485 (7400), 593–598. doi:10.1038/nature11044
Qin, H., Zhao, A.-D., Sun, M.-L., Ma, K., and Fu, X.-B. (2020). Direct conversion of human fibroblasts into dopaminergic neuron-like cells using small molecules and protein factors. Mil. Med. Res. 7 (1), 52. doi:10.1186/s40779-020-00284-2
Roh, H. C., Tsai, L. T. Y., Shao, M., Tenen, D., Shen, Y., Kumari, M., et al. (2018). Warming induces significant reprogramming of beige, but not Brown, adipocyte cellular identity. Cell Metab. 27 (5), 1121–1137. doi:10.1016/j.cmet.2018.03.005
Russo, G. L., Sonsalla, G., Natarajan, P., Breunig, C. T., Bulli, G., Merl-Pham, J., et al. (2021). CRISPR-mediated induction of neuron-enriched mitochondrial proteins boosts direct glia-to-neuron conversion. Cell Stem Cell 28 (3), 524–534.e7. doi:10.1016/j.stem.2020.10.015
Schwartzentruber, A., Boschian, C., Lopes, F. M., Myszczynska, M. A., New, E. J., Beyrath, J., et al. (2020). Oxidative switch drives mitophagy defects in dopaminergic parkin mutant patient neurons. Sci. Rep. 10 (1), 15485. doi:10.1038/s41598-020-72345-4
Shintaku, J., Peterson, J. M., Talbert, E. E., Gu, J.-M., Ladner, K. J., Williams, D. R., et al. (2016). MyoD regulates skeletal muscle oxidative metabolism cooperatively with alternative NF-κb. Cell Rep. 17 (2), 514–526. doi:10.1016/j.celrep.2016.09.010
Song, K., Nam, Y.-J., Luo, X., Qi, X., Tan, W., Huang, G. N., et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485 (7400), 599–604. doi:10.1038/nature11139
Soufi, A., Garcia, M. F., Jaroszewicz, A., Osman, N., Pellegrini, M., and Zaret, K. S. (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161 (3), 555–568. doi:10.1016/j.cell.2015.03.017
Stepper, P., Kungulovski, G., Jurkowska, R. Z., Chandra, T., Krueger, F., Reinhardt, R., et al. (2017). Efficient targeted DNA methylation with chimeric DCas9-dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res. 45 (4), 1703–1713. doi:10.1093/nar/gkw1112
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–872. doi:10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–676. doi:10.1016/j.cell.2006.07.024
Takahashi, K., and Yamanaka, S. (2016). A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 17 (3), 183–193. doi:10.1038/nrm.2016.8
Tapscott, S. J., Davis, R. L., Thayer, M. J., Cheng, P. F., Weintraub, H., and Lassar, A. B. (1988). MyoD1: a nuclear phosphoprotein requiring a myc homology region to convert fibroblasts to myoblasts. Science 242 (4877), 405–411. doi:10.1126/science.3175662
Taylor, M. V., and Hughes, S. M. (2017). Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 72, 33–44. doi:10.1016/j.semcdb.2017.11.020
Tercan, B., Aguilar, B., Huang, S., Dougherty, E. R., and Shmulevich, I. (2022). Probabilistic boolean networks predict transcription factor targets to induce transdifferentiation. iScience 25 (9), 104951. doi:10.1016/j.isci.2022.104951
Thorel, F., Népote, V., Avril, I., Kohno, K., Desgraz, R., Chera, S., et al. (2010). Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464 (7292), 1149–1154. doi:10.1038/nature08894
Tian, C., Li, Y., Huang, Y., Wang, Y., Chen, D., Liu, J., et al. (2015). Selective generation of dopaminergic precursors from mouse fibroblasts by direct lineage conversion. Sci. Rep. 5, 12622. doi:10.1038/srep12622
Victor, M. B., Richner, M., Olsen, H. E., Lee, S. W., Monteys, A. M., Ma, C., et al. (2018). Striatal neurons directly converted from huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat. Neurosci. 21 (3), 341–352. doi:10.1038/s41593-018-0075-7
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Südhof, T. C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463 (7284), 1035–1041. doi:10.1038/nature08797
Wada, R., Muraoka, N., Inagawa, K., Yamakawa, H., Miyamoto, K., Sadahiro, T., et al. (2013). Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. U. S. A. 110 (31), 12667–12672. doi:10.1073/pnas.1304053110
Wang, H., Keepers, B., Qian, Y., Xie, Y., Colon, M., Liu, J., et al. (2022a). Cross-lineage potential of Ascl1 uncovered by comparing diverse reprogramming regulatomes. Cell Stem Cell 29 (10), 1491–1504.e9. doi:10.1016/j.stem.2022.09.006
Wang, H., Yang, Y., Liu, J., and Qian, L. (2021). Direct cell reprogramming: approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 22 (6), 410–424. doi:10.1038/s41580-021-00335-z
Wang, R., Chen, F., Chen, Q., Wan, X., Shi, M., Chen, A. K., et al. (2022b). MyoD is a 3D genome structure organizer for muscle cell identity. Nat. Commun. 13 (1), 205. doi:10.1038/s41467-021-27865-6
Wapinski, O. L., Lee, Q. Y., Chen, A. C., Li, R., Corces, M. R., Ang, C. E., et al. (2017). Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep. 20 (13), 3236–3247. doi:10.1016/j.celrep.2017.09.011
Watson, J. L., Juergens, D., Bennett, N. R., Trippe, B. L., Yim, J., Eisenach, H. E., et al. (2023). De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100. doi:10.1038/s41586-023-06415-8
Weintraub, H., Dwarki, V. J., Verma, I., Davis, R., Hollenberg, S., Snider, L., et al. (1991). Muscle-specific transcriptional activation by MyoD. Genes Dev. 5 (8), 1377–1386. doi:10.1101/gad.5.8.1377
Weintraub, H., Tapscott, S. J., Davis, R. L., Thayer, M. J., Adam, M. A., Lassar, A. B., et al. (1989). Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. U. S. A. 86 (14), 5434–5438. doi:10.1073/pnas.86.14.5434
Weng, M., Hu, H., Graus, M. S., Tan, D. S., Gao, Y., Ren, S., et al. (2023). An engineered Sox17 induces somatic to neural stem cell fate transitions independently from pluripotency reprogramming. Sci. Adv. 9 (34), eadh2501. doi:10.1126/sciadv.adh2501
Yang, Y., Chen, R., Wu, X., Zhao, Y., Fan, Y., Xiao, Z., et al. (2019). Rapid and efficient conversion of human fibroblasts into functional neurons by small molecules. Stem Cell Rep. 13 (5), 862–876. doi:10.1016/j.stemcr.2019.09.007
Yu, X., Zhang, Y., Li, T., Ma, Z., Jia, H., Chen, Q., et al. (2017). Long non-coding RNA linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 8, 14016. doi:10.1038/ncomms14016
Yun, K., and Wold, B. (1996). Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr. Opin. Cell Biol. 8 (6), 877–889. doi:10.1016/s0955-0674(96)80091-3
Zangi, L., Lui, K. O., von Gise, A., Ma, Q., Ebina, W., Ptaszek, L. M., et al. (2013). Modified MRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 31 (10), 898–907. doi:10.1038/nbt.2682
Zhang, J., Chen, S., Zhang, D., Shi, Z., Li, H., Zhao, T., et al. (2016). Tet3-Mediated DNA demethylation contributes to the direct conversion of fibroblast to functional neuron. Cell Rep. 17 (9), 2326–2339. doi:10.1016/j.celrep.2016.10.081
Zhao, Y., Londono, P., Cao, Y., Sharpe, E. J., Proenza, C., O’Rourke, R., et al. (2015). High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 6, 8243. doi:10.1038/ncomms9243
Zhao, Y. T., Fallas, J. A., Saini, S., Ueda, G., Somasundaram, L., Zhou, Z., et al. (2021). F-domain valency determines outcome of signaling through the angiopoietin pathway. EMBO Rep. 22 (12), e53471. doi:10.15252/embr.202153471
Zheng, X., Boyer, L., Jin, M., Mertens, J., Kim, Y., Ma, L., et al. (2016). Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife 5, e13374. doi:10.7554/eLife.13374
Zhou, H., Liu, J., Zhou, C., Gao, N., Rao, Z., Li, H., et al. (2018). In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-DCas9-activator transgenic mice. Nat. Neurosci. 21 (3), 440–446. doi:10.1038/s41593-017-0060-6
Zhou, Y., Liu, Z., Welch, J. D., Gao, X., Wang, L., Garbutt, T., et al. (2019). Single-cell transcriptomic analyses of cell fate transitions during human cardiac reprogramming. Cell Stem Cell 25 (1), 149–164. doi:10.1016/j.stem.2019.05.020
Keywords: transdifferentiation, direct reprogramming, partial reprogramming, pioneer factors, aging, signaling, skeletal muscle, cardiac muscles
Citation: Keshri R, Detraux D, Phal A, McCurdy C, Jhajharia S, Chan TC, Mathieu J and Ruohola-Baker H (2024) Next-generation direct reprogramming. Front. Cell Dev. Biol. 12:1343106. doi: 10.3389/fcell.2024.1343106
Received: 23 November 2023; Accepted: 12 January 2024;
Published: 02 February 2024.
Edited by:
Jesus Chimal-Monroy, National Autonomous University of Mexico, MexicoReviewed by:
Luis Covarrubias, National Autonomous University of Mexico, MexicoYogesh Srivastava, University of Texas MD Anderson Cancer Center, United States
Copyright © 2024 Keshri, Detraux, Phal, McCurdy, Jhajharia, Chan, Mathieu and Ruohola-Baker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hannele Ruohola-Baker, aGFubmVsZUB1dy5lZHU=
†These authors have contributed equally to this work
 Riya Keshri
Riya Keshri Damien Detraux1,2†
Damien Detraux1,2† Julie Mathieu
Julie Mathieu Hannele Ruohola-Baker
Hannele Ruohola-Baker