
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 31 January 2024
Sec. Cancer Cell Biology
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1341999
The current situation of hepatocellular carcinoma (HCC) management is challenging due to its high incidence, mortality, recurrence and metastasis. Recent advances in gene genetic and expression regulation have unveiled the significant role of non-coding RNA (ncRNA) in various cancers. This led to the formulation of the competing endogenous RNA (ceRNA) hypothesis, which posits that both coding RNA and ncRNA, containing miRNA response elements (MRE), can share the same miRNA sequence. This results in a competitive network between ncRNAs, such as lncRNA and mRNA, allowing them to regulate each other. Extensive research has highlighted the crucial role of the ceRNA network in HCC development, impacting various cellular processes including proliferation, metastasis, cell death, angiogenesis, tumor microenvironment, organismal immunity, and chemotherapy resistance. Additionally, the ceRNA network, mediated by lncRNA or circRNA, offers potential in early diagnosis and prevention of HCC. Consequently, ceRNAs are emerging as therapeutic targets for HCC. The complexity of these gene networks aligns with the multi-target approach of traditional Chinese medicine (TCM), presenting a novel perspective for TCM in combating HCC. Research is beginning to show that TCM compounds and prescriptions can affect HCC progression through the ceRNA network, inhibiting proliferation and metastasis, and inducing apoptosis. Currently, the lncRNAs TUG1, NEAT1, and CCAT1, along with their associated ceRNA networks, are among the most promising ncRNAs for HCC research. However, this field is still in its infancy, necessitating advanced technology and extensive basic research to fully understand the ceRNA network mechanisms of TCM in HCC treatment.
Hepatocellular carcinoma (HCC) is among of the most prevalent malignant tumors, characterized by diverse etiologies, significant morbidity and mortality, and a poor survival prognosis. Over half of the global HCC cases are reported in Asia (Singal et al., 2020). The insidious nature of HCC often results in the absence of early-stage clinical manifestations, leading to its frequent oversight. Consequently, most HCC cases are diagnosed at an advanced stage, beyond the optimal timeframe for surgical intervention, and with limited treatment options. Despite recent advancements in early diagnosis and treatment, the clinical outcomes for HCC remain poor, largely due to high rates of recurrence and metastasis (Ghouri et al., 2017).
Addressing this challenge, it is critical to develop effective strategies for the early screening and monitoring of HCC. While Alpha-fetoprotein (AFP) is a widely used diagnostic biomarker for HCC, its limited sensitivity and specificity, as evidenced by the presence of AFP-negative HCC patients, indicates a need for improvement (Wang and Wei, 2020). The advancement of systems biology and bioinformatics technologies has led to the development of new biomarkers, such as alpha-fetoprotein lectin 3 (AFP-L3), des-gamma-carboxyprothrombin (DCP), and glypican-3 (GPC3). However, the clinical applicability of these biomarkers requires further evaluation (Pinero et al., 2020). Therefore, exploring novel potential biomarkers is essential to enhance diagnostic methods and improve treatment outcomes for HCC.
Non-coding RNA (ncRNA), including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA), constitute a major portion of the human transcriptome (Chan and Tay, 2018), with only 1%–2% being protein-coding mRNAs (Carninci et al., 2005; Djebali et al., 2012). This discovery has sparked significant interest in studying the functions of ncRNAs. These RNAs have been identified as key players in various biological processes, particularly in cancer development. MiRNA, a small ncRNA of approximately 22 nucleotides, post-transcriptionally regulates gene expression by binding to miRNA response elements (MRE), leading to gene degradation or translation inhibition (Bartel, 2004; 2018). Aberrant miRNA expression is closely linked to tumorigenesis (Di Leva et al., 2014), exhibiting both carcinogenic and tumor-suppressive effects in a dosage-dependent manner (Svoronos et al., 2016). Initial research on miRNA focused on their unidirectional regulation of target genes. However, further investigation has revealed intricate co-regulatory networks between non-coding and coding RNAs, leading to the competing endogenous RNA (ceRNA) hypothesis (Salmena et al., 2011). The ceRNA network, implicated in tumor cell proliferation, metastasis, apoptosis, angiogenesis, chemotherapy resistance, stem cell regulation, tumor immunity (Chen et al., 2022c). Comprehensive research on ceRNA function in HCC could identify potential biomarkers for early diagnosis and novel therapeutic targets. CeRNAs, encompassing both ncRNA and mRNA, present multifaceted targets. While Western medicine often faces challenges in achieving multi-target effects, traditional Chinese medicine (TCM) has shown significant advantages due to its complex composition and diverse targets. Thus, this study rigorously reviews the role of ceRNAs in HCC development and their potential as therapeutic targets, offering a novel perspective for researching TCM and non-TCM compound interventions in HCC (Supplementary Figure).
To our knowledge, protein-coding genes are predominantly responsible for genome functionality. Research indicates that the number of protein-coding genes in C. elegans (Caenorhabditis elegans) is analogous to that in humans (Baltimore, 2001). However, C. elegans remains a simpler organism, suggesting that protein-coding genes alone do not dictate the complexity of higher organisms. Additionally, the human genome contains approximately 30 times more genes than C. elegans, underscoring the crucial role of non-coding genome components in the sophistication and diversity of higher eukaryotes compared to simpler organisms (Costa, 2008; Mattick, 2009). This realization has intensified interest in non-coding genes within human genomics. Recent studies have elucidated notable changes in non-coding genes through comprehensive analyses of cancer genomes and transcriptomes (Futreal et al., 2004; Stratton et al., 2009; Beroukhim et al., 2010), fueling a surge in non-coding gene research in oncology. Despite evidence of a strong association between lncRNAs and certain cancer-related pathological processes, the broader implications of non-coding genes' functions and the potential non-coding roles of coding genes remain largely unexplored (Nagano and Fraser, 2011). Consequently, researchers are increasingly investigating the interactions between coding and non-coding genes. Emerging studies suggest that RNA molecules can modulate each other’s expression by competing for a limited supply of miRNA under specific conditions (Seitz, 2009; Poliseno et al., 2010). This led to the formulation of the ceRNA hypothesis, postulating that various RNA transcripts interact through a unique mechanism mediated by MRE, influencing the physiological or pathological progression of organisms (Figure 1).
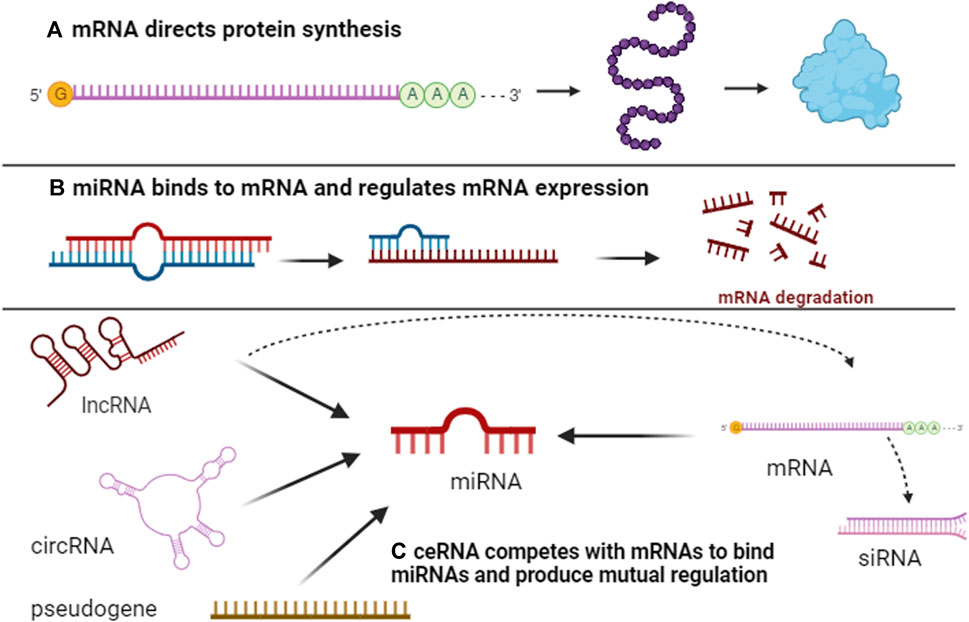
FIGURE 1. Overview of ceRNA networks. Initially, protein-coding gene mRNAs serve to carry genetic information, encode for protein synthesis, and execute biological functions (A). Subsequently, mRNAs interact with miRNAs through MRE, resulting in either mRNA degradation or inhibition of protein synthesis (B). Concurrently, ncRNAs, including lncRNAs, circRNAs, and pseudogenes, also possess MREs and establish a competitive dynamic with mRNAs. When ncRNAs bind more frequently to miRNAs, the latter’s binding to mRNAs is reduced, consequently mitigating mRNA degradation by miRNAs and facilitating gene interaction (C).
In ceRNA regulation, miRNAs are the focal point of competition, whereas coding genes' mRNAs or non-coding genes' lncRNA or circRNA with MRE are collectively identified as ceRNAs (Shi et al., 2021b; Chen et al., 2022c). These ceRNAs exhibit distinct biological characteristics and functions. While protein-coding genes' mRNAs are typically the downstream target genes in ceRNA networks and are primary targets for non-coding RNA interventions, they can also influence other mRNAs through MRE, creating a complex regulatory network and diversifying functions in biological and pathological contexts (Tay et al., 2014). MiRNA, approximately 22 nucleotides in length, regulate target genes at the transcriptional level by binding to MRE on target genes, leading to their downregulation or silencing (Bartel, 2009). MiRNA are known to modulate over 50% of mRNAs (Friedman et al., 2009; Thomas et al., 2010), playing a pivotal role in growth and the progression of various diseases, particularly cancer (Lu et al., 2008). LncRNAs, acting as sponges, guides, and regulators, influence transcription, chromatin structure, and histone modification, and are crucial in numerous biological processes, including epigenetic regulation, cell differentiation, motility, and apoptosis, thus drawing significant attention in gene regulation research (Abbasifard et al., 2020). CircRNAs, notable for their stable, ring-closed structure (Dong et al., 2023), are rich in MRE and excel as molecular "sponges", effectively countering miRNA-mediated inhibition (Kristensen et al., 2019). Pseudogenes, often disrupted by premature termination codons, frame-shifting mutations, insertions, or deletions, are traditionally regarded as "non-functional", "redundant", or "evolutionary remnants" (D'Errico et al., 2004). However, due to their high sequence similarity to miRNA-regulated known genes, pseudogenes actively compete in and contribute to ceRNA network regulation (Birney et al., 2007).
Advancements in epigenetic technologies have demonstrated that ncRNAs and ceRNA networks play a critical role in the pathogenesis of HCC. They are involved in regulating processes such as proliferation, invasion, metastasis, cell death, angiogenesis, the immune-tumor microenvironment, and chemoresistance (Figure 2).
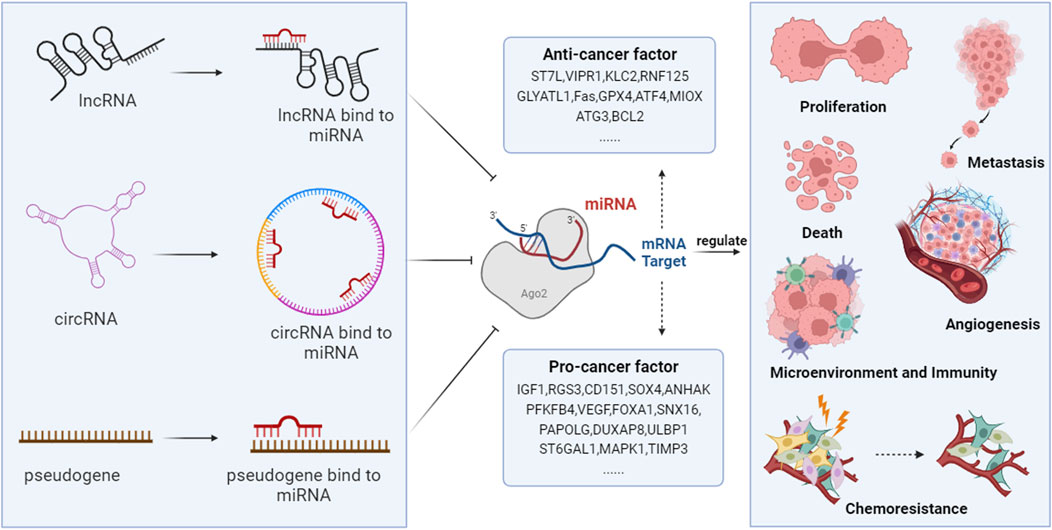
FIGURE 2. The ceRNA networks are involved in the development of HCC. NcRNAs, including lncRNAs, circRNAs, and pseudogenes, compete with coding RNAs for miRNA binding, as they all possess MRE. Through this mechanism, they regulate the degradation of miRNAs on target genes, thus modulating the expression of genes that are vital in controlling HCC’s proliferation, metastasis, angiogenesis, cell death, host immunity, tumor microenvironment, and chemotherapy resistance.
Uncontrolled proliferation is a primary characteristic of tumor cells, and numerous studied have indicated that ceRNA networks are crucial in regulating this proliferation. Non-coding RNAs regulate by competitively binding miRNAs to these proliferation-related genes, thereby affecting their transcription and expression. IGF1, a key cell growth-related factor, regulates growth, development, and the cell growth cycle, promoting cell proliferation, division, and regulating metabolism. Its expression was notably increased in various tumors (De Santi et al., 2016). LncRNA TUG1 acted as a ceRNA, sponging miR-1-3p to enhance IGF1 expression, thus facilitating HCC cell proliferation (Tang et al., 2022a). Transcription factors such as FOXA1 and E2F1 are known for their roles in carcinogenesis by modulating gene transcription and participating in processes like cell proliferation and differentiation (He et al., 2017). Studies showed that lncRNA MCM3AP-AS1 interacted with miR-194–5p, resulting in upregulated FOXA1, a target of miR-194–5p, which reduced HCC cell proliferation and cell cycle progression (Wang et al., 2019). Similarly, lncRNA APUE countered the inhibitory effect of miR-20b on E2F1, leading to increased E2F1 levels that foster cell cycle progression and tumor growth (Li et al., 2021c). Additionally, some ncRNAs induced protein signaling via the ceRNA network to boost HCC cell proliferation, such as UBE2MP1/miR-145–5p/RGS3 (Hao et al., 2022), lncRNA miat/miR-22–3p/Sirt1 (Zhao et al., 2019), and LINC00152/miR-143/KLC2 (Pellegrino et al., 2022). Furthermore, the pseudogene DUXAP8 encouraged HCC cell differentiation by modifying kinase function through DUXAP8/miR-490–5p/BUB1 (Zhang et al., 2020a). However, when certain lncRNAs target miRNAs of oncogenes, they often inhibit HCC proliferation, as observed with lncRNA MIR31HG (Yan et al., 2018) and lncRNA-AC079061.1 (Lin et al., 2022).
The aggressive and migratory behavior of tumors contributes to drug resistance and increased mortality rates. Tumor metastasis involves cell adhesion, extracellular matrix degradation, and phenotypic changes in tumor cells. NcRNAs can regulate HCC invasion by acting on relevant genes or signaling pathways through the ceRNA network. The epithelial-mesenchymal transition (EMT) is closely linked to HCC recurrence and metastasis, receiving increasing attention (Tam and Weinberg, 2013; Xie and Diehl, 2013). Signaling factors like CD151, SOX4, and FOSL2 are critical in EMT acquisition in HCC cells, enabling some ncRNAs to bind target miRNAs through sponging, thereby promoting HCC invasion and metastasis. Studies have demonstrated that PI3KC2 and LAMC1, as mRNAs, sequester miR-124, enhancing CD151 expression and facilitating HCC invasion and metastasis (Liu et al., 2016a). Circ-TLK1 and LncRNA HOSD-AS1 increased SOX4 expression by sponging miR-138–5p and miR-130a-3p, respectively, affecting EMT regulation and promoting aggressive HCC cell behavior (Wang et al., 2017; Lu Y. et al., 2022b). Circ0003998 bound miR-143–3p as a ceRNA both in vitro and in vivo, mitigating the inhibitory effect on EMT stimulator FOSL2 and advancing HCC invasion and metastasis (Song et al., 2020). Additionally, ANHAK influenced the EMT process through TGF-β, and circ0008194 acted as a sponge in the ceRNA network to promote HCC invasion (Liu et al., 2022c). RTKN2 and the Wnt/β-catenin signaling pathway contributed to tumor malignancy (Zhang et al., 2023). Has-circRNA-104348 indirectly regulated RTKN2 by binding miR-187–3p, promoting HCC lung metastasis in vivo (Huang et al., 2020). CeRNA networks are also involved in pathways like HCC cell metabolism and proliferation of metastatic foci, furthering HCC invasion and metastasis (Wei et al., 2020; Lai et al., 2021). Conversely, some ncRNA-mediated ceRNA networks inhibited HCC invasion by activating JAK1 polyubiquitination, increasing growth transcriptional repressors' expression, and enhancing glutamine metabolism (Lu et al., 2020; Bao et al., 2022; Peng et al., 2022b).
Cell death can be classified into programmed and non-programmed (necrosis) categories. Programmed cell death is an active process that can be inhibited by cell signaling inhibitors and includes apoptosis, autophagy, ferroptosis, mitotic catastrophe, bloat death, senescence-like cell death, and parapoptosis (D'Arcy, 2019; Kopeina and Zhivotovsky, 2022). Apoptosis, a well-researched programmed death mode, involves three signaling pathways, the death receptor, mitochondrial, and endoplasmic reticulum stress pathways, all culminating in caspase activation leading to apoptosis (Green, 2019). Autophagy, characterized by numerous autophagic vesicles in the cytoplasm, involves the fusion of these vesicles with lysosomal membranes for enzymatic degradation within lysosome (Kocaturk et al., 2019). Excessive autophagy results in autophagic cell death (Liu et al., 2023). Ferroptosis, a novel iron-dependent programmed cell death mode, primarily induces cell death through lipid peroxidation, catalyzed by divalent iron or ester oxygenase and affecting unsaturated fatty acids on cell membranes (Dixon et al., 2012). Conversely, necrosis, an uncontrolled cell death process unaffected by cell signaling inhibitors, occurs under physical or chemical environmental stimuli. Necrosis is marked by cell membrane disruption, cellular and organelle edema, absence of chromatin agglutination, and is often associated with inflammation and surrounding tissue damage (Park et al., 2023).
The ceRNA network in the apoptotic process of HCC cells exhibits varied roles depending on its target mRNAs, often leading to pro-apoptotic effects if targeting factors such as Fas (Zhang et al., 2017). When targeting apoptotic regulators, it inhibited apoptosis activation, as seen in the lncRNA SNHG6-003/miR-26a/b/TAK1 network (Cao et al., 2017). Similarly, acting on the FAK/Src/ERK pathway, it could inhibit apoptosis in HCC cells, exemplified by the lncRNA DBH-AS1/miR-138/FAK network (Bao et al., 2018).
Ferroptosis, characterized by iron-dependent lipid peroxidation, stands out as a distinct cell death process, differing from other regulated cell death forms in morphology, genetics, and biochemistry (Stockwell et al., 2017). The ceRNA network plays a significant role in the ferroptosis process in HCC cells. Recent studies found that ketamine induced ferroptosis in HCC cells through the Lnc-PVT1/miR-214–3p/GPX4 axis, reducing cell viability and proliferation (He et al., 2021). Knockdown of lncRNA HULC impacted HCC cell death, primarily by inducing ferroptosis via miR-3200-5p-mediated ATF4 targeting (Guan et al., 2022). LncRNA NEAT1 competitively bound to miR-362–3p, diminishing MIOX repression by miR-362–3p and increasing HCC cell susceptibility to ferroptosis (Zhang et al., 2022c). It has been suggested that lncRNA DNAJC27-AS1 and PPIF competitively bound miR-23b-3p, leading to an interaction that downregulates ferroptosis in HCC cells (Yang et al., 2023).
Autophagy involves engulfing cytoplasmic proteins or organelles and fusing them with lysosomes to form autophagic lysosomes, which degrade the contents, facilitating metabolism and cell renewal (Lee and Jang, 2015). Research indicated that autophagy was triggered in HCC under various stress conditions, such as hypoxia, nutrient deprivation, and chemotherapy, contributing to tumorigenesis (Codogno and Meijer, 2013; Cui et al., 2013). Autophagy-associated proteins ATG3 and ATG5 were involved in the lncRNA PVT1 and lncRNA HNF1A-AS1-mediated ceRNA network that promoted autophagy in HCC cells (Liu et al., 2016b; Yang et al., 2019).
Angiogenesis is essential for the growth and progression of HCC (Morse et al., 2019). This process occurs from the initial tumor stage through to metastasis. Vascular endothelial growth factor (VEGF) is a key angiogenic factor and is elevated in most malignant tumors (Han et al., 2021). The ceRNA network facilitates angiogenesis in HCC cells by targeting miRNAs that enhance VEGF and its receptor expression. For instance, lncRNA NEAT1 regulated angiogenesis in HCC by competing with VEGF for miR-125a-5p binding and modulating the AKT/mTOR and ERK pathways (Guo et al., 2022). Similarly, lncRNA MYLK-AS1 advanced HCC angiogenesis through the miR-424–5p/E2F7/VEGFR-2 pathway (Teng et al., 2020). In addition, ceRNA networks contributed to HCC angiogenesis by stimulating pro-angiogenic factors, such as the lncRNA MALAT1/miR-3064–5p/FOXA1 network (Zhang et al., 2020b). Moreover, research showed that cells lacking LINC00488 or with overexpressed miR-330–5p could suppress in vivo tumor growth and angiogenesis, highlighting LINC00488’s ceRNA role in HCC (Gao et al., 2019).
The tumor microenvironment (TME) is a dynamic internal environment that fosters tumor progression. Research indicates that altering the TME is a potential mechanism of action for anticancer drugs (Hanahan and Coussens, 2012). NcRNAs are pivotal in regulating innate and adaptive immunity (Carpenter et al., 2013). In HCC, the ceRNA network modulates the function of various immune cells, impacting host immunity and the TME (Zuo et al., 2021). For example, the lncRNA MIR4435-2HG/hsa-miR-1-3p/MMP9/hsa-miR-29–3p/DUXAP8 association with immune cell infiltration correlated with higher risk and worse outcomes in HCC (Zhang et al., 2021). Notably, NK cell infiltration is linked to tumor immune evasion. The LINC00638/miR-4732–3p/ULBP1 and lncRNA PART1/hsa-miR-125a-5p/CDK5R2 networks influenced NK cell function, fostering immune escape and vascular invasion in HCC (Qi et al., 2021; Cai et al., 2022). Additionally, the lncRNA MIR155HG enhanced tumor immune evasion induced by lipopolysaccharide through the miR-223/STAT1 axis, acting as a ceRNA (Peng et al., 2022a). Beyond NK cells, the ceRNA network also affects functional changes in B cells, T cells, and macrophages. For instance, LINC00261, by sponging miR105–5p, upregulated SELL expression in HCC, leading to altered B cell, T cell, and macrophage function (Song et al., 2022). Analysis of immune infiltration revealed that the lncRNA GSEC/miR-101–3p/SNX16/PAPOLG axis might influence macrophage polarization, a key factor in poor HCC prognosis (Hu et al., 2022). The ceRNA network related to CFHR4 regulated its expression, impacting the tumor immune microenvironment. High CFHR4 expression was significantly associated with increased dendritic cells, mast cells, neutrophils, NK cells, and T cells, underscoring its role in immune cell infiltration in HCC (Yu et al., 2022).
Chemotherapy is a pivotal treatment for tumors, yet its efficacy and side effects are influenced by patient-specific factors and tumor types. In HCC, characterized by high proliferation, metastasis, and drug resistance, some patients experience rapid deterioration post-chemotherapy. The role of ceRNA in cancer treatment resistance has become increasingly recognized.
Oxaliplatin, a primary chemotherapy agent for HCC, faces challenges with ceRNA-related resistance. Research indicated that lncRNA TINCR acted as a miR-195–3p sponge, reducing its inhibition of ST6GAL1. This triggers the NF-κB signaling pathway, thereby enhancing oxaliplatin resistance (Mei et al., 2022). Overexpression of lncRNA NR2F1-AS1 in oxaliplatin-resistant HCC tissues has been observed. It bound to miR-363, increasing ABCC1 expression and contributing to oxaliplatin resistance (Huang et al., 2018). Additionally, lncRNA KCNQ1OT1 regulated ABCC1 expression via miR-7-5p, promoting resistance in HepG2 and Huh7 cells (Hu et al., 2018). Thus, ABCC1 emerges as a crucial ceRNA target in combating oxaliplatin resistance in HCC. Moreover, lncRNA PCGEM1, HULC, and circFBXO11 have been implicated in oxaliplatin resistance through their respective ceRNA networks (Li et al., 2020; Chen J. et al., 2021a; Li et al., 2021b).
Cisplatin, another widely-used chemotherapy drug, has been the focus of studies on ceRNA network-related resistance in HCC. The lncRNA LEF1-AS1/miR-10a-5p/MSI1 network has been identified as a key player in cisplatin resistance. Through both in vitro and in vivo methods like gene knockdown or overexpression, it was found that LEF1-AS1 enhanced cisplatin resistance in HCC cells by targeting miR-10a-5p to elevate MSI1 expression and actuate the AKT signaling pathway (Gao et al., 2021).
Sorafenib is a leading drug for treating advanced HCC, extending the median survival of HCC patients by 12.3 months (Kudo et al., 2018). Yet, its effectiveness wanes due to resistance development. The upregulation of the pseudogene-derived lncRNA DUXAP8 has been linked to HCC progression and reduced sorafenib efficacy, principally through DUXAP8’s modulation of MAPK1 via miR-584–5p, thus activating the MAPK/ERK pathway (Liu et al., 2021). Additionally, circRNA-001241, by sequestering miR-21–5p, enhanced TIMP3 expression, contributing to sorafenib resistance in HCC (Yang and Wu, 2021). Furthermore, the lncRNAs TRERNA1, HANR, and TTN-AS1 were involved in sorafenib resistance, primarily via the RAS/Raf/MEK/ERK axis, promoting autophagy, and activating the PTEN/Akt pathway (Shi et al., 2020; Zhou et al., 2021c; Song et al., 2021). Conversely, circITCH and lncRNA FOXD2-AS1 may inhibit sorafenib resistance by targeting miR-20b-5p and miR-150–5p, underscoring a potential positive role of the ceRNA network in combating HCC chemotherapy resistance (Sui et al., 2019; Li et al., 2023). Further details about the ceRNA network in HCC are outlined in Table 1.
AFP is the most widely used serum biomarker for HCC diagnosis, crucial for early detection and disease monitoring. However, its diagnostic sensitivity is limited, specificity is suboptimal, and approximately 30% of early-stage HCC patients do not exhibit elevated AFP levels (Luo et al., 2020). Although advancements in liver ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI) have been significant, they are not suitable for routine screening in high-risk HCC individuals. Therefore, the identification of circulating biomarkers with high sensitivity, specificity, and ease of longitudinal monitoring is imperative.
With advancements in the study of the ceRNA network, its importance in the clinical diagnosis and prognosis of HCC has become evident. Abnormal expression of several ncRNAs and their associated ceRNA networks is often observed in HCC tissues and circulating fluids. One study utilized bioinformatics to construct a robust ceRNA network from public datasets, demonstrating the ZFAS1/hsa-miR-150–5p/GINS1 network’s significant contribution to HCC diagnosis and prognosis (Chen et al., 2022c). Additionally, research identified lncRNA MYCNOS, DLX6-AS1, LINC00221, CRNDE, and mRNAs CCNB1 and SHCBP1 as closely linked to HCC patient prognosis (Long et al., 2019). These findings underscore the potential of the ceRNA network in enhancing early diagnosis and predicting the outcome of HCC (Table 2).
Numerous studies have highlighted the significance of lncRNA in diagnosing and prognosticating HCC. The lncRNAs TUG1 and NEAT1, recently extensively researched, are emerging as novel biomarkers. TUG1, more effective than AFP for early HCC diagnosis, regulated AFP and served as a prognostic indicator for non-viral HCC (Lin et al., 2020). NEAT1, found to be diminished in hepatitis C and associated HCC, enhanced the diagnostic accuracy of HCC in hepatitis C patients when combined with TUG1, p53, and AFP (Mohyeldeen et al., 2020). TUG1 promoted SIX1 expression, increasing glycolysis and metastasis in HCC cells by interacting with miR-524–5p (Lu et al., 2022a) and activated the JAK/STAT3 pathway by targeting miR-144 (Lv et al., 2018). Additionally, NEAT1 influenced HCC progression by modulating the miR-9-BGH3 axis (Tripathi et al., 2022). Furthermore, the lncRNA CDKN2BAS/miR-153–5p/ARHGAP18 and RP11-386G11.10/miR-345–3p/HNRNPU networks, upregulated in HCC tissues, correlated with larger tumor size, advanced TNM stage, and poorer prognosis in patients (Chen et al., 2018; Xu et al., 2022). Also, a study identified four autophagy-related lncRNAs (LUCAT1, AC099850.3, ZFPM2-AS1, and AC009005.1) as prognostic markers in HCC. A predictive model developed using these lncRNAs was an independent prognostic factor and outperformed other clinicopathologic factors in predicting 1-, 3-, and 5-year survival rates (AUC 0.764, 0.738, and 0.717, respectively). LUCAT1 and ZFPM2-AS1 regulated autophagy in HCC cells through the miR-495–3p/DLC1 and miR-515–5p/DAPK2 axes, respectively (Wu et al., 2020). In summary, the ceRNA networks involving TUG1/miR-524–5p/SIX1, NEAT1/miR-9-BGH3, CDKN2BAS/miR-153–5p/MEK-ERK1/2, RP11-386G11.10/miR-345–3p/HNRNPU, and LUCAT1/miR-495–3p/DLC1 are promising for HCC diagnosis and prognosis.
CircRNAs also hold significant diagnostic value in HCC due to their stable expression, yielding numerous potential biomarkers for early HCC detection. Elevated levels of circ-0091579 in HCC tissues were strongly associated with reduced survival in patients (Zhang et al., 2018). Additionally, circ0091579 acted as an oncogenic factor in HCC development and progression by regulating ceRNA networks, including miR-136–5p/TRIM27 (Mao et al., 2022), miR-1287/PDK2 (Shu et al., 2021), miR-1225–5p/PLCB1 (Zhang et al., 2022a). The ceRNA networks of circ0016788, specifically circ0016788/miR-486/CDK4 and circ0016788/miR-506–3p/PAPR14, are known to significantly enhance HCC proliferation and invasion, presenting notable diagnostic potential for clinical research (Guan et al., 2019; Chen et al., 2021c). Similarly, the downregulated circGPR137B/miR-4739/FTO network indicated a poor prognosis for HCC patients (Liu et al., 2022b). Abnormal circRNA expression in the peripheral blood of HCC patients makes serum samples more conducive for early detection and diagnosis. A clinical study identified 12 distinct circRNAs in HCC patient blood, with three circRNAs (hsa_circ_0004001, hsa_circ_0004123, hsa_circ_0075792) demonstrating high sensitivity and specificity for HCC detection as evidenced by ROC analysis. These circRNAs regulated HCC progression by binding miRNAs and influencing VEGF/VEGFR, PI3K/AKt, mTOR, and Wnt signaling pathways (Sun et al., 2020). Furthermore, circ0000437 and circ0071662, aberrantly expressed in both HCC tissues and serum, are linked to tumor-node-metastasis (TNM) grade, differentiation degree, tumor size, and BCLC stage through their ceRNA networks circ0000437/miR-626/CDKN1B (Li and Liu, 2022) and circ0071662/miR-146b-3p/HPGD/NF2 (Abulizi et al., 2019) underlining their substantial clinical diagnostic relevance (Chen et al., 2022a; Wang et al., 2023). Therefore, the ceRNA network not only contributes to the pathogenesis of HCC but also plays a pivotal role in the clinical diagnosis and prognosis of HCC patients, offering biomarkers for early detection and ongoing monitoring.
Considering the pivotal role of the ceRNA network in HCC progression, it offers potential intervention targets for future HCC clinical treatments. As noted earlier, ncRNAs and their ceRNA networks are promising for drug-targeted therapy in HCC, attribute to their specific expression in HCC cells, reduced side effects, and regulatory targeting. TCM and modern molecular compounds have shown efficacy against HCC by influencing the ceRNA network (Figure 3). Currently, there is a scarcity of pharmacological research on treating HCC through ceRNA modulation. Research in TCM has mainly focused on active components rather than formulations, complemented by several studies on non-TCM products. With increasing insights into HCC’s epigenetic patterns, ceRNA networks are emerging as significant targets for novel drug development, particularly in TCM. Thus, the drugs have been categorized into two principal groups, TCM and non-TCM compounds, to elucidate their molecular mechanisms in HCC progression regulation via ceRNA networks.
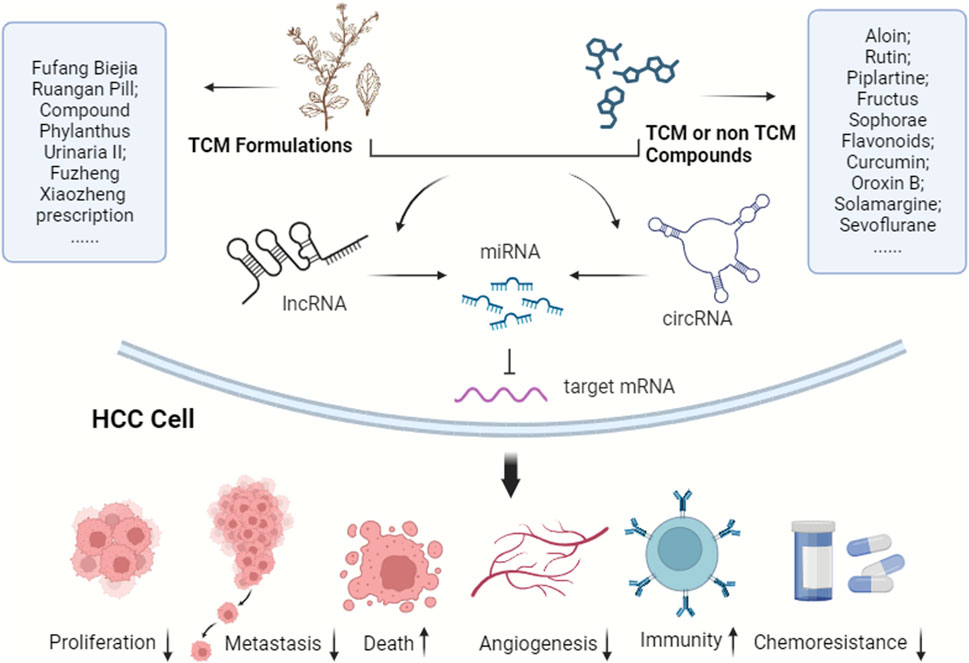
FIGURE 3. TCM and modern molecular compounds modulate the ceRNA network to inhibit HCC progression. These ingredients play crucial roles in various aspects, including suppressing HCC cell proliferation, invasion, and metastasis, inducing cell death, hindering angiogenesis, bolstering immunity, improving the tumor microenvironment, and reducing chemotherapy resistance.
The significant potential of TCM in HCC treatment necessitates an understanding of its mechanisms, particularly in complex herbal formulations. However, the complexity of these formulations and their multi-site, multi-target effects make it challenging to fully comprehend their therapeutic mechanisms through a single aspect or pathway. Advances in transcriptomics have revealed the involvement of ceRNA networks at various pathological stages of HCC, providing insights into HCC pathogenesis at the gene regulation level and presenting potential targets for TCM intervention. It has been established that ceRNA networks play a comprehensive role in TCM’s regulatory processes against HCC.
Chinese compounds, key constituents of TCM, have garnered significant attention in pharmacological research for their varied pharmacological properties. TCM employs heat-clearing and detoxifying agents for effective cancer cell elimination, inflammation reduction, and tumor growth inhibition. Scutellaria barbata D. Don (SB) and Oldenlandia diffusa (Willd.) Roxb (OD), commonly used in TCM to treat tumors by clearing heat and toxins, have been shown to inhibit hepatitis B virus-associated HCC growth in vitro and in vivo by affecting circRNAs and related ceRNA networks. Their active components, particularly total flavonoids, lignans, and apigenin, played a pivotal role in ceRNA network regulation (Yang et al., 2021). Aloin, a compound found in aloe vera plant, could inhibit HCC cell malignant phenotype such as proliferation, invasion, and promote apoptosis and autophagy by regulating the circ0011385/miR-149–5p/WT1 axis (Fu et al., 2021). Other studies, including those on Solasonine, Curcumin, and Piplartine, demonstrated the potential of these TCM-derived active ingredients against HCC through the ceRNA network regulation, lncRNA CCAT1/miR-375–3p/SP1 (Liu et al., 2019b), circ0078710/miR-378b/PRIM2 (Chen et al., 2022b), and circ100338/miR-141–3p/ZEB1 (Cheng et al., 2020), respectively. Moreover, the ceRNA network involving lncRNA BANCR/miR-590–5p/OLR1 was implicated in the mechanism by which Rutin, a TCM active ingredient, mitigated sorafenib resistance in HCC (Zhou et al., 2021a). Furthermore, TCM active compounds frequently interact with parts of ceRNA networks. LncRNA GAS5 contained miR-21 response elements and competed with miR-21, upregulating its target genes PDCD4 and PTEN for anticancer effects (Hu et al., 2016). Studies indicated that resveratrol (Li et al., 2014), matrine (Luo et al., 2017), oxymatrine (Huang et al., 2014), and ursolic acid (Liu et al., 2015) all reduced miR-21 expression in HCC cells, inhibiting proliferation and promoting apoptosis, suggesting their involvement in the lncRNA GAS5/miR-21/PDCD4 pathway for anti-HCC action. Sophorae Fructus flavonoids regulated the lncRNA FBXL19-AS1/miR-342–3p axis, inhibiting HCC cell proliferation and invasion (Jin et al., 2020). LncRNA ATB bound competitively to the miR-200 family, elevating ZEB1 and ZEB2 levels, inducing EMT, and promoting HCC cell invasion (Yuan et al., 2014). HOTAIR contained MRE of miR-1 and miR-218 and negatively regulated the expression of miR-1 and miR-218 (Su et al., 2016), increased the levels of their target gene EZH2 (Fu et al., 2015), which played a carcinogenic role. Research has suggested that Curcumin and Curcumol’s anti-HCC mechanism may be closely related to the inhibition of miR-200 expression (Liang et al., 2013) and the HOTAIR/miR-218-2/EZH2 pathway (Tian et al., 2021), respectively. Oroxin B, a flavonoid from Oroxylum indicum (L.) Vent, demonstrated anti-HCC activity and improved HCC pathology by reducing miR-221 levels and deactivating the PTEN/PI3K/Akt pathway (Li et al., 2019). Meanwhile, the tumor-suppressive lncRNA CASC2 bound miR-221, influencing Caspase-3 expression and affecting HCC cell growth (Sun et al., 2022a). These studies illustrate the extensive role of ceRNA networks as targets in the anti-HCC mechanisms of TCM’s active ingredients.
Compared to individual herbs or active compounds, TCM formulations can exert a more synergistic and comprehensive effect on the body. In TCM practice, these formulations are essential. Numerous clinical and preclinical studies have validated the significant efficacy of TCM formulations in treating HCC. The Jiedu Quyu Kangai Recipe, for instance, has been shown to improve clinical parameters following hepatic artery embolization and to modulate the immune system (Zhang et al., 2020c). The Dahuang Zhechong Pill, when combined with transcatheter arterial chemoembolization (TACE), enhanced liver function, boosted immunity, reduced metastasis risk, and alleviated adverse reactions in HCC treatment (Dai et al., 2021). Similarly, the Qinggan Huadu Zhuyu Decoction significantly ameliorated clinical symptoms, bolstered immune function, and extended survival time in post-TACE patients (Shi et al., 2021a). Mechanistic studies on TCM formulations have explored various pathways, including inducing tumor cell apoptosis, autophagy, and ferroptosis, arresting the cell cycle, reducing proliferation, impairing telomerase activity, counteracting multidrug resistance in tumors, and remodeling the tumor microenvironment and immune responses (Lin et al., 2017; Wang et al., 2020). With the advent of ceRNA research, some studies have begun to examine the gene network regulatory mechanisms of TCM formulations. For example, the Fufang Biejia Ruangan Pills have been found to potentially disrupt the malignant progression from liver fibrosis to liver cancer by regulating the lncRNA TUG1/miR-328–3p/SRSF9 ceRNA regulatory axis (Liu, 2022). The Compound Phylanthus urinaria Ⅱ has been shown to inhibit HepG2 cell proliferation by modulating the ceRNA network lncRNA CCAT1/miR-let-7a/HMGA2/Cyclin D1 axis, thereby enhancing the inhibitory effect of miR-let-7a on HMGA2 and Cyclin D1 expression, manifesting its anti-HCC properties (Chen et al., 2021b). In our previous research, transcriptomic analysis and qRT-PCR validation revealed that the Fuzheng Xiaozheng prescription could inhibit HCC proliferation and reduce hepatic inflammation via circRNA and associated ceRNA networks (Liu et al., 2022a). Overall, while the modulation of the ceRNA network by TCM formulations shows promise, research in this area is still sparse and warrants further exploration in the future.
In addition to TCM preparations, several natural compounds are known to regulate the ceRNA network. Three potential HCC therapeutic agents—decitabine, BW-B70C, and gefitinib—have been identified through big data mining and computational biology as promising for HCC treatment by modulating the circRNA-associated ceRNA network, though they require experimental validation (Xiong et al., 2018). Solamargine, a naturally occurring alkaloid, has been studied for its anti-HCC properties. It inhibited HCC growth and reversed sorafenib resistance via the ceRNA network HOTTIP-TUG1/miR-4726–5p/MUC1 (Tang et al., 2022b), and induced apoptosis and autophagy in HCC cells by suppressing the oncogenic factor LIF and modulating the miR-192–5p/CYR61/Akt signaling axis, repolarizing tumor-associated macrophages, and influencing other immune cells in the tumor microenvironment (Yin et al., 2022). Sevoflurane, a commonly used anesthetic, has been found to inhibit proliferation, invasion, and migration, and to promote apoptosis in HCC. Studies indicated that sevoflurane inhibited HCC progression by regulating the ceRNA networks circ0001649/miR-19a-3p/SGTB axis (Sun et al., 2023) and lncRNA KCNQ1OT1/miR-29a-3p CBX3 (Zhou et al., 2021b). Cyanidin-3-glucoside (C3G), extracted from Morus alba L., possesses notable antioxidant and antitumor activities. C3G could prevent diethylnitrosamine and acetamidofluorene-induced rat HCC precancerous lesions by regulating the autophagy protein ATG16L1 and its ceRNA network (circ0001345/miR-106b/ATG16L1), improving hepatic function and histopathological damage, thereby offering potential against HCC (Zabady et al., 2022). More detailed information on anti-HCC properties of TCM or non-TCM compounds is provided in Table 3.
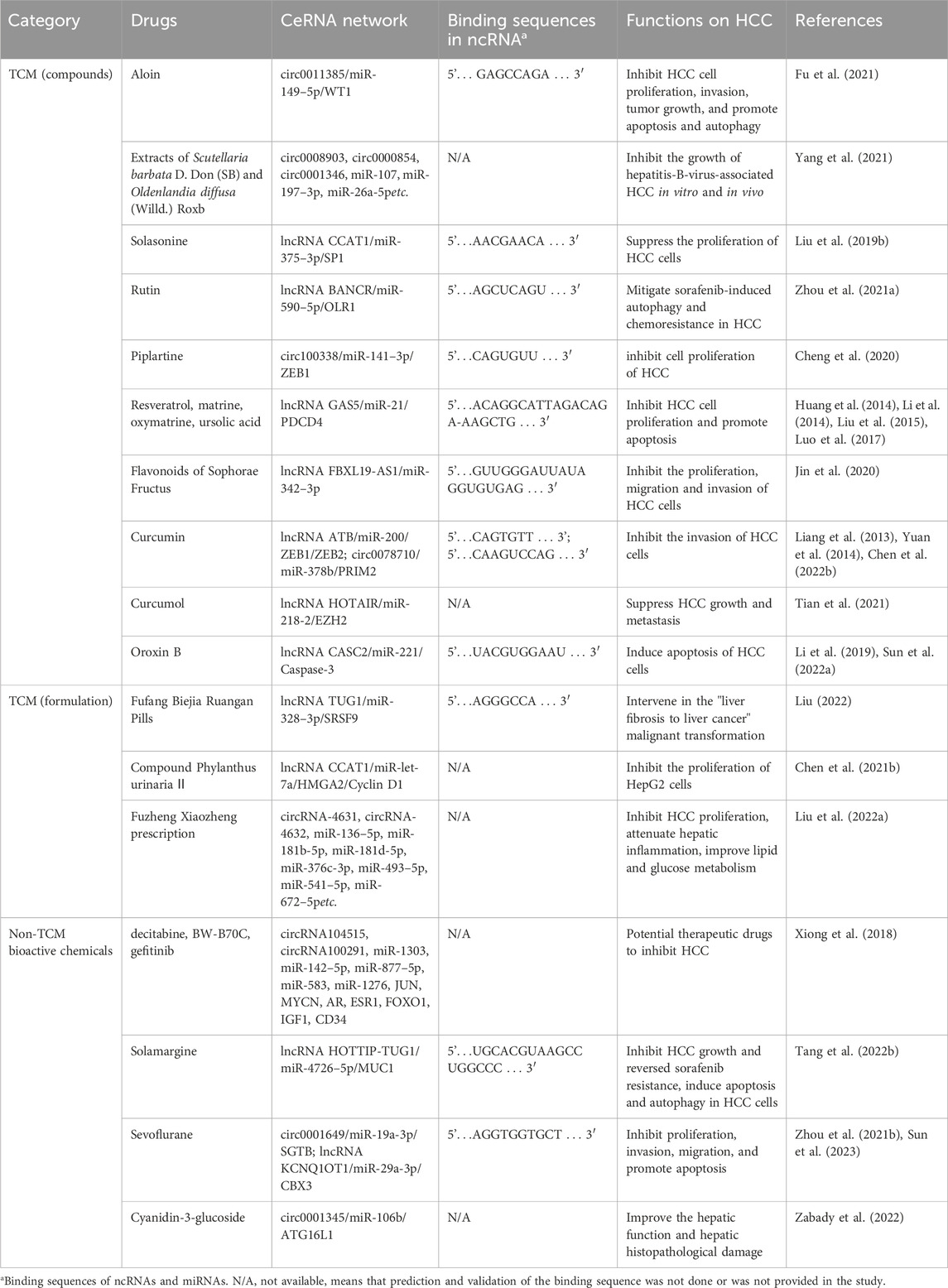
TABLE 3. TCM compounds and non-TCM compounds experimentally demonstrated to inhibit HCC by modulating the ceRNA network.
The progression from normal liver tissue to HCC is a lengthy and gradual process influenced by various factors, including hepatitis B or C virus infection, fat deposition, autoimmune disorders, alcoholism, and aflatoxin exposure. These factors can cause hepatocyte damage, followed by regeneration. However, persistent injury leads to continuous repair, eventually resulting in the development of fibrotic and proliferative nodules, precursors to HCC. As these hyperplastic nodules advance, characterized by liver lobular remodeling and abnormal cell proliferation, they may develop into overt HCC. Despite advancements in medical technology, the pathogenesis of HCC remains not fully understood. Therefore, investigating the regulatory mechanisms of HCC and identifying molecular targets for diagnosis, treatment, and prognosis are crucial for preventing and treating HCC.
As research into ncRNAs has deepened, genes like lncRNA and miRNA, once considered “noise”, have been increasingly recognized as pivotal in HCC development. The ceRNA network is extensively involved in various HCC pathological processes, including proliferation, migration, invasion, cell death, angiogenesis, and chemotherapy resistance. This review summarized the numerous ncRNAs and ceRNA networks involved in the development, diagnosis, and treatment of HCC, but most of them are only preliminary findings and the studies are superficial. Table 1, Table 2, Table 3 showed that certain ncRNAs played multiple roles, indicating that their roles have been widely confirmed and were better understood. LncRNAs such as TUG1 and NEAT1, along with their associated ceRNA networks, are extensively studied in HCC. They contributed significantly to HCC development, offering early diagnostic value and serving as targets in drug-regulatory mechanisms, potentially representing some of the most important ncRNAs in HCC (Bu et al., 2020; Farzaneh et al., 2022). TUG1, first identified in the retina of taurine-treated mice, is engaged in a variety of physiopathological processes involving the cardiovascular, respiratory, digestive, and nervous systems, and has important regulatory roles in a variety of tumor cells (Guo et al., 2020). TUG1 is located on chromosome 22q12.2 and includes 4 exons. Although TUG1 has 20 transcripts, 19 of them do not have the ability to encode proteins (Farzaneh et al., 2022). Previous studies have shown that TUG1 played an oncogenic role in tumors such as colorectal (Ibrahim et al., 2023), prostate (Yu et al., 2019), bladder (Tan et al., 2022), lung (Li et al., 2021a), oral squamous cell (Jiang et al., 2022), and ovarian cancers (Zhan et al., 2020), as well as being involved in the pathogenesis of non-cancerous diseases such as renal ischemia-reperfusion injury (Sun et al., 2022b), pulmonary fibrosis (Qi et al., 2023), and myocardial infarction (Dang et al., 2023). Thus, TUG1 could act as a pathogenic factor in a tissue-specific or environment-specific manner. Moreover, TUG1 performed a critical role in HCC by targeting multiple ceRNA networks. For example, TUG1/hsa-miR-582–5p/Siglec-15 contributed immunosuppression in HCC (Ren et al., 2022), TUG1/miR-137/AKT2 promoted the EMT process (Li et al., 2021d), TUG1/miR-335–5p/CXCR4 facilitated the activation of Wnt signaling pathway (Xie et al., 2020), TUG1/miR-328–3p/SRSF9 caused malignant progression of HCC (Liu Y. et al., 2022d) and was effectively inhibited as a target of prescription FufangBiejia Ruangan Pill. In brief, TUG1 could be a potential target for the diagnosis and prevention of HCC. As well, NEAT1, a lncRNA enriched in the nucleus, was discovered in 2007 in the paracrine cristae (Hutchinson et al., 2007). It has two transcripts, an unspliced non-coding transcript approximately 4 kb in length, and a lower-expression isoform greater than 17 kb. NEAT1 is localized to the pars compacta and has been extensively studied in a variety of cancers and its role as an oncogenic factor has been confirmed (Klec et al., 2019). Its mediated ceRNA network could promote the growth of HCC cells by inhibiting their senescence (Chen et al., 2023), regulating aerobic glycolysis (Zhang et al., 2022b), modulating sorafenib resistance (Niu et al., 2020), enhancing radioresistance in HCC (Chen and Zhang, 2019), and inhibiting the function of CD8+T cells (Yan et al., 2019). However, the most promising aspect of the current study is its early diagnosis and prognosis determination of HCC, and it has been elucidated in this review that the combined diagnostic model of NEAT1 in combination with TUG1 and AFP significantly outperformed the diagnostic efficiency of using AFP alone (Mohyeldeen et al., 2020). NEAT1 was significantly elevated in the serum of HCC patients, and ROC curve analysis showed AUC = 0.981, p < 0.001, sensitivity 100%, specificity 88.9% (Shaker et al., 2019). Additionally, The lncRNA CCAT1 is a typical pro-carcinogenic gene in HCC (Zhang et al., 2019), and is also repeatedly mentioned throughout this review. CCAT1 was first identified in colon cancer and mapped to chromosome 8q24.2. The CCAT1 gene encodes two isozymes, the short isoform CCAT1-S and the long isoform CCAT1-L. CCAT1-L is highly expressed mainly in the nucleus, whereas CCAT1-S is mainly found in the cytoplasm, and a positive correlation is likely to exist between the two (Liu et al., 2019a). The CCAT1-associated ceRNA network was involved in HCC cells autophagy (Guo et al., 2019), chemotherapeutic resistance (Xia et al., 2022), and promotion of tumor-associated macrophage polarization (Deng et al., 2020), and bound to C-Myc as an independent risk factor for HCC patients (Zhu et al., 2015). Furthermore, its related ceRNA network could be modulated by TCM and chemical drugs to produce anti-HCC effects, underscoring its research significance in HCC in this review. Alternatively, several target genes within the ceRNA network are noteworthy. The FOX gene family, regulated by various ncRNAs, and its related ceRNA networks such as lncRNA MCM3AP-AS1/miR-194–5p/FOXA1, lncRNA MFI2-AS1/miR-134/FOXM1, lncRNA MALAT1/miR-3064–5p/FOXA1, and circFBXO11/miR-605/FOXO3, played significant roles in HCC’s biological processes like proliferation, invasion, metastasis, and drug resistance, suggesting that FOX genes may be key targets for HCC prevention and treatment.
The discovery of key ncRNAs and ceRNA networks in HCC has broadened the scope of research beyond specific pathways, offering a comprehensive view of HCC’s malignant progression from a gene regulation standpoint. This paradigm shift has enhanced our understanding of HCC’s complex pathology, introducing new perspectives into its pathogenesis and enriching the research field. However, the study of ncRNAs is still nascent compared to that of protein-coding genes. While numerous specific ncRNAs and ceRNA networks have been identified in HCC, their functions are not fully understood, and their role remains confined to molecular expression levels. This limitation hinders the identification of ncRNAs with critical roles in HCC development. Furthermore, due to incomplete understanding, methods for detecting and characterizing functional ncRNAs are underdeveloped and need further refinement.
Research on the molecular mechanisms governing ceRNA network regulation in HCC is just beginning. Although the functions of ncRNAs and their role as ceRNA sponges are increasingly recognized, most studies are conducted at the cellular level using overexpression and knockdown assays. The variations in ceRNA interactions across different stages, populations, and their contributions to tumor aggressiveness and therapeutic resistance are yet to be clarified. Additionally, the human body’s complexity and various factors that can affect ceRNA activity, such as cellular localization, concentration of ceRNA components, interactions with RNA-binding proteins, RNA transcription, and ceRNA affinity in endogenous cellular environments, necessitate more preclinical experiments and clinical trials (Shi et al., 2021b). Moreover, while current research often depicts ceRNA interactions as singular, the existence of large, interconnected networks suggests significant secondary and indirect interactions that could substantially impact ceRNA regulation, warranting further investigation. Therefore, integrating advanced single-cell technologies, bioinformatics, and computer-aided modeling may elucidate the role of ceRNA interactions in tumor heterogeneity, offering profound clinical insights.
CeRNA networks, due to their impact on various aspects of HCC progression, present novel biomarkers or targets for HCC diagnosis and treatment. They have also elucidated the molecular mechanisms underlying the anticancer effects of drugs, particularly TCM, which is known for its multi-target, multi-process, and multi-effect approach. This insight offers a new perspective on the mechanistic study of TCM in HCC treatment, enhancing basic research and its application prospects. However, studies exploring drug modulation mechanisms within the ceRNA network are limited, possibly due to the nascent state of ceRNA research. Current investigations primarily focus on active compounds or specific segments of the ceRNA network. Concentrating solely on a single Chinese herb or an active component is inconsistent with the holistic principles of TCM, and examining a singular part of the ceRNA network overlooks the broader gene transcription interactions. Systematic research into TCM formulations through ceRNA networks remains underdeveloped. This gap can be attributed to the complexity of multi-ingredient herbal formulations, difficulty in identifying specific drug targets, and challenges inherent in TCM formulations, all of which pose significant challenges in TCM research. The diverse chemical components of TCM generate multiple biological effects through their interactions, complicating the identification of their material basis and action targets against HCC. Future research should, therefore, leverage modern techniques guided by TCM theory to unravel the ceRNA action mechanisms of TCM formulations against tumors. Employing genomics, transcriptomics, and bioinformatics will enable a comprehensive analysis of the ceRNA network mechanisms of TCM components in HCC intervention, systematically elucidating TCM’s material basis against HCC. As modern technology continues to advance, the roles of ceRNA regulation in HCC development and the mechanisms by which TCM treats HCC will become increasingly clear.
In conclusion, advancements in gene epigenetic technology have expanded research into ncRNAs, uncovering the regulatory mechanisms of the ceRNA network. Present research trends indicate that lncRNA TUG1, lncRNA NEAT1, and lncRNA CCAT1 are among the most extensively studied ncRNAs in HCC, owing to the significant role of their associated ceRNA networks, as confirmed by numerous studies. These insights not only enhance our understanding of HCC’s pathogenesis and progression from a gene transcriptional regulation standpoint but also pave the way for novel diagnostic, monitoring, and treatment approaches for HCC. Most notably, ceRNAs offer therapeutic targets for cancer treatment. Investigating the anticancer mechanisms of TCM or molecular compounds through the lens of the ceRNA network aids in systematically elucidating the material basis of TCM and its active components. However, it is imperative to integrate more contemporary medical technologies and undertake animal or clinical trials to explore the in vivo interactions of the ceRNA network and the systematic regulation of the ceRNA network by drugs, including TCM.
ZT: Conceptualization, Writing–review and editing. XuL: Data curation, Writing–review and editing. YZ: Methodology, Resources, Writing–review and editing. JL: Investigation, Visualization, Writing–review and editing. CL: Data curation, Funding acquisition, Writing–review and editing. XiL: Formal Analysis, Methodology, Writing–original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Sichuan Province (No.24NSFSC4811), the Special Support Less Developed Regions General Program of China Postdoctoral Science Foundation (No. 2021M693786), Program of Science and Technology Department of Sichuan Province (No. 2021JDRC0173).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1341999/full#supplementary-material
AFP, Alpha-fetoprotein; AFP-L3, fetoprotein lectin 3; ATG3, autophagy-related gene 3; ATG5, autophagy-related gene 5; C. elegans, Caenorhabditis elegans; ceRNA, competing endogenous RNA; CT, computer tomography; circRNA, circular RNA; C3G, cyanidin-3-glucoside; DCP, des-gamma-carboxyprothrombin; EMT, epithelial-mesenchymal transition; GPC3, glypican-3; HCC, hepatocellular carcinoma; lncRNA, long non-coding RNA; miRNA, microRNA; MRI, magnetic resonance imaging; MRE, miRNA response elements; ncRNA, non-coding RNA; NK cells, natural killer cells; NF-κB, nuclear factor-kappa B; OD, Oldenlandia diffusa (Willd.) Roxb; OLR1, oxidized low-density lipoprotein receptor 1; SB, Scutellaria barbata D. Don; TCM, traditional Chinese medicine; TME, tumor microenvironment; TACE, transcatheter arterial chemoembolization; VEGF, vascular endothelial growth factor; 3'UTR, 3' untranslated region.
Abbasifard, M., Kamiab, Z., Bagheri-Hosseinabadi, Z., and Sadeghi, I. (2020). The role and function of long non-coding RNAs in osteoarthritis. Exp. Mol. Pathol. 114, 104407. doi:10.1016/j.yexmp.2020.104407
Abulizi, R., Li, B., and Zhang, C. G. (2019). Circ_0071662, a novel tumor biomarker, suppresses bladder cancer cell proliferation and invasion by sponging miR-146b-3p. Oncol. Res. 2019, 88. doi:10.3727/096504019X15740729375088
Bao, J., Chen, X., Hou, Y., Kang, G., Li, Q., and Xu, Y. (2018). LncRNA DBH-AS1 facilitates the tumorigenesis of hepatocellular carcinoma by targeting miR-138 via FAK/Src/ERK pathway. Biomed. Pharmacother. 107, 824–833. doi:10.1016/j.biopha.2018.08.079
Bao, L., Li, P. F., Zhao, H. Y., Chen, L., Wang, Y. B., Liang, S., et al. (2022). Pseudogene PLGLA exerts anti-tumor effects on hepatocellular carcinoma through modulating miR-324-3p/GLYATL1 axis. Dig. Liver Dis. 54, 918–926. doi:10.1016/j.dld.2021.10.003
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi:10.1016/s0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi:10.1016/j.cell.2009.01.002
Beroukhim, R., Mermel, C. H., Porter, D., Wei, G., Raychaudhuri, S., Donovan, J., et al. (2010). The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905. doi:10.1038/nature08822
Bu, F. T., Wang, A., Zhu, Y., You, H. M., Zhang, Y. F., Meng, X. M., et al. (2020). LncRNA NEAT1: shedding light on mechanisms and opportunities in liver diseases. Liver Int. 40, 2612–2626. doi:10.1111/liv.14629
Cai, S., Du, R., Zhang, Y., Yuan, Z., Shang, J., Yang, Y., et al. (2022). Construction and comprehensive analysis of ceRNA networks and tumor-infiltrating immune cells in hepatocellular carcinoma with vascular invasion. Front. Bioinform 2, 836981. doi:10.3389/fbinf.2022.836981
Cao, C., Zhang, T., Zhang, D., Xie, L., Zou, X., Lei, L., et al. (2017). The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene 36, 1112–1122. doi:10.1038/onc.2016.278
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. doi:10.1126/science.1112014
Carpenter, S., Aiello, D., Atianand, M. K., Ricci, E. P., Gandhi, P., Hall, L. L., et al. (2013). A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792. doi:10.1126/science.1240925
Chan, J. J., and Tay, Y. (2018). Noncoding RNA:RNA regulatory networks in cancer. Int. J. Mol. Sci. 19, 1310. doi:10.3390/ijms19051310
Chen, D., Wang, J., Li, Y., Xu, C., Fanzheng, M., Zhang, P., et al. (2023). LncRNA NEAT1 suppresses cellular senescence in hepatocellular carcinoma via KIF11-dependent repression of CDKN2A. Clin. Transl. Med. 13, e1418. doi:10.1002/ctm2.1418
Chen, G. J., Xie, D. H., Zhang, P., and Zhou, H. K. (2022a). Circular RNA hsa_circ_0000437 may be used as a new indicator for the diagnosis and prognosis of hepatocellular carcinoma. Bioengineered 13, 14118–14124. doi:10.1080/21655979.2022.2081458
Chen, J., Yuan, D. Y., Hao, Q. Y., Zhu, D. M., and Chen, Z. (2021a). LncRNA PCGEM1 mediates oxaliplatin resistance in hepatocellular carcinoma via miR-129-5p/ETV1 axis in vitro. Adv. Clin. Exp. Med. 30, 831–838. doi:10.17219/acem/135533
Chen, J. Z., Huang, X. T., Wang, W. J., Xie, H. C., Li, J. F., Hu, Z. F., et al. (2018). LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging-Us 10, 3371–3381. doi:10.18632/aging.101645
Chen, L. W., Li, Q. M., Li, X. H., and Li, C. Q. (2021b). Effect of compound Phylanthus urinaria Ⅱ in regulating expression of lncRNA CCAT1 in hepatocellular carcinoma HepG2 cells. Chin. J. Exp. Tradit. Med. Formulae 27, 74–79. doi:10.13422/j.cnki.syfjx.20202123
Chen, M., Hu, G. S., Zhou, X., Peng, Z., and Wen, W. (2021c). Hsa_circ_0016788 regulates hepatocellular carcinoma progression via miR-506-3p/poly-adenosine diphosphate-ribose polymerase. J. Gastroen Hepatol. 36, 3457–3468. doi:10.1111/jgh.15635
Chen, Q., Guo, H., Zong, Y., and Zhao, X. (2022b). Curcumin restrains hepatocellular carcinoma progression depending on the regulation of the circ_0078710/miR-378b/PRIM2 axis. J. Recept Signal Transduct. Res. 42, 313–324. doi:10.1080/10799893.2021.1936554
Chen, S. S., Zhang, Y. C., Ding, X. Y., and Li, W. (2022c). Identification of lncRNA/circRNA-miRNA-mRNA ceRNA network as biomarkers for hepatocellular carcinoma. Front. Genet. 13, 838869. doi:10.3389/fgene.2022.838869
Chen, X., and Zhang, N. (2019). Downregulation of lncRNA NEAT1_2 radiosensitizes hepatocellular carcinoma cells through regulation of miR-101-3p/WEE1 axis. Cell Biol. Int. 43, 44–55. doi:10.1002/cbin.11077
Cheng, X. L., Tian, P., Zheng, W. Z., and Yan, X. T. (2020). Piplartine attenuates the proliferation of hepatocellular carcinoma cells via regulating hsa_circ_100338 expression. Cancer Med-Us 9, 4265–4273. doi:10.1002/cam4.3043
Codogno, P., and Meijer, A. J. (2013). Autophagy in the liver. J. Hepatol. 59, 389–391. doi:10.1016/j.jhep.2013.02.031
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigo, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi:10.1038/nature05874
Costa, F. F. (2008). Non-coding RNAs, epigenetics and complexity. Gene 410, 9–17. doi:10.1016/j.gene.2007.12.008
Cui, J. Z., Gong, Z. Y., and Shen, H. M. (2013). The role of autophagy in liver cancer: molecular mechanisms and potential therapeutic targets. Biochimica Biophysica Acta-Reviews Cancer 1836, 15–26. doi:10.1016/j.bbcan.2013.02.003
Dai, C. M., Jin, S., and Zhang, J. Z. (2021). Effect of Dahuang Zhechong Pills combined with TACE on VEGF, MMP-2, TGF-β1, and immune function of patients with primary liver cancer (blood stasis and collaterals blocking type). China J. Chin. Mater Med. 46, 722–729. doi:10.19540/j.cnki.cjcmm.20200716.501
Dang, Y., Hua, W., Zhang, X., Sun, H., Zhang, Y., Yu, B., et al. (2023). Anti-angiogenic effect of exo-LncRNA TUG1 in myocardial infarction and modulation by remote ischemic conditioning. Basic Res. Cardiol. 118, 1. doi:10.1007/s00395-022-00975-y
D'arcy, M. S. (2019). Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 43, 582–592. doi:10.1002/cbin.11137
Deng, L., Huang, S., Chen, B., Tang, Y., Huang, F., Li, D., et al. (2020). Tumor-linked macrophages promote HCC development by mediating the CCAT1/let-7b/HMGA2 signaling pathway. Onco Targets Ther. 13, 12829–12843. doi:10.2147/OTT.S283786
D'errico, I., Gadaleta, G., and Saccone, C. (2004). Pseudogenes in metazoa: origin and features. Brief. Funct. Genomic Proteomic 3, 157–167. doi:10.1093/bfgp/3.2.157
De Santi, M., Annibalini, G., Barbieri, E., Villarini, A., Vallorani, L., Contarelli, S., et al. (2016). Human IGF1 pro-forms induce breast cancer cell proliferation via the IGF1 receptor. Cell Oncol. (Dordr). 39, 149–159. doi:10.1007/s13402-015-0263-3
Di Leva, G., Garofalo, M., and Croce, C. M. (2014). MicroRNAs in cancer. Annu. Rev. Pathol. 9, 287–314. doi:10.1146/annurev-pathol-012513-104715
Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. doi:10.1016/j.cell.2012.03.042
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi:10.1038/nature11233
Dong, J. N., Zeng, Z. E., Huang, Y., Chen, C. P., Cheng, Z. E., and Zhu, Q. B. (2023). Challenges and opportunities for circRNA identification and delivery. Crit. Rev. Biochem. Mol. 58, 19–35. doi:10.1080/10409238.2023.2185764
Farzaneh, M., Ghasemian, M., Ghaedrahmati, F., Poodineh, J., Najafi, S., Masoodi, T., et al. (2022). Functional roles of lncRNA-TUG1 in hepatocellular carcinoma. Life Sci. 308, 120974. doi:10.1016/j.lfs.2022.120974
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi:10.1101/gr.082701.108
Fu, D. Z., Ji, Q., Wang, C. X., Yu, L., and Yu, R. (2021). Aloin decelerates the progression of hepatocellular carcinoma through circ_0011385/miR-149-5p/WT1 axis. Cell Cycle 20, 2476–2493. doi:10.1080/15384101.2021.1988227
Fu, W. M., Zhu, X., Wang, W. M., Lu, Y. F., Hu, B. G., Wang, H., et al. (2015). Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J. Hepatol. 63, 886–895. doi:10.1016/j.jhep.2015.05.016
Futreal, P. A., Coin, L., Marshall, M., Down, T., Hubbard, T., Wooster, R., et al. (2004). A census of human cancer genes. Nat. Rev. Cancer 4, 177–183. doi:10.1038/nrc1299
Gao, J., Dai, C., Yu, X., Yin, X. B., and Zhou, F. (2021). Long noncoding RNA LEF1-AS1 acts as a microRNA-10a-5p regulator to enhance MSI1 expression and promote chemoresistance in hepatocellular carcinoma cells through activating AKT signaling pathway. J. Cell Biochem. 122, 86–99. doi:10.1002/jcb.29833
Gao, J., Yin, X. B., Yu, X., Dai, C., and Zhou, F. (2019). Long noncoding RNA LINC00488 functions as a ceRNA to regulate hepatocellular carcinoma cell growth and angiogenesis through miR-330-5. Dig. Liver Dis. 51, 1050–1059. doi:10.1016/j.dld.2019.03.012
Ghouri, Y. A., Mian, I., and Rowe, J. H. (2017). Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J. Carcinog. 16, 1. doi:10.4103/jcar.JCar_9_16
Green, D. R. (2019). The coming decade of cell death research: five riddles. Cell 177, 1094–1107. doi:10.1016/j.cell.2019.04.024
Guan, L. L., Wang, F. F., Wang, M. J., Han, S. F., Cui, Z. H., Xi, S. M., et al. (2022). Downregulation of HULC induces ferroptosis in hepatocellular carcinoma via targeting of the miR-3200-5p/ATF4 Axis. Oxi Med. Cell. Longev. 2022, 9613095. doi:10.1155/2022/9613095
Guan, Z., Tan, J., Gao, W., Li, X., Yang, Y. D., Li, X. G., et al. (2019). Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J. Cell Physiol. 234, 500–508. doi:10.1002/jcp.26612
Guo, C., Qi, Y., Qu, J., Gai, L., Shi, Y., and Yuan, C. (2020). Pathophysiological functions of the lncRNA TUG1. Curr. Pharm. Des. 26, 688–700. doi:10.2174/1381612826666191227154009
Guo, J., Ma, Y., Peng, X., Jin, H., and Liu, J. (2019). LncRNA CCAT1 promotes autophagy via regulating ATG7 by sponging miR-181 in hepatocellular carcinoma. J. Cell Biochem. 120, 17975–17983. doi:10.1002/jcb.29064
Guo, J. Y., Yuan, Q., Fang, Y., Liao, J. M., and Zhang, Z. (2022). Long non-coding RNA NEAT1 promotes angiogenesis in hepatoma carcinoma via the miR-125a-5p/VEGF pathway. Open Life Sci. 17, 1229–1239. doi:10.1515/biol-2022-0498
Han, Z. Y., Dong, Y. C., Lu, J. Z., Yang, F., Zheng, Y. C., and Yang, H. Y. (2021). Role of hypoxia in inhibiting dendritic cells by VEGF signaling in tumor microenvironments: mechanism and application. Am. J. Cancer Res. 11, 3777–3793.
Hanahan, D., and Coussens, L. M. (2012). Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322. doi:10.1016/j.ccr.2012.02.022
Hao, F., Wang, N., Gui, H., Zhang, Y., Wu, Z., and Wang, J. (2022). Pseudogene UBE2MP1 derived transcript enhances in vitro cell proliferation and apoptosis resistance of hepatocellular carcinoma cells through miR-145-5p/RGS3 axis. Aging (Albany NY) 14, 7906–7925. doi:10.18632/aging.204319
He, G. N., Bao, N. R., Wang, S., Xi, M., Zhang, T. H., and Chen, F. S. (2021). Ketamine induces ferroptosis of liver cancer cells by targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des. Devel Ther. 15, 3965–3978. doi:10.2147/DDDT.S332847
He, X., Lindsay-Mosher, N., Li, Y., Molinaro, A. M., Pellettieri, J., and Pearson, B. J. (2017). FOX and ETS family transcription factors regulate the pigment cell lineage in planarians. Development 144, 4540–4551. doi:10.1242/dev.156349
Hu, H. Q., Yang, L., Li, L. J., and Zeng, C. C. (2018). Long non-coding RNA KCNQ1OT1 modulates oxaliplatin resistance in hepatocellular carcinoma through miR-7-5p/ABCC1 axis. Biochem. Bioph Res. Co. 503, 2400–2406. doi:10.1016/j.bbrc.2018.06.168
Hu, L. T., Ye, H., Huang, G. M., Luo, F., Liu, Y. W., Liu, Y., et al. (2016). Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumor Biol. 37, 2691–2702. doi:10.1007/s13277-015-4111-x
Hu, S. S., Zhang, J. Y., Guo, G. Q., Zhang, L., Dai, J., and Gao, Y. (2022). Comprehensive analysis of GSEC/miR-101-3p/SNX16/PAPOLG axis in hepatocellular carcinoma. Plos One 17, e0267117. doi:10.1371/journal.pone.0267117
Huang, G. Q., Liang, M., Liu, H. Y., Huang, J. H., Li, P. Q., Wang, C., et al. (2020). CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. 11, 1065. doi:10.1038/s41419-020-03276-1
Huang, H., Chen, J., Ding, C. M., Jin, X., Jia, Z. M., and Peng, J. (2018). LncRNA NR2F1-AS1 regulates hepatocellular carcinoma oxaliplatin resistance by targeting ABCC1 via miR-363. J. Cell Mol. Med. 22, 3238–3245. doi:10.1111/jcmm.13605
Huang, Z. S., Xiang, F. L., Zhou, X. H., Huang, Y. Q., Deng, Z. H., and Qiu, Y. Y. (2014). Effects of oxymatrine on the expression of microRNA-122 and microRNA-21 in hepatocellular carcinoma HepG2 cells. Chin. J. Gerontol. 34, 3079–3081.
Hutchinson, J. N., Ensminger, A. W., Clemson, C. M., Lynch, C. R., Lawrence, J. B., and Chess, A. (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39. doi:10.1186/1471-2164-8-39
Ibrahim, B. A., Gobran, M. A., Metwalli, A. M., Abd Elhady, W. A., Tolba, A. M., and Omar, W. E. (2023). Interplay of LncRNA TUG1 and TGF-β/P53 expression in colorectal cancer. Asian Pac J. Cancer Prev. 24, 3957–3968. doi:10.31557/APJCP.2023.24.11.3957
Jiang, L., Zhou, B., Fu, D., and Cheng, B. (2022). lncRNA TUG1 promotes the development of oral squamous cell carcinoma by regulating the MAPK signaling pathway by sponging miR-593-3p. Cell Cycle 21, 1856–1866. doi:10.1080/15384101.2022.2074624
Jin, H., Yu, J., Wang, Z. Y., Qiao, O., Wang, J., Han, J., et al. (2020). Mechanism of flavonoids of Sophaorae Fructus in inhibiting proliferation, migration and invasion of hepatocellular carcinoma cells by regulating LncRNA FBXL19-AS1/miR-342-3p pathway. China J. Chin. Mater Med. 45, 4440–4447. doi:10.19540/j.cnki.cjcmm.20200302.401
Klec, C., Prinz, F., and Pichler, M. (2019). Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol. Oncol. 13, 46–60. doi:10.1002/1878-0261.12404
Kocaturk, N. M., Akkoc, Y., Kig, C., Bayraktar, O., Gozuacik, D., and Kutlu, O. (2019). Autophagy as a molecular target for cancer treatment. Eur. J. Pharm. Sci. 134, 116–137. doi:10.1016/j.ejps.2019.04.011
Kopeina, G. S., and Zhivotovsky, B. (2022). Programmed cell death: past, present and future. Biochem. Biophys. Res. Commun. 633, 55–58. doi:10.1016/j.bbrc.2022.09.022
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
Kudo, M., Finn, R. S., Qin, S., Han, K. H., Ikeda, K., Piscaglia, F., et al. (2018). Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 391, 1163–1173. doi:10.1016/S0140-6736(18)30207-1
Lai, S. H., Quan, Z. P., Hao, Y. S., Liu, J., Wang, Z. Q., Dai, L., et al. (2021). Long non-coding RNA LINC01572 promotes hepatocellular carcinoma progression via sponging miR-195-5p to enhance PFKFB4-mediated glycolysis and PI3K/AKT activation. Front. Cell Devel Biol. 9, 783088. doi:10.3389/fcell.2021.783088
Lee, Y. J., and Jang, B. K. (2015). The role of autophagy in hepatocellular carcinoma. Int. J. Mol. Sci. 16, 26629–26643. doi:10.3390/ijms161125984
Li, J., Qin, X. P., Wu, R. S., Wan, L., Zhang, L., and Liu, R. (2020). Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J. Cell Mol. Med. 24, 5152–5161. doi:10.1111/jcmm.15162
Li, K., Niu, H., Wang, Y., Li, R., Zhao, Y., Liu, C., et al. (2021a). LncRNA TUG1 contributes to the tumorigenesis of lung adenocarcinoma by regulating miR-138-5p-HIF1A axis. Int. J. Immunopathol. Pharmacol. 35, 20587384211048265. doi:10.1177/20587384211048265
Li, N. N., Men, W. X., Zheng, Y. B., Wang, H. C., and Meng, X. S. (2019). Oroxin B induces apoptosis by down-regulating MicroRNA-221 resulting in the inactivation of the PTEN/PI3K/AKT pathway in liver cancer. Molecules 24, 4384. doi:10.3390/molecules24234384
Li, P., Li, Y. W., and Ma, L. T. (2021b). Long noncoding RNA highly upregulated in liver cancer promotes the progression of hepatocellular carcinoma and attenuates the chemosensitivity of oxaliplatin by regulating miR-383-5p/vesicle-associated membrane protein-2 axis. Pharmacol. Res. Perspe 9, e00815. doi:10.1002/prp2.815
Li, Q., Wang, W., Zhang, S., Xing, J., and Wang, Z. T. (2014). Effect of resveratrol on hepatocellular carcinoma by regulating STAT3 and miR-21. Chin. Pharmacol. Bull. 30, 186–191. doi:10.3969/j.issn.1001-1978.2014.02.010
Li, S. Y., Zhu, Y., Li, R. N., Huang, J. H., You, K., Yuan, Y. F., et al. (2021c). LncRNA lnc-APUE is repressed by HNF4α and promotes G1/S phase transition and tumor growth by regulating MiR-20b/E2F1 Axis. Adv. Sci. (Weinh) 8, 2003094. doi:10.1002/advs.202003094
Li, W., Ge, J., Xie, J., Yang, J., Chen, J., and He, T. (2021d). LncRNA TUG1 promotes hepatocellular carcinoma migration and invasion via targeting the miR-137/AKT2 Axis. Cancer Biother Radiopharm. 36, 850–862. doi:10.1089/cbr.2019.3297
Li, X., Yin, X., Bao, H., and Liu, C. (2023). Circular RNA ITCH increases sorafenib-sensitivity in hepatocellular carcinoma via sequestering miR-20b-5p and modulating the downstream PTEN-PI3K/Akt pathway. Mol. Cell Probes 67, 101877. doi:10.1016/j.mcp.2022.101877
Li, X. J., and Liu, Y. H. (2022). Hsa_circ_0000437 inhibits the development of endometrial carcinoma through miR-626/CDKN1B Axis. Protein Pept. Lett. 29, 611–620. doi:10.2174/0929866529666220622125016
Liang, H. H., Wei, P. L., Hung, C. S., Wu, C. T., Wang, W., Huang, M. T., et al. (2013). MicroRNA-200a/b influenced the therapeutic effects of curcumin in hepatocellular carcinoma (HCC) cells. Tumour Biol. 34, 3209–3218. doi:10.1007/s13277-013-0891-z
Lin, W. F., Lu, J. Y., Cheng, B. B., and Ling, C. Q. (2017). Progress in research on the effects of traditional Chinese medicine on the tumor microenvironment. J. Integr. Med. 15, 282–287. doi:10.1016/S2095-4964(17)60345-5
Lin, X. H., Zhang, D. Y., Liu, Z. Y., Tang, W. Q., Chen, R. X., Li, D. P., et al. (2022). lncRNA-AC079061.1/VIPR1 axis may suppress the development of hepatocellular carcinoma: a bioinformatics analysis and experimental validation. J. Transl. Med. 20, 379. doi:10.1186/s12967-022-03573-7
Lin, Y. H., Wu, M. H., Huang, Y. H., Yeh, C. T., and Lin, K. H. (2020). TUG1 is a regulator of AFP and serves as prognostic marker in non-hepatitis B non-hepatitis C hepatocellular carcinoma. Cells-Basel 9, 262. doi:10.3390/cells9020262
Liu, C., Huang, R., Yu, H., Gong, Y., Wu, P., Feng, Q., et al. (2022a). Fuzheng Xiaozheng prescription exerts anti-hepatocellular carcinoma effects by improving lipid and glucose metabolisms via regulating circRNA-miRNA-mRNA networks. Phytomedicine 103, 154226. doi:10.1016/j.phymed.2022.154226
Liu, K. M., Guo, L., and Xi, T. (2015). Mechanism study on apoptosis of hepatocellular carcinoma HepG2 cells induced by ursolic acid through inhibiting the expression of miR-21. J. China Pharm. Univ. 46, 745–750. doi:10.11665/j.issn.1000-5048.20150619
Liu, L. Y., Gu, M. J., Ma, J. H., Wang, Y., Li, M., Wang, H., et al. (2022b). CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol. Cancer 21, 149. doi:10.1186/s12943-022-01619-4
Liu, S., Yao, S., Yang, H., Liu, S., and Wang, Y. (2023). Autophagy: regulator of cell death. Cell Death Dis. 14, 648. doi:10.1038/s41419-023-06154-8
Liu, T., Zu, C. H., Wang, S. S., Song, H. L., Wang, Z. L., Xu, X. N., et al. (2016a). PIK3C2A mRNA functions as a miR-124 sponge to facilitate CD151 expression and enhance malignancy of hepatocellular carcinoma cells. Oncotarget 7, 43376–43389. doi:10.18632/oncotarget.9716
Liu, W., Pan, Y., Zhu, H. B., Zhou, Y., Zhang, H., Liu, L., et al. (2022c). CircRNA_0008194 functions as a ceRNA to promote invasion of hepatocellular carcinoma via inhibiting miR-190a/AHNAK signaling pathway. J. Clin. Lab. Anal. 36, e24286. doi:10.1002/jcla.24286
Liu, Y., Mao, X., Ma, Z., Chen, W., Guo, X., Yu, L., et al. (2022d). Aberrant regulation of LncRNA TUG1-microRNA-328-3p-SRSF9 mRNA Axis in hepatocellular carcinoma: a promising target for prognosis and therapy. Mol. Cancer 21, 36. doi:10.1186/s12943-021-01493-6
Liu, Y. D. (2022). Investigation on potential pharmacological mechanisms of Fufang Biejia ruangan Pills acting on the malignant transformation from liver fibrosis to hepatocellular carcinoma based on the regulatory network of competing endogenous RNAs. [Master 's thesis] (Fuzhou(IL): Fujian University of traditional Chinese Medicine).
Liu, Z., Chen, Q., and Hann, S. S. (2019a). The functions and oncogenic roles of CCAT1 in human cancer. Biomed. Pharmacother. 115, 108943. doi:10.1016/j.biopha.2019.108943
Liu, Z., Lu, J., Fang, H., Sheng, J., Cui, M., Yang, Y., et al. (2021). m6A modification-mediated DUXAP8 regulation of malignant phenotype and chemotherapy resistance of hepatocellular carcinoma through miR-584-5p/MAPK1/ERK pathway Axis. Front. Cell Dev. Biol. 9, 783385. doi:10.3389/fcell.2021.783385
Liu, Z., Ma, C. J., Tang, X. J., Tang, Q., Lou, L. J., Yu, Y. Y., et al. (2019b). The reciprocal interaction between LncRNA CCAT1 and miR-375-3p contribute to the downregulation of IRF5 gene expression by solasonine in HepG2 human hepatocellular carcinoma cells. Front. Oncol. 9, 1081. doi:10.3389/fonc.2019.01081
Liu, Z. Q., Wei, X. Q., Zhang, A. Q., Li, C. H., Bai, J., and Dong, J. H. (2016b). Long non-coding RNA HNF1A-AS1 functioned as an oncogene and autophagy promoter in hepatocellular carcinoma through sponging hsa-miR-30b-5p. Biochem. Bioph Res. Co. 473, 1268–1275. doi:10.1016/j.bbrc.2016.04.054
Long, J. Y., Bai, Y., Yang, X. B., Lin, J. Z., Yang, X., Wang, D. X., et al. (2019). Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 19, 90. doi:10.1186/s12935-019-0817-y
Lu, L., Huang, J. J., Mo, J. T., Da, X., Li, Q. X., Fan, M., et al. (2022a). Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol. Biol. Lett. 27, 17. doi:10.1186/s11658-022-00309-9
Lu, L., Li, S., Zhang, Y., Luo, Z., Chen, Y., Ma, J., et al. (2020). GFI1-Mediated upregulation of LINC00675 as a ceRNA restrains hepatocellular carcinoma metastasis by sponging miR-942-5p. Front. Oncol. 10, 607593. doi:10.3389/fonc.2020.607593
Lu, M., Zhang, Q., Deng, M., Miao, J., Guo, Y., Gao, W., et al. (2008). An analysis of human microRNA and disease associations. PLoS One 3, e3420. doi:10.1371/journal.pone.0003420
Lu, Y., Liu, Y., Zhang, K., and Jiang, L. (2022b). Circular RNA TLK1 exerts oncogenic functions in hepatocellular carcinoma by acting as a ceRNA of miR-138-5p. J. Oncol. 2022, 2415836. doi:10.1155/2022/2415836
Luo, P., Wu, S. Y., Yu, Y. L., Ming, X. L., Li, S., Zuo, X. L., et al. (2020). Current status and perspective biomarkers in AFP negative HCC: towards screening for and diagnosing hepatocellular carcinoma at an earlier stage. Pathol. Oncol. Res. 26, 599–603. doi:10.1007/s12253-019-00585-5
Luo, Y. L., Huang, Y. X., Lai, P., and Zhang, M. H. (2017). Effect of low expression of miR-21 on apoptosis of HepG2 cells induced by matrine. Chin. J. Pathophysiol. 33, 284–288. doi:10.3969/j.issn.1000-4718.2017.02.015
Lv, J., Kong, Y. K., Gao, Z. Q., Liv, Y. M., Zhu, P. F., and Yu, Z. J. (2018). LncRNA TUG1 interacting with miR-144 contributes to proliferation, migration and tumorigenesis through activating the JAK2/STAT3 pathway in hepatocellular carcinoma. Int. J. Biochem. Cell B 101, 19–28. doi:10.1016/j.biocel.2018.05.010
Mao, Y. T., Ding, Z. G., Jiang, M. Z., Yuan, B., Zhang, Y., and Zhang, X. B. (2022). Circ_0091579 exerts an oncogenic role in hepatocellular carcinoma via mediating miR-136- 5p/TRIM27. Biomed. J. 45, 883–895. doi:10.1016/j.bj.2021.12.009
Mattick, J. S. (2009). The genetic signatures of noncoding RNAs. PLoS Genet. 5, e1000459. doi:10.1371/journal.pgen.1000459
Mei, J., Lin, W., Li, S., Tang, Y., Ye, Z., Lu, L., et al. (2022). Long noncoding RNA TINCR facilitates hepatocellular carcinoma progression and dampens chemosensitivity to oxaliplatin by regulating the miR-195-3p/ST6GAL1/NF-κB pathway. J. Exp. Clin. Cancer Res. 41, 5. doi:10.1186/s13046-021-02197-x
Mohyeldeen, M., Ibrahim, S., Shaker, O., and Helmy, H. (2020). Serum expression and diagnostic potential of long non-coding RNAs NEAT1 and TUG1 in viral hepatitis C and viral hepatitis C-associated hepatocellular carcinoma. Clin. Biochem. 84, 38–44. doi:10.1016/j.clinbiochem.2020.06.005
Morse, M. A., Sun, W. J., Kim, R., He, A. R., Abada, P. B., Mynderse, M., et al. (2019). The role of angiogenesis in hepatocellular carcinoma. Clin. Cancer Res. 25, 912–920. doi:10.1158/1078-0432.CCR-18-1254
Nagano, T., and Fraser, P. (2011). No-nonsense functions for long noncoding RNAs. Cell 145, 178–181. doi:10.1016/j.cell.2011.03.014
Niu, Y., Tang, G., Wu, X., and Wu, C. (2020). LncRNA NEAT1 modulates sorafenib resistance in hepatocellular carcinoma through regulating the miR-149-5p/AKT1 axis. Saudi J. Gastroenterol. 26, 194–203. doi:10.4103/sjg.SJG_4_20
Park, W., Wei, S., Kim, B. S., Kim, B., Bae, S. J., Chae, Y. C., et al. (2023). Diversity and complexity of cell death: a historical review. Exp. Mol. Med. 55, 1573–1594. doi:10.1038/s12276-023-01078-x
Pellegrino, R., Castoldi, M., Ticconi, F., Skawran, B., Budczies, J., Rose, F., et al. (2022). LINC00152 drives a competing endogenous RNA network in human hepatocellular carcinoma. Cells-Basel 11, 1528. doi:10.3390/cells11091528
Peng, L. R., Pan, B. L., Zhang, X. X., Wang, Z. B., Qiu, J. C., Wang, X. Q., et al. (2022a). Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol. Toxicol. 38, 1159–1173. doi:10.1007/s10565-022-09718-0
Peng, R., Cao, J., Su, B. B., Bai, X. S., Jin, X., Wang, A. Q., et al. (2022b). Down-regulation of circPTTG1IP induces hepatocellular carcinoma development via miR-16-5p/RNF125/JAK1 axis. Cancer Lett. 543, 215778. doi:10.1016/j.canlet.2022.215778
Pinero, F., Dirchwolf, M., and Pessoa, M. G. (2020). Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells-Basel 9, 1370. doi:10.3390/cells9061370
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., and Pandolfi, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. doi:10.1038/nature09144
Qi, F., Du, X., Zhao, Z., Zhang, D., Huang, M., Bai, Y., et al. (2021). Tumor mutation burden-associated linc00638/miR-4732-3p/ULBP1 Axis promotes immune escape via PD-L1 in hepatocellular carcinoma. Front. Oncol. 11, 729340. doi:10.3389/fonc.2021.729340
Qi, F., Lv, Z. D., Huang, W. D., Wei, S. C., Liu, X. M., and Song, W. D. (2023). LncRNA TUG1 promotes pulmonary fibrosis progression via up-regulating CDC27 and activating PI3K/Akt/mTOR pathway. Epigenetics 18, 2195305. doi:10.1080/15592294.2023.2195305
Ren, Y., Lyu, J., Guo, Y., Yao, Y., and Hu, L. (2022). Long noncoding RNA TUG1 inhibits tumor progression through regulating siglec-15-related anti-immune activity in hepatocellular carcinoma. J. Immunol. Res. 2022, 9557859. doi:10.1155/2022/9557859
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. doi:10.1016/j.cell.2011.07.014
Seitz, H. (2009). Redefining microRNA targets. Curr. Biol. 19, 870–873. doi:10.1016/j.cub.2009.03.059
Shaker, O. G., Abdelwahed, M. Y., Ahmed, N. A., Hassan, E. A., Ahmed, T. I., Abousarie, M. A., et al. (2019). Evaluation of serum long noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma. IUBMB Life 71, 1571–1578. doi:10.1002/iub.2096
Shi, J., Zhou, Y. Q., Yuan, F., Xue, F., Tang, H. L., and Xue, B. Y. (2021a). Comparative study on the intervention effects of "Qinggan Huadu Zhuyu decoction" and huai Er granules on immune function and survival time of patients with liver cancer after TACE. Jiangsu J. Tradit. Chin. Med. 53, 41–44. doi:10.19844/j.cnki.1672-397X.2021.04.015
Shi, Y., Liu, J. B., Deng, J., Zou, D. Z., Wu, J. J., Cao, Y. H., et al. (2021b). The role of ceRNA-mediated diagnosis and therapy in hepatocellular carcinoma. Hereditas 158, 44. doi:10.1186/s41065-021-00208-7
Shi, Y., Yang, X. H., Xue, X. F., Sun, D., Cai, P., Song, Q. W., et al. (2020). HANR enhances autophagy-associated sorafenib resistance through miR-29b/atg9a Axis in hepatocellular carcinoma. Onco targets Ther. 13, 2127–2137. doi:10.2147/OTT.S229913
Shu, J. W., Du, J. Y., Wang, F. T., Cheng, Y., Chen, G. X., Xu, B., et al. (2021). Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis. Open Life Sci. 16, 69–83. doi:10.1515/biol-2021-0009
Singal, A. G., Lampertico, P., and Nahon, P. (2020). Epidemiology and surveillance for hepatocellular carcinoma: new trends. J. Hepatol. 72, 250–261. doi:10.1016/j.jhep.2019.08.025
Song, H., Huang, X. F., Hu, S. Y., Lu, L. L., and Yang, X. Y. (2022). The LINC00261/MiR105-5p/SELL axis is involved in dysfunction of B cell and is associated with overall survival in hepatocellular carcinoma. Peerj 10, e12588. doi:10.7717/peerj.12588
Song, L. N., Qiao, G. L., Yu, J., Yang, C. M., Chen, Y., Deng, Z. F., et al. (2020). Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J. Exp. Clin. Canc Res. 39, 114. doi:10.1186/s13046-020-01576-0
Song, W., Zheng, C. Q., Liu, M., Xu, Y., Qian, Y. Y., Zhang, Z. H., et al. (2021). TRERNA1 upregulation mediated by HBx promotes sorafenib resistance and cell proliferation in HCC via targeting NRAS by sponging miR-22-3p. Mol. Ther. 29, 2601–2616. doi:10.1016/j.ymthe.2021.04.011
Stockwell, B. R., Friedmann Angeli, J. P., Bayir, H., Bush, A. I., Conrad, M., Dixon, S. J., et al. (2017). Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171, 273–285. doi:10.1016/j.cell.2017.09.021
Stratton, M. R., Campbell, P. J., and Futreal, P. A. (2009). The cancer genome. Nature 458, 719–724. doi:10.1038/nature07943
Su, D. N., Wu, S. P., Chen, H. T., and He, J. H. (2016). HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol. Lett. 12, 4061–4067. doi:10.3892/ol.2016.5127
Sui, C. J., Dong, Z. T., Yang, C., Zhang, M. F., Dai, B. H., Geng, L., et al. (2019). LncRNA FOXD2-AS1 as a competitive endogenous RNA against miR-150-5p reverses resistance to sorafenib in hepatocellular carcinoma. J. Cell Mol. Med. 23, 6024–6033. doi:10.1111/jcmm.14465
Sun, J. C., Xu, H. B., Lei, Z., Li, Z. Q., Zhu, H. W., Deng, Z., et al. (2022a). The lncRNA CASC2 modulates hepatocellular carcinoma cell sensitivity and resistance to TRAIL through apoptotic and non-apoptotic signaling. Front. Oncol. 11, 726622. doi:10.3389/fonc.2021.726622
Sun, N., Gong, J., Zhang, W., Yang, X., and Liu, J. (2023). Sevoflurane suppresses hepatocellular carcinoma cell progression via circ_0001649/miR-19a-3p/SGTB axis. Histol. Histopathol. 38, 537–547. doi:10.14670/HH-18-484
Sun, X. H., Wang, Y. T., Li, G. F., Zhang, N., and Fan, L. (2020). Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 20, 226. doi:10.1186/s12935-020-01302-y
Sun, Z., Wu, J., Bi, Q., and Wang, W. (2022b). Exosomal lncRNA TUG1 derived from human urine-derived stem cells attenuates renal ischemia/reperfusion injury by interacting with SRSF1 to regulate ASCL4-mediated ferroptosis. Stem Cell Res. Ther. 13, 297. doi:10.1186/s13287-022-02986-x
Svoronos, A. A., Engelman, D. M., and Slack, F. J. (2016). OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 76, 3666–3670. doi:10.1158/0008-5472.CAN-16-0359
Tam, W. L., and Weinberg, R. A. (2013). The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 19, 1438–1449. doi:10.1038/nm.3336
Tan, J., Liu, B., Zhou, L., Gao, J., Wang, X. K., Liu, Y., et al. (2022). LncRNA TUG1 promotes bladder cancer malignant behaviors by regulating the miR-320a/FOXQ1 axis. Cell Signal 91, 110216. doi:10.1016/j.cellsig.2021.110216
Tang, K. K., Lv, D., Miao, L. L., Mao, Y. S., and Yu, X. Y. (2022a). LncRNA TUG1 functions as a ceRNA for miR-1-3p to promote cell proliferation in hepatic carcinogenesis. J. Clin. Lab. Anal. 36, e24415. doi:10.1002/jcla.24415
Tang, Q., Li, X. J., Chen, Y., Long, S. Q., Yu, Y. Y., Sheng, H. H., et al. (2022b). Solamargine inhibits the growth of hepatocellular carcinoma and enhances the anticancer effect of sorafenib by regulating HOTTIP-TUG1/miR-4726-5p/MUC1 pathway. Mol. Carcinog. 61, 417–432. doi:10.1002/mc.23389
Tay, Y., Rinn, J., and Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352. doi:10.1038/nature12986
Teng, F., Zhang, J. X., Chang, Q. M., Wu, X. B., Tang, W. G., Wang, J. F., et al. (2020). LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma (vol 39, 277, 2020). J. Exp. Clin. Canc Res. 39, 227. doi:10.1186/s13046-020-01780-y
Thomas, M., Lieberman, J., and Lal, A. (2010). Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17, 1169–1174. doi:10.1038/nsmb.1921
Tian, N. N., Zheng, Y. B., Li, Z. P., Zhang, F. W., and Zhang, J. F. (2021). Histone methylatic modification mediates the tumor-suppressive activity of curcumol in hepatocellular carcinoma via an Hotair/EZH2 regulatory axis. J. Ethnopharmacol. 280, 114413. doi:10.1016/j.jep.2021.114413
Tripathi, S. K., Pal, A., Ghosh, S., Goel, A., Aggarwal, R., Banerjee, S., et al. (2022). LncRNA NEAT1 regulates HCV- induced Hepatocellular carcinoma by modulating the miR-9-BGH3 axis. J. Gen. Virol. 103, 10. doi:10.1099/jgv.0.001809
Wang, H., Huo, X. S., Yang, X. R., He, J., Cheng, L. J., Wang, N., et al. (2017). STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 16, 136. doi:10.1186/s12943-017-0680-1
Wang, W., and Wei, C. (2020). Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 7, 308–319. doi:10.1016/j.gendis.2020.01.014
Wang, X., Zhang, J., Luo, F., and Shen, Y. (2023). Application of circular RNA Circ_0071662 in the diagnosis and prognosis of hepatocellular carcinoma and its response to radiotherapy. Dig. Dis. 41, 431–438. doi:10.1159/000527696
Wang, Y., Yang, L., Chen, T., Liu, X., Guo, Y., Zhu, Q., et al. (2019). A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol. Cancer 18, 28. doi:10.1186/s12943-019-0957-7
Wang, Y., Zhang, Q., Chen, Y., Liang, C. L., Liu, H., Qiu, F., et al. (2020). Antitumor effects of immunity-enhancing traditional Chinese medicine. Biomed. Pharmacother. 121, 109570. doi:10.1016/j.biopha.2019.109570
Wei, Y., Wang, Z., Zong, Y., Deng, D., Chen, P., and Lu, J. (2020). LncRNA MFI2-AS1 promotes HCC progression and metastasis by acting as a competing endogenous RNA of miR-134 to upregulate FOXM1 expression. Biomed. Pharmacother. 125, 109890. doi:10.1016/j.biopha.2020.109890
Wu, H., Liu, T. T., Qi, J. N., Qin, C. Y., and Zhu, Q. (2020). Four autophagy-related lncRNAs predict the prognosis of HCC through coexpression and ceRNA mechanism. Biomed. Res. Int-Uk 2020, 3801748. doi:10.1155/2020/3801748
Xia, C., Sun, Y., Li, Y., Ma, J., and Shi, J. (2022). LncRNA CCAT1 enhances chemoresistance in hepatocellular carcinoma by targeting QKI-5. Sci. Rep. 12, 7826. doi:10.1038/s41598-022-11644-4
Xie, F., Zhang, L., Yao, Q., Shan, L., Liu, J., Dong, N., et al. (2020). TUG1 promoted tumor progression by sponging miR-335-5p and regulating CXCR4-mediated infiltration of pro-tumor immunocytes in CTNNB1-mutated hepatoblastoma. Onco Targets Ther. 13, 3105–3115. doi:10.2147/OTT.S234819
Xie, G., and Diehl, A. M. (2013). Evidence for and against epithelial-to-mesenchymal transition in the liver. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G881–G890. doi:10.1152/ajpgi.00289.2013
Xiong, D. D., Dang, Y. W., Lin, P., Wen, D. Y., He, R. Q., Luo, D. Z., et al. (2018). A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J. Transl. Med. 16, 220. doi:10.1186/s12967-018-1593-5
Xu, K. Q., Xia, P., Gongye, X. D., Zhang, X., Ma, S. X., Chen, Z., et al. (2022). A novel lncRNA RP11-386G11.10 reprograms lipid metabolism to promote hepatocellular carcinoma progression. Mol. Metab. 63, 101540. doi:10.1016/j.molmet.2022.101540
Yan, K., Fu, Y., Zhu, N., Wang, Z., Hong, J. L., Li, Y., et al. (2019). Repression of lncRNA NEAT1 enhances the antitumor activity of CD8+T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int. J. Biochem. Cell Biol. 110, 1–8. doi:10.1016/j.biocel.2019.01.019
Yan, S., Tang, Z., Chen, K., Liu, Y., Yu, G., Chen, Q., et al. (2018). Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J. Exp. Clin. Cancer Res. 37, 214. doi:10.1186/s13046-018-0853-9
Yang, L., Peng, X. Q., Jin, H. Y., and Liu, J. G. (2019). Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene 697, 94–102. doi:10.1016/j.gene.2019.02.036
Yang, P. W., Chen, T. T., Zhao, W. X., Liu, G. W., Feng, X. J., Wang, S. M., et al. (2021). Scutellaria barbata D.Don and Oldenlandia diffusa (Willd.) Roxb crude extracts inhibit hepatitis-B-virus-associated hepatocellular carcinoma growth through regulating circRNA expression. J. Ethnopharmacol. 275, 114110. doi:10.1016/j.jep.2021.114110
Yang, Q., and Wu, G. (2021). CircRNA-001241 mediates sorafenib resistance of hepatocellular carcinoma cells by sponging miR-21-5p and regulating TIMP3 expression. Gastroenterol. Hepatol. 45, 742–752. doi:10.1016/j.gastrohep.2021.11.007
Yang, Z. L., He, K., Chen, W. G., and Chen, Y. (2023). A ferroptosis-related ceRNA network in hepatocellular carcinoma for potential clinical applications. Am. J. Transl. Res. 15, 3912–3927.
Yin, S. S., Jin, W. K., Qiu, Y. L., Fu, L. L., Wang, T., and Yu, H. Y. (2022). Solamargine induces hepatocellular carcinoma cell apoptosis and autophagy via inhibiting LIF/miR-192-5p/CYR61/Akt signaling pathways and eliciting immunostimulatory tumor microenvironment. J. Hematol. Oncol. 15, 32. doi:10.1186/s13045-022-01248-w
Yu, H. J., Wang, C. Q., Ke, S. J., Bai, M. Y., Xu, Y. A., Lu, S. A., et al. (2022). Identification of CFHR4 as a potential prognosis biomarker associated with lmmune infiltrates in hepatocellular carcinoma. Front. Immunol. 13, 892750. doi:10.3389/fimmu.2022.892750
Yu, J., Wang, Y., and Peng, S. (2019). The high expression of lncRNA TUG1 correlates with progressive tumor condition and less satisfying survival profiles in prostate cancer patients. Transl. Cancer Res. 8, 1817–1825. doi:10.21037/tcr.2019.08.33
Yuan, J. H., Yang, F., Wang, F., Ma, J. Z., Guo, Y. J., Tao, Q. F., et al. (2014). A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681. doi:10.1016/j.ccr.2014.03.010
Zabady, S., Mahran, N., Soltan, M. A., Alaa Eldeen, M., Eid, R. A., Albogami, S., et al. (2022). Cyanidin-3-Glucoside modulates hsa_circ_0001345/miRNA106b/ATG16L1 Axis expression as a potential protective mechanism against hepatocellular carcinoma. Curr. Issues Mol. Biol. 44, 1677–1687. doi:10.3390/cimb44040115
Zhan, F. L., Chen, C. F., and Yao, M. Z. (2020). LncRNA TUG1 facilitates proliferation, invasion and stemness of ovarian cancer cell via miR-186-5p/ZEB1 axis. Cell Biochem. Funct. 38, 1069–1078. doi:10.1002/cbf.3544
Zhang, C. X., Zhang, C. Y., Lin, J. M., and Wang, H. Y. (2018). Circular RNA Hsa_Circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma. Cell Physiol. Biochem. 51, 290–300. doi:10.1159/000495230
Zhang, D., Zhang, Y., Zhang, X. W., Zhai, H. J., Sun, X. L., and Li, Y. M. (2022a). Circ_0091579 serves as a tumor-promoting factor in hepatocellular carcinoma through miR-1225-5p/PLCB1 Axis. Dig. Dis. Sci. 67, 585–597. doi:10.1007/s10620-021-06861-2
Zhang, H., Chu, K., Zheng, C., Ren, L., and Tian, R. (2020a). Pseudogene DUXAP8 promotes cell proliferation and migration of hepatocellular carcinoma by sponging MiR-490-5p to induce BUB1 expression. Front. Genet. 11, 666. doi:10.3389/fgene.2020.00666
Zhang, H., Su, X., Burley, S. K., and Zheng, X. F. S. (2022b). mTOR regulates aerobic glycolysis through NEAT1 and nuclear paraspeckle-mediated mechanism in hepatocellular carcinoma. Theranostics 12, 3518–3533. doi:10.7150/thno.72581
Zhang, H., Wang, F., and Hu, Y. H. (2017). STARD13 promotes hepatocellular carcinoma apoptosis by acting as a ceRNA for Fas. Biotechnol. Lett. 39, 207–217. doi:10.1007/s10529-016-2253-6
Zhang, J., Cai, M., Jiang, D., and Xu, L. (2019). Upregulated LncRNA-CCAT1 promotes hepatocellular carcinoma progression by functioning as miR-30c-2-3p sponge. Cell Biochem. Funct. 37, 84–92. doi:10.1002/cbf.3375
Zhang, L., Hu, S. S., Chen, J. S., Ma, S. S., Liu, F. H., Liu, C. M., et al. (2021). Comprehensive analysis of the MIR4435-2HG/miR-1-3p/MMP9/miR-29-3p/DUXAP8 ceRNA network axis in hepatocellular carcinoma. Discov. Oncol. 12, 38. doi:10.1007/s12672-021-00436-3
Zhang, P., Ha, M., Li, L. B., Huang, X., and Liu, C. J. (2020b). MicroRNA-3064-5p sponged by MALAT1 suppresses angiogenesis in human hepatocellular carcinoma by targeting the FOXA1/CD24/Src pathway. Faseb J. 34, 66–81. doi:10.1096/fj.201901834R
Zhang, W. F., Huang, J. J., Lv, J. L., Huang, J. S., Huang, J. Z., Pan, Z., et al. (2020c). Effect of Jiedu Quyu Kangai Recipe on primary liver cancer after interventional therapy and its mechanism. J. Guangxi Uni Nat. Sci. Ed. 45, 1217–1225. doi:10.13624/j.cnki.issn.1001-7445.2020.1217
Zhang, X., Wang, J., and Li, H. (2023). RTKN2 knockdown alleviates the malignancy of breast cancer cells by regulating the Wnt/β-catenin pathway. Sci. Rep. 13, 23023. doi:10.1038/s41598-023-50153-w
Zhang, Y., Luo, M. Y., Cui, X. H., O'connell, D., and Yang, Y. F. (2022c). Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 29, 1850–1863. doi:10.1038/s41418-022-00970-9
Zhao, L. J., Hu, K. X., Cao, J. Z., Wang, P., Li, J., Zeng, K. W., et al. (2019). LncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging-Us 11, 7098–7122. doi:10.18632/aging.102240
Zhou, M., Zhang, G., Hu, J., Zhu, Y. Z., Lan, H. M., Shen, X. F., et al. (2021a). Rutin attenuates sorafenib-induced chemoresistance and autophagy in hepatocellular carcinoma by regulating BANCR/miRNA-590-5P/OLR1 Axis. Int. J. Biol. Sci. 17, 3595–3607. doi:10.7150/ijbs.62471
Zhou, W., Li, H., Shang, S., and Liu, F. (2021b). lncRNA KCNQ1OT1 reverses the effect of sevoflurane on hepatocellular carcinoma progression via regulating the miR-29a-3p/CBX3 axis. Braz J. Med. Biol. Res. 54, e10213. doi:10.1590/1414-431X2020e10213
Zhou, Y. P., Huang, Y. G., Dai, T., Hua, Z. Y., Xu, J., Lin, Y. T., et al. (2021c). LncRNA TTN-AS1 intensifies sorafenib resistance in hepatocellular carcinoma by sponging miR-16-5p and upregulation of cyclin E1. Biomed. Pharmacother. 133, 111030. doi:10.1016/j.biopha.2020.111030
Zhu, H. Q., Zhou, X., Chang, H., Li, H. G., Liu, F. F., Ma, C. Q., et al. (2015). Aberrant expression of CCAT1 regulated by c-myc predicts the prognosis of hepatocellular carcinoma. Asian Pac J. Cancer Prev. 16, 5181–5185. doi:10.7314/apjcp.2015.16.13.5181
Keywords: hepatocellular carcinoma, competing endogenous RNA, traditional Chinese medicine, progress, therapy
Citation: Tang Z, Li X, Zheng Y, Liu J, Liu C and Li X (2024) The role of competing endogenous RNA network in the development of hepatocellular carcinoma: potential therapeutic targets. Front. Cell Dev. Biol. 12:1341999. doi: 10.3389/fcell.2024.1341999
Received: 30 November 2023; Accepted: 16 January 2024;
Published: 31 January 2024.
Edited by:
Juanjuan Xiang, Central South University, ChinaReviewed by:
Yong Jiang, East Tennessee State University, United StatesCopyright © 2024 Tang, Li, Zheng, Liu, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Li, bGl4MTEwNkAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.