
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol., 15 May 2024
Sec. Cancer Cell Biology
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1332506
This article is part of the Research TopicState-of-the-art Insights into the Immuno-Oncological Effect of Aging and Circadian RhythmView all articles
 Kai Chen1,2†
Kai Chen1,2† Yaohui Wang3†
Yaohui Wang3† Dengxiong Li2†
Dengxiong Li2† Ruicheng Wu2†
Ruicheng Wu2† Jie Wang2
Jie Wang2 Wuran Wei2
Wuran Wei2 Wei Zhu1
Wei Zhu1 Wenhua Xie1
Wenhua Xie1 Dechao Feng2,4*
Dechao Feng2,4* Yi He1*
Yi He1*The Period (PER) gene family is one of the core components of the circadian clock, with substantial correlations between the PER genes and cancers identified in extensive researches. Abnormal mutations in PER genes can influence cell function, metabolic activity, immunity, and therapy responses, thereby promoting the initiation and development of cancers. This ultimately results in unequal cancers progression and prognosis in patients. This leads to variable cancer progression and prognosis among patients. In-depth studies on the interactions between the PER genes and cancers can reveal novel strategies for cancer detection and treatment. In this review, we aim to provide a comprehensive overview of the latest research on the role of the PER gene family in cancer.
With population aging, it is projected that by 2030, almost 20% of the world’s population will be over 65, and by 2050, this figure is expected to reach 1.6 billion (Feng et al., 2023a). Cancers, closely associated with aging, are characterized by aberrant cellular proliferation and differentiation, continuing to pose a significant threat to global health (Hanahan, 2022; Shen et al., 2022; Feng et al., 2023b; Wang et al., 2023). In 2020, approximately 19 million new cancer cases and over 10 million cancer-related deaths were recorded worldwide (Sung et al., 2021). Specifically, in China, there were 4.57 million new cancer cases and 3.00 million cancer-specific deaths in the same year (He and Ke, 2023). Traditional cancer treatments include radiation therapy, chemotherapy, and surgery (Jin et al., 2022; Sirhan et al., 2022; Xing et al., 2022; Association, 2023), while molecularly targeted therapies and immune checkpoint inhibitors have transformed oncology (Chen et al., 2022; Chan et al., 2023; Yin et al., 2023; Yu et al., 2023). Despite advancements, the adverse effects of treatments on patient survival and quality of life remain pressing challenges in cancer therapy (Zhang and Zhang, 2020; Peng et al., 2022), complicating patient management (Wang YH. et al., 2020; Mokhtari-Hessari and Montazeri, 2020). High-throughput sequencing, a pioneering molecular biology technique, has fueled new oncology research directions (Walter et al., 2022; Larson et al., 2023). Contrasting with the conventional American Joint Committee on Cancer staging, which relies on tumor size, lymph node status, and metastasis, new classifications based on tumor genetic expression patterns have emerged, correlating more closely with clinical outcomes and patient survival (Yin et al., 2020; Park et al., 2022; de Jong et al., 2023). This shift signifies a move towards personalized precision medicine in oncology, with the potential to uncover new therapeutic targets and prognostic biomarkers through a deeper understanding of molecular mechanisms in cancer (Hong et al., 2020; Bao et al., 2023).
The Earth’s rotation generates environmental variables with a 24-h periodicity, including temperature and light fluctuations (Hou et al., 2022; Laosuntisuk et al., 2023). Various life forms, including animals, plants, bacteria, fungi, have developed intricate internal timing mechanisms to synchronize their behavior and physiological processes with these cyclic environmental changes (Patton and Hastings, 2023). These mechanisms are known as “circadian rhythms,” a term derived from the Latin “circa diem”, meaning “about a day” (Allada and Bass, 2021). Thus, circadian rhythm denotes an organism’s inherent rhythm-regulating system. In humans and other mammals, this 24-h rhythm is orchestrated by the circadian clock (Huang et al., 2023). The suprachiasmatic nucleus (SCN) of the hypothalamus, acting as the central clock, utilizes neuroendocrine pathways to coordinate peripheral clocks throughout the body (Starnes and Jones, 2023; Zhang-Sun et al., 2023). This network of clocks ensures the consistency of vital functions and numerous physiological activities.
From a molecular perspective, the circadian clock in mammals is governed by a transcription-translation feedback loop (TTFL), involving circadian genes and associated proteins (Patke et al., 2020; Li et al., 2023). This process begins when Brain and Muscle ARNT-Like Protein 1 (BMAL1) and Circadian Locomotor Output Cycles Kaput (CLOCK) proteins form a heterodimer that binds to the E-box in the promoter regions of various genes (Cox and Takahashi, 2019). This includes clock-controlled genes (CCGs) and inhibitory elements like Period (PER) and Cryptochrome (CRY) proteins (Cao et al., 2021). As PER and CRY proteins levels rise, they eventually inhibit CLOCK-BMAL1 activity, thus regulating their own synthesis and setting the stage for a new circadian cycle. Additionally, RORα and REV-ERBs modulate BMAL1 expression through their interaction with REV-ERB-ROR response elements, creating a secondary loop (Preitner et al., 2002; Sato et al., 2004). D-box binding protein (DBP) and E4 promoter-binding protein 4 (E4BP4/NFIL3) also modulate gene expression and CCG activity via D-box promoters (Ripperger and Schibler, 2006). Furthermore, post-translational modifications (PTMs) by kinases/phosphatases and the ubiquitin-proteasome system, including Casein kinase Ⅰ epsilon (CKⅠε), β-transducin repeat-containing protein (β-TrCP), F-box and leucine-rich repeat protein 3 (FBXL3), and Tumor necrosis factor receptor-associated factor-2 (TRAF2) (Eide et al., 2005; Reischl et al., 2007; Siepka et al., 2007; Chen et al., 2018), maintain the stability and function of clock proteins. These mechanisms collectively create a delayed negative feedback loop, producing the roughly 24-h circadian rhythm. Our team has previously detailed this molecular mechanism in extensive studies (Feng et al., 2022; Zhu et al., 2023) (Figure 1).
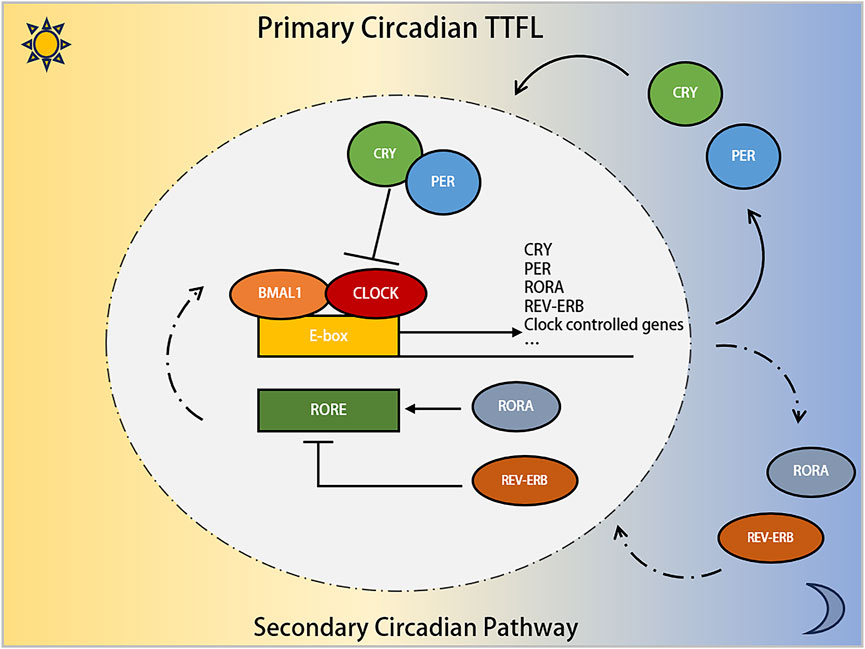
Figure 1. The involvement of the PER gene family in the molecular mechanism of Circadian Clock. The core circadian transcriptional machinery consists of the positive transcription factors (CLOCK and BMAL1 proteins), and the repressors factors (PER and CRY proteins). Additionally, the nuclear receptors RORα, and REV-ERB compose the secondary circadian pathway.
Given the pivotal role of circadian rhythm in regulating diverse biological functions, extensive research has shown that its disruption is associated with a heightened risk of various diseases, including cardiovascular disorders, neurological conditions, and cancers (Crnko et al., 2019; Sancar and Van Gelder, 2021; Niu et al., 2022). In 2007, the World Health Organization identified shift work that disrupts circadian rhythms as a Group 2A carcinogen (Straif et al., 2007). Epidemiological studies have indicated that long-term night shift workers face increased risks of certain cancers, such as breast, prostate, and lung cancer (Cordina-Duverger et al., 2022; Berge et al., 2023; Schernhammer et al., 2023). Remarkably, about 43% of protein-coding genes in the mammalian genome are regulated by circadian clock genes, which are integral to various critical physiological and pathological processes (Zhang et al., 2014). Alterations in circadian clock genes or their functions can lead to disrupted cellular activities linked with cancer hallmarks, including cell cycle disruption (Lévi et al., 2007; Qu et al., 2023), genomic instability (Zhang et al., 2023), metabolic reprogramming (Chun et al., 2022), and immune system dysregulation (Chen et al., 2020; Zhang et al., 2024). As a result, the link between circadian clock genes and cancer has become a focal point in oncological research. In this review, we concentrate on the PER gene family, a central component of the circadian clock, explore its connection to cancer and summarize current research findings in the area.
There are three homologous genes in mammals (PER1, PER2, and PER3) that are integral to the PER gene family, a key component of circadian clock genes (Deng and Yang, 2019). When Konopka and Benzer induced mutations in Drosophila melanogaster using ethyl methane sulfonate in 1971, they observed three distinct rhythm patterns in the mutants’ eclosion and locomotor activities. The per gene was subsequently located on the X chromosome of D. melanogaster, with its variants (pero, pers, and perl) associated with these rhythm changes (Konopka and Benzer, 1971). In 1984, the per gene was isolated and cloned by the teams of Jeffrey C. Hall (Zehring et al., 1984), Michael Rosbash (Reddy et al., 1984), and Michael Young (Bargiello and Young, 1984), who later received the 2017 Nobel Prize in Physiology or Medicine for their pioneering work on circadian rhythms. Further research by Hall and Rosbash demonstrated that the Per protein was a nuclear protein that oscillated between the cytoplasm and the nucleus, influencing the expression of its mRNA, leading to the hypothesis that the per gene acts as a transcription factor with feedback regulation (Zeng et al., 1994; Yildirim et al., 2022). Additionally, Young and his team found that the per gene’s oscillation was synchronized with another clock gene, Timeless (tim), and that mutations in tim significantly affected the Per protein’s synthesis, phosphorylation, and transport (Sehgal et al., 1995; Ahmad et al., 2021). These findings elucidated the interaction between Tim and Per proteins, supporting the TTFL model. Subsequent discoveries showed that CLOCK and BMAL1 proteins form a dimer that binds to the E-box to transcriptionally regulate the PER genes, confirming their role as positive regulators in the circadian rhythm (Vitaterna et al., 1994). The identification of other core clock genes, such as CLOCK (Allada et al., 1998), CYC (Rutila et al., 1998), and CRY (Todo et al., 1996), further elaborated the TTFL model, enhancing our understanding of circadian gene transcription regulation. These advancements propelled researches into circadian rhythms, underscoring the significance of clock genes in human physiology and diseases (Vitaterna et al., 2019) (Figure 2).
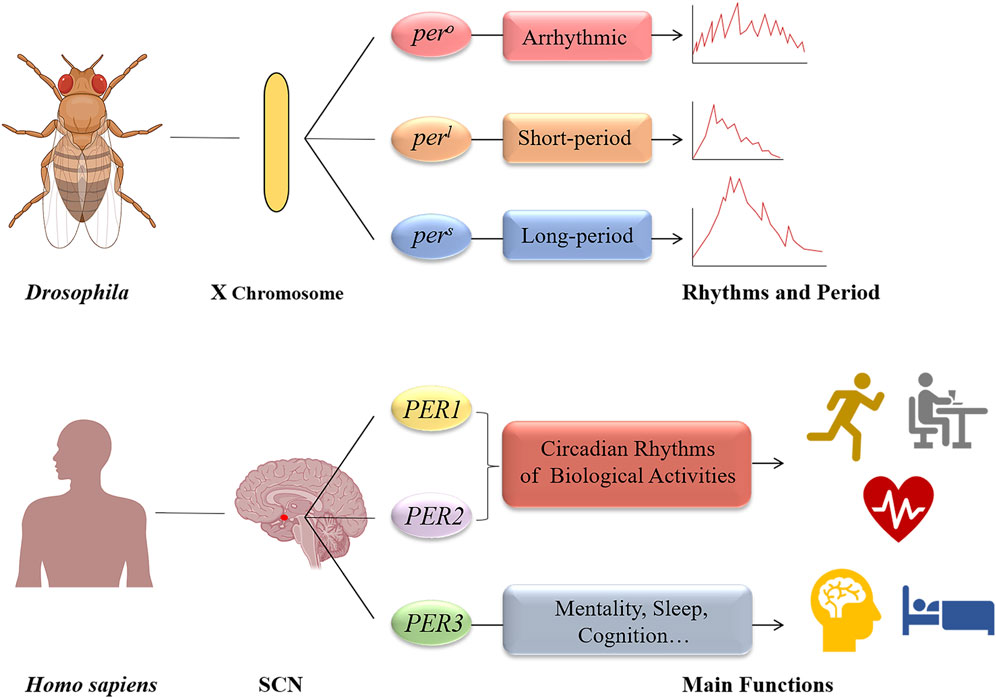
Figure 2. The period genes of Drosophila melanogaster and human. The discovery of the period gene can be traced back to 1971, when Konopka and Benzer identified three gene mutations (pero, pers, and perl) on the X chromosome of Drosophila melanogaster; As for human, there are three homologous genes (PER1, PER2, and PER3). The PER1 and PER2 genes are primarily responsible for maintaining circadian rhythms of basic biological activities. And PER3 gene play an important role in mentality, sleep, and cognition.
The PER gene family plays a pivotal role in maintaining circadian rhythms. Studies have demonstrated that Per2 homozygous mutant mice exhibit significantly shorter circadian rhythms than wild-type mice, which disappear in constant darkness (Zheng et al., 1999). Likewise, mice with homozygous Per1 mutations also display shorter rhythms, with Per2 influencing clock gene expression rhythm through transcriptional regulation (Zheng et al., 2001). Shiromani and colleagues found that Per1/Per2 double mutant mice quickly experienced a decrease in core body temperature rhythm and a reduced circadian period, while Per3 mutants showed no notable rhythm alterations (Shiromani et al., 2004). Nakamura et al. (Nakamura et al., 2023) observed that the triple knockout of Per1/Per2/Per3 severely disrupted the estrous cycle in C57BL/6J mice (melatonin deficient), potentially due to compromised SCN amplitude stability. While the contribution of PER3 to circadian rhythm maintenance is less pronounced than PER1 and PER2, it has been linked to nocturnal preferences, psychiatric disorders, sleep patterns, and cognitive functions (Chen et al., 2023). The PER3 gene exhibits polymorphism in humans, with a variable number of tandem repeats (VNTR) consisting of a 54-base pair sequence in the 18th exon (Barragan et al., 2022). Aytac et al. (Aytac et al., 2022) identified an association between the VNTR variant at the rs57875989 locus of the PER3 gene and bipolar affective disorder risk, combining clinical scale assessments and blood analysis. The 4R/4R genotype appears to be protective against bipolar disorder, whereas the 5R/5R genotype is linked to a higher incidence of moderate manic symptoms in a study of 121 patients and controls (Yegin et al., 2021). Additionally, a significant correlation was found between the rs228697 SNP in PER3 and increased vulnerability to sleep-wake disturbances in Alzheimer’s patients (Lozano-Tovar et al., 2023). Azevedo et al. (Azevedo et al., 2021) also noted a potential association between severe obesity and the rs228729 locus of the PER3 gene.
Emerging evidence suggests that alterations in clock genes, including the PER gene family (PER1, PER2, and PER3), play a significant role in cancer development and progression due to their regulatory functions in biological cycles and physiological processes. Researches have increasingly focused on comparing the PER genes expression levels in tumor and normal tissues to elucidate the potential link between PER genes aberrations and cancers. Downregulations of PER1, PER2, or PER3 have been observed in various cancers, including oral squamous cell carcinoma (Gong et al., 2021), head and neck squamous cell carcinoma (Li et al., 2019; Rahman et al., 2019), colorectal cancer (Orhan et al., 2019; Sahar et al., 2022), breast cancer (Liu et al., 2021; Liu et al., 2022a), ovarian cancer (Angelousi et al., 2019; Chen et al., 2021), melanoma (Lesicka et al., 2023), and hematological malignancies (Jiang et al., 2021). Studies have also highlighted the association between VNTR or SNPs polymorphism in the PER gene family and an increased risk of certain cancers, such as breast cancer (Fores-Martos et al., 2021; Song et al., 2023), colorectal cancer (Holipah and Kuroda, 2020), and prostate cancer (Wendeu-Foyet et al., 2019; Hinoura et al., 2021), through the analysis of tumor tissues or patient blood samples. To enhance our understanding of the prognostic significance of the PER gene family alterations in tumor progression, a growing body of research is exploring the relationship between these genetic changes and clinicopathological features.
A meta-analysis involving 7,476 cancer patients revealed that decreased PER1 gene expression was associated with poorer tumor differentiation and greater invasion depth, whereas reduced PER2 expression was correlated with advanced pathological stages and increased metastasis; furthermore, lower levels of both PER1 and PER2 were linked to shorter overall survivals (Zhang et al., 2020). Patients with advanced-stage head and neck squamous cell carcinoma exhibited markedly lower levels of PER1, PER2, and PER3 proteins compared to those with early-stage tumors, and higher levels of these proteins were associated with longer overall and recurrence-free survival. In oral squamous cell carcinoma, low PER2 expression was connected to poor prognosis, tumor grade progression, and lymph node metastasis (Xiong et al., 2018), whereas in lung cancer, increased PER2 expression was associated with less malignant differentiation and fewer lymph node metastases (Xiang et al., 2018). Transcriptome analysis from the GEO database indicated that PER3 gene expression could predict outcomes for ER+/HER2-breast cancer patients through multifactor Cox analysis (Cadenas et al., 2014). The link between VNTR or SNP variations in the PER gene family and cancer risk has also attracted significant research interest (Morales-Santana et al., 2019). Lesicka et al. (Lesicka et al., 2019) identified that the dominant phenotype of PER1 rs2735611 and the recessive phenotype of PER2 rs934945 were associated with increased breast cancer risk. Additionally, the higher prevalence of the PER2 VNTR 4R/3R and 3R/3R genotypes in pancreatic cancer patients compared to healthy controls suggested that a greater proportion of the 3R allele in the PER2 VNTR may be a risk factor for pancreatic cancer (Dagmura et al., 2021).
Overall, the majority of previous cancer studies have consistently demonstrated significant downregulation of the PER gene family, often linked to reduced patient survival, poorer prognosis, and clinicopathological factors such as low tumor differentiation, advanced tumor stage, lymph node metastasis, and more aggressive tumor characteristics. Furthermore, researches into VNTR or SNPs polymorphisms of the PER gene family have established a connection between their anomalies and an increased risk of certain cancers (Figure 3).
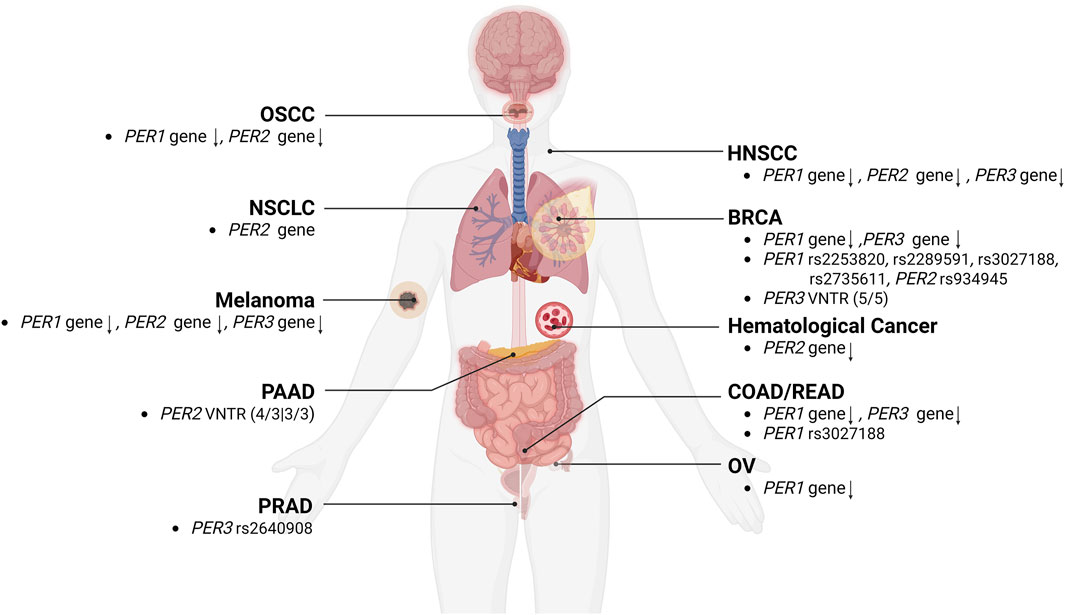
Figure 3. The relationships between alterations of the PER gene family and cancers. The aberrations, and VNTR or SNPs polymorphism of PER1, PER2, and PER3 gene were correlated with the development of various types of cancers, including BRCA (breast cancer), COAD/READ (colorectal cancer), HNSCC (head and neck squamous cell carcinoma), melanoma, NSCLC (non-small cell lung cancer), OSCC (oral squamous cell carcinoma), OV (ovarian cancer), PAAD (pancreatic adenocarcinoma), and PRAD (prostate cancer).
The cell cycle, which promotes cell division in a specific sequence and phases, exhibits a biological rhythm akin to the circadian clock. Cell cycle progression is governed by transient interactions between cyclins and cyclin-dependent kinases (CDKs), triggering phase transitions; While activities of the cyclin-CDK complexes are strictly regulated by CDK inhibitors like p16, p21, and WEE1, to halt the cell cycle under stress or damage conditions (Suski et al., 2021; Matthews et al., 2022). Consequently, there is growing interest in exploring the molecular interplay between the cell cycle and circadian rhythms, particularly regarding abnormal cell proliferation in cancers (Farshadi et al., 2020; Yao et al., 2021). For example, the PER1 protein has been shown to inhibit the cell cycle by interacting with checkpoint proteins ATM and CHK2 in colorectal cancer cells (Gery et al., 2006); similarly, NONO was found to influence the p16-Ink4A site by binding to PER1/PER2 proteins during the G1 phase, linking the cell cycle with the circadian rhythm (Kowalska et al., 2013). Conversely, the TP53 gene can inhibit PER2 gene activation by interfering with BMAL1/CLOCK-mediated E-box transcription, serving as a key cell cycle regulator (Miki et al., 2013; Zou et al., 2020; Engeland, 2022). Gao et al. (Gao et al., 2021) observed that IDH1 gene mutations in gliomas significantly reduced PER genes expression (PER1, PER2, and PER3) and altered the expression of cell cycle-related proteins like Cyclin A, CDK2, and Cyclin D3, resulting in increase of G1 phase cells and decrease of S phase cells. Overexpressing the PER2 gene in the chronic myelogenous leukemia cell line KCL22 led to G1 phase cell cycle arrest, while its downregulation expedited the transition to the S phase (Wang et al., 2020b; Basti et al., 2022). Recently, Han and colleagues integrated cell microarray analysis with experimental validation, revealing that overexpressing the PER1 gene affected the expression of cell cycle-related proteins such as Wee1, CRE-BP1, CDK1, and GADD45A, thereby inhibiting the cell cycle in cholangiocarcinoma cells (Han et al., 2016).
The epithelial-mesenchymal transition (EMT), marked by increased vimentin and decreased E-cadherin expression, is a critical process where cells transition from epithelial to mesenchymal traits (Bakir et al., 2020; Liu et al., 2022b; Huang et al., 2022; Lee et al., 2022). EMT is essential for tumor initiation, enhanced migration, metastasis, and treatment resistance (Pastushenko and Blanpain, 2019). Studies have shown that EMT in glioma C6 and breast cancer MCF-7 cells correlated with enhanced circadian rhythms and increased PER2 gene expression (De et al., 2020). Guo et al. (Guo et al., 2020) observed that elevated PER2 gene protected oral cancer cells from EMT via upregulation of TP53 protein. Conversely, PER2 knockdown in colorectal cancer cells activated the Snail/Slug-related EMT pathway, promoting proliferation and invasiveness of cancer cells (Xiong et al., 2022). Additionally, Lin et al. (Lin et al., 2020) reported that the mangiferin-induced EMT suppression in lung cancer cells was associated with decreased E-cadherin and increased PER1 protein expression. These findings suggest that the PER1 and PER2 genes play a role in inhibiting EMT, potentially reducing migration and invasion in cancer cells.
Metabolic reprogramming, a hallmark of cancer cells (Pavlova et al., 2022), involves altering cellular metabolism to manage different inputs and stressors (Wang et al., 2024). Cancer cells adapt their metabolic pathways to oncogenic mutations and external nutritional conditions, supporting their increased biosynthetic and energy needs while mitigating oxidative stress associated with survival and proliferation (Altea-Manzano et al., 2020; McGuirk et al., 2020; Schiliro and Firestein, 2021). An example is the Warburg effect, where cancer cells prefer glycolysis for energy production under aerobic conditions, highlighting their metabolic flexibility (Warburg et al., 1923; Warburg et al., 1924). Circadian rhythms regulate key biological processes associated with material and energy metabolism, including sleep-wake cycles (Daan et al., 1984; Wendrich et al., 2023), thermogenesis (Hasan et al., 2021), food intake (Teixeira et al., 2022), and glucose and lipid metabolism (Kalsbeek et al., 2010; Frazier et al., 2023; Small et al., 2023). Previous studies showed that mice lacking Bmal1 or Clock genes exhibit significant defects in gluconeogenesis, lipid, and glucose metabolism (Bolshette et al., 2023). In Drosophila, dietary restriction prolonged lifespan via promoting fat metabolism, which linked to increased oscillations and expressions of Per and Tim proteins (Katewa et al., 2016). Research on transgenic mice with the hPER2 S662G mutation revealed that derived lung cancer cells increased glucose, glutamine, and lactic acid consumption. Isotope tracing has also illustrated enhanced glucose utilization in cancer cell glycolysis and the tricarboxylic acid cycle (Papagiannakopoulos et al., 2016). Furthermore, Gong et al. (Gong et al., 2021) found that PER1 gene knockdown raised levels of key glycolytic enzymes, boosting glucose uptake and lactate production in oral squamous cell carcinoma cells. These studies indicate that disrupting PER1 or PER2 gene functions in cancer cells can lead to elevated metabolism and energy production, supporting their rapid proliferation and adaptability.
The tumor microenvironment (TME) comprises various cellular components (such as immune cells, fibroblasts, and endothelial cells) and acellular elements (including cytokines, growth factors, and the extracellular matrix) (Ribeiro Franco et al., 2020). Innate and adaptive immune cells within the TME interact with cancer cells directly or through chemokine and cytokine signaling (Liu et al., 2023a; Xiong et al., 2023; Zetrini et al., 2023). These interactions significantly influence the biological behavior and therapeutic responses of cancer cells, impacting patients’ clinical outcomes and prognosis (Neophytou et al., 2021). For example, a comprehensive pan-cancer analysis demonstrated a strong association between the upregulation of immunosuppressive molecules like PD-L1 and CTLA-4, and the downregulation of PER1, PER2, and PER3 genes, underscoring the impact of disrupted clock genes on T cell exhaustion and immune evasion in the TME (Wu et al., 2019). In ovarian cancer, B lymphocyte, macrophage, and neutrophil infiltration levels were inversely correlated with PER1 gene expression (Chen et al., 2021). Recently, a colon cancer research indicated that the epigenetic regulator CBX4 was negatively associated with myeloid-derived suppressor cells and cancer-associated fibroblasts, and showed coordinated expression with PER1 and PER3 genes (Wei et al., 2021). Yang et al. (Yang et al., 2019) found a specific circadian rhythm in PD-L1 expression and a positive correlation between the PER1 gene and CD4+ and CD8+ T cell infiltration on lung cancer. Furthermore, in endometrial cancer, the PER1 gene is linked to immunological factors like PD-1/PD-L1 and inflammatory markers such as TNF-α and IL-6 (Wang et al., 2020c). Additionally, chronic shift-lag-induced suppression of Per1 and Per2 genes impaired natural killer (NK) cell-mediated immunosurveillance and promoted tumorigenesis in mice, potentially due to decreased expression of immune functional receptors like Ly49D, Ly49G2, and Ly49H (Zeng et al., 2020). These studies robustly support the PER gene family’s regulatory role in immune cell function and tumor immune cell invasion.
To enhance patient prognosis, chemotherapy employs cytotoxic chemicals to eradicate cancer cells (van Stein et al., 2021). It may also serve as an adjuvant therapy alongside radiotherapy or surgery (Liu et al., 2022c). However, factors such as intratumor heterogeneity, adaptive mutations, epigenetic alterations, and metabolic changes enable some cancer cells to withstand clinical doses of medications, thereby escalating the challenge of medication resistance in cancer treatment (Vasan et al., 2019). Investigating the molecular mechanisms of drug resistance is crucial for improving therapeutic outcomes and introducing new treatment strategies (Nussinov et al., 2021; Liu et al., 2023b). Previous research revealed that oncogene-transformed mouse embryonic fibroblasts developed increased resistance to chemotherapeutic agents like methotrexate, gemcitabine, and etoposide, due to alterations of Per2 gene. Wang et al. (Wang et al., 2020d) found a link between the downregulation of PER2 gene expression in cisplatin-resistant ovarian cancer cells and the PI3K/AKT signaling pathway, which led to the activation of the multidrug resistance gene 1 (MDR1). Conversely, upregulating the PER2 gene in cervical cancer cells inhibited the PI3K/AKT pathway, diminishing multidrug resistance protein production and enhancing cisplatin’s lethal effect on cancer cells (Wang et al., 2022). Additionally, Cai et al. (Cai et al., 2018) noted a downregulation of PER3 gene in prostate cancer patients with paclitaxel-resistant, while the paclitaxel sensitivity of cancer cells could be rescued by overexpressing the PER3 gene. Moreover, Redondo and colleagues observed that lower PER2 gene expression increased the susceptibility of esophageal cancer cells to cisplatin, promoting higher rates of cell apoptosis (Redondo et al., 2021).
Radiation therapy is a prevalent cancer treatment modality. Increasing research focuses on the link between circadian rhythm and radiotherapy, given that specific circadian clock genes are vital for DNA repair and apoptosis induced by ionizing radiation and influence cell sensitivity to radiation at different cell cycle stages (Amiama-Roig et al., 2022). In glioma research, Zhu et al. (Zhu et al., 2019) observed that the downregulation of the PER1 gene in U343 cells diminished X-ray-induced DNA damage and cell death through the CHK2-TP53 pathway. A comprehensive cohort study of 1,690 breast cancer patients demonstrated a significant association between the genotype variations of PER3 gene and radiotherapy side-effects, suggesting the potential of the PER3 gene as a predictor for radiotherapy response (Webb et al., 2022). Furthermore, studies have explored the impact of radiation timing on long-term prognosis and adverse patient outcomes, along with the predictive capacity of clock gene profiles for radiotherapy efficacy (Jin et al., 2021; Sapienza et al., 2021; Kong et al., 2022; Tang et al., 2023).
Several studies have focused on the PER gene family in cell and animal models to augment cancer therapy. For instance, Yang et al. (Yang and Stockwell, 2008) demonstrated that IC261, a CKⅠε inhibitor, impedes fibrosarcoma cell proliferation by stabilizing PER2 protein. Oshima et al. (Oshima et al., 2019) conducted a proteomic analysis and identified that the CKⅡ inhibitor GO289 inhibited the PER2 phosphorylation sites, leading to extended circadian rhythms and suppressed growth in kidney cancer and acute myeloid leukemia cells. LY2857785, a CDK9 inhibitor, has been shown to decrease core clock protein levels including BMAL1 and PER2, by upregulating REV-ERBα expression (Ou et al., 2019). Additionally, mutations of PER2 could shorten circadian rhythms in mice induced by N-ethyl-N-nitrosourea (ENU) in the PAS domain, suggesting the PAS domain as target within the PER gene family for future interventions (Militi et al., 2016). These findings underscore the potential of targeting the PER gene family in cancer therapy. However, more clinical researches are necessary to elucidate their precise pharmacological effects and minimize unintended adverse impacts on healthy tissues.
As a crucial component of clock genes, the PER gene family in human (PER1, PER2, and PER3) plays a pivotal role in numerous significant pathological processes, including cancer initiation and development. These genes, by modulating downstream gene expression, are instrumental in regulating cell cycle and invasion processes, altering tumor cell metabolism, impacting the tumor immune environment, and influencing treatment responses (Figure 4; Table 1). This modulation leads to varied clinical and pathological features and prognostic outcomes in cancer patients. The dysregulated proliferative phenotype is a universally recognized hallmark of cancer. Consequently, research on the TP53 gene in cancer maintains a prominent and steadfast position, given its direct association with the cell cycle and proliferative phenotype, which may be intricately linked to circadian rhythms and the biological clock. Yet, the extent of the PER gene family’s influence on cancer cell cycles remains incompletely elucidated. In our review, we thoroughly reviewed the discovery history, fundamental biological functions, and the interplay between the PER gene family and various tumor characteristics. We also anticipate further studies on the relationship between the PER gene family and TP53 to elucidate their connections. Our review advocates for in-depth investigations into the PER gene family’s role in specific cancer types or stages, and their molecular mechanisms, to identify potential biomarkers for cancer risk and prognosis.
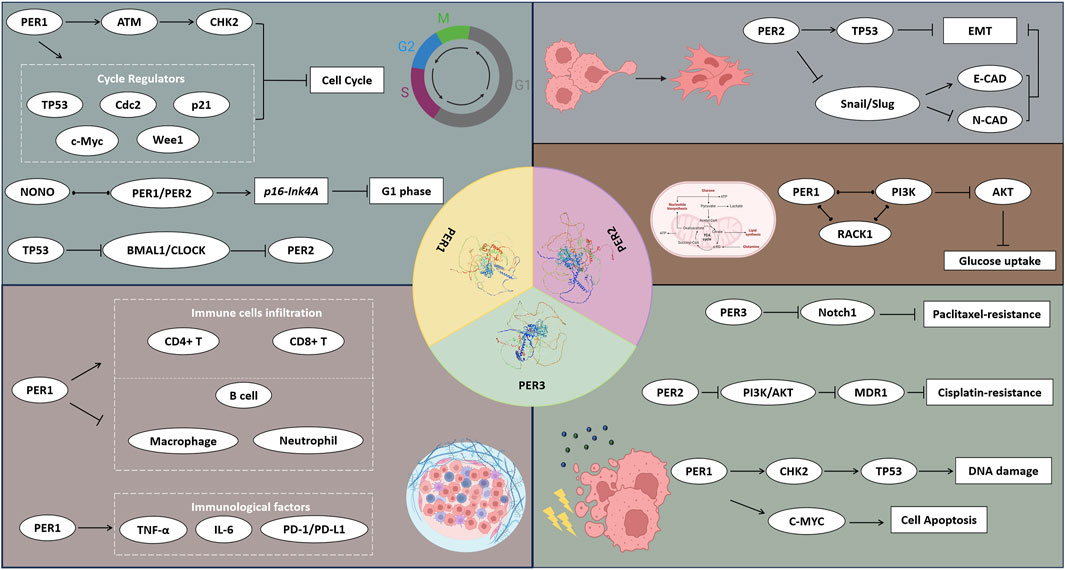
Figure 4. The potential mechanisms of PER1, PER2, and PER3 proteins affecting the hall markers of cancers. The PER1, PER2, and PER3 proteins influence the cell function, metabolism, TME, and therapy responses of cancers via a wide range of signal pathways and regulatory factors.
In future studies, our focus on the PER gene family will encompass several key areas (Feng et al., 2023a): Developing personalized treatment strategies: By harnessing a thorough comprehension of the PER genes’ expression and functional disparities across different cancer types, we aim to craft more targeted treatment methodologies to improve patient responses and survival rates (Hanahan, 2022); Integrating immunotherapy: This involves exploring how immunotherapy can enhance the immune system’s ability to detect and eliminate tumors, potentially including the creation of immunotherapeutic approaches directly linked to PER genes regulation (Shen et al., 2022); Deciphering treatment resistance mechanisms: Conducting detailed analyses of the resistance mechanisms associated with the PER genes to ascertain why some patients resist certain treatments, thereby providing insights for novel therapeutic development (Wang et al., 2023); Discovering early diagnostic and predictive biomarkers: Leveraging insights into the regulatory roles of PER genes in various biological processes to identify novel early diagnostic or predictive biomarkers, aiming to improve patient outcomes (Feng et al., 2023b); Fostering multidisciplinary collaboration and technological innovation: By enhancing collaboration across disciplines and integrating cutting-edge technologies from bioinformatics, molecular biology, and immunology, we plan to conduct an in-depth and comprehensive exploration of the mechanisms of action of PER genes in cancer.
KC: Conceptualization, Data curation, Formal Analysis, Investigation, Resources, Writing–original draft, Writing–review and editing. YW: Conceptualization, Data curation, Formal Analysis, Validation, Writing–original draft, Writing–review and editing. DL: Data curation, Formal Analysis, Investigation, Writing–review and editing. RW: Data curation, Formal Analysis, Investigation, Writing–review and editing. JW: Data curation, Writing–review and editing. WW: Data curation, Formal Analysis, Validation, Writing–review and editing. WZ: Data curation, Writing–review and editing. WX: Data curation, Writing–review and editing. DF: Project administration, Supervision, Validation, Writing–review and editing. YH: Project administration, Supervision, Validation, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This program was supported by the Chinese Scholarship Council (grant no. 202206240086), National Key Research and Development Program of China (2021YFC2009303), Project of Health Commission of Sichuan Province (21PJ041), the Key Research and Development Support Plan of Chengdu Science and Technology Bureau (2022-YF05-01568-SN), 2023 Jiaxing Key Discipline of Medicine-Urology (Supporting Subject) (2023-ZC-013) and 2020 Jiaxing Key Laboratory of Precision Diagnosis and Treatment of Urological Tumors (2020-MNZDSYS). The funders had no role in the study design, data collection or analysis, preparation of the manuscript, or the decision to publish.
We appreciated the Figdraw (www.figdraw.com), the Biorender (www.biorender.com/), the SWISS-MODEL (https://swissmodel.expasy.org/) and Chengdu Basebiotech Co., Ltd. for their assistance in drawing and data process.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, M., Li, W., and Top, D. (2021). Integration of circadian clock information in the Drosophila circadian neuronal network. J. Biol. Rhythms 36 (3), 203–220. Epub 2021/03/02. doi:10.1177/0748730421993953
Allada, R., and Bass, J. (2021). Circadian mechanisms in medicine. N. Engl. J. Med. 384 (6), 550–561. Epub 2021/02/11. doi:10.1056/NEJMra1802337
Allada, R., White, N. E., So, W. V., Hall, J. C., and Rosbash, M. (1998). A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell. 93 (5), 791–804. Epub 1998/06/18. doi:10.1016/s0092-8674(00)81440-3
Altea-Manzano, P., Cuadros, A. M., Broadfield, L. A., and Fendt, S. M. (2020). Nutrient metabolism and cancer in the in vivo context: a metabolic game of give and take. EMBO Rep. 21 (10), e50635. Epub 2020/09/24. doi:10.15252/embr.202050635
Amiama-Roig, A., Verdugo-Sivianes, E. M., Carnero, A., and Blanco, J. R. (2022). Chronotherapy: circadian rhythms and their influence in cancer therapy. Cancers (Basel) 14 (20), 5071. Epub 2022/10/28. doi:10.3390/cancers14205071
Angelousi, A., Kassi, E., Ansari-Nasiri, N., Randeva, H., Kaltsas, G., and Chrousos, G. (2019). Clock genes and cancer development in particular in endocrine tissues. Endocr. Relat. Cancer 26 (6), R305-R317–R17. Epub 2019/04/09. doi:10.1530/ERC-19-0094
Association, N. (2023). National health commission guidelines for Diagnosis and treatment of colorectal cancer 2023 in China (English version). Chin. J. Cancer Res. 35 (3), 197–232. Epub 2023/07/13. doi:10.21147/j.issn.1000-9604.2023.03.01
Aytac, H. M., Pehlivan, M., Oyaci, Y., and Pehlivan, S. (2022). Period3 (Per3) vntr variant associated with seasonal pattern and family history in bipolar disorder. Psychiatr. Danub 34 (4), 695–699. Epub 2022/12/23. doi:10.24869/psyd.2022.695
Azevedo, P. G., Miranda, L. R., Nicolau, E. S., Alves, R. B., Bicalho, M. A. C., Couto, P. P., et al. (2021). Genetic association of the period3 (Per3) clock gene with extreme obesity. Obes. Res. Clin. Pract. 15 (4), 334–338. Epub 2021/07/04. doi:10.1016/j.orcp.2021.06.006
Bakir, B., Chiarella, A. M., Pitarresi, J. R., and Emt, R. A. K. (2020). EMT, MET, plasticity, and tumor metastasis. Trends Cell. Biol. 30 (10), 764–776. Epub 2020/08/18. doi:10.1016/j.tcb.2020.07.003
Bao, H., Min, L., Bu, F., Wang, S., and Meng, J. (2023). Recent advances of liquid biopsy: interdisciplinary strategies toward clinical decision-making. Interdiscip. Med. 1 (4), e20230021. doi:10.1002/INMD.20230021
Bargiello, T. A., and Young, M. W. (1984). Molecular genetics of a biological clock in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 81 (7), 2142–2146. Epub 1984/04/01. doi:10.1073/pnas.81.7.2142
Barragan, R., Sorli, J. V., Coltell, O., Gonzalez-Monje, I., Fernandez-Carrion, R., Villamil, L. V., et al. (2022). Influence of DNA-polymorphisms in selected circadian clock genes on clock gene expression in subjects from the general population and their association with sleep duration. Med. Kaunas. 58 (9), 1294. Epub 2022/09/24. doi:10.3390/medicina58091294
Basti, A., Malhan, D., Dumbani, M., Dahlmann, M., Stein, U., and Relogio, A. (2022). Core-clock genes regulate proliferation and invasion via a reciprocal interplay with Macc1 in colorectal cancer cells. Cancers (Basel) 14 (14), 3458. Epub 2022/07/28. doi:10.3390/cancers14143458
Berge, L. A. M., Liu, F. C., Grimsrud, T. K., Babigumira, R., Støer, N. C., Kjærheim, K., et al. (2023). Night shift work and risk of aggressive prostate cancer in the Norwegian offshore petroleum workers (nopw) cohort. Int. J. Epidemiol. 52 (4), 1003–1014. Epub 2022/12/23. doi:10.1093/ije/dyac235
Bolshette, N., Ibrahim, H., Reinke, H., and Asher, G. (2023). Circadian regulation of liver function: from molecular mechanisms to disease pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 20, 695–707. Epub 2023/06/09. doi:10.1038/s41575-023-00792-1
Cadenas, C., van de Sandt, L., Edlund, K., Lohr, M., Hellwig, B., Marchan, R., et al. (2014). Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell. Cycle 13 (20), 3282–3291. Epub 2014/12/09. doi:10.4161/15384101.2014.954454
Cai, D. W., Chen, D., Sun, S. P., Liu, Z. J., Liu, F., Xian, S. Z., et al. (2018). Overexpression of Per3 reverses paclitaxel resistance of prostate cancer cells by inhibiting the notch pathway. Eur. Rev. Med. Pharmacol. Sci. 22 (9), 2572–2579. Epub 2018/05/18. doi:10.26355/eurrev_201805_14950
Cao, X., Yang, Y., Selby, C. P., Liu, Z., and Sancar, A. (2021). Molecular mechanism of the repressive phase of the mammalian circadian clock. Proc. Natl. Acad. Sci. U. S. A. 118 (2), e2021174118. Epub 2021/01/15. doi:10.1073/pnas.2021174118
Chan, K. I., Zhang, S., Li, G., Xu, Y., Cui, L., Wang, Y., et al. (2023). Myc oncogene: a druggable target for treating cancers with natural products. Aging Dis. 15, 640–697. Epub 2023/07/14. doi:10.14336/ad.2023.0520
Chen, H., Ma, X., Liu, J., Yang, Y., Fang, Y., Wang, L., et al. (2022). Clinical outcomes of atezolizumab in combination with etoposide/platinum for treatment of extensive-stage small-cell lung cancer: a real-world, multicenter, retrospective, controlled study in China. Chin. J. Cancer Res. 34 (4), 353–364. Epub 2022/10/07. doi:10.21147/j.issn.1000-9604.2022.04.04
Chen, H., Yang, P., Yang, D., Wang, D., Lu, M., Li, Y., et al. (2023). The per3(rs772027021) snp induces pigmentation phenotypes of dyschromatosis universalis hereditaria. J. Mol. Med. Berl. 101 (3), 279–294. Epub 2023/02/16. doi:10.1007/s00109-023-02288-6
Chen, M., Zhang, L., Liu, X., Ma, Z., and Lv, L. (2021). Per1 is a prognostic biomarker and correlated with immune infiltrates in ovarian cancer. Front. Genet. 12, 697471. Epub 2021/07/06. doi:10.3389/fgene.2021.697471
Chen, P., Hsu, W. H., Chang, A., Tan, Z., Lan, Z., Zhou, A., et al. (2020). Circadian regulator clock recruits immune-suppressive microglia into the gbm tumor microenvironment. Cancer Discov. 10 (3), 371–381. Epub 2020/01/11. doi:10.1158/2159-8290.Cd-19-0400
Chen, S., Yang, J., Yang, L., Zhang, Y., Zhou, L., Liu, Q., et al. (2018). Ubiquitin ligase Traf2 attenuates the transcriptional activity of the core clock protein Bmal1 and affects the maximal Per1 mrna level of the circadian clock in cells. FEBS J. 285 (16), 2987–3001. Epub 2018/06/24. doi:10.1111/febs.14595
Chun, S. K., Fortin, B. M., Fellows, R. C., Habowski, A. N., Verlande, A., Song, W. A., et al. (2022). Disruption of the circadian clock drives apc loss of heterozygosity to accelerate colorectal cancer. Sci. Adv. 8 (32), eabo2389. Epub 2022/08/11. doi:10.1126/sciadv.abo2389
Cordina-Duverger, E., Uchai, S., Tvardik, N., Billmann, R., Martin, D., Trédaniel, J., et al. (2022). Sleep traits, night shift work and lung cancer risk among women: results from a population-based case-control study in France (the welca study). Int. J. Environ. Res. Public Health 19 (23), 16246. Epub 2022/12/12. doi:10.3390/ijerph192316246
Cox, K. H., and Takahashi, J. S. (2019). Circadian clock genes and the transcriptional architecture of the clock mechanism. J. Mol. Endocrinol. 63 (4), R93-R102–R102. Epub 2019/09/27. doi:10.1530/JME-19-0153
Crnko, S., Du Pré, B. C., Sluijter, J. P. G., and Van Laake, L. W. (2019). Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat. Rev. Cardiol. 16 (7), 437–447. Epub 2019/02/24. doi:10.1038/s41569-019-0167-4
Daan, S., Beersma, D. G., and Borbély, A. A. (1984). Timing of human sleep: recovery process gated by a circadian pacemaker. Am. J. Physiol. 246 (2 Pt 2), R161–R183. Epub 1984/02/01. doi:10.1152/ajpregu.1984.246.2.R161
Dagmura, H., Yigit, S., Nursal, A. F., Duman, E., and Gumusay, O. (2021). Possible association of per2/per3 variable number tandem repeat polymorphism variants with susceptibility and clinical characteristics in pancreatic cancer. Genet. Test. Mol. Biomarkers 25 (2), 124–130. doi:10.1089/gtmb.2020.0179
De, A., Beligala, D. H., Sharma, V. P., Burgos, C. A., Lee, A. M., and Geusz, M. E. (2020). Cancer stem cell generation during epithelial-mesenchymal transition is temporally gated by intrinsic circadian clocks. Clin. Exp. Metastasis 37 (5), 617–635. Epub 2020/08/21. doi:10.1007/s10585-020-10051-1
de Jong, F. C., Laajala, T. D., Hoedemaeker, R. F., Jordan, K. R., van der Made, A. C. J., Boeve, E. R., et al. (2023). Non-muscle-invasive bladder cancer molecular subtypes predict differential response to intravesical Bacillus calmette-guerin. Sci. Transl. Med. 15 (697), eabn4118. Epub 2023/05/24. doi:10.1126/scitranslmed.abn4118
Deng, F., and Yang, K. (2019). Current status of research on the period family of clock genes in the occurrence and development of cancer. J. Cancer 10 (5), 1117–1123. Epub 2019/03/12. doi:10.7150/jca.29212
Eide, E. J., Woolf, M. F., Kang, H., Woolf, P., Hurst, W., Camacho, F., et al. (2005). Control of mammalian circadian rhythm by ckiepsilon-regulated proteasome-mediated Per2 Degradation. Mol. Cell. Biol. 25 (7), 2795–2807. Epub 2005/03/16. doi:10.1128/mcb.25.7.2795-2807.2005
Engeland, K. (2022). Cell cycle regulation: P53-P21-Rb signaling. Cell. Death Differ. 29 (5), 946–960. Epub 2022/04/02. doi:10.1038/s41418-022-00988-z
Farshadi, E., van der Horst, G. T. J., and Chaves, I. (2020). Molecular links between the circadian clock and the cell cycle. J. Mol. Biol. 432 (12), 3515–3524. Epub 2020/04/19. doi:10.1016/j.jmb.2020.04.003
Feng, D., Li, D., Wang, J., Wu, R., and Zhang, C. (2023b). Senescence-associated lncrnas indicate distinct molecular subtypes associated with prognosis and androgen response in patients with prostate cancer. Acta Mater. Medica 2 (3), 299–309. doi:10.15212/amm-2023-0025
Feng, D., Li, D., Xiao, Y., Wu, R., Wang, J., and Zhang, C. (2023a). Focal ablation therapy presents promising results for selectively localized prostate cancer patients. Chin. J. Cancer Res. 35 (4), 424–430. Epub 2023/09/11. doi:10.21147/j.issn.1000-9604.2023.04.08
Feng, D., Xiong, Q., Zhang, F., Shi, X., Xu, H., Wei, W., et al. (2022). Identification of a novel nomogram to predict progression based on the circadian clock and insights into the tumor immune microenvironment in prostate cancer. Front. Immunol. 13, 777724. Epub 2022/02/15. doi:10.3389/fimmu.2022.777724
Fores-Martos, J., Cervera-Vidal, R., Sierra-Roca, J., Lozano-Asencio, C., Fedele, V., Cornelissen, S., et al. (2021). Circadian performance in breast cancer: a germline and somatic genetic study of per3(vntr) polymorphisms and gene Co-expression. npj Breast Cancer 7 (1), 118. Epub 2021/09/12. doi:10.1038/s41523-021-00329-2
Frazier, K., Manzoor, S., Carroll, K., DeLeon, O., Miyoshi, S., Miyoshi, J., et al. (2023). Gut microbes and the liver circadian clock partition glucose and lipid metabolism. J. Clin. Investig. 133 (18), e162515. Epub 2023/09/15. doi:10.1172/jci162515
Gao, Y., Wu, Y., Zhang, N., Yuan, H., Wang, F., Xu, H., et al. (2021). Idh1 gene mutation activates smad signaling molecules to regulate the expression levels of cell cycle and biological rhythm genes in human glioma U87-Mg cells. Mol. Med. Rep. 23 (5), 354. Epub 2021/03/25. doi:10.3892/mmr.2021.11993
Gery, S., Komatsu, N., Baldjyan, L., Yu, A., Koo, D., and Koeffler, H. P. (2006). The circadian gene Per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell. 22 (3), 375–382. Epub 2006/05/09. doi:10.1016/j.molcel.2006.03.038
Gong, X., Tang, H., and Yang, K. (2021). Per1 suppresses glycolysis and cell proliferation in oral squamous cell carcinoma via the per1/rack1/pi3k signaling complex. Cell. Death Dis. 12 (3), 276. Epub 2021/03/17. doi:10.1038/s41419-021-03563-5
Guo, F., Tang, Q., Chen, G., Sun, J., Zhu, J., Jia, Y., et al. (2020). Aberrant expression and subcellular localization of Per2 promote the progression of oral squamous cell carcinoma. Biomed. Res. Int. 2020, 8587458. Epub 2020/03/19. doi:10.1155/2020/8587458
Han, Y., Meng, F., Venter, J., Wu, N., Wan, Y., Standeford, H., et al. (2016). Mir-34a-Dependent overexpression of Per1 decreases cholangiocarcinoma growth. J. Hepatol. 64 (6), 1295–1304. Epub 2016/03/01. doi:10.1016/j.jhep.2016.02.024
Hanahan, D. (2022). Hallmarks of cancer: new dimensions. Cancer Discov. 12 (1), 31–46. Epub 2022/01/14. doi:10.1158/2159-8290.CD-21-1059
Hasan, N., Nagata, N., Morishige, J. I., Islam, M. T., Jing, Z., Harada, K. I., et al. (2021). Brown adipocyte-specific knockout of Bmal1 causes mild but significant thermogenesis impairment in mice. Mol. Metab. 49, 101202. Epub 2021/03/07. doi:10.1016/j.molmet.2021.101202
He, Z., and Ke, Y. (2023). Challenge and future of cancer screening in China: insights from esophageal cancer screening practice. Chin. J. Cancer Res. 35 (6), 584–594. Epub 2024/01/11. doi:10.21147/j.issn.1000-9604.2023.06.03
Hinoura, T., Mukai, S., Kamoto, T., and Kuroda, Y. (2021). Per3 polymorphisms and their association with prostate cancer risk in Japanese men. J. Prev. Med. Hyg. 62 (2), E489–E495. Epub 2021/10/05. doi:10.15167/2421-4248/jpmh2021.62.2.1865
Holipah, H. T., and Kuroda, Y. (2020). Per1 Rs3027188 polymorphism and its association with the risk of colorectal cancer in the Japanese population. Gan Kagaku Ryoho 47 (6), 941–946. Epub 2020/06/17.
Hong, M., Tao, S., Zhang, L., Diao, L. T., Huang, X., Huang, S., et al. (2020). Rna sequencing: new technologies and applications in cancer research. J. Hematol. Oncol. 13 (1), 166. Epub 2020/12/06. doi:10.1186/s13045-020-01005-x
Hou, D., Lin, C., and Lin, Y. (2022). Diurnal circadian lighting accumulation model: a predictor of the human circadian phase shift phenotype. Phenomics 2 (1), 50–63. doi:10.1007/s43657-021-00039-6
Huang, W., Zong, J., Zhang, Y., Zhou, Y., Zhang, L., Wang, Y., et al. (2023). The role of circadian rhythm in neurological diseases: a translational perspective. Aging Dis. Epub 2023/10/10. doi:10.14336/AD.2023.0921
Huang, Y., Hong, W., and Wei, X. (2022). The molecular mechanisms and therapeutic strategies of emt in tumor progression and metastasis. J. Hematol. Oncol. 15 (1), 129. Epub 2022/09/09. doi:10.1186/s13045-022-01347-8
Jiang, H., Yang, X., Mi, M., Wei, X., Wu, H., Xin, Y., et al. (2021). Per2: a potential molecular marker for hematological malignancies. Mol. Biol. Rep. 48 (11), 7587–7595. Epub 2021/10/14. doi:10.1007/s11033-021-06751-w
Jin, F., Chen, Y., Jiang, Z., Li, Y., Zhao, C., Liu, L., et al. (2021). The correlation study of circadian clock gene Bmal1 regulates the biological behavior of human nasopharyngeal carcinoma cell after radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 111 (3, Suppl. ment), e229. doi:10.1016/j.ijrobp.2021.07.785
Jin, Y. H., Zeng, X. T., Liu, T. Z., Bai, Z. M., Dou, Z. L., Ding, D. G., et al. (2022). Treatment and surveillance for non-muscle-invasive bladder cancer: a clinical practice guideline (2021 edition). Mil. Med. Res. 9 (1), 44. Epub 2022/08/18. doi:10.1186/s40779-022-00406-y
Kalsbeek, A., Yi, C. X., La Fleur, S. E., and Fliers, E. (2010). The hypothalamic clock and its control of glucose homeostasis. Trends Endocrinol. Metab. 21 (7), 402–410. Epub 2010/03/23. doi:10.1016/j.tem.2010.02.005
Katewa, S. D., Akagi, K., Bose, N., Rakshit, K., Camarella, T., Zheng, X., et al. (2016). Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell. Metab. 23 (1), 143–154. Epub 2015/12/03. doi:10.1016/j.cmet.2015.10.014
Kong, D., Liu, Y., and Zhang, M. (2022). Expression of the circadian clock gene arntl associated with DNA repair gene and prognosis of patient with osteosarcoma. Mutat. Res. 825, 111801. Epub 2022/10/22. doi:10.1016/j.mrfmmm.2022.111801
Konopka, R. J., and Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 68 (9), 2112–2116. Epub 1971/09/01. doi:10.1073/pnas.68.9.2112
Kowalska, E., Ripperger, J. A., Hoegger, D. C., Bruegger, P., Buch, T., Birchler, T., et al. (2013). Nono couples the circadian clock to the cell cycle. Proc. Natl. Acad. Sci. U. S. A. 110 (5), 1592–1599. Epub 2012/12/26. doi:10.1073/pnas.1213317110
Laosuntisuk, K., Elorriaga, E., and Doherty, C. J. (2023). The game of timing: circadian rhythms intersect with changing environments. Annu. Rev. Plant Biol. 74, 511–538. Epub 2023/03/01. doi:10.1146/annurev-arplant-070522-065329
Larson, N. B., Oberg, A. L., Adjei, A. A., and Wang, L. (2023). A clinician's guide to bioinformatics for next-generation sequencing. J. Thorac. Oncol. 18 (2), 143–157. Epub 2022/11/16. doi:10.1016/j.jtho.2022.11.006
Lee, Y. I., Shim, J. E., Kim, J., Lee, W. J., Kim, J. W., Nam, K. H., et al. (2022). Wnt5a drives interleukin-6-dependent epithelial-mesenchymal transition via the jak/stat pathway in keloid pathogenesis. Burns Trauma 10, tkac023. Epub 2022/10/14. doi:10.1093/burnst/tkac023
Lesicka, M., Jablonska, E., Wieczorek, E., Peplonska, B., Gromadzinska, J., Seroczynska, B., et al. (2019). Circadian gene polymorphisms associated with breast cancer susceptibility. Int. J. Mol. Sci. 20 (22), 5704. doi:10.3390/ijms20225704
Lesicka, M., Nedoszytko, B., and Reszka, E. (2023). Disruptions of circadian genes in cutaneous melanoma-an in silico analysis of transcriptome databases. Int. J. Mol. Sci. 24 (12), 10140. Epub 2023/06/28. doi:10.3390/ijms241210140
Lévi, F., Filipski, E., Iurisci, I., Li, X. M., and Innominato, P. (2007). Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Cold Spring Harb. Symp. Quant. Biol. 72, 465–475. Epub 2008/04/19. doi:10.1101/sqb.2007.72.030
Li, W., Wang, Z., Cao, J., Dong, Y., and Chen, Y. (2023). Perfecting the life clock: the journey from pto to ttfl. Int. J. Mol. Sci. 24 (3), 2402. Epub 2023/02/12. doi:10.3390/ijms24032402
Li, Y. Y., Jin, F., Zhou, J. J., Yu, F., Duan, X. F., He, X. Y., et al. (2019). Downregulation of the circadian period family genes is positively correlated with poor head and neck squamous cell carcinoma prognosis. Chronobiol Int. 36 (12), 1723–1732. Epub 2019/10/15. doi:10.1080/07420528.2019.1648486
Lin, Y. S., Tsai, K. L., Chen, J. N., and Wu, C. S. (2020). Mangiferin inhibits lipopolysaccharide-induced epithelial-mesenchymal transition (emt) and enhances the expression of tumor suppressor gene Per1 in non-small cell lung cancer cells. Environ. Toxicol. 35 (10), 1070–1081. Epub 2020/05/19. doi:10.1002/tox.22943
Liu, H., Chen, X., Jia, Y., Chen, H., Wang, X., Liu, G., et al. (2023b). Facing inevitable parpis resistance: mechanisms and therapeutic strategies for breast cancer treatment. Interdiscip. Med. 1 (2), e20220013. doi:10.1002/INMD.20220013
Liu, L., Sun, Q., Davis, F., Mao, J., Zhao, H., and Ma, D. (2022b). Epithelial-mesenchymal transition in organ fibrosis development: current understanding and treatment strategies. Burns Trauma 10, tkac011. Epub 2022/04/12. doi:10.1093/burnst/tkac011
Liu, N., Chen, L., Yan, M., Tao, Q., Wu, J., Chen, J., et al. (2023a). Eubacterium rectale improves the efficacy of anti-Pd1 immunotherapy in melanoma via L-serine-mediated nk cell activation. Res. (Wash D C) 6, 0127. Epub 2023/05/24. doi:10.34133/research.0127
Liu, S., Chen, X., and Lin, T. (2022c). Emerging strategies for the improvement of chemotherapy in bladder cancer: current knowledge and future perspectives. J. Adv. Res. 39, 187–202. Epub 2022/07/02. doi:10.1016/j.jare.2021.11.010
Liu, Y., Hao, J., Yuan, G., Wei, M., Bu, Y., Jin, T., et al. (2021). Per1 as a tumor suppressor attenuated in the malignant phenotypes of breast cancer cells. Int. J. Gen. Med. 14, 7077–7087. Epub 2021/10/30. doi:10.2147/IJGM.S328184
Liu, Y., Wu, Z., Li, Y., Zhang, J., Gao, Y., Yuan, G., et al. (2022a). Per3 plays anticancer roles in the oncogenesis and progression of breast cancer via regulating mek/erk signaling pathway. J. Chin. Med. Assoc. 85 (11), 1051–1060. Epub 2022/10/25. doi:10.1097/JCMA.0000000000000781
Lozano-Tovar, S., Rodriguez-Agudelo, Y., Davila-Ortiz de Montellano, D. J., Perez-Aldana, B. E., Ortega-Vazquez, A., and Monroy-Jaramillo, N. (2023). Relationship between apoe, Per2, Per3 and Ox2r genetic variants and neuropsychiatric symptoms in patients with Alzheimer's disease. Int. J. Environ. Res. Public Health 20 (5), 4412. Epub 2023/03/12. doi:10.3390/ijerph20054412
Matthews, H. K., Bertoli, C., and de Bruin, R. A. M. (2022). Cell cycle control in cancer. Nat. Rev. Mol. Cell. Biol. 23 (1), 74–88. Epub 2021/09/12. doi:10.1038/s41580-021-00404-3
McGuirk, S., Audet-Delage, Y., and St-Pierre, J. (2020). Metabolic fitness and plasticity in cancer progression. Trends Cancer 6 (1), 49–61. Epub 2020/01/19. doi:10.1016/j.trecan.2019.11.009
Miki, T., Matsumoto, T., Zhao, Z., and Lee, C. C. (2013). P53 regulates Period2 expression and the circadian clock. Nat. Commun. 4, 2444. Epub 2013/09/21. doi:10.1038/ncomms3444
Militi, S., Maywood, E. S., Sandate, C. R., Chesham, J. E., Barnard, A. R., Parsons, M. J., et al. (2016). Early doors (edo) mutant mouse reveals the importance of period 2 (Per2) pas domain structure for circadian pacemaking. Proc. Natl. Acad. Sci. U. S. A. 113 (10), 2756–2761. Epub 2016/02/24. doi:10.1073/pnas.1517549113
Mokhtari-Hessari, P., and Montazeri, A. (2020). Health-related quality of life in breast cancer patients: review of reviews from 2008 to 2018. Health Qual. Life Outcomes 18 (1), 338. Epub 2020/10/14. doi:10.1186/s12955-020-01591-x
Morales-Santana, S., Morell, S., Leon, J., Carazo-Gallego, A., Jimenez-Lopez, J. C., and Morell, M. (2019). An overview of the polymorphisms of circadian genes associated with endocrine cancer. Front. Endocrinol. (Lausanne) 10, 104. Epub 2019/03/16. doi:10.3389/fendo.2019.00104
Nakamura, T. J., Takasu, N. N., Sakazume, S., Matsumoto, Y., Kawano, N., Pendergast, J. S., et al. (2023). Long days restore regular estrous cyclicity in mice lacking circadian rhythms. Heliyon 9 (6), e16970. Epub 2023/07/24. doi:10.1016/j.heliyon.2023.e16970
Neophytou, C. M., Panagi, M., Stylianopoulos, T., and Papageorgis, P. (2021). The role of tumor microenvironment in cancer metastasis: molecular mechanisms and therapeutic opportunities. Cancers (Basel) 13 (9), 2053. Epub 2021/05/01. doi:10.3390/cancers13092053
Niu, L., Zhang, F., Xu, X., Yang, Y., Li, S., Liu, H., et al. (2022). Chronic sleep deprivation altered the expression of circadian clock genes and aggravated Alzheimer's disease neuropathology. Brain Pathol. 32 (3), e13028. Epub 2021/10/21. doi:10.1111/bpa.13028
Nussinov, R., Tsai, C. J., and Jang, H. (2021). Anticancer drug resistance: an update and perspective. Drug Resist Updat 59, 100796. Epub 2021/12/27. doi:10.1016/j.drup.2021.100796
Orhan, T., Nielsen, P. B., Hviid, T. V. F., Rosen, A. W., and Gögenür, I. (2019). Expression of circadian clock genes in human colorectal cancer tissues using droplet digital pcr. Cancer Investig. 37 (2), 90–98. Epub 2019/02/09. doi:10.1080/07357907.2019.1571079
Oshima, T., Niwa, Y., Kuwata, K., Srivastava, A., Hyoda, T., Tsuchiya, Y., et al. (2019). Cell-based screen identifies a new potent and highly selective Ck2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci. Adv. 5 (1), eaau9060. Epub 2019/02/13. doi:10.1126/sciadv.aau9060
Ou, J., Li, H., Qiu, P., Li, Q., Chang, H. C., and Tang, Y. C. (2019). CDK9 modulates circadian clock by attenuating REV-ERBα activity. Biochem. Biophys. Res. Commun. 513 (4), 967–973. Epub 2019/04/22. doi:10.1016/j.bbrc.2019.04.043
Papagiannakopoulos, T., Bauer, M. R., Davidson, S. M., Heimann, M., Subbaraj, L., Bhutkar, A., et al. (2016). Circadian rhythm disruption promotes lung tumorigenesis. Cell. Metab. 24 (2), 324–331. Epub 2016/08/02. doi:10.1016/j.cmet.2016.07.001
Park, H., Tseng, S. C., Sholl, L. M., Hatabu, H., Awad, M. M., and Nishino, M. (2022). Molecular characterization and therapeutic approaches to small cell lung cancer: imaging implications. Radiology 305 (3), 512–525. Epub 2022/10/26. doi:10.1148/radiol.220585
Pastushenko, I., and Blanpain, C. (2019). Emt transition states during tumor progression and metastasis. Trends Cell. Biol. 29 (3), 212–226. Epub 2018/12/31. doi:10.1016/j.tcb.2018.12.001
Patke, A., Young, M. W., and Axelrod, S. (2020). Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell. Biol. 21 (2), 67–84. Epub 2019/11/27. doi:10.1038/s41580-019-0179-2
Patton, A. P., and Hastings, M. H. (2023). The mammalian circadian time-keeping system. J. Huntingt. Dis. 12 (2), 91–104. Epub 2023/05/01. doi:10.3233/JHD-230571
Pavlova, N. N., Zhu, J., and Thompson, C. B. (2022). The hallmarks of cancer metabolism: still emerging. Cell. Metab. 34 (3), 355–377. Epub 2022/02/07. doi:10.1016/j.cmet.2022.01.007
Peng, F., Liao, M., Qin, R., Zhu, S., Peng, C., Fu, L., et al. (2022). Regulated cell death (rcd) in cancer: key pathways and targeted therapies. Signal Transduct. Target Ther. 7 (1), 286. Epub 2022/08/14. doi:10.1038/s41392-022-01110-y
Preitner, N., Damiola, F., Lopez-Molina, L., Zakany, J., Duboule, D., Albrecht, U., et al. (2002). The orphan nuclear receptor rev-erbalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 110 (2), 251–260. Epub 2002/08/02. doi:10.1016/s0092-8674(02)00825-5
Qu, M., Zhang, G., Qu, H., Vu, A., Wu, R., Tsukamoto, H., et al. (2023). Circadian regulator bmal1::clock promotes cell proliferation in hepatocellular carcinoma by controlling apoptosis and cell cycle. Proc. Natl. Acad. Sci. U. S. A. 120 (2), e2214829120. Epub 2023/01/04. doi:10.1073/pnas.2214829120
Rahman, S., Kraljevic Pavelic, S., and Markova-Car, E. (2019). Circadian (De)Regulation in head and neck squamous cell carcinoma. Int. J. Mol. Sci. 20 (11), 2662. Epub 2019/06/04. doi:10.3390/ijms20112662
Reddy, P., Zehring, W. A., Wheeler, D. A., Pirrotta, V., Hadfield, C., Hall, J. C., et al. (1984). Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 38 (3), 701–710. Epub 1984/10/01. doi:10.1016/0092-8674(84)90265-4
Redondo, J. A., Bibes, R., Vercauteren Drubbel, A., Dassy, B., Bisteau, X., Maury, E., et al. (2021). Per2 circadian oscillation sensitizes esophageal cancer cells to chemotherapy. Biol. (Basel) 10 (4), 266. Epub 2021/04/04. doi:10.3390/biology10040266
Reischl, S., Vanselow, K., Westermark, P. O., Thierfelder, N., Maier, B., Herzel, H., et al. (2007). Beta-Trcp1-Mediated degradation of Period2 is essential for circadian dynamics. J. Biol. Rhythms 22 (5), 375–386. Epub 2007/09/19. doi:10.1177/0748730407303926
Ribeiro Franco, P. I., Rodrigues, A. P., de Menezes, L. B., and Pacheco, M. M. (2020). Tumor microenvironment components: allies of cancer progression. Pathol. Res. Pract. 216 (1), 152729. Epub 2019/11/19. doi:10.1016/j.prp.2019.152729
Ripperger, J. A., and Schibler, U. (2006). Rhythmic clock-bmal1 binding to multiple E-box motifs drives circadian dbp transcription and chromatin transitions. Nat. Genet. 38 (3), 369–374. Epub 2006/02/14. doi:10.1038/ng1738
Rutila, J. E., Suri, V., Le, M., So, W. V., Rosbash, M., and Hall, J. C. (1998). Cycle is a second bhlh-pas clock protein essential for circadian rhythmicity and transcription of Drosophila Period and timeless. Cell. 93 (5), 805–814. Epub 1998/06/18. doi:10.1016/s0092-8674(00)81441-5
Sahar, N. E., Qadir, J., Riaz, S. K., Bagabir, S. A., Muneer, Z., Sheikh, A. K., et al. (2022). Dysregulated expression of suppressor loop of circadian rhythm genes in colorectal cancer pathogenesis. Minerva Med. 113 (3), 497–505. Epub 2022/07/21. doi:10.23736/S0026-4806.22.07981-2
Sancar, A., and Van Gelder, R. N. (2021). Clocks, cancer, and chronochemotherapy. Science 371 (6524), eabb0738. Epub 2021/01/02. doi:10.1126/science.abb0738
Sapienza, L. G., Nasra, K., Berry, R., Danesh, L., Little, T., and Abu-Isa, E. (2021). Clinical effects of morning and afternoon radiotherapy on high-grade gliomas. Chronobiol Int. 38 (5), 732–741. Epub 2021/02/10. doi:10.1080/07420528.2021.1880426
Sato, T. K., Panda, S., Miraglia, L. J., Reyes, T. M., Rudic, R. D., McNamara, P., et al. (2004). A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron 43 (4), 527–537. Epub 2004/08/18. doi:10.1016/j.neuron.2004.07.018
Schernhammer, E., Bogl, L., Hublin, C., Strohmaier, S., Zebrowska, M., Erber, A., et al. (2023). The association between night shift work and breast cancer risk in the Finnish Twins cohort. Eur. J. Epidemiol. 38 (5), 533–543. Epub 2023/03/26. doi:10.1007/s10654-023-00983-9
Schiliro, C., and Firestein, B. L. (2021). Mechanisms of metabolic reprogramming in cancer cells supporting enhanced growth and proliferation. Cells 10 (5), 1056. Epub 2021/05/06. doi:10.3390/cells10051056
Sehgal, A., Rothenfluh-Hilfiker, A., Hunter-Ensor, M., Chen, Y., Myers, M. P., and Young, M. W. (1995). Rhythmic expression of timeless: a basis for promoting circadian cycles in Period Gene autoregulation. Science 270 (5237), 808–810. Epub 1995/11/03. doi:10.1126/science.270.5237.808
Shen, W., He, J., Hou, T., Si, J., and Chen, S. (2022). Common Pathogenetic mechanisms underlying aging and tumor and means of interventions. Aging Dis. 13 (4), 1063–1091. Epub 2022/07/21. doi:10.14336/ad.2021.1208
Shiromani, P. J., Xu, M., Winston, E. M., Shiromani, S. N., Gerashchenko, D., and Weaver, D. R. (2004). Sleep rhythmicity and homeostasis in mice with targeted disruption of mperiod genes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 287 (1), R47–R57. Epub 2004/03/20. doi:10.1152/ajpregu.00138.2004
Siepka, S. M., Yoo, S. H., Park, J., Song, W., Kumar, V., Hu, Y., et al. (2007). Circadian mutant overtime reveals F-box protein Fbxl3 regulation of Cryptochrome and Period Gene expression. Cell. 129 (5), 1011–1023. Epub 2007/04/28. doi:10.1016/j.cell.2007.04.030
Sirhan, Z., Thyagarajan, A., and Sahu, R. P. (2022). The efficacy of tucatinib-based therapeutic approaches for her2-positive breast cancer. Mil. Med. Res. 9 (1), 39. Epub 2022/07/13. doi:10.1186/s40779-022-00401-3
Small, L., Lundell, L. S., Iversen, J., Ehrlich, A. M., Dall, M., Basse, A. L., et al. (2023). Seasonal light hours modulate peripheral clocks and energy metabolism in mice. Cell. Metab. 35 (10), 1722–1735.e5. Epub 2023/09/10. doi:10.1016/j.cmet.2023.08.005
Song, S., Lei, L., Zhang, R., Liu, H., Du, J., Li, N., et al. (2023). Circadian disruption and breast cancer risk: evidence from a case-control study in China. Cancers (Basel) 15 (2), 419. Epub 2023/01/22. doi:10.3390/cancers15020419
Starnes, A. N., and Jones, J. R. (2023). Inputs and outputs of the mammalian circadian clock. Biol. (Basel) 12 (4), 508. Epub 2023/04/28. doi:10.3390/biology12040508
Straif, K., Baan, R., Grosse, Y., Secretan, B., El Ghissassi, F., Bouvard, V., et al. (2007). Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 8 (12), 1065–1066. Epub 2007/12/01. doi:10.1016/s1470-2045(07)70373-x
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. Epub 2021/02/05. doi:10.3322/caac.21660
Suski, J. M., Braun, M., Strmiska, V., and Sicinski, P. (2021). Targeting cell-cycle machinery in cancer. Cancer Cell. 39 (6), 759–778. Epub 2021/04/24. doi:10.1016/j.ccell.2021.03.010
Tang, Y., Li, Y., Zhao, C., Liu, L., He, Q., Li, Y., et al. (2023). Effects of biological clock gene Bmal1 and hypoxia-inducible factor hif-1α on proliferation, migration and radiotherapy sensitivity of nasopharyngeal carcinoma cells Hone1. Holist. Integr. Oncol. 2 (1), 26. doi:10.1007/s44178-023-00044-x
Teixeira, G. P., Guimarães, K. C., Soares, A., Marqueze, E. C., Moreno, C. R. C., Mota, M. C., et al. (2022). Role of chronotype in dietary intake, meal timing, and obesity: a systematic review. Nutr. Rev. 81 (1), 75–90. Epub 2022/07/01. doi:10.1093/nutrit/nuac044
Todo, T., Ryo, H., Yamamoto, K., Toh, H., Inui, T., Ayaki, H., et al. (1996). Similarity among the Drosophila (6-4)Photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science 272 (5258), 109–112. Epub 1996/04/05. doi:10.1126/science.272.5258.109
van Stein, R. M., Aalbers, A. G. J., Sonke, G. S., and van Driel, W. J. (2021). Hyperthermic intraperitoneal chemotherapy for ovarian and colorectal cancer: a review. JAMA Oncol. 7 (8), 1231–1238. Epub 2021/05/07. doi:10.1001/jamaoncol.2021.0580
Vasan, N., Baselga, J., and Hyman, D. M. (2019). A view on drug resistance in cancer. Nature 575 (7782), 299–309. Epub 2019/11/15. doi:10.1038/s41586-019-1730-1
Vitaterna, M. H., King, D. P., Chang, A. M., Kornhauser, J. M., Lowrey, P. L., McDonald, J. D., et al. (1994). Mutagenesis and mapping of a mouse gene, clock, essential for circadian behavior. Science 264 (5159), 719–725. Epub 1994/04/29. doi:10.1126/science.8171325
Vitaterna, M. H., Shimomura, K., and Jiang, P. (2019). Genetics of circadian rhythms. Neurol. Clin. 37 (3), 487–504. Epub 2019/07/02. doi:10.1016/j.ncl.2019.05.002
Walter, W., Pfarr, N., Meggendorfer, M., Jost, P., Haferlach, T., and Weichert, W. (2022). Next-generation diagnostics for precision oncology: Preanalytical considerations, technical challenges, and available technologies. Semin. Cancer Biol. 84, 3–15. Epub 2020/11/11. doi:10.1016/j.semcancer.2020.10.015
Wang, J., Wei, J. C., and Inuzuka, H. (2023). Aging and cancer hallmarks as therapeutic targets. Acta Mater. Medica 2 (3), 281–284. doi:10.15212/amm-2023-0018
Wang, N., Mi, M., Wei, X., and Sun, C. (2020b). Circadian clock gene Period2 suppresses human chronic myeloid leukemia cell proliferation. Exp. Ther. Med. 20 (6), 147. Epub 2020/10/24. doi:10.3892/etm.2020.9276
Wang, Y. H., Li, J. Q., Shi, J. F., Que, J. Y., Liu, J. J., Lappin, J. M., et al. (2020a). Depression and anxiety in relation to cancer incidence and mortality: a systematic review and meta-analysis of cohort studies. Mol. Psychiatry 25 (7), 1487–1499. Epub 2019/11/21. doi:10.1038/s41380-019-0595-x
Wang, Z., Li, F., He, S., Zhao, L., and Wang, F. (2022). Period circadian regulator 2 suppresses drug resistance to cisplatin by pi3k/akt pathway and improves chronochemotherapeutic efficacy in cervical cancer. Gene 809, 146003. Epub 2021/10/15. doi:10.1016/j.gene.2021.146003
Wang, Z., Wang, H., Guo, H., Li, F., Wu, W., Zhang, S., et al. (2020d). The circadian rhythm and core gene Period2 regulate the chemotherapy effect and multidrug resistance of ovarian cancer through the Pi3k signaling pathway. Biosci. Rep. 40 (11). Epub 2020/10/22. doi:10.1042/BSR20202683
Wang, Z., Wang, H., Wang, Z., He, S., Jiang, Z., Yan, C., et al. (2020c). Associated analysis of per1/tubb2b with endometrial cancer development caused by circadian rhythm disorders. Med. Oncol. 37 (10), 90. Epub 2020/09/15. doi:10.1007/s12032-020-01415-4
Wang, Z., Zhao, F., Xu, C., Zhang, Q., Ren, H., Huang, X., et al. (2024). Metabolic reprogramming in skin wound healing. Burns Trauma 12, tkad047. Epub 2024/01/05. doi:10.1093/burnst/tkad047
Warburg, O., Negelein, E., and Posener, K. (1923) Versuche an überlebendem Ca. USA: Gewebe Klin Wochenschr.
Warburg, O., Posener, K., and Negelein, E. (1924). Über den stoffwechsel der carcinomzelle. Naturwissenschaften 12 (50), 1131–1137. doi:10.1007/bf01504608
Webb, A. J., Harper, E., Rattay, T., Aguado-Barrera, M. E., Azria, D., Bourgier, C., et al. (2022). Treatment time and circadian genotype interact to influence radiotherapy side-effects. A prospective European validation study using the requite cohort. EBioMedicine 84, 104269. Epub 2022/09/22. doi:10.1016/j.ebiom.2022.104269
Wei, W., Zhao, W., and Zhang, Y. (2021). Cbx4 provides an alternate mode of colon cancer development via potential influences on circadian rhythm and immune infiltration. Front. Cell. Dev. Biol. 9, 669254. doi:10.3389/fcell.2021.669254
Wendeu-Foyet, M. G., Koudou, Y., Cenee, S., Tretarre, B., Rebillard, X., Cancel-Tassin, G., et al. (2019). Circadian genes and risk of prostate cancer: findings from the epicap study. Int. J. Cancer 145 (7), 1745–1753. Epub 2019/01/22. doi:10.1002/ijc.32149
Wendrich, K. S., Azimi, H., Ripperger, J. A., Ravussin, Y., Rainer, G., and Albrecht, U. (2023). Deletion of the circadian clock gene Per2 in the whole body, but not in neurons or astroglia, affects sleep in response to sleep deprivation. Clocks Sleep. 5 (2), 204–225. Epub 2023/04/24. doi:10.3390/clockssleep5020017
Wu, Y., Tao, B., Zhang, T., Fan, Y., and Mao, R. (2019). Pan-cancer analysis reveals disrupted circadian clock associates with T cell exhaustion. Front. Immunol. 10, 2451. Epub 2019/11/12. doi:10.3389/fimmu.2019.02451
Xiang, R., Cui, Y., Wang, Y., Xie, T., Yang, X., Wang, Z., et al. (2018). Circadian clock gene Per2 downregulation in non-small cell lung cancer is associated with tumour progression and metastasis. Oncol. Rep. 40 (5), 3040–3048. Epub 2018/09/19. doi:10.3892/or.2018.6704
Xing, S., Hu, K., and Wang, Y. (2022). Tumor immune microenvironment and immunotherapy in non-small cell lung cancer: update and New challenges. Aging Dis. 13 (6), 1615–1632. Epub 2022/12/06. doi:10.14336/ad.2022.0407
Xiong, D., Zhang, L., and Sun, Z. J. (2023). Targeting the epigenome to reinvigorate T cells for cancer immunotherapy. Mil. Med. Res. 10 (1), 59. Epub 2023/12/04. doi:10.1186/s40779-023-00496-2
Xiong, H., Yang, Y., Yang, K., Zhao, D., Tang, H., and Ran, X. (2018). Loss of the clock gene Per2 is associated with cancer development and altered expression of important tumor-related genes in oral cancer. Int. J. Oncol. 52 (1), 279–287. Epub 2017/11/09. doi:10.3892/ijo.2017.4180
Xiong, Y., Zhuang, Y., Zhong, M., Qin, W., Huang, B., Zhao, J., et al. (2022). Period 2 suppresses the malignant cellular behaviors of colorectal cancer through the epithelial-mesenchymal transformation process. Cancer control. 29, 10732748221081369. Epub 2022/03/01. doi:10.1177/10732748221081369
Yang, W. S., and Stockwell, B. R. (2008). Inhibition of Casein kinase 1-epsilon induces cancer-cell-selective, period2-dependent growth arrest. Genome Biol. 9 (6), R92. Epub 2008/06/04. doi:10.1186/gb-2008-9-6-r92
Yang, Y., Yuan, G., Xie, H., Wei, T., Zhu, D., Cui, J., et al. (2019). Circadian clock associates with tumor microenvironment in thoracic cancers. Aging (Albany NY) 11 (24), 11814–11828. Epub 2019/12/28. doi:10.18632/aging.102450
Yao, J., He, C., Zhao, W., Hu, N., and Long, D. (2021). Circadian clock and cell cycle: cancer and chronotherapy. Acta histochem. 123 (8), 151816. Epub 2021/11/21. doi:10.1016/j.acthis.2021.151816
Yegin, Z., Sarisoy, G., Erguner Aral, A., and Koc, H. (2021). For whom the circadian clock ticks? Investigation of period and clock gene variants in bipolar disorder. Chronobiol Int. 38 (8), 1109–1119. Epub 2021/06/12. doi:10.1080/07420528.2021.1917594
Yildirim, E., Curtis, R., and Hwangbo, D. S. (2022). Roles of peripheral clocks: lessons from the fly. FEBS Lett. 596 (3), 263–293. Epub 2021/12/05. doi:10.1002/1873-3468.14251
Yin, L., Duan, J. J., Bian, X. W., and Yu, S. C. (2020). Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22 (1), 61. Epub 2020/06/11. doi:10.1186/s13058-020-01296-5
Yin, Y., Yan, Y., Fan, B., Huang, W., Zhang, J., Hu, H. Y., et al. (2023). Novel combination therapy for triple-negative breast cancer based on an intelligent hollow carbon sphere. Res. (Wash D C) 6, 0098. Epub 2023/05/24. doi:10.34133/research.0098
Yu, A., Fu, J., Yin, Z., Yan, H., Xiao, X., Zou, D., et al. (2023). Continuous expression of interferon regulatory factor 4 sustains Cd8(+) T cell immunity against tumor. Res. (Wash D C) 6, 0271. Epub 2024/01/05. doi:10.34133/research.0271
Zehring, W. A., Wheeler, D. A., Reddy, P., Konopka, R. J., Kyriacou, C. P., Rosbash, M., et al. (1984). P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 39 (2 Pt 1), 369–376. Epub 1984/12/01. doi:10.1016/0092-8674(84)90015-1
Zeng, H., Hardin, P. E., and Rosbash, M. (1994). Constitutive overexpression of the Drosophila Period protein inhibits period mrna cycling. EMBO J. 13 (15), 3590–3598. Epub 1994/08/01. doi:10.1002/j.1460-2075.1994.tb06666.x
Zeng, X., Liang, C., and Yao, J. (2020). Chronic shift-lag promotes nk cell ageing and impairs immunosurveillance in mice by decreasing the expression of Cd122. J. Cell. Mol. Med. 24 (24), 14583–14595. Epub 2020/11/14. doi:10.1111/jcmm.16088
Zetrini, A. E., Lip, H., Abbasi, A. Z., Alradwan, I., Ahmed, T., He, C., et al. (2023). Remodeling tumor immune microenvironment by using polymer-lipid-manganese dioxide nanoparticles with radiation therapy to boost immune response of castration-resistant prostate cancer. Res. (Wash D C) 6, 0247. Epub 2023/10/05. doi:10.34133/research.0247
Zhang, C., Chen, L., Sun, L., Jin, H., Ren, K., Liu, S., et al. (2023). Bmal1 collaborates with clock to directly promote DNA double-strand break repair and tumor chemoresistance. Oncogene 42 (13), 967–979. Epub 2023/02/02. doi:10.1038/s41388-023-02603-y
Zhang, J., Lv, H., Ji, M., Wang, Z., and Wu, W. (2020). Low circadian clock genes expression in cancers: a meta-analysis of its association with clinicopathological features and prognosis. PLoS One 15 (5), e0233508. Epub 2020/05/22. doi:10.1371/journal.pone.0233508
Zhang, R., Lahens, N. F., Ballance, H. I., Hughes, M. E., and Hogenesch, J. B. (2014). A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U. S. A. 111 (45), 16219–16224. Epub 2014/10/29. doi:10.1073/pnas.1408886111
Zhang, X., Pant, S. M., Ritch, C. C., Tang, H. Y., Shao, H., Dweep, H., et al. (2024). Cell state dependent effects of Bmal1 on melanoma immunity and tumorigenicity. Nat. Commun. 15 (1), 633. Epub 2024/01/21. doi:10.1038/s41467-024-44778-2
Zhang, Y., and Zhang, Z. (2020). The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 17 (8), 807–821. Epub 2020/07/03. doi:10.1038/s41423-020-0488-6
Zhang-Sun, Z. Y., Xu, X. Z., Escames, G., Lei, W. R., Zhao, L., Zhou, Y. Z., et al. (2023). Targeting Nr1d1 in organ injury: challenges and prospects. Mil. Med. Res. 10 (1), 62. Epub 2023/12/11. doi:10.1186/s40779-023-00495-3
Zheng, B., Albrecht, U., Kaasik, K., Sage, M., Lu, W., Vaishnav, S., et al. (2001). Nonredundant roles of the Mper1 and Mper2 genes in the mammalian circadian clock. Cell. 105 (5), 683–694. Epub 2001/06/08. doi:10.1016/s0092-8674(01)00380-4
Zheng, B., Larkin, D. W., Albrecht, U., Sun, Z. S., Sage, M., Eichele, G., et al. (1999). The Mper2 gene encodes a functional component of the mammalian circadian clock. Nature 400 (6740), 169–173. Epub 1999/07/17. doi:10.1038/22118
Zhu, L., Wang, Q., Hu, Y., and Wang, F. (2019). The circadian gene Per1 plays an important role in radiation-induced apoptosis and DNA damage in glioma. Asian Pac J. Cancer Prev. 20 (7), 2195–2201. Epub 2019/07/28. doi:10.31557/APJCP.2019.20.7.2195
Zhu, W. Z., He, Q. Y., Feng, D. C., Wei, Q., and Yang, L. (2023). Circadian rhythm in prostate cancer: time to take notice of the clock. Asian J. Androl. 25 (2), 184–191. Epub 2022/09/09. doi:10.4103/aja202255
Keywords: period gene, circadian rhythm, carcinogenic effect, biological behavior, cancer therapy
Citation: Chen K, Wang Y, Li D, Wu R, Wang J, Wei W, Zhu W, Xie W, Feng D and He Y (2024) Biological clock regulation by the PER gene family: a new perspective on tumor development. Front. Cell Dev. Biol. 12:1332506. doi: 10.3389/fcell.2024.1332506
Received: 03 November 2023; Accepted: 30 April 2024;
Published: 15 May 2024.
Edited by:
Josefa Leon, Fundación para la Investigación Biosanitaria de Andalucía Oriental (FIBAO), SpainReviewed by:
Maria Grazia Totaro, IFOM—The FIRC Institute of Molecular Oncology, ItalyCopyright © 2024 Chen, Wang, Li, Wu, Wang, Wei, Zhu, Xie, Feng and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dechao Feng, ZmRjZmVuaXhAc3R1LnNjdS5lZHUuY24=, ZGVjaGFvLmZlbmdAdWNsLmFjLnVr; Yi He, ODQ3NDg0ODBAcXEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.