
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol. , 28 March 2024
Sec. Embryonic Development
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1280797
This article is part of the Research Topic Fertilization and Early Embryogenesis: From Research to Clinical Practice View all 14 articles
To study the genetic variation leading to the arrest phenotype of pronuclear (PN) zygotes. We recruited a family characterized by recurrent PN arrest during in vitro fertilization (IVF) and intracytoplasmic sperm injection cycles (ICSI) and performed whole-exome sequencing for 2 individuals. The transcriptome profiles of PN-arrest zygotes were assessed by single-cell RNA sequencing analysis. The variants were then validated by PCR amplification and Sanger sequencing in the affected individuals and other family members. A family characterized by recurrent PN arrest during IVF and ICSI cycles were enrolled after giving written informed consent. Peripheral blood samples were taken for DNA extraction. Three PN-arrest zygotes from patient III-3 were used for single-cell RNA-seq as described. This phenotype was reproduced after multiple cycles of egg retrieval and after trying different fertilization methods and multiple ovulation regimens. The mutant genes of whole exon sequencing were screened and verified. The missense variant c. C1630T (p.R544W) in RGS12 was responsible for a phenotype characterized by paternal transmission. RGS12 controls Ca2+ oscillation, which is required for oocyte activation after fertilization. Single-cell transcriptome profiling of PN-arrest zygotes revealed defective established translation, RNA processing and cell cycle, which explained the failure of complete oocyte activation. Furthermore, we identified proximal genes involved in Ca2+ oscillation–cytostatic factor–anaphase-promoting complex (Ca2+ oscillation–CSF–APC) signaling, including upregulated CaMKII, ORAI1, CDC20, and CDH1 and downregulated EMI1 and BUB3. The findings indicate abnormal spontaneous Ca2+ oscillations leading to oocytes with prolonged low CSF level and high APC level, which resulted in defective nuclear envelope breakdown and DNA replication. We have identified an RGS12 variant as the potential cause of female infertility characterized by arrest at the PN stage during multiple IVF and ICSI.
In vitro fertilization (IVF) is now routine for treating infertile women and has brought at least 8 million babies into the world. Indeed, the earliest embryonic development after fertilization is a complex process, including the formation of spermatozoa and oocyte pronuclei (two-pronuclear [2PN] zygote), cytoskeletal rearrangement, singamy, and initiation of cleavage of the zygote. After IVF trials, about 5% of fertilized human oocytes present early developmental arrest at the PN stage (Schmiady et al., 1987). Homozygous mutations in TLE6 (MIM: 612399) and PADI6 (MIM: 10363) have been reported to cause embryonic arrest at the 2- to 4-cell stage with normal cleavage in consanguineous families (Alazami et al., 2015; Xu et al., 2016). A patient presenting complete cleavage failure in 2PN oocytes after IVF carried the homozygous mutation c.322G > A (p.Glu108Lys) in TUBB8 (Yuan et al., 2018). However, the crucial gene responsible for PN-arrest zygotes from familial individuals remains largely unknown.
Oocyte activation is Ca2+-dependent, characterized by an increase in and spreading of intracellular Ca2+ waves. After formation of the male and female pronuclei, calcium signaling continues to play a crucial role in 2PN fusion, DNA synthesis, and initiation of the first cleavage. Therefore, optimal Ca2+ oscillation is required for both oocyte activation and 2PN fusion. Chemical artificial oocyte activation with the A23187 Ca2+ ionophore improved embryo development to the cleavage stage for zygotes with arrest at the PN stage. However, artificial oocyte activation with a Ca2+ ionophore resulted in partial success (Darwish and Magdi, 2015; Xi et al., 2020), so defects in an unknown mechanism can also contribute to Ca2+ oscillations causing 2PN fusion failure.
Here, we performed whole exome sequencing (WES) and single-cell RNA sequencing (RNA-seq) in a family characterized by recurrent PN arrest during IVF and intracytoplasmic sperm injection cycles. Our analysis implicated the causative variant in RGS12, a gene that controls Ca2+ oscillation, which is required for oocyte activation after fertilization.
This study was approved by the Ethics Committee of Guangdong Medical University Affiliated Hospital (YS2018010), and written informed consent was obtained from participants. Patients gave written informed consent for the use of abandoned zygotes and peripheral blood for research on the arrest mechanism of PN stage, with no monetary payment. All procedures used were performed in accordance with the relevant guidelines and regulations.
Families were recruited in the Reproductive Medicine Center at the Affiliated Hospital of Guangdong Medical University based on the observation of PN arrest during regular IVF treatment of 2 siblings. Eligible families and controls were enrolled after giving written informed consent. Peripheral blood samples were taken for DNA extraction.
Females III-2 and III-3 were 29 and 25 years old, with BMI of 18.82 and 18.92 kg/m2. They both had primary infertility due to unexplained infertility. In the family, the women’s ovarian function and sex hormones were normal, and the men’s semen was normal. The peripheral blood chromosomes of both woman and man were normal as well (Supplementary Table S1).
IVF and ICSI were performed according to the laboratory routine insemination procedures on the day of oocyte retrieval (Day 0). The presence of 2 pronuclei (2PN) was observed 16–18 h after insemination or injection, then zygotes were cultured in 25 µL pre-equilibrated cleavage medium at 37°C under 6% CO2. Embryo morphology was evaluated at 42–46 h (Day 2) and 68–72 h (Day 3) after insemination. Male and female pronuclei that continued to separate on Day 2 and 3 without fusion were defined as PN-arrest zygotes. More details of IVF/ICSI procedures are given in the following.
The ovulation promotion scheme and embryo development of patients with different egg retrieval cycles are described in detail in Supplementary Table S2.
Long agonist protocol: Briefly, 5–7 days after ovulation or after 15 days of being on pre-IVF Marvelon (N.V. Organon, Netherlands), gonadotropin-releasing hormone analogue (GnRH-a; single dose of 0.8–1.875 mg; Decapepty, Ferring, Germany) was administered. The GnRH-a administration phase ended on the human chorionic gonadotropin (hCG) administration day. The Gn phase of therapy, which started 14 days after starting GnRH-a delivery and finished on the hCG injection day, included a natural or synthetic follicle stimulating hormone (FSH) preparation (dose Gonal-F, Merck Serono, Germany), a luteinizing hormone (LH) preparation (dose Luveris, Merck Serono, Germany), and, finally, hCG (10,000 IU, Livzon Pharmaceutical Group, China). Eggs were harvested 34–36 h after hCG injection (generally, a single follicle with diameter of up to 19 mm, 2 follicles with diameter up to 18 mm, or 3 follicles with diameter up to 17 mm), when E2 levels were monitored (generally reaching a mean of 250–300 ng/L per dominant follicle [≥16 mm], and a proportion of dominant follicles as high as 60%).
Mild stimulation protocol: Without GnRH-a downregulation, a small number of ovulation-stimulating drugs is usually applied from the first 3–5 days of menstruation. Simple Gn was used to promote ovulation, or Gn was combined with oral ovulation-stimulating drugs to promote follicle growth After follicles grew and matured, after giving the hCG trigger, the egg was retrieved the next day.
Ejaculated samples were obtained after 3–5 days of abstinence. Semen was screened by the direct upstream method of fertilization culture fluid (G-IVF, Vitrolife Sweden AB). The pellet was resuspended in 0.3 mL fertilization culture fluid.
Transvaginal follicular aspiration was performed 36 h after recombinant hCG administration. After cumulus oocyte complexes were retrieved in 1 mL modified human tubal fluid medium (G-MOPS, Vitrolife Sweden AB) at 37°C, they were cultured in cleavage medium (G-1, Vitrolife Sweden AB) covered with light mineral oil (Vitrolife Sweden AB) at 37°C in 6% CO2 for 2–4 h before conventional IVF, ICSI or IVF + ICSI (half the eggs were fertilized as routine and half by microinjection).
All cumulus oocyte complexes were enzymatically treated with 80 µL hyaluronidase (Vitrolife Sweden AB) for 30–60 s with the aid of mechanical denudation to remove cumulus cells. The denuded oocytes were examined for maturity and integrity. Only metaphase II (MII) oocytes that had extruded the first polar body were injected by spermatozoa.
Fertilization was assessed 16–18 h after IVF or ICSI. On day 3 (68–72 h), cleaved embryos were morphologically graded by evaluating cell number, size, fragmentation and nucleation.
WES was performed for 2 affected individuals and 3 unaffected individuals, including an unaffected sibling and parents. Germline genomic DNA was subjected to exome capture (60 Mb) with the Agilent SureSelect Human All ExonV6 kit according to the manufacturer’s instructions (Agilent, Santa Clara, CA). Paired-end sequencing, resulting in 150 bases from each end of the fragments, involved using a HiSeq PE150 Genome Analyzer (Illumina) at Novogene Bioinformatics Technology (Beijing). Sequencing reads were mapped to the reference genome (GRCh37, UCSC hg19) by using the Burrow-Wheller Aligner and were analyzed by using the Genome Analysis Toolkit (GATK, v3.1) for calling single nucleotide variants, insertions and deletions. The 1,000 Genomes, Exome Sequencing Project (ESP6500), Exome Aggregation Consortium (ExAC) and an in-house database were used to annotate the minor allele frequency for each variant. In silico analysis, Sort Intolerant from Tolerant (SIFT), Polymorphism Phenotyping v2 (PolyPhen-2), MutationAssessor, and Genomic Evolutionary Rate Profiling (GERP++) were used to predict the impact of each non-synonymous variant.
The pipeline was designed to filter heterozygous variants 1) shared by both affected individuals; 2) absent in other unaffected family members; 3) not previously reported or reported to have a frequency <0.1% in the public databases 1,000 Genomes, ESP6500, and ExAC and the in-house database; and 4) frameshift, nonsense, splice-site and missense variants predicted to be damaging in at least 3 of the 4 algorithms SIFT, PolyPhen-2, MutationAssessor, and GERP++.
The variants were then validated by PCR amplification and Sanger sequencing in the affected individuals and other family members. The RGS12 gene-specific primers to generate the variation were 5′-CAGGTTCTGGGACCTAAACAAG-3′ (forward) and 5′-GACTGTGCAAGCTGGTGACT-3′ (reverse). Variants were evaluated for co-segregation based on an autosomal-dominant mode of inheritance.
Three PN-arrest zygotes from patient III-3 were used for single-cell RNA-seq as described (Zhang et al., 2018). Briefly, zygotes were transferred into lysate buffer by using a mouth pipette, and whole-cell lysates underwent reverse transcription reaction according to the manufacturer’s instructions. Terminal deoxynucleotidyl transferase was used to add a poly(A) tail to the 3ʹ end of the first-strand cDNA, then 20 + 10 cycles of PCR were used to amplify the single-cell cDNA. The libraries were sequenced on the HiSeq PE150 Genome Analyzer platform at Annoroad Gene Technology (Beijing; http://www.annoroad.com).
Overall read quality was checked by using FASTQC v.0.11.5. The raw sequence data, in the form of FASTQ files, were aligned to the human genome (GRCh38, Ensembl Homo_sapiens) by using HISAT2 (v. 2.1.0) and SAMTOOLS (v1.3.1). Read count and Fragments Per Kilobase Million (FPKM) mapped reads for each sample were generated by using HTSeq v0.6.0.
HTSeq read counts were uploaded into RNA-seq 2G (http://52.90.192.24:3838/rnaseq2g/) for DESeq2 analysis. Normalization involved default settings (“normalize count by DESeq/normalize logged by Loess”). P-values were adjusted with the Benjamini and Hochberg method for controlling the false discovery rate (FDR). Genes with FDR p < 0.05 and fold change >2 or <0.5 were considered differentially expressed.
RNA-seq normalized data (FPKM) were subjected to principal component analysis (PCA) by using an unsupervised approach to observe the whole clustering profile. Gene Ontology (GO, biological processes) and pathway enrichment involved using DAVID (http://david.abcc.ncifcrf.gov/) with the Benjamini and Hochberg FDR to adjust the p-value. Significantly enriched GO categories were visualized by using REVIGO (Supek et al., 2011) (http://revigo.irb.hr/). The network of enriched terms was evaluated by using Metascape (Zhou et al., 2019) (http://metascape.org/). To infer the transcription factor regulatory network of this study, we used all 1,665 human transcription factors in the human TFDB 3.0 database (http://bioinfo.life.hust.edu.cn/AnimalTFDB#!/).
All RNA-seq datasets generated in this study have been deposited in Gene Expression Omnibus. Human oocyte, pre-implantation embryo RNA-seq data were obtained from GSE44183 (Xue et al., 2013). The RNA-seq data for normal PN zygotes (n = 22) were downloaded from GSE6548 (Yanez et al., 2016). The list of differentially expressed genes from RNA-seq data for validation were from a previous publication (Suo et al., 2018).
Statistical analysis was performed on original data by using GraphPad Prism 8.0 and Student’s t-test with a significance threshold of p < 0.05.
To identify novel PN zygote arrest-specific genes, we recruited a family with multiple infertile individuals who presented recurrent visible PN zygotes with second polar-body emission that failed to complete PN fusion after 24–68 h during IVF (Figure 1A; Supplementary Table S2). The affected individuals had primary infertility with an unknown cause. Initially, the affected individuals presented complete cleavage failure in all 2PN fertilized oocytes after 2 cycles of IVF. Subsequently, the affected individuals underwent 2 cycles of ICSI, with similar outcomes. We performed WES for 2 affected individuals (III-2 and III-3), an unaffected individual (III-1), and their parents (II-2 and II-3) (Figure 1B). Given the pedigree structure, we used an autosomal-dominant inheritance pattern and identified heterozygous, rare, potential pathogenic variants co-segregated with PN zygote arrest. Initially, 13 candidate genes were filtered by WES (Supplementary Table S3), and only the missense variant c. C1630T (minor allele frequency = 0.00018 in ExAC database) resulting in a p. R544W of regulator of G protein signaling-12 (RGS12) was confirmed by Sanger sequencing results available for relatives and was characterized by paternal transmission (Figure 1D).
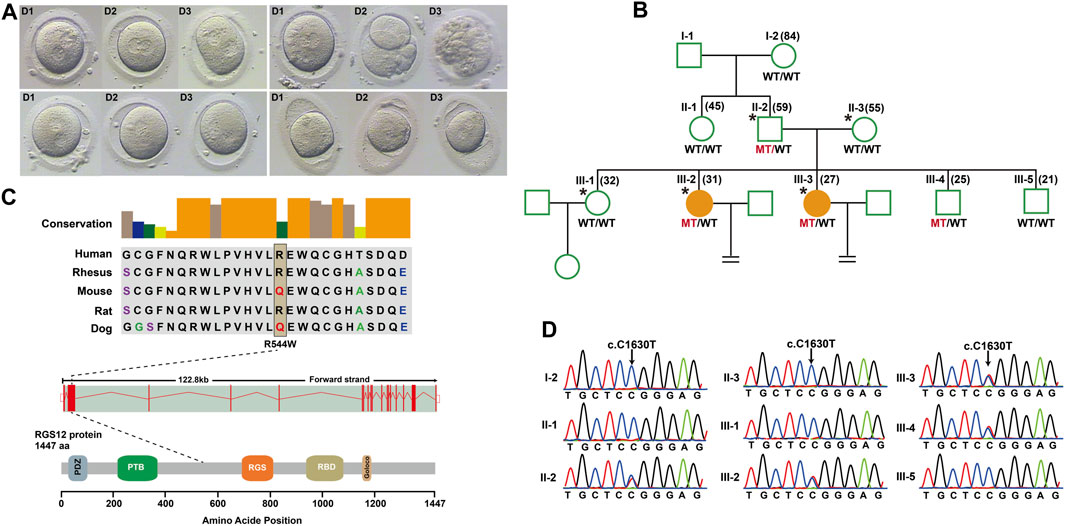
Figure 1. Structure of RGS12 and sites of pathogenic mutations associated with pronuclear (PN)-arrest zygotes. (A) Morphology of zygotes on days 1–3 after IVF. Most zygotes from affected individuals remained at the one-cell stage, and maternal and paternal pronuclei were separated. (B) Pedigrees of the family affected by infertility due to arrest at PN zygote stage. Squares denote male family members, circles denote female members, and solid symbols represent affected members. Equal signs indicate infertility. MT, mutant type; WT, wild type. (C) Conservation of amino acid residues affected by mutations in different species, structure of RGS12, and known domains of the gene product. Exons are red vertical bars, introns are dashed lines, and open rectangles at each end are noncoding exons. (D) Sanger sequencing confirmation of RGS12 variant (c.C1630T) in the family members.
RGS12 is the largest protein in the regulators of the G-protein signaling (RGS) family and is a negative regulator of specific G-protein–coupled receptor (GPCR) signals. RGS12 p. R544W is located between the RGS and PTB domains and presents a non-conserved pattern between human and mouse (Figure 1C), so it might have a species-specific functional effect. The predicted protein structure of R544W mutant RGS12 was further rendered with ChimeraX (28710774). We observed that the predicted structures of mutational intolerance corresponding to amino acids with inward-facing side chains (Figure 2). Consequently, mutant protein has decreased efficiency of PTB structural domain (PTB), which can bind to N-type calcium channels. RGS12 inhibits Ca2+ oscillations through PTB inhibition of G protein activity.
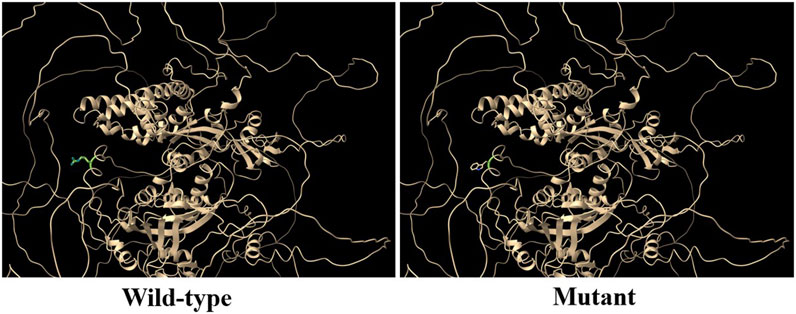
Figure 2. Three-dimensional structural analysis of wild-type and mutant RGS12 protein. Modeled structure of the RGS12 protein was painted by Chimera X software. Green indicated RGS protein structure in wild type, and the blue represent mutated RGS12 protein structure.
To describe the molecular landscape underlying the PN-arrest zygote (Figure 3A), we performed single-cell RNA-seq of PN-arrest zygotes (n = 3) from patient III-3 to explore the transcriptional profiles of PN-arrest zygotes by comparison with normal PN zygotes (n = 22, GSE65481) (Yanez et al., 2016). We found significant upregulation of 1,415 genes (fold change >2, p < 0.001) and downregulation of 1,545 genes (Figure 3B; Supplementary Table S4). Differentially expressed genes were enriched in the Gene Ontology (GO) terms (biological processes) RNA processing (FDR = 1.13 × 10−21), translational elongation (FDR = 1.25 × 10−16), intracellular transport (FDR = 1.63 × 10−15), and cell cycle (FDR = 4.11 × 10−13), so the oocyte-specific transcription and translation machinery is not completely established (Figures 3C, D). Pathway enrichment analysis revealed that differentially expressed genes in PN-arrest zygotes were also mainly involved in RNA processing and translation, such as ribosome (FDR = 6.40 × 10−22) and spliceosome (FDR = 1.52 × 10−5) (Figure 3F). The transition from oocyte to embryo is driven by a maternal stockpile of mRNA and translational machinery that is “packed” into the oocyte. Furthermore, oocyte activation after fertilization includes changes to oocyte coverings to prevent polyspermy, release of oocyte meiotic arrest, generation of haploid female and male pronuclei, changes in maternal mRNA and protein populations, and cytoskeletal rearrangements. Our transcriptional prolife results implied that the PN-arrest zygotes had properties of failure of complete oocyte activation after fertilization.
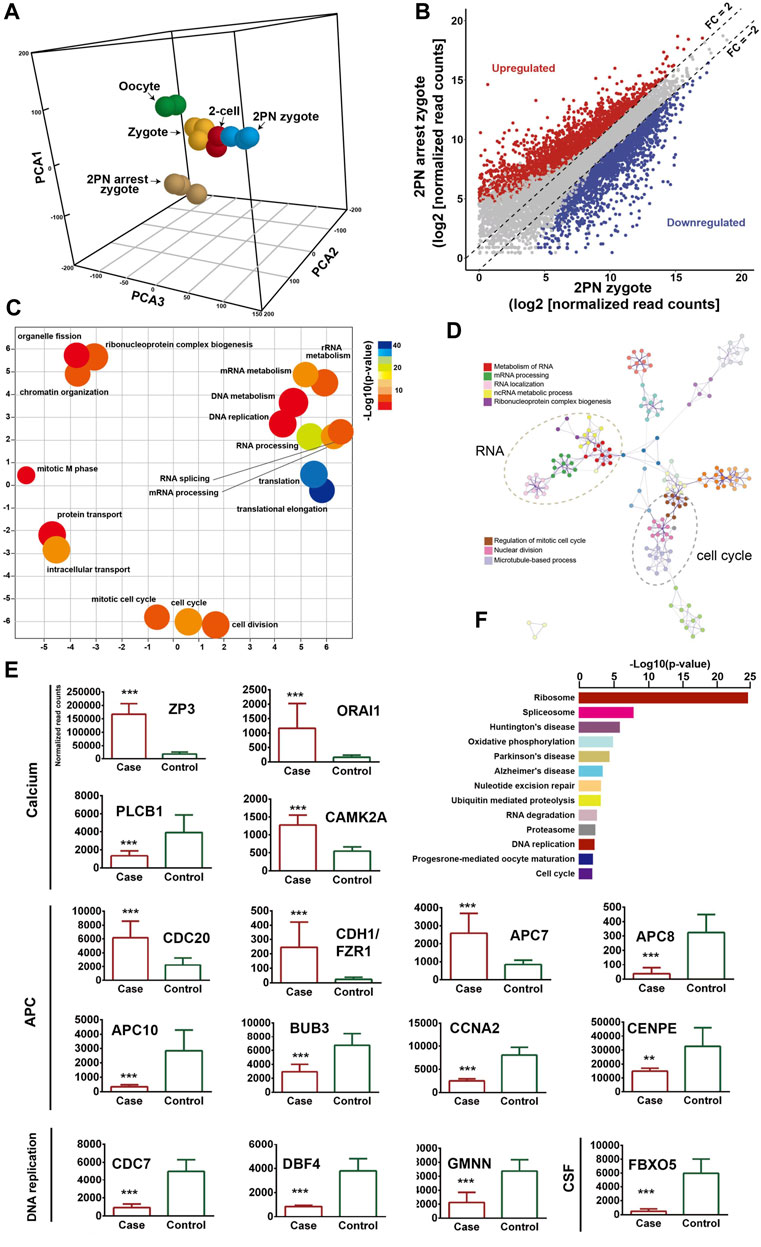
Figure 3. Transcriptome profile of PN arrest zygote by single-cell RNA-sequencing. (A) 3-D principal-component analysis (PCA) of the transcriptome of human oocytes, preimplantation embryos and PN-arrest zygotes. Cells of different preimplantation stages form distinct clusters, and PN-arrest zygotes had a specific transcriptome character. (B) Scatter plots comparing expression of genes with fold change >2, false discovery rate (FDR) < 0.05. (C) REVIGO scatter plot showing the cluster representatives in a 2-D space derived by applying multidimensional scaling to a matrix of semantic similarities for the Gene Ontology (GO) terms (biological processes). (D) Metascape enrichment network showing intra- and inter-cluster similarities of enriched GO terms. Cluster annotations are shown in color code. (E) Comparative analysis of genes in the Ca2+ oscillation–CSF–APC signaling pathway by average RNA-sequencing of normalized read counts for human PN and PN-arrest zygotes. FDR value, Data are mean ± SD. **, p < 0.001; ***, p < 0.0001. (F) Significantly enriched KEGG pathways in PN-arrest zygotes; length of column indicates the–log10 p-value.
Oocyte activation events present different Ca2+ requirements: 1) for cortical granules and blocking polyspermy; 2) inducing the resumption of meiosis including second meiotic polar body extrusion and initiating recruitment of maternal mRNAs; and 3) promoting pronuclear formation and initiation of embryonic mitosis. On oocyte activation, after PN formation, Ca2+ signaling continues to play a role in PN fusion and DNA synthesis for initiating embryonic mitosis during the oocyte-to-embryo transition.
Mature oocytes await fertilization while arrested at MII, which is maintained by the maturation promoting factor (MPF) consisting of cyclin B1/CDK1 subunits. Cytostatic factor (CSF) mediates MPF stabilization by inhibiting the anaphase-promoting complex (APC), which would otherwise destroy cyclin B. Fertilization breaks the MII arrest via cytoplasmic Ca2+ oscillation and triggers the APC, which mediates the degradation of cyclin B and thus inactivation of MPF. To confirm the spontaneous and abnormal Ca2+ oscillations, we traced the transcriptional change in genes that participate in vital processes from Ca2+ oscillation, CSF and APC (Figure 3E).
PN-arrest zygotes showed 9.54-fold increased ZP3 expression, which indicates sustained ZP3-evoked Ca2+ entry by Ca2+ influx and activation of G protein (Patel, 2004). ZP3-mediated calcium in-flow is important for fertilization, but abnormal calcium in-flow may be an important manifestation of PN arrest zygotes. Furthermore, plasma membrane Ca2+ channel ORAI1 mediates Ca2+ influx of oocytes after fertilization. In PN-arrest zygotes, ORAI1 showed 7.2-fold upregulation, which further confirmed the Ca2+ influx. These results indicate a spontaneous and abnormal Ca2+ oscillation in PN-arrest zygotes.
Next, we explored further evidence to support spontaneous and abnormal Ca2+ oscillation and its effects. CAMKII (CAMK2A) links Ca2+ oscillations and inactivates the MPF as well as translation and degradation of maternal mRNAs. PN-arrest zygotes showed upregulated CAMK2A, which implies the existence of prolonged Ca2+ oscillations. CaMKII activation by Ca2+ oscillations leads to activation of the APC by inhibiting CSF activity, which suppresses APC via EMI1 working with proto-oncogene, serine/threonine kinase (MOS). Because of abnormal Ca2+ oscillations, EMI1 (FBXO5) was significantly downregulated (12.25-fold) in PN-arrest zygotes, which indicates the lower CSF level and possible high APC level.
Activation of APC is regulated by 2 activators, CDC20 and CDH1. CDC20 and CDH1 expression was 2.7- and 10.3-fold increased, respectively, in PN-arrest zygotes. CDC20 and CDH1 bind to APC7, whose level was also increased in PN-arrest zygotes. The mitotic checkpoint complex (MCC), composed of CDC20, MAD2, BUBR1, CENPE and BUB3, acts as an APC inhibitor (Chao et al., 2012), but we found downregulated BUB3 and CENPE in PN-arrest zygotes. Downregulation of EMI1 and BUB3 and upregulation of CDC20, CDH1, and APC7 implied continually increased APC level in PN-arrest zygotes.
APC10 and cyclin A2 (CCNA2) were downregulated in PN-arrest zygotes, with no change in CCNB1 expression. We also found downregulation of GMNN (geminin), an APC substrate and essential for regulation of DNA replication for zygotes. In PN-arrest zygotes, CDC7/DBF4 expression was decreased ∼ five-fold. Therefore, continuous APC disrupted nuclear envelope breakdown (NEBD) and DNA replication in PN-arrest zygotes.
Increased expression of ZP3 and Orai1 in PN arrest zygotes may lead to disturbances in calcium inward flow and promote APC activity. Here we speculate that spontaneous and abnormal Ca2+ oscillation increased APC level by the mutant RGS12, thus leading to defective NEBD and DNA replication after 24–68 h in IVF trials. The trigger for the oocyte-to-embryo transition is oocyte activation. Therefore, our evidence supports that the PN arrest is due to spontaneous and abnormal Ca2+ oscillation causing prolonged APC activation. Hence, a precise pattern of Ca2+ oscillations after fertilization should be evaluated for further treating optimal oocyte activation.
We identified 589 common genes enriched in the GO terms translational elongation (FDR = 5.88 × 10−34) and translation (FDR = 2.45 × 10−17) and confirmed the incomplete oocyte activation. The key components of Ca2+ oscillation, CSF and APC signaling, EMI1, CCNA2, CDC7/DBF4, and GMNN were also identified (Figures 4A–D). To investigate the master regulators and construct the transcriptional regulatory network in the PN-arrest zygotes, we used the ARACNe method to analyze transcription factors. Only the transcription factor MAX was upregulated (Figure 4E), so MYC-MAX may play a critical role in the cell cycle entry of PN-arrest zygotes.
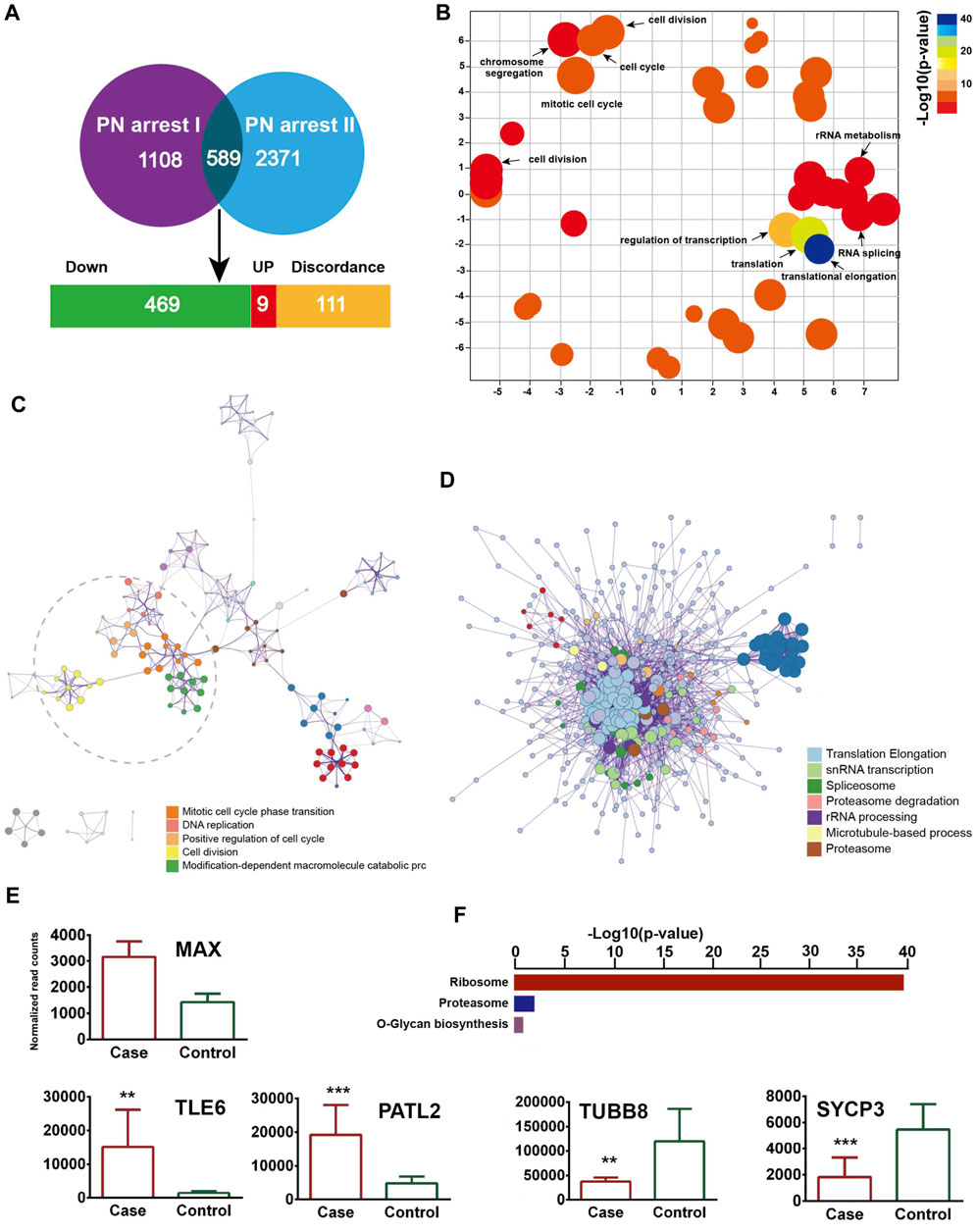
Figure 4. Validation of transcriptome profile in other PN-arrest zygotes. (A) Venn diagram shows overlapped differentially expressed genes between previous study by Suo et al. (2018) (PN arrest I) and our study (PN arrest II) for PN arrest groups each compared with their control group. (B) REVIGO scatterplot summarizes the overrepresented GO terms (biological processes) for representative subsets of terms. (C) Metascape enrichment network of the intra-cluster and inter-cluster similarities of enriched GO terms. (D) Metascape interactome network formed by all GO terms, confirming the defective RNA processing and translation in both PN-arrest zygote groups. (E) Relative expression of particularly interesting DEGs. FDR value, Data are mean ± SD. **, p < 0.001; ***, p < 0.0001. (F) Significantly enriched KEGG pathways shared in PN-arrest zygotes.
We found upregulation of 2- to 4-cell arrest-specific genes TLE6 and PATL2; hence, the RGS12 mutation caused earlier embryonic development arrest (Figure 4E). Furthermore, SYCP3 and TUBB8 were downregulated, which indicates defective cytoskeletal rearrangements. We did not find a disruption of zygotic arrest 1 (ZAR1), a oocyte-specific maternal-effect gene for mouse oocyte-to-embryo transition (Wu et al., 2003).
In this study, we used WES to explore mutations affecting familial individuals with recurrent PN arrest during IVF and ICSI cycles. We identified a heterozygous pathogenic variant in RGS12 that was responsible for the phenotype. Moreover, single-cell RNA-seq identified proximal genes involved in Ca2+ oscillation–CSF–APC signaling. The findings indicate that abnormal spontaneous Ca2+ oscillations led to oocytes with prolonged low CSF level and high APC level, which resulted in defective NEBD and DNA replication.
Maternal genes have a critical effect in the earliest stages of embryonic development. ZAR1 was first identified as an oocyte-specific maternal-effect gene that functions at the oocyte-to-embryo transition in mice (Wu et al., 2003), Novel synonymous variation (c.516C>T) and intron variation (c.964-55A>T) of ZAR1 were identified in individuals with recurrent uncleaved zygotes in IVF (Tian et al., 2020). Homozygous mutations in BTG4 caused zygotic cleavage failure in 4 independent affected females with infertility of unknown cause (Zheng et al., 2020). BTG4/CCR4-NOT–induced mRNA deadenylation is involved in regulating maternal mRNA stability (Sha et al., 2020). However, nothing is known about the genetic cause of the phenotype of human PN-arrest zygotes from familial individuals.
Here we describe a rare family with multiple infertile female members with phenotypes of human PN-arrest zygotes. Use of WES revealed that the affected members carried a paternally originated autosomal-dominant mutation (p.R544W) in RGS12, a member of the family of regulators of G-protein signaling. RGS12 variants have been considered the most promising candidates in multiple families affected by bipolar disorder identified by WES (Forstner et al., 2020). To the best of our knowledge, this is the first report to describe a variant in the RGS12 responsible for female infertility characterized by arrest at the PN stage during multiple IVF. The genetic basis for infertility characterized by abnormalities in human oocyte development and early embryogenesis (2- to 4-cell) has been described (Alazami et al., 2015; Feng et al., 2016; Xu et al., 2016). A variant in RGS12 extends the genetic causes of infertility. Therefore, our findings will facilitate genetic diagnoses for RGS12 mutation in identifying patients with infertility who are undergoing IVF and ICSI.
ZAR1 regulates mRNA translation during the oocyte-to-embryo transition (Tian et al., 2020). BTG4 variants impair the decay of maternal mRNA in zygotes of the affected individual with zygotic cleavage failure (Zheng et al., 2020). Unlike ZAR1 and BTG4, which affect maternal mRNA translation, RGS12 plays an important role in Ca2+ oscillations in osteoblasts, neurons and other cell types (Schiff et al., 2000; Richman et al., 2005; Li et al., 2019). Therefore RGS12 controls Ca2+ oscillations, which provides an important spatially restricted Ca2+ signal required for complete oocyte activation after fertilization and triggers CSF–APC signaling to switch from meiosis to mitotis. Our single-cell transcriptome sequencing data revealed unique features in translation, RNA processing and cell-cycle impairments of failure of complete oocyte activation and uncovered the Ca2+ oscillation–CSF–APC signaling pathway by which mutant RGS12 exerts its maternal effect on PN arrest. The genes involved in PN arrest of the Ca2+ oscillation–CSF–APC signaling pathway were validated in other studies (Suo et al., 2018). The partially activated oocytes not progressing further and becoming arrested again in the PN stage were described as a new MII arrest (Kubiak, 1989). The key genes underlying the PN-arrest zygote (Figure 5) improve our understanding of why and how RGS12 mutation causes the phenotype.
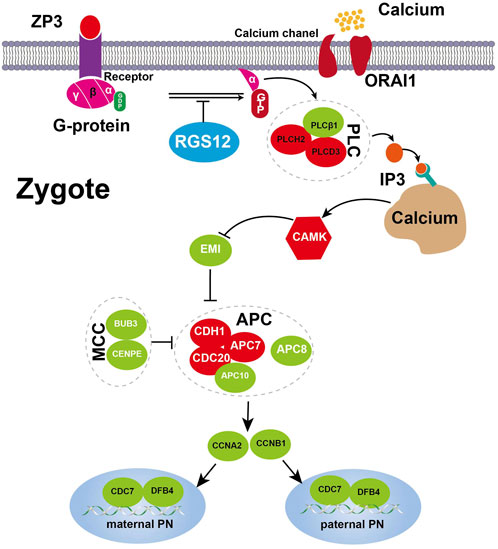
Figure 5. Expression changes of key genes involved Ca2+ oscillation–CSF–APC signaling. A proposed model for the RGS12-mutant oocyte with spontaneous and abnormal Ca2+ oscillations leading to prolonged low CSF and high APC level resulting in defective nuclear envelope breakdown and DNA replication. The key component genes are represented by green (downregulation) and red (upregulation), respectively. Detailed explanations are provided in the text.
Our validated single-cell transcriptional data support that a loss-of-function effect of RGS12 p. R544W causes spontaneous and abnormal Ca2+ oscillations in zygotes with 2PN arrest after fertilization. Our findings could explain why artificial oocyte activation using the A23187 Ca2+ ionophore improved embryo development only to the cleavage stage at a limited rate (one of 4 couples) (Darwish and Magdi, 2015). Spontaneous and abnormal Ca2+ oscillations in zygotes with 2PN arrest also raise the possibility of developing antagonists to block the excessive free Ca2+, such as Ca2+ chelators BAPTA and EGTA.
We have identified an RGS12 variant as the potential cause of female infertility characterized by arrest at the PN stage during multiple IVF and ICSI. This gene should be further screened in individuals with infertility caused by arrest at the PN stage during IVF and ICSI. These findings expand our knowledge of the genetic basis of human early embryonic arrest and provide the basis for genetic diagnoses of clinically infertile individuals with this phenotype.
Sequencing data have been deposited in public, open access repository of the Genome Sequence Archive for Human (http://bigd.big.ac.cn/gsa-human/) with the accession no. HRA006945.
The studies involving humans were approved by the Ethics Committee of Guangdong Medical University Affiliated Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The manuscript presents research on animals that do not require ethical approval for their study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
TM: Data curation, Funding acquisition, Resources, Writing–original draft, Writing–review and editing. SZ: Methodology, Data curation, Formal Analysis, Writing–review and editing. XX: Methodology, Data curation, Investigation, Formal Analysis, Writing–review and editing. JC: Methodology, Conceptualization, Data curation, Formal Analysis, Writing–review and editing. JW: Methodology, Data curation, Investigation, Formal Analysis, Writing–review and editing. GZ: Formal Analysis, Project administration, Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Nature Science Foundation of Guangdong Province (2022A1515010849) and the National Nature Science Foundation of China (81300484).
We acknowledge J. L. for excellent technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1280797/full#supplementary-material
APC, anaphase-promoting complex; BUB3, mitotic checkpoint protein; CaMKII, Calcium/calmodulin-dependent protein kinase II; CCNA2, cyclin A2; CDC, cell division cycle; CDH1, cadherin 1; CENPE, centromere protein E; CSF, cytostatic factor; EMI1, F-box protein 5; GMNN, geminin DNA replication inhibitor; ICSI, intracytoplasmic sperm injection; IVF, in vitro fertilization; ORAI1, calcium release-activated calcium modulator 1; PN, pronuclear; RGS12, regulator of G protein signaling 12.
Alazami, A. M., Awad, S. M., Coskun, S., Al-Hassan, S., Hijazi, H., Abdulwahab, F. M., et al. (2015). TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 16, 240. doi:10.1186/s13059-015-0792-0
Chao, W. C., Kulkarni, K., Zhang, Z., Kong, E. H., and Barford, D. (2012). Structure of the mitotic checkpoint complex. Nature 484 (7393), 208–213. doi:10.1038/nature10896
Darwish, E., and Magdi, Y. (2015). A preliminary report of successful cleavage after calcium ionophore activation at ICSI in cases with previous arrest at the pronuclear stage. Reprod. Biomed. Online 31 (6), 799–804. doi:10.1016/j.rbmo.2015.08.012
Feng, R., Sang, Q., Kuang, Y., Sun, X., Yan, Z., Zhang, S., et al. (2016). Mutations in TUBB8 and human oocyte meiotic arrest. N. Engl. J. Med. 374 (3), 223–232. doi:10.1056/NEJMoa1510791
Forstner, A. J., Fischer, S. B., Schenk, L. M., Strohmaier, J., Maaser-Hecker, A., Reinbold, C. S., et al. (2020). Whole-exome sequencing of 81 individuals from 27 multiply affected bipolar disorder families. Transl. Psychiatry 10 (1), 57. doi:10.1038/s41398-020-0732-y
Kubiak, J. Z. (1989). Mouse oocytes gradually develop the capacity for activation during the metaphase II arrest. Dev. Biol. 136 (2), 537–545. doi:10.1016/0012-1606(89)90279-0
Li, Z., Liu, T., Gilmore, A., Gómez, N. M., Fu, C., Lim, J., et al. (2019). Regulator of G Protein signaling protein 12 (Rgs12) controls mouse osteoblast differentiation via calcium channel/oscillation and gαi-ERK signaling. J. Bone Min. Res. 34 (4), 752–764. doi:10.1002/jbmr.3645
Patel, T. B. (2004). Single transmembrane spanning heterotrimeric g protein-coupled receptors and their signaling cascades. Pharmacol. Rev. 56 (3), 371–385. doi:10.1124/pr.56.3.4
Richman, R. W., Strock, J., Hains, M. D., Cabanilla, N. J., Lau, K. K., Siderovski, D. P., et al. (2005). RGS12 interacts with the SNARE-binding region of the Cav2.2 calcium channel. J. Biol. Chem. 280 (2), 1521–1528. doi:10.1074/jbc.M406607200
Schiff, M. L., Siderovski, D. P., Jordan, J. D., Brothers, G., Snow, B., De Vries, L., et al. (2000). Tyrosine-kinase-dependent recruitment of RGS12 to the N-type calcium channel. Nature 408 (6813), 723–727. doi:10.1038/35047093
Schmiady, H., Kentenich, H., and Stauber, M. (1987). Developmental arrest of fertilized human oocytes in the pronuclear stage after in vitro fertilization (IVF). Geburtshilfe Frauenheilkd 47 (6), 406–409. doi:10.1055/s-2008-1036145
Sha, Q. Q., Zheng, W., Wu, Y. W., Li, S., Guo, L., Zhang, S., et al. (2020). Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat. Commun. 11 (1), 4917. doi:10.1038/s41467-020-18680-6
Suo, L., Zhou, Y. X., Jia, L. L., Wu, H. B., Zheng, J., Lyu, Q. F., et al. (2018). Transcriptome profiling of human oocytes experiencing recurrent total fertilization failure. Sci. Rep. 8 (1), 17890. doi:10.1038/s41598-018-36275-6
Supek, F., Bosnjak, M., Skunca, N., and Smuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6 (7), e21800. doi:10.1371/journal.pone.0021800
Tian, Y., Yang, J., Peng, Y., Chen, T., Huang, T., Zhang, C., et al. (2020). Variation screening of zygote arrest 1(ZAR1) in women with recurrent zygote arrest during IVF/ICSI programs. Reprod. Sci. 27 (12), 2265–2270. doi:10.1007/s43032-020-00246-y
Wu, X., Viveiros, M. M., Eppig, J. J., Bai, Y., Fitzpatrick, S. L., and Matzuk, M. M. (2003). Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat. Genet. 33 (2), 187–191. doi:10.1038/ng1079
Xi, H., Fu, Y., Liu, C., Lu, X., Sui, L., Chen, Y., et al. (2020). Assisted oocyte activation with calcium ionophore 44 hours after intracytoplasmic sperm injection resulting in successful pregnancy. Gynecol. Endocrinol. 36 (11), 1035–1037. doi:10.1080/09513590.2020.1737667
Xu, Y., Shi, Y., Fu, J., Yu, M., Feng, R., Sang, Q., et al. (2016). Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am. J. Hum. Genet. 99 (3), 744–752. doi:10.1016/j.ajhg.2016.06.024
Xue, Z., Huang, K., Cai, C., Cai, L., Jiang, C. Y., Feng, Y., et al. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500 (7464), 593–597. doi:10.1038/nature12364
Yanez, L. Z., Han, J., Behr, B. B., Pera, R. A. R., and Camarillo, D. B. (2016). Human oocyte developmental potential is predicted by mechanical properties within hours after fertilization. Nat. Commun. 7, 10809. doi:10.1038/ncomms10809
Yuan, P., Zheng, L., Liang, H., Li, Y., Zhao, H., Li, R., et al. (2018). A novel mutation in the TUBB8 gene is associated with complete cleavage failure in fertilized eggs. J. Assist. Reprod. Genet. 35 (7), 1349–1356. doi:10.1007/s10815-018-1188-3
Zhang, Y., Yan, Z., Qin, Q., Nisenblat, V., Chang, H. M., Yu, Y., et al. (2018). Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol. Cell 72 (6), 1021–1034. doi:10.1016/j.molcel.2018.10.029
Zheng, W., Zhou, Z., Sha, Q., Niu, X., Sun, X., Shi, J., et al. (2020). Homozygous mutations in BTG4 cause zygotic cleavage failure and female infertility. Am. J. Hum. Genet. 107 (1), 24–33. doi:10.1016/j.ajhg.2020.05.010
Keywords: pronuclear arrest, zygote, RGS12, Ca2+ oscillation, case report
Citation: Ma T, Zhou S, Xie X, Chen J, Wang J and Zhang G (2024) A case report of a family with developmental arrest of human prokaryotic stage zygote. Front. Cell Dev. Biol. 12:1280797. doi: 10.3389/fcell.2024.1280797
Received: 21 August 2023; Accepted: 19 February 2024;
Published: 28 March 2024.
Edited by:
Katerina Komrskova, Institute of Biotechnology (ASCR), CzechiaReviewed by:
Renee J. Chosed, University of South Carolina, United StatesCopyright © 2024 Ma, Zhou, Xie, Chen, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guohong Zhang, Z19naHpoYW5nQHN0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.