
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell Dev. Biol., 23 February 2023
Sec. Morphogenesis and Patterning
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.971091
This article is part of the Research TopicSingle Cell Analysis to Study Cell Differentiation in Human DiseasesView all 6 articles
 Jieqi Wen†
Jieqi Wen† Rongsong Ling†
Rongsong Ling† Ruiyue Chen
Ruiyue Chen Siyan Zhang
Siyan Zhang Yarong Dai
Yarong Dai Tingtao Zhang
Tingtao Zhang Fanyu Guo
Fanyu Guo Qingxin Wang
Qingxin Wang Guixin Wang
Guixin Wang Yizhou Jiang*
Yizhou Jiang*Lipid metabolism disorder is the basis of atherosclerotic lesions, in which cholesterol and low-density lipoprotein (LDL) is the main factor involved with the atherosclerotic development. A high-fat and high-cholesterol diet can lead to this disorder in the human body, thus accelerating the process of disease. The development of single-cell RNA sequencing in recent years has opened the possibility to unbiasedly map cellular heterogeneity with high throughput and high resolution; alterations mediated by a high-fat and high-cholesterol diet at the single-cell transcriptomic level can be explored with this mean afterward. We assessed the aortic arch of 16-week old Apoe−/− mice of two control groups (12 weeks of chow diet) and two HFD groups (12 weeks of high fat, high cholesterol diet) to process single-cell suspension and use single-cell RNA sequencing to anatomize the transcripts of 5,416 cells from the control group and 2,739 from the HFD group. Through unsupervised clustering, 14 cell types were divided and defined. Among these cells, the cellular heterogeneity exhibited in endothelial cells and immune cells is the most prominent. Subsequent screening delineated ten endothelial cell subsets with various function based on gene expression profiling. The distribution of endothelial cells and immune cells differs significantly between the control group versus the HFD one. The existence of pathways that inhibit atherosclerosis was found in both dysfunctional endothelial cells and foam cells. Our data provide a comprehensive transcriptional landscape of aortic arch cells and unravel the cellular heterogeneity brought by a high-fat and high-cholesterol diet. All these findings open new perspectives at the transcriptomic level to studying the pathology of atherosclerosis.
As a chronic inflammatory disease, atherosclerosis is the major cause of myocardial and cerebral infarction and ischemia of the extremities, the underlying cause of about 50% of all deaths (Lusis, 2000). A disorder of lipid metabolism is the basis of atherosclerotic lesions, with cholesterol and low-density lipoprotein (LDL) acting as chief instigators (Lu and Daugherty, 2015). Several kinds of cells and cytokines are involved in the atherosclerotic progression.
Lesion of the diseased aorta originated from its intima. Minimally oxidized LDL diminishes nitric oxide to increase the permeability of endothelial layer and triggers endothelial cells to produce cytokines and chemokines, which helps monocytes pass through endothelial monolayer and convert into macrophages. Lipid accumulates here topically, and oxidation and modification of LDL happen under the presence of different enzymes and ROS. Macrophages then differentiate into foam cells with the help of highly oxidized LDL and gather. Under the regulation of cytokines, smooth muscle cells (SMCs) migrate into the inner lumen, and proliferate and secrete extracellular matrix (ECM) to promote the hyperplasia of fibrous cap and formation of plaque (Libby, 2000). Then macrophage and inflammatory T cells secrete various cytokines and enzymes to degrade matrix (Schönbeck et al., 2000), make the lesional plaque fragile, and induce calcification and precipitation. Finally the rupture of the plaque and the exposure of lipid core into blood recruit platelet initiate thrombosis. This blocks artery lumen and contributes to the ischemia or necrosis of organs or tissues supplied by the artery (Lusis, 2000).
It is clear that endothelial cells and macrophages play important roles in the development of atherosclerosis. Endothelial injury and repair are novel theories explaining pathogenesis of atherosclerosis (Mannarino and Pirro, 2008). Endothelial cells (ECs) are involved in a great range of homeostatic functions (Durand and Gutterman, 2013) through anti-coagulant, antithrombotic, and anti-inflammatory activity. Normally, endothelial cells regulate vascular tone, cell adhesion, and SMC proliferation (Paone et al., 2019). However, these functions lapse when pathological conditions appear and cause endothelial dysfunction (Werner et al., 2006). Most atherosclerotic risk factors can activate endothelial cells to secretes chemokines, cytokines, adhesion molecules, and intracellular adhesion molecules, hence aggregate immune cells (Davignon and Ganz, 2004). Macrophages play a vital role in the initiation of atherosclerosis and growth of plaque, while continuous inflammation leads to its apoptosis. In the absence of effective exocytosis, the accumulation of cell debris and apoptosis promotes the formation of a necrotic core in atherosclerotic plaques.
By thoroughly assessing the transcriptional landscape of aortic cells from mice administered with different diets, a panorama of the differences between normal and diseased endothelial cells can be looked at with the help of single-cell RNA sequencing. Western diet-mediated changes in immune cells, especially macrophages in the aortic arch, were also a focus in this study.
Our mice strain came from the Nanjing Model Animal Resource Information Platform. Apoe−/− male C57BL/6 mice (16 weeks old) were used to establish control and experimental group. APOE is often produced in monocytes and macrophages (Curtiss et al., 2000) and plays a critical role in blood lipid metabolism (Chen et al., 2017) as ligands for receptors that clear chylomicron and VLDL residue (Meir and Leitersdorf, 2004). So when APOE is knocked out, total cholesterol in plasma increases (Maganto-Garcia, Tarrio, and Lichtman, 2012), and the effect is multiplied especially under a high-fat and high-cholesterol diet. Female mice secrete estrogen, which lowers the content of LDL in plasma and enhances endovascular blood coagulation (Aryan et al., 2020). For the experimental group, to accelerate the progression of atherosclerosis, the mice were fed with high-fat and high-cholesterol food for about 12 weeks after they had been weaned (4 weeks old); this group is referred to as the Western diet (HFD) group for short. (Formula of high fat, high cholesterol diet: 20% sucrose, 15% lard, 1.2% cholesterol, 0.2% sodium cholate, 10% casein, 0.6% calcium hydrogen phosphate, 0.4% stone powder, 0.4% premix and 52.2% basal feed.) Meanwhile, another group of mice, the control group, was administered with a chow diet. Mice were euthanized after 12 weeks of being administered different diets. Animal studies were performed in compliance with ethical guidelines and use of animals, and the experimental protocol was approved by the Shenzhen University Animal Care and Use Committee.
After being euthanized, mice were locally perfused with cold PBS to remove the peripheral blood remaining in the aorta. The aorta were then harvested and incubated using the enzyme mix in the Lung Dissociated Kit (Cat# 130-095-927, Miltenyi Biotec). The aortic arch was cut into pieces before being immersed with the enzyme mix, and then the reaction was performed at 37°C for 40 min on a rotator. The product was then filtered through a 70 um strainer to remove the extra tissue, and the strainer was washed three times with Dulbecco’s modified Eagle’s medium (DMEM). The cell suspension was centrifuged for 5 min at 500×g, at 25°C. Pour off the supernatants and resuspend cells with DMEM again to obtain the final suspension.
Single-cell suspension of the two aortas from each group was pooled together as one sample. Single-cell RNA sequencing, library construction, and quality control were executed using the illumina-HiSeq3000 platform by Genergy Bio-technology (Shanghai) Co., Ltd. Dying cells with mitochondrial RNA above 30% and cells lacking information with UMI<200 were eliminated during the quality control, and 8,155 dissociated cells, with 5,416 cells from the control group and 2,739 from the HFD one, were filtered out thereafter. The median reads per cell were 3,955 for the control group and 3,421 for the HFD group, and transcripts detected per cell in the two groups were 1,364 and 1,069 respectively. (Figure 1A).
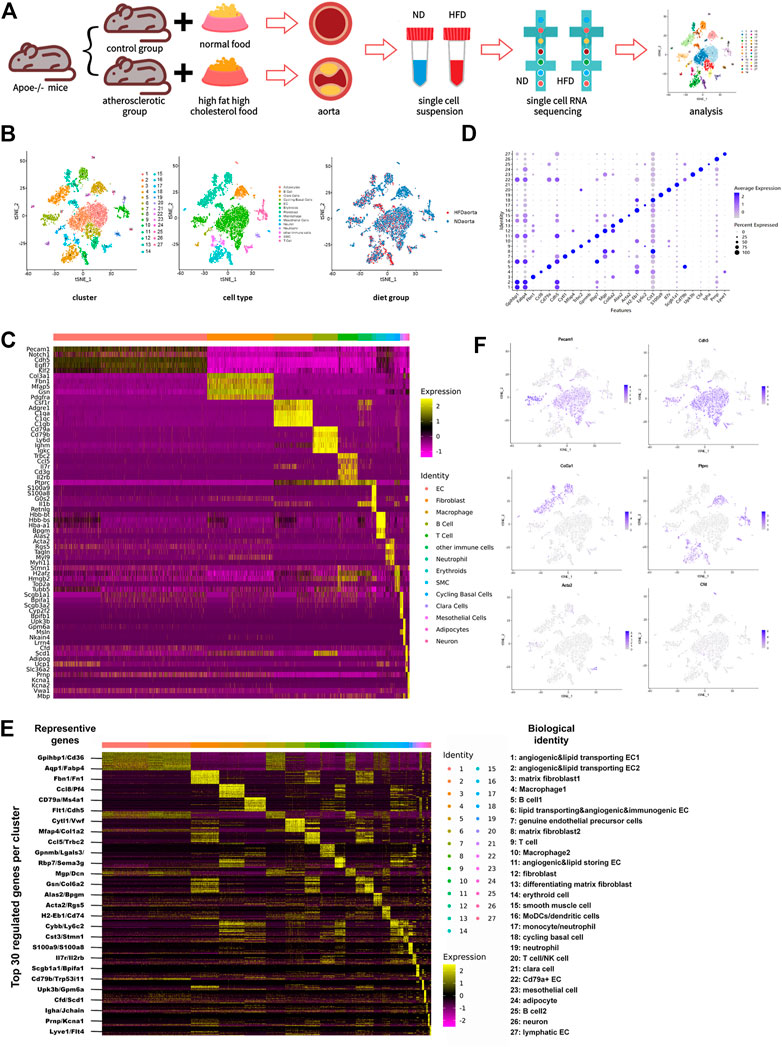
FIGURE 1. Transcriptome map of aortic cells extracted from both non-diseased and atherosclerotic Apoe−/− mice. (A) Schematic diagram of the experimental flow. (B) t-Stochastic neighbor embedding (t-SNE) representation of gene expression information in single cells from control (ND) group and Western diet (HFD) group. Colors denote different clusters (left), cell types (middle), and groups (right). (C) Heatmap of five representative genes for each aortic cell type. (D) Dot plot demonstrates distinct marker of each cluster. Size of dot corresponds to proportion of cells expressing each transcript, and dot color corresponds to expression level of transcript. (E) Heatmap showing the top 30 upregulated genes (ordered by decreasing P value) in each cluster, with representative genes selected for biological identification of each cluster marked on the left and biological identification on the right. The color from yellow to purple symbolizes the expression levels from high to low. (Scale: log2 fold change) (F) Gene expression patterns projected onto t-SNE plot profile expression of six significant cell markers. Endothelial cell: Pecam1 and Cdh5; Fibroblast: Col3a1; Immune cell: Ptprc; Smooth muscle cell: Acta2; Adipocyte: Cfd.
The Cell Ranger Single-cell Software Suite was used to demultiplex the experimental data; Illumina’s bcl2fastq was wrapped around by using the mkfastq command.
The calculations based on UMI-tools were used to control quality of RNA sequencing. Sample libraries balancing was carried out for the number of estimated reads per cell and then ran on the illumine-HiSeq300.
Based on the 10x Genomics documentation (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cellranger), demultiplexing, alignment filtering, barcode counting, UMI counting, and gene expression estimation were performed by Cell Ranger software on each sample. The LogNormalize algorithm worked in normalization. To compare experimental groups with normalized sequencing-depth and expression data, the IntegrateData (Seurat) was used to aggregate the gene expression estimates from each sample. Seurat (version 3.2.0) and R (version 4.0.0) package were used in downstream analysis. Cells with less than 200 genes were detected, and more than 30% of the mitochondrial gene count were filtered out as low quality or dying cells. Dimension reduction was then performed on normalized and logarithmized data by three stages of analysis, including the selection of variable genes, principal component analysis (nPCs = 50), and t-Distributed Stochastic Neighbor Embedding (t-SNE) with RunTSNE:dims = 1–30. Then the cell clustering was performed using the original Louvain algorithm (resolution = 0.9). We used the FindAllMarkers function to perform Wilcoxon rank-sum test based on the normalized data to identify gene markers in each cluster.
The Seurat package FindAllMarkers was used to analyze the differentially expressed genes (DEGs) between the two groups. The cowplot (version 1.1.1) and ggplot2 (version 3.3.5) were used for graphing. Sorting of endothelial cells were then performed with canonical markers Pecam1 and Cdh5 based on the estimated amount of EC in the total sample with resolution 0.4.
The mice to be sampled were individually isolated and kept in fasting conditions for 6 h, blood was collected below the jaw and stored in tubes without endotoxin. The samples were kept at 37°C for 1, 2 h to solidify the blood; blood was then left to clot overnight at 4°C. Afterwards, the serum was naturally precipitated, and centrifugation was performed for 10 min at 3,000 r/min at 4°C. Samples should be stored at −80°C if not used immediately.
The concentration of cholesterol and triglyceride were tested in serum with the Cholesterol Kit (CHOD-PAP Method) (Cat# 020080, Biosino) and Triglyceride Kit (GPO-PAP Method) (Cat# F001-2, Biosino). Prepare work solution. Mix work solution and cholesterol/triglyceride calibrator in different ratio to prepare cholesterol/triglyceride with different step concentrations. Mix work solution and sample in 100:1. The measured the concentration of cholesterol/triglyceride in serum using spectrophotometric method.
The statistical data of differentially expressed genes (DEGs) were calculated by Wilcoxon rank-sum test algorithm, and the threshold value of p-value was 0.05. The error bars in the bar graph represent standard error of mean (SEM). Two-tailed unpaired t-test and two-tailed Mann-Whitney test were used for statistical analysis unless stated otherwise. Statistical analysis was performed using GraphPad Prism version 8.0.2 or R version 3.2.0.
To prepare aortic cell suspension, fresh descending aorta and aortic root fragments, harvested from Apoe−/− male C57BL/6 mice (18 weeks old) fed with a Western diet and normal diet, three in each group, were incubated by an enzyme mix with 0.2 mg/mL Liberase (Roche, 5,401,054,001) and 2 U/mL Elastase (Sigma-Aldrich, E1250), with HBSS as the solvent. Digestion was done by rotating at 37°C in an oven for an hour. The product was filtered through the 35 um strainer and washed with HBSS. Cells were collected by centrifugation at room temperature, 500 xg for 5 min. The supernatant was discarded and the cells resuspended with staining buffer (3% BSA and 1%NaN in PBS).
Each group of cell suspension mentioned above was divided into four parts for incubation with the following four antibodies.
Incubation was performed for 1 h at 4°C in darkness. Separate isotype controls for each antibody were also prepared. Secondary antibodies labeled with fluorescent dye were diluted with 3% BSA and used to resuspend cells at room temperature for 30 min in darkness.
Cells were washed with PBS by centrifugation at 400 g for 5 min twice. Finally, cells were resuspended with cold staining buffer (3% BSA and 1%NaN in PBS), the cell number was counted, and flow cytometry was performed. Cells were sorted by flow cytometry (CytoFLEX, Beckman Coulter) and analyzed with flow cytometer (CytoFLEX, Beckman Coulter) (version 2.0).
Whole-cell map of the aorta arch mapped by single-cell sequencing techniques has often been mentioned in the previous studies (Kalluri et al., 2019; Zhao et al., 2021). This study provides a more comprehensive map of the transcriptional information of all aortic cells. Cells from control and HFD group were pooled together and distinguished into 27 cell clusters corresponding to 10 different cell types through unsupervised clustering done using Seurat and then were visualized by the dimension reduction via t-stochastic neighbor embedding (Figure 1B). To define the identity of each cell cluster, we performed differential expression analysis between each cluster and assigned a specific identity to each cluster based on the established lineage-specific marker genes (Figure 1C). Some of these marker genes may were only evenly and slightly upregulated or just topically upregulated in certain clusters (Figure 1D), but this could not exclude the role of these marker genes in identifying cell clusters. (Kalluri et al., 2019).
The largest population of cells in this study is endothelial cells, accounting for 43.2% (Supplementary Figure 1A). Under such high-resolution sequencing, seven endothelial cell clusters were distinguished despite the similarity of partial expression profiles among the clusters. Different types of immune cells were distributed in nine clusters unevenly and account for 28.8%, the second largest population.
Occupying a proportion of 18.7%, fibroblasts identified by particularly positive expression of genes encoding fibronectin (like Col3a1, Fbn1, Mfap5, Gsn, and Pdgfra) were divided into four clusters. Traditional matrix fibroblasts in cluster three and eight were characterized by notable expression of Fn1 and Mfap4 respectively. Enhanced expression of Mgp and Dcn indicates that fibroblasts in cluster 12 were involved in composition of ECM and collagen, while differentiating matrix fibroblasts in cluster 13 were featured with high expression of Gsn and Col6a2, two genes involved in ECM processing. Except fibroblasts in cluster 12, which exist exclusively in the control group, all other fibroblast subsets exist in both groups.
Smooth muscle cells are always defined by their usual marker Acta2. This population also showed a rise of Tagln, Rgs5, Myh9, and Myl11. Other cell types include erythroid cell (Hbb-bt, Hbb-bs, Hba-a1, Bpgm, and Alas2), cycling basal cells (Stmn1, H2afz, Hmgb2, Top2a, and Tubb5), clara cells (Scgb1a1, Bpifa1, Scgb3a2, Cyp2f2, and Bpifb1), mesothelial cells (Upk3b, Gpm6a, Msln, Nkain4, and Lrrn4), adipocytes (Cfd, Scd1, Adipoq, Ucp1, and Slc36a2), and neurons (Prnp, Kcna1, Kcna2, Vwa1, and Mbp). Among these, cycling basal cells, clara cells, mesothelial cells, and adipocytes have not been identified in similar studies (Figures 1C–F; Supplementary Figures 1A–C).
Sorting of specific types of cells was performed based on single-cell gene profile and dependent on identity genes or specific conditions. 2,785 cells with positive expression of endothelial canonical markers Pecam1 and Cdh5 were selected as endothelial cell lines from the total cell repository, with 1786 from the control group and 999 from the HFD group. These cells were divided into ten subpopulations with specific identification (Figures 2A, B; Supplementary Figure 2A).
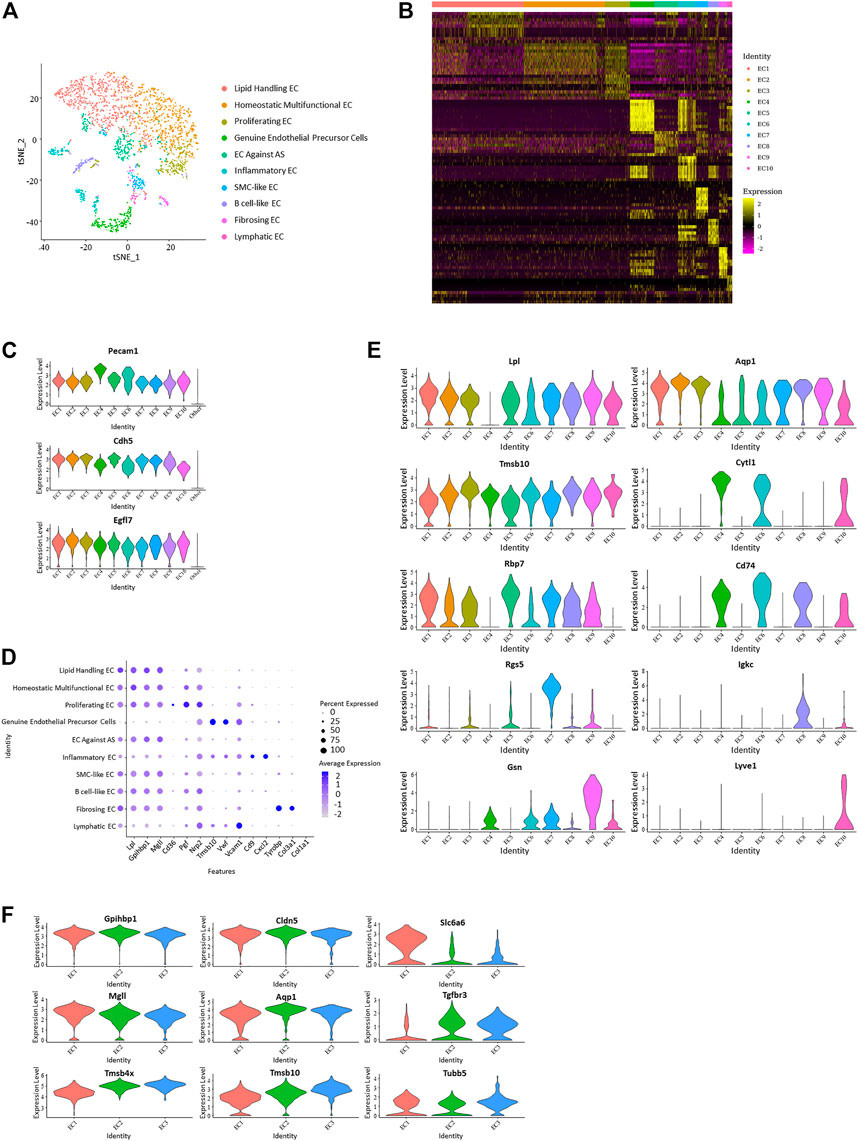
FIGURE 2. Differentiation of ten distinct vascular endothelial subpopulations. (A) t-Stochastic neighbor embedding representation of potential aortic endothelial cells extracted from total cell repertoire and separate them into 10 clusters. (B) Heatmap identifying top 10 upregulated genes of each endothelial subpopulation. (C) Violin plots of log-transformed gene expression of canonical endothelial markers distinguish these ten endothelial subpopulations from all other aortic cells. (D) Dot plot of functional markers showing the characteristics of each subpopulation. (Lipid handling: Lpl, Gpihbp1, Mgll, Cd36; Angiogenesis: Pgf, Nrp2, Tmsb10; Adhesion factor: Vwf, Vcam1, Cd9; Inflammation: Cxcl2, Tyrobp; Collagen forming: Col3a1, Col1a1) (E) Violin plots of log-transformed gene expression of selected markers demonstrating heterogeneity in each cluster. Violin plot y-axis demonstrates normalized transcript expression values. (F) Expression pattern of selected genes in endothelial subpopulation 1–3 showed heterogeneity and homogeneity between the three clusters. (Common: Gpihbp1, Cdh5; EC1 higher: Mgll, Slc6a6; EC2 higher: Aqp1; EC3 higher: Tgfbr3, Tmsb4x, Tmsb10, Tubb5).
Positive expression of EC canonical markers Nos3 (Knowles et al., 2000), Ptprb, and Notch1 in these selected cells distinguish them from other vascular cells and support the lineage assignment as ECs. Genes encoding proteins worked in endothelial adhesion and angiogenesis, such as Pecam1 (Sauteur et al., 2014; Lim et al., 2019), Cdh5, and Egfl7 (Charpentier et al., 2013; Usuba et al., 2019), which also showed enrichment in these cells (Figure 2C; Supplementary Figure 2B). Genes with average log-fold enrichment >2 and p-value <0.01 are the first choice to be used in distinguishing between individual endothelial cell subpopulations.
Expression of different functional genes demonstrates the functional multiplicity of these endothelial cells. Markers that have a role in the transporting and metabolism of lipids Lpl (Lutz et al., 2001), Gpihbp1 (Allan et al., 2017), Mgll (Nomura et al., 2010), and Cd36 (Jabs et al., 2018; Gerbod-Giannone et al., 2019) were upregulated in almost all clusters except EC 4, 6, and 10. Cells in EC 4 and 10 worked in cell adhesion (Vwf and Vcam1), while cells in EC six were inflammatory (Cxcl2 and Tyrobp). Of note, the gene uniquely expressed in endothelial colony-forming cells, Cytl1, showed a particular rise in EC 4, suggesting it is a genuine endothelial precursor cell; because endothelial colony-forming cells was thought to be a late endothelial precursor cell with a strong angiogenic function distinct from classical endothelial cells (d’Audigier et al., 2018). This type of endothelial cell has not been mentioned in similar studies. It uniquely performed an undetectable expression of Lpl and presented a higher expression of Nos3. Lyvel, a validated lymphatic endothelial cell marker that plays a role in lymphatic reactions such as leukocyte trafficking and helping to clear acute inflammatory response after myocardial infarction (Okuda et al., 2012; Vieira et al., 2018; Jackson, 2019), showed an exclusive positive expression in EC 10.
In addition to the transportation or metabolism of lipids, other endothelial subpopulations also present with different characteristics. Genes related to angiogenesis and cell proliferation, such as Pgf, Nrp2, and Tmsb10, have higher expression levels in EC 3. In VECs, PPARγ plays a protective role by increasing nitric oxide bioavailability and preventing oxidative stress. As a PPARγ target gene, RBP7 (Hu et al., 2017; Fang and Sigmund, 2020) is enriched in VECs and upregulated in EC5 in this study, which suggests it as EC against AS. Smooth muscle cell marker Myl9 exhibited a high expression in SMC-like EC in EC 7. Rgs5, which is involved in the induction of endothelial apoptosis (Lovschall et al., 2007), also performed significant upregulation in EC 7. Encoding proteins involved in B cell proliferation, Igkc, Ighm, and Cd79a (Kedmi et al., 2011), present a specific rise in EC eight and are identified as B cell-like EC. Fibrosing EC was identified with greatly increased Gsn, Dcn, and genes that play a role in collagen forming (Col1a2, Col3a1, Col1a1) (Figures 2B, D, E; Supplementary Figure 2C).
Among different gene markers, there exists a notable negative correlation between Cd36 and Vcam1. All endothelial subpopulations, except EC 4, 6, and 10, expressed higher Cd36 and have a reduced expression of Vcam1, while the three excluded subpopulations showed inverse expression. Interestingly, Pecam1 and Cdh5 also performed similar negative correlation in these endothelial cells.
From previous analysis, EC 1–3 were found to lack genes with log-fold enrichment >1, indicating that these clusters lack significant features. However, by further analysis, we observed that the genes involved in cell proliferation and angiogenesis presented progressively elevated expression levels from EC1 to EC3, suggesting that they may be continuous phenotypic gradients rather than conventionally different subpopulations (Figure 2F). Analysis focused on the top 30 upregulated genes of endothelial subpopulations 1–3 further revealed that the expression profiles of some cells in cluster EC1 overlap with those of EC2, while almost all of the cells in cluster EC3 have the expression features of EC2 (Supplementary Figure 2D).
Various endothelial subpopulations were distributed differently in the control group versus the HFD group. In total, the cells that exist preferentially in the HFD group indicated biological processes related with the development of mainly atherosclerosis. These subpopulations role in lipid clearance, utilization and storage, and exhibit property to against atherosclerosis but they also maintain role in inflammation and endothelial cell apoptosis. In contrast, those endothelial cells distributed mainly in the control group were involved in cell proliferating. Subpopulations EC1-3, which are assumed to have a developmental relationship, showed a distinct distribution between the two groups. EC1 has a biased distribution in the HFD group, while the other two were dominated by cells from the control group (Figures 3A, B).
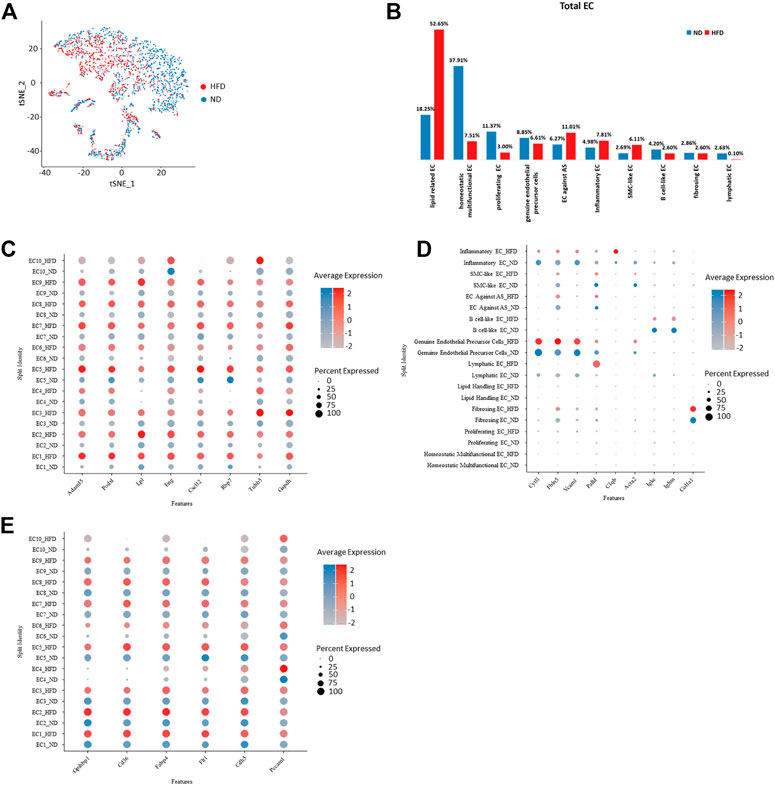
FIGURE 3. Homogeneity and heterogeneity between vascular endothelial cell of two origins uncover cellular variation respond to diet alteration. (A) t-Stochastic neighbor embedding representation of potential aortic endothelial cells extracted from total cell repertoire showing cellular origin in different colors. (B) Percentage of each endothelial population within total endothelial cells. Left: control (ND) group; Right: Western diet (HFD) group. (C–E) Transcriptional profiling of endothelial cells in ND versus HFD group notes diet-dependent genes with transcriptional upregulation in all HFD clusters. EC indicates endothelial cells (C) or diet dependent genes that are cluster specific (D) and conserved genes under regulation of diet. (E).
The transcriptional profiles of endothelial cells in the two experimental groups can help to reveal which biological processes are regulated by diet. In addition, the biological processes not influenced by diet were listed. We have summarized all endothelial cell markers that were used in recent single-cell studies of atherosclerosis (Table 1) as a reference.
All genes enhanced in the HFD group are related to atherosclerosis. Some indicate the progression of disease, such as Adam15 (Langer et al., 2005; Oksala et al., 2009), Lpl (Dugi et al., 1997), and Gadph (Perrotta et al., 2014), which play a role in pathological neovascularization, lipid utilization and storage, and innate immunity, respectively. Some play roles in the dysfunction of endothelial cells, such as Podxl which functions as an anti-adhesive molecule (Shoji et al., 2018) and Eng which regulates endothelial cell shape changes in response to blood flow and is required for normal structure (McAllister et al., 1994; Vicen et al., 2019). Some are anti-atherosclerotic, such as Cxcl12, which plays a protective role after myocardial infarction (Hartmann et al., 2015), and Rbp7, which helps to increase nitric oxide bioavailability (Figure 3C).
Cluster-specific markers also performed variously in the two groups and further confirmed the effect of diet. Cytl1 and Vcam1, markers of genuine endothelial precursor cells, showed unchanged expression under different types of diet, as does the marker of fibrosing EC, Col1a1. Fbln5, which is reinduced in atherosclerotic lesions, showed a rise in the HFD group. Similarly, canonical B cell marker Igkc and Ighm showed a reduced expression under the regulation of the Western diet (Figure 3D). The cells in the experimental group lost a lot of primary endothelial cell functions, such as cell formation, adhesion, and contraction, but inflammatory responses and some lipid-related functions (markers including Gpihbp1, Cd36, and Fabp4) were still retained (Figure 3E).
Immune cells are another emphasis in this study, and we define Ptprc + cells as immune cells. 1,615 cells were filtered out from the total cell population, with 1,083 from the control group and 532 from the HFD group. These cells were reclustered into 14 subpopulations. Identification of these cells was done based on the transcriptional profiles with the application of CIBERSORT (Figures 4A, B; Supplementary Figure 3A).
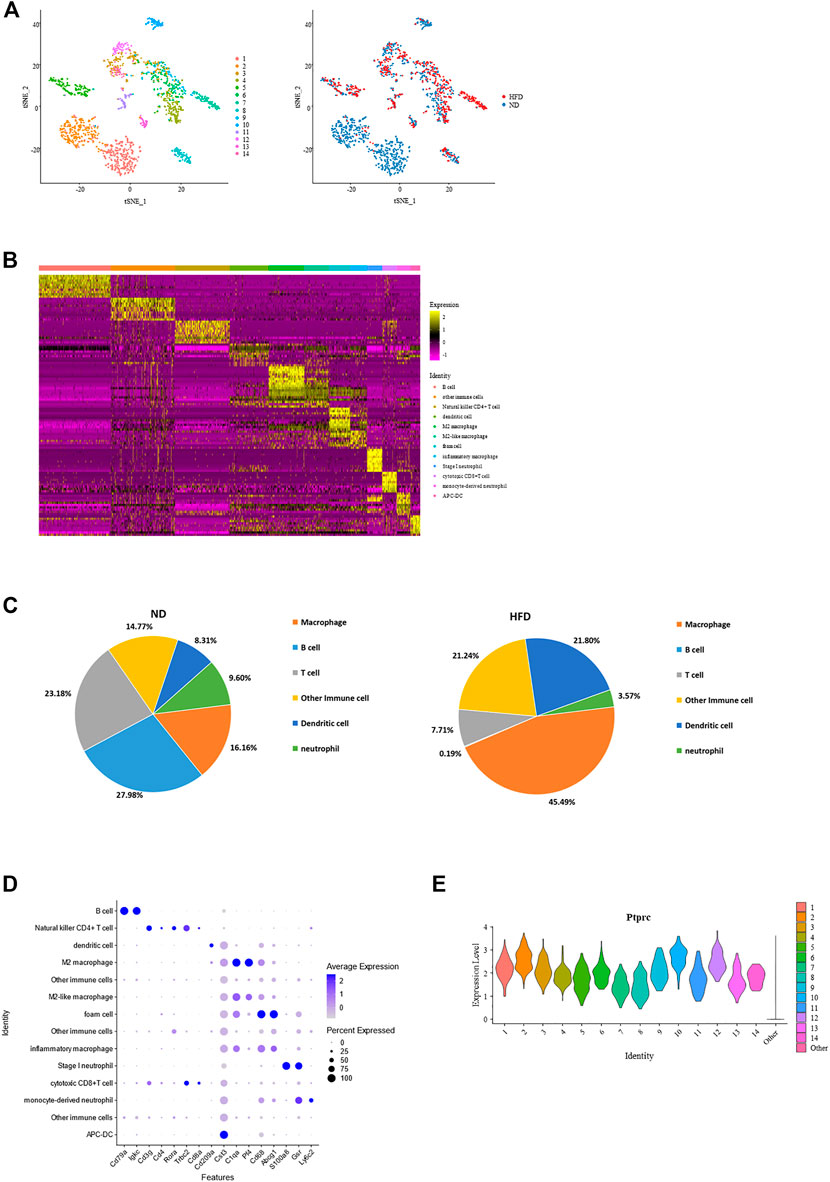
FIGURE 4. Identification of each immune cell cluster and their distribution reveal constitution of aortic immune cells and effect of diet. (A) T-Stochastic neighbor embedding (t-SNE) representation of gene expression information in aortic Ptptrc + immune cells. Cells from both control and Western diet group (right) are distinct into 14 clusters (left); CIBERSORT helps with their identification (middle). (B) Heatmap showing the top 10 upregulated genes in each cluster of immune cells, with biological identification of each cluster marked on the right. (C) Percentage of each immune cell type within total immune cells. Left: control (ND) group; Right: Western diet (HFD) group. (D) Dot plot showing the expression levels of representative marker genes for each immune cell type. (E) Violin plots of log-transformed gene expression of Ptprc distinguish these immune subpopulations from all other aortic cells.
Constitution of immune cells in the control group versus the HFD group reveals the great effect created by the Western diet. The most obvious heterogeneity brought by the Western diet was that B cells and T cells dominated the control group, accounting for 27.98% and 23.18% respectively, while macrophages had the largest proportion in HFD group, up to 45.49%. These 3 cell types dominate the immune cell. Another variance was the increase of dendritic cells and decrease of neutrophil (Figures 4A, C; Supplementary Figure 3A).
Macrophage had the largest proportion in all immune cells are were divided into four clusters, all showing a common expression of C1qa (De Couto et al., 2019). Pf4, an M2 macrophage marker, was expressed the highest in cluster4 and cluster6, while foam cell marker Cd68 and Abcg1 was enriched in cluster7. The proportion of B cell and T cell were similar. It should be noted that B cells (Cd79a, Cd79b, and Ly6d) existed exclusively in the control group. T cells (Trbc1, Trbc2 (Lefranc, 2014), Cd3d, and Cd3g) consisted of natural killer CD4+ T cells, Rora (Kästle et al., 2017), Il7r (Al-Mossawi et al., 2019), Icos (Niu et al., 2018) and Cd4, and cytotoxic CD8+ T cells, Cd8a and Cd8b1. The remainder of immune cell populations included two clusters of dendritic cells (DCs) with gene signatures Cd209a (cluster 3), Cst4 and Clec9a (cluster 14), two clusters of neutrophils (cluster 10: S100a8 and S100a9; cluster 10 and 12: Gsr (Yan et al., 2013); cluster 12: highly expressed Ly6c2, a marker of monocyte), and three other clusters of Ptprc + cells (Figures 4B, D; Supplementary Figure 3B). Regardless of type, all showed a high expression of Ptprc (Figure 4E).
As previously seen, four clusters were defined as macrophage. Common enrichment of C1qa (De Couto et al., 2019), C1qb (Giladi et al., 2018), C1qc (Zhao et al., 2020), and Mafb (Goudot et al., 2017) further confirmed the identity of macrophage (Figure 5A). The proportion of macrophage across all immune cells increased due to the appearance of foam cells according to the constitution of macrophage in the control group versus the HFD group (Supplementary Figure 4A). In the meantime, the proportion of M2 macrophage decreased. The remainder are M2-like macrophages and inflammatory macrophages (Figure 5B).
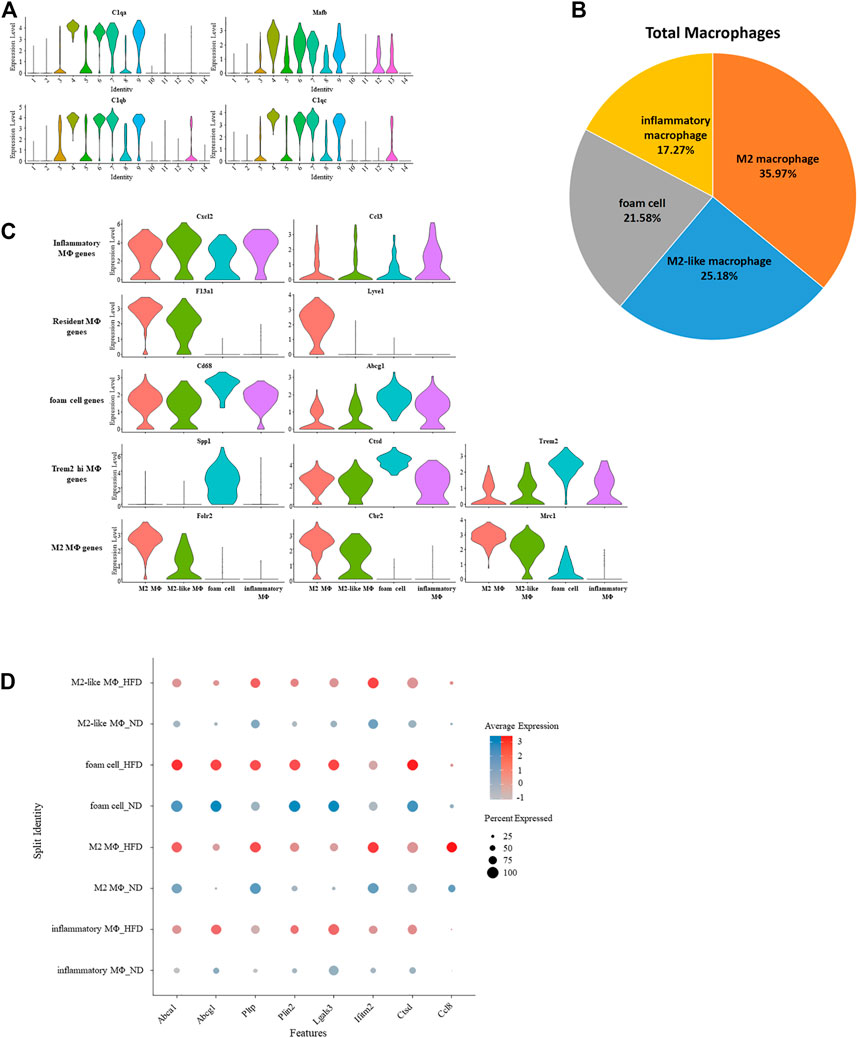
FIGURE 5. Gene expression signature of macrophage and its variation under western diet. (A) Violin plots of log-transformed gene expression of canonical macrophage markers in macrophages and all other aortic immune cells. (B) Percentage of each macrophage type. (C) Violin plots profile gene expression of markers for the five classes of macrophages in these selected macrophages. (D) Heterogeneity within macrophage of two origins demonstrated by dotplot.
Single-cell differential expression pattern helps differentiate these four macrophage subsets from one another, and specific enrichment of these genes in each subset helps further determine their identification. Previous studies established a gene pool for various macrophages. Despite greater expression of M2 macrophage markers Folr2, Mrc1, and Cbr2, M2 macrophage also had characteristics of resident macrophage (F13a1 (Beckers et al., 2017) and Lyve1 (Ensan et al., 2016)). The expression profile of M2-like macrophages is similar to M2 macrophages, inferred by a transitional relationship between them. Meanwhile, due to the high expression of the monocyte marker Ccr2, we speculate that it is the progenitor of M2 macrophages. Foam cells were identified because the relative upregulation of Cd68 and Abcg1 can promotes cholesterol accumulation, which matters in the formation of foam cells (Cheng et al., 2016). These cells also have an exclusively positive expression of Spp1. Spp1 encodes osteopontin, which is a factor related to the severity of lesions. Another related factor is cathepsin, encoded by Ctsb, Ctsd, and Ctsz (Cochain et al., 2018), which was expressed higher in the foam cell. This was also the case with Trem2, a putative factor in Trem2-high macrophages except osteopontin and cathepsin. With significant upregulation of several chemokines (Ccl3, Ccl4, Cxcl1, Cxcl2, and Cxcl16) that play a role in inflammation, inflammatory macrophages were identified (Figure 5C; Supplementary Figure 4B).
The effect of diet on macrophages can also be seen from genetic regulation under the Western diet. These genes were involved in biological pathways such as cholesterol transporting (Abca1, Abcg1, and Pltp), development of adipose tissue (Plin2), inflammation (Lgals3 and Ccl8), and cell apoptosis (Ifitm2 and Ctsd) (Figure 5D).
Pecam1 and Cdh5 are a pair of genes used to identify endothelial cells in this study. According to the scRNA sequencing data, an endothelial subpopulation, identified as genuine endothelial precursor cells specifically, was higher in Pecam1 and lower in Cdh5 compared to other endothelial subpopulations (Figure 1F). This subpopulation expressed Cytl1 exclusively (Figure 1D).
The Cytl1+ cells were found in three clusters in further analysis of sorted Pecam1+ Cdh5+ endothelial cells (Figure 2E), and retained the characteristics of high Pecam1 expression and low Cdh5 expression (Figure 2C). Upregulated Vcam1 expression level is another characteristic of these cells.
In the comparison of gene expression levels for endothelial cells isolated from a normal diet versus Western diet, Pecam1 and Cdh5 were listed in top 100 differentially expressed genes (DEGs), with Pecam1 lowered and Cdh5 enhanced in the Western diet group (Figure 3E).
Flow cytometry helped identify the distinct spatial location of high Pecam1 (Figure 6A) and low Cdh5 (Figure 6B) expression. Greater curvature of the aorta demonstrates less presence of high Pecam1-expressing cells and more presence of low Cdh5-expressing cells. The preference in location was found in both diet groups. Otherwise, the Western diet presented mild inhibition of the increase of low Cdh5-expressing cells.
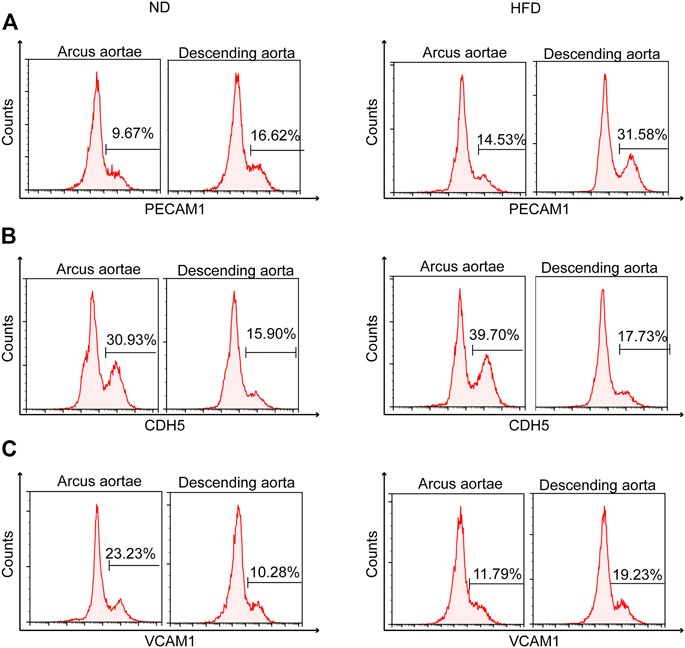
FIGURE 6. Identification of genetic heterogeneity in situ within endothelial cells. Flow cytometry of mouse aortic endothelial cells demonstrating heterogeneity of (A) Pecam1, (B) Cdh5, and (C) Vcam1 between greater (arcus aortae) and lesser (descending aorta) curvature of aorta, showing the influence of a Western diet.
Flow cytometry also helped identify the distinct spatial location of high Vcam1 expression (Figure 6C). High Vcam1-expressing cells showed a preference in descending aorta compared to the greater curvature of the aortic root under a high fat diet. A contrasting result was found under normal conditions.
Known as the region with disturbed blood flow, the aortic arch is composed of different cell types and is relevant in the progression of cardiovascular disease and is the research object in this study. With well-established single-cell RNA sequencing, we characterized different types of cells in the aortic arch and mining gene expression changes that occur within these cells under the influence of diet. We differentiated 14 classes of cells from the total sample, including endothelial cells, fibroblasts, smooth muscle cells, mesothelial cells, adipocytes, neurons, and immune cell populations, which were distributed in 27 clusters. Notably, the identification of cycling basal cell, clara cell, mesothelial cell, and adipocyte was new. Heterogeneous gene expression pattern distinguishes 10 subpopulations of endothelial cells with distinct functions and reveals which biological processes may be affected or remain unchanged under a Western diet. This study identifies genuine endothelial precursor cells for the first time in such studies, complementing the transcriptional information blank of this cell. Another class of cells focused on in this study was immune cells, especially macrophages. Seven immune cell types were distributed in 14 immune cell clusters, and four clusters of macrophages expressed different characteristics. A genes expression map uncovered the multiplicity of aortic cells and demonstrated that the Western diet accelerates atherosclerotic development by regulating the role of the genes in various biological processes.
In the total aortic cell pool, the seven endothelial subpopulations were similar in the expression levels of Pecam1 and Cdh5. The cell adhesion molecules encoded by Pecam1 and Cdh5 are essential for leukocyte trans-endothelial migration (Dasgupta et al., 2009) and the maintenance of vascular lumen homeostasis (Lampugnani et al., 2010). Negative expression correlation between these two genes was observed in each endothelial subpopulation. A genuine endothelial precursor cell exhibited higher Pecam1 and lower Cdh5 gene expression levels compared to the other endothelial cell clusters.
Another gene pair showing similar negative correlation between endothelial precursor cells and other endothelial cells is Vcam1 and Cd36. Based on the conclusions of previous single-cell studies, Vcam1 and Cd36 were relatively biased to be lower and higher expressed in classic endothelial cells and endothelial cell roles in lipid treating. A classic endothelial cell tends to be distributed in descending aorta with less curvature, while other endothelial cells tend to be distributed in the arterial root with greater curvature (Kalluri et al., 2019). Genuine endothelial precursor cells exhibited the same expression pattern as the classic endothelial cells mentioned previously. According to the relationship between the two gene pairs, the endothelial precursor cells involved in this study can be speculated to be like classic endothelial cells, possibly located at sites with smaller curvature. Sites with less curvature often suffer attenuated lipid accumulation and lower atherosclerotic risk rates. The result of flow cytometry supports the speculation on the location of genuine endothelial precursor cells and demonstrates the influence of the Western diet on this cell type. Nos3, known for its promotion effects in nitric oxide production and vasodilation, was significantly upregulated in these precursor cells, further indicating the lower atherosclerotic risk rate within it (Lusis, 2000). Data in the STRING database showed a co-expression relationship between Cdh5, Pecam1, and Vcam1, while Cd36 was not involved. However, whether the supposed relevance among these proteins are reliable and how to localize them remains to be addressed.
Flow cytometry was not used in this study, instead only algorithm was used in the cell sorting. Endothelial cells sorted this way in the study showed a similar proportion of genuine endothelial precursor cells in both experimental groups. Except genuine endothelial precursor cells, inflammatory endothelial cells also had a higher expression of Pecam1 and Vcam1 as well as a lower expression of Cdh5 and Cd36. The two endothelial cell subpopulations existing in both the control and HFD group indicated that some precursor cells and inflammatory cells from advanced atherosclerosis were clustered together with cells in a normal condition. It can be speculated that the transcriptional profiles of these cells in the late disease and normal states are similar, but whether they locate in atherosclerotic plaque remains to be determined.
Distribution of EC1-3 and expression profile of them indicates great probability that endothelial develops from the state of EC3 into EC1 during the disease progression. During the process, endothelial went through dysfunction and lost the function of angiogenesis and cell proliferation.
The Western diet can accelerate the progression of atherosclerosis (Supplementary Figures 5A–D) as well as endothelial dysfunction. Genes that showed an increased expression indicated the character of dysfunctional endothelial cells. These dysfunctional endothelial cells lost cell adhesion function (Podxl), which is one of the key functions in normal endothelial cells. Meanwhile, based on certain responses, based on certain responses, they appeared to inhibit the further dysfunction of endothelial cells (Hu et al., 2017), for example, changing shape in response to blood flow in order to keep normal structure (Eng) or promoting the production of nitric oxide (Rbp7) (Fang and Sigmund, 2020). In accordance with previous findings, Cxcl2 is enriched in endothelial cells in advanced atherosclerosis and suppresses atherosclerosis after myocardial infarction (F. Li et al., 2021). As a key enzyme in triglyceride metabolism, Tubb5 plays an important role in removing lipids from the blood, as well as in lipid utilization and storage (Weinstock et al., 1995). The enrichment of these biological processes suggests that in most cases, feedback inhibitory pathways for atherosclerosis development are activated in dysfunctional endothelial cells, potentially limiting further plaque expansion by inhibiting lipid accumulation and regulating vasodilation in the lesion.
Both Fbln5 and Vcam1 were upregulated in genuine endothelial precursor cells of mice with advanced atherosclerosis administered with a Western diet, implying that it induced more leukocytes that migrated to sites of inflammation. At the same time, lipid-related functions in these cells were not as active as in other endothelial cells, suggesting their absence in proatherogenic biological processes such as cholesterol transport, lipid utilization, or storage.
The results showed that some immune cells were absent in the HFD group, such as B cells and CD4 + T cells, indicating that the Western diet may inhibit the differentiation of aortic immune cells. A recent study pointed out that B1a and B1b lymphocytes produce IgM to inactivate oxidation-specific epitopes on LDL and thereby protect against atherosclerosis (Pattarabanjird et al., 2022). Therefore, it can be speculated that B cells initiate protective mechanisms in atherosclerosis, while the Western diet reduces the number of B cells, thus aggravating atherosclerosis. In accordance with a recent study published in Frontiers in Immunology, which indicated that neutrophils and APC-like neutrophils were dominant in the blood of hyperlipidemic patients rather than healthy patients (Zhao et al., 2022), neutrophils and dedicated antigen-presenting dendritic cells (APC-DC) were found to be higher in the HFD group than in the control group. However, in this study, there was no more research into the role of APC-DC in atherosclerosis. This could be further explored in the future.
The proportion of macrophages observed in Ptprc + cells increases with disease progression, similar to the findings of other researchers (Galkina et al., 2006; Cochain et al., 2018). In contrast, the proportional increase in macrophages observed in this study was caused by foam cells rather than resident macrophages or inflammatory macrophages. These foam cells were specifically enriched with the validated markers of Trem2 high macrophage. It can be inferred that the transition from macrophages to foam cells occurs in the late stages of disease and the cell of origin may be highly expressing Trem2. During the progression of atherosclerosis, the production of foam cells is inseparable from the high oxidation of LDL. High oxidization assists LDL recognition by the scavenger receptor Cd68 and thus uptake by macrophages occurs rapidly. Clearly, the significant upregulation of Cd68 with the above principle could explain the emergence of foam cells in this study.
The role of Abca1, Abcg1, and Pltp in foam cells is particularly apparent in the late stage of atherosclerosis. They help with the efflux of cholesterol in macrophages and uptake into HDL, hence promote the conversion of LDL to HDL. This facilitation of HDL synthesis predicts an inhibited conversion of macrophages to foam cells and can be one of the manifestations of feedback inhibition in advanced atherosclerosis, which slows down the further development of disease.
In conclusion, transcriptional information of aortic cells, especially VEC, was mapped using single-cell RNA sequencing. Significantly, we identified 10 endothelial subpopulations and established candidate marker genes of each subpopulation. In addition, all endothelial candidate markers identified in previous research of atherosclerosis with single-cell RNA sequencing were confirmed in this study. Notably, by observing its transcriptional signature, we found genuine endothelial precursor cells, which had not been mentioned in previous studies. Compared with previous studies, the results of this study broaden the understanding of endothelial cells and further enrich existing information about different endothelial cell markers.
In addition, comparison of gene expression profiles and constitutions of cell between the health and atherosclerotic group can help in understanding the post-modification induced by a high-fat high-cholesterol diet in various cells, especially endothelial cells and immune cells. Understanding the pathways involved in these modifications may contribute to further exploration of pathogenesis of atherosclerotic lesions.
There are some limitations to this study, which may comprise further areas of research. Validation of several newly demonstrated genes or pathways has not yet been carried out. Traditional methods such as immunohistochemistry and western blot could be used to validate the distribution of these genes and pathways more accurately.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE206239.
The animal study was reviewed and approved by Shenzhen University Animal Care and Use Committee.
JW designed the research and performed the experiments and RL performed data analysis; both contributed equally to this manuscript. JW wrote the article. RC conducted critical editing. SZ provided data support. YD, TZ, FG, QW, and GW helped in reagent and material preparation. All authors contributed to the article and approved the submitted version.
This work was supported by Shenzhen Science and Technology Innovation Commission grant JCYJ20180305124812444 and ZDSYS20190902092903237.
We thank Genergy Biotechnology (Shanghai) Co., Ltd. for the deep sequencing service.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.971091/full#supplementary-material
DC, Dendritic cell; EC, Endothelial cell; ECM, Extracellular matrix; HDL, High-density lipoprotein; HFD, Western diet; LDL, Low-density lipoprotein; SMC, Smooth muscle cell; t-SNE, t-stochastic neighbor embedding; VEC, Vascular endothelial cell.
Al-Mossawi, H., Yager, N., Taylor, C. A., Lau, E., Danielli, S., de Wit, J., et al. (2019). Context-specific regulation of surface and soluble IL7R expression by an autoimmune risk allele. Nat. Commun. 10 (1), 4575. doi:10.1038/s41467-019-12393-1
Allan, C. M., Jung, C. J., Larsson, M., Heizer, P. J., Tu, Y., Sandoval, N. P., et al. (2017). Mutating a conserved cysteine in GPIHBP1 reduces amounts of GPIHBP1 in capillaries and abolishes LPL binding. J. Lipid Res. 58 (7), 1453–1461. doi:10.1194/jlr.M076943
Aryan, L., Younessi, D., Zargari, M., Banerjee, S., Agopian, J., Rahman, S., et al. (2020). The role of estrogen receptors in cardiovascular disease. Int. J. Mol. Sci. 21 (12), 4314–4326. doi:10.3390/IJMS21124314
Beckers, C. M. L., Simpson, K. R., Griffin, K. J., Brown, J. M., Cheah, L. T., Smith, K. A., et al. (2017). Cre/lox studies identify resident macrophages as the major source of circulating coagulation factor XIII-A. Arteriosclerosis, Thrombosis, Vasc. Biol. 37 (8), 1494–1502. doi:10.1161/ATVBAHA.117.309271
Brandt, K. J., Burger, F., Baptista, D., Roth, A., Fernandes da Silva, R., Montecucco, F., et al. (2022). Single-cell analysis uncovers osteoblast factor growth differentiation factor 10 as mediator of vascular smooth muscle cell phenotypic modulation associated with plaque rupture in human carotid artery disease. Int. J. Mol. Sci. 23 (3), 1796. doi:10.3390/IJMS23031796
Burger, F., Baptista, D., Roth, A., Brandt, K. J., da Silva, R. F., Montecucco, F., et al. (2022). Single-cell RNA-seq reveals a crosstalk between hyaluronan receptor LYVE-1-expressing macrophages and vascular smooth muscle cells. Cells 11 (3), 411. doi:10.3390/CELLS11030411
Cai, J., Deng, J., Gu, W., Ni, Z., Liu, Y., Kamra, Y., et al. (2020). Impact of local alloimmunity and recipient cells in transplant arteriosclerosis. Circulation Res. 127 (8), 974–993. doi:10.1161/CIRCRESAHA.119.316470
Charpentier, M. S., Christine, K. S., Amin, N. M., Dorr, K. M., Kushner, E. J., Bautch, V. L., et al. (2013). CASZ1 promotes vascular assembly and Morphogenesis through the direct regulation of an EGFL7/RhoA-mediated pathway. Dev. Cell 25 (2), 132–143. doi:10.1016/j.devcel.2013.03.003
Chen, Y., Wen, S., Jiang, M., Zhu, Y., Ding, L., Shi, H., et al. (2017). Atherosclerotic dyslipidemia revealed by plasma lipidomics on ApoE−/− mice fed a high-fat diet. Atherosclerosis 262, 78–86. doi:10.1016/j.atherosclerosis.2017.05.010
Cheng, H. Y., Gaddis, D. E., Wu, R., McSkimming, C., Haynes, L. D., Taylor, A. M., et al. (2016). Loss of ABCG1 influences regulatory T cell differentiation and atherosclerosis. J. Clin. Investigation 126 (9), 3236–3246. doi:10.1172/JCI83136
Cochain, C., Vafadarnejad, E., Arampatzi, P., Pelisek, J., Winkels, H., Ley, K., et al. (2018). Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circulation Res. 122 (12), 1661–1674. doi:10.1161/CIRCRESAHA.117.312509
Curtiss, L. K., Boisvert, W. A., Mattot, V., Saubamea, B., Rossi, E., Guerin, C., et al. (2000). Apolipoprotein E and atherosclerosisEgfl7 represses the vasculogenic potential of human endothelial progenitor cells. Curr. Opin. lipidologyStem Cell Rev. Rep. 1114 (31), 24382–25191. doi:10.1097/00041433-200006000-00004
Dasgupta, B., Dufour, E., Mamdouh, Z., and Muller, W. A. (2009). A novel and critical role for tyrosine 663 in platelet endothelial cell adhesion molecule-1 trafficking and transendothelial migration. J. Immunol. 182 (8), 5041–5051. doi:10.4049/JIMMUNOL.0803192
Davignon, J., and Ganz, P. (2004). ‘Role of endothelial dysfunction in atherosclerosis’, Circulation. Lippincott Williams & Wilkins. doi:10.1161/01.cir.0000131515.03336.f8
De Couto, G., Jaghatspanyan, E., DeBerge, M., Liu, W., Luther, K., Wang, Y., et al. (2019). Mechanism of enhanced MerTK-Dependent macrophage efferocytosis by extracellular vesicles. Arteriosclerosis, Thrombosis, Vasc. Biol. 39 (10), 2082–2096. doi:10.1161/ATVBAHA.119.313115
Depuydt, M. A. C., Prange, K. H. M., Slenders, L., Ord, T., Elbersen, D., Boltjes, A., et al. (2020). Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circulation Res. 127 (11), 1437–1455. doi:10.1161/CIRCRESAHA.120.316770
Dugi, K. A., Feuerstein, I. M., Hill, S., Shih, J., Santamarina-Fojo, S., Brewer, H. B., et al. (1997). Lipoprotein lipase correlates positively and hepatic lipase inversely with calcific atherosclerosis in homozygous familial hypercholesterolemia. Arteriosclerosis, Thrombosis, Vasc. Biol. 17 (2), 354–364. doi:10.1161/01.ATV.17.2.354
Durand, M. J., and Gutterman, D. D. (2013). Diversity in mechanisms of endothelium-dependent vasodilation in health and disease. Microcirculation 20 (3), 239–247. doi:10.1111/micc.12040
Ensan, S., Li, A., Besla, R., Degousee, N., Cosme, J., Roufaiel, M., et al. (2016). Self-renewing resident arterial macrophages arise from embryonic CX3CR1 + precursors and circulating monocytes immediately after birth. Nat. Immunol. 17 (2), 159–168. doi:10.1038/ni.3343
Fang, S., and Sigmund, C. D. (2020). PPARγ and RhoBTB1 in hypertension. Curr. Opin. Nephrol. Hypertens. 29 (2), 161–170. doi:10.1097/MNH.0000000000000579
Galkina, E., Kadl, A., Sanders, J., Varughese, D., Sarembock, I. J., and Ley, K. (2006). Lymphocyte recruitment into the aortic wall before and during development of atherosclerosis is partially L-selectin dependent. J. Exp. Med. 203 (5), 1273–1282. doi:10.1084/JEM.20052205
Garrido, A. M., Kaistha, A., Uryga, A. K., Oc, S., Foote, K., Shah, A., et al. (2021). Efficacy and limitations of senolysis in atherosclerosis. Cardiovasc. Res. 118, 1713–1727. doi:10.1093/CVR/CVAB208
Gerbod-Giannone, M. C., Dallet, L., Naudin, G., Sahin, A., Decossas, M., Poussard, S., et al. (2019). Involvement of caveolin-1 and CD36 in native LDL endocytosis by endothelial cells. Biochimica Biophysica Acta - General Subj. 1863 (5), 830–838. doi:10.1016/j.bbagen.2019.01.005
Giladi, A., Paul, F., Herzog, Y., Lubling, Y., Weiner, A., Yofe, I., et al. (2018). Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20, 836–846. doi:10.1038/s41556-018-0121-4
Goudot, C., Coillard, A., Villani, A. C., Gueguen, P., Cros, A., Sarkizova, S., et al. (2017). Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 47 (3), 582–596.e6. doi:10.1016/j.immuni.2017.08.016
Gu, W., Ni, Z., Tan, Y. Q., Deng, J., Zhang, S. J., Lv, Z. C., et al. (2019). Adventitial cell atlas of wt (wild type) and ApoE (apolipoprotein E)-Deficient mice defined by single-cell RNA sequencing. Arteriosclerosis, thrombosis, Vasc. Biol. 39 (6), 1055–1071. doi:10.1161/ATVBAHA.119.312399
Hartmann, P., Schober, A., and Weber, C. (2015). ‘Chemokines and microRNAs in atherosclerosis’, cellular and molecular life sciences. New York: Birkhauser Verlag AG, 3253–3266. doi:10.1007/s00018-015-1925-z
Hu, C., Keen, H. L., Lu, K. T., Liu, X., Wu, J., Davis, D. R., et al. (2017). Retinol-binding protein 7 is an endothelium-specific PPAR γ cofactor mediating an antioxidant response through adiponectin. JCI insight 2 (6), e91738. doi:10.1172/JCI.INSIGHT.91738
Huang, X., Shen, W., Veizades, S., Liang, G., Sayed, N., and Nguyen, P. K. (2021). Single-cell transcriptional profiling reveals sex and age diversity of gene expression in mouse endothelial cells. Front. Genet. 12, 590377. doi:10.3389/FGENE.2021.590377
Jabs, M., Rose, A. J., Lehmann, L. H., Taylor, J., Moll, I., Sijmonsma, T. P., et al. (2018). Inhibition of endothelial notch signaling impairs fatty acid transport and leads to metabolic and vascular remodeling of the adult heart. Circulation 137 (24), 2592–2608. doi:10.1161/CIRCULATIONAHA.117.029733
Jackson, D. G. (2019). Hyaluronan in the lymphatics: The key role of the hyaluronan receptor LYVE-1 in leucocyte trafficking. J. Int. Soc. Matrix Biol. 78 (79), 219–235. doi:10.1016/J.MATBIO.2018.02.001
Kalluri, A. S., Vellarikkal, S. K., Edelman, E. R., Nguyen, L., Subramanian, A., Ellinor, P. T., et al. (2019). Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation 140 (2), 147–163. doi:10.1161/CIRCULATIONAHA.118.038362
Kästle, M., Bartel, S., Geillinger-Kastle, K., Irmler, M., Beckers, J., Ryffel, B., et al. (2017). microRNA cluster 106a∼363 is involved in T helper 17 cell differentiation. Immunology 152 (3), 402–413. doi:10.1111/imm.12775
Kedmi, M., Bar-Shira, A., Gurevich, T., Giladi, N., and Orr-Urtreger, A. (2011). Decreased expression of B cell related genes in leukocytes of women with Parkinson’s disease. Mol. Neurodegener. 6 (1), 66. doi:10.1186/1750-1326-6-66
Knowles, J. W., Reddick, R. L., Jennette, J. C., Shesely, E. G., Smithies, O., and Maeda, N. (2000). Enhanced atherosclerosis and kidney dysfunction in eNOS–/–Apoe–/– mice are ameliorated by enalapril treatment. J. Clin. Investigation 105 (4), 451–458. doi:10.1172/JCI8376
Lampugnani, M. G., Orsenigo, F., Rudini, N., Maddaluno, L., Boulday, G., Chapon, F., et al. (2010). CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J. Cell Sci. 123, 1073–1080. doi:10.1242/JCS.059329
Langer, H., May, A. E., Bultmann, A., and Gawaz, M. (2005). ADAM15 is an adhesion receptor for platelet GPIIb-IIIa and induces platelet activation. Thrombosis Haemostasis 94 (3), 555–561. doi:10.1160/TH04-12-0784
Lefranc, M. P. (2014). Immunoglobulin and T cell receptor genes: IMGT® and the birth and rise of immunoinformatics. Front. Immunol. 5 (FEB), 22. doi:10.3389/fimmu.2014.00022
Li, F., Yan, K., Wu, L., Zheng, Z., Du, Y., Liu, Z., et al. (2021). Single-cell RNA-seq reveals cellular heterogeneity of mouse carotid artery under disturbed flow. Cell death Discov. 7 (1), 180. doi:10.1038/S41420-021-00567-0
Li, S., Gao, Y., Ma, K., Li, Y., Liu, C., Yan, Y., et al. (2021). Lipid-related protein NECTIN2 is an important marker in the progression of carotid atherosclerosis: An intersection of clinical and basic studies. J. Transl. Intern. Med. 9 (4), 294–306. doi:10.2478/JTIM-2021-0044
Libby, P. (2000). “Changing concepts of atherogenesis,” in Journal of internal medicine (John Wiley & Sons), 349–358. doi:10.1046/j.1365-2796.2000.00654.x
Lim, R., Sugino, T., Nolte, H., Andrade, J., Zimmermann, B., Shi, C., et al. (2019). Deubiquitinase USP10 regulates Notch signaling in the endothelium. Sci. (New York, N.Y.) 364 (6436), 188–193. doi:10.1126/SCIENCE.AAT0778
Liu, Y. X., Yuan, P. Z., Wu, J. H., and Hu, B. (2021). Lipid accumulation and novel insight into vascular smooth muscle cells in atherosclerosis. J. Mol. Med. (Berlin, Ger. 99 (11), 1511–1526. doi:10.1007/S00109-021-02109-8
Lovschall, H., Mitsiadis, T. A., Poulsen, K., Jensen, K. H., and Kjeldsen, A. L. (2007). Coexpression of Notch3 and Rgs5 in the pericyte-vascular smooth muscle cell axis in response to pulp injury. Int. J. Dev. Biol. 51 (8), 715–721. doi:10.1387/IJDB.072393HL
Lu, H., and Daugherty, A. (2015). Atherosclerosis. ‘Atherosclerosis’, Arteriosclerosis, Thrombosis, Vasc. Biol. 35 (3), 485–491. doi:10.1161/ATVBAHA.115.305380
Lusis, A. J. (2000). ‘Atherosclerosis’, nature. England: Nature Publishing Group, 233–241. doi:10.1038/35025203
Lutz, E. P., Merkel, M., Kako, Y., Melford, K., Radner, H., Breslow, J. L., et al. (2001). Heparin-binding defective lipoprotein lipase is unstable and causes abnormalities in lipid delivery to tissues. J. Clin. Investigation 107 (9), 1183–1192. doi:10.1172/JCI11774
Ma, X., Deng, J., Han, L., Song, Y., Miao, Y., Du, X., et al. (2022). Single-cell RNA sequencing reveals B cell-T cell interactions in vascular adventitia of hyperhomocysteinemia-accelerated atherosclerosis. Protein & Cell 13, 540–547. doi:10.1007/S13238-021-00904-0
Maganto-Garcia, E., Tarrio, M., and Lichtman, A. H. (2012). Mouse models of atherosclerosis. Curr. Protoc. Immunol. 96 (1), 15.24.11–2415.24.23. doi:10.1002/0471142735.im1524s96
Mannarino, E., and Pirro, M. (2008). “Endothelial injury and repair: A novel theory for atherosclerosis,” in Angiology (Hyderabad: SAGE Publications Inc.), 69S–72S. doi:10.1177/0003319708320761
McAllister, K. A., Grogg, K. M., Johnson, D. W., Gallione, C. J., Baldwin, M. A., Jackson, C. E., et al. (1994). Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat. Genet. 8 (4), 345–351. doi:10.1038/NG1294-345
Meir, K. S., and Leitersdorf, E. (2004). Atherosclerosis in the apolipoprotein-E-deficient mouse: A decade of progress. Arteriosclerosis, thrombosis, Vasc. Biol. 24 (6), 1006–1014. doi:10.1161/01.ATV.0000128849.12617.F4
Ni, H., Liu, C., Chen, Y., Lu, Y., Ji, Y., Xiang, M., et al. (2022). MGP regulates perivascular adipose-derived stem cells DifferentiationToward smooth muscle cells via BMP2/SMAD pathway enhancing NeointimalFormation. Cell Transplant. 31, 9636897221075747. doi:10.1177/09636897221075747
Niu, L. L., Xuan, X., Wang, J., Li, L., Yang, D., Jing, Y., et al. (2018). Akt2 regulates the differentiation and function of NKT17 cells via FoxO-1-ICOS axis. Front. Immunol. 9 (SEP), 1940. doi:10.3389/fimmu.2018.01940
Nomura, D. K., Long, J. Z., Niessen, S., Hoover, H. S., Ng, S. W., and Cravatt, B. F. (2010). Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140 (1), 49–61. doi:10.1016/j.cell.2009.11.027
Oksala, N., Levula, M., Airla, N., Pelto-Huikko, M., Ortiz, R. M., Jarvinen, O., et al. (2009). ADAM-9, ADAM-15, and ADAM-17 are upregulated in macrophages in advanced human atherosclerotic plaques in aorta and carotid and femoral arteriesTampere vascular study. Ann. Med. 41 (4), 279–290. doi:10.1080/07853890802649738
Okuda, K. S., Astin, J. W., Misa, J. P., Flores, M. V., Crosier, K. E., and Crosier, P. S. (2012). Lyve1 expression reveals novel lymphatic vessels and new mechanisms for lymphatic vessel development in zebrafish. Dev. Camb. 139 (13), 2381–2391. doi:10.1242/dev.077701
Paone, S., et al. (2019). ‘Endothelial cell apoptosis and the role of endothelial cell-derived extracellular vesicles in the progression of atherosclerosis’, Cellular and Molecular Life Sciences. New York: Birkhauser Verlag AG, 1093–1106. doi:10.1007/s00018-018-2983-9
Pattarabanjird, T., Marshall, M., Upadhye, A., Srikakulapu, P., Garmey, J. C., Haider, A., et al. (2022). B-1b cells possess unique bHLH-driven P62-dependent self-renewal and atheroprotection. Circulation Res. 130, 981–993. doi:10.1161/CIRCRESAHA.121.320436
Perrotta, I., Aquila, S., and Mazzulla, S. (2014). ‘Expression profile and subcellular localization of GAPDH in the smooth muscle cells of human atherosclerotic plaque: An immunohistochemical and ultrastructural study with biological therapeutic perspectives’, Microscopy and Microanalysis. Cambridge University Press, 1145–1157. doi:10.1017/S1431927614001020
Pinheiro-de-Sousa, I., Fonseca-Alaniz, M. H., Teixeira, S. K., Rodrigues, M. V., and Krieger, J. E. (2022). Uncovering emergent phenotypes in endothelial cells by clustering of surrogates of cardiovascular risk factors. Sci. Rep. 12 (1), 1372. doi:10.1038/S41598-022-05404-7
Quiles-Jiménez, A., Gregersen, I., Segers, F. M., Skarpengland, T., Kroustallaki, P., Yang, K., et al. (2021). DNA glycosylase Neil3 regulates vascular smooth muscle cell biology during atherosclerosis development. Atherosclerosis 324, 123–132. doi:10.1016/J.ATHEROSCLEROSIS.2021.02.023
Sauteur, L., Krudewig, A., Herwig, L., Ehrenfeuchter, N., Lenard, A., Affolter, M., et al. (2014). Cdh5/VE-cadherin promotes endothelial cell interface elongation via cortical actin polymerization during angiogenic sprouting. Cell Rep. 9 (2), 504–513. doi:10.1016/j.celrep.2014.09.024
Schönbeck, U., Sukhova, G. K., Shimizu, K., Mach, F., and Libby, P. (2000). Inhibition of CD40 signaling limits evolution of established atherosclerosis in mice. Proc. Natl. Acad. Sci. U. S. A. 97 (13), 7458–7463. doi:10.1073/pnas.97.13.7458
Shoji, M., Takemoto, M., Kobayashi, K., Shoji, T., Mori, S., Sagara, J. I., et al. (2018). Serum podocalyxin levels correlate with carotid intima media thickness, implicating its role as a novel biomarker for atherosclerosis. Sci. Rep. 8 (1), 245. doi:10.1038/s41598-017-18647-6
Slenders, L., Landsmeer, L. P. L., Cui, K., Depuydt, M. A. C., Verwer, M., Mekke, J., et al. (2021). Intersecting single-cell transcriptomics and genome-wide association studies identifies crucial cell populations and candidate genes for atherosclerosis. Eur. heart J. open 2 (1), oeab043. doi:10.1093/EHJOPEN/OEAB043
Sorokin, V., Vickneson, K., Kofidis, T., Woo, C. C., Lin, X. Y., Foo, R., et al. (2020). Role of vascular smooth muscle cell plasticity and interactions in vessel wall inflammation. Front. Immunol. 11, 599415. doi:10.3389/FIMMU.2020.599415
Usuba, R., Pauty, J., Soncin, F., and Matsunaga, Y. T. (2019). EGFL7 regulates sprouting angiogenesis and endothelial integrity in a human blood vessel model. Biomaterials 197, 305–316. doi:10.1016/j.biomaterials.2019.01.022
Van Kuijk, K., Kuppe, C., Betsholtz, C., Vanlandewijck, M., Kramann, R., and Sluimer, J. C. (2019). Heterogeneity and plasticity in healthy and atherosclerotic vasculature explored by single-cell sequencing. Cardiovasc. Res. 115 (12), 1705–1715. doi:10.1093/CVR/CVZ185
Vicen, M., Vitverova, B., Havelek, R., Blazickova, K., Machacek, M., Rathouska, J., et al. (2019). Regulation and role of endoglin in cholesterol-induced endothelial and vascular dysfunction in vivo and in vitro. FASEB J. 33 (5), 6099–6114. doi:10.1096/fj.201802245R
Vieira, J. M., Norman, S., Villa Del Campo, C., Cahill, T. J., Barnette, D. N., Gunadasa-Rohling, M., et al. (2018). The cardiac lymphatic system stimulates resolution of inflammation following myocardial infarction. J. Clin. investigation 128 (8), 3402–3412. doi:10.1172/JCI97192
Weinstock, P. H., Bisgaier, C. L., Aalto-Setala, K., Radner, H., Ramakrishnan, R., Levak-Frank, S., et al. (1995). Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J. Clin. investigation 96 (6), 2555–2568. doi:10.1172/JCI118319
Werner, N., Wassmann, S., Ahlers, P., Kosiol, S., and Nickenig, G. (2006). Circulating CD31+/annexin V+ apoptotic microparticles correlate with coronary endothelial function in patients with coronary artery disease. Arteriosclerosis, Thrombosis, Vasc. Biol. 26 (1), 112–116. doi:10.1161/01.ATV.0000191634.13057.15
Williams, D., Mahmoud, M., Liu, R., Andueza, A., Kumar, S., Kang, D. W., et al. (2022). Stable flow-induced expression of KLK10 inhibits endothelial inflammation and atherosclerosis. eLife 11, e72579. doi:10.7554/ELIFE.72579
Yan, J., Ralston, M. M., Meng, X., Bongiovanni, K. D., Jones, A. L., Benndorf, R., et al. (2013). Glutathione reductase is essential for host defense against bacterial infection. Free Radic. Biol. Med. 61, 320–332. doi:10.1016/j.freeradbiomed.2013.04.015
Zhao, B., Wang, D., Liu, Y., Zhang, X., Wan, Z., Wang, J., et al. (2020). Six-gene signature associated with immune cells in the progression of atherosclerosis discovered by comprehensive bioinformatics analyses. Cardiovasc. Ther. 2020, 1230513. doi:10.1155/2020/1230513
Zhao, G., Lu, H., Liu, Y., Zhao, Y., Zhu, T., Garcia-Barrio, M. T., et al. (2021). Single-cell transcriptomics reveals endothelial plasticity during diabetic atherogenesis. Front. Cell Dev. Biol. 9, 689469. doi:10.3389/FCELL.2021.689469
Keywords: single-cell RNA sequencing (scRNAseq), western diet (WD), cellular heterogeneity, endothelial dysfunction (ED), atherosclerosis
Citation: Wen J, Ling R, Chen R, Zhang S, Dai Y, Zhang T, Guo F, Wang Q, Wang G and Jiang Y (2023) Diversity of arterial cell and phenotypic heterogeneity induced by high-fat and high-cholesterol diet. Front. Cell Dev. Biol. 11:971091. doi: 10.3389/fcell.2023.971091
Received: 16 June 2022; Accepted: 03 February 2023;
Published: 23 February 2023.
Edited by:
Jue Fan, Singleron Biotechnologies, ChinaReviewed by:
Laeticia Lichtenstein, University of Leeds, United KingdomCopyright © 2023 Wen, Ling, Chen, Zhang, Dai, Zhang, Guo, Wang, Wang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yizhou Jiang, amlhbmd5ekBzenUuZWR1LmNu
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.