
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 29 January 2024
Sec. Embryonic Development
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1327227
 Pranav S. Ramesh1,2,3
Pranav S. Ramesh1,2,3 Li-Fang Chu1,2,3*
Li-Fang Chu1,2,3*Somitogenesis is a hallmark feature of all vertebrates and some invertebrate species that involves the periodic formation of block-like structures called somites. Somites are transient embryonic segments that eventually establish the entire vertebral column. A highly conserved molecular oscillator called the segmentation clock underlies this periodic event and the pace of this clock regulates the pace of somite formation. Although conserved signaling pathways govern the clock in most vertebrates, the mechanisms underlying the species-specific divergence in various clock characteristics remain elusive. For example, the segmentation clock in classical model species such as zebrafish, chick, and mouse embryos tick with a periodicity of ∼30, ∼90, and ∼120 min respectively. This enables them to form the species-specific number of vertebrae during their overall timespan of somitogenesis. Here, we perform a systematic review of the species-specific features of the segmentation clock with a keen focus on mouse embryos. We perform this review using three different perspectives: Notch-responsive clock genes, ligand-receptor dynamics, and synchronization between neighboring oscillators. We further review reports that use non-classical model organisms and in vitro model systems that complement our current understanding of the segmentation clock. Our review highlights the importance of comparative developmental biology to further our understanding of this essential developmental process.
The canonical Notch signaling pathway is characterized by the activation of receptors on signal-receiving cells by ligands on signal-sending cells (Figure 1; Reviewed extensively in Kopan and Ilagan, 2009; Siebel and Lendahl, 2017; Zhou et al., 2022). In principle, the interacting extracellular domain of Notch ligands consists of a di-sulfide-rich DSL (Delta in mammals; Serrate in Drosophila; Lag-2 in C. elegans) domain and multiple tandem Epidermal Growth Factor (EGF) repeats. Notch receptors consist of a Notch Extracellular Domain (NECD), a Transmembrane Domain (TM), and a Notch Intracellular Domain (NICD) that are processed and fused in the endoplasmic reticulum and Golgi. Upon ligand-receptor binding, endocytosis is initiated in the signal-sending cell that exposes a characteristic S2 cleavage site in the NECD to the ADAM (A Disintegrin and Metalloprotease) family of proteases in the extra-cellular matrix (ECM). Subsequently, an S3 cleavage in the signal-receiving cell by the γ-secretase complex composed of Presenilin (Psen) 1/2, Nicastrin, APH1, and Presenilin enhancer (Psenen) releases the NICD to translocate into the nucleus. Here, NICD binds to a DNA-binding CSL complex (CBF1/RBPj in mammals, Suppressor of Hairless in Drosophila, Lag-1 in C. elegans) and a transcriptional coactivator mastermind-like (Maml 1–3) to initiate transcription of Notch target genes. In the absence of NICD, the CSL complex acts as a transcriptional repressor. Alternatively, a Notch ligand can bind the receptor intracellularly and prevent its trafficking to the cell surface thereby inhibiting the activation of this pathway (Figure 1). Both the activation and inhibition of this pathway are necessary for context-dependent biological processes (Reviewed extensively in Henrique and Schweisguth, 2019).
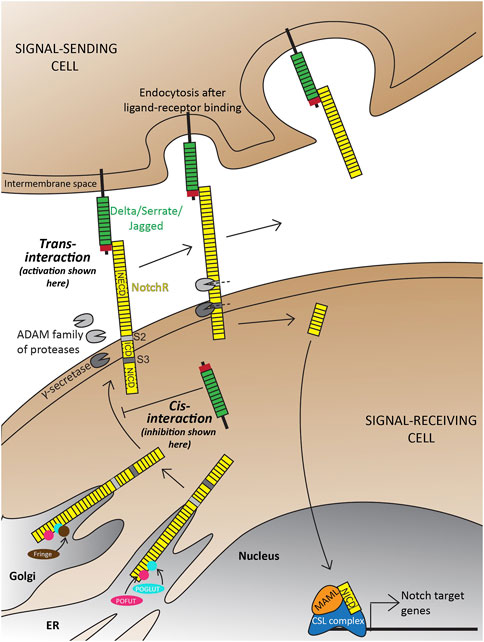
FIGURE 1. Overview of the Delta-Notch signaling pathway depicting trans-activation and cis-inhibition. Trans-activation is initiated when the DSL domain of the Delta/Serrate/Jagged ligands binds to the extracellular domain of Notch receptors. This initiates two crucial cleavages-the S2 cleavage by the ADAM protease; and subsequently an S3 cleavage by gamma-secretase. The two cleavages result in the production of free NICD that translocates into the nucleus to initiate transcription of target genes. Notch receptors, with are also a target of Notch signaling are subject to a wide range of post-translational modifications in the trans ER-Golgi space by Protein O-Fucosyltransferase (POFUT), Protein O-glucosyltransferase (POGLUT), Lunitic fringe (Lfng), etc., which subsequently affect the type of ligand-receptor interactions and downstream signaling. Illustrations adapted and modified from (Zhou et al., 2022).
Although the fundamental components of this pathway remain conserved across different species, the numbers and function of Notch receptors and ligands vary greatly across model organisms due to the differential degrees of genome duplications and loss across species (Table 1).
There are several layers of Notch signaling regulation, which have been extensively reviewed in the past (Kopan and Ilagan, 2009; Henrique and Schweisguth, 2019). Here, we will briefly introduce two, which will be complemented later in the review with a more focused view in the context of species-specificity and early development.
The first layer of regulation is at the level of Notch ligand-receptor interactions, which can happen in trans (ligand and receptors present on different cells; Figure 1), or cis (ligand and receptor on the same cell; Figure 1) (Jacobsen et al., 1998; Artavanis-Tsakonas et al., 1999; Hicks et al., 2000; Sprinzak et al., 2010; Nandagopal et al., 2019), depending on the Notch ligand-receptor pair interacting. For example, in vitro culture studies using immortalized cell lines have shown that Dll1 and Dll4 can act as trans-activating or cis-inhibiting ligands (Andrawes et al., 2013; Preuße et al., 2015), whereas Dll3 has been shown to act as a cis-inhibiting ligand by physically binding to Notch1 and blocking Notch signaling both in vitro in HEK293 cells and in vivo during both X. laevis and mouse neurogenesis (Ladi et al., 2005). Dll4 has been shown to have a strong cis-inhibitory effect on Notch1 receptors which can partially override Dll1-mediated trans-activation in vitro using HeLa cells and Chinese Hamster Ovary cells (Preuße et al., 2015). Another study, which also used immortalized HeLa cells in vitro, showed that Dll3 can act in cis, and potentiate the trans-activating capabilities of Dll1 (Bochter et al., 2022). While trans and cis-regulation have been mechanistically demonstrated, it is still challenging to generalize the activating/inhibiting effects of different ligands observed in the above studies to particular tissue- or species-specific contexts. For example, mutant mice carrying knock-in Dll4 in the endogenous Dll1 locus demonstrated that Dll1 and Dll4 are completely interchangeable during early retinal development, partially interchangeable during myogenesis, and not interchangeable during somitogenesis (Preuße et al., 2015).
The second layer of Notch signaling control arises through post-translational modifications of Notch receptors and ligands (Figure 1). Some of these include o-glucosylation, o-fucosylation, or N-Acetylglucosamination of consensus residues in the EGF repeats that allow a cell to distinguish between different ligand-receptor interactions (Kakuda and Haltiwanger, 2017). For example, o-glucosylation and o-fucosylation of the Notch receptors are carried out by Protein O-Glucosyl Transferase (POGLUT) and Protein O-Fucosyl Transferase (POFUT) respectively in the trans-Golgi network (Shi and Stanley, 2003; Acar et al., 2008). These modifications act as a base for N-Acetylglucosamination imparted by Notch signaling targets called the Fringe proteins which modulate the receptor’s ability to respond to ligands like Dll1 or Jag1 (Kakuda and Haltiwanger, 2017; Bochter et al., 2022). Seemingly, both Dll1 and Dll3 also have these conserved residues, making them amenable to these modifications (Bochter et al., 2022). Expression of one of the Fringe proteins- Lunatic Fringe (Lfng) in a signal-sending cell (containing the trans-activating ligand Dll1) has been shown to attenuate Notch-activation in vitro using HEK293T and NIH3T3 cells (Bochter et al., 2022). In vitro studies using immortalized cell lines have also shown that Lfng can potentiate Notch activation by Dll1, but inhibit activation by Jag1 (Brückner et al., 2000; Yang et al., 2005; Kakuda and Haltiwanger, 2017). This is intriguingly in contrast to in vivo data during somitogenesis between E8.75-E11.5, where lfng (the only Fringe gene expressed during mouse somitogenesis (Johnston et al., 1997)) can post-translationally inhibit NICD formation from the Notch1 receptor, thereby generating a cyclic NICD profile (Morimoto et al., 2005; Shifley et al., 2008). These results suggest that post-translational modifications imparted by Lfng on Notch receptors and ligands can be lineage-specific, and their consequences can be highly context-specific, for example, if Lfng is present in the signal-sending/receiving cell.
Notch signaling, and these different modes of regulation play crucial roles during animal development, but more importantly, are crucial for proper somitogenesis (Reviewed in Hubaud and Pourquié, 2014; Oates et al., 2012). In the following sections, we will review this developmental process, its history, and the complexity it involves at both the tissue and cellular levels.
The process of somitogenesis is an early developmental event occurring between the human embryo Carnegie stage 9 to stage 13 or within the first month post-fertilization (Orts-Llorca, 1981; O’Rahilly and Müller, 2001; O’Rahilly and Müller, 2010). However, knowledge about this process in human embryos has been largely lacking and most of what we have learned to date has been through classical model organisms like chick, mouse, and zebrafish. Most recently however, single-cell RNA sequencing data from early human gastrulating embryos has helped us infer critical information about human somite formation (Xiang et al., 2019; Tyser et al., 2021; Zeng et al., 2023).
Somitogenesis involves the sequential formation of transient, block-like structures called somites that eventually give rise to the bones, skeletal muscles, and dermis associated with the vertebrae. Somites differentiate from the presomitic mesoderm (PSM) tissue that spans along the anterior-posterior axis of a developing embryo. In this tissue, somites bud off of the anterior PSM, while new mesoderm cells are being generated by the mesoderm progenitors at the posterior end (or the tail bud) (Chalamalasetty et al., 2014; Row et al., 2016). The rates of these two processes at opposing ends of the PSM regulate the shrinkage of the tissue over time. Once the tissue is exhausted, somitogenesis is terminated, resulting in a species-specific duration of somitogenesis and somite numbers (Yoshikawa et al., 1997; Yoon and Wold, 2000; Ciruna and Rossant, 2001; Fior et al., 2012).
Pioneering work regarding the mechanisms underlying somitogenesis used chick as a model system with crucial speculations made regarding the origin of somites. One of the many views postulated the existence of a somitic ‘pre-pattern’ in the primitive streak that gets translated into the somite series produced by the PSM (Pasteels, 1937; Waddington, 1952). These views were backed by fate-mapping techniques in both chick and mouse that suggested the presence of stem-like cells in the primitive streak whereby their position along the A-P axis defined the future organization of the somitic tissue (Selleck and Stern, 1991; Psychoyos and Stern, 1996; Wilson and Beddington, 1996; Eloy-Trinquet and Nicolas, 2002).
More evidence for the existence of a pre-pattern came when a part of the chick PSM, if surgically inverted even as early as Hamburger-Hamilton (HH) stage 2 (15–20 somite stage), formed somites in an opposite posterior to anterior fashion (Figure 2A; Christ et al., 1974; Palmeirim et al., 1998; Menkes and Sandor, 1977). Additionally, when approximately one-fourth of a chick embryo at the gastrula stage was excised, or mutant mouse embryos with a shorter axis were analyzed around E10.5, the number of somites formed remained the same, with each new somite also being shorter containing fewer cells (Waddington and Deuchar, 1953; Flint et al., 1978). Landmark experiments performed using chick and snapping turtle embryos, where variable lengths of the segmental plate were excised and cultured in vitro showed that the number of somites formed from the intact segmental plate in the embryo and the explant, combined, always remained constant-about 10–12 somites (Figure 2B; Packard, 1978; Packard, 1980). Important work from Tam and others in the 1980s showed similar compensatory associations in mouse embryos. When one of the cells at the 2-cell blastula stage was surgically removed, or embryos were treated with DNA replication inhibitors resulting in smaller, half-sized embryos, but having the conserved number of somites (Figure 2C; Tam, 1981). Similar results have been reported in small-sized zebrafish embryos, which show a smaller somite size such that the somite number remains unchanged (Ishimatsu et al., 2018). Another recent study has also re-enforced the view of a pre-pattern by performing grafts of the primitive streak (PS) from older (HH8) to younger (HH4) chick embryos (Busby et al., 2023). While GFP+ grafts from an HH4 PS to an HH4 PS appeared in more anterior (first formed) somites, grafts from an HH8 PS transplanted into HH4 only populated the posterior (later formed) somites. This revealed that the progenitor addition to the PSM is regulated by an intrinsic timer or a definitive prepattern (Busby et al., 2023).
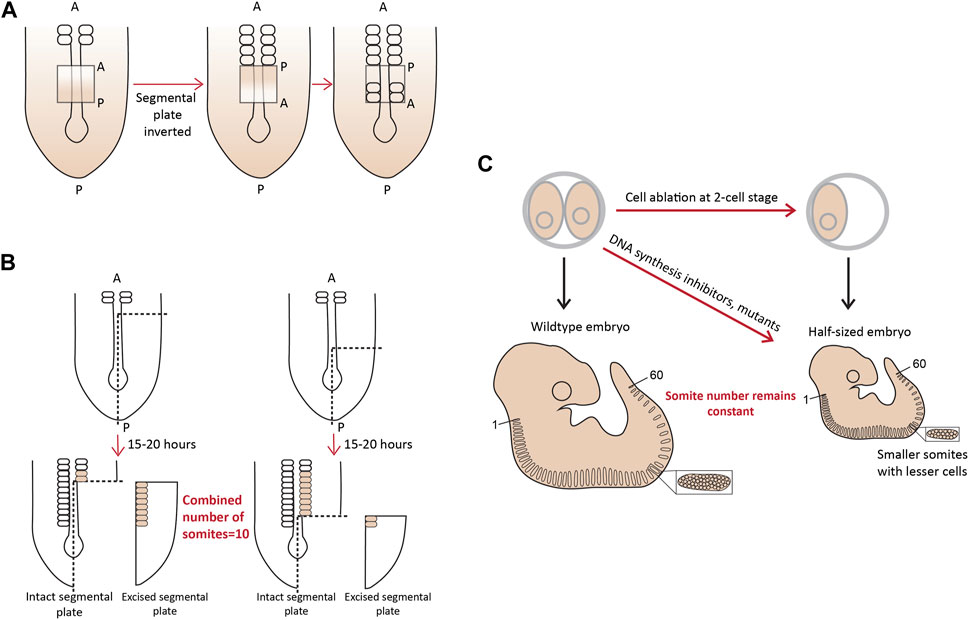
FIGURE 2. A molecular prepattern underlies the specification of PSM cells from the tail bud (A). Inversion of the unsegmented chick PSM results in this tissue forming somites in the opposite direction (B). An explant of chick segmental plate will form 10 somites irrespective of the site/timing of cutting (C). A smaller mouse embryo will form shorter somites, but maintain the mouse-specific somite number. Illustrations adapted and modified from (Pourquié, 2004).
These studies showed and strongly corroborated the existence of a molecular pre-pattern that regulates the metameric organization program of somites. To explain its molecular basis, Cooke and Zeeman in 1976 theoretically postulated the presence of the classical ‘Clock-and-Wavefront’ model (Cooke and Zeeman, 1976; Reviewed in Resende et al., 2014). It predicted that the formation of repeated patterns with high precision is possible if the developing tissue consists of a “clock” made up of individual phase-locked cellular oscillators that interact with a slowly regressing front generated by a signaling gradient to change the cell state (Figure 3). It was not until 1997 when the first experimental evidence for the clock surfaced in the chick embryo, and was termed the ‘Segmentation clock’ (Palmeirim et al., 1997). Notch targets c-hairy1/2 which belong to the Hairy/enhancer of split (Hes/her/Hey) group of basic-helix-loop-helix (bHLH) transcription factors (TFs) were shown to oscillate in the PSM. These oscillations occurred in a spatiotemporally coordinated manner resulting in “traveling” waves sweeping in a posterior-to-anterior direction once for every somite with a periodicity of 90 min (Palmeirim et al., 1997; Pourquié and Tam, 2001), matching the time taken to form one somite in chick, a process that begins at around HH stage 7. Oscillatory Hes/her/Hey genes are also present in mice and zebrafish that make up the clock during somitogenesis (Table 2) (Reviewed in detail in Kageyama et al., 2007; Maroto et al., 2008). These genes have orthologous and paralogous relationships to one another given the large number of gene duplications in zebrafish (Figure 4; Taylor et al., 2001), and are critical to the segmentation clock to varying degrees as we will discuss later below. Readers interested in a detailed analysis of these orthologous relationships and their evolution can refer to (Zhou et al., 2012; Kuretani et al., 2021).
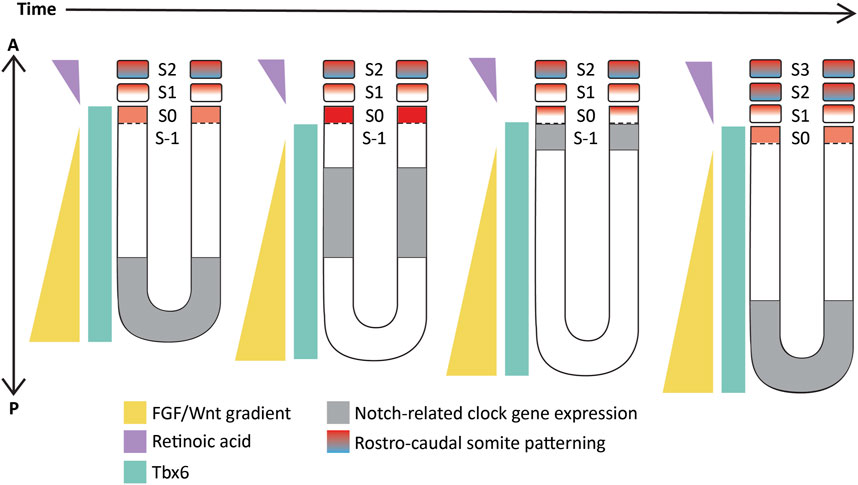
FIGURE 3. Schematic representation of the ‘Clock-and-wavefront’ model that shows the existence of a clock travelling from the posterior to the anterior PSM (in grey) coupled with a regressing wavefront (in yellow) which interact precisely with another T-box TF, Tbx6 (in cyan) to specify anterior PSM cells to differentiate into a somite. S-1: unsegmented PSM that will form the next somite; S0- The somite that is being specified; S1- The most recently formed somite; S2- The earliest formed somite that has completed rostro-caudal patterning (blue-red gradient). Illustrations adapted and modified from (Pourquié, 2011).
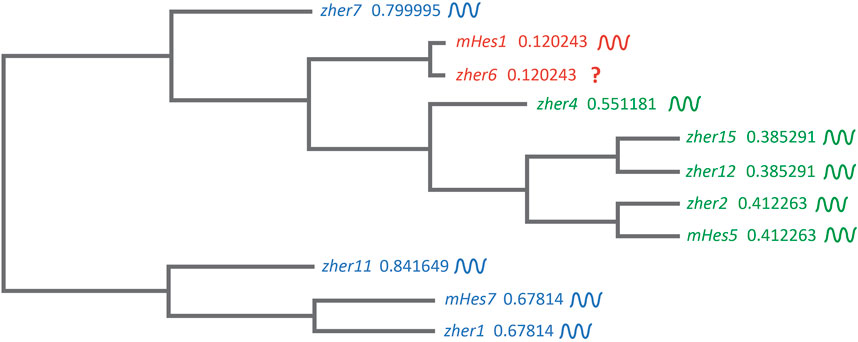
FIGURE 4. Clustal-based phylogenetic tree of mice (m) and zebrafish (z) Hes1/5/7 orthologs. Amino acid sequences of mHes7 (NP_149030.2), mHes5 (NP_034549.1), mHes1 (P35428) zher1 (Q90463), zher7 (Q9I9K1), zher11 (Q6W4T8), zher2 (Q90464), zher4 (Q90466), zher12 (Q6TA36), zher15 (Q7T3J0), and zher6 (Q6PBX3) were first aligned in the FASTA format using MUltiple Sequence Comparison by Log- Expectation (MUSCLE). The FASTA alignment file was input into the EMBL-EBI Simple Phylogeny tool. The tree was generated in a Clustal format with distance corrections and a UPGMA (Unweighted Pair Group Method with Arithmetic Mean) clustering method. The phylogenetic tree depicts the separation of the Hes1, Hes5, and Hes7 clusters. The symbol beside each gene shows their expression dynamics in the PSM, which is unclear specifically for her6 as it is expressed only during the 3-5 somite stage during fish segmentation.
Despite the elegance of the clock-and-wavefront model in explaining pattern formation during somitogenesis, certain properties of the segmentation clock make this process more complex than this proposed model.
Classically, the clock-and-wavefront model suggests the presence of regressing FGF/WNT gradients in the PSM that act as the ‘determination front’ determine the somite borders and the pace of somitogenesis (Figure 3; Sawada et al., 2001; Naiche et al., 2011; Akiyama et al., 2014). However, many studies have shown oscillating FGF and Wnt signaling targets in the caudal part of the mouse E8.5-10.5 PSM such that Notch and FGF targets oscillate in-phase, and out-of-phase of Wnt targets (Figure 5A; Dequéant et al., 2006; Hayashi et al., 2009). Furthermore, Notch and Wnt targets oscillate in-phase in the rostral E10.5 PSM when grown ex vivo as 2D cultures and disrupting this spatial relationship between the two pathways can disrupt somitogenesis (Sonnen et al., 2018). In addition, Hes7 has been shown to regulate the expression of FGF and Wnt target genes like Dusp6, Sprouty2, Axin2, and Snai1 during somitogenesis. For example, studies showed that Hes7 represses the expression of FGF targets, Sprouty2 and Dusp4, in the E10.5 mouse PSM (Niwa et al., 2007; Hayashi et al., 2009). One study showed that disrupting Hes7 cycling by changing gene length disrupted Axin2 and Dusp6 cyclic expression in the E10.5 PSM (Takashima et al., 2011). On the contrary, another study showed that Hes7−/− embryos still display dynamic Wnt (Axin2) and FGF (Sprouty2) target gene oscillations (Ferjentsik et al., 2009). This raises the question as to whether or not Wnt and FGF signaling oscillate or slowly regress as a gradient, or both (Figure 5A). Therefore, other models like the ‘two-phase model’ or the ‘phase-shift model’ have also been suggested that incorporate an interacting Notch, FGF/Erk, and Wnt oscillator (Reviewed in detail in Hubaud and Pourquié, 2014). Whether these expression patterns co-exist during the segmentation clock or are mutually exclusive remains to be resolved (Aulehla et al., 2003). It is possible to speculate that the wavefront is oscillatory under the control of Hes genes in the middle and anterior PSM but acts as a gradient in the more posterior tissue.
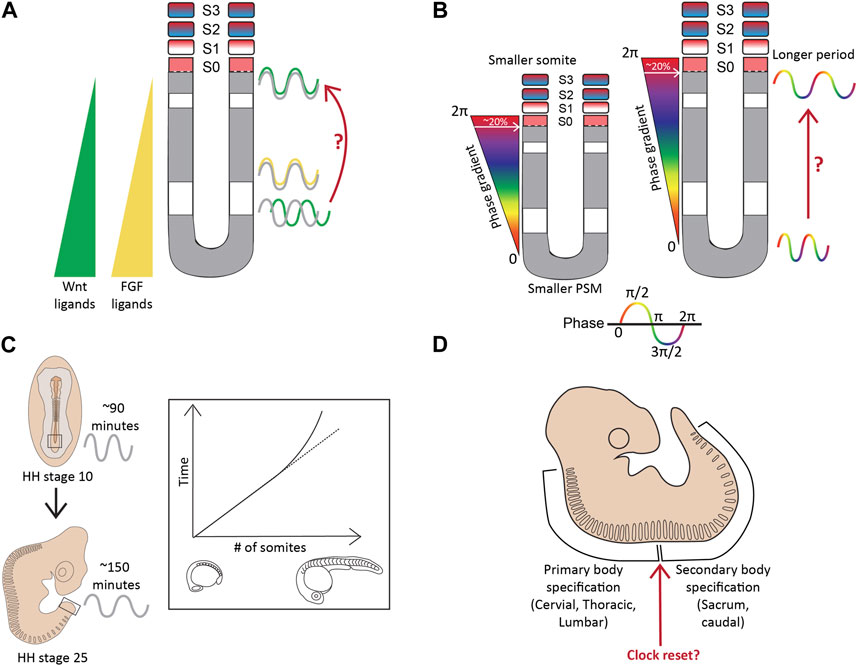
FIGURE 5. Complexities beyond the clock and wavefront (A). The regressing Wnt/FGF-based gradients from the posterior to anterior PSM specify the determination front, however, targets of these signaling pathways (green-Wnt; yellow-FGF) oscillate in the PSM, with changing phase relationships to Notch signaling targets (grey) (B). The period and the phase of cyclic gene oscillations change along the length of the PSM (C). The period of the segmentation clock changes temporally as somitogenesis proceeds (D). The anterior and posterior body segmentation are plausibly regulated by separate mechanisms. Illustrations adapted and modified from (Schröter et al., 2008; Lauschke et al., 2012).
More importantly, a recent study on the zebrafish posterior PSM showed that, unlike mice, Notch-target her7 and Erk signaling oscillate out of phase (Figure 6A; Simsek et al., 2023). This species-specific difference adds an additional layer of complexity to uncovering the mechanisms underlying these phase relationships between the different signaling pathways.
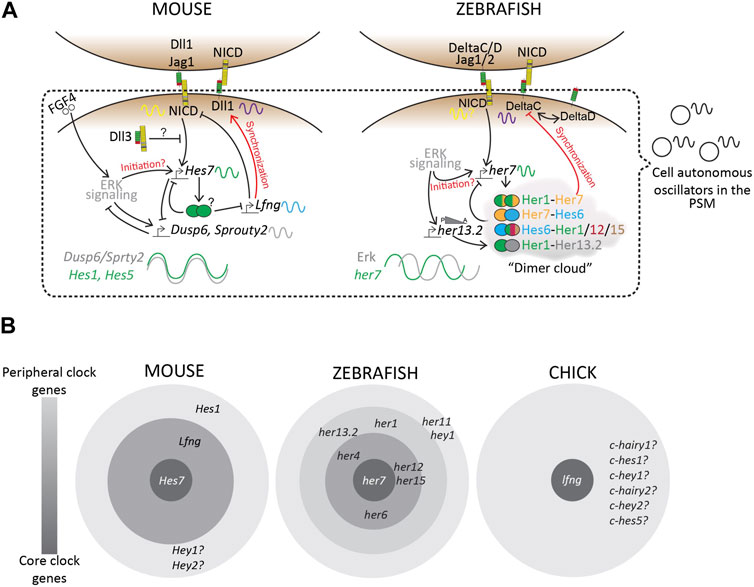
FIGURE 6. Transcriptional landscape of the segmentation clock. (A) A simplified schematic of the complex regulatory landscape underlying the mouse and zebrafish segmentation clocks. (B) “Core” and “Peripheral” bHLH genes and Lfng, amongst many others, occupy this regulatory landscape. Based on mutant phenotypes, these clock genes regulate the segmentation clock to varying degrees. Zebrafish consists of many more her/hey genes as a part of its segmentation clock, as compared to mouse or chick, plausibly making the zebrafish clock more robust to genetic or environmental perturbations compared to other species. Finally, we know very little about similar core vs. peripheral clock genes in other species including higher vertebrates like humans, resulting in a knowledge gap that needs to be studied to fully understand this biological clock.
Another deviation from the classic clock-and-wavefront model, that emerged in the decade following its discovery was that the clock is not constant, but rather highly dynamic at both the spatial and temporal axes.
Firstly, the period of the segmentation clock varies along the A-P axis of the PSM, slowing down in the anterior PSM, thereby generating waves of cyclic gene expression (Figure 5B). This phenomenon is apparent in the mouse and zebrafish PSM (Soroldoni et al., 2014; Shih et al., 2015; Liao et al., 2016; Tsiairis and Aulehla, 2016; Liao and Oates, 2017). In a 10–12 somite-stage zebrafish embryo, the her1 clock in the posterior PSM oscillates with a 1-somite periodicity (30 min), whereas with a slower 2-somite periodicity (∼60 min) in the anterior PSM (Shankaran et al., 2007; Shih et al., 2015). In mice, the period increases from ∼130 min in the posterior PSM to ∼170 min in the anterior PSM as assayed by oscillations of another clock gene Lfng in E10.5 2D PSM explants (Tsiairis and Aulehla, 2016). Alongside a period gradient, there also exists a ‘phase gradient’ such that somite formation in the anterior PSM always coincides with a 2π phase-shift of the clock compared to the posterior PSM (Lauschke et al., 2012). Essentially, this means that the cells in the anterior PSM are one cycle apart from the cells present posteriorly, reiterating the passing of one cycle for every somite formed. Additionally, a newly formed somite always spans ∼20% of this phase-gradient irrespective of PSM length (Figure 5B; Lauschke et al., 2012). In other words, a smaller PSM would form a “steeper” phase gradient to maintain this 2π phase shift, and as a result, 20% of this gradient results in smaller segments (Figure 5B). This presents a plausible mechanism underlying smaller-sized embryos forming the same number of (and smaller) somites.
One potential consequence of such a period/phase gradient has been hypothesized based on the existence of a “secondary” non-changing oscillator along the length of the PSM (Goodwin and Cohen, 1969; Lauschke et al., 2012). The phase of the segmentation clock changes as cells move along the PSM, and its relative phase difference with this secondary oscillator can help give cells the necessary spatial (alongside temporal) information to differentiate into a somite. One potential candidate for this secondary oscillator in mice is Wnt signaling. As mentioned in the previous sub-section, disrupting the phase relationship between Notch and Wnt oscillations results in disorganized somitogenesis. In zebrafish, one candidate is her12 whose phase relationship with her1 also changes along the PSM (Shankaran et al., 2007). However, whether or not these candidate oscillators also display a period/phase gradient along the PSM needs to be systematically characterized.
But what is the cause underlying this gradient? Chemical-based reduction in the levels of Wnt signaling in both mouse and chick PSM slows down the clock, forming one fewer somite compared to the untreated controls (Gibb et al., 2009). The Wnt3a ligand exists as a gradient in the PSM (Aulehla et al., 2003; 2008), and as a result, reducing levels of Wnt signaling along the length of the PSM could be one potential reason for the period gradient.
Perhaps, understanding the molecular and genetic mechanisms underlying the period gradient within a particular species could help us better understand how and why the clock time differs across different species.
The period of the clock also changes as segmentation proceeds, remaining rather constant for most of the duration of somite formation, but drastically slowing down by ∼1.5-fold for the last few somites in both chick and zebrafish (Figure 5C; Schröter et al., 2008; Gibb et al., 2009; Tenin et al., 2010). This layer of complexity has been less characterized during mouse segmentation. The time taken for somite formation in chick embryos between HH22-HH24 decelerates from 90 min to ∼150 min, and this slowing correlates with when Wnt3a levels change from a gradient in the PSM to being confined to the tip of the tail bud to finally disappearing (Gibb et al., 2009; Tenin et al., 2010). During this time, most if not all Notch activity is also lost in the PSM and the clock terminates before all somites are formed at HH25, suggesting a clock-independent regulation of somite formation towards the later stages of development (Tenin et al., 2010). Interestingly, something similar is observed in mouse embryos at E12.5 when all Wnt activity is restricted to the caudal tip and is completely lost at E13.5 before somitogenesis ceases at E14.5, suggesting a similar temporal alteration of the clock during mouse somitogenesis (Gibb et al., 2009). These results also suggest the intriguing possibility that the slower mouse clock (compared to chick) is due to the mouse PSM intrinsically expressing lower levels of Wnt ligands/effectors compared to the chick PSM.
Another layer of complexity in the process of somitogenesis arises from the speculations that the mechanisms regulating this process differ during anterior and posterior body specification (Figure 5D). The switch between primary and secondary body segmentation in mice occurs at the lumbosacral junction (Gossler and Tam, 2002).
Both Dll3−/− and Lfng−/− mice have highly disorganized thoracic and lumbar vertebrae, however, Lfng−/− mice can produce several normal sacral vertebrae before forming a disorganized sacrum and a short tail (Stauber et al., 2009; Williams et al., 2014; 2016; Bochter et al., 2022). Dll3−/−; Lfng−/− mice produced no normal sacral vertebrae, suggesting that while Lfng and Dll3 may work together during anterior body specification, they exhibit independent roles posteriorly. Furthermore, the deletion of an enhancer Fringe clock element 1 (FCE1) upstream of Lfng drastically reduced Lfng expression in the caudal E10.5 PSM while maintaining its anterior expression. LfngΔFCE1/ΔFCE1 embryos phenocopied Lfng−/− embryos during anterior segmentation including heavily disorganized and fused somites and missing ribs, but formed normal tail vertebrae (Shifley et al., 2008). qRT-PCR showed, unlike Lfng−/−, LfngΔFCE1/ΔFCE1 embryos still showed very low cyclic Lfng activity (Shifley et al., 2008; Williams et al., 2014). Similarly, while Dll1−/− mouse embryos fail to form somites throughout the body axis (De Angelis et al., 1997), embryos homozygous for a dominant negative allele of Dll1 can segment, but show fused/missing and split somites only during cervical and thoracic segmentation (Cordes et al., 2004). This suggests that while robust, high-level cyclic Lfng or Notch ligand function is crucial for primary body segmentation, this necessity is no longer present posteriorly, indicating a switch in mechanisms underlying the segmentation clock. Finally, FGF4 mouse mutants also show disorganized somite specification only during the 5-6 somite stage (E8.5) but show normal somitogenesis during the 24–26 somite stage (E9.5), corroborating this complexity (Anderson et al., 2020).
As we will also discuss later below, her1−/− zebrafish embryos, or her1-morpholino treated embryos show minor segmentation defects with diffuse somite borders only for the first 3-5 somites, while her7 knockdown resulted in severe somite border defects after a 5-somite delay (Henry et al., 2002). Furthermore, her1−/−;her7−/− embryos do not form somites throughout the embryonic axis (Henry et al., 2002). Inserting her1 and her7 transgenes in single chromosomes (different from the endogenous locations) in these double-knockout embryos resulted in partially rescued posterior, but not anterior segmentation (Keseroglu et al., 2023). This again suggests separate mechanisms regulating somitogenesis along the embryonic axis. These complexities arise as a result of intercommunication between the Notch, Wnt, and FGF signaling pathways.
In the following sections, we review the species-specific Notch signaling dynamics of the segmentation clock with a focus on bHLH-TFs, ligand-receptor functions, and how these dynamics are propagated by Notch-mediated cell synchronization. Readers interested in a detailed review specifically on the zebrafish segmentation clock can refer to (Venzin and Oates, 2020).
The Notch-responsive family of bHLH-TFs are transcriptional repressors that can bind to the E-box or N-box regions of certain promotors and suppress gene expression (Bessho et al., 2001b; Jones, 2004; Chen et al., 2005). Some of these transcription factors also bind to their own promoters to generate molecular oscillations via a negative feedback loop in the PSM (Holley et al., 2002; Bessho et al., 2003; Jones, 2004; Giudicelli et al., 2007). Mutating these binding sites on target promoters prevents this binding in vitro (Chen et al., 2005). Murine orthologs of Notch-responsive bHLH TFs c-hairy (Hes/Hey genes) include Hes1-7, Hey1-2, and HeyL, of which, Hes1, Hes5, Hes7, and Hey2 are expressed cyclically in the E8.5-E10.5 mouse PSM (Del Barco Barrantes et al., 1999; Jouve et al., 2000; Leimeister et al., 2000; Bessho et al., 2001b). Hey1 transcripts can also be detected in the mouse E9.5 embryos by in situ hybridization, but are not oscillatory (Leimeister et al., 1999). However, microarray experiments have shown that Hey1 indeed oscillates in the mouse E9.5 PSM (Krol et al., 2011). Similar to the first described expression patterns in chick embryos, mouse embryos also show autonomous traveling waves of cyclic gene expression sweeping along the PSM with a mouse-specific periodicity of 2 h (Aulehla et al., 2008). In fact, the Hes1 gene has also been shown to cycle with a conserved 2-h periodicity in other cell types like neural progenitor cells ex vivo, fibroblasts, myoblasts, and embryonic stem cells in vitro, suggesting that a species-specific ultradian rhythm of these bHLH-transcription factors is a conserved, vital feature of vertebrate development (Hirata et al., 2002; William et al., 2007; Ochi et al., 2020). This has been reviewed in detail (Carraco et al., 2022).
Interestingly, a high divergence has been observed in vivo and in silico between chick, mouse, and zebrafish when comparing oscillating genes in the PSM (Krol et al., 2011). Specifically, while the Notch, Wnt, and FGF signaling pathway targets oscillate in all three species, the cyclic expression patterns of orthologs of only two genes- Hes1 and Hes5 are conserved (Krol et al., 2011). We speculate that comparisons across more species could reveal additional conserved oscillators. As we will discuss in the following sections, a comparative analysis of the various Hes/her genes reveal diverging core clock genes and their importance in regulating the segmentation clock. We will review, in-depth, the complex regulatory web casted by these transcription factors during somitogenesis with the anticipation of detangling this species-specific divergence.
While Hes genes are cyclically expressed in the mouse PSM, not all of them seem to be essential for maintaining the clock. For example, Hes7−/− E8.5 and E9.5 PSM lose all Notch-responsive cyclic activity, including that of other clock genes like Lfng and Hes1, and show misexpression of somite maturation markers resulting in disfigured fused somites (Table 3; Bessho et al., 2001a; Ferjentsik et al., 2009; Hirata et al., 2004). However, the expression of upstream components like the Delta ligands and Notch receptors is unaffected suggesting the hierarchy of Notch signaling (Bessho et al., 2001b). On the other hand, Hes1−/− mice still show cyclic Lfng activity in the E10.5 PSM and somite formation is unaffected (Jouve et al., 2000). Lfng−/− E9.5-E10.5 PSM also show cyclic Hes7 and Notch activity, but form disfigured somites, a phenotype that is still less severe compared to Hes7−/− mice (Ferjentsik et al., 2009). Conceptually, this puts Hes7 as a core clock gene, Lfng as a peripheral clock gene, and Hes1 as a more peripheral gene (Figure 6B). In fact, the promoter of Lfng consists of N-box sequences to which Hes7 physically binds and represses expression (Chen et al., 2005). Interestingly, DNase-footprinting assays have shown that Hes1 only binds N-box sequences (and not E-boxes) to repress transcription (Sasai et al., 1992). Yet, Hes1 does not seem to affect Lfng transcription, indicating that these Notch signaling-related DNA-protein interactions are also highly lineage-specific. This kind of core vs. peripheral function may arise as a result of functional redundancy where the loss of Hes1 is fully compensated by Hes7. However, what makes one Hes gene a core player over another remains elusive.
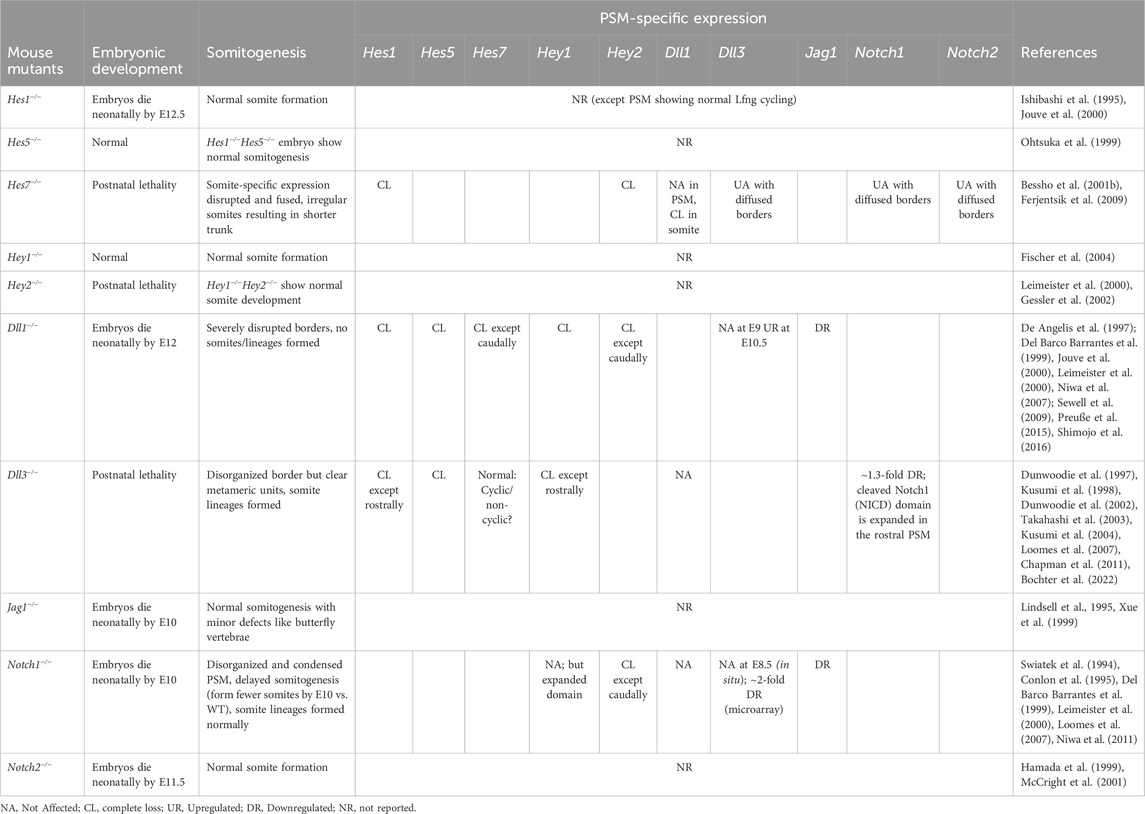
TABLE 3. Mice knockout phenotypes of PSM-specific Hes/her/Hey targets, Notch ligands, and receptors.
Despite this core function of Hes7, somites are formed nonetheless in Hes7−/− mice, albeit disfigured. This suggests the possibility of other core clock genes or the presence of signaling pathways like FGF or Wnt regulating the core clock circuit alongside Notch. Indeed, several Notch loss-of-function (LOF) mutant embryos that lose all cyclic Hes7 activity in the rostral and medial E9.5-E10.5 PSM, still show some caudal Hes7 expression, which is abrogated upon FGF inhibition (Ferjentsik et al., 2009). This suggests that FGF-induced Hes7 expression in the posterior PSM acts independently of Notch and possibly acts as a clock initiator. Whether or not this FGF-driven Hes7 is cyclic needs to be systemically characterized since one study has shown it to be non-cyclic (Ferjentsik et al., 2009), while another showed that this expression is dynamic (Niwa et al., 2007), despite both studies using in situ hybridization of the E9.5 caudal PSM. Furthermore, both Dll1−/− and Notch1−/− PSM at E9.5 show a caudally restricted Hey2, but not Hey1 expression, strengthening the idea of a caudally restricted Notch-independent clock centered around bHLH-transcription factors (Leimeister et al., 2000). More evidence of a peripheral role of Hey1 comes from the fact that its ubiquitous upregulated expression throughout the E9.5 PSM could still form up to ∼13 somites (Feller et al., 2008).
Surprisingly, the functioning of the core gene Hes7 appears to be RBPjk-independent despite its known canonical requirement for Notch target gene expression (Figure 1). RBPjk−/− embryos, which die at E9.5, still show cyclic Hes7 activity in the PSM (Ferjentsik et al., 2009). In agreement, a recent study used live light-sheet imaging of transgenic Lfng reporter carrying mice to study the onset of oscillatory dynamics in E6.5 mouse embryos and showed two interesting observations. First, Lfng shows a huge burst in its expression followed by two to three oscillations before the onset of somite formation. Second, and more importantly, RBPjk−/− embryos also show four to five Lfng oscillations prior to the onset of somitogenesis, but form disrupted somite borders subsequently (Falk et al., 2022). Although Hes7 acts upstream of Lfng and as a result, this burst of Lfng expression does not inform us about Hes7 dynamics, these results suggest an RBPJk-independent Notch activation. On the other hand, mice embryos mutant for Psen1/2, which lack γ-secretase activity do not form any somites (Huppert et al., 2005; Ferjentsik et al., 2009). These results could suggest a non-canonical Notch pathway downstream of Psen1 cleavage diverging before RBPjk function that still activates the core and peripheral clock genes to regulate the mouse segmentation clock.
In zebrafish, due to the high number of gene duplications (Taylor et al., 2001), there are approximately 23 her/hey genes of which 10 are expressed in the PSM (Table 2). Of these, 7 genes-her1, her7, her2, her4, her12, her15, and zhey1 oscillate (Table 2; Holley et al., 2000; Oates and Ho, 2002; Winkler et al., 2003; Sieger et al., 2004; Shankaran et al., 2007; Krol et al., 2011). One exception is her6 which is expressed in the PSM only between the 3 and 5-somite stage embryo, following which its expression is undetectable in the PSM and restricted only to newly formed somites (Pasini et al., 2004). Each of these different her genes are critical for regulating segmentation, but to different extents.
Embryos at the 5–10 somite stages in which her7 expression was disrupted either by morpholino-mediated knockdown or by its overexpression, resulted in a massive downregulation of its own transcription, that of her1 and other clock genes like deltaC, her12, and her15 (Oates and Ho, 2002; Gajewski et al., 2003; Giudicelli et al., 2007; Shankaran et al., 2007). Loss of her7 transcription also resulted in disrupted somite borders and fused somites throughout the embryonic axis after a 5-somite delay (Henry et al., 2002; Giudicelli et al., 2007). Loss of her1 expression by a genomic deletion, morpholino, or overexpression also resulted in a complete loss of its transcription, however, her7 and other cyclic gene transcripts including deltaC, her12, and her15 were attenuated but not fully lost, and were still cycling (Holley et al., 2002; Oates and Ho, 2002; Gajewski et al., 2003; Giudicelli et al., 2007; Shankaran et al., 2007). In agreement, neither her1-deletion nor her1-morpholino induced any severe segmentation phenotypes, other than deformed boundaries and elongated first few somites (Henry et al., 2002; Schröter and Oates, 2010). Furthermore, these defective boundaries were only observed in ∼85% of mutant embryos, suggesting incomplete penetrance which was not observed in her7 mutant embryos (Keseroglu et al., 2023).
These loss-of-function phenotypes suggest that her7 and her1 may behave as a core and peripheral clock gene respectively (Figure 6B), despite the fact that the two genes are genomically paired, are co-expressed, and show overlapping expression patterns in the PSM (Gajewski et al., 2003; Zinani et al., 2020). In addition, a combined loss of her1 and her7 led to a higher penetrance of the somitic defects by disrupting somite borders along the whole embryonic axis including the first few somites indicating a complementary function or redundancy between the two genes (Henry et al., 2002; Oates and Ho, 2002; Sieger et al., 2004).
Unlike her1 or her7, another bHLH-TF her13.2 (hes6 homolog) is independent of Delta-Notch signaling and under the direct control of FGF signaling. Due to this, her13.2 does not oscillate in the PSM, but is expressed in a posterior-to-anterior gradient (Kawamura et al., 2005). Potentially as a result of this differential expression, a complete loss of her13.2, either by a genomic deletion or morpholino-mediated knockdown did not result in any severe segmentation phenotypes, and instead slowed down the clock, resulting in fewer (six to seven fewer), but bigger somites (Schröter and Oates, 2010). Interestingly, the addition of a her1-morpholino in a her13.2−/− background or alongside her13.2-morpholino resulted in severe disruption of segmentation along the entire embryonic axis, indicating a genetic interaction between the two genes (Schröter and Oates, 2010). Morpholino-mediated knockdown of either her4 or her6 alone resulted in a severe disruption of her1 expression in the 10–12 somite stage embryo but showed minor segmentation defects with few somitic fusions (Takke and Campos-Ortega, 1999). This is particularly surprising given that her6 is not expressed in the PSM at this stage, indicating interesting somite-PSM genetic crosstalk which has not yet been seen in mice or chick. Similar to her13.2, double morpholino-knockdown of her1 and her4 resulted in severe defects with multiple fused somites (Takke and Campos-Ortega, 1999).
Four other PSM-specific her genes, her11, her12, her15, and zhey1 are only partially important for the maintenance of the segmentation clock. Misexpression of either her12 or her15 in the 10–12 somite stage embryos disrupted the cycling of clock genes and showed diffused somite borders in only ∼50% of the embryos (Shankaran et al., 2007). This low penetrance could be due to the wide range of homo/heterodimers that the Her proteins form to regulate gene expression (see below). An even lesser effect is seen when using MO-mediated knockdown of her11 or zhey1 at the 8–10 somite stage, where cyclic expression of her1/7 and other cyclic genes like deltaC is unaffected (Sieger et al., 2004).
Despite these diverging roles of individual bHLH-transcription factors, these results provide evidence that the hairy/Hes/her genes control their own expression, as well as that of other oscillating bHLH genes and other clock genes in the segmentation clock (Figures 6A, B; Bessho et al., 2001a; Ferjentsik et al., 2009; Shifley et al., 2008; Zhang and Gridley, 1998). Finally, although Lfng is expressed in the segmented somites, it does not oscillate in the zebrafish PSM (Mayburd et al., 2001; Prince et al., 2001). This emphasizes that, at least in zebrafish and mice, Hes7/her7 genes act as the core clock genes (Figure 6B).
Expression studies in chick have also shown a similar functional divergence between bHLH genes like c-hes1, c-hey1, c-hairy1, c-hey2, c-hairy2, and c-hes5, all of which show different expression pattern in the chick PSM at HH stage 9–10 (Palmeirim et al., 1997; Jouve et al., 2000; Leimeister et al., 2000; Krol et al., 2011).
Contrary to mouse or zebrafish, in chick, c-lfng may act as a core player (Figure 6B). Overexpression of c-lfng leads to a downregulation and loss of c-lfng, c-hairy1, and c-hairy2 cyclic mRNA expression in the PSM, which phenocopied Notch inhibition in the chick embryo (Dale et al., 2003). Additionally, microRNA-125a-5p-mediated post-transcriptional regulation of c-lfng in vivo is essential for chick segmentation suggesting that a tight control of its expression in the PSM is crucial for driving the segmentation clock (Riley et al., 2013). Interestingly, mir-125a-5p is dispensable for the mouse segmentation clock (Wahi et al., 2017), strengthening the view of a more core function of c-lfng in chick compared to the Hes genes in mice. In line with c-lfng being a core clock gene during chick somitogenesis, it also plays a critical role in somite polarization. In the anterior PSM, c-lfng is expressed as a tight band in the presumptive somite border. Mis-regulation of c-lfng expression results in disrupted somite borders and compartmentalization (Dale et al., 2003). Interestingly, when a group of PSM cells at the presumptive somite border were transplanted into an un-segmented PSM region, it induced ectopic fissure and border formation in the transplanted area (Sato et al., 2002). Essentially, border cells contained positional information about border formation which was encoded by a high c-lfng expression. This information was maintained even when transplanted elsewhere in the PSM resulting in ectopic somite borders.
A core function of hes/hairy genes in the chick segmentation clock cannot be ruled out; however, the lack of studies characterizing their roles through RNAi-mediated knockdowns makes it difficult to accurately predict these functions.
There are several features of the mouse segmentation clock that give it its characteristic 120-min period, one of them being delays associated with transcription and protein expression (Jensen et al., 2003; Monk, 2003; Hirata et al., 2004). One of the events that can affect these delays is splicing (Takashima et al., 2011; Alpert et al., 2017).
Introduction of an intronless Hes7 transgene fused to a luciferase showed that the luminescence preceded the endogenous protein expression domain by ∼21 min in E10.5 mouse PSM explants, suggesting a reduced delay in protein expression (Takashima et al., 2011). Surprisingly however, the period of the transgenic Hes7 did not significantly change, possibly due to interference from the endogenous Hes7. Mutant mice containing intronless Hes7 form highly disfigured, fused somites and a truncated body axis phenocopying Hes7−/− mice, indicating the importance of fine delays in maintaining correct patterning (Takashima et al., 2011). In fact, transgenic mice containing intronless Hes7, and not WT Hes7 showed severe segmentation defects resembling Hes7−/− mice (Harima et al., 2013). This suggests that intronic delay, but not copy number per se is essential to maintain proper Hes7 oscillations, and thereby somitogenesis. In contrast, another study reported kinked tails in Hes7+/− mice, indicating that the dose of Hes7, only when it drops below WT levels, could be important for somitogenesis (Bessho et al., 2001b).
Remarkably, introducing a transgene lacking two of the three Hes7 introns did not lead to any severe segmentation defects, but formed additional cervical and upper-thoracic segments, suggesting an acceleration of the clock. Furthermore, the removal of any one intron did not affect the clock or somitogenesis at all (Harima et al., 2013). These results show that while intronic delay is crucial, this criticality is dependent upon the number of introns present in a “dose-dependent” manner.
Once the Hes7 mRNA is spliced and the Hes7 protein is made, it needs to be degraded with rapid turnover to allow for the next cycle of expression to begin. Stabilizing the mouse Hes7 protein by mutating ubiquitination sites on Hes7 prevented its degradation and increased its half-life from 22 min (WT) to ∼30 min when measured in vitro (Hirata et al., 2004). In silico models showed that this small increase in Hes7 half-life severely dampened oscillations after 3-4 cycles. In agreement, knock-in mice containing this mutant Hes7 showed normal somitogenesis and cyclic Hes7 and Lfng expression until the 3-4 somite stage, following which the clock was fully disrupted resulting in defective segmentation (Hirata et al., 2004). These results show that protein turnover rates are central to maintaining proper cyclic gene expression, and therefore proper body segmentation.
In fact, a recent study used a transgenic Hes7 construct to show that the mouse Hes7 has an ∼2-fold faster decay rate compared to the human HES7, which correlates with the ∼2-fold faster mouse clock (Matsuda et al., 2020a). The authors also found that the human HES7 decayed at a mouse-specific rate when the transgene was expressed in the mouse PSM in vitro. Global translation inhibition in Zebrafish embryos at the 12–14-somite stage revealed that the half-life of Her7 is only ∼3.5 min (Giudicelli et al., 2007; Ay et al., 2013), compared to an ∼20 min and ∼40 min half-life of mouse Hes7 and human HES7 respectively (Matsuda et al., 2020a). These Hes/Her protein half-lives correlate with species-specific clock periods. These studies suggest that beyond the gene sequence itself, the surrounding transcriptional, translational, and degradation machinery plays a crucial role in regulating species-specific developmental timing.
However, this correlation may not extend to the chick segmentation clock, where blocking de-novo protein synthesis did not affect the dynamicity of either c-hairy1 or c-hey2 expression. This suggests a different mechanism for cyclic expression in chick (Palmeirim et al., 1997; McGrew et al., 1998; Leimeister et al., 2000). As a result, transcriptional/protein decay rates may only be one of the determinants of species-specific clock periods, and further comparative studies will be necessary to uncover the other ones.
bHLH-TFs like the Hes/her genes have the ability to form homo/hetero-dimerize with other bHLH genes via their helix-loop-helix structures (Jones, 2004). Altering dimerization parameters of Hes7 in silico can alter clock properties in the mouse PSM, however in vivo data for the same is lacking (Song et al., 2011).
In zebrafish however, the dimerization network of bHLH-TFs has been more extensively characterized. her genes form homo- and hetero-dimers in a “dimer cloud” (Figure 6A; Schröter et al., 2012), and this dimerization ability is essential for pattern formation in the fish embryo (Henry et al., 2002; Zinani et al., 2020; Zinani et al., 2022). her1 and her7 are present head-to-head on the same chromosome which enables their co-transcription and co-repression by a Her1/7 dimer. One study mutated the her1 and her7 genes on opposite sister chromatids resulting in reduced correlated gene expression dynamics and disrupted somitogenesis (Zinani et al., 2020). Her7 and Her13.2 also form heterodimers and enable autorepression of the her1 and her7, but also modulate the dimer topology within each cell crucial for the functioning of the clock (Figure 6A; Schröter et al., 2012). Her13.2 also affects the stability of the Her1/7 dimer via protein-protein competition and its loss results in increased stability of this dimer and a slower clock (Schröter et al., 2012). On the other hand, another study reported that Her7 acts as the critical node of the dimer topology and limits the availability of Hes13.2 to dimerize with the other cyclic Her proteins like Her1, Her12, and Her15 (Trofka et al., 2012). Indeed, electrophoretic mobility shift assays (EMSA) have shown that the cis-regulatory elements of her1 contain Her12 binding sites (Brend and Holley, 2009), and that Her12 plays an important role in regulating her1 expression (Shankaran et al., 2007).
Nonetheless, these studies suggest the complex interplay between bHLH-TFs necessary for somitogenesis, a feature not as extensive in mice or chick due to the limited number of Hes/Hey genes in the two species. This brings out an intriguing question- Does an extensive network of bHLH-TFs display redundancy in a way that makes zebrafish somitogenesis more robust to environmental/genetic aberrations? her1 knockout embryos show only ∼85% penetrance in segmentation defects suggesting some functional compensation by other her genes (Keseroglu et al., 2023). More comparative studies using different her mutants and across different species are necessary to address this question.
Upstream players of the Notch signaling pathway like the ligands and receptors play an important role in regulating and synchronizing this complex transcriptional landscape painted by bHLH-TFs, and the loss of these upstream components also disrupts somitogenesis.
The mouse PSM expresses three ligands- Dll1, Dll3, Jag1, and two Notch receptors- Notch1 and Notch2 (Swiatek et al., 1994; Lindsell et al., 1995; Shawber et al., 1996; De Angelis et al., 1997; Dunwoodie et al., 1997; Hamada et al., 1999). These different receptors and ligands play distinct roles during somitogenesis. While Dll1 and Dll3 null embryos show defects in somitogenesis, Jag1−/− embryos do not, except for minor defects like butterfly vertebrae (Table 3). Phenotypes of Dll1−/− and Dll3−/− embryos suggest that the expression of clock genes is independently regulated in the rostral and caudal PSM, and while Dll1 is required in both, Dll3 is only required in the caudal PSM (Del Barco Barrantes et al., 1999; Jouve et al., 2000; Dunwoodie et al., 2002). For example, Dll1−/− mice show a complete loss of Lfng in the E8.5 mouse PSM whereas Dll3−/− E9.5 embryos only lose the caudal Lfng expression (Del Barco Barrantes et al., 1999; Kusumi et al., 2004). This is also consistent with another study showing that the loss of Dll3 is epistatic to the loss of Lfng in E10.5 embryos (Riley et al., 2013; Bochter et al., 2022).
Furthermore, while Dll1−/− E9.5 embryos lose all Hes7 expression except in the caudal PSM, Dll3−/− E9.5 embryos still show normal cyclic Hes7 expression (Kusumi et al., 2004; Niwa et al., 2007; Sewell et al., 2009). Two other studies have challenged this by also using in situ hybridization showing that while Dll3−/− E9 PSM do not lose any Hes7 expression, they still become non-oscillatory (Chen et al., 2005; Bochter et al., 2022). On the other hand, Dll1−/− and Dll3−/− embryos both show complete loss of Hes1 and Hes5 expression (Del Barco Barrantes et al., 1999; Jouve et al., 2000; Dunwoodie et al., 2002).
These data suggest while Dll1 is crucial for regulating the expression of all Hes genes and Lfng, Dll3 is crucial only for Lfng and perhaps more peripheral bHLH gene expression. Potentially due to this “stronger” influence of Dll1, Dll1−/− mice are unable to establish any rostrocaudal compartmentalization, fail to make epithelial somites, show incomplete axis formation, and eventually die by E12 (De Angelis et al., 1997; Preuße et al., 2015; Shimojo et al., 2016). On the other hand, loss of Dll3 results in less severe phenotypes where although somite borders are not visible, clear metameric units are evident, albeit highly disorganized, which eventually also differentiate into different somitic lineages (Dunwoodie et al., 2002). Furthermore, Dll3−/− mice die postnatally within 10 days of birth (Dunwoodie et al., 2002). These phenotypes also reinforce the influence of a loss of core clock genes in Dll1−/− embryos versus the loss of more peripheral genes in Dll3−/− embryos. Interestingly, there are many more documented human DLL3 clinical mutations (versus DLL1) that result in a disorganized vertebral column. To date, there have been 28 reported DLL3 mutations resulting in a form of spondylocostal dysostosis (SCDO), SCDO1, which is a type of vertebral malformation (Kusumi et al., 2004; Umair et al., 2022). On the other hand, there has been only 1 DLL1 mutation causing another type of SCDO, SCDO7 (Barhoumi et al., 2019). One plausible reason for this could be that, similar to mice, mutations in DLL1 (as opposed to DLL3) are likely to be more embryonic lethal in humans, resulting in this disparity in reported cases.
Both Dll1−/− and Notch1−/− E8.5-E9.5 PSM completely lose Hey2 expression except in the caudal PSM (Leimeister et al., 2000; Niwa et al., 2011), but whereas Dll1−/− mice show a complete loss of Hey1, it is unaffected in Notch1−/− embryos (Table 3; Leimeister et al., 2000). This suggests that for Hey2 expression, Dll1 and Notch1 act linearly, but there may be additional ligand-receptor pairs regulating Hey1 expression in the mouse PSM. Additionally, while Dll1−/− PSM lose all Lfng expression and somite polarity, Notch1−/− PSM show only a reduced Lfng expressed with diffused borders (Leimeister et al., 2000). Somite formation is retarded in these embryos which, by E9.5, have only formed 14–16 somitic pairs compared to the 20–25 pairs in WT embryos, and eventually die by E11-11.5 (Swiatek et al., 1994; Conlon et al., 1995). The fact that Notch1−/− embryos can form somites is surprising given that Notch2−/− embryos also do not show any somitogenesis defects, but Psen1/2 −/− embryos show an absence of somites altogether (Hamada et al., 1999; Huppert et al., 2005; Ferjentsik et al., 2009). Notch1 likely serves as the core receptor during somitogenesis. In Notch1−/− embryos, partial compensation by Notch2 allows the retarded formation of 14–16 somites before embryonic death. On the other hand, when Notch2 is knocked out, Notch1 can fully compensate for its function. Alternatively, Psen1/2 may possess a Notch-independent role in regulating somitogenesis.
All of the above studies combined show that a complex, spatiotemporally regulated medley of Notch ligands, receptors, and their targets is of critical importance in regulating the mouse segmentation clock.
In zebrafish, deltaC (beamter; bea) and deltaD (after eight; aei) LOF mutant embryos show different segmentation phenotypes in the PSM, suggesting that they may serve separate roles in regulating the segmentation clock (Holley et al., 2000; Holley et al., 2002; Mara et al., 2007). While deltaD (aei) mutants display defective somitogenesis after 8-9 somites, deltaC (bea) mutants show disruption following just 3-4 somites indicating that, like dll1, deltaC acts as a more core regulator of segmentation (Van Eeden et al., 1996; 1998; Jiang et al., 2000). In fact, aei/bea double mutants show the same segmentation phenotypes as the bea mutants, reflecting the more primary function of deltaC (Jülich et al., 2005). This is in contrast to expression patterns of cyclic genes, where in situ hybridization in bea mutants display a salt-and-pepper expression of her1 and her7 throughout the 8–12 somite stage PSM, whereas her1 expression is completely lost (except in the anterior and posterior tips) in aei mutants (Holley et al., 2000; 2002). This phenotype is partially reminiscent of Hes7 expression in Dll1−/− mouse PSM. In contradiction, one study used high-resolution single-cell imaging and fluorescent in situ hybridization to show that deltaD−/− embryos show no posterior expression of her1 in the 12-somite stage PSM (Mara et al., 2007). As a result, it may be difficult to directly associate ligand similarities between different species based on mutant phenotypes alone. Nonetheless, these studies suggest a more “synchronization" role for DeltaC such that its loss results in a random, salt-and-pepper expression, but a more “driver” role for DeltaD such that its loss results in a complete loss of cyclic gene expression (Holley et al., 2002).
Irrespective of the ligand or receptor function lost, at least a few somite cycles must elapse before the blockade of Notch signaling and severe segmentation defects. This can happen if Notch signaling regulates synchronization primarily, but not the clock directly. Alternatively, it is also possible that Notch signaling in fish is important only for the posterior clock, but not the anterior one, where a different signaling pathway is a primary driver. In fact, a recent study showed that her1−/−;her7−/− fish can still show a proper segmentation profile if provided with exogenous pulses of FGF/Erk signaling inhibition between the 17–22 somite stage (Simsek et al., 2023). These results are complemented by the observation that deltaD;deltaC single and double knockout fish embryos show normal axial development, and are viable and fertile (Jülich et al., 2005), as opposed to ligand knockout mice which are embryonic lethal (Figure 7, Lower panel).
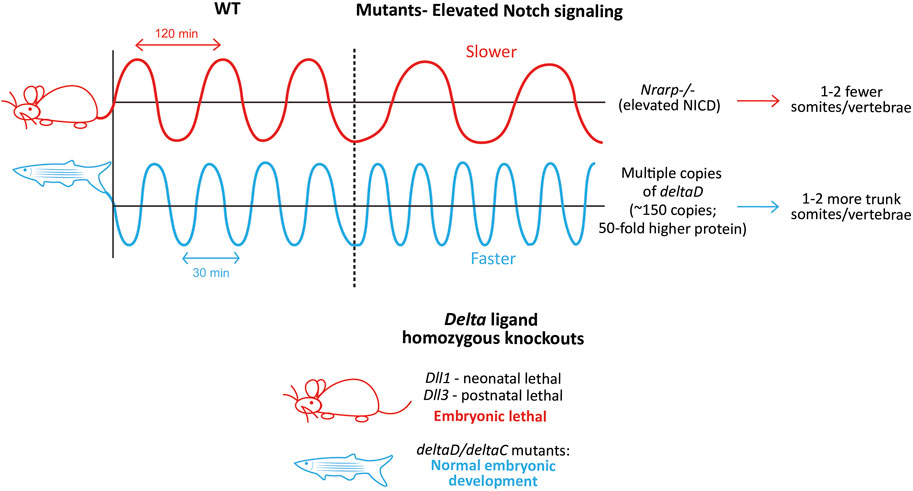
FIGURE 7. Species-specific differences in clock dynamics and Notch mutant phenotypes. Elevated levels of the Notch signaling pathway opposingly affect the mouse and zebrafish clock periods, and as a result, the final number of somites/vertebrae formed (Upper panel). Species-specific differences in the requirement of Delta ligands during embryonic development. Mouse-red; zebrafish-blue (Lower panel)..
While the presence of Notch signaling is necessary for the integrity of the mouse clock, the level of Notch signaling itself can also regulate the clock by changing its properties (Figure 7, Upper panel). For example, mice with a LOF mutation in the Notch regulated ankyrin repeat protein (Nrarp), a cyclic Notch target that represses NICD cleavage in the mouse PSM (Sewell et al., 2009), form ∼2 fewer vertebrae compared to WT mice, and ∼50% of homozygous mutant mice show a kinked tail (Kim et al., 2011). In line with the function of Nrarp, Nrarp−/− E10.5 PSM show ∼1.9-fold higher NICD levels. Furthermore, when WT pregnant mice were treated with LY-411575 (0.1 mg/kg; a γ-secretase inhibitor that inhibits Notch signaling) every 24 h starting at E7.5, NICD levels in E10.5 embryos dropped by 10%–20%, resulting in a slightly faster clock in ∼50% embryos that formed 1 extra somite compared to the untreated controls. Remarkably, the same dose of LY-411575 treatment to mice carrying Nrarp−/− embryos also resulted in a rescue in ∼50% of embryos forming one additional somite compared to the untreated mutants. This suggests that the loss of Nrarp results in an increase in NICD levels, in turn increasing the period. Interestingly however, the cyclic expression of clock genes like Hes7, Lfng, and that of NICD were unaffected in these mutant embryos. As a result, Nrarp may also be a more peripheral component, consistent with studies showing that the loss of Dll3 results in the complete loss of Nrarp expression (Sewell et al., 2009; Bochter et al., 2022). One plausible explanation for the increased period in Nrarp−/− embryos is that higher levels of Lfng may be necessary to build up to effectively repress the higher amount of NICD, which may take significantly more time, hence slowing the clock. However, increasing the half-life of NICD also slowed the clock (Wiedermann et al., 2015). This indicates that not just the levels of NICD, but the time spent by NICD in the cell also affects clock periodicity, suggesting a Lfng-independent mechanism.
The fish clock is also sensitive to the levels of Notch signaling, however in the direction opposite to that of mice. Transgenic zebrafish embryos containing multiple copies of deltaD produced more somites and vertebrae compared to WT embryos (Figure 7, Upper panel; Liao et al., 2016). Interestingly, the authors generate two lines, Dover with ∼10-fold higher DeltaD levels in the homozygotes (copy number ∼15), and Damascus with ∼50-fold higher DeltaD in the heterozygotes (copy number ∼150). While both the homozygous and heterozygous Dover mutants showed normal somitogenesis, none of the homozygous Damascus mutants survive. Surprisingly, the heterozygous Damascus mutants develop normally, and form an overall 2 additional trunk somites compared to WT siblings, with clock periods being ∼1.5 min faster specifically in the trunk region (4–19 somite stage). In contrast, neurogenesis was affected even with the slightest increase in DeltaD levels, suggesting the robustness of somitogenesis to tolerate gene expression changes compared to other lineages.
On the other hand, DAPT treatment (another γ-secretase inhibitor that inhibits Notch signaling) or homozygous aei/des embryos show a slower segmentation clock at the 3-4 somite stage (heterozygotes do not, even though protein levels are halved (Liao et al., 2016)), resulting in larger anterior somites before the onset of segmental defects (Herrgen et al., 2010). In the presence of DAPT, Damascus mutants showed no change in the clock period (Liao et al., 2016). This suggests that while no deltaD copies (aei embryos) confer segmental defects, moderately abundant copies do not affect somitogenesis, or even embryonic development. This could be because deltaD does not oscillate in the fish PSM, and as a result, too much of it does not disrupt its endogenous expression dynamics. Overexpression of Dll1 in mouse embryos can disrupt Dll1 oscillations and thereby somitogenesis (Shimojo et al., 2016). As a result, it is possible that the introduction of multiple copies of deltaC disrupts somitogenesis.
These results reinforce that while in mice, Notch signaling primarily drives the expression of oscillatory genes, in zebrafish, Notch signaling only synchronizes and propagates her1/7 oscillations. This is supported by the ‘delayed coupling theory’ where reduced Notch signaling results in weaker coupling strength and higher delays in information transfer between neighboring PSM cells, thereby resulting in a collective slower clock (Morelli et al., 2009; Herrgen et al., 2010; Liao et al., 2016; Yoshioka-Kobayashi et al., 2020). Due to the lack of genetic tools, similar Notch signaling manipulative studies have not been reported yet in chick embryos. It would be interesting to characterize the effects of in ovo electroporation of c-delta1 or c-notch1/c-NICD cDNA on the chick segmentation clock time.
The role of Notch signaling as a synchronizer in mouse embryos has evolved over many decades, since initially Notch signaling was only considered to be essential to maintain gene oscillations in individual PSM cells, based on Notch ligand and receptor LOF mutant phenotypes. Two studies showed that when mouse E10.5 or chick PSM tissue (14–18 somite stage) is dissected into multiple fragments, they are each able to oscillate maintaining the correct clock schedule compared to an intact tissue (Maroto et al., 2005; Masamizu et al., 2006). This showed that at least at the global level, groups of cells maintained synchrony despite being cultured separately. With the advent of single-cell resolution imaging, the role of Notch signaling as a cell synchronizer alongside being a ‘switch’ to maintain oscillations became easier to study. Masamizu et al., 2006 showed first that dissociated E9.5-10.5 transgenic mouse tailbud cells showed asynchronous and unstable Hes1 oscillations in vitro (Masamizu et al., 2006). Another study culturing dissociated E9.5 transgenic mouse tail bud explants showed similar dampened and weak Lfng oscillations (Hubaud et al., 2017). A similar experiment in 14–18 somite stage chick embryos showed that when the PSM was dissociated and cultured in vitro in separate pools, c-Lfng oscillated asynchronously across pools as assayed by in situ hybridization, suggesting that cell-cell communication is necessary to maintain stable, synchronous oscillations (Maroto et al., 2005). However, was this property Notch-dependent? Using an in vitro culture technique of mouse E10.5 tail bud explants, Tsiairis and Aulehla, (2016) showed that DAPT treatment affected the ability of dissociated and reassembled PSM cells to self-organize into spatiotemporal wave patterns upon reaggregation in vitro (Lauschke et al., 2012; Tsiairis and Aulehla, 2016).
A major obstacle in understanding the role of Notch as a synchronizer in mouse segmentation stems from the fact that Notch signaling directly controls Hes7 or Lfng oscillations throughout the PSM. As a result, the dissociation of the role of Notch as a driver of oscillations and as a synchronizer between oscillators is a challenge. The dampening and instability of Hes7 and Lfng oscillations in Notch pathway LOF mutants or DAPT-treated embryos may be because of reduced expression of these genes rather than due to a loss of synchrony. To combat this, one study used chimeric mouse embryos consisting of WT and Dll1−/− cells and showed that the synchronization rates in the E10.5 PSM reduced linearly with decreasing number of WT cells in the chimeras (Okubo et al., 2012). They also used chimeric mice containing WT and Lfng−/− cells to show that Lfng could regulate Hes7 oscillations non-cell autonomously. Another study also used immortalized cell lines to show that Lfng can repress Dll1 (alongside NICD) cell-autonomously to regulate levels of Notch signaling cell-autonomously (via NICD) and non-autonomously (via Dll1) (Okubo et al., 2012; Bochter et al., 2022). These data suggest a unique role of Lfng in being responsible for maintaining synchronization during somitogenesis (Figure 6A). A recent study supports the these results through single-cell tracking of Hes7-positive and negative cells in Lfng−/− PSM that showed individual oscillators lose synchrony (Yoshioka-Kobayashi et al., 2020).
In zebrafish Notch-LOF embryos, the first few somites develop normally and the progressive worsening of somitogenesis has been attributed to a loss of synchronization between individual oscillators. For example, in a ‘re-synchronization assay’, zebrafish embryos treated with DAPT that start to form disfigured somites after about 5 somites, promptly re-form normal somites after DAPT withdrawal due to the “resynchronization” of the clock (Liao et al., 2016; Uriu et al., 2021). As the tissue recovers following DAPT withdrawal, the defective and normal somites intermingle transiently showing that recovery occurs at multiple levels: At the cellular level with rapid resynchronization between individual cells via Delta-Notch signaling; and the tissue level with the transfer of this resynchronization “information” throughout the PSM (Uriu et al., 2021). deltaD/aei or notch1/des heterozygous mutants fail to resynchronize effectively after DAPT treatment, whereas deltaC/bea mutants recover similarly to WT embryos, suggesting ligand-specific roles in carrying out synchronization (Mara et al., 2007). In contrast, embryos with multiple deltaD copies can resynchronize faster than their WT siblings (Liao et al., 2016).
Similar to mice, advances in single-cell resolution imaging of Her1 oscillations in vivo showed that individual cells oscillate autonomously, which begin cycling out of phase in deltaD (aie), deltaC (beamter), and notch1a (des) embryos (Delaune et al., 2012). A more recent study corroborated this by culturing the caudal PSM cells in vitro at low densities and detecting cell-autonomous, but asynchronous and noisy her1 oscillations (Webb et al., 2016). This high background noise was absent in cells belonging to an intact PSM, suggesting a role for collective tissue-level processes in increasing the precision of the clock.
Interestingly, the suggested mechanism of synchronization in zebrafish is slightly different from mice (Figure 6A). The Her1/7 dimer is predicted to directly repress deltaC expression levels (Horikawa et al., 2006). One study showed that the deltaC expression pattern in her1-LOF fish is only altered in the anterior PSM but not posterior, suggesting that this predicted mechanism of synchronization differs along the embryonic axis (Choorapoikayil et al., 2012). However, this remains to be systematically characterized.
Much of the early knowledge about body segmentation before studies in chick embryos came from arthropods like Drosophila (Davis and Patel, 1999; Pourquie, 2003). However, unlike vertebrates, genetic studies have shown that Notch signaling does not have a link to fly segmentation (Johnston and Nüsslein-Volhard, 1992). Surprisingly, homologs of the genes delta, notch, and hairy all show a segmental expression pattern during early development of another arthropod, the spider Cupiennius salei (Stollewerk et al., 2003). RNAi-mediated knockdown of delta and notch resulted in disorganized hairy segmentation, similar to vertebrates. Similar segmented expression of Notch components and their roles in embryonic segmentation have been observed in other invertebrates like the cockroach Periplanata americana and the centipede Strigamia maritima, but not in the cricket Gryllus bimaculatus (Pueyo et al., 2008; Reviewed extensively in; Kainz et al., 2011; Brena and Akam, 2013; Clark et al., 2019). This suggests that a wider sampling of invertebrate species beyond classical models like Drosophila can reveal otherwise unidentified complex ancestral segmentation mechanisms. As a result, it is essential to expand our horizon on this developmental process by using non-classical in vivo and in vitro model systems, including other non-model vertebrates and potentially even invertebrate species to better understand this process.
Corn snake embryos form 200–300 somites with a timing of ∼100 min per somite (Table 4; Gomez et al., 2008). Interestingly, the house snake which has very similar developmental characteristics to the corn snake, displays a clock of ∼60 min suggesting that small genetic differences within a given genus can drastically affect clock timing (Gomez et al., 2008). This correlation is challenging to verify when only using classical models like mouse and fish, which inherently are very different. Reptiles like the green anole lizard and alligator have also been used to study somitogenesis which unlike traditional model species, do not express Lfng in the PSM. While the corn snake does, it exhibits significantly more stripes in the PSM compared to mice, suggesting a different mode of clock gene regulation (Gomez et al., 2008; Eckalbar et al., 2012). In contrast, similar to classical models, Dll1 displays oscillatory expression in the anole lizard (Eckalbar et al., 2012). Another example is Dll3, while expressed uniformly throughout the mouse PSM, is confined to the caudal-most PSM and displays static anterior bands in the anole lizard. These observations suggest possibly conserved and divergent mechanisms regulating the core versus peripheral clock players respectively in the reptilian segmentation clock.
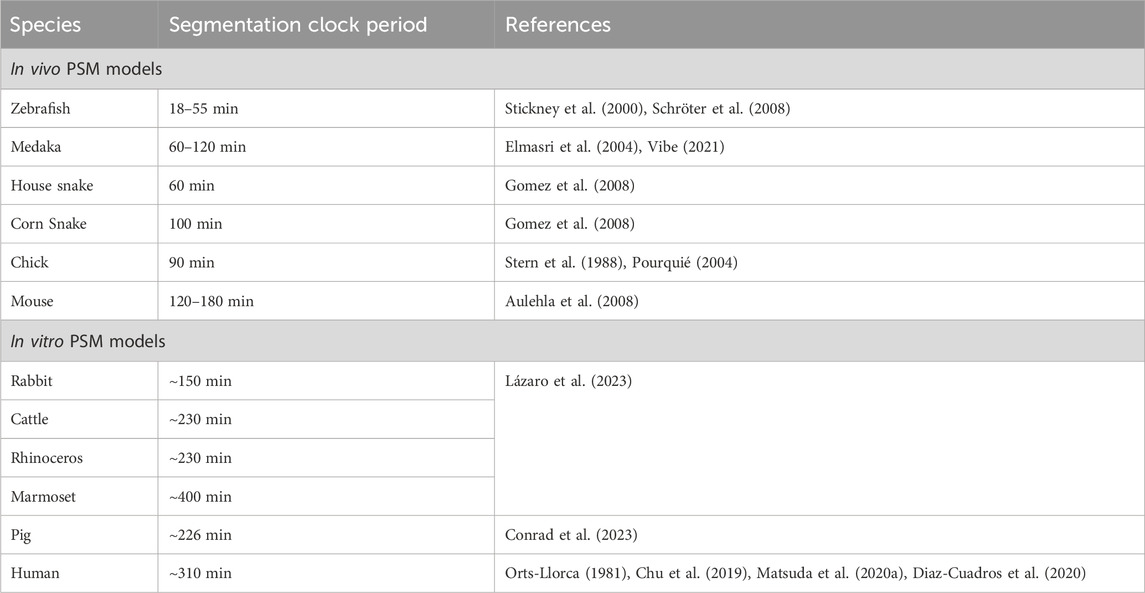
TABLE 4. Species-specific segmentation clock periods across various in vivo and in vitro model systems.
Studies on other teleost fish species like Oryzias latipes (Medaka) and Tagifuku rubripes (Japanese pufferfish) also show some interesting clock divergence from zebrafish (Danio rerio) (Elmasri et al., 2004; Gajewski et al., 2006). Medaka embryos show a clock timing of ∼60 min at 26°C, whereas zebrafish embryos tick approximately two-fold faster at 25–30 min at 26°C suggesting similar intra-species differences like snakes (Table 4; Elmasri et al., 2004; Schröter et al., 2008). In fact, 60 min is the clock speed at the posterior PSM which slows down by almost half to ∼120 min in the anterior PSM of a 3-5 somite-stage medaka embryo (Vibe, 2021). Given that the anterior speed of the medaka embryo (unlike zebrafish embryos) is very similar to the posterior PSM clock in mice, understanding why a period gradient arises in medaka could help decode the underlying mechanisms of species-specific clock times. Genetic knockout phenotypes have also shown that the Medaka her7 is functionally similar to the zebrafish her1 and her7 combined, and the Medaka her1/11 to the zebrafish her11 over her1 (Elmasri et al., 2004; Sieger et al., 2006). These phenotypes further support the idea that teleost embryos use her7 as a core clock driver and her1/11 genes as more peripheral ones.
Another advantage that Medaka has for decoding the genetics underlying species-specific clock times is that different inbred Medaka sub-species show different her7 clock periods (Seleit et al., 2023). They also have variable PSM and somite lengths, which positively correlate with clock periodicity. Crossbreeding species with different periods provides a useful approach to understand the genetic basis of clock periodicity. Accordingly, intercrossing these different Medaka species results in a wide range of meiotic recombination across the genome, yielding F2 progeny showing a wide range of clock periods (Seleit et al., 2023). The authors further show that while clock period scales with PSM size in parental fishes, these two parameters do not correlate in F2 progeny. On the other hand, somite size still scales with PSM size. This raises the possibility that there is a spectrum of somite numbers formed in the F2 embryos. However, this has not been shown. In addition, whole genome sequencing of F2 embryos revealed that the Medaka dll1 is responsible for controlling PSM size, but not somite number/border formation or clock period. This result is in contrast to Dll1−/− mice where no somite borders are formed, indicating a potential evolutionarily divergent role of Notch ligands.
One way to combat the inability to perform inter-species crosses in mammals is to carry out in vivo or in vitro gene-swapping experiments. One study showed that the species-specific oscillatory period of Hes7 is not entirely regulated by local genomic sequences (Matsuda et al., 2020a). In particular, the human segmentation clock ticks with a periodicity of ∼5 h (Chu et al., 2019; Matsuda et al., 2020b; Diaz-Cuadros et al., 2020) and when the mouse Hes7 locus was swapped with the human HES7 in vivo, it results in only an ∼20–30 min extension of the mouse period when measured in E10.5 mouse PSM explants (Matsuda et al., 2020a). This genomic manipulation caused minor, but significant segmentation defects in the form of a curved spine and a kinked short tail. Possibly, mutant embryos containing the human HES7 that showed a change in the clock time of >20–30 min may have been embryonic lethal, thereby only the embryos with the minimal clock perturbation survive. Furthermore, because this gene swapping was limited to a BAC construct extending only up to the 3′ UTR of the neighboring gene Per1, one cannot exclude the possibility of cis-elements downstream of Hes7 affecting its periodicity. While there are only limited reports on how the segmentation clock and the circadian clock could interact genetically (Umair et al., 2022), the PER1 gene also displays an in-phase gene oscillation phenotype with HES7 in human PSM cells in vitro (Matsuda et al., 2020b).
Aligning with the ideology of the importance of comparative developmental biology, recent studies have established embryonic stem cells or induced pluripotent stem cells (iPSCs) derived from various species of different sizes and gestational lengths such as marmosets, rhinoceros, cattle, rabbits (Lázaro et al., 2023) and pigs (Conrad et al., 2023) (Table 4). These species-specific PSCs display intrinsic developmental timing phenotypes when differentiated to a PSM and other lineages (Matsuda et al., 2020a; Rayon et al., 2020; Lázaro et al., 2023). The in vitro clock period observed across these different species also correlates with in vivo gestation times, body size, and biochemical reaction rates in vitro (Matsuda et al., 2020a; Conrad et al., 2023; Diaz-Cuadros et al., 2023; Lázaro et al., 2023). Therefore, carefully curated in vivo embryonic staging data can help determine how closely these in vitro systems can mimic somitogenesis. In addition, recent studies have showcased the power of both 2D and 3D mouse and human in vitro models in recapitulating hallmarks of somite formation, thereby giving us insights, especially into human development (Diaz-Cuadros and Pourquie, 2021; van den Brink et al., 2020; Budjan et al., 2022; Chu et al., 2019; Diaz-Cuadros et al., 2020; Matsuda et al., 2020b; Miao et al., 2022; Sanaki-Matsumiya et al., 2022; Veenvliet et al., 2020; Yamanaka et al., 2022). Multiple studies have shown human pluripotent stem cells (PSCs)-derived PSM cells capable of undergoing 3D morphological somite-like budding that can further our knowledge of how temporal information translates to spatial information in our species (Miao et al., 2022; Sanaki-Matsumiya et al., 2022; Yamanaka et al., 2022).
In this review, we summarized existing knowledge on the segmentation clock biology by leveraging the similarities and differences of the mouse segmentation clock with that of zebrafish and chick. Through insightful comparisons with non-classical model organisms and novel in vitro models, we illuminate the profound importance of these systems in advancing our understanding of the segmentation clock and its evolution. In the future, we anticipate that novel in vivo and in vitro approaches will provide complimentary knowledge to better understand vertebrate segmentation and embryonic development.
PSR: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing. L-FC: Conceptualization, Funding acquisition, Supervision, Validation, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was undertaken thanks to funding support from the Faculty of Veterinary Medicine, University of Calgary (UCVM), Alberta Children’s Hospital Research Institute (ACHRI), the Canada Research Chairs Program (CRC, L-FC, 950-232985), Canada Foundation for Innovation (CFI, L-FC, 40653), Natural Sciences and Engineering Research Council of Canada (NSERC, L-FC, RGPIN2021-02580), Government of Canada’s New Frontiers in Research Fund (NFRFE-2020-00446).
We are grateful to the many pioneers and scientists who have dedicated their work to elucidating the mechanisms underpinning somitogenesis. Due to space limitations, we apologize for not being able to mention all the important works contributing to our understanding of the segmentation clock in this article. We thank all members of the Chu Laboratory for their helpful comments on this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acar, M., Jafar-Nejad, H., Takeuchi, H., Rajan, A., Ibrani, D., Rana, N. A., et al. (2008). Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132, 247–258. doi:10.1016/J.CELL.2007.12.016
Akiyama, R., Masuda, M., Tsuge, S., Bessho, Y., and Matsui, T. (2014). An anterior limit of FGF/Erk signal activity marks the earliest future somite boundary in zebrafish. Development 141, 1104–1109. doi:10.1242/DEV.098905
Alpert, T., Herzel, L., and Neugebauer, K. M. (2017). Perfect timing: splicing and transcription rates in livingcells. Wiley Interdiscip. Rev. RNA 8. doi:10.1002/WRNA.1401
Anderson, M. J., Magidson, V., Kageyama, R., and Lewandoski, M. (2020). Fgf4 maintains Hes7 levels critical for normal somite segmentation clock function. Elife 9, e55608–e55622. doi:10.7554/ELIFE.55608
Andrawes, M. B., Xu, X., Liu, H., Ficarro, S. B., Marto, J. A., Aster, J. C., et al. (2013). Intrinsic selectivity of notch 1 for delta-like 4 over delta-like 1. J. Biol. Chem. 288, 25477–25489. doi:10.1074/jbc.M113.454850
Artavanis-Tsakonas, S., Rand, M. D., and Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Sci. (80-. ) 284, 770–776. doi:10.1126/SCIENCE.284.5415.770
Aulehla, A., Wehrle, C., Brand-Saberi, B., Kemler, R., Gossler, A., Kanzler, B., et al. (2003). Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4, 395–406. doi:10.1016/S1534-5807(03)00055-8
Aulehla, A., Wiegraebe, W., Baubet, V., Wahl, M. B., Deng, C., Taketo, M., et al. (2008). A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat. Cell Biol. 10, 186–193. doi:10.1038/NCB1679
Ay, A., Knierer, S., Sperlea, A., Holland, J., and Özbudak, E. M. (2013). Short-lived her proteins drive robust synchronized oscillations in the Zebrafish segmentation clock. Dev 140, 3244–3253. doi:10.1242/dev.093278
Barhoumi, T., Nashabat, M., Alghanem, B., Alhallaj, A. S., Boudjelal, M., Umair, M., et al. (2019). Delta Like-1 gene mutation: a novel cause of congenital vertebral malformation. Front. Genet. 10, 534. doi:10.3389/fgene.2019.00534
Bessho, Y., Hirata, H., Masamizu, Y., and Kageyama, R. (2003). Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456. doi:10.1101/gad.1092303
Bessho, Y., Miyoshi, G., Sakata, R., and Kageyama, R. (2001a). Hes7: a bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes cells. 6, 175–185. doi:10.1046/J.1365-2443.2001.00409.X
Bessho, Y., Sakata, R., Komatsu, S., Shiota, K., Yamada, S., and Kageyama, R. (2001b). Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15, 2642–2647. doi:10.1101/GAD.930601
Bettenhausen, B., Hrabe de Angelis, M., Simon, D., Guenet, J. L., and Gossler, A. (1995). Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development 121, 2407–2418. doi:10.1242/DEV.121.8.2407
Bierkamp, C., and Campos-Ortega, J. A. (1993). A zebrafish homologue of the Drosophila neurogenic gene Notch and its pattern of transcription during early embryogenesis. Mech. Dev. 43, 87–100. doi:10.1016/0925-4773(93)90027-U
Bochter, M. S., Servello, D., Kakuda, S., D’Amico, R., Ebetino, M. F., Haltiwanger, R. S., et al. (2022). Lfng and Dll3 cooperate to modulate protein interactions in cis and coordinate oscillatory Notch pathway activation in the segmentation clock. Dev. Biol. 487, 42–56. doi:10.1016/J.YDBIO.2022.04.004
Brena, C., and Akam, M. (2013). An analysis of segmentation dynamics throughout embryogenesis in the centipede Strigamia maritima. BMC Biol. 11, 112–118. doi:10.1186/1741-7007-11-112
Brend, T., and Holley, S. A. (2009). Expression of the oscillating gene her1 is directly regulated by Hairy/Enhancer of Split, T-box, and Suppressor of Hairless proteins in the zebrafish segmentation clock. Dev. Dyn. 238, 2745–2759. doi:10.1002/DVDY.22100
Brückner, K., Perez, L., Clausen, H., and Cohen, S. (2000). Glycosyltransferase activity of fringe modulates notch–delta interactions. Nat 406, 411–415. doi:10.1038/35019075
Budjan, C., Liu, S., Ranga, A., Gayen, S., Pourquie, O., and Hormoz, S. (2022). Paraxial mesoderm organoids model development of human somites. Elife 11, e68925. doi:10.7554/ELIFE.68925
Busby, L., Nájera, G. S., and Steventon, B. (2023). A population intrinsic timer controls Hox gene expression and cell dispersion during progenitor addition to the body axis. bioRxiv, 2023.05.10.540133. doi:10.1101/2023.05.10.540133
Caprioli, A., Goitsuka, R., Pouget, C., Dunon, D., and Jaffredo, T. (2002). Expression of Notch genes and their ligands during gastrulation in the chicken embryo. Mech. Dev. 116, 161–164. doi:10.1016/S0925-4773(02)00136-3
Carraco, G., Martins-Jesus, A. P., and Andrade, R. P. (2022). The vertebrate Embryo Clock: common players dancing to a different beat. Front. Cell Dev. Biol. 10, 944016. doi:10.3389/fcell.2022.944016
Chalamalasetty, R. B., Garriock, R. J., Dunty, W. C., Kennedy, M. W., Jailwala, P., Si, H., et al. (2014). Mesogenin 1 is a master regulator of paraxial presomitic mesoderm differentiation. Dev 141, 4285–4297. doi:10.1242/dev.110908
Chapman, G., Sparrow, D. B., Kremmer, E., and Dunwoodie, S. L. (2011). Notch inhibition by the ligand DELTA-LIKE 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum. Mol. Genet. 20, 905–916. doi:10.1093/HMG/DDQ529
Chen, J., Kang, L., and Zhang, N. (2005). Negative feedback loop formed by Lunatic fringe and Hes7 controls their oscillatory expression during somitogenesis. Genesis 43, 196–204. doi:10.1002/GENE.20171
Choorapoikayil, S., Willems, B., Ströhle, P., and Gajewski, M. (2012). Analysis of her1 and her7 mutants reveals a spatio temporal separation of the Somite clock module. PLoS One 7, e39073. doi:10.1371/journal.pone.0039073
Christ, B., Jacob, H. J., and Jacob, M. (1974). Die somitogenese beim huhnerembryo. zur determination der segmentierungsrichtung. Verh. Anat. Ges. 68, 573–579.
Chu, L. F., Mamott, D., Ni, Z., Bacher, R., Liu, C., Swanson, S., et al. (2019). An in vitro human segmentation clock model derived from embryonic stem cells. Cell Rep. 28, 2247–2255. doi:10.1016/j.celrep.2019.07.090
Ciruna, B., and Rossant, J. (2001). FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell 1, 37–49. doi:10.1016/S1534-5807(01)00017-X
Clark, E., Peel, A. D., and Akam, M. (2019). Arthropod segmentation. Dev 146, dev170480. doi:10.1242/dev.170480
Conlon, R. A., Reaume, A. G., and Rossant, J. (1995). Notch1 is required for the coordinate segmentation of somites. Development 121, 1533–1545. doi:10.1242/DEV.121.5.1533
Conrad, J. V., Meyer, S., Ramesh, P. S., Neira, J. A., Rusteika, M., Mamott, D., et al. (2023). Efficient derivation of transgene-free porcine induced pluripotent stem cells enables in vitro modeling of species-specific developmental timing. Stem Cell Rep. 18, 2328–2343. doi:10.1016/J.STEMCR.2023.10.009
Cooke, J., and Zeeman, E. C. (1976). A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 58, 455–476. doi:10.1016/S0022-5193(76)80131-2
Cordes, R., Schuster-Gossler, K., Serth, K., and Gossler, A. (2004). Specification of vertebral identity is coupled to Notch signalling and the segmentation clock. Development 131, 1221–1233. doi:10.1242/DEV.01030
Dale, J. K., Maroto, M., Dequeant, M. L., Malapert, P., McGrew, M., and Pourquie, O. (2003). Periodic Notch inhibition by Lunatic Fringe underlies the chick segmentation clock. Nat, 275–278. doi:10.1038/nature01244
Davis, G. K., and Patel, N. H. (1999). The origin and evolution of segmentation. Trends Cell Biol. 9, M68–M72. doi:10.1016/S0962-8924(99)01663-3
De Angelis, M. H., McIntyre, J., and Gossler, A. (1997). Maintenance of somite borders in mice requires the Delta homologue DII1. Nature 386, 717–721. doi:10.1038/386717A0
del Amo, F. F., Gendron-Maguire, M., Swiatek, P. J., Jenkins, N. A., Copeland, N. G., and Gridley, T. (1993). Cloning, analysis, and chromosomal localization of notch-1, a mouse homolog of drosophila notch. Genomics 15, 259–264. doi:10.1006/GENO.1993.1055
Delaune, E. A., François, P., Shih, N. P., and Amacher, S. L. (2012). Single-cell-resolution imaging of the impact of notch signaling and mitosis on segmentation clock dynamics. Dev. Cell 23, 995–1005. doi:10.1016/j.devcel.2012.09.009
Del Barco Barrantes, I., Elia, A. J., Wünsch, K., Hrabe de Angelis, M., Mak, T. W., Rossant, J., et al. (1999). Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9, 470–480. doi:10.1016/S0960-9822(99)80212-7
Dequéant, M. L., Glynn, E., Gaudenz, K., Wahl, M., Chen, J., Mushegian, A., et al. (2006). A complex oscillating network of signaling genes underlies the mouse segmentation clock. Sci. (80- 314, 1595–1598. doi:10.1126/science.1133141
Diaz-Cuadros, M., Miettinen, T. P., Skinner, O. S., Sheedy, D., Díaz-García, C. M., Gapon, S., et al. (2023). Metabolic regulation of species-specific developmental rates. Nature 613, 550–557. doi:10.1038/S41586-022-05574-4
Diaz-Cuadros, M., and Pourquie, O. (2021). In vitro systems: a new window to the segmentation clock. Dev. Growth Differ. 63, 140–153. doi:10.1111/DGD.12710
Diaz-Cuadros, M., Wagner, D. E., Budjan, C., Hubaud, A., Tarazona, O. A., Donelly, S., et al. (2020). In vitro characterization of the human segmentation clock. Nat, 113–118. doi:10.1038/s41586-019-1885-9
Dunwoodie, S. L., Clements, M., Sparrow, D. B., Sa, X., Conlon, R. A., and Beddington, R. S. P. (2002). Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129, 1795–1806. doi:10.1242/DEV.129.7.1795
Dunwoodie, S. L., Henrique, D., Harrison, S. M., and Beddington, R. S. P. (1997). Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development 124, 3065–3076. doi:10.1242/DEV.124.16.3065
Eckalbar, W. L., Lasku, E., Infante, C. R., Elsey, R. M., Markov, G. J., Allen, A. N., et al. (2012). Somitogenesis in the anole lizard and alligator reveals evolutionary convergence and divergence in the amniote segmentation clock. Dev. Biol. 363, 308–319. doi:10.1016/J.YDBIO.2011.11.021
Elmasri, H., Winkler, C., Liedtke, D., Sasado, T., Morinaga, C., Suwa, H., et al. (2004). Mutations affecting somite formation in the Medaka (Oryzias latipes). Mech. Dev. 121, 659–671. doi:10.1016/j.mod.2004.04.003
Eloy-Trinquet, S., and Nicolas, J. F. (2002). Cell coherence during production of the presomitic mesoderm and somitogenesis in the mouse embryo. Development 129, 3609–3619. doi:10.1242/DEV.129.15.3609
Falk, H. J., Tomita, T., Monke, G., McDole, K., and Aulehla, A. (2022). Imaging the onset of oscillatory signaling dynamics during mouse embryo gastrulation. Dev 149. doi:10.1242/dev.200083
Feller, J., Schneider, A., Schuster-Gossler, K., and Gossler, A. (2008). Noncyclic Notch activity in the presomitic mesoderm demonstrates uncoupling of somite compartmentalization and boundary formation. Genes Dev. 22, 2166–2171. doi:10.1101/gad.480408
Ferjentsik, Z., Hayashi, S., Dale, J. K., Bessho, Y., Herreman, A., De Strooper, B., et al. (2009). Notch is a critical component of the mouse somitogenesis oscillator and is essential for the formation of the somites. PLoS Genet. 5, e1000662. doi:10.1371/journal.pgen.1000662
Fiddes, I. T., Lodewijk, G. A., Mooring, M., Bosworth, C. M., Ewing, A. D., Mantalas, G. L., et al. (2018). Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369. doi:10.1016/j.cell.2018.03.051
Fior, R., Maxwell, A. A., Ma, T. P., Vezzaro, A., Moens, C. B., Amacher, S. L., et al. (2012). The differentiation and movement of presomitic mesoderm progenitor cells are controlled by mesogenin 1. Dev 139, 4656–4665. doi:10.1242/dev.078923
Fischer, A., Schumacher, N., Maier, M., Sendtner, M., and Gessler, M. (2004). The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18, 901–911. doi:10.1101/GAD.291004
Flint, O. P., Ede, D. A., Wilby, O. K., and Proctor, J. (1978). Control of somite number in normal and amputated mutant mouse embryos: an experimental and a theoretical analysis. Development 45, 189–202. doi:10.1242/DEV.45.1.189
Gajewski, M., Elmasri, H., Girschick, M., Sieger, D., and Winkler, C. (2006). Comparative analysis of her genes during fish somitogenesis suggests a mouse/chick-like mode of oscillation in medaka. Dev. Genes Evol. 216, 315–332. doi:10.1007/S00427-006-0059-6
Gajewski, M., Sieger, D., Alt, B., Leve, C., Hans, S., Wolff, C., et al. (2003). Anterior and posterior waves of cyclic her1 gene expression are differentially regulated in the presomitic mesoderm of zebrafish. Development 130, 4269–4278. doi:10.1242/DEV.00627
Gessler, M., Knobeloch, K. P., Helisch, A., Amann, K., Schumacher, N., Rohde, E., et al. (2002). Mouse gridlock: No aortic coarctation or deficiency, but fatal cardiac defects in Hey2 -/- mice. Curr. Biol. 12, 1601–1604. doi:10.1016/S0960-9822(02)01150-8
Gibb, S., Zagorska, A., Melton, K., Tenin, G., Vacca, I., Trainor, P., et al. (2009). Interfering with Wnt signalling alters the periodicity of the segmentation clock. Dev. Biol. 330, 21–31. doi:10.1016/J.YDBIO.2009.02.035
Giudicelli, F., Özbudak, E. M., Wright, G. J., and Lewis, J. (2007). Setting the tempo in development: an investigation of the zebrafish somite clock mechanism. PLoS Biol. 5, e150–e1323. doi:10.1371/journal.pbio.0050150
Gomez, C., Özbudak, E. M., Wunderlich, J., Baumann, D., Lewis, J., and Pourquié, O. (2008). Control of segment number in vertebrate embryos. Nat, 335–339. doi:10.1038/nature07020
Goodwin, B. C., and Cohen, M. H. (1969). A phase-shift model for the spatial and temporal organization of developing systems. J. Theor. Biol. 25, 49–107. doi:10.1016/S0022-5193(69)80017-2
Gossler, A., and Tam, P. P. L. (2002). Somitogenesis: segmentation of the paraxial mesoderm and the delineation of tissue compartments. Mouse Dev., 127–149. doi:10.1016/B978-012597951-1/50009-3
Haddon, C., Smithers, L., Schneider-Maunoury, S., Coche, T., Henrique, D., and Lewis, J. (1998). Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development 125, 359–370. doi:10.1242/dev.125.3.359
Hamada, Y., Kadokawa, Y., Okabe, M., Ikawa, M., Coleman, J. R., and Tsujimoto, Y. (1999). Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 126, 3415–3424. doi:10.1242/DEV.126.15.3415
Harima, Y., Takashima, Y., Ueda, Y., Ohtsuka, T., and Kageyama, R. (2013). Accelerating the tempo of the segmentation clock by reducing the number of introns in the Hes7 gene. Cell Rep. 3, 1–7. doi:10.1016/j.celrep.2012.11.012
Hayashi, S., Shimoda, T., Nakajima, M., Tsukada, Y., Sakumura, Y., Dale, J. K., et al. (2009). Sprouty4, an FGF inhibitor, displays cyclic gene expression under the control of the notch segmentation clock in the mouse PSM. PLoS One 4, e5603. doi:10.1371/JOURNAL.PONE.0005603
Henrique, D., Adam, J., Myat, A., Chitnis, A., Lewis, J., and Ish-Horowicz, D. (1995). Expression of a delta homologue in prospective neurons in the chick. Nature 375, 787–790. doi:10.1038/375787A0
Henrique, D., and Schweisguth, F. (2019). Mechanisms of notch signaling: a simple logic deployed in time and space. Dev 146, dev172148. doi:10.1242/dev.172148
Henry, C. A., Urban, M. K., Dill, K. K., Merlie, J. P., Page, M. F., Kimmel, C. B., et al. (2002). Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development 129, 3693–3704. doi:10.1242/DEV.129.15.3693
Herrgen, L., Ares, S., Morelli, L. G., Schröter, C., Jülicher, F., and Oates, A. C. (2010). Intercellular coupling regulates the period of the segmentation clock. Curr. Biol. 20, 1244–1253. doi:10.1016/j.cub.2010.06.034
Hicks, C., Johnston, S. H., DiSibio, G., Collazo, A., Vogt, T. F., and Weinmaster, G. (2000). Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat. Cell Biol. 2, 515–520. doi:10.1038/35019553
Higuchi, M., Kiyama, H., Hayakawa, T., Hamada, Y., and Tsujimoto, Y. (1995). Differential expression of Notch1 and Notch2 in developing and adult mouse brain. Mol. Brain Res. 29, 263–272. doi:10.1016/0169-328X(94)00257-F
Hirata, H., Bessho, Y., Kokubu, H., Masamizu, Y., Yamada, S., Lewis, J., et al. (2004). Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 36, 750–754. doi:10.1038/ng1372
Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K., et al. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843. doi:10.1126/SCIENCE.1074560
Holley, S. A., Geisler, R., and Nüsslein-Volhard, C. (2000). Control of her1 expression during zebrafish somitogenesis by a Delta-dependent oscillator and an independent wave-front activity. Genes Dev. 14, 1678–1690. doi:10.1101/gad.14.13.1678
Holley, S. A., Jülich, D., Rauch, G. J., Geisler, R., and Nüsslein-Volhard, C. (2002). her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129, 1175–1183. doi:10.1242/DEV.129.5.1175
Horikawa, K., Ishimatsu, K., Yoshimoto, E., Kondo, S., and Takeda, H. (2006). Noise-resistant and synchronized oscillation of the segmentation clock. Nature 441, 719–723. doi:10.1038/nature04861
Hubaud, A., and Pourquié, O. (2014). Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15, 709–721. doi:10.1038/NRM3891
Hubaud, A., Regev, I., Mahadevan, L., and Pourquié, O. (2017). Excitable dynamics and yap-dependent mechanical cues drive the segmentation clock. Cell 171, 668–682. doi:10.1016/j.cell.2017.08.043
Huppert, S. S., Ilagan, M. X. G., De Strooper, B., and Kopan, R. (2005). Analysis of notch function in presomitic mesoderm suggests a γ-secretase-independent role for presenilins in somite differentiation. Dev. Cell 8, 677–688. doi:10.1016/J.DEVCEL.2005.02.019
Ishibashi, M., Ang, S. L., Shiota, K., Nakanishi, S., Kageyama, R., and Guillemot, F. (1995). Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9, 3136–3148. doi:10.1101/GAD.9.24.3136
Ishimatsu, K., Hiscock, T. W., Collins, Z. M., Kartika Sari, D. W., Lischer, K., Richmond, D. L., et al. (2018). Size-reduced embryos reveal a gradient scaling-based mechanism for zebrafish somite formation. Dev 145, dev161257. doi:10.1242/dev.161257
Jacobsen, T., Brennan, K., Arias, A., and Muskavitch, M. A. (1998). Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development 125, 4531–4540. doi:10.1242/dev.125.22.4531
Jensen, M. H., Sneppen, K., and Tiana, G. (2003). Sustained oscillations and time delays in gene expression of protein Hes1. FEBS Lett. 541, 176–177. doi:10.1016/S0014-5793(03)00279-5
Jiang, Y. J., Aerne, B. L., Smithers, L., Haddon, C., Ish-Horowicz, D., and Lewis, J. (2000). Notch signalling and the synchronization of the somite segmentation clock. Nat 408, 475–479. doi:10.1038/35044091
Johnston, D. S., and Nüsslein-Volhard, C. (1992). The origin of pattern and polarity in the Drosophila embryo. Cell 68, 201–219. doi:10.1016/0092-8674(92)90466-P
Johnston, S. H., Rauskolb, C., Wilson, R., Prabhakaran, B., Irvine, K. D., and Vogt, T. F. (1997). A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development 124, 2245–2254. doi:10.1242/DEV.124.11.2245
Jones, S. (2004). An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226–6. doi:10.1186/gb-2004-5-6-226
Jouve, C., Palmeirim, I., Henrique, D., Beckers, J., Gossler, A., Ish-Horowicz, D., et al. (2000). Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127, 1421–1429. doi:10.1242/DEV.127.7.1421
Jülich, D., Chiaw, H. L., Round, J., Nicolaije, C., Schroeder, J., Davies, A., et al. (2005). beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev. Biol. 286, 391–404. doi:10.1016/J.YDBIO.2005.06.040
Kageyama, R., Ohtsuka, T., and Kobayashi, T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251. doi:10.1242/DEV.000786
Kainz, F., Ewen-Campen, B., Akam, M., and Extavour, C. G. (2011). Notch/Delta signalling is not required for segment generation in the basally branching insect Gryllus bimaculatus. Development 138, 5015–5026. doi:10.1242/DEV.073395
Kakuda, S., and Haltiwanger, R. S. (2017). Deciphering the Fringe-mediated Notch Code: identification of activating and inhibiting sites allowing discrimination between ligands. Dev. Cell 40, 193–201. doi:10.1016/J.DEVCEL.2016.12.013
Kawamura, A., Koshida, S., Hijikata, H., Sakaguchi, T., Kondoh, H., and Takada, S. (2005). Zebrafish Hairy/Enhancer of split protein links FGF signaling to cyclic gene expression in the periodic segmentation of somites. Genes Dev. 19, 1156–1161. doi:10.1101/gad.1291205
Keseroglu, K., Zinani, O. Q. H., Keskin, S., Seawall, H., Alpay, E. E., and Özbudak, E. M. (2023). Stochastic gene expression and environmental stressors trigger variable somite segmentation phenotypes. Nat. Commun. 14, 6497–6499. doi:10.1038/s41467-023-42220-7
Kim, W., Matsui, T., Yamao, M., Ishibashi, M., Tamada, K., Takumi, T., et al. (2011). The period of the somite segmentation clock is sensitive to Notch activity. Mol. Biol. Cell 22, 3541–3549. doi:10.1091/mbc.E11-02-0139
Kopan, R., and Ilagan, M. X. G. (2009). The canonical notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233. doi:10.1016/J.CELL.2009.03.045
Krol, A. J., Roellig, D., Dequéant, M. L., Tassy, O., Glynn, E., Hattem, G., et al. (2011). Evolutionary plasticity of segmentation clock networks. Development 138, 2783–2792. doi:10.1242/dev.063834
Kuretani, A., Yamamoto, T., Taira, M., and Michiue, T. (2021). Evolution of hes gene family in vertebrates: the hes5 cluster genes have specifically increased in frogs. BMC Ecol. Evol. 21, 147–215. doi:10.1186/s12862-021-01879-6
Kusumi, K., Mimoto, M. S., Covello, K. L., Beddington, R. S. P., Krumlauf, R., and Dunwoodie, S. L. (2004). Dll3 pudgy mutation differentially disrupts dynamic expression of somite genes. genesis 39, 115–121. doi:10.1002/GENE.20034
Kusumi, K., Sun, E. S., Kerrebrock, A. W., Bronson, R. T., Chi, D. C., Bulotsky, M. S., et al. (1998). The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat. Genet. 19, 274–278. doi:10.1038/961
Ladi, E., Nichols, J. T., Ge, W., Miyamoto, A., Yao, C., Yang, L. T., et al. (2005). The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 170, 983–992. doi:10.1083/JCB.200503113
Lardelli, M., Dahlstrand, J., and Lendahl, U. (1994). The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech. Dev. 46, 123–136. doi:10.1016/0925-4773(94)90081-7
Lauschke, V. M., Tsiairis, C. D., François, P., and Aulehla, A. (2012). Scaling of embryonic patterning based on phase-gradient encoding. Nat 493, 101–105. doi:10.1038/nature11804
Lázaro, J., Costanzo, M., Sanaki-Matsumiya, M., Girardot, C., Hayashi, M., Hayashi, K., et al. (2023). A stem cell zoo uncovers intracellular scaling of developmental tempo across mammals. Cell Stem Cell 30, 938–949.e7. doi:10.1016/J.STEM.2023.05.014
Leimeister, C., Dale, K., Fischer, A., Klamt, B., Hrabe De Angelis, M., Radtke, F., et al. (2000). Oscillating expression of c-hey2 in the presomitic mesoderm suggests that the segmentation clock may use combinatorial signaling through multiple interacting bHLH factors. Dev. Biol. 227, 91–103. doi:10.1006/DBIO.2000.9884
Leimeister, C., Externbrink, A., Klamt, B., and Gessler, M. (1999). Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech. Dev. 85, 173–177. doi:10.1016/S0925-4773(99)00080-5
Leslie, J. D., Ariza-McNaughton, L., Bermange, A. L., McAdow, R., Johnson, S. L., and Lewis, J. (2007). Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development 134, 839–844. doi:10.1242/dev.003244
Liao, B. K., Jörg, D. J., and Oates, A. C. (2016). Faster embryonic segmentation through elevated Delta-Notch signalling. Nat. Commun. 7, 11861–11912. doi:10.1038/ncomms11861
Liao, B. K., and Oates, A. C. (2017). Delta-Notch signalling in segmentation. Arthropod Struct. Dev. 46, 429–447. doi:10.1016/j.asd.2016.11.007
Lindsell, C. E., Shawber, C. J., Boulter, J., and Weinmaster, G. (1995). Jagged: a mammalian ligand that activates notch1. Cell 80, 909–917. doi:10.1016/0092-8674(95)90294-5
Loomes, K. M., Stevens, S. A., O’Brien, M. L., Gonzalez, D. M., Ryan, M. J., Segalov, M., et al. (2007). Dll3 and Notch1 genetic interactions model axial segmental and craniofacial malformations of human birth defects. Dev. Dyn. 236, 2943–2951. doi:10.1002/DVDY.21296
Lorent, K., Yeo, S. Y., Oda, T., Chandrasekharappa, S., Chitnis, A., Matthews, R. P., et al. (2004). Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development 131, 5753–5766. doi:10.1242/DEV.01411
Mara, A., Schroeder, J., Chalouni, C., and Holley, S. A. (2007). Priming, initiation and synchronization of the segmentation clock by deltaD and deltaC. Nat. Cell Biol. 9, 523–530. doi:10.1038/NCB1578
Maroto, M., Dale, J. K., Dequéant, M. L., Petit, A. C., and Pourquié, O. (2005). Synchronised cycling gene oscillations in presomitic mesoderm cells require cell-cell contact. Int. J. Dev. Biol. 49, 309–315. doi:10.1387/IJDB.041958MM
Maroto, M., Iimura, T., Dale, J. K., and Bessho, Y. (2008). BHLH proteins and their role in somitogenesis. Adv. Exp. Med. Biol. 638, 124–139. doi:10.1007/978-0-387-09606-3_7
Masamizu, Y., Ohtsuka, T., Takashima, Y., Nagahara, H., Takenaka, Y., Yoshikawa, K., et al. (2006). Real-time imaging of the somite segmentation clock: revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc. Natl. Acad. Sci. U. S. A. 103, 1313–1318. doi:10.1073/pnas.0508658103
Matsuda, M., Hayashi, H., Garcia-Ojalvo, J., Yoshioka-Kobayashi, K., Kageyama, R., Yamanaka, Y., et al. (2020a). Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Sci. (80-. ) 369, 1450–1455. doi:10.1126/SCIENCE.ABA7668
Matsuda, M., Yamanaka, Y., Uemura, M., Osawa, M., Saito, M. K., Nagahashi, A., et al. (2020b). Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580, 124–129. doi:10.1038/S41586-020-2144-9
Mayburd, A. L., Tan, Y., Kassner, R. J., and Tautz, D. (2001). Homologues of c-hairy1 (her9) and lunatic fringe in zebrafish are expressed in the developing central nervous system, but not in the presomitic mesoderm. Dev. Genes Evol. 211, 493–500. doi:10.1007/s00427-001-0181-4
McCright, B., Gao, X., Shen, L., Lozier, J., Lan, Y., Maguire, M., et al. (2001). Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128, 491–502. doi:10.1242/DEV.128.4.491
McGrew, M. J., Dale, J. K., Fraboulet, S., and Pourquié, O. (1998). The lunatic fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol. 8, 979–982. doi:10.1016/S0960-9822(98)70401-4
Menkes, B., and Sandor, S. (1977). Somitogenesis: regulation potencies, sequence determination and primordial interactions. Vertebrate limb somite Morphog., 405–419.
Miao, Y., Djeffal, Y., De Simone, A., Zhu, K., Lee, J. G., Lu, Z., et al. (2022). Reconstruction and deconstruction of human somitogenesis in vitro. Nat 614, 500–508. doi:10.1038/s41586-022-05655-4
Monk, N. A. M. (2003). Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr. Biol. 13, 1409–1413. doi:10.1016/S0960-9822(03)00494-9
Morelli, L. G., Ares, S., Herrgen, L., Schröter, C., Jülicher, F., and Oates, A. C. (2009). Delayed coupling theory of vertebrate segmentation. HFSP J. 3, 55–66. doi:10.2976/1.3027088
Morimoto, M., Takahashi, Y., Endo, M., and Saga, Y. (2005). The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature 435, 354–359. doi:10.1038/nature03591
Myat, A., Henrique, D., Ish-Horowicz, D., and Lewis, J. (1996). A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev. Biol. 174, 233–247. doi:10.1006/DBIO.1996.0069
Naiche, L. A., Holder, N., and Lewandoski, M. (2011). FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proc. Natl. Acad. Sci. U. S. A. 108, 4018–4023. doi:10.1073/pnas.1007417108
Nandagopal, N., Santat, L. A., and Elowitz, M. B. (2019). Cis-activation in the Notch signaling pathway. Elife 8, e37880. doi:10.7554/ELIFE.37880
Niwa, Y., Masamizu, Y., Liu, T., Nakayama, R., Deng, C. X., and Kageyama, R. (2007). The initiation and propagation of Hes7 oscillation are cooperatively regulated by fgf and notch signaling in the somite segmentation clock. Dev. Cell 13, 298–304. doi:10.1016/j.devcel.2007.07.013
Niwa, Y., Shimojo, H., Isomura, A., González, A., Miyachi, H., and Kageyama, R. (2011). Different types of oscillations in notch and Fgf signaling regulate the spatiotemporal periodicity of somitogenesis. Genes Dev. 25, 1115–1120. doi:10.1101/gad.2035311
Oates, A. C., and Ho, R. K. (2002). Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129, 2929–2946. doi:10.1242/DEV.129.12.2929
Oates, A. C., Morelli, L. G., and Ares, S. (2012). Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development 139, 625–639. doi:10.1242/DEV.063735
Ochi, S., Imaizumi, Y., Shimojo, H., Miyachi, H., and Kageyama, R. (2020). Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Dev 147, dev182204. doi:10.1242/DEV.182204
Ohtsuka, T., Ishibashi, M., Gé Rald Gradwohl, G., Nakanishi, S., Ois Guillemot, F., and Kageyama, R. (1999). Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207. doi:10.1093/EMBOJ/18.8.2196
Okubo, Y., Sugawara, T., Abe-Koduka, N., Kanno, J., Kimura, A., and Saga, Y. (2012). Lfng regulates the synchronized oscillation of the mouse segmentation clock via trans-repression of Notch signalling. Nat. Commun. 3, 1141–1149. doi:10.1038/ncomms2133
O’Rahilly, R., and Müller, F. (2010). Developmental stages in human embryos: revised and new measurements. Cells. Tissues. Organs 192, 73–84. doi:10.1159/000289817
O’Rahilly, R. R., and Müller, F. (2001). Human embryology and teratology. 3rd Edition, 520. Available at: https://openlibrary.org/works/OL1970822W/Human_embryology_teratology (Accessed September 20, 2023).
Orts-Llorca, F. (1981). La somitogenèse chez l’embryon humain: apparition des myotomes. Bull. Assoc. Anat. (Nancy) 65, 467–482.
Packard, D. S. (1978). Chick somite determination: the role of factors in young somites and the segmental plate. J. Exp. Zool. 203, 295–306. doi:10.1002/JEZ.1402030212
Packard, D. S. (1980). Somite formation in cultured embryos of the snapping turtle, Chelydra serpentina. Development 59, 113–130. doi:10.1242/DEV.59.1.113
Palmeirim, I., Dubrulle, J., Henrique, D., Ish-Horowicz, D., and Pourquié, O. (1998). Uncoupling segmentation and somitogenesis in the chick presomitic mesoderm. Dev. Genet. 23, 77–85. doi:10.1002/(SICI)1520-6408(1998)23:1<77::AID-DVG8>3.0.CO;2-3
Palmeirim, I., Henrique, D., Ish-Horowicz, D., and Pourquié, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. doi:10.1016/S0092-8674(00)80451-1
Pasini, A., Jing, Y. J., and Wilkinson, D. G. (2004). Two zebrafish Notch-dependent hairy/Enhancer-of-split-relatedgenes, her6 and her4, are required to maintain the coordination of cyclic gene expression in the presomitic mesoderm. Development 131, 1529–1541. doi:10.1242/DEV.01031
Pasteels, J. (1937). Etudes sur la gastrulation des vertébrés méroblastiques. III. Oiseaux. IV Conclusions générales. Arch. Biol. 48, 381–488.
Pourquie, O. (2003). Vertebrate somitogenesis: a novel paradigm for animal segmentation? Int. J. Dev. Biol. 47, 597–603. doi:10.1387/IJDB.14756335
Pourquié, O. (2004). The chick embryo: a leading model in somitogenesis studies. Mech. Dev. 121, 1069–1079. doi:10.1016/J.MOD.2004.05.002
Pourquié, O. (2011). Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663. doi:10.1016/J.CELL.2011.05.011
Pourquié, O., and Tam, P. P. L. (2001). A nomenclature for prospective somites and phases of cyclic gene expression in the presomitic mesoderm. Dev. Cell 1, 619–620. doi:10.1016/S1534-5807(01)00082-X
Preuße, K., Tveriakhina, L., Schuster-Gossler, K., Gaspar, C., Rosa, A. I., Henrique, D., et al. (2015). Context-dependent functional divergence of the notch ligands DLL1 and DLL4 in vivo. PLoS Genet. 11 (6), e1005328. doi:10.1371/journal.pgen.1005328
Prince, V. E., Holley, S. A., Bally-Cuif, L., Prabhakaran, B., Oates, A. C., Ho, R. K., et al. (2001). Zebrafish lunatic fringe demarcates segmental boundaries. Mech. Dev. 105, 175–180. doi:10.1016/S0925-4773(01)00398-7
Psychoyos, D., and Stern, C. D. (1996). Fates and migratory routes of primitive streak cells in the chick embryo. Development 122, 1523–1534. doi:10.1242/DEV.122.5.1523
Pueyo, J. I., Lanfear, R., and Couso, J. P. (2008). Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc. Natl. Acad. Sci. U. S. A. 105, 16614–16619. doi:10.1073/PNAS.0804093105
Rayon, T., Stamataki, D., Perez-Carrasco, R., Garcia-Perez, L., Barrington, C., Melchionda, M., et al. (2020). Species-specific pace of development is associated with differences in protein stability. Science 369, eaba7667. doi:10.1126/SCIENCE.ABA7667
Resende, T. P., Andrade, R. P., and Palmeirim, I. (2014). Timing embryo segmentation: dynamics and regulatory mechanisms of the vertebrate segmentation clock. Biomed. Res. Int. 2014, 718683. doi:10.1155/2014/718683
Riley, M. F., Bochter, M. S., Wahi, K., Nuovo, G. J., and Cole, S. E. (2013). Mir-125a-5p-mediated regulation of Lfng is essential for the avian segmentation clock. Dev. Cell 24, 554–561. doi:10.1016/J.DEVCEL.2013.01.024
Robbins, J., Blondel, B. J., Gallahan, D., and Callahan, R. (1992). Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J. Virol. 66, 2594–2599. doi:10.1128/JVI.66.4.2594-2599.1992
Row, R. H., Tsotras, S. R., Goto, H., and Martin, B. L. (2016). The zebrafish tailbud contains two independent populations of midline progenitor cells that maintain long-term germ layer plasticity and differentiate in response to local signaling cues. Dev 143, 244–254. doi:10.1242/dev.129015
Sanaki-Matsumiya, M., Matsuda, M., Gritti, N., Nakaki, F., Sharpe, J., Trivedi, V., et al. (2022). Periodic formation of epithelial somites from human pluripotent stem cells. Nat. Commun. 13, 2325. doi:10.1038/S41467-022-29967-1
Sasai, Y., Kageyama, R., Tagawa, Y., Shigemoto, R., and Nakanishi, S. (1992). Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 6, 2620–2634. doi:10.1101/GAD.6.12B.2620
Sato, Y., Yasuda, K., and Takahashi, Y. (2002). Morphological boundary forms by a novel inductive event mediated by Lunatic fringe and Notch during somitic segmentation. Development 129, 3633–3644. doi:10.1242/DEV.129.15.3633
Sawada, A., Shinya, M., Jiang, Y. J., Kawakami, A., Kuroiwa, A., and Takeda, H. (2001). Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128, 4873–4880. doi:10.1242/DEV.128.23.4873
Schröter, C., Ares, S., Morelli, L. G., Isakova, A., Hens, K., Soroldoni, D., et al. (2012). Topology and dynamics of the zebrafish segmentation clock core circuit. PLOS Biol. 10, e1001364. doi:10.1371/JOURNAL.PBIO.1001364
Schröter, C., Herrgen, L., Cardona, A., Brouhard, G. J., Feldman, B., and Oates, A. C. (2008). Dynamics of zebrafish somitogenesis. Dev. Dyn. 237, 545–553. doi:10.1002/DVDY.21458
Schröter, C., and Oates, A. C. (2010). Segment number and axial identity in a segmentation clock period mutant. Curr. Biol. 20, 1254–1258. doi:10.1016/j.cub.2010.05.071
Seleit, A., Brettell, I., Fitzgerald, T., Vibe, C., Loosli, F., Wittbrodt, J., et al. (2023). Modular control of time and space during vertebrate axis segmentation. bioRxiv. doi:10.1101/2023.08.30.555457
Selleck, M. A. J., and Stern, C. D. (1991). Fate mapping and cell lineage analysis of Hensen’s node in the chick embryo. Development 112, 615–626. doi:10.1242/DEV.112.2.615
Sewell, W., Sparrow, D. B., Smith, A. J., Gonzalez, D. M., Rappaport, E. F., Dunwoodie, S. L., et al. (2009). Cyclical expression of the Notch/Wnt regulator Nrarp requires modulation by Dll3 in somitogenesis. Dev. Biol. 329, 400–409. doi:10.1016/J.YDBIO.2009.02.023
Shankaran, S. S., Sieger, D., Schröter, C., Czepe, C., Pauly, M. C., Laplante, M. A., et al. (2007). Completing the set of h/E(spl) cyclic genes in zebrafish: her12 and her15 reveal novel modes of expression and contribute to the segmentation clock. Dev. Biol. 304, 615–632. doi:10.1016/J.YDBIO.2007.01.004
Shawber, C., Boulter, J., Lindsell, C. E., and Weinmaster, G. (1996). JAGGED2: a serrate-like gene expressed during rat embryogenesis. Dev. Biol. 180, 370–376. doi:10.1006/DBIO.1996.0310
Shi, S., and Stanley, P. (2003). Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 100, 5234–5239. doi:10.1073/PNAS.0831126100
Shifley, E. T., VanHorn, K. M., Perez-Balaguer, A., Franklin, J. D., Weinstein, M., and Cole, S. E. (2008). Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development 135, 899–908. doi:10.1242/dev.006742
Shih, N. P., François, P., Delaune, E. A., and Amacher, S. L. (2015). Dynamics of the slowing segmentation clock reveal alternating two-segment periodicity. Dev 142, 1785–1793. doi:10.1242/dev.119057
Shimojo, H., Isomura, A., Ohtsuka, T., Kori, H., Miyachi, H., and Kageyama, R. (2016). Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 30, 102–116. doi:10.1101/GAD.270785.115
Siebel, C., and Lendahl, U. (2017). Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97, 1235–1294. doi:10.1152/PHYSREV.00005.2017
Sieger, D., Ackermann, B., Winkler, C., Tautz, D., and Gajewski, M. (2006). her1 and her13.2 are jointly required for somitic border specification along the entire axis of the fish embryo. Dev. Biol. 293, 242–251. doi:10.1016/J.YDBIO.2006.02.003
Sieger, D., Tautz, D., and Gajewski, M. (2004). her11 is involved in the somitogenesis clock in zebrafish. Dev. Genes Evol. 214, 393–406. doi:10.1007/s00427-004-0427-z
Simsek, M. F., Chandel, A. S., Saparov, D., Zinani, O. Q. H., Clason, N., and Özbudak, E. M. (2023). Periodic inhibition of Erk activity drives sequential somite segmentation. Nature 613, 153–159. doi:10.1038/S41586-022-05527-X
Smithers, L., Haddon, C., Jiang, Y. J., and Lewis, J. (2000). Sequence and embryonic expression of deltaC in the zebrafish. Mech. Dev. 90, 119–123. doi:10.1016/S0925-4773(99)00231-2
Song, H., Yuan, Z., Zhang, J., and Zhou, T. (2011). Molecular level dynamics of genetic oscillator--the effect of protein-protein interaction. Eur. Phys. J. E. Soft Matter 34, 77. doi:10.1140/EPJE/I2011-11077-8
Sonnen, K. F., Lauschke, V. M., Uraji, J., Falk, H. J., Petersen, Y., Funk, M. C., et al. (2018). Modulation of phase shift between Wnt and notch signaling oscillations controls mesoderm segmentation. Cell 172, 1079–1090. doi:10.1016/j.cell.2018.01.026
Soroldoni, D., Jörg, D. J., Morelli, L. G., Richmond, D. L., Schindelin, J., Julicher, F., et al. (2014). Genetic oscillations. A Doppler effect in embryonic pattern formation. Science 345, 222–225. doi:10.1126/SCIENCE.1253089
Sprinzak, D., Lakhanpal, A., Lebon, L., Santat, L. A., Fontes, M. E., Anderson, G. A., et al. (2010). Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86–90. doi:10.1038/NATURE08959
Stauber, M., Sachidanandan, C., Morgenstern, C., and Ish-Horowicz, D. (2009). Differential axial requirements for lunatic fringe and Hes7 transcription during mouse somitogenesis. PLoS One 4, e7996. doi:10.1371/JOURNAL.PONE.0007996
Stern, C. D., Fraser, S. E., Keynes, R. J., and Primmett, D. R. N. (1988). A cell lineage analysis of segmentation in the chick embryo. Development 104 (Suppl. l), 231–244. doi:10.1242/DEV.104.SUPPLEMENT.231
Stickney, H. L., Barresi, M. J. F., and Devoto, S. H. (2000). Somite development in zebrafish. Dev. Dyn. 219 (3), 287–303. doi:10.1002/1097-0177(2000)9999:9999<::AID-DVDY1065>3.0.CO;2-A
Stollewerk, A., Schoppmeier, M., and Damen, W. G. M. (2003). Involvement of Notch and Delta genes in spider segmentation. Nat 423, 863–865. doi:10.1038/nature01682
Suzuki, I. K., Gacquer, D., Van Heurck, R., Kumar, D., Wojno, M., Bilheu, A., et al. (2018). Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370–1384. doi:10.1016/J.CELL.2018.03.067
Swiatek, P. J., Lindsell, C. E., Del Amo, F. F., Weinmaster, G., and Gridley, T. (1994). Notch1 is essential for postimplantation development in mice. Genes Dev. 8, 707–719. doi:10.1101/GAD.8.6.707
Takahashi, Y., Inoue, T., Gossler, A., and Saga, Y. (2003). Feedback loops comprising Dll1, Dll3 and Mesp2, and differential involvement of Psen1 are essential for rostrocaudal patterning of somites. Development 130, 4259–4268. doi:10.1242/DEV.00629
Takashima, Y., Ohtsuka, T., González, A., Miyachi, H., and Kageyama, R. (2011). Intronic delay is essential for oscillatory expression in the segmentation clock. Proc. Natl. Acad. Sci. U. S. A. 108, 3300–3305. doi:10.1073/pnas.1014418108
Takke, C., and Campos-Ortega, J. A. (1999). her1, a zebrafish pair-rule like gene, acts downstream of notch signalling to control somite development. Development 126, 3005–3014. doi:10.1242/DEV.126.13.3005
Tam, P. P. L. (1981). The control of somitogenesis in mouse embryos. Development 65, 103–128. doi:10.1242/DEV.65.SUPPLEMENT.103
Taylor, J. S., Van de Peer, Y., Braasch, I., and Meyer, A. (2001). Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. Ser. B 356, 1661–1679. doi:10.1098/RSTB.2001.0975
Tenin, G., Wright, D., Ferjentsik, Z., Bone, R., McGrew, M. J., and Maroto, M. (2010). The chick somitogenesis oscillator is arrested before all paraxial mesoderm is segmented into somites. BMC Dev. Biol. 10, 24–12. doi:10.1186/1471-213X-10-24
Trofka, A., Schwendinger-Schreck, J., Brend, T., Pontius, W., Emonet, T., and Holley, S. A. (2012). The Her7 node modulates the network topology of the zebrafish segmentation clock via sequestration of the Hes6 hub. Development 139, 940–947. doi:10.1242/dev.073544
Tsiairis, C. D., and Aulehla, A. (2016). Self-organization of embryonic genetic oscillators into spatiotemporal wave patterns. Cell 164, 656–667. doi:10.1016/j.cell.2016.01.028
Tyser, R. C. V., Mahammadov, E., Nakanoh, S., Vallier, L., Scialdone, A., and Srinivas, S. (2021). Single-cell transcriptomic characterization of a gastrulating human embryo. Nature 600, 285–289. doi:10.1038/S41586-021-04158-Y
Umair, M., Younus, M., Shafiq, S., Nayab, A., and Alfadhel, M. (2022). Clinical genetics of spondylocostal dysostosis: a mini review. Front. Genet. 13, 996364. doi:10.3389/FGENE.2022.996364
Uriu, K., Liao, B. K., Oates, A. C., and Morelli, L. G. (2021). From local resynchronization to global pattern recovery in the zebrafish segmentation clock. Elife 10, e61358–e61391. doi:10.7554/ELIFE.61358
van den Brink, S. C., Alemany, A., van Batenburg, V., Moris, N., Blotenburg, M., Vivié, J., et al. (2020). Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nat 582, 405–409. doi:10.1038/s41586-020-2024-3
Van Eeden, F. J. M., Granato, M., Schach, U., Brand, M., Furutani-Seiki, M., Haffter, P., et al. (1996). Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development 123, 153–164. doi:10.1242/DEV.123.1.153
Van Eeden, F. J. M., Holley, S. A., Haffter, P., Nu¨sslein, C., and Nu¨sslein-Volhard, N. (1998). Zebrafish segmentation and pair-rule patterning. Dev. Genet. 23, 65–76. doi:10.1002/(sici)1520-6408(1998)23:1<65::aid-dvg7>3.3.co;2-v(1998)
Vargesson, N., Patel, K., Lewis, J., and Tickle, C. (1998). Expression patterns of Notch1, Serrate1, Serrate2 and Delta1 in tissues of the developing chick limb. Mech. Dev. 77, 197–199. doi:10.1016/S0925-4773(98)00138-5
Veenvliet, J. V., Bolondi, A., Kretzmer, H., Haut, L., Scholze-Wittler, M., Schifferl, D., et al. (2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science 370, eaba4937. doi:10.1126/science.aba4937
Venzin, O. F., and Oates, A. C. (2020). What are you synching about? Emerging complexity of Notch signaling in the segmentation clock. Dev. Biol. 460, 40–54. doi:10.1016/J.YDBIO.2019.06.024
Vibe, C. B. (2021). The temperature response of the medakasegmentation clock and its link to robustness in embryonic patterning. doi:10.11588/HEIDOK.00028769
Waddington, C. H., and Deuchar, E. M. (1953). Studies on the mechanism of meristic SegmentationI. The dimensions of somites. Development 1, 349–356. doi:10.1242/DEV.1.4.349
Wahi, K., Friesen, S., Coppola, V., and Cole, S. E. (2017). Putative binding sites for mir-125 family miRNAs in the mouse Lfng 3’UTR affect transcript expression in the segmentation clock, but mir-125a-5p is dispensable for normal somitogenesis. Dev. Dyn. 246, 740–748. doi:10.1002/DVDY.24552
Webb, A. B., Lengyel, I. M., Jörg, D. J., Valentin, G., Jülicher, F., Morelli, L. G., et al. (2016). Persistence, period and precision of autonomous cellular oscillators from the zebrafish segmentation clock. Elife 5, e08438. doi:10.7554/ELIFE.08438
Westin, J., and Lardelli, M. (1997). Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev. Genes Evol. 207, 51–63. doi:10.1007/S004270050091
Wiedermann, G., Bone, R. A., Silva, J. C., Bjorklund, M., Murray, P. J., and Dale, J. K. (2015). A balance of positive and negative regulators determines the pace of the segmentation clock. Elife 4, e05842. doi:10.7554/ELIFE.05842
William, D. A., Saitta, B., Gibson, J. D., Traas, J., Markov, V., Gonzalez, D. M., et al. (2007). Identification of oscillatory genes in somitogenesis from functional genomic analysis of a human mesenchymal stem cell model. Dev. Biol. 305, 172–186. doi:10.1016/J.YDBIO.2007.02.007
Williams, D. R., Shifley, E. T., Braunreiter, K. M., and Cole, S. E. (2016). Disruption of somitogenesis by a novel dominant allele of Lfng suggests important roles for protein processing and secretion. Dev 143, 822–830. doi:10.1242/DEV.128538
Williams, D. R., Shifley, E. T., Lather, J. D., and Cole, S. E. (2014). Posterior skeletal development and the segmentation clock period are sensitive to Lfng dosage during somitogenesis. Dev. Biol. 388, 159–169. doi:10.1016/J.YDBIO.2014.02.006
Williams, R., Nelson, L., Dowthwaite, G. P., Evans, D. J. R., and Archer, C. W. (2009). Notch receptor and Notch ligand expression in developing avian cartilage. J. Anat. 215, 159–169. doi:10.1111/J.1469-7580.2009.01089.X
Wilson, V., and Beddington, R. S. P. (1996). Cell fate and morphogenetic movement in the late mouse primitive streak. Mech. Dev. 55, 79–89. doi:10.1016/0925-4773(95)00493-9
Winkler, C., Elmasri, H., Klamt, B., Volff, J. N., and Gessler, M. (2003). Characterization of hey bHLH genes in teleost fish. Dev. Genes Evol. 213, 541–553. doi:10.1007/s00427-003-0360-6
Xiang, L., Yin, Y., Zheng, Y., Ma, Y., Li, Y., Zhao, Z., et al. (2019). A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nat 577, 537–542. doi:10.1038/s41586-019-1875-y
Xue, Y., Gao, X., Lindsell, C. E., Norton, C. R., Chang, B., Hicks, C., et al. (1999). Embryonic lethality and vascular defects in mice lacking the notch ligand Jagged1. Hum. Mol. Genet. 8, 723–730. doi:10.1093/HMG/8.5.723
Yamanaka, Y., Hamidi, S., Yoshioka-Kobayashi, K., Munira, S., Sunadome, K., Zhang, Y., et al. (2022). Reconstituting human somitogenesis in vitro. Nat 614, 509–520. doi:10.1038/s41586-022-05649-2
Yang, L. T., Nichols, J. T., Yao, C., Manilay, J. O., Robey, E. A., and Weinmaster, G. (2005). Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol. Biol. Cell 16, 927–942. doi:10.1091/MBC.E04-07-0614
Yoneya, T., Tahara, T., Nagao, K., Yamada, Y., Yamamoto, T., Osawa, M., et al. (2001). Molecular cloning of Delta-4, a new mouse and human Notch ligand. J. Biochem. 129, 27–34. doi:10.1093/OXFORDJOURNALS.JBCHEM.A002832
Yoon, J. K., and Wold, B. (2000). The bHLH regulator pMesogenin1 is required for maturation and segmentation of paraxial mesoderm. Genes Dev. 14, 3204–3214. doi:10.1101/GAD.850000
Yoshikawa, Y., Fujimori, T., McMahon, A. P., and Takada, S. (1997). Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev. Biol. 183, 234–242. doi:10.1006/DBIO.1997.8502
Yoshioka-Kobayashi, K., Matsumiya, M., Niino, Y., Isomura, A., Kori, H., Miyawaki, A., et al. (2020). Coupling delay controls synchronized oscillation in the segmentation clock. Nat, 119–123. doi:10.1038/s41586-019-1882-z
Zeng, B., Liu, Z., Lu, Y., Zhong, S., Qin, S., Huang, L., et al. (2023). The single-cell and spatial transcriptional landscape of human gastrulation and early brain development. Cell Stem Cell 30, 851–866.e7. doi:10.1016/J.STEM.2023.04.016
Zhang, N., and Gridley, T. (1998). Defects in somite formation in lunatic fringe-deficient mice. Nature 394, 374–377. doi:10.1038/28625
Zhou, B., Lin, W., Long, Y., Yang, Y., Zhang, H., Wu, K., et al. (2022). Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 71 (7), 95–33. doi:10.1038/s41392-022-00934-y
Zhou, M., Yan, J., Ma, Z., Zhou, Y., Abbood, N. N., Liu, J., et al. (2012). Comparative and evolutionary analysis of the HES/HEY gene family reveal exon/intron loss and teleost specific duplication events. PLoS One 7, e40649. doi:10.1371/JOURNAL.PONE.0040649
Zinani, O. Q. H., Keseroğlu, K., Ay, A., and Özbudak, E. M. (2020). Pairing of segmentation clock genes drives robust pattern formation. Nat 589, 431–436. doi:10.1038/s41586-020-03055-0
Keywords: Notch signailing pathway, segmentation clock, gene oscillation, presomitic mesoderm (PSM), somitogenesis, somite
Citation: Ramesh PS and Chu L-F (2024) Species-specific roles of the Notch ligands, receptors, and targets orchestrating the signaling landscape of the segmentation clock. Front. Cell Dev. Biol. 11:1327227. doi: 10.3389/fcell.2023.1327227
Received: 24 October 2023; Accepted: 20 December 2023;
Published: 29 January 2024.
Edited by:
Aimin Liu, The Pennsylvania State University (PSU), United StatesReviewed by:
Ildiko M. L. Somorjai, University of St Andrews, United KingdomCopyright © 2024 Ramesh and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Fang Chu, bGlmYW5namFjay5jaHVAdWNhbGdhcnkuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.