
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 13 December 2023
Sec. Molecular and Cellular Pathology
Volume 11 - 2023 | https://doi.org/10.3389/fcell.2023.1321995
N6-methyladenosine (m6A) represents the most abundant modification of messenger RNA (mRNA) and is regulated by methyltransferases (writers), demethylases (erasers), and m6A-binding proteins (readers). A dynamic modification process is implicated in nearly every critical stage of RNA metabolism, including mRNA stability, transcription, translation, splicing, nuclear export, and decay. Notably, m6A methylation is significantly enriched in the brain and has recently been shown to be associated with neurodevelopmental disorders and the development of Parkinson’s disease (PD). In this review, we summarize the proteins involved in the process of m6A modification and elucidate the emerging role of m6A modification in PD, which could illuminate alternative strategies for the prevention and treatment of PD.
Epigenetics is a form of stable inheritance that does not change the basic sequence of the DNA and includes DNA methylation, histone modification, and RNA modification of both mRNA and non-coding RNA (ncRNA). Compared to DNA methylation and histone post-translational modification, RNA modification has been less thoroughly studied. However, the recent development of RNA sequencing technology has fostered increased research into RNA epigenetics, and more than 170 RNA modifications have been discovered (Wiener and Schwartz, 2021). These modifications are mainly m1A, m5C, m6A, m7G, etc. N6-methyladenosine (m6A) was initially identified in 1974. It is considered the most common internal transcriptional modification, especially in eukaryotic mRNA (Desrosiers et al., 1974; Dubin and Taylor, 1975; Fang et al., 2021).
The methylation of m6A describes the addition of a methyl group at the sixth nitrogen position of adenine and is recognized as a dynamic, reversible modification process (Jia et al., 2011). It is regulated by methyltransferases, demethylases, and m6A-binding proteins, called writers, erasers, and readers, respectively. M6A methylation occurs in various RNA species, including mRNA, tRNA, rRNA, small nuclear RNA, microRNA precursors, and long non-coding RNA (Punekar et al., 2013; Du et al., 2018; van Tran et al., 2019). The dynamic modification of m6A occurs in nearly all stages of RNA metabolism, including mRNA stability, transcription, translation, splicing, nuclear export, and decay (Alarcón et al., 2015a; Aguilo et al., 2015; Meyer et al., 2015; Yue et al., 2015; Du et al., 2016; Yoon et al., 2017; Zhao et al., 2017).
Parkinson’s disease (PD) is a chronic neurodegenerative disease that predominately affects the motor nervous system. Its key clinical symptoms include resting tremors, bradykinesia, myotonia, and postural imbalance. The pivotal pathological changes observed in PD are the degeneration and subsequent death of dopaminergic neurons in the substantia nigra (Lotankar et al., 2017).
M6A-specific methylated RNA immunoprecipitation (MeRIP) has revealed abundant m6A modifications in the brain (Meyer et al., 2012). As a result, an increasing number of studies have investigated the functional significance of m6A modification in the nervous system and its effects on normal physiology. In this review, we will summarize the enzyme proteins that contribute to the process of m6A modification and explore the emerging role and biological significance of m6A modification in PD. This will ultimately provide new insights into the diagnosis and treatment of PD.
M6A methylation is mainly catalyzed by a methyltransferase complex (MTC), which includes METTL3, METTL14, WTAP, VIRMA/KIAA1429, RBM15, ZC3H13, and Hakai (Figure 1). METTL3, which is highly conserved in eukaryotes, was the first m6A methyltransferase to be discovered and is the most critical core component of the MTC, serving a catalytic function (Bokar et al., 1997; Geula et al., 2015). METTL14 principally acts as an RNA binding scaffold, stabilizing the MTC structure and enhancing METTL3’s catalytic activity. WTAP, a regulatory subunit of the RNA methyltransferase complex, links METTL3 to METTL14 and facilitates the dimer’s localization (Wang Y. et al., 2014; Liu et al., 2014; Ping et al., 2014). VIRMA/KIAA1429 recruits and guides the catalytic core methyltransferase components (METTL3/METTL14/WTAP) to specific RNA regions for m6A methylation (Yue et al., 2018). RBM15 and RBM15B, although they lack catalytic function, can bind to METTL3 and WTAP, directing these two proteins to specific RNA sites for m6A modification (Patil et al., 2016). ZC3H13 primarily promotes MTC’s binding to RNA, and its interaction with WTAP can substantially enhance the MTC’s catalytic function, regulating RNA m6A methylation in the nucleus (Wen et al., 2018). Although Hakai is less well-studied, it is also part of the m6A biogenesis mechanism in vertebrates and plants. In mammalian cells, Hakai strongly interacts with WTAP, and studies indicate that it is a core member of the m6A-modified protein family and an essential component of the MTC in Drosophila and human cells (Horiuchi et al., 2013; Bawankar et al., 2021).
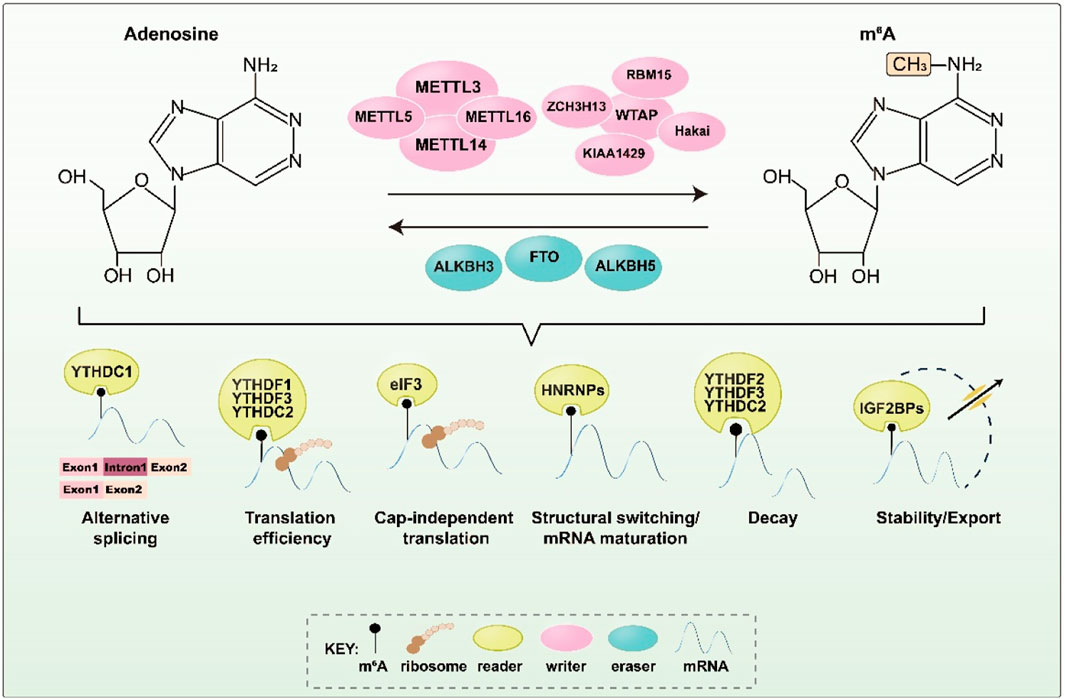
FIGURE 1. Key proteins and corresponding functions during m6A methylation. M6A methylation is regulated by m6A methyltransferase, demethylase, and m6A binding protein to perform a variety of biological functions. It affects critical stages of RNA metabolism and downstream gene expression regulation.
In addition, METTL16 and METTL5 perform m6A methylation in a non-MTC-dependent manner. METTL6, a conserved U6 snRNA methyltransferase, controls SAM homeostasis by post-transcriptionally regulating the expression of SAM synthase genes (Pendleton et al., 2017). METTL5 is mainly responsible for catalyzing m6A modification on 18S rRNA (van Tran et al., 2019; Lei et al., 2023).
The discovery of demethylases has proven that m6A methylation is a dynamic and reversible modification process. The currently recognized m6A demethylases are mainly FTO, ALKBH5, and ALKBH3.
FTO, also known as fat mass and obesity-associated protein, was the first m6A demethylase to be discovered and showed potential in regulating selective splicing and 3′ end mRNA processing and translation (Jia et al., 2011; Bartosovic et al., 2017; Zhang et al., 2019). FTO can catalyze the demethylation of both m6A and m6A.m., with preferences that are likely influenced by its subcellular localization (Mauer et al., 2017; Wei et al., 2018). In contrast, ALKBH5 (alkylation protein AlkB homolog 5), the second identified m6A demethylase, exhibits no activity toward m6A.m. substrates. It significantly influences mRNA output and RNA metabolism by reducing the level of m6A in nuclear speckles (Zheng et al., 2013). Both FTO and ALKBH5 catalyze the demethylation of m6A through an Fe(II)- and α-ketoglutarate-dependent mechanism, initially oxidizing m6A to N6-hydroxymethyladenosine (hm6A) before converting hm6A to N6-formyladenosine (f6A) and, eventually, transforming f6A into adenosine (A) to complete the demethylation process. Recent studies have reported that another AlkB family homolog, ALKBH3, also facilitates m6A demethylation in tRNA and enhances protein translation efficiency during cancer cell proliferation (Ueda et al., 2017).
The m6A reader mainly contributes to RNA recruitment. The YTH domain family, including YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2, are the first identified m6A readers. They bind directly to m6A through a common YTH domain (Dominissini et al., 2012). YTHDF1 promotes translation by elevating ribosome occupancy, recruiting translation initiation complex eukaryotic initiation factor 3 (eIF3), or working with YTHDF3 (Meyer et al., 2015; Wang et al., 2015; Shi et al., 2017; Lin et al., 2019). YTHDF2 contributes to mRNA stability and influences mRNA decay, while YTHDF3 affects the decay of methylated mRNA mediated by YTHDF2 (Wang X. et al., 2014; Shi et al., 2017; Fei et al., 2020). In the nucleus, YTHDC1 affects mRNA splicing and nuclear export, as well as mediates transcriptional repression by interacting with m6A-modified long non-coding RNA (lncRNA). Notably, YTHDC1 recognizes m6A-modified XIST to promote XIST-mediated X-chromosome silencing (Xu et al., 2014; Patil et al., 2016; Xiao et al., 2016; Roundtree et al., 2017). YTHDC2, which possesses 3'→5′ RNA helicase activity, selectively binds to m6A on its consensus motif and improves the translation efficiency of its target in spermatogenesis (Hsu et al., 2017; Wojtas et al., 2017).
In addition to the YTH domain family, heterogeneous nuclear ribonucleoproteins (HNRNPs), including HNRNPA2B1, HNRNPC, and HNRNPG, can also act as m6A readers. HNRNPA2B1 accelerates the processing of primary miRNA (pri-miRNA) by interacting with the DGCR8 protein in an m6A-dependent manner and regulates the alternative splicing of transcripts (Alarcón et al., 2015a; Alarcón et al., 2015b). Both HNRNPC and HNRNPG can modulate mRNA abundance and splicing after recognizing m6A (Zarnack et al., 2013; Liu et al., 2015; Liu et al., 2017). Moreover, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), including IGF2BP1/2/3, enhance mRNA stability and translation efficiency in an m6A-dependent manner by recognizing the GG (m6A) C motif (Huang et al., 2018). FMRP, another m6A reader, can specifically bind to m6A-modified RNA and interact with CRM1 to promote the nuclear export of these RNAs, thereby regulating gene expression and influencing the development of neural stem cells and the nervous system (Edens et al., 2019).
PD is a multifactorial neurodegenerative disease, the etiology of which has not been fully clarified to date, and no definitive and reliable clinical or testing tools currently exist to determine its cause. PD may be related to the interaction between age, environmental, and genetic factors.
Age is the greatest risk factor for developing PD. The prevalence of PD increases exponentially with age; 2%–3% of the population over 65 years of age is affected by PD (Pang et al., 2019; Kumar et al., 2022). In addition, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), can induce typical Parkinson’s disease in both humans and primates. Certain environmental substances, including pesticides, herbicides, and specific industrial chemicals, share structural similarities with MPTP. Consequently, the environment could be an etiological factor in PD (Herrero et al., 1993; Nonnekes et al., 2018). Moreover, genetic factors significantly influence the development of PD, with data suggesting that genetic variation is found in 5%–10% of people with PD. In a comprehensive analysis using a large population-based twin registry, the heritability of PD for an age of diagnosis of less than 50 years was estimated at 0.83 (Goldman et al., 2019; Uwishema et al., 2022). Recently, genome-wide analyses of clinical cases of PD patients have identified a new set of PD-associated genes, including ANK2, DNAH1, and STAB1 (Yang et al., 2023).
The two main hallmarks of PD pathophysiology are the accumulation of misfolded α-synuclein (α-Syn) and the decline of dopaminergic neurons in the substantia nigra (SN). α-Syn, a soluble protein that is predominantly found in the presynaptic and perinuclear regions of the central nervous system, is believed to be integral to cell membrane processes. Normal α-Syn exists as a single intrinsically disordered protein within the healthy brain (Fakhree et al., 2018). However, the misfolding and aggregation of α-Syn monomers results in the formation of pathological oligomers and protofibrils within neurons, which are associated with the progression of PD (Kalia et al., 2013).
Dopaminergic neurons are responsible for the storage and release of dopamine as a neurotransmitter and represent a widely distributed class of neurons within the brain that are crucially implicated in the regulation of significant physiological functions. The degeneration of nigrostriatal dopaminergic neurons is recognized as a central feature of PD, which is triggered by various factors, including mitochondrial dysfunction, Lewy body accumulation, neuroinflammation, excitotoxicity, and metal accumulation (Vallone et al., 2000).
Chen et al. established cellular and rat models of PD using 6-hydroxydopamine (6-OHDA) and observed significantly reduced m6A levels in the striatal regions of the PD rat model and the PD cellular model. They found that ALKBH5 was notably elevated in the striatum of the PD brain, while FTO remained unchanged. Conversely, the PD cellular model exhibited upregulated FTO expression but no significant difference in ALKBH5. FTO knockdown increased m6A levels, subsequently inhibiting GRIN1 expression and reducing glutamate binding to receptors, thereby mitigating neurotoxicity (Chen et al., 2019).
Similarly, Geng et al. observed reduced total m6A levels and increased FTO expression in both an MPTP-treated PD mouse model and an MPP + induced-PD MN9D cell model. They suggested that FTO affects ATM expression by influencing the stability of ATM mRNA in dopaminergic neurons. FTO knockdown inhibited ATM expression, suppressing the upregulation of α-Syn and the downregulation of tyrosine hydroxylase (TH), thereby alleviating dopaminergic neuron death in vitro in a PD model (Geng et al., 2023).
Furthermore, FTO regulation appears to be closely linked to the control of neurotransmitter dopamine transmission (Hess et al., 2013). FTO inactivation may impair dopamine receptor type 2 (D2R) and type 3 (D3R)-dependent control of neuronal activity and behavioral response. FTO is thought to target GNAO1, GRIN1, and SYN1 to facilitate the D2R–D3R–G protein-coupled inward rectifier potassium signaling (GIRK) cascade, thereby promoting dopamine signal transduction through D2R and D3R signals. In addition, Qiu et al. identified five m6A-SNPs associated with PD, three of which were located in the ALKBH5 gene, suggesting that m6A-SNPs may contribute to the risk of PD (Qiu et al., 2020).
He et al. compared m6A levels in peripheral blood mononuclear cells (PBMCs) from PD patients and healthy individuals. Their study revealed significantly reduced m6A levels in PD patients compared to healthy participants. The expression of m6a-related proteins, including METTL3, METTL14, and YTHDF2, was also notably downregulated. Further research indicated that METTL14 influences the stability of α-Syn mRNA and modulates α-Syn expression in an m6A-dependent manner, potentially offering valuable diagnostic information for PD (He et al., 2023).
Yu et al. established PD mice via MPTP and confirmed the aberrant expression of certain m6A-related proteins in the substantia nigra and striatum. They observed significantly reduced mRNA expression of RBM15b and YTHDF1 in the substantia nigra of PD mice, while IGF2BP1 expression was elevated. Moreover, in the striatum, the expression of RBM15, HNRNPG, METTL3, YTHDF1, HNRNPC, IGF2BP3, and RBM15 was markedly downregulated (Yu et al., 2022).
Furthermore, Gong et al. demonstrated that GLRX overexpression attenuated motor dysfunction and dopamine neuron degeneration in PD mice. They proposed that IGF2BP2 enhances GLRX mRNA stability in an m6A-dependent manner, whereas the knockdown of METTL3 substantially reduces the presence of GLRX mRNA enriched by an IGF2BP2-specific antibody. The study strongly associated m6A methylation modification with the progression of PD in mice (Gong et al., 2023).
Koranda et al. discovered that METTL14 deficiency leads to reduced m6A levels in the striatum without altering cell numbers or morphology. This deficiency also increased neuronal excitability and enhanced striatal sensitivity to dopamine agonist (DA) drugs, suggesting m6A’s essential role in maintaining striatal function and learning ability in adult mice (Koranda et al., 2018).
Additionally, Quan et al.’s data analysis suggested that HNRNPC might contribute to PD pathogenesis by inhibiting the proliferation of dopaminergic neurons, promoting their apoptosis, and inducing immune inflammation (Quan et al., 2021).
Overall, the emerging evidence indicates that m6A modifications play a pivotal role in the development of PD, presenting potential opportunities to prevent and treat this neurodegenerative disorder (Table 1).
As one of the most prevalent internal RNA modifications in eukaryotic mRNA, m6A methylation has emerged as a critical regulator of neuronal development and the pathogenesis of PD. This revelation offers a fresh perspective on the epigenetic regulation underlying Parkinson’s disease. Despite recent advancements, the significance of m6A modification in PD remains poorly understood, with several questions yet to be answered. For instance, the variations in m6A levels during different phases of PD development, the mechanisms responsible for these variations, and the correlations between m6A expression disparities in various tissue systems and the diversity of m6A functions remain largely unknown. Addressing these queries in future studies is crucial to unraveling the potential regulatory role of m6A in PD.
A comprehensive understanding of the role that m6A methylation plays in physiological homeostasis and disease, along with a deep exploration of the regulatory mechanisms governing the expression and function of m6A-associated proteins, is imperative. Components of these pathways may offer promising therapeutic targets for PD treatment. Delving into the intricacies of m6A modifications as PD progresses will also contribute to the investigation of PD-targeting therapies. The recognition of the critical role of m6A methylation in PD signifies a promising avenue for disease diagnosis and treatment, revealing new possibilities for future research.
JZ: Writing–original draft. YH: Writing–review and editing. RH: Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the National Natural Science Foundation of China (No. 32200466).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilo, F., Zhang, F., Sancho, A., Fidalgo, M., Di Cecilia, S., Vashisht, A., et al. (2015). Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17 (6), 689–704. doi:10.1016/j.stem.2015.09.005
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015a). HNRNPA2B1 is a mediator of m6a-dependent nuclear RNA processing events. Cell 162 (6), 1299–1308. doi:10.1016/j.cell.2015.08.011
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519 (7544), 482–485. doi:10.1038/nature14281
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 45 (19), 11356–11370. doi:10.1093/nar/gkx778
Bawankar, P., Lence, T., Paolantoni, C., Haussmann, I. U., Kazlauskiene, M., Jacob, D., et al. (2021). Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat. Commun. 12 (1), 3778. doi:10.1038/s41467-021-23892-5
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna 3 (11), 1233–1247.
Chen, X., Yu, C., Guo, M., Zheng, X., Ali, S., Huang, H., et al. (2019). Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 10 (5), 2355–2363. doi:10.1021/acschemneuro.8b00657
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. doi:10.1038/nature11112
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626. doi:10.1038/ncomms12626
Du, Y., Hou, G., Zhang, H., Dou, J., He, J., Guo, Y., et al. (2018). SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 46 (10), 5195–5208. doi:10.1093/nar/gky156
Dubin, D. T., and Taylor, R. H. (1975). The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 2 (10), 1653–1668. doi:10.1093/nar/2.10.1653
Edens, B. M., Vissers, C., Su, J., Arumugam, S., Xu, Z., Shi, H., et al. (2019). FMRP modulates neural differentiation through m6a-dependent mRNA nuclear export. Cell Rep. 28 (4), 845–854. doi:10.1016/j.celrep.2019.06.072
Fakhree, M. A. A., Nolten, I. S., Blum, C., and Claessens, M. (2018). Different conformational subensembles of the intrinsically disordered protein α-synuclein in cells. J. Phys. Chem. Lett. 9 (6), 1249–1253. doi:10.1021/acs.jpclett.8b00092
Fang, F., Wang, X., Li, Z., Ni, K., and Xiong, C. (2021). Epigenetic regulation of mRNA N6-methyladenosine modifications in mammalian gametogenesis. Mol. Hum. Reprod. 27 (5), gaab025. doi:10.1093/molehr/gaab025
Fei, Q., Zou, Z., Roundtree, I. A., Sun, H. L., and He, C. (2020). YTHDF2 promotes mitotic entry and is regulated by cell cycle mediators. PLoS Biol. 18 (4), e3000664. doi:10.1371/journal.pbio.3000664
Geng, Y., Long, X., Zhang, Y., Wang, Y., You, G., Guo, W., et al. (2023). FTO-targeted siRNA delivery by MSC-derived exosomes synergistically alleviates dopaminergic neuronal death in Parkinson's disease via m6A-dependent regulation of ATM mRNA. J. Transl. Med. 21 (1), 652. doi:10.1186/s12967-023-04461-4
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347 (6225), 1002–1006. doi:10.1126/science.1261417
Goldman, S. M., Marek, K., Ottman, R., Meng, C., Comyns, K., Chan, P., et al. (2019). Concordance for Parkinson's disease in twins: a 20-year update. Ann. Neurol. 85 (4), 600–605. doi:10.1002/ana.25441
Gong, X., Huang, M., and Chen, L. (2023). NRF1 mitigates motor dysfunction and dopamine neuron degeneration in mice with Parkinson's disease by promoting GLRX m6 A methylation through upregulation of METTL3 transcription. CNS Neurosci. Ther. doi:10.1111/cns.14441
He, H., Zhang, Q., Liao, J., Lei, J., Luo, M., Huang, J., et al. (2023). METTL14 is decreased and regulates m6 A modification of α-synuclein in Parkinson's disease. J. Neurochem. 166 (3), 609–622. doi:10.1111/jnc.15882
Herrero, M. T., Hirsch, E. C., Kastner, A., Ruberg, M., Luquin, M. R., Laguna, J., et al. (1993). Does neuromelanin contribute to the vulnerability of catecholaminergic neurons in monkeys intoxicated with MPTP? Neuroscience 56 (2), 499–511. doi:10.1016/0306-4522(93)90349-k
Hess, M. E., Hess, S., Meyer, K. D., Verhagen, L. A., Koch, L., Brönneke, H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16 (8), 1042–1048. doi:10.1038/nn.3449
Horiuchi, K., Kawamura, T., Iwanari, H., Ohashi, R., Naito, M., Kodama, T., et al. (2013). Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288 (46), 33292–33302. doi:10.1074/jbc.M113.500397
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27 (9), 1115–1127. doi:10.1038/cr.2017.99
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20 (3), 285–295. doi:10.1038/s41556-018-0045-z
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. doi:10.1038/nchembio.687
Kalia, L. V., Kalia, S. K., McLean, P. J., Lozano, A. M., and Lang, A. E. (2013). α-Synuclein oligomers and clinical implications for Parkinson disease. Ann. Neurol. 73 (2), 155–169. doi:10.1002/ana.23746
Koranda, J. L., Dore, L., Shi, H., Patel, M. J., Vaasjo, L. O., Rao, M. N., et al. (2018). Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99 (2), 283–292. doi:10.1016/j.neuron.2018.06.007
Kumar, S., Goyal, L., and Singh, S. (2022). Tremor and rigidity in patients with Parkinson's disease: emphasis on epidemiology, pathophysiology and contributing factors. CNS Neurol. Disord. Drug Targets 21 (7), 596–609. doi:10.2174/1871527320666211006142100
Lei, K., Lin, S., and Yuan, Q. (2023). N6-methyladenosine (m6A) modification of ribosomal RNAs (rRNAs): critical roles in mRNA translation and diseases. Genes Dis. 10 (1), 126–134. doi:10.1016/j.gendis.2021.10.005
Lin, X., Chai, G., Wu, Y., Li, J., Chen, F., Liu, J., et al. (2019). RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 10 (1), 2065. doi:10.1038/s41467-019-09865-9
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. doi:10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. doi:10.1038/nature14234
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. doi:10.1093/nar/gkx141
Lotankar, S., Prabhavalkar, K. S., and Bhatt, L. K. (2017). Biomarkers for Parkinson's disease: recent advancement. Neurosci. Bull. 33 (5), 585–597. doi:10.1007/s12264-017-0183-5
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature 541 (7637), 371–375. doi:10.1038/nature21022
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5' UTR m6A promotes cap-independent translation. Cell 163 (4), 999–1010. doi:10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149 (7), 1635–1646. doi:10.1016/j.cell.2012.05.003
Nonnekes, J., Post, B., Tetrud, J. W., Langston, J. W., and Bloem, B. R. (2018). MPTP-induced parkinsonism: an historical case series. Lancet Neurol. 17 (4), 300–301. doi:10.1016/s1474-4422(18)30072-3
Pang, S. Y., Ho, P. W., Liu, H. F., Leung, C. T., Li, L., Chang, E. E. S., et al. (2019). The interplay of aging, genetics and environmental factors in the pathogenesis of Parkinson's disease. Transl. Neurodegener. 8, 23. doi:10.1186/s40035-019-0165-9
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. doi:10.1038/nature19342
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169 (5), 824–835. doi:10.1016/j.cell.2017.05.003
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 (2), 177–189. doi:10.1038/cr.2014.3
Punekar, A. S., Liljeruhm, J., Shepherd, T. R., Forster, A. C., and Selmer, M. (2013). Structural and functional insights into the molecular mechanism of rRNA m6A methyltransferase RlmJ. Nucleic Acids Res. 41 (20), 9537–9548. doi:10.1093/nar/gkt719
Qiu, X., He, H., Huang, Y., Wang, J., and Xiao, Y. (2020). Genome-wide identification of m6A-associated single-nucleotide polymorphisms in Parkinson's disease. Neurosci. Lett. 737, 135315. doi:10.1016/j.neulet.2020.135315
Quan, W., Li, J., Liu, L., Zhang, Q., Qin, Y., Pei, X., et al. (2021). Influence of N6-methyladenosine modification gene HNRNPC on cell phenotype in Parkinson's disease. Park. Dis. 2021, 9919129. doi:10.1155/2021/9919129
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 6, e31311. doi:10.7554/eLife.31311
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27 (3), 315–328. doi:10.1038/cr.2017.15
Ueda, Y., Ooshio, I., Fusamae, Y., Kitae, K., Kawaguchi, M., Jingushi, K., et al. (2017). AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 7, 42271. doi:10.1038/srep42271
Uwishema, O., Onyeaka, H., Badri, R., Yücel, A. N., Korkusuz, A. K., Ajagbe, A. O., et al. (2022). The understanding of Parkinson's disease through genetics and new therapies. Brain Behav. 12 (5), e2577. doi:10.1002/brb3.2577
Vallone, D., Picetti, R., and Borrelli, E. (2000). Structure and function of dopamine receptors. Neurosci. Biobehav Rev. 24 (1), 125–132. doi:10.1016/s0149-7634(99)00063-9
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47 (15), 7719–7733. doi:10.1093/nar/gkz619
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161 (6), 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 (2), 191–198. doi:10.1038/ncb2902
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71 (6), 973–985. doi:10.1016/j.molcel.2018.08.011
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69 (6), 1028–1038. doi:10.1016/j.molcel.2018.02.015
Wiener, D., and Schwartz, S. (2021). The epitranscriptome beyond m6A. Nat. Rev. Genet. 22 (2), 119–131. doi:10.1038/s41576-020-00295-8
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., and Pillai, R. S. (2017). Regulation of m6A transcripts by the 3'→5' RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 68 (2), 374–387. doi:10.1016/j.molcel.2017.09.021
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61 (4), 507–519. doi:10.1016/j.molcel.2016.01.012
Xu, C., Wang, X., Liu, K., Roundtree, I. A., Tempel, W., Li, Y., et al. (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10 (11), 927–929. doi:10.1038/nchembio.1654
Yang, J., Wu, X., and Song, Y. (2023). Recent advances in novel mutation genes of Parkinson's disease. J. Neurol. 270 (8), 3723–3732. doi:10.1007/s00415-023-11781-4
Yoon, K. J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171 (4), 877–889. doi:10.1016/j.cell.2017.09.003
Yu, Z., Huang, L., Xia, Y., Cheng, S., Yang, C., Chen, C., et al. (2022). Analysis of m6A modification regulators in the substantia nigra and striatum of MPTP-induced Parkinson's disease mice. Neurosci. Lett. 791, 136907. doi:10.1016/j.neulet.2022.136907
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4, 10. doi:10.1038/s41421-018-0019-0
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29 (13), 1343–1355. doi:10.1101/gad.262766.115
Zarnack, K., König, J., Tajnik, M., Martincorena, I., Eustermann, S., Stévant, I., et al. (2013). Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 152 (3), 453–466. doi:10.1016/j.cell.2012.12.023
Zhang, X., Wei, L. H., Wang, Y., Xiao, Y., Liu, J., Zhang, W., et al. (2019). Structural insights into FTO's catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. U. S. A. 116 (8), 2919–2924. doi:10.1073/pnas.1820574116
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18 (1), 31–42. doi:10.1038/nrm.2016.132
Keywords: N6-methylAdenosine (m6A), RNA methylation, methyltransferase, epigenetics, neurodevelopment, Neurological disease
Citation: Zhou J, Han Y and Hou R (2023) Potential role of N6-methyladenosine modification in the development of Parkinson’s disease. Front. Cell Dev. Biol. 11:1321995. doi: 10.3389/fcell.2023.1321995
Received: 15 October 2023; Accepted: 04 December 2023;
Published: 13 December 2023.
Edited by:
Krishna Bhat, University of South Florida, United StatesReviewed by:
Partha Sarathi Sarkar, University of Texas Medical Branch at Galveston, United StatesCopyright © 2023 Zhou, Han and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruizhe Hou, aG91cnVpemhlamxkeEBqbHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.