- 1Department of Anatomy and Cell Biology, George Washington University, School of Medicine and Health Sciences, Washington, DC, United States
- 2Department of Veterinary and Animal Sciences, University of Massachusetts, Amherst, MA, United States
Introduction: The Six1 transcription factor plays important roles in the development of cranial sensory organs, and point mutations underlie craniofacial birth defects. Because Six1’s transcriptional activity can be modulated by interacting proteins, we previously screened for candidate interactors and identified zinc-finger MYM-containing protein 4 (Zmym4) by its inclusion of a few domains with a bona fide cofactor, Sine oculis binding protein (Sobp). Although Zmym4 has been implicated in regulating early brain development and certain cancers, its role in craniofacial development has not previously been described.
Methods: We used co-immunoprecipitation and luciferase-reporter assays in cultured cells to test interactions between Zmym4 and Six1. We used knock-down and overexpression of Zmym4 in embryos to test for its effects on early ectodermal gene expression, neural crest migration and craniofacial cartilage formation.
Results: We found no evidence that Zmym4 physically or transcriptionally interacts with Six1 in cultured cells. Nonetheless, knockdown of endogenous Zmym4 in embryos resulted in altered early cranial gene expression, including those expressed in the neural border, neural plate, neural crest and preplacodal ectoderm. Experimentally increasing Zmym4 levels had minor effects on neural border or neural plate genes, but altered the expression of neural crest and preplacodal genes. At larval stages, genes expressed in the otic vesicle and branchial arches showed reduced expression in Zmym4 morphants. Although we did not detect defects in neural crest migration into the branchial arches, loss of Zmym4 resulted in aberrant morphology of several craniofacial cartilages.
Discussion: Although Zmym4 does not appear to function as a Six1 transcriptional cofactor, it plays an important role in regulating the expression of embryonic cranial genes in tissues critical for normal craniofacial development.
1 Introduction
Six1 is a key transcriptional regulator of many developmental processes and several single nucleotide mutations are causative for Branchio-oto-renal syndrome (BOR) and Deafness, autosomal dominant 23 (DFNA23) (reviewed in (Moody and Saint-Jeannet, 2014; Moody and LaMantia, 2015; Riddiford and Schlosser, 2016; Smith, 2018; Streit, 2018; Schlosser, 2021)). Six1 can function as either a transcriptional activator or repressor depending upon the binding of different cofactors to its protein interaction domain (Silver et al., 2003; Brugmann et al., 2004; Anderson et al., 2012). Previously, we identified zinc-finger MYM-containing 4 (Zmym4) as a candidate Six1 cofactor (Neilson et al., 2010), based on structural similarities to Box2 and Box3 of Sine oculis-binding protein (Sobp), a cofactor discovered in Drosophila that also is expressed in frogs and mice (Kenyon et al., 2005; Tavares et al., 2021). While a closely related MYM-containing protein (Zmym2) interacts with Six4 during renal development (Connaughton et al., 2020), nothing is known about possible interactions between Zmym4 and Six transcription factors, including Six1, or whether Zmym4 is involved in craniofacial development.
The function of ZMYM4 has only recently begun to be investigated in human health. Mutations in ZMYM4 are associated with severe childhood obesity (Greenhill, 2020; Marenne et al., 2020; Regan and Shah, 2020), schizophrenia (Xiang et al., 2020), heritable substance use disorders (Zhang et al., 2022), heritable sleep disorders (Spada et al., 2016), and cancers (Goyal et al., 2017; Goyal et al., 2017; Goyal et al., 2019; Goyal et al., 2021; Moon et al., 2021; Yu et al., 2023). Despite keen interest in the newly identified role of ZMYM4 in human disease, little is known whether it has a role in early developmental processes, despite its abundant expression, particularly in craniofacial anlagen, during embryogenesis (Neilson et al., 2010).
During Xenopus development, Zmym4 is expressed in the neural plate and neural tube, preplacodal ectoderm (PPE) and cranial sensory placodes, neural crest (NC), branchial arches (BAs), somites and nephric mesoderm (Neilson et al., 2010). Similar expression patterns have been described at E10.5 in the mouse (Gray et al., 2004) and in the E15.5 mouse genitourinary tract (GUDMAP; www.gudmap.org, 2004). These tissues also express Six1 during development (reviewed in (Moody and Saint-Jeannet, 2014; Moody and LaMantia, 2015; Riddiford and Schlosser, 2016; Streit, 2018)), suggesting that Zmym4 and Six1 may have the opportunity to interact during critical periods of craniofacial development. Since Zmym4 contains two domains with homology to Drosophila Sobp (Box2 and Box3; (Kenyon et al., 2005)) and Xenopus Sobp interacts with Six1 and is required for craniofacial development (Tavares et al., 2021), we assessed whether Zmym4 also interacts with Six1 and/or modifies its transcriptional activity.
Herein, we show by assays in HEK293T cells that Zmym4 does not co-immunoprecipitate or transcriptionally interact with Six1. Nonetheless, knockdown of endogenous Zmym4 in embryos resulted in altered early cranial gene expression, including genes expressed in the neural border (NB), neural plate (NP), NC and PPE. Experimentally increasing Zmym4 levels had minor effects on NB or NP genes, but altered the expression of NC and PPE genes. At larval stages, genes expressed in the otic vesicle and BAs showed reduced expression in Zmym4 morphants. Although we did not detect defects in the migration of NC cells into the BAs, loss of Zmym4 resulted in aberrant morphology of several craniofacial cartilages. Thus, although Zmym4 does not appear to function as a Six1 transcriptional cofactor, herein we show that it plays an important role in regulating the expression of embryonic cranial genes in tissues critical for normal craniofacial development.
2 Materials and methods
2.1 Plasmid constructs
Two partial Xenopus laevis zmym4 (BC068960, BC108780) plasmids were purchased from Dharmacon. To generate a full-length zmym4 plasmid construct, the BC108780 ORF was subcloned into the BamHI (Thermo Fisher Scientific Cat# FD0054) and XbaI (New England Biolabs Cat# R0145S) sites of pCS2+ (pCS2+zmym4) using the Clone EZ PCR cloning kit (GenScript Gene Synthesis Cat# L00339). PCR was then used to amplify the region of the BC068960 plasmid spanning from the internal SpeI restriction site to the 3′ BamHI, removing the 3′ UTR present in the BC068960 plasmid. The PCR-generated second half of the zmym4 ORF was then ligated into the SpeI (New England Biolabs Cat# R0133S) and 3′ BamHI sites of the pCS2+zmym4 plasmid (pCS2+zmym4. L). To generate both morpholino (MO)-sensitive and MO-insensitive HA-tagged full-length zmym4 constructs, gBlock fragments were designed (Supplementary Figure S1A) and purchased from Integrative DNA Technologies. These contained the 5′ restriction site, a restriction site found within the ORF, and either the 5′UTR region containing the binding site for the translation blocking MO or an edited version of the 5′UTR removing the binding site for MO1 and editing the nucleotide sequence for MO2 to prevent MO binding but conserving the amino acid sequence. After confirming that these 5′ gBlock fragments were successfully ligated into pCS2+zmym4. L (pCS2+zmym4. L-5′UTR; pCS2+zmym4. L-5′MOinsensitive) and confirmed by full-length sequencing, the process was repeated at the 3′ end to insert a gBlock fragment containing a 5′ restriction site found within the ORF, the 3′ restriction site, and an HA-tag just before the TAA stop codon (pCS2+zmym4. L-3′HA; pCS2+zmym4. L-5′UTR3′HA; pCS2+zmym4. L-5′MOinsensitive3′HA). The inserted 3′HA sequence was confirmed in each construct by full-length sequencing and expression validated by Western blot (Supplementary Figure S1B).
2.2 Cell culture
HEK293T/17 cells (American Type Culture Collection (ATCC) CRL-11268, RRID:CVCL_0063) were cultured in Dulbecco’s modified Eagle medium (DMEM; Cytiva Life Sciences Cat# SH30022.01) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific Cat# 10437-028) and penicillin-streptomycin (PenStrep; Gibco; Thermo Fisher Scientific Cat# 15070-063). Cells were plated into 24-well plates for luciferase assays and into 6-well plates for co-immunoprecipitation (Co-IP) experiments. Cells were transfected according to the manufacturer’s protocol using Lipofectamine 3,000 (Invitrogen; Thermo Fisher Scientific Cat# L3000-015). Cells were processed for each assay 48 h after transfection.
2.3 Co-immunoprecipitation
HEK293T cells were transfected with 1.5 μg of pCS2+3′MycSix1, pCS2+zmym4. L-3′HA, or pCS2+3′MycSix1 and pCS2+zmym4. L-3′HA. Cytoplasmic proteins were extracted from cells 48 h after transfection using a volume of NE-PER Nuclear and Cytoplasmic Extraction Buffer CER I (Thermo Fisher Scientific Cat# 78833) according to the size of the cell pellet as per manufacturer instructions with added Halt Protease Inhibitor Cocktail with EDTA (Thermo Fisher Scientific Cat# 78430). After collecting the cytoplasmic fraction, the remaining nuclear cell pellet was lysed using NER buffer with added Halt and EDTA, according to the manufacturer protocol. The concentration of the cytoplasmic and nuclear supernatants was determined using the Pierce BCA Assay Kit (Thermo Fisher Scientific Cat# 23225). The same concentration of each sample was then subjected to immunoprecipitation using the Pierce anti-c-Myc magnetic beads (Thermo Fisher Scientific Cat# 88842) or the Pierce anti-HA magnetic beads (Thermo Fisher Scientific Cat# 88836). Proteins were washed according to the manufacturer protocol for each of the beads and eluted using the Pierce Lane Marker Non-Reducing Sample Buffer (Thermo Fisher Scientific Cat# 39001) for the anti-C-Myc beads and a 2 μg per ml concentration of HA peptide (Thermo Fisher Scientific Cat# 26184) following the gentle elution protocol in the manufacturer protocol for the anti-HA beads. The eluted proteins were reduced using 100 mM dithiothreitol (DTT; Thermo Fisher Scientific Cat# R0861) (anti-C-Myc beads) or Laemmli Sample Buffer (Bio-Rad Laboratories Cat# 161-0747 with 2% β-mercaptoethanol (BME; Bio-Rad Laboratories Cat# 161-0710) (anti-HA beads) at 100 °C for 10 min prior to SDS-PAGE and Western blot using 8%–16% mini-protean TGX gels (Bio-Rad Laboratories Cat# 456-1104). For control experiments, 30 μL of each lysate in CER or NER buffer was diluted in Laemmli Sample Buffer with 2% BME, incubated at 100 °C for 10 min prior to SDS-PAGE and Western blot. Immobilon-FL PVDF membranes (Fisher Scientific Cat# IPFL00010) were probed with mouse anti-Myc (9B11 at 1:1,000; Cell Signaling Technology Cat# 2040, RRID:AB_2148465), rabbit anti-HA (C29F4 at 1:1,000; Cell Signaling Technology Cat# 3724, RRID:AB_1549585), rabbit anti-Zmym4 (1:1,000; Lifespan Biosciences (LSBio) Cat# LS-C193170/200,169), rabbit anti-Six1 (D5S2S at 1:1,000; Cell Signaling Technology Cat# 16960, RRID:AB_2798773), or mouse anti-HA (6E2 at 1:1,000; Cell Signaling Technology Cat# 2999, RRID:AB_1264166). For all blots, mouse anti-αTubulin (DM1A at 1:5,000; Novus Biologicals Cat#NB110-93473, RRID:AB_1236645) was used as a control and the IRDye 680RD donkey anti-rabbit IgG (1:5,000; LI-COR Biosciences Cat# 925-68073, RRID:AB_2716687) and IRDye 800 CW goat anti-mouse IgG (1:5,000; LI-COR Biosciences Cat#925-32210, RRID:AB_2687825) were used as secondary antibodies. The Zmym4 antibody was validated by Western blot to generate a protein of the expected size and to overlap with the HA-antibody positive band generated by the pCS2+zmym4-3′HA construct. Experiments were repeated at least three independent times. Blots were scanned using the LI-COR Odyssey Classic Infrared Imaging System and analyzed using LI-COR Image Studio Software (RRID:SCR_015795).
2.4 Luciferase assay
HEK293T cells were transfected with 200 ng of pGL3-6xMEF3-Firefly luciferase, a Six1-inducible reporter containing 6 Six1 binding sites (Ford et al., 2000), and 100 ng of pRL-TK-Renilla luciferase reporter in addition to different combinations of pCS2+ (empty vector control), pCS2+3′FlagSix1, pCS2+5′MycEya1, and pCS2+zmym4. L (400 ng each). Plasmids encoding Six1 BOR variants are as previously reported (Shah et al., 2020). At 48 h post-transfection, cells were resuspended directly in 100 μL of Passive Lysis Buffer (from the Dual Luciferase Assay Kit; Promega Corporation Cat# E1980) for 1 h at room temperature and then frozen at −80 °C overnight. The following day, 20 μL of lysate was analyzed using the Dual Luciferase Assay kit following the manufacturer protocol. Experiments were repeated at least 5 times. ANOVA with Tukey post hoc multiple comparisons test was performed using GraphPad Prism 9 software (RRID:SCR_015795). Expression of exogenous proteins from the transfected plasmids was confirmed by standard Western blotting using the antibodies described for Co-IP with the exception of Eya1, which was identified using the mouse anti-Myc and anti-Eya1 antibodies (1:1,000; Proteintech Cat# 22658-1-AP, RRID:AB_2879145) and Six1, which was identified using the mouse anti-DYKDDDDK (9A3 at 1:1,000; Cell Signaling Technology Cat# 8146, RRID:AB_10950495) or rabbit anti-Six1 antibodies described above.
2.5 In vitro synthesis of RNAs
mRNAs encoding X. laevis zmym4 (pCS2+zmym4. L or pCS2+zmym4. L-5′MOins3′HA) and a nuclear-localized β-galactosidase (nβgal) lineage tracer were synthesized in vitro according to the manufacturer protocol (SP6 mMessage mMachine kit; Thermo Fisher Scientific Cat# AM1340). Antisense RNA DIG-labeled probes for in situ hybridization (ISH) were synthesized in vitro (MEGAscript SP6 Transcription Kit; Invitrogen, Thermo Fisher Scientific Cat# AM1330, as previously described (Yan et al., 2009).
2.6 Embryo microinjections
Fertilized X. laevis embryos were obtained as previously described and chosen at the 2-cell stage to accurately identify the dorsal and ventral animal blastomeres (Klein, 1987; Miyata et al., 1987; Moody, 2018). When selected embryos reached the 8-cell stage, the dorsal-animal and ventral-animal blastomeres that predominantly give rise to the neural crest and cranial placodes (Moody and Kline, 1990) were each microinjected with 1 nL of mRNA (100 pg mixed with 100 pg nβgal) or MOs (4.5 ng of an equimolar mixture of MO1+MO2) according to standard methods (Moody, 2018). Only one side of the embryo was microinjected rendering the uninjected side as an internal control. Embryos were cultured in diluted Steinburg’s solution at 16 °C until fixation.
2.7 Morpholino oligonucleotides
To knockdown endogenous levels of Zmym4 protein in vivo, two lissamine-tagged translation-blocking antisense morpholino oligonucleotides (MOs) were purchased (GeneTools, LLC) with the following sequences: Zmym4 MO1: 5′-GACTGTGATTAGTATCAGCATCCAT-3′ and Zmym4 MO2: 5′-TATTCCATCGCCCGGTCAGCCCGGT-3’. These MOs bind in the 5′UTR directly adjacent to the starting ATG or to the beginning of the coding region of both the L- and S-homoeolog of X. laevis zmym4 (Supplementary Figure S1A). To verify the ability of the MOs to block zmym4 translation, both cells of a 2-cell X. laevis embryo were injected with an equimolar mixture of the Zmym4 MOs. At stage 26 (Nieuwkoop and Faber, 1994), MO-injected and sibling control embryos were snap frozen on dry ice and stored overnight at −80 °C. They were lysed in 500 μL of RIPA buffer with added HALT and EDTA protease inhibitors and mechanically disrupted using a 1.5 mL pestle. To remove the yolk proteins, samples were sequentially centrifuged at 500 g, 750 x g, 1000 x g, and 1250 x g at 4 °C and the supernatant transferred to a clean 1.5 mL microcentrifuge tube between each centrifugation step. Expression of Zmym4 protein was determined Western blot, as described above (Supplementary Figure S1C) and protein levels normalized to αTubulin to determine knockdown efficacy (Supplementary Figure S1D). The specificity of the knockdown was determined by injecting embryos at the 8-cell stage with an equimolar mixture of Zmym4 MOs, as above, with or without mRNA encoding zmym4-5′MOins3′HA, which is insensitive to MO binding (Supplementary Figure S1A). These embryos were processed for in situ hybridization (see below) and assessed for the percentage of embryos showing reduced sox9 expression (Supplementary Figure S1E).
2.8 Histochemistry and in situ hybridization
Embryos were cultured to early neural plate (stage 13), neural fold (stages 16–18), or larval (stages 28–32) stages (Nieuwkoop and Faber, 1994), fixed in 4% paraformaldehyde (PFA; MP Biomedicals Cat# 150146), stained for βgal histochemistry (if mRNA-injected), and processed for in situ hybridization (ISH) as previously described (Yan et al., 2009). Only embryos in which the fluorescent MOs or nβGal lineage tracer were located in the appropriate tissue domains were analyzed for the expression of dlx5, foxd3, irx1, msx1, pax3, six1, sox2, sox9, sox11, tbx1, or tfap2α. For each gene, the staining intensity and size of the expression domain were compared between the injected, lineage-labeled side to the control, uninjected side of the same embryo, thus minimizing inter-embryo variation that might occur with differences in developmental stages or experimental batch. Embryos that showed a more intense staining pattern or larger expression domain on the injected side compared to control side were scored as “increased.” Embryos that showed a less intense staining pattern or smaller expression domain on the injected side compared to the control side were scored as “decreased.” The percentages of embryos that showed “increased” expression, “decreased” expression, or “no change” between injected and control sides of the same embryo were calculated for each gene, and differences in the frequency assessed for significance (p < 0.05) by the Chi-square test using GraphPad Prism 9. Embryos were collected from at least three different sets of outbred, wild-type parents to account for genetic diversity for each of the genes assessed in both MO and mRNA treatment conditions.
2.9 Neural crest migration assays
The extent of cranial NC migration was assessed using two approaches: in vivo and in vitro. In vivo studies were performed as previously described (Cousin et al., 2012). Briefly, the dorsal animal blastomere of the 8-cell embryo, which is the major precursor of the NC (Moody and Kline, 1990), was microinjected with 100 pg of membrane-associated GFP (mbGFP) mRNA alone or in combination with either 1.3 ng of Zmym4 MOs equimolar mixture (as above) or 1 ng of Sobp MOs (characterized in (Tavares et al., 2021)). When embryos reached neural crest migratory stages (stages 22-23), those expressing the GFP lineage marker in the dorsal anterior region were scored for the presence or absence of fluorescently labeled NC cells in the NC migration pathways into the BAs; partial migration was scored as presence. For each experiment, the results from embryos in which only lineage tracer was injected (control) were used as a normalizer and set to 100%. The control and morphant numbers were compared using a two-tailed, paired Student’s t-test. In vitro studies were performed as described in Cousin and Alfandari (2018). One cell of the embryo was injected at the 2-cell stage with either 200 pg of mbGFP mRNA alone or in combination with either 2.5 ng of Zmym4 MOs equimolar mixture (as above) or 2 ng of Sobp MOs. At stage 15, explants of fluorescently labeled cranial NC were dissected from the embryo, individually placed into a 96-well plate coated with 20 μg/mL of Fibronectin (FN), and cultured at 18 °C in Danilchik medium containing 50 μg/mL gentamicin. Once the explants attached to the plate (at ∼1 h post explant), time lapse images were taken using a Keyence microscope at a rate of one image every 3 min for ∼10–11 h. Movies were assembled using Keyence BZ-X Analyser software. The surface area occupied by the explant at the beginning and at the end of the movie were determined manually using Fiji software. The ratio of these areas (end/beginning) were calculated and normalized to the ratio obtained from control explants and compared using a two-tailed, paired Student’s t-test.
2.10 TUNEL staining
To determine if changes in NC gene expression might result from apoptosis, embryos were injected on one side with Zmym4 MOs as above and cultured to larval stages (stage 35–36). Embryos were fixed and processed for TUNEL staining as wholemounts according to the manufacturer protocol (ApopTag In Situ Apoptosis Detection Kit; EMD Millipore Cat# S7101) as adapted by (Zaghloul and Moody, 2007). After staining, the skin was removed with fine forceps and the number of TUNEL-positive cells in all four BAs counted on the control and MO-injected sides of each embryo. The numbers were compared using a two-tailed, paired t-test (p < 0.05) using GraphPad Prism 9.
2.11 Cartilage staining
Embryos were injected with Zmym4 MOs as above and grown to tadpole stages (stages 45–47) when the cranial cartilages have formed. Tadpoles were fixed in 4% PFA, dehydrated to 100% ethanol, stained overnight in Alcian blue (Sigma-Aldrich Cat# A5268) and cleared in a KOH (Emsure; EMD Millipore Cat# 1310-S8-3):glycerol (Fisher Scientific Cat# BP229-1) series, as previously described (Walker and Kimmel, 2007; Tavares et al., 2021). Morphological dysmorphologies in cartilages contributing to the lower jaw (infrarostral, Meckel’s, ceratohyal), otic capsule, and pharyngeal apparatus (ceratobranchial cartilages) (Svensson and Haas, 2005) were each visually scored as “normal”, “mildly defective”, or “severely defective” as previously described (Keer et al., 2022). Briefly, a defect was scored as “mild” if slightly displaced or reduced in size and “severe” if the cartilage was significantly displaced or significantly reduced or missing. The frequencies of each phenotype per cartilaginous element were calculated across three independent experiments using the Chi-squared test (p < 0.05). To quantify the extent of the disruption of the craniofacial cartilages, images of 76 Zmym4 morphants were taken at 32X using the Olympus cellSens software (RRID:SCR_014551) on an Olympus SZX16 stereomicroscope. These images were imported into Adobe Photoshop 2022 (RRID:SCR_014199) and the scale bar used to set the measurement scale for quantification (174 pixels = 200 μm). The length of both left and right Meckel’s, ceratohyal, and otic capsule cartilages, and the length of the unpaired infrarostral cartilage were measured as illustrated (Supplementary Figure S2). These measurements were then exported into GraphPad Prism 9 and the measurements for each element analyzed for normal distribution with the Shapiro-Wilks test. As the measurements for one of the elements were not normally distributed, each data set was analyzed for significant differences between the morphants and sibling controls using a Mann-Whitney test (p < 0.05).
3 Results
To identify novel Six1-interacting proteins that might be causative candidates for BOR, we previously screened the fly interactome for Sine oculis interacting proteins whose orthologs are also expressed in Xenopus embryos (Neilson et al., 2010). Zinc finger MYM-binding protein 4 (Zmym4) was identified as a candidate by its sequence similarity to a few domains found in Drosophila Sobp (Box2 and Box3) (Kenyon et al., 2005), which were confirmed to be present in Xenopus Sobp (Tavares et al., 2021). Xenopus Sobp contains two zinc fingers, a proline rich domain, two SUMO-interacting motifs (SIMs), and a functional nuclear localization signal (NLS) (Figure 1A). Box2 and Box3 span the two zinc finger domains and share sequence homology with two regions in Zmym4 (Figure 1B). Xenopus laevis Zmym4, which contains nine zinc finger domains, is highly conserved between the long and short homoeologs, the Xenopus tropicalis and human homologs, and the Drosophila ortholog Without children (Woc; Supplementary Figure S3). Zmym4 has a predicted glucocorticoid-like DNA binding domain that spans part of the first zinc finger domain as well as a DUF3504 domain in the C-terminal region (Figure 1B), which is related to the tyrosine recombinase element of Crypton DNA transposon elements suggesting it may function as a transcriptional activator and/or repressor (Kojima and Jurka, 2011). The DUF3504 domain also exhibits in silico structural similarity and likely evolutionary conservation with the BEN family DNA-binding domains (Pan et al., 2023), also suggesting a nuclear function. A putative NLS was detected within the DUF3504 domain (Figure 1B). Furthermore, Zmym4 has been shown to be a B-MYB binding protein and that it is SUMOylated, suggesting that it contains currently unannotated, non-canonical SIMs consistent with what has been described for ZMYM2 and ZMYM3 (Anderson, 2015; Cibis et al., 2020). These sequence similarities to other nuclear factors suggest that Zmym4 may function as a transcription factor.
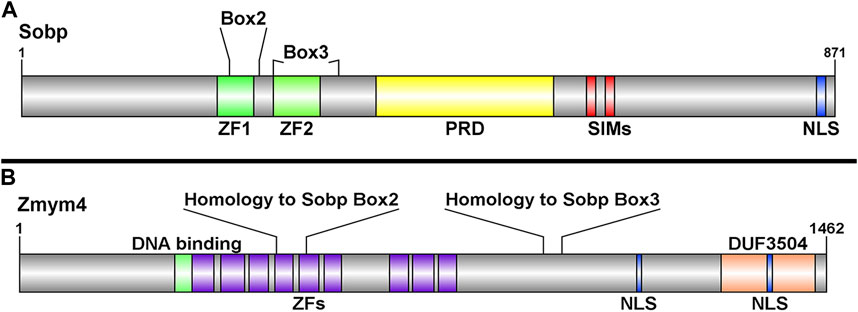
FIGURE 1. Schematic representations of the Sobp and Zmym4 proteins highlighting their different domains. (A) Sobp protein structure adapted from Tavares et al., 2021 highlighting Box2 and Box3 identified by Kenyon et al., 2005. Sobp contains two zinc fingers (ZF1, ZF2), a proline rich domain (PRD), two SUMO-interacting motifs (SIM), and a functional nuclear localization signal (NLS). (B) The Zmym4 protein contains nine zinc finger MYM-containing domains (purple boxes) identified in human ZMYM4 and aligned by sequence homology (Supplementary Figure S3). The areas of sequence homology with Sobp Box2 and Box3 are indicated. Zmym4 contains two putative NLSs, a glucocorticoid-like DNA binding domain (green box), and a C-terminal DUF3504 domain. The NLSs were identified using cNLS mapper (Kojima and Jurka, 2011), the DNA-binding and DUF3504 domains were identified using InterPro (https://www.ebi.ac.uk/interpro/) and the protein structures were generated using DOG2.0.1 (http://dog.biocuckoo.org/). Numbers on the right of each schematic indicate amino acid length of the protein.
3.1 Zmym4 is unlikely to function as a Six1 co-factor
A number of cofactors have been identified to regulate Six1 activity. Eya1 is required for Six1 to function as transcriptional activator, whereas Groucho (a.k.a., Grg4, Tle4) causes Six1 to function as a transcriptional repressor (Silver et al., 2003; Brugmann et al., 2004; Anderson et al., 2012). Other Six1-binding proteins (i.e., Pa2G4, Mcrs1, Sobp) act to reduce the activity of the Six1-Eya1 transcriptional complex (Neilson et al., 2017; Neilson et al., 2020; Tavares et al., 2021; Keer et al., 2022).
Neilson et al. (2010) showed that Zmym4 is expressed in similar embryonic tissues as both Six1 and Eya1, including the branchial arches (BAs), neural crest (NC) and cranial placodes. This co-expression, along with its sequence similarity to Sobp (Figure 1), suggest that Zmym4 may interact with Six1 during early cranial development. To determine whether Zmym4 can interact with Six1, HEK293T cells were transfected with either Myc-tagged Six1, HA-tagged Zmym4, or both and the lysates assayed by Co-IP using either anti-c-Myc tagged beads or anti-HA tagged beads. Despite performing multiple replicates under different conditions (i.e., performing the IP for either Myc- or HA-tag, different concentrations of input protein, and different elution conditions), we were unable to detect binding of Zmym4 to Six1 in this assay (Figure 2A). It is notable that the flowthrough from the Co-IP double transfection shows a lower level of Zmym4 compared to the flowthrough from the single Zmym4-3′HA lane. This may indicate that the co-expression of Six1 in the doubly transfected cells affects Zmym4 translation or stability. However, in previous reports we have seen this same reduction even when there is clear evidence for a Six1-cofactor interaction (Neilson et al., 2017; Neilson et al., 2020; Tavares et al., 2021). Therefore, we do not believe that the lack of Zmym4 binding to Six1 in the Co-IP lane is due to a reduction in Zmym4 levels. However, this requires further biochemical validation.
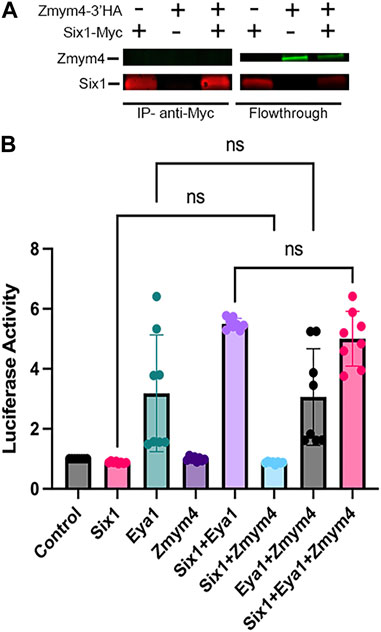
FIGURE 2. Zmym4 does not bind to Six1 or alter the transcriptional activity of Six1, Eya1, or the Six1-Eya1 transcriptional complex in cultured cells. (A) HEK293T cells were transfected with either Myc-tagged Six1, HA-tagged Zmym4, or both and were tested for co-immunoprecipitation (Co-IP) with anti-Myc tagged magnetic beads. In the Co-IP assay (IP—anti-Myc), when Six1 was transfected alone, it bound to the Myc-tagged beads (left lane: Zmym4-3′HA (−), Six1-Myc (+)), whereas when Zmym4 was transfected alone, it did not bind, as expected (middle lane: Zmym4-3′HA (+), Six1-Myc (−)). When both were transfected, (right lane: Zmym4-3′HA (+), Six1-Myc (+)), Six1 was immunoprecipitated by the beads but Zmym4 was not detected in the sample. The flowthrough lanes contained detectable Myc-tagged Six1 but no HA-tagged Zmym4 (left lane), detectable HA-tagged Zmym4 but no Myc-tagged Six1 (middle lane) and detectable Myc-tagged Six1 and HA-tagged Zmym4 (right lane). (B) Luciferase activity of the pGL3-6XMEF-luciferase reporter in HEK293T cells transfected with different combinations of empty vector control, Six1, Eya1, and Zmym4 plasmids. Data were normalized to Renilla expressed by a constitutively active promoter. There was no significant difference between the transcriptional activity of Six1 alone, Zmym4 alone and Six1+Zmym4, between Eya1 alone and Eya1+Zmym4, or between Six1+Eya1 and Six1+Eya1+Zmym4 (ns = p > 0.05).
Despite the lack of direct binding between Zmym4 and Six1, a recent publication identified that single nucleotide polymorphism (SNP) variants of Zmym4 are able to act on nearby genes (Spada et al., 2016); therefore, it was important to determine whether Zmym4 may have an indirect role in regulating the transcriptional activity of Six1, Eya1, or the Six1-Eya1 transcriptional complex. To assess whether Zmym4 could modulate Six1 transcriptional activity, HEK293T cells were co-transfected with a Six1-inducible reporter (Ford et al., 2000) and different combinations of Six1, Eya1, and/or Zmym4 encoding plasmids. As demonstrated previously (Patrick et al., 2009; Tavares et al., 2021), in the presence of Eya1, Six1 induced a significant increase in luciferase activity over control plasmid or Six1 alone (Figure 2B). In contrast, neither Zmym4 alone or in the presence of Six1 caused a significant change in luciferase activity over that of the control or Six1 alone (Figure 2B), indicating that Zmym4 does not alter Six1 transcriptional activity in this assay. Next, as the HEK293T cells have a low level of endogenous Six1 expression, reflected in the moderate activation of the Six1-inducible luciferase reporter with the transfection of only Eya1 (Tavares et al., 2021) (Figure 2B), we investigated whether Zmym4 was able to alter the transcriptional activity of Eya1. We again found that there was no significant difference in the transcriptional activity of the Six1-inducible luciferase reporter activity when assayed with Eya1 alone or in the presence of Zmym4 (Figure 2B). Finally, we assayed whether Zmym4 could alter the activity of the Six1-Eya1 transcriptional complex. We noted no significant difference in luciferase reporter activity in the presence of Six1+Eya1 with or without the addition of Zmym4 (Figure 2B). Consistent with a previous study that showed BOR variants of Six1 are transcriptionally deficient (Tavares et al., 2021), we did not detect any effect on transcriptional activity when Zmym4 was transfected with a Six1 BOR variant either alone or in the presence of Eya1 (Supplementary Figure S4). Together, these data show that despite some sequence similarity to Sobp, Zmym4 is unlikely to function as a Six1 cofactor as it neither binds to Six1 nor alters Six1 or Eya1 transcriptional activity in the in vitro assays we performed.
3.2 Zmym4 affects cranial neural crest and placode gene expression
Although we did not detect any evidence that Zmym4 could bind to Six1 or modulate its transcriptional activity in a cell line, its expression in the developing NC and cranial placodes suggests a cranial developmental function. Therefore, we assayed whether loss- or gain-of-function of Zmym4 in embryos could affect the expression of genes that regulate early cranial development. First, to determine whether Zmym4 is required for the formation of the cranial NC or PPE, we decreased the endogenous levels of Zmym4 with specific translation blocking MOs injected into their precursor blastomeres at the 8-cell stage (Moody and Kline, 1990). These embryos were then assessed for the expression of genes that are expressed in the NB (msx1, tfap2α, pax3), NP (sox2, sox11, irx1), cranial NC (foxd3, sox9), or PPE (six1, sox11, irx1, sox9). The size of the expression domain and/or intensity of the ISH color reaction were then compared between the MO-injected and control side of each embryo and scored as either “increased” (e.g., larger or more intense), “decreased” (e.g., smaller or less intense), or “no change”.
At early neural plate stages (stages 13–14), in the majority of embryos knockdown (KD) of Zmym4 decreased the expression domains of msx1 and tfap2α but expanded the pax3 domain (Figure 3A, A1). At neural fold stages (stages 16–18), Zmym4 KD primarily expanded three NP genes (sox2, sox11, irx1; Figure 3B, B1) and reduced two NC genes (foxd3, sox9; Figure 3C, C1). Zmym4 KD reduced the domains of three PPE genes - six1, sox9, irx1—in the majority of embryos. In contrast, the sox11 PP E domain was reduced in about a third of embryos, expanded in a third and unchanged in a third (Figure 3D, D1). Thus, loss of Zmym4 has a significant impact on the expression domains of genes required for the proper patterning of the embryonic ectoderm that gives rise to the central and peripheral nervous systems and several craniofacial sensory structures.
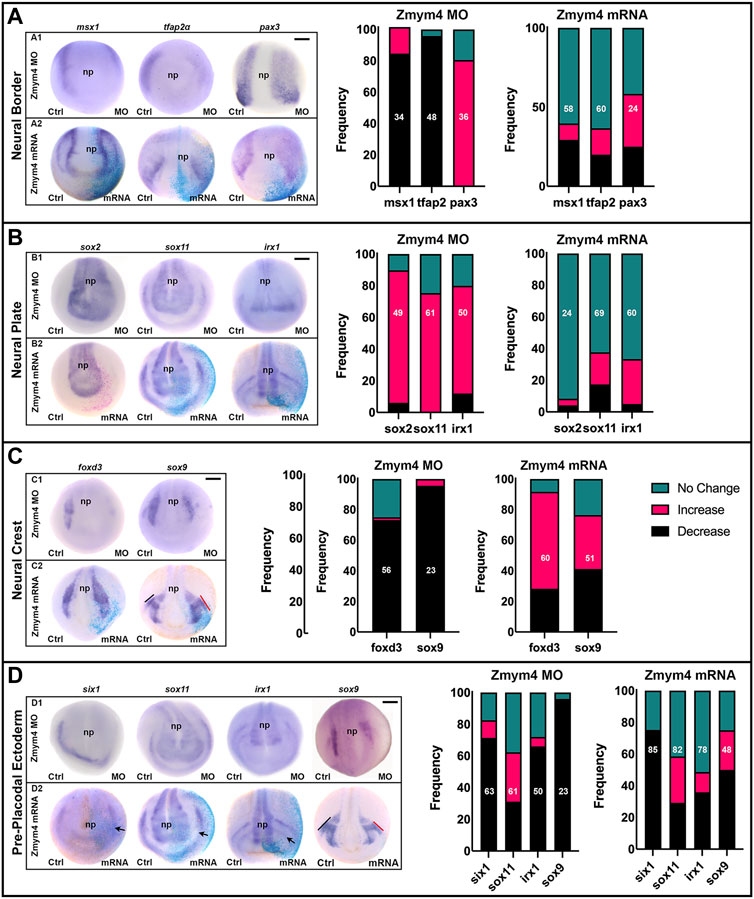
FIGURE 3. Altering the levels of Zmym4 in embryos differentially altered the expression of early cranial genes. (A) In the neural border zone, knockdown of Zmym4 by MO injection (A1) resulted in decreased expression of msx1 and tfap2α and broader expression of pax3 on the MO-injected side compared to the control (Ctrl) side of the same embryo. Increased expression of Zmym4 by mRNA injection (A2) caused no change in expression in most embryos. The percentages of embryos that showed “no change” (green), “increased” (pink), and “decreased” (black) expression on the injected side compared to the control side of the same embryo are reported in the bar graphs; the number of embryos examined for each gene is indicated within the bar. (B) In the neural plate, knockdown of Zmym4 (B1) resulted primarily in expanded expression of sox2, sox11, and irx1, whereas increased expression of Zmym4 (B2) caused no change in their expression in most embryos. Bar graphs are as in (A). (C) In the neural crest, knockdown of Zmym4 (C1) resulted primarily in reduced expression of foxd3 and sox9. Increased expression of Zmym4 (C2) primarily increased foxd3 expression, and caused broader sox9 expression in some embryos (∼35%; C2) and reduced expression in others (∼41%; D2). Bar graphs are as in (A). (D) In the preplacodal ectoderm, knockdown of Zmym4 (D1) primarily reduced the expression of six1, irx1, sox9; in contrast, nearly equal numbers of embryos showed no change, expanded or reduced expression of sox11. Increased expression of Zmym4 (D2) showed a minor decrease in six1, whereas for sox11, irx1 and sox9 some embryos showed broader expression while others showed reduced expression. In the D2 embryo images, PPE expression of six1 on mRNA-injected side is slightly reduced compared to control, sox11 is reduced and irx1 is reduced in the anterior placode (arrows). In D2, sox9 is reduced in the otic placode on the injected side (red bar) compared to control (black bar); in C2, sox9 is larger in the otic placode on the injected side (red bar) compared to control (black bar). Bar graphs are as in (A). All embryo images are anterior views with dorsal to the top. np, neural plate. Black scale bars in top right corners = 300 μm.
To determine whether increasing Zmym4 protein above endogenous levels altered early gene expression, zmym4 mRNA was microinjected as described for the MOs. At early neural plate stages, only a minority of embryos showed either an increase or decrease in NB gene expression (Figure 3A, A2). Similarly, the majority of embryos showed no effect on the NP genes (Figure 3B, B2). In contrast, increasing levels of Zmym4 primarily expanded the NC domain of foxd3 and about equally expanded or decreased the NC domain of sox9 (Figure 3C, C2). Increased levels of Zmym4, however, had variable effects on PPE genes. The six1 domain showed a minor decrease in most embryos (Figure 3D, D2). Although many embryos showed decreased levels of irx1 (36%) or sox9 (50%) PPE expression, other embryos showed expanded domains (Figure 3D, D2). Similar to the effects of its KD, increased Zmym4 protein resulted in about equal proportions of embryos with decreased, increased or no change in the sox11 PPE expression domain (Figure 3D, D2).
Together, these results suggest that loss of Zmym4 alters the expression of NB genes that results in expansion of the NP genes at the expense of NC and PPE genes. These data are consistent with Zmym4 being important for early brain development (Xiang et al., 2020), but also suggest a required function in the development of the NC and PPE. Increased expression of Zmym4, on the other hand, had only small effects on either the NB or NP, but instead expanded NC genes seemingly at the expense of PPE genes. One explanation could be that Zmym4 levels after 100 pg mRNA injections were not increased sufficiently for an effect. To test this, we doubled the mRNA concentration (200 pg) but detected no statistically significant differences from the 100 pg injections (p > 0.05; Chi-square test; data not shown). Therefore, we propose that Zmym4 targets may be sensitive to threshold amounts of protein and exceeding those levels may not affect the frequencies at which we observe the assessed phenotypes.
3.3 Loss of Zmym4 results in dysmorphology of the craniofacial cartilages
Since loss and gain of Zmym4 had opposite effects on NC gene expression and mostly reduced that of PPE genes, we asked whether those changes resulted in later disruptions of gene expression in the structures derived from these progenitor fields: the otic vesicle (OV), which is of placode origin, and the BAs, whose mesenchymal core is derived from cranial NC. Both the OV and BA express genes that are required for NC migration and chondrogenesis, including dlx5, sox9, and tbx1 (Acampora et al., 1999; Depew et al., 1999; Garg et al., 2001; Spokony et al., 2002; Mori-Akiyama et al., 2003; Saint-Germain et al., 2004; Brown et al., 2005; Moraes et al., 2005; Tazumi et al., 2010). Zmym4 KD decreased the intensity and domain size of dlx5 in both the OV and BAs (Figures 4A,B). In about 75% of these embryos all BA were affected and in 25% only the posterior BAs were affected, as illustrated in Figure 4A. Loss of Zmym4 had less of an effect on sox9 or tbx1 expression. sox9 was decreased in the OV in 25% of embryos and in the BA in 43% of embryos, whereas tbx1 was decreased in the OV in 19% of embryos and in the BA in 15% of embryos (Figures 4A,B). Because many deficiencies in cranial NC after loss of gene function can be attributed to increased apoptosis (e.g., Jones et al., 2008; Ross and Zarbalis, 2014), we performed a TUNEL analysis at late larval stages (∼stages 35–36), when NC migration into the BAs is complete. KD of Zmym4 caused no significant difference in the number of TUNEL positive cells between the control BAs (mean = 3.13 cells, N = 15) and the morphant BAs (mean = 3.53 cells; p > 0.05) (Figure 4C). These results indicate that the reduction of dlx5, sox9 or tbx1 BA expression domains were not due to cell death.
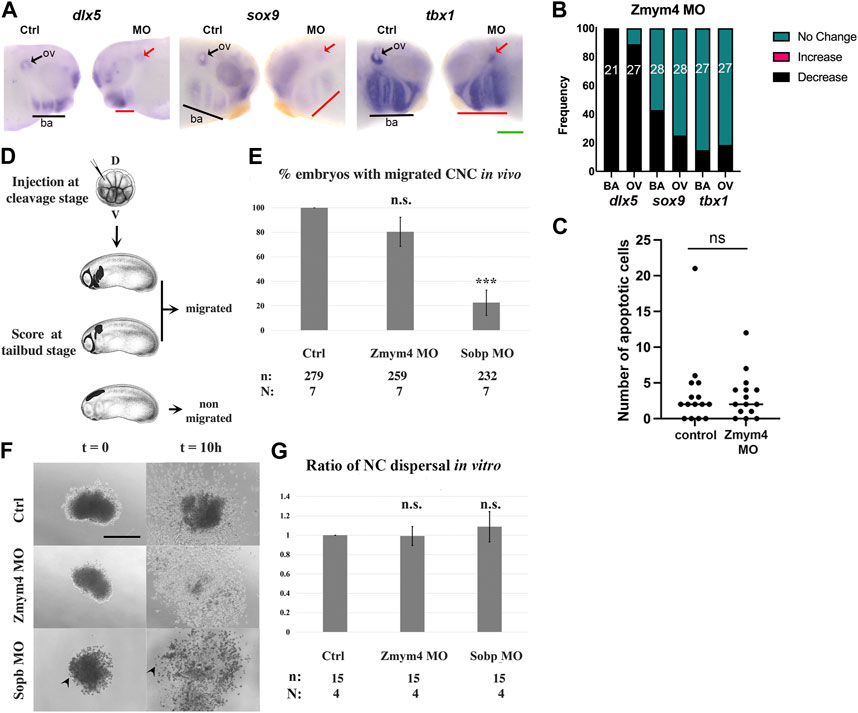
FIGURE 4. Loss of Zmym4 caused reduced otic vesicle and branchial arch gene expression. (A) Knockdown of Zmym4 reduced the expression dlx5, sox9, and tbx1 in larval otic vesicles (ov, arrows) and branchial arches (ba, underlined) on the MO-injected side compared to the control (Ctrl) side of the same embryo. Green scale bar in lower right corner indicates 200 μm. (B) The percentages of embryos that showed “no change” (green), “increased” (pink), or “decreased” (black) expression on the MO-injected side compared to the control side of the same embryo; the number of embryos examined for each gene is indicated within the bar. Zmym4 knockdown reduced the OV and BA expression of dlx5 in nearly every embryo, whereas it reduced sox9 or tbx1 expression only in small percentages of embryos. (C) Scatter plot of the number of apoptotic cells counted on the control and MO-injected sides of Zmym4 morphant larvae (n = 15). There was no significant difference (ns = p > 0.05) in the number of apoptotic cells after loss of Zmym4. (D) Blastomeres on the dorsal (D) animal side of cleavage stage embryos were microinjected with mbGFP mRNA ± Zmym4 MOs or Sobp MOs. At tailbud stages, embryos were scored as “migrated” if some fluorescently labeled NC entered a migratory pathway and as “non-migrated” if none of them entered any pathway. Embryo images downloaded from Xenbase (https://www.xenbase.org/xenbase/zahn.do). (E) The percentage of embryos with migrated NC cells in the assay shown in (D) n = the number of embryos analyzed; N = the number of independent trials. n. s. = not significant compared to controls. ***, p < 0.001, Student’s t-test (F) DIC images of typical cranial NC explants when they first adhered to the substrate (t = 0) and 10 h later (t = 10h). Arrowheads indicate Sobp morphant cells that are rounded rather than flattened on the FN substrate. Scale bar = 200 μm. (G) The ratio of the area the NC covered between the start and end of the culture period (end/beginning) (NC dispersal). n. s. = not significant compared to controls (p > 0.05, Student’s t-test). n = the number of embryos analyzed; N = the number of independent trials.
To determine whether the reduction in BA gene expression in Zmym4 morphants was due to delayed NC migration into the BAs, we performed in vivo and in vitro NC migration assays. For the in vivo assay, the presence or absence of GFP-labeled NC in the branchial arch migratory pathways was assessed at stage 25 in both Sobp and Zmym4 morphants, as described in Weir et al. (2021) (Figure 4D). We found that Sobp KD led to a significant decrease in the percentage of embryos that contained NC cells in any of the migratory streams (Figure 4E), as expected from the previously reported craniofacial defects (Tavares et al., 2021). In contrast, Zmym4 KD did not lead to a significant decrease compared to controls (Figure 4E). For the in vitro assay, explanted cranial NC derived from control, Zmym4 morphant or Sobp morphant embryos were assessed by time lapse microscopy. The area occupied by the cells of the explant at the beginning (Figure 4F t = 0) and at the end (Figure 4F t = 10) of the culture period was measured and the ratio (end/beginning) calculated and normalized to the ratio obtained from control embryo explants. In each of the three conditions, the initial explants and the cells derived from them attached to the FN substrate (Figure 4F). The only difference was that the control cells and the Zmym4 morphant cells flattened as they dispersed from the explant, whereas the Sobp morphant cells remained rounded throughout the time course of the assay (Figure 4F, arrowheads). Consistent with the in vivo assay, Zmym4 KD did not lead to a significant change in the area occupied by NC cells that migrated away from the explant (Figure 4G). Although Sobp KD cells did not flatten on the FN substrate, they also dispersed over an area that was not significantly different from controls (Figure 4G). Together, these NC migration assays show that Zmym4 KD does not appear to affect the ability of NCs to migrate into the BAs.
Since dlx5, sox9, and tbx1 also are required for cranial chondrogenesis (Depew et al., 1999; Garg et al., 2001; Mori-Akiyama et al., 2003; Tazumi et al., 2010), we assessed whether their altered BA expression at larval stages would lead to altered morphology of the craniofacial cartilages. To test this, Zmym4 morphants were grown to tadpole stages and their cartilages stained with Alcian blue. Loss of Zmym4 resulted in dysmorphology of the craniofacial cartilages, marked by compression of the infrarostral cartilage, thickening and shortening of Meckel’s cartilage, compression of the ceratohyal, and reduced size of the otic capsule and ceratobranchial cartilages (Figure 5A). When individual cartilages were measured and scored for severity of morphological defect, we found that the majority of tadpoles examined (n = 76) had at least a mild defect on the MO-injected side (Figure 5B). It is not surprising that many craniofacial cartilages were affected by Zmym4 KD since reduced dlx5 expression was observed in most of the branchial arches at tailbud stages (75%); perhaps those tadpoles in which the cartilages appeared normal (∼25–30%; Figure 5B) represent the larvae (∼25%) in which reduced dlx5 was only observed in the posterior BAs, as shown in Figure 4A. Taken together, these data confirm that Zmym4 is required for normal development of the NC-derived craniofacial cartilages.
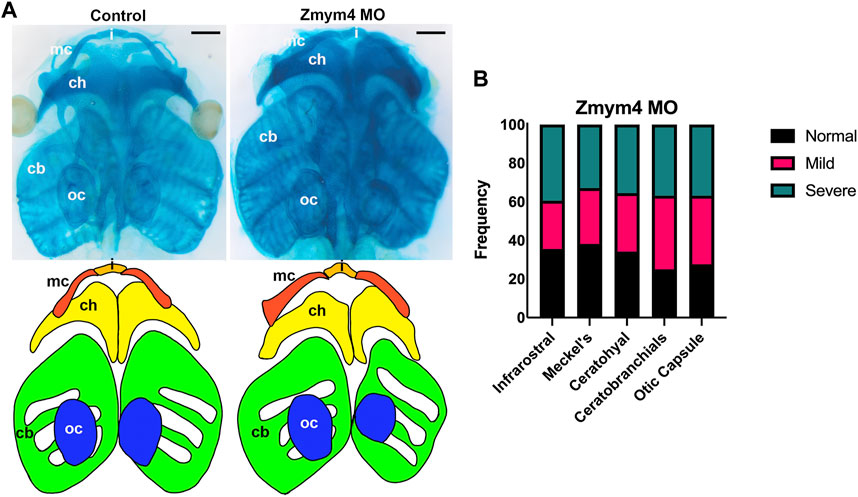
FIGURE 5. Loss of Zmym4 results in craniofacial cartilage dysmorphologies. (A) Top: ventral views of Alcian blue stained heads from a control tadpole and a sibling Zmym4 morphant (Zmym4 MO). Loss of Zmym4 on the right side of the tadpole resulted in compression of the infrarostral cartilage (I), thickening and shortening of the Meckel’s cartilage (mc), shortening and compression of the ceratohyal cartilage (ch) and reduced size of the ceratobranchial cartilages (cb). The otic capsule (oc) also was reduced in size on the injected side. Bottom: tracing of the craniofacial cartilages to better demonstrate the differences in morphology between the control and morphant infrarostral (orange), Meckel’s (red), ceratohyal (yellow), ceratobranchial (green) and otic capsule (blue) cartilages. Black scale bars in top right corners = 200 μm. (B) Bar graph depicting the percentage of Zmym4 morphants (n = 76) that displayed either “normal” (black), mildly affected (pink), or severely aberrant (green) morphology for each of the five craniofacial cartilages analyzed.
4 Discussion
Most of the cranial sensory and skeletal tissues are derived from neural tube, neural crest and preplacodal ectodermal stem cells during embryogenesis. It is hoped that by understanding the molecular and cellular interactions that result in the formation of these different stem populations, regenerative therapies may be developed to treat the large number of known syndromic and non-syndromic craniofacial dysmorphologies. Our long-standing interest in this quest has been to reveal the molecular interactions between the Six1 transcription factor and putative interacting partners that may account for variable craniofacial malformations found in Branchio-oto-renal syndrome (BOR) patients that include: the outer, middle, and inner ear; branchial fistulas and cysts; and in some cases renal abnormalities (Moody et al., 2015; Smith, 2018). Approximately 5% of patients presenting with BOR have a mutation in SIX1, and approximately 45% present with a mutation in EYA1, which encodes a SIX1 co-factor that is required for transcriptional activation (Buller et al., 2001; Ruf et al., 2004; Lee et al., 2007; Sanggaard et al., 2007; Patrick et al., 2009; Li et al., 2010; Moody and Saint-Jeannet, 2014; Moody and LaMantia, 2015; Klingbeil et al., 2017; Smith, 2018). We hypothesized that since nearly half of patients have a mutation in EYA1, it is likely that unidentified SIX1 interacting proteins may be causative (Moody et al., 2015). To this end, we previously screened the fly interactome for binding partners of the Drosophila homologue of Six1 whose orthologs are also expressed in Xenopus embryos (Neilson et al., 2010). These screens identified new Six1 cofactors, including Proliferation-associated 2G4 (Pa2G4), Microspherule protein 1 (Mcrs1), and Sine oculis binding protein (Sobp) (Neilson et al., 2017; Neilson et al., 2020; Tavares et al., 2021; Keer et al., 2022). This screen also identified zinc finger MYM-containing 4 (Zmym4) as a potential Six1 cofactor based on its structural similarities to Box2 and Box3 of Sobp (Kenyon et al., 2005; Neilson et al., 2010). In this report, we tested whether Zmym4 can interact with Six1, affect its transcriptional activity and/or is required for the development of the embryonic stem populations of important craniofacial structures.
4.1 Zmym4 does not interact directly with Six1 but may act as a pioneer or scaffold protein
We found no evidence in the Co-IP or luciferase assays that we used that Zmym4 binds to Six1 or alters its transcriptional activity, thus indicating it is unlikely to be a bona fide Six1 cofactor. Nonetheless, the KD experiments demonstrate that Zmym4 is required for the proper apportioning of gene expression domains that divide the embryonic ectoderm into neural plate, neural crest and cranial placodes. While there is no direct evidence yet for the manner by which Zmym4 regulates cranial gene expression, work on two highly similar proteins, ZMYM2 and ZMYM3, allow us to speculate that Zmym4 may have a developmental function in transcriptional control by remodeling chromatin.
ZMYM2 interacts with the fibronectin type-III (FNIII) domain of ATF7IP, which is required for the efficient transcriptional silencing of both genes and transposable elements by the SETDBI complex (Tsusaka et al., 2020). The FNIII domain is similar to an “ITEFSL” sequence found within MYC homology box II that enables histone acetylation and is essential for FNIII domain interactions (Kalkat et al., 2018; Tsusaka et al., 2020). ZMYM4 also possesses an ITEFSL-like sequence (Supplementary Figure S3), suggesting that it also may interact with the FNIII domain of ATF7IP (Tsusaka et al., 2020). ZMYM3 serves as a scaffolding protein that coordinates interactions between deacetylases, demethylases, and RNase H type enzymes (including HDAC and CoREST), and the eighth and ninth zinc fingers of ZMYM2 protein can bind directly to the LSD1-CoREST-HDAC complex (Shapson-Coe et al., 2019). As RNase H2 can interact with both ZMYM2 and ZMYM4, it has been suggested that ZMYM4 may also interact with the LSD1-CoREST-HDAC complex to mediate transcriptional regulation (Jabbari et al., 2018; Shapson-Coe et al., 2019). Furthermore, ZMYM2 and ZMYM3 contain a new class of SUMO-interacting motifs (SIMs) that can interact with SUMO2 in a 1:1 stoichiometry, and recent studies show that ZMYM4 is also SUMOylated (Anderson, 2015; Lamoliatte et al., 2017; Cibis et al., 2020). Both ZMYM2 and ZMYM4 are B-MYB binding proteins, although the interaction between ZMYM4 and B-MYB does not appear to be direct (Cibis et al., 2020). Nonetheless, a recent MYB chromatin immunoprecipitation mass spectrophotometry (ChIP-MS) assay identified that only MYB and seven other proteins were specifically enriched in this assay, one of which was ZMYM4 (Yong et al., 2023). Thus, given the similarities with ZMYM2 and ZMYM3, we predict that Zmym4 may accomplish its effects on cranial gene expression by modulating or stabilizing interactions between complexes that modify chromatin availability, perhaps allowing it to act as a pioneer factor during early development.
We found that loss of Zmym4 at neural border stages decreased the expression of NB genes (msx1, tfap2α) and neural plate genes (sox2, sox11, irx1), whereas its overexpression had minimal effects on them. These results are consistent with Zmym4 acting as a pioneer factor in the early neural ectoderm. Pioneer transcription factors implement new cell fates during development by binding to DNA target sites in a closed chromatin configuration and modulating active-chromatin histone modifications that remodel the chromatin into an open configuration, thus making critical sites accessible to other transcription factors (Balsalobre and Drouin, 2022). As such, loss of a pioneer factor typically causes significant changes in downstream gene expression, but overexpression of the same factor typically results in little to no change in later gene expression because the endogenous protein has already opened the chromatin, making critical binding sites already available. An important next step will be to biochemically determine whether Zmym4 acts as a chromatin remodeling/pioneer protein that regulates early cranial gene expression.
4.2 Zmym4 may act as a transcription factor in NC and PPE stem populations
Interestingly, the effects of loss- and gain-of-function of Zmym4 on NC and PPE genes were quite different from those on NB and NP genes. Loss of Zmym4 resulted in the reduction foxd3 and sox9 expression in the premigratory NC stem population, whereas its overexpression resulted primarily in broadening their expression domains. These data would suggest that Zmym4 may act as a NC-promoting transcription factor, which is consistent with our identification of a DUF3504 domain in the C-terminal region of Zmym4 (Figure 1; Supplementary Figure S3). DUF3504 domains recently were shown to exhibit in silico structural similarities and evolutionary conservation with BEN family DNA-binding domains, suggesting they have the ability to serve as a transcription factor (Pan et al., 2023). ZMYM4 also has downstream alternative initiation sites (dATI), which are characterized by having an AUG codon downstream of the canonical initiator methionine codon that can be used to generate multiple proteins from the same mRNA sequence and to traffic proteins to different cellular compartments (Kazak et al., 2013). In ZMYM4, these dATI sites have been predicted to function in transcriptional regulation, supporting the possibility that Zmym4 could function in this way in the NC stem population (Kazak et al., 2013).
The potential function of Zmym4 as a PPE-determining transcription factor is less clear. Both loss- and gain-of-function resulted primarily in a reduction in the expression of six1, whose protein is a key regulator of the other PPE genes. Zmym4 KD resulted primarily in reduced irx1 and sox9 domains; overexpression of Zmym4 caused reduction to a lesser extent and broader domains in a smaller percentage of embryos. Both loss- and gain-of-function resulted in nearly equal proportions of reduced and broadened expression of sox11. Since the formation of the PPE is regulated by a complex gene regulatory network that differentially effects each participating gene (Klein et al., 2002; Grocott et al., 2012; Moody and LaMantia, 2015; Riddiford and Schlosser, 2016; Hintze et al., 2017; Maharana and Schlosser, 2018), an important next goal will be to sort the direct and indirect effects of Zmym4 KD on each element in the network.
It is worth noting that ZMYM4 also has been shown to have numerous functions outside of direct transcriptional control, the details of which are only now being uncovered. These include: 1) ZMYM4-AS1, a long non-coding RNA that can be used as a competitive endogenous RNA to regulate gene expression levels and affect cell function by competing with microRNAs (Li et al., 2021), 2) a circular RNA form (i.e., circ_0011536) that may function as a microRNA sponge in colorectal and endometrial cancers (Cheng et al., 2018; Yin et al., 2022), and 3) fusion with a rolling-circle transposon that generates an alternative transcript of ZMYM4 that contains a cryptic splice site, an initiation codon, and a few codons in the predicted ORF from the transposon donor (Thomas et al., 2014). Therefore, the possible functions of Zmym4 during early cell fate decisions underlying the apportioning of the embryonic ectoderm into the various stem populations that contribute to craniofacial structures - the neural plate, neural crest, preplacodal ectoderm, and the non-neural ectoderm—remain to be determined by future in-depth studies.
4.3 Early loss of Zmym4 causes defects in craniofacial chondrogenesis
Given our finding that loss of Zmym4 results in reduction of NC and PPE gene expression, we investigated whether KD of Zmym4 had effects on later derivatives of these stem populations, the OV - derived from the otic placode - and the BA- derived from the cranial neural crest. Consistent with the gene expression data at neural fold stages, we found that loss of Zmym4 resulted in reduced expression of dlx5, sox9 and tbx1 in the OV and BA at larval stages. At tadpole stages, these gene expression changes persisted as dysmorphologies in the craniofacial cartilages. One potential cause of the reduction in gene expression domains and cartilage element is errors in migration of the NC into the BA, especially since Sox9 and Tbx1 are required for this migration (Spokony et al., 2002; Moraes et al., 2005). However, we did not detect any defects using either an in vivo or ex vivo assay. Another potential cause is increased apoptosis during BA formation; again, we did not detect a significant difference in the number of apoptotic cells in the BA between controls and Zmym4 morphants. A third potential cause is reduced cell proliferation either prior to or during migration of the NCs. Although we did not test this directly, KD of ZMYM4 previously was shown to have no obvious effects on cell cycle progression (Cibis et al., 2020). Therefore, the possible functions of Zmym4 during craniofacial chondrogenesis remain to be determined.
4.4 Conclusion
Our study is the first to identify a role for Zmym4 during early cranial gene expression and later craniofacial chondrogenesis. While very little is currently known about the function of ZMYM4 itself, its closely related proteins ZMYM2 and ZMYM3 have been shown to contain non-canonical SIMs and are important in chromatin remodeling either directly (ZMYM2) or by stabilizing the interactions between the LSD1-CoREST-HDAC repressive complex and chromatin by acting as scaffolding (ZMYM3). As ZMYM4 has been identified as being critical for early brain development (Xiang et al., 2020) and our data show that it is required for the proper apportioning of the embryonic ectoderm into neural plate, neural crest and PPE, it will be important to determine if Zmym4 accomplishes this by regulating chromatin accessibility during neural development, especially by facilitating the cell fate change from a multipotent state to a more restricted one. Data from human cell lines also indicate that ZMYM4 may have a direct transcriptional function (Kojima and Jurka, 2011; Pan et al., 2023), which we predict may impact the expression of neural crest and placode genes. Although we have yet to uncover the molecular mechanism(s) by which this multifunctional protein regulates these processes, taken together, the data presented herein demonstrate that Zmym4 plays an important role in regulating craniofacial development. They also identify exciting avenues of future study into the potential roles of this factor in the pathophysiology of craniofacial dysmorphologies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) of the George Washington University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
KJ: Conceptualization, Formal Analysis, Investigation, Methodology, Validation, Visualization, Data curation, Writing–original draft. KN: Conceptualization, Formal Analysis, Investigation, Methodology, Validation, Funding acquisition, Writing–review and editing. HC: Conceptualization, Formal Analysis, Investigation, Methodology, Writing–review and editing. AT: Conceptualization, Formal Analysis, Investigation, Methodology, Writing–review and editing. HM: Formal Analysis, Investigation, Writing–review and editing. DA: Formal Analysis, Investigation, Writing–review and editing, Conceptualization, Funding acquisition, Methodology, Project administration, Supervision. SM: Conceptualization, Formal Analysis, Investigation, Methodology, Writing–review and editing, Funding acquisition, Project administration, Supervision, Validation, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. NIH R01 DE022065 to SM; NIH R01 DE026434 to SM, KN and DA; NIH R01 DE016289 to DA.
Acknowledgments
The National Xenopus Resource (http://mbl.edu/xenopus/, RRID:SCR_013731) and Xenbase (http://www.xenbase.org, RRID:SCR_003280) provided invaluable information for carrying out this work. Embryo images used in Figure 4D were downloaded from Xenbase (https://www.xenbase.org/xenbase/zahn.do). We thank Richard Wargowsky for assistance in cloning the full length pCS2+zmym4. L. plasmid.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The reviewer (J-P S-J) declared a past co-authorship with the author (SM) to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.1274788/full#supplementary-material
References
Acampora, D., Merlo, G. R., Paleari, L., Zerega, B., Postiglione, M. P., Mantero, S., et al. (1999). Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 126, 3795–3809. doi:10.1242/dev.126.17.3795
Anderson, A. M., Weasner, B. M., Weasner, B. P., and Kumar, J. P. (2012). Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139, 991–1000. doi:10.1242/dev.077255
Anderson, O. (2015). Dissertation thesis. (Dundee, Scotland). UK, University of Dundee. Available at: https://discovery.dundee.ac.uk/en/studentTheses/the-identification-and-characterisation-of-proteins-interacting-w. The identification and characterisation of proteins interacting with SUMO-ubiquitin hybrid chains.
Balsalobre, A., and Drouin, J. (2022). Pioneer factors as master regulators of the epigenome and cell fate. Nat. Rev. Molec. Cell. Biol. 23, 449–464. doi:10.1038/s41580-022-00464-z
Brown, S. T., Wang, J., and Groves, A. K. (2005). Dlx gene expression during chick inner ear development. J. Comp. Neurol. 483, 48–65. doi:10.1002/cne.20418
Brugmann, S. A., Pandur, P. D., Kenyon, K. L., Pignoni, F., and Moody, S. A. (2004). Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development 131, 5871–5881. doi:10.1242/dev.01516
Buller, C., Xu, X., Marquis, V., Schwanke, R., and Xu, P. (2001). Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum. Molec. Gen. 10, 2775–2781. doi:10.1093/hmg/10.24.2775
Cheng, Q., Ning, D., Chen, J., Li, X., Chen, X., and Jiang, L. (2018). SIX1 and DACH1 influence the proliferation and apoptosis of hepatocellular carcinoma through regulating p53. Cancer Biol. Ther. 19, 381–390. doi:10.1080/15384047.2018.1423920
Cibis, H., Biyanee, A., Dörner, W., Mootz, H. D., and Klempnauer, K. (2020). Characterization of the zinc finger proteins ZMYM2 and ZMYM4 as novel B-MYB binding proteins. Sci. Rep. 10, 8390. doi:10.1038/s41598-020-65443-w
Connaughton, D. M., Dai, R., Owen, D. J., Marquez, J., Mann, N., Graham-Paquin, A. L., et al. (2020). Mutations of the transcriptional corepressor ZMYM2 cause syndromic urinary tract malformations. Amer. J. Hum. Genet. 107, 727–742. doi:10.1016/j.ajhg.2020.08.013
Cousin, H., Abbruzzese, G., McCusker, C., and Alfandari, D. (2012). ADAM13 function is required in the 3 dimensional context of the embryo during cranial neural crest cell migration in Xenopus laevis. Dev. Biol. 368, 335–344. doi:10.1016/j.ydbio.2012.05.036
Cousin, H., and Alfandari, D. (2018). Cranial neural crest explants”, Cold Spring Harb. Protoc. 2018, pdb.prot097394, doi:10.1101/pdb.prot097394
Depew, M. J., Liu, J. K., Long, J. E., Presley, R., Meneses, J. J., Pedersen, R. A., et al. (1999). Dlx5 regulates regional development of the branchial arches and sensory capsules. Development 126, 3831–3846. doi:10.1242/dev.126.17.3831
Ford, H. L., Landesman-Bollag, E., Dacwag, C. S., Stukenberg, P. T., Pardee, A. B., and Seldin, D. C. (2000). Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J. Biol. Chem. 275, 22245–22254. doi:10.1074/jbc.M002446200
Garg, V., Yamagishi, C., Hu, T., Kathiriya, I. S., Yamagishi, H., and Srivastava, D. (2001). Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev. Biol. 235, 62–73. doi:10.1006/dbio.2001.0283
Goyal, L., Kongpetch, S., Crolley, V. E., and Bridgewater, J. (2021). Targeting FGFR inhibition in cholangiocarcinoma. Cancer Treat. Rev. 95, 102170. doi:10.1016/j.ctrv.2021.102170
Goyal, L., Saha, S. K., Liu, L. Y., Siravegna, G., Leshchiner, I., Ahronian, L. G., et al. (2017). Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 7, 252–263. doi:10.1158/2159-8290.CD-16-1000
Goyal, L., Shi, L., Liu, L. Y., Fece de la Cruz, F., Lennerz, J. K., Raghavan, S., et al. (2019). TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov. 9, 1064–1079. doi:10.1158/2159-8290.CD-19-0182
Gray, P. A., Fu, H., Luo, P., Zhao, Q., Yu, J., Ferrari, A., et al. (2004). Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306, 2255–2257. doi:10.1126/science.1104935
Greenhill, C. (2020). Three more genes linked with childhood obesity. Nat. Rev. Endocrinol. 16, 402–403. doi:10.1038/s41574-020-0380-6
Grocott, T., Tambalo, M., and Streit, A. (2012). The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective. Dev. Biol. 370, 3–23. doi:10.1016/j.ydbio.2012.06.028
Hintze, M., Prajapati, R. S., Tambalo, M., Christophorou, N. A. D., Anwar, M., Grocott, T., et al. (2017). Cell interactions, signals and transcriptional hierarchy governing placode progenitor induction. Development 144, 2810–2823. doi:10.1242/dev.147942
Jabbari, K., Heger, P., Sharma, R., and Wiehe, T. (2018). The diverging routes of BORIS and CTCF: an interactomic and phylogenomic analysis. Life (Basel) 8, 4. doi:10.3390/life8010004
Jones, N. C., Lynn, M. L., Gaudenz, K., Sakai, D., Aoto, K., Rey, J., et al. (2008). Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 14, 125–133. doi:10.1038/nm1725
Kalkat, M., Resetca, D., Lourenco, C., Chan, P., Wei, Y., Shiah, Y., et al. (2018). MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell. 72, 836–848. doi:10.1016/j.molcel.2018.09.031
Kazak, L., Reyes, A., Duncan, A. L., Rorbach, J., Wood, S. R., Brea-Calvo, G., et al. (2013). Alternative translation initiation augments the human mitochondrial proteome. Nucleic Acids Res. 41, 2354–2369. doi:10.1093/nar/gks1347
Keer, S., Cousin, H., Jourdeuil, K., Neilson, K. M., Tavares, A. L. P., Alfandari, D., et al. (2022). Mcrs1 is required for branchial arch and cranial cartilage development. Dev. Biol. 489, 62–75. doi:10.1016/j.ydbio.2022.06.002
Kenyon, K. L., Li, D. J., Clouser, C., Tran, S., and Pignoni, F. (2005). Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev. Dyn. 234, 497–504. doi:10.1002/dvdy.20442
Klein, S. L., Strausberg, R. L., Wagner, L., Pontius, J., Clifton, S. W., and Richardson, P. (2002). Genetic and genomic tools for Xenopus research: the NIH Xenopus initiative. Dev. Dyn. 225, 384–391. doi:10.1002/dvdy.10174
Klein, S. L. (1987). The first cleavage furrow demarcates the dorsal-ventral axis in Xenopus embryos. Dev. Biol. 120, 299–304. doi:10.1016/0012-1606(87)90127-8
Klingbeil, K. D., Greenland, C. M., Arslan, S., Paneque, A. L., Gurkan, H., Ulusal, S. D., et al. (2017). Novel EYA1 variants causing Branchio-oto-renal syndrome. Inter. J. ped. Otorhinolaryngol. 98, 59–63. doi:10.1016/j.ijporl.2017.04.037
Kojima, K. K., and Jurka, J. (2011). Crypton transposons: identification of new diverse families and ancient domestication events. Mob. DNA 2, 12. doi:10.1186/1759-8753-2-12
Lamoliatte, F., McManus, F. P., Maarifi, G., Chelbi-Alix, M. K., and Thibault, P. (2017). Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 8, 14109. doi:10.1038/ncomms14109
Lee, K. Y., Kim, S., Kim, U. K., Ki, C., and Lee, S. H. (2007). Novel EYA1 mutation in a Korean branchio-oto-renal syndrome family. Inter. J. ped. Otorhinolaryngol. 71, 169–174. doi:10.1016/j.ijporl.2006.08.023
Li, M., Chen, Y., Chen, Z., Chen, M., Huo, H., and Bu, S. (2021). Identification of prognostic biomarkers and potential regulatory axes for acute myeloid leukemia based on a competitive endogenous RNA network.
Li, Y., Manaligod, J. M., and Weeks, D. L. (2010). EYA1 mutations associated with the branchio-oto-renal syndrome result in defective otic development in Xenopus laevis. Biol. Cell. 102, 277–292. doi:10.1042/BC20090098
Maharana, S. K., and Schlosser, G. (2018). A gene regulatory network underlying the formation of pre-placodal ectoderm in Xenopus laevis. BMC Biol. 16, 79. doi:10.1186/s12915-018-0540-5
Marenne, G., Hendricks, A. E., Perdikari, A., Bounds, R., Payne, F., Keogh, J. M., et al. (2020). Exome sequencing identifies genes and gene sets contributing to severe childhood obesity, linking PHIP variants to repressed POMC transcription. Cell. Metab. 31, 1107–1119.e12. doi:10.1016/j.cmet.2020.05.007
Miyata, S., Kageura, H., and Kihara, H. K. (1987). Regional differences of proteins in isolated cells of early embryos of Xenopus laevis. Cell. Differ. 21, 47–52. doi:10.1016/0045-6039(87)90447-7
Moody, S. A., and LaMantia, A. S. (2015). Transcriptional regulation of cranial sensory placode development. Curr. Top. Dev. Biol. 111, 301–350. doi:10.1016/bs.ctdb.2014.11.009
Moody, S. A. (2018). Lineage tracing and fate mapping in Xenopus embryos. Cold Spring Harb. Protoc. 12, prot097253. doi:10.1101/pdb.prot097253
Moody, S. A., Neilson, K. M., Kenyon, K. L., Alfandari, D., and Pignoni, F. (2015). Using Xenopus to discover new genes involved in Branchiootorenal spectrum disorders. Comp. Biochem. Physiol. Part C. Toxicol. Pharmacol. 178, 16–24. doi:10.1016/j.cbpc.2015.06.007
Moody, S. A., and Saint-Jeannet, J. (2014). in Development of the pre-placodal ectoderm and cranial sensory placodes” in Principles of Developmental Genetics. Editor S. A. Moody (NY (China: Elsevier), 331–356.
Moody, S., and Kline, M. (1990). Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol. 182, 347–362. doi:10.1007/BF02433495
Moon, S. W., Son, H. J., Chae, J., Yoo, N. J., An, C. H., and Lee, S. H. (2021). Expression and mutation alterations of ZMYM4 gene in gastric and colonic cancers. Appl. Immunohistochem. Mol. Morphol. 29, 570–575. doi:10.1097/PAI.0000000000000939
Moraes, F., Nóvoa, A., Jerome-Majewska, L. A., Papaioannou, V. E., and Mallo, M. (2005). Tbx1 is required for proper neural crest migration and to stabilize spatial patterns during middle and inner ear development. Mech. Dev. 122, 199–212. doi:10.1016/j.mod.2004.10.004
Mori-Akiyama, Y., Akiyama, H., Rowitch, D. H., and de Crombrugghe, B. (2003). Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc. Nat. Acad. Sci. U. S. A. 100, 9360–9365. doi:10.1073/pnas.1631288100
Neilson, K. M., Abbruzzesse, G., Kenyon, K., Bartolo, V., Krohn, P., Alfandari, D., et al. (2017). Pa2g4 is a novel Six1 co-factor that is required for neural crest and otic development. Dev. Biol. 421, 171–182. doi:10.1016/j.ydbio.2016.11.021
Neilson, K. M., Keer, S., Bousquet, N., Macrorie, O., Majumdar, H. D., Kenyon, K. L., et al. (2020). Mcrs1 interacts with Six1 to influence early craniofacial and otic development. Dev. Biol. 467, 39–50. doi:10.1016/j.ydbio.2020.08.013
Neilson, K. M., Pignoni, F., Yan, B., and Moody, S. A. (2010). Developmental expression patterns of candidate cofactors for vertebrate six family transcription factors. Dev. Dyn. 239, 3446–3466. doi:10.1002/dvdy.22484
Nieuwkoop, P. D., and Faber, J. (1994). Normal table of Xenopus laevis (daudin): A systematical & chronological survey of the development from the fertilized egg till the end of metamorphosis. New York: Garland Science.
Pan, A., Zeng, Y., Liu, J., Zhou, M., Lai, E. C., and Yu, Y. (2023). Unanticipated broad phylogeny of BEN DNA-binding domains revealed by structural homology searches. Curr. Biol. 33, 2270–2282.e2. doi:10.1016/j.cub.2023.05.011
Patrick, A. N., Schiemann, B. J., Yang, K., Zhao, R., and Ford, H. L. (2009). Biochemical and functional characterization of six SIX1 branchio-oto-renal syndrome mutations. J. Biol. Chem. 284, 20781–20790. doi:10.1074/jbc.M109.016832
Regan, J. A., and Shah, S. H. (2020). Obesity genomics and metabolomics: A nexus of cardiometabolic risk. Curr. Cardiol. Rep. 22, 174. doi:10.1007/s11886-020-01422-x
Riddiford, N., and Schlosser, G. (2016). Dissecting the pre-placodal transcriptome to reveal presumptive direct targets of Six1 and Eya1 in cranial placodes. eLife 5, e17666. doi:10.7554/eLife.17666
Ross, A. P., and Zarbalis, K. S. (2014). The emerging roles of ribosome biogenesis in craniofacial development. Front. Physiol. 5, 26. doi:10.3389/fphys.2014.00026
Ruf, R. G., Xu, P., Silvius, D., Otto, E. A., Beekmann, F., Muerb, U. T., et al. (2004). SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Nat. Acad. Sci. U. S. A. 101, 8090–8095. doi:10.1073/pnas.0308475101
Saint-Germain, N., Lee, Y. H., Zhang, Y., Sargent, T. D., and Saint-Jeannet, J. P. (2004). Specification of the otic placode depends on Sox9 function in Xenopus. Development 131, 1755–1763. doi:10.1242/dev.01066
Sanggaard, K. M., Rendtorff, N. D., Kjaer, K. W., Eiberg, H., Johnsen, T., Gimsing, S., et al. (2007). Branchio-oto-renal syndrome: detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses. Eur. J. Hum. Genet. 15, 1121–1131. doi:10.1038/sj.ejhg.5201900
Schlosser, G. (2021). Development of sensory and neurosecretory cell types. Boca Raton, FL: CRC Press.
Shah, A. M., Krohn, P., Baxi, A. B., Tavares, A. L. P., Sullivan, C. H., Chillakuru, Y. R., et al. (2020). Six1 proteins with human branchio-oto-renal mutations differentially affect cranial gene expression and otic development. Dis. Mod. Mech. 13, dmm043489. doi:10.1242/dmm.043489
Shapson-Coe, A., Valeiras, B., Wall, C., and Rada, C. (2019). Aicardi-Goutières Syndrome associated mutations of RNase H2B impair its interaction with ZMYM3 and the CoREST histone-modifying complex. PloS One 14, e0213553. doi:10.1371/journal.pone.0213553
Silver, S. J., Davies, E. L., Doyon, L., and Rebay, I. (2003). Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol 23, 5989–5999. doi:10.1128/mcb.23.17.5989-5999.2003
Smith, R. J. H. (2018). in Branchiootorenal spectrum disorders” in GeneReviews. Editors M. P. Adam, G. M. Mirzaa, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. W. Grippet al. (China: Univ Washington Press). Available at: https://www.ncbi.nlm.nih.gov/books/NBK1380/.
Spada, J., Scholz, M., Kirsten, H., Hensch, T., Horn, K., Jawinski, P., et al. (2016). Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE adult study. J. Sleep. Res. 25, 690–701. doi:10.1111/jsr.12421
Spokony, R. F., Aoki, Y., Saint-Germain, N., Magner-Fink, E., and Saint-Jeannet, J. P. (2002). The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development 129, 421–432. doi:10.1242/dev.129.2.421
Streit, A. (2018). Specification of sensory placode progenitors: signals and transcription factor networks. Inter. J. Dev. Biol. 62, 195–205. doi:10.1387/ijdb.170298as
Svensson, M. E., and Haas, A. (2005). Evolutionary innovation in the vertebrate jaw: A derived morphology in anuran tadpoles and its possible developmental origin. BioEssays 27, 526–532. doi:10.1002/bies.20224
Tavares, A. L. P., Jourdeuil, K., Neilson, K. M., Majumdar, H. D., and Moody, S. A. (2021). Sobp modulates the transcriptional activation of Six1 target genes and is required during craniofacial development. Development 148, dev199684. doi:10.1242/dev.199684
Tazumi, S., Yabe, S., and Uchiyama, H. (2010). Paraxial T-box genes, Tbx6 and Tbx1, are required for cranial chondrogenesis and myogenesis. Dev. Biol. 346, 170–180. doi:10.1016/j.ydbio.2010.07.028
Thomas, J., Phillips, C. D., Baker, R. J., and Pritham, E. J. (2014). Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Gen. Biol. Evol. 6, 2595–2610. doi:10.1093/gbe/evu204
Tsusaka, T., Fukuda, K., Shimura, C., Kato, M., and Shinkai, Y. (2020). The fibronectin type-III (FNIII) domain of ATF7IP contributes to efficient transcriptional silencing mediated by the SETDB1 complex. Epigenet. Chrom. 13, 52. doi:10.1186/s13072-020-00374-4
Walker, M. B., and Kimmel, C. B. (2007). A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82, 23–28. doi:10.1080/10520290701333558
Weir, E., McLinden, G., Alfandari, D., and Cousin, H. (2021). Trim-Away mediated knock down uncovers a new function for Lbh during gastrulation of Xenopus laevis. Dev. Biol. 470, 74–83. doi:10.1016/j.ydbio.2020.10.014
Xiang, B., Yang, J., Zhang, J., Yu, M., Huang, C., He, W., et al. (2020). The role of genes affected by human evolution marker GNA13 in schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 98, 109764. doi:10.1016/j.pnpbp.2019.109764
Yan, B., Neilson, K. M., and Moody, S. A. (2009). foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev. Biol. 329, 80–95. doi:10.1016/j.ydbio.2009.02.019
Yin, T., Du, S., Zhao, D., Sun, X., Zhou, Y., Wang, Q., et al. (2022). Identification of circ_0000375 and circ_0011536 as novel diagnostic biomarkers of colorectal cancer. World J. Clin. Cases 10, 3352–3368. doi:10.12998/wjcc.v10.i11.3352
Yong, W. K., Rane, G., Anuar, N. Z., Shao, X., Goh, C. Y., Khanchandani, V., et al. (2023). ChIP-MS reveals the local chromatin composition by label-free quantitative proteomics. bioRxiv. doi:10.1101/2023.01.27.525999
Yu, Y., Liu, S., Ren, B., Nelson, J., Jarrard, D., Brooks, J. D., et al. (2023). Fusion gene detection in prostate cancer samples enhances the prediction of prostate cancer clinical outcomes from radical prostatectomy through machine learning in a multi-institutional analysis. Amer. J. Pathol. 193, 392–403. doi:10.1016/j.ajpath.2022.12.013
Zaghloul, N. A., and Moody, S. A. (2007). Alterations of rx1 and pax6 expression levels at neural plate stages differentially affect the production of retinal cell types and maintenance of retinal stem cell qualities. Dev. Biol. 306, 222–240. doi:10.1016/j.ydbio.2007.03.017
Keywords: neural border, neural plate, preplacodal ectoderm, neural crest, otic vesicle, xenopus, Six1.
Citation: Jourdeuil K, Neilson KM, Cousin H, Tavares ALP, Majumdar HD, Alfandari D and Moody SA (2023) Zmym4 is required for early cranial gene expression and craniofacial cartilage formation. Front. Cell Dev. Biol. 11:1274788. doi: 10.3389/fcell.2023.1274788
Received: 08 August 2023; Accepted: 18 September 2023;
Published: 03 October 2023.
Edited by:
Lingyong Jiang, Shanghai Jiao Tong University, ChinaReviewed by:
Yuyao Tian, Harvard Medical School, United StatesJean-Pierre Saint-Jeannet, New York University, United States
Copyright © 2023 Jourdeuil, Neilson, Cousin, Tavares, Majumdar, Alfandari and Moody. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sally A. Moody, c2Ftb29keUBnd3UuZWR1
†Present address: Andre L. P. Tavares, Department of Biological Sciences, University of Delaware, Newark, DE, United States
 Karyn Jourdeuil1
Karyn Jourdeuil1 Dominique Alfandari
Dominique Alfandari Sally A. Moody
Sally A. Moody